Abstract
Background
Colon cancer is one of the most common cancers worldwide. IFN-stimulated gene 15 (ISG15), a ubiquitin-like molecule, is strongly up-regulated by type I interferon as a crucial response to a variety of microbial and cellular stress stimuli. However, the role of ISG15 in colon cancer remains unclear.
Methods
The expression of ISG15 in colon cancer tissues and cell lines was detected by using Western blotting and immunohistochemistry. ISG15 expression levels of colon cancer cells treated with trametinib was verified by using the data downloaded from the Gene Expression Omnibus (GEO) databases, quantitative real-time PCR analysis and Western blots assays. ISG15-siRNA was used to silence ISG15 in colon cancer cell line to determine the roles of ISG15 in colon cancer cell proliferation.
Results
ISG15 was highly expressed in colon cancer tissues and ISG15 upregulation was closely associated with poor prognoses in colon cancer patients. Enhanced ISG15 expression promoted the migration and proliferation of colon cancer cells in vitro, while ISG15 knockdown decreased cell proliferation and metastasis. In addition, we first found that the mRNA and protein expression of ISG15 were up-regulated following trametinib treatment. Further investigation showed that ISG15 knockdown could enhance the anti-cancer effect of trametinib in colon cancer cells.
Conclusion
We proposed an interesting possibility that ISG15 may be a prognostic bio-marker, and the combined targeting of ISG15 and MEK might be a promising therapeutic strategy for colon cancer.
Keywords: ISG15, MEK, Trametinib, GEO
Introduction
Colon cancer is one of the most common malignant cancers and among the leading causes of cancer-related deaths worldwide.1 The combination therapy of oxaliplatin and fluoropyrimidine has been the standard adjuvant therapy in patients with stage III/IV colon cancer. However, these chemotherapies are toxic and sometimes ineffective. Due to its heterogenicity, multiple genetic mutations were regarded as the underlying causes of this disease, for example, mutations in phosphatidylinositol-3 kinase (PI3K) and the mitogen activated protein kinase (MAPK) pathways. Deregulation of MAPK pathway genes have been found, including the amplification and mutation of KRAS, BRAF, and MEK1.2 Therefore, targeting the downstream MEK in the mutated tumors might be a new strategy for colon cancers, especially the patients with KRAS or BRAF mutations. Several MEK inhibitors have entered clinical trial evaluation. However, clinical activity was poor following the treatment with any single MEK inhibitor, and acquired drug resistance appears inevitable.3–8 Trametinib is a novel oral MEK inhibitor which has been approved by the FDA (Food and Drug Administration) for BRAF-mutated patients alone or in combination with dabrafenib.9–11 Combination remedies of MEK inhibitors, rather than single medicine therapy, was considered to be more effective in various tumors.12–14 Therefore, there is an urgent clinical demand for new synergic agents to cooperate with trametinib to enhance the survival of patients with colon cancer.
Interferon-Stimulated Gene 15 (ISG15), a ubiquitin-like protein (UBL),15 is an important oncoprotein and has become a potential diagnostic16 and therapeutic17 target for cancer treatment. The function of ISG15 was largely underestimated mainly due to its low expression in most human malignancies.18 Some studies have found that ISG15 expression was elevated and ISG15-conjugates in many malignant tumors, including melanoma19 and oral squamous cell carcinoma20,21 as well as malignancies of the breast,16 endometrium,18 and bladder.22 However, the roles of ISG15 in tumorigenesis and its responses to anticancer treatments in colon cancer remain largely unknown. In a recent study, Roulois et al23 showed that DNA methylation inhibitor 5-aza-2-deoxycytidine (5-AZACdR) could enhance the expression of ISG15 in LIM1215 colon cancer cells, which implied the nature of ISG15 as a tumor suppressor in colorectal cancer. Controversially, Desai et al18 discovered that ISG15 expression and ISG15-conjugated proteins in two colon cancer cases were up-regulated in comparison to normal colon tissues. Whether ISG15 acts as a tumor suppressor or promotor remains controversial.
In this study, we explored the function of ISG15 in colon cancer cell lines. Our results showed that high expression of ISG15 was an intrinsic feature for colon cancer, and ISG15 promoted the cell proliferation and metastasis. Trametinib could increase the expression of ISG15 shown by gene expression analysis. After treatment of a synergistic combination of trametinib with ISG15 siRNA, proliferation and colony formation was inhibited in vitro. Thus, combined targeting of ISG15 and MEK might be a promising therapeutic strategy for colon cancer treatment.
Materials and Methods
Tissue Samples and Study Cohort
Sixty-six pairs of tumor samples and matched adjacent non-tumor tissues were obtained from the Shanghai Outdo Biotech Co., Ltd. (Shanghai, China). All the patients signed informed consent forms. This study was approved by the Ethics Committee of Taizhou Hospital of Zhejiang Province. ISG15 expression was detected in all specimens. Two pathologists were appointed to evaluate the specimens separately without prior knowledge of the clinical statuses of the specimens.
Immunohistochemistry
Immunohistochemistry (IHC) was performed using the biotin-streptavidin HRP detection system according to the manufacturer’s instructions. The tissue chips were incubated with ISG15 antibody (1:100; Abcam, Cambridge, UK) in phosphate-buffered saline (PBS) overnight at 4°C in a humidified container. Biotinylated secondary antibodies (Zhongshan Golden Bridge Biotechnology Co. Ltd., China) were applied. The sections were incubated with HRP-streptavidin conjugates appropriate for detecting ISG15. Proper positive and negative controls were included in each IHC assay.
Western Blotting Analysis
Total proteins (30 µg) were subjected to fractionation by SDS polyacrylamide gel electrophoresis and immunoblotting assay. Antibodies used in the study included the following: anti-ISG15, anti-PARP, anti-c-Myc, anti-pERK, anti-ERK (all Abcam, Cambridge, UK), and anti-GAPDH (Zhongshan Golden Bridge Biotechnology Co. Ltd., China).
Cell Culture and RNA Interference
Human colon cancer cell lines were obtained from the State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, including RKO, HCT116 and SW480. And all the cells were tested and authenticated by an AmpFlSTR Identifiler PCR assays in the year of 2019 in TSINGKE Biological Technology Company. The results showed that RKO and SW480 were 100% exact matched, and HCT116 was 98% matched. All the cell lines were confirmed to be mycoplasma negative via a PCR method and directly thawed from the liquid nitrogen jar. Cells were grown in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum (Gibco, USA), 100mU/mL penicillin, and 100 µg/mL streptomycin in a 5% CO2 atmosphere at 37°C. Cells (1x10 5 cells/well) were seeded into six-well plates and transfected with the indicated constructs using Lipofectamine 2000 (Invitrogen/Life Sciences) according to the manufacturer’s instructions. After 72 h, the transfected cells were harvested for further analysis. The sequence of the siRNA sense strand was as follows: si-ISG15#1: 5-’TCCTGGTGAGGAATAACAA-3ʹ, si-ISG15#2: 5-’CCAUGUCGGUGUCAGAGCUTT-3ʹ.
CCK-8 and Colony Formation Assays
CCK-8 assay kit was used for colon cancer cell proliferation analysis. Colon cancer cells were seeded in 96-well plate at a density of 5x10^3 cells per well. After incubation, 10 µL CCK8 was added to the wells at different times. The absorbance was measured at 450 nm by a microplate reader (Bio-Rad, Hercules, CA, USA). Colony formation assay was performed to detect the capacity of cell proliferation. RKO, HCT116 and SW480 cells were seeded into 35 mm dishes with 3x10^3 cells per well and cultured for 10 days. Colonies were then fixed with 10% formaldehyde for 30 min and stained for 5 min with 0.5% crystal violet.
Wound Healing Assay
Colon cancer cells were cultured in 6-well plates and grown until 80–90% confluence. After that, the cell monolayer was scratched with a sterile pipette tip. An Olympus microscope was applied for taking pictures of cell morphology at different time intervals (0,48h). The results were analyzed by ImageJ software.
Matrigel Invasion Assays
The capacity invasiveness was determined in a 24-well transwell plate (8 up pore size; Costar). Briefly, 6x10^ 4 cells were suspended and added to the upper chamber of each insert coated with 200 mg/mL of Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). After 48 h, the cells that traversed the membrane and spread to the lower surface of the filters were stained with haematoxylin and counted.Each experiment was repeated three times independently.
RNA Extraction and qRT-PCR
Total RNA was isolated through the guanidinium thiocyanate-phenol-chloroform extraction method using TRIzolsolution (Invitrogen, USA), and cDNA synthesis was done using the Advantage RT-forPCRKit (Takara, Japan). The forward and reverse primers were designed to span two different exons for each gene.The primers for human ISG15 were forward:5ʹ-TGGACAAATGCGACGAACC-3ʹ; reverse: 5ʹ-TTCGTCGTTCACTCGCC-3ʹ.CFX96 Real-Time PCR detectionsystem (Bio-Rad, Hercules, CA, USA) was used to perform qRT-PCR. The threshold cycle number was determinedusing iCycler software version 3.0. Reactions were performed in duplicate and the threshold cycle numbers wereaveraged. The data were analysed using the 2^−ΔΔCT method.
Edu Assay
Quantification of cellular proliferative capacity was determined by 5-Ethynyl-2ʹ-deoxyuridine assay using a Cell-Light TM EdU Apollo®488 In Vitro Imaging Kit (Guangzhou RiboBio Co., LTD), according to the manufacturer’s instruction.
Identification of Differentially Expressed Genes from Public Microarray Data
GSE112282 gene expression profiles were downloaded from the Gene Expression Omnibusdatabase (GEO, http://www.ncbi.nlm.nih.gov/geo). These profiles, deposited by Wyce A et al in 2018, are comprised of two untreated replicates of human colon cells and two replicates of the same cells treated with 25 nM trametinib for 24 h and 96 h. The dataset was analyzed with R BIOCONDUCTOR packages, raw datasets were normalized and DEGs were screened out via the LIMMA package using the cut-off criteria of P-value < 0.05 and |log2(fold change)|>1.5.
Functional Enrichment Analysis
DAVID (https://david.ncifcrf.gov/) was used to perform functional and pathway enrichment analysis. Gene ontology (GO) analysis, including biological process (BP), cellular component (CC) and molecular function (MF)24 enrichment analysis, was carried out for the upregulated genes. P<0.001 was regarded as indicating statistical significance.
Protein-Protein Interaction Network Construction
In order to interpret the underlying molecular mechanisms, the online Search Tool for the Retrieval of Interacting Genes (STRING) database was utilized to construct a protein-protein interaction (PPI) network of the DEGs. An interaction score of not < 0.4 (medium confidence score) was deemed statistically significant, and the PPI was visualized. Subsequently, the hub genes were selected based on connection degree by CYTOSCAPE software.
Statistical Analysis
Assays for characterizing phenotypes of cells were analyzed by Student’s test or One-Way ANOVA, presented as the means ± SDs. In the clinical specimen study, the associations of ISG15expression with categorical variables were analyzed using the χ2 test or Fisher’s exact test as appropriate. And other variables were analyzed with Spearman correlation test. P values of <0.05 were deemed statistically significant.
Results
Up-Regulation of ISG15 Expression in Colon Cancer Samples
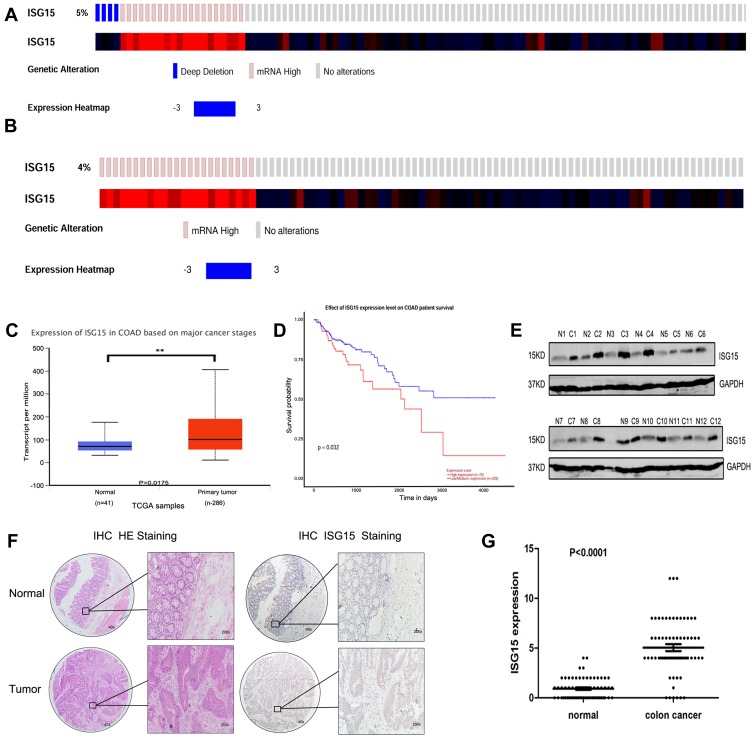
cBioPortal was applied for cancer genomics datasets analysis. ISG15 was amplified and the mutation frequency was 5% in colon cancers (Figure 1A). Among them, the ones with high expression of ISG15 mRNA accounted for 4% (Figure 1B). Moreover, ISG15 mRNA was over-expressed in colon cancer specimens compared to their non-tumor counterparts. (Figure 1C, P < 0.05). Kaplan-Meier analysis demonstrated that patients with high ISG15 expression had significantly shorter overall survival (OS) than patients with low ISG15 expression (Figure 1D, P < 0.05). Using Western blot, 83.3% (10/12) of the cases showed relatively higher ISG15 expression than their non-tumor counterpart, consistent with the results of mRNA expression analysis (Figure 1E). To evaluate whether the expression of ISG15 changes as colon cancer progresses, IHC was performed by using an independent formalin-fixed, paraffin-embedded-based (FFPE-based) tissue microarray (TMA) including 66 pairs of colon cancer patients. The results showed that ISG15 expression in colon cancer tissues was obviously higher than that in adjacent noncancerous colon tissues (Figure 1F). As indicated in Figure 1G, the mean immunoreactivity scores of the normal and cancer tissues were 1.02 and 5.01, respectively. Samples with scores above 5 were considered as high ISG15 expression ones, leading to the greatest number of tumors classified as having or not having a clinical outcome. In our study, high ISG15 levels were detected in 30/66 (45.5%) of colon cancer patients. The results showed ISG15 expression did not correlate with patient sex, age or lymph node status (P>0.05). It was, indeed, associated with poor histological classification (P < 0.01, Table 1). Overall, our data suggested that ISG15 level was higher in colon cancer samples.
Figure 1.
Upregulation of ISG15 expression in colon cancer samples. (A, B) ISG15 status in a colon cancer study obtained from http://www.cbioportal.org. (C) Expression of ISG15 in 286 TCGA cancer and 41 normal samples indicated that ISG15 was obviously upregulated in colon cancer. (D) Kaplan-Meier analysis indicated that high ISG15 expression predicted poorer prognosis in colon cancer. The error bars in all graphs represent SD, and each experiment was repeated three times (**p< 0.01 ). (E) The difference in protein expression levels of ISG15 in colon cancer tissues was compared with the corresponding adjacent tissues by Western blot in 12 pairs of human colon cancer specimens. (F, G) The differential expression of ISG15 in colon cancer samples (n = 66) and adjacent normal colon tissues (n =66) is shown by immunohistochemistry. ISG15 staining scores for tumor tissues and normal tissues are represented by a frequency distribution (0–4: low expression; 5–12: high expression).
Table 1.
Correlation Between ISG15 Expression and Clinicopathological Features of Colon Cancer Patients P-Value=0.0087*: the ISG15 Expression Was Associated with Poor Histological Classification
| Statistical Results of ISG15 Expression in 66 Colon Cancer Cells Specimens | ||||
|---|---|---|---|---|
| Variables | ISG15 Expression | |||
| N | Low Expression, n | High Expression, n | P-value | |
| All cases | 66 | 36 | 30 | |
| Age | ||||
| ≥ 50 | 43 | 21 | 22 | 0.2997a |
| < 50 | 23 | 15 | 8 | |
| Gender | ||||
| Male | 37 | 18 | 19 | 0.3254a |
| Female | 29 | 18 | 11 | |
| Pathology grade | ||||
| I (well) | 12 | 9 | 3 | 0.0087**,b |
| I-II (moderate) | 37 | 23 | 14 | |
| II-III (poor) | 17 | 4 | 13 | |
| Lymph node status | ||||
| N- | 40 | 21 | 19 | 0.8015a |
| N+ | 36 | 15 | 11 | |
Notes: **p<0.01, The ISG15 expression was associated with poor histological classification. aFisher’s exact test was used for statistical analyses. bPearson’s χ2 test was used for statistical analyses.
Knockdown of ISG15 Inhibits Cancerous Proliferation and Migration
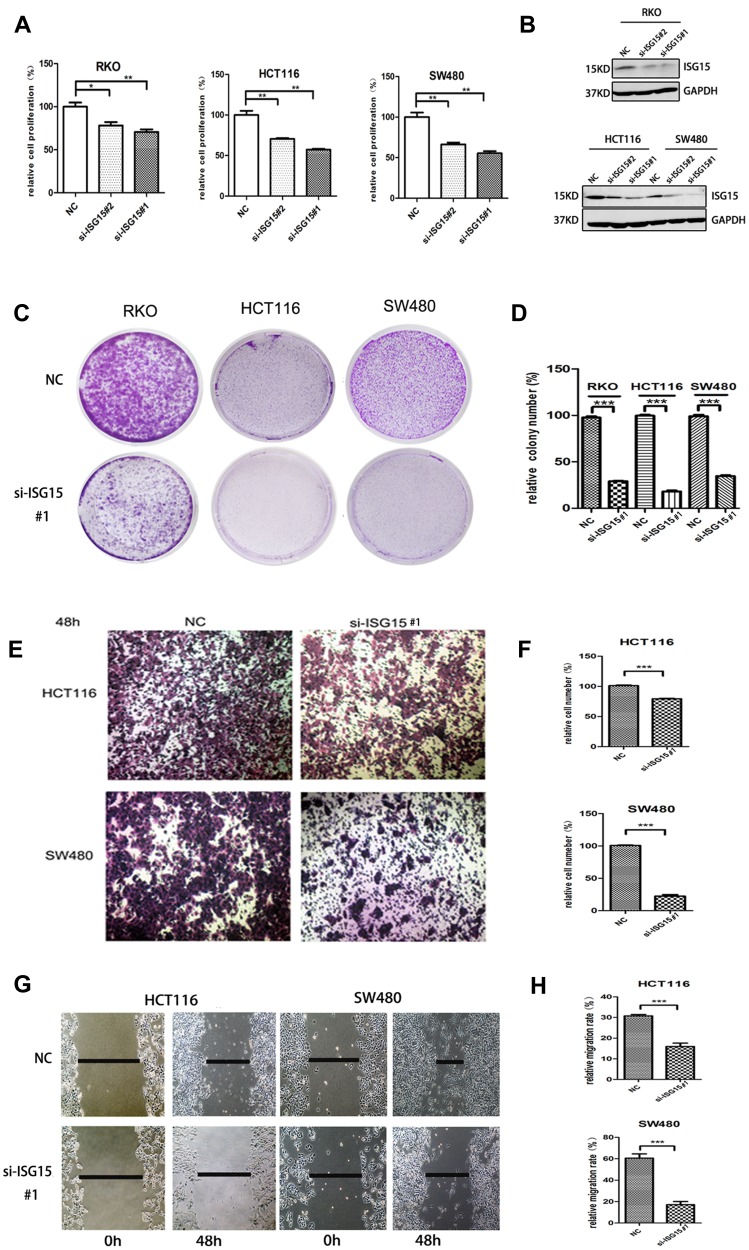
We investigated the effect of ISG15 suppression on the malignant phenotypes of colon cancer cells. Results showed that knockdown of ISG15 with si-SG15#1 and si-SG15#2 significantly inhibited cell proliferation in the RKO, HCT116 and SW480 cell lines (Figure 2A). As indicated in Figure 2B, knockdown by siRNAs resulted in reduced ISG15 protein in colon cancer cell lines. And we concluded that si-SG15#1 inhibitory effect was stronger than si-SG15#2. Colony formation assays and Edu assay also indicated that si-ISG15#1 could significantly inhibited the proliferation and growth of colon cancer cells (Figure 2C and D, Figure S1A and B). Next, we examined the migratory abilities of HCT116 and SW480 cells by using transwell migration assay. HCT116 and SW480 cells transfected with si-ISG15#1 displayed lower efficiency of cell migration compared to the vector control (Figure 2E and F, P<0.01). We also performed a wound healing assay by using RKO, HCT116 and SW480 human colon cancer cells. As shown in Figure 2G and H and Figure S1C, after 48 hrs of silence, si-ISG15#1 treatment significantly impeded gap closure compared to the vector control. These results indicated that suppression of ISG15 could inhibit the cell growth and migration in colon cancer cells.
Figure 2.
Knockdown ISG15 inhibits cancerous proliferation and migration. (A) Cell proliferation profiling in RKO, HCT116 and SW480 cells infected with si-ISG15#1 or si-ISG15#2 continuously for 3 days as analyzed by a CCK8 assay (*p< 0.05, **p< 0.01). (B) Protein levels of ISG15 in the RKO, HCT116 and SW480 cell lines infected with si-ISG15#1 or si-ISG15#2. GAPDH was used as an internal control. NC, Negative control; si-ISG15, siRNA of ISG15. (C, D) Colony formation in RKO, HCT116 and SW480 cells infected with si-NC or si-ISG15. The histogram shows the average colony numbers and represents the mean±SEM of three independent experiments (***p< 0.001). (E, F) Transwell invasion assay in si-NC or si-ISG15#1 transfected HCT116 and SW480 cells. The error bars in all graphs represent the SDs (***p< 0.001). (G, H) Wound-healing assay after transfecting with si-NC or si-ISG15#1 for 48 h in HCT116 and SW480 cells. Data are shown as the mean ± standard error of the mean (***p< 0.001).
Trametinib Treatment Activates the Expression of ISG15 by Identification DEGs
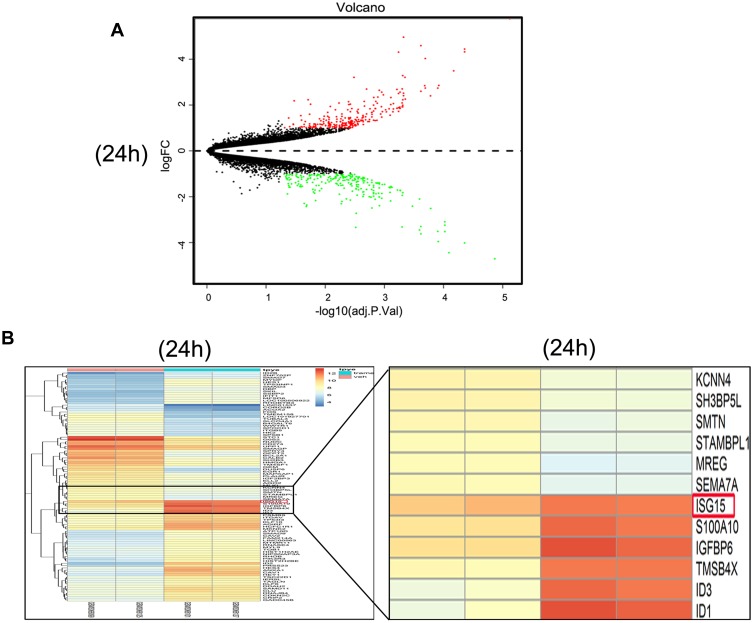
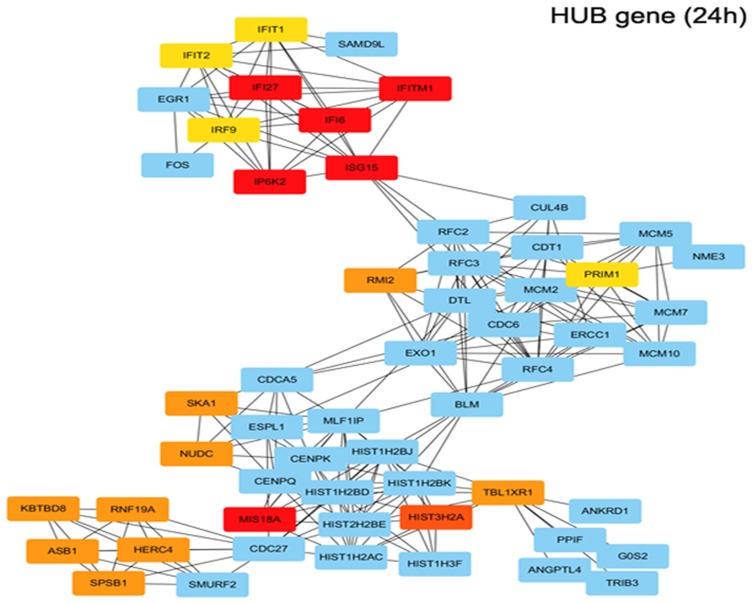
The gene expression profiles of GSE112282 were downloaded from the GEO database, to unravel molecular mechanisms underlying trametinib in the tumors of an experimental mouse model. The GSE112282 dataset contained following four groups: DMSO (control) or 30 nM trametinib for 24 or 96 hrs each in the RKO colon cancer cell line. We found 193 and 304 genes differentially expressed between the control and trametinib groups at 24 and 96 hrs, respectively (>1.5-fold, p<0.05, Figure 3A and B, Figure S2A and 2B). Gene ontology analysis were applied to discover the functions of the DEGs for 24 hrs (Figure S3) and 96 hrs (Figure S4). The DEGs of up-regulated genes were significantly enriched in biological processes related to ossification, transforming growth factor beta receptor signaling pathway (i.e., ISG15, TOB1, ID1) at 24 hrs (Table 2) and type I interferon signaling pathway, cellular response to type I interferon (i.e., ISG15, IRF9, IFITM3, IFI6, IFIT/12/3, and IP6K4) at 96 hrs (Table S1). The STRING database and Module analysis via Cytoscape software were constructed to identify gene interactions, the significant hub nodes of the network included IP6K2, ISG15, IFI6, IFI27 and IFITM1 at 24 hrs (Figure 4) and IFITM3, IRF9, ISG15, IP6K2 and HIST1H2BD at 96 hrs (Figure S5). Node degree and betweenness were calculated by the DMNC method to obtain hub nodes. The top 15 candidate hub genes were identified which may play a central role in this network (Table S2). And no matter which stage for the trametinib groups (24 hrs or 96 hrs), we found ISG15 always appears as a hub gene and was associated with type I interferon signaling pathway and cellular response to type I interferon. Therefore, the PPI network analysis and hub gene selection and analysis showed that treatment of colon cancer cells with trametinib could induced a dependency on ISG15 signaling especially in terms of type I interferon signaling pathway.
Figure 3.
Trametinib treatment activates the expression of ISG15 as shown by the identification of DEGs. (A) Volcanoplot of the 193 identified DEGs at 24 hrs. Red indicates DEGs with a |log2FC|>1.5. DEG, differentially expressed gene; FC, fold change. (B) Heat map showing the 193 differentially expressed genes betweenthe trametinib (n=2) and control groups at 24 hrs. (n=2; >1.5-fold, p<0.05). The most highly upregulated (red) and downregulated (green) genes in the trametinib group are listed.
Table 2.
Functional Enrichment Analysis of Up-Regulated Genes Were Significantly Enriched in Biological Process Terms (24h)
| Category | ID | Term/gene function | GeneID | Gene count | pvalue |
|---|---|---|---|---|---|
| GO_TERM_BP | GO:0001503 | Ossification | TOB1/FSTL3/ANO6/FGFR3/ID1/ID3/JUND/SMAD3/TP53INP2/BMPR1A/BMPR2/KLF10/TMSB4X/TUFT1/KAZALD1/CAT/IFITM1/DHRS3/EIF2AK3/ISG15 | 20 | 1.28E-07 |
| GO_TERM_BP | GO:0007179 | Transforming growth factor beta receptor signaling pathway | CDKN2B/ID1/SMAD3/SMAD7/SMAD9/PTPRK/SKIL/BMPR1A/KLF10/CAV1/CAV2/ADAM9 | 12 | 3.58E-06 |
| GO_TERM_BP | GO:0007178 | Transmembane receptor protein serine/threonine kinase signaling pathway | TOB1/FSTL3/CDKN2B/HES1/ID1/SMAD3/SMAD7/SMAD9/SKIL/BMPR1A/BMPR2/KLF10/CAV1/CAV2/ADAM9 | 16 | 6.40E-06 |
| GO_TERM_BP | GO:0030282 | Bone Mineralization | ANO6/FGFR3/SMAD3/BMPR1A/BMPR2/KLF10/TUFT1/EIF2AK3/ISG15 | 9 | 6.56E-06 |
| GO_TERM_BP | GO:0071560 | Cellular response to transforming growth factor beta stimulus | CDKN2B/ANKRD1/ID1/SMAD3/SMAD7/SMAD9/PTPRK/SKIL/BMPR1A/KLF10/CAV1/CAV2/ADAM9 | 13 | 8.08E-06 |
| GO_TERM_BP | GO:0071559 | Response to transforming growth factor beta | CDKN2B/ANKRD1/ID1/SMAD3/SMAD7/SMAD9/PTPRK/SKIL/BMPR1A/KLF10/CAV1/CAV2/ADAM9 | 13 | 9.80E-06 |
| GO_TERM_BP | GO:0050678 | Regulating of epithelial cell proliferation | CDKN1B/CDKN2B/ANG/HOXA5/HES1/ID1/MIR24-2/SMAD3/PTPRK/PEX2/BMPR1A/BMPR2/CAV1/CAV2/MAGED1 | 15 | 1.53E-05 |
| GO_TERM_BP | GO:0060337 | Type I interferon signaling pathway | IRF9/IFI6/IFI27/IFIT2/IFIT1/IP6K2/IFITM1/ISG15 | 8 | 2.15E-05 |
| GO_TERM_BP | GO:0071357 | Cellular response to type I interferon | IRF9/IFI6/IFI27/IFIT2/IFIT1/IP6K2/IFITM1/ISG15 | 8 | 2.15E-05 |
| GO_TERM_BP | GO:0050673 | Epithelial cell proliferation | CDKN1B/CDKN2B/ANG/HOXA5/HES1/ID1/ID2/MIR24-2/SMAD3/PTPRK/PEX2/BMPR1A/BMPR2/CAV1/CAV2/MAGED1 | 16 | 2.63E-05 |
| GO_TERM_BP | GO:0034340 | Repose to type I interferon | IRF9/IFI6/IFI27/IFIT2/IFIT1/IP6K2/IFITM1/ISG15 | 8 | 3.22E-05 |
| GO_TERM_BP | GO:0030278 | Regulation of ossification | TBO1/ANO6/ID1/ID3/JUND/SMAD3/BMPR1A/BMPR2/IFITM1/DHRS3/ISG15 | 11 | 3.49E-05 |
Figure 4.
Functional enrichment analysis of upregulated DEGs in trametinib treatment for 24h. Using the STRING online database and Cytoscape, upregulated genes in the trametinib groups were filtered into the DEG PPI network complex. (IP6K2, ISG15, IFI6, IFI27, IFITM1, MIS18A and so on are hub genes in red or orange; other linked genes in blue).
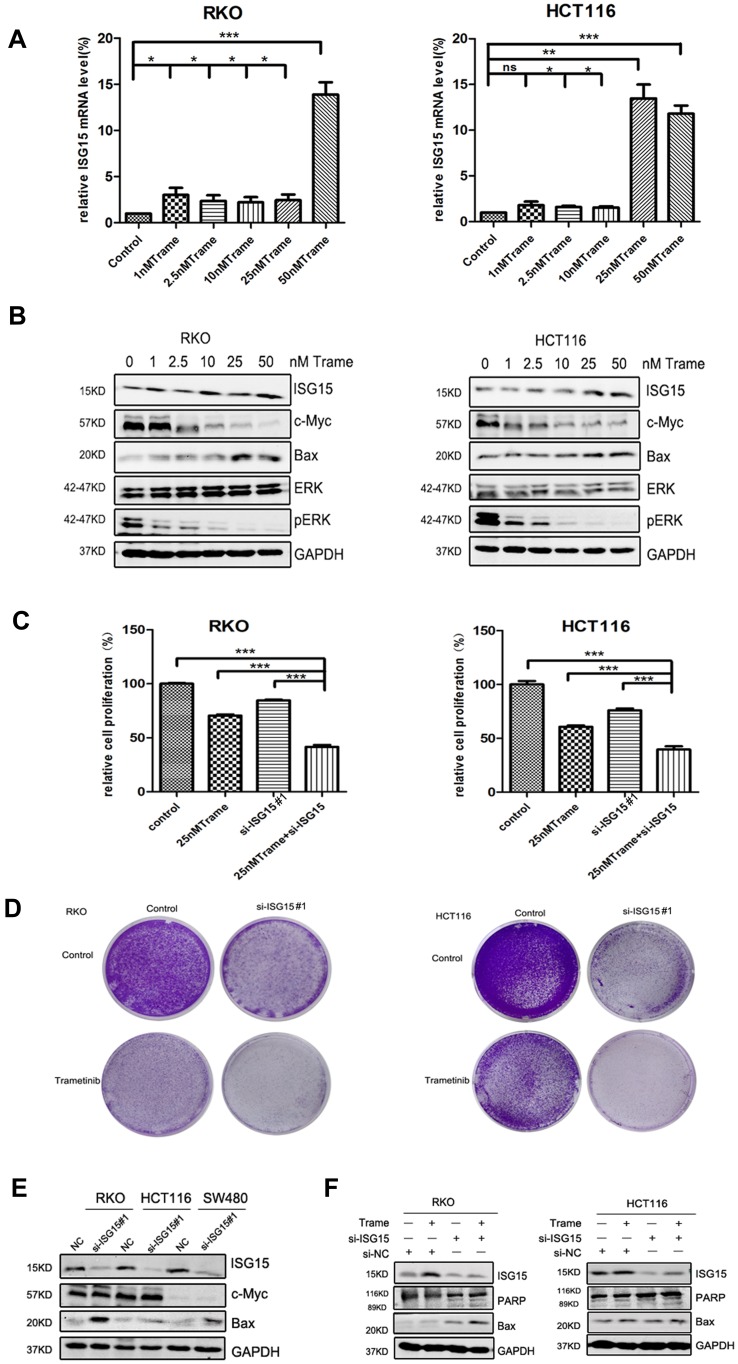
Inhibition of ISG15 Enhances the Anti-Cancer Effect of Trametinib in Colon Cancer Cells
We next tested whether combined therapeutic strategies with trametinib and ISG15 inhibition could be effective in treating colon cancer cells. As shown in Figure 5B, trametinib decreased the expression of c-Myc, a cell proliferation related protein, and improved the expression of Bax, an apoptosis inducing protein. Furthermore, trametinib increased the mRNA and protein expression of ISG15 (Figure 5A and B). Therefore, we treated cancer cells with ISG15 siRNA and trametinib and found that this combination had a synergistic effect that inhibited cell proliferation (Figure 5C, Figure S1D) and colony formation (Figures 5D, S1E and F). Next, we also examined the expression of c-Myc and Bax by Western blot following the knockdown of ISG15, and found that suppression of ISG15 had no effect on c-Myc, but promoted the upregulation of Bax (Figure 5E). Finally, we combined trametinib with si-ISG15 to investigate whether ISG15 inhibition enhances the cytotoxicity of trametinib against colon cancer at the protein levels. Our results showed that si-ISG15 synergized with trametinib to dramatically reduce the expression of ISG15 and increase the pro-apoptotic genes PARP and Bax (Figure 5F). Together, ISG15 inhibition combined with trametinib displayed synergistic inhibitory effects in colon cancer cells.
Figure 5.
ISG15 inhibition enhances the anti-cancer effect of trametinib in colon cancer cells. (A) The mRNA expression of ISG15 following treatment with the indicated concentrations of trametinib (0–50 nM) in colon cancer cells for 2 d by qPCR assay (*p< 0.05, **p< 0.01, ***p< 0.001). (B) WB for ISG15, c-Myc, Bax, pERK and ERK following treatment with the indicated concentrations of trametinib (0–50 nM) in colon cancer cells for 2 d. (C) Histograms shows the percentage of cell proliferation in colon cancer cells after transfection with si-ISG15 and/or 25 nM trametinib by CCK8 assays at day 3 (***p< 0.001). (D) Colony formation assays show that knockdown of ISG15 combined with 25 nM trametinib inhibited colon cancer cell proliferation at day 7. (E) The levels of ISG15, c-Myc and Bax following ISG15 silencing in colon cancer cells for 3 d as determined via Western blot. NC, Negative control; si-ISG15, siRNA of ISG15. (F) The levels of ISG15, cleaved PARP and Bax following treatment with si-ISG15 (and/or 25 nM trametinib) in RKO and HCT116 cells for 3 d. NC, Negative control; si-ISG15, siRNA of ISG15.
Discussion
ISG15, a 15 KDa protein, belongs to the ubiquitin-like (UBL) superfamily25 which is synthesized by several cells in response to various stimuli such as type-I IFNs, virus and genotoxicity.26 In addition, the function of ISG15 involves in diverse cellular pathways, including translation, RNA splicing, chromatin remodeling, polymerase II transcription, cytoskeleton organization, stress responses,27 autophagy28 and metabolism.29 Previous studies have discovered that ISG15 expression is associated with tumor responses to chemotherapy, radiotherapy and IFN-α treatment.30,31,32 Recently researchers have identified ISG15-deficient patients and suggested the underlying process of ISG15 regulation, these might cast some light on the intricacy of this protein. Also, there has been voices demanding a re-evaluation of ISG15.33 Several investigations has demonstrated the carcinogenicity of ISG15, which played an important role in cancer-associated inflammation.18,20,21,23 ISG15 is also associated with cytoskeleton disruption, cancer cell migration and poor prognosis.34 Recently, ISG15 is thought to function both as an oncogene and a tumor-suppressor gene in different tumors.35 For example, knockdown of ISG15 expression reduced breast cancer cell motility compared to the control in cell migration assays.36 In another study, Jeon et al37 reported that ISG15, as a possible tumor suppressor, provoked anchorage-dependent cell growth after doxorubicin treatment in mouse breast cancer model. Adam R.Brown38 purposed a process through which those select IFN-stimulated genes, especially ISG15, may facilitate colon cancer cell survival and growth by suppressing apoptosis, which was consistent with our findings. In addition, protein ISGylation could exacerbate experimental colitis and colitis-associated cancer. Jun-Bao Fana39 discovered an elevation of inflammation-related cytokines as well as the p38 MAP kinase activation induced by LPS in mouse. Above all, ISG15 plays an important role in the progression of various tumors. However, the pathological or physiological functions of ISG15 and their implications for targeted therapy have not been clearly elucidated in colon cancer.
In our study, immunohistochemical analysis and Western blot analysis were used to detect ISG15 expression in colon tumor tissues and pericancerous tissues. The results demonstrated that the expression of ISG15 protein was significantly higher in tumors than that in adjacent control tissues. Additionally, inhibition of ISG15 with siRNA could suppress tumor growth and migration in colon cancer. Using bioinformatics, it was also predicted that the ISG15 mRNA level of colon cancer patients was higher than that of normal patients, and the expression level of ISG15 was inversely proportional to the overall survival (OS) in colon cancer patients. Further investigation in gene expression indicated that trametinib treatment could induce the expression of ISG15. While suppression of ISG15 by siRNA could enhance the cytotoxicity of trametinib against colon cancer.
In summary, we have identified ISG15 as an oncogene in colon cancer, with a mechanism that mainly dealt with the proliferation and metastasis of colon cancer cells. Notably, our data point out the possibility that inhibition of ISG15 with small molecules which do not currently exist combined with trametinib treatment would be a novel therapeutic strategy for colon cancer.
Ethics Approval and Consent to Participate
The human colon cancer tissues as provided by the Ethics Committee of Taizhou Hospital of Zhejiang Province.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No.81572850).
Disclosure
No potential conflict of interest was reported by the authors.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.v61:2 [DOI] [PubMed] [Google Scholar]
- 2.Oddo D, Sennott EM, Barault L, et al. Molecular landscape of acquired resistance to targeted therapy combinations in BRAF-mutant colorectal cancer. Cancer Res. 2016;76:4504–4515. doi: 10.1158/0008-5472.CAN-16-0396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerji U, Camidge DR, Verheul HM, et al. The first-in-human study of the hydrogen sulfate (Hyd-Sulfate) capsule of the MEK1/2 Inhibitor AZD6244 (ARRY-142886): a phase I open-label multicenter trial in patients with advanced cancer. Clin Cancer Res. 2010;16:1613–1623. doi: 10.1158/1078-0432.CCR-09-2483 [DOI] [PubMed] [Google Scholar]
- 4.Lorusso PM, Adjei AA, Varterasian M, et al. Phase I and pharmacodynamic study of the oral MEK inhibitor CI-1040 in patients with advanced malignancies. J Clin Oncol. 2005;23:5281–5293. doi: 10.1200/JCO.2005.14.415 [DOI] [PubMed] [Google Scholar]
- 5.Rinehart J, Adjei AA, Lorusso PM, et al. Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. J Clin Oncol. 2004;22:4456–4462. doi: 10.1200/JCO.2004.01.185 [DOI] [PubMed] [Google Scholar]
- 6.Poulikakos PI, Solit DB. Resistance to MEK inhibitors: should we co-target upstream? Sci Signal. 2011;4:pe16. doi: 10.1126/scisignal.2001948 [DOI] [PubMed] [Google Scholar]
- 7.Turke AB, Song Y, Costa C, et al. MEK inhibition leads to PI3K/AKT activation by relieving a negative feedback on ERBB receptors. Cancer Res. 2012;72:3228–3237. doi: 10.1158/0008-5472.CAN-11-3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britten CD. PI3K and MEK inhibitor combinations: examining the evidence in selected tumor types. Cancer Chemother Pharmacol. 2013;71:1395–1409. doi: 10.1007/s00280-013-2121-1 [DOI] [PubMed] [Google Scholar]
- 9.Gilmartin AG, Bleam MR, Groy A, et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin Cancer Res. 2011;17:989–1000. doi: 10.1158/1078-0432.CCR-10-2200 [DOI] [PubMed] [Google Scholar]
- 10.Infante JR, Papadopoulos KP, Bendell JC, et al. A phase 1b study of trametinib, an oral mitogen-activated protein kinase kinase (MEK) inhibitor, in combination with gemcitabine in advanced solid tumours. Eur J Cancer. 2013;49:2077–2085. doi: 10.1016/j.ejca.2013.03.020 [DOI] [PubMed] [Google Scholar]
- 11.Tolcher AW, Bendell JC, Papadopoulos KP, et al. A phase IB trial of the oral MEK inhibitor trametinib (GSK1120212) in combination with everolimus in patients with advanced solid tumors. Ann Oncol. 2015;26:58–64. doi: 10.1093/annonc/mdu482 [DOI] [PubMed] [Google Scholar]
- 12.Levine BD, Cagan RL. Drosophila lung cancer models identify trametinib plus statin as candidate therapeutic. Cell Rep. 2016;14:1477–1487. doi: 10.1016/j.celrep.2015.12.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue Z, Vis DJ, Bruna A, et al. MAP3K1 and MAP2K4 mutations are associated with sensitivity to MEK inhibitors in multiple cancer models. Cell Res. 2018;28:719–729. doi: 10.1038/s41422-018-0044-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manchado E, Weissmueller S, Morris JT, et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature. 2016;534:647–651. doi: 10.1038/nature18600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haas AL, Ahrens P, Bright PM, Ankel H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J Biol Chem. 1987;262:11315–11323. [PubMed] [Google Scholar]
- 16.Bektas N, Noetzel E, Veeck J, et al. The ubiquitin-like molecule interferon-stimulated gene 15 (ISG15) is a potential prognostic marker in human breast cancer. Breast Cancer Res. 2008;10:R58. doi: 10.1186/bcr2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desai SD, Wood LM, Tsai YC, et al. ISG15 as a novel tumor biomarker for drug sensitivity. Mol Cancer Ther. 2008;7:1430–1439. doi: 10.1158/1535-7163.MCT-07-2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai SD, Haas AL, Wood LM, et al. Elevated expression of ISG15 in tumor cells interferes with the ubiquitin/26S proteasome pathway. Cancer Res. 2006;66:921–928. doi: 10.1158/0008-5472.CAN-05-1123 [DOI] [PubMed] [Google Scholar]
- 19.Padovan E, Terracciano L, Certa U, et al. Interferon stimulated gene 15 constitutively produced by melanoma cells induces e-cadherin expression on human dendritic cells. Cancer Res. 2002;62:3453–3458. [PubMed] [Google Scholar]
- 20.Laljee RP, Muddaiah S, Salagundi B, et al. Interferon stimulated gene-ISG15 is a potential diagnostic biomarker in oral squamous cell carcinomas. Asian Pac J Cancer Prev. 2013;14:1147–1150. doi: 10.7314/APJCP.2013.14.2.1147 [DOI] [PubMed] [Google Scholar]
- 21.Chi LM, Lee CW, Chang KP, et al. Enhanced interferon signaling pathway in oral cancer revealed by quantitative proteome analysis of microdissected specimens using 16O/18O labeling and integrated two-dimensional LC-ESI-MALDI tandem MS. Mol Cell Proteomics. 2009;8:1453–1474. doi: 10.1074/mcp.M800460-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen JB, Aaboe M, Borden EC, Goloubeva OG, Hassel BA, Orntoft TF. Stage-associated overexpression of the ubiquitin-like protein, ISG15, in bladder cancer. Br J Cancer. 2006;94:1465–1471. doi: 10.1038/sj.bjc.6603099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roulois D, Loo YH, Singhania R, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ontology CG. The gene ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pitha-Rowe I, Petty WJ, Feng Q, et al. Microarray analyses uncover UBE1L as a candidate target gene for lung cancer chemoprevention. Cancer Res. 2004;64:8109–8115. doi: 10.1158/0008-5472.CAN-03-3938 [DOI] [PubMed] [Google Scholar]
- 26.D’Cunha J, Ramanujam S, Wagner RJ, Witt PL, Knight EJ, Borden EC. In vitro and in vivo secretion of human ISG15, an IFN-induced immunomodulatory cytokine. J Immunol. 1996;157:4100–4108. [PubMed] [Google Scholar]
- 27.Zhao C, Denison C, Huibregtse JM, Gygi S, Krug RM. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc Natl Acad Sci USA. 2005;102:10200–10205. doi: 10.1073/pnas.0504754102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu D, Zhang T, Xiao J, et al. Modification of BECN1 by ISG15 plays a crucial role in autophagy regulation by type I IFN/interferon. Autophagy. 2015;11:617–628. doi: 10.1080/15548627.2015.1023982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baldanta S, Fernandez-Escobar M, Acin-Perez R, et al. ISG15 governs mitochondrial function in macrophages following vaccinia virus infection. PLoS Pathog. 2017;13:e1006651. doi: 10.1371/journal.ppat.1006651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pitha-Rowe I, Hassel BA, Dmitrovsky E. Involvement of UBE1L in ISG15 conjugation during retinoid-induced differentiation of acute promyelocytic leukemia. J Biol Chem. 2004;279:18178–18187. doi: 10.1074/jbc.M309259200 [DOI] [PubMed] [Google Scholar]
- 31.Weichselbaum RR, Ishwaran H, Yoon T, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci U S A. 2008;105:18490–18495. doi: 10.1073/pnas.0809242105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogunovic D, Byun M, Durfee LA, et al. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science. 2012;337:1684–1688. doi: 10.1126/science.1224026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Bogunovic D, Payelle-Brogard B, et al. Human intracellular ISG15 prevents interferon-alpha/beta over-amplification and auto-inflammation. Nature. 2015;517:89–93. doi: 10.1038/nature13801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hadjivasiliou A. ISG15 implicated in cytoskeleton disruption and promotion of breast cancer. Expert Rev Proteomics. 2012;9:7. doi: 10.1586/epr.12.36 [DOI] [PubMed] [Google Scholar]
- 35.Andersen JB, Hassel BA. The interferon regulated ubiquitin-like protein, ISG15, in tumorigenesis: friend or foe? Cytokine Growth Factor Rev. 2006;17:411–421. doi: 10.1016/j.cytogfr.2006.10.001 [DOI] [PubMed] [Google Scholar]
- 36.Desai SD, Reed RE, Burks J, et al. ISG15 disrupts cytoskeletal architecture and promotes motility in human breast cancer cells. Exp Biol Med (Maywood). 2012;237:38–49. doi: 10.1258/ebm.2011.011236 [DOI] [PubMed] [Google Scholar]
- 37.Jeon YJ, Jo MG, Yoo HM, et al. Chemosensitivity is controlled by p63 modification with ubiquitin-like protein ISG15. J Clin Invest. 2012;122:2622–2636. doi: 10.1172/JCI61762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown AR, Simmen RC, Raj VR, Van TT, MacLeod SL, Simmen FA. Kruppel-like factor 9 (KLF9) prevents colorectal cancer through inhibition of interferon-related signaling. Carcinogenesis. 2015;36:946–955. doi: 10.1093/carcin/bgv104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan JB, Miyauchi-Ishida S, Arimoto K, et al. Type I IFN induces protein ISGylation to enhance cytokine expression and augments colonic inflammation. Proc Natl Acad Sci U S A. 2015;112:14313–14318. doi: 10.1073/pnas.1505690112 [DOI] [PMC free article] [PubMed] [Google Scholar]