Abstract
Vitamin D deficiency is prevalent in pregnant women and is associated with adverse pregnancy outcomes, in particular disorders of malplacentation. The active form of vitamin D, 1,25-dihydroxyvitamin D3 (1,25(OH)2D3), is a potent regulator of innate and adaptive immunity, but it’s immune effects during pregnancy remain poorly understood. During early gestation, the predominant immune cells in maternal decidua are uterine natural killer cells (uNK), but the responsivity of these cells to 1,25(OH)2D3 is unknown despite high levels of 1,25(OH)2D3 in decidua. Transcriptomic responses to 1,25(OH)2D3 were characterised in paired donor uNK and peripheral natural killer cells (pNK) following cytokine (CK)-stimulation. RNAseq analyses indicated 911 genes were differentially expressed in CK-stimulated uNK versus CK-stimulated pNK in the absence of 1,25(OH)2D3, with predominant differentially-expressed pathways being associated with glycolysis and transforming growth factor β (TGFβ). RNAseq also showed that the vitamin D receptor (VDR) and its heterodimer partner retinoid X receptor were differentially-expressed in CK-stimulated uNK vs CK-stimulated pNK. Further analyses confirmed increased expression of VDR mRNA and protein, as well as VDR-RXR target in CK-stimulated uNK. RNAseq analysis showed that in CK-stimulated pNK, 1,25(OH)2D3 induced 38 and suppressed 33 transcripts, whilst in CK-stimulated uNK 1,25(OH)2D3 induced 46 and suppressed 19 genes. However, multiple comparison analysis of transcriptomic data indicated that 1,25(OH)2D3 had no significant overall effect on gene expression in either CK-stimulated pNK or uNK. These data indicate that CK-stimulated uNK are transcriptionally distinct from pNK and, despite expressing abundant VDR, neither pNK nor uNK are sensitive targets for vitamin D.
Keywords: vitamin D, natural killer cells, decidua, peripheral blood, pregnancy
Introduction
Human implantation and placental development present a unique challenge for the maternal immune system (Taglauer, et al. 2010). From embryonic implantation, semi-allogeneic fetal extravillous trophoblast cells invade the maternal decidua and inner myometrium (including remodelling the spiral arteries) facilitating normal haemochorial placentation. Throughout these actions, the materno-fetal immune function remains active, with diverse mechanisms mounted to support fetal development and placentation (Abumaree, et al. 2012; Erlebacher 2013). From early pregnancy a high proportion of resident leukocytes, displaying unique phenotypes and functions reside within the uteroplacental interface (Bulmer, et al. 2010). CD56bright uterine natural killer cells (uNK) (60–70%) are predominant, appearing phenotypically distinct from CD56dim NK cells in peripheral blood (pNK) (<10% total circulating immune cell population). In addition to the classical pro-cytotoxic, anti-tumorigenic and anti-viral effects associated with pNK (Koopman, et al. 2003), uNK have also been shown to play a pivotal role in embryonic implantation and haemochorial placental development (Lash, et al. 2010).
Regulation of uNK function involves a complex network of factors (Szekeres-Bartho 2008), and we have postulated that vitamin D plays a role in this process. Vitamin D is a secosteroid that exerts powerful immuno-regulatory responses on cells from both the innate and adaptive immune systems following immune activation (Adams and Hewison 2008; Hewison 2011). Early in pregnancy, maternal concentrations of the active form of vitamin D, 1,25-dihydroxyvitamin D3 (1,25(OH)2D3), increase dramatically (Tamblyn, et al. 2017). Moreover, the enzyme that catalyzes synthesis of 1,25(OH)2D3 from precursor 25-hydroxyvitamin D3 (25(OH)D3), 25-hydroxyvitamin D-1α-hydroxylase (CYP27B1) (Gray, et al. 1979; Weisman, et al. 1979; Zehnder, et al. 2002), as well as the nuclear vitamin D receptor (VDR) for 1,25(OH)2D3 (Zehnder et al. 2002), are abundantly expressed by both maternal decidua (stromal and immune cells) and fetal trophoblast (Evans, et al. 2004). The precise targets for decidual 1,25(OH)2D3 have yet to be defined, but may include effects upon VDR-expressing decidual immune cells (Zehnder et al. 2002). The aim of the current study was to characterise ex vivo the effects of 1,25(OH)2D3 on gene expression in activated uNK from first trimester decidual tissue, and to contrast these changes with effects of 1,25(OH)2D3 on paired pNK.
Materials and Methods
Ethical approval
Paired decidual and peripheral blood samples were obtained from healthy pregnant women undergoing 1st trimester elective surgical termination of pregnancy (TOP) (<12 weeks [w] gestational age [GA]; range 7–11+2 w) following written informed consent (2014–2017; NHS REC approval 06/Q2707/12 and 13/WM/0178). GA was determined by ultrasound measurement of crown rump length, and relevant anonymised patient demographics obtained as summarised (Supplemental Table 1).
Isolation of NK cells from peripheral blood and decidua
CD56+ cell-enriched isolates were prepared by modification of a previously published method (Lash, et al. 2006). Briefly, whole decidual tissue was minced and DNase/collagenase-treated. Lymphocyte purification was then carried out by density gradient centrifugation (Ficoll Plus, GE Healthcare Life Sciences, Little Chalfont, UK). For initial cell surface marker analysis, anti-CD56 reactive magnetic bead separation (MidiMACS, Miltenyi Biotec, Bisley, UK) was performed to obtain CD56+ cells.
Fluorescence Activated Cell Sorting (FACS)
For cell culture and RNAseq analysis, density-centrifuged cell populations were FACS sorted. Cells were surface stained with primary mAb as summarised and then washed in PBS and re-constituted in 500 μL MACS (PBS, pH 7.2, 0.5% bovine serum albumin (BSA), 2mM EDTA) for cell sorting. Propidium iodide (PI) was added to assess cell viability at the point of cell sorting, using a BD FACSAria Fusion Flow Cytometer (BD Biosciences, Wokingham, UK). The gating strategy for isolating live CD3-/CD56+ NK from peripheral blood and decidua is shown in Supplemental Figure 1. Live CD45+ CD3− CD14− CD56+ NKp46+ pNK and uNK were identified (overall purity ≥96%; pNK 99.0–99.2%, uNKs 96.0–98.7%). Monoclonal antibodies (mAb) used to gate live cells were: anti-CD56, anti-NKp46 (Miltenyi Biotec, Bisley, UK), anti-CD3, anti-CD8, anti-CD14 from BD Biosciences (San Jose, CA, USA). Isotype matched IgG and IgM controls conjugated to the appropriate fluorochrome (BD Bioscience, San Jose, CA, USA and eBioscience, Waltham, MA, USA) were used for non-specific binding.
Cell culture
Paired uNK and pNK were cultured in complete cell culture medium (Penstrep 100μg/ml, L-glutamine 2mM, RPMI 1640, fetal calf serum (10%) (Sigma-Aldrich, Irvine, UK) at 37°C and 5% CO2 for 24 hours (h), as either unstimulated controls or with cytokine (CK) stimulation using recognised NK cell co-activators: IL-2 [50 IU/ mL], IL-12 [10 ng/ mL], and IL-15 [10 ng/ mL]). Unstimulated and CK uNK and pNK were cultured in the presence and absence of 1,25(OH)2D3 (10nM)(Enzo Lifesciences, Exeter, UK) co-treatment.
Flow cytometric analysis of CD69 and VDR
Expression of VDR (D6 anti-VDR, Santa Cruz Biotechnology, Santa Cruz, CA, USA) and CD69 (anti-CD69, BD Biosciences, San Jose, CA, USA), a recognised surface marker of NK cell activation (Clausen, et al. 2003) was assessed in paired pNK and uNK following 24 h culture in the presence and absence of CK stimulation with or without 1,25(OH)2D3. Briefly, cultured cells were harvested, washed in PBS and incubated with live/dead fixable discrimination dye (Thermo Fisher, Waltham, MA, USA) on ice for 30 min. Cells were washed with PBS, incubated for 30 min with surface primary mAb at 4°C and then re-washed. For intracellular VDR analysis, a protocol optimised from Bendix et al was used (Bendix, et al. 2015). Briefly, following surface staining cells were fixed for 20 min in 100 μL 4% PFA, washed twice in 2 mL 2% FCS-PBS, and incubated for 30 min on ice in 75 μL 5% FCS 0.5% Triton X solution (Sigma-Aldrich, Irvine, UK). Cells were then incubated with anti-VDR mAb for 30 min, washed with 2% FCS-PBS and re-incubated with anti-mouse IgG2a APC secondary mAb under the same conditions. Cells were then re-washed twice in 2% FCS-PBS followed by PBS and stored at 4°C prior to analysis. Flow cytometry data were collected using a multi-channel Dako Cyan flow cytometer (Beckman Coulter, High Wycombe, UK). Data were analysed using Flow Jo V10 (Tree star, Inc., Ashland, USA), gating live CD45+ CD56+ NKp46+ cells (Supplemental Figure 2). The specificity of the D6 anti-VDR mAb in VDR positive control cells (activated T cells) was demonstrated using flow cytometry, immunoprecipitation and chromatin immunoprecipitation (Supplemental Figure 3).
Confocal Imaging of VDR in CD56+ pNKs
CD56+ pNKs were isolated and stimulated overnight with CK in the presence of 1,25(OH)2D3 as described above. Cells were surface-labelled for CD56 using FITC-anti-CD56 (clone REA196 Miltenyi Biotec), then fixed and permeabilised for VDR labelling with mouse anti-human VDR (D6 clone, Santa Cruz Biotechnology, Santa Cruz, CA, USA) or mouse IgG2a isotype control according to the protocol described above for flow cytometry but with goat anti-mouse IgG2a-594 and goat-anti-FITC-488 (molecular probes, Fisher Scientific) secondary labelling for 30 mins in the presence of Hoechst 33342 (10 μg/ml; Fisher Scientific). After a final wash, cells were re-suspended in ProlongR Gold antifade reagent (Molecular Probes, Cell Signalling Technologies, UK) and placed under coverslip on charged microscope slides. Edges were sealed with nail varnish and 22-slice Z stack images of cells at 63x magnification, resolution 1024 × 1024 pixels per image were captured using the Zeiss LSM 880, Axio Observer confocal microscope and studied with Zen2012 software (Supplemental Figure 4).
Quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted as described previously (Jeffery, et al. 2012). cDNA was prepared by reverse transcription Taqman Reverse Transcription Reagents Kit (Applied Biosystems, Roche, Nersey, USA) as per manufacturer’s instructions. qRT-PCR was performed on an ABI 7500 qPCR machine using Taqman assays from Applied Biosystems for VDR (Hs00172113_m1) and multiplexed with VIC labelled 18S rRNA (4319413E) (Applied Biosystems, Roche, Nersey, USA). Paired uNK and pNK gene expression following 24-hour culture in the presence and absence of CK-stimulation with and without 1,25(OH)2D3 co-treatment was measured. Relative expression compared to paired unstimulated pNK was assessed using fold-change calculated by the 2−ΔΔCt method. All statistical analyses were carried out using Sigmaplot 9.0 software (Systat Inc., San Jose, CA, USA). Experimental means were compared statistically using one-way ANOVA, with the Holm-Sidak method used as a post hoc multiple-comparison procedure. Statistical analyses for RT-qPCR analyses were carried out using raw ΔCt values.
RNA sequence analysis (RNA-seq)
Total RNA was extracted using the RNeasy Micro Kit and RNeasy MinElute spin columns (QIAGEN, Hilden Germany), as per manufacturer’s instructions, eluted in 15 μL RNAse-free water, and immediately stored at −80°C prior to library preparation and sequencing (Source Bioscience Facilities, Nottingham, UK). Following initial QC check using the Agilent BioAnalyzer 2100 (Agilent, Santa Clara, CA, USA) cDNA library preparation was performed using Switching Mechanism at the 5′ end of RNA Template (SMART)er Stranded Total RNA Seq Kit – Pico (Clontech, Mountain View, CA, USA) (RNA range = pg). Random primers and locked nucleic acid (LNA) technology were utilised to optimise total coverage. Illumina Adapters and Indexes (barcode markers) (Illumina, Inc., San Diego, USA) were added at PCR1 with rRNA cleaved by ZapR in the presence of R-Probes (mammalian-specific). The non-cleaved fragments underwent further enrichment in PCR2 cycle. All libraries were QC validated using the Agilent BioAnalyzer 2100 (Santa Clara, CA, USA) to confirm index samples amplified concentration and size distribution. The libraries were pooled and re-validated to assess molarity and size distribution prior to loading (1.8pM concentration) onto a High Output NextSeq 500 Flow Cell pv2. (Illumina) across 4 lanes for RNA-seq using 75-base-pair (bp) paired-end reading, generating ~800 million total reads with an average of 25 million per sample. Raw data was returned in the FastQ Phred+33 (Illumina 1.9) format.
Bioinformatics and Pathway analysis
Bioinformatics analysis of RNA-seq analyses was performed at University of Birmingham, UK using Partek Flow Software (Partek, St Louis, Missouri, USA). Data were trimmed for adapter sequences and pre-alignment QA/QC was performed before being mapped (aligned) using STAR-2.4.1d (Dobin, et al. 2013). Post-alignment QA/QC was performed with a coverage report generated. Reads were quantified to transcriptome hg38_RefSeq (Genome Reference Consortium GRCh38), and normalised using quantile normalisation. Median total aligned reads was 71567649 (IQR; 64250828–74820738), representing 85.7% (84.5 – 87.5%) coverage. Differential gene expression was defined by a cut-off fold-change of < −1.5 and > 1.5, p-value ≤ 0.05 and false discovery rate (FDR)-step up ≤ 0.05. P-values were calculated using F-statistics, which calculates variance within samples. Three analyses were assessed: (i) CK uNK versus (vs) CK pNK; (ii) CK uNK vs CK uNK + 1,25(OH)2D3; (iii) CK pNK vs CK pNK + 1,25(OH)2D3.
Characterisation of common biological processes in the CK-activated uNK and pNK groups was carried out using PathVisio (Version 3.2.4), an open-source pathway analysis and visualization software tool for biological data analysis, using previously described protocols (Munoz Garcia, et al. 2018). Briefly, analysis was performed using the 11164 raw gene counts obtained, with corresponding ENTREZ ID and symbol from the HUGO Gene Nomenclature Committee (HGNC) identifiers. A p-value <0.05 was applied to determine differentially-expressed genes. The Z-Score, which signifies whether the total number of genes in the pathway are over-represented (Z-score >1.96) compared to the complete data set, was then calculated. The curated human pathway collection from the online-pathway repository WikiPathways (Kutmon, et al. 2016) including the Reactome (Bohler, et al. 2016) collection (downloaded May 2017) was used.
Results
Comparison of gene expression in cytokine (CK)-stimulated pNK and uNK cells
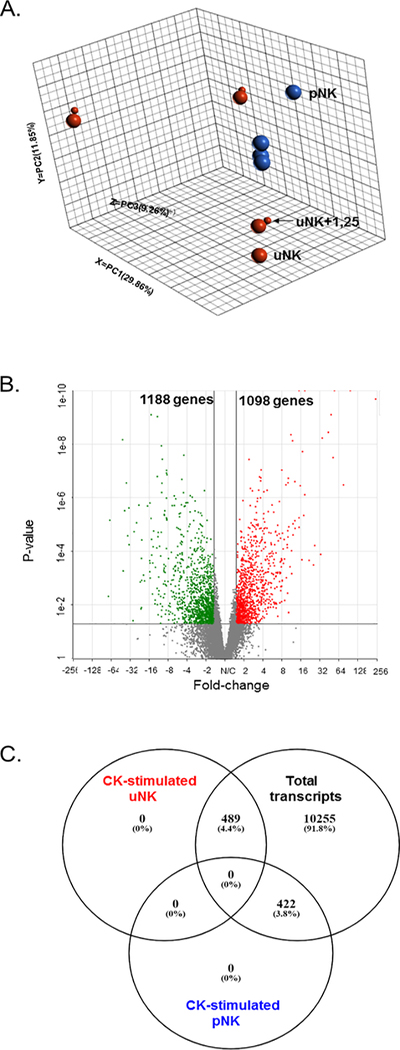
Principal Component Analysis (PCA) of unbiased genome-wide RNAseq analyses revealed wide variance in the transcriptional profiles of CK-stimulated uNK and pNK (Figure 1A). In both cell types, co-treatment with 1,25(OH)2D3 did not result in PCA separation of transcriptional profiles from their respective CK controls. Volcano plots of differentially expressed genes in CK-stimulated uNK relative to CK-stimulated pNK using a cut-off of p≤0.05 and fold change ≤−1.5 and ≥+1.5 showed 1098 up-regulated genes in CK-stimulated uNKs (red) and 1188 down-regulated genes (green), with those not significantly different in grey (n= 11163) (Figure 1B). Adjustment for false discovery rate (FDR) identified 911 transcripts to be differentially-expressed between CK-stimulated uNK and pNK, with 422 down-, and 489 up-regulated in CK-stimulated uNK relative to CK-stimulated pNK (p value ≤ 0.05; log2 fold-change ≤1.5 or ≥1.5, FDR step-up ≤ 0.05) (Figure 1C). Significantly regulated genes in CK-stimulated uNK relative to CK-stimulated pNK included up-regulation of NCAM1 (CD56) (fold-change 5.29), and down-regulation of FCG3RA (CD16A) (fold change −14.57), both of which are well-recognised differentially expressed markers for CD56bright NK versus CD56dim pNK subsets (Koopman et al. 2003; Poli, et al. 2009b). Interestingly, granzyme (GZMK) (fold-change (7.83)) and CD96 (2.53), which permits adhesive interactions and modulates responses between activated T and NK cells (Georgiev, et al. 2018), were both up-regulated in CK-stimulated uNK. CD276 (7.71), a regulator of T cell cytotoxicity (Picarda, et al. 2016) was also up-regulated in CK uNK.
Figure 1. Transcriptomic analysis of cytokine (CK) stimulated pNK and uNK cells.
A. Principal component analysis (PCA) analysis of CK-stimulated uNK and pNKs in the presence and absence of 1,25(OH)2D3. 3-dimensional dot-plot summarising the main sources of variance across the whole RNA-seq data set by principal components; PC1 29.9%, PC2 11.9%, PC3 9.3%. This includes pNK (red), uNK (blue) in the presence (small dot) and absence (large dot) of 1,25(OH)2D3 co-treatment. B. Volcano plot summary of differentially expressed genes in CK-stimulated uNK relative to CK-stimulated pNK. Significantly up-regulated genes in CK-stimulated uNKs are in red (n=1098), down-regulated genes in green (n=1188), with those not significantly different in grey (n= 11163). A cut-off of p≤0.05 and fold change ≤−1.5 or ≥+1.5 was used to determine significance. C. Venn diagram summarising the distribution of differentially-expressed genes in CK-stimulated uNK relative to CK-stimulated pNK compared to total transcripts in CK-stimulated uNK and CK pNK following adjustment for false discover rate (FDR): p value ≤ 0.05; log2 fold-change ≤1.5 or ≥1.5, FDR step-up ≤ 0.05.
To better delineate the differences in transcript expression between CK-stimulated uNK and pNK, complementary pathway analysis was performed on FDR-adjusted differentially-expressed data using WikiPathways (WP) workflows. A broad spectrum of enriched canonical pathways was identified, with significantly enriched pathways ranked from a high to low Z-score (Supplemental Dataset 1). Pathways identified as being significantly enriched based on a Z-score ≥1.96 are shown in Table 1. Overall, 29 WP pathways were significantly enriched in CK-stimulated uNK relative to CK-stimulated pNK, including pathways related to hypoxia-inducible factor (HIF)1α and peroxisomal proliferator-activated receptor (PPAR)γ regulation of glycolysis, and transforming growth factor (TGF)α signalling (Supplemental Figures 5A–5S).
Table 1. Pathway analysis of differential gene expression in CK uNK versus CK Pnk.
Summary of WikiPathways data-base analysis for CK uNK versus CK pNK. Frequency of significant differentially-expressed genes (p-value <0.05, Z-Score >1.96), total genes measured as a proportion of the total frequency of pathway enriched genes were ranked based on Z score.
| Pathway | positive | measured | total | % positive | Z Score | p-value |
|---|---|---|---|---|---|---|
| HIF1A and PPARG regulation of glycolysis | 6 | 6 | 19 | 100.00% | 3.64 | 0 |
| TGF-beta Signaling Pathway | 51 | 111 | 133 | 45.95% | 3.38 | 0.003 |
| Cori Cycle | 8 | 10 | 53 | 80.00% | 3.33 | 0 |
| Gastric Cancer Network 1 | 14 | 22 | 30 | 63.64% | 3.29 | 0.001 |
| Retinoblastoma Gene in Cancer | 38 | 80 | 98 | 47.50% | 3.16 | 0.001 |
| Extracellular vesicle-mediated signaling in recipient cells | 12 | 19 | 31 | 63.16% | 3.01 | 0 |
| Regulation of sister chromatid separation at metaphase-anaphase transition | 10 | 15 | 16 | 66.67% | 2.96 | 0.004 |
| Calcium Regulation in the Cardiac Cell | 31 | 65 | 164 | 47.69% | 2.88 | 0.003 |
| Primary Focal Segmental Glomerulosclerosis FSGS | 20 | 38 | 78 | 52.63% | 2.86 | 0.003 |
| Fluoropyrimidine Activity | 13 | 22 | 58 | 59.09% | 2.82 | 0.001 |
| Integrin-mediated Cell Adhesion | 30 | 64 | 102 | 46.88% | 2.72 | 0.004 |
| Focal Adhesion | 46 | 108 | 201 | 42.59% | 2.57 | 0.014 |
| Chemokine signaling pathway | 42 | 99 | 172 | 42.42% | 2.42 | 0.024 |
| Ebola Virus Pathway on Host | 39 | 91 | 141 | 42.86% | 2.41 | 0.018 |
| B Cell Receptor Signaling Pathway | 36 | 83 | 99 | 43.37% | 2.4 | 0.009 |
| Nanoparticle-mediated activation of receptor signaling | 12 | 22 | 36 | 54.55% | 2.36 | 0.016 |
| Lamin A-processing pathway | 4 | 5 | 8 | 80.00% | 2.35 | 0.015 |
| Signal Transduction of S1P Receptor | 11 | 20 | 26 | 55.00% | 2.3 | 0.022 |
| MFAP5-mediated ovarian cancer cell motility and invasiveness | 6 | 9 | 18 | 66.67% | 2.29 | 0.026 |
| Microglia Pathogen Phagocytosis Pathway | 15 | 30 | 44 | 50.00% | 2.22 | 0.027 |
| Histone Modifications | 27 | 61 | 68 | 44.26% | 2.21 | 0.024 |
| Association Between Physico-Chemical Features & Toxicity Associated Pathways | 22 | 48 | 78 | 45.83% | 2.19 | 0.026 |
| Human Thyroid Stimulating Hormone (TSH) signaling pathway | 24 | 54 | 67 | 44.44% | 2.1 | 0.032 |
| Mesodermal Commitment Pathway | 35 | 84 | 154 | 41.67% | 2.08 | 0.024 |
| Apoptosis-related network due to altered Notch3 in ovarian cancer | 20 | 44 | 54 | 45.45% | 2.04 | 0.047 |
| Regulation of Wnt/B-catenin Signaling by Small Molecule Compounds | 7 | 12 | 33 | 58.33% | 2.03 | 0.036 |
| T-Cell antigen Receptor (TCR) Signaling Pathway | 32 | 77 | 93 | 41.56% | 1.97 | 0.047 |
| Purine metabolism | 6 | 10 | 75 | 60.00% | 1.96 | 0.044 |
| G Protein Signaling Pathways | 25 | 58 | 97 | 43.10% | 1.96 | 0.047 |
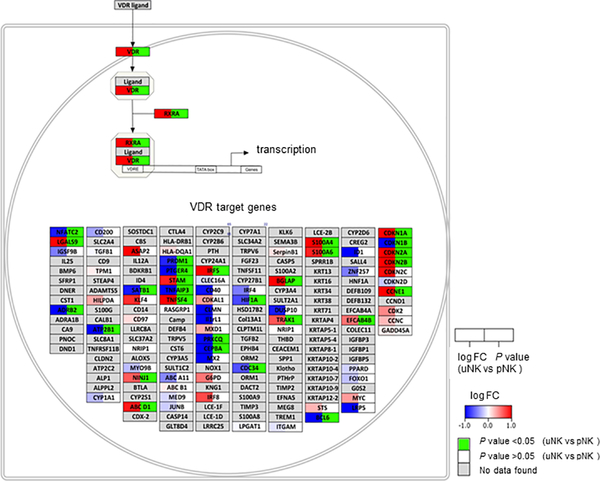
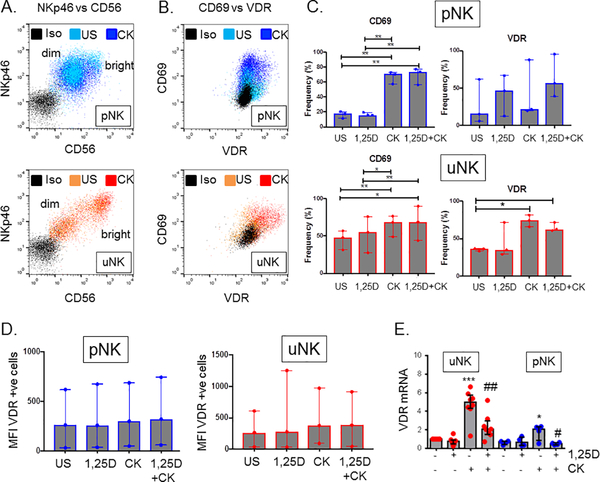
VDR expression in uNK is up-regulated following cytokine stimulation
RNA seq analysis revealed that VDR mRNA expression is significantly upregulated in CK stimulated uNK (fold change 8.56) relative to CK stimulated pNK. Analysis of differentially expressed pathways in CK-stimulated uNK versus CK-stimulated pNK in Supplemental Dataset 1 also showed enhanced expression of pathways associated with VDR and its heterodimer partner retinoid X receptor (RXR) (Z score 1.83, p=0.07) (Figure 2), as well as non-genomic actions of 1,25(OH)2D3 (Z score 1.88, p=0.062) (Supplemental Figure 5A). This suggests that capacity for 1,25(OH)2D3-VDR responses is elevated in CK-stimulated uNK relative to CK-stimulated pNK. Studies were therefore carried out to further characterise VDR expression in uNK and pNK. Flow cytometry showed that pNK and uNK expressed similar levels of the NK marker NKp46. Analysis of CD56 expression showed that uNK cells were characterised by more distinct populations of CD56bright and CD56dim cells than pNK (Figure 3A). Both CD56 and NKp46 showed no change in response to either CK stimulation or 1,25(OH)2D3 treatment in either uNK or pNK cells for median fluorescence intensity (MFI) (Figure 3A) or frequency of expression (data not shown). As shown in Figure 3B and 3C, CK-stimulation significantly up-regulated expression of the activation marker CD69 in both uNK (median 47.9%; IQR 31.5–56.6 to 68.1%; 48.9–76.4) and pNK (17.7%; 11.5–20.3 to 70.5%; 57.4–72.8), but treatment with 1,25(OH)2D3 had no significant effect on CD69 expression in either the unstimulated or CK-stimulated uNK or pNK populations (Figure 3C).
Figure 2. Pathway analysis of vitamin D receptor (VDR) signalling-related genes in CK-stimulated uNK versus CK-stimulated pNK.
Visualisation of genes with enhanced (red) or suppressed (blue) expression in CK-stimulated uNK versus CK-stimulated pNK. Individual genes are shown in boxes and box colour split into two parts, (1) the log2 fold-change in the left part of the box (blue down-regulated, white not changed, red up-regulated) and (2) p-value for statistical significance is shown in the right part of the box (green when significant). Pathway elements not assessed in the dataset are shown in grey.
Figure 3. Expression of VDR in paired peripheral blood and uterine natural killer cells.
A. Representative flow cytometry plots for expression of NKp46 and CD56 in isolated unstimulated (US) and cytokine (CK)-stimulated peripheral blood natural killer cells (pNK) and uterine natural killer cells (uNK) relative to isotype control (Iso). B. Representative flow cytometry plots for expression of VDR and CD69 in US and CK-stimulated CD3-CD56+ uNK and pNK cells relative to Iso control. C. Summary of VDR and CD69 surface protein expression in US- and CK-uNK and -pNK cells in the presence or absence of 1,25(OH)2D3 (1,25D). D. Median fluorescence intensity (MFI) for VDR expression in pNK and uNK cells. E. mRNA expression for VDR in isolated US and CK uNK and pNK cells in the presence and absence of 1,25(OH)2D3. Relative expression compared to US uNK cells; median value with bars denoting inter-quartile range. Effect of CK stimulation and 1,25(OH)2D3 was assessed by two-way ANOVA. Stars indicate level of significant difference between groups for which multiple comparisons analysis indicated significance *<0.05, **<0.01.
Without stimulation the frequency of NK cells that expressed VDR was low for both uNK (36.2%; 33.1–36.5) and pNK (15.8%; 5.81–61.6). CK stimulation significantly increased VDR frequency in uNK (74.2%; 65.6–81.3) although the absolute level of VDR expression by each expressing cell (MFI) was not significantly increased (Figure 3D). 1,25(OH)2D3 had no significant effect on VDR protein expression in either uNK or pNK in the presence or absence of CK-stimulation (Figure 3C). Expression of VDR mRNA was also significantly induced by CK-stimulation in uNK (p=0.0001) and pNK (p=0.017). Interestingly, this effect was suppressed in both cell types by co-treatment with 1,25(OH)2D3 (Figure 3D). Confocal imaging of CK-stimulated pNK cells (Supplemental Figure 4) showed co-localization of VDR protein with both DNA (Hoescht staining) and the plasma membrane (CD56), consistent with both membrane and nuclear localisation of VDR.
Effects of 1,25(OH)2D3 on gene expression in CK-stimulated pNK and uNK cells
When compared to CK-stimulated pNK, CK-stimulated pNK treated with 1,25(OH)2D3, showed only 71 genes differentially-expressed genes (fold-change ±1.5; p≤0.05). Of these, 33 (46.5%) were down- and 38 (53.5%) up-regulated by 1,25(OH)2D3. In CK-stimulated uNK treated with 1,25(OH)2D3 66 genes were differentially-expressed relative to CK-stimulation of uNK alone. Of these, 46 (69.7%) were down- and 19 (30.3%) up-regulated by 1,25(OH)2D3. In contrast to pNK, 1,25(OH)2D3 primarily targeted uNK genes implicated in cell processing, specifically cell adhesion, apoptosis, migration and angiogenesis (n=27; 41.5%) (Table 2). Due to the relatively small number of genes differentially-expressed in response to 1,25(OH)2D3 in either CK-stimulated pNK or uNK, adjustment of data to incorporate fold-change ±1.5, p-value ≤ 0.05, FDR-step up < 0.05 resulted in no significant effect of 1,25(OH)2D3 for either CK-stimulated uNK or CK-stimulated pNK. Furthermore, pathway analysis using non-adjusted differentially expressed gene data for either CK-stimulated uNK or CK-stimulated pNK cell treated with 1,25(OH)2D3 showed no significant enrichment of any specific pathways by 1,25(OH)2D3 (data not shown), and no commonality with any of the pathways that are known to be regulated by 1,25(OH)2D3 in other immune cells (Figure 4).
Table 2. Effect of 1,25(OH)2D3 on gene expression in CK pNK and uNK.
Summary of total genes differentially induced or suppressed by 1,25(OH)2D3 in CK-stimulated pNK and CK-stimulated uNK (fold change < −1.5 or > +1.5, p ≤ 0.05), with sub-categorisation according to transcript function.
| Functional classification | Genes expressed in pNK |
Genes expressed in uNK |
||||
|---|---|---|---|---|---|---|
| Total transcripts | Up-regulated by 1,25(OH)2D3 | Down-regulated by 1,25(OH)2D3 | Total transcripts | Up-regulated by 1,25(OH)2D3 | Down-regulated by 1,25(OH)2D3 | |
| Cell structure | 3 | TTLL1, FAM161A | CMYA5 | 2 | CAMSAP2, SOBP | |
| Cell survival, proliferation, invasion, adhesion, angiogenesis, trafficking | 15 | TMEM14A, MMP14, PIK3R6, ENG, RABL2A, DEPDC1B, TRIM35, CAB39L, AP5S1 | TSPAN4, SERPINI1, NRP2, RAPGEFL1, FAM195A, AP4M1 | 27 | TSPAN2, ARAP3, ITGAM,RARRES3, ADGRE5, FGL2, DYSF, NINI1, ZFP91-CNTF, DENND6B, BGLAP, DOCK3, RNF165, TAX1BP3, TRIM35, TAGLN2, RGS3 | ADGRG1, TMPRSS6, CYGB, C8orf44-SGK3, LZTS3, MARCKSL1, NACAD, TCF7L2, PHOSPHO2-KLH23, P2RY11 |
| Immune function | 3 | LXN, MTCP1, RSAD2 | 4 | SERPINB1, LGALS9, RELT | RSAD2 | |
| Metabolism and lipogenesis | 17 | ACOT1, ACSF2, CCBL1, MTHFD2L, PPP1R3F, AMPD3, GRHPR | PM20D2, TKFC, KDELC1, BDH2, GMPR, BCKDHB, NBR2, NPR2, ENPP1, NR2F6 | 7 | TMEM56-RWDD3, SARDH, CYP1A1, GDPGP1, ACSL1, GDE1, ADA | |
| Genetic | 10 | CBX8, DNAJC30, RPA3, EXOSC7, PPFIBP1 | TUT1, GTPBP3, MIR600HG, TCEANC, TYRO3P | 9 | TFEC, WWC2, ZBTB7A, HIC1, EFL1, HNRNPLL,TRAK1, CREG1 | BHLHB9 |
| Ion transport | 1 | SLC35E3 | 2 | SLC31A1 | SLC22A1 | |
| Unknown function | 10 | LOC101929767, TTC32, LOC100287042, PROB1 | ARMCX4, LOC441666, LOC100506804, PRRG4, CBWD6, THAP8 | 8 | ZSWIM5, LOC101928464, TNRC6C-AS1, POMGNT2, PLEKho2, NARS2, INTS6L | C1QTNF3-AMACR |
| Anti-sense, non-coding, snoRNAs | 11 | BRWD1-IT2, SNORA67, SNORA17B, LOC100129083, ADNP-AS1, RPL13P5 | C22orf34, SNHG25, TRIM47, MRPL42P5, GCSHP3 | 6 | SNORA17A, SIGLEC17P, NPPA-As1 | CCDC102A, ALMS1-ITI, LOC100505771 |
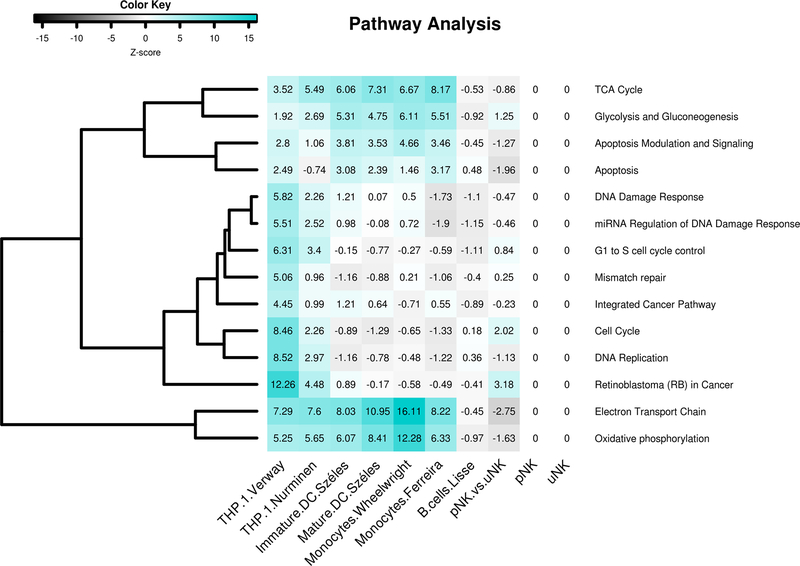
Figure 4. Pathway analysis of transcriptomic responses to 1,25(OH)2D3-treated in THP-1 cells, dendritic cells, monocytes and B cells compared to effects in pNK and uNK.
Heat map representing Z-scores for pathways significantly altered by 1,25(OH)2D3 comparing pathway analysis on datasets from THP-1 cells, dendritic cells (DC), monocytes and B cells with effects of 1,25(OH)2D3 on CK-stimulated pNK and uNK. Z-scores >1.96 indicate that more genes are significantly altered in this pathway compared to the complete dataset. Genes differentially expressed between CK-stimulated uNK and pNK in the absence of 1,25(OH)2D3 (pNK vs uNK) showed minimal overlap with the effect of 1,25(OH)2D3 on immune cells. The effects of 1,25(OH)2D3 on CK-stimulated pNK or uNK showed no commonality with the other immune cells.
Discussion
Previous studies using DNA microarrays have described transcriptomic variations in CD56bright pNK and CD56dim pNK cells, as well as CD56 bright uNK (Koopman et al. 2003). In the current study, pNK or uNK were not sub-categorized according to CD56 brightness, because CD56dim pNK predominate in peripheral blood (approximately 95% of total pNK cell population), whilst CD56bright uNK are the prevalent NK type in decidua (>95% of total uNK population). These differences imply that uNKs are constitutively active. In agreement with this, we observed that uNK were larger with abundant perforin and granzyme. Importantly, no change in CD56 or NKp46 activation receptor expression was measured in uNK or pNK in response to CK-stimulation or treatment with the active form of vitamin D, 1,25(OH)2D3. CD56bright NKs are considered efficient immunomodulatory cytokine producers, and only become cytotoxic following appropriate activation. However, in contrast to NK from most other sites, uNKs do not express the activating receptor CD16 which mediates antibody-dependent cellular cytotoxicity (Poli, et al. 2009a). It appears that whilst uNKs maintain an intrinsic capacity to exert cytotoxic functions when specifically challenged, this function is regulated in normal early pregnancy (Moffett and Colucci 2014). Further undefined local mechanisms, such as cytokine or hormone secretion, or crosstalk with other immune cell types within the decidual micro-environment are also thought to suppress the potential lytic effects of uNKs (Hanna, et al. 2006). In the current study, pro-inflammatory IL-2, IL-12 and IL-15 were utilised for CK challenge, as these are recognised NK cell activators, with both IL-12 and IL-15 constitutively expressed within the decidua from initial implantation (Kovacs, et al. 1992; Zourbas, et al. 2001). Although IL-2 concentrations appear low within normal decidua (Jokhi, et al. 1994), significantly elevated IL-2 has been reported in the setting of malplacentation (Garzia, et al. 2013), where vitamin D-deficiency is also more common (Robinson, et al. 2011).
The first aim of the current study was to compare the transcriptomic profiles of uNK versus pNK following immune activation by CK-stimulation. The second objective of the study was to determine if CK-stimulated uNK and pNK were responsive to 1,25(OH)2D3 in a similar manner to that reported for other components of the immune system such as macrophages and T cells (Adams and Hewison 2008). Since the effects of 1,25(OH)2D3 on NK cell function in decidua and maternal blood are still unclear, NK cell subsets were not further sub-classified.
In the current study approximately 900 genes were differentially-expressed in CK-stimulated uNK versus CK-stimulated pNK, similar to previous DNA array analyses for unstimulated CD56dim pNK versus unstimulated CD56bright uNK (Koopman et al. 2003). In CK-stimulated uNK a comparable number of up- (429) and down-regulated (422) genes were measured when adjusted for FDR. By contrast, in the absence of CK-stimulation, it was reported that uNK almost exclusively exhibited enhanced transcription relative to pNK (Koopman et al. 2003). uNK responses to CK stimulation appear highly distinct from paired pNK. In the circulation, pNK function as an early cytolytic response to tumors or viral infection (Chiossone, et al. 2018; Vivier, et al. 2011). By contrast, consistent with previous reports, we observed that uNK are poorly cytolytic and express low levels of pNK cytokines such as interferon-γ (IFN-γ) and tumour necrosis factor-α (TNF-α) (Kopcow, et al. 2005; Vacca, et al. 2006), and appear less reactive with low CD107 (degranulation marker) expression relative to pNKs in response to CK challenge (data not published). The unique phenotype of uNK may reflect their adaptation within this unique maternal tissue microenvironment to aid decidualisation, embryonic implantation, and subsequent fetal development (Pollheimer, et al. 2018).
Detailed pathway analysis verified that CK-stimulated uNK are phenotypically distinct from CK-stimulated pNK, with cell metabolism, signalling and processing, and cancer pathways consistently over-expressed in CK-stimulated uNK relative to CK-stimulated pNK. These observations are consistent with the known roles of tissue-resident uNK in placental development and fetal implantation. The significant inter-pathway transcript variance between CK-stimulated uNK and pNK underlines the complex diversity of uNK cell function. HIF1A and PPARγ regulation of glycolysis (Z score 3.64) was the most significantly differentially-expressed pathway in CK-stimulated uNK (Supplemental Figure 5B). The Cori cycle, a lactate-glucose carbon recycling system between muscle and liver that decreases circulating lactate was also enriched in CK-stimulated uNK (Z score 3.33) Supplemental Figure 5D). Genes associated with glycolysis and gluconeogenesis were also enhanced in CK-activated uNK (Z score 1.83, 14/30 genes, p=0.067), further underlining the active metabolic remodelling in uNK relative to pNK. During early pregnancy decidualisation is associated with enhanced glucose influx (Frolova and Moley 2011; Kommagani, et al. 2013). Recent studies have shown that the decidua acts in a manner similar to that reported for tumors by generating energy via a high rate of glycolysis, even under aerobic conditions rather, than conventional oxidative phosphorylation (Zuo, et al. 2015). This is referred to as a decidual Warburg-like glycolysis similar to that reported in tumors and was dependent on expression of HIF1α and the lactate transporter protein (MCT4) (Zuo et al. 2015). Collectively, these data suggest that a key feature of activated uNK is to regulate glucose metabolism and lactate transport within the decidua. Dysregulation of glucose metabolism in the human uterus is associated with subfertility and aberrant implantation (Kommagani et al. 2013). Glycolytic flux targets in uNK may therefore represent novel translational strategies to precisely time and/or therapeutically extend the window of receptivity in women at high risk for early pregnancy loss. Consistent with previous microarray analysis of 1st trimester uNK and adult non-paired pNK (Wang, et al. 2014), the current study showed that CK-stimulated uNK are enriched for pathways associated with transcriptional regulation, which may serve to differentially regulate NK development and function in the decidual microenvironment. Whether pNK undergo significant transcriptional alterations following recruitment to the decidua or, alternatively, uNK represent an entirely distinct subset of NK cells remains unclear (Hanna, et al. 2003; Vacca, et al. 2011).
Of the total transcripts measured in CK activated uNK and pNK, only 66 (0.59%) and 71 (0.64%) respectively were significantly regulated by 1,25(OH)2D3 (p≤0.05, fold change ± 1.5). Although transcriptomic analysis indicated that CK uNK and CK pNK respond differentially to 1,25(OH)2D3, this effect was statistically insignificant as the current analysis was not significantly powered to account for multiple comparisons (FDR step up < 0.05). Sample size and participant heterogeneity are potential reasons for this. Intra-participant variability for pNK was high, whereas the uNK demonstrated comparatively low variability. The heterogeneity in pNK gene expression may be in part attributable to differences in gestational age, maternal age, ethnicity and smoking status (Supplemental Table I), albeit n numbers are small. Cellular heterogeneity may also be a contributing factor to sample variability as recent single cell RNA sequencing data indicates 3 highly distinct decidual NK subsets to be resident within first trimester decidua (Vento-Tormo, et al. 2018).
Analysis of commonly regulated biological pathways, as detailed previously (Munoz Garcia et al. 2018), demonstrates that responses to 1,25(OH)2D3 in CK-stimulated uNK and pNK diverge significantly from other immune subsets including monocytes, dendritic cells and B cells (Figure 4). This may in part reflect different treatment regimens. Here a physiological treatment dose (10nM) of 1,25(OH)2D3 was utilised, whereas for monocytic THP-1 cells a 10-fold higher treatment dose (100nM) was applied (Seuter, et al. 2016). Temporal variations may also be a contributory factor. For THP-1 cells 46 genes were induced/ suppressed after 2.5 h, 288 at 4 h and 1204 at 24 h. In the current study we assessed mRNA expression in NK cells after 24 h to maximise potential variations in gene expression, however both pNK and uNK cells may show different patterns of gene regulation by 1,25(OH)2D3 at earlier or later time points. The distinct transcriptional responses to 1,25(OH)2D3 by NK cells may also reflect differential distribution of VDR.
Data reported here provide further evidence for the distinct phenotype of uNK cells relative to circulating pNK. By comparing the gene expression profile of these cells under conditions of immune activation, we confirmed enrichment of distinct functional pathways in uNK such as TGF-β signalling (Supplemental Figure 5C) which has previously been shown to promote uNK development (Keskin, et al. 2007). Our data have also revealed novel features of CK-stimulated uNK, notably enrichment of pathways associated with metabolism, suggesting a role for uNK cells in immunometabolic activity within the decidua. The lack of 1,25(OH)2D3 response by uNK and pNK contrasts the expression of VDR by both cell types and the enhancement of this receptor in CK-stimulated uNK vs pNK. Given the abundant levels of 1,25(OH)2D3 within decidual tissue (Tamblyn et al. 2017), and the potent immunomodulatory actions of 1,25(OH)2D3 (Chun, et al. 2014), the lack of response by NK cells is intriguing. Further studies are required to better define the role of the VDR in NK cells. Data in the current study suggest possible cell membrane expression of VDR and this may support non-genomic responses to 1,25(OH)2D3 (Hii and Ferrante 2016). In the case of uNK, it is also possible that 1,25(OH)2D3 is able to influence these cells indirectly via other immune cells such as macrophages that have an established transcriptional and functional response to 1,25(OH)2D3. It is now clear that uNK play a key role in mediating the cross-talk between immune cells within the decidua (Vento-Tormo et al. 2018), and vitamin D may therefore play a role in this process. Finally, the data presented in the current study have focused on transcriptional activity but it is also possible that 1,25(OH)2D3 exerts epigenetic effects on NK cells independent of transcription. In recent studies of uNK cells it has been shown that repeated pregnancies are associated with uNK cells that have a unique transcriptomic and epigenetic signature consistent with trained immune responses (Gamliel, et al. 2018). Based on its established epigenetic activity (Carlberg 2019), it is tempting to speculate that vitamin D may contribute to this facet of uNK function.
Supplementary Material
Acknowledgments
We would like to thank Matt MacKenzie for help with flow cytometry cell sorting, and Scott P. Davies and Jennifer L. Marshall for help with confocal imaging.
Study funding
This study was supported by funding from NIH (AR063910, MH), a Royal Society Wolfson Merit Award (MH), Mason Medical Trust (JAT), and Wellbeing of Women (RTF401, JAT).
Declaration of Interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported. Vitabiotics Pregnacare generously supported this work through the charity Wellbeing of Women, which independently selected the research through its AMRC accredited peer review process led by its Research Advisory Committee. Dr Tamblyn and her work were not impacted or influenced in any way by its sources of funding.
References
- Abumaree MH, Chamley LW, Badri M & El-Muzaini MF 2012. Trophoblast debris modulates the expression of immune proteins in macrophages: a key to maternal tolerance of the fetal allograft? J Reprod Immunol 94 131–141. [DOI] [PubMed] [Google Scholar]
- Adams JS & Hewison M 2008. Unexpected actions of vitamin D: new perspectives on the regulation of innate and adaptive immunity. Nat Clin Pract Endocrinol Metab 4 80–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendix M, Dige A, Deleuran B, Dahlerup JF, Peter Jorgensen S, Bartels LE, Husted LB, Harslof T, Langdahl B & Agnholt J 2015. Flow cytometry detection of vitamin D receptor changes during vitamin D treatment in Crohn’s disease. Clin Exp Immunol. 181 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohler A, Wu G, Kutmon M, Pradhana LA, Coort SL, Hanspers K, Haw R, Pico AR & Evelo CT 2016. Reactome from a WikiPathways Perspective. PLoS Comput Biol 12 e1004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer JN, Williams PJ & Lash GE 2010. Immune cells in the placental bed. Int J Dev Biol 54 281–294. [DOI] [PubMed] [Google Scholar]
- Carlberg C 2019. Nutrigenomics of Vitamin D. Nutrients 11 E676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiossone L, Dumas PY, Vienne M & Vivier E 2018. Natural killer cells and other innate lymphoid cells in cancer. Nat Rev Immunol 18 671–688. [DOI] [PubMed] [Google Scholar]
- Chun RF, Liu PT, Modlin RL, Adams JS & Hewison M 2014. Impact of vitamin D on immune function: Lessons learned from genome-wide analysis 5 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen J, Vergeiner B, Enk M, Petzer AL, Gastl G & Gunsilius E 2003. Functional significance of the activation-associated receptors CD25 and CD69 on human NK-cells and NK-like T-cells. Immunobiology 207 85–93. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M & Gingeras TR 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlebacher A 2013. Immunology of the maternal-fetal interface. Annu Rev Immunol 31 387–411. [DOI] [PubMed] [Google Scholar]
- Evans KN, Bulmer JN, Kilby MD & Hewison M 2004. Vitamin D and placental-decidual function. J Soc Gynecol Investig 11 263–271. [DOI] [PubMed] [Google Scholar]
- Frolova AI & Moley KH 2011. Quantitative analysis of glucose transporter mRNAs in endometrial stromal cells reveals critical role of GLUT1 in uterine receptivity. Endocrinology 152 2123–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamliel M, Goldman-Wohl D, Isaacson B, Gur C, Stein N, Yamin R, Berger M, Grunewald M, Keshet E, Rais Y, et al. 2018. Trained Memory of Human Uterine NK Cells Enhances Their Function in Subsequent Pregnancies. Immunity 48 951–962 e955. [DOI] [PubMed] [Google Scholar]
- Garzia E, Clauser R, Persani L, Borgato S, Bulfamante G, Avagliano L, Quadrelli F & Marconi AM 2013. Prolactin and proinflammatory cytokine expression at the fetomaternal interface in first trimester miscarriage. Fertil Steril 100 108–115 e101–102. [DOI] [PubMed] [Google Scholar]
- Georgiev H, Ravens I, Papadogianni G & Bernhardt G 2018. Coming of Age: CD96 Emerges as Modulator of Immune Responses. Front Immunol 9 1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray TK, Lester GE & Lorenc RS 1979. Evidence for extra-renal 1 alpha-hydroxylation of 25-hydroxyvitamin D3 in pregnancy. Science 204 1311–1313. [DOI] [PubMed] [Google Scholar]
- Hanna J, Goldman-Wohl D, Hamani Y, Avraham I, Greenfield C, Natanson-Yaron S, Prus D, Cohen-Daniel L, Arnon TI, Manaster I, et al. 2006. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat Med 12 1065–1074. [DOI] [PubMed] [Google Scholar]
- Hanna J, Wald O, Goldman-Wohl D, Prus D, Markel G, Gazit R, Katz G, Haimov-Kochman R, Fujii N, Yagel S, et al. 2003. CXCL12 expression by invasive trophoblasts induces the specific migration of CD16- human natural killer cells. Blood 102 1569–1577. [DOI] [PubMed] [Google Scholar]
- Hewison M 2011. Antibacterial effects of vitamin D. Nat Rev Endocrinol 7 337–345. [DOI] [PubMed] [Google Scholar]
- Hii CS & Ferrante A 2016. The Non-Genomic Actions of Vitamin D. Nutrients 8 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery LE, Wood AM, Qureshi OS, Hou TZ, Gardner D, Briggs Z, Kaur S, Raza K & Sansom DM 2012. Availability of 25-Hydroxyvitamin D3 to APCs Controls the Balance between Regulatory and Inflammatory T Cell Responses. J Immunol 189 5155–5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokhi PP, King A, Sharkey AM, Smith SK & Loke YW 1994. Screening for cytokine messenger ribonucleic acids in purified human decidual lymphocyte populations by the reverse-transcriptase polymerase chain reaction. J Immunol 153 4427–4435. [PubMed] [Google Scholar]
- Keskin DB, Allan DS, Rybalov B, Andzelm MM, Stern JN, Kopcow HD, Koopman LA & Strominger JL 2007. TGFbeta promotes conversion of CD16+ peripheral blood NK cells into CD16- NK cells with similarities to decidual NK cells. Proc Natl Acad Sci U S A 104 3378–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kommagani R, Szwarc MM, Kovanci E, Gibbons WE, Putluri N, Maity S, Creighton CJ, Sreekumar A, DeMayo FJ, Lydon JP, et al. 2013. Acceleration of the glycolytic flux by steroid receptor coactivator-2 is essential for endometrial decidualization. PLoS Genet 9 e1003900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman LA, Kopcow HD, Rybalov B, Boyson JE, Orange JS, Schatz F, Masch R, Lockwood CJ, Schachter AD & Park PJ 2003. Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. The Journal of experimental medicine 198 1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopcow HD, Allan DS, Chen X, Rybalov B, Andzelm MM, Ge B & Strominger JL 2005. Human decidual NK cells form immature activating synapses and are not cytotoxic. Proc Natl Acad Sci U S A 102 15563–15568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs E, Meichsner-Frauli M & Ludwig H 1992. Interleukin-2 receptor positive cells in human decidua during the first trimester of pregnancy and their association with macrophages. Arch Gynecol Obstet 251 93–100. [DOI] [PubMed] [Google Scholar]
- Kutmon M, Riutta A, Nunes N, Hanspers K, Willighagen EL, Bohler A, Melius J, Waagmeester A, Sinha SR, Miller R, et al. 2016. WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Res 44 D488–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lash GE, Otun HA, Innes BA, Percival K, Searle RF, Robson SC & Bulmer JN 2010. Regulation of extravillous trophoblast invasion by uterine natural killer cells is dependent on gestational age. Hum Reprod 25 1137–1145. [DOI] [PubMed] [Google Scholar]
- Lash GE, Schiessl B, Kirkley M, Innes BA, Cooper A, Searle RF, Robson SC & Bulmer JN 2006. Expression of angiogenic growth factors by uterine natural killer cells during early pregnancy. J Leukoc Biol 80 572–580. [DOI] [PubMed] [Google Scholar]
- Moffett A & Colucci F 2014 Uterine NK cells: active regulators at the maternal-fetal interface. J Clin Invest 124 1872–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz Garcia A, Kutmon M, Eijssen L, Hewison M, Evelo CT & Coort SL 2018. Pathway analysis of transcriptomic data shows immunometabolic effects of vitamin D. J Mol Endocrinol 60 95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picarda E, Ohaegbulam KC & Zang X 2016. Molecular Pathways: Targeting B7-H3 (CD276) for Human Cancer Immunotherapy. Clin Cancer Res 22 3425–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli A, Michel T, Theresine M, Andres E, Hentges F & Zimmer J 2009a. CD56bright natural killer (NK) cells: an important NK cell subset. Immunology 126 458–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli A, Michel T, Thérésine M, Andrès E, Hentges F & Zimmer J 2009b. CD56(bright) natural killer (NK) cells: an important NK cell subset. Immunology 126 458–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollheimer J, Vondra S, Baltayeva J, Beristain AG & Knofler M 2018. Regulation of Placental Extravillous Trophoblasts by the Maternal Uterine Environment. Front Immunol 9 2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CJ, Wagner CL, Hollis BW, Baatz JE & Johnson DD 2011. Maternal vitamin D and fetal growth in early-onset severe preeclampsia. Am J Obstet Gynecol 204 556 e551–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuter S, Neme A & Carlberg C 2016. Epigenome-wide effects of vitamin D and their impact on the transcriptome of human monocytes involve CTCF. Nucleic Acids Res 44 4090–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres-Bartho J 2008. Regulation of NK cell cytotoxicity during pregnancy. Reprod Biomed Online 16 211–217. [DOI] [PubMed] [Google Scholar]
- Taglauer ES, Adams Waldorf KM & Petroff MG 2010. The hidden maternal-fetal interface: events involving the lymphoid organs in maternal-fetal tolerance. The International journal of developmental biology 54 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamblyn JA, Susarla R, Jenkinson C, Jeffery LE, Ohizua O, Chun RF, Chan SY, Kilby MD & Hewison M 2017. Dysregulation of maternal and placental vitamin D metabolism in preeclampsia. Placenta 50 70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca P, Moretta L, Moretta A & Mingari MC 2011. Origin, phenotype and function of human natural killer cells in pregnancy. Trends Immunol 32 517–523. [DOI] [PubMed] [Google Scholar]
- Vacca P, Pietra G, Falco M, Romeo E, Bottino C, Bellora F, Prefumo F, Fulcheri E, Venturini PL, Costa M, et al. 2006. Analysis of natural killer cells isolated from human decidua: Evidence that 2B4 (CD244) functions as an inhibitory receptor and blocks NK-cell function. Blood 108 4078–4085. [DOI] [PubMed] [Google Scholar]
- Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park JE, Stephenson E, Polanski K, Goncalves A, et al. 2018. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM & Ugolini S 2011. Innate or adaptive immunity? The example of natural killer cells. Science 331 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhou Y, Fu B, Wu Y, Zhang R, Sun R, Tian Z & Wei H 2014. Molecular signatures and transcriptional regulatory networks of human immature decidual NK and mature peripheral NK cells. Eur J Immunol 44 2771–2784. [DOI] [PubMed] [Google Scholar]
- Weisman Y, Harell A, Edelstein S, David M, Spirer Z & Golander A 1979. 1 alpha, 25-Dihydroxyvitamin D3 and 24,25-dihydroxyvitamin D3 in vitro synthesis by human decidua and placenta. Nature 281 317–319. [DOI] [PubMed] [Google Scholar]
- Zehnder D, Evans KN, Kilby MD, Bulmer JN, Innes BA, Stewart PM & Hewison M 2002. The ontogeny of 25-hydroxyvitamin D(3) 1alpha-hydroxylase expression in human placenta and decidua. Am J Pathol 161 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourbas S, Dubanchet S, Martal J & Chaouat G 2001. Localization of pro-inflammatory (IL-12, IL-15) and anti-inflammatory (IL-11, IL-13) cytokines at the foetomaternal interface during murine pregnancy. Clin Exp Immunol 126 519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo RJ, Gu XW, Qi QR, Wang TS, Zhao XY, Liu JL & Yang ZM 2015. Warburg-like Glycolysis and Lactate Shuttle in Mouse Decidua during Early Pregnancy. J Biol Chem 290 21280–21291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.