Abstract
A causal factor in mammalian aging is the accumulation of senescent cells (SnCs). SnCs cause chronic inflammation, and removing SnCs decelerates aging in mice. Despite their importance, turnover rates of SnCs are unknown, and their connection to aging dynamics is unclear. Here we use longitudinal SnC measurements and induction experiments to show that SnCs turn over rapidly in young mice, with a half-life of days, but slow their own removal rate to a half-life of weeks in old mice. This leads to a critical-slowing-down that generates persistent SnC fluctuations. We further demonstrate that a mathematical model, in which death occurs when fluctuating SnCs cross a threshold, quantitatively recapitulates the Gompertz law of mortality in mice and humans. The model can go beyond SnCs to explain the effects of lifespan-modulating interventions in Drosophila and C. elegans, including scaling of survival-curves and rapid effects of dietary shifts on mortality.
Subject terms: Senescence, Ageing, Systems biology, Dynamical systems, Stochastic modelling
One of the underlying causes of aging is the accumulation of senescent cells, but their turnover rates and dynamics during ageing are unknown. Here the authors measure and model senescent cell production and removal and explore implications for mortality.
Introduction
Senescent cells (SnCs) accumulate with age in mice and humans in many tissues1–7, due in part to DNA damage, damaged telomeres, and oxidative stress5,8. These cells, characterized by high levels of p16 and SA-β-Gal5, enter permanent cell cycle arrest, and secrete a characteristic profile of molecules including pro-inflammatory signals9 and factors that slow regeneration9 (Fig. 1a). They have physiological roles in development, cancer prevention, and wound healing9–11. However, as organisms age, accumulating levels of SnC cause chronic inflammation and increase the risk of many age-related diseases, including osteoarthritis, neurodegeneration, and atherosclerosis12–24.
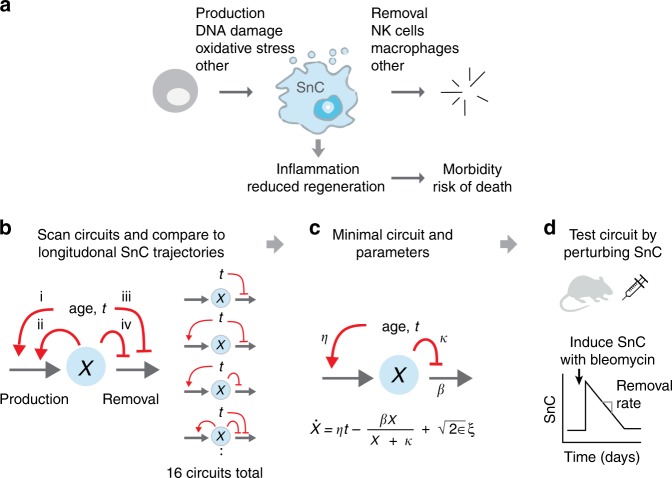
Fig. 1.
Approach for inferring SnC dynamics throughout adulthood. a Many processes, including DNA damage and developmental and paracrine signals, lead to SnC production. SnCs are cleared by immune mechanisms, and secrete factors that lead to morbidity and mortality. b–d We scanned a wide class of models for SnC dynamics, and compared them to longitudinal SnC data and direct SnC perturbation experiments to establish a minimal model for SnC stochastic dynamics and determine its rate constants. In the minimal model, η is the increase in SnC production rate with age, β is the removal rate, κ is the half-way saturation point for removal, and ϵ is the noise amplitude.
Accumulation of SnCs is known to be causal for aging in mice: continuous targeted elimination of whole-body SnCs increases mean lifespan by 25%, attenuates age-related deterioration of heart, kidney, and fat, delays cancer development25 and causes improvement in the above-mentioned diseases.
These studies indicate that SnC abundance is an important causal variable in the aging process. Despite their importance, however, the production and removal rates of SnCs are unknown9,26. For example, it is unclear whether SnCs passively accumulate or if they are turned over rapidly, and if so, whether their half-life changes with age. Since turnover affects the ability of a system to respond to fluctuations, information about these rates is crucial in order to mathematically test ideas about the possible role of SnCs in the age-dependent variations in morbidity and mortality between individuals.
Here, we address this experimentally and theoretically. To understand the dynamics of SnCs, we scanned a wide class of mathematical models of SnC dynamics, and compared these models to longitudinal SnC trajectories1 and direct SnC induction experiments in mice (Fig. 1b–d). The models all describe SnC production and removal. They differ from one another in the way that production and removal rates are affected by age and by SnC abundance. The models describe all combinations of four possible mechanisms for accumulation of SnCs (Fig 1b): (i) SnC production rate increases with age due to accumulation of mutations27, telomere damage, and other factors that trigger cellular senescence11, (ii) SnCs catalyze their own production by paracrine and bystander effects28, (iii) SnC removal decreases with age due to age-related decline in immune surveillance functions29, and (iv) SnCs reduce their own removal rate, which can be due to SnC-related signaling, such as SASP, downregulation of immune surveillance by SnCs, SnCs saturating immune surveillance mechanisms (similar to saturation of an enzyme by its substrate), or to disruption of tissue and extracellular matrix architecture that interferes with removal.
Mechanism (iv) is distinct from mechanism (iii) because the decline in removal rate in (iv) depends on SnC abundance, rather than on age directly. Although (iv) can arise from various biological processes, we denote it for simplicity ‘saturation of removal’. These four effects lead to 16 different circuits (Fig. 1b) with all combinations of whether or not each of effects (i–iv) occur. Additionally, each of the 16 models includes parameters for basal production and removal. The models have rate constants that are currently uncharacterized. We also tested models which incorporate additional non-linearities (Supplementary Note 1, Supplementary Fig. 1).
Results
SnC dynamics during ageing in mice
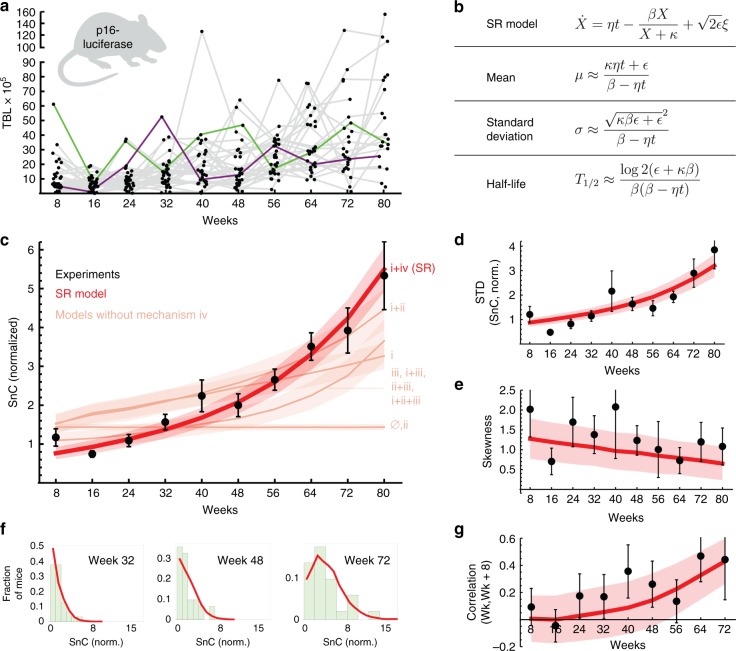
To find which of the model mechanisms best describes SnC dynamics, and with which rate constants, we compared the models to longitudinal data on SnC abundance in mice collected by Burd et al. 1. SnC abundance was measured using a luciferase reporter for the expression of p16INK4a, a biomarker for SnCs. Total body luminescence (TBL) was monitored every 8 weeks for 33 mice, from early age (8 weeks) to middle–late adulthood (80 weeks) (Fig. 2a).
Fig. 2.
Saturated-removal (SR) model captures longitudinal SnC trajectories in mice. a Total body luminescence (TBL) of p16-luciferase in mice (n = 33). Gray lines connect data from the same individual mice (green and purple lines are examples of individual trajectories). b SR model equations and their approximate analytical solutions. The SR model (red line) captures c the mean SnC abundance, d standard deviation of SnC abundance, e skewness, and f shape of the distributions among equal-aged individuals, and g correlation between subsequent measurements on the same individuals. TBL was normalized to give a mean abundance of 1 at young ages. Maximum-likelihood parameters for the SR model are: η = 0.15 day−1 year−1, β = 0.27 day−1, κ = 1.1, ε = 0.14 day−1. Pink lines in c: best-fit of all models without saturation mechanism iv, that have an age-related increase in SnCs, best-fit parameters are in Supplementary Note 1. Mean and standard error (shaded red, pink regions) are from bootstrapping. Source data are provided as a Source Data file.
The luciferase in these mice was introduced into one of the p16 loci, causing the mice to be heterozygous for p16, which may impair proper activation of the senescence program. We therefore also tested longitudinal measurements of SnCs based on another method. For this we obtained longitudinal data from Yamakoshi et al. 30, who measured SnC abundance by creating a transgenic mouse model with a human p16 gene tagged with luciferase, retaining the native p16 loci. Although this dataset has much fewer mice, it shows similar dynamics to the dataset of Burd et al.1 (Supplementary Note 1, Supplementary Fig. 2), suggesting a similar underlying dynamical process.
We tested how well each model describes the longitudinal SnC trajectories of Burd et al.1 by finding the maximum-likelihood parameters for each of the 16 models, adjusting for number of parameters (Supplementary Notes 1 and 2, Supplementary Tables 1–4). A principle emerges from this comparison: in order to capture the longitudinal dynamics, the mechanism must have rapid turnover of SnCs on the timescale of a few days in young mice, and it also must include mechanism (iv), which represents a decline in removal that depends on SnC abundance rather than directly on age. The simplest model that describes the data thus has only two interactions (Fig. 1c): SnC production rate increases linearly with age (mechanism i), and SnCs slow down their own removal rate (mechanism iv). We call this model the saturating removal model (SR model), whose equation is given in Fig. 2b.
The SR model captures the accelerating rise of mean SnC abundance with age in the longitudinal data (Fig. 2c and Supplementary Figs. 3, 4): as SnCs accumulate, they slow their own removal, leading to even higher SnC levels. The SR model also explains the SnC variability between individuals which accelerates with age (Fig. 2d), and the SnC distributions among equal-aged individuals (Fig. 2e), which are skewed to the right (Fig. 2f).
Importantly, the SR model captures the fact that SnC fluctuations become more persistent with age, as evidenced by an increasing correlation between subsequent measurements (Fig. 2g, F-test for linear regression, p-value 0.0047; F-statistic 16.5): individuals with higher (or lower) than average SnC levels stay higher (or lower) for longer periods with age. This increased persistence is due to the effect of SnCs on their own removal rate. Models without mechanism iv (saturation of removal) show a poor overall fit (pink lines in Fig. 2c, ΔBIC > 44.3).
SnC lifetime is days in young mice and weeks in old mice
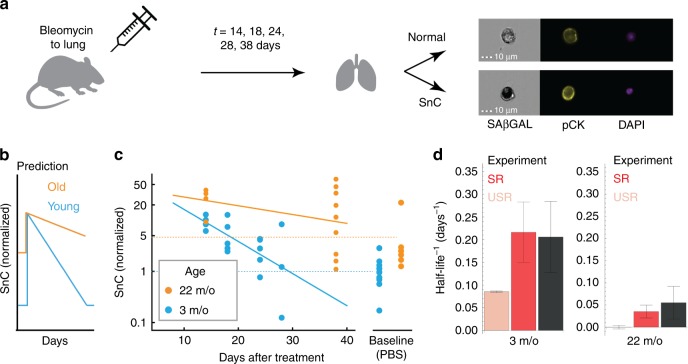
The maximum-likelihood parameters of the SR model (listed in the caption of Fig. 2) provide quantitative predictions for SnC half-lives: SnC turnover is rapid in young mice, with a half-life of about 5 ± 1 days at 3 months of age; Turnover slows with age, so that SnC half-life is about 25 ± 6 days at 22 months.
We tested these predictions using experiments in mice by inducing SnCs and analyzing their dynamics. To induce senescence in mice lungs we used intra-tracheal bleomycin administration (Fig. 3a), a DNA-damaging agent that induces cellular senescence in the lung epithelium a few days after treatment5,31.
Fig. 3.
SnC half-life measurements in mice support SR model predictions. a Bleomycin or PBS was introduced by intratracheal installation to mice on day 0. Lungs were analyzed on the indicated days thereafter. Representative images of lung cells analyzed by imaging flow cytometry show how senescent epithelial cells were identified, using SA-β-Gal, Pan-Cytokeratin (pCK), and DAPI staining. SnC removal rate was estimated by log-linear fit. b The SR model predicts that SnCs rapidly return to baseline in young mice and that removal is slower in old mice. c Fraction of SnCs in mouse lungs after treatment with bleomycin (1.5 U/kg). In young mice, SnC levels return to baseline with a half-life of about 5 days. In old mice, baseline SnC levels are about five-fold higher, and SnC removal rate is slower than in young mice . d SnC removal rates (half-life−1) for young and old mice (mean and standard error from bootstrapping, black) agree with the SR model predictions (red, mean and SE were calculated by bootstrapping, see the “Methods” section). The best-fit model without mechanism (iv), the USR model (mechanisms i + iii), shows a poor prediction (pink). For both ages, the USR prediction is different from the observed half-life with p < 0.01 from bootstrapping. Source data are provided as a Source Data file.
We quantified the fraction of senescent lung epithelial cells at different time points following bleomycin administration (Fig. 3a) using imaging flow cytometry. Epithelial SnCs were defined as cells positive for a senescent cell marker (SA-β-Gal) and an epithelial marker (pan-Cytokeratin, pCK). This cell population was also HMGB1 nuclear negative, as expected in SnCs5,32, and previously shown5 to correspond to non-proliferative cells (negative Ki67 assay, see Supplementary Note 3, Supplementary Fig. 6).
In 3-month-old mice, SnC levels decayed with a half-life of τ = 4.7 days (τ−1 = 0.21 +/− 0.07 days−1) and reached their baseline level within less than a month (Fig. 3b, c), as predicted. SnC levels in young mice lungs are thus in a rapid dynamic balance of production and removal.
To test the prediction that removal slows with age (Fig. 3b), we performed the bleomycin treatment in old mice (22-month old). In these mice, the baseline level of SnCs was about five-fold higher than in young mice (Fig. 3d). SnCs decayed with a half-life of τ = 18 days, τ−1 = 0.055 +/− 0.035 days−1), slower than that of young mice as predicted (p = 0.038 from bootstrapping, Fig. 3b).
These turnover measurements quantitatively agreed with the predictions of the SR model (Fig. 3d, Supplementary Note 4, Supplementary Fig. 7) with no additional fit. This agreement occurred despite the use of distinct SnC markers in the two data sets (SA-β-Gal in the bleomycin experiment vs. p16INK4A-luciferase in the longitudinal experiment), suggesting consistency between the measurement methods.
Our results suggest a core mechanism in which SnC production rate rises linearly with age, and SnCs slow their own removal (Supplementary Note 5, Supplementary Fig. 8). This slowdown of removal accelerates SnC accumulation with age. Slowdown of removal also amplifies fluctuations in SnC levels at old ages. This amplification, known as critical slowing down33,34, results in long-lasting differences among individuals at old ages. In other words, young mice have large spare removal capacity of SnC; old mice have much smaller spare removal capacity. This smaller removal capacity means that any addition of SnCs takes longer to remove, causing larger and more persistent variation in SnC levels among individuals (Fig. 2g).
The SR model quantitatively recapitulates the Gompertz law
In the remainder of the paper, we use mathematical analysis to explore the implications of these findings for the question of variability in mortality. Mortality times vary even in inbred organisms raised in the same conditions, demonstrating a non-genetic component to mortality35,36. In many species, including mice and humans, risk of death rises exponentially with age, a relation known as the Gompertz law37–39, and decelerates at very old ages. The Gompertz law has no known explanation at the cellular level.
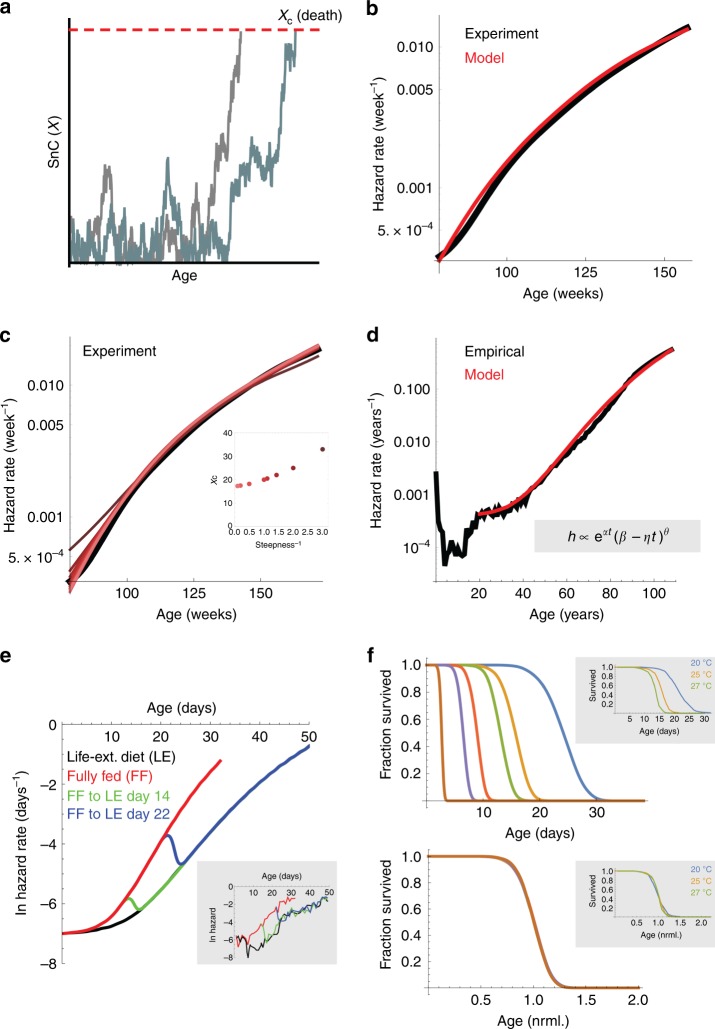
To connect SnC dynamics and mortality, we need to know the relationship between SnC abundance and risk of death1. The precise relationship is currently unknown. Clearly, SnC abundance is not the only cause for morbidity and mortality. It seems to be an important causal factor because removing SnCs from mice increases mean lifespan25, and adding SnCs to mice increases risk of death and causes age-related phenotypes23. We therefore explored the simple possibility that death can be modeled to occur when SnC abundance exceeds a threshold level XC, representing a collapse of an organ system or a tipping point such as sepsis (Fig. 4a). Thus, death is modeled as a first-passage time process, when SnC cross XC. We use this assumption to illustrate our approach, because it provides analytically solvable results. We also show that other dependencies between risk of death and SnC abundance, such as sigmoidal functions with various degrees of steepness, provide similar conclusions.
Fig. 4.
SR model can explain the variability in mortality between individuals. a To model the relation between risk of death and SnC levels, we assumed a simple threshold model where death occurs when SnC abundance exceeds a critical threshold XC. b Mouse mortality (C57BL/6J mice obtained from the Mouse Phenome Database60, black line) is well fit by the SR model (red line) with parameters consistent with the data of Figs. 1, 2, with death defined when SnC exceed a threshold (η = 0.084 day−1 year−1, β = 0.15 day−1, κ = 0.5, ε = 0.16 day−1, XC = 17). c Similar results are obtained by assuming a more general sigmoidal dependency between SnC abundance X and risk of death: . Parameters are the same as b, except that XC is adjusted according to the steepness parameter θ (inset). d The SR model with added age-independent extrinsic mortality of 0.4 × 10−3 year−1 (red) matches human mortality statistics61 (black). Inset: approximate analytical solution for the first passage time in the SR model shows the Gompertz law and deceleration at old ages. The parameters are similar to b, except a ~60-fold decrease in η: η = 0.00135 day−1 year−1, β = 0.15 day−1, κ = 0.5, ε = 0.142 day−1, XC = 17. e SR model describes rapid shifts in mortality when fully fed Drosophila transition to a lifespan-extending dietary intervention (LE), (inset: experimental data from Mair et al.45), with β = 1 h−1, κ = 1, ε = 1 h−1, η = 0.03 day -1 h−1 and XC = 15. LE was modeled by a decrease in η: η = 0.02 h−1 day−1 (changes in other parameters lead to similar conclusions, see Supplementary Note 8). f Lifespan of C. elegans raised at different temperatures varies by an order of magnitude, but survival curves collapse on a single curve when time is scaled by mean lifespan (inset: data from Stroustrup et al. 35). The SR model provides scaling for perturbations that affect η, but not other parameters (β = 1 h−1, κ = 1, and ε = 1 h−1, η = 0.07 h−1 day−1, XC = 20, Supplementary Note 9). Source data are provided as a Source Data file.
The SR model analytically reproduces the Gompertz law, including the observed deceleration of mortality rates at old ages (Fig. 4b–d, Supplementary Note 2, Supplementary Fig. 5, Supplementary Table 5). Notably, most models without both rapid turnover and slowdown of removal do not provide the Gompertz law (Supplementary Note 2). The deceleration of mortality rates at very old ages occurs in the model due to the increased persistence of SnC at old age. Those with high SnC have already died, whereas those with low SnC retain low SnC levels for long periods of time and avoid death. The SR model gives a good fit to the observed mouse mortality curve (Fig. 4b, c, Supplementary Note 1) using parameters that agree with the present experimental half-life measurements and longitudinal SnC data (Supplementary Note 1). Thus, turnover of days in the young and weeks in the old provides SnC variation such that individuals cross the death threshold at different times, providing the observed mortality curves.
The SR model can describe the observed increase in mean lifespan of mice in experiments in which a fraction of SnCs are continually removed (Supplementary Note 6). More generally, the SR model can address the use of drugs that eliminate SnCs, known as senolytics40. To reduce toxicity concerns, it is important to establish regimes of low dose and large inter-dose spacing41. The model provides a rational basis for scheduling senolytic drug administrations. Specifically, treatment should start at old age, and can be as infrequent as the SnC turnover time (~month in old mice) and still be effective (Supplementary Note 6).
We also adapted our results from the mouse data to study human mortality curves. In humans, mortality has a large non-heritable component42,43. A good description of human mortality data, corrected for extrinsic mortality, is provided by the same parameters as in mice, except for a 60-fold slower increase in SnC production rate with age in the human parameter set (Fig. 4d, Supplementary Note 7, Supplementary Table 6). This slower increase in SnC production rate can be due to improved DNA maintenance in humans compared to mice44. We conclude that the critical slowing-down described by the SR model provides a possible cellular mechanism for the variation in mortality between individuals.
SR-type dynamics and ageing of Drosophila and C. elegans
The generality of the SR model suggests that it might also apply to organisms where ageing may be driven by factors other than SnCs, such as Drosophila melanogaster and C. elegans, in which lifespan variation is well-studied35,45. In these organisms, the present approach can be extended by considering X as a causal factor for aging, that accumulates with age and has SR-type dynamics46, namely turnover that is much more rapid than the lifetime, increasing production and self-slowing removal. One clue for the identity of such factors may be gene-expression variations in young organisms that correlate with individual lifespan47–49, and the actions of genes that modulate lifespan39,50–53.
Work in C. elegans and Drosophila provides constraints to test the SR model. For example, Drosophila shows rapid switches between hazard curves when transitioned between normal and lifespan-extending diets (Fig. 4e, inset). These switches are well-described by the SR model, due to its rapid turnover property (Fig. 4e and Supplementary Note 8, Supplementary Fig. 9). The rapid turnover property entails that the level of X can adjust after a change in any of the parameters of the model. A model without rapid turnover could not explain these results.
We further tested whether the SR model can explain the survival curves of C. elegans under different life-extending genetic, environmental, and dietary perturbations35. These perturbations change mean lifespan by up to an order of magnitude. The survival curves show a remarkable feature called temporal scaling: the survival curves collapse onto approximately the same curve when age is scaled by mean lifespan (Fig. 4f insets). That is, the entire distribution of death times, including its mean and standard deviation, is determined by a single parameter, which depends on the perturbation. We find that the SR model provides the shape of the survival curves, as well as their temporal scaling feature. Temporal scaling is found in the SR model by assuming that the perturbations affect the accumulation rate η (Fig. 4f, Supplementary Note 9 and Supplementary Fig. 10A).
Temporal scaling cannot be explained by models without rapid turnover (Supplementary Fig. 10B), or by varying any other parameter except η in the SR model. Thus, we predict loss of temporal-scaling of survival curves when a perturbation affects other SR-model parameters, such as removal rate β or noise ∈ (Supplementary Fig. 10C, D). This prediction may apply to exceptional perturbations in which temporal scaling is not found, such as the eat-2 and nuo-6 mutations (Supplementary Fig. 10E, F). We conclude that the SR model of rapid turnover with critical-slowing down is a candidate explanation for the temporal scaling of survival curves in C. elegans.
Discussion
In this study, we propose a framework for the dynamics of SnCs based on rapid turnover that slows with age. Bleomycin-induced SnC half-life is days in young mice and weeks in old mice, causing critical slowing down, which greatly amplifies the differences between individual SnC levels at old age. We theoretically explore the implications of this slowdown in a model in which SnCs cause death when they exceed a threshold. The widening variation in SnC levels with age causes a mortality distribution that follows the Gompertz law of exponentially increasing risk of death. The mortality distribution of mice and humans is well-described by the SR model with the SnC half-lives measured here. Future work may test this proposed connection between SnC dynamics and mortality by experimentally measuring risk of death as a function of SnC abundance.
The rapid removal of SnCs that we observe following bleomycin-induced DNA damage is in line with studies that showed efficient removal of SnCs in vivo following liver fibrosis or induction by senescence by mutant Ras54–56. On the other hand, when senescence was induced in the skin by directly activating the cell-cycle inhibitor p14ARF, which was not associated with an increase in tissue cytokine expression or inflammation, the induced SnCs persisted in the tissue for several weeks57. Clearance may thus depend on the tissue, on the method of senescence induction, and on the presence of SASP.
The present analysis of longitudinal p16 trajectories suggests that SnC slow down their own removal rate. This effect may be due to several mechanisms, including SASP, disruption of tissue architecture, or SnC abundance exceeding immune capacity. For the latter effect, SnC abundance at old age needs to be comparable to the abundance of the immune cells that remove them, which make up on the order of 0.1% of the body’s cells58,59. Further research is needed to characterize these effects.
Our results suggest that treatments that remove SnCs can therefore have a double benefit: an immediate benefit from a reduced SnC load, and a longer-term benefit from increased SnC removal. Similarly, interventions that increase removal capacity, for example by augmenting the immune surveillance of SnC, are predicted to be an effective approach to reduce SnC levels. More generally, the present combination of experiment and theory can be extended to explore further stochastic processes in aging, in order to bridge between the population-level and molecular-level understanding of aging.
Methods
Stochastic model simulation
Simulations of the stochastic models were performed by using the ItoProcess function of Mathematica (V11.3), with a step size of 1 day. Negative X values were avoided using a reflecting boundary condition at X = 0. In simulations that included mortality, time of death was the first time-point where X exceeded XC.
Model comparison to p16INK-luciferase measurements
We sought for each model the parameters that maximize the log-likelihood of the measured trajectories (Fig. 1a). We calculated the log-likelihood of a model m with parameters θ as follows. Let Xi,j be the measured SnC level (SnC = TBL/9.63 to give SnC = 1 for young mice) of mouse j at time point i (with X0,j = 0). We denote by Probm,θ,i(a|b) the probability of reaching SnC level a at time point i given SnC level b at time point i−1. We call such a step from i−1 to i a sub-trajectory. We estimated this probability from simulations (4000 simulations for every such sub-trajectory). The log-likelihood is , n = 294 sub-trajectories. For each model, we sought the parameter set that maximizes the log-likelihood (see Supplementary Note 1 for more details). Confidence intervals for the best-fit parameters, as well as for estimates for SnC half-life, were calculated by bootstrapping (selecting mice at random with replacements). Modeling experimental noise (multiplicative noise with amplitude up to 30%) did not affect the best-fit parameters (Supplementary Note 1). To find parameters for the model that describe both the longitudinal trajectories as well as mouse mortality statistics, we scanned the subset of parameters that fit the mortality distribution of mice. Mortality statistics of WT (C57BL/6J) mice were obtained from the Mouse Phenome Database60. Because mortality in young mice appears to be unassociated with the accumulation of SnCs1, we only considered deaths that occurred after age one year (which make up 97% of the total deaths in the dataset). We performed a comprehensive scan of values of β0 and κ0 and constrained η,∈ to values that give a mean and standard deviation of the simulated mortality distributions that is within 2% of the empirical values for WT mice. The critical SnC level XC was set at XC = 17, which is the maximal SnC level in the Burd et al. dataset1. We then calculated maximum-likelihood parameters and confidence intervals as described above.
Population-level measures
The mean and variance at time-point i are the mean and variance of , where N is the number of mice. Autocorrelation is the Pearson correlation of the two vectors . The measures were calculated in the same manner from model simulations. Typical SnC removal rate (half-life−1) for the model at a given age i was estimated by , where is the mean SnC level at age i (see Supplementary Note 4 for discussion of alternative ways to estimate SnC half-life). For the USR model, typical SnC removal rate at age i is (β0−β1i) log(2)−1.
Quantification of SnCs in mouse lung epithelium
We subjected 3-month-old (young) and 22-month-old (aged) C57BL6 mice to intra-tracheal installation of 1.5 U/kg bleomycin (Sigma) solution in PBS (or PBS as a control treatment). We euthanized the mice at 14, 18, 24, and 28 days for the young mice and day 14 and day 38 for the aged mice. The quantification of senescent epithelial cells was performed as previously described5 with modification. Lung tissue was chopped into 2–5 mm pieces in HBSS (14025050, Gibco) on ice and incubated in the 5 ml dissociation buffer (1 mg/ml Collagenase Type IV (C9263, Sigma), 0.1 mg/ml DNase I (10104159001, Roche) in HBSS) at 37 °C for 50 min. Cells were washed with HBBS and then fixed with 4% PFA for 5 min. Post fixation, cells were washed and incubated with X-Gal-staining solution for 16 h at 37 °C. The X-Gal-staining solution consisted of 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6 × 3H2O and 2.5 mM X-Gal (Inalco) in PBS at pH 5.5 containing 1 mM MgCl2. Post X-Gal staining the cells were fixed with fixation buffer for 30 min at 4 °C and washed with permeabilization buffer (00-5223-56, eBioscience, San Diego, CA). The cells were then incubated with PE-conjugated pan-cytokeratin (ab52460, Abcam) and HMGB1 (ab18256, Abcam) antibodies for an hour at 4 °C. For visualization of HMGB1 antibody, we used Qdot605-labeled Goat Anti-Rabbit antibody (Q11402MP, ThermoFisher). Antibodies were diluted in the permeabilization buffer with the dilution of 1:100 of PE-conjugated pan-cytokeratin and primary HMGB1, and 1:50 of Qdot605. Before visualization, the cells were stained with DAPI and filtered through a 100 µm membrane. The resulting cells were analyzed by imaging flow-cytometry using ImageStreamX mark II (Amnis, Part of EMD milipore—Merck, Seattle, WA, USA, see Supplementary Note 3 for gating strategy summary). PE staining was collected at channel 3, the DAPI at channel 7 and the Qdot605 at channel 10, in addition to the bright-field images collected at channels 1 and 9. Analysis of the image data was performed using IDEAS 6.2 software. Cells were first gated according to their area (in μm2) and aspect ratio (ratio between width and length) of the bright field images, to eliminate debris and aggregates. Then, we gated on focused cells using the gradient RMS (which measures the sharpness quality of an image by using the average gradient of a pixel normalized for variations in intensity levels) and contrast (measures the sharpness quality of an image by detecting large changes of pixel values). Cropped cells were excluded by using the centroid X feature (the number of pixels in the horizontal axis from the upper, left corner of the image to the center of the image mask). To verify that only single cells were analyzed, cells were further gated for single nuclei using the area and aspect ratio of the nuclear image of the DAPI staining. SA-beta-Gal staining was quantified using the Mean pixel (the mean of the background-subtracted pixels) contained in the bright-field image5. Staining of pCK was quantified using the Intensity (the sum of the background subtracted pixel values within the image) and the Max pixel (the largest value of the background-subtracted pixels contained in the image) features of the corresponding channels. To quantify staining of HMGB1 specifically, its intensity was calculated. We first gated for pCK-positive cells then for HMGB1 negative, SA-β-Gal-positive cells to quantitate the SnCs in lung epithelium. Following the method, establishment of pCK positive, SA-β-Gal-positive cells were considered senescent in further experiments. In total, the mice analyzed were 3-month-olds treated with 1.5 U/kg (n = 17), 3-month-olds treated with PBS (n = 13), 22-month-olds treated with 1.5 U/kg (n = 13), and 22-month-olds treated with PBS (n = 6). We complied with all relevant ethical regulations for animal testing and research. The experiments were approved by the Weizmann IACUC committee.
Analysis of the bleomycin treatment time series
We estimated the turnover of SnCs by calculating the removal time after a perturbation with bleomycin. The removal rate is the slope of the log-linear regression model, which we fit for each experiment (with confidence intervals calculated by bootstrapping). We obtained the response time predicted by the model by bootstrapping and simulating the model after perturbation (see Supplementary Note 4 for details). Statistical significance tests were computed by bootstrapping.
Estimation of hazard and survival functions
We fit hazard and survival functions from mortality data by interpolation using the Mathematica (V11.3) function SmoothKernelDistribution and then applying the Mathematica functions HazardFunction and SurvivalFunction. For the mice survival data, the SmoothKernelDistribution was computed with a bandwidth of 80 days.
Simulation of Drosophila and C. elegans survival curves
We simulated the mortality trajectories of Drosophila and C. elegans using the SR model, by assuming a rapid turnover and saturation β = 1 h−1, κ = 1[au], and also set ε = 1 [au]2 h−1 where [au] is the mean level of X in young organisms. These parameters correspond to a turnover of X on the order of hours. For C. elegans, to fit the survival curve obtained by Stroustrup et al.35, we set η = 0.07[au] h−1 day−1 and assumed that death occurs when X > XC for XC = 20[au]. Similarly, to fit the Mair et al.45 data, we set the following parameters for the Drosophila simulations: η = 0.03[au] h−1 day−1 and XC = 15[au], and assumed a baseline mortality of ln hazard = −7 day−1.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
U.A. is the incumbent of the Abisch-Frenkel chair. O.K. is an Azrieli Fellow. This work was supported by grants to V.K. from the European Research Council under the European Union’s FP7 and H2020 Programs and the Israel Science Foundation.
Source Data
Author contributions
Conceptualization, O.K., U.A. and V.K.; Planning and setup of mouse experiments, A.A., V.K., Execution of experiments: A.A., Analysis of ImageStreamX data, A.A, Z.P.; Derivation of theory, O.K. and U.A.; Data analysis and modeling, O.K.; Writing manuscript, O.K. and U.A.; Review and editing: O.K., A.A., U.A. and V.K.
Data availability
The source data underlying Figs. 2a, c, d–g, 3c, 4b–d, and Supplementary Fig. 2A is available as a Source File. All other data are available from the corresponding author upon request.
Code availability
Custom code was written in Mathematica 11.3 (Wolfram) and can be found in the following link: https://github.com/omerka-weizmann/sncdynamics. The analysis file contains functions for simulating circuit topologies, obtaining mortality statistics and survival-curve scaling, computing log-likelihood of stochastic trajectories, and computing various summary statistics of stochastic trajectory data.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Omer Karin, Amit Agrawal.
Contributor Information
Valery Krizhanovsky, Email: valery.krizhanovsky@weizmann.ac.il.
Uri Alon, Email: uri.alon@weizmann.ac.il.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-019-13192-4.
References
- 1.Burd CE, et al. Monitoring tumorigenesis and senescence in vivo with a p16INK4a-Luciferase model. Cell. 2013;152:340–351. doi: 10.1016/j.cell.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y, et al. Expression of p16INK4a in peripheral blood T-cells is a biomarker of human aging. Aging Cell. 2009;8:439–448. doi: 10.1111/j.1474-9726.2009.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melk A, et al. Expression of p16INK4a and other cell cycle regulator and senescence associated genes in aging human kidney. Kidney Int. 2004;65:510–520. doi: 10.1111/j.1523-1755.2004.00438.x. [DOI] [PubMed] [Google Scholar]
- 4.Sofue T, et al. Tubular cell senescence in the donated kidney predicts allograft function, but not donor remnant kidney function, in living donor kidney transplantation. Am. J. Nephrol. 2018;47:8–17. doi: 10.1159/000485845. [DOI] [PubMed] [Google Scholar]
- 5.Biran A, et al. Quantitative identification of senescent cells in aging and disease. Aging Cell. 2017;16:661–671. doi: 10.1111/acel.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. Cellular senescence in aging primates. Science. 2006;311:1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- 7.Zhou S, et al. Age-related intrinsic changes in human bone-marrow-derived mesenchymal stem cells and their differentiation to osteoblasts. Aging Cell. 2008;7:335–343. doi: 10.1111/j.1474-9726.2008.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muñoz-Espín D, Serrano M. Cellular senescence: from physiology to pathology. Nat. Rev. Mol. Cell Biol. 2014;15:482–496. doi: 10.1038/nrm3823. [DOI] [PubMed] [Google Scholar]
- 9.He S, Sharpless NE. Senescence in health and disease. Cell. 2017;169:1000–1011. doi: 10.1016/j.cell.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demaria M, et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev. Cell. 2014;31:722–733. doi: 10.1016/j.devcel.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.d’Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat. Rev. Cancer. 2008;8:512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 12.Baar MP, et al. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell. 2017;169:132–147.e16. doi: 10.1016/j.cell.2017.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang J, et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 2016;22:78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yosef R, et al. Directed elimination of senescent cells by inhibition of BCL-W and BCL-XL. Nat. Commun. 2016;7:11190. doi: 10.1038/ncomms11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu M, et al. Targeting senescent cells enhances adipogenesis and metabolic function in old age. eLife. 2015;4:e12997. doi: 10.7554/eLife.12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roos CM, et al. Chronic senolytic treatment alleviates established vasomotor dysfunction in aged or atherosclerotic mice. Aging Cell. 2016;15:973–977. doi: 10.1111/acel.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Childs BG, et al. Senescent intimal foam cells are deleterious at all stages of atherosclerosis. Science. 2016;354:472–477. doi: 10.1126/science.aaf6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schafer MJ, et al. Cellular senescence mediates fibrotic pulmonary disease. Nat. Commun. 2017;8:14532. doi: 10.1038/ncomms14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogrodnik M, et al. Cellular senescence drives age-dependent hepatic steatosis. Nat. Commun. 2017;8:15691. doi: 10.1038/ncomms15691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeon OH, et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat. Med. 2017;23:775–781. doi: 10.1038/nm.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krishnamurthy J, et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 22.Sagiv A, et al. p53 in bronchial club cells facilitates chronic lung inflammation by promoting senescence. Cell Rep. 2018;22:3468–3479. doi: 10.1016/j.celrep.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 23.Xu, M. et al. Senolytics improve physical function and increase lifespan in old age. Nat. Med. 24, 1246–1256 (2018). [DOI] [PMC free article] [PubMed]
- 24.Musi N, et al. Tau protein aggregation is associated with cellular senescence in the brain. Aging Cell. 2018;17:e12840. doi: 10.1111/acel.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker DJ, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530:184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enge M, et al. Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell. 2017;171:321–330.e14. doi: 10.1016/j.cell.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Acosta JC, et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat. Cell Biol. 2013;15:978–990. doi: 10.1038/ncb2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aw D, Silva AB, Palmer DB. Immunosenescence: emerging challenges for an ageing population. Immunology. 2007;120:435–446. doi: 10.1111/j.1365-2567.2007.02555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamakoshi K, et al. Real-time in vivo imaging of p16Ink4a reveals cross talk with p53. J. Cell Biol. 2009;186:393–407. doi: 10.1083/jcb.200904105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aoshiba K, Tsuji T, Nagai A. Bleomycin induces cellular senescence in alveolar epithelial cells. Eur. Respir. J. 2003;22:436–443. doi: 10.1183/09031936.03.00011903. [DOI] [PubMed] [Google Scholar]
- 32.Davalos AR, et al. p53-dependent release of Alarmin HMGB1 is a central mediator of senescent phenotypes. J. Cell Biol. 2013;201:613–629. doi: 10.1083/jcb.201206006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Podolskiy, D. et al. Critical dynamics of gene networks is a mechanism behind ageing and Gompertz law. Phys. Q-Bio. Preprint at ArXiv:150204307 (2015).
- 34.Scheffer M, et al. Early-warning signals for critical transitions. Nature. 2009;461:53–59. doi: 10.1038/nature08227. [DOI] [PubMed] [Google Scholar]
- 35.Stroustrup N, et al. The temporal scaling of Caenorhabditis elegans ageing. Nature. 2016;530:103–107. doi: 10.1038/nature16550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finch, C. & Kirkwood, T. B. L. Chance, Development, and Aging (Oxford University Press, 2000).
- 37.Gompertz B. On the nature of the function expressive of the Law of Human Mortality, and on a new mode of determining the value of life contingencies. Philos. Trans. R. Soc. Lond. 1825;115:513–583. doi: 10.1098/rstl.1825.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirkwood TBL. Deciphering death: a commentary on Gompertz (1825) ‘On the nature of the function expressive of the law of human mortality, and on a new mode of determining the value of life contingencies’. Philos. Trans. R. Soc. B. 2015;370:20140379. doi: 10.1098/rstb.2014.0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson TE. Increased life-span of age-1 mutants in Caenorhabditis elegans and lower Gompertz rate of aging. Science. 1990;249:908–912. doi: 10.1126/science.2392681. [DOI] [PubMed] [Google Scholar]
- 40.Ovadya Y, Krizhanovsky V. Strategies targeting cellular senescence. J. Clin. Invest. 2018;128:1247–1254. doi: 10.1172/JCI95149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirkland JL, Tchkonia T, Zhu Y, Niedernhofer LJ, Robbins PD. The clinical potential of senolytic drugs. J. Am. Geriatr. Soc. 2017;65:2297–2301. doi: 10.1111/jgs.14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaplanis J, et al. Quantitative analysis of population-scale family trees with millions of relatives. Science. 2018;360:171–175. doi: 10.1126/science.aam9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruby JG, et al. Estimates of the heritability of human longevity are substantially inflated due to assortative mating. Genetics. 2018;210:1109–1124. doi: 10.1534/genetics.118.301613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hart RW, Setlow RB. Correlation between deoxyribonucleic acid excision-repair and life-span in a number of mammalian species. Proc. Natl Acad. Sci. USA. 1974;71:2169–2173. doi: 10.1073/pnas.71.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mair W, Goymer P, Pletcher SD, Partridge L. Demography of dietary restriction and death in Drosophila. Science. 2003;301:1731–1733. doi: 10.1126/science.1086016. [DOI] [PubMed] [Google Scholar]
- 46.Kikis, E. A., Gidalevitz, T. & Morimoto, R. I. Protein homeostasis in models of aging and age-related conformational disease. In Protein Metabolism and Homeostasis in Aging, Vol. 694 (ed. Tavernarakis, N.) 138–159 (Springer US, 2010). [DOI] [PMC free article] [PubMed]
- 47.Rea SL, Wu D, Cypser JR, Vaupel JW, Johnson TE. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat. Genet. 2005;37:894–898. doi: 10.1038/ng1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu D, Rea SL, Yashin AI, Johnson TE. Visualizing hidden heterogeneity in isogenic populations of C. elegans. Exp. Gerontol. 2006;41:261–270. doi: 10.1016/j.exger.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 49.Casanueva MO, Burga A, Lehner B. Fitness trade-offs and environmentally induced mutation buffering in isogenic C. elegans. Science. 2012;335:82–85. doi: 10.1126/science.1213491. [DOI] [PubMed] [Google Scholar]
- 50.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 51.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 52.Gems D, Partridge L. Genetics of longevity in model organisms: debates and paradigm shifts. Annu. Rev. Physiol. 2013;75:621–644. doi: 10.1146/annurev-physiol-030212-183712. [DOI] [PubMed] [Google Scholar]
- 53.Clancy DJ, et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- 54.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krizhanovsky V, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kang T-W, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479:547–551. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- 57.Tokarsky-Amiel R, et al. Dynamics of senescent cell formation and retention revealed by p14ARF induction in the epidermis. Cancer Res. 2013;73:2829–2839. doi: 10.1158/0008-5472.CAN-12-3730. [DOI] [PubMed] [Google Scholar]
- 58.Flindt, R. Amazing Numbers in Biology (Springer-Verlag, 2006).
- 59.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grubb SC, Bult CJ, Bogue MA. Mouse phenome database. Nucleic Acids Res. 2014;42:D825–D834. doi: 10.1093/nar/gkt1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barbieri M, et al. Data resource profile: the human mortality database (HMD) Int. J. Epidemiol. 2015;44:1549–1556. doi: 10.1093/ije/dyv105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source data underlying Figs. 2a, c, d–g, 3c, 4b–d, and Supplementary Fig. 2A is available as a Source File. All other data are available from the corresponding author upon request.
Custom code was written in Mathematica 11.3 (Wolfram) and can be found in the following link: https://github.com/omerka-weizmann/sncdynamics. The analysis file contains functions for simulating circuit topologies, obtaining mortality statistics and survival-curve scaling, computing log-likelihood of stochastic trajectories, and computing various summary statistics of stochastic trajectory data.