Abstract
Background
Traditional Chinese medicine (TCM) formulations have proven to be advantageous in clinical treatment and prevention of disease. LiuWei DiHuang Pill (LWDH Pill) is a TCM that was employed to treat type 2 diabetes mellitus (T2DM). However, a holistic network pharmacology approach to understanding the active ingredients and the therapeutic mechanisms underlying T2DM has not been pursued.
Methods
A network pharmacology approach including drug-likeness evaluation, oral bioavailability prediction, virtual docking, and network analysis has been used to predict the active ingredients and potential targets of LWDH Pill in the treatment of type 2 diabetes.
Results
The comprehensive network pharmacology approach was successfully to identify 45 active ingredients in LWDH Pill. 45 active ingredients hit by 163 potential targets related to T2DM. Ten of the more highly predictive components (such as :quercetin, Kaempferol, Stigmasterol, beta-sitosterol, Kadsurenone, Diosgenin, hancinone C, Hederagenin, Garcinone B, Isofucosterol) are involved in anti-inflammatory, anti-oxidative stress, and the reduction of beta cell damage. LWDH Pill may play a role in the treatment of T2DM and its complications (atherosclerosis and nephropathy) through the AGE-RAGE signaling pathway, TNF signaling pathway, and NF-kappa B signaling pathway.
Conclusion
Based on a systematic network pharmacology approach, our works successfully predict the active ingredients and potential targets of LWDH Pill for application to T2DM and helps to illustrate mechanism of action on a comprehensive level. This study provides identify key genes and pathway associated with the prognosis and pathogenesis of T2DM from new insights, which also demonstrates a feasible method for the research of chemical basis and pharmacology in LWDH Pill.
Keywords: LWDH Pill, type 2 diabetes mellitus, T2DM, network pharmacology
Introduction
Type 2 diabetes mellitus (T2DM) is a chronic disease, affecting about 90% of the people with diabetes. It would cause serious damages to the heart, blood vessels, eyes, kidneys, and nerves with long-time hyperglycemic status.1–4 Single target treatment of T2DM may be effective in controlling blood glucose levels in patients, but it is not effective in treating complications. Multiple pathways of treatment are often required to achieve and maintain long-term glycemic control.5,6 An economic cost estimate based on T2DM showed that per capita annual expenditures for treating diabetes in each country range from $242 to $11.917, which emerge a considerable impact in terms of costs to society, health systems, individuals and employers and in terms of a reduction in the productive workforce and productivity.7,8 Traditional Chinese medicine (TCM) herbal formula (FuFang in Chinese) was characterized by “multi-component, multichannel”, which has its own advantages and peculiarity in personalized treatment and early intervention.9
LiuWei DiHuang Pill (LWDH Pill), first mentioned during the Song Dynasty (AD 1119) writen by Qian Yi in his book “Pediatric Medicinals and Patterns”, is a traditional prescription for the treatment of many types of diseases in China clinically,such as asthenia of renal yin.10 It is described as good effect in T2DM with less toxicity and side effects, consisting six herbs: Rehmanniae radix preparata (Shu Di Huang: SDH), Cornus officinalis Sieb. (Shan Zhu Yu: SZY), Paeonia suffruticosa Andr. (Mu Dan Pi: MDP), Dioscorea opposite Thunb (Shan Yao: SY), Poria cocos (Schw.) Wolf (Fu Lin: FL), and Alisma orientale (Sam.) Juzep (Ze Xie: ZX), in the ratio of 8:4:4:3:3:3, respectively.11 Researches had been demonstrated that LWDH, as a safe and effective formula, were employed to improve T2DM and its complications, including diabetic nephropathy, diabetic encephalopathy, and diabetic muscle atrophy.11–13 Most of these correlations between LWDH and the corresponding treatment are derived from practical experience and long-term therapeutic observations. Thus, it is necessary to investigate scientific and technologic approaches to extend the understanding of the synergistic effects of LWDH Pill chemical compounds in treating T2DM.14,15
Recently, network pharmacology (NP) was proposed as a promising approach to discover TCM from a systems perspective and at the molecular level.16,17 Zeng et al, illuminated the molecular synergy of YHD for HER2-positive breast cancer (such as: reduces TGF-β1 secretion, regulating IGF-1receptors, down-regulating the expression of EGFR, et al.), by a network pharmacology approach.18 NP considers the contribution of each ingredient in TCMs to adequately explain the effects generated by an entire formula.19,20 Lee et al predicted 21 bioactive compounds and 57 genes linked to hyperlipidemia and atherosclerosis in Yijin-Tang by Network analysis.21 Pang et al, built constituent-target network, constituent-target-target network and target-biological pathway network indicate CDK5, MAOB, 5-HTR1A, GSK3-β, and found that COMT were important nodes for Naodesheng (NDS) formula in the treatment of Alzheimer’s disease (AD).22 These previous studies suggested that NP will be a good predictive tool for exploring the chemical compositions of LWDH and its relationships with T2DM. Our study is the first to identify potential bioactive compounds in LWDH Pill and elucidate its mechanisms in T2DM treatment by using the NP approach.
Materials And Methods
Ingredients Database Construction And ADME Screening
All candidate compounds of these six Chinese medicinal herbs in LWDH were collected from the three following databases: (1) Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) database (http://lsp.nwu.edu.cn/tcmsp.php, Ver.2.3).23 A total of 500 Chinese herbal medicines and 30,069 ingredients from the Chinese Pharmacopoeia (2010 edition) were registered in the TCMSP database.24 (2) Bioinformatics Analysis Tool for Molecular mechanism of Traditional Chinese Medicine (BATMAN) (http://bionet.ncpsb.org/batman-tcm/, updated January 2016), which consists of 46,914 formulas, 8159 herbs, and 25,210 ingredients through literature mining and database integration.25 (3) TCM Database@Taiwan (http://tcm.cmu.edu.tw/). A total of 351 compounds were identified in LWDH, including 77 in SDH, 231 in SZY, 59 in MDP, 75 in SY, 65 in FL, and 45 in ZX. Details of the 351 compounds are shown in Table S1.
Prior to target prediction, absorption, distribution, metabolism, and excretion (ADME) was employed to select bioactive components that contribute to its therapeutic effects, while those with poor pharmacological properties and bad drug ability compounds were removed.26 In order to obtain compounds with higher oral absorption, utilization, and biological properties for further analysis, we require that the candidate components meet two of the following parameters (1) Oral bioavailability (OB) ≥ 30%, (2) Drug-likeness (DL) ≥ 0.18.
The related genes of LWDH PILL bioactive components were obtained from TCMSP and DrugBank (https://www.drugbank.ca/, ver. 5.1.2) databases under the condition of Homo sapiens. UniProt KB (http://www.uniprot.org/), which prevents, for instance, over-annotation of similar proteins like paralogs and putative products of pseudogenes, was employed to standardize gene names and organisms.27 After that, these revised targets will lead to STRING (https://string-db.org/, ver. 11.0) predicting the associated protein–protein interaction (PPI) to obtain target genes directly and indirectly related to T2DM.
Predicting Putative Targets Of T2DM
The T2DM-related target proteins were screened from two sources: (1) GeneCards (https://www.genecards.org/, ver. 4.9.0) database; it is an online catalog of human genes and genetic diseases, which enables effectively navigate gene–disease linkages.28 2. OMIM (http://www.omim.org/, updated February 19, 2019) database possesses more than 15,500 gene entries and focuses on explaining gene–phenotype relationships.29 Only “Homo sapiens” proteins linked to T2DM were selected, and it should be considered whether there was a protein–ligand complex, crystal structure determination method, resolution, and so on. We got 7822 genes totally, and details are described in Table S2.
Protein–Protein Interaction (PPI)
The retrieved LWDH PILL active ingredient targets are correlated with T2DM targets by the STRING (https://string-db.org/, ver. 11.0) database.30 The condition was limited to “Homo sapiens”. In this study, a high confidence score with correlation degree ≥ 0.700, as the cut-off value, was set to obtain a network.
Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis For Targets
KEGG pathway analysis was performed to further probe the vital biological process of achieved targets and to validate the reliability of the integrated results.31–33 Finally, KEGG (http://www.genome.ad.jp/kegg/, updated February 1, 2019) terms with p-value ≤ 0.05 were selected for further research.
Network Construction And Analysis
Network construction was performed by the network visualization software Cytoscape (ver. 3.5.0) as follows: (1) Compound–compound target network; (2) Compound–T2DM PPI network; (3) LWDH PILL–T2DM network. Moreover, four topological properties of the hub network, including “Degree” and “Betweenness Centrality”, were calculated to screen the LWDH Pill candidate targets with topological importance.34,35
To obtain the links between network clusters and picked out hub genes with high degree of connectivity, “Molecular Complex Detection” (MCODE: a plug-in of Cytoscape) was employed to investigate node composition.36 Degree cutoff=2, Node Score Cutoff=0.2, and K-Core=2 were selected as the advanced options.37,38
Validation Of Compound–Target Interaction
Molecular Operating Environment (MOE 2015.10 software) was employed to validate the compound–target association in our study.26,39 The target proteins in all networks were obtained 3D structures from the RCSB PDB database (http://www.rcsb.org/). The proteins and ligands were prepared in the MOE tool prior to performing the docking process. The crystal structure of the target protein is pretreated, including removal of water molecules, Protonate 3D hydrogenation, correction of protein structure, energy optimization and retention of the active region of the target.40 The structure of ligands needs to meet to low-energy conformations. MOE-dock tool was used to semi-flexibly couple the active ingredient to the target protein. The placement function and scoring function use Triangle Matcher and London dG, respectively, and the rest use default parameters.
Results And Discussion
Compound–Compound Target Network Analysis
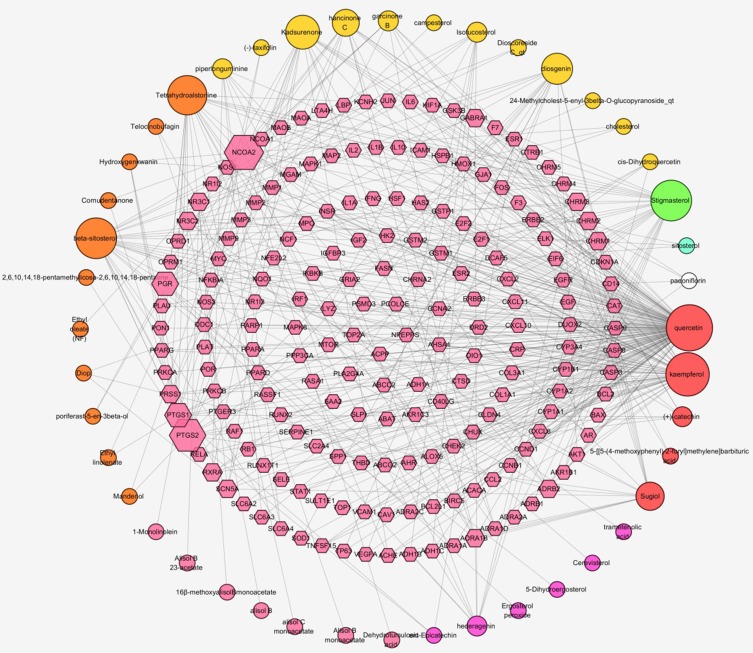
There are 229 nodes (185 compound target nodes and 45 compound nodes) and 418 edges composed compound–compound target network (Figure 1). Details of the 45 potentially bioactive ingredients and 185 target genes are provided in Table 1. In this network, we can find that some special compounds, interacting with multiple targets, participated in the regulation of multiple targets (central nodes, for instance, Quercetin, Kaempferol, Stigmasterol, Beta-Sitosterol, Kadsurenone, Diosgenin, Hancinone C, Hederagenin, Garcinone B, Isofucosterol).
Figure 1.
Compound–compound target network of LWDH Pill. Compound–compound target network of LWDH Pill consists of 185 compound target nodes and 45 compounds. The gene targets are described as a pink hexagon. Red, orange, yellow, pink, and purple circles represent MDP, SZY, SY, ZX, and FL, respectively. Green circle stands for common compound of SDH, SZY, and SY. Blue circle is employed to stand for common compound of SDH, SZY, MDP, and ZX. White circle represents common compound of MDP and FL. Node size of gene targets and compounds is proportional to the number of constituent. Lines stand for relation of compounds and target nodes.
Table 1.
Compound–Compound Target Network Information
| Mol ID | Compound | MW | OB (%) | DL | Medicine | Database Source |
|---|---|---|---|---|---|---|
| MOL000359 | Sitosterol | 414.79 | 36.91 | 0.75 | SDH, SZY, MDP, ZX | TCMSP |
| MOL000449 | Stigmasterol | 412.77 | 43.83 | 0.76 | SDH, SZY, SY, ZX | TCMSP |
| MOL001494 | Mandenol | 308.56 | 42 | 0.19 | SZY | TCMSP |
| MOL001495 | Ethyl linolenate | 306.54 | 46.1 | 0.2 | SZY | TCMSP |
| MOL001771 | Poriferast-5-en-3beta-ol | 414.79 | 36.91 | 0.75 | SZY | TCMSP |
| MOL002879 | Diop | 390.62 | 43.59 | 0.39 | SZY | TCMSP |
| MOL002883 | Ethyl oleate (NF) | 310.58 | 32.4 | 0.19 | SZY | TCMSP |
| MOL000358 | Beta-sitosterol | 414.79 | 36.91 | 0.75 | SZY | TCMSP |
| MOL005481 | 2,6,10,14,18-pentamethylicosa-2,6,10,14.18-pentaene | 342.67 | 33.4 | 0.24 | SZY | TCMSP |
| MOL005503 | Cornudentanone | 378.56 | 39.66 | 0.33 | SZY | TCMSP, Taiwan, BATMAN-TCM |
| MOL005530 | Hydroxygenkwanin | 300.28 | 36.47 | 0.27 | SZY | TCMSP |
| MOL005531 | Telocinobufagin | 402.58 | 69.99 | 0.79 | SZY | TCMSP, Taiwan, BATMAN-TCM |
| MOL008457 | Tetrahydroalstonine | 352.47 | 32.42 | 0.81 | SZY | TCMSP, Taiwan, BATMAN-TCM |
| MOL000098 | Quercetin | 302.25 | 46.43 | 0.28 | MDP | TCMSP |
| MOL000211 | Betulinic acid | 456.78 | 55.38 | 0.78 | MDP | TCMSP |
| MOL000422 | Kaempferol | 286.25 | 41.88 | 0.24 | MDP | TCMSP |
| MOL000492 | (+)-Catechin | 290.29 | 54.83 | 0.24 | MDP | TCMSP |
| MOL007374 | 5-[[5-(4-methoxyphenyl)-2-furyl]methylene]barbituric acid | 312.3 | 43.44 | 0.3 | MDP | TCMSP |
| MOL001924 | Paeoniflorin | 480.51 | 53.87 | 0.79 | MDP, FL | BATMAN-TCM |
| MOL002222 | Sugiol | 300.48 | 36.11 | 0.28 | MDP | BATMAN-TCM, Taiwan |
| MOL001559 | Piperlonguminine | 273.36 | 30.71 | 0.18 | SY | TCMSP |
| MOL001736 | (-)-Taxifolin | 304.27 | 60.51 | 0.27 | SY | TCMSP |
| MOL000322 | Kadsurenone | 356.45 | 54.72 | 0.38 | SY | TCMSP |
| MOL005430 | Hancinone C | 400.51 | 59.05 | 0.39 | SY | TCMSP |
| MOL005435 | 24-Methylcholest-5-enyl-3belta-O-glucopyranoside_qt | 400.76 | 37.58 | 0.72 | SY | TCMSP |
| MOL005438 | Campesterol | 400.76 | 37.58 | 0.71 | SY | TCMSP, BATMAN-TCM |
| MOL005440 | Isofucosterol | 412.77 | 43.78 | 0.76 | SY | TCMSP |
| MOL005458 | Dioscoreside C_qt | 444.72 | 36.38 | 0.87 | SY | TCMSP |
| MOL000546 | Diosgenin | 414.69 | 80.88 | 0.81 | SY | TCMSP, BATMAN-TCM |
| MOL005465 | Aids180907 | 394.45 | 45.33 | 0.77 | SY | TCMSP |
| MOL000953 | Clr | 386.73 | 37.87 | 0.68 | SY | TCMSP |
| MOL004580 | Cis-Dihydroquercetin | 304.27 | 66.44 | 0.27 | SY | BATMAN-TCM |
| MOL000275 | Trametenolic acid | 456.78 | 38.71 | 0.8 | FL | TCMSP |
| MOL000279 | Cerevisterol | 430.74 | 37.96 | 0.77 | FL | TCMSP |
| MOL000282 | 5-dihydroergosterol | 398.74 | 43.51 | 0.72 | FL | TCMSP |
| MOL000283 | Ergosterol peroxide | 430.74 | 40.36 | 0.81 | FL | TCMSP |
| MOL000285 | Dehydrotumulosic acid | 482.77 | 38.26 | 0.82 | FL | TCMSP |
| MOL000296 | Hederagenin | 414.79 | 36.91 | 0.75 | FL | TCMSP |
| MOL000073 | Ent-Epicatechin | 290.29 | 48.96 | 0.24 | FL | Taiwan |
| MOL000830 | Alisol B | 472.78 | 34.47 | 0.82 | FL, ZX | Taiwan |
| MOL000831 | Alisol B monoacetate | 514.82 | 35.58 | 0.81 | ZX | TCMSP, Taiwan, BATMAN-TCM |
| MOL000849 | 16β-methoxyalisol B monoacetate | 544.85 | 32.43 | 0.77 | ZX | TCMSP |
| MOL000856 | Alisol C monoacetate | 514.77 | 33.06 | 0.83 | ZX | TCMSP |
| MOL002464 | 1-monolinolein | 354.59 | 37.18 | 0.3 | ZX | TCMSP |
| MOL000862 | Alisol B 23-acetate | 514.82 | 35.58 | 0.81 | FL, ZX | TCMSP, Taiwan |
This suggests that LWDH Pill compounds may act synergistically on these targets, exerting pharmacological effects in T2DM and other diseases.
Compound–T2DM PPI Network Analysis
Compound–T2DM PPI Network
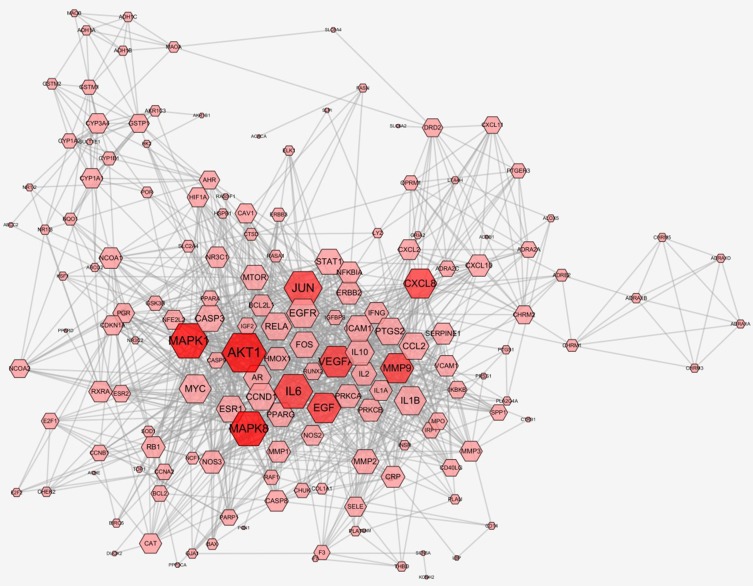
A total of 174 overlapping genes were obtained by looking for the intersection of the above compound target and the 7822 T2DM targets. The compound–disease co-targets information was exhibited in Figure S1 and Table S3. Import compound–disease co-targets information into STRING, we get compound–T2DM target PPI network with higher degree (degree ≥ 0.700) of connectivity (Figure 1), which contains 163 nodes and 2735 edges (Figure 2).
Figure 2.
Compound–Type 2 diabetes mellitus PPI network. Compound–T2DM target PPI network contains 163 gene nodes and 2735 edges. Pink hexagon represents gene targets. The red nodes (AKT1, MAPK1, MAPK8, IL6, JUN, VEGFA, EGF, MMP9, and CXCL8) have higher degrees. Node size of gene targets is proportional to the number of degrees.
In PPI network, some nodes (AKT1, MAPK1, MAPK8, IL6, JUN, VEGFA, EGF, MMP9, and CXCL8) have higher degrees. The number of edges of each node is quite large (66 in AKT1, 55 in MAPK1, 54 in MAPK8, 53 at IL6, 52 in JUN, 45 in VEGFA, 42 in EGF, and 41 in MMP9 and CXCL8).
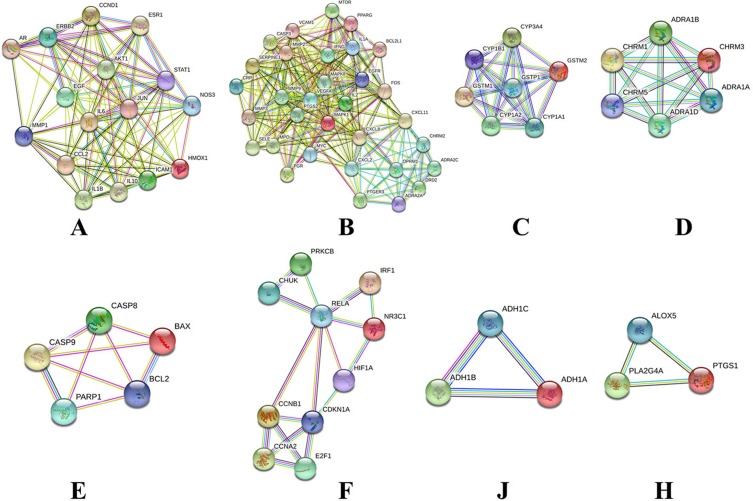
Cluster Of Compound–T2DM PPI Network
Targets information in PPI network queried from STRING protein database was analyzed by MCODE tool returned 8 central gene clusters (Figure 3 and Table 2). The top 3 modules with score > 6 were selected to detect significant modules in this PPI network. There are some T2DM-related targets in those clusters. Cluster 1 includes AR, HMOX1, AKT1, NOS3, ESR1, STAT1, IL1B, IL10, ICAM1, IL6, EGF, and MMP1. Cluster 2 gets CXCL8, EGFR, MAPK8, VEGFA, MMP9, MAPK1, PTGS2, MYC, MMP2, and FOS with high degree value. CYP1A2, CYP1A1, GSTM2, GSTP1, CYP1B1, CYP3A4, and GSTM1 were involved in Cluster 3.
Figure 3.
Cluster of compound–Type 2 diabetes mellitus PPI network. Eight clusters were identified in compound–Type 2 diabetes mellitus PPI network (A, B, C, D, E, F, G, and H) stand for clusters 1, 2, 3, 4, 5, 6, 7, and 8).
Table 2.
Cluster Of Compound–Type 2 Diabetes Mellitus PPI Network
| Cluster | Score (Density*#Nodes) | Nodes | Edges | Genes |
|---|---|---|---|---|
| 1 | 10.267 | 16 | 77 | JUN, AR, ERBB2, HMOX1, CCL2, AKT1, NOS3, CCND1, ESR1, STAT1, IL1B, IL10, ICAM1, IL6, EGF, MMP1 |
| 2 | 10.194 | 32 | 158 | MYC, MAPK1, VEGFA, SELE, MPO, MMP9, IFNG, CXCL11, OPRM1, DRD2, EGFR, CXCL2, PTGER3, FOS, ADRA2A, ADRA2C, MAPK8, MMP2, CHRM2, BCL2L1, IL2, MTOR, SERPINE1, PTGS2, CASP3, PPARG, MMP3, CRP, VCAM1, PGR, IL1A, CXCL8 |
| 3 | 6.667 | 7 | 20 | CYP1B1, GSTM2, CYP1A1, CYP1A2, GSTP1, CYP3A4, GSTM1 |
| 4 | 6 | 6 | 15 | CHRM1, CHRM5, ADRA1A, ADRA1B, ADRA1D, CHRM3 |
| 5 | 3.5 | 5 | 7 | PARP1, BCL2, CASP9, BAX, CASP8 |
| 6 | 3.111 | 10 | 14 | RELA, NR3C1, CHUK, E2F1, IRF1, CCNB1, HIF1A, CDKN1A, CCNA2, PRKCB |
| 7 | 3 | 3 | 3 | ADH1A, ADH1C, ADH1B |
| 8 | 3 | 3 | 3 | PLA2G4A, ALOX5, PTGS1 |
In retrospective analysis, T(−413)A single nucleotide polymorphism (SNP) of Heme oxygenase 1 (HMOX1) promoter was significantly more susceptible to albuminuria development than those carrying the A allele, especially in T2DM with longer duration and poor glycemic control.41–43 Yin et al, investigate that blocking of George et al, suggested that the role of Akt deficiency in human type 2 diabetes was underscored by the discovery of a family with type 2 diabetes associated with an inherited loss of function Akt2 mutation that seems to act in a dominant-negative manner to inhibit other Akt isoforms.44 Chen et al discovered that the haplodeficiency of Akt1 is sufficient to convert the mild insulin resistance in Akt2-/- mice to an overt type 2 diabetes, indicating a complementary role of Akt1 in the genesis of type 2 diabetes initiated by the deficiency of Akt2.45 Yin et al also investigated that blocking of AKT1 expression in pancreatic β-cells and peripheral tissues may lead to hyperglycemia and diabetes in Chinese Han population.46 Thus, these results showed that Akt1 and AKT2 are relatived to diabetes. The study reported inflammatory cytokines disorder was used as critical early biomarkers for estimating the T2DM risks and complications.47,48 T2DM patients had significantly higher levels of inflammatory markers interleukin-1B (IL-1B) and IL-6, while IL-10 showed lower levels, compared with non-diabetic patients.47,49 STAT1 is highly expressed in patients with T2DM. It can induce beta-cell injury by regulating IL-1B-activated beta-cell gene network in coordination with NF-kappaB.50,51 Yamada et al. showed that the 13989A→G(Ile118Val) polymorphism on CYP3A4 was significantly associated with T2DM sensitivity or drug resistance.52
The production of reactive oxygen species (ROS) abnormally increases in pancreatic β-cells when exposed to high concentrations of glucose and/or free fatty acids for a prolonged period of time, resulting in a burden of oxidative stress, further contributing to dysfunction of pancreatic β-cells.53 Oxidative stress increases intracellular matrix metalloproteinases including MMP1, MMP9, and MMP2 expression and intracellular activity, which response to induce impaired insulin secretion, declined glucose utilization, insulin resistance, and abnormal hepatic glucose production.54–56 Glutathione S-transferases (GSTs) are a family of multi-gene and multifunctional detoxification enzymes, which enable to remove ROS and S-thiolated protein regeneration caused by oxidative stress, metabolites, and chemicals.57–59 In Cluster 3, two families of related genes were found, mu and pi encoded by GSTM and GSTP genes. Tang et al confirmed by meta-analysis that 1354 T2DM from literature and database were significantly associated with Glutathione S-transferases mu 1 (GSTM1) polymorphism, with a pooled OR=1.66 (95% CI=1.17–2.37).60 Andrea et al illustrated that GSTM2 is one of the male-specific genes and plays an important role in liver metabolism, detoxification, and secretion.61 Adina’s data suggest that Val allele of the GSTP1 Ile105Val polymorphism (OR = 2.0, 95% CI=1.20–3.35, p=0.007) and GSTP1 Val/Val genotype (diabetic patients and controls: 13.1% vs 5.1%, resp.) may be associated with higher risk of T2DM in Romanian population.62
This suggests that LWDH Pill may be involved in the regulation of these genes, which are key or central genes in the development of T2DM.
Pathway Of Compounds–T2DM PPI Network
The KEGG pathway enrichment analysis was performed on the genes in the above mentioned three modules (Cluster 1, Cluster 2, and Cluster 3), displayed in Figure S2.
Clusters 1, 2, and 3 were enriched to 10 KEGG pathways, respectively, which were enriched respectively and mainly in AGE-RAGE signaling pathway in diabetic complications (p=5.08E−15), Pathways in cancer (p= 1.37E−12), Fluid shear stress and atherosclerosis (p= 4.42E−10), Chagas disease (p= 3.77E−9), TNF signaling pathway (p= 4.85E−9), Kaposi’s sarcoma-associated herpesvirus infection (p= 1.22E−11), IL-17 signaling pathway (p= 1.03E−13). The details are described in Table S4.
LWDH Pill–T2DM Network Analysis
LWDH Pill–T2DM Network
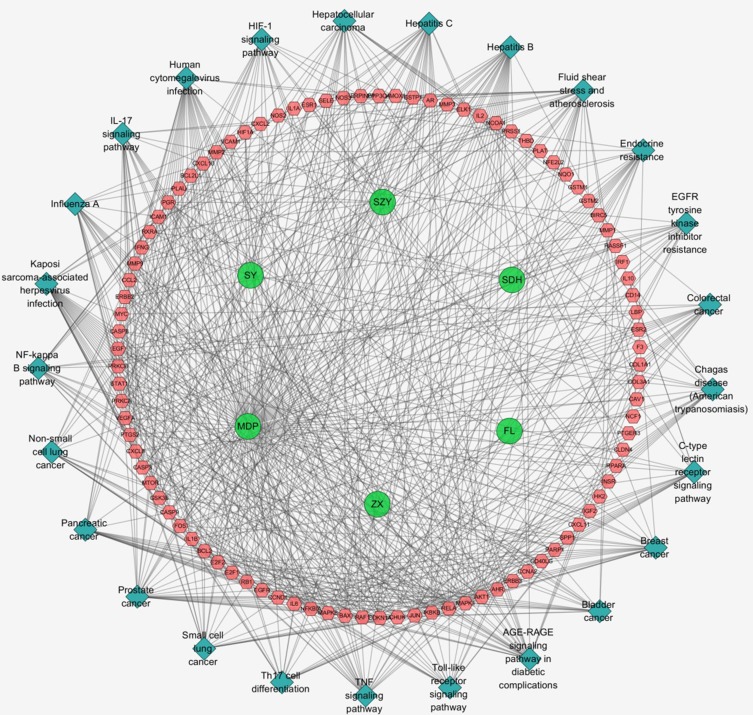
To shed light on the potential mechanisms of LWDH Pill acting on T2DM, the putative LWDH Pill target–T2DM-related target network consisting of LWDH herbs, compound–disease interactional proteins, and KEGG pathways was constructed based on PPI databases. Our pathway-act-network demonstrates interactions in multiple pathways including cross-talk of the pathway terms and the transitive relation. There are 125 nodes (6 herb nodes, 94 T2DM-related target nodes, and 25 pathway nodes) and 681 edges in LWDH Pill–T2DM network. The details of 25 signaling pathway nodes are shown in Figure 4 and Table 3.
Figure 4.
LWDH Pill–Type 2 diabetes mellitus network. Pathway of LWDH Pill–T2DM network includes 125 nodes (6 herb nodes represented by green circles, 94 T2DM-related target nodes represented by pink hexagon, and 25 pathway nodes described by blue diamond) and 681 edges.
Table 3.
KEGG Pathway Analysis Based On LWDH Pill–Type 2 Diabetes Mellitus Network
| No. | Pathway ID | Pathway Name | P value | Count | Gene Name |
|---|---|---|---|---|---|
| 1 | 05167 | Kaposi sarcoma-associated herpesvirus infection | 4.39E-20 | 31 | PTGS2, IL6, RELA, AKT1, VEGFA, CCND1, FOS, CDKN1A, BAX, CASP9, MAPK1, RB1, JUN, CASP3, NFKBIA, CASP8, RAF1, HIF1A, STAT1, MYC, ICAM1, CXCL8, CXCL2, CHUK, E2F1, E2F2, IKBKB, MAPK8, PPP3CA, GSK3B, MTOR |
| 2 | 05163 | Human cytomegalovirus infection | 1.35E-17 | 31 | PTGS2, IL6, RELA, EGFR, AKT1, VEGFA, CCND1, CDKN1A, BAX, CASP9, MAPK1, RB1, CASP3, ELK1, NFKBIA, CASP8, RAF1, PRKCA, MYC, IL1B, CCL2, PTGER3, CXCL8, PRKCB, CHUK, E2F1, E2F2, IKBKB, PPP3CA, GSK3B, MTOR |
| 3 | 05418 | Fluid shear stress and atherosclerosis | 1.11E-21 | 29 | RELA, AKT1, VEGFA, BCL2, FOS, MMP2, MMP9, JUN, HMOX1, CAV1, ICAM1, IL1B, CCL2, SELE, VCAM1, NOS3, PLAT, THBD, IFNG, IL1A, NCF1, GSTP1, NFE2L2, NQO1, CHUK, GSTM1, GSTM2, IKBKB, MAPK8 |
| 4 | 05161 | Hepatitis B | 1.23E-19 | 29 | IL6, RELA, AKT1, BCL2, FOS, CDKN1A, BAX, CASP9, MMP9, MAPK1, RB1, JUN, CASP3, ELK1, NFKBIA, CASP8, RAF1, PRKCA, STAT1, MYC, CXCL8, PRKCB, BIRC5, CHUK, E2F1, E2F2, IKBKB, MAPK8, CCNA2 |
| 5 | 04933 | AGE-RAGE signaling pathway in diabetic complications | 8.84E-25 | 28 | IL6, RELA, AKT1, VEGFA, CCND1, BCL2, BAX, MMP2, MAPK1, JUN, CASP3, PRKCA, STAT1, F3, ICAM1, IL1B, CCL2, SELE, VCAM1, CXCL8, PRKCB, NOS3, THBD, SERPINE1, COL1A1, IL1A, COL3A1, MAPK8 |
| 6 | 05160 | Hepatitis C | 5.23E-17 | 26 | RXRA, RELA, EGFR, AKT1, CCND1, CDKN1A, BAX, CASP9, MAPK1, EGF, RB1, CASP3, NFKBIA, CASP8, RAF1, STAT1, MYC, IFNG, CLDN4, PPARA, CXCL10, CHUK, E2F1, E2F2, IKBKB, GSK3B |
| 7 | 05215 | Prostate cancer | 3.33E-21 | 25 | AR, MMP3, RELA, EGFR, AKT1, CCND1, BCL2, CDKN1A, CASP9, PLAU, MMP9, MAPK1, EGF, RB1, NFKBIA, RAF1, ERBB2, PLAT, GSTP1, CHUK, E2F1, E2F2, IKBKB, GSK3B, MTOR |
| 8 | 05225 | Hepatocellular carcinoma | 3.91E-14 | 24 | EGFR, AKT1, CCND1, BCL2L1, CDKN1A, BAX, MAPK1, RB1, ELK1, RAF1, PRKCA, HMOX1, MYC, PRKCB, GSTP1, NFE2L2, NQO1, E2F1, E2F2, IGF2, GSTM1, GSTM2, GSK3B, MTOR |
| 9 | 04668 | TNF signaling pathway | 2.32E-17 | 23 | PTGS2, IL6, MMP3, RELA, AKT1, FOS, MMP9, MAPK1, JUN, CASP3, NFKBIA, CASP8, ICAM1, IL1B, CCL2, SELE, VCAM1, CXCL2, CXCL10, CHUK, IRF1, IKBKB, MAPK8 |
| 10 | 04657 | IL-17 signaling pathway | 7.05E-18 | 22 | PTGS2, IL6, MMP3, RELA, FOS, MMP9, MAPK1, JUN, CASP3, NFKBIA, CASP8, MMP1, IL1B, CCL2, CXCL8, IFNG, CXCL2, CXCL10, CHUK, IKBKB, MAPK8, GSK3B |
| 11 | 05164 | Influenza A | 4.33E-12 | 22 | IL6, PRSS1, RELA, AKT1, CASP9, MAPK1, JUN, NFKBIA, RAF1, PRKCA, STAT1, ICAM1, IL1B, CCL2, CXCL8, PRKCB, IFNG, IL1A, CXCL10, IKBKB, MAPK8, GSK3B |
| 12 | 05212 | Pancreatic cancer | 9.09E-19 | 21 | RELA, EGFR, AKT1, VEGFA, CCND1, BCL2L1, CDKN1A, BAX, CASP9, MAPK1, EGF, RB1, RAF1, STAT1, ERBB2, CHUK, E2F1, E2F2, IKBKB, MAPK8, MTOR |
| 13 | 04066 | HIF-1 signaling pathway | 5.70E-16 | 21 | IL6, RELA, EGFR, AKT1, VEGFA, BCL2, CDKN1A, MAPK1, EGF, PRKCA, HIF1A, ERBB2, HMOX1, PRKCB, NOS3, SERPINE1, IFNG, INSR, HK2, NOS2, MTOR |
| 14 | 05224 | Breast cancer | 1.75E-12 | 21 | PGR, ESR1, EGFR, AKT1, CCND1, FOS, CDKN1A, BAX, MAPK1, EGF, RB1, JUN, RAF1, ERBB2, MYC, E2F1, E2F2, GSK3B, NCOA1, ESR2, MTOR |
| 15 | 01522 | Endocrine resistance | 5.32E-15 | 20 | ESR1, EGFR, AKT1, CCND1, BCL2, FOS, CDKN1A, BAX, MMP2, MMP9, MAPK1, RB1, JUN, RAF1, ERBB2, E2F1, E2F2, MAPK8, ESR2, MTOR |
| 16 | 05222 | Small cell lung cancer | 2.61E-14 | 19 | PTGS2, RXRA, RELA, AKT1, CCND1, BCL2, BCL2L1, CDKN1A, BAX, CASP9, RB1, CASP3, NFKBIA, MYC, CHUK, E2F1, E2F2, NOS2, IKBKB |
| 17 | 05142 | Chagas disease (American trypanosomiasis) | 1.87E-13 | 19 | IL6, RELA, AKT1, FOS, MAPK1, IL10, JUN, NFKBIA, CASP8, IL1B, CCL2, CXCL8, IL2, SERPINE1, IFNG, CHUK, NOS2, IKBKB, MAPK8 |
| 18 | 04620 | Toll-like receptor signaling pathway | 2.25E-13 | 19 | IL6, CD14, LBP, RELA, AKT1, FOS, MAPK1, JUN, NFKBIA, CASP8, STAT1, IL1B, CXCL8, CXCL11, CXCL10, CHUK, SPP1, IKBKB, MAPK8 |
| 19 | 05210 | Colorectal cancer | 8.32E-14 | 18 | EGFR, AKT1, CCND1, BCL2, FOS, CDKN1A, BAX, CASP9, MAPK1, EGF, JUN, CASP3, RAF1, MYC, BIRC5, MAPK8, GSK3B, MTOR |
| 20 | 04064 | NF-kappa B signaling pathway | 5.17E-13 | 18 | PTGS2, CD14, LBP, RELA, BCL2, BCL2L1, PLAU, NFKBIA, ICAM1, IL1B, VCAM1, CXCL8, PRKCB, PARP1, CXCL2, CHUK, CD40LG, IKBKB |
LWDH–T2DM network analysis showed that six herbs in LWDH act synergistically to treat T2DM and its complications. The study by Yokozawa et al shows that the glucose content of the renal tissue in high blood glucose group was 3.89 mg/g, but this declined to 3.27 mg/g when treated with dried Rehmanniae Radix extract.63 SDH and SZY have been well evidenced to play a beneficial synergistic interaction in the ameliorating of renal injury in C57BL/KsJ-db/db diabetic mice by inhibiting AGEs/RAGE/SphK1 signaling pathway.64 Qian et al demonstrated that alcohol extract of SZY can increase GLUT4 mRNA and its protein expression in T2DM rats through promoting proliferation of islet and increasing postprandial secretion of insulin and therefore accelerate glucose transport.65 Qian et al demonstrated that alcohol extract of SZY can accelerate glucose transport by promoting islet proliferation and increasing postprandial secretion of insulin, thereby increasing GLUT4 mRNA and protein expression in T2DM rats.65 Zhang’s findings indicated that MDP decreased significantly the overexpression of IL-6, MCP-1, TGF-β1, ICAM-1, and RAGE in high-glucose-fat diet and streptozotocin (STZ)-induced diabetic nephropathy rats.66 In addition, previous studies showed that polysaccharides of SY, FL, and ZX perform the potential antidiabetic effect.67,68
Pathway Of LWDH Pill–T2DM Network
In LWDH Pill–T2DM network, some signaling pathways (such as AGE-RAGE signaling pathway in diabetic, IL-17 signaling pathway, TNF signaling pathway, HIF-1 signaling pathway, endocrine resistance, Toll-like receptor signaling pathway, NF-kappa B signaling pathway, and EGFR tyrosine kinase inhibitor resistance) with higher degrees are closely related to the development, metastasis, and drug resistance of T2DM.
Pro-inflammatory and/or oxidative stress mediators are directly interlinked with the pathogenesis of T2DM and development of insulin resistance.69,70 The AGE-RAGE signaling pathway plays a key role in the production of vascular complications (eg. atherosclerosis and kidney disease) that induce T2DM.71,72 The formation and accumulation of advanced glycation end products (AGEs) was accelerated upregulated with increased circulating glucose as well as other reducing sugars, such as galactose and fructose, in T2DM, and subsequently AGEs-their receptor RAGE interaction to a signaling cascade, stimulating NAD(P)H oxidase and increasing the production of ROS eventually lead to the vascular endothelial dysfunction.73–77 There are also accumulating evidence that the interaction of AGE/RAGE and tumor necrosis factor-alpha (TNF-α) signaling, perhaps even by amplifying one another, induces production of superoxide in T2DM.73,78–80 Nuclear factor-κB (NF-κB) was characterized as a transcription factor activated through inflammation and oxidative stress, and involved in cytokines (e.g., TNF-α, IL-6, and IL-1B) and adhesion molecules [e.g., intercellular adhesion molecule-1 (ICAM1)] expression.81 Akash et al proposed that the level of TNF-α disordered raised in adipocytes and/or peripheral tissues induces insulin resistance by impairing the insulin signaling through serine phosphorylation leading the development of T2DM.70 Zhang et al confirmed that PDTC pretreatment, a specific NF-kappa B inhibitor, was able to attenuate the translocation of NF-κBp65 protein into the nucleus of human endothelial cells after high glucose treatment.81 Apoptosis of β cell damage, caused by antigen-presenting cell activation mediated by the Toll-like receptor 2 (TLR2) pathway, was an initial event for the development of autoimmune diabetes.82,83 Additionally, previous reports have shown that vascular dysfunction of T2DM will also occur when EGFR tyrosine kinase activity is enhanced.84,85 Patricia et al have demonstrated that use of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors in T2DM can significantly improve glucose tolerance, reduce insulin resistance, reduce circulating levels of TNF-α, IL-6, and decrease macrophage infiltration in free fatty acids.86,87
Our research confirmed that LWDH Pill can achieve the potential characteristics of multi-target- multi-disease through the LWDH Pill-T2DM network.
Validation Of Compound–Target Interaction
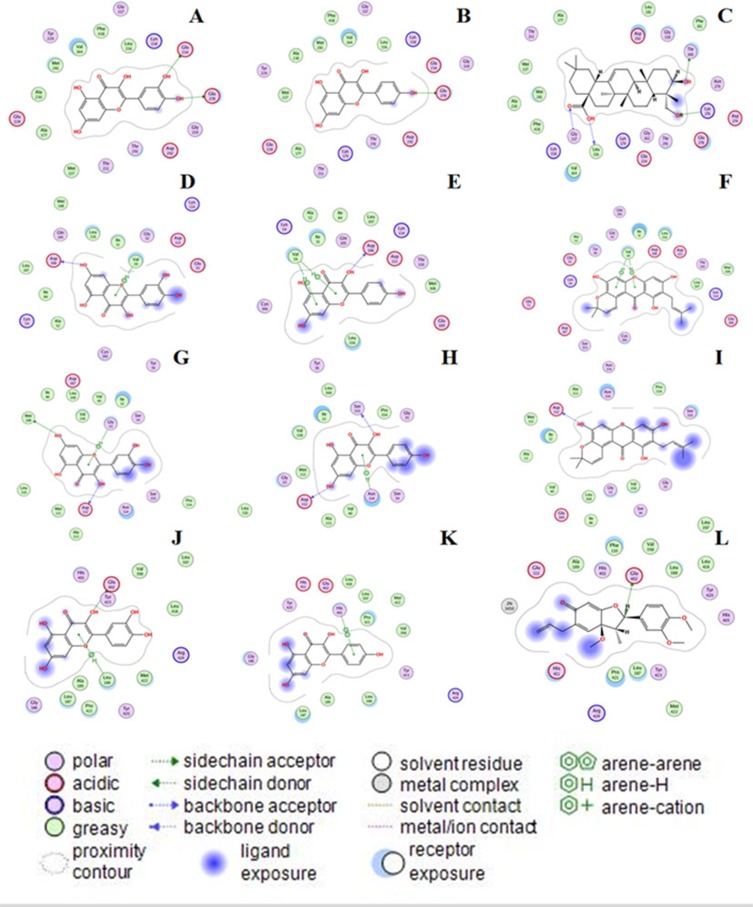
Molecular docking was further applied in our study to assess the interaction between components and targets, reducing complexity and improving the accuracy of the constituent-target network. Virtual screening using MOE was performed to determine the binding affinity between protein models and 10 potentially active compounds (including Quercetin, Kaempferol, Stigmasterol, Beta-Sitosterol, Kadsurenone, Diosgenin, Hancinone C, Hederagenin, Garcinone B, Isofucosterol) obtained from compound–compound target network. Molecular docking analyzed four targets (AKT1, MAPK1, MAPK8 and MMP9) and 10 compounds (Table 4 and Figure 5). The results of these signal transductions have been reported to be the possible mechanism for LWDH Pill in treating diabetic complications.
Table 4.
Virtual Docking Of Ten Bioactive Ingredients From LWDH Pill For Type 2 Diabetes Mellitus Targets
| No. | Compound | Structure | Binding Energy/(kcal·mol−1) | |||
|---|---|---|---|---|---|---|
| AKT1 | MAPK1 | MAPK8 | MMP9 | |||
| 1 | Quercetin | 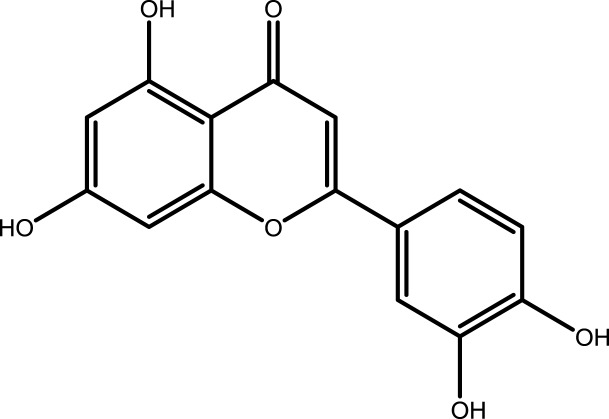 |
−6.8181 | −6.3394 | −7.1100 | −6.2667 |
| 2 | Kaempferol | 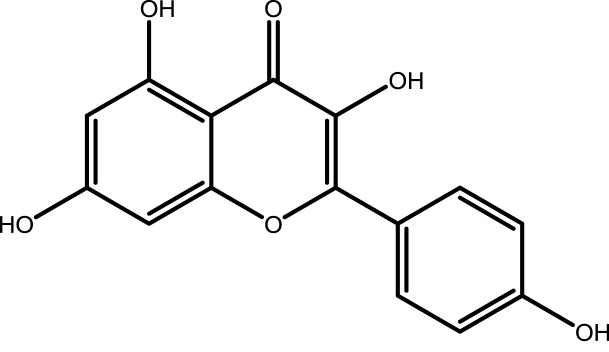 |
−6.5715 | −6.3819 | −6.3977 | −5.9005 |
| 3 | Stigmasterol | 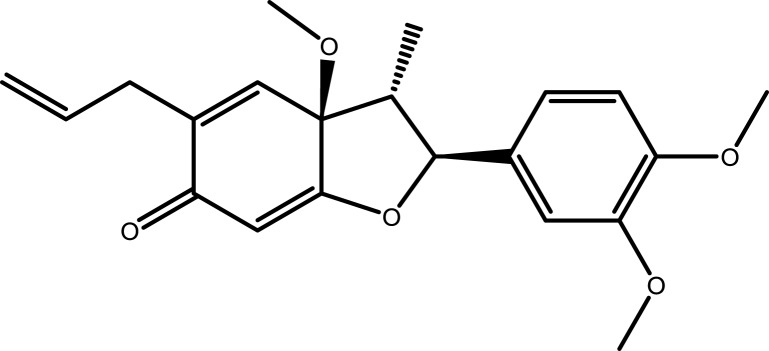 |
−6.8343 | −7.9176 | −5.5400 | −6.2369 |
| 4 | Beta-sitosterol | 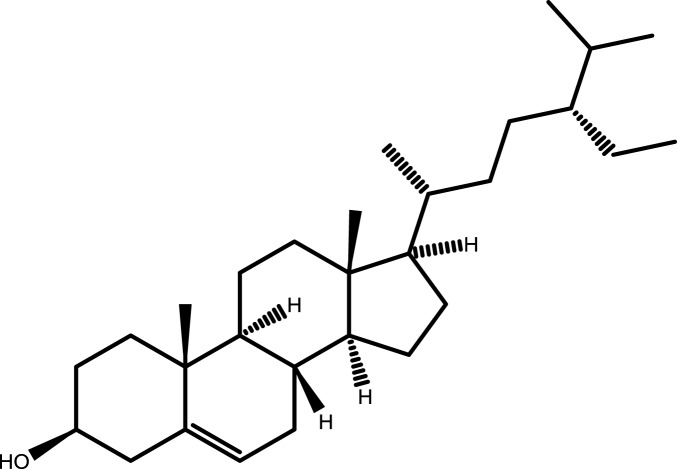 |
−6.8563 | −7.6574 | −5.5483 | −6.0089 |
| 5 | Kadsurenone | 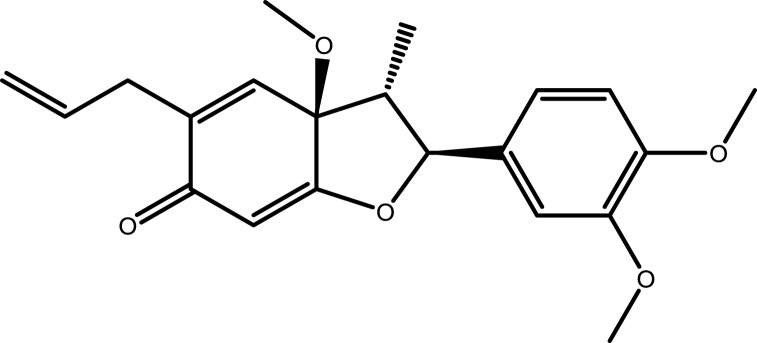 |
−7.0874 | −7.3170 | −6.1939 | −5.7586 |
| 6 | Diosgenin | 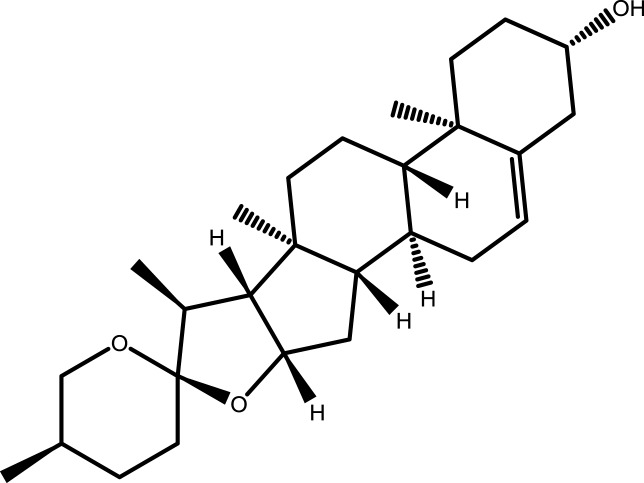 |
−5.5341 | −7.1899 | −5.3117 | −6.1969 |
| 7 | Hancinone C | 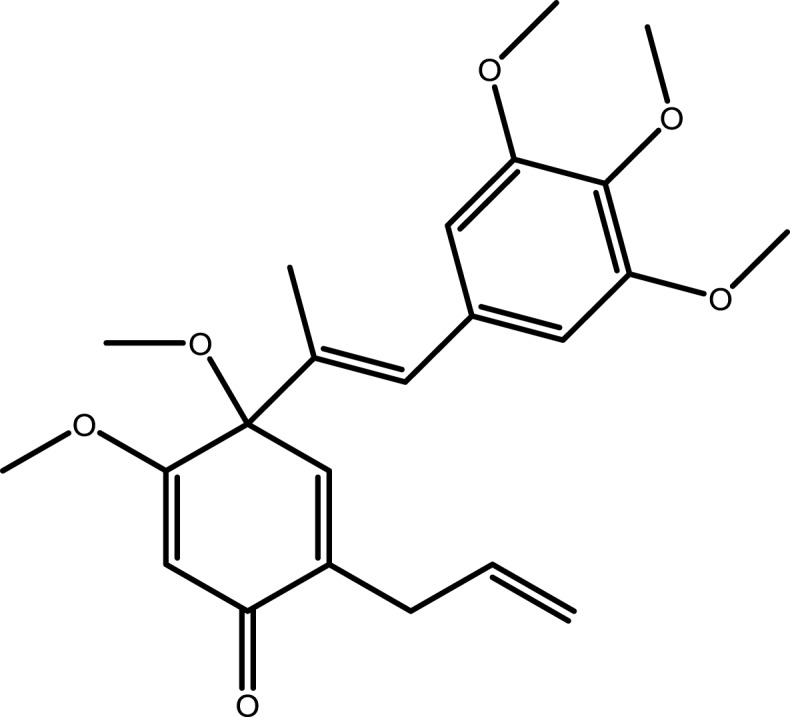 |
−7.7971 | −7.0791 | −5.6103 | −5.9619 |
| 8 | Hederagenin | 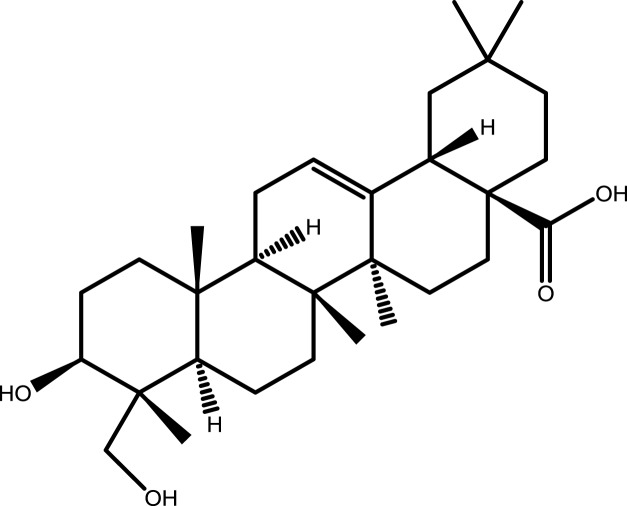 |
−3.7120 | −6.3780 | −5.3367 | −5.3195 |
| 9 | Garcinone B | 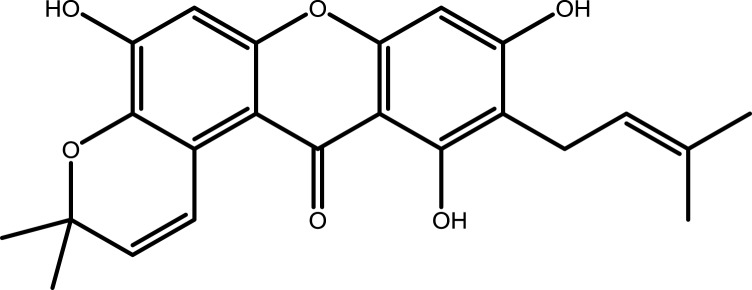 |
−7.1429 | −7.2783 | −6.7518 | −6.5401 |
| 10 | Isofucosterol | 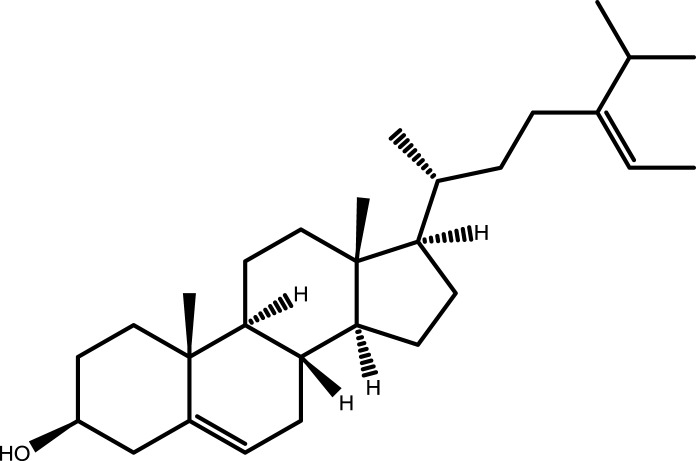 |
−7.4410 | −8.4075 | −5.8677 | −5.6977 |
Note: The combination with lower binding energy scores is more stable.
Figure 5.
Virtual docking of bioactive ingredients from LWDH Pill for Type 2 diabetes mellitus targets. The virtual docking of quercetin with AKT1, MAPK1, MAPK8, and MMP9 was represented by A, D, G, and J, respectively. C stands for the docking of AKTI and Hederagenin. The virtual docking of Kaempferol with AKT1, MAPK1, MAPK8, and MMP9 was represented by B, E, H, and K, respectively. The virtual docking of Garcinone B with MAKP1 and MAKP8 was represented by F and I, respectively. L stands for the virtual docking of MMP9 and Kadsurenone.
The three-dimensional structures of the proteins 4kl, 3w55, 2g01, and 1gkc were obtained by searching AKT1, MAPK1, MAPK8, and MMP9 from the PDB protein database, respectively. Quercetin has been demonstrated to downregulate the expression of RNA and protein levels of MAPK, TNF-α, IL-6, and IL-1B in rat pancreatic histopathological damage.88,89 The virtual docking of kaempferol with AKT1, MAPK1, MAPK8, and MMP9 was represented by B, E, H, and K, respectively. Kaempferol fixed the binding cavity of T2DM target AKT1 through H-bonds with residues Glu 278 (2.89 Å). Xu et al have validated that the anti-Diabetic retinopathy (DR) effect of Kaempferol (5–25 mM) was mediated via downregulating the expression of Erk1/2 and AKT1.90 The complex Kaempferol–MAPK1 was stabilized by one H-bond and two π-H bonds with residues including Asp 106 (2.94 Å), Val 39 (4.41 Å), and Val 39 (3.86 Å), respectively, which further verifies Kaempferol in LWDH Pill anti-T2DM effects. Same as the complex of Kaempferol–MAPK8 was stabilized by one π-H and two H-bonds with residues including Ser 155 (2.90 Å), Asp 112 (2.84 Å), and Asn 114 (4.23 Å), respectively, and Kaempferol–MMP9 was stabilized by π-π bonds with residues HIS 401 (3.96 Å).
Conclusion
In the past three decades, the prevalence of T2DM, a constellation of metabolic aberrations, has risen dramatically worldwide.91 The global consensus is that the health goal by 2025 is to stop the rise of diabetes and obesity.92 LWDH Pill is described as a commonly used drug for treatment and prevention of diabetes for thousands of years. Our research is the first report to explain the mechanisms of LWDH Pill and validate that LWDH Pill plays an active therapeutic role in T2DM, especially in diabetic complications (atherosclerosis and nephropathy). A total of 7822 genes were found related to T2DM by using bioinformatics analysis. Among them, 163 hub genes were selected; furthermore, AKT1, MAKP1, MAPK8, and MMP9 might be the core genes for LWDH Pill in treating T2DM and its complications. Although we have examined the possible role of LWDH Pill in treating T2DM, there are some limitations in the present study. Further in vitro and in vivo experimental validation is needed to support our research.
Acknowledgments
This work was supported by Natural Science Foundation of China (No.81603400, No.81673585), the China Postdoctoral Science Foundation (2019M652776); the National key R&D program of China (No: 2018YFC1703400; 2018YFC1704400); the Training Program for Excellent Young Innovators of Changsha (kq1802017); the Science Foundation of Hunan Province (2019JJ50345) and the Program of Survey of Chinese Medicines of China ([2017]66). The authors acknowledge the support of Hunan Province Traditional Chinese Medicine Preparation and Quality Traceability Engineering and Technology Center, and the 2011 Collaboration and Innovation Center for Digital Chinese Medicine in Hunan.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Adeshara KA, Diwan AG, Tupe RS. Diabetes and complications: cellular signaling pathways, current understanding and targeted therapies. Curr Drug Targets. 2016;17(11):1309–1328. doi: 10.2174/1389450117666151209124007 [DOI] [PubMed] [Google Scholar]
- 2.Feudtner C. Diabetes: the sweet irony of modern technology. Bull World Health Organ. 2011;8(2):90–91. doi: 10.2471/BLT.11.040211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarwar NGP, Seshasai SR, Gobin R, Kaptoge S, Di A. Diabetes mellitus, fasting blood glucose concentration, and risk of vascular disease: a collaborative meta-analysis of 102 prospective studies. Emerging Risk Factors Collab. 2010;365:2215–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sastre AA, Vernooij R, Harmand MG. Effect of the treatment of type 2 diabetes mellitus on the development of cognitive impairment and dementia. Cochrane Database Syst Rev. 2017;6:CD003804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vuylsteke V, Chastain LM, Maggu GA, Brown C. Imeglimin: a potential new multi-target drug for type 2 diabetes. Drugs R D. 2015;15(3):227–232. doi: 10.1007/s40268-015-0099-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ena J, Gomez-Huelgas R, Sanchez-Fuentes D, et al. Management of patients with type 2 diabetes and multiple chronic conditions: a Delphi consensus of the Spanish Society of Internal Medicine. Eur J Intern Med. 2016;27:31–36. doi: 10.1016/j.ejim.2015.10.015 [DOI] [PubMed] [Google Scholar]
- 7.Jacobs E, Hoyer A, Brinks R, Icks A, Kuß O, Rathmann W. Healthcare costs of type 2 diabetes in Germany. Diabetic Med. 2017;34(6):855–861. doi: 10.1111/dme.13336 [DOI] [PubMed] [Google Scholar]
- 8.Seuring T, Archangelidi O, Suhrcke M. The economic costs of type 2 diabetes – a global systematic review. PharmacoEconomics. 2015;33(8):811–831. doi: 10.1007/s40273-015-0268-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wenjuan X, Liangxiao Z, Yuhong H, Qianxu Y, Hongbin X, Deqin Z. Discrimination of type 2 diabetes mellitus corresponding to different traditional Chinese medicine syndromes based on plasma fatty acid profiles and chemometric methods. J Ethnopharmacol. 2012;143(2):463–468. doi: 10.1016/j.jep.2012.06.045 [DOI] [PubMed] [Google Scholar]
- 10.Wang P, Sun H, Lv H, et al. Thyroxine and reserpine-induced changes in metabolic profiles of rat urine and the therapeutic effect of Liu Wei Di Huang Wan detected by UPLC-HDMS. J Pharm Biomed Anal. 2010;53(3):631–645. doi: 10.1016/j.jpba.2010.04.032 [DOI] [PubMed] [Google Scholar]
- 11.Dai B, Wu Q, Zeng C, et al. The effect of Liuwei Dihuang decoction on PI3K/Akt signaling pathway in liver of type 2 diabetes mellitus (T2DM) rats with insulin resistance. J Ethnopharmacol. 2016;192:382–389. doi: 10.1016/j.jep.2016.07.024 [DOI] [PubMed] [Google Scholar]
- 12.Tseng YT, Chang WH, Lin CC, Chang FR, Wu PC, Lo YC. Protective effects of Liuwei dihuang water extracts on diabetic muscle atrophy. Phytomedicine. 2019;53:96–106. doi: 10.1016/j.phymed.2018.09.032 [DOI] [PubMed] [Google Scholar]
- 13.Xu ZJ, Shu S, Li ZJ, Liu YM, Zhang RY, Zhang Y. Liuwei Dihuang pill treats diabetic nephropathy in rats by inhibiting of TGF-beta/SMADS, MAPK, and NF-kB and upregulating expression of cytoglobin in renal tissues. Medicine. 2017;96(3):e5879. doi: 10.1097/MD.0000000000005879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Sun H, Zhang A, Sun W, Wang P, Wang Z. Potential role of metabolomics approaches in the area of traditional Chinese medicine: as pillars of the bridge between Chinese and Western medicine. J Pharm Biomed Anal. 2011;55(5):859–868. doi: 10.1016/j.jpba.2011.01.042 [DOI] [PubMed] [Google Scholar]
- 15.Li S, Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chin J Nat Med. 2013;11(2):110–120. doi: 10.1016/S1875-5364(13)60037-0 [DOI] [PubMed] [Google Scholar]
- 16.Zhang G, Li Q, Chen Q. Network pharmacology: a new approach for Chinese Herbal Medicine Research. Evid Based Complement Alternat Med. 2013;2013(8):621423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao Y, Hao J, Jin ZQ, et al. Network pharmacology-based and clinically relevant prediction of the active ingredients and potential targets of Chinese herbs in metastatic breast cancer patients. Oncotarget. 2017;8(16):27007–27021. doi: 10.18632/oncotarget.15351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng L, Yang K. Exploring the pharmacological mechanism of Yanghe Decoction on HER2-positive breast cancer by a network pharmacology approach. J Ethnopharmacol. 2017;199:68–85. doi: 10.1016/j.jep.2017.01.045 [DOI] [PubMed] [Google Scholar]
- 19.Zuo H, Zhang Q, Su S, Chen Q, Yang F, Hu Y. A network pharmacology-based approach to analyse potential targets of traditional herbal formulas: an example of Yu Ping Feng decoction. Sci Rep. 2018;8:1. doi: 10.1038/s41598-018-29764-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kibble M, Saarinen N, Tang J, Wennerberg K, Mäkelä S, Aittokallio T. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Nat Prod Rep. 2015;32(8):1249–1266. doi: 10.1039/c5np00005j [DOI] [PubMed] [Google Scholar]
- 21.Lee AY, Park W, Kang TW, Cha MH, Chun JM. Network pharmacology-based prediction of active compounds and molecular targets in Yijin-Tang acting on hyperlipidaemia and atherosclerosis. J Ethnopharmacol. 2018;221:151–159. doi: 10.1016/j.jep.2018.04.027 [DOI] [PubMed] [Google Scholar]
- 22.Kang D, Pang XC, Ying Z, et al. Network pharmacology-based analysis of Chinese herbal NaoDeSheng formula for application to Alzheimer disease. Chin J Nat Med. 2018;16(1):53–62. doi: 10.1016/S1875-5364(18)30029-3 [DOI] [PubMed] [Google Scholar]
- 23.Jinlong R, Peng L, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;16(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J, Cheung F, Tan HY, et al. Identification of the active compounds and significant pathways of yinchenhao decoction based on network pharmacology. Mol Med Rep. 2017;16(4):4583–4592. doi: 10.3892/mmr.2017.7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, Guo F, Wang Y, et al. BATMAN-TCM: a bioinformatics analysis tool for molecular mechanism of traditional Chinese medicine. Sci Rep. 2016;6:21146. doi: 10.1038/srep21146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Rui J, Ding X, Yang X. Exploring the multicomponent synergy mechanism of Banxia Xiexin Decoction on irritable bowel syndrome by a systems pharmacology strategy. J Ethnopharmacol. 2019;233:158–168. doi: 10.1016/j.jep.2018.12.033 [DOI] [PubMed] [Google Scholar]
- 27.Breuza L, Poux S, Estreicher A, et al. The UniProtKB guide to the human proteome. Database (Oxford). 2016;2016:bav120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stelzer G, Rosen N, Plaschkes I, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses: the GeneCards suite. Cur Protoc Bioinf. 54(1), 1–302016. [DOI] [PubMed] [Google Scholar]
- 29.Amberger JS, Hamosh A. Searching Online Mendelian Inheritance in Man (OMIM): a knowledgebase of human genes and genetic phenotypes. Cur Protoc Bioinf. 2017;58(1):1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneeweiss A, Ruckhäberle E, Huober J. Chemotherapy for metastatic breast cancer – an anachronism in the era of personalised and targeted oncological therapy? Geburtshilfe Frauenheilkd. 2015;75(6):574–583. doi: 10.1055/s-0035-1546150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minoru K. A database for post-genome analysis. Trends Genet. 1997;13 375–376. [DOI] [PubMed] [Google Scholar]
- 32.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu WM, Yang K, Jiang LJ, Hu JQ, Zhou XZ. Integrated modules analysis to explore the molecular mechanisms of phlegm-stasis cementation syndrome with ischemic heart disease. Front Physiol. 2018;9:7. doi: 10.3389/fphys.2018.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qian B, Junyang L, Chao H, et al. Topological, functional, and dynamic properties of the protein interaction networks rewired by benzo(a)pyrene. Toxicol Appl Pharmacol. 2015;283(2):83–91. doi: 10.1016/j.taap.2015.01.006 [DOI] [PubMed] [Google Scholar]
- 35.Guo R, Zhang X, Su J, et al. Identifying potential quality markers of Xin-Su-Ning capsules acting on arrhythmia by integrating UHPLC-LTQ-Orbitrap, ADME prediction and network target analysis. Phytomedicine. 2018;44:117–128. doi: 10.1016/j.phymed.2018.01.019 [DOI] [PubMed] [Google Scholar]
- 36.Gary D, Bader CWH. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4(2):1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ni M, Liu X, Wu J, et al. Identification of candidate biomarkers correlated with the pathogenesis and prognosis of non-small cell lung cancer via integrated bioinformatics analysis. Front Genet. 2018;9:469. doi: 10.3389/fgene.2018.00173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang E, Zhang X. Identification of breast cancer hub genes and analysis of prognostic values using integrated bioinformatics analysis. Cancer Biomark. 2017;21(1):373–381. doi: 10.3233/CBM-170550 [DOI] [PubMed] [Google Scholar]
- 39.Vilar S, Cozza G, Moro S. Medicinal chemistry and the Molecular Operating Environment (MOE): application of QSAR and molecular docking to drug discovery. Curr Top Med Chem. 2008;8(18):1555–1572. doi: 10.2174/156802608786786624 [DOI] [PubMed] [Google Scholar]
- 40.Maier JK, Labute P. Assessment of fully automated antibody homology modeling protocols in molecular operating environment. Proteins. 2014;82(8):1599–1610. doi: 10.1002/prot.24576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andrews M, Leiva E, Arredondo-Olguín M. Short repeats in the heme oxygenase 1 gene promoter is associated with increased levels of inflammation, ferritin and higher risk of type-2 diabetes mellitus. J Trace Elem Med Biol. 2016;37:25–30. doi: 10.1016/j.jtemb.2016.06.001 [DOI] [PubMed] [Google Scholar]
- 42.Lee EY, Lee YH, Kim SH, et al. Association between heme oxygenase-1 promoter polymorphisms and the development of albuminuria in type 2 diabetes: a case-control study. Medicine. 2015;94(43):e1825. doi: 10.1097/MD.0000000000000874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SE, Lee YB, Jun JE, et al. Increment of serum bilirubin as an independent marker predicting new-onset type 2 diabetes mellitus in a Korean population. Nutr Metab Cardiovasc Dis. 2017;27(3):234–240. doi: 10.1016/j.numecd.2016.10.003 [DOI] [PubMed] [Google Scholar]
- 44.George S. A family with severe insulin resistance and diabetes due to a mutation in AKT2. Science. 2004;304(5675):1325–1328. doi: 10.1126/science.1096706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen WS, Peng X-D, Wang Y, et al. Leptin deficiency and beta-cell dysfunction underlie type 2 diabetes in compound Akt knockout mice. Mol Cell Biol. 2009;29(11):3151. doi: 10.1128/MCB.01792-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin X, Xu Z, Zhang Z, et al. Association of PI3K/AKT/mTOR pathway genetic variants with type 2 diabetes mellitus in Chinese. Diabetes Res Clin Pract. 2017;128:127–135. doi: 10.1016/j.diabres.2017.04.002 [DOI] [PubMed] [Google Scholar]
- 47.Fathy SA, Mohamed MR, Ali MAM, El-Helaly AE, Alattar AT. Influence of IL-6, IL-10, IFN-gamma and TNF-alpha genetic variants on susceptibility to diabetic kidney disease in type 2 diabetes mellitus patients. Biomarkers. 2018;24:1–13. [DOI] [PubMed] [Google Scholar]
- 48.Senthilkumar GP, Anithalekshmi MS, Yasir M, Parameswaran S, Packirisamy RM, Bobby Z. Role of omentin 1 and IL-6 in type 2 diabetes mellitus patients with diabetic nephropathy. Diabetes Metab Syndr. 2018;12(1):23–26. doi: 10.1016/j.dsx.2017.08.005 [DOI] [PubMed] [Google Scholar]
- 49.Kologrivova IV, Suslova TE, Koshel’Skaya OA, Vinnitskaya IV, Trubacheva OA. System of matrix metalloproteinases and cytokine secretion in type 2 diabetes mellitus and impaired carbohydrate tolerance associated with arterial hypertension. Bull Exp Biol Med. 2014;156(5):635–638. doi: 10.1007/s10517-014-2413-4 [DOI] [PubMed] [Google Scholar]
- 50.EH CN K, Saha PK, Xiao L, et al. miR-30a remodels subcutaneous adipose tissue inflammation to improve insulin sensitivity in obesity. Diabetes. 2018;67(12):2541–2553. doi: 10.2337/db17-1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.M WN C, Jonas JC, Jörns A, Lenzen S, Eizirik DL. Mechanisms of pancreatic beta-cell death in type 1 and type 2 diabetes: many differences, few similarities. Diabetes. 2005;2:S97–107. [DOI] [PubMed] [Google Scholar]
- 52.Yamada YMH, Watanabe S, Kato K, et al. Association of a polymorphism of CYP3A4 with type 2 diabetes mellitus. Int J Mol Med. 2007;20(5):703–707. [PubMed] [Google Scholar]
- 53.Rehman K, Akash MSH. Mechanism of generation of oxidative stress and pathophysiology of type 2 diabetes mellitus: how are they interlinked? J Cell Biochem. 2017;118(11):3577–3585. doi: 10.1002/jcb.26097 [DOI] [PubMed] [Google Scholar]
- 54.Liu C, Wan X, Ye T, et al. Matrix metalloproteinase 2 contributes to pancreatic beta cell injury induced by oxidative stress. PLoS One. 2014;9(10):e110227. doi: 10.1371/journal.pone.0110227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kostov K, Blazhev A, Atanasova M, Dimitrova A. Serum concentrations of endothelin-1 and matrix metalloproteinases-2, -9 in pre-hypertensive and hypertensive patients with type 2 diabetes. Int J Mol Sci. 2016;17(8). doi: 10.3390/ijms17081182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma YZ, Jiang QY, Kong DQ. Association between matrix metallopeptidase 1 and type 2 diabetes mellitus coexisting with coronary heart disease in a Han Chinese population. Genet Mol Res. 2016;15(2). doi: 10.4238/gmr.15027938 [DOI] [PubMed] [Google Scholar]
- 57.Chasseaud LF. The role of glutathione and glutathione S-transferases in the metabolism of chemical carcinogens and other electrophilic agents. Adv Cancer Res. 1979;29(29):175–274. [DOI] [PubMed] [Google Scholar]
- 58.Antas P, Carneiro M, Reis B, et al. GST transcriptional changes induced by a toxic Microcystis aeruginosa strain in two bivalve species during exposure and recovery phases. Ecotoxicology. 2018;27(9):1272–1280. doi: 10.1007/s10646-018-1980-y [DOI] [PubMed] [Google Scholar]
- 59.Sheehan D, Meade G, Foley VM, Dowd CA. Structure, function and evolution of glutathione transferases implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem J. 2001;360(1):1–16. doi: 10.1042/0264-6021:3600001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang ST, Wang CJ, Tang HQ, Zhang Q, Wang Y. Evaluation of glutathione S-transferase genetic variants affecting type 2 diabetes susceptibility: a meta-analysis. Gene. 2013;530(2):301–308. doi: 10.1016/j.gene.2013.08.043 [DOI] [PubMed] [Google Scholar]
- 61.Babelova A, Burckhardt BC, Salinas-Riester G, Pommerenke C, Burckhardt G, Henjakovic M. Next generation sequencing of sex-specific genes in the livers of obese ZSF1 rats. Genomics. 2015;106(4):204–213. doi: 10.1016/j.ygeno.2015.07.006 [DOI] [PubMed] [Google Scholar]
- 62.Stoian A, Banescu C, Balasa RI, et al. Influence of GSTM1, GSTT1, and GSTP1 polymorphisms on type 2 diabetes mellitus and diabetic sensorimotor peripheral neuropathy risk. Dis Markers. 2015;2015:638693. doi: 10.1155/2015/105358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takako Yokozawa, Kim HY, Yamabe N. Amelioration of diabetic nephropathy by dried Rehmanniae Radix (Di Huang) extract. Am J Chin Med (Gard City N Y). 2004;32(6):829–839. doi: 10.1142/S0192415X04002442 [DOI] [PubMed] [Google Scholar]
- 64.Xing L, Dai G, Gaohong L, et al. Synergistic interaction of effective parts in Rehmanniae Radix and Cornus officinalis ameliorates renal injury in C57BL/KsJ-db/db diabetic mice: involvement of suppression of AGEs/RAGE/SphK1 signaling pathway. J Ethnopharmacol. 2016;185:110–119. doi: 10.1016/j.jep.2016.03.017 [DOI] [PubMed] [Google Scholar]
- 65.Qian DS, Zhu YF, Zhu Q. Effect of alcohol extract of cornus officinalis Sieb. et Zucc on GLUT4 expression in skeletal muscle in type 2 (non-insulin-dependent) diabetic mellitus rats. Zhongguo Zhong Yao Za Zhi. 2001;26(12):859–862. [PubMed] [Google Scholar]
- 66.Zhang M-H, Feng L, Zhu M-M, et al. The anti-inflammation effect of Moutan Cortex on advanced glycation end products-induced rat mesangial cells dysfunction and high-glucose-fat diet and streptozotocin-induced diabetic nephropathy rats. J Ethnopharmacol. 2014;151(1):591–600. doi: 10.1016/j.jep.2013.11.015 [DOI] [PubMed] [Google Scholar]
- 67.Li Q, Li W, Gao Q, Zou Y. Hypoglycemic effect of Chinese Yam (Dioscorea opposita rhizoma) polysaccharide in different structure and molecular weight. J Food Sci. 2017;82(10):2487–2494. doi: 10.1111/1750-3841.13919 [DOI] [PubMed] [Google Scholar]
- 68.Hsu PC, Tsai YT, Lai JN, Wu CT, Lin SK, Huang CY. Integrating traditional Chinese medicine healthcare into diabetes care by reducing the risk of developing kidney failure among type 2 diabetic patients: a population-based case control study. J Ethnopharmacol. 2014;156:358–364. doi: 10.1016/j.jep.2014.08.029 [DOI] [PubMed] [Google Scholar]
- 69.Akash MSH, Kanwal R, Shuqing C. Role of inflammatory mechanisms in pathogenesis of type 2 diabetes mellitus. J Cell Biochem. 2013;114(3):525–531. doi: 10.1002/jcb.24402 [DOI] [PubMed] [Google Scholar]
- 70.Akash MS, Rehman K, Liaqat A. Liaqat A tumor necrosis factor-alpha role in development of insulin resistance and pathogenesis of type 2 diabetes mellitus. J Cell Biochem. 2018;119(1):105–110. doi: 10.1002/jcb.26174 [DOI] [PubMed] [Google Scholar]
- 71.Chawla D, Bansal S, Banerjee BD, Madhu SV, Kalra OP, Tripathi AK. Role of advanced glycation end product (AGE)-induced receptor (RAGE) expression in diabetic vascular complications. Microvasc Res. 2014;95:1–6. doi: 10.1016/j.mvr.2014.06.010 [DOI] [PubMed] [Google Scholar]
- 72.Koulis C, Watson AM, Gray SP, Jandeleit-Dahm KA. Linking RAGE and Nox in diabetic micro- and macrovascular complications. Diabetes Metab. 2015;41(4):272–281. doi: 10.1016/j.diabet.2015.01.006 [DOI] [PubMed] [Google Scholar]
- 73.Gao X, Zhang H, Schmidt AM, Zhang C. AGE/RAGE produces endothelial dysfunction in coronary arterioles in type 2 diabetic mice. Am J Physiol Heart Circ Physiol. 2008;295(2):H491. doi: 10.1152/ajpheart.00464.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kay AM, Simpson CL, Stewart JA Jr. The role of AGE/RAGE signaling in diabetes-mediated vascular calcification. J Diabetes Res. 2016;2016:6809703. doi: 10.1155/2016/6809703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamagishi S, Matsui T, Nakamura K. Blockade of the advanced glycation end products (AGEs) and their receptor (RAGE) system is a possible mechanism for sustained beneficial effects of multifactorial intervention on mortality in type 2 diabetes. Med Hypotheses. 2008;71(5):749–751. doi: 10.1016/j.mehy.2008.05.039 [DOI] [PubMed] [Google Scholar]
- 76.Tan KCB, Chow WS, Ai VHG, Metz C, Bucala R, Lam KSL. Advanced glycation end products and endothelial dysfunction in type 2 diabetes. Diabetes Care. 2002;25(6):1055. doi: 10.2337/diacare.25.6.1055 [DOI] [PubMed] [Google Scholar]
- 77.Wautier MP, Chappey O, Corda S, Stern DM, Schmidt AM, Wautier JL. Activation of NADPH oxidase by AGE links oxidant stress to altered gene expression via RAGE. Am J Physiol Endocrinol Metab. 2001;280(5):E685–E694. doi: 10.1152/ajpendo.2001.280.5.E685 [DOI] [PubMed] [Google Scholar]
- 78.Csiszar AUZ. Endothelial dysfunction and vascular inflammation in type 2 diabetes interaction of AGE-RAGE and TNF-α signaling. Am J Physiol Heart Circ Physiol. 2008;295(2):H475. doi: 10.1152/ajpheart.00644.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xue G, Souad B, Andrea P, et al. Tumor necrosis factor-alpha induces endothelial dysfunction in Lepr(db) mice. Circulation. 2007;115(2):245–254. doi: 10.1161/CIRCULATIONAHA.106.650671 [DOI] [PubMed] [Google Scholar]
- 80.Andrea P, Xue G, Souad B, et al. Tumor necrosis factor-alpha induces endothelial dysfunction in the prediabetic metabolic syndrome. Circ Res. 2006;99(1):69–77. doi: 10.1161/01.RES.0000229685.37402.80 [DOI] [PubMed] [Google Scholar]
- 81.Zheng X, Zhu S, Chang S, et al. Protective effects of chronic resveratrol treatment on vascular inflammatory injury in streptozotocin-induced type 2 diabetic rats: role of NF-kappa B signaling. Eur J Pharmacol. 2013;720(1–3):147–157. [PubMed] [Google Scholar]
- 82.Kim HS, Han MS, Chung KW, et al. Toll-like receptor 2 senses beta-cell death and contributes to the initiation of autoimmune diabetes. Immunity. 2007;27(2):321–333. doi: 10.1016/j.immuni.2007.06.010 [DOI] [PubMed] [Google Scholar]
- 83.Dasu MR, Martin SJ. Toll-like receptor expression and signaling in human diabetic wounds. World J Diabetes. 2014;5(2):219–223. doi: 10.4239/wjd.v5.i2.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mali V, Haddox S, Hornersmith C, Matrougui K, Belmadani S. Essential role for EGFR tyrosine kinase and ER stress in myocardial infarction in type 2 diabetes. Pflugers Arch. 2018;470(3):471–480. doi: 10.1007/s00424-017-2097-5 [DOI] [PubMed] [Google Scholar]
- 85.Souad B, Palen DI, Gonzalez-Villalobos RA, Boulares HA, Khalid M. Elevated epidermal growth factor receptor phosphorylation induces resistance artery dysfunction in diabetic db/db mice. Diabetes. 2008;57(6):1629–1637. doi: 10.2337/db07-0739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Prada PO, Ropelle ER, Mourão RH, et al. EGFR tyrosine kinase inhibitor (PD153035) improves glucose tolerance and insulin action in high-fat diet-fed mice. Diabetes. 2009;58(12):2910. doi: 10.2337/db08-0506 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 87.Z LY L, Overstreet JM, Chung S, et al. Inhibition of epidermal growth factor receptor activation is associated with improved diabetic nephropathy and insulin resistance in type 2 diabetes. Diabetes. 2018;67(9):1847–1857. doi: 10.2337/db17-1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen S, Jiang H, Wu X, Fang J. Therapeutic effects of quercetin on inflammation, obesity, and type 2 diabetes. Mediators Inflamm. 2016;2016:9340637. doi: 10.1155/2016/9340637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zheng J, Wu J, Chen J, et al. Therapeutic effects of quercetin on early inflammation in hypertriglyceridemia-related acute pancreatitis and its mechanism. Pancreatology. 2016;16(2):200–210. doi: 10.1016/j.pan.2016.01.005 [DOI] [PubMed] [Google Scholar]
- 90.Xu XH, Zhao C, Peng Q, Xie P, Liu QH. Kaempferol inhibited VEGF and PGF expression and in vitro angiogenesis of HRECs under diabetic-like environment. Braz J Med Biol Res. 2017;50(3):e5396. doi: 10.1590/1414-431X20176071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.MB G, Rathmann W, Charbonnel B, et al. Treatment of type 2 diabetes mellitus worldwide: baseline patient characteristics in the global DISCOVER study. Diabetes Res Clin Pract. 2019;151:20–32. doi: 10.1016/j.diabres.2019.03.024 [DOI] [PubMed] [Google Scholar]
- 92.Health AGDo, Ageing. What is diabetes? Australian Government Department of Health & Ageing. 2006;1:1-2. Available from: http://www.health.gov.au/internet/main/publishing.nsf/Content/pq. [Google Scholar]