Abstract
The ATP-binding cassette subfamily B member 1 (ABCB1) multidrug transporter P-glycoprotein plays a central role in clearance of xenobiotics in humans and is implicated in cancer resistance to chemotherapy. We used double electron electron resonance spectroscopy to uncover the basis of stimulation of P-glycoprotein adenosine 5′-triphosphate (ATP) hydrolysis by multiple substrates and illuminate how substrates and inhibitors differentially affect its transport function. Our results reveal that substrate-induced acceleration of ATP hydrolysis correlates with stabilization of a high-energy, post-ATP hydrolysis state characterized by structurally asymmetric nucleotide-binding sites. By contrast, this state is destabilized in the substrate-free cycle and by high-affinity inhibitors in favor of structurally symmetric nucleotide binding sites. Together with previous data, our findings lead to a general model of substrate and inhibitor coupling to P-glycoprotein.
One Sentence Summary:
Transport substrates and inhibitors differentially interact with an asymmetric high energy post ATP hydrolysis state of the ABC transporter P-glycoprotein.
Efficient substrate extrusion by ATP-binding cassette (ABC) efflux transporters, including the mammalian P-glycoprotein (Pgp), entails the transduction of adenosine 5′-triphosphate (ATP) energy, harvested in nucleotide-binding domains (NBDs), to protein conformational motion in transmembrane domains (TMDs) (1–11). Transport models for Pgp have emerged from an expanding database of structures (12–14), the elucidation of its substrate-coupled conformational dynamics (8), and biochemical studies of ATP turnover (15–18). Convincing evidence supports a two-stroke model with alternating ATP hydrolysis at the two nucleotide-binding sites (NBSs) (16, 18, 19). Investigation of the substrate-coupled conformational cycle of Pgp by double electron electron resonance (DEER) spectroscopy (20–22) bolstered this model by uncovering a vanadate-trapped high-energy posthydrolysis state (HES), previously referred to as the transition state of ATP hydrolysis (23, 24), that results from hydrolysis of ATP molecules in structurally and catalytically asymmetric NBSs (8). Concomitant with the formation of the HES, the transporter samples occluded (OO) and outward-facing (OF) conformations, suggesting that the power stroke for transition between inward-facing (IF) and OF states requires ATP hydrolysis. By contrast, a recent cryo–electron microscopy (cryo-EM) structure of an ATP-bound Pgp mutant in an OF conformation (25) motivated a transport model wherein substrate extrusion precedes initiation of ATP hydrolysis. The mutant was impaired for ATP hydrolysis by glutamine substitution of two catalytic glutamate residues (Fig. 1A), substitutions that were previously demonstrated to abrogate NBS asymmetry and stabilize the OF conformation by ATP binding (8).
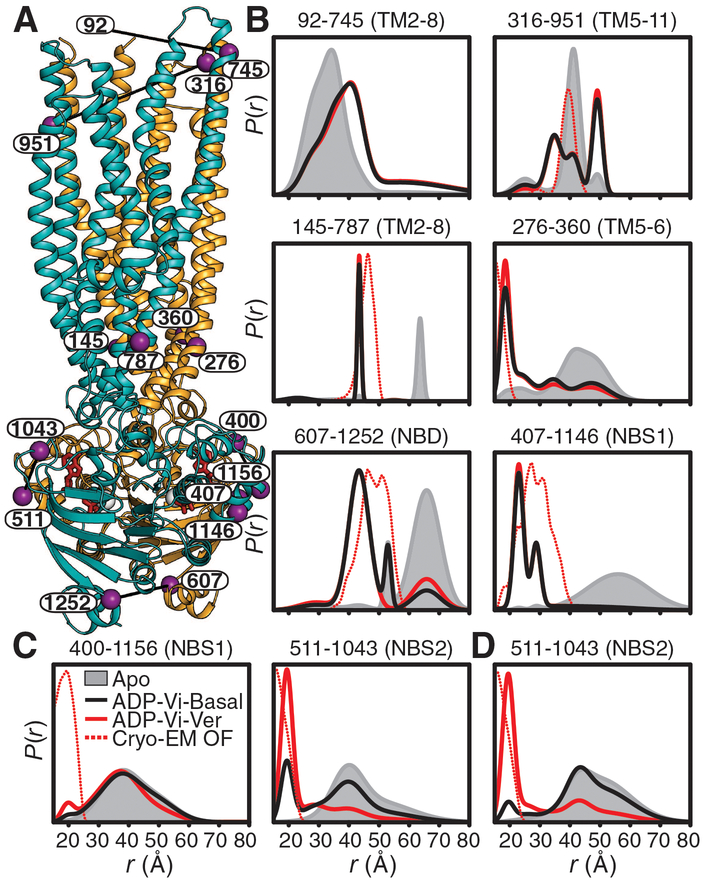
Fig. 1. HESs for basal and substrate-coupled cycles differ by the conformation of the A-loops.
(A) Ribbon representation of the OF Pgp [Protein Data Bank (PDB) code 6C0V] with the N- and C-terminal halves colored with orange and cyan, respectively, and highlighting the positions of spin-label pairs as purple spheres. (B) Distance distributions in the TMDs and NBDs and (C and D) the A-loops obtained in nucleotide-free Pgp (Apo) and the HES (ADP-Vi) in the presence and absence of the substrate Ver in nanodiscs. The corresponding distributions predicted from the cryo-EM OF structure are shown as dashed lines. (D) Distance distributions for NBS2 in mixed micelles are shown for reference. Residue 92 is not resolved in the cryo-EM structure, precluding prediction of distance distributions.
Resolving the discrepancy between the two models requires elucidation of how substrate binding in the TMD is allosterically coupled to ATP hydrolysis. For this purpose, we compared Pgp’s conformational cycles in the absence (basal cycle) and presence of the substrate verapamil (Ver), which accelerates ATP turnover (stimulated cycle). Distance distributions for selected spin-label pairs that were previously shown to fingerprint the IF-to-OF transition (8) were measured in lipid nanodiscs (Fig. 1) (26) and in mixed-detergent/lipid micelles (figs. S1 and S2). For both basal and stimulated cycles (Fig. 1B), we observed a pattern of distance changes between ligand-free (apo) Pgp (black traces in Fig. 1) and the HES (trapped by vanadate after ATP hydrolysis, ADP-Vi, red traces in Fig. 1) consistent with the model of alternating access described previously (8). Assembly of the NBD catalytic dimer in the HES (e.g., residue pair 607–1252), which brings together the Walker and ABC signature motifs to form the NBSs, is coupled to the homogeneous closure of the intracellular TMD, manifested by almost complete shift of the distributions to shorter distances relative to the corresponding IF, apo-Pgp distributions. At the extracellular side, the TMD undergoes an opening movement as evidenced by distinct longer-distance components. However, independent of substrate binding, a substantial fraction of apo-like distances persists in the extracellular distributions in contrast to the intracellular side of the TMD, which suggests the population of an OO conformation (extracellularly closed) in addition to an OF conformation (extracellularly open).
For most spin-label pairs, we observed minor differences (≅10%) between the basal and substrate-coupled HES distance distributions (see fig. S1), which is consistent with efficient Vi trapping and similar overall conformation. Substantial changes upon substrate binding were limited to the distributions of spin-label pairs monitoring the conserved A-loops (27), which are located in the NBSs and directly pack against the adenosine of bound ATP through a conserved tyrosine (Figs. 2A, 1C). A distinct short-distance component (~ 67%) is evident in the substrate-coupled HES (Fig. 1C) at the NBS2 A-loop (511–1043), but not at the equivalent pair at the NBS1 A-loop (400–1156). We previously interpreted the structural asymmetry between A-loops as a signature of high-affinity occlusion of ATP in NBS2 and its hydrolysis in NBS1 (8), which parallels the intrinsic catalytic asymmetry of substrate-coupled Pgp (16, 18). By contrast, for substrate-free HES, the short-distance component at NBS2 is a minor population (~ 25%) in nanodiscs (Fig. 1C) and negligible (~ 7%) in mixed micelles (Fig. 1D). Thus, the basal and stimulated cycles differ by the heterogeneity and asymmetry of the A-loops in otherwise structurally similar OO and OF conformations.
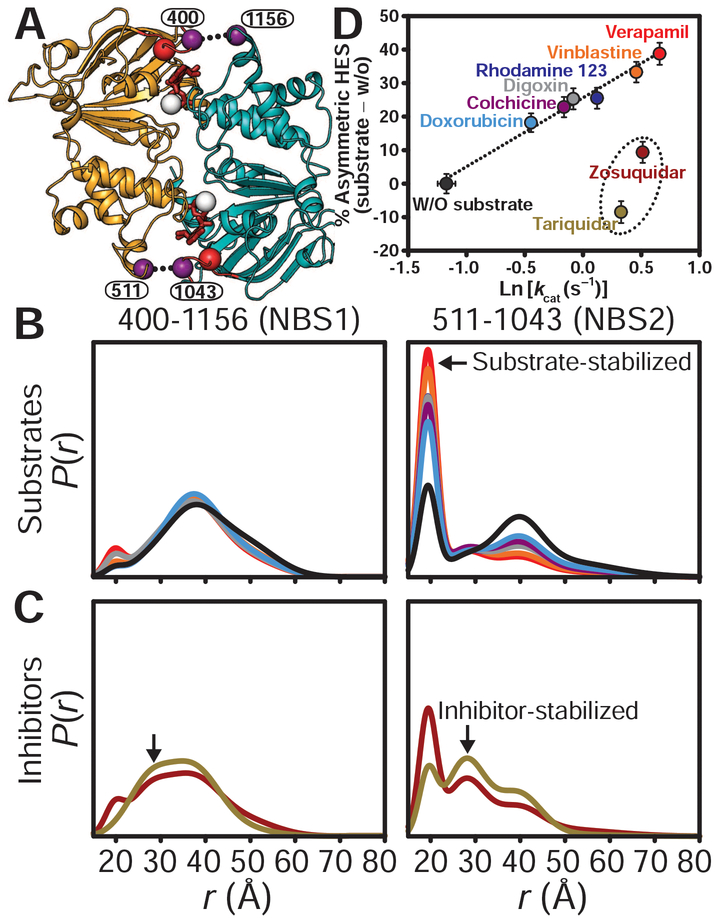
Fig. 2. Stimulation of ATP turnover by substrates entails stabilization of an asymmetric HES.
(A) Cytoplasmic view of the NBD dimer in the OF conformation in complex with ATP (red sticks) showing the A-loop (red spheres) spin-label pairs. (B) Distance distributions of the A-loop pairs highlighting the substrate dependence of the short-distance component (arrow) at NBS2. (C) Pgp inhibitors stabilize an HES with a distinct distance component (arrows) relative to substrates. (D) The asymmetric HES population for transport substrates is directly related to the activation energy of ATP turnover (Ln kcat). Experiments in (B) to (D) were performed in nanodiscs. Distance distributions were obtained in the trapped (ADP-Vi) HES.
Whereas DEER distance distributions in the NBD and the intracellular TMD were consistent with the cryo-EM structure of the double E/Q mutant, A-loop asymmetry was not observed in this presumed OF structure (Fig. 1 and fig. S2), thus predicting similar short-distance components for pairs monitoring the A-loops. This deviation is rationalized by the finding that A-loop asymmetry can be abrogated by the double E/Q mutation (8) and reversed by a single E/Q mutation (fig. S3). Another notable deviation is evident on the extracellular side, which is more closed (apo-like) in the cryo-EM structure relative to the HES (fig. S2). The cryo-EM density at the extracellular side was diffuse, suggesting a dynamic region (25), a finding consonant with the heterogeneous experimental DEER distributions.
To uncover the mechanistic implications of the asymmetric HES, we determined how a set of substrates, which display different transport efficiency (28) and ATP turnover rates (15) (figs. S4 to S6), modulate A-loop heterogeneity and asymmetry (Fig. 2 and fig. S7). Furthermore, Arrhenius analysis of ATP turnover for a subset of these substrates demonstrated a common rate-limiting step (15). Because transport substrates can inhibit ATP turnover at high concentrations, saturating stimulatory substrate concentrations were selected (determined in figs. S4B and S6 and tables S1 and S2). We found that substrates most efficient in stimulating ATP turnover (e.g., Ver) induced the largest population of the short component at NBS2. Indeed, a linear relationship emerged between this component’s population, representing the spectroscopic asymmetric HES, and the natural logarithm of stimulated ATP turnover rate constant (kcat). Other pairs in the NBD and TMD did not show substantial population changes (i.e., >10%), which exclude the possibility of heterogeneous Vi trapping and therefore link the allosteric effects of substrate binding to the conformation of A-loops. Given that Ln (kcat) reflects the activation energy of the hydrolytic transition state, we conclude that substrate-induced reduction in the activation energy requires stabilization of the catalytically and structurally asymmetric HES. Moreover, distinct kcat values and populations of HES asymmetry imply differential interaction of the substrates with this state, which has OO and OF conformations. Thus, substrate extrusion must occur simultaneously with or subsequent to ATP hydrolysis.
In contrast to transport substrates, the Pgp high-affinity inhibitors zosuquidar and tariquidar (12, 29) (Figs. 2C and 3 and fig. S8) induced broader distance distributions at NBS1, suggesting the population of an intermediate-distance component in the HES. A similar component was observed at NBS2 (black arrows in Fig. 2C), whereas the population of the short-distance component, stabilized by substrates, was similar to that of the basal cycle for zosuquidar and was reduced for tariquidar. The intermediate-distance components reveal a distinct, more symmetric conformation of the A-loops associated with high-affinity inhibitor binding.
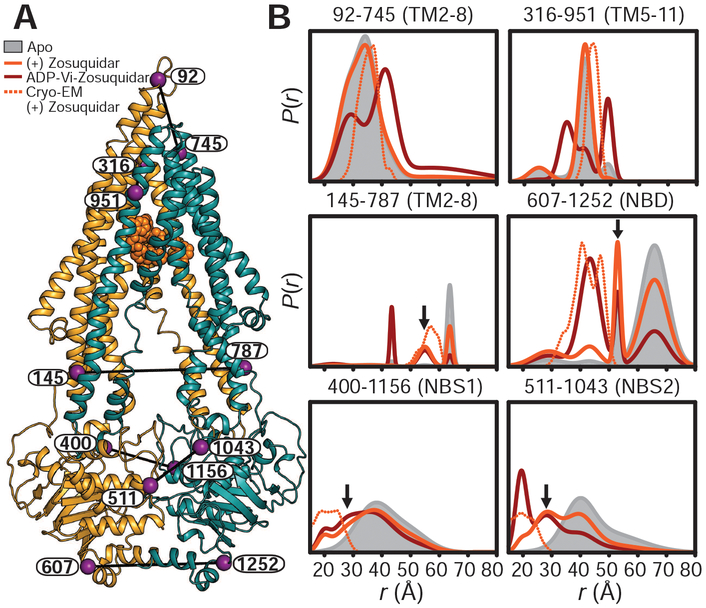
Fig. 3. The high-affinity inhibitor zosuquidar stabilizes a distinct IF conformation of Pgp.
(A) Ribbon representation of zosuquidar-bound Pgp cryo-EM structure (PDB code 6FN1) highlighting the positions of spin-label pairs as purple spheres. (B) Distance distributions obtained in nanodiscs for apo-Pgp, zosuquidar-bound nucleotide-free Pgp, and the HES (ADP-Vi). Predicted distributions are shown as dotted lines. Arrows highlight components on the intracellular side of the TMD and at the A-loops that are either not observed or are minor components in apo-Pgp. The y axes in Figs. 1 and 3 are identical.
To uncover how high-affinity inhibitors abrogate transport and destabilize the asymmetric HES while stimulating ATP turnover (30), we mapped structural changes in apo-Pgp induced by inhibitor binding (fig. S8, C and D). Although the distance distributions were heterogeneous, suggesting a conformational equilibrium, a distinct IF conformation was observed wherein the TMD intracellular side and NBDs were closer relative to apo-Pgp (solid orange traces, black arrows in Fig. 3 and fig. S8). By contrast, distance distributions were unchanged upon the addition of excess substrates to apo-Pgp (fig. S8B), indicating a lack of structural changes. Moreover, the IF conformation stabilized by inhibitors persisted in the HES (ADP-Vi, dark red traces in Fig. 3), suggesting that inhibitors not only reduce the asymmetry at the A-loops but also impair homogeneous closing of the intracellular TMD and the formation of an OF conformation (8). The direction of the distance changes in the inhibitor-stabilized IF agrees with a recent cryo-EM structure of zosuquidar-bound Pgp (Fig. 3) (12), with one distance component overlapping the distributions predicted from this structure (dashed orange traces in Fig. 3). However, the conformational equilibrium implied by heterogeneous DEER distributions is likely restricted in this structure by binding of an extracellular antibody and a disulfide bond introduced at the NBD interface.
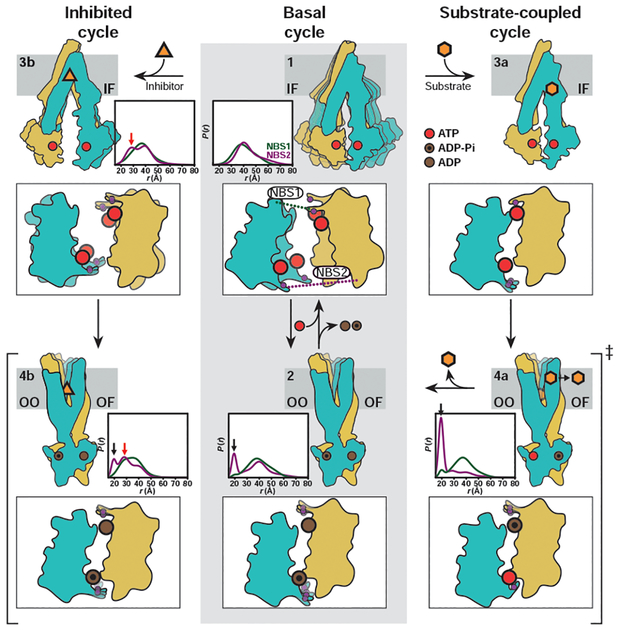
Our findings can be framed in a model that describes how inhibitors and substrates differentially couple to the Pgp transport cycle. The model (Fig. 4) invokes distinct HESs differing by the configuration of the NBSs but otherwise having almost identical OO/OF conformations. The basal cycle proceeds primarily through the NBS-symmetric HES in which both ATP molecules have been hydrolyzed presumably in random order (Fig. 4, step 2). Because Vi trapping of ADP, subsequent to ATP hydrolysis, can only occur in one NBS at a time (8, 16, 18), the symmetric HES indicates that the basal cycle has reduced catalytic asymmetry.
Fig. 4. Model of Pgp transport and inhibition.
The basal cycle (middle panels, steps 1 and 2) entails conformational sampling by ATP-bound Pgp to enable NBD dimerization followed by population of a symmetric HES. The substrate-coupled cycle (steps 1, 3a, 4a, and 2) is initiated by substrate binding. As previously postulated (8, 16, 18), a transient conformation with one tightly bound ATP molecule is likely populated. Hydrolysis of one ATP stabilizes the asymmetric HES (step 4a), which consists of OO/OF conformations. Hydrolysis of the second ATP leads to the symmetric HES as in the basal cycle (step 2). Pgp inhibition (steps 1, 3b, and 4b) is initiated by stabilization of an IF conformation in which the extracellular sides of TMD is apo-like, whereas the intracellular sides of TMDs and NBDs are closer than the apo state. ATP hydrolysis proceeds through a heterogeneous HES. For each intermediate, we show cytoplasmic views of the NBDs highlighting the conserved A-loop tyrosines as purple spheres along with associated DEER distributions in nanodiscs.
Stimulation of ATP hydrolysis by substrates proceeds through an NBS-asymmetric HES, in which one ATP molecule is hydrolyzed whereas the other is occluded with high affinity (Fig. 4, step 4a). We propose that asymmetric occlusion of ATP lowers the enthalpic barrier for hydrolysis and consequently accelerates the overall cycle, which is manifested by a larger kcat. The substrate-dependent asymmetric HES population is consistently less than 100%, implying that the symmetric HES is also populated in the substrate-stimulated cycle. Whereas it is possible that the substrate-coupled cycle involves a transient conformation bound to two ATP molecules similar to the cryo-EM structure (25), our findings establish that substrate extrusion cannot proceed until at least one ATP is hydrolyzed. Stabilization of the asymmetric HES for Pgp reconstituted in nanodiscs compared with mixed micelles (Fig. 1, C and D) rationalizes the stimulation of basal ATP hydrolysis (tables S1 and S2) (26) in the former and suggests that HES stability is lipid dependent. It also structurally contextualizes substrate-dependent Gibbs energy analysis of ATP turnover that postulated a single rate-limiting conformational step (15), i.e. transition to the OF conformation, and linked intrinsically different kcat of substrates to their affinities to this rate-limiting step as observed here for the asymmetric HES.
Inhibitors stabilize an NBS-heterogeneous HES (Fig. 4, step 4b) distinct from that of the basal cycle. We suggest that high-affinity inhibitors accelerate ATP hydrolysis by inducing an IF conformation in which the NBDs are closer relative to ligand-free Pgp, effectively reducing the conformational entropy of the IF ensemble. The energy of ATP hydrolysis is not sufficient to homogeneously close the intracellular side in the HES, as evidenced by persistent apo-like distance components (Fig. 3B, black arrow in panel 145–787). Accordingly, transport, requiring isomerization to an OF, is impaired by inhibitor binding.
In addition to illuminating the mechanism of allosteric modulation of Pgp by transport substrates and inhibitors, our approach of monitoring the conformational equilibrium of Pgp–specifically direct detection of the distinct HES populations–provides a strategy to identify allosteric modulators for multidrug ABC exporters in general, thereby accelerating the development of new inhibitors.
Supplementary Material
Acknowledgments:
We thank Brandy Verhalen for helpful discussions, Derek P. Claxton and Richard A. Stein for critical reading and editing of the manuscript.
Funding: This work was supported by National Institutes of Health grant U54-GM087519 (to H.S.M. and R.K.N.).
Footnotes
Competing interests: The authors declare there are no competing interests.
Data and materials availability: The generated data including the DEER experiments are available in the manuscript or the Supplementary Materials.
Supplementary Materials:
science.sciencemag.org/content/364/6441/689/suppl/DC1
Materials and Methods
Figs. S1 to S8
Tables S1 and S2
DEER Data Appendices
References and Notes:
- 1.Sharom FJ, Pharmacogenomics 9, 105–127 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Ferreira RJ, Bonito CA, Ferreira MJU, dos Santos DJVA, WIREs Comput. Mol. Sci 7, e1316 (2017). [Google Scholar]
- 3.Rees DC, Johnson E, Lewinson O, Nat. Rev. Mol. Cell. Biol 10, 218–227 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Locher KP, Nat. Struct. Mol. Biol 23, 487–493 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Szöllősi D, Rose-Sperling D, Hellmich UA, Stockner T, Biochim. Biophys. Acta Biomembr 1860, 818–832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor NMI, et al. , Nature 546, 504–509 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Johnson ZL, Chen J, Cell 168, 1075–1085 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Verhalen B, Dastvan R, et al. , Nature 543, 738–741 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barth K, et al. , J. Am. Chem. Soc 140, 4527–4533 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Zoghbi ME, et al. , J. Biol. Chem 292, 20412–20424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishra S, et al. , Elife, e02740 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alam A, et al. , Proc. Natl. Acad. Sci. U.S.A 115, E1973–E1982 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esser L, et al. , J. Biol. Chem 292, 446–461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Jaimes KF, Aller SG, Protein Sci 23, 34–46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Shawi MK, Polar MK, Omote H, Figler RA, J. Biol. Chem 278, 52629–52640 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Tombline G, Senior AE, J. Bioenerg. Biomembr 37, 497–500 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Tombline G, et al. , Biochemistry 47, 3294–3307 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Siarheyeva A, Liu R, Sharom FJ, J. Biol. Chem 285, 7575–7586 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janas E, et al. , J Biol Chem 278, 26862–9 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Jeschke G, Annu. Rev. Phys. Chem 63, 419–446 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Mchaourab HS, Steed PR, Kazmier K, Structure 19, 1549–1561 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schiemann O, Prisner TF, Q. Rev. Biophys 40, 1–53 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Urbatsch IL, Sankaran B, Bhagat S, Senior AE, J. Biol. Chem 270, 26956–26961 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Oldham ML, Chen J, Proc. Natl. Acad. Sci. U.S.A 108, 15152–15156 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim Y, Chen J, Science 359, 915–919 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Shukla S, Abel B, Chufan EE, Ambudkar SV, J. Biol. Chem 292, 7066–7076 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ambudkar SV, Kim IW, Xia D, Sauna ZE, FEBS Lett 580, 1049–1055 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Stein WD, Cardarelli C, Pastan I, Gottesman MM, Mol. Pharmacol 45, 763–772 (1994). [PubMed] [Google Scholar]
- 29.Chufan EE, Kapoor K, Ambudkar SV, Biochem. Pharmacol 101, 40–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loo TW, Clarke DM, Biochem. Pharmacol 92, 558–566 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Zou P, Mchaourab HS, Biophys. J 98, L18–L20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smriti, Zou P, Mchaourab HS, J. Biol. Chem 284, 13904–13913 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeschke G, Polyhach Y, Phys. Chem. Chem. Phys 9, 1895–1910 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Stein RA, Beth AH, Hustedt EJ, Methods Enzymol 563, 531–567 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hustedt EJ, Marinelli F, Stein RA, Faraldo-Gómez JD, Mchaourab HS, Biophys. J 115, 1200–1216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polyhach Y, Bordignon E, Jeschke G, Phys. Chem. Chem. Phys 13, 2356–2366 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.