Abstract
In recent years, mass spectrometry has emerged as a core component of fundamental discoveries in virology. As a consequence of their coevolution, viruses and host cells have established complex, dynamic interactions that function either in promoting virus replication and dissemination or in host defense against invading pathogens. Thus, viral infection triggers an impressive range of proteome changes. Alterations in protein abundances, interactions, posttranslational modifications, subcellular localizations, and secretion are temporally regulated during the progression of an infection. Consequently, understanding viral infection at the molecular level requires versatile approaches that afford both breadth and depth of analysis. Mass spectrometry is uniquely positioned to bridge this experimental dichotomy. Its application to both unbiased systems analyses and targeted, hypothesis-driven studies has accelerated discoveries in viral pathogenesis and host defense. Here, we review the contributions of mass spectrometry–based proteomic approaches to understanding viral morphogenesis, replication, and assembly and to characterizing host responses to infection.
Keywords: viral proteomics, virus-host interactions, AP-MS, secretome, posttranslational modifications
INTRODUCTION
As obligate parasites, viruses have coevolved with their hosts, developing finely tuned mechanisms to capture and manipulate host cellular processes for their replication and spread. Furthermore, in response to infection, cells deploy a range of intrinsic defense mechanisms to curb immediate viral replication and mobilize local and distal immune effectors. Consequently, viral infection triggers an impressive array of changes in the cell proteome, metabolome, and lipidome. These changes are starting to be established as key markers of the infection status. Mass spectrometry (MS) provides the unique ability to accurately define these diverse changes via either global unbiased analyses or targeted hypothesis-driven strategies. Here, we review the contribution of MS-based proteomic approaches to understanding viral and host protein functions and their regulation during infection.
The proteome—that is, the total protein content of a cell—is extraordinarily complex. Quantitative proteomic analyses have provided estimates of 0.5–3 × 106 proteins/μm3 for human cells (1). These proteins are encoded by ∼16,000–20,000 open reading frames, and all are subject to transcriptional, posttranscriptional, translational, and posttranslational regulation, which varies with cell and tissue types. Viruses co-opt and further expand the complexity of the proteome during infection. In fact, it is at the proteome level that most cellular pathways are modulated; viral or host genes exert most of their functions during infection at the protein level. Therefore, defining protein function during infection is challenging.
As with other research fields, virology has reached a stage when expanding research boundaries and answering long-standing questions benefit from the integration of technologies from diverse disciplines. Using modern quantitative MS, protein levels, localizations, interactions, and posttranslational modifications (PTMs) can be unambiguously and unbiasedly defined during infection. Thus, because they are both versatile and unique in the data they provide, MS-based proteomic strategies can readily be integrated with microscopy, biochemistry, genomic, and molecular virology approaches to generate a systems biology view of the viral replication cycle (Figure 1). As discussed in this review, MS studies have characterized virion composition and structure, virus-host protein interactions, global and subcellular proteome changes, infection-induced secretomes, and viral and cellular protein PTM functions. Our laboratory has extensively studied virus-host protein interactions that provide direct mechanistic insights regarding protein complexes and cellular pathways modulated during infection, thereby offering new targets for antiviral therapeutic intervention. Given the inherent dynamic nature of protein interactions, the study of virus-host interactions has benefited from the development of sensitive and quantitative MS approaches to quantify infection-driven changes in protein interactions, to capture stable and transient interactions, and to build functional interaction networks (Figure 2). To provide hands-on experience with analyzing proteomic data sets, we have developed two tutorials for generating protein interaction networks (Supplemental Tutorial 1) and determining protein interaction specificity (Supplemental Tutorial 2) (Figure 2). We also provide a list of prominent qualitative, quantitative, and structural MS-based approaches relevant to virology (Table 1). As MS and related computational strategies are continuously improving, we discuss several emerging approaches that show great promise for application to virology studies.
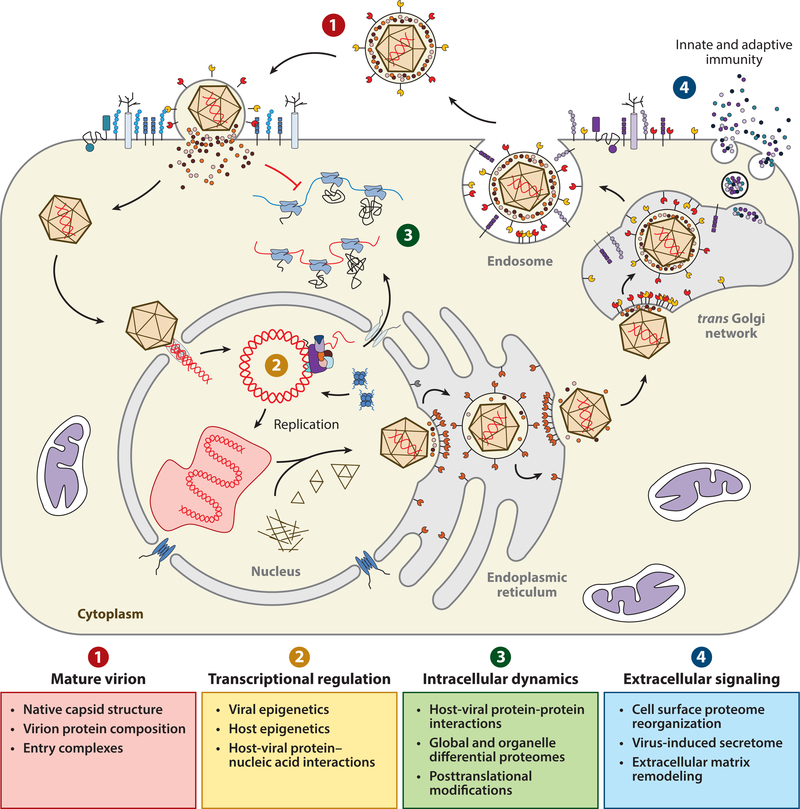
Figure 1.
Contribution of mass spectrometry (MS)-based proteomics to understanding viral infection. The productive infection cycle of the prototypic human virus herpes simplex virus 1 (HSV-1) is depicted within a susceptible host cell. Key aspects of the virus life cycle that can be studied using MS-based proteomics are designated by numbers. Briefly, the composition and structure of mature virions can be characterized by tandem MS and native MS, respectively (①). Affinity purification (AP)-MS can detail interactions between viral and host constituents at the cell surface mediating entry (①); in the nucleus regulating transcription, genome replication, and capsid assembly (②); and in the cytosol aiding egress and assembly (③). Quantitative MS can measure virus-induced, organelle-specific changes in protein expression and posttranslational modifications (③) and the secretion of antiviral mediators that initiate the innate and adaptive immune response (④). Viral proteins are shaded red and yellow; host proteins are shaded blue and purple.
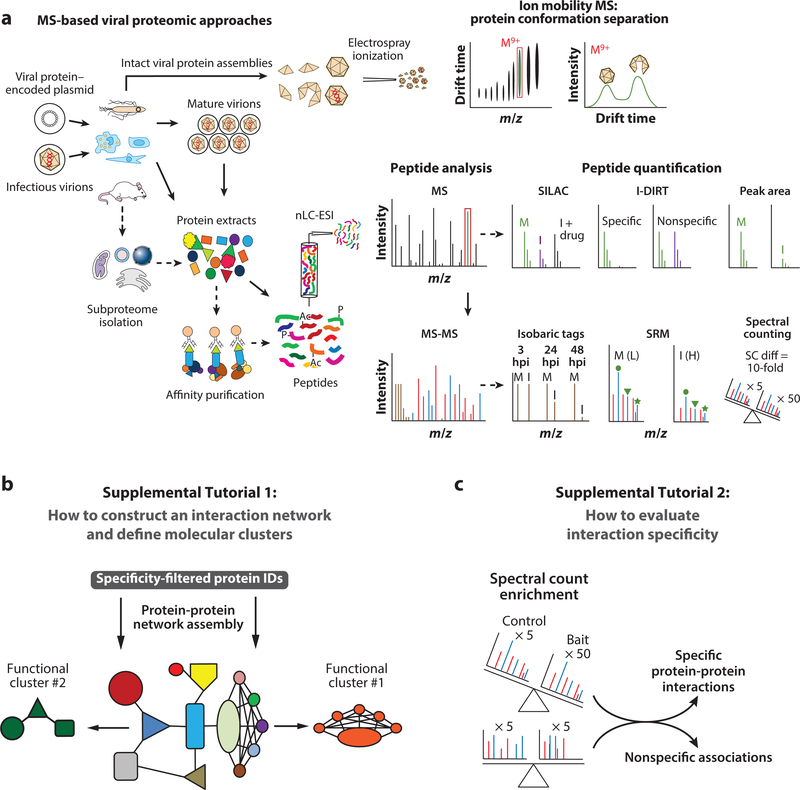
Figure 2.
(a) Multifaceted mass spectrometry (MS)-based analyses of viral structures, interactions, and pathogenesis. Orthogonal workflows illustrate biochemical and MS-based approaches that can be used to study recombinant viral proteins expressed in bacteria or mammalian cells, extracellular virions, or virus-infected mammalian cells. Masses of intact viral assemblies can be determined by electrospray ionization (ESI)–native MS (also called top-down MS), with an orthogonal ion mobility separation to assess molecular compactness (top). Peptide-based MS (also called bottom-up MS) analysis requires denatured protein extracts obtained from extracellular virions, infected cells, organelles, or virus-host complexes. Protein extracts can be directly digested to MS-amenable peptides, or they can first be separated by SDS-PAGE and then in-gel digested into peptides. Following nano–liquid chromatography and ESI (nLC-ESI), peptide sequencing is performed by successive rounds of intact and fragment peptide mass measurements (tandem MS) in the mass spectrometer. Peptide quantification strategies use MS or tandem MS spectra as the basis for protein-level quantification. Global quantitative techniques can use either isotope labeling—within the cell, for simultaneous comparison of usually up to three samples (SILAC), or at the peptide level, for comparison of currently up to ten samples (isobaric tags)—or label-free quantification (peak area or spectral counts). Selected reaction monitoring (SRM) is used for targeted analyses in isotopic or label-free workflows. (b,c) Supplemental Tutorials 1 and 2 provide hands-on experience in the computational analysis of AP-MS data sets. (b) In Supplemental Tutorial 1, a data set of high-confidence protein identifications from HDAC1-EGFP immunoisolations is provided as an example for construction and analysis of functional protein networks using the STRING database of known and predicted protein-protein interactions. (c) In Supplemental Tutorial 2, protein identifications from HDAC1-EGFP immunoisolations that have not been filtered for specificity are provided. The tutorial demonstrates how to use spectral counting to classify specific versus nonspecific associations. Abbreviations: H, heavy; hpi, hours postinfection; I, infected; L, light; M, mock; M9+, molecular ion with a charge state of 9; SC diff, spectral count difference.
Table 1.
A how-to guide for MS-based proteomic characterization of viral infection
| How to… | Technique | Description | Advantages | Drawbacks |
|---|---|---|---|---|
| Qualitative | ||||
| Confirm viral protein expression or purity from low-complexity samples | Mass fingerprint | MALDI-TOF detection of intact peptide masses (MS1); can be preceded by one- or two-dimensional electrophoresis | Rapid protein identification | Requires low protein complexity; limited sequence validation |
| Characterize viral and host proteomes | Peptide sequencing (tandem MS) | Predominant technology for viral proteomic studies; determination of peptide amino acid sequences using gas-phase fragmentation | Minimal sample handling and deep proteome coverage | Specialized computational approaches needed for large data sets |
| Quantitative | ||||
| Identify differences in relative protein expression between mock- and virus-infected conditions | Spectral counting (label-free) | Relative abundance differences inferred by comparing number of spectra per protein acquired by tandem MS | Facile; no modification to qualitative workflows | Subtle expression differences not detected |
| Quantify relative protein expression between different mock- and virus-infected conditions | Peak area (label-free) | Intact peptide abundances are integrated by area under the curve; relative or absolute (with external calibration) quantification | Greater dynamic range and precision than spectral counting | High-resolution instrumentation needed; computationally demanding |
| Quantify relative protein expression and posttranslational modifications (usually between two or three cellular conditions) | Metabolic labeling | Protein labeling in cells or animals with light or heavy isotopes (e.g., 15N salts or 13C/15N amino acids); relative peak areas of light and heavy peptides in MS1 spectra are used for quantification | Minimizes experimental variation; accurate measurement of small changes in relative expression | Increased cost, spectrum complexity, and computational demands; limited/expensive use in animals |
| Simultaneously quantify relative protein expression at different time points postinfection | Isobaric tagging | Peptide labeling (following digestion) using amine-reactive reagents; simultaneous relative quantification of up to 10 samples | High-throughput multiplexing of time points or biological replicates | Reduced linear quantification range |
| Determine the absolute abundance of proteins in virus-infected cells | Selected reaction monitoring (SRM) | Targeted analysis of selected peptides (e.g., <100) by tandem MS for relative or absolute quantification; often paired with isotope-labeled peptide/protein standards (e.g., AQUA or QconCAT) | Higher sensitivity and quantitative precision (<10%) than MS1-based methods; identified signature peptides are universal among labs | Each method requires separate optimization and validation |
| Determine viral capsid assembly states and subunit arrangements under different salt and pH conditions | Native ion mobility MS | Analysis of intact proteins or noncovalent protein assemblies at lower complexity using a high-resolution mass detector (e.g., TOF or Orbitrap); ion mobility separates by structural “compactness” | Distinguish among structural isoforms of large protein complexes | Currently requires micromolar concentrations; technically challenging |
| Determine interprotein contacts within capsids or other complexes; map immune cell epitopes for antivirals and vaccines | Hydrogen/deuterium exchange | Quantify the differential uptake or release of deuterium from amide hydrogen depending on solvent accessibility of a protein surface and hydrogen bonding | Detect shifts in protein conformations; resolution of several amino acids | Technically challenging; back-exchange and bias toward fast-exchanging regions |
| Determine protein interaction topologies in a cellular context | Cross-linking MS (XL-MS) | Covalent linking of proteins; stabilizes weak interactions in AP-MS; exact cross-linked residues identified by tandem MS | Determine direct interactions at single-residue resolution | Cross-linked species are of low abundance; technically challenging |
Abbreviations: AP, affinity purification; AQUA, absolute quantification; MALDI, matrix-assisted laser desorption ionization; MS, mass spectrometry; QConCAT, quantification concatemer; TOF, time of flight.
VIRION COMPOSITION AND STRUCTURE
Mature viral particles are a microcosm of their host, composed of cell-derived amino acids, lipids, and nucleotides. The intricate assembly of these components is required to produce an infectious virus. Although many complementary analytical methods have contributed to understanding the composition of extracellular virions (2, 3), MS has provided a powerful means for characterizing their detailed protein composition and macromolecular structures (Figure 2). For compositional analysis, most studies have focused on viruses with high proteomic complexity, including those in the families Herpesviridae (4–9) and Poxviridae (10–12). A noteworthy exception was the comparison of adenovirus subtypes using isotope-labeled (18O) water by the Fenselau laboratory (13), an early application of 18O for comparative quantitative proteomics of complex biological samples. MS analysis of virion composition has also offered proof of evolutionary origins. In a pioneering study, MS analysis of purified channel catfish virus virions provided amino acid sequence data to support the common evolutionary origin of this virus with other herpesviruses (14), a hypothesis previously based only on capsid and virion morphology. Subsequently, deeper proteomic analyses have been conducted on mammalian herpesviruses, such as human (8) and murine (5) cytomegaloviruses, herpes simplex viruses (7, 9, 15), Epstein–Barr virus (EBV) (4), and pseudorabies virus (PRV) (6). Technological advances in MS instrumentation have improved the sensitivity of detection, providing near-complete proteome coverage and access to low-abundance virion components (6, 7, 16). These developments have helped establish that virions contain host proteins and have triggered the expansion of MS studies to the proteomes of filoviruses, HIV, influenza, and respiratory syncytial virus (16–22). Nevertheless, defining virion composition still presents challenges and is a developing area of research. Purifying homogeneous populations of virions is difficult; for example, the presence of nonenveloped capsids in preparations of mature herpesvirus virions is common. Therefore, virion preparation purity must be assessed using microscopy techniques prior to MS analysis. The identification of virion-associated host proteins also presents challenges in distinguishing those that are specifically and functionally incorporated. It is critical that host factors are screened to eliminate spurious candidates. Current approaches involve determining proteinase sensitivity (6, 9) and/or performing functional RNA interference–based virus propagation assays (16, 22).
Recent technology developments have allowed MS to also contribute insights into the structure of virions. An emerging biomolecular technique, termed native MS, enables the detection of complexes of several million daltons (23,24).When combined with ion mobility, a gas phase–based separation, native MS becomes a valuable technique for structural virologists to determine the size and shape of intermediate and mature capsid structures (Figure 2) (25–27). As many capsid proteins self-assemble in vitro, native ion mobility MS can monitor the dynamics of capsid morphology and stability as a function of pH and ionic strength (28, 29). For example, native MS and atomic force microscopy have demonstrated that the single-stranded RNA genome of the picorna-like Triatoma virus stabilizes or triggers uncoating of the capsid in a pH-dependent fashion through electrostatic genome-capsid interactions (29). In addition to native ion mobility MS, hydrogen/deuterium (H/D) exchange MS has been used as a complementary strategy for characterizing the assembly of viral capsids (30). In this technique, the rates of amide proton exchange (deuterium versus protium) are used as a measure of relative solvent accessibility. For the study of macromolecular virus structures, this technique allows determination of amino acid sequence regions that undergo changes in conformation and/or modification of intermolecular contacts during infection. For example, the Prevelige laboratory (31) compared H/D exchange rates for immature, mature, and Gag/CA5 mutant capsids. H/D exchange rates supported increased protection in the β-hairpin and the NTD-CTD interface in mature virions, which interestingly had yet to form in the Gag/CA5 structural intermediate. Overall, these approaches rely on the stability of complexes in solution and in gas phase (for native MS) and therefore are technically demanding; however, with increases in the mass resolution and acquisition speed of the instruments used, analysis of intermediate virion structures with increased heterogeneity and mass will become feasible.
VIRUS-HOST PROTEIN INTERACTIONS
Dynamic Protein-Protein Interactions During Infection
The progression of viral infection depends on the establishment of numerous finely tuned virus-host and virus-virus protein interactions. Through these dynamic protein-protein interactions, complex cellular pathways are modulated to promote viral replication or act in host defense. Understanding which host factors are targeted by viral proteins provides direct insight into the molecular details of infection and identifies regulatory hubs for antiviral intervention. The identification of virus-host interactions has benefited greatly from the use and continuous development of antibody-based affinity purification (AP; also known as immunoprecipitation) strategies (reviewed in 32). Their integration with MS (AP-MS) (Figure 2) has allowed a protein’s interacting partners to be identified with little to no a priori knowledge (33). Most studies have focused on interactions at one given time point of infection. For example, the Knipe laboratory (34, 35) used AP-MS to study virus-host protein complexes containing the immediate-early proteins ICP27 or ICP8 at an early stage of herpes simplex virus 1 (HSV-1) infection in human Hep2 cells. Interestingly, components of SWI/SNF and ISWI chromatin-remodeling complexes were coisolated with the single-stranded DNA–binding protein ICP8. The identification of chromatin-remodeling enzymes was a key finding, because at that time, their role in productive HSV-1 infection was not clear. This study clearly stimulated further investigations, as chromatin-remodeling complexes are now established as important players during infection (reviewed, e.g., in 36).
Although the spatial and temporal regulation of virus-host interactions is at the core of their functions during the progression of an infection, it was not until the use of fluorescent probes for AP by Cristea and coworkers (37) that the applicability of AP-MS was demonstrated for the isolation of protein complexes at different stages of a viral infection (38). In this initial study, the Sindbis virus nonstructural protein nsP3 was affinity-tagged at its native genomic locus with green fluorescent protein (GFP), while maintaining the virus’s replication competence and infectivity in fibroblasts (38). The GFP tag afforded the temporal examination of protein localization and interactions (37) at a given moment in infected cells (38, 39). Further advancements introduced through this AP-MS strategy were cryogenic cell lysis of infected cells and fast isolations on magnetic beads, which helped preserve complexes close to their original state in the cell, maximized viral bait isolation, and reduced nonspecific interactions (38). These strategies offered a temporal distinction of virus-host interactions, such as with G3BP1 throughout the infection and with heterogeneous nuclear ribonucleoproteins at early and 14–3-3 proteins at late stages of Sindbis virus infection. The power of these approaches for the discovery and functional characterization of various virus-host interactions (33) was demonstrated in our studies of diverse viruses, including Sindbis virus (38, 40), human cytomegalovirus (HCMV) (41–44), HSV-1 (45), PRV (46, 47), and West Nile virus (48).
Another significant advancement to increase AP specificity was the development of the tandem affinity purification (TAP) tag system, originally applied to characterizing multiprotein complexes inyeast(49)and subsequently extended to mammalian systems(50).As the name implies, TAP uses a dual epitope tag for successive immunoaffinity isolations and has been applied to interactions within virus-infected cells (51–54). For example, a streptavidin-and calmodulin-binding TAP system was used to isolate the vaccinia virus A56 and K2 proteins, which localize to the cell surface and are required for interaction with the entry fusion complex (51). Overall, the two-step TAP system reduces background contaminants and identifies stable interactions, but unlike single-step AP, it often cannot retain transient interactions.
Although studying virus-protein interactions in the context of an infection by using full-length, replication-competent viruses is ideal, in vitro or cellular overexpression of individual affinity-tagged viral proteins has also shown utility for studying interactions (55–60), particularly when the generation of recombinant viruses has proved difficult. For example, in a large-scale interactome study, all the viral proteins encoded by the HIV genome were individually expressed in 293 and Jurkat cells and subjected to TAP AP-MS analysis (61). Almost 200 interactions were identified in both cell types. Moreover, 97 of 127 selected interactions were confirmed by reciprocal isolations, highlighting the confidence of this data set. In general, one drawback of the expression of single viral proteins within cellular systems is that it cannot fully recapitulate the diversity and temporality of signaling pathway modulation during an infection. Furthermore, virus-host interactions that depend on the expression of other viral proteins are missed. Nonetheless, this approach can isolate interactions of biological significance, as evidenced by the aforementioned HIV interactome, which stimulated further biological insights into HIV pathogenesis. Ultimately, these studies accelerate insights into how viral proteins could directly alter host cell functions.
In combination with advances in molecular virology, AP-MS studies have significantly improved our understanding of the molecular mechanisms that underlie the virus life cycle. As the number of studies and the diversity of human viruses are sizable, we focus on selected works in which MS-based proteomics has facilitated insights into key aspects of virus biology, namely virus–host cell signaling, genome replication, and assembly and maturation.
Elegant AP-MS studies have described protein interactions early during infection that permit virus entry and promote cell survival (62–64). For example, after initial binding to cell surface receptors, Kaposi sarcoma–associated herpesvirus (KSHV) is rapidly translocated in a c-Cbl-dependent manner to lipid rafts and internalized into macropinosomes (65). To define signaling molecules that stimulate c-Cbl recruitment, KSHV entry complexes were isolated via α3β1 integrin at a remarkably early time of infection (5 min postinfection) in human endothelial cells (64). EphA2 was found in a functional complex with integrins, c-Cbl, and myosin IIA and was further shown to impact macropinocytic entry and KSHV trafficking (64). In another study, Gabaev et al. (66) identified the HCMV pUL11—a glycoprotein expressed at the cell surface—as an interaction partner and ligand of the receptor tyrosine phosphatase CD45. Importantly, the authors determined that this interaction generated an immunosuppressive phenotype, as pUL11 binding reduced tyrosine phosphorylation, leading to inhibition of T cell proliferation (66).
Once inside the host cells, the virus initiates programs aimed at replication of its genome. Genome replication mechanisms vary widely across virus families, yet all viruses leverage host protein functions. AP-MS has been employed to determine the identity of such host proteins, for instance, proteins in replication complexes at viral promoters in herpesviruses (67–69), viral polymerases of influenza virus (53), and preintegration complexes in HIV-1 (70). In HCMV infection, the virion protein pUL83 was found to induce transcription from the viral major immediate-early promoter (MIEP). To explore the molecular mechanism of this function, we defined pUL83 interactions, identifying the interferon-inducible protein IFI16 (43). Using chromatin immunoprecipitation, pUL83 and IFI16 were simultaneously localized to the MIEP. Interestingly, pUL83 not only sequestered IFI16 but also co-opted it for regulation of immediate-early gene expression. The subsequent identification of IFI16 as the first known nuclear DNA sensor (70a, 71, 72) and of the pUL83-dependent inhibition of this sensing function (73) further highlights the ability of AP-MS approaches to drive discoveries in molecular virology.
Following genome replication and viral protein synthesis, a coordinated program assembles the final infectious particle. Global proteomic approaches have characterized virion compositions (see above), but the cellular factors that coordinate virion assembly are less well understood. Integration of AP-MS with imaging tools can help determine how viruses hijack host vesicular transport and secretory pathways (74–76). In Ebola virus infection, because VP40 expression alone and its transport to the plasma membrane (PM) lead to release of filamentous virus-like particles (77), Yamayoshi et al. (54) examined its host interactions in 293 cells. The COPII complex component Sec24C was identified and validated by colocalization and reciprocal isolations, and its contribution to virus-like particle formation was demonstrated by knockdown studies (54). For viruses with greater virion proteome complexity, such as herpesviruses, trafficking and assembly of >50 distinct virion components are required. To investigate the spatial-temporal relationship of virion components during HCMV virion assembly, we combined AP-MS, molecular virology, and confocal microscopy (42). During HCMV infection, GFP-expressing fusions of pUL99 or pUL32—viral proteins required for virion assembly—were found to associate with distinct host factors, including ubiquitin and clathrin. The existing functional knowledge regarding these factors led to the model that pUL99 associates with the ESCRT/TGN pathway, whereas pUL32 traffics in clathrin-coated vesicles. Interestingly, at the same time, the viral glycoprotein gB localized to endosomes, and it was not until late in infection that all three proteins merged within a large virion assembly structure (42).
Independent of virus type or stage in the life cycle, the true value of AP-MS lies in its complementarity with biochemistry, molecular biology, and optical approaches. AP-MS has the unique ability to identify and functionally profile previously unknown interactions from a complex biological state. Quantitative MS approaches and continuous developments in bioinformatics (see Computational Strategies for Protein Interaction Analysis, below) can provide a precise definition of temporal changes in virus-host interactions and are expected to continue expanding the range of AP-MS applications to virology studies.
Viral RNA–Host Protein Interactions
As many viruses do not encode their own transcription machinery, regulation of viral gene transcription is achieved via recruitment of host polymerases and host factors. Although protein AP can target host proteins to identify unknown effectors of viral transcription, an alternative approach is to target the viral genome itself (78, 79). AP-MS approaches have been adapted to identify proteins that bind viral RNA (vRNA), most often to the 5′ or 3′ untranslated regions thought to function in the initial stages of vRNA replication and translation (80). Similar to protein-based AP-MS, RNA-based AP-MS employs epitope tagging, with biotin as a tag of choice due to its ease of conjugation to nucleotides and high affinity with avidin (Kd = ∼10−14 M). For example, the first RNA-based AP-MS studies investigated cellular factors associated with the 3′ untranslated region of hepatitis C virus RNA using biotinylated oligonucleotides complementary to the vRNA (81, 82). However, nonspecific binding can be an issue given the negatively charged phosphate backbone of oligonucleotides. To address this issue, a DNA mimic, termed peptide nucleic acid, was developed to replace the phosphate backbone with amide bonds, providing reduced nonspecific binding (83). For viruses such as poliovirus that do not use host polymerases to replicate, we have shown that UV-sensitive nucleotide analogs can be incorporated selectively into the vRNA, which is ideal for studying vRNA-protein associations directly from infected cells (84). Because associations in close apposition are cross-linked, nonspecific binding is reduced through high-stringency isolation. Overall, although nucleotide-based AP-MS is not as technologically refined as its protein-based sibling, future studies targeting virus-induced transcriptional effectors will benefit greatly from these affinity-and MS-based strategy developments.
Computational Strategies for Protein Interaction Analysis
The sensitivity of AP-MS methodologies has allowed detection of lower-abundance species, which include indirect specific and nonspecific associations. Although this expands the pool of candidate binding partners, often it results in data sets too large to evaluate with functional assays. Recently developed computational platforms address this challenge, evaluating interaction specificity and functional relationships for diverse AP-MS data sets (85–87). A major advancement was the development of scoring algorithms for filtering nonspecific interactions (61, 88–90). Although these tools have yet to be used routinely for virus-host AP-MS data sets, we have used the Significance Analysis of INTeractome (SAINT) scoring algorithm (90), which models protein spectral counts between experimental and control affinity purifications (Figure 2), in our study of HSV-1 pUL46 tegument protein interactions during infection (45). One notable pUL46 interaction was the viral E3 ubiquitin ligase ICP0, which triggered a proteasome-dependent degradation of pUL46—a first example of virus-induced degradation of a viral protein. Another interaction specificity predictor, MiST, distinguished high-confidence virus-host protein interactions of epitope-tagged HIV proteins individually expressed in human cells (61). These computational approaches are effective at predicting nonspecific associations to the resin and tag, but contaminants that bind to the isolated proteins are more difficult to discern. To address this issue, an approach using metabolic labeling with stable isotopes was developed (I-DIRT; see Figure 2) (91, 92).
As proteins usually exist within multiple complexes with distinct functions, high-scoring candidate interactions can be further evaluated through functional classification and network-based relationships. Bioinformatic network tools, including STRING and GeneMania (93, 94), can score direct and indirect protein functional relationships using public database repositories, such as IntAct and Reactome (95, 96). Virus-centric repositories have also been established, including ViralZone(97) and VirusMINT (98), as well as GPS-Prot, which focuses on HIV-host interactions (99). As bioinformatic strategies are a critical aspect of interaction studies, this review includes two interactive tutorials for construction of functional protein interaction networks (Supplemental Tutorial 1) and evaluation of protein interaction specificity (Supplemental Tutorial 2).
To supplement interaction networks, recent studies have demonstrated the utility of overlaying annotation and quantitative information to identify high-value candidates for focused biological validation (92). Future AP-MS studies will benefit from increased use of quantitative MS (100), particularly for determining complex stability and absolute abundances within viral complexes. The continued refinement of computational tools will ensure reliable cataloging of virus-host interactions and streamlined web-based interfaces to fuel the cycle of hypothesis generation and testing.
MASS SPECTROMETRY–BASED ANALYSIS OF GLOBAL PROTEOMES DURING VIRAL INFECTION
Intracellular Proteome Expression and Reorganization
Viral infection triggers a drastic transformation in intracellular proteomes, including de novo expression of viral proteins and reorganization of organelles. With the ability of MS-based proteomics to simultaneously quantify thousands of proteins, an unbiased proteome profiling of infected cells permits the discovery of new viral gene products not previously predicted or observed. Recently, MS and ribosome occupancy profiling were used to identify and temporally resolve unique, previously unreported HCMV polypeptides (>50) and transcripts (>550) during infection (101). Sequences of uncharacterized proteins and transcripts could be mapped to nonannotated or overlapping annotated HCMV genomic loci and corresponded with alternative splicing or translation sites within known HCMV gene transcripts. This study highlighted the necessity of MS approaches to reveal the true genetic complexity and protein diversity of even well-studied viruses.
Moreover, as alterations in protein localizations and abundances within subcellular compartments reflect host defense responses and viral morphogenesis processes, several studies have utilized biochemical fractionation or affinity isolation techniques in conjunction with MS to characterize target organelle subproteomes. These enrichment strategies confer increased sensitivity, providing greater molecular details of host and viral processes. For instance, metabolic labeling using stable isotope labeling in cell culture (SILAC; see Figure 2) was employed to quantify virus-induced expression changes in Golgi-enriched fractions from mouse hepatitis virus (MHV)-infected hepatocytes (102). Here, mock-and virus-infected cells were labeled with light-or heavy-isotopic amino acids and then mixed 6 h postinfection, allowing enrichment of Golgi membranes from a single isolation. Overall, SILAC is well suited for minimizing quantification variance from multistep proteomic approaches. Following MS quantification, selected differentially regulated Golgi-resident and vesicle trafficking factors, such as SEC22B and the Ragulator complex protein LAMTOR1, were shown to alter MHV virion assembly using RNA interference and overexpression assays. Other noteworthy studies have coupled nuclear-cytoplasmic biochemical fractionation with quantitative MS to delineate modulation of compartment-specific pathway functions during HSV-1 (103), influenza A (104), adenovirus (105), and respiratory syncytial virus (106) infection.
Subproteome studies have also been extended to the PM. Functioning at the interface between the cell and the extracellular space, the PM is a site of intercellular signal integration and propagation. Furthermore, viruses that derive their envelopes from cellular membranes capture integral viral and cellular proteins necessary for subsequent infection. Due to their roles in viral morphogenesis and their accessibility to small molecules, PM constituents are promising targets for antivirals. To profile cell surface proteins by MS, PM enrichment by chemical derivatization–affinity purification confers reduced organelle contamination and improved detection of low-abundance membrane receptors (107, 108). Berro et al. (109) used cell surface biotinylation coupled with streptavidin affinity chromatography to investigate PM changes in HIV-1 latently infected lymphocytes. Differentially expressed integral and membrane-associated proteins, including the antiapoptotic proteins XIAP and Mcl-1, were identified and validated by orthogonal techniques. Interestingly, drug inhibition of XIAP and Mcl-1 expression sensitized latently infected cells to apoptosis. More recently, during latent HCMV infection, the degradation of the PM drug transporter MRP1 by the viral protein pUL138 was discovered by SILAC-MS after PM enrichment (110). Loss of MRP1 conferred sensitivity to cytotoxic MRP1 drug substrates in latently infected monocytes and hematopoietic progenitors. Therefore, these studies propose therapeutic strategies for the specific elimination of cells latently infected with HIV or HCMV. Our lab used affinity isolation and label-free MS quantification (Figure 2) to investigate HCMV-induced PM changes across productive immediate-early, early, and late stages of infection (111). The low-density lipoprotein receptor LRP1, which negatively regulates cholesterol influx into cells (15), was strongly upregulated early in infection. Disruption of LRP1 activity increased intracellular and progeny HCMV envelope cholesterol levels, which was correlated with greater virion infectivity. Similar to our HCMV studies, depletion of cholesterol was observed to compromise infectivity of other enveloped viruses, such as HIV-1, hepatitis B and C viruses, and influenza (112–115). Thus, the LRP1 PM recruitment could reflect a general antiviral response that starves the infected cell of cholesterol, promoting the production of infection-incompetent virions. Altogether, quantitative MS–based subproteomic profiling heightens our understanding of host-virus dynamics by providing sensitivity and spatial resolution during infection.
Cellular Secretomes During Infection
Viral infection also significantly alters the proteome within the extracellular space. This is critical from the host perspective, as the virus-induced secretion of signaling factors mediates the onset of innate and adaptive immunity (116). Secreted cytokines, inflammation effectors, and cell adhesion molecules recruit and activate circulating blood leukocytes and alert neighboring cells. These events initiate tissue destruction and antiviral cellular programs at the infection epicenter, abating viral dissemination. Importantly, approximately 10% of the human genome open reading frames encode proteins that are secreted (117). Considering their physiological importance and protein richness, the characterization of cellular secretomes by MS can drive key insights into the molecular mechanisms of host immunological responses.
Due to their roles in immunosurveillance, monocyte-derived macrophages have been a focus for many MS-based secretome analyses (118–121). In one study, isobaric tagging and tandem MS (Figure 2 and Table 1) quantified the secretion of >400 proteins from HSV-1-infected macrophages (121). Interestingly, bioinformatic analyses revealed that 80% of these are exosomal proteins, suggesting export through nonconventional, vesicle-mediated pathways. Furthermore, though the HSV-1-induced secretome was composed of known secreted danger signals and immunomodulators, interferon-inducible proteins not known to be secreted were also detected, including IFIT1, STAT1, and MxA. Finally, the authors found no secretion of the proinflammatory cytokine interleukin-1β, a process known to depend on the inflammasome—a caspase-1-containing complex (122). These results implicated an HSV-1 mechanism for antagonizing inflammasome activity, which was subsequently demonstrated (123). Hence, in a single MS-based secretome study, the authors were able to unbiasedly confirm and discover a variety of cellular secretion responses to viral infection and to predict an immunoevasive function of HSV-1. Interestingly, the influenza A–induced secretome (120) exhibited significant overlap with those of HSV-1 (121) and HCMV (124) in cytokines, cathepsin proteases, and danger-associated signals, suggesting the activation of common downstream signaling pathways in response to different viruses. Secreted cathepsin protease activity was shown to be required for both inflammasome stimulation and apoptosis in response to influenza (120). These studies illustrate the ability of quantitative MS to complement traditional antibody array–based analyses of secretomes and reveal details of cellular immune responses to viral infection.
MS-based secretome analyses have also identified factors that contribute to virus-associated pathologies. Accruing data suggest that chronic inflammation triggered by persistent, lifelong HCMV infection contributes to vascular pathologies, including atherosclerosis and allograft rejection (125–127). Considering recent evidence implicating angiogenesis in HCMV-associated vasculopathies (128), Dumortier et al. (124) profiled the secretome of HCMV-infected human dermal fibroblasts. Pathway analysis revealed that the HCMV-specific secretome was enriched for proangiogenic and wound healing factors. Indeed, supernatants from HCMV-infected cells induced angiogenesis and wound healing responses in primary human endothelial cells. Additionally, other MS-based studies have investigated the mechanisms of HIV-associated neurodegeneration through secretome profiling of HIV-infected phagocytes (118, 119). Together, these studies substantiate the effectiveness of MS in understanding the immunological and pathophysiological consequences of viral infection through characterization of cellular secretion patterns.
ROLES OF PROTEIN POSTTRANSLATIONAL MODIFICATIONS IN VIRUS INFECTION
In addition to changes in abundances and subcellular localizations, proteins are subject to regulation by posttranslational modifications (PTMs), which, during an infection, can be modulated by either cell-or virus-encoded factors. PTMs have been recognized to regulate an expanding array of protein properties, for example, stability, localizations, activities, and interactions. Furthermore, the list of existing PTMs is growing faster than our understanding of the functions they mediate. Without a doubt, the temporal and spatial definition of PTMs during infection can provide critical mechanistic knowledge regarding important viral and cellular processes. To this end, MS-based proteomic strategies conferring high sensitivity have been developed for the unambiguous identification, quantification, and localization of PTMs within protein structures.
Several MS studies have focused on intracellular PTM dynamics on viral proteins during infection. For example, Bell et al. (129) discovered 95 phosphorylations and 10 ubiquitinations within HSV-1 proteins enriched for cellular trafficking and immune evasion functions. Although this study did not functionally assess the identified PTMs, it highlights the sensitivity and breadth of modern MS-based proteomics with respect to PTM identification and the scarcity of information regarding viral PTMs in the context of infection. Analysis of these phosphorylation sites revealed great sequence degeneracy, suggesting that the kinases mediating these phosphorylation events either have altered behavior during infection or have not yet been characterized. Global PTM studies are, therefore, effective ways of building comprehensive databases for future functional characterization of modifications and the pathways that regulate them.
The ability of global PTM studies to access lower-abundance modifications has benefited from a variety of affinity enrichment strategies (as reviewed in 130), including PTM-specific antibodies and immobilized metals, such as Fe and TiO2. Recently, TiO2 phospho-enrichment was successfully applied for mapping the phosphoproteome of influenza A–infected human cell lines (131). Here, an identified phosphorylation within the influenza A nucleoprotein was found to be necessary for efficient viral replication. As an alternative to PTM-specific enrichment, immunoaffinity isolation of a protein of interest allows the simultaneous detection of various PTMs within the target protein, aiding the investigation of PTM-dependent protein function. For instance, the HCMV immunosuppressant protein pUL83 (73) was known for several decades to be heavily phosphorylated (132); however, the sites and functions of these phosphorylations remained unclear. Recently, we identified eight phosphosites within immunoaffinity-isolated pUL83 and established that one phosphorylation, under the control of host kinases, functions to block pUL83-dependent dispersal of cellular innate immunity effectors (73). Similarly, several identified phosphorylations within the multifunctional HSV-1 protein ICP0 were found to modulate its ubiquitin ligase, transactivation, and localization activities using an AP-MS/mutagenesis approach (133). In addition to studying intracellular viral PTMs, several studies have profiled modifications within purified mature virions (6, 131, 134, 135). This strategy may distinguish PTMs important for virion structure, entry, and dissemination from those added de novo to regulate intracellular viral protein functions over the course of infection.
Dynamic PTMs on host proteins have also been demonstrated to mediate critical cellular functions related to viral infection. For instance, the DNA sensor IFI16 is an effector of innate immune responses during viral infection, detecting viral DNA in both the nucleus and the cytoplasm (71–73, 136, 137). Using AP-MS, we identified two acetylations within its nuclear localization signal that modulate IFI16 nuclear-cytoplasmic distribution, effectively expanding its range of immunosurveillance to both subcellular compartments (71). Interestingly, multiple acetylations within other innate immunity signaling components—OAS2, TRIM25, IRAK4, and RIG-I—were identified using anti-acetyllysine antibody enrichment and MS (138). As drug inhibition of cellular deacetylases strongly attenuates pathogen-and immunogen-induced immune responses (139–141), it is tempting to speculate that acetylation plays a key role in modulating immune signaling pathways. Further studies of acetylation-dependent functions are required to strengthen this hypothesis.
PTM-dependent pathways have also been characterized by investigating the enzymes that regulate these modifications using MS-based chemoproteomic approaches. As PTM-modifying pathways are commonly implicated in disease pathogenesis, a plethora of small-molecule inhibitors have been developed to target them. Taking advantage of this concept, metabolic labeling was effectively coupled with AP of cellular kinases via immobilized inhibitors to quantify cellular kinome changes during HCMV infection (142). The authors demonstrated that drug inhibition of AMPK, a kinase found to be upregulated during infection, decreased virus growth, in agreement with a previous RNA interference screen of kinases during HCMV infection (143). Whereas this chemoproteomic strategy assessed relative protein abundances, other MS-amenable techniques have been designed to measure enzyme activity. For instance, hemagglutinin-affinity-tagged ubiquitin derivatives were developed as active site–directed probes for deubiquitinating enzymes (DUBs) (144). By using AP of active DUBs and MS analysis, the activities of several DUBs were found to be strongly upregulated in EBV-infected lymphocytes (145). Unique DUB activity was additionally identified following EBV establishment of latency and correlated with changes in cell proliferation rates. Altogether, these studies provide just a glimpse of the value that defining PTMs in the context of infection can provide for understanding the molecular details of viral infection.
CONCLUSIONS AND FUTURE PERSPECTIVES
Intimate virus-host coevolution has shaped viruses in their every facet. Their genomic organization, virion structure, and protein arsenal are all finely tuned to exploit and manipulate the molecular processes of their permissive host cells. As a consequence, viruses must induce complex alterations in intracellular protein abundance and activity to dismantle inhibitory host functions and promote their own propagation. For this reason, modern MS-based proteomics, in conjunction with orthogonal approaches, is perfectly suited for acquiring a systems understanding of virus-host dynamics. The sensitivity, speed, accuracy, and analytical breadth conferred by recent advances in MS instrumentation, sample preparation, and bioinformatics have allowed researchers to successfully elucidate the molecular details of viruses and their interactions with hosts.
Thus far, global proteome studies have provided a wealth of quantitative information regarding intracellular protein identifications and abundances during viral infection. Paired with MS detection, parallel genomic and bioinformatic analyses have enabled functional categorization of host response pathways and identification of previously uncharacterized viral gene products. However, the dynamic range and spatial-temporal insight of current MS-based proteomic approaches are still limiting factors. These issues can be partly mitigated using organelle enrichment strategies performed across multiple time points of infection, which offer not only protein temporal kinetics and subcellular localizations but also increased sensitivity for subtle proteomic changes otherwise undetected in global analyses. These fractionation approaches come with their own challenges, but organelle enrichment techniques are continuously improving and show promise for uncovering mechanisms of virion assembly. Apart from sample fractionation, targeted detection using selected reaction monitoring (SRM) (Figure 2 and Table 1) also holds great potential for accurate quantification of low-abundance proteins or PTMs (146). Future incorporation of SRM in viral studies can provide relative and absolute quantification of critical factors, and possibly stoichiometry within infection-induced structures; in addition, the application of SRM to high-throughput identification of biomarkers of infection in clinical settings is a particularly exciting future direction.
Apart from global and subproteome studies, immunoaffinity isolation coupled with MS has provided invaluable insights into the dynamic functions and components of viral and cellular protein complexes during infection. These MS-based analyses come with their own challenges, including distinguishing specific from nonspecific associations. The incorporation of computational analyses to predict interaction specificity into proteomic workflows represents a technical milestone in resolving these issues. Integration of these strategies with the metabolic labeling strategy I-DIRT can offer additional validation of interaction specificity and, importantly, a measure of the relative stability of a protein (92). Furthermore, because functional interaction networks constructed from single affinity purifications cannot differentiate direct and indirect protein interactions within complexes, improvements in chemical cross-linking reagents and bioinformatic tools will accelerate the definition of protein interface topologies, as elegantly demonstrated for virus-plant interactions (147). Finally, characterizing virus-host protein interactions at multiple stages of infection can be laborious and expensive (due to extensive instrument analysis time). Isobaric tag labeling (Figure 2 and Table 1) after immunoisolation of virus-host protein complexes may provide an effective multiplexing strategy to simultaneously analyze multiple stages of infection. Altogether, the future incorporation of the above techniques into viral studies will help expand our knowledge of the viral and host protein complexes that mediate critical processes during infection.
In recent years, it has become increasingly evident that viral infection modulates the epigenetic landscape of the cell. This is accomplished via interactions of viral proteins with chromatin-remodeling enzymes and virus-induced changes in histone PTMs and DNA methylation profiles. Furthermore, the discovery that nuclear-replicating DNA viral genomes associate with core histones implicates cellular effectors of chromatinization as important regulators of viral gene expression. Integration of epigenetic studies with MS technologies will aid in delineating the mechanisms by which viruses exploit cellular chromatin structural components and transcriptional machinery.
In summary, MS-based proteomics has already significantly contributed to multifaceted discoveries in virology. Ongoing improvements in MS and bioinformatics are expected to further expand the range of applications and establish MS as a routine tool in virology studies.
Supplementary Material
SUMMARY POINTS.
The integration of mass spectrometry–based proteomic platforms with orthogonal techniques and research disciplines provides mechanistic insights into viral infection that are hard to obtain by traditional approaches.
Tandem mass spectrometry–based peptide sequencing and native ion mobility mass spectrometry are powerful tools for elucidating virion protein composition and intermediate capsid structures, respectively.
The development of diverse affinity purification–mass spectrometry workflows, including fluorescent affinity probes, quantitative mass spectrometry, and bioinformatic analyses, has made it possible to define spatially and temporally dynamic virus-host protein interactions during viral infections.
Comprehensive and unbiased whole-cell proteome studies offer a systems view of infection and delineate previously uncharacterized cellular response pathways and viral gene products.
Biochemical and affinity-based subcellular fractionation techniques coupled with mass spectrometry can sensitively analyze organelle-specific host and viral processes during infection, pointing to infection biomarkers and new targets for antiviral therapeutics.
Mass spectrometry–based secretome analyses of virus-infected immune cells can ascertain secreted immunomodulatory factors that contribute to host immunological responses and virus-associated pathologies.
Viral and host posttranslational modifications that modulate a vast range of protein and pathway functions can be conclusively identified and quantified using various mass spectrometry approaches.
FUTURE ISSUES.
Affinity purification–mass spectrometry in conjunction with label-free and metabolic labeling approaches can reveal protein interaction specificity, as well as the relative stability of interactions within virus-host protein complexes during infection.
The use of chemical cross-linking reagents within affinity purification–mass spectrometry workflows can determine direct virus-host protein interactions, identifying intermolecular protein interfaces to reconstruct protein complex topology.
For hypothesis-driven viral proteomics, targeted mass spectrometry approaches, such as selected reaction monitoring, allow sensitive quantification of proteins or posttranslational modifications within complex biological samples, making them promising candidates for clinical applications.
Mass spectrometry approaches provide the opportunity to begin defining the virus-induced changes in the host epigenetic landscape by mapping histone modifications and characterizing the functions of chromatin-remodeling enzymes in the context of infection.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants (NIDA DP1DA026192 and NIAID R21AI102187) to I.M.C., an HFSPO award (RGY0079/2009-C) to I.M.C., a New Jersey Commission on Cancer Research postdoctoral fellowship to T.M.G., and an American Heart Association scholarship to B.A.D.
Glossary
- Posttranslational modifications (PTMs):
functional groups that modify amino acids in proteins to regulate localization and function
- Affinity purification–mass spectrometry (AP-MS):
enrichment of a cellular structure or complex and analysis by mass spectrometry
- Significance Analysis of INTeractome (SAINT):
computational approach that uses label-free data to predict specific protein interactions
- I-DIRT:
a metabolic labeling AP-MS method for distinguishing specific from nonspecific interactions and relative interaction stability
- Stable isotope labeling of amino acids in cell culture (SILAC):
metabolic labeling of proteins for relative MS-based quantification
- Selected reaction monitoring (SRM):
targeted fragmentation of specific peptides, often used in quantitative applications
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
The Annual Review of Virology is online at virology.annualreviews.org
LITERATURE CITED
- 1.Milo R 2013. What is the total number of protein molecules per cell volume? A call to rethink somepublished values. BioEssays 35:1050–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grunewald K, Cyrklaff M. 2006. Structure of complex viruses and virus-infected cells by electron cryotomography. Curr. Opin. Microbiol 9:437–42 [DOI] [PubMed] [Google Scholar]
- 3.Kuznetsov YG, McPherson A. 2011. Atomic force microscopy in imaging of viruses and virus-infectedcells. Microbiol. Mol. Biol. Rev 75:268–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johannsen E, Luftig M, Chase MR, Weicksel S, Cahir-McFarland E, et al. 2004. Proteins of purified Epstein–Barr virus. Proc. Natl. Acad. Sci. USA 101:16286–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kattenhorn LM, Mills R, Wagner M, Lomsadze A, Makeev V, et al. 2004. Identification of proteins associated with murine cytomegalovirus virions. J. Virol 78:11187–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kramer T, Greco TM, Enquist LW, Cristea IM. 2011. Proteomic characterization of pseudorabies virus extracellular virions. J. Virol 85:6427–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loret S, Guay G, Lippé R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol 82:8605–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, et al. 2004. Identification of proteinsin human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol 78:10960–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vidick S, Leroy B, Palmeira L, Machiels B, Mast J, et al. 2013. Proteomic characterization of murid herpesvirus 4 extracellular virions. PLoS ONE 8:e83842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung CS, Chen CH, Ho MY, Huang CY, Liao CL, Chang W. 2006. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol 80:2127–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manes NP, Estep RD, Mottaz HM, Moore RJ, Clauss TRW, et al. 2008. Comparative proteomics of human monkeypox and vaccinia intracellular mature and extracellular enveloped virions. J. Proteome Res 7:960–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoder JD, Chen TS, Gagnier CR, Vemulapalli S, Maier CS, Hruby DE. 2006. Pox proteomics: mass spectrometry analysis and identification of vaccinia virion proteins. Virol. J 3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. 2001. Proteolytic 18O labeling for comparative proteomics: model studies with two serotypes of adenovirus. Anal. Chem 73:2836–42 [DOI] [PubMed] [Google Scholar]
- 14.Davison AJ, Davison MD. 1995. Identification of structural proteins of channel catfish virus by mass spectrometry. Virology 206:1035–43 [DOI] [PubMed] [Google Scholar]
- 15.Terrand J, Bruban V, Zhou L, Gong W, El Asmar Z, et al. 2009. LRP1 controls intracellular cholesterol storage and fatty acid synthesis through modulation of Wnt signaling. J. Biol. Chem 284:381–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spurgers KB, Alefantis T, Peyser BD, Ruthel GT, Bergeron AA, et al. 2010. Identification of essential filovirion-associated host factors by serial proteomic analysis and RNAi screen. Mol. Cell. Proteomics 9:2690–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, et al. 2006. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol 80:9039–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linde ME, Colquhoun DR, Ubaida Mohien C, Kole T, Aquino V, et al. 2013. The conserved set of host proteins incorporated into HIV-1 virions suggests a common egress pathway in multiple cell types. J. Proteome Res 12:2045–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radhakrishnan A, Yeo D, Brown G, Myaing MZ, Iyer LR, et al. 2010. Protein analysis of purified respiratory syncytial virus particles reveals an important role for heat shock protein 90 in virus particle assembly. Mol. Cell. Proteomics 9:1829–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saphire AC, Gallay PA, Bark SJ. 2006. Proteomic analysis of human immunodeficiency virus using liquid chromatography/tandem mass spectrometry effectively distinguishes specific incorporated host proteins. J. Proteome Res 5:530–38 [DOI] [PubMed] [Google Scholar]
- 21.Shaw ML, Stone KL, Colangelo CM, Gulcicek EE, Palese P. 2008. Cellular proteins in influenza virusparticles. PLoS Pathog. 4:e1000085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stegen C, Yakova Y, Henaff D, Nadjar J, Duron J, Lippé R. 2013. Analysis of virion-incorporated host proteins required for herpes simplex virus type 1 infection through a RNA interference screen. PLoS ONE 8:e53276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uetrecht C, Barbu IM, Shoemaker GK, van Duijn E, Heck AJR. 2011. Interrogating viral capsid assembly with ion mobility–mass spectrometry. Nat. Chem 3:126–32 [DOI] [PubMed] [Google Scholar]
- 24.Pease LF III. 2012. Physical analysis of virus particles using electrospray differential mobility analysis. Trends Biotechnol. 30:216–24 [DOI] [PubMed] [Google Scholar]
- 25.Alexander CG, Jürgens MC, Shepherd DA, Freund SMV, Ashcroft AE, Ferguson N. 2013. Thermo-dynamic origins of protein folding, allostery, and capsid formation in the human hepatitis B virus core protein. Proc. Natl. Acad. Sci. USA 110:E2782–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vörös J, Urbanek A, Rautureau GJP, O’Connor M, Fisher HC, et al. 2014. Large-scale production,¨ structural and biophysical characterizations of the human hepatitis B viruspolymerase.J.Virol 88:2584–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shepherd DA, Holmes K, Rowlands DJ, Stonehouse NJ, Ashcroft AE. 2013. Using ion mobility spectrometry–mass spectrometry to decipher the conformational and assembly characteristics of the hepatitis B capsid protein. Biophys. J 105:1258–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shoemaker GK, van Duijn E, Crawford SE, Uetrecht C, Baclayon M, et al. 2010. Norwalk virus assembly and stability monitored by mass spectrometry. Mol. Cell. Proteomics 9:1742–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snijder J, Uetrecht C, Rose RJ, Sanchez-Eugenia R, Marti GA, et al. 2013. Probing the biophysical interplay between a viral genome and its capsid. Nat. Chem 5:502–9Native MS and atomic force microscopy reveal the constraints of Triatoma virus capsid packaging and uncoating.
- 30.Tuma R, Coward LU, Kirk MC, Barnes S, Prevelige PE Jr. 2001. Hydrogen-deuterium exchange as aprobe of folding and assembly in viral capsids. J. Mol. Biol 306:389–96 [DOI] [PubMed] [Google Scholar]
- 31.Monroe EB, Kang S, Kyere SK, Li R, Prevelige PE Jr. 2010. Hydrogen/deuterium exchange analysis of HIV-1 capsid assembly and maturation. Structure 18:1489–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miteva YV, Budayeva HG, Cristea IM. 2013. Proteomics-based methods for discovery, quantification, and validation of protein-protein interactions. Anal. Chem 85:749–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rowles DL, Terhune SS, Cristea IM. 2013. Discovery of host-viral protein complexes during infection. Methods Mol. Biol 1064:43–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fontaine-Rodriguez EC, Taylor TJ, Olesky M, Knipe DM. 2004. Proteomics of herpes simplex virus infected cell protein 27: association with translation initiation factors. Virology 330:487–92 [DOI] [PubMed] [Google Scholar]
- 35.Taylor TJ, Knipe DM. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol 78:5856–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guise AJ, Budayeva HG, Diner BA, Cristea IM. 2013. Histone deacetylases in herpesvirus replication and virus-stimulated host defense. Viruses 5:1607–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cristea IM, Williams R, Chait BT, Rout MP. 2005. Fluorescent proteins as proteomic probes. Mol. Cell. Proteomics 4:1933–41 [DOI] [PubMed] [Google Scholar]
- 38.Cristea IM, Carroll JW, Rout MP, Rice CM, Chait BT, MacDonald MR. 2006. Tracking and elucidating Alphavirus-host protein interactions. J. Biol. Chem 281:30269–78The first temporal AP-MS study of virus-host protein interactions during the progression of an infection.
- 39.Frolova E, Gorchakov R, Garmashova N, Atasheva S, Vergara LA, Frolov I. 2006. Formation of nsP3-specific protein complexes during Sindbis virus replication. J. Virol 80:4122–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cristea IM, Rozjabek H, Molloy KR, Karki S, White LL, Rice CM, et al. 2010. Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: role for G3BP1 and G3BP2 in virus replication. J. Virol 84:6720–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moorman NJ, Cristea IM, Terhune SS, Rout MP, Chait BT, Shenk T. 2008. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3:253–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moorman NJ, Sharon-Friling R, Shenk T, Cristea IM. 2010. A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Mol. Cell. Proteomics 9:851–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cristea IM, Moorman NJ, Terhune SS, Cuevas CD, O’Keefe ES, et al. 2010. Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J. Virol 84:7803–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terhune SS, Moorman NJ, Cristea IM, Savaryn JP, Cuevas-Bennett C, et al. 2010. Human cytomegalovirus UL29/28 protein interacts with components of the NuRD complex which promote accumulation of immediate-early RNA. PLoS Pathog. 6:e1000965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin AE, Greco TM, Döhner K, Sodeik B, Cristea IM. 2013. A proteomic perspective of inbuilt viral¨ protein regulation: pUL46 tegument protein is targeted for degradation by ICP0 during herpes simplex virus type 1 infection. Mol. Cell. Proteomics 12:3237–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kramer T, Greco TM, Taylor MP, Ambrosini AE, Cristea IM, Enquist LW. 2012. Kinesin-3 mediates axonal sorting and directional transport of alphaherpesvirus particles in neurons. Cell Host Microbe 12:806–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kratchmarov R, Kramer T, Greco TM, Taylor MP, Ch’ng TH, et al. 2013. Glycoproteins gE and gIare required for efficient KIF1A-dependent anterograde axonal transport of alphaherpesvirus particles in neurons. J. Virol 87:9431–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Youn S, Li T, McCune BT, Edeling MA, Fremont DH, et al. 2012. Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of West Nile virus. J. Virol 86:7360–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol 17:1030–32 [DOI] [PubMed] [Google Scholar]
- 50.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, et al. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463–73 [DOI] [PubMed] [Google Scholar]
- 51.Wagenaar TR, Moss B. 2007. Association of vaccinia virus fusion regulatory proteins with the multicomponent entry/fusion complex. J. Virol 81:6286–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka M, Sata T, Kawaguchi Y. 2008. The product of the herpes simplex virus 1 UL7 gene interacts with a mitochondrial protein, adenine nucleotide translocator 2. Virol. J 5:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayer D, Molawi K, Martínez-Sobrido L, Ghanem A, Thomas S, et al. 2007. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J. Proteome Res 6:672–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamayoshi S, Noda T, Ebihara H, Goto H, Morikawa Y, et al. 2008. Ebola virus matrix protein VP40 uses the COPII transport system for its intracellular transport. Cell Host Microbe 3:168–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jäger S, Kim DY, Hultquist JF, Shindo K, LaRue RS, et al. 2012. Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection. Nature 481:371–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naji S, Ambrus G, Cimermančič P, Reyes JR, Johnson JR, et al. 2012. Host cell interactome of HIV-1 Rev includes RNA helicases involved in multiple facets of virus production. Mol. Cell. Proteomics 11:M111.015313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chi YH, Semmes OJ, Jeang KT. 2011. A proteomic study of TAR-RNA binding protein (TRBP)-associated factors. Cell Biosci. 1:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee SJ, Shim HY, Hsieh A, Min JY, Jung G. 2009. Hepatitis B virus core interacts with the host cell nucleolar protein, nucleophosmin 1. J. Microbiol 47:746–52 [DOI] [PubMed] [Google Scholar]
- 59.Mukerji J, Olivieri KC, Misra V, Agopian KA, Gabuzda D. 2012. Proteomic analysis of HIV-1 Nef cellular binding partners reveals a role for exocyst complex proteins in mediating enhancement of intercellular nanotube formation. Retrovirology 9:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smirnova EV, Collingwood TS, Bisbal C, Tsygankova OM, Bogush M, et al. 2008. TULA proteins bind to ABCE-1, a host factor of HIV-1 assembly, and inhibit HIV-1 biogenesis in a UBA-dependent fashion. Virology 372 [DOI] [PubMed] [Google Scholar]
- 61.Jäger S, Cimermančič P, Gulbahce N, Johnson JR, McGovern KE, et al. 2012. Global landscape of HIV-human protein complexes. Nature 481:365–70First interactome of all proteins encoded by a viral genome using individually expressed viral proteins.
- 62.Gordón-Alonso M, Sala-Valdés M, Rocha-Perugini V, Pérez-Hernández D, López-Martín S, et al. 2012. EWI-2 association with α-actinin regulates T cell immune synapses and HIV viral infection. J. Immunol. 189:689–700 [DOI] [PubMed] [Google Scholar]
- 63.Tran K, Kamil JP, Coen DM, Spector DH. 2010. Inactivation and disassembly of the anaphase-promoting complex during human cytomegalovirus infection is associated with degradation of the APC5 and APC4 subunits and does not require UL97-mediated phosphorylation of Cdh1. J. Virol 84:10832–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chakraborty S, Veettil MV, Bottero V, Chandran B. 2012. Kaposi’s sarcoma–associated herpesvirus interacts with EphrinA2 receptor to amplify signaling essential for productive infection. Proc. Natl. Acad. Sci. USA 109:E1163–72 Mechanistic insight into macropinocytic entry facilitated by AP-MS of viral entry complexes at 5 min postinfection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raghu H, Sharma-Walia N, Veettil MV, Sadagopan S, Chandran B. 2009. Kaposi’s sarcoma–associated herpesvirus utilizes an actin polymerization–dependent macropinocytic pathway to enter human dermal microvascular endothelial and human umbilical vein endothelial cells. J. Virol 83:4895–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gabaev I, Steinbrück L, Pokoyski C, Pich A, Stanton RJ, et al. 2011. The human cytomegalovirus UL11 protein interacts with the receptor tyrosine phosphatase CD45, resulting in functional paralysis of T cells. PLoS Pathog. 7:e1002432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao Y, Colletti K, Pari GS. 2008. Identification of human cytomegalovirus UL84 virus-and cell-encoded binding partners by using proteomics analysis. J. Virol 82:96–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kagele D, Rossetto CC, Tarrant MT, Pari GS. 2012. Analysis of the interactions of viral and cellular factors with human cytomegalovirus lytic origin of replication, oriLyt. Virology 424:106–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lester JT, DeLuca NA. 2011. Herpes simplex virus 1 ICP4 forms complexes with TFIID and mediatorin virus-infected cells. J. Virol 85:5733–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raghavendra NK, Shkriabai N, Graham RL, Hess S, Kvaratskhelia M, Wu L. 2010. Identification of host proteins associated with HIV-1 preintegration complexes isolated from infected CD4+ cells. Retrovirology 7:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70a.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, et al. 2011. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma–associated herpesvirus infection. Cell Host Microbe 9:363–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li T, Diner BA, Chen J, Cristea IM. 2012. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc. Natl. Acad. Sci. USA 109:10558–63Acetylation modulates the localization of the first viral DNA sensor that functions in the nucleus.
- 72.Orzalli MH, DeLuca NA, Knipe DM. 2012. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc. Natl. Acad. Sci. USA 109:E3008–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li T, Chen J, Cristea IM. 2013. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe 14:591–99 Host-mediated phosphorylation of a viral protein during infection inhibits its function in viral immune evasion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Emmott E, Munday D, Bickerton E, Britton P, Rodgers MA, et al. 2013. The cellular interactome of the coronavirus infectious bronchitis virus nucleocapsid protein and functional implications for virus biology. J. Virol 87:9486–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Greninger AL, Knudsen GM, Betegon M, Burlingame AL, DeRisi JL. 2012The3A protein from multiple picornaviruses utilizes the Golgi adaptorproteinACBD3 to recruit PI4KIIIβ. J. Virol 86:3605–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lemnitzer F, Raschbichler V, Kolodziejczak D, Israel L, Imhof A, et al. 2013. Mouse cytomegalovirus egress protein pM50 interacts with cellular endophilin-A2. Cell Microbiol. 15:335–51 [DOI] [PubMed] [Google Scholar]
- 77.Noda T, Sagara H, Suzuki E, Takada A, Kida H, Kawaoka Y. 2002. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J. Virol 76:4855–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vidalain PO, Tangy F.2010. Virus-host protein interactions in RNA viruses. Microbes Infect. 12:1134–43 [DOI] [PubMed] [Google Scholar]
- 79.Vashist S, Urena L, Chaudhry Y, Goodfellow I. 2012. Identification of RNA-protein interaction networks involved in the norovirus life cycle. J. Virol 86:11977–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Galán C, Sola I, Nogales A, Thomas B, Akoulitchev A, et al. 2009. Host cell proteins interacting with the 3′ end of TGEV coronavirus genome influence virus replication. Virology 391:304–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harris D, Zhang Z, Chaubey B, Pandey VN. 2006. Identification of cellular factors associated with the 3′-nontranslated region of the hepatitis C virus genome. Mol. Cell. Proteomics 5:1006–18 [DOI] [PubMed] [Google Scholar]
- 82.Tingting P, Caiyun F, Zhigang Y, Pengyuan Y, Zhenghong Y. 2006. Subproteomic analysis of the cellular proteins associated with the 3′ untranslated region of the hepatitis C virus genome in human liver cells. Biochem. Biophys. Res. Commun 347:683–91 [DOI] [PubMed] [Google Scholar]
- 83.Upadhyay A, Dixit U, Manvar D, Chaturvedi N, Pandey VN. 2013. Affinity capture and identification of host cell factors associated with hepatitis C virus (+) strand subgenomic RNA. Mol. Cell. Proteomics 12:1539–52A novel peptide nucleic acid–neamine conjugate for in situ capturing of viral RNA–host interactions.
- 84.Lenarcic EM, Landry DM, Greco TM, Cristea IM, Thompson SR. 2013. Thiouracil cross-linking mass spectrometry: a cell-based method to identify host factors involved in viral amplification. J. Virol 87:8697–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koh GC, Porras P, Aranda B, Hermjakob H, Orchard SE. 2012Analyzing protein-protein interaction networks. J. Proteome Res 11:2014–31 [DOI] [PubMed] [Google Scholar]
- 86.Nesvizhskii AI. 2012. Computational and informatics strategies for identification of specific protein interaction partners in affinity purification mass spectrometry experiments. Proteomics 12:1639–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sardiu ME, Washburn MP. 2011. Building protein-protein interaction networks with proteomics and informatics tools. J. Biol. Chem 286:23645–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sowa ME, Bennett EJ, Gygi SP, Harper JW. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138:389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sardiu ME, Cai Y,Jin J, Swanson SK, Conaway RC, et al. 2008. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc. Natl. Acad. Sci. USA 105:1454–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, et al. 2011. SAINT: probabilistic scoring of affinity purification–mass spectrometry data. Nat. Methods 8:70–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tackett AJ, DeGrasse JA, Sekedat MD, Oeffinger M, Rout MP, Chait BT. 2005. I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J. Proteome Res 4:1752–56 [DOI] [PubMed] [Google Scholar]
- 92.Joshi P, Greco TM, Guise AJ, Luo Y, Yu F, et al. 2013. The functional interactome landscape of the human histone deacetylase family. Mol. Syst. Biol 9:672.Integrated label-free and metabolic labelling approaches measure the relative stability and specificity of protein interactions.
- 93.Zuberi K, Franz M, Rodriguez H, Montojo J, Lopes CT, et al. 2013. GeneMANIA prediction server 2013 update. Nucleic Acids Res. 41:W115–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, et al. 2013. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41:D808–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, et al. 2014. The Reactome pathway knowledgebase. Nucleic Acids Res. 42:D472–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, et al. 2010. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 38:D525–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hulo C, de Castro E, Masson P, Bougueleret L, Bairoch A, et al. 2011ViralZone: a knowledge resource to understand virus diversity. Nucleic Acids Res. 39:D576–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chatr-aryamontri A, Ceol A, Peluso D, Nardozza A, Panni S, et al. 2009. VirusMINT: a viral protein interaction database. Nucleic Acids Res. 37:D669–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fahey ME, Bennett MJ, Mahon C, Jäger S, Pache L, et al. 2011. GPS-Prot: a web-based visualization platform for integrating host-pathogen interaction data. BMC Bioinform. 12:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gingras AC, Raught B. 2012. Beyond hairballs: the use of quantitative mass spectrometry data to understand protein-protein interactions. FEBS Lett. 586:2723–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, et al. 2012. Decoding human cytomegalovirus. Science 338:1088–93Combination of transcriptomics and proteomics identifies novel viral translation products during HCMV infection.
- 102.Vogels MW, van Balkom BW, Kaloyanova DV, Batenburg JJ, Heck AJ, et al. 2011. Identification of host factors involved in coronavirus replication by quantitative proteomics analysis. Proteomics 11:64–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Santamaría E, Mora MI, Potel C, Fernández-Irigoyen J, Carro-Roldan E, et al. 2009. Identification of replication-competent HSV-1 Cgal+ strain signaling targets in human hepatoma cells by functional organelle proteomics. Mol. Cell. Proteomics 8:805–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Emmott E, Wise H, Loucaides EM, Matthews DA, Digard P, Hiscox JA. 2010. Quantitative proteomics using SILAC coupled to LC-MS/MS reveals changes in the nucleolar proteome in influenza A virus–infected cells. J. Proteome Res 9:5335–45 [DOI] [PubMed] [Google Scholar]
- 105.Lam YW, Evans VC, Heesom KJ, Lamond AI, Matthews DA. 2010. Proteomics analysis of the nucleolus in adenovirus-infected cells. Mol. Cell. Proteomics 9:117–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Munday DC, Emmott E, Surtees R, Lardeau CH, Wu W, et al. 2010. Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus. Mol. Cell. Proteomics 9:2438–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Peirce MJ, Wait R, Begum S, Saklatvala J, Cope AP. 2004. Expression profiling of lymphocyte plasma membrane proteins. Mol. Cell. Proteomics 3:56–65 [DOI] [PubMed] [Google Scholar]
- 108.Wollscheid B, Bausch-Fluck D, Henderson C, O’Brien R, Bibel M, et al. 2009. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat. Biotechnol 27:378–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Berro R, de la Fuente C, Klase Z, Kehn K, Parvin L, et al. 2007. Identifying the membrane proteome of HIV-1 latently infected cells. J. Biol. Chem 282:8207–18 [DOI] [PubMed] [Google Scholar]
- 110.Weekes MP, Tan SY, Poole E, Talbot S, Antrobus R, et al. 2013. Latency-associated degradation of the MRP1 drug transporter during latent human cytomegalovirus infection. Science 340:199–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gudleski-O’Regan N, Greco TM, Cristea IM, Shenk T. 2012. Increased expression of LDL receptor–related protein 1 during human cytomegalovirus infection reduces virion cholesterol and infectivity. Cell Host Microbe 12:86–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pollock S, Nichita NB, Böhmer A, Radulescu C, Dwek RA, Zitzmann N. 2010. Polyunsaturated liposomes are antiviral against hepatitis B and C viruses and HIV by decreasing cholesterol levels in infected cells. Proc. Natl. Acad. Sci. USA 107:17176–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sun X, Whittaker GR. 2003. Role for influenza virus envelope cholesterol in virus entry and infection. J. Virol 77:12543–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Campbell SM, Crowe SM, Mak J. 2002. Virion-associated cholesterol is critical for the maintenance of HIV-1 structure and infectivity. AIDS 16:2253–61 [DOI] [PubMed] [Google Scholar]
- 115.Guyader M, Kiyokawa E, Abrami L, Turelli P, Trono D. 2002. Role for human immunodeficiency virus type 1 membrane cholesterol in viral internalization. J. Virol 76:10356–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bonjardim CA, Ferreira PC, Kroon EG. 2009. Interferons: signaling, antiviral and viral evasion. Immunol. Lett 122:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pavlou MP, Diamandis EP. 2010. The cancer cell secretome: a good source for discovering biomarkers? J. Proteomics 73:1896–906 [DOI] [PubMed] [Google Scholar]
- 118.Ciborowski P, Enose Y, Mack A, Fladseth M, Gendelman HE. 2004. Diminished matrix metalloproteinase 9 secretion in human immunodeficiency virus–infected mononuclear phagocytes: modulation of innate immunity and implications for neurological disease. J. Neuroimmunol 157:11–16 [DOI] [PubMed] [Google Scholar]
- 119.Ciborowski P, Kadiu I, Rozek W, Smith L, Bernhardt K, et al. 2007. Investigating the human immunodeficiency virus type 1–infected monocyte-derived macrophage secretome. Virology 363:198–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lietzén N, Ohman T, Rintahaka J, Julkunen I, Aittokallio T, et al. 2011. Quantitative subcellular proteome and secretome profiling of influenza A virus–infected human primary macrophages. PLoS Pathog. 7:e1001340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Miettinen JJ, Matikainen S, Nyman TA. 2012. Global secretome characterization of herpes simplex virus 1–infected human primary macrophages. J. Virol 86:12770–78Multiplexed, quantitative MS analysis of the HSV-1-stimulated macrophage secretome identifies interferon-inducible proteins.
- 122.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, et al. 2010. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol 11:395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Johnson KE, Chikoti L, Chandran B. 2013. Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. J. Virol 87:5005–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dumortier J, Streblow DN, Moses AV, Jacobs JM, Kreklywich CN, et al. 2008. Human cytomegalovirus secretome contains factors that induce angiogenesis and wound healing. J. Virol 82:6524–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vliegen I, Duijvestijn A, Grauls G, Herngreen S, Bruggeman C, Stassen F. 2004. Cytomegalovirus infection aggravates atherogenesis in apoE knockout mice by both local and systemic immune activation. Microbes Infect. 6:17–24 [DOI] [PubMed] [Google Scholar]
- 126.Merigan TC, Renlund DG, Keay S, Bristow MR, Starnes V, et al. 1992. A controlled trial of ganciclovir to prevent cytomegalovirus disease after heart transplantation. N. Engl. J. Med 326:1182–86 [DOI] [PubMed] [Google Scholar]
- 127.Vliegen I, Herngreen SB, Grauls GE, Bruggeman CA, Stassen FR. 2005. Mouse cytomegalovirus antigenic immune stimulation is sufficient to aggravate atherosclerosis in hypercholesterolemic mice. Atherosclerosis 181:39–44 [DOI] [PubMed] [Google Scholar]
- 128.Streblow DN, Dumortier J, Moses AV, Orloff SL, Nelson JA. 2008. Mechanisms of cytomegalovirus-accelerated vascular disease: induction of paracrine factors that promote angiogenesis and wound healing. Curr. Top. Microbiol. Immunol 325:397–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bell C, Desjardins M, Thibault P, Radtke K. 2013. Proteomics analysis of herpes simplex virus type 1–infected cells reveals dynamic changes of viral protein expression, ubiquitylation, and phosphorylation. J. Proteome Res 12:1820–29 [DOI] [PubMed] [Google Scholar]
- 130.Zhao Y, Jensen ON. 2009. Modification-specific proteomics: strategies for characterization of posttranslational modifications using enrichment techniques. Proteomics 9:4632–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hutchinson EC, Denham EM, Thomas B, Trudgian DC, Hester SS, et al. 2012. Mapping the phosphoproteome of influenza A and B viruses by mass spectrometry. PLoS Pathog. 8:e1002993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Roby C, Gibson W. 1986. Characterization of phosphoproteins and protein kinase activity of virions, noninfectious enveloped particles, and dense bodies of human cytomegalovirus. J. Virol 59:714–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Davido DJ, von Zagorski WF, Lane WS, Schaffer PA. 2005. Phosphorylation site mutations affect herpes simplex virus type 1 ICP0 function. J. Virol 79:1232–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fang CY, von Zagorski WF, Lane WS, Schaffer PA. 2010. Global analysis of modifications of the human BK virus structural proteins by LC-MS/MS. Virology 402:164–76 [DOI] [PubMed] [Google Scholar]
- 135.Bergström Lind S, Artemenko KA, Elfineh L, Zhao Y, Bergquist J, Pettersson U. 2013. Post translational modifications in adenovirus type 2. Virology 447:104–11 [DOI] [PubMed] [Google Scholar]
- 136.Jakobsen MR, Bak RO, Andersen A, Berg RK, Jensen SB, et al. 2013. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc. Natl. Acad. Sci. USA 110:E4571–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, et al. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol 11:997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, et al. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834–40 [DOI] [PubMed] [Google Scholar]
- 139.Halili MA, Andrews MR, Labzin LI, Schroder K, Matthias G,et al. 2010. Differential effects of selective HDAC inhibitors on macrophage inflammatory responses to the Toll-like receptor 4 agonist LPS. J. Leukoc. Biol 87:1103–14 [DOI] [PubMed] [Google Scholar]
- 140.Roger T, Lugrin J, Le Roy D, Goy G, Mombelli M, et al. 2011. Histone deacetylase inhibitors impair innate immune responses to Toll-like receptor agonists and to infection. Blood 117:1205–17 [DOI] [PubMed] [Google Scholar]
- 141.Mombelli M, Lugrin J, Rubino I, Chanson AL, Giddey M, et al. 2011. Histone deacetylase inhibitors impair antibacterial defenses of macrophages. J. Infect. Dis 204:1367–74 [DOI] [PubMed] [Google Scholar]
- 142.Hutterer C, Wandinger SK, Wagner S, Muller R, Stamminger T, et al. 2013. Profiling of the kinome of cytomegalovirus-infected cells reveals the functional importance of host kinases Aurora A, ABL and AMPK. Antivir. Res 99:139–48 [DOI] [PubMed] [Google Scholar]
- 143.Terry LJ, Vastag L, Rabinowitz JD, Shenk T. 2012. Human kinome profiling identifies a requirement for AMP-activated protein kinase during human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 109:3071–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, et al. 2002. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol 9:1149–59 [DOI] [PubMed] [Google Scholar]
- 145.Ovaa H, Kessler BM, Rolén U, Galardy PJ, Ploegh HL, Masucci MG. 2004. Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc. Natl. Acad. Sci. USA 101:2253–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Picotti P, Aebersold R. 2012. Selected reaction monitoring–based proteomics: workflows, potential, pitfalls and future directions. Nat. Methods 9:555–66 [DOI] [PubMed] [Google Scholar]
- 147.Chavez JD, Cilia M, Weisbrod CR, JuH J, Eng JK, et al. 2012. Cross-linking measurements of the Potato leafroll virus reveal protein J. Proteome Res 11:2968–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.