Roy et al. highlight a harmful cross-talk that can arise between spindle assembly checkpoint silencing and chromosome biorientation due to the involvement of protein phosphatase 1 in both the processes.
Abstract
Accurate chromosome segregation during cell division requires the spindle assembly checkpoint (SAC), which detects unattached kinetochores, and an error correction mechanism that destabilizes incorrect kinetochore–microtubule attachments. While the SAC and error correction are both regulated by protein phosphatase 1 (PP1), which silences the SAC and stabilizes kinetochore–microtubule attachments, how these distinct PP1 functions are coordinated remains unclear. Here, we investigate the contribution of PP1, docked on its conserved kinetochore receptor Spc105/Knl1, to SAC silencing and attachment regulation. We find that Spc105-bound PP1 is critical for SAC silencing but dispensable for error correction; in fact, reduced PP1 docking on Spc105 improved chromosome segregation and viability of mutant/stressed states. We additionally show that artificially recruiting PP1 to Spc105/Knl1 before, but not after, chromosome biorientation interfered with error correction. These observations lead us to propose that recruitment of PP1 to Spc105/Knl1 is carefully regulated to ensure that chromosome biorientation precedes SAC silencing, thereby ensuring accurate chromosome segregation.
Introduction
During cell division, chromosomes often form syntelic attachments, wherein both sister kinetochores establish end-on attachments with microtubules from the same spindle pole (Fig. 1 A). For accurate chromosome segregation, these erroneous attachments must be corrected before the cell enters anaphase. However, recent studies show that end-on kinetochore–microtubule attachments, whether they are monopolar, syntelic, or bipolar, can silence the spindle assembly checkpoint (SAC; Etemad et al., 2015; Tauchman et al., 2015). To prevent chromosome missegregation, the kinetochore must allow SAC silencing only after bipolar attachments form (Fig. 1 A). How the kinetochore meets this requirement is unclear, because the same enzyme, protein phosphatase 1 (PP1), antagonizes both the SAC and the error correction machinery. PP1 silences the SAC by dephosphorylating the kinetochore protein KNL1/Spc105 to enable anaphase onset (London et al., 2012; Meadows et al., 2011; Nijenhuis et al., 2014; Rosenberg et al., 2011). It stabilizes kinetochore–microtubule attachments by dephosphorylating microtubule-binding kinetochore components such as the Ndc80 complex (Liu et al., 2010; Posch et al., 2010). This dual role of PP1 creates the possibility of a harmful cross-talk between SAC silencing and error correction: if PP1 is recruited for SAC silencing before chromosome biorientation, it can inadvertently stabilize syntelic attachments and thus cause chromosome missegregation. Therefore, it is important to understand how the kinetochore ensures that the correction of syntelic attachments and chromosome biorientation precedes SAC silencing.
Figure 1.
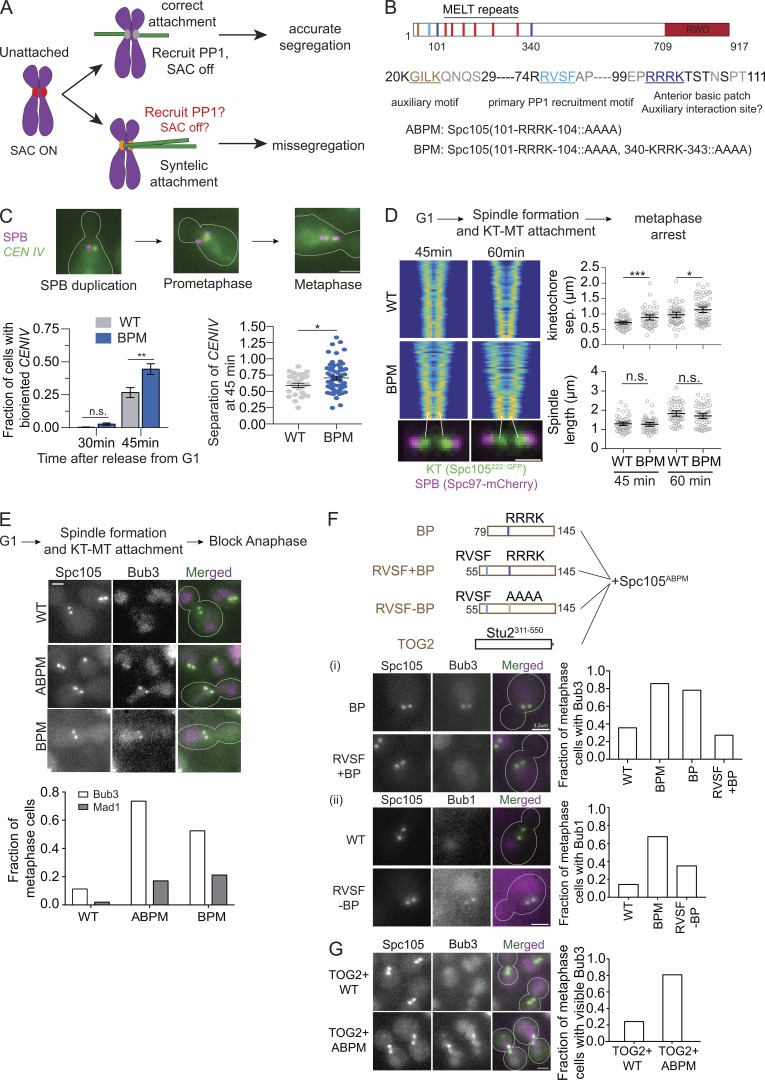
The basic patch near the N-terminus of Spc105 contributes to Glc7 recruitment. (A) Model of how cross-talk between SAC silencing and error correction can interfere with the correction of syntelic attachments and promote chromosome missegregation. (B) Functional domains of Spc105 and the amino acid sequence of its N-terminus. The mutations in Spc105 used in this study are noted at the bottom. (C) Representative micrographs of TetO-TetR-GFP spots. CENIV achieves biorientation faster in cells expressing Spc105BPM compared with WT cells (data presented as mean + SEM; P = ∼0.0041 at 45 min using two-way ANOVA). Sister centromere separation is higher in cells expressing Spc105BPM compared with WT cells, even though the spindle length is not. Scale bar: ∼3.2 µm. The measurements were pooled from three experiments; for WT, n = 273 and 342 at 30 and 45 min, respectively; for BPM, n = 176 and 281 at 30 and 45 min; **, P < 0.01 for the fraction of cells with bioriented CENIV at 45 min; *, P < 0.05 for sister centromere separation at 45 min. (D) Left: V-plots display the normalized distribution of kinetochores along the spindle axis for the indicated strains (n > 50 for each time point). Each row of pixels in the plot represents the symmetrized distribution of Spc105222∷GFP or Spc105BPM,222∷GFP along the spindle axis in one cell. Rows are ranked according to spindle length (see Materials and methods and Marco et al. [2013]). Scale bar: 1.6 µm. Right, top: Average sister kinetochore separation (data presented as mean + SEM; P = 0.0005 [***] and 0.0121 [*] for 45 and 60 min, respectively, using unpaired t test). Right, bottom: Distance between two spindle poles remains unchanged (data presented as mean + SEM; P = 0.6523 and 0.1932 for 45 and 60 min, respectively, using unpaired t test, from two experiments). (E) Top: Workflow. Middle: Representative micrographs of yeast cells expressing the indicated proteins. Scale bar: ∼3.2 µm. Bottom: Frequency of metaphase cells with visible Bub3 and Mad1 at the kinetochores (pooled from two experiments; for Bub3-mCherry, n = 204, 196, and 179, respectively; for Mad1-mCherry, n = 101, 94, and 123). In this and subsequent assays yielding two-category (presence or absence of visible recruitment) scoring data for WT and mutant Spc105, we used Fisher’s exact test for the fractions calculated from the total number of observations. P < 0.0001 for Bub3-mCherry and P < 0.0003 for Mad1-mCherry recruitment. (F) Fusion of an extra basic patch with or without RVSF and their effect on Bub3-Bub1 localization in metaphase cells. Top: Schematics of the fragments fused to the N-terminus of Spc105ABPM. (F, i) Left: Representative micrographs. Scale bar: ∼3.2 µm. Right: Bar graph shows the fraction of cells with visible Bub3-mCherry recruitment (n = 90, 70, 96, and 114, respectively, pooled from two experiments; P < 0.0001 using Fisher’s exact test). (F, ii) Left: Representative micrographs of cells expressing the indicated proteins. Scale bar: ∼3.2 µm. Right: Bar graph shows the fraction of cells with visible Bub1-mCherry recruitment (n = 42, 120, and 103, respectively, pooled from two experiments; P < 0.0152 using Fisher’s exact test). (G) The effect of TOG2-Spc105 fusions on Bub3-mCherry recruitment to bioriented kinetochores. Left: Representative micrographs. Scale bar: ∼3.2 µm. Right: Bar graph showing fraction of metaphase cells with visible Bub3-mCherry recruitment (n = 112 and 144 for TOG2-Spc105 and TOG2-Spc105ABPM, respectively, pooled from two experiments). n.s., not significant.
To study the coordination between the SAC silencing and error correction activities, we investigated the importance of regulated PP1 recruitment by Spc105. PP1 recruitment by Spc105 is necessary for SAC silencing, and it is also suggested to play a role in stabilizing kinetochore–microtubule attachments (Hendrickx et al., 2009; Liu et al., 2010; London et al., 2012; Nijenhuis et al., 2014; Rosenberg et al., 2011). Interestingly, in human cells, Aurora B, the kinase responsible for error correction, down-regulates the interaction between KNL1/Spc105 and PP1. In principle, this regulation can minimize the cross-talk discussed above, but this Aurora B–mediated regulation is thought to be important mainly for robust SAC in unattached kinetochores (Liu et al., 2010; Nijenhuis et al., 2014). Using cell biological and genetic experiments in budding yeast, we find that PP1 is recruited by Spc105 via the combined activities of the conserved RVSF motif and a patch of basic residues downstream from it. However, this PP1 is required only for SAC silencing; it is not required for stabilizing kinetochore–microtubule attachments. In fact, we find that when PP1 is recruited by Spc105 prematurely, i.e., before sister kinetochore biorientation, it inadvertently stabilizes syntelic attachments and interferes with error correction. These observations suggest that the PP1 recruited to the kinetochore via Spc105 can interfere with chromosome biorientation, and that this recruitment is likely to be regulated to minimize this effect.
Results and discussion
The loss of a conserved basic patch near the N-terminus of Spc105 leads to defective SAC silencing and also improves kinetochore biorientation
The N-terminus of Spc105 contains two known PP1 binding sites: the RVSF motif (the primary binding site) and the G/SILK motif (the secondary site, Fig. 1 B). The Aurora B kinase, known as Ipl1 in budding yeast, phosphorylates the RVSF motif, but the abrogation of this phosphoregulation does not appear to have detectable effects on chromosome segregation in budding yeast (Rosenberg et al., 2011). The secondary PP1 binding site in budding yeast lacks a phosphorylatable residue. This suggests the existence of a third element involved in the regulation of PP1 recruitment. Indeed, mutation of a basic patch consisting of four residues in the N-terminus of the Caenorhabditis elegans homologue of Spc105 results in defective SAC silencing, raising the possibility that the basic patch could regulate PP1 activity directly or indirectly (Espeut et al., 2012). The budding yeast Spc105 contains two basic patches: an anterior one spanning residues 101–104 and a posterior one spanning residues 340–343. Notably, the anterior conserved, four-residue basic patch with the sequence RRRK may be subject to phosphoregulation, because the serine and threonine residues located immediately downstream from the basic patch are phosphorylated by mitotic and S-phase kinases (pSYT repository of phosphorylated peptides hosted at the Global Proteome Machine Database; http://gpmdb.thegpm.org/psyt/index.html; original observations from Kanshin et al. [2017] and Smolka et al. [2007]). Therefore, to understand the activity of the basic patch, we created the basic patch mutant (BPM) of Spc105, or Spc105BPM, wherein the basic residues in both basic patches are replaced with nonpolar alanine residues (101-RRRK-104::AAAA, 340-KRRK-343::AAAA). Mutation of either the anterior basic patch (101-RRRK-104::AAAA; referred to as Spc105ABPM) or both basic patches showed no adverse effects on cell growth or viability (Fig. S1 A). This mutation also did not affect the number of Spc105 molecules in the kinetochore (Fig. S1 B).
The earlier study of the function of the basic patch in CeKNL1 suggested that it binds to microtubules (Espeut et al., 2012). Therefore, we first tested how the basic patches of Spc105 contribute to microtubule binding. Single molecules of recombinant Spc105 phosphodomain (residues 2–455 as a part of 6xHIS-MBP-Spc1052-455,222::GFP) did not detectably interact with taxol-stabilized porcine microtubules based on total internal reflection fluorescence (TIRF) microscopy observations (not depicted). Therefore, we tested the binding of Spc105- and Spc105BPM-coated microspheres to microtubules. WT 6xHIS-MBP-Spc1052-455,222::GFP–coated microspheres readily bound to microtubules, and whereas 6xHIS-MBP-Spc1052-455,BPM,222::GFP–coated microspheres did not bind (Fig. S1 C). These results indicate that the basic patches of Spc105 can interact with microtubules. However, since we did not observe the binding of single molecules of the phosphodomain with the microtubule, we expected that the basic patches may contribute only a weak microtubule-binding activity.
We analyzed the effect of Spc105BPM on kinetochore biorientation and force generation in vivo using a centromere proximal TetO array to visualize centromere IV (CENIV). We quantified the fraction of cells that achieve biorientation 30 and 45 min after release from G1 arrest. After 45 min, CENIV achieved biorientation in a significantly larger fraction of cells expressing Spc105BPM compared with WT cells (Fig. 1 C). Interestingly, the separation between the bioriented CENIV was also significantly larger in cells expressing Spc105BPM (Fig. 1 C). We confirmed these results by examining the kinetics of biorientation of all 16 pairs of sister kinetochores. In this experiment, we used GFP-tagged Spc105 to visualize all kinetochores and quantified their distribution over the spindle using a previously described method (Marco et al., 2013). Cells expressing Spc105BPM bioriented their kinetochores faster than cells expressing WT Spc105 (compare the V-plots in Fig. 1 D, left, which displays kinetochore distribution of >50 spindles imaged at the indicated times after release from G1 arrest). These results show that Spc105BPM accelerates kinetochore biorientation. The average distance between the centroids of sister kinetochore clusters was again larger in cells expressing Spc105BPM, even though the spindle length was the same (scatterplots in Fig. 1 D). This increased separation is suggestive of higher force generation by the yeast kinetochore, even though the microtubule-binding activity of the basic patch is absent. In C. elegans and human cells, the basic patch does not affect kinetochore force generation (Espeut et al., 2012; Zhang et al., 2014). Neither the increased sister kinetochore separation nor the accelerated chromosome biorientation can be easily explained by the loss of the microtubule binding activity contributed by the basic patches. They suggest that the basic patches contribute to a function other than force generation.
We next assessed whether Spc105BPM impairs SAC silencing, similar to the reported function of the basic patch in CeKNL1. For this, we quantified the recruitment of Bub3- or Bub1-mCherry to bioriented kinetochores in metaphase-arrested cells (using CDC20 repression; see Materials and methods). Bub3 is an excellent reporter of active SAC signaling. It binds only to phosphorylated MELT motifs in Spc105 as a part of the Bub3-Bub1 complex (Aravamudhan et al., 2015; Hiruma et al., 2015; Ji et al., 2015; London et al., 2012; Primorac et al., 2013). Once stable kinetochore–microtubule attachment forms, PP1, known as Glc7 in yeast, dephosphorylates the MELT motifs to suppress Bub3 recruitment (London et al., 2012). Therefore, in WT cells, only a small minority of metaphase-arrested yeast cells show detectable Bub3 recruitment at bioriented kinetochores (Fig. 1 E). In contrast, under the same conditions in a majority of yeast cells expressing either Spc105BPM or Spc105ABPM, Bub3-mCherry colocalized with bioriented kinetochores (Fig. 1 E). We next examined the recruitment of Mad1 to kinetochores under the same conditions. Mad1 binding to the kinetochore is required to activate the SAC. In contrast to Bub3, visible Mad1 localization was seen in a small fraction of the cells expressing either Spc105BPM or Spc105ABPM (Fig. 1 E, micrographs and quantification shown in Fig. S1 D). Consistent with this observation, the growth rate of Spc105BPM cultures is similar to the growth rate of WT cells (Fig. S1 F).
Our current understanding of the kinetochore suggests two different mechanisms to explain the abnormal Bub3-mCherry recruitment to kinetochores in metaphase-arrested cells. First, the basic patch solely provides microtubule binding activity, and the loss of this activity in Spc105BPM releases the Spc105 phosphodomain from the microtubule lattice, thereby bringing the MELT motifs into proximity of the Mps1 kinase and promoting their phosphorylation (Aravamudhan et al., 2015). Alternatively, Spc105BPM reduces Glc7 recruitment or its activity, and thus suppresses the dephosphorylation of MELT motifs and increases Bub3 recruitment. To test whether the loss of microtubule-binding activity of the basic patch is responsible for abnormal Bub3 recruitment, we asked whether reintroduction of the basic patch at a different location suppresses the abnormal Bub3 recruitment. We fused a fragment of Spc105 containing the basic patch and its surrounding residues to the N-terminus of Spc105ABPM (residues 79–145 [BP]; Fig. 1 Fi). This did not suppress the abnormal Bub3 recruitment. As an alternative approach, we fused the TOG2 domain (Stu2311–550) from Stu2/XMAP215, a known microtubule-binding domain, to the N-terminus of Spc105ABPM (Ayaz et al., 2012). Bioriented kinetochores in the majority of the cells expressing TOG2-Spc105ABPM, but not TOG2-Spc105, recruited Bub3-mCherry, suggesting that microtubule binding by the Spc105 phosphodomain is not sufficient to suppress the Bub3 recruitment/retention (Fig. 1 G). The basic patch likely acts via a novel mechanism.
We next considered whether the basic patch suppresses Bub3-mCherry recruitment by promoting Glc7 recruitment or activity. To test this, we appended a fragment of Spc105 (residues 55–145) containing the RVSF motif either with (RVSF + BP) or without (RVSF – BP) the basic patch to the N-terminus of Spc105ABPM (see schematics at the top of Fig. 1 F). The abnormal Bub3-Bub1 recruitment was suppressed only when the extra RVSF motif was introduced along with the downstream basic patch (RVSF + BP, Fig. 1 F, i and Fii). These data strongly suggest that the RVSF motif and the basic patch act together to suppress Bub3 recruitment, likely through recruiting or activating Glc7 at the yeast kinetochore.
The basic patch promotes Glc7 recruitment to the kinetochore
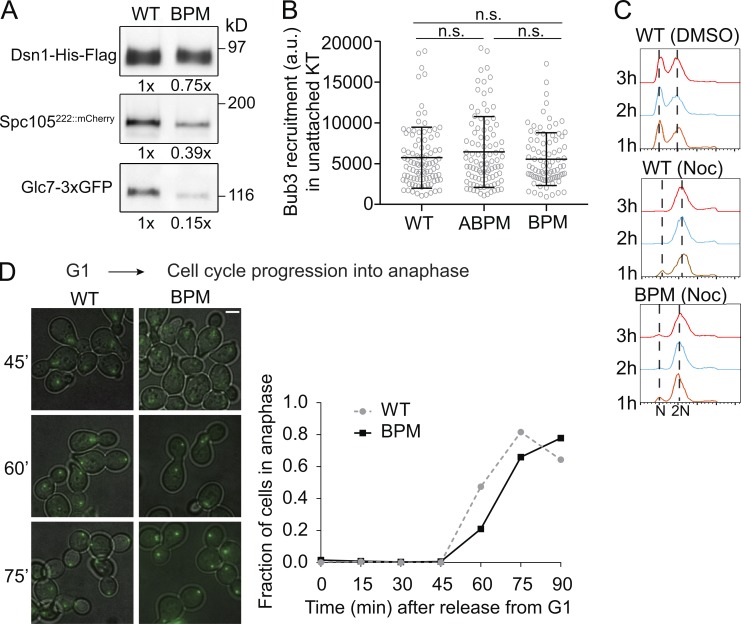
To directly test whether the basic patch contributes to Glc7 binding to Spc105 in vivo, we used the kinetochore particle pulldown assay to assess the interaction of Glc7 with kinetochores in cells expressing Spc105BPM (Gupta et al., 2018). Briefly, we immunoprecipitated Dsn1-Flag to pull down kinetochore particles from yeast cells expressing either Spc105222::mCherry or Spc105BPM,222::mCherry and quantified the amount of Spc105 and Glc7-3xGFP coprecipitating with the kinetochore particles (see Materials and methods). Even though we equalized total protein concentration in each lysate before immunoprecipitation, the amount of Spc105222::mCherry coprecipitating with Dsn1-Flag varied (see Materials and methods for details regarding the imaging and quantification methodology). Therefore, we normalized the amount of Glc7-3xGFP for the level of Spc105222::mCherry in each coprecipitate. Upon this normalization, we found that the level of Glc7 associating with kinetochore particles containing Spc105BPM,222::mCherry was reduced by ∼62–85% (Fig. 2 A; Western blot with loading volumes adjusted for coprecipitated Spc105 amounts from another trial is shown in Fig. S1 E). Thus, the basic patch mutation significantly reduces the interaction of Glc7 with yeast kinetochores. This experiment indicates that the basic patch cooperates with the RVSF motif to achieve normal levels of Glc7 recruitment in yeast kinetochores.
Figure 2.
Spc105BPM does not enhance SAC signaling. (A) Coprecipitation of Glc7-3xGFP with kinetochore particles containing either WT Spc105 or Spc105BPM. Numbers below each lane indicate the band intensity relative to the band intensity for the WT strain. The reduced coprecipitation of Spc105BPM reflects experimental variation and not a reduction in the number of kinetochore-bound molecules (see Fig. S1 B). (B) Bub3-mCherry recruitment by unattached kinetochore clusters in strains expressing the indicated Spc105 variant (mean ± SD; P = 0.2501 obtained from one-way ANOVA, n = 97, 97, and 88, respectively, pooled from two experiments). (C) Flow cytometry of the DNA content from cultures treated with nocodazole (Noc). The 1n and 2n peaks correspond to G1 and G2/M cell populations, respectively (representative histograms from two trials). (D) Left: Representative overlays of transmitted light and fluorescence micrographs of yeast cells at indicated time points after release from G1 arrest. The chart shows the fraction of cells entering anaphase at the indicated time after release from G1 arrest (data pooled from two experiments). It should be noted that the drop in the fraction of anaphase cells in the WT culture at the last time point is because many cells entered the next cell cycle. Scale bar: ∼2.0 µm. Total number of cells analyzed at each time point: WT, n = 206, 182, 385, 276, 274, 250, and 129 at 0, 15, 30, 45, 60, 75, and 90 min, respectively; BPM, n = 331, 353, 286, 306, 414, 408, and 260 at 0, 15, 30, 45, 60, 75, and 90 min, respectively. n.s., not significant.
Spc105BPM does not enhance SAC signaling from unattached kinetochores, but causes a minor delay in metaphase-to-anaphase transition
The lack of Mad1 recruitment to bioriented kinetochores in cells expressing Spc105BPM and normal growth rate indicated that the defect in SAC silencing does not translate into a significant cell cycle delay (Fig. 1 E and S1 F). However, because of the involvement of the basic patch in Glc7 recruitment, we analyzed in detail whether Spc105BPM affects SAC signaling and its silencing. We treated cells expressing either WT Spc105 or Spc105BPM with the microtubule poison nocodazole to depolymerize the spindle and quantified the amount of Bub3-mCherry recruited by unattached kinetochores. In both cases, unattached kinetochores recruited similar amounts of Bub3, indicating that SAC signaling was unaffected by the mutation (Fig. 2 B). This is consistent with our prior finding that Glc7 activity has little influence on Bub3 recruitment in unattached kinetochores (Aravamudhan et al., 2016). We next used flow cytometry to quantify changes in the DNA content of cells treated with nocodazole over time. Spc105BPM had no detectable effect on the DNA content, indicating that the strength of the SAC was not detectably compromised by the mutation (Fig. 2 C). Finally, we tested whether Spc105BPM delays the metaphase-to-anaphase transition by monitoring its timing in a synchronized cell population. Anaphase onset was marginally delayed in cells expressing Spc105BPM, similar to the effect of a similar mutation in C. elegans (Fig. 2 D). Together, these results show that Spc105BPM does not affect SAC signaling from unattached kinetochores but causes a small delay in anaphase onset.
Spc105BPM suppresses benomyl sensitivity of chromosome segregation mutants in an Sgo1-independent manner
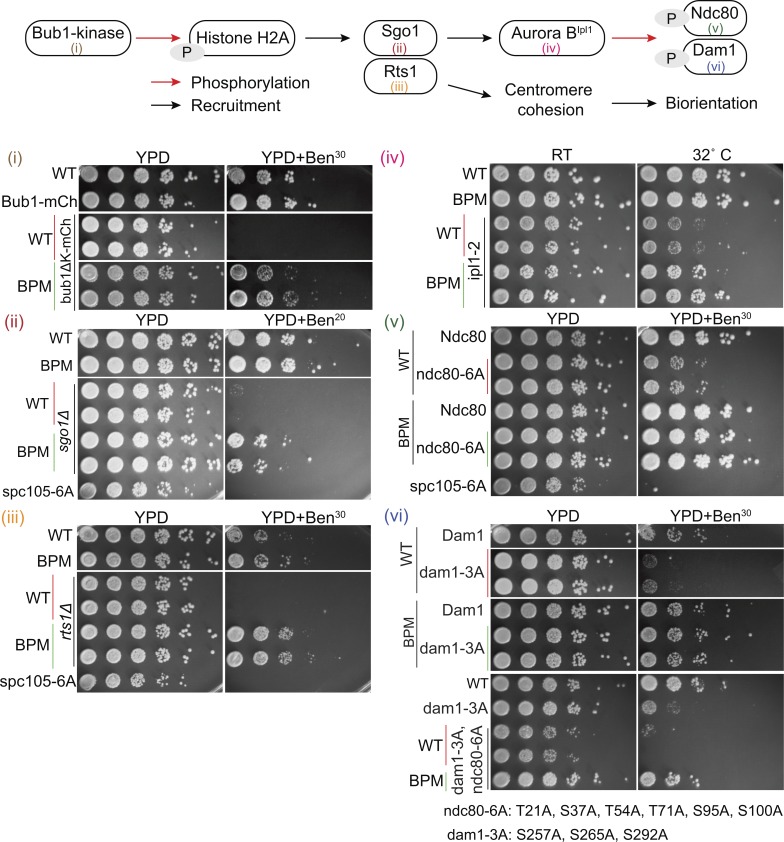
According to the current understanding, reduced Glc7 recruitment by Spc105BPM should reduce the dephosphorylation of microtubule-binding kinetochore proteins, and therefore destabilize kinetochore–microtubule attachments. This should in turn delay kinetochore biorientation and reduce sister kinetochore separation. The faster kinetics of kinetochore biorientation in cells expressing Spc105BPM (Fig. 1 C) is inconsistent with this model. One potential explanation for the observed phenotype is that the higher Bub1 level at the kinetochore promotes Shugoshin (Sgo1) recruitment (Fig. S2 A) and enhances sister centromere cohesion and Aurora B activity, and thus promotes sister kinetochore biorientation ( Salic et al., 2004; Kawashima et al., 2010; Peplowska et al., 2014). To test this possibility, we examined interactions between the genes involved in the Sgo1-mediated error correction pathway and Spc105BPM using the benomyl sensitivity assay (Fig. 3, top panel, schematic of the error correction pathway). In this assay, yeast cells are grown on medium containing low doses of the drug benomyl. In the presence of benomyl, microtubule dynamicity increases, and as a result, the spindle becomes short, oscillatory movements of bioriented kinetochores dampen, and centromeric tension is significantly reduced (Pearson et al., 2003). To proliferate under these conditions, yeast cells must possess a robust SAC signaling and error correction mechanism. Mutant strains that are defective in either SAC signaling (e.g., mad2Δ) or error correction (e.g., sgo1Δ) grow poorly or are inviable specifically on benomyl-containing medium (Fig. 3).
Figure 3.
Spc105BPM suppresses defects in chromosome biorientation and error correction independently of Sgo1. Top: Schematic of the sister centromere cohesion and biorientation pathway in budding yeast. Bottom: Suppression of benomyl sensitivity by Spc105 BPM. Serial dilutions of yeast cells spotted on either rich medium or medium containing either 20 or 30 µg/ml benomyl. WT at the top of each plating indicates WT strain included as a positive control. WT, BPM, or ABPM in other rows refers to the Spc105 allele. spc105-6A, wherein all six MELT motifs are rendered nonphosphorylatable, was used as the negative control.
Yeast strains carrying either the bub1Δkinase mutation, wherein Bub1 lacks its kinase domain, or sgo1Δ are impaired in error correction but competent in SAC signaling (Fig. S2 B). Because of the error correction defect, they grow on normal medium, but they cannot grow on benomyl-containing medium (Fig. 3, i and ii). To test the role of Glc7 binding in error correction, we examined the genetic interactions of the Spc105 mutants and chimeras with bub1Δkinase or sgo1Δ. Strikingly, spc105BPM displayed a positive genetic interaction with both these strains: the double mutants spc105BPM bub1Δkinase and spc105BPM sgo1Δ grew robustly on benomyl-containing medium (Fig. 3, i and ii). These results indicate that reduced PP1 recruitment to the kinetochore via Spc105 results in improved chromosome segregation in cells with an impaired error correction mechanism. Using the positive genetic interaction between spc105BPM and sgo1Δ as a functional readout, we confirmed that the mutation of the anterior basic patch (Spc105ABPM), but not the posteriorly located basic patch (340-KRRK-343::AAAA, referred to as Spc105PBPM), suppresses the benomyl lethality of sgo1Δ cells (Fig. S2 C). Moreover, consistent with the model that the anterior basic patch works in concert with the RVSF motif, the fusion of RVSF – BP, but not RVSF + BP, to the N-terminus of Spc105 suppressed the benomyl sensitivity of bub1Δkinase (Fig. S2 D; see Fig. 1 F for schematics). These results further confirm that the RVSF motif and the basic patch work together to recruit Glc7.
To further elucidate how Spc105BPM promotes accurate chromosome segregation, we studied genetic interactions of spc105BPM and genes involved in kinetochore biorientation and error correction (see the pathway schematic in Fig. 3). spc105BPM suppressed the benomyl lethality of deletion of the gene RTS1 (Fig. 3 iii), which encodes a regulatory subunit of protein phosphatase 2A involved in sister chromatid cohesion (Peplowska et al., 2014; Xu et al., 2009). spc105BPM also suppressed the temperature sensitivity of a strain expressing ipl1-2 (Ipl1-H352Y) that has a weaker kinase activity (Fig. 3 iv). Finally, spc105BPM suppressed the benomyl sensitivity of strains expressing ndc80-6A and dam1-3A, alleles of the microtubule-binding proteins Ndc80 and Dam1, wherein all but one of the known Ipl1 phosphorylation sites in the respective strain are rendered nonphosphorylatable (Fig. 3, v and vi, mutated phosphosites indicated at the bottom; Cheeseman et al., 2002; Lampson et al., 2004; Pinsky et al., 2006b; Tanaka et al., 2002). These mutations normally result in hyperstable kinetochore–microtubule attachments, which interfere with error correction (Akiyoshi et al., 2009). Strikingly, Spc105BPM suppressed these defects, indicating that the accuracy of chromosome segregation is significantly improved in these strains.
The suppression of the benomyl sensitivity of the mutant strains with either reduced Ipl1 kinase activity or nonphosphorylatable Ipl1 substrates by spc105BPM is similar to the well-known compensation of changes in Ipl1 activity with reciprocal changes in Glc7 activity in yeast (Francisco et al., 1994; Pinsky et al., 2006a; Robinson et al., 2012). However, the suppression of the benomyl-sensitivity of the rts1Δ mutant by spc105BPM cannot be explained by this simple model. rts1Δ impairs chromosome biorientation by impairing sister centromere cohesion; it does not affect centromeric recruitment of either Sgo1 or Ipl1, indicating that Ipl1 activity is unaffected by this mutation (Peplowska et al., 2014). Yet, the reduced binding of Glc7 to Spc105BPM leads to significant improvement in chromosome segregation in the rts1Δ mutant. This observation suggests that spc105BPM reduces the reliance of the kinetochore on the error correction pathway.
Spc105BPM improves the accuracy of chromosome segregation in sgo1Δ cells
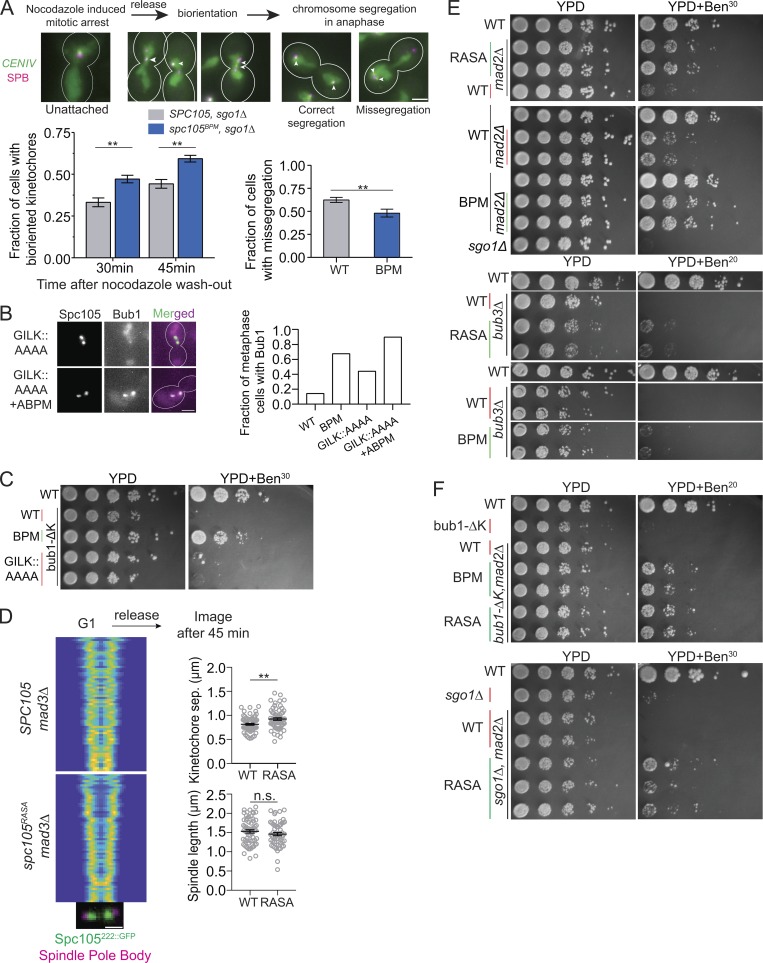
The benomyl lethality of yeast strains carrying mutations in centromeric, kinetochore, and SAC proteins is due to lower accuracy of chromosome segregation. Therefore, the suppression of this benomyl lethality by spc105BPM of these mutants suggests that spc105BPM improves the accuracy of chromosome segregation in these mutant strains. To test this prediction, we monitored chromosome segregation by fluorescently marking CENIV using TetO repeats in sgo1Δ cells. We used this mutant strain background because chromosome missegregation is rare and not easily quantified in WT cells under normal growing conditions (Verzijlbergen et al., 2014). We first destroyed the spindle by treating cells with nocodazole, and then monitored the kinetics of CENIV biorientation. Consistent with our earlier observations, TetO spots marking CENIV separated from one another faster in cells expressing Spc105BPM compared with WT Spc105 (Fig. 4 A, right). Importantly, the frequency of CENIV missegregation was significantly reduced in the sgo1Δ cells expressing Spc105BPM. Following this result, we assessed whether Spc105BPM reduced the incidence of chromosome missegregation compared with WT Spc105 when chromosome biorientation was challenged by benomyl treatment. When mutant and WT cells were synchronized in G1 and then released into the cell cycle in benomyl-containing medium, we found that CENIV marked with TetO repeats missegregated more frequently in WT cells compared with the spc105BPM mutants (Fig. S2 F). Moreover, the spc105BPM cells grew significantly faster in benomyl-containing medium compared with WT cells (growth measured by monitoring OD600, Fig. S2 F). The reduced incidence of chromosome missegregation and significantly faster growth of the spc105BPM mutant compared with even WT cells in medium containing benomyl strongly suggests that the weakening of Spc105-Glc7 interaction results in improved chromosome biorientation.
Figure 4.
Glc7 recruited by the RVSF motif in Spc105 is not required for chromosome biorientation. (A) Top: Workflow used to study chromosome biorientation. Arrowheads show the positions of CENIV foci within the spindle axis in each cell. Scale bar: ∼3.2 µm. Bottom, left: Fraction of cells with bioriented CENIV at the indicated time after nocodazole washout (data presented as mean + SEM; two-way ANOVA revealed P = 0.0066 at 30 min, P = 0.0043 at 45 min; for Spc105WT, n = 434 and 464 at 30 and 45 min, respectively; for Spc105BPM, n = 458 and 411 at 30 and 45 min, accumulated from three repeats). Bottom, right: Fraction of anaphase cells with chromosome IV missegregation quantified 45 min after wash (data presented as mean + SEM; *, P = 0.0079 according to unpaired t test). (B) The fraction of metaphase cells with detectable Bub1-mCherry recruitment to bioriented kinetochores in cells expressing the indicated Spc105 variant (P < 0.0015 for all mutant-WT comparisons using Fisher’s exact test. n = 42, 120, 133, and 87 cells for WT, BPM, GILK::AAAA, and GILK::AAAA+ABPM, respectively, accumulated from two repeats). Scale bar: ∼3.2 µm. (C) Spotting assay of the indicated strains on benomyl-containing medium. (D) Separation between sister kinetochore clusters in the indicated strains and at the indicated time after release from a G1 arrest (analysis performed as in Fig. 1 C, >48 cells displayed). Scale bar: 1.6 µm. Right, top: Separation between two sister kinetochores in metaphase (data presented as mean + SEM; *, P = 0.0014, unpaired t test). Right, bottom: Distance between two sister spindle pole bodies in metaphase (data presented as mean + SEM; P = 0.2285, obtained from unpaired t test). (E and F) Spotting assay of the indicated strains on benomyl-containing medium. n.s., not significant.
Glc7 recruited via the RVSF motif in Spc105 is not required for stabilization of kinetochore–microtubule attachments
The results so far led us to two main conclusions: (a) Spc105BPM recruits a significantly lower amount of Glc7 to the kinetochore, and (b) Spc105BPM accelerates the kinetics of chromosome biorientation under normal growth conditions and improves the accuracy of chromosome segregation when the process of chromosome biorientation is challenged. Together, these conclusions imply that a weakened Spc105–Glc7 interaction impairs SAC silencing but improves the accuracy of chromosome segregation when chromosome biorientation is challenged either by mutations in a wide range of genes involved in the process or by microtubule destabilization due to benomyl. To test this further, we determined whether mutations of the canonical Glc7 binding sites in Spc105, the GILK and RVSF motif, result in phenotypes similar to Spc105BPM. Cells expressing Spc105GILK::AAAA (21-GILK-24::AAAA) showed abnormal recruitment of Bub3-Bub1 to bioriented kinetochores, similar to Spc105BPM (Fig. 4 B). Furthermore, mutation of the anterior basic patch and the GILK motif together (21-GILK-24::AAAA and 101-RRRK-104::AAAA) produced an additive effect: the fraction of metaphase cells recruiting Bub3-Bub1 increased in this double mutant (Fig. 4 B). However, spc105GILK::AAAA by itself did not suppress the benomyl sensitivity of bub1Δkinase strains (Fig. 4 C), suggesting that the GILK motif makes a relatively small contribution to Glc7 recruitment compared with the anterior basic patch.
To determine if Glc7 recruitment via the RVSF motif in Spc105 is required for kinetochore biorientation, we used the spc105RASA allele, which completely blocks Glc7 binding to Spc105. spc105RASA cells arrest in metaphase and cannot grow. Therefore, we used the spc105RASA mad3Δ double mutant to create viable cells (Rosenberg et al., 2011). As before, we analyzed the kinetics of kinetochore biorientation by imaging the distribution of fluorescently labeled kinetochores over the spindle as cells progressed from G1 into the cell cycle. Similar to Spc105BPM, kinetochore clusters in spc105RASA mad3Δ cells were separated by a larger distance compared with mad3Δ cells (Fig. 4 D). This confirms that the recruitment of Glc7 via Spc105 is not required for kinetochore biorientation (Rosenberg et al., 2011).
A weakened interaction between Glc7 and PP1 may improve kinetochore biorientation simply by delaying SAC silencing, and thus providing additional time for kinetochore biorientation (Muñoz-Barrera et al., 2015). Therefore, we exploited the SAC-deficient spc105RASA strains and the benomyl sensitivity assay to directly test whether the accuracy of chromosome segregation was improved in the absence of a functional SAC. Because the SAC is inactive in these strains (Fig. S2 E), any improvement in the accuracy of chromosome segregation must occur because of improved sister kinetochore biorientation and error correction. Strikingly, spc105RASA mad2Δ grew robustly on benomyl-containing medium, even though mad2Δ grew poorly under the same conditions (Fig. 4 E). spc105RASA also partially suppressed the benomyl lethality of bub3Δ. The milder rescue likely reflects the additional defects due to bub3Δ that include lower Sgo1 recruitment to the centromere and defects in APC/C function (Fig. 4 E; Yang et al., 2015). spc105BPM also similarly suppressed the benomyl sensitivity due to mad2Δ and mildly suppressed the benomyl lethality due to bub3Δ. Most strikingly, the triple mutants spc105RASA mad2Δ bub1Δkinase and spc105RASA mad2Δ sgo1Δ also grew on benomyl-containing medium (Fig. 4 F). Thus, the improved accuracy of chromosome segregation in these strains is not because of delayed SAC silencing.
Taken together, our results suggest that Glc7 recruitment by the RVSF motif of Spc105 is not required for stabilizing kinetochore–microtubule attachments; on the contrary, our results imply that the Glc7 recruited by Spc105 may stabilize syntelic kinetochore–microtubule attachments and interfere with the error correction mechanism. A surprising and important observation supporting this view is that inactivation of the RVSF motif improves chromosome segregation in strains harboring mutations in genes involved in chromosome biorientation subjected to stress by low concentrations of benomyl (Fig. 3).
Artificial tethering of PP1 to Spc105 before biorientation has a deleterious effect on chromosome segregation
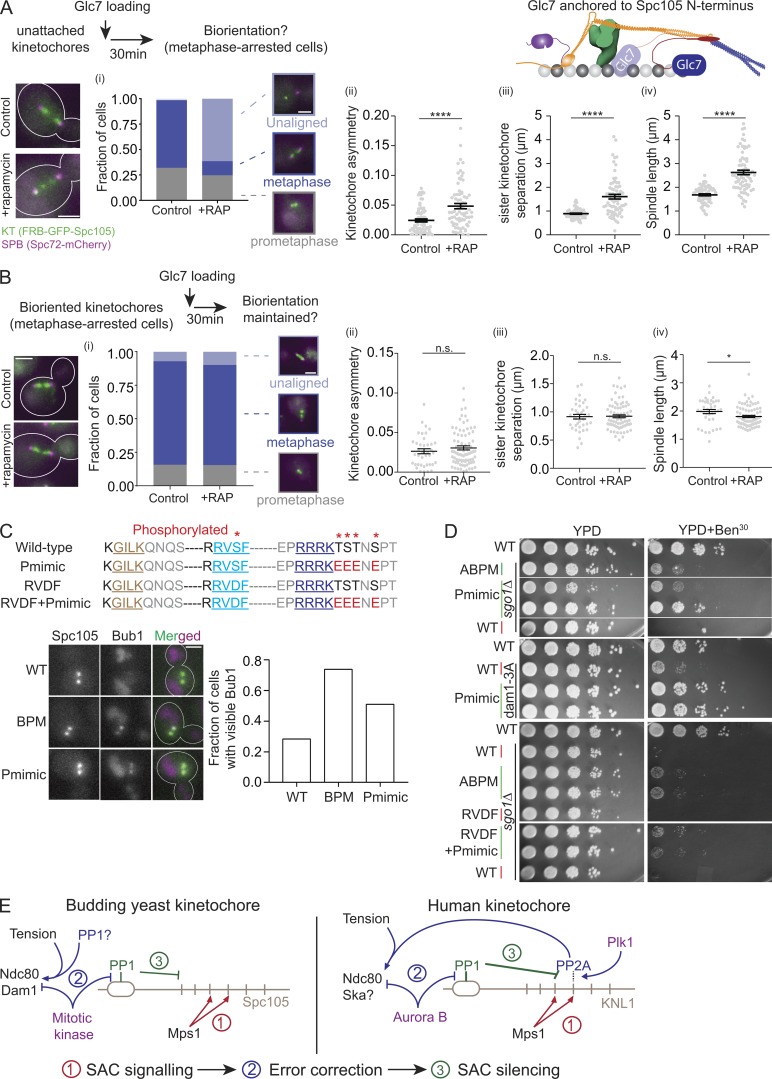
To directly test whether Glc7 recruitment via Spc105 interferes with chromosome biorientation, we used rapamycin-induced dimerization of the Fkbp12 and Frb domains to artificially tether Glc7 at the N-terminus of Spc105 (Haruki et al., 2008). In this experiment, we tethered Glc7 close to its normal binding sites in Spc105 by adding rapamycin to the growth medium either before or after sister kinetochore biorientation, and then monitored its effect on kinetochore biorientation and attachment. These cells express both Spc105 and Frb-Spc105 to avoid tethering abnormally high amounts of Glc7.
We first monitored the effect of Glc7 tethering to Spc105 before chromosome biorientation. For this, we arrested cells in mitosis by treating them with nocodazole, and then tethered Glc7 to Spc105 by treating the cells with rapamycin (workflow depicted at the top of Fig. 5 A). In nocodazole-treated cells, Bub3 recruitment to unattached kinetochores remained unchanged even after rapamycin treatment, indicating that SAC signaling remained unaffected by the treatment (Fig. S3 A). We next released these cells from the mitotic block and examined kinetochore distribution over the mitotic spindle 30 min after nocodazole washout. To ensure that the potentially faster SAC silencing by the tethered Glc7 did not induce premature anaphase onset, we also blocked anaphase onset in these cells by depleting Cdc20. After 30 min, the majority of untreated cells displayed normal spindle morphology, with two bioriented kinetochore clusters (Fig. 5 A, left). In contrast, most rapamycin-treated cells showed abnormal kinetochore–microtubule attachment and spindle morphology (Fig. 5 A, bar graph). The morphological defects included (a) unequal distribution of kinetochores between the two spindle pole bodies (kinetochore asymmetry, Fig. 5 A, i, quantified by calculating the absolute difference between the normalized intensity of the brightest pixel in each spindle half), (b) unaligned kinetochores along the spindle axis, (c) reduced separation between kinetochore clusters and spindle pole bodies (0.27 ± 0.02 µm instead of the normal 0.43 ± 0.11 µm; mean ± SEM; Fig. S3 B), (d) increased separation between sister kinetochores (Fig. 5 A, iii), and (e) abnormally long spindles (Fig. 5 A, iv). The unequal distribution of kinetochores between the two spindle pole bodies is a hallmark of defective chromosome biorientation (Marco et al., 2013). The significant increase in the spindle length also indicates that there is a smaller number of bioriented chromosomes opposing the outward forces generated within the spindle (Bouck and Bloom, 2007).
Figure 5.
Delayed recruitment of Glc7 for SAC silencing is necessary for chromosome biorientation. (A) Top: Workflow used to tether Glc7 at the N-terminus of Spc105 under prophase-like conditions. Cartoon at the right: 2D schematic of the yeast kinetochore. Bottom left: Micrographs depicting the indicated proteins. Scale bar: ∼3.2 µm. (A, i) Bar graph depicts the scoring of cells based on spindle morphology (key provided on the right, n = 126 and 89 for control and rapamycin-treated samples, respectively, pooled from three technical repeats performed on two biological replicates). Scale bar in magnified inset images: ∼3.2 µm. (A, ii–iv) Scatter plots display the indicated quantity in untreated control and rapamycin-treated cells. The data are presented as mean ± SEM. For ii, n = 88 and 91 control and rapamycin-treated cells, respectively; data were pooled from three experimental repeats, ****, P < 0.0001 by unpaired t test); for iii, n = 87 control and rapamycin-treated cells pooled from three experimental repeats, ****, P < 0.0001 by unpaired t test; and for iv, n = 127 and 153 control and rapamycin-treated cells pooled from three experimental repeats, ****, P < 0.0001 by unpaired t test. (B) Workflow for tethering Glc7 to the N-terminus of Spc105 in metaphase-arrested cells. Left: Micrographs depicting the indicated proteins. Scale bar: ∼3.2 µm. (B, i) Bar graph indicates the scoring of cells based on spindle morphology (key provided on the right; n = 70 and 142 for control and rapamycin-treated cells, respectively, pooled from three technical repeats). Scale bar for magnified inset images: ∼3.2 µm. (B, ii–iv) Scatter plots display the indicated quantity in untreated control and rapamycin-treated cells. The data are shown as mean ± SEM. For ii, n = 39 and 80 control and rapamycin-treated cells pooled from two experimental repeats, P = 0.3298 by unpaired t test. In iii and iv, n = 38 and 80 control and rapamycin-treated cells pooled from two experimental repeat; for iii, P = 0.8844 by unpaired t test; for iv, *, P = 0.0244 by unpaired t test. (C) Top: Residues downstream from the basic patch that are known to be phosphorylated are marked with an asterisk at the top. Lower sequences display the mutants used in this study. Bottom: Representative images and quantification of the fraction of metaphase cells with detectable Bub1-mCherry recruitment to bioriented kinetochores. P < 0.0005 for the comparisons displayed on the graph by Fisher’s exact test. We pooled the data from two experimental repeats where we analyzed 169, 103, and 133 cells for WT, BPM, and Pmimic, respectively. Scale bar: ∼3.2 µm. (D) Spotting assay of the indicated Spc105 mutants on benomyl-containing medium. (E) Left: Model mechanisms that mitigate harmful cross-talk between the SAC silencing and error correction in budding yeast kinetochore. Right: Schematic of the proposed model for the human kinetochore. Dashed line indicates indirect recruitment of PP2A via Mad3/BubR1, which is promoted by the Plk1 kinase. n.s., not significant.
We next studied the effects of Glc7 tethering to Spc105 after the process of chromosome biorientation is complete (Fig. 5 B, schematic at the top). For this, we first repressed CDC20 expression to block anaphase onset, providing sister kinetochores with sufficient time to form bipolar attachments, and then added rapamycin to tether Glc7 to the N-terminus of Spc105. In this case, Glc7 tethering had no discernible effects on the metaphase spindle (scatterplots in Fig. 5 B): the spindle morphology was nearly identical with or without rapamycin treatment, kinetochores were symmetrically distributed in two distinct clusters, and spindle length decreased slightly. Thus, Glc7 tethering to Spc105 after kinetochore biorientation does not adversely affect kinetochore–microtubule attachments.
It should be noted that a prior study has shown that Glc7 fusion to the N-terminus of Spc105RASA generates a viable strain with intact SAC signaling, which is inconsistent with the results above (Rosenberg et al., 2011). However, we found that chromosome biorientation was significantly delayed in these cells compared with a WT strain of the same strain background, likely because a significant number of cells showed unaligned and unattached kinetochores (Fig. S3 C). This strain is also benomyl sensitive, likely because of defects in chromosome segregation.
These experiments demonstrate that a premature recruitment of Glc7, before kinetochore biorientation, interferes with kinetochore biorientation by stabilizing syntelic attachments. However, Glc7 recruitment after kinetochore biorientation does not detectably affect kinetochore–microtubule attachment. These data strongly suggest that the recruitment of Glc7 via Spc105 must be delayed until sister kinetochores establish bipolar attachments.
Putative phosphoregulation adjacent to the basic patch may reduce its activity
As mentioned previously, the serine and threonine residues immediately downstream from the basic patch are phosphorylated by mitotic kinases (pSYT; also see Materials and methods). This phosphorylation may interfere with the activity of the basic patch. To test this idea, we replaced the reported phosphorylated residues (marked with an asterisk in Fig. 5 C) with negatively charged residues (Fig. 5 C, top panel). Similar to Spc105BPM, Bub1-mCherry recruitment to bioriented kinetochore clusters was elevated in cells expressing the phosphomimic allele (Fig. 5 C, bottom panel). This allele suppressed the benomyl sensitivity of sgo1Δ as well as the nonphosphorylatable dam1-3A allele (Fig. 5 D). A phosphomimic mutation in the RVSF motif alone did not rescue the benomyl sensitivity of sgo1Δ cells (Fig. 5 D, lower panel).
In summary, our results indicate that Spc105-bound PP1 is critical for SAC silencing but dispensable for the stabilization of kinetochore–microtubule attachments. Surprisingly, we find that reduced PP1 docking on Spc105 improves chromosome biorientation and viability in mutant strains with impaired chromosome biorientation subjected to experimental challenges. In addition, artificial tethering of PP1 to Spc105 before chromosome biorientation had a severe negative effect on chromosome segregation. Thus, our observations highlight the potential for harmful cross-talk between SAC silencing and chromosome biorientation and error correction due to their common regulation by PP1. We propose that this deleterious cross-talk is avoided by regulating the delivery of PP1 until after biorientation. We additionally suggest that phosphoregulation of the Spc105-Glc7 interaction may be important for this regulated delivery (Fig. 5 E, left panel).
In addition to highlighting the importance of tightly regulated PP1 delivery, our results suggest that the PP1 activity required for attachment stabilization is not derived from the PP1 bound to the RVSF motif of Spc105 (Espeut et al., 2012; Rosenberg et al., 2011; Zhang et al., 2014). A recent study suggests that the motor protein Cin8 transports PP1/Glc7 to the kinetochore (Suzuki et al., 2018). However, it is unclear how this mechanism would deliver Glc7 preferentially to kinetochores with correct, but not incorrect, attachments. It is also possible that this activity comes from diffusive interactions between PP1 and the kinetochore. A dedicated Glc7 recruitment mechanism may not even be necessary, as the intercentromeric tension generated by sister kinetochores selectively stabilizes bipolar attachments (Akiyoshi et al., 2010; Franck et al., 2007; Miller et al., 2016).
To achieve accurate chromosome segregation in human cells, similar mechanisms can be expected to bring about the ordered execution of chromosome biorientation first, and then SAC silencing. Even though a more complex network of three kinases regulates these two processes (Fig. 5 E, right panel), their targets, including KNL1, are similar (Etemad et al., 2015; Hiruma et al., 2015; London et al., 2012; Nijenhuis et al., 2014; Suijkerbuijk et al., 2012). Aurora B regulates PP1 recruitment by phosphorylating the RVSF motif and the SILK motif, which, interestingly, lie directly adjacent to the basic patch in KNL1 (Liu et al., 2010; Welburn et al., 2010). A recent in vitro study found that Knl1 binding to the microtubule and PP1 is mutually exclusive and proposed a model wherein microtubule binding contributes to regulated PP1 binding to KNL1 (Bajaj et al., 2018). However, this model does not explain why microtubule binding by KNL1 is needed in the first place if (a) PP1 binding is inhibited by phosphorylation in unattached kinetochores, where microtubules are absent, and (b) the stronger PP1 binding is expected to displace the microtubule from KNL1 anyway, when microtubules are present. Thus, the functional significance of the microtubule-binding activity of basic patch in KNL1, which does not contribute to kinetochore force generation, needs to be better understood (Espeut et al., 2012; Zhang et al., 2014). Our findings suggest a new model (Fig. 5 E, right panel), wherein Aurora B suppresses PP1 recruitment by KNL1 to ensure that error correction is completed before SAC silencing takes place. In line with this, kinetochore recruitment of PP1 peaks after chromosome biorientation (Liu et al., 2010).
Materials and methods
Plasmid and strain construction
Budding yeast strains and plasmids used in this study are listed in Tables S1 and S2, respectively. Strains containing multiple genetic modifications were constructed using standard yeast genetics. GFP(S65T)- and mCherry-tagged proteins were used to localize kinetochores by microscopy. The C-terminal tags and gene deletion cassettes were introduced at the endogenous locus through homologous recombination of PCR amplicons (Bähler et al., 1998). A 7–amino acid linker (sequence RIPGLIN) separates the tag (GFP and mCherry) from the C-terminus of the tagged protein. We have previously observed that the intensity of mCherry-tagged kinetochore proteins varies significantly from strain to strain due to inherent variability of the brightness of mCherry. Therefore, we created all Bub3-mCherry and Mad1-mCherry strains by crossing a specific transformant of Bub3-mCherry or Mad1-mCherry with strains expressing WT Spc105 or its mutants. Mad1-mCherry is always accompanied by nup60Δ to prevent it from localizing to the nuclear envelopes (Scott et al., 2005).
Every Spc105 chimera contains a 397-bp upstream and a 250-bp downstream sequence as promoter (prSPC105) and terminator (trSPC105), respectively. We inserted 711-bp GFP (S65T) fragment (237 amino acids) at the 222nd amino acid position of Spc105 by subcloning, introducing an extra BamHI site (Gly-Ser) upstream of the GFP fragment and a NheI site (Ala-Ser) downstream of the GFP fragment. To insert mCherry using the same BamHI and NheI sites, we used a 705-bp mCherry fragment (235 amino acids) that was codon-optimized for yeast. We introduced mutations in basic patches (amino acids 101–104 RRRK and 340–343 KRRK) and phosphorylation sites (S77, amino acids 105–107, and S109) by subcloning as well. We incorporated anterior basic patch mutation by initially incorporating an extra BspEI site that was later removed by site-directed mutagenesis to create pAJ525. To create posterior basic patch mutation, we introduced a silent mutation that created a MluI site, which aided us in introducing the mutation at amino acids 340–343. Likewise, we introduced a silent mutation creating a SacII site 9 bp upstream of phosphorylation sites (amino acids 105–107 TST and S109) which helped to build the phosphomimetic chimeras. To construct Spc105 chimeras with N-termini fusion (Spc105 79–145, Spc105 55–145, and TOG2), we introduced an extra BspEI site (Ser-Gly) upstream and a MluI site (Thr-Arg) downstream, so that we could integrate the fragments after the start codon of Spc105. The TOG2 fragment also contains a C-terminal linker peptide of AGGA. FRB-GFP fusion in plasmid pAJ603 consists of 279-bp FRB (93 amino acids) and 711-bp GFP, which are linked with each other by a 30-bp fragment which codes for LESSGSGSGS. The whole fragment is ligated within BamHI–NheI sites after the start codon of Spc105.
To build haploid strains expressing Spc105222::GFP alleles (WT or mutant with GFP inserted at amino acid 222), first we deleted a WT genomic copy of SPC105 in a diploid strain of YEF473 to form the strain AJY3278 (SPC105/spc105Δ::NAT1). Then we transformed this strain with the chimeras with pRS305, after linearizing the plasmid with BstEII. We sporulated the correct transformants to obtain nourseothricin-resistant and leucine prototroph segregants. We linearized the plasmids based on pRS306 by StuI before transformation. To visualize centromere 4 segregation, first we built a diploid strain that is homozygous for CENIV-tetO, tetR-GFP, and SPC29-mCherry but heterozygous for SPC105 (AJY5160). We digested pAJ817 (WT) and pAJ818 (BPM) by SacII–KpnI before transforming them in this strain. After that, we sporulated the correct transformants to obtain the haploid strains.
We obtained the pSB148 from the Biggins laboratory (Fred Hutchinson Cancer Research Center, Seattle, WA). In this plasmid, the IPL1 ORF is flanked by a 1.0-kb upstream sequence as promoter (prIPL1) and a 654-bp downstream sequence as terminator (trIPL1). We performed site-directed mutagenesis with the QuikChange kit (Agilent Technologies) to introduce the ipl1-2 (H352Y) mutation in the IPL1 ORF. We acquired sgo1Δ and rts1Δ from the yeast deletion library (Giaever et al., 2002). The strains with sgo1Δ demonstrated variable growth. Hence, we backcrossed them with YEF473 to remove any background mutations. We tested both normal and slow-growing segregants for growth assays on yeast extract peptone dextrose (YPD; yeast extract 1%, peptone 2%, and dextrose 2%) and YPD + benomyl. We confirmed the strains with bub1-ΔK (kinase-deleted bub1) using Western blot with anti-mCherry antibody and localization at unattached kinetochores.
To build the strains containing ndc80-6A or dam1-3A, first we deleted one allele of NDC80 or DAM1 ORF with TRP1 marker. Next, we replaced the deleted allele with ndc80-6A (KAN) or dam1-3A (KAN) by transforming the heterozygous deletion strains of NDC80 and DAM1 with SacII–ApaI digest of pAJ108 (ndc80-6A) and SacII–SmaI digest of pSB617 (dam1-3A), respectively. After confirming the integration of the cassettes, we sporulated the diploids to obtain tryptophan auxotroph and G418-resistant segregants. We also confirmed the haploid strains with ndc80-6A and dam1-3A by colony check PCR, followed by Sanger sequencing.
To construct the plasmids containing 6XHIS-MBP-Spc1052-455,222::GFP, we used a pET28a chimera in which maltose-binding protein (MBP) is already cloned within NheI–BamHI sites. 6xHIS and MBP are separated by a linker of 13 amino acids (SSGLVPRGSHMAS), and a TEV protease cleavage site is present at the downstream of MBP. We cloned fragments containing SPC1052-455,222::GFP of WT and BPM within SalI–EagI sites that are present downstream of MBP-TEV. Hence translation of the whole chimera will result in expression of a 1,123–amino-acid-long protein in which 6xHIS-MBP-TEV is linked with the phosphodomain by a 9–amino-acid-long peptide fragment (GSEFELRRP).
Phosphorylation of the N-terminus of Spc105
The posttranslational modifications of Spc105 are cataloged in the pSYT database. The entry for Spc105 within this database can be viewed at the Global Protein Machine Database (Craig et al., 2004) at http://psyt.thegpm.org/~/dblist_pep_modmass/label=YGL093W&modmass=80@STY&display=0.
Cell culture
We grew yeast strains in YPD at RT (25°C) and 32°C. For strains with W303 background, we supplemented the medium with 0.1 mg/ml adenine. To form zygotes for constructing diploid strains, we mixed overnight-grown cultures of two strains of opposite mating types and spotted them on YPD plates, which were incubated for 3–4 h at 32°C. To induce sporulation, we grew diploid yeast cells in YPD overnight to stationary phase. Then the cells were pelleted down and resuspended with starvation medium (yeast extract 0.1% and potassium acetate 1%) and incubated for 4–5 d at RT. We prepared YPD liquid supplemented with benomyl (30 µg/ml) as described previously (Gupta et al., 2018).
Metaphase arrest by Cdc20 depletion
We synchronized the strains expressing Cdc20 from prMET3 by treating them with α factor (2 µg/ml) for 2 h in synthetic dextrose medium lacking methionine. After that, they were washed to remove α factor and released in YPD supplemented with 2 M methionine to knock down Cdc20 expression for 1–2 h. To release from Cdc20 repression-mediated metaphase arrest, we washed the cells with synthetic medium lacking methionine and incubated them in the same.
Kinetochore particle pulldown experiments
We used strains expressing Dsn1-His-Flag, Glc7-3XGFP, and either Spc105222::mcherry or Spc105BPM,222::mCherry. We purified the kinetochores as previously described (Gupta et al., 2018). Briefly, we harvested and lysed cells in a blender in the presence of liquid nitrogen and prepared clear lysate by ultracentrifugation at 24,000 rpm for 90 min at 4°C. Equal amounts of precleared lysates were incubated with α-Flag-M2–conjugated magnetic Dynabeads at 4°C for 150 min. The beads were then washed, and Flag-tagged protein was eluted from the beads by incubation with 0.5 mg/ml 3xFlag peptide solution at RT for 30 min. Western blotting was performed using commercial antibodies (α-Flag M2 [Sigma-Aldrich], 1:5,000; α-Ds-Red [Santa Cruz Biotechnologies], 1:2,000; α-GFP, JL-8 [Living Colors], 1:3,000). The primary antibodies were detected using HRP-conjugated secondary antibodies per the manufacturer’s instructions. The resulting bioluminescence was detected and quantified using the C600 imager from Azure Biosystems. The band intensities from the Western blot were measured using ImageJ (National Institutes of Health).
Treatment of cells to study spindle pole and CENIV separation
For this experiment, we used strains with TetO-CENIV, TetR-GFP, and Spc29-mCherry. For the assay mentioned in Fig. 1 C, we synchronized them at G1 stage with α factor (2 µg/ml) for 105 min. They were then washed and released in YPD, and cell samples were collected at 30, 45, and 60 min. For the assay mentioned in Fig. 4 A, we used strains of 5267 (Spc105WT, sgo1Δ) and 5273 (Spc105BPM, sgo1Δ) and synchronized them at G1 stage with α factor (2 µg/ml) for 105 min. They were then washed and released in nocodazole-supplemented (15 µg/ml) medium to disrupt the spindle and activate the spindle checkpoint for 2 h. After that, we again washed them to remove nocodazole and released them in fresh YPD. Cell samples were collected and imaged at 0, 30, and 45 min.
To set a time course for imaging experiments involving benomyl supplementation, starting from overnight inoculums, we grew the cells in fresh YPD for 1-1.5 h before adding α factor (2 µg/ml) to synchronize the cells at G1 for 105 min. After that, we washed the cells to remove α factor and released them from G1 by resuspending them in YPD + benomyl liquid medium. We collected aliquots of cell samples at 105 and 135 min to image them.
Benomyl sensitivity assay
We prepared 10-fold serial dilutions of log-phase cultures starting from 0.1 OD600 and spotted them on YPD or YPD containing 20 or 30 µg/ml benomyl (YPD + ben20 and YPD + ben30, respectively). Plates were incubated at 32°C, and plate images were taken after 2 d (on YPD) or 3 d (YPD + benomyl). All benomyl spotting assays were performed with at least two or three biological replicates wherever possible and two technical replicates on both YPD + ben20 and YPD + ben30, because benomyl is not readily soluble in medium, and it also has relatively poor thermal stability. Both these factors are problematic when making agar medium, because benomyl must be dissolved in the agar once it reaches ∼60°C. Thus, we find that the effect of benomyl in YPD plates on control strains varies slightly from one batch to another. Therefore, we performed each spotting experiment at two different benomyl concentrations. Furthermore, we included a WT positive control that should grow and a negative control strain that should grow either poorly or not at all in YPD + benomyl. If the strains behave as expected at both benomyl concentrations, we show the results from the higher concentration.
For the spotting assay shown in Fig. 4 F, we incubated the ben30 plate for 5 d at 30°C, as the growth rates of sgo1Δ and mad2Δ double mutant are poorer than that of sgo1Δ single mutant even in YPD. One should compare the growth of the double mutant of mad2Δ and bub1-ΔK (Fig. 4 F, top panel) or the double mutant of mad2Δ and sgo1Δ (bottom panel) in the background of WT and BPM or RASA in benomyl-containing medium.
96-well plate liquid culture assay
We prepared this experiment as described previously (Hung et al., 2018). To set a 96-well plate liquid culture, we used log-phase cultures of 0.05 OD600 as determined by a standard spectrophotometer. The cells were then pelleted down, resuspended, and diluted with YPD + benomyl liquid or YPD control. We dispensed 160 µl of medium in each well of a sterile 96-well plate, diluting the cells by fivefold. For each strain, we set three technical repeats in YPD or YPD + benomyl. To measure OD600 continuously, we placed the 96-well plate in a SpectraMax 340PC plate reader, incubated it for 36 h at 30°C without shaking, and measured the absorbance of the samples every 20 min.
Microscopy and image acquisition and analyses
The cells were imaged at RT in the presence of synthetic dextrose medium supplemented with essential amino acids. We added nocodazole and methionine to the mounting medium to image the nocodazole-arrested cells and Cdc20-depleted cells, respectively. Fluorescence imaging was conducted on a Nikon Ti-E inverted microscope with a 1.4-NA, 100×, oil-immersion objective (Joglekar et al., 2013). A 10-plane Z-stack was acquired (200-nm separation between adjacent planes). To measure Bub3 and Mad1-mCherry, the 1.5× opto-var lens was used. Total fluorescence of kinetochore clusters (16 kinetochores in metaphase) was measured by integrating the intensities over a 6 × 6-pixel region centered on the maximum-intensity pixel. The median intensity of pixels immediately surrounding the 6 × 6-pixel area was used to correct for background fluorescence. The calculations were performed using ImageJ or semi-automated Matlab programs as described previously (Joglekar et al., 2006).
Analysis of kinetochore biorientation was conducted using a Matlab GUI according to the scheme developed previously (Marco et al., 2013). Briefly, a region of interest containing the two spindle poles was manually selected from each cell. The script then identified the locations of the spindle poles and rotated the image so that the spindle axis (defined by the two spindle poles) coincided with the horizontal axis. Next, the fluorescence intensity in the GFP channel (used to record Spc105222∷GFP) over a rectangular region centered on the spindle axis and extending 4 pixels on either side of the spindle axis was summed to define the kinetochore distribution at each pixel along the spindle axis. The kinetochore distribution was then normalized by the total fluorescence to obtain the fluorescence line scan for each cell. Line scans from >50 cells were then displayed as a heat map in Figs. 1 D and 4 D. G1-arrested yeast cells expressing either WT Spc105 or Spc105BPM were released into the cell cycle, and the distribution of kinetochores along the spindle axis (defined by the locations of the two spindle poles) was analyzed at 15-min intervals and visualized as a heat-map. Because yeast cells enter mitosis after ∼30 min following release from a G1 arrest, only 45- and 60-min time points are shown in Fig. 1 D, and the 45-min time point is shown in Fig. 4 D. In this visualization, each pixel row represents the kinetochore fluorescence line scan for a single cell, and the pixel rows have been sorted according to spindle length (longest to shortest from top to bottom, >50 cells for each cell for each time point).
Quantitative assessment of the relative distribution of kinetochores in the two spindle halves was performed on the basis of the normalized kinetochore distribution as calculated above. Briefly, the brightest pixel in each spindle half was identified from the fluorescence line scan, and the absolute value of the intensity difference between these pixels was used as the measure of asymmetry in kinetochore distribution.
Statistical analyses
To quantify the frequency with which metaphase cells visibly recruited SAC proteins (Bub1, Bub3, or Mad1) at bioriented kinetochores in a cell population, we performed imaging of each strain at least twice to obtain a significant number of metaphase cells (>50). The scoring analysis divided the metaphase cell population into two groups: cells with visible SAC protein localization and the cells without detectable localization. The variation in the fraction of cells with visible SAC protein localization from individual experiments did not vary significantly. Therefore, we pooled observations from all experiments. To ascertain the statistical significance of these categorical scoring data, we applied Fisher’s exact test in GraphPad Prism (version 8). The number of cells analyzed for each strain is noted in the figure legends. To ascertain the statistical significance of the rest of the data, we applied two-way ANOVA using GraphPad Prism. The P values from these tests are noted in the figure legends.
Flow cytometry
We performed flow cytometry as described previously (Baum and Clarke, 2000). Starting from overnight inoculum, we grew the yeast strains to mid-log phase. Then we added nocodazole (final concentration 15 µg/ml) to the cultures to depolymerize the spindle and activate the spindle checkpoint (Gillett et al., 2004). We collected the cell samples of 0.1 OD600 at designated time points (0, 1, 2, and 3 h) and fixed the samples by pelleting the cells and resuspending them in 70% ethanol. We stored them at 4°C overnight. The next day, we pelleted the cells to remove ethanol and treated them with bovine pancreatic RNase (Millipore Sigma, final concentration 170 ng/µl) in RNase buffer (10 mM Tris, pH 8.0, and 15 mM NaCl) at 37°C for ≥6 h. After that, we pelleted the cells to remove RNase and resuspended them in 1× PBS. On the day of the experiment, we treated the cells with propidium iodide (Millipore Sigma, final concentration 5 µg/ml) in 1× PBS for 1 h at RT before subjecting them to the Synergy Head Cell Sorter at the Biomedical Research Core Facility, University of Michigan Medical School.
Spc105 phosphodomain purification
The 6xHIS-MBP-Spc1052-455,222::GFP fusion construct was expressed in BL21-Rosetta 2 (DE3; Novagen) or T7 expression Escherichia coli cells (NEB) using 0.25 mM IPTG for 16 h at 18°C. Cells were then harvested and resuspended in the lysis buffer (20 mM Tris, pH 7.5, 300 mM NaCl, 5 mM imidazole, 0.1% Triton X-100 [Sigma-Aldrich], 5% glycerol, and 1 mM PMSF) supplemented with complete EDTA-free protease inhibitor mix (Roche). Cells were lysed by sonication (3× for 3 min with 30-s pulse on and 30-s pulse off). Cleared supernatant was incubated with Ni-NTA agarose beads (Invitrogen) for 2 h at 4°C. Beads were washed with lysis buffer containing 500 mM NaCl and 30 mM imidazole, and protein was eluted with lysis buffer containing 150 mM NaCl and 250 mM imidazole. Subsequently, the protein was loaded onto a 16/60 Superdex 200 size exclusion column (GE) equilibrated with gel filtration buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 5% glycerol, and 1 mM DTT). Fractions were analyzed by SDS-PAGE and Coomassie staining, and peak fractions were aliquoted and stored at −80°C.
Preparation of X-rhodamine–labeled microtubules
X-rhodamine (Cytoskeleton)–labeled microtubules were prepared by using a mixture of X-rhodamine (1.25 mg/ml) and unlabeled tubulins (10mg/ml) in BRB80 buffer (80 mM Pipes/KOH, pH 6.8, 1 mM MgCl2, and 1 mM EGTA) containing 1 mM GTP and 4 mM MgCl2 at 37°C. Microtubules were further stabilized by the addition of 10 µM taxol and incubation at 37°C for 15 min. Microtubules were pelleted down by ultracentrifugation (55,000 rpm for 10 min at 37°C) to get rid of unpolymerized tubulin.
Decoration of polystyrene beads with recombinant Spc105 phosphodomain
The assay was performed as previously described with small modifications (Espeut et al., 2012). In brief, decoration of Spc105 proteins on polystyrene beads was achieved using streptavidin-biotin system. 100 nm each of WT and mutant Spc105 proteins was incubated with 10 µl of 0.1% (wt/vol) streptavidin polystyrene beads (Spherotech) conjugated with anti-penta-his biotin antibody (Qiagen) for 90 min at 4°C. Before imaging, the beads were sonicated for 3 min in a water bath containing ice cubes. Subsequently, the beads were introduced inside the flow chamber coated with taxol-stabilized microtubules. The images were acquired using a TIRF microscope.
Sample preparation for TIRF imaging
Flow cells were created as described previously (Verma et al., 2015). Flow cells were first incubated with 30 µl of 1:100 dilution of monoclonal anti–β-tubulin antibody (Sigma-Aldrich) for 10 min in a humidified chamber, followed by another incubation of 0.5% (wt/vol) Pluronic F-127 (Sigma-Aldrich) for 10 min and 30 µl of X-rhodamine–labeled microtubules for 20 min. The flow cells were then incubated with 30 µl of 2.5 mg/ml casein for 5 min to block nonspecific sites. After blocking, flow cells were incubated with 30 µl of polystyrene beads decorated with WT or mutant Spc105 proteins and a scavenging system (40 nM d-glucose, 250 nM glucose oxidase, 64 nM catalase, and 1% [vol/vol] β-mercaptoethanol in BRB80 containing 50 mM NaCl, 80 mM Pipes, 1 mM MgCl2, 1 mM EGTA, and 50 mM NaCl, pH 6.8). Subsequently, chambers were sealed, and samples were immediately imaged using a TIRF microscope.
TIRF microscopy
All images were acquired as described previously (Verma et al., 2015) with small modifications. In brief, images were acquired on a Nikon Ti-E microscope equipped with a 100×, 1.4-NA CFI-Apo oil-immersion objective, an EMCCD camera (iXon+ DU 897; Andor), a 3-line (488, 561, and 640 nm) monolithic laser combiner with an acousto-optic tunable filter laser system (Agilent) and Nikon NIS-Elements software. Images of X-rhodamine–labeled tubulin and Spc105-GFP were acquired at the following settings: 600 frames at 50-ms exposure time, conversion gain 1×, EM multiplier gain setting 288, 561 laser power 40%, and 488 laser power 40%. For TIRF image analysis, the length of the microtubule was calculated with Fiji software, and intensity of the Spc105-GFP conjugated with streptavidin polystyrene beads was measured using custom software in Matlab as described previously (Joglekar et al., 2006).
Artificial tethering of Glc7 to the N-terminus of Spc105
For these experiments, we used the yeast strains wherein Glc7-Fkbp can be tethered to the N-terminus of Spc105 (FRB-GFP-Spc105) by adding rapamycin to the growth medium. To investigate Glc7 tethering in prophase, haploid strains were arrested in G1 by α factor treatment (2 µg/ml) for 105 min. They were then washed and released in YPD medium supplemented with methionine (to repress CDC20 expression and block anaphase onset). We added nocodazole to the medium (15 µg/ml) to disrupt the spindle structure and activate the SAC 30 min after release from the G1 arrest (at which time cells are in S phase, with duplicated spindle pole bodies, and are in the process of kinetochore biorientation). After 30 min, the cells were washed again to remove nocodazole and released in rapamycin- and methionine-supplemented growth medium for 30 min before imaging. To tether Glc7 in metaphase cells, we repressed CDC20 expression for 70 min. Then we supplemented the medium with rapamycin and incubated for 30 min before imaging.
Online supplemental material
Tables S1 and S2 contain the lists of strains and plasmid constructs used in this study. Fig. S1 shows that the Mad1 localization in bioriented kinetochores of BPM remains similar to that of WT, and there was no significant alteration in growth rate of BPM, supporting Figs. 1 and 2. Fig. S2, which supports Figs. 3 and 4, demonstrates that the RVSF and anterior basic patch work together to regulate PP1/Glc7 binding. Mutating the anterior basic patch results in significant enhancement of the error correction process, which in turn leads to the rescue of cell growth in the presence of microtubule poison benomyl. Fig. S3 supports Fig. 5 by revealing that biorientation is significantly affected if PP1 is constitutively bound to N-Spc105, although regulated tethering of PP1 to N-termini of Spc105 in an unattached kinetochore does not interfere with SAC signaling.
Supplementary Material
Acknowledgments
We thank Mara Duncan (University of Michigan Medical School, Ann Arbor, MI) for sharing reagents. We thank Amitabha Gupta, Rena Evans, and Sue Biggins (Fred Hutchinson Cancer Research Center, Seattle, WA) for guidance and training for performing yeast kinetochore particle pull-down experiments and for providing key reagents. We thank members of the Joglekar laboratory and the Duncan laboratory for their support and constructive comments.
This work was funded by National Institutes of Health grants RO1-GM-088908 and GM-095005 to A.P. Joglekar.
The authors declare no competing financial interests.
Author contributions: Conceptualization: A.P. Joglekar and B. Roy. Funding acquisition: A.P. Joglekar. Resources development: B. Roy, J. Sim, and A. Fontan. Investigation: B. Roy, J. Sim, A. Fontan, and V. Verma. Visualization, data analysis and data validation: B. Roy, J. Sim, A. Fontan, and V. Verma. Writing – original draft: A.P. Joglekar. Writing – review and editing: B. Roy and A. P. Joglekar.
References
- Akiyoshi B., Nelson C.R., Ranish J.A., and Biggins S.. 2009. Analysis of Ipl1-mediated phosphorylation of the Ndc80 kinetochore protein in Saccharomyces cerevisiae. Genetics. 183:1591–1595. 10.1534/genetics.109.109041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi B., Sarangapani K.K., Powers A.F., Nelson C.R., Reichow S.L., Arellano-Santoyo H., Gonen T., Ranish J.A., Asbury C.L., and Biggins S.. 2010. Tension directly stabilizes reconstituted kinetochore–microtubule attachments. Nature. 468:576–579. 10.1038/nature09594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravamudhan P., Chen R., Roy B., Sim J., and Joglekar A.P.. 2016. Dual mechanisms regulate the recruitment of spindle assembly checkpoint proteins to the budding yeast kinetochore. Mol. Biol. Cell. 27:3405–3417. 10.1091/mbc.e16-01-0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravamudhan P., Goldfarb A.A., and Joglekar A.P.. 2015. The kinetochore encodes a mechanical switch to disrupt spindle assembly checkpoint signalling. Nat. Cell Biol. 17:868–879. 10.1038/ncb3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz P., Ye X., Huddleston P., Brautigam C.A., and Rice L.M.. 2012. A TOG:αβ-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science. 337:857–860. 10.1126/science.1221698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A. III, Steever A.B., Wach A., Philippsen P., and Pringle J.R.. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951. [DOI] [PubMed] [Google Scholar]
- Bajaj R., Bollen M., Peti W., and Page R.. 2018. KNL1 Binding to PP1 and Microtubules Is Mutually Exclusive. Structure. 26:1327–1336.e4. 10.1016/j.str.2018.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M., and Clarke L.. 2000. Fission yeast homologs of human CENP-B have redundant functions affecting cell growth and chromosome segregation. Mol. Cell. Biol. 20:2852–2864. 10.1128/MCB.20.8.2852-2864.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck D.C., and Bloom K.. 2007. Pericentric chromatin is an elastic component of the mitotic spindle. Curr. Biol. 17:741–748. 10.1016/j.cub.2007.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Anderson S., Jwa M., Green E.M., Kang J., Yates J.R. III, Chan C.S., Drubin D.G., and Barnes G.. 2002. Phospho-regulation of kinetochore–microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172. 10.1016/S0092-8674(02)00973-X [DOI] [PubMed] [Google Scholar]
- Craig R., Cortens J.P., and Beavis R.C.. 2004. Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 3:1234–1242. 10.1021/pr049882h [DOI] [PubMed] [Google Scholar]
- Espeut J., Cheerambathur D.K., Krenning L., Oegema K., and Desai A.. 2012. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J. Cell Biol. 196:469–482. 10.1083/jcb.201111107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad B., Kuijt T.E., and Kops G.J.. 2015. Kinetochore-microtubule attachment is sufficient to satisfy the human spindle assembly checkpoint. Nat. Commun. 6:8987 10.1038/ncomms9987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco L., Wang W., and Chan C.S.. 1994. Type 1 protein phosphatase acts in opposition to IpL1 protein kinase in regulating yeast chromosome segregation. Mol. Cell. Biol. 14:4731–4740. 10.1128/MCB.14.7.4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck A.D., Powers A.F., Gestaut D.R., Gonen T., Davis T.N., and Asbury C.L.. 2007. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat. Cell Biol. 9:832–837. 10.1038/ncb1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., et al. . 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 418:387–391. 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- Gillett E.S., Espelin C.W., and Sorger P.K.. 2004. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 164:535–546. 10.1083/jcb.200308100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Evans R.K., Koch L.B., Littleton A.J., and Biggins S.. 2018. Purification of kinetochores from the budding yeast Saccharomyces cerevisiae. Methods Cell Biol. 144:349–370. 10.1016/bs.mcb.2018.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H., Nishikawa J., and Laemmli U.K.. 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell. 31:925–932. 10.1016/j.molcel.2008.07.020 [DOI] [PubMed] [Google Scholar]
- Hendrickx A., Beullens M., Ceulemans H., Den Abt T., Van Eynde A., Nicolaescu E., Lesage B., and Bollen M.. 2009. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem. Biol. 16:365–371. 10.1016/j.chembiol.2009.02.012 [DOI] [PubMed] [Google Scholar]
- Hiruma Y., Sacristan C., Pachis S.T., Adamopoulos A., Kuijt T., Ubbink M., von Castelmur E., Perrakis A., and Kops G.J.. 2015. Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science. 348:1264–1267. 10.1126/science.aaa4055 [DOI] [PubMed] [Google Scholar]
- Hung C.W., Martínez-Márquez J.Y., Javed F.T., and Duncan M.C.. 2018. A simple and inexpensive quantitative technique for determining chemical sensitivity in Saccharomyces cerevisiae. Sci. Rep. 8:11919 10.1038/s41598-018-30305-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z., Gao H., and Yu H.. 2015. Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science. 348:1260–1264. 10.1126/science.aaa4029 [DOI] [PubMed] [Google Scholar]
- Joglekar A., Chen R., and Lawrimore J.. 2013. A Sensitized Emission Based Calibration of FRET Efficiency for Probing the Architecture of Macromolecular Machines. Cell. Mol. Bioeng. 6:369–382. 10.1007/s12195-013-0290-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A.P., Bouck D.C., Molk J.N., Bloom K.S., and Salmon E.D.. 2006. Molecular architecture of a kinetochore–microtubule attachment site. Nat. Cell Biol. 8:581–585. 10.1038/ncb1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanshin E., Giguère S., Jing C., Tyers M., and Thibault P.. 2017. Machine Learning of Global Phosphoproteomic Profiles Enables Discrimination of Direct versus Indirect Kinase Substrates. Mol. Cell. Proteomics. 16:786–798. 10.1074/mcp.M116.066233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima S.A., Yamagishi Y., Honda T., Ishiguro K., and Watanabe Y.. 2010. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 327:172–177. 10.1126/science.1180189 [DOI] [PubMed] [Google Scholar]
- Lampson M.A., Renduchitala K., Khodjakov A., and Kapoor T.M.. 2004. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 6:232–237. 10.1038/ncb1102 [DOI] [PubMed] [Google Scholar]
- Liu D., Vleugel M., Backer C.B., Hori T., Fukagawa T., Cheeseman I.M., and Lampson M.A.. 2010. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 188:809–820. 10.1083/jcb.201001006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N., Ceto S., Ranish J.A., and Biggins S.. 2012. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 22:900–906. 10.1016/j.cub.2012.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco E., Dorn J.F., Hsu P.H., Jaqaman K., Sorger P.K., and Danuser G.. 2013. S. cerevisiae chromosomes biorient via gradual resolution of syntely between S phase and anaphase. Cell. 154:1127–1139. 10.1016/j.cell.2013.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows J.C., Shepperd L.A., Vanoosthuyse V., Lancaster T.C., Sochaj A.M., Buttrick G.J., Hardwick K.G., and Millar J.B.. 2011. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev. Cell. 20:739–750. 10.1016/j.devcel.2011.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.P., Asbury C.L., and Biggins S.. 2016. A TOG Protein Confers Tension Sensitivity to Kinetochore-Microtubule Attachments. Cell. 165:1428–1439. 10.1016/j.cell.2016.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Barrera M., Aguilar I., and Monje-Casas F.. 2015. Dispensability of the SAC Depends on the Time Window Required by Aurora B to Ensure Chromosome Biorientation. PLoS One. 10:e0144972 10.1371/journal.pone.0144972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijenhuis W., Vallardi G., Teixeira A., Kops G.J., and Saurin A.T.. 2014. Negative feedback at kinetochores underlies a responsive spindle checkpoint signal. Nat. Cell Biol. 16:1257–1264. 10.1038/ncb3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C.G., Maddox P.S., Zarzar T.R., Salmon E.D., and Bloom K.. 2003. Yeast kinetochores do not stabilize Stu2p-dependent spindle microtubule dynamics. Mol. Biol. Cell. 14:4181–4195. 10.1091/mbc.e03-03-0180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peplowska K., Wallek A.U., and Storchova Z.. 2014. Sgo1 regulates both condensin and Ipl1/Aurora B to promote chromosome biorientation. PLoS Genet. 10:e1004411 10.1371/journal.pgen.1004411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky B.A., Kotwaliwale C.V., Tatsutani S.Y., Breed C.A., and Biggins S.. 2006a Glc7/protein phosphatase 1 regulatory subunits can oppose the Ipl1/aurora protein kinase by redistributing Glc7. Mol. Cell. Biol. 26:2648–2660. 10.1128/MCB.26.7.2648-2660.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky B.A., Kung C., Shokat K.M., and Biggins S.. 2006b The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat. Cell Biol. 8:78–83. 10.1038/ncb1341 [DOI] [PubMed] [Google Scholar]
- Posch M., Khoudoli G.A., Swift S., King E.M., Deluca J.G., and Swedlow J.R.. 2010. Sds22 regulates aurora B activity and microtubule-kinetochore interactions at mitosis. J. Cell Biol. 191:61–74. 10.1083/jcb.200912046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primorac I., Weir J.R., Chiroli E., Gross F., Hoffmann I., van Gerwen S., Ciliberto A., and Musacchio A.. 2013. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. eLife. 2:e01030 10.7554/eLife.01030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson L.C., Phillips J., Brou L., Boswell E.P., and Tatchell K.. 2012. Suppressors of ipl1-2 in components of a Glc7 phosphatase complex, Cdc48 AAA ATPase, TORC1, and the kinetochore. G3 (Bethesda). 2:1687–1701. 10.1534/g3.112.003814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg J.S., Cross F.R., and Funabiki H.. 2011. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr. Biol. 21:942–947. 10.1016/j.cub.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A., Waters J.C., and Mitchison T.J.. 2004. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 118:567–578. 10.1016/j.cell.2004.08.016 [DOI] [PubMed] [Google Scholar]
- Scott R.J., Lusk C.P., Dilworth D.J., Aitchison J.D., and Wozniak R.W.. 2005. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 16:4362–4374. 10.1091/mbc.e05-01-0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka M.B., Albuquerque C.P., Chen S.H., and Zhou H.. 2007. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. USA. 104:10364–10369. 10.1073/pnas.0701622104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk S.J., Vleugel M., Teixeira A., and Kops G.J.. 2012. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore–microtubule attachments. Dev. Cell. 23:745–755. 10.1016/j.devcel.2012.09.005 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Gupta A., Long S.K., Evans R., Badger B.L., Salmon E.D., Biggins S., and Bloom K.. 2018. A Kinesin-5, Cin8, Recruits Protein Phosphatase 1 to Kinetochores and Regulates Chromosome Segregation. Curr. Biol. 28:2697–2704.e3. 10.1016/j.cub.2018.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T.U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J., and Nasmyth K.. 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 108:317–329. 10.1016/S0092-8674(02)00633-5 [DOI] [PubMed] [Google Scholar]
- Tauchman E.C., Boehm F.J., and DeLuca J.G.. 2015. Stable kinetochore–microtubule attachment is sufficient to silence the spindle assembly checkpoint in human cells. Nat. Commun. 6:10036 10.1038/ncomms10036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma V., Mallik L., Hariadi R.F., Sivaramakrishnan S., Skiniotis G., and Joglekar A.P.. 2015. Using Protein Dimers to Maximize the Protein Hybridization Efficiency with Multisite DNA Origami Scaffolds. PLoS One. 10:e0137125 10.1371/journal.pone.0137125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzijlbergen K.F., Nerusheva O.O., Kelly D., Kerr A., Clift D., de Lima Alves F., Rappsilber J., and Marston A.L.. 2014. Shugoshin biases chromosomes for biorientation through condensin recruitment to the pericentromere. eLife. 3:e01374 10.7554/eLife.01374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J.P.I., Vleugel M., Liu D., Yates J.R. III, Lampson M.A., Fukagawa T., and Cheeseman I.M.. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore–microtubule interface. Mol. Cell. 38:383–392. 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Cetin B., Anger M., Cho U.S., Helmhart W., Nasmyth K., and Xu W.. 2009. Structure and function of the PP2A-shugoshin interaction. Mol. Cell. 35:426–441. 10.1016/j.molcel.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Tsuchiya D., and Lacefield S.. 2015. Bub3 promotes Cdc20-dependent activation of the APC/C in S. cerevisiae. J. Cell Biol. 209:519–527. 10.1083/jcb.201412036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Lischetti T., and Nilsson J.. 2014. A minimal number of MELT repeats supports all the functions of KNL1 in chromosome segregation. J. Cell Sci. 127:871–884. 10.1242/jcs.139725 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.