HAMLET (human alpha-lactalbumin made lethal to tumor cells) is a protein-lipid complex derived from human milk that was first described for its tumoricidal activity. Later studies showed that HAMLET also has direct bactericidal activity against select species of bacteria, with highest activity against Streptococcus pneumoniae.
Keywords: HAMLET, alpha-lactalbumin, antibiotic resistance, sensitization, potentiation, streptococci, Streptococcus, group A Streptococcus, group B Streptococcus, Pneumococcus
ABSTRACT
HAMLET (human alpha-lactalbumin made lethal to tumor cells) is a protein-lipid complex derived from human milk that was first described for its tumoricidal activity. Later studies showed that HAMLET also has direct bactericidal activity against select species of bacteria, with highest activity against Streptococcus pneumoniae. Additionally, HAMLET in combination with various antimicrobial agents can make a broad range of antibiotic-resistant bacterial species sensitive to antibiotics. Here, we show that HAMLET has direct antibacterial activity not only against pneumococci but also against Streptococcus pyogenes (group A streptococci [GAS]) and Streptococcus agalactiae (group B streptococci [GBS]). As with pneumococci, HAMLET treatment of GAS and GBS resulted in depolarization of the bacterial membrane, followed by membrane permeabilization and death, which was able to be inhibited by calcium and sodium transport inhibitors. Treatment of clinical antibiotic-resistant isolates of S. pneumoniae, GAS, and GBS with sublethal concentrations of HAMLET in combination with antibiotics decreased the MICs of the antibiotics into the sensitive range. This effect could also be blocked by ion transport inhibitors, suggesting that HAMLET’s bactericidal and combination treatment effects used similar mechanisms. Finally, we show that HAMLET potentiated the effects of erythromycin against erythromycin-resistant bacteria more effectively than penicillin G potentiated killing bacteria resistant to erythromycin. These results show that HAMLET effectively (i) kills three different species of pathogenic streptococci by similar mechanisms and also (ii) potentiates the activities of macrolides and lincosamides more effectively than combination treatment with beta-lactams. These findings suggest a potential therapeutic role for HAMLET in repurposing antibiotics currently causing treatment failures in patients.
INTRODUCTION
Antibiotic resistance is a serious and growing threat to global health. The problem was addressed by the WHO in a 2014 report, aimed to develop a global action plan to combat further resistance development. The report proclaimed that if nothing is done to counteract antibiotic resistance, we will enter a postantibiotic era within this century (1). Although this has led to increased surveillance of resistance levels in all member countries and increased efforts to address and combat resistance development, antibiotic resistance is still increasing.
Drug-resistant strains of certain bacterial species are considered especially problematic due to their current impact on global health, and therefore, a pathogen priority list for intervention was presented by the WHO in 2017 (2). Non-penicillin-susceptible Streptococcus pneumoniae is among the 13 priority pathogens listed. Additionally, penicillin-resistant S. pneumoniae is categorized as a serious threat, and macrolide-resistant group A and B streptococci (GAS and GBS, respectively) are considered concerning threats to global health by the Centers for Disease Control and Prevention in the United States. Novel strategies to combat antibiotic resistance in these organisms are thus acutely needed.
HAMLET (human alpha-lactalbumin made lethal to tumor cells) is a protein-lipid complex from human milk with direct bactericidal activity against a select set of Gram-positive and Gram-negative bacteria, such as S. pneumoniae, Haemophilus influenzae, and Mycobacterium tuberculosis (3–5). HAMLET-induced death in S. pneumoniae requires binding of the complex to the bacterial membrane, resulting in a sodium-dependent calcium transport and membrane depolarization that can be inhibited by calcium and sodium transport inhibitors (4). Additionally, HAMLET was shown to require kinase activity for full bactericidal activity, as kinase inhibition partially blocked bacterial death (6). However, HAMLET has no bactericidal activity against Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, or Enterococcus faecalis (3). Yet, HAMLET treatment of methicillin-resistant S. aureus (MRSA) decreased the MICs of methicillin for MRSA strains to within the sensitive range, and combination treatment with HAMLET and methicillin was able to eliminate MRSA nasopharyngeal colonization (7). The ability of HAMLET to make antibiotic-resistant bacteria more sensitive to the antibiotic to which they are resistant is true also for HAMLET-resistant, Gram-negative organisms, such as E. coli and A. baumannii (7), and recent information suggests that combination treatment with sublethal concentrations of HAMLET and antibiotics can increase the activities of antibiotics against antibiotic- or drug-resistant strains of S. pneumoniae and M. tuberculosis (5, 6).
In this study, we first evaluated the antibacterial effect of HAMLET against a panel of pneumococcal clinical isolates with various antibiotic resistance patterns and show that HAMLET kills all strains equally well, regardless of their antibiotic resistance mechanism. We next investigated the antibacterial activity of HAMLET against macrolide-resistant clinical isolates of group A and group B streptococci and showed (i) that HAMLET had direct antibacterial activity that required the same signaling mechanisms as seen in pneumococci and (ii) that HAMLET at sublethal concentrations in combination with macrolides/lincosamides exhibited better bacterial killing than combination treatment with penicillin G (PcG) and erythromycin (Erm). These results show the potential for the future use of HAMLET-antibiotic combination therapy against streptococcal infections caused by resistant organisms.
RESULTS
Antibacterial activity of HAMLET against antibiotic-resistant S. pneumoniae clinical strains.
Previous studies have revealed that HAMLET has bactericidal activity against a select set of Gram-positive and Gram-negative bacteria but fails to directly kill a number of other species, including E. coli, S. aureus, P. aeruginosa, or E. faecalis (3–5). Furthermore, HAMLET has been shown to kill various strains of S. pneumoniae, some of which were resistant to antibiotics, but no comprehensive analysis of HAMLET’s activity against pneumococcal isolates with various antibiotic resistance characteristics currently circulating in the population has been done. Additionally, no investigation of HAMLET’s activity against other pathogenic streptococci has been performed.
To address this, we first tested HAMLET’s activity against a panel of clinical pneumococcal strains with various antibiotic resistance profiles collected and characterized by the European Committee on Antimicrobial Susceptibility Testing (EUCAST; see Materials and Methods). The specific antibiotic resistance mechanism for the clinical strains was determined by PCR amplification of known resistance cassettes, when possible (see Table S1 in the supplemental material for details). MIC broth dilution assays were performed according to the CSLI standard, and the assay was validated using the EUCAST reference and quality control strain S. pneumoniae ATCC 49619. Using this strain, we obtained MICs of 0.125 to 0.25 μg/ml for penicillin G and 0.06 μg/ml for erythromycin, in accordance with the quality control limits determined by Jorgensen et al. (8).
HAMLET had direct antibacterial activity against all S. pneumoniae strains tested, irrespective of their serotype or antibiotic resistance pattern (Table 1). HAMLET showed similar MIC values of between 20 and 40 μg/ml (i.e., between 1.2 and 2.4 μM and within 2 dilutions) for all strains tested. Furthermore, HAMLET showed a bactericidal concentration (BC) of at least a 3-log10 reduction in the number of viable organisms after 3 h of incubation in the broth dilution assay at concentrations between 40 and 80 μg/ml (i.e., between 2.4 and 4.8 μM and within 2 dilutions), suggesting that HAMLET had similar antibacterial activities against all strains of pneumococci, regardless of genetic background or antibiotic resistance mechanism.
TABLE 1.
HAMLET MICs and BCs for S. pneumoniae clinical isolates
| Isolate (serotype) | Antibiotic resistance (gene[s] or resistance)a | HAMLET MIC or range (μg/ml) | HAMLET BC or range (μg/ml)b |
|---|---|---|---|
| ATCC 49619 | None | 20 | 40 |
| 3974 (14) | Erythromycin (ermB) | 20 | 40 |
| 7545 (14) | Erythromycin (mef + msrD) | 20 | 80 |
| 12627 (6B) | Erythromycin (mef + msrD) | 20–40 | 40 |
| 13331 (14) | Erythromycin (mef + msrD) | 40 | 40 |
| 17476 (23A) | Erythromycin (mef + msrD) | 40 | 40 |
| 1947 (19F) | Trimethoprim-sulfamethoxazole (R) | 20 | 80 |
| 16467 (6A) | Trimethoprim-sulfamethoxazole (R) | 20 | 40 |
| 17446 (16A/F) | Trimethoprim-sulfamethoxazole (I) | 40 | 40 |
| 18091 (23F) | Trimethoprim-sulfamethoxazole (I) | 40 | 40 |
| 4269 (3) | Fluoroquinolone (R) | 20 | 40 |
| 13985 (19F) | Tetracycline (R) | 20 | 40 |
| 17144 (19F) | Tetracycline (R) | 20 | 40 |
| 6701 (4) | Penicillin G (I) | 20 | 40 |
| 16998 (6B) | Penicillin G (I) | 20–40 | 40 |
| 12116 (35B) | Penicillin G (I), cephalosporin-ampicillin (R) | 20–40 | 40 |
| 16031 (35A) | Penicillin G and cephalosporin (I or R) | 40 | 40 |
| 19000 (35B) | Penicillin G and cephalosporin (I or R) | 40 | 40 |
| 18120 (6A) | Penicillin G and cephalosporin (I or R) | 40 | 40 |
| 4393 (23F) | Penicillin G and cephalosporin (I or R) | 40 | 40–80 |
I, intermediate resistance; R, resistant.
BC, bactericidal concentration, as defined by the concentration of HAMLET resulting in at least a 3-log10 reduction in viability in the MIC assay 3 h postinoculation.
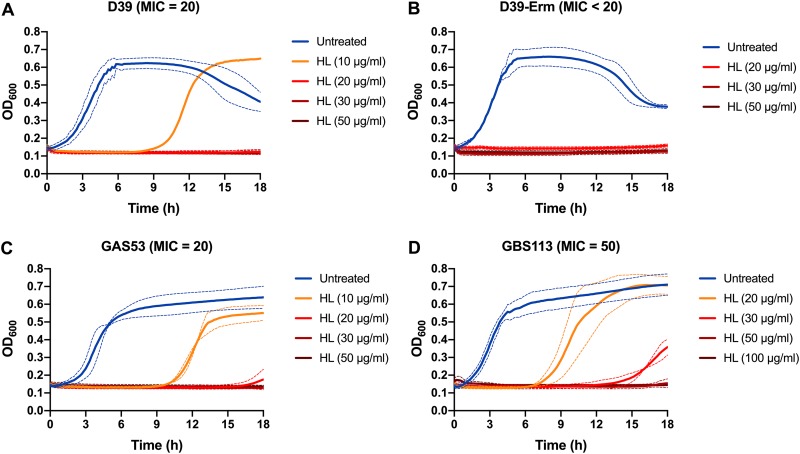
To obtain more-specific MIC values for some strains, further MIC assays were conducted using additional HAMLET concentrations, as exemplified by the wild-type strain SPN-D39 and its Erm-resistant mutant in Fig. 1A and B, and direct bactericidal activity was confirmed in short-term killing assays (1-h treatment), where HAMLET concentrations of 75 to 100 μg/ml (4 to 6 μM) resulted in more than a 3-log10 decrease in the number of viable pneumococci (Fig. S1A).
FIG 1.
MIC growth curves of pathogenic streptococci. Combined MIC growth curves for D39 (A), D39-C0832 (Erm resistant) (B), GAS clinical isolate 53 (C), and GBS clinical isolate 113 (D) in the presence of increasing concentrations of HAMLET (HL). Bacteria were grown in the presence of HAMLET for 18 h at 37°C, with the absorbance at 600 nm (OD600) recorded every 10 min. The lowest concentration of HAMLET at which no growth was detected over 18 h was considered the MIC. The figure shows combined graphs for each strain based on two (D39-C0832), three (for the GAS and GBS strains), or four (for D39) separate experiments run in duplicate wells. The OD600 is represented by a solid line, and dashed lines of the same color represent the standard deviation.
Antibacterial activity of HAMLET against Erm-resistant GAS and GBS clinical isolates.
As HAMLET’s activity against other pathogenic streptococci has not been determined, we performed MIC assays for HAMLET also against Erm-resistant group A streptococci (GAS) and group B streptococci (GBS) expressing different Erm resistance mechanisms (as listed in Table 2).
TABLE 2.
HAMLET MICs and BCs for erythromycin-resistant S. pyogenes and S. agalactiae clinical isolates
| Species and isolate | Erythromycin resistance gene(s) | HAMLET MIC or range (μg/ml) | HAMLET BC (μg/ml)a |
|---|---|---|---|
| S. pyogenes | |||
| 6 | mefA and msrD | 25–50 | 100 |
| 8 | mefA and msrD | 25–50 | 100 |
| 138 | mefA and msrD | 25–50 | 100 |
| 53 | ermB | 25 | 100 |
| 128 | ermB | 25–50 | 100 |
| 125 | ermTR | 50 | 100 |
| S. agalactiae | |||
| 114 | ermB | 100 | 400 |
| 126 | ermB | 50 | 200 |
| 129 | ermB | 100 | 400 |
| 51 | ermTR | 50–100 | 400 |
| 113 | ermTR | 50–100 | 400 |
| 76 | Undetermined | 100 | 400 |
| 150 | Undetermined | 100 | 400 |
BC, bactericidal concentration, as defined by the concentration of HAMLET resulting in at least a 3-log10 reduction in viability in the MIC assay 3 h postinoculation.
HAMLET-induced antibacterial activity was observed for all clinical GAS and GBS strains (listed in Table 2 and exemplified visually for two strains in Fig. 1C and D). The GAS strains were almost as sensitive to HAMLET as pneumococci, with MIC values ranging from 20 to 50 μg/ml (1.2 to 3 μM) and a 3-log10 reduction in bacterial viability over 3 h reached at 100 μg/ml (6 μM) for all 6 strains. The GBS strains required slightly higher HAMLET concentrations, with MIC values ranging from 50 to 100 μg/ml (3 to 12 μM), and a 3-log10 reduction in bacterial viability over 3 h was reached at 200 to 400 μg/ml (12 to 24 μM) for all seven strains. As for pneumococci, this was confirmed in short-term killing assays where HAMLET concentrations of 75 to 100 μg/ml (4 to 6 μM) resulted in a 3-log10 or higher decrease in the number of viable GAS within 1 h (Fig. S1B), whereas a concentration of 250 μg/ml (15 μM) was required to obtain the same level of killing in GBS (Fig. S1C). This indicated that higher concentrations of HAMLET were required to kill GBS than S. pneumoniae and GAS strains but that the mechanism was potentially the same. The data also show that HAMLET’s activities did not differ between strains within each species and were not affected by intraspecies genetic background or antimicrobial resistance mechanisms.
Bacterial growth inhibition and death by HAMLET in streptococci require membrane depolarization through sodium and calcium transport.
Earlier studies have shown that the direct bactericidal activity of HAMLET in pneumococci requires a sodium-dependent influx of calcium, resulting in membrane depolarization followed by membrane disruption, which can be inhibited by the calcium transport inhibitor ruthenium red (RuR) and the sodium/calcium exchange inhibitor dichlorobenzamil (DCB) (4, 9). Whether HAMLET-induced growth inhibition uses the same mechanism(s) and whether membrane depolarization and calcium influx are also involved in the bacteriostatic and direct bactericidal activity of other streptococci have not been determined.
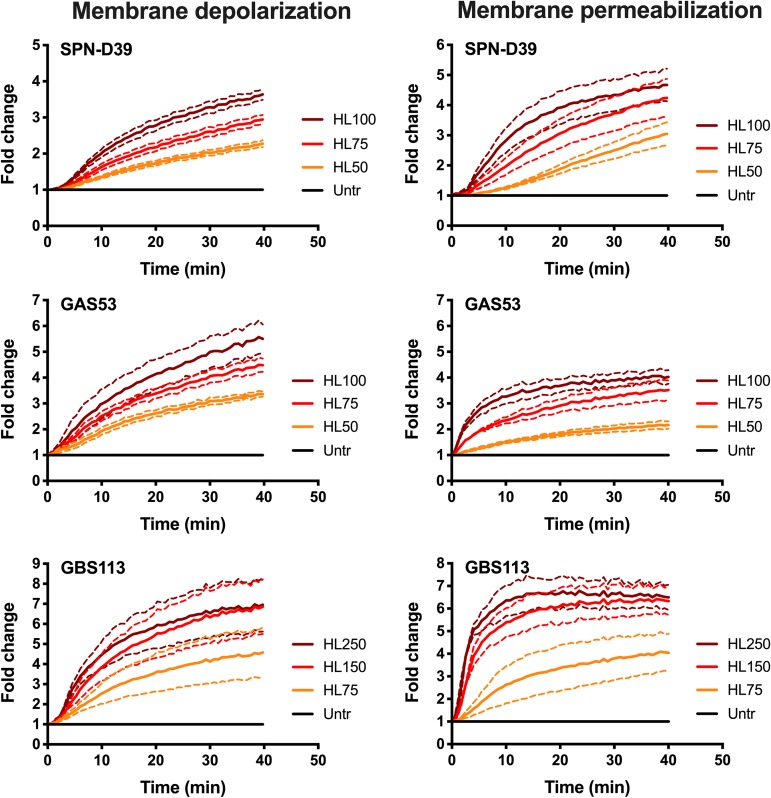
We therefore used the pneumococcal strain D39 as a positive control, as it has been used in previous experiments delineating these mechanisms, and we compared the results using two strains each of GAS and GBS. We first measured membrane depolarization and disruption/permeabilization in S. pneumoniae, GAS, and GBS after exposure to increasing concentrations of HAMLET and showed that all three species responded with a concentration-dependent increase in depolarization (Fig. 2, left panels) and subsequent membrane permeability (Fig. 2, right panels). Analogously with the results in the MIC assays above, a higher concentration of HAMLET was required to obtain a similar level of depolarization and membrane disruption in the slightly more HAMLET-resistant GBS strains.
FIG 2.
HAMLET-induced membrane depolarization and permeabilization in S. pneumoniae, GAS, and GBS. Bacteria were grown in THY, washed in PBS, and resuspended in PBS with 25 mM glucose to energize the cells. DiBAC4(3) and propidium iodide were added to the bacterial suspension, and the cells were allowed to equilibrate for 40 min at 37°C before the experiment was started. At 0 min, HAMLET was added at 50, 75, and 100 μg/ml (labeled HL50, HL75, and HL100, respectively, in the figure) for SPN-D39 and GAS 53 and at 75, 150, and 250 μg/ml (labeled HL75, HL150, and HL250, respectively, in the figure) for GBS 113. (Left panels) Membrane polarity was measured every 30 s using a 485/20-nm excitation and 528/20-nm emission filter combination for 40 min. Depolarization of the membrane is detected through an increased DiBAC4(3) fluorescence over time. (Right panels) Membrane integrity was recorded every 30 s using a 528/20-nm excitation and 605/20-nm emission filter combination for 40 min. Membrane disruption was detected through increased propidium (PI) fluorescence intensity over time. The graphs show the average of results from at least 3 experiments for each strain as a solid line, with dashed lines of the same color representing the standard deviations. Untr, untreated.
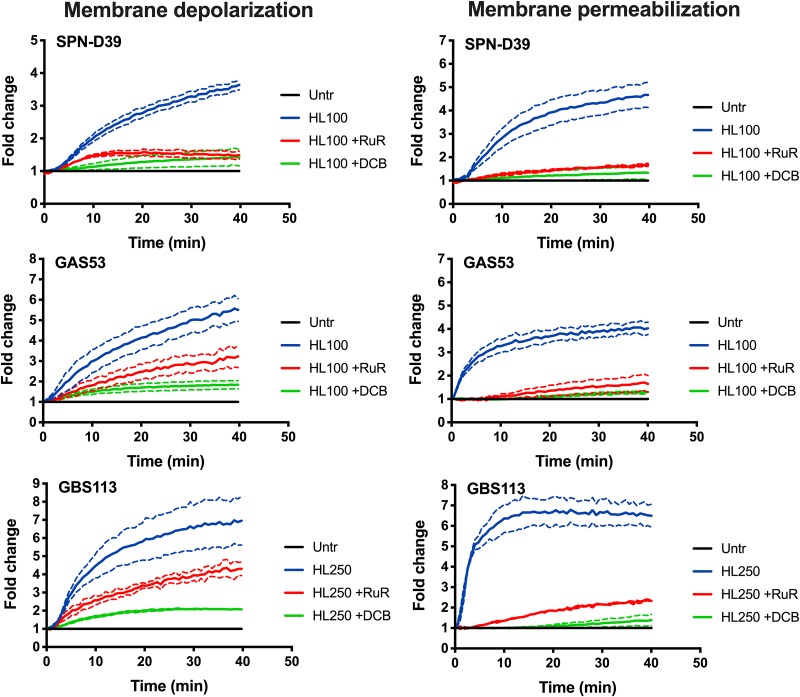
Depolarization and permeabilization of the bacterial cell membrane were effectively inhibited in the presence of RuR and DCB in all three species (Fig. 3), suggesting that the mechanisms involved in HAMLET-induced death leading to membrane disruption and bacterial death were similar in pneumococci, GAS, and GBS. Interestingly, although both inhibitors reduced membrane depolarization, the inhibitory activity of DCB was generally stronger than the inhibition seen in the presence of RuR (Fig. 3, left panels). The significant, albeit varied, reduction in membrane depolarization induced by either inhibitor still resulted in an almost complete inhibition of membrane disruption (Fig. 3, right panels).
FIG 3.
Inhibition of membrane depolarization and permeabilization by calcium and sodium transport inhibitors. Bacteria were grown in THY, washed, and resuspended in PBS with 25 mM glucose to energize the cells. DiBAC4(3) and propidium iodide were added to the bacterial suspension, and the cells were allowed to equilibrate for 40 min at 37°C before the experiment was started. At 0 min, the bacterial cells were pretreated with inhibitors (30 μM [final concentration] ruthenium red [RuR] and 25 μM [final concentration] dichlorobenzamil [DCB]), after which 100 μg/ml (6 μM; for SPN-D39 and GAS strains) and 250 μg/ml (15 μM; for GBS) of HAMLET was added. The samples were immediately read in a fluorescence plate reader every 30 s for 40 min. (Left panels) Membrane polarity was measured using a 485/20-nm excitation and 528/20-nm emission filter combination, and depolarization of the membrane was detected through increased DiBAC4(3) fluorescence over time. (Right panels) Membrane integrity was recorded using a 528/20-nm excitation and 605/20-nm emission filter, and membrane disruption was detected through an increased propidium (PI) fluorescence intensity over time. The graphs show the average of results from 3 experiments for each strain as a solid line, with dashed lines of the same color representing the standard deviation.
Inhibition of sodium and calcium transport by DCB and RuR also resulted in an increased MIC value of HAMLET for all three species tested, suggesting a role for ion transport in HAMLET’s antibacterial activity during cell growth (Fig. S2). The HAMLET MIC for S. pneumoniae strain D39 increased from less than 20 μg/ml in the absence of an inhibitor to >50 μg/ml in the presence of RuR and to 50 μg/ml in the presence of DCB. For both GAS strains, the HAMLET MIC was less than 20 μg/ml in the absence of inhibitors, which increased to over 50 μg/ml in the presence of either RuR or DCB. For GBS strains, the HAMLET MIC was 50 μg/ml for both strains, which increased 4-fold to 200 μg/ml in the presence of inhibitors.
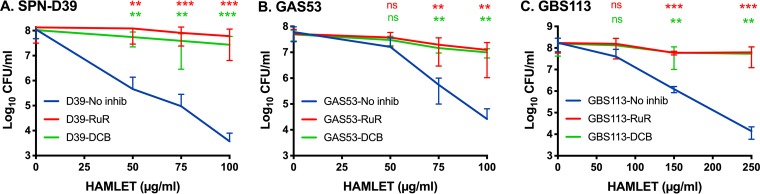
Inhibition of calcium and sodium transport had an even more pronounced effect on HAMLET-induced bacterial death in all three species. Exposure of bacteria to HAMLET concentrations used to induce membrane depolarization and disruption for 1 h resulted in almost complete rescue of bacterial viability (Fig. 4). Both inhibitors significantly and almost completely blocked HAMLET-induced bacterial death in all five strains tested. Taken together, our results reveal that the bactericidal effect of HAMLET on GAS and GBS, in addition to pneumococci, is related to sodium and calcium transport systems in the bacterial cell.
FIG 4.
Inhibition of HAMLET-induced death. Bacteria were grown in THY and washed and resuspended in PBS with 25 mM glucose to keep the bacteria energized. The bacterial suspensions were prepared to obtain a starting concentration of approximately 1 × 108 CFU/ml. The bacteria were preincubated for 5 min at 37°C in the presence or absence of ruthenium red (RuR; 30 μM; red line) or dichlorobenzamil (DCB; 25 μM; green line). Then, increasing concentrations of HAMLET were added to wells of each bacterial strain, and the bacteria were allowed to incubate for 1 h at 37°C. Bacteria were then serially diluted, dilutions were plated onto agar, and colonies were allowed to grow for 24 to 48 h. Viable CFU were counted, and the concentration in numbers of CFU per milliliter was calculated and is depicted in the graphs. (A) S. pneumoniae D39, (B) GAS 53, and (C) GBS 113. The results represent the mean data from at least 5 separate experiments, with standard deviations. Statistical comparison of groups was performed using Welch’s ANOVA, with Dunnett’s multiple-comparison test used for comparisons of individual groups. P values are presented from Dunnett’s comparison of experiments with no inhibitor (No inhib) versus RuR (red asterisks) and no inhibitor versus DCB (green asterisks). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significant difference.
HAMLET enhances the activities of antibiotics against S. pneumoniae.
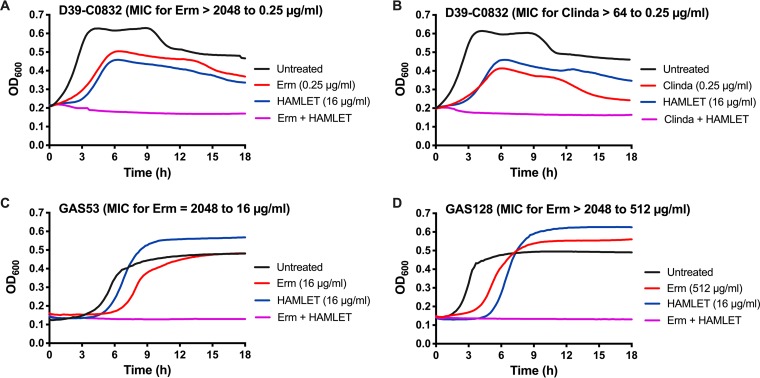
Previous studies have shown that HAMLET, besides having bactericidal activity against certain species, enhances the activities of some antibiotics, such as penicillin, erythromycin, and methicillin, both against strains sensitive to HAMLET-induced killing, such as S. pneumoniae, and against strains resistant to HAMLET’s direct killing activity, such as S. aureus (6, 7). To better understand the scope of HAMLET’s ability to increase bacterial killing or growth inhibition in the presence of antibiotics, we first tested strains with antibiotic cassettes inserted in the genome of the well-defined lab strain D39. These experiments indicated that sublethal or subinhibitory concentrations of HAMLET were able to act in concert with penicillin, a macrolide (Erm), a lincosamide (clindamycin), and kanamycin, as shown by a decrease in the MICs of these antibiotics, whereas no combination effect was observed using tetracycline, chloramphenicol, or streptomycin (Table 3). HAMLET had its strongest effect in combination with Erm and clindamycin, which resulted in an >8,000-fold decrease in the MIC compared with that of erythromycin alone and a >256-fold decrease in the MIC compared with that of clindamycin alone against the erythromycin-resistant strain D39-C0832 carrying an ermB cassette (Table 3; Fig. 5A and B). Combination effects of HAMLET and penicillin against a penicillin-resistant strain and HAMLET and kanamycin against a kanamycin-resistant strain resulted in 4-fold or higher decreases in the respective MIC values measured in the absence of HAMLET.
TABLE 3.
Combination treatment of HAMLET with various antibiotics against isogenic strains carrying resistance cassettes
| Strain name | Antibiotic resistance | Antibiotic MIC (μg/ml) |
Fold reduction MIC | |
|---|---|---|---|---|
| Without HAMLET | With HAMLETb | |||
| SP670a | Penicillin G | 4 | 0.25 | 16 |
| D39-C0832 | Erythromycin (ermB) | 2,048 | 0.25 | 8,192 |
| Clindamycin (ermB) | >64 | 0.25 | >256 | |
| D39-StkP | Kanamycin (aphA3) | >1,024 | 256 | >4 |
| D39-PspC | Tetracycline (tetM) | 8 | 8 | None |
| D39-PitB | Chloramphenicol (cat) | >16 | >16 | None |
| D39-SmR | Streptomycin (rpsL) | >16 | >16 | None |
Clinical isolate SP670 was used, as no penicillin G-resistant strain in the D39 background was available.
The MICs of HAMLET for the strains were similar between the isogenic mutants (30 μg/ml for the HAMLET batch used). For the combination experiments, a sub-MIC concentration of HAMLET, representing 50 to 75% of the MIC (15 to 22 μg/ml) was used.
FIG 5.
HAMLET and erythromycin or clindamycin combination treatment of erythromycin-resistant streptococci. Each bacterial strain was grown in THY in the presence of increasing concentrations of erythromycin (1 μg/ml to 2,048 μg/ml) or clindamycin (0.125 μg/ml to 64 μg/ml) in a 2-fold dilution series with or without the addition of sub-MICs of HAMLET. For each graph, the untreated bacterial growth curve is shown with a black line, the specific erythromycin or clindamycin concentration that resulted in no growth in the presence of HAMLET is shown with a red line, the sub-MIC HAMLET concentration used is shown with a blue line, and the combination treatment (erythromycin [Erm] or clindamycin [Clinda] and HAMLET) is shown with a purple line. In the title of each graph is the name of each strain, the MIC of erythromycin without HAMLET present, and the resulting MIC in the presence of HAMLET. (A and B) D39-C0832 (D39 carrying an ermB cassette) treated with erythromycin (A) or clindamycin (B) alone or in combination with HAMLET. (C and D) GAS 53 (C) and GBS 113 (D) treated with erythromycin alone or in combination with HAMLET. The figure shows a representative graph for each strain and the MIC shift after combination treatment was determined from at least 3 separate experiments in duplicate. The complete data set is presented in Tables 3 and 4.
HAMLET enhances the activities of macrolides against GAS and GBS.
Similarly, subinhibitory concentrations of HAMLET in combination with erythromycin resulted in a decreased MIC for all GAS and GBS strains compared with the MIC of erythromycin alone (Table 4). Our results show that treatment of GAS strains with erythromycin in the presence or absence of sub-MICs of HAMLET revealed a 4- to 256-fold decrease in the erythromycin MIC when HAMLET was present. For the GBS strains, a >2- to 1,024-fold decrease in the erythromycin MIC was observed in the presence of HAMLET. For both GAS and GBS strains, the highest reduction in the MIC value was observed for those strains with the greatest erythromycin resistance, which was most often associated with carrying the ermB gene. Representative growth curves for one GAS strain and one GBS strain are shown in Fig. 5C and D, and they show that the MIC of erythromycin is decreased in the presence of subinhibitory concentrations of HAMLET, similarly to what was observed for S. pneumoniae. This effect was also inhibited by addition of the ion transport inhibitors RuR and DCB. Using GAS 53 and GBS 113 as examples, in the absence of inhibitors, addition of subinhibitory concentrations of HAMLET resulted in a shift of the MIC from 2,048 μg/ml to 16 μg/ml (128-fold) or 8 μg/ml to 1 μg/ml (8-fold) for GAS 53 and GBS 113, respectively (Fig. 5C and D). However, in the presence of RuR or DCB, the shift in the MIC for GAS 53 in the presence of HAMLET was only 2-fold (2,048 to 1,024 μg/ml and 4-fold (2,048 to 512 μg/ml), respectively (data not shown). For strain GBS 113, the MIC did not shift in the presence of RuR and was shifted only 2-fold (from 8 to 4 μg/ml) in the presence of DCB. These results reveal that HAMLET was able to effectively enhance the activity of erythromycin against macrolide-resistant GAS and GBS strains and that ion transport plays an important role also in HAMLET’s antibiotic-potentiating effect.
TABLE 4.
Combination treatment of HAMLET and erythromycin against erythromycin-resistant S. pyogenes and S. agalactiae clinical isolates
| Species and isolate | Erythromycin resistance gene(s) | Erythromycin MIC (μg/ml) |
Fold reduction (MIC) | |
|---|---|---|---|---|
| Without HAMLET | With HAMLETa | |||
| S. pyogenes | ||||
| 6 | mefA and msrD | 8 | 2 | 4 |
| 8 | mefA and msrD | 8 | 0.5 | 16 |
| 138 | mefA and msrD | 8 | 0.5 | 16 |
| 53 | ermB | 2,048 | 8 | 256 |
| 128 | ermB | 2,048 | 512 | 4 |
| 125 | ermTR | 32 | 2 | 16 |
| S. agalactiae | ||||
| 114 | ermB | >2,048 | 4 | >512 |
| 126 | ermB | >2,048 | 8 | >256 |
| 129 | ermB | >2,048 | 1,024 | >2 |
| 51 | ermTR | 16 | 1 | 16 |
| 113 | ermTR | 8 | 1 | 8 |
| 76 | Undetermined | 1,024 | 1 | 1,024 |
| 150 | Undetermined | 32 | 0.5 | 64 |
The MICs of HAMLET against the strains were similar between the isogenic mutants (50 and 125 μg/ml for GAS and GBS strains, respectively, with the HAMLET batch used). For the combination experiments, a sub-MIC concentration of HAMLET, representing 40 to 60% of the MIC (20 to 30 μg/ml for GAS strains and 30 to 75 μg/ml for GBS strains) was used.
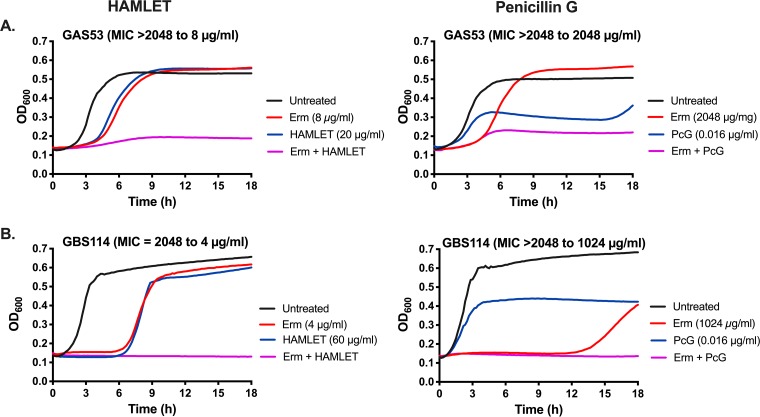
HAMLET potentiates the effect of Erm against Erm-resistant bacteria more effectively than PcG.
Combination treatments, using two or more antimicrobial drugs to treat an infection, are commonly used in clinics, with HIV and tuberculosis treatment regimens as good examples (10–12). As shown above, combination treatment with HAMLET and antibiotics can increase bacterial death and decrease the MIC of an antibiotic to the sensitive range also for bacteria resistant to this antibiotic. To investigate how HAMLET, as an antibacterial agent, compares with another antibiotic, such as penicillin G (PcG; to which these bacteria are sensitive, with MIC values ranging from 0.016 to 0.032 μg/ml) in potentiating the effect of erythromycin against erythromycin-resistant bacteria, combination treatments were performed. Macrolide-resistant GAS and GBS were treated with various concentrations of erythromycin in the presence or absence of sub-MICs of either HAMLET or PcG. In these experiments, the presence of sub-MICs of HAMLET was able to decrease the MIC of Erm 256- and 16-fold for GAS 53 and 125, respectively, and 512- and 1,024-fold for GBS 114 and 76, respectively (Table 4; see also Fig. 6 for MIC graphs for strains GAS 53 and GBS 114). The corresponding effects of combination treatment of Erm with sub-MICs of PcG (i.e., the highest concentration of PcG at which any growth was detected) were only 2- and 4-fold for GAS 53 and 125, respectively, and 4- and 2-fold for GBS 114 and 76, respectively (Fig. 6 presents representative MIC graphs for strains GAS 53 and GBS 114). These results suggest that, at least for erythromycin, combination treatment with sub-MIC HAMLET concentrations resulted in a much greater reduction of the erythromycin MICs for the resistant strains than combination treatment with penicillin G.
FIG 6.
Combination treatment of erythromycin-resistant streptococci with erythromycin and HAMLET or penicillin G. Each bacterial strain was grown in THY in the presence of increasing concentrations of erythromycin (1 μg/ml to 2,048 μg/ml) in a 2-fold dilution series with or without the addition of sub-MICs of HAMLET or penicillin G (PcG). For each graph, the untreated bacterial growth curve is shown with a black line, the specific erythromycin concentration that resulted in no growth in the presence of HAMLET or PcG is shown with a red line, the sub-MIC HAMLET or PcG concentrations used are shown with a blue line, and the combination treatment (erythromycin and HAMLET or PcG) is shown with a purple line. In the title of each graph is the name of each strain, the MIC of erythromycin without HAMLET or PcG, and the resulting MIC in the presence of HAMLET or PcG at the concentration depicted in the graph legend. (A) GAS 53 treated with erythromycin alone or in combination with either HAMLET (left) or PcG (right); (B) GBS 114 treated with erythromycin alone or in combination with either HAMLET (left) or PcG (right). The figure shows a representative graph for each strain and the MIC shift after combination treatment was determined from at least 3 separate experiments in duplicate; the full data set is presented in Results.
DISCUSSION
In 2014, the WHO ratified a resolution to combat antimicrobial resistance and determined this problem to be one of the main threats to human health (1). A recent study that showed that 33,000 deaths from antibiotic-resistant bacteria occurred within the European Economic Area in 2015, an increase of almost 250% since 2007 (13). Data from other parts of the world have shown similarly significant increases in resistance patterns in bacteria over the last few years (14–16). These and other studies show that resistance levels vary between bacterial organisms in different parts of the world and also between various countries or regions within the same economic area and are often linked to antibiotic use in the community (13, 17–19). Actions with regard to both antibiotic use and novel strategies to treat infections with antibiotic-resistant organisms are therefore urgently needed.
In this study, we showed for the first time that HAMLET, a protein-lipid complex from human milk consisting of α-lactalbumin and oleic acid, has bactericidal activity against the pathogenic streptococci S. pyogenes and S. agalactiae, as previously shown for other bacteria (3, 6, 20). Interestingly, the mechanism of bacterial death was associated with both membrane depolarization and membrane disruption, similar to what has been observed for S. pneumoniae (4, 9), suggesting this to be a general mode of action for HAMLET-induced bacterial death. As other agents, including antimicrobial peptides, with direct effects on membrane function and integrity are being explored and tested in clinical studies (21–23), this suggests a potential for the use of HAMLET in future therapy. Similarly, trials have begun to explore the use of lytic bacteriophages as novel antibacterial treatments (24). The potential advantage of HAMLET and other similar strategies is that the target, the bacterial membrane, is difficult for the bacteria to change, resulting in less risk for resistance development.
Besides its bactericidal activity, HAMLET added in combination with antibiotics to which the treated bacteria are resistant resulted in a reversal of antibiotic resistance for both GAS and GBS strains resistant to macrolides and S. pneumoniae strains resistant to various antibiotics. HAMLET’s reversal of antibiotic resistance was strongest for macrolides and lincosamides, whereas no significant effect was observed for other intracellular antibiotics, such as tetracycline and chloramphenicol. Previous and preliminary data from our laboratory has shown that treatment of S. pneumoniae with sublethal concentrations of HAMLET results in an increased association of fluorescent gentamicin, an intracellular aminoglycoside, with the bacterial cells (6), potentially explaining the increased activity of gentamicin in the presence of HAMLET. Thus, increased activities of intracellular antibiotics may well be related to their intracellular accumulation based on the decreased function of energy-dependent export of antibiotics due to a decreased production of ATP in HAMLET-treated cells. However, to what extent such an accumulation occurs for different antibiotics is, at present, unknown and will be an interesting question for future studies. Treatment with HAMLET in combination with an antibiotic resulted in the elimination of bacteria at a concentration that was in the sensitive range based on the EUCAST breakpoints. This is similar to what has been observed using HAMLET combination treatment against other bacterial species, including important, serious-threat, priority pathogens, such as M. tuberculosis, S. aureus, and A. baumannii (5–7). These three organisms, together with penicillin-resistant S. pneumoniae, are listed on the WHO global priority pathogen list, which includes the 13 most critical antibiotic-resistant organisms threatening human health (2). Interestingly, so far, HAMLET has been shown to have either a direct effect or a combination effect with relevant antibiotics on 7 of these 13 organisms, and further research is in progress to investigate the effect of HAMLET directly or in combination with antibiotics for the remaining ones.
Combination treatment regimens or other sensitization strategies are currently being used successfully in clinics or are being explored as promising future strategies (25–27). Classical examples are the regimen of multiple drugs in treating tuberculosis, HIV, and malaria based on the problem with antimicrobial resistance development in these organisms (10, 12, 28–30). Unfortunately, resistance development in both Mycobacterium tuberculosis (10, 31, 32) and HIV (33) is still increasing. Another successful strategy used in clinics is the inclusion of beta-lactamase inhibitors, such as clavulanic acid, to improve the efficacy of beta-lactam treatment; however, strains expressing a novel class of beta-lactamases (metallo-beta-lactamases, causing resistance to carbapenems) are emerging (34).
With the advent of multidrug-resistant bacteria, combination treatment with two or more antibiotics of different classes has also been explored against bacterial infections (25). In vitro and animal infection experiments exploring the activities of combinations of different antibiotic classes have shown synergistic and improved treatment effects against multiple Gram-positive and Gram-negative organisms (35–38), including pneumococci (35, 39, 40). Additionally, clinical efficacy has been shown, especially for colistin or fosfomycin in combination with various antibiotics, in the treatment of infections caused by multidrug-resistant Gram-negative organisms (41, 42). In this study, treatment of macrolide-resistant GAS and GBS with HAMLET and macrolides showed a significantly higher synergistic effect than combination treatment of the same bacteria with macrolides and penicillin. Similarly, our previous results with HAMLET have shown that HAMLET in combination with antibiotics also has efficacy in vivo. HAMLET-antibiotic combination therapy against colonization with antibiotic-resistant S. pneumoniae or S. aureus caused a significant and almost-complete eradication of the bacteria (6, 7). Although HAMLET was superior to penicillin G in killing resistant GAS and GBS strains in combination with macrolides, two aspects need to be considered. First, although macrolides and beta-lactams have different targets that may potentially cause additive or synergistic effects, that was not observed to any higher degree in this study. However, this does not mean that other combinations of antibiotics will not work better for combination treatment against macrolide-resistant streptococci. Second, despite the fact that HAMLET was superior to penicillin G in potentiating killing in the presence of macrolides, future experiments evaluating in vivo efficacy will be required.
These data imply that monotherapy using agents, such as HAMLET, that target molecules and mechanisms to which the bacteria have a hard time becoming resistant (43, 44) should be explored further. Additionally, combination treatments with optimal combinations of antimicrobial agents, especially including agents that are in themselves not antibiotics or have effects that have the potential to escape the evolutionary pressure necessary for the bacteria to initiate resistance development, are potentially viable avenues to future treatment strategies to combat antibiotic resistance.
MATERIALS AND METHODS
Reagents.
Oligonucleotides for PCR were purchased from Invitrogen/ThermoFisher Scientific (for primer sequences [see Table S2 in the supplemental material]). Bacterial growth medium (Todd-Hewitt broth and brain heart infusion broth), yeast extract, and remaining chemicals and inhibitors were purchased from Sigma-Aldrich at the highest purity level available. Penicillin G, tetracycline, kanamycin, clindamycin, and streptomycin stocks were suspended in water, while erythromycin and chloramphenicol stocks were dissolved in ethanol. Antibiotic stocks were diluted at least 25-fold in phosphate-buffered saline (PBS; 30 mM Na2HPO4, 10 mM KH2PO4, 120 mM NaCl, pH 7.4), or growth medium before use in the assays.
Production and characterization of HAMLET from human milk.
Human alpha-lactalbumin purified from human milk was converted into HAMLET by complexing apo-protein (treated with EDTA) with oleic acid (C18:1; Sigma) on a DEAE-containing ion exchange matrix, and HAMLET complex was eluted with salt as described previously (32). The eluted HAMLET was dialyzed with water to remove salt, and the desalted protein-lipid complex was lyophilized and saved at –20°C until use. HAMLET concentrations presented in this paper represent the weight of the total complex per milliliter of solution.
Bacterial strain characteristics and growth conditions.
All pneumococcal strains used in this study are listed in Table S1 in the supplemental material, and the characteristics of the GAS and GBS strains are presented in Table 2. The use of all clinical, antibiotic-resistant isolates for this study was kindly authorized by Gunnar Kahlmeter, Infectious Disease Clinic, Växjö, Sweden, the curator of the collection of strains from the European Committee on Antimicrobial Susceptibility Testing (EUCAST). Streptomycin-resistant D39 was generated by growing D39 on blood agar plates containing 200 μg/ml of streptomycin and isolating spontaneous mutants after overnight growth carrying mutations in the rpsL gene, as described previously (45, 46). Pneumococcal GAS and GBS strains were grown in Todd-Hewitt medium with addition of 0.5% yeast extract (THY) as described previously (47), sterile glycerol was added to a final concentration of approximately 15%, and 1-ml aliquots of each strain were saved at –80°C as starter cultures for each experiment. The genetic basis for erythromycin resistance was determined by PCR using primers listed in Table S2 based on the general protocols presented in references 48, to ,50, with slight modifications and optimization of primer sequences and annealing temperatures. The PCR was performed using reagents from Thermo Fisher Scientific, and the annealing temperature ranged from 50 to 57°C for the various primer pairs, with colonies from plate-grown bacteria serving as the source of template DNA. For ermB, primers designed for the pneumococcal ermB gene were initially used, and for ermTR and mefA, published primer sequences (all listed in Table S2) were initially used to detect the genes in the strain. New primers were designed to optimize the PCR for GAS and GBS strains from which no product was obtained. Despite specifically designing primers for strains GBS 76 and GBS 150, no product was obtained.
In vitro susceptibility tests.
MICs were determined in 96-well microtiter plates using the microdilution method according to approved standards of the CLSI (51), with some modifications. Rather than using the prescribed Mueller-Hinton broth, Todd-Hewitt broth with 0.5% yeast extract (THY) was used, as it is a common medium used for streptococcal growth. As indicated in prior studies, THY produced reliable MIC values for streptococci and required approximately 15% less HAMLET to obtain activity, similar to that on Mueller-Hinton broth (6). To test antibiotic or HAMLET susceptibility alone, a 2-fold dilution series of antibiotics of interest or HAMLET was prepared in duplicate wells. For HAMLET, an initial 2-fold dilution series ranging from 320 μg/ml to 10 μg/ml was used to assess the MICs for pneumococci and a series ranging from 400 μg/ml to 25 μg/ml was used to assess the MICs for GAS and GBS strains. Each well was then seeded with a bacterial concentration of approximately 105 CFU/ml in a total volume of 250 μl THY, and the plate was incubated for 18 h at 37°C in a Synergy 2 microplate reader (BioTek, Winooski, VT). The optical density at 600 nm (OD600) was recorded every 10 min to monitor bacterial growth.
For combination treatments, a 2-fold dilution series of antibiotics of choice was prepared to determine the MIC of each agent as delineated above. At the same time, sub-MICs of HAMLET or penicillin G (the highest concentration of HAMLET or penicillin G that still resulted in bacterial growth) was added to dilution series of erythromycin or clindamycin. Finally, the bacterial suspension was added, and bacterial growth was recorded by monitoring the OD600 as described above. However, when using an inoculum of 105 CFU/ml in these experiments, we observed that addition of sub-MICs of erythromycin resulted in a significant right shift of the bacterial growth curve, which was not observed after addition of, e.g., penicillin G (Fig. S3A, C, and E). This made it difficult to clearly discern the added effects of HAMLET in these assays over the 18-h time period studied. We therefore performed the MIC assays with increased bacterial inoculum concentrations. When we used 107 CFU/ml, the growth curves were left shifted enough to produce more discernible results, without resulting in an effect on the MIC of erythromycin for either the erythromycin-sensitive reference strain ATCC 49619 or the erythromycin-resistant GBS 76 strain (Fig. S3B and D). Thus, all experiments with HAMLET-antibiotic or antibiotic-antibiotic in combination were conducted with this inoculum. The MIC was defined as the lowest concentration of antimicrobial agent solution at which no increase in OD600 was detected over 18 h. For MIC assays involving ion inhibitors, ruthenium Red (RuR; 30 μM) or dichlorobenzamil (DCB; 25 μM) was added to the bacterial suspension 5 min before addition of the antibiotic dilutions.
Associated with the MIC assay, bactericidal concentrations (BCs) were determined as described previously (51) from a separate plate set up identically to the MIC plate. Viable organisms were determined by plating bacterial dilutions from all wells at 3 h after the initiation of the MIC assay. The BC was defined as the lowest concentration of antimicrobials yielding colony counts that were <0.1% (3 log10 reduction) of the initial inoculum (as determined by colony counts from the growth control well immediately after inoculation) after overnight growth on blood agar plates at 37°C (51).
Short-time bactericidal assay.
To assess the bactericidal activity of HAMLET, bacteria were grown in THY to late logarithmic phase (OD600 of approximately 0.6), harvested by centrifugation at 9,000 × g for 2 min in a microcentrifuge, washed once in PBS, and resuspended to the original volume in PBS. The bacterial suspension was energized with 25 mM glucose and treated with the concentrations of HAMLET and/or antibiotics indicated in the figures for various times. In experiments examining ion transport, the ion transport inhibitors RuR (30 μM) and DCB (25 μM) were added to the bacterial suspension 5 min before the addition of HAMLET and antibiotics. Bacterial viability was assessed by plating 10-fold serial dilutions of treated and untreated bacterial suspensions on brain heart infusion agar plates. CFU were counted after overnight growth at 37°C.
Measuring bacterial membrane depolarization and membrane permeabilization.
Membrane polarization and integrity were measured as described in detail in reference 9, with minor modifications. Streptococci were grown to late log phase (OD600 of approximately 0.6) in THY, pelleted by centrifugation at 9,000 × g for 2 min in a microcentrifuge, and washed twice by resuspension in PBS. The bacterial pellet was resuspended in PBS to the original volume and energized by the addition of 25 mM glucose. To measure membrane potential changes of the streptococcal membrane, a final concentration of 250 nM DiBAC4(3) [bis-(1,3-dibutylbarbituric acid) trimethine oxonol; Invitrogen] from a 200× stock in dimethyl sulfoxide (DMSO) was added. Simultaneously, a final concentration of 20 μg/ml propidium iodide (PI; Sigma) from a 50× stock in PBS was added to monitor the integrity of the bacterial membrane. In a 96-well plate, 180 μl of this bacterial suspension was then added to each well of a microtiter plate, and the plate was then placed into a prewarmed (37°C) Synergy 2 multimode microplate reader (BioTek), where fluorescence readings from DiBAC4(3) (485/20-nm excitation, 528/20-nm emission) and PI (528/20-nm excitation, 605/20-nm emission) were recorded every 30 s for 40 min to allow the bacteria to energize and equilibrate. The plate was then programmed to exit the plate reader, and 20 μl of PBS containing vehicle alone or HAMLET and antibiotic combinations was added in the presence or absence of a specific ion transport inhibitor. Triton X-100 (0.1%; Sigma-Aldrich) was used as a positive control for membrane depolarization and rupture to validate each assay (data not shown). The plate was then placed immediately into the fluorescence reader, where fluorescence readings were recorded for another 40 min. The ratio of the fluorescence intensity (in arbitrary units) of the untreated control to that of the HAMLET-antibiotic-treated sample was calculated for the no-inhibitor samples and for the inhibitor samples using all values over the final 40 min and plotted.
Statistical analysis.
GraphPad (Prism 8 software) was used for graph development and statistical analysis. Where applicable, statistical significance was determined by Welch’s analysis of variance (ANOVA) test (assuming nonidentical standard deviations), with Dunnett’s multiple-comparison test for comparisons of individual groups. P values of >0.05 were considered statistically significant. Data are presented as mean values, with error bars representing standard deviations, i.e., the errors of mean values of multiple biological data points, as described in detail in the figure legend for each experiment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Anki Mossberg, Lund University, Lund, Sweden, for purification of the HAMLET batches used in this study, for reading the manuscript, and for providing helpful comments, Gunnar Kahlmeter, Infectious Disease Clinic, Växjö, Sweden, for providing pneumococcal strains from the EUCAST collection, and Birgitta Andersson, Lund University, Malmö, Sweden, for performing the serotyping of these pneumococcal strains.
This study was supported by research grants K2015-99X-22878-01-6 and 2018-05947 from the Swedish Medical Research Council (A.P.H.), by grants from the Alfred Österlund Foundation, Malmö, Sweden (A.P.H.), The Royal Physiographic Society, Lund, Sweden (F.A.), and the Anna and Edwin Berger Foundation, Stockholm, Sweden (K.R.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01193-19.
REFERENCES
- 1.WHO. 2014. Antimicrobial resistance: global report of surveillance. WHO, Geneva, Switzerland. [Google Scholar]
- 2.Tacconelli E, Carmeli Y, Harbarth S, Kahlmeter G, Kluytman J, Mendelson M, Pulcini C, Singh N, Theuretzbacker U. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. WHO, Geneva, Switzerland. [Google Scholar]
- 3.Håkansson A, Svensson M, Mossberg AK, Sabharwal H, Linse S, Lazou I, Lönnerdal B, Svanborg C. 2000. A folding variant of alpha-lactalbumin with bactericidal activity against Streptococcus pneumoniae. Mol Microbiol 35:589–600. doi: 10.1046/j.1365-2958.2000.01728.x. [DOI] [PubMed] [Google Scholar]
- 4.Clementi EA, Marks LR, Duffey ME, Hakansson AP. 2012. A novel initiation mechanism of death in Streptococcus pneumoniae induced by the human milk protein-lipid complex HAMLET and activated during physiological death. J Biol Chem 287:27168–27182. doi: 10.1074/jbc.M112.371070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meikle V, Mossberg AK, Mitra A, Hakansson AP, Niederweis M. 2018. A protein complex from human milk enhances the activity of antibiotics and drugs against Mycobacterium tuberculosis. Antimicrob Agents Chemother 63:e01846-18. doi: 10.1128/AAC.01846-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marks LR, Clementi EA, Hakansson AP. 2012. The human milk protein-lipid complex HAMLET sensitizes bacterial pathogens to traditional antimicrobial agents. PLoS One 7:e43514. doi: 10.1371/journal.pone.0043514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marks LR, Clementi EA, Hakansson AP. 2013. Sensitization of Staphylococcus aureus to methicillin and other antibiotics in vitro and in vivo in the presence of HAMLET. PLoS One 8:e63158. doi: 10.1371/journal.pone.0063158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jorgensen JH, Doern GV, Ferraro MJ, Knapp CC, Swenson JM, Washington JA. 1992. Multicenter evaluation of the use of Haemophilus test medium for broth microdilution antimicrobial susceptibility testing of Streptococcus pneumoniae and development of quality control limits. J Clin Microbiol 30:961–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clementi EA, Marks LR, Roche-Hakansson H, Hakansson AP. 2014. Monitoring changes in membrane polarity, membrane integrity, and intracellular ion concentrations in Streptococcus pneumoniae using fluorescent dyes. J Vis Exp 84:e51008. doi: 10.3791/51008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith T, Wolff KA, Nguyen L. 2013. Molecular biology of drug resistance in Mycobacterium tuberculosis. Curr Top Microbiol Immunol 374:53–80. doi: 10.1007/82_2012_279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corbett EL, Watt CJ, Walker N, Maher D, Williams BG, Raviglione MC, Dye C. 2003. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch Intern Med 163:1009–1021. doi: 10.1001/archinte.163.9.1009. [DOI] [PubMed] [Google Scholar]
- 12.Melo R, Lemos A, Preto AJ, Bueschell B, Matos-Filipe P, Barreto C, Almeida JG, Silva RDM, Correia JDG, Moreira I. 4 September 2018. An overview of antiretroviral agents for treating HIV infection in paediatric population. Curr Med Chem doi: 10.2174/0929867325666180904123549. [DOI] [PubMed] [Google Scholar]
- 13.Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, Colomb-Cotinat M, Kretzschmar ME, Devleesschauwer B, Cecchini M, Ouakrim DA, Oliveira TC, Struelens MJ, Suetens C, Monnet DL, the Burden of AMR Collaborative Group. 2019. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis 19:56–66. doi: 10.1016/S1473-3099(18)30605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zafar A, Hasan R, Nizamuddin S, Mahmood N, Mukhtar S, Ali F, Morrissey I, Barker K, Torumkuney D. 2016. Antibiotic susceptibility in Streptococcus pneumoniae, Haemophilus influenzae and Streptococcus pyogenes in Pakistan: a review of results from the Survey of Antibiotic Resistance (SOAR) 2002–15. J Antimicrob Chemother 71(Suppl 1):i103–i109. doi: 10.1093/jac/dkw076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torumkuney D, Chaiwarith R, Reechaipichitkul W, Malatham K, Chareonphaibul V, Rodrigues C, Chitins DS, Dias M, Anandan S, Kanakapura S, Park YJ, Lee K, Lee H, Kim JY, Lee Y, Lee HK, Kim JH, Tan TY, Heng YX, Mukherjee P, Morrissey I. 2016. Results from the Survey of Antibiotic Resistance (SOAR) 2012–14 in Thailand, India, South Korea and Singapore. J Antimicrob Chemother 71(Suppl 1):i3–i19. doi: 10.1093/jac/dkw073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherazard R, Epstein M, Doan TL, Salim T, Bharti S, Smith MA. 2017. Antimicrobial resistant Streptococcus pneumoniae: prevalence, mechanisms, and clinical implications. Am J Ther 24:e361–e369. doi: 10.1097/MJT.0000000000000551. [DOI] [PubMed] [Google Scholar]
- 17.Alividza V, Mariano V, Ahmad R, Charani E, Rawson TM, Holmes AH, Castro-Sánchez E. 2018. Investigating the impact of poverty on colonization and infection with drug-resistant organisms in humans: a systematic review. Infect Dis Poverty 7:76. doi: 10.1186/s40249-018-0459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang KL, Caffrey NP, Nóbrega DB, Cork SC, Ronksley PE, Barkema HW, Polachek AJ, Ganshorn H, Sharma N, Kellner JD, Ghali WA. 2017. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: a systematic review and meta-analysis. Lancet Planet Health 1:e316–e327. doi: 10.1016/S2542-5196(17)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mölstad S, Löfmark S, Carlin K, Erntell M, Aspevall O, Blad L, Hanberger H, Hedin K, Hellman J, Norman C, Skoog G, Stålsby-Lundborg C, Tegmark Wisell K, Åhrén C, Cars O. 2017. Lessons learnt during 20 years of the Swedish strategic programme against antibiotic resistance. Bull World Health Organ 95:764–773. doi: 10.2471/BLT.16.184374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hakansson AP, Roche-Hakansson H, Mossberg AK, Svanborg C. 2011. Apoptosis-like death in bacteria induced by HAMLET, a human milk lipid-protein complex. PLoS One 6:e17717. doi: 10.1371/journal.pone.0017717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldberg K, Sarig H, Zaknoon F, Epand RF, Epand RM, Mor A. 2013. Sensitization of gram-negative bacteria by targeting the membrane potential. FASEB J 27:3818–3826. doi: 10.1096/fj.13-227942. [DOI] [PubMed] [Google Scholar]
- 22.Balakrishna R, Wood SJ, Nguyen TB, Miller KA, Suresh Kumar EV, Datta A, David SA. 2006. Structural correlates of antibacterial and membrane-permeabilizing activities in acylpolyamines. Antimicrob Agents Chemother 50:852–861. doi: 10.1128/AAC.50.3.852-861.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor PL, Rossi L, De Pascale G, Wright GD. 2012. A forward chemical screen identifies antibiotic adjuvants in Escherichia coli. ACS Chem Biol 7:1547–1555. doi: 10.1021/cb300269g. [DOI] [PubMed] [Google Scholar]
- 24.Vazquez R, Garcia E, Garcia P. 2018. Phage lysins for fighting bacterial respiratory infections: a new generation of antimicrobials Front Immunol 9:2252. doi: 10.3389/fimmu.2018.02252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Worthington RJ, Melander C. 2013. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol 31:177–184. doi: 10.1016/j.tibtech.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gill EE, Franco OL, Hancock RE. 2015. Antibiotic adjuvants: diverse strategies for controlling drug-resistant pathogens. Chem Biol Drug Des 85:56–78. doi: 10.1111/cbdd.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright GD. 2016. Antibiotic adjuvants: rescuing antibiotics from resistance. Trends Microbiol 24:862–871. doi: 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Richman DD. 2001. HIV chemotherapy. Nature 410:995–1001. doi: 10.1038/35073673. [DOI] [PubMed] [Google Scholar]
- 29.Mitchison D, Davies G. 2012. The chemotherapy of tuberculosis: past, present and future. Int J Tuber Lung Dis 16:724–732. doi: 10.5588/ijtld.12.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nosten F, White NJ. 2007. Artemisinin-based combination treatment of falciparum malaria. Am J Trop Med Hyg 77:181–192. doi: 10.4269/ajtmh.2007.77.181. [DOI] [PubMed] [Google Scholar]
- 31.Trofimov V, Costa-Gouveia J, Hoffmann E, Brodin P. 2017. Host-pathogen systems for early drug discovery against tuberculosis. Curr Opin Microbiol 39:143–151. doi: 10.1016/j.mib.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 32.Rojano B, Caminero JA, Hayek M. 2019. Curving tuberculosis: current trends and future needs. Ann Glob Health 85:5. doi: 10.5334/aogh.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aves T, Tambe J, Siemieniuk RA, Mbuagbaw L. 2018. Antiretroviral resistance testing in HIV-positive people. Cochrane Database Syst Rev 11:CD006495. doi: 10.1002/14651858.CD006495.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dortet L, Poirel L, Nordmann P. 2014. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res Int 2014:249856. doi: 10.1155/2014/249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Desbiolles N, Piroth L, Lequeu C, Neuwirth C, Portier H, Chavanet P. 2001. Fractional maximal effect method for in vitro synergy between amoxicillin and ceftriaxone and between vancomycin and ceftriaxone against Enterococcus faecalis and penicillin-resistant Streptococcus pneumoniae. Antimicrob Agents Chemother 45:3328–3333. doi: 10.1128/AAC.45.12.3328-3333.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Entenza JM, Moreillon P. 2009. Tigecycline in combination with other antimicrobials: a review of in vitro, animal and case report studies. Int J Antimicrob Agents 34:8.e1–8.e9. doi: 10.1016/j.ijantimicag.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 37.Entenza JM, Giddey M, Vouillamoz J, Moreillon P. 2010. In vitro prevention of the emergence of daptomycin resistance in Staphylococcus aureus and enterococci following combination with amoxicillin/clavulanic acid or ampicillin. Int J Antimicrob Agents 35:451–456. doi: 10.1016/j.ijantimicag.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 38.Kastoris AC, Rafailidis PI, Vouloumanou EK, Gkegkes ID, Falagas ME. 2010. Synergy of fosfomycin with other antibiotics for Gram-positive and Gram-negative bacteria. Eur J Clin Pharmacol 66:359–368. doi: 10.1007/s00228-010-0794-5. [DOI] [PubMed] [Google Scholar]
- 39.Drago L, Nicola L, Rodighiero V, Larosa M, Mattina R, De Vecchi E. 2011. Comparative evaluation of synergy of combinations of beta-lactams with fluoroquinolones or a macrolide in Streptococcus pneumoniae. J Antimicrob Chemother 66:845–849. doi: 10.1093/jac/dkr016. [DOI] [PubMed] [Google Scholar]
- 40.Cottagnoud P, Acosta F, Cottagnoud M, Neftel K, Tauber MG. 2000. Synergy between trovafloxacin and ceftriaxone against penicillin-resistant pneumococci in the rabbit meningitis model and in vitro. Antimicrob Agents Chemother 44:2179–2181. doi: 10.1128/aac.44.8.2179-2181.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petrosillo N, Ioannidou E, Falagas ME. 2008. Colistin monotherapy vs. combination therapy: evidence from microbiological, animal and clinical studies. Clin Microbiol Infect 14:816–827. doi: 10.1111/j.1469-0691.2008.02061.x. [DOI] [PubMed] [Google Scholar]
- 42.Bassetti M, Righi E. 2015. New antibiotics and antimicrobial combination therapy for the treatment of gram-negative bacterial infections. Curr Opin Crit Care 21:402–411. doi: 10.1097/MCC.0000000000000235. [DOI] [PubMed] [Google Scholar]
- 43.Czaplewski L, Bax R, Clokie M, Dawson M, Fairhead H, Fischetti VA, Foster S, Gilmore BF, Hancock RE, Harper D, Henderson IR, Hilpert K, Jones BV, Kadioglu A, Knowles D, Ólafsdóttir S, Payne D, Projan S, Shaunak S, Silverman J, Thomas CM, Trust TJ, Warn P, Rex JH. 2016. Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect Dis 16:239–251. doi: 10.1016/S1473-3099(15)00466-1. [DOI] [PubMed] [Google Scholar]
- 44.Allen HK, Trachsel J, Looft T, Casey TA. 2014. Finding alternatives to antibiotics. Ann N Y Acad Sci 1323:91–100. doi: 10.1111/nyas.12468. [DOI] [PubMed] [Google Scholar]
- 45.Lipsitch M, Dykes JK, Johnson SE, Ades EW, King J, Briles DE, Carlone GM. 2000. Competition among Streptococcus pneumoniae for intranasal colonization in a mouse model. Vaccine 18:2895–2901. doi: 10.1016/S0264-410X(00)00046-3. [DOI] [PubMed] [Google Scholar]
- 46.Sung CK, Li H, Claverys JP, Morrison DA. 2001. An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl Environ Microbiol 67:5190–5196. doi: 10.1128/AEM.67.11.5190-5196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tyx RE, Roche-Hakansson H, Hakansson AP. 2011. Role of dihydrolipoamide dehydrogenase in regulation of raffinose transport in Streptococcus pneumoniae. J Bacteriol 193:3512–3524. doi: 10.1128/JB.01410-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woodbury RL, Klammer KA, Xiong Y, Bailiff T, Glennen A, Bartkus JM, Lynfield R, Van Beneden C, Beall BW, Active Bacterial Core Surveillance Team. 2008. Plasmid-borne erm(T) from invasive, macrolide-resistant Streptococcus pyogenes strains. Antimicrob Agents Chemother 52:1140–1143. doi: 10.1128/AAC.01352-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daly MM, Doktor S, Flamm R, Shortridge D. 2004. Characterization and prevalence of MefA, MefE, and the associated msr(D) gene in Streptococcus pneumoniae clinical isolates. J Clin Microbiol 42:3570–3574. doi: 10.1128/JCM.42.8.3570-3574.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seppälä H, Skurnik M, Soini H, Roberts MC, Huovinen P. 1998. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob Agents Chemother 42:257–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fuchs PC, Barry AL, Brown SD. 2002. In vitro bactericidal activity of daptomycin against staphylococci. J Antimicrob Chemother 49:467–470. doi: 10.1093/jac/49.3.467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.