Lemmens and Lindqvist discuss how DNA replication and mitosis are coordinated and propose a cell cycle model controlled by brakes.
Abstract
The core function of the cell cycle is to duplicate the genome and divide the duplicated DNA into two daughter cells. These processes need to be carefully coordinated, as cell division before DNA replication is complete leads to genome instability and cell death. Recent observations show that DNA replication, far from being only a consequence of cell cycle progression, plays a key role in coordinating cell cycle activities. DNA replication, through checkpoint kinase signaling, restricts the activity of cyclin-dependent kinases (CDKs) that promote cell division. The S/G2 transition is therefore emerging as a crucial regulatory step to determine the timing of mitosis. Here we discuss recent observations that redefine the coupling between DNA replication and cell division and incorporate these insights into an updated cell cycle model for human cells. We propose a cell cycle model based on a single trigger and sequential releases of three molecular brakes that determine the kinetics of CDK activation.
Introduction
Pioneering autoradiography studies in the 1960s revealed that the events preceding cell division involved a sequence of distinct metabolic states, called cell cycle phases. These were characterized by a period of DNA synthesis (S-phase) surrounded by periods in which no DNA synthesis was detected (G1- and G2-phase; Baserga, 1965). Decades later, molecular activities that drive cell cycle progression were identified, and a central role for cyclins in complex with cyclin-dependent kinases (CDKs) was established (Nurse, 1990; Stern and Nurse, 1996). D-type cyclins (which assemble with CDK4 or CDK6) and E-type cyclins (which assemble with CDK2) function as molecular triggers for cell cycle entry (Sherr, 1993). D- and E-type cyclins generate positive feedback loops that increase cyclin expression, thereby producing a surge in cyclin-CDK activity that irreversibly leads to cell cycle commitment (Bertoli et al., 2013). Overexpression of D- or E-type cyclins alleviates growth factor dependence, overrides G1 arrest, and advances S-phase entry (Brewer et al., 1999; Ohtsubo and Roberts, 1993; Pagano et al., 1994; Quelle et al., 1993; Resnitzky et al., 1994). The kinetics of cell cycle commitment is influenced by external stimuli (e.g., growth factors), genetic context (e.g., oncogenes), and factors inherited from previous cell cycles (e.g., DNA damage signals; Arora et al., 2017; Barr et al., 2017; Moser et al., 2018; Pardee, 1989; Schwarz et al., 2018; Spencer et al., 2013; Yang et al., 2017).
Once committed to the cell cycle, E- and A-type cyclins complex with CDK2 to drive DNA replication. These cyclin-CDK2 complexes are later joined in by cyclin A–CDK1 and cyclin B–CDK1, ensuring a rise in CDK activity. As the cyclin-CDK complexes transit from low activity to high activity, they phosphorylate thousands of residues in various target proteins to initiate mitosis (Dephoure et al., 2008; Ly et al., 2017; Ohta et al., 2016; Olsen et al., 2010; Swaffer et al., 2018). Cyclin-CDK activity is controlled at multiple levels, involving complex formation of cyclin-CDKs, direct binding of accessory/inhibitory proteins, posttranslational modifications, and regulated activity of phosphatases that reverse cyclin-CDK–mediated phosphorylation (Hégarat et al., 2016; Malumbres, 2014; Nilsson, 2019). Many regulators of cyclin-CDK activity are also direct or indirect targets of cyclin-CDK activity, creating positive feedback loops that ensure that cyclin-CDK activity continues to rise once activated (Hégarat et al., 2016; Lindqvist et al., 2009; Pomerening, 2009). One such regulator is the kinase PLK1, which, apart from activating cyclin-CDK, plays a key role in mitotic entry and progression (Combes et al., 2017; Joukov and De Nicolo, 2018; Pintard and Archambault, 2018). Here we discuss how spiraling cyclin-CDK activities are kept in check and coordinated with genome duplication. We incorporate these insights into an updated cell cycle model based on three molecular brakes that control the level of CDK activation and thus the timing of cell division.
DNA replication and mitosis: From a gap to a link
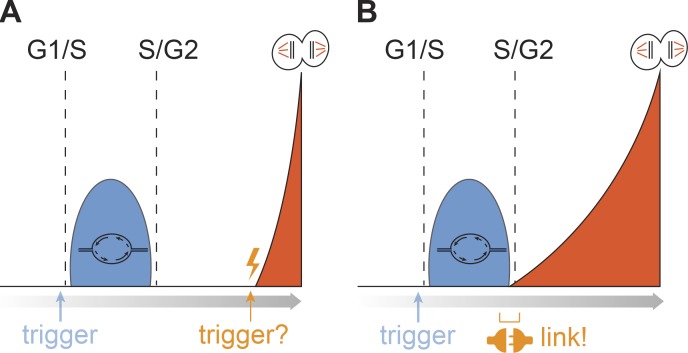
What triggers cell division has remained a key unanswered question for cell cycle research (Mchedlishvili et al., 2015). In eukaryotic cells, DNA replication and cell division are separated by the intervening G2-phase. Due to the several-hour duration of G2 in somatic cells, DNA replication and cell division were seen as largely independent events, which suggested that the trigger of cell division was not directly coupled to DNA replication (Fig. 1 A). Accordingly, much effort has gone into identifying what triggers cell division at the end of G2-phase. Although many proteins are involved (Hégarat et al., 2016; Lindqvist et al., 2009), recent data suggest a particular involvement of cyclin A–CDK and PLK1 in initiating mitotic entry. Apart from directly phosphorylating targets that promote mitosis, cyclin A in complex with CDK1 or CDK2 phosphorylates the Aurora A cofactor Bora to stimulate activation of PLK1 (Gheghiani et al., 2017; Lindqvist et al., 2009; Macůrek et al., 2008; Seki et al., 2008; Thomas et al., 2016; Vigneron et al., 2018). However, cyclin A–CDK complexes are active from the onset of S-phase (Fotedar et al., 1996; Girard et al., 1991; Minshull et al., 1990), raising the question, what determines the timing of mitosis?
Figure 1.
Models on DNA replication and mitotic triggers. Gray arrow depicts time toward mitosis, and dashed lines indicate G1/S and S/G2 transitions. (A) Model 1 is based on two independent triggers: one trigger for DNA replication (blue) and one trigger for mitotic kinase activation (orange). (B) Model 2 is based on a single trigger for DNA replication (blue) and mitotic kinase activation (orange), yet a signaling cascade links both processes and separates the two events in time. Kinases that drive mitotic entry are activated when bulk DNA replication is completed at the S/G2 transition.
Recent advances have shown that while DNA replication and cell division occur at different times, from a signaling perspective, both processes are heavily connected. While CDK1 and PLK1 are classified as mitotic kinases, both are active before mitosis. In fact, mass spectrometry studies indicated that >100 mitotic phosphorylation sites are already phosphorylated in G2-phase (Ly et al., 2017). Quantitative immunofluorescence approaches also showed that Lamin A/C and TCTP (substrates of CDK1 and PLK1, respectively) are increasingly phosphorylated through G2-phase. Importantly, these phosphorylated residues are first detected at the S/G2 transition, implying a molecular link between DNA replication and mitotic entry (Akopyan et al., 2014; Cucchi et al., 2010; Lemmens et al., 2018). A live-cell sensor that responds to PLK1 activity confirmed these observations and showed nuclear PLK1 target phosphorylation at the moment cells completed DNA replication (Akopyan et al., 2014). Similarly, a live-cell sensor based on the CDK1 phosphorylation motif in SLBP was degraded the moment cells finished bulk DNA synthesis (Bajar et al., 2016). Thus, both PLK1 and CDK1 start to phosphorylate target proteins at the S/G2 transition, suggesting a direct link between S-phase completion and mitotic kinase activation (Fig. 1 B).
Mitotic entry: From a trigger to a brake
The tight temporal connection between completion of DNA replication and activation of mitotic kinases raises several fundamental questions. What triggers the onset of mitotic kinase activities at the S/G2 transition? And if mitotic kinases can be activated hours before mitosis, why are they not activated earlier? In fact, inhibition of phosphatases can induce mitosis in S-phase without production of new proteins (Yamashita et al., 1990). Similarly, inhibition of WEE1, a negative regulator of CDK1 and CDK2, can promote premature mitotic entry in S-phase (Aarts et al., 2012; Sørensen and Syljuåsen, 2012). Thus, mitotic kinases can, in principle, be activated in S-phase, but are kept suppressed until the S/G2 transition.
To explain the notion that eukaryotic cells are able to proliferate with high fidelity, Hartwell and Weinert (1989) theorized that human cells might possess surveillance mechanisms that allow cell division only when DNA replication is complete. While this idea was postulated decades ago, direct empirical evidence proved hard to obtain. Recent data show that human cells that cannot initiate DNA replication prematurely activate PLK1 and CDK1 and promptly enter mitosis, demonstrating that DNA replication itself is a key regulator of mitotic kinases (Lemmens et al., 2018). The absence of DNA replication causes immediate CDK hyperactivation upon S-phase entry and is sufficient to trigger premature mitotic entry, showing that the process of DNA replication generates a brake signal that controls the timing of mitosis (Lemmens et al., 2018).
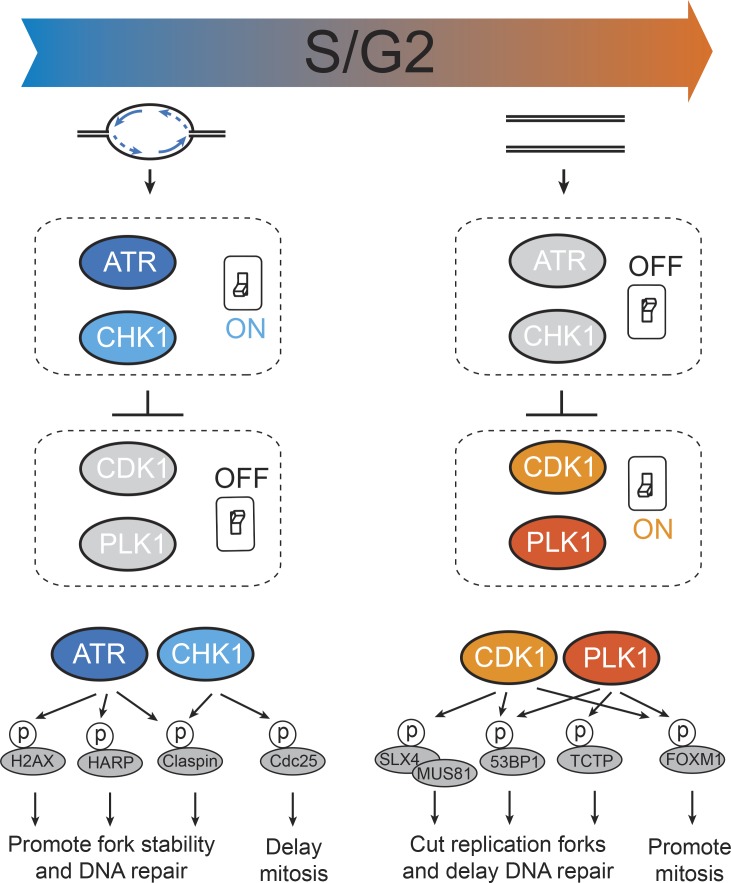
While the absence of DNA replication advances mitotic entry (Lemmens et al., 2018), perturbation of ongoing replication postpones mitotic entry (Bajar et al., 2016; Boddy et al., 1998; Darzynkiewicz et al., 2011; Lemmens et al., 2018; Sørensen et al., 2003; Zineldeen et al., 2017). The latter could be explained by delayed completion of DNA replication, but altered replication fork progression typically induces immediate DNA damage, which obscures the role of unperturbed DNA replication in cell cycle regulation (Darzynkiewicz et al., 2011). DNA damage is a potent and well-studied inhibitor of mitotic entry (Harrison and Haber, 2006; Sancar et al., 2004). Stalled replication forks halt the cell cycle primarily through the action of ATR and CHK1 kinases, and several studies show that the ATR–CHK1 axis also plays an important role during an unperturbed S-phase (Petermann and Caldecott, 2006; Saldivar et al., 2017; Sørensen and Syljuåsen, 2012). ATR and CHK1 are essential proteins with highly conserved functions in cell cycle regulation (Brown and Baltimore, 2000; Cliby et al., 1998; Eykelenboom et al., 2013; Hekmat-Nejad et al., 2000; Niida et al., 2005; Petermann et al., 2006; Schmitt et al., 2006; Sørensen et al., 2003; Sørensen et al., 2004). In particular, multiple targets of ATR and CHK1 can stabilize DNA replication forks and promote DNA repair (Fig. 2; Kumagai and Dunphy, 2003; Scorah and McGowan, 2009; Uto et al., 2004; Ward and Chen, 2001; Yuan et al., 2009). However, although inhibition of ATR is toxic to cells, it can be rescued by simultaneous depletion of the CDK-activating phosphatase Cdc25A, suggesting that a key function of ATR is to counteract CDK activity (Ruiz et al., 2016). In line with this finding, inhibition of CHK1 was shown to lead to premature activation of CDK1 and PLK1 in S-phase (Lemmens et al., 2018; Saldivar et al., 2018; Syljuåsen et al., 2005). Importantly, a CDK1-dependent transcriptional program of mitotic regulators was found to be prematurely initiated upon inhibition of ATR in S-phase (Saldivar et al., 2018). These observations point toward a model in which mitotic kinases are actively suppressed during DNA replication (Sørensen and Syljuåsen, 2012). As long as DNA replication is ongoing, ATR-CHK1 inhibits CDK1 and PLK1, ensuring that DNA replication and mitosis are separated in time (Fig. 2). The phosphorylation events triggered by ATR-CHK1 are likely complemented by other pathways, including ubiquitin-mediated processes. For instance, the SCF E3 ligase component cyclin F shows synthetic lethality with Chk1 inhibition (Burdova et al., 2019) and, similar to ATR, can attenuate the transcriptional response that drives mitosis (Burdova et al., 2019; Clijsters et al., 2019; Klein et al., 2015; Mavrommati et al., 2018).
Figure 2.
Molecular switches at the S/G2 transition. The S/G2 transition is dictated by DNA replication status. DNA replication in S-phase activates ATR/Chk1 signaling, which represses CDK1 and PLK1 activity and promotes fork stabilization and DNA repair. Completion of bulk DNA replication allows CDK1 and PLK1 activation, which promotes mitotic entry and processing of persistent replication intermediates. Lower panel depicts examples of phosphorylation targets of ATR/CHK1 or CDK1/PLK1. The ATR/CHK1 axis inhibits mitotic entry (e.g., via CDC25) and promotes fork stability (e.g., via H2AX, HARP, and Claspin), while the CDK1/PLK1 axis promotes mitotic entry (e.g., via FOXM1) and alters DNA repair (e.g., via 53BP1, SLX4, and TCTP).
Although the exact nature of the replication intermediate controlling CDK activity during an unperturbed S-phase is unknown, a recent study suggests it involves single-stranded DNA (Saldivar et al., 2018). Whether the presence of single-stranded DNA is complemented by another DNA replication intermediate, signaling from active replication forks, DNA topological stress, and/or low levels of stochastic DNA damage, the process of DNA replication constitutes an integral signaling component of the human cell cycle. Completion of DNA replication removes the substrates for ATR-CHK1 activity and hence allows the immediate activation of mitotic kinases at the S/G2 transition (Lemmens et al., 2018; Saldivar et al., 2018). Thus, changing from an S-phase state to a G2-phase state requires release of a brake, rather than the generation of a new trigger (Fig. 2).
Better to brake than to break
Mitosis is sensitive to persisting replication intermediates that can prevent sister chromatid separation. Hence, persisting replication intermediates are cut by structure-specific nucleases to promote faithful sister chromatid disjunction (Chan and West, 2018; Naim et al., 2013; Ying et al., 2013). While cleavage of a few remaining replication intermediates during mitosis might be beneficial, such activities in S-phase would cause genome-wide havoc. Different mechanisms have evolved to restrict endonucleases to mitosis. Whereas the GEN1 nuclease depends on nuclear envelope breakdown for access to DNA (Chan and West, 2018), the Mus81-Slx4 nuclease depends on PLK1 and CDK1 activity for assembly (Svendsen et al., 2009; Cucchi et al., 2010; Duda et al., 2016; Wyatt et al., 2017). Interestingly, PLK1 and CDK1 also actively suppress DNA double-strand repair during mitosis (Benada et al., 2015; Orthwein et al., 2014), possibly preventing immediate religation of cut chromosomes and thus deleterious chromosome entanglements. Reinstating double-strand break repair during mitosis is highly toxic and causes telomere fusions (Orthwein et al., 2014).
The fact that CDK1 and PLK1 suppress DNA repair and activate endonucleases makes them tailored for mitosis but dangerous in S-phase (Fig. 2; Chen et al., 2009; Cucchi et al., 2010; Duda et al., 2016; van Vugt et al., 2010; Zhang et al., 2012). In addition, CDK deregulation can lead to various DNA replication stresses, including faulty origin firing, nucleotide imbalances, and RPA exhaustion (Gaillard et al., 2015; Hills and Diffley, 2014; Toledo et al., 2017). Indeed, inhibition or depletion of the WEE1 kinase, a negative regulator of CDK1 and CDK2, leads to activation of mitotic kinases in S-phase, DNA damage, and altered DNA repair (Beck et al., 2010, 2012; Heald et al., 1993; Krajewska et al., 2013). Mutation of CDK1 so that it can no longer be targeted by WEE1 leads to embryonic lethality in mice, where cells arrest in S-phase with high levels of damaged DNA (Szmyd et al., 2019). Similarly, inhibition of CHK1 causes immediate DNA damage in replicating cells (Lemmens et al., 2018; Syljuåsen et al., 2005). In all three cases, DNA damage could be prevented by simultaneous CDK inhibition, revealing unrestrained CDK activity as the cause of genome instability. A brake on PLK1 and CDK1 activity during S-phase is therefore instrumental for maintaining DNA integrity.
When cells experience DNA damage, an elaborate DNA damage response (DDR) is triggered, which inhibits mitotic kinase activities and cell cycle progression (Harrison and Haber, 2006; Sancar et al., 2004). The DDR buys time for DNA repair and prevents propagation of damaged genomes. A common form of DNA damage during S-phase is replication stress, which is due to faulty DNA replication. Replication stress can be caused by many factors, including deregulation of CDK activity by oncogenes such as cyclin E and Cdc25A (Beck et al., 2010; Gaillard et al., 2015; Macheret and Halazonetis, 2015; Toledo et al., 2017). As treatments that induce premature CDK activation cause DNA damage, they are expected to trigger a feedback loop in which the DDR counteracts the rising CDK activities. Indeed, fluctuation of CDK2 activity has been observed upon spontaneous and ectopic DNA replication stress, and a subset of cells fails to enter mitosis upon WEE1/CHK1 inhibition (Daigh et al., 2018; Lemmens et al., 2018). Given that PLK1 and CDK1 control structure-specific DNA nucleases (Chan and West, 2018; Wyatt et al., 2017), DNA replication intermediates can act as downstream substrates as well as upstream regulators of mitotic kinase activity. Interestingly, forced mitotic entry by WEE1 inhibition requires the structure-specific DNA nuclease Mus81, suggesting that stalled forks need to be processed to allow premature mitotic entry (Duda et al., 2016). Thus, high CDK activity during S-phase leads to replication stress, which in turn limits CDK activity, revealing a delicate balancing mechanism that brakes CDK activities in S-phase.
A cell cycle model based on brakes
The quantitative threshold model for cell cycle progression, originally introduced by Stern and Nurse (1996) to explain that a single cyclin-CDK can drive the fission yeast cell cycle, has for decades stood the test of time (Coudreuse and Nurse, 2010; Swaffer et al., 2016). The quantitative model focuses on the amount of CDK activity rather than the identity of different cyclin-CDK complexes. In this model, S-phase requires relatively low cyclin-CDK activity, whereas mitosis only occurs above a much higher threshold of cyclin-CDK activity. Although likely complemented with different specificities of various cyclin-CDK complexes in other eukaryotes, this model explains why DNA replication commences before cell division (Hochegger et al., 2008; Uhlmann et al., 2011). However, it raises the question of how the cell cycle control system is wired to ensure that cyclin-CDK activity does not rise too quickly. As discussed earlier, DNA replication is sensitive to high CDK activity and needs to be completed before cell division.
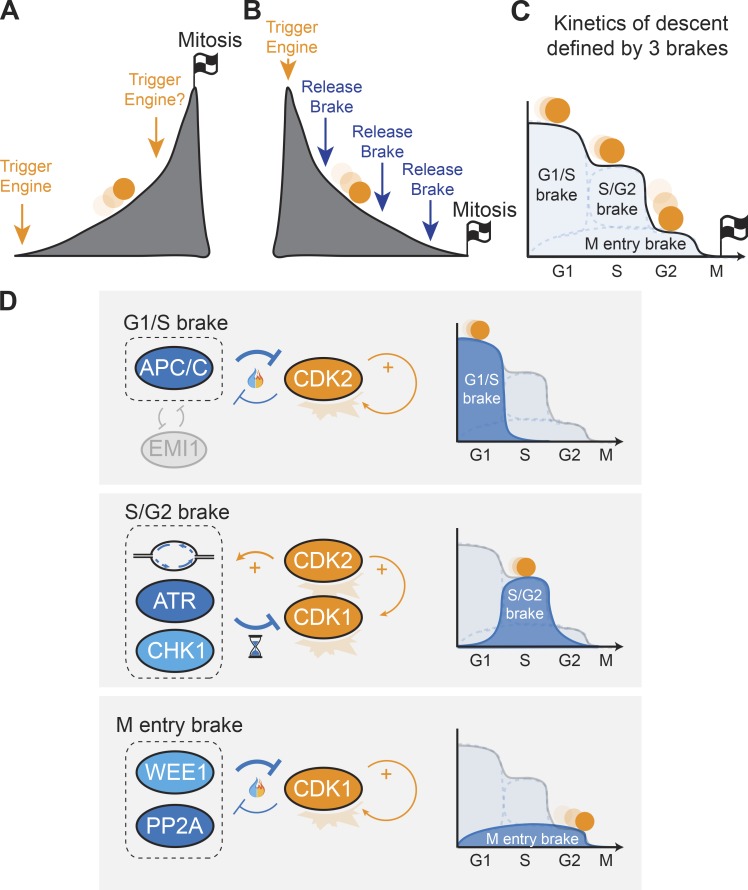
Increasing cyclin-CDK activity can in principle be accomplished by triggering additional sets of engines that push cyclin-CDK activity to higher levels. Accordingly, the road to mitosis is sometimes envisioned as a mountain. Approaching mitosis, the slope of the mountain is very steep, reflecting the high levels of CDK activity required for cell division (Fig. 3 A). The increasing CDK activity observed in cycling cells is primarily due to the presence of multiple positive feedback loops (Lindqvist et al., 2009; Medema and Lindqvist, 2011; Pomerening, 2009). These feedback loops ensure that once CDK activation is initiated, it will autonomously continue to increase in an exponential fashion, similar to a ball rolling down a mountain picking up speed without the need for extra engines. As discussed, progressing from an S-phase state to a G2-phase state requires the release of a brake (DNA replication), rather than the generation of a new trigger. Similarly, deficiencies in WEE1 or ATR/CHK1 advance mitotic entry, revealing a poised state toward cell division that needs to be restrained. Thus, mitotic entry per se does not require an extra engine or trigger in G2-phase, but instead might involve a single trigger in G1-phase and a set of counteracting brakes. These molecular brakes ensure that CDK activation occurs in a stepwise manner and on par with the processes required for faithful cell division. For instance, CDK-dependent FOXM1 phosphorylation is initiated early but plateaus during S-phase, which postpones full activation of the promitotic transcriptional program until after genome duplication (Saldivar et al., 2018). We believe recent discoveries fit a model in which the road to mitotic entry, instead of a steep climb, resembles a well-monitored descent (Fig. 3 B). We here propose a model in which the descent is controlled by three brake modules that collectively determine CDK activity output and thus the timing of mitosis (Fig. 3 C).
Figure 3.
A brake model for cell cycle progression. (A) Model 1 is based on multiple independent triggers that initiate cell cycle engines and push the cell into mitosis, akin to pushing a ball up a mountain. (B) Model 2 is based on a single trigger and multiple brakes that restrain cell cycle engines and mitotic entry, akin to braking a ball rolling down a mountain. (C) At least three brake modules collectively determine cell cycle progression. Visualized as an energy landscape, the brakes define the slope of the descent and thus the speed of the ball rolling toward mitosis. (D) Basic signaling circuits of the three brake modules controlling CDK activity (left) and the timing of the respective brakes during the cell cycle (right). The G1/S brake and M entry brake are wired to CDK via double-negative feedback loops, and eventually are overruled by self-amplifying CDK activity. The S/G2 brake is wired to CDK via a transient incoherent feed-forward loop: CDK2 activity triggers DNA replication, which inhibits CDK1, but the brake is inherently transient because it resolves the moment the cell completes genome replication.
Instead of focusing solely on the source and amount of CDK activity, we propose that it is more informative to think of the cell cycle in terms of an energy landscape, as is frequently done for chemical reactions, and for cell biology has been made popular by the cellular route to differentiation (Takahashi, 2012; Waddington, 1957). Because the signaling landscape in which a given CDK activity acts is critical to its outcome, we propose that the kinetics of the cell cycle relies on the condition of the road and strength of the brakes rather than the power of the engine. We discriminate a G1/S brake, a S/G2 brake, and an M-entry brake. While all three brakes regulate CDK activity, they have different effectors and act at different stages of the cell cycle. In unchallenged conditions, the release of each brake corresponds to a cell cycle transition.
G1/S brake
The G1/S brake prevents G1/S transition and is mainly controlled by the anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase. Activation of APC/C in mitosis triggers the separation of duplicated chromosomes and resets the cell cycle. The APC/C complex triggers the degradation of many CDK activators (including cyclin A, cyclin B, and PLK1) and thus constitutes a major brake to cell cycle progression (Alfieri et al., 2017; Watson et al., 2019). Recent data indicate that inactivation of the APC/C complex occurs in a switch-like manner and precedes S-phase entry (Barr et al., 2016; Cappell et al., 2016, 2018; Yuan et al., 2014). Rising CDK activity in G1-phase, driven by D- and E-type cyclins that are not targeted by APC/C, ultimately shuts off the G1/S brake. CDK inactivates APC/C both directly (through inactivating phosphorylations on CDH1, an activator of APC/C) and indirectly (through induced expression of EMI1, a negative regulator of APC/C; Hsu et al., 2002; Kramer et al., 2000; Lukas et al., 1999). A dual negative feedback between APC/C and EMI1 generates a bistable switch: once a threshold level of CDK activity is reached, the G1/S brake is turned off in an irreversible manner (Barr et al., 2016; Cappell et al., 2016, 2018). Linking spiraling CDK activation to dual negative feedback loops ensures full commitment into S-phase (Fig. 3 D).
S/G2 brake
The S/G2 brake requires DNA replication (Lemmens et al., 2018) and acts via various substrates of ATR/CHK1 that counteract CDK activation (Lemmens et al., 2018; Saldivar et al., 2018; Sørensen et al., 2003). While the G1/S brake is controlled by shifting double-negative feedback loops, the S/G2 brake is manifested through a transient incoherent feed-forward loop. Rising CDK2 activity promotes CDK1 activity and triggers the initiation of DNA replication, which by itself constitutes a brake on CDK1 activation. In other words, the cell pushes the gas and brake at the same time, yet the brake is inherently transient. Completion of DNA replication at the S/G2 transition resolves the brake signal and allows activation of CDK1 and PLK1 (Fig. 3 D).
Current models suggest that ATR/CHK1 postpones the activation of CDK1 through Cdc25/Wee1 regulation, similar to the response to DNA damage (Goto et al., 2019; Sørensen and Syljuåsen, 2012). How PLK1 is suppressed during unperturbed proliferation is not clear, but after DNA damage, PLK1 activity is limited by dephosphorylation, disruption of Aurora A–Bora interaction, and direct methylation (Bruinsma et al., 2017; Hu et al., 2018; Li et al., 2019). In addition, PLK1 and CDK1 activity are tightly interlinked, and suppression of one can affect the other (Gheghiani et al., 2017; Lindqvist et al., 2009; Macůrek et al., 2008; Seki et al., 2008; Thomas et al., 2016; Vigneron et al., 2018). How quickly mitotic kinase activity builds up at the S/G2 transition is likely linked to the kinetics of DNA replication completion: a cell that completes DNA replication efficiently is expected to activate mitotic kinases promptly, whereas a cell sustaining stalled forks is expected to activate CDK1 and PLK1 relatively slowly, providing extra time to resolve DNA replication issues before mitotic entry.
M-entry brake
Finally, cells are equipped with an M-entry brake that prevents premature mitosis throughout interphase and is mostly controlled by WEE1 and PP1/PP2A activities. The M-entry brake is a composite brake module that acts both on CDK itself and on its substrates. Similar to the G1/S brake, the M-entry brake is controlled by dual negative feedback loops (Fig. 3 D). Both WEE1 and the PP1/PP2A phosphatases counteract CDK activity but are also restrained by CDK activity. WEE1 inhibits CDK1 by direct phosphorylation, but elevated levels of CDK1 activity (together with PLK1 activity) target WEE1 for degradation (Parker and Piwnica-Worms, 1992; Watanabe et al., 2005). Similarly, the PP2A-B55 phosphatase effectively counteracts CDK1 target phosphorylation, yet elevated levels of CDK1 activity reduce PP2A-B55 activity through the Greatwall kinase and the PP1 phosphatase (Dohadwala et al., 1994; Gharbi-Ayachi et al., 2010; Grallert et al., 2015; Kwon et al., 1997; Mochida et al., 2010). The M-entry brake thus functions as a constitutive counterweight to the spiraling CDK activity, which raises the threshold for mitotic entry and supports full commitment after the G2/M transition.
Together, these three brakes ensure a controlled stepwise descent toward mitosis: the G1/S brake restricts CDK activity in G1-phase, the S/G2 brake restricts CDK activity in S-phase, and the M-entry brake suppresses CDK activity throughout interphase and determines the final kinetics of mitotic entry (Fig. 3). Defects in any of these brakes cause unrestrained CDK activation, which leads to hasty cell cycle progression and thus severe errors in DNA replication and/or chromosome segregation (Aarts et al., 2012; Beck et al., 2012; Brown and Baltimore, 2000; Cappell et al., 2016; Labib and De Piccoli, 2011; Lemmens et al., 2018; Niida et al., 2005; Sørensen and Syljuåsen, 2012; Yamashita et al., 1990).
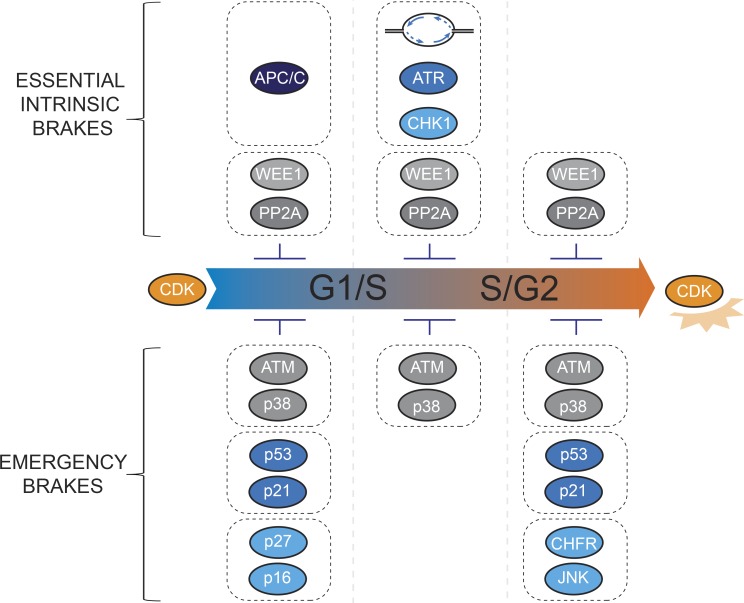
From “intrinsic checkpoint” to “intrinsic brake”
Given that the G1/S brake, S/G2 brake, and M-entry brake are required for cell survival and rely on cues inherent to cell proliferation (i.e., DNA replication and CDK activity oscillations), we refer to these brakes as “essential” or “intrinsic” brakes. These intrinsic brakes can also be called “checkpoints” by the original definition given by Hartwell and Weinert (1989) or as redefined by Elledge (1996). The word “checkpoint,” however, has led to confusion in the field because it traditionally refers to a predefined point or barrier on a street or path that can only be passed if certain conditions are fulfilled. Using “checkpoints” to describe cell cycle regulation thus implies that cellular status is checked at defined moments (e.g., just before cell cycle transitions) and that the approach leading up to these checkpoints is not monitored. For example, the terms “S-M checkpoint” or “S/G2 checkpoint” could imply that DNA replication status is monitored just before mitotic entry or S/G2 transition, respectively (Eykelenboom et al., 2013; Saldivar et al., 2018). However, the factors that enforce the cell cycle delay (i.e., ATR/CHK1) are active throughout S-phase and cause oscillating CDK activities well before S/G2 transition (Daigh et al., 2018; Lemmens et al., 2018; Michelena et al., 2019; Saldivar et al., 2018). We therefore favor the use of “brake” terminology, instead of “checkpoint” terminology. The timing of cell cycle transitions can be explained by the sequential release of three essential brake signals that counteract CDK activation (Fig. 3).
Extra brakes in case of emergency
Thus far we have mainly focused on the intrinsic signaling circuits that determine cell cycle progression, but cell proliferation is highly sensitive to external cues. A multitude of developmental or experimental conditions can slow down or arrest the cell cycle (Edgar and Lehner, 1996; Elledge, 1996; Kipreos and van den Heuvel, 2019). As a result, the term “checkpoint” has gained a second meaning in cell biology, referring to a halt in cell cycle progression due to external stresses. Most famous among this second class of checkpoints are the DNA damage checkpoints, which prevent G1/S and G2/M transition upon DNA damage (Cuadrado et al., 2009; Falck et al., 2001; Gutierrez et al., 2010; Jackson and Bartek, 2009; Mirzayans et al., 2012; Privette and Petty, 2008; Zarubin and Han, 2005). Examples of effectors of DNA damage checkpoints are ATM and the p53-p21 axis, which are critical for maintaining genome integrity but are not required for cell cycle progression per se (Bartkova et al., 2006; Sancar et al., 2004; Zhou and Elledge, 2000). Although a DNA damage checkpoint can be induced by a single DNA break (van den Berg et al., 2018), low levels of DNA damage are often not sufficient to sustain a cell cycle arrest (Deckbar et al., 2007, 2010; Syljuåsen et al., 2006). In G2-phase, this depends on the self-amplifying properties of promitotic signaling, which eventually overcomes checkpoint signaling (Jaiswal et al., 2017; Liang et al., 2014). Apart from strengthening CDK activity, in particular PLK1 can by multiple means counteract checkpoint kinases (Mailand et al., 2006; Mamely et al., 2006; Peschiaroli et al., 2006; Syljuåsen et al., 2006; van Vugt et al., 2004, 2010). The G2 DNA damage checkpoint therefore delays rather than blocks mitosis, arguing that also DNA damage checkpoints can function as brakes rather than strict checkpoint barriers. The cell cycle thus is controlled by two distinct classes of brakes: (a) “essential” brakes that are inherent to the cell cycle and (b) “emergency” brakes that are conditional and come into play upon stress. Examples of brake effectors for each class are depicted in Fig. 4. The process of DNA replication can modulate the timing and amplitude of both types of brakes. For instance, DNA replication triggers ATR/CHK1 signaling (Lemmens et al., 2018; Saldivar et al., 2018; Sørensen et al., 2003) but also p21 degradation (Abbas et al., 2008; Coleman et al., 2015), substantiating a tight link between genome duplication and cell cycle progression.
Figure 4.
Two types of brakes control cell cycle timing. Intrinsic brakes are essential to life and ensure cell cycle phase dependence, while emergency brakes delay or arrest cell cycle progression upon external cues and stresses. The APC/C complex, ATR/CHK1, and WEE1/PP2A represent intrinsic brakes required to restrain CDK activation during the unchallenged cell cycle (top). In contrast, signaling factors such as ATM, p38, p27, p16, p53, p21, CHFR, and JNK are not essential for cell proliferation, per se, but effectively inhibit CDK activation upon stress (lower panel).
Concluding remarks
The activities capable of copying a living cell have intrigued scientists for decades, and through the years many fundamental concepts of the cell cycle have been exposed (Baserga, 1965; Dephoure et al., 2008; Nurse, 1990; Ohta et al., 2016; Stern and Nurse, 1996). One key feature of cycling cells is the distinct surge of DNA incorporation in S-phase, in which the cell duplicates its genome before cell division. It is now clear that DNA replication is not just an output of the cell cycle, but in fact feeds back into the signaling networks controlling mitotic entry. Linking DNA replication to the mitotic entry network dismisses the need for separate triggers while allowing temporal separation. We propose a cell cycle model based on a single trigger, which together with a set of three molecular brakes generates distinct waves of protein activities. This brake model can explain gradual cyclin-CDK activation and distinct phase transitions, as well as commitment to complete the cell cycle once initiated.
Many aspects of this model require further study. Are there situations in which additional triggers are needed? What is the molecular identity of the DNA replication intermediate postponing mitosis? How can DNA replication impose differential regulation on different cyclin-CDK complexes? What systems properties are emerging for the S/G2 transition, and how does that affect signaling sensitivity and stability? Moreover, while we propose three basic brake circuits controlling CDK activation, more work is needed to determine their exact wiring. Understanding the nature of these brake modules will help explain and predict the kinetics and behaviors of the different cell cycle transitions. The current pace of discoveries in the cell cycle field is encouraging, and the continuous development of time-resolved technologies at single-cell resolution will surely further our understanding of how cells establish concerted waves of protein activities to control the road to cell division.
Acknowledgments
This work was supported by grants from the Swedish Cancer Foundation (to B. Lemmens and A. Lindqvist), Strategic Research Program in Cancer at Karolinska Institutet (grant number 2201; B. Lemmens), and the Swedish Research Council (A. Lindqvist).
The authors declare no competing financial interests.
References
- Aarts M., Sharpe R., Garcia-Murillas I., Gevensleben H., Hurd M.S., Shumway S.D., Toniatti C., Ashworth A., and Turner N.C.. 2012. Forced mitotic entry of S-phase cells as a therapeutic strategy induced by inhibition of WEE1. Cancer Discov. 2:524–539. 10.1158/2159-8290.CD-11-0320 [DOI] [PubMed] [Google Scholar]
- Abbas T., Sivaprasad U., Terai K., Amador V., Pagano M., and Dutta A.. 2008. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 22:2496–2506. 10.1101/gad.1676108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopyan K., Silva Cascales H., Hukasova E., Saurin A.T., Müllers E., Jaiswal H., Hollman D.A., Kops G.J., Medema R.H., and Lindqvist A.. 2014. Assessing kinetics from fixed cells reveals activation of the mitotic entry network at the S/G2 transition. Mol. Cell. 53:843–853. 10.1016/j.molcel.2014.01.031 [DOI] [PubMed] [Google Scholar]
- Alfieri C., Zhang S., and Barford D.. 2017. Visualizing the complex functions and mechanisms of the anaphase promoting complex/cyclosome (APC/C). Open Biol. 7:170204 10.1098/rsob.170204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora M., Moser J., Phadke H., Basha A.A., and Spencer S.L.. 2017. Endogenous Replication Stress in Mother Cells Leads to Quiescence of Daughter Cells. Cell Reports. 19:1351–1364. 10.1016/j.celrep.2017.04.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajar B.T., Lam A.J., Badiee R.K., Oh Y.H., Chu J., Zhou X.X., Kim N., Kim B.B., Chung M., Yablonovitch A.L., et al. 2016. Fluorescent indicators for simultaneous reporting of all four cell cycle phases. Nat. Methods. 13:993–996. 10.1038/nmeth.4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr A.R., Cooper S., Heldt F.S., Butera F., Stoy H., Mansfeld J., Novák B., and Bakal C.. 2017. DNA damage during S-phase mediates the proliferation-quiescence decision in the subsequent G1 via p21 expression. Nat. Commun. 8:14728 10.1038/ncomms14728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr A.R., Heldt F.S., Zhang T., Bakal C., and Novák B.. 2016. A Dynamical Framework for the All-or-None G1/S Transition. Cell Syst. 2:27–37. 10.1016/j.cels.2016.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L.V., Kolettas E., Niforou K., Zoumpourlis V.C., et al. 2006. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 444:633–637. 10.1038/nature05268 [DOI] [PubMed] [Google Scholar]
- Baserga R. 1965. The Relationship of the Cell Cycle to Tumor Growth and Control of Cell Division: A Review. Cancer Res. 25:581–595. [PubMed] [Google Scholar]
- Beck H., Nähse V., Larsen M.S., Groth P., Clancy T., Lees M., Jørgensen M., Helleday T., Syljuåsen R.G., and Sørensen C.S.. 2010. Regulators of cyclin-dependent kinases are crucial for maintaining genome integrity in S phase. J. Cell Biol. 188:629–638. 10.1083/jcb.200905059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck H., Nähse-Kumpf V., Larsen M.S., O’Hanlon K.A., Patzke S., Holmberg C., Mejlvang J., Groth A., Nielsen O., Syljuåsen R.G., and Sørensen C.S.. 2012. Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Mol. Cell. Biol. 32:4226–4236. 10.1128/MCB.00412-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benada J., Burdová K., Lidak T., von Morgen P., and Macurek L.. 2015. Polo-like kinase 1 inhibits DNA damage response during mitosis. Cell Cycle. 14:219–231. 10.4161/15384101.2014.977067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoli C., Skotheim J.M., and de Bruin R.A.. 2013. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 14:518–528. 10.1038/nrm3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M.N., Furnari B., Mondesert O., and Russell P.. 1998. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 280:909–912. 10.1126/science.280.5365.909 [DOI] [PubMed] [Google Scholar]
- Brewer J.W., Hendershot L.M., Sherr C.J., and Diehl J.A.. 1999. Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proc. Natl. Acad. Sci. USA. 96:8505–8510. 10.1073/pnas.96.15.8505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E.J., and Baltimore D.. 2000. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 14:397–402. [PMC free article] [PubMed] [Google Scholar]
- Bruinsma W., Aprelia M., García-Santisteban I., Kool J., Xu Y.J., and Medema R.H.. 2017. Inhibition of Polo-like kinase 1 during the DNA damage response is mediated through loss of Aurora A recruitment by Bora. Oncogene. 36:1840–1848. 10.1038/onc.2016.347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdova K., Yang H., Faedda R., Hume S., Chauhan J., Ebner D., Kessler B.M., Vendrell I., Drewry D.H., Wells C.I., et al. 2019. E2F1 proteolysis via SCF-cyclin F underlies synthetic lethality between cyclin F loss and Chk1 inhibition. EMBO J. 38:e101443 10.15252/embj.2018101443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappell S.D., Chung M., Jaimovich A., Spencer S.L., and Meyer T.. 2016. Irreversible APC(Cdh1) Inactivation Underlies the Point of No Return for Cell-Cycle Entry. Cell. 166:167–180. 10.1016/j.cell.2016.05.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappell S.D., Mark K.G., Garbett D., Pack L.R., Rape M., and Meyer T.. 2018. EMI1 switches from being a substrate to an inhibitor of APC/CCDH1 to start the cell cycle. Nature. 558:313–317. 10.1038/s41586-018-0199-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.W., and West S.C.. 2018. GEN1 Endonuclease: Purification and Nuclease Assays. Methods Enzymol. 600:527–542. 10.1016/bs.mie.2017.11.020 [DOI] [PubMed] [Google Scholar]
- Chen Y.J., Dominguez-Brauer C., Wang Z., Asara J.M., Costa R.H., Tyner A.L., Lau L.F., and Raychaudhuri P.. 2009. A conserved phosphorylation site within the forkhead domain of FoxM1B is required for its activation by cyclin-CDK1. J. Biol. Chem. 284:30695–30707. 10.1074/jbc.M109.007997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliby W.A., Roberts C.J., Cimprich K.A., Stringer C.M., Lamb J.R., Schreiber S.L., and Friend S.H.. 1998. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 17:159–169. 10.1093/emboj/17.1.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clijsters L., Hoencamp C., Calis J.J.A., Marzio A., Handgraaf S.M., Cuitino M.C., Rosenberg B.R., Leone G., and Pagano M.. 2019. Cyclin F Controls Cell-Cycle Transcriptional Outputs by Directing the Degradation of the Three Activator E2Fs. Mol. Cell. 74:1264–1277.e7. 10.1016/j.molcel.2019.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman K.E., Grant G.D., Haggerty R.A., Brantley K., Shibata E., Workman B.D., Dutta A., Varma D., Purvis J.E., and Cook J.G.. 2015. Sequential replication-coupled destruction at G1/S ensures genome stability. Genes Dev. 29:1734–1746. 10.1101/gad.263731.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combes G., Alharbi I., Braga L.G., and Elowe S.. 2017. Playing polo during mitosis: PLK1 takes the lead. Oncogene. 36:4819–4827. 10.1038/onc.2017.113 [DOI] [PubMed] [Google Scholar]
- Coudreuse D., and Nurse P.. 2010. Driving the cell cycle with a minimal CDK control network. Nature. 468:1074–1079. 10.1038/nature09543 [DOI] [PubMed] [Google Scholar]
- Cuadrado M., Gutierrez-Martinez P., Swat A., Nebreda A.R., and Fernandez-Capetillo O.. 2009. p27Kip1 stabilization is essential for the maintenance of cell cycle arrest in response to DNA damage. Cancer Res. 69:8726–8732. 10.1158/0008-5472.CAN-09-0729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucchi U., Gianellini L.M., De Ponti A., Sola F., Alzani R., Patton V., Pezzoni A., Troiani S., Saccardo M.B., Rizzi S., et al. 2010. Phosphorylation of TCTP as a marker for polo-like kinase-1 activity in vivo. Anticancer Res. 30:4973–4985. [PubMed] [Google Scholar]
- Daigh L.H., Liu C., Chung M., Cimprich K.A., and Meyer T.. 2018. Stochastic Endogenous Replication Stress Causes ATR-Triggered Fluctuations in CDK2 Activity that Dynamically Adjust Global DNA Synthesis Rates. Cell Syst. 7:17–27.e3. 10.1016/j.cels.2018.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz Z., Halicka H.D., Zhao H., and Podhorecka M.. 2011. Cell synchronization by inhibitors of DNA replication induces replication stress and DNA damage response: analysis by flow cytometry. Methods Mol. Biol. 761:85–96. 10.1007/978-1-61779-182-6_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckbar D., Birraux J., Krempler A., Tchouandong L., Beucher A., Walker S., Stiff T., Jeggo P., and Löbrich M.. 2007. Chromosome breakage after G2 checkpoint release. J. Cell Biol. 176:749–755. 10.1083/jcb.200612047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckbar D., Stiff T., Koch B., Reis C., Löbrich M., and Jeggo P.A.. 2010. The limitations of the G1-S checkpoint. Cancer Res. 70:4412–4421. 10.1158/0008-5472.CAN-09-3198 [DOI] [PubMed] [Google Scholar]
- Dephoure N., Zhou C., Villén J., Beausoleil S.A., Bakalarski C.E., Elledge S.J., and Gygi S.P.. 2008. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA. 105:10762–10767. 10.1073/pnas.0805139105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohadwala M., da Cruz e Silva E.F., Hall F.L., Williams R.T., Carbonaro-Hall D.A., Nairn A.C., Greengard P., and Berndt N.. 1994. Phosphorylation and inactivation of protein phosphatase 1 by cyclin-dependent kinases. Proc. Natl. Acad. Sci. USA. 91:6408–6412. 10.1073/pnas.91.14.6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda H., Arter M., Gloggnitzer J., Teloni F., Wild P., Blanco M.G., Altmeyer M., and Matos J.. 2016. A Mechanism for Controlled Breakage of Under-replicated Chromosomes during Mitosis. Dev. Cell. 39:740–755. 10.1016/j.devcel.2016.11.017 [DOI] [PubMed] [Google Scholar]
- Edgar B.A., and Lehner C.F.. 1996. Developmental control of cell cycle regulators: a fly’s perspective. Science. 274:1646–1652. 10.1126/science.274.5293.1646 [DOI] [PubMed] [Google Scholar]
- Elledge S.J. 1996. Cell cycle checkpoints: preventing an identity crisis. Science. 274:1664–1672. 10.1126/science.274.5293.1664 [DOI] [PubMed] [Google Scholar]
- Eykelenboom J.K., Harte E.C., Canavan L., Pastor-Peidro A., Calvo-Asensio I., Llorens-Agost M., and Lowndes N.F.. 2013. ATR activates the S-M checkpoint during unperturbed growth to ensure sufficient replication prior to mitotic onset. Cell Reports. 5:1095–1107. 10.1016/j.celrep.2013.10.027 [DOI] [PubMed] [Google Scholar]
- Falck J., Mailand N., Syljuåsen R.G., Bartek J., and Lukas J.. 2001. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 410:842–847. 10.1038/35071124 [DOI] [PubMed] [Google Scholar]
- Fotedar A., Cannella D., Fitzgerald P., Rousselle T., Gupta S., Dorée M., and Fotedar R.. 1996. Role for cyclin A-dependent kinase in DNA replication in human S phase cell extracts. J. Biol. Chem. 271:31627–31637. 10.1074/jbc.271.49.31627 [DOI] [PubMed] [Google Scholar]
- Gaillard H., García-Muse T., and Aguilera A.. 2015. Replication stress and cancer. Nat. Rev. Cancer. 15:276–289. 10.1038/nrc3916 [DOI] [PubMed] [Google Scholar]
- Gharbi-Ayachi A., Labbé J.C., Burgess A., Vigneron S., Strub J.M., Brioudes E., Van-Dorsselaer A., Castro A., and Lorca T.. 2010. The substrate of Greatwall kinase, Arpp19, controls mitosis by inhibiting protein phosphatase 2A. Science. 330:1673–1677. 10.1126/science.1197048 [DOI] [PubMed] [Google Scholar]
- Gheghiani L., Loew D., Lombard B., Mansfeld J., and Gavet O.. 2017. PLK1 Activation in Late G2 Sets Up Commitment to Mitosis. Cell Reports. 19:2060–2073. 10.1016/j.celrep.2017.05.031 [DOI] [PubMed] [Google Scholar]
- Girard F., Strausfeld U., Fernandez A., and Lamb N.J.. 1991. Cyclin A is required for the onset of DNA replication in mammalian fibroblasts. Cell. 67:1169–1179. 10.1016/0092-8674(91)90293-8 [DOI] [PubMed] [Google Scholar]
- Goto H., Natsume T., Kanemaki M.T., Kaito A., Wang S., Gabazza E.C., Inagaki M., and Mizoguchi A.. 2019. Chk1-mediated Cdc25A degradation as a critical mechanism for normal cell cycle progression. J. Cell Sci. 132:jcs223123 10.1242/jcs.223123 [DOI] [PubMed] [Google Scholar]
- Grallert A., Boke E., Hagting A., Hodgson B., Connolly Y., Griffiths J.R., Smith D.L., Pines J., and Hagan I.M.. 2015. A PP1-PP2A phosphatase relay controls mitotic progression. Nature. 517:94–98. 10.1038/nature14019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez G.J., Tsuji T., Cross J.V., Davis R.J., Templeton D.J., Jiang W., and Ronai Z.A.. 2010. JNK-mediated phosphorylation of Cdc25C regulates cell cycle entry and G(2)/M DNA damage checkpoint. J. Biol. Chem. 285:14217–14228. 10.1074/jbc.M110.121848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J.C., and Haber J.E.. 2006. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 40:209–235. 10.1146/annurev.genet.40.051206.105231 [DOI] [PubMed] [Google Scholar]
- Hartwell L.H., and Weinert T.A.. 1989. Checkpoints: controls that ensure the order of cell cycle events. Science. 246:629–634. 10.1126/science.2683079 [DOI] [PubMed] [Google Scholar]
- Heald R., McLoughlin M., and McKeon F.. 1993. Human wee1 maintains mitotic timing by protecting the nucleus from cytoplasmically activated Cdc2 kinase. Cell. 74:463–474. 10.1016/0092-8674(93)80048-J [DOI] [PubMed] [Google Scholar]
- Hégarat N., Rata S., and Hochegger H.. 2016. Bistability of mitotic entry and exit switches during open mitosis in mammalian cells. BioEssays. 38:627–643. 10.1002/bies.201600057 [DOI] [PubMed] [Google Scholar]
- Hekmat-Nejad M., You Z., Yee M.C., Newport J.W., and Cimprich K.A.. 2000. Xenopus ATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Curr. Biol. 10:1565–1573. 10.1016/S0960-9822(00)00855-1 [DOI] [PubMed] [Google Scholar]
- Hills S.A., and Diffley J.F.. 2014. DNA replication and oncogene-induced replicative stress. Curr. Biol. 24:R435–R444. 10.1016/j.cub.2014.04.012 [DOI] [PubMed] [Google Scholar]
- Hochegger H., Takeda S., and Hunt T.. 2008. Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nat. Rev. Mol. Cell Biol. 9:910–916. 10.1038/nrm2510 [DOI] [PubMed] [Google Scholar]
- Hsu J.Y., Reimann J.D., Sørensen C.S., Lukas J., and Jackson P.K.. 2002. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APC(Cdh1). Nat. Cell Biol. 4:358–366. 10.1038/ncb785 [DOI] [PubMed] [Google Scholar]
- Hu X., Li Z., Ding Y., Geng Q., Xiahou Z., Ru H., Dong M.-Q., Xu X., and Li J.. 2018. Chk1 modulates the interaction between myosin phosphatase targeting protein 1 (MYPT1) and protein phosphatase 1cβ (PP1cβ). Cell Cycle. 17:421–427. 10.1080/15384101.2017.1418235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.P., and Bartek J.. 2009. The DNA-damage response in human biology and disease. Nature. 461:1071–1078. 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal H., Benada J., Müllers E., Akopyan K., Burdova K., Koolmeister T., Helleday T., Medema R.H., Macurek L., and Lindqvist A.. 2017. ATM/Wip1 activities at chromatin control Plk1 re-activation to determine G2 checkpoint duration. EMBO J. 36:2161–2176. 10.15252/embj.201696082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukov V., and De Nicolo A.. 2018. Aurora-PLK1 cascades as key signaling modules in the regulation of mitosis. Sci. Signal. 11:eaar4195 10.1126/scisignal.aar4195 [DOI] [PubMed] [Google Scholar]
- Kipreos E.T., and van den Heuvel S.. 2019. Developmental Control of the Cell Cycle: Insights from Caenorhabditis elegans. Genetics. 211:797–829. 10.1534/genetics.118.301643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D.K., Hoffmann S., Ahlskog J.K., O’Hanlon K., Quaas M., Larsen B.D., Rolland B., Rösner H.I., Walter D., Kousholt A.N., et al. 2015. Cyclin F suppresses B-Myb activity to promote cell cycle checkpoint control. Nat. Commun. 6:5800 10.1038/ncomms6800 [DOI] [PubMed] [Google Scholar]
- Krajewska M., Heijink A.M., Bisselink Y.J., Seinstra R.I., Silljé H.H., de Vries E.G., and van Vugt M.A.. 2013. Forced activation of Cdk1 via wee1 inhibition impairs homologous recombination. Oncogene. 32:3001–3008. 10.1038/onc.2012.296 [DOI] [PubMed] [Google Scholar]
- Kramer E.R., Scheuringer N., Podtelejnikov A.V., Mann M., and Peters J.M.. 2000. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol. Biol. Cell. 11:1555–1569. 10.1091/mbc.11.5.1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A., and Dunphy W.G.. 2003. Repeated phosphopeptide motifs in Claspin mediate the regulated binding of Chk1. Nat. Cell Biol. 5:161–165. 10.1038/ncb921 [DOI] [PubMed] [Google Scholar]
- Kwon Y.G., Lee S.Y., Choi Y., Greengard P., and Nairn A.C.. 1997. Cell cycle-dependent phosphorylation of mammalian protein phosphatase 1 by cdc2 kinase. Proc. Natl. Acad. Sci. USA. 94:2168–2173. 10.1073/pnas.94.6.2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K., and De Piccoli G.. 2011. Surviving chromosome replication: the many roles of the S-phase checkpoint pathway. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366:3554–3561. 10.1098/rstb.2011.0071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmens B., Hegarat N., Akopyan K., Sala-Gaston J., Bartek J., Hochegger H., and Lindqvist A.. 2018. DNA Replication Determines Timing of Mitosis by Restricting CDK1 and PLK1 Activation. Mol. Cell. 71:117–128.e3. 10.1016/j.molcel.2018.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wang H.Y., Zhao X., Duan H., Cheng B., Liu Y., Zhao M., Shu W., Mei Y., Wen Z., et al. 2019. A methylation-phosphorylation switch determines Plk1 kinase activity and function in DNA damage repair. Sci. Adv. 5:eaau7566 10.1126/sciadv.aau7566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Esposito A., De S., Ber S., Collin P., Surana U., and Venkitaraman A.R.. 2014. Homeostatic control of polo-like kinase-1 engenders non-genetic heterogeneity in G2 checkpoint fidelity and timing. Nat. Commun. 5:4048 10.1038/ncomms5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist A., Rodríguez-Bravo V., and Medema R.H.. 2009. The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J. Cell Biol. 185:193–202. 10.1083/jcb.200812045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C., Sørensen C.S., Kramer E., Santoni-Rugiu E., Lindeneg C., Peters J.M., Bartek J., and Lukas J.. 1999. Accumulation of cyclin B1 requires E2F and cyclin-A-dependent rearrangement of the anaphase-promoting complex. Nature. 401:815–818. 10.1038/44611 [DOI] [PubMed] [Google Scholar]
- Ly T., Whigham A., Clarke R., Brenes-Murillo A.J., Estes B., Madhessian D., Lundberg E., Wadsworth P., and Lamond A.I.. 2017. Proteomic analysis of cell cycle progression in asynchronous cultures, including mitotic subphases, using PRIMMUS. eLife. 6:e27574 10.7554/eLife.27574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macheret M., and Halazonetis T.D.. 2015. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 10:425–448. 10.1146/annurev-pathol-012414-040424 [DOI] [PubMed] [Google Scholar]
- Macůrek L., Lindqvist A., Lim D., Lampson M.A., Klompmaker R., Freire R., Clouin C., Taylor S.S., Yaffe M.B., and Medema R.H.. 2008. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 455:119–123. 10.1038/nature07185 [DOI] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Bartek J., and Lukas J.. 2006. Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell. 23:307–318. 10.1016/j.molcel.2006.06.016 [DOI] [PubMed] [Google Scholar]
- Malumbres M. 2014. Cyclin-dependent kinases. Genome Biol. 15:122 10.1186/gb4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamely I., van Vugt M.A., Smits V.A., Semple J.I., Lemmens B., Perrakis A., Medema R.H., and Freire R.. 2006. Polo-like kinase-1 controls proteasome-dependent degradation of Claspin during checkpoint recovery. Curr. Biol. 16:1950–1955. 10.1016/j.cub.2006.08.026 [DOI] [PubMed] [Google Scholar]
- Mavrommati I., Faedda R., Galasso G., Li J., Burdova K., Fischer R., Kessler B.M., Carrero Z.I., Guardavaccaro D., Pagano M., and D’Angiolella V.. 2018. β-TrCP- and Casein Kinase II-Mediated Degradation of Cyclin F Controls Timely Mitotic Progression. Cell Reports. 24:3404–3412. 10.1016/j.celrep.2018.08.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mchedlishvili N., Jonak K., and Saurin A.T.. 2015. Meeting report--Getting Into and Out of Mitosis. J. Cell Sci. 128:4035–4038. 10.1242/jcs.181107 [DOI] [PubMed] [Google Scholar]
- Medema R.H., and Lindqvist A.. 2011. Boosting and suppressing mitotic phosphorylation. Trends Biochem. Sci. 36:578–584. 10.1016/j.tibs.2011.08.006 [DOI] [PubMed] [Google Scholar]
- Michelena J., Gatti M., Teloni F., Imhof R., and Altmeyer M.. 2019. Basal CHK1 activity safeguards its stability to maintain intrinsic S-phase checkpoint functions. J. Cell Biol. 218:2865–2875. 10.1083/jcb.201902085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshull J., Golsteyn R., Hill C.S., and Hunt T.. 1990. The A- and B-type cyclin associated cdc2 kinases in Xenopus turn on and off at different times in the cell cycle. EMBO J. 9:2865–2875. 10.1002/j.1460-2075.1990.tb07476.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzayans R., Andrais B., Hansen G., and Murray D.. 2012. Role of p16(INK4A) in Replicative Senescence and DNA Damage-Induced Premature Senescence in p53-Deficient Human Cells. Biochem. Res. Int. 2012:951574 10.1155/2012/951574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida S., Maslen S.L., Skehel M., and Hunt T.. 2010. Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science. 330:1670–1673. 10.1126/science.1195689 [DOI] [PubMed] [Google Scholar]
- Moser J., Miller I., Carter D., and Spencer S.L.. 2018. Control of the Restriction Point by Rb and p21. Proc. Natl. Acad. Sci. USA. 115:E8219–E8227. 10.1073/pnas.1722446115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim V., Wilhelm T., Debatisse M., and Rosselli F.. 2013. ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat. Cell Biol. 15:1008–1015. 10.1038/ncb2793 [DOI] [PubMed] [Google Scholar]
- Niida H., Tsuge S., Katsuno Y., Konishi A., Takeda N., and Nakanishi M.. 2005. Depletion of Chk1 leads to premature activation of Cdc2-cyclin B and mitotic catastrophe. J. Biol. Chem. 280:39246–39252. 10.1074/jbc.M505009200 [DOI] [PubMed] [Google Scholar]
- Nilsson J. 2019. Protein phosphatases in the regulation of mitosis. J. Cell Biol. 218:395–409. 10.1083/jcb.201809138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. 1990. Universal control mechanism regulating onset of M-phase. Nature. 344:503–508. 10.1038/344503a0 [DOI] [PubMed] [Google Scholar]
- Ohta S., Kimura M., Takagi S., Toramoto I., and Ishihama Y.. 2016. Identification of Mitosis-Specific Phosphorylation in Mitotic Chromosome-Associated Proteins. J. Proteome Res. 15:3331–3341. 10.1021/acs.jproteome.6b00512 [DOI] [PubMed] [Google Scholar]
- Ohtsubo M., and Roberts J.M.. 1993. Cyclin-dependent regulation of G1 in mammalian fibroblasts. Science. 259:1908–1912. 10.1126/science.8384376 [DOI] [PubMed] [Google Scholar]
- Olsen J.V., Vermeulen M., Santamaria A., Kumar C., Miller M.L., Jensen L.J., Gnad F., Cox J., Jensen T.S., Nigg E.A., et al. 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3:ra3 10.1126/scisignal.2000475 [DOI] [PubMed] [Google Scholar]
- Orthwein A., Fradet-Turcotte A., Noordermeer S.M., Canny M.D., Brun C.M., Strecker J., Escribano-Diaz C., and Durocher D.. 2014. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science. 344:189–193. 10.1126/science.1248024 [DOI] [PubMed] [Google Scholar]
- Pagano M., Theodoras A.M., Tam S.W., and Draetta G.F.. 1994. Cyclin D1-mediated inhibition of repair and replicative DNA synthesis in human fibroblasts. Genes Dev. 8:1627–1639. 10.1101/gad.8.14.1627 [DOI] [PubMed] [Google Scholar]
- Pardee A.B. 1989. G1 events and regulation of cell proliferation. Science. 246:603–608. 10.1126/science.2683075 [DOI] [PubMed] [Google Scholar]
- Parker L.L., and Piwnica-Worms H.. 1992. Inactivation of the p34cdc2-cyclin B complex by the human WEE1 tyrosine kinase. Science. 257:1955–1957. 10.1126/science.1384126 [DOI] [PubMed] [Google Scholar]
- Peschiaroli A., Dorrello N.V., Guardavaccaro D., Venere M., Halazonetis T., Sherman N.E., and Pagano M.. 2006. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell. 23:319–329. 10.1016/j.molcel.2006.06.013 [DOI] [PubMed] [Google Scholar]
- Petermann E., and Caldecott K.W.. 2006. Evidence that the ATR/Chk1 pathway maintains normal replication fork progression during unperturbed S phase. Cell Cycle. 5:2203–2209. 10.4161/cc.5.19.3256 [DOI] [PubMed] [Google Scholar]
- Petermann E., Maya-Mendoza A., Zachos G., Gillespie D.A., Jackson D.A., and Caldecott K.W.. 2006. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol. Cell. Biol. 26:3319–3326. 10.1128/MCB.26.8.3319-3326.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L., and Archambault V.. 2018. A unified view of spatio-temporal control of mitotic entry: Polo kinase as the key. Open Biol. 8:180114 10.1098/rsob.180114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerening J.R. 2009. Positive-feedback loops in cell cycle progression. FEBS Lett. 583:3388–3396. 10.1016/j.febslet.2009.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privette L.M., and Petty E.M.. 2008. CHFR: A Novel Mitotic Checkpoint Protein and Regulator of Tumorigenesis. Transl. Oncol. 1:57–64. 10.1593/tlo.08109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quelle D.E., Ashmun R.A., Shurtleff S.A., Kato J.Y., Bar-Sagi D., Roussel M.F., and Sherr C.J.. 1993. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 7:1559–1571. 10.1101/gad.7.8.1559 [DOI] [PubMed] [Google Scholar]
- Resnitzky D., Gossen M., Bujard H., and Reed S.I.. 1994. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol. Cell. Biol. 14:1669–1679. 10.1128/MCB.14.3.1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S., Mayor-Ruiz C., Lafarga V., Murga M., Vega-Sendino M., Ortega S., and Fernandez-Capetillo O.. 2016. A Genome-wide CRISPR Screen Identifies CDC25A as a Determinant of Sensitivity to ATR Inhibitors. Mol. Cell. 62:307–313. 10.1016/j.molcel.2016.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldivar J.C., Cortez D., and Cimprich K.A.. 2017. Publisher correction: The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 18:783 10.1038/nrm.2017.116 [DOI] [PubMed] [Google Scholar]
- Saldivar J.C., Hamperl S., Bocek M.J., Chung M., Bass T.E., Cisneros-Soberanis F., Samejima K., Xie L., Paulson J.R., Earnshaw W.C., et al. 2018. An intrinsic S/G2 checkpoint enforced by ATR. Science. 361:806–810. 10.1126/science.aap9346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Lindsey-Boltz L.A., Unsal-Kaçmaz K., and Linn S.. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73:39–85. 10.1146/annurev.biochem.73.011303.073723 [DOI] [PubMed] [Google Scholar]
- Schmitt E., Boutros R., Froment C., Monsarrat B., Ducommun B., and Dozier C.. 2006. CHK1 phosphorylates CDC25B during the cell cycle in the absence of DNA damage. J. Cell Sci. 119:4269–4275. 10.1242/jcs.03200 [DOI] [PubMed] [Google Scholar]
- Schwarz C., Johnson A., Koivomagi M., Zatulovskiy E., Kravitz C.J., Doncic A., and Skotheim J.M.. 2018. A Precise Cdk Activity Threshold Determines Passage through the Restriction Point. Mol. Cell. 69:253–264.e5. 10.1016/j.molcel.2017.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorah J., and McGowan C.H.. 2009. Claspin and Chk1 regulate replication fork stability by different mechanisms. Cell Cycle. 8:1036–1043. 10.4161/cc.8.7.8040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki A., Coppinger J.A., Jang C.Y., Yates J.R., and Fang G.. 2008. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 320:1655–1658. 10.1126/science.1157425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C.J. 1993. Mammalian G1 cyclins. Cell. 73:1059–1065. 10.1016/0092-8674(93)90636-5 [DOI] [PubMed] [Google Scholar]
- Sørensen C.S., and Syljuåsen R.G.. 2012. Safeguarding genome integrity: the checkpoint kinases ATR, CHK1 and WEE1 restrain CDK activity during normal DNA replication. Nucleic Acids Res. 40:477–486. 10.1093/nar/gkr697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen C.S., Syljuåsen R.G., Falck J., Schroeder T., Rönnstrand L., Khanna K.K., Zhou B.B., Bartek J., and Lukas J.. 2003. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell. 3:247–258. 10.1016/S1535-6108(03)00048-5 [DOI] [PubMed] [Google Scholar]
- Sørensen C.S., Syljuåsen R.G., Lukas J., and Bartek J.. 2004. ATR, Claspin and the Rad9-Rad1-Hus1 complex regulate Chk1 and Cdc25A in the absence of DNA damage. Cell Cycle. 3:939–945. 10.4161/cc.3.7.972 [DOI] [PubMed] [Google Scholar]
- Spencer S.L., Cappell S.D., Tsai F.C., Overton K.W., Wang C.L., and Meyer T.. 2013. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell. 155:369–383. 10.1016/j.cell.2013.08.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern B., and Nurse P.. 1996. A quantitative model for the cdc2 control of S phase and mitosis in fission yeast. Trends Genet. 12:345–350. 10.1016/S0168-9525(96)80016-3 [DOI] [PubMed] [Google Scholar]
- Svendsen J.M., Smogorzewska A., Sowa M.E., O’Connell B.C., Gygi S.P., Elledge S.J., and Harper J.W.. 2009. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 138:63–77. 10.1016/j.cell.2009.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaffer M.P., Jones A.W., Flynn H.R., Snijders A.P., and Nurse P.. 2016. CDK Substrate Phosphorylation and Ordering the Cell Cycle. Cell. 167:1750–1761.e16. 10.1016/j.cell.2016.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaffer M.P., Jones A.W., Flynn H.R., Snijders A.P., and Nurse P.. 2018. Quantitative Phosphoproteomics Reveals the Signaling Dynamics of Cell-Cycle Kinases in the Fission Yeast Schizosaccharomyces pombe. Cell Reports. 24:503–514. 10.1016/j.celrep.2018.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syljuåsen R.G., Jensen S., Bartek J., and Lukas J.. 2006. Adaptation to the ionizing radiation-induced G2 checkpoint occurs in human cells and depends on checkpoint kinase 1 and Polo-like kinase 1 kinases. Cancer Res. 66:10253–10257. 10.1158/0008-5472.CAN-06-2144 [DOI] [PubMed] [Google Scholar]
- Syljuåsen R.G., Sørensen C.S., Hansen L.T., Fugger K., Lundin C., Johansson F., Helleday T., Sehested M., Lukas J., and Bartek J.. 2005. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol. Cell. Biol. 25:3553–3562. 10.1128/MCB.25.9.3553-3562.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szmyd R., Niska-Blakie J., Diril M.K., Renck Nunes P., Tzelepis K., Lacroix A., van Hul N., Deng L.W., Matos J., Dreesen O., et al. 2019. Premature activation of Cdk1 leads to mitotic events in S phase and embryonic lethality. Oncogene. 38:998–1018. 10.1038/s41388-018-0464-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K. 2012. Cellular reprogramming--lowering gravity on Waddington’s epigenetic landscape. J. Cell Sci. 125:2553–2560. 10.1242/jcs.084822 [DOI] [PubMed] [Google Scholar]
- Thomas Y., Cirillo L., Panbianco C., Martino L., Tavernier N., Schwager F., Van Hove L., Joly N., Santamaria A., Pintard L., and Gotta M.. 2016. Cdk1 Phosphorylates SPAT-1/Bora to Promote Plk1 Activation in C. elegans and Human Cells. Cell Reports. 15:510–518. 10.1016/j.celrep.2016.03.049 [DOI] [PubMed] [Google Scholar]
- Toledo L., Neelsen K.J., and Lukas J.. 2017. Replication Catastrophe: When a Checkpoint Fails because of Exhaustion. Mol. Cell. 66:735–749. 10.1016/j.molcel.2017.05.001 [DOI] [PubMed] [Google Scholar]
- Uhlmann F., Bouchoux C., and López-Avilés S.. 2011. A quantitative model for cyclin-dependent kinase control of the cell cycle: revisited. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366:3572–3583. 10.1098/rstb.2011.0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uto K., Inoue D., Shimuta K., Nakajo N., and Sagata N.. 2004. Chk1, but not Chk2, inhibits Cdc25 phosphatases by a novel common mechanism. EMBO J. 23:3386–3396. 10.1038/sj.emboj.7600328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg J., Manjón A.G., Kielbassa K., Feringa F.M., Freire R., and Medema R.H.. 2018. A limited number of double-strand DNA breaks is sufficient to delay cell cycle progression. Nucleic Acids Res. 46:10132–10144. 10.1093/nar/gky786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vugt M.A., Brás A., and Medema R.H.. 2004. Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol. Cell. 15:799–811. 10.1016/j.molcel.2004.07.015 [DOI] [PubMed] [Google Scholar]
- van Vugt M.A., Gardino A.K., Linding R., Ostheimer G.J., Reinhardt H.C., Ong S.E., Tan C.S., Miao H., Keezer S.M., Li J., et al. 2010. A mitotic phosphorylation feedback network connects Cdk1, Plk1, 53BP1, and Chk2 to inactivate the G(2)/M DNA damage checkpoint. PLoS Biol. 8:e1000287 10.1371/journal.pbio.1000287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron S., Sundermann L., Labbe J.C., Pintard L., Radulescu O., Castro A., and Lorca T.. 2018. Cyclin A-cdk1-Dependent Phosphorylation of Bora Is the Triggering Factor Promoting Mitotic Entry. Dev. Cell. 45:637–650.e7. 10.1016/j.devcel.2018.05.005 [DOI] [PubMed] [Google Scholar]
- Waddington C.H. 1957. The Strategy of the Genes; a Discussion of Some Aspects of Theoretical Biology. Allen & Unwin, London, 262 pp. [Google Scholar]
- Ward I.M., and Chen J.. 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276:47759–47762. 10.1074/jbc.C100569200 [DOI] [PubMed] [Google Scholar]
- Watanabe N., Arai H., Iwasaki J., Shiina M., Ogata K., Hunter T., and Osada H.. 2005. Cyclin-dependent kinase (CDK) phosphorylation destabilizes somatic Wee1 via multiple pathways. Proc. Natl. Acad. Sci. USA. 102:11663–11668. 10.1073/pnas.0500410102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E.R., Brown N.G., Peters J.M., Stark H., and Schulman B.A.. 2019. Posing the APC/C E3 Ubiquitin Ligase to Orchestrate Cell Division. Trends Cell Biol. 29:117–134. 10.1016/j.tcb.2018.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt H.D., Laister R.C., Martin S.R., Arrowsmith C.H., and West S.C.. 2017. The SMX DNA Repair Tri-nuclease. Mol. Cell. 65:848–860.e11. 10.1016/j.molcel.2017.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita K., Yasuda H., Pines J., Yasumoto K., Nishitani H., Ohtsubo M., Hunter T., Sugimura T., and Nishimoto T.. 1990. Okadaic acid, a potent inhibitor of type 1 and type 2A protein phosphatases, activates cdc2/H1 kinase and transiently induces a premature mitosis-like state in BHK21 cells. EMBO J. 9:4331–4338. 10.1002/j.1460-2075.1990.tb07882.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.W., Chung M., Kudo T., and Meyer T.. 2017. Competing memories of mitogen and p53 signalling control cell-cycle entry. Nature. 549:404–408. 10.1038/nature23880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S., Minocherhomji S., Chan K.L., Palmai-Pallag T., Chu W.K., Wass T., Mankouri H.W., Liu Y., and Hickson I.D.. 2013. MUS81 promotes common fragile site expression. Nat. Cell Biol. 15:1001–1007. 10.1038/ncb2773 [DOI] [PubMed] [Google Scholar]
- Yuan J., Ghosal G., and Chen J.. 2009. The annealing helicase HARP protects stalled replication forks. Genes Dev. 23:2394–2399. 10.1101/gad.1836409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Srividhya J., De Luca T., Lee J.H., and Pomerening J.R.. 2014. Uncovering the role of APC-Cdh1 in generating the dynamics of S-phase onset. Mol. Biol. Cell. 25:441–456. 10.1091/mbc.e13-08-0480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarubin T., and Han J.. 2005. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 15:11–18. 10.1038/sj.cr.7290257 [DOI] [PubMed] [Google Scholar]
- Zhang J., de Toledo S.M., Pandey B.N., Guo G., Pain D., Li H., and Azzam E.I.. 2012. Role of the translationally controlled tumor protein in DNA damage sensing and repair. Proc. Natl. Acad. Sci. USA. 109:E926–E933. 10.1073/pnas.1106300109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.B., and Elledge S.J.. 2000. The DNA damage response: putting checkpoints in perspective. Nature. 408:433–439. 10.1038/35044005 [DOI] [PubMed] [Google Scholar]
- Zineldeen D.H., Shafik N.M., and Li S.F.. 2017. Alternative Chk1-independent S/M checkpoint in somatic cells that prevents premature mitotic entry. Med. Oncol. 34:70 10.1007/s12032-017-0932-3 [DOI] [PubMed] [Google Scholar]