Abstract
PTEN is a major tumor-suppressor protein whose expression and biological activity are frequently diminished in sporadic or inherited cancers. PTEN gene deletion or loss-of-function mutations favor tumor cell growth and are commonly found in clinical practice. In addition, diminished PTEN protein expression is also frequently observed in tumor samples from cancer patients in the absence of PTEN gene alterations. This makes PTEN protein levels a potential biomarker parameter in clinical oncology, which can guide therapeutic decisions. The specific detection of PTEN protein can be achieved by using highly defined anti-PTEN monoclonal antibodies (mAbs), characterized with precision in terms of sensitivity for the detection technique, specificity for PTEN binding, and constraints of epitope recognition. This is especially relevant taking into consideration that PTEN is highly targeted by mutations and posttranslational modifications, and different PTEN protein isoforms exist. The precise characterization of anti-PTEN mAb reactivity is an important step in the validation of these reagents as diagnostic and prognostic tools in clinical oncology, including their routine use in analytical immunohistochemistry (IHC). Here, we review the current status on the use of well-defined anti-PTEN mAbs for PTEN immunodetection in the clinical context and discuss their potential usefulness and limitations for a more precise cancer diagnosis and patient benefit.
Biomarker detection constitutes one of the key parameters in precision oncology providing information to stratify cancer patients in terms of diagnosis, prognosis, and response to therapies (Salgado et al. 2017; Bode and Dong 2018). The analysis of genomic data from both tumor and normal-tissue samples is becoming one of the major sources of clinically relevant information for cancer patient stratification and therapy optimization. However, other molecular variables, such as the presence of biomarker proteins in biological samples, are more informative than genetic data in some types of neoplasias (Schmidt et al. 2016; Twomey et al. 2017). Analytical immunohistochemistry (IHC), which relies on the use of sensitive monoclonal antibodies (mAbs) targeting specific biomarker proteins, is one of the most universal diagnostic techniques in modern clinical oncology. IHC provides local information at the single-cell level on the biomarker expression in the whole tumor and its microenvironment, guiding the pathologist in the diagnosis and prognosis of most solid malignancies (Leong et al. 2010; Matos et al. 2010). Power-resolution advantages of IHC include the monitoring of biomarker expression without disrupting the architecture of the tissue, which has especial relevance in tumors displaying high intratumor heterogeneity (ITH) (McGranahan and Swanton 2015; López and Angulo 2018), the opportunity of detecting specific protein posttranslational modifications that are functionally and clinically relevant (Sperinde et al. 2010; Bodo and Hsi 2011), and the possibility of monitoring the subcellular localization of the biomarker protein (Cheuk and Chan 2004). Alternative techniques to monitor the presence of cancer biomarker proteins using mAbs exist, including radioimmunoassay, enzyme-linked immunosorbent assays (ELISAs), and membrane- and bead-based protein arrays, although their implementation in the clinic is more limited (Zhang et al. 2014).
The PTEN tumor-suppressor protein plays a major and unique role in the control of cell growth and survival in all tissues, and PTEN protein is partially or totally lost in a large number of human tumors, which makes it an excellent biomarker and therapeutic target candidate in clinical oncology (McCabe et al. 2016; McLoughlin et al. 2018; Bazzichetto et al. 2019). PTEN is encoded by a single gene, but distinct PTEN protein isoforms exist, generated by alternative initiation of messenger RNA (mRNA) translation or mRNA alternative splicing (Sharrard and Maitland 2000; Malaney et al. 2017). The more abundant PTEN protein isoform contains 403 amino acids (PTEN 1–403), and constitutes the PTEN form referenced in most of the studies. Several commercial anti-PTEN mAbs are available and have been extensively used in cancer research, but their clinical use in diagnostic or predictive tests measuring PTEN presence in tumors by IHC is still pending analytical validation. This, in part, can be because, of the difficulties to standardize the IHC preanalytical and analytical conditions of mAb usage, as well as variations in the PTEN immunostaining scoring and the performance attributed to each anti-PTEN mAb in different laboratories (Eritja et al. 2015). In addition, the lack of precision in the definition of the epitopes recognized by the distinct anti-PTEN mAbs can also be a caveat in the accurate interpretation of their IHC reactivity patterns in tumor samples. In this review, we summarize the current status of the use of anti-PTEN mAbs as diagnostic and prognostic tools in human neoplasias and highlight the importance of the precise characterization of anti-PTEN mAbs in providing a more accurate assistance to precision oncology.
PTEN GENE, mRNA, AND PROTEIN EXPRESSION ALTERATIONS IN HUMAN TUMORS
The human PTEN gene was originally identified as a protein tyrosine phosphatase–encoding tumor-suppressor gene frequently deleted in multiple human advanced cancers, including brain, breast, and prostate cancer (Li and Sun 1997; Li et al. 1997; Steck et al. 1997). In more recent years, it has been determined that the PTEN gene and its protein product are recurrently altered in most human cancers because of the key role played by PTEN PI(3,4,5)P3 (PIP3) phosphatase activity in the negative regulation of the prosurvival PI3K/AKT/mTOR pathway, as well as by PTEN PIP3 phosphatase-independent tumor-suppressive functions (Worby and Dixon 2014; Pulido 2015; Lee et al. 2018). PTEN distributes in a highly regulated manner at different cell compartments, and the tumor-suppressive functional consequences of PTEN partitioning between membranes, cytoplasm, and nucleus have been well documented (Vazquez and Devreotes 2006; Gil et al. 2007; Bassi and Stambolic 2013; Kreis et al. 2014; Bononi and Pinton 2015). PTEN functional deficiency associates with hyperactivation of the PI3K/AKT/mTOR pathway. This makes the determination of PTEN gene loss by molecular or FISH analysis, or the assessment of PTEN protein expression by IHC, relevant to stratify patients who could benefit from therapies based on PI3K, AKT, or mTOR inhibition by small molecule drugs (Dillon and Miller 2014; Papa and Pandolfi 2019). In this respect, the global analysis of genetically characterized human cancer cell lines revealed that genetic alterations in PTEN are associated with increased sensitivity to PI3K/AKT/mTOR inhibitors and decreased sensitivity to receptor-tyrosine kinase (RTK) inhibitors (Yang et al. 2013; Dillon and Miller 2014), a concept that can be extended to tumor resistance to RTK inhibitors and chemotherapeutic agents (Shoman et al. 2005; Mellinghoff et al. 2007; Steelman et al. 2008). In addition, PTEN loss has also been associated with resistance to PD-1-inhibitor T-cell-mediated immunotherapy in several cancer types (Rieth and Subramanian 2018). On the other hand, because of the positive role of nuclear PTEN in DNA-damage repair, PTEN loss sensitizes cancer cells to poly (ADP-ribose) polymerase (PARP) inhibitors by a synthetic lethality mechanism (Mendes-Pereira et al. 2009; Dedes et al. 2010; McEllin et al. 2010).
PTEN gene mutations are found in ∼5% of human tumor samples, ranking in the top group of genes most commonly mutated in human cancer (Tan et al. 2015). Importantly, a copy of the PTEN gene is also mutated in the germline of patients with PTEN hamartoma tumor syndrome (PHTS) (Yehia and Eng 2018), making determination of PTEN protein levels and function critical in the follow-up of their disease. From a global perspective, PTEN gene or PTEN protein loss is associated with cancer progression and resistance to therapies in most human tumors, although divergent findings have been reported (Table 1). For instance, in glioblastoma, ∼30% of studies revealed a positive prognostic value for PTEN gene or protein expression, including a study showing a positive predictive value for response to the erlotinib RTK inhibitor (Table 1; Mellinghoff et al. 2005; Montano et al. 2016). In other malignancies, such as breast cancer, most of the meta-analysis studies indicated a positive prognostic value for PTEN expression, including studies with a positive predictive value for response to trastuzumab anti-human epidermal growth receptor 2 (HER2) therapy (Table 1; Nagata et al. 2004; Wang et al. 2013; Zhang et al. 2019).
Table 1.
Global clinical significance of PTEN loss
| Cancer type | Clinical significance of PTEN loss | Reference |
|---|---|---|
| Endometrial | Low diagnostic accuracy in EHa | Raffone et al. 2019a |
| Association with increased risk of EC in EH | Raffone et al. 2019b | |
| Glioblastoma | Prognostic value only in 30% of studies | Montano et al. 2016 |
| Prostate | ↑ GS; ↑ capsular penetration | Wang and Dai 2015 |
| ↑ GS; ↑ recurrence | Gao et al. 2016 | |
| ↓ PFS | Xie et al. 2017 | |
| Prognostic value | Wise et al. 2017 | |
| Prognostic value | Jamaspishvili et al. 2018 | |
| Prognostic and predictive (recurrence) value | Carneiro et al. 2018 | |
| Lung | ↓ OS; ↓ PFS | Gu et al. 2016 |
| ↑ Stage; ↑ LNM | Ji et al. 2018 | |
| ↑ Stage; ↑ distant metastasis; ↓ OS | Zhao et al. 2017 | |
| ↓ OS | Xiao et al. 2016 | |
| Gastric | ↓ OS | Chen et al. 2014 |
| Breast | Resistance to TZMB in recurrent or metastatic patients | Wang et al. 2013 |
| No predictive or prognostic value | Wang et al. 2015c | |
| ↓ OS | Yang et al. 2016 | |
| ↑ Stage; ↑ LNM; ↓ PFS; ↓ OS | Xu et al. 2017 | |
| ↑ Stage; ↑ LNM; ↓ PFS; ↓ OS | Li et al. 2017 | |
| Resistance to TZMB | Zhang et al. 2019 | |
| Ovarian | No prognostic value | Cai et al. 2014 |
| Colorectal | Resistance to anti-EGFR; unclear prognostic value | Lo Nigro et al. 2016 |
| Resistance to anti-EGFR | Therkildsen et al. 2014 | |
| Renal | ↓ DSS | Tang et al. 2017 |
| ↑ Stage; ↑ distant and LNM | Que et al. 2018 |
Note that this is not a comprehensive list. Selected reviews and meta-analysis studies with information on the clinical significance of PTEN gene or PTEN protein loss are denoted.
DSS, Disease-specific survival; EC, endometrial carcinoma; EGFR, epidermal growth factor receptor; EH, endometrial hyperplasia; GS, Gleason score; LNM, lymph node metastasis; OS, overall survival; PFS, progression-free survival; TZMB, trastuzumab.
aText in italics indicates studies in which the PTEN status does not show prognostic or predictive values.
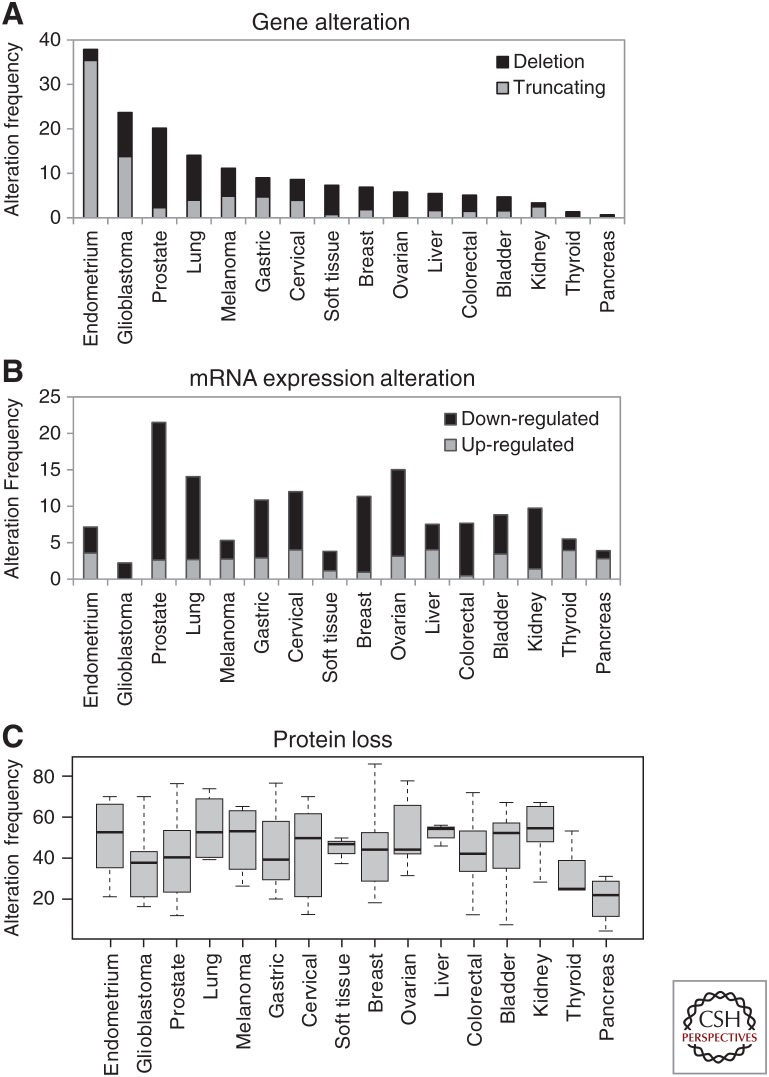
Regulation of PTEN expression, abundance, and function is exerted at multiple genetic and nongenetic levels under physiologic conditions, and aberrant PTEN alterations in cancer are caused by a wide variety of mechanisms (Hollander et al. 2011; Leslie and Foti 2011; Boosani and Agrawal 2013; Correia et al. 2014; Milella et al. 2015; Li et al. 2018b; Alvarez-Garcia et al. 2019). PTEN has been proposed as an obligate haploinsufficient tumor suppressor, in which partial loss of expression, rather than complete loss, renders maximal tumorigenicity (Berger and Pandolfi 2011). This is relevant in clinical practice because weak PTEN protein expression is often observed in tumors, as compared with normal tissues. PTEN gene alterations resulting in PTEN protein loss are evident and differential in distinct cancer types. For instance, advanced prostate cancers and lung cancers frequently show PTEN gene deletion, whereas endometrial cancers and glioblastomas often show PTEN mutations generating PTEN truncated proteins, including premature termination codon (PTC), frameshift small insertions, and frameshift small deletions (Fig. 1A; www.cbioportal.org; Cerami et al. 2012). Alterations in PTEN mRNA levels are also different in distinct types of cancer with prostate, ovarian, lung, and breast cancers displaying a higher frequency of PTEN mRNA down-regulation (Fig. 1B). This is the result of a combination of factors, including tissue-specific gene promoter hypermethylation, alterations in the activity of PTEN transcription factors, and pathologic modifications in the balance between microRNAs (miRNAs) and long noncoding RNAs targeting PTEN (Hollander et al. 2011; Boosani and Agrawal 2013; Taulli et al. 2013; Lu et al. 2016; Li et al. 2018b).
Figure 1.
PTEN gene and protein alterations in human tumors. (A) Frequency of PTEN gene deletion in human tumors and PTEN mutations causing loss of PTEN protein (premature termination codon [PTC] mutations and frameshift small deletions or insertions). (B) Frequency of PTEN messenger RNA (mRNA) down- or up-regulation in human tumors (z-score threshold ±2). In both cases, the alteration frequencies are indicated for 16 different human cancers from data generated by the TCGA Research Network and using the cBioPortal database (cBioPortal for Cancer Genomics; Cerami et al. 2012; Gao et al. 2013). Cancer types are as follows: endometrial, uterine corpus endometrial carcinoma; gliobastoma, glioblastoma multiforme; prostate, prostate adenocarcinoma; lung, lung squamous cell carcinoma; melanoma, skin cutaneous melanoma; gastric, stomach adenocarcinoma; cervical, cervical squamous cell carcinoma; soft tissue, sarcoma; breast, breast invasive carcinoma; ovarian, ovarian serous cystadenocarcinoma; liver, liver hepatocellular carcinoma; colorectal, colorectal adenocarcinoma; bladder, bladder urothelial carcinoma; kidney, kidney renal clear cell carcinoma; thyroid, thyroid carcinoma; pancreas, pancreatic adenocarcinoma. (C) Boxplot of the frequency of PTEN protein loss in human tumors as detected by immunohistochemistry (IHC). Whiskers represent the minimum and maximum of all of the data; boxes represent the values between quartiles 1 and 3, and bands inside the boxes represent the median. Data are compilations from Tables 2–4. Note that the cancer categories in C have a wider coverage of cancer subtypes than the categories in A and B.
A more homogeneous global pattern of PTEN expression alteration in human neoplasms is observed when analyzing PTEN protein levels in tumors using IHC. Although there are important variations among different studies, most of the cancer types show, on average and independently of the anti-PTEN mAb used, total or partial loss of PTEN protein expression in 30%–50% of cases (Fig. 1C). This is in accordance with the multicenter study by Millis et al. (2016), using about 20,000 samples from diverse solid tumors, which revealed 30% of samples with PTEN protein loss as determined by IHC with the anti-PTEN 6H2.1 mAb. Interestingly, this study also documented the coexistence of PTEN loss or PTEN mutations with PIK3CA mutations in tumors, supporting the notion that PTEN PIP3 phosphatase-independent tumor-suppressive functions have clinical relevance. These findings illustrate the high incidence of PTEN protein loss in human tumors, even in the absence of PTEN genetic alterations, which has important clinical implications in prognostic and therapy-response prediction. Heterogeneous PTEN gene deletion or focal loss of PTEN protein expression in tumors is a common event in some human cancers, which makes examination of PTEN expression in neoplastic tissues by IHC very important (Garg et al. 2012; Zaldumbide et al. 2016; Yun et al. 2019). For instance, heterogeneity of PTEN immunostaining in glioblastoma tumors was associated with poor patient outcome (Idoate et al. 2014). Moreover, alterations in PTEN subcellular compartmentation, such as dynamic changes in PTEN nuclear-cytoplasmic distribution during oncogenesis, may also have clinical relevance, which is revealed by IHC (Gil et al. 2015; Nosaka et al. 2017; Mingo et al. 2018; Mukherjee et al. 2018). Thus, detection of local PTEN protein expression in tumor samples by analytical IHC constitutes a fundamental methodology whose accuracy to assist clinical precision oncology needs to be optimized. A number of ongoing clinical trials based on PI3K/AKT/mTOR, PARP, or immune checkpoint inhibitors are using PTEN gene or PTEN protein loss as a stratifying patient criterion (clinicaltrials.gov). Optimization of PTEN protein IHC would benefit these trials, as well as the potential implementation of novel PTEN-dependent anticancer therapies.
PTEN PROTEIN DETECTION BY IHC USING DEFINED ANTI-PTEN mAbs
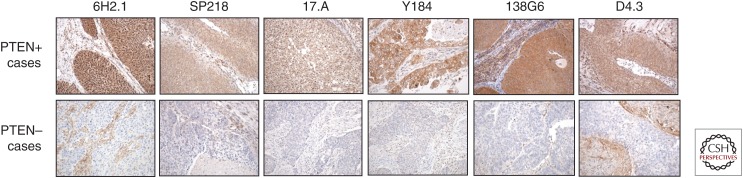
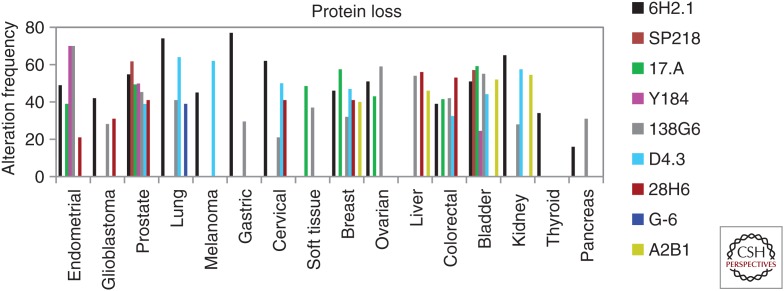
A major factor influencing the accurate determination of PTEN protein expression in tumors by IHC is the sensitivity and specificity of the mAb. In Table 2, a list is provided of commercial anti-PTEN mAbs available for the analysis of formalin-fixed paraffin-embedded (FFPE) tissues by IHC, as reported in the literature or indicated by the manufacturer. Some of these mAbs have been experimentally validated, and their IHC use conditions, efficiency, and reliability have been, in some cases, evaluated and compared (Pallares et al. 2005; Sakr et al. 2010; Sangale et al. 2011; Carvalho et al. 2014; Maiques et al. 2014; Eritja et al. 2015; Lavorato-Rocha et al. 2015; Ágoston et al. 2016; Castillo-Martin et al. 2016; Gil et al. 2016; Lotan et al. 2016a; Guedes et al. 2019). In Table 3 and Figure 2, the comparative staining of samples from prostate and bladder urothelial carcinomas with six experimentally validated anti-PTEN mAbs is shown, illustrating PTEN-positive and PTEN-negative cases. Table 4 is a compilation of IHC studies addressing the frequency of PTEN protein loss in FFPE samples from different cancer types, with indications of the specific anti-PTEN mAbs used and the clinical associations found in each study. As shown, there are differences in the frequency of usage of the distinct mAb within each cancer type, which makes it difficult to perform appropriate comparisons. There are also variations in the results obtained with the same antibody in a given cancer type, which in some cases can be a result of clinical differences in the analyzed cohorts. The clustering of studies taking into consideration the mAb used (same mAb for each cancer type) provided average values of PTEN loss between 35% and 65% of cases (Fig. 3). In spite of these extensive analyses, a consensus on which anti-PTEN mAb is more appropriate for IHC evaluation of PTEN expression in tumors does not exist yet. It is likely that this choice will depend on several factors, such as the tissue of interest and PTEN subcellular localization (cytoplasm vs. nucleus) under scrutiny. In addition, the identity of the recognized epitope in PTEN protein, and whether the detection of noncanonical PTEN isoforms is part of the study, could be important in the adequate selection of anti-PTEN mAbs as IHC biomarker tools (see below). In this respect, most of the IHC experimentally validated anti-PTEN mAbs recognize short linear epitopes at the PTEN carboxyl terminus, which reflects both the frequent usage of PTEN carboxy-terminal peptides as the immunogens and the antigenic immunodominance of the PTEN carboxyl terminus unstructured region (Fig. 4; Mingo et al. 2019). Finally, a group of anti-PTEN mAbs suitable for IHC analysis, in some cases targeting non-carboxy-terminal PTEN regions, is pending experimental evaluation and validation (Table 2).
Table 2.
Selected commercial anti-PTEN mAbs suitable for IHC
| mAb | Isotype | Host | Immunogena | Epitopeb | Referencec |
|---|---|---|---|---|---|
| 6H2.1 | IgG | Mouse | PTEN 304–403 | 392–398 | Perren et al. 1999 |
| SP218 | IgG | Rabbit | Carboxy-terminal PTEN peptide | 394–402 | Castillo-Martin et al. 2016 |
| 17.A (Ab-4) | IgM | Mouse | PTEN 2–403 | 392–402 | Torres et al. 2001 |
| Y184 | IgG | Rabbit | Carboxy-terminal PTEN peptide | 386–394 | Sangale et al. 2011 |
| 138G6 | IgG | Rabbit | Carboxy-terminal PTEN peptide | 388–394 | Bedolla et al. 2007 |
| D4.3 | Rabbit | Carboxy-terminal PTEN peptide | 388–394 | Schultz et al. 2010 | |
| 217702 | IgG1 | Mouse | PTEN 2–403 | 392–400 | Carvalho et al. 2014 |
| 28H6d | IgG1 | Mouse | PTEN-203–403 | Kimura et al. 2004 | |
| G-6 | IgG1 | Mouse | PTEN 1–403 | Wang et al. 2015b | |
| A2B1 | IgG1 | Mouse | PTEN 388–403 | Depowski et al. 2001 | |
| 11G8.11 | IgG | Mouse | PTEN 1–403 | Cascade BioScience | |
| EPR9941-2 | IgG | Rabbit | PTEN 1–403 | Abcam | |
| SP170 | IgG | Rabbit | PTEN 200–300 | Abcam; Sigma-Aldrich | |
| SP227 | IgG | Rabbit | PTEN 250–350 | Abcam; Sigma-Aldrich | |
| PTN-18 | IgG2a | Mouse | PTEN 386–403 | Sigma-Aldrich | |
| RM265 | Rabbit | Carboxy-terminal PTEN peptide | Sigma-Aldrich | ||
| H-3 | IgG2b | Mouse | PTEN 2–28 | Santa Cruz Biotechnology | |
| F-1 | IgG1 | Mouse | PTEN 3–29 | Santa Cruz Biotechnology | |
| 9E8 | IgG1 | Mouse | PTEN 320–400 | Abbkine | |
| 2C10 | IgG1 | Mouse | PTEN 320–400 | Abbkine | |
| EP2138Ye | IgG | Rabbit | PTEN phospho-Ser380 peptide | Roy and Dittmer 2011 | |
| EP229e | IgG | Rabbit | PTEN phospho-Thr366 peptide | Abcam |
Commercial anti-PTEN mAbs suitable for IHC, as reported or indicated by the supplier, are listed.
mAb, Monoclonal antibody; IHC, immunohistochemistry.
aPTEN amino acid numbering is indicated according to NP_000305.3.
bPrecise epitope mapping is provided, indicating the residues encompassing the epitope, according to Mingo et al. (2019) and our unpublished results.
cThe reference in which the mAb was first described (to the best of our knowledge) is indicated. When no reference is indicated, the supplier is indicated. Note that suppliers may change with time.
dThe anti-PTEN 28H6 mAb only stains nuclear PTEN in tissues.
eThese mAbs recognize PTEN phosphorylated at the indicated residues.
Table 3.
Comparative IHC staining of anti-PTEN mAbs of FFPE samples from prostate and bladder urothelial carcinomas
| mAb | Prostate (n = 81) | Bladder (n = 49) | ||
|---|---|---|---|---|
| Negative/positive | % Negative | Negative/positive | % Negative | |
| 6H2.1 | 62/19 | 76.5 | 25/24 | 51 |
| SP218 | 50/31 | 62 | 28/21 | 57 |
| 17.A (Ab-4) | 41/40 | 49 | 29/20 | 59 |
| Y184 | 33/48 | 41 | 12/37 | 24.5 |
| 138G6 | 45/36 | 56 | 27/22 | 55 |
| D4.3 | 42/39 | 52 | 33/16 | 67 |
Data are as reported in Mingo et al. (2019).
FFPE, Formalin-fixed paraffin-embedded.
Figure 2.
Immunohistochemistry (IHC) staining of formalin-fixed paraffin-embedded (FFPE) tumor tissue sections with different anti-PTEN monoclonal antibodies (mAbs). Bladder urothelial carcinoma samples, immunostained with six different anti-PTEN mAbs, are shown. For each mAb, a positive (+) and a negative (–) case are shown. Magnification, ×200.
Table 4.
PTEN IHC detection in human cancers using defined anti-PTEN mAbs
| Cancer type | Antibody | Frequency of PTEN protein loss (%)a | Clinical associations of PTEN protein loss | Reference |
|---|---|---|---|---|
| Endometrial | 6H2.1 | 61 | — | Mutter et al. 2000 |
| 6H2.1 | 68 | ↓ EIN versus EC | Monte et al. 2010 | |
| 6H2.1 | 64 | ↑ Endometroid versus nonendometroid | Djordjevic et al. 2012 | |
| 6H2.1 | 50 | — | Sangale et al. 2011 | |
| 6H2.1 | 40.5 | — | Garg et al. 2012 | |
| 6H2.1 | 31 | — | Pallares et al. 2005 | |
| 6H2.1 | 55 | ↑ PFS in obese patientsb | Westin et al. 2015 | |
| 6H2.1 | 24 | ↓ NPE versus EIN | Norimatsu et al. 2007 | |
| 17.A | 39 | ↓ NPE versus EC | Erkanli et al. 2006 | |
| Y184 | 70 | — | Sangale et al. 2011 | |
| 138G6 | 70 | — | Sangale et al. 2011 | |
| 28H6 | 21 | — | Pallares et al. 2005 | |
| Glioblastoma | 6H2.1 | 50 | ↑ Resistance to anti-EGFR | Mellinghoff et al. 2005 |
| 6H2.1 | 43 | — | Brown et al. 2008 | |
| 6H2.1 | 33 | — | Kreisl et al. 2009 | |
| 6H2.1 | 70 | ↓ OS heterogeneous versus homogeneous | Idoate et al. 2014 | |
| 6H2.1 | 16 | — | Ballester et al. 2017 | |
| 28H6 | 21 | — | Kim et al. 2010 | |
| 28H6 | 41 | — | Montano et al. 2011 | |
| 138G6 | 37.5 | — | Thiessen et al. 2010 | |
| 138G6 | 19 | — | Lv et al. 2012 | |
| Prostate | 6H2.1 | 27 | ↓ PFS combined with p27 loss | Halvorsen et al. 2003 |
| 6H2.1 | 39 | — | Verhagen et al. 2006 | |
| Y184 | 59 | ↑ Tumor grade | Al Bashir et al. 2019 | |
| 138G6 | 21.5 | ↓ PFS combined with high pAKT | Bedolla et al. 2007 | |
| 138G6 | 18 | ↑ PC death | Cuzick et al. 2013 | |
| 138G6 | 40 | ↓ PFS combined with ERG+ | Leinonen et al. 2013 | |
| 138G6 | 40 | ↓ OS | Ferraldeschi et al. 2015 | |
| 138G6 | 55 | — | Mehra et al. 2018 | |
| D4.3 | 38 | ↑ Pathologic features; ↓ metastasis | Lotan et al. 2011 | |
| D4.3 | 61 | ↓ PFS | Antonarakis et al. 2012 | |
| D4.3 | 52 | — | Gumuskaya et al. 2013 | |
| D4.3 | 13.5 | ↓ PFS | Chaux et al. 2012a | |
| D4.3 | 11.5 | ↑ Upgrading from biopsy to rp | Lotan et al. 2015 | |
| D4.3 | 25 | ↑ PC death combined with ERG– | Ahearn et al. 2016 | |
| D4.3 | 15.5 | ↑ Upgrading from G6 to G7 | Trock et al. 2016 | |
| D4.3 | 27 | ↑ Upgrading from biopsy (G7) to rp (nonorgan confined disease) | Guedes et al. 2017 | |
| D4.3 | 24 | ↓ PFS | Lotan et al. 2016b | |
| D4.3 | 55 | ↓ PFS combined with ERG– | Lahdensuo et al. 2016 | |
| D4.3 | 22 | ↓ PFS | Lotan et al. 2017 | |
| D4.3 | 14 | ↓ PFS | Lokman et al. 2018 | |
| 28H6 | 41 | ↓ PFS combined with ERG– | Kim et al. 2015 | |
| Lung | 6H2.1 | 74 | ↑ Well- versus poorly differentiated; | Marsit et al. 2005 |
| ↑ Stages I and II versus III and IV | ||||
| 138G6 | 41 | ↑ LSCC versus LAC; ↓ PFS in LAC | ||
| D4.3 | 64 | ↑ LSCC versus LAC; associated with smoking | Yanagawa et al. 2012 | |
| G-6 | 39 | ↓ Well- versus poorly differentiated; | Hlaing et al. 2018 | |
| ↓ stages I-II versus III-IV; ↓ OS | Wang et al. 2015b | |||
| Melanoma | 6H2.1 | 65 | — | Zhou et al. 2000 |
| 6H2.1 | 44 | ↑ BM and ↓ OS in BRAFV600 patients | Bucheit et al. 2014 | |
| 6H2.1 | 26 | ↑ Cadherin switch; ↓ PFS | Lade-Keller et al. 2013 | |
| D4.3 | 62 | ↓ OS | Giles et al. 2019 | |
| Gastric | 6H2.1 | 77 | — | Bamias et al. 2010 |
| 138G6 | 39 | — | Tran et al. 2013 | |
| 138G6 | 20 | — | Kim et al. 2016 | |
| Cervical | 6H2.1 | 62 | ↑ Pelvic lymph node metastasis | Eijsink et al. 2010 |
| 138G6 | 21 | — | Tinker et al. 2013 | |
| D4.3 | 50 | — | Ueno et al. 2013 | |
| 28H6 | 12 | — | El-Mansi and Williams 2006 | |
| 28H6 | 70 | ↓ CIN versus ICC | Vázquez-Ulloa et al. 2011 | |
| Soft tissue | 17.A | 50 | — | Torres et al. 2001 |
| 17.A | 47 | ↓ OS | Teng et al. 2011 | |
| 138G6 | 37 | — | Valkov et al. 2011 | |
| Breast | 6H2.1 | 33 | Association with ER– and PR– | Perren et al. 1999 |
| 6H2.1 | 86 | — | Yonemori et al. 2009 | |
| 6H2.1 | 43 | ↑ Resistance to TZMB; ↑ response to lapatinib (nuclear staining evaluation) | Dave et al. 2011 | |
| 6H2.1 | 27.5 | Discordance between primary tumors and metastases | Gonzalez-Angulo et al. 2011 | |
| 6H2.1 | 55.5 | ↓ OS | Razis et al. 2011 | |
| 6H2.1 | 30 | ↓ Resistance to TZMB | Gschwantler-Kaulich et al. 2017 | |
| 17.A | 50 | — | Torres et al. 2001 | |
| 17.A | 77 | — | Panigrahi et al. 2004 | |
| 17.A | 45.5 | ↓ Stages I and II versus stages III and IV; associated with TNB tumors | Siddiqui et al. 2016 | |
| 138G6 | 52 | ↑ Resistance to TZMB; ↓ OS | Esteva et al. 2010 | |
| 138G6 | 26 | — | Perez et al. 2013 | |
| 138G6 | 18 | — | Beelen et al. 2014 | |
| D4.3 | 81 | ↑ Resistance to TZMB + anthracycline-taxane-based chemotherapy | Loibl et al. 2016 | |
| D4.3 | 24 | ↓ Five-year survival in LNM patients | Wang et al. 2017 | |
| D4.3 | 37 | ↑ Resistance to TZMB + lapatinib | Rimawi et al. 2018 | |
| 28H6 | 26 | — | Bakarakos et al. 2010 | |
| 28H6 | 52 | ↑ Resistance to TZMB | Fabi et al. 2010 | |
| 28H6 | 45 | — | Duman et al. 2013 | |
| A2B1 | 48 | ↑ BC death; ↑ LNM | Depowski et al. 2001 | |
| A2B1 | 32 | ↓ PFS | Capodanno et al. 2009 | |
| Ovarian | 6H2.1 | 78 | — | Kurose et al. 2001 |
| 6H2.1 | 31 | ↑ PFS in stage I and II patients and in nondifferentiated SC | de Graeff et al. 2008 | |
| 6H2.1 | 37.5 | — | Ho et al. 2009 | |
| 6H2.1 | 65.5 | — | Roh et al. 2010 | |
| 6H2.1 | 42 | ↑ PFS in high-grade SC | Bakkar et al. 2015 | |
| 17.A | 41.5 | — | Wang et al. 2005 | |
| 17.A | 44 | ↓ OS in TP53+ patients | Kolasa et al. 2006 | |
| 138G6 | 69 | ↓ PFS | Lee and Park 2009 | |
| 138G6 | 49 | ↓ OS in high-grade SC | Martins et al. 2014 | |
| Liver | 138G6 | 54 | ↑ Liver function grading | Zhou and Li 2018 |
| 28H6 | 56 | — | Bassullu et al. 2012 | |
| A2B1 | 46 | ↓ PFS; ↓ OS | Su et al. 2016 | |
| Colorectal | 6H2.1 | 28 | ↑ MSI+ versus MSI– tumors | Zhou et al. 2002 |
| 6H2.1 | 71 | ↑ Tumor stage | Nassif et al. 2004 | |
| 6H2.1 | 42 | — | Goel et al. 2004 | |
| 6H2.1 | 45 | — | Hocking et al. 2014 | |
| 6H2.1 | 34 | ↑ Tumor stage | Lin et al. 2015 | |
| 17.A | 41.5 | ↑ Resistance to anti-EGFR + irinotecan in metastatic tumors | Loupakis et al. 2009 | |
| 138G6 | 12 | ↓ OS in LM patients | Atreya et al. 2013 | |
| 138G6 | 72 | — | Karapetis et al. 2014 | |
| D4.3 | 32.5 | ↓ OS in anti-EGFR therapy | Sood et al. 2012 | |
| 28H6 | 56 | ↑ LM versus nonLM patients; ↑ LM versus pt; ↓ five-year survival in LM patients | Sawai et al. 2008 | |
| 28H6 | 50 | ↓ PFS; ↓ OS | Jang et al. 2010 | |
| Bladder | D4.3 | 35 | ↑ Pathologic features | Schultz et al. 2010 |
| D4.3 | 7.5 | ↑ Pathologic features; ↑ LNM; ↓ OS | Rieken et al. 2017 | |
| A2B1 | 52 | — | Litlekalsoy et al. 2012 | |
| Renal | 6H2.1 | 65 | — | Zaldumbide et al. 2016 |
| 138G6 | 28 | — | Figlin et al. 2009 | |
| D4.3 | 48 | — | Chaux et al. 2012b | |
| D4.3 | 67 | — | Chaux et al. 2013 | |
| A2B1 | 54.5 | — | He et al. 2007 | |
| Thyroid | 6H2.1 | 24 | — | Alvarez-Nuñez et al. 2006 |
| 6H2.1 | 53 | ↑ LNM | Min et al. 2013 | |
| 6H2.1 | 24.5 | Associated with follicular variant of papillary thyroid cancer | Beg et al. 2015 | |
| Pancreas | 6H2.1 | 4 | — | Perren et al. 2000 |
| 6H2.1 | 18 | ↑ Resistance to anti-EGFR | Boeck et al. 2013 | |
| 6H2.1 | 26 | ↑ Recurrence/metastasis; ↓ OS | Foo et al. 2013 | |
| 138G6 | 31 | Association with invasive carcinoma; ↓ OS | Garcia-Carracedo et al. 2013 |
Note that this is not a comprehensive list. Selected studies with specific information on the anti-PTEN mAb used and the percentage of cases with PTEN protein loss are denoted. Studies addressing differential expression of PTEN in cytoplasm and in the nucleus of tumor cells are not included.
BC, Breast cancer; BM, brain metastasis; CIN, cervical intraepithelial neoplasia; ER, estrogen receptor; ICC, invasive cervical carcinoma; LAC, lung adenocarcinoma; LM, liver metastasis; LNM, lymph node metastasis; LSCC, lung squamous cell carcinoma; NPE, normal proliferative endometrium; PC, prostate cancer; pCR, pathological complete response; PR, progesterone receptor; pt, primary tumor; rp, radical prostatectomy; SC, serous carcinoma; TNB, triple negative breast.
aPTEN loss includes partial loss (weak or focal expression) or total loss (absence of expression) of PTEN protein detection.
bText in italics indicates studies in which the indicated association with PTEN loss is in contradiction with PTEN tumor-suppressor function. Empty lines indicate no clinical associations reported in the study.
Figure 3.
Frequency of PTEN protein loss in human tumors, as detected by IHC using defined anti-PTEN mAbs. Data are compilations from Tables 3 and 4 and are represented as the average of samples with PTEN protein loss clustered by cancer type and the anti-PTEN mAb used. Note that the distinct mAbs have not been used in all cancer types.
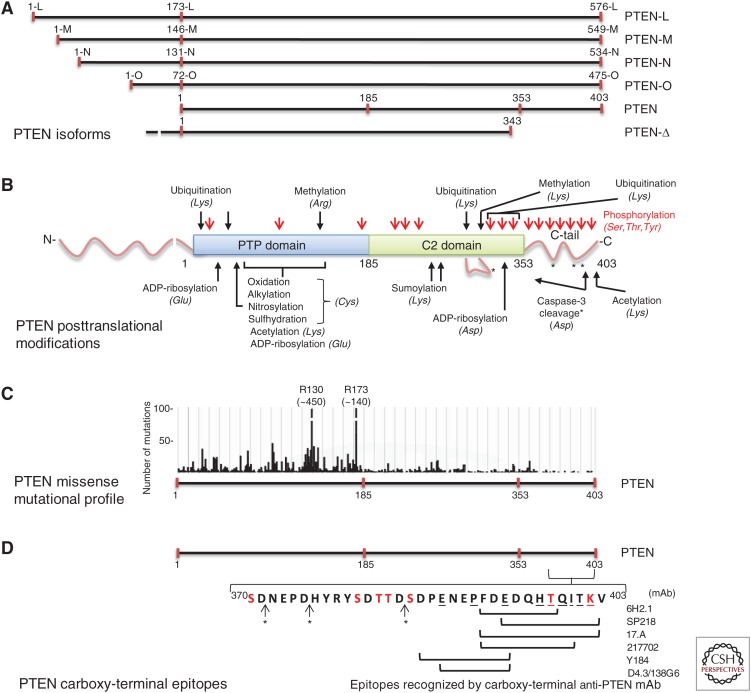
Figure 4.
Variability of PTEN amino acid composition and posttranslational modifications in relation with PTEN carboxy-terminal epitopes. (A) Schematic depiction of PTEN isoforms. PTEN long isoforms (PTEN-L, PTEN-M, PTEN-N, and PTEN-O), generated by alternative translation initiation, are shown in the upper part. Amino acid numbering and nomenclature are according to Pulido et al. (2014) and Tzani et al. (2016). PTEN-Δ isoform (PTEN 1-343-Ser), generated by alternative splicing, is shown in the bottom. PTEN (1–403) is shown in the middle with indication of the residues flanking the PTP and C2 domains and the carboxy-terminal intrinsically disordered region (C-tail). (B) Schematic of PTEN posttranslational modifications. The distinct posttranslational modifications undergone in the different PTEN domains are denoted with indications of the identity of the residues modified as reported for PTEN (1–403; see Table 5 for more information). The disordered region in the C2 domain (residues 286–310) is shown as a line loop. Note that the existence of these modifications in the PTEN long isoforms has not been reported. (C) Number of PTEN missense mutations along PTEN protein found in human tumor samples as annotated in the COSMIC database (Catalogue of Somatic Mutations in Cancer, Wellcome Trust Sanger Institute; Forbes et al. 2017). Note that the y-axis is scaled to allow visualization of low-frequency mutations. The total number of missense mutations for residues R130 and R173 is in parentheses. (D) Delimitation of PTEN carboxy-terminal epitopes recognized by the indicated commercial anti-PTEN mAb. PTEN carboxy-terminal amino acid sequence (residues 370–403) is indicated with a one-letter code. Amino acids in red are subjected to phosphorylation or acetylation (K402). Amino acids underlined are targeted by disease-associated mutations (see Table 6 for more information). *, caspase-3 cleavage sites as shown in B. Epitope mapping is from Mingo et al. (2019) and our unpublished observations.
PTEN PROTEIN IMMUNODETECTION BY OTHER TECHNIQUES
In addition to IHC, other techniques based on the use of anti-PTEN mAbs, such as immunoblot, ELISA, and flow cytometry, have been used to monitor PTEN expression in human biological samples. Some of these techniques have the advantage of a higher qualitative or quantitative resolution in terms of PTEN molecular properties, which could be important when determination of well-defined PTEN protein levels during the evolution of patient disease is desired, as could be the case in PHTS patients. Immunoblot is the standard technique to validate the specificity of anti-PTEN mAbs using cell lysates from PTEN-positive and PTEN-negative cells, and provides information on the relative molecular size of the detected PTEN proteins, which is important when addressing the expression of PTEN isoforms (Wang et al. 2015a). Immunoblot has been successfully used to semiquantitatively monitor the PTEN protein levels in PHTS patient-derived lymphoblast cell lines, and its use has been proposed as a predictor of PTEN germline mutations (Ngeow et al. 2012). Other membrane-based antibody approaches, including reverse phase protein array (RPPA), have been used in high-throughput monitoring of parallel expression of PTEN protein and other biomarkers in human cancer cell lines and tumor clinical samples (Stemke-Hale et al. 2008; Calderaro et al. 2014; Wiegand et al. 2014; Aslan et al. 2018). Flow cytometry allows single-cell quantitative analysis of molecular markers from cells in solution, although in the case of intracellular markers, such as PTEN protein, flow cytometry requires cell fixation and permeabilization. Flow cytometry has been mainly applied to monitor PTEN protein expression in hemopoietic cells (Yang et al. 2007; Woolley and Salmena 2016; Wu and Song 2018). Finally, examples of ELISA as a technique to monitor PTEN concentration from patient samples include its use in the determination of circulating PTEN protein levels in serum from cancer or diabetic patients (Li et al. 2015b; Razavi et al. 2017; Wu and Song 2018). These tests could provide diagnostic or predictive information, but further standardization and analytical and clinical validation are required.
PRECISE DEFINITION OF THE REACTIVITY OF ANTI-PTEN mAbs
Several PTEN protein variants are generated by the alternative initiation of PTEN mRNA translation, resulting in PTEN proteins with amino-terminal extensions in their amino acid sequence and distinct subcellular localizations (Fig. 4A; Hopkins and Parsons 2014; Pulido et al. 2014; Tzani et al. 2016; Malaney et al. 2017). Although a variety of functions have been proposed for some of these amino-terminal-extended PTEN variants, little is known about their expression in human cancer and their specific tumor-suppressive roles (Hopkins et al. 2013; Liang et al. 2014, 2017; Wang et al. 2015a,b, 2018; Cao et al. 2018; Li et al. 2018a; Jochner et al. 2019). The longer PTEN variant, PTEN-L (573 amino acids; also described as PTEN-α), can be secreted to the extracellular environment and internalized to acceptor cells, where it can execute growth-restrictive functions (Hopkins et al. 2013; Wang et al. 2015a). In addition, PTEN-L is also found located in mitochondria, where it regulates mitophagy and mitochondrial function (Liang et al. 2014; Li et al. 2018a; Wang et al. 2018). The generation, characterization, and validation of anti-PTEN mAbs that specifically recognize the distinct amino-terminal-extended PTEN isoforms are necessary to monitor the expression and function of these longer PTEN forms during oncogenic processes. Alternative splicing of the unique PTEN precursor mRNA renders a PTEN isoform lacking the carboxy-terminal PTEN amino acids encoded in exon 9 (PTEN-Δ, PTEN 1-343-Ser; Sharrard and Maitland 2000). Distinct to other PTEN aberrant nonfunctional isoforms generated by alternative splicing (Agrawal et al. 2005; Agrawal and Eng 2006; Sarquis et al. 2006), PTEN-Δ may behave as a functional PTEN protein, although its specific involvement in human oncogenesis has not been disclosed (Breuksch et al. 2018). As mentioned above, most of the anti-PTEN mAbs currently used recognize carboxy-terminal PTEN epitopes (Table 2; Fig. 4; Mingo et al. 2019), which precludes the detection of PTEN-Δ isoforms using these reagents, regardless of PTEN mRNA translational initiation.
PTEN protein is heavily targeted by diverse regulatory posttranslational modifications, including phosphorylation, ubiquitination, sumoylation, and proteolysis, among others (Fig. 4B; Table 5; Aronchik et al. 2014; Correia et al. 2014; Xu et al. 2014; Kim et al. 2017; Lee et al. 2018). Phosphorylation of the PTEN carboxy-terminal region regulates PTEN protein stability, nuclear-cytoplasmic shuttling, and function. These modifications affect not only PTEN function, but also the antigenicity of the PTEN modified protein. For instance, the PTEN regulatory carboxy-terminal tail is proteolytically cleaved by caspase-3 during apoptosis (Torres et al. 2003; Singh et al. 2013), which generates PTEN truncated proteins with reduced stability and displaying enhanced membrane binding and nuclear accumulation (Georgescu et al. 1999; Gil et al. 2006), but lacking the immunodominant carboxy-terminal region. As occurs with the PTEN-Δ isoforms, this precludes the use of most of the current anti-PTEN mAbs to detect the PTEN caspase-3 cleaved forms. In addition, the PTEN carboxy-terminal tail is enriched in Ser/Thr residues that can be phosphorylated by multiple protein kinases, including CK2, CK1, GSK3β, and LKB1, among others (Fig. 4B; Table 5; Hopkins et al. 2014; Fragoso and Barata 2015; Lee et al. 2018). These phosphorylations regulate PTEN inter- and intramolecular interactions that result in the fine-tuning of PTEN subcellular location and activity in cells, which is highly relevant in the context of PTEN protein immunodetection in clinical samples. A model is proposed in which carboxy-terminal phosphorylated PTEN adopts a stable compact conformation with reduced catalytic activity and impaired membrane binding and nuclear accumulation (Vazquez et al. 2000, 2001; Gil et al. 2007; Odriozola et al. 2007; Rahdar et al. 2009; Bolduc et al. 2013; Chia et al. 2015; Masson et al. 2016). In line with this model, direct inhibition of CK2 has been proposed as a feasible approach to reconstitute PTEN function in human cancer (Shehata et al. 2010; Barata 2011). Our PTEN epitope mapping analysis suggests that most of PTEN carboxy-terminal phosphorylations do not affect the binding of anti-PTEN carboxy-terminal mAbs to PTEN. One exception is PTEN Thr398 phosphorylation by the ataxia telangiectasia-mutated (ATM) kinase during the DNA-damage response, a process that favors PTEN nuclear exclusion (Bassi et al. 2013). Phosphorylation of PTEN at Thr398 is likely to impede the binding of the anti-PTEN mAbs recognizing the more carboxy-terminally located PTEN epitopes (Mingo et al. 2019). Thus, in cells exposed to DNA-damaging chemotherapy drugs, the recognition of PTEN by some anti-PTEN carboxy-terminal mAbs could be reduced. Phospho-specific anti-PTEN mAbs (antiphospho PTEN) targeting PTEN carboxy-terminal phosphorylated residues exist, including some mAbs suitable for IHC (Table 2), which could provide relevant complementary information to the PTEN protein expression status in clinical samples. For instance, a positive correlation was found in Kaposi's sarcoma tumor specimens between antiphospho PTEN (Ser380) and antiphospho Akt (Thr308) IHC staining, suggesting the Ser 380 phosphorylation-mediated inactivation of PTEN in these tumors (Roy and Dittmer 2011). However, the effective use of antiphospho PTEN mAbs in the clinical setting is still limited, mainly because of the multiplicity of clustered PTEN carboxy-terminal phosphorylation sites, which hampers the accurate detection of the PTEN phosphorylated residues, and to the complexity of PTEN phosphorylation/dephosphorylation events in different tissues. Together, these observations point out the necessity of validation of anti-PTEN mAbs recognizing non-carboxy-terminal PTEN regions, which could be used in PTEN immunodetection analyses in parallel with the currently used anti-PTEN mAbs (Andrés-Pons et al. 2005).
Table 5.
PTEN amino acids posttranslationally modified
| Amino acida | Posttranslational modification | Referenceb |
|---|---|---|
| Lys13 | Ubiquitination | Trotman et al. 2007 |
| Tyr27 | Phosphorylation | Liu et al. 2014 |
| Glu40 | ADP-ribosylation | Li et al. 2015a |
| Lys66 | Ubiquitination | Gupta and Leslie 2016 |
| Cys71 | Oxidation | Lee et al. 2002 |
| Alkylation | Covey et al. 2010 | |
| Nitrosylation | Kwak et al. 2010 | |
| Sulfhydration | Ohno et al. 2015 | |
| Cys83 | Nitrosylation | Kwak et al. 2010 |
| Ser113 | Phosphorylation | Chen et al. 2015 |
| Cys124 | Oxidation | Lee et al. 2002 |
| Alkylation | Covey et al. 2010 | |
| Nitrosylation | Kwak et al. 2010 | |
| Sulfhydration | Ohno et al. 2015 | |
| Lys125, Lys128 | Acetylation | Okumura et al. 2006 |
| Glu150 | ADP-ribosylation | Li et al. 2015a |
| Arg159 | Methylation | Feng et al. 2019 |
| Lys 163 | Acetylation | Meng et al. 2016 |
| Tyr174 | Phosphorylation | Liu et al. 2014 |
| Ser229, Thr232 | Phosphorylation | Li et al. 2005 |
| Tyr240 | Phosphorylation | Fenton et al. 2012 |
| Lys254, Lys266 | Sumoylation | Huang et al. 2012 |
| Lys289 | Ubiquitination | Trotman et al. 2007 |
| Asp301 | Cleavage | Torres et al. 2003 |
| Lys313 | Methylation | Nakakido et al. 2015 |
| Thr319, Thr 321 | Phosphorylation | Li et al. 2005 |
| Asp326 | ADP-ribosylation | Li et al. 2015a |
| Lys327, Lys330 | Ubiquitination | Lee et al. 2013 |
| Tyr336 | Phosphorylation | Yim et al. 2009 |
| Lys 342, Lys344 | Ubiquitination | Lee et al. 2013 |
| Ser362, Thr366 | Phosphorylation | Al-Khouri et al. 2005 |
| Ser370 | Phosphorylation | Torres and Pulido 2001 |
| Asp371, Asp375 | Cleavage | Torres et al. 2003 |
| Ser380, Thr382 Thr383 | Phosphorylation | Torres and Pulido 2001 |
| Asp384 | Cleavage | Torres et al. 2003 |
| Ser385 | Phosphorylation | Torres and Pulido 2001 |
| Thr398 | Phosphorylation | Bassi et al. 2013 |
| Lys402 | Acetylation | Ikenoue et al. 2008 |
aAmino acids are indicated by the three-letter code.
bThe reference in which the posttranslational modification was first described (to the best of our knowledge) is indicated. Additional potential posttranslationally modified residues, as identified by proteomic studies, can be found at www.phosphosite.org.
Regulated mono- and polyubiquitination occurs in a variety of PTEN Lys residues, controlling PTEN protein stability, dimerization, subcellular localization, and tumor-suppressive functions (Leslie et al. 2016; Lee et al. 2018, 2019). This may have an important impact in the clinic because pharmacological reactivation of PTEN in cancer treatment has been proposed using direct inhibitors of E3 ubiquitin ligases targeting PTEN for degradative or nondegradative ubiquitination (Aronchik et al. 2014; Lee et al. 2019). In addition, ubiquitination constitutes another important source of antigenic variability in several regions of PTEN protein, which should be considered in the case of anti-PTEN mAbs recognizing these amino acid regions. Finally, the amino-terminal extensions of PTEN long isoforms present potential posttranslational modification motifs, including abundant phosphorylation and O-glycosylation motifs, and PTEN-L binds to the carbohydrate-binding protein concanavalin A (Hopkins et al. 2013; Malaney et al. 2013). It will be important to precisely characterize the posttranslational modifications of PTEN long isoforms, and relate these modifications with the function and antigenic properties of these PTEN proteoforms.
In addition to mutations causing PTEN protein loss (Fig. 1A), the PTEN gene is frequently targeted in tumors and PHTS patients by missense mutations resulting in single amino acid substitutions, which have variable consequences in PTEN protein stability, subcellular localization, and function (Rodríguez-Escudero et al. 2011; Gil et al. 2015; Leslie and Longy 2016). The distribution of PTEN somatic missense mutations along the PTEN protein is shown in Figure 4C, showing some of the hotspot mutations at the PTP domain. Note that the frequency of missense mutations in the distinct PTEN domains follows the pattern PTP domain >C2 domain >carboxy-terminal tail. These mutations have the potential to affect PTEN tumor-suppressor functions, and they may also introduce antigenic modifications in PTEN protein, which can affect the binding of anti-PTEN mAbs and the interpretation of PTEN immunodetection analyses, especially when the mutation has no clear pathologic effect. This is shown in Table 6 for mutations targeting the PTEN carboxy-terminal tail and the currently used anti-PTEN carboxyl terminus mAbs. As shown, most of the disease-associated mutations at the PTEN carboxyl terminus affect the binding of distinct anti-PTEN mAbs in concordance with their antigen specificity (Table 6; Fig. 4D), although these mutations were found not to affect the inhibitory function of PTEN on the PI3K/AKT pathway (Mingo et al. 2019). The low relative frequency of somatic or inherited mutations in the PTEN gene segment encoding the PTEN carboxy-terminal tail makes this region a good recognition target for reliable anti-PTEN mAbs in clinical practice, although the physiologic or pathologic conditions in which the PTEN carboxy-terminal epitopes are lost (including PTEN alternative splicing, caspase-3 cleavage, posttranslational modifications, and targeting by carboxy-terminal PTC mutations) needs to be addressed in the interpretation of the results.
Table 6.
Reactivity of anti-PTEN mAbs with carboxy-terminal PTEN amino-acid substitution variants associated to disease
| Mutation | Cancer type/disease | mAb | ||||||
|---|---|---|---|---|---|---|---|---|
| 6H2.1 | SP218 | 17.A | Y184 | 138G6 | D4.3 | |||
| WT | + | + | + | + | + | + | ||
| E388Q | Renal carcinomaa | + | + | + | +/− | +/− | +/− | |
| P391L | PHTSb | + | + | + | +/− | +/− | +/− | |
| P391H | PHTSb | + | + | + | +/− | +/− | +/− | |
| P391S | ASDc | + | + | + | − | + | +/− | |
| E394K | Urothelial carcinomad | + | +/− | − | − | +/− | +/− | |
| H397Y | Gliomaa | |||||||
| Stomach carcinomaa | − | − | +/− | + | + | + | ||
| T398S | Gliomaa | |||||||
| Ovarian carcinomaa | + | + | +/− | + | + | + | ||
| Q399H | Lung carcinomaa | + | + | − | + | + | + | |
| I400V | PHTSb | + | + | + | + | + | + | |
| T401I | Gliomaa | |||||||
| Leiomyosarcomaa | + | − | +/− | + | + | + | ||
| K402E | PHTSb | + | + | − | + | + | + | |
| K402N | PHTSb | + | + | − | + | + | + | |
Binding results are from Mingo et al. (2019) and our unpublished observations.
+, Binding equivalent to PTEN wild type (WT); +/−, diminished binding; –, no binding.
aCOSMIC (Wellcome Trust Sanger Institute; Forbes et al. 2017).
bClinVar (NCBI; Landrum et al. 2016).
cHGMD Professional (Stenson et al. 2014).
dcBioPortal (Cerami et al. 2012).
As a summary, when searching for optimal non-carboxy-terminal PTEN amino acid sequences as epitopic regions to obtain an anti-PTEN mAb, it would be ideal to choose PTEN protein peptides with low mutational load and posttranslational modifications. In this regard, the PTEN PTP domain is more heavily targeted by mutations than the C2 domain (Fig. 4C), which makes the PTEN C2 domain a good target for the development of clinically robust anti-PTEN mAbs. Moreover, the PTEN PTP domain displays 48% amino acid identity with the PTP domains from the testis-specific proteins TPTE and TPIP (Walker et al. 2001; Tapparel et al. 2003), which could favor cross-reactivity of mAbs raised against PTEN PTP domain peptides. The rational design of anti-PTEN mAbs recognizing PTEN precise epitopes, with potential use in clinical oncology, requires detailed knowledge about the antigenicity and specificity of the chosen immunogen peptide, its isoform-dependent location in the PTEN polypeptide chain, and its targeting by posttranslational modifications or mutations.
CONCLUDING REMARKS
The decrease in the amount of PTEN tumor-suppressor protein in tissues, independently of the acute functional regulation of its catalysis, is known to have pathological consequences. This makes the mAb-based determination of PTEN protein expression levels in patients and human tumors a relevant undertaking in clinical trials and routine clinical practice in the near future, including routine analytical IHC in clinical pathology. It would be important to set up a consensus on the use of specific anti-PTEN mAbs in clinical practice and how to score the expression and subcellular localization of PTEN protein in the analysis of tumor samples. The complexity in the physiologic regulation of PTEN function, including the abundance of PTEN posttranslational modifications, the existence of distinct PTEN isoforms, and the high frequency of PTEN gene mutations in tumors, are issues that need to be considered in the interpretation of PTEN expression results obtained with specific anti-PTEN mAbs. The use in parallel of more than one anti-PTEN mAb, recognizing different PTEN epitopes, could help to obtain relevant information in this regard. An important requirement for the clinical validation of anti-PTEN mAbs, and their optimal use as prognostic and predictive informative tools in clinical oncology, is the precise definition of their reactivity in terms of epitope specificity and epitope recognition constraints. The availability of a well-defined panel of anti-PTEN mAbs recognizing different PTEN regions, suitable to monitor with accuracy the expression of PTEN proteins in biological samples and tumor tissue sections, will be helpful for the efficacy of precision cancer therapies dependent on PTEN tumor-suppressor function.
ACKNOWLEDGMENTS
This work was partially supported by Grants SAF2013-48812-R (to R.P.) and SAF2016-79847-R (to R.P. and J.I.L.) from Ministerio de Economía y Competitividad (Spain and Fondo Europeo de Desarrollo Regional), Grant 2013111011 from Gobierno Vasco, Departamento de Salud (Basque Country, Spain), and 239813 from The Norwegian Research Council, Norway (to C.E.N-X.). J.M. is the recipient of a predoctoral fellowship (PRE_2014_1_285) from Gobierno Vasco, Departamento de Educación (Basque Country, Spain). L.T. is the recipient of an oncology predoctoral fellowship from Asociación Española Contra el Cáncer (AECC, Junta Provincial de Bizkaia, Spain). We thank Ikerbasque, the Basque Foundation for Science, for their help and support.
Footnotes
Editors: Charis Eng, Joanne Ngeow, and Vuk Stambolic
Additional Perspectives on The PTEN Family available at www.perspectivesinmedicine.org
REFERENCES
- Ágoston EI, Micsik T, Ács B, Fekete K, Hahn O, Baranyai Z, Dede K, Bodoky G, Bursics A, Kulka J, et al. 2016. In depth evaluation of the prognostic and predictive utility of PTEN immunohistochemistry in colorectal carcinomas: Performance of three antibodies with emphasis on intracellular and intratumoral heterogeneity. Diagn Pathol 11: 61 10.1186/s13000-016-0508-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal S, Eng C. 2006. Differential expression of novel naturally occurring splice variants of PTEN and their functional consequences in Cowden syndrome and sporadic breast cancer. Hum Mol Genet 15: 777–787. 10.1093/hmg/ddi492 [DOI] [PubMed] [Google Scholar]
- Agrawal S, Pilarski R, Eng C. 2005. Different splicing defects lead to differential effects downstream of the lipid and protein phosphatase activities of PTEN. Hum Mol Genet 14: 2459–2468. 10.1093/hmg/ddi246 [DOI] [PubMed] [Google Scholar]
- Ahearn TU, Pettersson A, Ebot EM, Gerke T, Graff RE, Morais CL, Hicks JL, Wilson KM, Rider JR, Sesso HD, et al. 2016. A prospective investigation of PTEN loss and ERG expression in lethal prostate cancer. J Natl Cancer Inst 108: djv346 10.1093/jnci/djv346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Bashir S, Alzoubi A, Alfaqih MA, Kheirallah K, Smairat A, Haddad H, Al-Dwairy A, Fawwaz BAB, Alzoubi M, Trpkov K. 2019. PTEN loss in a prostate cancer cohort from Jordan. Appl Immunohistochem Mol Morphol 10.1097/PAI.0000000000000732 [DOI] [PubMed] [Google Scholar]
- Al-Khouri AM, Ma Y, Togo SH, Williams S, Mustelin T. 2005. Cooperative phosphorylation of the tumor suppressor phosphatase and tensin homologue (PTEN) by casein kinases and glycogen synthase kinase 3β. J Biol Chem 280: 35195–35202. 10.1074/jbc.M503045200 [DOI] [PubMed] [Google Scholar]
- Alvarez-Garcia V, Tawil Y, Wise HM, Leslie NR. 2019. Mechanisms of PTEN loss in cancer : It's all about diversity. Semin Cancer Biol. 10.1016/j.semcancer.2019.02.001 [DOI] [PubMed] [Google Scholar]
- Alvarez-Nuñez F, Bussaglia E, Mauricio D, Ybarra J, Vilar M, Lerma E, de Leiva A, Matias-Guiu X, Thyroid Neoplasia Study Group. 2006. PTEN promoter methylation in sporadic thyroid carcinomas. Thyroid 16: 17–23. 10.1089/thy.2006.16.17 [DOI] [PubMed] [Google Scholar]
- Andrés-Pons A, Valiente M, Torres J, Gil A, Roglá I, Ripoll F, Cervera J, Pulido R. 2005. Functional definition of relevant epitopes on the tumor suppressor PTEN protein. Cancer Lett 223: 303–312. 10.1016/j.canlet.2004.09.047 [DOI] [PubMed] [Google Scholar]
- Antonarakis ES, Keizman D, Zhang Z, Gurel B, Lotan TL, Hicks JL, Fedor HL, Carducci MA, De Marzo AM, Eisenberger MA. 2012. An immunohistochemical signature comprising PTEN, MYC, and Ki67 predicts progression in prostate cancer patients receiving adjuvant docetaxel after prostatectomy. Cancer 118: 6063–6071. 10.1002/cncr.27689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronchik I, Kundu A, Quirit JG, Firestone GL. 2014. The antiproliferative response of indole-3-carbinol in human melanoma cells is triggered by an interaction with NEDD4-1 and disruption of wild-type PTEN degradation. Mol Cancer Res 12: 1621–1634. 10.1158/1541-7786.MCR-14-0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan O, Cremona M, Morgan C, Cheung LW, Mills GB, Hennessy BT. 2018. Preclinical evaluation and reverse phase protein array-based profiling of PI3K and MEK inhibitors in endometrial carcinoma in vitro. BMC Cancer 18: 168 10.1186/s12885-018-4035-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atreya CE, Sangale Z, Xu N, Matli MR, Tikishvili E, Welbourn W, Stone S, Shokat KM, Warren RS. 2013. PTEN expression is consistent in colorectal cancer primaries and metastases and associates with patient survival. Cancer Med 2: 496–506. 10.1002/cam4.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakarakos P, Theohari I, Nomikos A, Mylona E, Papadimitriou C, Dimopoulos AM, Nakopoulou L. 2010. Immunohistochemical study of PTEN and phosphorylated mTOR proteins in familial and sporadic invasive breast carcinomas. Histopathology 56: 876–882. 10.1111/j.1365-2559.2010.03570.x [DOI] [PubMed] [Google Scholar]
- Bakkar RM, Xie SS, Urbauer DL, Djordjevic B, Vu K, Broaddus RR. 2015. Intact PTEN expression by immunohistochemistry is associated with decreased survival in advanced stage ovarian/primary peritoneal high-grade serous carcinoma. Int J Gynecol Pathol 34: 497–506. 10.1097/PGP.0000000000000205 [DOI] [PubMed] [Google Scholar]
- Ballester LY, Fuller GN, Powell SZ, Sulman EP, Patel KP, Luthra R, Routbort MJ. 2017. Retrospective analysis of molecular and immunohistochemical characterization of 381 primary brain tumors. J Neuropathol Exp Neurol 76: 179–188. [DOI] [PubMed] [Google Scholar]
- Bamias A, Karina M, Papakostas P, Kostopoulos I, Bobos M, Vourli G, Samantas E, Christodoulou C, Pentheroudakis G, Pectasides D, et al. 2010. A randomized phase III study of adjuvant platinum/docetaxel chemotherapy with or without radiation therapy in patients with gastric cancer. Cancer Chemother Pharmacol 65: 1009–1021. 10.1007/s00280-010-1256-6 [DOI] [PubMed] [Google Scholar]
- Barata JT. 2011. The impact of PTEN regulation by CK2 on PI3K-dependent signaling and leukemia cell survival. Adv Enzyme Regul 51: 37–49. 10.1016/j.advenzreg.2010.09.012 [DOI] [PubMed] [Google Scholar]
- Bassi C, Stambolic V. 2013. PTEN, here, there, everywhere. Cell Death Differ 20: 1595–1596. 10.1038/cdd.2013.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi C, Ho J, Srikumar T, Dowling RJ, Gorrini C, Miller SJ, Mak TW, Neel BG, Raught B, Stambolic V. 2013. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science 341: 395–399. 10.1126/science.1236188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassullu N, Turkmen I, Dayangac M, Yagiz Korkmaz P, Yasar R, Akyildiz M, Yaprak O, Tokat Y, Yuzer Y, Bulbul Dogusoy G. 2012. The predictive and prognostic significance of c-erb-B2, EGFR, PTEN, mTOR, PI3K, p27, and ERCC1 expression in hepatocellular carcinoma. Hepat Mon 12: e7492 10.5812/hepatmon.7492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzichetto C, Conciatori F, Pallocca M, Falcone I, Fanciulli M, Cognetti F, Milella M, Ciuffreda L. 2019. PTEN as a prognostic/predictive biomarker in cancer: An unfulfilled promise? Cancers 11: E435 10.3390/cancers11040435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedolla R, Prihoda TJ, Kreisberg JI, Malik SN, Krishnegowda NK, Troyer DA, Ghosh PM. 2007. Determining risk of biochemical recurrence in prostate cancer by immunohistochemical detection of PTEN expression and Akt activation. Clin Cancer Res 13: 3860–3867. 10.1158/1078-0432.CCR-07-0091 [DOI] [PubMed] [Google Scholar]
- Beelen K, Opdam M, Severson TM, Koornstra RH, Vincent AD, Wesseling J, Muris JJ, Berns EM, Vermorken JB, van Diest PJ, et al. 2014. PIK3CA mutations, phosphatase and tensin homolog, human epidermal growth factor receptor 2, and insulin-like growth factor 1 receptor and adjuvant tamoxifen resistance in postmenopausal breast cancer patients. Breast Cancer Res 16: R13 10.1186/bcr3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg S, Siraj AK, Jehan Z, Prabakaran S, Al-Sobhi SS, Al-Dawish M, Al-Dayel F, Al-Kuraya KS. 2015. PTEN loss is associated with follicular variant of Middle Eastern papillary thyroid carcinoma. Br J Cancer 112: 1938–1943. 10.1038/bjc.2015.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AH, Pandolfi PP. 2011. Haplo-insufficiency: a driving force in cancer. J Pathol 223: 137–146. 10.1002/path.2800 [DOI] [PubMed] [Google Scholar]
- Bode AM, Dong Z. 2018. Recent advances in precision oncology research. NPJ Precis Oncol 2: 11 10.1038/s41698-018-0055-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodo J, Hsi ED. 2011. Phosphoproteins and the dawn of functional phenotyping. Pathobiology 78: 115–121. 10.1159/000296015 [DOI] [PubMed] [Google Scholar]
- Boeck S, Jung A, Laubender RP, Neumann J, Egg R, Goritschan C, Vehling-Kaiser U, Winkelmann C, Fischer von Weikersthal L, Clemens MR, et al. 2013. EGFR pathway biomarkers in erlotinib-treated patients with advanced pancreatic cancer: Translational results from the randomised, crossover phase 3 trial AIO-PK0104. Br J Cancer 108: 469–476. 10.1038/bjc.2012.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc D, Rahdar M, Tu-Sekine B, Sivakumaren SC, Raben D, Amzel LM, Devreotes P, Gabelli SB, Cole P. 2013. Phosphorylation-mediated PTEN conformational closure and deactivation revealed with protein semisynthesis. Elife 2: e00691 10.7554/eLife.00691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bononi A, Pinton P. 2015. Study of PTEN subcellular localization. Methods 77-78: 92–103. 10.1016/j.ymeth.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boosani CS, Agrawal DK. 2013. PTEN modulators: A patent review. Expert Opin Ther Pat 23: 569–580. 10.1517/13543776.2013.768985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuksch I, Welter J, Bauer HK, Enklaar T, Frees S, Thüroff JW, Hasenburg A, Prawitt D, Brenner W. 2018. In renal cell carcinoma the PTEN splice variant PTEN-Δ shows similar function as the tumor suppressor PTEN itself. Cell Commun Signal 16: 35 10.1186/s12964-018-0247-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PD, Krishnan S, Sarkaria JN, Wu W, Jaeckle KA, Uhm JH, Geoffroy FJ, Arusell R, Kitange G, Jenkins RB, et al. 2008. Phase I/II trial of erlotinib and temozolomide with radiation therapy in the treatment of newly diagnosed glioblastoma multiforme: North Central Cancer Treatment Group Study N0177. J Clin Oncol 26: 5603–5609. 10.1200/JCO.2008.18.0612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheit AD, Chen G, Siroy A, Tetzlaff M, Broaddus R, Milton D, Fox P, Bassett R, Hwu P, Gershenwald JE, et al. 2014. Complete loss of PTEN protein expression correlates with shorter time to brain metastasis and survival in stage IIIB/C melanoma patients with BRAFV600 mutations. Clin Cancer Res 20: 5527–5536. 10.1158/1078-0432.CCR-14-1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Xu L, Tang H, Yang Q, Yi X, Fang Y, Zhu Y, Wang Z. 2014. The role of the PTEN/PI3K/Akt pathway on prognosis in epithelial ovarian cancer: A meta-analysis. Oncologist 19: 528–535. 10.1634/theoncologist.2013-0333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderaro J, Rebouissou S, de Koning L, Masmoudi A, Herault A, Dubois T, Maille P, Soyeux P, Sibony M, de la Taille A, et al. 2014. PI3K/AKT pathway activation in bladder carcinogenesis. Int J Cancer 134: 1776–1784. 10.1002/ijc.28518 [DOI] [PubMed] [Google Scholar]
- Cao Y, Wang H, Yang L, Zhang Z, Li C, Yuan X, Bu L, Chen L, Chen Y, Li CM, et al. 2018. PTEN-L promotes type I interferon responses and antiviral immunity. Cell Mol Immunol 15: 48–57. 10.1038/cmi.2017.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capodanno A, Camerini A, Orlandini C, Baldini E, Resta ML, Bevilacqua G, Collecchi P. 2009. Dysregulated PI3K/Akt/PTEN pathway is a marker of a short disease-free survival in node-negative breast carcinoma. Hum Pathol 40: 1408–1417. 10.1016/j.humpath.2009.02.005 [DOI] [PubMed] [Google Scholar]
- Carneiro A, Barbosa ARG, Takemura LS, Kayano PP, Moran NKS, Chen CK, Wroclawski ML, Lemos GC, da Cunha IW, Obara MT, et al. 2018. The role of immunohistochemical analysis as a tool for the diagnosis, prognostic evaluation and treatment of prostate cancer: A systematic review of the literature. Front Oncol 8: 377 10.3389/fonc.2018.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho KC, Maia BM, Omae SV, Rocha AA, Covizzi LP, Vassallo J, Rocha RM, Soares FA. 2014. Best practice for PTEN gene and protein assessment in anatomic pathology. Acta Histochem 116: 25–31. 10.1016/j.acthis.2013.04.013 [DOI] [PubMed] [Google Scholar]
- Castillo-Martin M, Thin TH, Collazo Lorduy A, Cordon-Cardo C. 2016. Immunopathologic assessment of PTEN expression. Methods Mol Biol 1388: 23–37. 10.1007/978-1-4939-3299-3_3 [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. 2012. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaux A, Peskoe SB, Gonzalez-Roibon N, Schultz L, Albadine R, Hicks J, De Marzo AM, Platz EA, Netto GJ. 2012a. Loss of PTEN expression is associated with increased risk of recurrence after prostatectomy for clinically localized prostate cancer. Mod Pathol 25: 1543–1549. 10.1038/modpathol.2012.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaux A, Schultz L, Albadine R, Hicks J, Kim JJ, Allaf ME, Carducci MA, Rodriguez R, Hammers HJ, Argani P, et al. 2012b. Immunoexpression status and prognostic value of mammalian target of rapamycin and hypoxia-induced pathway members in papillary cell renal cell carcinomas. Hum Pathol 43: 2129–2137. 10.1016/j.humpath.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaux A, Albadine R, Schultz L, Hicks J, Carducci MA, Argani P, Allaf M, Netto GJ. 2013. Dysregulation of the mammalian target of rapamycin pathway in chromophobe renal cell carcinomas. Hum Pathol 44: 2323–2330. 10.1016/j.humpath.2013.05.014 [DOI] [PubMed] [Google Scholar]
- Chen J, Li T, Liu Q, Jiao H, Yang W, Liu X, Huo Z. 2014. Clinical and prognostic significance of HIF-1α, PTEN, CD44v6, and survivin for gastric cancer: A meta-analysis. PLoS ONE 9: e91842 10.1371/journal.pone.0091842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JH, Zhang P, Chen WD, Li DD, Wu XQ, Deng R, Jiao L, Li X, Ji J, Feng GK, et al. 2015. ATM-mediated PTEN phosphorylation promotes PTEN nuclear translocation and autophagy in response to DNA-damaging agents in cancer cells. Autophagy 11: 239–252. 10.1080/15548627.2015.1009767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheuk W, Chan JK. 2004. Subcellular localization of immunohistochemical signals: Knowledge of the ultrastructural or biologic features of the antigens helps predict the signal localization and proper interpretation of immunostains. Int J Surg Pathol 12: 185–206. 10.1177/106689690401200301 [DOI] [PubMed] [Google Scholar]
- Chia YC, Catimel B, Lio DS, Ang CS, Peng B, Wu H, Zhu HJ, Cheng HC. 2015. The C-terminal tail inhibitory phosphorylation sites of PTEN regulate its intrinsic catalytic activity and the kinetics of its binding to phosphatidylinositol-4,5-bisphosphate. Arch Biochem Biophys 587: 48–60. 10.1016/j.abb.2015.10.004 [DOI] [PubMed] [Google Scholar]
- Correia NC, Gírio A, Antunes I, Martins LR, Barata JT. 2014. The multiple layers of non-genetic regulation of PTEN tumour suppressor activity. Eur J Cancer 50: 216–225. 10.1016/j.ejca.2013.08.017 [DOI] [PubMed] [Google Scholar]
- Covey TM, Edes K, Coombs GS, Virshup DM, Fitzpatrick FA. 2010. Alkylation of the tumor suppressor PTEN activates Akt and β-catenin signaling: A mechanism linking inflammation and oxidative stress with cancer. PLoS ONE 5: e13545 10.1371/journal.pone.0013545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick J, Yang ZH, Fisher G, Tikishvili E, Stone S, Lanchbury JS, Camacho N, Merson S, Brewer D, Cooper CS, et al. 2013. Prognostic value of PTEN loss in men with conservatively managed localised prostate cancer. Br J Cancer 108: 2582–2589. 10.1038/bjc.2013.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave B, Migliaccio I, Gutierrez MC, Wu MF, Chamness GC, Wong H, Narasanna A, Chakrabarty A, Hilsenbeck SG, Huang J, et al. 2011. Loss of phosphatase and tensin homolog or phosphoinositol-3 kinase activation and response to trastuzumab or lapatinib in human epidermal growth factor receptor 2-overexpressing locally advanced breast cancers. J Clin Oncol 29: 166–173. 10.1200/JCO.2009.27.7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedes KJ, Wetterskog D, Mendes-Pereira AM, Natrajan R, Lambros MB, Geyer FC, Vatcheva R, Savage K, Mackay A, Lord CJ, et al. 2010. PTEN deficiency in endometrioid endometrial adenocarcinomas predicts sensitivity to PARP inhibitors. Sci Transl Med 2: 53ra75 10.1126/scitranslmed.3001538 [DOI] [PubMed] [Google Scholar]
- de Graeff P, Crijns AP, Ten Hoor KA, Klip HG, Hollema H, Oien K, Bartlett JM, Wisman GB, de Bock GH, de Vries EG, et al. 2008. The ErbB signalling pathway: Protein expression and prognostic value in epithelial ovarian cancer. Br J Cancer 99: 341–349. 10.1038/sj.bjc.6604471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depowski PL, Rosenthal SI, Ross JS. 2001. Loss of expression of the PTEN gene protein product is associated with poor outcome in breast cancer. Mod Pathol 14: 672–676. 10.1038/modpathol.3880371 [DOI] [PubMed] [Google Scholar]
- Dillon LM, Miller TW. 2014. Therapeutic targeting of cancers with loss of PTEN function. Curr Drug Targets 15: 65–79. 10.2174/1389450114666140106100909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic B, Hennessy BT, Li J, Barkoh BA, Luthra R, Mills GB, Broaddus RR. 2012. Clinical assessment of PTEN loss in endometrial carcinoma: Immunohistochemistry outperforms gene sequencing. Mod Pathol 25: 699–708. 10.1038/modpathol.2011.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman BB, Sahin B, Acikalin A, Ergin M, Zorludemir S. 2013. PTEN, Akt, MAPK, p53 and p95 expression to predict trastuzumab resistance in HER2 positive breast cancer. J BUON 18: 44–50. [PubMed] [Google Scholar]
- Eijsink JJ, Noordhuis MG, ten Hoor KA, Kok M, Hollema H, de Bock GH, Nijman HW, Schuuring E, Wisman GB, van der Zee AG. 2010. The epidermal growth factor receptor pathway in relation to pelvic lymph node metastasis and survival in early-stage cervical cancer. Hum Pathol 41: 1735–1741. 10.1016/j.humpath.2010.04.017 [DOI] [PubMed] [Google Scholar]
- El-Mansi MT, Williams AR. 2006. Evaluation of PTEN expression in cervical adenocarcinoma by tissue microarray. Int J Gynecol Cancer 16: 1254–1260. 10.1136/ijgc-00009577-200605000-00046 [DOI] [PubMed] [Google Scholar]
- Eritja N, Santacana M, Maiques O, Gonzalez-Tallada X, Dolcet X, Matias-Guiu X. 2015. Modeling glands with PTEN deficient cells and microscopic methods for assessing PTEN loss: Endometrial cancer as a model. Methods 77-78: 31–40. 10.1016/j.ymeth.2014.11.001 [DOI] [PubMed] [Google Scholar]
- Erkanli S, Kayaselcuk F, Kuscu E, Bagis T, Bolat F, Haberal A, Demirhan B. 2006. Expression of survivin, PTEN and p27 in normal, hyperplastic, and carcinomatous endometrium. Int J Gynecol Cancer 16: 1412–1418. 10.1136/ijgc-00009577-200605000-00071 [DOI] [PubMed] [Google Scholar]
- Esteva FJ, Guo H, Zhang S, Santa-Maria C, Stone S, Lanchbury JS, Sahin AA, Hortobagyi GN, Yu D. 2010. PTEN, PIK3CA, p-AKT, and p-p70S6K status: Association with trastuzumab response and survival in patients with HER2-positive metastatic breast cancer. Am J Pathol 177: 1647–1656. 10.2353/ajpath.2010.090885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabi A, Metro G, Di Benedetto A, Nisticò C, Vici P, Melucci E, Antoniani B, Perracchio L, Sperduti I, Milella M, et al. 2010. Clinical significance of PTEN and p-Akt co-expression in HER2-positive metastatic breast cancer patients treated with trastuzumab-based therapies. Oncology 78: 141–149. 10.1159/000312656 [DOI] [PubMed] [Google Scholar]
- Feng J, Dang Y, Zhang W, Zhao X, Zhang C, Hou Z, Jin Y, McNutt MA, Marks AR, Yin Y. 2019. PTEN arginine methylation by PRMT6 suppresses PI3K-AKT signaling and modulates pre-mRNA splicing. Proc Natl Acad Sci 116: 6868–6877. 10.1073/pnas.1811028116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton TR, Nathanson D, Ponte de Albuquerque C, Kuga D, Iwanami A, Dang J, Yang H, Tanaka K, Oba-Shinjo SM, Uno M, et al. 2012. Resistance to EGF receptor inhibitors in glioblastoma mediated by phosphorylation of the PTEN tumor suppressor at tyrosine 240. Proc Natl Acad Sci 109: 14164–14169. 10.1073/pnas.1211962109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraldeschi R, Nava Rodrigues D, Riisnaes R, Miranda S, Figueiredo I, Rescigno P, Ravi P, Pezaro C, Omlin A, Lorente D, et al. 2015. PTEN protein loss and clinical outcome from castration-resistant prostate cancer treated with abiraterone acetate. Eur Urol 67: 795–802. 10.1016/j.eururo.2014.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figlin RA, de Souza P, McDermott D, Dutcher JP, Berkenblit A, Thiele A, Krygowski M, Strahs A, Feingold J, Boni J, et al. 2009. Analysis of PTEN and HIF-1α and correlation with efficacy in patients with advanced renal cell carcinoma treated with temsirolimus versus interferon-α. Cancer 115: 3651–3660. 10.1002/cncr.24438 [DOI] [PubMed] [Google Scholar]
- Foo WC, Rashid A, Wang H, Katz MH, Lee JE, Pisters PW, Wolff RA, Abbruzzese JL, Fleming JB, Wang H. 2013. Loss of phosphatase and tensin homolog expression is associated with recurrence and poor prognosis in patients with pancreatic ductal adenocarcinoma. Hum Pathol 44: 1024–1030. 10.1016/j.humpath.2012.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al. 2017. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res 45: D777–D783. 10.1093/nar/gkw1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso R, Barata JT. 2015. Kinases, tails and more: Regulation of PTEN function by phosphorylation. Methods 77-78: 75–81. 10.1016/j.ymeth.2014.10.015 [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Sign 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, Mei Y, Sun H, Nie Z, Liu X, Wang S. 2016. The association of phosphatase and tensin homolog (PTEN) deletion and prostate cancer risk: A meta-analysis. Biomed Pharmacother 83: 114–121. 10.1016/j.biopha.2016.06.020 [DOI] [PubMed] [Google Scholar]
- Garcia-Carracedo D, Turk AT, Fine SA, Akhavan N, Tweel BC, Parsons R, Chabot JA, Allendorf JD, Genkinger JM, Remotti HE, et al. 2013. Loss of PTEN expression is associated with poor prognosis in patients with intraductal papillary mucinous neoplasms of the pancreas. Clin Cancer Res 19: 6830–6841. 10.1158/1078-0432.CCR-13-0624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg K, Broaddus RR, Soslow RA, Urbauer DL, Levine DA, Djordjevic B. 2012. Pathologic scoring of PTEN immunohistochemistry in endometrial carcinoma is highly reproducible. Int J Gynecol Pathol 31: 48–56. 10.1097/PGP.0b013e3182230d00 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu MM, Kirsch KH, Akagi T, Shishido T, Hanafusa H. 1999. The tumor-suppressor activity of PTEN is regulated by its carboxyl-terminal region. Proc Natl Acad Sci 96: 10182–10187. 10.1073/pnas.96.18.10182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A, Andrés-Pons A, Fernández E, Valiente M, Torres J, Cervera J, Pulido R. 2006. Nuclear localization of PTEN by a Ran-dependent mechanism enhances apoptosis: Involvement of an N-terminal nuclear localization domain and multiple nuclear exclusion motifs. Mol Biol Cell 17: 4002–4013. 10.1091/mbc.e06-05-0380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A, Andrés-Pons A, Pulido R. 2007. Nuclear PTEN: A tale of many tails. Cell Death Differ 14: 395–399. 10.1038/sj.cdd.4402073 [DOI] [PubMed] [Google Scholar]
- Gil A, Rodríguez-Escudero I, Stumpf M, Molina M, Cid VJ, Pulido R. 2015. A functional dissection of PTEN N-terminus: Implications in PTEN subcellular targeting and tumor suppressor activity. PLoS ONE 10: e0119287 10.1371/journal.pone.0119287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A, López JI, Pulido R. 2016. Assessing PTEN subcellular localization. Methods Mol Biol 1388: 169–186. 10.1007/978-1-4939-3299-3_12 [DOI] [PubMed] [Google Scholar]
- Giles KM, Rosenbaum BE, Berger M, Izsak A, Li Y, Illa Bochaca I, Vega-Saenz de Miera E, Wang J, Darvishian F, Zhong H, et al. 2019. Revisiting the clinical and biologic relevance of partial PTEN loss in melanoma. J Invest Dermatol 139: 430–438. 10.1016/j.jid.2018.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A, Arnold CN, Niedzwiecki D, Carethers JM, Dowell JM, Wasserman L, Compton C, Mayer RJ, Bertagnolli MM, Boland CR. 2004. Frequent inactivation of PTEN by promoter hypermethylation in microsatellite instability-high sporadic colorectal cancers. Cancer Res 64: 3014–3021. 10.1158/0008-5472.CAN-2401-2 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Angulo AM, Ferrer-Lozano J, Stemke-Hale K, Sahin A, Liu S, Barrera JA, Burgues O, Lluch AM, Chen H, Hortobagyi GN, et al. 2011. PI3K pathway mutations and PTEN levels in primary and metastatic breast cancer. Mol Cancer Ther 10: 1093–1101. 10.1158/1535-7163.MCT-10-1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschwantler-Kaulich D, Tan YY, Fuchs EM, Hudelist G, Köstler WJ, Reiner A, Leser C, Salama M, Attems J, Deutschmann C, et al. 2017. PTEN expression as a predictor for the response to trastuzumab-based therapy in Her-2 overexpressing metastatic breast cancer. PLoS ONE 12: e0172911 10.1371/journal.pone.0172911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Ou W, Huang L, Wu J, Li S, Xu J, Feng J, Liu B, Zhou Y. 2016. PTEN expression is associated with the outcome of lung cancer: Evidence from a meta-analysis. Minerva Med 107: 342–351. [PubMed] [Google Scholar]
- Guedes LB, Tosoian JJ, Hicks J, Ross AE, Lotan TL. 2017. PTEN loss in Gleason score 3 + 4 = 7 prostate biopsies is associated with nonorgan confined disease at radical prostatectomy. J Urol 197: 1054–1059. 10.1016/j.juro.2016.09.084 [DOI] [PubMed] [Google Scholar]
- Guedes LB, Morais CL, Fedor H, Hicks J, Gurel B, Melamed J, Lee P, Gopalan A, Knudsen BS, True LD, et al. 2019. Effect of preanalytic variables on an automated PTEN immunohistochemistry assay for prostate cancer. Arch Pathol Lab Med 143: 338–348. 10.5858/arpa.2018-0068-OA [DOI] [PubMed] [Google Scholar]
- Gumuskaya B, Gurel B, Fedor H, Tan HL, Weier CA, Hicks JL, Haffner MC, Lotan TL, De Marzo AM. 2013. Assessing the order of critical alterations in prostate cancer development and progression by IHC: Further evidence that PTEN loss occurs subsequent to ERG gene fusion. Prostate Cancer Prostatic Dis 16: 209–215. 10.1038/pcan.2013.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Leslie NR. 2016. Controlling PTEN (phosphatase and tensin homolog) stability: A dominant role for lysine 66. J Biol Chem 291: 18465–18473. 10.1074/jbc.M116.727750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvorsen OJ, Haukaas SA, Akslen LA. 2003. Combined loss of PTEN and p27 expression is associated with tumor cell proliferation by Ki-67 and increased risk of recurrent disease in localized prostate cancer. Clin Cancer Res 9: 1474–1479. [PubMed] [Google Scholar]
- He L, Fan C, Gillis A, Feng X, Sanatani M, Hotte S, Kapoor A, Tang D. 2007. Co-existence of high levels of the PTEN protein with enhanced Akt activation in renal cell carcinoma. Biochim Biophys Acta 1772: 1134–1142. 10.1016/j.bbadis.2007.07.001 [DOI] [PubMed] [Google Scholar]
- Hlaing AM, Furusato B, Udo E, Kitamura Y, Souda M, Masutani M, Fukuoka J. 2018. Expression of phosphatase and tensin homolog and programmed cell death ligand 1 in adenosquamous carcinoma of the lung. Biochem Biophys Res Commun 503: 2764–2769. 10.1016/j.bbrc.2018.08.037 [DOI] [PubMed] [Google Scholar]
- Ho CM, Lin MC, Huang SH, Huang CJ, Lai HC, Chien TY, Chang SF. 2009. PTEN promoter methylation and LOH of 10q22-23 locus in PTEN expression of ovarian clear cell adenocarcinomas. Gynecol Oncol 112: 307–313. 10.1016/j.ygyno.2008.09.040 [DOI] [PubMed] [Google Scholar]
- Hocking C, Hardingham JE, Broadbridge V, Wrin J, Townsend AR, Tebbutt N, Cooper J, Ruszkiewicz A, Lee C, Price TJ. 2014. Can we accurately report PTEN status in advanced colorectal cancer? BMC Cancer 14: 128 10.1186/1471-2407-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander MC, Blumenthal GM, Dennis PA. 2011. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer 11: 289–301. 10.1038/nrc3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins BD, Parsons RE. 2014. Molecular pathways: Intercellular PTEN and the potential of PTEN restoration therapy. Clin Cancer Res 20: 5379–5383. 10.1158/1078-0432.CCR-13-2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins BD, Fine B, Steinbach N, Dendy M, Rapp Z, Shaw J, Pappas K, Yu JS, Hodakoski C, Mense S, et al. 2013. A secreted PTEN phosphatase that enters cells to alter signaling and survival. Science 341: 399–402. 10.1126/science.1234907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins BD, Hodakoski C, Barrows D, Mense SM, Parsons RE. 2014. PTEN function: The long and the short of it. Trends Biochem Sci 39: 183–190. 10.1016/j.tibs.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Yan J, Zhang J, Zhu S, Wang Y, Shi T, Zhu C, Chen C, Liu X, Cheng J, et al. 2012. SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nat Commun 3: 911 10.1038/ncomms1919 [DOI] [PubMed] [Google Scholar]
- Idoate MA, Echeveste J, Diez-Valle R, Lozano MD, Aristu J. 2014. Biological and clinical significance of the intratumour heterogeneity of PTEN protein expression and the corresponding molecular abnormalities of the PTEN gene in glioblastomas. Neuropathol Appl Neurobiol 40: 736–746. 10.1111/nan.12117 [DOI] [PubMed] [Google Scholar]
- Ikenoue T, Inoki K, Zhao B, Guan KL. 2008. PTEN acetylation modulates its interaction with PDZ domain. Cancer Res 68: 6908–6912. 10.1158/0008-5472.CAN-08-1107 [DOI] [PubMed] [Google Scholar]
- Jamaspishvili T, Berman DM, Ross AE, Scher HI, De Marzo AM, Squire JA, Lotan TL. 2018. Clinical implications of PTEN loss in prostate cancer. Nat Rev Urol 15: 222–234. 10.1038/nrurol.2018.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang KS, Song YS, Jang SH, Min KW, Na W, Jang SM, Jun YJ, Lee KH, Choi D, Paik SS. 2010. Clinicopathological significance of nuclear PTEN expression in colorectal adenocarcinoma. Histopathology 56: 229–239. 10.1111/j.1365-2559.2009.03468.x [DOI] [PubMed] [Google Scholar]
- Ji ZY, Li HF, Lei Y, Rao YW, Sun ML, Wang XW, Zhang YL. 2018. Phosphatase and tensin homolog protein may be linked to lymph node metastasis and tumor node metastasis staging in nonsmall cell lung cancer. J Cancer Res Ther 14: S138–S144. 10.4103/0973-1482.165865 [DOI] [PubMed] [Google Scholar]
- Jochner MCE, An J, Lättig-Tünnemann G, Kirchner M, Dagane A, Dittmar G, Dirnagl U, Eickholt BJ, Harms C. 2019. Unique properties of PTEN-L contribute to neuroprotection in response to ischemic-like stress. Sci Rep 9: 3183 10.1038/s41598-019-39438-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karapetis CS, Jonker D, Daneshmand M, Hanson JE, O'Callaghan CJ, Marginean C, Zalcberg JR, Simes J, Moore MJ, Tebbutt NC, et al. 2014. PIK3CA, BRAF, and PTEN status and benefit from cetuximab in the treatment of advanced colorectal cancer—Results from NCIC CTG/AGITG CO.17. Clin Cancer Res 20: 744–753. 10.1158/1078-0432.CCR-13-0606 [DOI] [PubMed] [Google Scholar]
- Kim B, Myung JK, Seo JH, Park CK, Paek SH, Kim DG, Jung HW, Park SH. 2010. The clinicopathologic values of the molecules associated with the main pathogenesis of the glioblastoma. J Neurol Sci 294: 112–118. 10.1016/j.jns.2010.03.019 [DOI] [PubMed] [Google Scholar]
- Kim SH, Kim SH, Joung JY, Lee GK, Hong EK, Kang KM, Yu A, Nam BH, Chung J, Seo HK, et al. 2015. Overexpression of ERG and wild-type PTEN are associated with favorable clinical prognosis and low biochemical recurrence in prostate cancer. PLoS ONE 10: e0122498 10.1371/journal.pone.0122498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Shin SJ, Beom SH, Jung M, Choi YY, Son T, Kim HI, Cheong JH, Hyung WJ, Noh SH, et al. 2016. Comprehensive expression profiles of gastric cancer molecular subtypes by immunohistochemistry: Implications for individualized therapy. Oncotarget 7: 44608–44620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Suh J, Surh YJ, Na HK. 2017. Regulation of the tumor suppressor PTEN by natural anticancer compounds. Ann NY Acad Sci 1401: 136–149. 10.1111/nyas.13422 [DOI] [PubMed] [Google Scholar]
- Kimura F, Watanabe J, Hata H, Fujisawa T, Kamata Y, Nishimura Y, Jobo T, Kuramoto H. 2004. PTEN immunohistochemical expression is suppressed in G1 endometrioid adenocarcinoma of the uterine corpus. J Cancer Res Clin Oncol 130: 161–168. 10.1007/s00432-003-0517-8 [DOI] [PubMed] [Google Scholar]
- Kolasa IK, Rembiszewska A, Janiec-Jankowska A, Dansonka-Mieszkowska A, Lewandowska AM, Konopka B, Kupryjańczyk J. 2006. PTEN mutation, expression and LOH at its locus in ovarian carcinomas. Relation to TP53, K-RAS and BRCA1 mutations. Gynecol Oncol 103: 692–697. 10.1016/j.ygyno.2006.05.007 [DOI] [PubMed] [Google Scholar]
- Kreis P, Leondaritis G, Lieberam I, Eickholt BJ. 2014. Subcellular targeting and dynamic regulation of PTEN: Implications for neuronal cells and neurological disorders. Front Mol Neurosci 7: 23 10.3389/fnmol.2014.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisl TN, Lassman AB, Mischel PS, Rosen N, Scher HI, Teruya-Feldstein J, Shaffer D, Lis E, Abrey LE. 2009. A pilot study of everolimus and gefitinib in the treatment of recurrent glioblastoma (GBM). J Neurooncol 92: 99–105. 10.1007/s11060-008-9741-z [DOI] [PubMed] [Google Scholar]
- Kurose K, Zhou XP, Araki T, Cannistra SA, Maher ER, Eng C. 2001. Frequent loss of PTEN expression is linked to elevated phosphorylated Akt levels, but not associated with p27 and cyclin D1 expression, in primary epithelial ovarian carcinomas. Am J Pathol 158: 2097–2106. 10.1016/S0002-9440(10)64681-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak YD, Ma T, Diao S, Zhang X, Chen Y, Hsu J, Lipton SA, Masliah E, Xu H, Liao FF. 2010. NO signaling and S-nitrosylation regulate PTEN inhibition in neurodegeneration. Mol Neurodegener 5: 49 10.1186/1750-1326-5-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lade-Keller J, Riber-Hansen R, Guldberg P, Schmidt H, Hamilton-Dutoit SJ, Steiniche T. 2013. E- to N-cadherin switch in melanoma is associated with decreased expression of phosphatase and tensin homolog and cancer progression. Br J Dermatol 169: 618–628. 10.1111/bjd.12426 [DOI] [PubMed] [Google Scholar]
- Lahdensuo K, Erickson A, Saarinen I, Seikkula H, Lundin J, Lundin M, Nordling S, Butzow A, Vasarainen H, Bostrom PJ, et al. 2016. Loss of PTEN expression in ERG-negative prostate cancer predicts secondary therapies and leads to shorter disease-specific survival time after radical prostatectomy. Mod Pathol 29: 1565–1574. 10.1038/modpathol.2016.154 [DOI] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, et al. 2016. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44: D862–D868. 10.1093/nar/gkv1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavorato-Rocha AM, Anjos LG, Cunha IW, Vassallo J, Soares FA, Rocha RM. 2015. Immunohistochemical assessment of PTEN in vulvar cancer: Best practices for tissue staining, evaluation, and clinical association. Methods 77–78: 20–24. 10.1016/j.ymeth.2014.12.017 [DOI] [PubMed] [Google Scholar]
- Lee YK, Park NH. 2009. Prognostic value and clinicopathological significance of p53 and PTEN in epithelial ovarian cancers. Gynecol Oncol 112: 475–480. 10.1016/j.ygyno.2008.11.031 [DOI] [PubMed] [Google Scholar]
- Lee SR, Yang KS, Kwon J, Lee C, Jeong W, Rhee SG. 2002. Reversible inactivation of the tumor suppressor PTEN by H2O2. J Biol Chem 277: 20336–20342. 10.1074/jbc.M111899200 [DOI] [PubMed] [Google Scholar]
- Lee JT, Shan J, Zhong J, Li M, Zhou B, Zhou A, Parsons R, Gu W. 2013. RFP-mediated ubiquitination of PTEN modulates its effect on AKT activation. Cell Res 23: 552–564. 10.1038/cr.2013.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YR, Chen M, Pandolfi PP. 2018. The functions and regulation of the PTEN tumour suppressor: New modes and prospects. Nat Rev Mol Cell Biol 19: 547–562. [DOI] [PubMed] [Google Scholar]
- Lee YR, Chen M, Lee JD, Zhang J, Lin SY, Fu TM, Chen H, Ishikawa T, Chiang SY, Katon J, et al. 2019. Reactivation of PTEN tumor suppressor for cancer treatment through inhibition of a MYC-WWP1 inhibitory pathway. Science 364: eaau0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen KA, Saramaki OR, Furusato B, Kimura T, Takahashi H, Egawa S, Suzuki H, Keiger K, Ho Hahm S, Isaacs WB, et al. 2013. Loss of PTEN is associated with aggressive behavior in ERG-positive prostate cancer. Cancer Epidemiol Biomarkers Prev 22: 2333–2344. 10.1158/1055-9965.EPI-13-0333-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong TY, Cooper K, Leong AS. 2010. Immunohistology–past, present, and future. Adv Anat Pathol 17: 404–418. 10.1097/PAP.0b013e3181f8957c [DOI] [PubMed] [Google Scholar]
- Leslie NR, Foti M. 2011. Non-genomic loss of PTEN function in cancer: Not in my genes. Trends Pharmacol Sci 32: 131–140. 10.1016/j.tips.2010.12.005 [DOI] [PubMed] [Google Scholar]
- Leslie NR, Longy M. 2016. Inherited PTEN mutations and the prediction of phenotype. Semin Cell Dev Biol 52: 30–38. 10.1016/j.semcdb.2016.01.030 [DOI] [PubMed] [Google Scholar]
- Leslie NR, Kriplani N, Hermida MA, Alvarez-Garcia V, Wise HM. 2016. The PTEN protein: Cellular localization and post-translational regulation. Biochem Soc Trans 44: 273–278. 10.1042/BST20150224 [DOI] [PubMed] [Google Scholar]
- Li DM, Sun H. 1997. TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor β. Cancer Res 57: 2124–2129. [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. 1997. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275: 1943–1947. 10.1126/science.275.5308.1943 [DOI] [PubMed] [Google Scholar]
- Li Z, Dong X, Wang Z, Liu W, Deng N, Ding Y, Tang L, Hla T, Zeng R, Li L, et al. 2005. Regulation of PTEN by Rho small GTPases. Nat Cell Biol 7: 399–404. 10.1038/ncb1236 [DOI] [PubMed] [Google Scholar]
- Li N, Zhang Y, Han X, Liang K, Wang J, Feng L, Wang W, Songyang Z, Lin C, Yang L, et al. 2015a. Poly-ADP ribosylation of PTEN by tankyrases promotes PTEN degradation and tumor growth. Genes Dev 29: 157–170. 10.1101/gad.251785.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Xiao R, Li CP, Huangfu J, Mao JF. 2015b. Increased plasma levels of FABP4 and PTEN is associated with more severe insulin resistance in women with gestational diabetes mellitus. Med Sci Monit 21: 426–431. 10.12659/MSM.892431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Shen Y, Wang M, Yang J, Lv M, Li P, Chen Z, Yang J. 2017. Loss of PTEN expression in breast cancer: Association with clinicopathological characteristics and prognosis. Oncotarget 8: 32043–32054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Yang J, Yang C, Zhu M, Jin Y, McNutt MA, Yin Y. 2018a. PTENα regulates mitophagy and maintains mitochondrial quality control. Autophagy 14: 1742–1760. 10.1080/15548627.2018.1489477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhang T, Guo L, Huang L. 2018b. Regulation of PTEN expression by noncoding RNAs. J Exp Clin Cancer Res 37: 223 10.1186/s13046-018-0898-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, He S, Yang J, Jia X, Wang P, Chen X, Zhang Z, Zou X, McNutt MA, Shen WH, et al. 2014. PTENα, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab 19: 836–848. 10.1016/j.cmet.2014.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Chen X, Yin Q, Ruan D, Zhao X, Zhang C, McNutt MA, Yin Y. 2017. PTENβ is an alternatively translated isoform of PTEN that regulates rDNA transcription. Nat Commun 8: 14771 10.1038/ncomms14771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PC, Lin JK, Lin HH, Lan YT, Lin CC, Yang SH, Chen WS, Liang WY, Jiang JK, Chang SC. 2015. A comprehensive analysis of phosphatase and tensin homolog deleted on chromosome 10 (PTEN) loss in colorectal cancer. World J Surg Oncol 13: 186 10.1186/s12957-015-0601-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litlekalsoy J, Hostmark JG, Costea DE, Illemann M, Laerum OD. 2012. Time-dependent biological differences in molecular markers of high-grade urothelial cancer over 7 decades (ras proteins, pTEN, uPAR, PAI-1 and MMP-9). Virchows Arch 461: 541–551. 10.1007/s00428-012-1323-y [DOI] [PubMed] [Google Scholar]
- Liu J, Visser-Grieve S, Boudreau J, Yeung B, Lo S, Chamberlain G, Yu F, Sun T, Papanicolaou T, Lam A, et al. 2014. Insulin activates the insulin receptor to downregulate the PTEN tumour suppressor. Oncogene 33: 3878–3885. 10.1038/onc.2013.347 [DOI] [PubMed] [Google Scholar]
- Loibl S, Darb-Esfahani S, Huober J, Klimowicz A, Furlanetto J, Lederer B, Hartmann A, Eidtmann H, Pfitzner B, Fasching PA, et al. 2016. Integrated analysis of PTEN and p4EBP1 protein expression as predictors for pCR in HER2-positive breast cancer. Clin Cancer Res 22: 2675–2683. 10.1158/1078-0432.CCR-15-0965 [DOI] [PubMed] [Google Scholar]
- Lokman U, Erickson AM, Vasarainen H, Rannikko AS, Mirtti T. 2018. PTEN loss but not ERG expression in diagnostic biopsies is associated with increased risk of progression and adverse surgical findings in men with prostate cancer on active surveillance. Eur Urol Focus 4: 867–873. 10.1016/j.euf.2017.03.004 [DOI] [PubMed] [Google Scholar]
- Lo Nigro C, Ricci V, Vivenza D, Granetto C, Fabozzi T, Miraglio E, Merlano MC. 2016. Prognostic and predictive biomarkers in metastatic colorectal cancer anti-EGFR therapy. World J Gastroenterol 22: 6944–6954. 10.3748/wjg.v22.i30.6944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López JI, Angulo JC. 2018. Pathological bases and clinical impact of intratumor heterogeneity in clear cell renal cell carcinoma. Curr Urol Rep 19: 3 10.1007/s11934-018-0754-7 [DOI] [PubMed] [Google Scholar]
- Lotan TL, Gurel B, Sutcliffe S, Esopi D, Liu W, Xu J, Hicks JL, Park BH, Humphreys E, Partin AW, et al. 2011. PTEN protein loss by immunostaining: Analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin Cancer Res 17: 6563–6573. 10.1158/1078-0432.CCR-11-1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan TL, Carvalho FL, Peskoe SB, Hicks JL, Good J, Fedor H, Humphreys E, Han M, Platz EA, Squire JA, et al. 2015. PTEN loss is associated with upgrading of prostate cancer from biopsy to radical prostatectomy. Mod Pathol 28: 128–137. 10.1038/modpathol.2014.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan TL, Wei W, Ludkovski O, Morais CL, Guedes LB, Jamaspishvili T, Lopez K, Hawley ST, Feng Z, Fazli L, et al. 2016a. Analytic validation of a clinical-grade PTEN immunohistochemistry assay in prostate cancer by comparison with PTEN FISH. Mod Pathol 29: 904–914. 10.1038/modpathol.2016.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan TL, Wei W, Morais CL, Hawley ST, Fazli L, Hurtado-Coll A, Troyer D, McKenney JK, Simko J, Carroll PR, et al. 2016b. PTEN loss as determined by clinical-grade immunohistochemistry assay is associated with worse recurrence-free survival in prostate cancer. Eur Urol Focus 2: 180–188. 10.1016/j.euf.2015.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan TL, Heumann A, Rico SD, Hicks J, Lecksell K, Koop C, Sauter G, Schlomm T, Simon R. 2017. PTEN loss detection in prostate cancer: Comparison of PTEN immunohistochemistry and PTEN FISH in a large retrospective prostatectomy cohort. Oncotarget 8: 65566–65576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loupakis F, Pollina L, Stasi I, Ruzzo A, Scartozzi M, Santini D, Masi G, Graziano F, Cremolini C, Rulli E, et al. 2009. PTEN expression and KRAS mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. J Clin Oncol 27: 2622–2629. 10.1200/JCO.2008.20.2796 [DOI] [PubMed] [Google Scholar]
- Lu YM, Cheng F, Teng LS. 2016. The association between phosphatase and tensin homolog hypermethylation and patients with breast cancer, a meta-analysis and literature review. Sci Rep 6: 32723 10.1038/srep32723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S, Teugels E, Sadones J, De Brakeleer S, Duerinck J, Du Four S, Michotte A, De Grève J, Neyns B. 2012. Correlation of EGFR, IDH1 and PTEN status with the outcome of patients with recurrent glioblastoma treated in a phase II clinical trial with the EGFR-blocking monoclonal antibody cetuximab. Int J Oncol 41: 1029–1035. 10.3892/ijo.2012.1539 [DOI] [PubMed] [Google Scholar]
- Maiques O, Santacana M, Valls J, Pallares J, Mirantes C, Gatius S, Garcia Dios DA, Amant F, Pedersen HC, Dolcet X, et al. 2014. Optimal protocol for PTEN immunostaining; role of analytical and preanalytical variables in PTEN staining in normal and neoplastic endometrial, breast, and prostatic tissues. Hum Pathol 45: 522–532. 10.1016/j.humpath.2013.10.018 [DOI] [PubMed] [Google Scholar]
- Malaney P, Uversky VN, Davé V. 2013. The PTEN long N-tail is intrinsically disordered: Increased viability for PTEN therapy. Mol Biosyst 9: 2877–2888. 10.1039/c3mb70267g [DOI] [PubMed] [Google Scholar]
- Malaney P, Uversky VN, Davé V. 2017. PTEN proteoforms in biology and disease. Cell Mol Life Sci 74: 2783–2794. 10.1007/s00018-017-2500-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit CJ, Zheng S, Aldape K, Hinds PW, Nelson HH, Wiencke JK, Kelsey KT. 2005. PTEN expression in non-small-cell lung cancer: Evaluating its relation to tumor characteristics, allelic loss, and epigenetic alteration. Hum Pathol 36: 768–776. 10.1016/j.humpath.2005.05.006 [DOI] [PubMed] [Google Scholar]
- Martins FC, Santiago I, Trinh A, Xian J, Guo A, Sayal K, Jimenez-Linan M, Deen S, Driver K, Mack M, et al. 2014. Combined image and genomic analysis of high-grade serous ovarian cancer reveals PTEN loss as a common driver event and prognostic classifier. Genome Biol 15: 526 10.1186/s13059-014-0526-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson GR, Perisic O, Burke JE, Williams RL. 2016. The intrinsically disordered tails of PTEN and PTEN-L have distinct roles in regulating substrate specificity and membrane activity. Biochem J 473: 135–144. 10.1042/BJ20150931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos LL, Trufelli DC, de Matos MG, da Silva Pinhal MA. 2010. Immunohistochemistry as an important tool in biomarkers detection and clinical practice. Biomark Insights 5: 9–20. 10.4137/BMI.S2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe N, Kennedy RD, Prise KM. 2016. The role of PTEN as a cancer biomarker. Oncoscience 3: 54–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEllin B, Camacho CV, Mukherjee B, Hahm B, Tomimatsu N, Bachoo RM, Burma S. 2010. PTEN loss compromises homologous recombination repair in astrocytes: Implications for glioblastoma therapy with temozolomide or poly(ADP-ribose) polymerase inhibitors. Cancer Res 70: 5457–5464. 10.1158/0008-5472.CAN-09-4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N, Swanton C. 2015. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 27: 15–26. 10.1016/j.ccell.2014.12.001 [DOI] [PubMed] [Google Scholar]
- McLoughlin NM, Mueller C, Grossmann TN. 2018. The therapeutic potential of PTEN modulation: Targeting strategies from gene to protein. Cell Chem Biol 25: 19–29. [DOI] [PubMed] [Google Scholar]
- Mehra R, Salami SS, Lonigro R, Bhalla R, Siddiqui J, Cao X, Spratt DE, Palapattu GS, Palanisamy N, Wei JT, et al. 2018. Association of ERG/PTEN status with biochemical recurrence after radical prostatectomy for clinically localized prostate cancer. Med Oncol 35: 152 10.1007/s12032-018-1212-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, et al. 2005. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med 353: 2012–2024. 10.1056/NEJMoa051918 [DOI] [PubMed] [Google Scholar]
- Mellinghoff IK, Cloughesy TF, Mischel PS. 2007. PTEN-mediated resistance to epidermal growth factor receptor kinase inhibitors. Clin Cancer Res 13: 378–381. 10.1158/1078-0432.CCR-06-1992 [DOI] [PubMed] [Google Scholar]
- Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, Kim JS, Waldman T, Lord CJ, Ashworth A. 2009. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med 1: 315–322. 10.1002/emmm.200900041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z, Jia LF, Gan YH. 2016. PTEN activation through K163 acetylation by inhibiting HDAC6 contributes to tumour inhibition. Oncogene 35: 2333–2344. 10.1038/onc.2015.293 [DOI] [PubMed] [Google Scholar]
- Milella M, Falcone I, Conciatori F, Cesta Incani U, Del Curatolo A, Inzerilli N, Nuzzo CCMA, Vaccaro V, Vari S, Cognetti F, et al. 2015. PTEN: Multiple functions in human malignant tumors. Front Oncol 5: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millis SZ, Ikeda S, Reddy S, Gatalica Z, Kurzrock R. 2016. Landscape of phosphatidylinositol-3-kinase pathway alterations across 19 784 diverse solid tumors. JAMA Oncol 2: 1565–1573. 10.1001/jamaoncol.2016.0891 [DOI] [PubMed] [Google Scholar]
- Min HS, Lee C, Jung KC. 2013. Correlation of immunohistochemical markers and BRAF mutation status with histological variants of papillary thyroid carcinoma in the Korean population. J Korean Med Sci 28: 534–541. 10.3346/jkms.2013.28.4.534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingo J, Rodríguez-Escudero I, Luna S, Fernández-Acero T, Amo L, Jonasson AR, Zori RT, López JI, Molina M, Cid VJ, et al. 2018. A pathogenic role for germline PTEN variants which accumulate into the nucleus. Eur J Hum Genet 26: 1180–1187. 10.1038/s41431-018-0155-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingo J, Luna S, Gaafar A, Nunes-Xavier CE, Torices L, Mosteiro L, Ruiz R, Guerra I, Llarena R, Angulo JC, et al. 2019. Precise definition of PTEN C-terminal epitopes and its implications in clinical oncology. NPJ Precis Oncol 3: 11 10.1038/s41698-019-0083-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montano N, Cenci T, Martini M, D'Alessandris QG, Pelacchi F, Ricci-Vitiani L, Maira G, De Maria R, Larocca LM, Pallini R. 2011. Expression of EGFRvIII in glioblastoma: Prognostic significance revisited. Neoplasia 13: 1113–1121. 10.1593/neo.111338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montano N, D'Alessandris QG, Izzo A, Fernandez E, Pallini R. 2016. Biomarkers for glioblastoma multiforme: Status quo. J Clin Transl Res 2: 3–10. [PMC free article] [PubMed] [Google Scholar]
- Monte NM, Webster KA, Neuberg D, Dressler GR, Mutter GL. 2010. Joint loss of PAX2 and PTEN expression in endometrial precancers and cancer. Cancer Res 70: 6225–6232. 10.1158/0008-5472.CAN-10-0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Patterson AL, George JW, Carpenter TJ, Madaj ZB, Hostetter G, Risinger JI, Teixeira JM. 2018. Nuclear PTEN localization contributes to DNA damage response in endometrial adenocarcinoma and could have a diagnostic benefit for therapeutic management of the disease. Mol Cancer Ther 17: 1995–2003. 10.1158/1535-7163.MCT-17-1255 [DOI] [PubMed] [Google Scholar]
- Mutter GL, Lin MC, Fitzgerald JT, Kum JB, Baak JP, Lees JA, Weng LP, Eng C. 2000. Altered PTEN expression as a diagnostic marker for the earliest endometrial precancers. J Natl Cancer Inst 92: 924–930. 10.1093/jnci/92.11.924 [DOI] [PubMed] [Google Scholar]
- Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, Sahin AA, Klos KS, Li P, Monia BP, Nguyen NT, et al. 2004. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell 6: 117–127. 10.1016/j.ccr.2004.06.022 [DOI] [PubMed] [Google Scholar]
- Nakakido M, Deng Z, Suzuki T, Dohmae N, Nakamura Y, Hamamoto R. 2015. Dysregulation of AKT pathway by SMYD2-mediated lysine methylation on PTEN. Neoplasia 17: 367–373. 10.1016/j.neo.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif NT, Lobo GP, Wu X, Henderson CJ, Morrison CD, Eng C, Jalaludin B, Segelov E. 2004. PTEN mutations are common in sporadic microsatellite stable colorectal cancer. Oncogene 23: 617–628. 10.1038/sj.onc.1207059 [DOI] [PubMed] [Google Scholar]
- Ngeow J, He X, Mester JL, Lei J, Romigh T, Orloff MS, Milas M, Eng C. 2012. Utility of PTEN protein dosage in predicting for underlying germline PTEN mutations among patients presenting with thyroid cancer and Cowden-like phenotypes. J Clin Endocrinol Metab 97: E2320–E2327. 10.1210/jc.2012-2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norimatsu Y, Moriya T, Kobayashi TK, Sakurai T, Shimizu K, Tsukayama C, Ohno E. 2007. Immunohistochemical expression of PTEN and β-catenin for endometrial intraepithelial neoplasia in Japanese women. Ann Diagn Pathol 11: 103–108. 10.1016/j.anndiagpath.2006.06.009 [DOI] [PubMed] [Google Scholar]
- Nosaka R, Yamasaki F, Saito T, Takayasu T, Kolakshyapati M, Amatya VJ, Takeshima Y, Sugiyama K, Kurisu K. 2017. Role for loss of nuclear PTEN in a harbinger of brain metastases. J Clin Neurosci 44: 148–154. 10.1016/j.jocn.2017.06.004 [DOI] [PubMed] [Google Scholar]
- Odriozola L, Singh G, Hoang T, Chan AM. 2007. Regulation of PTEN activity by its carboxyl-terminal autoinhibitory domain. J Biol Chem 282: 23306–23315. 10.1074/jbc.M611240200 [DOI] [PubMed] [Google Scholar]
- Ohno K, Okuda K, Uehara T. 2015. Endogenous S-sulfhydration of PTEN helps protect against modification by nitric oxide. Biochem Biophys Res Commun 456: 245–249. 10.1016/j.bbrc.2014.11.066 [DOI] [PubMed] [Google Scholar]
- Okumura K, Mendoza M, Bachoo RM, DePinho RA, Cavenee WK, Furnari FB. 2006. PCAF modulates PTEN activity. J Biol Chem 281: 26562–26568. 10.1074/jbc.M605391200 [DOI] [PubMed] [Google Scholar]
- Pallares J, Bussaglia E, Martínez-Guitarte JL, Dolcet X, Llobet D, Rue M, Sanchez-Verde L, Palacios J, Prat J, Matias-Guiu X. 2005. Immunohistochemical analysis of PTEN in endometrial carcinoma: A tissue microarray study with a comparison of four commercial antibodies in correlation with molecular abnormalities. Mod Pathol 18: 719–727. 10.1038/modpathol.3800347 [DOI] [PubMed] [Google Scholar]
- Panigrahi AR, Pinder SE, Chan SY, Paish EC, Robertson JF, Ellis IO. 2004. The role of PTEN and its signalling pathways, including AKT, in breast cancer; an assessment of relationships with other prognostic factors and with outcome. J Pathol 204: 93–100. 10.1002/path.1611 [DOI] [PubMed] [Google Scholar]
- Papa A, Pandolfi PP. 2019. The PTEN–PI3K axis in cancer. Biomolecules 9: 153 10.3390/biom9040153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez EA, Dueck AC, McCullough AE, Chen B, Geiger XJ, Jenkins RB, Lingle WL, Davidson NE, Martino S, Kaufman PA, et al. 2013. Impact of PTEN protein expression on benefit from adjuvant trastuzumab in early-stage human epidermal growth factor receptor 2-positive breast cancer in the North Central Cancer Treatment Group N9831 trial. J Clin Oncol 31: 2115–2122. 10.1200/JCO.2012.42.2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perren A, Weng LP, Boag AH, Ziebold U, Thakore K, Dahia PL, Komminoth P, Lees JA, Mulligan LM, Mutter GL, et al. 1999. Immunohistochemical evidence of loss of PTEN expression in primary ductal adenocarcinomas of the breast. Am J Pathol 155: 1253–1260. 10.1016/S0002-9440(10)65227-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perren A, Komminoth P, Saremaslani P, Matter C, Feurer S, Lees JA, Heitz PU, Eng C. 2000. Mutation and expression analyses reveal differential subcellular compartmentalization of PTEN in endocrine pancreatic tumors compared to normal islet cells. Am J Pathol 157: 1097–1103. 10.1016/S0002-9440(10)64624-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido R. 2015. PTEN: A yin-yang master regulator protein in health and disease. Methods 77-78: 3–10. 10.1016/j.ymeth.2015.02.009 [DOI] [PubMed] [Google Scholar]
- Pulido R, Baker SJ, Barata JT, Carracedo A, Cid VJ, Chin-Sang ID, Dave V, den Hertog J, Devreotes P, Eickholt BJ, et al. 2014. A unified nomenclature and amino acid numbering for human PTEN. Sci Signal 7: pe15 10.1126/scisignal.2005560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que WC, Qiu HQ, Cheng Y, Liu MB, Wu CY. 2018. PTEN in kidney cancer: A review and meta-analysis. Clin Chim Acta 480: 92–98. 10.1016/j.cca.2018.01.031 [DOI] [PubMed] [Google Scholar]
- Raffone A, Travaglino A, Saccone G, Campanino MR, Mollo A, De Placido G, Insabato L, Zullo F. 2019a. Loss of PTEN expression as diagnostic marker of endometrial precancer: A systematic review and meta-analysis. Acta Obstet Gynecol Scand 98: 275–286. 10.1111/aogs.13513 [DOI] [PubMed] [Google Scholar]
- Raffone A, Travaglino A, Saccone G, Viggiani M, Giampaolino P, Insabato L, Mollo A, De Placido G, Zullo F. 2019b. PTEN expression in endometrial hyperplasia and risk of cancer: A systematic review and meta-analysis. Arch Gynecol Obstet 299: 1511–1524. 10.1007/s00404-019-05123-x [DOI] [PubMed] [Google Scholar]
- Rahdar M, Inoue T, Meyer T, Zhang J, Vazquez F, Devreotes PN. 2009. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc Natl Acad Sci 106: 480–485. 10.1073/pnas.0811212106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razavi SA, Modarressi MH, Yaghmaei P, Tavangar SM, Hedayati M. 2017. Circulating levels of PTEN and KLLN in papillary thyroid carcinoma: Can they be considered as novel diagnostic biomarkers? Endocrine 57: 428–435. 10.1007/s12020-017-1368-4 [DOI] [PubMed] [Google Scholar]
- Razis E, Bobos M, Kotoula V, Eleftheraki AG, Kalofonos HP, Pavlakis K, Papakostas P, Aravantinos G, Rigakos G, Efstratiou I, et al. 2011. Evaluation of the association of PIK3CA mutations and PTEN loss with efficacy of trastuzumab therapy in metastatic breast cancer. Breast Cancer Res Treat 128: 447–456. 10.1007/s10549-011-1572-5 [DOI] [PubMed] [Google Scholar]
- Rieken M, Shariat SF, Karam JA, Foerster B, Khani F, Gust K, Abufaraj M, Wood CG, Weizer AZ, Raman JD, et al. 2017. Frequency and prognostic value of PTEN loss in patients with upper tract urothelial carcinoma treated with radical nephroureterectomy. J Urol 198: 1269–1277. 10.1016/j.juro.2017.06.096 [DOI] [PubMed] [Google Scholar]
- Rieth J, Subramanian S. 2018. Mechanisms of intrinsic tumor resistance to immunotherapy. Int J Mol Sci 19: E1340 10.3390/ijms19051340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimawi MF, De Angelis C, Contreras A, Pareja F, Geyer FC, Burke KA, Herrera S, Wang T, Mayer IA, Forero A, et al. 2018. Low PTEN levels and PIK3CA mutations predict resistance to neoadjuvant lapatinib and trastuzumab without chemotherapy in patients with HER2 over-expressing breast cancer. Breast Cancer Res Treat 167: 731–740. 10.1007/s10549-017-4533-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Escudero I, Oliver MD, Andrés-Pons A, Molina M, Cid VJ, Pulido R. 2011. A comprehensive functional analysis of PTEN mutations: Implications in tumor- and autism-related syndromes. Hum Mol Genet 20: 4132–4142. 10.1093/hmg/ddr337 [DOI] [PubMed] [Google Scholar]
- Roh MH, Yassin Y, Miron A, Mehra KK, Mehrad M, Monte NM, Mutter GL, Nucci MR, Ning G, McKeon FD, et al. 2010. High-grade fimbrial-ovarian carcinomas are unified by altered p53, PTEN and PAX2 expression. Mod Pathol 23: 1316–1324. 10.1038/modpathol.2010.119 [DOI] [PubMed] [Google Scholar]
- Roy D, Dittmer DP. 2011. Phosphatase and tensin homolog on chromosome 10 is phosphorylated in primary effusion lymphoma and Kaposi's sarcoma. Am J Pathol 179: 2108–2119. 10.1016/j.ajpath.2011.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakr RA, Barbashina V, Morrogh M, Chandarlapaty S, Andrade VP, Arroyo CD, Olvera N, King TA. 2010. Protocol for PTEN expression by immunohistochemistry in formalin-fixed paraffin-embedded human breast carcinoma. Appl Immunohistochem Mol Morphol 18: 371–374. 10.1097/PAI.0b013e3181d50bd5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado R, Moore H, Martens JWM, Lively T, Malik S, McDermott U, Michiels S, Moscow JA, Tejpar S, McKee T, et al. 2017. Societal challenges of precision medicine: Bringing order to chaos. Eur J Cancer 84: 325–334. 10.1016/j.ejca.2017.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangale Z, Prass C, Carlson A, Tikishvili E, Degrado J, Lanchbury J, Stone S. 2011. A robust immunohistochemical assay for detecting PTEN expression in human tumors. Appl Immunohistochem Mol Morphol 19: 173–183. 10.1097/PAI.0b013e3181f1da13 [DOI] [PubMed] [Google Scholar]
- Sarquis MS, Agrawal S, Shen L, Pilarski R, Zhou XP, Eng C. 2006. Distinct expression profiles for PTEN transcript and its splice variants in Cowden syndrome and Bannayan–Riley–Ruvalcaba syndrome. Am J Hum Genet 79: 23–30. 10.1086/504392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai H, Yasuda A, Ochi N, Ma J, Matsuo Y, Wakasugi T, Takahashi H, Funahashi H, Sato M, Takeyama H. 2008. Loss of PTEN expression is associated with colorectal cancer liver metastasis and poor patient survival. BMC Gastroenterol 8: 56 10.1186/1471-230X-8-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt KT, Chau CH, Price DK, Figg WD. 2016. Precision oncology medicine: The clinical relevance of patient-specific biomarkers used to optimize cancer treatment. J Clin Pharmacol 56: 1484–1499. 10.1002/jcph.765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz L, Albadine R, Hicks J, Jadallah S, DeMarzo AM, Chen YB, Nielsen ME, Gonzalgo ML, Sidransky D, Schoenberg M, et al. 2010. Expression status and prognostic significance of mammalian target of rapamycin pathway members in urothelial carcinoma of urinary bladder after cystectomy. Cancer 116: 5517–5526. 10.1002/cncr.25502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrard RM, Maitland NJ. 2000. Alternative splicing of the human PTEN/MMAC1/TEP1 gene. Biochim Biophys Acta 1494: 282–285. 10.1016/S0167-4781(00)00210-4 [DOI] [PubMed] [Google Scholar]
- Shehata M, Schnabl S, Demirtas D, Hilgarth M, Hubmann R, Ponath E, Badrnya S, Lehner C, Hoelbl A, Duechler M, et al. 2010. Reconstitution of PTEN activity by CK2 inhibitors and interference with the PI3-K/Akt cascade counteract the antiapoptotic effect of human stromal cells in chronic lymphocytic leukemia. Blood 116: 2513–2521. 10.1182/blood-2009-10-248054 [DOI] [PubMed] [Google Scholar]
- Shoman N, Klassen S, McFadden A, Bickis MG, Torlakovic E, Chibbar R. 2005. Reduced PTEN expression predicts relapse in patients with breast carcinoma treated by tamoxifen. Mod Pathol 18: 250–259. 10.1038/modpathol.3800296 [DOI] [PubMed] [Google Scholar]
- Siddiqui S, Akhter N, Deo SV, Shukla NK, Husain SA. 2016. A study on promoter methylation of PTEN in sporadic breast cancer patients from North India. Breast Cancer 23: 922–931. 10.1007/s12282-015-0665-0 [DOI] [PubMed] [Google Scholar]
- Singh M, Chaudhry P, Fabi F, Asselin E. 2013. Cisplatin-induced caspase activation mediates PTEN cleavage in ovarian cancer cells: A potential mechanism of chemoresistance. BMC Cancer 13: 233 10.1186/1471-2407-13-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood A, McClain D, Maitra R, Basu-Mallick A, Seetharam R, Kaubisch A, Rajdev L, Mariadason JM, Tanaka K, Goel S. 2012. PTEN gene expression and mutations in the PIK3CA gene as predictors of clinical benefit to anti-epidermal growth factor receptor antibody therapy in patients with KRAS wild-type metastatic colorectal cancer. Clin Colorectal Cancer 11: 143–150. 10.1016/j.clcc.2011.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperinde J, Jin X, Banerjee J, Penuel E, Saha A, Diedrich G, Huang W, Leitzel K, Weidler J, Ali SM, et al. 2010. Quantitation of p95HER2 in paraffin sections by using a p95-specific antibody and correlation with outcome in a cohort of trastuzumab-treated breast cancer patients. Clin Cancer Res 16: 4226–4235. 10.1158/1078-0432.CCR-10-0410 [DOI] [PubMed] [Google Scholar]
- Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, et al. 1997. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet 15: 356–362. 10.1038/ng0497-356 [DOI] [PubMed] [Google Scholar]
- Steelman LS, Navolanic PM, Sokolosky ML, Taylor JR, Lehmann BD, Chappell WH, Abrams SL, Wong EW, Stadelman KM, Terrian DM, et al. 2008. Suppression of PTEN function increases breast cancer chemotherapeutic drug resistance while conferring sensitivity to mTOR inhibitors. Oncogene 27: 4086–4095. 10.1038/onc.2008.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, Carey M, Hu Z, Guan Y, Sahin A, et al. 2008. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res 68: 6084–6091. 10.1158/0008-5472.CAN-07-6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. 2014. The Human Gene Mutation Database: Building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 133: 1–9. 10.1007/s00439-013-1358-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su R, Nan H, Guo H, Ruan Z, Jiang L, Song Y, Nan K. 2016. Associations of components of PTEN/AKT/mTOR pathway with cancer stem cell markers and prognostic value of these biomarkers in hepatocellular carcinoma. Hepatol Res 46: 1380–1391. 10.1111/hepr.12687 [DOI] [PubMed] [Google Scholar]
- Tan H, Bao J, Zhou X. 2015. Genome-wide mutational spectra analysis reveals significant cancer-specific heterogeneity. Sci Rep 5: 12566 10.1038/srep12566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Li X, Gao Y, Chen L, Gu L, Chen J, Lyu X, Zhang Y, Zhang X. 2017. Phosphatase and tensin homolog (PTEN) expression on oncologic outcome in renal cell carcinoma: A systematic review and meta-analysis. PLoS ONE 12: e0179437 10.1371/journal.pone.0179437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapparel C, Reymond A, Girardet C, Guillou L, Lyle R, Lamon C, Hutter P, Antonarakis SE. 2003. The TPTE gene family: Cellular expression, subcellular localization and alternative splicing. Gene 323: 189–199. 10.1016/j.gene.2003.09.038 [DOI] [PubMed] [Google Scholar]
- Taulli R, Loretelli C, Pandolfi PP. 2013. From pseudo-ceRNAs to circ-ceRNAs: A tale of cross-talk and competition. Nat Struct Mol Biol 20: 541–543. 10.1038/nsmb.2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng HW, Wang HW, Chen WM, Chao TC, Hsieh YY, Hsih CH, Tzeng CH, Chen PC, Yen CC. 2011. Prevalence and prognostic influence of genomic changes of EGFR pathway markers in synovial sarcoma. J Surg Oncol 103: 773–781. 10.1002/jso.21852 [DOI] [PubMed] [Google Scholar]
- Therkildsen C, Bergmann TK, Henrichsen-Schnack T, Ladelund S, Nilbert M. 2014. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: A systematic review and meta-analysis. Acta Oncol 53: 852–864. 10.3109/0284186X.2014.895036 [DOI] [PubMed] [Google Scholar]
- Thiessen B, Stewart C, Tsao M, Kamel-Reid S, Schaiquevich P, Mason W, Easaw J, Belanger K, Forsyth P, McIntosh L, et al. 2010. A phase I/II trial of GW572016 (lapatinib) in recurrent glioblastoma multiforme: Clinical outcomes, pharmacokinetics and molecular correlation. Cancer Chemother Pharmacol 65: 353–361. 10.1007/s00280-009-1041-6 [DOI] [PubMed] [Google Scholar]
- Tinker AV, Ellard S, Welch S, Moens F, Allo G, Tsao MS, Squire J, Tu D, Eisenhauer EA, MacKay H. 2013. Phase II study of temsirolimus (CCI-779) in women with recurrent, unresectable, locally advanced or metastatic carcinoma of the cervix. A trial of the NCIC Clinical Trials Group (NCIC CTG IND 199). Gynecol Oncol 130: 269–274. 10.1016/j.ygyno.2013.05.008 [DOI] [PubMed] [Google Scholar]
- Torres J, Pulido R. 2001. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J Biol Chem 276: 993–998. 10.1074/jbc.M009134200 [DOI] [PubMed] [Google Scholar]
- Torres J, Navarro S, Roglá I, Ripoll F, Lluch A, Garcia-Conde J, Llombart-Bosch A, Cervera J, Pulido R. 2001. Heterogeneous lack of expression of the tumour suppressor PTEN protein in human neoplastic tissues. Eur J Cancer 37: 114–121. 10.1016/S0959-8049(00)00366-X [DOI] [PubMed] [Google Scholar]
- Torres J, Rodriguez J, Myers MP, Valiente M, Graves JD, Tonks NK, Pulido R. 2003. Phosphorylation-regulated cleavage of the tumor suppressor PTEN by caspase-3: Implications for the control of protein stability and PTEN–protein interactions. J Biol Chem 278: 30652–30660. 10.1074/jbc.M212610200 [DOI] [PubMed] [Google Scholar]
- Tran TN, Brettingham-Moore K, Duong CP, Mitchell C, Clemons NJ, Phillips WA. 2013. Molecular changes in the phosphatidylinositide 3-kinase (PI3K) pathway are common in gastric cancer. J Surg Oncol 108: 113–120. 10.1002/jso.23357 [DOI] [PubMed] [Google Scholar]
- Trock BJ, Fedor H, Gurel B, Jenkins RB, Knudsen BS, Fine SW, Said JW, Carter HB, Lotan TL, De Marzo AM. 2016. PTEN loss and chromosome 8 alterations in Gleason grade 3 prostate cancer cores predicts the presence of un-sampled grade 4 tumor: Implications for active surveillance. Mod Pathol 29: 764–771. 10.1038/modpathol.2016.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, Pavletich NP, Carver BS, Cordon-Cardo C, Erdjument-Bromage H, et al. 2007. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 128: 141–156. 10.1016/j.cell.2006.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey JD, Brahme NN, Zhang B. 2017. Drug-biomarker co-development in oncology—20 years and counting. Drug Resist Updat 30: 48–62. 10.1016/j.drup.2017.02.002 [DOI] [PubMed] [Google Scholar]
- Tzani I, Ivanov IP, Andreev DE, Dmitriev RI, Dean KA, Baranov PV, Atkins JF, Loughran G. 2016. Systematic analysis of the PTEN 5′ leader identifies a major AUU initiated proteoform. Open Biol 6: 150203 10.1098/rsob.150203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno S, Sudo T, Oka N, Wakahashi S, Yamaguchi S, Fujiwara K, Mikami Y, Nishimura R. 2013. Absence of human papillomavirus infection and activation of PI3K-AKT pathway in cervical clear cell carcinoma. Int J Gynecol Cancer 23: 1084–1091. 10.1097/IGC.0b013e3182981bdc [DOI] [PubMed] [Google Scholar]
- Valkov A, Kilvaer TK, Sorbye SW, Donnem T, Smeland E, Bremnes RM, Busund LT. 2011. The prognostic impact of Akt isoforms, PI3K and PTEN related to female steroid hormone receptors in soft tissue sarcomas. J Transl Med 9: 200 10.1186/1479-5876-9-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Devreotes P. 2006. Regulation of PTEN function as a PIP3 gatekeeper through membrane interaction. Cell Cycle 5: 1523–1527. 10.4161/cc.5.14.3005 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Ramaswamy S, Nakamura N, Sellers WR. 2000. Phosphorylation of the PTEN tail regulates protein stability and function. Mol Cell Biol 20: 5010–5018. 10.1128/MCB.20.14.5010-5018.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Grossman SR, Takahashi Y, Rokas MV, Nakamura N, Sellers WR. 2001. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. J Biol Chem 276: 48627–48630. 10.1074/jbc.C100556200 [DOI] [PubMed] [Google Scholar]
- Vázquez-Ulloa E, Lizano M, Avilés-Salas A, Alfaro-Moreno E, Contreras-Paredes A. 2011. Abnormal distribution of hDlg and PTEN in premalignant lesions and invasive cervical cancer. Gynecol Oncol 122: 663–668. 10.1016/j.ygyno.2011.05.017 [DOI] [PubMed] [Google Scholar]
- Verhagen PC, van Duijn PW, Hermans KG, Looijenga LH, van Gurp RJ, Stoop H, van der Kwast TH, Trapman J. 2006. The PTEN gene in locally progressive prostate cancer is preferentially inactivated by bi-allelic gene deletion. J Pathol 208: 699–707. 10.1002/path.1929 [DOI] [PubMed] [Google Scholar]
- Walker SM, Downes CP, Leslie NR. 2001. TPIP: A novel phosphoinositide 3-phosphatase. Biochem J 360: 277–283. 10.1042/bj3600277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dai B. 2015. PTEN genomic deletion defines favorable prognostic biomarkers in localized prostate cancer: A systematic review and meta-analysis. Int J Clin Exp Med 8: 5430–5437. [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kristensen GB, Helland A, Nesland JM, Børresen-Dale AL, Holm R. 2005. Protein expression and prognostic value of genes in the erb-b signaling pathway in advanced ovarian carcinomas. Am J Clin Pathol 124: 392–401. 10.1309/BL7EMW66LQX6GFRP [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu Y, Du Y, Yin W, Lu J. 2013. The predictive role of phosphatase and tensin homolog (PTEN) loss, phosphoinositol-3 (PI3) kinase (PIK3CA) mutation, and PI3K pathway activation in sensitivity to trastuzumab in HER2-positive breast cancer: A meta-analysis. Curr Med Res Opin 29: 633–642. 10.1185/03007995.2013.794775 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang P, Lin C, Yu Q, Wu J, Wang L, Cui Y, Wang K, Gao Z, Li H. 2015a. Relevance and therapeutic possibility of PTEN-long in renal cell carcinoma. PLoS ONE 10: e114250 10.1371/journal.pone.0114250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen H, Liao Y, Chen N, Liu T, Zhang H, Zhang H. 2015b. Expression and clinical evidence of miR-494 and PTEN in non–small cell lung cancer. Tumour Biol 36: 6965–6972. 10.1007/s13277-015-3416-0 [DOI] [PubMed] [Google Scholar]
- Wang Y, Qu X, Shen HC, Wang K, Liu Q, Du JJ. 2015c. Predictive and prognostic biomarkers for patients treated with anti-EGFR agents in lung cancer: A systemic review and meta-analysis. Asian Pac J Cancer Prev 16: 4759–4768. 10.7314/APJCP.2015.16.11.4759 [DOI] [PubMed] [Google Scholar]
- Wang LL, Hao S, Zhang S, Guo LJ, Hu CY, Zhang G, Gao B, Zhao JJ, Jiang Y, Tian WG, et al. 2017. PTEN/PI3K/AKT protein expression is related to clinicopathological features and prognosis in breast cancer with axillary lymph node metastases. Hum Pathol 61: 49–57. 10.1016/j.humpath.2016.07.040 [DOI] [PubMed] [Google Scholar]
- Wang L, Cho YL, Tang Y, Wang J, Park JE, Wu Y, Wang C, Tong Y, Chawla R, Zhang J, et al. 2018. PTEN-L is a novel protein phosphatase for ubiquitin dephosphorylation to inhibit PINK1-Parkin-mediated mitophagy. Cell Res 28: 787–802. 10.1038/s41422-018-0056-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin SN, Ju Z, Broaddus RR, Krakstad C, Li J, Pal N, Lu KH, Coleman RL, Hennessy BT, Klempner SJ, et al. 2015. PTEN loss is a context-dependent outcome determinant in obese and non-obese endometrioid endometrial cancer patients. Mol Oncol 9: 1694–1703. 10.1016/j.molonc.2015.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand KC, Hennessy BT, Leung S, Wang Y, Ju Z, McGahren M, Kalloger SE, Finlayson S, Stemke-Hale K, Lu Y, et al. 2014. A functional proteogenomic analysis of endometrioid and clear cell carcinomas using reverse phase protein array and mutation analysis: protein expression is histotype-specific and loss of ARID1A/BAF250a is associated with AKT phosphorylation. BMC Cancer 14: 120 10.1186/1471-2407-14-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise HM, Hermida MA, Leslie NR. 2017. Prostate cancer, PI3K, PTEN and prognosis. Clin Sci 131: 197–210. 10.1042/CS20160026 [DOI] [PubMed] [Google Scholar]
- Woolley JF, Salmena L. 2016. Measurement of PTEN by flow cytometry. Methods Mol Biol 1388: 39–51. 10.1007/978-1-4939-3299-3_4 [DOI] [PubMed] [Google Scholar]
- Worby CA, Dixon JE. 2014. Pten. Annu Rev Biochem 83: 641–669. 10.1146/annurev-biochem-082411-113907 [DOI] [PubMed] [Google Scholar]
- Wu J, Song Y. 2018. Expression and clinical significance of serum MMP-7 and PTEN levels in patients with acute myeloid leukemia. Oncol Lett 15: 3447–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Hu CP, He BX, Chen X, Lu XX, Xie MX, Li W, He SY, You SJ, Chen Q. 2016. PTEN expression is a prognostic marker for patients with non–small cell lung cancer: A systematic review and meta-analysis of the literature. Oncotarget 7: 57832–57840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Xie B, Liu C, Wang J, Xu Y. 2017. Association of PTEN expression with biochemical recurrence in prostate cancer: Results based on previous reports. OncoTargets Ther 10: 5089–5097. 10.2147/OTT.S132653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang Z, Zhou SF, Lu N. 2014. Posttranslational regulation of phosphatase and tensin homolog (PTEN) and its functional impact on cancer behaviors. Drug Design Dev Ther 8: 1745–1751. 10.2147/DDDT.S71061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Zhang C, Cui J, Liu J, Li J, Jiang H. 2017. The prognostic value and potential drug target of phosphatase and tensin homolog in breast cancer patients: A meta-analysis. Medicine 96: e8000 10.1097/MD.0000000000008000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa N, Leduc C, Kohler D, Saieg MA, John T, Sykes J, Yoshimoto M, Pintilie M, Squire J, Shepherd FA, et al. 2012. Loss of phosphatase and tensin homolog protein expression is an independent poor prognostic marker in lung adenocarcinoma. J Thorac Oncol 7: 1513–1521. 10.1097/JTO.0b013e3182641d4f [DOI] [PubMed] [Google Scholar]
- Yang J, Liu J, Zheng J, Du W, He Y, Liu W, Huang S. 2007. A reappraisal by quantitative flow cytometry analysis of PTEN expression in acute leukemia. Leukemia 21: 2072–2074. 10.1038/sj.leu.2404740 [DOI] [PubMed] [Google Scholar]
- Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, et al. 2013. Genomics of drug sensitivity in cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 41: D955–D961. 10.1093/nar/gks1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZY, Yu YY, Yuan JQ, Shen WX, Zheng DY, Chen JZ, Mao C, Tang JL. 2016. The prognostic value of phosphatase and tensin homolog negativity in breast cancer: A systematic review and meta-analysis of 32 studies with 4393 patients. Crit Rev Oncol Hematol 101: 40–49. 10.1016/j.critrevonc.2016.01.013 [DOI] [PubMed] [Google Scholar]
- Yehia L, Eng C. 2018. 65 years of the double helix: one gene, many endocrine and metabolic syndromes: PTEN-opathies and precision medicine. Endocr Relat Cancer 25: T121–T140. 10.1530/ERC-18-0162 [DOI] [PubMed] [Google Scholar]
- Yim EK, Peng G, Dai H, Hu R, Li K, Lu Y, Mills GB, Meric-Bernstam F, Hennessy BT, Craven RJ, et al. 2009. Rak functions as a tumor suppressor by regulating PTEN protein stability and function. Cancer Cell 15: 304–314. 10.1016/j.ccr.2009.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemori K, Tsuta K, Shimizu C, Hatanaka Y, Hashizume K, Ono M, Kouno T, Ando M, Tamura K, Katsumata N, et al. 2009. Immunohistochemical expression of PTEN and phosphorylated Akt are not correlated with clinical outcome in breast cancer patients treated with trastuzumab-containing neo-adjuvant chemotherapy. Med Oncol 26: 344–349. 10.1007/s12032-008-9127-2 [DOI] [PubMed] [Google Scholar]
- Yun JW, Lee S, Ryu D, Park S, Park WY, Joung JG, Jeong J. 2019. Biomarkers associated with tumor heterogeneity in prostate cancer. Transl Oncol 12: 43–48. 10.1016/j.tranon.2018.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaldumbide L, Erramuzpe A, Guarch R, Pulido R, Cortes JM, Lopez JI. 2016. Snail heterogeneity in clear cell renal cell carcinoma. BMC Cancer 16: 194 10.1186/s12885-016-2237-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Soori G, Dobleman TJ, Xiao GG. 2014. The application of monoclonal antibodies in cancer diagnosis. Exp Rev Mol Diagn 14: 97–106. 10.1586/14737159.2014.866039 [DOI] [PubMed] [Google Scholar]
- Zhang C, Guo Y, Li J, Tian X, Duan X. 2019. The role of the phosphatase and tensin homolog status in predicting pathological complete response to neoadjuvant anti-HER2 therapies in HER2-positive primary breast cancer: A meta-analysis. Medicine 98: e14261 10.1097/MD.0000000000014261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zheng R, Li J, Lin F, Liu L. 2017. Loss of phosphatase and tensin homolog expression correlates with clinicopathological features of non-small cell lung cancer patients and its impact on survival: A systematic review and meta-analysis. Thorac Cancer 8: 203–213. 10.1111/1759-7714.12425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Li X. 2018. Association of PTEN expression with liver function and inflammatory changes in patients with liver cancer after chemotherapy. Oncol Lett 16: 6633–6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XP, Gimm O, Hampel H, Niemann T, Walker MJ, Eng C. 2000. Epigenetic PTEN silencing in malignant melanomas without PTEN mutation. Am J Pathol 157: 1123–1128. 10.1016/S0002-9440(10)64627-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XP, Loukola A, Salovaara R, Nystrom-Lahti M, Peltomäki P, de la Chapelle A, Aaltonen LA, Eng C. 2002. PTEN mutational spectra, expression levels, and subcellular localization in microsatellite stable and unstable colorectal cancers. Am J Pathol 161: 439–447. 10.1016/S0002-9440(10)64200-9 [DOI] [PMC free article] [PubMed] [Google Scholar]