Abstract
Ion mobility spectrometry (IMS) is a widely used analytical technique providing rapid gas phase separations. IMS alone is useful, but its coupling with mass spectrometry (IMS–MS) and various front-end separation techniques has greatly increased the molecular information achievable from different omic analyses. IMS–MS analyses are specifically gaining attention for improving metabolomic, lipidomic, glycomic, proteomic and exposomic analyses by increasing measurement sensitivity (e.g. S/N ratio), reducing the detection limit, and amplifying peak capacity. Numerous studies including national security-related analyses, disease screenings and environmental evaluations are illustrating that IMS–MS is able to extract information not possible with MS alone. Furthermore, IMS–MS has shown great utility in salvaging molecular information for low abundance molecules of interest when high concentration contaminant ions are present in the sample by reducing detector suppression. This review highlights how IMS–MS is currently being used in omic analyses to distinguish structurally similar molecules, isomers, molecular classes and contaminant ions.
Keywords: Ion Mobility Spectrometry, Mass Spectrometry, Omics, Proteomics, Lipidomics, Metabolomics, Glycomics, Exposomics
1. Introduction
Mass spectrometry (MS)-based analyses have become one of the most informative methods for studying complex mixtures in the 21st century. However, evaluating complex biological and environmental systems with MS-based measurements can be extremely challenging due to many molecule types (e.g. proteins, lipids, small molecules, metals, etc.) present in the different samples and the wide range of concentrations they exist at. To address these challenges and enable more effective MS measurements, advanced sample preparation techniques are often employed to separate or fractionate the diverse molecular classes. These methods included biphasic and triphasic extractions, depletion of the most abundant proteins in blood plasma and serum analyses, and extensive solid phase extractions to remove salts and clean up the samples prior to their injection into the MS platform. Unfortunately, even the most advanced procedures are not always able to completely clean up the samples and limit the detector suppression that can occur when highly concentrated species are present with those at much lower abundance. Isomeric molecules of the same elemental composition and similar structure can even more challenges for analyses when they cannot be distinguished by their fragmentation pattern or with chromatography. Ion mobility spectrometry (IMS) is therefore being increasingly utilized to address some of these limitations.
Over the last decade, the use of IMS in analytical measurements has rapidly increased with applications in national security-related analyses, patient screening, environmental monitoring and numerous other areas (1–5). IMS is a gas phase technique that provides rapid structural separations based on the balance of two forces that impact the movement of an ion, the electric field and the drag force from the collision with buffer gas molecules (6). In IMS, compounds of different sizes, shapes, and charges are separated as they travel through a buffer gas. Variations on the electric field and stationary state of the buffer gas have given rise to multiple IMS-based platforms. Drift tube IMS (DTIMS) (7–9), traveling wave IMS (TWIMS) (10), trapped IMS (TIMS) (11), field asymmetric IMS (FAIMS; also called differential mobility spectrometry (DMS) or differential ion mobility spectrometry (DIMS)) (12, 13) and differential mobility analyzers (DMA) (14, 15) are five of the most common forms of IMS used in current analyses of complex samples. DTIMS and TWIMS allow all ions to be analyzed simultaneously in the measurements, while TIMS, FAIMS and DMA devices are scanned to evaluate specific ions or classes separately. Additionally, the measured mobilities for molecular species in DTIMS, TWIMS, TIMS and DMA can be converted into rotationally averaged collision cross sections (CCS) (16, 17), which provide useful descriptors of the corresponding ions’ 3- dimensional gas-phase structures under the given experimental conditions. In DTIMS and DMA, CCS values are calculated directly from the measured arrival times to provide structural insight and remove pressure, temperature and length dependencies. However, calibration procedures for the CCS values are required with TWIMS and TIMS measurements. Due to the alternating high and low fields used in FAIMS, no current approach exists to allow CCS values to be obtained, but the use of alternating high and low fields does allow FAIMS to separate species that presently challenge other IMS methods.
DTIMS, TWIMS, TIMS, DMA and FAIMS have all shown great utility in separating molecular classes and interfering ions of various forms. Additionally, they can be interfaced with mass spectrometers (18) to permit simultaneous acquisition of IMS structural and MS mass information on a rapid millisecond to second timescale. The CCS values from IMS separations are then used to support molecular identifications from the IMS–MS analyses as they provide structural insight. Although CCS and m/z are not truly orthogonal identifiers due to some correlation between the two characteristics, different molecular classes still exhibit distinct ‘trend lines’. This allows potential insight to be garnered about unknown analytes based on their position in the m/z versus CCS trend line space (19). Molecular dynamics and density function theory (DFT) candidate structures have also been utilized for comparison of the experimental CCS values and showed excellent agreement with errors often <2%. This agreement is due to the actual molecule properties such as bond types, charge position and molecular compaction, which can used be theoretically predict structures and further explain the different trend lines. Since IMS is able to quickly separate distinct chemical species such as carbohydrates, peptides, lipids, and organic material from other components, it enables better analyses for the molecule of choice in complex samples (Figure 1) (19, 20). These measurements can then be used to quickly screen conditions while gathering information about the classes of molecular species present, differentiating molecular isomers and separating contaminates from the compounds of interest. Furthermore, these separations can be coupled with liquid chromatography (LC) separations, enabling multi-dimensional measurements that address highly complex samples without increasing the analysis time (21–23). The additional IMS separation can also be used to significantly reduce the LC separation times while maintaining the depth of coverage characteristic of much longer traditional LC-MS analyses (24). Examples of how IMS has been used to separate molecular structures, isomers, chemical classes and reduce detector suppression caused by contaminant ions are illustrated in this review to show its utility in omic measurements.
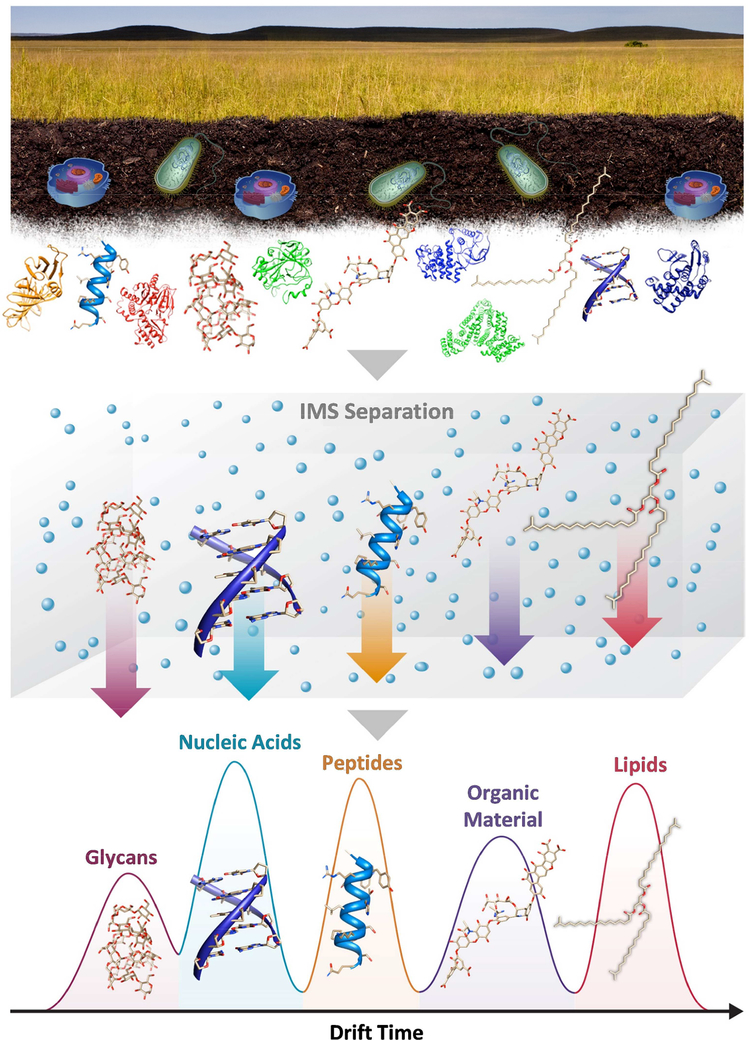
Figure 1.
The various molecule types present in environmental and biological samples greatly hinder comprehensive MS measurements. IMS provides a way of separating these molecule types such as glycans, nucleotides, peptides, organic material and lipids based on their structures. In this IMS evaluation, glycans traversing the IMS drift cell fastest due to their ring-based structures, while lipids are slowest due to their rigid linear backbone.
2. IMS–MS in Proteomic Analyses
In proteomics, measurements with high throughput and sensitivity are essential for improving the characterization of complex mixtures in both discovery and validation studies. Proteomic measurements are typically divided into bottom-up, middle-down and top-down approaches. In top-down approaches, intact proteins are separated, detected and identified; whereas both bottom-up and middle-down strategies utilize digestion, where the intact proteins are broken down and identified by their characteristic peptides. Middle-down and bottom-up approaches are differentiated based on the endopeptidase used in the digestion, as middle-down analyses often utilize endopeptidase that results in larger peptides (e.g. Lys–C or OMP–T) than bottom-up techniques which commonly utilize trypsin or pepsin.
Since IMS–MS provides relative structural information not easily obtained using MS alone, it has been utilized in top down proteomic studies since the 1990s to investigate structural changes in proteins that could result in different diseases (e.g. misfolding and aggregation) (25–27). IMS–MS has been specifically used to study intact proteins related to intrinsically disordered proteins implicated in neurodegenerative diseases like Alzheimer’s (amyloid–β protein) and Parkinson’s (α–synuclein protein) (28–30). The urge to understand how native protein structures change upon stress is also promoting collision induced unfolding (CIU) studies, where energy is imparted on proteins or protein complexes to measure their structural changes (31). In most CIU analyses, proteins tend to adopt compact conformations at the lowest energies, however, as the energy is increased the proteins and complexes slowly begin to unfold. Recent examples of CIU have probed the conformational stability of protein complexes and protein-ligand interactions (32, 33), serving as an invaluable tool in the repertoire of structural biology techniques.
In both bottom-up and middle-down approaches, extensive LC times (often >100 minutes) and high-resolution mass spectrometers with tandem MS capabilities are used to distinguish the multiply charged isomeric species. However, even these LC–MS/MS methodologies still lack the ability to fully resolve all peptide components, especially when many of these important components are at low abundance. Performing LC–MS analyses provides obvious benefits in improving the resolution of proteomic measurements without sacrificing analysis time and even has the capability to speed up the LC times (24, 34). Furthermore, IMS enables a way of separating peptide and contaminant ions if they are extracted together and cannot be removed using cleanup methods such as when DTIMS was utilized to separate CHAPS and SDS from peptides of interest (35, 36). FAIMS devices have also shown utility in separating 1+ contaminant ions from higher charge state proteins and peptides of interest so to not fill up the linear ion trap or orbitrap (37). This capability has been extremely important in avoiding biases in trap-based mass analyzers, which require automated gain control (AGC). Specifically, AGC is used in linear ion traps and orbitraps to limit the number of ion charges accumulated to prevent excessive space charge effects and allow high measurement accuracy to be achieved. However, any high concentration contaminant causes the AGC to greatly reduce trapping time so that lower abundance ions that might be of more interest do not have enough time to collect in the trap and provide a reasonable signal for detection. LC-IMS- MS measurements have also greatly aided the analysis of complex environmental water, soil, and plant material samples which have many diverse types of molecular contaminants (36). In processing these environmental samples, organic material is typically extracted with the proteins. The three-dimensional measurements are therefore particularly useful for separating the peptides from the high concentrations of organic material (e.g., humic acid substances in soil and polyphenols in plants), natural contaminants (e.g., abundant salts or polymers), and detergents (35, 36). The IMS separation is able to move the organic material to a different arrival time area to greatly improve the coverage of environmental samples (Figure 2). LC–IMS–MS proteomic measurements have also been used to reduce the number of false positives resulting from complex samples due to the extra dimension for. Characterization(38).Advantages such as higher sensitivity and throughput than LC-MS measurements alone have also been noted in LC-IMSMS studies (39, 40).
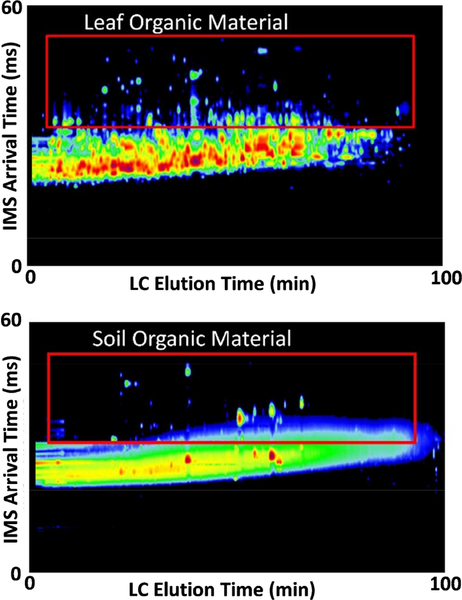
Figure 2.
The LC and IMS separation of the organic material contaminant from peptides in an Arabidopsis leaf (top) and a soil sample (bottom). The IMS separation was able to move most of the organic material to a different drift space due to the different molecular structures. All molecular species are normalized to the highest concentration molecules in these plots, illustrating that the leaf peptides did not have as much interference from the organic material as the soil did.
The ability of LC-IMS–MS to provide high throughput sensitive analyses and separate peptides from contaminants that can arise from glassware and the multiple proteomic extraction steps is also extremely important in biological studies. In one study (41), plasma was collected from Ebola patients in a field biosafety level-4 laboratory (BSL–4) setting in the affected area of Sierra Leone. During the immunodepletion and viral inactivation steps of Ebola infected plasma samples, polymers were unavoidably introduced causing problems for ion trap-based MS platforms. In the analyses, the AGC function of the QExactive quickly terminated the accumulation of ions due to signal from the polymers dominating the samples. However, during the corresponding LC–IMS–MS analyses of the same samples, the IMS separation moved the polymers to a different spectral region. Since TOF instruments do not require an AGC due to their larger charge capacity, they do not have the space-charge issues that can be observed in trap instruments. However, during IMS separations when the ions are bunched in the axial direction, TOF detector saturation can occur due to the larger number of ions-per-push. This mainly happens with TDC detectors and is why many manufactures have moved to ADC-based detectors. Due to the AGC bias and sensitivity improvements due to IMS, a drastic increase in the number of proteins detected with LC-IMS–MS versus the LC-QExactive was observed (Figure 3A). The number of observed enzymes that mapped to the KEGG atlas also greatly increased with the LC-IMS–MS analysis (Figure 3B), allowing additional information to be uncovered about the mechanistic role these proteins play in the Ebola disease (41).
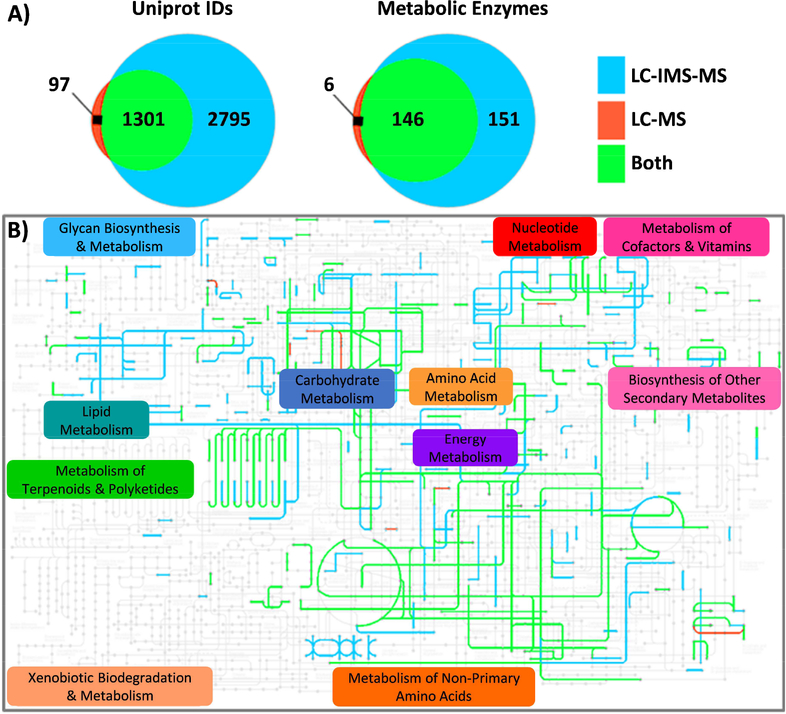
Figure 3.
Proteins identified in plasma samples contaminated with polymers from Ebola patients were compared for a LC-QExactive MS (noted as LC-MS (41)) and LC-IMS-QTOF MS (noted as LC-IMS- MS). A) Venn diagrams for the Uniprot protein IDs (left) and those that mapped to human metabolic pathways in the KEGG database (right) and identified by LC-MS only (red), LC-IMS-MS only (blue), and the overlap of both (green). All proteins were identified with at least two peptides and a requirementthat one peptide must be unique (i.e., the case of a single peptide matching only one protein in thereference database). B) The specific enzymes that mapped to the KEGG atlas using the color schemefrom A), illustrating the much higher coverage with the LC-IMS-MS analysis.
High throughput proteomic analyses have also been of great interest for analyzing the numerous patients necessary to address human biodiversity. Recently, IMS was even used to combine both the discovery and validation steps to simultaneously increase coverage and throughput. In this discovery and targeted monitoring (DTM) approach (42, 43), heavy-labeled peptide standards are spiked into a tryptic digest for highly sensitive and precise relative quantitation of the selected target peptides. Global analyses are also performed in these studies so that unknown peptides can be characterized based on their LC elution times, CCS values, and accurate masses. Results leveraging DTM strategies can provide better overall protein sequence coverage and detection of lower abundance peptides.
3. IMS–MS in Small Molecule Metabolomic and Exposomic Studies
The evaluation of small molecules with IMS–MS is also of great interest due to the many isomers and nominal mass isobars that co-elute and fragment together when selection windows of 0.5 Da or greater are utilized. In metabolomic measurements, small molecule metabolites, intermediates and products of metabolism are studied in environmental and biological systems (44). The presence of xenobiotic molecules in these metabolomic evaluations is also possible as they can be introduced through various sources such as chemical manufacturing plants, consumption of foods and drugs, or contact with personal-care products. The study of these xenobiotic molecules is often called exposomics, but can be coupled with metabolomic measurements depending on the polarity of the xenobiotic extraction and ion source needed for its detection (e.g. polyaromatic hydrocarbons will not be found in polar exactions and analyses with electrospray ionization). The complexity of the different biological and environmental matrices and the occurrence of thousands of different small molecules therefore requires analytical methods able to screen and identify many compounds in a single run. The application of LC–IMS–MS has therefore shown great promise for the untargeted analysis of small molecules in biological and environmental samples due to its speed and multidimensional analysis dimensions. IMS–MS and LC–IMS–MS have been utilized in numerous studies to improve both selectivity and coverage of the metabolome by providing additional structural information compared to routine MS or LC–MS-based methods (45). Since different molecular classes fall on distinct trend lines based on their CCS and m/z values, IMS has been extremely valuable for separating out many different types of small molecules (Figure 4). To date there have been various CCS databases released for different substance classes such as peptides (46–48), N-glycans (49), drug-like compounds (48), metabolites (50, 51), lipids (52–54), environmental toxins (55) and various biomolecules (56), ultimately aiding in their classification. There is also great effort going into the standardization of the CCS values from the same type of instruments as well as different IMS techniques (57–59). Considerations into standardizations have related to the type of ion mobility spectrometry performed, buffer gas composition, calibration procedures, and instrumental parameters such as electric fields, temperature and pressure. Recent reviews have demonstrated that addressing these questions requires communal consensus between both academic and industrial researchers to advance the application of IMS–MS technology and provide a frame of reference for reporting CCS measurements (60).
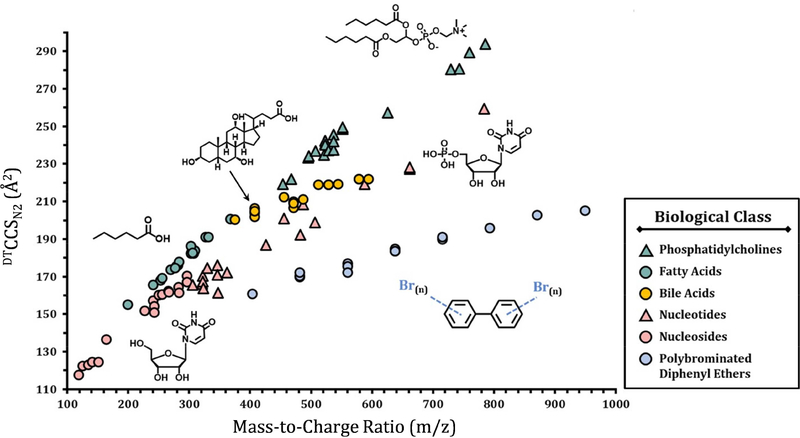
Figure 4.
The distinct CCS values verses m/z trend lines for the different classes of small molecules examined with DTIMS using N2 as the drift gas. The depronated values are shown for all molecules except the phosphatidylcholines ([M+H]+) and PBDEs ([M–Br–O]−).
Previously, LC–IMS–MS small molecule evaluations have been performed for biological matrices and environmental samples including wastewater (48). Wastewater, in particular, provides many challenges for analytical measurements as anthropogenic compounds such as pharmaceuticals, pesticides, or personal-care products can be found in the water cycle. The application of LC–IMS–MS for the untargeted analysis of water samples was found to be a very useful tool due to its speed and multiple analysis dimensions, which provided better identifications than LC–MS alone. Two-dimensional LC systems have even been combined with IMS–MS to perform four dimensional separations in the different omic analyses to increase the possible peak capacity even further (61). Additionally, for qualitative analysis of unknown molecules, the ability to empirically measure CCS values offers the possibility to possibly identify substances by the combination of their exact mass and their specific size. Molecular modeling (62) and machine learning (63, 64) also provide the capability to annotate molecules present in samples and predict possible identities or molecular classification for unknown molecules by mapping them to specific trend lines. These theoretical approaches have the potential to enable the annotation and identification of many small molecules, even though only ~30,000 in databases to date have measured MS/MS values.
4. IMS–MS in the Analysis of Lipidomic and Glycomic Isomers
Both lipidomic and glycomics measurements are also proving increasingly important for understanding biological and environmental systems. However, both molecule types challenge current techniques due to their many possible isomers. Lipid isomers often occur from different fatty acyl positions (sn–1/sn–2 vs. sn–2/sn–1), sn-backbones connectivities, double bond positions (positional and conformational), similar fatty acyl groups (14:0/14:0 vs. 16:0/12:0), S versus R orientations, and distinct but structurally similar headgroups. Glycan evaluations are also exceedingly difficult due to the high abundance of isomeric species resulting from isomeric monosaccharides (glucose, galactose, mannose, etc.), anomericity (α- versus β-linkages), glycosidic linkage position (e.g. α(1–4) linkage, α(1–6) linkage, or β(1–4)), and branching versus linear glycan forms in which the monosaccharide composition is identical but the connectivity along the monosaccharide chain differs. Because of the abundance of structural variability for both lipids and glycans, IMS–MS has emerged as a potential solution. While many of the isomeric glycan and lipid groups can be separated using specific chromatographic methods, the large variability in chemical properties such as polarity can greatly complicate both global lipidomic and glycomic analyses. Furthermore, LC parameter optimization can be very time consuming and costly. Since obtaining better resolving power with IMS only consists of buffer gas optimization (e.g. lower temperatures or addition of mobile phase modifiers) and it is amenable to a range of pre-separation techniques and ionization methods, it is an ideal complement to existing methods (65).
To date, numerous instances of IMS–MS have been published demonstrating improvements in lipid identification. In one study IMS–MS/MS analyses were utilized to provide a high level of structural information for phosphatidylcholines (PCs), including differentiation of fatty acyl substituents at the sn–1 versus sn–2 position, and location of fatty acyl double bond(s) for PCs in plasma (66). A different evaluation showed that glycero- and phospholipid isomers can be separated using high resolution FAIMS–MS/MS with ~75% success when forming silver adducts of the target lipids (67, 68). To increase lipid identifications, many research groups are compiling their own CCS libraries for lipids, similar to what has been done for small molecules/metabolites (56, 69). The work to date has illustrated greatly improved selectivity in various lipidomics approaches, including LC–IMS–MS shotgun approaches (58). Machine- learning algorithm-based CCS prediction has also been implemented to generate large-scale CCS libraries in support of lipidomics (69). These values are also being utilized with LC elution times, MS vales and MS/MS fragmentation patterns for identification, so that the multiple characteristics enable the reduction of false positives (52).
To date, elucidating detailed information about glycans and glycoconjugates has been limited due to their structural complexity (70). The earliest IMS applications involved separation and CCS measurement of small oligosaccharides such as tetrasaccharides, hexasaccharides, and cyclic oligosaccharides (cyclodextrins) (26, 71). More challenging glycans were then successfully resolved using IMS–MS include anomeric trisaccharides (72), high mannose N- glycans (73), and even glycopeptides (74–76). Most IMS–MS studies of glycans to date have mainly focused on positive ion mode characterization due to its higher ionization efficiency (77–80), but recent reports on negative ion mode are also showing great promise (81). Additionally, various salt and metal ions adducts have been shown to enhance isomeric separations and provide further opportunities for better glycan separations (81, 82). Thus, future evaluations of lipids and glycans are expected to rely heavily on IMS analyses for the rapid and sensitive separation their isomers, greatly improving their characterization and identification (83).
5. Conclusions
While IMS–MS was once characterized as an emerging technique in the omics field, it is being increasingly integrated into modern omic measurements. Its utility in separating peptides and proteins with only minor structural differences has shown to be extremely powerful in bottom-up, middle-down and top-down studies. It has also greatly enabled the evaluation of isomers, which were previously inseparable or required long chromatography. Additionally, the ability of IMS–MS to distinguishing contaminant ions and molecular classes is proving to be essential in all omic analyses of complex environmental and biological samples.
The CCS values from IMS measurements are also proving to be important in many omic areas. By measuring CCS values for available standards and compiling databases, rapid targeted analyses and identification of small molecules, lipids, glycans and peptides are possible. These CCS values are also being incorporated into computational methods such as theoretical modeling of structures and machine-based approaches, enabling molecular insight when no standards are available. Undoubtedly, the increasing utility of IMS–MS hinges on increasing its resolving power and successfully coupling it with other pre-separation, fragmentation and software tools. Fortunately, developments in all these areas are taking place to increase the capabilities of IMS–MS measurements. Higher resolution IMS and MS instrument platforms such as those possible with structures for lossless ion manipulations (SLIM) are becoming available, providing even more confidence in global measurements of complex biological samples (84). New fragmentation approaches such as ECD are being added to various instrumental IMS platforms and software tools enabling rapid analyses are also being developed to push the technology further. Since the analysis of the multi-dimensional LC–IMS–MS data has proven to be difficult due to the many dimensions (LC, IMS, MS and DIA or DDA MS/MS), the future seems promising with its incorporation into numerous software packages including the open source Skyline software (85). Thus, IMS–MS capabilities are only expected to increase in the future, making it an essential tool for high quality omic analyses.
Highlights:
IMS is a widely used analytical technique providing rapid gas phase separations
IMS coupled with MS is rapidly gaining attention for improving omic analyses
IMS–MS is able to extract information not possible with MS alone in complex samples
IMS–MS distinguishes isomers, isobars, molecular classes and contaminant ions
Acknowledgements
The authors would like to thank Nathan Johnson for help with the figures. Portions of this research were supported by grants from the NIH National Institute of Environmental Health Sciences (R01 ES022190 and P42 ES027704), National Institute of General Medical Sciences (P41 GM103493), National Institute of Allergy and Infectious Disease (U19 AI106772), National Institute of Child Health and Human Development (R21 HD084788) and the Laboratory Directed Research and Development Program at Pacific Northwest National Laboratory. This research used capabilities developed by the Pan-omics program (funded by the U.S. Department of Energy Office of Biological and Environmental Research Genome Sciences Program). This work was performed in the W. R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a DOE national scientific user facility at the Pacific Northwest National Laboratory (PNNL). PNNL is operated by Battelle for the DOE under contract DE-AC05–76RL0 1830.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Borsdorf H, Eiceman GA. Ion Mobility Spectrometry: Principles and Applications. Applied Spectroscopy Reviews. 2006;41(4):323–75. doi: 10.1080/05704920600663469. [DOI] [Google Scholar]
- 2.Lapthorn C, Pullen F, Chowdhry BZ. Ion mobility spectrometry-mass spectrometry (IMS–MS) of small molecules: Separating and assigning structures to ions. Mass Spectrometry Reviews. 2013;32(1):43–71. doi: 10.1002/mas.21349. [DOI] [PubMed] [Google Scholar]
- 3.Lanucara F, Holman SW, Gray CJ, Eyers CE. The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat Chem. 2014;6(4):281–94. doi: 10.1038/nchem.1889. [DOI] [PubMed] [Google Scholar]
- 4.Chouinard CD, Wei MS, Beekman CR, Kemperman RHJ, Yost RA. Ion Mobility in Clinical Analysis: Current Progress and Future Perspectives. Clinical Chemistry. 2016;62(1):124–33. doi: 10.1373/clinchem.2015.238840. [DOI] [PubMed] [Google Scholar]
- 5.May JC, Gant-Branum RL, McLean JA. Targeting the untargeted in molecular phenomics with structurally-selective ion mobility-mass spectrometry. Current opinion in biotechnology. 2016;39:192–7. doi: 10.1016/j.copbio.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mason E, McDaniel E. Transport Properites of Ions in Gases. New York: Wiley; 1988. [Google Scholar]
- 7.Cohen MJ, Karasek FW. Plasma Chromatography™—A New Dimension for Gas Chromatography and Mass Spectrometry. Journal of Chromatographic Science. 1970;8(6):330–7. doi: 10.1093/chromsci/8.6.330. [DOI] [Google Scholar]
- 8.Hoaglund CS, Valentine SJ, Sporleder CR, Reilly JP, Clemmer DE. Three-Dimensional Ion Mobility/TOFMS Analysis of Electrosprayed Biomolecules. Analytical Chemistry. 1998;70(11):2236–42. doi: 10.1021/ac980059c. [DOI] [PubMed] [Google Scholar]
- 9.Wyttenbach T, Kemper PR, Bowers MT. Design of a new electrospray ion mobility mass spectrometer. International Journal of Mass Spectrometry. 2001;212(1–3):13–23. doi: 10.1016/S1387-3806(01)00517-6. [DOI] [Google Scholar]
- 10.Giles K, Pringle SD, Worthington KR, Little D, Wildgoose JL, Bateman RH. Applications of a travelling wave-based radio-frequency-only stacked ring ion guide. Rapid Commun Mass Spectrom. 2004;18(20):2401–14. Epub 2004/09/24. doi: 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- 11.Michelmann K, Silveira JA, Ridgeway ME, Park MA. Fundamentals of Trapped Ion Mobility Spectrometry. J Am Soc Mass Spectrom. 2015;26(1):14–24. doi: 10.1007/s13361-014-0999-4. [DOI] [PubMed] [Google Scholar]
- 12.Brown LJ, Creaser CS. Field Asymmetric Waveform Ion Mobility Spectrometry Analysis of Proteins and Peptides: A Review. Current Analytical Chemistry. 2013;9(2):192–8. [Google Scholar]
- 13.Guevremont R High-field asymmetric waveform ion mobility spectrometry: A new tool for mass spectrometry. J Chromatogr A. 2004;1058(1–2):3–19. doi: 10.1016/j.chroma.2004.08.119. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez de la Mora J. Mobility Analysis of Proteins by Charge Reduction in a Bipolar Electrospray Source. Anal Chem. 2018;90(20):12187–90. Epub 2018/09/11. doi: 10.1021/acs.analchem.8b03296. [DOI] [PubMed] [Google Scholar]
- 15.Rawat VK, Buckley DT, Kimoto S, Lee MH, Fukushima N, Hogan CJ. Two dimensional size-mass distribution function inversion from differential mobility analyzer-aerosol particle mass analyzer (DMA-APM) measurements. J Aerosol Sci. 2016;92:70–82. doi: 10.1016/j.jaerosci.2015.11.001. [DOI] [Google Scholar]
- 16.Mason EA, Homer W. Schamp J. Mobility of Gaseous Ions in Weak Electric Fields. Annals of Physics. 1958;4(3):233–70. [Google Scholar]
- 17.Revercomb HE, Mason EA. Theory of Plasma Chromatography/Gaseous Electrophoresis - A Reviews. Analytical Chemistry. 1975;47(7):970–83. [Google Scholar]
- 18.Guevremont R, Siu KW, Wang J, Ding L. Combined Ion Mobility/Time-of-Flight Mass Spectrometry Study of Electrospray-Generated Ions. Anal Chem. 1997;69(19):3959–65. Epub 1997/10/01. doi: 10.1021/ac970359e. [DOI] [PubMed] [Google Scholar]
- 19.May JC, Goodwin CR, Lareau NM, Leaptrot KL, Morris CB, Kurulugama RT, Mordehai A, Klein C, Barry W, Darland E, Overney G, Imatani K, Stafford GC, Fjeldsted JC, McLean JA. Conformational ordering of biomolecules in the gas phase: nitrogen collision cross sections measured on a prototype high resolution drift tube ion mobility-mass spectrometer. Anal Chem. 2014;86(4):2107–16. Epub 2014/01/23. doi: 10.1021/ac4038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metz TO, Page JS, Baker ES, Tang KQ, Ding J, Shen YF, Smith RD. High-resolution separations and improved ion production and transmission in metabolomics. Trac-Trend Anal Chem. 2008;27(3):205–14. doi: 10.1016/j.trac.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suhr H Plasma Chromatography. New York: Plenum Press; 1984. [Google Scholar]
- 22.Henderson SC, Valentine SJ, Counterman AE, Clemmer DE. ESI/ion trap/ion mobility/time-of-flight mass spectrometry for rapid and sensitive analysis of biomolecular mixtures. Anal Chem. 1999;71(2):291–301. [DOI] [PubMed] [Google Scholar]
- 23.Valentine SJ, Counterman AE, Hoaglund CS, Reilly JP, Clemmer DE. Gas-phase separations of protease digests. J Am Soc Mass Spectrom. 1998;9(11):1213–6. doi: 10.1016/S1044-0305(98)00101-9. [DOI] [PubMed] [Google Scholar]
- 24.Baker ES, Livesay EA, Orton DJ, Moore RJ, Danielson WF, 3rd, Prior DC, Ibrahim YM, LaMarche BL, Mayampurath AM, Schepmoes AA, Hopkins DF, Tang K, Smith RD, Belov ME. An LC-IMS–MS platform providing increased dynamic range for high-throughput proteomic studies. J Proteome Res. 2010;9(2):997–1006. Epub 2009/12/17. doi: 10.1021/pr900888b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clemmer DE, Hudgins RR, Jarrold MF. Naked Protein Conformations: Cytochrome c in the Gas Phase. J Am Chem Soc. 1995;117:10141–2. doi: 10.1021/ja00145a037‚. [DOI] [Google Scholar]
- 26.Lee S, Wyttenbach T, Bowers MT. Gas phase structures of sodiated oligosaccharides by ion mobility/ion chromatography methods. International Journal of Mass Spectrometry and Ion Processes. 1997;167/168:605–14. [Google Scholar]
- 27.Hoaglund CS, Valentine SJ, Sporleder CR, Reilly JP, Clemmer DE. Three-Dimensional Ion Mobility/TOFMS Analysis of Electrosprayed Biomolecules. Analytical Chemistry. 1998;70:2236–42. [DOI] [PubMed] [Google Scholar]
- 28.Bleiholder C, Dupuis NF, Wyttenbach T, Bowers MT. Ion mobility-mass spectrometry reveals a conformational conversion from random assembly to beta-sheet in amyloid fibril formation. Nat Chem. 2011;3(2):172–7. Epub 2011/01/25. doi: 10.1038/nchem.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith DP, Radford SE, Ashcroft AE. Elongated oligomers in beta2-microglobulin amyloid assembly revealed by ion mobility spectrometry-mass spectrometry. Proc Natl Acad Sci U S A. 2010;107(15):6794–8. Epub 2010/03/31. doi: 10.1073/pnas.0913046107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phillips AS, Gomes AF, Kalapothakis JM, Gillam JE, Gasparavicius J, Gozzo FC, Kunath T, MacPhee C, Barran PE. Conformational dynamics of alpha-synuclein: insights from mass spectrometry. Analyst. 2015;140(9):3070–81. Epub 2015/03/11. doi: 10.1039/c4an02306d. [DOI] [PubMed] [Google Scholar]
- 31.Dixit SM, Polasky DA, Ruotolo BT. Collision induced unfolding of isolated proteins in the gas phase: past, present, and future. Curr Opin Chem Biol. 2018;42:93–100. Epub 2017/12/06. doi: 10.1016/j.cbpa.2017.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eschweiler JD, Martini RM, Ruotolo BT. Chemical Probes and Engineered Constructs Reveal a Detailed Unfolding Mechanism for a Solvent-Free Multidomain Protein. J Am Chem Soc. 2017;139(1):534–40. Epub 2016/12/14. doi: 10.1021/jacs.6b11678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allison TM, Reading E, Liko I, Baldwin AJ, Laganowsky A, Robinson CV. Quantifying the stabilizing effects of protein-ligand interactions in the gas phase. Nat Commun. 2015;6:8551 Epub 2015/10/07. doi: 10.1038/ncomms9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valentine SJ, Plasencia MD, Liu X, Krishnan M, Naylor S, Udseth HR, Smith RD, Clemmer DE. Toward Plasma Proteome Profiling with Ion Mobility-Mass Spectrometry. Journal of Proteome Research. 2006;5:2977–84. [DOI] [PubMed] [Google Scholar]
- 35.Hengel SM, Floyd E, Baker ES, Zhao R, Wu S, Pasa-Tolic L. Evaluation of SDS depletion using an affinity spin column and IMS–MS detection. Proteomics. 2012;12(21):3138–42. Epub 2012/09/01. doi: 10.1002/pmic.201200168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baker ES, Burnum-Johnson KE, Ibrahim YM, Orton DJ, Monroe ME, Kelly RT, Moore RJ, Zhang X, Theberge R, Costello CE, Smith RD. Enhancing bottom-up and top-down proteomic measurements with ion mobility separations. Proteomics. 2015;15(16):2766–76. Epub 2015/06/06. doi: 10.1002/pmic.201500048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abbatiello SE, Cardasis H, Belford M, Sarracino D, Neil J, Stephenson J, Blackburn M. Improved Detection of Proteins in Complex Sample Matrices by Infusion Using FAIMS. Poster Presented at ASMS 2015. [Google Scholar]
- 38.Crowell KL, Baker ES, Payne SH, Ibrahim YM, Monroe ME, Slysz GW, LaMarche BL, Petyuk VA, Piehowski PD, Danielson WF, 3rd, Anderson GA, Smith RD. Increasing Confidence of LC-MS Identifications by Utilizing Ion Mobility Spectrometry. Int J Mass Spectrom. 2013;354–355:312–7. Epub 2014/08/05. doi: 10.1016/j.ijms.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker ES, Burnum-Johnson KE, Jacobs JM, Diamond DL, Brown RN, Ibrahim YM, Orton DJ, Piehowski PD, Purdy DE, Moore RJ, Danielson WF 3rd, Monroe ME, Crowell KL, Slysz GW, Gritsenko MA, Sandoval JD, Lamarche BL, Matzke MM, Webb-Robertson BJ, Simons BC, McMahon BJ, Bhattacharya R, Perkins JD, Carithers RL Jr., Strom S, Self SG, Katze MG, Anderson GA, Smith RD. Advancing the high throughput identification of liver fibrosis protein signatures using multiplexed ion mobility spectrometry. Mol Cell Proteomics. 2014;13(4):1119–27. Epub 2014/01/10. doi: 10.1074/mcp.M113.034595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X, Plasencia M, Ragg S, Valentine SJ, Clemmer DE. Development of high throughput dispersive LC-ion mobility-TOFMS techniques for analysing the human plasma proteome. Briefings in functional genomics & proteomics. 2004;3(2):177–86. [DOI] [PubMed] [Google Scholar]
- 41.Eisfeld AJ, Halfmann PJ, Wendler JP, Kyle JE, Burnum-Johnson KE, Peralta Z, Maemura T, Walters KB, Watanabe T, Fukuyama S, Yamashita M, Jacobs JM, Kim YM, Casey CP, Stratton KG, Webb-Robertson BJM, Gritsenko MA, Monroe ME, Weitz KK, Shukla AK, Tian MY, Neumann G, Reed JL, van Bakel H, Metz TO, Smith RD, Waters KM, N’jai A, Sahr F, Kawaoka Y. Multi-platform ‘Omics Analysis of Human Ebola Virus Disease Pathogenesis. Cell Host Microbe. 2017;22(6):817-+. doi: 10.1016/j.chom.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burnum-Johnson KE, Nie S, Casey CP, Monroe ME, Orton DJ, Ibrahim YM, Gritsenko MA, Clauss TR, Shukla AK, Moore RJ, Purvine SO, Shi T, Qian W, Liu T, Baker ES, Smith RD. Simultaneous Proteomic Discovery and Targeted Monitoring using Liquid Chromatography, Ion Mobility Spectrometry, and Mass Spectrometry. Mol Cell Proteomics. 2016;15(12):3694–705. Epub 2016/09/28. doi: 10.1074/mcp.M116.061143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garabedian A, Benigni P, Ramirez CE, Baker ES, Liu T, Smith RD, Fernandez-Lima F. Towards Discovery and Targeted Peptide Biomarker Detection Using nanoESI-TIMS-TOF MS. J Am Soc Mass Spectrom. 2018;29(5):817–26. Epub 2017/09/11. doi: 10.1007/s13361-017-1787-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fiehn O Metabolomics- The link between genotypes and phenotypes. Plant Molecular Biology. 2002;48:155–71. [PubMed] [Google Scholar]
- 45.Ortmayr K, Causon TJ, Hann S, Koellensperger G. Increasing selectivity and coverage in LC-MS based metabolome analysis. TrAC Trends in Analytical Chemistry. 2016;82:358–66. doi: 10.1016/j.trac.2016.06.011. [DOI] [Google Scholar]
- 46.Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Collision Cross Sections of Proteins and Their Complexes: A Calibration Framework and Database for Gas-Phase Structural Biology. Analytical Chemistry. 2010;82(22):9557–65. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- 47.Valentine SJ, Counterman AE, Clemmer DE. A database of 660 peptide ion cross sections: Use of intrinsic size parameters for bona fide predictions of cross sections. J Am Soc Mass Spectr. 1999;10(11):1188–211. doi: Doi 10.1016/S1044-0305(99)00079-3. [DOI] [PubMed] [Google Scholar]
- 48.Stephan S, Hippler J, Kohler T, Deeb AA, Schmidt TC, Schmitz OJ. Contaminant screening of wastewater with HPLC-IM-qTOF-MS and LC+LC-IM-qTOF-MS using a CCS database. Anal Bioanal Chem. 2016;408(24):6545–55. Epub 2016/08/09. doi: 10.1007/s00216-016-9820-5. [DOI] [PubMed] [Google Scholar]
- 49.Hofmann J, Struwe WB, Scarff CA, Scrivens JH, Harvey DJ, Pagel K. Estimating collision cross sections of negatively charged N-glycans using traveling wave ion mobility-mass spectrometry. Anal Chem. 2014;86(21):10789–95. Epub 2014/10/01. doi: 10.1021/ac5028353. [DOI] [PubMed] [Google Scholar]
- 50.Zheng X, Aly NA, Zhou Y, Dupuis KT, Bilbao A, Paurus VL, Orton DJ, Wilson R, Payne SH, Smith RD, Baker ES. A structural examination and collision cross section database for over 500 metabolites and xenobiotics using drift tube ion mobility spectrometry. Chem Sci. 2017;8(11):7724–36. Epub 2018/03/24. doi: 10.1039/c7sc03464d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paglia G, Angel P, Williams JP, Richardson K, Olivos HJ, Thompson JW, Menikarachchi L, Lai S, Walsh C, Moseley A, Plumb RS, Grant DF, Palsson BO, Langridge J, Geromanos S, Astarite G. Ion Mobility-Derived Collision Cross Section As an Additional Measure for Lipid Fingerprinting and Identification. Analytical Chemistry. 2015;87(2):1137–44. doi: 10.1021/ac503715v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Z, Shen X, Chen X, Tu J, Xiong X, Zhu ZJ. LipidIMMS Analyzer: Integrating multi-dimensional information to support lipid identification in ion mobility - mass spectrometry based lipidomics. Bioinformatics. 2018. Epub 2018/07/28. doi: 10.1093/bioinformatics/bty661. [DOI] [PubMed] [Google Scholar]
- 53.Kyle JE, Aly N, Zheng XY, Burnum-Johnson KE, Smith RD, Baker ES. Evaluating lipid mediator structural complexity using ion mobility spectrometry combined with mass spectrometry. Bioanalysis. 2018;10(5):279–89. doi: 10.4155/bio-2017-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paglia G, Kliman M, Claude E, Geromanos S, Astarita G. Applications of ion-mobility mass spectrometry for lipid analysis. Analytical and Bioanalytical Chemistry. 2015;407(17):4995–5007. doi: 10.1007/s00216-015-8664-8. [DOI] [PubMed] [Google Scholar]
- 55.Zheng XY, Dupuis KT, Aly NA, Zhou YX, Smith FB, Tang KQ, Smith RD, Baker ES. Utilizing ion mobility spectrometry and mass spectrometry for the analysis of polycyclic aromatic hydrocarbons, polychlorinated biphenyls, polybrominated diphenyl ethers and their metabolites. Analytica Chimica Acta. 2018;1037:265–73. doi: 10.1016/j.aca.2018.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.May JC, Goodwin CR, Lareau NM, Leaptrot KL, Morris CB, Kurulugama RT, Mordehai A, Klein C, Barry W, Darland E, Overney G, Imatani K, Stafford GC, Fjeldsted JC, McLean JA. Conformational Ordering of Biomolecules in the Gas Phase: Nitrogen Collision Cross Sections Measured on a Prototype High Resolution Drift Tube Ion Mobility-Mass Spectrometer. Analytical Chemistry. 2014;86(4):2107–16. doi: 10.1021/ac4038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stow SM, Causon TJ, Zheng X, Kurulugama RT, Mairinger T, May JC, Rennie EE, Baker ES, Smith RD, McLean JA, Hann S, Fjeldsted JC. An Interlaboratory Evaluation of Drift Tube Ion Mobility-Mass Spectrometry Collision Cross Section Measurements. Anal Chem. 2017;89(17):9048–55. Epub 2017/08/02. doi: 10.1021/acs.analchem.7b01729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paglia G, Angel P, Williams JP, Richardson K, Olivos HJ, Thompson JW, Menikarachchi L, Lai S, Walsh C, Moseley A, Plumb RS, Grant DF, Palsson BO, Langridge J, Geromanos S, Astarita G. Ion mobility-derived collision cross section as an additional measure for lipid fingerprinting and identification. Anal Chem. 2015;87(2):1137–44. Epub 2014/12/17. doi: 10.1021/ac503715v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paglia G, Williams JP, Menikarachchi L, Thompson JW, Tyldesley-Worster R, Halldorsson S, Rolfsson O, Moseley A, Grant D, Langridge J, Palsson BO, Astarita G. Ion mobility derived collision cross sections to support metabolomics applications. Anal Chem. 2014;86(8):3985–93. Epub 2014/03/20. doi: 10.1021/ac500405x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gabelica V, Shvartsburg AA, Afonso C, Barran P, Benesch JLP, Bleiholder C, Bowers MT, Bilbao A, Bush MF, Campbell JL, Campuzano IDG, Causon T, Clowers BH, Creaser CS, De Pauw E, Far J, Fernandez-Lima F, Fjeldsted JC, Giles K, Groessl M, Hogan CJ Jr., Hann S, Kim HI, Kurulugama RT, May JC, McLean JA, Pagel K, Richardson K, Ridgeway ME, Rosu F, Sobott F, Thalassinos K, Valentine SJ, Wyttenbach T. Recommendations for reporting ion mobility Mass Spectrometry measurements. Mass Spectrom Rev. 2019. Epub 2019/02/02. doi: 10.1002/mas.21585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stephan S, Jakob C, Hippler J, Schmitz OJ. A novel four-dimensional analytical approach for analysis of complex samples. Anal Bioanal Chem. 2016;408(14):3751–9. Epub 2016/04/03. doi: 10.1007/s00216-016-9460-9. [DOI] [PubMed] [Google Scholar]
- 62.Zheng X, Renslow RS, Makola MM, Webb IK, Deng L, Thomas DG, Govind N, Ibrahim YM, Kabanda MM, Dubery IA, Heyman HM, Smith RD, Madala NE, Baker ES. Structural Elucidation of cis/trans Dicaffeoylquinic Acid Photoisomerization Using Ion Mobility Spectrometry-Mass Spectrometry. J Phys Chem Lett. 2017;8(7):1381–8. Epub 2017/03/08. doi: 10.1021/acs.jpclett.6b03015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Z, Xiong X, Zhu ZJ. MetCCS predictor: a web server for predicting collision cross-section values of metabolites in ion mobility-mass spectrometry based metabolomics. Bioinformatics. 2017;33(14):2235–7. Epub 2017/03/24. doi: 10.1093/bioinformatics/btx140. [DOI] [PubMed] [Google Scholar]
- 64.Soper-Hopper MT, Petrov AS, Howard JN, Yu SS, Forsythe JG, Grover MA, Fernandez FM. Collision cross section predictions using 2-dimensional molecular descriptors. Chem Commun (Camb). 2017;53(54):7624–7. Epub 2017/06/24. doi: 10.1039/c7cc04257d. [DOI] [PubMed] [Google Scholar]
- 65.Silveira JA, Servage KA, Gamage CM, Russell DH. Cryogenic Ion Mobility-Mass Spectrometry Captures Hydrated Ions Produced During Electrospray Ionization. Journal of Physical Chemistry A. 2013;117(5):953–61. doi: 10.1021/jp311278a. [DOI] [PubMed] [Google Scholar]
- 66.Castro-Perez J, Roddy TP, Nibbering NM, Shah V, McLaren DG, Previs S, Attygalle AB, Herath K, Chen Z, Wang SP, Mitnaul L, Hubbard BK, Vreeken RJ, Johns DG, Hankemeier T. Localization of fatty acyl and double bond positions in phosphatidylcholines using a dual stage CID fragmentation coupled with ion mobility mass spectrometry. J Am Soc Mass Spectrom. 2011;22(9):1552–67. Epub 2011/09/29. doi: 10.1007/s13361-011-0172-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bowman AP, Abzalimov RR, Shvartsburg AA. Broad Separation of Isomeric Lipids by High-Resolution Differential Ion Mobility Spectrometry with Tandem Mass Spectrometry. J Am Soc Mass Spectrom. 2017;28(8):1552–61. Epub 2017/05/04. doi: 10.1007/s13361-017-1675-2. [DOI] [PubMed] [Google Scholar]
- 68.Berry KA, Barkley RM, Berry JJ, Hankin JA, Hoyes E, Brown JM, Murphy RC. Tandem Mass Spectrometry in Combination with Product Ion Mobility for the Identification of Phospholipids. Anal Chem. 2017;89(1):916–21. Epub 2016/12/14. doi: 10.1021/acs.analchem.6b04047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou Z, Tu J, Xiong X, Shen X, Zhu ZJ. LipidCCS: Prediction of Collision Cross-Section Values for Lipids with High Precision To Support Ion Mobility-Mass Spectrometry-Based Lipidomics. Anal Chem. 2017;89(17):9559–66. Epub 2017/08/03. doi: 10.1021/acs.analchem.7b02625. [DOI] [PubMed] [Google Scholar]
- 70.Bertozzi CR, Rabuka D, Varki A. Essentials of Glycobiology. 2009. [PubMed]
- 71.Liu Y, Clemmer DE. Characterizing Oligosaccharides Using Injected-Ion Mobility/Mass Spectrometry. Analytical Chemistry. 1997;69:2504–9. [DOI] [PubMed] [Google Scholar]
- 72.Hofmann J, Hahm HS, Seeberger PH, Pagel K. Identification of carbohydrate anomers using ion mobility-mass spectrometry. Nature. 2015;526(7572):241–4. Epub 2015/09/30. doi: 10.1038/nature15388. [DOI] [PubMed] [Google Scholar]
- 73.Harvey DJ, Scarff CA, Edgeworth M, Struwe WB, Pagel K, Thalassinos K, Crispin M, Scrivens J. Travelling-wave ion mobility and negative ion fragmentation of high-mannose N-glycans. J Mass Spectrom. 2016;51(3):219–35. Epub 2016/03/10. doi: 10.1002/jms.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hinneburg H, Hofmann J, Struwe WB, Thader A, Altmann F, Varon Silva D, Seeberger PH, Pagel K, Kolarich D. Distinguishing N-acetylneuraminic acid linkage isomers on glycopeptides by ion mobility-mass spectrometry. Chem Commun (Camb). 2016;52(23):4381–4. Epub 2016/03/02. doi: 10.1039/c6cc01114d. [DOI] [PubMed] [Google Scholar]
- 75.Creese AJ, Cooper HJ. Separation and identification of isomeric glycopeptides by high field asymmetric waveform ion mobility spectrometry. Anal Chem. 2012;84(5):2597–601. Epub 2012/01/28. doi: 10.1021/ac203321y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu F, Trinidad JC, Clemmer DE. Glycopeptide Site Heterogeneity and Structural Diversity Determined by Combined Lectin Affinity Chromatography/IMS/CID/MS Techniques. J Am Soc Mass Spectrom. 2015;26(7):1092–102. Epub 2015/04/05. doi: 10.1007/s13361-015-1110-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu M, Bendiak B, Clowers B, Hill HH Jr. Ion mobility-mass spectrometry analysis of isomeric carbohydrate precursor ions. Anal Bioanal Chem. 2009;394:1853–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fenn LS, McLean JA. Structural resolution of carbohydrate positional and structural isomers based on gas-phase ion mobility-mass spectrometry. Phys Chem Chem Phys. 2011;13:2196–205. [DOI] [PubMed] [Google Scholar]
- 79.Both P, Green AP, Gray CJ, Šardzík R, Voglmeir J, Fontana C, Austeri M, Rejzek M, Richardson D, Field RA, Widmalm G, Flitsch SL, Eyers CE. Discrimination of epimeric glycans and glycopeptides using IM-MS and its potential for carbohydrate sequencing. Nat Chem. 2014;6(1):65–74. doi: 10.1038/nchem.1817 http://www.nature.com/nchem/journal/v6/n1/abs/nchem.1817.html#supplementary-information. [DOI] [PubMed] [Google Scholar]
- 80.Hofmann J, Hahm HS, Seeberger PH, Pagel K. Identification of carbohydrate anomers using ion mobility-mass spectrometry. Nature. 2015;526(7572):241–4. doi: 10.1038/nature1538810.1038/nature15388http://www.nature.com/nature/journal/v526/n7572/abs/nature15388.html#supplementary-informationhttp://www.nature.com/nature/journal/v526/n7572/abs/nature15388.html#supplementary-information . [DOI] [PubMed] [Google Scholar]
- 81.Zheng X, Zhang X, Schocker NS, Renslow RS, Orton DJ, Khamsi J, Ashmus RA, Almeida IC, Tang K, Costello CE, Smith RD, Michael K, Baker ES. Enhancing glycan isomer separations with metal ions and positive and negative polarity ion mobility spectrometry-mass spectrometry analyses. Anal Bioanal Chem. 2017;409(2):467–76. Epub 2016/09/09. doi: 10.1007/s00216-016-9866-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang Y, Dodds ED. Discrimination of Isomeric Carbohydrates as the Electron Transfer Products of Group II Cation Adducts by Ion Mobility Spectrometry and Tandem Mass Spectrometry. Anal Chem. 2015;87(11):5664–8. Epub 2015/05/09. doi: 10.1021/acs.analchem.5b00759. [DOI] [PubMed] [Google Scholar]
- 83.Nagy G, Attah IK, Garimella SVB, Tang KQ, Ibrahim YM, Baker ES, Smith RD. Unraveling the isomeric heterogeneity of glycans: ion mobility separations in structures for lossless ion manipulations. Chem Commun. 2018;54(83):11701–4. doi: 10.1039/c8cc06966b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ibrahim YM, Hamid AM, Deng L, Garimella SV, Webb IK, Baker ES, Smith RD. New frontiers for mass spectrometry based upon structures for lossless ion manipulations. Analyst. 2017;142(7):1010–21. Epub 2017/03/07. doi: 10.1039/c7an00031f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.MacLean BX, Pratt BS, Egertson JD, MacCoss MJ, Smith RD, Baker ES. Using Skyline to Analyze Data-Containing Liquid Chromatography, Ion Mobility Spectrometry, and Mass Spectrometry Dimensions. J Am Soc Mass Spectrom. 2018;29(11):2182–8. Epub 2018/07/27. doi: 10.1007/s13361-018-2028-5. [DOI] [PMC free article] [PubMed] [Google Scholar]