Abstract
All organisms must protect their genome from constantly occurring DNA damage. To this end, cells have evolved complex pathways for repairing sites of DNA lesions, and multiple in vitro and in vivo techniques have been developed to study these processes. In this review, we discuss the commonly used laser microirradiation method for monitoring the accumulation of repair proteins at DNA damage sites in cells, and we outline several strategies for deriving kinetic models from such experimental data. We discuss an example of how in vitro measurements and in vivo microirradation experiments complement each other to provide insight into the mechanism of PARP1 recruitment to DNA lesions. We also discuss a strategy to combine data obtained for the recruitment of many different proteins in a move toward fully quantitating the spatiotemporal relationships between various damage responses, and we outline potential venues for future development in the field.
Keywords: Laser microirradiation, kinetic modelling, accumulation, recruitment to DNA damage sites, DNA repair protein, PARP1, Q-FADD
1. Introduction
Throughout the lifetime of an organism, its genetic material is constantly assaulted by many endogenous and exogenous DNA-damaging agents. Damage to DNA can result in simple base substitutions or more complex changes such as formation of single or double strand DNA breaks (SSBs or DSBs). SSBs commonly arise due to the spontaneous oxidative action of endogenously-formed free radicals, ultimately resulting in a break in one of the two strands of the DNA double helix [1,2]. Such DNA lesions can cause obstruction of the transcription and/or replication machineries. Indeed, if SSBs are missed by the surveillance machinery and remain unrepaired in cycling cells, DNA replication forks can collapse in S-phase cells, resulting in the conversion of SSBs to DSBs. Another direct cause of DSBs is exposure to ionizing radiation. When cells fail to repair DSBs, untoward consequences such as genomic instability and cell death will occur. All living organisms have evolved highly coordinated mechanisms for repairing damaged DNA, thereby safeguarding genome integrity [3–6]. These processes are the subject of intense study.
Early in the process of SSB repair (SSBR), poly (ADP-ribose) polymerase 1 (PARP1) rapidly accumulates at sites of DNA damage. PARP1 is a molecular sensor of various forms of damaged DNA and is enzymatically activated upon binding to damage sites. This results in the rapid synthesis of negatively charged poly-ADP ribose (PAR) chains onto itself and other target proteins [7, 8]. These PAR chains, consisting of up to several hundred ADP-ribose molecules in a branched conformation, recruit other repair proteins such as X-ray repair cross-complementing protein 1 (XRCC1), ligase 3, polynucleotide kinase 3’-phosphate (PNKP), aprataxin, and DNA polymerase β to mediate SSBR [9–11]. DSB repair (DSBR) occurs via one of two main pathways: homologous recombination (HR), or classical non-homologous end joining (c-NHEJ). DSBR usually activates a process known as ‘end resection’, wherein the DNA ends undergo nucleolytic cleavage from the 5’ to 3’ end to form a long ssDNA segment on the 3’ strand. Repair of DSBs by HR is favored when DNA end resection occurs since the long 3’ ssDNA can invade the homologous DNA strand. This process occurs predominantly in the late S and G2 phases of the cell cycle, when a sister chromatid becomes available [12]. HR is initiated by recruitment of meiotic recombination 11 (MRE11) and ataxia telangiectasia mutated (ATM) to DSBs. The recombinase RAD51 is responsible for strand invasion, ultimately leading to the completion of the repair process by RAD52, RAD54 and Polδ [13]. Alternatively, in the absence of a homologous sequence or end resection, the c-NHEJ pathway is favored. In this process, the DNA ends are directly ligated, which requires factors such as Ku70/80, DNA-PKcs and DNA ligase IV [14–16]. The absence of Ku70/80 is compensated by PARP1 recruitment to locations of DSBs [17]. In vitro and in vivo studies conducted over the past fifty years have revealed that most of the above listed repair proteins often participate in multiple DNA repair pathways; however, the exact mechanisms of how the requisite cohort of proteins assemble to perform their activities to effectively repair diverse DNA lesions is yet to be fully determined.
A number of in vitro and in vivo strategies are currently utilized to dissect DNA damage repair, and a complete understanding of these mechanisms requires the use of multiple experimental approaches. In vitro approaches include, but are not limited to, the use of fluorescence polarization, Fluorescence Resonance Energy Transfer (FRET), and stopped-flow anisotropy assays to study the mechanisms governing association (and dissociation) of repair factors with model DNA fragments, plasmids, or nucleosomes [18, 19]. These experiments provide detailed insights into the molecular behavior of individual or small subsets of proteins that participate in DNA repair. Several in vivo methods such as PCR-based assays, the ‘comet assay’, and TUNEL assays can quantitate the degree of cellular DNA damage [20]. Fluorescence techniques, e.g. Fluorescence Recovery After Photobleaching (FRAP), Fluorescence Loss In Photobleaching (FLIP), Fluorescence Correlation Spectroscopy (FCS), and laser microirradiation, can be used to monitor repair protein kinetics in a nuclear context and can be interpreted in conjunction with the aforementioned in vitro measurements. In FRAP and FLIP, a small region within the cell is rapidly photobleached by a short pulse of high intensity radiation, and the recovery of fluorescently-tagged protein to the region (FRAP) or depletion of fluorescence outside the region (FLIP) is examined by time-lapse microscopy. On the other hand, FCS measures the change in fluorescence intensity as the protein of interest traverses a femtoliter volume within the cell [21–30]. While FRAP, FLIP, and FCS are typically designed to quantitate protein movement independent of DNA damage, laser microirradiation specifically focuses on the process of repair protein accumulation at DNA lesions [31–34].
Recent massive efforts have been placed on using laser microirradiation to understand the spatiotemporal details of repair protein accumulation at sites of DNA damage [35, 36]. In this approach, damage foci are induced by short wavelength (355 nm – 400 nm) light, and the accumulation of repair proteins to the irradiated site is detected either by immunofluorescence or by live-cell tracking of the fluorescently-tagged protein(s) of interest. Cells can be pre-sensitized by the addition of halogenated nucleotide analogs, such as bromodeoxyuridine (BrdU), or DNA-intercalating Hoechst dyes, which enables the use of 405 nm laser lines typically available on standard laser scanning confocal microscopes. Laser microirradiation is a powerful method for monitoring the kinetics of recruitment and the sequence of assembly of repair protein complexes on damaged DNA [16, 35–37]. However, a major limitation for this approach to reach its full potential has been the lack of reliable methods to derive meaningful quantitative information from imaging data. In this review, we highlight the widespread use of laser microirradiation over the last four decades, and we describe various methods currently in use for analyzing the accumulation of DNA repair proteins. In addition, we suggest approaches by which quantitation could be improved.
2. A Brief History of laser-induced DNA Damage Studies
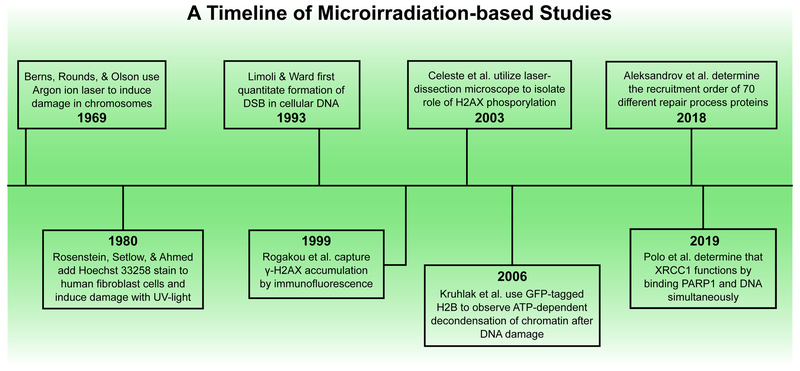
One of the earliest reports for the use of intense light to induce DNA damage in vivo dates back to 1969 (Fig. 1). Here, an argon ion laser was used to induce lesions on pre-determined sites on chromosomes of salamander lung cells stained with acridine orange, and it was found that cells could survive localized doses of microirradiation and continue to divide [38]. In 1978, another group used a mercury arc lamp to generate 313 nm ultraviolet (UV) rays to introduce DSBs in bromouracil-substituted cellular DNA in E. coli cells. Sensitivity of E. coli cells to photolysis increased linearly with the number of bromouracil molecules incorporated into the DNA [39]. In a modification of this method reported two years later, Hoechst 33258 stain was used to enhance the photolytic effect of BrdU treatment in human fibroblasts irradiated with 365 nm light. This wavelength of UV-light presented an advantage over the earlier approach that used 313 nm light since many aromatic ring-containing carcinogens form DNA adducts that strongly absorb 313 nm light [40]. In 1993, Limoli and Ward quantitated the formation of DSBs in cellular DNA subjected to photolysis using a combination of BrdU, Hoechst 33258, and near-visible UV light using the alkaline filter elution method [41]. In the following years, multiple groups continued to make improvements to this approach. These efforts enabled the visualization of γ-H2AX foci generated in human cells by indirect immunofluorescence by Rogakou et al. Here, the irradiating laser was guided along a pre-drawn path using an analog joystick [42]. To understand the significance of γ-H2AX in the DNA damage signaling process, Celeste et al. introduced DSBs in Hoechst-stained mouse embryonic fibroblasts (MEFs) using a laser-dissecting microscope (337 nm). This study suggested that H2AX phosphorylation was not required for formation of damage foci but was necessary for the concentration of repair factors like NBS1, 53BP1, and BRCA1 at DNA lesions. These efforts together established γ-H2AX as a DSB marker in the DNA repair field [43].
Figure 1.
Timeline of DNA microirradiation studies. While this timeline is not exhaustive, it highlights a few of the many significant microirradiation experiments over the 50-year history of the field.
In following years, the use of a laser scanning microscope to generate a precise path of DNA damage followed by real time live-cell imaging became a widely used approach in the field of DNA repair. Concomitantly, the biological functions of many repair proteins including NBS1, XRCC1, PARP1, and PARP2 were uncovered, resulting in a clearer understanding of the DNA repair signaling process [31,44, 45]. Since then, longer wavelengths of light (365-405 nm) have been utilized for introducing DSBs in mammalian cells stained with Hoechst dyes to compare the functions of repair proteins that participate in different repair mechanisms [31, 46, 47]. Some groups also began using laser microirradiation without prior chemical pre-sensitization in an attempt to investigate particular kinds of DNA lesions [48, 49]. In fact, DNA sensitization is not required when high-energy multiphoton near-infrared (NIR) lasers are used to inflict DNA damage; however, such high energies may ultimately cause premature cell death [48–51]. It is important to note that the type of DNA lesions induced and the way these lesions are processed could vary with the choice of the chemical sensitizer, the wavelength of light, and the duration of the pulse. These variations result in formation of a poorly understood mixture of DNA damage types such as pyrimidine dimers, inappropriate base modifications and inter-strand crosslinks [48, 52]. This heterogeneity has deterred researchers from comparing their findings to those of other groups, thereby limiting the ability to comprehend the interplay between different repair pathways.
Laser microirradiation has also been used as a tool to investigate remodeling of chromatin structure that accompanies the DNA repair process [53, 54]. The dynamic alteration of chromatin architecture that occurs as a consequence of disruption of DNA double-helical structure has been visualized using a photoactivatable version of GFP-tagged histone H2B (PAGFP-H2B) [55]. The 364 nm laser line was used to simultaneously induce DSBs and photoconvert the PAGFP to the active form. This study concluded that local chromatin undergoes rapid ATP-dependent decondensation immediately after the DNA damaging event [55], and another group independently established that this process was mediated by PARP1-dependent PARylation [56].
The last five years have witnessed an insurgence of information regarding the recruitment of several DNA damage response (DDR) proteins to laser-induced damage sites. In 2015, Izhar et al. performed a focused screen for proteins that arrived at DNA lesions post UV laser microirradiation [37]. Their work revealed that multiple families of transcription factors localized at damage sites in a DNA binding domain-dependent and/or a PARP-dependent manner [37]. In another study, Kochan et al. revisited numerous DSBR-related reports and performed a systematic analysis of laser-microirradiation data generated for 79 repair proteins to obtain insight into the spatiotemporal details of DSBR [36]. More recently, in 2018, Aleksandrov et al. carried out the mammoth task of measuring and mathematically modeling the recruitment kinetics of 70 proteins at laser-induced damage sites both in the presence and absence of a PARP inhibitor in living HeLa Kyoto cell lines [35]. This work provided information on the timing and sequence of arrival and removal of repair proteins at complex DNA lesions under similar experimental conditions [35]. Since then, many more research groups have relied on the powerful approach of laser microirradiation to probe the kinetics of recruitment of several repair proteins and their mutant counterparts [18, 33, 57–60].
3. Methods for quantitative analysis of laser microirradiation data
Laser microirradiation experiments are most frequently described by a fluorescence intensity time series for the region of interest after the damage event (Fig. 2). Such graphs depict the accumulation (and subsequent dissipation) of fluorescently labeled repair proteins at laser-induced damage sites. In practice, intensity values are generated from raw images using proprietary (Nikon Elements, ZEN) or publicly available (ImageJ) [61] imaging software packages, and each point within the timeseries is generally normalized to pre-irradiation values and corrected for background fluorescence. Furthermore, the fluorescence loss that occurs over the course of the experiment (photobleaching) can also be accounted for. A common practice in the field is to average data from 10-20 independently damaged nuclei and report a mean value with error for each time point within the curve [31,62, 63].
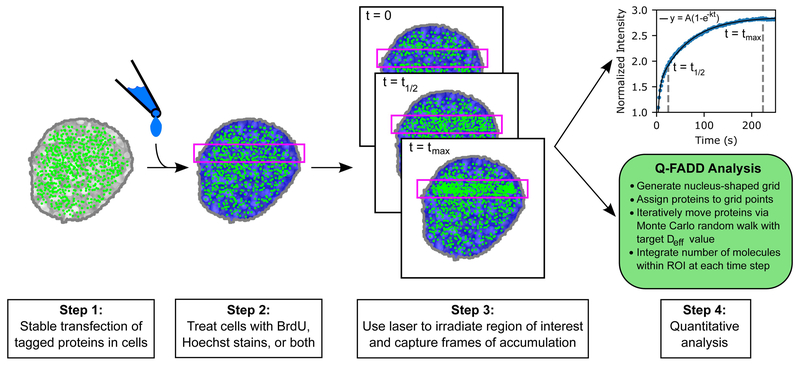
Figure 2.
A schematic of the microirradiation measurement and analysis workflow. First, cells are transfected with a vector containing the fluorescently labeled protein of interest. Then, cells may be pre-sensitized for DNA damage by the introduction of BrdU, Hoechst dyes, or both. After pre-sensitization, cells are placed in a confocal microscope and microirradiated with a laser beam, producing a region-of-interest (ROI) across the length of the cell (as shown above). Immediately preceding and following microirradiation, accumulation of labeled repair proteins to DNA lesions in the ROI is captured by live images over a set course of time. These images are then analyzed and converted to a fluorescence intensity timeseries, which is fit using the techniques described in Section 3.
Currently, one of the most widely used methods for quantitating accumulation timescales for various DNA repair proteins is to report the time required for half of the maximum accumulation to occur (t1/2). This is the quickest form of analysis currently in use, and it is a reliable method for comparing the accumulation of multiple repair proteins within a single nucleus. However, when accumulation kinetics are being compared between distinct nuclei, it is important to note that t1/2 is strongly influenced by the shape and size of the nucleus under examination [64] . Therefore, averaging multiple timeseries as part of a single kinetics population is not necessarily correct, as populations with diverse features (such as bimodality) cannot be solely assessed from a single mean and standard deviation [64–66]. Moreover, while t1/2 can be used to swiftly assess the speed of protein accumulation, it provides little information on the underlying physics of the protein transport process (simple, facilitated or anomalous diffusion).
An additional layer of quantitation can be obtained from accumulation data by fitting it to an exponential model [I(t) = A(1−e−kt)], where the accumulation rate constants (k) can be derived directly from the equation of best fit. Many proteins can be described by a first-order exponential fit [36, 67], but some proteins may require additional exponential terms [35, 68]. In these cases, the first-order term may describe a phase of rapid accumulation, whereas a second-order exponential would describe accumulation at longer timescales. Another method based in exponential fitting is the “Consecutive Chain Reactions” (CRC) model employed by Aleksandrov et al. to rationalize both protein accumulation and removal as a series of consecutive reactions, each with its own characteristic rate [35]. These approaches provide quantitative information about the underlying kinetic rates of accumulation, allowing for potential comparisons between the activation of different repair proteins. If these experiments are performed under similar conditions, one can actually determine the sequence of arrival to damage sites for a collection of proteins [35]. Nonetheless, exponential fitting still suffers from similar restrictions as does t1/2 evaluation, since these models are both affected by nuclear size and shape [64]. As a result, it is difficult to deduce a direct and meaningful physical interpretation of the diffusion process involved, especially when multiple nuclei are averaged to provide a single intensity timeseries [64]. Furthermore, it is important to exercise caution and avoid the spurious use of multiple exponentials for fitting accumulation curves, as the inclusion of unwarranted extra parameters to the model could lead to arbitrarily strong fits to the experimental data and lead to a potential misinterpretation of the underlying biological processes [69].
Recent efforts in our own lab have focused on developing a quantitative method to explain repair protein accumulation at DNA lesions that recognizes the role of nuclear shape and size. Quantitation of Fluorescence Accumulation after DNA Damage (Q-FADD) assumes that repair proteins move within the nucleus by simple free diffusion [64], as previous studies have shown that many proteins may navigate to sites of DNA damage through a free diffusion process [70–73]. Q-FADD utilizes a Monte Carlo simulation to describe molecular motion within a two-dimensional, nucleus-shaped grid to track protein accumulation within the defined region-of-interest (ROI), i. e. the DNA damage site. Originally, Q-FADD was developed to explain the diffusive behaviors of PARP1, PARP2, and Histone Parylation Factor 1 (HPF1), a new player in the DNA repair field [64, 74, 75]. Notably, the effective diffusion coefficient (Deff) derived from Q-FADD can be directly compared to Deff from other methods, such as FRAP and FCS [76]. While Q-FADD is able to provide Deff values in agreement with these other methods, levels of information such as dwell times and binding kinetics are integrated into a mobility fraction parameter, and derivation of these underlying properties requires orthogonal information from separate techniques.
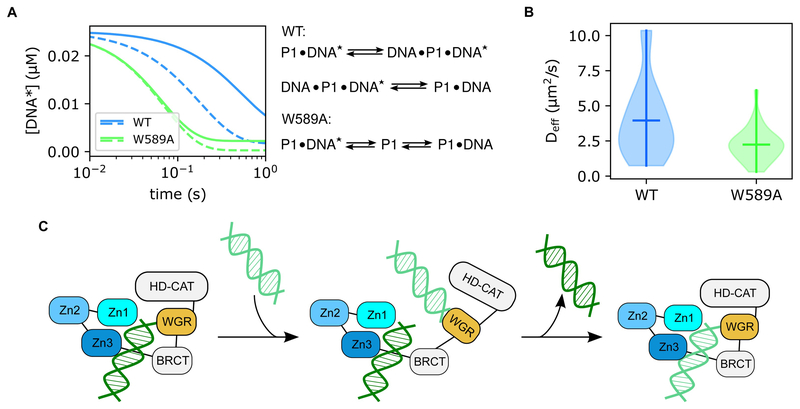
We have employed Q-FADD to elucidate the dynamic mechanism of PARP1 recruitment in vivo [18]. Indeed, PARP1 is among the fastest proteins recruited to sites of DNA damage [35], but the means by which PARP1 so rapidly seeks out sections of DSBs was largely unknown. In vitro kinetics work showed that purified PARP1 rapidly binds to DNA ends and that the release of bound DNA requires formation of a ternary complex with a second piece of DNA [18]. These data suggested that PARP1 navigates DNA through an intersegment transfer scheme, similar to how a child navigates playground monkey bars by a hand-over-hand exchange. Mutation studies identified the WGR domain of PARP1 as responsible for this action, as W589A mutation or WGR domain deletion prevented ternary complex formation. Similarly, Q-FADD analysis showed that W589A mutation significantly decreased the observed Deff value describing PARP1 accumulation (3.7 ± 0.6 μm2/s vs 2.1 ± 0.2 μm2/s, p = 0.0094), providing in vivo evidence for the “monkey-bar” mechanism of PARP1 mobility and confirming the important role of the WGR domain for biological efficacy (Fig. 3). As the mechanism was proposed by in vitro observations and confirmed through in vivo microirradiation quantitation, this study provides a classic example for the enhanced level of biological insight that can be gained from the combined use of quantitative in vivo and in vitro techniques.
Figure 3.
The “monkey bar” mechanism of PARP1 mobility. (A) Stopped-flow kinetics models show that wild-type PARP1 (blue) releases pre-bound DNA fragments ([DNA*]) at a significantly slower rate than the W589A mutant (green). The W589A mutant showed no difference in off-rate observation between low A Timeline of Microirradiation-based Studies(0.4 μM, solid line) and high (4 μM, dashed line) concentrations of competitor DNA, but wild-type PARP1 displays faster release of the initially bound strand when exposed to a higher concentration of competitor DNA. In addition, fitting of W589A kinetics were adequately described by a two-state equation, whereas wild-type data required formation of an intermediate state. (B) Violin plots of modeled Deff values from Q-FADD analyses on wild-type (blue) and W589A (green) microirradiation data. These models show that wild-type PARP1 diffuses significantly more quickly to DNA lesions than the W589A mutant (p-value = 0.0094). (C) Graphical representation of the “monkey bar” mechanism derived from the combination of these data. The WGR domain of PARP1 releases from the currently bound DNA strand and binds to a neighboring strand of DNA, then the original strand of DNA is released and the complex collapses around the newly bound DNA fragment. Figures were adapted from our published (free-access) data [18].
Until now, we have focused on the importance of quantitating microirradiation data for individual proteins. However, comparative analysis of multiple repair proteins also benefits from more quantitative approaches. For example, the recent study by Aleksandrov et al. used a divisive clustering algorithm (DIANA) to group a wide range of proteins in different arrival and removal clusters according to their measured t1/2 values [35]. From their results, they were able to show that when proteins are clustered according to their accumulation timescales, members of the same response pathway can be found within the same arrival cluster. By comparing to a library of known protein-protein interactions, they were also able to identify proteins that regulate DNA damage repair between pathways and timescales. Upon addition of PARP inhibitors, it was observed that proteins from PARP-independent pathways experienced changes in both their accumulation and depletion rates. Thus, inhibiting one repair pathway can cause the cell to alter priority in favor of another. Moreover, these results suggest that complex crosstalk may exist between various repair responses. As such, Aleksandrov et al. have demonstrated the importance of using several levels of quantitation to promote better understanding of the cellular response to DNA damage.
4. Perspective/Future Directions
Here, we have revisited the 50-year history of the laser microirradiation field, and we have highlighted several techniques that are currently used for quantitating microirradiation observations. Recent advancements in fluorescent labeling [77, 78]. as well as superior imaging systems and user-friendly computational tools have resulted in the ability to derive deeper biophysical knowledge from laser microirradiation experiments. Since robust modeling of irradiation data can provide insight into protein transport mechanisms, distribution of mobile and immobile protein populations, as well as (dis)association constants, continued emphasis should be placed on using these enhanced resources to develop even more robust quantitative modeling approaches, similar to current developments in methods such as FRAP and FCS [72, 79–81].
While the quantitation methods discussed in this review provide a range of insights to biological processes, differences in results obtained from various experimental approaches are still difficult to interpret [24] . First, the presence of the fluorescence tag may affect diffusion kinetics in a protein-specific manner [82]. Additionally, proteins could have smaller Deff values or accumulation rate constants because they bind to other proteins or immobile chromatin structures, whereas proteins with a larger accumulation rate or Deff value may encounter fewer binding and unbinding events. In this way, quantitative models should still be interpreted with caution and supplemented with other measurements, although future developments in modeling techniques could potentially be extended to include prediction of protein residence times on chromatin and thereby recover this information.
Improvements to quantitative in vivo modeling should also not stop at the per-protein or single timeseries level. While the previously discussed study by Aleksandrov et al. was conducted on t1/2 values [35], it is important to note that methods such as DIANA clustering are not restricted to t1/2 measures alone. This type of analysis can be used to compare accumulation rates from exponential fits, as well as Deff values from Q-FADD analyses. Moreover, there are a variety of clustering algorithms one might employ. The DIANA method is a divisive form of hierarchical clustering, where clusters are formed by iteratively separating proteins from a master family, but one could also cluster proteins in an agglomerative manner, where proteins are iteratively merged from individuals in to clusters instead of divided from a larger pool [83]. Beyond hierarchical schemes, centroid-based measures like the k-means method [84] or density-based methods such as DBSCAN [85] provide alternative approaches to characterizing pathways of DNA damage response from microirradiation data. In this way, through rigorous quantitation at multiple levels, the cellular response to DNA damage events can ultimately be fully understood.
Acknowledgement
We thank Johannes Rudolph for critical reading of the manuscript. JM and KL are supported by National Institutes of Health – National Cancer Institute (R01 CA218255) and the University of Colorado Cancer Center (Pilot Funding Grant ST63501792). Members of the Luger Lab are also supported by the Howard Hughes Medical Institute.
ABBREVIATIONS
- SSBs
Single strand breaks
- SSBR
Single strand break repair
- DSBs
Double strand breaks
- DSBR
Double strand break repair
- PARP1/2
Poly ADP-ribose polymerase 1/2
- PAR
Poly ADP-ribose
- XRCC1
X-ray repair cross-complementing protein 1
- PNKP
polynucleotide kinase 3’-phosphate
- HR
homologous recombination
- c-NHEJ
classical non-homologous end joining
- ssDNA
single stranded DNA
- DNA-PKcs
DNA-dependent protein kinase, catalytic subunit
- FRET
Fluorescence Resonance Energy Transfer
- MRE11
meiotic recombination 11
- ATM
ataxia telangiectasia mutated
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick-end labeling
- FRAP
Fluorescence Recovery After Photobleaching
- FLIP
Fluorescence Loss In Photobleaching
- FCS
Fluorescence Correlation Spectroscopy
- BrdU
bromodeoxyuridine
- UV
ultraviolet
- NBS1
Nibrin
- NIR
near-infrared
- PAGFP-H2B
photoactivatable GFP-tagged histone H2B
- DDR
DNA damage response
- CRC
Consecutive Chain Reactions
- Q-FADD
Quantitation of Fluorescence Accumulation after DNA Damage
- ROI
region-of-interest
- HPF1
Histone Parylation Factor 1
- Deff
effective diffusion coefficient
- DIANA
Divisive Analysis
- DBSCAN
density-based scan
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement: None declared
References
- 1.Caldecott KW, Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays, 2001. 23(5): p. 447–55. [DOI] [PubMed] [Google Scholar]
- 2.Abbotts R and Wilson DM 3rd, Coordination of DNA single strand break repair. Free Radic Biol Med, 2017. 107: p. 228–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou BB and Elledge SJ, The DNA damage response: putting checkpoints in perspective. Nature, 2000. 408(6811): p. 433–9. [DOI] [PubMed] [Google Scholar]
- 4.Sancar A, et al. , Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem, 2004. 73: p. 39–85. [DOI] [PubMed] [Google Scholar]
- 5.Sirbu BM and Cortez D, DNA damage response: three levels of DNA repair regulation. Cold Spring Harb Perspect Biol, 2013. 5(8): p. a012724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleck O and Nielsen O, DNA repair. J Cell Sci, 2004. 117(Pt 4): p. 515–7. [DOI] [PubMed] [Google Scholar]
- 7.de Murcia G and de Murcia J. Menissier, Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem Sci, 1994. 19(4): p. 172–6. [DOI] [PubMed] [Google Scholar]
- 8.Ray Chaudhuri A and Nussenzweig A, The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol, 2017. 18(10): p. 610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caldecott KW, DNA single-strand break repair. Exp Cell Res, 2014. 329(1): p. 2–8. [DOI] [PubMed] [Google Scholar]
- 10.Caldecott KW, Single-strand break repair and genetic disease. Nat Rev Genet, 2008. 9(8): p. 619–31. [DOI] [PubMed] [Google Scholar]
- 11.Kim MY, Zhang T, and Kraus WL, Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev, 2005. 19(17): p. 1951–67. [DOI] [PubMed] [Google Scholar]
- 12.Karanam K, et al. , Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol Cell, 2012. 47(2): p. 320–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright WD, Shah SS, and Heyer WD, Homologous recombination and the repair of DNA double-strand breaks. J Biol Chem, 2018. 293(27): p. 10524–10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiruvella KK, Liang Z, and Wilson TE, Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol, 2013. 5(5): p. a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ceccaldi R, Rondinelli B, and D’Andrea AD, Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol, 2016. 26(1): p. 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bekker-Jensen S, et al. , Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol, 2006. 173(2): p. 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M, et al. , PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res, 2006. 34(21): p. 6170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudolph J, et al. , Poly(ADP-ribose) polymerase 1 searches DNA via a ‘monkey bar’ mechanism. Elife, 2018. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muthurajan UM, et al. , Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. Proc Natl Acad Sci U S A, 2014. 111(35): p. 12752–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figueroa-Gonzalez G and Perez-Plasencia C, Strategies for the evaluation of DNA damage and repair mechanisms in cancer. Oncol Lett, 2017. 13(6): p. 3982–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phair RD and Misteli T, Kinetic modelling approaches to in vivo imaging. Nat Rev Mol Cell Biol, 2001. 2(12): p. 898–907. [DOI] [PubMed] [Google Scholar]
- 22.Lippincott-Schwartz J, Snapp E, and Kenworthy A, Studying protein dynamics in living cells. Nat Rev Mol Cell Biol, 2001. 2(6): p. 444–56. [DOI] [PubMed] [Google Scholar]
- 23.Reits EA and Neefjes JJ, From fixed to FRAP: measuring protein mobility and activity in living cells. Nat Cell Biol, 2001. 3(6): p. E145–7. [DOI] [PubMed] [Google Scholar]
- 24.Mueller F, et al. , FRAP and kinetic modeling in the analysis of nuclear protein dynamics: what do we really know? Curr Opin Cell Biol, 2010. 22(3): p. 403–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Axelrod D, et al. , Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys J, 1976. 16(9): p. 1055–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dundr M and Misteli T, Measuring dynamics of nuclear proteins by photobleaching. Curr Protoc Cell Biol, 2003. Chapter 13: p. Unit 13 5. [DOI] [PubMed] [Google Scholar]
- 27.Koster M, Frahm T, and Hauser H, Nucleocytoplasmic shuttling revealed by FRAP and FLIP technologies. Curr Opin Biotechnol, 2005. 16(1): p. 28–34. [DOI] [PubMed] [Google Scholar]
- 28.Dittrich P, et al. , Accessing molecular dynamics in cells by fluorescence correlation spectroscopy. Biol Chem, 2001. 382(3): p. 491–4. [DOI] [PubMed] [Google Scholar]
- 29.Pramanik A, Ligand-receptor interactions in live cells by fluorescence correlation spectroscopy. Curr Pharm Biotechnol, 2004. 5(2): p. 205–12. [DOI] [PubMed] [Google Scholar]
- 30.Maiti S, Haupts U, and Webb WW, Fluorescence correlation spectroscopy: diagnostics for sparse molecules. Proc Natl Acad Sci U S A, 1997. 94(22): p. 11753–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortusewicz O, et al. , Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res, 2007. 35(22): p. 7665–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong J, et al. , RBM45 competes with HDAC1 for binding to FUS in response to DNA damage. Nucleic Acids Res, 2017. 45(22): p. 12862–12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mani RS, et al. , Domain analysis of PNKP-XRCC1 interactions: Influence of genetic variants of XRCC1. J Biol Chem, 2019. 294(2): p. 520–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Q, et al. , PARP2 mediates branched poly ADP-ribosylation in response to DNA damage. Nat Commun, 2018. 9(1): p. 3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aleksandrov R, et al. , Protein Dynamics in Complex DNA Lesions. Mol Cell, 2018. 69(6): p. 1046–1061 e5. [DOI] [PubMed] [Google Scholar]
- 36.Kochan JA, et al. , Meta-analysis of DNA double-strand break response kinetics. Nucleic Acids Res, 2017. 45(22): p. 12625–12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Izhar L, et al. , A Systematic Analysis of Factors Localized to Damaged Chromatin Reveals PARP-Dependent Recruitment of Transcription Factors. Cell Rep, 2015. 11(9): p. 1486–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berns MW, Rounds DE, and Olson RS, Effects of laser micro-irradiation on chromosomes. Exp Cell Res, 1969. 56(2): p. 292–8. [DOI] [PubMed] [Google Scholar]
- 39.Krasin F and Hutchinson F, Double-strand breaks from single photochemical events in DNA containing 5-bromouracil. Biophys J, 1978. 24(3): p. 645–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenstein BS, Setlow RB, and Ahmed FE, Use of the dye Hoechst 33258 in a modification of the bromodeoxyuridine photolysis technique for the analysis of DNA repair. Photochem Photobiol, 1980. 31(3): p. 215–22. [DOI] [PubMed] [Google Scholar]
- 41.Limoli CL and Ward JF, A new method for introducing double-strand breaks into cellular DNA. Radiat Res, 1993. 134(2): p. 160–9. [PubMed] [Google Scholar]
- 42.Rogakou EP, et al. , Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol, 1999. 146(5): p. 905–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Celeste A, et al. , Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol, 2003. 5(7): p. 675–9. [DOI] [PubMed] [Google Scholar]
- 44.Lukas C, et al. , Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol, 2003. 5(3): p. 255–60. [DOI] [PubMed] [Google Scholar]
- 45.Lan L, et al. , In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc Natl Acad Sci U S A, 2004. 101(38): p. 13738–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mortusewicz O, et al. , Recruitment of RNA polymerase II cofactor PC4 to DNA damage sites. J Cell Biol, 2008. 183(5): p. 769–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bradshaw PS, Stavropoulos DJ, and Meyn MS, Human telomeric protein TRF2 associates with genomic double-strand breaks as an early response to DNA damage. Nat Genet, 2005. 37(2): p. 193–7. [DOI] [PubMed] [Google Scholar]
- 48.Dinant C, et al. , Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. J Cell Sci, 2007. 120(Pt 15): p. 2731–40. [DOI] [PubMed] [Google Scholar]
- 49.Kim JS, et al. , Specific recruitment of human cohesin to laser-induced DNA damage. J Biol Chem, 2002. 277(47): p. 45149–53. [DOI] [PubMed] [Google Scholar]
- 50.Mari PO, et al. , Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci U S A, 2006. 103(49): p. 18597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen BP, et al. , Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J Biol Chem, 2005. 280(15): p. 14709–15. [DOI] [PubMed] [Google Scholar]
- 52.Kong X, et al. , Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nucleic Acids Res, 2009. 37(9): p. e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bao Y and Shen X, Chromatin remodeling in DNA double-strand break repair. Curr Opin Genet Dev, 2007. 17(2): p. 126–31. [DOI] [PubMed] [Google Scholar]
- 54.Peterson CL and Cote J, Cellular machineries for chromosomal DNA repair. Genes Dev, 2004. 18(6): p. 602–16. [DOI] [PubMed] [Google Scholar]
- 55.Kruhlak MJ, et al. , Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol, 2006. 172(6): p. 823–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strickfaden H, et al. , Poly(ADP-ribosyl)ation-dependent Transient Chromatin Decondensation and Histone Displacement following Laser Microirradiation. J Biol Chem, 2016. 291(4): p. 1789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kong X, et al. , Biphasic recruitment of TRF2 to DNA damage sites promotes non-sister chromatid homologous recombination repair. J Cell Sci, 2018. 131(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith R and Timinszky G, Monitoring Poly(ADP-Ribosyl)ation in Response to DNA Damage in Live Cells Using Fluorescently Tagged Macrodomains. Methods Mol Biol, 2018. 1813: p. 11–24. [DOI] [PubMed] [Google Scholar]
- 59.Morotomi-Yano K, et al. , Dynamic behavior of DNA topoisomerase IIbeta in response to DNA double-strand breaks. Sci Rep, 2018. 8(1): p. 10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Polo LM, et al. , Efficient Single-Strand Break Repair Requires Binding to Both Poly(ADP-Ribose) and DNA by the Central BRCT Domain of XRCC1. Cell Rep, 2019. 26(3): p. 573–581 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tampere M and Mortusewicz O, DNA Damage Induction by Laser Microirradiation. Bioprotocol, 2016. 6(23): p. e2039. [Google Scholar]
- 62.Xie S, et al. , Timeless Interacts with PARP-1 to Promote Homologous Recombination Repair. Mol Cell, 2015. 60(1): p. 163–76. [DOI] [PubMed] [Google Scholar]
- 63.Sellou H, et al. , The poly(ADP-ribose)-dependent chromatin remodeler Alc1 induces local chromatin relaxation upon DNA damage. Mol Biol Cell, 2016. 27(24): p. 3791–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mahadevan J, et al. , Q-FADD: A Mechanistic Approach for Modeling the Accumulation of Proteins at Sites of DNA Damage. Biophys J, 2019. 116(11): p. 2224–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karanam K, Loewer A, and Lahav G, Dynamics of the DNA damage response: insights from live-cell imaging. Brief Funct Genomics, 2013. 12(2): p. 109–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fay DS and Gerow K, A biologist’s guide to statistical thinking and analysis. WormBook, 2013: p. 1–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lukas C, et al. , Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J, 2004. 23(13): p. 2674–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bekker-Jensen S, et al. , Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J Cell Biol, 2005. 170(2): p. 201–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mayer J, Khairy K, and Howard J, Drawing an elephant with four complex parameters. American Journal of Physics, 2010. 78(6): p. 648–649. [Google Scholar]
- 70.Kong M, et al. , Single-Molecule Imaging Reveals that Rad4 Employs a Dynamic DNA Damage Recognition Process. Mol Cell, 2016. 64(2): p. 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Friis I and Solov’yov IA, Activation of the DNA-repair mechanism through NBS1 and MRE11 diffusion. PLOS Computational Biology, 2018. 14(7): p. e1006362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mazza D, et al. , A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res, 2012. 40(15): p. e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hinow P, et al. , The DNA binding activity of p53 displays reaction-diffusion kinetics. Biophys J, 2006. 91(1): p. 330–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gibbs-Seymour I, et al. , HPF1/C4orf27 Is a PARP-1-Interacting Protein that Regulates PARP-1 ADP-Ribosylation Activity. Mol Cell, 2016. 62(3): p. 432–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bonfiglio JJ, et al. , Serine ADP-Ribosylation Depends on HPF1. Mol Cell, 2017. 65(5): p. 932–940 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kozlowski M, The molecular mechanism of PARP1 activation and its downstream roles in ALC1-regulated transcription. 2014. [Google Scholar]
- 77.Toseland CP, Fluorescent labeling and modification of proteins. J Chem Biol, 2013. 6(3): p. 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crivat G and Taraska JW, Imaging proteins inside cells with fluorescent tags. Trends Biotechnol, 2012. 30(1): p. 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carrero G, et al. , Using FRAP and mathematical modeling to determine the in vivo kinetics of nuclear proteins. Methods, 2003. 29(1): p. 14–28. [DOI] [PubMed] [Google Scholar]
- 80.Phair RD, Gorski SA, and Misteli T, Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol, 2004. 375: p. 393–414. [DOI] [PubMed] [Google Scholar]
- 81.Kaufman EN and Jain RK, Quantification of transport and binding parameters using fluorescence recovery after photobleaching. Potential for in vivo applications. Biophys J, 1990. 58(4): p. 873–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nenninger A, Mastroianni G, and Mullineaux CW, Size dependence of protein diffusion in the cytoplasm of Escherichia coli. J Bacteriol, 2010. 192(18): p. 4535–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leonard Kaufman PJR, Finding Groups in Data: An Introduction to Cluster Analysis. 2008. [Google Scholar]
- 84.MacQueen J Some methods for classification and analysis of multivariate observations In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Volume 1: Statistics. 1967. Berkeley, Calif.: University of California Press. [Google Scholar]
- 85.Ester M, et al. , A density-based algorithm for discovering clusters a density-based algorithm for discovering clusters in large spatial databases with noise, in Proceedings of the Second International Conference on Knowledge Discovery and Data Mining 1996, AAAI Press: Portland, Oregon p. 226–231. [Google Scholar]