Abstract
Fungi of the genus Trichoderma are a rich source of enzymes, such as cellulases and hemicellulases, that can degrade lignocellulosic biomass and are therefore of interest for biotechnological approaches seeking to optimize biofuel production. The essential transcription factor ACE3 is involved in cellulase production in Trichoderma reesei; however, the mechanism by which ACE3 regulates cellulase activities is unknown. Here, we discovered that the nominal ace3 sequence in the T. reesei genome available through the Joint Genome Institute is erroneously annotated. Moreover, we identified the complete ace3 sequence, the ACE3 Zn(II)2Cys6 domain, and the ACE3 DNA-binding sites containing a 5′-CGGAN(T/A)3-3′ consensus. We found that in addition to its essential role in cellulase production, ace3 is required for lactose assimilation and metabolism in T. reesei. Transcriptional profiling with RNA-Seq revealed that ace3 deletion down-regulates not only the bulk of the major cellulase, hemicellulase, and related transcription factor genes, but also reduces the expression of lactose metabolism–related genes. Additionally, we demonstrate that ACE3 binds the promoters of many cellulase genes, the cellulose response transporter gene crt1, and transcription factor–encoding genes, including xyr1. We also observed that XYR1 dimerizes to facilitate cellulase production and that ACE3 interacts with XYR1. Together, these findings uncover how two essential transcriptional activators mediate cellulase gene expression in T. reesei. On the basis of these observations, we propose a model of how the interactions between ACE3, Crt1, and XYR1 control cellulase expression and lactose metabolism in T. reesei.
Keywords: Trichoderma reesei, transcription factor, protein-protein interaction, cellulase, DNA binding protein, protein-DNA interaction, gene expression, fungi, Hypocrea jecorina, gene knockout, ace3, protein-protein interaction, transcription factor, Trichoderma reesei (Hypocrea jecorina), Zn(II)2Cys6 protein, renewables, gene regulation, hydrolase, transcriptomics
Introduction
Reducing fossil fuel consumption and replacing fossil fuels with alternate energy sources presents a major global challenge in the coming decades (1, 2). Lignocellulosic biomass has long been known to be the most abundant sustainable resource for biorefineries and the production of biofuels (3). Lignocellulosic biomass must be hydrolyzed by cellulases and hemicellulase synergistically into simple sugars to facilitate fermentation (4, 5). Enzymes required for the degradation of lignocellulosic biomass and continuation of the carbon cycle are mainly produced by microorganisms, especially by fungi belonging to the Trichoderma genus (6). The saprophytic filamentous fungus Trichoderma reesei (teleomorph Hypocrea jecorna) acts as a dominant workhorse for the industrial production of extracellular cellulase and hemicellulase, where up to 100 g/liter extracellular proteins can be obtained using certain industrial strains (7, 8).
The majority of the cellulase and hemicellulase genes are regulated by the fine-tuned cooperation between several transcription factors in T. reesei (9–11). Two Zn(II)2Cys6 type TFs, XYR1 (12) and ACE34 (13), are essential for cellulase production. In 2006, XYR1 was identified as a general and essential transcriptional activator for the genetic expression of cellulases, including the four main cellulases, CBHI, CBHII, EGLI, and EGLII, and hemicellulases, including the two major xylanases, XYNI and XYNII (14–16). This protein is also required for d-xylose metabolism in T. reesei (12). The deletion of xyr1 resulted in the absence of cellulase and hemicellulase production (17). In 2014, another essential transcription factor, ACE3, was identified from transcriptional profiling data from T. reesei cultures grown in different lignocellulose-derived cellulase inducers (13). Additionally, the deletion of ace3 completely abrogates the expression of cellulase, whereas this deletion merely reduces expression of xylanase (13). XYR1 can bind to a 5′-GGC(T/A)3-3′ motif (12, 17); however, the DNA binding motifs of ACE3 remain unknown.
An uptake system for cellulase inducers was also reported to be essential for cellulase production in T. reesei (18). Trire2:3405 was first considered to be a putative lactose permease after BLAST analysis was performed in 2013, and its deletion was thought to completely abolish the growth of T. reesei in a lactose-based medium (18). After its initial identification, Trire2:3405 was named as the cellulose response transporter Crt1, and this protein is thought to act as an essential receptor or transceptor involved in cellulase gene induction that functions without the need for intracellular sugar delivery activity (19). The absence of Crt1 resulted in no cellulase production following lactose-, cellulose-, cellobiose-, or sophorose-mediated induction (19).
These phenomena indicate that the expression of cellulase genes requires the participation of two master regulators, ACE3 and XYR1, and the cellulose response transporter Crt1 in T. reesei. ACE3, XYR1, and Crt1 are all essential for cellulase production; however, the DNA-binding motifs and the mechanism by which ACE3 regulates cellulase expression remain unknown. The regulatory mechanism underlying the master activators XYR1 and ACE3, which synergistically mediate cellulase production in T. reesei, is completely unknown. The molecular mechanism controlling the interdependence of ACE3, XYR1, and Crt1 for regulating cellulase expression is also unclear.
In this study, we found that ace3 is essential for lactose metabolism, we identified the appropriate ace3 sequence for analysis, and we further analyzed the DNA-binding sites of ACE3. ACE3 can bind to the promoters of two factors essential for cellulase expression, crt1 and xyr1, and this binding indicated that the cross-talk between ACE3, Crt1, and XYR1 is responsible for mediating cellulase expression and lactose metabolism in T. reesei. We also demonstrated that protein–protein interactions occur between two Zn(II)2Cys6 type master regulators, XYR1 and ACE3. Our research demonstrates the direct and indirect mechanisms by which ACE3 regulates cellulase induction, and our findings shed light on the novel ACE3–XYR1 interactions that may mediate cellulase gene expression in T. reesei. These findings provide insights into the regulatory mechanisms for cellulase and hemicellulase production and enable us to highlight the potential applications for glucoside hydrolase in the production of biofuels and biorefineries through the use of fungi.
Results
Transcriptional profile of the Δace3 mutant cultured in the presence of lactose
A chance discovery (see Text S1 and Figs. S1–S3) showed that an 8-amino acid FLAG tag (20) fused to the C terminus of ACE3 (to express an ACE3-FLAG fusion) or XYR1 (to express an XYR1-FLAG fusion) completely inactivates their cellulase-inducing function. The ACE3-FLAG mutants (6aACE3-FLAG strains) and ACE3 deletion mutants (Δace3 strains) were also found to be unable to grow in medium containing lactose as the sole carbon source (Fig. S1B). Interestingly, cellulase is not involved in the utilization of lactose in T. reesei (21). Therefore, we hypothesize that ACE3 could also be involved in the primary mechanism for the metabolism of lactose in T. reesei.
To address why ace3 inactivation strains could not grow in lactose and to further explore the function of ACE3, the full genomic responses of the parental strain QM6a (WT) and ACE3 deletion strain (Δace3) and were assessed by RNA sequencing (RNA-Seq) to profile the genome-wide mRNA abundance observed following lactose addition. Twelve hours after transfer into a 2% lactose solution, 4128 genes (Fig. S4) were identified as significantly different compared with those of the WT strain in the presence of lactose (p-adjust < 0.05 and |log2FC (Δace3/WT)| > = 1).
Genes that were highly down-regulated (log2FC (Δace3/WT) ≤ −2) in the Δace3 mutant in the presence of lactose are listed in Table S1. Twenty-two cellulase-related genes were down-regulated, including major cellulase genes such as cbh1, cbh2, egl1, egl2, egl3, and bgl2 (Table S1). This result is consistent with the absence of cellulase production in the Δace3 strain. Fifteen hemicellulase-related genes were down-regulated, including major hemicellulase genes such as xyn1, xyn2, xyn3, and bxl1 (Table S1). This result is consistent with the reduced xylanase production in the Δace3 strain. Seven transcription factor genes were down-regulated, including important transcription factors regulating cellulase expression, such as xyr1, ace3, clr-2, yellow pigment regulator 1 ypr1, and some transcription factors of unknown function (Table S1). The down-regulated transcriptional levels of xyr1 observed in the Δace3 strain indicated that ACE3 could indirectly regulate cellulase gene expression through the main factor XYR1. The significantly down-regulated expression of ypr1 in the Δace3 strain is in accord with the remarkably decreased level of yellow pigment production (Fig. S5). Also, seven putative lactose metabolism-related genes were significantly down-regulated, and these included the putative lactose transporter gene Trire2:3405 (18) that was provisionally named crt1, as it was identified as a cellulose response transporter (19), and the bga1 gene (22), which enables the first step of lactose utilization to occur (Table S1). The down-regulation of bga1 is likely the reason why Δace3 cannot grow in lactose.
Characterization of the binding specificity of ACE3
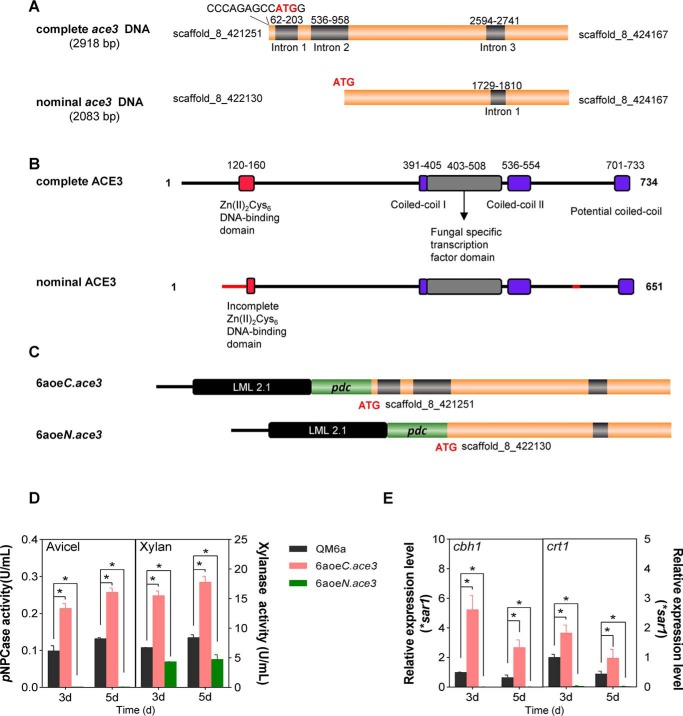
We discovered that the nominal ace3 sequence (Trire2:77513, estExt_GeneWisePlus.C_80307) in the T. reesei genome available through the Joint Genome Institute is erroneously annotated (Fig. 1, A and B), and this sequence encoded a truncated Zn(II)2Cys6 DNA-binding domain that was neither functional (Fig. 1, C–E) nor commonly observed for a natural transcription factor (23). The complete ORF of ace3, which encodes a 734-amino acid protein (Fig. 1), was manually corrected (Text S2 and Fig. S6). Phylogenetic analysis indicates that ACE3 is a genus-specific transcriptional activator in Trichoderma and is essential for cellulase production (Fig. S7).
Figure 1.
Complete ACE3 structure and structural consequences of the nominal ACE3 sequence. A, complete and nominal ace3 sequences. B, the DNA-binding domain (red) of complete ACE3 is orthologous to that of Gal4 from Saccharomyces cerevisiae. Two coiled-coil (25) domains of ACE3 are indicated in purple. The potential coiled-coil domain of ACE3 is located at the C-terminal end of ACE3 (701–733 amino acids). The filamentous fungi-specific transcription factor domain (FSTFD; gray) regulates target gene expression. Hence, the nominal ACE3 sequence harbors the incomplete Zn(II)2Cys6 DNA-binding domain and erroneous N-terminal and C-terminal sequences (red line). The cDNA sequence of the complete ace3 sequence was submitted to GenBankTM (submission ID: BankIt2146245 ace3 MH819249). C, construction of two ace3 overexpression mutants. pdc is the strong constitutive promoter of pyruvate decarboxylase in T. reesei. LML 2.1 is the erasable hygromycin selection marker in T. reesei. Strain 6aoeC.ace3 is the complete ace3 overexpression mutant. Strain 6aoeN.ace3 is the nominal ace3 overexpression mutant. D, pNPCase and xylanase activities of T. reesei mutants 6aoeC.ace3 and 6aoeN.ace3 in Avicel and xylan, respectively. E, relative transcriptional levels of cbh1 and crt1 in T. reesei mutants in Avicel. The mRNA levels were normalized to the SAR1 housekeeping gene in each sample. Using the pdc promoter overexpressing the nominal ACE3 sequence totally blocked cellulase production compared with the ACE3 deletion strain (Fig. S2D). Overexpression of the complete ACE3 structure can increase the cellulase production (such as cbh1) and transcriptional levels of crt1 in T. reesei. Values are the mean ± S.D. (error bars) of the results from three independent experiments. Asterisks, significant difference compared with the untreated strain (*, p < 0.05, Student's t test).
As a transcription factor, ACE3 should regulate cellulase by directly binding to its promoter. To verify this possibility, we chose cbh1, which encodes CBH I, the principal component of the cellulase mixture secreted by T. reesei, and analyzed it via an electrophoretic mobility shift assay (EMSA).
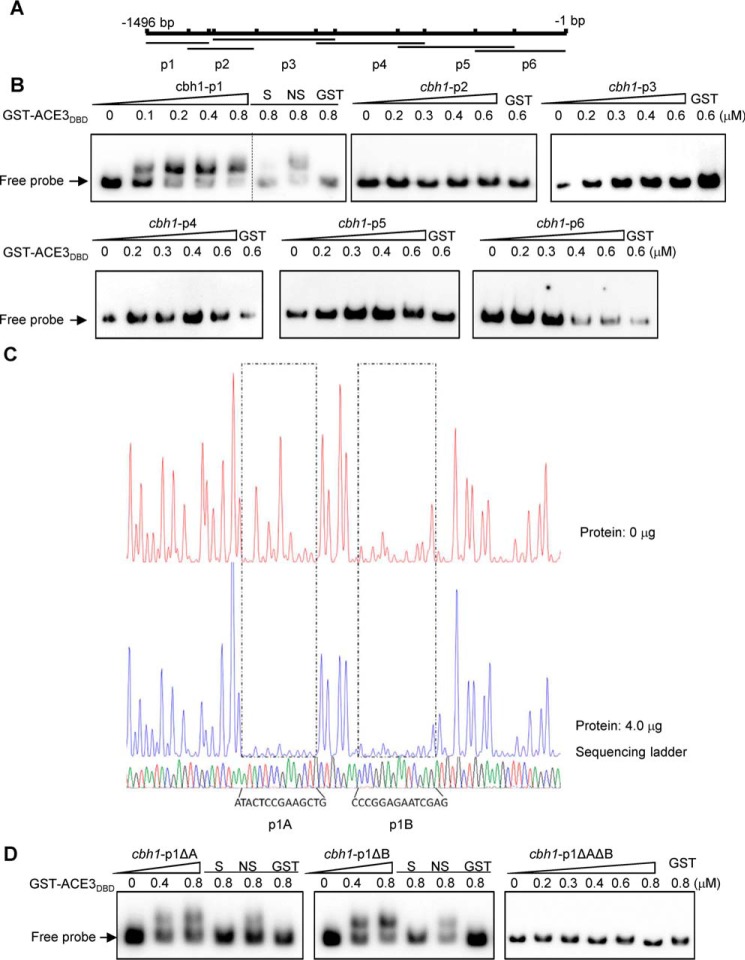
In the EMSAs, the peptide containing the Zn(II)2Cys6 domain (DNA-binding domain (DBD)) of ACE3 (amino acids 79–222) that was fused to a GST tag was expressed in Escherichia coli Rosetta (DE3) and purified using GST affinity chromatography. The purified peptide GST-ACE3DBD was incubated with the biotin-labeled cbh1-p1 (−1496 to −1272), cbh1-p2 (−1344 to −1102), cbh1-p3 (−1253 to −821), cbh1-p4 (−890 to −503), cbh1-p5 (−599 to −183), and cbh1-p6 (−322 to −1) fragments of the cbh1 promoter (Fig. 2A). EMSA results revealed that GST-ACE3DBD was bound only to cbh1-p1 (−1496 to −1272), as only the mobility of cbh1-p1 clearly shifted among six DNA probes (Fig. 2B).
Figure 2.
ACE3 binds to cbh1 promoter in vivo and vitro. A, the cbh1 promoter was divided into the six parts, cbh1-p1, -p2, -p3, -p4, -p5, and -p6 fragments of the cbh1 promoter, and there was a 50–150-bp overlap between adjacent fragments, each of which were ∼200 to 450 bp long. B, the binding of GST-ACE3DBD (GST-ACE379–222) to the different cbh1 promoter fragments. EMSAs were performed by incubating biotin-labeled fragments cbh1-p1 (−1496 to −1272), cbh1-p2 (−1344 to −1102), cbh1-p3 (−1253 to −821), cbh1-p4 (−890 to −503), cbh1-p5 (−599 to −183), and cbh1-p6 (−322 to −1) of the cbh1 promoter with the GST tag–fused Zn(II)2Cys6 domain of ACE3 (aa 79–222) expressed by E. coli and purified by GST affinity chromatography. For each EMSA, a 10 nm biotin-labeled fragment and 0, 0.1, 0.2, 0.4, 0.6, and 0.8 μm (the increase amount is indicated by a right-pointing triangle) recombinant GST-ACE379–222 were added. EMSAs with only the GST tag (GST), a 200-fold excess of unlabeled specific fragments (S), or nonspecific competitor fragment (sperm DNA) (NS) were conducted as controls. C, electropherograms of the DNase I digest of the FAM-labeled cbh1-p1 fragment incubated without proteins (top of each panel) or with 4 μg of GST-ACE3DBD. The respective nucleotide sequences bound by GST-ACE3DBD are indicated as p1A and p1B (bottom of the panel). D, binding characteristics of GST-ACE3DBD to mutant cbh1-p1 fragments. EMSAs were identified by incubating biotin-labeled fragments cbh1-p1ΔA (deleting p1A), cbh1-p1ΔB (deleting p1B), and cbh1-p1ΔAΔB (deleting p1A and p1B). The dashed line indicates the splicing site of gels.
A DNase I footprinting assay was conducted using the above cbh1-p1 fragment to determine the binding regions of ACE3 on the fragment. The results of this assay indicated that GST-ACE3DBD could bind to two regions on the cbh1-p1 fragment, specifically p1A (−1444 to −1428 of cbh1) and p1B (−1423 to −1410 of cbh1) (Fig. 2C).
To verify the DNase I footprinting assay in vitro, three DNA probes (cbh1-p1ΔA, cbh1-p1ΔB, and cbh1-p1ΔAΔB) were constructed by deleting p1A, p1B, or both, respectively. EMSA results showed that ACE3 could not bind to cbh1-p1ΔAΔB (Fig. 2D). Both regions (p1A and p1B) are independently in contact with the ACE3 DNA-binding domain (Fig. 2D).
The two binding sites (p1A and p1B) were also verified in vivo by knockout at the cbh1 promoter in T. reesei. We obtained the 6aDcbh1p1AB strains with knocked out p1AB sequences (−1444 to −1410 of cbh1). Compared with the parental strain QM6a, the pNPCase activity in 6aDcbh1p1AB was significantly decreased by 30–40% (Fig. S8A). The qPCR data showed that cbh1 transcript levels were markedly decreased (50–60%) in the 6aDcbh1p1AB strains (Fig. S8B), and this is consistent with the enzyme activity data. These results indicated that the two binding sites of ACE3 in the cbh1 promoter are functional and regulate the expression of cbh1.
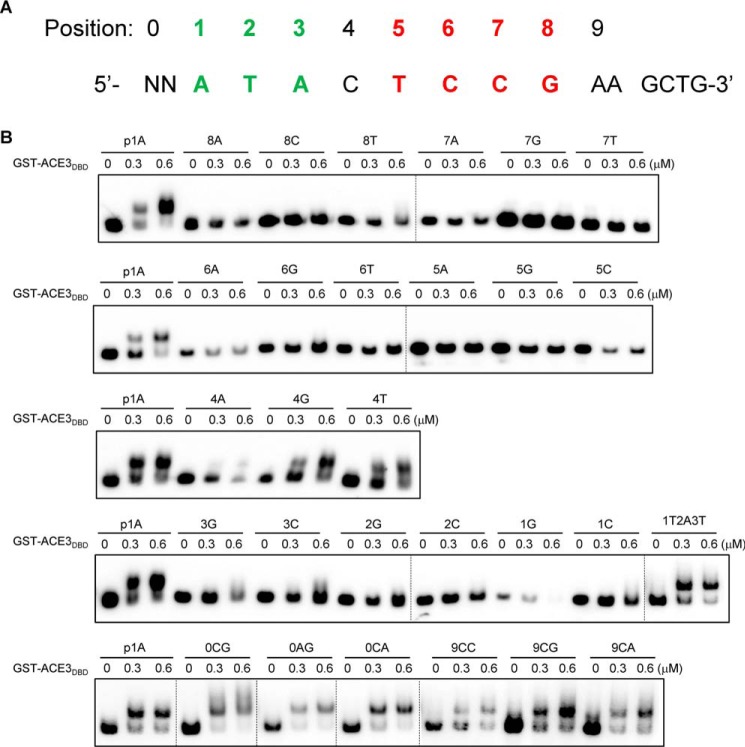
To accurately confirm the ACE3 binding motif sequences and address whether ACE3 could exhibit binding affinity for the somewhat different oligonucleotide motif, we needed to define the important nucleotides that affect the specificity of binding of ACE3. For this analysis, the 5′-ATACTCCGAAGCTG-3′ p1A sequence was used as an original probe for mutagenesis.
The effect of mutations within p1A on ACE3 binding was examined by quantitative EMSA. The mutational analyses at positions 0–9 are summarized in Fig. 3A. When any T or A was mutated to G or C in positions 1–3, except for during an A and T interchange, this resulted in a complete loss of ACE3 binding (Fig. 3B). Additionally, any mutations in positions 5–8 resulted in a complete loss of ACE3 binding (Fig. 3B). The C at position 4 can be mutated to any of the other three bases (A, G, or T) (Fig. 3B). Positions before 1 and after 8 do not belong to the binding motif, as they are not conserved by in silico analysis and can be mutated to any other bases (Fig. 3B). These results illustrated that functional ACE3-binding sequences are 5′-CGGAN(T/A)3-3′ motifs (the reverse complement motifs, 5′-(A/T)3NTCCG-3′).
Figure 3.
Determination of binding motif sequences of ACE3 in 14-bp p1A fragments. A, positions (0–9) of each base on the 14-bp p1A fragments. The core sites of the ACE3-binding motif are shown in red (cannot be mutated) and green (A or T). B, binding characteristics of GST-ACE3DBD (GST-ACE379–222) to mutant p1A fragments. EMSAs were performed by incubating biotin-labeled fragments p1A and its mutants (designated as position number + base) with the GST tag–fused Zn(II)2Cys6 domain of ACE3 (aa 79–222) expressed by E. coli and purified by GST affinity chromatography. For each EMSA, a 10 nm biotin-labeled fragment and 0–0.6 μm recombinant GST-ACE379–222 were added. Dashed lines indicate the splicing sites of gels.
To test the ability of ACE3 to bind to promoters of other down-regulated genes in Δace3 (Table S1) and to assess whether ACE3 mediated the expression of these genes directly or indirectly, scanning was performed to identify ACE3-binding motifs in the 5′-upstream regions (about 2000 bp) of all 51 target genes listed in Table S1. A large number of the 5′-CGGAN(T/A)3-3′ motifs were detected in the 5′-upstream regions of 41 target genes (Table S1). We also found two ACE3 binding sites in its own promoter, indicating the transcriptional self-activation effect of ACE3 on ace3, which is in accordance with the down-regulated transcriptional level of ace3 in Δace3 (Table S1). No binding site was found in the promoters of several well-known cellulase genes, including cbh2, egl2, and Trire2:50215, indicating that ACE3 regulated these genes indirectly. Also, we did not identify any binding sites in the 5′-upstream regions of xyn1, one of two main xylanase genes from T. reesei, suggesting that ACE3 could not regulate this gene directly. Therefore, the Δace3 mutant exhibits a detectable level of xylanase activity and transcription instead of the abolished activity observed with cellulase. Interestingly, bga1, which is essential for achieving growth in lactose, showed no ACE3-binding sites in its promoter, illustrating that ACE3 indirectly regulated its expression. Notably, another essential gene involved in lactose metabolism and cellulase gene induction, crt1, can be directly controlled by ACE3 via four ACE3-binding sites in its promoter.
EMSA analyses were also performed using probes from 17 selected promoter regions (Table S1). As shown in Fig. S9, the specific binding of ACE3 was observed for oligonucleotide probes containing 5′-CGGAN(T/A)3-3′ from promoters of five cellulase-related genes (cbh1, egl1, bgl2, cel3c, and cip2), three hemicellulase-related genes (bxl1, agl1, and man1), six transcription factor genes (xyr1, ace3, ypr1, clr-2, tre70351, and tre108381), and three lactose metabolism–related genes (crt1, tr56684, and tre104072). Based on these results, it is inferred that the 5′-CGGAN(T/A)3-3′ motifs that exist in numerous promoters function as ACE3-binding sites. ACE3 can regulate the expression of important cellulase and xylanase genes directly and indirectly through other factors, such as the key transcription factor XYR1, the cellulose response transporter Crt1, and others.
Truncation of the C terminus of ACE3 and XYR1 inhibits the function of these proteins
To confirm the effects of the C-terminal domain of ACE3 and XYR1 on the production of cellulase and xylanase in T. reesei, C-terminal truncation of these proteins was performed in situ (Fig. S10). As shown in Fig. S10A, a native ACE3 cassette named A8, which contained all the 734 amino acids of ACE3, was used as the positive control. The A7 cassette (amino acids 1–700 of ACE3) was constructed by deleting the 34 amino acids at the C terminus of ACE3. The T. reesei transformants 6aA8 and 6aA7 were obtained. T. reesei 6aA8 exhibited levels of cellulase and xylanase production that were similar to those observed with the parental strain QM6a; however, 6aA7 exhibited a cellulase-negative, xylanase-decreased phenotype (Fig. S10, B and C). Also, the conidia of 6aA7 germinate poorly and grow when lactose is used as the sole source of carbon, as observed with the conidia of the ace3 deletion strain. Truncating the 34 amino acids present at the C terminus of ACE3 completely abolishes protein function, as observed with the deletion of ace3.
As shown in Fig. S11A, a native XYR1 cassette named X8, which contained all 940 amino acids of XYR1, was used as the positive control. XYR1 truncated analogues X7 (amino acids 1–909 of XYR1), X6 (amino acids 1–865 of XYR1), X5 (amino acids 1–775 of XYR1), X3 (amino acids 1–700 of XYR1), and X2 (amino acids 1–368 of XYR1) were constructed (Fig. S11A), and the T. reesei transformants TRX8, TRX7, TRX6, TRX5, TRX3, and TRX2 were obtained. T. reesei TRX8 exhibits levels of cellulase and xylanase production that are similar to those observed in the parental strain; however, cellulase activity was significantly decreased in the TRX7 strain (Fig. S11B). Additionally, TRX6, TRX5, and TRX3 exhibit a cellulase-negative phenotype (Fig. S11, B and C). The xylanase production and the expression levels of the two main xylanase genes xyn1 and xyn2 in TRX6, TRX5, and TRX3 remain unaffected, as compared with those of the positive control TRX8 (Figs. S11D and S12). Despite this, TRX2 showed no cellulase or xylanase production, similar to that observed upon the deletion of xyr1 (Fig. S11, B–D). These results indicate that the C-terminal amino acids 866–940 of XYR1 are essential for cellulase induction. Truncating amino acids 866–940 of XYR1 (to construct X6) completely abolished cellulase production; however, this domain is not essential for xylanase induction. Even truncating the 240 C-terminal amino acids of XYR1 (to construct X3) resulted in the xylanase production remaining entirely unaffected. Truncating C-terminal amino acids 368–700 of X3 (to construct X2) resulted in a xylanase-negative phenotype; therefore, amino acids 368–700 of XYR1 are essential for xylanase production. These results illustrated that the 240 C-terminal amino acids of XYR1 are necessary for cellulase expression but not completely necessary for xylanase expression. This indicates that the regulation of cellulase is more complex than that of xylanase, as the expression of cellulase requires not only ACE3, but also the complete XYR1 sequence, whereas xylanase does not.
The C-terminal domain of XYR1 mediates homodimerization and cellulase production
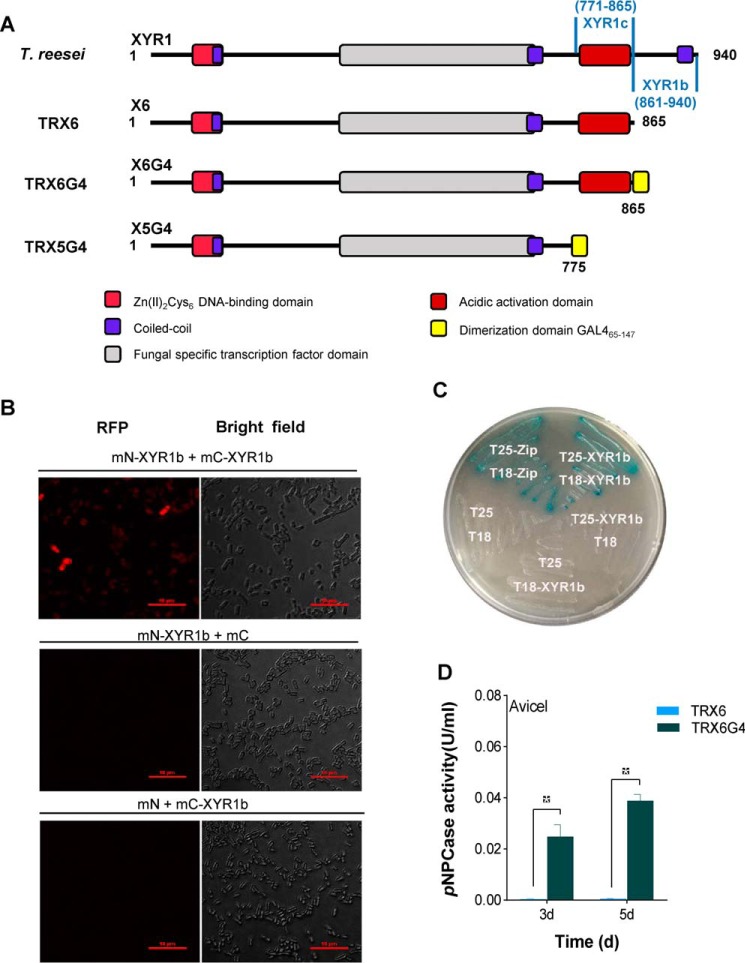
The presence of C-terminal amino acids 866–940 of XYR1 is essential for cellulase induction. Lichius et al. (24) predicted a coiled-coil domain (amino acids 910–935 of XYR1) using COILS (25, 26), and they hypothesized that it mediated the homodimerization of XYR1.
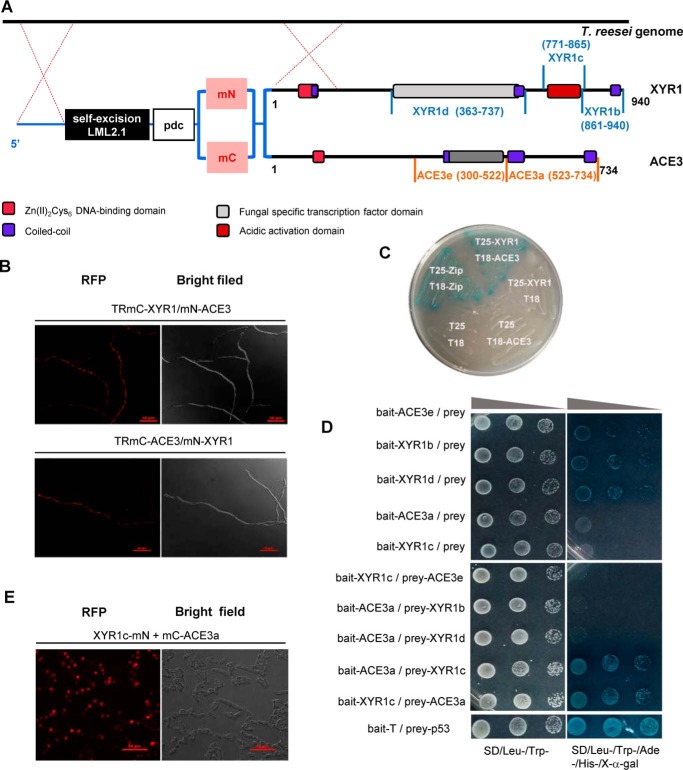
To confirm whether the C terminus of XYR1 can be formed via self-assembly, we detected homodimer formation in two XYR1b domains (amino acids 861–940 of XYR1; Fig. 4A) via a bimolecular fluorescence complementation (BiFC) assay, a yeast two-hybrid (Y2H) assay, and a bacterial two-hybrid (B2H) assay. As shown in Fig. 4B, the red fluorescence produced during the BiFC assay revealed positive interactions occurring between two XYR1b domains, which is indicative of dimerization. In contrast, no red fluorescence was observed in negative controls (Fig. 4B). We also used the classical Y2H assay to further confirm interactions occurring within the two XYR1b domains. Autoactivation of pGBKT7-XYR1b will interfere with our Y2H experiment, so B2H was selected instead of Y2H. The B2H assay confirmed that self-assembly occurred between the two XYR1b domains, and this resulted in transformed E. coli that could grow on maltose and that expressed the β-gal gene (Fig. 4C). The results of the BiFC and B2H assays confirmed that the two XYR1b domains could interact to form homodimers, which partially proved that homodimerization occurs in XYR1.
Figure 4.
C-terminal domain of XYR1 mediates homodimerization. A, construction of T. reesei TRX6G4 and TRX5G4. The XYR1b domain is amino acids 861–940 of XYR1; the XYR1c domain is amino acids 771–865 of XYR1; and GAL465–147 is a well-known dimerization domain of GAL4 (amino acids 65–147) from S. cerevisiae. The structure of XYR1 is from Lichius et al. (24). B, BiFC assay showing the protein–protein interactions in the XYR1b domain in E. coli. Strains expressing protein partners fused with N-terminal aa 1–159 of a fluorescent protein mCherry (mN) or C-terminal aa 160–237 of mCherry (mC) are indicated on each panel. Negative controls for the BiFC assay of the XYR1b domain were performed (mN-XYR1b + XYR1b and mN + mC-XYR1b). Positive interactions resulted in red fluorescence. C, B2H assay reveals the protein–protein interactions of the XYR1b domain. Strains with proteins that could bind with each other showed up as a blue colony on M63 plates supplemented with 5-bromo-4-chloro-3-indolyl β-d-galactoside and isopropyl-β-d-thiogalactopyranoside. T18 and T25 are two complementary fragments of adenylate cyclase (CyaA), obtained from Bordetella pertussis. Zip, the leucine zipper motif of GCN4. Strains are indicated by their expressed T18 or T25 fusion proteins. A strain expressing T25-zip and T18-zip fusion proteins was used as a positive control. It was observed as a blue colony (Cya+ phenotype) because of the dimerization of leucine zipper motifs, which were appended to the T25 and T18 fragments, respectively. A strain expressing T18 and T25 was used as the negative control. The strain expressing T25-XYR1b and T18-XYR1b fusion proteins was observed as a blue colony, which indicates a positive protein–protein interaction. D, pNPCase activities of T. reesei strains TRX6G4 and TRX6 on Avicel.
To confirm whether the homodimerization of XYR1b is responsible for cellulase expression, we performed X6 reconstitution using a well-known dimerization domain (amino acids 65–147) of GAL4 (23, 27) from Saccharomyces cerevisiae to replace the XYR1b domain of XYR1 to construct a chimeric factor X6G4. The X6G4 cassette was transformed into T. reesei QM6a as a substitute for xyr1, which resulted in the T. reesei transformant TRX6G4 (Fig. 4A).
Interestingly, TRX6G4 strains exhibited a remarkably compensatory pNPCase activity, as compared with that of the TRX6 strains that exhibited a cellulase-negative phenotype (Fig. 4D). Additionally, the four main cellulase genes cbh1, cbh2, egl1, and egl2 were also markedly expressed in the TRX6G4 strains, as compared with their expression in TRX6 strains (Fig. S13). This is consistent with the results observed for pNPCase activity. These results indicated that a functional dimerization domain was fused to the C terminus of X6, which truncated the C-terminal XYR1b domain from XYR1, and could restore cellulase expression. It is likely that the XYR1b domain possesses a function similar to that of the functional dimerization domain of GAL465–147. Hence, GAL465–147 can replace the XYR1b domain to reconstitute a functional chimeric cellulase transcription factor X6G4 to induce cellulase expression in the same manner as WT XYR1. Thus, the cellulase compensatory mechanism observed in X6G4 and the results of the BiFC and B2H assays described above indicated that 80 amino acids at the C terminus of XYR1 mediate the homodimerization of XYR1, a process essential for cellulase expression in T. reesei.
Another XYR1 reconstitution strain (TRX5G4), however, could not recover cellulase activity or facilitate cellulase gene transcription. These results indicated that the XYR1c domain (amino acids 771–865 of XYR1; Fig. 4A), which was lost in X5G4 but retained in X6G4, has some unknown function that is also necessary for cellulase expression to occur.
ACE3 interacts with XYR1
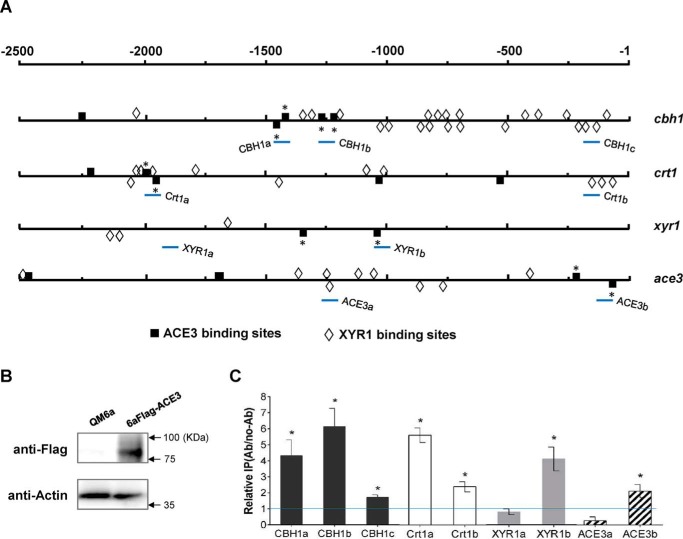
ACE3 binds to two regions (−1444 to −1428 and −1423 to −1410) in the 5′-upstream region of cbh1, including a pair of everted repeat motifs and a pair of direct repeat motifs (Fig. 5A). Also, four ACE3 binding sites were found in the ace3 promoter, indicating that ACE3 was self-activated. Two ACE3-binding sites located distantly from three XYR1-binding sites were found in the promoter of xyr1 (Fig. 5A). Using ChIP, we demonstrated that ACE3 binds to these key regions in vivo (Fig. 5, B and C).
Figure 5.
ACE3 binds to promoters in vitro and in vivo. A, schematic representation of the localization of 5′-GGC(A/T)3–3′ and 5′-CGGAN(T/A)3–3′ motifs within the 2.5-kb upstream region of cbh1, crt1, xyr1, and ace3 genes. The 5′-CGGAN(T/A)3–3′ motifs analyzed in the electrophoretic mobility shift assay (Fig. S9) are indicated with an asterisk. The promoter regions for ChIP analysis are shown as CBH1-a, CBH1-b, CBH1-c, Crt1-a, Crt1-b, XYR1-a, XYR1-b, ACE3-a, and ACE3-b (bottom lines). B, design of the ChIP studies. FLAG-tagged ACE3 is generated in the 6aFLAG-ACE3 strain. Expression is confirmed via Western blot analysis. Parental strain QM6a was used as a negative control. Thirty micrograms of total protein were loaded into each lane, and the expression of β-actin was used as a positive reference. C, a ChIP assay was performed with 6aFLAG-ACE3 cells grown in lactose. Immunoprecipitation was conducted using anti-FLAG antibody. Relative IP levels were normalized to Ab/no-Ab at the PSAR1 region, which used as a reference. Ratios higher than 1 were considered as the enrichment of the DNAs due to ACE3 binding. Error bars, S.D. of three biological replicates. Asterisks, significant difference compared with the untreated strain (*, p < 0.05, Student's t test). ACE3 binds to the promoters of CBH1, Crt1, XYR1, and ACE3 with higher binding activity near the 5′-GGC(A/T)3–3′ and 5′-CGGAN(T/A)3–3′ motifs. Error bars, S.D.
We identified at least 22 GGC(A/T)3 motifs (17) that exist in the 2.5-kb upstream region of cbh1, of which six motifs were confirmed to participate in XYR1 binding by a previous study (17). The binding sites of XYR1 and ACE3 are adjacent and interlaced in the cbh1 promoter (Fig. 5A). This is similar to what was observed in the crt1 promoter (Fig. 5A). The coexistent ACE3- and XYR1-binding motifs showed that XYR1 and ACE3 co-regulated cbh1 and crt1 expression directly. The presence of adjacent tandem ACE3-binding motifs (everted repeat, direct repeat, and inverted repeat motifs) suggested that ACE3 can bind as homodimers (28). Both XYR1- and ACE3-binding sites were found in their own promoters, which illustrated the complex interactive process underlying the activation of ace3 and xyr1.
It is essential to investigate the need for two master regulators, XYR1 and ACE3, to mediate cellulase expression in T. reesei. It is also of interest to determine why XYR1 and ACE3 bind so closely to sites within the promoters of downstream genes (Fig. 5A). Given this, we postulated that there is a protein–protein interaction between ACE3 and XYR1 to form heterodimers. To test this, we investigated whether ACE3 interacted with XYR1 using the B2H and BiFC assays (Fig. 6A). The mCherry fused XYR1 and ACE3 were under the control of the pdc promoter and transformed successively into T. reesei to construct the co-expression strains TmC-XYR1/mN-ACE3 and TmC-ACE3/mN-XYR1 (Fig. 6A). In the co-expression strains, red fluorescence was distributed in the mycelia of T. reesei (Fig. 6B). In contrast, no red fluorescence was observed in single fusion protein transformants or the parental strain (Fig. S14). Next, the B2H assay indicated that the complete sequence of ACE3 interacts with XYR1 to form blue colonies on a maltose M63 plate (Fig. 6C). Thus, our results confirmed that ACE3 interacts with XYR1.
Figure 6.
ACE3 interacts with XYR1. A, construction of co-expression strains RmC-XYR1/mN-ACE3 and TRmC-ACE3/mN-XYR1. The two domains of mCherry (mN and mC) were fused with the N terminus of XYR1 or ACE3. The mCherry-fused XYR1 and ACE3 were under the control of the pdc promoter and were transformed into T. reesei. The structure of XYR1 is from Lichius et al. (24). B, BiFC assay results revealed the protein–protein interactions of XYR1 and ACE3. Positive interactions resulted in fluorescence. C, B2H assay results revealed the protein–protein interactions of XYR1 and ACE3. The strain expressing T25-XYR1 and T18-ACE3 fusion proteins was observed as a blue colony, which indicates a positive protein–protein interaction. D, Y2H assay results revealed the protein–protein interactions of XYR1c and ACE3a domains. The vectors pGBKT7 and pGADT7 were used for the expression of bait and prey proteins, respectively. XYR1b (amino acids 861–940 of XYR1), XYR1c (amino acids 771–865 of XYR1), and XYR1d (amino acids 363–737 of XYR1), ACE3a (amino acids 523–734 of ACE3), and ACE3e (amino acids 171–522 of ACE3) were all cloned into bait or prey plasmids. The yeast strains Y187 and Y2HGold were used as hosts for transforming bait- and prey-relating vectors, respectively. The positive isolates observed after yeast mating were selected on S.D. plates lacking tryptophan and leucine (Trp−, Leu−). Then the concentration of the positive isolates was adjusted until OD600 values of 2, 0.2, and 0.02 were obtained, and 5 μl of each solution with a specific concentration was spotted on plates lacking tryptophan and leucine (Trp−, Leu−) and on selection plates lacking tryptophan, leucine, and histidine (Trp−, Leu−, His−) containing 125 ng/ml aureobasidin A (AbA) and 40 μg/ml X-α-galactosidase (X-α-gal). Plates were grown at 30 °C for 3–5 days. Positive interactions resulted in blue spots. Bait-T/prey-p53 was used as the positive control. E, BiFC assay results revealed the protein–protein interactions of XYR1c and ACE3a domains. Strains expressing protein partners fused with mN or mC are indicated in each panel. Positive interactions resulted in fluorescence.
Given that ACE3 interacts with XYR1, we examined which domains in these two proteins interact with each other. To investigate the areas of XYR1 and ACE3 that participate in these interactions, three domains of XYR1, including XYR1b (amino acids 861–940 of XYR1), XYR1c (amino acids 771–865 of XYR1), and XYR1d (amino acids 363–737 of XYR1), and two domains of ACE3, including ACE3a (amino acids 523–734 of ACE3) and ACE3e (amino acids 300–522 of ACE3), were intercepted for the Y2H assay (Fig. 6, A and D). The baits for autoactivation were tested, and the results showed that XYR1b, XYR1d, and ACE3e are autoactivated (Fig. 6D), supporting their role in transcriptional activation. Additionally, there are strong protein–protein interactions between XYR1c and ACE3a (Fig. 6D). These results were again confirmed by performing a BiFC assay (Figs. 4 and 6E). Thus, our results indicated that ACE3 interacted with XYR1 and that protein–protein interactions occur between XYR1c and ACE3a domains to form a heterodimer or heteromultimer.
Discussion
In this study, we serendipitously discovered that ACE3, in addition to its essential role in cellulase production, is also required for lactose assimilation and metabolism. To verify the role of ACE3 in lactose metabolism, the transcriptional profile of the Δace3 mutant grown in lactose was assessed. The data indicated that cellulose response transporter gene crt1 and β-gal gene bga1 were significantly down-regulated. Crt1 functions as an essential receptor or transceptor of lactose signals (18, 19), and BGAI exerts certain functions during the first step of lactose utilization, and this explains why Δace3 cannot grow in lactose. EMSAs revealed that the regulator ACE3 can bind to the promoter of crt1, indicating that the direct regulation of crt1 expression is one of the molecular mechanisms by which ACE3 mediates lactose metabolism (Fig. S9). Still, ACE3 may regulate bga1 expression indirectly, as no ACE3 binding site is located in its promoter. The precise mechanism of regulation of bga1 by ACE3, however, needs to be examined further.
We identified that the functional ACE3-binding sequence was 5′-CGGAN(T/A)3-3′. ACE3-binding motifs include the core CGG triplets, which are commonly reported in fungal Zn(II)2Cys6 proteins (29–32). Zn(II)2Cys6 proteins can bind as homodimers to CGG triplets that are oriented in everted, inverted, or direct repeats (28); however, certain Zn(II)2Cys6 proteins, such as ALCR from Aspergillus nidulans and XYR1 from T. reesei, bind to an asymmetrical target sequence as a monomer (17, 33). EMSA and ChIP assays demonstrated that ACE3 binds to the promoters of these key genes in vitro and in vivo (Fig. 5).
The deletion of two ACE3 binding sites (−1444 to −1410 of cbh1) causes the pNPCase activity to decrease significantly by 30–40% (Fig. S8). Even the deletion of five ACE3 binding sites (deleting −2446 to −2238 and −1444 to −1225 of cbh1) reduced cbh1 transcription by ∼50–80% and did not totally abolish cbh1 expression in the manner observed for the ace3 deletion strain. There are two explanations for this difference. First, some sites that did not exactly match the 5′-CGGAN(T/A)3-3′ sequence may have been present in the cbh1 promoter (−1225 to −1 of cbh1) and were likely still functional for enabling ACE3 binding as a homodimer. Second, ACE3 can indirectly mediate cbh1 transcription through other factors, such as XYR1 and Crt1 (Fig. S9), in addition to directly activating cbh1 by binding to its promoter. The deletion of ace3 totally abolishes the direct and indirect regulation of cellulase expression by ace3, whereas the deletion of ACE3 binding sites only interrupts its direct regulation of gene expression.
We first demonstrated that the C-terminal domain of XYR1 mediated the homodimerization of XYR1 via in vitro and in vivo experiments (Fig. 4). The C-terminal domain of XYR1, XYR1b, regulates homodimerization, and the interaction between two XYR1 molecules is required for cellulase production. Therefore, a FLAG label fused to the final C-terminal encoding codon of XYR1 can inactivate this protein, as the fused FLAG may hinder the homodimerization of XYR1.
We first identified the interactions between ACE3 and XYR1 (Fig. 6). Additionally, the binding sites of XYR1 and ACE3 are adjacent and interlaced into the promoters of cellulase-related genes (Fig. 5A). Consequently, we can speculate that XYR1 and ACE3 bind as heterodimers to the promoters of downstream genes, as Zn(II)2Cys6 proteins can interact with DNA as monomers, homodimers, or heterodimers (34–36). Also, putative heterodimers of XYR1 and ACE3 may be essential for cellulase production. Therefore, fusing a FLAG label to the final C-terminal encoding codon of ACE3 can inactivate this protein, as the fused FLAG may hinder the heterodimerization of ACE3 and XYR1.
Strong protein–protein interactions exist between XYR1c (amino acids 771–865 of XYR1) and ACE3a (amino acids 523–734 of ACE3) (Fig. 6, D and E). These interactions can explain why X5G4 could not recover its cellulase activity whereas X6G4 could—the XYR1c domain is lost in the X5G4 structure. Interestingly, one mutation, A824V, in XYR1 has been reported to result in a highly elevated basal level of cellulase expression in T. reesei (37). This point mutation occurs in the XYR1c domain, which interacts with ACE3 in our study. The mutation A824V may increase the interactions occurring between ACE3 and XYR1. Further research is required to study these interactions to obtain more effective mutants of ACE3 or XYR1 to improve cellulase production.
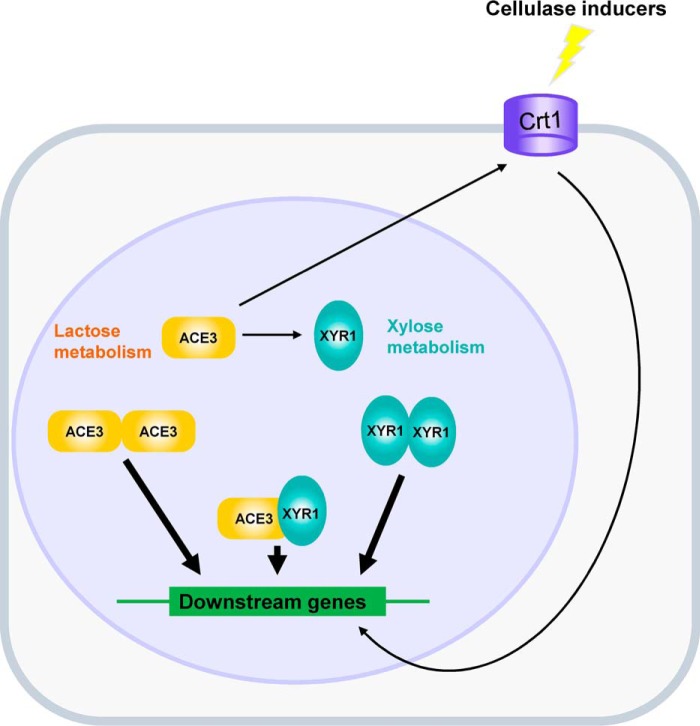
Based on our findings, a regulatory model showing a summary of the interactions between ACE3, Crt1, and XYR1 for mediating cellulase expression and lactose metabolism in T. reesei was constructed (Fig. 7). ACE3 is essential for lactose metabolism. ACE3 mediates lactose metabolism by regulating crt1 and bga1. ACE3 can bind to the promoters of many cellulase genes to directly mediate cellulase production, and ACE3 can bind to crt1 and transcriptional factor genes such as xyr1 to indirectly regulate cellulase production. XYR1 can dimerize, and this is essential for cellulase production. Also, ACE3 interacts with XYR1, a finding that sheds light on the artificial evolution of two essential transcriptional activators for the purpose of enhancing cellulase gene expression in T. reesei.
Figure 7.
Generating a model of ACE3 for mediating cellulase production in T. reesei. ACE3 is essential for lactose metabolism. XYR1 is essential for xylose metabolism (11, 12). ACE3 can bind to the promoters of many cellulase genes to directly mediate cellulase production. ACE3 can bind to crt1 (19) and transcriptional factor genes such as xyr1 to indirectly regulate cellulase production. XYR1 can bind to the promoters of cellulase genes to form homodimers that are essential for cellulase production. Moreover, ACE3 interacts with XYR1, which illustrated that heterodimers of ACE3 and XYR1 were formed during cellulase gene expression in T. reesei.
The regulation of xylanases is less complex than that of cellulases in T. reesei, because ACE3 and the dimerization domain of XYR1 are not necessary for xylanase production (Figs. S2D and S11D). ACE3 only acts as a transcriptional enhancer to improve XYNI expression levels indirectly by binding to the xyr1 promoter to activate its expression (Table S1, Fig. 5, and Fig. S9). X3 (Fig. S11A) is the core structure of XYR1, which includes a DNA-binding domain and a transcription factor domain, both of which are enough for xyn1 and xyn2 expression (Figs. S11D and S12). These results led us to construct the enhanced transcriptional activator XYR1VP (38) and X3VP5 to improve xylanase production in T. reesei. Further research will focus on ACE3–XYR1 interactions to aid in the design and construct of better mutants and/or to redesign ACE3 and XYR1 for improving hydrolytic enzyme production in T. reesei. Additionally, the interaction between two essential fungi-specific Zn(II)2Cys6 type transcriptional activators in T. reesei will shed new light on the molecular mechanisms underlying the expression of hydrolytic enzymes in other fungi.
Experimental procedures
Microbial strains and cultivation conditions
T. reesei QM6a (ATCC 13631) and transformants were maintained on potato dextrose agar (Difco) for the generation of conidia. All strains were cultivated at 28 °C (200 rpm) in MA medium (0.1% polypeptone, 0.05% yeast extract, 0.14% (NH4)2SO4, 0.03% urea, 0.2% KH2PO4, 0.03% CaCl2·2H2O, 0.03% MgSO4·7H2O, 0.05% Tween 80, 0.005‰ FeSO4·7H2O, 0.0016‰ MnSO4·H2O, 0.0014‰ ZnSO4·7H2O, and 0.002‰ CoCl2·6H2O, in which 2% (w/v) glucose, avicel, lactose, or xylan was used as a carbon source).
E. coli DH5α was used for plasmid propagation and cultured at 37 °C in Luria–Bertani medium. The E. coli transformants were obtained by kanamycin-resistant screening. The Agrobacterium tumefaciens AGL-1 strain was used for the transformation of T. reesei under standard electroporation protocols (39). E. coli Rosetta (DE3) was used as a host for the expression of recombinant proteins, and these stains were grown in the TB medium.
Plasmid construction and transformation
The primers, plasmids, and transformants used in this study are listed in Table S2. LML2.1 (40) was used as the skeleton for all plasmids, and the 5′-arms and 3′-arms of the homology double exchange were ligated using the PacI/XbaI and SwaI sites of linearized LML2.1, respectively, using the One Step Cloning Kit (Vazyme Biotech, Nanjing, China). The identity of resultant plasmids was confirmed by DNA sequencing. After Agrobacterium-mediated transformation, transformants were verified by diagnostic PCR, DNA sequencing, and qPCR for copy number (41). The hygromycin marker gene of transformants was rescued using xylose-induced cre recombinase expression (40) prior to further analysis. Additional information on materials and methods can be found in Text S3.
Biomass, cellulase, and xylanase production
These assays were performed as described previously (42, 43) with modifications. Conidia (2 × 106) were cultivated at 28 °C (200 rpm) in 50 ml of MA medium supplemented with 2% carbon source. One milliliter of culture liquid was collected at various intervals to perform the biomass concentration assay. Biomass concentrations from different carbon sources were indirectly measured by calculating the total amount of intracellular proteins (42). The supernatant was used for cellulase and xylanase assays. Mycelia were collected for RNA extraction. The activity of the produced cellulase and xylanase was measured against that of p-nitrophenyl-d-cellobioside or xylan at a pH of 5.0, respectively (43).
RT-qPCR analysis
RNA extraction, cDNA synthesis, and RT-qPCR experiments were performed as described previously (43). The transcriptional levels of the sar1 gene and the RNA of the parental strain were measured for reference calculation and data normalization. All samples were analyzed in at least three independent biological experiments with three RT-qPCR replicates in each assay. The primers used for real-time PCR are listed in Table S2.
Expression and purification of the ACE3 DNA-binding domain
The expression and purification were performed as described previously (17) with modifications. The DBD of ACE3 (amino acids 79–222) was cloned and ligated to the BamHI/XhoI sites of linearized vector pGEX4T-2 (GE Healthcare) using the One Step Cloning Kit, which encoded the fused DNA-binding domain of ACE3 with N-terminal GSH-S-transferase (GST-ACE3DBD). The resulting expression plasmid pGEX4T-2-ACE3DBD was transformed into Rosetta (DE3) Competent E. coli (Tiangen, China), and the transformed stains were grown at 37 °C in TB medium. A final isopropyl-β-d-thiogalactopyranoside concentration of 0.1 mm was then added to induce the production of the target protein at an A600 of ∼0.5–0.6, and the cultures were incubated at 18 °C (180 rpm) for 8 h. Then bacteria were collected by centrifugation and lysed by sonication in 20 mm PBS buffer containing 1 mm DTT and 1 mm phenylmethylsulfonyl fluoride. Next, GST-ACE3DBD was purified on a GST-TrapTM (Senhui Microsphere Tech Co., Ltd), according to the manufacturer's instructions. The Bradford method was used for the determination of protein concentration, with BSA serving as a standard. All purified proteins were stored with 40% (w/v) glycerol at −80 °C.
EMSAs
EMSAs were performed as described previously (17) with modifications. The PCR products or dsDNA obtained after annealing were directly ligated with the pEASY®-Blunt cloning vector (TransGen Biotech, Beijing) and acted as templates of probes. Then a universal 5′-biotin–labeled primer pair (5′-TTAACCCTCACTAAAGGGACTA-3′ and 5′-CGACTCACTATAGGGCGAAT-3′) was used for amplification, and the above templates were used to obtain biotin-labeled DNA fragments. The biotinylated PCR products were purified for use as EMSA probes. EMSA was performed according to the manufacturer's protocol (Beyotime, Shanghai, China). Quantification was performed by densitometry using a Tanon 6200 chemiluminescence image analysis system (Shanghai, China).
DNase I footprinting assay
The fluorescent 6-carboxyfluorescein (FAM)-labeled probes of cbh1-p1 (−1496 to −1272 of cbh1) were PCR-amplified using Dpx DNA polymerase (TOLO Biotech, Shanghai, China) and the primers FAM-labeled M13F and M13R. DNase I footprinting assays were performed using a method similar to that described by Zianni et al. (44). The assay was repeated three times to achieve stability.
BiFC assay
BiFC assays were performed as described previously (45) with modifications. First, the N terminus of mCherry (encoding amino acids 1–159 of mCherry) was fused to XYR1b (amino acids 861–940 of XYR1) and ACE3a (amino acids 523–734 of ACE3) using a flexible linker (GGGGSGGGGS). The fused protein genes mN159-ACE3a and mN159-XYR1b were then inserted into the NdeI/HindIII sites of pET21a to construct pET21a-mN159-ACE3a and pET21a-mN159-XYR1b, respectively. Then the C terminus of mCherry (encoding amino acids 160–236 of mCherry) was fused to XYR1b (amino acids 861–940 of XYR1) and XYR1c (amino acids 771–865 of XYR1) using the same flexible linker (GGGGSGGGGS). The fused protein genes mC160-XYR1b and mC160-XYR1c were then inserted into the NcoI/HindIII sites of pET28a to obtain the plasmids pET28a-mC160-XYR1b and pET28a-mC160-XYR1c, respectively. The two pairs of resulting plasmids (pET28a-mC160-XYR1c/pET21a-mN159-ACE3a and pET28a-mC160-XYR1b/pET21a-mN159-XYR1b) were co-transformed into E. coli Rosetta (DE3) to form E. coli mC160-XYR1c/mN159-ACE3a and E. coli mC160-XYR1b/mN159-XYR1b, respectively. It should be noted that two mCherry fragment genes were fused to both the N-terminal and C-terminal ends of tested protein domains (XYR1b, XYR1c, and ACE3a) to find the best means of achieving fusion and for protein interactions to occur effectively. Positive interactions resulted in red fluorescence. The primers, plasmids, and strains used in the BiFC assay are listed in Table S2.
BiFC assay in T. reesei
The mC160-XYR1/mN159-ACE3 and mC160-ACE3/mN159-XYR1 plasmid pairs were co-transformed into T. reesei to construct co-expression strains TRmC-XYR1/mN-ACE3 and TRmC-ACE3/mN-XYR1. It should be noted that two mCherry fragment genes were fused to both the N-terminal and C-terminal ends of tested protein (XYR1 and ACE3) to find the best means of achieving fusion and for protein interactions to occur effectively. Positive interactions resulted in red fluorescence. The primers, plasmids, and strains used in the BiFC assay are listed in Table S2.
B2H assay
B2H assays were performed as described previously (46) with modifications. The Euromedex bacterial two-hybrid (BACTH) system was used to detect protein–protein interactions and was performed in accordance with manufacturer protocols. The entire xyr1 and ace3 genes were fused to pKT25 and pUT18C, respectively, using the One Step Cloning Kit (Vazyme Biotech, Nanjing). To detect the self-dimerization of the XYR1b domain (amino acids 861–940 of XYR1), XYR1b was fused simultaneously to pKT25 and pUT18C to determine the interactions occurring at the XYR1 C terminus. The B2H assay was performed according to the manufacturer's protocol (Euromedex, Souffelweyersheim, France). The primers, plasmids, and strains used in the B2H assay are listed in Table S2.
Y2H assays
Y2H assays were performed as described previously (47) with modifications. The vectors pGBKT7 and pGADT7 were used for achieving the expression of bait and prey proteins, respectively. Three domains of XYR1, including XYR1b (amino acids 861–940 of XYR1), XYR1c (amino acids 771–865 of XYR1), and XYR1d (amino acids 363–737 of XYR1), and two domains of ACE3, including ACE3a (amino acids 523–734 of ACE3) and ACE3e (amino acids 300–522 of ACE3), were all cloned into pGBKT7 and pGADT7 to test their “bait” or “prey” functions, respectively, in Gold Yeast one- and two-hybrid screening systems (Clontech). All constructs were produced using the One Step Cloning Kit (Vazyme Biotech). Y2H assays were performed according to manufacturer's protocol (Clontech). The primers, plasmids, and strains used in the Y2H assay are listed in Table S2.
RNA-Seq
Parental and Δace3 strains were cultured in MA medium containing 1% (w/v) glycerol for 48 h. Then the mycelia with the same wet weights were collected, washed with sterile water, and transferred to a fresh MA medium containing 1% (w/v) lactose for 12 h. Total RNA was extracted from the mycelia of each sample using TRIzol® reagent, according the manufacturer's instructions (Invitrogen), and genomic DNA was removed using DNase I (TaKara). Then RNA quality was determined using the 2100 Bioanalyzer (Agilent) and quantified using the ND-2000 (NanoDrop Technologies). Only high-quality RNA samples (OD260/280 = 1.8–2.2, OD260/230 ≥ 2.0, RNA Integrity Number ≥ 6.5, 28S/18S ≥ 1.0) were used to construct the sequencing library. The six RNA samples, which included three high-quality biological replicates of parental and Δace3 strains grown in lactose, were used for library preparation, by using the TruSeq RNA sample preparation kit from Illumina (San Diego, CA), using 1 μg of total RNA. Libraries were paired-end–sequenced using the Illumina HiseqTM 2000 (125 PE) instrument developed by Shanghai Majorbio Biotechnology Co., Ltd. (Shanghai, China). Quality-filtered reads were mapped against the T. reesei 2.0 reference genome (http://fungi.ensembl.org/Trichoderma_reesei/Info/Index)6 using the TopHat2 software, which allowed no more than a two-nucleotide mismatch (48). The expression level of each transcript was calculated using the fragments per kilobase of exon per million mapped reads (FPKM) method, used for identifying differential expression genes (DEGs) between two different samples. RSEM (http://deweylab.github.io/RSEM/)6 (49) and R statistical package software EdgeR (Empirical analysis of Digital Gene Expression in R, http://www.bioconductor.org/packages/2.12/bioc/html/edgeR.html)6 (50) were used for quantifying the abundance of genes and for differential expression analysis. The FPKM method was used to identify DEGs based on a false discovery rate of <0.05 and estimated absolute log2 -fold change of >1 between different genotypes. In addition, GO functional enrichment analysis was performed to identify which DEGs were significantly enriched in GO terms, if the Bonferroni-corrected p value was ≤0.05, as compared with that of the whole-transcriptome background (see File S7). GO functional enrichment were carried out by Goatools (https://github.com/tanghaibao/Goatools)6 (53). The raw whole transcriptome shotgun sequencing data have been submitted to the NCBI SRA website (SUB5285647).
Fluorescence analysis
mCherry was visualized using a laser-scanning confocal microscope (A1R, Nikon, Japan) composed of a Texas Red filter (500–620-nm band pass excitation filter and emission filter of 670 nm). The recombinant E. coli were observed using a ×100 oil immersion objective, and mycelia were examined using a ×60 air objective. Images were processed using the NIS Elements software.
Protein extraction and Western blot analysis
Conidia (2 × 106) of 6aFLAG-ACE3 were cultivated at 28 °C (200 rpm) in 50 ml of MA medium supplemented with 2% lactose for 72 h. A total of 20 ml of mycelium were collected and dried by filtration paper. The cell lysate was obtained by bead-beating in lysis buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1% Nonidet P-40 (Beyotime), 0.1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor mixture set I (Merck Millipore) (51). One-milliliter aliquots were mixed with 1.8 g of glass beads (Biospec Products, Bartlesville, OK) in a 2.0-ml screw-cap tube followed by disruption with a bead disrupter (FastPrep-24, MP) for 20 cycles (1 min vibrating at 6.0 m/s and 5 min resting on ice for each cycle). The supernatant was collected by centrifugation at 20,000 × g for 30 min. The protein concentration of the supernatant was measured using the BCA Protein Assay kit (Beyotime), and 30 μg of total protein was loaded in each lane. After SDS-denaturing gel electrophoresis, the protein was transferred to a polyvinylidene difluoride membrane (Millipore). The membranes were then washed and blotted with either anti-FLAG polyclonal antibody (Beyotime) or anti-β-actin antibody (Beyotime). Protein was visualized using anti-mouse or anti-rabbit IgG antibody conjugate to horseradish peroxidase (Beyotime), and ECL reagents (Beyotime). The chemiluminescent image was analyzed using the Tanon 6200 Chemiluminescence image analysis system (Shanghai, China).
ChIP assay
Conidia (2 × 106) of 6aFLAG-ACE3 were cultivated at 28 °C (200 rpm) in 50 ml of MA medium supplemented with 2% lactose for 72 h. The mycelium were cross-linked with 1% formaldehyde for 30 min at room temperature and stopped by adding glycine to a final concentration of 125 mm. A total of 20 ml of mycelium were collected by filtration, washed twice with cold TBS (20 mm Tris-HCl, pH 7.5, 150 mm NaCl), and dried. Cells were lysed by bead-beating in 1 ml of FA-140 lysis buffer (50 mm HEPES, 140 mm NaCl, 1% Triton X-100, 1 mm EDTA, 0.1% sodium deoxycholate, 0.1 mm phenylmethylsulfonyl fluoride, 1× protease inhibitor mixture set I) (52). The cell lysate was drawn off the beads and centrifuged at a maximum speed (20,000 × g) for 30 min at 4 °C. The chromatin pellet was resuspended in 1 ml of FA-140 lysis buffer and sonicated on ice six times with 30-s pulses using a Bioruptor (Diagenode s.a. BELGIUM) at the low-power setting to shear chromatin to an average length of ∼450 bp. Sonicated chromatin solution was centrifuged twice at 10,000 × g for 10 min at 4 °C. The supernatant was then aliquoted into three tubes (labeled “input,” “IP,” and “no-Ab”). The IP samples were incubated overnight at 4 °C with anti-FLAG polyclonal antibody at a dilution of 1:50. Both IP and no-Ab samples were incubated with 20 μl of ChIP-grade protein A/G beads from the EZ-Magna ChIP A/G kit, (Millipore) overnight at 4 °C and then washed as per the manufacturer's instructions. DNA was then eluted from the beads according the protocol. The purified DNA samples were analyzed by qPCR, and results were compared with a standard curve prepared from input DNA. The amount of immunoprecipitated specific promoter DNA was determined relative to no-Ab DNA corresponding to the amplicon for the promoter of SAR1. S.D. values were calculated from the results of three independent biological replicates.
The specific primer pairs SAR1-F/SAR1-R for SAR1 promoter (PSAR1) and CBH1–1F/CBH1–1R, CBH1–2F/CBH1–2R, CBH1–3F/CBH1–3R, Crt1–1F/Crt1–1R, Crt1–2F/Crt1–2R, XYR1–1F/XYR1–1R, XYR1–2F/XYR1–2R, ACE3–1F/ACE3–1R, and ACE3–2F/ACE3–2R for promoters are listed in Table S2.
Statistical analysis
All data were obtained from at least three biological replicates (each with three technical replicates) assayed in duplicate and presented as mean ± S.D. values. Student's t test was performed to determine the differences among grouped data. The statistical significance was assessed if p was <0.05.
Author contributions
J. Z., C. W., and W. W. data curation; J. Z. and Y. C. formal analysis; J. Z. visualization; J. Z., C. W., and W. W. methodology; J. Z. and W. W. writing-original draft; Y. C. and W. W. conceptualization; Y. C., W. W., and D. W. supervision; P. L. and D. W. resources; P. L. software; W. W. validation; W. W. investigation; W. W. project administration; W. W. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Menghao Cai and Qiyao Wang for providing the B2H system and Yushu Ma for providing the Y2H and BiFc systems.
Note added in proof
In the version of this article that was published as a Paper in Press on September 9, 2019, the XYR1 gene model referred to an older one with 920 amino acids. This error has now been corrected and does not affect the results or conclusions of this work.
This work was supported by the Fundamental Research Funds for the Central Universities (Grant 222201714053) and the Open Funding Project of the State Key Laboratory of Bioreactor Engineering (Grant 2018). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1 and S2, Figs. S1–S14, and Text S1–S3.
Raw whole-transcriptome shotgun sequencing data have been submitted to the NCBI SRA website with BioProject ID PRJNA526091.
J. Zhang, Y. Chen, C. Wu, P. Liu, W. Wang, and D. Wei, unpublished data.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- ACE3
- activator of cellulase expression 3
- XYR1
- xylanase regulator 1
- Crt1
- cellulose response transporter 1
- aa
- amino acid
- FAM
- 6-carboxyfluorescein
- BiFC
- bimolecular fluorescence complementation
- B2H
- bacterial two-hybrid
- Y2H
- yeast two-hybrid
- EMSA
- electrophoretic mobility shift assay
- DBD
- DNA-binding domain
- GST
- glutathione S-transferase
- qPCR
- quantitative PCR
- OD
- optical density
- DEG
- differential expression gene
- FPKM
- fragments per kilobase of exon per million mapped reads
- pNPCase
- p-nitrophenylcellobiosidase.
References
- 1. Solomon B. D. (2010) Biofuels and sustainability. Ann. N.Y. Acad. Sci. 1185, 119–134 10.1111/j.1749-6632.2009.05279.x [DOI] [PubMed] [Google Scholar]
- 2. Stern P. C., Janda K. B., Brown M. A., Steg L., Vine E. L., and Lutzenhiser L. (2016) Opportunities and insights for reducing fossil fuel consumption by households and organizations. Nat. Energy 1, 16043 10.1038/nenergy.2016.43 [DOI] [Google Scholar]
- 3. Baeyens J., Kang Q., Appels L., Dewil R., Lv Y., and Tan T. (2015) Challenges and opportunities in improving the production of bio-ethanol. Prog. Energ. Combust. 47, 60–88 10.1016/j.pecs.2014.10.003 [DOI] [Google Scholar]
- 4. Himmel M. E., Ding S. Y., Johnson D. K., Adney W. S., Nimlos M. R., Brady J. W., and Foust T. D. (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315, 804–807 10.1126/science.1137016 [DOI] [PubMed] [Google Scholar]
- 5. Soni S. K., Sharma A., and Soni R. (2018) Cellulases: role in lignocellulosic biomass utilization. Methods Mol. Biol. 1796, 3–23 10.1007/978-1-4939-7877-9_1 [DOI] [PubMed] [Google Scholar]
- 6. Gemishev O., Zapryanov S., Blagoev A., Markova M., and Savov V. (2014) Effect of multiple short highly energetic X-ray pulses on the synthesis of endoglucanase by a mutant strain of Trichoderma reesei-M7. Biotechnol. Biotechnol. Equip. 28, 850–854 10.1080/13102818.2014.966543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martinez D., Berka R. M., Henrissat B., Saloheimo M., Arvas M., Baker S. E., Chapman J., Chertkov O., Coutinho P. M., Cullen D., Danchin E. G., Grigoriev I. V., Harris P., Jackson M., Kubicek C. P., et al. (2008) Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 26, 553–560 10.1038/nbt1403 [DOI] [PubMed] [Google Scholar]
- 8. Gusakov A. V. (2011) Alternatives to Trichoderma reesei in biofuel production. Trends Biotechnol. 29, 419–425 10.1016/j.tibtech.2011.04.004 [DOI] [PubMed] [Google Scholar]
- 9. Saloheimo A., Aro N., Ilmén M., and Penttilä M. (2000) Isolation of the ace1 gene encoding a Cys2-His2 transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei. J. Biol. Chem. 275, 5817–5825 10.1074/jbc.275.8.5817 [DOI] [PubMed] [Google Scholar]
- 10. Aro N., Saloheimo A., Ilmén M., and Penttilä M. (2001) ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276, 24309–24314 10.1074/jbc.M003624200 [DOI] [PubMed] [Google Scholar]
- 11. Cziferszky A., Mach R. L., and Kubicek C. P. (2002) Phosphorylation positively regulates DNA binding of the carbon catabolite repressor Cre1 of Hypocrea jecorina (Trichoderma reesei). J. Biol. Chem. 277, 14688–14694 10.1074/jbc.M200744200 [DOI] [PubMed] [Google Scholar]
- 12. Stricker A. R., Grosstessner-Hain K., Würleitner E., and Mach R. L. (2006) Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and d-xylose metabolism in Hypocrea jecorina. Eukaryot. Cell 5, 2128–2137 10.1128/EC.00211-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Häkkinen M., Valkonen M. J., Westerholm-Parvinen A., Aro N., Arvas M., Vitikainen M., Penttilä M., Saloheimo M., and Pakula T. M. (2014) Screening of candidate regulators for cellulase and hemicellulase production in Trichoderma reesei and identification of a factor essential for cellulase production. Biotechnol. Biofuels 7, 14 10.1186/1754-6834-7-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zeilinger S., Mach R. L., Schindler M., Herzog P., and Kubicek C. P. (1996) Different inducibility of expression of the two xylanase genes xyn1 and xyn2 in Trichoderma reesei. J. Biol. Chem. 271, 25624–25629 10.1074/jbc.271.41.25624 [DOI] [PubMed] [Google Scholar]
- 15. Carle-Urioste J. C., Escobar-Vera J., El-Gogary S., Henrique-Silva F., Torigoi E., Crivellaro O., Herrera-Estrella A., and El-Dorry H. (1997) Cellulase induction in Trichoderma reesei by cellulose requires its own basal expression. J. Biol. Chem. 272, 10169–10174 10.1074/jbc.272.15.10169 [DOI] [PubMed] [Google Scholar]
- 16. Zeilinger S., Mach R. L., and Kubicek C. P. (1998) Two adjacent protein binding motifs in the cbh2 (Cellobiohydrolase II-encoding) promoter of the fungus Hypocrea jecorina (Trichoderma reesei) cooperate in the induction by cellulose. J. Biol. Chem. 273, 34463–34471 10.1074/jbc.273.51.34463 [DOI] [PubMed] [Google Scholar]
- 17. Furukawa T., Shida Y., Kitagami N., Mori K., Kato M., Kobayashi T., Okada H., Ogasawara W., and Morikawa Y. (2009) Identification of specific binding sites for XYR1, a transcriptional activator of cellulolytic and xylanolytic genes in Trichoderma reesei. Fungal Genet. Biol. 46, 564–574 10.1016/j.fgb.2009.04.001 [DOI] [PubMed] [Google Scholar]
- 18. Ivanova C., Bååth J. A., Seiboth B., and Kubicek C. P. (2013) Systems analysis of lactose metabolism in Trichoderma reesei identifies a lactose permease that is essential for cellulase induction. PLoS One 8, e62631 10.1371/journal.pone.0062631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang W., Kou Y., Xu J., Cao Y., Zhao G., Shao J., Wang H., Wang Z., Bao X., Chen G., and Liu W. (2013) Two major facilitator superfamily sugar transporters from Trichoderma reesei and their roles in induction of cellulase biosynthesis. J. Biol. Chem. 288, 32861–32872 10.1074/jbc.M113.505826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hopp T. P., Prickett K. S., Price V. L., Libby R. T., March C. J., Cerretti D. P., Urdal D. L., and Conlonet P. J. (1988) A short polypeptide marker sequence useful for recombinant protein identification and purification. Nat. Biotechnol. 6, 1204–1210 10.1038/nbt1088-1204 [DOI] [Google Scholar]
- 21. Seiboth B., Hartl L., Salovuori N., Lanthaler K., Robson G. D., Vehmaanperä J., Penttilä M. E., and Kubicek C. P. (2005) Role of the bga1-encoded extracellular β-galactosidase of Hypocrea jecorina in cellulase induction by lactose. Appl. Environ. Microbiol. 71, 851–857 10.1128/AEM.71.2.851-857.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gamauf C., Marchetti M., Kallio J., Puranen T., Vehmaanperä J., Allmaier G., Kubicek C. P., and Seiboth B. (2007) Characterization of the bga1-encoded glycoside hydrolase family 35 β-galactosidase of Hypocrea jecorina with galacto-β-d-galactanase activity. FEBS J. 274, 1691–1700 10.1111/j.1742-4658.2007.05714.x [DOI] [PubMed] [Google Scholar]
- 23. Hong M., Fitzgerald M. X., Harper S., Luo C., Speicher D. W., and Marmorstein R. (2008) Structural basis for dimerization in DNA recognition by Gal4. Structure 16, 1019–1026 10.1016/j.str.2008.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lichius A., Bidard F., Buchholz F., Le Crom S., Martin J., Schackwitz W., Austerlitz T., Grigoriev I. V., Baker S. E., Margeot A., Seiboth B., and Kubicek C. P. (2015) Genome sequencing of the Trichoderma reesei QM9136 mutant identifies a truncation of the transcriptional regulator XYR1 as the cause for its cellulase-negative phenotype. BMC Genomics 16, 326 10.1186/s12864-015-1526-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lupas A., Van Dyke M., and Stock J. (1991) Predicting coiled coils from protein sequences. Science 252, 1162–1164 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- 26. Lupas A. (1996) Coiled coils: new structures and new functions. Trends Biochem. Sci. 21, 375–382 10.1016/S0968-0004(96)10052-9 [DOI] [PubMed] [Google Scholar]
- 27. Wang X., Chen X., and Yang Y. (2012) Spatiotemporal control of gene expression by a light-switchable transgene system. Nat. Methods 9, 266–269 10.1038/nmeth.1892 [DOI] [PubMed] [Google Scholar]
- 28. MacPherson S., Larochelle M., and Turcotte B. (2006) A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 70, 583–604 10.1128/MMBR.00015-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Micheli M., Bille J., Schueller C., and Sanglard D. (2002) A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol. Microbiol. 43, 1197–1214 10.1046/j.1365-2958.2002.02814.x [DOI] [PubMed] [Google Scholar]
- 30. Axelrod J. D., Majors J., and Brandriss M. C. (1991) Proline-independent binding of PUT3 transcriptional activator protein detected by footprinting in vivo. Mol. Cell Biol. 11, 564–567 10.1128/MCB.11.1.564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vashee S., Xu H., Johnston S. A., and Kodadek T. (1993) How do “Zn2Cys6” proteins distinguish between similar upstream activation sites? Comparison of the DNA-binding specificity of the GAL4 protein in vitro and in vivo. J. Biol. Chem. 268, 24699–24706 [PubMed] [Google Scholar]
- 32. Larsen L. A., Armstrong J. S., Grønskov K., Hjalgrim H., Macpherson J. N., Brøndum-Nielsen K., Hasholt L., Nørgaard-Pedersen B., and Vuust J. (2000) Haplotype and AGG-interspersion analysis of FMR1.CGG)(n) alleles in the Danish population: implications for multiple mutational pathways towards fragile X alleles. Am. J. Med. Genet. 93, 99–106 [DOI] [PubMed] [Google Scholar]
- 33. Nikolaev I., Lenouvel F., and Felenbok B. (1999) Unique DNA binding specificity of the binuclear zinc AlcR activator of the ethanol utilization pathway in Aspergillus nidulans. J. Biol. Chem. 274, 9795–9802 10.1074/jbc.274.14.9795 [DOI] [PubMed] [Google Scholar]
- 34. Lohr D., Venkov P., and Zlatanova J. (1995) Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 9, 777–787 10.1096/fasebj.9.9.7601342 [DOI] [PubMed] [Google Scholar]
- 35. Schjerling P., and Holmberg S. (1996) Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 24, 4599–4607 10.1093/nar/24.23.4599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Todd R. B., and Andrianopoulos A. (1997) Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet. Biol. 21, 388–405 10.1006/fgbi.1997.0993 [DOI] [PubMed] [Google Scholar]
- 37. Derntl C., Gudynaite-Savitch L., Calixte S., White T., Mach R. L., and Mach-Aigner A. R. (2013) Mutation of the xylanase regulator 1 causes a glucose blind hydrolase expressing phenotype in industrially used Trichoderma strains. Biotechnol. Biofuels 6, 62 10.1186/1754-6834-6-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang J., Wu C., Wang W., Wang W., and Wei D. (2018) Construction of enhanced transcriptional activators for improving cellulase production in Trichoderma reesei RUT C30. Bioresour. Bioprocess. 5, 40 10.1186/s40643-018-0226-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Michielse C. B., Hooykaas P. J., van den Hondel C. A., and Ram A. F. (2008) Agrobacterium-mediated transformation of the filamentous fungus Aspergillus awamori. Nat. Protoc. 3, 1671–1678 10.1038/nprot.2008.154 [DOI] [PubMed] [Google Scholar]
- 40. Zhang L., Zhao X., Zhang G., Zhang J., Wang X., Zhang S., Wang W., and Wei D. (2016) Light-inducible genetic engineering and control of non-homologous end-joining in industrial eukaryotic microorganisms: LML 3.0 and OFN 1.0. Sci. Rep. 6, 20761 10.1038/srep20761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li C., Lin F., Zhou L., Qin L., Li B., Zhou Z., Jin M., and Chen Z. (2017) Cellulase hyper-production by Trichoderma reesei mutant SEU-7 on lactose. Biotechnol. Biofuels 10, 228 10.1186/s13068-017-0915-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen Y., Shen Y., Wang W., and Wei D. (2018) Mn2+ modulates the expression of cellulase genes in Trichoderma reesei Rut-C30 via calcium signaling. Biotechnol. Biofuels 11, 54 10.1186/s13068-018-1055-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang J., Zhang G., Wang W., Wang W., and Wei D. (2018) Enhanced cellulase production in Trichoderma reesei RUT C30 via constitution of minimal transcriptional activators. Microb. Cell Fact. 17, 75 10.1186/s12934-018-0926-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zianni M., Tessanne K., Merighi M., Laguna R., and Tabita F. R. (2006) Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J. Biomol. Tech. 17, 103–113 [PMC free article] [PubMed] [Google Scholar]
- 45. Fan J. Y., Cui Z. Q., Wei H. P., Zhang Z. P., Zhou Y. F., Wang Y. P., and Zhang X. E. (2008) Split mCherry as a new red bimolecular fluorescence complementation system for visualizing protein-protein interactions in living cells. Biochem. Biophys. Res. Commun. 367, 47–53 10.1016/j.bbrc.2007.12.101 [DOI] [PubMed] [Google Scholar]
- 46. Wang X., Wang Q., Wang J., Bai P., Shi L., Shen W., Zhou M., Zhou X., Zhang Y., and Cai M. (2016) Mit1 transcription factor mediates methanol signaling and regulates the alcohol oxidase 1 (AOX1) promoter in Pichia pastoris. J. Biol. Chem. 291, 6245–6261 10.1074/jbc.M115.692053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang M., Wu Z., and Fields S. (1995) Protein-peptide interactions analyzed with the yeast two-hybrid system. Nucleic Acids Res. 23, 1152–1156 10.1093/nar/23.7.1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S. L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li B., and Dewey C. N. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Robinson M. D., McCarthy D. J., and Smyth G. K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. James Theoga Raj C., Croft T., Venkatakrishnan P., Groth B., Dhugga G., Cater T., and Lin S. J. (2019) The copper-sensing transcription factor Mac1, the histone deacetylase Hst1, and nicotinic acid regulate de novo NAD+ biosynthesis in budding yeast. J. Biol. Chem. 294, 5562–5575 10.1074/jbc.RA118.006987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li M., Petteys B. J., McClure J. M., Valsakumar V., Bekiranov S., Frank E. L., and Smith J. S. (2010) Thiamine biosynthesis in Saccharomyces cerevisiae is regulated by the NAD+-dependent histone deacetylase Hst1. Mol. Cell Biol. 30, 3329–3341 10.1128/MCB.01590-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Klopfenstein D. V., Zhang L., Pedersen B. S., Ramírez F., Warwick Vesztrocy A., Naldi A., Mungall C. J., Yunes J. M., Botvinnik O., Weigel M., Dampier W., Dessimoz C., Flick P., and Tang H. (2018) GOATOOLS: a Python library for Gene Ontology analyses. Sci. Rep. 8, 10872 10.1038/s41598-018-28948-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.