Abstract
Objective
To investigate compositional differences in the gut microbiota associated with bone homeostasis and fractures in a cohort of older adults.
Methods
Faecal microbiota profiles were determined from 181 individuals with osteopenia (n = 61) or osteoporosis (n = 60), and an age- and gender-matched group with normal BMD (n = 60). Analysis of the 16S (V3-V4 region) amplicon dataset classified to the genus level was used to identify significantly differentially abundant taxa. Adjustments were made for potential confounding variables identified from the literature using several statistical models.
Results
We identified six genera that were significantly altered in abundance in the osteoporosis or osteopenic groups compared with age- and gender-matched controls. A detailed study of microbiota associations with meta-data variables that included BMI, health status, diet and medication revealed that these meta-data explained 15–17% of the variance within the microbiota dataset. BMD measurements were significantly associated with alterations in the microbiota. After controlling for known biological confounders, five of the six taxa remained significant. Overall microbiota alpha diversity did not correlate to BMD in this study.
Conclusion
Reduced BMD in osteopenia and osteoporosis is associated with an altered microbiota. These alterations may be useful as biomarkers or therapeutic targets in individuals at high risk of reductions in BMD. These observations will lead to a better understanding of the relationship between the microbiota and bone homeostasis.
Keywords: osteoporosis, gut microbiota, bone mineral density, elderly, osteopenia
Rheumatology key messages
Reduced BMD is associated with taxon-specific signatures in the gut microbiota.
Medication, anthropometric measures, nutrition and gender are associated with gut microbiota composition.
Confounders do not explain the microbiota–bone density interactions observed here.
Introduction
Osteoporosis, characterized by reduced BMD and degradation of the micro-architectural structure of bone, affects over 27.5 million people in Europe [1]. Over the age of 50 years, one in three women and one in five men, worldwide, will experience an osteoporotic fracture in their lifetime, representing a significant burden for patients and health care providers [2]. The aetiology of osteoporosis and its precursor, osteopenia, is multi-factorial. Contributing factors include oestrogen and vitamin D deficiency, and genetic modification in regulatory genes such as vitamin D receptors and TGF-β [3]. Osteoporosis occurrence is accelerated in patients with immune-mediated inflammatory conditions, where excessive production of pro-inflammatory cytokines leads to increased osteoclastic bone resorption (e.g. IBD, RA and AS) [4–6]. The gut microbiome is known to modulate immune cell activities and alterations in the microbiome have previously been associated with these inflammatory conditions [7].
The gut microbiome shares a complex relationship with the host. Development and maturation of the innate and adaptive immunity in the host is dependent on appropriate exposure to the gut microbiota [8]. Alterations in the microbiota may result in immune system modulation or activation. Circulating osteoclastogenic cytokines may be increased in a T-cell-dependent mechanism by the microbiota, which can drive bone resorption in inflammatory conditions [9]. Several investigations have identified microbes that regulate the production of hormones or improve uptake of vitamins that are integral to bone health [10, 11].
Studies with germ-free and antibiotic-treated animals have indicated the possibility of gut microbial influence on both bone mass accumulation and turnover. These animals have shown a reduction in osteoclastic precursor cell number [12], an increase in bone mass [13], and improvement in bone strength and material properties [14].
We have previously identified significant microbiota alterations associated with inflamm-aging and frailty in an elderly cohort [15]. Other studies have demonstrated that the absence of gut microbiota leads to a reduction in bone mechanical strength [16] and inversely, long term colonization of pathogen-free gut microbiota increases bone formation [17]. In contrast, another recent study suggested that microbiota restoration in germ-free mice does not affect bone loss [18]. These conflicting findings may, in part, be due to different animal genotypes, the anti-microbials administered and the absence or presence of particular taxa in their baseline microbiota.
Our aim in the present study was to determine whether gut microbiota features are associated with BMD in a cohort of individuals at high risk of reduced BMD and fractures. In addition to this, any genus-level taxa associated with altered BMD would be identified by comparing the gut microbiota composition of osteopenic and osteoporotic patients with those of age- and gender-matched controls with normal BMD. Our hypothesis was that intestinal microbiota composition was different in the osteoporotic subjects. Furthermore, we developed and applied a rigorous statistical regime to remove the effect of potentially confounding variables.
Methods
Subject recruitments and clinical information
Ethical approval was granted by the Clinical Research Ethics Committee of the Cork Teaching Hospitals before recruitment. Adult female and male subjects, aged 55–75 years, were recruited from the bone densitometry unit at Cork University Hospital, Cork, Ireland. The indications for referral for BMD assessment by dual-energy X-ray absorptiometry were varied, with referrals from primary, secondary and tertiary care. No single specific referral criterion was used, and request for assessment was at the discretion of the attending clinician and not the study investigators. Written informed consent was obtained from the participants. Individuals with a known history of alcohol abuse, participation in an investigational drug trial in the 30 days before enrolment, use of antibiotics in the 3 months prior to bone density measurement, and previous partial or total colectomy were excluded. No measure was taken to exclude participants with co-existing OA, aortic calcification or fractures. Altogether, stool samples were collected from 193 participants. Due to lack of vitamin D information from 12 samples, they were excluded from the analysis, resulting in the final dataset comprising of 181 participants.
Patients underwent dual-energy X-ray absorptiometry assessment of BMD (g/cm2) at the femoral neck and antero-posterior lumbar spine (L1-L4) with a GE Healthcare Lunar iDXA machine (GE Healthcare, Madison, WI) and enCORE software (V.13.4, 2010) using standardized methodology [19]. T-score threshold was used to define three groups based on their BMD. These were normal BMD (n = 60) with a T-score of ⩾–1, patients with osteopenia (n = 61) with a T-score between –1 and –2.5, and patients with osteoporosis (n = 60) were defined as having a T-score of ⩽–2.5 [20, 21]. The detailed procedure of recording anthropometric, clinical, dietary and medications information is recorded in the supplementary material, available at Rheumatology online.
Molecular methods and bioinformatics
Genomic DNA was extracted from 0.25 g of each of the faecal samples based on a modified Yu and Morrison protocol [22]. The V3-V4 region of the 16S rRNA gene was amplified and sequenced [23] on the Illumina MiSeq platform at Moorepark Teagasc Food Research Centre, Fermoy, Ireland. The reads were merged using FLASH (v1.2.8) [24]. The forward adapters were removed using cutadapt (v1.8.3). The quality filtering of reads and removal of reverse primers were carried out using the QIIME (v1.9.1) [25] pipeline with default settings. The removal of chimeric sequences and generation of operational taxonomic units at 97% identity threshold was done using USEARCH (v8.1) [26]. Representative operational taxonomic units were classified using the Ribosomal Database Project (RDP) database (v11.4) [27] implemented in mothur (v1.34.4) [28]. α- and β-diversity measures were produced from a rarefied dataset (10 613 reads per sample).
Statistical analysis
All statistical analyses were carried out in the R statistical software (v3.4.0) [29]. Significance was determined by a cut-off P-value ⩽0.05 and P-adjusted ⩽0.05 (Benjamini-Hochberg procedure) unless stated otherwise. P-adjusted for pairwise comparison is based on the P-values obtained from all the pairwise comparisons for each variable.
Analysis of meta-data
Kruskal–Wallis, Dunn’s test (v1.3.4) [30] and/or χ2 tests were carried out to identify anthropometric, clinical, dietary and medications significantly different between the groups. For χ2 testing, at least seven participants were present across the whole dataset for that factor.
Analysis of microbiota data
Kruskal–Wallis test was used to determine significant difference in α-diversity measures between the groups. Co-inertia analysis was used to explore the covariance between the dietary dataset and microbiota dataset. DESeq2 (v1.16.1) [31] was used to identify differentially abundant taxa from the microbiota dataset. The dataset was filtered to retain only those taxa that were present in at least 20% of the samples across the whole dataset. A DESeq2 model adjusted for BMI and gender was used to identify genera that were significantly differentially abundant.
Identification of meta-data variables associated with beta-diversity
Meta-data variables significantly associated with variations in global microbiota profiles were identified using permutational multivariate analysis of variance. A nominal P-value of ⩽0.05 was used as the analysis was a confirmation of previously established associations. Subjects with diseases such as coeliac disease, diverticulitis and inflammatory arthritis conditions were present within the dataset and were tested separately. Inflammatory and non-inflammatory diseases can alter the microbiota with a common dysbiosis signature [32]. To investigate the common signature of microbiota-associated inflammatory diseases, we created an inflammatory disease index, where the presence of any one of the microbiota-associated conditions (coeliac, diverticulitis, arthritis, IBD and multiple sclerosis) was considered. Nominally significant meta-data variables were added to a single permutational multivariate analysis of variance model to identify overall effect sizes. The cumulative effect was calculated based on these pre-defined groups of variables.
Analysis of confounding variables
Clinical variables that have been reported to interact with the microbiota were identified from the literature (supplementary Table S1, available at Rheumatology online). These included diet [Healthy Food Diversity (HFD) index] [33], Barthel score [34], Godin leisure time activity score [35], Mini-Mental State Examination scores [36], Mini Nutritional Assessment [37] and Carlson co-morbidity index [38]. Secondly, the meta-data identified as significantly different between the subject groups were confirmed by a literature search (Table 1, supplementary Table S2, available at Rheumatology online; P-adjusted ⩽0.05) and were added to the analysis as potential confounders.
Table 1.
Significant characteristics of the participants in the final dataset
| Meta-data | Healthy (n = 60) | Osteopenia (n = 61) | Osteoporosis (n = 60) | Significance |
|---|---|---|---|---|
| Gender (male/female) | 13/47 | 7/54 | 11/49 | NS |
| Age (years) | 63.57 ± 5.73 | 64.84 ± 5.28 | 65.07 ± 5.58 | NS |
| BMI | 29.09 ± 4.57 | 27.20 ± 4.80 | 23.96 ± 3.31 | *** |
| Weight (kg) | 78.86 ± 13.60 | 70.96 ± 14.44 | 61.65 ± 9.44 | *** |
| Waist circumference (cm) | 95.71 ± 11.95 (13/46) | 89.81 ± 12.40 (6/54) | 81.81 ± 9.36 | *** |
| Hip circumference (cm) | 106.71 ± 9.83 (13/46) | 103.63 ± 10.45 (6/53) | 96.66 ± 7.26 | *** |
| Waist–hip ratio | 0.90 ± 0.08 (13/46) | 0.87 ± 0.06 (6/53) | 0.85 ± 0.07 | ** |
| Mid arm circumference (cm) | 30.98 ± 3.62 (12/47) | 28.85 ± 3.97 | 26.80 ± 2.91 | *** |
| Calf circumference (cm) | 37.69 ± 3.73 (11/47) | 35.76 ± 4.28 | 33.93 ± 2.76 | *** |
| AP spine T-score | 0.28 ± 1.02 | –1.16 ± 0.87 | –2.86 ± 0.74 | *** |
| AP spine BMD (g/cm2) | 1.22 ± 0.13 | 1.04 ± 0.11 | 0.84 ± 0.09 | *** |
| Neck-femur T-score | –0.54 ± 0.35 | –1.27 ± 0.53 | –1.95 ± 0.80 | *** |
| Neck-femur BMD (g/cm2) | 0.98 ± 0.09 | 0.84 ± 0.07 | 0.84 ± 0.68 | *** |
| Vitamin D3 [25(OH)D3] (nmol/L) | 60.49 ± 20.84 | 69.98 ± 25.27 | 75.96 ± 26.43 | ** |
| Total Vitamin D [25(OH)D)] (nmol/L) | 63.68 ± 20.57 | 72.40 ± 25.36 | 79.18 ± 26.07 | ** |
| Calcium supplements (yes/no) | 10/50 | 31/30 | 35/25 | *** |
| Bisphosphonate medication (yes/no) | 4/56 | 6/55 | 17/43 | *** |
Group-wise comparisons of the clinical variables. Kruskal–Wallis or χ2 statistic was used to determine significance. The values represent mean ± s.d. or number of samples per group. 25(OH)D3: vitamin D3; Total Vitamin D [25(OH)D]: total vitamin D. Significance: P-adjusted.
≤0.0005;
≤0.005;
NS: not significant. Values in brackets for circumference measures and waist–hip ratio represents different sample size. The complete list of sample characteristics along with pairwise comparisons is available in supplementary Table S2, available at Rheumatology online. AP: anterior-posterior.
Confounding factors were modelled using a general linear mixed-effect model using the negative binomial distribution, and the sequencing depth was controlled for by categorizing the number of reads into four quartiles and adding this information as a random effect to the model. Firstly, univariate general linear mixed-effect models were generated with individual confounding factors as the predictor and the significant taxa as the response. The confounders identified as significant for individual taxa were controlled for in a bivariate model. To maximize the number of known confounders identified, a nominal P-value was regarded as significant. In this model, the effect of group category was evaluated after adjustment for the individual significant confounders. Summary reports were generated for both the univariate and bivariate general linear mixed-effect models to explain the contribution of the predictors.
An expanded methodology is available in the supplementary material, available at Rheumatology online.
Results
Descriptive statistics of the study population
In the present study, samples and clinical information for 181 individuals were analysed. These patients were evenly divided between those with normal BMD (n = 60), osteopenia (n = 61) and osteoporosis (n = 60) groups. Clinical, physiological, biomedical and dietary measures were investigated and significant differences between normal BMD, osteopenia and osteoporosis participants were detected. Differences in bone density measurements (T-score and BMD of the anterior-posterior spine and neck of femur) were confirmed and differences in BMI, weight, circumference measures, vitamin D levels, and the use of calcium and bisphosphonate supplements were noted (Table 1, supplementary Table S2 and Figs S1 and S2A and B, available at Rheumatology online). Due to the recruitment by clinical referral of this high risk cohort, there was a high rate of fractures in all groupings, with percentages for one or more fractures being 40% (24/60), 59% (36/61) and 42% (25/60) for normal BMD, osteopenia and osteoporosis groups, respectively, and percentages for two or more fractures being 7% (4/60), 23% (14/61) and 15% (9/60), respectively.
Microbiota characterization
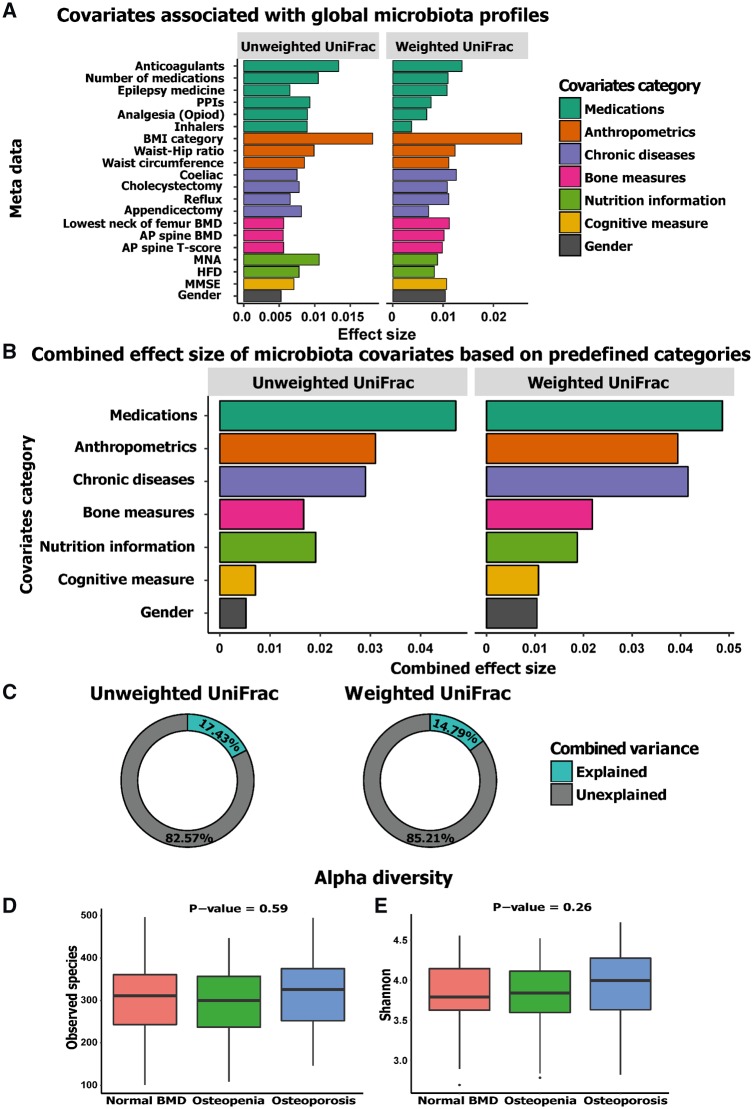
The microbiota composition of the samples analysed was dominated by phylum Firmicutes, with a mean abundance of 78.9% across the whole dataset, followed in rank abundance order by Bacteroidetes, accounting for 14.9%. Other phyla accounted for 5.8%, while 0.4% were unclassified (supplementary Fig. S3A, available at Rheumatology online). The core microbiota consisted of 23 genera that were found in at least 90% of the samples. The top five genera with mean relative abundance in the whole dataset were Faecalibacterium (11.7%), Bacteroides (9.4%), Roseburia (7.9%), Blautia (7.6%) and Coprococcus (3.2%) (supplementary Fig. S3B, available at Rheumatology online). Based on principal coordinate analysis on different β-diversity measures, Axes 1 and 2 explained 11–17% and 8–13% of variance, respectively (supplementary Fig. S4A and Table S3, available at Rheumatology online). The relationship of BMD measures with global microbiota profile was visualized using distance-based redundancy analysis, testing anterior-posterior spine BMD measure with Bray–Curtis distance (supplementary Fig. S4B, available at Rheumatology online). With regard to α-diversity, an average richness of 308.7 ± 84.2 was observed and extrapolated richness (chao1) was estimated at 406.8 ± 122 (Fig. 1D, supplementary Fig. S4C, available at Rheumatology online) No significant difference was observed in any of the alpha diversity indices among the three clinical groups (Fig. 1D and E, supplementary Fig. S4C and D, available at Rheumatology online).
Fig. 1.
Effect size of covariates significantly associated with global microbiota profiles
Significance was defined as a P-value of 0.05. (A) A total of 20 factors were identified to be nominally significantly associated with β-diversity. The bar plot shows the variation explained by each factor individually on microbiota composition (weighted and unweighted UniFrac). The factors are sorted based on their mean cumulative (grouped into predefined categories) and individual effect size from both distance measures. (B) The combined variance explained by the predefined categories. (C) The donut plot shows the portion of combined variance explained by the nominally significant factors on weighted and unweighted UniFrac measures, respectively. (D and E) The lack of significant difference in observed species diversity measure and Shannon index, respectively. PPIs: proton pump inhibitors; MNA: Mini Nutritional Assessment; HFD: Healthy Food Diversity; MMSE: Mini-Mental State Examination.
Association of gut microbiota with covariates
Both sets of bone density measurements and one of the T-scores tested explained a significant amount of microbiota variance (P-value ⩽0.05), verifying the original hypothesis that BMD is associated with alterations in the microbiota (Fig. 1, supplementary Table S4, available at Rheumatology online). We extended this beta-diversity analysis to known microbiota-associated putative meta-data variables to measure their effect on the microbiota (supplementary Table S4, available at Rheumatology online). This analysis identified 20 meta-data variables to be associated with the global microbiota profile (Fig. 1A), with BMI having the largest effect size individually (2.1%). An inflammatory disease index was created indicating the presence or absence of a disease, disorder or condition. This index showed a significant association with the β-diversity (Bray-Curtis P-value 0.042, R2 = 0.009).
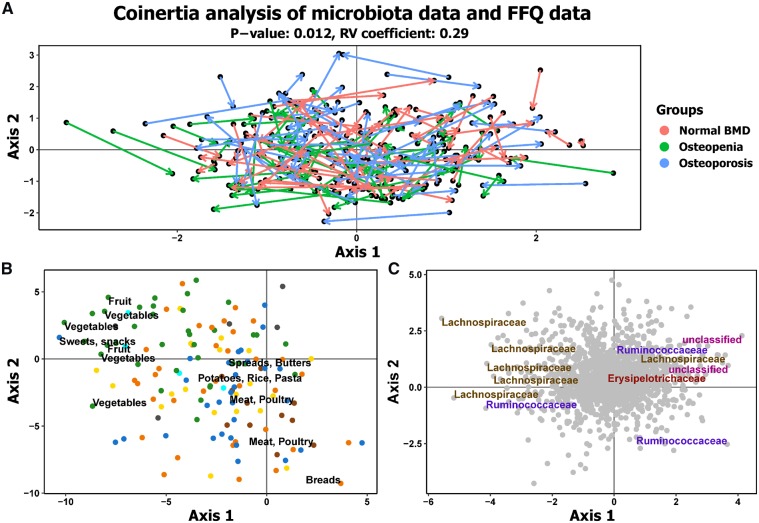
Among the significant variables, the combined effect-size of the different medications explained the most variance (4.8%), followed by anthropometric measures (3.5%). Chronic diseases explained 3.5% and BMD measurement was the fourth largest contributor to effect size (2%). Nutritional information (HFD and Mini Nutritional Assessment), cognitive measures (Mini-Mental State Examination) and gender explained 1.4, 1 and 0.6% of variance, respectively (Fig. 1B). Overall, a cumulative total range of 15–17% of the variance in our dataset was explained, which indicates that stochastic factors explain the majority of the variance in global microbiota composition (Fig. 1C). Analysis of the Food Frequency Questionnaire data and diet quality as measured by the HFD index revealed no significant difference in diet composition or HFD across the three groups. Co-inertia analysis of the Food Frequency Questionnaire dataset with the microbiota dataset (Fig. 2A) graphically confirmed a significant co-variation between the two datasets, which was independent of the defined bone health groups.
Fig. 2.
Food profile is significantly associated with microbiota profile based on the CIA
(A) The CIA of the FFQ PCA and microbiota PCA, where the arrows relate the position of the samples in the FFQ dataset in relation to the microbiota dataset. (B) The FFQ item category associated with the visualized trends. Green dots represent fruits and vegetables, orange represents grains, cereals and bread, brown represents meats, cyan represents fish, yellow represents dairy products, blue represents sweets, cakes and alcohol, and grey represents vitamins, minerals and tea. The food items on the most extreme ends are labelled. (C) The microbial taxa at family level associated with visualized trends. The taxa present at the extreme ends are labelled. CIA: co-inertia analysis; FFQ: Food Frequency Questionnaire; PCA: principal component analysis.
Identification of significantly differentially abundant taxa in patients with osteopenia and osteoporosis
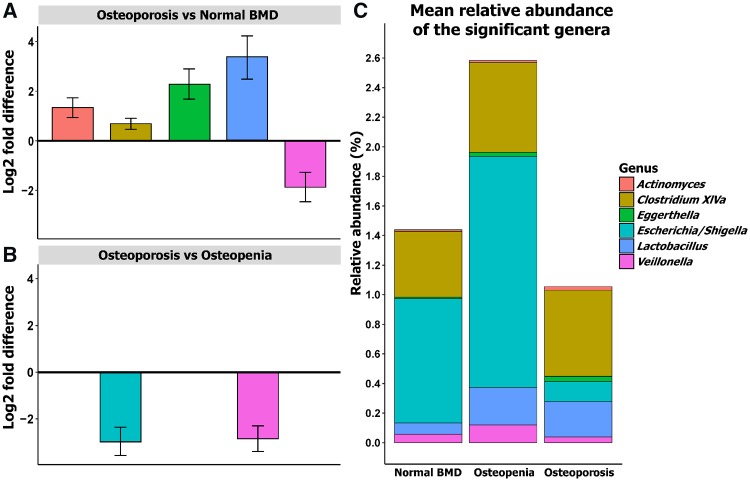
DESeq2 statistical analysis was used to identify genera that were differentially abundant across the groups with adjustment for BMI and gender (Fig. 3A and B, supplementary Table S5, available at Rheumatology online). In summary, we found that Escherichia/Shigella and Veillonella were more abundant in subjects with osteopenia compared with those with osteoporosis. Actinomyces, Eggerthella, Clostridium Cluster XlVa and Lactobacillus were more abundant in subjects with osteoporosis compared with the normal BMD group. We did not identify any taxa significantly differentially abundant in osteopenia compared with the normal BMD group. The relative abundance of these taxa is shown in Fig. 3C.
Fig. 3.
Taxa with differential abundance across the BMD groups
Plot of the log2-fold difference from the significantly differentially abundant genera in pairwise analysis between the groups from the DESeq2 analysis when the model is adjusted for BMI and gender. Based on the log2-fold difference, (A) shows the genera that are significantly higher in osteoporosis compared with normal BMD, (B) represents the genera that are significantly more abundant in osteoporosis compared with osteopenia and (C) represents the relative abundance of the significant genera in the three groups identified in DESeq2.
Alterations at taxonomic levels are not associated with confounding factors
It is well established that many confounding factors may affect the intestinal microbiota [39]. Therefore, it is important to account for confounders potentially affecting the significant taxa identified. We implemented an in-depth statistical analysis to control for potential cofounders based on a combination of previously published approaches [39, 40]. Each significant taxon was tested against the confounding meta-data factors as outlined in the Methods section. A total of 29 factors and the inflammatory disease index were analysed and based on the results of the univariate models (supplementary Table S6, available at Rheumatology online), the bivariate models explaining the associations with each significant genus were generated (supplementary Table S7, available at Rheumatology online).
Significant associations with the different significantly differentially abundant genera were explained by a range of factors including diet, frailty variables, levels of physical activity, medications, weight, BMI, gender and bone density measurements, including the osteopenic and osteoporotic groups (supplementary Table S6, available at Rheumatology online) based on the univariate models. Based on the bivariate models, five of the six previously identified genera remained significantly differentially abundant after adjustment for known confounding factors (supplementary Table S7, available at Rheumatology online). The inflammatory disease index did not show any significant association with these significant taxa in the bivariate models. Lactobacillus abundance was not significantly associated with any of the bone density measurements in the univariate and bivariate models unless BMI was included in the model and therefore was no longer considered.
Our analysis shows that BMI is significantly associated with anterior-posterior spine BMD measures but not with lowest neck of femur BMD values (supplementary Tables S8a and S9a, available at Rheumatology online). The removal of the effect of BMI, medications and vitamin D levels (supplementary Tables S8b–e and S9b–e, available at Rheumatology online) retained all but two of the results, with Clostridium XlVa and Veillonella losing significance (supplementary Tables S8f and S9f, available at Rheumatology online).
Discussion
This is the largest study to-date to investigate associations between the microbiota and reduced bone density in a human cohort including individuals suffering from osteopenia and osteoporosis. We have identified significant associations between different gut microbial genera and reduced bone density in this well-characterized cohort. Extensive investigation by considering the potential influence of various confounders clearly established that the taxonomic differences observed are not explained by the confounders.
It has been observed that the microbiome field suffers from a proliferation of small datasets that show associations of the microbiome with particular diseases or states, without the ability to adequately control for confounding variables. Here we show that global alterations in the gut microbiota are associated with BMD measures, and these interactions explain a similar amount of variance compared with other known microbiota-associated diseases and disorders. This confirms our hypothesis of the association of the gut microbiota alterations with a reduction in BMD in the elderly.
Diseases, disorders and medical conditions are associated with smaller effect sizes compared with medications [39, 41]. In-depth analysis of confounding variables revealed that bisphosphonate and calcium supplements show no significant association with the global microbiota profile. This is consistent with previous reports that bisphosphonates are not significantly associated with gut microbiota markers and the evidence for microbiota alteration in association with calcium intake is weak [42]. We identified six individual gut microbial taxa that may affect bone metabolism. This modest result contrasts with a small cohort study that identified a large number of alterations associated with osteoporosis and osteopenia patients in the microbiota at the global and genus level [43]. The lack of replication of these global alterations in this cohort shows the importance of adequate sample sizes and controlling for multiple testing when investigating possible new associations.
A loss of microbiota diversity is associated with a wide range of disease states, and microbiota diversity is widely considered as an important indicator of health. Within this context, the lack of significant differences in the within-sample diversity measures is interesting. However, it has been observed previously that despite loss of commensal population with the elderly microbiota and noticeable differences in microbiome composition and other host-associated factors (e.g. inflammation, dietary patterns), there was no significant observable difference in overall diversity in ageing individuals [44] and between frail and non-frail elderly individuals [40].
The taxa identified resonate well with the bone density–microbiome literature. Actinomyces abundance in the osteoporosis group here is in concordance with findings that Actinomyces is involved in the development of bisphosphonate-related osteonecrosis of the jaw [45], and it has been proposed that prolonged courses of antimicrobial therapy targeting this organism may lead to better clinical outcomes [46]. The increase in Clostridium XlVa in the osteoporotic group represents a means by which the gut microbiota may influence bone state acting through several differentiating mechanisms [47]. Clostridium XlVa induces accumulation and differentiation of T-regulator cells, which in turn are responsible for bone homeostasis [48]. Clostridium XlVa is an important producer of butyrate, a short chain fatty acid known to stimulate bone formation [49]. Further functional analysis of this group of microorganisms may provide insight into how the gut microbiota affects BMD through modulation of the host’s immune system and metabolism.
Vitamin D receptor polymorphisms are associated with increased osteoporotic fracture risk [50]. The increase in Eggerthella abundance in the osteoporotic group is of interest, as absence of the vitamin D receptor leads to increased Eggerthella abundance and other unfavourable alterations in the intestinal microbiota in murine models [51]. The current investigation also found that vitamin D concentration is associated with a decrease in the relative abundance of Escherichia/Shigella (supplementary Tables S6 and S7, available at Rheumatology online), mirroring other findings looking at vitamin D supplementation [52]. The high relative abundance of this genus in osteopenic but not in osteoporotic patients may be partially due to the greater use of oral vitamin D supplementation among the patients with osteoporosis.
A number of microbes belonging to the phylum Firmicutes are known metabolizers of isoflavone diadzin to equol, which is an oestrogen analogue [53]. This includes species from the genus Veillonella, which we have observed to be decreased in osteoporotic patients. This suggests that a reduction in Veillonella would lead to lower production of equol, which in turn leads to a lack of inhibition of bone resorption.
An analysis of the meta-data revealed that diet and BMI were large contributors to variance in the dataset, with BMI being the largest single contributor, in line with numerous reports linking gut microbiota with obesity [41]. Our study investigated and confirmed the effect of these variables that can alter the microbiota as reported by previous studies. These included various medications that have a profound effect on the microbiota profiles such as proton pump inhibitors and the general term of polypharmacy [42, 54]. Thus, the current study corroborates previous reports which show that cumulative medication use has the largest effect size on global microbiota profiles [39, 41]. However, neither these alterations nor chronic diseases [41, 55] or anthropometric measures explained the observed microbiota alterations.
The relationships between BMI and BMD and the microbiota is complex. Although lower BMI has been associated with a higher fracture rate [56], a high amount of fat mass may provide no beneficial effect on bone health [57]. Within this study, individuals with a higher BMI tended to have higher BMD, which is consistent with the literature [58]. BMI is known to be associated with microbiota alterations. Our analysis has considered both of these BMI associations. Of the taxa related to BMD, Lactobacillus and Veillonella were significantly related (P-value <0.05) to both the obese category and BMD, while Clostridium XlVa showed trends of associations with the obese category (P-value <0.1). However, the Lactobacillus correlation was not significant without adjustment for BMI and so was considered a false positive. Further analysis showed that with removal of variance associated with BMI and medications from the BMD measures results in Veillonella and Clostridium XlVa losing significance. Other results were unaffected, showing that the associations are independent of BMI. Therefore, the association of Clostridium XlVa and Veillonella with BMD should be interpreted with caution.
This is the first investigation of the intestinal microbiota in a large well-characterized human adult cohort with respect to BMD, with one previous study having a limited sample size [43]. Nevertheless, the current study has certain limitations. Due to the recruitment of individuals through consultant referral, the normal BMD cohort are not truly representative of the general population, as highlighted by the high fracture rate in this group. However, a history of fractures was not associated with a detectable alteration in the microbiota, and controlling for this variable confirmed the BMD results but did not improve the analysis. Due to the incomplete information of the standalone vitamin supplements, we included serum vitamin D levels to use directly measured concentrations to account for vitamin D. The number of variables that can be tested in the identification of confounding factors through statistical analyses is limited by the sample size. However, this analysis was not dependent on the statistical identification of confounding variables, with the majority of the variables being identified from the literature before the commencement of the analysis, and with all additional variables being supported by the literature. The reported study is also observational and the association with BMD does not imply direct causation. However, the literature supports the notation that these taxa may have functional links to bone health and this microbial contribution to bone health may represent a modifiable environmental factor in the prevention and treatment of osteoporosis. Despite the limitations discussed, changes in gut bacterial composition with respect to bone health suggest that further exploration and mechanistic studies are warranted.
In conclusion, we identified taxa-specific differences in the gut microbiota profiles associated with normal BMD, osteopenic and osteoporotic subjects. These genera could be potential biomarkers and therapeutic targets in high risk cohorts. These differences support the concept that specific genera within the gut exert influence on bone metabolism in the host, subsequently affecting bone health in adulthood.
Supplementary Material
Acknowledgements
We would like to acknowledge the contribution of Jennifer Connolly. We kindly acknowledge the academic support infrastructure of the APC Microbiome Ireland, University College Cork, Cork. M.D. carried out the bioinformatics analysis, compilation and interpretation of the work, and drafting and revising of the manuscript submitted. O.C., D.M.K. and M.G.M. contributed to the concept and design of the work, acquisition of data and reviewed the manuscript before submission. E.M.C. carried out the compilation of data, DNA extraction and performed the library preparation for next-generation sequencing. H.N. and M.N. carried out the acquisition and compilation of the data. C.M. carried out the acquisition of data and reviewed the manuscript before submission. P.W.O’T. contributed to the concept and design of the work, interpretation of the work, and drafting and revising of the manuscript submitted. F.S. contributed to the concept and design of the work and revising of the manuscript submitted. I.B.J. contributed to the concept and design of the work, interpretation of the work, and drafting and revising of the manuscript submitted.
Funding: This work was partly supported by the Irish Centre for Arthritis Research and Education (ICARE). The authors are supported in part by Science Foundation Ireland (SFI) in the form of a research centre grant (SFI/12/RC/2273). I.B.J. and M.D. are funded by an SFI Starting Investigator Research Grant (SIRG) (13/SIRG/2128).
Disclosure statement : The authors have declared no conflicts of interest.
Supplementary data
Supplementary data are available at Rheumatology online.
References
- 1. Hernlund E, Svedbom A, Ivergård M. et al. Osteoporosis in the European Union: medical management, epidemiology and economic burden. A report prepared in collaboration with the International Osteoporosis Foundation (IOF) and the European Federation of Pharmaceutical Industry Associations (EFPIA). Arch Osteoporos 2013;8:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sozen T, Ozisik L, Basaran NC.. An overview and management of osteoporosis. Eur J Rheumatol 2017;4:46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eastell R, O’Neill TW, Hofbauer LC. et al. Postmenopausal osteoporosis. Nat Rev Dis Primers 2016;2:16069. [DOI] [PubMed] [Google Scholar]
- 4. Ali T, Lam D, Bronze MS, Humphrey MB.. Osteoporosis in inflammatory bowel disease. Am J Med 2009;122:599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sambrook PN, Spector TD, Seeman E. et al. Osteoporosis in rheumatoid arthritis. A monozygotic co-twin control study. Arthritis Rheum 1995;38:806–9. [DOI] [PubMed] [Google Scholar]
- 6. Donnelly S, Doyle DV, Denton A. et al. Bone mineral density and vertebral compression fracture rates in ankylosing spondylitis. Ann Rheum Dis 1994;53:117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clemente JC, Manasson J, Scher JU.. The role of the gut microbiome in systemic inflammatory disease. BMJ 2018;360:j5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peterson CT, Sharma V, Elmén L, Peterson SN.. Immune homeostasis, dysbiosis and therapeutic modulation of the gut microbiota. Clin Exp Immunol 2015;179:363–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hsu E, Pacifici R.. From osteoimmunology to osteomicrobiology: how the microbiota and the immune system regulate bone. Calcif Tissue Int 2018;102:512–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baker JM, Al-Nakkash L, Herbst-Kralovetz MM.. Estrogen-gut microbiome axis: physiological and clinical implications. Maturitas 2017;103:45–53. [DOI] [PubMed] [Google Scholar]
- 11. Jones ML, Martoni CJ, Prakash S.. Oral supplementation with probiotic L. reuteri NCIMB 30242 increases mean circulating 25-hydroxyvitamin D: a post hoc analysis of a randomized controlled trial. J Clin Endocrinol Metab 2013;98:2944–51. [DOI] [PubMed] [Google Scholar]
- 12. Sjögren K, Engdahl C, Henning P. et al. The gut microbiota regulates bone mass in mice. J Bone Miner Res 2012;27:1357–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nobel YR, Cox LM, Kirigin FF. et al. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat Commun 2015;6:7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pytlik M, Folwarczna J, Janiec W.. Effects of doxycycline on mechanical properties of bones in rats with ovariectomy-induced osteopenia. Calcif Tissue Int 2004;75:225–30. [DOI] [PubMed] [Google Scholar]
- 15. Claesson MJ, Jeffery IB, Conde S. et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012;488:178–84. [DOI] [PubMed] [Google Scholar]
- 16. Guss JD, Horsfield MW, Fontenele FF. et al. Alterations to the gut microbiome impair bone strength and tissue material properties. J Bone Miner Res 2017;32:1343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yan J, Herzog JW, Tsang K. et al. Gut microbiota induce IGF-1 and promote bone formation and growth. Proc Natl Acad Sci USA 2016;113:E7554–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Quach D, Collins F, Parameswaran N, McCabe L, Britton RA.. Microbiota reconstitution does not cause bone loss in germ-free mice. mSphere 2018;3:e00545-17. 10.1128/mSphereDirect.00545-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hind K, Oldroyd B, Truscott JG.. In vivo precision of the GE Lunar iDXA densitometer for the measurement of total-body, lumbar spine, and femoral bone mineral density in adults. J Clin Densitom 2010;13:413–17. [DOI] [PubMed] [Google Scholar]
- 20. Kanis JA, Melton LJ 3rd, Christiansen C, Johnston CC, Khaltaev N.. The diagnosis of osteoporosis. J Bone Miner Res 1994;9:1137–41. [DOI] [PubMed] [Google Scholar]
- 21. Kanis JA, McCloskey EV, Johansson H. et al. European guidance for the diagnosis and management of osteoporosis in postmenopausal women. Osteoporos Int 2013;24:23–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu Z, Morrison M.. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 2004;36:808–12. [DOI] [PubMed] [Google Scholar]
- 23. Klindworth A, Pruesse E, Schweer T. et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 2013;41:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Magoc T, Salzberg SL.. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011;27:2957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Caporaso JG, Kuczynski J, Stombaugh J. et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460–1. [DOI] [PubMed] [Google Scholar]
- 27. Cole JR, Wang Q, Fish JA. et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 2014;42(Database issue):D633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schloss PD, Westcott SL, Ryabin T. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009;75:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2017.
- 30.Dinno A. dunn.test: Dunn’s Test of Multiple Comparisons Using Rank Sums. R package version 1.3.4. 2017. https://cran.r-project.org/web/packages/dunn.test/dunn.test.pdf (26 April 2017, date last accessed).
- 31. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Duvallet C, Gibbons SM, Gurry T, Irizarry RA, Alm EJ.. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat Commun 2017;8:1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Drescher LS, Thiele S, Mensink GB.. A new index to measure healthy food diversity better reflects a healthy diet than traditional measures. J Nutr 2007;137:647–51. [DOI] [PubMed] [Google Scholar]
- 34. Mahoney FI, Barthel DW.. Functional evaluation: the Barthel Index. Md State Med J 1965;14:61–5. [PubMed] [Google Scholar]
- 35. Godin G, Shephard RJ.. A simple method to assess exercise behavior in the community. Can J Appl Sport Sci 1985;10:141–6. [PubMed] [Google Scholar]
- 36. Molloy DW, Alemayehu E, Roberts R.. Reliability of a standardized Mini-Mental State Examination compared with the traditional Mini-Mental State Examination. Am J Psychiatry 1991;148:102–5. [DOI] [PubMed] [Google Scholar]
- 37. Guigoz Y, Vellas B.. The Mini Nutritional Assessment (MNA) for grading the nutritional state of elderly patients: presentation of the MNA, history and validation. Nestle Nutr Workshop Ser Clin Perform Programme 1999;1:3–11; discussion 11–2. [DOI] [PubMed] [Google Scholar]
- 38. Charlson ME, Pompei P, Ales KL, MacKenzie CR.. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 1987;40:373–83. [DOI] [PubMed] [Google Scholar]
- 39. Zhernakova A, Kurilshikov A, Bonder MJ. et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016;352:565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jeffery IB, Lynch DB, O’Toole PW.. Composition and temporal stability of the gut microbiota in older persons. ISME J 2016;10:170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Falony G, Joossens M, Vieira-Silva S. et al. Population-level analysis of gut microbiome variation. Science 2016;352:560–4. [DOI] [PubMed] [Google Scholar]
- 42. Jackson MA, Verdi S, Maxan ME. et al. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat Commun 2018;9:2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang J, Wang Y, Gao W. et al. Diversity analysis of gut microbiota in osteoporosis and osteopenia patients. PeerJ 2017;5:e3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bian G, Gloor GB, Gong A. et al. The gut microbiota of healthy aged Chinese is similar to that of the healthy young. mSphere 2017;2:e00327-17. 10.1128/mSphere.00327-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arranz Caso JA, Flores Ballester E, Ngo Pombe S. et al. [Bisphosphonate related osteonecrosis of the jaw and infection with Actinomyces]. Med Clin (Barc) 2012;139:676–80. [DOI] [PubMed] [Google Scholar]
- 46. De Ceulaer J, Tacconelli E, Vandecasteele SJ.. Actinomyces osteomyelitis in bisphosphonate-related osteonecrosis of the jaw (BRONJ): the missing link? Eur J Clin Microbiol Infect Dis 2014;33:1873–80. [DOI] [PubMed] [Google Scholar]
- 47. Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A.. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog 2013;5:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bozec A, Zaiss MM.. T regulatory cells in bone remodelling. Curr Osteoporos Rep 2017;15:121–5. [DOI] [PubMed] [Google Scholar]
- 49. Lucas S, Omata Y, Hofmann J. et al. Short-chain fatty acids regulate systemic bone mass and protect from pathological bone loss. Nat Commun 2018;9:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu J, Shang DP, Yang S. et al. Association between the vitamin D receptor gene polymorphism and osteoporosis. Biomed Rep 2016;5:233–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jin D, Wu S, Zhang YG. et al. Lack of vitamin D receptor causes dysbiosis and changes the functions of the murine intestinal microbiome. Clin Ther 2015;37:996–1009.e7. [DOI] [PubMed] [Google Scholar]
- 52. Bashir M, Prietl B, Tauschmann M. et al. Effects of high doses of vitamin D3 on mucosa-associated gut microbiome vary between regions of the human gastrointestinal tract. Eur J Nutr 2016;55:1479–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rafii F. The role of colonic bacteria in the metabolism of the natural isoflavone daidzin to equol. Metabolites 2015;5:56–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ticinesi A, Milani C, Lauretani F. et al. Gut microbiota composition is associated with polypharmacy in elderly hospitalized patients. Sci Rep 2017;7:11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang W, Wang J, Li J. et al. Cholecystectomy damages aging-associated intestinal microbiota construction. Front Microbiol 2018;9:1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. De Laet C, Kanis JA, Odén A. et al. Body mass index as a predictor of fracture risk: a meta-analysis. Osteoporos Int 2005;16:1330–8. [DOI] [PubMed] [Google Scholar]
- 57. Zhao LJ, Liu YJ, Liu PY. et al. Relationship of obesity with osteoporosis. J Clin Endocrinol Metab 2007;92:1640–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Beck TJ, Petit MA, Wu G. et al. Does obesity really make the femur stronger? BMD, geometry, and fracture incidence in the women's health initiative-observational study. J Bone Miner Res 2009;24:1369–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.