Abstract
DNA accessibility is thought to be of major importance in regulating gene expression. We test this hypothesis using a restriction enzyme as a probe of chromatin structure and as a proxy for transcription factors. We measured the digestion rate and the fraction of accessible DNA at almost all genomic AluI sites in budding yeast and mouse liver nuclei. Hepatocyte DNA is more accessible than yeast DNA, consistent with longer linkers between nucleosomes, suggesting that nucleosome spacing is a major determinant of accessibility. DNA accessibility varies from cell to cell, such that essentially no sites are accessible or inaccessible in every cell. AluI sites in inactive mouse promoters are accessible in some cells, implying that transcription factors could bind without activating the gene. Euchromatin and heterochromatin have very similar accessibilities, suggesting that transcription factors can penetrate heterochromatin. Thus, DNA accessibility is not likely to be the primary determinant of gene regulation.
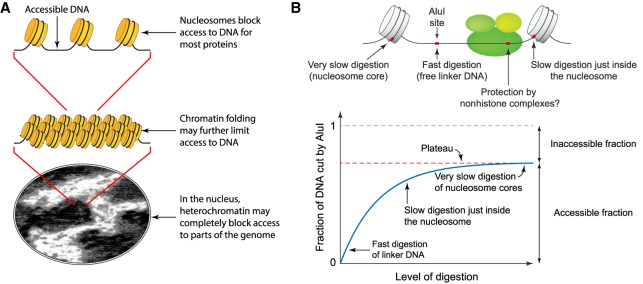
Genomic DNA is packaged into chromatin, which is composed of regularly spaced nucleosomes. Human and mouse cells contain relatively open euchromatin (similar to yeast chromatin) and extremely compact heterochromatin (Becker et al. 2016; Allshire and Madhani 2018). Most genes located in heterochromatin are completely repressed (Becker et al. 2016). Controlling the accessibility of DNA to transcription factors is thought to be of major importance in regulating gene activation, primarily through precise positioning of a nucleosome over regulatory elements such as promoters and enhancers, blocking access to transcription factors (Fig. 1A). Activation is thought to occur when an ATP-dependent chromatin remodeler removes the blocking nucleosome, allowing transcription factors to bind, although precisely how remodelers are targeted to regulatory elements is still unclear (Voss and Hager 2014; Venkatesh and Workman 2015). DNA accessibility may also be modulated at higher levels of chromatin structure (Fig. 1A): Linker histone-dependent condensation of the chromatin filament may limit access to linker DNA between nucleosomes. Furthermore, large-scale compaction may occlude transcription factors from heterochromatin domains, perhaps involving liquid droplet phase separation (Larson et al. 2017). If DNA accessibility is the primary determinant of gene regulation, then it is crucial for repression that critical regulatory elements are blocked in all cells in a population. Otherwise, there would be inappropriate gene activation in some cells.
Figure 1.
A fully quantitative assay for DNA accessibility in chromatin. (A) DNA accessibility may depend on the degree of chromatin compaction. (B) Principle of the restriction enzyme protection assay.
The accessibility model described above is appealing, but it has not yet been tested using a fully quantitative genome-wide assay. MNase-seq data are difficult to quantify because micrococcal nuclease destroys the DNA as digestion proceeds. Furthermore, nucleosomes are digested at different rates depending on the sequences they contain, resulting in apparently different relative occupancies as digestion proceeds (Chereji et al. 2017; Schwartz et al. 2019). MNase-seq data are typically normalized to the genomic average, and relative nucleosome occupancies are estimated, although they are still subject to the caveat above. Three other valuable methods—DNase-seq (John et al. 2011), ATAC-seq (which uses a transposase) (Schep et al. 2015), and RED-seq/NA-Seq (which use restriction enzymes) (Gargiulo et al. 2009; Chen et al. 2014)—report the relative accessibilities of open regions in chromatin. Small DNA fragments excised from accessible DNA sequences (typically regulatory elements) are isolated and sequenced. However, the rest of the genome is excluded from the analysis because it is still present as very long DNA molecules. Consequently, the signal and relative amounts of each accessible element depend on the extent of digestion or, in the case of ATAC-seq, on the extent of transposition, allowing only relative measurements. Because these methods sequence only the small fraction of accessible DNA fragments, they are only semiquantitative.
We have adapted the restriction enzyme protection assay to measure accessibility (Linxweiler and Hörz 1984; Fascher et al. 1990; Jack and Eggert 1990; Archer et al. 1991; Verdin et al. 1993; Wallrath and Elgin 1995; Shen et al. 2001). This assay measures both the absolute accessibility of the DNA and the rate at which accessible sites are cut. It has been used in vitro to monitor nucleosome reconstitution (Zheng and Hayes 2003), to detect nucleosome shifts (Studitsky et al. 1994), and to measure the activities of chromatin remodeling enzymes (Tsukiyama and Wu 1995). It depends on the fact that restriction enzymes are essentially unable to cut their cognate sites within a nucleosome (Linxweiler and Hörz 1984; Polach and Widom 1995). Restriction enzymes cut nucleosomal DNA 102–105 times slower than linker DNA, with faster rates for DNA just inside the nucleosome, as it is more likely to dissociate transiently from the histone octamer (Polach and Widom 1995; Chereji and Morozov 2014). These large rate differences result in a plateau in the digestion, corresponding to the fraction of accessible DNA (Fig. 1B; Supplemental Methods). Here, we use the restriction enzyme protection assay to measure the accessibility of a large number of specific sites throughout the genome in nuclei from budding yeast and mouse liver.
Results
A fully quantitative measure of DNA accessibility: qDA-seq
We used the restriction enzyme AluI to measure both the absolute DNA accessibility (i.e., the fraction of the DNA that is accessible to AluI) and the initial rate at which these accessible sites are cut. This simple method, which we term “quantitative DNA accessibility” assay (qDA-seq), involves treating nuclei with a restriction enzyme at different concentrations and sonicating the DNA into small fragments, followed by paired-end sequencing (Supplemental Fig. S1). The sonication step is necessary because the AluI digest contains many long DNA fragments derived from protected chromatin, which are not suitable for Illumina sequencing.
AluI cuts the sequence AG|CT to yield blunt ends. The yeast genome has approximately 40,000 AluI sites; the mouse genome has approximately 12.6 million sites. After sequencing, we calculate the fraction of DNA molecules cut at each AluI site as a function of AluI concentration up to ∼50 nM. The accessible fraction is measured by counting the number of DNA molecules starting or ending at a specific genomic AluI site as a fraction of all DNA molecules containing the same site. A direct comparison of accessible fractions in different cell types is possible; critically, no normalization is necessary. We note that although most of the protection observed is likely to be owing to nucleosomes, the nucleosome may not be the only complex that is resistant to restriction enzymes. Such complexes would have to be stable enough to protect an AluI site during the 20-min incubation.
DNA accessibility in yeast varies from cell to cell
To avoid potential complications owing to increased accessibility of replicating DNA, we arrested haploid yeast cells in the G1 phase of the cell cycle using α-factor. Nuclei were digested with increasing concentrations of AluI, and the expected plateau was observed at essentially all AluI sites (see below). It is important to note that only one copy of a unique genomic sequence is present in each cell because the cells are haploid. Therefore, the plateau value indicates the fraction of cells in which a particular unique AluI site is accessible. Each site is accessible in some cells and inaccessible in the other cells.
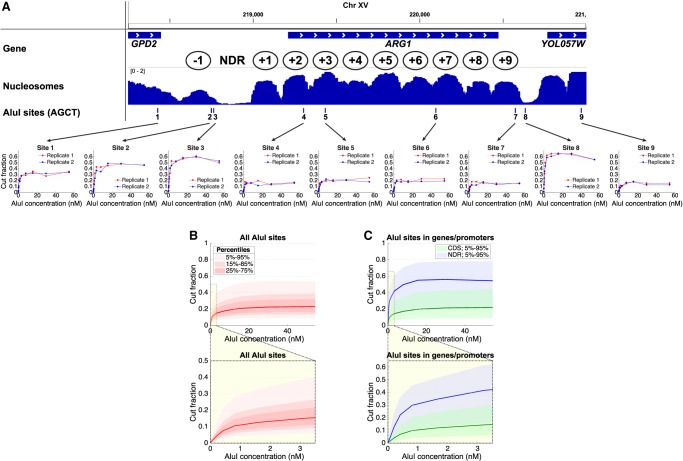
We present the ARG1 gene as an example (Fig. 2A). Digestion at an AluI site (site 2; Fig. 2A) just inside the −1 nucleosome reaches a plateau at ∼45% cut, indicating that this site is accessible in ∼45% of the cells and inaccessible in the remaining ∼55% of cells. A neighboring AluI site (site 3) located close to the upstream border of the nucleosome-depleted region (NDR) at the ARG1 promoter is more accessible, at ∼50%. In contrast, all three AluI sites in the ARG1 coding region (sites 4, 5, and 6; located within the +2, +3, and +7 nucleosomes, respectively) have lower accessibilities (∼15%–20%), suggesting that nucleosome occupancy is higher on the coding region (∼75%–80%), consistent with MNase-seq data (Fig. 2A), and indicating that these sites are accessible in only one in four or five cells. The AluI site in the YOL057W promoter downstream from ARG1 (site 8) is much more accessible, but the digestion still reaches a plateau at ∼60%, indicating that this site is protected in ∼40% of cells, even though the MNase-seq data show that it is located within a deep NDR, predicting a nucleosome occupancy close to zero (Fig. 2A). Instead, we attribute this protection to nonhistone proteins stably bound at the YOL057W promoter in ∼40% of cells (Chereji et al. 2017). A plateau is reached at all AluI sites, indicating that each site is accessible in some cells and protected in the remaining cells. We obtained similar data for ARG1 sites 5 and 8 using duplex qPCR (Supplemental Fig. S2).
Figure 2.
DNA accessibility in yeast varies from cell to cell. (A) AluI accessibility of the ARG1 gene in arrested haploid yeast cells. (Upper panel) Nucleosome occupancy (MNase-seq data) (Ocampo et al. 2016) in wild-type cells normalized to the genomic average (=1). Ovals indicate approximate nucleosome positions. (Lower panel) AluI digestion at each of nine sites (data for two independent experiments are shown). The plateau value is a measure of the fraction of cells in which the AluI site is accessible. Each site is accessible in some cells and inaccessible in the rest. (B) Digestion kinetics for all approximately 40,000 AluI sites as a function of [AluI] (11 digestion points) for haploid cells arrested with α-factor. Red line indicates median level of digestion; pink shading, data ranges. The lightest pink area includes 90% of the AluI sites (i.e., the 5%–95% data range, which excludes the 5% of AluI sites that are the least cut and the 5% of sites that are the most cut). Below the main panel: an expansion of the plot to show the initial stages of digestion. (C) Kinetics for AluI sites in gene bodies (between start and stop codons) and promoter NDRs defined using the positions of the +1 and −1 nucleosomes (Chereji et al. 2018). Blue line indicates median level of digestion in NDRs; green line, median level of digestion in gene bodies.
For genome-wide analysis of the data, we superimposed the plots for all approximately 40,000 AluI sites (Fig. 2B). A plateau is reached at a median accessibility of ∼22%, indicating that the median AluI site is inaccessible in ∼78% of cells. The data range shows that 90% of AluI sites are cut in only ∼9%–55% of cells (Fig. 2B). This observation indicates that yeast cells are very heterogeneous in DNA accessibility. In particular, we note that none of the approximately 40,000 AluI sites are accessible in >95% of cells. Moreover, of the 96 AluI sites that are apparently inaccessible in both replicate experiments (0% cut), at least 78% are sites present in the S288C genome sequence but not in our strain (which is derived from W303); that is, these AluI sites are absent in our strain owing to polymorphisms and deletions. To gain more insight, we divided the AluI sites into gene body sites and promoter sites (Fig. 2C). The median accessibility in gene bodies is very similar to that for all sites (∼20%), because gene bodies account for a very large fraction of the yeast genome. If we make the simple assumption that a nucleosome protects 147 bp of every 165 bp (the average nucleosome spacing in yeast) (Thomas and Furber 1976), the predicted protection is 89% (147/165), which is higher than observed (∼80%), suggesting that there may be some digestion just inside the nucleosome as observed in vitro (Polach and Widom 1995) or that there may be occasional gaps in the nucleosomal arrays. In the former case, a value of 80% is consistent with a protected inner nucleosome core of 132 bp (80% of 165 bp), suggesting that the outer ∼7 bp on both sides of the nucleosome is vulnerable to AluI.
The median accessibility of promoters, defined by their NDRs, is ∼53% (range, 90% of sites cut in 22%–78% of cells), which is much higher than in gene bodies and consistent with nucleosome depletion. However, digestion within the NDR still reaches a plateau, indicating the presence of stable complexes protecting the NDR in about half of the cells, presumably corresponding to nonhistone barrier complexes (Chereji et al. 2017). Estimation of AluI digestion rates at accessible sites in promoter NDRs and genes, assuming first order kinetics (see Supplemental Material), indicates that NDR sites are digested only about 1.3 times faster than linker DNA sites in gene bodies (Fig. 2B,C; Supplemental Fig. S3).
Imperfect nucleosome positioning can account for cell-to-cell heterogeneity in DNA accessibility
We plotted the mean AluI accessibility for all approximately 5000 yeast genes as a function of distance from the first (+1) nucleosome on the gene, which typically covers the transcription start site (TSS) in yeast (Fig. 3A; Mavrich et al. 2008). The extent of digestion as a function of AluI concentration is shown. In the absence of AluI, there is a background of ∼1% cut, corresponding to random fragmentation of the DNA at an AluI site by sonication. Digestion increases with increasing AluI concentration up to ∼15 nM, beyond which there is no more digestion. There is a strong peak at the NDR, with a maximum mean value of ∼55% cut. Promoters are more accessible than gene bodies, but a resistant complex is present in about half of the cells.
Figure 3.
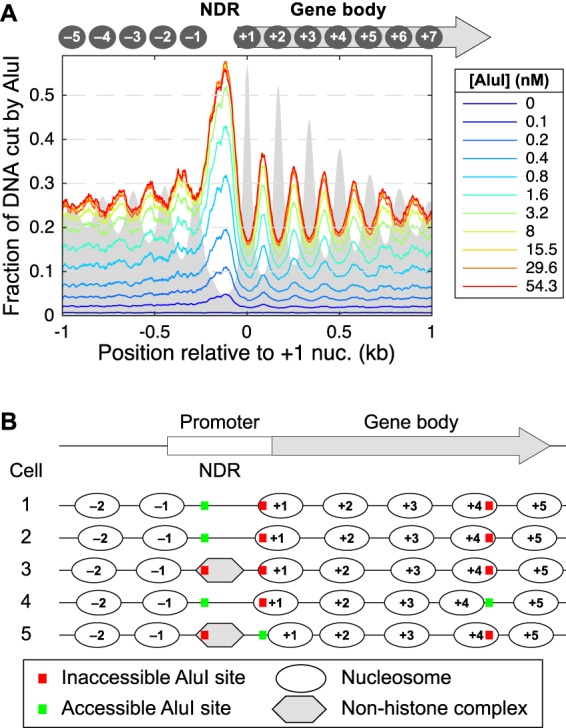
Genomic analysis of AluI accessibility reveals imperfect nucleosome phasing in yeast. (A) Mean accessibility as a function of distance from the center of the +1 nucleosome (defined by Chereji et al. 2018) on all approximately 5000 yeast genes. (B) Heterogeneous nucleosome positioning model to explain the AluI accessibility data. On a typical gene, nucleosomes are positioned slightly differently in each cell such that a particular AluI site is inside a nucleosome in one cell and in a linker in another cell. The cartoon shows the nucleosome positions on a gene in five different cells. An AluI site in the coding region is in the linker (accessible) in only one cell out of five (20% accessibility), whereas an AluI site in the promoter NDR is accessible in three out of five cells (60% accessibility). The observed average values are ∼25% in the coding region and ∼55% in the NDR (see A).
In gene bodies, an oscillatory pattern is observed around a mean value of ∼25% accessibility, anticorrelated with phased nucleosomes observed in MNase-seq data, such that the AluI peaks coincide with linkers and the AluI troughs coincide with nucleosomes. The amplitude of this oscillation provides a quantitative estimate of the degree of phasing. Perfectly phased nucleosomal arrays (i.e., each nucleosome occupies an identical position in every cell) predict 100% cutting at AluI sites in linkers (i.e., cut in all cells) and 0% cutting at nucleosomal sites (i.e., blocked in all cells). In fact, the oscillations are relatively weak: The average probability of cutting an AluI site located at the +1 nucleosome position is ∼15%, compared with ∼35% for AluI sites in linkers. Thus, the +1 nucleosome is shifted or absent in ∼15% of cells. These data can be explained by a model in which regularly spaced nucleosomes are positioned slightly differently in different cells, such that an AluI site is protected in ∼80% of cells and in an accessible linker in ∼20% of cells (Fig. 3B). Similarly, promoters are blocked by a stable complex in about half of the cells.
Heavy transcription correlates with increased DNA accessibility of yeast gene bodies
We treated exponentially growing yeast cells with 3-aminotriazole (3AT), which induces the amino acid starvation response mediated by the Gcn4 transcription factor (Hinnebusch and Natarajan 2002). We have shown previously that 3AT induces heavy transcription of ARG1, HIS4, and a few other genes, resulting in chromatin disruption and loss of nucleosome occupancy over the coding region and flanking regions (Cole et al. 2014). As expected, the AluI accessibility of the ARG1 and HIS4 gene bodies increases after 3AT treatment, whereas the accessibility of AluI sites in GAL1, which is not induced by 3AT, is unaffected (Supplemental Fig. S4). We also note that growing cells and α-factor-arrested cells have similar DNA accessibilities at the global level, suggesting that replication does not have a strong effect on global accessibility (Supplemental Fig. S4; cf. with Fig. 3A).
The mouse hepatocyte genome is more accessible than the yeast genome
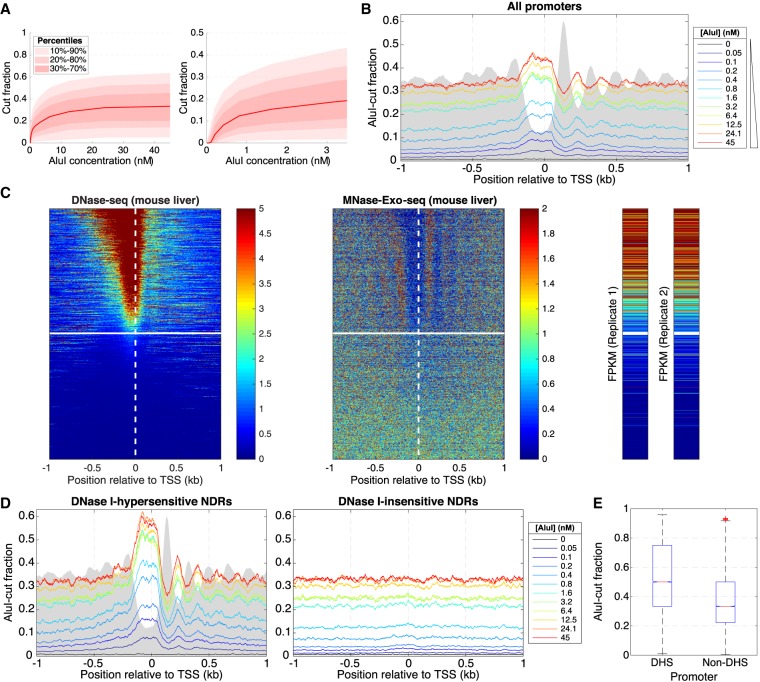
We performed the same experiment using mouse liver nuclei (Supplemental Fig. S1). AluI digestion resulted in a plateau at a median accessibility of ∼34% (Fig. 4A). Thus, the mouse hepatocyte genome is more accessible than the yeast genome (∼22%) (Fig. 2B). Although this observation seems counter-intuitive, given that the yeast genome is very active and lacks heterochromatin, it is consistent with the longer average nucleosome spacing in hepatocytes (∼195 bp) (van Holde 1989) relative to yeast (∼165 bp) (Supplemental Fig. S1). More insight is obtained by examining the chromatin structure in the vicinity of the average mouse promoter after alignment of all approximately 24,000 genes on the TSS (Fig. 4B). AluI digestion in the promoter NDR just upstream of the TSS reaches a plateau at ∼45% accessibility, which is higher than in the flanking regions (∼32% accessible/ ∼68% protected). The protection of genic DNA is consistent with a protected inner nucleosome core of 133 bp (68% of 195 bp), which is essentially the same as that observed for yeast genes (132 bp). Weak nucleosome phasing is apparent downstream.
Figure 4.
Inactive gene promoters are accessible to AluI in some mouse liver cells. (A) AluI digestion of mouse hepatocyte nuclei (12 digestion points): (left) all data; (right) initial digestion. The data range shows that 80% of AluI sites (with at least five reads) are cut in 5%–60% of cells. (B) Average AluI accessibility plotted as a function of distance from the TSS on all approximately 24,000 mouse genes. Gray area indicates MNase-Exo-seq data (nucleosome dyads) (Cole et al. 2016) on an arbitrary scale. (C) Heat map analysis of all approximately 24,000 mouse genes sorted according to the DNase I hypersensitivity of their promoters in mouse hepatocytes (data from ENCODE) and aligned on the TSS: (left) DNase I cut density; (middle) nucleosome dyad positions (Cole et al. 2016); (right) RNA-seq data (two biological replicates from ENCODE). The white line divides hypersensitive and insensitive promoters (defined in Supplemental Fig. S5). (D) Average AluI accessibility plotted as a function of distance from the TSS for DNase I–hypersensitive and DNase I–insensitive promoters defined in C. (E) Distribution of AluI-cut fractions corresponding to all AluI sites located in promoters (region [TSS − 185 bp; TSS + 85 bp]), separated by DNase I hypersensitivity.
Inactive mouse gene promoters are accessible in some cells
We sorted the genes according to the DNase I hypersensitivity of their promoters in mouse hepatocytes. This analysis revealed two classes of promoter: hypersensitive and insensitive (Fig. 4C; Supplemental Fig. S5A; Chereji and Clark 2018). After sorting using the same gene order, nucleosome positioning (MNase-Exo-seq) data (Cole et al. 2016) and hepatocyte gene expression data show that genes with hypersensitive promoters are mostly active, with an NDR and phased nucleosomes, whereas genes with DNase I–insensitive promoters are inactive, lack phasing, and have no NDR (Fig. 4C; Supplemental Fig. S5B). A finer analysis of these data, using deciles, is presented in Supplemental Figure S6.
Analysis of the AluI data indicates that active genes show better phasing and higher NDR accessibility (∼58%) than all genes (Fig. 4, cf. D and B). On the other hand, DNA accessibility on both sides of the NDR is unchanged (∼32%) (Fig. 4D). In contrast, DNase I–insensitive genes are uniformly accessible (∼32%), including promoters, with no evidence for an NDR or nucleosome phasing, consistent with the nucleosome positioning data (Fig. 4C). Although inactive promoters have a lower mean accessibility (∼32%) than active promoters (∼58%), they are at least partly accessible in approximately one in three haploid genomes (i.e., on at least one allele in half of these diploid cells). The unexpected accessibility of AluI sites in inactive promoters probably reflects the lack of nucleosome phasing, such that regularly spaced, but unphased nucleosomal arrays result in protection of promoter AluI sites in cells in which they are nucleosomal and exposure in the other cells in which they are in linker DNA (cf. Fig. 3B).
Euchromatin and heterochromatin have very similar DNA accessibilities
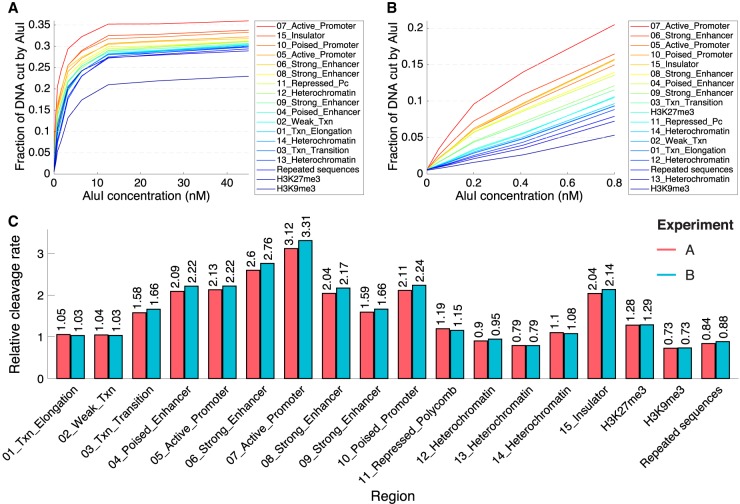
We compared the accessibilities of euchromatin and heterochromatin using a 15-state epigenetic model for mouse hepatocyte chromatin derived from histone modification patterns and ChIP-seq data for Pol II and CTCF (Fig. 5; Bogu et al. 2016). The median absolute AluI-accessible fraction is similar for all 15 chromatin states (the plateau values range from ∼29% to ∼36%), indicating that all states are accessible, including all heterochromatin states and Polycomb-repressed regions (Fig. 5A). Active promoters (states 5 and 7) and strong enhancers (states 6 and 8) were defined in the model of Bogu et al. (2016) primarily by the presence of the H3K4me1, H3K4me3, and H3K27ac histone marks and Pol II, whereas insulators were defined primarily by CTCF binding (state 15). All three of these regulatory elements are more accessible to AluI than the euchromatin and heterochromatin states, because they are short and dominated by an NDR, which has a higher average accessibility than the flanking chromatin (Fig. 4D, left). Note that the accessibility of active promoters (∼36%) (Fig. 5A) averages lower than at promoter NDRs (58%) (Fig. 4D), because the epigenetic state model includes both the NDR and its modified flanking nucleosomes. Most importantly, the curves for the euchromatin states (1 and 2), defined by the H3K36me3 mark (Bogu et al. 2016), track with those for the heterochromatin states, defined by the H3K27me3 mark (Polycomb-repressed; state 11) or by the absence of active marks (states 12–14), indicating that the differences between them are negligible (Fig. 5A).
Figure 5.
Heterochromatin and euchromatin have very similar DNA accessibilities in mouse hepatocytes. (A) AluI digestion kinetics for sites in the 15 epigenetic chromatin states in mouse hepatocytes defined by Bogu et al. (2016), for sites in repeated sequences (defined by RepeatMasker) and for sites marked by H3K9me3 (constitutive heterochromatin) or H3K27me3 (facultative heterochromatin) (data from Nicetto et al. 2019). (B) Initial rates of AluI digestion. (C) Quantitative comparison of initial digestion rates for the 15 epigenetic states defined by Bogu et al. (2016), repeated sequences, and regions marked by H3K9me3 or H3K27me3. Data for biological replicate experiments A and B are shown. See Supplemental Material for details of the analysis.
The hidden Markov model used by Bogu et al. (2016) to define the various chromatin states did not include the histone marks typical of constitutive heterochromatin (H3K9me2 and H3K9me3). We confirmed that heterochromatin has a similar accessibility to euchromatin using two independent additional analyses. First, we analyzed H3K9me3 ChIP-seq data for adult mouse hepatocytes (Nicetto et al. 2019). These data indicate that constitutive heterochromatin defined by the H3K9me3 mark has a somewhat lower but still quite similar average absolute accessibility (plateau at ∼23%) than the heterochromatin states defined by Bogu et al. (∼29%) (Fig. 5A; Bogu et al. 2016). In the case of Polycomb-repressed/-facultative heterochromatin, analysis of ChIP-seq data for H3K27me3 from the same source (Nicetto et al. 2019) indicates a very similar average absolute accessibility (∼28%) to that obtained for the same mark in the Bogu model (state 11; ∼29%) (Fig. 5A). Second, we used mouse genome annotations to determine the absolute AluI accessibilities of different annotated regions (Supplemental Fig. S7). We observed that all annotated regions have similar accessibilities except promoters (because of their NDRs, as discussed above). Repeated sequences, which are strongly enriched in constitutive heterochromatin, have similar average absolute accessibility (∼29%) to that of the heterochromatin states defined by Bogu et al. (∼29%) (Fig. 5A; Bogu et al. 2016).
Although the absolute AluI accessibilities (plateau values) are similar for euchromatin and heterochromatin, it seemed possible that they might be digested at very different rates, reflecting their very different degrees of compaction. Accordingly, we analyzed the initial AluI digestion rates for accessible sites in mouse chromatin (Fig. 5B,C). Regulatory elements containing NDRs (active promoters, insulators, and strong enhancers) are digested about three times faster than the other chromatin states (cf. yeast NDRs) (Supplemental Fig. S3). However, accessible sites in heterochromatin (states 11–14) are digested at virtually the same rate as those in euchromatin (active genes; states 1 and 2) (Fig. 5B,C). Similarly, the relative rate of AluI digestion of facultative heterochromatin as defined by Bogu et al. (2016) (state 11; approximately 1.2-fold) is very similar to that defined by H3K27me3 ChIP-seq data (approximately 1.3-fold) (Nicetto et al. 2019). The rate of digestion of constitutive heterochromatin defined by the H3K9me3 mark (approximately 0.7-fold) is a little slower than for the various heterochromatin states of Bogu et al. (2016) (states 12, 13, and 14, which range from approximately 0.8-fold to approximately 1.1-fold), but this is a very small effect. If heterochromatin really blocks accessibility, a very large difference in AluI digestion rates for euchromatin and heterochromatin is expected, but it is not observed. Thus, DNA accessibility in mouse hepatocytes does not depend strongly on epigenetic state.
Discussion
Nucleosome spacing, phasing, and DNA accessibility in chromatin
Our data indicate that access to most of the genome is blocked by nucleosomes in yeast (∼78%) and in mouse hepatocytes (∼68%), consistent with average nucleosome spacings of ∼165 bp and ∼195 bp, respectively, and a protected inner core of ∼133 bp. Thus, nucleosome spacing is the major determinant of absolute DNA accessibility. The inner nucleosome core completely blocks AluI, but because nucleosomes can occupy different positions in different cells (Shen et al. 2001; Cole et al. 2011; Small et al. 2014), all AluI sites are accessible in some cells. Thus, DNA accessibility varies from cell to cell. In yeast, most nucleosomes are phased, but nucleosome positioning is not strong enough to guarantee the inaccessibility of specific sites (Fig. 3). In mouse cells, most nucleosomes are regularly spaced but are only well positioned (phased) in the vicinity of active regulatory elements. Although inactive promoters have no NDR or phased nucleosomes, the nucleosomes are still regularly spaced, such that the probability of an AluI site being in the linker is determined by the average spacing.
The plateau in AluI digestion indicates that protection is stable during the 20-min digestion period, which is inconsistent with widespread nucleosome mobility in isolated nuclei, which would predict continued digestion if nucleosomes slide back and forth, alternately exposing and burying AluI sites. Nucleosomes may be more mobile in vivo owing to the activities of various ATP-dependent chromatin remodelers capable of moving nucleosomes. It is likely that isolation of nuclei “freezes” the chromatin structure in the absence of ATP. Nucleosome mobility in vivo would be expected to increase the accessibility of DNA in chromatin.
Accessibility of promoter NDRs
The typical promoter NDR in yeast and mouse is inaccessible to AluI in ∼40% of cells. What is the source of this protection? Some protection can be attributed to residual nucleosome occupancy, as shown by MNase-seq data. On the other hand, many NDRs are almost nucleosome free but still protected in ∼40% of cells (e.g., ARG1 site 8 in Fig. 2A). Transcription factors are unlikely to provide much protection because they typically have fast on/off kinetics, resulting in windows of opportunity for AluI. Instead, based on our previous work (Nagarajavel et al. 2013; Chereji et al. 2017), we propose that NDR protection is largely owing to stable complexes resembling the TFIIIB–TFIIIC complex found at tRNA genes, which has a stability similar to that of a nucleosome. Barrier complexes are thought to be responsible for nucleosome phasing, but their composition is unknown and may differ from one promoter to the next. Such barrier complexes must be stably bound during the 20-min digestion to explain the observed plateau. Unlike the nucleosome, which bestows almost uniform protection to the DNA wrapped around its central core, barrier complexes may contain stretches of accessible DNA. In the case of the TFIIIB–TFIIIC complex, there is an MNase-sensitive site located between TFIIIB and TFIIIC (Nagarajavel et al. 2013). Thus, the location of an AluI site within a barrier complex could determine whether it is accessible or not.
DNase I hypersensitivity, ATAC-seq, and AluI digestion at promoters
The hypersensitivity of active promoters to DNase I and the contrast between active and inactive promoters (Fig. 4C, cf. top and bottom halves of the heat map) suggest a very large difference in promoter accessibility between active and inactive promoters. In contrast, our AluI data indicate that the difference in absolute accessibility is quite small: ∼58% at the average active promoter NDR compared with ∼32% at inactive promoters (which have no NDR) (Fig. 4D). Similarly, our data also reveal that the difference in initial AluI digestion rates between active promoters and chromatin lacking NDRs is only approximately threefold (Fig. 5B,C). To reconcile these apparently different results, we note that DNase I hypersensitivity correlates with the presence of a promoter NDR (MNase-seq data) (Fig. 4C) and that DNase I data derive from short DNA fragments released at a very early stage in digestion and so are heavily enriched for open chromatin states (NDRs); the rest of the genome is not sequenced. DNA fragments from open promoters are therefore amplified relative to the rest of the genome, resulting in a large artificial difference in accessibility between active and inactive promoters. Similar considerations apply to all methods that sequence only the initial digestion products, including typical ATAC-seq experiments. In our qDA-seq method, all of the DNA fragments are sequenced. We also perform an enzyme titration to prove that the accessibility limit (plateau) has been reached, but this is not possible with DNase-seq, because DNase I is not completely blocked by nucleosomes.
More generally, the DNase I hypersensitivity and Tn5 transposase (ATAC-seq) sensitivity of a promoter depend on two factors. First is the fraction of accessible promoters: An AluI site in a specific active, open promoter is accessible in some cells but not in the others (information not provided by DNase I or ATAC-seq data). The higher this fraction is, the more DNase I or transposase cutting there would be. Second is the width of the NDR (target size): The wider the NDR, the higher the probability of DNase I or transposase cleavage, because they are nonspecific nucleases (sequence preference may be another important factor). This is clear from the heat maps in Figure 4, in which the promoters with the most DNase I cleavage are also the ones with the widest NDRs (cf. tops of the DNase I and MNase-seq heat maps). A promoter is unlikely to contain more than one AluI site, and so, the target size is a much less important factor.
Alternative methods for quantitative measurement of DNA accessibility involve the use of DNA methyltransferases. These enzymes can be used to methylate cytosines in accessible CpG or GpC dinucleotides, which can then be identified by their resistance to bisulfite conversion after sequencing (e.g., NOMe-seq [Kelly et al. 2012] or MAPit [Nabilsi et al. 2014]). The methylation pattern reveals the footprints of nucleosomes and other stably bound proteins and therefore has a much higher resolution than qDA-seq (which is limited by the distribution of restriction sites), although it is not as good as MNase-seq. This approach requires considerably more sequence coverage than qDA-seq, and the bioinformatic analysis is much more complex.
Restriction enzymes as proxies for sequence-specific transcription factors
Because restriction enzymes are sequence specific, they may be considered proxies for transcription factors. Sequence-specific transcription factors must search the DNA sequence to find their cognate binding sites. The search process is facilitated by one-dimensional diffusion of the transcription factor along the DNA in a nonspecific binding mode, with occasional dissociation and reassociation events (Halford and Marko 2004; Woringer and Darzacq 2018). Both transcription factors and restriction enzymes find their cognate sites using this type of mechanism. When a transcription factor locates a cognate site, it remains bound for a relatively long time and may recruit other factors. A restriction enzyme locates a cognate site in the same way but then cuts the DNA instead, providing a record of that binding event, which we detect and quantify in our experiment. We note that the AluI concentration range used in our experiments is in the expected range for transcription factors (up to ∼50 nM).
Mouse promoter accessibility and gene repression
The accessibility of AluI sites in inactive hepatocyte promoters implies that transcription factors can bind their cognate sites in inactive promoters in some cells, depending on whether the site is in a linker or not, and that gene inactivity is not primarily because of binding site occlusion. Thus, our data are inconsistent with a simple repression model in which positioned nucleosomes prevent transcription factors from binding their cognate sites. Although cognate sites in inactive promoters are accessible in some cells because they are located in linker DNA, an NDR is not created, suggesting that transcription factor access to DNA is insufficient for gene activation. Thus, events downstream from initial transcription factor binding determine whether a gene is active or not (Chen and Widom 2005). We propose that the key is the formation of the stable complex represented by the NDR. Pioneer factors (defined as sequence-specific transcription factors capable of binding nucleosomal sites with high affinity) (Zaret and Mango 2016) may be critical, because they have the potential to bind their sites in all cells in a population, whether or not they are occupied by a nucleosome. If so, gene activation would depend on whether the pioneer factor is expressed. However, for reasons that are unclear, some pioneer factors cannot access all of their sites in vivo (Donaghey et al. 2018). An alternative model is that the key to NDR formation may be the clustering of transcription factor binding sites at promoters and enhancers; several specific transcription factors may have to be expressed and bind in concert to form an NDR before a gene can be activated. In this cooperative multisite model (Adams and Workman 1995; Mirny 2010), single-factor binding events at cognate sites in linker DNA would not be sufficient for NDR formation; all of the factors involved would need to be expressed to initiate NDR formation and gene activation.
Heterochromatin and euchromatin have similar DNA accessibilities
Our data indicate that heterochromatin is not generally less accessible than euchromatin. This conclusion is consistent with a recent quantitative analysis of MNase-seq data for human cells (Schwartz et al. 2019). We note that some accessibility is expected given that constitutive heterochromatin must be transcribed to produce the RNA required for its repression (Grewal 2010). Our data indicate that transcription factors would be expected to penetrate heterochromatin, even in its extremely compact state. However, the size of the transcription factor may be critical. Although AluI, which is a monomer with a relative molecular mass of about 46,000, is similar in size to many transcription factors, theoretical modeling suggests that much larger complexes may be excluded from compact heterochromatin (Maeshima et al. 2015). This is a distinct possibility given that many transcription factors are associated with large complexes.
Methods
AluI digestion of yeast nuclei
Yeast strain YDC111 (MATa ade2-1 can1-100 leu2-3,112 trp1-1 ura3-1) (Kim et al. 2006) was grown at 30°C in synthetic complete (SC) medium to A600 = ∼0.2 and arrested in G1 by addition of α-factor (FDA Core Facility) to 10 µg/mL. Arrest was monitored by observing the appearance of the “shmoo” phenotype in a light microscope. After 2 h, the cells were harvested by filtration and stored at −80°C. Spheroplasts were prepared from ∼100 A600 units of cells in 15 mL SM buffer (SC medium with 1 M sorbitol, 50 mM Tris-HCl at pH 8.0, 20 mM 2-mercaptoethanol) by digestion with ∼26,000 units of lyticase (Sigma-Aldrich, L-2524) for ≤5 min at 30°C. Digestion of the cell wall was monitored by measuring the A600 of 30 µL cell suspension in 1 mL 1% SDS and was considered complete when the A600 decreased to <10% of the initial value. For 3AT experiments, YDC111 cells were grown to midlog phase at 30°C either in SC medium lacking histidine, followed by addition of 3AT (Sigma-Aldrich, 61-82-5) to 10 mM for 20 min, or in SC medium (control) and stored as above. Spheroplasting of 3AT-treated cells was performed in SM medium lacking histidine. Spheroplasts were centrifuged in a precooled Sorvall SA600 rotor (7500 rpm for 5 min at 4°C) and washed once with 25 mL cold ST buffer (1 M sorbitol, 50 mM Tris-HCl at pH 8.0). Spheroplasts were lysed by resuspension in 20 mL cold F buffer (18% w/v Ficoll-PM400 [GE Healthcare 17-0300-50], 40 mM potassium phosphate [pH 6.5], 1 mM magnesium chloride; protease inhibitors [Roche 05056489001] and 5 mM 2-mercaptoethanol were added just before use). The lysate was applied to a step gradient of 15 mL cold FG buffer (7% w/v Ficoll-PM400, 20% glycerol, 40 mM potassium phosphate [pH 6.5], 1 mM magnesium chloride, with protease inhibitors and 5 mM 2-mercaptoethanol as above) and centrifuged in an SA600 rotor (12,500 rpm for 20 min at 4°C). The pellets (crude nuclei) were resuspended in 4.4 mL prewarmed AluI digestion buffer (10 mM HEPES at pH 7.5, 35 mM NaCl, 5 mM MgCl2, with protease inhibitors and 5 mM 2-mercaptoethanol) and divided into 11 400-µL aliquots. AluI (New England Biolabs, R0137 at 0.015 mg/mL; MW = 46,000) was added (0, 1, 2.5, 5, 10, 20, 40, 100, 200, 400, 800 units), mixed thoroughly, and incubated for 20 min at 25°C. Digestion was stopped by adding 50 µL 90 mM EDTA, 9% SDS. Aliquots (180 µL) were removed from each digest to ascertain the level of digestion; the remainders were stored at −20°C before sonication. For gel analysis, the DNA was purified by addition of 10 µL 20% SDS, mixing, and the addition of 50 µL 5 M potassium acetate, followed by two extractions with an equal volume of chloroform, precipitation with 0.7 vol. isopropanol, and one wash with 75% ethanol. The purified DNA was dissolved in 20 µL 10 mM Tris-HCl (pH 8.0), 0.1 mM EDTA, 0.5 mg/mL RNase A and incubated at 37°C for 1 h. The DNA was analyzed in a 1% agarose gel stained with SYBR Gold (Invitrogen S11494). For sonication, the samples were adjusted to 450 µL with 180 µL AluI digestion buffer, transferred to 15-mL Sumilon TPX tubes (Diagenode C30010009), and sonicated using a Diagenode bioruptor 300 at 4°C and high power: 20 cycles of 30 sec on and 30 sec off. The DNA was purified, treated with RNase as above, purified again using QIAGEN PCR purification columns (QIAGEN, 28106), and eluted in 50 µL 10 mM Tris-HCl at pH 8.0, 0.1 mM EDTA (TE[0.1]). Concentrations were determined by measuring A260. The degree of sonication was checked by analysis in a 2% agarose gel stained with SYBR Gold; DNA sizes ranged from ∼100 to ∼700 bp. Before library preparation, the DNA was treated with repair enzymes (New England Biolabs, PreCR kit M0309) and purified using QIAGEN PCR purification columns as above.
AluI digestion of mouse liver nuclei
Livers were dissected from pregnant (E13.5) female NIH/Swiss mice and stored at −80°C. For each experiment, a liver was thawed on ice and gently disrupted in a glass homogenizer containing 12 mL cold buffer A per gram liver (buffer A: 0.34 M sucrose, 60 mM KCl, 15 mM NaCl, 15 mM Tris-HCl at pH 8.0, 0.5 mM spermidine-HCl, 0.15 mM spermine, 1 mM Na-EDTA, 15 mM 2-mercaptoethanol, and protease inhibitors as above). The homogenate was filtered through four layers of cheesecloth into a 50-mL tube on ice. Crude nuclei were collected by applying the filtrate to two 4-mL step gradients of buffer A with 1 M sucrose in 15-mL tubes and spinning in a Sorvall SA600 rotor at 12,500 rpm for 15 min at 4°C. The supernatants were decanted, and solid material on the tube sides was removed with a tissue. The nuclei were washed by gentle resuspension of both pellets in a total of 5 mL buffer A and centrifuged for 5 min as above. The supernatant was removed, the pellet resuspended in 1 mL buffer A, and placed on ice. The DNA concentration was estimated by measuring the A260 of 2 µL nuclei in 1 mL 1 M NaOH. The volume of nuclei corresponding to 50 A260 units was transferred to a 1.5-mL microfuge tube and centrifuged for 5 min at 14,000 rpm at 4°C. The supernatant was removed, and the nuclei were resuspended in 1 mL mouse AluI digestion buffer (0.34 M sucrose, 60 mM KCl, 15 mM NaCl, 15 mM Tris-HCl at pH 8.0, 5 mM MgCl2, 15 mM 2-mercaptoethanol, with protease inhibitors as above). The A260 of the diluted nuclei (10 µL) was measured as above. Twelve aliquots of carefully resuspended nuclei, each containing 20 µg DNA in 200 µL digestion buffer, were titrated with AluI as follows: 0, 0.3, 0.6, 1.3, 2.5, 5, 10, 20, 40, 80, 160, 320 units. The samples were mixed gently but thoroughly with a 1-mL pipette and incubated for 20 min at 37°C. Digestion was stopped by adding 200 µL 2% SDS, 20 mM EDTA, 10 mM Tris-HCl (pH 8.0); mixing thoroughly; and incubating for 40 min at room temperature to ensure complete protein removal from DNA. The extent of digestion was determined by analysis of DNA purified from 50 µL of each digest in an agarose gel, after RNase treatment as above. The remaining 350 µL was stored at −20°C before sonication. The samples were warmed to room temperature to dissolve precipitated SDS; the volumes were adjusted to 450 µL with 100 µL 10 mM TE(0.1) and sonicated as above. The DNA was purified as above and dissolved in 45 µL 50 mM Tris-HCl (pH 8.0), 5 mM EDTA, 0.1 mg/mL RNase A and incubated for 1 h at 37°C. The salt concentration was adjusted by addition of 5 µL 10× NEB buffer 4, and the DNA was purified using QIAGEN PCR columns. DNA was eluted in 50 µL TE(0.1). Concentrations were measured by A260. The degree of sonication was checked by analysis in a 2% agarose gel; DNA sizes ranged from ∼100 to ∼700 bp.
Illumina paired-end library preparation
The Illumina paired-end adaptor was ligated to ∼500 ng purified sonicated AluI-digested DNA using the NEBNext ultra DNA library kit for Illumina (New England Biolabs, E7370) according to the manufacturer's instructions. The ligated DNA samples were purified without size selection using AMPure XP beads at a 1:1 ratio (Beckman, A63880). The DNA (50–100 ng) was amplified using the Phusion Hi-Fi PCR master mix with HF buffer (New England Biolabs, M0531) or the Q5 Hot Start HiFi PCR master mix (7–10 cycles; New England Biolabs, E6625AA). Library quality was checked in an agarose gel. Sequencing was performed using either an Illumina HiSeq 2500 or an Illumina NextSeq 500.
Bioinformatics and data analysis
Paired-end reads were aligned to the Saccharomyces cerevisiae reference genome sacCer3 or to the Mus musculus reference genome mm10, using Bowtie 2 (Langmead and Salzberg 2012) with the parameters -X 5000 --very-sensitive, to map sequences up to 5 kb with maximum accuracy. Coverage information is given in Supplemental Figure S8. For every AluI motif (AGCT) found in the genome, we estimated the fraction of nuclei in which the given motif was cleaved by AluI, fcut = Ncut/(Ncut + Nuncut), by counting the number of reads that were cut at this site (with an end at this site), Ncut, and the number of reads that were not cut at this site (overlapping the motif), Nuncut, using the Bioinformatics toolbox from MATLAB. The fact that a cut site will generate two fragment ends was accounted for as follows: AluI cleavage produces a left fragment ending with AG and a right fragment starting with CT. For every site, the number of fragments ending at AG, , the number of fragments starting at CT, , and the number of fragments containing intact sites, Nuncut, were counted. Two estimations of the cut fraction, and , are obtained, which theoretically should be equal. Because we sequenced 50 nt from both ends of the DNA fragments, we discarded the AluI sites that were separated by <50 bp apart, as the reads originating from cleavages at both sites were underrepresented in the properly aligned reads. Duplicate reads were not removed for two reasons: (1) They occur at low frequency in the sonicated “0 nM AluI” controls (∼2% of reads in the yeast data after exclusion of the rDNA, for which the probability of genuine duplicate reads is much higher than for unique sequences), and (2) the probability of a genuine duplicate read corresponding to fragments cut with AluI at both ends is much higher than random (∼9% of reads in the most digested AluI samples [54 nM] are duplicates; rDNA excluded), and it is therefore important not to remove duplicates to avoid biasing the results. We also note that the probability of a genuine duplicate increases with the number of reads. GEO database data used are MNase-Exo-seq (GSE65889), DNase-seq (GSM1014183 in GSE37074), and RNA-seq (GSM2071423 and GSM2071424 in GSE78391). The theoretical framework for modeling the kinetics of restriction enzyme digestion is presented in Supplemental Methods (in “Restriction enzyme digestion kinetics”; Supplemental Figs. S9, S10).
Data access
The sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE115693. Custom MATLAB code for analyzing AluI cleavage data is available at GitHub (https://github.com/rchereji/AluI_accessibility_analysis) and as Supplemental Code.
Supplementary Material
Acknowledgments
We thank Gary Felsenfeld, Gordon Hager, Alan Hinnebusch, Tom Johnson, and Diego Presman for comments on the manuscript; Valya Russanova for technical support; and Phil Lee, Will Huffman, and Doug Fields for mouse livers. We thank the NHLBI Sequencing Core Facility (Yan Luo, Poching Liu, and Yuesheng Li) for paired-end sequencing. This study used the computational resources of the NIH HPC Biowulf cluster. This work was supported by the Intramural Research Program of the National Institutes of Health (NICHD).
Author contributions: The study was designed by R.V.C., J.O., P.R.E., and D.J.C. Experiments were performed by J.O., P.R.E., H.K.P., and D.J.C. R.V.C. wrote the scripts and performed the bioinformatic analysis. Writing, reviewing, and editing were done by R.V.C., J.O., P.R.E., and D.J.C.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.249326.119.
References
- Adams CC, Workman JL. 1995. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol Cell Biol 15: 1405–1421. 10.1128/MCB.15.3.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Madhani HD. 2018. Ten principles of heterochromatin formation and function. Nat Rev Mol Cell Biol 19: 229–244. 10.1038/nrm.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer TK, Cordingley MG, Wolford RG, Hager GL. 1991. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol 11: 688–698. 10.1128/MCB.11.2.688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JS, Nicetto D, Zaret KS. 2016. H3K9me3-dependent heterochromatin: barrier to cell fate changes. Trends Genet 32: 29–41. 10.1016/j.tig.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogu GK, Vizán P, Stanton LW, Beato M, Di Croce L, Marti-Renom MA. 2016. Chromatin and RNA maps reveal regulatory long noncoding RNAs in mouse. Mol Cell Biol 36: 809–819. 10.1128/MCB.00955-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Widom J. 2005. Mechanism of transcriptional silencing in yeast. Cell 120: 37–48. 10.1016/j.cell.2004.11.030 [DOI] [PubMed] [Google Scholar]
- Chen PB, Zhu LJ, Hainer SJ, McCannell KN, Fazzio TG. 2014. Unbiased chromatin accessibility profiling by RED-seq uncovers unique features of nucleosome variants in vivo. BMC Genomics 15: 1104 10.1186/1471-2164-15-1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereji RV, Clark DJ. 2018. Major determinants of nucleosome positioning. Biophys J 114: 2279–2289. 10.1016/j.bpj.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereji RV, Morozov AV. 2014. Ubiquitous nucleosome crowding in the yeast genome. Proc Natl Acad Sci USA 111: 5236–5241. 10.1073/pnas.1321001111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereji RV, Ocampo J, Clark DJ. 2017. MNase-sensitive complexes in yeast: nucleosomes and non-histone barriers. Mol Cell 65: 565–577.e3. 10.1016/j.molcel.2016.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereji RV, Ramachandran S, Bryson TD, Henikoff S. 2018. Precise genome-wide mapping of single nucleosomes and linkers in vivo. Genome Biol 19: 19 10.1186/s13059-018-1398-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole HA, Howard BH, Clark DJ. 2011. Activation-induced disruption of nucleosome position clusters on the coding regions of Gcn4-dependent genes extends into neighbouring genes. Nucleic Acids Res 39: 9521–9535. 10.1093/nar/gkr643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole HA, Ocampo J, Iben JR, Chereji RV, Clark DJ. 2014. Heavy transcription of yeast genes correlates with differential loss of histone H2B relative to H4 and queued RNA polymerases. Nucleic Acids Res 42: 12512–12522. 10.1093/nar/gku1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole HA, Cui F, Ocampo J, Burke TL, Nikitina T, Nagarajavel V, Kotomura N, Zhurkin VB, Clark DJ. 2016. Novel nucleosomal particles containing core histones and linker DNA but no histone H1. Nucleic Acids Res 44: 573–581. 10.1093/nar/gkv943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaghey J, Thakurela S, Charlton J, Chen JS, Smith ZD, Gu H, Pop R, Clement K, Stamenova EK, Karnik R, et al. 2018. Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat Genet 50: 250–258. 10.1038/s41588-017-0034-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fascher KD, Schmitz J, Hörz W. 1990. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J 9: 2523–2528. 10.1002/j.1460-2075.1990.tb07432.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargiulo G, Levy S, Bucci G, Romanenghi M, Fornasari L, Beeson KY, Goldberg SM, Cesaroni M, Ballarini M, Santoro F, et al. 2009. NA-Seq: a discovery tool for the analysis of chromatin structure and dynamics during differentiation. Dev Cell 16: 466–481. 10.1016/j.devcel.2009.02.002 [DOI] [PubMed] [Google Scholar]
- Grewal SI. 2010. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev 20: 134–141. 10.1016/j.gde.2010.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford SE, Marko JF. 2004. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res 32: 3040–3052. 10.1093/nar/gkh624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Natarajan K. 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryotic Cell 1: 22–32. 10.1128/EC.01.1.22-32.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack RS, Eggert H. 1990. Restriction enzymes have limited access to DNA sequences in Drosophila chromosomes. EMBO J 9: 2603–2609. 10.1002/j.1460-2075.1990.tb07442.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung M-H, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. 2011. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet 43: 264–268. 10.1038/ng.759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TK, Liu Y, Lay FD, Liang G, Berman BP, Jones PA. 2012. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res 22: 2497–2506. 10.1101/gr.143008.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, McLaughlin N, Lindstrom K, Tsukiyama T, Clark DJ. 2006. Activation of Saccharomyces cerevisiae HIS3 results in Gcn4p-dependent, SWI/SNF-dependent mobilization of nucleosomes over the entire gene. Mol Cell Biol 26: 8607–8622. 10.1128/MCB.00678-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ. 2017. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547: 236–240. 10.1038/nature22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linxweiler W, Hörz W. 1984. Reconstitution of mononucleosomes: characterization of distinct particles that differ in the position of the histone core. Nucleic Acids Res 12: 9395–9413. 10.1093/nar/12.24.9395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima K, Kaizu K, Tamura S, Nozaki T, Kokubo T, Takahashi K. 2015. The physical size of transcription factors is key to transcriptional regulation in chromatin domains. J Phys Condens Matter 27: 064116 10.1088/0953-8984/27/6/064116 [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18: 1073–1083. 10.1101/gr.078261.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA. 2010. Nucleosome-mediated cooperativity between transcription factors. Proc Natl Acad Sci USA 107: 22534–22539. 10.1073/pnas.0913805107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabilsi NH, Deleyrolle LP, Darst RP, Riva A, Reynolds BA, Kladde MP. 2014. Multiplex mapping of chromatin accessibility and DNA methylation within targeted single molecules identifies epigenetic heterogeneity in neural stem cells and glioblastoma. Genome Res 24: 329–339. 10.1101/gr.161737.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajavel V, Iben JR, Howard BH, Maraia RJ, Clark DJ. 2013. Global ‘bootprinting’ reveals the elastic architecture of the yeast TFIIIB–TFIIIC transcription complex in vivo. Nucleic Acids Res 41: 8135–8143. 10.1093/nar/gkt611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicetto D, Donahue G, Jain T, Peng T, Sidoli S, Sheng L, Montavon T, Becker JS, Grindheim JM, Blahnik K, et al. 2019. H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science 363: 294–297. 10.1126/science.aau0583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo J, Chereji RV, Eriksson PR, Clark DJ. 2016. The ISW1 and CHD1 ATP-dependent chromatin remodelers compete to set nucleosome spacing in vivo. Nucleic Acids Res 44: 4625–4635. 10.1093/nar/gkw068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polach KJ, Widom J. 1995. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol 254: 130–149. 10.1006/jmbi.1995.0606 [DOI] [PubMed] [Google Scholar]
- Schep AN, Buenrostro JD, Denny SK, Schwartz K, Sherlock G, Greenleaf WJ. 2015. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res 25: 1757–1770. 10.1101/gr.192294.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz U, Németh A, Diermeier S, Exler JH, Hansch S, Maldonado R, Heizinger L, Merkl R, Längst G. 2019. Characterizing the nuclease accessibility of DNA in human cells to map higher order structures of chromatin. Nucleic Acids Res 47: 1239–1254. 10.1093/nar/gky1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CH, Leblanc BP, Alfieri JA, Clark DJ. 2001. Remodeling of yeast CUP1 chromatin involves activator-dependent repositioning of nucleosomes over the entire gene and flanking sequences. Mol Cell Biol 21: 534–547. 10.1128/MCB.21.2.534-547.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small EC, Xi L, Wang J-P, Widom J, Licht JD. 2014. Single-cell nucleosome mapping reveals the molecular basis of gene expression heterogeneity. Proc Natl Acad Sci USA 111: E2462–E2471. 10.1073/pnas.1400517111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studitsky VM, Clark DJ, Felsenfeld G. 1994. A histone octamer can step around a transcribing polymerase without leaving the template. Cell 76: 371–382. 10.1016/0092-8674(94)90343-3 [DOI] [PubMed] [Google Scholar]
- Thomas JO, Furber V. 1976. Yeast chromatin structure. FEBS Lett 66: 274–280. 10.1016/0014-5793(76)80521-2 [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. 1995. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell 83: 1011–1020. 10.1016/0092-8674(95)90216-3 [DOI] [PubMed] [Google Scholar]
- van Holde KE. 1989. Chromatin. Springer New York, New York. [Google Scholar]
- Venkatesh S, Workman JL. 2015. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol 16: 178–189. 10.1038/nrm3941 [DOI] [PubMed] [Google Scholar]
- Verdin E, Paras P, Van Lint C. 1993. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J 12: 3249–3259. 10.1002/j.1460-2075.1993.tb05994.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, Hager GL. 2014. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet 15: 69–81. 10.1038/nrg3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath LL, Elgin SC. 1995. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev 9: 1263–1277. 10.1101/gad.9.10.1263 [DOI] [PubMed] [Google Scholar]
- Woringer M, Darzacq X. 2018. Protein motion in the nucleus: from anomalous diffusion to weak interactions. Biochem Soc Trans 46: 945–956. 10.1042/BST20170310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Mango SE. 2016. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev 37: 76–81. 10.1016/j.gde.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Hayes JJ. 2003. Intra- and inter-nucleosomal protein–DNA interactions of the core histone tail domains in a model system. J Biol Chem 278: 24217–24224. 10.1074/jbc.M302817200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.