Abstract
Genetic conflicts arise when the evolutionary interests of two genetic elements are not aligned. Conflicts between genomes (e.g. pathogen versus host) or within the same genome (e.g internal parasitic DNA sequences versus the rest of the host genome) can both foster ‘molecular arms races’ in which genes on both sides of the conflict rapidly evolve due to bouts of adaptation and counter-adaptation. Importantly, a source of genetic novelty is needed to fuel these arms races. In this review, we highlight gene conversion as a major force in generating the novel alleles on which selection can act. Using examples from both intergenomic and intragenomic conflicts, we feature the mechanisms by which gene conversion facilitates the rapid evolution of genes in conflict.
Rapid evolution and genetic conflicts [antagonistic coevolution]
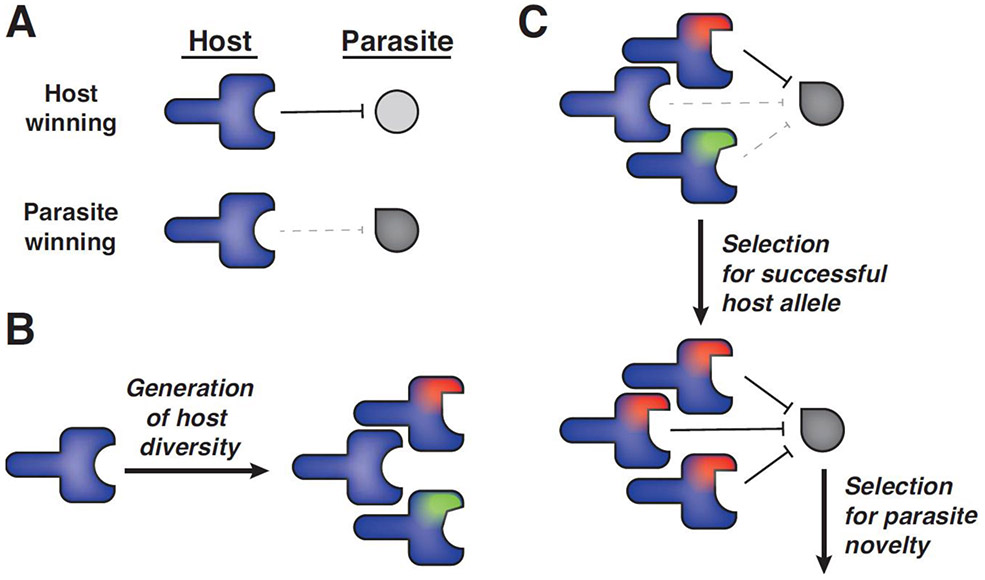
Biologists sometimes confound the conservation of a gene with its importance. In these cases, well conserved genes are considered essential and those that are poorly conserved genes, or rapidly evolving, are considered to be less important for organismal fitness. However, genes involved in genetic conflicts can be amongst the most rapidly evolving while also being critically important for the fitness of an organism. For example, toll or toll-like receptors (TLRs) are critical pathogen recognition molecules in both vertebrates and invertebrates, yet show a large number of lineage-specific gene births, gene losses and signatures of diversifying selection on individual codons [1-3]. The rapid evolution of genes in conflict is often due to the fact that there is not a single stable fitness optimum for these genes (Figure 1) [4,5].
Figure 1: Molecular arms races between hosts and parasites continually select for evolutionary innovations.
A) Hosts can encode genes that suppress a specific parasite. However, parasite variants may exist that escape host-mediated inhibition and result in a fitness disadvantage to the host. B) Genetic mutations arise randomly in populations, as indicated by differences in shapes and colors, which contribute to the diversity of host alleles. C) Host variants that suppress a common parasite will have a selective advantage and spread in a population. This now gives a selective advantage to novel or mutant parasites that can evade the common host variant. These cycles of adaptation and counter-adaptation can persist as long as the conflict is present and can lead to recurrent and rapid diversifying evolution. The generation of novel alleles is critical in these arms races and the gene conversion mechanisms shown in figure 2 can generate the types of novelty shown in panel B and facilitate the spread of adaptive variants as shown in panel C.
The molecular arms races driven by genetic conflicts require genetic diversity. Any genetic mutation will increase the variation present within a population, giving selection an opportunity to act. Here, we highlight the contribution of gene conversion in facilitating rapid evolution in host-parasite genetic conflicts. We draw our examples from two disparate types of genetic conflicts to illustrate the breadth of this topic: host immunity genes that rapidly evolve to defend against invading pathogens and parasitic meiotic drive genes that bias their own transmission into more than half of the gametes generated by a heterozygote.
The outcomes of gene conversion: changes in gene copy number and the generation of variation within paralogs
Broadly defined, gene conversion describes when an allele at one locus (the recipient) is changed by copying sequence from a different genomic locus (the donor) (Figure 2). The donor and recipient loci can be different alleles of the same gene or loci of different sizes and at nonallelic sites. Gene conversion occurs as a result of homologous recombination to repair a DNA double strand break (DSB) [6]. For example, site A could sustain a DSB. During the repair of that break, sequence information could be copied from a different site (e.g. site B) that shares some homology to site A. Through this process, site A was gene converted [7].
Figure 2: Mechanisms by which gene conversion generates genetic novelty.
A) The black and green lines represent nonallelic loci, the yellow arrowheads represent a repeated sequence and the blue box represents a gene. A DNA DSB in the green locus is repaired by copying sequence from the black locus, including the blue gene. The recombination event was seeded by homology shared between the yellow arrows. B) The blue boxes represent members of a gene family. Over time, the red variant can spread to all members of the blue gene family via gene conversion. C) The blue, red, and light blue boxes represent diverged members of a gene family. Over time, gene conversion can shuffle the variation between members of the family to generate novel alleles.
One way that gene conversion can contribute to adaptive evolution in an arms race scenario is by generating duplicate genes (Figure 2A). This can happen if a DSB is repaired by copying a nonallelic sequence that contains a gene. Often called ‘segmental duplication,’ this process can be seeded by a stretch of homology shared between the donor and receptor sites. As the sites are nonallelic, the homology is often provided by distributed repetitive sequences found throughout genomes, such as those derived from transposable elements (Figure 2A) [6,8,9]. It is important to note that duplicate genes do not always arise as a result of gene conversion. For example, uneven crossing over between homologous chromosomes or sister chromatids can generate tandem gene duplications and retroposition can generate dispersed gene duplicates. However, even in these cases, gene conversion can play an important role in the probability of fixation of these newly duplicated paralogs [10].
Once there is more than one variant of a gene in the genome, whether they are different alleles of a gene found on homologous chromosomes or paralogous copies born by gene duplication, gene conversion can play a further role in the evolution of these copies [10]. In many cases, gene conversion promotes the homogenization of the duplicates, effectively reducing the amount of genetic diversity (Figure 2B) [11,12]. This can allow beneficial variants to spread quickly throughout a family of DNA elements into higher copy, or help purge deleterious alleles [10,13].
Gene conversion can also shuffle variants within the duplicates to generate novel chimeras of preexisting alleles (Figure 2C). In fact, even pseudogenized or silenced gene copies can facilitate diversification of intact homologous genes [14,15]. This can be favored by selection if diversity within the gene family is beneficial [10]. For example, diversity could broaden the number of potential targets of a gene family, especially amongst proteins with modular domains.
Overall, gene conversion can expand adaptive potential by generating duplicate genes, by spreading adaptive variants within a gene family, and by shuffling existing variation to generate novel alleles [10]. It is important to highlight, however, that not all mutations generated by gene conversion will be adaptive, even those within genes in conflict. Like any type of mutation, variants generated by gene conversion can be deleterious or even neutral.
Examples of gene conversion in host-pathogen conflicts.
The generation of diversity is critical for host defense genes to be able to adapt to rapidly evolving pathogens. While a great deal of prior literature has focused on amino acid substitutions and their role in molecular arms races [16,17], host immune genes can adapt via numerous mechanisms including those mediated by gene conversion. For instance, changes in gene repertoire, via gene duplication and expansion of gene families, is becoming increasingly appreciated for its role in host defense against pathogens. In invertebrates, which lack an adaptive immune response, changes in gene copy number generates much of the diversity in immune genes between and within species [18]. Commonly expanded gene families include those that are involved in recognition of pathogen-associated molecular patterns (PAMPs) such as bacterial peptidoglycan molecules, as well as small antimicrobial peptides that are effectors of the anti-pathogen response [18,19].
Recent studies have similarly revealed the importance of evolving large repertoires and functional diversity within several gene families that are involved in the mammalian innate antiviral gene response, including APOBECs [20], TRIM genes [21,22], AIM2-like receptors [23], and IFITs [24]. A recent cross-species analysis highlighted this trend on a genomic level: mammalian genes that are involved in conflicts with pathogens are much more likely to be part of variable copy gene families [25]. In many cases, the duplicate genes become subfunctionalized and specialized such that they recognize or combat distinct pathogens. This allows the host to avoid potential evolutionary tradeoffs that could occur if a single gene needs to adapt to restrict multiple pathogens [16].
Beyond copy number expansion, gene conversion also serves to rapidly change the sequences of immunity genes. For instance, some of the key signaling cytokines in the innate immune system, type I interferons, are encoded by a multigene family that has extensively diversified in mammalian species by both gene duplication and gene conversion between paralogs [26]. Likewise, paralogous families of antimicrobial peptides in both Drosophila and Caenorhabditis species show evidence for repeated gene conversion and homogenization [27,28]. In all of these cases, the whole gene conversion that has occurred completely ‘overwrites’ another allele, leaving two identical copies in the genome and complicating the inference of the ancestry of these genes.
In many other cases, however, the gene conversion is only partial, creating new chimeras that are shuffled versions of the ancestral alleles. For instance, in two families of mammalian innate immune effectors, the IFIT and Mx genes, there has been repeated partial gene conversion between paralogs [24,29], generating new combinations of domains that are predicted to confer unique functionality to these proteins. Likewise, the NAIP family of inflammasome sensors is characterized by extensive regions of recombination that have shuffled regions from several different rodent paralogs [30].
This shuffling is reminiscent of the genetic innovation that occurs during development of the adaptive immune systems of vertebrates. In the adaptive immune system of most vertebrates, as well as the evolutionary distinct adaptive immune system of jawless fish, recombination between defined loci occurs in somatic cells to generate a tremendous diversity of specialized immune molecules [31]. Although mechanistically distinct from gene conversion, the result of all of these genetic processes is the reshuffling of homologous regions that have independently diverged in order to generate novelty that can provide larger genetic changes than individual mutations. Indeed, this gene-conversion based shuffling of immune proteins is likely a primary mechanism by which diversity is generated in the immune system of plants, whereby paralogous Nod-like receptor (NLR) proteins undergo partial gene conversion to generate new anti-pathogen specificity [32]. Interestingly, such a mechanism for rapid generation of diversity may also contribute to rapid evolution of speciation barriers, as nearly all examples of hybrid incompatibility genes in a recent study in plants were mapped to NLR genes [33]. Given the many independent instances where gene conversion or recombination underlies genetic innovation in immunity proteins, there is a clear selective advantage to shuffling regions of pre-existing alleles in order to create new ways to recognize and defeat pathogens.
Examples of gene conversion amongst meiotic drive genes.
Meiotic drive genes cheat during gametogenesis to give themselves a transmission advantage into the next generation. These genes are often considered selfish, but they are widespread in eukaryotes [34]. Instead of being transmitted to 50% of offspring like normal alleles, drivers can promote their own transmission into up to 100% of offspring [34]. This cheating is generally costly to fitness, so meiotic drivers can be suppressed by unlinked regions of the genome, thereby setting up a genetic conflict between loci in the same genome [35].
Gene conversion is emerging as a driver of innovation amongst meiotic drive genes. First, gene conversion has likely contributed to the expansion and/or birth of meiotic driver genes [36-40]. This may be especially true when important components of drive systems consist of segments of DNA that can be multicopy within a genome. When these duplicate, the new copy can be selected because if it can cause drive and/or confer immunity against other drivers in the gene family [37-39,41].
The Spok3 and Spok4 meiotic drive genes of the fungus Podospora anserina, for example, are found within homologous 74-167 kb segments of DNA that may have moved to different chromosomal locations in different lineages via gene conversion [39]. Other Spok genes are flanked by transposon sequences that may have facilitated their expansion via gene conversion within filamentous fungi to as many as 11 copies in one lineage [38].
Novel duplicates of meiotic drive elements can also be selected if increased copy number of a sequence can establish drive or helps a drive system escape suppression [42]. For example, high copy number of the R2d sequence promotes meiotic drive in female house mice [43,44]. Around 2 million years ago, the ancestral R2d1 locus duplicated to generate the R2d2 locus (~6 Mb away), which subsequently expanded to multicopy [43,45]. The endpoints of the R2d2 locus are not resolved, so it is currently unclear if there is shared homology flanking the two R2d loci. Gene conversion is a likely mechanism of segmental duplication because the copied region is not a tandem duplication that could be generated by unequal crossing over. In addition, the duplication is 127 kb long and spans several genes, which would be unlikely for a retroposed fragment as these are generally made from processed transcripts of single genes [43,45].
Gene conversion is also thought to act on duplicated meiotic drive genes beyond a potential role in generating the duplicates [36,39,40,45]. For example, gene conversion has occurred within the duplicated Kindr genes that comprise one part of the knob meiotic driver found on the abnormal chromosome 10 (Ab10) in Maize. The gene conversion has contributed to the homogenization of the Kindr genes, which encode kinesin proteins that are required for the drive of the Ab10 knob and other distributed knob drivers found in Mazie [46]. The functional impact of Kindr gene homogenization is unclear, but it may facilitate maintenance of multiple functional copies of the genes, analogous to the mechanism thought to maintain functional gene copies on primate Y chromosomes [12,47].
Gene conversion has also been implicated in promoting the diversification of duplicated meiotic drive genes within a genome. The wtf genes, for example, comprise a family of meiotic drive genes found in the fission yeast S. pombe [40,48]. Different isolates contain between 25-38 wtf genes–some of which are meiotic drivers, some are suppressors of drive, and some have unknown functions. Many of these genes are adjacent to meiotic DSB hotspots and extensive gene conversion between wtf genes shuffles variation between them and facilitates their rapid divergence [36]. This is known to generate novel alleles with novel functions. For example, the wtf18-2 suppressor of the wtf13 meiotic driver was born via gene conversion of wtf13 sequence onto the wtf18 locus [41]. The wtf genes thus illustrate how gene conversion can facilitate innovation on both side of the genetic conflict between meiotic drivers and their suppressors.
Concluding thoughts and future perspectives
Most genes in eukaryotic genomes are well-suited to their current tasks and mutations that alter their functions are often purged by negative selection. On the contrary, mutations that generate new functionality are often selected for in genes that are involved in genetic conflicts.
Increasingly, gene conversion has been discovered to play a key role in generating such functional innovation by both producing duplicate copies of genes within a genome, and by homogenizing or shuffling those duplicate copies.
In this review, we describe several diverse examples of gene conversion playing a role in intergenomic and intragenomic conflicts. However, these instances are likely just the tip of the iceberg. Nonallelic gene conversion often occurs within the context of duplicate or multicopy gene families. Even with next generation whole-genome sequencing, accurately assembling these multicopy sequences was challenging or even impossible. Now, with third generation long read sequencing technologies, these previously inaccessible regions can more easily be deciphered. In the future, increased numbers of assembled genomes promise to facilitate higher-resolution analyses of entire gene families and even repetitive noncoding sequences like centromeres that may be hotspots for gene conversion and genetic conflict [49,50]. These data sets will undoubtedly uncover many more instances in which gene conversion has diversified genomes between species and even within a population of individuals. With a greater appreciation of the extent to which gene conversion is acting, it is likely that other instances of adaptive genetic novelty generated by gene conversion will be discovered, perhaps especially amongst genes in conflict where generation and diversification of gene duplicates may be particularly advantageous.
Acknowledgements:
Research in the Zanders lab is supported by the Stowers Institute for Medical Research, The March of Dimes, a Searle Scholars award, and the National Institutes of Health (NIH; award numbers R00GM114436 and DP2GM132936). Research in the Daugherty lab is supported by the UCSD Division of Biological Sciences, a Pew Biomedical Scholars award, and the National Institutes of Health (NIH; award numbers K22AI119017 and R35GM133633).
Footnotes
Conflict of Interest: SEZ is an inventor on a patent application based on wtf killers (patent application serial 62/491,107). MDD declares no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aderem A, Ulevitch RJ: Toll-like receptors in the induction of the innate immune response. Nature 2000, 406:782–787. [DOI] [PubMed] [Google Scholar]
- 2.Lewis SH, Obbard DJ: Recent insights into the evolution of innate viral sensing in animals. Curr Opin Microbiol 2014, 20:170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roach JC, Glusman G, Rowen L, Kaur A, Purcell MK, Smith KD, Hood LE, Aderem A: The evolution of vertebrate Toll-like receptors. Proc Natl Acad Sci U S A 2005, 102:9577–9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Queller DC, Strassmann JE: Evolutionary Conflict. Annual Review of Ecology, Evolution, and Systematics 2018, 49:73–93.•• This is an excellent review of genetic conflict including molecular arms races.
- 5.Burt A, Trivers R: Genes in conflict: the biology of selfish genetic elements. Cambridge, Mass.: Belknap Press of Harvard University Press; 2006. [Google Scholar]
- 6.Sasaki M, Lange J, Keeney S: Genome destabilization by homologous recombination in the germ line. Nat Rev Mol Cell Biol 2010, 11:182–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen JM, Cooper DN, Chuzhanova N, Ferec C, Patrinos GP: Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet 2007, 8:762–775. [DOI] [PubMed] [Google Scholar]
- 8.Dennis MY, Eichler EE: Human adaptation and evolution by segmental duplication. Curr Opin Genet Dev 2016, 41:44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bailey JA, Liu G, Eichler EE: An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet 2003, 73:823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Innan H, Kondrashov F: The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 2010, 11:97–108. [DOI] [PubMed] [Google Scholar]
- 11.Eickbush TH, Eickbush DG: Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics 2007, 175:477–485.• This work assembled all members of the wtf gene family from multiple natural isolates and shows that the gene family is rapidly evolving. Much of the evolution is driven by extensive gene conversion across most wtf genes.
- 12.Trombetta B, Cruciani F: Y chromosome palindromes and gene conversion. Hum Genet 2017, 136:605–619. [DOI] [PubMed] [Google Scholar]
- 13.Ellison CE, Bachtrog D: Non-allelic gene conversion enables rapid evolutionary change at multiple regulatory sites encoded by transposable elements. Elife 2015, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bittihn P, Tsimring LS: Gene Conversion Facilitates Adaptive Evolution on Rugged Fitness Landscapes. Genetics 2017, 207:1577–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takuno S, Nishio T, Satta Y, Innan H: Preservation of a pseudogene by gene conversion and diversifying selection. Genetics 2008, 180:517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daugherty MD, Malik HS: Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet 2012, 46:677–700. [DOI] [PubMed] [Google Scholar]
- 17.Duggal NK, Emerman M: Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol 2012, 12:687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchmann K: Evolution of Innate Immunity: Clues from Invertebrates via Fish to Mammals. Front Immunol 2014, 5:459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh J, Lun CM, Majeske AJ, Sacchi S, Schrankel CS, Smith LC: Invertebrate immune diversity. Dev Comp Immunol 2011, 35:959–974. [DOI] [PubMed] [Google Scholar]
- 20.Munk C, Willemsen A, Bravo IG: An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol Biol 2012, 12:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tareen SU, Sawyer SL, Malik HS, Emerman M: An expanded clade of rodent Trim5 genes. Virology 2009, 385:473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han K, Lou DI, Sawyer SL: Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet 2011, 7:e1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brunette RL, Young JM, Whitley DG, Brodsky IE, Malik HS, Stetson DB: Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J Exp Med 2012, 209:1969–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daugherty MD, Schaller AM, Geballe AP, Malik HS: Evolution-guided functional analyses reveal diverse antiviral specificities encoded by IFIT1 genes in mammals. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaw AE, Hughes J, Gu Q, Behdenna A, Singer JB, Dennis T, Orton RJ, Varela M, Gifford RJ, Wilson SJ, et al. : Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biol 2017, 15:e2004086.• This work reveals the diversity of innate immunity protein repertoires across several mammalian species, underscoring the importance of gene duplication in the evolution of mammalian immunity.
- 26.Woelk CH, Frost SD, Richman DD, Higley PE, Kosakovsky Pond SL: Evolution of the interferon alpha gene family in eutherian mammals. Gene 2007, 397:38–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lazzaro BP, Clark AG: Evidence for recurrent paralogous gene conversion and exceptional allelic divergence in the Attacin genes of Drosophila melanogaster. Genetics 2001, 159:659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas JH: Concerted evolution of two novel protein families in Caenorhabditis species. Genetics 2006, 172:2269–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell PS, Young JM, Emerman M, Malik HS: Evolutionary Analyses Suggest a Function of MxB Immunity Proteins Beyond Lentivirus Restriction. PLoS Pathog 2015, 11:e1005304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tenthorey JL, Kofoed EM, Daugherty MD, Malik HS, Vance RE: Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes. Mol Cell 2014, 54:17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boehm T: Design principles of adaptive immune systems. Nat Rev Immunol 2011, 11:307–317. [DOI] [PubMed] [Google Scholar]
- 32.Borrelli GM, Mazzucotelli E, Marone D, Crosatti C, Michelotti V, Vale G, Mastrangelo AM: Regulation and Evolution of NLR Genes: A Close Interconnection for Plant Immunity. Int J Mol Sci 2018, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chae E, Bomblies K, Kim ST, Karelina D, Zaidem M, Ossowski S, Martin-Pizarro C, Laitinen RA, Rowan BA, Tenenboim H, et al. : Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell 2014, 159:1341–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindholm AK, Dyer KA, Firman RC, Fishman L, Forstmeier W, Holman L, Johannesson H, Knief U, Kokko H, Larracuente AM, et al. : The Ecology and Evolutionary Dynamics of Meiotic Drive. Trends Ecol Evol 2016, 31:315–326. [DOI] [PubMed] [Google Scholar]
- 35.Crow JF: Why is Mendelian segregation so exact? Bioessays 1991, 13:305–312. [DOI] [PubMed] [Google Scholar]
- 36.Eickbush MTY JM; Zanders SE : Killer meiotic drive and dynamic evolution of the wtf gene family. Mol Biol Evol 2019, In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tao Y, Araripe L, Kingan SB, Ke Y, Xiao H, Hartl DL: A sex-ratio meiotic drive system in Drosophila simulans. II: an X-linked distorter. PLoS Biol 2007, 5:e293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grognet P, Lalucque H, Malagnac F, Silar P: Genes that bias Mendelian segregation. PLoS Genet 2014, 10:e1004387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogan AAA-V SL; Granger-Farbos A; Svedberg J; Bastiaans E; Debets AJM; Coustou V; Yvanne H; Clave C; Saupe SJ; Johannesson H: Combinations of Spok genes create multiple meiotic drivers in Podospora. bioRxiv 2019.• This work identified that the Spok family of meiotic drive genes underlie all known, but unmapped, meiotic drivers in Podospora anserina. The authors also describe how the Spok3 and Spok4 genes are found within a haplotype termed the Spok block that has moved around within the genome in different isolates.
- 40.Hu W, Jiang ZD, Suo F, Zheng JX, He WZ, Du LL: A large gene family in fission yeast encodes spore killers that subvert Mendel's law. Elife 2017, 6.•• This manuscript identifies wtf genes as meiotic drivers in S. pombe and shows that the evolution of the wtf gene family has been shaped by gene conversion.
- 41.Bravo Nunez MA, Lange JJ, Zanders SE: A suppressor of a wtf poison-antidote meiotic driver acts via mimicry of the driver's antidote. PLoS Genet 2018, 14:e1007836.• This manuscript identifies the wtf18-2 gene as a suppressor of the wtf13 meiotic driver. The wtf18-2 allele arose by a gene conversion event that overwrote part of the wtf18 locus with wtf13 sequence. This demonstrated that the wtf gene family includes both meiotic drivers and suppressors of drive.
- 42.Shen R, Wang L, Liu X, Wu J, Jin W, Zhao X, Xie X, Zhu Q, Tang H, Li Q, et al. : Genomic structural variation-mediated allelic suppression causes hybrid male sterility in rice. Nat Commun 2017, 8:1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Didion JP, Morgan AP, Clayshulte AM, McMullan RC, Yadgary L, Petkov PM, Bell TA, Gatti DM, Crowley JJ, Hua K, et al. : A multi-megabase copy number gain causes maternal transmission ratio distortion on mouse chromosome 2. PLoS Genet 2015, 11:e1004850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Didion JP, Morgan AP, Yadgary L, Bell TA, McMullan RC, Ortiz de Solorzano L, Britton-Davidian J, Bult CJ, Campbell KJ, Castiglia R, et al. : R2d2 Drives Selfish Sweeps in the House Mouse. Mol Biol Evol 2016, 33:1381–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morgan AP, Holt JM, McMullan RC, Bell TA, Clayshulte AM, Didion JP, Yadgary L, Thybert D, Odom DT, Flicek P, et al. : The Evolutionary Fates of a Large Segmental Duplication in Mouse. Genetics 2016, 204:267–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dawe RK, Lowry EG, Gent JI, Stitzer MC, Swentowsky KW, Higgins DM, Ross-Ibarra J, Wallace JG, Kanizay LB, Alabady M, et al. : A Kinesin-14 Motor Activates Neocentromeres to Promote Meiotic Drive in Maize. Cell 2018, 173:839–850 e818.•• This manuscript shows that Kindr, a multicopy gene encoding a kinesin motor protein, is required for meiotic drive of maize knobs.
- 47.Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Page DC: Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 2003, 423:873–876. [DOI] [PubMed] [Google Scholar]
- 48.Nuckolls NL, Bravo Nunez MA, Eickbush MT, Young JM, Lange JJ, Yu JS, Smith GR, Jaspersen SL, Malik HS, Zanders SE: wtf genes are prolific dual poison-antidote meiotic drivers. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang CH, Chavan A, Palladino J, Wei X, Martins NMC, Santinello B, Chen CC, Erceg J, Beliveau BJ, Wu CT, et al. : Islands of retroelements are major components of Drosophila centromeres. PLoS Biol 2019, 17:e3000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malik HS, Henikoff S: Major evolutionary transitions in centromere complexity. Cell 2009, 138:1067–1082. [DOI] [PubMed] [Google Scholar]