SUMMARY
Genetic screens are critical for the systematic identification of genes underlying cellular phenotypes. Pooling gene perturbations greatly improves scalability, but is not compatible with imaging of complex and dynamic cellular phenotypes. Here, we introduce a pooled approach for optical genetic screens in mammalian cells. We use targeted in situ sequencing to demultiplex a library of genetic perturbations following image-based phenotyping. We screened a set of 952 genes across millions of cells for involvement in NF-κB signaling by imaging the translocation of RelA (p65) to the nucleus. Screening at a single time point across 3 cell lines recovered 15 known pathway components, while repeating the screen with live-cell imaging revealed a role for Mediator complex subunits in regulating the duration of p65 nuclear retention. These results establish a highly multiplexed approach to image-based screens of spatially and temporally defined phenotypes with pooled libraries.
In Brief
A screening approach that combines high-content imaging with in situ sequencing can identify genes that affect spatially and temporally defined phenotypes like morphology and subcellular localization, expanding the list of scientific questions that can be asked with CRISPR-based tools.
Graphical Abstract

INTRODUCTION
Genetic perturbation screens are powerful tools for establishing causal links between genes and phenotypes in mammalian systems. Pooling genetic perturbations has greatly improved the ability to perform screens of many genetic elements but has been limited to enrichment-based phenotypic assays (Gilbert et al., 2014; Parnas et al., 2015; Shalem et al., 2014; Wang et al., 2014) or single-cell molecular profiling (Adamson et al., 2018; Datlinger et al., 2017; Dixit et al., 2016; Jaitin et al., 2016; Rubin et al., 2019). Conversely, imaging can capture a rich set of phenotypes at scale, including dynamics, but has not been compatible with pooled approaches, limiting the ability to systematically screen for genes that impact cell function.
Pooling genetic perturbations offers several advantages, making it practical to conduct comprehensive, genome-scale screens to find relevant components in a biological process. Pooled libraries can be constructed, introduced into cells, and read out as single samples, dramatically reducing the cost, effort and time needed to perform large-scale screens. Moreover, combining all perturbations into a single sample reduces batch effects and improves statistical power since all perturbed and control cells experience the same conditions. These advantages make pooled screening a preferred method in the biological community.
Historically, pooled screens have relied on enriching populations of cells for a phenotype of interest, followed by next-generation sequencing (NGS) to measure changes in perturbation abundance. Common enrichment-based phenotypes include differential cell fitness (e.g., under drug selection) (Shalem et al., 2014; Wang et al., 2014) and differential fluorescence of a marker (e.g., a genetic reporter or immunostained protein), followed by separation of a target population via fluorescence-activated cell sorting (FACS) (Parnas et al., 2015). Although highly scalable, these approaches are limited to population-level measurement, often rely on indirect reporters for the biological activity of interest, and necessarily limit phenotypes to one or a few parameters. Recently, pooled perturbations were integrated with single cell RNA-seq (Adamson et al., 2016; Datlinger et al., 2017; Dixit et al., 2016; Jaitin et al., 2016), and subsequently with chromatin accessibility profiling by ATAC-seq and protein detection by mass cytometry (Rubin et al., 2019; Wroblewska et al., 2018) to achieve high-dimensional readout for pooled screens. These molecular profiling screens capture high-dimensional representations of cell state, yielding more information about perturbation effects. However, the relationship between RNA or other molecular profiles and cellular functions is often unknown. Moreover, existing single cell molecular profiling approaches are destructive and thus cannot be used to directly monitor dynamic processes over time in individual cells.
Imaging offers an attractive alternative as it can collect many spatially and temporally resolved parameters from millions of individual cells. A multitude of optical assays have been optimized for genetic screens of protein localization, enzyme activity, metabolic state, molecular/cellular dynamics and cellular morphology in a variety of biological contexts, including mitosis (Moffat et al., 2006; Neumann et al., 2010), endocytosis (Collinet et al., 2010), viral infection (Karlas et al., 2010), differentiation (Chia et al.,2010), metabolism (Guo et al., 2008), DNA damage (Floyd et al., 2013), autophagy (Orvedahl et al., 2011) and synaptogenesis (Linhoff et al., 2009). However, these and other high-content screens required expensive and laborious testing of arrayed perturbations. Image-based screening of pooled perturbations has thus far only been demonstrated with fluorescence in situ hybridization (FISH)-based approaches that required high-magnification imaging with limited scalability for mammalian applications (Lawson et al., 2017; Wang et al., 2019).
Here, we report an optical pooled screening method that greatly expands the range of phenotypes amenable to large-scale pooled perturbation screens in mammalian cells. Our approach uses microscopy to determine first the phenotype and then the perturbation identity in each cell. We identify perturbations by targeted in situ sequencing (Ke et al., 2013) of either compact perturbations (e.g., CRISPR sgRNAs) or short associated barcodes. In situ sequencing uses enzymatic amplification to generate high signal levels, permitting imaging at low magnification, which made it feasible for us to screen millions of cells from a pooled library. We first vetted this approach by screening a synthetic reporter activated by CRISPR-induced mutations to estimate the accuracy of mapping perturbations to phenotypes in situ. Next, we performed optical pooled CRISPR loss-of-function screens across three cell lines to study p65 nuclear translocation in response to pro-inflammatory signals, identifying core NF-κB pathway members and additional factors as hits. We used optical pooled screens with time-lapse imaging to uncover a role for Mediator components as regulators of p65 nuclear retention time and subsequently found that MED12/MED24 knockouts led to sustained expression of key NF-κB target genes. The approach demonstrated here paves the way to pooled image-based screens of genome-scale libraries.
RESULTS
A single integrated genetic perturbation can be identified by in situ RNA sequencing
We developed an optical barcoding strategy to enable pooled screens of microscopy-based phenotypes (Figure 1A). In our approach, perturbations are identified via targeted in situ sequencing of an expressed barcode (Figure 1B). As pooled screens in mammalian cells generally use single-copy lentiviral integration to deliver perturbations, we focused on establishing reliable in situ sequencing of barcode transcripts in this format. To optically barcode CRISPR perturbations, we modified an existing lentiviral CRISPR guide RNA (sgRNA) expression vector (LentiGuide-Puro) to express both an sgRNA and a 12-nt barcode, and termed the resulting vector LentiGuide-BC. The barcodes and constant flanking sequences were inserted into the 3’ UTR of the Pol Il-transcribed antibiotic resistance gene, a highly expressed mRNA suitable for in situ detection.
Figure 1. Optical pooled genetic screens.
(A) In pooled screens, a library of genetic perturbations is introduced, typically at a single copy per target cell. In existing approaches, cellular phenotypes are evaluated by bulk NGS of enriched cell populations or single-cell molecular profiling (e.g. single-cell RNA-seq). In optical pooled screens, high-content imaging assays are used to extract rich spatiotemporal information from the sample prior to enzymatic amplification and in situ detection of RNA barcodes, enabling linkage between the phenotype and perturbation genotype of each cell.
(B) Targeted in situ sequencing is used to read out RNA barcodes expressed from a single genomic integration. Barcode transcripts are fixed in place, reverse transcribed, and hybridized with single-stranded DNA padlock probes, which bind to common sequences flanking the barcode. The 3’ arm of the padlock is extended and ligated, copying the barcode into a circularized ssDNA molecule, which then undergoes rolling circle amplification. The barcode sequence is then read out by multiple rounds of in situ sequencing-by-synthesis. See also Figure S1A.
To test in situ identification of perturbations, we transduced a LentiGuide-BC library containing 40 sgRNA- barcode pairs into HeLa-TetR-Cas9 cells at low multiplicity of infection (MOI < 0.01). We prepared samples for targeted in situ sequencing using a padlock-based approach (Ke et al., 2013) in which variable sequences within an RNA transcript are converted to single-stranded DNA and enzymatically amplified via in situ reverse transcription, padlock extension/ligation, and rolling circle amplification (RCA) (Figures 1B, 2A and S1A). Sequencing the amplified DNA with a 4-color sequencing-by-synthesis chemistry over 12 cycles generated high quality reads with excellent uniformity among barcodes (Figure 2B and S1B-G). After image segmentation and base calling analysis, more than 85% of sequence reads mapped exactly to one of the 40 expected barcodes out of the 412 = 16.7 million possible 12-nt sequences (Figure 2C, STAR Methods).
Figure 2. Identification of perturbation barcodes by in situ sequencing.
(A) Schematic of perturbation detection by in situ sequencing. Barcodes representing perturbations are expressed on a Pol II transcript and enzymatically converted into cDNA and amplified by RCA. RCA products serve as templates for sequencing-by-synthesis, in which barcodes are read out by multiple cycles of fluorescent nucleotide incorporation, imaging and dye cleavage.
(B) A 125-nt oligo pool encoding perturbations (sgRNAs) and associated 12-nt barcodes was cloned into a lentiviral vector (lentiGuide-BC) and delivered into HeLa cells. Expressed barcode sequences were read out by padlock detection, rolling circle amplification, and 12 cycles of sequencing-by-synthesis. A linear filter (Laplacian-of-Gaussian, kernel width σ = 1 pixel) was applied to sequencing channels to enhance spot-like features (scale bar 10 μm; composite image of DAPI and four sequencing channels). See also Movie S1.
(C) >80% of barcodes map to 40 designed sequences out of 16.7 million possible 12-nt sequences. See also Figure S1B-D.
(D) Most cells contain multiple barcode reads that map to the designed library.
(E) Cellular read distribution further categorized by read identity for 77% of cells containing at least one read.
(F) The number of possible barcodes scales geometrically with barcode length. Sufficient 12-nt barcodes can be designed to cover a genome-scale perturbation library while maintaining the ability to detect and reject single or double sequencing errors (minimum pairwise Levenshtein distance d = 2 or 3, respectively).
Perturbation detection in situ is compatible with the demands of large screens
Next, we demonstrated that an in situ sequencing approach can meet the demands of large pooled screens: (1) measurement across millions of cells quickly; (2) high efficiency and accuracy of perturbation mapping per cell; (3) detection of one or more perturbations delivered independently; and (4) compatibility with a large number of perturbations.
We showed that the in situ readout step can process millions of cells within a few days to provide the high coverage (typically 100–1,000 cells/perturbation) needed for high-throughput optical screening. High signal intensity allows accurate sequence data to be obtained at low optical magnification across large fields of view, each containing thousands of cells (Figure 2B, Movie S1, Table S1). We maximized fluorescence signal-to-background by optimizing the barcode amplification protocol, including the post-fixation step that follows reverse transcription and the conditions for padlock extension/ligation (Figure S2A). Using the optimized protocol, in situ sequencing spots were readily visible at 10X optical magnification, with at least one exactly mapped read detected in more than 77% of transduced cells (Figures 2B, 2D and 2E).
Next, we constructed two types of large barcoded perturbation libraries using oligo microarray synthesis. In the first approach, we designed a set of 83,314 12-nt barcodes using criteria that balanced GC content, minimized homopolymer repeats, and maintained a minimum edit distance between barcodes of three (STAR Methods). This allows rejection of reads containing up to two single base insertion/deletion/substitution errors arising from oligo synthesis or in situ processing (Figure 2F, STAR Methods) (Buschmann and Bystrykh, 2013). We then used a two-step procedure to clone a library of sgRNAs and associated barcodes into LentiGuide-BC (STAR Methods). In contrast to perturbation barcoding by random pairing (Wang et al., 2019), this approach pairs sgRNAs with specific barcodes in silico, ensuring efficient use of available barcodes. We addressed a known problem whereby barcode-sgRNA associations are swapped due to reverse transcription-mediated recombination during lentiviral infection (Adamson et al., 2018; Hill et al., 2018; Sack et al., 2016; Xie et al., 2018) by using a modified lentiviral packaging protocol (Feldman et al., 2018) that reduced the frequency of cells exhibiting swapped barcodes from >28% to <5%.
In the second approach, we used the CROP-seq vector to directly sequence a Pol II-transcribed copy of the sgRNA, rather than relying on auxiliary barcodes (Datlinger et al., 2017). We observed accurate sequencing of sgRNAs in HeLa-TetR-Cas9 cells, but with reduced fluorescent signal intensity compared to LentiGuide-BC. We then systematically tested a set of 84 CROP-seq-targeting padlocks with a range of padlock binding arm sequences, padlock backbone lengths and backbone sequences (STAR Methods, Figure S2B, Table S2) to yield a top-performing CROP-seq padlock with a 4-fold increase in reads per cell and 1.7-fold increase in signal intensity relative to the median padlock.
Moreover, we showed that in situ sequencing could be used to read out multiple perturbations within the same cell. To this end, we transduced HeLa-TetR-Cas9 cells with one CROP-seq library carrying a puromycin selection marker, followed by a second CROP-seq library carrying a zeocin selection marker. After serial transduction and antibiotic selection, we performed in situ sequencing on both libraries simultaneously. Most cells (81%) contained reads mapping to two sgRNAs (Figure S2C,D). The ability to independently sequence perturbations delivered by separate vectors suggests a straightforward and general route to higher-order combinatorial screens.
Accurate mapping of phenotype to genotype in an optical pooled screen
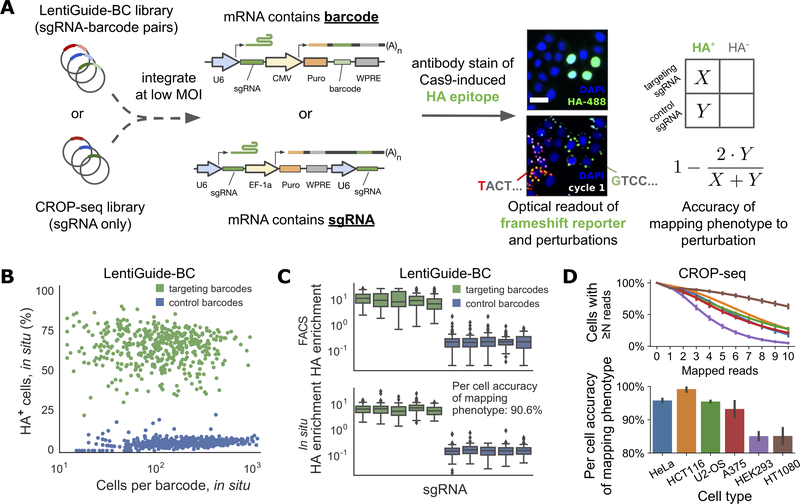
We next showed that our approach can correctly map genetic perturbations to cell phenotypes in situ by performing a reporter imaging screen, in which a lentiviral reporter produces an HA-tagged, nuclear- localized H2B protein after a Cas9-induced +1 frameshift in a target region (Figures 3A and S3A, STAR Methods). Cells expressing the reporter can either be screened in situ or by FACS. The reporter is highly specific and sensitive, with a mean in situ activation across 5 targeting sgRNAs of 65 ± 2.7% and background of <0.001 % in the absence of a targeting sgRNA (Figures 3B and S3B). We transduced cells stably expressing the frameshift reporter with a LentiGuide-BC library containing 972 barcodes redundantly encoding 5 targeting and 5 control sgRNAs (average of 97 barcodes per sgRNA) (Table S3). We then induced Cas9 expression, measured reporter activation by immunofluorescence, and determined barcode sequences by in situ sequencing.
Figure 3. Accuracy of phenotype-to-genotype mapping assessed with a fluorescent reporter.
(A) Workflow for CRISPR-Cas9 knockout-based screening of a genetically-encoded frameshift reporter. A library of targeting and non-targeting guides was cloned into either LentiGuide-BC or the CROP-seq vector and transduced into cells at low MOI. Cas9 expression generates indels at the frameshift reporter target locus in cells with a targeting guide and leads to expression of a nuclear-localized HA epitope. HA expression was assayed by immunofluorescence and correlated with sgRNAs detected by in situ sequencing. Frameshift reporter accuracy was estimated using the relative abundances of HA+ cells mapped to targeting and non-targeting guides (X and Y, respectively). Scale bar is 30 μm.
(B) Targeting and control barcodes expressed from LentiGuide-BC in HeLa-TetR-Cas9 cells were well separated by fraction of HA+ cells.
(C) The same cell library was screened by flow sorting cells into HA+ and HA- bins and performing next-generation sequencing of the genomically integrated barcode. See also Figure S3D.
(D) The experiment was repeated across a panel of cell lines using the CROP-seq library and an optimized padlock detection protocol, yielding a similar distribution of mapped reads (top) and frameshift reporter accuracies (bottom). Error bars indicate the range between two replicate sequencing experiments. Cell types are indicated by the same colors in both plots.
All barcodes encoding targeting sgRNAs were distinguishable from control sgRNAs by HA+ fraction, with a per-cell identification accuracy of 90.6% (based on false positive events in which HA+ cells were assigned control sgRNAs, STAR Methods). Indeed, there were no errors in barcode-to-phenotype assignment even for barcodes represented by very few cells (Figures 3B and S3C, STAR Methods). Screening the same cell library by FACS with NGS readout showed similar enrichment of targeting sgRNAs (Figures 3C and S3D), with consistent representation of most barcodes in both contexts (95% within 5-fold abundance, Figure S1). We achieved comparably robust mapping of CRISPR sgRNAs to the frameshift reporter phenotype with the CROP-seq vector in HeLa cells as well as U2-OS, HCT116, A375, HT1080 and HEK293 cells (Figure 3D), with per-cell identification accuracy ranging from 83–98%. Thus, the combined errors that may arise from oligonucleotide synthesis, library cloning, lentiviral delivery, barcode diffusion during in situ processing, barcode readout by in situ sequencing, or incorrect assignment of reads to cells during image processing are infrequent enough to permit pooled functional screens.
An optical pooled screen in millions of cells for regulators of NF-κB activation
After demonstrating the ability to screen genetic knockouts for optical phenotypes, we sought to identify genes required for activation of NF-κB, a family of transcription factors (p50, p52, p65, RelB and c-Rel) that translocate to the nucleus in response to a host of stimuli (Gewurz et al., 2012; Hayden and Ghosh, 2012; Pahl, 1999). At baseline, NF-κB dimers are maintained in an inactive state in the cytoplasm by inhibitory IκB proteins, which mask nuclear localization signals to prevent NF-κB translocation into the nucleus. Upon stimulation of cell surface receptors, a signal cascade is initiated, leading to downstream activation of the IKK complex. IKK-β then phosphorylates IκB proteins, triggering their phosphorylation- dependent ubiquitination and subsequent degradation by the proteasome, releasing NF-κB from inhibition. Free NF-κB dimers may then translocate to the nucleus and induce the expression of genes promoting cell proliferation, survival, and pro-inflammatory responses. It is known that post-translational modifications play a key role in regulating the baseline state and activation of NF-κB. In addition to its role in proteolytic degradation, ubiquitin has been shown to function in IKK activation and the activity of many core NF-κB pathway members is controlled by alterations in ubiquitination state.
We used an established nuclear translocation assay to measure the localization of a p65-mNeonGreen reporter in HeLa cells following stimulation with either IL-1β or TNFα, cytokines that activate NF-κB via different pathways (Figure 4A). We screened 3,063 sgRNAs targeting 963 genes using the LentiGuide- BC design. We included all GO-annotated ubiquitin ligase and deubiquitinase enzymes, as well as 425 immune-related genes in the library, hypothesizing that ubiquitin signaling may play as-yet unrecognized roles in NF-κB activation and relaxation (return to the baseline cellular distribution) (Chen, 2005) (Figure 4A, Table S3). After stimulation with either IL-1β or TNFα, we imaged p65-mNeonGreen translocation, and then performed in situ sequencing to identify sgRNAs. A total of 3,037,909 cells were retained for analysis after filtering cells based on reporter expression, nuclear morphology, and exact barcode mapping (952 out of 963 genes retained for analysis) (Figure S3E). We scored the degree of p65-mNeonGreen translocation in each cell by cross-correlation with a DAPI nuclear stain. We ranked the perturbations by the difference of their translocation score distribution from negative control sgRNAs to identify gene knockouts that led to defects in response to IL-1β and/or TNFα (Figures 4B, 4C and S4A, Tables S1 and S3, STAR Methods).
Figure 4. A screen for regulators of NF-κB signaling.
(A) Workflow for CRISPR-Cas9 knockout-based screening using a fluorescently tagged reporter cell line. Screen hits were identified by the failure of p65-mNeonGreen to translocate to the nucleus following stimulation with IL-1β or TNFα cytokines.
(B) Known NF-κB regulators were identified as high-ranking screen hits. Cells were assigned translocation scores based on the pixelwise correlation between mNeonGreen fluorescence and a DAPI nuclear stain; thus a score 1 indicates maximum translocation while a score of −1 indicates maximum cytoplasmic localization. The translocation defect for a gene was defined based on the integrated difference in the distribution of translocation scores relative to non-targeting control sgRNAs across three replicate screens. See also Tables S1 and S3.
(C) Cumulative distributions of translocation scores (second-ranked guide) of known NF-κB regulators in response to both cytokines. The shaded areas depict the difference between the translocation score distributions for targeting sgRNAs and non-targeting control sgRNAs (gray).
(D) NF-κB pathway map (KEGG HSA04064) color-coded as in (B). KEGG pathway members colored gray did not show a translocation defect when individually knocked out in HeLa cells.
(E) Top-ranked genes were validated with arrayed CRISPR-Cas9 knockouts (scale bar 10 μm). Histograms show the cumulative distributions of IL-1β and TNFα-induced translocation scores (averaged over two guides) for each gene knockout compared to wild type cells (gray). See also Figure S4B.
The screen recovered most known NF-κB regulators and uncovered novel candidates
The hits in our screen included known pathway components annotated by KEGG (Kanehisa and Goto, 2000) for NF-κB activation by IL-1β signaling (5/5 genes), TNFα signaling (4/7 genes) and downstream components (5/7 genes), including cytokine-specific receptors, adaptor proteins, and factors that activate the shared regulator MAP3K7 (Figure 4D) (Gewurz et al., 2012). Hits common to both cytokine stimulations included MAP3K7 and its target, the IKK complex (CHUK, IKBKB, IKBKG), as well as components of the SKP1-CUL1-F-box ubiquitin ligase complex and proteasome subunits, which together promote degradation of the inhibitor NFKBIA/IκBα and nuclear translocation of p65.
We confirmed the results of the pooled screen by individual CRISPR knockouts, with 19 out of 20 top-ranked hits validated (z-score threshold 3.75, Figure 4E and Table S3). Phenotype strength was well correlated between the primary screening and validation ranks (Spearman’s ρ = 0.84 and 0.73 for IL-1β and TNFα, respectively), emphasizing the quantitative nature of the primary screen (Figure S4B). The p65-mNeonGreen screen showed high sensitivity, detecting genes known to be involved in NF-κB activation that show only modest translocation defects in our model system. This set includes genes such as RBCK1/HOIL1 (LUBAC complex, poly-ubiquitination of IKBKG/NEMO and RIPK1), DCUN1D1 (neddylation of CUL1) and COPS5 (COP9 signalosome, regulates neddylation).
Interestingly, the set of screening hits validated by arrayed knockout using multiple individual sgRNAs included known negative regulators of NF-κB activation, such as NFKBIA/κBα and CYLD. Knockout of these negative regulators may lead to permanent baseline activation of NF-κB signaling, which in turn causes negative feedback activation (Jäättelä et al., 1996), potentially rendering perturbed cells refractory to induced NF-κB activation. The screen also identified several candidate regulators, including BAP1, HCFC1, and KCTD5. Among these, BAP1 has been previously described to deubiquitinate HCFC1 (Machida et al., 2009), with relevance for controlling metabolism (Bononi et al., 2017), ER-stress signaling (Dai et al., 2017), cell-cycle progression (Misaghi et al., 2009), and viral gene expression (Johnson et al., 1999).
In order to investigate cell line-dependent wiring of the NF-κB signaling pathway, we repeated the same screen with an antibody against endogenous p65 in HeLa, A549 and HCT116 cells. The cytokine concentration for each cell line was determined by imaging p65 translocation in wild type cells 40 minutes after stimulation and using the lowest concentration that saturated translocation (STAR Methods). This titration was found to improve screen sensitivity, but the remainder of the screening protocol was identical across cell lines. The antibody screens detected many of the KEGG-annotated regulators found in the HeLa p65-mNeonGreen screen (17/19 in HeLa, 18/19 in A549 and 11/19 in HCT116, Table S3). Notably, some annotated regulators were not detected as hits in any of the three cell lines. Among these, TRAF5 and BIRC3 have functionally redundant counterparts (TRAF2 and BIRC2) (Mahoney et al., 2008; Tada et al., 2001). Indeed, TRAF2 scored as a positive regulator in all three cell lines, while BIRC2 was only detected in HeLa, suggesting its role may vary among the different cell lines. Meanwhile, knockdown of TAB1 was previously shown not to impact IKK activation (Chen, 2005; Wang et al., 2001), consistent with our finding that TAB1 was not detected as a hit in any cell line. Comparing high-scoring genes across cell lines revealed that the E2 ubiquitin-conjugating enzyme UBE2N was strongly involved in IL-1β-dependent p65 activation in A549 cells, while being largely dispensable in the other cell lines tested (Figure S4C). Similarly, BIRC2 was detected only in HeLa cells to be involved in TNFα-dependent p65 translocation. The robustness and scalability of the optical pooled screening protocol makes it possible to conduct in-depth analyses of biological pathways across cellular backgrounds.
High-content analysis of morphology distinguishes regulators by function
In addition to the translocation defect measured in knockouts of NF-κB positive regulators, we observed that certain classes of genes led to similar morphological changes in cells. For example, proteasomal knockouts induced a distinct morphology consisting of rounded cells (low eccentricity) with enlarged nuclei. We hypothesized that loss of genes with related functions might induce similar morphological changes, providing additional dimensions to interpret our screening results, and thus re-scored each cell in the primary HeLa p65-mNeonGreen screen for cell and nuclear morphology and nuclear stain intensity (STAR Methods). Members of the CUL1-RBX1-SKP1-FBXW11 complex and its substrate NEDD8 showed increased cell and nuclear area, with 4 out of 5 genes grouped together by PCA (Figures S4D-H). UFD1, a member of the VCP/p97-UFD1-NPL4 complex that mediates post-ubiquitination degradation of IκBα, shared a similar morphological profile to COPS5, which interacts directly with VCP/p97 as part of the COP9 signalosome (Cayli et al., 2009; Li et al., 2014). Disruption of the chromatin remodeler INO80, as well as NOP53/GLTSCR2, and UBA52, genes with roles in ribosome biogenesis, showed a decrease in cell and nuclear area. The ability to group classify knockouts into functional categories based on morphological changes is a key benefit of image-based screening, which could be further enhanced by staining additional cellular markers to extract more information from each cell (Gustafsdottir et al., 2013).
A live-cell imaging screen identifies MED12 and MED24 as regulators of NF-κB translocation kinetics
Activation of NF-κB involves a cascade of signaling events and feedback loops whose kinetics determine the dynamic response of the pathway. As optical pooled screens can be readily combined with live-cell imaging, we screened a CROP-seq library of sgRNAs targeting the same 952 genes (5,638 sgRNAs) detected in the initial activation screen for variability in the timing of p65 activation and relaxation (434,505 cells analyzed, Table S3). Cells were stimulated with IL-1β or TNFα and imaged at 23 minute intervals for 6 hours post-stimulation. Nuclei were tracked using a cell-permeable DNA stain and p65-mNeonGreen nuclear translocation was assessed at each time point. Following live-cell analysis, cells were fixed and the perturbation in each cell was read out by in situ sequencing of the sgRNA sequences. The live-cell screen closely reproduced hits from the fixed-cell activation screen described above, with 15 out of the 20 top positive regulators shared when the live cell analysis was restricted to a single matched time point (STAR Methods).
Hierarchical clustering of mean translocation time profiles revealed distinct populations of positive and negative regulators (Figure 5A). To quantify changes in translocation kinetics, we defined a cumulative defect metric for each gene. First, we integrated over time the difference between each cell’s translocation score and the mean translocation score of non-targeting controls. Then, for each gene, the distribution of cell-level cumulative defects was tested for statistically significant deviations from the non-targeting control population (STAR Methods, Table S4). By integrating differences in translocation over time, we were able to identify known key negative regulators of the pathway with effects on p65 relaxation that did not score in the initial static screen, including TNFAIP3 (an NF-κB target gene that provides negative feedback by deubiquitinating multiple upstream signaling components) (Chen, 2005), KEAP1 (a ubiquitin ligase involved in degradation of IKKβ) (Lee et al., 2009) and USP7 (a deubiquitinase that slows ubiquitination and proteasomal degradation of NF-κB) (Colleran et al., 2013).
Figure 5. A live-cell screen identifies a role for Mediator components in regulating p65 kinetics.
(A) The initial 952 gene translocation screen was repeated using live-cell imaging to monitor p65 translocation kinetics (n = 361,587 analyzed cells). Hierarchical clustering of the time-dependent translocation difference between each gene and the non-targeting controls grouped together KEGG-annotated regulators, as well as other positive and negative regulators with distinct cytokine-specific kinetic signatures. See also Table S4.
(B) and (C) A validation live-cell screen was performed to improve sampling of regulators identified by the primary screen and helped group regulators with distinct p65 translocation kinetics. Individual traces represent different sgRNAs for each gene. See also Figure S5 and Table S4.
(D) Clonal knockouts of MED12 and MED24 recapitulated the increased retention time phenotype seen in the primary screen. Each trace represents a different knockout clone (scale bar 10 μm).
(E) Cytokine stimulation of Mediator clonal knockouts led to differential activation of NF-κB target genes, including increased expression of chemokines after stimulation with TNFα (1 ng/mL) or IL-1β (30 ng/mL). Error bars show the range among knockout clones or wild type biological replicates, clones same as in (D).
(F) IL1B expression was induced in the Mediator clonal knockouts, but not in wild type cells, by TNFα stimulation.
Next, we performed a pooled validation screen for 61 genes that showed evidence for a kinetic phenotype (10 sgRNAs/gene). We optimized conditions for live-cell imaging and cell tracking by using faster microscope hardware, allowing us to link time-resolved p65 translocation data to perturbation barcodes from 2,595,514 cells (median of 3,611 cells/sgRNA) at 30 minute intervals. At this high level of sampling, individual sgRNAs targeting a given gene showed excellent concordance (Figure 5B and Figure S5). The pooled validation screen confirmed several genes that were initially detected as lower-ranked hits in the activation screen, while adding additional kinetic details. For example, at later time points, RIPK1 showed an increased translocation defect while KCTD5 and HCFC1 were seen to negatively regulate p65.
The validation screen also confirmed MED12 and MED24 as previously unknown negative regulators of p65 nuclear translocation (Figures 5B, 5C and S5). Clonal homozygous knockouts of each of these Mediator complex subunits confirmed delayed relaxation relative to wild type cells (Figure 5D). Of note, a mono-allelic MED24 knockout clone showed a much weaker phenotype, underscoring the high sensitivity of the primary optical screen despite the heterogeneity of alleles generated by Cas9 cleavage (Figure 5D). We performed RNA sequencing of clonal knockout lines after stimulation with either TNFα or IL-1β. Among highly induced genes (>50-fold increase in wild type cells after 4 hours of stimulation with either cytokine, adjusted p-value < 10−3), we identified 6 genes (CCL2, CCL20, CXCL1, CXCL2, CXLC3, CXCL8) that were differentially expressed in either MED12 or MED24 clonal knockout cells (>8-fold change versus wild type cells at 4 hours post-stimulation with either cytokine, adjusted p-value < 10−3) (Figure 5E, STAR Methods), all of which encode for chemokines that attract immune cells to sites of inflammation. Even more strikingly, pro-IL-1β, which is a well-established NF-κB target gene in the context of Toll-like receptor activation (Sims and Smith, 2010), was transcriptionally induced in MED12 or MED24 deficient HeLa cells but not in wild type cells upon TNFα stimulation (Figure 5F). These results suggest that, apart from the well-studied function of the Mediator complex in recruiting active RNA polymerase-II to DNA-bound transcription factors (Malik and Roeder, 2005) including to p65 (Guermah et al., 1998), Mediator components may be involved in restricting the transcriptional response of induced genes during pro-inflammatory NF-κB signaling.
DISCUSSION
Optical pooled screens enable systematic analysis of the genetic components underpinning a wide range of spatially and temporally defined phenotypes, including subcellular localization, live-cell dynamics, and high-content morphological profiling. The in situ sequencing framework is compatible with any perturbation that can be identified by a short expressed sequence and readily scales to millions of cells and genome-scale libraries.
We applied pooled CRISPR loss-of-function screening targeting 952 genes in multiple cell lines to identify regulators of p65 translocation, recovering nearly all annotated regulators of TNFα- and IL-1β -induced NF-κB activation. Moreover, by performing a pooled live-cell imaging screen, we discovered two components of the Mediator complex as previously unknown negative regulators of pro-inflammatory signaling that we validated in clonal knockout cell lines. Although the Mediator complex has been implicated in p65-mediated gene transcription (Essen et al., 2009), Mediator components have not previously been shown to be involved in negative regulation of either the nuclear translocation or the transcriptional response of NF-κB. As one of the most potent pyrogens in the human body, IL-1β is secreted only upon transcriptional (NF-κB) as well as post-transcriptional (Caspase 1) licensing. Our results indicated that Mediator components play a role in suppressing pro-IL-1β induction by TNFα signaling, which might provide a mechanism for preventing aberrant feedback activation of immune signaling pathways (Bauernfeind et al., 2016). The ability to screen dynamic responses, such as p65 translocation kinetics, adds a rich temporal dimension to the profiling of gene knockouts, permitting direct measurement of response onset and duration in addition to strength, directionality (positive/negative) and cytokine specificity.
The use of in situ sequencing to read out perturbations has several advantages over alternative approaches. Amplification by RCA enables fast in situ sequencing at low magnification (10X), greatly increasing throughput. The 12-nt sequences used here can robustly distinguish >80,000 perturbations, sufficient to encode genome-scale perturbation libraries. Existing CRISPR sgRNA libraries can be read out directly (using the CROP-seq vector) while other perturbations (e.g., ORFs, non-coding sequences) can be paired with short barcodes (Melnikov et al., 2012; Yang et al., 2011). By comparison, reported methods for highly multiplexed FISH require higher imaging magnification (60X) and barcodes longer than 200 bp, precluding cost-effective direct oligo array synthesis. Epitope-based protein barcodes are a promising method for enzyme-free decoding of pooled elements, but are currently limited in scale to ~100 barcodes (Wroblewska et al., 2018).
Despite the rich phenotypic information provided by imaging assays, optical phenotyping has been underused for screening applications due to the substantial cost, time and labor required to execute large-scale arrayed perturbation screens. In recent years, there has been wide use of pooled genome-scale CRISPR screens based on enriching cells via fitness or reporter fluorescence in various model systems, but only one large-scale (2,281 sgRNA) CRISPR imaging screen (Groot et al., 2018). Pooling improves data quality and dramatically reduces the cost and labor required, making genome-scale screens with image-based phenotyping accessible to many laboratories.
Optical pooled screening serves as an important complement to single-cell molecular profiling, which also provides high-content data but is not yet able to deliver dynamic or spatially resolved information. The LentiGuide-BC and CROP-seq libraries described in this study both generate mRNA encoding perturbation identity, so the same libraries of perturbed cells can be screened in situ to read dynamic and/or spatially resolved information and by complementary molecular profiling methods, such as single cell RNA-seq. This approach could assist in establishing functional relationships between molecular profiles and many cellular phenotypes (e.g., morphology, motility, electrical depolarization, cell-to-cell interactions).
Our approach is broadly applicable across many settings. We demonstrate the identification of multiple perturbations within the same cell, providing a straightforward route to study higher order genetic interactions. The potential to integrate optical screening with high-dimensional morphological profiling (Gustafsdottir et al., 2013) and in situ multiplexed gene expression analysis (Chen et al., 2015; Ke et al., 2013; Lee et al., 2014; Lubeck et al., 2014; Wang et al., 2014) raises the prospect of learning phenotypes from high-content data rather than pre-specifying phenotypes of interest. Libraries of endogenous or engineered protein variants could be screened for effects on cell structure or other optically-defined phenotypes. Optical pooled screening in a 2D or 3D co-culture system could be used to analyze non-cell autonomous phenotypes based on physical contact (e.g., formation of adhesion complexes, direct contact signaling, neurotransmission). Existing protocols for in situ sequencing in tissue samples (Ke et al., 2013; Wang et al., 2018) highlight the exciting possibility of perturbing cells in vivo and measuring the resulting phenotypes within the native spatial context.
STAR ★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Paul Blainey (pblainey@broadinstitute.org)
Plasmids generated in this study have been deposited to Addgene (additional details provided in the Key Resources Table).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-HA antibody | Cell Signaling Technologies | 3724 |
| rabbit anti-p65 antibody | Cell Signaling Technologies | 8242 |
| goat anti-rabbit IgG Alexa Fluor 488 | Cell Signaling Technologies | 4412 |
| Bacterial and Virus Strains | ||
| Endura Electrocompetent Cells | Lucigen | 69242 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| TNFα | Invivogen | rcyc-htnfa |
| IL-1β | Invivogen | rcyec-hil1b |
| Critical Commercial Assays | ||
| Revertaid H minus RT | Thermo Fisher Scientific | EP0452 |
| Ribolock RNase inhibitor | Thermo Fisher Scientific | EO0384 |
| RNase H | Enzymatics / Qiagen Beverly | Y9220L |
| TaqlT DNA polymerase | Enzymatics / Qiagen Beverly | P7620L |
| Stoffel Fragment | Blirt SA / DNA Gdansk | RP810 |
| Ampligase | Lucigen | A3210K |
| Phi29 DNA polymerase | Thermo Fisher Scientific | EP0092 |
| MiSeq 500 cycle Nano kit | Illumina | MS-103-1003 |
| Deposited Data | ||
| RNA-seq of clonal knockouts | Gene Expression Omnibus | GSE132704 |
| Screening image data | Image Data Resource | Accession number pending |
| Experimental Models: Cell Lines | ||
| HeLa-TetR-Cas9 | Iain Cheeseman Lab, Whitehead Institute | N/A |
| Oligonucleotides | ||
| oRT_LentiGuide-BC (for use with LentiGuide-BC): G+AC+GT+GT+GC+TT+AC+CCAAAGG | This paper | N/A |
| oPD_LentiGuide-BC (for use with LentiGuide-BC): /5Phos/actggctattcattcgcCTCCTGTTCGA CAGTCAGCCGCATCTGCGTCTA1 1 IAGTGGAGCCC TT Gtgttcaatcaacattcc | This paper | N/A |
| oSBS_LentiGuide-BC (for use with oPD_LentiGuide- BC): TTCGACAGTCAGCCGCATCTGCGTCTATTTAGTGG AGCCCTT Gtgttcaatcaacattcc | This paper | N/A |
| oRT_CROPseq (for use with CROPseq-puro and CROPseq-zeo): G+AC+TA+GC+CT+TA+TT+TTAACTTGCTAT | This paper | N/A |
| oPD_CROPseq (for use with CROPseq-puro and CROPseq-zeo, optimized padlock): /5Phos/gttttagagctagaaatagcaagCTCCTGTTCGACACC TACCCACCTCATCCCACTCTTCAaaaggacgaaacaccg | This paper | N/A |
| oSBS_CROPseq (for use with oPD_CROPseq): CACCTCATCCCACTCTTCAaaaggacgaaacaccg | This paper | N/A |
| oRT_CROPseq-v2 (for use with CROPseq-puro-v2): A+CT+CG+GT+GC+CA+CT+TTT | This paper | N/A |
| oPD_CROPseq-v2 (for use with CROPseq-puro-v2, optimized padlock):/5Phos/GTTTCAGAGCTATGCTGGCTCCTGT TCGCTTCTCCCTTACCTCCTTCCCTTCCATCCTATAT CCTCCACTCATAaaaggacgaaaCACCg | This paper | N/A |
| oSBS_CROPseq-v2 (for use with oPD_CROPseq-v2): TCCATCCTATATCCTCCACTCATAaaaggacgaaaCACCg | This paper | N/A |
| oPD_323 (previously used with CROPseq-puro; not optimized): /5Phos/gttttagagctagaaatagcCTCCTGTTCGACAGTCA GCCGCATCTGCGTCTATTTAGTGGAGCCCTTGaagga cgaaacaccg | This paper | N/A |
| oSBS_394 (previously used with oPD_323; not optimized): TCAGCCGCATCTGCGTCTATTTAGTGGAGCCCTTGa aggacgaaacaccg | This paper | N/A |
| Recombinant DNA | ||
| LentiCas9-blast | Sanjana et al, 2014 | Addgene Plasmid #52962 |
| LentiGuide-Puro | Sanjana et al, 2014 | Addgene Plasmid #52963 |
| pXPR_011 | Doench et al, 2014 | Addgene Plasmid #59702 |
| CROPseq-Guide-Puro | Datlinger et al., 2017 | Addgene Plasmid #86708 |
| pR_LG | Feldman et al., 2018 | Addgene Plasmid #112895 |
| LentiGuide-BC | This paper | Addgene Plasmid #127168 |
| LentiGuide-BC-CMV-Puro | This paper | Addgene Plasmid #127169 |
| LentiGuide-BC-CMV-Puro | This paper | Addgene Plasmid #127170 |
| pL_FR_Hygro | This paper | Addgene Plasmid #127171 |
| pR14_p65-mNeonGreen | This paper | Addgene Plasmid #127172 |
| CROPseq-Zeo | This paper | Addgene Plasmid #127173 |
| CROPseq-Puro-v2 | This paper | N/A |
| Software and Algorithms | ||
| Snakemake | Köster et al., 2012 | https://snakemake.readthedocs.io/en/stable/ |
| Sample in situ sequencing data and analysis pipeline from data to sequencing read table | This paper | https://github.com/blaineylab/OpticalPooledScreens |
| Other | ||
| Microscope light source | Lumencor | Sola SE365 FISH |
| Camera | Hamamatsu | ORCA-Flash 4.0 v3 |
| Objective Lens | Nikon | CFI Plan Apochromat Lambda 10X/0.45 |
| DAPI filter set | Semrock | LED-DAPI-A-NTE-ZERO |
| GFP filter set | Semrock | GFP-1828A-NTE-ZERO |
| Cy3 filter set (MiSeq G) | Semrock | FF01-534/20-25 FF552-Di02-25×36 FF01-572/28-25 |
| Alexa Fluor 594 filter set (MiSeq T) | Semrock | FF03-575/25-25 FF596-Di01-25×36 FF01-615/24-25 |
| Cy5 filter set (MiSeq A) | Semrock | FF01-635/18-25 FF652-Di01-25×36 FF01-680/42-25 |
| Cy7 filter set (MiSeq C) | Semrock | FF01-661/20-25 FF695-DI01-25X36 FF01-732/68-25 |
| Sample in situ sequencing data and analysis pipeline from data to sequencing read table | This paper | https://github.com/blaineylab/OpticalPooledScreens |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Tissue culture
HEK293FT cells (Thermo Fisher Scientific R70007) were cultured in DMEM with sodium pyruvate and GlutaMAX (Life 10569044) supplemented with heat-inactivated fetal bovine serum (Seradigm 97068–085) and 100 U/mL penicillin-streptomycin (Thermo Fisher Scientific 15140163). HeLa cells were cultured in the same media with serum substituted for 10% tetracycline-screened fetal bovine serum (Hyclone SH30070.03T). All other cell lines (A549, HCT116, HT1080, A375) were cultured in the same media with 10% heat-inactivated fetal bovine serum (Sigma F4135).
In preparation for in situ analysis, cells were seeded onto glass-bottom plates (6-well: Cellvis P06–1.5H-N, 24-well: Greiner Bio-one 662892, 96-well: Greiner Bio-one 655892) at a density of 50,000 cells/cm2 and incubated for 2 days to permit proper cell attachment, spreading, and colony formation.
Selection of inducible HeLa Cas9 clone
Parental HeLa-TetR-Cas9 cells were a gift from Iain Cheeseman. In order to select a clone with optimal induction and Cas9 activity for further experiments, single cells were sorted into a 96-well plate (Sony SH800), clonally expanded, and screened for Cas9 activity after 8 days with and without 1 μg/mL doxycycline induction. Cas9 activity was assessed by transducing each clone with pXPR_011 (Addgene #59702), a reporter vector expressing GFP and an sgRNA targeting GFP (Doench et al., 2014), and using FACS to read out efficiency of protein knockdown. Additionally, gene editing was directly assessed by transducing HeLa-TetR-Cas9 clones with a guide targeting TFRC (sgRNA:CTATACGCCACATAACCCCC). Genomic DNA was extracted from both uninduced and induced clones by resuspending in cell lysis buffer (10 mM Tris pH 7.5, 1 mM CaCl2, 3 mM MgCl2, 1 mM EDTA, 1% Triton X-100, and 0.2 mg/mL Proteinase K), and heating for 10 minutes at 65º0 and 15 minutes at 95º0. The guide target region was amplified by PCR (P5 primer: CTCTTTCCCTACACGACGCTCTTCCGATCTATGACCTTAGGCTTATTTTAACTTAATC, P7 primer: CTGGAGTTCAGACGTGTGCTCTTCCGATCTGAGTCTCATGCACTGTTTTGC) and sequenced on an Illumina MiniSeq. The best clones showed efficient indel generation (≥ 97%) in the presence of doxycycline and minimal cutting (≤ 2%) in its absence.
METHOD DETAILS
Lentivirus production
HEK293FT cells were seeded into 15-cm plates or multi-well plates at a density of 100,000 cells/cm2. After 20 hours, cells were transfected with pMD2.G (Addgene #12259), psPAX2 (Addgene #12260), and a lentiviral transfer plasmid (2:3:4 ratio by mass) using Lipofectamine 3000 (Thermo Fisher Scientific L3000015). Media was exchanged after 4 hours and supplemented with 2 mM caffeine 20 hours post-transfection to increase viral titer. Viral supernatant was harvested 48 hours after transfection and filtered through 0.45 μm cellulose acetate filters (Corning 431220).
Lentiviral transduction
Cells were transduced by adding viral supernatant supplemented with polybrene (8 μg/mL) and centrifuging at 1000g for 2 hours at 33º0. At 5 hou rs post-infection, media was exchanged. At 24 hours post-infection, cells were passaged into media containing selection antibiotic at the following concentrations: 1 μg/mL puromycin (Thermo Fisher Scientific A1113802), 300 μg/mL hygromycin (Invivogen ant-hg-1), 30 μg/mL blasticidin (Thermo Fisher Scientific A1113903), and 300 μg/mL zeocin (Thermo Fisher Scientific R25001).
For lentiviral transduction of LentiGuide-BC libraries, a carrier plasmid was utilized to minimize recombination between distant genetic elements (e.g., sgRNA and associated barcode). Libraries were packaged following the above protocol, with the library transfer plasmid diluted in integration-deficient carrier vector pR_LG (1:10 mass ratio of library to carrier, Addgene #112895) (Feldman et al., 2018).
Library cell line validation
For library transductions, multiplicity of infection (Table S1) was estimated by counting colonies after sparse plating and antibiotic selection. Genomic DNA was also extracted for NGS validation of library representation.
Next generation sequencing of libraries
Genomic DNA was extracted using an extraction mix as described above. Barcodes and sgRNAs were amplified by PCR from a minimum of 106 genomic equivalents per library using NEBNext 2X Master Mix (initial denaturation for 5 minutes at 98ºC, follow ed by 28 cycles of annealing for 10 seconds at 65ºC , extension for 25 seconds at 72ºC, and denaturation for 20 seconds at 98ºC).
Library design and cloning
A set of 12-nt barcodes was designed by selecting 83,314 barcodes from the set of 16.7 million possible 12-nt sequences by filtering for GC content between 25% and 75%, no more than 4 consecutive repeated bases, and minimum substitution and insertion/deletion edit distance (Levenshtein distance) of 3 between any pair of barcodes. Ensuring a minimum edit distance is useful for detecting and correcting errors, which arise mainly from DNA synthesis and in situ reads with low signal-to-background ratios. The E-CRISP web tool was used to select sgRNA sequences targeting genes of interest (Heigwer et al., 2014). Barcode-sgRNA pairs were randomly assigned and co-synthesized on a 125-nt 90K oligo array (CustomArray/Genscript) (Table S3). Individual libraries were amplified from the oligo pool by dial-out PCR (Schwartz et al., 2012) and cloned into LentiGuide-BC or LentiGuide-BC-CMV (the latter contains the CMV promoter instead of the EF1a promoter) via two steps of Golden Gate assembly using BsmBI restriction sites. Then, the sgRNA scaffold sequence and desired resistance cassette were inserted using BbsI restriction sites. Libraries were transformed in electrocompetent cells (Lucigen Endura) and grown in liquid culture for 18 hours at 30ºC before extracti ng plasmid DNA. The sgRNA-barcode association was validated by Sanger sequencing individual colonies from the final library.
Padlock-based RNA detection
Preparation of targeted RNA amplicons for in situ sequencing was adapted from published protocols with modifications to improve molecular detection efficiency and amplification yield (Ke et al., 2013; Larsson et al., 2010). Cells were seeded onto glass-bottom dishes two days prior to in situ processing. For D458 cells, glass bottom plates were coated with poly-L-lysine (0.01% w/v in water, Sigma Aldrich) for 30 minutes and washed with PBS prior to seeding. Cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences 15714) for 30 minutes, washed with PBS, and permeabilized with 70% ethanol for 30 minutes. Permeabilization solution was carefully exchanged with PBS-T wash buffer (PBS + 0.05% Tween-20) to minimize sample dehydration. Reverse transcription mix (1× RevertAid RT buffer, 250 μM dNTPs, 0.2 mg/mL BSA, 1 μM RT primer, 0.8 U/μL Ribolock RNase inhibitor, and 4.8 U/μL RevertAid H minus reverse transcriptase) was added to the sample and incubated for 16 hours at 37ºC. Following reverse transcription, cells were washed 5 times with PBS-T and post-fixed with 3% paraformaldehyde and 0.1% glutaraldehyde for 30 minutes at room temperature, then washed with PBS-T 5 times. After this step, cells expressing p65-mNeonGreen were imaged. Samples were thoroughly washed again with PBS-T, incubated in a padlock probe and extension-ligation reaction mix (1× Ampligase buffer, 0.4 U/μL RNase H, 0.2 mg/mL BSA, 100 nM padlock probe, 0.02 U/μL TaqIT polymerase, 0.5 U/μL Ampligase and 50 nM dNTPs) for 5 minutes at 37C and 90 minutes a t 45C, and then washed 2 times with PBS-T. Circularized padlocks were amplified with rolling circle amplification mix (1× Phi29 buffer, 250 μM dNTPs, 0.2 mg/mL BSA, 5% glycerol, and 1 U/μL Phi29 DNA polymerase) at 300ºC for either 3 hours or overnight.
In situ sequencing
Rolling circle amplicons were prepared for sequencing by hybridizing a mix containing sequencing primer oSBS_LentiGuide-BC or oSBS_CROP-seq (1 μM primer in 2X SSC + 10% formamide) for 30 minutes at room temperature. Barcodes were read out using sequencing-by-synthesis reagents from the Illumina MiSeq 500 cycle Nano kit (Illumina MS-103–1003). First, samples were washed with incorporation buffer (Nano kit PR2) and incubated for 3 minutes in incorporation mix (Nano kit reagent 1) at 60ºC. Samples were then thoroughly washed with PR2 at 60ºC (6 was hes for 3 minutes each) and placed in 200 ng/mL DAPI in 2x SSC for fluorescence imaging. Following each imaging cycle, dye terminators were removed by incubation for 6 minutes in Illumina cleavage mix (Nano kit reagent 4) at 60ºC, and samples were thoroughly washed with PR2.
Padlock optimization
Improved padlock sequences were found for the CROP-seq and CROP-seq-v2 (contains a modified sgRNA scaffold for increased CRISPR efficiency) vectors by pooled testing of a set of 84 padlocks (per vector) for in situ RNA detection efficiency and RCA amplification yield. Sequences varied in the length of the cDNA binding region (melting temperature from 45ºC, 49ºC, or 54ºC), and in overall length (72, 77, 83, or 89 nt for CROP-seq and 59, 69, 79, or 90 nt for CROP-seq-v2). The padlock backbone sequence is not functionally constrained; here, backbone sequences were randomly generated with G nucleotides excluded to reduce secondary structure. The backbone sequences were then filtered to have C content of 50±5% and no homopolymers exceeding 3 nt. Padlock sequences with the most positive minimum free energies were selected from these filtered sequences using mfold’s zipfold utility (Zuker, 2003). For compatibility with pooled testing, the backbone sequences for each vector were required to begin with a unique 5-mer barcode. Padlocks were individually synthesized (Integrated DNA Technologies) and pooled prior to phosphorylation with T4 PNK (New England Biolabs) according to the manufacturer’s instructions. Padlock-based RNA detection and in situ sequencing of the padlock backbones were performed as previously described in HeLa-TetR-Cas9 cells, using a sequencing primer that targeted the 5-mer padlock barcode. After identifying padlocks by 5 cycles of in situ sequencing, the padlocks were hybridized with a fluorescently labeled detection oligo to give a barcode-independent estimate of relative RCA yield. Relative RNA detection efficiency (fraction of sequencing reads from a given padlock) was used as a proxy for absolute RNA detection efficiency.
Fluorescence microscopy
All in situ sequencing images were acquired using a Ti-E Eclipse inverted epifluorescence microscope (Nikon) with automated XYZ stage control and hardware autofocus. An LED light engine (Lumencor Sola SE FISH II) was used for fluorescence illumination and all hardware was controlled using Micromanager software (Edelstein et al., 2010). In situ sequencing cycles were imaged using a 10X 0.45 NA CFI Plan Apo Lambda objective (Nikon) with the following filters (Semrock) and exposure times for each base: G (excitation 534/20 nm, emission 572/28 nm, dichroic 552 nm, 200 ms); T (excitation 575/25 nm, emission 615/24 nm, dichroic 596 nm, 200 ms); A (excitation 635/18 nm, emission 680/42 nm, dichroic 652 nm, 200 ms); C (excitation 661/20 nm, emission 732/68 nm, dichroic 695 nm, 800 ms).
Frameshift reporter screen
HeLa-TetR-Cas9 cells were stably transduced at MOI > 2 with pL_FR_Hygro and selected with hygromycin for 7 days to generate the HeLa-TetR-Cas9-FR cell line. Cells transduced with the pL_FR_Hygro lentiviral vector express an open reading frame consisting of a 50-nt frameshift reporter target sequence, followed by an H2B histone coding sequence with C-terminus HA epitope tag (+1 frameshift), followed by a second H2B sequence with C-terminus myc tag (+0 frameshift) and hygro antibiotic resistance cassette (+0 frameshift). The H2B-HA, H2B-myc, and hygromycin resistance sequences are preceded by self-cleaving 2A peptides in the same reading frame. Before generation of Cas9-mediated indel mutations, cells express the coding sequences with +0 frameshift. Subsequent activation of the reporter by co-expression of Cas9 and a targeting sgRNA leads to mutations in the target sequence, which may alter the downstream reading frame. A frameshift of +1 leads to expression of the H2B-HA protein, which can be visualized by immunofluorescence and detected by microscopy or flow cytometry. Integration of multiple copies of reporter per cell increases the likelihood of generating a +1 frameshift in at least one copy.
HeLa-TetR-Cas9-FR cells were used to screen targeting and control sgRNAs. A barcoded sgRNA perturbation library with 972 barcodes, each encoding one of 5 targeting or 5 control sgRNAs, was synthesized and cloned into LentiGuide-BC-CMV. This library was transduced into HeLa-TetR-Cas9-FR cells at MOI < 0.05 in three replicates, which were independently cultured and screened. Following 4 days of puromycin selection, cells were collected to validate library representation by NGS. Cas9 expression was induced by supplementing the culture media with 1 μg/mL doxycycline for 6 days. Cells were then split for screening either via in situ sequencing or by FACS.
For in situ screening, 500,000 cells were seeded into each well of a glass-bottom 6-well plate (CellVis). After two days of culture, in situ padlock detection and sequencing were carried out as above, with the modification that prior to sequencing-by-synthesis, cells were immunostained to detect frameshift reporter activation by blocking and permeabilizing with 3% BSA + 0.5 % Triton X-100 for 5 minutes, incubating in rabbit anti-HA (1:1000 dilution in 3% BSA) for 30 minutes, washing with PBS-T and incubating with goat anti-rabbit F(ab’)2 fragment Alexa 488 (CST 4412S, 1:1000 dilution in 3% BSA) for 30 minutes. Samples were changed into imaging buffer (200 ng/mL DAPI in 2X SSC) and phenotype images were acquired.
FACS screening was carried out by fixing cells with 4% PFA, permeabilizing with 70% ice-cold ethanol, and immunostaining with the same anti-HA primary and secondary antibodies and dilutions used for in situ analysis. Cells were sorted into HA+ and HA- populations (Sony SH800) and genomically integrated perturbations were sequenced as described above. The enrichment for each barcoded perturbation was defined as the HA+/HA- ratio of normalized read counts.
Frameshift reporter screens in U2-OS, A375, HT1080, HCT116 and HEK293T cell types were performed by first transducing these cells with lentiCas9-blast (Addgene #52962) (Sanjana et al., 2014) and selecting with blasticidin for 4 days. Cas9-expressing cells were then transduced with pL_FR_Hygro and selected with hygromycin for 7 days to generate reporter lines. These reporter lines were transduced with a CROPseq-puro library consisting of the same 5 targeting and 5 non-targeting sgRNAs used above. Cell libraries were selected with puromycin for 4 days and cells were seeded onto glass-bottom dishes 2 days prior to in situ sequencing.
NF-kB activation screens
HeLa-TetR-Cas9 cells were transduced with pR14_p65-mNeonGreen, a C-terminal fusion of p65 with a bright monomeric green fluorescent protein (Allele Biotechnology). Fluorescent cells were sorted by FACS (Sony SH800) and re-sorted to select for cells with stable expression. This reporter cell line was further transduced with a 4,063-barcode sgRNA library (962 genes targeted and 1,000 barcodes assigned to non-targeting sgRNAs) in LentiGuide-BC-CMV. Cells were selected with puromycin for 4 days and library representation was validated by NGS.
Cas9 expression was induced with 1 μg/mL doxycycline and cells were seeded onto 6-well cover glass-bottom plates 2 days prior to translocation experiments. The total time between Cas9 induction and performing the NF-κB activation assay was 7 days. Cells were stimulated with either 30 ng/mL TNFα or 30 ng/mL IL-1β (Invivogen) for 40 minutes prior to fixation with 4% paraformaldehyde for 30 minutes and initiation of the in situ sequencing protocol. Translocation phenotypes were recorded after the post-fixation step by exchanging for imaging buffer (2X SSC + 200 ng/mL DAPI) and imaging the nuclear DAPI stain and p65-mNeonGreen. After phenotyping, the remainder of the in situ sequencing protocol (gap-fill and rolling circle amplification) and 12 bases of sequencing-by-synthesis were completed.
HeLa Cas9-blast, A549 Cas9-blast and HCT116 Cas9-blast cells were transduced with a CROPseq-puro library consisting of 5,738 sgRNAs (952 genes targeted, 100 non-targeting sgRNAs). Cells were selected for 4 days and seeded onto 6-well plates 2 days prior to translocation experiments. The total time between library transduction and the screen was between 7 and 14 days. Cells were stimulated with TNFα or IL-1β (30 ng/mL for HCT116, 3 ng/mL for HeLa and A549) for 40 minutes prior to fixation with 4% paraformaldehyde for 30 minutes. Cells were then processed for in situ sequencing as in the HeLa p65-mNeonGreen activation screen, except for the following differences. Cells were stained after the post-fixation step by incubating for 1 hour with a rabbit antibody against p65 (CST 8242S, 1:400 dilution in 3% BSA), washing with PBS-T, then incubating with goat anti-rabbit F(ab’)2 fragment Alexa 488 (CST 4412S, 1:1000 dilution in 3% BSA) for 45 minutes and washing with PBS-T before proceeding to the gap- fill step. After RCA, cells were exchanged into imaging buffer (2X SSC + 200 ng/mL DAPI) and imaged with DAPI and 488 (p65) filters. After phenotyping, sequencing primer was hybridized and 10 bases of sequencing-by-synthesis were acquired.
NF-κB live-cell screens
HeLa-TetR-Cas9 stably expressing the p65-mNeonGreen reporter were transduced with the 952-gene CROPseq-puro library described above. Cells were selected with puromycin for 4 days and library representation was validated by NGS.
Cas9 expression was induced for 7 days with 1 μg/mL doxycycline and cells were seeded onto 6-well cover glass-bottom plates 2 days prior to translocation experiments. The total time between the start of Cas9 induction and performing the NF-κB live-cell assay was 14 days. Prior to the experiment, culture media was exchanged for imaging media: 0.1 ng/mL Hoechst 33342 in phenol red-free DMEM with HEPES and L-glutamine (Thermo Fisher Scientific 21063029), supplemented with 10% FBS and 100 U/mL penicillin-streptomycin. Cells were returned to the incubator for 2 hours, then stimulated with either 30 ng/mL TNFα or 30 ng/mL IL-1β and immediately loaded onto an automated live-cell microscope with environmental control (Zeiss CellDiscoverer 7). Images of the Hoechst nuclear stain and p65- mNeonGreen were acquired at 5X magnification at 23 minute intervals over 6 hours. Immediately after stopping live-cell imaging, cells were fixed and the in situ sequencing protocol was carried out as above.
Pooled validation was conducted with a new CROPseq-puro library targeting 65 candidate genes from the primary screen (10 sgRNAs per gene, 100 non-targeting sgRNAs). The validation screen was conducted following the same protocol as the primary live-cell screen, except that imaging was carried out on a Nikon Ti2 inverted microscope equipped with a live-cell incubator (Okolab). Images were acquired at 10X magnification in 30 minute intervals.
NF-κB arrayed validation
Top-ranking genes from the primary pooled screen were validated with individual sgRNAs. For each gene, 2 or 3 sgRNAs were tested, including at least one sgRNA not used in the primary screen. HeLa-TetR-Cas9 cells expressing p65-mNeonGreen were prepared and assayed as in the pooled screen, except that cells were transduced with the LentiGuide-Puro sgRNA expression vector (Addgene #52963) (Sanjana et al., 2014), and the translocation assay was carried out in 96-well cover glass plates. Image acquisition and data analysis were performed with the same hardware and software settings as in the pooled screen.
Hit validation using clonal knockout cell lines
Hela-TetR-Cas9 cells expressing p65-mNeonGreen were transfected with synthetic crRNA/tracrRNA (IDT; MED12 sgRNA: GGATCTTGAGCTACGAACAC; MED24 sgRNA: GCGCTGGAGTGACTACCAAT) using Lipofectamine 2000 (Thermo Fisher Scientific). Two days after transfection, single cell clones were seeded at a density of 0.6 cells/well into 480 96-wells per target gene. After two weeks, live clones were identified by microscopy and were expanded to a 6-well format. Clones were genotyped by replacing media with direct lysis buffer (1 mM CaCl2, 3 mM MgCl2, 1 mM EDTA, 1% Triton X-100, 10 mM Tris pH 7.5, 0.2 mg/ml Proteinase K), heating to 65º0 for 1 0 minutes, and incubating at 95º0 for 15 minutes to inactivate Proteinase K. PCR amplification of target loci, deep sequencing (Illumina MiSeq), and data analysis using the OutKnocker.org software were performed as described (Schmid-Burgk, Genome Research 2014). Clones bearing out-of-frame mutations in all alleles were further expanded, and analyzed by automated live cell microscopy (Nikon Ti2 inverted microscope with Okolab live-cell incubator) for 6 hours after stimulation with TNFα or IL-1β as described above.
RNA-seq analysis of clonal knockout cell lines
Clonal cell lines and wild type control cells were plated in 24-well format at a density of 50,000 cells/well. The next day, cells were stimulated for 0, 1, or 4 hours with TNFα or IL-1β (3 ng/mL) prior to lysis. Cells were washed with PBS, and lysed at room temperature in 150 μl TCL buffer (Qiagen, supplemented with 1% beta-mercaptoethanol) per well, then stored at −80°C. Smart-seq2 was performed as described (Trombetta et al., 2014). Libraries were sequenced on a NextSeq 500 (Illumina) using the High Output Kit v2 75-cycle kit (paired end, 36 cycles forward, 36 cycles reverse).
QUANTIFICATION AND STATISTICAL ANALYSIS
Image analysis
Images of cell phenotype and in situ sequencing of perturbations were coarsely aligned during acquisition using nuclear masks to calibrate plate position, and finely aligned during analysis using cross-correlation of DAPI or in situ sequencing signal between cycles. Nuclei were detected using local thresholding and watershed-based segmentation. Cells were segmented using local thresholding of cytoplasmic background from in situ sequencing signal and assignment of pixels to the nearest nucleus by the fast-marching method. Frameshift reporter and NF-κB translocation phenotypes were quantified by calculating pixel-wise correlations between the nuclear DAPI channel and 488 channel (HA stain or p65-mNeonGreen, respectively). Mitotic cells and cells with abnormally high or low reporter expression were filtered out based on maximum DAPI signal and mean p65-mNeonGreen signal (Figure S4A).
Sequencing reads were detected by applying a Laplacian-of-Gaussian linear filter (kernel width σ = 1 pixel) to subtract low-frequency background and enhance the point-like sequencing spots. Peaks were detected by calculating the per-pixel, per-channel standard deviation over sequencing cycles, averaging over color channels, and finding local maxima in the resulting image. The base intensity at each cycle was defined as the maximum value in a 3×3 pixel window centered on the read. A linear transformation to correct for optical cross-talk and intensity differences between color channel was then estimated from the data and applied. Finally, each base was called according to the channel with maximum corrected intensity, and a per-base quality score was defined as the ratio of intensity for the maximum channel to total intensity for all channels. The output of the sequencing image analysis was a table recording each sequencing read along with the identity of the overlapping cell, quality score per base, and spatial location.
Data analysis functions were written in Python, using Snakemake for workflow control (Köster and Rahmann, 2012).
Frameshift reporter misidentification rate estimation
To estimate the rate at which cells are assigned an incorrect perturbation barcode, we first assumed all HA+ cells mapped to a non-targeting control sgRNA (4.7%) were false positive events due to incorrect barcode assignment (supported by the very low false positive rate (<0.001 %) of the frameshift reporter itself, measured for a single perturbation in arrayed format). However, as incorrect barcode assignments were equally likely to map an HA+ cell to a targeting or control sgRNA, we estimated the misidentification rate to be twice as large, or 9.4%.
NF-κB activation screen analysis
Nuclei of individual cells were segmented by thresholding background-subtracted DAPI signal and separating the resulting regions using the watershed method. Cells with at least one read exactly matching a library barcode were retained for analysis. In order to remove mitotic cells and cells with abnormally high or low reporter expression, cells were further filtered based on nuclear area, maximum DAPI signal, and mean p65-mNeonGreen signal. Pixel-wise DAPI-mNeonGreen correlation within the segmented nuclear region, described above, was used to define the translocation score for each cell as it most clearly separated perturbations against known NF-κB genes from non-targeting controls. The phenotypic effects of perturbations targeting known NF-κB genes ranged from a large increase in fully untranslocated cells (e.g., MAP3K7) to more subtle negative shifts in the distribution of scores (e.g., IKBKB).
To capture a broad range of effect sizes, we calculated an sgRNA translocation defect in a given replicate by computing the difference in translocation score distribution compared to non-targeting controls (the shaded area in Fig. 4, C and E). We found this metric performed better at separating known genes from controls than the often-used Kolmogorov-Smirnov distance. We defined the gene translocation defect as the second-largest sgRNA translocation defect for sgRNAs targeting that gene. This statistic helps reduce the false positive rate due to clonal effects (integration of an sgRNA into a cell that is defective in translocation) which are independent among sgRNAs and screen replicates, as well as false negatives due to inefficient sgRNAs.
A permutation test was used to calculate p-values for the gene translocation defects. Random subsets of sgRNA translocation defects were sampled from non-targeting controls to build a null distribution (3 sgRNAs per replicate, repeated 100,000 times). The cumulative null distribution was used to determine p-values for the gene translocation defects. Hits at an estimated FDR <10% and <20% were identified using the Benjamini-Hochberg procedure (Table S3). KEGG-annotated genes were defined as members of KEGG pathway HSA04064 (NF-kappa B signaling pathway) between IL-1β or TNFα and p65/p50.
Cellular morphology analysis
Cellular morphology was analyzed by calculating image features from DAPI intensity as well as cell and nuclear geometry. We summarized the gene-level (across all guides and cells) distribution of morphological features (cell area, nucleus area, DAPI max, DAPI mean, cell eccentricity and nucleus eccentricity) by its first, second and third quartiles, producing an 18-dimensional vector for each gene. We performed dimensionality reduction by Principal Components Analysis (PCA) and visualized the first two principal components, finding distinct clusters for regulators downstream of IKK (Figure S4).
Analysis of NF-κB live-cell screens
Image analysis was conducted as for the initial NF-κB screen, with additional preprocessing steps to (a) track cells through the live-cell time course, and (b) align the final time point of live-cell with the first cycle of sequencing-by-synthesis. For each cell, the translocation score at each time point was subtracted from a baseline translocation score, interpolated in time from control cells imaged in the same well. For each gene, an integrated translocation score was calculated by integrating each cell’s baseline-subtracted translocation scores from 45 to 345 minutes post-stimulation. Statistical significance for deviation in the integrated translocation score between perturbed and control cells was quantified using the non- parametric Mann-Whitney U test.
NF-κB arrayed validation
The translocation defect for each sgRNA and cytokine translocation was assessed by computing the difference in translocation score distribution compared to the average of at least 3 non-targeting control sgRNAs assayed on the same plate. For each cytokine, the translocation defects were standardized using the mean and standard deviation of the translocation defects for non-targeting control guides. These standardized values were averaged over replicate sgRNAs for a given gene and cytokine pair to obtain validation Z-scores (Table S3).
RNA-seq quantification
Transcript abundance was quantified using kallisto and differential gene expression was analyzed using DEBrowser as a graphical interface to edgeR, with default edgeR parameters: TMM-based normalization, dispersion estimated using the edgeR function estimateDisp, and p-values adjusted with the Benjamini-Hochberg procedure (Bray et al., 2016; Kucukural et al., 2019; Robinson et al., 2010). Highly induced NF-κB genes were defined as genes with >50-fold increase after 4 hours of stimulation by either cytokine in wild type cells (adjusted p-value < 10−3). MED12 or MED24-dependent genes were defined as genes with >8-fold change between knockout and wild type conditions after 4 hours of stimulation with either cytokine. All pairwise comparisons were performed using the edgeR function exactTest (n = 3 or 4 biological replicates). Biological replicates were defined as replicate clonal knockout cell lines (for MED12 and MED24 knockouts) or replicate stimulations (for wild type control cells).
Supplementary Material
(A) In order to determine the identity of the lentiviral vector integrated in each cell, all cellular RNAs are first fixed in place by formaldehyde treatment. A reverse transcription primer containing locked nucleic acid (LNA) bases is hybridized to the mRNA containing the barcode sequence. Complementary DNA (cDNA) is generated using a reverse transcriptase lacking RNase activity, producing an RNA-DNA hybrid. The cells are then fixed again (post-fixed) with a mixture of formaldehyde and glutaraldehyde to improve cDNA retention.
A single reaction mix containing RNase H, a DNA polymerase lacking 5’ to 3’ exonuclease activity, a DNA ligase, and a padlock DNA oligonucleotide is then added. Digestion of the RNA strand exposes the cDNA bases, allowing the padlock to hybridize to the cDNA at sites flanking the barcode. The DNA polymerase extends the padlock, copying the barcode sequence until it reaches the annealed 5’ padlock arm. Once extended, the padlock is then ligated into a single-stranded DNA circle. This reaction can potentially be inhibited by strand-displacing activity of the polymerase, which may prevent padlock circularization (Chen et al., 2018). During this step, the cDNA is retained in place via hybridization to the RNA strand at the LNA-modified bases within the RT primer, which inhibit RNase H digestion.
Phi29 polymerase is used to perform rolling circle amplification of the circularized padlock. The 3’ exonuclease activity of Phi29 polymerase digests the single-stranded portion of the cDNA strand, generating a primer for rolling circle amplification. The amplified single-stranded DNA product contains tandem repeats of the padlock backbone sequences and barcode, which can be read out by sequencing-by-synthesis.
The overall protocol provides a high level of sequence specificity, conferred by hybridization of the RT primer to a unique priming site, hybridization of the padlock to the flanking sites, the preference of the ligase to act only on exactly matched DNA, and sequencing-by-synthesis of the cell-derived barcode sequence itself.
(B) Read-level intensity comparison across cycles. Each point represents the intensity in a given cycle (2–12) on the y-axis relative to the intensity in the same channel in the first cycle on the x-axis. For each plot, 300 reads were randomly sampled from all reads with minimum quality score of 0.2.
(C) Example compensation matrix used to correct for relative intensity and spectral crosstalk, from the dataset shown in Figure 2. Corrected intensities are calculated by multiplying the raw channel intensities by the compensation matrix.
(D) Fraction of reads that map (edit distance = 0) and nearly map (edit distance > 0) to a barcode expected in the 40-plex pool.
(E) Comparison of barcode abundances measured by in situ sequencing or NGS (R2 = 0.55). The relative abundance of 95% of barcodes was within 5-fold (indicated by dashed lines).
(F) In situ sequencing was carried out on HeLa or D458 medulloblastoma cells expressing CROP-seq-BFP (cell segmentation outlined, 10X magnification, scale bar 50 μm). D458 cells, normally grown in suspension, were adhered by poly-L-lysine treatment (STAR Methods).
(G) The number of reads detected in HeLa and D458 cells increased with BFP intensity before plateauing, due in part to difficulty segmenting individual reads at density above ~1 read / 10 pixels. Box plot indicates median, 25th and 75th quartiles, and twice the interquartile range (n = 13,000 cells per cell line).
(A) Padlock detection efficiency was increased more than two-fold compared to literature protocols (Chen et al., 2018; Ke et al., 2013) by optimizing the dNTP concentration and polymerase used for the padlock extension-ligation reaction. A striking improvement in detection efficiency was observed when using Stoffel fragment with a dNTP concentration 1000-fold less than previously published (Ke et al., 2013). It is possible that reducing dNTP concentrations decreases the strand displacement activity of the polymerase, improving padlock circularization. Although Stoffel fragment has been discontinued by its manufacturer, we obtained similar results with another commercially available truncation mutant of Taq polymerase (Qiagen TaqIT). While optimizing post-fixation conditions for detection efficiency, we observed that modifying the standard 4% formaldehyde fixative to 3% formaldehyde and 0.1% glutaraldehyde (“glutaraldehyde postfix”) led to a dramatic increase in the yield of overall fluorescence signal from each spot. We presume the improvement was due to an increase in the efficiency of rolling circle amplification, although no specific mechanism was identified. The protocol comparison was performed on a single multi-well plate, using HeLa-TetR-Cas9 cells transduced with LentiGuide-BC. Each data point represents a technical replicate of the in situ protocol.
(B) A set of 84 padlock probes was synthesized with binding sites flanking the sgRNA sequence in the CROP-seq vector. Padlock probe length, 5’ binding sequence (i.e., binding site on sgRNA scaffold), and non-binding sequence content were varied (STAR Methods). Padlocks contained a barcode in the non-binding sequence so they could be pooled and tested in a single in situ reaction, using in situ sequencing to demultiplex the padlock identity. The relative detection efficiency (count) and RCA yield (intensity) were quantified using a dye-labeled hybridization probe complementary to the 3’ binding site, which was common across all padlock probes. Data points are colored by either padlock length (not including the 20 nt added during the gap-fill step) or Tm of the padlock 5’ binding arm. The optimized padlock is identified by a black circle.
(C) Multiple perturbations can be delivered via separate lentiviral vectors and detected in the same cell. HeLa-TetR-Cas9 cells were sequentially transduced with CROPseq-puro and CROPseq-zeo libraries (each containing a pool of 95 sgRNAs). The in situ padlock detection protocol was the same as for a single vector library. The cumulative fraction of cells with at least N reads of different sgRNAs can be calculated. For example, a total of 70% of all cells imaged had 2 or more reads for two independent sgRNAs (heatmap position 2,2).
(D) Images were acquired at 10X magnification and could still resolve most barcode spots per cell (scale bar 200 μm; composite image of four sequencing channels; white outlines indicate segmented cells). Reads that failed to map to a known barcode (e.g., due to overlapping signal) are indicated by grey squares in the “base calls” panels of S2D.).
(A) Schematic of a frameshift reporter that converts CRISPR-Cas9-induced indel mutations into a positive fluorescent signal.
(B) The frameshift reporter was read out by microscopy in HeLa-TetR-Cas9-FR cells in the absence of a targeting sgRNA. A myc epitope tag in the original, unedited frame was stained to confirm expression levels. The reporter was found to have a very low background, with zero false positives observed among >400,000 cells (dashed line indicates threshold for defining HA+ cells).
(C) Phenotype data for the frameshift reporter screen (random subset of 10,000 cells plotted). HA+ cells were defined by thresholding based on HA-488 fluorescence intensity and pixel-wise correlation with DAPI nuclear stain.
(D) Flow analysis for the FACS-based frameshift reporter screen. Cells from the HA+ and HA- gates were sorted (top left) and sgRNA abundance was compared by NGS. Sorted cells from the HA+ (top middle) and HA- (top right) gates were re-analyzed to verify sorting accuracy. The ratio of HA+ to HA- cells in the re-analyzed populations sets an upper bound for relative sgRNA enrichment of ~300X.
(E) Schematic of image analysis pipeline showing intermediate images (LoG-transformed data, nuclei masks, cell masks max-filtered data and peak locations) and tables (summarizing bases, reads, cells and phenotypes). Example data and code provided at https://github.com/feldman4/OpticalPooledScreens.).
(A) Phenotype filtering criteria for the primary screen used to exclude mitotic cells and cells with low p65-mNeonGreen reporter expression.
(B) For both IL-1β and TNFα, primary screen gene rankings correlate well (Spearman’s p > 0.73) with rankings in validation screen of single-gene CRISPR-Cas9 knockouts. Proteasome subunits are not shown as they exhibited severe negative fitness effects in arrayed validation experiments, likely biasing the surviving cells to those with incomplete protein knockout.
(C) Translocation distributions for sgRNAs targeting UBE2N and BIRC2 across the HeLa p65-mNeonGreen reporter and HeLa, A549 and HCT116 antibody screens.
(D) Cells from the primary screen were analyzed based on morphological features. Dimensionality reduction by Principal Components Analysis grouped a subset of genes by known function in the NF-kB pathway. Genes are plotted by PCA component 1 (46% of variance explained) and component 2 (24%).
(E) PCA was carried out based on the first (Q1), second (Q2) and third (Q3) quartiles of the per-gene distribution for each morphological parameter. A percentile-based statistic was used instead of a population mean in order to minimize sensitivity to outliers while still capturing changes to the distribution. Plotted are the average values for each quartile after standardization across gene categories (color codes same as in (D)).
(F) Fraction of variance explained by the top principal components.
(G) Weights of the top principal components.
(H) Example cells randomly sampled from the dataset used in (D) and (E). DAPI nuclear stain is shown in gray and the segmented boundary for each cell is outlined (scale bar 6 μm).).
Time traces of translocation score (correlation between p65-mNeonGreen and Hoechst nuclear stain) from the live-cell validation screen. Each trace averages over all cells for a single sgRNA, with each gene represented by 10 sgRNAs. The average trace over 50 non-targeting sgRNAs is shown in grey. ).
Summary of scale and throughput of the NF-κB screen performed, as well as projections for two hypothetical screens using either low-density cell culture or a genome-scale perturbation library, assuming a cell mapping rate of 80% (typical for EF1a in HeLa cells, Figure 2E) and 600 cells screened per perturbation. The screening approach described in this study uses fully pooled protocols for library construction, cloning, and cell culture. As a result, practical limitations to the scale of optical pooled screens come mainly from the time required to image large numbers of cells and the cost of reagents for padlock detection and in situ sequencing. The key figure determining both throughput and cost is the total surface area processed, which affects the volume of reagents used and the time per sequencing cycle. The total readout time may be substantially reduced by using a two-color sequencing chemistry (e.g., Illumina MiniSeq/NextSeq) and optimized microscope hardware.
Principal reagent costs (direct) for in situ amplification of barcodes and sequencing-by-synthesis. One 6-well plate corresponds to approximately 2–4 million mapped cells for the cell lines used in this study. For example, the activation screen and static p65 screens listed in Table S1 each employed one 6-well plate.
Summary of screen metrics, including number of cells passing a phenotyping filter (e.g. non-mitotic with reporter expression above threshold) and number of those cells with ≥ 1 or 2 mapped reads.).
Spot counts and intensities for competitive padlock detection assays for LentiGuide-BC, CROPseq and CROPseq-v2.).
List of oligo sequences used for pooled cloning of perturbation libraries. Libraries of paired barcodes and sgRNAs were amplified from oligo pools and cloned into LentiGuide-BC or LentiGuide-BC-CMV (STAR Methods).
Summary of translocation defects and p-values for HeLa p65-mNeonGreen reporter screen. Each gene was assigned a score for lack of p65-mNeonGreen translocation in response to IL-1β and TNFα stimulation. Gene p-values were calculated by repeatedly sampling sets of three permuted control sgRNAs to generate gene-level null translocation scores. Genes scored as hits at estimated FDR <10% were identified by the Benjamini-Hochberg procedure (STAR Methods).
Screen ranks for p65 translocation defects and a summary table for KEGG genes detected across HeLa, A549 and HCT116 cells, color-coded based on FDR threshold for each screen (genes with FDR < 10% and rank < 100 labeled in green).
Table of translocation defects, adjusted p-values and screen ranks for all genes across HeLa, A549 and HCT116 cells. Translocation defect for each gene was calculated as the integrated difference between the translocation score distributions for an sgRNA and the non-targeting population, averaged over all sgRNAs for a gene.).
Table of integrated translocation defects and p-values for primary kinetic screen. Genes were assigned p-values based on comparing the translocation either at 45 minutes post-stimulation (as in the initial screen) or from 45 minutes to 345 minutes post-stimulation (STAR Methods).).
Example data showing a complete 10X magnification field of view with 12 cycles of sequencing, taken from the same dataset used in Fig. 2.
Highlights.
In situ sequencing of perturbations or barcodes enables image-based pooled screens
p65 translocation is assayed by imaging in fixed and live cell pools
Pooled live-cell screen identifies MED12 and MED24 as negative regulators of NF-κB
ACKNOWLEDGEMENTS
We thank Rhiannon Macrae, Iain Cheeseman, Aviv Regev, Fei Chen, Arjun Raj, Nir Hacohen, Benjamin Gewurz, Sara Jones, Emily Botelho, and members of the Blainey and Zhang labs for critical feedback and discussions. We thank Leslie Gaffney for assistance with figures. We gratefully acknowledge Linyi Gao for the gift of the integration-deficient co-packaging plasmid pR_LG. We thank Zohar Bloom-Ackermann for assistance with live-cell imaging and FuNien Tsai for preparing in situ sequencing of D458 cells.
This work was supported by a Simons Center Seed Grant from MIT, the Broad Institute through startup funding (to P.C.B) and the BN10 program, and two grants from the National Human Genome Research Institute (HG009283 and HG006193). P.C.B. is supported by a Career Award at the Scientific Interface from the Burroughs Wellcome Fund. F.Z. is a New York Stem Cell Foundation–Robertson Investigator. F.Z. is supported by NIH grants (1R01-HG009761, 1R01-MH110049, and 1DP1-HL141201); the Howard Hughes Medical Institute; the New York Stem Cell, Simons, Paul G. Allen Family, and Vallee Foundations; the Poitras Center for Affective Disorders Research at MIT; the Hock E. Tan and K. Lisa Yang Center for Autism Research at MIT; J. and P. Poitras; and R. Metcalfe. A.M. is supported by the Swedish Research Council (grant 2015–06403). R.J.C. is supported by a Fannie and John Hertz Foundation Fellowship and an NSF Graduate Research Fellowship. J.L.S.-B. is supported by an EMBO Long-Term Fellowship (ALTF 199–2017).
The Broad Institute and MIT may seek to commercialize aspects of this work, and related applications for intellectual property have been filed.
Footnotes
DATA AND CODE AVAILABILITY
Python scripts, including a complete pipeline from example data to sequencing reads, will be made available, along with plasmid sequences and updated protocols at https://github.com/blaineylab/OpticalPooledScreens. RNA-seq data is deposited in the Gene Expression Omnibus (GSE132704). Raw and processed image data is available on request.
DECLARATION OF INTERESTS
J.S.B and A.M. declare no competing interests. A.M is an employee at AstraZeneca. D.F. is an employee at insitro. A.J.G. is an employee of and holds equity in Arbor Biotechnologies, Inc. P.C.B is a consultant to and holds equity in 10X Genomics, General Automation Lab Technologies, Celsius Therapeutics, and Next Gen Diagnostics, LLC. P.C.B is a consultant to insitro. F.Z. is a founder, consultant to and holds equity in Arbor Biotechnologies, Beam Therapeutics, Editas Medicine, Pairwise Plants, and Sherlock Biosciences. The Broad Institute and MIT have filed U.S. patent applications on work described in this manuscript and may seek to license the technology. P.C.B, D.F., R.J.C., A.S. are listed as inventors.
REFERENCES
- Adamson B, Norman TM, Jost M, Cho MY, Nuñez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, et al. (2016). A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell 167, 1867–1882.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson B, Norman TM, Jost M, and Weissman JS (2018). Approaches to maximize sgRNA-barcode coupling in Perturb-seq screens. BioRxiv 298349. [Google Scholar]
- Bauernfeind F, Niepmann S, Knolle PA, and Hornung V. (2016). Aging-Associated TNF Production Primes Inflammasome Activation and NLRP3-Related Metabolic Disturbances. The Journal of Immunology 197, 2900–2908. [DOI] [PubMed] [Google Scholar]
- Bononi A, Yang H, Giorgi C, Patergnani S, Pellegrini L, Su M, Xie G, Signorato V, Pastorino S, Morris P, et al. (2017). Germline BAP1 mutations induce a Warburg effect. Cell Death Differ 24, 1694–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Buschmann T, and Bystrykh LV (2013). Levenshtein error-correcting barcodes for multiplexed DNA sequencing. BMC Bioinformatics 14, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ (2005). Ubiquitin Signaling in the NF-kB Pathway. Nat. Cell Biol 7, 758–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, and Zhuang X. (2015). Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Sun Y-C, Church GM, Lee JH, and Zador AM (2018). Efficient in situ barcode sequencing using padlock probe-based BaristaSeq. Nucleic Acids Res 46, e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia N-Y, Chan Y-S, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau M-S, et al. (2010). A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 468, 316–320. [DOI] [PubMed] [Google Scholar]
- Colleran A, Collins PE, O’Carroll C, Ahmed A, Mao X, McManus B, Kiely PA, Burstein E, and Carmody RJ (2013). Deubiquitination of NF-κB by Ubiquitin-Specific Protease-7 promotes transcription. Proc. Natl. Acad. Sci. U. S. A 110, 618–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinet C, Stöter M, Bradshaw CR, Samusik N, Rink JC, Kenski D, Habermann B, Buchholz F, Henschel R, Mueller MS, et al. (2010). Systems survey of endocytosis by multiparametric image analysis. Nature 464, 243–249. [DOI] [PubMed] [Google Scholar]
- Dai F, Lee H, Zhang Y, Zhuang L, Yao H, Xi Y, Xiao Z-D, You MJ, Li W, Su X, et al. (2017). BAP1 inhibits the ER stress gene regulatory network and modulates metabolic stress response. Proc. Natl. Acad. Sci. U. S. A 114, 3192–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Jia G, Choi J, Ma H, Anaya E, Ye C, Shankar P, and Wu H. (2015). Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol 16, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datlinger P, Rendeiro AF, Schmidl C, Krausgruber T, Traxler P, Klughammer J, Schuster LC, Kuchler A, Alpar D, and Bock C. (2017). Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, et al. (2016). Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167, 1853–1866.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, and Root DE (2014). Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol 32, 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essen D van, Engist B, Natoli G, and Saccani S. (2009). Two Modes of Transcriptional Activation at Native Promoters by NF-kB p65. PLOs Biol 7, e1000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman D, Singh A, Garrity AJ, and Blainey PC (2018). Lentiviral co-packaging mitigates the effects of intermolecular recombination and multiple integrations in pooled genetic screens. BioRxiv 262121. [Google Scholar]
- Floyd SR, Pacold ME, Huang Q, Clarke SM, Lam FC, Cannell IG, Bryson BD, Rameseder J, Lee MJ, Blake EJ, et al. (2013). The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature 498, 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewurz BE, Towfic F, Mar JC, Shinners NP, Takasaki K, Zhao B, Cahir-McFarland ED, Quackenbush J, Xavier RJ, and Kieff E. (2012). Genome-wide siRNA screen for mediators of NF-κB activation. Proc. Natl. Acad. Sci. U. S. A 109, 2467–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. (2014). Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot R. de, Lüthi J, Lindsay H, Holtackers R, and Pelkmans L. (2018). Large-scale image-based profiling of single-cell phenotypes in arrayed CRISPR-Cas9 gene perturbation screens. Mol. Syst. Biol 14, e8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guermah M, Malik S, and Roeder RG (1998). Involvement of TFIID and USA components in transcriptional activation of the human immunodeficiency virus promoter by NF-kappaB and Sp1. Mol. Cell. Biol 18, 3234–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, and Farese RV (2008). Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453, 657–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsdottir SM, Ljosa V, Sokolnicki KL, Wilson JA, Walpita D, Kemp MM, Seiler KP, Carrel HA, Golub TR, Schreiber SL, et al. (2013). Multiplex Cytological Profiling Assay to Measure Diverse Cellular States. PlOs ONE 8, e80999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, and Ghosh S. (2012). NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev 26, 203–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F, Kerr G, and Boutros M. (2014). E-CRISP: fast CRISPR target site identification. Nat. Methods 11, 122–123. [DOI] [PubMed] [Google Scholar]
- Hill AJ, McFaline-Figueroa JL, Starita LM, Gasperini MJ, Matreyek KA, Packer J, Jackson D, Shendure J, and Trapnell C. (2018). On the design of CRISPR-based single-cell molecular screens. Nat. Methods 15, 271–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäättelä M, Mouritzen H, Elling F, and Bastholm L. (1996). A20 zinc finger protein inhibits TNF and IL-1 signaling. J. Immunol. Baltim. Md 1950 156, 1166–1173. [PubMed] [Google Scholar]
- Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, David E, Salame TM, Tanay A, van Oudenaarden A, and Amit I. (2016). Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell 167, 1883–1896.e15. [DOI] [PubMed] [Google Scholar]
- Johnson KM, Mahajan SS, and Wilson AC (1999). Herpes simplex virus transactivator VP16 discriminates between HCF-1 and a novel family member, HCF-2. J. Virol 73, 3930–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, and Goto S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlas A, Machuy N, Shin Y, Pleissner K-P, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, et al. (2010). Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463, 818–822. [DOI] [PubMed] [Google Scholar]
- Ke R, Mignardi M, Pacureanu A, Svedlund J, Botling J, Wählby C, and Nilsson M. (2013). In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods 10, 857–860. [DOI] [PubMed] [Google Scholar]
- Köster J, and Rahmann S. (2012). Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 28, 2520–2522. [DOI] [PubMed] [Google Scholar]
- Kucukural A, Yukselen O, Ozata DM, Moore MJ, and Garber M. (2019). DEBrowser: interactive differential expression analysis and visualization tool for count data. BMC Genomics 20, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson C, Grundberg I, Söderberg O, and Nilsson M. (2010). In situ detection and genotyping of individual mRNA molecules. Nat. Methods 7, 395–397. [DOI] [PubMed] [Google Scholar]
- Lawson MJ, Camsund D, Larsson J, Baltekin Ö, Fange D, and Elf J. (2017). In situ genotyping of a pooled strain library after characterizing complex phenotypes. Mol. Syst. Biol 13, 947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D-F, Kuo H-P, Liu M, Chou C-K, Xia W, Du Y, Shen J, Chen C-T, Huo L, Hsu M-C, et al. (2009). KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol. Cell 36, 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, Terry R, Jeanty SSF, Li C, Amamoto R, et al. (2014). Highly multiplexed subcellular RNA sequencing in situ. Science 343, 1360–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhoff MW, Lauren J, Cassidy RM, Dobie FA, Takahashi H, Nygaard HB, Airaksinen MS, Strittmatter SM, and Craig AM (2009). An unbiased expression screen for synaptogenic proteins identifies the LRRTM protein family as synaptic organizers. Neuron 61, 734–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, and Cai L. (2014). Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 11, 360–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, and Dutta A. (2009). The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J. Biol. Chem 284, 34179–34188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, and Roeder RG (2005). Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci 30, 256–263. [DOI] [PubMed] [Google Scholar]
- Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, Feizi S, Gnirke A, Callan CG, Kinney JB, et al. (2012). Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol 30, 271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misaghi S, Ottosen S, Izrael-Tomasevic A, Arnott D, Lamkanfi M, Lee J, Liu J, O’Rourke K, Dixit VM, and Wilson AC (2009). Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol. Cell. Biol 29, 2181–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. (2006). A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298. [DOI] [PubMed] [Google Scholar]
- Nakatake Y, Fujii S, Masui S, Sugimoto T, Torikai-Nishikawa S, Adachi K, and Niwa H. (2013). Kinetics of drug selection systems in mouse embryonic stem cells. BMC Biotechnol 13, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche J-K, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. (2010). Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464, 721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, Sumpter R, Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, et al. (2011). Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature 480, 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahl HL (1999). Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 18, 6853–6866. [DOI] [PubMed] [Google Scholar]
- Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, et al. (2015). A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell 162, 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin AJ, Parker KR, Satpathy AT, Qi Y, Wu B, Ong AJ, Mumbach MR, Ji AL, Kim DS, Cho SW, et al. (2019). Coupled Single-Cell CRiSpR Screening and Epigenomic Profiling Reveals Causal Gene Regulatory Networks. Cell 176, 361–376.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack LM, Davoli T, Xu Q, Li MZ, and Elledge SJ (2016). Sources of Error in Mammalian Genetic Screens. G3 Genes Genomes Genet 6, 2781–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F. (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JJ, Lee C, and Shendure J. (2012). Accurate gene synthesis with tag-directed retrieval of sequence-verified DNA molecules. Nat. Methods 9, 913–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JE, and Smith DE (2010). The IL-1 family: regulators of immunity. Nat. Rev. Immunol 10, 89–102. [DOI] [PubMed] [Google Scholar]
- Wang C, Lu T, Emanuel G, Babcock HP, and Zhuang X. (2019). Imaging-based pooled CRISPR screening reveals regulators of lncRNA localization. Proc. Natl. Acad. Sci 116, 10842–10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, and Lander ES (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, et al. (2018). Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewska A, Dhainaut M, Ben-Zvi B, Rose SA, Park ES, Amir E-AD, Bektesevic A, Baccarini A, Merad M, Rahman AH, et al. (2018). Protein Barcodes Enable High-Dimensional Single-Cell CRISPR Screens. Cell 175, 1141–1155.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Cooley A, Armendariz D, Zhou P, and Hon GC (2018). Frequent sgRNA-barcode recombination in single-cell perturbation assays. PloS One 13, e0198635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Boehm JS, Yang X, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T, et al. (2011). A public genome-scale lentiviral expression library of human ORFs. Nat. Methods 8, 659–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) In order to determine the identity of the lentiviral vector integrated in each cell, all cellular RNAs are first fixed in place by formaldehyde treatment. A reverse transcription primer containing locked nucleic acid (LNA) bases is hybridized to the mRNA containing the barcode sequence. Complementary DNA (cDNA) is generated using a reverse transcriptase lacking RNase activity, producing an RNA-DNA hybrid. The cells are then fixed again (post-fixed) with a mixture of formaldehyde and glutaraldehyde to improve cDNA retention.
A single reaction mix containing RNase H, a DNA polymerase lacking 5’ to 3’ exonuclease activity, a DNA ligase, and a padlock DNA oligonucleotide is then added. Digestion of the RNA strand exposes the cDNA bases, allowing the padlock to hybridize to the cDNA at sites flanking the barcode. The DNA polymerase extends the padlock, copying the barcode sequence until it reaches the annealed 5’ padlock arm. Once extended, the padlock is then ligated into a single-stranded DNA circle. This reaction can potentially be inhibited by strand-displacing activity of the polymerase, which may prevent padlock circularization (Chen et al., 2018). During this step, the cDNA is retained in place via hybridization to the RNA strand at the LNA-modified bases within the RT primer, which inhibit RNase H digestion.
Phi29 polymerase is used to perform rolling circle amplification of the circularized padlock. The 3’ exonuclease activity of Phi29 polymerase digests the single-stranded portion of the cDNA strand, generating a primer for rolling circle amplification. The amplified single-stranded DNA product contains tandem repeats of the padlock backbone sequences and barcode, which can be read out by sequencing-by-synthesis.
The overall protocol provides a high level of sequence specificity, conferred by hybridization of the RT primer to a unique priming site, hybridization of the padlock to the flanking sites, the preference of the ligase to act only on exactly matched DNA, and sequencing-by-synthesis of the cell-derived barcode sequence itself.
(B) Read-level intensity comparison across cycles. Each point represents the intensity in a given cycle (2–12) on the y-axis relative to the intensity in the same channel in the first cycle on the x-axis. For each plot, 300 reads were randomly sampled from all reads with minimum quality score of 0.2.
(C) Example compensation matrix used to correct for relative intensity and spectral crosstalk, from the dataset shown in Figure 2. Corrected intensities are calculated by multiplying the raw channel intensities by the compensation matrix.
(D) Fraction of reads that map (edit distance = 0) and nearly map (edit distance > 0) to a barcode expected in the 40-plex pool.
(E) Comparison of barcode abundances measured by in situ sequencing or NGS (R2 = 0.55). The relative abundance of 95% of barcodes was within 5-fold (indicated by dashed lines).
(F) In situ sequencing was carried out on HeLa or D458 medulloblastoma cells expressing CROP-seq-BFP (cell segmentation outlined, 10X magnification, scale bar 50 μm). D458 cells, normally grown in suspension, were adhered by poly-L-lysine treatment (STAR Methods).
(G) The number of reads detected in HeLa and D458 cells increased with BFP intensity before plateauing, due in part to difficulty segmenting individual reads at density above ~1 read / 10 pixels. Box plot indicates median, 25th and 75th quartiles, and twice the interquartile range (n = 13,000 cells per cell line).
(A) Padlock detection efficiency was increased more than two-fold compared to literature protocols (Chen et al., 2018; Ke et al., 2013) by optimizing the dNTP concentration and polymerase used for the padlock extension-ligation reaction. A striking improvement in detection efficiency was observed when using Stoffel fragment with a dNTP concentration 1000-fold less than previously published (Ke et al., 2013). It is possible that reducing dNTP concentrations decreases the strand displacement activity of the polymerase, improving padlock circularization. Although Stoffel fragment has been discontinued by its manufacturer, we obtained similar results with another commercially available truncation mutant of Taq polymerase (Qiagen TaqIT). While optimizing post-fixation conditions for detection efficiency, we observed that modifying the standard 4% formaldehyde fixative to 3% formaldehyde and 0.1% glutaraldehyde (“glutaraldehyde postfix”) led to a dramatic increase in the yield of overall fluorescence signal from each spot. We presume the improvement was due to an increase in the efficiency of rolling circle amplification, although no specific mechanism was identified. The protocol comparison was performed on a single multi-well plate, using HeLa-TetR-Cas9 cells transduced with LentiGuide-BC. Each data point represents a technical replicate of the in situ protocol.
(B) A set of 84 padlock probes was synthesized with binding sites flanking the sgRNA sequence in the CROP-seq vector. Padlock probe length, 5’ binding sequence (i.e., binding site on sgRNA scaffold), and non-binding sequence content were varied (STAR Methods). Padlocks contained a barcode in the non-binding sequence so they could be pooled and tested in a single in situ reaction, using in situ sequencing to demultiplex the padlock identity. The relative detection efficiency (count) and RCA yield (intensity) were quantified using a dye-labeled hybridization probe complementary to the 3’ binding site, which was common across all padlock probes. Data points are colored by either padlock length (not including the 20 nt added during the gap-fill step) or Tm of the padlock 5’ binding arm. The optimized padlock is identified by a black circle.
(C) Multiple perturbations can be delivered via separate lentiviral vectors and detected in the same cell. HeLa-TetR-Cas9 cells were sequentially transduced with CROPseq-puro and CROPseq-zeo libraries (each containing a pool of 95 sgRNAs). The in situ padlock detection protocol was the same as for a single vector library. The cumulative fraction of cells with at least N reads of different sgRNAs can be calculated. For example, a total of 70% of all cells imaged had 2 or more reads for two independent sgRNAs (heatmap position 2,2).
(D) Images were acquired at 10X magnification and could still resolve most barcode spots per cell (scale bar 200 μm; composite image of four sequencing channels; white outlines indicate segmented cells). Reads that failed to map to a known barcode (e.g., due to overlapping signal) are indicated by grey squares in the “base calls” panels of S2D.).
(A) Schematic of a frameshift reporter that converts CRISPR-Cas9-induced indel mutations into a positive fluorescent signal.
(B) The frameshift reporter was read out by microscopy in HeLa-TetR-Cas9-FR cells in the absence of a targeting sgRNA. A myc epitope tag in the original, unedited frame was stained to confirm expression levels. The reporter was found to have a very low background, with zero false positives observed among >400,000 cells (dashed line indicates threshold for defining HA+ cells).
(C) Phenotype data for the frameshift reporter screen (random subset of 10,000 cells plotted). HA+ cells were defined by thresholding based on HA-488 fluorescence intensity and pixel-wise correlation with DAPI nuclear stain.
(D) Flow analysis for the FACS-based frameshift reporter screen. Cells from the HA+ and HA- gates were sorted (top left) and sgRNA abundance was compared by NGS. Sorted cells from the HA+ (top middle) and HA- (top right) gates were re-analyzed to verify sorting accuracy. The ratio of HA+ to HA- cells in the re-analyzed populations sets an upper bound for relative sgRNA enrichment of ~300X.
(E) Schematic of image analysis pipeline showing intermediate images (LoG-transformed data, nuclei masks, cell masks max-filtered data and peak locations) and tables (summarizing bases, reads, cells and phenotypes). Example data and code provided at https://github.com/feldman4/OpticalPooledScreens.).
(A) Phenotype filtering criteria for the primary screen used to exclude mitotic cells and cells with low p65-mNeonGreen reporter expression.
(B) For both IL-1β and TNFα, primary screen gene rankings correlate well (Spearman’s p > 0.73) with rankings in validation screen of single-gene CRISPR-Cas9 knockouts. Proteasome subunits are not shown as they exhibited severe negative fitness effects in arrayed validation experiments, likely biasing the surviving cells to those with incomplete protein knockout.
(C) Translocation distributions for sgRNAs targeting UBE2N and BIRC2 across the HeLa p65-mNeonGreen reporter and HeLa, A549 and HCT116 antibody screens.
(D) Cells from the primary screen were analyzed based on morphological features. Dimensionality reduction by Principal Components Analysis grouped a subset of genes by known function in the NF-kB pathway. Genes are plotted by PCA component 1 (46% of variance explained) and component 2 (24%).
(E) PCA was carried out based on the first (Q1), second (Q2) and third (Q3) quartiles of the per-gene distribution for each morphological parameter. A percentile-based statistic was used instead of a population mean in order to minimize sensitivity to outliers while still capturing changes to the distribution. Plotted are the average values for each quartile after standardization across gene categories (color codes same as in (D)).
(F) Fraction of variance explained by the top principal components.
(G) Weights of the top principal components.
(H) Example cells randomly sampled from the dataset used in (D) and (E). DAPI nuclear stain is shown in gray and the segmented boundary for each cell is outlined (scale bar 6 μm).).
Time traces of translocation score (correlation between p65-mNeonGreen and Hoechst nuclear stain) from the live-cell validation screen. Each trace averages over all cells for a single sgRNA, with each gene represented by 10 sgRNAs. The average trace over 50 non-targeting sgRNAs is shown in grey. ).
Summary of scale and throughput of the NF-κB screen performed, as well as projections for two hypothetical screens using either low-density cell culture or a genome-scale perturbation library, assuming a cell mapping rate of 80% (typical for EF1a in HeLa cells, Figure 2E) and 600 cells screened per perturbation. The screening approach described in this study uses fully pooled protocols for library construction, cloning, and cell culture. As a result, practical limitations to the scale of optical pooled screens come mainly from the time required to image large numbers of cells and the cost of reagents for padlock detection and in situ sequencing. The key figure determining both throughput and cost is the total surface area processed, which affects the volume of reagents used and the time per sequencing cycle. The total readout time may be substantially reduced by using a two-color sequencing chemistry (e.g., Illumina MiniSeq/NextSeq) and optimized microscope hardware.
Principal reagent costs (direct) for in situ amplification of barcodes and sequencing-by-synthesis. One 6-well plate corresponds to approximately 2–4 million mapped cells for the cell lines used in this study. For example, the activation screen and static p65 screens listed in Table S1 each employed one 6-well plate.
Summary of screen metrics, including number of cells passing a phenotyping filter (e.g. non-mitotic with reporter expression above threshold) and number of those cells with ≥ 1 or 2 mapped reads.).
Spot counts and intensities for competitive padlock detection assays for LentiGuide-BC, CROPseq and CROPseq-v2.).
List of oligo sequences used for pooled cloning of perturbation libraries. Libraries of paired barcodes and sgRNAs were amplified from oligo pools and cloned into LentiGuide-BC or LentiGuide-BC-CMV (STAR Methods).
Summary of translocation defects and p-values for HeLa p65-mNeonGreen reporter screen. Each gene was assigned a score for lack of p65-mNeonGreen translocation in response to IL-1β and TNFα stimulation. Gene p-values were calculated by repeatedly sampling sets of three permuted control sgRNAs to generate gene-level null translocation scores. Genes scored as hits at estimated FDR <10% were identified by the Benjamini-Hochberg procedure (STAR Methods).
Screen ranks for p65 translocation defects and a summary table for KEGG genes detected across HeLa, A549 and HCT116 cells, color-coded based on FDR threshold for each screen (genes with FDR < 10% and rank < 100 labeled in green).
Table of translocation defects, adjusted p-values and screen ranks for all genes across HeLa, A549 and HCT116 cells. Translocation defect for each gene was calculated as the integrated difference between the translocation score distributions for an sgRNA and the non-targeting population, averaged over all sgRNAs for a gene.).
Table of integrated translocation defects and p-values for primary kinetic screen. Genes were assigned p-values based on comparing the translocation either at 45 minutes post-stimulation (as in the initial screen) or from 45 minutes to 345 minutes post-stimulation (STAR Methods).).
Example data showing a complete 10X magnification field of view with 12 cycles of sequencing, taken from the same dataset used in Fig. 2.