Ticks are important vectors of infectious emerging diseases and tick-borne phleboviruses represent a growing threat to humans globally. We employed here a high-resolution, label-free mass spectrometry and RNA interference screen approach to reveal the host cell protein GBF1 as a proviral factor, not only for tick-borne phleboviruses, but also for many other important zoonotic RNA viruses. This study lays the basis for the development of innovative antiviral strategies against a broad range of human pathogenic viruses.
Keywords: Viruses, glycoproteins, affinity proteomics, label-free quantification, peptide mass fingerprinting, assembly, egress, GBF1, replication, Uukuniemi
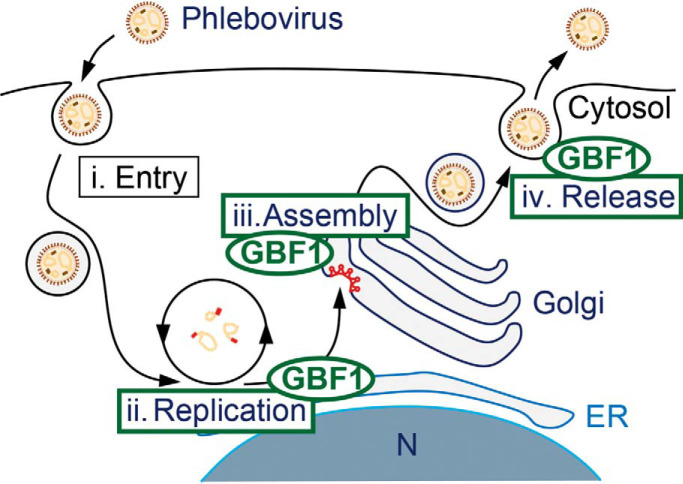
Graphical Abstract
Highlights
Quantitative proteomics reveals GBF1 to interact with Uukuniemi virus glycoproteins.
GBF1 is a proviral host factor for phleboviruses and other zoonotic RNA viruses.
GBF1 supports phlebovirus replication and assembly and release of particles.
The GBF1 inhibitor Golgicide A blocks infection with zoonotic RNA viruses.
Abstract
Novel tick-borne phleboviruses in the Phenuiviridae family, which are highly pathogenic in humans and all closely related to Uukuniemi virus (UUKV), have recently emerged on different continents. How phleboviruses assemble, bud, and exit cells remains largely elusive. Here, we performed high-resolution, label-free mass spectrometry analysis of UUKV immunoprecipitated from cell lysates and identified 39 cellular partners interacting with the viral envelope glycoproteins. The importance of these host factors for UUKV infection was validated by silencing each host factor by RNA interference. This revealed Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (GBF1), a guanine nucleotide exchange factor resident in the Golgi, as a critical host factor required for the UUKV life cycle. An inhibitor of GBF1, Golgicide A, confirmed the role of the cellular factor in UUKV infection. We could pinpoint the GBF1 requirement to UUKV replication and particle assembly. When the investigation was extended to viruses from various positive and negative RNA viral families, we found that not only phleboviruses rely on GBF1 for infection, but also Flavi-, Corona-, Rhabdo-, and Togaviridae. In contrast, silencing or blocking GBF1 did not abrogate infection by the human adenovirus serotype 5 and immunodeficiency retrovirus type 1, the replication of both requires nuclear steps. Together our results indicate that UUKV relies on GBF1 for viral replication, assembly and egress. This study also highlights the proviral activity of GBF1 in the infection by a broad range of important zoonotic RNA viruses.
Enveloped viruses are surrounded by a host cell derived lipid bilayer into which the viral glycoproteins are embedded (1). To assemble an infectious particle, virus glycoproteins undergo processing and maturation steps in the host cell secretory pathway and possibly in the extracellular space (2). After initial protein synthesis at the endoplasmic reticulum (ER)1 membrane, viral glycoproteins are transported through the cellular secretory pathway to the plasma membrane (3). During this process they are glycosylated and oftentimes proteolytically processed to acquire a metastable structure needed for infection of new cells (3). The timing and subcellular location of assembly, i.e. when the viral genome and capsid become enveloped by the virus glycoprotein-containing membrane, differs among enveloped viruses. Some viruses bud from intracellular membranes such as the ER or the Golgi, whereas others bud from the plasma membrane (4, 5). In the former two cases, the processing of the glycoproteins occurs during transport of the assembled particle through the secretory pathway. For several viruses it remains elusive, which cellular components of the trafficking machinery are essential for particle release.
We focus here on Uukuniemi virus (UUKV), which belongs to the genus Phlebovirus in the family Phenuiviridae (6). As such, UUKV is closely related to Rift Valley fever virus (RVFV), an important pathogen in both human and livestock (7). UUKV is moreover the viral model to study the highly pathogenic tick-borne human phleboviruses that have recently emerged in different parts of the world such as severe fever with thrombocytopenia syndrome virus (SFTSV) in China and Heartland virus (HRTV) in the United States (8, 9). Like other phleboviruses, UUKV has a tri-segmented single-stranded mainly negative-sense RNA genome, which is exclusively replicated in the cytosol of infected cells (9). The viral genome encodes four structural proteins, namely the nucleoprotein N, the RNA-dependent RNA polymerase L, and a polypeptide precursor that is further processed into the two transmembrane glycoproteins GN and GC. Cleavage, folding, and maturation of the polypeptide precursor into GN and GC take place in the ER and Golgi (10). At the Golgi membrane viral particles acquire their lipid bilayer envelope and bud into the Golgi lumen. The pathway used by the virus to exit cells remains poorly characterized. Extracellular UUKV particles are enveloped, roughly spherical with an icosahedral shape of t = 12, a diameter of about 125 nm and spike-like projections of 5–10 nm (11). The spikes are composed of the two envelope glycoproteins GN and GC, responsible for the attachment and entry of the virus into the target cells. UUKV penetrates host cells by acid-activated membrane fusion from late endosomal compartments, and therefore, belongs to the large group of late-penetrating viruses, which depend on late endosomal cues for infection (12, 13). However, many aspects of the virus exit, replication, and entry programs remain to be elucidated at the molecular and cellular levels.
Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (GBF1), the orthologue of the Drosophila protein Gartenzwerg (14), is a ubiquitously expressed guanine nucleotide exchange factor (GEF), which activates the ADP-ribosylation factor (ARF) family of GTPases (15). It resides at the cis-Golgi and is important for intracellular retrograde trafficking in the early secretory pathway (16). GBF1 regulates ARF and coat protein I (COPI) dependent Golgi - ER trafficking (17). During this process GBF1 cycles between a membrane bound and cytosolic state (18).
Many RNA viruses, which replicate in the cytoplasm, reshape intracellular membranes to generate shielded replication compartments. GBF1 aids in replication complex formation for several enveloped plus strand RNA viruses including yellow fever virus (YFV), hepatitis C virus (HCV), human coronavirus 229E (HCoV-229E), and dengue virus (DENV) (19–21). Notably, also non-enveloped viruses hijack GBF1. Poliovirus reshapes intracellular membranes to form shielded replication compartments and recruits GBF1 through its non-structural protein 3A. Interestingly, the GEF activity of GBF1 is dispensable for poliovirus RNA replication (22, 23). The negative strand RNA virus vesicular stomatitis virus (VSV), but also the non-enveloped RNA viruses Coxsackievirus B and hepatitis E virus, similarly depend on GBF1 for their genome replication (24–26). Even though a range of RNA viruses hijacks GBF1 for genome replication complex formation, we currently lack knowledge whether GBF1 plays a role in replication or assembly and release of negative strand RNA viruses of the Phenuiviridae family in the Bunyavirales order.
Here, we use an integrative virology-proteomics approach and identify GBF1 as an interaction partner of the GN/GC glycoproteins of the Phenuiviridae family member UUKV. We show that GBF1 is required for a post-entry step in the UUKV life cycle. Knockdown and pharmacological inhibition of GBF1 demonstrates that negative strand RNA viruses of the Phlebovirus genus including the emerging Toscana virus (TOSV) and RVFV rely on GBF1 for efficient infection of epithelial cells. DNA viruses and retroviruses in contrast replicate independently of GBF1, whereas other unrelated RNA viruses also rely on GBF1 for replication. This work demonstrates that GBF1 is a broad proviral factor for plus and negative strand RNA viruses.
EXPERIMENTAL PROCEDURES
Cells
All products used for cell culture were purchased from Thermo Fisher Scientific (Waltham, MA). Human lung epithelial A549 and embryonic kidney 293T cells as well as green monkey epithelial Vero cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 2 mm glutamine. Human hepatoma Huh-7.5 cells stably expressing Firefly luciferase (Huh-7.5 FLuc) were cultured in DMEM supplemented with 10% FBS and 2 mm glutamine and non-essential amino acids. 293T cells, TZM-bl cells and PM-1 cells were obtained from and cultured as recommended by the NIH AIDS Program. Indicator TZM-bl cells (previously named JC53BL cells) are human epithelial HeLa cell derivatives engineered to express CD4 and CCR5, in addition to the already endogenously expressed CXCR4. In addition, they carry a Tat-responsive luciferase gene under the control of the human immunodeficiency virus 1 (HIV-1) long terminal repeat promoter. PM-1 cells are Hut78-human T cell derivatives. The hamster kidney BHK-21 cells were grown in Glasgow's minimal essential medium (GMEM) supplemented with 10% tryptose phosphate broth (TPB) and 5% FBS.
Plasmids
The expression plasmids pUUK-N and pUUK-L as well as the plasmids pRF108-S and pRF108-L code for the UUKV nucleoprotein N and polymerase L, and the anti-genomic full length UUKV S and L RNA segments respectively (9, 10). The pRF108-S.ΔNSsGFP was obtained as follow. The cDNAs corresponding to the 5′ noncoding region (NCR)-N sequence and green fluorescent protein (GFP) sequence-3′ NCR were synthesized by PCR from plasmid DNA encoding the UUKV S segment and GFP, respectively, using Herculase II fusion DNA polymerase (Agilent, Santa Clara, CA). The primers AATCGTCTCTAGGTACACAAAGACCTCCAACTTAGCTATCG and AATCGTCTCTGGGCCGAAGCCCTTTTAGAGTCC were used for the amplification of the first cDNA, and the primers AATCGTCTCTGCCCAAACTAGAGTCCGGACTTGTACAGCTCG and AATCGTCTCTGGGACACAAAGACCCTCCAACATTAAGCATGGTGAGCAAGGGCGAGGAGC for the second. Both PCR products were digested with the enzyme BsmBI (New England Biolabs, Ipswich, MA) and subcloned together into the pRF108 plasmid vector.
Plasmids coding for HIV-1 NL4.3, LAI, and YU-2 as well as VSV-G were obtained from O. Keppler (LMU, Germany) whereas the plasmid encoding the transmitted/founder strain CHO.77t (T/F) was obtained from F. Kirchhoff (Ulm University, Germany).
Viruses
All viruses were amplified from the cell lines most efficient for production according to well-established, standard protocols. Briefly, the prototype strains of UUKV S23, TOSV ISS, and Semliki Forest virus (SFV) as well as the engineered Chikungunya virus (CHIKV) encoding GFP or Nano-luciferase have been described previously (27–31). UUKV S23, TOSV ISS, and SFV were produced and titrated in BHK-21 cells (12). GFP- and Nano-luciferase-recombinant CHIKV were generated by electroporation of full-length RNA into BHK-21 cells and viral titers determined on 293T cells. The Langat virus (LGTV) TP21, tick-borne encephalitis virus (TBEV) Hypr 71, and Japanese encephalitis virus (JEV) Nakayama strains were kind gifts of G. Dobler (the Bundeswehr Institute of Microbiology, Germany) and S. Vene (Folkhälsoinstitutet, Sweden), respectively. The GFP-recombinant RVFV strain ZH548 was previously described (32). Production and titration of RVFV, LGTV, TBEV, and JEV were performed in Vero cells (33–35). Human adenovirus serotype 5 (HAdV-5, strain Ad75) was produced from and tittered on A549 cells as described previously (36), with the exception that the virions were eluted in sterile phosphate buffered saline (PBS) when desalting on a NAP column (GE Healthcare, Chicago, IL). HCV full length RNA encoding a Renilla luciferase was in vitro transcribed, electroporated into Huh-7.5 cells, and cell supernatants were harvested 48 h, 72 h, and 96 h after electroporation. HCV stocks were tittered on Huh-7.5 cells. The various infectious HIV-1 strains, i.e. NL4.3, LAI, YU-2, and T/F, were generated by calcium phosphate DNA precipitation of 293T cells with plasmids encoding the respective HIV-1 strains. HIV-1 Ba-L was obtained from the NIH AIDS Reagent Program and passaged on PM-1 cells. Recombinant VSV expressing GFP and Renilla luciferase-expressing recombinant human coronavirus strain HCoV-229E were kindly provided by G. Zimmer (University of Bern, Switzerland) (37) and V. Thiel (University of Bern, Switzerland) (38), respectively. Although VSV was produced in BHK-21 cells and tittered on Vero cells, coronavirus was amplified and tittered on Huh-7.5 cells. The multiplicity of infection (MOI) is given for each virus according to the titer determined on the respective cell lines mentioned above.
Small Interfering RNAs (siRNAs), Antibodies, and Reagents
siRNAs (see the complete list in Table I) were all obtained from Thermo Fisher Scientific except for the AllStars negative-control siRNA (si_Ctrl) that was purchased from Qiagen (Hilden, Germany). The mouse monoclonal antibody (mAb) 8B11A3 and 6G9E5 are directed against the UUKV nucleoprotein N and GN, respectively (kind gift from Ludwig Institute for Cancer Research, Sweden) (39). The mouse immune ascitic fluid recognizes all TOSV structural proteins (generous gift from R.B. Tesh, University of Texas) (34). The mouse mAb E2–1 targets the SFV glycoprotein E2 and was kind gift from M.C. Kielian (Albert Einstein College of Medicine) (40). The mouse mAbs anti-GBF1 (sc-136240) and anti-actin (sc-58679) were both obtained from Santa Cruz. Stock of Golgicide A (GCA, Merck, Darmstadt, Germany) was dissolved in methanol or dimethyl sulfoxide (DMSO) respectively.
Table I. siRNA sequences of GN/GC interaction partners.
| Gene control | siRNA (ID) ctrl | Catalogue number AllStars negative | siRNA sense sequence |
|---|---|---|---|
| SAAL1 | 1 | s41430 | CAUCCAGCUAUUUAUGAUAtt |
| 2 | s41431 | GCAAGAGUAUCUAAAGAUAtt | |
| HEATR3 | 1 | s30019 | GUCUUUCAGUGCUACAGCAtt |
| 2 | s30020 | GUGACGCAUUUAUGGAGAAtt | |
| VDAC2 | 1 | s14771 | AUCAAGUCUUCUUACAAGAtt |
| 2 | s14772 | GGAGGAUCAAUUUAUCAGAtt | |
| GOLPH3 | 1 | s34400 | GUACGGGAACGAUUAGCUAtt |
| 2 | s34401 | CUAUUAACAAGAAAGGUAAtt | |
| TRAPPC2L | 1 | s28534 | AGCCCUUCGAGACAACGAAtt |
| 2 | s28536 | AGGUGAAGUUUGUCAUGGUtt | |
| TNPO3 | 1 | s24030 | GGGACUCAUUGCUAACCCAtt |
| 2 | s24031 | CCUUACGAAUUGGAGCUAAtt | |
| ERLEC1 | 1 | s26043 | CCUACAGAAUUGAGUCUUAtt |
| 2 | s26044 | GGACUUACGAAGUAUGUCAtt | |
| GBF1 | 1 | s16633 | CAACCACAAUGUUCGUAAAtt |
| 2 | s16634 | GCAUAGUUUCGGUCAUCUAtt | |
| 3 | s16635 | GAGCACUACUUGUACAUGAtt | |
| SEL1L | 1 | s12674 | GGCUUAUGACUGCCUAUAAtt |
| 2 | s12675 | GCACCGAUGUAGAUUAUGAtt | |
| SURF4 | 1 | s13649 | UCAUAGCUCUGCAGACGAUtt |
| 2 | s13650 | AGUUCCUCCGUGUCACAAAtt | |
| GOLPH3L | 1 | s30429 | CCGCCUUACUCUUAUGGAAtt |
| 2 | s30431 | GAGAAACAGCGACUAGUGAtt | |
| MARS | 1 | s8517 | GGAGCUGAGGAUAACUAUAtt |
| 2 | s8518 | CAGAGCAAGUGGACCUGUAtt |
Fluorescently Labeled Viral Particles
Labeling of UUKV with Alexa Fluor (AF) 488-succinimidyl esters (Thermo Fisher Scientific) was performed in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, 20 mm) with three molecules of dye for one molecule of UUKV glycoproteins according to a previously established procedure (34, 41).
siRNA-mediated Gene Silencing, CRISPR/Cas9 Knockout of GBF1, and GCA Inhibition
siRNA reverse transfections were performed using Lipofectamine RNAiMAX reagent according to the manufactures recommendations (Thermo Fisher Scientific). Briefly, 25,000 of the indicated cells were transfected with a 20 nm final concentration of siRNAs in FBS-free medium and seeded in a 24-well dish 3 days before exposition to the viruses. For GBF1 knockout, 293T cells were transfected with three GBF1-targeting clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9) guide RNAs (gRNAs), or a non-targeting CRISPR/Cas9 gRNA as a control, using the system pSpCas9(BB)-2A-GFP (PX458) that was a gift from F. Zhang (Broad Institute) (Addgene, Watertown, MA, plasmid # 48138; http://n2t.net/addgene:48138; RRID:Addgene_48138) (42). All three target plasmids were co-transfected at a ratio of 1:1:1 and 72 h later, cells were re-seeded and transfected again following the same procedure (43). GFP-expression was used as an indicator of GBF1 knockdown. Infection was performed 3 days later as described below. For GBF1 inhibition experiments, cells were pretreated with indicated concentrations of GCA or solvent for up to 60 min before infection.
Immunofluorescence-based Infection Assays (UUKV, TOSV, SFV, HAdV-5, and LGTV)
Following silencing, knockout, and block of GBF1, cells were infected with UUKV, TOSV, SFV, HAdV-5, or LGTV at the indicated MOIs as previously shown (9). Briefly, for siRNA-based infection assays, transfected A549 cells were washed 1 h before infection, exposed to UUKV, TOSV, and SFV, washed again, and fixed 8 h post-infection. For GBF1 knockout-based infection assays, transfected 293T cells were infected with HAdV-5 and LGTV for 48 h and fixed. To assess the capacity of GCA to block infection, A549 or BHK-21 cells were first exposed to the drug for up to 60 min and subsequently to UUKV, TOSV, SFV, and HAdV-5 in the presence of the drug for 1 h. The input virus was then washed away, and cells incubated in the continuous presence of the drug for 7 h before fixation in the case of UUKV, TOSV, and SFV. In the case of HAdV-5, cells were freeze-thawed three times 24 h post-infection in order to release progeny virus. The supernatant was subsequently used to infect newly seeded A549 cells and the number of progeny viruses was determined by fluorescence-focus assay. Therefore, A549 cells were fixed 48 h later, permeabilized and intracellular viral antigens stained with primary antibodies for 1 h, washed, and subsequently exposed to AF647-conjugated secondary antibodies (Molecular Probes) for 45 min. The mAbs 8B11A3 and 2E-1 were used to detect the UUKV nucleoprotein N and the SFV glycoprotein E2, respectively. Infection with TOSV was detected with mouse immune ascitic fluid whereas that by HAdV-5 and LGTV with the primary antibodies clone 20/11 (Merck) and 1786,3 (44), respectively. Infection of cells by HAdV-5 produced from GCA-pretreated cells was quantified by imaging with a Trophos system (Luminy Biotech Enterprises, Luminy, France). For the other immunofluorescence-based assays, the infection index was determined by flow cytometry analysis with a FACS Celesta or FACSLSRII flow cytometer (Becton Dickinson, Franklin Lakes, NJ) using either FlowJo (Treestar, Franklin Lakes, NJ) or FACSDiva (Becton Dickinson) software. In the case of 293T cells knocked-out for GBF1 and exposed to HAdV-5 or LGTV, infection was analyzed in cell populations positive for GFP, which corresponded to cells knocked-out for GBF1.
GFP-based Infection Assays (VSV, CHIKV, and RVFV)
siRNA-transfected or GCA-pretreated A549 or Huh-7.5 FLuc cells were exposed to GFP-expressing VSV, CHIKV, or RVFV at the indicated MOI and GFP-positive cells were quantified up to 40 h post-infection with a FACS Celesta or an Accuri C6 flow cytometer (Becton Dickinson).
Focus Forming Unit-based Infection Assays with TBEV and JEV
A549 cells were first exposed to TBEV and JEV for 1 h, virus inoculum removed, and GCA added. 24 h post-infection, cells were washed twice to remove GCA and cell viability in GCA treated, uninfected cells was analyzed using CellTiter-Glo® (Promega, Madison, WI). Supernatants containing virions were collected and tittered by immunostaining and focus forming assay on Vero cells as previously described (45).
Luminometry-based Infection Assays (CHIKV, HCV, HCoV-229E, and HIV-1)
siRNA-transfected or GCA-pretreated Huh-7.5 FLuc or TZM-bl cells were infected with HIV-1, CHIKV, HCV-JcR2a, or HCoV-229E and the infection stopped after up to 48 h. GCA-pretreated TZM-bl cells were infected with HIV-1 strains according to a standard procedure (46). On HIV-1 challenge and Tat production, TZM-bl cells indicate infection by expressing luciferase (47, 48). Luciferase reporter gene activity in cell lysates, which correlates with infectivity, was measured by luminometry using a Centro LB 960 Microplate luminometer and Ascent 2.0 software.
Co-immunoprecipitation (Co-IP)
A549 cells (1,5 × 108 cells per experimental replicate) were incubated with sucrose cushion purified UUKV or mock supernatants at MOI 2 for 2 h in the cold in DMEM, 20 mm HEPES, 1 mm CaCl2, 0.2% human serum albumin, pH 7.4, followed by extensive washes and snap freezing of virus bound cells. Cells were thawed and lysed for 30 min on ice shortly before IP using 50 mm HEPES, pH 7.4, 150 mm NaCl, 10% glycerol, 1% NP-40, 1 mm CaCl2. For mass spectrometry (MS) analysis, one-step IPs from quadruplicate samples of A549-mock, A549-UUKV and virus only samples, containing the equivalent amount of virus particles as the A549-UUKV inoculum, were performed using aminolink plus protein A/G resin (Pierce, Dallas, TX) with covalently bound antibodies against GN/GC (rabbit U2 antibody, (49)) as described in (50). To capture intracellular interaction partners of GN/GC antibodies specific to GN/GC were incubated with cell lysates for 16 h. GN/GC and associated proteins were eluted from the resin using glycine buffer (pH 2.8) and precipitated with ethanol, sodium acetate (pH 5), glycogen as described in (51). To identify proteins which bind non-specifically to the antibody matrix, we performed affinity purifications from cells that had been incubated with virus-free cell culture supernatants and used the same experimental conditions as for the UUKV containing samples. Experiments were conducted in four biological replicates from four independent A549 cell passages. Efficiency of bait enrichment was determined by immunoblotting. All IP eluates were used for liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis.
Co-IPs of GN/GC mutants were performed by transfecting pcDNA UUK GN/GC and mutants, with alanines in position 46–50 or 23–24 in the GN cytoplasmic tail (52, 53), into 293T cells. Cells were lysed 24 h post transfection and the GN was immunoprecipitated with the mAb 6G9E5 (39). Proteins were separated on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blot analysis. Proteins were detected with rabbit polyclonal GBF1 (Abcam, Cambridge, United Kingdom), rabbit polyclonal GN/GC (Goliat) and beta-tubulin (rabbit polyclonal; Abcam) antibodies.
Experimental Design and Statistical Rationale
To generate the MS dataset, four biological replicates of each of the following three conditions were analyzed
GN/GC co-IP from A549 cells incubated with purified UUKV supernatant (A549 + UUKV)
GN/GC co-IP from A549 cells incubated with mock supernatant (A549 + Mock)
Purified UUKV supernatant only (UUKV only).
A549 + Mock and UUKV only samples served as negative controls to identify bona fide host cell interaction partners of UUKV GN/GC glycoproteins. Biological quadruplicates of each condition were considered as sufficient for the statistical analysis because the replicates showed a high correlation und clustered together as shown by unsupervised hierarchical clustering. Because independent samples with normal distributions and unequal variances were compared, a parametric two-tailed Welch's t test was used for the statistical analysis. A permutation-based false discovery rate (FDR) of 1% and a S0 parameter of 1 was employed to identify proteins that significantly differed in abundance (54). Detailed t test results of the pairwise comparisons A549 + UUKV versus A549 + Mock and UUKV only versus A549 + Mock are shown in supplemental Table S1.
Sample Preparation for MS
For MS analysis, protein samples were reduced, alkylated and tryptically digested as described in (55). In brief, protein samples were reduced with 10 mm dithiotreitol for 30 min at room temperature (RT) followed by alkylation with 55 mm iodoacetamide for 20 min at RT in the dark. Remaining iodoacetamide was quenched with 100 mm thiourea. Proteins were digested with 1 μg LysC (Wako Chemicals, Tokyo, Japan) at RT for 3 h and 1 μg trypsin (Sigma, Saint Louis, MO) at RT overnight. Protein digestion was stopped with 0.6% (v/v) trifluoroacetic acid and 2% (v/v) acetonitrile before peptides were loaded onto reversed phase C18 StageTips (3MTM EmporeTM, IVA Analysentechnik, Düsseldorf, Germany). Peptides were desalted using 0.5% (v/v) acetic acid and subsequently eluted from the C18 StageTips with 50 μl 80% (v/v) acetonitrile in 0.5% (v/v) acetic acid. After concentrating and drying in a SpeedVac (Thermo Scientific), peptides were resuspended in 10 μl 2% (v/v) acetonitrile, 0.1% (v/v) trifluoroacetic acid in 0.5% (v/v) acetic acid and stored at −20 °C until mass spectrometric analysis.
Mass Spectrometric Analysis Including Database Search Parameters and Acceptance Criteria for Identifications
Peptide mixtures were analyzed in a single-run liquid chromatography LC-MS/MS format as described in (55). In brief, peptides were separated using a nanoflow UHPLC instrument (EASY-nLC 1200, Thermo Scientific). Separated peptides were analyzed on a benchtop quadrupole-Orbitrap instrument (Q Exactive HF mass spectrometer, Thermo Scientific) with a nanoelectrospray ion source (Thermo Scientific), which was coupled on-line to the liquid chromatography instrument. MS data were acquired as described previously using the Xcalibur software (Thermo Scientific).
Protein identification and quantification from MS raw files was performed using the computational proteomics platform MaxQuant (software version 1.5.5.2). Match-between runs was enabled and additional GN/GC co-IP samples from HeLa cells incubated with purified UUKV or mock supernatants were acquired, which served as peptide matching reference. The MS/MS spectra were searched against the human UniProtKB FASTA database (92,013 forward entries including isoforms; version from December 2015) and a common contaminant database (247 entries) by the implemented Andromeda search engine with a FDR of 1% at peptide and protein level (56, 57). To identify the UUKV viral proteins L, N, GN and GC in the peptide mixtures, the MS/MS spectra were additionally searched against the corresponding FASTA files for these four viral proteins. Additionally, hits were checked against the Crapome contaminant repository (58). The protease configuration in the Andromeda search engine was set to “Trypsin/P” allowing cleavage at the carboxyl side of the amino acids lysine or arginine (also if a proline follows) with a maximum of two missed cleavages. Carbamidomethylation was set as a fixed modification and acetylation (N terminus) as well as methionine oxidation were set as variable modifications. Initial precursor mass deviation of up to 4.6 ppm and fragment mass deviation up to 20 ppm were allowed. Inclusion criteria were set to at least one unique or razor peptide with a minimum length of 7 amino acids per protein group. Protein identification required at least one razor peptide and proteins that could not be discriminated based on unique peptides were grouped into protein groups. For protein quantification, MaxLFQ algorithms were used (59).
Bioinformatics Analysis
Proteomics data was analyzed with the statistical software environment R and R Studio, respectively (60, 61), and the Perseus computational platform (62). Proteins matching to the reversed or contaminants database as well as peptides only identified by side modification were excluded from the analysis. MaxLFQ values were used for protein quantification and the data were filtered to contain at least three valid values per protein identification in at least one group of replicates. 350 protein groups were derived from only one unambiguously identified peptide. The annotated spectra and MS/MS information for these single peptide identifications are provided in supplemental Fig. S4 and supplemental Table S3. The base 2 logarithm of the MaxLFQ intensity was calculated for each protein group in each sample before comparing relative protein abundances. The number of identified protein groups per condition and the Pearson correlation between biological replicates were determined and subsequently missing values were replaced. Missing values were imputed by a Gaussian distribution with a 30% width relative to the standard deviation of the measured values and a downshift of the mean by 1.8 standard deviations of the valid data. In addition, principal component analysis (PCA) of the individual MS samples was performed. Detailed information for all protein identifications that remained after data processing including information about the number of distinct peptides, sequence coverage, identification type, non-imputed/imputed MaxLFQ values, intensity values, iBAQ values, GO annotations is provided in supplemental Table S2. A parametric two-tailed Welch's t test with a permutation-based FDR of 1% and a S0 parameter of 1 was employed to identify proteins that significantly differed in abundance (54). Proteins significantly enriched in GN/GC co-IPs from A549 cells incubated with purified UUKV versus A549 cells incubated with mock supernatants were determined (A549 + UUKV versus A549 + Mock) and 43 significantly enriched proteins identified (39 host proteins and the four viral protein L, N, GN and GC). Significant proteins were functionally characterized by annotation enrichment analysis using annotations from the UniProtKB Keywords (63). All annotations with intersection sizes greater than 2 and p values less than 0.05 were considered. Enrichment factors were calculated by Fisher's exact test.
As a control, significantly enriched proteins in virus only samples versus GN/GC co-IPs from A549 cells incubated with mock supernatants were determined (UUKV only versus A549 + Mock) and 47 significantly enriched proteins identified (43 host proteins and the four viral protein L, N, GN and GC). The correlation of the t test differences of the pairwise comparisons A549 + UUKV versus A549 + Mock and UUKV only versus A549 + Mock was determined.
All identified proteins, significant proteins of the A549 + UUKV versus A549 + Mock comparison and significant proteins in the UUKV only versus A549 + Mock comparison were analyzed for annotated membrane association. Proteins were classified as membrane-associated if they were annotated as “membrane,” “plasma membrane,” or “intracellular membrane-bounded organelle” according to the Gene Ontology (GO) category cellular component (GOCC) (64). For functional follow up of GN/GC associated proteins, 12 proteins were selected, of which 9 additionally were classified as membrane-associated.
Molecular network enrichment analysis was performed using the Ingenuity Pathway Analysis software package (Qiagen).
Virus-like Particle (VLP) Assay
pUUK-GN/GC and pUUK-N plasmids expressing the UUKV glycoprotein precursor (p110) and nucleoprotein under the control of a cytomegalovirus (CMV) promoter (10). Transfection of BHK-21 or 293T cells was performed using GeneJuice (Novagen, Darmstadt, Germany) following the manufacturer's protocol using 8 μg of DNA/sample in total. Briefly, the transfection medium was removed 24 h post transfection and fresh medium containing 10 mm GCA (Sigma) or DMSO was added. Cells and VLPs were collected 16–18 h after the addition of GCA. Cells were lysed (0.5 m Tris-HCl pH8, 1 m NaCl, 1% Triton X-100) and supernatant of transfected cells was collected, and concentrated by ultracentrifugation (100,000 × g, 90 min 4 °C, SW41, Beckman Coulter, Brea, CA). The pellet was resuspended in non-reducing Laemmli SDS-PAGE sample buffer. Proteins were separated with SDS-PAGE and Western blot analysis.
Protein Analysis
Proteins were separated by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes (iBlot Transfer Stacks, Invitrogen, Waltham, MA), and analyzed by Western blotting. Briefly, PVDF membranes were first incubated with the primary mouse mAbs against GBF1 (1:500), actin (1:1,000), or primary rabbit polyclonal Ab recognizing both the UUK GN and GC proteins all diluted in Tris-buffered saline containing 0.1% Tween and 5% milk, and then with an anti-mouse horseradish peroxidase-conjugated secondary antibody (1:10,000; Santa Cruz, Dallas, TX). Bound antibodies were detected by exposure to enhanced chemiluminescence reagents (SuperSignal, Thermo Fisher Scientific or SuperSignal West Pico kit (Pierce)) according to manufactures recommendations. For semi quantitative analysis Fiji/Image J Gel analyze was used.
Binding and Internalization Assay
Fluorescent UUKV-AF488 (MOI ∼20) was bound to siRNA-transfected A549 cells on ice in binding buffer [DMEM (pH ∼7.4) containing 0.2% BSA] for 2 h as described before (65). In internalization assays, fluorescent virus-bound A549 cells were washed and rapidly warmed to 37 °C to trigger endocytosis for up to 30 min. To distinguish between internalized and external particles, samples were treated with trypan blue (0.01%; Sigma) for 15 s at RT (49). Both virus binding and internalization were quantified by flow cytometry using a FACS Celesta (Becton Dickinson).
Reverse Genetics System
The anti-genomic segments S.ΔNSsGFP and L from UUKV were rescued by transfecting 1.6 × 105 BHK-21 cells with the expression plasmids pUUK-L (0.3 μg) and pUUK-N (0.09 μg) together with 0.15 μg each of pRF108- S.ΔNSsGFP and pRF108-L, as described previously (9). Cells were overlaid with GMEM containing the indicated concentrations of GCA and supplemented with 5% FBS and 10% TPB 1 h post-transfection. Samples were harvested, fixed, and analyzed by flow cytometry 24 h later using a FACS Celesta (Becton Dickinson).
RESULTS
Quantitative Proteomics Identifies GBF1 as UUKV Glycoprotein Interaction Partner
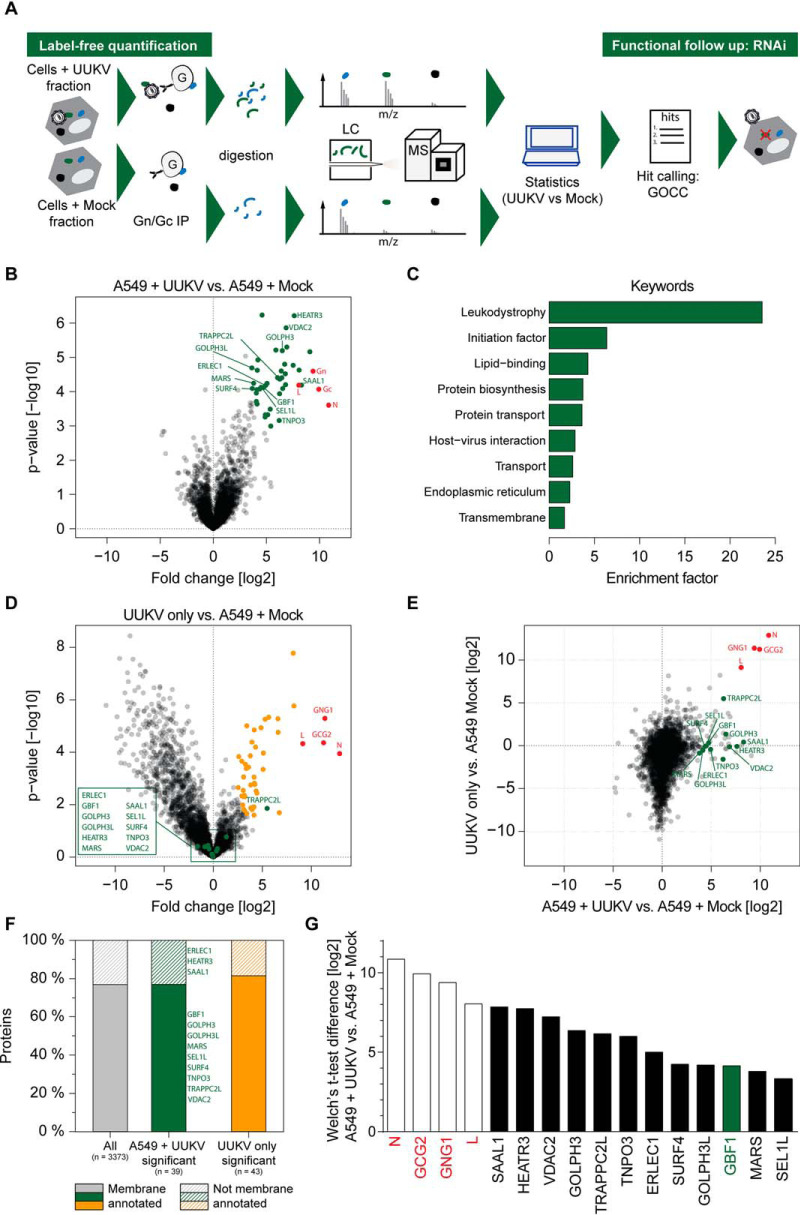
We have previously characterized UUKV GN/GC glycoprotein interactions with the C-type lectins DC-SIGN and l-SIGN, which are critical for virus entry (34, 49). However, interactions of GN/GC during late stages of the UUKV life cycle, i.e. during assembly and release of progeny particles, remained enigmatic. To better understand UUKV glycoprotein-host cell interactions, we performed affinity purification - mass spectrometry (AP-MS) experiments from permissive human epithelial cells (A549). These cells constitute a gold standard model system to study UUKV and other phleboviruses (12). To enrich for virus particles capable of binding to cellular attachment factors we incubated purified UUKV particles or mock cell supernatants at a MOI of 2 with cell suspensions for 2 h on ice, washed away unbound virus particles extensively and lysed the cells for 30 min in the cold. In order to not only allow interactions of GN/GC with cell surface attachment factors, but also with intracellular proteins contained in the lysate, we carried out the affinity purification of GN/GC and associating host proteins for 16 h in the cold. To control for host proteins associated with UUKV, we performed GN/GC affinity purifications from the UUKV preparation in the absence of cells. Moreover, we used lysates from cells incubated with virus-free mock cell supernatants to control for nonspecific binding of proteins to the antibody matrix. Samples were then processed for and subjected to label-free MS analysis (Fig. 1A).
Fig. 1.
Label-free quantitative proteomics identifies GBF1 as UUKV GN/GC interaction partner. A, Scheme of the label-free quantification approach to identify UUKV GN/GC interaction partners in A549 cells. B, Volcano plot of UUKV GN/GC affinity - purification mass spectrometry (AP-MS) comparing A549 cells incubated with purified UUKV and A549 cells incubated with mock supernatants. Proteins significantly enriched with UUKV GN/GC in A549 lysates are labeled in green and viral proteins are labeled in red (FDR<0.01, S0 = 1). Follow-up hits are indicated by protein names. Median protein intensity differences and -log10 p values of quadruplicates are shown. C, Enrichment analysis of significant UUKV GN/GC interaction partners using UniProt Keywords. Enrichment factors were calculated by Fisher's exact test (annotations with intersection sizes > 2 and p value < 0.05 are shown). D, Volcano plot of UUKV GN/GC AP-MS comparing UUKV particle preparation and A549 cells incubated with mock supernatants. Proteins significantly enriched with UUKV GN/GC in the particle preparation are labeled in orange, follow-up hits are labeled in green and indicated by protein name and viral proteins are labeled in red (Welch's t test, FDR < 0.01, S0 = 1). Median protein intensity differences and -log 10 p values of quadruplicates are shown. E, Correlation of protein enrichment (t test differences) with GN/GC from UUKV in A549 cells (x axis) versus UUKV particle preparation (y axis). Labels as in (B). F, Number of membrane annotated and not membrane annotated proteins of all identified proteins, significantly with GN/GC enriched proteins in A549 lysates incubated with UUKV and significantly with GN/GC enriched proteins in UUKV particle preparation. The four viral proteins L, N, GN and GC were excluded from the analysis. G, Median intensity differences of selected top twelve cellular GN/GC interaction partners in UUKV versus mock samples from quadruplicate samples. Enriched viral proteins are shown in red.
We next determined specific UUKV GN/GC interactions by comparing biological quadruplicates and using the MaxLFQ algorithm (59). Thirty-nine host proteins of a total of 3377 unambiguously identified proteins (supplemental Fig. S1A) passed the threshold of Welch's t test significance when comparing A549 cells incubated with and without UUKV (FDR < 0.01, S0 = 1) (Fig. 1B, supplemental Table S1). Experimental replicates clustered as shown by unsupervised hierarchical clustering (supplemental Fig. S1B) and showed a high correlation (A549 without and with UUKV: correlation coefficients 0.86 and 0.88; UUKV only: 0.77) (supplemental Fig. S1C). All three experimental conditions, i.e. UUKV only, A549 without UUKV and A549 with UUKV, clearly segregated, whereas the segregation between the A549 without UUKV and A549 with UUKV samples was less prominent (supplemental Fig. S1D). We found the viral bait glycoproteins GN and GC as well as the two UUKV structural proteins N and L enriched in the virus containing samples. Keyword enrichment analysis of the 39 hits from the A549 UUKV samples revealed that two categories of cellular “transport” as well as “host-virus interactions” were among the top nine keywords (Fig. 1C). To distinguish proteins, which were associated with the UUKV particle preparation, either as integral components of the particle or as contaminant of the UUKV preparation, we also determined UUKV associated proteins in the absence of cells. As expected, we identified the UUKV structural proteins GN, GC, N, and L (Fig. 1D, Table II). From the host proteins identified in the A549 UUKV sample, only TRAPPC2L was also identified in the UUKV particle preparation (Fig. 1D and 1E). Because particle associated proteins may play a crucial role in the UUKV life cycle, we did not exclude TRAPPC2L from follow up analyses. GOCC term analysis of the 39 GN/GC co-purifying host proteins demonstrated a strong enrichment of membrane-associated proteins (77%) (Fig. 1F). When performing pathway analyses, we identified the 'Cellular Assembly and Organization' network as a top scoring network. For functional testing, we concentrated here on proteins within this network. As UUKV traffics within intracellular vesicles to enter and egress host cells, we selected nine proteins with clearly reported membrane association in this network (GBF1, GOLPH3, GOLPH3L, MARS, SEL1L, SURF4, TNPO3, TRAPPC2L and VDAC2) and additionally three proteins with high significance in this network (ERLEC1, HEATR3, SAAL1) for follow up analysis (Fig. 1G).
Table II. Selected MS hits used for follow-up infection study.
| Gene names | Protein names | Welch's t-test difference | Welch's t-test p value | Welch's t-test p value [-log10] |
|---|---|---|---|---|
| N | 10.86 | 2.49E-04 | 3.60 | |
| GCG2 | 9.93 | 8.54E-05 | 4.07 | |
| GNG1 | 9.39 | 2.54E-05 | 4.59 | |
| L | 8.05 | 6.50E-05 | 4.19 | |
| SAAL1 | Serum amyloid A like 1 | 8.30 | 6.46E-05 | 4.19 |
| HEATR3 | HEAT repeat containing 3 | 7.62 | 6.12E-07 | 6.21 |
| VDAC2 | Voltage dependent anion channel 2 | 6.86 | 1.38E-06 | 5.86 |
| GOLPH3 | Golgi phosphoprotein 3 | 6.49 | 6.39E-06 | 5.19 |
| TRAPPC2L | Trafficking protein particle complex 2 like | 6.26 | 4.33E-05 | 4.36 |
| TNPO3 | Transportin 3 | 6.20 | 7.70E-04 | 3.16 |
| ERLEC1 | Endoplasmic reticulum lectin 1 | 4.94 | 6.57E-05 | 4.18 |
| GBF1 | Golgi brefeldin A resistant guanine nucleotide exchange factor 1 | 4.73 | 7.57E-05 | 4.12 |
| SEL1L | SEL1L ERAD E3 ligase adaptor subunit | 4.50 | 7.50E-05 | 4.12 |
| SURF4 | Surfeit 4 | 4.39 | 8.37E-05 | 4.08 |
| GOLPH3L | Golgi phosphoprotein 3 like | 4.14 | 2.39E-05 | 4.62 |
| MARS | Methionyl-tRNA synthetase | 3.80 | 5.70E-05 | 4.24 |
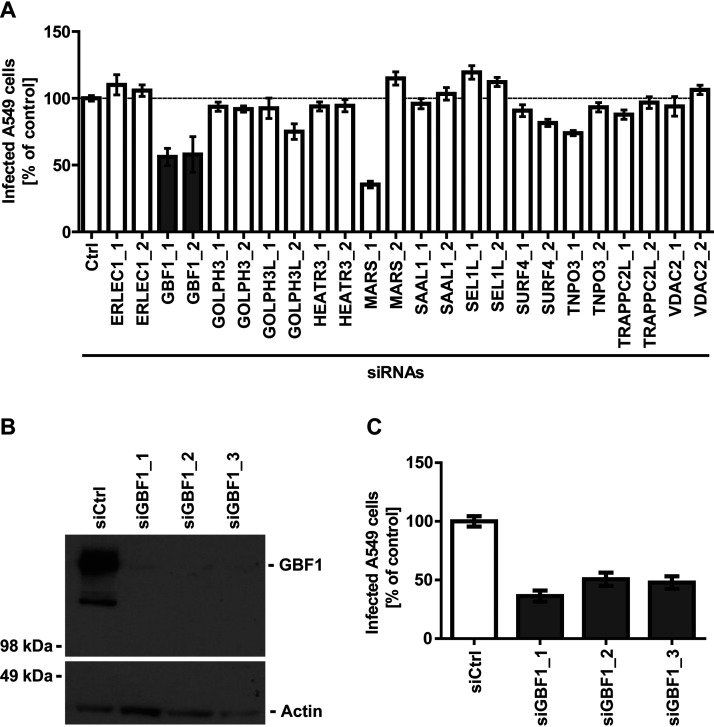
siRNA Screen Identifies GBF1 as Host Factor with a Role in UUKV Infection
Based on the mass spectrometric significance score and the GO analysis, we selected the 12 UUKV GN/GC interacting host factors ERLEC1, GBF1, GOLPH3, GOLPH3L, HEATR3, MARS, SAAL1, SEL1L, SURF4, TNOP3, TRAPPC2L, and VDAC2 for functional analysis. In a contaminant repository the 12 host factors were found in less than six out of 411 unspecific pullout experiments except for MARS (107/411), SURF4 (35/411), and VDAC2 (58/411) (58). To determine whether UUKV relies on these host factors for productive infection, we used a procedure combining siRNA silencing and a flow cytometry-based infection assay recently established in our group (65). Briefly, each gene was silenced by two single, non-overlapping siRNAs before infection. Infected cells were then subjected to immunofluorescence staining against newly synthetized UUKV nucleoprotein N and analysis by flow cytometry. This means that the N protein was used in this assay as a readout of infectious entry, i.e. the assay includes all steps from virus binding to genome release into the cytosol and viral replication until N expression. In this approach, and all the further experiments involving UUKV, we used the N protein to monitor infection and limited our assay to a single infectious cycle by harvesting cells 8 h after exposure to the virus. In A549 cells, a complete UUKV cycle lasts about 12 h.
In our siRNA screen, we found only one gene, GBF1, for which silencing by the two non-overlapping siRNAs resulted in a minimum 50% decrease in UUKV infection (Fig. 2A). Silencing of SEL1L led to a slight increase in infection, which was however not statistically significant (>0.05). Therefore, we pursued our investigation with GBF1, a GEF that governs recruitment of proteins to membranes and intracellular vesicle trafficking by promoting guanosine diphosphate to guanosine triphosphate exchange.
Fig. 2.
GBF1 silencing reduces UUKV infection in A549 cells. A, A549 cells treated with the indicated siRNAs (20 nm) were infected with UUKV (MOI ∼1) and harvested 8 h later. Infection was analyzed by flow cytometry following immunofluorescence staining against the viral nucleoprotein N, and results were normalized to the infection of cells treated with a negative-control siRNA (si_Ctrl). Arithmetic mean ± S.E. of the mean (S.E.) from three independent infection experiments shown. B, Efficiency of GBF1 silencing assessed by Western blotting. GBF1 levels in A549 cells treated with siRNAs targeting GBF1 were compared with GBF1 levels in cells treated with negative-control siRNAs (si_Ctrl). C, A549 cells were treated with three siRNAs against GBF1 (20 nm) and exposed to UUKV (MOI ∼5). Infection was analyzed via flow cytometry, after immunofluorescence staining against N, 8 h later. The values were normalized to the infection level in samples treated with si_Ctrl. Arithmetic means ± S.E. from three independent infection experiments shown.
To further address the potential involvement of GBF1 in UUKV infection, we first depleted A549 cells of GBF1 by using three siRNAs with non-overlapping target sites. GBF1 expression was reduced to a level at which it was nearly not detectable as assayed by western blotting (Fig. 2B). In the cells silenced for the GBF1 protein, UUKV infection was reduced to 40–50% of that in cells transfected with control siRNAs (Fig. 2C). The results indicate that GBF1 is, indeed, needed for efficient UUKV infection.
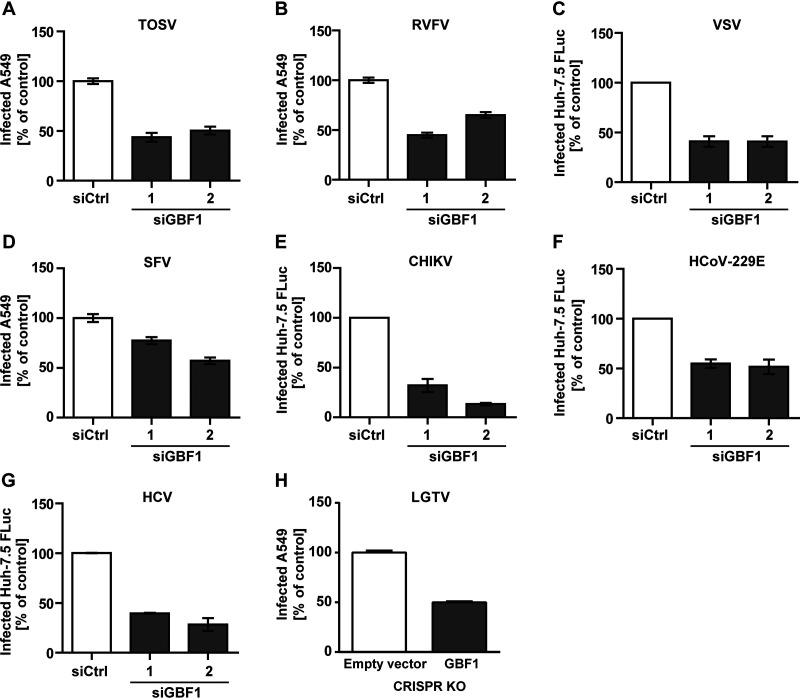
GBF1 Is A Proviral Host Factor For Of A Wide Range Of Enveloped RNA Viruses
As GBF1 has not previously been described to promote infection with phleboviruses but with other viruses, we next sought to systematically analyze which virus families rely on GBF1. To test this, we included in our flow cytometry-based infection investigation the two closely related phleboviruses TOSV and RVFV. To detect TOSV infection, we used primary antibodies that recognize all the viral structural proteins. RVFV was previously genetically modified to express GFP instead of the viral non-structural protein NSs (32). GFP was therefore used to monitor RVFV replication by flow cytometry rather than immunofluorescence staining. Using this approach, we found that silencing of GBF1 by two non-overlapping siRNAs resulted in a 40–60% decrease in infection of A549 cells by TOSV and RVFV (Fig. 3A and 3B).
Fig. 3.
GBF1 silencing reduced infection of a wide range of enveloped RNA viruses. Cells were treated with siRNAs targeting GBF1 and then exposed to (A) TOSV (MOI ∼2, A549 cells), (B) RVFV (MOI ∼3, A549 cells), (C) VSV (MOI 0.01, Huh-7.5 FLuc cells), (D) SFV (MOI ∼25, A549 cells), (E) CHIKV (MOI 1, Huh-7.5 FLuc cells), (F) HCoV-229E (MOI ∼0.1, Huh-7.5 FLuc cells), and (G) HCV (MOI ∼0.1, Huh-7.5 FLuc cells). To detect TOSV and SFV infected cells, primary antibodies targeting the envelope glycoprotein E2 and all viral structural proteins, respectively were used. For RVFV, VSV and CHIKV, GFP reporter viruses were used to infect target cells and GFP fluorescence was monitored. Infection was quantified by flow cytometry 8 h post inoculation for RVFV, SFV, and TOSV, and 16 h for VSV and CHIKV. Infection with HCoV-229E and HCV Renilla-luciferase reporters was determined 24 h post inoculation and 48 h post inoculation, respectively, by luminometry. H, 293T cells were transfected with three different GBF1 specific Cas9 guide RNAs (gRNAs) or a non-targeting control gRNA. Cells were then infected with LGTV at MOI of 1 and infection of Cas9 transfected cells was quantified by flow cytometry 48 h later. Data were normalized to infection of cells treated with non-targeting control siRNA or gRNA. Arithmetic means ± S.E. from three independent infection experiments shown.
We next expanded our investigation to nine viruses from different viral families. These viruses display differential tropism and a differential ability to amplify in cell monolayers in vitro. The choice of the cellular model system for each virus was therefore made based on the capacity of the cell lines to productively replicate each virus. For this reason, we extended the study from A549 cells to four other lines, namely BHK-21, Huh-7.5 FLuc, 293T, and TZM-bl cells.
In addition to viruses from the Phenuiviridae family, we first included VSV, as another negative strand RNA virus. We used a recombinant VSV strain expressing a GFP reporter and determined infectivity by flow cytometric quantification of GFP expression. In line with the results obtained with the different bunyaviruses and published work (26), Huh-7.5 FLuc cells treated with GBF1 targeting siRNAs were less susceptible to VSV as compared to cells treated with a negative control siRNA (Fig. 3C). These data demonstrate that GBF1 is involved in infection of negative strand RNA viruses of different families.
We then expanded our research to positive strand RNA viruses and their dependence on GBF1. We included two viruses belonging to the Togaviridae family: SFV and CHIKV. For SFV we used primary antibodies targeting the viral envelope glycoprotein E2 and determined infectivity by immunofluorescence. The knockdown of GBF1 expression led to a decrease in infection of A549 cells by SFV up to 40% (Fig. 3D). For CHIKV infection of Huh-7.5 FLuc cells we used a reporter virus expressing GFP from a subgenomic reporter (30) and determined infectivity by flow cytometry. The two siRNAs targeting GBF1 led to a minimum 60% reduction in GFP-positive cells, the second leading to an 80% reduction compared to the control cells (Fig. 3E). Further we tested HCoV-229E, HCV and Langat virus (LGTV) representing positive strand RNA viruses of the Coronaviridae and Flaviviridae family, respectively. HCoV-229E and HCV harbored a luciferase reporter gene, thus we determined infectivity by luciferase activity quantification. siRNA knockdown of GBF1 resulted in a 60 and 50% reduced infectivity for HCV and CoV in Huh-7.5 FLuc cells, respectively without measurable cytotoxicity (Fig. 3F and 3G). GBF1 was knocked down in 293T cells by expression of Cas9 with three different GBF1 specific gRNAs or a non-targeting control gRNA, followed by LGTV infection. The viral E protein was stained, and infection rate was measured in Cas9 transfected (GFP-positive) cells by flow cytometry. CRISPR knockdown of GBF1 resulted in 50% reduction of LGTV infection, confirming a role of GBF1 in the life cycle of enveloped RNA viruses (Fig. 3H).
Golgicide A (GCA) is an Inhibitor for a Range of Enveloped RNA Viruses
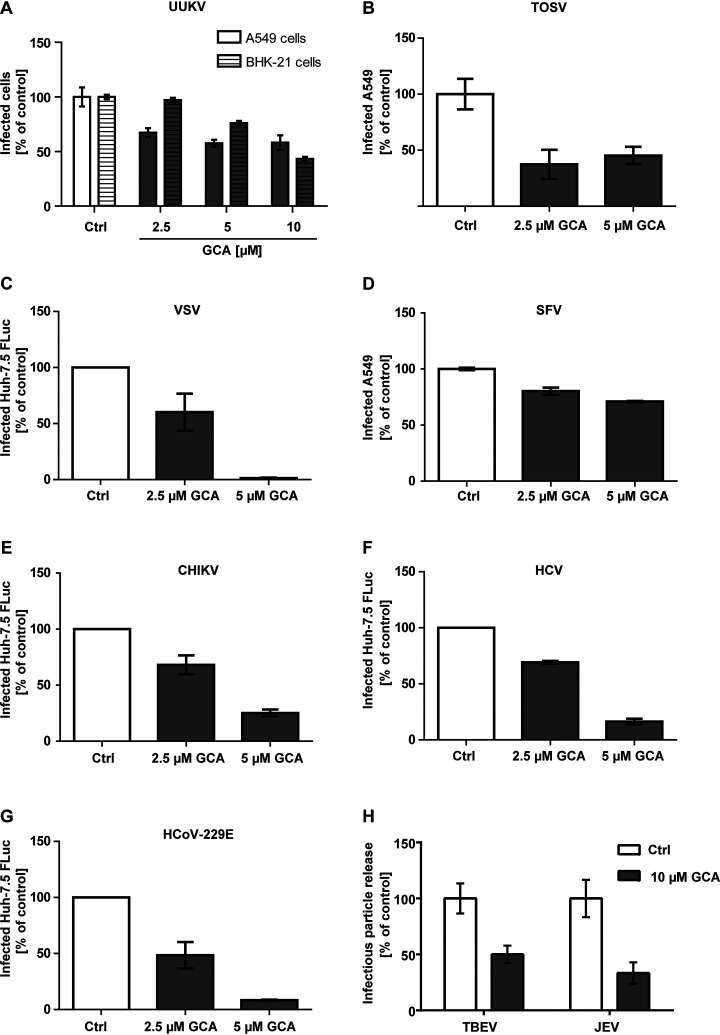
In an complementary approach, we tested the effect of GCA, a quinoline compound that specifically targets GBF1 and impairs GBF1-mediated intracellular vesicle trafficking (66), on infection with our panel of viruses. When A549 cells or BHK-21 cells were exposed to UUKV in the continuous presence of GCA, we observed a dose-dependent reduction of infection (Fig. 4A). Similarly, GCA reduced infection with TOSV in A549 cells (Fig. 4B), and VSV infection in Huh-7.5 FLuc cells (Fig. 4C).
Fig. 4.
The GBF1 inhibitor Golgicide A (GCA) blocks infection with RNA viruses. Cells were pretreated with GCA, an inhibitor of GBF1 activity, and then infected with (A) UUKV (MOI ∼5, A549 and MOI ∼0.5, BHK-21 cells), (B) TOSV (MOI ∼2, A549 cells), (C) VSV (MOI 0.01, Huh-7.5 FLuc cells), (D) SFV (MOI ∼15, A549 cells), (E) CHIKV (MOI ∼1, Huh-7.5 FLuc cells), (F) HCV (MOI ∼0.1, Huh-7.5 FLuc cells), and (G) HCoV-229E (MOI ∼0.1, Huh-7.5 FLuc cells) in the continuous presence of the drug. Infection was performed and quantified as described in Fig. 3, except for CHIKV where a Nano-luciferase reporter virus was used, and infection was determined by luminometry. H, A549 cells were infected with TBEV and JEV (MOI ∼1), and after 1 h, the inoculum was removed and GCA added. 24 h post-infection the supernatant was harvested, and viral titers were quantified by focus forming assay. Data are normalized to samples where the inhibitor had been omitted. Arithmetic means ± S.E. from three independent infection experiments shown.
To confirm that GBF1 is required for the life cycle of positive strand RNA viruses, we tested SFV in A549 cells, and Renilla- and Nano-luciferase reporter strains of CHIKV, HCV and CoV in Huh-7.5 FLuc cells. The constitutive expression of luciferase in Huh-7.5 cells enabled us to normalize the effect of GCA on infectivity (Renilla and Nano luciferase activity) to its effect on cell viability (Firefly luciferase activity). Pretreatment and infection in presence of 5 μm GCA led to a reduction of SFV, CHIKV, HCV and CoV infection to 20% of the DMSO control, whereas the lower concentration of 2.5 μm GCA only mildly decreased CHIKV, HCV and CoV infection (Fig. 4D–4G). Addition of 10 μm GCA directly after TBEV and JEV infection led to a reduction in release of infectious progeny particles of 50 and 66% respectively (Fig. 4H).
Altogether the data indicate that GBF1 promotes productive infection by a wide range of enveloped RNA viruses including UUKV, TOSV, VSV, SFV, CHIKV, HCV, CoV, TBEV and JEV. The results also imply that the GBF1 inhibitor GCA has a broad antiviral activity, not limited to UUKV.
GBF1 Is Dispensable for DNA Virus and Retrovirus Infection
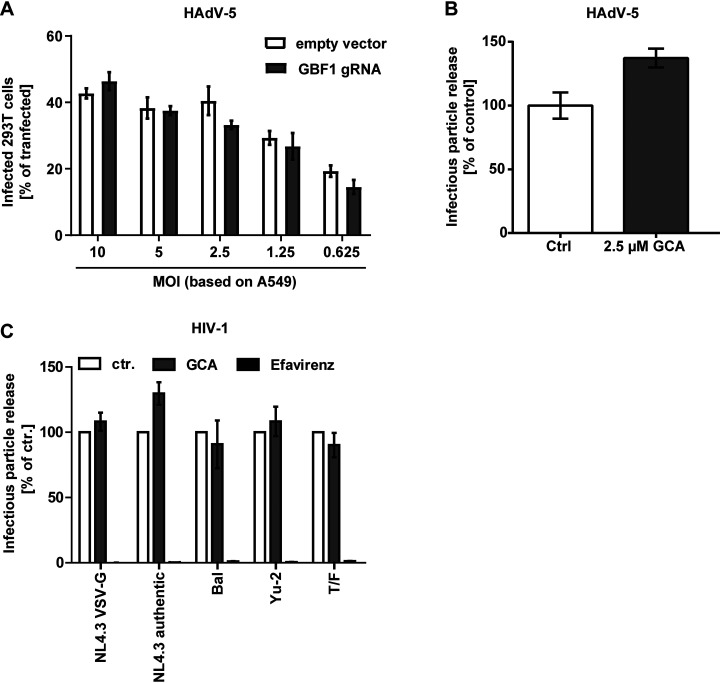
Our results indicate that many RNA viruses rely on GBF1 for productive infection. To investigate the role of GBF1 in DNA virus infection, we used HAdV-5. We knocked-down GBF1 in 293T cells by transient expression of Cas9 with three different GBF1 specific gRNAs or a non-targeting control gRNA, and measured HAdV-5 infection in Cas9 transfected (GFP-positive) cells by flow cytometry. No difference in HAdV-5 infectivity was observed whether GBF1 was present or not (Fig. 5A).
Fig. 5.
GBF1 is dispensable for adenovirus and HIV-1 infection. A, 293T cells were transfected with three different GBF1 specific Cas9 gRNAs or a non-targeting control gRNA. Cells were then infected with HAdV-5 at different MOIs and infection of Cas9 transfected cells was quantified by flow cytometry 48 h later. B, A549 cells were pretreated with GCA, an inhibitor of GBF1 activity, and then infected with HAdV-5 (MOI ∼0.1) in the continuous presence of the drug for 24 h. Thereafter number of progeny viruses was quantified by fluorescent-focus assay. C, GBF1 does not influence HIV-1 late steps of infection. TZM-bl cells were infected with VSV-G-pseudotyped HIV-1 NL4.3 GFP, HIV-1 NL4.3, HIV-1 BaL, HIV-1 YU-2, and the transmitted/founder strain CHO.77t (T/F). Cells were washed 16 h post-infection, and fresh medium containing either DMSO or GCA (each 2.5 μm) was added to the cells. In addition, cells were treated, in a separate condition, with the reverse transcriptase inhibitor efavirenz (20 nm). 24 h later, virus supernatant was collected and used for inoculation of TZM-bl cells, followed by determination of secreted infectivity by either luminometry or flow cytometry. Arithmetic means ± S.E. from three independent infection experiments shown.
To confirm that GBF1 was dispensable for DNA virus infection, we also investigated the impact of GCA. Specifically, we tested if GCA affects HAdV-5 replication and production of new adenovirus particles by infecting A549 cells in presence of GCA and subsequent analysis of release of infectious progeny viruses by fluorescent-focus assay. At 2.5 μm GCA, which we determined not to affect cell viability, progeny HAdV-5 particles production increased to 135% of solvent controls (Fig. 5B). This indicates that GBF1 is dispensable for HAdV-5 infection.
To unravel if GBF1 was required by all enveloped viruses independently of their replication site, we investigated if the lentivirus human immunodeficiency virus 1 (HIV-1) required GBF1 for infection. HIV-1, as all enveloped viruses, relies on the ER and Golgi for its envelope glycoprotein biosynthesis and maturation. In contrast to the above tested enveloped viruses, HIV-1 does not replicate in the cytoplasm. Interestingly, the efficiency of HIV-1 early entry steps was not affected by GCA in TZM-bl cells, observed by similar infectivity levels regardless of the presence of the drug (supplemental Fig. S2). Like the observation at the level of HIV-1 entry, we could not observe any alterations in the efficiency of secretion of HIV-1 infectivity from TZM-bl cells treated with GCA (Fig. 5C). Thus, in contrast to many RNA viruses GBF1 seems dispensable for DNA virus and lentivirus infection.
GBF1 Is Critical for UUKV Replication and Infectious Particle Release
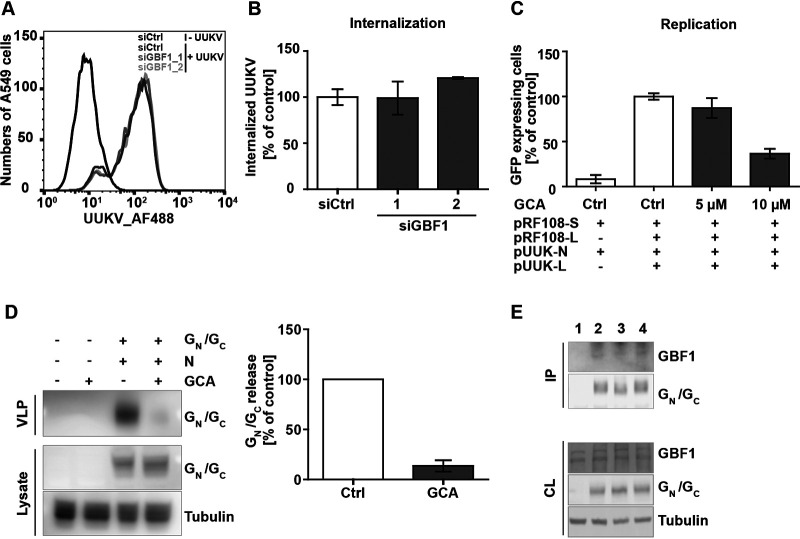
GBF1 is an important RNA replication factor for several plus strand RNA viruses including HCV, CoV, YFV, Coxsackievirus B and Sindbis virus (19). We therefore sought to determine which UUKV life cycle step was impaired by GBF1 silencing. Using AF488-labeled UUKV particles (UUKV-AF488) and flow cytometry analysis, we found that virus binding to the A549 cell surface was unaffected by GBF1 silencing (Fig. 6A). To assess virus uptake into A549 cells, a flow cytometry-based assay was used to distinguish virus particles at the plasma membrane from those which were internalized (41). This assay relies on the biochemical property of the membrane-impermeable dye, trypan blue, that quenches the fluorescence emitted by UUKV-AF488 at the cell surface while leaving intracellular viruses unquenched. When the amount of trypan blue-resistant fluorescence of cell-associated AF488-conjugated UUKV was analyzed 30 min after temperature shift, no significant difference was observed between GBF1 silenced and the control cells (Fig. 6B). This indicates that internalization of viral particles by endocytosis is functional in the absence of GBF1. Together the results show that GBF1 is playing a role in UUKV life cycle steps succeeding virus binding and uptake.
Fig. 6.
GBF1 depletion impairs UUKV replication and particle production. A, AF488-labeled UUKV (UUKV-AF488) was bound to A549 cells silenced for GBF1 expression (si_GBF1_1 and _2) or transfected with a negative-control siRNA (si_Ctrl) for 2 h on ice before fixation and flow cytometry analysis. B, UUKV-AF488 (MOI ∼20) was bound to A549 cells treated with si_Ctrl or siRNAs against GBF1 on ice before warming to 37 °C for 30 min. Internalization was quantified by flow cytometry after cell fixation and trypan blue treatment to quench fluorescence of cell surface-bound viruses. Internalization was normalized to internalization in cells treated with si_Ctrl. Arithmetic means ± S.E. from three independent infection experiments shown. C, BHK-21 cells were transfected with Pol I-driven plasmid DNAs encoding the anti-genomic L and S.ΔNSsGFP UUKV RNA segments, and then, subjected to GCA. Cells were harvested 24 h post-transfection, and GFP expression was analyzed by flow cytometry. D, BHK-21 cells were transfected with UUK-GN/GC and UUK-N expression plasmids media was exchanged after 24 h to media containing GCA (10 μm) and cells and media were collected 16–18 h post media exchange. Supernatant were concentrated by ultracentrifugation before protein analysis with Western blotting. Left panel shows representative blots and right panel show quantification, mean values and standard deviation (n = 3) of the released GN/GC normalized to intracellular GN/GC levels. E, 293T cells were transfected with plasmids expressing wt UUK-GN/GC (lane 2), UUK-GN/GC mutant where position 46–50 (lane 3) in the cytoplasmic tail is exchanged to alanine and the cytoplasmic tail UUK-GN/GC mutant 23–24 (lane 4). 24 h post-transfection IP was performed using the mAb 6G9E5 against GN, proteins in IPs and cell lysates (CL) were separated and detected by Western blotting. Representative blots are shown from three independent experiments.
To test whether GCA impairs the replication of UUKV, we used an assay derived from the reverse genetics system that we have recently developed for this virus (9). In this approach, the anti-genomic full-length RNA segments from UUKV rely on the cellular Pol I promoter for transcription (supplemental Fig. S3A). The M segment that encodes GN and GC was missing to prevent viral assembly and propagation, and the S segment was genetically engineered to express GFP instead of the viral non-structural protein NSs (supplemental Fig. S3B). BHK-21 cells were co-transfected with plasmids coding for the viral polymerase L and nucleoprotein N, whose expression depends on the CMV promoter. This allowed the transcription of the anti-genomic UUKV RNAs into mRNAs, and in turn, GFP expression. Transfected cells were imaged by flow cytometry and GFP served as readout to monitor viral transcription and replication (Fig. 6C). No signal was observed in the absence of plasmids coding for the viral polymerase L, confirming that GFP expression was specific to UUKV replication. In the presence of all constructs, including that encoding the polymerase L, we observed that about 20% of transfected cells expressed GFP. When these cells were subjected to treatment with GCA, the GFP signal decreased 3-fold with the highest concentration of the drug (Fig. 6C), suggesting that GBF1 was required for UUKV RNA genome transcription and replication.
We identified GBF1 to interact with UUKV GN/GC, which points toward an involvement of GBF1 in GN/GC trafficking or UUKV particle assembly. To uncouple viral replication from particle assembly, we performed a VLP assay (10), which scores for assembly and release of UUKV. In this assay, plasmids expressing the GN/GC and N protein were transfected into BHK-21 cells, 24 h post transfection media was exchanged and GCA was added, cells and released particles were analyzed by Western blotting 16–18 h post GCA addition. A strong reduction in released glycoprotein was detected in the GCA treated samples compared to the control, indicating that GBF1 is important for assembly and release of UUKV (Fig. 6D).
The GN contains a cytoplasmic tail (1–81 aa) which harbors signals necessary for intracellular localization (46–50 aa), packaging of viral nucleoproteins (76–81 aa) and initiation of budding (23–24 aa) (52, 53). GN/GC trafficking and budding mutants (aa changed to alanines), which we previously characterized (53), were therefore analyzed for their ability to bind to GBF1. To that end, co-IP analysis between wt and mutants of GN/GC with GBF1 was performed. Two different mutants were chosen; the alanine mutant 46–50 which is retained in the ER, and the mutant with alanine in positions 23–24 which is retained in the Golgi and unable to release particles (52, 53). 293T cells were transfected with GN/GC expressing plasmids and co-IP between GN/GC and endogenous expressed GBF1 was performed. Wt UUKV glycoproteins were able to interact with endogenous GBF1 (Fig. 6E), confirming the MS data. Analyzing the interactions between mutant glycoprotein and GBF1 showed no difference in coprecipitation compared to wt (Fig. 6E). Together, the results indicate that interactions between GBF1 and GN/GC already occur in the ER.
DISCUSSION
Phleboviruses replicate and assemble in cytosolic intracellular compartments (8). Virus genomes, together with the viral nucleoprotein and polymerase, form ribonucleoproteins that bud into the lumen of the Golgi to form enveloped progeny viruses. Viral particles are believed to be exported out of the cell through the secretory pathway. In this study, we identify the cis-Golgi resident GEF GBF1 as a critical host factor, which promotes phlebovirus replication, assembly, and release through interactions with the viral envelope glycoproteins GN/GC.
Phlebovirus glycoproteins are synthesized at the ER and transported to the cis-Golgi, where virus budding occurs. We observed that UUKV GN/GC glycoproteins bind to GBF1. In addition, we identified another ER - Golgi transport protein (TRAPPC2L) and two Golgi - plasma membrane transport proteins (GOLPH3, GOLPH3L) as putative GBF1 interaction partners. Silencing of GBF1, but not of the additional Golgi proteins, reproducibly reduced UUKV infection. GBF1 silencing or inhibition did not seem to affect UUKV cell entry up to the endocytosis step, but inhibited UUKV genome replication as well as particle assembly and release.
None of the identified UUKV associated proteins, except TRAPPC2L, were identified in the UUKV preparation used in this study. This suggests that the interaction with GBF1 and the additional 38 proteins occurs during infection. In this study we did not follow up on the UUKV associated proteins, which may be integral parts of the virions or contaminants of the virus preparation. Distinguishing between these two options and investigating the role of UUKV particle integrated host proteins is part of future work. Of note, the silencing efficiency of candidate genes other than GBF1 was not assessed in our siRNA-based middle-throughput screen. A role of the 11 other host factors in UUKV infection, tested in this screen, can thus not be formally excluded. Therefore, these factors should not be dismissed by others in future studies.
The fact that GBF1 is a cis-Golgi resident protein (67) is in line with our finding of a role of GBF1 in UUKV particle assembly and egress. Our work suggests a role for GBF1 in budding of UUKV into the Golgi. We find that GBF1 also interacts with the GN mutant aa 46–50 which is retained in the ER (52, 53). Thus GN/GC complex formation with GBF1 can occur at the ER and at the cis-Golgi. This result also indicates that GBF1 does not rely on the aa motif 46–50 to bind GN and interacts with another domain in the viral glycoprotein. Additional cellular factors are most likely involved, and essential, in the GN/GC maturation and progeny assembly processes. Moreover, our results show that the binding motif between GBF1 and GN/GC does not overlap with the budding motif aa 23–24 (53) in the cytoplasmic tail of GN. A role of GBF1 in infectious particle release was previously demonstrated for flaviviruses (43). Whether GBF1 binds to flavivirus glycoproteins to exert its function remains elusive. Here we show that GBF1 is a host factor important for infectious phlebovirus particle release.
Our study reports an interaction of GBF1 with UUKV glycoproteins. Previously, GBF1 interactions with DENV non-structural protein 5 and HCV non-structural protein 3 have been reported. Both viruses are members of the Flaviviridae family of enveloped positive strand RNA viruses and extensively reshape intracellular membranes to form their replication complexes. For DENV, GBF1 is essential for the formation of replication complexes (68). The protein further supports transport of DENV capsid protein to lipid droplets (21). HCV requires GBF1 rather for genome replication than for formation of replication complexes (20, 69). Thus, GBF1 seems to have a broad spectrum of functions in virus life cycle steps, which involve the transport of viral proteins between cytoplasmic membrane compartments and the function of membrane associated viral proteins.
Our study also shows an apparent dependence of UUKV on GBF1 for viral replication. Infection with isolates from the Peribunyaviridae family, closely related to the phleboviruses, have been shown to induce a dramatical reorganization of the ER/Golgi membrane network, which is believed to be critical for the replication of these viruses (70, 71). It is realistic to postulate that UUKV also triggers membrane rearrangements in infected cells and that GBF1 plays an important role in the process. UUKV glycoproteins have indeed been reported to cause morphological changes of the Golgi complex in the absence of virus maturation (72). In this model, GBF1 would have two functions in the UUKV life cycle, one rather early, during the establishment of viral transcription/replication, and a second occurring later, during virus replication and assembly. It remains to be determined whether the two molecular functions are interdependent.
Additional RNA viruses with (classical swine fever virus, mouse hepatitis virus, VSV) and without an envelope (poliovirus, Coxsackievirus B, hepatitis E virus) require GBF1 for RNA replication (19, 22–25, 73, 74). For several of these viruses, GBF1 is necessary for reshaping of intracellular membranes to build protected virus replication compartments. All these viruses have in common that they replicate in cytoplasmic compartments. Interestingly, influenza virus, a RNA virus that replicates in the nucleus of infected cells, was shown to rely on GBF1, not for viral replication, but for infectious particle assembly (75). Clearly, we could establish that non-enveloped DNA viruses such as adenoviruses and lentiviruses replicate and form particles independently of GBF1. The envelope glycoprotein of HIV-1, gp120, is synthesized in the ER and then transported through the Golgi to the plasma membrane, from where the virus assembles and is released in the outer space. Our observations therefore suggest that the loss of GBF1 function did not impair the general trafficking through the Golgi. Whether enveloped DNA viruses that replicate and assemble in the cytoplasm, e.g. poxviruses, and enveloped DNA viruses that replicate in the nucleus and have cytoplasmic assembly steps, e.g. herpes simplex virus, require GBF1 will be interesting to assess. Currently, our data demonstrate a specific role of GBF1 as a host factor essential for RNA viruses. Whether GBF1 is required for both viral assembly and replication in the case of most these RNA viruses will need further work.
Effectors of the antiviral innate immune response target all steps of the virus life cycle including replication and release of infectious particles. GBF1 is targeted by the interferon inducible antiviral protein viperin and this interaction blocks release of infectious flavivirus particles (43). Interestingly, viperin was shown to restrict Bunyamwera virus, which is a member of the Bunyavirales order like UUKV (76). Whether GBF1 binding to viperin causes the antiviral effect of viperin in this context and whether viperin similarly inhibits phlebovirus infection is a possibility that we currently test.
The broad function of GBF1 in infection with human pathogenic RNA viruses from diverse families including flaviviruses, phleboviruses, and alphaviruses make the protein an attractive target for broad spectrum antiviral therapies. GCA, the GBF1 specific inhibitor used in this study, is a reversible inhibitor, suggesting that short term treatment with minimal side effects would be feasible. In the absence of alternative antiviral agents, transient GBF1 inhibition could represent a treatment strategy for highly pathogenic phleboviruses such as SFTSV, HRTV, and RVFV.
DATA AVAILABILITY
The mass spectrometry data have been deposited at the ProteomeXchange Consortium (http://www.proteomexchange.org) (77) via the PRIDE partner repository (78) with the dataset identifier PXD015194. The protein interactions from this publication have been submitted to the IMEx (http://www.imexconsortium.org) consortium through IntAct and assigned the identifier IM-27097 (79).
Supplementary Material
Acknowledgments
We thank the NIH AIDS Reagent Program for providing reagents. We acknowledge C.M. Rice for Huh-7.5 cells and R. Bartenschlager for HCV infectious clones, V. Thiel for CoV 229E Renilla reporter viruses, G. Zimmer for the VSV GFP reporter virus, and G. Simmons for the CHIKV GFP reporter strain. We thank S. Kunz for critical reading of the manuscript. We are also grateful to Thomas Pietschmann and Thomas Schulz for constant support.
Footnotes
* This work was funded by the Knut and Alice Wallenberg Foundation, the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) - Projektnummer 158989968 - SFB 900 project C7 and DFG project GE-2145/3-2 to GG; by grants from CellNetworks Research Group funds, Heidelberg, and from DFG (LO-2338/1-1 and LO-2338/3-1) to PYL; by DFG funding (Collaborative Research Centre SFB900, project C8, Projektnummer 158989968; Priority Programme 1923), funding from the Helmholtz Center for Infection Research (HZI) and the Berlin Institute of Health (BIH) to CG; by the international Infection Biology Ph.D. program of Hannover Biomedical Research School to RM and VP; by the GABBA PhD Program to VP; and by the Swedish research council (vetenskapsrådet) dnr 2018–05851 and the Laboratory for molecular infection medicine Sweden (MIMS) to AKÖ.
 This article contains supplemental material. No author has an actual or perceived conflict of interest with the contents of this article.
This article contains supplemental material. No author has an actual or perceived conflict of interest with the contents of this article.
1 The abbreviations used are:
- ER
- endoplasmic reticulum
- HEPES
- 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- ARF
- ADP-ribosylation factor
- AP-MS
- Affinity purification-mass spectrometry
- AF
- Alexa Fluor
- CHIKV
- Chikungunya virus
- CRISPR
- clustered regularly interspaced short palindromic repeats
- COPI
- coat protein I
- Cas9
- CRISPR associated protein 9
- CMV
- cytomegalovirus
- DENV
- dengue virus
- DMEM
- Dulbecco's Modified Eagle's Medium
- DMSO
- dimethyl sulfoxide
- GFP
- green fluorescence protein
- FDR
- false discovery rate
- FBS
- fetal bovine serum
- GMEM
- Glasgow's minimal essential medium
- GBF1
- Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1
- GCA
- Golgicide A
- GEF
- guanine nucleotide exchange factor
- GOCC
- Gene Ontology category cellular component
- HCV
- hepatitis C virus
- HRTV
- Heartland virus
- HAdV-5
- human adenovirus serotype 5
- HCoV-229E
- human coronavirus 229E
- HIV-1
- human immunodeficiency virus 1
- gRNA
- guide RNA
- IP
- immunoprecipitation
- JEV
- Japanese encephalitis virus
- LGTV
- Langat virus
- LC-MS/MS
- liquid chromatography tandem mass spectrometry
- MS
- mass spectrometry
- MOI
- multiplicity of infection
- NCR
- noncoding region
- PBS
- Phosphate Buffered Saline
- PVDF
- polyvinylidene difluoride
- RVFV
- Rift Valley fever virus
- RT
- room temperature
- SFV
- Semliki Forest virus
- SFTSV
- severe fever with thrombocytopenia syndrome virus
- siRNA
- small interfering RNA
- si_Ctrl
- siRNA control
- SDS-PAGE
- sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SEM
- standard error of the mean
- TBEV
- tick-borne encephalitis virus
- TOSV
- Toscana virus
- TPB
- tryptose phosphate broth
- UUKV
- Uukuniemi virus
- VSV
- vesicular stomatitis virus
- VLP
- virus-like particle
- YFV
- yellow fever virus.
REFERENCES
- 1. Helenius, A. (2018) Virus entry: looking back and moving forward. J. Mol. Biol. 430, 1853–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lozach, P. Y. (2018) Early virus-host cell interactions. J. Mol. Biol. 430, 2555–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Griffiths, G., and Rottier, P. (1992) Cell biology of viruses that assemble along the biosynthetic pathway. Semin. Cell Biol. 3, 367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bedi, S., and Ono, A. (2019) Friend or foe: the role of the cytoskeleton in influenza A virus assembly. Viruses 11, pii: E46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matsuoka, Y., Chen, S. Y., and Compans, R. W. (1991) Bunyavirus protein transport and assembly. Curr. Top. Microbiol. Immunol. 169, 161–179 [DOI] [PubMed] [Google Scholar]
- 6. Uckeley, Z. M., Koch, J., Tischler, N. D., Léger, P., Lozach, P. Y. (2019) Cell biology of phlebovirus entry. Virologie 23, 176–187 [DOI] [PubMed] [Google Scholar]
- 7. Albornoz, A., Hoffmann, A. B., Lozach, P. Y., and Tischler, N. D. (2016) Early Bunyavirus-host cell interactions. Viruses 8, pii: E143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Léger, P., and Lozach, P. Y. (2015) Bunyaviruses:. from transmission by arthropods to virus entry into the mammalian host first-target cells. Future Virol. 10, 859–881 [Google Scholar]
- 9. Mazelier, M., Rouxel, R. N., Zumstein, M., Mancini, R., Bell-Sakyi, L., and Lozach, P. Y. (2016) Uukuniemi virus as a tick-borne virus model. Virol. J. 90, 6784–6798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Overby, A. K., Popov, V., Neve, E. P. A., and Pettersson, R. F. (2006) Generation and analysis of infectious virus-like particles of uukuniemi virus (bunyaviridae): a useful system for studying bunyaviral packaging and budding. J. Virol. 80, 10428–10435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Overby, A. K., Pettersson, R. F., Grünewald, K., and Huiskonen, J. T. (2008) Insights into bunyavirus architecture from electron cryotomography of Uukuniemi virus. Proc. Natl. Acad. Sci. U.S.A. 105, 2375–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lozach, P.-Y., Mancini, R., Bitto, D., Meier, R., Oestereich, L., Overby, A. K., Pettersson, R. F., and Helenius, A. (2010) Entry of bunyaviruses into mammalian cells. Cell Host Microbe 7, 488–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lozach, P. Y., Huotari, J., and Helenius, A. (2011) Late-penetrating viruses. Curr. Opin. Virol. 1, 35–43 [DOI] [PubMed] [Google Scholar]
- 14. Wang, S., Meyer, H., Ochoa-Espinosa, A., Buchwald, U., Onel, S., Altenhein, B., Heinisch, J. J., Affolter, M., and Paululat, A. (2012) GBF1 (Gartenzwerg)-dependent secretion is required for Drosophila tubulogenesis. J. Cell Sci. 125, 461–472 [DOI] [PubMed] [Google Scholar]
- 15. Claude, A., Zhao, B. P., Kuziemsky, C. E., Dahan, S., Berger, S. J., Yan, J. P., Armold, A. D., Sullivan, E. M., and Melançon, P. (1999) GBF1: A novel Golgi-associated BFA-resistant guanine nucleotide exchange factor that displays specificity for ADP-ribosylation factor 5. J. Cell Biol. 146, 71–84 [PMC free article] [PubMed] [Google Scholar]
- 16. Kawamoto, K., Yoshida, Y., Tamaki, H., Torii, S., Shinotsuka, C., Yamashina, S., and Nakayama, K. (2002. Jul.) GBF1, a guanine nucleotide exchange factor for ADP-ribosylation factors, is localized to the cis-Golgi and involved in membrane association of the COPI coat. Traffic 3, 483–495 [DOI] [PubMed] [Google Scholar]
- 17. García-Mata, R., Szul, T., Alvarez, C., and Sztul, E. (2003) ADP-ribosylation factor/COPI-dependent events at the endoplasmic reticulum-Golgi interface are regulated by the guanine nucleotide exchange factor GBF1. Mol. Biol. Cell 14, 2250–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Szul, T., Garcia-Mata, R., Brandon, E., Shestopal, S., Alvarez, C., and Sztul, E. (2005) Dissection of membrane dynamics of the ARF-guanine nucleotide exchange factor GBF1. Traffic 6, 374–385 [DOI] [PubMed] [Google Scholar]
- 19. Ferlin, J., Farhat, R., Belouzard, S., Cocquerel, L., Bertin, A., Hober, D., Dubuisson, J., and Rouillé, Y. (2018) Investigation of the role of GBF1 in the replication of positive-sense single-stranded RNA viruses. J. Gen. Virol. 99, 1086–1096 [DOI] [PubMed] [Google Scholar]
- 20. Goueslain, L., Alsaleh, K., Horellou, P., Roingeard, P., Descamps, V., Duverlie, G., Ciczora, Y., Wychowski, C., Dubuisson, J., and Rouillé, Y. (2010) Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J. Virol. 84, 773–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iglesias, N. G., Mondotte, J. A., Byk, L. A., De Maio, F. A., Samsa, M. M., Alvarez, C., and Gamarnik, A. V. (2015) Dengue virus uses a non-canonical function of the host GBF1-Arf-COPI system for capsid protein accumulation on lipid droplets. Traffic 16, 962–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Belov, G. A., Kovtunovych, G., Jackson, C. L., and Ehrenfeld, E. (2010) Poliovirus replication requires the N-terminus but not the catalytic Sec7 domain of ArfGEF GBF1. Cell Microbiol. 12, 1463–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Belov, G. A., Altan-Bonnet, N., Kovtunovych, G., Jackson, C. L., Lippincott-Schwartz, J., and Ehrenfeld, E. (2007) Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J. Virol. 81, 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lanke, K. H. W., van der Schaar, H. M., Belov, G. A., Feng, Q., Duijsings, D., Jackson, C. L., Ehrenfeld, E., and van Kuppeveld, F. J. (2009) GBF1, a guanine nucleotide exchange factor for Arf, is crucial for coxsackievirus B3 RNA replication. J. Virol. 83, 11940–11949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Farhat, R., Ankavay, M., Lebsir, N., Gouttenoire, J., Jackson, C. L., Wychowski, C., Moradpour, D., Dubuisson, J., Rouillé, Y., and Cocquerel, L. (2018) Identification of GBF1 as a cellular factor required for hepatitis E virus RNA replication. Cell Microbiol. 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Panda, D., Das, A., Dinh, P. X., Subramaniam, S., Nayak, D., Barrows, N. J., Pearson, J. L., Thompson, J., Kelly, D. L., Ladunga, I., and Pattnaik, A. K. (2011) RNAi screening reveals requirement for host cell secretory pathway in infection by diverse families of negative-strand RNA viruses. Proc. Natl. Acad. Sci. U.S.A. 108, 19036–19041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pettersson, R., and Kääriäinen, L. (1973) The ribonucleic acids of Uukuniemi virus, a noncubical tick-borne arbovirus. Virology 56, 608–619 [DOI] [PubMed] [Google Scholar]
- 28. Giorgi, C., Accardi, L., Nicoletti, L., Gro, M. C., Takehara, K., Hilditch, C., Morikawa, S., and Bishop, D. H. (1991) Sequences and coding strategies of the S RNAs of Toscana and Rift Valley fever viruses compared to those of Punta Toro, Sicilian Sandfly fever, and Uukuniemi viruses. Virology 180, 738–753 [DOI] [PubMed] [Google Scholar]
- 29. Helenius, A., Kartenbeck, J., Simons, K., and Fries, E. (1980) On the entry of Semliki forest virus into BHK-21 cells. J. Cell Biol. 84, 404–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vanlandingham, D. L., Tsetsarkin, K., Hong, C., Klingler, K., McElroy, K. L., Lehane, M. J., and Higgs, S. (2005) Development and characterization of a double subgenomic chikungunya virus infectious clone to express heterologous genes in Aedes aegypti mosquitoes. Insect Biochem. Mol. Biol. 35, 1162–1170 [DOI] [PubMed] [Google Scholar]
- 31. Jin, J., Sherman, M. B., Chafets, D., Dinglasan, N., Lu, K., Lee, T. H., Carlson, L. A., Muench, M. O., Simmons, G. (2018) An attenuated replication-competent chikungunya virus with a fluorescently tagged envelope. PLoS Negl. Trop. Dis. 12, e0006693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Billecocq, A., Gauliard, N., Le May, N., Elliott, R. M., Flick, R., and Bouloy, M. (2008. Sep 1) RNA polymerase I-mediated expression of viral RNA for the rescue of infectious virulent and avirulent Rift Valley fever viruses. Virology 378, 377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Billecocq, A., Vialat, P., and Bouloy, M. (1996. Dec.) Persistent infection of mammalian cells by Rift Valley fever virus. J. Gen. Virol. 77, 3053–3062 [DOI] [PubMed] [Google Scholar]
- 34. Léger, P., Tetard, M., Youness, B., Cordes, N., Rouxel, R. N., Flamand, M., and Lozach, P. Y. (2016) Differential use of the C-type lectins L-SIGN and DC-SIGN for phlebovirus endocytosis. Traffic 17, 639–656 [DOI] [PubMed] [Google Scholar]
- 35. Lindqvist, R., Mundt, F., Gilthorpe, J. D., Wölfel, S., Gekara, N. O., Kröger, A., and Overby, A. K. (2016) Fast type I interferon response protects astrocytes from flavivirus infection and virus-induced cytopathic effects. J. Neuroinflammation 13, 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Johansson, S. M. C., Nilsson, E. C., Elofsson, M., Ahlskog, N., Kihlberg, J., and Arnberg, N. (2007) Multivalent sialic acid conjugates inhibit adenovirus type 37 from binding to and infecting human corneal epithelial cells. Antiviral Res. 73, 92–100 [DOI] [PubMed] [Google Scholar]
- 37. Hoffmann, M., Wu, Y. J., Gerber, M., Berger-Rentsch, M., Heimrich, B., Schwemmle, M., and Zimmer, G. (2010) Fusion-active glycoprotein G mediates the cytotoxicity of vesicular stomatitis virus M mutants lacking host shut-off activity. J. Gen. Virol. 91, 2782–2793 [DOI] [PubMed] [Google Scholar]
- 38. Pfefferle, S., Schöpf, J., Kögl, M., Friedel, C. C., Müller, M. A., Carbajo-Lozoya, J., Stellberger, T., von Dall'Armi, E., Herzog, P., Kallies, S., Niemeyer, D., Ditt, V., Kuri, T., Züst, R., Pumpor, K., Hilgenfeld, R., Schwarz, F., Zimmer, R., Steffen, I., Weber, F., Thiel, V., Herrler, G., Thiel, H.J., Schwegmann-Wessels, C., Pöhlmann, S., Haas, J., Drosten, C., and von Brunn, A. (2011) The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 7, e1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Persson, R., and Pettersson, R. F. (1991. Jan.) Formation and intracellular transport of a heterodimeric viral spike protein complex. J. Cell Biol. 112, 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kielian, M., Jungerwirth, S., Sayad, K. U., and DeCandido, S. (1990. Oct) Biosynthesis, maturation, and acid activation of the Semliki Forest virus fusion protein. J. Virol. 64, 4614–4624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hoffmann, A. B., Mazelier, M., Léger, P., and Lozach, P. Y. (2018) Deciphering Virus Entry with Fluorescently Labeled Viral Particles. Methods Mol. Biol. 1836, 159–183 [DOI] [PubMed] [Google Scholar]
- 42. Ran, F. A., Hsu, P. D., Wright, J., Agarwala, V., Scott, D. A., and Zhang, F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vonderstein, K., Nilsson, E., Hubel, P., Nygård Skalman, L., Upadhyay, A., Pasto, J., Pichlmair, A., Lundmark, R., and Överby, A. K. (2018) Viperin targets flavivirus virulence by inducing assembly of noninfectious capsid particles. J. Virol. 92, pii, e01751–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Niedrig, M., Klockmann, U., Lang, W., Roeder, J., Burk, S., Modrow, S., Pauli, G. (1994) Monoclonal antibodies directed against tick-borne encephalitis virus with neutralizing activity in vivo. Acta Virol. 38, 141–149 [PubMed] [Google Scholar]
- 45. Overby, A. K., Popov, V. L., Niedrig, M., and Weber, F. (2010. Sep.) Tick-borne encephalitis virus delays interferon induction and hides its double-stranded RNA in intracellular membrane vesicles. J. Virol. 84, 8470–8483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lodermeyer, V., Ssebyatika, G., Passos, V., Ponnurangam, A., Malassa, A., Ewald, E., Stürzel, C. M., Kirchhoff, F., Rotger, M., Falk, C. S., Telenti, A., Krey, T., and Goffinet, C. (2018) The antiviral activity of the cellular glycoprotein LGALS3BP/90K is species specific. J. Virol. 92, pii: e00226–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Platt, E. J., Wehrly, K., Kuhmann, S. E., Chesebro, B., and Kabat, D. (1998) Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72, 2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wei, X., Decker, J. M., Liu, H., Zhang, Z., Arani, R. B., Kilby, J. M., Saag, M. S., Wu, X., Shaw, G. M., and Kappes, J. C. (2002) Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46, 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lozach, P. Y., Kühbacher, A., Meier, R., Mancini, R., Bitto, D., Bouloy, M., and Helenius, A. (2011) DC-SIGN as a receptor for phleboviruses. Cell Host Microbe 10, 75–88 [DOI] [PubMed] [Google Scholar]
- 50. Bruening, J., Lasswitz, L., Banse, P., Kahl, S., Marinach, C., Vondran, F. W., Kaderali, L., Silvie, O., Pietschmann, T., Meissner, F., and Gerold, G. (2018) Hepatitis C virus enters liver cells using the CD81 receptor complex proteins calpain-5 and CBLB. PLoS Pathog. 14, e1007111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schneider, C., Newman, R. A., Sutherland, D. R., Asser, U., and Greaves, M. F. (1982. Sep 25) A one-step purification of membrane proteins using a high efficiency immunomatrix. J. Biol. Chem. 257, 10766–10769 [PubMed] [Google Scholar]
- 52. Overby, A. K., Pettersson, R. F., and Neve, E. P. A. (2007) The glycoprotein cytoplasmic tail of Uukuniemi virus (Bunyaviridae) interacts with ribonucleoproteins and is critical for genome packaging. J. Virol. 81, 3198–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Overby, A. K., Popov, V. L., Pettersson, R. F., and Neve, E. P. A. (2007) The cytoplasmic tails of Uukuniemi Virus (Bunyaviridae) G(N) and G(C) glycoproteins are important for intracellular targeting and the budding of virus-like particles. J. Virol. 81, 11381–11391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tusher, V. G., Tibshirani, R., and Chu, G. (2001. Apr 24) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gerold, G., Meissner, F., Bruening, J., Welsch, K., Perin, P. M., Baumert, T. F., Vondran, F. W., Kaderali, L., Marcotrigiano, J., Khan, A. G., Mann, M., Rice, C. M., and Pietschmann, T. (2015) Quantitative proteomics identifies serum response factor binding protein 1 as a host factor for hepatitis C virus entry. Cell Rep. 12, 864–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cox, J., Neuhauser, N., Michalski, A., Scheltema, R. A., Olsen, J. V., and Mann, M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 57. Cox, J., and Mann, M. (2008. Dec.) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 58. Mellacheruvu, D., Wright, Z., Couzens, A. L., Lambert, J. P., St-Denis, N. A., Li, T., Miteva, Y. V., Hauri, S., Sardiu, M. E., Low, T. Y., Halim, V. A., Bagshaw, R. D., Hubner, N. C., Al-Hakim, A., Bouchard, A., Faubert, D., Fermin, D., Dunham, W. H., Goudreault, M., Lin, Z. Y., Badillo, B. G., Pawson, T., Durocher, D., Coulombe, B., Aebersold, R., Superti-Furga, G., Colinge, J., Heck, A. J., Choi, H., Gstaiger, M., Mohammed, S., Cristea, I. M., Bennett, K. L., Washburn, M. P., Raught, B., Ewing, R. M., Gingras, A. C., and Nesvizhskii, A. I. (2013) The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cox, J., Hein, M. Y., Luber, C. A., Paron, I., Nagaraj, N., and Mann, M. (2014. Sep.) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Team RC. (2016) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 61. Team RS (2016) RStudio: Integrated development environment for R. Boston, MA: RStudio [Google Scholar]
- 62. Tyanova, S., Temu, T., Sinitcyn, P., Carlson, A., Hein, M. Y., Geiger, T., Mann, M., and Cox, J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 [DOI] [PubMed] [Google Scholar]
- 63. Magrane, M., and UniProt Consortium (2011) UniProt Knowledgebase: a hub of integrated protein data. Database 2011, bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gene Ontology Consortium (2015) Gene Ontology Consortium: going forward. Nucleic Acids Res. 43, D1049–D1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Meier, R., Franceschini, A., Horvath, P., Tetard, M., Mancini, R., von Mering, C., Helenius, A., and Lozach, P. Y. (2014. Aug) Genome-wide small interfering RNA screens reveal VAMP3 as a novel host factor required for Uukuniemi virus late penetration. J. Virol. 88, 8565–8578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sáenz, J. B., Sun, W. J., Chang, J. W., Li, J., Bursulaya, B., Gray, N. S., and Haslam, D. B. (2009) Golgicide A reveals essential roles for GBF1 in Golgi assembly and function. Nat. Chem. Biol. 5, 157–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Szul, T., Grabski, R., Lyons, S., Morohashi, Y., Shestopal, S., Lowe, M., and Sztul, E. (2007) Dissecting the role of the ARF guanine nucleotide exchange factor GBF1 in Golgi biogenesis and protein trafficking. J. Cell Sci. 120, 3929–3940 [DOI] [PubMed] [Google Scholar]
- 68. Carpp, L. N., Rogers, R. S., Moritz, R. L., and Aitchison, J. D. (2014. Nov.) Quantitative proteomic analysis of host-virus interactions reveals a role for Golgi brefeldin A resistance factor 1 (GBF1) in dengue infection. Mol. Cell. Proteomics 13, 2836–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lebsir, N., Goueslain, L., Farhat, R., Callens, N., Dubuisson, J., Jackson, C. L., and Rouillé, Y. (2019. Mar 15) Functional and physical interaction between the Arf activator GBF1 and Hepatitis C Virus NS3 protein. J. Virol. 93, pii: e01459–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fontana, J., López-Montero, N., Elliott, R. M., Fernández, J. J., and Risco, C. (2008. Oct.) The unique architecture of Bunyamwera virus factories around the Golgi complex. Cell Microbiol. 10, 2012–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sanz-Sánchez, L., and Risco, C. (2013) Multilamellar structures and filament bundles are found on the cell surface during bunyavirus egress. PLoS ONE 8, e65526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gahmberg, N., Kuismanen, E., Keränen, S., and Pettersson, R. F. (1986) Uukuniemi virus glycoproteins accumulate in and cause morphological changes of the Golgi complex in the absence of virus maturation. J. Virol. 57, 899–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Verheije, M. H., Raaben, M., Mari, M., Te Lintelo, E. G., Reggiori, F., van Kuppeveld, F. J. M., Rottier, P. J., and de Haan, C. A. (2008) Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 4, e1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Liang, W., Zheng, M., Bao, C., and Zhang, Y. (2017) CSFV proliferation is associated with GBF1 and Rab2. J. Biosci. 42, 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Watanabe, T., Kawakami, E., Shoemaker, J. E., Lopes, T. J. S., Matsuoka, Y., Tomita, Y., Kozuka-Hata, H., Gorai, T., Kuwahara, T., Takeda, E., Nagata, A., Takano, R., Kiso, M., Yamashita, M., Sakai-Tagawa, Y., Katsura, H., Nonaka, N., Fujii, H., Fujii, K., Sugita, Y., Noda, T., Goto, H., Fukuyama, S., Watanabe, S., Neumann, G., Oyama, M., Kitano, H, and Kawaoka, Y. (2014) Influenza virus-host interactome screen as a platform for antiviral drug development. Cell Host Microbe. 16, 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Carlton-Smith, C., and Elliott, R. M. (2012) Viperin, MTAP44, and protein kinase R contribute to the interferon-induced inhibition of Bunyamwera Orthobunyavirus replication. J. Virol. 86, 11548–11557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Vizcaíno, J. A., Deutsch, E. W., Wang, R., Csordas, A., Reisinger, F., Ríos, D., Dianes, J. A., Sun, Z., Farrah, T., Bandeira, N., Binz, P. A., Xenarios, I., Eisenacher, M., Mayer, G., Gatto, L., Campos, A., Chalkley, R. J., Kraus, H. J., Albar, J. P., Martinez-Bartolomé, S., Apweiler, R., Omenn, G. S., Martens, L., Jones, A. R., and Hermjakob, H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Vizcaíno, J. A., Csordas, A., Del-Toro, N., Dianes, J. A., Griss, J., Lavidas, I., Mayer, G., Perez-Riverol, Y., Reisinger, F., Ternent, T., Xu, Q. W., Wang, R., and Hermjakob, H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, 11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Orchard, S., Ammari, M., Aranda, B., Breuza, L., Briganti, L., Broackes-Carter, F., Campbell, N.H., Chavali, G., Chen, C., del-Toro, N., Duesbury, M., Dumousseau, M., Galeota, E., Hinz, U., Iannuccelli, M., Jagannathan, S., Jimenez, R., Khadake, J., Lagreid, A., Licata, L., Lovering, R.C., Meldal, B., Melidoni, A.N., Milagros, M., Peluso, D., Perfetto, L., Porras, P., Raghunath, A., Ricard-Blum, S., Roechert, B., Stutz, A., Tognolli, M., van Roey, K., Cesareni, G., and Hermjakob, H. (2014) The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 42, D358–D363 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data have been deposited at the ProteomeXchange Consortium (http://www.proteomexchange.org) (77) via the PRIDE partner repository (78) with the dataset identifier PXD015194. The protein interactions from this publication have been submitted to the IMEx (http://www.imexconsortium.org) consortium through IntAct and assigned the identifier IM-27097 (79).