Chloroplast lysophosphatidylcholine acyltransferases are involved in PC acyl editing with newly synthesized fatty acids, and not in lysophosphatidylcholine transport for galactolipid synthesis.
Abstract
The eukaryotic pathway of galactolipid synthesis involves fatty acid synthesis in the chloroplast, followed by assembly of phosphatidylcholine (PC) in the endoplasmic reticulum (ER), and then turnover of PC to provide a substrate for chloroplast galactolipid synthesis. However, the mechanisms and classes of lipids transported between the chloroplast and the ER are unclear. PC, PC-derived diacylglycerol, phosphatidic acid, and lyso-phosphatidylcholine (LPC) have all been implicated in ER-to-chloroplast lipid transfer. LPC transport requires lysophosphatidylcholine acyltransferase (LPCAT) activity at the chloroplast to form PC before conversion to galactolipids. However, LPCAT has also been implicated in the opposite chloroplast-to-ER trafficking of newly synthesized fatty acids through PC acyl editing. To understand the role of LPC and LPCAT in acyl trafficking we produced and analyzed the Arabidopsis (Arabidopsis thaliana) act1 lpcat1 lpcat2 triple mutant. LPCAT1 and LPCAT2 encode the major lysophospholipid acyltransferase activity of the chloroplast, and it is predominantly for incorporation of nascent fatty acids exported form the chloroplast into PC by acyl editing. In vivo acyl flux analysis revealed eukaryotic galactolipid synthesis is not impaired in act1 lpcat1 lpcat2 and uses a PC pool distinct from that of PC acyl editing. We present a model for the eukaryotic pathway with metabolically distinct pools of PC, suggesting an underlying spatial organization of PC metabolism as part of the ER–chloroplast metabolic interactions.
INTRODUCTION
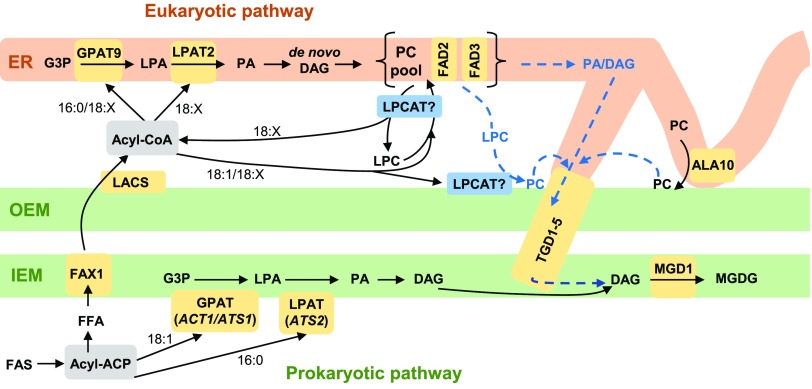
Membranes that encompass, subdivide, and provide scaffolds for protein localization are essential to all living cells. In plant leaf tissue, the thylakoid membranes within the chloroplast are composed predominantly from the galactolipids monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG) and are the essential structures that hold the photosynthetic apparatuses. The metabolic pathways of photosynthetic membrane production have been studied for over 40 years through biochemical, genetic, and molecular biology approaches and have been extensively reviewed across the decades (Roughan and Slack, 1982; Browse and Somerville, 1991; Ohlrogge and Browse, 1995; Moreau et al., 1998; Kelly and Dörmann, 2004; Benning, 2008, 2009; Shimojima et al., 2009; Li-Beisson et al., 2013, 2017; Boudière et al., 2014; Hurlock et al., 2014; Block and Jouhet, 2015; Bastien et al., 2016; Botella et al., 2017; LaBrant et al., 2018). This research has led to a complicated metabolic model that requires the trafficking of lipid substrates from the chloroplast, to the endoplasmic reticulum (ER), and then back into the chloroplast to produce galactolipids (Figure 1). Most of the biosynthetic enzymes and associated genes of fatty acid synthesis, glycerolipid assembly, and fatty acid desaturation crucial to produce galactolipids have been identified. However, a major area of uncertainty surrounds the process of lipid transfer between the chloroplast and the ER, and even the lipid species that is transferred from the ER back into the plastid is still unclear (LaBrant et al., 2018).
Figure 1.
Model of Wild-Type Arabidopsis Acyl Trafficking Within Leaf Glycerolipid Synthesis.
The model focuses on the trafficking of acyl groups between the chloroplast and the ER for MGDG synthesis. The model is centered around the ER “PC pool,” which is involved in de novo PC synthesis, desaturation, acyl editing, and turnover to produce the substrate for MGDG production. Chloroplast OEM PC produced from transfer of LPC or PC may also be a substrate for MGDG synthesis. Key enzymes/transporters are in yellow; uncertain reactions are in blue and have dashed lines.
Figure 1 displays the current state of the two-pathway model of leaf MGDG synthesis (for more detailed comprehensive recent reviews please see Li-Beisson et al., 2013, 2017; Hurlock et al., 2014; Botella et al., 2017; LaBrant et al., 2018). Fatty acids are synthesized up to 18-carbon saturated or monounsaturated fatty acids while esterified to acyl carrier protein (ACP) within the chloroplast stroma. In some plants these acyl-ACPs can be used by the acyl selective glycerol-3-phosphate acyltransferase (GPAT) and lysophosphatidic acid acyltransferase (LPAT) of the “prokaryotic pathway” (ACT1 or ATS1, and ATS2, respectively; Kunst et al., 1988; Nishida et al., 1993; Yu et al., 2004) to produce a molecular species of phosphatidic acid (PA) containing oleate (18:1, number carbons: number double bonds) at the sn-1 position, and palmitate (16:0) at the sn-2 position. Dephosphorylation produces the diacylglycerol (DAG) substrate of MGDG synthase (MGD1; Jarvis et al., 2000). Subsequent desaturation of the 18:1/16:0 molecular species to 18:3/16:3 produces the abundant polyunsaturated molecular species of MGDG characteristic of chloroplast membranes.
Plants that produce up to half of the MGDG through the prokaryotic pathway are known as 16:3 plants in reference to the 16:3 that accumulates at sn-2 of MGDG. Only ∼12% of Angiosperms [including Arabidopsis (Arabidopsis thaliana)] are 16:3 plants (Mongrand et al., 1998). In contrast with 16:3 plants, 18:3 plants accumulate 18:3 at the sn-2 position of MGDG through glycerolipid assembly by the ER-localized “eukaryotic pathway” where sn-2 acyltransferases are selective for 18-carbon unsaturated fatty acids. In all plants, the eukaryotic pathway GPAT and LPAT (AtGPAT9 and AtLPAT2, respectively; Kim et al., 2005; Shockey et al., 2016; Singer et al., 2016) use cytosolic acyl-CoA to produce PA, and DAG in a parallel pathway to that in chloroplast (Figure 1). The de novo synthesized DAG is used to produce ER phospholipids such as phosphatidylcholine (PC) and phosphatidylethanolamine (PE). PC is also the extra-plastidic site for desaturation of 18:1, to 18:2 and 18:3 through the FAD2 and FAD3 enzymes, respectively (Arondel et al., 1992; Okuley et al., 1994). Eukaryotic MGDG is synthesized from a polyunsaturated DAG backbone derived from PC (Slack et al., 1977). However, the mechanism and location of PC turnover, and the lipid class transferred from the ER to the chloroplast, are still unclear (Figure 1, blue dashed lines). A key aspect of the eukaryotic pathway is that extensive trafficking of acyl groups between the chloroplast and the ER is required.
Fatty acid export from the chloroplast begins with hydrolysis of acyl-ACPs by fatty acid thioesterases (Figure 1; Bates et al., 2013). The subsequent free fatty acids (FFA) are transported across the chloroplast inner envelope membrane (IEM) by FAX1 (Li et al., 2015), and likely other members of the FAX family. The mechanism of FFA transfer across the chloroplast outer envelope membrane (OEM) is not clear but may involve vectoral diffusion driven by activation of FFA to acyl-CoA by long chain acyl-CoA synthetases on the cytoplasmic side of the OEM and/or the ER (Schnurr et al., 2002; Koo et al., 2004; Zhao et al., 2010; Jessen et al., 2015), which prevents FFA diffusion back into the plastid. Original models of the eukaryotic pathway assumed acyl-CoA-containing newly synthesized fatty acids were used by the ER-localized GPAT and LPAT to produce initial molecular species of glycerolipids containing 16:0 or 18:1 at sn-1, and 18:1 at sn-2 before further desaturation on PC (e.g., Ohlrogge and Browse, 1995). However, metabolic tracing experiments in various plant tissues have demonstrated that the majority of newly synthesized fatty acids exported from the plastid are initially rapidly incorporated into PC by a process known as acyl editing, which is essentially an acyl-CoA:PC fatty acid exchange cycle (Williams et al., 2000; Bates et al., 2007, 2009, 2012; Tjellström et al., 2012; Yang et al., 2017). This exchange between PC and the acyl-CoA pool produces a mixture of nascent fatty acids exported from the plastid with previously synthesized fatty acids derived from PC, some of which may have been desaturated to 18:2, or 18:3. This mixed acyl-CoA pool is thus the substrate for the eukaryotic GPAT and LPAT reactions for de novo glycerolipid assembly. In particular, short time point (≤1 min) [14C]acetate labeling of fatty acid synthesis in pea (Pisum sativum) leaves and Arabidopsis cells was crucial to demonstrating that nascent fatty acids are predominantly incorporated into the sn-2 position of PC by a lysophosphatidylcholine acyltransferase (LPCAT)-type reaction faster than incorporation into de novo DAG of the eukaryotic pathway (Bates et al., 2007; Tjellström et al., 2012). These results suggest that the lysophosphatidylcholine (LPC) pool and LPCAT enzymes involved in acyl editing may be part of the acyl trafficking of nascent fatty acids from the chloroplast to the ER (Figure 1).
The lipids DAG, PA, PC, and LPC have all been suggested as the species transported from the ER to the chloroplast in the eukaryotic pathway (Figure 1). DAG was first suggested to be transferred from the ER to the plastid after metabolic labeling experiments indicated that both the glycerol and fatty acids of PC were incorporated into MGDG together (Slack et al., 1977), and analysis of Arabidopsis PA hydrolase mutants (pah1 and pah2) under phosphate starvation supported this conclusion (Nakamura et al., 2009). However, combining the pah1 and pah2 mutants with the act1 mutation, which eliminates the prokaryotic pathway, did not appear to affect eukaryotic galactolipid synthesis under phosphate replete conditions (Fan et al., 2014). Recent work characterizing the Arabidopsis tgd1-5 mutants has indicated a transporter system involved in transferring the lipid substrate for MGDG synthesis from the chloroplast OEM to the IEM where MGDG synthesis occurs (Xu et al., 2003, 2005; Awai et al., 2006; Lu et al., 2007; Wang et al., 2012b, 2013; Fan et al., 2015). The mutants have impaired eukaryotic MGDG production and accumulate unusual trigalactosyldiacylglycerols as a phenotype. The TGD2 and TGD4 components of this transporter system bind to PA, and isolated chloroplasts from the tdg1 mutant effectively convert exogenous DAG into MGDG but exogenous PA conversion to MGDG is partially reduced. Therefore, PA has been proposed as the molecule transported from the ER to the plastid (Xu et al., 2005; Lu and Benning, 2009; Wang et al., 2012b, 2013). However, it is also been suggested that the role of PA is to destabilize membranes to reduce the energy barrier to transport of a different lipid species (LaBrant et al., 2018). A recent mathematical modeling approach suggested DAG was a better substrate to transport than PA, but limited PA transport was also required to activate MGDG synthesis (Maréchal and Bastien, 2014). Both DAG and PA can be produced by lipases in the ER, and thus could be transferred from the ER to the chloroplast and then into the IEM for MGDG synthesis. A different approach would be to first move PC from the ER to the chloroplast, and then derive PA or DAG from PC for transport to the IEM by the mechanisms discussed above.
PC is highly abundant in the outer leaflet of the OEM but is not present in other chloroplast membranes (Dorne et al., 1985), whereas the ER lipid PE is absent from chloroplasts. PC can be selectively transferred over PE from liposomes to isolated chloroplasts, a process which is dependent on the proteins within the chloroplast OEM (Yin et al., 2015), suggesting PC could be directly transferred from the ER, possibly through ER–chloroplast membrane contact sites (Andersson and Dörmann, 2009; Mueller-Schuessele and Michaud, 2018). Recent characterization of the flippase ALA10 (Botella et al., 2016) suggests it could be involved in enriching PC in ER membrane contact sites before transfer to the plastid (Botella et al., 2017). Rather than trafficking of a whole membrane lipid, LPC is a more water-soluble derivative of PC that could more easily traverse an aqueous space between the two compartments. The abundant LPCAT activities associated with the exterior of the chloroplast would then regenerate PC at the OEM (Bessoule et al., 1995; Tjellström et al., 2012). The transfer of LPC from the ER to plastid was supported by long time point (2–100 h) pulse-chase metabolic labeling studies in leek (Allium porrum) seedlings, which demonstrated a loss of fatty acids from sn-1/sn-2 PC and the subsequent accumulation in mostly sn-1 MGDG (Mongrand et al., 1997, 2000). The authors concluded that it was labeled sn-1-acyl-LPC that was transferred to the chloroplast and then reacylated with unlabeled fatty acids by LPCAT at the OEM during the chase in route to MGDG production. These results suggest that LPC and chloroplast LPCAT activity may be part of the ER-to-chloroplast lipid transfer reactions of the leaf eukaryotic pathway.
The discussion of previous research above indicates that LPC and LPCAT activity may have roles in both trafficking of fatty acids from the chloroplast to the ER, and from the ER to the chloroplast. Arabidopsis has four enzymes with demonstrated in vitro LPCAT activity: AtLPCAT1 and AtLPCAT2 (Ståhl et al., 2008; Wang et al., 2012a, 2014; Lager et al., 2013), and the lysophosphatidylethanolmine acyltransferases AtLPEAT1 and AtLPEAT2 (Stålberg et al., 2009; Jasieniecka-Gazarkiewicz et al., 2016). AtLPCAT1 and AtLPCAT2 have a strong preference for 18-carbon unsaturated acyl-CoAs over 16:0-CoA, and thus could produce the sn-1/2 18-carbon molecular species of PC (and subsequent MGDG) characteristic of the eukaryotic pathway (Lager et al., 2013). Short time point metabolic tracing of lipid metabolism in developing seeds of the LPCAT1 LPCAT2 double mutant (lpcat1 lpcat2) indicated the initial incorporation of nascent fatty acids into PC through sn-2 acyl editing was abolished (Bates et al., 2012). Instead the acyl groups are rerouted and initially esterified to G3P through the GPAT and LPAT reactions of the eukaryotic pathway before de novo PC synthesis. This result suggests that the Arabidopsis LPCAT1 and LPCAT2 enzymes are involved in the flux of acyl groups from the plastid to the ER in developing seeds, and that LPEAT1 and LPEAT2 cannot compensate for the loss of LPCATs in the acyl-editing cycle. However, these previous results are not directly applicable to leaves for two reasons: (1) In developing seeds LPCAT1 and LPCAT2 are expressed at two- to threefold higher levels than LPEAT1 and LPEAT2 (Supplemental Figure 1), which may explain the lack of compensatory acyl-editing activity by LPEAT1 and LPEAT2 in the lpcat1 lpcat2 mutant seeds. In leaves LPEAT1 and LPEAT2 are expressed at similar or even higher levels than LPCAT1 and LPCAT2 (Supplemental Figure 1). Therefore, it is possible that LPCAT1/LPCAT2 and LPEAT1/LPEAT2 may both contribute the acyl-editing LPCAT activity in leaves. (2) Quantitative acyl flux between the plastid and the ER is distinctly different between developing leaves and developing oilseed tissues. In leaves of 18:3 plants >60% of all fatty acids exported to the ER are reincorporated into the plastid for chloroplast membrane production. However, in oilseeds the major flux of acyl groups is for ER-localized triacylglycerol (TAG) synthesis such that >95% of acyl groups accumulate in extra-plastidial oil bodies and membranes, with very little flux back into the plastid (Li-Beisson et al., 2013). Thus, previous acyl flux studies on developing seeds of lpcat1 lpcat2 are not appropriate for measuring the role of LPCATs in the flux of acyl groups from the ER to the plastid for galactolipid production in leaves.
Therefore, to understand the roles of LPC and LPCAT activity in acyl flux to and from the ER and chloroplast we crossed the Arabidopsis lpcat1 lpcat2 double mutant (Bates et al., 2012) with the act1 mutant (Kunst et al., 1988). ACT1 (also called ATS1; Nishida et al., 1993) encodes the chloroplast GPAT. The act1 mutant eliminates prokaryotic pathway MGDG synthesis and enhances acyl flux through the eukaryotic pathway, similar to 18:3 plants. Our analysis of lipid accumulation, chloroplast-associated LPCAT activity, and in vivo acyl fluxes through both short time point metabolic tracing and long time point pulse-chase experiments further clarify the role of LPCATs within leaf PC acyl editing, and distinguish a separate metabolically active pool of PC involved in providing the substrate for chloroplast lipid synthesis.
RESULTS
Production and Characterization of the act1 lpcat1 lpcat2 Triple Mutant
In Arabidopsis leaves both the eukaryotic pathway and prokaryotic pathway contribute approximately equally to MGDG production (Browse et al., 1986). To better understand the roles of LPCAT1 and LPCAT2 specifically in eukaryotic pathway galactolipid production, we crossed the lpcat1 lpcat2 double mutant (Bates et al., 2012) with the act1 mutant, which essentially eliminates prokaryotic pathway galactolipid production (Kunst et al., 1988). The act1 allele is partially leaky, and residual GPAT activity remaining in the chloroplast is used for phosphatidylglycerol (PG) production (Xu et al., 2006). Previous crosses of act1 with tgd1-1 (a component of the transporter complex that imports the eukaryotic pathway lipid substrate into the chloroplast) was embryo lethal (Xu et al., 2005). We reasoned if the transport of LPC to chloroplasts and its subsequent conversion to PC by LPCAT at the chloroplast OEM was a key part of the eukaryotic pathway, then the act1 lpcat1 lpcat2 triple mutant may also demonstrate developmental or vegetative growth defects. Crossed F1 seeds were grown to maturity, and seed was collected and re-sown. The segregating F2 plants were screened for homozygosity of the lpcat1 lpcat2 double T-DNA mutation by PCR, and for homozygosity of the act1 mutation by the lack of 16:3 in leaf lipids by gas chromatography (Kunst et al., 1988). During the initial screening no growth phenotypes were observed. Homozygous act1 lpcat1 lpcat2 triple mutants were subsequently grown side by side with parental lines and wild-type Col-0. Across vegetative growth, the size of the triple mutants was within the plant-to-plant variation range of the parental lines (Supplemental Figure 2). Therefore, the act1 lpcat1 lpcat2 triple mutation has minimal effects on plant vegetative growth.
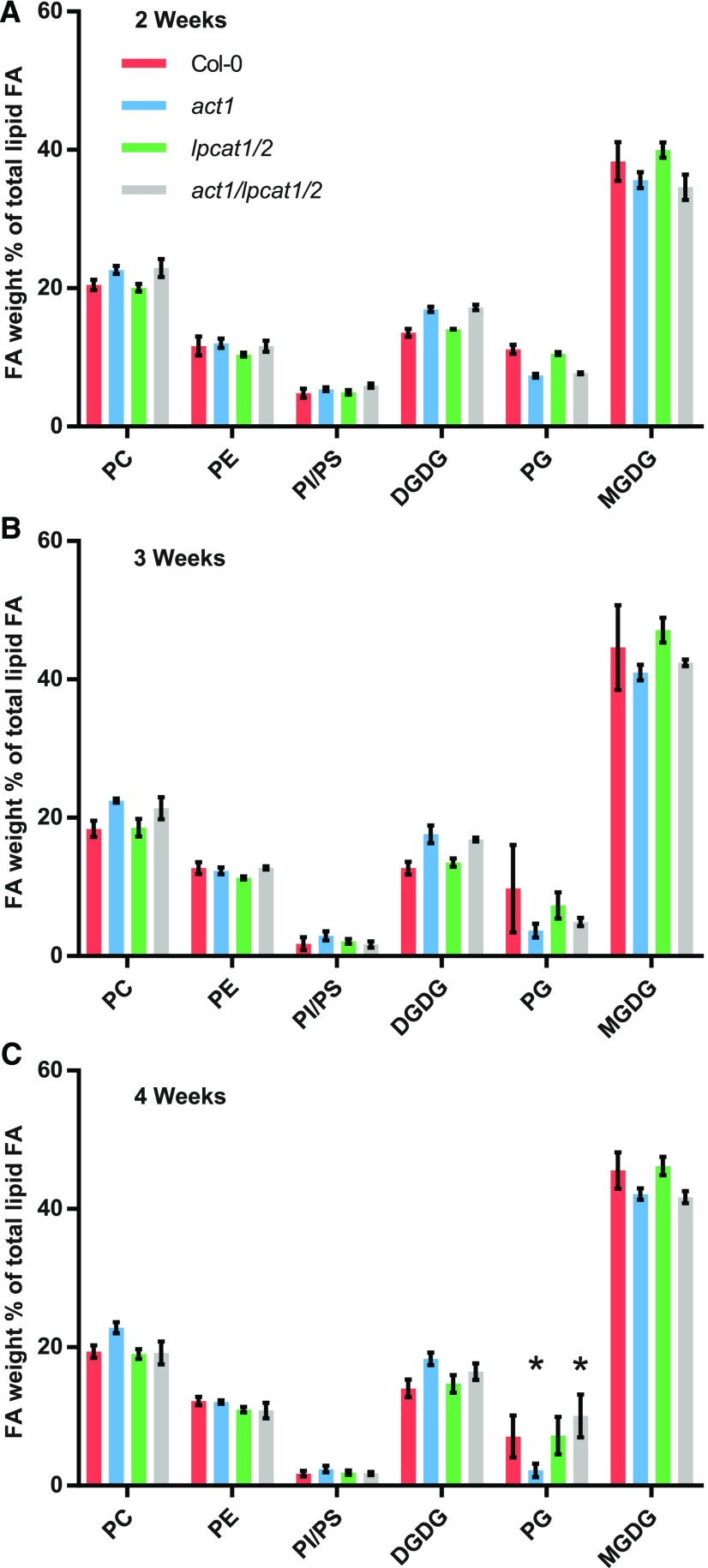
To determine if the act1 lpcat1 lpcat2 triple mutant had defects in leaf lipid production, we measured the relative accumulation of leaf membrane lipids at three developmental stages (2, 3, and 4 weeks after germination) in four lines of Arabidopsis: wild-type Col-0, act1, lpcat1 lpcat2 double mutant, and the act1 lpcat1 lpcat2 triple mutant (Figure 2). In general the relative accumulation of leaf membrane lipids in the lpcat1 lpcat2 mutant was similar to Col-0; however, as previously characterized, the act1 mutant has a significant change from Col-0 with less MGDG and PG, and a corresponding increase in PC and DGDG (Figures 2A to 2C; Kunst et al., 1988). Therefore, to understand the effect of the lpcat1 lpcat2 mutation when acyl flux through the eukaryotic pathway is enhanced in the act1 background, our main comparison is between act1 and the act1 lpcat1 lpcat2 triple mutant. At 2 and 3 weeks there were no significant changes in membrane lipid abundance between the two lines (Figures 2A and 2B). At 4 weeks (Figure 2C), only PG demonstrated a significant increase from 2.2% ± 1.0% in the act1 line to 10.1 ± 3.1% in the act1 lpcat1 lpcat2 line (P-value = 0.0013).
Figure 2.
Membrane Lipid Composition of Leaves across Development from Wild-Type and Mutant Lines.
The relative abundance of leaf membrane lipids was determined at three developmental stages: 2 weeks (A), 3 weeks (B), and 4 weeks (C) after germination. The data represent the average and se of 2–4 biological replicates. Significant (P-value < 0.05, Two-way ANOVA with multiple comparisons) differences within individual lipid abundances between act1 and act1 lpcat1 lpcat2 are indicated by asterisks above the bars. FA, fatty acid.
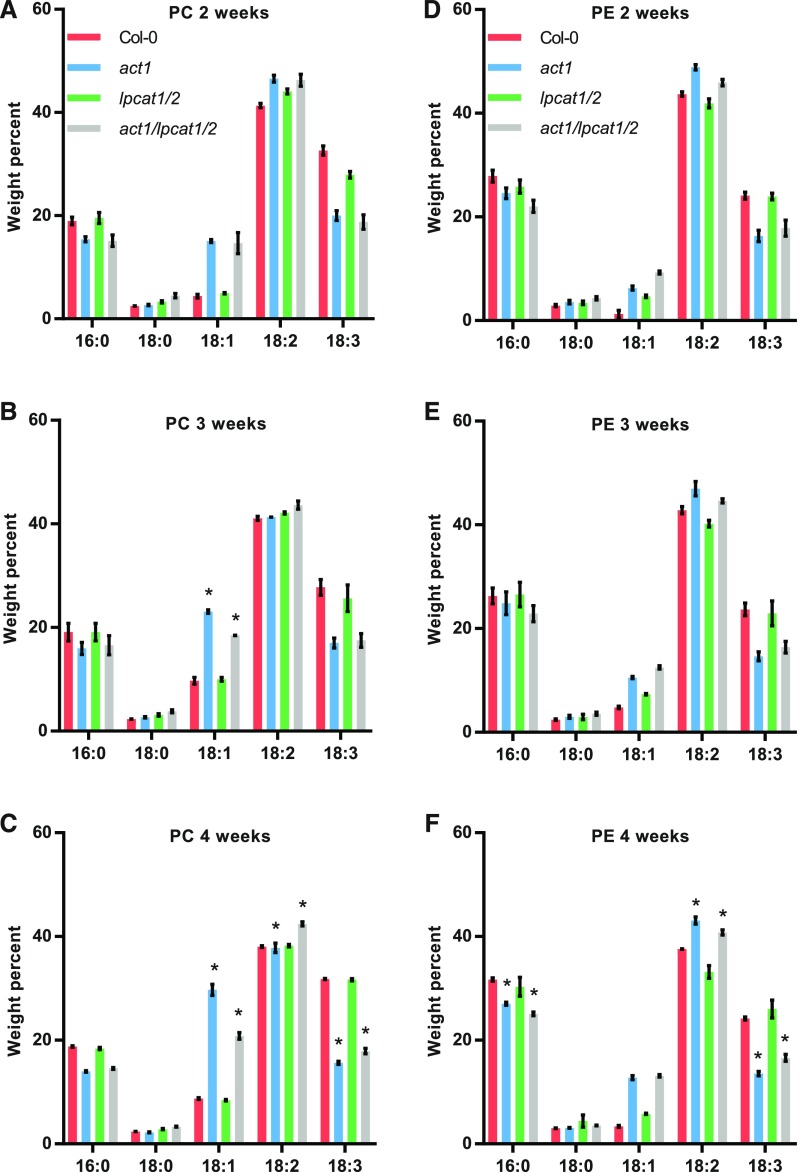
Even though the amount of PC or PE did not significantly change between act1 and act1 lpcat1 lpcat2, there were significant changes to their fatty acid compositions, especially at later stages in development (Figure 3). There was no change in either PC or PE at 2 weeks after germination (Figures 3A and 3D). At 3 weeks PC 18:1 content decreased from 23.1% ± 0.3% in the act1 line to 18.5% ± 0.05% in the act1 lpcat1 lpcat2 line (Figure 3B). At 4 weeks PC 18:1 decreased from 29.7% ± 1.1% in act1 to 20.8% ± 0.6% in act1 lpcat1 lpcat2, which was mostly compensated for by significant increases in 18:2 (from 37.8% ± 0.9% to 42.4% ± 0.4%) and 18:3 (from 15.6% ± 0.3% to 17.8% ± 0.5%). The only significant change in PE between act1 and act1 lpcat1 lpcat2 was at 4 weeks (Figure 3F), where 16:0 decreased (from 27.0% ± 0.3% to 25.1% ± 0.3%), 18:2 decreased (from 43.1% ± 0.7% to 40.7% ± 0.5%), and 18:3 increased (from 13.6% ± 0.4% to 16.6% ± 0.7%). Supplemental Figures 3, 4, and 5 report the fatty acid composition for all other lipids measured in Figure 2 at 2, 3, and 4 weeks, respectively. The major galactolipid products of the eukaryotic pathway (MGDG and DGDG) did not have any significant changes in fatty acid composition between act1 and act1 lpcat1 lpcat2. The change in abundance of PG measured at 4 weeks (Figure 2C) had only a limited effect on its fatty acid composition with a significant increase in 18:3 in the in act1 lpcat1 lpcat2 line (from 30.7% ± 0.3% to 32.3% ± 0.1%).
Figure 3.
Fatty Acid Composition of PC and PE Across Leaf Development From Wild-Type And Mutant Lines.
The fatty acid composition of PC [(A) to (C)] and PE [(D) to (F)] were determined at three developmental stages: 2 weeks [(A) and (D)], 3 weeks [(B) and (E)], and 4 weeks [(C) and (F)] after germination. The data represent the average and se of 2–4 biological replicates. Significant (p-value < 0.05, Two-way ANOVA with multiple comparisons) differences within individual lipid abundances between act1 and act1 lpcat1 lpcat2 are indicated by asterisks above the bars.
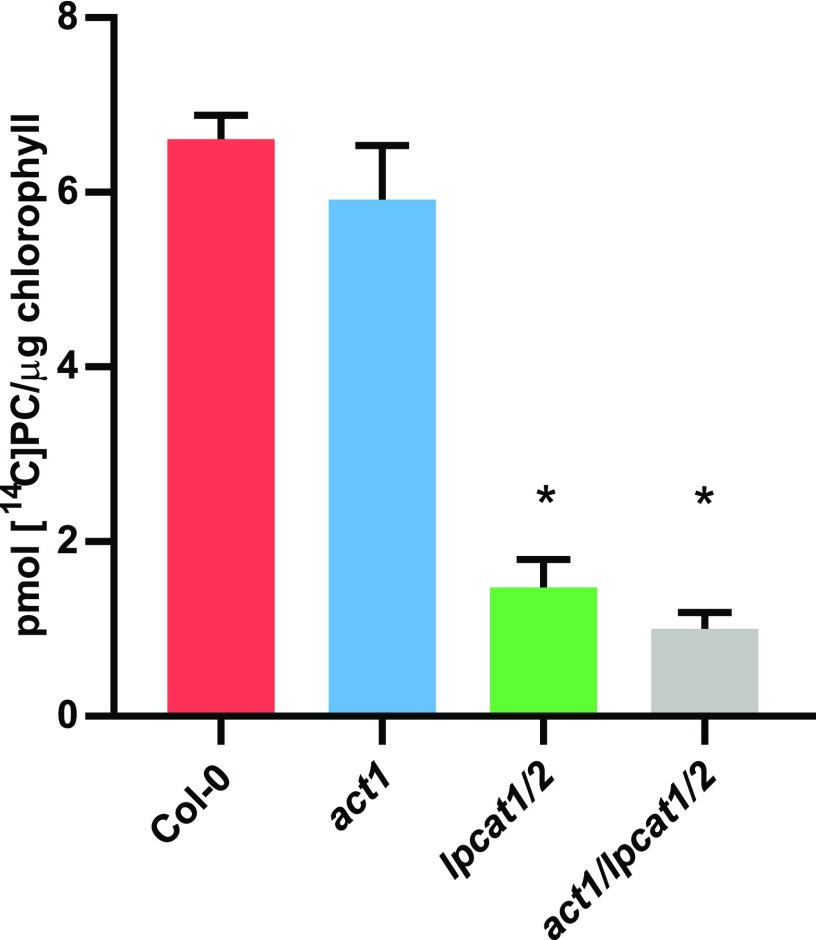
Together, the limited effect of the act1 lpcat1 lpcat2 triple mutation compared with the act1 mutation alone on plant growth and lipid accumulation across leaf development suggests that LPCAT1 and LPCAT2 are not essential for eukaryotic pathway lipid metabolism when the prokaryotic pathway is limiting. However, other lysophospholipid acyltransferases (such as LPEAT1 and LPEAT2) have demonstrated LPCAT activity in vitro (Stålberg et al., 2009; Jasieniecka-Gazarkiewicz et al., 2016) and are expressed at similar levels to LPCAT1 LPCAT2 in leaves (Supplemental Figure 1). Thus, it is possible that other lysophospholipid acyltransferases may compensate for the loss of LPCAT activity in the act1 lpcat1 lpcat2 background. In vitro LPCAT activity has also been associated with multiple subcellular membrane fractions (Bessoule et al., 1995; Tjellström et al., 2012; Wang et al., 2014). To measure if the chloroplast-associated LPCAT activity that has been hypothesized to be involved in the eukaryotic pathway of galactolipid synthesis (Mongrand et al., 1997, 2000; Moreau et al., 1998) is actually reduced in the lpcat1 lpcat2 mutants, we performed LPCAT assays on chloroplasts isolated from each plant line (Figure 4). The controls Col-0 and act1 did not exhibit a significant difference in the LPCAT activity that produced [14C]PC from the addition of LPC and [14C]oleoyl-CoA to the isolated chloroplasts. However, [14C]PC production was reduced ∼85% in act1 lpcat1 lpcat2 from wild-type levels (Figure 4).
Figure 4.
LPCAT Activity in Isolated Chloroplasts.
Isolated chloroplasts from Col-0, act1, lpcat1 lpcat2, and act1 lpcat1 lpcat2 were incubated with 1 mM soy LPC, and 13.6 µM [14C]oleoyl-CoA for 30 min at 30°C and radioactivity incorporated into PC measured. The data represent the average and se of three independent assays from chloroplasts isolated from each line. Significant (p-value < 0.05, Student’s t test) differences from the Col-0 control are indicated by asterisks above the bars.
There was also no significant difference in [14C]PC synthesis between lpcat1 lpcat2 and act1 lpcat1 lpcat2 lines. It is not clear if the residual [14C]PC synthesis within the lpcat1 lpcat2 backgrounds is chloroplast-localized LPCAT activity, or if it is due to the activity of other lysophospholipid acyltransferases from partial contamination of the isolated chloroplasts with other cellular membrane fractions (Larsson et al., 2007; Stålberg et al., 2009; Bulat and Garrett, 2011; Jasieniecka-Gazarkiewicz et al., 2016). Nevertheless, the major chloroplast-associated LPCAT activity in the act1 lpcat1 lpcat2 triple mutant was mostly eliminated (Figure 4), and it had little to no effect on growth or leaf galactolipid accumulation (Figure 2; Supplemental Figure 2), suggesting LPCAT activity is not an essential part of eukaryotic pathway galactolipid synthesis. However, the mass accumulation of MGDG and DGDG does not indicate the metabolic pathway of synthesis. To better understand how the loss of the major chloroplast LPCAT activity affects acyl flux out of the chloroplast and through the eukaryotic pathway into galactolipids of the act1 lpcat1 lpcat2 triple mutant, we moved on to an in vivo metabolic labeling approach during the stage of rapid leaf growth (3-week-old plants).
Rapid In Vivo Metabolic Labeling to Characterize the Effect of lpcat1 lpcat2 on the Entry of Nascent Fatty Acids into the Eukaryotic Pathway through Acyl Editing
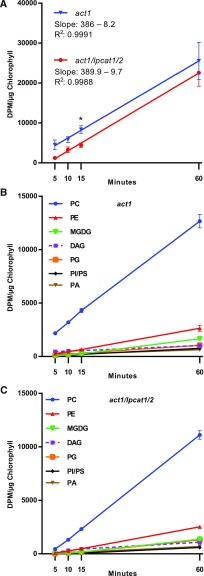
Newly synthesized fatty acids produced in the stroma of the plastid are exported as FFA and esterified to coenzyme A in the ER for use by the various acyltransferases of the eukaryotic pathway (Li-Beisson et al., 2013). Rapid metabolic labeling experiments in leaves, seeds, and plant suspension cells have demonstrated that nascent fatty acids exported from the plastid are predominantly directly incorporated into PC through an LPCAT-type reaction within the acyl-editing cycle, rather than first esterified to glycerol-3-phosphate through de novo glycerolipid biosynthesis (Bates et al., 2007, 2009, 2012; Tjellström et al., 2012; Yang et al., 2017). This rapid incorporation into PC may take place on the chloroplast surface where significant LPCAT activity resides and ER-plastid connection sites are present (Andersson et al., 2007; Tjellström et al., 2012; Botella et al., 2017). We demonstrated that the lpcat1 lpcat2 knockout eliminates most of the chloroplast-associated LPCAT activity (Figure 4). To determine if the knockout of lpcat1 lpcat2 also affects the initial incorporation of nascent fatty acids into the eukaryotic pathway through PC acyl editing, we followed the continual incorporation of [14C]acetate into fatty acid synthesis and lipid assembly (Allen et al., 2015) in developing leaves of act1 and act1 lpcat1 lpcat2 over a short time course from 5 to 60 min (Figure 5). [14C]acetate incorporation into total lipids was linear and had the same rate of accumulation in each line (Figure 5A). However, on average act1 had more total disintegrations per minute per microgram chlorophyll than act1 lpcat1 lpcat2. Higher accumulation of label but with the same rate may be due to a difference in [14C]acetate concentration in the incubation medium for each line, or possibly a small difference in total chlorophyll content used for normalization. Accumulation of 14C labeled fatty acids into different glycerolipids was also linear, indicating continuous biosynthesis of each lipid over the time course in each line (Figures 5B and 5C). Together these results indicate similar rates of total fatty acid biosynthesis in each line, and no indication of lipid degradation during the time course.
Figure 5.
Initial Incorporation of [14C]Acetate-Labeled Nascent Fatty Acids into Leaf Lipids.
Continuous [14C]acetate labeling of 3-week-old leaves over a 1-h time course.
(A) Total 14C accumulation in organic extractable lipids, and linear regression. All data points are average and se from three sets of independently labeled plants. Significant differences (Student’s t test, p-value <0.05) in lipid labeling between lines at each time point are indicated by asterisks above the data points.
(B) and (C) Incorporation of [14C]acetate into major labeled membrane lipids and DAG in the act1 and act1 lpcat1 lpcat2 lines, respectively. All data points are average and se from three sets of independently labeled plants.
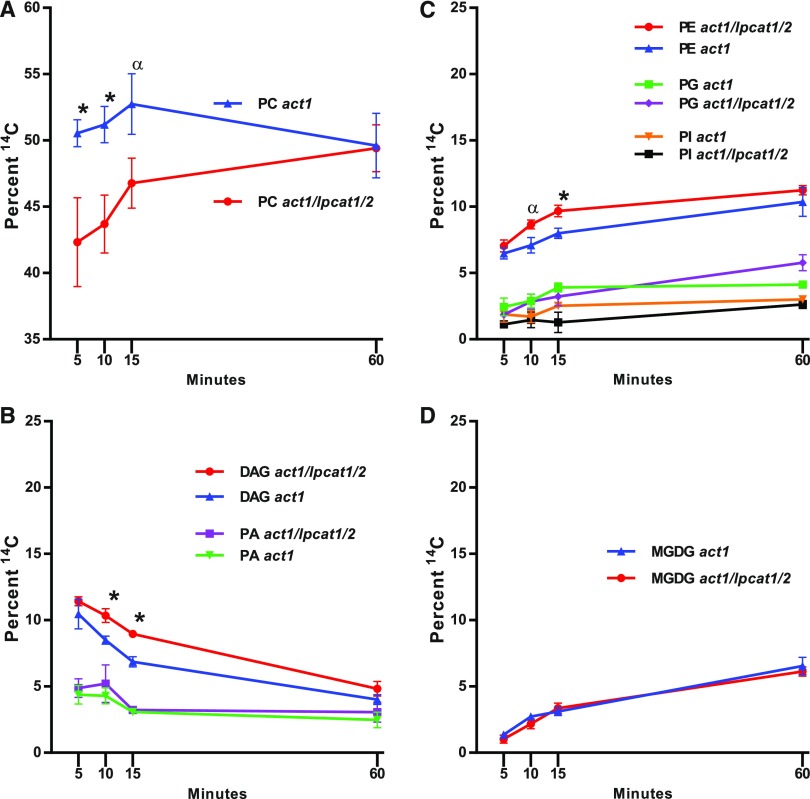
The relative accumulation of labeled lipids in act1 and act1 lpcat1 lpcat2 from Figure 5 is shown in Figure 6. At 60 min the relative labeling between lipids was similar in both lines; however, the initial incorporation of nascent fatty acids into PC of the act1 lpcat1 lpcat2 line was delayed compared with act1 (Figure 6A). The decrease in PC was mostly compensated for by significant increases in DAG and PE (Figures 6B and 6C). There was no significant difference measured between the lines for labeling of PA, PG, and phosphatidylinositol (PI) and phosphatidylserine (PS) together (Figures 6B and 6C), and notably MGDG (Figure 6D).
Figure 6.
Relative Accumulation of [14C]Acetate-Labeled Nascent Fatty Acids into Leaf Lipids.
The relative labeling of individual lipids to the total labeled lipids in each line compared between lines.
(A) PC.
(B) DAG and PA.
(C) PE, PG, and PI/PS.
(D) MGDG.
All data points are average and se from three sets of independently labeled plants. Significant differences (Student’s t test, p-value <0.05) in lipid labeling between lines at each time point are indicated by asterisks above the data points. An α above the data indicates a p-value <0.07.
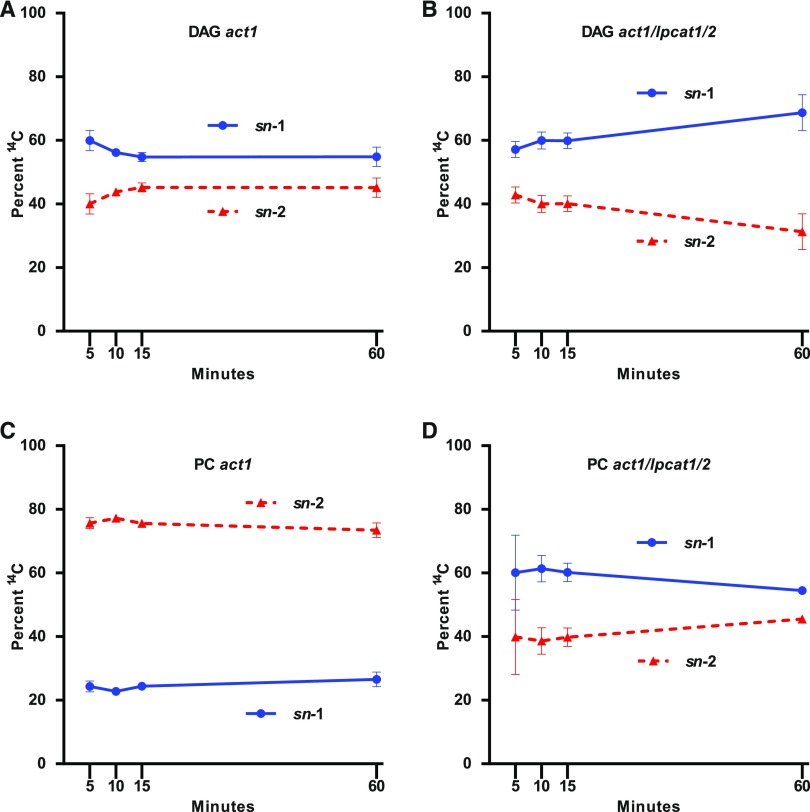
To gain a better understanding of the mechanisms of newly synthesized fatty acid incorporation into the eukaryotic pathway, we characterized the positional distribution of the 14C-labeled fatty acids in PC and DAG across the labeling time course for both act1 and act1 lpcat1 lpcat2 (Figure 7). In act1 DAG contained similar amounts of nascent fatty acids at both stereochemical positions with a slight preference for sn-1 (55–60%) over sn-2 (40–45%) across the time course (Figure 7A). In the PC of act1 most of the nascent fatty acids accumulated at the sn-2 position (∼75%) at all time points (Figure 7C). In act1 lpcat1 lpcat2, DAG stereochemical labeling was similar to that of DAG in act1 with a slight preference for the sn-1 position (Figure 7B). However, PC of act1 lpcat1 lpcat2 was distinctly different than act1 PC, with the stereochemical labeling demonstrating a preference for sn-1 labeling (∼60%) across the time course, similar to DAG from both lines. The rapid incorporation of nascent fatty acids predominantly in sn-2 of PC of act1 (Figures 5B and 7C) is characteristic of newly synthesized fatty acids first entering eukaryotic glycerolipids through PC acyl editing. However, in act1 lpcat1 lpcat2 the initial delay of PC labeling and increase of DAG labeling (Figures 5C, 6A, and 6B), combined with the similar stereochemical labeling of PC and DAG across the time course (Figures 7B and 7D) is consistent with an elimination of nascent fatty acid entry into eukaryotic glycerolipids through PC acyl editing and a reorientation of acyl flux to first move through DAG then into PC. These results indicate that LPCAT1 and LPCAT2 are involved in the direct incorporation of nascent fatty acids into PC through acyl editing as the fatty acids exit the chloroplast, and that other lysophospholipid acyltransferases do not compensate for the loss of LPCAT1 and LPCAT2 activity in the leaf acyl editing cycle.
Figure 7.
Stereochemical Analysis of [14C]Acetate-Labeled Nascent Fatty Acids Incorporation into DAG and PC.
Continuous [14C]acetate-labeled DAG and PC from Figures 5 and 6 were collected and subjected to lipase-based regiochemical analysis of 14C fatty acid locations in the sn-1 or sn-2 position of the glycerol backbone.
(A) act1 DAG.
(B) act1 lpcat1 lpcat2 DAG.
(C) act1 PC.
(D) act1 lpcat1 lpcat2 PC. All data points are average and se from three sets of independently labeled plants.
Pulse-Chase Metabolic Labeling to Characterize the Effect of lpcat1 lpcat2 on the PC–MGDG Precursor–Product Relationship of the Eukaryotic Pathway
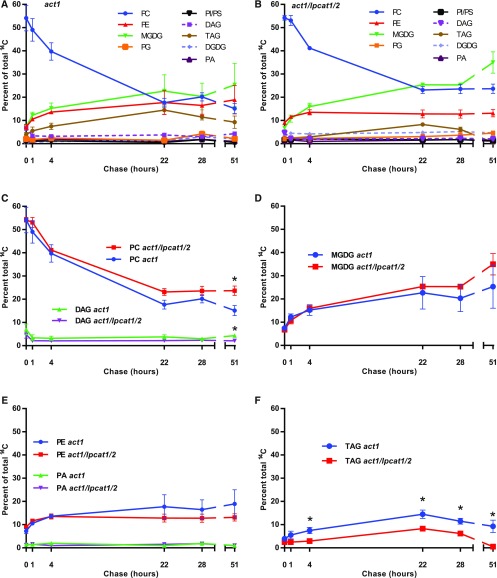
To measure acyl flux through the longer-term precursor–product relationships within leaf lipid metabolism, we performed a pulse-chase metabolic tracing experiment. Three-week-old rosettes from both the act1 and act1 lpcat1 lpcat2 lines were pulsed with [14C]acetate for 15 min, the radioisotope was washed off, and then the samples were chased without radiolabel for an additional 51 h to measure the redistribution of fatty acids as eukaryotic pathway intermediates turn over with time (Figure 8). Similar to the short time point continuous [14C]acetate labeling experiment (Figures 5 and 6), at the end of the pulse most of the newly synthesized radioactive fatty acids were in PC in both the act1 (Figure 8A) and act1 lpcat1 lpcat2 (Figure 8B) lines. During the chase the radioactivity in PC rapidly declined, and the fatty acids were redistributed predominantly into MGDG, followed by PE, and then TAG. All other measured lipids (PG, PA, DAG, PI, PS, and DGDG) contained only minor amounts of radioactivity over the chase period (Figures 8A and 8B). When the relative labeling of individual lipids was compared between the plant lines, there was no statistical difference in the labeling pattern for the major labeled lipids PC, MGDG, and PE. PC levels differed only at the 51-h time point where there was more labeled PC in act1 lpcat1 lpcat2 than in act1 (Figures 8C to 8E). In addition, there was no difference between the lines over the time course for labeling of DAG (Figure 8C) and PA (Figure 8E), which are intermediates of glycerolipid synthesis. This result suggests that the quantitative turnover of PC to provide the substrate for MGDG synthesis within the eukaryotic pathway is not affected by the lpcat1 lpcat2 mutations.
Figure 8.
Pulse-Chase [14C]Acetate Tracking of Leaf Lipid Precursor–Product Relationships.
A 15-min [14C]acetate pulse of 3-week-old whole rosettes was followed by a chase up to 51 h.
(A) and (B) Relative labeling of lipids within act1 (A), or act1 lpcat1 lpcat2 (B).
(C) to (F) Relative labeling of major labeled individual lipids to the total labeled lipids in each line compared between lines.
(C) PC and DAG.
(D) MGDG.
(E) PE and PA.
(F) TAG.
All data points are average and se from three sets of independently labeled plants, except for PA, which had 1–3 replicates. In (C) to (F), significant differences (Student’s t test, p-value <0.05) in lipid labeling between lines at each time point are indicated by asterisks above the data points.
Interestingly, TAG labeling was significantly different between the two plant lines (Figure 8F). TAG accumulation is very minor in leaf tissue under normal circumstances (typically <1% of total lipid mass), but a dynamic small pool of TAG that is constantly synthesized and degraded can be measured through radiolabeling (Fan et al., 2014; Tjellström et al., 2015). Both lines demonstrated the same trend, with the continual increase of labeled fatty acid in TAG until the 22-h time point, and then a decrease through the remaining time course. However, starting at the 4-h time point the act1 line had a significantly higher proportion of labeled fatty acids in TAG than the act1 lpcat1 lpcat2 line did (Figure 8F).
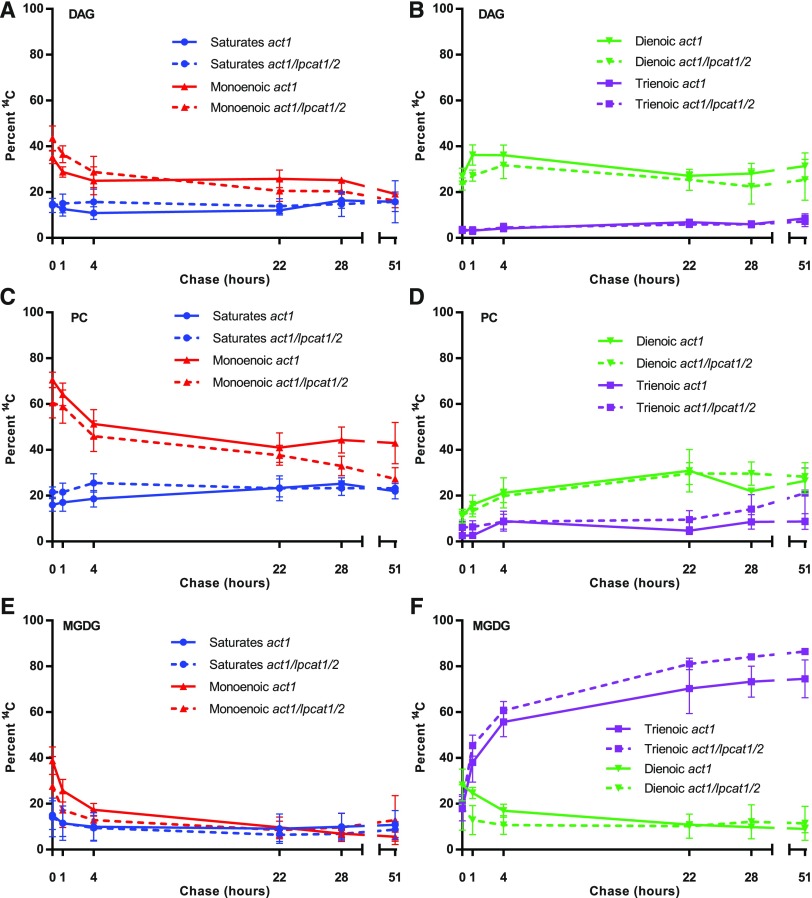
To gain a better understanding of the role of the LPCAT1 LPCAT2 enzymes in leaf eukaryotic pathway metabolism, we analyzed the changes in both the labeled fatty acid composition (Figure 9), and their stereochemical location (Figure 10) within DAG, PC, and MGDG across the [14C]acetate pulse-chase time course (Figure 8). DAG, PC, and MGDG each had a unique profile of 14C-fatty acid accumulation. However, when the composition of labeled fatty acids within each lipid was compared between the act1 and act1 lpcat1 lpcat2 lines there was no significant differences between the lines (Figure 9). There was limited change in the composition of DAG over the 51-h chase period (Figures 9A to 9B), consistent with its role as an intermediate of lipid metabolism that is not a substrate of fatty acid desaturases. However, PC and MGDG had larger changes in composition as both lipid classes are substrates for desaturases, and the acyl groups that are initially incorporated into PC eventually accumulate in MGDG (Figure 8). In PC the major change in composition was a decrease in monoenoic fatty acids, with a concomitant increase in both dienoic and trienoic fatty acids as a proportion of the labeled fatty acids remaining in PC (Figures 9C and 9D). The major change in MGDG was a large increase in trienoic fatty acids (Figure 9F). When the precursor–product relationship of acyl transfer between PC and MGDG (Figure 8) is considered, these results are consistent with the loss of nascent 14C-18:1 initially incorporated into PC (through both desaturation and acyl transfer) and its subsequent accumulation as 14C-18:3 in MGDG. These results are consistent with the current understanding of the eukaryotic pathway (Figure 1). Together with Figure 8, this result suggests the lpcat1 lpcat2 mutations have little to no effect on the flux of total acyl groups (Figure 8) or select fatty acids (Figure 9) from PC to MGDG within the eukaryotic pathway.
Figure 9.
Radiolabeled Fatty Acid Composition of DAG, PC, and MGDG over the [14C]Acetate Pulse-Chase Time Course.
The radiolabeled fatty acids in different lipids from Figure 8 are represented as total saturated fatty acids (e.g., 16:0, 18:0), monoenoic fatty acids (e.g., 18:1), dienoic (e.g., 18:2), and trienoic (e.g., 18:3). The proportion of each fatty acid within each lipid is compared between plant lines with act1 as solid lines, and act1 lpcat1 lpcat2 as dashed lines.
(A) and (B) DAG.
(C) and (D) PC.
(E) and (F) MGDG.
All data points are average and se from three sets of independently labeled plants from Figure 8. Significant differences (Student’s t test, p-value <0.05) in lipid labeling between lines at each time point are indicated by asterisks above the data points.
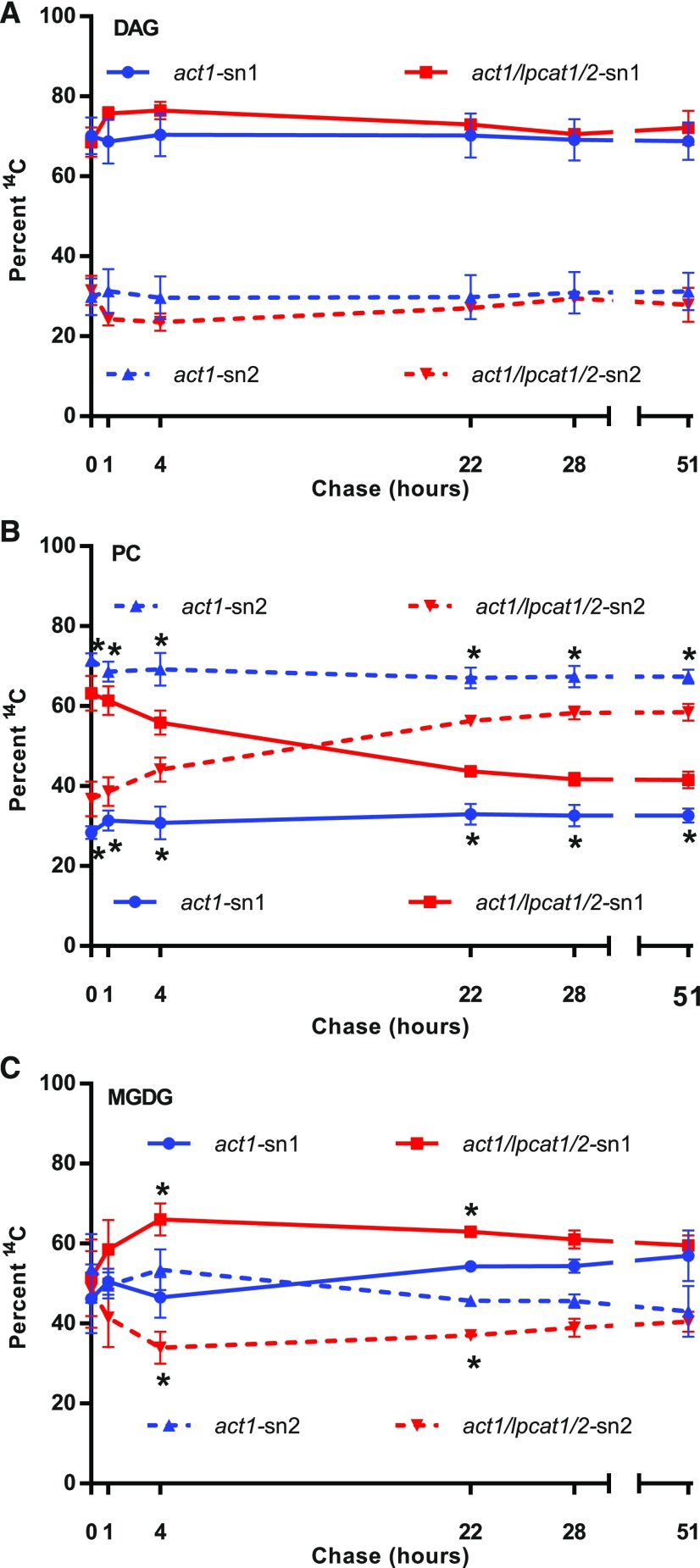
Figure 10.
Stereochemical Analysis of [14C]Acetate-Labeled Fatty Acids within DAG, PC, and MGDG over the Pulse-Chase Time Course.
The sn-1 position is solid lines; the sn-2 position is dashed lines. The act1 samples are blue lines; the act1 lpcat1 lpcat2 samples are red lines.
(A) DAG.
(B) PC.
(C) MGDG.
All data points are average and se from three sets of independently labeled plants from Figure 8. Significant differences (Student’s t test, p-value <0.05) in lipid labeling between act1 and act1 lpcat1 lpcat2 stereochemical positions at each time point are indicated by asterisks next to the act1 blue lines in (B), and next to the act1 lpcat1 lpcat2 red lines in (C).
Despite the similarities of acyl accumulation and composition over the pulse-chase between the act1 and act1 lpcat1 lpcat2 lines, the stereochemical location of the labeled acyl groups revealed significant differences between the lines (Figure 10). During the pulse-chase experiment labeled DAG can represent de novo DAG at early time points. At later time points labeled DAG can represent de novo DAG synthesized with fatty acids removed from other lipids, and DAG derived from membrane lipid or TAG turnover. The 14C-fatty acid stereochemistry in DAG throughout the pulse-chase time course indicated more labeled fatty acids at the sn-1 position than the sn-2 position (Figure 10A), similar to the short time point continuous [14C]acetate labeling experiment (Figures 7A and 7B). There was no significant difference between act1 or act1 lpcat1 lpcat2 at any time point, indicating the loss of LPCAT1 LPCAT2 does not affect initial or prolonged DAG metabolism. The act1 PC stereochemistry was similar to the continuous labeling experiment (Figure 7C) throughout the pulse-chase with more 14C-fatty acids at sn-2 than sn-1 (Figure 10B), consistent with the incorporation of nascent fatty acids into sn-2 PC by LPCAT-mediated acyl editing. The act1 lpcat1 lpcat2 PC stereochemistry was initially similar to the short time point continuous labeling experiment (Figure 7D) with more label at sn-1 than sn-2, consistent with the loss of rapid incorporation of nascent fatty acids into PC through acyl editing. However, after 4 h of chase there was a switch in stereochemistry, with more labeled fatty acids in the sn-2 position rather than the sn-1 (Figure 10B), more similar to PC from the act1 mutant. At all time points the labeled fatty acid stereochemistry in PC was significantly different between the act1 and act1 lpcat1 lpcat2 lines. For MGDG, the early time points indicated similar stereochemical localization of the 14C-fatty acids in both positions; at later time points in the chase more sn-1 labeled fatty acids accumulated in both lines (Figure 10C). It is important to point out that MGDG accumulated as the major labeled lipid by the end of the chase period (Figures 8A and 8B), and that the stereochemistry of labeled acyl groups was similar to that of DAG throughout the time course (Figure 10A) and that of initial PC of the act1 lpcat1 lpcat2 line (Figures 7D and 10B), but not similar to the labeled PC, which remains near the end of the time course in both lines (Figure 10B).
DISCUSSION
Biochemical, genetic, and molecular biology research on plant membrane lipid assembly over the past 40 years has indicated a complicated metabolic network of reactions (Figure 1) that requires the trafficking of intermediates between multiple subcellular compartments to produce the diverse molecular species of lipids crucial to cellular function. While many of the acyltransferases and desaturases involved in lipid assembly have been identified the pathways of acyl trafficking, the identity of lipid intermediates, and the trafficking proteins involved in the eukaryotic pathway, have remained more elusive. Major advances over the past 15 years include the identification of a free fatty acid transporter for export of nascent fatty acids from the plastid (Li et al., 2015), the determination that nascent fatty acids exported from the chloroplast in leaves are predominantly first incorporated into PC by an LPCAT-type reaction of acyl editing rather than the initial attachment to G3P through de novo glycerolipid assembly (Bates et al., 2007), and the characterization of a protein complex involved in the transport of the eukaryotic pathway assembled lipid intermediate into the plastid for galactolipid production (Xu et al., 2005). However, the exact lipid species transported from the ER to the chloroplast has remained unclear; DAG, PA, PC, and LPC have all been suggested. Eukaryotic pathway-derived lipids are characterized by 18-carbon fatty acids at the sn-2 position, whereas the sn-2 acyltransferases of the prokaryotic pathway in the plastid use 16-carbon fatty acids. If LPC is transported to the outer membrane of the plastid, it must be acylated to PC by an LPCAT activity before turnover to DAG or PA to produce the correct molecular species of the eukaryotic pathway. Because LPCAT activity has been implicated in both the trafficking of fatty acids from the chloroplast to the ER in seeds (Bates et al., 2012) and from the ER to the chloroplast in leaves (Mongrand et al., 1997, 2000; Moreau et al., 1998), we sought to gain a better understanding of the roles of LPCAT1 and LPCAT2 in leaf acyl trafficking by analyzing the lipid accumulation and acyl fluxes within the act1 lpcat1 lpcat2 mutant background.
LPCAT1 and LPCAT2 Encode Chloroplast-Localized LPCATs that Are Involved in the Direct Incorporation of Newly Synthesized Fatty Acids into PC through Acyl Editing
The rapid incorporation of nascent fatty acids into predominantly the sn-2 position of PC through acyl editing, rather than through de novo glycerolipid synthesis, was originally characterized in developing pea leaves, an 18:3 plant (Bates et al., 2007). In this study the act1 mutation (Kunst et al., 1988) was used to essentially convert Arabidopsis into an 18:3 plant. Here we demonstrate that in act1 leaves nascent fatty acids are also predominantly incorporated initially into the sn-2 position of PC, consistent with a highly active PC acyl-editing cycle in Arabidopsis leaves (Figures 5B, 6A, and 7C). When LPCAT1 and LPCAT2 were additionally mutated in the act1 lpcat1 lpcat2 line, the chloroplast-associated LPCAT activity was reduced at least 85% (Figure 4), the initial incorporation of nascent fatty acids into PC was reduced concomitantly with an increase into DAG (Figures 6A and 6B), and the stereochemistry of incorporation into PC was completely switched to favor sn-1 in a proportion similar to the rapidly synthesized de novo DAG (Figures 7B and 7D). These results suggest that without chloroplast-associated LPCAT1 and LPCAT2 activity the newly synthesized fatty acids are rerouted to enter PC through the GPAT and LPAT reactions of de novo glycerolipid synthesis rather than LPCAT-based acyl editing in leaves (Figure 11). It is not clear if the residual ∼15% of wild-type LPCAT activity measured in the act1 lpcat1 lpcat2 isolated chloroplasts is due to other chloroplast-associated lysophospholipid acyltransferases (LPLAT; Larsson et al., 2007; Stålberg et al., 2009; Bulat and Garrett, 2011; Jasieniecka-Gazarkiewicz et al., 2016), or due to partial contamination of the isolated chloroplasts with other cellular membrane fractions containing LPLATs (Larsson et al., 2007; Tjellström et al., 2012). However, the complete switch in the in vivo–labeled PC stereochemistry suggests that any other putative chloroplast-associated LPLATs do not compensate for the lack of LPCAT1 and LPCAT2 in the direct flux of nascent fatty acids into PC through acyl editing.
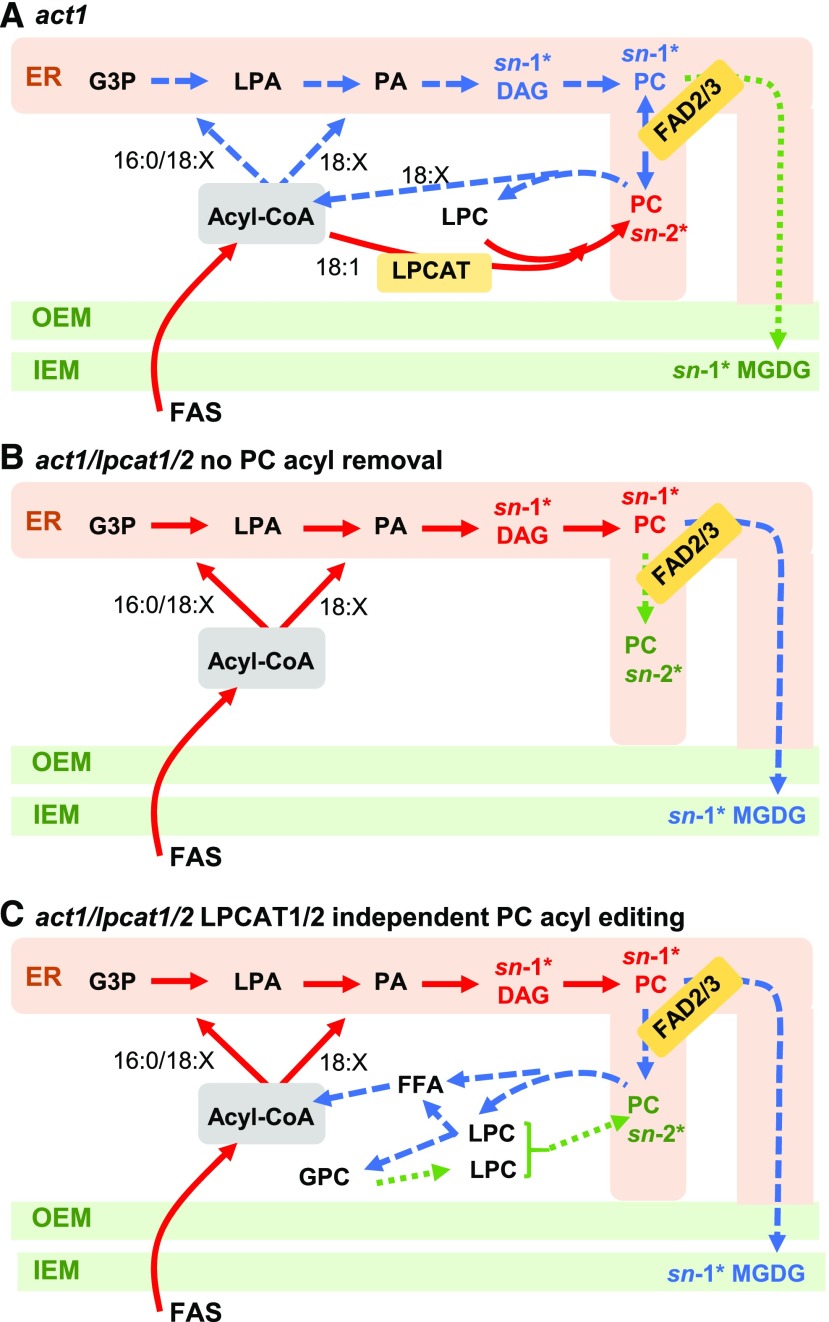
Figure 11.
Models of [14C]Acetate Pulse-Chase Labeling of MGDG Synthesis in act1 and act1 lpcat1 lpcat2 Leaves.
The models indicate the relative rate of labeled fatty acid flux through the eukaryotic pathway of MGDG synthesis within the pulse-chase experiment. Red solid lines represent initial reactions, blue large dashed lines represent the next set of reactions labeled over time, and the green small dashed lines represent the slowest set of reactions labeled over time within each model. Likewise, for the DAG, PC, and MGDG pools, the major labeled stereochemical position at various time points is indicated by the position noted with an asterisk and color coding the same as the lines. No specific time points are intended, and each model color coding is independent from the others, representing only relative labeling within each model.
(A) act1.
(B) act1 lpcat1 lpcat2 with no PC acyl chain removal from residual acyl-editing mechanisms.
(C) act1 lpcat1 lpcat2 with compensating acyl-editing reactions that lead to acyl chain removal from PC and incorporation into the acyl-CoA pool, and the switching of PC labeled stereochemistry from sn-1 to sn-2.
Roles of PC Acyl Editing in Leaves
Here we demonstrate a role for LPCAT1 and LPCAT2 in the direct incorporation of newly synthesized fatty acids into PC as they exit the chloroplast in leaves and show that this role is dispensable in the lpcat1 lpcat2 background (Figures 11A and 11B). However, the role of LPCAT1 and LPCAT2 in leaves likely extends beyond trafficking of nascent fatty acids to PC. PC is the site of ER-localized fatty acid desaturation (Li-Beisson et al., 2013). Previously, LPCAT1 and LPCAT2 were demonstrated to be involved in acyl flux through PC to provide polyunsaturated fatty acids (PUFA) for seed triacylglycerol biosynthesis (Bates et al., 2012; Wang et al., 2012a). Acyl flux through PC for PUFA production is likely also a key role for acyl editing in leaves. Recent work has indicated that the amount of PUFA that accumulate in ER lipids is related to both desaturase activity and the rate of acyl flux through PC. When acyl flux slows down, more PUFA accumulate due to enhanced residence time on PC for desaturation (Maatta et al., 2012; Meï et al., 2015; Botella et al., 2016). While young leaves are expanding, acyl flux through PC is high for membrane lipid production, and little change in PC fatty acid composition was observed in the act1 lpcat1 lpcat2 mutant (Figure 3A). However, as leaves matured, more 18:2 and 18:3 and less 18:1 accumulated in PC as compared with act1 (Figures 3B and 3C), and a similar change was observed in PE (Figure 3F). This result suggests that LPCAT1- and LPCAT2-based acyl editing has homeostatic roles likely involving distribution of PUFA to other lipids across the leaf life cycle. In the lpcat1 lpcat2 mutant background, the plant may compensate for the loss of acyl editing by increasing other mechanisms of acyl flux through PC as indicated in seeds (Bates et al., 2012; Wang et al., 2012a), or providing PUFA from chloroplast sources. Recently, a PG lipase was implicated in the export of PUFA from the chloroplast for seed oil biosynthesis (Wang et al., 2017; Aulakh and Durrett, 2019). The only lipid with a change in abundance in the act1 lpcat1 lpcat2 line was PG (Figure 2C). It is possible that the loss of LPCAT1- and LPCAT2-based acyl editing has activated this or other mechanisms of chloroplast-to-ER trafficking of PUFA. However, the in vivo metabolic labeling experiments (Figures 6 and 8) did not measure a significant difference in PG labeling, suggesting acyl flux through PG may be a minor contribution to ER PUFA content.
The only lipid that had significant differences in [14C]fatty acid accumulation between lines across the pulse-chase time course was TAG (Figure 8F). The act1 lpcat1 lpcat2 line accumulated less labeled TAG than did the act1 line. TAG does not accumulate to high mass levels in leaves, but a small metabolically active pool that is constantly synthesized and turned over is believed to act as a FFA buffer during times of high rates of fatty acid synthesis or stress (Xu and Shanklin, 2016). Recently, phospholipid:diacylglycerol acyltransferase (PDAT) was demonstrated to be a key part of TAG production in Arabidopsis leaves (Fan et al., 2013a, 2013b, 2014). PDAT transfers a fatty acid from the sn-2 position of PC to DAG, producing TAG and LPC. LPCAT works in tandem with PDAT to regenerate PC from the coproduced LPC (Xu et al., 2012). Together, PDAT and LPCAT could lead to channeling of nascent fatty acids exported from the plastid into PC and then TAG during high rates of fatty acid synthesis. The reduced TAG labeling in the act1 lpcat1 lpcat2 mutant is likely due to inefficient PDAT activity without an LPCAT to regenerate the PC substrate. Together, these results suggest a variety of possible roles for LPCAT1- and LPCAT2-based acyl editing in leaves.
MGDG Production from PC Is Independent of LPCAT1 and LPCAT2
LPCAT activity has been implicated in the acylation of LPC transported from the ER to the chloroplast as part of eukaryotic pathway trafficking of substrates for MGDG synthesis. This previous conclusion originally came from in vitro experiments demonstrating the transfer of LPC from isolated microsomes to isolated chloroplasts from leek seedlings, and its acylation to PC by the chloroplast-associated LPCAT activity (Bessoule et al., 1995). Further in vivo metabolic labeling pulse-chase experiments in leek seedlings demonstrated that PC containing predominantly sn-2 labeled fatty acids gave rise to MGDG labeled mostly at sn-1 (Mongrand et al., 1997, 2000). The conclusion was that only the sn-1 fatty acid was transferred to the chloroplast and, combined with the previous in vitro experiments, suggested that LPC was the molecule transferred from the ER to the chloroplast.
The act1 mutation eliminates the prokaryotic pathway of MGDG synthesis. When this mutant was crossed with the tgd1-1 mutant (a part of the OEM to IEM transporter that provides substrate for chloroplast lipid synthesis), no viable double mutants were recovered (Xu et al., 2005), indicating that disruptions of the eukaryotic pathway in the act1 background are lethal. However, we demonstrate that the act1 lpcat1 lpcat2 triple mutation causes at least an 85% reduction in chloroplast LPCAT activity (Figure 4), little to no growth alteration (Supplemental Figure 2), and no effect on the accumulation of galactolipids (Figure 2). Therefore, we conclude that LPCAT1 and LPCAT2, and LPC trafficking are not a key part of eukaryotic pathway galactolipid synthesis. However, we cannot rule out that the residual ∼15% of wild-type LPLAT activity associated with isolated act1 lpcat1 lpcat2 chloroplasts may represent a minimal flow of LPC transported from the ER for other purposes, such as incorporation of PC into the outer leaflet of the chloroplast OEM. If this minimal flow of LPC occurs, the formation of PC must be through a LPLAT other than LPCAT1 and LPCAT2.
To gain a better understanding of the mechanisms involved in the PC–MGDG precursor–product relationship we performed a long-term [14C]acetate pulse-chase experiment. The act1 mutant and act1 lpcat1 lpcat2 triple mutant showed little difference in quantitative turnover of initially labeled PC or in the subsequent incorporation of the labeled fatty acids into MGDG (Figure 8). In addition, the labeled fatty acid composition of DAG, PC, and MGDG was the same between the two lines across the pulse-chase time course (Figure 9). These results further suggest that the lpcat1 lpcat2 mutation does not affect the ER-to-chloroplast trafficking of the eukaryotic pathway. When the stereochemistry of labeled fatty acids in PC and MGDG of act1 were analyzed, we found a similar result to that of the leek seedlings (Mongrand et al., 2000) where PC was mostly sn-2 labeled and the labeled MGDG that accumulated from turnover of PC was mostly sn-1 labeled (Figure 10). From the act1 labeling data alone (in an essentially 18:3 plant, similar to leek), the transfer of LPC would make sense. However, the stereochemical analysis of DAG, PC, and MGDG of the act1 lpcat1 lpcat2 line revealed a different underlying mechanism.
In both the short continuously labeling time course and at the end of the pulse (time 0), the stereochemistry of DAG and PC from act1 lpcat1 lpcat2 were very similar, with more nascent labeled fatty acids at sn-1 than sn-2 (Figures 7 and 10). Therefore, the lack of LPCAT1- and LPCAT2-based acyl editing leads to nascent fatty acid incorporation into PC though eukaryotic de novo glycerolipid assembly, which dictates the stereochemical distribution of fatty acids in DAG and PC. The labeled fatty acid stereochemical distribution that accumulates in MGDG over time in both lines (Figure 10C) is also very similar to the de novo synthesized DAG and PC (Figures 10A and 10B). Therefore, we conclude that the DAG backbone used to synthesize eukaryotic MGDG is derived from a PC pool produced from de novo eukaryotic glycerolipid assembly and is distinct from the pool of PC undergoing LPCAT1- and LPCAT2-based acyl editing.
Figure 11 incorporates the [14C]acetate pulse-chase data onto new models of eukaryotic pathway acyl flux that demonstrate the metabolically distinct pools of PC involved in acyl editing and eukaryotic pathway MGDG production. When LPCAT1 and LPCAT2 are present in act1 the labeling of PC is dominated by the rapid sn-2 acyl editing (Figure 11A). Acyl editing is a constant exchange of acyl groups in PC with the acyl-CoA pool, and it allows the PUFA produced on PC to be used by the GPAT and LPAT reactions of the eukaryotic pathway (Bates et al., 2007, 2009, 2012; Bates, 2016). Therefore, during the pulse-chase the labeled acyl groups can leave PC by acyl editing and are assembled into DAG with more sn-1 labeling than sn-2, which is then used for PC synthesis. If this de novo synthesized pool of PC is rapidly turned over to produce the substrate for MGDG synthesis, MGDG will have the same sn-1-labeled stereochemistry, and it would not have much effect on the stereochemistry of “total labeled PC,” which is dominated by the separate highly labeled acyl-editing PC pool. It is only when LPCAT activity is removed in the act1 lpcat1 lpcat2 triple mutant that the flux through de novo PC to MGDG can be measured separately from the acyl-edited PC pool (Figure 10), which reveals a clear PC–MGDG precursor–product relationship (Figures 11B and 11C). Therefore, the glycerol backbone and both fatty acids (derived mostly from PC acyl editing) that are assembled onto de novo PC are ultimately the “DAG backbone” used for chloroplast MGDG synthesis. This model is also supported by recent characterization of an unusual Δ6 desaturated fatty acid produced transgenically in Arabidopsis leaves at only the sn-2 position of PC. However, the Δ6D fatty acid was redistributed approximately equally to the sn-1 and sn-2 positions of MGDG (Hurlock et al., 2018). Removal of the Δ6D fatty acid from PC by acyl editing (Figure 11A) and its subsequent incorporation into both positions of de novo DAG by GPAT/LPAT activities of the eukaryotic pathway before MGDG synthesis is consistent with our new model of acyl flux.
Changes to Eukaryotic Pathway Acyl Flux within the act1 lpcat1 lpcat2 Background
In wild-type and act1 leaves, the PC acyl-editing cycle may occur by at least three mechanisms (Bates, 2016): (1) both the forward and reverse reactions of LPCAT (Lager et al., 2013; Jasieniecka-Gazarkiewicz et al., 2016); (2) a phospholipase A2 (PLA2) hydrolysis of PC to LPC and a FFA, FFA activation to acyl-CoA by long chain acyl-CoA synthetases, and LPC conversion to PC by the forward LPCAT reaction using a different acyl-CoA [also known as the Lands Cycle (Lands, 1965)]; (3) either mechanism 1 or 2 plus a LPC:LPC transacylase (LPCT; Lager et al., 2015) and a glycerophosphocholine acyltransferase (GPCAT; Lager et al., 2015; Gląb et al., 2016). LPCT transfers an acyl group from one LPC to another producing PC and glycerophosphocholine, which is then converted back to PC by the combined action of GPCAT and LPCAT. In relation to the multiple possible enzymatic mechanisms for acyl editing, two important details from the [14C]acetate pulse-chase experiment must be pointed out. (1) In both act1 and act1 lpcat1 lpcat2, DAG has ∼30% 14C-PUFA at the end of the pulse (time 0 chase), indicating newly synthesized 18:1 is rapidly incorporated into PC for desaturation, and then incorporated into DAG. (2) The act1 lpcat1 lpcat2 PC stereochemical labeling within the pulse-chase experiment switches from more sn-1 label at time 0 to more sn-2 label at the end of the time course. Based on the mechanisms of acyl editing, multiple possible scenarios could explain both the act1 lpcat1 lpcat2 DAG PUFA content and the stereochemistry switch in PC (Figures 11B and 11C).
First, in model Figure 11B the lpcat1 lpcat2 mutation eliminates acyl chain removal from PC by acyl editing (e.g., eliminating acyl-editing mechanism 1). Therefore, the PUFA-labeled DAG represents PC-derived DAG after desaturation. In model 11B, the switch in labeled PC stereochemistry may be through selective molecular species trafficking. Not all PC that is synthesized de novo is turned over for chloroplast lipid synthesis. Some PC has a structural role within various cellular endomembrane systems. It is possible that the turnover of mostly sn-1 labeled molecular species for chloroplast lipid synthesis has left behind a majority of molecular species that contain sn-2 labeled fatty acids.
Second, the lpcat1 lpcat2 mutation eliminates the LPCAT portion of a Lands Cycle, but not the continual generation of LPC by PLA2. In model Figure 11C red and blue arrows only, PLA1- or PLAB-based turnover of LPC generated by the PLA2 would completely remove the fatty acids from PC, leading to complete PC turnover. Considering PC is also undergoing desaturation (Figure 9), the [14C]18:1 originally incorporated into PC will be converted to [14C]18:2 and [14C]18:3 over time. When these fatty acids are removed and then reused for de novo glycerolipid synthesis, it will produce de novo DAG containing PUFA, and the labeled fatty acid stereochemistry in PC will then be determined by the acyl selectivity of GPAT/LPAT. This will lead to different labeled stereochemical molecular species of PC produced from the 14C-PUFA and newly synthesized 12C-18:1 during the chase. In support of this hypothesis, increased expression of various lipases with as-yet uncharacterized functions was measured in developing seeds of the lpcat1 lpcat2 mutant, suggesting the possibility of a modified method to remove PUFA from PC in the lpcat1 lpcat2 background (Wang et al., 2012a).
Third (model Figure 11C, all arrows), PLA2 activity in act1 lpcat1 lpcat2 would produce LPC, which could be converted back to PC by LPCT (as in acyl-editing mechanism 3). This type of reaction would transfer an sn-1 acyl group from one LPC to a second LPC and thus could move a labeled fatty acid from the sn-1 to sn-2 position in PC. The glycerophosphocholine also produced can be reacylated to LPC by GPCAT, and thus could produce a cycle of sn-1/sn-2 acyl switching within the lpcat1 lpcat2 background. From the current experiments it is not clear which of these three possibilities may be occurring, but it is likely that a pool of PC that remains in the ER (model 1, Figure 11B) may be undergoing acyl turnover (models 2 and 3, Figure 11C), which leads to a different stereochemistry of the labeled fatty acids in PC over time (Figure 10B).
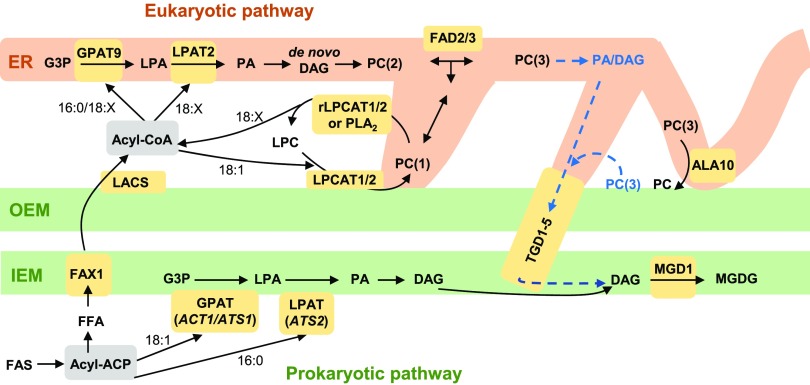
Current Model for Leaf Glycerolipid Synthesis and Trafficking in Wild-Type Arabidopsis Leaves
Figure 12 is a modification of Figure 1 based on our current results and displays the current areas of uncertainty in the eukaryotic pathway of leaf glycerolipid synthesis. The model no longer indicates that LPC can be transferred from the ER to the chloroplast for galactolipid production. It also has three metabolically distinct pools of PC. PC(1) is involved in LPCAT1- and LPCAT2-based acyl editing and may be located at the chloroplast surface or a ER-chloroplast membrane contact site. A membrane contact site might make the most sense because it would allow PC containing newly synthesized 18:1 to diffuse through the ER to the FAD2 and FAD3 enzymes for desaturation. PC(2) is the pool that is produced by de novo PC synthesis within the eukaryotic pathway; it is a substrate for desaturases and provides PC that migrates through the ER to other locations. PC(3) is the pool that is turned over to produce the substrate for MGDG synthesis, and it may have been further desaturated by FAD2 and FAD3 than PC(2). In addition, the exact PC(3)-derived intermediate (DAG or PA) transported to the IEM by the TGD1-5 complex is still unclear. The location of PC(3) turnover is also unclear; it could be the ER, the chloroplast surface, or a ER-chloroplast membrane contact site. Recent characterization of the ALA10 flippase mutant suggests the likely involvement of a membrane contact site for ER-to-chloroplast trafficking (Botella et al., 2016). When the multiple possible roles for membrane contact sites in acyl trafficking are considered, it may be plausible that there are ER-chloroplast contact sites with specific functions in acyl export from the chloroplast involving LPCAT1- and LPCAT2-based acyl editing and separate contact sites with specific function in eukaryotic lipid import into the chloroplast for galactolipid production.
Figure 12.
Updated Model of Wild-Type Arabidopsis Acyl Trafficking within Leaf Glycerolipid Assembly Clarifying the Role of LPCAT1 and LPCAT2.
The model focuses on the trafficking of acyl groups between the chloroplast and the ER for MGDG synthesis. Here the model separates PC involved in acyl editing “PC(1)” from PC synthesized de novo “PC(2)” and PC that provides the substrate for MGDG synthesis “PC(3).” The model also allows that PC acyl editing may take place in the ER or at the chloroplast surface, which could be a way to move acyl groups into the ER by PC movement through membrane contact sites. The PC(3) pool is derived from de novo synthesized PC(2), which may have been further desaturated by FAD2 and FAD3. The substrate for MGDG synthesis may come from turnover of the PC(3) pool in the ER, or turnover of the PC(3) at the chloroplast surface. Key enzymes/transporters are in yellow; uncertain reactions are in blue and have dashed lines. rLPCAT, reverse LPCAT reaction.
The findings presented here strongly enhance our understanding of eukaryotic pathway of membrane lipid production in leaves by demonstrating that (1) LPCAT1 and LPCAT2 encode the major chloroplast-associated LPCAT activities; (2) the major role of LPCAT1 and LPCAT2 is for direct incorporation of newly synthesized fatty acids into PC through acyl editing as the fatty acids are transported out of the chloroplast; (3) LPCAT1 and LPCAT2 activity is not involved in the transfer of LPC from the ER to the chloroplast within eukaryotic pathway galactolipid production; (4) the PC–MGDG precursor–product relationship of acyl flux involves removal of acyl chains from PC by acyl editing before de novo PC synthesis and the subsequent turnover of PC for MGDG production; and (5) PC acyl editing and PC turnover for MGDG production involve metabolically distinct pools of PC. This last result suggests that an underlying spatial organization of distinct PC metabolism may be a key part of the efficient acyl trafficking through the eukaryotic pathway. While there is still uncertainty regarding which PC-derived lipid is trafficked from the ER to the chloroplast, the Arabidopsis act1 lpcat1 lpcat2 line characterized here may be particularly useful for future studies because it allows for metabolic tracing of the PC–MGDG precursor–product relationship without the complications of acyl flux around the PC acyl-editing cycle.
METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) lines used in this study include wild-type Columbia-0 (Col-0), act1 mutant (ACT1, At1G32200; Kunst et al., 1988), lpcat1 lpcat2 double mutant (LPCAT1, AT1G12640; LPCAT2, AT1G63050; Bates et al., 2012), and the act1 lpcat1 lpcat2 triple mutant generated here.
Plant Germination and Growth
Seeds were sterilized in aqueous 10% (v/v) bleach, 27% (v/v) ethanol, and 0.1% (w/v) SDS, rinsed with water 5 times, and applied to germination plates (1× Murashige and Skoog salts, 0.05% [w/v] MES free acid, 1% [w/v] Suc, and 0.8% [w/v] Agar, pH 5.7) in a 0.1% (w/v) agar solution. The plates were incubated at 4°C for 3 d, then placed in a growth chamber under ∼150 µmol photons m−2 s−1 white light using 14 h/10 h day/night cycle at 23°C constant temperature until all lines germinated and produced two true leaves (∼7–10 d). The seedlings were then transferred to soil and placed back into the growth chambers. All plants were watered 3 times a week with on watering consisting of Peter’s NPK 20-20-20 (0.957 g/l) fertilizer solution. During the crossing and harvest of seeds, the plants were grown at the same growth condition but with constant light.
Production of act1 lpcat1 lpcat2 Triple Mutant
The act1 and lpcat1 lpcat2 were crossed via cross-pollination by hand. The screening of act1 was done by identifying absence of hexadecatrienoic acid (16:3) in whole leaf fatty acid methyl esters (FAME) by gas chromatography. The screening of lpcat1 lpcat2 was done by PCR of leaf tissue with primers previously described by Bates et al., (2012) and Xu et al., (2012) with the Phire Plant Direct PCR Mastermix (Thermo Fisher Scientific) as per the manufacturer’s instructions.
Production of FAME and Gas Chromatography
Plant tissue and collected lipid samples were converted to FAME with an internal 17:0 TAG standard by heating to 85°C for 1.5 h in 5% sulfuric acid in methanol. After forcing a phase separation by adding hexane and 0.88% (w/v) potassium chloride, the hexane phase containing the FAME was analyzed by gas chromatography with flame ionization detection on a Shimadzu GC-2010 with a RESTEK Rtx-65 column (30 m, 0.25 mm ID, df = 0.25 µm), with method run parameters of 190°C before for 2 min, and then the temperature increased to 270°C at 10°C/min and held at 270°C for 2 min. The detector was set at 275°C.
Lipid Extraction
The lipid extraction is based off of Hara and Radin (1978). Plant tissues were quenched in 80-85°C isopropanol with 0.01% (w/v) butylated hydroxytoluene (BHT) for 10 min. The tissue was homogenized with polytron and moved to new glass tubes. The polytron was washed with isopropanol and hexane to recover all remaining sample and combined with the ground tissue to a final proportion of hexane:isopropanol:water of 6:4:0.2 (v/v/v). The polytron was washed further between samples to avoid cross-contamination. Lipids were collected into the hexane phase by adding half of the sample volume of 6.6% sodium sulfate. The aqueous phase was back extracted using hexane:isopropanol (7:2, v/v) and combined with the previous hexane extract. The combined organic sample was dried down under nitrogen and resuspended in known volume of toluene. For radiolabeled samples, the lipids were resuspended in chloroform:methanol (2:1, v/v) and subjected to a second phase separation by the addition of 0.88% (w/v) potassium chloride to remove any excess radiolabel. The organic phase was collected, dried down in nitrogen, and resuspended in known volume of toluene. Chlorophyll was measured as in Arnon (1949). Each replicate was a separate extraction of enough leaf material from many plants to make ∼0.3 g fresh weight.
Chloroplast Isolation and LPCAT Assays
Arabidopsis lines were grown on soil in a growth chamber set to 12/12-h day/night cycle, 25°C, and 150 μmol photons m−2 s−1 light. Leaf tissue (25 g) was harvested at 36 d (Col-0, lpcat1 lpcat2) and 44 d (act1, act1 lpcat1 lpcat2) after 16 h of dark treatment. Chloroplast isolation was as described by Kubis et al., (2008), with a modified concentration of 0.33 M sorbitol (instead of 0.3 M) for the isolation and resuspension buffers. The chlorophyll content of isolated chloroplasts was measured as in Arnon (1949) with a Genesys 50 UV-Vis spectrophotometer (Thermo Fisher Scientific).
LPCAT assays were performed on chloroplasts equivalent to 150 µg chlorophyll, in 0.3 mL in a 1.5-mL tube containing 1 mM soy LPC, and 13.6 µM [14C]oleoyl-CoA 55 mCi/mmol (American Radiolabeled Chemicals, Inc.), at 30°C, with 300 rpm mixing for 30 min on a Thermomixer (Eppendorf). The reaction was stopped by adding 1.2 mL CHCl3:methanol:Formic Acid (2:1:0.1, v/v/v) and vigorous vertexing. The assay mixture was transferred to 8-mL glass tubes, and the assay vessel was washed once with a second aliquot of CHCl3:methanol:formic acid and then combined with the previous extract. Addition of 0.3 mL KCl to the mixture and centrifugation at 2000 g produced phase separation, and the lower organic phase was removed to a new 8-mL glass tube. The aqueous phase was washed with 1 mL of CHCl3 and combined with the previous organic phase. The CHCl3 extract was evaporated under a stream of N2 and resuspended in 100 μL CHCl3. Two aliquots of 5 μL were dissolved in 5 mL Eco-Scint liquid scintillation cocktail (National Diagnostics) and radioactivity measured with a Packard 2200CA Liquid Scintillation Counter to quantify the radioactivity in the whole extract. The remaining extract was loaded onto Millipore-Sigma Silica gel 60 thin layer chromatography (TLC) plates in 1-cm bands with nonradioactive lipid standards in adjacent lanes. The TLC plate was developed in CHCl3:methanol:acetic acid:acetone/water (35:25:4:14:2, v/v/v/v/v). After development, the TLC plate was air dried and stained with iodine vapor for visualization of lipid mass bands, and the standards were marked with a radioactive dot. The TLC plate was placed against phosphor imaging screen for 24 h and developed by a GE Typhoon FLA 7000 phosphor imager. Identification of radioactive lipids from the assays was based on comigration with lipid standards. Relative quantification of all radioactive bands was by ImageQuant software version 7.0.
In vivo [14C]Acetate Metabolic Labeling
For both continuous and pulse-chase labeling 3-week-old plant tissue was floated on incubation medium consisting of 20 mM MES, 0.1× Murashige and Skoog salts, and 0.01% (v/v) Tween 20 at pH 5.5, in a shaking water bath at 23°C under ∼150 µmol photons m−2 s−1 white light.
For the continuous labeling, leaves were harvested into the incubation media and placed in the shaking water bath to equilibrate temperature. To start the time course the medium was removed and replaced with incubation medium containing 0.255 mM [1-14C]acetate sodium salt 55 mCi/mmol (American Radiolabeled Chemicals Inc.) at 12.75 µCi/ml. The leaves were incubated for different time points (5, 10, 15, and 60 minutes) after which they were removed from the medium and quenched in isopropanol with 0.01% (w/v) BHT at 85°C before lipid extraction. Fifteen leaves were used per time point replicate. The labeled medium was reused between different time points of same replicate/plant line. Each time point was harvested/radiolabeled separately in replicates of three per plant line.
For the pulse-chase labeling, whole rosettes were harvested by removing the roots, and immediately placed into the incubation medium. Once rosettes for all time points for each replicate/plant line were collected, the incubation medium was removed and replaced with medium containing 0.153 mM [14C]acetate at 7.65 µCi/ml. After 15 min of pulse, the radiolabeled medium was removed, and the tissues were washed 5 times using incubation medium. The 0–time point was immediately collected, and the remaining rosettes were incubated in incubation medium for 1, 4, 22, 28, and 51 h. The collected tissues were quenched in isopropanol with 0.01% (w/v) BHT at 85°C before lipid extraction. Three to four whole rosettes were used per time point replicate. Three replicate pulse-chase time courses were performed per line, and the radiolabeled medium for the pulse was reused for the three replicates within each line.
Analysis of radioactivity of extracts in disintegrations per minute by liquid scintillation counting in EcoScint Original scintillation fluid (National Diagnostics) was on a Beckman Coulter LS 6500 liquid scintillation counter. Relative radioactivity of lipids separated on TLC plates was measured using phosphor imaging on a GE Typhoon FLA7000 and ImageQuant analysis software.
Glycerolipid and FAME Separations by TLC
Polar lipids were separated using TLC plates (20 × 20 cm Analtech Silica gel HL 250 µM thickness) pretreated with 0.15 M ammonium sulfate and baked at 120°C for 3 h. Less than 250 µg lipid was loaded per centimeter and separated in toluene:acetone:water (30:91:7, v/v/v). Neutral lipids were loaded directly onto the untreated EMD Millipore silica gel 60 20 × 20 cm TLC plates and separated in hexane:ether:acetic acid (70:30:1, v/v/v). FAME were loaded onto EMD Millipore plates treated in 7.5% (w/v) silver nitrate in acetonitrile and baked at 100°C for 5 min before use. The FAME were first separated to 75% of plate height in hexane:ether (1:1, v/v) and then fully developed in hexane:ether (9:1, v/v).
Stereochemical Analysis of 14C Labeled Lipids
Lipids were separated by TLC, stained with 0.005% (w/v) primulin in acetone:water (4:1, v/v), and visualized under UV light. Polar lipids were eluted from silica gel with chloroform:methanol:water (5:5:1, v/v/v), and the chloroform phase was collected after phase partitioning with 0.88% (w/v) KCl. Neutral lipids were eluted from silica gel with chloroform/methanol (9:1, v/v). All lipids were dried under nitrogen before being suspended in diethyl ether for lipase digestion.
Stereochemical analysis of DAG and MGDG was done by enzymatic digest with lipase from Rhizomucor miehei (Sigma). Buffer consisting of 50 mM boric acid and 5 mM calcium chloride at 7.8 pH and the lipase were added at 4:1 ratio for DAG and 39:1 ratio for MGDG. The reaction was performed for 15 min for both DAG and MGDG with a goal of 50 to 60% and 20 to 30% digestion, respectively. The reaction was stopped by adding chloroform:methanol (1:1, v/v), and the chloroform phase was collected for TLC. The digested lipids were separated using hexane:diethyl ether:acetic acid (35:70:1.5, v/v/v) for DAG and acetone:toluene:water (91:30:7.5, v/v/v) for MGDG on nontreated silica TLC plates.
Stereochemical analysis of PC was done with Phospholipase A2 from bee venom (Apis mellifera; Sigma). Buffer containing 50 mM Tris-HCl and 5 mM calcium chloride at 8.7 pH and PLA2 were added such that the enzyme was ∼0.25 units. The reaction was performed for 5 min, and the reaction mixture was dried down under nitrogen. The digested lipids were extracted by adding chloroform:methanol:0.15 M acetic acid (38:19:15, v/v/v) and collecting the organic phase. The lipids extracted were separated using chloroform:methanol:acetic acid:water (50:30:8:4, v/v/v/v) on silica TLC plates.
Data Analysis
All calculations from raw data were done in Microsoft Excel. Graphing and statistical analysis done with GraphPad Prism version 7.04.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers LPCAT1 (At1g12640), LPCAT2 (At1g63050), and ACT1/ATS1 (At1g32200).
Supplemental Data
Supplemental Figure 1. Relative gene expression of LPCAT1, LPCAT2, LPEAT1, LPEAT2 in leaves and seeds of wild-type Arabidopsis.
Supplemental Figure 2. Pictures of growth of Col-0, act1, lpcat1 lpcat2, and act1 lpcat1 lpcat2.
Supplemental Figure 3. Lipid fatty acid composition at 2 weeks.
Supplemental Figure 4. Lipid fatty acid composition at 3 weeks.
Supplemental Figure 5. Lipid fatty acid composition at 4 weeks.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Jay Shockey for critical reading of the article and helpful discussions. This work was supported by the National Science Foundation (grant 1930559), and the Agriculture and Food Research Initiative from the USDA National Institute of Food and Agriculture (grant 2017-67013-29481). In addition, this work used the Mississippi IDeA Networks of Biomedical Research Excellence facilities, funded by the National Institutes of Health (grant P20GM103476).
Author Contributions
N.K., B.S.J., and P.D.B. designed the research, performed research, analyzed data, and wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Allen D.K., Bates P.D., Tjellström H. (2015). Tracking the metabolic pulse of plant lipid production with isotopic labeling and flux analyses: Past, present and future. Prog. Lipid Res. 58: 97–120. [DOI] [PubMed] [Google Scholar]
- Andersson M., Dörmann P. (2009). Chloroplast membrane lipid biosynthesis and transport In The Chloroplast: Interactions with the Environment, Sandelius A.S., and Aronsson H., eds (Springer, Berlin, Heidelberg; ), pp. 125–158. [Google Scholar]
- Andersson M.X., Goksör M., Sandelius A.S. (2007). Optical manipulation reveals strong attracting forces at membrane contact sites between endoplasmic reticulum and chloroplasts. J. Biol. Chem. 282: 1170–1174. [DOI] [PubMed] [Google Scholar]
- Arnon D.I. (1949). Copper enzymes in isolated chloroplasts - polyphenoloxidase in Beta-Vulgaris. Plant Physiol. 24: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arondel V., Lemieux B., Hwang I., Gibson S., Goodman H.M., Somerville C.R. (1992). Map-based cloning of a gene controlling omega-3 fatty acid desaturation in Arabidopsis. Science 258: 1353–1355. [DOI] [PubMed] [Google Scholar]
- Aulakh K., Durrett T.P. (2019). The plastid lipase PLIP1 is critical for seed viability in diacylglycerol acyltransferase1 mutant seed. Plant Physiol. 180: 1962–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awai K., Xu C., Tamot B., Benning C. (2006). A phosphatidic acid-binding protein of the chloroplast inner envelope membrane involved in lipid trafficking. Proc. Natl. Acad. Sci. USA 103: 10817–10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien O., Botella C., Chevalier F., Block M.A., Jouhet J., Breton C., Girard-Egrot A., Maréchal E. (2016). Chapter One - New Insights on Thylakoid Biogenesis in Plant Cells In International Review of Cell and Molecular Biology, Jeon K.W., ed (Academic Press; ), pp. 1–30. [DOI] [PubMed] [Google Scholar]
- Bates P.D. (2016). Understanding the control of acyl flux through the lipid metabolic network of plant oil biosynthesis. Biochim. Biophys. Acta 1861 (9 Pt B): 1214–1225. [DOI] [PubMed] [Google Scholar]
- Bates P.D., Durrett T.P., Ohlrogge J.B., Pollard M. (2009). Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol. 150: 55–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates P.D., Fatihi A., Snapp A.R., Carlsson A.S., Browse J., Lu C. (2012). Acyl editing and headgroup exchange are the major mechanisms that direct polyunsaturated fatty acid flux into triacylglycerols. Plant Physiol. 160: 1530–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates P.D., Ohlrogge J.B., Pollard M. (2007). Incorporation of newly synthesized fatty acids into cytosolic glycerolipids in pea leaves occurs via acyl editing. J. Biol. Chem. 282: 31206–31216. [DOI] [PubMed] [Google Scholar]
- Bates P.D., Stymne S., Ohlrogge J. (2013). Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 16: 358–364. [DOI] [PubMed] [Google Scholar]
- Benning C. (2008). A role for lipid trafficking in chloroplast biogenesis. Prog. Lipid Res. 47: 381–389. [DOI] [PubMed] [Google Scholar]
- Benning C. (2009). Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu. Rev. Cell Dev. Biol. 25: 71–91. [DOI] [PubMed] [Google Scholar]
- Bessoule J.J., Testet E., Cassagne C. (1995). Synthesis of phosphatidylcholine in the chloroplast envelope after import of lysophosphatidylcholine from endoplasmic reticulum membranes. Eur. J. Biochem. 228: 490–497. [PubMed] [Google Scholar]
- Block M.A., Jouhet J. (2015). Lipid trafficking at endoplasmic reticulum-chloroplast membrane contact sites. Curr. Opin. Cell Biol. 35: 21–29. [DOI] [PubMed] [Google Scholar]
- Botella C., Jouhet J., Block M.A. (2017). Importance of phosphatidylcholine on the chloroplast surface. Prog. Lipid Res. 65: 12–23. [DOI] [PubMed] [Google Scholar]
- Botella C., Sautron E., Boudiere L., Michaud M., Dubots E., Yamaryo-Botté Y., Albrieux C., Marechal E., Block M.A., Jouhet J. (2016). ALA10, a phospholipid flippase, controls FAD2/fad3 desaturation of phosphatidylcholine in the ER and affects chloroplast lipid composition in Arabidopsis thaliana. Plant Physiol. 170: 1300–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudière L., Michaud M., Petroutsos D., Rébeillé F., Falconet D., Bastien O., Roy S., Finazzi G., Rolland N., Jouhet J., Block M.A., Maréchal E. (2014). Glycerolipids in photosynthesis: composition, synthesis and trafficking. Biochim. Biophys. Acta 1837: 470–480. [DOI] [PubMed] [Google Scholar]
- Browse J., Somerville C. (1991). Glycerolipid synthesis - biochemistry and regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42: 467–506. [Google Scholar]
- Browse J., Warwick N., Somerville C.R., Slack C.R. (1986). Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the ‘16:3’ plant Arabidopsis thaliana. Biochem. J. 235: 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulat E., Garrett T.A. (2011). Putative N-acylphosphatidylethanolamine synthase from Arabidopsis thaliana is a lysoglycerophospholipid acyltransferase. J. Biol. Chem. 286: 33819–33831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorne A.J., Joyard J., Block M.A., Douce R. (1985). Localization of phosphatidylcholine in outer envelope membrane of spinach chloroplasts. J. Cell Biol. 100: 1690–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Yan C., Xu C. (2013a). Phospholipid:diacylglycerol acyltransferase-mediated triacylglycerol biosynthesis is crucial for protection against fatty acid-induced cell death in growing tissues of Arabidopsis. Plant J. 76: 930–942. [DOI] [PubMed] [Google Scholar]
- Fan J., Yan C., Zhang X., Xu C. (2013b). Dual role for phospholipid:diacylglycerol acyltransferase: enhancing fatty acid synthesis and diverting fatty acids from membrane lipids to triacylglycerol in Arabidopsis leaves. Plant Cell 25: 3506–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Zhai Z., Yan C., Xu C. (2015). Arabidopsis TRIGALACTOSYLDIACYLGLYCEROL5 interacts with TGD1, TGD2, and TGD4 to facilitate lipid transfer from the endoplasmic reticulum to plastids. Plant Cell 27: 2941–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Yan C., Roston R., Shanklin J., Xu C. (2014). Arabidopsis lipins, PDAT1 acyltransferase, and SDP1 triacylglycerol lipase synergistically direct fatty acids toward β-oxidation, thereby maintaining membrane lipid homeostasis. Plant Cell 26: 4119–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Głąb B., Beganovic M., Anaokar S., Hao M.-S., Rasmusson A.G., Patton-Vogt J., Banaś A., Stymne S., Lager I. (2016). Cloning of glycerophosphocholine acyltransferase (GPCAT) from fungi and plants; a novel enzyme in phosphatidylcholine synthesis. J. Biol. Chem. 291: 25066–25076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara A., Radin N.S. (1978). Lipid extraction of tissues with a low-toxicity solvent. Anal. Biochem. 90: 420–426. [DOI] [PubMed] [Google Scholar]
- Hurlock A.K., Roston R.L., Wang K., Benning C. (2014). Lipid trafficking in plant cells. Traffic 15: 915–932. [DOI] [PubMed] [Google Scholar]
- Hurlock A.K., Wang K., Takeuchi T., Horn P.J., Benning C. (2018). In vivo lipid ‘tag and track’ approach shows acyl editing of plastid lipids and chloroplast import of phosphatidylglycerol precursors in Arabidopsis thaliana. Plant J. 95: 1129–1139. [DOI] [PubMed] [Google Scholar]
- Jarvis P., Dörmann P., Peto C.A., Lutes J., Benning C., Chory J. (2000). Galactolipid deficiency and abnormal chloroplast development in the Arabidopsis MGD synthase 1 mutant. Proc. Natl. Acad. Sci. USA 97: 8175–8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasieniecka-Gazarkiewicz K., Demski K., Lager I., Stymne S., Banaś A. (2016). Possible role of different yeast and plant lysophospholipid:Acyl-CoA Acyltransferases (LPLATs) in acyl remodelling of phospholipids. Lipids 51: 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen D., Roth C., Wiermer M., Fulda M. (2015). Two activities of long-chain acyl-coenzyme A synthetase are involved in lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis. Plant Physiol. 167: 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.A., Dörmann P. (2004). Green light for galactolipid trafficking. Curr. Opin. Plant Biol. 7: 262–269. [DOI] [PubMed] [Google Scholar]
- Kim H.U., Li Y., Huang A.H.C. (2005). Ubiquitous and endoplasmic reticulum-located lysophosphatidyl acyltransferase, LPAT2, is essential for female but not male gametophyte development in Arabidopsis. Plant Cell 17: 1073–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo A.J.K., Ohlrogge J.B., Pollard M. (2004). On the export of fatty acids from the chloroplast. J. Biol. Chem. 279: 16101–16110. [DOI] [PubMed] [Google Scholar]
- Kubis S.E., Lilley K.S., Jarvis P. (2008). Isolation and Preparation of Chloroplasts from Arabidopsis thaliana Plants In 2D PAGE: Sample Preparation and Fractionation, Posch A., ed (Totowa, NJ: Humana Press; ), pp. 171–186. [DOI] [PubMed] [Google Scholar]
- Kunst L., Browse J., Somerville C. (1988). Altered regulation of lipid biosynthesis in a mutant of Arabidopsis deficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proc. Natl. Acad. Sci. USA 85: 4143–4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBrant E., Barnes A.C., Roston R.L. (2018). Lipid transport required to make lipids of photosynthetic membranes. Photosynth. Res. 138: 345–360. [DOI] [PubMed] [Google Scholar]
- Lager I., Glab B., Eriksson L., Chen G., Banas A., Stymne S. (2015). Novel reactions in acyl editing of phosphatidylcholine by lysophosphatidylcholine transacylase (LPCT) and acyl-CoA:glycerophosphocholine acyltransferase (GPCAT) activities in microsomal preparations of plant tissues. Planta 241: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lager I., et al. (2013). Plant acyl-CoA:lysophosphatidylcholine acyltransferases (LPCATs) have different specificities in their forward and reverse reactions. J. Biol. Chem. 288: 36902–36914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lands W.E.M. (1965). Lipid metabolism. Annu. Rev. Biochem. 34: 313–346. [DOI] [PubMed] [Google Scholar]
- Larsson K.E., Kjellberg J.M., Tjellström H., Sandelius A.S. (2007). LysoPC acyltransferase/PC transacylase activities in plant plasma membrane and plasma membrane-associated endoplasmic reticulum. BMC Plant Biol. 7: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y., Neunzig J., Lee Y., Philippar K. (2017). Plant membrane-protein mediated intracellular traffic of fatty acids and acyl lipids. Curr. Opin. Plant Biol. 40: 138–146. [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y., et al. (2013). Acyl-lipid metabolism. The Arabidopsis Book 11: e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Gügel I.L., Giavalisco P., Zeisler V., Schreiber L., Soll J., Philippar K. (2015). FAX1, a novel membrane protein mediating plastid fatty acid export. PLoS Biol. 13: e1002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Benning C. (2009). A 25-amino acid sequence of the Arabidopsis TGD2 protein is sufficient for specific binding of phosphatidic acid. J. Biol. Chem. 284: 17420–17427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Xu C., Awai K., Jones A.D., Benning C. (2007). A small ATPase protein of Arabidopsis, TGD3, involved in chloroplast lipid import. J. Biol. Chem. 282: 35945–35953. [DOI] [PubMed] [Google Scholar]
- Maatta S., Scheu B., Roth M.R., Tamura P., Li M., Williams T.D., Wang X., Welti R. (2012). Levels of Arabidopsis thaliana leaf phosphatidic acids, phosphatidylserines, and most trienoate-containing polar lipid molecular species increase during the dark period of the diurnal cycle. Front. Plant Sci. 3: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal E., Bastien O. (2014). Modeling of regulatory loops controlling galactolipid biosynthesis in the inner envelope membrane of chloroplasts. J. Theor. Biol. 361: 1–13. [DOI] [PubMed] [Google Scholar]
- Meï C., Michaud M., Cussac M., Albrieux C., Gros V., Maréchal E., Block M.A., Jouhet J., Rébeillé F. (2015). Levels of polyunsaturated fatty acids correlate with growth rate in plant cell cultures. Sci. Rep. 5: 15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongrand S., Bessoule J.J., Cabantous F., Cassagne C. (1998). The C-16: 3/C-18: 3 fatty acid balance in photosynthetic tissues from 468 plant species. Phytochemistry 49: 1049–1064. [Google Scholar]
- Mongrand S., Bessoule J.J., Cassagne C. (1997). A re-examination in vivo of the phosphatidylcholine-galactolipid metabolic relationship during plant lipid biosynthesis. Biochem. J. 327: 853–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongrand S., Cassagne C., Bessoule J.J. (2000). Import of lyso-phosphatidylcholine into chloroplasts likely at the origin of eukaryotic plastidial lipids. Plant Physiol. 122: 845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau P., Bessoule J.J., Mongrand S., Testet E., Vincent P., Cassagne C. (1998). Lipid trafficking in plant cells. Prog. Lipid Res. 37: 371–391. [DOI] [PubMed] [Google Scholar]
- Mueller-Schuessele S.J., Michaud M. (2018). Plastid Transient and Stable Interactions with Other Cell Compartments In Plastids: Methods and Protocols, E. Maréchal, New York N.Y., ed (Springer, US: ), pp. 87–109. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Koizumi R., Shui G., Shimojima M., Wenk M.R., Ito T., Ohta H. (2009). Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc Natl Acad Sci U S A 106: 20978–20783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida I., Tasaka Y., Shiraishi H., Murata N. (1993). The gene and the RNA for the precursor to the plastid-located glycerol-3-phosphate acyltransferase of Arabidopsis thaliana. Plant Mol. Biol. 21: 267–277. [DOI] [PubMed] [Google Scholar]
- Ohlrogge J., Browse J. (1995). Lipid biosynthesis. Plant Cell 7: 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuley J., Lightner J., Feldmann K., Yadav N., Lark E., Browse J. (1994). Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6: 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughan P.G., Slack C.R. (1982). Cellular-organization of glycerolipid metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 33: 97–132. [Google Scholar]
- Schnurr J.A., Shockey J.M., de Boer G.J., Browse J.A. (2002). Fatty acid export from the chloroplast. Molecular characterization of a major plastidial acyl-coenzyme A synthetase from Arabidopsis. Plant Physiol. 129: 1700–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima M., Ohta H., Nakamura Y. (2009). Biosynthesis and function of chloroplast lipids In Lipids in Photosynthesis. (Springer; ), pp. 35–55. [Google Scholar]
- Shockey J., Regmi A., Cotton K., Adhikari N., Browse J., Bates P.D. (2016). Identification of Arabidopsis GPAT9 (At5g60620) as an essential gene involved in triacylglycerol biosynthesis. Plant Physiol. 170: 163–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer S.D., Chen G., Mietkiewska E., Tomasi P., Jayawardhane K., Dyer J.M., Weselake R.J. (2016). Arabidopsis GPAT9 contributes to synthesis of intracellular glycerolipids but not surface lipids. J. Exp. Bot. 67: 4627–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack C.R., Roughan P.G., Balasingham N. (1977). Labelling studies in vivo on the metabolism of the acyl and glycerol moieties of the glycerolipids in the developing maize leaf. Biochem. J. 162: 289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhl U., Stålberg K., Stymne S., Ronne H. (2008). A family of eukaryotic lysophospholipid acyltransferases with broad specificity. FEBS Lett. 582: 305–309. [DOI] [PubMed] [Google Scholar]
- Stålberg K., Ståhl U., Stymne S., Ohlrogge J. (2009). Characterization of two Arabidopsis thaliana acyltransferases with preference for lysophosphatidylethanolamine. BMC Plant Biol. 9: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjellström H., Strawsine M., Ohlrogge J.B. (2015). Tracking synthesis and turnover of triacylglycerol in leaves. J. Exp. Bot. 66: 1453–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjellström H., Yang Z., Allen D.K., Ohlrogge J.B. (2012). Rapid kinetic labeling of Arabidopsis cell suspension cultures: Implications for models of lipid export from plastids. Plant Physiol. 158: 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Froehlich J.E., Zienkiewicz A., Hersh H.L., Benning C. (2017). A plastid phosphatidylglycerol lipase contributes to the export of acyl groups from plastids for seed oil biosynthesis. Plant Cell 29: 1678–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Kazachkov M., Shen W., Bai M., Wu H., Zou J. (2014). Deciphering the roles of Arabidopsis LPCAT and PAH in phosphatidylcholine homeostasis and pathway coordination for chloroplast lipid synthesis. Plant J. 80: 965–976. [DOI] [PubMed] [Google Scholar]
- Wang L., Shen W., Kazachkov M., Chen G., Chen Q., Carlsson A.S., Stymne S., Weselake R.J., Zou J. (2012a). Metabolic interactions between the Lands cycle and the Kennedy pathway of glycerolipid synthesis in Arabidopsis developing seeds. Plant Cell 24: 4652–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Anderson N.S., Benning C. (2013). The phosphatidic acid binding site of the Arabidopsis trigalactosyldiacylglycerol 4 (TGD4) protein required for lipid import into chloroplasts. J. Biol. Chem. 288: 4763–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Xu C., Benning C. (2012b). TGD4 involved in endoplasmic reticulum-to-chloroplast lipid trafficking is a phosphatidic acid binding protein. Plant J. 70: 614–623. [DOI] [PubMed] [Google Scholar]
- Williams J.P., Imperial V., Khan M.U., Hodson J.N. (2000). The role of phosphatidylcholine in fatty acid exchange and desaturation in Brassica napus L. leaves. Biochem. J. 349: 127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Fan J., Froehlich J.E., Awai K., Benning C. (2005). Mutation of the TGD1 chloroplast envelope protein affects phosphatidate metabolism in Arabidopsis. Plant Cell 17: 3094–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Fan J., Riekhof W., Froehlich J.E., Benning C. (2003). A permease-like protein involved in ER to thylakoid lipid transfer in Arabidopsis. EMBO J. 22: 2370–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Shanklin J. (2016). Triacylglycerol metabolism, function, and accumulation in plant vegetative tissues. Annu. Rev. Plant Biol. 67: 179–206. [DOI] [PubMed] [Google Scholar]
- Xu C., Yu B., Cornish A.J., Froehlich J.E., Benning C. (2006). Phosphatidylglycerol biosynthesis in chloroplasts of Arabidopsis mutants deficient in acyl-ACP glycerol-3- phosphate acyltransferase. Plant J. 47: 296–309. [DOI] [PubMed] [Google Scholar]
- Xu J., Carlsson A.S., Francis T., Zhang M., Hoffman T., Giblin M.E., Taylor D.C. (2012). Triacylglycerol synthesis by PDAT1 in the absence of DGAT1 activity is dependent on re-acylation of LPC by LPCAT2. BMC Plant Biol. 12: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Wang G., Li J., Bates P.D., Wang X., Allen D.K. (2017). Phospholipase Dζ enhances diacylglycerol flux into triacylglycerol. Plant Physiol. 174: 110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C., Andersson M.X., Zhang H., Aronsson H. (2015). Phosphatidylcholine is transferred from chemically-defined liposomes to chloroplasts through proteins of the chloroplast outer envelope membrane. FEBS Lett. 589: 177–181. [DOI] [PubMed] [Google Scholar]
- Yu B., Wakao S., Fan J., Benning C. (2004). Loss of plastidic lysophosphatidic acid acyltransferase causes embryo-lethality in Arabidopsis. Plant Cell Physiol. 45: 503–510. [DOI] [PubMed] [Google Scholar]
- Zhao L., Katavic V., Li F., Haughn G.W., Kunst L. (2010). Insertional mutant analysis reveals that long-chain acyl-CoA synthetase 1 (LACS1), but not LACS8, functionally overlaps with LACS9 in Arabidopsis seed oil biosynthesis. Plant J. 64: 1048–1058. [DOI] [PubMed] [Google Scholar]