Abstract
Background
Retinoid X Receptor Gamma (RXRG) is a member of the nuclear receptor superfamily and plays a role in tumour suppression. This study aims to explore the prognostic significance of RXRG in breast cancer.
Methods
Primary breast cancer tissue microarrays (n = 923) were immuno-stained for RXRG protein and correlated with clinicopathological features, and patient outcome.
Results
Nuclear RXRG expression was significantly associated with smaller tumour size (p = 0.036), lower grade (p < 0.001), lobular histology (p = 0.016), lower Nottingham Prognostic Index (p = 0.04) and longer breast cancer-specific survival (p < 0.001), and longer time to distant metastasis (p = 0.002). RXRG expression showed positive association with oestrogen receptor (ER)-related biomarkers: GATA3, FOXA1, STAT3 and MED7 (all p < 0.001) and a negative correlation with the Ki67 proliferation marker. Multivariate analysis demonstrated RXRG protein as an independent predictor of longer breast cancer-specific survival and distant metastasis-free survival. In the external validation cohorts, RXRG expression was associated with improved patients’ outcome (p = 0.025). In ER-positive tumours, high expression of RXRG was associated with better patient outcome regardless of adjuvant systemic therapy. ER signalling pathway was the top predicted master regulator of RXRG protein expression (p = 0.005).
Conclusion
This study provides evidence for the prognostic value of RXRG in breast cancer particularly the ER-positive tumours.
Subject terms: Breast cancer, Prognostic markers
Introduction
Breast cancer is the most common cancer among women worldwide.1 Oestrogen receptor (ER) and progesterone receptor (PR), which are members of the nuclear receptor superfamily of transcription factors, are important in predicting prognosis and establishing therapeutic strategies for breast cancer treatment. Recent studies have revealed growing evidence of the involvement of nuclear receptors, other than ER and PR, in breast cancer development and progression.2,3 Drugs targeting nuclear receptors are widely used in the clinic for treating patients.4 Expression levels of some nuclear receptors, such as thyroid hormone receptor beta (THRb), COUP transcription factor 2 (COUP-TF2), peroxisome proliferator-activated receptor gamma (PPARG) and liver receptor homologue-1 (LRH-1), are associated with clinicopathological variables and can predict outcome in tamoxifen-treated patients.5 The glucocorticoid receptor (GR) in breast cancer exerts anti-proliferative and anti-apoptotic activities and its overexpression is associated with features characteristic of longer survival.6,7 Moreover, in tamoxifen-treated ER-positive breast cancer, androgen receptor (AR; also a member of the nuclear receptor superfamily) status has prognostic value and it is reported to be a crucial factor in deciding treatment regime.8 With these important roles in breast cancer, other nuclear receptors could therefore provide additional therapeutic targets for breast cancer management.9–11
Retinoids derived from vitamin A are signalling molecules that play important roles in cell differentiation and proliferation12 and act via retinoic acid receptors (RARs) and retinoid X receptors (RXRs), which are members of the nuclear receptors superfamily. Retinoids are well documented for their ability to induce differentiation and arrest proliferation in cancer.12,13 The RXR family are known to form heterodimers with other nuclear receptors, including the vitamin D receptor (VDR), peroxisome proliferator-activated receptors (PPARs) and RARs.11 There are three subtypes of the Retinoid X Receptor (RXR), namely RXR Alpha (RXRα; NR2B1), RXRβ (NR2B2) and RXRγ (NR2B3).14 These receptors have tumour suppressor properties, particularly as their ligand 9-cis-retinoic acid,12 and impede cellular proliferation.15 Moreover, the RXR family are involved in mediating the anti-proliferative effects of retinoic acid (RA) as partners of RARB and RARA.12
RXRG has been demonstrated to modulate cellular differentiation and apoptosis in different tumour types. For example, enhanced expression of RXRG was associated with increased apoptosis in ovarian cancer.16 Epigenetic silencing of RXRG correlated with decreased overall survival in lung cancer.17 In ovarian cancer tumour models, RXRG activation re-sensitises ovarian carcinoma cells to apoptosis. However, the mechanism by which this occurs is still unclear. With minimal toxicity both in vitro and in vivo, novel RXR family members (rexinoids), have been reported to suppress breast cancer development in several animal models and have been extensively evaluated either alone or in combination with selective ER modulators.18 One RXRG partner, RARA was shown to influence the ERα transcriptional complex in oestrogen-treated MCF-7 breast cancer cells.19,20 Altogether, these findings indicate that RXRG could have a function in tumour pathogenesis and could potentially be promising cancer therapeutics.
Therefore, this study aimed to investigate the potential prognostic role of RXRG in breast cancer with a focus on the luminal ER-positive class.
Methods
Study cohort
This study was conducted on a large cohort (n = 923) of primary breast cancer from patients who presented to Nottingham City Hospital with available clinicopathological data, as previously described.21–23 Treatment and outcome data, including breast cancer-specific survival and distant metastasis-free interval was maintained on a prospective basis. Breast cancer-specific survival was defined as the duration (in months) from the date of primary surgery to the time of death because of breast cancer. Distant metastasis-free interval was defined as the duration (in months) from primary surgical treatment to the occurrence of first distant recurrence.
Evaluation of RXRG protein expression
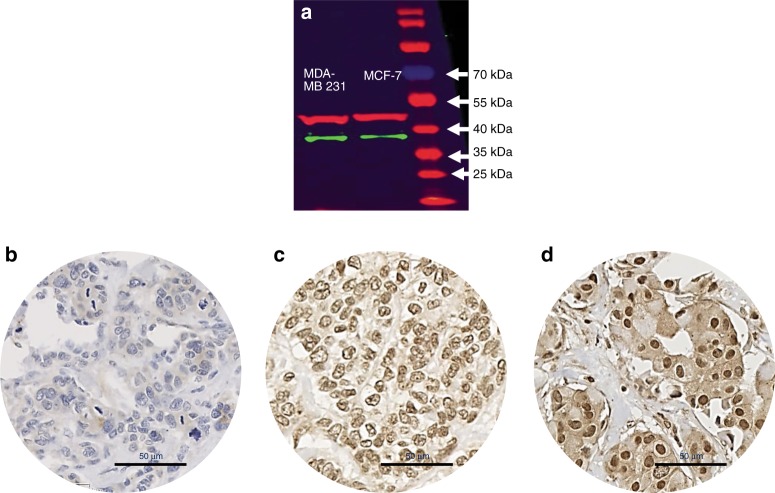
RXRG protein expression was evaluated using immunohistochemistry (IHC) preceded by validation of the rabbit RXRG antibody (Abcam, ab15518) specificity using western blot. For the latter, cell lysates of MDA-MB-231 and MCF-7 cell lines (obtained from the American Type Culture Collection; Rockville, MD, USA) were incubated with the primary antibody at 1:700 dilution. The specificity of the antibody was validated with a single specific band at the predicted molecular weight (39 kDa, Fig. 1a).
Fig. 1.
Western blot and immunohistochemical expression of RXRG in breast cancer. a Western blotting results for RXRG expression in MCF-7 and MDA-MB231 breast cancer cell lines using rabbit polyclonal antibody (Abcam, ab15518). Green and red bands represent RXRG and the house-keeping Beta-Actin, respectively. RXRG protein expression in breast cancer tissue microarrays cores. b Negative/no staining c showing low expression and d showing high immunoreactivity. Images are at x40 magnification
For evaluation of the morphological pattern of protein expression and suitability of tissue microarrays for its assay, immunohistochemistry was assessed in full-face breast cancer tissue sections (n = 10). Tumour samples were arrayed onto tissue microarrays as previously described.21 Four micrometre sections from the tissue microarrays and full-face sections were immunohistochemically stained using the Novolink Max Polymer Detection system (Leica, Newcastle, UK). The antibody was incubated 24 h at the concentration of 1:300.
The modified Histo-score (H-score) method was used in assessing immunohistochemistry staining, taking the staining intensity and percentage positivity into account.24 High-resolution digital images were generated via scanning the stained slides using Nanozoomer (Hamamatsu Photonics, Welwyn Garden City, UK) at x20 magnification to facilitate the scoring of the tissue microarrays cores. The sections were blindly double scored by two researchers, including a consultant histopathologist for ~25% cores to assess inter-observer concordance. Inter-observer agreement was determined, and the intra-class correlation co-efficient was 0.83, indicating an excellent concordance between scorers. Moreover, the discordant cases were re-scored by the both observers and a consensus score was agreed and assigned. Biomarkers closely relevant to breast cancer carcinogenesis, progression and outcome were also available for this cohort of patients (See Tables 2 and 3). Immunohistochemistry staining and dichotomisation of these biomarkers were used as per previous publications.6,22,25–33
Table 2.
Associations between RXRG expression and other biomarkers in the whole series, in ER-positive and ER-negative breast cancer series
| Parameters | RXRG expression whole cohort | RXRG expression ER-positive cohort | RXRG expression ER-negative cohort | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Negative/low expression N (%) | High expression N (%) | p-value (χ2) | Negative/low expression N (%) | High expression N (%) | p-value (χ2) | Negative/low expression N (%) | High expression N (%) | p-value (χ2) | |
| Oestrogen (ER) status | |||||||||
| Negative | 138 (63.0) | 81 (37.0) | <0.0001 (20.142) | ||||||
| Positive | 319 (48.7) | 380 (54.4) | |||||||
| Progesterone (PR) status | |||||||||
| Negative | 201 (56.3) | 156 (43.7) | 0.018 (7.726) | 67 (45.0) | 82 (55.0) | 0.780 (0.137) | 134 (65.0) | 73.0 (35.0) | 1.00 (0.543) |
| Positive | 247 (46.8) | 281 (53.2) | 246 (46.7) | 281 (53.3) | 1 (100.0) | 0 (0.00) | |||
| Human epidermal growth factor receptor 2 (HER2) | |||||||||
| Negative | 371 (48.4) | 395 (51.6) | 0.057 (3.612) | 269 (44.2) | 339 (55.8) | 0.102 5.750) | 102 (66.0) | 53 (34.0) | 0.016 (1.928) |
| Positive | 72 (57.6) | 53 (42.4) | 41 (59.4) | 28 (40.6) | 31 (55.0) | 25 (45.0) | |||
| Forkhead box protein A1 (FOXA1) | |||||||||
| Negative | 235 (65.1) | 126 (34.9) | <0.00001 (33.053) | 133 (61.0) | 85 (39.0) | <0.0001 (19.026) | 102 (71.0) | 41 (29.0) | 0.194 (2.220) |
| Positive | 103 (41.5) | 145 (58.5) | 92 (40.4) | 136 (59.6) | 11 (55.0) | 9 (45.0) | |||
| GATA-binding protein 3 (GATA3) | |||||||||
| Negative | 266 (62.3) | 161 (37.7) | <0.00001 (36.024) | 169 (58.9) | 118 (41.1) | 0.0001 (23.251 | 97 (69.3) | 43 (30.7) | 0.312 (2.220) |
| Positive | 43 (32.6) | 89 (67.4) | 43 (3.3) | 86 (66.7) | 0 (0.00) | 1(100) | |||
| Brain-expressed X-linked protein 1(BEX1) | |||||||||
| Negative | 149 (70.0) | 64 (30.0) | <0.00001 (31.812) | 99 (67.3) | 48 (32.7) | <0.00001 (24.131) | 50 (77.0) | 15 (23.0) | 0.032 (5.610) |
| Positive | 184 (46.1) | 215 (53.9) | 133 (42.8) | 178 (57.2) | 51 (59.0) | 36 (41.0) | |||
| Cluster of differentiation 71 (CD71) | |||||||||
| Negative | 139 (50.2) | 138 (49.8) | 0.049 (4.891) | 115 (47.1) | 129 (52.9) | 0.496 (2.396) | 25 (71.0) | 10 (29.0) | 0.838 (0.114) |
| Positive | 218 (58.9) | 152 (41.1) | 130 (54.2) | 110 (45.8) | 89 (69.0) | 41 (32.0) | |||
| Ki67 | |||||||||
| Negative | 120 (41.0) | 173 (59.0) | 0.0004 (15.903) | 104 (39.4) | 160 (60.6) | 0.014 (9.660) | 15 (56.0) | 12 (44.0) | 0.678 (0.590) |
| Positive | 240 (56.1) | 188 (43.9) | 150 (25.6) | 135 (47.4) | 90 (63.0) | 52 (37.0) | |||
| Cytokeartin5/6 (CK5/6) | |||||||||
| Negative | 298 (49.1) | 309 (50.9) | 0.020 (7.883) | 242 (46.7) | 276 (53.3) | 1.63 (0.157) | 56 (63.0) | 32 (37.0) | 0.623 (0.402) |
| Positive | 70 (63.6) | 40 (36.4) | 8 (42.1) | 11 (57.9) | 62 (68.0) | 29 (32.0) | |||
| Phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha (PIK3CA) | |||||||||
| Negative | 71 (40.1) | 106 (59.9) | 0.0004 (15.545) | 60 (38.7) | 95 (61.3) | 0.012 (10.045) | 11 (55.0) | 9 (45.0) | 0.458 (0.832) |
| Positive | 307 (57.2) | 230 (42.8) | 205 (53.8) | 176 (46.2) | 102 (65.0) | 54 (35.0) | |||
| N-cadherin | |||||||||
| Negative | 66 (34.2) | 127 (65.8) | <0.00001 (32.387) | 53 (32.3) | 111 (67.7) | <0.00001 (20.774) | 13 (46.0) | 15 (54.0) | 0.034 (6.434) |
| Positive | 286 (58.4) | 204 (41.6) | 194 (53.7) | 167 (46.3) | 92 (71.0) | 37 (29.0) | |||
| Signal transducer and activator of transcription 3 (STAT3) | |||||||||
| Negative | 283 (59.7) | 191 (40.3) | <0.00001 (35.589) | 197 (57.3) | 147 (42.7) | <0.00001 (28.678) | 86 (66.0) | 44 (34.0) | 0.210 (1.734) |
| Positive | 61 (34.3) | 117 (65.7) | 45 (30.8) | 101 (69.2) | 16 (53.0) | 14 (47.0) | |||
| Mediator of RNA polymerase II transcription subunit 7 (MED7) | |||||||||
| Negative | 275 (67.7) | 131 (32.3) | <0.00001 (105.75) | 117 (63.4) | 102 (36.6) | <0.00001 (68.053) | 97 (79.0) | 26 (21.0) | <0.00001 (32.610) |
| Positive | 105 (30.2) | 243 (69.8) | 81 (28.7) | 201 (71.3) | 24 (37.0) | 41 (63.0) | |||
Significant p-values are highlighted in bold
Table 3.
Associations between RXRG expression and other nuclear receptors in the whole series, ER-positive and ER-negative breast cancer series
| Parameters | Whole cohort | ER-positive cohort | ER-negative cohort | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Negative/ low expression N (%) | High expression N (%) | p-value (χ2) | Negative/ low expression N (%) | High expression N (%) | p-value (χ2) | Negative/ low expression N (%) | High expression N (%) | p-value (χ2) | |
| Androgen receptor (AR) | |||||||||
| Negative | 253 (70.7) | 105 (29.3) | <0.0001 (105.72) | 156 (70.0) | 69 (30.0) | <0.0001 (77.25) | 97 (74.0) | 34 (26.0) | 0.0003(15.66) |
| Positive | 103 (31.4) | 225 (68.6) | 88 (30.3) | 202 (69.7) | 14 (39.0) | 22 (61.0) | |||
| Glucocorticoid receptor (GR) | |||||||||
| Negative | 184 (71.0) | 75 (29.0) | <0.0001 (67.10) | 108 (66.0) | 57 (34.0) | <0.0001 (36.88) | 76 (82.0) | 17 (18.0) | 0.00001(22.52) |
| Positive | 129 (37.4) | 216 (62.6) | 100 (36.0) | 180 (64.0) | 28 (45.0) | 34 (55.0) | |||
| Liver receptor homologue-1(LRH-1) | |||||||||
| Negative | 220 (65.5) | 116 (34.5) | <0.0001 (45.94) | 142 (63.0) | 85 (37.0) | <0.0001 (34.53) | 77 (73.0) | 29 (27.0) | 0.039 (5.13) |
| Positive | 135 (39.5) | 207 (60.5) | 103 (36.0) | 180 (64.0) | 32 (55.2) | 26 (44.8) | |||
| Peroxisome proliferator-activated receptor beta (PPARβ) | |||||||||
| Negative | 227 (67.0) | 112 (33.0) | <0.00001 (59.84) | 142 (64.0) | 80 (36.0) | <0.0001 (40.83) | 85 (74.0) | 30 (26.0) | 0.004 (10.556) |
| Positive | 94 (35.3) | 172 (64.7) | 78 (34.0) | 152 (66.0) | 15 (44.0) | 19 (56.0) | |||
| Peroxisome proliferator-activated receptor gamma (PPARγ) | |||||||||
| Negative | 267 (69.0) | 120 (31.0) | <0.00001 (107.54) | 175 (67.0) | 86 (33.0) | <0.0001 (77.30) | 92 (74.0) | 33 (26.0) | 0.00001 (24.55) |
| Positive | 51 (25.0) | 15 7 (75.0) | 437(25.0) | 141 (75.0) | 3 (15.8) | 16 (84.2) | |||
| Retinoid A receptor alpha (RARa) | |||||||||
| Negative | 238 (68.0) | 114 (32.0) | <0.00001 (72.29) | 193 (50.0) | 194 (50.0) | <0.00001 (24.13) | 85 (80.0) | 21 (20.0) | <0.00001 (22.46) |
| Positive | 117 (35.0) | 216 (65.0) | 52 (37.0) | 88 (63.0) | 26 (44.0) | 33 (56.0) | |||
| Retinoic acid-related orphan receptor gamma (RORγ) | |||||||||
| Negative | 294 (55.0) | 244 (45.0) | 0.002 (13.58) | 115 (47.1) | 129 (52.9) | 0.033 (6.69) | 100 (68.0) | 47 (32.0) | p = 0.22 (2.979) |
| Positive | 60 (38.0) | 98 (62.0) | 130 (54.2) | 110 (45.8) | 8 (47.1) | 9 (52.9) | |||
| Vitamin D receptor (VDR) | |||||||||
| Negative | 216 (59.0) | 153 (41.0) | 0.004 (12.85) | 133 (52.0) | 121 (48.0) | 0.090 (3.16) | 82 (72.6) | 31 (27.4) | 0.014 (10.309) |
| Positive | 145 (45.0) | 178 (55.0) | 119 (45.0) | 148 (55.0) | 26 (47.3) | 29 (52.7) | |||
| Photoreceptor cell-specific nuclear receptor (PNR) | |||||||||
| Negative | 206 (56.0) | 161 (44.0) | 0.030 (5.09) | 148 (52.0) | 138 (48.0) | 0.042 (4.80) | 57 (73.0) | 21 (27.0) | 0.22 (2.334) |
| Positive | 162 (48.0) | 178 (52.0) | 103 (43.0) | 141 (57.0) | 59 (62.0) | 36 (38.0) | |||
Significant p-values are highlighted in bold
Gene expression cohorts
The clinicopathological significance of RXRG mRNA expression was assessed using a subset (n = 150) of the Nottingham series that was included in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) data set.34 The aim of this investigation is to understand the molecular biology of RXRG protein expression as an end product, therefore, the analysis was completed utilising cases with RXRG protein expression. The definition of cases into low versus high groups was based on RXRG protein expression.
External validation was performed using the Breast Cancer Gene-Expression Miner v4.0 (bc-GenExMiner v4.0),35 as previously described.33,36 Breast cancer cases data set (n = 818) within The Cancer Genome Atlas (TCGA)37 was also used for external validation of RXRG mRNA expression. Patient outcome following systemic treatment was further validated using KM Plotter (n = 3951).38
Pathway analysis
Differential gene expression between RXRG negative and positive cases was assessed using the Robina implementation of EdgeR.39 Differential expression with >2-fold difference and a false discovery rate of q < 0.05 between RXRG-negative and -positive cases were considered significant. Webgestalt (http://www.webgestalt.org) was used to annotate the differential gene expression list and to identify over-represented gene ontologies and pathways.40
Statistical analysis
IBM SPSS 22.0 (Chicago, IL, USA) software was used for statistical analysis. The H-scores of expressions of nuclear RXRG did not follow a normal distribution. For this reason, expression of RXRG protein was used to define patient groups based on prediction of breast cancer-specific survival using X-tile (http://tissuearray.org; Yale University, USA).41 Chi-squared test was used to evaluate the association between expression of other biomarkers and the clinicopathological parameters. Correlation between RXRG and ER/PR expression was analysed using Spearman’s correlation co-efficient test. Association between RXRG expression, clinicopathological parameters and other related biomarkers using the continuous H-score were evaluated.
Kaplan–Meier analysis with log-rank test for significance was performed to assess breast cancer-specific survival and distant metastasis-free interval. Interaction between RXRG and ER was evaluated using Cox regression model, which was also used for multivariate survival analysis with adjustment of covariates to test independence from standard prognostic factors in breast cancer (nodal stage, tumour grade, tumour size, ER level of expression (defined as percentage of positive tumour cells), and Ki67. The STRING database (http://string-db.org)42 was used to evaluate the interaction with RXRG and other nuclear receptors in steroid signalling pathways. The p-values were adjusted using Bonferroni correction for multiple testing. A p-value of <0.05 was considered significant.
This study obtained ethics approval by the North West–Greater Manchester Central Research Ethics Committee under the title: Nottingham Health Science Biobank (NHSB), reference number 15/NW/0685. All samples from Nottingham used in this study were pseudo-anonymised and collected prior to 2006 and stored in compliance with the UK Human Tissue Act.
Results
RXRG protein expression
Full-face tissue sections (Supplementary Fig. 1a–c) showed high RXRG expression in the normal glandular epithelium (Supplementary Fig. 1b). In contrast, low RXRG immunopositivity was observed in the nuclei of invasive cancer cells (Supplementary Fig. 1c), with some malignant cells additionally featuring cytoplasmic staining. On tissue microarrays, RXRG protein expression levels varied from absent to high (Fig. 1b–d). In the 923 scorable cores, the cutoff points of the RXRG nuclear H-score was set at 175 by X-tile analysis, where low expression is defined as H-scores <175 and high expression as H-scores ≥175. Low RXRG nuclear expression was observed in 458/923 (49.6%) cases and high expression was observed in 465/923 (50.4%) cases. Low RXRG mRNA expression was found in 73/150 (49%), whereas high RXRG mRNA expression was observed in 77/150 (51%) cases.
Relationship between RXRG protein expression and clinicopathological variables
In the whole cohort and ER-positive sub-cohort, RXRG was associated with features of favourable prognosis, including smaller tumour size (p = 0.036), lower histological grade (p < 0.00001), less pleomorphism (p = 0.042), lower mitotic scores (p < 0.00001), lobular and special tumour types of excellent prognosis (p = 0.016), and lower Nottingham Prognostic Index (p < 0.05; Table 1). Moreover, significant association was observed with breast cancer molecular intrinsic subtypes (p < 0.00001 and p = 0.009), for the whole series and ER-positive tumours, respectively (Table 1). High RXRG expression was primarily observed in luminal A tumours (136/214, 63.6%), while it was less expressed in HER2+ and triple-negative breast cancer.
Table 1.
Associations between RXRG expression and clinicopathological features in the whole series, ER-positive and ER-Negative breast cancer series
| Parameters | RXRG expression whole cohort | RXRG expression ER-positive cohort | RXRG expression ER-negative cohort | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Negative/ low expression N (%) | High expression N (%) | p-value (χ2) | Negative/ low expression N (%) | High expression N (%) | p-value (χ2) | Negative/ low expression N (%) | High expression N (%) | p-value (χ2) | |
| Age at diagnosis (years) | |||||||||
| <50 | 167 (51.2) | 159 (48.8) | 1.473 (0.520) | 94 (43.3) | 123 (56.7) | 1.239 (0.682) | 72 (68.6) | 33 (31.4) | 0.123 (2.673) |
| ≥50 | 291 (48.7) | 306 (51.3) | 225 (46.7) | 257 (53.3) | 66 (57.9) | 48 (42.1) | |||
| Histological grade | |||||||||
| 1 | 52 (35.6) | 94 (64.4) | <0.00001 (44.423) | 49 (35.8) | 88 (64.2) | <0.00001 (25.929) | 2 (40.0) | 3 (60.0) | 0.530 (1.271) |
| 2 | 130 (40.8) | 189 (59.2) | 122 (39.9) | 184 (60.1) | 8 (61.5) | 5 (38.5) | |||
| 3 | 273 (60.9) | 175 (39.1) | 145 (58.5) | 103 (41.5) | 128 (63.6) | 71 (35.7) | |||
| Tubules | |||||||||
| 1 | 11 (26.2) | 31 (73.8) | 0.004 (13.895) | 11(27.5) | 29 (72.5) | 0.172 (6.284) | 0 (0.0) | 1 (100.0) | 0.376 (1.959) |
| 2 | 140 (46.1) | 164 (53.9) | 123 (44.2) | 155 (55.8) | 17 (68.0) | 8 (32.0) | |||
| 3 | 289 (53.4) | 252 (46.6) | 169 (48.0) | 183 (52.0) | 120 (63.5) | 69 (36.5) | |||
| Pleomorphism | |||||||||
| 1 | 5 (23.8) | 16 (76.2) | <0.00001 (23.960) | 5 (26.3) | 14 (73.7) | 0.042 (10.294) | 0 (0.0) | 1 (100.0) | 0.406 (1.803) |
| 2 | 144 (41.4) | 204 (58.6) | 136 (40.6) | 199 (59.4) | 8 (66.7) | 4 (33.3) | |||
| 3 | 291 (56.2) | 227 (43.8) | 162 (51.3) | 154 (48.7) | 129 (63.9) | 73 (36.1) | |||
| Mitosis | |||||||||
| 1 | 111 (36.0) | 197 (64.0) | <0.00001 (53.653) | 107 (36.0) | 190 (64.0) | <0.00001 (22.597) | 4 (44.4) | 5 (55.6) | 0.170 (3.452) |
| 2 | 77 (43.3) | 101 (56.7) | 67 (42.4) | 91 (57.6) | 10 (50.0) | 10 (50.0) | |||
| 3 | 252 (62.8) | 149 (37.2) | 129 (60.0) | 86 (40.0) | 123 (66.1) | 63 (33.9) | |||
| Stage | |||||||||
| I | 280 (50.5) | 275 (49.5) | 1.69 (0.337) | 203 (47.6) | 221 (52.4) | 1.064 (2.200) | 80 (60.6) | 52 (39.4) | 0.522 (1.300) |
| II | 141 (49.1) | 146 (50.9) | 97 (43.7) | 125 (56.3) | 43 (68.3) | 20 (31.7) | |||
| III | 34 (47.2) | 38 (52.8) | 19 (38.0) | 31 (62.0) | 15 (68.2) | 7 (31.8) | |||
| Tumour size | |||||||||
| <2.0 cm | 182 (42.8) | 243 (57.2) | 0.0005 (15.355) | 143 (40.6) | 209 (59.4) | 0.036 (7.550) | 38 (54.3) | 32 (45.7) | 0.071 (3.609) |
| ≥2.0 cm | 274 (55.8) | 217 (44.2) | 174 (51.0) | 167 (49.0) | 100 (67.6) | 48 (32.4) | |||
| Histological type | |||||||||
| Ductal | 403 (53.3) | 353 (46.7) | 0.0001 (29.455) | 277 (49.5) | 283 (50.5) | 0.016 (19.281) | 125 (64.8) | 68 (35.2) | 0.071 (10.161) |
| Lobular | 32 (32.3) | 67 (67.7) | 32 (33.0) | 65 (67.0) | 0 (0.00) | 2 (100.0) | |||
| Medullary-like | 12 (57.1) | 9 (42.9) | 1 (50.0) | 1 (50.0) | 11 (57.9) | 8 (42.1) | |||
| Special typea | 8 (22.2) | 28 (77.8) | 6 (18.8) | 26 (81.3) | 2 (100.0) | 0 (0.0) | |||
| IHC subtypes | |||||||||
| ER+/HER2– low proliferation | 78 (36.4) | 136 (63.6) | <0.00001 (37.474) | 78 (36.4) | 136 (63.6) | 0.009 (14.564) | 0.103 (2.849) | ||
| ER+/HER2 – high proliferation | 147 (50.3) | 145 (49.7) | 147 (50.3) | 145 (49.7) | |||||
| Triple negative | 102 (68.0) | 48 (32.0) | 102 (68.0) | 48 (32.0) | |||||
| HER2 + | 71 (57.3) | 53 (42.7) | 31 (55.4) | 25 (44.6) | |||||
| Nottingham Prognostic Index | |||||||||
| GPG | 105 (39.2) | 163 (60.8) | 0.0004 (19.294) | 101 (39.8) | 153 (60.2) | 0.040 (6.538) | 3 (30.0) | 7 (70.0) | 0.051 (5.943) |
| MPG | 260 (52.5) | 235 (47.5) | 165 (48.1) | 178 (51.9) | 95 (62.9) | 56 (37.1) | |||
| PPG | 91 (59.5) | 62 (40.5) | 51 (45.7) | 45 (46.9) | 40 (70.2) | 17 (29.8) | |||
Significant p-values are highlighted in bold;
GPG good prognostic group, MPG moderate prognostic group, PPG poor prognostic group
aSpecial types of excellent prognosis (invasive tubular, invasive cribriform, invasive mucinous, invasive papillary carcinoma)
There was a significant positive linear correlation between RXRG and ER expression in the whole cohort and in ER-positive tumours (r = 0.30, p < 0.0001 and r = 0.20, p = 0.016, respectively). Similar results were observed with PR expression (r = 0.20, p = 0.014 and r = 0.17, p = 0.016; respectively). High-nuclear RXRG expression showed significant positive association with ER and PR positivity (p < 0.0001 and p = 0.018, respectively), while negative association was observed with basal cytokeratin CK5/6 (p = 0.020; Table 2). High expression of RXRG was positively associated with luminal subtype-related biomarkers in both the whole cohort and ER-positive tumours, including ER-chromatin interaction regulator Forkhead box protein A1 (FOXA1; p < 0.00001) and human brain-expressed X-linked1 (BEX1; p < 0.00001). Significant positive associations were observed with cell cycle regulatory proteins such as GATA3 (p = 0.0001), and STAT3 (p < 0.00001); markers also known to be over-expressed in ER-positive breast cancer and associated with favourable outcome.21,43 By contrast, negative associations were observed with the proliferation marker Ki67 (p = 0.014), epithelial–mesenchymal transition markers such as N-cadherin (p < 0.00001) and phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha (PIK3CA; p = 0.012). In addition, the mediator subunit MED7 was positively associated (p < 0.00001) with RXRG (Table 2; both whole and ER-positive cohort). In ER-negative tumours, only MED7 (p < 0.00001), BEX1 (p = 0.032) and N-cadherin (p = 0.034) showed significant association with RXRG (Table 2).
Positive associations were observed between the nuclear expression of RXRG and the expression of nuclear receptors, including PPARγ, PPARβ, AR, RARα, glucocorticoid receptor and liver receptor homologue-1 (p for all <0.001) (Table 3; in the whole cohort, ER-positive and ER-negative cohort). Moreover, using the continuous H-score to assess the association between RXRG expression and the clinicopathological parameters, as well as other breast cancer-related biomarkers revealed similar significant association to those obtained with the categorised RXRG (Supplementary Table 1).
Association between RXRG protein expression and patients’ outcome
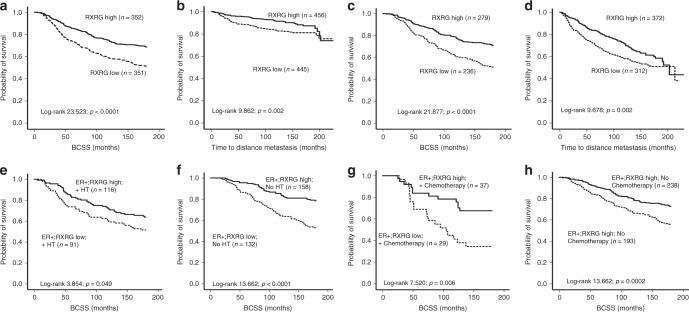
High expression of RXRG was associated with longer breast cancer-specific survival (p < 0.0001; Fig. 2a). Regarding distant metastasis, high RXRG expression was associated with a lower probability of distant metastasis (p = 0.002; Fig. 2b). Cox proportional multivariate analysis showed that RXRG expression was an independent indicator of both longer breast cancer-specific survival and distant metastasis-free interval in the whole cohort (HR = 0.6; 95% CI = 0.4–0.8; p = 0.04 and HR = 0.7; 95% CI = 0.6–0.9; p = 0.025, respectively) independent of the standard prognostic parameters of breast cancer, including tumour size, histological grade, nodal stage, ER status and proliferative fraction as assessed by Ki67. Comparable results were obtained when we included the ER level of expression as a continuous variable to the multivariate analysis of the ER-positive cohort (Table 4).
Fig. 2.
Kaplan–Meier plot for the association of RXRG nuclear expression. Whole series: a Breast cancer-specific survival, b distant metastasis-free survival. In ER-positive tumours. c Breast cancer-specific survival, d distant metastasis-free survival. Kaplan–Meier analysis of breast cancer-specific survival showing the impact of treatment on RXRG nuclear protein expression in ER-positive cohort; e in patients who did receive hormone therapy f in patients that did not receive hormone therapy g in patients who did receive chemotherapy and h in patients who did not receive chemotherapy with significance determined using the log-rank test
Table 4.
Univariate and multivariate analysis of RXRG expression compared with tumour stage, grade, size, Ki67and ER status for breast cancer-specific survival and distant metastasis-free survival
| Variable | Breast cancer-specific survival | Distant metastasis-free interval | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | |||||||||
| HR | 95% CI | p-value | HR | 95% CI | p-value | HR | 95% CI | p-value | HR | 95% CI | p-value | |
| Whole cohort | ||||||||||||
| Stage | 2.1 | 1.9–2.4 | <0.0001 | 2.2 | 1.7– 2.8 | <0.0001 | 2.3 | 2.1–2.5 | <0.0001 | 2.0 | 1.6–2.4 | <0.0001 |
| Grade | 2.3 | 2.0–2.6 | <0.0001 | 1.7 | 1.3– 2.5 | <0.0001 | 1.7 | 1.6–2.0 | <0.0001 | 1.3 | 1.1–1.6 | 0.039 |
| Tumour size | 2.1 | 1.8–2.5 | <0.0001 | 1.6 | 1.1–2.2 | 0.006 | 1.9 | 1.6–2.2 | <0.0001 | 1.4 | 1.1–1.9 | 0.005 |
| ERa | 0.9 | 0.9–1.1 | <0.0001 | 1.1 | 0.9–1.2 | 0.558 | 0.9 | 0.8–1.1 | <0.0001 | 1.6 | 1.1–2.3 | 0.026 |
| Ki67 | 2.6 | 2.1–3.1 | <0.0001 | 1.5 | 1.1–2.3 | 0.027 | 2.1 | 1.7–2.5 | <0.0001 | 1.6 | 1.2–2.2 | 0.004 |
| RXRG | 0.6 | 0.4–0.7 | <0.0001 | 0.6 | 0.4–0.8 | 0.040 | 0.8 | 0.6–0.9 | 0.003 | 0.7 | 0.6–0.9 | 0.025 |
| ER+cohort | ||||||||||||
| Stage | 2.0 | 1.8–2.4 | <0.0001 | 2.1 | 1.6–2.7 | <0.0001 | 2.2 | 1.9–2.4 | <0.0001 | 2.0 | 1.6–2.4 | <0.0001 |
| Grade | 2.4 | 2.1–2.8 | <0.0001 | 1.6 | 1.2–2.3 | 0.004 | 1.9 | 1.6–2.1 | <0.0001 | 1.3 | 0.9–1.7 | 0.084 |
| Tumour size | 2.3 | 1.9–2.9 | <0.0001 | 1.6 | 1.1–2.4 | 0.025 | 2.2 | 1.8–2.6 | <0.0001 | 1.5 | 1.1–2.1 | 0.024 |
| ERa | 0.9 | 0.9–1.0 | 0.101 | 0.9 | 0.8–1.1 | 0.428 | 1.0 | 0.9–1.2 | 0.002 | 0.9 | 0.8–1.1 | 0.456 |
| Ki67 | 2.9 | 2.3–3.7 | <0.0001 | 1.8 | 1.2–2.9 | 0.005 | 2.4 | 1.9–3.0 | <0.0001 | 1.8 | 1.2–2.6 | 0.002 |
| RXRG | 0.5 | 0.4–0.7 | <0.0001 | 0.5 | 0.4–0.7 | 0.004 | 0.7 | 0.5–0.9 | 0.002 | 0.7 | 0.5–0.9 | 0.036 |
Significant p-values highlighted in bold
aER used as a continuous variable (percentage of positive tumour cells)
Similarly, in ER-positive tumours, high RXRG levels were predictive of longer breast cancer-specific survival (p < 0.0001; Fig. 2c) and longer distant metastasis-free interval (p = 0.002; Fig. 2d). The Cox regression model demonstrated that RXRG was an independent predictor of both breast cancer-specific survival and longer distant metastasis-free interval (HR = 0.5; 95% CI = 0.4–0.7; p = 0.004 and HR = 0.7; 95% CI = 0.5–0.9; p = 0.036, respectively, Table 4). In triple-negative breast cancer and HER2+ phenotypes, RXRG expression was neither associated with breast cancer-specific survival nor with distant metastasis-free interval.
RXRG positivity was associated with a significant survival advantage in patients with ER-positive tumours irrespective of hormonal therapy (p = 0.049 and p < 0.0001, respectively, Fig. 2e, f). Similarly, in ER-positive patients who either received or did not receive adjuvant chemotherapy, the prognostic advantage of positive RXRG expression was maintained (p = 0.006 and p = 0.002, respectively) (Fig. 2g, h). Supporting this, evaluation of the interaction between RXRG and ER level of expression (RXRG*ER) using the Cox regression model showed significant association with longer breast cancer-specific survival and distant metastasis-free interval (both p = 0.001).
There was a trend towards a positive linear correlation between RXRG mRNA and protein expression in the subset of Nottingham cases within the METABRIC study (n = 150), that has data on both mRNA and protein expression, however, the association did not reach statistical significance (r = 0.20, p = 0.077).
Genomic study and pathway analysis
We next identified differential gene expression between patients with low versus high RXRG mRNA expression in the Nottingham primary operable breast cancer series, which were included in the METABRIC34 study (n = 150). This analysis identified 1048 significant differentially expressed genes (p < 0.05), comprises of 554 over-expressed and 494 downregulated genes, associated with reduced RXRG expression. Analysis of the differential gene expression list identified over-represented pathways, including dysregulation of genes regulating ER signalling pathway (Supplementary Table 2; p = 0.0053; FOS and AP-1 transcription factor subunit). Other relevant pathways involved in regulating RXRG protein expression included cAMP signalling pathway (p = 0.001; ADORA1), protein digestion and absorption pathway (p = 0.001; COL4A2 and SLC7A7 and the ABC transport pathway (p = 0.002; ABCB9 and ABCD3). Interaction with RXRG and other nuclear receptors in steroid signalling pathways are summarised in Supplementary Fig. 2.
RXRG genomic profiling
Expression analysis for RXRG mRNA using Breast Cancer Gene-Expression Miner v4.0 showed that high RXRG expression was associated with older age at diagnosis (n = 3600; Supplementary Fig. 3a; p = 0.0082), lower histological tumour grade (n = 3518; p = 0.0024; Supplementary Fig. 3b), ER-positive status (n = 5558; Supplementary Fig. 3c; p = 0.029). Among PAM50 subtypes, RXRG mRNA was associated with luminal subtypes (n = 5607; p = 0.0024; Supplementary Fig. 3d) and non-triple-negative status (n = 1275; p = 0.014; Supplementary Fig. 3e). Targeted prognostic analyses for RXRG with nodal status and positive ER status patients (n = 33 data sets, 3941 patients) indicated that high gene expression correlated with adverse event-free survival (HR = 0.88; 95% CI = 0.79–0.98; p = 0.025; Supplementary Fig. 3f). Consistent with this, Kaplan–Meier analysis38 indicates high RXRG expression showed significant survival advantage irrespective of systemic treatment in (n = 3951; p < 0.0001; Supplementary Fig. 3g). To confirm this, we examined the TCGA-BRCA44,45 data set and found high RXRG mRNA expression was associated with longer disease-free intervals, post-menopausal status, and differential ER, PR and HER2 expression (Supplementary Fig. 4a–f).
Discussion
Understanding the mechanisms by which RXRs exert their effects in breast cancer remains incomplete.12 To our knowledge, this is the first study to define the prognostic role RXRG in breast cancer using a large clinical data set with long-term follow up. Results from the current study provide evidence that high expression of RXRG protein was significantly associated good long-term clinical outcome. Our study shows that high-nuclear RXRG was associated with ER-positive tumours, and is consistent with previous reports, which shows it confers a better prognostic impact.46 Indeed, the positive correlation between RXRG and ER expression, and association of higher RXRG with improved patient outcome independent of ER expression, suggest that RXRG could be a potential surrogate marker for ER expression in our cohort. Moreover, RXRG expression is significantly higher in breast cancer histologic subtypes with better prognosis such as invasive lobular carcinoma,46,47 in contrast to ductal or medullary-like tumours, which typically are associated with poorer outcomes.
In this study, ER-positive breast cancer showed the highest expression of RXRG compared to HER2+ and triple-negative breast cancer. Moreover, elevated expression of RXRG was associated with ER associated markers, such as GATA3,48 FOXA1,49 BEX,30 STAT343 and MED7.33 As noted earlier, RXRs and RARs form heterodimeric complexes, which bind DNA at specific retinoid responsive elements and regulate the various transcriptional processes.12 In breast cancer, functional interactions between retinoic acid and oestrogen signalling are complex and well documented.2,19,20
In this study pathway, analyses were conducted to explore the differentially enriched pathways associated with increased expression levels of RXRG protein. Results on pathway analysis confirmed our IHC findings reinforcing the importance of RXRG expression and ER status, where it revealed a positive association between high RXRG expression and ER positivity, and on patients’ survival. Our results indicated that the ER enriched pathway was the top master regulator of RXRG. Thus, we exposed a positive correlation between the genes regulating the ER pathway and RXRG protein expression, suggesting that suppressed expression of those indicators may inhibit signalling via the ER pathway and consequently affecting RXRG expression. For instance, dimerised ER directly binds to DNA sequences called oestrogen response elements (EREs) in relevant activated genes and activate gene transcription. However, ER is also known to use non-classical pathways via Activator protein 1 (AP-1) or via Specificity protein 1 (Sp-1).50 In ER-positive, breast cancer cell lines, ER enhanced ADORA1 mRNA and protein levels. Inhibition of ADORA1 reduced the binding activity of ER to its target gene indicating its role for the transcriptional activity of ER on oestrogen stimulation.51 By decreasing COL4A2 mRNA levels through miR-29b may be contribute to the tumorigenicity in ER-positive BC cells.52 The aforementioned studies have revealed the potential role of these biomarkers in ER-related pathways and may affect RXRG expression. However, it is important to note that the role of RXRG within ER-related pathways may be quite complex, depending on the specific interacting partners. For example, in this study, RXRG expression was negatively associated with PIK3CA. PIK3CA mutations are strongly associated with ER-positive tumours with better prognostic characteristics.53 Thus, its inverse relationship to PIK3CA warrants further investigation in the context of ER-associated pathways. Interestingly, in the MNU-induced rat mammary tumour models, the RXR-selective retinoid bexarotene (Targretin), suppressed ER-positive tumour development with minimal toxicity.54
In this study, the negative correlation with N-cadherin, CK5/6 and Ki67 indicates that RXRG expression is not associated with aggressive breast cancers. Elevated N-cadherin expression is associated with epithelial–mesenchymal transition (EMT) and tumour aggressiveness.55 In thyroid carcinoma, administration of ligands selective for RXRG resulted in a 30% reduction in cell proliferation,56 which is in agreement with low proliferation index and high RXRG expression. High molecular weight cytokeratin are strongly associated with high histological grade, and worse patient outcome31 and their negative association with RXRG further reinforces its role as a good prognostic indicator.
Nuclear RXRG expression displayed strong positive associations with other nuclear receptors. Studies have shown that RXRs form heterodimers with many nuclear receptors, including RARs, VDRs, PPARs, liver-x receptor (LXRs) and farnesoid X receptors (FXRs),57 suggesting that the positive correlations in our study could be due to heterodimer formation with one or more of these nuclear receptors. For instance, in breast cancer cells treated with ligands specific for PPARγ and RXR/RAR, troglitazone and 9-cis-retinoic acid, respectively, a reduction in proliferation was observed,58 and low doses of PPARγ and RXR ligands also promoted apoptosis.59 This suggests that RXRs have an anti-tumorigenic role, potentially through heterodimer formation with PPARγ. Treatment of thyroid cancer cells containing both RXRG and PPARγ with their ligands resulted in a synergistic increase in apoptotic activity.56 This suggests that RXRγ-PPARγ heterodimer may be present, and that the activation of this heterodimer leads to a synergistic increase in apoptosis. For this reason, we propose that increased expression of RXRG could potentiate heterodimer formation and activation of other nuclear receptors (e.g., VDR, RAR and PPARγ) thereby enhancing their anti-tumorigenic functions.
Regarding the association with patient outcome, high-nuclear RXRG expression was associated with improved breast cancer-specific survival and a longer time to distant metastasis in the whole series and in ER-positive breast cancer. However, in other breast cancer subtypes RXRG did not show any association with patient outcome. This might be due to the smaller sample size of ER-negative, HER2+ and triple-negative breast tumours in this cohort. Further investigation of larger cohorts of ER-negative, HER2+, and triple-negative breast tumours is therefore warranted. Our findings are consistent with previous reports in breast and renal cancer.60,61 In our study, these outcome associations were independent of other well-established prognostic variables. Interestingly, increased RXRG expression showed improved outcome regardless of adjuvant hormonal therapy or chemotherapy status. Hence, in chemotherapy-intolerant patients, therapeutic manipulation of RXRG on its own, or in combination with other therapies, may be helpful in improving the existing treatment regimen, particularly as next-generation RXR subtype-selective rexinoids enter clinical testing and use. Furthermore, assessment of RXRG mRNA levels using bc-GenExMiner and TCGA demonstrated that high RXRG mRNA expression is significantly associated with better tumour characteristics and longer event-free survival of breast cancer patients, which corroborates with RXRG protein expression. RARA mRNA expression levels in breast cancer patients treated with hormonal therapy predicted positive outcome,19 which is in agreement with our findings.
In summary, high RXRG expression in breast cancer is associated with favourable prognostic parameters and is an independent prognostic factor with prolonged patient survival. The interaction between RXRG, ER, and other nuclear receptors may explain the prognostic effect of RXRG in breast cancer. There is evidence that rexinoids are more effective anti-cancer agents than retinoids in preclinical models and show minimal toxicity.62 Therefore, further studies to validate the potential of RXRG as a therapeutic target in breast cancer are therefore warranted.
Supplementary information
Acknowledgements
We thank the Nottingham Health Science Biobank and Breast Cancer Now Tissue Bank for the provision of tissue samples.
Author contributions
C.J. participated in its conception, design, experimentation, analysis, interpretation and manuscript drafting. S.A. conducted the immunohistochemical studies and participated in the analysis and interpretation. M.S.T. helped with pathology review and manuscript drafting; M.A., F.Q.G. and I.A. helped in immune-histochemical analysis and interpretation; M.A., S.K., I.A., M.A.A., S.A., N.P.M., I.O.E. and A.R.G. participated in interpretation and manuscript drafting. E.A.R. conceived and supervised the study, participated in its design, interpretation and analysis, including drafting. All authors contributed to drafting and reviewing the manuscript and approved the submitted and final version.
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
All patients included in this study were consented. This work obtained ethics approval by the North West–Greater Manchester Central Research Ethics Committee under the title:Nottingham Health Science Biobank (NHSB), reference number 15/NW/0685. We can declare that this study is complying with Helsinki declaration.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Data availability
The authors confirm the data that has been used in this work is available on reasonable request.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41416-019-0589-0.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Hua S, Kittler R, White KP. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 2009;137:1259–1271. doi: 10.1016/j.cell.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ni M, Chen Y, Lim E, Wimberly H, Bailey ST, Imai Y, et al. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer Cell. 2011;20:119–131. doi: 10.1016/j.ccr.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muscat GE, Eriksson NA, Byth K, Loi S, Graham D, Jindal S, et al. Research resource: nuclear receptors as transcriptome: discriminant and prognostic value in breast cancer. Mol. Endocrinol. 2013;27:350–365. doi: 10.1210/me.2012-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doan TB, Graham JD, Clarke CL. Emerging functional roles of nuclear receptors in breast cancer. J. Mol. Endocrinol. 2017;58:R169–R190. doi: 10.1530/JME-16-0082. [DOI] [PubMed] [Google Scholar]
- 6.Abduljabbar R, Negm OH, Lai CF, Jerjees DA, Al-Kaabi M, Hamed MR, et al. Clinical and biological significance of glucocorticoid receptor (GR) expression in breast cancer. Breast Cancer Res. Treat. 2015;150:335–346. doi: 10.1007/s10549-015-3335-1. [DOI] [PubMed] [Google Scholar]
- 7.Goya L, Maiyar AC, Ge Y, Firestone GL. Glucocorticoids induce a G1/G0 cell cycle arrest of Con8 rat mammary tumor cells that is synchronously reversed by steroid withdrawal or addition of transforming growth factor-alpha. Mol. Endocrinol. 1993;7:1121–1132. doi: 10.1210/mend.7.9.8247014. [DOI] [PubMed] [Google Scholar]
- 8.Hilborn E, Gacic J, Fornander T, Nordenskjold B, Stal O, Jansson A. Androgen receptor expression predicts beneficial tamoxifen response in oestrogen receptor-alpha-negative breast cancer. Br. J. Cancer. 2016;114:248–255. doi: 10.1038/bjc.2015.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abedin SA, Thorne JL, Battaglia S, Maguire O, Hornung LB, Doherty AP, et al. Elevated NCOR1 disrupts a network of dietary-sensing nuclear receptors in bladder cancer cells. Carcinogenesis. 2009;30:449–456. doi: 10.1093/carcin/bgp005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Battaglia S, Maguire O, Thorne JL, Hornung LB, Doig CL, Liu S, et al. Elevated NCOR1 disrupts PPARalpha/gamma signaling in prostate cancer and forms a targetable epigenetic lesion. Carcinogenesis. 2010;31:1650–1660. doi: 10.1093/carcin/bgq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long MD, Campbell MJ. Pan-cancer analyses of the nuclear receptor superfamily. Nucl. Receptor Res. 2015;2:101182. doi: 10.11131/2015/101182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang XH, Gudas LJ. Retinoids, retinoic acid receptors, and cancer. Annu. Rev. Pathol. 2011;6:345–364. doi: 10.1146/annurev-pathol-011110-130303. [DOI] [PubMed] [Google Scholar]
- 13.Dawson MI, Xia Z. The retinoid X receptors and their ligands. Biochim. Biophys. Acta. 2012;1821:21–56. doi: 10.1016/j.bbalip.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nuclear Receptors Nomenclature C. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- 15.Conzen SD. Minireview: nuclear receptors and breast cancer. Mol. Endocrinol. 2008;22:2215–2228. doi: 10.1210/me.2007-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalra RS, Bapat SA. Expression proteomics predicts loss of RXR-gamma during progression of epithelial ovarian cancer. PLoS ONE. 2013;8:e70398. doi: 10.1371/journal.pone.0070398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SM, Lee JY, Choi JE, Lee SY, Park JY, Kim DS. Epigenetic inactivation of retinoid X receptor genes in non-small cell lung cancer and the relationship with clinicopathologic features. Cancer Genet. Cytogenet. 2010;197:39–45. doi: 10.1016/j.cancergencyto.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 18.Zanardi S, Serrano D, Argusti A, Barile M, Puntoni M, Decensi A. Clinical trials with retinoids for breast cancer chemoprevention. Endocr. Relat. Cancer. 2006;13:51–68. doi: 10.1677/erc.1.00938. [DOI] [PubMed] [Google Scholar]
- 19.Ross-Innes CS, Stark R, Holmes KA, Schmidt D, Spyrou C, Russell R, et al. Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes Dev. 2010;24:171–182. doi: 10.1101/gad.552910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kittler R, Zhou J, Hua S, Ma L, Liu Y, Pendleton E, et al. A comprehensive nuclear receptor network for breast cancer cells. Cell Rep. 2013;3:538–551. doi: 10.1016/j.celrep.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, et al. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int. J. Cancer. 2005;116:340–350. doi: 10.1002/ijc.21004. [DOI] [PubMed] [Google Scholar]
- 22.Aleskandarany MA, Green AR, Benhasouna AA, Barros FF, Neal K, Reis-Filho JS, et al. Prognostic value of proliferation assay in the luminal, HER2-positive, and triple-negative biologic classes of breast cancer. Breast Cancer Res. 2012;14:R3. doi: 10.1186/bcr3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blamey RW, Ellis IO, Pinder SE, Lee AH, Macmillan RD, Morgan DA, et al. Survival of invasive breast cancer according to the Nottingham Prognostic Index in cases diagnosed in 1990-1999. Eur. J. Cancer. 2007;43:1548–1555. doi: 10.1016/j.ejca.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 24.McCarty KS, Jr., KS McCarty., Sr. Histochemical approaches to steroid receptor analyses. Semin. Diagn. Pathol. 1984;1:297–308. [PubMed] [Google Scholar]
- 25.Aleskandarany MA, Rakha EA, Ahmed MA, Powe DG, Ellis IO, Green AR. Clinicopathologic and molecular significance of phospho-Akt expression in early invasive breast cancer. Breast Cancer Res. Treat. 2011;127:407–416. doi: 10.1007/s10549-010-1012-y. [DOI] [PubMed] [Google Scholar]
- 26.Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109:25–32. doi: 10.1002/cncr.22381. [DOI] [PubMed] [Google Scholar]
- 27.Abduljabbar R, Al-Kaabi MM, Negm OH, Jerjees D, Muftah AA, Mukherjee A, et al. Prognostic and biological significance of peroxisome proliferator-activated receptor-gamma in luminal breast cancer. Breast Cancer Res. Treat. 2015;150:511–522. doi: 10.1007/s10549-015-3348-9. [DOI] [PubMed] [Google Scholar]
- 28.Aleskandarany MA, Abduljabbar R, Ashankyty I, Elmouna A, Jerjees D, Ali S, et al. Prognostic significance of androgen receptor expression in invasive breast cancer: transcriptomic and protein expression analysis. Breast Cancer Res. Treat. 2016;159:215–227. doi: 10.1007/s10549-016-3934-5. [DOI] [PubMed] [Google Scholar]
- 29.Habashy HO, Powe DG, Staka CM, Rakha EA, Ball G, Green AR, et al. Transferrin receptor (CD71) is a marker of poor prognosis in breast cancer and can predict response to tamoxifen. Breast Cancer Res. Treat. 2010;119:283–293. doi: 10.1007/s10549-009-0345-x. [DOI] [PubMed] [Google Scholar]
- 30.Naderi A, Teschendorff AE, Beigel J, Cariati M, Ellis IO, Brenton JD, et al. BEX2 is overexpressed in a subset of primary breast cancers and mediates nerve growth factor/nuclear factor-kappaB inhibition of apoptosis in breast cancer cell lines. Cancer Res. 2007;67:6725–6736. doi: 10.1158/0008-5472.CAN-06-4394. [DOI] [PubMed] [Google Scholar]
- 31.Alshareeda AT, Soria D, Garibaldi JM, Rakha E, Nolan C, Ellis IO, et al. Characteristics of basal cytokeratin expression in breast cancer. Breast Cancer Res. Treat. 2013;139:23–37. doi: 10.1007/s10549-013-2518-x. [DOI] [PubMed] [Google Scholar]
- 32.Habashy HO, Powe DG, Rakha EA, Ball G, Paish C, Gee J, et al. Forkhead-box A1 (FOXA1) expression in breast cancer and its prognostic significance. Eur. J. Cancer. 2008;44:1541–1551. doi: 10.1016/j.ejca.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 33.Joseph C, Macnamara O, Craze M, Russell R, Provenzano E, Nolan CC, et al. Mediator complex (MED) 7: a biomarker associated with good prognosis in invasive breast cancer, especially ER+ luminal subtypes. Br. J. Cancer. 2018;118:1142–1151. doi: 10.1038/s41416-018-0041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rakha EA, Martin S, Lee AH, Morgan D, Pharoah PD, Hodi Z, et al. The prognostic significance of lymphovascular invasion in invasive breast carcinoma. Cancer. 2012;118:3670–3680. doi: 10.1002/cncr.26711. [DOI] [PubMed] [Google Scholar]
- 35.Jezequel P, Campone M, Gouraud W, Guerin-Charbonnel C, Leux C, Ricolleau G, et al. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res. Treat. 2012;131:765–775. doi: 10.1007/s10549-011-1457-7. [DOI] [PubMed] [Google Scholar]
- 36.Kurozumi S, Joseph C, Sonbul S, Gorringe KL, Pigera M, Aleskandarany MA, et al. Clinical and biological roles of Kelch-like family member 7 in breast cancer: a marker of poor prognosis. Breast Cancer Res. Treat. 2018;170:525–533. doi: 10.1007/s10549-018-4777-z. [DOI] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 39.Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, et al. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucl. Acids Res. 2012;40:W622–W627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucl. Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin. Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 42.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucl. Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aleskandarany MA, Agarwal D, Negm OH, Ball G, Elmouna A, Ashankyty I, et al. The prognostic significance of STAT3 in invasive breast cancer: analysis of protein and mRNA expressions in large cohorts. Breast Cancer Res. Treat. 2016;156:9–20. doi: 10.1007/s10549-016-3709-z. [DOI] [PubMed] [Google Scholar]
- 44.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hennigs A, Riedel F, Gondos A, Sinn P, Schirmacher P, Marme F, et al. Prognosis of breast cancer molecular subtypes in routine clinical care: a large prospective cohort study. BMC Cancer. 2016;16:734. doi: 10.1186/s12885-016-2766-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rakha EA, El-Sayed ME, Menon S, Green AR, Lee AH, Ellis IO. Histologic grading is an independent prognostic factor in invasive lobular carcinoma of the breast. Breast Cancer Res. Treat. 2008;111:121–127. doi: 10.1007/s10549-007-9768-4. [DOI] [PubMed] [Google Scholar]
- 48.Hosoda M, Yamamoto M, Nakano K, Hatanaka KC, Takakuwa E, Hatanaka Y, et al. Differential expression of progesterone receptor, FOXA1, GATA3, and p53 between pre- and postmenopausal women with estrogen receptor-positive breast cancer. Breast Cancer Res. Treat. 2014;144:249–261. doi: 10.1007/s10549-014-2867-0. [DOI] [PubMed] [Google Scholar]
- 49.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Safe S, Kim K. Non-classical genomic estrogen receptor (ER)/specificity protein and ER/activating protein-1 signaling pathways. J. Mol. Endocrinol. 2008;41:263–275. doi: 10.1677/JME-08-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Z, Yin P, Reierstad S, O’Halloran M, Coon VJ, Pearson EK, et al. Adenosine A1 receptor, a target and regulator of estrogen receptoralpha action, mediates the proliferative effects of estradiol in breast cancer. Oncogene. 2010;29:1114–1122. doi: 10.1038/onc.2009.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang C, Gao C, Zhuang JL, Ding C, Wang Y. A combined approach identifies three mRNAs that are down-regulated by microRNA-29b and promote invasion ability in the breast cancer cell line MCF-7. J. Cancer Res. Clin. Oncol. 2012;138:2127–2136. doi: 10.1007/s00432-012-1288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dumont AG, Dumont SN, Trent JC. The favorable impact of PIK3CA mutations on survival: an analysis of 2587 patients with breast cancer. Chin. J. Cancer. 2012;31:327–334. doi: 10.5732/cjc.012.10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gottardis MM, Bischoff ED, Shirley MA, Wagoner MA, Lamph WW, Heyman RA. Chemoprevention of mammary carcinoma by LGD1069 (Targretin): an RXR-selective ligand. Cancer Res. 1996;56:5566–5570. [PubMed] [Google Scholar]
- 55.Hulit J, Suyama K, Chung S, Keren R, Agiostratidou G, Shan W, et al. N-cadherin signaling potentiates mammary tumor metastasis via enhanced extracellular signal-regulated kinase activation. Cancer Res. 2007;67:3106–3116. doi: 10.1158/0008-5472.CAN-06-3401. [DOI] [PubMed] [Google Scholar]
- 56.Klopper JP, Hays WR, Sharma V, Baumbusch MA, Hershman JM, Haugen BR. Retinoid X receptor-gamma and peroxisome proliferator-activated receptor-gamma expression predicts thyroid carcinoma cell response to retinoid and thiazolidinedione treatment. Mol. Cancer Ther. 2004;3:1011–1020. [PubMed] [Google Scholar]
- 57.Shulman AI, Mangelsdorf DJ. Retinoid x receptor heterodimers in the metabolic syndrome. N. Engl. J. Med. 2005;353:604–615. doi: 10.1056/NEJMra043590. [DOI] [PubMed] [Google Scholar]
- 58.Elstner E, Williamson EA, Zang C, Fritz J, Heber D, Fenner M, et al. Novel therapeutic approach: ligands for PPARgamma and retinoid receptors induce apoptosis in bcl-2-positive human breast cancer cells. Breast Cancer Res. Treat. 2002;74:155–165. doi: 10.1023/a:1016114026769. [DOI] [PubMed] [Google Scholar]
- 59.Bonofiglio D, Cione E, Qi H, Pingitore A, Perri M, Catalano S, et al. Combined low doses of PPARgamma and RXR ligands trigger an intrinsic apoptotic pathway in human breast cancer cells. Am. J. Pathol. 2009;175:1270–1280. doi: 10.2353/ajpath.2009.081078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heublein S, Mayr D, Meindl A, Kircher A, Jeschke U, Ditsch N. Vitamin D receptor, Retinoid X receptor and peroxisome proliferator-activated receptor gamma are overexpressed in BRCA1 mutated breast cancer and predict prognosis. J. Exp. Clin. Cancer Res. 2017;36:57. doi: 10.1186/s13046-017-0517-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Obara W, Konda R, Akasaka S, Nakamura S, Sugawara A, Fujioka T. Prognostic significance of vitamin D receptor and retinoid X receptor expression in renal cell carcinoma. J. Urol. 2007;178:1497–1503. doi: 10.1016/j.juro.2007.05.119. [DOI] [PubMed] [Google Scholar]
- 62.Uray IP, Dmitrovsky E, Brown PH. Retinoids and rexinoids in cancer prevention: from laboratory to clinic. Semin. Oncol. 2016;43:49–64. doi: 10.1053/j.seminoncol.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm the data that has been used in this work is available on reasonable request.