Abstract
Peroxisome proliferator-activated receptor γ (PPARγ) is the central regulator of adipogenesis, and its dysregulation is linked to obesity and metabolic diseases. Identification of the factors that regulate PPARγ expression and activity is therefore crucial for combating obesity. Aryl hydrocarbon receptor (AhR) is a ligand-activated transcription factor with a known role in xenobiotic detoxification. Recent studies have suggested that AhR also plays essential roles in energy metabolism. However, the detailed mechanisms remain unclear. We previously reported that experiments with adipocyte-specific Cullin 4b (Cul4b)-knockout mice showed that CUL4B suppresses adipogenesis by targeting PPARγ. Here, using immunoprecipitation, ubiquitination, real-time PCR, and GST-pulldown assays, we report that AhR functions as the substrate receptor in CUL4B-RING E3 ubiquitin ligase (CRL4B) complex and is required for recruiting PPARγ. AhR overexpression reduced PPARγ stability and suppressed adipocyte differentiation, and AhR knockdown stimulated adipocyte differentiation in 3T3-L1 cells. Furthermore, we found that two lysine sites on residues 268 and 293 in PPARγ are targeted for CRL4B-mediated ubiquitination, indicating cross-talk between acetylation and ubiquitination. Our findings establish a critical role of AhR in regulating PPARγ stability and suggest that the AhR–PPARγ interaction may represent a potential therapeutic target for managing metabolic diseases arising from PPARγ dysfunction.
Keywords: aryl hydrocarbon receptor (AhR) (AHR), peroxisome proliferator-activated receptor (PPAR), E3 ubiquitin ligase, protein degradation, ubiquitylation (ubiquitination), adipocyte differentiation, cullin 4B, peroxisome proliferator-activated receptor γ (PPARγ)
Introduction
Exorbitant body fat in obese people is thought to be a major reason for insulin resistance, cardiovascular diseases, and diabetes (1). Obesity-related insulin resistance is associated with dysregulation of lipid storage and chronic inflammation in adipose tissue (2). Insulin resistance is likely to be induced by inappropriate regulation of gene expression required for adipocyte differentiation or functions. Adipocyte differentiation from preadipocytes is controlled by a number of transcriptional cascades, particularly PPARγ2 (3). PPARγ proteins are expressed in two isoforms, PPARγ1 and PPARγ2. PPARγ1 is expressed in a number of tissues. PPARγ2 is only observed in adipocytes and is crucial in maintaining normal insulin sensitivity (4).
PPARγ protein is post-translationally regulated by several modifications. Phosphorylation on Ser-273 of PPARγ is modified by Cdk5 to dysregulate the expression of specific genes, such as adiponectin (5). Acetylation on Lys-268 and Lys-293 of PPARγ is a signal of lipid storage and cell proliferation, whereas deacetylation of these two sites results in energy expenditure and promotes insulin sensitivity (6). PPARγ degradation is linked to the regulation of its transcriptional activity. Recently, several E3s have been identified in adipocytes, including seven in absentia homolog 2 (SIAH2) (7), makorin ring finger protein 1 (MKRN1) (8), tripartite motif protein 23 (TRIM23) (9), and neural precursor cell–expressed developmentally down-regulated protein 4 (NEDD4) (10). These E3s are located predominantly in the cytoplasm, and not all of them target PPARγ toward proteasomal degradation.
Cullin 4B (CUL4B) acts as a scaffold protein that assembles DDB1, ROC1, and DDB1–CUL4 association factor (DCAF) to form CUL4B-RING E3 ubiquitin ligases (CRL4B). CRL4B uses a variety of DCAFs to assemble different E3 ligases to specifically target substrate (11). Thus, DCAF is correlated with substrate selection and specificity. By targeting different substrates for Ub-dependent degradation or modification, CRL4B has been shown to participate in the regulation of diverse physiologically and developmentally controlled processes. Patients with CUL4B mutations manifest mental and growth retardation as well as central obesity (12–15). Using adipocyte-specific Cul4b-knockout mice, we previously showed that CUL4B functions as a negative regulator of adipogenesis (16). However, the DCAF in CRL4B complex that recruits PPARγ remains unknown.
AhR, also known as dioxin receptor, was reported to play essential roles in xenobiotic and energy metabolism (17–20). In this study, we identified AhR as a novel factor that negatively regulates PPARγ protein stability via forming the CRL4B E3 ligase complex. The physiological function of AhR in adipocyte differentiation was further demonstrated using AhR-overexpressing and -knockdown 3T3-L1 cell lines. Our study provides a direct link between AhR and PPARγ and indicates that AhR–PPARγ interaction is a potential therapeutic target in PPARγ-related diseases.
Results
AhR negatively regulates adipocyte differentiation
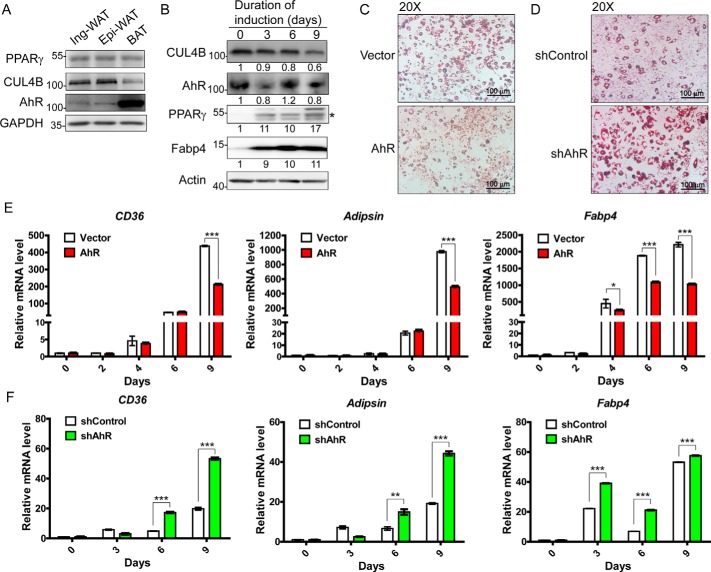
To investigate the role of AhR in adipocyte differentiation, we first examined the expression level of AhR in mouse adipose tissues. Similar to CUL4B and PPARγ, AhR was detected in white adipocyte tissues, including inguinal (subcutaneous) and epididymal (visceral) fat pads. AhR was expressed highly in brown adipose tissue (Fig. 1A). We also examined the mRNA and protein levels of AhR, CUL4B, and PPARγ as well as its target genes at different time points during differentiation. mRNA levels and protein levels of PPARγ and adipogenic markers were increased dramatically from day 0 postinduction. The transcriptional level of Cul4b was eventually reduced, and its protein level was also decreased dramatically at late stages of adipogenic differentiation, which is consistent with our previous study (16). Notably, the mRNA level of Ahr was also ultimately decreased during the process of differentiation, consistent with its protein level (Figs. S1 and 1B).
Figure 1.
Adipogenesis is negatively regulated by AhR. A, the levels of the indicated proteins in mouse adipose tissues. B, protein levels of CUL4B and its related proteins were determined in 3T3-L1 cells. Upon differentiation with induction medium, cells were harvested and lysed for Western blotting. The protein band of interest is marked with a star. Signals from the Western blots were analyzed by Volume Analysis of Quantity One software with volume background subtraction. C, adipogenesis of AhR-overexpressing 3T3-L1 cells. Cells stably expressing empty vector or AhR were constructed using a lentivirus system. At day 9 postinduction, the cells were stained for lipid droplets using Oil Red O. D, adipogenesis of AhR-knockdown 3T3-L1. Stable cell lines were constructed using a lentivirus expressing shRNA (sh) for scrambled sequences (control) or mouse Ahr. On day 9 after induction of differentiation, cells were stained with Oil Red O. E, in AhR-overexpressing 3T3-L1 cells, total RNA samples were extracted at different time points upon induction. Samples were subjected to quantitative PCR analysis of adipogenic markers (CD36, Adipsin, and Fabp4). F, in AhR-knockdown 3T3-L1 cells, total RNA samples were extracted at different time points. Samples were subjected to quantitative PCR analysis of adipogenic markers (CD36, Adipsin, and Fabp4). Data are presented as mean ± S.D. (error bars); n = 4 with * representing p < 0.05, ** representing p < 0.01, and *** representing p < 0.001 by Student's t test. Ing-WAT, inguinal white adipose tissue; Epi-WAT, epididymal white adipose tissue; BAT, brown adipose tissue.
We next studied whether AhR negatively regulates adipocyte differentiation like CUL4B does. Overexpression of AhR using a retrovirus suppressed the adipocyte differentiation of 3T3-L1 cells, as demonstrated with Oil Red O staining (Fig. 1C and S2). In contrast, knockdown of AhR expression led to increased lipid accumulation (Fig. 1D and S2). Consistently, overexpressed AhR led to decreased expression of adipogenic markers, such as CD36, Adipsin, and Fabp4 (Fig. 1E), whereas knockdown of AhR induced increased expression of adipocyte-specific genes (Fig. 1F).
AhR decreases the stability of PPARγ protein via a proteasome-dependent mechanism
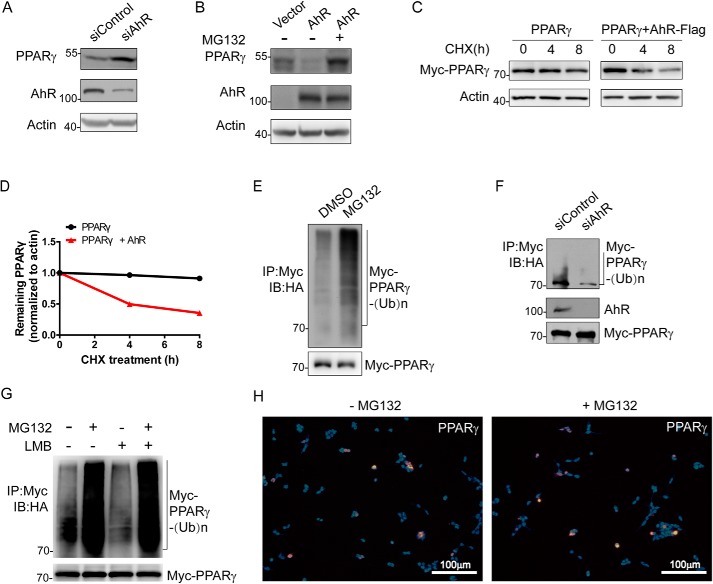
To determine whether AhR regulates adipogenesis via modulating the PPARγ level, we examined the expression of PPARγ when AhR expression was knocked down or overexpressed. Although transcriptional levels of PPARγ in 3T3-L1 cells were not obviously changed by either AhR knockdown or overexpression (Fig. S3), knockdown of AhR expression significantly increased the endogenous protein level of PPARγ (Fig. 2A). Conversely, overexpression of AhR in 3T3-L1 cells induced a decrease of endogenous PPARγ. Importantly, the decreased PPARγ led by the overexpression of AhR was efficiently blocked by the administration of MG132, a proteasome inhibitor, suggesting that AhR may down-regulate PPARγ by a proteasome-dependent degradation mechanism (Fig. 2B). To further confirm this notion, we measured the half-life of PPARγ in AhR-overexpressing HEK293T cells. As expected, overexpression of AhR significantly increased PPARγ decay (Fig. 2, C and D). Consistently, the administration of MG132 significantly increased the accumulation of polyubiquitinated PPARγ in HEK293T cells transfected with AhR and PPARγ plasmids (Fig. 2E). Knockdown of AhR in 3T3-L1 preadipocytes also resulted in a reduction of polyubiquitinated PPARγ (Fig. 2F). Furthermore, in the presence or absence of leptomycin B (LMB) or MG132, we observed that PPARγ was ubiquitinated and targeted for proteasomal degradation in the nucleus (Fig. 2G). Immunofluorescence analysis results also showed that PPARγ accumulated in the nucleus upon MG132 treatment (Fig. 2H). Taken together, these results suggest that AhR decreases the stability of PPARγ protein via a proteasome-dependent mechanism.
Figure 2.
AhR affects PPARγ protein level and stability. A, effect of AhR ablation on PPARγ protein level. 3T3-L1 cells were transfected with the indicated siRNAs (si) for 8 h. Cells were harvested, lysed, and immunoprecipitated with the indicated antibodies followed by Western blotting. B, effect of AhR overexpression on PPARγ protein level. 3T3-L1 cells were transfected with the indicated plasmids expressing AhR-Flag-His in the absence or presence of MG132. Cells were harvested, lysed, and immunoprecipitated with the indicated antibodies followed by Western blotting. C, effect of AhR on the protein stability of PPARγ. HEK293T cells were transfected with the indicated plasmids expressing Myc-PPARγ in the absence or presence of AhR-Flag-His for CHX-chase assay. Cells were harvested and lysed. The cell lysate was then detected with the indicated antibodies. D, turnover of PPARγ was determined by Western blotting. Signals from immunoblots were analyzed using Quantity One. PPARγ protein signals were normalized with the actin protein signals, and the percentage of PPARγ protein remaining was plotted against time. E, ubiquitination product mediated by CUL4B–AhR-associated complex was accumulated in the presence of proteasome inhibitor MG132. HEK293T cells were transfected with plasmids expressing Flag-Cul4B, Myc-PPARγ, AhR-Flag-His, and HA-Ub. Cells were then treated with DMSO or MG132 for 3 h. F, effect of CUL4B–AhR-associated complex on PPARγ ubiquitination. 3T3-L1 cells were transfected with the indicated siRNA for 5 h. The cells were then transfected with plasmids expressing Flag-Cul4B, Myc-PPARγ, and HA-Ub. After MG132 treatment for 4 h, cells were harvested, lysed, and immunoprecipitated with the indicated antibodies followed by Western blotting. G, HEK293T cells were transfected with plasmids expressing Flag-Cul4B, AhR-Flag-His, HA-Ub, and Myc-PPARγ. Cells were treated with MG132 for 3 h and LMB for 4 h followed by immunoprecipitation and Western blotting. Input (5%) was used for Western blotting. H, HEK293T cells were transfected with plasmids expressing Myc-PPARγ and AhR-Flag-His. Cells were treated with MG132 or DMSO for 6 h. Immunofluorescence against PPARγ was performed. Images were collected using a fluorescence microscope. IP, immunoprecipitation; IB, immunoblotting.
AhR functions as a substrate receptor for CRL4B-mediated PPARγ degradation
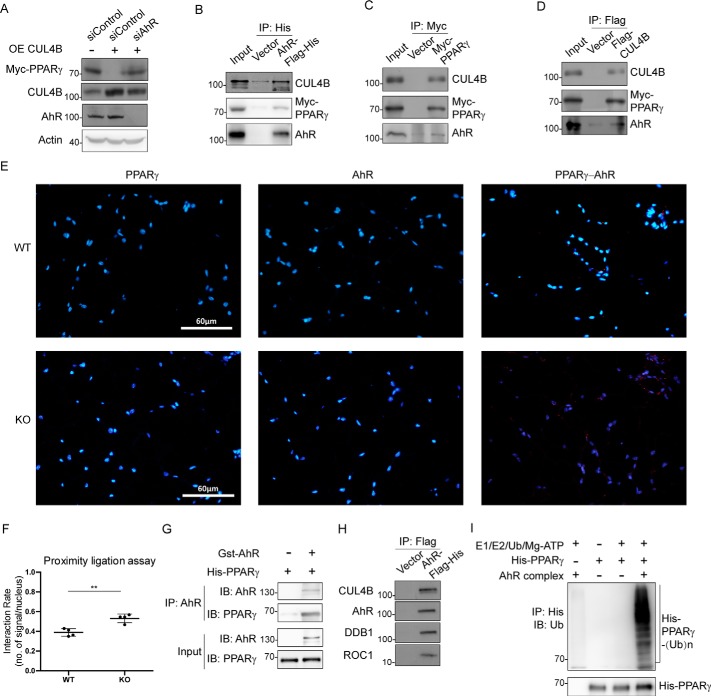
The fact that AhR decreases the stability of PPARγ protein prompted us to determine whether AhR acts as a substrate receptor for CRL4B-mediated PPARγ degradation. When overexpression of CUL4B decreased PPARγ in 3T3-L1 cells, knockdown of AhR could efficiently block the reduction of PPARγ caused by CUL4B overexpression (Fig. 3A), suggesting that AhR is required for the CRL4B-mediated PPARγ degradation. To further strengthen this notion, we performed coimmunoprecipitation assays in 3T3-L1 cells to determine possible physical association among AhR, CUL4B, and PPARγ. As shown in the figure, when AhR was immunoprecipitated from 3T3-L1 cells, both CUL4B and PPARγ were brought down as well (Fig. 3B). Consistently, AhR was also coimmunoprecipitated with antibodies against PPARγ or CUL4B (Fig. 3, C and D). Interactions between PPARγ and AhR were also confirmed in mouse adipose tissue using proximity ligation assay (PLA). Consistent with increased PPARγ level led by CUL4B deletion, more positive signals showing PPARγ–AhR interactions were detected in adipose tissues of adipocyte-specific knockout mice (Fig. 3, E and F). GSH S-transferase (GST) pulldown experiments were performed to further examine whether AhR binds PPARγ. Our results showed that AhR directly interacts with PPARγ (Fig. 3G).
Figure 3.
AhR directly interacts with PPARγ and functions as a substrate receptor in the E3 ligase complex. A, CUL4B affects PPARγ stabilization dependent on AhR. 3T3-L1 cells were transfected with the indicated siRNA and plasmids. Cells were harvested, lysed, and immunoprecipitated with the indicated antibodies followed by Western blotting. B–D, interactions among CUL4B, AhR, and PPARγ. 3T3-L1 cells were transfected with the plasmids as indicated. The transfected cells were then harvested and immunoprecipitated with the indicated antibodies individually. E, interactions between PPARγ and AhR were tested in epididymal white adipose tissues from WT and knockout mice. PLA (Duolink, Sigma) was performed, and images were collected using a fluorescence microscope. F, number of signals per nucleus was calculated and analyzed using Prism software. Data are presented as mean ± S.D. (error bars); n = 4 with ** representing p < 0.01 by Student's t test. G, direct interactions between AhR and PPARγ. GST-AhR and His-PPARγ were expressed in E. coli individually followed by protein purification and immunoprecipitation and pulldown. The proteins were detected using the indicated antibodies. Input (5%) was used for Western blotting. H, HEK293T cells were transfected with AhR-Flag-His or Flag-His vector. Cells were then harvested and sonicated followed by Flag pulldown. The coimmunoprecipitated products used in the ubiquitination assay were detected using the indicated antibodies. I, ubiquitination of PPARγ targeted by the AhR complex in vitro. Purified human recombinant His-PPARγ (full-length) was incubated with E1, E2, Ub, and ATP in the absence and presence of the AhR complex and the coimmunoprecipitated products obtained in F. Reactions were performed at 37 °C for 1 h followed by Flag pulldown. Ubiquitination of His-PPARγ was analyzed by Western blotting using anti-Ub antibody. IP, immunoprecipitation; IB, immunoblotting; OE, overexpressing.
We next determined whether CUL4B–AhR complex represented an E3 ligase for PPARγ. To this end, His-Flag-AhR was expressed in HEK293T cells, and cellular extracts were prepared by affinity purification. As expected, the components of AhR complex, including endogenous CUL4B, DDB1, and ROC1, were detected in the affinity-purified fractions, thereby forming CRL4BAhR E3 complex (Fig. 3H). Importantly, the affinity-purified AhR complex significantly increased the amount of polyubiquitinated PPARγ, as indicated by an in vitro ubiquitination assay (Fig. 3I). Taken together, these results suggest that AhR functions as the substrate receptor in CRL4BAhR complex by recruiting PPARγ and facilitating its ubiquitination.
Domain–domain interactions between PPARγ and AhR
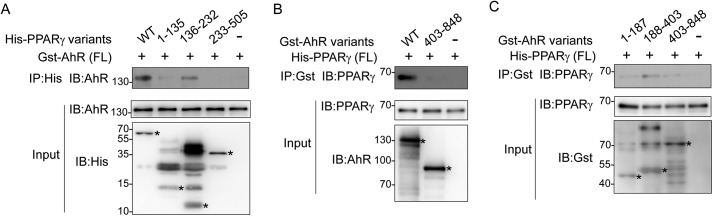
To map out the region where PPARγ (isoform 2) binds AhR, we designed several plasmids expressing truncated PPARγ. Full-length AhR and PPARγ truncations were expressed in Escherichia coli. Our results showed that PPARγ binds AhR mainly on its DNA-binding domain (136–232aa). The N-terminal region of PPARγ (1–135aa) seemed to assist in binding AhR, whereas the C-terminal region of PPARγ, including its ligand-binding domain (233–505aa), did not contribute to binding (Fig. 4A).
Figure 4.
AhR recruits PPARγ upon domain–domain interactions. A, mapping of the PPARγ domain responsible for binding AhR. GST-AhR (full length (FL)) and His-PPARγ truncations were expressed in E. coli individually followed by protein purification. Proteins were mixed for Ni2+ pulldown followed by Western blotting. Input (5%) was detected using the indicated antibodies. Protein bands of interest are marked with stars. B and C, mapping of the AhR domain responsible to interact with PPARγ. His-PPARγ (full length) and GST-AhR truncations were expressed in E. coli individually followed by protein purification. Proteins were mixed followed by immunoprecipitation with the indicated antibodies and Western blotting. Input (5%) was detected using the indicated antibodies. Protein bands of interest are marked with stars. IP, immunoprecipitation; IB, immunoblotting.
Meanwhile, we investigated the region of AhR that contributes in binding PPARγ. All truncated AhRs were expressed in E. coli and purified separately. Because neither AhR nor GST antibodies recognize all of the AhR truncations, we performed two sets of experiments. AhR 403–848aa was assayed in both experiments. Our results demonstrated that the C-terminal fragment of AhR (403–848aa) does not pull down PPARγ (Fig. 4B). Instead of full-length AhR, AhR fragment (188–403aa), including its ligand-binding domain, tends to pull down more PPARγ, suggesting that this region mainly contributes to recruiting PPARγ (Fig. 4C).
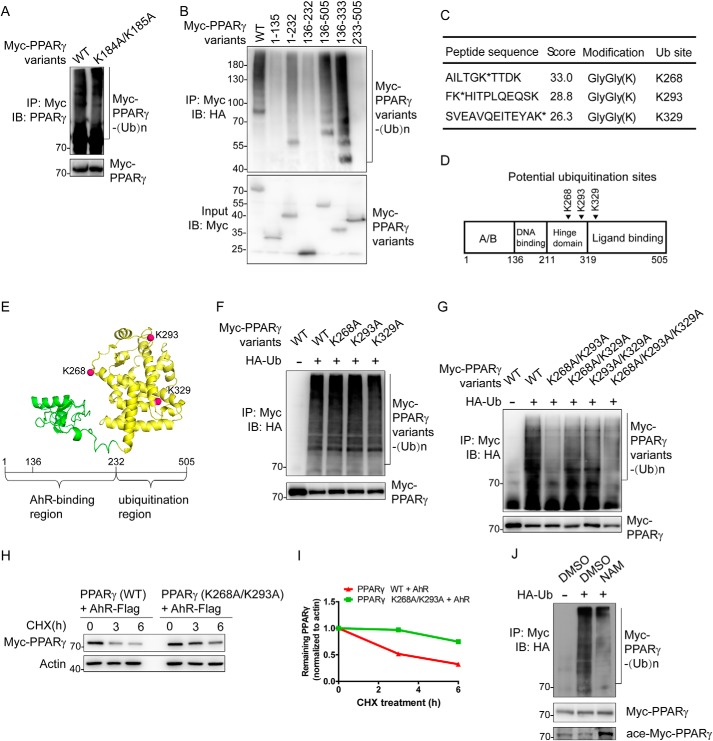
Two lysines located on the hinge domain of PPARγ are targeted for ubiquitination by CRL4BAhR
Lys-184 and Lys-185 on PPARγ were previously reported as ubiquitination sites mediated by MKRN1 (8). To examine whether CRL4BAhR targets the same residues, we mutated Lys-184/185 to alanines and performed a ubiquitination assay. Our results showed that K184A/K185A mutant exhibited a level of ubiquitination similar to that of WT PPARγ, suggesting that CRL4BAhR catalyzes PPARγ ubiquitination on novel site(s) (Fig. 5A). PPARγ is a lysine-rich protein. To identify the amino acid residue(s) targeted by CRL4BAhR, we first mapped the ubiquitinated region using different PPARγ truncations. Truncations 1–135aa, 136–232aa, and 233–505aa were barely ubiquitinated, suggesting that the targeted lysine was not located within these regions. Truncations 136–333aa and 136–505aa that contain the major AhR-binding region (136–232aa) were both ubiquitinated with levels similar to that of WT PPARγ, indicating that the 233–333aa region of PPARγ is required for ubiquitination by CRL4BAhR. Additionally, retaining the ability to bind AhR, truncation 1–232aa was also ubiquitinated but with a lower level compared with WT PPARγ, possibly due to nonspecific modifications (Fig. 5B).
Figure 5.
Identification of ubiquitination site(s) in PPARγ. A, CRL4BAhR targets PPARγ for ubiquitination at a novel site(s). HEK293T cells were transfected with plasmids expressing Flag-Cul4B, AhR-Flag-His, HA-Ub, and Myc-PPARγ variants. Cells were treated with MG132 for 3 h and harvested for the following immunoprecipitation and Western blotting. Input (5%) was used for Western blotting. B, domain–domain interactions are required for PPARγ ubiquitination. HEK293T cells were transfected with plasmids expressing Flag-Cul4B, AhR-Flag-His, HA-Ub, and Myc-PPARγ truncations. Cells were treated with MG132 for 6 h and harvested for the following immunoprecipitation and Western blotting. Input (5%) was used for Western blotting. C, list of PPARγ ubiquitination sites determined by MS. HEK293T cells were transfected with plasmids expressing Flag-Cul4B, AhR-Flag-His, HA-Ub, and Myc-PPARγ. Cells were treated with MG132 for 10 h and harvested for immunoprecipitation using anti-Myc antibody. D, schematic structure of PPARγ. Arrowheads indicate potential ubiquitinated sites. E, the model of PPARγ isoform 2 (created from Protein Data Bank code 1PRG (65)) is demonstrated in cartoon. The AhR-binding region is colored green, and the ubiquitination region is colored yellow. Potential ubiquitination sites are shown as red spheres. F, single mutations on PPARγ barely affect ubiquitination. HEK293T cells were transfected with plasmids expressing Flag-Cul4B, AhR-Flag-His, HA-Ub, and Myc-PPARγ variants. Cells were treated with MG132 for 3 h and harvested for the following immunoprecipitation and Western blotting. Input (5%) was used for Western blotting. G, effect of dual mutations on PPARγ ubiquitination. HEK293T cells were transfected with plasmids expressing Flag-Cul4B, AhR-Flag-His, HA-Ub, and Myc-PPARγ variants. Cells were treated with MG132 for 3 h and harvested for the following immunoprecipitation and Western blotting. Input (5%) was used for Western blotting. H, effect of dual mutations on PPARγ stability. HEK293T cells were transfected with the indicated plasmids expressing AhR-Flag-His and Myc-PPARγ variants for CHX-chase assay. The final concentration of CHX used in the treatment was 60 μg/ml. The cell lysate was then detected with the indicated antibodies. I, turnover of WT and mutated PPARγ was determined by Western blotting. Signals from immunoblots were analyzed using Quantity One software. Protein signals of PPARγ variants were normalized with the actin protein signals, and the percentage of PPARγ variants remaining was plotted against time. J, SirT1 inhibitor NAM reduced PPARγ ubiquitination level. HEK293T cells were transfected with plasmids expressing Flag-Cul4B, AhR-Flag-His, HA-Ub, and Myc-PPARγ. Cells were treated with MG132 (20 μm; 1.5 h) in the absence or presence of NAM (100 μm; 48 h). Cells were then harvested for the following immunoprecipitation and Western blotting. Input (5%) was used for Western blotting. IP, immunoprecipitation; IB, immunoblotting.
We next characterized the specific ubiquitination site(s) on PPARγ by mass spectrometry (MS). This can be accomplished by affinity purification of Myc-tagged PPARγ, isolating the captured proteins by SDS-PAGE, extracting the high-molecular-weight modified protein, and performing in-gel tryptic digestion. According to MS results, three peptide fragments gave GG–K ubiquitin-modification signals (a mass shift of +114.0429 Da) with high scores (experiments were done by Applied Protein Technology Co.). These peptides contain Lys-268, Lys-293, and Lys-329 individually (Fig. 5C), which are also located in the 233–333aa region of PPARγ (Fig. 5D). Lys-268 and Lys-293 were reported as two evolutionally conserved residues in the hinge domain (6) (Fig. 5E). We then used point mutation analysis to confirm the residue(s) that is polyubiquitinated. As shown in the figure, mutation on a single site of these three residues did not affect the substrate ubiquitination level (Fig. 5F). However, the ubiquitination levels of K268A/K293A double mutant or K268A/K293A/K329A triple mutant were significantly decreased, suggesting that Lys-268 and Lys-293 were both targeted by CRL4BAhR (Fig. 5G). Consistently, in the presence of AhR, mutations on Lys-268 and Lys-293 could remarkably prolong PPARγ half-life in HEK293T cells (Fig. 5, H and I). SirT1 was previously reported to deacetylate PPARγ on its Lys-268 and Lys-293 (6). In the presence of NAM, an inhibitor of SirT1 (21), acetylation of PPARγ was increased, whereas ubiquitination of PPARγ was decreased (Fig. 5J), suggesting that acetylation on these two sites competes against ubiquitination. However, inhibiting ubiquitination by knockdown of AhR does not increase the acetylation level of PPARγ (Fig. S4).
Discussion
In adipocytes, PPARγ protein has a short half-life (22). Several E3s were found to target PPARγ for proteasomal degradation. MKRN1 and SCFFBXO9 promote PPARγ proteasomal degradation in the cytoplasm (8, 23). SIAH2 has been shown to be localized in the nucleus and to promote PPARγ ubiquitination dependent on thiazolidinedione (24). Some E3s were not associated with PPARγ degradation. TRIM23 regulates PPARγ ubiquitination and stabilizes it (9). NEDD4 has been reported to induce both Lys-48– and Lys-63–linked polyubiquitination of PPARγ (10, 25). Although WDTC1 binds PPARγ in adipocytes, CRL4BWDTC1 complex promotes histone H2A Lys-119 monoubiquitination and plays a role in transcriptional repression (26). However, our previous study demonstrated that knockdown of CUL4B and DDB1 resulted in a significant increase in the half-life of PPARγ, indicating that CRL4B complex might use a different DCAF rather than WDTC1 to promote PPARγ ubiquitination and degradation. AhR, a nuclear receptor, has been shown to recruit estrogen receptor-α for proteasomal degradation in a ligand-dependent manner (27). In this study, we have shown here that AhR functions as the substrate receptor and recruits PPARγ onto CRL4BAhR E3 complex. Up-regulation of AhR shortens the PPARγ protein half-life via forming the CRL4BAhR E3 complex to facilitate Lys-268– and Lys-293–linked polyubiquitination of PPARγ, thereby blocking adipocyte differentiation. Knockdown of AhR stabilizes PPARγ and promotes adipocyte differentiation in 3T3-L1 cells.
PPARγ plays a central role during adipocyte differentiation (28, 29). Increasing attention has been paid to PPARγ post-translational modifications (30–32). Despite the fact that PPARγ binds its E3 ligases through its N-terminal (8) or C-terminal region (10, 23), our data showed that PPARγ interacts with AhR mainly on its DNA-binding domain and part of the hinge region. The hinge region has been reported to regulate subcellular distribution (33) and to interact with many nuclear receptors, such as androgen receptor (34, 35), estrogen receptor (36, 37), glucocorticoid receptor (38), and PPARα (39).
PPARγ is a nuclear receptor with a nuclear localization signal region between amino acids 181 and 224 (40, 41). The nuclear localization signal in CUL4B is located in its N terminus between amino acids 37 and 40 (42). Upon binding with ligand and interaction with HSP90, AhR translocates to the nucleus (43). Other components in CRL4BAhR E3 complex were all reported to localize mainly in the nucleus (44–46). Although ubiquitin, E1, E2, E3s, and proteasomal subunits were found in the nucleus, some studies suggest that nuclear export is required for the degradation of nuclear substrates (43, 47–49). However, other studies indicated that a nuclear ubiquitin–proteasome system may also be responsible for the degradation of several transcription factors (50–53). In our case, PPARγ was ubiquitinated and targeted for proteasomal degradation in the nucleus.
Acetylation, which modifies the lysine residue of target proteins, including histone and nonhistone proteins, is now recognized as a critical step in transcriptional regulation (54). Interestingly, many of these identified acetylated substrates are involved in ubiquitin-dependent proteolysis. Several identified acetylated lysine residues are also potential ubiquitination sites in proteins (55, 56). For example, the same lysine residues at the C terminus of p53 can be modified by both acetylation and ubiquitination, implicating a role of acetylation in regulating p53 stability (57). Acetylation is also integrated with other post-translational modifications to regulation PPARγ activity (58, 59). PPARγ acetylation on Lys-268 and Lys-293 is a signal of lipid storage and cell proliferation. Conversely, deacetylation of PPARγ induces energy expenditure and promotes insulin sensitivity (6). We used MS analysis on trypsin-digested peptides to identify three ubiquitinated lysines at residues 268, 293, and 329. Among them, two evolutionally conserved residues, Lys-268 and Lys-293, were main ubiquitinated sites verified by the following ubiquitination and cycloheximide (CHX)-chase assays. In some cases, acetylation competes with ubiquitination for the same lysine sites to prevent protein degradation (60). Deacetylation of PPARγ makes Lys-268 and Lys-293 available for ubiquitination by CRL4BAhR and subsequent proteasomal degradation. Therefore, it is important to explore the mechanism of post-translational modifications on PPARγ.
Previously, we reported that scaffold protein CUL4B of CRL4B E3 ligase complex functions as a negative regulator of adipogenesis. We observed that depletion of CUL4B improves adipose function and protects against glucose intolerance and insulin resistance (16). A similar phenotype was observed in AhR-knockout mice. Exposing AhR−/− mice to a high-fat diet showed lower fasting glucose levels and improved glucose tolerance (61). In addition to AhR−/− mice, AhR+/− mice, which express about 30% of the AhR levels, also exhibited significantly improved glucose tolerance and insulin sensitivity (62).
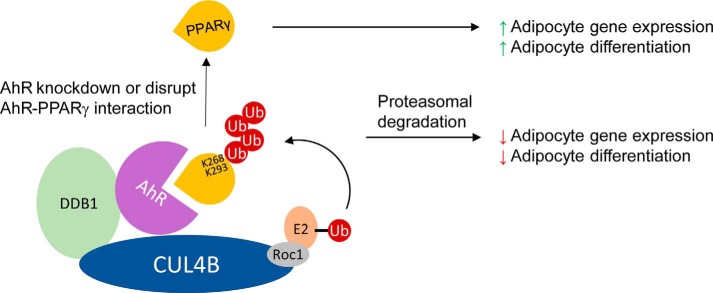
Recent studies revealed that AhR plays critical roles in energy metabolism (17, 18). Suppression of AhR activity can improve metabolic function to avoid obesity (62). Unlike the low-affinity direct or indirect interaction between SIAH2 and PPARγ (24), AhR functions as a substrate receptor, directly recruits substrate PPARγ, facilitates its ubiquitination, and promotes subsequent proteasomal degradation (Fig. 6). Further studies are certainly required to reveal the precise mechanism of substrate selection, cross-talk between acetylation and ubiquitination, and detailed AhR–PPARγ interactions for the treatment of obesity and metabolic diseases.
Figure 6.
CRL4BAhR-mediated PPARγ ubiquitination regulates adipocyte differentiation.
Experimental procedures
Plasmids
Full-length cDNAs of AhR and PPARγ were inserted into either pcDNA3.1 (Invitrogen), pGEX4T1 (GE Healthcare), or pRsfduet-1 (Novagen) vectors as indicated. Truncated mutants of AhR (1–187aa, 188–403aa, and 403–848aa) and truncated mutants of PPARγ (1–135aa, 136–232aa, 1–232aa, 136–505aa, 233–505aa, and 136–333aa) were amplified by PCR and cloned into vectors as indicated. Different point mutations in PPARγ or AhR were generated by site-directed mutagenesis. Other plasmids were generated and described previously (16, 63).
Antibodies and chemicals
For Western blotting, the following antibodies were used: PPARγ (C26H12, Cell Signaling Technology), AhR (ab2769, Abcam), CUL4B (HPA011880, Sigma), HA (rabbit D110004 and mouse D199961, BBI Life Sciences Corp. (BBI), and 600401384, Rockland), Myc (rabbit D110006 and mouse D153566, BBI), β-actin (sc-69879, Santa Cruz Biotechnology), and acetylated lysine (9441, Cell Signaling Technology). The following reagents were used to culture cells or perform experiments: X-tremeGene HP DNA transfection reagent (Roche Applied Science), X-tremeGene siRNA transfection reagent (Roche Applied Science), polyethylenimine (23966, Polysciences Inc.), CHX (HY-12320, MedChemExpress (MCE)), MG132 (HY-13259, MCE), LMB (S1726, Beyotime), N-ethylmaleimide (NEM; EB0450, BBI), DMSO (A503039, Sangon Biotech), 4′,6-diamidino-2-phenylindole (ab104139, Abcam), GSH reduced (A600229, BBI), and phenylmethylsulfonyl fluoride (PB0425, BBI).
Cell culture and adipocyte differentiation
HEK293T and 3T3-L1 cells were grown in DMEM (Gibco) with 10% bovine serum (Sigma-Aldrich). Adipocytes differentiated from 3T3-L1 cells were first maintained in DMEM with 10% bovine serum (Gibco) for 2–3 days. Cells were then differentiated in DMEM containing 10% fetal bovine serum, 1 μm dexamethasone, 520 μm isobutylmethylxanthine, and 1 μg/ml insulin for 7 days. Differentiated cells were stained using Oil Red O (Sigma-Aldrich). The stained cells were photographed using a camera-connected microscope (Olympus).
Generation of AhR knockdown and overexpression stable cell lines
AhR shRNA construct (LV3(H1/GFP&Puro)) and AhR overexpression plasmid (LV5(EF-1aF/GFP&Puro)) were purchased from GenePharma. Lentivirus packaging was done at GenePharma. Lentiviral supernatants of shRNA and AhR overexpression plasmids were used to infect 3T3-L1 cells following the manufacturer's instructions. Infected cells were selected by puromycin (1.5 μg/ml) treatment for 6 days.
Protein stability assay and analysis
HEK293T cells were cotransfected with Myc-PPARγ (2 μg) and AhR-Flag-His (4 μg) or the empty vector (4 μg). After 20 h, cells were treated with 60 μg/ml CHX to inhibit protein synthesis. CHX-treated cells were harvested at different time points (0, 4, and 8 h) and processed for immunoblotting with anti-PPARγ antibody. Anti-actin antibody was used as an internal control. Signals from the Western blots were analyzed by Volume Analysis of Quantity One software with volume background subtraction (Bio-Rad).
Protein purification and binding assays
pGEX4T1-AhR and pRsfduet-PPARγ (isoform 2) were expressed in E. coli and purified using GSH-Sepharose 4B (GE Healthcare) or Ni2+-NTA (GE Healthcare) separately. The purified GST-AhR and His-PPARγ were further purified by size-exclusion chromatography (Superdex 200 or Superdex 75, GE Healthcare) and stored in protein buffer containing 25 mm Tris, pH 7.6, 150 mm NaCl, and 1 mm DTT. The binding assays were performed by mixing His-PPARγ and GST-AhR. The mixture was then incubated with AhR antibody and protein A/G-Sepharose (Santa Cruz Biotechnology) for 2 h at 4 °C. Immunoprecipitates were boiled in sample loading buffer for 5 min.
Immunoprecipitation
HEK293T cells were lysed using lysis buffer (50 mm Tris-HCl, pH 7.6, 150 mm NaCl, 0.5% sodium desoxycholate, and 0.5% Nonidet-P40) with protease inhibitor mixture (Roche Applied Science) followed by sonication (8 s, 12 cycles). Cell lysate supernatants were incubated with the indicated antibodies and protein A/G-Sepharose (Santa Cruz Biotechnology) for 2 h at 4 °C. Immunoprecipitates were boiled in sample loading buffer for 5 min.
PLA
Adult male mice (WT and adipocyte-specific Cul4b knockout) were housed at the animal unit at the Department of Genetics, Shandong University. Mouse adipose tissues were extracted and fixed for embedding in paraffin. 4-μm samples of the paraffin tissue array were stained against the indicated antibodies according to the Duolink® in situ PLA (Sigma) protocol. Antibodies used in this experiment include PPARγ (mouse 95128S, Cell Signaling Technology; 1:400), AhR (rabbit 83200S, Cell Signaling Technology; 1:50), and CUL4B (HPA011880, Sigma; 1:600). Images were collected using a fluorescence microscope (Olympus BX51).
Ubiquitination assay
HEK293T or 3T3-L1 cells were first transfected with the indicated plasmids or siRNAs and treated with the indicated chemicals, including MG132 (20 μm), NAM (100 μm), and LMB (10 ng/ml). Cells were then harvested with PBS containing 10 μm NEM to prevent deubiquitination. Cells were lysed in 2% SDS by boiling the samples for 10 min followed by sonication (8 s, one cycle). Lysed-sample supernatants were incubated with Myc antibody (D153566, BBI) and mixed with protein A/G-Sepharose (Santa Cruz Biotechnology) for 2 h at 4 °C followed by Western blotting or Coomassie Blue staining for MS.
In vitro ubiquitination assay
His-PPARγ was expressed in E. coli and purified with Ni2+-NTA (GE Healthcare) followed by size-exclusion chromatography (Superdex 75, GE Healthcare). His-PPARγ was incubated with 200 ng of E1 (UBE1), 500 ng of E2 (UbcH5c), 10 μg of His-Ub, and 2 mm ATP (Enzo Life Sciences) as described previously (8). The reaction was performed in the absence and presence of AhR-Flag-His coimmunoprecipitation products. After 1-h incubation at 37 °C, samples were quenched in 6 m guanidinium HCl, pH 8, containing 5 mm NEM. His-ubiquitinated proteins were pulled down with Ni2+-NTA (GE Healthcare) followed by washing and elution in sample buffer (64). The mixture was then boiled in loading dye at 95 °C for 10 min to disrupt protein–protein interactions.
Mass spectrometry
Mass spectrometry experiments were done by Applied Protein Technology Co. LC-MS/MS analysis was performed on a Q Exactive mass spectrometer (Thermo Fisher Scientific) for 60 min. MS data were acquired using a data-dependent top 10 method dynamically choosing the most abundant precursor ions from the survey scan (300–1800 m/z) for higher-energy collisional-dissociation fragmentation. Survey scans were acquired at a resolution of 70,000 at m/z 200, the resolution for higher-energy collisional-dissociation spectra was set to 17,500 at m/z 200, and the isolation width was 2 m/z. The instrument was run with peptide recognition mode enabled. The raw data are available in the supporting information.
The MS search parameters used were as follows. The peak list–generating software and search engine used was Mascot2.2. The UniProt database was used for sequence searching. We actually searched 161,584 entries in the database. Trypsin was used in our experiments with specificity sites Lys/Arg. Two missed and/or nonspecific cleavages were permitted. The fixed and variable modifications (including residue specificity) used include carbamidomethyl (Cys) and oxidation (Met) and Gly-Gly (Lys), respectively. Mass tolerance for precursor ions was 20 ppm, and that for fragment ions was 0.1 Da. The threshold score/expectation value for accepting individual spectra was no less than 20.
Immunofluorescence analysis
HEK293T cells were cotransfected with Myc-PPARγ (2 μg) and AhR-Flag-His (4 μg) and treated with DMSO or MG132 for 6 h. Cells were fixed (P0098, Beyotime) for 30 min at room temperature and washed with PBS buffer three times. Cells were stained with anti-PPARγ antibody (1:100). Images were collected using a fluorescence microscope (Olympus BX51).
RT-PCR analysis
RNAs were extracted using TRIzol (Invitrogen) according to the manufacturer's directions. cDNAs were synthesized from total RNA using reverse transcriptase (EP0441, Thermo), amplified, and analyzed using a SYBR Green PCR kit and real-time PCR. The primers used in this study are described in the supporting information. Each gene expression level was normalized by the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene.
Statistical analysis
Statistical analyses on RT-PCR and PLA were performed using Prism software. Measurement data were expressed as mean ± S.D. A paired, two-tailed Student's t test was used to determine the significance between two groups. p < 0.05 was regarded as the threshold value for statistical significance. Statistical analyses on protein level and stability were performed using Quantity One software.
Author contributions
H. D. formal analysis; H. D. and Y. G. supervision; H. D. and Y. G. funding acquisition; H. D., Y. D., X. Z., Q. Y., and Q. D. investigation; H. D. writing-original draft; H. D. and Y. G. project administration; H. D. and Y. G. writing-review and editing; Y. S. and P. L. resources; Y. S. and P. L. methodology; Y. G. conceptualization.
Supplementary Material
Acknowledgments
We thank Associate Prof. Jiangxia Li and Liping Qin for technical help. We also acknowledge Prof. Chengjiang Gao for advice.
This work was supported by the National Natural Science Foundation of China (Grants 31872810 and 81401634), Shandong Province Science and Technology Development Project (Grant 2017GSF221013), and Young Scholars Program of Shandong University (Grant 21300075614018). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4, Table S1, and supporting information.
The mass spectrometric raw data and spectral libraries associated with this manuscript are available from ProteomeXchange with the accession number PXD015830.
- PPAR
- peroxisome proliferator-activated receptor
- AhR
- aryl hydrocarbon receptor
- CUL4B
- Cullin 4B
- SIAH2
- seven in absentia homolog 2
- MKRN1
- makorin ring finger protein 1
- TRIM23
- tripartite motif protein 23
- NEDD4
- neural precursor cell–expressed developmentally down-regulated protein 4
- DCAF
- DDB1–CUL4 association factor
- CRL4B
- CUL4B-RING E3 ubiquitin ligase
- Ub
- ubiquitin
- LMB
- leptomycin B
- PLA
- proximity ligation assay
- GST
- GSH S-transferase
- aa
- amino acids
- NAM
- nicotinamide
- CHX
- cycloheximide
- HA
- hemagglutinin
- NEM
- N-ethylmaleimide
- DMEM
- Dulbecco's modified Eagle's medium
- NTA
- nitrilotriacetic acid.
References
- 1. Rocha V. Z., and Libby P. (2009) Obesity, inflammation, and atherosclerosis. Nat. Rev. Cardiol. 6, 399–409 10.1038/nrcardio.2009.55 [DOI] [PubMed] [Google Scholar]
- 2. Jung U. J., and Choi M. S. (2014) Obesity and its metabolic complications: the role of adipokines and the relationship between obesity, inflammation, insulin resistance, dyslipidemia and nonalcoholic fatty liver disease. Int. J. Mol. Sci. 15, 6184–6223 10.3390/ijms15046184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cristancho A. G., and Lazar M. A. (2011) Forming functional fat: a growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol. 12, 722–734 10.1038/nrm3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Semple R. K., Chatterjee V. K., and O'Rahilly S. (2006) PPARγ and human metabolic disease. J. Clin. Investig. 116, 581–589 10.1172/JCI28003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choi J. H., Banks A. S., Estall J. L., Kajimura S., Boström P., Laznik D., Ruas J. L., Chalmers M. J., Kamenecka T. M., Blüher M., Griffin P. R., and Spiegelman B. M. (2010) Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARγ by Cdk5. Nature 466, 451–456 10.1038/nature09291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qiang L., Wang L., Kon N., Zhao W., Lee S., Zhang Y., Rosenbaum M., Zhao Y., Gu W., Farmer S. R., and Accili D. (2012) Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell 150, 620–632 10.1016/j.cell.2012.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kilroy G., Carter L. E., Newman S., Burk D. H., Manuel J., Möller A., Bowtell D. D., Mynatt R. L., Ghosh S., and Floyd Z. E. (2015) The ubiquitin ligase Siah2 regulates obesity-induced adipose tissue inflammation. Obesity 23, 2223–2232 10.1002/oby.21220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim J. H., Park K. W., Lee E. W., Jang W. S., Seo J., Shin S., Hwang K. A., and Song J. (2014) Suppression of PPARγ through MKRN1-mediated ubiquitination and degradation prevents adipocyte differentiation. Cell Death Differ. 21, 594–603 10.1038/cdd.2013.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Watanabe M., Takahashi H., Saeki Y., Ozaki T., Itoh S., Suzuki M., Mizushima W., Tanaka K., and Hatakeyama S. (2015) The E3 ubiquitin ligase TRIM23 regulates adipocyte differentiation via stabilization of the adipogenic activator PPARγ. eLife 4, e05615 10.7554/eLife.05615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li J. J., Wang R., Lama R., Wang X., Floyd Z. E., Park E. A., and Liao F. F. (2016) Ubiquitin ligase NEDD4 regulates PPARγ stability and adipocyte differentiation in 3T3-L1 cells. Sci. Rep. 6, 38550 10.1038/srep38550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jackson S., and Xiong Y. (2009) CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 34, 562–570 10.1016/j.tibs.2009.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zou Y., Liu Q., Chen B., Zhang X., Guo C., Zhou H., Li J., Gao G., Guo Y., Yan C., Wei J., Shao C., and Gong Y. (2007) Mutation in CUL4B, which encodes a member of cullin-RING ubiquitin ligase complex, causes X-linked mental retardation. Am. J. Hum. Genet. 80, 561–566 10.1086/512489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang B., Zhao W., Yuan J., Qian Y., Sun W., Zou Y., Guo C., Chen B., Shao C., and Gong Y. (2012) Lack of Cul4b, an E3 ubiquitin ligase component, leads to embryonic lethality and abnormal placental development. PLoS One 7, e37070 10.1371/journal.pone.0037070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tarpey P. S., Raymond F. L., O'Meara S., Edkins S., Teague J., Butler A., Dicks E., Stevens C., Tofts C., Avis T., Barthorpe S., Buck G., Cole J., Gray K., Halliday K., et al. (2007) Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am. J. Hum. Genet. 80, 345–352 10.1086/511134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hannah J., and Zhou P. (2015) Distinct and overlapping functions of the cullin E3 ligase scaffolding proteins CUL4A and CUL4B. Gene 573, 33–45 10.1016/j.gene.2015.08.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li P., Song Y., Zan W., Qin L., Han S., Jiang B., Dou H., Shao C., and Gong Y. (2017) Lack of CUL4B in adipocytes promotes PPARγ-mediated adipose tissue expansion and insulin sensitivity. Diabetes 66, 300–313 10.2337/db16-0743 [DOI] [PubMed] [Google Scholar]
- 17. Wada T., Sunaga H., Miyata K., Shirasaki H., Uchiyama Y., and Shimba S. (2016) Aryl hydrocarbon receptor plays protective roles against high fat diet (HFD)-induced hepatic steatosis and the subsequent lipotoxicity via direct transcriptional regulation of Socs3 gene expression. J. Biol. Chem. 291, 7004–7016 10.1074/jbc.M115.693655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu P., Yan J., Liu K., Garbacz W. G., Wang P., Xu M., Ma X., and Xie W. (2015) Activation of aryl hydrocarbon receptor dissociates fatty liver from insulin resistance by inducing fibroblast growth factor 21. Hepatology 61, 1908–1919 10.1002/hep.27719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Taylor K. W., Novak R. F., Anderson H. A., Birnbaum L. S., Blystone C., Devito M., Jacobs D., Köhrle J., Lee D. H., Rylander L., Rignell-Hydbom A., Tornero-Velez R., Turyk M. E., Boyles A. L., Thayer K. A., et al. (2013) Evaluation of the association between persistent organic pollutants (POPs) and diabetes in epidemiological studies: a national toxicology program workshop review. Environ. Health Perspect. 121, 774–783 10.1289/ehp.1205502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Abbott B. D., Schmid J. E., Pitt J. A., Buckalew A. R., Wood C. R., Held G. A., and Diliberto J. J. (1999) Adverse reproductive outcomes in the transgenic Ah receptor-deficient mouse. Toxicol. Appl. Pharmacol. 155, 62–70 10.1006/taap.1998.8601 [DOI] [PubMed] [Google Scholar]
- 21. Bitterman K. J., Anderson R. M., Cohen H. Y., Latorre-Esteves M., and Sinclair D. A. (2002) Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277, 45099–45107 10.1074/jbc.M205670200 [DOI] [PubMed] [Google Scholar]
- 22. Hauser S., Adelmant G., Sarraf P., Wright H. M., Mueller E., and Spiegelman B. M. (2000) Degradation of the peroxisome proliferator-activated receptor γ is linked to ligand-dependent activation. J. Biol. Chem. 275, 18527–18533 10.1074/jbc.M001297200 [DOI] [PubMed] [Google Scholar]
- 23. Lee K. W., Kwak S. H., Koo Y. D., Cho Y. K., Lee H. M., Jung H. S., Cho Y. M., Park Y. J., Chung S. S., and Park K. S. (2016) F-box only protein 9 is an E3 ubiquitin ligase of PPARγ. Exp. Mol. Med. 48, e234 10.1038/emm.2016.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kilroy G., Kirk-Ballard H., Carter L. E., and Floyd Z. E. (2012) The ubiquitin ligase Siah2 regulates PPARγ activity in adipocytes. Endocrinology 153, 1206–1218 10.1210/en.2011-1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Han L., Wang P., Zhao G., Wang H., Wang M., Chen J., and Tong T. (2013) Upregulation of SIRT1 by 17β-estradiol depends on ubiquitin-proteasome degradation of PPAR-γ mediated by NEDD4-1. Protein Cell 4, 310–321 10.1007/s13238-013-2124-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Groh B. S., Yan F., Smith M. D., Yu Y., Chen X., and Xiong Y. (2016) The antiobesity factor WDTC1 suppresses adipogenesis via the CRL4WDTC1 E3 ligase. EMBO Rep. 17, 638–647 10.15252/embr.201540500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ohtake F., Baba A., Takada I., Okada M., Iwasaki K., Miki H., Takahashi S., Kouzmenko A., Nohara K., Chiba T., Fujii-Kuriyama Y., and Kato S. (2007) Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature 446, 562–566 10.1038/nature05683 [DOI] [PubMed] [Google Scholar]
- 28. Chawla A., Schwarz E. J., Dimaculangan D. D., and Lazar M. A. (1994) Peroxisome proliferator-activated receptor (PPAR) γ: adipose-predominant expression and induction early in adipocyte differentiation. Endocrinology 135, 798–800 10.1210/endo.135.2.8033830 [DOI] [PubMed] [Google Scholar]
- 29. Hamm J. K., el Jack A. K., Pilch P. F., and Farmer S. R. (1999) Role of PPARγ in regulating adipocyte differentiation and insulin-responsive glucose uptake. Ann. N.Y. Acad. Sci. 892, 134–145 10.1111/j.1749-6632.1999.tb07792.x [DOI] [PubMed] [Google Scholar]
- 30. van Beekum O., Fleskens V., and Kalkhoven E. (2009) Posttranslational modifications of PPAR-γ: fine-tuning the metabolic master regulator. Obesity 17, 213–219 10.1038/oby.2008.473 [DOI] [PubMed] [Google Scholar]
- 31. Park H. S., Ju U. I., Park J. W., Song J. Y., Shin D. H., Lee K. H., Jeong L. S., Yu J., Lee H. W., Cho J. Y., Kim S. Y., Kim S. W., Kim J. B., Park K. S., and Chun Y. S. (2016) PPARγ neddylation essential for adipogenesis is a potential target for treating obesity. Cell Death Differ. 23, 1296–1311 10.1038/cdd.2016.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jennewein C., Kuhn A. M., Schmidt M. V., Meilladec-Jullig V., von Knethen A., Gonzalez F. J., and Brüne B. (2008) Sumoylation of peroxisome proliferator-activated receptor γ by apoptotic cells prevents lipopolysaccharide-induced NCoR removal from κB binding sites mediating transrepression of proinflammatory cytokines. J. Immunol. 181, 5646–5652 10.4049/jimmunol.181.8.5646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pawlak M., Lefebvre P., and Staels B. (2012) General molecular biology and architecture of nuclear receptors. Curr. Top. Med. Chem. 12, 486–504 10.2174/156802612799436641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Buchanan G., Ricciardelli C., Harris J. M., Prescott J., Yu Z. C., Jia L., Butler L. M., Marshall V. R., Scher H. I., Gerald W. L., Coetzee G. A., and Tilley W. D. (2007) Control of androgen receptor signaling in prostate cancer by the cochaperone small glutamine rich tetratricopeptide repeat containing protein α. Cancer Res. 67, 10087–10096 10.1158/0008-5472.CAN-07-1646 [DOI] [PubMed] [Google Scholar]
- 35. Jin F., Claessens F., and Fondell J. D. (2012) Regulation of androgen receptor-dependent transcription by coactivator MED1 is mediated through a newly discovered noncanonical binding motif. J. Biol. Chem. 287, 858–870 10.1074/jbc.M111.304519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yan J., Kim Y. S., Yang X. P., Albers M., Koegl M., and Jetten A. M. (2007) Ubiquitin-interaction motifs of RAP80 are critical in its regulation of estrogen receptor α. Nucleic Acids Res. 35, 1673–1686 10.1093/nar/gkl1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burns K. A., Li Y., Liu L., and Korach K. S. (2014) Research resource: comparison of gene profiles from wild-type ERα and ERα hinge region mutants. Mol. Endocrinol. 28, 1352–1361 10.1210/me.2014-1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chodankar R., Wu D. Y., Schiller B. J., Yamamoto K. R., and Stallcup M. R. (2014) Hic-5 is a transcription coregulator that acts before and/or after glucocorticoid receptor genome occupancy in a gene-selective manner. Proc. Natl. Acad. Sci. U.S.A. 111, 4007–4012 10.1073/pnas.1400522111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Georgiakaki M., Chabbert-Buffet N., Dasen B., Meduri G., Wenk S., Rajhi L., Amazit L., Chauchereau A., Burger C. W., Blok L. J., Milgrom E., Lombès M., Guiochon-Mantel A., and Loosfelt H. (2006) Ligand-controlled interaction of histone acetyltransferase binding to ORC-1 (HBO1) with the N-terminal transactivating domain of progesterone receptor induces steroid receptor coactivator 1-dependent coactivation of transcription. Mol. Endocrinol. 20, 2122–2140 10.1210/me.2005-0149 [DOI] [PubMed] [Google Scholar]
- 40. Agostini M., Schoenmakers E., Mitchell C., Szatmari I., Savage D., Smith A., Rajanayagam O., Semple R., Luan J., Bath L., Zalin A., Labib M., Kumar S., Simpson H., Blom D., et al. (2006) Non-DNA binding, dominant-negative, human PPARγ mutations cause lipodystrophic insulin resistance. Cell Metab. 4, 303–311 10.1016/j.cmet.2006.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Iwamoto F., Umemoto T., Motojima K., and Fujiki Y. (2011) Nuclear transport of peroxisome-proliferator activated receptor α. J. Biochem. 149, 311–319 10.1093/jb/mvq144 [DOI] [PubMed] [Google Scholar]
- 42. Zou Y., Mi J., Cui J., Lu D., Zhang X., Guo C., Gao G., Liu Q., Chen B., Shao C., and Gong Y. (2009) Characterization of nuclear localization signal in the N terminus of CUL4B and its essential role in cyclin E degradation and cell cycle progression. J. Biol. Chem. 284, 33320–33332 10.1074/jbc.M109.050427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Davarinos N. A., and Pollenz R. S. (1999) Aryl hydrocarbon receptor imported into the nucleus following ligand binding is rapidly degraded via the cytoplasmic proteasome following nuclear export. J. Biol. Chem. 274, 28708–28715 10.1074/jbc.274.40.28708 [DOI] [PubMed] [Google Scholar]
- 44. Wittschieben B. Ø., Iwai S., and Wood R. D. (2005) DDB1-DDB2 (xeroderma pigmentosum group E) protein complex recognizes a cyclobutane pyrimidine dimer, mismatches, apurinic/apyrimidinic sites, and compound lesions in DNA. J. Biol. Chem. 280, 39982–39989 10.1074/jbc.M507854200 [DOI] [PubMed] [Google Scholar]
- 45. Scrima A., Konícková R., Czyzewski B. K., Kawasaki Y., Jeffrey P. D., Groisman R., Nakatani Y., Iwai S., Pavletich N. P., and Thomä N. H. (2008) Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell 135, 1213–1223 10.1016/j.cell.2008.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Furukawa M., Zhang Y., McCarville J., Ohta T., and Xiong Y. (2000) The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell. Biol. 20, 8185–8197 10.1128/MCB.20.21.8185-8197.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tomoda K., Kubota Y., and Kato J. (1999) Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature 398, 160–165 10.1038/18230 [DOI] [PubMed] [Google Scholar]
- 48. Diehl J. A., Cheng M., Roussel M. F., and Sherr C. J. (1998) Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12, 3499–3511 10.1101/gad.12.22.3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rodriguez M. S., Thompson J., Hay R. T., and Dargemont C. (1999) Nuclear retention of IκBα protects it from signal-induced degradation and inhibits nuclear factor κB transcriptional activation. J. Biol. Chem. 274, 9108–9115 10.1074/jbc.274.13.9108 [DOI] [PubMed] [Google Scholar]
- 50. Lo R. S., and Massagué J. (1999) Ubiquitin-dependent degradation of TGF-β-activated smad2. Nat. Cell Biol. 1, 472–478 10.1038/70258 [DOI] [PubMed] [Google Scholar]
- 51. Floyd Z. E., Trausch-Azar J. S., Reinstein E., Ciechanover A., and Schwartz A. L. (2001) The nuclear ubiquitin-proteasome system degrades MyoD. J. Biol. Chem. 276, 22468–22475 10.1074/jbc.M009388200 [DOI] [PubMed] [Google Scholar]
- 52. Blondel M., Galan J. M., Chi Y., Lafourcade C., Longaretti C., Deshaies R. J., and Peter M. (2000) Nuclear-specific degradation of Far1 is controlled by the localization of the F-box protein Cdc4. EMBO J. 19, 6085–6097 10.1093/emboj/19.22.6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lenk U., and Sommer T. (2000) Ubiquitin-mediated proteolysis of a short-lived regulatory protein depends on its cellular localization. J. Biol. Chem. 275, 39403–39410 10.1074/jbc.M006949200 [DOI] [PubMed] [Google Scholar]
- 54. Drazic A., Myklebust L. M., Ree R., and Arnesen T. (2016) The world of protein acetylation. Biochim. Biophys. Acta 1864, 1372–1401 10.1016/j.bbapap.2016.06.007 [DOI] [PubMed] [Google Scholar]
- 55. Martínez-Balbás M. A., Bauer U. M., Nielsen S. J., Brehm A., and Kouzarides T. (2000) Regulation of E2F1 activity by acetylation. EMBO J. 19, 662–671 10.1093/emboj/19.4.662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Grönroos E., Hellman U., Heldin C. H., and Ericsson J. (2002) Control of Smad7 stability by competition between acetylation and ubiquitination. Mol. Cell 10, 483–493 10.1016/S1097-2765(02)00639-1 [DOI] [PubMed] [Google Scholar]
- 57. Li M., Luo J., Brooks C. L., and Gu W. (2002) Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 277, 50607–50611 10.1074/jbc.C200578200 [DOI] [PubMed] [Google Scholar]
- 58. Tian L., Wang C., Hagen F. K., Gormley M., Addya S., Soccio R., Casimiro M. C., Zhou J., Powell M. J., Xu P., Deng H., Sauve A. A., and Pestell R. G. (2014) Acetylation-defective mutant of Pparγ is associated with decreased lipid synthesis in breast cancer cells. Oncotarget 5, 7303–7315 10.18632/oncotarget.2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mayoral R., Osborn O., McNelis J., Johnson A. M., Oh D. Y., Izquierdo C. L., Chung H., Li P., Traves P. G., Bandyopadhyay G., Pessentheiner A. R., Ofrecio J. M., Cook J. R., Qiang L., Accili D., et al. (2015) Adipocyte SIRT1 knockout promotes PPARγ activity, adipogenesis and insulin sensitivity in chronic-HFD and obesity. Mol. Metab. 4, 378–391 10.1016/j.molmet.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu Y., Dentin R., Chen D., Hedrick S., Ravnskjaer K., Schenk S., Milne J., Meyers D. J., Cole P., Yates J. 3rd, Olefsky J., Guarente L., and Montminy M. (2008) A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature 456, 269–273 10.1038/nature07349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Korecka A., Dona A., Lahiri S., Tett A. J., Al-Asmakh M., Braniste V., D'Arienzo R., Abbaspour A., Reichardt N., Fujii-Kuriyama Y., Rafter J., Narbad A., Holmes E., Nicholson J., Arulampalam V., et al. (2016) Bidirectional communication between the Aryl hydrocarbon Receptor (AhR) and the microbiome tunes host metabolism. NPJ Biofilms Microbiomes 2, 16014 10.1038/npjbiofilms.2016.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xu C. X., Wang C., Zhang Z. M., Jaeger C. D., Krager S. L., Bottum K. M., Liu J., Liao D. F., and Tischkau S. A. (2015) Aryl hydrocarbon receptor deficiency protects mice from diet-induced adiposity and metabolic disorders through increased energy expenditure. Int. J. Obes. 39, 1300–1309 10.1038/ijo.2015.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hu H., Yang Y., Ji Q., Zhao W., Jiang B., Liu R., Yuan J., Liu Q., Li X., Zou Y., Shao C., Shang Y., Wang Y., and Gong Y. (2012) CRL4B catalyzes H2AK119 monoubiquitination and coordinates with PRC2 to promote tumorigenesis. Cancer Cell 22, 781–795 10.1016/j.ccr.2012.10.024 [DOI] [PubMed] [Google Scholar]
- 64. Dou H., Buetow L., Hock A., Sibbet G. J., Vousden K. H., and Huang D. T. (2012) Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat. Struct. Mol. Biol. 19, 184–192 10.1038/nsmb.2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nolte R. T., Wisely G. B., Westin S., Cobb J. E., Lambert M. H., Kurokawa R., Rosenfeld M. G., Willson T. M., Glass C. K., and Milburn M. V. (1998) Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 395, 137–143 10.1038/25931 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.