Abstract
Objective
Phosphatidylethanolamine methyltransferase (PEMT) generates phosphatidylcholine (PC), the most abundant phospholipid in the mitochondria and an important acyl chain donor for cardiolipin (CL) biosynthesis. Mice lacking PEMT (PEMTKO) are cold-intolerant when fed a high-fat diet (HFD) due to unclear mechanisms. The purpose of this study was to determine whether PEMT-derived phospholipids are important for the function of uncoupling protein 1 (UCP1) and thus for maintenance of core temperature.
Methods
To test whether PEMT-derived phospholipids are important for UCP1 function, we examined cold-tolerance and brown adipose (BAT) mitochondria from PEMTKO mice with or without HFD feeding. We complemented these studies with experiments on mice lacking functional CL due to tafazzin knockdown (TAZKD). We generated several conditional mouse models to study the tissue-specific roles of PEMT, including mice with BAT-specific knockout of PEMT (PEMT-BKO).
Results
Chow- and HFD-fed PEMTKO mice completely lacked UCP1 protein in BAT, despite a lack of difference in mRNA levels, and the mice were accordingly cold-intolerant. While HFD-fed PEMTKO mice exhibited reduced mitochondrial CL content, this was not observed in chow-fed PEMTKO mice or TAZKD mice, indicating that the lack of UCP1 was not attributable to CL deficiency. Surprisingly, the PEMT-BKO mice exhibited normal UCP1 protein levels. Knockout of PEMT in the adipose tissue (PEMT-AKO), liver (PEMT-LKO), or skeletal muscle (PEMT-MKO) also did not affect UCP1 protein levels, suggesting that lack of PEMT in other non-UCP1-expressing cells communicates to BAT to suppress UCP1. Instead, we identified an untranslated UCP1 splice variant that was triggered during the perinatal period in the PEMTKO mice.
Conclusions
PEMT is required for UCP1 splicing that yields functional protein. This effect is derived by PEMT in nonadipocytes that communicates to BAT during embryonic development. Future research will focus on identifying the non-cell-autonomous PEMT-dependent mechanism of UCP1 splicing.
Keywords: Brown adipose tissue, UCP1, PEMT, Phosphatidylcholine, Cardiolipin, Alternative splicing
Highlights
-
•
Whole-body ablation of PEMT results in a complete loss of UCP1 protein.
-
•
While PEMT-derived PC may participate in CL biosynthesis, changes in mitochondrial PC or CL do not explain the loss of UCP1 protein.
-
•
PEMT regulates brown adipose UCP1 in a non-cell-autonomous manner.
-
•
PEMT deletion promotes the use of an alternative 3’ splice junction in exon 5 of UCP1, preventing UCP1 protein expression.
Abbreviations
- ADP
adenosine diphosphate
- ATP
adenosine triphosphate
- BAT
brown adipose tissue
- CI
complex I
- CII
complex II
- CIII
complex III
- CIV
complex IV
- CL
cardiolipin
- CS
citrate synthase
- CV
complex V
- EM
electron microscopy
- ER
endoplasmic reticulum
- ETS
electron transport system
- GDP
guanosine diphosphate
- HFD
high-fat diet
- IL-33
interleukin-33
- IMM
inner mitochondrial membrane
- MAM
mitochondrial-associated membrane
- MIM
mitochondrial isolation medium
- NADP
nicotinamide adenine dinucleotide phosphate
- NADPH
nicotinamide adenine dinucleotide phosphate reduced
- Nt
nucleotide
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PEMT
phosphatidylethanolamine methyltransferase
- PEMT +/−
PEMT heterozygous knockout
- PEMT-AKO
PEMT adipose-specific knockout
- PEMT-BKO
PEMT brown adipose-specific knockout
- PEMTcKO
PEMT conditional knockout
- PEMTKO
PEMT knockout
- PEMT-LKO
PEMT liver-specific knockout
- PEMT-MKO
PEMT muscle-specific knockout
- ST2
interleukin 1 receptor-like 1
- TAZ
tafazzin
- TAZKD
tafazzin knockdown
- TLC
thin-layer chromatography
- UCP1
uncoupling protein 1
- WT
wild-type
1. Introduction
Brown adipose tissue (BAT) generates heat as a result of the inefficient coupling of mitochondrial oxidative phosphorylation [1]. Mitochondrial thermogenesis in adipose tissue is largely driven by uncoupling protein 1 (UCP1), which resides in the inner mitochondrial membrane (IMM) [2]. UCP1 dissipates the proton gradient across the IMM, independent of complex V, thereby uncoupling O2 consumption from adenosine triphosphate (ATP) synthesis [[3], [4], [5], [6]]. UCP1-dependent thermogenesis represents a potential mechanism to increase energy expenditure and protect from obesity through the futile cycling of substrates derived from carbohydrates and lipids [[7], [8], [9]]. While much is known about the transcriptional control of UCP1, little is known about how the IMM lipid composition affects the activity of UCP1 to facilitate uncoupled respiration.
Phosphatidylcholine (PC) is the most abundant phospholipid in the mitochondria and is generated in part by phosphatidylethanolamine methyltransferase (PEMT) [[10], [11], [12]]. PEMT resides in the endoplasmic reticulum and mitochondrial-associated membranes (MAM) and synthesizes PC by tri-methylation of phosphatidylethanolamine (PE) [13,14]. Mitochondria do not possess an enzyme for the autonomous synthesis of PC and rely on PC import through the MAM [15]. Additionally, PC is an important acyl-chain donor for cardiolipin (CL), a phospholipid unique to mitochondrial membranes that is required for thermogenic respiration [[16], [17], [18]]. This study was driven by a previous observation that mice with a whole-body deletion of PEMT (PEMTKO) become cold-intolerant when fed a high-fat diet (HFD) [19], which was attributed to insufficient glucose supply. Based on findings that PEMT provides mitochondrial PC in the liver [12], we investigated the possibility that PEMT intrinsically alters cellular bioenergetics and UCP1-dependent thermogenesis by altering mitochondrial phospholipids.
2. Materials and methods
2.1. Animals
All mice used in this study were of the C57BL/6 J background. The PEMTKO mice were a generous gift from Dr. Dennis Vance at the University of Alberta [20]. We generated PEMT conditional knockout mice (PEMTcKO, with exon 3 of the Pemt gene flanked with loxP sites) [21] that were crossed to UCP1-Cre mice (Jackson Laboratory, stock #: 024670), albumin-Cre mice (Jackson Laboratory, stock #: 003574), HSA-MerCreMer mice (a gift from Dr. Karyn Esser, University of Florida), or adiponectin-Cre mice (Jackson Laboratory, stock #: 028020) to obtain tissue-specific knockout mice. The tafazzin knockdown (TAZKD) mice were obtained from Jackson Laboratory (stock #: 014648). The mice were either fed a standard chow diet (Teklad 2020X) or a 42% HFD (Teklad 88137). At 2–4 months of age, the PEMT-deficient mice were studied for a chow-fed condition or placed on a HFD for 10 weeks. The TAZKD mice were fed a 625 mg/kg doxycycline chow diet (Teklad 09628) to induce TAZ knockdown as previously described [ 22,23]. The TAZKD mice were given doxycycline containing chow at 2 months of age for 4 months. No sex-dependent differences were observed in the experimental mice used in this study. All the mice were fasted for 4 h prior to euthanasia and tissue collection. Unless otherwise noted, the data presented are from mice housed at an ambient temperature of 22 °C. All the animal experiments were performed with the approval of the Institutional Animal Care and Use Committee at East Carolina University and the University of Utah.
2.2. Cell culture
SV40T preadipocytes were a gift from Dr. Kai Ge from the NIDDK. SV40T preadipocytes were differentiated to brown adipocytes as previously described [24]. Briefly, preadipocytes were grown to confluency in growth media (10% fetal bovine serum and high-glucose Dulbecco's modified Eagle medium containing glutamine). Induction media (growth media with 20 nM insulin, 1 nM T3, 0.5 mM 3-isobutyl-1-methl-xanthine, 2 μg/ml dexamethasone, and 0.125 mM indomethacin) was added to confluent cells for 48 h and then replaced with differentiation media (growth media with 20 nM insulin and 1 nM T3). Differentiation media were refreshed every 48 h for 6 days. The lentivirus system was used to infect the preadipocytes with plasmids coding for shRNAs against PEMT and TAZ. Infected preadipocytes were then differentiated to brown adipocytes after puromycin selection to ensure the death of noninfected cells.
2.3. Metabolic phenotyping
Body composition was measured using a Bruker MiniSpec NMR. Whole-body VO2, RER, and activity levels were measured using a CLAMS system (Columbus Instruments). Cold-tolerance testing was carried out in a 4 °C cold room. Prior to cold-tolerance testing, the mice were injected with a temperature-sensitive transponder (Bio Medic Data Systems, IPTT 300). One week after the injections, the mice were transferred to a 4 °C cold room for 6–8 h, and their core temperature was assessed using a Reader-Programmer (Bio Medic Data Systems, DAS 8007). The mice were single-housed in cages containing bedding with access to food and water throughout the cold-tolerance test. For long-term cold-exposure experiments, the mice were acclimated to the cold following a protocol used to acclimate UCP1 null mice to the cold [25]. Briefly, the mice were housed in a rodent incubator at 18°C for 2 weeks, after which the temperature was lowered to 6.5°C for 7 days. The mice were euthanized after a 4 h fast and their tissues were harvested. For glucose-tolerance testing, their blood glucose levels were measured following an IP injection of 20% glucose (5 μL/g body mass) at time points 0, 15, 30, 60, and 120 min.
2.4. Mitochondrial assays
BAT tissue was finely minced in mitochondrial isolation medium (MIM, 300 mM sucrose, 10 mM HEPES, and 1 mM EGTA) and homogenized in MIM with 1 mg/ml BSA. Homogenates were centrifuged at 10,000×g, and the supernatant was discarded. The pellet was resolubilized in MIM and spun at 200×g, and the supernatant was transferred to a new tube leaving behind the pellet. The supernatant was then centrifuged again at 200×g and transferred to a new tube. The supernatant was then centrifuged at 10,000×g and discarded. The mitochondrial pellet was resolubilized in MIM and used for the mitochondrial assays. Mitochondrial O2 consumption was measured using Oroboros oxygraphs https://advances.sciencemag.org/content/5/9/eaax8352. UCP1-dependent respiration was stimulated with 0.5 mM malate and 5 mM pyruvate. UCP1 was then inhibited using 4 mM guanosine diphosphate (GDP) [26]. ATP production was measured using a FluoroMax-4 (Horiba Scientific). ATP production was coupled enzymatically to reduced nicotinamide adenine dinucleotide phosphate (NADPH) production as previously described [27]. Briefly, ATP synthesis was stimulated using 0.5 mM malate, 5 mM pyruvate, 5 mM glutamate, and 5 mM succinate in the presence of 20 μM, 200 μM, or 2000 μM adenosine diphosphate (ADP). NADPH fluorescence was measured every 2 s (excitation 340 and emission 460). The ATP/O ratio was determined by dividing ATP production by O2 consumption using the same conditions for ATP production.
2.5. Mass spectrometry and thin-layer chromatography (TLC)
Lipids were extracted from mitochondria using a modified Matyash lipid extraction [28]. Briefly, a mixture of cold methyl-tert-butyl ether, methanol, and internal standards (SPLASH Mix Avanti Polar Lipids 330707 and Cardiolipin Mix I Avanti Polar Lipids LM6003) was added to 150 μg of mitochondrial protein. The samples were vortexed and then sonicated for 1 min. The samples were then incubated on ice for 15 min and vortexed every 5 min. Phase separation was induced by adding 300 μL of H2O and centrifuging at 15,000×g, and the organic phase was then dried using a SpeedVac. The dried lipids were reconstituted in a 9:1 methanol:toluene mixture. Liquid chromatography-mass spectrometry (LC-MS) was then performed on the reconstituted lipids using an Agilent 6490 UPLC-QQQ mass spectrometer (PC species) and an Agilent 6530 UPLC-QTOF mass spectrometer (CL species). For TLC, the lipids were extracted from isolated mitochondria using a Bligh-Dyer extraction. TLC plates were developed with a chloroform:glacial acetic acid:methanol:water mobile phase (85:25:5:2, all values in mL). The plates were then dried and sprayed with a charring reagent (4% phosphoric acid and 5% copper sulfate) and heated at 190 °C for approximately 15 min. The intensity of the lipid spots was measured using an Odyssey Infrared Imager.
2.6. Electron microscopy (EM)
BAT was minced in an ice-cold fixative (2.5% glutaraldehyde and 1% paraformaldehyde) and subsequently incubated in fixative for 48 h at 4 °C. After fixation, the minced tissues were washed 3 × 10 min in cold 0.1 M phosphate buffer. The tissues were then post-fixed in 1% osmium tetroxide in 0.1 M phosphate buffer at 4 °C for 1 h, after which the tissues were washed 3 × 10 min in 0.1 M phosphate buffer. The tissues were then dehydrated in 25%, 50%, 75%, and 100% EtOH and embedded in Spurr's resin. The tissues were then sectioned and stained with uranyl acetate and imaged using a JEOL 1200EX transmission electron microscope with a Soft Imaging Systems MegaView III CCD camera.
2.7. Histology
A small piece of BAT was fixed in 4% paraformaldehyde in PBS for 48 h and subsequently placed in 70% ETOH for 48 h. The tissue was then paraffin embedded, cut into 5 μm sections, and stained with hematoxylin and eosin. The sectioned tissues were imaged using an Axio Scan.Z1 (Zeiss).
2.8. Western blotting
Frozen BAT was homogenized in lysis buffer and centrifuged at 12,000×g. The supernatant was taken, and the protein concentration was quantified using the Pierce BCA protein assay kit. Equal amounts of protein were mixed with Laemmli sample buffer and loaded onto a gradient gel (Bio-Rad). The proteins were then transferred onto nitrocellulose membranes and blocked with 5% BSA in TBST. The membranes were treated with primary antibodies overnight (Table S1). The membranes were then washed 5 times with TBST and incubated in secondary antibody with 5% milk. The membranes were then again washed 5 times in TBST and 2 times in TBS before being treated with ECL (PerkinElmer) and imaged with a FluorChem E imager (ProteinSimple).
2.9. Quantitative PCR and DNA sequencing
BAT was homogenized in 1 ml of TRIzol, treated with 200 μL of chloroform, and mixed by several inversions. The mixture was then centrifuged at 12,000×g, and the aqueous layer was transferred to a new Eppendorf tube containing 500 μL of 100% isopropanol and centrifuged at 12,000×g. The resulting pellet was washed with 80% ETOH, which was then aspirated. The RNA pellet was then solubilized in Tris–EDTA buffer. The RNA was then reverse transcribed to the cDNA using the iScript cDNA Synthesis Kit (Bio-Rad). cDNA was added to a mixture of SYBR Green (Thermo Fisher Scientific) and primers (Table S2). The sample mixtures were pipetted onto a 384 plate and analyzed with QuantStudio 12K Flex (Life Technologies). For DNA sequencing, the RNA was reverse transcribed to the cDNA, and UCP1 cDNA was amplified via PCR. The resulting PCR product was purified using ExoSAP-IT reagent according to the manufacturer's instructions (Thermo Fisher Scientific). The sequence of the PCR product was determined using Sanger sequencing by the University of Utah DNA Sequencing Core.
2.10. Statistical analyses
All data presented herein are expressed as mean ± SEM. The level of significance was set at p < 0.05. Student's t-tests were used to determine the significance between the experimental groups and two-way ANOVAs were used where appropriate. The sample size (n) for each experiment is shown in the figure legends and corresponds to the sample derived from the individual mice or for cell culture experiments on an individual batch of cells. Statistical analyses were performed using GraphPad Prism software.
3. Results
3.1. PEMTKO mice lack UCP1 protein in BAT
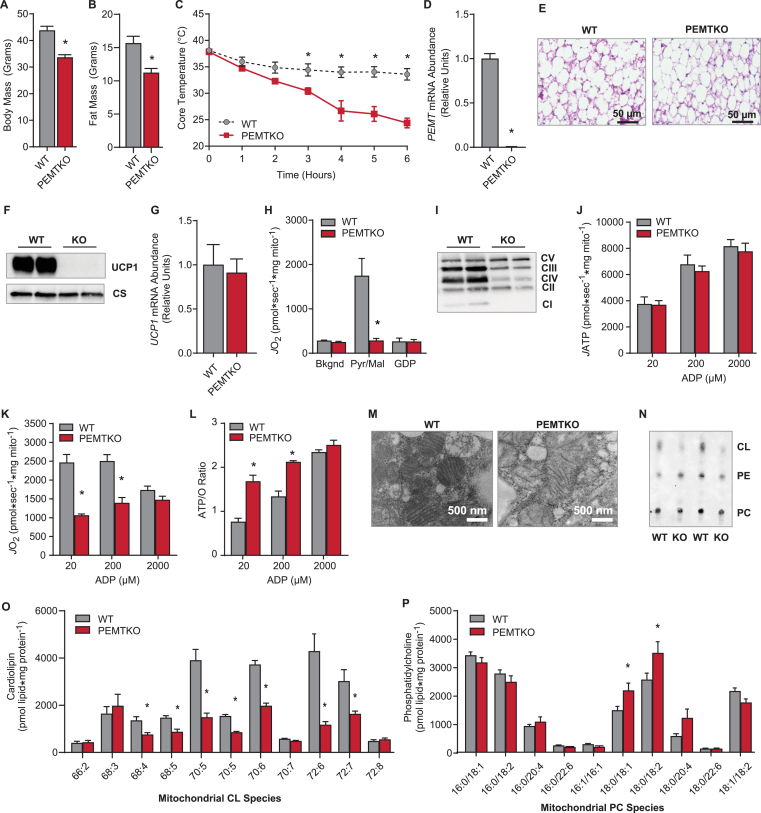
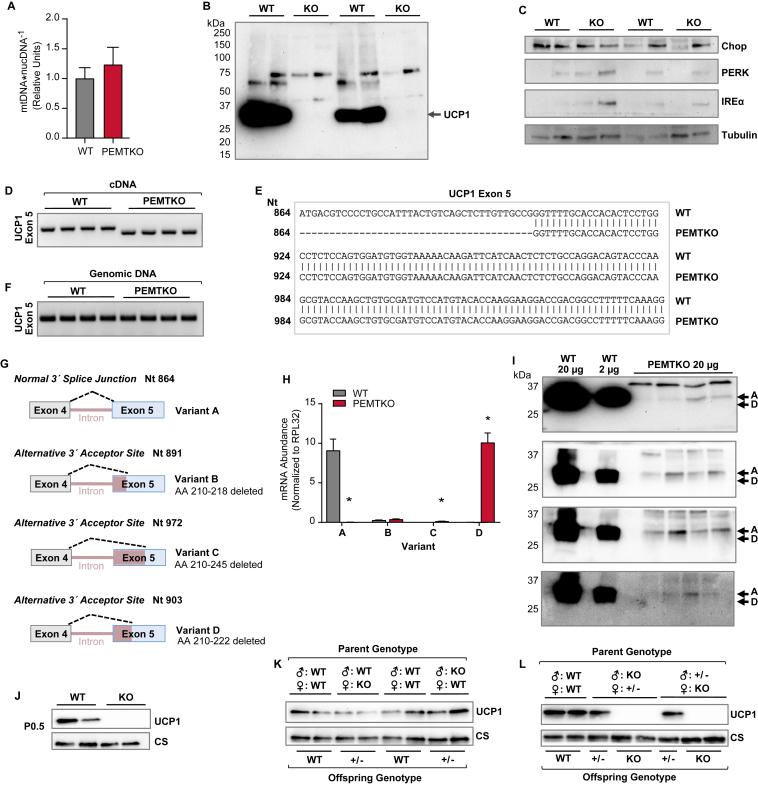
Whole-body deletion of PEMT results in diet-induced cold-intolerance [19], despite reports of increased metabolic rate [21 ,29]. Consistent with previous reports, following 10 weeks of high-fat feeding, the PEMTKO mice had lower body mass and fat mass than the wild-type controls (WT) (Figure 1A, B) [21,29]. Nevertheless, the PEMTKO mice were more prone to hypothermia during an acute cold challenge (Figure 1C). The absence of PEMT in BAT was confirmed by qPCR (Figure 1D). BAT from the PEMTKO mice was similarly engorged with lipids compared with the WT mice despite the differences in body and fat mass (Figure 1E). Strikingly, Western blotting of BAT revealed a complete loss of UCP1 protein in the PEMTKO mice (Figure 1F), which would promote cold-intolerance [30]. Remarkably, the loss of UCP1 protein occurred despite the apparently normal UCP1 mRNA levels (Figure 1G), suggesting mechanisms independent of transcription. In agreement with the loss of UCP1 protein, mitochondria from PEMTKO BAT completely lacked UCP1-dependent respiration (Figure 1H). Lack of PEMT also promoted substantial decreases in complexes I, III, and IV of the electron transport system (ETS), an increase in complex V, and no change in complex II (Figure 1I), very similar to what is found in UCP1 knockout mice [31,32]. The altered levels of ETS complexes did not affect ATP production (Figure 1J); however, ATP was produced at a much lower O2 cost (Figure 1K), resulting in the highly efficient coupling of oxidative phosphorylation (Figure 1L).
Figure 1.
PEMTKO mice fed HFD lack UCP1 protein concomitant with decreased CL. (A) Body mass in the WT or PEMTKO mice fed HFD for 10 weeks, n = 8. (B) Fat mass, n = 8. (C) Cold-tolerance test in the PEMTKO mice. The mice were single-housed and placed in a temperature-controlled room at 4 °C for 6 h, n = 3–5. (D) mRNA levels of PEMT in BAT, n = 7–8. (E) Images of BAT sections stained with hematoxylin and eosin. (F) Protein levels of UCP1 and citrate synthase (CS). (G) mRNA levels of UCP1, n = 4. (H) UCP1-dependent respiration in mitochondria isolated from BAT. Respiration was stimulated with 0.2 mM malate and 5 mM pyruvate. UCP1 was inhibited using 4 mM GDP, n = 6. (I) Protein levels of ETS complexes in whole tissue lysates. (J) ATP production in isolated BAT mitochondria. ATP production was measured in the presence of 0.2 mM malate, 5 mM glutamate, 5 mM pyruvate, and 5 mM succinate at increasing concentrations of ADP, n = 3–4. (K) O2 consumption using the same conditions described for ATP production, n = 3–4. (L) ATP/O ratio calculated from the ATP produced per oxygen consumed, n = 3–4. (M) EM images of BAT mitochondria. (N) Detection of phospholipids extracted from the BAT mitochondria by TLC. (O) Analysis of CL species via LC-MS, n = 9. (P) Analysis of PC species via LC-MS, n = 4–5. Data are expressed as mean ± SEM, *p < 0.05.
3.2. The role of CL in BAT UCP1 deficiency
Mitochondria from PEMTKO BAT had abnormal morphology and lacked distinct cristae (Figure 1M), suggesting alterations of the lipid membranes. Analysis of mitochondrial phospholipids revealed a substantial decrease in mitochondrial CL in PEMTKO BAT without a change in mitochondrial PC (Figure 1N–P). PCs are the preferred acyl-chain donors for CL remodeling via the enzyme tafazzin (TAZ) [16,18], the final step of CL biosynthesis. Recent research has demonstrated that CL is necessary for both the stability and function of UCP1 [17,33]. We therefore hypothesized that the loss of CL was responsible for the lack of UCP1 protein in the PEMTKO mice.
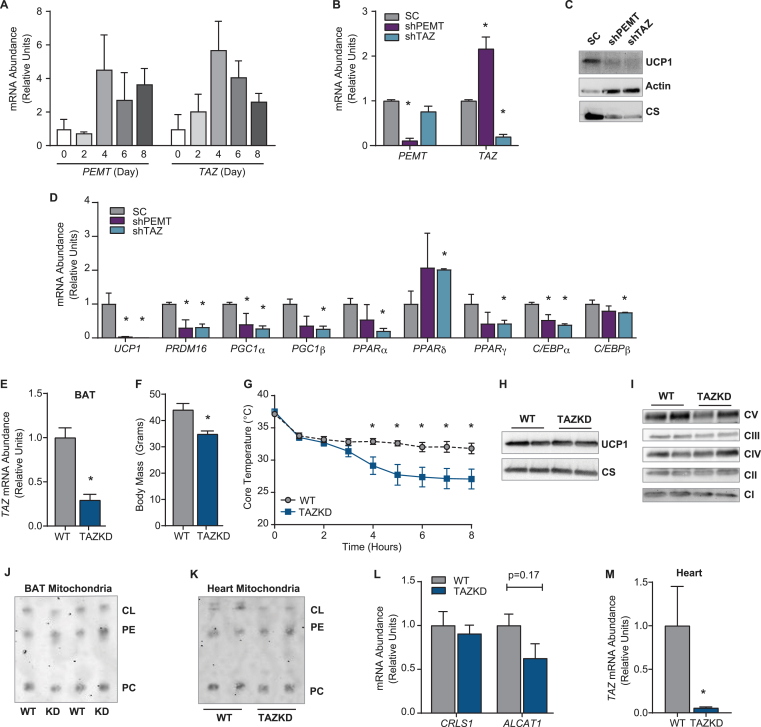
To determine whether the loss of TAZ function would phenocopy PEMT deletion, we used immortalized brown adipocytes [24]. Both PEMT and TAZ mRNA were induced during differentiation (Figure 2A), and silencing PEMT or TAZ dramatically decreased UCP1 protein (Figure 2B, C). However, this was likely a secondary consequence due to impaired brown adipogenesis, as the mRNA levels of UCP1 and genes involved in adipogenesis were substantially decreased (Figure 2D). This was different from our findings in vivo, as BAT from the PEMTKO mice exhibited normal UCP1 mRNA levels (Figure 1G). While these in vitro data are informative, we further investigated the effect of CL on UCP1 protein in fully differentiated BAT in vivo. Doxycycline-inducible TAZKD mice have been used extensively to study the role of TAZ in heart tissue, and we used this mouse model to examine the function of TAZ in BAT. Upon doxycycline treatment, the TAZKD mice express an shRNA against TAZ to silence its expression presumably in all tissues. Doxycycline treatment reduced the TAZ mRNA levels by 71% in BAT (Figure 2E). As previously reported, WT mice gained more weight on doxycycline chow than TAZKD littermates (Figure 2F) [ 22,23,34]. Similar to the PEMTKO mice, the TAZKD mice exhibited decreased tolerance to cold exposure (Figure 2G). However, BAT from the TAZKD mice had normal UCP1 and ETS protein or activity compared to the WT controls (Figure 2H&I, S1A-D), suggesting that different mechanisms drive cold sensitivity in TAZKD mice. These observations may be interpreted to mean that CL is dispensable for UCP1 protein. However, the BAT mitochondrial CL content in was not lower in the TAZKD mice compared to the WT mice (Figure 2J), suggesting that the TAZKD mouse model is an imperfect model for CL deficiency in BAT. This was somewhat surprising due to a robust decrease in CL in heart mitochondria from these mice (Figure 2K). A previous study showed that lack of TAZ in the liver triggers activation of compensatory CL acylation pathways, but this was not the case in BAT [34] (Figure 2L). Rather, the lack of changes in BAT CL may be attributable to an imperfect knockdown of TAZ in BAT compared to heart (Figure 2M), despite our use of high-dose doxycycline (625 mg/kg), which has precedence for effectively inducing doxycycline-dependent expression [22 ,35].
Figure 2.
TAZKD mice exhibit normal UCP1 and CL levels in BAT. (A) mRNA levels of PEMT and TAZ during differentiation of preadipocytes to brown adipocytes, n = 2. (B) Knockdown confirmation of PEMT and TAZ mRNA following silencing using the lentivirus system. shRNA against PEMT (shPEMT) or TAZ (shTAZ) administered to preadipocytes via infection with viral media for 48 h. Control cells received a scrambled virus (SC). After infection, preadipocytes were differentiated, n = 2. (C) Protein levels of UCP1, actin, and CS. (D) mRNA levels of UCP1 and other genes involved in adipogenesis, n = 2. (E) TAZ mRNA abundance in BAT, n = 4. (F) Body mass, n = 5–6. (G) Cold-tolerance test, n = 10. (H) Protein levels of UCP1 and CS. (I) Protein abundance of ETS complexes. (J) Mitochondrial phospholipids in BAT measured by TLC. (K) Mitochondrial phospholipids isolated from heart mitochondria. (L) mRNA levels of cardiolipin synthase (CRLS1) and acyl-CoA:lysocardiolipin acyltransferase-1 (ALCAT1), n = 3–5. (M) TAZ mRNA levels in heart, n = 4. Data are expressed as mean ± SEM, *p < 0.05.
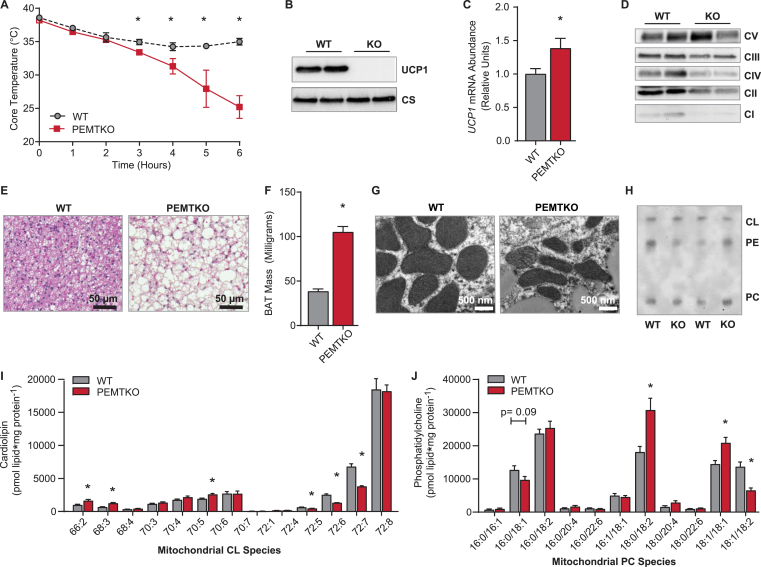
To gain further insights into the relationship between mitochondrial CL and UCP1 protein, we studied PEMTKO mice fed a standard chow diet, which have previously been reported have normal cold tolerance [19]. We reasoned that if the loss of mitochondrial CL was promoting the loss of UCP1 and cold-intolerance, then the chow-fed PEMTKO mice that were not cold-sensitive may have undisturbed mitochondrial CL levels. To our surprise, these mice also exhibited cold-intolerance and loss of UCP1 protein in BAT, despite elevated UCP1 mRNA (Figure 3A–C). The PEMTKO mice fed a chow diet also had decreased levels of ETS complexes I, II, III, and IV, but not V (Figure 3D), essentially mirroring the mitochondrial phenotype observed in the HFD-fed animals (Figs. S1E–H). BAT from the WT mice was devoid of large lipid droplets, but BAT from the PEMTKO mice was engorged with more and larger lipid droplets (Figure 3E, F), consistent with the loss of UCP1 and reduced thermogenic capacity. From these observations, we expected that the BAT mitochondria from the standard chow diet-fed PEMTKO mice would appear similarly abnormal as in the HFD-fed condition. However, the mitochondrial morphology demonstrated by EM revealed strikingly normal-appearing cristae (Figure 3G). Surprisingly, the BAT mitochondrial CL and PC content was largely unchanged between the WT and PEMTKO mice (Figure 3H–J). Together, these observations indicate that a lack of CL is not the mechanism that mediates the loss of UCP1 protein in BAT from the PEMTKO mice.
Figure 3.
PEMTKO mice fed a standard chow diet lack UCP1 despite normal CL levels. (A) Cold-tolerance in the PEMTKO mice fed a standard chow diet, n = 3–5. (B) Protein levels of UCP1 and CS. (C) UCP1 mRNA levels, n = 4. (D) Protein levels of ETS complexes in whole tissue lysates. (E) Images of BAT sections stained with hematoxylin and eosin from the PEMTKO mice fed a chow diet. (F) Wet weight of BAT, n = 4. (G) EM images of BAT mitochondria. (H) TLC analysis of BAT mitochondrial phospholipids. (I) Levels of CL species measured by LC-MS, n = 8. (J) Mitochondrial PC species by measured by LC-MS, n = 8. Data are expressed as mean ± SEM, *p < 0.05.
3.3. UCP1-cre-dependent deletion of PEMT does not ablate UCP1
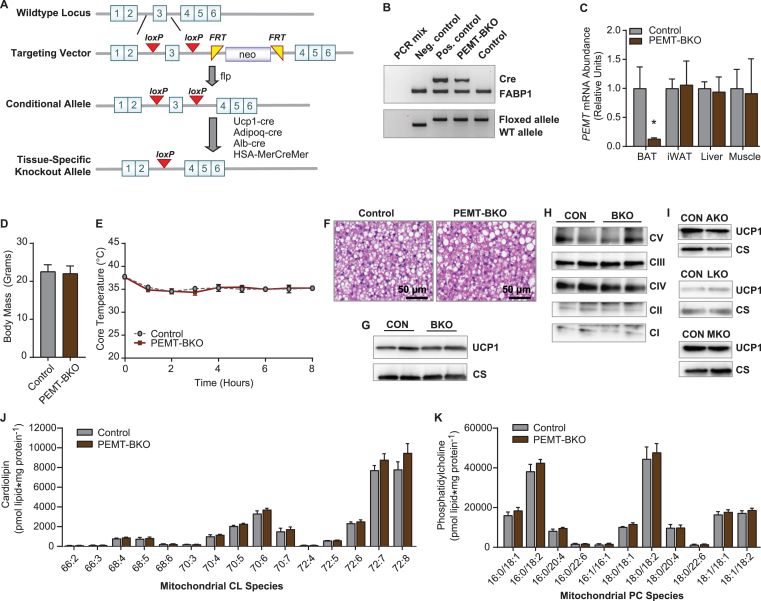
We also studied the tissue-specific effect of PEMT on UCP1 protein in BAT. This was accomplished by crossing our newly generated PEMTcKO mice [21] with UCP1-Cre mice (PEMT-BKO) (Figure 4A, B). This strategy successfully yielded mice with PEMT deficient BAT, but not white adipose tissue, liver, or muscle (Figure 4C). The PEMT-BKO mice were not different from the loxP flanked controls in terms of body weight (Figure 4D). We first studied these mice with standard chow feeding, as the effect of whole-body PEMT knockout on the loss of BAT UCP1 protein was present under this condition. Surprisingly, the PEMT-BKO mice maintained core temperature in response to cold exposure (Figure 4E) and displayed normal gross morphology (Figure 4F), unlike the PEMTKO mice. Furthermore, BAT from the PEMT-BKO mice had unchanged protein levels of UCP1 and ETS complexes (Figure 4G, H). These data provide striking evidence that the effect of PEMT on regulating UCP1 protein does not occur in a cell-autonomous manner. We also crossed the PEMTcKO mice with adiponectin-Cre mice, albumin-Cre mice, and HSA-MerCreMer mice, but the loss of PEMT in these tissue-specific models did not promote the loss of UCP1 protein in BAT (Figure 4I).
Figure 4.
BAT-specific deletion of PEMT does not cause loss of UCP1. (A) Schematic of the targeting vector used to generate the PEMTcKO mice. (B) Genotyping PCR identifying the PEMT-BKO mice and loxP flanked controls. (C) PEMT mRNA levels in BAT, inguinal white adipose (iWAT), liver, and muscle, n = 3–4. (D) Body mass, n = 14–21. (E) Cold-tolerance test in the PEMT-BKO mice, n = 3. (F) Histology images of BAT stained with hematoxylin and eosin. (G) Protein levels of UCP1 and CS. (H) Protein levels of ETS complexes. (I) UCP1 and CS protein levels in adipose-specific PEMT knockouts (PEMT-AKO), liver-specific knockouts (PEMT-LKO), and skeletal muscle-specific knockouts (PEMT-MKO). (J) CL species in mitochondria isolated from BAT from the PEMT-BKO mice, n = 4–5. (K) PC species in isolated mitochondria from BAT from the PEMT-BKO mice, n = 4–5. Data are expressed as mean ± SEM, *p < 0.05.
The PEMT-BKO mice were studied in greater detail. The BAT mitochondrial PC or CL content was not different between the control and PEMT-BKO mice (Figure 4J, K). These findings are analogous to those in the whole-body PEMTKO mice, suggesting that PEMT in BAT does not contribute to mitochondrial phospholipids under standard chow diet-fed conditions. We then placed the control and PEMT-BKO mice on a HFD for 10 weeks to determine whether the lack of PEMT compromised BAT's role in adaptations to obesity. However, the PEMT-BKO mice gained weight similarly to the control mice and were metabolically similar in all aspects except for a small but significant decrease in their core temperature during cold exposure (Fig. S2A–P). Thus, PEMT plays only a marginal role in BAT homeostasis. Similar to the HFD-fed whole-body PEMTKO mice, mitochondrial CL was lower in the HFD-fed PEMT-BKO mice compared to the HFD-fed control mice (Fig. S2Q), suggesting that PEMT contributes to BAT mitochondrial CL under HFD-fed (but not standard chow diet-fed) conditions. Similar to the HFD-fed whole-body PEMTKO mice, the mitochondrial PC content was not different between the HFD-fed control and PEMT-BKO mice (Fig. S2R). Overall, the cell-autonomous effect of PEMT on BAT was marginal. Rather, PEMT in another cell type regulates BAT UCP1 substantially. To explore these mechanisms, we returned to the PEMTKO mouse model.
3.4. PEMT-dependent alternative splicing of UCP1
What is the mechanism by which UCP1 protein is absent in BAT from the PEMTKO mice? The mitochondrial content of BAT was largely unchanged (Figure 5A), and together with the lack of changes in the mitochondrial matrix protein citrate synthase (Figure 1F), it is highly unlikely that the loss of PEMT targets entire populations of mitochondria for lysosomal degradation. UCP1 protein may be ubiquitinated and targeted for proteolysis [36], but UCP1 showed no sign of ubiquitin conjugation (Figure 5B). A lack of PEMT is known to induce ER stress in the liver [37], which, if true in BAT, could impair the translation of UCP1. However, we found no evidence of ER stress in BAT from the PEMTKO mice (Figure 5C). Odegaard et al. recently demonstrated that UCP1 mRNA contains alternative 3′ acceptor sites in exon 5, which results in a loss of UCP1 protein [32]. Similar to the present study, the loss of UCP1 protein observed by Odegaard et al. also appears to occur in a non-cell-autonomous fashion. To test this possibility, we reverse transcribed UCP1 mRNA from BAT and PCR-amplified a region that includes exon 5 splice sites. The size of the PCR products was lower in the transcripts from the PEMTKO mice compared to the WT mice, suggesting potential splicing (Figure 5D). Sanger sequencing of BAT UCP1 cDNA revealed a 39 nucleotide (Nt) excision (Nt 864–903) at the 5′ end of exon 5 (Figure 5E) in the PEMTKO mice, a region coding for amino acids 210–222. We similarly PCR-amplified exon 5 of UCP1 from genomic DNA using primers overlapping with the excised region in the mRNA and found no difference in the PCR products (Figure 5F), excluding the possibility of a genomic mutation. These data demonstrate that the loss of PEMT promotes the utilization of an alternative 3′ splice acceptor site in exon 5 of UCP1 instead of the normal splice junction at Nt 864.
Figure 5.
Whole-body ablation of PEMT results in alternative splicing of UCP1 mRNA perinatally. (A) Mitochondrial DNA abundance normalized to nuclear DNA, n = 4. (B) Whole-gel UCP1 immunoblot to detect potentially ubiquitinated UCP1. (C) Abundance of proteins involved in the ER stress response. (D) Amplicon of UCP1 exon 5 in cDNA reverse transcribed from mRNA. (E) Sequence of UCP1 exon 5 in cDNA reverse transcribed from mRNA in the WT and PEMTKO mice, n = 3. (F) Amplicon of UCP1 exon 5 in genomic DNA isolated from the PEMTKO mice. (G) Schematic of alternative 3′ splice junctions in exon 5 of UCP1. (H) mRNA levels of UCP1 splice variants in the PEMTKO mice, n = 4. (I) Measurement of UCP1 protein using commercially available antibodies whose epitopes lie outside the deleted amino acid (AA) sequence in variant D. The antibodies and their epitopes from top to bottom: Alpha Diagnostic UCP11-A, epitope: C-terminus; Abcam ab23841, epitope: AA100-200; Abcam ab10983, epitope: AA145-159; and Cell Signaling 14670S, epitope: C-terminus. All of the antibodies were predicted to detect all of the UCP1 splice variants based on the epitopes. (J) UCP1 protein levels in BAT from the PEMTKO mice at birth. (K) UCP1 protein levels in BAT from 2-week-old PEMT+/- offspring from breeder pairs of either male WT, PEMTKO female, or male PEMTKO, female WT. (L) UCP1 protein levels in BAT from 2-week-old PEMTKO and PEMT+/- mice that were offspring of breeder pairs of either male PEMTKO, female PEMT+/-, or male PEMT+/-, female PEMTKO. Data are expressed as mean ± SEM, *p < 0.05.
Odegaard et al. previously reported the presence of two alternatively spliced forms of UCP1 mRNA resulting from alternate 3′ acceptor sites in exon 5 [32]. The authors designated the WT variant of UCP1 mRNA as variant A (splice junction at Nt 864), and their newly discovered variants as variants B and C (splice junctions at Nt 891 and Nt 972, respectively). Our splice variant was different from these other two variants, which we designated as variant D (splice junction at Nt 903) (Figure 5G). Using variant-specific primers, we verified that PEMT deletion promotes a near complete loss of WT variant A (Figure 4H). Variants B and C were similarly barely detectable in BAT from the PEMTKO mice, although there was a small but significant increase in variant C compared to the WT mice. Variant D was not detectable in BAT from the WT mice, but the transcript level in the PEMTKO mice was similar to the content of variant A in BAT from the WT mice (Figure 5H). This explains the lack of differences in the mRNA quantified in Figure 1G, which utilized PCR primers that would detect all four variants.
To determine whether splice variant D UCP1 mRNA produces a functional protein, we used four commercially available UCP1 antibodies whose epitopes did not overlap with the missing 13 amino acids of variant D. None of these antibodies detected a protein product for splice variant D, which would be predicted to produce a band ∼1.5 kDa lower than WT UCP1 (Figure 5I), suggesting that a functional protein was not produced from splice variant D. Indeed, UCP1 protein for variant A appeared in BAT from the PEMTKO mice despite its transcript being over 1000-fold lower compared to variant D. We also examined these mice under prolonged cold exposure to determine whether UCP1 protein could be induced under this condition. After an acclimation protocol previously described for UCP1 null mice [ 25 ], we successfully performed 7-day cold exposure on the WT and PEMTKO mice. As expected, prolonged cold exposure increased BAT UCP1 protein content in the WT mice compared to those housed at room temperature (Fig. S3A). In contrast, prolonged cold exposure had no effect on BAT UCP1 protein content in the PEMTKO mice (Fig. S3AFig. S3A). Similarly, cold exposure induced higher UCP1 protein content in inguinal white adipose tissue (iWAT) in the WT mice compared with room temperature controls (Fig. S3B), but not in iWAT from the PEMTKO mice. Taken together, our data demonstrate that PEMT is required for UCP1 protein expression via splicing of UCP1 transcripts to produce variant A. The important caveat is that this mechanism does not operate in a cell-autonomous manner, as demonstrated by our findings in the PEMT-BKO mice.
Odegaard et al. reported that a lack of IL-33 or its receptor, ST2, promoted the expression of UCP1 splice variants B and C during the perinatal period, thus abrogating UCP1 protein from birth [32]. Similarly, the PEMTKO mice lacked UCP1 from birth (Figure 5J), suggesting that PEMT is required during perinatal activation of the normal UCP1 3′ splice acceptor site for exon 5. Although the UCP1 splice variants reported in the IL-33/ST2-deficient models were different from those we found in the PEMTKO mice, we examined how this system may have been affected in our mouse model. Surprisingly, circulating IL-33 was higher, not lower, in the PEMTKO mice compared to the WT mice (Fig. S3C), suggesting that UCP1 splicing in the PEMTKO mice was not due to the lack of IL-33. Our observations on circulating ST2 were more complex. ST2 is expressed as a membrane-bound receptor for IL-33. When IL-33 levels rise (as observed), a soluble form of ST2 (sST2) becomes expressed and released into circulation, intercepting IL-33 and preventing an excessive immune response [38]. Compared to the WT mice, blood from the PEMTKO mice contained lower levels of ST2, while the sST2 levels were greater. Lower levels of ST2 likely indicate reduced ST2 signaling, and thus this could have potentially contributed to UCP1 splicing in the PEMTKO mice. However, PEMT knockout also coincided with the increased sST2 content, which is usually a signature of upregulated IL-33/ST2 signaling. It is also noteworthy that the membrane-bound, active form of ST2 was present (∼30% of WT) in the PEMTKO mice. Thus, it is highly likely that ST2 signaling was intact in the PEMTKO mice, unlike the ST2 knockout mice. We also could not detect IL-33 or ST2 in BAT in the WT or PEMTKO mice (Fig. S3D).
Given that a lack of PEMT promotes alternative splicing in UCP1 during development in a non-cell-autonomous manner, PEMT in the embryo or the mother could be required to appropriately express UCP1. To determine if a lack of PEMT in the mother alone was sufficient to induce alternative splicing in UCP1, we crossed female PEMTKO mice with male WT mice and male PEMTKO mice with female WT mice. PEMT heterozygous knockouts (PEMT+/-) born to either set of parents had normal UCP1 levels (Figure 5K), indicating that the loss of PEMT in the mother or the partial loss of PEMT in the offspring was not sufficient to induce splicing. We further pursued a possibility that the loss of UCP1 protein may have been induced by a lack of PEMT in both the mother and offspring and tested this by crossing either PEMTKO males with PEMT+/- females or PEMTKO females with PEMT+/- males. Regardless of the combination, the only factor that produced the UCP1-deficient phenotype was when the offspring were PEMTKO (Figure 5L). Together, these data indicate that the loss of PEMT in the offspring mediates UCP1 splicing during perinatal development regardless of the parents’ genotype.
4. Discussion
We initially sought to determine the cellular mechanism by which mice with whole-body PEMT deletion are cold-intolerant. We found that BAT from the PEMTKO mice completely lacked UCP1 protein. Furthermore, this effect was not mediated by PEMT in BAT, as the mice with UCP1-Cre or adiponectin-Cre driven deletions of PEMT had normal UCP1 protein levels. Rather, the lack of PEMT in other tissue induced alternative splicing of UCP1, resulting in a truncated mRNA that did not yield UCP1 protein.
A previous study that documented cold sensitivity in PEMTKO mice attributed insufficient glucose supply as the mechanism for reduced thermogenesis [19]. We report that this phenotype is also the result of their lack of intrinsic ability for UCP1-dependent thermogenesis. Curiously, the prior study reported normal UCP1 protein content in PEMTKO mice. Hence, we utilized four different UCP1 antibodies, including the one used by Gao et al. [19](Abcam #ab10983) to verify that BAT from the PEMTKO mice did not contain detectable UCP1 protein. The root of this discrepancy is unclear. We acquired the PEMTKO mice from Gao et al. less than a year after their study was published, making it unlikely that these mice were different. While there were some minor experimental differences between the present study and that of Gao et al. (length of fasting, cold-tolerance duration, etc.), these are unlikely to account for the discrepancy in the UCP1 phenotype. One possible explanation is that the Abcam UCP1 antibody utilized in their study was a different batch than the one used in our study (GR3188478-13). Polyclonal antibodies generated from different rabbits are not identical, so it is possible that the batch used by Gao et al. produced a non-specific band that was mistaken for UCP1. Gao et al. also reported normal cold sensitivity in standard chow diet-fed PEMTKO mice. We note that the cold sensitivity in our chow-fed PEMTKO mice was modestly delayed compared to our HFD-fed PEMTKO mice, such that they began to display low temperatures after the 3 h mark, which was the duration of the cold exposure experiments employed by Gao et al. One important finding by Gao et al. was that cold sensitivity in PEMTKO mice was rescuable by dietary choline supplementation. Circulating choline is reduced in PEMTKO mice, and its dietary supplementation is known to rescue other phenotypes found in PEMTKO mice [ 29 ]. Reduced circulating choline is thought to be due to the lack of PEMT in the liver, as PC generated by this enzyme is cleaved to produce choline that is secreted into circulation. However, BAT from our PEMT-LKO mice exhibited normal UCP1 protein content, suggesting that choline's effect to rescue cold sensitivity was likely due to an effect of choline in other tissues. Intracellular choline can supply headgroup for PC, thus potentially rescuing the lack of PEMT through this mechanism.
We initially hypothesized that the loss of UCP1 was due to the loss of mitochondrial CL in the HFD-fed PEMTKO mice, as CL is known to be required for thermogenic respiration. Indeed, the PEMTKO mice fed a HFD exhibited a robust decrease in CL, suggesting a potential connection between CL and the loss of UCP1. However, the loss of UCP1 protein was not due to a lack of CL, as the standard chow diet-fed PEMTKO mice had no UCP1 protein despite normal CL levels. Although previous work indicated that PEMT supplies mitochondria with PC in the liver [12], our data in the PEMTKO and PEMT-BKO mice indicate that PEMT was dispensable for maintaining mitochondrial PC or CL in BAT with a standard chow diet. Meanwhile, PEMT in BAT appeared to modestly contribute to the CL levels in a diet-dependent manner, although this had no robust effects on BAT function.
Our results are strikingly similar to the findings of Odegaard et al. who discovered two alternatively spliced UCP1 mRNAs arising from alternative 3′ acceptor sites in exon 5 of IL-33 and ST2 null mice during the perinatal period [32]. Similarly, we demonstrated that the loss of PEMT resulted in alternative splicing of UCP1 mRNA during development. Nevertheless, PEMT-dependent UCP1 splicing occurred at a third unique splice site, producing a 39 Nt deletion that we called variant D. Variant D, similar to variants B and C reported by Odegaard et al. does not produce functional protein, leading to a loss of UCP1-dependent thermogenic respiration and compromised whole-body thermoregulation.
What is the mechanism by which PEMT promotes UCP1 splicing? In our present study, we were unable to identify the cell type whose lack of PEMT prompted this process. Neither UCP1-Cre nor adiponectin-Cre mediated deletion of PEMT phenocopied whole-body PEMTKO mice, indicating that PEMT acts non-cell-autonomously and is independent of PEMT in white adipose tissues. We also tested albumin-Cre mice, as liver PEMT is known to robustly alter circulating cytokines and lipid milieu [29], and HSA-MerCreMer mice, as myokines can regulate adipose thermogenic programming [[39], [40], [41]] However, neither of these interventions promoted the loss of UCP1 protein in BAT. Odegaard et al. were similarly unable to identify the cell type that mediated splicing in their study; however, they were able to rule out IL-5-expressing cells, adaptive immune cells, and canonical IL-33/ST2 signaling through MyD88. Given the difference in the splice variants between our studies, the mechanism that produces UCP1 variant D is likely different than the one that produced variant C. Nevertheless, it is important to note that ST2 found in circulation was lower (∼30% of WT), but not absent, in the PEMTKO mice compared to the WT mice. The change coincided with greater IL-33 and sST2, likely suggesting that the IL-33/ST2 axis was intact in the PEMTKO mice. While it remains possible that the IL-33/ST2 axis may be involved in PEMT-dependent mechanisms, it is difficult to assess the exact nature of this interaction without understanding the non-canonical signaling of ST2 that regulates UCP1 splicing. It also remains possible that PEMT-dependent splicing of UCP1 is mediated by some population of immune cells. Investigation of these possibilities are currently ongoing but is beyond the scope of the current study.
Regardless of the identity of the cell type whose lack of PEMT promotes loss of UCP1 protein in BAT, these cells likely communicate with brown adipocytes either through direct cell-to-cell interactions or via the secretion of cytokines. There are plausible scenarios on how PEMT may robustly affect these cell behaviors. For example, as a consumer of S-adenosylmethionine, PEMT may affect histone methylation to induce changes in transcription. Indeed, deletion of PEMT can alter S-adenosylmethionine utilization, leading to hypermethylation of histones and robust effects on gene transcription [42]. Another possible mechanism arises from PEMT's role in maintaining ER homeostasis. PEMT ablation is known to cause ER stress [37], which may alter protein translation and other cellular processes in cells that mediate changes in BAT via signaling events.
5. Conclusion
In summary, PEMT plays a vital role in thermogenesis by regulating UCP1 protein expression via alternative splicing. Surprisingly, PEMT within BAT does not mediate this effect, nor does it substantially affect BAT function. Rather, PEMT's action occurs in a non-cell-autonomous manner, suggesting that a lack of PEMT prevents cell-to-cell communication required to induce properly spliced UCP1 transcripts. While the precise mechanisms remain enigmatic, our work helps reveal an exciting avenue of UCP1 regulation that may have profound implications for whole-body metabolism.
Author contributions
The experiments and data analysis were conducted by J.M.J, A.R.P.V, P.J.F., C.T.L., J.A.M, K.A.K, and J.E.C. The experimental design and manuscript review were performed by J.M.J, K.A.K, P.D.N., I.J.L, and K.F. The manuscript was written by J.M.J and K·F.
Acknowledgments
We thank Dr. Dennis Vance from the University of Alberta and Dr. Karyn Esser from the University of Florida for donating the PEMTKO mice and HSA-MerCreMer mice, respectively. We also thank Dr. Kai Ge from the NIDDK for donating the SV40T brown preadipocytes for this study. We thank Diana L. Lim for her assistance with the figures. This project was funded by NIH grants DK095774, DK107397, DK109888, and AG063077 to K·F., DK115867 and DK118333 to I.J.L., DK110656 to P.D.N., American Heart Association Predoctoral fellowships 19PRE34380991 to J.M.J. and 18PRE33960491 to A.R.P·V, and the Larry H. & Gail Miller Family Foundation to P.J.F. The University of Utah Metabolomics Core Facility is supported by S10 OD016232, S10 OD021505, and U54 DK110858.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.10.007.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiological Reviews. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 2.Rousset S., Alves-Guerra M.C., Mozo J., Miroux B., Cassard-Doulcier A.M., Bouillaud F. The biology of mitochondrial uncoupling proteins. Diabetes. 2004;53(Suppl 1):S130–S135. doi: 10.2337/diabetes.53.2007.s130. [DOI] [PubMed] [Google Scholar]
- 3.Divakaruni A.S., Brand M.D. The regulation and physiology of mitochondrial proton leak. Physiology. 2011;26:192–205. doi: 10.1152/physiol.00046.2010. [DOI] [PubMed] [Google Scholar]
- 4.Garlid K.D., Jaburek M., Jezek P. The mechanism of proton transport mediated by mitochondrial uncoupling proteins. FEBS Letters. 1998;438:10–14. doi: 10.1016/s0014-5793(98)01246-0. [DOI] [PubMed] [Google Scholar]
- 5.Klingenberg M., Winkler E. The reconstituted isolated uncoupling protein is a membrane potential driven H+ translocator. The EMBO Journal. 1985;4:3087–3092. doi: 10.1002/j.1460-2075.1985.tb04049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker N., Crichton P.G., Vidal-Puig A.J., Brand M.D. Uncoupling protein-1 (UCP1) contributes to the basal proton conductance of brown adipose tissue mitochondria. Journal of Bioenergetics and Biomembranes. 2009;41:335–342. doi: 10.1007/s10863-009-9232-8. [DOI] [PubMed] [Google Scholar]
- 7.Kontani Y., Wang Y., Kimura K., Inokuma K.I., Saito M., Suzuki-Miura T. UCP1 deficiency increases susceptibility to diet-induced obesity with age. Aging Cell. 2005;4:147–155. doi: 10.1111/j.1474-9726.2005.00157.x. [DOI] [PubMed] [Google Scholar]
- 8.Kozak L.P., Anunciado-Koza R. UCP1: its involvement and utility in obesity. International Journal of Obesity. 2008;32(Suppl 7):S32–S38. doi: 10.1038/ijo.2008.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nedergaard J., Golozoubova V., Matthias A., Asadi A., Jacobsson A., Cannon B. UCP1: the only protein able to mediate adaptive non-shivering thermogenesis and metabolic inefficiency. Biochimica et Biophysica Acta. 2001;1504:82–106. doi: 10.1016/s0005-2728(00)00247-4. [DOI] [PubMed] [Google Scholar]
- 10.Heden T.D., Neufer P.D., Funai K. Looking beyond structure: membrane phospholipids of skeletal muscle mitochondria. Trends in Endocrinology and Metabolism. 2016;27:553–562. doi: 10.1016/j.tem.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath S.E., Daum G. Lipids of mitochondria. Progress in Lipid Research. 2013;52:590–614. doi: 10.1016/j.plipres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 12.van der Veen J.N., Lingrell S., da Silva R.P., Jacobs R.L., Vance D.E. The concentration of phosphatidylethanolamine in mitochondria can modulate ATP production and glucose metabolism in mice. Diabetes. 2014;63:2620–2630. doi: 10.2337/db13-0993. [DOI] [PubMed] [Google Scholar]
- 13.Ridgway N.D., Vance D.E. Phosphatidylethanolamine N-methyltransferase from rat liver. Methods in Enzymology. 1992;209:366–374. doi: 10.1016/0076-6879(92)09045-5. [DOI] [PubMed] [Google Scholar]
- 14.Vance D.E., Ridgway N.D. The methylation of phosphatidylethanolamine. Progress in Lipid Research. 1988;27:61–79. doi: 10.1016/0163-7827(88)90005-7. [DOI] [PubMed] [Google Scholar]
- 15.Flis V.V., Daum G. Lipid transport between the endoplasmic reticulum and mitochondria. Cold Spring Harbor Perspectives in Biology. 2013;5 doi: 10.1101/cshperspect.a013235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abe M., Sawada Y., Uno S., Chigasaki S., Oku M., Sakai Y. Role of acyl chain composition of phosphatidylcholine in tafazzin-mediated remodeling of cardiolipin in liposomes. Biochemistry. 2017;56:6268–6280. doi: 10.1021/acs.biochem.7b00941. [DOI] [PubMed] [Google Scholar]
- 17.Sustarsic E.G., Ma T., Lynes M.D., Larsen M., Karavaeva I., Havelund J.F. Cardiolipin synthesis in Brown and beige fat mitochondria is essential for systemic energy homeostasis. Cell Metabolism. 2018;28:159–174. doi: 10.1016/j.cmet.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y., Malhotra A., Ren M., Schlame M. The enzymatic function of tafazzin. Journal of Biological Chemistry. 2006;281:39217–39224. doi: 10.1074/jbc.M606100200. [DOI] [PubMed] [Google Scholar]
- 19.Gao X., van der Veen J.N., Fernandez-Patron C., Vance J.E., Vance D.E., Jacobs R.L. Insufficient glucose supply is linked to hypothermia upon cold exposure in high-fat diet-fed mice lacking PEMT. The Journal of Lipid Research. 2015;56:1701–1710. doi: 10.1194/jlr.M059287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walkey C.J., Donohue L.R., Bronson R., Agellon L.B., Vance D.E. Disruption of the murine gene encoding phosphatidylethanolamine N-methyltransferase. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:12880–12885. doi: 10.1073/pnas.94.24.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verkerke A.R.P., Ferrara P.J., Lin C.-T., Johnson J.M., Ryan T.E., Maschek J.A. Phospholipid methylation regulates muscle metabolic rate through Ca2+ transport efficiency. Nature Metabolism. 2019;1:876–885. doi: 10.1038/s42255-019-0111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acehan D., Vaz F., Houtkooper R.H., James J., Moore V., Tokunaga C. Cardiac and skeletal muscle defects in a mouse model of human Barth syndrome. Journal of Biological Chemistry. 2011;286:899–908. doi: 10.1074/jbc.M110.171439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson J.M., Ferrara P.J., Verkerke A.R.P., Coleman C.B., Wentzler E.J., Neufer P.D. Targeted overexpression of catalase to mitochondria does not prevent cardioskeletal myopathy in Barth syndrome. Journal of Molecular and Cellular Cardiology. 2018;121:94–102. doi: 10.1016/j.yjmcc.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai B., Lee J.E., Jang Y., Wang L., Peng W., Ge K. MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis. Nucleic Acids Research. 2017;45:6388–6403. doi: 10.1093/nar/gkx234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golozoubova V., Hohtola E., Matthias A., Jacobsson A., Cannon B., Nedergaard J. Only UCP1 can mediate adaptive nonshivering thermogenesis in the cold. The FASEB Journal. 2001;15:2048–2050. doi: 10.1096/fj.00-0536fje. [DOI] [PubMed] [Google Scholar]
- 26.Nigro M., Santos A.T., Barthem C.S., Louzada R.A., Fortunato R.S., Ketzer L.A. A change in liver metabolism but not in brown adipose tissue thermogenesis is an early event in ovariectomy-induced obesity in rats. Endocrinology. 2014;155:2881–2891. doi: 10.1210/en.2013-1385. [DOI] [PubMed] [Google Scholar]
- 27.Lark D.S., Torres M.J., Lin C.T., Ryan T.E., Anderson E.J., Neufer P.D. Direct real-time quantification of mitochondrial oxidative phosphorylation efficiency in permeabilized skeletal muscle myofibers. American Journal of Physiology - Cell Physiology. 2016;311:C239–C245. doi: 10.1152/ajpcell.00124.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matyash V., Liebisch G., Kurzchalia T.V., Shevchenko A., Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. The Journal of Lipid Research. 2008;49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs R.L., Zhao Y., Koonen D.P., Sletten T., Su B., Lingrell S. Impaired de novo choline synthesis explains why phosphatidylethanolamine N-methyltransferase-deficient mice are protected from diet-induced obesity. Journal of Biological Chemistry. 2010;285:22403–22413. doi: 10.1074/jbc.M110.108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enerback S., Jacobsson A., Simpson E.M., Guerra C., Yamashita H., Harper M.E. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature. 1997;387:90–94. doi: 10.1038/387090a0. [DOI] [PubMed] [Google Scholar]
- 31.Kazak L., Chouchani E.T., Stavrovskaya I.G., Lu G.Z., Jedrychowski M.P., Egan D.F. UCP1 deficiency causes brown fat respiratory chain depletion and sensitizes mitochondria to calcium overload-induced dysfunction. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:7981–7986. doi: 10.1073/pnas.1705406114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Odegaard J.I., Lee M.W., Sogawa Y., Bertholet A.M., Locksley R.M., Weinberg D.E. Perinatal licensing of thermogenesis by IL-33 and ST2. Cell. 2016;166:841–854. doi: 10.1016/j.cell.2016.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee Y., Willers C., Kunji E.R., Crichton P.G. Uncoupling protein 1 binds one nucleotide per monomer and is stabilized by tightly bound cardiolipin. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:6973–6978. doi: 10.1073/pnas.1503833112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cole L.K., Mejia E.M., Vandel M., Sparagna G.C., Claypool S.M., Dyck-Chan L. Impaired cardiolipin biosynthesis prevents hepatic steatosis and diet-induced obesity. Diabetes. 2016;65:3289–3300. doi: 10.2337/db16-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cawthorne C., Swindell R., Stratford I.J., Dive C., Welman A. Comparison of doxycycline delivery methods for Tet-inducible gene expression in a subcutaneous xenograft model. Journal of Biomolecular Techniques. 2007;18:120–123. [PMC free article] [PubMed] [Google Scholar]
- 36.Clarke K.J., Adams A.E., Manzke L.H., Pearson T.W., Borchers C.H., Porter R.K. A role for ubiquitinylation and the cytosolic proteasome in turnover of mitochondrial uncoupling protein 1 (UCP1) Biochimica et Biophysica Acta. 2012;1817:1759–1767. doi: 10.1016/j.bbabio.2012.03.035. [DOI] [PubMed] [Google Scholar]
- 37.Gao X., Van der Veen J.N., Vance J.E., Thiesen A., Vance D.E., Jacobs R.L. Lack of phosphatidylethanolamine N-methyltransferase alters hepatic phospholipid composition and induces endoplasmic reticulum stress. Biochimica et Biophysica Acta. 2015;1852:2689–2699. doi: 10.1016/j.bbadis.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 38.Griesenauer B., Paczesny S. The ST2/IL-33 Axis in immune cells during inflammatory diseases. Frontiers in Immunology. 2017;8:475. doi: 10.3389/fimmu.2017.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bostrom P., Wu J., Jedrychowski M.P., Korde A., Ye L., Lo J.C. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao R.R., Long J.Z., White J.P., Svensson K.J., Lou J., Lokurkar I. Meteorin-like is a hormone that regulates immune-adipose interactions to increase beige fat thermogenesis. Cell. 2014;157:1279–1291. doi: 10.1016/j.cell.2014.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts L.D., Bostrom P., O'Sullivan J.F., Schinzel R.T., Lewis G.D., Dejam A. beta-Aminoisobutyric acid induces browning of white fat and hepatic beta-oxidation and is inversely correlated with cardiometabolic risk factors. Cell Metabolism. 2014;19:96–108. doi: 10.1016/j.cmet.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye C., Sutter B.M., Wang Y., Kuang Z., Tu B.P. A metabolic function for phospholipid and histone methylation. Molecular Cell. 2017;66:180–193. doi: 10.1016/j.molcel.2017.02.026. e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.