Abstract
Purpose
One in eight women will develop breast cancer, 15–20% of whom will have triple-negative breast cancer (TNBC), an aggressive breast cancer with no current targeted therapy. We have demonstrated that riluzole, an FDA-approved drug for treating amyotrophic lateral sclerosis, inhibits growth of TNBC. In this study, we explore potential synergism between riluzole and paclitaxel, a chemotherapeutic agent commonly used to treat TNBC, in regulating TNBC proliferation, cell cycle arrest, and apoptosis.
Methods
TNBC cells were treated with paclitaxel and/or riluzole and synergistic effects on cell proliferation were quantified via MTT assay and Compusyn analysis. Apoptosis was observed morphologically and by measuring cleaved PARP/caspase three products. Microarray analysis was performed using MDA-MB-231 cells to examine cell cycle genes regulated by riluzole and any enhanced effects on paclitaxel-mediated cell cycle arrest, determined by FACS analysis. These results were confirmed in vivo using a MDA-MB-231 xenograft model.
Results
Strong enhanced or synergistic effects of riluzole on paclitaxel regulation of cell cycle progression and apoptosis was demonstrated in all TNBC cells tested as well as in the xenograft model. The MDA-MB-231, SUM149, and SUM229 cells, which are resistant to paclitaxel treatment, demonstrated the strongest synergistic or enhanced effect. Key protein kinases were shown to be upregulated in this study by riluzole as well as downstream cell cycle genes regulated by these kinases.
Conclusions
All TNBC cells tested responded synergistically to riluzole and paclitaxel strongly suggesting the usefulness of this combinatorial treatment strategy in TNBC, especially for patients whose tumors are relatively resistant to paclitaxel.
Keywords: Riluzole, Triple-negative breast cancer, cell cycle, Apoptosis, Paclitaxel
Introduction
According to the American Cancer Society, one in eight women in the U.S. will develop breast cancer [1], and approximately 15% of these cases will be triple-negative breast cancer (TNBC), an aggressive subtype of breast cancer that does not respond to drugs targeting the estrogen receptor (ER) or HER2 [2]. Although newer regimens are becoming more accepted, standard-of-care chemotherapy for TNBC still typically consists of concurrent doxorubicin and cyclophosphamide, followed by a taxane [2]. Unfortunately, these drugs are not targeted therapies, and they often fail to completely eradicate the tumor, resulting in recurrence [3–5]. In addition, this multidrug chemotherapy regimen induces toxic side effects. As such, a targeted therapy, such as tamoxifen for the ER+ breast cancer subtype and trastuzumab for the HER2+ breast cancer subtype, would be highly desirable for TNBC.
To identify a targeted agent for TNBC, in a previous study we reported that TNBC expresses higher levels of metabotropic glutamate receptor-1 (mGluR1) than non-cancerous epithelium and other breast cancer subtypes [6, 7]. Additionally, we identified mGluR1 as a pro-tumor and pro-angiogenic factor [6–8]. Studies in melanoma have shown that riluzole, an FDA-approved orally available drug used to treat amyotrophic lateral sclerosis [9, 10], functions as an mGluR1 inhibitor resulting in reduced cell proliferation and tumor growth [11, 12]. Our results showed that riluzole inhibits TNBC proliferation, invasion, and colony formation, strongly suggesting a role for riluzole in the systemic therapy of TNBC [7, 8, 13]. However, the correlation between mGluR1 levels in TNBC cells and inhibition of cell growth using riluzole was not as tight as demonstrated to be in melanoma. Additionally, mGluR1 knockdown and overexpression studies found little effect on cell sensitivity to riluzole [13], suggesting that in TNBC riluzole also works through mGluR1-independent pathways, which may involve inhibition of various signaling pathways involving PI3′K, Akt, or PKC [14–17].
The goal of this study is to determine whether combining riluzole with paclitaxel, currently part of the standard of care for TNBC, in preclinical models of TNBC results in synergistic or additive anti-tumor effects. Paclitaxel is known to inhibit cell growth by inhibiting spindle function attributed to its suppression of microtubule dynamics [18]. However, resistance remains a significant problem when using paclitaxel to treat TNBC [19, 20]. Our results demonstrate a strong synergistic or enhanced effect of riluzole and paclitaxel on cell growth and apoptosis in both TNBC cell lines and a TNBC xenograft model.
Materials and methods
Reagents and cell culture
Cell culture reagents were purchased from Life Technologies (Carlsbad, CA) except fetal bovine serum (FBS), purchased from Thermo Fisher Scientific (Waltham, MA). Human SUM TNBC cell lines were a kind gift from Dr. Stephen P. Ethier. All other cell lines (MDA-MB-231, MDA-MB-468, BT549) were purchased from ATCC. Cell lines were authenticated via cytogenetic analysis and used within 6 months of purchase or stored in liquid nitrogen for future use.
Cell proliferation
To determine whether riluzole and paclitaxel synergistically inhibit proliferation of TNBC cells, MTT assays were performed as previously described [13]. Briefly, cells were treated with riluzole (Sigma-Aldrich, 1–50 μM) and paclitaxel (Invitrogen, 0.5–25 nM), at constant ratio of 1:2000 (riluzole:paclitaxel). Cell viability was determined on day 3 by MTT assay [13]. In some experiments, cell numbers were determined in parallel with the MTT assay by counting manually on a hemocytometer. To assess the interaction between riluzole and paclitaxel, Compusyn 1.0 software was used to generate isoboles. Using this method of isoboles [21], the dose-effect data of individual drugs measured above was used to determine the expected combination and then statistically compared to the actual combination effect measured to determine synergism, additivity, or anti-additive interactions. The combination index (CI) at different Fa levels was also determined using this software based on the following equation:
Western blot analysis of cleaved PARP and caspase 3 proteins
Cells were treated with riluzole (5 or 10 μM) and/or paclitaxel (5–10 nM) and collected by scraping in RIPA lysis buffer (Santa Cruz, CA). Protein (10–30 μg) was separated by SDS-PAGE and transferred to PVD membranes. Detection of PARP and caspase 3 cleavage products was performed using respective primary and secondary antibodies (Cell Signaling Technology, Danvers, MA) and detected by chemiluminescence. Primary blots were reprobed with anti-GAPDH antibody (Novus Biologicals, Littleton, CO).
Live cell analysis
SUM149 cells were treated with riluzole (15 μM) and/or paclitaxel (7.5 nM) for 72 h. Cell images were captured on a Nikon Ti E-Series inverted microscope and morphologically analyzed.
Cell cycle analysis
MDA-MB-231 and SUM149 cells were treated with riluzole and/or paclitaxel and stored in PBS/ethanol at 4 °C before incubation with anti-phospho-Ser/Thr-Pro MPM-2 (EMD Millipore Corp, Temecula, CA), that recognizes proteins phosphorylated by M-phase promoting factor followed by anti-mouse IgG (Sigma-Aldrich). Cells were washed, incubated in 10 μg/ml RNase A and 20 μg/ml PI (Sigma-Aldrich), and analyzed by FACS. Cells were treated with fixable viability dye eFluor 450 (eBioscience; San Diego, CA) before ethanol fixation.
Microarray analysis of riluzole mediated pathways
MDA-MB-231 cells were treated with riluzole (25 μM) or vehicle and RNA isolated using RNeasy Plus Mini Kit (Qiagen, Valencia, CA) including an extra DNase step. RNA was quality assessed using the 2100 Bioanalyzer System and hybridized to the Illumina® Human HT-12v4 array then washed, stained, and scanned. The data generated were uploaded to BeadStudio, background-corrected, and normalized using rank invariant algorithm. Differentially expressed genes were identified using the Illumina Custom Error Model and genes differentially expressed were uploaded to Genomatix software suite to determine over-represented canonical pathways. The online DAVID tool was used to determine Gene Ontology Biological Process terms over-represented by the data.
RT-PCR analysis of M-phase regulators
TNBC cells were treated with riluzole (37.5 or 15 μM) or vehicle (0.1% DMSO) and RNA extracted using RNeasy Plus Mini Kit (Qiagen). Reverse transcription was performed with 2 μg RNA using High-capacity cDNA Reverse Transcription Kit (Life Technologies). PCR was performed using ABsolute QPCR Mix (Thermo Scientific) with the following sense/anti-sense oligonucleotide primers:
| PLK1 | Sense | 5′-TACCTTGTTAGTGGGCAAACC-3′ |
| Anti-sense | 5′-GGGTTGATGTGCTTGGGAATA-3′ | |
| CDC25B | Sense | 5′-GTGCTTGGTCTGTTTGACTTTAC-3′ |
| Anti-sense | 5′-GACCGAGTGGGTAACTGATATTT-3′ | |
| CCNB1 | Sense | 5′-GATGCAGAAGATGGAGCTGAT-3′ |
| Anti-sense | 5′-TCCCGACCCAGTAGGTATTT-3′ | |
| CCNB2 | Sense | 5′-GATCCCTCAGCTGAACTCAAA-3′ |
| Anti-sense | 5′-GGCACAATGAAGCACACATC-3′ | |
| CDC25C | Sense | 5′-CATCCACAAGAGAGGAAGGAAG-3′ |
| Anti-sense | 5′-GACATCTGGACAGACGGTAAAG-3′ | |
| GAPDH | Sense | 5′-ACA ACT TTG GTA TCG TGG AAG G-3′ |
| Anti-sense | 5′-CAG TAG AGG CAG GCA TGA TGT TC-3′ |
For tissue samples, RNA was isolated from tissue (50 mg) using the RNeasy Plus Universal Mini Kit (Qiagen) and RT-PCR performed as described above.
Xenografts
MDA-MB-231 cells (1 × 106) in Matrigel (1:1) were injected into mammary fat pads of female SCID/beige mice, 6 and 8 weeks old (Envigo; Haslett, MI) and allowed to grow until xenografts reached a mean size of 40–50 mm3 (approximately 2 weeks), at which point they were divided into eight experimental groups, consisting of ten mice per group, such that the means did not vary more than 10%. Treatment then began with paclitaxel (2.7, 4.5, 7.2 mg/kg) alone or together with 18 mg/kg riluzole or vehicle (DMSO). Paclitaxel was administered i.v. three times a week and riluzole was given i.p. five times per week. Tumor size was measured three times a week using a Vernier caliper and calculated using the following formula: length × width × depth/2. Treatment continued until tumors in control group either reached a volume of 1000 mm3 or ulcerated, whichever came first. For cell cycle gene analysis, a second xenograft study was performed as described above but mice only received 1 week of treatment with either riluzole (18 mg/kg) or vehicle (DMSO). After treatment, tumors were harvested and snap frozen in liquid nitrogen until gene analysis.
Statistical analyses
Numerical data were analyzed using GraphPad Prism (v.7.0) for Macintosh. Unless otherwise indicated, all numerical results are expressed as mean ± SEM and statistical analysis performed by one-way or two-way repeated measures analysis of variance (ANOVA) followed by multiple comparison procedure with Student–Newman Keuls method. A value of p ≤ 0.05 or p ≤ 0.01 was considered significant. For Compusyn analyses, the conformity of data to the mass action law was confirmed for all treatment groups by p ≤ 0.05. Differentially expressed genes using Illumina platform were identified using the Illumina Custom Error Model. A p value was associated with every differential call and genes with a p value more than 0.05 were discarded. In addition, genes were discarded if fold-change in expression was less than 1.3.
Results
Riluzole and paclitaxel act synergistically to inhibit cell proliferation of various TNBC cells
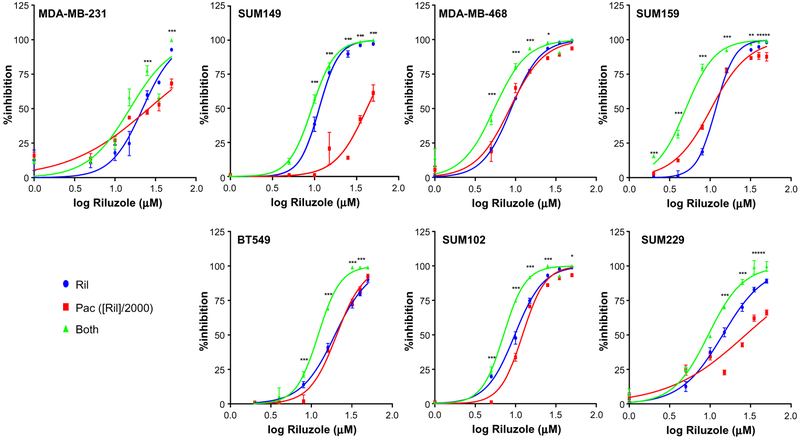
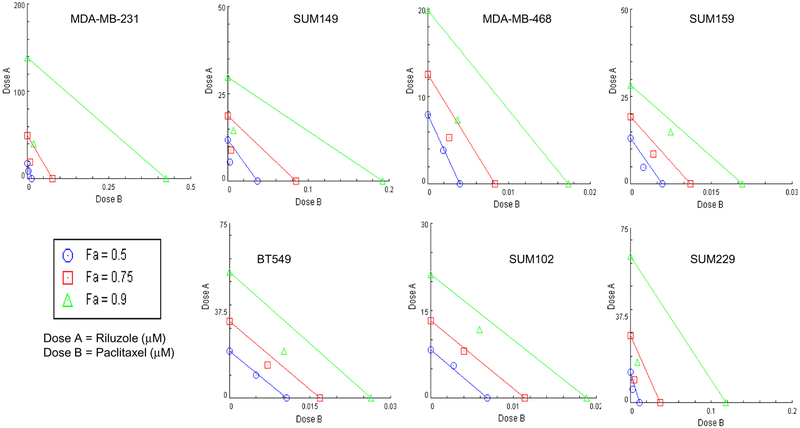
Cell proliferation in various TNBC cell lines was measured after treatment with riluzole and/or paclitaxel. As expected, riluzole significantly inhibited cell proliferation in a dose-dependent manner in all TNBC cell lines tested, with ED50 values ranging from 5 to 20 μM (Fig. 1), consistent with previous studies [13]. Paclitaxel also significantly inhibited cell proliferation in all TNBC cells but with a wider range of ED50 values, ranging from 4 to 40 nM. MDA-MB-231, SUM149, and SUM229 cell lines had higher ED50 values and never reached 75% inhibition suggesting resistance to paclitaxel compared to other cell lines. With the combined dose, growth inhibition was significantly enhanced in all TNBC cells compared to paclitaxel treatment alone (Fig. 1). Isobologram analysis using Compusyn software determined that the enhanced effect of the combined treatment in all cell lines was synergistic for at least one of the fractional effect (Fa) doses demonstrated in the isobologram and determined by CI values (Fig. 2 and Table 1). Interestingly, the strongest synergistic effect (i.e., synergism at all Fa doses) was observed in the more resistant cell lines (MDA-MB-231, SUM149, and SUM229).
Fig. 1.
Riluzole and paclitaxel inhibit cell proliferation of various TNBC cells. Cells were plated in RPMI containing 5% FBS at 1 × 104 cells/well in 96-well plates and treated with varying concentrations of riluzole and paclitaxel at a constant ratio of 1:2000 (paclitaxel to riluzole). Cell proliferation was determined on day three using MTT assay and initial absorbance on day of treatment was subtracted from absorbance on day 3 and results expressed as percentage of vehicle (0.05% DMSO)-treated control. Results represent three experiments performed in triplicate and graphed using GraphPad Prism software. Two-way ANOVA test was performed on the data where *p < 0.05, **p < 0.01, ***p < 0.001 when comparing combined treatment to paclitaxel alone. For all cell lines tested, inhibition of cell proliferation was significantly greater in the presence of paclitaxel and riluzole together compared to paclitaxel treatment alone
Fig. 2.
Riluzole and paclitaxel inhibit cell proliferation in a synergistic manner. Isobolograms of the data generated in Fig. 1 demonstrating synergism in all cell lines tested. Isobolograms were generated using Compusyn 1.0 software. Using this method, the dose–effect data of the individual drugs measured above were used to determine the expected combination and then statistically compared to the actual combination effect measured to determine whether there was synergism, additivity, or anti-additive interactions. These results are expressed in an isobologram that graphs the effective doses of inhibition at 50% (Fa 0.5), 75% (Fa 0.75), and 90% (Fa 0.9) for the individual drugs as x- and y-intercept values. Synergism is demonstrated in all cell lines by the dose pair plotting as a point (symbol) below their respective Fa isobole or line
Table 1.
Combination Index (CI) values for riluzole/paclitaxel combination treatment
| Cell line | CI values | ||
|---|---|---|---|
| Fa = 0.50 | Fa = 0.75 | Fa = 0.90 | |
| MDA-MB-231 | 0.86 | 0.52 | 0.34 |
| SUM149 | 0.54 | 0.53 | 0.53 |
| MDA-MB-468 | 0.98 | 0.75 | 0.58 |
| SUM159 | 0.78 | 0.82 | 0.89 |
| BT549 | 0.96 | 0.85 | 0.76 |
| SUM102 | 1.07 | 0.97 | 0.87 |
| SUM229 | 0.68 | 0.48 | 0.35 |
Synergistic (CI < 1); Additive (CI = 1); Antagonistic (CI > 1)
Riluzole and paclitaxel together enhance cell apoptosis of various TNBC cells
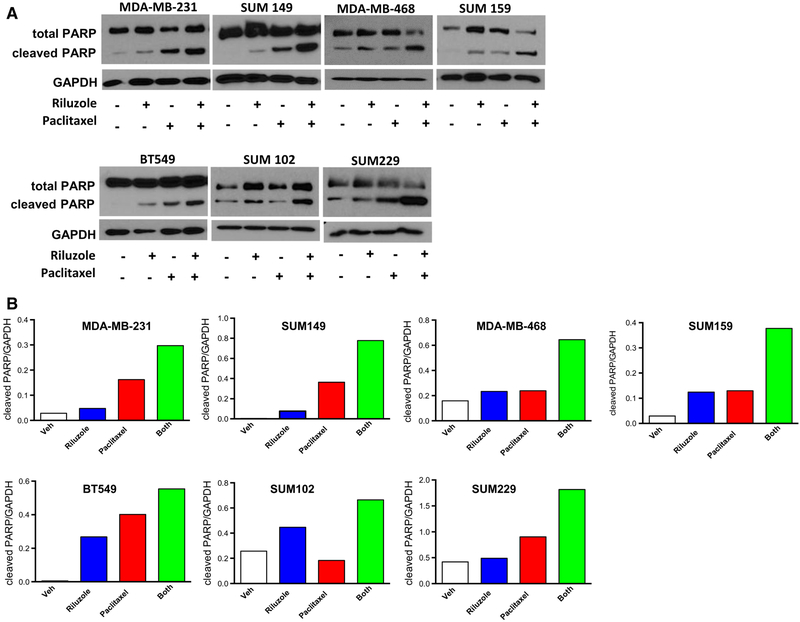
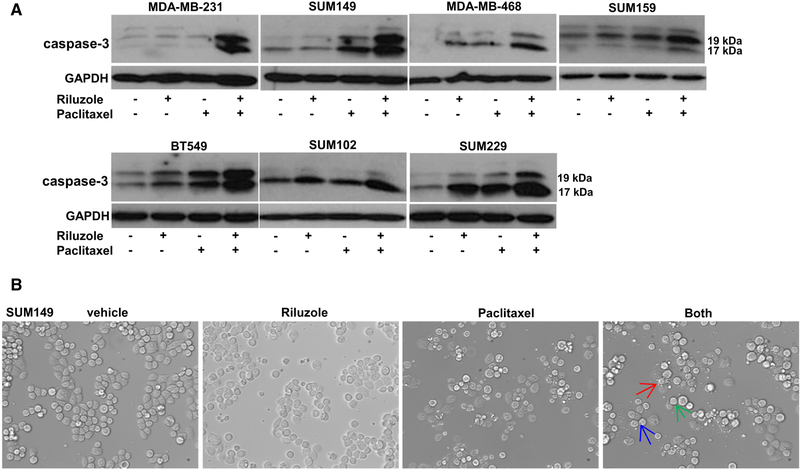
Synergy between riluzole and paclitaxel inhibiting TNBC cell growth suggested that apoptosis might also be enhanced with combined treatment. To test this hypothesis, we incubated TNBC cells with riluzole (5–10 μM) and paclitaxel (5–10 nM) together or alone and assessed cells for proteolytic cleavage of PARP and caspase-3. Even at low doses of riluzole or paclitaxel, PARP and caspase-3 p17 and p19 cleavage products were detected 24 h after treatment (Figs. 3, 4a). Enhanced levels of both PARP and caspase-3 cleavage products with the combined treatment were also detected as early as 24 h in all cell lines tested with the strongest enhanced effect demonstrated in MDAMB-231, SUM149, and SUM229 cells. Live cell imaging of SUM149 cells revealed condensation of chromatin (pyknosis) and nuclear fragmentation (karyorrhexis), both signs of apoptosis/necrosis, after treatment with riluzole or paclitaxel, which was also enhanced by combined treatment (Fig. 4b).
Fig. 3.
Riluzole and paclitaxel together enhance cell apoptosis. a Cells were plated in RPMI containing 5% FBS at 1 × 106 cells per 100 mm dish and allowed to grow for 48 h before treatment with riluzole (5–10 μM) and/or paclitaxel (5–10 nM). After 24 h treatment, cells were collected, protein isolated, and PARP cleavage detected by Western analysis. Blots are representative of at least two experiments. b Representative density graph of the blots in (a) where PARP cleavage values are normalized to their respective GAPDH values
Fig. 4.
Confirmation of enhanced apoptosis by riluzole/paclitaxel in TNBC cells. a Various TNBC cells were plated at 1 × 106 cells per 100 mm dish and allowed to grow for 48 h before treatment with riluzole (5–10 μM) and/or paclitaxel (5–10 nM). After 24 h treatment, cells were collected and the caspase 3 cleavage products, p17 and p19, were detected by Western analysis. Blots are representative of at least two experiments. b Microscopic images of live SUM149 cells from (a) imaged before collecting for Western analysis. Images confirm enhanced apoptotic cells in the combined treatment demonstrated by arrows which indicate either increased pyknosis (blue), karyorrhexis (green), or membrane blebbing and apoptotic body formation (red)
Riluzole increases M-phase proteins and arrests cells in M-phase
To determine how riluzole mediates cell cycle arrest leading to apoptosis, cDNA microarray analysis was performed with vehicle- and riluzole-treated MDA-MB-231 cells. After 24 h treatment, 290 genes were found differentially expressed in the riluzole-treated group (see Online Resource 1 for complete gene list) [22]. A majority of these genes fall into six major canonical pathways that play key roles in cell cycle regulation (Table 2). Further analysis of these differentially regulated genes using the DAVID tool show these genes map to categories associated with mitosis, cell division, and cell cycle progression (Table 3). qPCR analysis of TNBC cells treated for 24 h with riluzole or vehicle confirmed differential expression of up to five of these genes (PLK1, CDC25B, CDC25C, CCNB1, CCNB2) which are major regulators of mitosis (Fig. 5). All of these genes were significantly upregulated by riluzole (37.5 μM) in the MDA-MB-231 cells compared to vehicle-treated cells except for PLK1 which was upregulated by 24% but not significantly different from vehicle cells. In the other TNBC cells, only two or three of the genes were significantly upregulated. However, these cells were stimulated with a much lower concentration of riluzole (15 μM) because of low EC50 values determined in MTT assays.
Table 2.
Canonical pathways and associated genes in MDA-MB-231 cells regulated by riluzole
| Canonical pathway | p value | Observed genes |
|---|---|---|
| PLK1 signaling events | 5.75E–12 | CCNB1, CDC25B, KIF20A, AURKA, TPX2, CENPE, NDC80, PLK1 |
| Aurora B signaling | 4.16E–10 | KIF23, KIF20A, AURKA, NDC80, KIF2C, CENPA, RACGAP1 |
| Aurora A signaling | 1.72E–07 | CDC25B, AURKA, DLGAP5, TPX2, TACC3, CENPA, NFKBIA, BIRC5 |
| FOXM1 transcription factor network | 2.13E–06 | CCNB1, CCNE1, CDC25B, NEK2, PLK1, CENPA, CCNB2, BIRC5 |
| CDK regulation of DNA replication | 4.95E–04 | CCNE1, MCM2, MCM5, MCM6 |
| PLK3 signaling events | 9.09E–03 | CCNE1, CDC25C |
Table 3.
Gene Ontology Biological Process terms over-represented in riluzole-treated MDA231 cells
| Biological process term | Bonferroni p value |
|---|---|
| M-phase | 5.25E-25 |
| Mitosis | 1.77E-23 |
| M-phase of mitotic cell cycle | 2.83E-23 |
| Regulation of mitotic cell cycle | 4.15E-07 |
| Spindle organization | 1.57E-06 |
| Regulation of cell cycle process | 3.91E-06 |
| Regulation of nuclear division | 5.82E-06 |
| Regulation of mitosis | 5.82E-06 |
| Spindle checkpoint | 1.09E-04 |
| Meiosis | 1.43E-04 |
| M-phase of meiotic cell cycle | 1.43E-04 |
| Establishment of chromosome localization | 2.21E-04 |
| Mitotic spindle organization | 2.21E-04 |
| Mitotic cell cycle checkpoint | 3.99E-04 |
| Centrosome cycle | 4.61E-04 |
| Cellular amino acid metabolic process | 5.02E-04 |
| Amino acid activation | 5.18E-04 |
| tRNA aminoacylation for protein translation | 5.18E-04 |
| Regulation of mitotic metaphase/anaphase transition | 7.19E-04 |
| Cellular amine metabolic process | 8.34E-04 |
| Cell cycle checkpoint | 8.62E-04 |
| Establishment of spindle orientation | 9.62E-04 |
| Establishment of mitotic spindle orientation | 9.62E-04 |
| Establishment of mitotic spindle localization | 1.78E-03 |
| Centrosome organization | 2.18E-03 |
| Establishment of spindle localization | 2.83E-03 |
| Microtubule organizing center organization | 2.83E-03 |
| Mitotic sister chromatid segregation | 3.07E-03 |
| Negative regulation of mitotic metaphase/anaphase transition | 3.44E-03 |
| Mitotic cell cycle spindle assembly checkpoint | 3.44E-03 |
| Negative regulation of nuclear division | 4.10E-03 |
| Negative regulation of mitosis | 4.10E-03 |
| Oocyte maturation | 4.82E-03 |
| Protein-DNA complex assembly | 6.42E-03 |
Fig. 5.
Riluzole increases M-phase proteins in TNBC cells. Various TNBC cells were treated overnight with riluzole and mitotic cell cycle genes (PLK1, CDC25B, CDC25C, CCNB1, CCNB2) detected by RT-QPCR and normalized to GAPDH as the reference gene. Results are representative of two experiments, performed in triplicate where *p < 0.05 compared to vehicle-treated cells
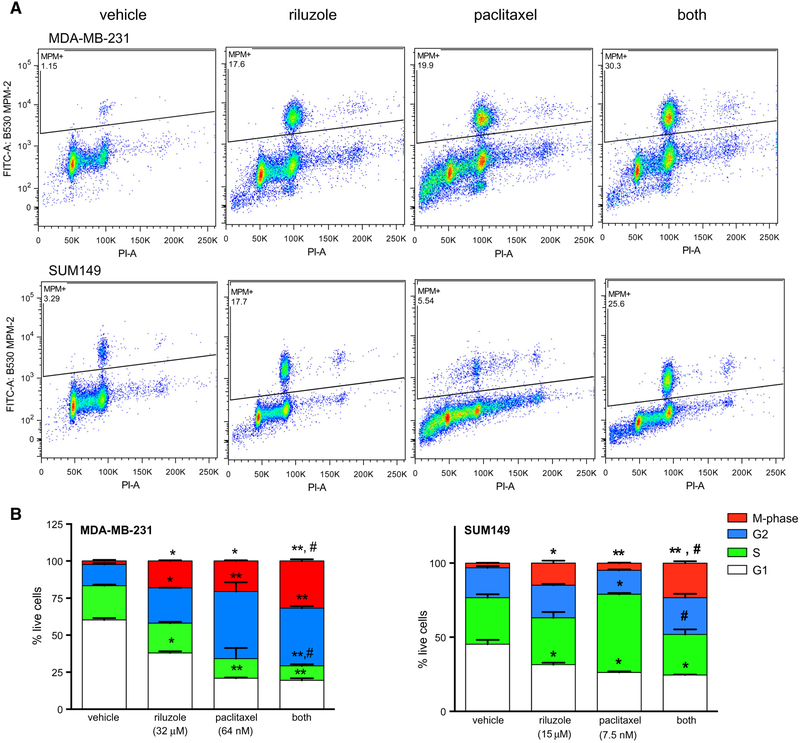
Cell cycle FACS analysis using anti-MPM2 antibody in conjunction with standard propidium iodide DNA staining demonstrated a dramatic and significant increase in the percentage of cells in M-phase after 24 h riluzole treatment in both MDA-MB-231 and SUM149 cells (seven- and fourfold, respectively) compared to control (Fig. 6), associated with a significant decrease in the percentage of G1-phase cells in both cell lines by 33.7 and 32.3%, respectively. Paclitaxel also significantly increased M-phase cells in both cell lines compared to vehicle-treated cells but the effect was not as dramatic in SUM149 cells as in MDAMB-231 cells (60% vs. eightfold, respectively). This difference is probably due to treatment of SUM149 cells with a lower concentration of paclitaxel (7.5 nM compared to 64 nM for MDA-MB-231), based on MTT EC50 values. Similar to riluzole treatment, this increase in M-phase cells in both MDA-MB-231 and SUM149 cell lines correlated with a significant decrease in the number of cells in G1-phase by 67 and 50%, respectively. Treatment of both cell lines with riluzole and paclitaxel together significantly increased the percentage of cells in M-phase compared to paclitaxel treatment alone, although not as dramatically in the MDA-MB-231 cells as in SUM149 cells (55% and 3.7-fold increase, respectively). This is not surprising since paclitaxel alone induced an eightfold increase in the percentage of M-phase cells in MDA-MB-231 cells. This increase in M-phase cells with the combined treatment correlated with a significant decrease in S-phase cells by greater than 50% in both cell lines when compared to paclitaxel treatment alone.
Fig. 6.
Riluzole and paclitaxel together enhance M-phase arrest in TNBC cells. MDA-MB-231 and SUM149 cells were treated overnight with riluzole and/or paclitaxel and the number of cells in M-phase, G2, S, or G1 were determined by FACS analysis. a Representative scatter plots of live cells staining positive for MPM-2 proteins (PI and FITC positive), proteins that are phosphorylated by M-phase promoting factor during mitosis. Riluzole increases and number of cells in M-phase for both MDA-MB-231 and SUM149 cells and riluzole and paclitaxel together (both) induce a higher percentage of cells expressing MPM-2 proteins compared to their respective vehicle control or paclitaxel-treated cells. b The data generated from the scatterplots above and from standard propidium iodide DNA staining were analyzed and graphed using prism software. G2 is calculated by subtracting percent of cells in M-phase from the G2/M values. The results are expressed as the percent of total live cells and are the average mean ± SEM of two experiments, performed in duplicate where *p < 0.05 and **p < 0.01 compared to vehicle control cells and #p < 0.05 compared to paclitaxel treatment alone. Riluzole significantly increases the percentage of cells in M-phase compared to vehicle-treated alone in both cell types and the percent of cells in M-phase is significantly greater in the combined treatment groups compared to paclitaxel treatment alone
Riluzole enhances tumor growth inhibition by paclitaxel in vivo
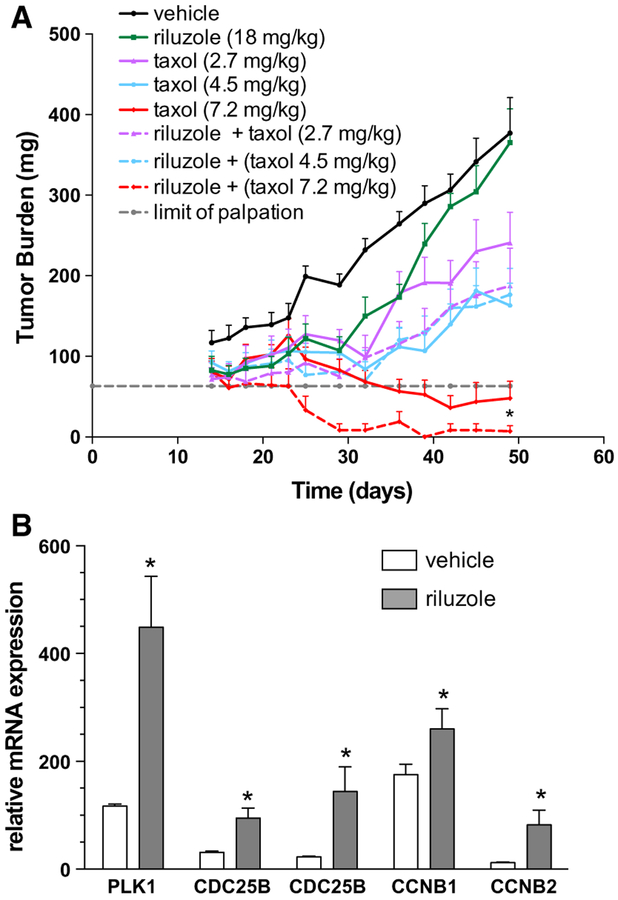
Our results thus far suggest that riluzole and paclitaxel together could act synergistically in patients. To validate this hypothesis in a preclinical model, we used an MDAMB-231 TNBC xenograft model in which tumor-bearing mice were treated with riluzole (18 mg/kg) or paclitaxel at three concentrations (2.7, 4.5, 7.2 mg/kg), alone or together, and tumor growth inhibition (TGI) was determined and compared between groups (Fig. 7a). The two-drug combination produced at their respective highest non-toxic total doses a TGI of 100% with eight out of the nine mice below the limit of palpation (i.e., below 63 mg) on day 49 (day of harvest). This result was superior to either agent at equivalent dose monotherapy (riluzole: 1% TGI; paclitaxel: 92% TGI with only four out of eight mice below tumor palpation limit). Lower dose combination groups more closely tracked the efficacy effects of single agent paclitaxel at equivalent doses over riluzole and were found to be sub-therapeutic (TGI values <58%). Despite this, of note is the observation that the two lower combination doses each produced 1–2 mice below tumor palpation limit by day 49, suggesting there was at minimum, an additive or potentiating effect of riluzole on paclitaxel efficacy.
Fig. 7.
Riluzole enhances tumor growth inhibition by paclitaxel in vivo and arrests cells in M-phase. a MDA-MB-231 cells (1 × 106)were injected into the mammary fat pads of SCID/beige mice and allowed to grow until the xenografts reached a mean size of 40–50 mm3 (day 14) at which point they were divided into experimental groups of ten mice each and treated with paclitaxel (2.7, 4.5, 7.2 mg/kg) alone or together with 18 mg/kg riluzole or vehicle (DMSO). Treatment continued until tumors reached a mean size of 1000 mm3 or began to ulcerate. Results represent the mean tumor weight ± SEM where *p < 0.05 compared to tumors treated with paclitaxel alone. b Cell cycle gene analysis was performed by RT-QPCR on tumors from a second xenograft study performed as described above with four mice per group but mice only received 1 week of treatment with either riluzole (18 mg/kg) or vehicle (DMSO) treatment. Results represent the mean mRNA-expressed ± SEM for each cell cycle gene normalized using GAPDH as the reference gene. *p < 0.05 compared to vehicle-treated tumor genes
A second animal study was undertaken to ascertain whether riluzole could arrest tumor cells in M-phase cell cycle. For this study, once xenografts reached a mean size of 40–50 mm3, they were treated for only 1 week with riluzole or vehicle at their respective highest non-toxic total doses and RNA isolated and analyzed for various cell cycle genes. Similar to in vitro results, riluzole-treated mouse tissue expressed significantly more cell cycle genes associated with mitosis compared to vehicle-treated tissue (Fig. 7b). This effect of riluzole was more dramatic than the in vitro results with a greater than twofold increase in all genes analyzed except CCNB1, which was still significantly increased by 75%.
Discussion
TNBC makes up a minority of human breast cancers but is responsible for a disproportionate number of deaths [23]. TNBC responds well initially to taxane-containing chemotherapy regimens but rapidly develops resistance. Thus, finding new combinatorial treatments to optimize their clinical usefulness is imperative. In this study, we demonstrate an enhanced or synergistic effect of riluzole and paclitaxel on cell cycle progression and apoptosis in TNBC cell lines as well as the MDA-MB-231 tumor model. Some TNBC cell lines, such as MDA-MB-231, SUM149, and SUM229 cells, responded more strongly whereas responses in other cell lines were less dramatic. This is expected given that TNBC does not have a single dominant driver of tumorigenicity, hampering efforts to develop targeted drugs against this subtype. That all TNBC cell lines tested responded synergistically to this combination treatment strongly suggests the usefulness of this combinatorial treatment strategy in TNBC.
Surprisingly, the effect of riluzole monotherapy on xenograft growth was modest with tumors in these mice becoming refractory to continuous riluzole monotherapy, a different result compared to our previous studies in nu/nu mice or the 4T1 syngeneic tumor model [6–8]. Both these animal models have a fully intact immune system except for functional T-cells, in the case of the nu/nu mice. In the current study, SCID beige mice were used which lack both T- and B-cells and have impaired NK cell activity. This suggests that the immune system may be playing a role in mediating riluzole’s anti-tumor effect in mice and could be responsible for refractoriness to riluzole demonstrated in this immune-deficient animal model. In support of this, riluzole has been shown to increase survival of CD8 T-cells in HIV-1-infected individuals and enhance proliferation of anti-CD3/CD28-stimulated T-cells [24].
Key serine/threonine protein kinases (PLK1, Aurora A-B, CDK, and PLK3) were upregulated by riluzole as well as their downstream cell cycle genes. These serine/threonine kinase receptors play key roles in regulating mitosis, and upregulation of these kinases suggests either a direct effect of riluzole on the regulation of these protein kinase pathways or an indirect effect resulting from riluzole’s effects on other key aspects of cell proliferation or cell survival. In melanoma, a majority of riluzole’s anti-tumor effect is mediated through glutamate signaling, specifically mGluR1 [11, 12]. However, we previously determined that a significant proportion of riluzole’s anti-tumor activity in breast cancer likely derives from mechanisms other than mGluR1-mediated signaling [13]. In the current study, we show significant down-regulation of genes regulating lipid metabolism in MDA-MB-231 cells. Further analysis of these genes and their role(s) in mediating cell growth and proliferation may provide insight into the mechanism by which riluzole regulates growth of TNBC.
The strongest synergism between riluzole and paclitaxel was observed in SUM229, MDA-MB-231, and SUM149 cells, with increased synergism corresponding with the relative resistance of these cells to paclitaxel. Interestingly, other studies have reported that SUM149 and SUM229 cells are resistant to paclitaxel [19] and that this resistance is associated with overexpression of cell surface EGFR and loss of PI3 K regulation resulting from PTEN loss [25, 26]. This constitutive EGF-PTEN-independent EGFR signaling pathway drives inflammatory and anti-apoptotic pathways [19, 27, 28] and has been demonstrated as a common pathway associated with a drug-resistant subset of TNBC [20, 29]. The strong synergistic effect of riluzole and paclitaxel demonstrated in SUM229 and SUM149 cells suggests that this combinatorial treatment could be effective in treating this of TNBC with a dysregulated EGFR subgroup.
Recently, studies in melanoma have demonstrated an inhibitory effect of riluzole on glutamate-mediated PI3 K/AKT signaling, an observation that has been tentatively confirmed in a human phase 0 trial [14, 16, 17]. Interestingly, PI3 K is dysregulated in EGFR-PTEN null TNBC which results in high constitutive AKT expression and is the driving force regulating tumor growth in these cells [30]. This observation has been confirmed in another study demonstrating that AKT inhibition can effectively kill this subset of TNBC [31]. Therefore, it appears that riluzole, through inhibition of AKT activity, may inhibit tumor growth in the EGFR-PTEN null subgroup of TNBC patients as well as other AKT-driven breast cancer subtypes [29, 32, 33]. Further studies of how riluzole regulates AKT phosphorylation and the specificity of the AKT family, will be useful in the development of therapeutics agents for treating paclitaxel-resistant tumors.
In addition to AKT, riluzole inhibits PKC alpha activity in mixed mouse cortical cultures by directly binding to the catalytic domain [15]. PKC alpha is known to regulate EGFR-mediated tumor growth in prostate cancer [34] where riluzole is an effective anti-tumor agent [35]. This suggests in TNBC that riluzole may mediate its anti-tumor effects through inhibition of either PKC alpha, AKT activity, or both since PKC alpha is a known positive regulator of AKT activity [36].
These results demonstrate synergistic and enhanced effects of riluzole and paclitaxel on growth and apoptosis in both TNBC cells as well as in vivo in a MDA-MB-231 xenograft TNBC tumor model. In addition, we also identify novel key protein kinases upregulated by riluzole and downstream cell cycle genes regulated by these kinases. These results suggest that riluzole will be useful for treating TNBC, tumors resistant to paclitaxel. Further studies into signaling pathways and targets effected by riluzole in TNBC will be critical to the design of clinical trials employing this drug as well as in the future design of new drugs targeting these pathways.
Supplementary Material
Acknowledgements
We are grateful to Dr. Stephen Ethier for kindly providing us with his SUM cell lines. We are also grateful to the Animal Model and Therapeutics Evaluation core for their help with the animal portion of this study, the Genomics core for microarray analysis and the MICR core for FACS analysis.
Funding This study was supported by Breakthrough Award Level 1 Grant# BC142052, funded by the Department of Defense through its Breast Cancer Research Program, a part of its Congressionally Directed Medical Research Program. Animal work was supported by a combination of institutional research funds and a grant from the DMC Foundation. The Microscopy, Imaging, and Cytometry Resources (MICR) Core is supported, in part, by NIH Center grant P30CA022453 to Karmanos Cancer Institute, Wayne State University and the Perinatology Research Branch of the National Institutes of Child Health and Development, Wayne State University. The Genomics Core is supported, in part, by NIH Center grant P30 CA022453 to the Karmanos Cancer Institute at Wayne State University. A portion of the in vivo xenograft study was funded by the Committee for Student Research at Oakland University William Beaumont School of Medicine and under the guidance of Dr. David M. Thomas.
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
Ethical approval All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted. Animal studies were approved by the local Institutional Animal Care and Use Committee (IACUC) at Wayne State University which is structured and operated in accordance with NIH’s Office of Laboratory Animal Welfare (OLAW) Public Health Service Policy on Humane Care and Use of Laboratory Animals (Public Health Service NIH Assurance Number D16–00198).
Research involving human and animal rights This article does not contain any studies with human participants performed by any of the authors.
Availability of data and materials: The dataset supporting the conclusions of this article is available in the GEO repository, (Accession #GSE96653: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE96653) and is also included within the article (and in Online Resource 1). Additional file 1 is an excel (.xls) file listing the 290 genes found to be differentially expressed by riluzole.
Electronic supplementary material The online version of this article (doi:10.1007/s10549-017-4435-x) contains supplementary material, which is available to authorized users.
References
- 1.American Cancer Society I: breast cancer facts & figures (2015–2016). American Cancer Society, Inc, Atlanta [Google Scholar]
- 2.Aysola K, Desai A, Welch C, Xu J, Qin Y, Reddy V, Matthews R, Owens C, Okoli J, Beech DJ et al. (2013) Triple negative breast cancer—an overview. Hereditary Genet. doi: 10.4172/2161-1041.S2-001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA (2007) Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 13(15 Pt 1):4429–4434 [DOI] [PubMed] [Google Scholar]
- 4.Mancini P, Angeloni A, Risi E, Orsi E, Mezi S (2014) Standard of care and promising new agents for triple negative metastatic breast cancer. Cancers 6(4):2187–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newman LA, Reis-Filho JS, Morrow M, Carey LA, King TA (2015) The 2014 Society of Surgical Oncology Susan G. Komen for the Cure Symposium: triple-negative breast cancer. Ann Surg Oncol 22(3):874–882 [DOI] [PubMed] [Google Scholar]
- 6.Banda M, Speyer CL, Semma SN, Osuala KO, Kounalakis N, Torres Torres KE, Barnard NJ, Kim HJ, Sloane BF, Miller FR et al. (2014) Metabotropic glutamate receptor-1 contributes to progression in triple negative breast cancer. PLoS ONE 9(1):e81126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Speyer CL, Smith JS, Banda M, DeVries JA, Mekani T, Gorski DH (2012) Metabotropic glutamate receptor-1: a potential therapeutic target for the treatment of breast cancer. Breast Cancer Res Treat 132(2):565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Speyer CL, Hachem AH, Assi AA, Johnson JS, DeVries JA, Gorski DH (2014) Metabotropic glutamate receptor-1 as a novel target for the antiangiogenic treatment of breast cancer. PLoS ONE 9(3):e88830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellingham MC (2011) A review of the neural mechanisms of action and clinical efficiency of riluzole in treating amyotrophic lateral sclerosis: what have we learned in the last decade? CNS Neurosci Ther 17(1):4–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wokke J (1996) Riluzole. Lancet 348(9030):795–799 [DOI] [PubMed] [Google Scholar]
- 11.Khan AJ, Wall B, Ahlawat S, Green C, Schiff D, Mehnert JM, Goydos JS, Chen S, Haffty BG (2011) Riluzole enhances ionizing radiation-induced cytotoxicity in human melanoma cells that ectopically express metabotropic glutamate receptor 1 in vitro and in vivo. Clin Cancer Res 17(7):1807–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wall BA, Wangari-Talbot J, Shin SS, Schiff D, Sierra J, Yu LJ, Khan A, Haffty B, Goydos JS, Chen S (2014) Disruption of GRM1-mediated signalling using riluzole results in DNA damage in melanoma cells. Pigment Cell Melanoma Res 27(2):263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speyer CL, Nassar MA, Hachem AH, Bukhsh MA, Jafry WS, Khansa RM, Gorski DH (2016) Riluzole mediates anti-tumor properties in breast cancer cells independent of metabotropic glutamate receptor-1. Breast Cancer Res Treat 157(2):217–228 [DOI] [PubMed] [Google Scholar]
- 14.Le MN, Chan JL, Rosenberg SA, Nabatian AS, Merrigan KT, Cohen-Solal KA, Goydos JS (2010) The glutamate release inhibitor Riluzole decreases migration, invasion, and proliferation of melanoma cells. J Invest Dermatol 130(9):2240–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noh KM, Hwang JY, Shin HC, Koh JY (2000) A novel neuro-protective mechanism of riluzole: direct inhibition of protein kinase C. Neurobiol Dis 7(4):375–383 [DOI] [PubMed] [Google Scholar]
- 16.Rosenberg SA, Niglio SA, Salehomoum N, Chan JL, Jeong BS, Wen Y, Li J, Fukui J, Chen S, Shin SS et al. (2015) Targeting glutamatergic signaling and the PI3 kinase pathway to halt melanoma progression. Transl Oncol 8(1):1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yip D, Le MN, Chan JL, Lee JH, Mehnert JA, Yudd A, Kempf J, Shih WJ, Chen S, Goydos JS (2009) A phase 0 trial of riluzole in patients with resectable stage III and IV melanoma. Clin Cancer Res 15(11):3896–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jordan MA, Wilson L (2004) Microtubules as a target for anti-cancer drugs. Nat Rev Cancer 4(4):253–265 [DOI] [PubMed] [Google Scholar]
- 19.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA (2011) Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 121(7):2750–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuda H, Baggerly KA, Wang Y, Zhang Y, Gonzalez-Angulo AM, Meric-Bernstam F, Valero V, Lehmann BD, Pietenpol JA, Hortobagyi GN et al. (2013) Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin Cancer Res 19(19):5533–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loewe S (1953) The problem of synergism and antagonism of combined drugs. Arzneimittelforschung 3(6):285–290 [PubMed] [Google Scholar]
- 22.Speyer CL, Gorski DH (2017) Genome-wide analysis of differential gene expression in MDA-MB-231 cells after overnight treatment with riluzole. GEO Accession: GSE96653 [Google Scholar]
- 23.Cleere DW (2010) Triple-negative breast cancer: a clinical update. Commun Oncol 7:203–211 [Google Scholar]
- 24.Achour A, M’Bika JP, Biquard JM (2009) Enhanced endogenous type I interferon cell-driven survival and inhibition of spontaneous apoptosis by Riluzole. Virology 386(1):160–167 [DOI] [PubMed] [Google Scholar]
- 25.Berquin IM, Dziubinski ML, Nolan GP, Ethier SP (2001) A functional screen for genes inducing epidermal growth factor autonomy of human mammary epithelial cells confirms the role of amphiregulin. Oncogene 20(30):4019–4028 [DOI] [PubMed] [Google Scholar]
- 26.Rao GS, Murray S, Ethier SP (2000) Radiosensitization of human breast cancer cells by a novel ErbB family receptor tyrosine kinase inhibitor. Int J Radiat Oncol Biol Phys 48(5):1519–1528 [DOI] [PubMed] [Google Scholar]
- 27.Martin V, Botta F, Zanellato E, Molinari F, Crippa S, Mazzucchelli L, Frattini M (2012) Molecular characterization of EGFR and EGFR-downstream pathways in triple negative breast carcinomas with basal like features. Histol Histopathol 27(6):785–792 [DOI] [PubMed] [Google Scholar]
- 28.Streicher KL, Willmarth NE, Garcia J, Boerner JL, Dewey TG, Ethier SP (2007) Activation of a nuclear factor kappaB/inter-leukin-1 positive feedback loop by amphiregulin in human breast cancer cells. Mol Cancer Res 5(8):847–861 [DOI] [PubMed] [Google Scholar]
- 29.Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She QB, Maurer M, Koujak S, Ferrando AA, Malmstrom P, Memeo L et al. (2007) Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci USA 104(18):7564–7569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kappler CS, Guest ST, Irish JC, Garrett-Mayer E, Kratche Z, Wilson RC, Ethier SP (2015) Oncogenic signaling in amphiregulin and EGFR-expressing PTEN-null human breast cancer. Mol Oncol 9(2):527–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.She QB, Chandarlapaty S, Ye Q, Lobo J, Haskell KM, Leander KR, DeFeo-Jones D, Huber HE, Rosen N (2008) Breast tumor cells with PI3 K mutation or HER2 amplification are selectively addicted to Akt signaling. PLoS ONE 3(8):e3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hellyer NJ, Cheng K, Koland JG (1998) ErbB3 (HER3) interaction with the p85 regulatory subunit of phosphoinositide 3-kinase. Biochem J 333(Pt 3):757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holbro T, Beerli RR, Maurer F, Koziczak M, Barbas CF 3rd, Hynes NE (2003) The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: Erbb2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci USA 100(15):8933–8938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart JR, O’Brian CA (2005) Protein kinase C-{alpha} mediates epidermal growth factor receptor transactivation in human prostate cancer cells. Mol Cancer Ther 4(5):726–732 [DOI] [PubMed] [Google Scholar]
- 35.Akamatsu K, Shibata MA, Ito Y, Sohma Y, Azuma H, Otsuki Y (2009) Riluzole induces apoptotic cell death in human prostate cancer cells via endoplasmic reticulum stress. Anticancer Res 29(6):2195–2204 [PubMed] [Google Scholar]
- 36.Li L, Sampat K, Hu N, Zakari J, Yuspa SH (2006) Protein kinase C negatively regulates Akt activity and modifies UVC-induced apoptosis in mouse keratinocytes. J Biol Chem 281(6):3237–3243 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.