Parkinson’s disease–associated LRRK2 kinase is activated on the Golgi by Rab29 but phosphorylates Rab10 there and on other compartments. This study shows that phosphorylation is restricted to membrane surfaces but need not take place on the Golgi.
Abstract
LRRK2 kinase mutations cause familial Parkinson’s disease and increased phosphorylation of a subset of Rab GTPases. Rab29 recruits LRRK2 to the trans-Golgi and activates it there, yet some of LRRK2’s major Rab substrates are not on the Golgi. We sought to characterize the cell biology of LRRK2 activation. Unlike other Rab family members, we show that Rab29 binds nucleotide weakly, is poorly prenylated, and is not bound to GDI in the cytosol; nevertheless, Rab29 only activates LRRK2 when it is membrane bound and GTP bound. Mitochondrially anchored, GTP-bound Rab29 is both a LRRK2 substrate and activator, and it drives accumulation of active LRRK2 and phosphorylated Rab10 on mitochondria. Importantly, mitochondrially anchored LRRK2 is much less capable of phosphorylating plasma membrane–anchored Rab10 than soluble LRRK2. These data support a model in which LRRK2 associates with and dissociates from distinct membrane compartments to phosphorylate Rab substrates; if anchored, LRRK2 can modify misdelivered Rab substrates that then become trapped there because GDI cannot retrieve them.
Introduction
Although the majority of Parkinson’s disease cases are idiopathic, mutations in the LRRK2 gene are the most common cause of familial Parkinson’s disease (Simón-Sánchez et al., 2009; Alessi and Sammler, 2018). LRRK2 is a large, 2,527-residue, multidomain protein kinase that also contains a GTPase “ROC-COR” domain and other protein scaffolding domains. Pathogenic mutations in both the ROC-COR and kinase domains increase LRRK2’s basal kinase activity, which has made LRRK2 an attractive pharmaceutical target. The pathogenic R1441G mutation in the ROC GTPase domain shows enhanced GTP binding and increased kinase activity (Lewis et al., 2007; Ito et al., 2016); these features make it useful for understanding the consequences of LRRK2 phosphorylation on cell physiology that underlie Parkinson's disease. WT and R1441G LRRK2 are largely cytosolic but a pool (∼20%) associates with membranes (Berger et al., 2010; Purlyte et al., 2018) and colocalizes with Rab8 and Rab10 GTPases (Purlyte et al., 2018). Membrane-associated LRRK2 shows a higher level of kinase activity than cytosolic LRRK2 (Berger et al., 2010; Purlyte et al., 2018), but little is known about the molecular basis for membrane association or activation.
Recent phospho-proteomic analyses identified a subset of Rab GTPases as LRRK2 substrates (Steger et al., 2017), including Rab8, Rab10, and Rab29. Rab proteins are master regulators of all membrane trafficking events in eukaryotic cells (Wandinger-Ness and Zerial, 2014; Pfeffer, 2017). Rabs are membrane anchored by stable geranylgeranylation of their C-termini; when GTP bound, they recruit specific effector proteins to membrane surfaces to mediate transport vesicle formation, motility, and target recognition (Pfeffer, 2017). So-called switch regions report the identity of the Rab protein–bound nucleotide; they change conformation between GTP- and GDP-bound states (Stroupe and Brunger, 2000; Itzen and Goody, 2011). LRRK2 phosphorylates Rabs on their switch II regions, which interferes with binding to most of their binding partners, including GDI (Rab GDP dissociation inhibitor) that mediates Rab membrane delivery and extraction (Steger et al., 2016; Liu et al., 2018). Phosphorylated Rab8 and Rab10 (pRab10) proteins also bind with higher affinity to certain effectors, leading to their relocalization to the mother centriole (Dhekne et al., 2018; Madero-Pérez et al., 2018a).
The Rab29 gene is located within the PARK16 locus linked to Parkinson’s disease (Simón-Sánchez et al., 2009); however, it is much less clear how mutations in the PARK16 locus influence Parkinson’s disease. Rab29’s normal function and effector proteins remain elusive, although depletion of Rab29 has been suggested to alter Golgi structure, T cell receptor recycling, mannose 6-phosphate receptor recycling, and cilia formation (Wang et al., 2014; Onnis et al., 2015). Rab29 and LRRK2 double-knockout mice exhibit a nonadditive enlarged kidney phenotype, implying that these genes act in a common pathway (Kuwahara et al., 2016). Additionally, in Caenorhabditis elegans, Rab29 and LRRK2 orthologues function together in a signaling pathway controlling axon termination (Kuwahara et al., 2016).
Rab29 has recently been shown to activate pathogenic LRRK2 kinase activity and, at least when overexpressed, to recruit LRRK2 to the Golgi complex (Purlyte et al., 2018). In this study, we show that Rab29 is an unusual Rab protein in that it binds nucleotide poorly and is inefficiently prenylated in cells. Nevertheless, GTP binding by Rab29 and membrane association are required for its ability to trigger LRRK2 activation and substrate phosphorylation. Surprisingly, the identity of the membrane to which Rab29 or LRRK2 associates is not important for either of these processes.
Results
Rab29 has unique biochemical properties
To characterize Rab29-mediated activation of LRRK2, we sought to understand Rab29’s biochemical properties. Previous work suggested that Rab29 binds nucleotide more poorly than other Rab GTPases, based upon its rapid release of prebound, labeled nucleotides (Delprato et al., 2004; Beilina et al., 2014). In addition, the Q67L mutant that in other Rabs would be expected to lock the protein in an active conformation was not localized to the Golgi and bound nucleotide more weakly than Rab29 WT protein (Beilina et al., 2014). Additionally, a small pool of T21N Rab29 localizes to the Golgi (Wang et al., 2014; Madero-Pérez et al., 2018b). This suggested that canonical mutations cannot be used to study Rab29 and that additional mutagenesis and careful characterization would be required.
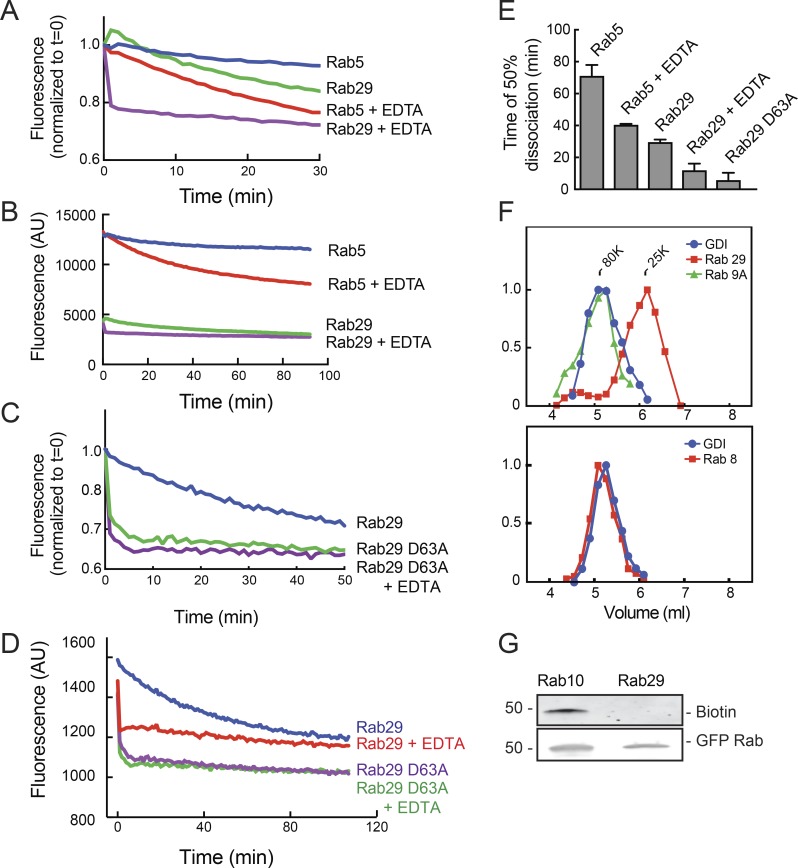
We used fluorescent 2′-(3′)-bis-O-(N-methylanthraniloyl)-GDP (MANT-GDP; Delprato et al., 2004) to monitor the dissociation of nucleotide from Rab29 and its mutant derivatives (Fig. 1, A–E). Purified Rab29 released MANT-GDP twofold faster than the well-characterized Rab5 GTPase (Fig. 1, A, B, and E). Upon addition of EDTA to chelate Mg2+, Rab29 released nucleotide instantaneously, much quicker than Rab5 protein (compare red and purple lines, Fig. 1, A and B).
Figure 1.
Rab29 binds nucleotide weakly and does not bind GDI in the cytosol. (A–D) Purified His-Rab5, His-Sumo Rab29, or His-Sumo Rab29 D63A was loaded with MANT-GDP, and the release of MANT-GDP fluorescence was followed after addition of an excess of unlabeled GDP. Shown are examples of experiments performed at least three times. A and C are normalized to time zero; B and D are unnormalized, and a longer time course is shown. (E) The curves from B and D were fitted to a nonlinear regression one-phase decay function. Times of 50% dissociation from three experiments performed in duplicate are shown. Error bars represent SEM. (F) Cytosol collected from 293T cells transfected with either Myc-Rab8A or Myc-Rab29 were applied onto an Sephacryl S100 size exclusion column and fractions immunoblotted using antibodies to GDI, Myc, or endogenous Rab9. (G) HEK392T cells were transfected with GFP-Rab10 or GFP-Rab29. Cell lysates were incubated with biotin-labeled geranyl pyrophosphate for 4 h at RT. GFP-Rabs were immunoprecipitated using beads to which GFP-binding nanobody was attached and immunoblotted for incorporated biotin geranyl pyrophosphate using streptavidin-IR800 and chicken anti-GFP antibody for total Rab protein. Shown is an example of an experiment performed three times. The molecular mass is shown at left in kilodaltons in this and all subsequent figures.
Rab29 D63A was created to generate a protein that binds nucleotide extremely poorly for experiments to test the importance of Rab29 nucleotide-binding capacity. The D63 residue is conserved throughout the Ras superfamily and coordinates a water molecule adjacent to the Mg2+ ion that holds the nucleotide in place. D63 is also involved in GDI binding (Rak et al., 2003). Although Rab29 D63A could be loaded with similar amounts of MANT-GDP compared with WT Rab29 (in reactions containing nucleotide at high concentrations; Fig. 1 D), it released the bound nucleotide very rapidly (Figs. 1, C and D), and EDTA had no additional effect, suggesting that this mutant binds magnesium poorly. Given that Rab29 D63A is entirely cytosolic upon expression in HEK293T cells (see below), it cannot be functional.
Cytosolic Rab29 is not GDI bound
Essentially all Rab GTPases studied to date exist in two pools: in cytosol or membrane bound. When present in cytosol, Rabs are complexed with GDI, which solubilizes these (mostly) doubly prenylated Rab GTPases (Araki et al., 1990). Thus, to date, gel filtration of cytosol has shown that Rabs chromatograph as an ∼80-kD complex with GDI (Soldati et al., 1993), as shown for Rab9A and Rab8A (Fig. 1 F). Rab29 is ∼35% membrane bound, which is comparable to other Rab GTPases (Overmeyer et al., 2001). Unexpectedly, unlike other Rabs, cytosolic Rab29 eluted after GDI as an ∼25-kD monomer upon S100 gel filtration of cytosol from HEK293T cells (Fig. 1 F, red line in top panel).
GDI binding depends on Rab GTPase prenylation; thus, it is possible that Rab29 fails to bind GDI in the cytosol because it is less efficiently prenylated compared with other Rab proteins. We compared the abilities of GFP-Rab29 or GFP-Rab10 to be prenylated in vitro using HEK293T cytosol and a biotinylated geranyl pyrophosphate precursor (Nguyen et al., 2009). As shown in Fig. 1 G, in vitro Rab29 prenylation was not detected under conditions in which Rab10 prenylation was readily observed. Although prenylation of endogenous Rab29 likely occurs in cells, Rab29’s weak nucleotide binding and inefficient prenylation could lead to decreased GDI binding, possibly explaining the pool of monomeric Rab29 in the cytosol.
Rab prenylation is needed for LRRK2 activation and substrate phosphorylation
At least 80% of LRRK2 is in the cytosol (Biskup et al., 2006; Berger et al., 2010; Purlyte et al., 2018); the remainder is on membranes. We showed previously that exogenously expressed Rab29 recruits cytosolic LRRK2 onto membranes, where it becomes preferentially activated (Purlyte et al., 2018). Given that cytosolic Rab29 is not bound to GDI, it is possible that it is available to activate LRRK2 in the cytosol. We thus first tested whether Rab29 prenylation is needed for LRRK2 activation by depleting cells of the prenylation precursor geranylgeranyl diphosphate by treatment with the HMG-CoA reductase inhibitor lovastatin. LRRK2 activation was then monitored using an anti-LRRK2 pS1292 antibody that detects activated LRRK2 protein.
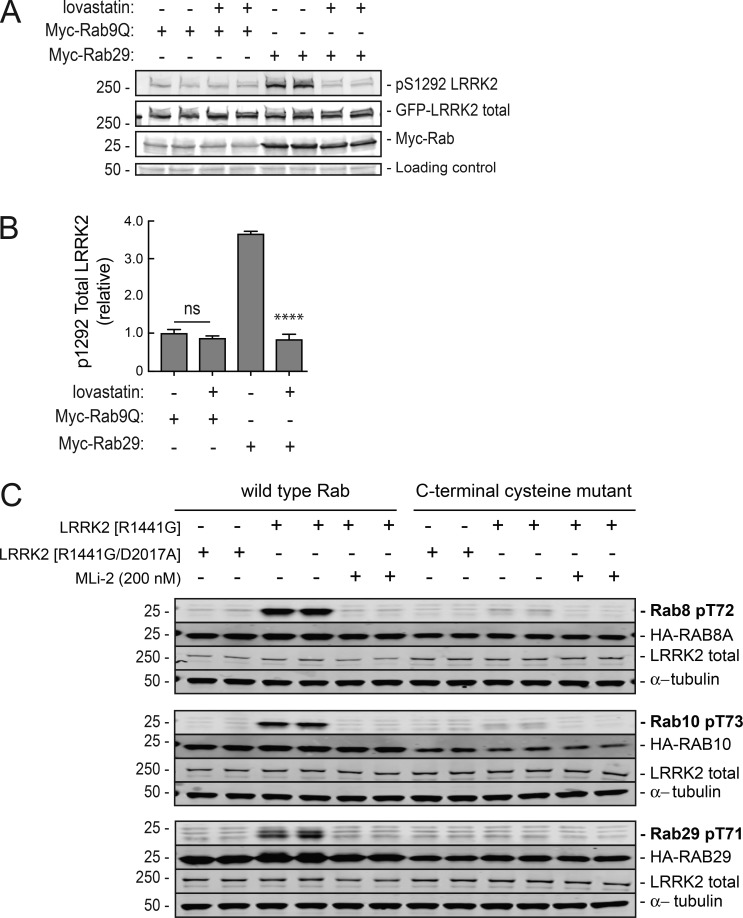
As shown in Fig. 2 (A and B), lovastatin treatment of Myc-Rab29– and R1441G LRRK2–transfected HEK293T cells strongly inhibited LRRK2 activation, as monitored by pS1292 antibody staining, consistent with a previous report (Liu et al., 2018). In Rab9-containing control reactions, pS1292 LRRK2 levels were unchanged by lovastatin treatment. Thus, despite the presence of both free Rab29 and LRRK2 in cytosol, Rab29 activation of LRRK2 is sensitive to lovastatin treatment. This finding is likely explained by a requirement for Rab29 prenylation and membrane association; however, lovastatin will also interfere with prenylation of other proteins, so such a conclusion would be premature.
Figure 2.
Membrane association enhances Rab29 activation of pathogenic LRRK2 and Rab phosphorylation. (A) HEK293T cells were transfected with R1441G LRRK2 and Myc-Rab29 or Myc-Rab9 and treated with either DMSO or 10 µM lovastatin to inhibit prenylation. Samples were immunoblotted 24 h later for pS1292 LRRK2, total LRRK2 (UDD3), and Myc. An ∼50-kD nonspecific band is shown at bottom as a loading control. (B) Quantitation of A. Error bars represent SEM from two experiments; P < 0.0001 from two-tailed unpaired t test. (C) HA-tagged Rab8A, Rab10, and Rab29 were coexpressed with FLAG-R1441G LRRK2 (±200 nM MLi-2, 1.5 h) or FLAG-R1441G/D2017A LRRK2 in HEK293T cells. Mutant Rab8A(C204S), Rab10(C199/200S), and Rab29(C202/203S) were used along with the corresponding WT Rabs. Samples were immunoblotted for the respective phosphorylated Rabs, HA, total LRRK2, and tubulin.
LRRK2 phosphorylation of its substrates, Rab8A, Rab10, and Rab29, also required that these Rab proteins gain membrane association by C-terminal prenylation, since deletion of their C-terminal cysteine residues that become prenyl modified strongly decreased their phosphorylation by LRRK2 kinase, as monitored using phospho-specific anti-Rab antibodies (Fig. 2 C). Important controls included LRRK2 R1441G/D2017A, a mutant LRRK2 that is defective in kinase activity (Jaleel et al., 2007), and cell treatment with MLi-2, a LRRK2-specific inhibitor (Scott et al., 2017) that also abolished Rab protein phosphorylation. Together, these data suggest that Rabs and LRRK2 only interact in association with membranes.
Membrane identity is not important for Rab29 activation of LRRK2
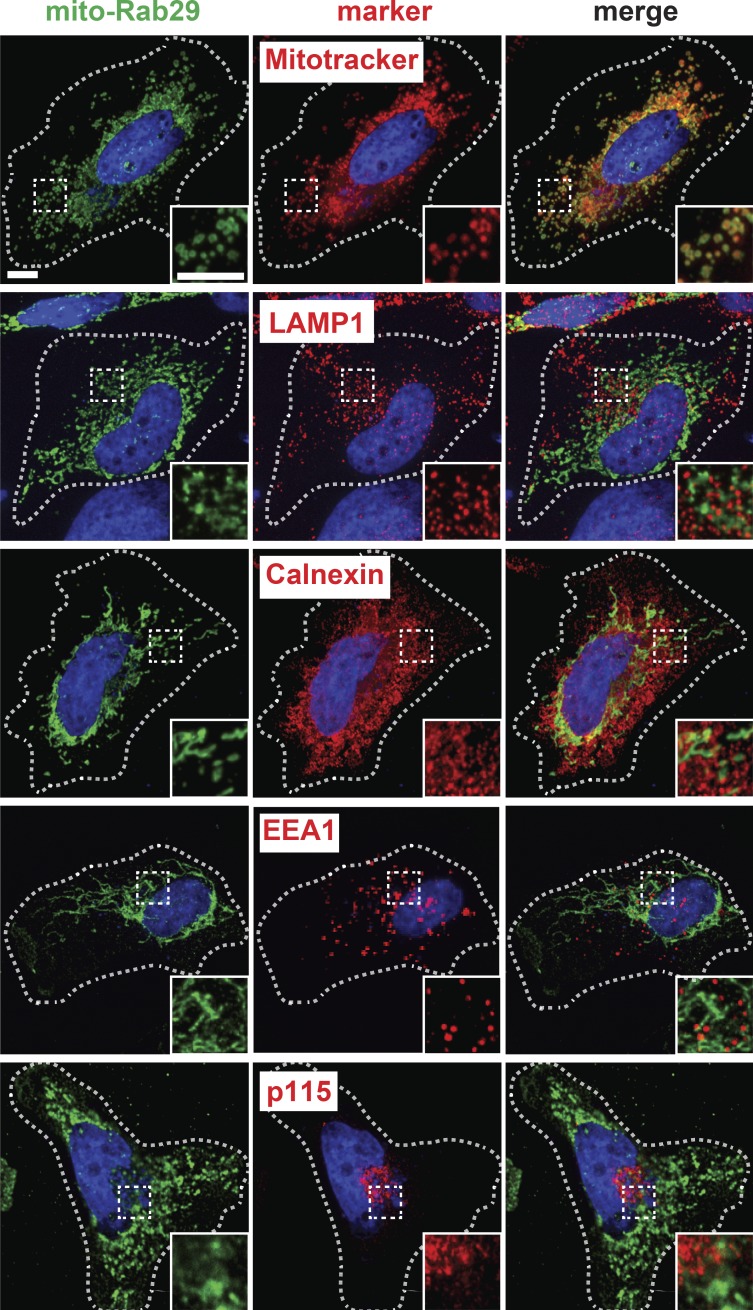
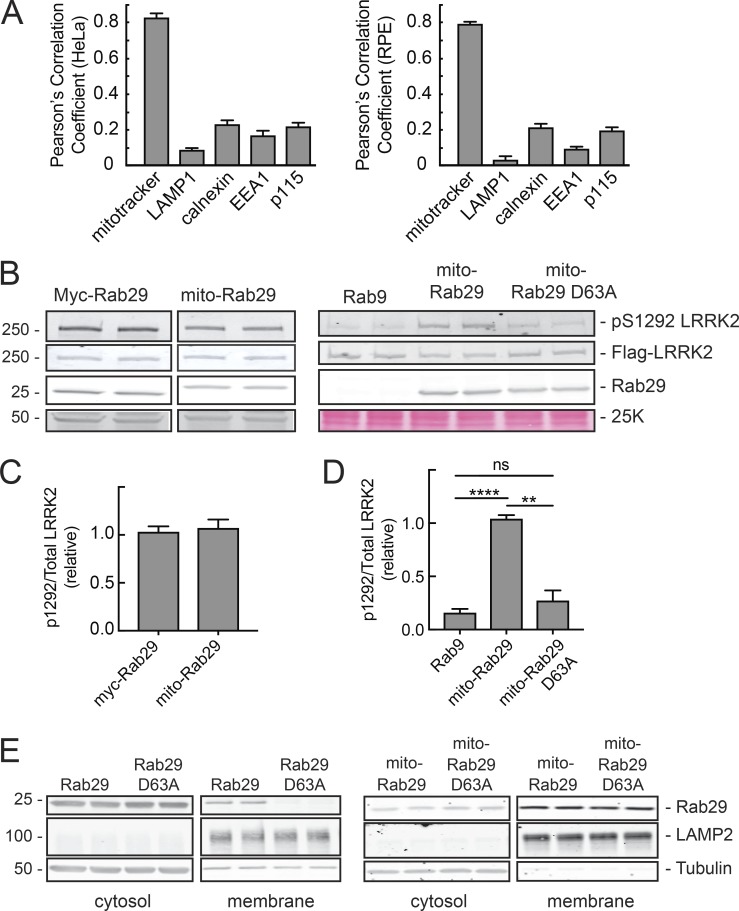
Because membrane association is required for both Rab29-mediated LRRK2 activation and LRRK2-mediated Rab substrate phosphorylation, we next tested whether the identity of the membrane housing the Rab is important. For these experiments, we first relocalized Rab29 to mitochondria by replacing Rab29’s two C-terminal cysteines with an amphipathic helix derived from monoamine oxidase A (Wong and Munro, 2014); this hybrid protein will be referred to as mito-Rab29. Upon expression in HeLa and retinal pigment epithelial (RPE) cells, mito-Rab29 localized predominantly to mitochondria, as monitored by colocalization with MitoTracker in both HeLa and RPE cells (Figs. 3 and 4 A). Mito-Rab29 did not colocalize with markers of lysosomes, the endoplasmic reticulum, early endosomes, or the Golgi apparatus (Figs. 3 and 4 A). Compared with Golgi-localized Myc-Rab29 (Purlyte et al., 2018), mito-Rab29 was comparable in its ability to activate LRRK2, as monitored by the generation of pS1292 LRRK2 (Fig. 4, B [top row, left panels] and C).
Figure 3.
Mitochondrially targeted Rab29 localizes to mitochondria but not other compartments. HeLa cells transfected with mito-Rab29 were stained with MitoTracker 488, anti-Myc, and anti-marker protein antibodies: LAMP1, Calnexin, EEA1, and Golgi protein p115. Scale bars, 10 µm. All insets represent a 2.2× magnification.
Figure 4.
Mitochondrially anchored Rab29-GTP activates LRRK2 as efficiently as Golgi-associated Rab29. (A) Quantitation of Fig. 3 for HeLa (left) and RPE cells (right). Pearson’s correlation coefficients calculated from ≥20 cells are shown. Error bars represent SEM. (B) HEK293T cells were transfected with LRRK2-R1441G and either Myc-Rab29 or mito-Rab29, Rab9, or mito-Rab29 D63A. Samples were immunoblotted for pS1292 LRRK2, total LRRK2 (UDD3), and Myc-tag after 24 h of expression. An ∼50-kD nonspecific band is shown at left as a loading control; a portion of the Ponceau S–stained filter (∼25 kD) is shown at bottom right as a loading control. (C) Quantitation of left panel of B. Amount of active (pS1292) LRRK2 normalized to the amount of total LRRK2 and Rab29. Error bars represent SEM from four independent experiments; P > 0.5 from two-tailed unpaired t test. (D) Quantitation of right panel of B. Amount of active, pS1292 LRRK2 normalized to the amount of total LRRK2 and Rab29. Error bars represent SEM from three experiments with duplicate samples in each; P values from unpaired t test: **, P = 0.0017; ****, P < 0.0001. (E) Immunoblot of membrane proteins and the equivalent volumes of cytosolic proteins from HEK293T cells transfected with Myc-Rab29 WT, Myc-Rab29 D63A, mito-Rab29 WT, or mito-Rab29 D63A. Blots were probed with anti-Myc antibodies for Myc-Rab29 or mito-Rab29 anti-LAMP2 as a membrane marker and anti-tubulin as a cytosolic marker.
Until now, there was no established Rab29 mutant that was certain to have lost its nucleotide-binding capacity. To explore the importance of bound nucleotide on Rab29’s ability to activate LRRK2 once present on membranes, we took advantage of the D63A Rab29 mutant. Upon expression as a Myc-tagged protein in cells, this protein failed to associate with membranes (Fig. 4 E, left panels), likely because it cannot interact with the prenylation machinery that requires nucleotide binding. However, the protein efficiently associated with membranes in its corresponding mitochondrial targeting form (Fig. 4 E, right panels).
Mito-Rab29 D63A failed to activate R1441G LRRK2 (Fig. 4, B [right panels] and D), which demonstrates the requirement for Rab29 nucleotide binding for LRRK2 activation. Importantly, these data show that the identity of the membrane upon which Rab29 sits does not influence its ability to activate LRRK2. Moreover, Rab29 must be both membrane and nucleotide bound to accomplish LRRK2 activation.
Membrane identity is not important for LRRK2 Rab substrate phosphorylation
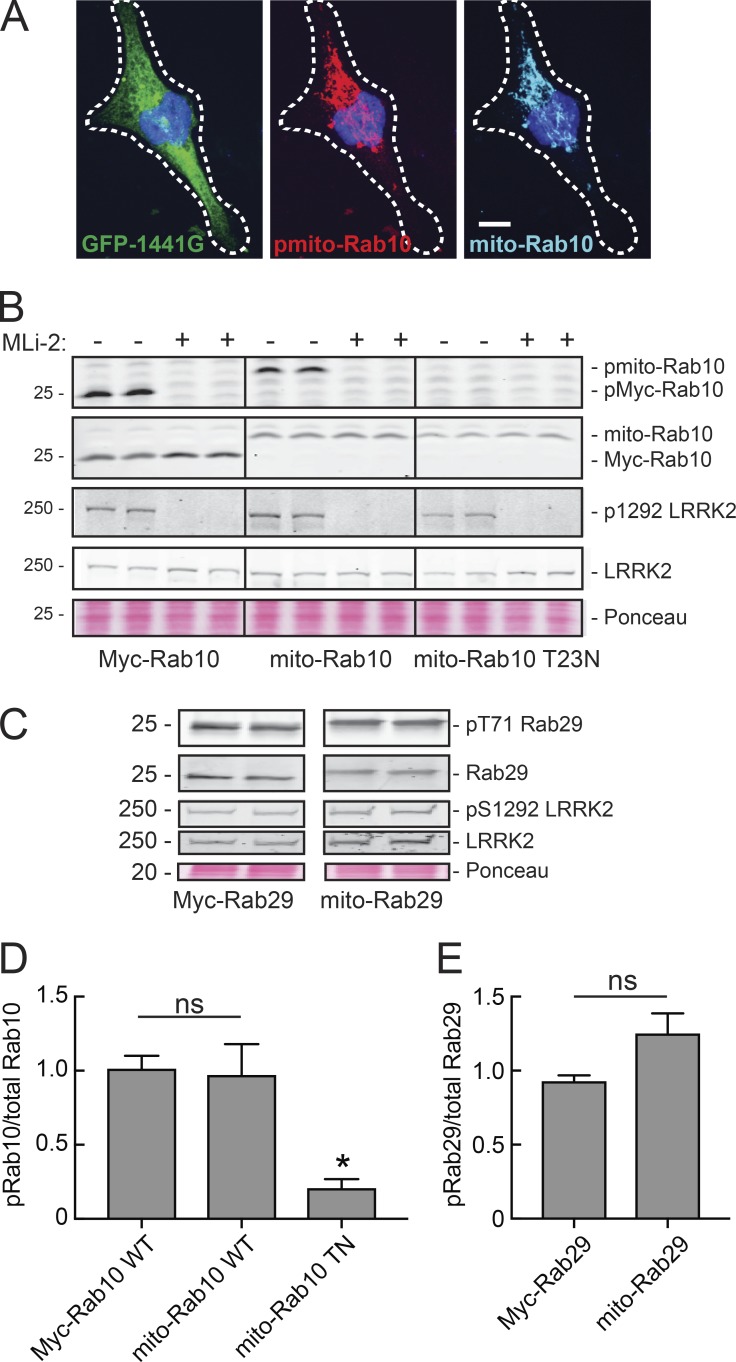
Rab10 colocalizes with R1441G LRRK2 on both perinuclear and peripheral membrane vesicles (Purlyte et al., 2018). We anchored Rab10 onto mitochondria to assess whether it is still a substrate for LRRK2 phosphorylation when localized to a nonnative membrane surface (Fig. 5 A). The protein was specifically localized to mitochondria, as determined by a Pearson’s correlation coefficient with the mitochondrial marker Mitofilin of 0.81 (20 cells measured). Although GFP-1441G LRRK2 was cytosolic, mito-Rab10 was phosphorylated on mitochondria (pRab10 shown in red, Fig. 5 A). Fig. 5 B (top row) shows the levels of pRab10 detected in cells expressing R1441G LRRK2 and Myc-Rab10 (left column), mito-Rab10 (center column), or GDP-preferring mito-Rab10 T23N (right column) in reactions with or without LRRK2 MLi-2 inhibitor. Even when relocalized to mitochondria, Rab10 was phosphorylated by LRRK2 to an extent similar to that seen for WT Myc-Rab10 protein (Fig. 5, B and D); GDP-preferring Rab10 T23N was not phosphorylated by LRRK2 on mitochondria. Thus, LRRK2 can phosphorylate membrane-associated Rab10 that is capable of efficient GTP binding, consistent with a recent report on Rab10 phosphorylation (Liu et al., 2018).
Figure 5.
Artificially anchored, nucleotide-bound Rab29 and Rab10 are substrates for pathogenic LRRK2. (A) Light microscopy of A549 Rab10 knockout cells transfected with GFP-R1441G LRRK2 (green) and mito-Rab10 (cyan). pRab10 is shown in red. Scale bar, 10 µm. (B) HEK293T cells transfected with GFP-R1441G LRRK2 and mito-Rab10, Myc-Rab10, or Myc-Rab29 T23N. Lysates were immunoblotted for pRab10, Myc-tag, pS1292 LRRK2, GFP for total LRRK2, and tubulin. A portion of the Ponceau S–stained filter (∼25 kD) is shown as a loading control. (C) HEK293T cells transfected with R1441G LRRK2, mito-Rab29, or Myc-Rab29 expressed for 24 h. Lysates were immunoblotted for pThr71 Rab29, GFP for total LRRK2, PS1292, and Myc-tag. A portion of the Ponceau S–stained filter (∼20 kD) is shown as a loading control. (D) Quantitation of B. Error bars represent SEM from three independent experiments. P values calculated from unpaired t test. *, P = 0.02 (two tailed). (E) Quantitation of C. pRab10 or Rab29 levels normalized to total Rab and p1292/total LRRK2 levels. In D and E, error bars represent SEM from three independent experiments with duplicate samples. Two-tailed P values were calculated using an unpaired t test.
Mito-Rab29 was also phosphorylated by R1441G LRRK2 (Fig. 5, C and E). This suggests that there are no integral membrane effector proteins on Rab10- or Rab29-native membranes that are needed for Rab phosphorylation by LRRK2. However, GTP binding is required for LRRK2-Rab substrate recognition.
LRRK2 activated by mito-Rab29 causes local Rab10 accumulation
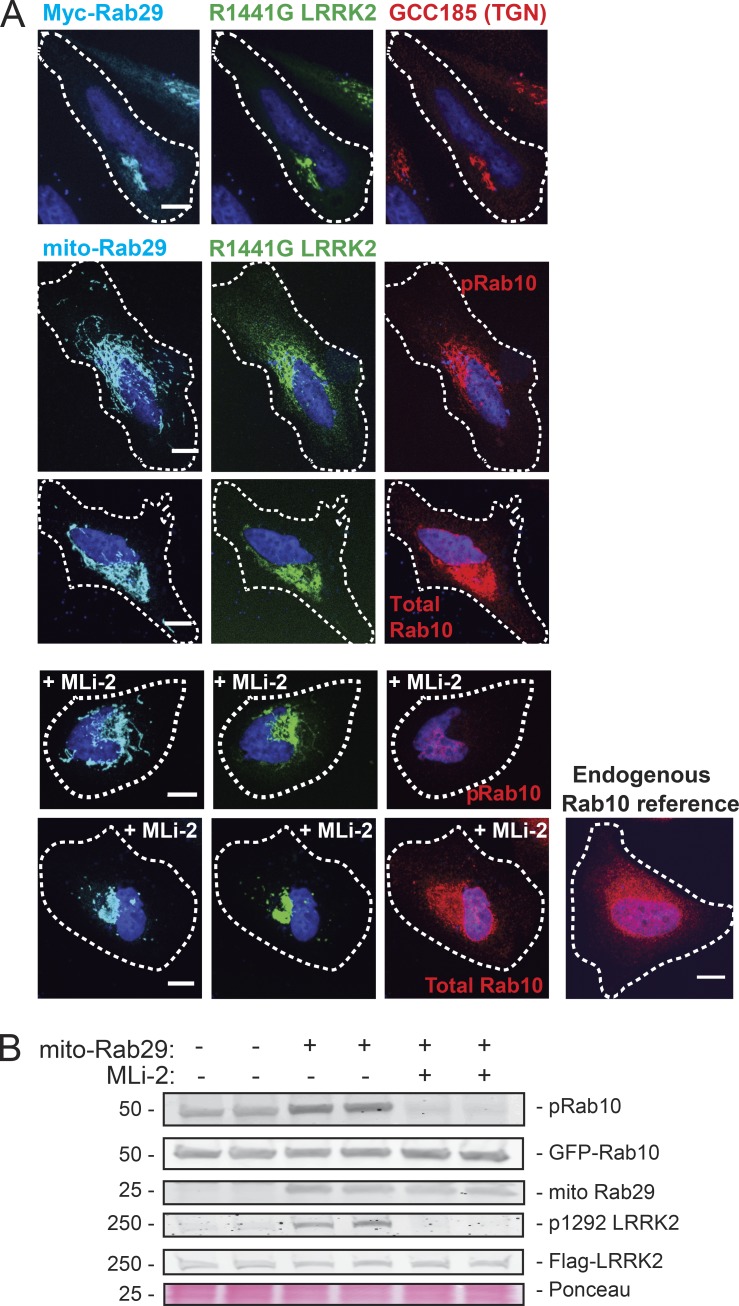
Because Rab29 can activate and recruit LRRK2 onto the Golgi complex (Purlyte et al., 2018) and mito-Rab29 can also activate LRRK2, as determined by immunoblot, we tested whether mito-Rab29 recruits LRRK2 onto mitochondria. In cells expressing both R1441G LRRK2 and mito-Rab29, LRRK2 was recruited onto mitochondria (Fig. 6 A). Quantitation of the colocalization of mito-Rab29 with R1441G LRRK2 by Pearson's correlation coefficient confirmed a high level of colocalization for the two proteins (Fig. 7 A).
Figure 6.
Mitochondrially anchored Rab29 recruits pathogenic LRRK2, pRab10, and total Rab10 to mitochondria. (A) Top row: Light microscopy of HeLa cells transfected with Myc-Rab29 (cyan) and GFP-R1441G LRRK2 (green); the TGN marker GCC185 is shown in red. Second row: Light microscopy of HeLa cells transfected with mito-Rab29 (cyan) and GFP-R1441G LRRK2 (green) expressed for 24 h. Endogenous pRab10 is shown in red. Third row: Mito-Rab29 (cyan) and GFP-R1441G LRRK2 (green); endogenous total Rab10 is shown in red. Fourth row: Mito-Rab29 (cyan) and GFP-R1441G LRRK2 (green). 24 h after transfection, cells were treated with 200 nM MLi-2 for 45 min, fixed, and stained. Endogenous pRab10 is shown in red. Fifth row: Mito-Rab29 (cyan) and GFP-R1441G LRRK2 (green); 24 h after transfection, cells were treated with 200 nM MLi-2 for 45 min, fixed, and stained. Endogenous total Rab10 is shown in red. Reference image of Rab10 without R1441G LRRK2 or mito-Rab29 expression is shown. Scale bars, 10 µm. (B) Immunoblot of HEK293T cells expressing mito-Rab29, 1441G LRRK2, and GFP-Rab10 for 24 h. Blot was probed for pRab10, Myc-tag, GFP, pS1292 LRRK2, and total LRRK2 (UDD3). MLi-2 treatment was 200 nM at the time of transfection. A portion of the Ponceau S–stained filter (∼25 kD) is shown as a loading control.
Figure 7.
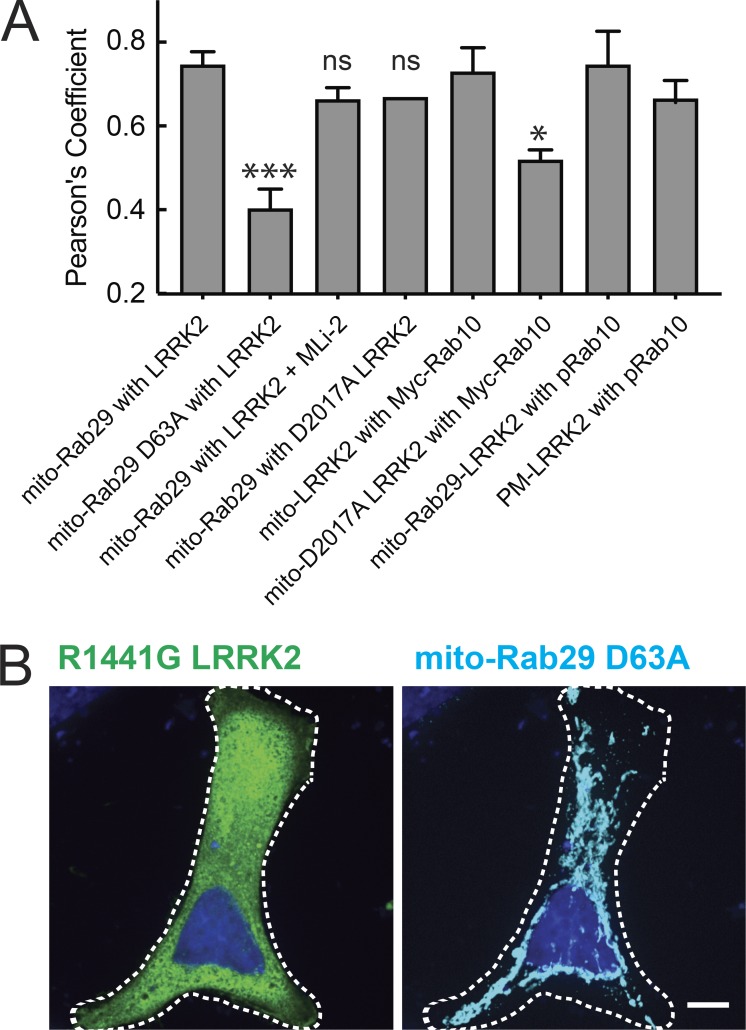
Mitochondrially anchored Rab29-GTP recruits LRRK2. (A) First four bars: Pearson’s correlation coefficients calculated from three independent experiments with ≥40 cells per condition between LRRK2 and Rab29 in HeLa cells expressing GFP-R1441G LRRK2 or D2017A LRRK2 and either mito-Rab29 WT or mito-Rab29 D63A after 24 h of expression with or without MLi-2. MLi-2 treatment was 200 nM at the time of transfection (first four bars). P values were calculated from unpaired t tests; *, P = 0.03; ***, P = 0.0007 (two tailed). Second four bars: Pearson’s correlation coefficients calculated from three independent experiments with ≥20 cells each; total Myc-Rab10 and mito-R1441G LRRK2 or mito-D2017A LRRK2 (fifth and sixth bars); LRRK2 recruited to mitochondria by mito-Rab29 and pRab10 (seventh bar); PM-LRRK2 and pRab10 (eighth bar). Error bars represent SEM from three experiments. (B) Light microscopy of a HeLa cell transfected with GFP-R1441G (green) and mito-Rab29 D63A (cyan). Scale bar, 10 µm.
We expected that LRRK2 recruited and activated on the surface of mitochondria would then release from that compartment to phosphorylate Rab10 on peripheral vesicles. To our surprise, when mito-Rab29 recruited LRRK2 onto mitochondria, pRab10 also relocalized to that compartment (Figs. 6 A and 7 A). This is surprising, as Rab proteins usually localize to membranes where they encounter their cognate guanine nucleotide exchange factors and are stabilized by subsequent, GTP-dependent effector binding (Aivazian et al., 2006; Barr, 2013). We hypothesize that GDI can be promiscuous in its delivery of Rabs to membranes (Barr, 2013); because Rab phosphorylation blocks interaction with GDI (Steger et al., 2016), a mistakenly delivered Rab on a mitochondrial surface will become trapped there by a mechanism that depends on LRRK2 kinase activity.
A prediction of this model is that Rab10 should be retrieved from mitochondria by GDI upon LRRK2 inhibition with MLi-2. Control experiments showed that phosphorylation of Rab10 trapped on mitochondria was reversed upon MLi-2 treatment (Fig. 6 B). Under these conditions, mito-Rab29 and LRRK2 retained their mitochondrial localizations, while total Rab10 reverted to a more general perinuclear staining (Fig. 6 A, bottom two rows). Loss of Rab10 accumulation on mitochondria was seen in as little as 15 min of inhibitor addition (45 min shown; Fig. 6 A, bottom panel), supporting the conclusion that this accumulation is phosphorylation dependent. Thus, when driven onto mitochondria by Rab29 recruitment and activation, most LRRK2 traps substrates at that location in an activity-dependent manner.
Active Rab29 recruits LRRK2
Just as active Rab29 was needed to activate LRRK2 when anchored on mitochondrial membranes (Fig. 4, B and D), active Rab29 was also more capable of recruiting cytosolic R1441G LRRK2 to that compartment, despite similar levels of protein expression of the WT and mutant Rab proteins (Figs. 4 E, 6 A, and 7). Fig. 7 B shows cytosolic localization of LRRK2 upon coexpression with mito-Rab29 D63A; this is in stark contrast with the complete relocalization of LRRK2 to mitochondria when coexpressed with mito-Rab29 WT (Fig. 6 A). Mito-Rab29 (WT) recruited LRRK2 to mitochondria with a Pearson’s correlation coefficient of >0.7, while mito-Rab29 D63A and LRRK2 had a Pearson’s correlation coefficient of 0.4 (Fig. 7 A). Thus, recruitment and activation by Rab29 are linked and both depend on Rab29’s ability to bind GTP.
We also tested whether LRRK2 recruitment was dependent on LRRK2 kinase activity. HeLa cells were transfected with mito-Rab29 and either R1441G LRRK2 or D2017A (kinase-dead) LRRK2. The Pearson’s correlation coefficient between LRRK2 and mito-Rab29 was unchanged when kinase-dead D2017A LRRK2 was used or when cells were treated with MLi-2 (Fig. 7 A). This indicates that the Rab29-LRRK2 interaction is insensitive to any conformational changes associated with LRRK2 activity changes.
A test of the importance of LRRK2 membrane–cytosol equilibrium
While trans-Golgi–localized Rab29 activates LRRK2 and recruits it to the Golgi, most of LRRK2’s primary substrates, Rab8A, Rab10, and Rab12, show broader distribution across the Golgi stack and also elsewhere (compare Figs. 6 A and 8 A; Purlyte et al., 2018). One possibility is that activated LRRK2 releases from the trans-Golgi to phosphorylate Rabs on nearby compartments. To explore this, we anchored GFP-LRRK2 on mitochondria using a mitochondrially anchored, camelid GFP-nanobody (GBP) that displays an ∼1-nM Kd for GFP (Kubala et al., 2010).
Figure 8.
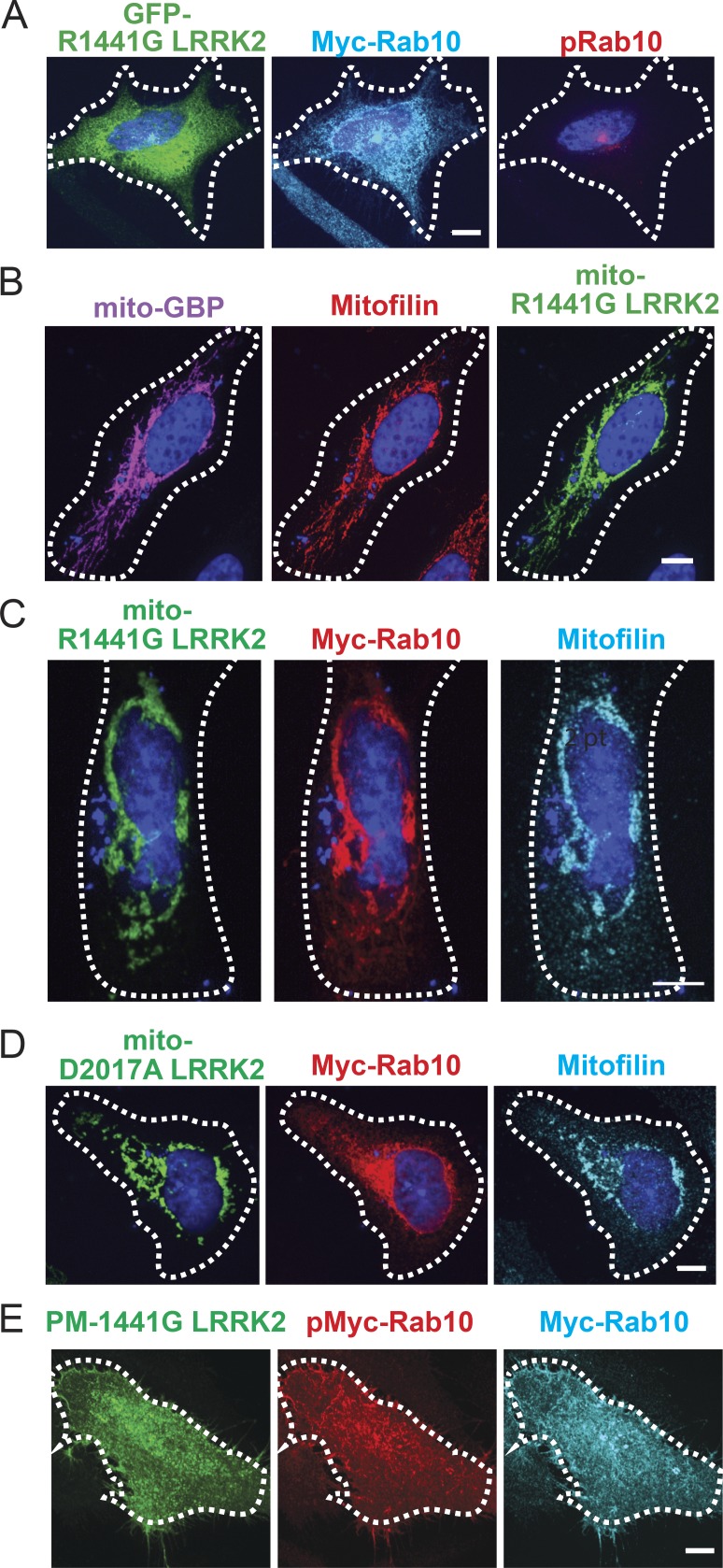
Mislocalized LRRK2 phosphorylates and traps pRab10. (A) Light microscopy of an A549 Rab10 knockout cell expressing untagged GBP, GFP-R1441G LRRK2 (green), and Myc-Rab10 (cyan). pRab10 is shown in red. (B) Light microscopy of a HeLa cell stained for expressed mito-GBP (magenta) and “mito” GFP-R1441G LRRK2 (green). Mitofilin staining shown in red. (C) Light microscopy of a HeLa cell expressing mito-GBP, GFP-R1441G LRRK2 (green), and Myc-Rab10 (red). Mitofilin is shown in cyan. (D) Light microscopy of a HeLa cell expressing mito-GBP, GFP-D2017A LRRK2 (green), and Myc-Rab10 (red). Mitofilin is shown in cyan. (E) Light microscopy of a HeLa cell expressing PM-GBP, GFP-R1441G LRRK2 (green), and Myc-Rab10 (cyan); pMyc-Rab10 shown in red. The entire Z-stack series of GFP in this cell is shown in Video 1. Scale bars, 10 µm.
As shown in Fig. 8 B, mito-GBP was accurately targeted to mitochondria, as determined by its colocalization with the protein Mitofilin (Fig. 8 B). In addition, expression of mito-GBP relocalized GFP-R1441G LRRK2 to the surface of mitochondria (Pearson’s correlation coefficient = 0.82 for 20 cells); we will refer to GBP-bound, mitochondrially localized GFP-R1441G LRRK2 as mito-R1441G LRRK2.
To test whether mito-LRRK2 can phosphorylate Rab10, we expressed either soluble Myc-GBP or mito-GBP and GFP-R1441G LRRK2 with Myc-Rab10 and stained cells for pRab10 in A549 Rab10 knockout cells (Dhekne et al., 2018; Figs. 8, A and C). When LRRK2 was soluble in cells expressing Myc-GBP, Myc-Rab10 was seen on perinuclear membranes, and pRab10 accumulated over the mother centriole as a tight spot in the perinuclear region (Fig. 8 A), as we showed previously (Dhekne et al., 2018). When mito-GBP was used to anchor LRRK2 on mitochondria, total Myc-Rab10 accumulated on mitochondria (Fig. 8 C and quantitation in Fig. 7 A), analogous to what was observed in cells expressing mito-Rab29 to recruit LRRK2 (Fig. 6 A). Recruitment of Rab10 to mitochondria by mito-LRRK2 was phosphorylation dependent; kinase-dead mito-D2017A LRRK2 did not recruit significant amounts of Rab10 onto mitochondria (Figs. 8 D and quantitation in Fig. 7 A).
As an independent test of R1441G LRRK2’s ability to trap Rab10 on a membrane surface, we also anchored LRRK2 to the inner leaflet of the plasma membrane using GBP containing the C-terminal polybasic region that targets K-Ras to the plasma membrane (Apolloni et al., 2000). We refer to this construct as PM-LRRK2 (Video 1). PM-LRRK2 relocalized pRab10 and total Rab10 to the plasma membrane (Fig. 8 E and quantitation in Fig. 7 A). These data provide further evidence that LRRK2 can drive the relocalization and accumulation of Rab10 to inappropriate membranes via phosphorylation.
Mito-LRRK2 rarely phosphorylates plasma membrane–targeted Rab10
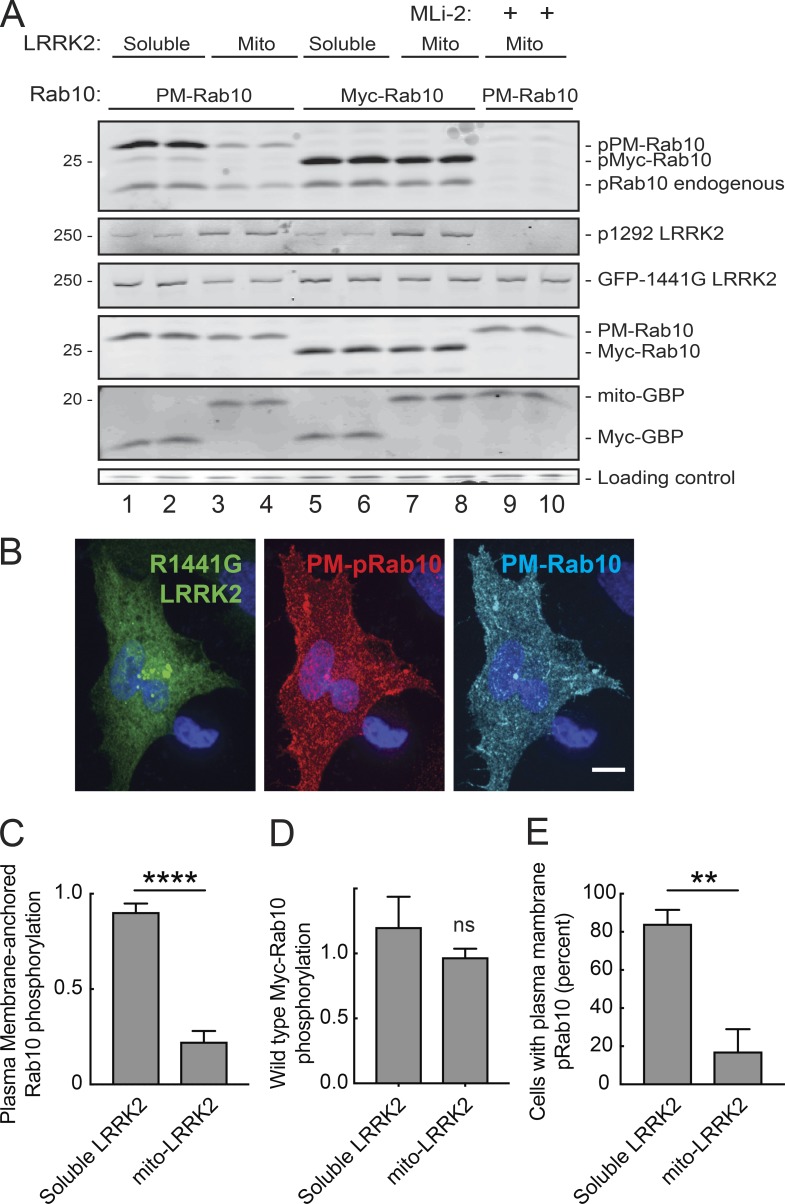
In these experiments thus far, it was possible for Rab10 to find LRRK2 on mitochondria or the plasma membrane (via GDI; compare Figs. 6 A, 8 C, and 9) or cytoplasmic LRRK2 to find Rabs on their native (or foreign) membrane compartments. Can an anchored LRRK2 phosphorylate a Rab anchored on another membrane surface? To study this, we anchored both LRRK2 and Rab10 on different membranes and monitored subsequent, LRRK2-mediated Rab10 phosphorylation. For these experiments, Rab10 was anchored onto the plasma membrane by replacing its C-terminal cysteines with the polybasic C-terminus of K-Ras. This construct is referred to here as PM-Rab10.
Figure 9.
Soluble LRRK2 phosphorylates plasma membrane–targeted Rab10 more efficiently than mitochondrially anchored LRRK2. (A) Representative immunoblot of HeLa cells transfected with either soluble GFP-R1441G LRRK2 (with soluble Myc-GBP; lanes 1, 2, 5, and 6) or mitochondrially anchored R1441G-LRRK2 (with mito-GBP; lanes 3, 4, and 7–10) and Myc-Rab10 (lanes 5–8) or PM-Rab10 (lanes 1–4, 9, and 10). One set of samples was treated with 200 nM Mli-2 at the time of transfection (lanes 9 and 10). Blots were probed for pRab10, pS1292 LRRK2, GFP, and Myc-tag. An ∼37-kD nonspecific band is shown below as a loading control. (B) Light microscopy of an A549 Rab10 knockout cell expressing GFP-R1441G LRRK2 (green), Myc-GBP, and PM-Rab10 (cyan). pRab10 shown in red. See also Video 2. Scale bar, 10 µm. (C) Quantitation of data shown in A. Ratio of pMyc-Rab10/total Myc-Rab10 signal normalized to the amount of total LRRK2 in the sample. Error bars represent SEM from four independent experiments. ****, P < 0.0001, unpaired t test two-tailed. (D) Quantitation of A. Ratio of phosphorylated PM-Rab10/total PM-Rab10 signal normalized to total LRRK2. Error bars represent SEM from three independent experiments. Unpaired t test two-tailed P value = ns (E) Quantitation of the percentage of A549 Rab10 knockout cells transfected with GFP-R1441G LRRK2, Myc- or mito-GBP, and phosphorylated PM-Rab10 positive. Scored based on presence or absence of phosphorylated PM-Rab10. Error bars represent SEM from four independent experiments with >30 cells scored per experiment; **, P = 0.0029, two-tailed unpaired t test.
A significant fraction of PM-Rab10 accurately localized to the cytoplasmic surface of the plasma membrane of A549 Rab10 knockout cells (Fig. 9 B and Video 2). As shown in Fig. 9 A (lanes 1–4, top row), soluble LRRK2 was much more effective at phosphorylating plasma membrane–anchored Rab10 than LRRK2 anchored to mitochondria (Fig. 9, A and C). Upon coexpression with GFP-R1441G LRRK2 and the soluble control protein Myc-GBP, PM-Rab10 was phosphorylated and pRab10 localized to the plasma membrane in ∼80% of cells (Fig. 9, B and E). Upon coexpression with mito-LRRK2 instead of soluble LRRK2, PM-Rab10 phosphorylation was detected in <20% of cells (Fig. 9 E). It is possible that a small amount of phosphorylated PM-Rab10 was modified before it reached the plasma membrane. (Note that this protein could not use GDI for membrane delivery because it is not prenylated.) Collectively, these data support the conclusion that the ability of LRRK2 to move between membranes and the cytosol is crucial for efficient Rab phosphorylation. Rab phosphorylation by immobilized LRRK2 relies on misdelivery of Rab10 to mitochondria; a much lower percentage of phosphorylation seen with anchored enzyme and substrate might reflect access via membrane contact sites between mitochondria and the plasma membrane.
Finally, Fig. 9 A (second row) shows the relative activity of soluble R1441G LRRK2 (cells expressing Myc-GBP) compared with mitochondrially anchored R1441G LRRK2 in cells expressing mito-GBP. Although total LRRK2 expression levels were comparable (Fig. 9 A, third row), LRRK2 was more highly autophosphorylated upon mitochondrial membrane anchoring, as detected by increased anti-pS1292 staining (Fig. 9 A, second row). Despite higher autophosphorylation in these samples, Myc-Rab10 substrate phosphorylation was comparable when catalyzed by either soluble or mitochondrially localized LRRK2 protein (Fig. 9, A [lanes 5–8, top row] and D). It is possible that membrane anchoring concentrates the kinase in a mode that self activates without necessarily providing adequate substrate access to achieve greater levels of substrate phosphorylation. Future work will be needed to explore the basis of LRRK2 activation on membrane surfaces.
Discussion
We have presented here data indicating that LRRK2 is optimally able to access membrane-associated Rab substrates from a cytosolic pool. A key experiment involved anchoring LRRK2 on mitochondria and Rab10 at the plasma membrane; soluble or plasma membrane–anchored LRRK2 had no problem phosphorylating WT Rab10, but mitochondrially anchored LRRK2 was much less able to phosphorylate plasma membrane–anchored Rab10. Mitochondria-associated LRRK2 was nevertheless active; when it was recruited onto mitochondria by mitochondrially anchored Rab29, it could be activated there for both auto- and substrate phosphorylation; it was also active when anchored to mitochondria without exogenous Rab29
Because of its ability to bind and activate pathogenic LRRK2, Rab29 is an important Rab protein to understand in relation to the molecular basis of familial Parkinson’s disease (MacLeod et al., 2013; Beilina et al., 2014; Purlyte et al., 2018). Our data confirm that Rab29 is an unusual Rab; it binds nucleotide poorly, appears to be less efficiently prenylated, and is not bound to GDI in cytosol. Rab29 is most closely related to the proteins Rab32 and Rab38, and there are no obvious sequence differences that would suggest that it would behave differently from other Rab proteins; its structure has not yet been determined. Future studies of Rab29 must be designed with great care, as canonical Rab29 mutations such as Q67L (Beilina et al., 2014) do not function as would be expected (locked in the active conformation) and should not be used (this protein binds nucleotide more poorly than WT). Additionally, Rab29 D63A can serve in future experiments as an appropriate, inactive Rab29 control protein because it is entirely cytosolic and displays nucleotide binding that is EDTA insensitive. Note that a small fraction of this mutant may bind GTP in cells where the GTP concentration approaches 0.5 mM.
We have shown that Rabs are phosphorylated on membranes and their prenylation is required for LRRK2 modification, confirming and expanding a previous report (Liu et al., 2018). Rab8, Rab10, and Rab29 all require prenylation-conferred membrane localization to be phosphorylated by LRRK2, and Rab29 needs to be membrane localized to activate LRRK2. If Rab29 is not GDI bound in cytosol, then why is it unable to activate the LRRK2 that is mostly cytosolic? It is possible that membrane association confers a local concentration effect and increases the local Rab concentration on a two-dimensional surface; membrane association may also influence LRRK2 conformation.
For the first time, we have been able to dissociate the importance of Rab nucleotide binding from the process of membrane association. When the Rab10 GDP-preferring mutant Rab10 T23N is membrane anchored, it is not a substrate for LRRK2 kinase. Additionally, when Rab29 is membrane anchored but unable to bind nucleotide (D63A mutant), it does not activate LRRK2. These data demonstrate that LRRK2 binds Rab29 as a bona fide Rab effector in cells, and for Rab substrates, GTP binding likely orders the switch II domain for efficient recognition by LRRK2.
Our ability to relocalize LRRK2 activation and substrate phosphorylation to the mitochondrial surface or plasma membrane inner leaflet demonstrates that the identity of the membrane supporting this reaction does not matter; there are no integral membrane Golgi proteins required for Rab29 activation of LRRK2 kinase. Similarly, Rab10 is also phosphorylated on mitochondria or the plasma membrane, suggesting that LRRK2 does not require an activating or binding protein on endogenous Rab10-containing membranes. We cannot rule out the possibility that artificially anchored Rab29 or Rab10 recruits a soluble binding partner to the new compartment that enhances activation. In vitro reconstitution of Rab29 activation will clarify this issue. Another remaining mystery is why LRRK2 is activated upon membrane association, even independent of Rab29 binding.
One perhaps surprising finding in this study is that pRab10 accumulates at the site of LRRK2 activation; when mitochondrially anchored Rab29 recruits LRRK2, both total and pRab10 accumulate on the surface of those mitochondria, an abnormal localization for Rab10. Similarly, plasma membrane–anchored LRRK2 led to the accumulation of pRab10 at that location. Our experimental setup traps mislocalized Rabs at the site of phosphorylation, because this modification blocks their ability to interact with GDI protein (Steger et al., 2016). Normally, GDI delivers GDP-bound Rabs to membranes; they become stabilized on specific membranes upon encountering their cognate guanine nucleotide exchange factors to convert them to their GTP-bound forms, enabling subsequent effector binding (Barr, 2013). When delivered to the wrong membrane, GDP-Rabs are usually removed by GDI, which recognizes GDP-bound Rab proteins. When LRRK2 phosphorylates mislocalized Rab proteins, GDI can no longer bind and relocalize them. Note that some of our assays used overexpressed proteins, which may have a higher tendency for mislocalization if endogenous effector interactions become saturated; nevertheless, we did detect relocalization of endogenous Rab10 protein (Fig. 6). Mislocalization of Rab10 to mitochondria was due to LRRK2 action, because it was reversed upon addition of the LRRK2 kinase inhibitor MLi-2. Additionally, kinase-dead mito-D2017A LRRK2 did not relocalize Rab10 to mitochondria (Fig. 8), supporting our contention that accumulation is phosphorylation dependent. A yet-to-be-identified phosphatase must act on pRab10 to permit subsequent Rab10 extraction by GDI. These experiments reveal a significant level of GDI-Rab mistargeting that has been previously underappreciated.
The large accumulation of pRab10 and total Rab10 on LRRK2-containing membrane surfaces suggests that Rab phosphorylation (and subsequent membrane trapping) could have large consequences on membrane trafficking events, as shown recently for pRab8A (Rivero-Ríos et al., 2019). Upon LRRK2 phosphorylation, native Rabs will be unable to be recognized by GDI protein, thereby interfering with their retrieval and delivery to donor membranes. Thus, in addition to losing the capacity to bind effector proteins, LRRK2 Rab phosphorylation is also predicted to interfere with the GDI retrieval process needed for optimal Rab GTPase functioning (Steger et al., 2016).
We have shown that LRRK2 associates preferentially with GTP-bearing Rab29 in cells, apparently independent of LRRK2 kinase activity. This suggests that Rab29 binds to a portion of the protein that is insensitive to conformational changes associated with kinase activation. This is a somewhat surprising finding, as Rab29 is better able to activate hyperactive, pathogenic LRRK2 compared with WT LRRK2 protein. It is possible that under the conditions of exogenous protein expression, our experiments were not able to distinguish more subtle affinity differences. Future work will identify more precisely the Rab29-binding site on LRRK2 protein.
Finally, if Rab29 recruits LRRK2 to the Golgi, then why would that not be sufficient to trap pRab10 at the Golgi under normal conditions? It is important to note that most LRRK2 is not present on the Golgi unless Rab29 is overexpressed; Rab29 is normally a poorly abundant protein in most cultured cells, and although Rab29 can activate LRRK2 at the Golgi, pathogenic LRRK2 mutants are nevertheless active in the absence of Rab29. All of these features indicate that there is still much to be learned about LRRK2 cell biology and the significance of Rab29 activation in vivo.
In summary, active LRRK2 is a bona fide Rab29 effector that recognizes GTP-bound Rab GTPase substrates and phosphorylates them in the context of native and artificial membrane surfaces. Beyond LRRK2 activation, Rab29’s additional physiological roles remain poorly characterized and warrant further analysis. At least under conditions of LRRK2 overexpression, Rab phosphorylation can drive Rab accumulation at the site of phosphorylation, which could lead to broad trafficking defects. Greater understanding of the activities of phosphorylated Rabs will add much to our understanding of the etiology of Parkinson’s disease.
Materials and methods
General cloning and plasmids
DNA constructs were amplified in Escherichia coli DH5α and purified using mini spin columns (Econospin). All sequences were verified by sequencing (http://www.sequetech.com). Plasmids encoding His-Sumo-Rab29 (DU50295), HA-Rab 8A (DU35414), HA-Rab10 (DU44250), and HA-Rab29 (DU5022) were obtained from the Medical Research Council unit at Dundee, and GBP was obtained from Addgene (49172). Myc-Rab 9Q, Myc-Rab29, Myc-Rab10, and GFP-binding protein were cloned into pcDNA 3.1 with an N-terminal 3X-Myc tag between the BamHI and NotI sites. Rab9 was cloned into pcDNA 3.1 lacking an N-terminal 3X-Myc tag. Rab29 and Rab10 mutants were generated by site-directed mutagenesis using Pfu Turbo DNA polymerase (Agilent Technologies). A mitochondrial targeting tag was added to the C-termini of Myc-Rab29 (residues 1–201), Myc-Rab10 (residues 1–198), and Myc-GBP following a 5′-GAGAGAAA-3′ linker between the NotI and ApaI sites. Mito-Rab29 primers were used to amplify Rab29 with the linker (5′-ATTATGGATCCATGGGCAGCCGCGAC-3′, 5′-ATAATGCGGCCGCACCGGCGCCAGCACCGGACCAGCTGGAGGACTT-3′), or to amplify the mitochondrial targeting tag (5′-ATTATGCGGCCGCCTTCTGGGAAAGGAACCTGC-3′, 5′-ATAATGGGCCCTCAAGACCGTGGCAGGAG-3′). The mitochondrial targeting tag sequence was composed of human monoamine oxygenase A amino acids 490–527. Plasma membrane–targeted Rab10 contained K-Ras residues 178–188 appended onto the C-terminus of Rab10 residues 1–198 and GBP using the NotI and ApaI restriction sites in pcDNA 3.1. Primers to amplify K-ras tail included 5′-ATTATGCGGCCGCCAAAGATGGTAAAAAGAAG-3′ and 5′-ATAATGGGCCCTTACATAATTACACACTTTGT-3′. C-terminal EGFP-LRRK2-R1441G was cloned into modified pSLQ1371 with EGFP at the N-terminus. All constructs are for human genes.
Cell culture
HEK293T, A549, RPE, and HeLa cells (ATCC) were cultured at 37°C and under 5% CO2 in Dulbecco’s modified Eagle’s medium containing 8% fetal bovine serum, 2 mM Glutamine, and penicillin (100 U/ml)/streptomycin (100 µg/ml). HEK293T and HeLa cells were transfected with polyethylenimine HCl MAX 4000 (Polysciences). A549 and RPE cells were transfected using lipofectamine 3000 according to the manufacturer’s instructions.
Immunoblotting
SDS-PAGE gels were transferred onto nitrocellulose membranes using a Bio-Rad Trans-turbo blot system. Membranes were blocked with either 5% skim milk or 3% BSA in Tris-buffered saline with Tween-20 for 30 or 60 min at RT, respectively. Primary antibodies used diluted in blocking buffer were rabbit anti-GDI (Soldati et al., 1993) 1:1,000, mouse anti-Rab9 (Lombardi et al., 1993) 1:1,000, mouse anti-Myc (9E10 Hybridoma culture supernatant undiluted), 1.2 µg/ml rabbit anti-pT73Rab10 (230261; Abcam), rabbit anti-pT71 Rab29 (ab241062; Abcam) 1:500, rabbit anti-phospho-Ser1292 LRRK2 (ab203181; Abcam) 1:1,000, chicken anti-GFP (GFP-1010; Aves) 1:1,000, rabbit anti-LRRK2 (ab133518; Abcam), rabbit antitubulin (11224-1-AP; Proteintech) 1:2,000, and mouse anti-LAMP2 culture supernatant (H4B4; Developmental Studies Hybridoma Bank). Primary antibody incubations were either 1 h at RT or overnight at 4°C. LI-COR secondary antibodies diluted in blocking buffer used were 680 nm donkey anti-mouse (1:10,000), 800 nm donkey anti-mouse (1:5,000), 680 nm donkey anti-rabbit (1:10,000), 800 nm donkey anti-rabbit (1:5,000), 680 nm donkey anti-chicken (1:10,000), and 800 nm streptavidin (1:5,000). Secondary antibody incubations were for 1 h at RT. Blots were imaged using an Odyssey Infrared scanner (LI-COR) and quantified using ImageJ software.
Membrane fractionation and gel filtration chromatography
HEK293T cells were transfected with Myc-Rab29 or Myc-Rab8 plasmids. 24 h after transfection, cells were chilled, washed, resuspended in PBS, and then pelleted at 1,000 rpm for 5 min in a swinging bucket rotor. Cell pellets were resuspended in hypotonic buffer (10 mM Hepes, pH 7.4) and incubated on ice for 15 min. Buffer (5×) was added to achieve a final concentration of 1× resuspension buffer (50 mM Hepes, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 0.5 mM DTT, and 100 nM GDP and protease inhibitor cocktail; Sigma). The suspension was passed 20 times through a 25G syringe. Nuclei were pelleted by centrifugation at 1,000 ×g for 5 min at 4°C. The post-nuclear supernatant was spun at 100,000 ×g for 15 min in a TLA100.2 rotor in a table-top ultracentrifuge. The resulting cytosolic supernatant was removed, and the membrane pellet was solubilized in resuspension buffer containing 0.5% Triton X-100. Protein was determined by Bradford assay (Bio-Rad). Cytosol was snap frozen in liquid nitrogen and stored at −80°C. Thawed cytosol was applied onto a 10-ml Sephacryl S100 column (GE Healthcare) equilibrated in buffer containing 50 mM Hepes, pH 7.4, 100 mM NaCl, 5 mM MgCl2, 0.2 mM DTT, 10 µM GDP, and 10 mM PMSF. Fractions were collected and weighed for volume, and protein was determined. Fractions were analyzed by immunoblotting as described above.
Determination of LRRK2 activation by Rab29
HEK293T cells were transfected with indicated plasmids; 24 h after transfection, cells were chilled, washed, and resuspended in PBS. Where applicable, cells were treated with 10 µM lovastatin (Abcam) at the time of transfection. Cell pellets were resuspended in lysis buffer (50 mM Hepes, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 0.5 mM DTT, and 100 nM GTP, protease inhibitor cocktail, 0.1 µg/ml microcystin [Calbiochem], 1.15 mM NaF, and 5 mM sodium pyrophosphate). Lysates were incubated at 4°C with rotation for 15 min and spun at 16,873 relative centrifugal force in a microcentrifuge for 15 min. Lysate (120 µg) was analyzed by immunoblot after resolution on a 4–20% gradient gel (Bio-Rad). Relative activation of LRRK2 was determined using the ratio of pS1292/total LRRK2, normalizing for Rab29 levels.
Determination of Rab phosphorylation
HEK293T or HeLa cells were transfected with the indicated plasmids using polyethylenimine HCl MAX 4000; 24 h later, cells were chilled and washed with PBS. Cells were scraped into buffer containing 50 mM Tris/HCl, pH 7.5, 1% Triton X-100, 1 mM EGTA, 1 mM sodium orthovanadate, 50 mM NaF, 10 mM β-glycerophosphate, 5 mM sodium pyrophosphate, 0.1 µg/ml Microcystin-LR, 270 mM sucrose, and protease inhibitor cocktail (Sigma). Samples were immunoblotted on 15% gels or 4–20% precast gradient gels (Bio-Rad).
In vitro prenylation
Cytosol was incubated with 5 µM biotin geranyl pyrophosphate (Jena Biosciences). Samples were rotated for 4 h at RT and incubated with GFP-binding protein coupled to NHS-activated Sepharose 4 fast flow resin (GE Healthcare) for 1 h rotating end over end at 4°C. Resin was washed three times with 1 ml PBS, eluted with 50 µl 2× SDS sample buffer, and boiled for 5 min. Samples were resolved on a 12% SDS-PAGE gel for immunoblot analysis.
Rab purification and MANT-GDP assay
His-Sumo-Rab29 WT and D63A were purified from a 1L culture of Rosetta II cells induced at OD600 = 0.5–0.7 with 0.4 mM IPTG (Gold Biotechnology) overnight at 17°C. Cells were resuspended in 50mM Hepes pH 7.4, 200mM NaCl, 5mM MgCl2, 5mM imidazole, 0.5mM DTT, 20 µM GTP, 10% (vol/vol) glycerol, and protease inhibitors (Sigma) by one passage through an Emulsiflex-C5 apparatus (Avestin) at 10,000 lb/in2 followed by centrifugation at 13,000 rpm for 25 min in a FiberLite F15 rotor (ThermoFisher). Clarified lysates were incubated with 30 µl cOmplete His-Tag resin (Sigma) for 2 h rotating at 4°C. Resin was washed three times with 1ml lysis buffer and eluted in 150 mM imidazole-containing lysis buffer. Protein was immediately desalted using a PD MiniTrap G-25 column (GE Healthcare) to remove imidazole. His-tagged Rab5 was used as a control and purified as described previously (Aivazian et al., 2006).
For the MANT-GDP binding assay, Rab proteins were loaded with 25-fold molar excess of MANT-GDP (Invitrogen) in buffer containing 20 mM Hepes, 150 mM NaCl, 8% glycerol, and 25 mM EDTA in a 25°C water bath for 2 h. The reaction was quenched with addition of MgCl2 to achieve a final concentration of 40 mM. Proteins were desalted using a 0.5-ml 7K molecular weight cutoff Zeba spin desalting column (ThermoFisher) into 30 mM Hepes, pH 7.4, 150 mM NaCl, and 5 mM MgCl2. Samples were diluted to 1 µM in a final volume of 98 µl of either desalting buffer or EDTA containing buffer with 30 mM Hepes, pH 7.4, 150 mM NaCl, 5 mM MgCl2, and 20 mM EDTA. Samples were loaded into a black 94-well plate. Fluorescence was monitored in a Tecan infinite 200Pro plate reader with excitation at 360 nm and monitoring 440 nm emission every minute. After an equilibration period, 2 µl of 10 mM unlabeled GDP was added to achieve a final concentration of 200 µM. Observed pseudo-first-order rate constants were extracted from a nonlinear least-squares fit.
Light microscopy
Cells were plated on glass coverslips and the next day transfected with indicated plasmids. Cells were fixed with 3.5% paraformaldehyde in PBS for 15 min, permeabilized for 3 min in 0.1% Triton X-100, and blocked with 1% BSA in PBS. Antibodies were diluted as follows: mouse anti-Myc (9E10 hybridoma culture supernatant 1:3), rabbit anti-pRab10 (MJF-R21-22-5, 1 µg/ml; Abcam), rabbit anti-total Rab10 (MJF-R23 1:500; Abcam), sheep polyclonal anti-Rab29 (S984D; 2 µg/ml; University of Dundee), rabbit anti-Mitofilin (NB100, 1:500; Novus), mouse anti GFP (1:1,000; Neuromab), rabbit anti-calnexin (1:1,000; gift from Ron Kopito, Stanford University, Palo Alto, CA), mouse anti EEA1 (1:200; BD Biosciences), rabbit anti-GCC185, and mouse anti-P115 (ascites). Primary and secondary antibody incubations were for 1 h at RT. Highly cross-absorbed H+L secondary antibodies (Life Technologies) conjugated to Alexa Fluor 488, 568, or 647 were used at 1:2,000. MitoTracker Green FM was used at 500 nM before fixation for 30 min at 37°C. Glass coverslips were mounted onto slides using mowiol. All images were obtained using Metamorph software with a spinning disk confocal microscope (Yokogawa) with an electron-multiplying charge coupled device camera (Andor) and a 100× 1.4-NA oil-immersion objective at RT. Typical exposure times of 100–300 ms were used. Nuclei were stained with 0.1 µg/ml DAPI (Sigma). Pearson’s correlation coefficients were calculated by analyzing maximum intensity projection images with CellProfiler software (Carpenter et al., 2006).
Statistics
Graphs were made using GraphPad Prism 6 software. Error bars indicate SEM. A Student’s unpaired t test was used to test significance. Two-tailed P values <0.05 were considered statistically significant. Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Video 1 shows a confocal Z-seriesof the HeLa cell shown in Fig. 8 E expressing PM-GBP, GFP-R1441G LRRK2, and Myc-Rab10 to illustrate the localization of plasma membrane–anchored LRRK2 detected in the GFP channel. Video 2 shows a confocal Z-series of the A549 Rab10 knockout cell shown in Fig. 9 B expressing GFP-R1441G LRRK2, Myc-GBP, and PM-Rab10 to illustrate the localization of plasma membrane–anchored Rab10 distribution detected in the 647-nm channel.
Supplementary Material
Acknowledgments
We thank Dr. David Lambright for his help designing the MANT nucleotide release experiments.
This work was supported by grants from the National Institutes of Health (DK37332 to S.R. Pfeffer), the Michael J. Fox Foundation for Parkinson’s Research (to S.R. Pfeffer and D.R. Alessi), and the Medical Research Council (MC_UU_12016/2 to D.R. Alessi). R.C. Gomez was supported by a National Science Foundation Graduate Research Fellowship (DGE-114747) and a Stanford Graduate Fellowship.
The authors declare no competing financial interests.
Author contributions: R.C. Gomez did all the experiments, except for those shown in Fig. 2 C (P. Lis), Fig. 2 (A and B) and Figs. 3 and 4 A (P. Wawro). The project was overseen by S.R. Pfeffer and D.R. Alessi, who also co-wrote the manuscript with R.C. Gomez.
References
- Aivazian D., Serrano R.L., and Pfeffer S.. 2006. TIP47 is a key effector for Rab9 localization. J. Cell Biol. 173:917–926. 10.1083/jcb.200510010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessi D.R., and Sammler E.. 2018. LRRK2 kinase in Parkinson’s disease. Science. 360:36–37. 10.1126/science.aar5683 [DOI] [PubMed] [Google Scholar]
- Apolloni A., Prior I.A., Lindsay M., Parton R.G., and Hancock J.F.. 2000. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell. Biol. 20:2475–2487. 10.1128/mcb.20.7.2475-2487.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki S., Kikuchi A., Hata Y., Isomura M., and Takai Y.. 1990. Regulation of reversible binding of smg p25A, a ras p21-like GTP-binding protein, to synaptic plasma membranes and vesicles by its specific regulatory protein, GDP dissociation inhibitor. J. Biol. Chem. 265:13007–13015. [PubMed] [Google Scholar]
- Barr F.A. 2013. Review series: Rab GTPases and membrane identity: causal or inconsequential? J. Cell Biol. 202:191–199. 10.1083/jcb.201306010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilina A., Rudenko I.N., Kaganovich A., Civiero L., Chau H., Kalia S.K., Kalia L.V., Lobbestael E., Chia R., Ndukwe K., et al. North American Brain Expression Consortium . 2014. Unbiased screen for interactors of leucine-rich repeat kinase 2 supports a common pathway for sporadic and familial Parkinson disease. Proc. Natl. Acad. Sci. USA. 111:2626–2631. 10.1073/pnas.1318306111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger Z., Smith K.A., and Lavoie M.J.. 2010. Membrane localization of LRRK2 is associated with increased formation of the highly active LRRK2 dimer and changes in its phosphorylation. Biochemistry. 49:5511–5523. 10.1021/bi100157u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biskup S., Moore D.J., Celsi F., Higashi S., West A.B., Andrabi S.A., Kurkinen K., Yu S.W., Savitt J.M., Waldvogel H.J., et al. 2006. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann. Neurol. 60:557–569. 10.1002/ana.21019 [DOI] [PubMed] [Google Scholar]
- Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delprato A., Merithew E., and Lambright D.G.. 2004. Structure, exchange determinants, and family-wide rab specificity of the tandem helical bundle and Vps9 domains of Rabex-5. Cell. 118:607–617. 10.1016/j.cell.2004.08.009 [DOI] [PubMed] [Google Scholar]
- Dhekne H.S., Yanatori I., Gomez R.C., Tonelli F., Diez F., Schüle B., Steger M., Alessi D.R., and Pfeffer S.R.. 2018. A pathway for Parkinson’s Disease LRRK2 kinase to block primary cilia and Sonic hedgehog signaling in the brain. eLife. 7:e40202 10.7554/eLife.40202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito G., Katsemonova K., Tonelli F., Lis P., Baptista M.A., Shpiro N., Duddy G., Wilson S., Ho P.W., Ho S.L., et al. 2016. Phos-tag analysis of Rab10 phosphorylation by LRRK2: a powerful assay for assessing kinase function and inhibitors. Biochem. J. 473:2671–2685. 10.1042/BCJ20160557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzen A., and Goody R.S.. 2011. GTPases involved in vesicular trafficking: structures and mechanisms. Semin. Cell Dev. Biol. 22:48–56. 10.1016/j.semcdb.2010.10.003 [DOI] [PubMed] [Google Scholar]
- Jaleel M., Nichols R.J., Deak M., Campbell D.G., Gillardon F., Knebel A., and Alessi D.R.. 2007. LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson’s disease mutants affect kinase activity. Biochem. J. 405:307–317. 10.1042/BJ20070209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubala M.H., Kovtun O., Alexandrov K., and Collins B.M.. 2010. Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci. 19:2389–2401. 10.1002/pro.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara T., Inoue K., D’Agati V.D., Fujimoto T., Eguchi T., Saha S., Wolozin B., Iwatsubo T., and Abeliovich A.. 2016. LRRK2 and RAB7L1 coordinately regulate axonal morphology and lysosome integrity in diverse cellular contexts. Sci. Rep. 6:29945 10.1038/srep29945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P.A., Greggio E., Beilina A., Jain S., Baker A., and Cookson M.R.. 2007. The R1441C mutation of LRRK2 disrupts GTP hydrolysis. Biochem. Biophys. Res. Commun. 357:668–671. 10.1016/j.bbrc.2007.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Bryant N., Kumaran R., Beilina A., Abeliovich A., Cookson M.R., and West A.B.. 2018. LRRK2 phosphorylates membrane-bound Rabs and is activated by GTP-bound Rab7L1 to promote recruitment to the trans-Golgi network. Hum. Mol. Genet. 27:385–395. 10.1093/hmg/ddx410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi D., Soldati T., Riederer M.A., Goda Y., Zerial M., and Pfeffer S.R.. 1993. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 12:677–682. 10.1002/j.1460-2075.1993.tb05701.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod D.A., Rhinn H., Kuwahara T., Zolin A., Di Paolo G., McCabe B.D., Marder K.S., Honig L.S., Clark L.N., Small S.A., and Abeliovich A.. 2013. RAB7L1 interacts with LRRK2 to modify intraneuronal protein sorting and Parkinson’s disease risk. Neuron. 77:425–439. 10.1016/j.neuron.2012.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madero-Pérez J., Fdez E., Fernández B., Lara Ordóñez A.J., Blanca Ramírez M., Gómez-Suaga P., Waschbüsch D., Lobbestael E., Baekelandt V., Nairn A.C., et al. 2018a Parkinson disease-associated mutations in LRRK2 cause centrosomal defects via Rab8a phosphorylation. Mol. Neurodegener. 13:3 10.1186/s13024-018-0235-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madero-Pérez J., Fernández B., Lara Ordóñez A.J., Fdez E., Lobbestael E., Baekelandt V., and Hilfiker S.. 2018b RAB7L1-Mediated Relocalization of LRRK2 to the Golgi Complex Causes Centrosomal Deficits via RAB8A. Front. Mol. Neurosci. 11:417 10.3389/fnmol.2018.00417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen U.T., Guo Z., Delon C., Wu Y., Deraeve C., Fränzel B., Bon R.S., Blankenfeldt W., Goody R.S., Waldmann H., et al. 2009. Analysis of the eukaryotic prenylome by isoprenoid affinity tagging. Nat. Chem. Biol. 5:227–235. 10.1038/nchembio.149 [DOI] [PubMed] [Google Scholar]
- Onnis A., Finetti F., Patrussi L., Gottardo M., Cassioli C., Spanò S., and Baldari C.T.. 2015. The small GTPase Rab29 is a common regulator of immune synapse assembly and ciliogenesis. Cell Death Differ. 22:1687–1699. 10.1038/cdd.2015.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmeyer J.H., Wilson A.L., and Maltese W.A.. 2001. Membrane targeting of a Rab GTPase that fails to associate with Rab escort protein (REP) or guanine nucleotide dissociation inhibitor (GDI). J. Biol. Chem. 276:20379–20386. 10.1074/jbc.M101511200 [DOI] [PubMed] [Google Scholar]
- Pfeffer S.R. 2017. Rab GTPases: master regulators that establish the secretory and endocytic pathways. Mol. Biol. Cell. 28:712–715. 10.1091/mbc.e16-10-0737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purlyte E., Dhekne H.S., Sarhan A.R., Gomez R., Lis P., Wightman M., Martinez T.N., Tonelli F., Pfeffer S.R., and Alessi D.R.. 2018. Rab29 activation of the Parkinson’s disease-associated LRRK2 kinase. EMBO J. 37:1–18. 10.15252/embj.201798099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak A., Pylypenko O., Durek T., Watzke A., Kushnir S., Brunsveld L., Waldmann H., Goody R.S., and Alexandrov K.. 2003. Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase. Science. 302:646–650. 10.1126/science.1087761 [DOI] [PubMed] [Google Scholar]
- Rivero-Ríos P., Romo-Lozano M., Madero-Pérez J., Thomas A.P., Biosa A., Greggio E., and Hilfiker S.. 2019. The G2019S variant of leucine-rich repeat kinase 2 (LRRK2) alters endolysosomal trafficking by impairing the function of the GTPase RAB8A. J. Biol. Chem. 294:4738–4758. 10.1074/jbc.RA118.005008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J.D., DeMong D.E., Greshock T.J., Basu K., Dai X., Harris J., Hruza A., Li S.W., Lin S.I., Liu H., et al. 2017. Discovery of a 3-(4-Pyrimidinyl) Indazole (MLi-2), an Orally Available and Selective Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitor that Reduces Brain Kinase Activity. J. Med. Chem. 60:2983–2992. 10.1021/acs.jmedchem.7b00045 [DOI] [PubMed] [Google Scholar]
- Simón-Sánchez J., Schulte C., Bras J.M., Sharma M., Gibbs J.R., Berg D., Paisan-Ruiz C., Lichtner P., Scholz S.W., Hernandez D.G., et al. 2009. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet. 41:1308–1312. 10.1038/ng.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati T., Riederer M.A., and Pfeffer S.R.. 1993. Rab GDI: a solubilizing and recycling factor for rab9 protein. Mol. Biol. Cell. 4:425–434. 10.1091/mbc.4.4.425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger M., Tonelli F., Ito G., Davies P., Trost M., Vetter M., Wachter S., Lorentzen E., Duddy G., Wilson S., et al. 2016. Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. eLife. 5:e12813 10.7554/eLife.12813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger M., Diez F., Dhekne H.S., Lis P., Nirujogi R.S., Karayel O., Tonelli F., Martinez T.N., Lorentzen E., Pfeffer S.R., et al. 2017. Systematic proteomic analysis of LRRK2-mediated Rab GTPase phosphorylation establishes a connection to ciliogenesis. eLife. 6:e31012 10.7554/eLife.31012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroupe C., and Brunger A.T.. 2000. Crystal structures of a Rab protein in its inactive and active conformations. J. Mol. Biol. 304:585–598. 10.1006/jmbi.2000.4236 [DOI] [PubMed] [Google Scholar]
- Wandinger-Ness A., and Zerial M.. 2014. Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb. Perspect. Biol. 6:a022616 10.1101/cshperspect.a022616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Ma Z., Xu X., Wang Z., Sun L., Zhou Y., Lin X., Hong W., and Wang T.. 2014. A role of Rab29 in the integrity of the trans-Golgi network and retrograde trafficking of mannose-6-phosphate receptor. PLoS One. 9:e96242 10.1371/journal.pone.0096242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M., and Munro S.. 2014. Membrane trafficking. The specificity of vesicle traffic to the Golgi is encoded in the golgin coiled-coil proteins. Science. 346:1256898 10.1126/science.1256898 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.