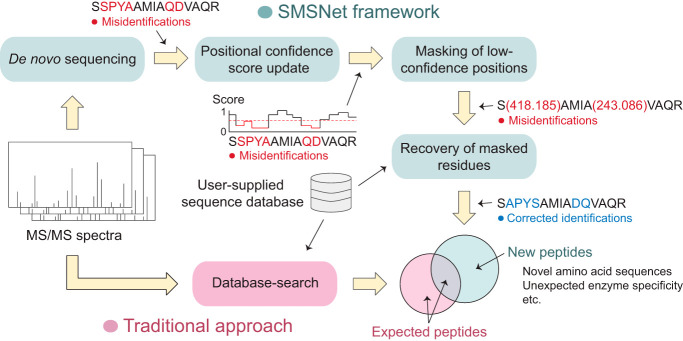
De novo peptide sequencing is a promising approach for discovering new peptides. However, its performance is hindered by the fact that most MS/MS spectra do not contain complete amino acid sequence information. Here, we present a deep learning-based de novo sequencing model, SMSNet, together with a post-processing strategy that pinpoints misidentified residues and utilizes user-provided sequence database to revise the identifications. The results show that many new HLA antigens and phosphopeptides can be discovered with our strategy.
Keywords: De novo sequencing, mass spectrometry, bioinformatics searching, phosphoproteome, peptides, deep learning, software
Graphical Abstract
Highlights
Deep learning-based hybrid de novo sequencing with database search strategy.
Accurate identifications via ability to revise confidence scores and amino acids.
Discovery of >10,000 potential new HLA antigens and human phosphopeptides.
A dataset of >26 million annotated HCD spectra from Q Exactive instruments.
Abstract
Typical analyses of mass spectrometry data only identify amino acid sequences that exist in reference databases. This restricts the possibility of discovering new peptides such as those that contain uncharacterized mutations or originate from unexpected processing of RNAs and proteins. De novo peptide sequencing approaches address this limitation but often suffer from low accuracy and require extensive validation by experts. Here, we develop SMSNet, a deep learning-based de novo peptide sequencing framework that achieves >95% amino acid accuracy while retaining good identification coverage. Applications of SMSNet on landmark proteomics and peptidomics studies reveal over 10,000 previously uncharacterized HLA antigens and phosphopeptides, and in conjunction with database-search methods, expand the coverage of peptide identification by almost 30%. The power to accurately identify new peptides of SMSNet would make it an invaluable tool for any future proteomics and peptidomics studies, including tumor neoantigen discovery, antibody sequencing, and proteome characterization of non-model organisms.
Typical analyses of mass spectrometry-based proteomics and peptidomics data rely on database-search approaches that provide the best known answers and cannot identify unexpected peptides and proteins. In contrast, de novo peptide sequencing attempts to determine the amino acid sequences directly from observed mass spectra but at times suffers from low accuracy. The capability of de novo approaches to identify new peptides is crucial for studying non-model organisms with incomplete databases (1), for identifying uncharacterized mutations or polymorphisms, and for discovering peptides derived from complex processing of RNA and proteins, such as proteasome-mediated splicing (2–4) and translation of canonically non-coding regions of the genome (5).
Despite advancements in de novo peptide sequencing (6–9), use of these methods in routine tandem mass spectrometry (MS/MS) data remains challenging. Recently, DeepNovo (6, 10) has shown that deep learning can be effectively applied to the de novo peptide sequencing problem, which resulted in clear improvements over its predecessors. Nonetheless, there is still a huge performance gap in the accuracy and number of identified peptides between de novo approaches and standard database-search approaches. Key parts of this limitation lie in the nature of peptide MS/MS spectra which are noisy and sometimes lack crucial information. When interpreting an MS/MS spectrum, evidence for certain amino acid sequence comes in the form of a series of observed ions whose sequential mass differences match to masses of specific amino acids within an error threshold. However, most MS/MS spectra do not contain a complete series of ions that would enable definite deduction of every amino acid position within the original sequence (11). In these cases, without prior information such as a database of expected amino acid sequences, it would be very challenging or even impossible to arrive at the correct answers.
Here, we introduce SMSNet, a hybrid de novo peptide sequencing approach that leverages a multi-step Sequence-Mask-Search strategy to address the issue of missing ions in MS/MS spectra. SMSNet adopts the encoder-decoder deep learning architecture that has been widely used in machine translation (12), basically formulating peptide sequencing as a spectra-to-peptide language translation problem. At the initial sequence step, SMSNet determines the full amino acid sequence as well as positional confidence scores for each input MS/MS spectrum. Then, during the mask step, identified amino acid positions with low confidence scores are converted into mass tags that indicate the total masses of masked amino acids. Finally, in the search step, all identifications are compared against an input amino acid sequence database in order to recover the exact amino acid sequences from mass tags. Our approach essentially combines the strengths of modern, machine learning-based methods (6, 8), which are able to determine the whole amino acid sequence, and sequence tag methods (13, 14), which use high-confidence partial sequences as seeds for retrieving full sequences from databases. The current version of SMSNet can identify unmodified forms of all twenty amino acids plus oxidized Methionine and phosphorylated Serine, Threonine, and Tyrosine, but the model can be re-trained to recognize additional post-translational modifications.
Application of SMSNet on large-scaled studies of human leukocyte antigen (HLA)1 peptidomes and epidermal growth factor (EGF)-treated glioblastoma's phosphoproteomes reveals over 10,000 previously uncharacterized HLA antigens and over 4000 previously uncharacterized phosphopeptides. SMSNet's identifications are in almost perfect agreement with results from database searches and exhibit known characteristics of HLA antigens or could be traced to known phosphoproteins and phosphosites. Furthermore, more than 6000 of newly identified HLA antigens have not been reported in the Immune Epitope Database (15) and should contribute to the growing interests in neoantigen discovery for immunotherapy. The power to accurately identify new peptides of SMSNet would make it an invaluable tool for any future proteomics and peptidomics studies.
EXPERIMENTAL PROCEDURES
Data Acquisition
A combined mass spectrometry dataset consisting of more than 27 million peptide-spectrum matches (PSM) was obtained from the Proteomics and Metabolomics Core Facility at The Wistar Institute (Philadelphia, PA). All MS/MS spectra were acquired on Q Exactive HF or Q Exactive Plus mass spectrometers (Thermo Fisher Scientific, Bremen, Germany) and processed using MaxQuant (16) by scientists at the Core Facility. Peptide level false discovery rate was set at 1%. Multiple sets of variable modifications and multiple protein databases were used depending on the goals and scopes of individual mass spectrometry experiments. A partial list of species and the corresponding numbers of peptides identified in this dataset is included in supplemental Table S1. More information on MaxQuant search settings can be found in Supplementary Methods. Importantly, all metadata have been removed to safeguard the identity of principal investigators and the details of their research projects.
From these 27 million PSMs, we constructed three individual training datasets: (1) WCU-MS-M, which consists of 25,174,942 MS/MS spectra that correspond to unmodified peptides and peptides containing oxidized Methionine, (2) WCU-MS-P, which consists of 26,943,975 MS/MS spectra that correspond to unmodified peptides, peptides containing oxidized Methionine, and peptides containing phosphorylated Serine, Threonine, or Tyrosine, and (3) WCU-MS-BEST, which consists of 1,239,045 MS/MS spectra that were assigned the highest quality scores by MaxQuant (the score column in evidence output file) for each unique unmodified peptide and charge state. There is no cutoff on the quality score. In other words, the WCU-MS-BEST data set contains the highest quality MS/MS spectrum for each unmodified peptide at each charge state.
We also acquired three external datasets for evaluating SMSNet's performance on peptides from diverse species and on MS/MS data from multiple laboratories. For direct comparison with DeepNovo, we downloaded 1,422,793 PSMs from 9 studies of distinct species (PRIDE accessions PXD005025, PXD004948, PXD004325, PXD004565, PXD004536, PXD004947, PXD003868, PXD004467, and PXD004424) that were previously curated by DeepNovo's authors (high-resolution spectra only). For evaluating SMSNet's ability to discover new peptides, we downloaded 83 raw files consisting of more than 3.5 million MS/MS spectra from an HLA peptidome study of mono-allelic cell lines (17) (MassIVE accession MSV000080527). Finally, for testing SMSNet's ability to identify phosphorylated peptides, we downloaded 12 raw files consisting of more than 676,000 MS/MS spectra from a comprehensive phosphoproteome study of control and epidermal growth factor-treated glioblastoma cells (18) (PRIDE accession PXD009227). All of these datasets were acquired on Q Exactive mass spectrometers with high-resolution MS/MS and HCD fragmentation method. High-quality MS/MS spectra of synthetic peptides were downloaded from the ProteomeTools HCD Spectral Library (19). It should be noted that this data set was acquired on Orbitrap Fusion Lumos mass spectrometer with high-resolution MS/MS and HCD fragmentation method.
Data Preprocessing
MS/MS spectra in the WCU-MS datasets were extracted from raw files and centroided using Thermo Fisher Scientific's MSFileReader version 3.0. MS/MS spectra in the HLA peptidome and phosphoproteome datasets were extracted from raw files into mgf format using ProteoWizard version 3.0.11133 (20) with the following filter parameters: Peak Picking = Vendor for MS1 and MS2, Zero Samples = Remove for MS2, MS Level = 2–2, and the default Title Maker. De-noising and de-isotoping of MS/MS spectra were not performed. Essentially, SMSNet model allows each MS/MS peak to be of any charge state during the analysis.
For inputting into SMSNet, MS/MS spectra were truncated at 5000 Da and the observed m/z were discretized at 0.1 Da and 0.01 Da resolutions to produce vector representations with length of 50,000 and 500,000, respectively. The lower resolution vector provides an overview of the spectrum for the encoder whereas the higher resolution vector is used by the candidate ion stack. The details of each component are described in the next section. MS/MS peak intensities were also normalized so that they sum to 1.0.
Model Architecture
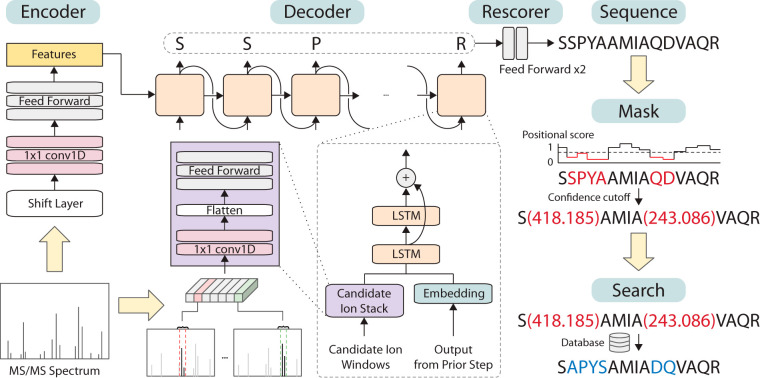
Inspired by DeepNovo, we developed our deep learning model focusing on integrating domain knowledge to create a specialized model for de novo peptide sequencing, which we called SMSNet. Both SMSNet and DeepNovo utilize the encoder-decoder architecture, a specialized module to extract only the relevant part of MS/MS spectra for consideration at each step (the ion-CNN in DeepNovo and the Candidate Ion Stack in SMSNet), and a knapsack approach for checking whether certain mass values can be explained by some amino acid combinations. However, SMSNet diverges from DeepNovo in two aspects: first, the shift layer in SMSNet's encoder module (Fig. 1) helps the model learn long-range relationships between MS/MS peaks whose mass difference matches to some amino acid, and second, SMSNet accepts the possibility that some peptide identifications cannot be fully resolved and thus includes extensive post-processing.
Fig. 1.
Overview of the Sequence-Mask-Search framework. SMSNet encodes the input MS/MS spectrum and passes the information to the decoder module which outputs amino acids sequentially. During the sequencing process, relevant m/z regions from the input MS/MS spectrum are extracted and fed to the decoder. Post-processing steps involve the adjustment of positional confidence scores, the replacement of low confidence positions by mass tags, and the recovery of exact amino acid sequences in masked segments through database search.
Overall, by viewing a peptide sequence as a list of amino acids, we can view the peptide sequencing problem as a problem of identifying amino acid one by one until termination. Let X be an input mass spectrum data, the model can be written as:
where yi's is the identified amino acid at position i, y0 is a special start token, and N is the peptide length. Our model consists of three main components: an encoder, a decoder, and an ion stack. In general, the encoder tries to capture an overview of the input mass spectrum and uses it to initialize the decoder. Then, conditioning on the prefix, the ion stack focuses on the relevant part of the spectrum and uses it to compute features for identifying the next amino acid. Finally, the decoder calculates probabilities for the next amino acid using its previous output and features from the ion stack. The model architecture is illustrated in Fig. 1. Every layer in the networks used rectified linear unit (ReLU) as the activation function unless specified otherwise.
Encoder
The encoder was designed to encode an overview of the input spectrum vector into a feature vector of size 1024 which will be used to initialize the hidden state and cell state of the decoder. To integrate the knowledge from the peptide fragmentation process into the model, we restructured the input to make it more likely for the encoder to capture the relationship between positions that could be used to determine amino acid presences. First, the input vector of length 50,000 was duplicated A times, where A is the number of possible amino acids, into a tensor of shape (50000, A). A is 21 when training on datasets with 20 amino acids plus oxidized Methionines and 24 when training on datasets with 20 amino acids plus oxidized Methionines and phosphorylated Serines, Threonines, and Tyrosines. Each copy of the original input vector is shifted to the left according to each amino acid mass, and then padded with zeros. For example, with the resolution of 0.1 Da, the vector representing Alanine is shifted to the left by floor(71.037 × 10) = 710. The first 710 values in the vector are discarded, and 710 zeros are padded to the right. Secondly, we created another vector of values from 0 to 49,999 to indicate the index of each positions on the spectrum, and then normalized it to have zero mean and unit variance. The index vector was then concatenated to the input to provide the information regarding the position, resulting in a tensor of shape (50000, A + 1).
The restructured input was then passed to the encoder neural networks consisting of three 1 × 1 convolution layers, followed by three fully connected layers. Each of the 1 × 1 convolution layers applies the same transformation to every input position separately and computes features along the second dimension of the input tensor. This forces the encoder to learn about the structure at each location. The three kernels have shape (1, 32), (1, 64), and (1, 2) that would produce a tensor of shape (50000, 32), (50000, 64), and (50000, 2) respectively after each layer. After that, the feature vector was flattened and passed through three fully connected layers with dimension 512, 512, and 1024, finally resulting in a vector of size 1024. For regularization, a dropout layer with dropout rate of 0.4 was used between the first and second fully connected layer.
Decoder
The decoder is a type of recurrent neural network that receives the feature vector from the encoder and uses it to generate a sequence of amino acids by outputting amino acids one by one. This is similar to the technique used in training neural networks for image captioning (21) or machine translation (12, 22, 23) where the input information (an image or a sentence in one language) is encoded into a vector representation, then passed to a decoder to generate the intended output (a caption or a sentence in a different language). Normally, the decoder for image captioning takes only the previously outputted word as input. In SMSNet, the decoder also takes as input a feature vector calculated by the candidate ion stack based on previous outputs for each step. This additional input was designed to provide the model more context about the next amino acid.
In the decoder, we used two layers of long short-term memory (LSTM) of size 512 with layer normalization (24) on top of each layer and a residual connection (25) around the second layer. The same encoded vector of length 1,024 was split into two halves and used as initial values for the hidden state and memory in both layers. At each step, the LSTMs take as input a vector of length 544, a concatenated vector between a feature vector of length 512 from the candidate ion stack and an embedding vector of size 32 of the previous amino acid. Then, the output from LSTMs is passed through a fully connected layer with a softmax activation function to produce probabilities for each amino acid. The shape of the last output depends on the number of possible amino acids (20, 21, or 24 depending on the number of modified amino acids considered).
The decoder always outputs one of the predefined modified or unmodified amino acids. Our current implementation does not allow gap in the output sequence at this stage but instead allows conversion of portions of output sequence into gaps in the post-processing stage described below.
Candidate Ion Stack
Given the total mass of the current prefix sequence, the candidate ion stack retrieves relevant m/z sections of the mass spectrum to compute a feature vector for the decoder. For each possible amino acid, 9 ion types were considered: a, b, b(2+), b-H2O, b-NH3, y, y(2+), y-H2O, and y-NH3. Suppose there are 21 different amino acids, for each of the 9 ions, we sliced a small window of size 0.2 Da (corresponding to 20 elements at 0.01 resolution) from the original input vector of size 500,000, resulting in 189 20-element vectors. These vectors were stacked together to form an input of shape (189, 20). It should be noted that because of high computational cost for checking whether a neutral loss is expected based on amino acid composition of an ion, our current implementation allows both NH3 and H2O neutral losses on all b-ions and y-ions. Additionally, the current implementation does not identify nor make use of charge state information for each MS/MS peak. Peak intensities were not explicitly included in the scoring as we expect the model to learn to distinguish signals from noises in a purely data-driven manner.
The candidate ion stack consists of two 1 × 1 convolution layers followed by two fully-connected layers. The idea is to force the model to first learn the peak patterns of each ion, then learn the relationship between ions based on the calculated features. The two 1 × 1 convolution layers have 32 and 64 filters respectively, whereas both fully connected layers have 512 dimensions. The output feature tensor was then used as input for the decoder.
Inference
During inference, we used beam search with beam size of 20 to explore and find the most likely sequence of amino acids. At each step, all remaining hypotheses are ranked by the following formulas (26):
where P(Y | X) is the product of the previously identified amino acid probabilities. The length penalty term is used to compensate for the fact that longer sequences usually have lower score values than the shorter ones. Additionally, during each step, we filtered out hypotheses where the difference between current prefix mass and the precursor mass did not match any possible amino acid combinations using the knapsack search algorithm.
The beam search decoding would continue until a special ending token is produced or a maximum length of 50 is reached for every remaining beam. After the decoding process ended, the amino acid sequence with the best score according to the provided formulas was selected as the final output. If the correct peptide contains residues with unexpected modifications, the beam search process has the capability to replace those sections of the peptide sequence with isobaric combinations of predefined residues, if ones exist, and proceed to identify the correct amino acid sequences in the remaining sections of the peptide. The incorrect isobaric sections should have low confidence scores and subsequently be masked in the post-processing stage described below. It should be noted that if the appropriate sequence database and amino acid modifications are considered, these peptides can be correctly identified during the post-processing stage.
Training, Validation, and Test Sets Partitioning
To ensure that training, validation, and testing sets do not share a common peptide, we first partitioned unique peptides into three sets, then constructed training, validation, and testing sets from mass spectrum data associated with these peptides. Accounting for the fact that some peptides appear in the datasets much more often than the others, we kept only one random data entry per peptide in validation and testing sets. The validation set was used for choosing the model architecture, determining the number of training steps, as well as setting all hyper-parameters. For WCU-MS-M and WCU-MS-P, we used validation and test sets of size 50,000. During training, peptides longer than 30 amino acids were ignored. Details of data set sizes can be found in supplemental Table S2.
Model Training
We modeled the peptide sequencing task as a series of amino acid identifications where each identification is a multi-class classification problem. We chose the focal loss (27), which is a dynamically scaled cross-entropy loss, as a loss function for our model. For binary classification tasks, the focal loss is defined as:
where pt = p for the class with y = 1 and pt = 1- p otherwise, p is the model's estimated probability for the class with label y = 1, and α and γ are hyper-parameters for balancing the importance of positive/negative examples and easy/hard examples, respectively. We set α to 0.25 and γ to 1.0 as it performed best on the validation set. The focal loss is chosen instead of normal cross-entropy loss because we suspected that there is an imbalance between easy examples with complete mass spectrum evidence and hard examples with missing peaks.
To extend the focal loss to multi-class classification, we can view a multi-class classification problem as many binary classification problems. Concretely, we can pass the output of the last layer of the model through multiple sigmoid functions to obtain binary probabilities of being each class, then use the provided formula to calculate the focal loss. The loss function is thus the summation of the focal loss of each class:
where N is the number of possible amino acid classes, pt(k) is pt of class k. The values of α and γ are the same for every class. For inference, the sigmoid function was substituted with a softmax function to compute probability scores which sum to 1.
We initialized all parameters by drawing from a uniform distribution between −0.1 and 0.1, and trained the model using stochastic gradient descent with learning rate decay. An initial learning rate of 0.01 was used until two-thirds of the maximum training step. Afterward, the learning rate was halved every one-twelfth of the maximum training steps. The gradient of the loss was normalized so that its L2-norm was less than or equal to 5. With a batch size of 32, the models were trained for 4,000,000 steps on WCU-MS-M and WCU-MS-P datasets, which took roughly one month on Nvidia GeForce GTX 1080 Ti.
Ablation Study
To evaluate the impact of each component to the performance of SMSNet, we performed ablation studies by making some modifications to the model, and then measuring the performance degradation caused by those modifications. The following modifications were tested:
Not using layer normalization after LSTM layers in the decoder
Using normal cross-entropy loss instead of focal loss
Not considering neutral losses, b-H2O, b-NH3, y-H2O, and y-NH3 ions, in the candidate ion stack.
Removing the shift mechanism in the encoder. In this variation, we removed the 1 × 1 convolution layers and fed the low-resolution input vector of size 50,000 directly to the fully-connected layer.
Removing the encoder entirely and initializing the decoder with a vector of zeros
Every modified model was trained for 20 epochs from new initialization on the WCU-MS-BEST dataset. This is the same number of epochs that was used to train the main model until convergence.
Data Preprocessing for the Rescorer
Unlike the main model, the rescorer model operates solely on the level of amino acids. For each hypothesized amino acid, it predicts the confidence level of the identification. The following features were used: peptide length, numbers of amino acids with probability more than 0.7, 0.8, and 0.9, a geometric mean of amino acid probabilities in the peptide, the position of the amino acid normalized by the peptide length, probabilities of amino acids at index t - 1 to t + 2 for current index t. We chose these features on the basis that they are not taken into consideration by the main model during the decoding process, and they gave the lowest loss on the validation set. The label for each data point is 1 if the given de novo amino acid matches the true label and 0 otherwise.
As the rescorer is designed to evaluate amino acids labels identified by the main model, we could not use the original training set that the main model was trained on. Therefore, the original validation set was split into rescorer training and validation sets with ratio of a 90:10. The test set were still the same for both tasks. It should be noted that because the rescorer module is trained at amino acid level, each peptide-spectrum match in the validation set gives rise to multiple data points for training this module. In fact, there are more than 700,000 data points for training the rescorer module in most cases (supplemental Table S3).
Rescorer
We designed the post-processing model to be a shallow neural network consisting of two fully connected layers of size 64. To train the model, we used binary cross-entropy loss and the Adam optimizer with default parameters of β1 = 0.9 and β2 = 0.999. The rescoring validation set was used for early stopping.
Database Search for Resolving Ambiguous Identifications
To determine the exact amino acid sequences for identifications with mass tags or ambiguous Leucine/Isoleucine positions, we search for possible matches within a given protein sequence database. For analyzing HLA peptidome and human phosphoproteome dataset, the UniProt (28) reference human proteome was used (downloaded April 2019). For evaluating whether each of SMSNet's identifications could be matched to a unique possibility, the UniProt reference proteome for either human, mouse (Mus musculus), budding yeast (Saccharomyces cerevisiae), Escherichia coli strain K12, or a combination of all four species was used (downloaded April 2019). All databases include isoforms and predicted proteins. An amino acid sequence within the database is considered a match to an ambiguous identification if (1) all non-Isoleucine positions in both sequences match, (2) all Isoleucines in the identification match to either Leucine or Isoleucine in the database sequence, and (3) all mass tags in the identification match to amino acid substrings in the database sequence whose weights differ less than 20 ppm from the corresponding mass tags. Only amino acid modifications recognized by the corresponding SMSNet model were considered during the search. For example, when evaluating the SMSNet model that can identify phosphorylated Serine, every Serine in the database is allowed to be in either the unmodified form or the phosphorylated form.
Performance Comparison with DeepNovo
To compare the performance of our model with DeepNovo, we trained both DeepNovo and SMSNet on three data sets, one from nine species used in Deep-Novo, one from our new data set, and one from ProteomeTools spectral library. Both models used the same training, validation, and test sets. For DeepNovo, we used the code provided together with their publication.
The first data set is constructed by combining all high-resolution data sets in DeepNovo publication. As we only focused on amino acid with Methionine-oxidation, any peptide that contains deamidated Asparagine or Glutamine in the original data set was discarded. The remaining data consist of 1,422,793 mass spectra from 256,200 unique peptides. Because of its lower number of unique peptides, instead of using 50,000 unique peptides as validation and testing sets as in other experiments, we sampled only 20,000 unique peptides from the data set and used all of their associated spectra, resulting in validation and test sets of size 111,365 and 112,995, respectively.
The second data set, called WCU-MS-BEST, is a subset of WCU-MS-M data set that contained only peptide with no amino acid modification. We selected only the spectra with the best quality score according to MaxQuant for each unique peptide and charge state to form an easy but diverse dataset. In total, there are 1,239,045 spectra of 869,206 unique peptides. The validation and test set each contains 50,000 unique peptide spectra (two spectra were later removed from the test set because of mismatches between their precursor masses and the labels, resulting in the test set of size 49,998). For peptides with many charge states, we randomly chose one charge state and discarded the rest.
The third data set corresponds to the high-quality spectral library of 202,648 unique synthetic peptides. Because of its low number of unique peptides and spectra, we sampled only 20,000 unique peptides each to create the validation and test sets. The summary of all data sets can be found in supplemental Table S2.
The amino acid vocabulary was set according to the data set for both models, with 21 possible amino acids for the first data set, and 20 for the second data set, and 21 for the third data set. Apart from the amino acid vocabulary, our model settings were the same as in other experiments. For DeepNovo, we set the spectrum resolution to 0.02 Da and kept other default parameters. At inference time, both models used beam search with beam size 20 to find the most probable peptide for each input.
Definition of Amino Acid's Evidence
Given a mass spectrum, we determined that an amino acid has supporting evidence if it follows our defined criteria. First, for an amino acid with mass Maa and prefix mass Mprefix, there must be ions with mass Mprefix and Mprefix + Maa present in the spectrum. Secondly, a fragmented ion is said to be present in the spectrum if there is at least a peak with any intensity within 0.1 Da of its theoretical a, b, b-NH3, b-H2O, b(2+), y, y-NH3, y-H2O, or y(2+) ions. The first and last amino acids in a peptide only require one ion mass presence.
Evaluation of Newly Identified HLA Antigens
Because the HLA peptidome data sets considered here (17) were derived from cell cultures that contain fetal bovine serum and immunoprecipitated using protein G, several MS/MS spectra may correspond to these contaminants. We searched SMSNet's identifications against a combined database of UniProt reference human proteome (downloaded April 2019), UniProt reference bovine proteome (downloaded August 2019), and Streptococcus Immunoglobulin G-binding protein G (UniProt accessions: P19909 and P06654). Isoforms and predicted proteins are included. This revealed that the overwhelming majority (57,394 out of 58,565) of identified peptides are from human, and only 1080 and 91 peptides are from bovine and G protein, respectively. For the comparison to Spectrum Mill and all subsequent analyses, only peptides mapped to human proteins are considered.
As most HLA antigens are non-tryptic whereas most peptides in SMSNet's training data sets are tryptic, there may be some systematic errors in SMSNet's identified peptides. One possibility is that SMSNet mis-assigned b-ions as y-ions and vice versa. If this is the case, among identifications where SMSNet disagrees with prior study, we expected peptides identified by prior study to be more like the reversed sequences of SMSNet's identifications than to SMSNet's identifications. However, the Levenshtein distances between peptides identified by prior study and SMSNet's identifications are significantly lower than the distances between peptides identified by prior study and the reversed sequences of SMSNet's identifications (mean distances are 5.27 and 8.23, respectively, Wilcoxon sign test p value = 2.64e-31). Overall, SMSNet appears to be able to identify non-tryptic peptides.
To check the novelty of newly identified antigens, we downloaded the latest list of known HLA antigens from the Immune Epitope Database (IEDB) (downloaded April 2019) and searched whether peptide-HLA pairs that were identified by SMSNet have been previously reported. It should be noted that negative entries and entries with ambiguous HLA allele names in the IEDB database were excluded from consideration. To verify whether newly identified antigens possess the expected properties, NetMHCPan version 4.0 (29) was used to predict the binding affinity (in percentile rank) and the 9-residue core binding motif for each identified peptide-HLA pair. The profiles of allele-specific core binding motifs were visualized using WebLogo (30).
Determination of the Origins of Identified HLA Antigens
We iteratively determined the origins of HLA antigens identified by SMSNet by (1) searching all identified amino acid sequences against a reference human proteome from UniProt (isoforms and predicted proteins included, downloaded April 2019), (2) searching the amino acid sequences without any hit in the first step against hypothetical open reading frames extracted from published RNA sequencing data of the cell lines used (17) and from reference human non-coding RNAs downloaded from RefSeq (GRCh38) (31), and (3) searching the amino acid sequences without any hit in the two prior steps against hypothetical spliced peptides generated by joining two peptides from the same protein. RNA sequencing data were aligned to the GRCh38 human reference genome using HISAT2 (32). Sequence variants were called using GATK version 4 (33). Reference non-coding RNAs were extracted from GRCh38's reference transcriptome based on the “ncRNA,” “non-coding RNA,” and “long non-coding RNA” tags. Hypothetical spliced peptides were generated by joining two peptides, each with length at least 3, from non-overlapping regions of the same protein.
Evaluation of Newly Identified Phosphopeptides
We compared SMSNet's identification for each MS/MS spectrum in the dataset to previously reported result (18) by considering only the actual amino acid sequences without any modification. This is because many predicted peptides contain multiple Serine, Threonine, and Tyrosine residues located near each other and so it is often difficult to pinpoint the exact location of phosphorylation(s). To evaluate the contribution of SMSNet to human phosphoproteome study, we appended newly identified amino acid sequences to a reference human proteome downloaded from UniProt (isoforms and predicted proteins included, downloaded April 2019) and reanalyzed raw mass spectrometry data using MaxQuant. MaxQuant analysis settings were set as described in the prior study (18). Namely, variable modifications include Oxidation (M), Acetyl (protein N-term), and Phosphorylation (STY). Enzyme specificity was set as Trypsin/P with 2 maximum missed cleavages. Fixed modification includes only Carbamidomethyl (C). Precursor mass tolerances was set at 4.5 ppm for the main search and MS/MS mass tolerance was set at 20 ppm. Other settings were left as default. The Match Between Runs and Second Peptide search functionalities were disabled as we want to focus on whether newly identified amino acid sequences would be selected by MaxQuant as the main identification for previously unannotated MS/MS spectra. False discovery rates were set at 1% for both PSM and protein levels. Minimum peptide length was set at 7. Newly identified phosphopeptides from the re-analysis that were not reported in prior study were searched against the PhosphoSitePlus database (34) (downloaded April 2019). Annotated spectra for newly identified phosphopeptides were produced using PDV version 1.5.3 (35) with MS/MS mass tolerance of 20 ppm and are available on Figshare (DOI: 10.6084/m9.figshare.9784814).
RESULTS
SMSNet Model Training
We acquired a large collection of >26 million anonymized MS/MS spectra from the Proteomics and Metabolomics Facility at The Wistar Institute for developing SMSNet. This dataset consists of about 1 million unique peptides from diverse species (supplemental Table S1). Four versions of SMSNet were developed based on distinct training datasets for evaluation on external data sets and comparison with DeepNovo. To compare with DeepNovo, we trained both SMSNet and DeepNovo on a high-resolution MS/MS data set curated by DeepNovo's authors (1,422,793 spectra) and on a collection of 1,239,045 highest quality spectra each representing a peptide in our dataset (named WCU-MS-BEST, see Experimental Procedures). For further evaluations, SMSNet was trained on a set of 25,174,942 spectra of unmodified peptides and peptides containing oxidized methionine (named WCU-MS-M) and on spectra in WCU-MS-M dataset plus additional 1,769,033 spectra of peptides containing phosphorylated serine, threonine, or tyrosine (named WCU-MS-P).
SMSNet employs the encoder-decoder architecture which has been widely used in machine translation to identify amino acid sequence from input MS/MS spectra sequentially from the N terminus to the C terminus of the peptide (Fig. 1). The encoder embeds the input MS/MS spectrum into a fixed-length vector representation through multiple 1D-convolutional and feed forward layers. The decoder, consisting of long short-term memory (LSTM) layers, predicts the likelihood distribution of the next amino acid based on the current state of the model and evidence from the corresponding m/z regions in the input spectrum (the candidate ion windows in Fig. 1). Once the entire amino acid sequence has been identified, SMSNet adjusts the confidence score for each position in the identification through feed forward layers (the rescorer in Fig. 1). These steps comprise the sequence phase of the Sequence-Mask-Search framework. Then, during the mask phase, identified amino acid positions whose confidence scores lie below a user-specified cutoff were replaced by mass tags that reflect their combined masses. Here, we set the confidence score cutoff so that the false discovery rate at amino acid level is less than 5%. Finally, during the search phase, SMSNet attempts to recover the exact amino acid sequences from masked positions by searching all identifications against a reference amino acid sequence database.
Performance Evaluation On Held-out MS/MS Data
We evaluated SMSNet's performance against DeepNovo by training and testing both tools on the same high-resolution MS/MS spectra from three separate datasets (see Experimental Procedures). In each test, MS/MS spectra were split so that the train and test sets contain different peptides (supplemental Table S2). At each threshold on the positional confidence score, an identified amino acid whose score is above the threshold is considered correct only if its mass and prefix mass (the combined mass of earlier amino acid positions in the sequence) differs less than 0.0001 Da and 0.03 Da from the ground truth, respectively. Identified amino acids whose scores are lower than the threshold are considered in the calculation of recall but excluded from the calculation of accuracy. At the peptide level, an identified peptide is considered correct only if all of its amino acid positions whose scores are higher than the threshold are correct and that it contains at least 4 identified amino acid positions. Peptides with less than 4 identified amino acid positions after applying the score threshold are removed from consideration. To provide fair comparisons, because DeepNovo does not perform post-processing, the performance of SMSNet with and without re-scoring (the outputs prior to and after the Rescorer module in Fig. 1) are considered together here.
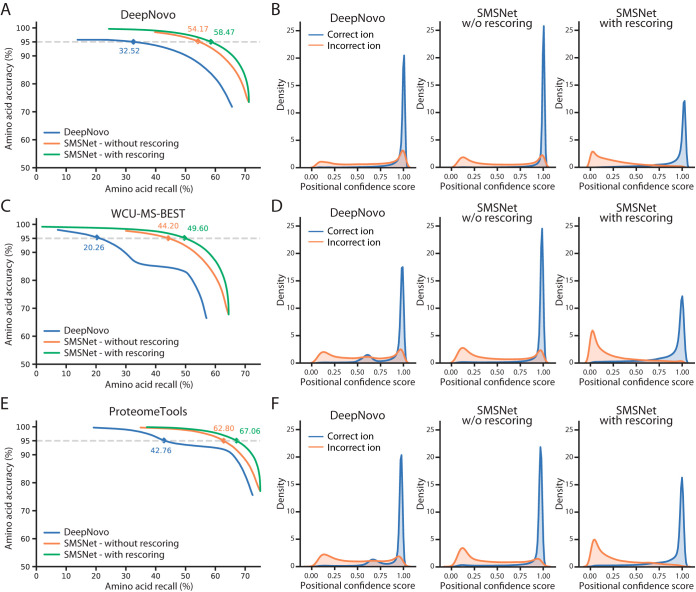
SMSNet consistently outperforms DeepNovo, achieving 21.65% higher amino acid recall than DeepNovo at 5% amino acid false discovery rates on the dataset curated by DeepNovo's authors (Fig. 2A). If all identified amino acids are considered, SMSNet achieves 71.24% amino acid recall and 47.11% peptide recall whereas DeepNovo achieves 65.57% and 44.41% recall, respectively. These differences resulted from the fact that SMSNet's output positional confidence scores are slightly better at distinguishing between correct and incorrect positions than DeepNovo's do (Fig. 2B). Similar difference in performance was observed when both tools were evaluated on our WCU-MS-BEST dataset (Fig. 2C). Here, if all identified amino acids are considered, SMSNet achieves 44.73% amino acid recall and 64.45% peptide recall whereas DeepNovo achieves 37.57% and 57.02% recall, respectively. Interestingly, the distribution of confidence scores for the correctly identified positions produced by DeepNovo became bi-modal with a new mode at around 0.6 in this latter test (Fig. 2D) whereas SMSNet's outputs remain unaffected. We suspect that DeepNovo's model may have overfit to some coincidental patterns in this dataset. Similar results could be observed when DeepNovo and SMSNet were evaluated on high-quality MS/MS spectra of synthetic peptides acquired from ProteomeTools database (Fig. 2E–2F).
Fig. 2.
SMSNet outperforms state-of-the-art de novo peptide sequencing tool. To provide fair comparisons, SMSNet and DeepNovo were retrained using the same high-resolution MS/MS spectra from the indicated datasets. Furthermore, as DeepNovo does not include post-processing, the performance of SMSNet with and without re-scoring are shown together here. A, Amino acid-level performances for SMSNet and DeepNovo when evaluated on the dataset curated by DeepNovo's authors. The corresponding recalls at 5% amino acid false discovery rate are indicated. B, Histograms showing the distributions of positional confidence scores produced by SMSNet and DeepNovo when evaluated on the dataset curated by DeepNovo's authors. C–D, Similar plots showing performances of SMSNet and DeepNovo when evaluated on our WCU-MS-BEST dataset. E–F, Similar plots showing performances of SMSNet and DeepNovo when evaluated on high-quality MS/MS spectra of synthetic peptides acquired from ProteomeTools database.
We also evaluated the contribution of each key component of SMSNet's architecture by removing individual component from the model, retraining the model, and determining the degradation in performance (Table I). This reveals that the normalization of recurrent neural network layers (24) and the use of focal loss (27), which conceptually makes the model gives higher weights to data points that are difficult to fit, contribute up to 1.32% to the recall at peptide level and 0.66% at amino acid level. In comparison, the inclusion of neutral loss ions to the model, which is one of the standard considerations in any peptide sequencing approaches, contribute 1.72% to the recall at peptide level and 1.45% at the amino acid level. Interestingly, the shift layer alone (Fig. 1) is almost as important as the whole encoder architecture to the performance (4.50% versus 4.67% contribution to the recall at peptide level and 2.55% versus 2.64% at amino acid level).
Table I. Ablation analysis of SMSNet's main components. Percentages are shown.
| Components | Peptide recall (%) | Difference | Amino acid recall (%) | Difference |
|---|---|---|---|---|
| Final architecture | 44.73 | – | 64.45 | – |
| Without layer normalization | 44.10 | −0.63 | 63.89 | −0.56 |
| Cross-entropy loss instead of focal loss | 43.41 | −1.32 | 63.79 | −0.66 |
| Not considering neutral loss ions | 43.01 | −1.72 | 63.00 | −1.45 |
| Without shift layer in encoder | 40.23 | −4.50 | 61.90 | −2.55 |
| Without the entrie encoder | 40.06 | −4.67 | 61.81 | −2.64 |
Impact of the Sequence-Mask-Search Framework
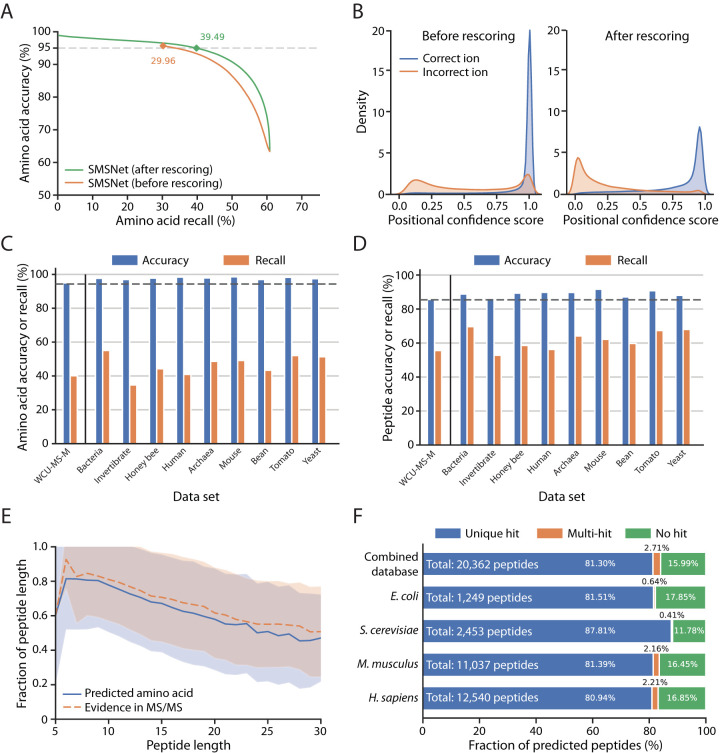
By design, the underlying architecture of SMSNet, which sequentially output the likelihood of the next amino acid based on the model's current state, assumes that the identifications for prior positions are correct and is unable to adjust the confidence scores of prior positions even if later identifications drastically change the context of the sequence. Thus, we trained a neural network module to adjust the resulting positional confidence score based on the information of the whole output amino acid sequence (the rescorer in Fig. 1, see Experimental Procedures). The objective of this rescorer is to maximize the separation in confidence score between correctly identified and incorrectly identified amino acid positions. This post-processing step improves the recalls at amino acid level by 9.53% and 9.08% when SMSNet was evaluated on the WCU-MS-M and WCU-MS-P datasets, respectively (Fig. 3A–3B and supplemental Fig. S1).
Fig. 3.
The Sequence-Mask-Search framework significantly improves de novo peptide sequencing accuracy. The WCU-MS-M dataset was used to train and evaluate the performance of SMSNet here. A, Amino acid-level performances for SMSNet before and after the positional confidence score adjustment step. The corresponding recalls at 5% amino acid false discovery rate are indicated. B, Histograms showing the distributions of positional confidence scores produced by SMSNet before and after score adjustment. C, Bar plots showing amino acid-level accuracies and recalls of SMSNet on a test set derived from WCU-MS-M and on MS/MS spectra of nine species that comprise the dataset curated by DeepNovo's authors. The threshold on positional confidence score was selected so that 5% amino acid false discovery rate was achieved on the WCU-MS-M test set (the leftmost bars). Dashed line indicates the expected 95% accuracy based on the applied score threshold. D, Similar bar plots showing the results at peptide-level. Dashed line indicates the expected accuracy level based on the applied score threshold. E, Line plots comparing the fraction of identified amino acid positions that pass the same score threshold used in C–D in peptides of various lengths (blue line) to the fraction of amino acid positions that can be definitely determined based on observed ions in the MS/MS spectra (orange dashed line). Shaded area indicates the ±1 standard deviation range. F, Stacked bar plots showing the fraction of identified peptides that could be matched to various protein sequence databases. Amino acid sequence database for each species was downloaded from UniProt (see Experimental Procedures). Combined database integrates amino acid sequences from all four species considered. In each bar, only identifications whose ground truths exist within the corresponding database were counted. “Unique hit” means that the identified sequence matches to exactly one possibility in the database. “Multi-hit” means that the identified sequence matches to multiple possibilities. “No hit” means the identified sequence does not match to anything in the database.
We examined the impact of masking identified positions with low confidence scores on the performance of SMSNet and whether the correct amino acid for each masked position could be recovered by searching against a reference amino acid sequence database. The key concern here is that in order to achieve high accuracy, so many amino acid positions may be masked that the resulting identified peptides are no longer informative. Here, we trained SMSNet and the rescorer using the WCU-MS-M dataset, identified the adjusted confidence score threshold that corresponds to 5% amino acid false discovery rate on this dataset, and then determined the accuracy and recall achieved by SMSNet at the same threshold on the dataset curated by DeepNovo's authors. This revealed that SMSNet with the rescore module performs consistently on MS/MS spectra from diverse species and laboratories at both amino acid and peptide levels (Fig. 3C–3D). Furthermore, the amount of masked amino acid positions at 5% false discovery rate threshold closely matches the actual number of amino acid positions whose corresponding ions are missing from the corresponding MS/MS spectra (Fig. 3E). In other words, SMSNet did not apply too many masks more than the amount required to cover all missing data.
Although the masking step effectively improves the accuracy of SMSNet without too much sacrifice in recall, most uses of proteomics and peptidomics data require fully identified amino acid sequences where mass tags and the Leucine/Isoleucine ambiguity have been resolved. Therefore, we explored whether the correct amino acids that correspond to masked positions could be recovered if an appropriate amino acid sequence database is provided. There are three possible outcomes here. If the identified peptide is incorrect or contain unexpected amino acid sequence, then there would be no match in the database. On the other hand, if the identified peptide is correct, but too many masks were introduced, then there may be multiple matches with distinct sequences in the database. Finally, if the identified peptide is correct and contains a small number of masked positions, a unique sequence hit could be recovered from the database. Our results show that the correct amino acid sequences could be unambiguously recovered for more than 80% of SMSNet's identifications, even when amino acid sequences from multiple species were used at once (Fig. 3F).
SMSNet Discovers More Than 10,000 New HLA Antigens
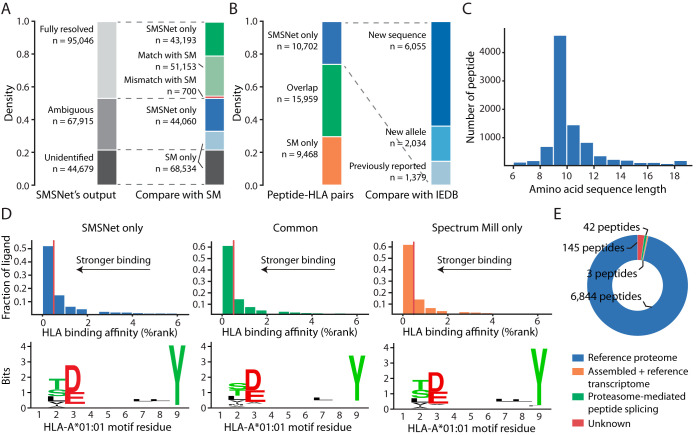
To evaluate the utility of SMSNet on real-life mass spectrometry dataset that contains MS/MS spectra of unexpected peptides, we analyzed a large-scale HLA class I peptidome dataset of mono-allelic human B lymphoblastoid cell lines (17) which consists of more than 35 million MS/MS spectra. The SMSNet model trained on WCU-MS-M dataset was used here because most peptides should not be post-translationally modified. At 5% amino acid level false discovery rate, SMSNet made full-sequence identifications for 95,046 MS/MS spectra, 43,193 of which were not annotated by the prior study. SMSNet's identifications also matched prior study on 51,153 out of 51,853 MS/MS spectra (Fig. 4A, 98.65% agreement rate). Even though SMSNet was trained primarily using tryptic peptides, it was able to produce highly concordant identifications compared with database search approach and there was no systematic error, such as reversal of sequence because of misidentification of b-ions as y-ions and vice versa, among 213 mismatches between SMSNet and Spectrum Mill (see Experimental Procedures). SMSNet uncovered 10,702 unreported peptide-HLA pairs, 8089 of which are new antigens according to the Immune Epitope Database (Fig. 4B). Newly identified antigens are of the right lengths (8–12 amino acids, Fig. 4C), predicted to bind strongly to their corresponding HLA molecules, and contain the expected core binding motifs (Fig. 4D and supplemental Fig. S2). Altogether, these evidences strongly suggest that SMSNet's identifications are true HLA class I antigens.
Fig. 4.
SMSNet uncovers many new HLA antigens. A, Stacked bar plots showing the numbers of MS/MS spectra identified by SMSNet and the overlaps between SMSNet and prior study (17), which utilized Spectrum Mill (SM) software (Agilent Technologies, Inc., Santa Clara, CA) for MS/MS data interpretation. B, Stacked bar plots showing the overlap between peptide-HLA pairs identified by SMSNet and prior study or the Immune Epitope Database (IEDB). “New sequence” indicates that the identified peptide contains amino acid sequence that has not been reported as antigen for any specific HLA allele in IEDB. “New allele” indicates that the identified sequence has been reported to bind to HLA alleles other than the ones considered here. There are 7034 distinct peptides among 10,702 newly identified peptide-HLA pairs. C, The length distribution of 7034 peptides newly identified by SMSNet. D, Histograms and sequence logos comparing the predicted binding affinities and core sequence motifs between peptide-HLA pairs identified by only SMSNet (left), by both SMSNet and prior study (middle), or by prior study only (right). Binding affinities and core motifs were predicted using NetMHCPan. Vertical red lines designate the 0.5% rank threshold typically used to select strong binders. E, Pie chart showing the origins of 7,034 peptides newly identified by SMSNet. Peptide sources were determined by searching amino acid sequences against human proteome, transcriptome, and a database of theoretically possible spliced peptides (see Experimental Procedures).
Additionally, as recent reports indicated that a fraction of HLA antigens may originate from proteasome-mediated peptide splicing (2–4) and non-coding regions of the genome (5), we explored whether SMSNet discovered any new antigen whose amino acid sequence does not match known human proteins in the UniProt database. Among 7034 peptides newly identified by SMSNet, 6844 peptides (97.30%) were mapped to known proteins and only a handful were mapped to non-coding RNAs or explained as possible products of proteasome-mediated peptide splicing (Fig. 4E). The high proportion of matches to known proteins here is expected because the majority of SMSNet's identifications contain mass tags and ambiguous Leucine/Isoleucine residues that must be resolved through searching against reference database. If we also include 68,159 MS/MS spectra with partial sequence identifications (Ambiguous in Fig. 4A) in this analysis, then the number of peptides that could be mapped to non-coding RNAs or explained as possible products of proteasome-mediated splicing rose to 592 and 1154, respectively. Compared to the number of peptides mapped to known protein, the small number of potential spliced peptides identified here is in concordance with recent analysis of HLA peptidomes which attributed around 2–6% of identified antigens as spliced peptides (4). Overall, we estimated that as many as 43.60% of all unique identifications made by SMSNet still remain unresolved. All of SMSNet's fully resolved and partial identifications are available as Supplementary Excel Table.
SMSNet Improves the Coverage of Phosphoproteome
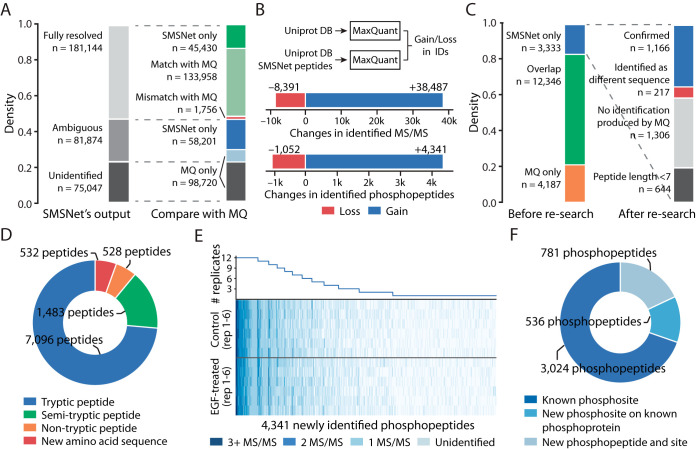
Lastly, we evaluated the power of SMSNet to identify post-translationally modified peptides by analyzing a phosphoproteome dataset of control and epidermal growth factor (EGF)-treated glioblastoma cells (18) that was previously analyzed with MaxQuant. Phosphorylation was selected because of its importance in biology and because our WCU-MS-P dataset contains a sizeable number of phosphorylated peptides (1,769,033 MS/MS spectra) for the model to learn. At 5% amino acid level false discovery rate, SMSNet made full-sequence identifications for 181,144 MS/MS spectra, 133,958 of which are in agreement with previous study (Fig. 5A, see Experimental Procedures). Further, SMSNet assigned the same phosphorylation sites as previous study in 78,609 out of 81,440 sites (96.52%) on 68,344 phosphopeptides which contain multiple Serine, Threonine, and Tyrosine residues.
Fig. 5.
SMSNet improves the coverage of human phosphoproteome. A, Stacked bar plots showing the numbers of MS/MS spectra identified by SMSNet and the overlaps between SMSNet and prior study (18). B, The gains and losses in number of identified MS/MS spectra and phosphopeptides after adding SMSNet's identifications to the human proteome database and re-analyzing with MaxQuant (see Experimental Procedures). C, Stacked bar plots showing the numbers of phosphopeptides identified by MaxQuant and SMSNet and the numbers of new phosphopeptides that could be confirmed by re-analyzing SMSNet's identifications with MaxQuant. Peptides shorter than 7 amino acids were not considered during the search. “No identification produced by MQ” indicates that none of the MS/MS spectra of a phosphopeptide were identified as peptides by MaxQuant. “Identified as different sequence” indicates that none of the MS/MS spectra of a phosphopeptide were confirmed by MaxQuant as the same identification produced by SMSNet and at least one MS/MS spectrum was identified as a different peptide. D, Pie chart showing the composition of all newly identified peptides gained by adding SMSNet's identifications to MaxQuant's search. This includes phosphopeptide and non-phosphopeptide identifications from all MS/MS spectra that were not previously identified by prior study. It should be noted that because MaxQuant searches were performed with full-tryptic enzyme specificity, all identifications of semi-tryptic peptides, non-tryptic peptides, and peptides with new amino acid sequences were possible because of sequences supplied by SMSNet. E, Heatmap and line plot showing the reproducibility of 4,341 newly identified phosphopeptides after re-analysis using MaxQuant across 6 control and 6 epidermal growth factor (EGF)-treated replicates. Each row in the heatmap corresponds to one mass spectrometry experiment. F, Pie chart showing the overlap between newly identified phosphopeptides and known phosphoproteins and phosphosites in the PhosphoSitePlus database. An identified phosphopeptide was counted as “Known phosphosites” only if all identified phosphorylation sites on that peptide are reported in the database. Identified phosphopeptides that contain unreported phosphosites were grouped based on whether they could be mapped to known phosphoproteins in the database.
Next, we appended SMSNet's identifications to a human protein database and re-searched the MS/MS spectra using MaxQuant. This results in a net gain of 30,096 identified MS/MS spectra and 3,289 identified phosphopeptides over previous study's result (Fig. 5B). Among 3333 new phosphopeptides identified by SMSNet, 1166 were confirmed by MaxQuant whereas only 217 resulted in conflicting identifications (Fig. 5C). 644 phosphopeptides were shorter than 7 amino acids and out of scope of the MaxQuant analysis. Overall, by supplementing MaxQuant's search with both phosphopeptide and non-phosphopeptide sequences from SMSNet, we were able to gain 532 peptides with new amino acid sequences and 2001 semi-tryptic and non-tryptic peptides that would not be discovered in typical full-tryptic search (Fig. 5C). Furthermore, the majority of newly identified phosphopeptides could be observed in multiple replicate samples (Fig. 5D) and mapped to known phosphosites and phosphoproteins in the PhosphoSitePlus database (Fig. 5E). All SMSNet's fully-resolved and partial identifications are available as Supplementary Excel Table.
DISCUSSION
SMSNet incorporates several recent computer vision and machine translation techniques (24–27) as well as key considerations from earlier de novo peptide sequencing approaches (6, 14), all of which contribute to its strong performance. The structure of the recurrent neural network in the decoder module was improved from DeepNovo's design so that more of the relevant information are exposed to the network. To better capture the notion that some amino acid positions are easy to determine from evidences in MS/MS spectra whereas some could be extremely difficult to do so, we implemented the focal loss (27). Coincidentally, this feature has also been introduced in a later version of DeepNovo (10). Interestingly, one of the most impactful features in SMSNet turns out to be the shift layer (Fig. 1), which was implemented to help the encoder module detect pairs of MS/MS peaks whose mass differences match to some amino acids. When we removed this layer, the performance of SMSNet dropped by almost as much as removing the whole encoder module. This indicates that incorporating domain-specific knowledge is highly critical for handling complex data generated in the field of biotechnology.
Generalizability is always a major concern in any machine learning study. We have shown that even though SMSNet was trained primarily on MS/MS data of tryptic peptides (>95% of peptides in the WCU-MS dataset end with a Lysine or an Arginine) that were acquired in a single laboratory, the model consistently performs well on MS/MS data from diverse species and laboratories (Fig. 3C–3D) and is able to identify a large number of HLA antigens that end with non-Lysine/non-Arginine residues. Therefore, SMSNet should be able to identify any peptide regardless of the protease enzyme used. Additionally, although our evaluations of SMSNet mainly involved MS/MS spectra from Q Exactive mass spectrometers with higher-energy collisional dissociation (HCD), it is possible to adapt the framework of SMSNet for data from different mass spectrometers and peptide fragmentation methods. Part of SMSNet's capability to adapt to new MS/MS dataset can be seen through its high performance on MS/MS spectra acquired on Orbitrap Fusion Lumos (19) (Fig. 2E).
One limitation of SMSNet is that the effectiveness of the final search step, which attempts to recover unique amino acid possibilities for the masked positions, depends on the quality and completeness of the database provided. Our result shows that multiple sequence databases could be given to SMSNet at once without apparent deterioration in performance (the combined database in Fig. 3F). Hence, one can potentially incorporate known DNA polymorphisms or disease-associated mutations into the protein database to improve the identification coverage of SMSNet. Another possibility is to include all amino acid arrangements that fit the mass tags and perform a scoring-based database search to select the best matches. In this regard, a recent advancement in database search using deep learning-assisted prediction of fragment ion intensity (36) would help improve SMSNet's search step.
Most importantly, we have demonstrated the power of SMSNet to uncover new peptides in large-scale proteomics and peptidomics datasets. High level of agreement between SMSNet and MaxQuant (Fig. 5A), together with the findings that SMSNet's identifications exhibit expected characteristics of HLA antigens (Fig. 4B–4C) and contain known phosphosites (Fig. 5E), indicates that newly identified peptides are true positives. Our results also show that SMSNet can improve the coverage of peptide identification by almost 30% (Fig. 4A) and identify new amino acid sequences that better explain observed MS/MS spectra than those in the database do (supplemental Fig. S3). Although many of SMSNet's new identifications are semi-tryptic and non-tryptic forms of known peptides (Fig. 5C) which could theoretically be discovered through database search, SMSNet can process 50,000 MS/MS spectra in 1.14 h whereas partial or no enzyme specificity search would take much longer and may result in many false positives. Altogether, SMSNet should become an invaluable tool for future proteomics and peptidomics studies, such as neoantigen discovery, antibody sequencing, and characterization of non-model organisms, as well as for mining novel peptides and detecting potential contaminants in well-studied samples.
DATA AVAILABILITY
SMSNet is available open-source on GitHub (https://github.com/cmb-chula/SMSNet). The trained models (on WCUMS-M and WCU-MS-P) are available on Figshare (https://figshare.com/articles/SMSNet_s_trained_models/8259122). Processed mass spectrometry datasets used for evaluating SMSNet against DeepNovo (the WCU-MS-BEST, DeepNovo, and ProteomeTools datasets) are available on Figshare (https://figshare.com/articles/SMSNet_s_training_datasets_WCU-MS-BEST_DeepNovo_ProteomeTools_/9734105). The whole WCU-MS dataset which contain peptides without modification, peptides with Methionine oxidation, and peptides with phosphorylation is available on Figshare (https://figshare.com/articles/SMSNet_s_training_datasets_WCU-MS-ALL_/9738839). RIDE accessions for the DeepNovo datasets are PXD005025, PXD004948, PXD004325, PXD004565, PXD004536, PXD004947, PXD003868, PXD004467, and PXD004424. MassIVE accession for the HLA peptidome dataset is MSV000080527. PRIDE accession for the human phosphoproteome dataset is PXD009227. The ProteomeTools spectral library is available on ProteomeTools website (http://www.proteometools.org/index.php?id=53). SMSNet's identification results for the HLA peptidome and human phosphoproteome studies as well as MaxQuant's re-search result for the human phosphoproteome dataset are provided as supplementary Excel tables and are also available on Figshare (https://figshare.com/articles/SMSNet_s_predictions_for_HLA_peptidome_and_human_phosphoproteome_datasets/8259134). Annotated spectra for newly identified phosphopeptides are available on Figshare (https://figshare.com/articles/Annotated_spectra_of_potential_new_human_phosphopeptides_PXD009227_EGF_stimulate_dataset_/9784814).
Supplementary Material
Acknowledgments
We gratefully acknowledge the contribution of mass spectrometry dataset from the Proteomics and Metabolomics Core Facility at The Wistar Institute, the support of the Chulalongkorn Academic Advancement into Its 2nd Century Project, and the donation of TITAN Xp graphic card used in this research by the NVIDIA Corporation. We especially thank Mark A. Knepper (the Epithelial Systems Biology Laboratory, National Heart, Lung, and Blood Institute, National Institute of Health) and Trairak Pisitkul (Systems Biology Center, Chulalongkorn University, Thailand) for facilitating access to computing resources at the National Institute of Health, and for providing critical advice on the manuscript. This work used high performance computing resources of the Biowulf cluster, National Institute of Health (http://hpc.nih.gov) and the Center of Excellence for Medical Genomics, Faculty of Medicine, Chulalongkorn University, Thailand.
Footnotes
* This work was supported by the Ratchadapisek Sompoch Endowment Fund, Faculty of Medicine, Chulalongkorn University grant RA62/037 (to S.S.) and the Grant for Special Task Force for Activating Research, Ratchadapisek Sompoch Endowment Fund, Chulalongkorn University (to E.C. and S.S.). The authors declare no competing interest.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- HLA
- human leukocyte antigen
- EGF
- epidermal growth factor
- PSM
- peptide-spectrum matches.
REFERENCES
- 1. Muth, T., Hartkopf, F., Vaudel, M., and Renard, B. Y. (2018) A potential golden age to come - current tools, recent use cases, and future avenues for de novo sequencing in proteomics. Proteomics 18, 1700150. [DOI] [PubMed] [Google Scholar]
- 2. Liepe, J., Marino, F., Sidney, J., Jeko, A., Bunting, D. E., Sette, A., Kloetzel, P. M., Stumpf, M. P., Heck, A. J., and Mishto, M. (2016) A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science 354, 354–358 [DOI] [PubMed] [Google Scholar]
- 3. Faridi, P., Chen, L., Ramarathinam, S. H., Vivian, J. P., Illing, P. T., Mifsud, N. A., Ayala, R., Song, J., Gearing, L. J., Hertzog, P. J., Ternette, N., Rossjohn, J., Croft, N. P., and Purcell, A. W. (2018) A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands. Sci. Immunol. 3, pii: eaar3947. [DOI] [PubMed] [Google Scholar]
- 4. Mylonas, R., Beer, I., Iseli, C., Chong, C., Park, H.-S., Gfeller, D., Coukos, G., Xenarios, I., Muller, M., and Bassani-Sternberg, M. (2018) Estimating the contribution of proteasomal spliced peptides to the HLA-I ligandome. Mol. Cell. Proteomics 17, 2347–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Laumont, C. M., Vincent, K., Hesnard, L., Audemard, E., Bonneil, E., Laverdure, J.-P., Gendron, P., Courcelles, M., Hardy, M.-P., Cote, C., and others. (2018) Noncoding regions are the main source of targetable tumor-specific antigens. Sci. Transl. Med. 10, eaau5516. [DOI] [PubMed] [Google Scholar]
- 6. Tran, N. H., Zhang, X., Xin, L., Shan, B., and Li, M. (2017) De novo peptide sequencing by deep learning. Proc. Natl. Acad. Sci. 114, 8247–8252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frank, A., and Pevzner, P. (2005) PepNovo: de novo peptide sequencing via probabilistic network modeling. Anal. Chem. 77, 964–973 [DOI] [PubMed] [Google Scholar]
- 8. Ma, B., Zhang, K., Hendrie, C., Liang, C., Li, M., Doherty-Kirby, A., and Lajoie, G. (2003) PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 17, 2337–2342 [DOI] [PubMed] [Google Scholar]
- 9. Ma, B. (2015) Novor: real-time peptide de novo sequencing software. Journal of the Am. Soc. Mass Spectrom. 26, 1885–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tran, N. H., Qiao, R., Xin, L., Chen, X., Liu, C., Zhang, X., Shan, B., Ghodsi, A., and Li, M. (2019) Deep learning enables de novo peptide sequencing from data-independent-acquisition mass spectrometry. Nat. Methods 16, 63–66 [DOI] [PubMed] [Google Scholar]
- 11. Ma, B., and Johnson, R. S. (2012) De Novo Sequencing and Homology Searching. Mol. Cell. Proteomics 11, O111.014902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sutskever, I., Vinyals, O., and Le, Q. V. (2014) Sequence to sequence learning with neural networks. Adv. Neural Information Processing Syst. 27, 3104–3112 [Google Scholar]
- 13. Mann, M., and Wilm, M. (1994) Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal. Chem. 66, 4390–4399 [DOI] [PubMed] [Google Scholar]
- 14. Johnson, R. S., and Taylor, J. A. (2002) Searching sequence databases via de novo peptide sequencing by tandem mass spectrometry. Mol. Biotechnol. 22, 301–315 [DOI] [PubMed] [Google Scholar]
- 15. Vita, R., Overton, J. A., Greenbaum, J. A., Ponomarenko, J., Clark, J. D., Cantrell, J. R., Wheeler, D. K., Gabbard, J. L., Hix, D., Sette, A., and Peters, B. (2015) The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 43, D405–D412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cox, J., and Mann, M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 17. Abelin, J. G., Keskin, D. B., Sarkizova, S., Hartigan, C. R., Zhang, W., Sidney, J., Stevens, J., Lane, W., Zhang, G. L., Eisenhaure, T. M., Clauser, K. R., Hacohen, N., Rooney, M. S., Carr, S. A., and Wu, C. J. (2017) Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity 46, 315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Humphrey, S. J., Karayel, O., James, D. E., and Mann, M. (2018) High-throughput and high-sensitivity phosphoproteomics with the EasyPhos platform. Nat. Protocols 13, 1897–1916 [DOI] [PubMed] [Google Scholar]
- 19. Zolg, D. P., Wilhelm, M., Schnatbaum, K., Zerweck, J., Knaute, T., Delanghe, B., Bailey, D. J., Gessulat, S., Ehrlich, H. C., Weininger, M., Yu, P., Schlegl, J., Kramer, K., Schmidt, T., Kusebauch, U., Deutsch, E. W., Aebersold, R., Moritz, R. L., Wenschuh, H., Moehring, T., Aiche, S., Huhmer, A., Reimer, U., and Kuster, B. (2017) Building ProteomeTools based on a complete synthetic human proteome. Nat. Methods 14, 259–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chambers, M. C., MacLean, B., Burke R. a. A. D,. Ruderman D. L,. Neumann S, Gatto L., Fischer, B., Pratt, B., Egertson, J., Hoff, K., Kessner, D., Tasman, N., Shulman, N., Frewen, B., Baker, T. A., Brusniak, M.-Y., Paulse, C., Creasy, D., Flashner, L., Kani, K., Moulding, C., Seymour, S. L., Nuwaysir, L. M., Lefebvre, B., Kuhlmann, F., Roark, J., Rainer, P., Detlev, S., Hemenway, T., Huhmer, A., Langridge, J., Connolly, B., Chadick, T., Holly, K., Eckels, J., Deutsch, E. W., Moritz, R. L., Katz, J. E., Agus, D. B., MacCoss, M., Tabb, D. L., and Mallick, P. (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vinyals, O., Toshev, A., Bengio, S., and Erhan, D. (2015) Show and tell: A neural image caption generator. Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 3156–3164 [Google Scholar]
- 22. Bahdanau, D., Cho, K., and Bengio, Y. (2014) Neural machine translation by jointly learning to align and translate. arXiv preprint arXiv,1409.0473 [Google Scholar]
- 23. Cho, K., Van Merriënboer. Bart Gulcehre, C., Bahdanau, D., Bougares, F., Schwenk, H., and Bengio, Y. (2014) Learning phrase representations using RNN encoder-decoder for statistical machine translation. arXiv preprint arXiv,1406.1078 [Google Scholar]
- 24. Ba, J. L., Kiros, J. R., and Hinton, G. E. (2016) Layer normalization. arXiv preprint arXiv,1607.06450 [Google Scholar]
- 25. He, K., Zhang, X., Ren, S., and Sun, J. (2016) Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 770–778 [Google Scholar]
- 26. Wu, Y., Schuster, M., Chen, Z., Le, Q. V., Norouzi, M., Macherey, W., Krikun, M., Cao, Y., Gao, Q., Macherey, K., and others. (2016) Google's neural machine translation system: Bridging the gap between human and machine translation. arXiv preprint arXiv,1609.08144 [Google Scholar]
- 27. Lin, T.-Y., Goyal, P., Girshick, R., He, K., and Dollá, r, Piotr. (2017) Focal loss for dense object detection. Proceedings of the IEEE international conference on computer vision, pp. 2980–2988 [Google Scholar]
- 28. Consortium, UniProt (2018) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jurtz, V., Paul, S., Andreatta, M., Marcatili, P., Peters, B., and Nielsen, M. (2017) NetMHCpan-4.0: Improved peptide–MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol. 199, 3360–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Crooks, G. E., Hon, G., Chondonia, J. M., and Brenner, S. E. (2004) WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O'Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., Rajput, B., Robbertse, B., Smith-White, B., Ako-Adjei, D., Astashyn, A., Badretdin, A., Bao, Y., Blinkova, O., Brover, V., Chetvernin, V., Choi, J., Cox, E., Ermolaeva, O., Farrell, C. M., Goldfarb, T., Gupta, T., Haft, D., Hatcher, E., Hlavina, W., Joardar, V. S., Kodali, V. K., Li, W., Maglott, D., Masterson, P., McGarvey, K. M., Murphy, M. R., O'Neill, K., Pujar, S., Rangwala, S. H., Rausch, D., Riddick, L. D., Schoch, C., Shkeda, A., Storz, S. S., Sun, H., Thibaud-Nissen, F., Tolstoy, I., Tully, R. E., Vatsan, A. R., Wallin, C., Webb, D., Wu, W., Landrum, M. J., Kimchi, A., Tatusova, T., DiCuccio, M., Kitts, P., Murphy, T. D., and Pruitt, K. D. (2015) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim, D., Langmead, B., and Salzberg, S. L. (2015) HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., Garimella, K., Altshuler, D., Gabriel, S., Daly, M., and DePristo, M. A. (2010) The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hornbeck, P. V., Zhang, B., Murray, B., Kornhauser, J. M., Latham, V., and Skrzypek, E. (2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 34, D512–D520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li, K., Vaudel, M., Zhang, B., Ren, Y., and Wen, B. (2019) PDV: an integrative proteomics data viewer. Bioinformatics 35, 1249–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gessulat, S., Schmidt, T., Zolg, D. P., Samaras, P., Schnatbaum, K., Zerweck, J., Knaute, T., Rechenberger, J., Delanghe, B., Huhmer, A., Reimer, U., Ehrlich, H.-C., Aiche, S., Kuster, B., and Wilhelm, M. (2019) Prosit: proteome-wide prediction of peptide tandem mass spectra by deep learning. Nat. Methods 16, 509–518 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SMSNet is available open-source on GitHub (https://github.com/cmb-chula/SMSNet). The trained models (on WCUMS-M and WCU-MS-P) are available on Figshare (https://figshare.com/articles/SMSNet_s_trained_models/8259122). Processed mass spectrometry datasets used for evaluating SMSNet against DeepNovo (the WCU-MS-BEST, DeepNovo, and ProteomeTools datasets) are available on Figshare (https://figshare.com/articles/SMSNet_s_training_datasets_WCU-MS-BEST_DeepNovo_ProteomeTools_/9734105). The whole WCU-MS dataset which contain peptides without modification, peptides with Methionine oxidation, and peptides with phosphorylation is available on Figshare (https://figshare.com/articles/SMSNet_s_training_datasets_WCU-MS-ALL_/9738839). RIDE accessions for the DeepNovo datasets are PXD005025, PXD004948, PXD004325, PXD004565, PXD004536, PXD004947, PXD003868, PXD004467, and PXD004424. MassIVE accession for the HLA peptidome dataset is MSV000080527. PRIDE accession for the human phosphoproteome dataset is PXD009227. The ProteomeTools spectral library is available on ProteomeTools website (http://www.proteometools.org/index.php?id=53). SMSNet's identification results for the HLA peptidome and human phosphoproteome studies as well as MaxQuant's re-search result for the human phosphoproteome dataset are provided as supplementary Excel tables and are also available on Figshare (https://figshare.com/articles/SMSNet_s_predictions_for_HLA_peptidome_and_human_phosphoproteome_datasets/8259134). Annotated spectra for newly identified phosphopeptides are available on Figshare (https://figshare.com/articles/Annotated_spectra_of_potential_new_human_phosphopeptides_PXD009227_EGF_stimulate_dataset_/9784814).