Abstract
Several antidepressant drugs activate tropomyosin-related kinase B (TRKB) receptor, but it remains unclear whether these compounds employ a common mechanism for TRKB activation. Here, using MS, we found that a single intraperitoneal injection of fluoxetine disrupts the interaction of several proteins with TRKB in the hippocampus of mice. These proteins included members of adaptor protein complex-2 (AP-2) involved in vesicular endocytosis. The interaction of TRKB with the cargo-docking μ subunit of the AP-2 complex (AP2M) was confirmed to be disrupted by both acute and repeated fluoxetine treatments. Of note, fluoxetine disrupted the coupling between full-length TRKB and AP2M, but not the interaction between AP2M and the TRKB C-terminal region, indicating that the fluoxetine-binding site in TRKB lies outside the TRKB:AP2M interface. ELISA experiments revealed that in addition to fluoxetine, other chemically diverse antidepressants, such as imipramine, rolipram, phenelzine, ketamine, and its metabolite 2R,6R-hydroxynorketamine, also decreased the interaction between TRKB and AP2M in vitro. Silencing the expression of AP2M in a TRKB-expressing mouse fibroblast cell line (MG87.TRKB) increased cell-surface expression of TRKB and facilitated its activation by brain-derived neurotrophic factor (BDNF), observed as levels of phosphorylated TRKB. Moreover, animals haploinsufficient for the Ap2m1 gene displayed increased levels of active TRKB, along with enhanced cell-surface expression of the receptor in cultured hippocampal neurons. Taken together, our results suggest that disruption of the TRKB:AP2M interaction is a common mechanism underlying TRKB activation by several chemically diverse antidepressants.
Keywords: brain-derived neurotrophic factor (BDNF), clathrin, drug action, receptor tyrosine kinase, molecular pharmacology, neurotrophic receptor tyrosine kinase 2 (NTRK2), adaptor protein complex-2 (AP-2), neuroplasticity
Introduction
Compromised plasticity has been suggested as one of the major causes of depression (1). Supporting this idea, morphological and functional deficits have been observed in the brains of patients with mood disorders (2–4). Reduced neurotrophic support, such as by brain-derived neurotrophic factor (BDNF)2 signaling via TRKB, has been linked to the atrophy observed in these patients in the neuronal networks regulating mood and cognition (5, 6).
Antidepressant drugs (AD) have been suggested to act by improving neuronal connectivity, plasticity, and information processing mainly by targeting monoamine neurotransmission, such as serotonin (5-HT) and noradrenaline (4). In line with this, an intact BDNF-TRKB signaling system is crucial for the efficacy of antidepressant compounds, as mice overexpressing a dominant negative TRKB isoform (TRKB.T1) or haploinsufficient for BDNF (Bdnf+/−) are not responsive to AD (7). Furthermore, the activation of TRKB receptor signaling (and putatively the reinstatement of plasticity) is associated with the response to AD (7–9).
The binding of BDNF to TRKB initiates dimerization and phosphorylation of tyrosine residues in the intracellular portion of the receptor (10), which allows docking of adaptor proteins to these sites. Phosphorylated tyrosine 515, for example, serves as a docking site for SHC adaptor protein, whereas phospholipase Cγ (PLC-γ1) binds to the phosphorylated Tyr-816 residue (11). Interestingly, the phosphorylation of TRKB receptors at Tyr-706/707 and Tyr-816 residues are enhanced by several antidepressants in adult mouse hippocampus, whereas no change was detected at the Tyr-515 residue (7, 9). Despite the evidence on the necessity of the BDNF/TRKB system for antidepressant responses, the mechanism by which these drugs might trigger TRKB activation remains unclear.
TRKB is mainly localized intracellularly in vesicles that are translocated to the neuronal surface through neuronal activity (12–14). In this scenario, proteins able to modulate TRKB transit to and from the cell surface, therefore modulating its exposure to BDNF, provide an interesting opportunity to understand the antidepressant-induced effects. In line with this idea, our MS-based screen for TRKB interactors that change upon fluoxetine treatment identified the AP-2 complex as a TRKB interactor. AP-2 is crucial for clathrin-dependent endocytosis and participates in the removal of several transmembrane receptors and their ligands from the cell surface (15). AP-2 is a heterotetrameric complex that consists of two large chains (α- and β-adaptin), one medium chain (μ, AP2M), and one small chain (σ) (16). TRK receptors have been identified among the targets for rapid clathrin-mediated endocytosis upon binding to their ligands (17). Earlier studies demonstrated that BDNF-induced phosphorylation of TRKB receptors recruits AP-2 complex and clathrin to the plasma membrane in cultured hippocampal neurons (18). More recently, a novel relationship has been established between TRKB receptors and AP-2, suggesting that autophagosomes containing activated TRKB are transported to the cell soma in an AP-2–dependent manner (19).
In the present study, we identified a putative binding site on the TRKB receptor for AP2M, which provides a known interface for interaction between these proteins. Thus, we aimed at investigating whether the interface between TRKB and AP2M is a site of action for AD. Specifically, we tested the effects of several AD, such as fluoxetine, imipramine, rolipram, phenelzine, ketamine, and RR-HNK, as well as nonantidepressants with psychoactive effects (diazepam and chlorpromazine) on the interaction of TRKB receptors with AP2M.
Results
AP2M is an interaction partner of TRKB receptors
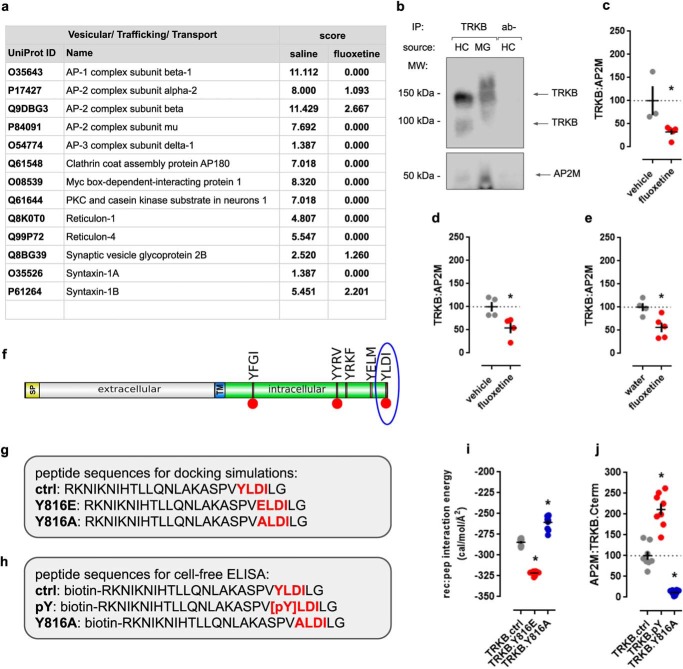
Samples from mouse hippocampus (vehicle (n = 3) and fluoxetine-treated (30 mg/kg, single i.p. injection, n = 3)) were processed for co-IP with TRKB-specific antibody and analyzed by MS to investigate the TRKB interactome and the changes in the interactome induced by acute fluoxetine administration. The complete list of interactors can be found in FigShare (Table S1). The identified interactors that are components of the synaptic and endocytic machineries are listed with their score from MS in Fig. 1a. In the present study, we decided to focus on the interaction with AP-2 as a putative modulator of TRKB surface localization and activation because three of four subunits of the complex were found associated with the TRKB receptor (Fig. 1a). Moreover, acute fluoxetine administration induced uncoupling of TRKB from AP-2, which is evidenced by the reduced scores of the AP-2 complex subunits α-2 (P17427), β (Q9DBG3), and μ (P84091) in the samples from fluoxetine-treated mice compared with the vehicle-treated control animals (Fig. 1a).
Figure 1.
Interaction between TRKB and AP-2. a, list of TRKB interactors involved in vesicle trafficking with MS scores in vehicle- and fluoxetine-treated groups. For a full list of interacting proteins and the effects of fluoxetine administration, see Tables S1 and S2 in the FigShare repository, respectively. b, co-IP of TRKB (145 or 100 kDa) and AP2M (50 kDa; IP: TRKB, WB: AP2M, TRKB) in samples from MG87.TRKB cells and hippocampus of 7-week-old male mouse. Acute fluoxetine administration (30 min) decreases the TRKB:AP2M interaction in mouse hippocampus measured by Western blotting (IP: AP2M, WB: TRKB) (c) or ELISA (d), respectively. e, repeated administration of fluoxetine (7 days) also disrupts the TRKB:AP2M interaction, as measured by ELISA. f, the C-terminal region of TRKB exhibits AP2M-binding motifs identified by the Eukaryotic Linear Motif library (TRG_ENDOCYTIC_2): Y515FGI, Y705YRV, Y726RKF, Y782ELM, Y816LDI (blue circle). Yellow, signal peptide (SP); gray, extracellular portion (N-terminal); blue, transmembrane (TM); green, intracellular portion (C-terminal). Red circles, sites of tyrosine phosphorylation in TRKB. g, the docking simulations were performed using sequences of the peptides spanning the last 26 amino acids of TRKB receptor. h, synthetic peptides (0.1 μg/ml = 34 μm) were used in cell-free ELISA to measure their binding to AP2M protein in MG.87.TRKB cell lysate. Red residues represent the TRG_ENDOCYTIC_2-positive motif and mutated sites. i, the phosphomimetic peptide (TRKB.Y816E, red circles) shows lower free energy for the receptor:peptide docking simulations than WT peptide (TRKB.ctrl, gray circles), whereas TRKB.Y816A mutant (blue circles) displayed the highest free energy values (n = 10/group). j, phosphorylated peptide (TRKB.pY, red circles) showed a higher interaction with AP2M compared with control peptide (TRKB.ctrl, gray circles), whereas TRKB.Y816A mutant (blue circles) displayed the lowest interaction levels, as measured by cell-free ELISA (n = 8/group). *, p < 0.05 from the WT peptide TRKB.ctrl. Black crosses, mean ± S.E.
Several cargo proteins docked to the AP-2 complex are bound via recognition of single or multiple tyrosine-based motifs by the AP2M subunit. These motifs in the cytosolic tails of transmembrane receptors consist of four amino acid residues (20). The initial tyrosine residue of the motif is followed by two random amino acids, and the signal ends with one of the hydrophobic residues leucine, isoleucine, phenylalanine, valine, or methionine: YXXϕ (27). We searched for such tyrosine-based motifs in mature full-length TRKB. The Eukaryotic Linear Motif library (21) indicated the presence of multiple putative tyrosine-based sorting signals (classified as TRG ENDOCYTIC 2 motifs, TRG2) in the cytosolic domain of the rat and mouse canonical TRKB sequence: Y515FGI, Y705YRV, Y726RKF, Y782ELM, Y816LDI, as seen in Fig. 1f. Interestingly, one of the presumed AP2M-binding sites, Y816LDI, which is located within the C-terminal portion of TRKB, is phosphorylated following antidepressant administration (9). Thus, we focused on the TRKB interaction with the cargo-docking μ subunit of the AP-2 complex (AP2M). We further confirmed the interaction between TRKB and AP2M in lysates of mouse hippocampus and the TRKB-overexpressing MG87 cell line by co-IP and Western blotting (Fig. 1b).
The acute administration of fluoxetine (30 mg/kg) decreased the TRKB:AP2M interaction, as measured by Western blotting (t(6) = 2.805, p = 0.0310, vehicle n = 3, fluoxetine n = 5) or ELISA (t(6) = 2.966, p = 0.0251, vehicle n = 4, fluoxetine n = 4) (Fig. 1, c and d). To find out whether the drug-induced disruption persists long-term with extended drug treatment, we examined the effect of 7-day exposure to fluoxetine in mice (in the drinking water (15 mg/kg)). This regimen was chosen based on evidence indicating the effectiveness of fluoxetine in a learned helplessness model in mice (22). Indeed, the decrease in the TRKB:AP2M coupling was also found in mouse hippocampus (t(7) = 3.115; p = 0.0170, vehicle n = 4, fluoxetine n = 5), as analyzed by ELISA (Fig. 1e). Furthermore, we validated the assay based on the decrease in the TRKB:AP2M interaction following the partial depletion of AP2M in MG87.TRKB cells (data and plot found in the FigShare repository; mean/S.D./n of percentage from ctrl: 100.0/34.24/11; siAP2M: 47.40/25.62/10; t(19) = 3.952, p = 0.0009).
To assess the interaction mechanism between AP2M and TRKB, we generated a model of full-length AP2M in the RaptorX server (23, 24). For the docking simulations, the AP2M model and the last 26 amino acid (aa) residues from the C-terminal portion of TRKB (Fig. 1g), including variants with either a phospho-dead (Y816A) or a phosphomimetic mutation (Y816E) of the critical tyrosine, were uploaded to the CABS-dock server (25). We obtained the Gibbs' free energy for the AP2M:TRKB peptide interaction, and the lowest 10 values from each group were used in statistical analysis. The interaction energy of the AP2M:TRKB.Y816E complex was lower than that for the AP2M:TRKB.ctrl complex, suggesting a more stable interaction, whereas it was significantly higher for the AP2M:TRKB.Y816A complex compared with AP2M:TRKB.ctrl (Kruskal–Wallis = 25.83; p < 0.0001, n = 10/group) (Fig. 1i). Based on these in silico results, we proceeded to analyze the AP2M:TRKB C terminus interaction in vitro by ELISA. For this purpose, three variants of N-biotinylated synthetic peptides of the last 26 aa of rat TRKB were used (ctrl, pY, or Y816A) (Fig. 1h). The interaction between AP2M and the phosphorylated TRKB peptide (TRKB.pY) was stronger compared with control peptide. In line with our in silico data, the AP2M:TRKB.Y816A interaction was weaker compared with the control peptide (Kruskal–Wallis = 20.48; p < 0.0001, n = 8/group) (Fig. 1j).
Role of AP2M in TRKB function
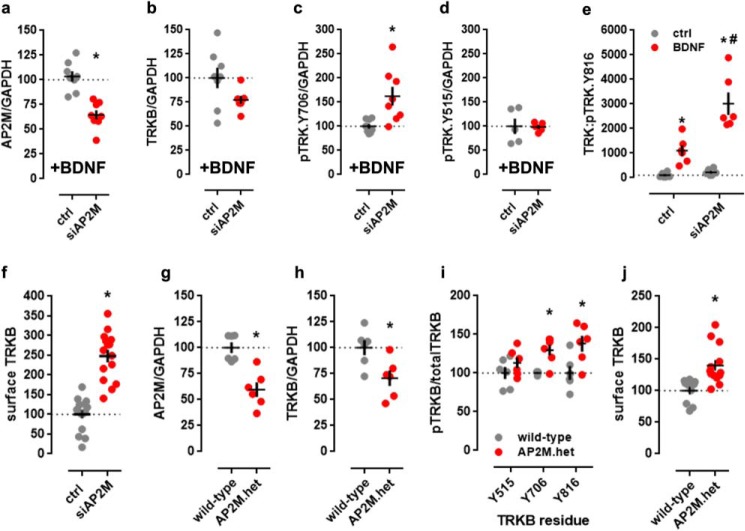
Previous studies indicate that AP2M depletion in mammalian cells prevents the assembly of functional AP-2 complexes (26) and results in a loss of AP-2 function (17). Thus, we investigated the impact of AP2M down-regulation on activation of TRKB receptor in MG87.TRKB cells overexpressing TRKB receptor. To induce TRKB signaling, the cells were challenged with BDNF following the silencing of AP2M, as there was no detectable phosphorylation of the receptor without BDNF. The depletion of AP2M (t(14) = 5.521; p < 0.0001, n = 8/group) (Fig. 2a) was associated with a marginal decrease in TRKB levels (t(13) = 1.901; p = 0.0797, ctrl n = 8, siAP2M n = 7) (Fig. 2b). However, silencing of AP2M enhanced the effect of BDNF on TRKB phosphorylation at residues Tyr-706/707 (t(14) = 3.099; p = 0.0078, n = 8/group) (Fig. 2c) and Tyr-816 (F(1, 20) = 12.21; p = 0.0023, n = 6/group) (Fig. 2e), but not at residue Tyr-515 (t(8) = 0.092; p = 0.928, n = 5/group) (Fig. 2d). The plasma membrane localization of TRKB receptors was also increased (t(28) = 7.71; p < 0.0001, n = 15/group) (Fig. 2f) by the down-regulation of AP2M in the absence of BDNF.
Figure 2.
AP2M regulates TRKB cell-surface localization and response to BDNF. Cultured MG87.TRKB cells were transfected with shRNA to silence the expression of AP2M and challenged with BDNF (0 or 25 ng/ml/15 min). Silencing of AP2M (50 kDa) (a) was associated with decrease in total TRKB (145 kDa) levels (b) and increase in phosphorylation of TRKB at pY706 (145 kDa) (c) but not in pY515 (145 kDa) (d) as measured by Western blotting (n = 5–8/group). e, phosphorylation of the pY816 site was augmented in siAP2M samples following the BDNF treatment as measured in ELISA (n = 6/group). f, the silencing of AP2M expression in MG87.TRKB led to an increased exposure of TRKB at the cell surface, as measured by surface ELISA (n = 6/group). The Western blot analysis of protein samples from AP2M.het animals showed decreased levels of AP2M (g) and total TRKB (h) in the hippocampus, associated with increased levels of phosphorylated TRKB at Tyr-706 and Tyr-816 residues (n = 5–6/group) (i). j, surface TRKB levels were also increased in primary cultures of hippocampal cells from AP2M.het mice in surface ELISA. *, p < 0.05 for comparison with ctrl/WT group; #, p < 0.05 for comparison with ctrl/siAP2M group. Black crosses, mean ± S.E.
Consistently, the reduced level of AP2M in the hippocampus of haploinsufficient AP2M.het mice (t(10) = 4.63; p = 0.0009, n = 6/group) (Fig. 2g) was associated with reduced levels of total TRKB in the hippocampus of these animals (t(10) = 2.794; p = 0.019, n = 6/group) (Fig. 2h). However, we observed increased levels of pTRKB (genotype: F(1,29) = 17.59; p = 0.0002, n = 6/group) at residues Tyr-706/707 and Tyr-816 (Fisher's LSD, p = 0.015 and 0.0016, respectively) (Fig. 2i). Similarly to the observed effects in MG87.TRKB cells, hippocampal cultures of AP2M.het animals (P0) showed increased levels of surface TRKB (t(21) = −3.603, p = 0.002, WT n = 10, AP2M.het n = 13) (Fig. 2j).
Antidepressants and BDNF differentially affect the TRKB:AP2M interaction
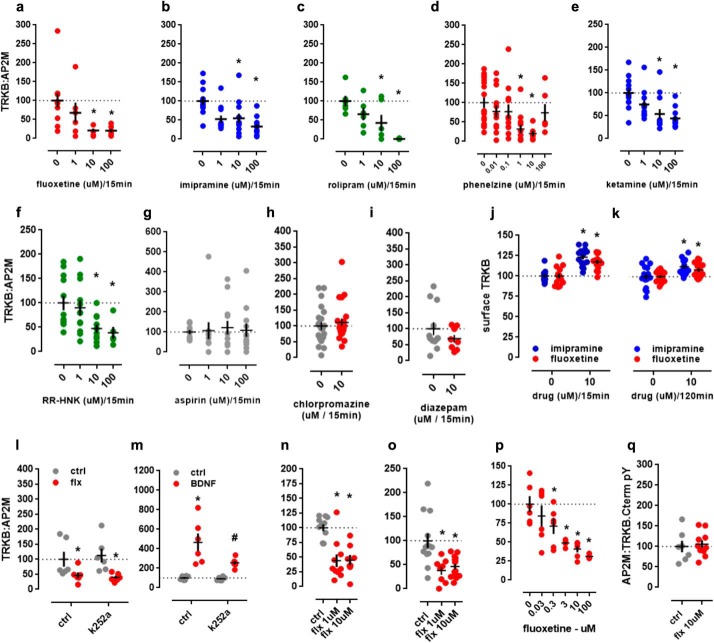
The systemic administration of fluoxetine (30 mg/kg, i.p., 30 min) has a disruptive effect on the TRKB:AP2M interaction. We next tested whether other antidepressants similarly disrupt the TRKB:AP2M interaction. Indeed, the AD fluoxetine, imipramine, rolipram, phenelzine, ketamine, and RR-HNK reduced the TRKB:AP2M interaction in MG87.TRKB cells as measured by ELISA (fluoxetine: F(3, 26) = 4.554, p = 0.011, n = 12, 6, 6, 6; imipramine: F(3, 43) = 7.883, p = 0.0003, n = 12, 11, 12, 12; rolipram: F(3, 19) = 6.78, p = 0.0027, n = 6, 6, 6, 5; phenelzine: Kruskal–Wallis = 19.34, p = 0.0017, n = 18, 12, 12, 12, 6, 6; ketamine: F(3, 44) = 7.421, p = 0.0004, n = 12, 12, 12, 12; RR-HNK: F(3, 35) = 4.022, p = 0.0147, n = 12, 12, 10, 5) (Fisher's LSD p < 0.05) (Fig. 3, a–f). However, the negative control aspirin did not induce a disruption (aspirin: F(3, 40) = 0.082, p = 0.96, n = 10, 10, 12, 12) (Fig. 3g). Furthermore, the administration of non-AD with a psychoactive profile, such as chlorpromazine or diazepam, failed to affect the TRKB:AP2M interaction (for chlorpromazine: t(42) = 0.6143; p = 0.5423, n = 22, 22; for diazepam: t(18) = 1.167, p = 0.2583, n = 12, 8) (Fig. 3, h and i).
Figure 3.
Antidepressant-induced disruption of the TRKB:AP2M interaction. MG87.TRKB cell cultures were treated with antidepressant fluoxetine (a), imipramine (b), rolipram (c), phenelzine (d), ketamine (e), or RR-HNK (f), and the TRKB:AP2M interaction was determined by ELISA. The compound aspirin (g), chlorpromazine (h), or diazepam (i) was used as a negative control (n = 6–22/group). j, surface levels of TRKB increased following fluoxetine or imipramine treatment, as measured by surface ELISA (10 μm/15 min; n = 15/group), and surface levels of TRKB (k) were still elevated 2 h after the administration of these drugs. ELISA measurements showed that the acute treatment of MG87.TRKB cells with k252a (10 μm) did not prevent fluoxetine (10 μm)-induced effects (l), but did prevent BDNF (25 ng/ml)-induced effects (m) on the TRKB:AP2M interaction (n = 6–12/group). Cortical (n) and hippocampal (o) cells (8–10 DIV) were treated with fluoxetine (1–10 μm/15 min), and the levels of TRKB:AP2M were determined by ELISA as described (n = 8–13/group). As shown by a cell-free ELISA, fluoxetine was able to disrupt the AP2M interaction with full-length (p), but not with TRKB C-terminal (q) peptide in lysates from MG87.TRKB cells (n = 6–10/group). *, p < 0.05 from ctrl/ctrl, water, or 0 μm; #, p < 0.05 from ctrl/k252a group. Black crosses, mean ± S.E.
The administration of fluoxetine or imipramine increased the levels of surface exposed TRKB in MG87.TRKB cells as measured by ELISA (15 min: F(1, 54) = 62.54, p < 0.0001, ctrl/fluoxetine n = 14/14, ctrl/imipramine n = 15/15; 120 min: F(1, 56) = 15.35, p = 0.0002, n = 15/group). Interestingly, the disruptive effect of fluoxetine on the TRKB:AP2M interaction is not dependent on TRKB activation, because pretreatment with the TRK inhibitor k252a was not able to prevent this effect (interaction effect: F(1,20) = 0.3066, p = 0.586, n = 6/group) (Fig. 3l). BDNF administration (25 ng/ml) also resulted in an increased interaction of TRKB receptors with AP2M, but contrary to what was observed for antidepressants, this increase depended on TRKB activation, because pretreatment with k252a prevented this effect (interaction effect: F(1, 20) = 4.528, p = 0.046, n = 6/group) (Fig. 3m). Similarly to the observations for MG87.TRKB cells, fluoxetine was able to disrupt the AP2M:TRKB interaction in primary cultures of cortical (F(2, 26) = 14.75, p < 0.0001, n = 11, 10, 8) and hippocampal cells (F(2, 32) = 9.562, p = 0.0006, n = 13, 8, 13) of rat embryos (Fig. 3, n and o).
Finally, we observed that fluoxetine disrupted the interaction between full-length TRKB and AP2M in MG87.TRKB cell lysates as measured in a cell-free ELISA (F(5, 30) = 9.71; p < 0.0001, n = 6/group) with effective doses ranging from 0.3 to 100 μm (Fisher's LSD, p < 0.05 for all) (Fig. 3p). However, no effect of fluoxetine (10 μm) was observed on the interaction between AP2M and the TRKB C-terminal peptide (t(15) = 0.35; p = 0.73, ctrl n = 7, fluoxetine n = 10) (Fig. 3q), suggesting additional binding sites for fluoxetine outside of this region.
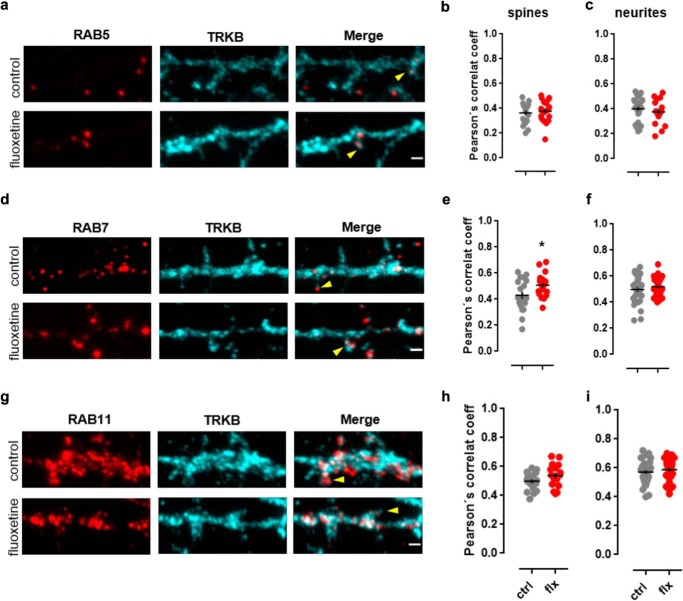
To the present point, it is plausible to consider that disruption of TRKB:AP2M occurs in the cell surface as well as in endocytic vesicles. Therefore, to address the fate of TRKB receptors after an acute fluoxetine challenge, we determined the colocalization of TRKB with early, late, and recycling endosome markers RAB5, RAB7, and RAB11, respectively (27, 28) (Fig. 4, a, d, and g). Student's t test indicates no significant effect of fluoxetine administration and the colocalization of TRKB and RAB proteins in neurites (RAB5: t(41) = 0.8490, p = 0.4008, ctrl n = 26, fluoxetine n = 17; RAB7: t(56) = 0.7730, p = 0.4428 ctrl n = 30, fluoxetine n = 28; RAB11: t(64) = 0.8233, p = 0.4134, ctrl n = 37, fluoxetine n = 29) of cortical neurons (Fig. 4, c, f, and i). No effect of fluoxetine treatment was observed in spines for TRKB:RAB5 (t(36) = 0.5053, p = 0.6164; ctrl n = 19/group) (Fig. 4b) or TRKB:RAB11 (t(31) = 1.561, p = 0.1288, ctrl n = 17, fluoxetine n = 16) (Fig. 4h) colocalization. However, a significant effect of fluoxetine was observed on the colocalization of TRKB and RAB7 (t(36) = 2.230, p = 0.0321, ctrl n = 19/group) (Fig. 4e).
Figure 4.
Effect of fluoxetine on TRKB colocalization with endosomal markers. The colocalization (indicated by the arrows) of TRKB with early (RAB5) (a–c), late (RAB7) (d–f), and recycling (RAB11) (g–i) endosome markers was determined in the spines and neurites of cultured cortical neurons (14–16 DIV) treated with fluoxetine (1 μm/15 min) (n = 16–37 spines or neurite shafts/group). *, p < 0.05 from ctrl group. Black crosses, mean ± S.E. Scale bars, 1 μm.
Discussion
Sub-chronic (29) and chronic fluoxetine treatment (30, 31) can induce changes in the proteomics profile in rodent brains. This compound altered the expression of proteins involved in neuroprotection, serotonin biosynthesis, and axonal transport in hippocampus and frontal cortex (30), as well as in endocytosis and transport in visual cortex (31).
The present study identified the endocytic complex AP-2 as a target whose interaction with TRKB is reduced following acute fluoxetine treatment. In line with a robust interaction between these proteins, we found three of four subunits of the AP-2 complex interacting with TRKB, whereas in the case of related complexes (i.e. AP-3 and AP-1), only one subunit was co-precipitated for each: AP3D1 and AP1B1. Furthermore, we identified tyrosine-based motifs in the TRKB receptor as putative binding sites for the AP2M subunit of the AP-2 adaptor complex. Especially the region around the Tyr-816 residue of TRKB conformed to the classical consensus for an AP2M-interacting motif. In line with its classical role as an endocytic adaptor complex, AP-2 regulates the surface localization of TRKB. Decreased expression of AP2M resulted in increased surface levels of TRKB, facilitating its activation by its ligand BDNF. We also found increased levels of activated TRKB in the hippocampus of AP2M haploinsufficient mice, as well as an increased exposure of TRKB at the surface of cultured hippocampal neurons from these animals. Finally, we observed that classical AD, such as fluoxetine, imipramine, rolipram, and phenelzine, as well as the fast acting antidepressants ketamine and RR-HNK are able to disrupt the coupling between AP2M and TRKB, resulting in increased exposure of TRKB at the cell surface, at concentrations found in the central nervous system after systemic administration (32–34).
It has been established that activation, endocytosis, and trafficking of TRKB receptors regulate the propagation of the downstream signaling response (35). There are three main cascades downstream of TRKB receptor activation. MAPK/ERK signaling regulating neuronal differentiation and growth is activated by recruitment of adaptors to the Tyr-515 site of the receptor. Phosphorylation of the Tyr-515 residue also induces a PI3K/Akt cascade promoting cell survival and growth. Finally, phosphorylation of the Tyr-816 residue is the key step for docking PLC-γ1, which regulates protein kinase C isoforms and induces the release of Ca2+ from internal sources to activate Ca2+/calmodulin (11, 36). Eventually, the post-endocytic sorting machinery plays a major role during the fate-determination process of the receptor signaling (37). In this scenario, some of the vesicles carrying the receptors are destined for retrograde transport toward the cell body (38, 39), whereas some are recycled back to the cell surface (40). For instance, Pincher, a membrane-trafficking protein, regulates the formation of TRK-carrying endosomes from plasma membrane ruffles in soma, axons, and dendrites (41). Pincher-mediated retrograde transport of signaling TRK receptors is rescued from lysosomal degradation (42). Moreover, BDNF-induced survival signaling in hippocampal neurons has been shown to be regulated by endophilin-A, which is involved in endosomal sorting of active TRKB receptors (27). Evidence suggests that the maintenance of long-term potentiation in hippocampal slices depends on the recycling and re-secretion of BDNF, thus emphasizing the necessity of TRKB receptor recycling back to the cell surface (43). Another study indicates that Slitrk5, by coupling to TRKB receptors, promotes its recycling to the cell surface in striatal neurons (44).
AP-2, as a member of the endocytic machinery (45), contributes to the formation of clathrin-coated vesicles around cargo proteins for endocytosis (46). In line with this idea, our results indicate a BDNF-induced enhancement of the TRKB:AP2M interaction, suggesting that the receptor is endocytosed upon activation. Although acute treatment with AD (e.g. 30 min) can induce a rapid phosphorylation of TRKB receptor in the mouse brain (7), these drugs disrupted the interaction of TRKB:AP2M. Thus, we suggest that the differential regulation of TRKB:AP2M by BDNF and AD may underlie the differential surface expression of the receptor. Zheng et al. (47), as well as Sommerfeld et al. (48), demonstrated that BDNF quickly reduces the TRKB receptor surface level, whereas our data indicate that antidepressants increase such levels. Interestingly, AMPA receptors, also committed to AP-2 complex-dependent endocytosis in the brain (49, 50), are internalized upon activation by glutamate. However, whereas the activation of NMDA receptors results in rapid reinsertion of AMPAR subunits to the cell surface, AMPARs activated in the absence of NMDAR activation are destined for degradation in lysosomes (51). Thus, the source of the activation signal recruits alternative machineries to differentially regulate the fate of the target receptor.
Although the AP-2 complex is best known for its major role in endocytosis (52), it remains doubtful whether the main purpose of the identified AP2M:TRKB interaction is facilitated endocytosis of TRKB. A recent study performing endocytosis assays with WT and AP2M null neurons overexpressing EGFP-TRKB and treated with BDNF could not detect any significant defect in the uptake of TRKB upon loss of AP-2. Instead, the authors showed that the retrograde axonal transport of autophagosomes containing TRKB receptor depends on AP-2 linking the autophagy adaptor LC3 to p150.Glued, an activator of the dynein motor required for autophagosome transport (19). This led us to conclude that the action of the TRKB–AP-2 complex is not necessarily restricted to the neuronal surface, but can take place in different neuronal compartments. Therefore, antidepressants could disrupt the interaction of TRKB receptors with AP2M at the cell surface, as well as in the autophagosomes.
Disruption of the AP2M:TRKB interaction in internal vesicles could prevent the interaction of the complex with the members of the autophagy-lysosome pathway that can lead to degradation of TRKB receptors (19). The delayed delivery of the signaling endosomes to the soma may locally enhance the specific signaling pathways that can recruit more plasticity-related proteins to the synapses. The two above-mentioned scenarios are not exclusive, although our TRKB:RAB colocalization data point toward changes that take place mainly at the cell-surface level. RAB11-positive endosomes have been shown to accumulate in dendrites of hippocampal neurons after BDNF challenge to support dendritic TRKB recycling (28). As we failed to observe an enhancement of TRKB:RAB11 colocalization in spines and neurites under fluoxetine, we suggest that the drug-induced regulation of TRKB surface exposure takes place in the cell membrane, at least shortly after the drug administration. On the other hand, the increase of TRKB:RAB7 colocalization only in spines but not in neurites suggests that TRKB receptors directed toward degradation could be stalled, which could contribute to the maintenance of previously activated and internalized receptors.
In the present study, we combined in vivo, in vitro, and in silico methods to gain insight into putative interaction domains between TRKB and AP-2. The in silico techniques allowed us to obtain a priori knowledge about possible interactions, especially involving the Tyr-816 region and the AP2M subunit. In fact, in the present study, the in silico results on the AP2M interaction with the TRKB C terminus matched the in vitro observations. As predicted by the CABS-dock server, AP2M displayed an increased interaction with phosphorylated TRKB (simulated using the phosphomimetic mutation Y816E). Interestingly, the Y816A mutation led to a decrease below the levels of control peptide (WT, nonphosphorylated) observed by both in silico (interpreted as a less stable complex) and in vitro assays. This suggests that there is some level of interaction between AP2M and nonactivated TRKB that is further potentiated upon TRKB phosphorylation. However, the role of such interaction, if any, or its occurrence in vivo remains to be investigated.
At this point, we speculated that the interaction of AP2M and TRKB could be a putative target for AD. Supporting this idea, cell-free assays indicated that the disruption of the TRKB:AP2M complex by fluoxetine happens in a dose-dependent manner. However, the same effect was not replicated in the AP2M:TRKB.peptide interaction assay. Therefore, although our data support the idea of a binding site for AD in the TRKB complex, it is unlikely that this binding site is between the receptor C-terminal portion and the AP2M subunit. Moreover, this site displays a low affinity for fluoxetine, given that effective doses are found above 0.3 μm. In fact, we have recently identified a low-affinity binding site for antidepressants in the transmembrane domain of TRKB (53). However, it is presently unclear how the conformational changes triggered by antidepressant binding to TRKB could alter the interaction of TRKB with AP-2.
Altogether, we suggest a novel mechanism where the antidepressant-induced disruption of the TRKB:AP2M interaction promotes TRKB cell-surface exposure and consequently BDNF signaling. However, further investigation will be necessary to address the precise mechanism and the long-term consequences of such a disruption.
Materials and methods
Animals
Adult (7-week-old) male C57BL/6RccHsd mice from Harlan (Horst aan de Maas, Netherlands), kept with free access to food and water, were used for TRKB immunoprecipitation and MS analysis. Briefly, the animals received a single i.p. injection of fluoxetine (30 mg/kg) and were euthanized by CO2. An independent cohort of female and another one of male mice from the same supplier (12–18 weeks old) received fluoxetine for 7 days or single i.p. injection, respectively. The hippocampi were collected and processed for ELISA and co-IP as described below. All procedures were in accordance with international guidelines for animal experimentation and the County Administrative Board of Southern Finland (ESLH-2007-09085/Ym-23, ESAVI/7551/04.10.07/2013).
The generation of floxed AP2M mice carrying loxP sites in front of exon 2 and behind exon 3 of the AP2M encoding gene Ap2m1 has been described (54). These conditional knockout (KO) mice were crossed with CMV-Cre deleter mice to achieve ubiquitous excision of exons 2 and 3. The resulting AP2M.het animals were crossed with AP2M.wt mice, thus excluding the generation of AP2M homozygous KO, which are embryonically lethal (55). All animals were genotyped prior to experiments using PCR on genomic DNA extracted from tissue biopsies and confirmed by Western blot analysis of hippocampus. The KO allele was detected using the forward primer GCTCTAAAGGTTATGCCTGGTGG and the reverse primer CCAAGGGACCTACAGGACTTC, which generate a fragment of 404 bp (PCR at 58 °C, 25 cycles). All experiments involving AP2M.het mice were reviewed and approved by the ethics committee of the “Landesamt für Gesundheit und Soziales” (LAGeSo) Berlin and were conducted according to the committee's guidelines. For brain dissection, animals (15 weeks old) were anesthetized with isoflurane prior to cervical dislocation. The hippocampus was dissected on ice and processed as described below.
Drugs
Acetylsalicylic acid (aspirin, #A2093, Sigma–Aldrich), fluoxetine (#H6995, Bosche Scientific), imipramine (#I7379-5G, Sigma–Aldrich), phenelzine (#P6777, Sigma–Aldrich), rolipram (#R6520, Sigma–Aldrich), ketamine (#3131, Tocris), 2R,6R-hydroxynorketamine (#6094, Tocris), diazepam (#D0899, Sigma–Aldrich), chlorpromazine (#C8138, Sigma–Aldrich), recombinant human BDNF (#450-02, Peprotech), and k252a (TRK inhibitor, #K2015, Sigma–Aldrich) were used (56, 57). All compounds were dissolved in DMSO for in vitro experiments, except BDNF, diluted in PBS. For in vivo administration, fluoxetine was diluted in sterile saline for intraperitoneal single injection (7) or in the drinking water for a repeated-administration regimen (8).
Immunoprecipitation and mass spectrometry
For TRKB proteome quantitative MS analysis, adult mouse hippocampal samples (n = 3) were mechanically homogenized in NP lysis buffer (20 mm Tris-HCl, 137 mm NaCl, 10% glycerol, 50 mm NaF, 1% Nonidet P-40, 0.05 mm Na3VO4, containing a mixture of protease and phosphatase inhibitors (#P2714 and #P0044, respectively, Sigma–Aldrich)). The samples were centrifuged (16,000 × g) at 4 °C for 15 min, and the supernatant was subjected to preclearing (control) and TRKB immunoprecipitation using Protein G–Sepharose–containing spin columns (#69725, Pierce Spin Columns Snap Cap). Briefly, 30 μl of Protein G–Sepharose were added to the columns and washed twice with 200 μl of NP buffer. Samples (400 μg of total protein) were cleared through the columns without antibody (1 h, under rotation at 4 °C). Unbound samples were collected (1000 × g, 1 min at 4 °C), preincubated with 5 μl of TRKB antibody (goat anti-mouse TRKB antibody, #AF1494, R&D Systems), and cleared through new Protein G–Sepharose tubes. After 1 h of rotation in the cold, the columns were washed twice with NP buffer and twice with TBS buffer (1000 × g, 1 min at 4 °C). The proteins were then eluted from the column with 0.1 m glycine (3 × 1000 × g, 1 min, 4 °C) or with Laemmli buffer for samples submitted to Western blotting and stored at −80 °C until further analysis. Cysteine bonds of the proteins were reduced with tris(2-carboxyethyl)-phosphine hydrochloride salt (#C4706, Sigma–Aldrich) and alkylated with iodoacetamide (#57670, Fluka, Sigma–Aldrich). Samples were digested overnight with trypsin (sequencing grade modified trypsin, #V5111, Promega) that specifically cleaves the carboxylic side of lysine and arginine amino acids. The digested peptides were then purified with C18 microspin columns (Harvard Apparatus) (58).
Mass spectrometry analysis was performed on an Orbitrap Elite hybrid mass spectrometer (Thermo Fisher Scientific) coupled to EASY-nLC II system (Xcalibur version 2.7.0 SP1, Thermo Fisher Scientific). The peptides were separated with a 60-min linear gradient from 5 to 35% of buffer B (98% acetonitrile and 0.1% formic acid in MS grade water). The analysis was performed in data-dependent acquisition: a high-resolution (60,000) FTMS full scan (m/z 300–1700) was followed by top20 CID-MS2 scans in an ion trap (Energy 35). The maximum fill time was 200 ms for both Fourier-transform ion cyclotron resonance MS (full AGC target 1,000,000) and the ion trap (MSn AGC target of 50,000). The MS/MS spectra were searched against the mouse component of the UniProtKB database (release 2012_08, 16,943 entries) using the SEQUEST search engine in Proteome DiscovererTM software (version 1.4, Thermo Fisher Scientific). Carbamidomethylation (+57.021464 Da) of cysteine residues was used as static modification, and oxidation (+15.994491 Da) of methionine was used as dynamic modification. Precursor mass tolerance and fragment mass tolerance were set to <15 ppm and <0.8 Da, respectively. A maximum of two missed cleavages was allowed, and the results were filtered to a maximum false discovery rate of 0.05 that was calculated by Target Decoy PSM validator, a tool of the Proteome Discoverer software. Raw MS data files can be found in FigShare.
For each protein-protein interaction, besides spectral counting values, a DR-score was calculated, and the specific interactions were filtered as in PMID (59). The full list of identified interactions is presented in Table S1. Proteins differing between saline- and fluoxetine-treated samples are highlighted in Table S2.
Cell culture and silencing of AP2M expression
The cell line 3T3 (mouse fibroblasts) stably transfected to express TRKB (MG87.TRKB) were used for in vitro assays (57). The MG87.TRKA cell line was used for antibody validation. The cells were maintained at 5% CO2, 37 °C in Dulbecco's modified Eagle's medium (containing 10% fetal calf serum, 1% penicillin/streptomycin, 1% l-glutamine, and 400 μg/ml G418 (only for MG87.TRKB)).
For silencing AP2M expression, MG87.TRKB cells were transfected with a mix of four AP2M-specific shRNA sequences (#TG712191 (Origene) or scrambled sequence as control) using Lipofectamine 2000 (#11668019, Thermo Fisher Scientific) as transfecting agent according to the manufacturer's instructions. The expression of GFP reporter was confirmed by microscopy 24 h after transfection, and AP2M expression levels were determined by Western blotting. The cells were challenged with BDNF (0 or 25 ng/ml/15 min) and lysed for ELISA or Western blotting as described below.
Primary cultures of cortical and hippocampal cells from rat embryos (embryonic day 18) or postnatal day 0 mice (for AP2M.het and wt) were prepared as described in the literature (60) and maintained in NeuroBasal medium (Thermo Fisher Scientific) supplemented with 1% B-27 supplement (Thermo Fisher Scientific), 1% penicillin/streptomycin, and 1% l-glutamine at 37 °C (60).
Sample processing and Western blotting
Samples from hippocampi of male and female mice were sonicated in NP lysis buffer and centrifuged (16,000 × g at 4 °C for 15 min). The supernatants were either submitted to TRKB:AP2M ELISA, or processed for immunoprecipitation of TRKB or AP2M, and submitted to SDS-PAGE. Supernatant of samples from AP2M.wt and AP2M.het mice, as well as samples from MG87.TRKB cells, were also submitted to SDS-PAGE. For cell-free ELISAs described below, the samples from MG87.TRKB cells were acidified with 1 m HCl (pH ∼2) at room temperature (RT) for 15 min, and the pH was restored to 7.4 with 1 m NaOH. This procedure aims to disrupt noncovalent interactions between proteins in a given complex.
The resolved proteins were transferred to polyvinylidene difluoride membranes, blocked with 3% BSA in TBST (20 mm Tris-HCl, 150 mm NaCl, 0.1% Tween 20, pH 7.6) for 2 h at RT and incubated overnight (ON) at 4 °C with antibodies against TRKB (1:1000, #AF1494, R&D Systems), AP2M (1:1000, #sc-515920, Santa Cruz Biotechnology, Inc.), pTRKB.Y515 (1:1000, #9141, Cell Signaling), pTRKB.Y706/7 (1:1000, #4621, Cell Signaling), pTRKB.Y816 (1:1000, #4168, Cell Signaling), or GAPDH (1:5000, #2118 (Cell Signaling) or #ab8245 (Abcam)). The membranes were then incubated with HRP-conjugated secondary antibodies (1:5000, anti-Gt, #61-1620, Invitrogen; anti-Rb, #170-5046, Bio-Rad; or mouse IgG κ–binding protein, #516102, Santa Cruz Biotechnology), and the HRP activity was detected by incubation with ECL and exposure to a CCD camera. The signal from each target was subtracted from background, normalized by GAPDH or TRKB (145 kDa), and expressed as a percentage of AP2M.wt, scrambled, or vehicle-treated groups.
AP2M:TRKB interaction and surface TRKB ELISAs
MG87.TRKB cells were incubated with fluoxetine, imipramine, rolipram, aspirin, ketamine, RR-HNK (1–100 μm), phenelzine (0.01–100 μm), chlorpromazine (10 μm), or diazepam (10 μm). An independent group received k252a (10 μm), followed 10 min later by fluoxetine (10 μm) or BDNF (25 ng/ml). The cells were lysed 15 min after the last drug administration. For comparative reasons, primary cultures of cortical and hippocampal cells from rat embryo (8–10 DIV) were incubated with fluoxetine (1–10 μm/15 min). The concentration ranges were chosen based on the literature (57). Finally, mice received fluoxetine at 15 mg/kg for 7 days in drinking water (8) or at 30 mg/kg as a single i.p. injection 30 min (9) before sample collection, and the TRKB:AP2M interaction was analyzed by a sandwich ELISA method and/or co-IP using hippocampal lysates.
The AP2M:TRKB interaction was determined by ELISA based on the general method described in the literature (57, 61), with minor adjustments to detect TRK:AP2M interaction. Briefly, white 96-well plates (OptiPlate 96F-HB, PerkinElmer Life Sciences) were coated with capturing anti-TRK antibody (1:1000, #92991S (Cell Signaling) or #AF1494 (R&D Systems)) in carbonate buffer (pH 9.8) ON at 4 °C. Following a blocking step with 3% BSA in PBST (137 mm NaCl, 10 mm phosphate, 2.7 mm KCl, 0.1% Tween 20, pH 7.4) (2 h, RT), samples were incubated ON at 4 °C. The incubation with antibody against AP2M (1:2000, #sc-515920, Santa Cruz Biotechnology; ON, 4 °C) was followed by HRP-conjugated mouse IgG κ–binding protein (1:5000, #516102, Santa Cruz Biotechnology; 2 h, RT). Finally, the chemiluminescent signal generated by the reaction with ECL was analyzed in a plate reader (Varioskan Flash, Thermo Fisher Scientific).
The surface levels of TRKB were also determined by ELISA (47) in MG87.TRKB cells treated with fluoxetine or imipramine (10 μm; 15 min or 2 h) and also after AP2M down-regulation. Hippocampal neurons (10 DIV) of haploinsufficient Ap2m mice were also tested for their TRKB surface expression. Briefly, cells cultivated in clear-bottom 96-well plates (ViewPlate 96, PerkinElmer Life Sciences) were washed with ice-cold PBS and fixed with 100 μl of 4% paraformaldehyde per well. After washing with PBS and blocking with PBS containing 5% nonfat dry milk and 5% BSA, the samples were incubated with primary anti-TRKB antibody (1:1000 in blocking buffer, #AF1494, R&D Systems) ON at 4 °C. Following a brief washing with PBS, the samples were incubated with HRP-conjugated anti-Gt antibody (1:5000 in blocking buffer, #61-1620, Invitrogen) for 1 h at RT. The cells were washed four times with 200 μl of PBS for 10 min each. Finally, the chemiluminescent signal generated by reaction with ECL was acquired in a plate reader.
The cell-free assays were performed in white 96-well plates. The plates were precoated with anti TRKB antibody (1:1000, #AF1494, R&D Systems) in carbonate buffer (pH 9.8) ON at 4 °C. Following blocking with 3% BSA in PBST buffer (2 h at RT), 120 μg of total protein from each sample (treated as for cell-free ELISA) were added and incubated ON at 4 °C under agitation. The plates were then washed three times with PBST buffer, and fluoxetine (0.01–100 μm) was added for 15 min. Following the washing protocol, the secondary antibody against AP2M was added (1:2000, #sc-515920, Santa Cruz Biotechnology) ON at 4 °C under agitation. The plates were then incubated with HRP-conjugated mouse IgG κ–binding protein (1:5000, #516102, Santa Cruz Biotechnology) for 2 h at RT. The HRP activity was detected by luminescence following incubation with ECL on a plate reader.
For the cell-free AP2M:TRKB C-terminal interaction, the plates were precoated with antibody against AP2M (1:500, #516102, Santa Cruz Biotechnology) in carbonate buffer, and then samples (treated as for cell-free ELISA) from MG87.TRKB cells were added ON at 4 °C. N-Biotinylated synthetic peptides (ctrl, pY, or Y816A; Genscript; sequences described in Fig. 1h) of the last 26 aa of rat TRKB (0.1 μg/ml = 34 nm, in PBST) were added for 1 h at RT, followed by HRP-conjugated streptavidin (1:10,000, #21126, Thermo Fisher Scientific) for 1 h at RT. The luminescence was determined upon incubation with ECL as described. In an independent experiment, immobilized AP2M from MG87.TRKB lysates was incubated with pY TRKB C-terminal peptide and fluoxetine (0 or 10 μm) for 1 h at RT, and the luminescence was determined via HRP-conjugated streptavidin activity reaction with ECL by a plate reader. The background-subtracted signal from each sample in each of the described assays was expressed as a percentage of the control group (vehicle-treated or WT peptide).
Immunofluorescence staining and image analysis
The colocalization experiments of TRKB and the endosome markers RAB5, RAB7, and RAB11 (62) were performed in cultured cortical cells. Briefly, 15 DIV cells were fixed in 4% paraformaldehyde (in PBS) after being treated with fluoxetine (1 μm, 15 min). The coverslips were washed in PBS and incubated in blocking buffer (5% normal donkey serum, 1% BSA, 0.1% gelatin, 0.1% Triton X-100, 0.05% Tween 20 in PBS) for 1 h at RT. The cells were stained with anti-TRKB antibody (1:500, #AF1494, R&D Systems) and anti-RAB5 antibody (1:500, #NB120-13253, Novus Biologicals), anti-RAB7 antibody (1:500, #D95F2, Cell Signaling), or anti-RAB11A antibody (5 μg/ml, #71-5300, Thermo Fisher Scientific) ON at 4 °C under agitation. Next, Alexa Fluor–conjugated secondary antibodies (Thermo Fisher Scientific) were diluted in PBS (1:1000). After a brief wash in PBS, the coverslips were incubated in donkey anti-goat-647 for TRKB and donkey anti-rabbit-568 for the RAB-specific antibodies for 45 min at RT and fixed in Dako Fluorescence Mounting Medium (S3023, Dako North America, Inc.). Control coverslips without primary antibody staining were also included.
Imaging was performed with a Leica TCS SP8 X inverted confocal microscope, ×63 oil objective (1.40 numeric aperture) at 1024 × 1024-pixel resolution. At least 6–10 Z-stack steps were acquired with 0.30-μm intervals from each image. The ImageJ FIJI Coloc2 plug-in was used for the colocalization analysis. The experimenters selected and analyzed the regions of interest (spines or neurites) blindly to the treatments. The regions of interest that passed the requirement of Costes' p value > 0.95 were selected for further statistical analysis of Pearson's correlation coefficient values from ctrl and fluoxetine groups.
The antibodies used in Western blotting, ELISA, or immunostaining were validated in previous studies (63–65) (e.g. against pY residues of TRKB) (7, 9, 57). The specificity of AP2M antibody was assessed in this study in transgenic mice and AP2M-silenced cells. For the antibody used to detect total TRKB, we performed Western blotting with samples from MG87 cells expressing TRKB or TRKA, and the results can be found on the FigShare repository.
Structural bioinformatics data mining and docking assay
To address a putative interaction between AP2M and TRKB, we submitted the full-length, canonical sequence of rat and mouse TRKB (Uniprot: Q63604 or P15209, respectively) to the Eukaryotic Linear Motif library (21). Several TRG_ENDOCYTIC_2 motifs (TRG2; a tyrosine-based sorting signal responsible for the interaction with AP2M subunit) were found for both mouse and rat TRKB. Of particular interest, the Tyr-816 residue is also activated by BDNF and antidepressants. Thus, the sequence of the last 26 aa residues of rat TRKB (WT, Y/E, or Y/A) and the full-length AP2M model generated through the RaptorX server (23) were submitted to docking simulations using the CABS-dock server (25).
Statistical analysis
Data were analyzed by Student's t test and one- or two-way analysis of variance (or their nonparametric equivalents) followed by Fisher's LSD post hoc test, when appropriate. Values of p < 0.05 were considered significant. All data generated in the present study, including MS raw files, are stored in FigShare under a CC-BY license, DOI: 10.6084/m9.figshare.7976528.
Author contributions
S. M. F. and P. C. C. conceptualization; S. M. F., C. A. B., L. V., H. G., I. C., R. M., T. M., M. V., and P. C. C. investigation; S. M. F. and L. L. methodology; S. M. F., L. L., H. G., and P. C. C. writing-original draft; H. G. data curation; H. G. visualization; T. M., M. V., P. C. C., and E. C. writing-review and editing; P. C. C. and E. C. supervision; E. C. project administration.
Supplementary Material
Acknowledgments
We thank Sulo Kolehmainen and Seija Lågas (University of Helsinki), Claudia Schmidt (Leibniz-Forschungsinstitut für Molekulare Pharmakologie), and the Biomedicum Imaging Unit for technical assistance, as well as Dr. Henri Huttunen for RAB5 and RAB7 antibodies. We also thank Drs. Tomi Rantamaki, Cassiano R. A. Faria Diniz, and Caroline Biojone for helpful comments on the manuscript.
This work was supported by European Research Council (ERC) Grant 322742 (iPLASTICITY), EU Joint Programme-Neurodegenerative Disease Research (JPND) project CircProt Grants 1301225 and 643417, Academy of Finland Grants 294710 and 307416, the Sigrid Juselius Foundation, the Jane and Eljas Erkko Foundation, and Deutsche Forschungsgemeinschaft (DFG) Grant SFB958/A01. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1 and S2.
- BDNF
- brain-derived neurotrophic factor
- TRKB
- tropomyosin-related kinase B
- AP-2
- adaptor protein complex-2
- AP2M
- μ subunit of the AP-2 complex
- AD
- antidepressant drugs
- PLC
- phospholipase C
- i.p.
- intraperitoneal
- IP
- immunoprecipitation
- ctrl
- control
- aa
- amino acid(s)
- LSD
- least significant difference
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal–regulated kinase
- KO
- knockout
- AGC
- automatic gain control
- RT
- room temperature
- ON
- overnight
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- HRP
- horseradish peroxidase
- DIV
- days in vitro
- PI3K
- phosphatidylinositol 3-kinase
- AMPA
- α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- AMPAR
- AMPA receptor
- NMDA
- N-methyl-d-aspartate
- NMDAR
- NMDA receptor.
References
- 1. Castrén E. (2005) Is mood chemistry? Nat. Rev. Neurosci. 6, 241–246 10.1038/nrn1629 [DOI] [PubMed] [Google Scholar]
- 2. Stockmeier C. A., Mahajan G. J., Konick L. C., Overholser J. C., Jurjus G. J., Meltzer H. Y., Uylings H. B. M., Friedman L., and Rajkowska G. (2004) Cellular changes in the postmortem hippocampus in major depression. Biol. Psychiatry 56, 640–650 10.1016/j.biopsych.2004.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Drevets W. C. (2001) Neuroimaging and neuropathological studies of depression: implications for the cognitive-emotional features of mood disorders. Curr. Opin. Neurobiol. 11, 240–249 10.1016/S0959-4388(00)00203-8 [DOI] [PubMed] [Google Scholar]
- 4. Castrén E. (2004) Neurotrophic effects of antidepressant drugs. Curr. Opin. Pharmacol. 4, 58–64 10.1016/j.coph.2003.10.004 [DOI] [PubMed] [Google Scholar]
- 5. Rantamäki T., and Castrén E. (2008) Targeting TrkB neurotrophin receptor to treat depression. Expert Opin. Ther. Targets 12, 705–715 10.1517/14728222.12.6.705 [DOI] [PubMed] [Google Scholar]
- 6. Neto F. L., Borges G., Torres-Sanchez S., Mico J. A., and Berrocoso E. (2011) Neurotrophins role in depression neurobiology: a review of basic and clinical evidence. Curr. Neuropharmacol. 9, 530–552 10.2174/157015911798376262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saarelainen T., Hendolin P., Lucas G., Koponen E., Sairanen M., MacDonald E., Agerman K., Haapasalo A., Nawa H., Aloyz R., Ernfors P., and Castrén E. (2003) Activation of the TrkB neurotrophin receptor is induced by antidepressant drugs and is required for antidepressant-induced behavioral effects. J. Neurosci. 23, 349–357 10.1523/JNEUROSCI.23-01-00349.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Karpova N. N., Pickenhagen A., Lindholm J., Tiraboschi E., Kulesskaya N., Agústsdóttir A., Antila H., Popova D., Akamine Y., Bahi A., Sullivan R., Hen R., Drew L. J., and Castrén E. (2011) Fear erasure in mice requires synergy between antidepressant drugs and extinction training. Science 334, 1731–1734 10.1126/science.1214592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rantamäki T., Hendolin P., Kankaanpää A., Mijatovic J., Piepponen P., Domenici E., Chao M. V., Männistö P. T., and Castrén E. (2007) Pharmacologically diverse antidepressants rapidly activate brain-derived neurotrophic factor receptor TrkB and induce phospholipase-Cγ signaling pathways in mouse brain. Neuropsychopharmacology 32, 2152–2162 10.1038/sj.npp.1301345 [DOI] [PubMed] [Google Scholar]
- 10. Cunningham M. E., and Greene L. A. (1998) A function-structure model for NGF-activated TRK. EMBO J. 17, 7282–7293 10.1093/emboj/17.24.7282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Minichiello L. (2009) TrkB signalling pathways in LTP and learning. Nat. Rev. Neurosci. 10, 850–860 10.1038/nrn2738 [DOI] [PubMed] [Google Scholar]
- 12. Haapasalo A., Sipola I., Larsson K., Akerman K. E. O., Stoilov P., Stamm S., Wong G., and Castren E. (2002) Regulation of TRKB surface expression by brain-derived neurotrophic factor and truncated TRKB isoforms. J. Biol. Chem. 277, 43160–43167 10.1074/jbc.M205202200 [DOI] [PubMed] [Google Scholar]
- 13. Du J., Feng L., Yang F., and Lu B. (2000) Activity- and Ca2+-dependent modulation of surface expression of brain-derived neurotrophic factor receptors in hippocampal neurons. J. Cell Biol. 150, 1423–1434 10.1083/jcb.150.6.1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meyer-Franke A., Wilkinson G. A., Kruttgen A., Hu M., Munro E., Hanson M. G. Jr., Reichardt L. F., and Barres B. A. (1998) Depolarization and cAMP elevation rapidly recruit TrkB to the plasma membrane of CNS neurons. Neuron 21, 681–693 10.1016/S0896-6273(00)80586-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doherty G. J., and McMahon H. T. (2009) Mechanisms of endocytosis. Annu. Rev. Biochem. 78, 857–902 10.1146/annurev.biochem.78.081307.110540 [DOI] [PubMed] [Google Scholar]
- 16. Kirchhausen T. (1993) Coated pits and coated vesicles—sorting it all out. Curr. Opin. Struct. Biol. 3, 182–188 10.1016/S0959-440X(05)80150-2 [DOI] [Google Scholar]
- 17. Grimes M. L., Beattie E., and Mobley W. C. (1997) A signaling organelle containing the nerve growth factor-activated receptor tyrosine kinase, TrkA. Proc. Natl. Acad. Sci. U.S.A. 94, 9909–9914 10.1073/pnas.94.18.9909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beattie E. C., Howe C. L., Wilde A., Brodsky F. M., and Mobley W. C. (2000) NGF signals through TrkA to increase clathrin at the plasma membrane and enhance clathrin-mediated membrane trafficking. J. Neurosci. 20, 7325–7333 10.1523/JNEUROSCI.20-19-07325.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kononenko N. L., Classen G. A., Kuijpers M., Puchkov D., Maritzen T., Tempes A., Malik A. R., Skalecka A., Bera S., Jaworski J., and Haucke V. (2017) Retrograde transport of TrkB-containing autophagosomes via the adaptor AP-2 mediates neuronal complexity and prevents neurodegeneration. Nat. Commun. 8, 14819 10.1038/ncomms14819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ohno H., Stewart J., Fournier M. C., Bosshart H., Rhee I., Miyatake S., Saito T., Gallusser A., Kirchhausen T., and Bonifacino J. S. (1995) Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science 269, 1872–1875 10.1126/science.7569928 [DOI] [PubMed] [Google Scholar]
- 21. Dinkel H., Van Roey K., Michael S., Kumar M., Uyar B., Altenberg B., Milchevskaya V., Schneider M., Kühn H., Behrendt A., Dahl S. L., Damerell V., Diebel S., Kalman S., Klein S., et al. (2016) ELM 2016–data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res. 44, D294–D300 10.1093/nar/gkv1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Holanda V. A. D., Medeiros I. U., Asth L., Guerrini R., Calo' G., and Gavioli E. C. (2016) Antidepressant activity of nociceptin/orphanin FQ receptor antagonists in the mouse learned helplessness. Psychopharmacology 233, 2525–2532 10.1007/s00213-016-4310-1 [DOI] [PubMed] [Google Scholar]
- 23. Källberg M., Wang H., Wang S., Peng J., Wang Z., Lu H., and Xu J. (2012) Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 7, 1511–1522 10.1038/nprot.2012.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peng J., and Xu J. (2011) RaptorX: exploiting structure information for protein alignment by statistical inference. Proteins 79, Suppl. 10, 161–171 10.1002/prot.23175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kurcinski M., Jamroz M., Blaszczyk M., Kolinski A., and Kmiecik S. (2015) CABS-dock web server for the flexible docking of peptides to proteins without prior knowledge of the binding site. Nucleic Acids Res. 43, W419–W424 10.1093/nar/gkv456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Deborde S., Perret E., Gravotta D., Deora A., Salvarezza S., Schreiner R., and Rodriguez-Boulan E. (2008) Clathrin is a key regulator of basolateral polarity. Nature 452, 719–723 10.1038/nature06828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Burk K., Murdoch J. D., Freytag S., Koenig M., Bharat V., Markworth R., Burkhardt S., Fischer A., and Dean C. (2017) EndophilinAs regulate endosomal sorting of BDNF-TrkB to mediate survival signaling in hippocampal neurons. Sci. Rep. 7, 2149 10.1038/s41598-017-02202-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lazo O. M., Gonzalez A., Ascaño M., Kuruvilla R., Couve A., and Bronfman F. C. (2013) BDNF regulates Rab11-mediated recycling endosome dynamics to induce dendritic branching. J. Neurosci. 33, 6112–6122 10.1523/JNEUROSCI.4630-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cecconi D., Mion S., Astner H., Domenici E., Righetti P. G., and Carboni L. (2007) Proteomic analysis of rat cortical neurons after fluoxetine treatment. Brain Res. 1135, 41–51 10.1016/j.brainres.2006.12.008 [DOI] [PubMed] [Google Scholar]
- 30. Carboni L., Vighini M., Piubelli C., Castelletti L., Milli A., and Domenici E. (2006) Proteomic analysis of rat hippocampus and frontal cortex after chronic treatment with fluoxetine or putative novel antidepressants: CRF1 and NK1 receptor antagonists. Eur. Neuropsychopharmacol. 16, 521–537 10.1016/j.euroneuro.2006.01.007 [DOI] [PubMed] [Google Scholar]
- 31. Ruiz-Perera L., Muniz M., Vierci G., Bornia N., Baroncelli L., Sale A., and Rossi F. M. (2015) Fluoxetine increases plasticity and modulates the proteomic profile in the adult mouse visual cortex. Sci. Rep. 5, 12517 10.1038/srep12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bolo N. R., Hodé Y., Nédélec J. F., Lainé E., Wagner G., and Macher J. P. (2000) Brain pharmacokinetics and tissue distribution in vivo of fluvoxamine and fluoxetine by fluorine magnetic resonance spectroscopy. Neuropsychopharmacology 23, 428–438 10.1016/S0893-133X(00)00116-0 [DOI] [PubMed] [Google Scholar]
- 33. Lumsden E. W., Troppoli T. A., Myers S. J., Zanos P., Aracava Y., Kehr J., Lovett J., Kim S., Wang F.-H., Schmidt S., Jenne C. E., Yuan P., Morris P. J., Thomas C. J., Zarate C. A. Jr., et al. (2019) Antidepressant-relevant concentrations of the ketamine metabolite (2R,6R)-hydroxynorketamine do not block NMDA receptor function. Proc. Natl. Acad. Sci. U.S.A. 116, 5160–5169 10.1073/pnas.1816071116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zanos P., Moaddel R., Morris P. J., Georgiou P., Fischell J., Elmer G. I., Alkondon M., Yuan P., Pribut H. J., Singh N. S., Dossou K. S. S., Fang Y., Huang X.-P., Mayo C. L., Wainer I. W., et al. (2016) NMDAR inhibition-independent antidepressant actions of ketamine metabolites. Nature 533, 481–486 10.1038/nature17998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harrington A. W., and Ginty D. D. (2013) Long-distance retrograde neurotrophic factor signalling in neurons. Nat. Rev. Neurosci. 14, 177–187 10.1038/nrn3253 [DOI] [PubMed] [Google Scholar]
- 36. Huang E. J., and Reichardt L. F. (2001) Neurotrophins: roles in neuronal development and function. Annu. Rev. Neurosci. 24, 677–736 10.1146/annurev.neuro.24.1.677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cosker K. E., and Segal R. A. (2014) Neuronal signaling through endocytosis. Cold Spring Harb. Perspect. Biol. 6, a020669 10.1101/cshperspect.a020669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ehlers M. D., Kaplan D. R., Price D. L., and Koliatsos V. E. (1995) NGF-stimulated retrograde transport of trkA in the mammalian nervous system. J. Cell Biol. 130, 149–156 10.1083/jcb.130.1.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bhattacharyya A., Watson F. L., Bradlee T. A., Pomeroy S. L., Stiles C. D., and Segal R. A. (1997) Trk receptors function as rapid retrograde signal carriers in the adult nervous system. J. Neurosci. 17, 7007–7016 10.1523/JNEUROSCI.17-18-07007.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen Z.-Y., Ieraci A., Tanowitz M., and Lee F. S. (2005) A novel endocytic recycling signal distinguishes biological responses of Trk neurotrophin receptors. Mol. Biol. Cell 16, 5761–5772 10.1091/mbc.e05-07-0651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Valdez G., Akmentin W., Philippidou P., Kuruvilla R., Ginty D. D., and Halegoua S. (2005) Pincher-mediated macroendocytosis underlies retrograde signaling by neurotrophin receptors. J. Neurosci. 25, 5236–5247 10.1523/JNEUROSCI.5104-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shao Y., Akmentin W., Toledo-Aral J. J., Rosenbaum J., Valdez G., Cabot J. B., Hilbush B. S., and Halegoua S. (2002) Pincher, a pinocytic chaperone for nerve growth factor/TrkA signaling endosomes. J. Cell Biol. 157, 679–691 10.1083/jcb.200201063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Santi S., Cappello S., Riccio M., Bergami M., Aicardi G., Schenk U., Matteoli M., and Canossa M. (2006) Hippocampal neurons recycle BDNF for activity-dependent secretion and LTP maintenance. EMBO J. 25, 4372–4380 10.1038/sj.emboj.7601303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Song M., Giza J., Proenca C. C., Jing D., Elliott M., Dincheva I., Shmelkov S. V., Kim J., Schreiner R., Huang S.-H., Castrén E., Prekeris R., Hempstead B. L., Chao M. V., Dictenberg J. B., et al. (2015) Slitrk5 mediates BDNF-dependent TrkB receptor trafficking and signaling. Dev. Cell 33, 690–702 10.1016/j.devcel.2015.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pearse B. M., and Robinson M. S. (1990) Clathrin, adaptors, and sorting. Annu. Rev. Cell Biol. 6, 151–171 10.1146/annurev.cb.06.110190.001055 [DOI] [PubMed] [Google Scholar]
- 46. Collins B. M., McCoy A. J., Kent H. M., Evans P. R., and Owen D. J. (2002) Molecular architecture and functional model of the endocytic AP2 complex. Cell 109, 523–535 10.1016/S0092-8674(02)00735-3 [DOI] [PubMed] [Google Scholar]
- 47. Zheng J., Shen W.-H., Lu T.-J., Zhou Y., Chen Q., Wang Z., Xiang T., Zhu Y.-C., Zhang C., Duan S., and Xiong Z.-Q. (2008) Clathrin-dependent endocytosis is required for TrkB-dependent Akt-mediated neuronal protection and dendritic growth. J. Biol. Chem. 283, 13280–13288 10.1074/jbc.M709930200 [DOI] [PubMed] [Google Scholar]
- 48. Sommerfeld M. T., Schweigreiter R., Barde Y. A., and Hoppe E. (2000) Down-regulation of the neurotrophin receptor TrkB following ligand binding: evidence for an involvement of the proteasome and differential regulation of TrkA and TrkB. J. Biol. Chem. 275, 8982–8990 10.1074/jbc.275.12.8982 [DOI] [PubMed] [Google Scholar]
- 49. Lee S. H., Liu L., Wang Y. T., and Sheng M. (2002) Clathrin adaptor AP2 and NSF interact with overlapping sites of GluR2 and play distinct roles in AMPA receptor trafficking and hippocampal LTD. Neuron 36, 661–674 10.1016/S0896-6273(02)01024-3 [DOI] [PubMed] [Google Scholar]
- 50. Kastning K., Kukhtina V., Kittler J. T., Chen G., Pechstein A., Enders S., Lee S. H., Sheng M., Yan Z., and Haucke V. (2007) Molecular determinants for the interaction between AMPA receptors and the clathrin adaptor complex AP-2. Proc. Natl. Acad. Sci. U.S.A. 104, 2991–2996 10.1073/pnas.0611170104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ehlers M. D. (2000) Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron 28, 511–525 10.1016/S0896-6273(00)00129-X [DOI] [PubMed] [Google Scholar]
- 52. Robinson M. S. (1987) 100-kD coated vesicle proteins: molecular heterogeneity and intracellular distribution studied with monoclonal antibodies. J. Cell Biol. 104, 887–895 10.1083/jcb.104.4.887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Casarotto P. C., Girych M., Fred S. M., Moliner R., Enkavi G., Biojone C., Cannarozzo C., Brunello C. A., Steinzeig A., Winkel F., Patil S., Vestring S., Serchov T., Laukkanen L., Cardon I., et al. (2019) Antidepressants act by binding to the cholesterol-interaction site at TRKB neurotrophin receptor. bioRxiv 10.1101/757989 [DOI] [Google Scholar]
- 54. Kononenko N. L., Puchkov D., Classen G. A., Walter A. M., Pechstein A., Sawade L., Kaempf N., Trimbuch T., Lorenz D., Rosenmund C., Maritzen T., and Haucke V. (2014) Clathrin/AP-2 mediate synaptic vesicle reformation from endosome-like vacuoles but are not essential for membrane retrieval at central synapses. Neuron 82, 981–988 10.1016/j.neuron.2014.05.007 [DOI] [PubMed] [Google Scholar]
- 55. Mitsunari T., Nakatsu F., Shioda N., Love P. E., Grinberg A., Bonifacino J. S., and Ohno H. (2005) Clathrin adaptor AP-2 is essential for early embryonal development. Mol. Cell Biol. 25, 9318–9323 10.1128/MCB.25.21.9318-9323.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Koizumi S., Contreras M. L., Matsuda Y., Hama T., Lazarovici P., and Guroff G. (1988) K-252a: a specific inhibitor of the action of nerve growth factor on PC 12 cells. J. Neurosci. 8, 715–721 10.1523/JNEUROSCI.08-02-00715.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Antila H., Autio H., Turunen L., Harju K., Tammela P., Wennerberg K., Yli-Kauhaluoma J., Huttunen H. J., Castrén E., and Rantamäki T. (2014) Utilization of in situ ELISA method for examining Trk receptor phosphorylation in cultured cells. J. Neurosci. Methods 222, 142–146 10.1016/j.jneumeth.2013.11.001 [DOI] [PubMed] [Google Scholar]
- 58. Varjosalo M., Keskitalo S., Van Drogen A., Nurkkala H., Vichalkovski A., Aebersold R., and Gstaiger M. (2013) The protein interaction landscape of the human CMGC kinase group. Cell Rep. 3, 1306–1320 10.1016/j.celrep.2013.03.027 [DOI] [PubMed] [Google Scholar]
- 59. Sowa M. E., Bennett E. J., Gygi S. P., and Harper J. W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 10.1016/j.cell.2009.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sahu M. P., Nikkilä O., Lågas S., Kolehmainen S., and Castrén E. (2019) Culturing primary neurons from rat hippocampus and cortex. Neuronal Signal. 3, NS20180207 10.1042/NS20180207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Antila H., Casarotto P., Popova D., Sipila P., Guirado R., Kohtala S., Ryazantseva M., Vesa L., Lindholm J., Yalcin I., Sato V., Goos H., Lempriere S., Cordeira J., Autio H., et al. (2016) TrkB signaling underlies the rapid antidepressant effects of isoflurane. bioRxiv 10.1101/084525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bucci C., Alifano P., and Cogli L. (2014) The role of rab proteins in neuronal cells and in the trafficking of neurotrophin receptors. Membranes 4, 642–677 10.3390/membranes4040642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ranieri R., Ciaglia E., Amodio G., Picardi P., Proto M. C., Gazzerro P., Laezza C., Remondelli P., Bifulco M., and Pisanti S. (2018) N6-Isopentenyladenosine dual targeting of AMPK and Rab7 prenylation inhibits melanoma growth through the impairment of autophagic flux. Cell Death Differ. 25, 353–367 10.1038/cdd.2017.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Brunello C. A., Yan X., and Huttunen H. J. (2016) Internalized Tau sensitizes cells to stress by promoting formation and stability of stress granules. Sci. Rep. 6, 30498 10.1038/srep30498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Eisfeld A. J., Kawakami E., Watanabe T., Neumann G., and Kawaoka Y. (2011) RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J. Virol. 85, 6117–6126 10.1128/JVI.00378-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.