Abstract
Parasesarma liho Koller, Liu & Schubart, 2010 and P. cognatum Rahayu & Li, 2013 from East and Southeast Asia are similar species that have been confused in several studies. Here, we re-examined the type specimens of both species and found identical main characters, which is supported by the molecular identity of the mitochondrial cytochrome oxidase subunit I gene. As a result, we treat P. cognatum as a junior subjective synonym of P. liho. We also show that the male paratype of P. paucitorum Rahayu & Ng, 2009 is conspecific with P. liho, although P. paucitorum s. str. remains a distinct but allied species. The distribution of P. liho is updated to include Japan (Ryukyus), Taiwan, Philippines (Cebu) and Indonesia (Sulawesi).
Keywords: Parasesarma, P. liho, P. cognatum, P. paucitorum, Morphology, Mitochondrial COI
BACKGROUND
Crabs of the family Sesarmidae Dana, 1851 form part of the dominant macrofauna of the Indo-West Pacific mangrove ecosystems, playing an especially important ecological role as “ecosystem engineers” (Lee 1998; Kristensen 2008). They inhabit landward regions of mangrove fringes and can tolerate high temperature and salinity fluctuations (Theurkauff et al. 2018). With 69 recognised species, Parasesarma De Man, 1895 is the most speciose genus of this family, especially after most species of Perisesarma De Man, 1895 have been moved to Parasesarma and the recent descriptions of more new species (see Ng et al. 2008; Shahdadi et al. 2017 2018a 2019; Li et al. 2018; Shahdadi and Schubart 2017; Fratini et al. 2019). In Taiwan, 12 species of this genus have been reported, of which eight, viz. P. cognatum Rahayu & Li, 2013, P. corallicum Ng, Davie & Li, 2016, P. kuekenthali (De Man, 1902), P. kui Li, Rahayu & Ng, 2018, P. lepidum (Tweedie, 1950), P. liho Koller, Liu & Schubart, 2010, P. macaco Li, Rahayu & Ng, 2018, and P. ungulatum (H. Milne Edwards, 1853), were added after the latest revision of Taiwanese crabs in 2001 (Ng et al. 2001 2016 2017; Koller et al. 2010; Rahayu and Li 2013; Li 2015; Hsu and Shih 2018; Li et al. 2018 2019).
Recently, several taxonomic studies of crabs have used molecular evidence to support the descriptions or recognition of new or reinstated species (cf. Chu et al. 2015); this is also the case within the family Sesarmidae (Schubart et al. 1998 2009; Gillikin and Schubart 2004; Koller et al. 2010; Naderloo and Schubart 2010; Ragionieri et al. 2012; Thiercelin and Schubart 2014; Cannicci et al. 2017; Shahdadi et al. 2017 2018a b). To help identify species of Taiwanese sesarmids, a DNA barcoding approach using the cytochrome oxidase subunit I (COI) marker (Hebert et al. 2003a b) was undertaken. A preliminary result of the COI analyses showed that only one clade was obtained for specimens labelled as P. liho and P. cognatum from Taiwan; and this required further investigation.
Parasesarma liho Koller, Liu & Schubart, 2010 (type locality: Hualien, Taiwan) is distributed in Hualien and Taitung, eastern Taiwan (Koller et al. 2010), as well as the Ryukyus, Japan (Okinawa, Miyako and Ishigaki; Maenosono and Naruse 2015). Parasesarma cognatum Rahayu & Li, 2013 (type locality: Pingtung, Taiwan) is distributed in Pingtung (southern Taiwan) and Hualien (eastern Taiwan), as well as in Cebu (the Philippines) according to Rahayu and Li (2013). Rahayu and Li (2013: 639) mentioned that P. cognatum is different from P. liho in the proportions of ambulatory propodi, structure of the male first gonopod, number and shape of dactylar tubercles of male chela, and coloration. However, the similarity between the two species has been remarked by Maenosono and Naruse (2015: 22), who questioned their identities.
In the present study, the types of these species, as well as more specimens of different sizes, were examined and their COI sequences compared, including specimens from various localities. The types of the allied P. paucitorum Rahayu & Ng, 2009 from Indonesia were also studied.
MATERIALS AND METHODS
New specimens with the appearance of Parasesarma liho and P. cognatum were collected from their type localities and other areas in southern and eastern Taiwan, as well as from Cebu, Philippines. The holotypes and paratypes of P. liho, P. cognatum and P. paucitorum were also included. Individuals of other related species (see below) were also studied for comparison (Table 1). Those specimens were deposited in the Biodiversity Research Museum, Academia Sinica, Taipei, Taiwan (ASIZ); the Muséum national d’Histoire naturelle, Paris (MNHN); the Museum Zoologi Bogor, Indonesian Institute of Sciences, Indonesia (MZB); the Zoological Collections of the Department of Life Science, National Chung Hsing University, Taichung, Taiwan (NCHUZOOL); the National Museum of Marine Biology and Aquarium, Pingtung, Taiwan (NMMBA); National Museum of Natural Science, Taichung, Taiwan (NMNS); the Senckenberg Museum, Frankfurt am Main, Germany (SMF); Zoological Reference Collection of the Lee Kong Chian Natural History Museum, National University of Singapore (ZRC); and the Zoologische Staatssammlung, München (Munich), Germany (ZSM).
Table 1.
Found haplotypes of the COI gene of Parasesarma liho, P. cognatum and P. paucitorum, as well as the outgroups. For abbreviations of museums and universities see MATERIALS AND METHODS
| Locality | sample size | Catalogue no. | Haplotype of COI | Access. no. of COI |
| P. liho or P. cognatum | ||||
| Taiwan | ||||
| Hualien: Meilun R. estuary | 1 | SMF 36266 (holotype of P. liho) | —a | LC490879 |
| 1 | ZSM A20100040 (paratype of P. liho) | —a | LC490880 | |
| 1 | NCHUZOOL 15027 | PRL-C1 | LC490881 | |
| Taitung: Dulanwan | 2 | NCHUZOOL 15025 | PRL-C1 | LC490881 |
| Pingtung: Niou R. estuary | 1 | NCHUZOOL 15031 | PRL-C1 | LC490881 |
| Pingtung: Gangkou R. estuary | 1 | NMMBCD 3975 (holotype of P. cognatum) | PRL-C1 | LC490881 |
| 2 | NMMBCD 3976 (paratypes of P. cognatum) | PRL-C1 | LC490881 | |
| 1 | NMMBCD 3976 (paratype of P. cognatum) | PRL-C2 | LC490882 | |
| 1 | NCHUZOOL 15028 | PRL-C1 | LC490881 | |
| 1 | ZRC 2013.1757 | —a | LC490883 | |
| 1 | ZRC 2013.1757 | —a | LC490883 | |
| Pingtung: Houwan | 3 | NCHUZOOL 15022 | PRL-C1 | LC490881 |
| 1 | NCHUZOOL 15425 | PRL-C1 | LC490881 | |
| Pingtung: Baoli R. | 1 | NCHUZOOL 15024 | PRL-C1 | LC490881 |
| Philippines | ||||
| Cebu: Kawasan | 1 | ASIZCR | PRL-C1 | LC490881 |
| 1 | ASIZCR | PRL-C1 | LC490881 | |
| 1 | NCHUZOOL 15034 | PRL-C2 | LC490882 | |
| Indonesia | ||||
| Sulawesi: Manado | 1 | ZRC 2019.0578 (male paratype of P. paucitorum) | —a | LC490884 |
| Others | ||||
| P. paucitorum: Sulawesi, Indonesia | 1 | MZB Cru 2243 (holotype) | LC490885 | |
| 1 | ZRC 2008.0869 (female paratype) | LC490886 | ||
| P. kui: Gangkou R. estuary, Pingtung, Taiwan | 1 | NMNS 7779-015 (holotype) | LC490887 | |
| P. macaco: Baoli R. estuary, Pingtung, Taiwan | 1 | NMNS-7779-005 (holotype) | LC490888 | |
| P. tripectinis: Dajia R. estuary, Taichung, Taiwan | 1 | NCHUZOOL 15428 | LC490889 | |
| P. pictum: Nangan, Matsu, Taiwan | 1 | NCHUZOOL 15427 | LC490890 | |
| P. affine: Danshuei, New Taipei, Taiwan | 1 | NCHUZOOL 15426 | LC490891 | |
| P. dumacense: Cebu, Philippines | ZRC 2008.0833 | KX400929 |
asequences are shorter and not included for further analyses (see RESULTS).
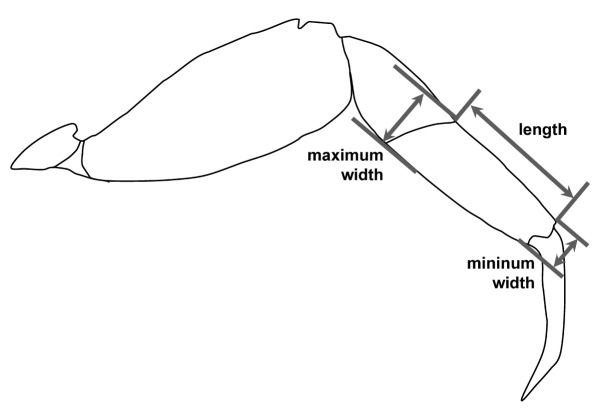
The abbreviations P4 is used for the fourth pereiopods (third ambulatory legs), and G1 for male first gonopods. Measurements, all in millimeters (mm), are of the maximum carapace width (CW) and carapace length (CL). The G1s of different sized specimens were compared to determine whether there is a size effect on the morphology of G1. The ratios of length/width of the P4 propodus for different-sized specimens were calculated, as it was used to distinguish P. liho and P. cognatum by Rahayu and Li (2013). Because different authors could measure different positions from the photograph (Shih and Do 2014), both the proximal and distal widths of the propodi were measured to obtain the range of width in our study (Fig. 1). The morphology of the upper margin of the cheliped merus of the holotype was also examined to confirm the presence of a large subdistal spine, as mentioned in Rahayu and Li (2013: 637) and Maenosono and Naruse (2015: 22, fig. 7B).
Fig. 1.
Schematic drawing showing the measurement of the length, as well as the maximum and minimum widths of the propodi of the fourth pereiopods (third ambulatory leg, P4) used in this study.
Genomic DNA was isolated from the muscle tissue of the pleon or walking leg with different kits (Shih et al. 2016; Shahdadi and Schubart 2017). A portion of the COI gene was amplified with PCR using the primers LCO1490, HCO2198 (Folmer et al. 1994) or COL6 and COH6 (Schubart 2009). The PCR conditions for the above primers were: denaturation for 50 s at 94°C, annealing for 70 s at 45–47°C (45 s at 48–50°C for COL6-COH6), and extension for 60 s at 72°C (40 cycles), followed by extension for 10 min at 72°C. Sequences were obtained by automated sequencing (Applied Biosystems 3730) and verified with the complementary strand. Sequences of the different haplotypes were deposited in the DNA Data Bank of Japan (DDBJ) (accession numbers in Table 1).
For comparative purposes, we included the species or species complexes used in the study of P. liho, P. cognatum and P. paucitorum in Koller et al. (2010), Rahayu and Li (2013) and Rahayu and Ng (2009), respectively, using the following as outgroups: P. affine (De Haan, 1837), P. dumacense (Rathbun, 1914), P. leptosoma (Hilgendorf, 1869) (now corresponding to P. kui Li, Rahayu & Ng, 2018, P. macaco Li, Rahayu & Ng, 2018 of this species complex; see Li et al. 2018), P. pictum (De Haan, 1835), and P. tripectinis (Shen, 1940) (Table 1). The COI sequences of P. dumacense was downloaded from GenBank (accession number: KX400929).
The best-fitting model for sequence evolution was determined by Partition Finder (vers. 2.1.1; Lanfear et al. 2017), selected by the Bayesian information criterion (BIC). The obtained best model (GTR + I) was subsequently used for a Bayesian inference (BI) analysis. The BI analysis was performed with MrBayes (vers. 3.2.3, Ronquist et al. 2012). The phylogenetic analyses were run with four chains for 10 million generations and four independent runs, with trees sampled every 1000 generations. The convergence of chains was determined by the average standard deviation of split frequency values below the recommended 0.01 (Ronquist et al. 2019) and the first 3000 trees were discarded as burnin. The maximum likelihood (ML) analysis was conducted in RAxML (vers. 7.2.6, Stamatakis 2006). Because RAxML does not accept the GTR + I model, the second best model, GTR + G (i.e., GTRGAMMA), was used with 100 runs, and the best ML tree was found by comparing the likelihood scores. The robustness of the ML tree was evaluated by 1000 bootstrap pseudoreplicates under the model GTRGAMMA. The relationships of the COI haplotypes among P. liho and other related species were examined by using the program PopART (vers. 1.7, Leigh and Bryant 2015). Basepair (bp) differences and the pairwise estimates of Kimura 2-parameter (K2P) distances (Kimura 1980) for genetic diversities between haplotypes were calculated with MEGA (vers. 10.0.5, Kumar et al. 2018).
Material examined (see Table 1): Holotype of P. liho: male (13.0 × 12.3 mm) (SMF 36266), Meilun R. (= River) estuary, Hualien, Taiwan, coll. H.-C. Liu, 31 October 2009; paratypes: 1 male (12.7 × 11.5 mm) (ZSM A20100040), same data as holotype; 1 male (14.2 × 13.1 mm) (SMF 36269), 1 male (14.5 × 13.1 mm) (MNHN B32312), same locality as holotype, coll. H.-C. Liu, 6 November 2000.
Holotype of P. cognatum: male (14.3 × 13.1 mm) (NMMBCD 3975), Gangkou R. estuary, Manjhou, Pingtung, Taiwan, coll. J.-J. Li, 1 September 2012; paratypes: 2 females (11.5 × 10.3 mm; 14.4 × 12.6 mm) (NMMBCD 3506), same locality as holotype, coll. J.-J. Li, 8 June 2012 (the data is different from that in Rahayu and Li 2013).
Others: Taiwan: 1 female (10.8 mm) (NCHUZOOL 15024), Baoli R. estuary, Pingtung, coll. P.-Y. Hsu et al., 11 July 2017; 1 male (14.2 mm) (NCHUZOOL 15030), Houwan, Pingtung, coll. P.-Y. Hsu, 26 June 2012; 2 males (11.1–16.2 mm), 5 females (12.6–16.0 mm) (NCHUZOOL 15022), Houwan, Pingtung, coll. R.-H. Lee, 19 September 2013; 3 males (9.7–14.9 mm), 1 female (9.3 mm) (NCHUZOOL 15425), Houwan, Pingtung, coll. P.-Y. Hsu and C.-Y. Chi, 3 December 2016; 1 female (16.2 mm) (NCHUZOOL 15029), Leidashih, Kenting, Pingtung, coll. J.-H. Lee, 18 August 2012; 1 male (16.7 mm) (NCHUZOOL 15031), Niou R. estuary, Pingtung, coll. P.-Y. Hsu, 19 January 2016; 2 males (13.3–13.0 mm), 1 female (11.8 mm) (ZRC 2013.1757), Gangkou R. estuary, Pingtung, coll. J.-J. Li, 19 February 2013; 2 females (11.7–12.6 mm) (NCHUZOOL 15023), Fushuei Bridge, Gangkou R. estuary, Pingtung, coll. P.-Y. Hsu et al., 12 July 2017; 2 females (8.6–13.9 mm) (NCHUZOOL 15028), Gangkou R. estuary, Pingtung, coll. P.-Y. Hsu et al., 4 September 2017; 8 males (4.8–12.4 mm), 2 females (5.6–6.0 mm) (NCHUZOOL 15025), Dulanwan, Taitung, coll. P.-Y. Hsu et al., 9 August 2017; 1 male (6.1 mm), 2 females (10.9–10.9 mm) (NCHUZOOL 15032), Jihuei Fishing Port, Taitung, coll. P.-Y. Hsu et al., 10 August 2017; 1 male (11.6 mm) (NCHUZOOL 15027), Meilun R. estuary, Hualien, coll. J.-H. Lee, 18 May 2012; 1 male (11.6 mm), 3 females (10.2–11.3 mm) (NCHUZOOL 15026), Meilun R. estuary, Hualien, coll. P.-Y. Hsu et al., 10 August 2017. Philippines: 1 male (11.2 mm), 1 female (16.4 mm) (ASIZCR), Kawasan, Cebu, coll. H.-C. Liu, 4 December 2001; 2 males (13.0–13.3 mm) (NCHUZOOL 15034), Kawasan, Cebu, coll. J.-J. Li, 6 September 2018. Indonesia: 1 male (15.5 mm) (ZRC 2019.0578, ex ZRC 2008.0869 partim) (paratype of P. paucitorum Rahayu and Ng, 2009), Manado, northern Sulawesi, Indonesia, coll. P. K. L. Ng, 17 July 2003.
Comparative material: Parasesarma paucitorum: 1 male (19.7 mm) (MZB Cru 2243, holotype), 1 female (19.2 mm) (ZRC 2008.0869, paratype), Manado, northern Sulawesi, Indonesia, coll. P. K. L. Ng, 17 July 2003. P. affine: 1 male (29.6 mm) (NCHUZOOL 15426), Danshuei, New Taipei City, Taiwan, 30 June 2006. P. dumacense: 1 male (20.4 mm) (ZRC 2008.0833), Kawasan Waterfall, Cebu, Philippines, coll. H.-C. Liu, 25 November 2001. P. pictum: 1 male (15.0 mm) (NCHUZOOL 15427), Nangan, Matsu, Taiwan, coll. P.-Y. Hsu et al., 26 August, 2011. P. tripectinis: 1 male (9.7 mm) (NCHUZOOL 15428), Dajia R. estuary, Taichung, Taiwan, 31 May 2014.
RESULTS
Morphology
The CWs of the holotypes of Parasesarma liho (SMF 36266) and P. cognatum (NMMBCD 3975), as well as the paratypes of P. liho (SMF 36269; MNHN B32312) used for description and/or drawing are 13.0 mm, 14.3 mm, 14.2 mm and 14.5 mm, respectively. Different sized specimens with CW from 4.8 to 16.7 mm of males (n = 19) and 5.6 to 16.4 mm of females (n = 22) were included in this study.
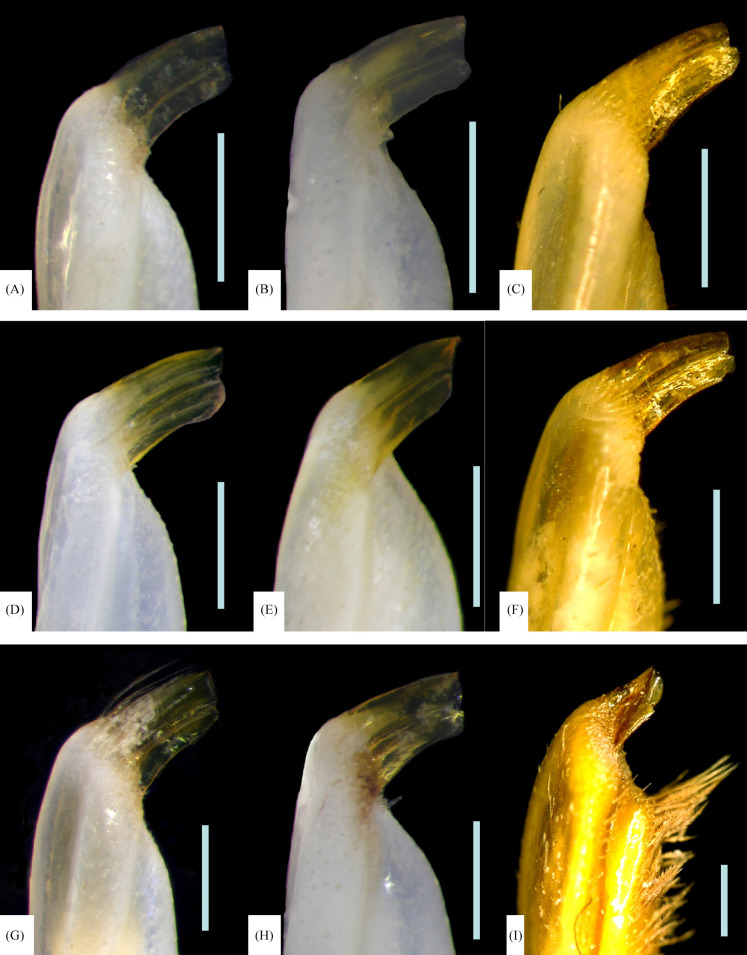
The distal part of the G1s of the specimens of P. liho and P. cognatum, with CW ranging from 11.1 to 16.7 mm, are quite similar in the form of the apical processes (Fig. 2A–E, G, H). The morphology of G1 of the holotype (SMF 36266) was examined and also agrees with other G1s shown in figure 2.
Fig. 2.
The morphological variation of distal part the right G1s of Parasesarma liho (A–H), and P. paucitorum (I). A, CW 11.1 mm (NCHUZOOL 15022), Pingtung, Taiwan; B, CW 11.6 mm (NCHUZOOL 15027), Hualien, Taiwan; C, CW 12.7 mm (ZSM A20100040, paratype of P. liho), Hualien, Taiwan; D, CW 13.29 mm (NCHUZOOL 15034), Cebu, Philippines; E, CW 14.3 mm (NMMBCD 3975, holotype of P. cognatum), Pingtung, Taiwan; F, CW 15.5 mm (ZRC 2019.0578, paratype of P. paucitorum), Sulawesi, Indonesia; G, CW 16.2 mm (NCHUZOOL 15022), Pingtung, Taiwan; H, CW 16.7 mm (NCHUZOOL 15031), Pingtung, Taiwan; I, CW 19.7 mm (MZB Cru 2243, holotype of P. paucitorum), Sulawesi, Indonesia. Scales bars = 0.5 mm.
The length/width ratios of the P4 propodi are shown in table 2; the differences in the maximum and minimum ratios are large, ranging from 2.67 to 4.76, for different sizes of CW (7.0–16.7 mm), including the paratype of P. liho (SMF 36269 and MNHN B32312; ratios: 2.79 and 2.67, respectively; based on Koller et al. 2010: figs. 2f, 3c) and the holotype of P. cognatum (NMMBCD 3975; ratio: 2.79).
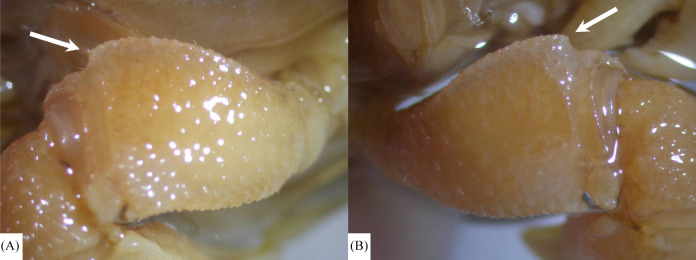
The chelar morphology of the types of P. liho and P. cognatum are similar, including the 11–12 elongate tubercles on the dorsal margins (Fig. 3A–D). The holotype of P. cognatum (NMMBCD 3975) was re-examined, confirming only a subdistal angle, not a spine, on the outer margin of the cheliped merus (Fig. 4).
Table 2.
The ratios of length/maximum width and length/minimum width of P4 propodi of specimens of Parasesarma liho with different size. The ratios of paratypes of P. liho was measured by Koller et al. (2010: figs. 2f, 3c). For abbreviations of museums and universities see MATERIALS AND METHODS
| CW (mm) | cat. no. | Length/maximum width | length/minimum width |
| 7.0 | NCHUZOOL 15025 | 3.19 | 4.76 |
| 11.1 | NCHUZOOL 15022 | 2.89 | 4.17 |
| 11.6 | NCHUZOOL 15027 | 2.81 | 4.15 |
| 11.6 | NCHUZOOL 15026 | 2.67 | 3.79 |
| 14.2 | SMF 36269 (paratype of P. liho) | 2.79 | 4.01 |
| 14.5 | MNHN B32312 (paratype of P. liho) | 2.67 | 3.88 |
| 14.3 | NMMBCD 3975 (holotype of P. cognatum) | 2.79 | 4.17 |
| 16.7 | NCHUZOOL 15031 | 3.36 | 4.53 |
Fig. 3.
The morphology of left chela of types of Parasesarma liho (A–B), P. cognatum (C–D) and P. paucitorum (E–H). A, C, E, G, outer view; B, D, F, H, upper view. A, B, holotype of P. liho (CW 13.0 mm, SMF 36266); C, D, holotype of P. cognatum (CW 14.3 mm, NMMBCD 3975); E, F, paratype of P. paucitorum (CW 15.5 mm, ZRC 2019.0578); G, H, holotype of P. paucitorum (CW 19.7 mm, MZB Cru 2243). Scales bars = 2 mm.
Fig. 4.
The outer surface of chelipedal meri of Parasesarma cognatum (holotype, NMMBCD 3975). A, left cheliped; B, right cheliped. Arrow indicates a subdistal angle on the upper margin of chelipedal merus.
For the coloration of P. liho in the field, the carapace is light brown with dark brown blotches, whereas the chelipeds and legs are uniform yellow to brownish yellow (Fig. 5).
Fig. 5.
The coloration of Parasesarma liho in the field in Taiwan. A, specimen (not captured) from Gangkou R. estuary, Pingtung; B, specimen (not captured) from Meilun R. estuary, Hualien.
The male paratype of P. paucitorum (ZRC 2019.0578) was also found to resemble other specimens of P. liho (and P. cognatum), but somewhat different from the holotype of P. paucitorum (MZB Cru 2243), including the G1s (Fig. 2F, I), chelae (Fig. 3E–H), as well as in the fresh coloration of carapace and legs (Rahayu and Ng 2009: fig. 1).
DNA analysis
The molecular results comprised 22 P. liho-like and P. cognatum-like specimens, including the holotype and paratypes of P. liho and P. cognatum (Table 1). Available COI sequences for the holotype of P. liho, the paratype of P. cognatum and the male paratype of P. paucitorum were a bit shorter (614–635 bp see below), but otherwise identical to most other sequences with 658 bp. Therefore, they were omitted from further analyses. The male paratype of P. paucitorum (ZRC 2019.0578) has a similar genetic sequence as P. liho (see above) and is also referred to as being in the P. liho clade (see below). In total, three haplotypes of P. liho (including P. cognatum) are found from among the studied specimens (Table 1).
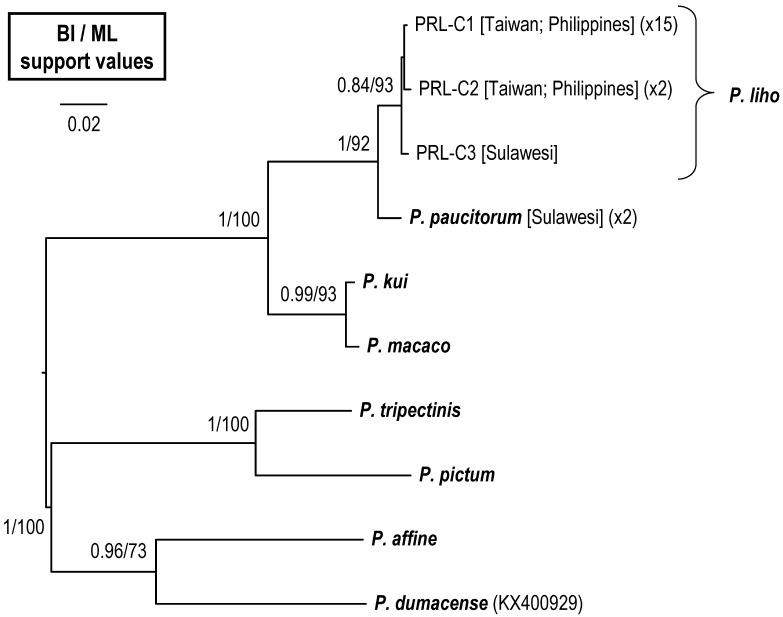
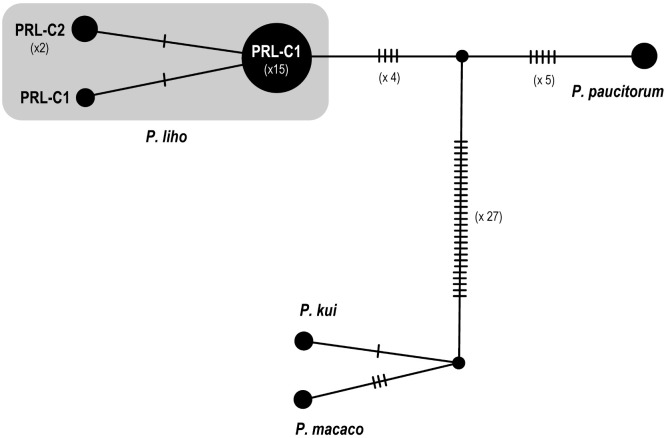
The reconstructed COI phylogenetic tree (Fig. 6) shows that the three haplotypes of P. liho (including the male paratype of P. paucitorum) form a distinct clade, sister to another clade including real P. paucitorum s. str. The mean pairwise nucleotide divergence with the K2P distances and bp differences of haplotypes of the two clades are shown in table 3. The intraspecific K2P nucleotide divergence within P. liho (including P. cognatum) is 0.05% (0–0.32%), and the interspecific K2P distance between P. liho and the P. paucitorum clade is 1.49% (1.46%–1.64%). Molecular data shows that the male paratype of P. paucitorum belongs to the P. liho clade and is different from the other types of P. paucitorum (Figs. 6, 7, Table 3). The haplotype network based on COI haplotypes (Fig. 7) further shows P. liho and P. paucitorum to be separated by 11 steps, and both species to be different from P. kui and P. macaco by 34–38 steps.
Fig. 6.
A Bayesian inference (BI) tree of Parasesarma liho, as well as the outgroups, based on the cytochrome oxidase subunit I genes (COI). Probability values at the nodes represent support values for BI and maximum likelihood (ML). For haplotype names, see table 1.
Table 3.
Matrix of percentage pairwise K2P nucleotide divergences (lower left) and mean number of differences (upper right) based on COI within the clade of Parasesarma liho (including types of P. cognatum and the male paratype of P. paucitorum) and other related species (see Table 1). Values of range are shown in parentheses
| Intraspecific |
Interspecific |
|||||
| Nucleotide divergence | Mean nucleotide difference | P. liho | P. paucitorum | P. kui | P. macaco | |
| P. liho | 0.05 (0-0.32) | 0.32 (0-2) | 9.17 (9-10) | 33.11 (33-34) | 35.11 (35-36) | |
| P. paucitorum | 0 | 0 | 1.49 (1.46-1.64) | 33 (33-33) | 33 (33-33) | |
| P. kui | 5.26 (5.22-5.58) | 5.5 (5.5-5.5) | 4 (4-4) | |||
| P. macaco | 5.6 (5.56-5.94) | 5.5 (5.5-5.5) | 0.61 (0.61-0.61) | |||
Fig. 7.
Genealogical network for the COI haplotypes observed within the clades of Parasesarma liho and other related species. Unlabelled hatches indicate inferred haplotypes not found in the sampled population. For haplotype names, see table 1.
DISCUSSION
In this study, we confirm that Parasesarma cognatum Rahayu & Li, 2013 is a synonym of Parasesarma liho Koller, Liu & Schubart, 2010 based on the evidence from morphology (Figs. 2–4, Table 2) and mitochondrial DNA (Figs. 6–7, Table 3).
The main differences between the two species was supposedly the morphology of G1 and the ratio of length/width of P4 propodus (Rahayu and Li 2013). The apical processes of the presently examined G1s are quite different from that of the paratype of P. liho (SMF 36269), which has a very tapering tip (Koller et al. 2010: fig. 2e). After comparing the line drawings of G1 in Koller et al. (2010), the tapering tip of the apical process in figure 2e (left) was likely due to breakage before or after it was denuded because it appeared broader when it was still with setae (Fig. 2d); which agrees with the specimens examined (Fig. 2) and the holotype of P. cognatum (Rahayu and Li 2013: fig. 7A-D). The morphology of both G1 structures of the holotype (SMF 36266) and the other paratypes (see Material examined) were also examined and agrees with the other G1s of this species (see Fig. 2).
Rahayu and Li (2013) mentioned the length/width ratio of P4 propodi in P. liho being 2.8 and that of P. cognatum 3.6, but our ratios for the paratype of P. liho are 2.79 and 2.67 (based on Koller et al. 2010: figs. 2f, 3c); and the holotype of P. cognatum is 2.79 (Table 2). The different values are probably caused by different positions used for measurement by different authors (see Shih and Do 2014), but the values are still variable, even if measured in a standardised manner (Table 2). The character of the length/width ratio of P4 propodi (Koller et al. 2010; Rahayu and Li 2013; Maenosono and Naruse 2015) is here considered not useful to distinguish the two species.
Rahayu and Li (2013: 637) described the merus of the chelipeds of P. cognatum as “outer margin tuberculate, with large subdistal spine”, but this character was not clearly shown in the corresponding figures. Maenosono and Naruse (2015: 22, fig. 7B) identified the species in the Ryukyus as P. liho because the specimens did not have such a large subdistal spine on the upper (= outer) margin of the merus (Koller et al. 2010: fig. 2a). In our study, it is clear that only a subdistal angle (not really a spine) is present on the upper margin of the merus of the holotype of P. cognatum (Fig. 4), a character that agrees with other specimens of P. liho (see Koller et al. 2010; Maenosono and Naruse 2015).
In the phylogenetic tree and haplotype network (Figs. 5, 6), only three haplotypes with only 1 or 2 bp difference were found in the clade of P. liho, including the haplotypes identical to the shorter sequences of the types of P. liho and P. cognatum. This supports the hypotheses that the two species should now be considered as conspecific. The two trees (Figs. 5, 6) also show that the male paratype of P. paucitorum should be identified as P. liho. The minimum interspecific divergence between the P. liho clade and the sister species, P. paucitorum, is 1.49%, which is small compared with those of most families in the Thoracotremata (Varunidae, Mictyridae, Ocypodidae, Dotillidae and Sesarmidae; see Chu et al. 2015). The small interspecific divergence between P. liho and P. paucitorum implies that these species are quite young, with an estimated divergence time of 0.88 mya (with an uncorrected p-distance divergence of 1.47%, based on the substitution rate of 1.66% per millions of years for COI for marine Sesarma in Schubart et al. 1998). The possibility remains that the small interspecific divergence could also be caused by mitochondrial introgression (e.g., Cannicci et al. 2017), but this needs further study by using nuclear evidence.
Koller et al. (2010) mentioned that P. liho is morphologically similar to P. paucitorum Rahayu & Ng, 2009 (type locality Manado, Sulawesi, Indonesia). The holotype and male paratype of P. paucitorum are, however, quite differently coloured (Rahayu and Ng 2009: figs. 1–2), with the colour of the paratype being very close to that of P. liho (Fig. 4; see Li and Chiu 2013: 82; Rahayu and Li 2013: fig. 4; Maenosono and Naruse 2015: figs. 2F–H, 8). Our study finds that the male paratype of P. paucitorum (ZRC 2019.0578) actually belongs to P. liho instead (Table 3, Figs. 5, 6). The holotype and paratype female of P. paucitorum remain genetically different.
Even if the interspecific genetic distance is small, adult P. liho and P. paucitorum can still be distinguished by the chelae (Fig. 3A–F vs. Fig. 3G–H), G1s (Fig. 2A–H vs. Fig. 2I), as well as the coloration of the adult carapace and ambulatory legs (Fig. 5 vs. Rahayu and Ng 2009: fig. 1A). That these two closely allied species occur together in one location in Sulawesi is somewhat surprising (all the specimens of P. paucitorum were collected together at the same time), but not without precedence in other sesarmids.
As P. cognatum is from now on synonymized with P. liho, and the male paratype of P. paucitorum also turned out to belong to P. liho, the updated distribution of P. liho is Japan (Okinawa, Miyako and Ishigaki), Taiwan (Hualien, Taitung, Pingtung), Philippines (Cebu) and Indonesia (Sulawesi).
CONCLUSIONS
In our study, the type specimens of Parasesarma liho and P. cognatum were re-examined and their main characters were found to be identical, which are also supported by the evidence from COI. Parasesarma cognatum is treated as a junior subjective synonym of P. liho accordingly. In addition, the male paratype of P. paucitorum is confirmed to be conspecific with P. liho. As a result, the updated distribution of P. liho is Japan (Ryukyus), Taiwan, Philippines (Cebu) and Indonesia (Sulawesi).
Acknowledgments
This study was supported by a grant from the Ministry of Science and Technology (MOST 105-2621-B-005-002-MY3), Executive Yuan, Taiwan, to HTS. We wish to thank Chia-Wei Lin for the type specimens of P. cognatum in NMMBCD; Kristin Arnold and Nicolas Noli for the pictures of type specimens of P. liho in SMF; and the member of HTS’s laboratory for collecting specimens. We acknowledge Peter K. L. Ng, one anonymous referee and the editor Benny K. K. Chan, all of whom greatly improved the manuscript.
Footnotes
Authors’ contributions: HTS conceived this study, the molecular analysis, and drafted the manuscript. PYH and AS performed the morphological comparison, measurements and the molecular work. CDS contributed to the discussion and drafted the manuscript. JJL collected and processed the samples, performed the ecological observation, and drafted the manuscript. All authors read and approved the final manuscript.
Competing interests: The authors declare that they have no conflict of interest.
Availability of data and materials: Sequences generated in the study have been deposited into the DNA Data Bank of Japan (DDBJ) database (accession numbers in Table 1 in manuscript).
Consent for publication: Not applicable.
Ethics approval consent to participate: Not applicable.
References
- Cannicci S, Schubart CD, Innocenti G, Dahdouh-Guebas F, Shahdadi A, Fratini S. 2017. A new species of the genus Parasesarma De Man 1895 from East African mangroves and evidence for mitochondrial introgression in sesarmid crabs. Zool Anz 269:89– 99. doi:10.1016/j.jcz.2017.08.002.
- Chu KH, Schubart CD, Shih HT, Tsang LM. 2015. Genetic diversity and evolution of Brachyura. In: Castro P, Davie PJF, Guinot D, Schram FR, von Vaupel Klein JC (eds) Treatise on zoology – anatomy, taxonomy, biology – The Crustacea, complementary to the volumes translated from the French of the Traité de Zoologie. Brill. Leiden, 9(C)(II), Decapoda: Brachyura (Part 2), pp. 775– 820.
- Gillikin DP, Schubart CD. 2004. Ecology and systematics of mangrove crabs of the genus Perisesarma (Crustacea: Brachyura: Sesarmidae) from East Africa. Zool J Linn Soc 141:435–445. doi:10.1111/j.1096-3642.2004.00125.x.
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299. [PubMed]
- Fratini S, Cannicci S, Porri F, Innocenti G. 2019. Revision of the Parasesarma guttatum species complex reveals a new pseudocryptic species in south-east African mangroves. Invertebr Syst 33:208–224. doi:10.1071/is18028.
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. 2003a. Biological identifications through DNA barcodes. Proc R Soc Lond B 270:313–321. doi:10.5772/49967. [DOI] [PMC free article] [PubMed]
- Hebert PDN, Ratnasingham S, deWaard RJ. 2003b. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond B 270:S96–S99. doi:10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed]
- Hsu PY, Shih HT. 2018. A new record crab of Parasesarma lepidum (Decapoda: Brachyura: Sesarmidae) from southern Taiwan. Coll Res 31:85–90.
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi:10.1007/bf01731581. [DOI] [PubMed]
- Koller P, Liu HC, Schubart CD. 2010. A new semiterrestrial species of the genus Parasesarma De Man, 1895, from Taiwan (Crustacea: Decapoda: Brachyura: Sesarmidae). Crust Monogr 14:357–368. doi:10.1163/9789047427759_024.
- Kristensen E. 2008. Mangrove crabs as ecosystem engineers; with emphasis on sediment processes. J Sea Res 59:30–43. doi:10.1016/j.seares.2007.05.004.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi:10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol 34:772–773. doi:10.1093/molbev/msw260. [DOI] [PubMed]
- Lee S Y. 1998. Ecological role of grapsid crabs in mangrove ecosystems: a review. Mar Freshw Res 49:335–343. doi:10.1071/mf97179.
- Leigh JW, Bryant D. 2015. PopART: Full-feature software for haplotype network construction. Methods Ecol Evol 6:1110– 1116. doi:10.1111/2041-210x.12410.
- Li JJ. 2015. Two new records of Parasesarma De Man, 1895 and Ptychognathus Stimpson, 1858 (Decapoda: Brachyura: Grapsoidea) from Taiwan. Taiwan J Biodivers 17:49–57. (in Chinese)
- Li JJ, Chiu YW. 2013. Land Crabs of Hengchun Peninsula. National Museum of Marine Biology & Aquarium, Pingtung, 91 pp. (in Chinese)
- Li JJ, Hsu JW, Ng NK, Shih HT. 2019. Eight new records of grapsoid crabs (Decapoda: Brachyura: Sesarmidae, Varunidae) from the coasts of Taiwan. Crustaceana 92. (in press).
- Li JJ, Rahayu DL, Ng PKL. 2018. Identity of the tree-spider crab, Parasesarma leptosoma (Hilgendorf, 1869) (Decapoda: Brachyura: Sesarmidae), with descriptions of seven new species from the Western Pacific. Zootaxa 4482:451–490. doi:10.11646/zootaxa.4482.3.2. [DOI] [PubMed]
- Maenosono T, Naruse T. 2015. Morphological characteristics and taxonomical problems of the genus Parasesarma De Man, 1895 (Crustacea: Decapoda: Brachyura: Sesarmidae) from the Ryukyu Archipelago, Japan. Fauna Ryukyuana 23:1–41. (in Japanese)
- Naderloo R, Schubart CD. 2010. Description of a new species of Parasesarma (Crustacea; Decapoda; Brachyura; Sesarmidae) from the Persian Gulf, based on morphological and genetic characteristics. Zool Anz 249:33–43. doi:10.1016/j.jcz.2010.01.003.
- Ng PKL, Davie PJF, Li JJ. 2016. On the identities of Parasesarma carolinense (Rathbun, 1907) and Parasesarma sigillatum (Tweedie, 1950), with description of a new species from Taiwan (Crustacea: Brachyura: Sesarmidae). Raffles B Zool 64:257–268.
- Ng PKL, Guinot D, Davie PJF. 2008. Systema Brachyurorum: Part I. An annotated checklist of extant brachyuran crabs of the world. Raffles B Zool Suppl 17:1–296.
- Ng PKL, Shih HT, Ho PH, Wang CH. 2017. An updated annotated checklist of brachyuran crabs from Taiwan (Crustacea: Decapoda). J Nat Taiwan Mus 70:1–208. doi:10.6532/JNTM.201712_70(3;4).01.
- Ng PKL, Wang CH, Ho PH, Shih HT. 2001. An annotated checklist of brachyuran crabs from Taiwan (Crustacea: Decapoda). Nat Taiwan Mus Spec Publ Ser 11:1–86.
- Ragionieri L, Fratini S, Schubart CD. 2012. Revision of the Neosarmatium meinerti species complex (Decapoda: Brachyura: Sesarmidae), with descriptions of three pseudocryptic Indo-West Pacific species. Raffles B Zool 60:71–87.
- Rahayu DL, Li JJ. 2013. A new species of the genus Parasesarma (Crustacea: Brachyura: Sesarmidae) from Taiwan and the Philippines, and redescription of P. jamelense (Rathbun, 1914). Raffles B Zool 61:633-639.
- Rahayu DL, Ng PKL. 2009. Two new species of Parasesarma De Man, 1895, from Southeast Asia (Crustacea: Decapoda: Brachyura: Sesarmidae). Zootaxa 1980:29–40. doi:10.5281/zenodo.185266.
- Ronquist F, Huelsenbeck JP, van der Mark P. 2005. MrBayes 3.1 manual. Available at http://mrbayes.csit.fsu.edu/manual.php. Accessed 9 September 2019.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MRBAYES 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi:10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed]
- Schubart CD. 2009. Mitochondrial DNA and decapod phylogenies: the importance of pseudogenes and primer optimization. In: Martin JW, Crandall KA, Felder DL (eds) Decapod Crustacean Phylogenetics. Crustacean Issues. CRC Press, Taylor & Francis Group. Boca Raton, London, New York, 18, pp. 47–65.
- Schubart CD, Diesel R, Hedges SB. 1998. Rapid evolution to terrestrial life in Jamaican crabs. Nature 393:363–365. doi:10.1038/30724.
- Schubart CD, Liu HC, Ng PKL. 2009. Revision of Selatium Serène & Soh, 1970 (Crustacea: Brachyura: Sesarmidae), with description of a new genus and two new species. Zootaxa 2154:1–29. doi:10.5281/zenodo.188831.
- Schubart CD, Reimer J, Diesel R. 1998. Morphological and molecular evidence for a new endemic freshwater crab, Sesarma ayatum sp. n., (Grapsidae, Sesarminae) from eastern Jamaica. Zool Scrip 27:373–380. doi:10.1111/j.1463-6409.1998.tb00468.x.
- Shahdadi A, Davie PJF, Schubart CD. 2017. Perisesarma tuerkayi, a new species of mangrove crab from Vietnam (Decapoda, Brachyura, Sesarmidae), with an assessment of its phylogenetic relationships. Crustaceana 90:1155–1175. doi:10.1163/15685403-00003663.
- Shahdadi A, Davie PJF, Schubart CD. 2018b. Systematics and phylogeography of the Australasian mangrove crabs Parasesarma semperi and P. longicristatum (Decapoda: Brachyura: Sesarmidae) based on morphological and molecular data. Invertebr Syst 32:196–214. doi:10.1071/is17040.
- Shahdadi A, Davie PJF, Schubart CD. 2019. A new species of Parasesarma (Decapoda: Brachyura: Sesarmidae) from northern Australian mangroves and its distinction from morphologically similar species. Zool Anz 279:116–125. doi:10.1016/j.jcz.2019.01.005.
- Shahdadi A, Ng PKL, Schubart CD. 2018a. Morphological and phylogenetic evidence for a new species of Parasesarma De Man, 1895 (Crustacea: Decapoda: Brachyura: Sesarmidae) from the Malay Peninsula, previously referred to as Parasesarma indiarum (Tweedie, 1940). Raffles B Zool 66:739–762.
- Shahdadi A, Schubart CD. 2017. Taxonomic review of Perisesarma (Decapoda: Brachyura: Sesarmidae) and closely related genera based on morphology and molecular phylogenetics: new classification, two new genera and the questionable phylogenetic value of the epibranchial tooth. Zool J Linn Soc 182:517–548 (printed in 2018). doi:10.1093/zoolinnean/zlx032.
- Shih HT, Do VT. 2014. A new species of Tiwaripotamon Bott, 1970, from northern Vietnam, with notes on T. vietnamicum (Dang & Ho, 2002) and T. edostilus Ng & Yeo, 2001 (Crustacea, Brachyura, Potamidae). Zootaxa 3764:26–38. doi:10.11646/zootaxa.3764.1.2. [DOI] [PubMed]
- Shih HT, Ng PKL, Davie PJF, Schubart CD, Türkay M, Naderloo R, Jones DS, Liu MY. 2016. Systematics of the family Ocypodidae Rafinesque, 1815 (Crustacea: Brachyura), based on phylogenetic relationships, with a reorganization of subfamily rankings and a review of the taxonomic status of Uca Leach, 1814, sensu lato and its subgenera. Raffles B Zool 64:139–175.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihoodbased phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi:10.1093/bioinformatics/btl446. [DOI] [PubMed]
- Theuerkauff D, Rivera-Ingraham GA, Roques JAC, Azzopardi L, Bertini M, Lejeune M, Farcy E, Lignot J, Sucré E. 2018. Salinity variation in a mangrove ecosystem: a physiological investigation to assess potential consequences of salinity disturbances on mangrove crabs. Zool Stud 57:36. doi:10.6620/ZS.2018.57-36. [DOI] [PMC free article] [PubMed]
- Thiercelin N, Schubart CD. 2014. Transisthmian differentiation in the tree-climbing mangrove crab Aratus H. Milne Edwards, 1853 (Crustacea, Brachyura, Sesarmidae), with description of a new species from the tropical eastern Pacific. Zootaxa 3793:545–560. doi:10.11646/zootaxa.3793.5.3. [DOI] [PubMed]