Abstract
Genome-wide association studies (GWAS) have recurrently associated sequence variation nearby LIN28B with pubertal timing, growth and disease. However, the biology linking LIN28B with these traits is still poorly understood. With our study, we sought to elucidate the mechanisms behind the LIN28B associations, with a special focus on studying LIN28B function at the hypothalamic-pituitary (HP) axis that is ultimately responsible for pubertal onset. Using CRISPR-Cas9 technology, we first generated lin28b knockout (KO) zebrafish. Compared to controls, the lin28b KO fish showed both accelerated growth tempo, reduced adult size and increased expression of mitochondrial genes during larval development. Importantly, data from the knockout zebrafish models and adult humans imply that LIN28B expression has potential to affect gene expression in the HP axis. Specifically, our results suggest that LIN28B expression correlates positively with the expression of ESR1 in the hypothalamus and POMC in the pituitary. Moreover, we show how the pubertal timing advancing allele (T) for rs7759938 at the LIN28B locus associates with higher testosterone levels in the UK Biobank data. Overall, we provide novel evidence that LIN28B contributes to the regulation of sex hormone pathways, which might help explain why the gene associates with several distinct traits.
Subject terms: Genetics, Transgenic organisms, Computational biology and bioinformatics, Heritable quantitative trait
Introduction
Pubertal timing is a complex trait that varies extensively in the general population. Individuals at the extreme tails of the distribution show an increased risk for many disease and socially adverse consequences including type 2 diabetes and depression1. Although several monogenic syndromes that affect pubertal timing have been identified, we still understand relatively little about the factors that control the timing of pubertal onset when development proceeds normally2–5. To shed more light on these mechanisms, our project focused on understanding the function of a pubertal timing-associated gene LIN28B, which has been robustly shown to affect developmental timing and growth in several species6–8.
While high-impact mutations causing rare monogenic forms of disturbed pubertal timing have been identified already a few decades ago, LIN28B presents one of the first genetic loci associated with puberty in genome-wide association studies (GWAS), affecting pubertal timing in the general population9,10. Recently, we showed that the same genetic variants nearby LIN28B that delay pubertal onset, also associate with increased LIN28B expression in the hypothalamus and the pituitary, the key structures regulating sexual maturation11. Since the original GWAS-studies linking LIN28B to puberty, the pleiotropic potential of the gene has become evident: currently, in humans, variants in the LIN28B region have been associated with several hormonally regulated traits ranging from depression and bone mineral density (BMD) to finger length ratios and height12–15.
Potentially, the pleiotropic effects of LIN28B might be a consequence of the gene’s local actions in different tissues during development, might occur due to systemic effects induced by the gene’s expression in the hypothalamus and the pituitary, or be a combination of both. At the molecular level, the LIN28-genes are known to control cell division, growth and differentiation - mainly through the well-established interaction with let7 microRNAs16,17. Many models suggest LIN28B and a homologous gene LIN28A contribute to cellular metabolism and proliferation: for example, both genes have been robustly implicated in several cancers and shown to affect metabolism in cells and mice18,19. Since LIN28B expression promotes cell proliferation, it is not surprising that the gene is both ubiquitously and prominently expressed during embryogenesis20,21. However, data from adult humans indicate that the genetic variants linked to pubertal timing affect LIN28B expression mainly in the hypothalamus and the pituitary, thus indicating that LIN28B primarily may impact on pubertal timing and related phenotypes through these tissues11.
The development of the hypothalamic-pituitary (HP) axis is well-conserved in vertebrates22. In this study, we have combined zebrafish knockout models with human data to address the genetic mechanisms whereby LIN28B might affect growth and development, with a special focus on LIN28B function at the HP axis. We demonstrate that in zebrafish CRISPR-Cas9 knockdown of lin28b accelerates the tempo of fish growth, simultaneously reducing the size of adult fish. Moreover, the lin28b KO zebrafish show evidence of altered expression of mitochondrial genes during larval development, as well as changes in estrogen signalling. We also show that increased LIN28B expression correlates with increased expression of several genes in the hypothalamus and the pituitary in humans, including ESR1 and POMC. Finally, we report how the pubertal timing advancing alleles in the LIN28B locus associate with higher serum testosterone levels in the UK Biobank data, highlighting a general mechanism whereby LIN28B could affect several of the phenotypes associated with the gene.
Results
Generation of the lin28b KO zebrafish
In order to assess the impacts of permanent lin28b dysregulation on zebrafish development, we generated lin28b mutant zebrafish strains with the help of CRISPR-Cas9 technology23. To create mutants by inducing double stranded breaks (DSB) into DNA, we designed guide RNA’s targeting exon1 and exon3 of zebrafish lin28b and co-injected these with Cas9 mRNA into embryos, resulting in mosaic fish (F0). These fish were subsequently outcrossed with the in-house strain, and the resulting offspring (F1) was screened for potential protein truncating mutations in lin28b. The generated protein truncating mutations that were chosen for this study either deleted the initiation codon (exon1) or introduced a premature stop codon to the canonical zebrafish lin28b transcript (exon3) (Supplementary data). Finally, two heterozygous fish were bred together to generate lin28b homozygous fish and their sibling controls (F2). The fish from these breedings generally showed the expected 1:2:1 genotypic ratio. However, the sex ratio of the fish appeared consistently distorted: despite the genotype of the F2 fish, the great majority of the fish (~85–90%) were male. As the sex determination in zebrafish appears complex, the result may reflect the fact that the fish which were used in this study were enriched for genetic factors promoting male development of that the growth conditions favoured the development of male fish. Hence, our results are largely based on data from male fish. Both exon1 and exon3 mutants were viable in the homozygous state, and showed no obvious phenotypes upon superficial visual inspection, despite a significant decrease in the amount of lin28b mRNA compared to controls (Fig. 1A–C). Since the different mutational strains were phenotypically indistinguishable, both exon1 and exon3 mutants (collectively referred to as the lin28b KO zebrafish) were used in parallel to produce data for the study. cDNA sequencing further confirmed lin28b RNA expressed in the mutants likely yields truncated protein that is prone to undergo non-sense mediated decay (NMD) (Supplementary data 1 and Supplementary Fig. 1).
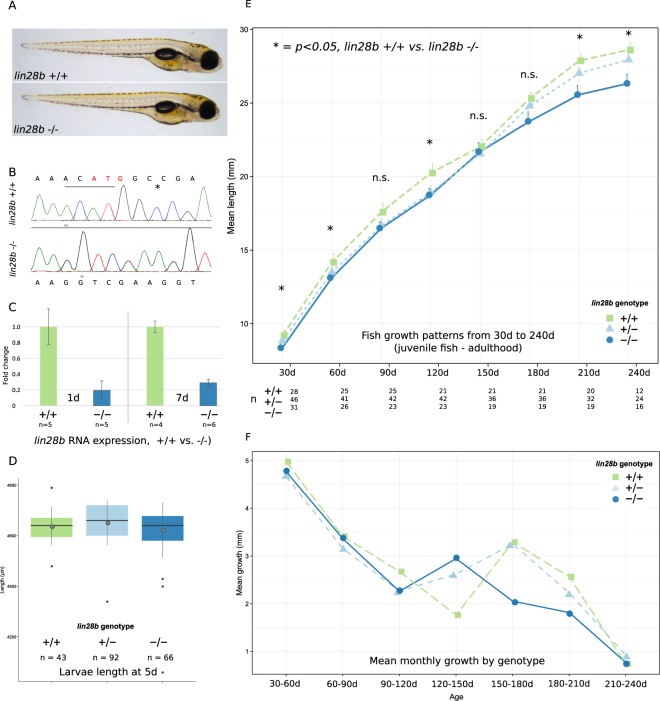
Figure 1.
Characteristics and growth patterns of the lin28b KO fish. (A) Images of 5d old lin28b control (+/+) and KO (−/−) larvae (B) Sequencing traces showing the four deleted bases (underlined) + one mismatch (asterisk) in exon1 mutant fish. The bases deleted in the mutants overlap the translation initiation codon for lin28b (in red). (C) Full length lin28b mRNA expression in control vs lin28b KO RNA-Seq samples at 1d and 7d. The expression of lin28b RNA appears downregulated in the KO fish, likely due to NMD related mechanisms. (D) Comparison of the lin28b KO and the control zebrafish size at 5d. No significant size differences were observed at this stage (One-Way-ANOVA with post-hoc Tukey HSD, Error bars = SEM). (E) Growth curves for the lin28b KO and the control fish. The growth of the KO fish was reduced, but the size difference temporarily vanishedaround the time of sexual maturation (One-Way-ANOVA with post-hoc Tukey HSD, Error bars = SEM, *P < 0.05). (F) Mean monthly growth by genotype. Around the period when the fish undergo sexual maturation the growth rate peaks transiently. The lin28b KO fish seem to undergo this transient peak in growth earlier than controls.
The lin28b KO fish show both advanced growth tempo and reduced growth
The LIN28 gene family has been robustly associated with vertebrate growth, as the genes have been shown to contribute to the tempo and amplitude of growth in various species ranging from humans to nematodes. Previously, we analysed zebrafish models where lin28b was either transiently knocked down or overexpressed during the embryogenesis, showing how short-term, non-physiological lin28b overexpression can increase both larval and adulthood fish length11. In the current study, we wanted to test the consequences of permanent lin28b KO on fish growth and assessed these effects over the fish life-course from larval stages to adulthood.
We began by assessing whether the lin28b KO affected larval fish size, genotyping and measuring 5-day old zebrafish from heterozygote crosses (N = 201). However, at this stage, the length of the KO zebrafish appeared not significantly reduced (−/−) compared to the heterozygotes (+/−) and the wild-type siblings (+/+) (Fig. 1D).
Secondly, targeting the growth between the juvenile period and adulthood, we wanted to assess whether the knockouts exhibited similar alterations in the tempo and amplitude of longitudinal growth as previously reported for humans and mice. We designed an experiment where we tracked the growth of F2 zebrafish from 30 days post fertilisation (dpf) (juvenile fish) into 240 dpf (adult fish) by 30d intervals (Fig. 1E). Already by 30 dpf, the lin28b KO fish appeared on average significantly smaller than their lin28b +/+ siblings (p < 0.05). The KO fish remained smaller also at the end of the experiment at 240 dpf, matching the results from humans and mice linking lower LIN28B expression with smaller adult size. Although on average the size of the heterozygote fish appeared to fall between the +/+ and −/− groups, suggesting additive genetic effects, this group did not show statistically significant differences in growth compared to the other genotypes at any time point.
Besides causing smaller juvenile and adult size (growth amplitude), tracking the growth between these two time points allowed us to observe how the growth progresses throughout this period (growth tempo). Data from humans indicates that the individuals carrying alleles associated with lower LIN28B expression are born slightly smaller and show reduced adult height. Nonetheless, since the timing of their pubertal growth spurt is advanced, they temporarily appear taller than their peers in their early teens6. In fish stocks, growth patterns between the individual fish are known to vary extensively, but similarly as in humans, before sexual maturation, many zebrafish seem to experience transient slowing of growth followed by a period of more rapid growth24. Remarkably, we observed a similar trend in the growth patterns of the lin28b KO fish as seen in the human data (Fig. 1F). Around the timing of sexual maturation, (120–150 dpf in our strain) the size difference between the lin28b KO and the control fish temporarily disappeared, only to reappear later. Thus, mirroring the growth patterns previously reported in humans, our data suggest that in addition to affecting zebrafish size, lin28b may regulate the temporal pattern of fish growth, with the lin28b KO advancing the growth velocity around the timing of sexual maturation.
The lin28b KO fish show advanced HP axis development, links to estrogen signalling and upregulation of mitochondrial pathways
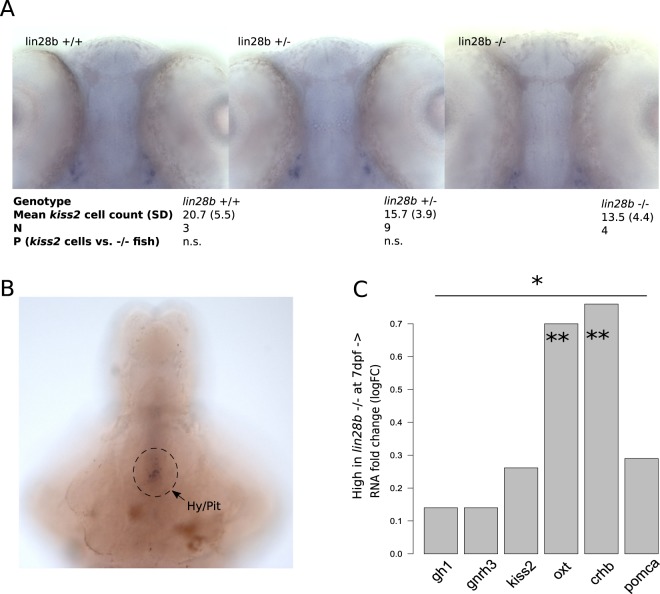
After discovering that our zebrafish model replicated some of the growth phenotypes previously associated with LIN28B, we turned our focus towards lin28b function at the HP axis. We first wanted to evaluate whether the lin28b KO fish showed changes in the structure and function of the HP axis already at the larval stages. We previously detected that knocking down lin28b temporarily in zebrafish embryos interfered with the development of the hypothalamic kiss2 neurons, which are known to participate to gonadotropin release across species11. In the lin28b KO fish, the development of the kiss2 neurons was however not dramatically affected at 3 dpf (Fig. 2A).
Figure 2.
Evaluation of the lin28b KO effects on the HP axis in zebrafish larvae. (A) Representative images of kiss2 expression at 3 dpf in the control (+/+), the heterozygous (+/−), and the lin28b KO (−/−) zebrafish. Though this varied between individual samples, the staining location and intensity was comparable between the groups. Despite a trend towards less kiss2 expressing cells in the KO vs. control fish, we did not detect significant differences in the neuron numbers between the samples (Welch Two Sample t-test). The analysis was complicated by the fact that the +/+ group consists of WT fish from the exon3 crosses, whereas 3 out of 4 −/− fish were obtained from exon1 crosses. As the gene expression data from other experiments indicated that kiss2 transcripts are not significantly up- or downregulated in the KO fish, we concluded that the lin28b KO does not prevent normal development of the kiss2 neurons (e.g. C, Supplementary data 2, and Fig. 3). (B) Image of 7 dpf +/+ zebrafish brain showing weak staining by lin28b anti-sense mRNA in the hypothalamic-pituitary region. (C) HP axis RNA expression was slightly upregulated in the KO fish at 7 dpf. The analysis was restricted to hormonal genes that were expressed in high enough quantities to pass filtering in the RNA-seq experiment. Oxytocin (oxt) and corticotrophin-releasing-hormone beta (chrb) appeared upregulated in the KO samples (**p < 0.005, and overall the expression of the studied HP axis genes appeared upregulated in the lin28b KO zebrafish based on the RNAseq data from 7 dpf (*p < 0.05) (Welch two-sample t-test).
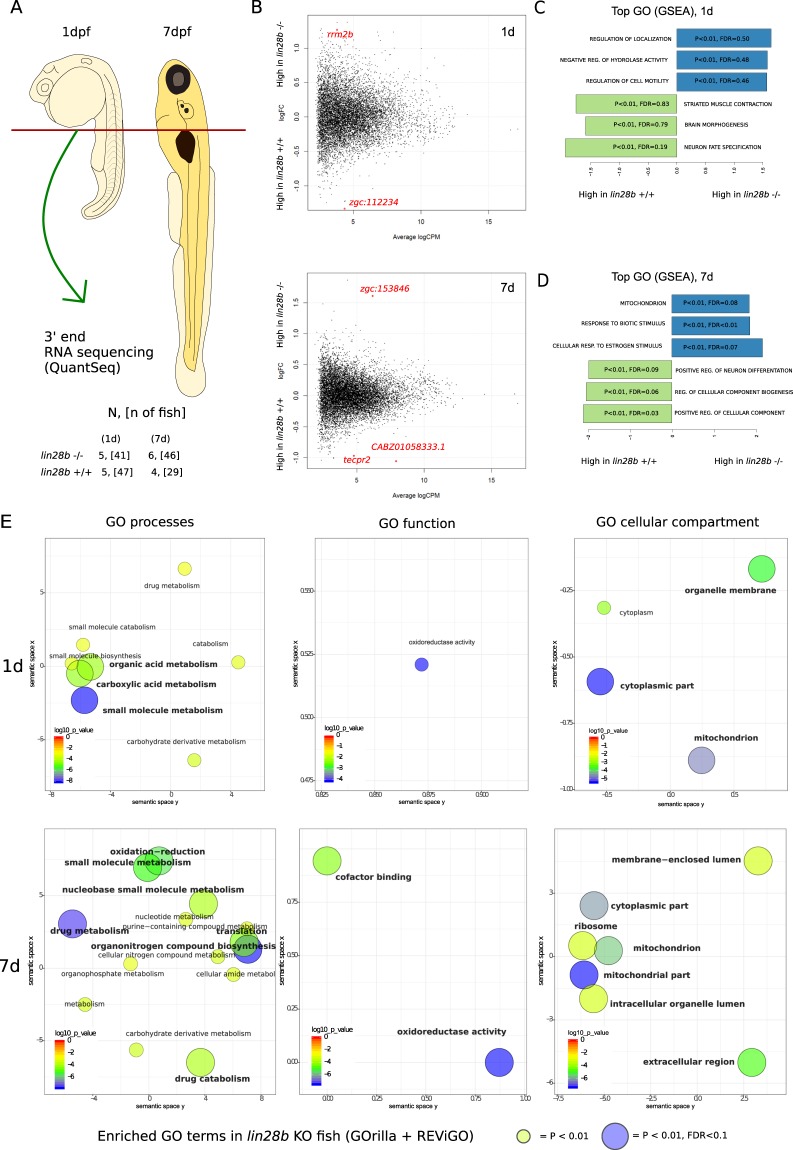
To probe the HP axis development further, we performed RNA-seq analysis of embryonic and larval zebrafish. At 7 dpf, lin28b expression in the zebrafish brain seems already restricted to specific neuronal networks including the hypothalamus and pituitary (Fig. 2B)11. We first screened the RNA-seq data for expression of peptide hormones belonging to growth-hormone, gonadotropin, oxytocin or proopiomelanocortin pathways, and found out that six genes (gh1, gnrh3, kiss2, oxt, crhb and pomca) were expressed in high enough quantities to be subjected for detailed analysis. Although kiss2 mRNA levels were not significantly affected, the lin28b KO larvae showed increased expression of oxt and crhb mRNAs (P < 0.005) at 7 dpf. When analysing the expression of all the six HP axis genes together, we noticed that the expression of these genes was collectively upregulated in the KO zebrafish at 7 dpf (P < 0.05, Welch Two Sample t-test, Fig. 2C). Besides these links to the function of the HP axis, we however saw no evidence for example for steroidogenic enzymes being affected at this stage (Supplementary Table 1). We then turned in to assessing the global gene expression patterns between the lin28b KO and the control fish. In the global analysis of the RNA-seq data, only a couple of transcripts passed false discovery rate (FDR), potentially owing to a relatively small sample size (Fig. 3 and Supplementary Fig. 2). These included rrm2b and zgc:153846 that were upregulated, and zgc:112234, tecpr2 and ENSDARG00000090575, downregulated in the lin28b KO samples. The gene upregulated in 1-day-old knockouts, rrm2b is a part of the ribonucleotide reductase complex, regulating DNA repair and mtDNA synthesis in non-proliferating cells, whereas zgc:153846, upregulated in 7d KO larvae encodes for a lens protein, γ-Crystallin25,26. The genes downregulated in the knockouts include a gene encoding for a histone 2B (zgc:112234), another one involved in the regulation of autophagy (tecpr2) and finally a transcript that has been annotated as a rnf213 orthologue (ENSDARG00000090575)27. However, the gene ontology (GO) analyses using GSEA (Gene Set Enrichment Analysis) and GOrilla (Gene Ontology enRIchment anaLysis and visualization) highlighted several interesting pathways that appeared to differ between the KOs and controls28,29. In the GSEA analysis the top GO terms enriched for the lin28b KOs at 7 dpf included “mitochondrion”, “response to biotic stimulus” and “cellular response to estrogen stimulus” (P < 0.01 and FDR < 0.1), and the GOrilla results also highlighted the mitochondrial pathways (Fig. 3).
Figure 3.
Summary of the RNA-Seq experiment examining the global RNA expression in the lin28b mutant fish at 1d and 7d. (A) Sampling and study design. After dissecting the anterior part from embryos/larvae, we pooled 6–10 samples together. The resulting 4–6 biological replicates per group were subjected to RNA sequencing. (B) Plots of mean logFC differences between the lin28b −/− and the +/+ fish at 1dpf (upper panel), and at 7dpf (lower panel). The genes showing significantly different expression in the global analysis (p < 0.00005, FDR < 0.1) are highlighted in red. (C,D) Results from the gene ontology analysis by GSEA, showing the GO categories differentiating between the lin28b −/− and the +/+ groups. E) Enriched GO terms in the lin28b −/− fish from the GOrilla analysis. Upper panels shows the results for the 1dpf fish and the lower panel for the 7dpf zebrafish.
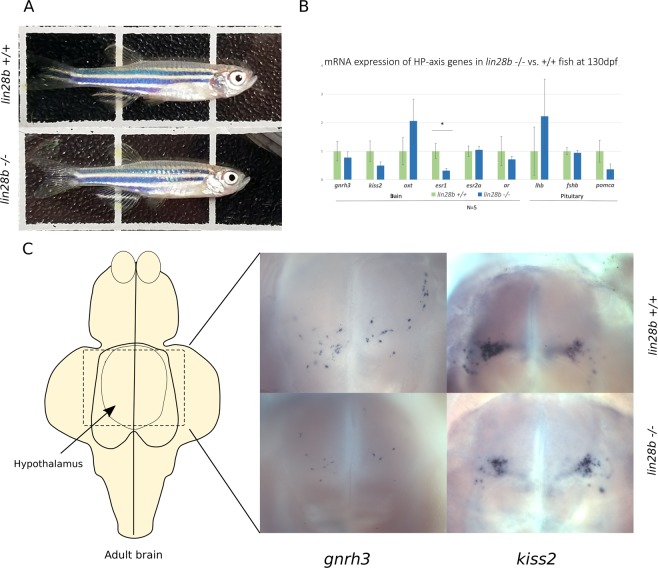
After screening the RNA-seq data, to see whether the potential changes in the HP axis gene expression and estrogen signalling might last till adulthood, we then assessed the expression levels of estrogen signalling- and pubertal timing-related genes in the KO and control fish at 130 dpf by qPCR. We observed that esr1 expression appeared downregulated in the KO-fish brain compared to the control fish (P = 0.04) (Fig. 4B). Although the expression of pomca, gnrh3 and kiss2 also were on average lower in the lin28b KO fish than the controls, the differences were not statistically significant. Finally, we estimated whether the lin28b KO would impose changes in the organisation of hypothalamic Kiss2 or Gnrh3 neurons in adult fish. With in situ hybridisation we located respective RNA expression in adult lin28b KO and control zebrafish brains. We however could not detect any consistent differences in the location or staining intensity of kiss2 and gnrh3 expressing neurons between the KO and control fish (Fig. 4C).
Figure 4.
Inspection of the HP axis of the lin28b KO fish after sexual maturation. (A) Images of 130 day old fish (lin28b KO =−/−, lower panel). The KO fish behaved normally and showed no obvious phenotypic differences compared to controls at this stage. (B) qPCR results from 130d old fish evaluating gene expression at the HP axis. The results suggest that esr1 expression is downregulated in the KO brain compared to controls, although for example gnrh3 expression was not significantly different between the groups in the analysis. N = 5 per group, One-Way-ANOVA with post-hoc Tukey HSD, Error bars = SEM, *P < 0.05. Size of the fish used in the qPCR experiment is shown in the Supplementary Table 2. (C) Left: Schematic diagram of adult zebrafish brain (ventral view) with the hypothalamic region in the middle (arrow). Right: visualization of gnrh3 and kiss2 expression in the hypothalami of 210d old lin28b KO (−/−) and control (+/+) fish by in situ hybridization. The localization of gnrh3/kiss2 expression appeared roughly similar, although the number of stained neurons varied between individual samples within each group (2–4 samples per group). The variation appeared potentially related to the size differences of the fish (for samples shown: gnrh3, +/+ size = 32 mm/300 mg; −/−, 27 mm/200 mg; kiss2+/+30 mm/230 mg, −/− 29 mm/190 mg).
LIN28B expression correlates positively with the expression of several HP axis genes, including ESR1 and POMC in adult humans
The results from the lin28b KO fish prompted us to further assess the connection between LIN28B, estrogen signalling and HP axis function. In adult humans LIN28B expression is relatively strong in the hypothalamus and the pituitary compared to most tissues, and many of the phenotypes associated with LIN28B such as growth and puberty are regulated by the HP axis. Previous analyses by us and others further show that sequence variants in the LIN28B region affect the gene’s expression primarily in the hypothalamus and the pituitary, and a hypothesis has been proposed that decreasing LIN28 expression in the hypothalamus might be permissive for pubertal onset11,30. To further explore LIN28B function at the HP axis, we leveraged the Genotype-Tissue Expression (GTEx) database, containing tissue specific gene expression data from hundreds of adult humans, looking for correlations between LIN28B and selected hormonal genes expressed in the HP axis (Fig. 5). The current release of GTEx (v7) consists of 11688 post mortem samples from mostly white adults, with median age over 50 yrs31. We included in this analysis pubertal timing genes belonging to kisspeptin-GNRH-gonadotropin pathway (KISS, KISS1R, GNRH1, GNRHR, LH, FSHB), and genes that were considered relevant based on the zebrafish data, including steroid hormone receptors that regulate the activity of pubertal timing pathway (ESR1, ESR2, AR) and proopiomelanocortin (POMC), the most abundant hormonal mRNA expressed in the pituitary. We observed that in the hypothalami of adult humans, higher LIN28B expression correlated with higher expression of ESR1, ESR2, AR, KISS1, KISS1R and GNRH1 (unadjusted P-values ranging from 8e-10 to 3e-19). Compared to other genes expressed in the hypothalamus at a similar level, the correlation of LIN28B with ESR1 and AR was among the top 5% of most significant results (permuted P-value < 0.05). Thus, contrasting our expectations, higher LIN28B expression in the hypothalamus, which previously has been linked with later pubertal onset, correlated with higher expression of genes that are known to promote sexual maturation.
Figure 5.
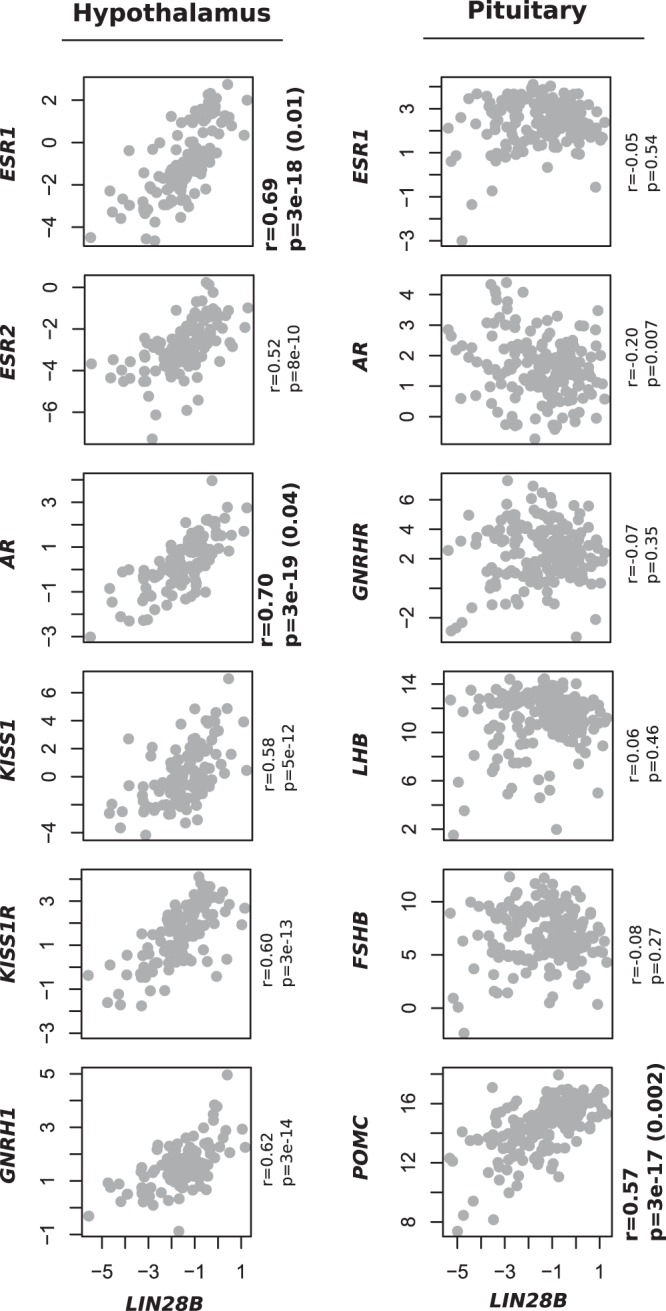
LIN28B correlations with hormonal genes in the hypothalamus and pituitary in GTEx. The figures show relative LIN28B expression on the X-axis compared to the expression of selected genes from the hypothalamus in the Y-axis (log2 TPM + 1). LIN28B expression correlated positively with all the tested genes in the hypothalamus (R = 0.58–0.70, P < 8e-10))). Particularly, the expression of ESR1 and AR was upregulated: these genes showed statistically more robust correlations with LIN28B than the majority of the other ~50000 transcripts (p-value in parenthesis). Although GNRHR, LHB and FSHB showed no correlations with LIN28B levels in the pituitary, we observed a statistically significant positive correlation between LIN28B and POMC expression (R = 0.57, P = 3e-17). The correlation with POMC was among the most robust correlations for LIN28B in the pituitary.
The hypothalamic pathway including KISS1 and GNRH1 stimulates pituitary gonadotropin secretion through gonadotropin releasing hormone receptors (GNRHR). Therefore, we expected that higher LIN28B expression would correlate with higher mRNA levels of GNRHR, lutenizing hormone (LH) and follicle-stimulating hormone (FSH). Nonetheless, GNRHR, LH and FSH levels did not correlate with LIN28B expression in the pituitary (Fig. 5). Surprisingly, although LIN28B levels did not correlate with gonadotrophin mRNA expression, we noted that these correlated positively with the expression of POMC, coding a precursor for several peptides including b-endorphin, melanin, and adrenocorticotrophic hormone (ACTH) (unadjusted P = 3e-17, permuted P = 0.002, Fig. 5). While the current GTEx data include over 50,000 transcripts from the pituitary, we found that POMC was among the three most abundant transcripts in that tissue. Moreover, it was among the top 20 genes whose expression correlates with LIN28B31. The strong association with POMC further suggests that LIN28B may – either directly or indirectly – contribute to hormonal dynamics at the HP axis.
Pubertal timing advancing alleles at the LIN28B locus associate with higher serum testosterone levels
Collectively, the results from our zebrafish models and the GTEx-database suggest that LIN28B may affect the expression of hormonal genes at the HP axis in an unexpected manner. Therefore, we finally wanted to explore the possibility that the gene might affect serum hormone levels in adults. Previously, we have shown that the puberty-delaying allele (C) at rs7759938 nearby LIN28B associates with higher LIN28B expression at the HP axis. For the current study, we performed an association analysis to detect whether the same variant would correlate with serum testosterone levels in the UK biobank database containing over 350,000 individuals with testosterone data available. Remarkably, we detected that the C-allele at rs7759938 was robustly associated with significantly lower serum testosterone levels (P = 9.2e-16, Table 1). The result was mostly driven by the signal in males (P = 2.5E-37) whereas it did not reach genome-wide significance (P < 5e-8) in females (P = 0.0067). The data, nonetheless, strongly link higher LIN28B expression with lower serum testosterone levels in adult humans.
Table 1.
Association of rs7759938 with serum testosterone levels in the UK biobank data. A1 = effect allele, A2 = other allele, EAF = effect allele frequency.
| SNP | N | Chr | Pos | A1/A2 | EAF | Beta | SE | P | |
|---|---|---|---|---|---|---|---|---|---|
| Males | rs7759938 | 176212 | 6 | 105378954 | C/T | 0.32 | −0.044 | 0.003 | 1.5E-37 |
| Females | rs7759938 | 174850 | 6 | 105378954 | C/T | 0.32 | −0.010 | 0.003 | 0.0067 |
| M + F combined | rs7759938 | 351062 | 6 | 105378954 | C/T | 0.32 | −0.020 | 0.002 | 9.2e-16 |
Discussion
Over the past years, GWAS have linked sequence variants at the LIN28B locus with multiple traits. In the current study, we focused on exploring the mechanisms behind these associations. Our data show that lin28b affects growth and metabolism in zebrafish, and links the gene with estrogen signalling. Importantly, analyses of gene expression data from humans indicate that LIN28B may affect hormonal dynamics at the hypothalamus and pituitary, and based on data from ~350,000 individuals from the UK Biobank we report a novel association with LIN28B and serum testosterone levels. Altogether, these results provide important insights into the potential mechanisms whereby LIN28B may contribute to human phenotypes.
Despite the variable physical features of different vertebrates, many basic mechanisms regulating growth have been conserved during the course of evolution. For example, genetic variation in VGLL3 has been shown to affect timing of sexual maturation in humans as well as in fish, and LIN28 genes have been shown to contribute to growth and developmental timing in a wide range of metazoans ranging from C. elegans to humans6,11,32–34. In this study, we report that knocking out lin28b accelerates the growth tempo but leads to reduced adult size in zebrafish. The growth pattern of the lin28b KO zebrafish is strikingly similar to humans who carry SNPs that associate with lower LIN28B expression6. The result suggests that the LIN28B-related genetic mechanisms control ontogeny across different vertebrate species.
With our study, we specifically aimed to investigate whether LIN28B contributes to the development and function of the HP axis. Our results suggest that while the development of the HP axis might be slightly advanced by the lin28b KO, the gene is not essential for the formation of the HP axis in zebrafish. Since zebrafish have been shown to activate various compensatory mechanisms to ensure normal HP axis development, even when critical genes such as kiss2 or gnrh3 have been knocked out, the finding that the lin28b KO does not dramatically affect the development of the HP axis is not surprising35,36. In fact, gene loss by knockout may be generally quite well compensated in zebrafish, which potentially could explain the discrepancy between the current and our previous study where transient knockdown of lin28b with MOs significantly interfered with the development of the hypothalamic kiss2 neurons11,37,38. For example, it remains to be studied to which extent the homologous gene lin28a might compensate for the loss of lin28b. However, in the current study we saw evidence that knocking out lin28b might induce subtle changes in hormonal signalling throughout development: genes which respond to estrogen signalling were upregulated in the KO larvae and esr1a appeared downregulated in the adult lin28b KO brains. In our study we did not assess whether these changes might be attributable for example to changes in the expression of the let7 microRNAs, which are the known regulatory targets of the LIN28-genes.
Besides the links between LIN28B and estrogen signalling, our lin28b KO zebrafish showed an increased expression of mitochondrial genes during the first days of development. Loss of LIN28 function has been shown to increase mitochondrial activity and prolong mitochondrial mRNA half-lives also in murine pluripotent stem cell models39. Since mitochondrial metabolism is the preferred means of energy production in differentiated cells, theoretically, this might be a reflection of the lin28b KO zebrafish having less dividing, and more differentiated cells than controls at this stage. As speculated in the case of LIN28A and mice, this could lead to a reduction in the total amount cells in these fish, providing a potential mechanistic explanation for the association between LIN28B and vertebrate body size40.
In addition to studying lin28b expression in zebrafish, we also leveraged the GTEx database and showed that LIN28B affects gene expression in the HP axis of adult humans. The finding that LIN28B correlates positively with genes such as KISS1 and GNRH1 was somewhat unexpected, since the activity of these genes is known to promote sexual maturation – a phenotype related to lower LIN28B expression. However, given that the LIN28B expression-increasing allele correlates negatively with serum testosterone levels, one may speculate that LIN28B indirectly, via reduced testosterone levels, might promote the expression of these steroid hormone receptors through allostatic regulation. Moreover, one may further speculate that sex hormones binding ER an AR, including testosterone, might be the potential mediators whereby LIN28B could affect many of the phenotypes associated with the gene, including for example pubertal timing, finger length ratios, and increased bone mineral density (BMD). Although our study does not directly address causation, considering the known functions of testosterone (promotion of pubertal development, contribution to finger length ratios, and increasing BMD), in theory we could assume that pubertal development might be triggered earlier, finger length ratios are likely masculinized and BMD increased for individuals carrying testosterone-increasing genetic variants in the LIN28B locus14,41,42. Nonetheless, the molecular mechanisms underlying the association between LIN28B and testosterone levels remain unknown and therefore warrant further studies.
Besides correlating with testosterone levels, we saw evidence that LIN28B may affect also estrogen signalling. While this might be directly related to changes in androgen levels, in theory estrogen might also have an independent effect on several LIN28B associated phenotypes. Studies in mice have shown that estrogen receptor alpha (ERα) signalling in Kiss1 neurons is crucial for the regulation of pubertal onset: the ablation of ERα from these neurons dramatically advanced the pubertal development in mice, though stalling it later43. On the other hand, ERα knockout in medial basal hypothalamus leads to increased bone mineral density (BMD) in female mice via an effect on Kiss1 neurons44. In principle, the positive correlation between LIN28B and ESR1 expression in the hypothalamus thus might be directly relevant for example to LIN28B associations with puberty and BMD.
Further underscoring the widespread effects that LIN28B might have on hormonal genes expressed at the HP axis, we additionally observed a strong positive correlation between LIN28B and POMC expression in the pituitary. Notably, also increased POMC-activity has been associated with lower prenatal testosterone exposure45. POMC gives rise to several peptides that regulate human physiology, like β-endorphin, which might be relevant for example to LIN28B association with depression46. Overall, the results from our study suggest that LIN28B has widespread effects on gene expression at the HP axis, and LIN28B-induced changes in sex steroid signalling might contribute to many of the phenotypes associated with the gene in the human studies.
Conclusion
Taken together, our results suggest that LIN28B genes regulate growth and development across vertebrates in an evolutionarily conserved manner. Besides showing how lin28b knockout affects zebrafish growth, our results indicate that LIN28B can contribute to gene expression dynamics at the HP axis, highlighting especially the ESR1- and POMC-related pathways. Importantly, the sequence variants linked to higher LIN28B expression also associate with lower serum testosterone levels in adult humans, which might mediate the gene’s association with various phenotypes.
Materials and Methods
Generation of the lin28b KO zebrafish
Zebrafish for the experiments were obtained from a breeding line that has been maintained in the Panula laboratory for more than 10 years (Kaslin and Panula, 2001; Kaslin et al., 2004). Permits for the experiments were attained from the Office of the Regional Government of Southern Finland, in agreement with the ethical guidelines of the European convention for the protection of vertebrate animals used for experimental and other scientific purposes.
We designed two CRISPR-Cas9 models targeting exon1 and exon3 of zebrafish lin28b. The target site for exon1 was 5′-GGAGGGTCCGCGGGGAAACA-3′ and for exon3 5′-GGCTTCAGGAGCCTGCGGGA-3′. To create mutants, 30 zebrafish embryos were injected with 600 pg Cas9 mRNA and 50 pg exon1 or 25 pg exon3 gRNA. (Supplementary Table 3). Mutations in lin28b were induced in approximately 80–90% of F0 fish. Fish from the two mutational strains, carrying mutations either in exon1 or exon3 of lin28b were mated with the in-house strain to obtain F1 zebrafish heterozygous for lin28b mutations. These two strains of F1 fish were subsequently crossed to obtain F2 control (+/+), heterozygous (+/−) and KO (−/−) zebrafish used in the analyses. Details of the induced mutations are shown in Supplementary data 1 and Supplementary Fig. 1.
Fish growth analysis
We characterized the growth of the lin28b KO fish in two separate experiments. First, we targeted the larval fish size (at 5d) and in another experiment followed the growth of juvenile fish up to adulthood (from 30 dpf to 240 dpf). In the experiment including larval fish we measured altogether 201 5d old fish that were imaged, genotyped, and analyzed for their body length and head circumference. All the fish in this experiment were obtained from lin28b exon1 F1 crosses.
In the from-juvenile-to-adult experiment, fish length and weight were measured at 30d intervals (weight starting from 120dpf). Fish were genotyped from tail fin samples either by sequencing (exon1) or by High-Resolution-Melting curve analysis (HRM) (exon3) at 30d intervals. Sequencing and HRM primers and conditions are shown in Supplementary Table 3. lin28b +/+, +/− and −/− fish were grown together. Two of the tanks contained exon1 KOs and controls, (starting N = 32 and 34 fish per 5 liter tank), and one contained exon3 KOs and controls N = 38). The researcher measuring and sequencing the fish (JP) was always blinded for their genotype information.
RNA isolation, RNAseq and qPCR
Total RNA was extracted using miRNeasy mini Kit (Qiagen Inc., Valencia, CA) according to the instructions of the manufacturer, For the RNA-seq experiment, we collected altogether 163 genotyped lin28b −/− and +/+ fish at 1dpf and 7 dpf. Pooled samples each containing RNA from 6–11 embryos were subsequently subjected to 3′ end RNA sequencing (Fig. 4A), producing an average read count of >10 million reads per sample. After quality control and filtering, the differential expression (DE) analyses included altogether 9304 and 11262 genes for 1dpf samples and 7 dpf samples, respectively (Supplementary data 1). P-value distribution of the data suggested that notable differences in the gene expression profiles between the lin28b KO and the control samples were likely to exist: however, only a couple of transcripts passed the false discovery rate (FDR) (Fig. 4B, Supplementary Fig. 3A and supplementary data 1). Out of five pooled 1d KO samples subjected to RNA-seq, two were based on exon1 mutant fish, and three were based on exon3 mutants. Out of six 7d KO samples, 3 were based on exon1 mutants and other 3 on exon3 mutants. The 1d and 7d samples subjected to RNA sequencing were DNAse treated, showing median RNA Integrity Number (RIN) > 9.5. Quality and quantity of the the extracted RNA samples were analyzed with 2100 Bioanalyzer RNA 6000 Nano Kit (Agilent, Santa Clara, CA, USA) and Qubit RNA BR kit (Thermo Fisher Scientific, Waltham, MA, USA). For genomic DNA contamination measurement, Qubit DNA BR kit (Thermo Fisher Scientific, Waltham, MA, USA) was used. mRNA libraries were prepared from 80 ng of extracted RNA with QuantSeq3′ mRNA-Seq Library Prep Kit (Lexogen GmbH, Vienna, Austria) according to user guide version 015UG009V0221. ERCC RNA spike-in mix (Life Technologies, Carlsbad, CA, USA) was added as control to each sample according to manufacturer’s instructions. Libraries were quantified and pooled for sequencing using 2100 Bioanalyzer DNA High Sensitivity Kit (Agilent, Santa Clara, CA, USA). Sequencing was performed with Illumina HiSeq2500 system in HiSeq high output mode using v4 chemistry (Illumina, San Diego, CA, USA). Read length for the paired-end run was 2 × 101 bp and target coverage 5 M reads for each library. Using Chipster analysis platform, sequencing reads were preprocessed with FastX and PRINSEQ, aligned to zebrafish genome build GRCz10 with TopHat2, and read count tables used in the analysis were produced by HTSeq. The final differential expression (DE) analysis between the KO and control samples was made in R version 3.4.3 using edgeR47. Gene ontology (GO) pathway analyses of the RNA-seq data were performed with Gene Set Enrichment Analysis (GSEA) and Gene Ontology enRIchment anaLysis and visuaLizAtion (GORILLA) tools in combination with REViGO29,48.
For qPCR experiments, RNA was extracted from a single brain/pituitary (130d). All samples in the 130 dpf experiment were obtained by breeding F1 lin28b exon1 mutants. 0.1–0.2 μg of RNA was reverse-transcribed using SuperScript® VILO cDNA Synthesis Kit. The experiments were run with the Light Cycler® 480 instrument using Roche Light Cycler 480 SYBR Green I master mix as instructed (F. Hoffmann-La Roche Ltd, Switzerland). 0.75 μl of 5 mM primers and 2 μl of 1:10 dilution of the cDNA was used per 15 μl reaction. Primer sequences and cycling conditions are shown in (Supplementary Table 3) Fluorescence changes were monitored after every cycle. Dissociation curve analysis was performed to ensure only a single amplicon was obtained. Reactions were run as duplicates and relative expression levels were calculated based on b-actin, using the 2-δδCT method.
In situ hybridisation
WISH of gnrh3 and kiss2 probes was performed on 4% paraformaldehyde (PFA)-fixed brain from 240d old fish based on the protocol by Thisse & Thisse, as previously described11.
GTEx
The Genotype-Tissue Expression (GTEx) project provides the scientific community with a resource of gene expression and genotype data from hundreds of individuals and thousands of samples from a diverse set of human tissues, with written informed consent for the use of samples obtained directly from the living participants, or their next-of-kin in case of deceased individuals28. We queried this database to examine whether LIN28B expression correlates with the expression of key genes involved in the regulation of pubertal timing in the hypothalamus and the pituitary.
For the current analysis release (GTEx Analysis V7 release, dbGaP Accession phs000424.v6.p1), the GTEx project has collected tissue samples from 714 postmortem donors (244 females, 470 males; age range 20–70), produced RNA sequencing data from 11688 tissue samples spanning 53 unique tissue types and generated genotype data for up to 635 donors. The data production and analysis procedures are available through the GTEx Portal (http://gtexportal.org) and described in detail in28.
Genetic association analysis
For the current study, we utilised the UK Biobank database containing over 500,000 individuals, aged between 40 and 69 at the time of recruitment, described in detail by Bycroft et al.49. At recruitment, participants provided electronic signed consent. Ethics approval for the UK Biobank study was obtained from the North West Centre for Research Ethics Committee (11/NW/0382). All experiments were performed in accordance to relevant guidelines and regulations. Under UK Biobank application number 22627, we run an association analysis against serum testosterone for pubertal timing associated marker rs7759938, which was previously shown to correlate with LIN28B expression levels at the hypothalamus and the pituitary. Analysis was performed using linear mixed model association testing implemented in BOLT-LMM v2.3 software50. Sex-specific log-transformed testosterone levels were adjusted for age and BMI, inverse normalised, and the analyses were run using 10 PCs as covariates. Additional covariates included menopause for the analysis involving females and sex in the combined analysis. The analyses were restricted on individuals from white British ancestry, with testosterone levels between+/−5SD from the population mean for each sex.
Statistical analyses
All analyses were performed with R version 3.4.3. Fish genotype ratios were assessed with Chi-square test. Growth of fish was compared with ANOVA using post-hoc Tukey HSD. Statistics for qPCR results from 130d fish were calculated using one-way ANOVA with post-hoc Tukey HSD.
Supplementary information
Acknowledgements
The authors would like to acknowledge the contribution of the Zebrafish core facility supported by University of Helsinki and Biocenter Finland and Academy of Finland. Special thanks to Henri Koivula and Riikka Pesonen for maintenance of the fish. Anu Suoranta and Pirkko Mattila are warmly acknowledged for their contributions to the RNA-seq experiment which was performed at the FIMM Technology Centre. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal and dbGaP accession number phs000424.v6.p1. 393. This research has been conducted using the UK Biobank Resource, and we want to specifically acknowledge Matti Pirinen for the access to the data. Jake Lin is acknowledged for help with the grammar of the manuscript. Samuli Ripatti as the head of the Centre of Excellence in Complex Disease Genetics (CoECDG) is warmly thanked for providing support for this study. This work has been funded by the Academy of Finland grant No. 267561 and JTL was supported by the Finnish Cultural Foundation.
Author contributions
J.T.L., Y.C.C., J.P., T.T., L.L. and N.J. performed experiments and analyzed data. J.T.L. and Y.C.C. generated reagents. E.W. and P.P. supervised the study. J.T.L. drafted the manuscript. J.T.L. and E.W. edited the manuscript. All authors critically evaluated and revised the manuscript.
Data availability
The gene expression datasets analysed during the current study are available from the GTEx portal (https://gtexportal.org/). The UK biobank data is available for researches upon request from the UK Biobank (https://www.ukbiobank.ac.uk/researchers/). The results from the RNA-seq dataset from 1d and 7d old zebrafish generated and analysed during the current study are included in the supplementary files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-54475-6.
References
- 1.Day, F. R., Elks, C. E., Murray, A., Ong, K. K. & Perry, J. R. B. Puberty timing associated with diabetes, cardiovascular disease and also diverse health outcomes in men and women: the UK Biobank study. Sci. Rep. 5, 11208, 10.1038/srep11208, https://www.nature.com/articles/srep11208#supplementary-information (2015). [DOI] [PMC free article] [PubMed]
- 2.Gajdos ZK, Hirschhorn JN, Palmert MR. What controls the timing of puberty? An update on progress from genetic investigation. Curr Opin Endocrinol Diabetes Obes. 2009;16:16–24. doi: 10.1097/MED.0b013e328320253c. [DOI] [PubMed] [Google Scholar]
- 3.Day FR, Perry JRB, Ong KK. Genetic Regulation of Puberty Timing in Humans. Neuroendocrinology. 2015;102:247–255. doi: 10.1159/000431023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plant TM. Neuroendocrine control of the onset of puberty. Front. Neuroendocrinol. 2015;38:73–88. doi: 10.1016/j.yfrne.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herbison AE. Control of puberty onset and fertility by gonadotropin-releasing hormone neurons. Nature Reviews. Endocrinology. 2016;12:452. doi: 10.1038/nrendo.2016.70. [DOI] [PubMed] [Google Scholar]
- 6.Widén E, et al. Distinct Variants at LIN28B Influence Growth in Height from Birth to Adulthood. The American Journal of Human Genetics. 2010;86:773–782. doi: 10.1016/j.ajhg.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elks, C. E. et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat. Genet. 42, 1077–1085, http://www.nature.com/ng/journal/v42/n12/abs/ng.714.html#supplementary-information (2010). [DOI] [PMC free article] [PubMed]
- 8.Corre C, et al. Sex-specific regulation of weight and puberty by the Lin28/let-7 axis. The Journal of endocrinology. 2016;228:179–191. doi: 10.1530/JOE-15-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ong, K. K. et al. Genetic variation in LIN28B is associated with the timing of puberty. Nat. Genet. 41, 729–733, http://www.nature.com/ng/journal/v41/n6/suppinfo/ng.382_S1.html (2009). [DOI] [PMC free article] [PubMed]
- 10.Sulem Patrick, Gudbjartsson Daniel F, Rafnar Thorunn, Holm Hilma, Olafsdottir Elinborg J, Olafsdottir Gudridur H, Jonsson Thorvaldur, Alexandersen Peter, Feenstra Bjarke, Boyd Heather A, Aben Katja K, Verbeek Andre L M, Roeleveld Nel, Jonasdottir Aslaug, Styrkarsdottir Unnur, Steinthorsdottir Valgerdur, Karason Ari, Stacey Simon N, Gudmundsson Julius, Jakobsdottir Margret, Thorleifsson Gudmar, Hardarson Gudmundur, Gulcher Jeffrey, Kong Augustine, Kiemeney Lambertus A, Melbye Mads, Christiansen Claus, Tryggvadottir Laufey, Thorsteinsdottir Unnur, Stefansson Kari. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nature Genetics. 2009;41(6):734–738. doi: 10.1038/ng.383. [DOI] [PubMed] [Google Scholar]
- 11.Leinonen JT, Chen Y-C, Tukiainen T, Panula P, Widén E. Transient modification of lin28b expression - permanent effects on zebrafish growth. Mol. Cell. Endocrinol. 2019;479:61–70. doi: 10.1016/j.mce.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Hyde CL, et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat. Genet. 2016;48:1031–1036. doi: 10.1038/ng.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lango Allen Hana, Estrada Karol, Lettre Guillaume, Berndt Sonja I., Weedon Michael N., Rivadeneira Fernando, Willer Cristen J., Jackson Anne U., Vedantam Sailaja, Raychaudhuri Soumya, Ferreira Teresa, Wood Andrew R., Weyant Robert J., Segrè Ayellet V., Speliotes Elizabeth K., Wheeler Eleanor, Soranzo Nicole, Park Ju-Hyun, Yang Jian, Gudbjartsson Daniel, Heard-Costa Nancy L., Randall Joshua C., Qi Lu, Vernon Smith Albert, Mägi Reedik, Pastinen Tomi, Liang Liming, Heid Iris M., Luan Jian’an, Thorleifsson Gudmar, Winkler Thomas W., Goddard Michael E., Sin Lo Ken, Palmer Cameron, Workalemahu Tsegaselassie, Aulchenko Yurii S., Johansson Åsa, Carola Zillikens M., Feitosa Mary F., Esko Tõnu, Johnson Toby, Ketkar Shamika, Kraft Peter, Mangino Massimo, Prokopenko Inga, Absher Devin, Albrecht Eva, Ernst Florian, Glazer Nicole L., Hayward Caroline, Hottenga Jouke-Jan, Jacobs Kevin B., Knowles Joshua W., Kutalik Zoltán, Monda Keri L., Polasek Ozren, Preuss Michael, Rayner Nigel W., Robertson Neil R., Steinthorsdottir Valgerdur, Tyrer Jonathan P., Voight Benjamin F., Wiklund Fredrik, Xu Jianfeng, Hua Zhao Jing, Nyholt Dale R., Pellikka Niina, Perola Markus, Perry John R. B., Surakka Ida, Tammesoo Mari-Liis, Altmaier Elizabeth L., Amin Najaf, Aspelund Thor, Bhangale Tushar, Boucher Gabrielle, Chasman Daniel I., Chen Constance, Coin Lachlan, Cooper Matthew N., Dixon Anna L., Gibson Quince, Grundberg Elin, Hao Ke, Juhani Junttila M., Kaplan Lee M., Kettunen Johannes, König Inke R., Kwan Tony, Lawrence Robert W., Levinson Douglas F., Lorentzon Mattias, McKnight Barbara, Morris Andrew P., Müller Martina, Suh Ngwa Julius, Purcell Shaun, Rafelt Suzanne, Salem Rany M., Salvi Erika, Sanna Serena, Shi Jianxin, Sovio Ulla, Thompson John R., Turchin Michael C., Vandenput Liesbeth, Verlaan Dominique J., Vitart Veronique, White Charles C., Ziegler Andreas, Almgren Peter, Balmforth Anthony J., Campbell Harry, Citterio Lorena, De Grandi Alessandro, Dominiczak Anna, Duan Jubao, Elliott Paul, Elosua Roberto, Eriksson Johan G., Freimer Nelson B., Geus Eco J. C., Glorioso Nicola, Haiqing Shen, Hartikainen Anna-Liisa, Havulinna Aki S., Hicks Andrew A., Hui Jennie, Igl Wilmar, Illig Thomas, Jula Antti, Kajantie Eero, Kilpeläinen Tuomas O., Koiranen Markku, Kolcic Ivana, Koskinen Seppo, Kovacs Peter, Laitinen Jaana, Liu Jianjun, Lokki Marja-Liisa, Marusic Ana, Maschio Andrea, Meitinger Thomas, Mulas Antonella, Paré Guillaume, Parker Alex N., Peden John F., Petersmann Astrid, Pichler Irene, Pietiläinen Kirsi H., Pouta Anneli, Ridderstråle Martin, Rotter Jerome I., Sambrook Jennifer G., Sanders Alan R., Oliver Schmidt Carsten, Sinisalo Juha, Smit Jan H., Stringham Heather M., Bragi Walters G., Widen Elisabeth, Wild Sarah H., Willemsen Gonneke, Zagato Laura, Zgaga Lina, Zitting Paavo, Alavere Helene, Farrall Martin, McArdle Wendy L., Nelis Mari, Peters Marjolein J., Ripatti Samuli, van Meurs Joyce B. J., Aben Katja K., Ardlie Kristin G., Beckmann Jacques S., Beilby John P., Bergman Richard N., Bergmann Sven, Collins Francis S., Cusi Daniele, den Heijer Martin, Eiriksdottir Gudny, Gejman Pablo V., Hall Alistair S., Hamsten Anders, Huikuri Heikki V., Iribarren Carlos, Kähönen Mika, Kaprio Jaakko, Kathiresan Sekar, Kiemeney Lambertus, Kocher Thomas, Launer Lenore J., Lehtimäki Terho, Melander Olle, Mosley Jr Tom H., Musk Arthur W., Nieminen Markku S., O’Donnell Christopher J., Ohlsson Claes, Oostra Ben, Palmer Lyle J., Raitakari Olli, Ridker Paul M., Rioux John D., Rissanen Aila, Rivolta Carlo, Schunkert Heribert, Shuldiner Alan R., Siscovick David S., Stumvoll Michael, Tönjes Anke, Tuomilehto Jaakko, van Ommen Gert-Jan, Viikari Jorma, Heath Andrew C., Martin Nicholas G., Montgomery Grant W., Province Michael A., Kayser Manfred, Arnold Alice M., Atwood Larry D., Boerwinkle Eric, Chanock Stephen J., Deloukas Panos, Gieger Christian, Grönberg Henrik, Hall Per, Hattersley Andrew T., Hengstenberg Christian, Hoffman Wolfgang, Mark Lathrop G., Salomaa Veikko, Schreiber Stefan, Uda Manuela, Waterworth Dawn, Wright Alan F., Assimes Themistocles L., Barroso Inês, Hofman Albert, Mohlke Karen L., Boomsma Dorret I., Caulfield Mark J., Adrienne Cupples L., Erdmann Jeanette, Fox Caroline S., Gudnason Vilmundur, Gyllensten Ulf, Harris Tamara B., Hayes Richard B., Jarvelin Marjo-Riitta, Mooser Vincent, Munroe Patricia B., Ouwehand Willem H., Penninx Brenda W., Pramstaller Peter P., Quertermous Thomas, Rudan Igor, Samani Nilesh J., Spector Timothy D., Völzke Henry, Watkins Hugh, Wilson James F., Groop Leif C., Haritunians Talin, Hu Frank B., Kaplan Robert C., Metspalu Andres, North Kari E., Schlessinger David, Wareham Nicholas J., Hunter David J., O’Connell Jeffrey R., Strachan David P., Wichmann H.-Erich, Borecki Ingrid B., van Duijn Cornelia M., Schadt Eric E., Thorsteinsdottir Unnur, Peltonen Leena, Uitterlinden André G., Visscher Peter M., Chatterjee Nilanjan, Loos Ruth J. F., Boehnke Michael, McCarthy Mark I., Ingelsson Erik, Lindgren Cecilia M., Abecasis Gonçalo R., Stefansson Kari, Frayling Timothy M., Hirschhorn Joel N. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warrington NM, et al. Genome-wide association study identifies nine novel loci for 2D:4D finger ratio, a putative retrospective biomarker of testosterone exposure in utero. Hum. Mol. Genet. 2018;27:2025–2038. doi: 10.1093/hmg/ddy121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris JA, et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet. 2019;51:258–266. doi: 10.1038/s41588-018-0302-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heo I, et al. Lin28 Mediates the Terminal Uridylation of let-7 Precursor MicroRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 17.Viswanathan SR, Daley GQ. Lin28: A MicroRNA Regulator with a Macro Role. Cell. 2010;140:445–449. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Balzeau J, Menezes MR, Cao S, Hagan JP. The LIN28/let-7 Pathway in Cancer. Frontiers in genetics. 2017;8:31–31. doi: 10.3389/fgene.2017.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu H, et al. The Lin28/let-7 Axis Regulates Glucose Metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang D-H, Moss EG. Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse. Gene Expression Patterns. 2003;3:719–726. doi: 10.1016/S1567-133X(03)00140-6. [DOI] [PubMed] [Google Scholar]
- 21.Yokoyama S, et al. Dynamic gene expression of Lin-28 during embryonic development in mouse and chicken. Gene Expression Patterns. 2008;8:155–160. doi: 10.1016/j.gep.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machluf Y, Gutnick A, Levkowitz G. Development of the zebrafish hypothalamus. Ann. N. Y. Acad. Sci. 2011;1220:93–105. doi: 10.1111/j.1749-6632.2010.05945.x. [DOI] [PubMed] [Google Scholar]
- 23.Li M, Zhao L, Page-McCaw PS, Chen W. Zebrafish Genome Engineering Using the CRISPR–Cas9 System. Trends Genet. 2016;32:815–827. doi: 10.1016/j.tig.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Developmental dynamics: an official publication of the American Association of Anatomists. 2009;238:2975–3015. doi: 10.1002/dvdy.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shang H, Li Q, Feng G, Cui Z. Molecular analysis and functions of p53R2 in zebrafish. Gene. 2011;475:30–38. doi: 10.1016/j.gene.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Greiling TMS, Houck SA, Clark JI. The zebrafish lens proteome during development and aging. Mol. Vis. 2009;15:2313–2325. [PMC free article] [PubMed] [Google Scholar]
- 27.Oz-Levi D, et al. Mutation in TECPR2 reveals a role for autophagy in hereditary spastic paraparesis. Am. J. Hum. Genet. 2012;91:1065–1072. doi: 10.1016/j.ajhg.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mootha Vamsi K, Lindgren Cecilia M, Eriksson Karl-Fredrik, Subramanian Aravind, Sihag Smita, Lehar Joseph, Puigserver Pere, Carlsson Emma, Ridderstråle Martin, Laurila Esa, Houstis Nicholas, Daly Mark J, Patterson Nick, Mesirov Jill P, Golub Todd R, Tamayo Pablo, Spiegelman Bruce, Lander Eric S, Hirschhorn Joel N, Altshuler David, Groop Leif C. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genetics. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 29.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sangiao-Alvarellos S, et al. Changes in Hypothalamic Expression of the Lin28/let-7 System and Related MicroRNAs During Postnatal Maturation and After Experimental Manipulations of Puberty. Endocrinology. 2013;154:942–955. doi: 10.1210/en.2012-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science348, 648–660, 10.1126/science.1262110%J Science (2015). [DOI] [PMC free article] [PubMed]
- 32.Barson Nicola J., Aykanat Tutku, Hindar Kjetil, Baranski Matthew, Bolstad Geir H., Fiske Peder, Jacq Céleste, Jensen Arne J., Johnston Susan E., Karlsson Sten, Kent Matthew, Moen Thomas, Niemelä Eero, Nome Torfinn, Næsje Tor F., Orell Panu, Romakkaniemi Atso, Sægrov Harald, Urdal Kurt, Erkinaro Jaakko, Lien Sigbjørn, Primmer Craig R. Sex-dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature. 2015;528(7582):405–408. doi: 10.1038/nature16062. [DOI] [PubMed] [Google Scholar]
- 33.Ambros V, Horvitz H. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226(4673):409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 34.Cousminer DL, et al. Genome-wide association and longitudinal analyses reveal genetic loci linking pubertal height growth, pubertal timing and childhood adiposity. Hum. Mol. Genet. 2013;22:2735–2747. doi: 10.1093/hmg/ddt104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marvel Miranda, Spicer Olivia Smith, Wong Ten-Tsao, Zmora Nilli, Zohar Yonathan. Knockout of the Gnrh genes in zebrafish: effects on reproduction and potential compensation by reproductive and feeding-related neuropeptides†. Biology of Reproduction. 2018;99(3):565–577. doi: 10.1093/biolre/ioy078. [DOI] [PubMed] [Google Scholar]
- 36.Tang H, et al. The kiss/kissr systems are dispensable for zebrafish reproduction: evidence from gene knockout studies. Endocrinology. 2015;156:589–599. doi: 10.1210/en.2014-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El-Brolosy MA, Stainier DYR. Genetic compensation: A phenomenon in search of mechanisms. PLOS Genetics. 2017;13:e1006780. doi: 10.1371/journal.pgen.1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El-Brolosy MA, et al. Genetic compensation triggered by mutant mRNA degradation. Nature. 2019 doi: 10.1038/s41586-019-1064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, et al. LIN28 Regulates Stem Cell Metabolism and Conversion to Primed Pluripotency. Cell Stem Cell. 2016;19:66–80. doi: 10.1016/j.stem.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Zhu Hao, Shah Samar, Shyh-Chang Ng, Shinoda Gen, Einhorn William S, Viswanathan Srinivas R, Takeuchi Ayumu, Grasemann Corinna, Rinn John L, Lopez Mary F, Hirschhorn Joel N, Palmert Mark R, Daley George Q. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nature Genetics. 2010;42(7):626–630. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swerdloff RS, Odell WD. Hormonal mechanisms in the onset of puberty. Postgrad. Med. J. 1975;51:200–208. doi: 10.1136/pgmj.51.594.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snyder Peter J., Peachey Helen, Hannoush Peter, Berlin Jesse A., Loh Louise, Holmes John H., Dlewati Abdallah, Staley Janet, Santanna Jill, Kapoor Shiv C., Attie Maurice F., Haddad John G., Strom Brian L. Effect of Testosterone Treatment on Bone Mineral Density in Men Over 65 Years of Age1. The Journal of Clinical Endocrinology & Metabolism. 1999;84(6):1966–1972. doi: 10.1210/jcem.84.6.5741. [DOI] [PubMed] [Google Scholar]
- 43.Mayer C., Acosta-Martinez M., Dubois S. L., Wolfe A., Radovick S., Boehm U., Levine J. E. Timing and completion of puberty in female mice depend on estrogen receptor -signaling in kisspeptin neurons. Proceedings of the National Academy of Sciences. 2010;107(52):22693–22698. doi: 10.1073/pnas.1012406108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herber CB, et al. Estrogen signaling in arcuate Kiss1 neurons suppresses a sex-dependent female circuit promoting dense strong bones. Nature Communications. 2019;10:163. doi: 10.1038/s41467-018-08046-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tiano, J. P. et al. Early-Life Exposure to Testosterone Programs the Hypothalamic Melanocortin System. Endocrinology152, 1661–1669, 10.1210/en.2010-1288%J Endocrinology (2011). [DOI] [PMC free article] [PubMed]
- 46.Hegadoren KM, O’Donnell T, Lanius R, Coupland NJ, Lacaze-Masmonteil N. The role of β-endorphin in the pathophysiology of major depression. Neuropeptides. 2009;43:341–353. doi: 10.1016/j.npep.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 47.Robinson MD, McCarthy DJ, Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loh Po-Ru, Tucker George, Bulik-Sullivan Brendan K, Vilhjálmsson Bjarni J, Finucane Hilary K, Salem Rany M, Chasman Daniel I, Ridker Paul M, Neale Benjamin M, Berger Bonnie, Patterson Nick, Price Alkes L. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nature Genetics. 2015;47(3):284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The gene expression datasets analysed during the current study are available from the GTEx portal (https://gtexportal.org/). The UK biobank data is available for researches upon request from the UK Biobank (https://www.ukbiobank.ac.uk/researchers/). The results from the RNA-seq dataset from 1d and 7d old zebrafish generated and analysed during the current study are included in the supplementary files.