Abstract
Transcriptional regulation by nuclear hormone receptors (NRs) requires multiple coregulators that modulate chromatin structures by catalyzing a diverse array of posttranslational modifications of histones. Different combinations of these modifications yield dynamic functional outcomes, constituting an epigenetic histone code. This code is inscribed by histone-modifying enzymes and decoded by effector proteins that recognize specific covalent marks. One important modification associated with active chromatin structures is methylation of histone H3-lysine 4 (H3K4). Crucial roles for this modification in NR transactivation have been recently highlighted through our purification and subsequent characterization of a steady-state complex associated with ASC-2, a coactivator of NRs and other transcription factors. This complex, designated ASCOM for ASC-2 complex, contains H3K4-methyltransferase MLL3/HALR or its paralogue MLL4/ALR and represents the first Set1-like H3K4-methyltransferase complex to be reported in vertebrates. This review focuses on recent progress in our understanding of how ASCOM-MLL3 and ASCOM-MLL4 influence NR-mediated gene transcription and of their physiological function.
I. Introduction
Nuclear hormone receptors (NRs) share a common modular structure comprised an N-terminal variable domain, a central DNA-binding domain (DBD), and a C-terminal ligand-binding domain (LBD).1 While the LBD harbors a ligand-dependent activation function referred to as AF2, the N-terminal domains of some NRs harbor AF1, a constitutive activation function. A variety of endocrine hormones, fatty acids, cholesterol, and lipid metabolites act as ligands for various NRs. During ligand-dependent transcriptional regulation by NRs, multiple transcriptional coregulators have been demonstrated to operate in a ligand-dependent, combinatorial manner, as dictated by the context of the individual target gene and cell type.2–4
Importantly, genes are normally compacted in closed chromatin structures in a transcriptionally inactive state.5 Thus, the establishment of open chromatin structures is an essential step toward successful transactivation. Consistent with this notion, NR transactivation depends on the ability of NR to recruit multiple coactivators whose primary function is to remodel or modify chromatin structures. In particular, histone-modifying enzymes catalyze a diverse array of posttranslational modifications of core and linker histones within chromatin.6,7 Interestingly, different combinations of these modifications appear to yield dynamic functional outcomes, constituting an epigenetic histone code. While this code is inscribed by histone-modifying enzymes catalyzing site-selective modifications, it is proposed to be decoded by effector proteins that recognize specific covalent marks.6,7
Methylation of histone H3-lysine 4 (H3K4) is one particular histone modification that has been recently highlighted for its roles in NR transactivation through our characterization of ASCOM, a novel NR coactivator complex associated with H3K4-methyltransferase (H3K4MT) MLL3/HALR or its paralogue MLL4/ALR.8,9 ASCOM belongs to a family of Set1-like H3K4MT complexes (Table I). It has been proposed that ASC-2 may play crucial roles in reproduction and a variety of endocrine functions.10 Our recent results also implicate ASCOM as a Set1-like complex specialized to regulate genes involved with metabolic homeostasis.11,12 Moreover, ASCOM contains another histone modifier, UTX, that removes trimethylated-H3K27, a mark for inactive chromatin structures.13–17 Thus, ASCOM is associated with two histone modifiers linked to transactivation and should serve as an excellent model system to study the potential cross talk between H3K4 and H3K27-methylation events. Interestingly, ASCOM appears to have a unique function as a platform for integrating the activities of several other coactivators,10 including the ATPase-dependent Swi/Snf chromatin remodeling complexes (see below), during NR transactivation.
TABLE I.
A Family of Set1-Like Complexes
| ySet1_C | hSet1_C | MLL1_C | MLL2_C | ASCOM |
|---|---|---|---|---|
| Set1 | hSet1α/β | MLL1 | MLL2 | MLL3/4 |
| Bre2 | ASH2L | ASH2L | ASH2L | ASH2L |
| Swd1 | RbBP5 | RbBP5 | RbBP5 | RbBP5 |
| Swd3 | WDR5 | WDR5 | WDR5 | WDR5 |
| Sdc1 | hDPY-30 | hDPY-30 | hDPY-30 | hDPY-30 |
| Swd2 | hSwd2 | |||
| SPP1 | CXXC1 | |||
| HCF1 | Menin | Menin | ASC-2 | |
| HCF1/HCF2 | PTIP | |||
| PA1 | ||||
| UTX | ||||
| α/β-Tubulins |
The yeast complex (left) is aligned with six human complexes. Three common subunits, which assemble an independent subcomplex, are shaded. This subcomplex forms a functional core H3K4MT complex, along with each individual H3K4MT enzyme. Unique subunits in each complex are shown in bold. Notably, additional subunit proteins may remain unidentified in each complex.
Here, we present an overview of current literature on the function of ASCOM-MLL3 and ASCOM-MLL4 in NR transactivation and further discuss future challenges in fully elucidating their physiological functions.
II. Activating Signal Cointegrator-2 (ASC-2)
NRs bind to specific response elements in target genes and regulate transcriptional initiation in a ligand-dependent manner.1 In the unliganded state, a subset of the NRs repress transcription by recruiting corepressors.2–4 Upon ligand binding, the conserved C-terminal LBD of NRs undergoes a dramatic conformational change, which is recognized by an α-helical LXXLL motif, named the NR box, which is often found associated with transcriptional coactivators.18,19
ASC-2, also named NCOA6 (nuclear receptor coactivator-6), TRBP (thyroid hormone receptor-binding protein), RAP250 (nuclear receptor-activating protein-250), NRC (nuclear receptor coregulator), and PRIP (peroxisome proliferator-activated receptor-interacting protein), has been shown to function as a coactivator of many NRs.10 In addition, AIB3 (amplified in breast cancer-3) has been identified as a human ASC-2 isoform in which the N-terminal 26 amino acids replace the first 88 amino acids of ASC-2. Of note, ASC-2 has two NR interaction boxes10 (Fig. 1A). While NR1 binds multiple NRs, NR2 interacts primarily with the liver X receptors (LXRs).20 The physiological importance of ASC-2 and its two NR boxes in transactivation by ASC-2-interacting NRs has been documented by studies with various ASC-2 mouse models.21–27 Notably, ASC-2 also functions as an important coactivator for an array of other classes of transcription factors.10
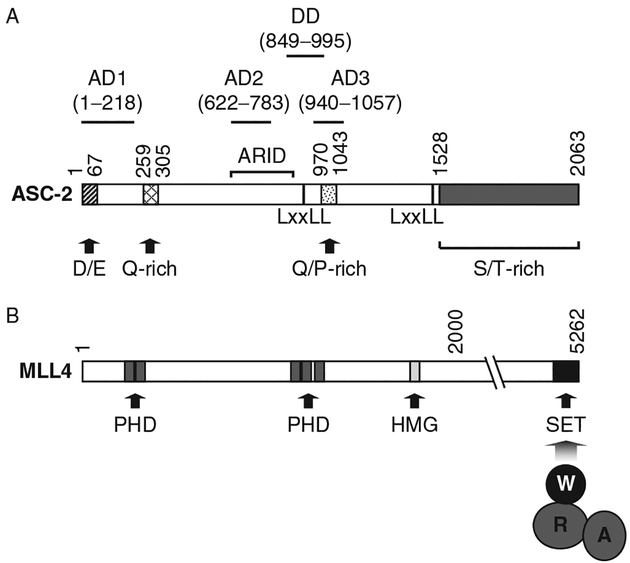
Fig. 1.
Schematic representations of ASC-2 and MLL4. Various domains that include the AD1–3, DD (dimerization domain), and ARID (AR-interacting domain) in ASC-2, as well as the PHD finger, HMG box, and SET domains in MLL4, are as indicated. The SET domain in MLL3/4 also serves as a docking site for a subcomplex consisting of WDR5 (W)-RbBP5 (R)-ASH2L (A).
A. Expression of ASC-2 and Its Isoforms
ASC-2 is a 250 kDa protein of 2063 amino acids that is expressed from a ~8–9 kb mRNA in different human tissues.28 ASC-2 mRNAs of 6.8, 4.5, and 3.6 kb also have been described.28 While the 3.6 kb transcript is the predominant ASC-2 mRNA species detected in heart and skeletal muscle, the 4.5 kb transcript is the major form in testes.28 The latter transcript has been identified in human testis as an alternatively spliced form of ASC-2 mRNA that encodes an ASC-2 isoform of 1070 amino acids.29 This isoform lacks the NR box 2 and a large internal region (amino acids 972–1964) toward the C-terminus. Mouse ASC-2 mRNA (~8–9 kb) encodes a protein of 2068 amino acids.28 A partial rat ASC-2 cDNA encoding the NR box 1 and AD3 followed by a stop codon and a poly A tail has also been isolated from a rat pituitary GH4C1 cell cDNA library.28
The mouse ASC-2 gene, containing 13 exons, is localized on chromosome 2,28 while rat ASC-2 resides on chromosome 3. The organization of human ASC-2 on chromosome 20q11 predicts a single gene, with 15 exons and 14 introns, that spans ~111 kb.30 Thus, the various sized ASC-2 mRNAs described above likely represent alternative splicing products of the ASC-2 gene. The sequence of the ASC-2 gene predicts a TATA-less promoter with four binding sites for Sp1, as well as sites for C/EBP and Myc/Max.30 However, the functional significance of these binding sites in regulating ASC-2 gene expression remains to be established.
Human, mouse, and rat ASC-2 proteins show over 90% sequence identity at the amino acid level. The distribution of the ASC-2 protein in mice has been studied by immunochemistry using antibody against ASC-2. These studies indicate relatively higher levels of ASC-2 in cells of endocrine target tissues, including testicular sertoli cells, follicular granulosa cells, and epithelial cells of the prostate, uterus, mammary gland, kidney tubules and urothelia23,31 (PNAS 2009;106:8513–8, and our unpublished results). ASC-2 protein expression has also been detected in thyroid and parathyroid cells and in the pancreatic islets of langerhans.23 Although medium to low expression is reported throughout a variety of tissues, the relatively higher levels of ASC-2 observed in reproductive and many types of endocrine tissues suggest that ASC-2 supports NR functions in reproduction and regulation of the endocrine system.23,31 ASC-2 also is relatively abundant in a variety of metabolic tissues and cell types, including the liver, pancreas, adipose tissues, and hypothalamic metabolic neurons23,31 (our unpublished results). These results are consistent with our proposed roles for ASCOM in metabolic homeostasis (see below).
B. Autonomous Transactivation Domains of ASC-2
Structure-function analysis of human ASC-2 has revealed a number of functional domains. First, we have mapped three autonomous transactivation domains, AD1, AD2, and AD3, that in human ASC-2 comprise residues 1–218, 622–849, and 849–1057, respectively.20 Mahajan and Samuels have also mapped two autonomous transactivation domains in ASC-2 to residues 302–783 and 940–1124.28 Thus, the boundaries of AD2 and AD3 are likely to be refined to ASC-2 residues 622–783 and 940–1057, respectively (Fig. 1A). Interestingly, while AD2 is dispensable for NR transactivation, the AD3 domain appears to play crucial roles for NR signaling and AF2-dependent enhancement of NR transactivation by ASC-2.32
The ~600 residue ASC-2 C-terminal region that is rich in Ser, Thr, and Leu (Fig. 1A) inhibits the autonomous transactivation function of ASC-2 in the context of full length ASC-2, as its deletion enhances the intrinsic activation potential of AD1, AD2, and AD3 in ASC-2.28 Notably, this region contains an array of putative phosphorylation sites for different protein kinases, raising the interesting possibility that the activity of ASC-2 may potentially be modulated by kinases and phosphatases. These results also suggest that this C-terminal region may function to control the transcriptional output of ASC-2 through inter- and intramolecular interactions. One interesting possibility is that the activation properties of ASC-2 could be modulated through exposure of the cryptic activation domain in response to liganded NRs, as previously exemplified by the finding that PGC-1 undergoes a conformational change upon NR binding.33 In support of this notion, a LexA fusion protein containing ASC-2 AD2 and AD3 domains is transcriptionally mild in yeast but shows a dramatically increased activity when coexpressed with RXR in the presence of its ligand. Similar results are also observed in mammalian cells. When an ASC-2 region containing the NR1 and AD2 regions is expressed in mammalian cells, it is moderately active when expressed alone. However, its activity is markedly enhanced when coexpressed with the liganded TR.28 These results lead to a proposal that the NR-coactivator function of ASC-2 is masked until it associates with liganded NRs.28
C. Two NR Boxes in ASC-2
ASC-2 contains three distinct NR-interacting surfaces. Two distinct NR boxes mediate direct interactions of ASC-2 with a variety of NRs.10 In addition, we have shown that ASC-2 contains a binding site for the retinoblastoma (Rb) tumor suppressor protein,34 which is known to interact with androgen receptor (AR).35 We have further shown that this region indeed functions as an indirect binding site for AR, as it recruits AR via Rb.34
1. NR1
Two functional NR boxes have been identified in ASC-2 (Fig. 1A). The NR box 1 shows ligand-dependent interactions with a wide variety of NRs that include RARs, RXRs, TRs, GR, ERs, VDR, and the PPARs.28,29,36 Accordingly, mutation of the leucine residues in NR1 abolishes the association of ASC-2 with NRs. Through the use of combinatorial peptide libraries, the NR boxes have been characterized as Class 1, 2, or 3 depending on the nature of the amino acid residue at position −1 or −2 of the LXXLL motif.37 The Class 2 NR box with Pro at −2 has been shown to interact with various NRs. The NR box 1 region of ASC-2 (LTSPLLVNLLQSDIS) resembles the Class 2 NR box and contains a Pro at the −2 position. Like other NR box-dependent coactivators, helix 12 of the NR AF2 domain is pivotal in the formation of an interface that contacts the ASC-2 NR1. Thus, either a point mutation (L398R) in helix 12 of chicken TRα or a deletion of helix 12 abolishes the association with ASC-2.28 Deletion of helix 12 of PPARγ also disrupts the interaction with ASC-2.38 We have found that the interactions of TRβ with the NR1 of ASC-2 are abolished by specific mutations in the TRβ helices 3, 5, and 6,28 which are known to form the binding pocket for SRC-1.39,40 Overall, these results indicate that the ASC-2:NR interactions are similar to those of other coactivators.
The Ser at the −3 position (Ser 884) of the ASC-2 NR1 has been demonstrated to be important for binding to LBDs of NRs, particularly for ERβ, TR, and RXR.41 Although in vitro phosphorylation of Ser 884 by MAPK reduces its interaction with NRs, it remains to be determined whether such regulation by phosphorylation occurs in vivo.41 Interestingly, the ASC-2 NR1 has been shown to associate more strongly with mouse ERβ1 than with ERβ2, consistent with the notion that mouse ERβ1 is a more potent estradiol-induced transcriptional activator than mouse ERβ2.42 Studies with a ~300 amino acid rat ASC-2 region that contains NR1 and AD3 indicate that NR1 enhances both the ligand-dependent activity and the intrinsic basal activity of AD3.32 Thus, the NR1 of ASC-2 embedded in AD3 appears to influence not only the association of ASC-2 with NRs, but also the activation potential of AD3. Correspondingly, the AD3 region has been shown to be necessary for transcriptional activation by NRs.32
2. NR2
ASC-2 contains a second NR box (Fig. 1A) that does not contain a conserved hydrophobic amino acid residue at the −1 position (EAPTSLSQLLDNSGA).28 While NR2 strongly interacts with LXRs,20 it does not recognize most NRs other than ERα, which shows ~10-fold lower affinity toward NR2 compared with NR1.28 The ASC-2 NR2 does not resemble any of the known NR boxes identified thus far in various NR coactivators or from combinatorial phage peptide libraries.37
RXR has been shown to enhance LXR transactivation, even in the absence of an LXR ligand, via a unique mechanism of allosteric regulation.43 Interestingly, we have found that LXR binding to the ASC-2 NR2 is enhanced by RXR and even more substantially by liganded RXR.44 We also have identified specific NR2 region residues that are involved both in its interaction with LXR and in the ASC-2-mediated transactivation of LXR in mammalian cells. Using these mutants, we have demonstrated that the NR2–LXR interaction surface is not altered by the presence of RXR and RXR ligand and that Ser 1490 is the critical determinant for the LXR-specific interaction of NR2.44 Notably, NR2, but not NR1, is essential for ASC-2-mediated transactivation of LXR in vivo and for the interaction between LXR–RXR and ASC-2 in vitro. These results indicate that RXR does not interact directly with the NR1 of ASC-2, but functions as an allosteric activator of LXR binding to the ASC-2 NR2.44
D. Homodimerization Domain in ASC-2
Most NRs activate transcription from target genes either as homodimers or as heterodimers with RXR.1 SRC family coactivators contain multiple NR boxes, such that a single SRC coactivator could interact simultaneously, through distinct NR boxes, with various NR dimmers.45 Similarly, the TRAP220/MED1 Mediator subunit has two NR boxes. One NR box interacts with RXR, while the other interacts preferentially with VDR or TR.46 Thus, a TRAP220/MED1 monomer has the potential to bind to NR–RXR heterodimers through two interfaces. However, ASC-2 contains a single NR box that is involved in interactions with multiple NRs. Potentially resolving the issue of how ASC-2 with a single functional NR box binds to and activates NR dimers, ASC-2 has been found to homodimerize.28 The homodimerization region (DD) of ASC-2 (Fig. 1A) has been mapped to a region of human ASC-2 containing amino acid 849–995.28,32 Embedded in this region is the NR1 motif, which is involved in NR interactions, but not homodimerization of ASC-2. Thus, a homodimer of ASC-2 with two functional NR box motifs has the potential to bind NR homo- and heterodimers with high affinity.28,32 However, in vivo evidence for formation of an ASC-2 homodimer has yet to be established.
III. Set1 -Like H3K4MT Complexes
A. Methylation of H3K4
H3K4-methylation is an evolutionarily conserved mark that is linked to transcriptionally active chromatin and has been proposed to counter the generally repressive chromatin environment imposed by H3K9/H3K27-methylation in higher eukaryotes.5 H3K4-methylation has been demonstrated to be associated with transcriptional activation in a variety of eukaryotic species.47,48 Lysine residues can be mono-, di-, or trimethylated at the ζ-amine in vivo. In particular, H3K4-trimethylation is tightly associated with the 5′ regions of transcriptionally active genes, as it shows a strong positive correlation with transcription rates, active polymerase II occupancy, and histone acetylation.49–54 Interestingly, the patterns of H3K4-dimethylation differ significantly between yeast and vertebrates. In Saccharomyces cerevisiae, dimethylated H3K4 is found throughout genes that, transcriptionally, are either active or poised and peaks around the middle of the coding region, whereas monomethylation is most abundant at the 3′ end of genes.49,50,54 In vertebrates, the majority of H3K4-dimethylation is coupled to H3K4-trimethylation in discrete zones about 5–20 nucleosomes in length proximate to highly transcribed genes.51,52
B. Multiple Set1-Like H3K4MT Complexes in Vertebrates
The majority of histone lysine methyltransferases contain a SET domain, which catalyzes the addition of methyl groups to the specific lysine residues.48 The catalytic SET domains of H3K4MTs can be arbitrarily grouped into either Set1-like H3K4MTs, which are related to the yeast Set1 and Drosophila Trx, or non-Set1-like H3K4MTs, which include ASH1, SET7/9, SMYD3, and Mei-setz.55–59 The first H3K4MT to be identified is S. cerevisiae Set1, a single enzyme responsible for all H3K4-methylation in yeast.60,61 Interestingly, this enzyme is found as a component of a large steady-state Set1 complex62,63 (Table I). We subsequently described a similar mammalian complex containing mixed lineage leukemia 3 (MLL3, also named HALR) or MLL4 (also named MLL2/ALR),8,9 and others later described homologous vertebrate complexes containing MLL1, MLL2, SET1α, or SET1β64–68 (Table I). These evolutionarily conserved complexes are collectively named “Set1-like H3K4MT complexes” (Table I). The multiplicity of Set1-like H3K4MT complexes in vertebrate genomes suggests that these complexes are not redundant in their function. In strong support of this notion, MLL1, MLL2, and MLL3 mutant mice display distinguishable phenotypes.9,11,12,64,69 The potential functional specialization of these enzymes likely results from their differential expression patterns, recruitment to different target genes, and/or methylation of distinct nonhistone substrates.
C. A Subcomplex of WDR5, RbBP5, and ASH2L in Set1-Like Complexes
The Set1-like H3K4MT complexes share at least three common subunits: WDR5, RbBP5, and ASH2L8,65–8,70 (Table I). Our recent biochemical reconstitution of a functional four-component MLL1 core complex reveals that recombinant WDR5, RbBP5, ASH2L, and MLL1 are sufficient for recapitulating H3K4MT activity comparable to that of the MLL1 holocomplex purified from human cells.71 Importantly, the subcomplex of WDR5, RbBP5, and ASH2L associates with the MLL1 SET domain (Fig. 1B) but can exist independently of the catalytic subunit, thereby serving as a structural platform that can associate with the SET domains of different MLL-family members8,71 (Fig. 2A). All three subcomplex components are required for H3K4-methylation by MLL1 in vitro and in vivo,71,72 with different components affecting methyl states to different degrees.71,73,74 In particular, WDR5 mediates interactions of the MLL1 catalytic unit both with the common structural platform and with the histone substrate71 (Fig. 1B).
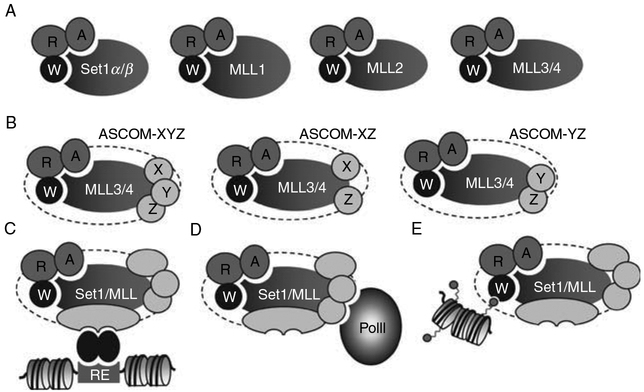
Fig. 2.
Working models for Set1-like complexes. (A) An independent subcomplex of WDR5–RbBP5–ASH2L is tethered to each Set1/MLL H3K4MT to form a functional core complex for H3K4MT activity. (B) ASCOM may represent a heterogeneous pool of similar complexes containing slightly different noncore subunits. (C) Transcription factor functions as a primary adaptor to recruit each Set1-like complex to its target genes. Interactions with the basal machinery (D) and modified histone tails (E) provide further stabilization of the Set1-like complex on its target genes.
D. H3K4 Methyl-Binding Effectors
Although H3K4 methylation has been proposed to function through countering the generally repressive chromatin environment imposed by H3K9/H3K27-methylation in higher eukaryotes,5 its precise role in transcription remains poorly understood. Like other histone modifications, histone methylation is proposed to act through the recruitment of downstream effector proteins, which in turn carry out specific independent functions on the chromatin template. Indeed, recent work has uncovered a remarkably wide variety of domains that have the capacity to recognize methylated H3-lysine residues.75 In particular, at least two distinct motifs have been shown to bind H3K4-trimethyl residues: the royal superfamily (including the chromodomains of CHD1 and tudor domains of JMJD2A) and the PHD-finger superfamily (including the PHD fingers of BPTF and ING proteins).76,77 Interestingly, many of these H3K4-methyl-binding proteins reside within protein complexes associated with chromatin remodeling and modifications. For example, CHD1 and BPTF are involved in ATP-dependent chromatin remodeling,78 ING proteins associate with and modulate the activity of histone acetyltransferase (HAT) and deacetylase (HDAC) complexes,79 and JMJD2A carries out histone demethylation.80 Thus, H3K4-methylation can be directly coupled to further chromatin remodeling and modification mechanisms necessary to carry out specific biological functions. The recruitment of remodeling machinery, such as the BPTF-containing NURF remodeling complex, may facilitate transcriptional activation by increasing the accessibility of the chromatin template to the transcriptional machinery.81 The association of H3K4-methyl with ING3–5 and yeast Yng1 containing acetyltransferase complexes is also consistent with the role of H3K4-methylation in transcriptional activation.82–84 Histone acetylation directly promotes more accessible chromatin structure and can further recruit bromodomain-containing transcriptional regulators.85 Interplay between H3K4-methylation and acetylation is evidenced by a high correlation of hyperacetylation and H3K4-methylation patterns in genomic location analyses.52–4,83 In further support of this notion, we have recently demonstrated that loss of the H3K4-methyl mark on RAR-target gene RAR-β2 in ASC-2-null cells is accompanied by loss of H3/H4-acetylation marks.9 Intriguingly, recent findings demonstrate that upon DNA damage ING2 recruits the Sin3/HDAC complex to silence transcription of cell proliferation genes, thus implicating H3K4-trimethylation in active gene repression as well.82 In support of this idea, H3K4-methyl-binding PHD fingers exist in well-established HDAC complexes in organisms ranging from yeast to humans.79 Interestingly, human CHD1 recognition of trimethylated H3K4 has been shown to function through the recruitment of factors implicated in transcriptional elongation and pre-mRNA processing.86 A PHD finger within RAG2, a protein involved in V(D)J recombination, has been shown to recognize trimethylated H3K4 and these interactions subsequently have been shown to play important roles in V(D)J recombination.87–89 The latter two sets of results expand the functional scope for H3K4-methylation beyond its well-defined role in transcriptional initiation.
IV. ASCOM in NR-Mediated Transactivation
A. ASCOM-MLL3 and ASCOM-MLL4
Our proteomic analysis of ASC-2 in HeLa nuclei has led to the surprising finding that ASC-2 does not exist as a single polypeptide but belongs to a large (~2 MDa) steady-state H3K4MT complex that is highly homologous to the yeast Set1 complex.8 This complex, which we named ASCOM, was the first mammalian Set1-like H3K4MT complex to be described.8,9 ASCOM, which contains either MLL3 or MLL4 as the H3K4MT, shares the RbBP5, ASH2L, and WDR5 subcomplex with other Set1-like complexes (Table I). The components unique to ASCOM include PTIP, PTIP-associated protein 1 (PA1), α- and β-tubulins, UTX, and possibly other additional proteins8,90,91 (Table I). Like Set1 and MLL1/2, the C-termini of MLL3 and MLL4 have a SET domain (Fig. 1B) that is associated with an intrinsic histone lysine-specific methyltransferase activity.5 We and others have shown that ASCOM is indeed a genuine H3K4MT complex.8,90,91 Interestingly, UTX has been found to be a H3K27-specific demethylase.13–17 Because H3K27-trimethylation is a repressive chromatin mark critical for maintaining embryonic stem (ES) cell pluripotency and plasticity in developing embryos, polycomb-mediated gene silencing, and X chromosome inactivation,92 ASCOM has two distinct histone modifiers linked to active chromatin. Virtually identical complexes have also been purified as MLL491 and PTIP90 complexes from K562 and HeLa cells, respectively. Notably, in our original purification, we have failed to determine the identity of UTX, PTIP, PA1, and WDR5 due to technical problems.8 In addition, a similar complex enriched in MLL4 has also been reported as a coactivator complex of ERα from DU4475 cells.93 This complex has been claimed to be distinct from ASCOM based on the relatively unaltered amount of RbBP5, ASH2L, and MLL4 in lysates immunodepleted for ASC-2.93 However, the reported purification includes ASC-2, although it is less abundant than MLL4.93 Moreover, RbBP5 and ASH2L are associated with other Set1-like complexes (Table I)94 and thus are not readily coimmunodepleted with ASC-2. In addition, currently it is not clear whether MLL4 can also be found outside of ASCOM, either as a single polypeptide or as a subunit of other distinct complexes. If MLL4 also exists unassociated with ASC-2, it would not be coimmunodepleted with ASC-2. Thus, it needs to be further examined whether this MLL4 complex93 is indeed distinct from ASCOM. Considering all the relevant studies, we favor a model in which a shared subcomplex of RbBP5, ASH2L, and WDR5 forms a core complex with MLL3 or MLL4 that may also form a heterogeneous population of complexes with ASC-2 and other proteins depending on cell types and/or target genes (Fig. 2B).
B. ASCOMs as Crucial H3K4MT Complexes for a Subset of NRs
Transactivation by RXR, RAR, TR, and PPARγ has been shown to be compromised in ASC-2−/− mouse embryo fibroblast (MEF) cells relative to wild-type cells.24,25,27,95 Moreover, we have found that RAR transactivation is correlated with RA-induced H3K4-trimethylation, and that this modification is redundantly mediated by ASCOM-MLL3 and ASCOM-MLL4 but not by related menin-containing MLL1/2-complexes (Table I), primarily due to the ability of ASC-2 to function as a specific linker for RARs to recruit ASCOM.9
ASC-2−/− MEF cells are refractory to PPARγ-mediated adipogenesis and fail to express adipogenic markers such as aP2,95 suggesting that ASC-2 is required for the adipogenic program by PPARγ. In addition, our recent results reveal that MLL3 also plays crucial roles in adipogenesis.12 First, MLL3Δ/Δ mice expressing an H3K4MT inactivated mutant of MLL3 have significantly less white fat. Second, MLL3Δ/Δ MEFs are mildly but consistently less responsive to inducers of adipogenesis than wild-type MEFs. Third, ASC-2, MLL3, and MLL4 are recruited to the PPARγ-activated aP2 gene during adipogenesis, and PPARγ is shown to interact directly with the purified ASCOM. Moreover, while H3K4-methylation of aP2 is readily induced in wild-type MEFs, it is not induced in ASC-2−/− MEFs and only partially induced in MLL3Δ/Δ MEFs.12 These results suggest that ASCOM-MLL3 and ASCOM-MLL4 likely function as crucial but redundant H3K4MT complexes for PPARγ-dependent adipogenesis.
ASC-2 similarly functions as a key adaptor for LXR recruitment of MLL3 and MLL4.11 Moreover, H3K4-trimethylation of a subset of metabolic target genes of LXRs, mediated redundantly by MLL3 and MLL4, is correlated with their expression.11 We also have found that ASCOM-MLL3 and ASCOM-MLL4 play crucial roles in bile acid homeostasis as specific H3K4MT coactivator complexes for FXR (Mol Endo 2009, in press, and our unpublished results). Overall, our results expand the roles for H3K4-trimethylation to transcriptional regulation of metabolic genes and suggest that ASCOM, among Set1-like complexes, functions as a Set1-like H3K4MT complex specialized for a subset of NRs that regulate metabolic genes.
Interestingly, MLL4 has been proposed to play crucial roles in ERα transactivation via direct interactions with ERα.93 Moreover, MLL1/2-complexes have also been shown to function with ERα via interactions between ERα and menin.96 These results highlight the important caveat that ASC-2 is not a general adaptor for NRs and that other subunits of ASCOM may also function as adaptors for some NRs (e.g., MLL4 for ERα),93 and that ASCOM is not the only Set1-like H3K4MT complex involved in NR functions.
C. Recruitment of Set1-Like H3K4MT Complexes
In S. cerevisiae, Set1 is thought to be recruited to chromatin by the phosphorylated form of RNA polymerase II carboxy-terminal domain (CTD), the histone chaperone FACT, and the Paf1 elongation complex. Moreover, its H3K4-trimethylation activity is dependent on H2B-monoubiquitination.5,47 However, this model fails to explain several observations in higher eukaryotes. First, H3K4-trimethylation is enriched in promoter regions. Second, interactions of methylated H3K4 and/or H3K4MTs with chromatin modifiers involved in transcription initiation, such as NURF81 and p300,97 occur at the promoter. Third, although direct interactions among FACT, the PAF complex, and the H2B-ubiquitination machinery are well documented and although Paf1 interacts (directly or indirectly) with Set1 in yeast,98 direct interactions of these components with H3K4MTs are not yet established in mammalian cells. Finally, several reports demonstrate that genetic or siRNA-mediated knockdown of components of the H2B ubiquitination machinery does not affect H3K4 mono- and dimethylation at target genes, suggesting intact recruitment of H3K4MTs.99,100
These results lead to our favored model in which the primary mechanisms of recruitment of at least some H3K4MTs to their target genes involve direct or indirect associations with transcription factors (Fig. 2C). In support of this notion, diverse site-specific transcription factors have been shown to associate with MLL-family complexes. For example, ERα associates with MLL2 and MLL4,93,96 β-catenin associates with MLL1 and MLL2,101 E2F6 associates with MLL1,102 and SET1α associates with the viral transcription factor VP16.66 In addition, apart from binding directly to p53 and effecting p53-dependent H3K4 methylation of chromatin templates independently of H2B ubiquitylation, the MLL1 complex stimulates p53-dependent transcription from a chromatin template in a reconstituted in vitro transcription system.102 We have also found that RARs and LXRs recruit MLL3 and MLL4 to the target genes of RARs and LXRs via interactions of these NRs with ASC-2, an integral subunit of MLL3/4 complexes.9,11 Importantly, associations with transcription factors likely function to determine the target specificity of Set1-like H3K4MT complexes. For instance, ASCOM, but not the related Set1-like MLL1/2 complexes, is targeted by RAR since ASCOM is capable of interacting with RAR via ASC-2 while MLL1/2 complexes show no interactions with RAR.9
The mechanisms of recruitment of Set1-like complexes to their target genes may also involve at least two additional types of interactions. First, interactions with the basal machinery have been proposed (Fig. 2D). Similar to yeast, Set1, MLL1, and MLL2 associate with the phosphorylated CTD of Pol II and components of the basal machinery,102,103 albeit not necessarily directly, and colocalize with the Pol II binding sites.104 This association with the basal machinery appears to be also conserved with Drosophila Trx.105 Second, interactions with modified histones may serve to further stabilize the association of Set1-like complexes with chromatin, as suggested by the discovery that WDR5 presents the H3K4 side chain for further methylation by MLL171,73 (Fig. 2E). Because H3K4MTs often contain a series of distinct modified histone binding domains (e.g., PHD fingers in MLL4 in Fig. 1B), future studies may uncover roles for additional interactions with modified histones in recruiting Set1-like complexes to their targets.
Taken together, we propose that recruitment of mammalian Set1-like H3K4MT complexes to their target genes may generally involve a two-step mechanism. In this model, the primary recruitment event is proposed to be carried out through interactions with specific transcription factors (Fig. 2C), followed by further stabilization of the complex on chromatin through interactions with basal transcription machinery and modified histones (Fig. 2D and E). The secondary stabilization might allow for efficient association of the complex with chromatin over a larger domain than dictated by the presence of the transcription factor on its DNA regulatory element. For example, both NURF and ING2 complexes could be recruited by the site-specific transcription factors and also be stabilized on chromatin by the specific recognition of H3 trimethylated at K4.82,106
V. Cross Talk of ASCOMs with Other Coactivators
Although NR transactivation involves multiple coactivators, it is unclear how these factors are functionally integrated. Interestingly, ASC-2 has been reported to bind both to HATs CBP and p30020,28,36 and, although direct interactions and functions were not established, to the TRAP/DRIP/ARC Mediator complex that links NRs to the basal transcription machinery.36 ASC-2 has also been linked to other coregulators that function in transcriptional initiation and subsequent mRNA processing steps (see below). Our recent results further indicate that the NR coactivator function of ASCOM requires a novel interplay with Swi/Snf that is likely mediated through specific interactions of the SET domain of MLL3/4 with INI1, a core subunit of Swi/Snf.107 These results raise the interesting possibility that ASCOM may serve as a novel platform for NR transactivation by integrating the functions of multiple coactivators through mutual interactions. Moreover, the two histone-modifying activities in ASCOM (i.e., MLL3/4 and UTX) may directly and/or indirectly affect these integration processes.
A. CBP/p300
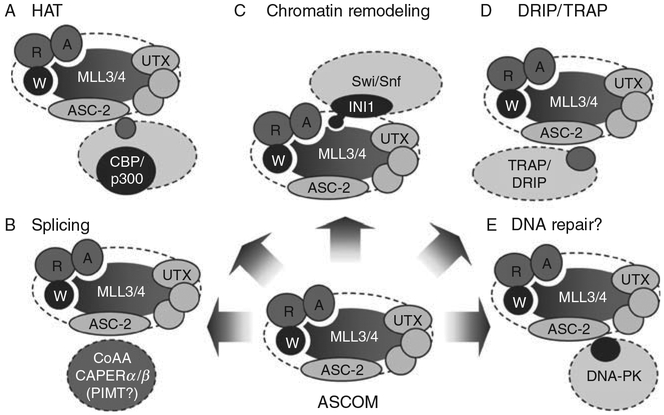
ASC-2 has been shown to form a high-affinity complex with CBP.20,28,36 Thus, expression of ASC-2 in 293 cells followed by extraction, affinity adsorption, and immunoblotting have indicated that a significant amount of CBP in the cell is associated with ASC-2. This association is likely an indirect event, as no direct interaction between full length ASC-2 and full length CBP was detected in yeast.108 Other in vitro binding studies have demonstrated that the C-terminal region of p300 (amino acids 1661–2414) can associate with the C-terminal region of ASC-2.36 Whether association of ASC-2 with CBP/p300 plays essential roles for ligand-dependent activation by NRs or other transcription factors activated by ASC-2 has not been directly addressed using CBP−/−/p300−/− cells. However, ligand-dependent activation of NRs by ASC-2 is completely blocked by expression of E1A, which is known to inactivate CBP/p300.20,28 We also have found that not only H3K4-trimethylation, but also H3/H4-acetylation of the RAR-target gene RAR-β2, is ablated in ASC-2-null cells.9 These results implicate ASC-2 in recruitment of a complex containing CBP/p300 and associated factors to ligand-bound NRs on gene promoters.20,28 However, because CBP/p300 appears to associate with primary coactivators, such as SRC-1, that directly interact with NRs, the recruitment of CBP by NRs may not necessarily require ASCOM. Moreover, recruitment of CBP/p300 and ASCOM to NR-target genes could be facilitated not only by mutual interactions but also by the possible presence of binding modules that recognize methylated H3K4 (by MLL3/4), demethylated H3K27 (by UTX), or acetylated H3/H4 (by CBP/p300). CBP/p300 complexes may also modify components of ASCOM and vice versa, resulting in enhanced function for these complexes. More studies are needed to distinguish between these possibilities. Nonetheless, these results are consistent with a possibility that ASCOM and CBP/p300 are functionally integrated during NR transactivation through interactions between these two complexes (Fig. 3A).
Fig. 3.
Interplays of ASCOM with different complexes (see text).
B. RNA-Binding Proteins
Increasing evidence indicates that transcription and pre-mRNA processing are functionally coupled to modulate gene expression. Interestingly, three proteins containing RNA recognition motifs (RRMs) have been identified as ASC-2-interacting proteins through yeast two-hybrid screenings; CAPER (for coactivator protein for AP-1 and ER receptor), PIMT (PRIP-interacting methyltransferase), and CoAA (for coactivator activator). The recent results demonstrate that CAPER and CoAA are clearly involved with both transcription and splicing, suggesting that ASCOM may play important roles in functionally linking these two processes (Fig. 3B).
We have isolated CAPER from a mouse liver cDNA library using the C-terminal region (amino acids 1172–1729) of ASC-2 as bait.109 This protein has been subsequently renamed CAPERα upon identification of its paralogue CAPERβ.110 CAPERα is identical to the nuclear autoantigens HCC1.3 and HCC1.4 reported in hepatocarcinoma.111 HCC1.3 and HCC1.4 are identical except for six additional amino acids in HCC1.4. Mouse CAPERα (HCC1.3) enhances transactivation by ERα and c-Jun.109 CAPERα contains three RRMs and associates with c-Jun and the liganded ERα via RRM3, while the C-terminus of CAPERα binds to ASC-2.109 Interestingly, CAPERα does not appear to enhance activation of other NRs such as RARs, RXRs, TR, and GR. The basis for ERα specificity is unclear, but this specificity implies that CAPERα may not enhance ERα function through association with ASC-2, which interacts with and enhances activation of many other NRs. Both CAPERα and CAPERβ coactivate the progesterone receptor (PR) in luciferase transcription reporter assays and alter alternative splicing of a calcitonin/calcitonin gene-related peptide minigene in a hormone-dependent manner.110 The importance of CAPER coactivators in the regulation of alternative RNA splicing of an endogenous cellular gene (VEGF) has been substantiated by siRNA knockdown of CAPERα.110 Mutational analysis of CAPERβ indicates that the transcriptional and splicing functions are located in distinct and separable domains of the protein.110 Further implicating CAPERs as potential splicing factors, CAPERα has been found in a purified spliceosome complex.112 Overall, these results indicate that NR-regulated transcription and pre-mRNA splicing can be directly linked via ASCOM and dual function coactivator molecules such as CAPERα and CAPERβ (Fig. 3B).
CoAA is another RRM-containing factor that was isolated from a GC cell cDNA library in a yeast two-hybrid screen using the C-terminal region (amino acids 1641–2063) of ASC-2 as bait.113 The 669 amino acid CoAA contains two RRMs near its N-terminus and the C-terminal auxiliary domain that interacts with ASC-2. Expression of CoAA enhances activation by a number of transcription factors that include NF-κB, CREB, AP-1, PR, TR, ER, and GR.113,114 Using transcriptional and splicing reporter genes driven by different promoters, CoAA has been shown to mediate transcriptional and splicing effects in a promoter-preferential manner.114 CoAA also associates with DNA-PK regulatory subunit Ku86 and poly (ADP-ribose) polymerase (PARP) from GH3 cells, raising the interesting possibility that it may act to functionally link ASCOM to DNA repair machinery in vivo.113
PIMT has been isolated in a yeast two-hybrid screen from a human liver cDNA library using a large C-terminal region (amino acids 773–2067) of ASC-2 as bait.115 PIMT is a ubiquitously expressed putative RNA methyltransferase that contains an invariant GXXGXXI motif near its N-terminus found in K-homology motifs of many RNA-binding proteins. Expression of PIMT enhances PPARγ and RXR activity, and this activity is further enhanced by expression of ASC-2. The putative methyltransferase activity of PIMT does not appear to be involved in its role as an activator since deletion of the methyltransferase domain does not affect its activating function.115 Like ASC-2, PIMT homodimerizes in vitro and may also form homo-oligomers.115 PIMT does not appear to methylate histones, but associates with CBP/p300 and PBP/TRAP220/MED1.116 However, whether PIMT is also involved in splicing remains to be tested.
C. Swi/Snf
Three lines of our recent results suggest the presence of a novel cross talk between ASCOM and the ATPase-dependent chromatin remodeling complex Swi/Snf during NR-mediated transactivation.107 First, these two complexes are colocalized in the nuclear matrix.117 Although transcriptionally active DNA has been suggested to be tightly associated with the nuclear matrix,118 the relevance of the nuclear matrix colocalization of ASCOM and Swi/Snf to their function remains to be further determined. Second, Swi/Snf promotes binding of ASC-2 to NR-target genes, and ASCOM assists binding of Swi/Snf to NR-target genes.107 Finally, we have discovered that the C-terminal SET domains of MLL3 and MLL4 directly interact with INI1, an integral subunit of Swi/Snf, and that these interactions play important roles in the mutually facilitated recruitment of ASCOM and Swi/Snf to NR-target genes107 (Fig. 3C). The involvement of these interactions in the cross talk between ASCOM and Swi/Snf is clearly demonstrated by our identification of a specific INI1 point mutant that affects the MLL3/4-SET interactions, but not the known hBRM and c-Myc interactions, of INI1.107 Both ASCOM and Swi/Snf are known to be recruited to NR-target genes through well-defined interaction interfaces; for example, ligand-dependent interactions of NRs and ASC-2 in recruiting ASCOM to NRs10 and interactions between various subunits of Swi/Snf and NRs in tethering Swi/Snf to NR-target genes.119 Thus, our results suggest a model in which the MLL3/4-SET:INI1 interactions provide “additional” interactions that can lead to mutual facilitation of ASCOM and Swi/Snf in recruitment to NR-target genes. Importantly, this study provides the molecular basis for a novel integration of two enzymatic complexes in NR transactivation, H3K4MT in ASCOM and the ATPase-dependent chromatin remodeler Swi/Snf.
These results, along with the possible interplay of ASCOM with CBP/p30020,28,36 and the TRAP/DRIP/ARC Mediator complex36 (Fig. 3D), further support the possibility that ASCOM has a distinct platform function in facilitating the recruitment of multiple coactivators to NRs. As these include the chromatin regulators Swi/Snf and CBP/p300, and as ASCOM itself is a H3K4MT complex, ASCOM is expected to play crucial roles in establishing transcriptionally active chromatin on NR-target genes. In addition, these chromatin remodelers and modifiers may “indirectly” affect recruitment of other coactivators. For instance, ASCOM-mediated H3K4-methylation may create a direct docking site for other chromatin remodeling/modifying complexes that contribute to the generation of a proper environment for transcription. Interestingly, despite the conserved nature of the SET domains throughout evolution, the SET–INI1 interactions, which were originally described in MLL1,120 are not shared with the SET domains of Ash1, E(Z), and SU(VAR)3–9.121 Given that MLL1 and MLL3/4 belong to the Set1-like family of complexes, these results raise the interesting possibility that Set1α/β and MLL2, the remaining members of the Set1-like complexes, may also communicate with Swi/Snf through INI1 interactions. This possibility is supported by the results that ASH2L and RbBP5, two common subcomplex subunits of all the Set1-like complexes (Table I), are also found associated with the nuclear matrix107 and that SNR1, a Drosophila homologue of INI1 that interacts with the C-terminal SET domain of Trx (a Drosophila orthologue of MLL1), has been shown to colocalize with approximately one-half of the TRX binding sites on larval salivary gland polytene chromosomes.120
VI. Physiological Roles of Key Subunits of ASCOM
A. ASC-2
1. DN1 and DN2
We have shown that an 80 amino acid ASC-2 peptide (DN1) containing NR1 blocks transactivation by NRs when expressed in transgenic mice. DN1 transgenic lines display pathological abnormalities in many different organs that include heart, pituitary, adrenal glands, brain, spleen, liver, and lung. In particular, we have observed eyes with microphthalmia and posterior lenticonus with cataract.22 We have also found that transgenic expression of another ASC-2 region (DN2; amino acids 1431–1511) containing NR2 blocks the activity of LXRs in vivo.21 Livers from DN2 transgenic mice show changes similar to those seen in livers from LXRα−/− mice, suggesting that ASC-2 plays a role in cholesterol and lipid metabolism in the liver, presumably through regulation of LXRs.21 On a high fat cholesterol diet, DN2 transgenic lines display rapid accumulation of large amounts of cholesterol and down-regulation of known lipid metabolizing target genes of LXR.21 Unexpectedly, DN1 and DN2 selectively block the activity of endogenous ASC-2, but not that of other coactivators such as TRAP220/MED1 or SRC-1.21,22 Since DN1 and DN2 would be expected to compete with the binding of other coactivators for liganded NRs, the mechanism underlying this specificity is unclear and requires further study. Importantly, recent studies of a conditional knockout of ASC-2 in liver122 suggest that the phenotypes observed in DN1 transgenic mice123 may not be simply ASC-2-dependent. Thus, while these transgenic studies have provided the initial clues to the probable biological role of ASC-2 in vivo, they should be interpreted with caution.
2. ASC-2 Mutant Mice
The ASC-2 gene has been knocked out in mice in a mixed C57BL/6–129S6 (C57/129) genetic background.24–27 These studies reveal that the ASC-2-null mutation is embryonic lethal and that mutant embryos die in utero between E8.5 and E12.5, while C57/129 ASC-2+/− mutant mice appear normal and grow similar to wild-type mice (see below). The cause of lethality in ASC-2−/− embryos is attributed to placental dysfunction and a number of developmental defects involving the heart, liver, and brain.24,25,27 ASC-2−/− embryos are growth retarded and half the size of wild-type or ASC-2+/− embryos.26 Consistent with these results, ASC-2−/− MEFs derived from E12.5 ASC-2−/− embryos exhibit growth retardation in culture compared to wild-type MEFs. ASC-2−/− MEFs also undergo apoptosis as they enter into the late log phase of growth,26 while wild-type MEFs are resistant to apoptosis under the same growth conditions. RNAi-mediated knockdown of ASC-2 in wild-type MEFs leads to a level of apoptosis similar to that found with ASC-2−/− MEFs.26 Apoptosis of the ASC-2−/− cells seems caspase-mediated since zVAD-fmk, a pan-caspase inhibitor, blocks apoptosis in ASC-2−/− MEFs.26 Overall, these findings suggest that ASC-2 likely regulates antiapoptotic and or prosurvival genes required for cell growth and development.
Interestingly, older ASC-2+/− C57/129 mice have been found to exhibit a wound healing phenotype26 (Table II). They spontaneously develop skin lesions or ulcers around the neck, ears, snout, and facial area. These regions correspond to the areas where mice groom and likely scratch themselves. This occurs in ~25% of both male and female ASC-2+/− C57/129 mice. These spontaneous lesions develop as early as 4–6 months of age in the ASC-2+/−C57/129 mice.26 Histopathology of the affected and normal regions of the skin show thickening of the epidermis, increased sebaceous glands, and a reduction or total loss of hair follicles. In addition, there is no leading edge of keratinocyte migration, which is seen in normal wound healing. This lack of keratinocyte migration has been reproduced using ex vivo skin explant cultures from 2-day-old C57/129 ASC-2+/− mice.26 Wild-type keratinocytes show robust migration when incubated with epidermal growth factor (EGF), whereas keratinocytes from the ASC-2+/− explants show no response to EGF. The molecular mechanism by which a reduction of ASC-2 in the skin of ASC-2+/− mice leads to chronic skin wounds remains to be further defined.
TABLE II.
Physiological Functions for ASC-2 and MLL3
| Defects in | Genotype | Comments |
|---|---|---|
| Wound heeling | ASC-2+/− | Defects occur in some old mice26 |
| Insulin secretion | ASC-2+/− | Apoptosis and decreased proliferation of islets23 |
| Growth | ASC-2+/− (129S6) | Stunted growth26 |
| MLL3Δ/Δ | Stunted growth9 | |
| Fertility | ASC-2+/− (129S6) | 26 |
| MLL3Δ/Δ | 9 | |
| Ductal branching of mammary glands | ASC-2−/− in mammary glands | No milk production during lactation124 |
| WAT (PPARγ-signaling) | MLL3Δ/Δ | MEFs show poorer adipogenic potential than wild-type MEFs in vitro,12 while ASC-2−/− MEFsdo not undergo adipogenesis95 |
| Hepatic lipid synthesis (LXR-signaling) | MLL3Δ/Δ | Less fats deposited to hepatocytes even with a high fat diet11 |
| Bile acid homeostasis (FXR-signaling) | MLL3Δ/Δ | Enlarged gall bladder (Mol Endo 2009, in press, and our unpublished results) |
| Mammary tumor formation suppression | ASC-2+/− | Polyoma middle-T antigen expressed in ASC-2+/− mammary glands133 |
| Urothelia tumor formation suppression | MLL3Δ/Δ | At least in part due to defects in p53- signaling (PNAS 2009;106:8513–8, and our unpublished results) |
References and additional remarks are included as comments.
In the pancreas, we have found that ASC-2 is expressed in the endocrine cells of islets of langerhans.23 In addition, our results reveal that overexpressed ASC-2 increases glucose-elicited insulin secretion, whereas insulin secretion is decreased in islets from ASC-2+/− mice. Moreover, primary rat islets ectopically expressing DN1 or DN2 exhibit decreased insulin secretion. The mass and number of islets also decrease in heterozygous mice, likely due to increased apoptosis and decreased proliferation of ASC-2+/− islets.23
3. 129S6 Isogenic ASC-2+/− Mice
In contrast to ASC-2+/− mice in a C57/129 mixed genetic background, isogenic ASC-2+/− 129S6 mice exhibit a neonatal growth phenotype and newborn pups are ~10–15% smaller than their wild-type littermates.26 However, within 2 months after weaning, ASC-2+/− 129S6 mice become similar in size to their wild-type littermates. Interestingly, ~3% of the ASC-2+/− 129S6 newborn pups are extremely growth stunted, weighing 70% less than their wild-type littermates. These mice often die before weaning. However, a small number of these mice survive and also become equivalent to wild-type littermates in weight.26 Interestingly, this phenotype seems distinct from MLL3Δ/Δ newborn mice whose uniformly smaller size and weight deficit persist through adulthood.12 The molecular mechanisms underlying impaired growth in MLL3Δ/Δ and isogenic 129S6 ASC-2+/− mice have yet to be determined.
In contrast to the absence of reproductive phenotypes in ASC-2+/− mice in C57/129 mixed genetic background, the fertility of both sexes of isogenic ASC-2+/− mice in 129S6 background is compromised.26 Both male and female mice are hypofertile and ~20% of isogenic ASC-2+/− females are sterile. These infertile females appear to mate based on the formation of vaginal plugs and exhibit normal estrus cycles. These results suggest abnormalities in oogenesis, implantation of fertilized ova, or a defect in placental function, as no embryos are detected between E8.5 and E12.5. The number of newborn ASC-2+/− pups obtained from crosses between ASC-2+/− 129S6 hypofertile males and females is far less than expected based on Mendelian distribution.26 This appears to result from a significant number of ASC-2+/− embryos dying in utero. Those ASC-2−/− 129S6 female mice which are hypofertile exhibit a progressive decline in fertility as they age and their newborn pups have a high rate of neonatal mortality. The reproductive phenotypes in isogenic ASC-2+/− 129S6 mice are similar to those found for MLL3Δ/Δ mice.9 Future studies should be directed at elucidating the molecular mechanism underlying male and female hypofertility in MLL3Δ/Δ and isogenic 129S6 ASC-2+/− mice.
4. Phenotypes from Conditional Knockouts
To overcome the early embryonic lethality of ASC-2−/− mice and to study the role of ASC-2 in select tissues of adult mice, conditional knockout mice for ASC-2 have been constructed. In ASC-2-deficient mammary glands,124 the elongation of ducts during puberty is not affected, but the number of ductal branches is decreased. Moreover, during pregnancy, the null mammary glands exhibit decreased alveolar density. Interestingly, the lactating ASC-2-deficient glands contain scant lobuloalveoli with many adipocytes, while the wild-type glands lack adipocytes. The null mammary glands fail to produce enough milk to nurse all the pups during lactation. These results suggest that ASC-2 contributes to efficient ductal branching of mammary glands in response to estrogen. A slight increase in apoptosis has been observed in the terminal buds from ASC-2-deficient glands, raising the possibility that abnormal apoptosis contributes to the impaired ductal branching.124
ASC-2 liver conditional knockout mice have been generated to study the in vivo role of ASC-2 as a coactivator for PPARα and CAR.122 We have reported that ASC-2 interacts with CAR and enhances the transcriptional activation by CAR in transfection assays.123 However, the in vivo data obtained from conditional knockout studies show that ASC-2 deficiency in the liver fails to alter CAR function, as both wild-type and ASC-2 liver conditional knockout mice are susceptible to acetaminophen-induced liver damage mediated by CAR and upregulate expression of CAR-target genes (e.g., CYP1A2, CYP2B10, CYP3A11, and CYP2E1) in response to CAR ligand. CAR mRNA and protein levels between these mice are similar, suggesting that ASC-2 does not alter hepatic CAR expression.122 Notably, these in vivo findings are somewhat contradictory to our earlier studies with transgenic mice expressing DN1, a fragment of ASC-2 containing the NR1, as these mice fail to show acetaminophen-induced hepatic necrosis.123 Similarly, ASC-2 has been shown to activate PPARs in vitro.29,38 However, in conditional knockout mice, the degree of PPARα ligand-mediated peroxisome proliferation in liver cells has been found to be essentially similar to that seen in wild-type liver cells, implying that, in the liver, ASC-2 is not involved for PPARα-mediated proliferation. Accordingly, the PPARα-specific target genes encoding fatty acyl-CoA oxidase, enoyl-CoA hydratase/L-3 hydoxyacyl-CoA hydrogenase (L-bifunctional enzyme; L-PBE), peroxisomal thiolase (PTL), and CYP4A1 increase markedly in both wild-type and ASC-2 liver conditional knockout mice, following treatment with the PPARα ligand. PPARα mRNA levels remain essentially similar in control and PPARα ligand-treated mice.122 Based on these results, it has been suggested that ASC-2 is dispensable in the liver for activating PPARα- and CAR-mediated gene expression.122 However, it needs to be further determined whether MLL3/4 complexes are also dispensable for the hepatic function of PPARα and CAR, as it is possible that MLL3/4 complexes still form in the absence of ASC-2 and that another subunit of ASCOM may function to tether ASCOM to PPARα and CAR. Alternatively, other Set1-like complex(es) may function redundantly with ASCOM, and inactivation of ASCOM alone may not be sufficient to alter PPARα and CAR functions in the liver. Further analyses of MLL3/4 mutant mice should clarify these issues.
B. Metabolic Phenotypes of MLL3
H3K4-trimethylation is an evolutionarily conserved mark for transcriptionally active chromatin.47,48 Interestingly, whereas yeast has a single enzyme responsible for this modification (H3K4MT), higher eukaryotes carry a number of H3K4MTs.94 This suggests that individual H3K4MTs in higher eukaryotes may have distinct target genes. However, the physiological roles for higher eukaryotic H3K4MTs and their target genes remain poorly understood. We have generated MLL3Δ/Δ mutant mice that express an MLL3 with a deletion of the Set catalytic domain that is involved in H3K4-methylation.9 The MLL3Δ/Δ mutation in a mixed C57/129 genetic background leads to partial embryonic lethality,9 while isogenic C57BL/6 MLL3Δ/Δ mice are completely embryonic lethal (our unpublished results). Interestingly, MLL3Δ/Δ mice share some similar phenotypes with isogenic 129S6 ASC-2+/− mice,26 providing genetic evidence for the assembly of MLL3 into ASCOM. Similar to isogenic 129S6 ASC-2+/− mice,26 MLL3Δ/Δ mice are stunted in their overall growth and weigh 30–40% less at birth. MLL3Δ/Δ females also exhibit a range of reproductive phenotypes from infertility to hypofertility, while males are generally hypofertile. In addition, like ASC-2−/− MEFs, MEFs generated from MLL3Δ/Δ mice divide at approximately half the rate of wild-type MEFs.9,26 Our phenotypic analyses of MLL3Δ/Δ mice reveal that MLL3 has multiple functions in vivo, likely as a component of ASCOM. In particular, our results reveal that MLL3, and possibly MLL4, may be specialized for regulating genes involved in metabolic homeostasis, thus revealing key roles for specific H3K4MTs in metabolism.
1. WAT Phenotypes of MLL3Δ/Δ Mice
ASC-2-null MEFs are refractory to PPARγ-stimulated adipogenesis and fail to express the PPARγ-responsive, adipogenic marker gene aP2.95 However, the specific roles for MLL3 and MLL4 in adipogenesis remain undefined. We recently have found that MLL3 plays crucial roles in adipogenesis.12 First, MLL3Δ/Δ mice have a significantly decreased amount of WAT with a favorable overall metabolic profile, including improved insulin sensitivity and increased energy expenditure. Second, MLL3Δ/Δ MEFs are mildly but consistently less responsive to inducers of adipogenesis than wild-type MEFs. Third, ASC-2, MLL3, and MLL4 are recruited to the PPARγ-activated aP2 gene during adipogenesis.12 Consistent with earlier demonstrations that ASC-2 binds directly to PPARγ and C/EBPα,10 two factors important for adipogenesis,125 ASCOM directly interacts with both free and promoter-bound PPARγ·RXRα heterodimers.12 Thus, it is possible that ASC-2 plays a similar role in facilitating ASCOM function through PPARγ and C/EBPα during adipogenesis. Moreover, while H3K4-methylation of aP2 is readily induced in wild-type MEFs, it is not induced in ASC-2−/− MEFs and only partially induced in MLL3Δ/Δ MEFs.12 These results suggest that ASCOM-MLL3 and ASCOM-MLL4 likely function as crucial but redundant H3K4MT complexes for PPARγ-dependent adipogenesis, uncovering an interesting connection between H3K4-trimethylation and adipogenesis.
Interestingly, we have observed that PPARγ exhibits both ligand-dependent and ligand-independent interactions with different components of the ASCOM complex,12 uncovering a previously unappreciated complexity in recruiting ASCOM to NRs. This could reflect, for example, the presence of a dynamic equilibrium distribution of ASC-2-containing and ASC-2 free complexes and an associated ligand-independent recruitment of the core complex, possibly through interactions with a PPARγ domain other than AF2, followed by a ligand- and AF2-dependent recruitment or stabilization of ASC-2. Another possibility is the presence in our purified PPARγ of an active AF2 conformation, due either to an endogenous ligand or to a natural equilibrium between active and inactive AF2 states,45 and an AF2-dependent recruitment of ASCOM through LXXLL motifs in MLL3/411,93 or ASC-2 with further stabilization of MLL3/4 or ASC-2 by the potent ectopic ligands.
2. Lipid and Bile Acid Phenotypes of MLL3Δ/Δ Mice
In further support of the selective involvement of ASCOM in metabolism, we have discovered two additional lines of metabolic phenotypes, in MLL3Δ/Δ mice that are ascribed to the roles of ASCOM as a H3K4MT coactivator of LXRs and FXR.
The second NR box of ASC-2 has been shown to specifically recognize LXRs.20,44 However, no exact role for either ASC-2 or MLL3/4 in LXR transactivation has been clearly defined. Our recent results reveal that the key function of ASC-2 in LXR transactivation is to present MLL3 and MLL4 to LXRs.11 Thus, ASC-2 is required for ligand-induced recruitment of MLL3 and MLL4 to LXRs, and LXR ligand T1317 induces not only expression of LXR-target genes but also their H3K4-trimethylation. Strikingly, both of these ligand effects are ablated in ASC-2-null cells but only partially suppressed in cells expressing an enzymatically inactivated mutant MLL3.11 Our results also show that LXR transactivation does not appear to require certain other Set1-like complexes, because, while ASC-2 is required for LXR transactivation, menin, an integral component of MLL1/2 Set1-like complexes, is dispensable.11 In comparison, targeted deletion of menin or siRNA-mediated reduction of MLL4 alone has been shown to significantly impair ERα transactivation.93,96 Taken together, these results suggest that ASCOM-MLL3 and ASCOM-MLL4 play redundant but essential roles in ligand-dependent H3K4-trimethylation and expression of LXR-target genes and that ASC-2 is likely a key determinant for LXR function through ASCOM (but not other Set1-like complexes). We have proposed that a subunit of Set1-like complexes determines the target specificity of each complex via direct protein–protein interactions with target transcription factors9 (Fig. 2C). The current study supports this model and validates the previously proposed specialized function for ASC-2 as a key adaptor to present ASCOM to LXRs.11,20,21,44,126
In further support of the role of ASCOM as a coactivator of LXRs, our analysis of hepatic mRNAs shows that the hepatic lipogenic LXR-target genes FAS and SREBP-1c are suppressed in MLL3Δ/Δ mice.11 Intriguingly, expression of another direct LXR-target gene Cyp7A1, that encodes a key enzyme in the conversion of cholesterol into bile acids is not downregulated.11 These results suggest that clearance of cholesterol via bile acid synthesis could be intact in MLL3Δ/Δ mice, which, along with the impaired expression of lipogenic genes, might have contributed to our failure to observe accumulation of oil-red-O positive lipid droplets in the livers of MLL3Δ/Δ mice even with a high fat diet.11 Thus, these results raise the interesting possibility that MLL3 may have some selectivity toward a subset of LXR-target genes involved in hepatic de novo lipogenesis. Future studies should be directed toward elucidation of the molecular basis underlying this interesting selectivity.
Our recent results reveal that expression of two FXR-target genes, BSEP and SHP, is downregulated in MLL3Δ/Δ mice (Mol Endo 2009, in press, and our unpublished results). SHP encodes an atypical orphan nuclear receptor that lacks a DBD.127 SHP forms a novel regulatory loop with FXR, which functions to maintain bile acid homeostasis.127 SHP expression is induced in response to increased hepatic levels of bile acids that bind and activate FXR, and SHP in turn suppresses expression of Cyp7A1 by antagonizing the activity of another NR, “liver receptor homolog 1 (LRH-1),” that functions as an inducer of Cyp7A1. This leads to a blockage in the further production of bile acids from cholesterol.127 Consistent with the decreased expression of SHP, we have found that Cyp7A1 is significantly upregulated in MLL3Δ/Δ mice (Mol Endo 2009, in press, and our unpublished results). In further support of these results, MLL3Δ/Δ mice exhibit a much higher level of plasma bile acid when fed a control diet supplemented with 0.5% (w/w) cholic acid (CA, a ligand for FXR) (Mol Endo 2009, in press, and our unpublished results), similar to FXR-null mice.128 In further indication for impaired bile acid homeostasis, MLL3Δ/Δ mice fed a control diet supplemented with 0.5% (w/w) CA also have a significantly enlarged gallbladder filled with bile acids and thus the total pool of bile acids in MLL3Δ/Δ mice is much larger than that in wild-type mice (Mol Endo 2009, in press, and our unpublished results). These results indicate that bile acid homeostasis is seriously disturbed in MLL3Δ/Δ mice. Interestingly, expression of BSEP and SHP is still induced and expression of Cyp7A1 is still suppressed in response to a control diet supplemented with 0.5% (w/w) CA in MLL3Δ/Δ mice, suggesting that another H3K4MT, likely MLL4, functions in the livers of MLL3Δ/Δ mice. Consistent with these results, both MLL3 and MLL4 are recruited to SHP via FXR, and suppression of SHP expression and SHP H3K4-trimethylation is much greater with siRNA-mediated downregulation of both MLL3 and MLL4 than with downregulation of MLL3 or MLL4 alone (Mol Endo 2009, in press, and our unpublished results). Overall, these results suggest that ASCOM-MLL3 and ASCOM-MLL4 act as redundant but crucial coactivators of FXR. Our newly established mice with a mutant MLL4 should help clarify whether MLL4 indeed functions redundantly with MLL3 in mediating FXR transactivation.
C. ASCOM in Cancers
The ASC-2 gene is amplified and overexpressed in human breast and other cancers.129–131 However, it has been difficult to determine whether ASCOM is involved in tumorigenesis or antitumorigenesis. This is mainly because ASC-2 and MLL3/4 target a complex array of transcription factors that include both tumorigenic and tumor suppressive proteins.10 However, as these ASCOM target factors have been identified mostly through cell transfection and in vitro studies,10 the roles for ASCOM in their transactivation remain to be validated in more physiological settings. The results discussed below, along with the finding that MLL3 maps to 7q36, a chromosome region that is frequently deleted in myeloid disorders,132 are in favor of tumor suppressive roles for ASCOM (see below). Given the multifunctionality of ASCOM, however, it is possible that ASCOM may also function to trigger tumorigenesis in different cell types.
1. Amplification and Overexpression of ASC-2 in Human Cancers
Human ASC-2 is localized on chromosome 20 (20q11). The ASC-2 gene was initially mapped to the 20q11 segment of chromosome 20 during a search for amplified and overexpressed genes mapping to chromosome 20q in breast cancer.130 Subsequently, we have found, using FISH analysis, that the ASC-2 copy number is increased to a moderate level (4–6 copies) in 14 out of 335 (4.2%) cases of breast cancer and to a high level (>6 copies) in 15 out of 335 (4.5%) cases of breast cancer.131 ASC-2 mRNA is also detected in 11 different breast cancer cell lines, with the highest expression in BT-474 cells.131 The ASC-2 gene is also found to be amplified in lung and colon cancers.131 Notably, the high level of ASC-2 expression in breast cancer cell lines does not correlate with the level of ER expression.131
2. Mammary Tumor Suppressive Function of ASC-2
To study the roles of ASC-2 in mammary tumorigenesis, ASC-2+/− mice have been crossed with mice carrying polyoma middle-T antigen (pyMT).133 This study has revealed that mammary tumor development in ASC-2+/− pyMT female and male mice is substantially accelerated compared with that in wild-type pyMT mice. Correspondingly, tumor formation in nude mice that receive premalignant ASC-2+/− pyMT mammary tissue is much faster than in nude mice that receive transplants from premalignant wild-type pyMT mammary tissue.133 This tumor acceleration is reported to result from increased cell proliferation and ductal hyperplasia and mammary intraepithelial neoplasia. Although the mechanism of pyMT-induced tumorigenesis is proposed to reflect a partial impairment of activated PPARγ/RXR,133 it is important to note that ASC-2 is a multifunctional coactivator and, thus, that the accelerated tumor phenotypes in ASC-2+/− pyMT mice may also involve ASC-2 coactivator functions for other tumor suppressive transcription factors such as p53 (see below).
3. ASCOM as a Coactivator of the Tumor Suppressor p53 and Kidney Phenotypes of MLL3Δ/Δ Mice
Further implicating ASCOM in tumor suppression pathways, we have found that ASCOM-MLL3 and ASCOM-MLL4 function as redundant but crucial coactivator complexes for the tumor suppressor p53 (PNAS 2009;106:8513–8, and our unpublished results). Our results reveal, first, that ASC-2 acts as a coactivator of p53 in reporter assays and is required for H3K4-trimethyation and expression of endogenous p53-target genes in response to the DNA damaging agent doxorubicin and, second, that ureter epithelial tumors result from targeted inactivation of MLL3 H3K4-methylation activity in the mouse (PNAS 2009;106:8513–8, and our unpublished results). Interestingly, this latter phenotype is exacerbated in a p53+/− background and the tumorigenic cells are heavily immunostained for γH2AX (PNAS 2009;106:8513–8, and our unpublished results), indicating a contribution of MLL3 to the DNA damage response pathway through p53. In support of redundant functions for MLL3 and MLL4 in this process, siRNA-mediated downregulation of both MLL3 and MLL4 is required to suppress doxorubicin-inducible expression of p53-target genes (PNAS 2009;106:8513–8, and our unpublished results). Importantly, this study identifies a physiologically relevant, specific H3K4-trimethytransferase coactivator complex for p53. Notably, and related, independent siRNA-based studies have indicated that p53 is regulated by ASC-2.32
Interestingly, PTIP, which has been proposed to play important roles in cellular responses to DNA damage,134–136 has recently been identified as an additional component of ASCOM.90,91 The tumor suppressor p53 plays a key role countering the adverse effects of DNA damage,137,138 which otherwise can be lethal or lead to oncogenic transformation. DNA damage induces the transcriptional activity of p53 via damage sensors such as ATM. Interestingly, PTIP appears to be required for ATM-mediated phosphorylation of p53 at Ser 15 and for DNA damage-induced upregulation of the cyclin-dependent kinase inhibitor p21.135 Correspondingly, the loss-of-function studies in mice indicate that PTIP is essential for the maintenance of genomic stability.134 One intriguing future challenge is to test whether this function of PTIP occurs in the context of ASCOM, particularly because ASC-2-null cells show increased phosphorylation of p53 at Ser 15 (PNAS 2009;106:8513–8, and our unpublished results). Regardless, as our results suggest that ASCOM acts as a tumor suppressive coactivator complex of p53, ASCOM, like PTIP, may also play crucial roles in the maintenance of genomic stability. Indeed, we show that targeted inactivation of MLL3 H3K4-methylation activity in the mouse results in ureter epithelial tumors accompanied by an increased level of damaged DNA and that ASC-2 appears to be important for DNA damage-induced expression of p53-target genes (PNAS 2009;106:8513–8, and our unpublished results).
ASC-2 appears to play a crucial role in effecting MLL3/4 function on p53-target genes, as our results reveal that MLL3 and MLL4 are recruited to p53 and carry out H3K4-trimehtylation of p53-target genes in an ASC-2-dependent manner (PNAS 2009;106:8513–8, and our unpublished results). The precise roles of ASC-2 in p53 transactivation remain unclear, as ASC-2 does not directly interact with p53 (PNAS 2009;106:8513–8, and our unpublished results). Our further search for the mechanistic basis underlying the associations between p53 and ASCOM reveal that 53BP1, a protein originally identified based on its interaction with p53,139,140 is recruited to p21-p53REs. Moreover, the timing of this recruitment precisely overlaps that of ASC-2 (PNAS 2009;106:8513–8, and our unpublished results), suggesting corecruitment of these two proteins. These results support the previously proposed role for 53BP1 as a coactivator of p53.140 Our proposal for an adaptor role for 53BP1 between p53 and ASCOM is further supported by our finding that 53BP1 and ASC-2 are readily coimmunopurified and that 53BP1 appears to directly interact not only with p53139,140 but also with ASC-2 (PNAS 2009;106:8513–8, and our unpublished results). However, because a significant level of ASC-2/MLL3 recruitment to p21-p53REs is still observed in 53BP1-null cells, additional mechanisms are expected to exist (PNAS 2009;106:8513–8, and our unpublished results). Thus, the detailed molecular basis underlying the recruitment of ASCOM to p53 remains to be further delineated.
Because the loss of p53 function is generally associated with most tumors, the seeming propensity of MLL3Δ/Δ mice to selectively develop urothelial tumors (PNAS 2009;106:8513–8, and our unpublished results) may appear paradoxical. On the one hand, this may reflect a redundancy between MLL3 and other MLLs in most tissues and, on the other hand, a more careful examination may yet uncover other types of tumors. In fact, ASCOM may guard against a broad range of epithelial tumors, and acceleration of pyMT-induced mammary tumorigenesis by haploid inactivation of ASC-2133 may similarly involve genomic instability caused by the impaired ability of ASCOM to support p53 transactivation. Nonetheless, it is possible that, along with a general redundancy between ASCOM-MLL3 and ASCOM-MLL4, an as-yet-to-be-characterized specific function of MLL3 in urothelium may selectively lead to urothelial tumors in MLL3Δ/Δ mice.
VII. Future Challenges
The studies discussed in this review suggest that ASCOM likely serves as an essential H3K4-methylation complex of multiple NRs and other transcription factors and accordingly regulates a diverse array of physiological processes. Most striking is the possibility that ASCOM may have a specialized function for a selective set of NRs involved in metabolism. However, more surprises for the physiological function of ASCOM are likely to be encountered as we continue to dissect the phenotypes of ASC-2/MLL3/MLL4 mutant mice. To fully decipher the physiological function of ASCOM, many outstanding questions remain to be answered, and a few immediate challenges are summarized below.
ASCOM is a unique Set1-like complex, because it also contains UTX, an H3K27-demethylase. H3K27-methylation is a posttranslational modification that is highly correlated with genomic silencing. Thus, ASCOM may serve as an excellent model system to study the potential interplay between H3K4- and H3K27-methylation events. Indeed, during retinoic acid signaling events, the recruitment of the UTX complex to HOX genes has been shown to result in H3K27-demethylation and a concomitant methylation of H3K4.16 Our preliminary results also indicate that these two modifications are inversely coregulated in RAR-target gene, RAR-β2, through MLL3/4 and UTX (our unpublished results). These results suggest a concerted mechanism for transcriptional activation in which cycles of H3K4-methylation by ASCOM are linked with the demethylation of H3K27 through UTX. It will be interesting to investigate whether these two distinct enzymes residing in the same complex modulate each other’s activity “directly.” Importantly, further studies of ASCOM may open new possibilities for pharmacological interventions of various disorders and diseases, because UTX and MLL3/4 should be chemically modulatable.
In our studies, MLL3 and MLL4 appear to function redundantly in regulating target genes for RAR, LXRs, FXR, and PPARγ9,11,12 (our unpublished results), likely due to the central adaptor role of ASC-2 in recruiting ASCOM-MLL3 and ASCOM-MLL4 to these NRs. Accordingly, the phenotypes of MLL3Δ/Δ mice elicited by these NRs may also be observed in MLL4 mutant animals. One important future challenge is to determine whether MLL3 and MLL4 also have their own unique sets of target genes. There are at least two related scenarios which would lead us to observe distinct functions between MLL3 and MLL4. First, if any component of ASCOM, which functions as an adaptor for specific NRs and/or transcription factors, is found only in ASCOM-MLL3 or ASCOM-MLL4, we would observe differences between MLL3 and MLL4 in regulating genes targeted by these NRs and transcription factors. Secondly, it is noted that MLL3 and MLL4 are gigantic in size but their overall homology is merely 30%.8,132 Thus, it is possible that these proteins may contain distinct interaction surfaces for some NRs and or transcription factors and thereby act as specific adaptors for selective ASCOM-MLL3 or ASCOM-MLL4 recruitment. Given our recent success in establishing MLL4 mutant mice (our unpublished results), we should be able to investigate these and other possibilities. Also, we should carefully examine whether any subunit of ASCOM is found selectively associated with MLL3 or MLL4.
Our unpublished results indicate that multiple subunits of ASCOM undergo specific proteolysis during adipogenesis. We also have found that MEKK1 relieves the cryptic autonomous activation function of full-length ASC-2 (our unpublished results), suggesting that ASC-2 is subjected to MAPK regulation through direct or indirect phosphorylation. Overall, ASC-2 is likely to be subject to diverse posttranslational modifications, which may serve as a focal point to communicate and integrate regulatory events mediated by NRs or other transcription factors and the signaling cascades generated by cell surface receptors.
Our finding of a coactivator role for ASCOM in p53 transactivation is consistent with some recent studies suggesting that ASC-2 could be involved in DNA repair. The ASC-2 C-terminal region has been shown to interact with components of the DNA-dependent protein kinase complex, including the catalytic subunit (DNA–PKc), regulatory subunits (Ku70 and Ku80), PARP, NuMA (p200), and DNA topoisomerase I36 (Fig. 2E). Although the role of DNA–PK in recombination and DNA repair has been well characterized,141 the involvement of DNA–PK in transcription is less clear. However, studies of ASC-2 activity in cells deficient in DNA–PK reveal a marked reduction in ASC-2-mediated enhancement of GR transactivation.141 Moreover, DNA–PK phosphorylates an ASC-2 fragment containing amino acids 714–999 in a DNA-dependent manner and a fragment containing amino acids 1237–2063 in a DNA-independent manner.142 In addition, ASC-2 stimulates the phosphorylation activity of DNA–PK in vitro, generating a phosphorylation pattern that differs from that of its DNA-mediated activation form. Interestingly, PTIP, a component of ASCOM,90,91 and 53BP1, a putative adaptor protein between p53 and ASCOM (PNAS 2009;106:8513–8, and our unpublished results), have also been shown to modulate DNA repair,134–136,143 although it remains to be determined whether PTIP functions through ASCOM in DNA repair. ASC-2 has also been shown to localize to a pS2 promoter nucleosome (NucE) that contains an ER binding site and TopoIIβ-mediated double strand breaks.144
Finally, with regard to the tumor suppressive function of ASCOM and its interplay with Swi/Snf, it is noted that at least two subunits of Swi/Snf, BRG1, and INI1, have been linked to tumor suppression pathways and that mice heterozygous for mutations at SNF5 and BRG1 are cancer-prone.145–147 Thus, one interesting future challenge is to investigate whether the MLL3/4: INI1 interactions that play crucial roles in the cross talk between ASCOM and Swi/Snf107 play any roles in integrating the two tumor suppressive pathways directed by ASCOM and Swi/Snf, respectively.
Acknowledgments
Work from authors’ laboratories was supported by NIH/NIDDK grants to J.W.L. (DK064678) and to R.G.R. (DK071900).
References
- 1.Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev 2001;81:1269–304. [DOI] [PubMed] [Google Scholar]
- 2.Lonard DM, Lanz RB, O’Malley BW. Nuclear receptor coregulators and human disease. Endocr Rev 2007;28:575–87. [DOI] [PubMed] [Google Scholar]
- 3.Nettles KW, Greene GL. Ligand control of coregulator recruitment to nuclear receptors. Annu Rev Physiol 2005;67:309–33. [DOI] [PubMed] [Google Scholar]
- 4.Rosenfeld MG, Glass CK. Coregulator codes of transcriptional regulation by nuclear receptors. J Biol Chem 2001;276:36865–8. [DOI] [PubMed] [Google Scholar]
- 5.Shilatifard A Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 2006;75:243–69. [DOI] [PubMed] [Google Scholar]
- 6.Jenuwein T, Allis CD. Translating the histone code. Science 2001;293:1074–80. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Fischle W, Cheung W, Jacobs S, Khorasanizadeh S, Allis CD. Beyond the double helix: writing and reading the histone code. Novartis Found Symp 2004;259:3–17; discussion 17–21, 163–9. [PubMed] [Google Scholar]
- 8.Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol 2003;23:140–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S, Lee DK, Dou Y, Lee J, Lee B, Kwak E, et al. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc Natl Acad Sci USA 2006;103:15392–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahajan MA, Samuels HH. Nuclear hormone receptor coregulator: role in hormone action, metabolism, growth, and development. Endocr Rev 2005;26:583–97. [DOI] [PubMed] [Google Scholar]
- 11.Lee S, Lee J, Lee SK, Lee JW. Activating signal cointegrator-2 is an essential adaptor to recruit histone H3 lysine 4 methyltransferases MLL3 and MLL4 to the liver X receptors. Mol Endocrinol 2008;22:1312–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, Saha PK, Yang QH, Lee S, Park JY, Suh Y, et al. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc Natl Acad Sci USA 2008;105:19229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 2007;449:731–4. [DOI] [PubMed] [Google Scholar]
- 14.Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA 2007;104:18439–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 2007;449:689–94. [DOI] [PubMed] [Google Scholar]
- 16.Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 2007;318:447–50. [DOI] [PubMed] [Google Scholar]
- 17.Smith ER, Lee MG, Winter B, Droz NM, Eissenberg JC, Shiekhattar R, et al. Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol Cell Biol 2008;28:1041–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional coactivators mediates binding to nuclear receptors. Nature 1997;387:733–6. [DOI] [PubMed] [Google Scholar]
- 19.Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, et al. The transcriptional coactivator p CIP binds CBP and mediates nuclear-receptor function. Nature 1997;387:677–84. [DOI] [PubMed] [Google Scholar]
- 20.Lee SK, Jung SY, Kim YS, Na SY, Lee YC, Lee JW. Two distinct nuclear receptor-interaction domains and CREB-binding protein-dependent transactivation function of activating signal cointegrator-2. Mol Endocrinol 2001;15:241–54. [DOI] [PubMed] [Google Scholar]
- 21.Kim SW, Park K, Kwak E, Choi E, Lee S, Ham J, et al. Activating signal cointegrator 2 required for liver lipid metabolism mediated by liver X receptors in mice. Mol Cell Biol 2003;23:3583–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SW, Cheong C, Sohn YC, Goo YH, Oh WJ, Park JH, et al. Multiple developmental defects derived from impaired recruitment of ASC-2 to nuclear receptors in mice: implication for posterior lenticonus with cataract. Mol Cell Biol 2002;22:8409–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeom SY, Kim GH, Kim CH, Jung HD, Kim SY, Park JY, et al. Regulation of insulin secretion and beta-cell mass by activating signal cointegrator 2. Mol Cell Biol 2006;26:4553–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antonson P, Schuster GU, Wang L, Rozell B, Holter E, Flodby P, et al. Inactivation of the nuclear receptor coactivator RAP250 in mice results in placental vascular dysfunction. Mol Cell Biol 2003;23:1260–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuang SQ, Liao L, Zhang H, Pereira FA, Yuan Y, DeMayo FJ, et al. Deletion of the cancer-amplified coactivator AIB3 results in defective placentation and embryonic lethality. J Biol Chem 2002;277:45356–60. [DOI] [PubMed] [Google Scholar]
- 26.Mahajan MA, Das S, Zhu H, Tomic-Canic M, Samuels HH. The nuclear hormone receptor coactivator NRC is a pleiotropic modulator affecting growth, development, apoptosis, reproduction, and wound repair. Mol Cell Biol 2004;24:4994–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu YJ, Crawford SE, Stellmach V, Dwivedi RS, Rao MS, Gonzalez FJ, et al. Coactivator PRIP, the peroxisome proliferator-activated receptor-interacting protein, is a modulator of placental, cardiac, hepatic, and embryonic development. J Biol Chem 2003;278:1986–90. [DOI] [PubMed] [Google Scholar]
- 28.Mahajan MA, Samuels HH. A new family of nuclear receptor coregulators that integrate nuclear receptor signaling through CREB-binding protein. Mol Cell Biol 2000;20:5048–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caira F, Antonson P, Pelto-Huikko M, Treuter E, Gustafsson JA. Cloning and characterization of RAP250, a novel nuclear receptor coactivator. J Biol Chem 2000;275:5308–17. [DOI] [PubMed] [Google Scholar]
- 30.Antonson P, Al-Beidh F, Matthews J, Gustafsson JA. The human RAP250 gene: genomic structure and promoter analysis. Gene 2004;327:233–8. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Liao L, Kuang SQ, Xu J. Spatial distribution of the messenger ribonucleic acid and protein of the nuclear receptor coactivator, amplified in breast cancer-3, in mice. Endocrinology 2003;144:1435–43. [DOI] [PubMed] [Google Scholar]
- 32.Mahajan MA, Murray A, Levy D, Samuels HH. Nuclear receptor coregulator (NRC): mapping of the dimerization domain, activation of p53 and STAT-2, and identification of the activation domain AD2 necessary for nuclear receptor signaling. Mol Endocrinol 2007;21:1822–34. [DOI] [PubMed] [Google Scholar]
- 33.Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O’Malley B, et al. Activation of PPARgamma coactivator-1 through transcription factor docking. Science 1999;286:1368–71. [DOI] [PubMed] [Google Scholar]
- 34.Goo YH, Na SY, Zhang H, Xu J, Hong S, Cheong J, et al. Interactions between activating signal cointegrator-2 and the tumor suppressor retinoblastoma in androgen receptor transactivation. J Biol Chem 2004;279:7131–5. [DOI] [PubMed] [Google Scholar]
- 35.Lu J, Danielsen M. Differential regulation of androgen and glucocorticoid receptors by retinoblastoma protein. J Biol Chem 1998;273:31528–33. [DOI] [PubMed] [Google Scholar]
- 36.Ko L, Cardona GR, Chin WW. Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proc Natl Acad Sci USA 2000;97:6212–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang C, Norris JD, Gron H, Paige LA, Hamilton PT, Kenan DJ, et al. Dissection of the LXXLL nuclear receptor-coactivator interaction motif using combinatorial peptide libraries: discovery of peptide antagonists of estrogen receptors alpha and beta. Mol Cell Biol 1999;19:8226–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y, Kan L, Qi C, Kanwar YS, Yeldandi AV, Rao MS, et al. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. J Biol Chem 2000;275:13510–6. [DOI] [PubMed] [Google Scholar]
- 39.Hu X, Lazar MA. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature 1999;402:93–6. [DOI] [PubMed] [Google Scholar]
- 40.Perissi V, Staszewski LM, McInerney EM, Kurokawa R, Krones A, Rose DW, et al. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev 1999;13:3198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ko L, Cardona GR, Iwasaki T, Bramlett KS, Burris TP, Chin WW. Ser-884 adjacent to the LXXLL motif of coactivator TRBP defines selectivity for ERs and TRs. Mol Endocrinol 2002;16:128–40. [DOI] [PubMed] [Google Scholar]
- 42.Zhao C, Toresson G, Xu L, Koehler KF, Gustafsson JA, Dahlman-Wright K. Mouse estrogen receptor beta isoforms exhibit differences in ligand selectivity and coactivator recruitment. Biochemistry 2005;44:7936–44. [DOI] [PubMed] [Google Scholar]
- 43.Willy PJ, Mangelsdorf DJ. Unique requirements for retinoid-dependent transcriptional activation by the orphan receptor LXR. Genes Dev 1997;11:289–98. [DOI] [PubMed] [Google Scholar]
- 44.Son YL, Park OG, Kim GS, Lee JW, Lee YC. RXR heterodimerization allosterically activates LXR binding to the second NR box of activating signal co-integrator-2. Biochem J 2008;410:319–30. [DOI] [PubMed] [Google Scholar]
- 45.Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, et al. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 1998;395:137–43. [DOI] [PubMed] [Google Scholar]
- 46.Ito M, Roeder RG. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol Metab 2001;12:127–34. [DOI] [PubMed] [Google Scholar]
- 47.Dehe PM, Geli V. The multiple faces of Set1. Biochem Cell Biol 2006;84:536–48. [DOI] [PubMed] [Google Scholar]
- 48.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 2005;6:838–49. [DOI] [PubMed] [Google Scholar]
- 49.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, et al. Active genes are tri-methylated at K4 of histone H3. Nature 2002;419:407–11. [DOI] [PubMed] [Google Scholar]
- 50.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 2003;11:709–19. [DOI] [PubMed] [Google Scholar]
- 51.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol 2004;6:73–7. [DOI] [PubMed] [Google Scholar]
- 52.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 2005;120:169–81. [DOI] [PubMed] [Google Scholar]
- 53.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev 2004;18:1263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 2005;122:517–27. [DOI] [PubMed] [Google Scholar]
- 55.Beisel C, Imhof A, Greene J, Kremmer E, Sauer F. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 2002;419:857–62. [DOI] [PubMed] [Google Scholar]
- 56.Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, et al. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev 2002;16:479–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, et al. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell 2001;8:1207–17. [DOI] [PubMed] [Google Scholar]
- 58.Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol 2004;6:731–40. [DOI] [PubMed] [Google Scholar]
- 59.Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 2005;438:374–8. [DOI] [PubMed] [Google Scholar]
- 60.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev 2001;15:3286–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, et al. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J 2001;20:7137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, et al. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci USA 2001;98:12902–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roguev A, Schaft D, Shevchenko A, Pim Pijnappel WWM, Wilm M, Aasland R, et al. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J 2001;20:7137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Glaser S, Schaft J, Lubitz S, Vintersten K, van der Hoeven F, Tufteland KR, et al. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development 2006;133:1423–32. [DOI] [PubMed] [Google Scholar]
- 65.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 2002;10:1107–17. [DOI] [PubMed] [Google Scholar]
- 66.Wysocka J, Herr W. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem Sci 2003;28:294–304. [DOI] [PubMed] [Google Scholar]
- 67.Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell 2004;13:587–97. [DOI] [PubMed] [Google Scholar]
- 68.Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, et al. Leukemia protooncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol 2004;24:5639–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 1995;378:505–8. [DOI] [PubMed] [Google Scholar]
- 70.Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem 2005;280:41725–31. [DOI] [PubMed] [Google Scholar]
- 71.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol 2006;13:713–9. [DOI] [PubMed] [Google Scholar]
- 72.Steward MM, Lee J, O’Donovan A, Wyatt M, Bernstein BE, Shilatifard A. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol 2006;13:852–4. [DOI] [PubMed] [Google Scholar]
- 73.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 2005;121:859–72. [DOI] [PubMed] [Google Scholar]
- 74.Steward MM, Lee JS, O’Donovan A, Wyatt M, Bernstein BE, Shilatifard A. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol 2006;13:852–4. [DOI] [PubMed] [Google Scholar]
- 75.Sims III RJ, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev 2006;20:2779–86. [DOI] [PubMed] [Google Scholar]
- 76.Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain “Royal Family”: tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci 2003;28:69–74. [DOI] [PubMed] [Google Scholar]
- 77.Bienz M The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci 2006;31:35–40. [DOI] [PubMed] [Google Scholar]
- 78.Neely KE, Workman JL. Histone acetylation and chromatin remodeling: which comes first? Mol Genet Metab 2002;76:1–5. [DOI] [PubMed] [Google Scholar]
- 79.Shi X, Gozani O. The fellowships of the INGs. J Cell Biochem 2005;96:1127–36. [DOI] [PubMed] [Google Scholar]
- 80.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell 2007;25:1–14. [DOI] [PubMed] [Google Scholar]
- 81.Mizuguchi G, Tsukiyama T, Wisniewski J, Wu C. Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin. Mol Cell 1997;1:141–50. [DOI] [PubMed] [Google Scholar]
- 82.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 2006;442:96–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell 2006;24:785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martin DG, Baetz K, Shi X, Walter KL, MacDonald VE, Wlodarski MJ, et al. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol Cell Biol 2006;26:7871–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang XJ. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays 2004;26:1076–87. [DOI] [PubMed] [Google Scholar]
- 86.Sims III RJ, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 2007;28:665–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 2007;450:1106–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity 2007;27:561–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ramon-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, et al. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci USA 2007;104:18993–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 2007;282:20395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol 2007;27:1889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res 2008;647:21–9. [DOI] [PubMed] [Google Scholar]
- 93.Mo R, Rao SM, Zhu YJ. Identification of the MLL2 complex as a coactivator for estrogen receptor alpha. J Biol Chem 2006;281:15714–20. [DOI] [PubMed] [Google Scholar]
- 94.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 2007;25:15–30. [DOI] [PubMed] [Google Scholar]
- 95.Qi C, Surapureddi S, Zhu YJ, Yu S, Kashireddy P, Rao MS, et al. Transcriptional coactivator PRIP, the peroxisome proliferator-activated receptor gamma (PPARgamma)-interacting protein, is required for PPARgamma-mediated adipogenesis. J Biol Chem 2003;278:25281–4. [DOI] [PubMed] [Google Scholar]
- 96.Dreijerink KM, Mulder KW, Winkler GS, Hoppener JW, Lips CJ, Timmers HT. Menin links estrogen receptor activation to histone H3K4 trimethylation. Cancer Res 2006;66:4929–35. [DOI] [PubMed] [Google Scholar]
- 97.Cho H, Orphanides G, Sun X, Yang XJ, Ogryzko V, Lees E, et al. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol 1998;18:5355–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 2003;11:721–9. [DOI] [PubMed] [Google Scholar]
- 99.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 2006;125:703–17. [DOI] [PubMed] [Google Scholar]
- 100.Shahbazian MD, Zhang K, Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol Cell 2005;19:271–7. [DOI] [PubMed] [Google Scholar]
- 101.Sierra J, Yoshida T, Joazeiro CA, Jones KA. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev 2006;20:586–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 2005;121:873–85. [DOI] [PubMed] [Google Scholar]
- 103.Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci USA 2005;102:14765–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, et al. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci USA 2005;102:8603–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Smith ST, Petruk S, Sedkov Y, Cho E, Tillib S, Canaani E, et al. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat Cell Biol 2004;6:162–7. [DOI] [PubMed] [Google Scholar]
- 106.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 2006;442:86–90. [DOI] [PubMed] [Google Scholar]
- 107.Lee S, Kim D, Goo Y, Lee YC, Lee SK, Lee JW. Crucial roles for interactions between MLL3/4 and INI1 in nuclear receptor transactivation. Mol Endocrinol 2009;23:610–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tian H, Mahajan MA, Wong CT, Habeos I, Samuels HH. The N-Terminal A/B domain of the thyroid hormone receptor-beta2 isoform influences ligand-dependent recruitment of coactivators to the ligand-binding domain. Mol Endocrinol 2006;20:2036–51. [DOI] [PubMed] [Google Scholar]
- 109.Jung DJ, Na SY, Na DS, Lee JW. Molecular cloning and characterization of CAPER, a novel coactivator of activating protein-1 and estrogen receptors. J Biol Chem 2002;277:1229–34. [DOI] [PubMed] [Google Scholar]
- 110.Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, et al. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol Cell 2005;17:429–39. [DOI] [PubMed] [Google Scholar]
- 111.Imai H, Chan EK, Kiyosawa K, Fu XD, Tan EM. Novel nuclear autoantigen with splicing factor motifs identified with antibody from hepatocellular carcinoma. J Clin Invest 1993;92:2419–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res 2002;12:1231–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Iwasaki T, Chin WW, Ko L. Identification and characterization of RRM-containing coactivator activator (CoAA) as TRBP-interacting protein, and its splice variant as a coactivator modulator (CoAM). J Biol Chem 2001;276:33375–83. [DOI] [PubMed] [Google Scholar]
- 114.Auboeuf D, Dowhan DH, Li X, Larkin K, Ko L, Berget SM, et al. CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol Cell Biol 2004;24:442–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhu Y, Qi C, Cao WQ, Yeldandi AV, Rao MS, Reddy JK. Cloning and characterization of PIMT, a protein with a methyltransferase domain, which interacts with and enhances nuclear receptor coactivator PRIP function. Proc Natl Acad Sci USA 2001;98:10380–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Misra P, Qi C, Yu S, Shah SH, Cao WQ, Rao MS, et al. Interaction of PIMT with transcriptional coactivators CBP, p300, and PBP differential role in transcriptional regulation. J Biol Chem 2002;277:20011–9. [DOI] [PubMed] [Google Scholar]
- 117.Reyes JC, Muchardt C, Yaniv M. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J Cell Biol 1997;137:263–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Razin SV, Iarovaia OV, Sjakste N, Sjakste T, Bagdoniene L, Rynditch AV, et al. Chromatin domains and regulation of transcription. J Mol Biol 2007;369:597–607. [DOI] [PubMed] [Google Scholar]
- 119.McKenna NJ, Xu J, Nawaz Z, Tsai SY, Tsai MJ, O’Malley BW. Nuclear receptor coactivators: multiple enzymes, multiple complexes, multiple functions. J Steroid Biochem Mol Biol 1999; 69:3–12. [DOI] [PubMed] [Google Scholar]
- 120.Rozenblatt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, et al. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI SNF complex. Proc Natl Acad Sci USA 1998;95:4152–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rozovskaia T, Rozenblatt-Rosen O, Sedkov Y, Burakov D, Yano T, Nakamura T, et al. Self-association of the SET domains of human ALL-1 and of Drosophila TRITHORAX and ASH1 proteins. Oncogene 2000;19:351–7. [DOI] [PubMed] [Google Scholar]
- 122.Sarkar J, Qi C, Guo D, Ahmed MR, Jia Y, Usuda N, et al. Transcription coactivator PRIP, the peroxisome proliferator-activated receptor (PPAR)-interacting protein, is redundant for the function of nuclear receptors PPARalpha and CAR, the constitutive androstane receptor, in mouse liver. Gene Exp 2007;13:255–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Choi E, Lee S, Yeom SY, Kim GH, Lee JW, Kim SW. Characterization of activating signal cointegrator-2 as a novel transcriptional coactivator of the xenobiotic nuclear receptor constitutive androstane receptor. Mol Endocrinol 2005;19:1711–9. [DOI] [PubMed] [Google Scholar]
- 124.Qi C, Kashireddy P, Zhu YT, Rao SM, Zhu YJ. Null mutation of peroxisome proliferator-activated receptor-interacting protein in mammary glands causes defective mammopoiesis. J Biol Chem 2004;279:33696–701. [DOI] [PubMed] [Google Scholar]
- 125.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem 2008;77:289–312. [DOI] [PubMed] [Google Scholar]
- 126.Li Q, Chu MJ, Xu J. Tissue- and nuclear receptor-specific function of the C-terminal LXXLL motif of coactivator NCoA6 AIB3 in mice. Mol Cell Biol 2007;27:8073–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Davis RA, Miyake JH, Hui TY, Spann NJ. Regulation of cholesterol-7alpha-hydroxylase: barely missing a SHP. J Lipid Res 2002;43:533–43. [PubMed] [Google Scholar]
- 128.Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR BAR impairs bile acid and lipid homeostasis. Cell 2000;102:731–44. [DOI] [PubMed] [Google Scholar]
- 129.Tanner MM, Tirkkonen M, Kallioniemi A, Isola J, Kuukasjarvi T, Collins C, et al. Independent amplification and frequent co-amplification of three nonsyntenic regions on the long arm of chromosome 20 in human breast cancer. Cancer Res 1996;56:3441–5. [PubMed] [Google Scholar]
- 130.Guan XY, Xu J, Anzick SL, Zhang H, Trent JM, Meltzer PS. Hybrid selection of transcribed sequences from microdissected DNA: isolation of genes within amplified region at 20q11-q13.2 in breast cancer. Cancer Res 1996;56:3446–50. [PubMed] [Google Scholar]
- 131.Lee SK, Anzick SL, Choi JE, Bubendorf L, Guan XY, Jung YK, et al. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J Biol Chem 1999;274:34283–93. [DOI] [PubMed] [Google Scholar]
- 132.Ruault M, Brun ME, Ventura M, Roizès G, De Sario A. MLL3, a new human member of the TRX/MLL gene family, maps to 7q36, a chromosome region frequently deleted in myeloid leukaemia. Gene 2002;284:73–81. [DOI] [PubMed] [Google Scholar]
- 133.Zhang H, Kuang SQ, Liao L, Zhou S, Xu J. Haploid inactivation of the amplified-in-breast cancer 3 coactivator reduces the inhibitory effect of peroxisome proliferator-activated receptor gamma and retinoid X receptor on cell proliferation and accelerates polyoma middle-T antigen-induced mammary tumorigenesis in mice. Cancer Res 2004;64:7169–77. [DOI] [PubMed] [Google Scholar]
- 134.Cho EA, Prindle MJ, Dressler GR. BRCT domain-containing protein PTIP is essential for progression through mitosis. Mol Cell Biol 2003;23:1666–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jowsey PA, Doherty AJ, Rouse J. Human PTIP facilitates ATM-mediated activation of p53 and promotes cellular resistance to ionizing radiation. J Biol Chem 2004;279:55562–9. [DOI] [PubMed] [Google Scholar]
- 136.Munoz M, Jowsey PA, Toth R, Rouse J. Phospho-epitope binding by the BRCT domains of hPTIP controls multiple aspects of the cellular response to DNA damage. Nucleic Acids Res 2007;35:5312–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene 2007;26:7773–9. [DOI] [PubMed] [Google Scholar]
- 138.Riches LC, Lynch AM, Gooderham NJ. Early events in the mammalian response to DNA double-strand breaks. Mutagenesis 2008;23:331–9. [DOI] [PubMed] [Google Scholar]
- 139.Iwabuchi K, Bartel PL, Li B, Marraccino R, Fields S. Two cellular proteins that bind to wild-type but not mutant p53. Proc Natl Acad Sci USA 1994;91:6098–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Iwabuchi K, Li B, Massa HF, Trask BJ, Date T, Fields S. Stimulation of p53-mediated transcriptional activation by the p53-binding proteins, 53BP1 and 53BP2. J Biol Chem 1998;273:26061–8. [DOI] [PubMed] [Google Scholar]
- 141.Nishino T, Morikawa K. Structure and function of nucleases in DNA repair: shape, grip and blade of the DNA scissors. Oncogene 2002;21:9022–32. [DOI] [PubMed] [Google Scholar]
- 142.Ko L, Chin WW. Nuclear receptor coactivator thyroid hormone receptor-binding protein (TRBP) interacts with and stimulates its associated DNA-dependent protein kinase. J Biol Chem 2003;278:11471–9. [DOI] [PubMed] [Google Scholar]
- 143.Zgheib O, Huyen Y, DiTullio RA Jr, Snyder A, Venere M, Stavridi ES, et al. ATM signaling and 53BP1. Radiother Oncol 2005;76:119–22. [DOI] [PubMed] [Google Scholar]
- 144.Ju BG, Rosenfeld MG. A breaking strategy for topoisomerase IIbeta/PARP-1-dependent regulated transcription. Cell Cycle 2006;5:2557–60. [DOI] [PubMed] [Google Scholar]
- 145.Medina PP, Sanchez-Cespedes M. Involvement of the chromatin-remodeling factor BRG1/SMARCA4 in human cancer. Epigenetics 2008;3:64–8. [DOI] [PubMed] [Google Scholar]
- 146.Sansam CG, Roberts CW. Epigenetics and cancer: altered chromatin remodeling via Snf5 loss leads to aberrant cell cycle regulation. Cell Cycle 2006;5:621–4. [DOI] [PubMed] [Google Scholar]
- 147.Roberts CW, Orkin SH. The SWI/SNF complex—chromatin and cancer. Nat Rev Cancer 2004;4:133–42. [DOI] [PubMed] [Google Scholar]