Abstract
Purpose
Genomic reprogramming and cellular dedifferentiation are critical to the success of de novo tissue regeneration in lower vertebrates such as zebrafish and axolotl. In tissue regeneration following injury or disease, differentiated cells must retain lineage while assuming a progenitor-like identity in order to repopulate the damaged tissue. Understanding the epigenetic regulation of programmed cellular dedifferentiation provides unique insights into the biology of stem cells and cancer and may lead to novel approaches for treating human degenerative conditions.
Methods
Using a zebrafish in vivo model of adult muscle regeneration, we utilized chromatin immunoprecipitation followed by massively parallel DNA sequencing (ChIP-seq) to characterize early changes in epigenetic signals, focusing on three well-studied histone modifications—histone H3 trimethylated at lysine 4 (H3K4me3), and histone H3 trimethylated or acetylated at lysine 27 (H3K27me3 and H3K27Ac, respectively).
Results
We discovered that zebrafish myocytes undergo a global, rapid, and transient program to drive genomic remodeling. The timing of these epigenetic changes suggests that genomic reprogramming itself represents a distinct sequence of events, with predetermined checkpoints, to generate cells capable of de novo regeneration. Importantly, we uncovered subsets of genes that maintain epigenetic marks paradoxical to changes in expression, underscoring the complexity of epigenetic reprogramming.
Conclusions
Within our model, histone modifications previously associated with gene expression act for the most part as expected, with exceptions suggesting that zebrafish chromatin maintains an easily editable state with a number of genes paradoxically marked for transcriptional activity despite downregulation.
Keywords: H3K4me3, H3K27me3, H3K27Ac, histone, chromatin, myocyte, reprogramming, dedifferentiation, stem cell, extraocular muscle
Regeneration of functional tissue following severe injury or disease is a longstanding goal of regenerative medicine. Skeletal muscle disorders represent a particularly high burden on human health and quality of life.1–4 Though mammalian muscle tissue is capable of local tissue repair, such repair cannot reverse the effects of significant volumetric muscle loss or fibrosis. In contrast, piscine and amphibian models of regeneration provide hope that with improved mechanistic understanding of the biology, the ability to regenerate tissue might be translatable to mammals.
Our laboratory has developed a novel model of de novo adult muscle regeneration using zebrafish extraocular muscles (EOMs). Within 24 hours post injury (hpi), residual myocytes dedifferentiate to form myoblast-like cells. These cells are capable of proliferating and regenerating the lost muscle mass through extensive genomic and cellular reprogramming.5,6 We hypothesized that the ability to reprogram “postmitotic” cells must involve epigenetic alterations that can persist through multiple cell divisions. Indeed, our transcriptome analysis identified significant changes in genes encoding epigenetic regulators, including the DNA methylation-related dnmt1, dnmt3aa, tet2, and tdg, the histone H3K27 trimethylation-related kdm6ba and ezh2, and the NuRD complex genes chd4a, hdac1, rbb4, mta2, chd4a, chd3, and mbd2.6
In order to test our hypothesis, we performed chromatin immunoprecipitation followed by massively parallel DNA sequencing (ChIP-seq) to examine global changes in modifications to histone H3 lysine 27 tri-methylation and acetylation, and histone H3 lysine 4 tri-methylation (H3K27me3, H3K27Ac, and H3K4me3, respectively). We found that changes in H3K27Ac and H3K4me3 correlated both with each other and with changes in gene expression at 9 and 18 hpi when compared with control. Conversely, H3K27me3 levels do not correlate with changes in gene expression. Interestingly, we discovered a subset of genes that are significantly downregulated but maintain epigenetic marks of active transcription, that is, paradoxical changes in epigenetic regulation of gene expression.
Methods
Zebrafish (Danio rerio) Rearing
All animal work was performed in compliance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and approved by the University of Michigan Committee on the Use and Care of Animals, protocol 06034. Sexually mature adult (4–18 months of age) wild-type and transgenic (α-actin::EGFP) zebrafish were spawned in our fish facility and raised according to standard protocol at 28°C with a 14-hour light to 10-hour dark cycle.
Surgery and Preparation of Tissue for ChIP-Seq
We anesthetized adult zebrafish using 0.05% tricaine methanesulfonate (Tricaine-S; Western Chemical, Ferndale, WA, USA) with 0.05% sodium bicarbonate buffer and placed them on moist Kimwipes (category number 06-666; Fisher Scientific, Hampton, NH, USA) under a stereomicroscope with green fluorescent protein (GFP) fluorescence illumination. We adducted both eyes, in turn, with a thin blunt probe, and excised a 0.5- to 1.0-mm segment (approximately 50%) of lateral rectus (LR) muscle (Fig. 1). This manipulation resulted in an immediate loss of LR muscle function.5 Uninjured LR muscles were harvested as control tissue and injured LR muscles harvested at 9 and 18 hpi. Muscles were accumulated from a group of fish (control: 119; 9 hpi: 192; 18 hpi: 200) to attain a final volume of 100 to 300 mg of per histone mark, per time point.
Figure 1.
Experimental design. LR muscle tissue (green) that was either uninjured (control, A) or allowed to recover for either 9 or 18 hpi (B) was harvested and immediately fixed in 37% formaldehyde in chilled PBS (final formaldehyde concentration 1%). (C) Prepared sample tissue was immunoprecipitated for ChIP-seq with antibodies against H3K4me3, H3K27me3, and H3K27Ac. Reads were aligned to the danRer10 genome and used to measure coverage and detect peaks (examples shown; see Methods).
Immediately upon removal from the orbit, the tissue was fixed by immersion in 37% formaldehyde in chilled PBS on ice (final formaldehyde concentration 1%) for 15 minutes to cross-link, per guidelines for chromatin immunoprecipitation (Active Motif, Carlsbad, CA, USA; personal communication). Fixation was quenched using 1:20 volume of 2.5 M Glycine Solution and the solution rotated at room temperature for 5 minutes. The mixture was centrifuged at 1250g for 5 minutes at 4°C. The supernatant was decanted, and the pellet was frozen at −90°C to −70°C.
ChIP-Seq Sample Preparation and Sequencing
Chromatin immunoprecipitation, library preparation, sequencing, and initial data processing were performed by Active Motif. Lysis buffer was added to the tissue sample and the chromatin was disrupted using a Dounce homogenizer. The product was sonicated, and the DNA sheared to fragments of 300 to 500 base pairs. Input genomic DNA was prepared by treating with RNase, proteinase K with heat, and ethanol precipitation. Other aliquots with 2 to 4 μg chromatin were precleared with protein A agarose beads (Invitrogen, Carlsbad, CA, USA), and regions of interest were isolated using 4 μg antibody against H3K27me3 (07-449; Millipore, Burlington, MA, USA), H3K27Ac (39133; Active Motif), or 3 μg antibody against H3K4me3 (39159; Active Motif). Similar to the input samples, the resulting complexes were washed, eluted with SDS buffer, and treated with RNase and proteinase K. They were incubated overnight at 65°C to reverse crosslinks, and the ChIP DNA products were purified with phenol-chloroform extraction and ethanol precipitation. ChIP and Input DNA were put through standard preparation steps of end-polishing, dA-addition, and adaptor ligation before sequencing on the Illumina NextSeq 500, generating 75-nt single-end reads.
ChIP-Seq Data Analysis
Reads were aligned to the danRer10 zebrafish reference genome using the Burrows-Wheeler Aligner (BWA) algorithm (default settings). Duplicate reads were removed and only uniquely mapped reads with mapping quality ≥ 25 were used for further analysis. Reads were extended to 150- to 250-nt in silico based on estimated average fragment length and used to generate coverage files (32-nt bins; bigWig format). Peak calling was performed using the model-based analysis of ChIP-seq (MACS) algorithm v2.1.07 with cutoff P value = 1 × 10−7. For H3K27me3, identification of enriched regions was performed using spatial clustering for identification of ChIP-enriched regions (SICER) algorithm v1.18 with false discovery rate (FDR) cutoff = 1e–10 and gap size of 3× fragment size (200 bp).
Samples were normalized by down-sampling to the smallest library. For each histone mark, overlapping peaks were merged into common regions (referred to as active regions). Statistical transformations and analyses were performed in Excel, Prism, and using locally curated R programs.
Reads within active regions were counted using BEDTools (v2.20.1)9 and normalized via the trimmed mean of M values (TMM) method (using edgeR v3.18.1)10 and reported as counts per million (CPM) for given samples (Fig. 2). Log2 fold-change values for active regions were obtained from edgeR (Figs. 3, 4). Each active region was assigned a closest gene based on proximity to the gene transcription start site (TSS), for genes < 10 kb away; to obtain unique 1:1 relationships between genes and active regions, a best match was determined for genes claimed multiple times as closest to active regions. For H3K27Ac, genes matching multiple active regions were assigned the one with the highest CPM, thus favoring distal regulatory elements (e.g., enhancers) where possible. For H3K4me3 and H3K27me3, priority is given to marks closer to the promoter. Thus, the gene was assigned to the active region that was strongest but within 2500 bp of the TSS. This 2500-bp limit was designed to be inclusive, especially with H3K27me3 peaks, which are much broader than H3K4me3. Correlation coefficients were calculated using log10(x + 0.001) transformations to include all points (Figs. 2–4). Data used to produce Figures 2, 3, and 4 can be found in Supplementary Data File S1. Summaries of the scatterplots in Figures 2 to 4, including correlation coefficients and P values, are in Supplementary Data File S1.
Figure 2.
Over the injury time course, H3K27Ac and H3K4me3 levels, but not H3K27me3, correlate with gene expression. Each point represents a unique gene and its most representative histone active region per modification indicated for control (A–C), 9 hpi (D–F), and 18 hpi (G–I). Gene expression is represented as FPKM and histone modification active regions are in CPM, both measures log10-transformed. Correlation is indicated by Pearson's r for log10-transformed values. Blue lines indicate linear regressions.
Figure 3.
Postinjury gene expression changes correlate with histone modification levels for H3K27Ac and H3K4me3, but not H3K27me3, for significant DE genes. (A–I) These scatterplots represent significantly DEGs with associated active regions for each histone modification. Changes for both genes and histone modifications are in LFC. Correlation is represented by Pearson's r and P values are from correlation tests (0.05 significance level). Blue lines represent linear regressions. Genes must have FDR < 0.05 and fold-change > 1.5 or < −1.5 in at least one comparison (9 hpi/control, 18 hpi/control, or 18 hpi/9 hpi).
Figure 4.
Comparison of changes among H3K27Ac, H3K4me3, and H3K27me3. LFCs were compared among histone modifications for all genes in the (A) control to 9 hpi and (B) control to 18 hpi periods. Histone modification changes within the subset of genes that were (C) upregulated and (D) downregulated were also compared for the time period of control to 9 hpi. Gene TSSs and flanking regions for expressed genes were analyzed at (E) control, (F) 9 hpi, and (G) 18 hpi. Black lines represent H3K27Ac, red lines represent H3K4me3, and gray lines represent input data. Correlations are represented by Pearson's r and linear regressions by blue lines.
For each given annotated gene, the mean peak densities from active regions within 10 kb of the gene are represented as an average peak value. These values are used to represent genes in Figures 5 and 6. Data for genes were obtained for two pathways and two functional subsets of interest from Saera-Vila et al.5 Within these groups, log10(RNA fragments per kilobase per million [FPKM] + 1.1) was plotted against average peak height for each histone mark at each time point (Fig. 5).
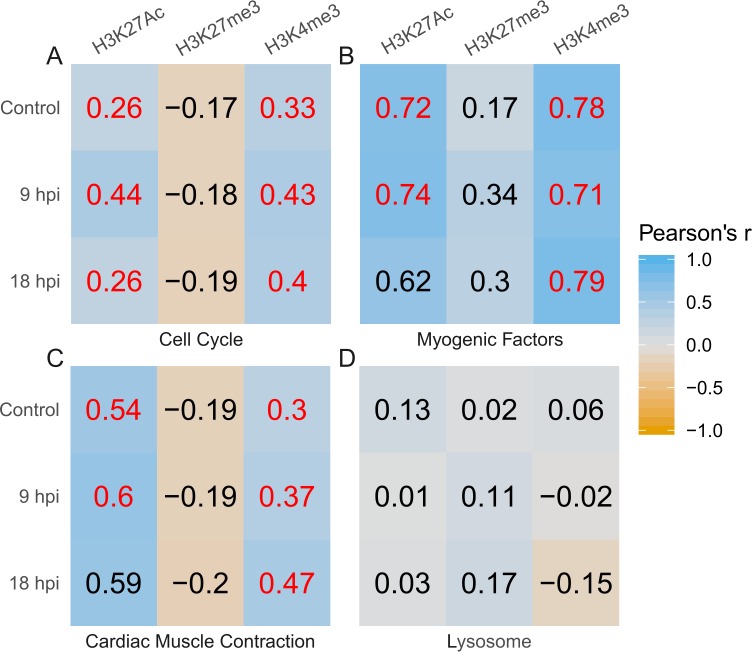
Figure 5.
Correlation of RNA-seq and ChIP-seq data for the subsets of genes related to cell cycle (A) and myogenic factors (B), and DEGs within the cardiac muscle contraction KEGG pathway (dre04260) (C) and lysosome KEGG pathway (dre04142) (D). Color gradient represents Pearson's correlation coefficient r, numerically expressed within each square. Red indicates if the value had P < 0.05 significance. Coefficients were obtained by plotting gene expression (log10 of FPKM) against the average peak value for each histone mark at each time point. Statistical significance is reported using Pearson's r value of correlation for data sets, which also have a P < 0.05 (all correlation and corresponding P values can be found in Supplementary Data File S2). A version of this plot that displays individual points in represented in Supplementary Figure S5.
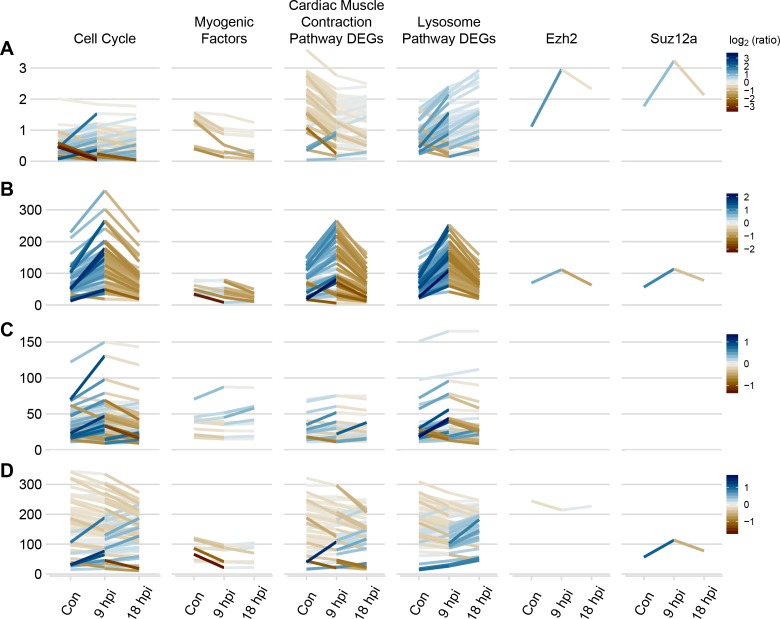
Figure 6.
RNA and chromatin patterns in select subsets of genes. Temporal changes in histone modification levels for genes related to cell cycle, myogenic factors, genes involved in cardiac muscle contraction KEGG pathway (dre04260), lysosome KEGG pathway (dre04142), and for individual genes ezh2 and suz12a (each subset takes one column of the plot). (A) Expression profiles of the genes over the time course (control, 9 hpi, 18 hpi). Measured in FPKM. Average peak value in active regions of H3K27Ac (B), H3K27me3 (C), and H3K4me3 (D), from control to 9 hpi to 18 hpi. Neither ezh2 nor suz12a had nearby H3K27me3 peaks, hence the plots for these genes are blank. For each of A to D, a divergent color scale represents the log2 ratio of signal between two time points (both 9 hpi/control and 18 hpi/9 hpi).
Gene lists for Lysosome and Cardiac Muscle Contraction (hereafter referred to as “muscle contraction” or, simply, “contractile”) include differentially expressed genes (DEGs) only, as defined in a previous publication.6 Myogenic factors are all genes related to myogenic differentiation, myocytes, or myoblasts. Cell cycle genes are all genes characterized as cyclins or cyclin-dependent kinases.
Within these groups, temporal changes were analyzed by plotting RNA FPKM and ChIP-seq average peak values for each histone mark, at each time point in succession (Fig. 6). The plots were also made using two individual genes of interest: ezh2 and suz12a.
Data Sources
All ChIP-seq data presented in this study are available at the Gene Expression Omnibus (GSE137304). RNA-sequencing (RNA-seq) data used in this study were previously published and publicly available (GEO: GSE92489).6 Briefly, TruSeq Stranded Total RNA libraries were sequenced using the Illumina HiSeq platform (50-base, stranded, single-end reads), aligned to danRer10 with TopHat/Bowtie2, expression values (FPKM) calculated using Cufflinks, and DEGs selected by testing with CuffDiff and a minimum fold change (significant with FDR < 0.05 and fold change > 1.5 up or down).
Results
We previously described global transcriptional changes using RNA-seq of uninjured LR muscle and muscle tissue collected from adult zebrafish at 9 and 18 hpi collected using laser microdissection.6 These results support the hypothesis that zebrafish muscle regeneration involves transient de-differentiation driven by alterations in epigenetic and transcriptional programs. At the time that the RNA-seq data were generated, we had not yet developed the microsurgical method used in this study. Due to a need for drastically higher concentrations of chromatin to perform ChIP-seq as compared with RNA-seq (approximately six times as many fish were killed), the collection strategy was modified for expediency and integrity of the muscle material. We utilized quantitative PCR (qPCR) to compare gene expression between RNA isolated using our surgical method versus the methods used to generate our RNA-seq data, and based on those results we conclude there are no significant differences in the populations of genes being correlated within this study (Supplementary Figs. S1A–D).
Using ChIP-seq on excised muscle tissue, we tested whether myocyte reprogramming involves set alterations in chromatin. We assessed three modifications: H3K4me3 for its known role in active promoters, H3K27Ac, which is present in super-enhancers and regulates “open” chromatin, and H3K27me3, which is found in heterochromatin and associated with decreased transcription.11 Because dedifferentiation and proliferation initiate during the first 20 hpi,5 we focused our analysis at 9 and 18 hpi, comparing changes to uninjured control.
“Activating” Histone Modifications Correlate With Gene Expression
Overlapping ChIP-seq peaks were merged into common regions across the control, 9 hpi, and 18 hpi time points for comparison purposes. We then counted the reads within these selected active regions and normalized using the TMM method (edgeR).10 For further analysis, to represent expressed genes in a 1:1 manner, we selected the most promoter-proximal active regions (or strong distal active regions, in the case of H3K27Ac). Proximal was defined as +/− 2500 bp of gene TSSs, as most of the signal densities around TSSs fell within this region (Figs. 4E–G). Genes with FPKM > 0 (in FPKM) were considered to be “expressed” for this purpose. We asked whether H3K27Ac, H3K27me3, or H3K4me3 levels (in CPM) were related to gene expression levels and found that both histone marks that putatively associate with transcribing genes (H3K27Ac and H3K4me3) were positively correlated to gene expression at all three time points (Figs. 2A, D, G, and 2C, F, I, respectively). In contrast, the putative repressive modification, H3K27me3, did not correlate with gene expression (Figs. 2B, E, H).
Changes in Histone Modification Levels Correlate With Changes in Gene Expression in Control EOMs Versus 9 and 18 hpi
Given the overall correlation of two histone marks with gene expression, we tested the correlation between reprogramming time points. To do this, we used our time series model to perform three comparisons—9 hpi/control, 18 hpi/control, and 18 hpi/9 hpi—within both sets of data: histone modifications and gene expression. We log transformed the results and compared the two sets, first using only the significantly DEGs teased out from our prior transcriptome analysis (Fig. 3). As expected, both H3K27Ac and H3K4me3 tracked with gene expression between the control and postinjury time points (9 and 18 hpi; Figs. 3A, D, and 3C, F, respectively), but not between the 9 and 18 hpi time points (Figs. 3G, I). Additionally, H3K27me3 does not correlate in any comparison (Figs. 3B, E, H). The lack of correlation between 9 and 18 hpi lead us to believe that the majority of chromatin changes that associate with changes in gene expression likely occur within the first 9 hpi. We also asked if similar behaviors could be observed in the entire gene set. Indeed, a positive correlation was observed between gene log fold change (LFC) and H3K27Ac and H3K4me3 LFC, but not H3K27me3 and not for the 18 hpi/9 hpi comparison (Supplementary Fig. S2). Although many points lay within quadrants I and III (direction of change matches between genes and histone marks), a notable number of H3K27Ac regions are increasing when the corresponding genes are downregulated (1249 out of 2084 downregulated differential expressed genes; H3K27Ac 9 hpi/control LFC > 1), indicating a class of acetylation that is increasing regardless of nearby gene expression changes (quadrant II; Supplementary Fig. S2A), suggesting that these regions might exert their effects at a distance.
H3K27Ac and H3K4me3 Are Correlated; H3K27Ac Transiently Increases
Because both H3K27Ac and H3K4me3 are associated with active/transcribing genes,12,13 which we also observed in our analysis, we asked whether they correlated directly with each other within the entire gene set. For both the 9 hpi/control and 18 hpi/control comparisons, H3K27Ac LFC did indeed correlate with H3K4me3 LFC (Figs. 4A, B). Conversely, there was no correlation between either activating mark and changes in H3K27me3 (Supplementary Fig. S3). For significantly DEGs, both upregulated and downregulated classes, H3K27Ac and H3K4me3 were positively correlated overall (Figs. 4C, D). However, a clear subset of genes with both marks show increasing H3K27Ac but not H3K4me3, suggesting a trend for increasing H3K27Ac independent of nearby gene expression changes. Again, there was little to no correlation between H3K27me3 and either activating mark in either upregulated or downregulated genes (Supplementary Fig. S4). Analysis of gene TSSs and flanking regions for expressed genes revealed a global increase of H3K27Ac between control and 9 hpi (Figs. 4E, F), which then reverts back to control levels at 18 hpi (Fig. 4G), reflecting a transient acetylation of H3K27 that may be critical for the reprogramming process.
Most Gene Groupings Exhibit Expected Correlation Between Histone Modification and Gene Expression
Four key groups of genes were selected for further analysis based on their relevance to muscle regeneration and findings from our previous work6 (Figs. 5A–D; gene lists in Supplementary Data File S2). Cell cycle genes included all available paralogs of cyclins and genes related to the cell cycle. Myogenic factors include available paralogs of mef2 and myf, as well as myod1 and myogenin. The lysosome and muscle contraction pathways were selected based on gene ontology (GO) terms, as described.6
For each group of genes, we compared gene expression to average peak value. For cell cycle, myogenic factors, and muscle contraction gene sets, both H3K27Ac and H3K4me3 were positively correlated to gene expression in at least two of the time points, though for some the correlation was weak (Fig. 5). H3K27me3 was not correlated to gene expression anywhere. In three of the four groups, H3K27Ac correlation with gene expression increased at 9 hpi and decreased by 18 hpi. H3K27me3 did not experience significant or observable change in correlation with expression. Correlation of gene expression and H3K4me3 was consistent throughout the time course (correlation values in Supplementary Data File S2). H3K4me3 correlation was strongest in plots for myogenic factors (Fig. 5B) and weakest in those for lysosome pathway DEGs (Fig. 5D).
H3K4me3 Is Not an Indication of Change in Gene Expression for the First 18 hpi
We examined the gene expression and histone modification profiles for the above gene sets as well as two individual genes—the methyltransferase ezh2 and its associated gene suz12a, both members of the polycomb repressive complex 2 (Fig. 6). Neither H3K4me3 nor H3K27me3 levels changed for any of these smaller gene groups, despite clear gene expression level changes in myogenic factors, muscle contraction genes, and lysosomal genes. Additionally, we found that although most gene expression changes were reflected in H3K4me3, such as suz12a, where H3K4me3 levels rise and fall with gene expression levels, others were not, such as ezh2. Hence, although overall expression levels correlate with H3K4me3 at each time point, there are clearly exceptions in some critical epigenetic genes within the timeframe of the study.
Genes Are Marked for Transcriptional Activity Despite Downregulation
Analysis revealed a particular subset of 46 genes that are downregulated despite persistent levels of histone modifications H3K27Ac and H3K4me3, which are usually associated with regions and promoters of active genes, respectively (gene LFC 9 hpi/control < 0, significant LFC 9 hpi/control = “yes”, −0.9 < histone modification LFC < 0.1 for both H3K27Ac and H3K4me3). More broadly, a group of approximately 250 significantly downregulated genes experience either stable or increasing levels of both histone modifications (gene LFC 9 hpi/control < 0, significant LFC 9 hpi/control = “yes”, mod LFC > −0.05; lists can be found in Supplementary Data File S3). We suggest that these genes may be bookmarked for reactivating transcriptional activity, as part of a tightly controlled transient reprogramming process.
Discussion
In this study, we tested the hypothesis that cellular dedifferentiation in adult zebrafish includes significant alterations in chromatin. We used an in vivo model of myocyte dedifferentiation to test for three modifications of histone H3. This model is notable for its ability to achieve the relatively synchronized and enriched cell population required for genome-wide mapping of chromatin modifications. To investigate early steps of epigenetic reprogramming (from 0 to 18 hpi, prior to onset of proliferation), we chose three modifications—H3K27Ac, H3K27me3, and H3K4me3—because of their known and conserved roles in regulating chromatin in mammals, yeast (Saccharomyces cerevisiae), and Tetrahymena thermophile.14–20
Global increases in H3K27Ac at 9 hpi followed by global decreases by 18 hpi indicate a rapid but transient activation across the chromatin landscape, as would be expected of a tightly regulated dedifferentiation process. This finding, accompanied by the relatively static levels of H3K27me3, leads us to hypothesize that zebrafish retain a relatively “open” chromatin structure compared with other species.21 Furthermore, the finding that most of the activating modifications occurred in the first 9 hours suggest that even a highly differentiated cell such as a multinucleated syncytial myocyte is poised for epigenetic reprogramming, supporting an active and highly regulated process. Indeed, studies have revealed that in certain cell types, high levels of histone H3 acetylation help to maintain cells in a less differentiated, pluripotent state.22,23 Combining this knowledge with our finding of a group of genes that are downregulated despite increasing levels of this modification lead us to conclude that some genes are bookmarked for future transcriptional activity. Further, after showing that H3K27Ac levels return to normal by 18 hpi, we hypothesize that the chromatin remodeling/reprogramming process reflects a tightly regulated program in and of itself. A better understanding of this process may lead to important insights about the potential for dedifferentiation-driven regeneration in other organisms, including mammals, as well as about the biology of stem cells and cancer.
An important aspect of our study was the finding that in the early time points of EOM regeneration, transcriptional changes were indeed concurrent with the activating histone markers H3K27Ac and H3K4me3. To further explore this finding, we selected gene sets of particular interest and assessed the epigenetic landscape therein. Indeed, analysis of gene groups involved in the cell cycle, myogenesis, lysosome, and muscle contraction, as well as the two members of the polycomb repressive complex 2, revealed a strong correlation between epigenetic modifications and changes in gene expression. However, we also uncovered paradoxical epigenetic marks. Particularly notable was the finding that, in certain subsets of genes, H3K4me3 remains relatively unchanged despite a significant reduction of gene expression. One such example can be seen within myogenic genes. Shortly after the proliferative burst, myogenic gene expression resumes, along with muscle regeneration. This suggests that H3K4me3 may mark promoters “poised” for resumption of expression—possibly a component of maintaining lineage restriction. Recently published work found that in mammals, promoters of many key oligopotent retinal neuronal genes can be held in a permissive chromatin state with high levels of histone activation markers combined with DNA methylation.24,25 Thus, further studies of DNA methylation states during zebrafish muscle regeneration could explain the inconsistency between expression and histone modification for some genes. A key feature of myocyte dedifferentiation is its transience and the ability to quickly return to the differentiated state. This suggests that the dedifferentiated myocyte is fundamentally different from a primary myoblast, which might be reflected in a paradoxical epigenetic signature, that is, genes that exhibit downregulation coincident with increasing levels of markers traditionally associated with active transcription.
Within these paradoxically regulated genes, we did not find pathway enrichment that could be considered unique from the larger pool containing all significantly downregulated genes (regardless of epigenetic patterns), nor were any particular genomic loci overrepresented. Instead, these genes represent a relatively small group with a unique epigenetic state—downregulated but marked for reactivation. Such paradoxical marks suggest that genomes have the ability to bookmark genes for rapid reactivation. Future studies will pursue the role of such paradoxical epigenetic changes in the regulation of transient/reversible genomic reprogramming and cellular dedifferentiation.
In summary, our results reveal that modifications known to be associated with activation of gene expression, that is, H3K27Ac and H3K4me3, correlate with changes in gene expression during the early reprogramming process. On the other hand, H3K27me3, a modification associated with chromatin silencing and transcriptional downregulation, is relatively nondynamic during myocyte reprogramming, with poor correlation to both gene expression changes and the other histone marks we explored. We hypothesize that zebrafish chromatin may be predisposed to an “open” conformation, potentially explaining why dedifferentiation appears to be a universal mechanism for zebrafish tissue regeneration.21,26,27 This possibility is consistent with the recent finding that the zebrafish genome is relatively hypo-methylated in at least one cell type during tissue regeneration.21 Finally, our findings reveal a number of genes that are paradoxically marked for transcriptional activity despite being downregulated. Future studies will explore mechanisms of chromatin reprogramming during cellular dedifferentiation, including regulation of super-enhancers28 and “poised” promoters.
Supplementary Material
Acknowledgments
The authors thank Shelby Unsworth, PhD, for helpful discussions and critical reading of the manuscript, and Ethan Kahana for his technical assistance with microsurgical tissue collection.
Supported by the University of Michigan Forbes Cancer Research Institute, a Physician-Scientist Award from Research to Prevent Blindness, Inc. (RPB), Grant R01 EY022633 from the National Eye Institute of the National Institutes of Health (NIH; AK), and an unrestricted grant from RPB to the Department of Ophthalmology and Visual Sciences. This research utilized the Vision Research Core (P30 EY007003), and the Cancer Center Research Core (P30 CA046592). AK is supported by the Mrs. William Davidson Emerging Scholar Award from the A. Alfred Taubman Medical Research Institute. The Zebrafish International Resource Center is supported by Grant P40 RR012546 from the NIH National Center for Research Resources. The authors alone are responsible for the content and writing of the paper.
Disclosure: C.F. Tingle, None; B. Magnuson, None; Y. Zhao, None; C.J. Heisel, None; P.E. Kish, None; A. Kahana, None
References
- 1.Grogan BF, Hsu JR, Skeletal Trauma Research Consortium Volumetric muscle loss. J Am Acad Orthop Surg. 2011;19(suppl 1):S35–S37. doi: 10.5435/00124635-201102001-00007. [DOI] [PubMed] [Google Scholar]
- 2.Grasman JM, Zayas MJ, Page RL, Pins GD. Biomimetic scaffolds for regeneration of volumetric muscle loss in skeletal muscle injuries. Acta Biomater. 2015;25:2–15. doi: 10.1016/j.actbio.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasukonis B, Kim J, Brown L, et al. Codelivery of infusion decellularized skeletal muscle with minced muscle autografts improved recovery from volumetric muscle loss injury in a rat model. Tissue Eng Part A. 2016;22:1151–1163. doi: 10.1089/ten.tea.2016.0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garg K, Ward CL, Hurtgen BJ, et al. Volumetric muscle loss: persistent functional deficits beyond frank loss of tissue. J Orthop Res. 2015;33:40–46. doi: 10.1002/jor.22730. [DOI] [PubMed] [Google Scholar]
- 5.Saera-Vila A, Kasprick DS, Junttila TL, et al. Myocyte dedifferentiation drives extraocular muscle regeneration in adult zebrafish. Invest Ophthalmol Vis Sci. 2015;56:4977–4993. doi: 10.1167/iovs.14-16103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Louie KW, Saera-Vila A, Kish PE, Colacino JA, Kahana A. Temporally distinct transcriptional regulation of myocyte dedifferentiation and Myofiber growth during muscle regeneration. BMC Genomics. 2017;18:854. doi: 10.1186/s12864-017-4236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 12.Lawrence M, Daujat S, Schneider R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 2016;32:42–56. doi: 10.1016/j.tig.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Huang H, Sabari BR, Garcia BA, Allis CD, Zhao Y. SnapShot: histone modifications. Cell. 2014;159:458–458. doi: 10.1016/j.cell.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia BA, Hake SB, Diaz RL, et al. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282:7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- 15.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim D-H, Tang Z, Shimada M, et al. Histone H3K27 trimethylation inhibits H3 binding and function of SET1-like H3K4 methyltransferase complexes. Mol Cell Biol. 2013;33:4936–4946. doi: 10.1128/MCB.00601-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 18.Jiao L, Liu X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science. 2015;350:aac4383. doi: 10.1126/science.aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 20.Jin W, Peng J, Jiang S. The epigenetic regulation of embryonic myogenesis and adult muscle regeneration by histone methylation modification. Biochem Biophys Rep. 2016;6:209–219. doi: 10.1016/j.bbrep.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powell C, Grant AR, Cornblath E, Goldman D. Analysis of DNA methylation reveals a partial reprogramming of the Müller glia genome during retina regeneration. Proc Natl Acad Sci U S A. 2013;110:19814–19819. doi: 10.1073/pnas.1312009110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J-H, Hart SRL, Skalnik DG. Histone deacetylase activity is required for embryonic stem cell differentiation. Genes N Y N 2000. 2004;38:32–38. doi: 10.1002/gene.10250. [DOI] [PubMed] [Google Scholar]
- 23.De Felice L, Tatarelli C, Mascolo MG, et al. Histone deacetylase inhibitor valproic acid enhances the cytokine-induced expansion of human hematopoietic stem cells. Cancer Res. 2005;65:1505–1513. doi: 10.1158/0008-5472.CAN-04-3063. [DOI] [PubMed] [Google Scholar]
- 24.Dvoriantchikova G, Seemungal RJ, Ivanov D. DNA methylation dynamics during the differentiation of retinal progenitor cells into retinal neurons reveal a role for the DNA demethylation pathway. Front Mol Neurosci. 2019;12:182. doi: 10.3389/fnmol.2019.00182. Available at: https://www.frontiersin.org/articles/10.3389/fnmol.2019.00182/full Accessed August 27, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dvoriantchikova G, Seemungal RJ, Ivanov D. Development and epigenetic plasticity of murine Müller glia. Biochim Biophys Acta BBA - Mol Cell Res. 2019;1866:1584–1594. doi: 10.1016/j.bbamcr.2019.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jopling C, Sleep E, Raya M, Martí M, Raya A, Belmonte JCI. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knopf F, Hammond C, Chekuru A, et al. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev Cell. 2011;20:713–724. doi: 10.1016/j.devcel.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 28.Kang J, Hu J, Karra R, et al. Modulation of tissue repair by regeneration enhancer elements. Nature. 2016;532:201–206. doi: 10.1038/nature17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.