To the Editor:
Acquired resistance during treatment will remain a principal driver of the extensively drug-resistant tuberculosis (XDR-TB) epidemic into the foreseeable future (1). Conventional culture-based phenotypic drug-susceptibility testing (DST) for Mycobacterium tuberculosis (M.tb) as conceived in the 1960s is laborious, scarce in many regions, and too slow for clinical decision-making. Existing commercial molecular TB diagnostic tests provide results within days but can only detect resistant subpopulations greater than 5–65% of the total M.tb population (2–4). We recently developed a novel targeted next-generation sequencing (NGS) approach (Single Molecule-Overlapping Reads [SMOR]) that is able to discern low-level minority resistant populations between 0.1% and 5% (i.e., microheteroresistance) for isoniazid, rifampin, fluoroquinolones (FQs), second-line injectables (SLIs), and pyrazinamide (genes katG and inhA/inhA promoter; rpoB; gyrA/B; rrs; and pncA, respectively) (5, 6). We hypothesized that genotypic microheteroresistance may precede the acquisition of phenotypic drug resistance in clinical practice.
Methods
We applied SMOR to sets of serial primary isolates, at a total of 145 time points, collected from 19 patients with at least rifampin-resistant TB who acquired additional FQ and/or SLI resistance (i.e., progressing to pre-XDR or XDR-TB). The patients were treated under programmatic conditions in the Western Cape, South Africa, 2014–2016, with each patient contributing a median of eight serial cultures (interquartile range [IQR], 5–9.5 cultures) over 26 months (IQR, 16–40 mo). Treatment records were available for eight patients, seven of whom (88%) were known to have been treated with contemporary standardized multidrug-resistant-TB regimens, including FQs and SLIs. This study was approved by institutional review boards at Stellenbosch University (#N09/11/296) and the University of California, San Francisco (#14-15090).
National Health Laboratory Service (NHLS) second-line DST was performed on Middlebrook 7H11 slants (Becton Dickinson) containing 2 μg/ml and 4 μg/ml of ofloxacin and amikacin, respectively. The control slants for all NHLS isolates found to be resistant to either drug were stored at Stellenbosch University, and additional second-line phenotypic DST was performed via the indirect proportion method in a BD BACTEC MGIT 960 system using identical drug concentrations. SMOR sequencing was performed on crude DNA extracts (generated by incubating ∼200 μl cells at 100°C for 30 min) from the original NHLS DST 7H11 agar, and DNA specimens were coded, blinded, amplified, and prepared as described previously (5). All SLI resistance target alleles in rrs (1401G, 1402T, and 1484T) and FQ resistance target alleles in gyrA, stratified by high-level (gyrA 94AAC, 94CAC, 94GGC, and 94TAC, and all gyrA88 mutations) and low-level (gyrA 90GTG, 91CCG, and 94GCC) resistance-associated variants (7), were covered with a minimum of 10 SMOR reads (i.e., 20 or more standard reads, a pair of reads for each sequenced amplicon molecule), resulting in at least 10× the total needed reads to quantify a proportion (e.g., 1,000 total amplicon reads to detect a 1% minor population). In one phenotypically resistant isolate without a genotypic correlate in gyrA, SMOR was also applied to gyrB (496CTC and 500CAC). Numerous no-template controls were used to ensure the lack of well-to-well sample or amplicon contamination. All sequencing read files were deposited in the National Institutes of Health Short Read Archive (SRA BioProject #PRJNA503635). The SMOR percent mutant values for each sampling point and patient were used as predictors at a range of prespecified cut-points (0.1%, 0.5%, 1%, 5%, and 20%), with phenotypic DST as the outcome. Because repeated measures of the SMOR percent mutant were used as predictors, confidence intervals (CIs) were obtained using bootstrap resampling by participant. Statistical analysis was conducted in R 3.4.1, RStudio 1.0.153, and Stata (v15.1).
Results
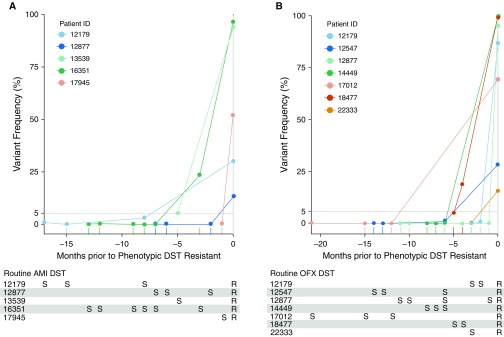
The association between preceding microheteroresistance and subsequent phenotypic resistance to SLIs and FQs was assessed independently. Among the patients who were phenotypically susceptible to SLIs at baseline (n = 16/19, 84%) and acquired SLI phenotypic resistance during follow-up (n = 14/16, 88%), six (43%; 95% CI, 18–71%) had preceding microheteroresistance for a median of 8.0 months (IQR, 5.5–9.75 mo) before they developed phenotypic resistance (Figure 1). Two patients did not acquire phenotypic resistance (follow-up time 23 and 37 mo) but had documented microheteroresistance. Among the patients who were phenotypically susceptible to FQs at baseline (n = 18/19, 95%) and developed phenotypic resistance during follow-up (n = 17/18, 94%), seven (41%; 95% CI, 18–67%) had preceding microheteroresistance for a median of 8.0 months (IQR, 3.0–12 mo) before phenotypic resistance was documented. The single patient who did not acquire resistance had no preceding microheteroresistance (followed for 5 mo). A receiver operating characteristic analysis using all heteroresistance measures over time as a continuous measure indicated that 89.7% (95% CI, 80–99.4%) and 90.9% (95% CI, 83.3–98.6%) of the patients who eventually developed reference-standard resistance to SLIs and FQs, respectively, had preceding heteroresistance of at least 0.5%.
Figure 1.
Mycobacterium tuberculosis genotypic microheteroresistance/phenotypic susceptibility in months preceding the first documented phenotypic resistance to (A) aminoglycosides or (B) fluoroquinolones among patients with rifampin-resistant tuberculosis. Second-line drug-susceptibility testing (DST) on Middlebrook 7H11 media containing 2 μg/ml and 4 μg/ml for ofloxacin (OFX) and amikacin (AMI), respectively, and Single Molecule-Overlapping Reads sequencing of crude DNA extracts from original cultures were performed at each time point from cultures obtained for routine care. All patients were assumed to be on standardized multidrug-resistant-tuberculosis regimens (this was confirmed for seven of eight patients with available treatment records). (A) Variant frequency refers to resistance-associated variants (RAVs) at rrs 1401G, except for patient 12877, where initial microheteroresistance was noted at rrs 1484T and subsequently became fixed at rrs 1401G. (B) Variant frequency refers to RAVs at gyrA 94GGC, gyrA 94GCC, and gyrB 496CTC/500CAC for patients 14449, 22333, and 18477, respectively. For patients 12179 and 17012, initial microheteroresistance was noted at gyrA 94GGC and subsequently became fixed at gyrA 90GTG and gyrA 91CCG, respectively. For patients 12547 and 12877, initial microheteroresistance was noted at gyrA 94CAC and gyrA 88TGC, respectively, and subsequently became fixed at gyrA 94GGC. Additional RAVs occurred concurrently, but to preserve clarity, not all are represented. Patients without acquired phenotypic resistance (i.e., those who remained susceptible through the study period or were resistant at baseline; AMI, n = 5; OFX, n = 3) or had no preceding microheteroresistance (i.e., either heteroresistance >5% or no preceding RAV detected; AMI, n = 8; OFX, n = 9) were excluded from the figure. R = resistant; S = susceptible.
Discussion
In this retrospective proof-of-concept study, we used a novel NGS procedure to demonstrate a high prevalence of preexisting microheteroresistance among individuals with amplified aminoglycoside and FQ resistance that preceded established phenotypic resistance by a median of 8 months. Furthermore, minority genotypic subpopulations were frequently documented at levels beneath Canetti and colleagues’ canonical 1% cut-point (8). These findings may have important implications for the detection and monitoring of drug-resistant TB.
The expanded use of targeted NGS has a number of clinical advantages over conventional techniques, including shorter turnaround times, more complete resistance profiles, and easier storage and transportation of samples. Minor resistant M.tb subpopulations identified through deep sequencing may indicate a transitory stage marked by selection and clonal interference before the development of fixed resistance (9, 10), whose emergence is a function of drug pressure and spatial heterogeneity due to granuloma communication with the airways. The replacement of gyrA 88TGC with gyrA 94GGC in our study could be representative of this phenomenon. Within this paradigm, more temporary reductions in cumulative drug pressure might account for the appearance and subsequent disappearance of microheteroresistance. The ultimate clinical significance of these subpopulations will depend on a pretest probability defined by drug efficacy and barrier to resistance, companion regimen, adherence, fitness cost, and patient predictors. An improved understanding of M.tb microheteroresistance within this context could have a significant impact on the clinical management of drug-resistant TB.
A limitation of our analysis is the purposeful selection of cases based on acquisition of aminoglycoside and/or FQ resistance, which precluded us from determining specificity. However, the much higher prevalence of microheteroresistance in our study relative to that noted in a previous study of drug-susceptible TB, where further acquisition of drug resistance did not occur (11), may be meaningful. Second, the 145 sampled time points in our study were retrospectively assessed and occurred at programmatically relevant intervals. Thus, although the sampled time points were likely associated with clinical events, they also represent times when clinical guidance was most needed. Third, the full genetic complement of resistance-associated variants continues to be elucidated, and loci other than those studied here may be determined in the future to confer phenotypic resistance.
In conclusion, previously undetectable microheteroresistant subpopulations frequently occur before fixed genotypic or phenotypic drug resistance develops in patients with baseline rifampin resistance. The incremental clinical utility of the detection and kinetics of these subpopulations as bacteriologic measures should be assessed in prospective trials including patients who develop or do not develop amplification of drug resistance.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank Tanja Dolby from the NHLS for processing the samples used in this study.
Footnotes
Supported by the National Institute of Allergy and Infectious Diseases R01AI131939 (J.Z.M. and D.M.E.), a National Research Foundation (NRF) Research Career Advancement Award (E.M.S.), NRF Incentive Funding (G.T. and R.M.W.), and the EDCTP2 program (G.T.), and the South African Medical Research Council (R.M.W.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NRF or the South African Medical Research Council.
Author Contributions: Conception or design: D.M.E., E.M.S., G.T., R.M.W., and J.Z.M. Acquisition of data: E.M.S. and R.M.W. Analysis and interpretation of data, D.M.E., E.M.S., E.J.K., C.J.A., K.W., D.J., D.L., E.V., G.T., F.A.S., R.M.W., and J.Z.M. Drafting of the manuscript: D.M.E., E.M.S., E.J.K., and J.Z.M. Revision of the manuscript for important intellectual content: G.T. and R.M.W.
Author disclosures are available with the text of this letter at www.atsjournals.org.
References
- 1.Sharma A, Hill A, Kurbatova E, van der Walt M, Kvasnovsky C, Tupasi TE, et al. Global Preserving Effective TB Treatment Study Investigators. Estimating the future burden of multidrug-resistant and extensively drug-resistant tuberculosis in India, the Philippines, Russia, and South Africa: a mathematical modelling study. Lancet Infect Dis. 2017;17:707–715. doi: 10.1016/S1473-3099(17)30247-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Folkvardsen DB, Svensson E, Thomsen VO, Rasmussen EM, Bang D, Werngren J, et al. Can molecular methods detect 1% isoniazid resistance in Mycobacterium tuberculosis? J Clin Microbiol. 2013;51:1596–1599. doi: 10.1128/JCM.00472-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakravorty S, Simmons AM, Rowneki M, Parmar H, Cao Y, Ryan J, et al. The new Xpert MTB/RIF Ultra: improving detection of Mycobacterium tuberculosis and resistance to rifampin in an assay suitable for point-of-care testing. MBio. 2017;8:e00812–e00817. doi: 10.1128/mBio.00812-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blakemore R, Story E, Helb D, Kop J, Banada P, Owens MR, et al. Evaluation of the analytical performance of the Xpert MTB/RIF assay. J Clin Microbiol. 2010;48:2495–2501. doi: 10.1128/JCM.00128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colman RE, Schupp JM, Hicks ND, Smith DE, Buchhagen JL, Valafar F, et al. Detection of low-level mixed-population drug resistance in Mycobacterium tuberculosis using high fidelity amplicon sequencing. PLoS One. 2015;10:e0126626. doi: 10.1371/journal.pone.0126626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Metcalfe JZ, Streicher E, Theron G, Colman RE, Allender C, Lemmer D, et al. Cryptic micro-heteroresistance explains Mycobacterium tuberculosis phenotypic resistance. Am J Respir Crit Care Med. 2017;196:1191–1201. doi: 10.1164/rccm.201703-0556OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miotto P, Tessema B, Tagliani E, Chindelevitch L, Starks AM, Emerson C, et al. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. Eur Respir J. 2017;50:1701354. doi: 10.1183/13993003.01354-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canetti G, Froman S, Grosset J, Hauduroy P, Langerova M, Mahler HT, et al. Mycobacteria: laboratory methods for testing drug sensitivity and resistance. Bull World Health Organ. 1963;29:565–578. [PMC free article] [PubMed] [Google Scholar]
- 9.de Vos M, Ley SD, Wiggins KB, Derendinger B, Dippenaar A, Grobbelaar M, et al. Bedaquiline microheteroresistance after cessation of tuberculosis treatment. N Engl J Med. 2019;380:2178–2180. doi: 10.1056/NEJMc1815121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trauner A, Liu Q, Via LE, Liu X, Ruan X, Liang L, et al. The within-host population dynamics of Mycobacterium tuberculosis vary with treatment efficacy. Genome Biol. 2017;18:71. doi: 10.1186/s13059-017-1196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin SS, Modongo C, Baik Y, Allender C, Lemmer D, Colman RE, et al. Mixed Mycobacterium tuberculosis-strain infections are associated with poor treatment outcomes among patients with newly diagnosed tuberculosis, independent of pretreatment heteroresistance. J Infect Dis. 2018;218:1974–1982. doi: 10.1093/infdis/jiy480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.