Genetic studies revealed that Abl family kinases interact functionally with microtubules, but the mechanism by which Abl kinases regulate microtubules remains unclear. Hu et al. provide the first evidence that the Abl family kinase Abl2 directly binds microtubules to regulate microtubule dynamics.
Abstract
Abl family kinases are essential regulators of cell shape and movement. Genetic studies revealed functional interactions between Abl kinases and microtubules (MTs), but the mechanism by which Abl family kinases regulate MTs remains unclear. Here, we report that Abl2 directly binds to MTs and regulates MT behaviors. Abl2 uses its C-terminal half to bind MTs, an interaction mediated in part through electrostatic binding to tubulin C-terminal tails. Using purified proteins, we found that Abl2 binds growing MTs and promotes MT polymerization and stability. In cells, knockout of Abl2 significantly impairs MT growth, and this defect can be rescued via reexpression of Abl2. Stable reexpression of an Abl2 fragment containing the MT-binding domain alone was sufficient to restore MT growth at the cell edge. These results show Abl2 uses its C-terminal half to bind MTs and directly regulate MT dynamics.
Introduction
Abl nonreceptor tyrosine kinases, Abl1 and Abl2 in vertebrates, play essential roles in the development and function of the heart, vasculature, brain, and immune system, and inappropriate activation of these kinases causes leukemias and promotes solid tumor progression (Qiu et al., 2010; Koleske et al., 1998; Srinivasan and Plattner, 2006; Chislock and Pendergast, 2013; Rizzo et al., 2015; Zipfel et al., 2004). Adhesion and growth factor receptors signal through Abl1 and Abl2 to activate several cytoskeletal effectors and coordinate changes in actin cytoskeletal structure (Plattner et al., 1999, 2004; Wang et al., 2001; Woodring et al., 2002; Miller et al., 2004; Van Etten et al., 1994). For example, in response to growth factor or integrin receptor activation, Abl2 phosphorylates the Arp2/3 complex regulators cortactin and N-WASp to promote actin-based cell edge protrusions, as well as the RhoA GTPase inhibitor p190RhoGAP to regulate cell:matrix adhesion dynamics and attenuate actomyosin contractility (Bradley et al., 2006; Oser et al., 2009; Lapetina et al., 2009; Boyle et al., 2007). Perturbations of these mechanisms disrupt cell migration, chemotaxis, and endocytosis in multiple cell types (Kain and Klemke, 2001; Peacock et al., 2007; Wetzel et al., 2012; Li et al., 2015), impair breast cancer cell invasion and metastasis (Mader et al., 2011), impede epithelial cell:cell adhesion (Grevengoed et al., 2001; Fox and Peifer, 2006; Zandy et al., 2007; Zandy and Pendergast, 2008), and compromise normal neuronal axon and dendrite development (Wills et al., 1999a,b; Giniger, 1998; Crowner et al., 2003; Moresco et al., 2005; Sfakianos et al., 2007).
To date, the majority of known Abl interactors control aspects of actin assembly, but key observations indicate that Abl family kinases also interact functionally with microtubules (MTs) to regulate cell morphogenesis. Genetic studies in Drosophila melanogaster indicate that Abl acts upstream of the MT-associated protein (MAP) CLASP to regulate neuronal axon pathfinding (Lee et al., 2004). Abl1 can phosphorylate CLASP in vitro, but the physiological consequences are unclear (Engel et al., 2014). Genetic and proteomic experiments in flies indicate that minispindles, the orthologue of the vertebrate MT polymerase XMAP215, and the MT motor protein kinesin-1 also interact genetically with abl (Martin et al., 2005; Lowery et al., 2010). Despite this compelling data that Abl family kinases interact functionally with MTs, the physical and mechanistic basis by which Abl family kinases regulate MTs is still unknown.
In addition to their kinase and kinase-regulatory Src homology (SH) 3 and SH2 domains, Abl family kinases contain large 600 amino acid C-terminal extensions. Here, we show that this Abl2 C-terminal half directly binds MTs and regulates MT dynamics. Abl2 binding to MTs is significantly impaired, but not completely disrupted, by increasing ionic strength or removing tubulin C-terminal tails. We show that Abl2 or the Abl2 C-terminal half is sufficient to increase the MT elongation rate, decrease the shortening rate, and reduce the catastrophe frequency in vitro. Knockout of Abl2 in both fibroblasts and COS-7 cells impairs MT growth, which can be rescued with reexpression of Abl2 or the Abl2 C-terminal half. Together, these data indicate that direct binding of Abl2 regulates MT dynamics both in vitro and in cells.
Results and discussion
The Abl2 C-terminal half binds MTs
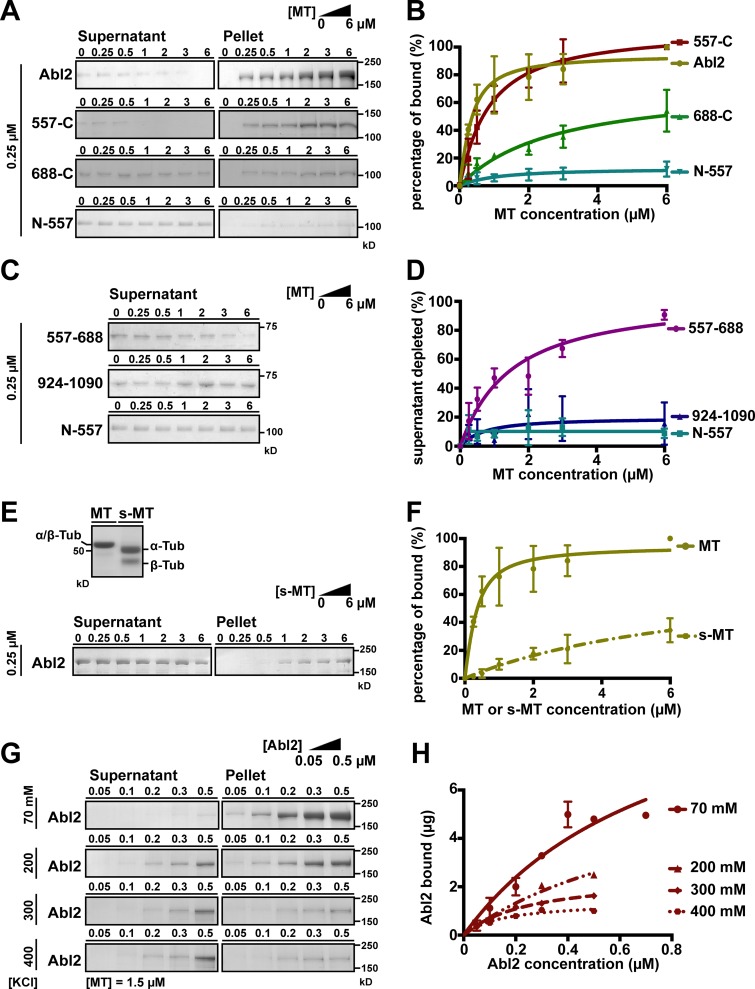
Abl2, an Abl family kinase, contains N-terminal tandem SH3, SH2, tyrosine kinase domains, and a large C-terminal half that mediates interactions with other proteins including actin and MTs (Lapetina et al., 2009; Miller et al., 2004; Wang et al., 2001; MacGrath and Koleske, 2012). The Abl2 actin-binding domains have been well defined, and these domains are required for inducing actin-rich dynamic protrusions at the cell periphery (Wang et al., 2001; Miller et al., 2004). Previous work indicated that a dimerized GST fusion to Abl2 amino acids 924–1090 can bind MTs (Miller et al., 2004), but our subsequent work showed that monomeric maltose binding protein (MBP) fusion to Abl2-924-1090 did not bind detectably to MTs (Fig. 1 D and Fig. S1 I). This led us to reexamine the features of Abl2 required for high-affinity MT binding. We purified Abl2 and Abl2 fragments via an N-terminal MBP fusion and used a MT cosedimentation assay to measure their binding to MTs (Table 1). 0.25 µM Abl2 was mixed with increasing concentrations of taxol-stabilized MTs and the relative amount of Abl2 pelleted after centrifugation was measured. When incubated alone, Abl2 remained in the supernatant, while increasing amounts of Abl2 were recovered in the pellet fraction as MT concentrations were increased. MBP-tagged Abl2 bound MTs with a Dissociation constant (Kd ) = 0.21 ± 0.05 µM (mean ± SD; Fig. 1, A and B; and Fig. S1 A). MBP alone did not bind MTs, and cleavage of MBP from Abl2 did not impact MT-binding affinity (Kd = 0.17 ± 0.06 µM; Fig. S1, G, Q, and R), indicating MBP did not impact Abl2 binding to MTs. Hence, we did not remove MBP tags from Abl2 fragments. While the Abl2 N-terminal half (Abl2-N-557) did not bind MTs, the Abl2 C-terminal half (Abl2-557-C) bound MTs strongly (Kd = 0.70 ± 0.13 µM; Fig. 1, A and B; and Fig. S1, B and D). We further truncated the Abl2 C-terminal half to identify the region necessary and sufficient for MT binding. Abl2-668-C, which lacks the central proline-rich domain, exhibited significantly weaker affinity than Abl2-557-C and did not saturate binding to MTs within the tested concentrations (Kd > 3 µM; Fig. 1, A and B; and Fig. S1 C). In contrast, the Abl2 N-terminal half plus the central proline-rich region (Abl2-N-688) bound MTs with an affinity (Kd = 0.24 ± 0.07 µM) similar to full-length Abl2 (Fig. S1 E). As MBP-tagged Abl2-557-688 comigrates with MTs on SDS-PAGE (Fig. S1 F), we measured its MT binding by monitoring its depletion from the supernatant, obtaining a Kd = 1.30 ± 0.30 µM, while Abl2-N-557 and Abl2-924-1090 did not detectably bind MTs (Fig. 1, C and D; and Fig. S1, D, H, and I). Together, these data reveal that Abl2 contains a primary MT-binding region, Abl2-557-688, that is necessary and sufficient for high-affinity MT binding. Binding of this domain to MTs is augmented by determinants in the N-terminal half, as Abl2-N-688 binds MTs with higher affinity than Abl2-557-688, and in Abl2-688-C, which mediates low-affinity binding to MTs.
Figure 1.
Abl2 used C-terminal regions to bind MTs and removal of the tubulin E-hook impairs Abl2:MT interaction. (A) A fixed concentration of 0.25 µM Abl2, Abl2-557-C, Abl2-688-C, or Abl2-N-557 was mixed with 0–6 µM MTs. The mixture was pelleted by high-speed centrifugation, and all of the pellet and one third of the supernatant were separated by SDS-PAGE. The amounts of Abl2 or Abl2 fragments in the supernatant and pellet were quantified by densitometry. Results are shown in B. The original gels are shown in Fig. S1, A–D. (C) Supernatant depletion of Abl2 fragments was used to infer MT binding. 0.25 µM Abl2-557-688 or Abl2-924-1090 was mixed with 0–6 µM MTs, then treated as in A. Results are shown in D. The original gels are shown in Fig. S1, H and I. (E) Equal amounts (16 µg) of MT or s-MT are shown on the SDS-PAGE. A fixed concentration of 0.25 µM Abl2 was mixed with 0–6 µM s-MTs, then treated as in A. Results are shown in F. The original gels are shown in Fig. S1 J. (G) A fixed concentration of 1.5 µM MT was mixed with 0.05–0.5 µM Abl2 and 70–400 mM KCl, then treated as in A. The amount of Abl2 recovered from pellet fraction versus Abl2 concentrations used in the assay is shown in H. The original gels are shown in Fig. S1, K–P. Error bars are presented as mean ± SD 5 ≤ n ≤ 7. The R2 values of curve fitting for Abl2 = 0.88, 557-C = 0.95, 688-C = 0.85, N-688 = 0.84, and 557–688 = 0.89. Tub, tubulin.
Table 1. Summary of MT binding of Abl2 and Abl2 fragments.
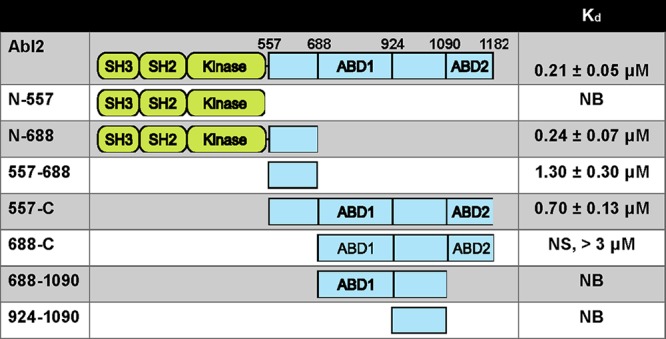 |
Abl2 contains tandem SH3, SH2, and kinase domains and a C-terminal half containing actin binding domains (ABD). Data are presented as mean ± SD; NB, not binding; NS, not saturating.
Abl2 binding to MTs is partly mediated by electrostatic interactions with the tubulin E-hook
Abl2 does not share sequence homology with any known MT-binding domains, but the amino acids in 557–688 are basic (isoelectric point [pI] = 10.24) and proline rich (12 prolines/131 residues), similar to the MT-binding regions of MAP2 and Tau, which bind to the negatively charged α/β-tubulin C termini known as the “E-hook” (Al-Bassam et al., 2002; Hinrichs et al., 2012; Lee et al., 1988). We hypothesized that Abl2 utilizes a similar electrostatic mechanism to bind MTs. We treated polymerized MTs with subtilisin to cleave the E-hook and measured Abl2 binding to subtilisin-treated MTs (s-MTs; Fig. 1 E and Fig. S1 J). Abl2 bound s-MTs weakly, and binding did not saturate within the tested concentrations (Kd > 3 µM; Fig. 1 F). We also probed the electrostatic nature of the Abl2:MT interaction by measuring the concentration dependence of Abl2 binding to MTs as a function of ionic strength. Abl2 bound MTs with a Kd = 0.36 ± 0.25 µM, similar to the affinity measured using a fixed Abl2 concentration and increasing MT concentrations. Abl2:MT binding was significantly reduced as the KCl concentration increased (Fig. 1, G and H; and Fig. S1, K–P). These data strongly suggest that electrostatic interactions between Abl2 and the tubulin E-hook contribute to Abl2:MT binding. Our finding that subtilisin or KCl treatment weakens but does not abolish Abl2:MT binding suggests that, in addition to the E-hook, Abl2 binds MTs via other interfaces.
The Abl2 C-terminal half binds to stabilized and growing MTs
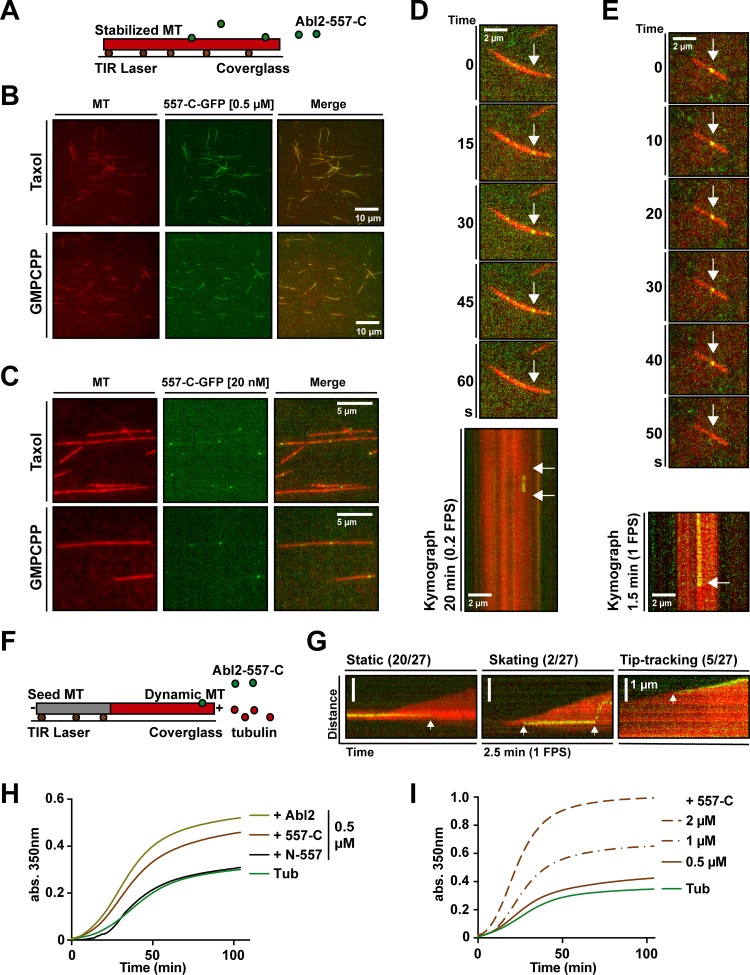
We used total internal reflection fluorescence (TIRF) microscopy to examine where Abl2-557-C bound to MTs. We stabilized rhodamine-MTs with Guanosine-5′-[(β,γ)-methyleno]triphosphate (GMPCPP) or taxol and monitored binding of 0.5 µM Abl2-557-C-GFP (Fig. 2 A). Abl2-557-C-GFP bound densely along the GMPCPP- or taxol-stabilized MTs (Fig. 2 B). Control experiments containing rhodamine-MTs alone showed that this apparent Abl2-557-C-GFP binding did not result from rhodamine bleed-through in the GFP channel (Fig. S2 A). To visualize the binding and/or possible movement of Abl2-557-C-GFP along stabilized MTs, GMPCPP-stabilized MTs were incubated with 20 nM Abl2-557-C-GFP, and images were captured at 0.2 frames per second (FPS; Fig. 2 C). The majority of Abl2-557-C-GFP puncta (64%) fell within a single low-intensity peak, and we further analyzed their behaviors (Fig. S2 B). Some Abl2-557-C-GFP puncta were observed to appear on the MT lattice, followed by single-step disappearance, due to either unbinding or photobleaching (Fig. 2 D). This finding strongly suggested that the low-intensity fluorescence puncta on the MT lattice were single Abl2-557-C-GFP molecules. To address possible transient movements of Abl2-557-C-GFP on MTs, we performed imaging at a faster capture rate (1 FPS) and confirmed that Abl2-557-C-GFP bound stably to stabilized MTs (Fig. 2 E). To visualize how Abl2-557-C-GFP bind to growing MTs, 50 nM Abl2-557-C-GFP and 7 µM rhodamine-tubulin were incubated with nonfluorescent GMPCPP-stabilized biotin-MTs. Abl2-557-C-GFP molecules that bound to the newly formed dynamic rhodamine-labeled MTs were imaged at 1 FPS (Fig. 2 F). Most Abl2-557-C-GFP (20/27) bound stably to the lattice of the growing MTs, while a minority of Abl2-557-C-GFP (5/27) puncta remained associated with the growing MT tips. In rare cases, Abl2-557-C-GFP puncta bound the MT lattice and skated toward the MT growing end (2/27; Fig. 2 G).
Figure 2.
Abl2-557-C-GFP binds on growing and stabilized MTs and Abl2 or Abl2-557-C promotes tubulin assembly. (A) Abl2-557-C-GFP (green) was incubated with rhodamine-labeled GMPCPP- or taxol-stabilized MTs (red) bound to the coverslip via anti-rhodamine antibodies (brown). Stabilized rhodamine-MTs were incubated with (B) 0.5 µM or (C) 20 nM Abl2-557-C-GFP. Kymographs of 20 nM Abl2-557-C-GFP on GMPCPP-stabilized MTs plotted from videos (D) at 0.2 FPS or (E) 1 FPS. (F) 50 nM Abl2-557-C-GFP and 7 µM rhodamine-tubulin was incubated with GMPCPP-stabilized biotin-MTs (gray) bound to the coverslip via neutravidin (brown). (G) Kymographs of Abl2-557-C-GFP on growing rhodamine-MTs. Events are categorized as static, skating, or tip-tracking. Tubulin assembly was monitored by measuring turbidity (A350). Representative time series of A350 measurements are shown. 18 µM tubulin and 1 mM GTP incubated alone or with (H) 0.5 µM Abl2, Abl2-557-C, or Abl2-N-557; (I) 0.5, 1, or 2 µM Abl2-557-C. n = 3. Arrows in D and E (top) show the position of single Abl2-557-C-GFP molecules binding to MTs. In the kymographs on D, E, and G, arrows mark the appearance and/or disappearance of the Abl2-557-C-GFP molecule binding and unbinding to MTs. TIR, total internal reflection.
Abl2 promotes tubulin assembly
We next sought to determine if the direct Abl2:MT interactions we observed were responsible for regulating MT dynamics. We measured whether Abl2 impacts tubulin assembly by monitoring turbidity at Absorbance 350 nm (A350). Tubulin polymerization resulting from incubation of 18 µM tubulin with GTP alone led to a time-dependent increase in A350. Inclusion of 0.5 µM Abl2 or Abl2-557-C led to a steeper increase in A350 that plateaued at a significantly higher level (Fig. 2 H). Abl2-557-C promoted tubulin assembly in a concentration-dependent manner (Fig. 2 I). Abl2-N-557, containing the kinase domain, did not impact tubulin assembly over tubulin alone. Control experiments showed that MBP did not affect tubulin assembly, and Abl2 or MBP alone did not impact A350 (Fig. S2, C and D). Negative stain EM images taken from samples of the turbidity experiments revealed that tubulin was assembled into MTs (Fig. S2 E), and we did not observe any significant MT bundles, as noted previously (Miller et al., 2004).
We next measured the physiological Abl2 concentration in cells. Using purified recombinant Abl2 as blotting standards, we found that Abl2 represents 0.19% of total protein in WT mouse 3T3 fibroblast and COS-7 cell extracts. Assuming that total cytoplasmic protein concentration is 100 mg/ml (Finka and Goloubinoff, 2013; Albe et al., 1990) and the molecular weight of Abl2 is 134 kD, we estimate the physiological concentration of Abl2 to be 1.4 µM (Fig. S3 A). This is within the concentration range at which Abl2 or Abl2 C-terminal half promotes tubulin assembly.
Abl2 increases MT elongation rate
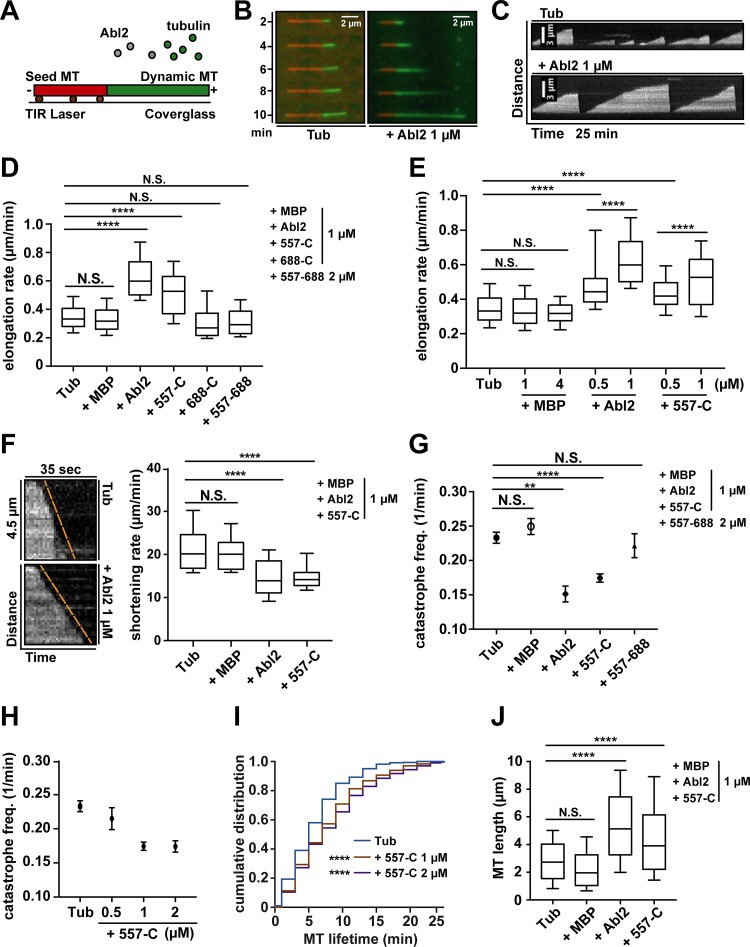
Given that Abl2 promotes tubulin assembly at physiological concentrations, we used TIRF microscopy to measure how Abl2 impacts MT dynamics at single MT resolution. GMPCPP-stabilized rhodamine-MT seeds were incubated with 7 µM Alexa 488–tubulin and varying concentrations of Abl2 or Abl2 fragments imaged over 25 min at 0.2 FPS (Fig. 3 A). MT elongation rate was quantified from kymographs of single MT behavior over time (Fig. 3, B and C; and Fig. S2 F). Under baseline conditions, MTs grew at 0.35 ± 0.01 µm/min. Inclusion of Abl2 or Abl2-557-C increased the MT elongation rate in a concentration-dependent manner, while MBP did not affect MT elongation (Fig. 3 E). 1 µM Abl2 or Abl2-557-C increased the elongation rate to 0.62 ± 0.02 µm/min or 0.52 ± 0.01 µm/min. In contrast, neither Abl2-557-688 nor Abl2-688-C impacted MT elongation (0.32 ± 0.01 µm/min or 0.31 ± 0.01 µm/min; Fig. 3 D). These observations indicate the Abl2 C-terminal half is sufficient to increase MT elongation rate and requires both the strong (557–688) and weak (688-C) MT-binding regions.
Figure 3.
Abl2 or Abl2-557-C is sufficient to regulate MT dynamics in vitro. (A) Alexa 488–labeled tubulin (green) polymerizes onto the ends of rhodamine-labeled GMPCPP-stabilized MTs (red) bound to the coverslip via anti-rhodamine antibodies (brown). (B and C) Time-lapse shots of MT growth and kymographs of MT dynamics. (D) MT elongation rates were measured alone or with 1 µM Abl2, Abl2-557-C, Abl2-688-C, MBP, or 2 µM Abl2-557-688. (E) MT elongation rates were measured alone or with 0.5 or 1 µM Abl2, 0.5 or 1 µM Abl2-557-C, or 1 or 4 µM MBP. (F) Kymographs of MT shrinkage for tubulin alone or with 1 µM Abl2. MT shortening rates were measured alone or with 1 µM Abl2, Abl2-557-C, or MBP. (G) MT catastrophe frequencies were measured alone or with 1 µM Abl2, Abl2-557-C, MBP, or 2 µM Abl2-557-688. (H) MT catastrophe frequencies were measured alone or with 0.5, 1, or 2 µM Abl2-557-C. (I) Cumulative frequencies of individual MT growth times were plotted based on kymographs of tubulin with 1 or 2 µM Abl2-557-C. Comparisons were made to tubulin control. (J) MT growth length at catastrophe was measured alone or with 1 µM Abl2, Abl2-557-C, or MBP. Error bars are presented as mean ± SEM n ≥ 100. **, P < 0.01; ****, P < 0.0001. freq., frequency; N.S., not significant.
Abl2 slows MT shrinkage and reduces catastrophe frequency
We also measured how Abl2 binding impacts MT shortening and catastrophe frequency. MT shortening rate with 7 µM Alexa 488–tubulin was monitored at 1 FPS. Under baseline conditions, MTs shrank at a rate of 20.9 ± 0.9 µm/min. Inclusion of 1 µM Abl2 or Abl2-557-C slowed MT shortening significantly to 13.5 ± 1.8 µm/min or 13.6 ± 0.4 µm/min, while MBP did not affect MT shortening (Fig. 3 F). MT catastrophe frequency was monitored for 25 min at 0.2 FPS. Under baseline conditions, MTs exhibit a catastrophe frequency of 0.23 min−1. Inclusion of 1 µM Abl2 or Abl2-557-C reduced catastrophe frequency to 0.15 min−1 or 0.17 min−1 (Fig. 3 G). Abl2-557-C reduced catastrophe frequency in a concentration-dependent manner (Fig. 3 H). 2 µM Abl2-557-688 did not affect MT catastrophe frequency (0.22 min−1; Fig. 3 G). Since MT catastrophe did not follow a normal distribution, a cumulative frequency of MT lifetimes was plotted to capture the change in overall population. Addition of 1 or 2 µM Abl2-557-C significantly shifted the distribution toward longer lifetimes (Fig. 3 I), while MBP did not affect the lifetime distribution of MTs (Fig. S2, G and H). We also recorded videos with an extended time scale (40 min) with 1 µM Abl2-557-C to address the possibility that the MTs with longer lifetimes were not adequately documented. Results showed MTs in 25- or 40-min videos had a similar lifetime distribution (Fig. S2, G and I), indicating 25-min recording time was acceptable for capturing catastrophe frequency. We also measured the MT growth length for each growing period to integrate the effects of Abl2 on MT elongation rate and catastrophe frequency. When incubated with 7 µM tubulin alone, MT growth length was 2.8 ± 0.1 µm. 1 µM Abl2 or Abl2-557-C significantly increased the MT growth length to 5.4 ± 0.3 µm or 4.5 ± 0.1 µm, respectively. MBP did not affect MT growth length (Fig. 3 J). Together, these results indicate that Abl2 uses its C-terminal half to stabilize MTs in vitro by slowing shrinkage and reducing catastrophe frequency.
Abl2 is required for normal MT growth in cells
Having demonstrated that purified Abl2 regulates MT dynamics in vitro, we examined how loss of Abl2 function impacts MT behaviors in cells. We measured MT growth in mouse 3T3 fibroblast and COS-7 cells by tracing the extension of MT plus-tip tracker mCherry-MACF43 (van de Willige et al., 2016; Honnappa et al., 2009; Slater et al., 2019). Because MT dynamics differ at the cell periphery compared with the cell center (Matov et al., 2010), we separately analyzed MT growth events in the inner 50% (cell center) and the outermost 25% of the cell (cell edge), based on distance from the observed MT organization center (MTOC) to the cell membrane. MT growth rates were quantified based on kymographs of MACF43 tracks (Fig. 4, A, B, and E; and Fig. S3 D).
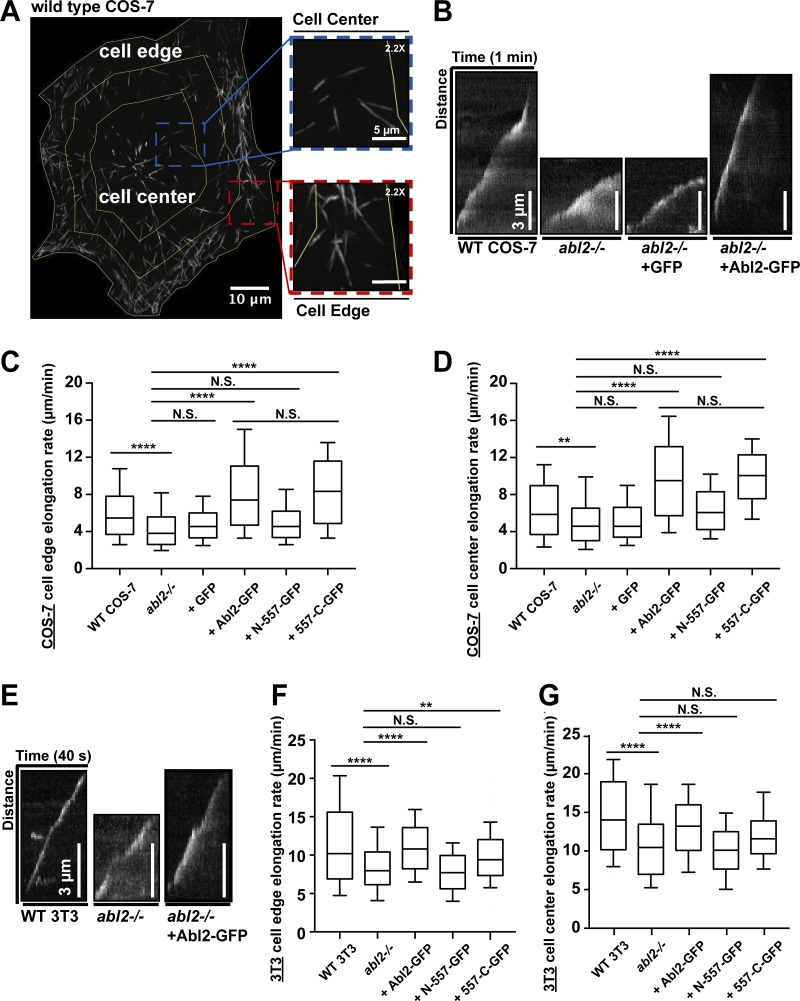
Figure 4.
Abl2 is required for normal MT growth in cells. Maximum intensity projection of mCherry-MACF43 time-lapse showed single MT tracks in a COS-7 cell. MT growth events in the inner one half of the cell, centered at the MTOC, were categorized as being within the cell center, and those in the outermost one quarter region were categorized as being within the cell edge. (B) Kymographs of the cell edge MT plus-tip growth in WT and abl2−/− COS-7 cells, and abl2−/− COS-7 cells re-expressing GFP or Abl2-GFP; (E) in WT and abl2−/− 3T3 cells, and abl2−/− 3T3 cells re-expressing Abl2-GFP, which are also shown in Fig. S3 E as references. Quantifications of MT plus-tip growth at the cell center and the cell edge in (C and D) COS-7 or (F and G) 3T3 WT, abl2−/−, and abl2−/− re-expressing Abl2-, N-557-, 557-C-GFP cells. n ≥ 150. **, P < 0.01; ****, P < 0.0001. N.S., not significant.
In control WT COS-7 cells, MTs grew at a rate of 6.1 ± 0.1 µm/min at the cell edge and 6.5 ± 0.2 µm/min at the cell center. In abl2−/− COS-7 cells, MT growth slowed by 25% at the cell edge (4.6 ± 0.2 µm/min) and 21% at the cell center (5.2 ± 0.2 µm/min; Fig. 4, B–D). Re-expression of Abl2-GFP in abl2−/− COS-7 cells increased MT growth rates to 8.3 ± 0.2 µm/min at the cell edge and 9.7 ± 0.2 µm/min at the cell center. Re-expression of Abl2-557-C-GFP also increased MT growth rates to 8.5 ± 0.2 µm/min at the cell edge and 10.0 ± 0.3 µm/min at the cell center (Fig. 4, C and D). The elevated rates of MT growth over those of WT cells likely resulted from the overexpression of Abl2-GFP (14-fold) or Abl2-557-C-GFP (18-fold; Fig. S3 B). That said, this effect was specific to the re-expression of Abl2 or Abl2-557-C, as similar overexpression of Abl2-N-557-GFP (12-fold), containing the tyrosine kinase domain but not the MT-binding regions, did not rescue MT growth at the cell edge (5.1 ± 0.2 µm/min) or the cell center (6.5 ± 0.2 µm/min). GFP expression also did not impact MT growth in abl2−/− COS-7 cells (edge: 4.9 ± 0.1 µm/min; center: 5.4 ± 0.2 µm/min; Fig. 4, C and D). Using dual-color TIRF imaging, we also observed EB3-GFP-positive MT plus-tips approaching and traversing though Abl2-RFP puncta at the cell edge in COS-7 cells (Fig. S3 E).
The COS-7 cell experiments demonstrate that reduction of Abl2 levels slowed MT growth, and overexpression of Abl2 or Abl2-557-C rescued this defect. We extended these results to 3T3 fibroblasts, where we could achieve more physiological re-expression levels of Abl2 or Abl2-557-C in abl2−/− 3T3 cells (Fig. S3 C). In WT 3T3 cells, MTs grew at a rate of 11.5 ± 0.4 µm/min at the cell edge and 14.6 ± 0.4 µm/min at the cell center. In abl2−/− 3T3 cells, MT growth was reduced by 26% at the cell edge (8.4 ± 0.3 µm/min) and 26% at the cell center (10.8 ± 0.4 µm/min; Fig. 4, E–G). Stable re-expression of Abl2-GFP (0.25-fold of endogenous level) in abl2−/− 3T3 cells restored MT growth to levels observed in WT cells (edge: 11.1 ± 0.2 µm/min; center: 13.1 ± 0.2 µm/min). Stable re-expression of Abl2-557-C-GFP (0.34-fold) was sufficient to restore MT growth at the cell edge, but not at the cell center (Fig. 4, F and G). This may reflect the preferential targeting of Abl2-557-C to the periphery in fibroblasts (Wang et al., 2001; Miller et al., 2004). Re-expression of Abl2-N-557-GFP (21-fold) did not rescue MT growth defect in abl2−/− 3T3 cells (edge: 7.9 ± 0.2 µm/min; center: 10.2 ± 0.3 µm/min).
Knowing that re-expression of Abl2-557-C rescued MT growth in cells, we next sought to test if Abl2-557-C regulates cell migration. Previously our laboratory showed that abl2−/− 3T3 cells had significantly higher migration speeds compared with WT 3T3 fibroblasts and that Abl2-GFP re-expression in abl2−/− 3T3 cells slowed migration to WT levels (Peacock et al., 2007). As before, we found that abl2−/− 3T3 cells showed a twofold increase in migration speed compared with WT 3T3 cells (WT: 11.88 ± 4.50 µm/h; abl2−/−: 22.18 ± 8.14 µm/h). Re-expression of Abl2-557-C-GFP in abl2−/− 3T3 cells partially slowed migration by 22.5% to 17.19 ± 9.01 µm/h, but unlike full-length Abl2, it did not slow migration to WT speeds (Fig. S3 F). GFP alone did not impact the accelerated abl2−/− migratory behavior (20.00 ± 7.56 µm/h). These data indicate that the Abl2-557-C contributes to normal cell migration, likely in combination with other activities of the protein.
A key finding was that the Abl2-557-C is sufficient to promote MT polymerization. We speculate that Abl2 may use its MT-binding regions to bridge adjacent protofilaments, providing longitudinal or lateral structural support to enhance MT stability and promote assembly. We demonstrate that Abl2 kinase activity is not absolutely required for the effects of Abl2 binding on MT elongation and stability. Given Abl family kinases can phosphorylate and regulate MAPs, including kinesin, CLASP, and Tau (Engel et al., 2014; Tremblay et al., 2010; Martin et al., 2005), it remains possible that these events may also contribute to MT-dependent processes in certain cellular contexts.
The primary interaction site of Abl2 with MTs, amino acids 557–668, share 43% sequence identity with the corresponding region in Abl1, which is also both basic (pI = 10.2) and proline-rich (12/122). We anticipate that Abl1 might also bind and/or regulate MTs. Given that Drosophila and Caenorhabditis elegans Abl shared much less sequence identity with amino acids 557–668 (25% and 29%, respectively), it is less clear if the invertebrate Abl kinases will bind or regulate MTs.
Previous work from our laboratory has demonstrated that Abl2 binds cooperatively to actin and promotes formation of actin-based protrusions, including lamellipodia in fibroblasts and invadopodia in invasive cancer cells (Wang et al., 2001; Miller et al., 2004; Peacock et al., 2007; Simpson et al., 2015). In these contexts, Abl2 localizes primarily with actin, and we did not observe significant Abl2 decoration on isolated MTs. However, a subset of MTs do preferentially target the Abl2:actin-rich protrusions in fibroblasts (Miller et al., 2004). These observations, in combination with the ability of Abl2 to regulate MT extension, suggest that actin-bound Abl2 may help to promote and steer MT growth into specific cell regions.
In summary, our experiments demonstrate that the Abl family nonreceptor tyrosine kinase Abl2 binds directly to MTs and controls MT dynamic behaviors. This represents a new cytoskeletal regulatory mechanism for an Abl family kinase.
Materials and methods
Molecular cloning, purification, and labeling of recombinant proteins
Murine Abl2 cDNAs were cloned with an MBP tag into the pFastBac1 vector (Invitrogen). Abl2 proteins engineered to start with the first common exon (glutamic acid, E74) of full-length Abl2, N-557, N-688, 557-C, 688-C, 557–688, 688–1090, 924–1090, and 557-C-GFP were generated using PCR-based amplification followed by cloning into pFastBac. Abl2, N-557, and 557-C cDNA were also cloned into pN1-EGFP expression vector for mammalian cell expression. Recombinant baculoviruses expressing Abl2 or Abl2 fragments in pFastBac vector constructs were generated using the Bac-to-Bac expression system in Sf9 insect cells according to the manufacturer’s instructions (Thermo Fisher Scientific). After expression in Hi5 insect cells for 48 h, cells were lysed in Hi5 Lysis Buffer (50 mM Hepes, pH 7.25, 500 mM KCl, 1% Triton X-100, 5% glycerol, 1 mM DTT, and protease inhibitors [benzamidine, aprotinin, leupeptin, chymostatin, pepstatin A, and PMSF]). Abl2 proteins were affinity-purified on Amylose resin (New England Biolabs) and eluted with 10 mM maltose. Abl2-557-688 cDNA was cloned into the pMAL-TEV vector (New England Biolabs) for protein expression in bacteria. Abl2-557-688 and MBP proteins were affinity-purified on Amylose resin and eluted with 10 mM maltose followed by chromatography on a Fast-SP Column with an elution gradient of 50 mM to 500 mM KCl. Porcine brain tubulin was purified as described previously (Castoldi and Popov, 2003). Tubulin is purified from porcine brain by two cycles of polymerization/depolymerization with high-molarity Pipes polymerization buffer (80 mM Pipes, pH 6.8, 10 mM MgCl2, and 20 mM EGTA) and depolymerization buffer (50 mM MES, pH 6.6, and 1 mM CaCl2). The purified tubulin was stored in BRB80 buffer (80 mM Pipes, pH 6.8, 1 mM MgCl2, and 1 mM EGTA). Cycled tubulin was labeled with Alexa 488 (Alexa Fluor 488 succinimidyl ester; Invitrogen) and rhodamine (TAMRA (5(6)-carboxytetramethylrhodamine succinimidyl ester); Invitrogen) as described (Brouhard et al., 2008). Cycled tubulin labeling was done by two cycles of polymerization/depolymerization. Tubulin was polymerized in BRB80 buffer and GTP, then labeled with labeling buffer (Hepes 100 mM, pH 8.6, 4 mM MgCl2, 1 mM EGTA, and glycerol 40% [vol/vol]) with 10% DMSO (vol/vol). Depolymerization was done by resuspending MTs in ice-cold BRB80 buffer. Biotinylated porcine brain tubulin was obtained from Cytoskeleton (T333P). GMPCPP (Jena Bioscience) stabilized MTs were grown as described (Yajima et al., 2012).
MT cosedimentation assays and quantification
Cosedimentation assays were performed as previously described (Campbell and Slep, 2011; Miller et al., 2004). Tubulin was polymerized at a final concentration of 100 µM at 37°C in polymerization buffer (80 mM Pipes, pH 6.8, 1 mM MgCl2, 1 mM EGTA, 1 mM GTP, and 15 nM paclitaxel [taxol]). The C-terminal 10–20 amino acids of α- and β-tubulin were removed by subtilisin treatment as previously described (Kerssemakers et al., 2006). Subtilisin (10 µg/ml) was incubated with preassembled MTs at 37°C for 1 h, and the reaction was terminated with 4 mM PMSF and 100 µg/ml aprotinin. The s-MTs were then pelleted and resuspended in BRB80. The taxol-stabilized MTs/s-MTs were set aside for cosedimentation. For MT cosedimentation assays, 0.25 µM Abl2 or Abl2 fragments were mixed with increasing concentrations of MTs or s-MTs (0 to 6 µM) at room temperature for 20 min in binding buffer (80 mM Pipes, pH 6.8, 70 mM KCl, 1 mM GTP, and 5 nM taxol [100 µl reaction volume]). For electrostatic interference assays, 1.5 µM MTs was mixed with increasing concentration of full-length Abl2 (0.05–0.7 µM) and KCl (70–400 mM) at room temperature for 20 min in binding buffer (80 mM Pipes, pH 6.8, 1 mM GTP, and 5 nM taxol [100 µl reaction volume]). Mixtures were pelleted by high-speed centrifugation at 120,000 × g for 20 min at 30°C. As a control, Abl2 or Abl2 fragments were centrifuged alone. Pellet and supernatant fractions were recovered and separated by SDS-PAGE, stained with Coomassie Blue G-250 (Bio-Rad Laboratories), and then destained in water. The SDS-PAGE gels were then scanned with a Canon LiDE-120 scanner and quantified by densitometry using ImageJ software. Binding affinity was quantified either as the percentage of Abl2/Abl2 fragments bound to MTs over the total amount of Abl2/Abl2 fragments in the reaction for each concentration, or as the amount of Abl2 bound to MTs for each concentration of Abl2. Experiments were repeated at least five times for each experimental condition (n ≥ 5). A specific binding curve with equation Y = Bmax * {c + Kd + X − sqrt[(c + Kd + X) *(c + Kd + X) − 4 * c * X]}/(2 * c) was used to fit the curve, where c = [Abl2]total, X = [MT]total, Y = [Abl2]bound, Kd is the dissociation constant of the Abl2-MT complex, and Bmax is the saturation percentage (%) of Abl2 that can bind to MT (Pollard, 2010). Binding curves, affinities (Kd), and R2 value for curve fitting were calculated using Prism8 (GraphPad).
In vitro imaging assays and quantification
MT dynamic imaging assays were performed as described preciously (Zanic et al., 2009; Gell et al., 2010; Brouhard et al., 2008). Imaging buffer consisting of BRB80 (80 mM Pipes, 1 mM MgCl2, and 1 mM EGTA, pH 6.8) was supplemented with 40 mM glucose, 40 µg/ml glucose oxidase, 16 µg/ml catalase, 0.1 mg/ml casein, 1 mM DTT, and 100 mM KCl. For measurements of MT elongation rate and catastrophe frequency, 20% rhodamine-labeled GMPCPP-stabilized MT seeds were anchored to coverslips via anti-rhodamine antibodies (Invitrogen). Reaction chambers were then perfused with 7 µM of 15% labeled Alexa 488–tubulin in imaging buffer with 1 mM GTP. Images were acquired every 5 s or at 0.2 FPS for 25 min on a Nikon Ti-E microscope with a 100×/1.49 oil objective, an Andor Zyla 4.2 sCMOS camera, and Nikon Elements software. An objective heater was used to keep the reaction chamber at 35°C. For MT shortening rate, images were acquired every 1 s or at 1 FPS for 2 min. Data were analyzed using kymographs from acquired images with ImageJ software. Elongation and shortening rates were determined by measuring the change in length of single MT filaments over time. Catastrophe frequency was expressed as the reciprocal of MT growth time. The quantification of kymographs was performed as described (Zanic et al., 2009). At least 100 MT tracks were quantified for each assay condition (n ≥ 100). For near simultaneous imaging of Abl2-557-C-GFP and stabilized rhodamine-MTs, 20% rhodamine-labeled GMPCPP- or taxol-stabilized MTs were anchored to coverslips via anti-rhodamine antibodies. The reaction chamber was then perfused with 0.5 µM or 20 nM Abl-557-C-GFP in imaging buffer, and images were acquired every 1 s (1 FPS) for 2 min or every 5 s (0.2 FPS) for 20 min. For near simultaneous imaging of Abl2-557-C-GFP and growing rhodamine-MTs, 10% biotin-labeled GMPCPP-stabilized MTs were anchored to coverslips via biotinylated BSA (Biotin-BSA; Thermo Fisher Scientific) and NeutrAvidin (Thermo Fisher Scientific) scaffolds. The GMPCPP-stabilized biotin-labeled short MTs were not fluorescently labeled. Reaction chambers were then perfused with 7 µM of 15% labeled rhodamine-tubulin and 50 nM Abl-557-C-GFP in imaging buffer and 1 mM GTP. Images were acquired every 1 s (1 FPS) for 3 min.
Turbidimetric measurement of tubulin assembly
18 µM porcine brain tubulin in BRB80 was incubated with 1 mM of GTP alone or in the presence of Abl2, Abl2 fragments, or MBP at 37°C. Tubulin assembly was monitored by measuring turbidity at 350 nm (A350) for 2 h using a SpectraMax M6 Multi-Mode Microplate Reader recording spectrophotometer. Control experiments were done by monitoring A350 for buffer alone (BRB80 + 1 mM GTP), buffer with 0.5 µM MBP, or buffer with 0.5 µM Abl2.
Cells, cell culture, and construct transfection
Mycoplasma free COS-7 cell lines (ATCC) were grown in DMEM supplemented with 10% FBS, 100 units/ml penicillin, 100 µg/ml streptomycin, and 2 mM L-glutamine. abl2−/− COS-7 cells were generated using CRISPR/Cas9. A guide sequence of 5′-GAGAAAGTGAGAGTAGCCCT-3′ with an adjacent protospacer adjacent motif, PAM (GGG) targeting the fourth exon of Abl2 was inserted into lentiCRISPR plasmid, then transfected into HEK293T cells to generate lentivirus. COS-7 cells were infected with the generated lentivirus then selected with 2 µg/ml puromycin for 72 h. abl2−/− COS-7 cells were transfected with Abl2, N-557, or 557-C in pN1-EGFP and polyethylenimine (PEI). Transfection was performed 24 to 48 h before imaging according to the manufacturer’s instructions. The same strategy was used to inactivate Abl2 in WT mouse 3T3 fibroblasts, using the guide sequence of 5′-CATGTAAAGTAACACGACGG-3′ with an adjacent PAM (CGG). Resistant Abl2-GFP, N-557-GFP, or 557-C-GFP was cloned into pLXSN vector and then transfected into HEK293T cells to generate retrovirus. abl2−/− 3T3 cells were infected with the retrovirus then selected with 400 µg/ml G418 for 7 d. Abl2- or Abl2 variants–expressing cells were obtained by fluorescence-activated cell sorting using abl2−/− 3T3 cells as a negative reference. Cell lysates were collected to determine the expression levels of Abl2 or Abl2 variants via immunoblotting.
Western blot analysis
Cells were lysed with 1× Laemmli Sample Buffer (LSB) buffer (8% SDS, 20% glycerol, 100 mM Tris, pH 6.8, 8% 2-mercaptoethanol, and complete protease inhibitors) at 95°C. Lysates were run on SDS-PAGE, then transferred to nitrocellulose, blocked using 5% milk, and immunoblotted with Ar11, which specifically recognizes Abl2 C-terminal half (residues 766–1182), or Ar19, which recognizes Abl2 N-terminal half (Ar11 and Ar19 were gifts from P. Davies, Albert Einstein Medical College, Bronx, NY). The intensity of each band was quantified using ImageJ and controlled by the intensity of Ponceau S stain image for each lane. abl2−/− COS-7 cells exhibited >92% loss of Abl2 signal and abl2−/− 3T3 fibroblasts exhibited >95% loss of Abl2 signal by blotting with Ar11 and Ar19.
Time-lapse live-cell microscopy and quantification
Cells were imaged on 30-mm no. 1.5 coverslips in an interchangeable dish (Bioptechs). Coverslips were plasma-cleaned for 2 min with H2/O2. Coverslips were coated with 50 µg/ml Poly-D-Lysine (Sigma-Aldrich) for 20 min at room temperature. Cells were seeded at 50,000 cells per coverslip. Cell dishes were maintained at 34°C in phenol red–free DMEM supplemented with 10% FBS and 20 mM Hepes (pH 7.3) while imaging. Images were acquired in TIRF mode every 1 s (1 FPS) for 2 min. Cell areas were divided by 15° radial lines centered at the observed MTOC. Cell centers were defined as the area within the inner half region of a cell determined by connecting the midpoints of all radial lines. Cell edges were defined as the outermost one quarter of a cell. Kymographs were generated by plotting the track of MT plus-tip marker fluorescence, mCherry-MACF43, as a function of time. MT plus-end growth rates were determined by measuring the displacement of MT plus-tip tracker over time. At least 150 tracks of mCherry-MACF43 were quantified for each assay condition (n ≥ 150).
Single-cell migration assay and quantification
WT and abl2−/− 3T3 fibroblasts, or abl2−/− 3T3 cells reexpressing GFP, or Abl2-557-C-GFP were plated at a subconfluence on six-well plates coated with 10 µg/ml fibronectin (Sigma-Aldrich). Phase-contrast images were acquired every 40 min for 6 h on a Keyence BZ-X710 All-in-One Fluorescence Microscope with a 4×/0.1 objective. Videos were analyzed with the Manual Tracking function in ImageJ, using one of the nucleoli to track the cells in each frame. At least 30 cells were tracked for each experimental condition (n ≥ 30). The cell migration tracking data were analyzed as described (Gorelik and Gautreau, 2014).
Statistical analyses
Comparisons of MT elongation, shrinkage, catastrophe frequency, and growth length were made with unpaired, two-tailed Student’s t tests, as appropriate. Comparisons of MT lifetime distribution were made with the Kolmogorov–Smirnov test. Significance was defined as *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. For MT catastrophe frequency analyses, error bars are presented as SEM (mean ± SEM). For MT cosedimentation experiments, values of Kd are expressed as mean ± SD, not binding, or not saturating. Error bars of the binding curves are presented as SD (mean ± SD). Calculations were performed in Prism7 (GraphPad).
Online supplemental material
Fig. S1 includes SDS-PAGE gel images for MT cosedimentation assays, including Abl2/Abl2 fragments with increasing MT or s-MT concentrations, MTs with increasing Abl2 concentrations, MTs with increasing Abl2 concentrations and increasing buffer ionic strength, tag-removed Abl2, or MBP with increasing MT concentrations. Fig. S2 includes fluorescence bleed-through control for in vitro imaging; single particle fluorescence intensity analyses; turbidity measurements for tubulin assembly of free-tubulin with Abl2 or Abl2 fragments; control turbidity measurements for buffer with Abl2 or MBP; negative stain images of MTs assembled in turbidity experiments; additional MT kymographs for in vitro imaging; and in vitro imaging control experiments for MBP and recording time. Fig. S3 includes Western blot images for determining physiological Abl2 concentration in WT 3T3 fibroblast and WT COS-7 cells; Abl2 or Abl2 fragments reexpression levels in abl2−/− 3T3 fibroblast and COS-7 cells; representative 3T3 cell images and MT-tracking kymographs; dual-color imaging experiments of MT plus-tip tracker and Abl2 puncta in COS-7 cells; and cell migration analyses.
Supplementary Material
Acknowledgments
We thank M. Mooseker, J. Howard, M. Podolski, and M. Mahamdeh for advice on experimental design and troubleshooting, G. Cammarata for experimental design assistance, and Koleske laboratory members for critical feedback on the project.
This work was supported by the National Institutes of Health, National Institute of Neurological Disorders and Stroke grants R01-NS-089662, R01-NS105640 (to A.J. Koleske), and National Institutes of Health, National Institute of Mental Health grant R01-MH115939 (to A.J. Koleske). Additional support was from the Randall W. Reyer Scholarship Fund (to Y. Hu), an American Heart Association Predoctoral Fellowship (to W. Lyu), and National Institute of Mental Health grant R01-MH109651 (to L.A. Lowery).
The authors declare no competing financial interests.
Author contributions: Y. Hu: conceptualization, investigation, methodology, visualization, and writing (original draft and editing); W. Lyu: conceptualization, investigation, methodology, visualization, and writing (review and editing); L.A. Lowery: resource, methodology, and writing (review and editing); and A.J. Koleske: conceptualization, methodology, supervision, funding acquisition, and writing (original draft and editing).
References
- Al-Bassam J., Ozer R.S., Safer D., Halpain S., and Milligan R.A.. 2002. MAP2 and tau bind longitudinally along the outer ridges of microtubule protofilaments. J. Cell Biol. 157:1187–1196. 10.1083/jcb.200201048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albe K.R., Butler M.H., and Wright B.E.. 1990. Cellular concentrations of enzymes and their substrates. J. Theor. Biol. 143:163–195. 10.1016/S0022-5193(05)80266-8 [DOI] [PubMed] [Google Scholar]
- Boyle S.N., Michaud G.A., Schweitzer B., Predki P.F., and Koleske A.J.. 2007. A critical role for cortactin phosphorylation by Abl-family kinases in PDGF-induced dorsal-wave formation. Curr. Biol. 17:445–451. 10.1016/j.cub.2007.01.057 [DOI] [PubMed] [Google Scholar]
- Bradley W.D., Hernández S.E., Settleman J., and Koleske A.J.. 2006. Integrin signaling through Arg activates p190RhoGAP by promoting its binding to p120RasGAP and recruitment to the membrane. Mol. Biol. Cell. 17:4827–4836. 10.1091/mbc.e06-02-0132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard G.J., Stear J.H., Noetzel T.L., Al-Bassam J., Kinoshita K., Harrison S.C., Howard J., and Hyman A.A.. 2008. XMAP215 is a processive microtubule polymerase. Cell. 132:79–88. 10.1016/j.cell.2007.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J.N., and Slep K.C.. 2011. αβ-Tubulin and microtubule-binding assays. Methods Mol. Biol. 777:87–97. 10.1007/978-1-61779-252-6_6 [DOI] [PubMed] [Google Scholar]
- Castoldi M., and Popov A.V.. 2003. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr. Purif. 32:83–88. 10.1016/S1046-5928(03)00218-3 [DOI] [PubMed] [Google Scholar]
- Chislock E.M., and Pendergast A.M.. 2013. Abl family kinases regulate endothelial barrier function in vitro and in mice. PLoS One. 8:e85231 10.1371/journal.pone.0085231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowner D., Le Gall M., Gates M.A., and Giniger E.. 2003. Notch steers Drosophila ISNb motor axons by regulating the Abl signaling pathway. Curr. Biol. 13:967–972. 10.1016/S0960-9822(03)00325-7 [DOI] [PubMed] [Google Scholar]
- Engel U., Zhan Y., Long J.B., Boyle S.N., Ballif B.A., Dorey K., Gygi S.P., Koleske A.J., and Vanvactor D.. 2014. Abelson phosphorylation of CLASP2 modulates its association with microtubules and actin. Cytoskeleton (Hoboken). 71:195–209. 10.1002/cm.21164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finka A., and Goloubinoff P.. 2013. Proteomic data from human cell cultures refine mechanisms of chaperone-mediated protein homeostasis. Cell Stress Chaperones. 18:591–605. 10.1007/s12192-013-0413-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox D.T., and Peifer M.. 2006. Abelson kinase (Abl) and RhoGEF2 regulate actin organization during cell constriction in Drosophila. Development. 134:567–578. 10.1242/dev.02748 [DOI] [PubMed] [Google Scholar]
- Gell C., Bormuth V., Brouhard G.J., Cohen D.N., Diez S., Friel C.T., Helenius J., Nitzsche B., Petzold H., Ribbe J., et al. 2010. Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. 95. First edition Elsevier; 221–245. [DOI] [PubMed] [Google Scholar]
- Giniger E. 1998. A role for Abl in notch signaling. Neuron. 20:667–681. 10.1016/S0896-6273(00)81007-7 [DOI] [PubMed] [Google Scholar]
- Gorelik R., and Gautreau A.. 2014. Quantitative and unbiased analysis of directional persistence in cell migration. Nat. Protoc. 9:1931–1943. 10.1038/nprot.2014.131 [DOI] [PubMed] [Google Scholar]
- Grevengoed E.E., Loureiro J.J., Jesse T.L., and Peifer M.. 2001. Abelson kinase regulates epithelial morphogenesis in Drosophila. J. Cell Biol. 155:1185–1198. 10.1083/jcb.200105102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs M.H., Jalal A., Brenner B., Mandelkow E., Kumar S., and Scholz T.. 2012. Tau protein diffuses along the microtubule lattice. J. Biol. Chem. 287:38559–38568. 10.1074/jbc.M112.369785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S., Gouveia S.M., Weisbrich A., Damberger F.F., Bhavesh N.S., Jawhari H., Grigoriev I., van Rijssel F.J.A., Buey R.M., Lawera A., et al. 2009. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 138:366–376. 10.1016/j.cell.2009.04.065 [DOI] [PubMed] [Google Scholar]
- Kain K.H., and Klemke R.L.. 2001. Inhibition of cell migration by Abl family tyrosine kinases through uncoupling of Crk-CAS complexes. J. Biol. Chem. 276:16185–16192. 10.1074/jbc.M100095200 [DOI] [PubMed] [Google Scholar]
- Kerssemakers J., Howard J., Hess H., and Diez S.. 2006. The distance that kinesin-1 holds its cargo from the microtubule surface measured by fluorescence interference contrast microscopy. Proc. Natl. Acad. Sci. USA. 103:15812–15817. 10.1073/pnas.0510400103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleske A.J., Gifford A.M., Scott M.L., Nee M., Bronson R.T., Miczek K.A., and Baltimore D.. 1998. Essential roles for the Abl and Arg tyrosine kinases in neurulation. Neuron. 21:1259–1272. 10.1016/S0896-6273(00)80646-7 [DOI] [PubMed] [Google Scholar]
- Lapetina S., Mader C.C., Machida K., Mayer B.J., and Koleske A.J.. 2009. Arg interacts with cortactin to promote adhesion-dependent cell edge protrusion. J. Cell Biol. 185:503–519. 10.1083/jcb.200809085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G., Cowan N., and Kirschner M.. 1988. The primary structure and heterogeneity of tau protein from mouse brain. Science. 239:285–288. 10.1126/science.3122323 [DOI] [PubMed] [Google Scholar]
- Lee H., Engel U., Rusch J., Scherrer S., Sheard K., and Van Vactor D.. 2004. The microtubule plus end tracking protein Orbit/MAST/CLASP acts downstream of the tyrosine kinase Abl in mediating axon guidance. Neuron. 42:913–926. 10.1016/j.neuron.2004.05.020 [DOI] [PubMed] [Google Scholar]
- Li R., Knight J.F., Park M., and Pendergast A.M.. 2015. Abl kinases regulate HGF/Met signaling required for epithelial cell scattering, tubulogenesis and motility. PLoS One. 10:e0124960 10.1371/journal.pone.0124960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery L.A., Lee H., Lu C., Murphy R., Obar R.A., Zhai B., Schedl M., Van Vactor D., and Zhan Y.. 2010. Parallel genetic and proteomic screens identify Msps as a CLASP-Abl pathway interactor in Drosophila. Genetics. 185:1311–1325. 10.1534/genetics.110.115626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGrath S.M., and Koleske A.J.. 2012. Arg/Abl2 modulates the affinity and stoichiometry of binding of cortactin to F-actin. Biochemistry. 51:6644–6653. 10.1021/bi300722t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader C.C., Oser M., Magalhaes M.A.O., Bravo-Cordero J.J., Condeelis J., Koleske A.J., and Gil-Henn H.. 2011. An EGFR-Src-Arg-cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer Res. 71:1730–1741. 10.1158/0008-5472.CAN-10-1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Ahern-Djamali S.M., Hoffmann F.M., and Saxton W.M.. 2005. Abl tyrosine kinase and its substrate Ena/VASP have functional interactions with kinesin-1. Mol. Biol. Cell. 16:4225–4230. 10.1091/mbc.E05-02-0116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matov A., Applegate K., Kumar P., Thoma C., Krek W., Danuser G., and Wittmann T.. 2010. Analysis of microtubule dynamic instability using a plus-end growth marker. Nat. Methods. 7:761–768. 10.1038/nmeth.1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A.L., Wang Y., Mooseker M.S., and Koleske A.J.. 2004. The Abl-related gene (Arg) requires its F-actin-microtubule cross-linking activity to regulate lamellipodial dynamics during fibroblast adhesion. J. Cell Biol. 165:407–419. 10.1083/jcb.200308055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresco E.M.Y., Donaldson S., Williamson A., and Koleske A.J.. 2005. Integrin-mediated dendrite branch maintenance requires Abelson (Abl) family kinases. J. Neurosci. 25:6105–6118. 10.1523/JNEUROSCI.1432-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oser M., Yamaguchi H., Mader C.C., Bravo-Cordero J.J., Arias M., Chen X., Desmarais V., van Rheenen J., Koleske A.J., and Condeelis J.. 2009. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J. Cell Biol. 186:571–587. 10.1083/jcb.200812176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock J.G., Miller A.L., Bradley W.D., Rodriguez O.C., Webb D.J., and Koleske A.J.. 2007. The Abl-related gene tyrosine kinase acts through p190RhoGAP to inhibit actomyosin contractility and regulate focal adhesion dynamics upon adhesion to fibronectin. Mol. Biol. Cell. 18:3860–3872. 10.1091/mbc.e07-01-0075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plattner R., Kadlec L., DeMali K.A., Kazlauskas A., and Pendergast A.M.. 1999. c-Abl is activated by growth factors and Src family kinases and has a role in the cellular response to PDGF. Genes Dev. 13:2400–2411. 10.1101/gad.13.18.2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plattner R., Koleske A.J., Kazlauskas A., and Pendergast A.M.. 2004. Bidirectional signaling links the Abelson kinases to the platelet-derived growth factor receptor. Mol. Cell. Biol. 24:2573–2583. 10.1128/MCB.24.6.2573-2583.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D. 2010. A guide to simple and informative binding assays. Mol. Biol. Cell. 21:4061–4067. 10.1091/mbc.e10-08-0683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z., Cang Y., and Goff S.P.. 2010. c-Abl tyrosine kinase regulates cardiac growth and development. Proc. Natl. Acad. Sci. USA. 107:1136–1141. 10.1073/pnas.0913131107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo A.N., Aman J., van Nieuw Amerongen G.P., and Dudek S.M.. 2015. Targeting Abl kinases to regulate vascular leak during sepsis and acute respiratory distress syndrome. Arterioscler. Thromb. Vasc. Biol. 35:1071–1079. 10.1161/ATVBAHA.115.305085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfakianos M.K., Eisman A., Gourley S.L., Bradley W.D., Scheetz A.J., Settleman J., Taylor J.R., Greer C.A., Williamson A., and Koleske A.J.. 2007. Inhibition of Rho via Arg and p190RhoGAP in the postnatal mouse hippocampus regulates dendritic spine maturation, synapse and dendrite stability, and behavior. J. Neurosci. 27:10982–10992. 10.1523/JNEUROSCI.0793-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson M.A., Bradley W.D., Harburger D., Parsons M., Calderwood D.A., and Koleske A.J.. 2015. Direct interactions with the integrin β1 cytoplasmic tail activate the Abl2/Arg kinase. J. Biol. Chem. 290:8360–8372. 10.1074/jbc.M115.638874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater P., Cammarata G.M., Monahan C., Bowers J.T., Yan O., Lee S., and Lowery L.. 2019. Characterization of Xenopus laevis Guanine Deaminase reveals new insights for its expression and function in the embryonic kidney. Dev. Dyn. 248:296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan D., and Plattner R.. 2006. Activation of Abl tyrosine kinases promotes invasion of aggressive breast cancer cells. Cancer Res. 66:5648–5655. 10.1158/0008-5472.CAN-06-0734 [DOI] [PubMed] [Google Scholar]
- Tremblay M.A., Acker C.M., and Davies P.. 2010. Tau phosphorylated at tyrosine 394 is found in Alzheimer’s disease tangles and can be a product of the Abl-related kinase, Arg. J. Alzheimers Dis. 19:721–733. 10.3233/JAD-2010-1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Willige D., Hoogenraad C.C., and Akhmanova A.. 2016. Microtubule plus-end tracking proteins in neuronal development. Cell. Mol. Life Sci. 73:2053–2077. 10.1007/s00018-016-2168-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten R.A., Jackson P.K., Baltimore D., Sanders M.C., Matsudaira P.T., and Janmey P.A.. 1994. The COOH terminus of the c-Abl tyrosine kinase contains distinct F- and G-actin binding domains with bundling activity. J. Cell Biol. 124:325–340. 10.1083/jcb.124.3.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Miller A.L., Mooseker M.S., and Koleske A.J.. 2001. The Abl-related gene (Arg) nonreceptor tyrosine kinase uses two F-actin-binding domains to bundle F-actin. Proc. Natl. Acad. Sci. USA. 98:14865–14870. 10.1073/pnas.251249298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel D.M., McMahon-Pratt D., and Koleske A.J.. 2012. The Abl and Arg kinases mediate distinct modes of phagocytosis and are required for maximal Leishmania infection. Mol. Cell. Biol. 32:3176–3186. 10.1128/MCB.00086-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills Z., Bateman J., Korey C.A., Comer A., and Van Vactor D.. 1999a The tyrosine kinase Abl and its substrate enabled collaborate with the receptor phosphatase Dlar to control motor axon guidance. Neuron. 22:301–312. 10.1016/S0896-6273(00)81091-0 [DOI] [PubMed] [Google Scholar]
- Wills Z., Marr L., Zinn K., Goodman C.S., and Van Vactor D.. 1999b Profilin and the Abl tyrosine kinase are required for motor axon outgrowth in the Drosophila embryo. Neuron. 22:291–299. 10.1016/S0896-6273(00)81090-9 [DOI] [PubMed] [Google Scholar]
- Woodring P.J., Litwack E.D., O’Leary D.D.M., Lucero G.R., Wang J.Y.J., and Hunter T.. 2002. Modulation of the F-actin cytoskeleton by c-Abl tyrosine kinase in cell spreading and neurite extension. J. Cell Biol. 156:879–892. 10.1083/jcb.200110014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima H., Ogura T., Nitta R., Okada Y., Sato C., and Hirokawa N.. 2012. Conformational changes in tubulin in GMPCPP and GDP-taxol microtubules observed by cryoelectron microscopy. J. Cell Biol. 198:315–322. 10.1083/jcb.201201161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandy N.L., and Pendergast A.M.. 2008. Abl tyrosine kinases modulate cadherin-dependent adhesion upstream and downstream of Rho family GTPases. Cell Cycle. 7:444–448. 10.4161/cc.7.4.5452 [DOI] [PubMed] [Google Scholar]
- Zandy N.L., Playford M., and Pendergast A.M.. 2007. Abl tyrosine kinases regulate cell-cell adhesion through Rho GTPases. Proc. Natl. Acad. Sci. USA. 104:17686–17691. 10.1073/pnas.0703077104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanic M., Stear J.H., Hyman A.A., and Howard J.. 2009. EB1 recognizes the nucleotide state of tubulin in the microtubule lattice. PLoS One. 4:e7585 10.1371/journal.pone.0007585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel P.A., Zhang W., Quiroz M., and Pendergast A.M.. 2004. Requirement for Abl kinases in T cell receptor signaling. Curr. Biol. 14:1222–1231. 10.1016/j.cub.2004.07.021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.