The accessory protein Nef of human immunodeficiency virus (HIV) is a primary determinant of viral pathogenesis. Nef is abundantly expressed during infection and reroutes a variety of cell surface proteins to disrupt host immunity and promote the viral replication cycle. Nef counteracts host defenses by sequestering and/or degrading its targets via the endocytic and secretory pathways. Nef does this by physically engaging a number of host trafficking proteins.
KEYWORDS: antiviral restriction factors, host-pathogen, immune receptors, lentiviruses, Nef, human immunodeficiency virus, protein structure, simian immunodeficiency virus, trafficking
ABSTRACT
The accessory protein Nef of human immunodeficiency virus (HIV) is a primary determinant of viral pathogenesis. Nef is abundantly expressed during infection and reroutes a variety of cell surface proteins to disrupt host immunity and promote the viral replication cycle. Nef counteracts host defenses by sequestering and/or degrading its targets via the endocytic and secretory pathways. Nef does this by physically engaging a number of host trafficking proteins. Substantial progress has been achieved in identifying the targets of Nef, and a structural and mechanistic understanding of Nef’s ability to command the protein trafficking machinery has recently started to coalesce. Comparative analysis of HIV and simian immunodeficiency virus (SIV) Nef proteins in the context of recent structural advances sheds further light on both viral evolution and the mechanisms whereby trafficking is hijacked. This review describes how advances in cell and structural biology are uncovering in growing detail how Nef subverts the host immune system, facilitates virus release, and enhances viral infectivity.
INTRODUCTION
Human immunodeficiency virus (HIV) efficiently manipulates host protein trafficking machinery to promote viral replication, evade immune killing, and circumvent antiviral restriction factors (1, 2). These mechanisms of viral pathogenesis are primarily orchestrated by the abundantly expressed HIV protein Nef within the endocytic and late secretory pathways. Nef is a nonenzymatic accessory protein encoded by the HIV type 1 (HIV-1), HIV-2, and simian immunodeficiency virus (SIV) genomes (3). Nef associates with the cytosolic leaflet of various cell membranes via N-myristoylation at G2 of the Nef anchor domain (PDB identifier [ID] 1QA5) (4, 5). Though a significant portion of Nef remains cytosolic during infection (6–10), membrane association is essential for Nef’s engagement with the host cell trafficking machinery, in particular, the clathrin-coated vesicle (CCV) machinery. Nef hijacks the CCV pathway to redirect specific transmembrane proteins away from the cell surface via either sequestration or lysosomal degradation mechanisms (Fig. 1). The end product is the effective downregulation of specific restriction factors, the cell surface proteins that signal infection to the adaptive immune system, and proteins interfering with nascent virion release.
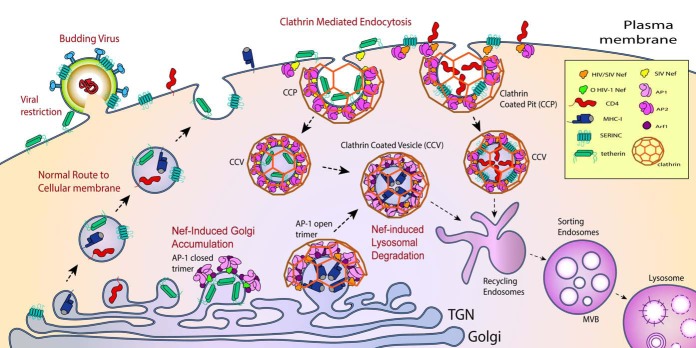
FIG 1.
Nef downregulates host factors. Schematic representation of the clathrin machinery hijacked for CD4 (red), tetherin (green), MHC-I (blue), and SERINC3/5 (teal) downregulation by HIV-1 Nef. These host factors are synthesized in the ER and transported to the Golgi apparatus and then the TGN. From the TGN, these proteins are shuttled to the plasma membrane (PM) via the constitutive secretory pathway where they engage in viral restriction and immune signaling. For both SIV and HIV, Nef-induced downregulation of CD4 and SERINCs occurs at the PM by linking these host factors to AP-2. For the case of CD4, it is believed that a tripartite assembly of Nef-CD4-AP-2 is formed. For SERINCs, this is not known. The Nef-AP-2 assembly triggers clathrin recruitment and budding of CD4 and SERINC3/5 into clathrin-coated pits (CCPs) and eventual budding into clathrin-coated vesicles (CCVs). These CCVs are then routed into a lysosomal degradation pathway. SIVs utilize the same pathway to downregulate simian tetherin. Both SIV and HIV Nefs downregulate newly synthesized MHC-I molecules by rerouting them into the endolysosomal system. This involves the formation of a tripartite assembly of Nef-MHC-I-AP-1 at the TGN membrane, which sequesters MHC-I molecules into CCVs. These CCVs are targeted to MVBs that will eventually fuse with the lysosomes, leading to MHC-I degradation. O-type HIV-1 Nef also targets human tetherin at the TGN similarly to targeting MHC-1, but the result is a Golgi accumulation mechanism of downregulation where Nef traps tetherin at the TGN in an AP-1-dependent manner. Nef-induced downregulation at the TGN is dependent on the formation of Arf1-mediated open and closed trimer of AP-1 for engagement with MHC-I and tetherin, respectively.
The three-dimensional structure of the 27- to 35-kDa Nef protein has been determined by both X-ray crystallography (11, 12) and nuclear magnetic resonance (5, 13–15) (Table 1). Nef consists of a folded core (residues 79 to 203, HIV-1 NL4-3 numbering), flexible N-terminal (residues 1 to 78) and C-terminal (residues 204 to 206) loops, and a central dileucine motif-bearing loop (residues 149 to 179) extending from the core. Depending on the structure determination, the dileucine loop can be either ordered or disordered (Table 1). Nef’s flexible regions are both easily accessible and capable of undergoing conformational changes, making them ideal for binding an array of host cell proteins (3, 16). To date, more than 70 Nef-interacting proteins in the human proteome have been identified (17, 18). The remarkable conformational plasticity of Nef has been explored in detail through X-ray crystallography (19, 20) and cryo-electron microscopy (cryo-EM) (21, 22) structural determinations of Nef in complexes with various cellular binding partners (Table 1).
TABLE 1.
PDBs containing Nef
| Structure composition | PDB ID(s) | Reference or source |
|---|---|---|
| Nef PDBs without dileucine motif resolved | ||
| HIV-1 Nef alone | 1AVV | 11 |
| 2Xl1 | 183 | |
| 6B72 | 154 | |
| 2NEF | 14 | |
| 3TB8 | Unpublished dataa | |
| HIV-1 Nef anchor domain | 5 | |
| Unmyristoylated | 1QA4 | |
| Myristoylated | 1QA5 | |
| HIV-1 Nef in complex with FYN SH3 | 1AVZ | 11 |
| 1EFN | 12 | |
| 4D8D | Unpublished datab | |
| HIV-1 Nef in complex with Hck SH3 | 3REA, 3REB | 184 |
| 4U5W | 185 | |
| HIV-1 Nef in complex with MHC-1 cytoplasmic domain and AP-1 μ1 CTD | 4EMZ, 4EN2 | 19 |
| SIVmac239 Nef in complex with TCR ξ peptide | 3IK5 | 125 |
| HIV-1 Nef protein in complex with antibody sdAb19 and an Hck SH3 domain | 4ORZ | 186 |
| Nef PDBs with dileucine motif resolved | ||
| HIV-1 Nef in complex with Hck SH3 | 3RBB | 187 |
| SIVmac239 Nef in complex with Hck SH3 | 5NUH | 83 |
| SIVmac239 Nef in complex with CD4-like peptide | 5NUI | 83 |
| SIVsmm Nef in complex with AP-2 | 6OWT | 113 |
| HIV-1 Nef bound AP-2 α-σ2 complex | 4NEE | 20 |
| HIV-1 Nef bound to AP-1-Arf1-tetherin complex | 6CM9 | 21 |
C. A. Dennis, M. Harris, and J. Jaeger, unpublished data.
A. Lugani, C. C. Lin, X. Shi, F. Hoh, L. Ponchon, C. Ktori, C. Dumas, J. E. Ladbury, Y. Collette, X. Morelli, and S. T. Arold, unpublished data.
The Nef protein is not essential for virus replication in vitro but is critical for viral replication and infectivity in vivo. Patients infected with a strain of HIV-1 lacking nef do not progress to AIDS, or they do so very slowly (23–25). To combat increasing drug resistance, new antiretroviral targets are being sought against HIV-1-interacting host proteins essential for immune evasion and proliferation. Thus, Nef’s host interactors are attractive pharmacological targets, as they are not subject to viral evolution and drug resistance, so long as sites can be found that are nonessential for normal host functions. In our experience working at the interface of HIV and SIV virology, membrane traffic, and structural biology, we have found that the information transfer between these three fields can be rate limiting for progress. This review is intended to synthesize information across these disciplines for the benefit of those working in all three areas.
TARGETS OF Nef COOPTATION
Clathrin-coated vesicle machinery.
Clathrin-mediated endocytosis (CME) is the primary mechanism by which transmembrane proteins, integral membrane proteins, and lipids are routed from the plasma membrane to the endosomal system in CCVs. CME thus plays a pivotal role in regulating plasma membrane proteostasis. In CME, the heterotetrameric clathrin adaptor protein complex (AP-2) connects clathrin on the one hand to membrane protein substrates and to lipids on the other. AP-2 is a heterotetramer composed of α, β2, μ2, and σ2 subunits. The N-terminal solenoidal “trunk” domains of α and β2, together with the whole μ2 and σ2 subunits, constitute the core of the complex, whereas the C-terminal “hinge” and “ear” domains of α and β2 subunits form long projections extending from the core (26).
AP-2 coordinates clathrin-coated pit (CCP) formation in CME and binds cargoes, which are normally integral membrane proteins, containing acidic dileucine (D/E)xxxL[L/I] (27) and YxxΦ (where Φ is a bulky hydrophobic residue) (28) endocytic motifs. The dileucine binding site is located on the α-σ2 hemicomplex and the tyrosine motif binding site is on the C-terminal domain (CTD) of the μ2 subunit. While Nef is not a transmembrane protein, its dileucine-based motif is a major determinant of its ability to interact with AP-2 (29). AP-2 cargo binding is initiated by a conformational change from the locked (inactive) cytosolic state to an unlocked (active) state. Unlocking is initiated through binding membranes containing phosphoinositide phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] (30) (Fig. 2A). PtdIns(4,5)P2 is enriched at the plasma membrane relative to other cellular compartments and is enriched further during CME. In the locked state (PDB ID 2VGL) (30), the μ2 CTD is nestled between the trunk domains of the α and β2 subunits. In the unlocked state (PDB ID 2XA7) (31), the μ2 CTD undergoes a very large motion in the course of which it is dislodged from the center of the complex and becomes poised to bind to membranes. The remainder of the complex relaxes and opens up to a lesser degree. Only when unlocked are the (D/E)xxxL[L/I] and YxxΦ endocytic cargo binding sites and the canonical clathrin box motif (LLNLD) exposed, resulting in the recruitment of clathrin and the initiation of CME (32). All Nefs interact with the unlocked state of AP-2 to downregulate host cell factors, including cluster of differentiation 4 (CD4), CD8, CD28, CD3, serine incorporator 3 (SERINC3), and SERINC5, while SIV, but not HIV, Nefs also downregulate tetherin in this way (33–41) (Fig. 1, 2A, and 3). Most lentiviral Nef proteins reduce the cell surface expression of various chemokine receptors, such as CXCR4 and CCR5. A conserved DRY motif in the second intracellular loop of these receptors is critical for Nef-dependent downregulation, but the mechanism is not understood at the structural level (42, 43).
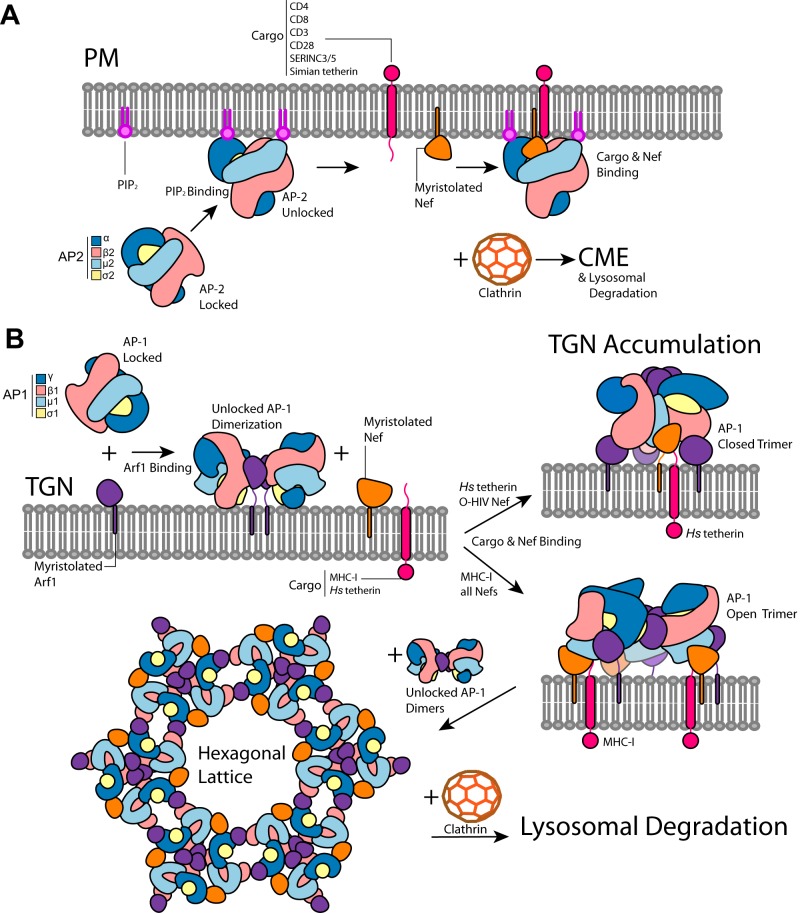
FIG 2.
Nef-dependent downregulation of host factors is dependent on clathrin adaptor proteins AP-2 and AP-1. (A) Locked AP-2 interacts with phosphatidylinositol 4,5-bisphosphate (PIP2) at the plasma membrane (PM) where it unlocks to expose its tyrosine (YxxΦ) and dileucine (ExxxLL) cargo binding sites. Nef can interact with unlocked AP-2 and specific host factors at these sites to force the downregulation of the targeted host factor. The host factors targeted by Nef include CD4, CD8, CD3, CD28, SERINC3/5, and simian tetherin. Nef accomplishes this by inducing clathrin-mediated endocytosis (CME) and shuttling the host factor into the lysosomal degradation pathway. (B) Locked AP-1 interacts with Arf1 at the trans-Golgi network (TGN), where it unlocks and forms dimers. Upon binding cargo and Nef at its cargo binding sites, AP-1 can form either closed or open trimers, depending on the specific cargo and the type of Nef, which is centered at a trimeric Arf1 interface. Host factors targeted at the TGN by Nef include MHC-I and human (Hs) tetherin. Closed and open trimers represent the distinct Nef-induced downregulation mechanisms of TGN accumulation and lysosomal degradation, respectively. In the case of the Nef-induced lysosomal degradation mechanism, with additional engagement of unlocked AP-1 dimers, Nef-bound open trimers can form a large hexagonal lattice which matches the hexagonal lattice of clathrin. This lattice binds clathrin to form CCVs and shuttles the bound cargo into the lysosomal degradation pathway.
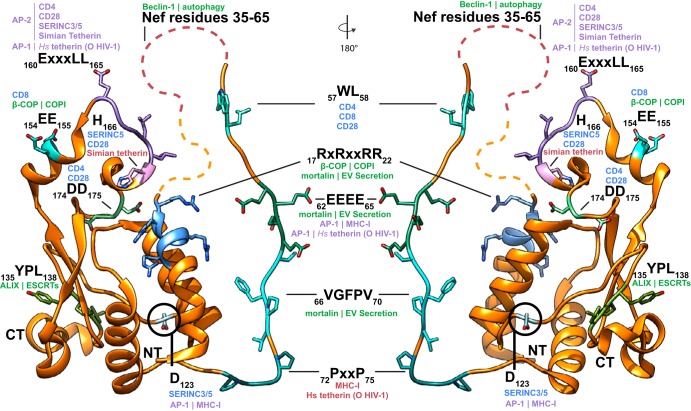
FIG 3.
Nef sites of interaction responsible for host factor downregulation and engagement with host trafficking machinery. Ribbon diagram of a composite structure of HIV-1 NL4-3 (orange). Residues 5 to 24 and 55 to 79 are from PDB ID 4EMZ, and residues 80 to 203 are from PDB ID 4NEE. Composite structure was built by aligning the Nef core of the two PDBs and combining them into a single PDB in Chimera. Where no structural data are available, the disordered regions are represented as dashed lines. For clarity, Nef residues 35 to 65 are represented by a single disordered region though they overlap the 57WL58 and 62EEEE65 sites of interaction. Sites of host protein engagement are highlighted with various colors, and their interacting residues are modeled. Human is abbreviated Hs. Sites that directly bind host factors, resulting in their downregulation, are in red text. Direct AP-binding sites required for host factor downregulation are in purple text; the labels indicate which AP binds at the highlighted site, followed by which host factors are downregulated as a result. Sites implicated in cargo binding, scaffolding, and other non-AP interactions in host substrate downregulation are in blue text. Putative Nef engagement sites of nonclathrin trafficking machinery, direct or indirect, are indicated in green text; the interacting protein is listed, followed by the trafficking machinery affected by binding. The N terminus (NT) and C terminus (CT) of Nef are labeled for reference.
AP-1 is the heterotetrameric clathrin adaptor complex devoted to the biogenesis of CCVs at the trans-Golgi network (TGN) (44, 45) (Fig. 1). AP-1 consists of the large subunits β1 and γ, the medium subunit μ1, and the small subunit σ1 (26, 46). Both β1 and γ bind to the Golgi-residing small G protein Arf1 (47). Like AP-2, AP-1 binds to cargo bearing either tyrosine or dileucine-based motifs (Fig. 2B). Like AP-2, AP-1 is in a locked conformation in the absence of activation (PDB ID 1W63) (48). Unlike AP-2, AP-1 is not responsive to PtdIns(4,5)P2. Instead, Arf1-GTP triggers the recruitment of AP-1 to the TGN membrane, in the course of which it unlocks AP-1 and promotes its assembly into dimers and trimers (PDB IDs 4HMY and 6CM9) (Fig. 2B) (21, 22, 47). Unlocked AP-1 can enter the same open conformation seen for unlocked AP-2, but unlike AP-2, it has also been observed in an even more open conformation known as the hyperunlocked state (PDB ID 4P6Z) (49). In the hyperunlocked state, Arf1-linked dimers and trimers of AP-1 can assemble into an extended hexagonal coat whose symmetry and dimensions match those of clathrin (Fig. 2B) (22). While a similar type of Arf1-dependent coat assembly can be seen in coat protein I (COPI) (PDB ID 5A1V) (50), no evidence has emerged that Arf1 interacts with AP-2 in CME. Thus, Arf1-stabilized coat formation is an additional complication pertaining to AP-1- but not AP-2-dependent sorting. All Nefs hijack AP-1 to downregulate major histocompatibility complex class I (MHC-I), while HIV-1 group O Nefs appear to be unique in their use of AP-1 to target tetherin in humans (51) (Fig. 3).
The AP-3 clathrin adaptor is required for cargo-selective transport to late endosomes. Its δ-σ3 hemicomplex has the conserved binding site for the dileucine motif, which was mapped by a yeast two-hybrid assay in vitro (52, 53). AP-3 is not known to function in Nef-dependent host factor downmodulation. However, Nef proteins colocalize with AP-3 in HeLa cells and mediate AP-3 membrane recruitment in an Arf1-independent manner (54), suggesting that an as-yet-unidentified role for Nef-recruited AP-3 is still possible.
Coatomer.
Nef has many other host partners involved in intracellular trafficking in addition to the AP complexes. These range from the endolysosomal system to autophagy (55–57). Nef binds to β-COP, a component of the COPI protein coat. COPI-coated vesicles mediate the retrograde trafficking of Golgi-derived vesicles to the endoplasmic reticulum (ER) and have also been proposed to participate in the maturation of multivesicular bodies (MVBs) and late endosomes (58, 59). A subset of COPI, also referred as endosomal COPs, containing β-, β′-, and ζ-COP, are found on endosomes (60). The Nef-β-COP interaction was first identified in a yeast two-hybrid screen for cellular proteins that interacted with Nef. The study revealed binding between the C termini of β-COP and Nef, which was later validated through in vitro and in vivo pulldowns with rhesus SIV Nef (61). Further studies have shown that the 154EE155 (diacidic) and 17RxR19 motifs of Nef are important for maintaining this binding interface (Fig. 3) (62, 63). However, in seemingly contradictory experimentation, mutations in the diacidic motif did not disrupt Nef and β-COP binding or CD4 downregulation (64). One possible explanation is that Nef can still bind via the 17RxR19 motif. This model is supported by the functional separation of MHC-I and CD4 degradation between the 17RxR19 and diacidic motifs of Nef, respectively (63). The specificity of these interfaces may help explain the destinations of cargo regulated by Nef as well. The Nef-β-COP interaction is a promising area for the further study of Nef-mediated hijacking of the host trafficking system and the multifaceted functions of the COPI in the endogenous context.
ESCRTs.
Nef is thought to affect endolysosomal trafficking by binding to components of the endosomal sorting complex required for transport (ESCRT) machinery. The ESCRT proteins play an essential role in the maturation and budding of intraluminal vesicles (ILVs) into endosomes, exosome biogenesis and release, and the release of many enveloped viruses, including HIV (65, 66). Endosomes possessing ILVs are referred to as multivesicular bodies (MVBs). The ILVs and their contents are degraded when the MVBs fuse with lysosomes. ESCRT proteins direct physiological cargo sorting into these MVBs, so it is natural to suppose that interactions between Nef and ESCRTs could be the mechanism for directing Nef cargoes to degradation in lysosomes. The main reported interaction between Nef and the ESCRT machinery occurs through the protein ALIX (67). ALIX functions in both ubiquitin-dependent and -independent cargo sorting and recruits the ESCRT-III protein CHMP4, which is directly responsible for ILV release from the endosome-limiting membrane (68). The V domain of ALIX binds tyrosine-based motifs (YPXnL) in late domains from HIVGag and SIVGag (69–74). Nef is reported to interact with ALIX using a 135YPL138 sequence that is conserved among HIV-1 and SIV strains (Fig. 3) (67). Though this motif is partially buried within the Nef core (75), functional studies have shown that eliminating this motif from Nef decreases viral yield from HIV-1-infected macrophages and that knockdown of ALIX reduces CD4 degradation without perturbing the degradation of epidermal growth factor (EGF), an established MVB cargo (67, 76). In the case of SIVs, it is an N-terminal flexible loop tyrosine-based motif (Y28GRL) in SIV Nef that ALIX binds (75). Immunofluorescence studies have also shown that the Nef-ALIX interaction occurs at late endosomes and lysosomes, providing microscopic evidence for the suspected role of ALIX in trafficking Nef-bound cargo for lysosomal degradation (76).
Exosomes.
Exosomes, an extracellular vesicle (EV) of endosomal origin, are heterogeneous membrane-enclosed structures released by cells into the cellular milieu which can transfer material from one cell to another (77). Nefs from both HIV and SIV are secreted from HIV-1-infected cells into EVs, providing a potential mechanism for cellular nonautonomous host-virus interactions and unconventional transmission of infection (78–80). Nef-positive EVs are capable of membrane fusion and depositing their contents, including Nef, into recipient cells (80). Further, evidence suggests that Nef facilitates its own secretion by increasing the production of components of the endosomal/exosomal pathway, particularly multivesicular bodies (81, 82). This could represent a novel mechanism whereby lentiviruses can influence CD4+ T cells as well as naive and noninfectible (CD4-negative) cells (80). Cell-to-cell transfer of Nef would consequently manipulate the cellular environment of cells surrounding infection. A Nef Arg cluster (R17, R19, R21, and R22) on helix 0 (H0; denoted helix 1 in reference 83), which overlaps the 17RxR19 motif, and the sequence 62EEEEVGFPV70 (secretion modification region [SMR] motif), which overlaps the 62EEEE65 acid patch, are required for Nef secretion in EVs (Fig. 3) (78, 84). Mortalin (mitochondrial Hsp70) interacts with the Nef SMR motif and is involved in Nef secretion into EVs (85, 86). Mortalin is a multipotent chaperone found in multiple subcellular locations and has been implicated in cellular processes ranging from neurodegeneration to exocytosis (86–88). The Nef myristoylation site and C-terminal core (residues 71 to 206) are reported to be dispensable for EV packaging (84). In addition to Nef, exosomes package viral RNA, microRNAs, and other proteins, which contributes to the HIV pathogenesis and apoptosis in bystander CD4+ T cells (80, 89–91). What aspect of the EV biogenesis machinery is hijacked or influenced by Nef remains to be determined. ALIX and other ESCRTs would be the most obvious candidates, as exosome biogenesis is in many cases ESCRT mediated (77).
Autophagy.
Autophagy is a cytoplasmic degradative pathway by which a double-membrane vesicle called the autophagosome engulfs cytosol, damaged organelles, or intracellular pathogens for lysosomal degradation (92, 93). Autophagy has many connections to innate and adaptive immunity (94). Autophagy can be controlled by immune receptor and cytokine signaling and is stimulated upon microbial recognition by innate immunity pattern recognition receptors (95). HIV-1 Nef functions in preventing the destruction of HIV components in autolysosomes and thus shields HIV from autophagy and its role as a cell-autonomous antimicrobial defense (96). Nef colocalizes with beclin-1 (BECN1), a key regulatory protein for controlling the activity of the autophagic lipid kinase complex phosphatidylinositol 3-kinase catalytic subunit type 3 (PI3KC3) and thus the phosphatidylinositol 3-phosphate (PI3P) content of autophagosomes. This interaction results in a Nef-dependent increase in viral yields from autophagic cells (96). Nef is capable of binding a peptide fragment of beclin-1 (residues 257 to 337 [57]). Administering this peptide in vivo was shown to activate phosphatidylinositol 3-kinase (PI3K) and induce autophagy, suggesting a mechanistic connection of the peptide to Nef-mediated inhibition of autophagy (57). Further, Nef binding with PI3KC3-C2, the form of PI3KC3 involved in autophagosome maturation, inhibits PI3KC2 activity in vitro (55). Residues 35 to 65 in the Nef protein share sequence homology with Rubicon, and Nef thus inhibits PI3KC3-C2 membrane binding by a similar mechanism (55). In addition to being in a highly disordered region of the protein (Fig. 3), the more N-terminal part of this sequence (35–54) is poorly conserved beyond the HIV-1 M-Nefs.
TARGETS OF Nef SUPPRESSION
CD4.
Cluster of differentiation 4 (CD4) is a type 1 transmembrane glycoprotein found on the surface of immune cells, such as T helper cells, monocytes, and macrophages, and is the major cellular receptor for HIV. Downregulation of CD4 is a conserved function of SIV and HIV Nefs (33, 97, 98), which accelerate the AP-2- and clathrin-dependent endocytosis of cell surface CD4 and direct CD4 to lysosomal degradation (99–101). Consistent with this, the depletion of AP-2 (102–104) or clathrin (102) impedes Nef-induced CD4 downregulation. Nef and AP-2 recruit CD4 to CCPs (105–107), resulting in CD4 internalization and delivery to the lysosome for degradation via endosomes and MVBs (99, 100, 108).
Nef binds directly to AP-2 at the plasma membrane via its α-σ2 hemicomplex (20, 53, 102, 103, 109, 110). This leads to the cooperative assembly of a tripartite CD4-Nef-AP-2 complex (103). CD4 contains a noncanonical dileucine-based sorting motif (408SQIKRLL414) in its C-terminal cytoplasmic tail. In the normal nondegradative endocytosis of CD4 in uninfected cells, the S408 residue upstream of the LL motif is phosphorylated such that it effectively mimics the glutamate or aspartate residues of the classical (D/E)xxxL(L/I) dileucine motif, which allows CD4 to interact with AP-2 directly (111). In infection, Nef uses its canonical dileucine motif to adapt CD4 to clathrin-mediated endocytosis in the absence of serine phosphorylation (33).
The crystal structure of Nef bound to the α-σ2 hemicomplex provided a model for AP-2 hijacking by Nef to downregulate CD4 via binding the full AP-2 tetramer in the unlocked conformation (PDB ID 4NEE) (20). The myristoylated N-terminal anchor domain of Nef is not involved in this complex formation, thus making it accessible for putative membrane interactions. The structure confirmed that the key leucine pair of the dileucine loop of Nef binds to AP-2 in the same way as physiological cargo (PDB ID 2JKR) (27). Two other motifs within the dileucine loop of Nef have been implicated as crucial in mediating interaction with AP-2. The NA7 Nef hydrophobic motif M168/L170 (112), corresponding to V168/L170 in NL4-3 Nef, had been proposed to bind to the σ2 subunit, while an acidic motif, 174DD175, had been suggested to bind to a basic patch on the α subunit (103, 110). The structure confirmed the importance of these regions, while revealing that the diacidic motif 174DD175 was actually involved in scaffolding the appropriate AP-2-binding conformation of the dileucine loop rather than direct contact with AP-2. In SIVs, Nef W203 just outside the dileucine loop is important for Nef binding to the α-σ2 hemicomplex. When mutated (W203S), Nef was deficient in downregulating not only simian CD4 but also CD28, CD8, SERINC5, and simian tetherin (113, 114). A conserved His (196 in SIV sooty mangabey [SIVsmm]) on the back side of the dileucine motif is important for downmodulation of CD4, CD8, CD28, SERINC5, and simian tetherin (114).
Nef directly binds to the C terminus (CT) of CD4 (15, 115, 116), albeit with low affinity in the context of the soluble binary complex. The membrane-proximal cluster of hydrophobic amino acids (M407 and/or I410) and the dileucine motif (413LL414) are known to affect CD4-Nef association and endocytosis (99, 115, 117). The N-terminal motif 57WL58 of Nef was proposed to mediate the low-affinity binding to the CT of CD4 (13, 118). Recently, a structure of an asymmetric SIV macaque (SIVmac) Nef dimer showed how the dileucine motif on one copy of Nef could bind in a hydrophobic pocket between strand β1 and helix H2 of a second copy (PDB ID 5NUI) (Fig. 4) (83), suggesting how the CD4 dileucine motif might bind. These predictions were borne out through mutational experiments, lending good confidence to the mapping of the CD4 site.
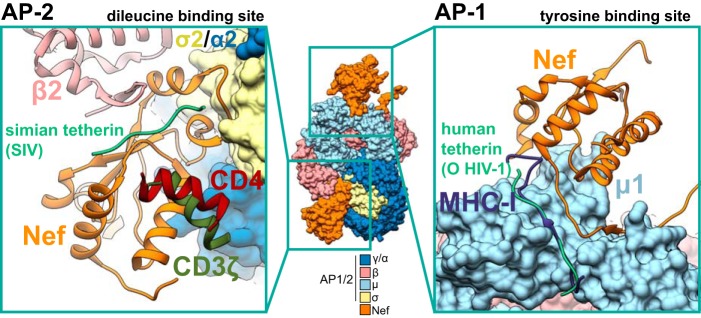
FIG 4.
Structures of Nef bound to AP-2 or AP-1 and host factors targeted for downregulation. Central composite structures of AP-2 and AP-1 were composed by aligning PDB IDs 6CM9, 4EMZ, 4NEE, and 6OWT. In this composite structure, Nef is bound at both the tyrosine and dileucine binding sites. The sites where Nef binds host factors CD4 (PDB ID 5NUI), CD3ζ (PDB ID 3IK5), and simian tetherin (6OWT) are indicated for the dileucine binding site of AP-2. The AP-2 β2 subunit is indicated to highlight the formation of the N-terminal β-hairpin upon binding to simian tetherin. The sites where Nef binds host factors MHC-I (PDB ID 4EMZ) and human tetherin (6CM9) are indicated for the tyrosine binding site of AP-1. All positions of the host factors were determined by aligning the Nef core of the respective PDBs to the Nef cores of PDB ID 4NEE (AP-2 interacting partners) and PDB ID 4EMZ (AP-1-interacting partners).
CD3.
Though the function was largely lost in HIV-1 and its direct precursor from chimpanzees (SIVcpz), the Nef proteins of HIV-2 and most SIVs efficiently internalize CD3, an essential cofactor in T-cell receptor (TCR) signaling (36). Nef-mediated internalization of CD3 allows HIV-2 and most SIVs to disrupt the formation of the immune synapse between infected CD4+ T cells and primary antigen-presenting cells (APCs) (119). By downregulating CD3, SIV and HIV-2 are thought to disrupt TCR-dependent T-cell activation (119), explaining why these infections do not lead to the hyperactivation of the immune system seen in HIV-1-infected individuals (120).
CD3 downmodulation depends on interactions between its ζ chain and the core of SIV and HIV-2 Nefs (121, 122). The ζ chain of the CD3 complex contains three copies of an immunoreceptor tyrosine-based activation motif (ITAM) with the consensus Yxx(L/I)x6–9Yxx(L/I). The tyrosines of the ITAMs are phosphorylated upon TCR ligation by Src family kinases and mediate downstream signal transduction and T-cell activation (123). Lentiviral Nefs downmodulating CD3 directly interact with the cytoplasmic tail of the ζ chain. The interaction sites overlap the ITAM regions and were mapped to the tyrosine-containing sequence motifs 72YNELNL77 and 123YSEIGM128, termed SIV Nef interaction domains 1 and 2 (SNID1 and SNID2), respectively (124). A crystal structure of SIVmac Nef bound to SNID1 has been determined (PDB ID 3IK5), which shows binding to the same hydrophobic pocket between strand β1 and helix H2 of Nef, as seen for CD4 (PDB ID 5NUI) (Fig. 4) (125). Functional studies showed that SIVmac Nef interacts with the ζ subunit of the CD3 complex, and both cooperate to bind AP-2 and consequently to suppress T-cell activation by downmodulating the TCR-CD3 complex (83, 126). Downregulation of CD3 is AP-2-dependent but does not require the Nef dileucine motif (126). Supporting this observation, a gain-of-function mutation of an HIV-1-based Nef allele capable of CD3 downregulation required the introduction of a tyrosine-based sorting motif in the N-terminal anchor domain of Nef (83). Thus, Nef not only uses distinct but overlapping domains to interact with the cytoplasmic portions of cellular receptors but also targets different AP-2 interaction surfaces to accelerate their endocytosis.
CD28.
CD28-initiated costimulatory signaling is critical for normal antigen-specific T-cell responses, and interference with the CD28 signaling pathway suppresses immune responses and anergy (127). HIV-1 and SIV Nef proteins downregulate cell surface expression of CD28 (38, 128, 129). CD28 downregulation by Nef involves direct interactions between Nef and CD28. Furthermore, HIV and SIV Nefs use overlapping but distinct target sites in the membrane-proximal region of the CD28 cytoplasmic domain (38). CD28 downregulation requires H196 of SIVmac239 Nef (114, 128). Nef-dependent downregulation of CD28 from the cell surface has been shown to involve both AP-1 and AP-2 (38, 130, 131). Downregulation of cell surface CD28 depends on the diacidic motif (174DD175), dileucine motif (164LL165), and CD4-interacting site (57WL58) of HIV-1 NL4-3 Nef (Fig. 3) (131). These interactions are consistent with the canonical mode of engagement of Nef to the AP-2 σ2-α hemicomplex, which would lead to a Nef-AP-2-CD28 ternary complex. Given these dependencies, engagement with AP-2 at the cell membrane to induce CME is probably the primary mode of Nef antagonism of CD28. Further to this point, HIV-1 Nef uses a combination of distinct and overlapping surfaces to interact with CD28 as it does with CD4, consistent with its AP-2 dependence (38).
CD8.
Host cellular immune responses mediated by CD8+ T lymphocytes (CTLs) play a pivotal role in controlling HIV/SIV viral proliferation (132, 133). However, during viral infection, a degradation of CTL function accompanies progression to symptomatic disease (134, 135). Though the major cellular target of HIV/SIV is the CD4 lymphocyte, both in vivo (136–139) and in vivo (136, 140–143) evidence show that CTLs are also susceptible to infection. Once CTLs are activated during HIV/SIV infection, they induce the expression of CD4 (136, 137, 144, 145), thus allowing for the infection of the CD8+ T lymphocytes and the subsequent downregulation of CD8 by Nef. CD8 is a transmembrane glycoprotein and a coreceptor of the T-cell receptor (TCR). HIV-1, HIV-2, and most SIV Nefs downregulate cell surface expression of the CD8αβ receptor in T cells in an AP-1-dependent manner (130, 146). The CD8α- and β-chain cytoplasmic tails contain no canonical AP-binding sorting signals. Nef-mediated downmodulation depends on the presence of the amino acid sequence FMK in the CD8 β-chain cytoplasmic tail (37). Nefs interact specifically with the cytoplasmic tail of CD8β, and downmodulation is dependent on the dileucine motif (164LL165) and its flanking diacidic motifs (154EE155 and 174DD175). The 57WL58 sequence, previously described as a binding site of the CD4 receptor (118), also has a role in CD8αβ downregulation (Fig. 3) (37, 114). These interactions are all consistent with a CD4-like downmodulation mechanism even though CD8 does not contain a CD4-like dileucine motif.
SERINC3/5.
It has long been known that Nef has a profound effect on enhancing HIV-1 infectivity (147). However, only recently was the Nef target most important to infectivity enhancement identified as SERINC5 (35, 39). The five human SERINC proteins are polytopic integral membrane proteins which are distributed through various cell compartments. Although there has been one early report that SERINCs are involved in connecting serine and lipid synthesis in yeast (148), this has not been corroborated. SERINC restriction of HIV infectivity does not seem to involve modulation of the lipid content of the virions (149). Neither the normal physiological function of this ancient and conserved family of proteins nor their mechanism of antagonizing HIV infectivity is fully known. Although SERINC3 is a Nef target and has also been implicated in suppressing viral infectivity (35, 39), SERINC5 has the strongest antiviral activity of the five. SERINC5 is thought to interact with Env such that its fusogenicity with target cells is impaired (150).
Clathrin and AP-2 are required for Nef to downregulate SERINC5 (35). The ability of Nef to counteract SERINC3/5 depends on the Nef dileucine motif, consistent with a dependence on AP-2 (Fig. 3) (35, 39). In SIV, SERINC5 downmodulation has been shown to be dependent on H196 of the dileucine loop (113, 114). SERINC3/5, however, lack canonical AP-2- or Nef-interacting motifs. Sensitivity of SERINC5 to Nef downregulation maps to the largest intracellular loop (loop 4) of SERINC5 (151). Two hydrophobic residues in loop 4 (L350 and I352) are crucial for susceptibility to Nef counteraction. Nef association with SERINC5 can be detected in cells using bimolecular fluorescence complementation (152). The most rigorous evidence for a direct physical linkage between Nef, SERINC5, and AP-2 would be the reconstitution of the ternary complex, something that has yet to be reported for either full-length SERINC3 or SERINC5 or a peptide based on loop 4.
It has been suggested counteraction of SERINCs by Nef is also dependent on dynamin-2 (Dyn2), based on a mutation at Nef D123 (35). Dyn2 coimmunoprecipitates with Nef and is required for the enhancement of HIV-1 infectivity by Nef (153). To date, a direct Dyn2-Nef interaction has not been corroborated structurally or biochemically with purified proteins. The ability to counteract SERINC5 was impaired by the Nef D123 mutation, which might suggest involvement of Dyn2, based on the original 2007 study (153). However, Nef D123 has been implicated structurally in AP-1 μ1-associated downregulation of MHC-I (19), as well as the dimerization of Nef (154), suggesting that the dependence of SERINC5 antagonism on a direct interaction with Dyn2 cannot be fully established without more evidence.
Following internalization, SERINC5 is localized in Rab5-positive early endosomes, Rab7-positive late endosomes, and Rab11-positive recycling endosomes (35, 152). The former two are consistent with shunting to lysosomal degradation and with the observation that SERINC5 is ultimately targeted to lysosomes for destruction (152). As with other Nef cargoes, the details of the hand-off from CCV into the degradative lysosomal sorting pathway are not well established, although it seems likely that ALIX and other ESCRTs are involved.
Tetherin.
Tetherin (BST2 or CD317) is an interferon-inducible cellular restriction factor. Tetherin functions by physically trapping budding progeny virions at the cell surface of infected cells, preventing their release (155–159) and promoting immune detection of infected cells (160). SIVs use Nef to downregulate tetherin from the cell surface (34, 41, 161, 162). SIV Nefs remove tetherin from the cell surface via AP-2 (163, 164). It has been reported that the total cellular tetherin levels remain unaffected during SIV infection, suggesting that Nef sequesters it to intracellular compartments rather than inducing its degradation (163, 164). However, the distribution of tetherin in SIV-infected cells revealed colocalization with TGN46 (TGN marker) and lysosome-associated membrane protein 1 (LAMP-1; lysosome marker) but not with CD63 (endosome marker), suggesting that tetherin accumulates in the TGN and in lysosomes while in the presence of Nef (163). However, tetherin also localizes to the TGN in uninfected cells; thus, SIV Nef’s contribution to this phenotype is hard to determine (165). Furthermore, the Nef-induced trafficking of MHC-I and CD4 to lysosomes for subsequent lysosomal degradation has been well established (63), and it also seems possible that endocytosed simian tetherin could follow this pathway.
The Nef dileucine loop residues adjacent to the LL residues themselves are critical for the antitetherin activity of SIV Nefs (163, 164, 166). Some of these mutations specifically disrupted downmodulation of tetherin but not other Nef and AP-2 dependent cargoes, such as CD4 (163, 166). Thus, residues surrounding the AP-2-binding site may be involved in the direct binding of tetherin rather than AP-2 recruitment, implying that the binding sites could be in close proximity and mutually influence one another. A direct physical interaction between Nef and the N-terminal cytoplasmic tail of tetherin has been proposed (163). The sensitivity of simian tetherin to SIV Nefs maps to a (G/D)DIWKK motif that is missing in its human ortholog (34, 41, 161). Thus, human tetherin is resistant to SIV Nef, and this is thought to have been one of the major hurdles to cross-species transmission of SIV to humans (161, 167).
Recently, the cryo-EM structure of the simian tetherin from sooty mangabey (SMM tetherin) and its SIV Nef (SIVsmm Nef) was determined in complex with AP-2 (PDB ID 6OWT) (113). Nef refolds the first α-helix of the β2 subunit of AP-2 to a β-hairpin, creating a binding site for the DIWK sequence on the SIVsmm Nef dileucine (190ExxxLV195) loop bound to the dileucine binding site of the AP-2 α-σ2 hemicomplex (Fig. 4). The DIWK binding site sits in a hydrophobic pocket centered around H192 and H196 of the SIVsmm Nef dileucine loop (Fig. 3). At this site, the DIWK motif is sandwiched between the newly formed AP-2 β2 β-hairpin and the SIVsmm Nef helix 0 (H0). H0 engages the same helix binding site implicated in CD4 and CD3ζ binding to HIV Nef (Fig. 4). Using in vivo mutational analysis, it was determined that the tetherin binding site in Nef is distinct from those of most other Nef substrates, including MHC-I, CD3, and CD4, but overlaps the site for SERINC5 restriction of viral infectivity (113). The structure explains the dependence of SIVs on the host tetherin DIWK sequence and the consequent barrier to human transmission.
During the 12 or more independent zoonotic transmissions of simian immunodeficiency viruses to humans, multiple adaptations occurred to overcome human tetherin’s insensitivity to SIV Nefs (161, 167, 168). While evidence suggests that some M-type HIV-1 strains can downregulate tetherin (168), it is largely agreed upon that the pandemic HIV-1 group M Vpu effectively counteracts human tetherin (157, 159). However, HIV-1 group O uses Nef for this purpose (51). The mechanism whereby O-Nefs downregulate tetherin is completely different than that utilized by SIV Nefs, as it must be since human tetherin lacks the DIWK motif. HIV-1 NL4-3 Nef, when bacterially expressed as a dephosphorylated protein such that it is an in vitro surrogate for an O-Nef, forms closed trimers containing three copies of hyperunlocked AP-1 and six copies of Arf1-GTP in the presence of the human tetherin tail (21). This assembly is centered on a trimer of Arf1 in which each molecule of Arf1 is bound to the γ subunit of AP-1 (PDB ID 6CM9). The edges of the trimer are bridged by Nef molecules (Fig. 2). Nef interacts through its dileucine motif with the γ-σ1 hemicomplex of one AP-1 tetramer. Nef binds to the tyrosine binding site of the μ1 domain of a second tetramer in trans in a ternary complex with the tetherin tail (Fig. 4). These bridges hold this trimer together and define it as a purely Nef-dependent assembly, which is nonexistent as far as is known in normal physiology. In contrast to MHC-I-bound open trimers of AP-1 described below, these tetherin-bound closed trimers do not appear to be capable of promoting CCV formation. The closed trimers appear to be a dead-end state, unique to infection, whose purpose is to sequester tetherin in the TGN. In this context, the function of AP-1 is not to package tetherin into CCVs, but to actually inhibit the anterograde sorting of tetherin. It is not clear what the benefit is to the virus of downmodulating activity via sequestration as opposed to the more permanent mechanisms of lysosomal degradation.
The ability of Nefs to form closed trimers with AP-1 appears to be regulated by phosphorylation within the dileucine loop. Casein kinase 1δ (CK1δ) localizes at the TGN (169). Phosphorylation of S169 in the dileucine loop of the NL4-3 M-Nef disrupts its interaction with the AP-1 γ-σ1 hemicomplex, preventing closed trimer formation (21). The mutation of Ser to Ala partially restores the ability of NL4-3 Nef to target tetherin, while the reciprocal mutation of the corresponding amino acids, Cys to Ser, in O-Nef impairs tetherin counteraction (21).
MHC-I.
MHC-I molecules deliver short endogenous peptides to the surface of antigen-presenting cells, allowing these epitopes to be recognized by CD8+ (cytotoxic) T lymphocytes (CTLs). In infected cells, viral peptides are processed and presented to the cell surface, activating CTLs for viral clearance. Therefore, the antigen presentation mediated by MHC-I is an essential pathway in adaptive immunity (170, 171). Nef disrupts antigen presentation both by sequestering MHC-I molecules in intracellular compartments and by sorting them for lysosomal degradation instead of plasma membrane presentation. Antigenic peptides are 8 to 10 amino acids in length and are mainly generated by proteasome in the cytosol (172). They are translocated into the ER lumen by the transporter associated with antigen processing (TAP). Nascent MHC-I molecules fold within ER lumen. ER resident chaperones facilitate MHC-I-TAP interactions and transfer peptides to MHC-I (173). After peptide loading, the MHC-I-peptide complex dissociates from TAP and then traffics through the ER and Golgi apparatus to the plasma membrane. It is at the TGN that HIV Nef targets MHC-I for downregulation.
MHC-I does not contain either of the canonical tyrosine-based YxxΦ or dileucine-based D/ExxxLL signals for AP sorting. In normal physiology, MHC-I endocytosis is mediated by a clathrin-independent pathway (174), and MHC-I does not interact with clathrin adaptors in the absence of Nef. In HIV-infected primary T cells, Nef promotes a physical interaction between the Golgi adaptor complex AP-1 and MHC-I (175). In particular, the AP-1 μ1 subunit is required to sequester the MHC-I-Nef complex at the TGN and into sorting MHC-I lysosomes (118, 176). The crystal structures of HIV-I Nef, MHC-I cytoplasmic tail (CD), and the μ1-C-terminal domain (CTD) complex show that Nef and MHC-I cooperatively assemble so as to mimic physiological Tyr motif binding to the μ1-CTD (19). In this structure, residues 314 to 332 of the MHC-I tail were sandwiched by a μ1-CTD β sheet and secured by Nef 62EEEE65 and 72PxxP75 (Fig. 3 and 4). The combined MHC-I and Nef interactions with μ1 are thus equivalent to those of a physiological cargo (TGN38) bound to the μ2-CTD of AP-2 (PDB ID 1BW8) (28). The AP-2 μ2 subunit lacks the equivalent of the Tyr374 in μ1 that stacks onto Nef Pro72 during binding, partially explaining why MHC-I does not bind to AP-2 even in the presence of Nef. Moreover, two extensive electrostatic interactions were identified in this structure. The Nef acidic sequence 62EEEE65 binds to the μ1 basic patch (K274, K298, K302, and R303) (Fig. 3) (19). This basic patch does not exist in AP-2 μ2. MHC-I D327 and Nef D123 together bind to a μ1 basic patch (R211, R225, R246, R393, and K396). Nef D123 is thus critical for binding to AP-1 μ1 (Fig. 3). Other functions have also been imputed to D123. This residue mediates lattice contacts in a crystallographic dimer of Nef and the SH3 domain of the tyrosine kinase Hck (PDB ID 5NUH) (83). However, most crystal structures involving Nef are not dimers, and there are even other structures of dimeric SH3 complexes of Nef that show D123 to be surface exposed and uninvolved in dimerization (Table 1). Another role imputed to D123 is in the interaction with dynamin-2 (153). Following the initial reports, there has been no structure or other substantiation of this complex. Thus, the only well-established function of D123 is in μ1 binding. It seems possible that some of the phenotypes attributed to dimerization or dynamin-2 binding could actually be reinterpreted in terms of a role for μ1.
AP-1 and the Golgi small GTPase Arf1 are capable of forming dimeric and open and closed trimeric assemblies in the absence of cargo, and Nef more efficiently promotes trimer formation (PDB IDs 4HMY and 6CM9) (Fig. 2B) (22). The dimeric and open trimeric assemblies can in turn form a hexagonal lattice whose symmetry and dimensions are a nearly perfect match for the clathrin lattice. It appears that even in normal physiology, AP-1 and Arf1 can form an ordered inner coat beneath clathrin so as to promote CCV formation and onward trafficking. The role of Nef here is to accelerate the process and to steer targeted cargo into the CCVs thus formed (63, 130).
FUTURE PERSPECTIVES
There is still limited direct information about the conformational arrangement of membrane-bound Nef in complex with clathrin adaptors and cargo receptors and how they assemble in clathrin vesicles. Some information on the conformation of Nef on membranes is available from time-resolved fluorescence kinetics on synthetic liposomes, neutron reflectometry, and hydrogen deuterium exchange coupled to mass spectrometry (HDX-MS) on a Langmuir monolayer (177–180). When Nef alone associates with a low-density lipid monolayer, its N-terminal region and the C-terminal unstructured loop undergo conformational changes. Ideally, it will be possible to use cryo-electron tomography (cryo-ET) to study their assembly in clathrin-coated vesicles. The highly interconnected COPI coat structure (50, 181) has been determined by cryo-ET from reconstituted COPI-coated vesicles in vitro and in cells (182). It should similarly be feasible to determine structurally how Nef, clathrin adaptor and cargo package inside the clathrin coats by using cryo-ET of these reconstituted systems.
Understanding Nef at the levels of structural biology and molecular virology has moved faster than our understanding at the cellular level. With new methods, such as lattice-light sheet microscopy and genome-edited cell lines, real-time visualization of Nef and its substrates throughout the trafficking pathway should change this situation. While an understanding of Nef’s role in endocytosis and at the TGN has advanced considerably, its roles at other internal membranes, including endosomes and autophagosomes, need to be further explored. Nef is a structurally plastic protein with extensive disordered regions and no unique active site. Targeting the Nef protein itself for antiretrovirals would seem to be challenging. On the other hand, structural analysis is showing how Nef hijacks a combination of host-mimetic and Nef-unique sites on its host partners, with the latter comprising an attractive set of targets for antiretroviral therapeutics.
ACKNOWLEDGMENTS
This research was supported by NIH grants R01 AI120691 (to X.R.), P50 GM082250 (to J.H.H.), and F32 GM125209 (to C.Z.B.), and by the Andrew Dougherty Vision Foundation (to C.Z.B.).
REFERENCES
- 1.Collins DR, Collins KL. 2014. HIV-1 accessory proteins adapt cellular adaptors to facilitate immune evasion. PLoS Pathog 10:e1003851. doi: 10.1371/journal.ppat.1003851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malim MH, Emerman M. 2008. HIV-1 accessory proteins–ensuring viral survival in a hostile environment. Cell Host Microbe 3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Geyer M, Fackler OT, Peterlin BM. 2001. Structure-function relationships in HIV-1 Nef. EMBO Rep 2:580–585. doi: 10.1093/embo-reports/kve141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fackler OT, Kienzle N, Kremmer E, Boese A, Schramm B, Klimkait T, Kucherer C, Mueller-Lantzsch N. 1997. Association of human immunodeficiency virus Nef protein with actin is myristoylation dependent and influences its subcellular localization. Eur J Biochem 247:843–851. doi: 10.1111/j.1432-1033.1997.00843.x. [DOI] [PubMed] [Google Scholar]
- 5.Geyer M, Munte CE, Schorr J, Kellner R, Kalbitzer HR. 1999. Structure of the anchor-domain of myristoylated and non-myristoylated HIV-1 Nef protein. J Mol Biol 289:123–138. doi: 10.1006/jmbi.1999.2740. [DOI] [PubMed] [Google Scholar]
- 6.Bentham M, Mazaleyrat S, Harris M. 2006. Role of myristoylation and N-terminal basic residues in membrane association of the human immunodeficiency virus type 1 Nef protein. J Gen Virol 87:563–571. doi: 10.1099/vir.0.81200-0. [DOI] [PubMed] [Google Scholar]
- 7.Breuer S, Gerlach H, Kolaric B, Urbanke C, Opitz N, Geyer M. 2006. Biochemical indication for myristoylation-dependent conformational changes in HIV-1 Nef. Biochemistry 45:2339–2349. doi: 10.1021/bi052052c. [DOI] [PubMed] [Google Scholar]
- 8.Fackler OT, Moris A, Tibroni N, Giese SI, Glass B, Schwartz O, Krausslich HG. 2006. Functional characterization of HIV-1 Nef mutants in the context of viral infection. Virology 351:322–339. doi: 10.1016/j.virol.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 9.Giese SI, Woerz I, Homann S, Tibroni N, Geyer M, Fackler OT. 2006. Specific and distinct determinants mediate membrane binding and lipid raft incorporation of HIV-1(SF2) Nef. Virology 355:175–191. doi: 10.1016/j.virol.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Coates K, Cooke SJ, Mann DA, Harris MP. 1997. Protein kinase C-mediated phosphorylation of HIV-I Nef in human cell lines. J Biol Chem 272:12289–12294. doi: 10.1074/jbc.272.19.12289. [DOI] [PubMed] [Google Scholar]
- 11.Arold S, Franken P, Strub MP, Hoh F, Benichou S, Benarous R, Dumas C. 1997. The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling. Structure 5:1361–1372. doi: 10.1016/s0969-2126(97)00286-4. [DOI] [PubMed] [Google Scholar]
- 12.Lee CH, Saksela K, Mirza UA, Chait BT, Kuriyan J. 1996. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell 85:931–942. doi: 10.1016/s0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- 13.Grzesiek S, Bax A, Clore GM, Gronenborn AM, Hu JS, Kaufman J, Palmer I, Stahl SJ, Wingfield PT. 1996. The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nat Struct Biol 3:340–345. doi: 10.1038/nsb0496-340. [DOI] [PubMed] [Google Scholar]
- 14.Grzesiek S, Bax A, Hu JS, Kaufman J, Palmer I, Stahl SJ, Tjandra N, Wingfield PT. 1997. Refined solution structure and backbone dynamics of HIV-1 Nef. Protein Sci 6:1248–1263. doi: 10.1002/pro.5560060613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grzesiek S, Stahl SJ, Wingfield PT, Bax A. 1996. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry 35:10256–10261. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
- 16.Geyer M, Peterlin BM. 2001. Domain assembly, surface accessibility and sequence conservation in full length HIV-1 Nef. FEBS Lett 496:91–95. doi: 10.1016/s0014-5793(01)02394-8. [DOI] [PubMed] [Google Scholar]
- 17.Jäger S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, Hernandez H, Jang GM, Roth SL, Akiva E, Marlett J, Stephens M, D'Orso I, Fernandes J, Fahey M, Mahon C, O'Donoghue AJ, Todorovic A, Morris JH, Maltby DA, Alber T, Cagney G, Bushman FD, Young JA, Chanda SK, Sundquist WI, Kortemme T, Hernandez RD, Craik CS, Burlingame A, Sali A, Frankel AD, Krogan NJ. 2011. Global landscape of HIV-human protein complexes. Nature 481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mailler E, Waheed AA, Park SY, Gershlick DC, Freed EO, Bonifacino JS. 2019. The autophagy protein ATG9A promotes HIV-1 infectivity. Retrovirology 16:18. doi: 10.1186/s12977-019-0480-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia X, Singh R, Homann S, Yang H, Guatelli J, Xiong Y. 2012. Structural basis of evasion of cellular adaptive immunity by HIV-1 Nef. Nat Struct Mol Biol 19:701–706. doi: 10.1038/nsmb.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren X, Park SY, Bonifacino JS, Hurley JH. 2014. How HIV-1 Nef hijacks the AP-2 clathrin adaptor to downregulate CD4. Elife 3:e01754. doi: 10.7554/eLife.01754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris KL, Buffalo CZ, Sturzel CM, Heusinger E, Kirchhoff F, Ren X, Hurley JH. 2018. HIV-1 Nefs are cargo-sensitive AP-1 trimerization switches in tetherin downregulation. Cell 174:659–671.e14. doi: 10.1016/j.cell.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen QT, Ren X, Zhang R, Lee IH, Hurley JH. 2015. HIV-1 Nef hijacks clathrin coats by stabilizing AP-1:Arf1 polygons. Science 350:aac5137. doi: 10.1126/science.aac5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deacon NJ, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker DJ, McPhee DA, Greenway AL, Ellett A, Chatfield C, Lawson VA, Crowe S, Maerz A, Sonza S, Learmont J, Sullivan JS, Cunningham A, Dwyer D, Dowton D, Mills J. 1995. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 24.Gorry PR, McPhee DA, Verity E, Dyer WB, Wesselingh SL, Learmont J, Sullivan JS, Roche M, Zaunders JJ, Gabuzda D, Crowe SM, Mills J, Lewin SR, Brew BJ, Cunningham AL, Churchill MJ. 2007. Pathogenicity and immunogenicity of attenuated, Nef-deleted HIV-1 strains in vivo. Retrovirology 4:66. doi: 10.1186/1742-4690-4-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. 1995. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med 332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 26.Owen DJ, Collins BM, Evans PR. 2004. Adaptors for clathrin coats: structure and function. Annu Rev Cell Dev Biol 20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- 27.Kelly BT, McCoy AJ, Spate K, Miller SE, Evans PR, Honing S, Owen DJ. 2008. A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature 456:976–979. doi: 10.1038/nature07422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Owen DJ, Evans PR. 1998. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science 282:1327–1332. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirchhausen T. 1999. Adaptors for clathrin-mediated traffic. Annu Rev Cell Dev Biol 15:705–732. doi: 10.1146/annurev.cellbio.15.1.705. [DOI] [PubMed] [Google Scholar]
- 30.Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. 2002. Molecular architecture and functional model of the endocytic AP2 complex. Cell 109:523–535. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- 31.Jackson LP, Kelly BT, McCoy AJ, Gaffry T, James LC, Collins BM, Honing S, Evans PR, Owen DJ. 2010. A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell 141:1220–1229. doi: 10.1016/j.cell.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly BT, Graham SC, Liska N, Dannhauser PN, Honing S, Ungewickell EJ, Owen DJ. 2014. Clathrin adaptors. AP2 controls clathrin polymerization with a membrane-activated switch. Science 345:459–463. doi: 10.1126/science.1254836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia JV, Miller AD. 1991. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature 350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- 34.Jia B, Serra-Moreno R, Neidermyer W Jr, Rahmberg A, Mackey J, Fofana IB, Johnson WE, Westmoreland S, Evans DT. 2009. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog 5:e1000429. doi: 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, Santoni FA, Pizzato M. 2015. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526:212–217. doi: 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schindler M, Munch J, Kutsch O, Li H, Santiago ML, Bibollet-Ruche F, Muller-Trutwin MC, Novembre FJ, Peeters M, Courgnaud V, Bailes E, Roques P, Sodora DL, Silvestri G, Sharp PM, Hahn BH, Kirchhoff F. 2006. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell 125:1055–1067. doi: 10.1016/j.cell.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 37.Stove V, Van de Walle I, Naessens E, Coene E, Stove C, Plum J, Verhasselt B. 2005. Human immunodeficiency virus Nef induces rapid internalization of the T-cell coreceptor CD8αβ. J Virol 79:11422–11433. doi: 10.1128/JVI.79.17.11422-11433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swigut T, Shohdy N, Skowronski J. 2001. Mechanism for down-regulation of CD28 by Nef. EMBO J 20:1593–1604. doi: 10.1093/emboj/20.7.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Usami Y, Wu Y, Gottlinger HG. 2015. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526:218–223. doi: 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willard-Gallo KE, Furtado M, Burny A, Wolinsky SM. 2001. Down-modulation of TCR/CD3 surface complexes after HIV-1 infection is associated with differential expression of the viral regulatory genes. Eur J Immunol 31:969–979. doi:. [DOI] [PubMed] [Google Scholar]
- 41.Zhang F, Wilson SJ, Landford WC, Virgen B, Gregory D, Johnson MC, Munch J, Kirchhoff F, Bieniasz PD, Hatziioannou T. 2009. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6:54–67. doi: 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michel N, Ganter K, Venzke S, Bitzegeio J, Fackler OT, Keppler OT. 2006. The Nef protein of human immunodeficiency virus is a broad-spectrum modulator of chemokine receptor cell surface levels that acts independently of classical motifs for receptor endocytosis and Gαi signaling. Mol Biol Cell 17:3578–3590. doi: 10.1091/mbc.e06-02-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venzke S, Michel N, Allespach I, Fackler OT, Keppler OT. 2006. Expression of Nef downregulates CXCR4, the major coreceptor of human immunodeficiency virus, from the surfaces of target cells and thereby enhances resistance to superinfection. J Virol 80:11141–11152. doi: 10.1128/JVI.01556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stamnes MA, Rothman JE. 1993. The binding of AP-1 clathrin adaptor particles to Golgi membranes requires ADP-ribosylation factor, a small GTP-binding protein. Cell 73:999–1005. doi: 10.1016/0092-8674(93)90277-w. [DOI] [PubMed] [Google Scholar]
- 45.Traub LM, Ostrom JA, Kornfeld S. 1993. Biochemical dissection of AP-1 recruitment onto Golgi membranes. J Cell Biol 123:561–573. doi: 10.1083/jcb.123.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Traub LM, Bonifacino JS. 2013. Cargo recognition in clathrin-mediated endocytosis. Cold Spring Harb Perspect Biol 5:a016790. doi: 10.1101/cshperspect.a016790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ren X, Farias GG, Canagarajah BJ, Bonifacino JS, Hurley JH. 2013. Structural basis for recruitment and activation of the AP-1 clathrin adaptor complex by Arf1. Cell 152:755–767. doi: 10.1016/j.cell.2012.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heldwein EE, Macia E, Wang J, Yin HL, Kirchhausen T, Harrison SC. 2004. Crystal structure of the clathrin adaptor protein 1 core. Proc Natl Acad Sci U S A 101:14108–14113. doi: 10.1073/pnas.0406102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia X, Weber E, Tokarev A, Lewinski M, Rizk M, Suarez M, Guatelli J, Xiong Y. 2014. Structural basis of HIV-1 Vpu-mediated BST2 antagonism via hijacking of the clathrin adaptor protein complex 1. Elife 3:e02362. doi: 10.7554/eLife.02362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dodonova SO, Diestelkoetter-Bachert P, von Appen A, Hagen WJ, Beck R, Beck M, Wieland F, Briggs JA. 2015. Vesicular transport. A structure of the COPI coat and the role of coat proteins in membrane vesicle assembly. Science 349:195–198. doi: 10.1126/science.aab1121. [DOI] [PubMed] [Google Scholar]
- 51.Kluge SF, Mack K, Iyer SS, Pujol FM, Heigele A, Learn GH, Usmani SM, Sauter D, Joas S, Hotter D, Bibollet-Ruche F, Plenderleith LJ, Peeters M, Geyer M, Sharp PM, Fackler OT, Hahn BH, Kirchhoff F. 2014. Nef proteins of epidemic HIV-1 group O strains antagonize human tetherin. Cell Host Microbe 16:639–650. doi: 10.1016/j.chom.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janvier K, Kato Y, Boehm M, Rose JR, Martina JA, Kim BY, Venkatesan S, Bonifacino JS. 2003. Recognition of dileucine-based sorting signals from HIV-1 Nef and LIMP-II by the AP-1 γ-σ1 and AP-3 δ-σ3 hemicomplexes. J Cell Biol 163:1281–1290. doi: 10.1083/jcb.200307157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mattera R, Boehm M, Chaudhuri R, Prabhu Y, Bonifacino JS. 2011. Conservation and diversification of dileucine signal recognition by adaptor protein (AP) complex variants. J Biol Chem 286:2022–2030. doi: 10.1074/jbc.M110.197178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Janvier K, Craig H, Hitchin D, Madrid R, Sol-Foulon N, Renault L, Cherfils J, Cassel D, Benichou S, Guatelli J. 2003. HIV-1 Nef stabilizes the association of adaptor protein complexes with membranes. J Biol Chem 278:8725–8732. doi: 10.1074/jbc.M210115200. [DOI] [PubMed] [Google Scholar]
- 55.Chang C, Young LN, Morris KL, von Bulow S, Schoneberg J, Yamamoto-Imoto H, Oe Y, Yamamoto K, Nakamura S, Stjepanovic G, Hummer G, Yoshimori T, Hurley JH. 2019. Bidirectional control of autophagy by BECN1 BARA domain dynamics. Mol Cell 73:339–353.e6. doi: 10.1016/j.molcel.2018.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pereira EA, daSilva LL. 2016. HIV-1 Nef: taking control of protein trafficking. Traffic 17:976–996. doi: 10.1111/tra.12412. [DOI] [PubMed] [Google Scholar]
- 57.Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, Huerta C, Virgin HW, Helms JB, Eerland R, Tooze SA, Xavier R, Lenschow DJ, Yamamoto A, King D, Lichtarge O, Grishin NV, Spector SA, Kaloyanova DV, Levine B. 2013. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 494:201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aniento F, Gu F, Parton RG, Gruenberg J. 1996. An endosomal beta COP is involved in the pH-dependent formation of transport vesicles destined for late endosomes. J Cell Biol 133:29–41. doi: 10.1083/jcb.133.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu F, Aniento F, Parton RG, Gruenberg J. 1997. Functional dissection of COP-I subunits in the biogenesis of multivesicular endosomes. J Cell Biol 139:1183–1195. doi: 10.1083/jcb.139.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitney JA, Gomez M, Sheff D, Kreis TE, Mellman I. 1995. Cytoplasmic coat proteins involved in endosome function. Cell 83:703–713. doi: 10.1016/0092-8674(95)90183-3. [DOI] [PubMed] [Google Scholar]
- 61.Benichou S, Bomsel M, Bodeus M, Durand H, Doute M, Letourneur F, Camonis J, Benarous R. 1994. Physical interaction of the HIV-1 Nef protein with beta-COP, a component of non-clathrin-coated vesicles essential for membrane traffic. J Biol Chem 269:30073–30076. [PubMed] [Google Scholar]
- 62.Piguet V, Gu F, Foti M, Demaurex N, Gruenberg J, Carpentier JL, Trono D. 1999. Nef-induced CD4 degradation: a diacidic-based motif in Nef functions as a lysosomal targeting signal through the binding of β-COP in endosomes. Cell 97:63–73. doi: 10.1016/s0092-8674(00)80715-1. [DOI] [PubMed] [Google Scholar]
- 63.Schaefer MR, Wonderlich ER, Roeth JF, Leonard JA, Collins KL. 2008. HIV-1 Nef targets MHC-I and CD4 for degradation via a final common β-COP-dependent pathway in T cells. PLoS Pathog 4:e1000131. doi: 10.1371/journal.ppat.1000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Janvier K, Craig H, Le Gall S, Benarous R, Guatelli J, Schwartz O, Benichou S. 2001. Nef-induced CD4 downregulation: a diacidic sequence in human immunodeficiency virus type 1 Nef does not function as a protein sorting motif through direct binding to β-COP. J Virol 75:3971–3976. doi: 10.1128/JVI.75.8.3971-3976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hurley JH, Cada AK. 2018. Inside job: how the ESCRTs release HIV-1 from infected cells. Biochem Soc Trans 46:1029–1036. doi: 10.1042/BST20180019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCullough J, Frost A, Sundquist WI. 2018. Structures, functions, and dynamics of ESCRT-III/Vps4 membrane remodeling and fission complexes. Annu Rev Cell Dev Biol 34:85–109. doi: 10.1146/annurev-cellbio-100616-060600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Costa LJ, Chen N, Lopes A, Aguiar RS, Tanuri A, Plemenitas A, Peterlin BM. 2006. Interactions between Nef and AIP1 proliferate multivesicular bodies and facilitate egress of HIV-1. Retrovirology 3:33. doi: 10.1186/1742-4690-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bissig C, Gruenberg J. 2014. ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol 24:19–25. doi: 10.1016/j.tcb.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 69.Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. 2007. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 70.Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 71.Zhai Q, Fisher RD, Chung HY, Myszka DG, Sundquist WI, Hill CP. 2008. Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV. Nat Struct Mol Biol 15:43–49. doi: 10.1038/nsmb1319. [DOI] [PubMed] [Google Scholar]
- 72.Fujii K, Munshi UM, Ablan SD, Demirov DG, Soheilian F, Nagashima K, Stephen AG, Fisher RJ, Freed EO. 2009. Functional role of Alix in HIV-1 replication. Virology 391:284–292. doi: 10.1016/j.virol.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. 2007. Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol 14:194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Munshi UM, Kim J, Nagashima K, Hurley JH, Freed EO. 2007. An Alix fragment potently inhibits HIV-1 budding: characterization of binding to retroviral YPXL late domains. J Biol Chem 282:3847–3855. doi: 10.1074/jbc.M607489200. [DOI] [PubMed] [Google Scholar]
- 75.Jesus da Costa L, Lopes Dos Santos A, Mandic R, Shaw K, Santana de Aguiar R, Tanuri A, Luciw PA, Peterlin BM. 2009. Interactions between SIVNef, SIVGagPol and Alix correlate with viral replication and progression to AIDS in rhesus macaques. Virology 394:47–56. doi: 10.1016/j.virol.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Amorim NA, da Silva EML, de Castro RO, da Silva-Januário ME, Mendonça LM, Bonifacino JS, da Costa LJ, daSilva LLP. 2014. Interaction of HIV-1 Nef protein with the host protein Alix promotes lysosomal targeting of CD4 receptor. J Biol Chem 289:27744–27756. doi: 10.1074/jbc.M114.560193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Niel G, D'Angelo G, Raposo G. 2018. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 78.Ali SA, Huang MB, Campbell PE, Roth WW, Campbell T, Khan M, Newman G, Villinger F, Powell MD, Bond VC. 2010. Genetic characterization of HIV type 1 Nef-induced vesicle secretion. AIDS Res Hum Retroviruses 26:173–192. doi: 10.1089/aid.2009.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Raymond AD, Campbell-Sims TC, Khan M, Lang M, Huang MB, Bond VC, Powell MD. 2011. HIV rype 1 Nef is released from infected cells in CD45+ microvesicles and is present in the plasma of HIV-infected individuals. AIDS Res Hum Retroviruses 27:167–178. doi: 10.1089/aid.2009.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McNamara RP, Costantini LM, Myers TA, Schouest B, Maness NJ, Griffith JD, Damania BA, MacLean AG, Dittmer DP. 2018. Nef secretion into extracellular vesicles or exosomes is conserved across human and simian immunodeficiency viruses. mBio 9:e02344-17. doi: 10.1128/mBio.02344-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Février B, Raposo G. 2004. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol 16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 82.Sanfridson A, Hester S, Doyle C. 1997. Nef proteins encoded by human and simian immunodeficiency viruses induce the accumulation of endosomes and lysosomes in human T cells. Proc Natl Acad Sci U S A 94:873–878. doi: 10.1073/pnas.94.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Manrique S, Sauter D, Horenkamp FA, Lulf S, Yu H, Hotter D, Anand K, Kirchhoff F, Geyer M. 2017. Endocytic sorting motif interactions involved in Nef-mediated downmodulation of CD4 and CD3. Nat Commun 8:442. doi: 10.1038/s41467-017-00481-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Campbell PE, Isayev O, Ali SA, Roth WW, Huang MB, Powell MD, Leszczynski J, Bond VC. 2012. Validation of a novel secretion modification region (SMR) of HIV-1 Nef using cohort sequence analysis and molecular modeling. J Mol Model 18:4603–4613. doi: 10.1007/s00894-012-1452-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shelton MN, Huang MB, Ali S, Johnson K, Roth W, Powell M, Bond V. 2013. Peptide-based identification of functional motifs and their binding partners. J Vis Exp 76:e50362. doi: 10.3791/50362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shelton MN, Huang MB, Ali SA, Powell MD, Bond VC. 2012. Secretion modification region-derived peptide disrupts HIV-1 Nef’s interaction with mortalin and blocks virus and Nef exosome release. J Virol 86:406–419. doi: 10.1128/JVI.05720-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Flachbartová Z, Kovacech B. 2013. Mortalin - a multipotent chaperone regulating cellular processes ranging from viral infection to neurodegeneration. Acta Virol 57:3–15. doi: 10.4149/av_2013_01_3. [DOI] [PubMed] [Google Scholar]
- 88.Londono C, Osorio C, Gama V, Alzate O. 2012. Mortalin, apoptosis, and neurodegeneration. Biomolecules 2:143–164. doi: 10.3390/biom2010143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pužar Dominkus P, Ferdin J, Plemenitas A, Peterlin BM, Lenassi M. 2017. Nef is secreted in exosomes from Nef.GFP-expressing and HIV-1-infected human astrocytes. J Neurovirol 23:713–724. doi: 10.1007/s13365-017-0552-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aqil M, Mallik S, Bandyopadhyay S, Maulik U, Jameel S. 2015. Transcriptomic analysis of mRNAs in human monocytic cells expressing the HIV-1 Nef protein and their exosomes. Biomed Res Int 2015:492395. doi: 10.1155/2015/492395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lenassi M, Cagney G, Liao M, Vaupotic T, Bartholomeeusen K, Cheng Y, Krogan NJ, Plemenitas A, Peterlin BM. 2010. HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic 11:110–122. doi: 10.1111/j.1600-0854.2009.01006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gatica D, Lahiri V, Klionsky DJ. 2018. Cargo recognition and degradation by selective autophagy. Nat Cell Biol 20:233–242. doi: 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morishita H, Mizushima N. 2019. Diverse cellular roles of autophagy, in press. Annu Rev Cell Dev Biol 35:453–475. doi: 10.1146/annurev-cellbio-100818-125300. [DOI] [PubMed] [Google Scholar]
- 94.Levine B, Kroemer G. 2019. Biological functions of autophagy genes: a disease perspective. Cell 176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Levine B, Mizushima N, Virgin HW. 2011. Autophagy in immunity and inflammation. Nature 469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, Wu L, Kominami E, Ueno T, Yamamoto A, Federico M, Panganiban A, Vergne I, Deretic V. 2009. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol 186:255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guy B, Kieny MP, Riviere Y, Le Peuch C, Dott K, Girard M, Montagnier L, Lecocq JP. 1987. HIV F/3′ orf encodes a phosphorylated GTP-binding protein resembling an oncogene product. Nature 330:266–269. doi: 10.1038/330266a0. [DOI] [PubMed] [Google Scholar]
- 98.Mariani R, Skowronski J. 1993. CD4 down-regulation by nef alleles isolated from human immunodeficiency virus type 1-infected individuals. Proc Natl Acad Sci U S A 90:5549–5553. doi: 10.1073/pnas.90.12.5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aiken C, Konner J, Landau NR, Lenburg ME, Trono D. 1994. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell 76:853–864. doi: 10.1016/0092-8674(94)90360-3. [DOI] [PubMed] [Google Scholar]
- 100.Rhee SS, Marsh JW. 1994. Human immunodeficiency virus type 1 Nef-induced down-modulation of CD4 is due to rapid internalization and degradation of surface CD4. J Virol 68:5156–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gondim MV, Wiltzer-Bach L, Maurer B, Banning C, Arganaraz E, Schindler M. 2015. AP-2 is the crucial clathrin adaptor protein for CD4 downmodulation by HIV-1 Nef in infected primary CD4+ T cells. J Virol 89:12518–12524. doi: 10.1128/JVI.01838-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chaudhuri R, Lindwasser OW, Smith WJ, Hurley JH, Bonifacino JS. 2007. Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J Virol 81:3877–3890. doi: 10.1128/JVI.02725-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chaudhuri R, Mattera R, Lindwasser OW, Robinson MS, Bonifacino JS. 2009. A basic patch on α-adaptin is required for binding of human immunodeficiency virus type 1 Nef and cooperative assembly of a CD4-Nef-AP-2 complex. J Virol 83:2518–2530. doi: 10.1128/JVI.02227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jin YJ, Cai CY, Zhang X, Zhang HT, Hirst JA, Burakoff SJ. 2005. HIV Nef-mediated CD4 down-regulation is adaptor protein complex 2 dependent. J Immunol 175:3157–3164. doi: 10.4049/jimmunol.175.5.3157. [DOI] [PubMed] [Google Scholar]
- 105.Burtey A, Rappoport JZ, Bouchet J, Basmaciogullari S, Guatelli J, Simon SM, Benichou S, Benmerah A. 2007. Dynamic interaction of HIV-1 Nef with the clathrin-mediated endocytic pathway at the plasma membrane. Traffic 8:61–76. doi: 10.1111/j.1600-0854.2006.00512.x. [DOI] [PubMed] [Google Scholar]
- 106.Foti M, Mangasarian A, Piguet V, Lew DP, Krause KH, Trono D, Carpentier JL. 1997. Nef-mediated clathrin-coated pit formation. J Cell Biol 139:37–47. doi: 10.1083/jcb.139.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Greenberg ME, Bronson S, Lock M, Neumann M, Pavlakis GN, Skowronski J. 1997. Co-localization of HIV-1 Nef with the AP-2 adaptor protein complex correlates with Nef-induced CD4 down-regulation. EMBO J 16:6964–6976. doi: 10.1093/emboj/16.23.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.daSilva LL, Sougrat R, Burgos PV, Janvier K, Mattera R, Bonifacino JS. 2009. Human immunodeficiency virus type 1 Nef protein targets CD4 to the multivesicular body pathway. J Virol 83:6578–6590. doi: 10.1128/JVI.00548-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Doray B, Lee I, Knisely J, Bu G, Kornfeld S. 2007. The gamma/sigma1 and α/σ2 hemicomplexes of clathrin adaptors AP-1 and AP-2 harbor the dileucine recognition site. Mol Biol Cell 18:1887–1896. doi: 10.1091/mbc.e07-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lindwasser OW, Smith WJ, Chaudhuri R, Yang P, Hurley JH, Bonifacino JS. 2008. A diacidic motif in human immunodeficiency virus type 1 Nef is a novel determinant of binding to AP-2. J Virol 82:1166–1174. doi: 10.1128/JVI.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pitcher C, Honing S, Fingerhut A, Bowers K, Marsh M. 1999. Cluster of differentiation antigen 4 (CD4) endocytosis and adaptor complex binding require activation of the CD4 endocytosis signal by serine phosphorylation. Mol Biol Cell 10:677–691. doi: 10.1091/mbc.10.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jin YJ, Cai CY, Mezei M, Ohlmeyer M, Sanchez R, Burakoff SJ. 2013. Identification of a novel binding site between HIV type 1 Nef C-terminal flexible loop and AP2 required for Nef-mediated CD4 downregulation. AIDS Res Hum Retroviruses 29:725–731. doi: 10.1089/AID.2012.0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Buffalo CZ, Sturzel CM, Heusinger E, Kmiec D, Kirchhoff F, Hurley JH, Ren X. 2019. Structural basis for tetherin antagonism as a barrier to zoonotic lentiviral transmission. Cell Host Microbe 26:359–368.e8. doi: 10.1016/j.chom.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schouest B, Weiler AM, Janaka SK, Myers TA, Das A, Wilder SC, Furlott J, Baddoo M, Flemington EK, Rakasz EG, Evans DT, Friedrich TC, Maness NJ. 2018. Maintenance of AP-2-dependent functional activities of Nef restricts pathways of immune escape from CD8 T lymphocyte responses. J Virol 92:e01822-17. doi: 10.1128/JVI.01822-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Preusser A, Briese L, Baur AS, Willbold D. 2001. Direct in vitro binding of full-length human immunodeficiency virus type 1 Nef protein to CD4 cytoplasmic domain. J Virol 75:3960–3964. doi: 10.1128/JVI.75.8.3960-3964.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rossi F, Gallina A, Milanesi G. 1996. Nef-CD4 physical interaction sensed with the yeast two-hybrid system. Virology 217:397–403. doi: 10.1006/viro.1996.0130. [DOI] [PubMed] [Google Scholar]
- 117.Salghetti S, Mariani R, Skowronski J. 1995. Human immunodeficiency virus type 1 Nef and p56lck protein-tyrosine kinase interact with a common element in CD4 cytoplasmic tail. Proc Natl Acad Sci U S A 92:349–353. doi: 10.1073/pnas.92.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mangasarian A, Piguet V, Wang JK, Chen YL, Trono D. 1999. Nef-induced CD4 and major histocompatibility complex class I (MHC-I) down-regulation are governed by distinct determinants: N-terminal alpha helix and proline repeat of Nef selectively regulate MHC-I trafficking. J Virol 73:1964–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Arhel N, Lehmann M, Clauss K, Nienhaus GU, Piguet V, Kirchhoff F. 2009. The inability to disrupt the immunological synapse between infected human T cells and APCs distinguishes HIV-1 from most other primate lentiviruses. J Clin Invest 119:2965–2975. doi: 10.1172/JCI38994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kirchhoff F. 2010. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 8:55–67. doi: 10.1016/j.chom.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 121.Iafrate AJ, Bronson S, Skowronski J. 1997. Separable functions of Nef disrupt two aspects of T cell receptor machinery: CD4 expression and CD3 signaling. EMBO J 16:673–684. doi: 10.1093/emboj/16.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schaefer TM, Bell I, Pfeifer ME, Ghosh M, Trible RP, Fuller CL, Ashman C, Reinhart TA. 2002. The conserved process of TCR/CD3 complex down-modulation by SIV Nef is mediated by the central core, not endocytic motifs. Virology 302:106–122. doi: 10.1006/viro.2002.1628. [DOI] [PubMed] [Google Scholar]
- 123.Osman N, Lucas S, Cantrell D. 1995. The role of tyrosine phosphorylation in the interaction of cellular tyrosine kinases with the T cell receptor ξ chain tyrosine-based activation motif. Eur J Immunol 25:2863–2869. doi: 10.1002/eji.1830251023. [DOI] [PubMed] [Google Scholar]
- 124.Schaefer TM, Bell I, Fallert BA, Reinhart TA. 2000. The T-cell receptor ξ chain contains two homologous domains with which simian immunodeficiency virus Nef interacts and mediates down-modulation. J Virol 74:3273–3283. doi: 10.1128/jvi.74.7.3273-3283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim WM, Sigalov AB, Stern LJ. 2010. Pseudo-merohedral twinning and noncrystallographic symmetry in orthorhombic crystals of SIVmac239 Nef core domain bound to different-length TCRξ fragments. Acta Crystallogr D Biol Crystallogr 66:163–175. doi: 10.1107/S090744490904880X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Swigut T, Greenberg M, Skowronski J. 2003. Cooperative interactions of simian immunodeficiency virus Nef, AP-2, and CD3-ξ mediate the selective induction of T-cell receptor-CD3 endocytosis. J Virol 77:8116–8126. doi: 10.1128/jvi.77.14.8116-8126.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rudd CE, Taylor A, Schneider H. 2009. CD28 and CTLA-4 coreceptor expression and signal transduction. Immunol Rev 229:12–26. doi: 10.1111/j.1600-065X.2009.00770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bell I, Schaefer TM, Trible RP, Amedee A, Reinhart TA. 2001. Down-modulation of the costimulatory molecule, CD28, is a conserved activity of multiple SIV Nefs and is dependent on histidine 196 of Nef. Virology 283:148–158. doi: 10.1006/viro.2001.0872. [DOI] [PubMed] [Google Scholar]
- 129.Khalid M, Yu H, Sauter D, Usmani SM, Schmokel J, Feldman J, Gruters RA, van der Ende ME, Geyer M, Rowland-Jones S, Osterhaus AD, Kirchhoff F. 2012. Efficient Nef-mediated downmodulation of TCR-CD3 and CD28 is associated with high CD4+ T cell counts in viremic HIV-2 infection. J Virol 86:4906–4920. doi: 10.1128/JVI.06856-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Leonard JA, Filzen T, Carter CC, Schaefer M, Collins KL. 2011. HIV-1 Nef disrupts intracellular trafficking of major histocompatibility complex class I, CD4, CD8, and CD28 by distinct pathways that share common elements. J Virol 85:6867–6881. doi: 10.1128/JVI.00229-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pawlak EN, Dirk BS, Jacob RA, Johnson AL, Dikeakos JD. 2018. The HIV-1 accessory proteins Nef and Vpu downregulate total and cell surface CD28 in CD4+ T cells. Retrovirology 15:6. doi: 10.1186/s12977-018-0388-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Miura T, Brockman MA, Schneidewind A, Lobritz M, Pereyra F, Rathod A, Block BL, Brumme ZL, Brumme CJ, Baker B, Rothchild AC, Li B, Trocha A, Cutrell E, Frahm N, Brander C, Toth I, Arts EJ, Allen TM, Walker BD. 2009. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte [corrected] recognition. J Virol 83:2743–2755. doi: 10.1128/JVI.02265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chopera DR, Woodman Z, Mlisana K, Mlotshwa M, Martin DP, Seoighe C, Treurnicht F, de Rosa DA, Hide W, Karim SA, Gray CM, Williamson C, CAPRISA 002 Study Team . 2008. Transmission of HIV-1 CTL escape variants provides HLA-mismatched recipients with a survival advantage. PLoS Pathog 4:e1000033. doi: 10.1371/journal.ppat.1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hoffenbach A, Langlade-Demoyen P, Dadaglio G, Vilmer E, Michel F, Mayaud C, Autran B, Plata F. 1989. Unusually high frequencies of HIV-specific cytotoxic T lymphocytes in humans. J Immunol 142:452–462. [PubMed] [Google Scholar]
- 135.Klein MR, van Baalen CA, Holwerda AM, Kerkhof Garde SR, Bende RJ, Keet IP, Eeftinck-Schattenkerk JK, Osterhaus AD, Schuitemaker H, Miedema F. 1995. Kinetics of Gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. J Exp Med 181:1365–1372. doi: 10.1084/jem.181.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Flamand L, Crowley RW, Lusso P, Colombini-Hatch S, Margolis DM, Gallo RC. 1998. Activation of CD8+ T lymphocytes through the T cell receptor turns on CD4 gene expression: implications for HIV pathogenesis. Proc Natl Acad Sci U S A 95:3111–3116. doi: 10.1073/pnas.95.6.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kitchen SG, Korin YD, Roth MD, Landay A, Zack JA. 1998. Costimulation of naive CD8+ lymphocytes induces CD4 expression and allows human immunodeficiency virus type 1 infection. J Virol 72:9054–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Saha K, Zhang J, Gupta A, Dave R, Yimen M, Zerhouni B. 2001. Isolation of primary HIV-1 that target CD8+ T lymphocytes using CD8 as a receptor. Nat Med 7:65–72. doi: 10.1038/83365. [DOI] [PubMed] [Google Scholar]
- 139.Zloza A, Sullivan YB, Connick E, Landay AL, Al-Harthi L. 2003. CD8+ T cells that express CD4 on their surface (CD4dimCD8bright T cells) recognize an antigen-specific target, are detected in vivo, and can be productively infected by T-tropic HIV. Blood 102:2156–2164. doi: 10.1182/blood-2002-07-1972. [DOI] [PubMed] [Google Scholar]
- 140.Brenchley JM, Hill BJ, Ambrozak DR, Price DA, Guenaga FJ, Casazza JP, Kuruppu J, Yazdani J, Migueles SA, Connors M, Roederer M, Douek DC, Koup RA. 2004. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J Virol 78:1160–1168. doi: 10.1128/jvi.78.3.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Imlach S, McBreen S, Shirafuji T, Leen C, Bell JE, Simmonds P. 2001. Activated peripheral CD8 lymphocytes express CD4 in vivo and are targets for infection by human immunodeficiency virus type 1. J Virol 75:11555–11564. doi: 10.1128/JVI.75.23.11555-11564.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Livingstone WJ, Moore M, Innes D, Bell JE, Simmonds P. 1996. Frequent infection of peripheral blood CD8-positive T-lymphocytes with HIV-1. Edinburgh Heterosexual Transmission Study Group. Lancet 348:649–654. doi: 10.1016/S0140-6736(96)02091-0. [DOI] [PubMed] [Google Scholar]
- 143.McBreen S, Imlach S, Shirafuji T, Scott GR, Leen C, Bell JE, Simmonds P. 2001. Infection of the CD45RA+ (naive) subset of peripheral CD8+ lymphocytes by human immunodeficiency virus type 1 in vivo. J Virol 75:4091–4102. doi: 10.1128/JVI.75.9.4091-4102.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cochrane A, Imlach S, Leen C, Scott G, Kennedy D, Simmonds P. 2004. High levels of human immunodeficiency virus infection of CD8 lymphocytes expressing CD4 in vivo. J Virol 78:9862–9871. doi: 10.1128/JVI.78.18.9862-9871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sullivan YB, Landay AL, Zack JA, Kitchen SG, Al-Harthi L. 2001. Upregulation of CD4 on CD8+ T cells: CD4dimCD8bright T cells constitute an activated phenotype of CD8+ T cells. Immunology 103:270–280. doi: 10.1046/j.1365-2567.2001.01243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Heigele A, Schindler M, Gnanadurai CW, Leonard JA, Collins KL, Kirchhoff F. 2012. Down-modulation of CD8αβ is a fundamental activity of primate lentiviral Nef proteins. J Virol 86:36–48. doi: 10.1128/JVI.00717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chowers MY, Spina CA, Kwoh TJ, Fitch NJ, Richman DD, Guatelli JC. 1994. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J Virol 68:2906–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Inuzuka M, Hayakawa M, Ingi T. 2005. Serinc, an activity-regulated protein family, incorporates serine into membrane lipid synthesis. J Biol Chem 280:35776–35783. doi: 10.1074/jbc.M505712200. [DOI] [PubMed] [Google Scholar]
- 149.Trautz B, Wiedemann H, Luchtenborg C, Pierini V, Kranich J, Glass B, Krausslich HG, Brocker T, Pizzato M, Ruggieri A, Brugger B, Fackler OT. 2017. The host-cell restriction factor SERINC5 restricts HIV-1 infectivity without altering the lipid composition and organization of viral particles. J Biol Chem 292:13702–13713. doi: 10.1074/jbc.M117.797332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sood C, Marin M, Chande A, Pizzato M, Melikyan GB. 2017. SERINC5 protein inhibits HIV-1 fusion pore formation by promoting functional inactivation of envelope glycoproteins. J Biol Chem 292:6014–6026. doi: 10.1074/jbc.M117.777714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dai W, Usami Y, Wu Y, Gottlinger H. 2018. A long cytoplasmic loop governs the sensitivity of the anti-viral host protein SERINC5 to HIV-1 Nef. Cell Rep 22:869–875. doi: 10.1016/j.celrep.2017.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shi J, Xiong R, Zhou T, Su P, Zhang X, Qiu X, Li H, Li S, Yu C, Wang B, Ding C, Smithgall TE, Zheng YH. 2018. HIV-1 Nef antagonizes SERINC5 restriction by downregulation of SERINC5 via the endosome/lysosome system. J Virol 92:e00196-18. doi: 10.1128/JVI.00196-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pizzato M, Helander A, Popova E, Calistri A, Zamborlini A, Palu G, Gottlinger HG. 2007. Dynamin 2 is required for the enhancement of HIV-1 infectivity by Nef. Proc Natl Acad Sci U S A 104:6812–6817. doi: 10.1073/pnas.0607622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wu M, Alvarado JJ, Augelli-Szafran CE, Ptak RG, Smithgall TE. 2018. A single beta-octyl glucoside molecule induces HIV-1 Nef dimer formation in the absence of partner protein binding. PLoS One 13:e0192512. doi: 10.1371/journal.pone.0192512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Evans DT, Serra-Moreno R, Singh RK, Guatelli JC. 2010. BST-2/tetherin: a new component of the innate immune response to enveloped viruses. Trends Microbiol 18:388–396. doi: 10.1016/j.tim.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jouvenet N, Neil SJ, Zhadina M, Zang T, Kratovac Z, Lee Y, McNatt M, Hatziioannou T, Bieniasz PD. 2009. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J Virol 83:1837–1844. doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 158.Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, Bieniasz PD. 2009. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Galão RP, Le Tortorec A, Pickering S, Kueck T, Neil SJ. 2012. Innate sensing of HIV-1 assembly by tetherin induces NFκB-dependent proinflammatory responses. Cell Host Microbe 12:633–644. doi: 10.1016/j.chom.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sauter D, Schindler M, Specht A, Landford WN, Munch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, Takehisa J, Ogando Y, Ochsenbauer C, Kappes JC, Ayouba A, Peeters M, Learn GH, Shaw G, Sharp PM, Bieniasz P, Hahn BH, Hatziioannou T, Kirchhoff F. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Schmökel J, Sauter D, Schindler M, Leendertz FH, Bailes E, Dazza MC, Saragosti S, Bibollet-Ruche F, Peeters M, Hahn BH, Kirchhoff F. 2011. The presence of a vpu gene and the lack of Nef-mediated downmodulation of T cell receptor-CD3 are not always linked in primate lentiviruses. J Virol 85:742–752. doi: 10.1128/JVI.02087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Serra-Moreno R, Zimmermann K, Stern LJ, Evans DT. 2013. Tetherin/BST-2 antagonism by Nef depends on a direct physical interaction between Nef and tetherin, and on clathrin-mediated endocytosis. PLoS Pathog 9:e1003487. doi: 10.1371/journal.ppat.1003487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhang F, Landford WN, Ng M, McNatt MW, Bieniasz PD, Hatziioannou T. 2011. SIV Nef proteins recruit the AP-2 complex to antagonize tetherin and facilitate virion release. PLoS Pathog 7:e1002039. doi: 10.1371/journal.ppat.1002039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Habermann A, Krijnse-Locker J, Oberwinkler H, Eckhardt M, Homann S, Andrew A, Strebel K, Kräusslich H-G. 2010. CD317/tetherin is enriched in the HIV-1 envelope and downregulated from the plasma membrane upon virus infection. J Virol 84:4646–4658. doi: 10.1128/JVI.02421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Götz N, Sauter D, Usmani SM, Fritz JV, Goffinet C, Heigele A, Geyer M, Bibollet-Ruche F, Learn GH, Fackler OT, Hahn BH, Kirchhoff F. 2012. Reacquisition of Nef-mediated tetherin antagonism in a single in vivo passage of HIV-1 through its original chimpanzee host. Cell Host Microbe 12:373–380. doi: 10.1016/j.chom.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sauter D, Specht A, Kirchhoff F. 2010. Tetherin: holding on and letting go. Cell 141:392–398. doi: 10.1016/j.cell.2010.04.022. [DOI] [PubMed] [Google Scholar]
- 168.Arias JF, Colomer-Lluch M, von Bredow B, Greene JM, MacDonald J, O’Connor DH, Serra-Moreno R, Evans DT. 2016. Tetherin antagonism by HIV-1 group M Nef proteins. J Virol 90:10701–10714. doi: 10.1128/JVI.01465-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang J, Davis S, Menon S, Zhang J, Ding J, Cervantes S, Miller E, Jiang Y, Ferro-Novick S. 2015. Ypt1/Rab1 regulates Hrr25/CK1δ kinase activity in ER-Golgi traffic and macroautophagy. J Cell Biol 210:273–285. doi: 10.1083/jcb.201408075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Blander JM. 2016. The comings and goings of MHC class I molecules herald a new dawn in cross-presentation. Immunol Rev 272:65–79. doi: 10.1111/imr.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Blander JM. 2018. Regulation of the cell biology of antigen cross-presentation. Annu Rev Immunol 36:717–753. doi: 10.1146/annurev-immunol-041015-055523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hewitt EW. 2003. The MHC class I antigen presentation pathway: strategies for viral immune evasion. Immunology 110:163–169. doi: 10.1046/j.1365-2567.2003.01738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Blees A, Januliene D, Hofmann T, Koller N, Schmidt C, Trowitzsch S, Moeller A, Tampe R. 2017. Structure of the human MHC-I peptide-loading complex. Nature 551:525–528. doi: 10.1038/nature24627. [DOI] [PubMed] [Google Scholar]
- 174.Blagoveshchenskaya AD, Thomas L, Feliciangeli SF, Hung CH, Thomas G. 2002. HIV-1 Nef downregulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell 111:853–866. doi: 10.1016/s0092-8674(02)01162-5. [DOI] [PubMed] [Google Scholar]
- 175.Roeth JF, Williams M, Kasper MR, Filzen TM, Collins KL. 2004. HIV-1 Nef disrupts MHC-I trafficking by recruiting AP-1 to the MHC-I cytoplasmic tail. J Cell Biol 167:903–913. doi: 10.1083/jcb.200407031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wonderlich ER, Williams M, Collins KL. 2008. The tyrosine binding pocket in the adaptor protein 1 (AP-1) μ1 subunit is necessary for Nef to recruit AP-1 to the major histocompatibility complex class I cytoplasmic tail. J Biol Chem 283:3011–3022. doi: 10.1074/jbc.M707760200. [DOI] [PubMed] [Google Scholar]
- 177.Kent MS, Murton JK, Sasaki DY, Satija S, Akgun B, Nanda H, Curtis JE, Majewski J, Morgan CR, Engen JR. 2010. Neutron reflectometry study of the conformation of HIV Nef bound to lipid membranes. Biophys J 99:1940–1948. doi: 10.1016/j.bpj.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Akgun B, Satija S, Nanda H, Pirrone GF, Shi X, Engen JR, Kent MS. 2013. Conformational transition of membrane-associated terminally acylated HIV-1 Nef. Structure 21:1822–1833. doi: 10.1016/j.str.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pirrone GF, Emert-Sedlak LA, Wales TE, Smithgall TE, Kent MS, Engen JR. 2015. Membrane-associated conformation of HIV-1 Nef investigated with hydrogen exchange mass spectrometry at a Langmuir monolayer. Anal Chem 87:7030–7035. doi: 10.1021/acs.analchem.5b01725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gerlach H, Laumann V, Martens S, Becker CF, Goody RS, Geyer M. 2010. HIV-1 Nef membrane association depends on charge, curvature, composition and sequence. Nat Chem Biol 6:46–53. doi: 10.1038/nchembio.268. [DOI] [PubMed] [Google Scholar]
- 181.Faini M, Prinz S, Beck R, Schorb M, Riches JD, Bacia K, Brugger B, Wieland FT, Briggs JA. 2012. The structures of COPI-coated vesicles reveal alternate coatomer conformations and interactions. Science 336:1451–1454. doi: 10.1126/science.1221443. [DOI] [PubMed] [Google Scholar]
- 182.Bykov YS, Schaffer M, Dodonova SO, Albert S, Plitzko JM, Baumeister W, Engel BD, Briggs JA. 2017. The structure of the COPI coat determined within the cell. Elife 6:e32493. doi: 10.7554/eLife.32493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Singh P, Yadav GP, Gupta S, Tripathi AK, Ramachandran R, Tripathi RK. 2011. A novel dimer-tetramer transition captured by the crystal structure of the HIV-1 Nef. PLoS One 6:e26629. doi: 10.1371/journal.pone.0026629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Breuer S, Schievink SI, Schulte A, Blankenfeldt W, Fackler OT, Geyer M. 2011. Molecular design, functional characterization and structural basis of a protein inhibitor against the HIV-1 pathogenicity factor Nef. PLoS One 6:e20033. doi: 10.1371/journal.pone.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Alvarado JJ, Tarafdar S, Yeh JI, Smithgall TE. 2014. Interaction with the Src homology (SH3-SH2) region of the Src-family kinase Hck structures the HIV-1 Nef dimer for kinase activation and effector recruitment. J Biol Chem 289:28539–28553. doi: 10.1074/jbc.M114.600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lülf S, Matz J, Rouyez MC, Jarviluoma A, Saksela K, Benichou S, Geyer M. 2014. Structural basis for the inhibition of HIV-1 Nef by a high-affinity binding single-domain antibody. Retrovirology 11:24. doi: 10.1186/1742-4690-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Horenkamp FA, Breuer S, Schulte A, Lulf S, Weyand M, Saksela K, Geyer M. 2011. Conformation of the dileucine-based sorting motif in HIV-1 Nef revealed by intermolecular domain assembly. Traffic 12:867–877. doi: 10.1111/j.1600-0854.2011.01205.x. [DOI] [PubMed] [Google Scholar]