Ribosome profiling combined with biochemical studies demonstrates that chloroplast SRP54 contacts ribosomal uL4 and enhances cotranslational insertion of thylakoid membrane proteins.
Abstract
Key proteins of the photosynthetic complexes are encoded in the chloroplast genome and cotranslationally inserted into the thylakoid membrane. However, the molecular details of this process are largely unknown. Here, we demonstrate by ribosome profiling that the conserved chloroplast signal recognition particle subunit (cpSRP54) is required for efficient cotranslational targeting of several central photosynthetic proteins, such as the PSII PsbA (D1) subunit, in Arabidopsis (Arabidopsis thaliana). High-resolution analysis of membrane-associated and soluble ribosome footprints revealed that the SRP-dependent membrane targeting of PsbA is already initiated at an early translation step before exposure of the nascent chain from the ribosome. In contrast to cytosolic SRP, which contacts the ribosome close to the peptide tunnel exit site, analysis of the cpSRP54/ribosome binding interface revealed a direct interaction of cpSRP54 and the ribosomal subunit uL4, which is not located at the tunnel exit site but forms a part of the internal peptide tunnel wall by a loop domain. The plastid-specific C-terminal tail region of cpSRP54 plays a crucial role in uL4 binding. Our data indicate a novel mechanism of SRP-dependent membrane protein transport with the cpSRP54/uL4 interaction as a central element in early initiation of cotranslational membrane targeting.
INTRODUCTION
The chloroplast genome encodes roughly half of the subunits of PSI, PSII, the cytochrome b6/f complex, the NADH dehydrogenase-like complex (NDH), and the ATP synthase in the thylakoid membrane, which mediate electron transport and ATP synthesis during photosynthesis. Among the subunits are many integral membrane proteins that are synthesized on thylakoid-bound ribosomes and cotranslationally inserted into the membrane (Jagendorf and Michaels, 1990; Kim et al., 1991; Zhang et al., 1999; Zoschke and Barkan, 2015; Króliczewski et al., 2016). In eukaryotic and prokaryotic cells, cotranslational targeting of membrane proteins to the endoplasmic reticulum or plasma membrane, respectively, depends on the signal recognition particle (SRP) system, which is conserved across all kingdoms of life and has been extensively studied (Akopian et al., 2013; Voorhees and Hegde, 2016; Steinberg et al., 2018). Cytosolic SRPs are ribonucleoproteins, and their minimal functional core is formed by two components, an SRP RNA and a conserved protein subunit, which is named SRP54 in eukaryotes and Fifty-four homolog (Ffh) in prokaryotes. Bacterial SRP binds to ribosome nascent chains (RNCs) and the SRP-RNC complex is then guided to the membrane-bound SRP receptor (FtsY) and finally to the SecYEG/YidC translocon machinery. The ribosomal subunit uL23, which surrounds the exit point of the ribosomal peptide tunnel, acts as a docking site for SRP (Pool et al., 2002; Gu et al., 2003; Ullers et al., 2003; Halic et al., 2006; Schaffitzel et al., 2006). SRP recruitment to the ribosome is triggered at an early stage of translation by short nascent chains within the exit tunnel (Houben et al., 2005; Bornemann et al., 2008; Berndt et al., 2009; Mercier et al., 2017). Recently, it has been shown that the early sensing of the nascent chain is mediated by the C terminus of the uL23-bound SRP protein (Ffh), which inserts into the ribosomal tunnel and receives information about the nascent chain (Jomaa et al., 2016; Denks et al., 2017). In the presence of a substrate protein, the C-terminal methionine-rich (M)-domain of Ffh binds the emerging signal sequence and the SRP-RNC complex is delivered to the membrane (Bornemann et al., 2008).
Chloroplasts of seed plants (spermatophytes) contain a unique SRP system that comprises chloroplast homologs of bacterial SecY (cpSecY1), the Ffh subunit of SRP (cpSRP54), the SRP receptor (cpFtsY), and YidC (Alb3) but differs from cytosolic SRP in several striking aspects (Akopian et al., 2013; Celedon and Cline, 2013; Ziehe et al., 2017). (1) During the evolution of higher plant chloroplasts from their cyanobacterial ancestor, the SRP RNA component was lost (Rosenblad and Samuelsson, 2004; Träger et al., 2012). (2) Chloroplasts contain a posttranslationally active SRP system mediating the transport of the nuclear-encoded light-harvesting chlorophyll a/b binding proteins to the thylakoid membrane (Schuenemann et al., 1998; Klimyuk et al., 1999; Groves et al., 2001; Holdermann et al., 2012). In this system, cpSRP54 forms a high-affinity complex with the plant-specific cpSRP43 protein. This interaction is mediated by a highly conserved streptophyte (land plants and charophyte algae)-specific C-terminal extension of cpSRP54 that harbors the positively charged cpSRP43 binding motif ARRKR (Funke et al., 2005; Holdermann et al., 2012; Dünschede et al., 2015). (3) In addition to the cpSRP43-associated pool of cpSRP54, chloroplasts contain a second pool of cpSRP54 that is associated with ribosomes (Franklin and Hoffman, 1993; Schuenemann et al., 1998). The ability of cpSRP54 to interact with nascent chains of PsbA (the reaction center subunit of PSII) and PetB (cytochrome b6; Nilsson et al., 1999; Nilsson and van Wijk, 2002; Piskozub et al., 2015) and the observation that young Arabidopsis (Arabidopsis thaliana) mutants lacking cpSRP54 (ffc) exhibit reduced levels of PsbA (Amin et al., 1999), PsaA, and PsaB (the reaction center proteins of PSI; Amin et al., 1999; Rutschow et al., 2008) suggested that the ribosome-associated pool of cpSRP54 plays a role in the cotranslational transport of at least some plastid-encoded thylakoid membrane proteins.
However, surprisingly little is known about the molecular details of cotranslational protein targeting in chloroplasts, an essential process for the biogenesis and maintenance of the photosynthetically active thylakoid membrane. We aimed to identify substrate proteins for cpSRP54-dependent transport in a chloroplast genome-wide approach and to elucidate the molecular function of ribosome-associated cpSRP54. Ribosome profiling of the Arabidopsis wild type and the cpSRP54-deficient ffc mutant demonstrates that a large set of the subunits of the photosynthetic apparatus require cpSRP54 for efficient transport and that membrane targeting can be triggered by short nascent chains from inside the peptide tunnel. Different to the bacterial SRP system, our analysis of the cpSRP54/ribosome binding interface did not point to a location of cpSRP54 at the exit point of the ribosomal peptide tunnel. However, we demonstrate a direct interaction with the surface-exposed globular domain of the ribosomal protein uL4, which forms part of the internal peptide tunnel by an extended loop. Our data show that chloroplasts contain a unique mechanism of the SRP-dependent formation of thylakoid membrane-associated RNC complexes and suggest that the unexpected contact between cpSRP54 and uL4 might allow the transfer of information about the nascent chain from the peptide tunnel to the surface-bound cpSRP54 to initiate targeting.
RESULTS
The cpSRP54-Deficient Arabidopsis ffc Mutant Exhibits an Altered Distribution of Soluble and Membrane-Associated Ribosome Footprint Abundances
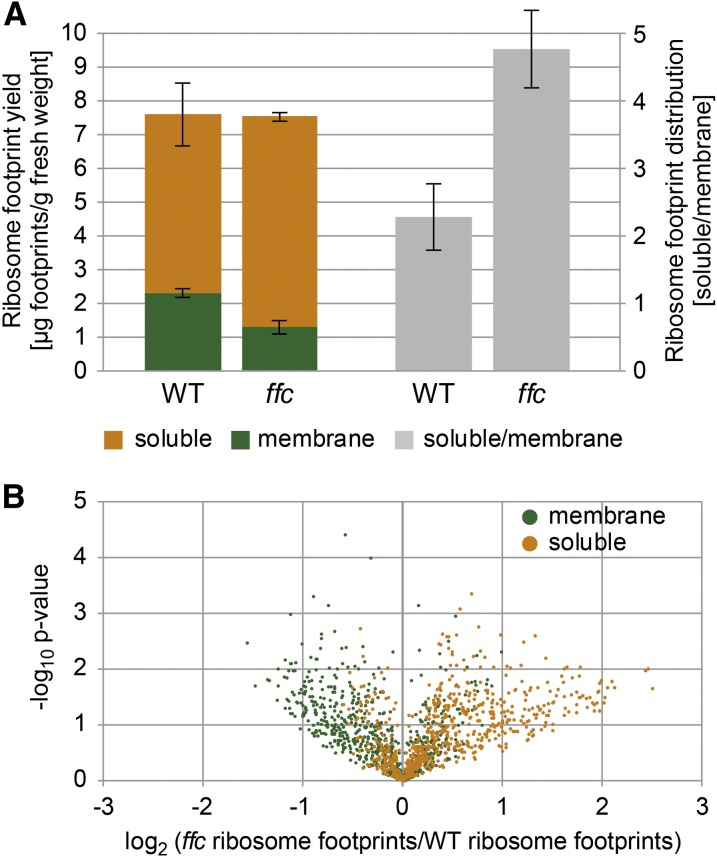
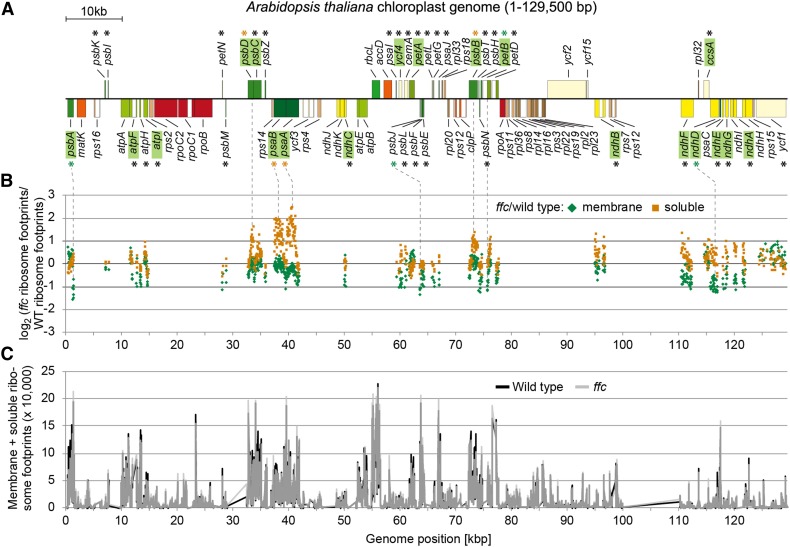
To elucidate the role of ribosome-bound cpSRP54 in translation or cotranslational targeting of plastid-encoded thylakoid membrane proteins, a previously described ribosomal profiling approach was used that distinguishes between membrane-associated and soluble footprints and identifies the regions of cotranslational membrane engagement in nascent peptides (Zoschke and Barkan, 2015; Zoschke et al., 2017). To this end, ribosome footprints from separated (thylakoid) membrane and soluble (stroma) fractions of the wild-type Arabidopsis plants and a mutant lacking cpSRP54 (ffc; Amin et al., 1999) were isolated and analyzed by hybridization to microarrays with highly tiled probes covering all chloroplast reading frames. Our data from the wild-type Arabidopsis showed the same set of proteins to be integrated co- or posttranslationally into the thylakoid membrane that was previously identified in maize (Zea mays; Zoschke and Barkan, 2015). While the general footprint yield and the relative translation output of all plastid open reading frames (ORFs) were unaltered between the ffc mutant and the wild-type plants, the ffc mutant showed a clear decrease in membrane footprint yield, resulting in a more than twofold increased ratio of soluble-to-membrane footprint yield (Figure 1A; Supplemental Figure 1). For each probe in transmembrane segment (TMS)–encoding ORFs, the change in abundances of membrane-associated and soluble footprints in ffc compared with the wild type was visualized against significances of changes in a volcano plot (Figure 1B). This revealed substantial alterations in the spatial distribution of ribosome footprints in the mutant plants with increased soluble and decreased membrane-associated footprints for many probes. These alterations in ffc compared with the wild type were plotted against the genome position of the probes located in TMS-encoding ORFs, most of which represent integral thylakoid membrane proteins (Figure 2B). Major changes in membrane–stroma distributions of ribosome footprints in the ffc mutant (more than twofold change in membrane or soluble abundance of at least two consecutive probes) were observed for several cotranslationally targeted proteins, including the PSI subunits PsaA and PsaB, the PSII subunits PsbA, PsbB, and PsbD, the cytochrome b6f complex subunit PetB, and the NADH dehydrogenase-like complex subunit NdhD (Figures 2A and 2B). By contrast, the overall translation behavior (i.e., footprint abundance and spreading) was not substantially altered for any chloroplast gene (Figure 2C; Supplemental Figure 1).
Figure 1.
Spatial Redistribution of Translation in the ffc Mutant.
(A) Left, ribosome footprint yield was determined for membrane (green) and soluble (orange) fractions in the wild-type (WT) and ffc mutant plants and normalized to the amount of fresh weight used as starting material. Vertical lines illustrate sds that are derived from three independent biological replicates. Right, ratios were calculated between soluble and membrane footprint yields shown on the left to illustrate the redistribution of ribosome footprints in the ffc mutant (gray).
(B) Volcano plot illustrating the relocation of ribosome footprints in the ffc mutant. For all probes located in reading frames with TMS(s), P-values were calculated to demonstrate the significance of the changes between ribosome footprint abundances in the wild-type (WT) and ffc mutant plants at the membrane and in the soluble fraction (green and orange dots, respectively). These P-values were plotted in log10 scale against log2 scale ratios of membrane-associated and soluble ribosome footprint abundances in the WT and ffc mutant plants, respectively.
Figure 2.
Genome-Wide Overview of the Spatial Ribosome Footprint Distribution and General Translation Output in the ffc Mutant.
(A) Map of the Arabidopsis chloroplast genome created with OGDraw (Lohse et al., 2013) and displaying protein-coding regions. For better visualization, only one of the two large inverted repeats is shown. Genes encoding proteins that contain TMS(s) are labeled with asterisks. Genes whose names are highlighted in green encode cotranslationally thylakoid-targeted proteins. Green and orange asterisks mark genes that display substantially altered membrane-associated and soluble ribosome footprint abundances in the ffc mutant compared with the wild type, respectively.
(B) Ribosome footprints were isolated from membrane and soluble fractions and analyzed by microarray hybridization (for details, see “Methods”). For both the membrane and soluble fraction, the log2-transformed ratio of ribosome footprint abundances in the ffc mutant relative to the wild type was calculated for all probes located in TMS-containing reading frames and plotted as green diamonds and orange squares, respectively, against the genome position according to the map shown in (A). Dotted lines connect genes in the map with substantially altered spatial ribosome footprint distribution in the ffc mutant compared with the wild type (>2-fold or <0.5-fold change for at least two consecutive probes). Data represent the average values of three independent biological replicates (see the Supplemental Data Set).
(C) Based on the ribosome footprint distributions shown in Figure 1A, weighted sums of membrane and soluble ribosome footprint abundances were calculated for each individual probe located in protein-coding regions (for details, see “Methods”) and plotted for wild type (black line) and ffc mutant (transparent gray line) against the genome position according to the map shown in (A). Data are based on average values from three independent biological replicates (see the Supplemental Data Set).
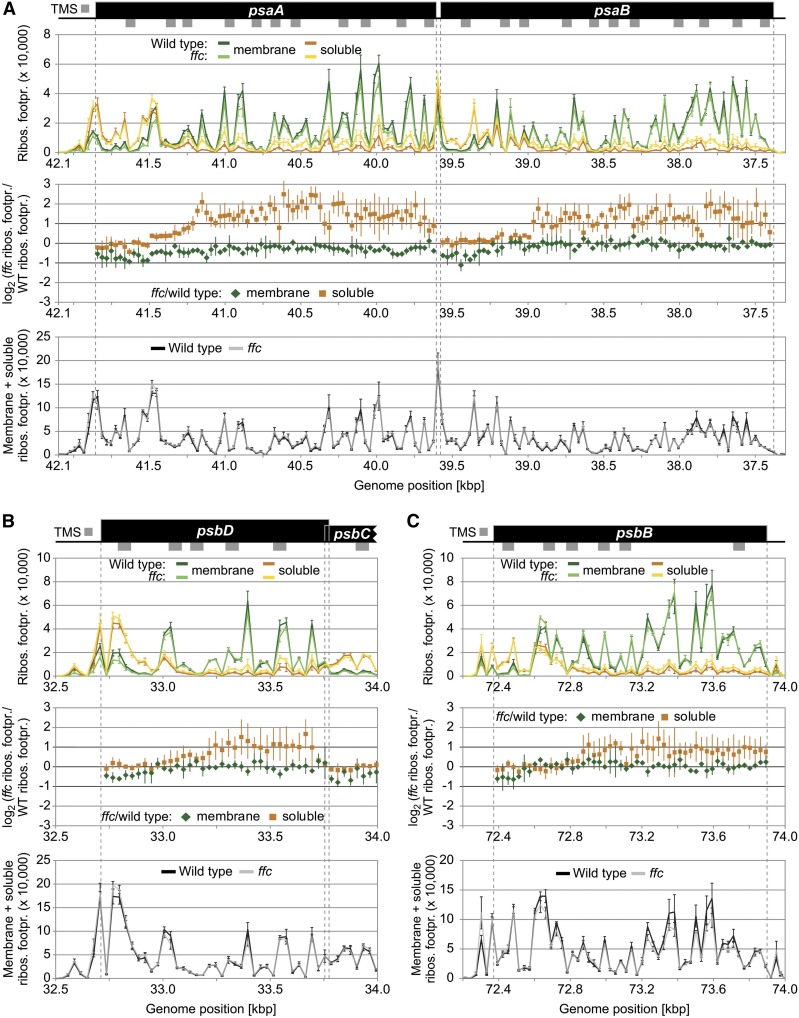
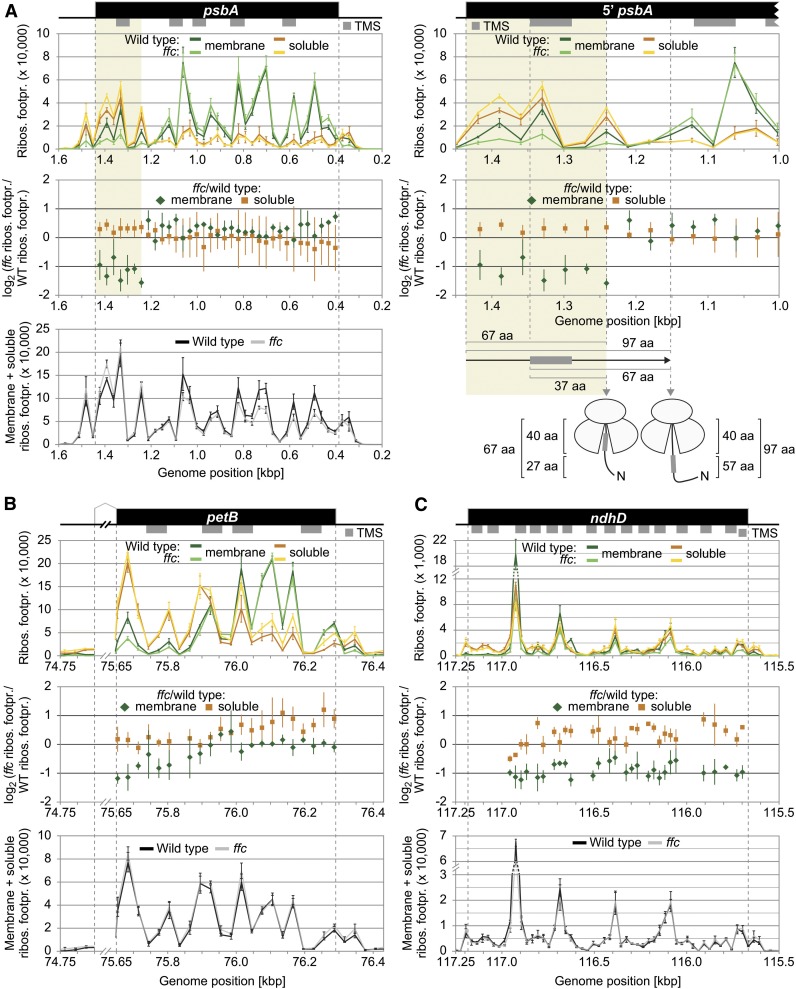
Other cotranslationally targeted proteins such as AtpI, AtpF, Ycf4, and PsbC or the NDH subunits A, B, C, E, F, and G are less affected in membrane–stroma distribution of their ribosome footprints (Figures 2A and 2B; Supplemental Figures 2A and 2B and 3). Clearly, no difference in spatial footprint distribution was observed for PetA, which supports previous results describing a cotranslational cpSecA-dependent transport of PetA (Voelker and Barkan, 1995; Röhl and van Wijk, 2001; Zoschke and Barkan, 2015) that can be considered to be independent of cpSRP54 (Supplemental Figure 2C). High-resolution views of the affected ORFs together with the corresponding footprint distributions in the wild type and ffc are shown in Figures 3 and 4 Consistent with previous data from maize (Zoschke and Barkan, 2015), the wild-type Arabidopsis displayed the typical footprint pattern for cotranslational transport with a clear transition from soluble to stable membrane-associated translation after emergence of the first TMS outside the ribosome exit tunnel. The ffc plants showed overall similar footprint patterns but with clear increases (more than twofold) in soluble footprints for two or more consecutive probes in the psaA, psaB, psbD, and psbB reading frames (Figure 3) or with clear decreases (more than twofold) in membrane-associated footprints for two or more consecutive probes in the psbA, petB, and ndhD reading frames (Figure 4). The increase of soluble footprints in psaA, psaB, psbD, and psbB in ffc occurs after exposure of the first TMS and therefore indicates a role of cpSRP54 in facilitating the formation or maintenance of stably thylakoid membrane-associated RNC complexes (Figure 3).
Figure 3.
Zoom-In Images for Reading Frames That Display Substantially Altered Soluble Ribosome Footprint Abundances in the ffc Mutant.
(A) to (C) Zoom-in images of ribosome footprint distributions and translation output in the psaA/B (A), psbD (B), and psbB (C) coding regions. From top to bottom: Gene maps with TMSs represented by gray rectangles below the coding regions (black boxes). TMS positions are based on the Aramemnon plant membrane protein database (Schwacke et al., 2003). (Top) Ribosome footprint abundances from membrane and soluble fractions (green and orange lines, respectively) from the wild-type and ffc mutant plants (dark and light colors, respectively) plotted against genome position according to the gene map. (Middle) Log2-transformed ratio of ribosome footprint abundances in the ffc mutant compared with the wild type for membrane-associated and soluble footprints (green diamonds and orange squares, respectively) as shown in Figure 2B. Note that these ratios were only calculated for probes that are located in protein-coding regions and that gave a ribosome footprint signal that passed the threshold of 200 above background. (Bottom) Weighted sums of membrane and soluble ribosome footprint abundances for the wild type (black line) and ffc mutant (transparent gray line) as shown in Figure 2C. In all diagrams, vertical lines represent sds that are based on three independent biological replicates. Dashed gray vertical lines mark the boundaries of protein-coding regions. The psbD and psbC reading frames are overlapping. Note that several regions in the shown reading frames display substantially increased levels of soluble ribosome footprints in the ffc mutant (i.e., more than twofold change for at least two consecutive probes), while the translation output and ribosome distribution within the reading frames are unaltered. Footpr., footprint; Ribos., ribosome; WT, wild type.
Figure 4.
Zoom-In images for Reading Frames That Display Substantially Altered Membrane-Associated Ribosome Footprint Abundances in the ffc Mutant.
(A) to (C) Zoom-in images of ribosome footprint distributions and translation output in the psbA (A), petB (B), and ndhD (C) coding regions. Data representation and annotations are as in Figure 3. The petB reading frame is interrupted by an intron whose size was reduced for better illustration by breaking the x axes. Note that several regions in the shown reading frames display substantially decreased levels of membrane-associated ribosome footprints in the ffc mutant (i.e., <0.5-fold change for at least two consecutive probes). The region that shows substantially decreased levels of membrane-associated ribosome footprints in psbA is shaded in ocher, and a zoom in of this region is shown on the right for the map and the two diagrams on top. Vertical gray dashed lines in this zoom in illustrate from left to right the start codon of psbA, the start of the first TMS, the end of the region that shows decreased membrane-associated ribosome footprints, and the point at which psbA translation shows stable engagement with the thylakoid membrane. Distances from the start codon or the start of the first TMS to the end of the region whose translation is spatially altered and the point of stable engagement are given in amino acids (aa) below the diagrams. The model illustrates that the region that shows decreased membrane-associated ribosome footprints is translated before exposure of the first TMS outside of the ribosome exit tunnel, which accommodates ∼40 amino acids. Footpr., footprint; Ribos., ribosome; WT, wild type; N, N-terminus.
Notably, the footprints of petB and especially psbA in ffc show a pattern with a prominent decrease of membrane-associated footprints at early stages of translation (Figures 4A and 4B). A similar tendency, but less pronounced, is also observed for psaA, psaB, psbD, and psbB (Figure 3). A close-up view of the footprint distribution in psbA shows that in the region between genome position 1444 (start codon) and 1242 (first 67 residues of the PsbA chain) membrane-associated footprints is substantially reduced (Figure 4A, right). At this translational stage, the first TMS of PsbA (amino acids 31 to 51) is fully buried inside the ribosomal exit tunnel as the tunnel accommodates ∼40 residues (Figure 4A, right). These data indicate that cpSRP54 facilitates at least for the central photosynthetic subunit of PSII an initial step of membrane association of ribosomes before the first TMS is exposed to the ribosomal surface. Similarly, our data showed substantially reduced membrane-associated footprints in psbJ, a reading frame whose single TMS is located too close to the stop codon to confer stable engagement of the translating ribosome with the thylakoid membrane (Supplemental Figure 4). This finding suggests that cpSRP54 might enhance the probably posttranslational transfer of PsbJ to the membrane insertase by anchoring the ribosome at the membrane close to the insertion site.
Altogether, our ribosome profiling data revealed substantially changed spatial ribosome distributions during translation elongation of chloroplast reading frames encoding central photosynthetic proteins, indicating that cpSRP54 enhances the efficiency of cotranslational thylakoid targeting for these substrate proteins.
The Level of Many Cotranslationally Targeted Thylakoid Membrane Proteins Is Largely Reduced and the Transport Machinery Is Upregulated in the ffc Mutant
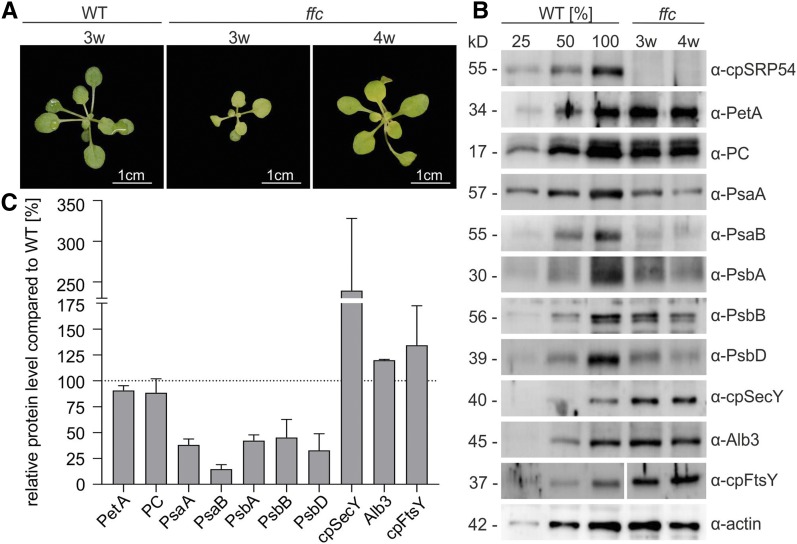
Previously, it was shown that the reaction center proteins of PSI and PSII (PsaA, PsaB, PsbA, and PsbD) were reduced in young ffc seedlings (10 d old) but reached normal levels in older plants (24 d old; Amin et al., 1999). Another study observed an ∼50% reduction in PSI core subunits but only a slight reduction (∼10%) of the PSII core components in the ffc plants (Rutschow et al., 2008). As our ribosome profiling data demonstrated that the spatial distribution of the translation of a large set of chloroplast-encoded thylakoid membrane proteins, including the central PSI and PSII subunits, is affected in 3-week-old ffc, we reexamined previous data. To this end, the levels of PSI and PSII subunits in leaf material of 3-week-old ffc and the wild-type plants were analyzed immunologically (Figures 5A to 5C). The actin level served as loading control. Equal loading was also evident by nearly the same level (90%) of PetA and plastocyanin (PC) in ffc compared with the wild type. Both proteins were targeted to thylakoids independently of cpSRP54, as shown previously by Knott and Robinson, (1994), Voelker and Barkan, (1995), Röhl and van Wijk, (2001), and in this work. The ffc plants exhibited a reduction of the PSI subunits PsaA and PsaB by >60%. The PSII subunits PsbA, PsbB, and PsbD were also substantially reduced (>50%). As ffc plants are developmentally delayed (Rutschow et al., 2008), we also analyzed 4-week-old ffc seedlings that show a similar developmental stage as the 3-week-old wild-type plants (Figure 5A). However, no difference in protein levels was observed between 3- and 4-week-old plants. The level of cpSecY is elevated in ffc, suggesting that an increased level of the cpSec translocase might compensate the loss of cpSRP54 , previously described by Amin et al., (1999). In line with these data, we observed a more than twofold upregulation of cpSecY in the ffc plants. Furthermore, the ffc plants showed elevated levels of the SRP receptor cpFtsY (134%) and Alb3 (120%). Immunoblot analysis of thylakoid membranes and stroma prepared from ffc chloroplasts verified that the affected proteins are integrated in the thylakoid membrane and do not accumulate in the stroma (Supplemental Figure 5).
Figure 5.
Levels of PSI and PSII Subunits and of Components of the Transport Machinery in the Thylakoid Membrane of the Wild Type and the ffc Mutant.
(A) Phenotypes of 3- and 4-week-old (3w, 4w) ffc mutants compared with the 3-week-old wild-type (WT) Arabidopsis plants grown on soil.
(B) Total protein extracts from ffc (3w and 4w) and the wild-type (WT, 3w) fresh leaf material were separated by SDS-PAGE and blotted for immunodetection with the indicated antibodies (PC). The WT samples corresponded to 25, 50, and 100% of total protein.
(C) Quantification of protein levels of 3w ffc mutants based on immunoblot band intensity (ImageJ) in relation to the 100% wild-type (WT) total protein extract (dotted line). The average intensities and corresponding sds were calculated from three to seven independent replicates.
Taken together, the clearly reduced level of core components of PSI and PSII in ffc confirms that cpSRP54 is required for an efficient targeting mechanism to the thylakoid membrane. However, cpSRP54 is not essential for this process in vivo, and its absence can probably be at least partially compensated by an upregulation of other components of the transport machinery.
Mapping the cpSRP54/Ribosome Binding Interface by Isotope-Coded Crosslinking Combined with Mass Spectrometry
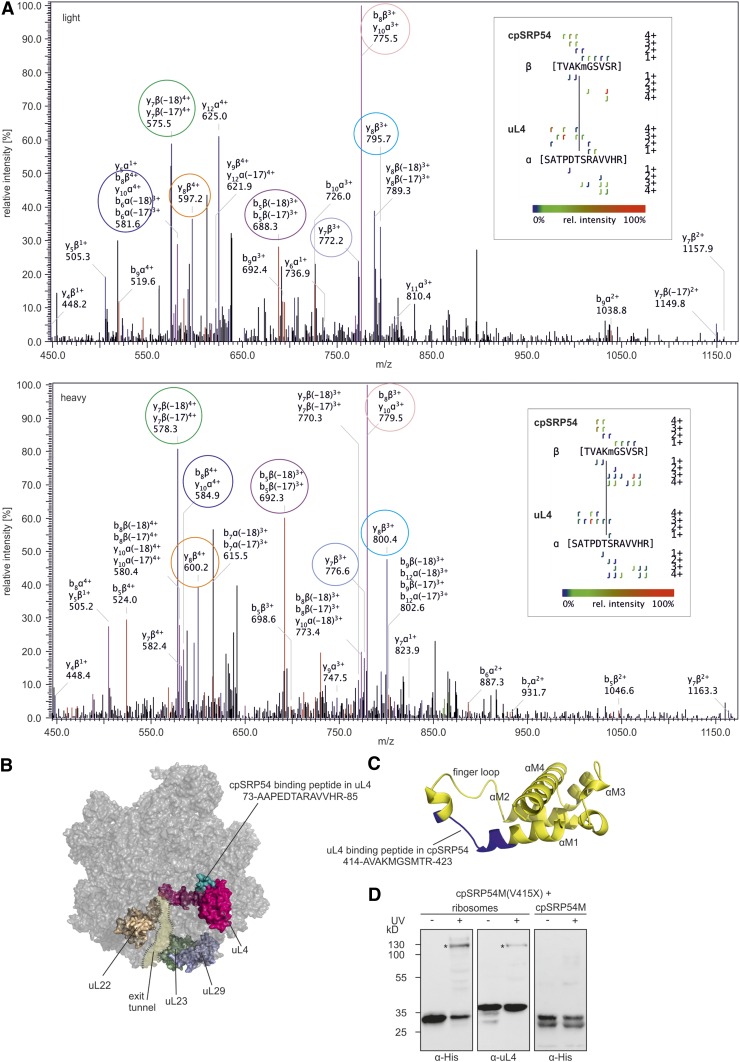
To investigate the cpSRP54-dependent initiation of cotranslational membrane targeting, we examined the cpSRP54/ribosome binding interface. To directly identify the interacting peptides, enriched ribosomes from pea (Pisum sativum), containing endogenous cpSRP54, were crosslinked by incubation with the isotope-coded crosslinker BS3-H12/D12, and tryptic peptides were analyzed by mass spectrometry. Crosslinks were identified based on the spectra of the light (H12) and heavy (D12) forms of BS3 and only considered as positive identification if a BS3-H12 crosslink corresponded to a BS3-D12 crosslink of identical retention time with similar fragment ion pattern. Furthermore, the mass difference of 12 D between the light and heavy form of BS3 was crucial for the validation of potential crosslinks (Cormann et al., 2016). This analysis led to the identification of a crosslink between cpSRP54 and the ribosomal protein uL4 (Figure 6A). The crosslinked peptides SATPDTSRAVVHR of uL4 and TVAKMGSVSR (crosslinked residues are indicated in bold) of cpSRP54 of P. sativum (Figure 6A) correspond to residues 73 to 85 (AAPEDTARAVVHR) and 414 to 423 (AVAKMGSMTR) of Arabidopsis uL4 and cpSRP54, respectively. According to the cryoelectron microscopy structure of spinach (Spinacia oleracea) chloroplast ribosomes (Ahmed et al., 2016; Bieri et al., 2017; Perez Boerema et al., 2018), uL4 is a globular protein with an extended loop forming part of the peptide tunnel. The cpSRP54 binding peptide in uL4 is located at the ribosomal surface (Figure 6B). The corresponding uL4 binding peptide in cpSRP54 is located in the M-domain (Figure 6C). To visualize this region in cpSRP54, a homology model of the M-domain of cpSRP54 was generated using the Phyre2 web portal based on the structure of the Thermus aquaticus Ffh (protein data bank [PDB]: 2FFH), which is predicted to most closely represent the structure of cpSRP54 (Henderson et al., 2016), as template. Similar to the M-domain of T. aquaticus Ffh (Keenan et al., 1998), the cpSRP54M-domain is predicted to consist of four amphipathic helices and a flexible finger loop that connects helix 1 to helix 2, forming a hydrophobic core. The uL4 binding peptide is located in the region connecting helix 1 and the finger loop (Figure 6C).
Figure 6.
Identification of Crosslinks between cpSRP54 and uL4.
(A) Fragmentation pattern of the crosslink. Chloroplast ribosomes of P. sativum were incubated with isotope-coded BS3 (1:1 mixture of H12-BS3 and D12-BS3). The sample was analyzed via liquid chromatography-electrospray ionization mass spectrometry. Assigned b-ions are depicted in red, y-ions in blue, and nonidentified ions in black. Ions that can be assigned to both b- and y-ions are depicted in pink. Fragment ion pairs of the light and heavy form that show the characteristic mass shift of 12 D are encircled in the same color. The crosslinked peptide-fragment is shown on the right side of each spectrum.
(B) Ribosomal location of uL4 (pink) and the cpSRP54 binding peptide in uL4 (green) is visualized using the cryoelectron microscopy structure of the 50S large subunit of the spinach chloroplast ribosome (PDB: 5X8T). Ribosomal proteins that form the exit tunnel or the tunnel exit point are depicted in brown (uL22), light green (uL23), and light blue (uL29).
(C) Location of the uL4 binding peptide (blue) in a homology model of cpSRP54M (yellow). The homology model was generated using the Phyre2 web portal (Kelley et al. 2015) and the signal sequence binding subunit of Ffh from T. aquaticus (PDB: 2FFH) as template.
(D) Recombinant cpSRP54M-His(V415X) containing the UV light-inducible crosslinker pBpa (X) was incubated with chloroplast ribosomes or His-cpSRP54M in the absence (−) or presence (+) of UV light. The assay was analyzed immunologically using antibodies against the His-Tag (α-His) and the ribosomal protein uL4 (α-uL4). Specific crosslinking products are indicated with an asterisk (*).
The Direct Contact between the Ribosomal Binding Region in cpSRP54 and uL4 Is Confirmed by Site-Specific Photo-Crosslinking
To independently test for a direct interaction between the uL4 binding region 414-AVAKMGSMTR-423 in cpSRP54 and uL4, the site-specific photoinducible crosslinker p-benzoyl-l-phenylalanine (pBpa) was incorporated into cpSRP54M-His at positions V415, M418, and M421. The recombinantly expressed labeled constructs were used for crosslinking studies with chloroplast ribosomes from P. sativum. To control for the formation of crosslinked dimers or oligomers of cpSRP54M, the labeled constructs were incubated with unlabeled His-cpSRP54M instead of ribosomes. A strong UV light-induced and ribosome-dependent specific crosslinking product with an apparent molecular weight of ∼120 kD was detected in immunoblots using α-His and α-uL4 antibodies for the cpSRP54M construct containing pBpa at position 415 [cpSRP54M(V415X); Figure 6D].
The C-Terminal Tail Region of cpSRP54 Contains a Second Ribosomal Binding Motif
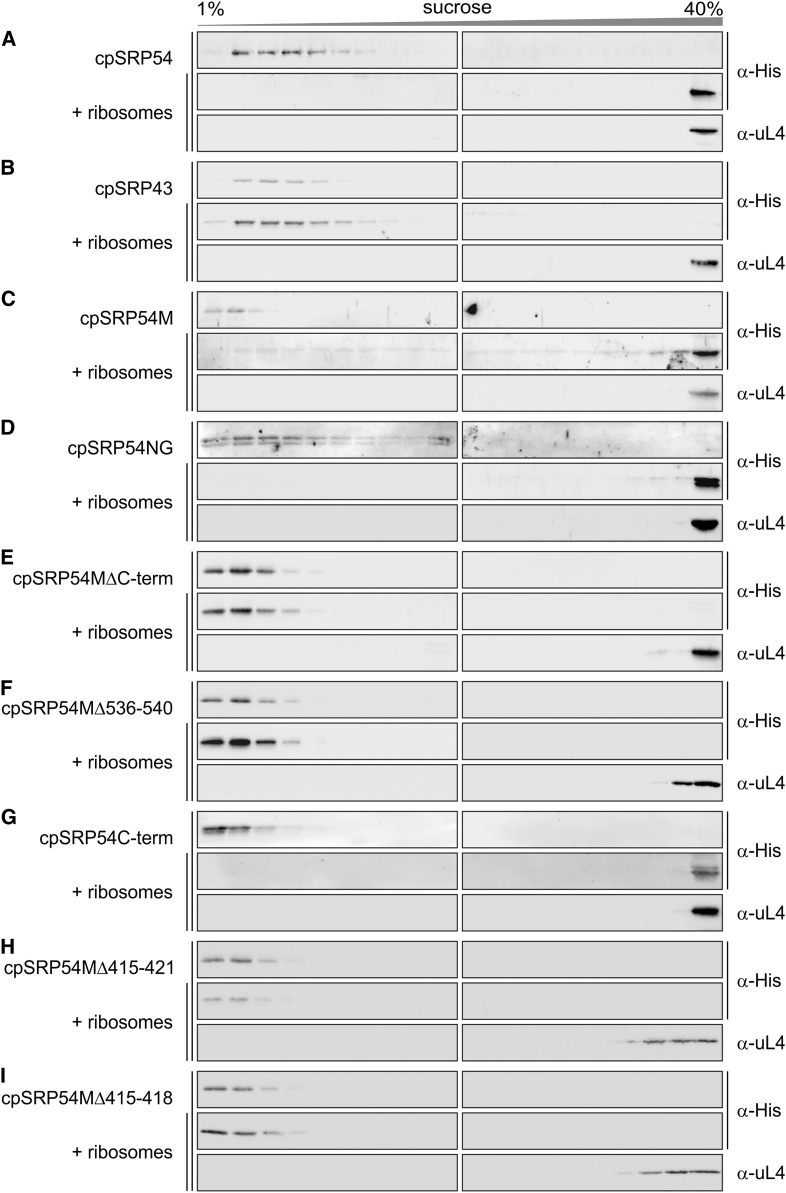
To further analyze the cpSRP54/ribosome interaction, the cofractionation of His-cpSRP54 and various truncation constructs with P. sativum chloroplast ribosomes on Suc gradients was determined. The sedimentation pattern of the His-cpSRP54 constructs and the ribosomes was analyzed by immunoblot analysis of the Suc gradient fractions using antibodies directed against the His-tag and uL4, respectively (Figure 7). As shown in Figure 7A, full-length cpSRP54 cosedimented with the ribosomes to the bottom of the gradient, while it remained in the top fractions of a control gradient performed in the absence of ribosomes. In agreement with previous results showing that the ribosome association is specific for the 54-kD subunit of the heterodimeric chloroplast SRP complex (Schuenemann et al., 1998; Nilsson et al., 1999), no cosedimentation of His-cpSRP43 with ribosomes was detected (Figure 7B). As expected, a clear cofractionation was observed for the M-domain of cpSRP54 including the C-terminal tail region (cpSRP54M; Figure 7C). However, the cpSRP54/ribosome interaction is not exclusively mediated by the M-domain as we also observed a cofractionation of a construct comprising the N-terminal NG-domain (cpSRP54NG) with ribosomes (Figure 7D). The presence of stromal cpSRP54 in two pools, one bound to cpSRP43 and the other bound to ribosomes, and the observation that recombinant cpSRP43 can displace stromal cpSRP54 from the ribosomes (Supplemental Figure 6) prompted us to analyze whether the C-terminal tail region of cpSRP54 (amino acids 530 to 564), which contains the cpSRP43 binding motif, plays a role in ribosome binding. Indeed, cpSRP54M lacking the C-terminal tail (cpSRP54MΔC-term) or the positively charged cpSRP43 binding motif 536-RRKRK-540 (cpSRP54MΔ536-540) did not cofractionate with ribosomes (Figures 7E and 7F). Furthermore, a cosedimentation of the recombinant C-terminal tail region (cpSRP54C-term) with ribosomes was observed (Figure 7G), indicating a direct contact between these components.
Figure 7.
Interaction of Recombinant cpSRP54 Constructs and Chloroplast Ribosomes.
(A) to (I) Recombinant His-tagged mature cpSRP54 (A), cpSRP43 (B), and the indicated cpSRP54 constructs (see [C] to [I]) were incubated with chloroplast ribosomes and loaded onto a Suc gradient. After ultracentrifugation, the gradient fractions were analyzed immunologically using antibodies against the His-tag (α-His) and the ribosomal protein uL4 (α-uL4). Sucrose density gradient centrifugation of the recombinant proteins in the absence of ribosomes was used as negative controls. cpSRP54M, M-domain (amino acids 371 to 564); cpSRP54NG, NG-domain (amino acids 77 to 370); Cterm, C-terminal tail region (amino acids 530 to 564).
To confirm the role of the uL4 binding region 414-AVAKMGSMTR-423 in ribosome binding, His-cpSRP54M constructs exhibiting deletions in this region were generated (cpSRP54Δ415-VAKMGSM-421 and cpSRP54Δ415-VAKM-418) and binding to ribosomes was analyzed using sucrose density gradient centrifugation. Neither of the deletion constructs cosedimented with ribosomes, remaining in the top fractions of the gradient (Figures 7H and 7I).
Therefore, our data indicate that cpSRP54M contains two ribosome binding sites: 415-VAKM-418 in the hydrophobic groove and 536-RRKRK-540 in the C-terminal tail region. The location of these binding sites in cpSRP54 is schematically presented in Figure 9A.
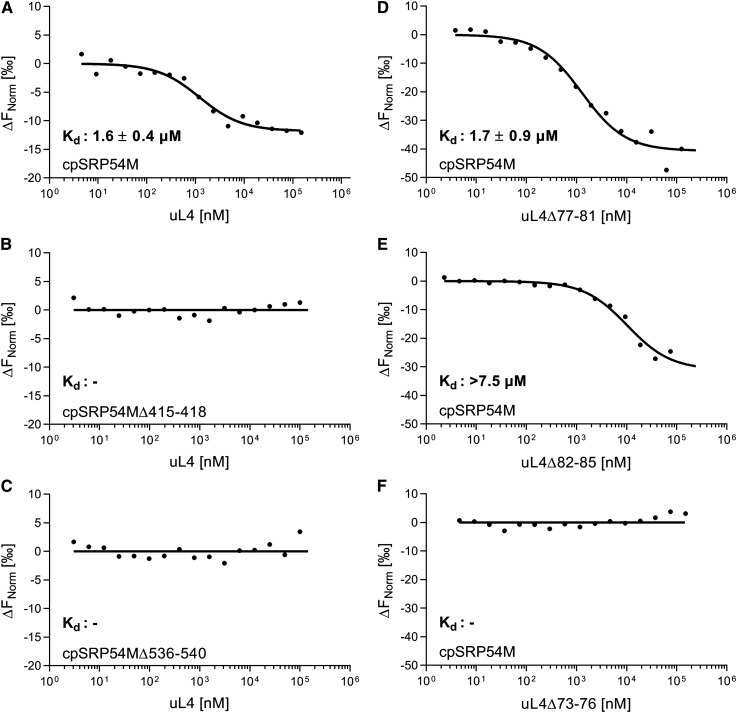
The Purified Recombinant Proteins cpSRP54M and uL4 Interact as Shown by Microscale Thermophoresis
To validate the direct interaction between cpSRP54 and uL4 and to quantitatively characterize this interaction the binding affinity of the recombinant proteins cpSRP54M and uL4 was assessed using microscale thermophoresis. To this end, fluorescently labeled His-cpSRP54M was mixed with increasing amounts of His-tagged uL4 and the thermophoretic movement of the fluorescent cpSRP54M was monitored. The results demonstrated that cpSRP54M binds uL4 directly and a Kd of 1.6 ± 0.4 µM was determined for the interaction (Figure 8A). No interaction was detected when cpSRP54M constructs with deletions of the uL4 binding motifs VAKM (amino acids 415 to 418) and RRKRK (amino acids 536 to 540) were used (Figures 8B and 8C). To confirm the cpSRP54 binding motif AAPEDTARAVVHR (amino acids 73 to 85) in uL4, the uL4 deletion constructs uL4Δ73-76, uL4Δ77-81, and uL4Δ82-85 were generated. As shown in Figure 8D, removal of residues 77 to 81 did not affect cpSRP54M binding (Kd of 1.7 ± 0.9 µM). However, deletion of residues 82 to 85 in uL4 led to an at least fivefold reduced affinity to cpSRP54M (Kd > 7.5 µM; Figure 8E) and the deletion of uL4 residues 73 to 76 resulted in a complete loss of cpSRP54M binding (Figure 8F).
Figure 8.
Determination of the Binding Affinity of cpSRP54/uL4 Complex Formation Using Microscale Thermophoresis.
(A) to (F) Fluorescently labeled His-cpSRP54M (see [A], and [D] to [F]), His-cpSRP54MΔ415-418 (B), and His-cpSRP54MΔ536-540 (C) (20 nM) were titrated with His-uL4 (see [A] to [C]) or the indicated His-uL4 constructs (see [D] to [F]). The difference in normalized fluorescence was plotted against the concentration of the indicated uL4 constructs. Raw microscale thermophoresis traces are shown in Supplemental Figure 7. The Kd values and corresponding sds were calculated from three independent replicates. The recombinant proteins used for microscale thermophoresis are shown in Supplemental Figure 8.
Taken together, these data clearly demonstrate a direct interaction of cpSRP54 and uL4. Furthermore, the data provide additional evidence for an important role of the uL4 binding motifs 415-VAKM-418 and 536-RRKRK-540 in cpSRP54 and the cpSRP54 binding motif 73-AAPE-76 in uL4 for the formation of the binding interface.
DISCUSSION
The interaction between cpSRP54 and chloroplast ribosomes was discovered approximately two decades ago; yet, the nature of this interaction and the functional role of ribosome-associated cpSRP54 remained largely unknown. Our study revealed that cpSRP54 binds to the surface-exposed globular domain of the ribosomal protein uL4 and is required for the efficient cotranslational insertion of a large subset of chloroplast-encoded thylakoid membrane proteins. The cpSRP54/uL4 interaction is an unexpected result, as cytosolic bacterial and eukaryotic SRP binds to ribosomal components surrounding the exit point of the polypeptide tunnel, where the M-domain of SRP interacts with the signal sequence of SRP-dependent substrate proteins (Pool et al., 2002; Gu et al., 2003; Ullers et al., 2003; Halic et al., 2006; Schaffitzel et al., 2006; Akopian et al., 2013; Steinberg et al., 2018). Although cpSRP54 is highly conserved, it differs from SRP particles of all three domains of life as it is not associated with an SRP RNA in higher plants. In cytosolic SRP systems, the SRP RNA plays a crucial role in the transfer of nascent chains to the translocon. In bacteria, the conserved apical tetraloop of the RNA is important for the initial recruitment of the SRP-receptor FtsY to the SRP-RNC complex (Jagath et al., 2001; Shen and Shan, 2010). Furthermore, the SRP RNA activates the GTP hydrolysis of the SRP/FtsY complex after the dimerized NG-domains of SRP and FtsY moved ∼100 Å to the distal end of the SRP RNA, which finally leads to disassembly of the SRP/FtsY complex (Ataide et al., 2011; Shen et al., 2012). Additionally, the repositioning of the NG heterodimer on the SRP RNA results in the exposure of the contact sites between the ribosome and the Sec translocase (Kuhn et al., 2015; Jomaa et al., 2016). In view of the critical role of the SRP RNA in cytosolic SRP systems, it is an intriguing question how cotranslational transport functions in chloroplasts in the absence of an SRP RNA component. The cpSRP54/uL4 interaction may contribute to the compensation of the evolutionary loss of the RNA by providing a different platform for cpFtsY interaction. As uL4 is not located at the tunnel exit point, the cpSRP54/cpFtsY interaction may occur without the steric interference of the ribosome/translocon binding interface that necessitates the large repositioning of the dimerized NG-domains along an RNA. The cpSRP54/uL4 interaction may also enable the early initiation of membrane targeting of ribosomes with short nascent chains inside the exit tunnel that we clearly detected for PsbA nascent chains. In cytosolic SRP systems, several reports show that SRP-dependent protein transport is initiated from nascent chains inside the ribosomal tunnel (Houben et al., 2005; Bornemann et al., 2008; Berndt et al., 2009; Mercier et al., 2017). In bacteria, the early sensing of the nascent chain is mediated by the C terminus of the bacterial SRP protein (Ffh), which contacts the intra-tunnel loop of ribosomal protein uL23 inside the ribosomal tunnel and receives information about the nascent peptide (Jomaa et al., 2016; Denks et al., 2017). We propose that in chloroplasts uL4 senses information about the nascent chain inside the tunnel and transduces it to the ribosomal surface to the uL4-bound cpSRP54 to initiate cpFtsY recruitment at the membrane. In this model the sensing is mediated by the tunnel-exposed loop of uL4 that has been shown to contact nascent chains in the most constricted part of the tunnel in bacteria and yeast (Saccharomyces cerevisiae; Houben et al., 2005; Zhang et al., 2013). The described transient interaction between cpSRP54 and the first transmembrane domain of the D1 nascent chain after emergence from the polypeptide tunnel (Nilsson et al., 1999; Nilsson and van Wijk, 2002) might occur after the initial establishment of an early membrane-associated targeting complex by repositioning of the M-domain of cpSRP54 to the exit point of the tunnel to support transfer of the nascent chain to the translocon. A stable membrane association of the translating ribosome occurs at a later step during insertion after further elongation of the nascent chain into the translocon (Figure 9B).
Figure 9.
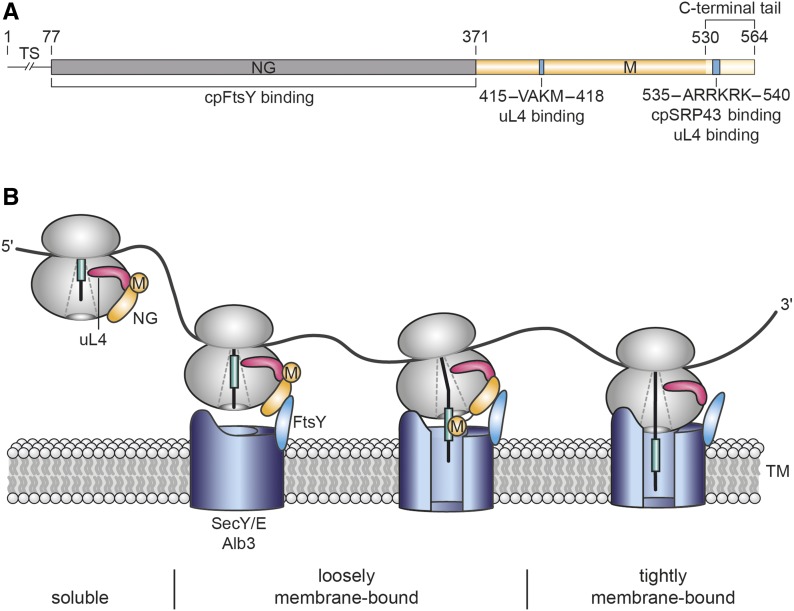
Chloroplast SRP54 and Its Role in Cotranslational Insertion of Thylakoid Membrane Proteins.
(A) Mature cpSRP54 is composed of an N-terminal N-domain (N, gray), a central G-domain with GTPase activity (G, gray), and a C-terminal Met-rich M-domain (M, yellow). CpSRP54 proteins differ from the bacterial homologs by a C-terminal tail region (light yellow) containing a positively charged cluster (blue) that mediates cpSRP43 binding (Funke et al., 2005; Holdermann et al., 2012) and binding to the chloroplast ribosomal protein uL4 (this work). Binding to uL4 is also mediated by a second binding motif within the M-domain (blue; this work). Amino acid positions (numbers above the diagram) refer to cpSRP54 from Arabidopsis. Residues 1 to 76 comprise the transit sequence (TS), which is cleaved off after chloroplast import.
(B) Model of cotranslational protein insertion combining previously published data and results of this work. CpSRP54 binds to ribosomes by a direct contact between its C-terminal M-domain and the ribosomal subunit uL4. uL4 is a globular protein with an extended loop forming part of the internal peptide tunnel at a narrow constriction site of the tunnel. The tip of the loop might sense information about the translational state of the ribosome and transduce it to the surface-bound cpSRP54, initiating early transfer of the ribosome-nascent chain to the thylakoid membrane prior to the emergence of the nascent chain from the exit tunnel. Initial docking to the membrane is mediated by heterodimer formation between cpSRP54NG and the homologous NG-domain of the SRP receptor, cpFtsY, and additional contacts between the ribosome and components of the cpSecY/Alb3 translocon that was shown to be involved in PsbA insertion previously (Walter et al., 2015a). Upon emergence of the hydrophobic targeting sequence, cpSRP54M repositions to the tunnel exit and mediates the transfer of the nascent chain into the translocase. GTP hydrolysis leads to disassembly of the cpSRP54/cpFtsY GTPase complex (Akopian et al., 2013). Ribosomes become tightly associated with membranes by insertion of the nascent chain into the translocon and, depending on the length of the nascent chain, by an additional direct contact with the lipid bilayer.
The clear, but not drastic visible and molecular phenotype of the ffc plants indicates that cpSRP54 mediates the efficient cotranslational protein transport but is not essential for this process. This finding is in line with results from Escherichia coli SRP, which indicate that SRP enhances the kinetics of cytoplasmic membrane proteome biogenesis (Wickström et al., 2011). As shown previously, ffc plants are only slightly affected in the PSII repair cycle (Walter et al., 2015b); thus, it is likely that cpSRP54 functions primarily in the de novo biogenesis of PSII during plant development. Notably, plant mutants lacking cpFtsY or the translocon components cpSecY or Alb3 exhibit strong chlorotic or even seedling-lethal phenotypes (Sundberg et al., 1997; Roy and Barkan, 1998; Asakura et al., 2004,2008; Tzvetkova-Chevolleau et al., 2007), indicating a critical role of these components in the tethering of RNCs at the translocon and subsequent insertion into the membrane. As our data show an upregulation of cpSecY, cpFtsY, and Alb3 in the ffc mutant, it is likely that the increased level of the transport machinery in the thylakoid membrane compensates at least partially the absence of cpSRP54.
METHODS
Plant Material and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) mutant lacking cpSRP54 (ffc 1-2, referred to as ffc) was described previously by Amin et al. (1999). Pea (Pisum sativum) cv Kelvedon Wonder and Arabidopsis plants (ecotype Columbia-0 and ffc) were grown on soil under cool-white light (Philips Master TL-D 58W/840 Reflex Eco; 8 h of light, 120 µmol photons m−2 s−1, 22°C, 65% humidity and 16 h of dark, 19,5°C, 65% humidity).
Ribosome Profiling
The spatial examination of stromal and thylakoid membrane–associated ribosomes of the 3-week-old wild-type and ffc plants was performed as described previously by Zoschke and Barkan, (2015) in three biological replicates (separate leaf material; reproducibility between replicates is shown in Supplemental Figure 9A and the Supplemental Data Set). A micrococcal nuclease pre-treatment was included to remove mRNA-tethered ribosomes from thylakoid membranes before pelleting the membranes (Zoschke and Barkan, 2015). Differing from the published protocol, thylakoid membranes were pelleted by centrifugation at 20,000g at 4°C for 15 min. The Arabidopsis custom microarrays (Supplemental Data Set) used contain four technical replicate spots for each probe, described previously by Trösch et al., (2018).
Data were initially normalized to eliminate general alterations introduced by technical variations (e.g., unequal labeling efficiencies; described previously by Trösch et al., [2018]). Subsequently, membrane and soluble signals were normalized to each other as described previously by Zoschke and Barkan, (2015). In brief, the ribosome distribution (indicated by signal intensities) in the first 200 nucleotides relative to the last 200 nucleotides of the psbD, psbC, psaA, psaB, and psbB ORFs (excluding the psbC/D overlap) were calculated from three normalized published data sets from the unfractionated wild type (Trösch et al., 2018) and used to normalize the soluble signal for the 5′ region and the membrane signal for the 3′ region, respectively, for each of the three biological replicates.
General ribosome coverage was calculated as a weighted sum of soluble and membrane-associated ribosome footprints in wild-type and ffc mutant plants. To this end, membrane and soluble footprints were summed up for each probe (reproducibility between biological replicates is shown in Supplemental Figure 9B and the Supplemental Data Set). In this calculation, soluble footprints were weighted different than membrane footprints by multiplication with a specific factor for the ffc (4.75) mutant and the wild type (2.28) according to determined spatial footprint distribution (Figure 1A). Averaged translation outputs were calculated from these values as described previously (Trösch et al., 2018).
Protein Extraction and Immunoblot Analyses
Total protein extracts of 100 mg of Arabidopsis leaves from 3- and 4-week-old plants were prepared using the TRIzol reagent (Life Technologies). Proteins were separated on 12 to 15% SDS-polyacrylamide gels and subsequently blotted onto nitrocellulose membrane (Macherey-Nagel). Transferred proteins were detected by specific antibodies against the His-tag (Penta·His HRP Conjugate, Qiagen, ID 34460, lot no. 157051123, dilution 1:4000), chloroplast uL4 (Agrisera, AS153076, lot no. 1512, dilution 1:5000), PsbA (Agrisera, AS05084, lot no. 1712, dilution 1:10,000), PsbB (Agrisera, AS04038, lot no. 1802, dilution 1:2000), PsbD (Agrisera, AS06146, lot no. 0905, dilution 1:2000), PsaA (Agrisera, AS03025, lot no. 0611, dilution 1:500), PsaB (Agrisera, AS10695, lot no. 1807, dilution 1:2000), PC (Agrisera, AS06141, lot no. 1511, dilution 1:5000), PetA (Agrisera, AS06119, lot no. 1611, dilution 1:2000), actin (Sigma-Aldrich, A0480, lot no. 127M4890V, dilution 1:2000), Rbcs (Agrisera, AS07259, dilution 1:5000), cpSRP54 (Walter et al. 2015a), Alb3 (Bals et al. 2010), cpSecY (Schuenemann et al., 1999), and cpFtsY (Tu et al. 1999) and enhanced chemiluminescence reaction (Thermo Fisher Scientific).
Isolation and Fractionation of Arabidopsis Chloroplasts
Arabidopsis plants (50 g of fresh weight, 3 to 4 weeks old) were homogenized in 500 mL of 300 mM sorbitol, 5 mM magnesium chloride, 5 mM EGTA, 5 mM EDTA, 10 mM NaHCO3, and 20 mM HEPES, pH 7.7 (isolation buffer), using an ULTRA-TURRAX T25 homogenizer. The homogenate was filtered through two layers of Miracloth and centrifuged for 5 min at 1000g and 4°C. The pellets were resuspended in remaining supernatant and loaded on two preformed Percoll gradients (50% Percoll, 0.5 mM reduced l-glutathione in isolation buffer). The preformation of the Percoll gradients was done by centrifugation for 30 min at 40,000g and 4°C with brake off. After centrifugation of the loaded Percoll gradients (at 7800g and 4°C, for 10 min with brake off), the lower green band including the intact chloroplasts was removed and washed one time in 300 mM sorbitol, 3 mM magnesium chloride, and 50 mM HEPES, pH 8.0. Intact chloroplasts were centrifuged (at 1000g and 4°C, for 5 min), lysed at 1 to 2 mg chlorophyll/mL in 10 mM magnesium chloride, and 50 mM HEPES, pH 8.0 (HM buffer), and separated in thylakoids and stroma by centrifugation (at 14,000 rpm and 4°C, for 10 min). Thylakoids were washed in HM buffer with 0.5 M NaCl and adjusted in HM buffer according to the chlorophyll concentration of chloroplasts.
Plasmid Construction
All constructs used for sucrose density gradient centrifugation and microscale thermophoresis were cloned into the pETDUET vector for expression with N-terminal His-tags. The constructs encoding Arabidopsis cpSRP43 (amino acids 61 to 376), cpSRP54 (amino acids 77 to 564), cpSRP54M (amino acids 372 to 564), and cpSRP54ΔC-term (amino acids 371 to 529) were described previously (Bals et al., 2010; Dünschede et al., 2015). The constructs cpSRP54NG (amino acids 77 to 371) and cpSRP54C-term (amino acids 529 to 564) were cloned using the restriction enzymes BamHI and SalI (forward [for] 5′-CCACAGCCAGGATCCGATGTTTGGTCAGTTGACTG-3′, reverse [rev] 5′-CGCAAGCTTGTCGACTTATCCTAGAATTCGTCCAGCCA-3′) and BamHI and HindIII, respectively (for 5′-TTTGGATCCGAAGGCTCCACCTGGA-3′, rev 5′-GTAAGCTTTTAGTTACCAGAGCCGAAGCC-3′). The cDNA encoding mature Arabidopsis chloroplast uL4 was PCR amplified from cDNA and cloned into the pETDUET vector via SalI/NdeI (for 5′-TTTGTCGACGTTTCAAAACTCGGCTC-3′, rev 5′-AAACATATGTTAAGCTTCCTCTGACCCTTCCGTCTCATCTTCATCGT-3′). The constructs cpSRP54MΔ415-421, cpSRP54MΔ415-418, cpSRP54MΔ536-540, uL4Δ73-76, uL4Δ77-81, and uL4Δ82-85 were generated using the QuikChange Lightning site-directed mutagenesis kit (Agilent) using pETDUET-cpSRP54M and pETDUET-uL4 as templates. For site-specific photo-crosslinking cpSRP54M was cloned into pET29b using NcoI and SalI for expression with a C-terminal His-tag (for 5′-CTAACCATGGGAATGGGAGATGTGCTT-3′, rev 5′-AAAGTCGACGTTACCAGAGCCGAAGCC-3′). Amber stop codons were introduced into the pET29b-cpSRP54M plasmid at the indicated sites using site-directed mutagenesis.
Protein Expression and Purification
His-tagged recombinant proteins were overexpressed in Escherichia coli strain BL21 (DE3) or strain Rosetta2 (DE3) and purified under native conditions using nickel-nitrilotriacetic acid resin or HisTrap columns (Qiagen, GE Healthcare). After purification, proteins used in microscale thermophoresis were stored in thermophoresis buffer (300 mM sodium chloride, 2.7 mM potassium chloride, 10 mM disodium phosphate, 1.8 mM dipotassium phosphate, 5 mM magnesium chloride, and 2 mM DTT, pH 7.3). Otherwise, proteins were purified in 20 mM HEPES, 300 mM sodium chloride, and 2 mM DTT, pH 7.5.
For the overexpression of proteins containing the site specific photo-crosslinker pBpA, E. coli BL21 (DE3), cells carrying the plasmid pEVOL, and the indicated pET29b-cpSRP54M plasmid were cultured overnight (37°C in terrific broth medium containing 25 µg/mL kanamycin and 34 µg/mL chloramphenicol). The main culture was supplemented with pBpA (dissolved in 1 M sodium hydroxide) to a final concentration of 0.2 mM. When the cells reached an OD600 of 0.7, l-Ara was added to a final concentration of 0.2% (w/v). After an incubation of 60 min at 30°C, expression of the cpSRP54M constructs was induced with 1 mM isopropyl-β-d-thiogalactopyranoside. The cells were harvested after a 16-h incubation at 30°C, and the His-tagged proteins were purified as described above. All steps were conducted in the dark or under red light. A Coomassie Brilliant Blue R 250–stained SDS-polyacrylamide gel of the purified recombinant proteins used in microscale thermophoresis and in vitro photo-crosslinking is shown in Supplemental Figure 8. The quality of recombinant cpSRP54M was checked by size exclusion chromatography as described previously (Dünschede et al., 2015). The quality of the uL4 constructs was checked by size exclusion chromatography (Superose 12 10/30) at a flow rate of 0.4 mL/min in thermophoresis buffer.
Isolation of Chloroplast 70S Ribosomes
Chloroplasts isolated from 9-d-old pea plants were adjusted to a chlorophyll concentration of 2 mg/mL in 20 mM HEPES, 50 mM potassium acetate, 6 mM magnesium acetate, and 2 mM DTT, pH 7.5 (LSB buffer) with 0.5 U of Ribolock RNase Inhibitor, 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, 1 µg/mL antipain, and 1 µg/mL leupeptin and incubated for 30 min on ice. The lysed chloroplasts were centrifuged (at 20,000g and 4°C, for 10 min), and 1 mL of the clear supernatant (stroma) was loaded onto a 2.5-mL Suc cushion (1 M Suc in LSB buffer). After centrifugation (at 70,000 rpm and 4°C, for 90 min; Rotor TLA 100.3), the ribosomal pellet was resuspended in LSB buffer.
Isotope-Coded Crosslinking
Enriched ribosomes (20 µg) in LSB plus 300 mM sodium chloride were incubated with BS3-H12/D12 in a ratio of 1:1 (Creative Molecules) containing the crosslinker in a final concentration of 2 mM and incubated on ice for 30 min. The reaction was stopped with ammonium carbonate with a final concentration of 100 mM and incubated for 15 min on ice. After tryptic digestion, the samples were purified and analyzed via mass spectrometry as described in Cormann et al. (2016). Data were evaluated using the StavroX v.3.5.1 software (Götze et al. 2012) according to Cormann et al. (2016).
Position-Specific in Vitro Photo-Crosslinking
Fifty micrograms of the indicated cpSRP54M-constructs were incubated with 1 mg of chloroplast ribosomes in 200 μL of LSB buffer. Crosslinks were induced by exposure to UV light for 5 h at 4°C. Before and after UV-light exposure, samples were taken and analyzed by immunoblot.
Sucrose Density Gradient Centrifugation
Enriched chloroplast ribosomes (1.6 mg) were incubated with 1.2 nmol of the indicated recombinant His-tagged protein constructs in LSB buffer for 30 min on ice and loaded onto a Suc gradient (layers of 15, 25, 35, and 45% [w/v] Suc in LSB buffer). After centrifugation (overnight, at 155,000g and 4°C), the gradient was fractionated and analyzed by immunoblot.
Microscale Thermophoresis
Recombinant proteins (His-cpSRP54M, His-cpSRP54MΔ415-418, and His-cpSRP54Δ536-540) were labeled using the Monolith NT.115 protein labeling kit RED-NHS. The interaction studies between the indicated cpSRP54M and His-uL4 constructs were performed in thermophoresis buffer supplemented with 0.05% Tween 20 in Monolith NT.115 MST premium coated capillaries. A dilution series of the indicated His-uL4 constructs in the micromolar range was created, while the concentration of the labeled interacting protein remained constant in the nanomolar range. Experiments were performed in the Monolith NT.115 instrument, and the data were evaluated using the MO.Affinity Analysis software (Nano Temper Technologies).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers cpSRP54 (At5g03940); chloroplast uL4 (At1g07320); psbA (AtCg00020).
Supplemental Data
Supplemental Figure 1. Minimal alterations of relative translation outputs in the ffc mutant.
Supplemental Figure 2. Zoom-in images for representative reading frames which do not display substantially altered spatial ribosome footprint distributions in the ffc mutant.
Supplemental Figure 3. Zoom-in image for the psbC reading frame which does not show a substantially altered spatial ribosome footprint distribution in the ffc mutant.
Supplemental Figure 4. Zoom-in image for the psbJ reading frame which displays substantially altered membrane-associated ribosome footprint abundances in the ffc mutant.
Supplemental Figure 5. Immunoblot analysis of PS I and PS II subunits and of components of the transport machinery in chloroplasts, thylakoids and stroma of the ffc mutant.
Supplemental Figure 6. Recombinant cpSRP43 displaces cpSRP54 from chloroplast ribosomes.
Supplemental Figure 7. Raw microscale thermophoresis data.
Supplemental Figure 8. Purified recombinant proteins used for microscale thermophoresis and in vitro photocrosslinking experiments.
Supplemental Figure 9. Reproducibility of ribosome footprint data between representative biological replicates.
Supplemental Data Set. Data sets of spatially resolved and recalculated standard ribosome profiling experiments.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Sabine Mölleken and Jennifer Ortelt for experimental support, Bernd Ackermann for help with the software PyMol and Phyre2 and figure preparation, and Maja Schuster for help with statistical analyses. We thank Klaus Hagemann and Gabriele Frenßen-Schenkel for graphical contributions and Silke Funke for technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft (SCHU 1163/6-2 within Research Unit FOR2092 to D.S.S., ZO 302/4-1 and SFB-TRR 175 A04 to R.Z., and funding within the Cluster of Excellence EXC 1069 RESOLV to M.M.N.).
AUTHOR CONTRIBUTIONS
A.H., B.D., M.M.N., R.Z., and D.S. designed the experiments. A.H., I.G., D.S.S., J.N., and A.B. performed the experiments. All authors contributed to the data analysis. A.H., M.M.N., R.Z., and D.S. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Ahmed T., Yin Z., Bhushan S. (2016). Cryo-EM structure of the large subunit of the spinach chloroplast ribosome. Sci. Rep. 6: 35793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopian D., Shen K., Zhang X., Shan S.O. (2013). Signal recognition particle: An essential protein-targeting machine. Annu. Rev. Biochem. 82: 693–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin P., Sy D.A., Pilgrim M.L., Parry D.H., Nussaume L., Hoffman N.E. (1999). Arabidopsis mutants lacking the 43- and 54-kilodalton subunits of the chloroplast signal recognition particle have distinct phenotypes. Plant Physiol. 121: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y., Hirohashi T., Kikuchi S., Belcher S., Osborne E., Yano S., Terashima I., Barkan A., Nakai M. (2004). Maize mutants lacking chloroplast FtsY exhibit pleiotropic defects in the biogenesis of thylakoid membranes. Plant Cell 16: 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y., Kikuchi S., Nakai M. (2008). Non-identical contributions of two membrane-bound cpSRP components, cpFtsY and Alb3, to thylakoid biogenesis. Plant J. 56: 1007–1017. [DOI] [PubMed] [Google Scholar]
- Ataide S.F., Schmitz N., Shen K., Ke A., Shan S.O., Doudna J.A., Ban N. (2011). The crystal structure of the signal recognition particle in complex with its receptor. Science 331: 881–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bals T., Dünschede B., Funke S., Schünemann D. (2010). Interplay between the cpSRP pathway components, the substrate LHCP and the translocase Alb3: An in vivo and in vitro study. FEBS Lett. 584: 4138–4144. [DOI] [PubMed] [Google Scholar]
- Berndt U., Oellerer S., Zhang Y., Johnson A.E., Rospert S. (2009). A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc. Natl. Acad. Sci. USA 106: 1398–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri P., Leibundgut M., Saurer M., Boehringer D., Ban N. (2017). The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J. 36: 475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornemann T., Jöckel J., Rodnina M.V., Wintermeyer W. (2008). Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat. Struct. Mol. Biol. 15: 494–499. [DOI] [PubMed] [Google Scholar]
- Celedon J.M., Cline K. (2013). Intra-plastid protein trafficking: How plant cells adapted prokaryotic mechanisms to the eukaryotic condition. Biochim. Biophys. Acta 1833: 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormann K.U., Möller M., Nowaczyk M.M. (2016). Critical assessment of protein cross-linking and molecular docking: An updated model for the interaction between photosystem II and Psb27. Front. Plant Sci. 7: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denks K., Sliwinski N., Erichsen V., Borodkina B., Origi A., Koch H.G. (2017). The signal recognition particle contacts uL23 and scans substrate translation inside the ribosomal tunnel. Nat. Microbiol. 2: 16265. [DOI] [PubMed] [Google Scholar]
- Dünschede B., Träger C., Schröder C.V., Ziehe D., Walter B., Funke S., Hofmann E., Schünemann D. (2015). Chloroplast SRP54 was recruited for posttranslational protein transport via complex formation with chloroplast SRP43 during land plant evolution. J. Biol. Chem. 290: 13104–13114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin A.E., Hoffman N.E. (1993). Characterization of a chloroplast homologue of the 54-kDa subunit of the signal recognition particle. J. Biol. Chem. 268: 22175–22180. [PubMed] [Google Scholar]
- Funke S., Knechten T., Ollesch J., Schünemann D. (2005). A unique sequence motif in the 54-kDa subunit of the chloroplast signal recognition particle mediates binding to the 43-kDa subunit. J. Biol. Chem. 280: 8912–8917. [DOI] [PubMed] [Google Scholar]
- Götze M., Pettelkau J., Schaks S., Bosse K., Ihling C.H., Krauth F., Fritzsche R., Kühn U., Sinz A. (2012). StavroX--A software for analyzing crosslinked products in protein interaction studies. J. Am. Soc. Mass Spectrom. 23: 76–87. [DOI] [PubMed] [Google Scholar]
- Groves M.R., Mant A., Kuhn A., Koch J., Dübel S., Robinson C., Sinning I. (2001). Functional characterization of recombinant chloroplast signal recognition particle. J. Biol. Chem. 276: 27778–27786. [DOI] [PubMed] [Google Scholar]
- Gu S.Q., Peske F., Wieden H.J., Rodnina M.V., Wintermeyer W. (2003). The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA 9: 566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M., Blau M., Becker T., Mielke T., Pool M.R., Wild K., Sinning I., Beckmann R. (2006). Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature 444: 507–511. [DOI] [PubMed] [Google Scholar]
- Henderson R.C., Gao F., Jayanthi S., Kight A., Sharma P., Goforth R.L., Heyes C.D., Henry R.L., Suresh Kumar T.K. (2016). Domain organization in the 54-kDa subunit of the chloroplast signal recognition particle. Biophys. J. 111: 1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdermann I., Meyer N.H., Round A., Wild K., Sattler M., Sinning I. (2012). Chromodomains read the arginine code of post-translational targeting. Nat. Struct. Mol. Biol. 19: 260–263. [DOI] [PubMed] [Google Scholar]
- Houben E.N., Zarivach R., Oudega B., Luirink J. (2005). Early encounters of a nascent membrane protein: Specificity and timing of contacts inside and outside the ribosome. J. Cell Biol. 170: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagath J.R., Matassova N.B., de Leeuw E., Warnecke J.M., Lentzen G., Rodnina M.V., Luirink J., Wintermeyer W. (2001). Important role of the tetraloop region of 4.5S RNA in SRP binding to its receptor FtsY. RNA 7: 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagendorf A.T., Michaels A. (1990). Rough thylakoids: Translation on photosynthetic membranes. Plant Sci. 71: 137–145. [Google Scholar]
- Jomaa A., Boehringer D., Leibundgut M., Ban N. (2016). Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon. Nat. Commun. 7: 10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan R.J., Freymann D.M., Walter P., Stroud R.M. (1998). Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell 94: 181–191. [DOI] [PubMed] [Google Scholar]
- Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Klein P.G., Mullet J.E. (1991). Ribosomes pause at specific sites during synthesis of membrane-bound chloroplast reaction center protein D1. J. Biol. Chem. 266: 14931–14938. [PubMed] [Google Scholar]
- Klimyuk V.I., Persello-Cartieaux F., Havaux M., Contard-David P., Schuenemann D., Meiherhoff K., Gouet P., Jones J.D., Hoffman N.E., Nussaume L. (1999). A chromodomain protein encoded by the Arabidopsis CAO gene is a plant-specific component of the chloroplast signal recognition particle pathway that is involved in LHCP targeting. Plant Cell 11: 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott T.G., Robinson C. (1994). The secA inhibitor, azide, reversibly blocks the translocation of a subset of proteins across the chloroplast thylakoid membrane. J. Biol. Chem. 269: 7843–7846. [PubMed] [Google Scholar]
- Króliczewski J., Piskozub M., Bartoszewski R., Króliczewska B. (2016). ALB3 insertase mediates cytochrome b6 co-translational import into the thylakoid membrane. Sci. Rep. 6: 34557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn P., Draycheva A., Vogt A., Petriman N.A., Sturm L., Drepper F., Warscheid B., Wintermeyer W., Koch H.G. (2015). Ribosome binding induces repositioning of the signal recognition particle receptor on the translocon. J. Cell Biol. 211: 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M., Drechsel O., Kahlau S., Bock R. (2013). OrganellarGenomeDRAW--A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41: W575-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier E., Holtkamp W., Rodnina M.V., Wintermeyer W. (2017). Signal recognition particle binds to translating ribosomes before emergence of a signal anchor sequence. Nucleic Acids Res. 45: 11858–11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R., Brunner J., Hoffman N.E., van Wijk K.J. (1999). Interactions of ribosome nascent chain complexes of the chloroplast-encoded D1 thylakoid membrane protein with cpSRP54. EMBO J. 18: 733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R., van Wijk K.J. (2002). Transient interaction of cpSRP54 with elongating nascent chains of the chloroplast-encoded D1 protein; ‘cpSRP54 caught in the act’. FEBS Lett. 524: 127–133. [DOI] [PubMed] [Google Scholar]
- Perez Boerema A., Aibara S., Paul B., Tobiasson V., Kimanius D., Forsberg B.O., Wallden K., Lindahl E., Amunts A. (2018). Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor. Nat. Plants 4: 212–217. [DOI] [PubMed] [Google Scholar]
- Piskozub M., Króliczewska B., Króliczewski J. (2015). Ribosome nascent chain complexes of the chloroplast-encoded cytochrome b6 thylakoid membrane protein interact with cpSRP54 but not with cpSecY. J. Bioenerg. Biomembr. 47: 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool M.R., Stumm J., Fulga T.A., Sinning I., Dobberstein B. (2002). Distinct modes of signal recognition particle interaction with the ribosome. Science 297: 1345–1348. [DOI] [PubMed] [Google Scholar]
- Röhl T., van Wijk K.J. (2001). In vitro reconstitution of insertion and processing of cytochrome f in a homologous chloroplast translation system. J. Biol. Chem. 276: 35465–35472. [DOI] [PubMed] [Google Scholar]
- Rosenblad M.A., Samuelsson T. (2004). Identification of chloroplast signal recognition particle RNA genes. Plant Cell Physiol. 45: 1633–1639. [DOI] [PubMed] [Google Scholar]
- Roy L.M., Barkan A. (1998). A SecY homologue is required for the elaboration of the chloroplast thylakoid membrane and for normal chloroplast gene expression. J. Cell Biol. 141: 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutschow H., Ytterberg A.J., Friso G., Nilsson R., van Wijk K.J. (2008). Quantitative proteomics of a chloroplast SRP54 sorting mutant and its genetic interactions with CLPC1 in Arabidopsis. Plant Physiol. 148: 156–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffitzel C., Oswald M., Berger I., Ishikawa T., Abrahams J.P., Koerten H.K., Koning R.I., Ban N. (2006). Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature 444: 503–506. [DOI] [PubMed] [Google Scholar]
- Schuenemann D., Amin P., Hartmann E., Hoffman N.E. (1999). Chloroplast SecY is complexed to SecE and involved in the translocation of the 33-kDa but not the 23-kDa subunit of the oxygen-evolving complex. J. Biol. Chem. 274: 12177–12182. [DOI] [PubMed] [Google Scholar]
- Schuenemann D., Gupta S., Persello-Cartieaux F., Klimyuk V.I., Jones J.D.G., Nussaume L., Hoffman N.E. (1998). A novel signal recognition particle targets light-harvesting proteins to the thylakoid membranes. Proc. Natl. Acad. Sci. USA 95: 10312–10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W.B., Flügge U.I., Kunze R. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K., Arslan S., Akopian D., Ha T., Shan S.O. (2012). Activated GTPase movement on an RNA scaffold drives co-translational protein targeting. Nature 492: 271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K., Shan S.O. (2010). Transient tether between the SRP RNA and SRP receptor ensures efficient cargo delivery during cotranslational protein targeting. Proc. Natl. Acad. Sci. USA 107: 7698–7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg R., Knüpffer L., Origi A., Asti R., Koch H.G. (2018). Co-translational protein targeting in bacteria. FEMS Microbiol. Lett. 365. [DOI] [PubMed] [Google Scholar]
- Sundberg E., Slagter J.G., Fridborg I., Cleary S.P., Robinson C., Coupland G. (1997). ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell 9: 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Träger C., et al. (2012). Evolution from the prokaryotic to the higher plant chloroplast signal recognition particle: The signal recognition particle RNA is conserved in plastids of a wide range of photosynthetic organisms. Plant Cell 24: 4819–4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trösch R., Barahimipour R., Gao Y., Badillo-Corona J.A., Gotsmann V.L., Zimmer D., Mühlhaus T., Zoschke R., Willmund F. (2018). Commonalities and differences of chloroplast translation in a green alga and land plants. Nat. Plants 4: 564–575. [DOI] [PubMed] [Google Scholar]
- Tu C.J., Schuenemann D., Hoffman N.E. (1999). Chloroplast FtsY, chloroplast signal recognition particle, and GTP are required to reconstitute the soluble phase of light-harvesting chlorophyll protein transport into thylakoid membranes. J. Biol. Chem. 274: 27219–27224. [DOI] [PubMed] [Google Scholar]
- Tzvetkova-Chevolleau T., et al. (2007). Canonical signal recognition particle components can be bypassed for posttranslational protein targeting in chloroplasts. Plant Cell 19: 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullers R.S., Houben E.N., Raine A., ten Hagen-Jongman C.M., Ehrenberg M., Brunner J., Oudega B., Harms N., Luirink J. (2003). Interplay of signal recognition particle and trigger factor at L23 near the nascent chain exit site on the Escherichia coli ribosome. J. Cell Biol. 161: 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker R., Barkan A. (1995). Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J. 14: 3905–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees R.M., Hegde R.S. (2016). Toward a structural understanding of co-translational protein translocation. Curr. Opin. Cell Biol. 41: 91–99. [DOI] [PubMed] [Google Scholar]
- Walter B., Hristou A., Nowaczyk M.M., Schünemann D. (2015a). In vitro reconstitution of co-translational D1 insertion reveals a role of the cpSec-Alb3 translocase and Vipp1 in photosystem II biogenesis. Biochem. J. 468: 315–324. [DOI] [PubMed] [Google Scholar]
- Walter B., Pieta T., Schünemann D. (2015b). Arabidopsis thaliana mutants lacking cpFtsY or cpSRP54 exhibit different defects in photosystem II repair. Front. Plant Sci. 6: 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickström D., Wagner S., Baars L., Ytterberg A.J., Klepsch M., van Wijk K.J., Luirink J., de Gier J.-W. (2011). Consequences of depletion of the signal recognition particle in Escherichia coli. J. Biol. Chem. 286: 4598–4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Paakkarinen V., van Wijk K.J., Aro E.M. (1999). Co-translational assembly of the D1 protein into photosystem II. J. Biol. Chem. 274: 16062–16067. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wölfle T., Rospert S. (2013). Interaction of nascent chains with the ribosomal tunnel proteins Rpl4, Rpl17, and Rpl39 of Saccharomyces cerevisiae. J. Biol. Chem. 288: 33697–33707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziehe D., Dünschede B., Schünemann D. (2017). From bacteria to chloroplasts: Evolution of the chloroplast SRP system. Biol. Chem. 398: 653–661. [DOI] [PubMed] [Google Scholar]
- Zoschke R., Barkan A. (2015). Genome-wide analysis of thylakoid-bound ribosomes in maize reveals principles of cotranslational targeting to the thylakoid membrane. Proc. Natl. Acad. Sci. USA 112: E1678–E1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R., Chotewutmontri P., Barkan A. (2017). Translation and co-translational membrane engagement of plastid-encoded chlorophyll-binding proteins are not influenced by chlorophyll availability in maize. Front. Plant Sci. 8: 385. [DOI] [PMC free article] [PubMed] [Google Scholar]