Abstract
Porcine parvovirus (PPV) is one of many pathogens responsible for reproductive failure in pregnant sows. Several studies have reported the appearance of new PPV strains that differ from previous isolates both genetically and antigenically. Thus, the protective effects of commercially inactivated vaccines could not be complete. In South America, the information about PPV is limited. Thus, the aim of the present study was to detect and characterize the PPV strains present in 131 mummies or stillbirths from normal deliveries in sows from a commercial swine farm of Argentina that uses the commercial vaccine. PCR results showed that 17/131 were positive to PPV. Ten of these viruses were isolated and sequenced. All viruses were related to the PPV1 sequence (NADL-2), maintaining the amino acid differences in positions 436 (S–P) and 565 (R–K). This study is the first to report the isolation of PPV in Argentina and the results suggest that PPV can cross the placenta even in vaccinated sows, thus affecting some of the fetuses and being able to cause fetal death in sows without reproductive failure. The results also suggest that vaccination only reduces clinical signs and reproductive disorders and may thus not be a perfect tool to manage PPV infection. This study provides information that needs to be studied in depth to improve strategies to prevent and control PPV infection in swine farms.
Keywords: Biotechnology, Immunology, Microbiology, Molecular biology, Veterinary medicine, Porcine parvovirus, Reproductive failures, Molecular characterization
Biotechnology; Immunology; Microbiology; Molecular Biology; Veterinary Medicine; porcine parvovirus, reproductive failures, molecular characterization.
1. Introduction
Porcine parvovirus (PPV) is one of many pathogens responsible for reproductive failure in pregnant sows. Epidemiological studies and diagnostic surveys have demonstrated that PPV is the main causative agent responsible for embryonic and fetal death in swine, causing mummification, stillbirths and delayed return to estrus (Mengeling, 2006). Parvoviruses are taxonomically classifiable into two subfamilies based on their host range: the Parvovirinae, which infect vertebrates, and the Densovirinae, which infect mainly insects and other arthropods (Ni et al., 2014). Viruses that infect swine are classified into Protoparvovirus, Bocaparvovirus, Copiparvovirus and Tetraparvovirus and the most representatives are: porcine parvovirus 1 (PPV1), PPV2, PPV3, PPV4, PPV5, PPV6, porcine bocavirus 1 (PBoV1), PBoV2, PBoV3A, PBoV3B, PBoV3C, PBoV3D and PBoV3E (Cotmore et al., 2019; Gava et al., 2015).
A typical parvovirus is non-enveloped, possesses a small and single-stranded DNA genome of 4–6.3 kb in length, and generally contains two open reading frames (ORFs): ORF1, encoding for a non-structural protein (NS1), and ORF2, encoding at least 2 capsid proteins -VP1 and VP2- (Allander et al., 2005). An additional ORF, named ORF3, which has been identified in some parvoviruses, encodes the accessory protein NP1, a non-structural protein found only in bocaviruses and PPV4 (Cheng et al., 2010; Cheung et al., 2010).
The capsid protein VP2 harbors major antigenic domains of PPV, which can induce PPV neutralizing antibodies. Thus, VP2 is the main target protein to neutralizing antibodies and plays a key role in PPV diagnosis and immune prophylaxis (Xu and Li, 2007; Kong et al., 2014). Despite the high level of conservation in the parvovirus sequence, PPV strains can be distinguished by their different pathogenicity. Some residue substitutions of the VP2 gene between the attenuated strain NADL-2 and the virulent Kresse strain (D378 -G, H383 –Q and S436 –P) have been found to play a crucial role in the function and properties of VP2. These amino acid differences are associated with the pathogenic phenotype of the Kresse strain, and all are located on the surface of the capsid. Besides, five differences have been found to be consistently present in the sequences of the Kresse strain and field isolates (I215 -T, D378 -G, H383 -Q, S436 –P and R565 -K) (Bergeron et al., 1996; Simpson et al., 2002; Shangjin et al., 2009).
Several studies have reported the appearance of new PPV strains that differ antigenically from previous isolates (Soares et al., 2003; Zimmermann et al., 2006). Furthermore, the protective effects of commercially available vaccines have shown lower neutralization activity against some of these new strains (Zeeuw et al., 2007; Józwik et al., 2009). Vaccines against PPV can be attenuated vaccines, inactivated vaccines, subunit vaccines and DNA vaccines. Attenuated vaccines, which have been used in some countries for several years, are expensive, cause side effects, and may revert to a pathogenic strain. However, they are likely to be more reactogenic and immunogenic than inactivated, subunit and DNA vaccines (Lima et al., 2004; Chen et al., 2010). Subunit vaccines elicit only moderate levels of protection and inactivated vaccines show deficiencies of specific cellular immune efficacy and failure to increase immunogenicity of weak antigens. However, despite these latter limitations of inactivated vaccines, they are the most common type of PPV vaccines used (Wang et al., 2012). In Argentina, only commercial inactivated PPV vaccines are available and are thus widely used to control reproductive failure due to PPV.
In South America, information about the subfamily Parvovirinae is limited. Soares et al. (2003) reported an analysis of the diversity of PPV strains in Brazil, by using the partial sequencing of the VP2 encoding gene. In Argentina, only an analysis on the diversity and characterization of canine parvovirus has been reported (Gallo Calderón et al., 2012). It is expected that PPV is present in most of the pig intensive commercial farms as an endemic disease as it occurs in other parts of the world; however, no report about this disease has yet been published in our country.
Based on the above, the objective of this study was to detect and characterize the strains of PPV present in mummies and stillbirth piglets collected from a subclinically infected farm in Argentina.
2. Material and methods
2.1. Sample collection and DNA extraction
A 2500-sow multi-site farrow-to-finish farm located in Santa Fe province, Argentina, was selected for the study. In this farm, the vaccination program against reproductive diseases includes commercial PPV (NADL-2) and leptospira inactivated vaccine during acclimation (170–190 days old) and 14 days before every mating. The farm is free of brucellosis, Aujeszky disease virus, porcine reproductive and respiratory stress syndrome virus and classical swine fever virus. The age of first mating is 230–240 days old.
Stillbirths and mummies from randomly selected normal deliveries (<2% of stillbirths and >11 born alive) were collected. From September to December 2016, a total of 131 mummies and stillborn (that represent 1.5% of the total of mummies and stillborn of the time frame studied) belonging to 74 sows (with more than one parity) were analyzed. The complete fetuses were placed in individual sterile bags, preserved at 4 °C, and taken to our Laboratory (Laboratorio de Virología, Facultad de Ciencias Veterinarias, Universidad Nacional de La Plata, La Plata, Argentina). Samples were preserved at -20 °C until analysis. At the laboratory, the crown-rump length of each fetus was measured and the following formula: days of gestation = length (in mm) x 3 + 21 was applied to estimate the fetal death age (Kirkwood et al., 2012). Thereafter, fetuses were classified into mummies, Type I and Type II stillbirths, according to Christianson (1992). Samples of tonsil, lung, liver, heart and kidney were individually collected from each fetus and immediately pooled and processed for routine virus isolation and PCR detection. Total DNA was extracted from sample homogenates by using the Wizard Genomic DNA Purification Kit (Promega-USA) according to the manufacturer's instructions.
2.2. PPV detection and sequence analysis
PCR detection was used to test all samples to amplify the highly conserved NS1 partial gene by using the PPVm Fw 5′- CTTGGAGCCGTGGAGCGAGC-3′ and PPVm Rv 5′- TGCACAGTTTTCACCAAAGCAGGC-3′ primers. The reaction was carried out in a final volume of 25 μl mixture containing 5X PCR buffer, 10 pmol of dNTPs, 10 pmol of each primer, 1 Units of Go Taq DNA polymerase (Promega) and 7% of DMSO. The reaction conditions were as follows: pre-denaturation at 94 °C for 5 min, followed by 35 cycles of 95 °C for 45 s, 60 °C for 45 s, 72 °C for 45 s, and a final elongation step at 72 °C for 7 min. PCR products were electrophoresed in 2% agarose gels in standard TBE buffer and stained with ethidium bromide. The analytical sensitivity of the PCR was determined using dilutions in base 10 of DNA positive control.
For sequence analysis, the VP2 gene was amplified as described by Soares et al. (2003). The PCR products were purified according to the manufacturer's protocols by using the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA). Sequencing reactions were performed in both directions with the same primers for amplification by PCR, using an automated sequencer (3130xl/3500xl Genetic Analyzer, Applied Biosystems, USA), at the Unidad Genómica of the National Institute of Agricultural Technology (INTA Castelar), Argentina.
The sequences were edited using BioEdit software version 7.2.1. Homology analyses were performed with the BLASTN program (National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov/BLAST/]). For PPV analysis, the partial sequences of VP2 were aligned in the MEGA program version 7.0, using the ClustalW algorithm. The phylogenetic dataset included 10 sequences obtained in this study and 53 sequences from GenBank, including PPV1, PPV2, PPV3 and PPV4 types and several PPV geographically related strains (Table 1). The phylogenetic trees were constructed using MEGA program. The evolutionary history was inferred by using the Maximum Likelihood (ML) method based on the Kimura 2-parameter model. Initial tree(s) for the heuristic search were obtained automatically by applying the Neighbor-Joining and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The nucleotide sequences obtained in this study were submitted to GenBank and accession numbers are shown in Table 1.
Table 1.
Details of the sequences used in this study.
2.3. PPV isolation
The methodology used was adapted according to Trindade et al. (2011) and Soares et al. (1999). Virus isolation was attempted with cloned porcine kidney (CPK) cells maintained in minimal essential medium (MEM) supplemented with L-glutamine and 10% fetal bovine serum. Infection of cells was performed simultaneously with cell disruption using trypsin. Confluent monolayer cultures of CPK cells grown in six-well plates were inoculated with serial decimal dilutions (10−1 to 10−3) of a positive PCR tissue pool in MEM. The plates were incubated at 37 °C for 60 min in an atmosphere of 5% CO2. After removing the inoculum, trypsin solution was added and incubated for 5 min. The cells were collected and transferred into T25 cell culture flasks. The cell culture was reincubated at 37 °C and examined daily for cytopathic effects until the fifth day. Then, two more repetitions using the same method were made for each sample and finally presence of PPV was evaluated by PCR and then, positive supernatants were titrated by the hemagglutination test. Guinea pig bloods were collected into Alsever's solution and stored at 4 °C. Erythrocytes were washed 3 times and final suspensions made up to 0.6% using phosphate buffered saline (PBS) at pH 7.2 was used as diluent throughout. Positive virus suspensions were stored at -70 °C.
3. Results
3.1. Samples and PCR
A total of 131 fetuses belonging to 74 sows (with more than one parity) were analyzed. Thirty-two percent (42/131) of the fetuses were classified as mummies, 38.17% (50/131) as type I stillbirths and 29.77% as a type II stillbirths (39/131). The mean age of the fetuses was 92.1 days old (range = 48–116). PCR amplification products were observed in 17 out of the 131 fetuses analyzed. The sensitivity of PCR for PPV was determined in 4 × 10−6 ng/μl.
3.2. Phylogenetic analysis
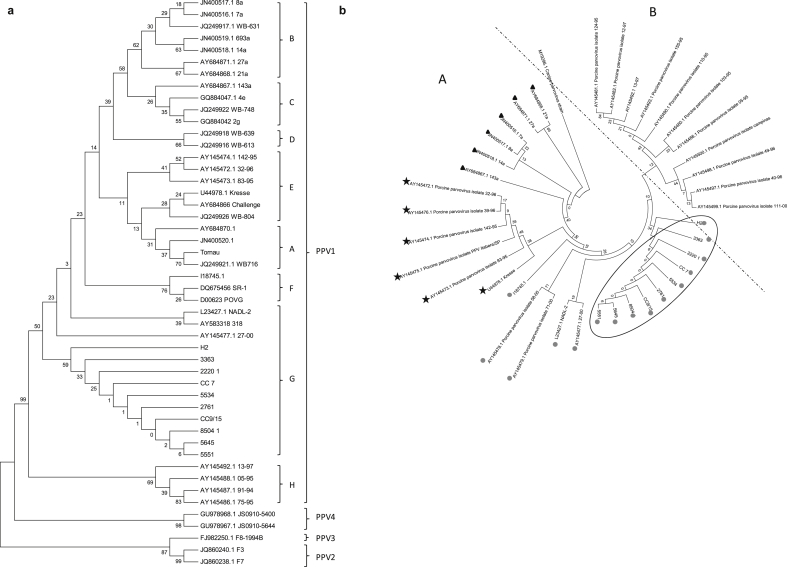
Phylogenetic analysis demonstrated that all Argentinean strains showed no differences between them and had high similarity to the PPV1 sequences. The phylogram obtained by ML analysis of the partial VP2 gene (494bp) showed the grouping of the PPV1, PPV2, PPV3 and PPV4 sequences (Fig. 1a). In accordance with previous reports by Cadar et al. (2012), the following clusters were observed within the PPV1 strains: cluster A, containing two highly virulent strains (Tornau AY684870.1 and JN400520.1) and one wild boar strain (WB-716); cluster B, including several highly virulent strains from Germany, one domestic pig strain from Austria, and one wild boar strain from Romania (WB-631); cluster C, grouping domestic pig strains from the USA, Germany and Europe and one wild boar strain from Romania; cluster D, grouping only two wild boar strains (WB-613 and WB-639); cluster E, grouping the highly virulent Kresse strain together with the challenge UK strain separately from a wild boar strain (WB-804) and another group of sequences represented by Brazilian domestic pig strains; cluster F, including a vaccine strain (I18745.1) and other strains from Spain and China; cluster G, represented by one Brazilian strain, one Chinese strain and the NADL-2 strain and all the Argentinean strains obtained in this study; and cluster H, including only Brazilian domestic pig strains. The PPV2 sequences (JQ860238.1 and JQ860240.1), the PPV3 sequence named F8–1994B, and the PPV4 strains (GU978968.1 and GU978967.1) grouped separately. A new phylogram was obtained when including the Brazilian sequences and the VP2 sequence of canine parvovirus (M19296.1) as outgroup. The phylogenetic tree showed two defined groups of sequences named Groups A and B, with similar distributions, as described by Soares et al. (2003) (Fig. 1b). Group A included three subgroups: subgroup a, including the Argentinean strains forming a separate group, NADL-2 and I18745.1 vaccine strains, and three Brazilian strains associated with mummification as clinical presentation (sequences represented as  ); subgroup b, represented by the highly virulent Kresse strain together with Brazilian strains, forming a group Kresse as described by Soares et al. (2003), and two other Brazilian strains named 39–96 and Itabera/SP in this study (sequences represented as
); subgroup b, represented by the highly virulent Kresse strain together with Brazilian strains, forming a group Kresse as described by Soares et al. (2003), and two other Brazilian strains named 39–96 and Itabera/SP in this study (sequences represented as  ); and subgroup c (sequences represented as
); and subgroup c (sequences represented as  ), formed by virulent German strains represented as Cluster B in Fig. 1a. In contrast to that described by Soares et al., when including the Argentinean strains and other Brazilian strains, some Brazilian strains (103–95, 05–95, 49–96 and 111–00) clustered within group B and not within group A.
), formed by virulent German strains represented as Cluster B in Fig. 1a. In contrast to that described by Soares et al., when including the Argentinean strains and other Brazilian strains, some Brazilian strains (103–95, 05–95, 49–96 and 111–00) clustered within group B and not within group A.
Fig. 1.
(a) Phylogenetic tree constructed by the Maximum Likelihood (ML) method based on the Kimura 2-parameter model based on partial DNA sequence of the VP2 gene. The tree shows the grouping of the PPV1, PPV2, PPV3 and PPV4 sequences. (b) Phylogenetic tree constructed by the Maximum Likelihood (ML) method based on the Kimura 2-parameter model based on partial DNA sequence of the VP2 gene. This tree included 22 Brazilian sequences and the VP2 sequence of canine parvovirus as outgroup. The phylogenetic tree shows two defined groups, A and B. Group A included three subgroups: subgroups a (sequences represented as ); subgroup b (sequences represented as
); subgroup b (sequences represented as  ); and subgroup c (sequences represented as
); and subgroup c (sequences represented as  ). The Argentinean strains are marked with a circular line.
). The Argentinean strains are marked with a circular line.
The phylogenetic tree from the amino acid sequence analysis maintained the same distribution of groups, differing only in the branch positions (data not shown). The analysis of differences between the NADL-2 and Kresse strains described by Bergeron et al. (1996) and Simpson et al. (2002) allowed observing that all Argentinean strains the as NADL-2 strain maintained the amino acid differences in the amino acid positions 436 (S–P) and 565 (R–K).
3.3. PPV isolation
Virus isolation was attempted with CPK cells. Only one strain (CC7) showed evident cytopathic effect. Besides, nine other viruses were isolated but showed no cytopathic characteristics. They were confirmed as PPV by PCR and the hemagglutination test with titer of hemagglutination between 1:64 and 1:256.
4. Discussion
PPV is found in almost all pig-breeding countries and PPV1 is considered endemic in swine herds worldwide. The main clinical sign of PPV infection in non-immune sows is reproductive failure, characterized by embryonic and fetal infection and death, usually without any clinical signs in the dam (Mengeling et al., 1979). Since 80′ parvovirus vaccination is widespread in Argentina swine farms and serological evidence of PPV infection were frequently observed. However, this is the first report of PPV isolation and characterization. In recent years there are numerous studies that showed genetic variation of PPV. Moreover, a constant increase in mummies and stillborn are observed in several parts of the world and also in Argentina. In this context it is imperative to study PPV isolates to identify risk of vaccine failure associate to evolution of PPV and the field efficacy of currently used vaccines. This study explored the molecular characteristics of PPV strains in Argentina and of the presence of PPV from fetuses and stillbirths delivered by sows without reproductive failure in Argentina. All the samples analyzed in this study were from fetal tissues of normal deliveries of vaccinated sows. The detection of PPV in fetal tissues (17/131) suggests that PPV can cross the placenta in vaccinated sows, thus affecting some of the fetuses and causing fetal death. This result is in agreement with previous experimental studies showing that an inactivated whole-virus vaccine did not protect against PPV infection (Józwik et al., 2009; Foerster et al., 2016). Differences in PCR-positive samples from fetal tissues between these previous studies an our present study could be associated with the type of samples analyzed; Józwik et al. (2009) and Foerster et al. (2016) analyzed only lung and kidney samples, whereas we studied tonsil, heart and liver samples, according to other studies (Streck et al., 2013). In this study, the detection of PPV in mummies and stillbirths collected from a subclinically infected farm which uses the PPV inactivated vaccine suggests, as expressed by other authors, that vaccination may not be a perfect tool to manage PPV infection and that it only reduces clinical signs and reproductive disorders (Józwik et al., 2009; Foerster et al., 2016). It is thus necessary to continue exploring the complete genome of PPV isolates and perform new analyses that can demonstrate this hypothesis.
The biological significance of the sequence variation seen among the isolates and vaccine strains is not clear yet (Zimmerman et al., 2006), and the origin and role of the PPV vaccine strains associated with mummification are unknown. The vaccines against PPV currently available are based on inactivated virus preparations of PPV1 strains isolated some 30 years ago. Experimental infection was demonstrated by the high mortality among the fetuses of sows infected with the PPV-27a strain compared with sows infected with other strains as well as with NADL-2 (Zeeuw et al., 2007; Józwik et al., 2009; Foerster et al., 2016).
Since field strains of PPV are difficult to be propagated in conventional cell lines, molecular studies of DNA have been carried out directly on clinical material. In the present study, we were able to isolate 10/17 PPV PCR-positive samples. Then, we analyzed the partial VP2 gene from the 10 PPV strains isolated and other published sequences (Table 1). The phylogram obtained of the partial VP2 gene showed the grouping of the PPV1, PPV2, PPV3 and PPV4 sequences (Fig. 1a). Cadar et al. (2013) described that comparative phylogeny of PPVs has placed PPV2 phylogenetically closer to PPV1, and PPV3 and PPV4 more distantly. In our analysis, the two sequences considered as PPV2 were topologically more distant from PPV1 sequences, probably due to the few sequences included. As shown by Cadar et al. (2013), PPV3 and PPV4 sequences grouped separately and different clusters were observed within PPV1 strains. The Argentinean strains grouped with NADL-2 vaccine strains together with the 27-00 Brazilian strain in the same cluster, as previously reported also by Soares et al. (2003), and they gave origin to group G. Cotmore et al. (2014) described that the genomic organization of PPV6 is similar to that of PPV5 but not to that of other PPV types, which contain an ORF3 in the middle of the viral genome. Phylogenetic analysis demonstrated that PPV6 and PPV5 form a distinct branch that is genetically different from viruses of previously defined genera in the subfamily Parvovirinae, and might be classified into the novel genus Copiparvovirus. For this reason, in our study, we did not include the PPV5 and PPV6 sequences. Rooting the ML tree based on nucleotide sequences using the reference VP2 sequence of canine parvovirus as outgroup, as seen in Fig. 1b, allowed the identification of two groups named Groups A and B, whose distribution was similar to that described by Soares et al. (2003). This analysis allowed defining various subgroups of sequences within group A, where the Argentinean strains formed a separate subgroup associated with the NADL-2 strain and Brazilian strains related to reproductive clinical presentation. Whether this similarity is associated with the detection of PPV in mummies and stillbirths or depends on other factors should be further analyzed. Few differences regarding the groups were detected when including the Argentinean and new Brazilian strains. Four Brazilian strains were within group B, in disagreement with that reported by Soares et al. (2003). In this study, all German strains included formed the subgroup c, all represented by virulent strains. Germany has a long PPV infection history and the genetic variability is complicated. Zimmermann et al. (2006) described a phylogenetic analysis of the full-length VP1 nucleotide sequences of German isolates and showed two defined clusters. In the present study, when we analyzed the amino acid sequence, we also analyzed the differences of residues at the positions described between Kresse and NADL-2 strains. We found that all field strains detected and analyzed have the same residues of NADL-2 at the amino acid positions 436 and 565 in the VP2 fragment. Similar results were obtained by Soares et al. (2003), who found that position 436 is one of the responsible ones for differences in tropism (allotropic determinant) between virulent PPV isolates and the NADL-2 strain. Like Soares, we can conclude that amino acid 436 is the one responsible for tissue tropism, and, based on the results of the present study, we can also conclude that it seems to be the main residue involved in the differentiation of the clusters.
This study showed the presence of PPV DNA in fetal tissues (mummies and stillbirths) of sows without reproductive failure, a fact that suggests a subclinical effect of PPV infection. Moreover, the 10 isolates reported here represent the first report of PPV in Argentina and are related to the strains isolated in Brazil.
This study provides information that needs to be studied in depth so as to improve prevention and control strategies for PPV infection in swine farms.
5. Conclusions
The results provides information about the PPV strains isolated from a 2500-sow multi-site farrow-to-finish farm located in Santa Fe province, Argentina with a vaccination program against reproductive diseases includes commercial PPV.
Declarations
Author contribution statement
M. S. Serena: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
J. A. Cappuccio: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
G.E. Metz, C. G. Aspitia: Performed the experiments.
M. Dibárbora: Analyzed and interpreted the data.
M. Gallo Calderón: Analyzed and interpreted the data; Wrote the paper.
M.G. Echeverría: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by Proyectos de Incentivos Docentes V221 and V260 from Universidad Nacional de La Plata, Argentina; PICT 2015-1232 nd PICT 2017-1075 from Agencia de Promocion Cientifica y Tecnologica, Argentina and PE INTA 1115057 from Instituto Nacional de Tecnologia Agropecuaria.
Competing interest statement
The authors declare no conflict of interest.
Additional information
Data associated with this study has been deposited at GenBank (see Table 1 for accession numbers).
Acknowledgements
The technical assistance of Ms MJ. Vazzano, Mr C. Leguizamón and Mr T. Gilead are gratefully acknowledged. MS Serena and JA Cappuccio contributed equally to this work.
References
- Allander T., Tammi M., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron J., Hébert B., Tijssen P. Genome organization of the Kresse strain of porcine parvovirus: identification of the allotropic determinant and comparision with those of NADL-2 and field isolates. J. Virol. 1996;70:2508–2515. doi: 10.1128/jvi.70.4.2508-2515.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadar D., Cságola A., Kiss T., Tuboly T. Capsid protein evolution and comparative phylogeny of novel porcine parvoviruses. Mol. Phylogenetics Evol. 2013;66:243–253. doi: 10.1016/j.ympev.2012.09.030. [DOI] [PubMed] [Google Scholar]
- Cadar D., Dán A., Tombácz K., Lorincz M., Kiss T., Becskei Z., Spînu M., Tuboly T., Cságola A. Phylogeny and evolutionary genetics of porcine parvovirus in wild boars. Infect. Genet. Evol. 2012;12:1163–1171. doi: 10.1016/j.meegid.2012.04.020. [DOI] [PubMed] [Google Scholar]
- Chen H.Y., Zhao L., Wei Z.Y., Cui B.A., Wang Z.Y., Li X.S., Xia P.A., Liu J.P. Enhancement of the immunogenicity of an infectious laryngotracheitis virus DNA vaccine by a bicistronic plasmid encoding glycoprotein B and interleukin-18. Antivir. Res. 2010;87:235–241. doi: 10.1016/j.antiviral.2010.05.009. [DOI] [PubMed] [Google Scholar]
- Cheng W.X., Li J.S., Huang C.P., Yao D.P., Liu N., Cui S.X., Jin Y., Duan Z.J. Identification and nearly full-length genome characterization of novel porcine bocaviruses. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.K., Wu G., Wang D., Bayles D.O., Lager K.M., Vincent A.L. Identification and molecular cloning of a novel porcine parvovirus. Arch. Virol. 2010;155:801–806. doi: 10.1007/s00705-010-0646-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson W.T. Stillbirths, mummies, abortions, and early embryonic death. Vet. Clin. N. Am. Food Anim. Pract. 1992;8:623–639. doi: 10.1016/s0749-0720(15)30708-8. [DOI] [PubMed] [Google Scholar]
- Cotmore S.F., Agbandje-McKenna M., Canuti M., Chiorini J.A., Eis-Hubinger A., Hughes J., Mietzsch M., Modha S., Ogliastro M., Pénzes J.J., Pintel D.J., Qiu J., Soderlund-Venermo M., Tattersall P., Tijssen P., ICTV Report Consortium ICTV virus taxonomy profile: parvoviridae. J. Gen. Virol. 2019;100:367–368. doi: 10.1099/jgv.0.001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore S.F., Agbandje-McKenna M., Chiorini J.A., Mukha D.V., Pintel D.J., Qiu J., Soderlund-Venermo M., Tattersall P., Tijssen P., Gatherer D., Davison A.J. The family Parvoviridae. Arch. Virol. 2014;159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerster T., Streck A.F., Speck S., Selbitz H.J., Lindner T., Truyen U. An inactivated whole-virus porcine parvovirus vaccine protects pigs against disease but does not prevent virus shedding even after homologous virus challenge. J. Gen. Virol. 2016;97:1408–1413. doi: 10.1099/jgv.0.000446. [DOI] [PubMed] [Google Scholar]
- Gallo Calderón M., Wilda M., Boado L., Keller L., Malirat V., Iglesias M., Mattion N., La Torre J. Study of canine parvovirus evolution: comparative analysis of full-length VP2 gene sequences from Argentina and international field strains. Virus Genes. 2012;44:32–39. doi: 10.1007/s11262-011-0659-8. [DOI] [PubMed] [Google Scholar]
- Gava D., Souza C.K., Schaefer R., Vincent A.L., Cantão M.E., Coldebella A., Ciacci-Zanella J.R. A TaqMan-based real-time PCR for detection and quantification of porcine parvovirus 4. J. Virol Methods. 2015;219:14–17. doi: 10.1016/j.jviromet.2015.03.011. [DOI] [PubMed] [Google Scholar]
- Huang L., Zhai S.L., Cheung A.K., Zhang H.B., Long J.X., Yuan S.S. Detection of a novel porcine parvovirus, PPV4, in Chinese swine herds. Virol. J. 2010;7:333. doi: 10.1186/1743-422X-7-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Józwik A., Manteufel J., Selbitz H.J., Truyen U. Vaccination against porcine parvovirus protects against disease, but does not prevent infection and virus shedding after challenge infection with a heterologous virus strain. J. Gen. Virol. 2009;90:2437–2441. doi: 10.1099/vir.0.012054-0. [DOI] [PubMed] [Google Scholar]
- Kirkwood R.N., Althouse G.C., Yaeger M.J., Carr J., Almond G.W. Diseases of Swine. tenth ed. John Wiley and Sons; West Sussex, UK: 2012. Diseases of the reproductive System; pp. 329–347. [Google Scholar]
- Kong M., Peng Y., Cui Y., Chang T., Wang X., Liu Z., Liu Y., Zhu Y., Luo Y., Tang Q., Feng L., Cui S. Development and evaluation of the rVP-ELISA for detection of antibodies against porcine parvovirus. J. Virol Methods. 2014;206:115–118. doi: 10.1016/j.jviromet.2014.06.008. [DOI] [PubMed] [Google Scholar]
- Lima K.M., dos Santos S.A., Rodrigues J.M., Jr., Silva C.L. Vaccine adjuvant: it makes the difference. Vaccine. 2004;22:2374–2379. doi: 10.1016/j.vaccine.2003.12.030. [DOI] [PubMed] [Google Scholar]
- Mengeling W.L. Diseases of Swine. ninth ed. Blackwell Publishing; Ames, IA, USA: 2006. Porcine parvovirus; pp. 373–385. [Google Scholar]
- Mengeling W.L., Brown T.T., Paul P.S., Gutekunst D.E. Efficacy of an inactivated virus vaccine for prevention of porcine parvovirus-induced reproductive failure. Am. J. Vet. Res. 1979;40:204–207. [PubMed] [Google Scholar]
- Ni J., Qiao C., Han X., Han T., Kang W., Zi Z., Cao Z., Zhai X., Cai X. Identification and genomic characterization of a novel porcine parvovirus (PPV6) in China. Virol. J. 2014;11:203. doi: 10.1186/s12985-014-0203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing L., Lv J., Li H., Tan Y., Hao H., Chen Z., Zhao J., Chen H. The recombinant nonstructural polyprotein NS1 of porcine parvovirus (PPV) as diagnostic antigen in ELISA to differentiate infected from vaccinated pigs. Vet. Res. Commun. 2006;30:175–190. doi: 10.1007/s11259-006-3212-9. [DOI] [PubMed] [Google Scholar]
- Ranz A.I., Manclus J.J., Diaz-Aroca E., Casal J.I. Porcine parvovirus: DNA sequence and genome organization. J. Gen. Virol. 1989;70:2541–2553. doi: 10.1099/0022-1317-70-10-2541. [DOI] [PubMed] [Google Scholar]
- Reed A.P., Jones E.V., Miller T.J. Nucleotide sequence and genome organization of canine parvovirus. J. Virol. 1988;62:266–276. doi: 10.1128/jvi.62.1.266-276.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shangjin C., Cortey M., Segalés J. Phylogeny and evolution of the NS1 and VP1/VP2 gene sequences from porcine parvovirus. Virus Res. 2009;140:209–215. doi: 10.1016/j.virusres.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Simpson A.A., Hébert B., Sullivan G.M., Parrish C.R., Zádori Z., Tijssen P., Rossmann M.G. The structure of porcine parvovirus: comparison with related viruses. J. Mol. Biol. 2002;315:1189–1198. doi: 10.1006/jmbi.2001.5319. [DOI] [PubMed] [Google Scholar]
- Soares M.R., Durigon E.L., Bersano J.G., Richtzenhain L.J. Detection of porcine parvovirus DNA by the polymerase chain reaction assay using primers to the highly conserved nonstructural protein gene, NS-1. J. Virol Methods. 1999;78:191–198. doi: 10.1016/s0166-0934(98)00177-3. [DOI] [PubMed] [Google Scholar]
- Soares M.R., Cortez A., Heinemann M.B., Sakamoto S.M., Martins V.G., Bacci M., Jr., De Campos Fernandes F.M., Richtzenhain L.J. Genetic variability of porcine parvovirus isolates revealed by analysis of partial sequences of the structural coding gene VP2. J. Gen. Virol. 2003;84:1505–1515. doi: 10.1099/vir.0.19011-0. [DOI] [PubMed] [Google Scholar]
- Streck A.F., Homeier T., Foerster T., Fischer S., Truyen U. Analysis of porcine parvoviruses in tonsils and hearts from healthy pigs reveals high prevalence and genetic diversity in Germany. Arch. Virol. 2013;158:1173–1180. doi: 10.1007/s00705-013-1603-0. [DOI] [PubMed] [Google Scholar]
- Szelei J., Liu K., Li Y., Fernandes S., Tijssen P. Parvovirus 4-like virus in blood products. Emerg. Infect. Dis. 2010;16:561–564. doi: 10.3201/eid1603.090746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade I.M.S., Soares Campos G., de Cassia Pimentel R., de Andrade Moura J.C., Sardi S.I. Vol. 5. PUBVET; Londrina: 2011. (Pesquisa sorológica do parvovirus suíno em reprodutoras de rebanhos suínos da Bahia). N. 25, Ed. 172, Art. 1164. [Google Scholar]
- Wang R.N., Wang Y.B., Geng J.W., Guo D.H., Liu F., Chen H.Y., Zhang H.Y., Cui B.A., Wei Z.Y. Enhancing immune responses to inactivated porcine parvovirus oil emulsion vaccine by co-inoculating porcine transfer factor in mice. Vaccine. 2012;30:5246–5252. doi: 10.1016/j.vaccine.2012.05.077. [DOI] [PubMed] [Google Scholar]
- Xu Y.G., Li Y.J. Induction of immune responses in mice after intragastricadministration of Lactobacillus casei producing porcine parvovirus VP2 protein. Appl. Environ. Microbiol. 2007;73:7041–7047. doi: 10.1128/AEM.00436-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeuw E.J., Leinecker N., Herwig V., Selbitz H.J., Truyen U. Study of the virulence and cross-neutralization capability of recent porcine parvovirus field isolates and vaccine viruses in experimentally infected pregnant gilts. J. Gen. Virol. 2007;88:420–427. doi: 10.1099/vir.0.82302-0. [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Ritzmann M., Selbitz H.J., Heinritzi K., Truyen U. VP1 sequences of German porcine parvovirus isolates define two genetic lineages. J. Gen. Virol. 2006;87:295–301. doi: 10.1099/vir.0.81086-0. [DOI] [PubMed] [Google Scholar]