Summary
BRCA1 or BRCA2 inactivation drives hereditary breast and ovarian cancer, but also creates vulnerability to Parp inhibitors. To search for additional targets whose inhibition is synthetically lethal in BRCA2-deficient backgrounds, we screened two pairs of BRCA2 isogenic cell lines with DNA repair-focused shRNA and CRISPR-based libraries. We found BRCA2-deficient cells are selectively dependent on multiple pathways including base excision repair, ATR signaling, and splicing. We identified APEX2 and FEN1 as synthetic lethal genes with both BRCA1 and BRCA2 loss-of-function. BRCA2-deficient cells require the apurinic endonuclease activity and PCNA binding domain of Ape2 (APEX2), but not Ape1 (APEX1). Furthermore, BRCA2-deficient cells require the 5’ flap endonuclease but not the 5’−3’ exonuclease activity of Fen1, and chemically inhibiting Fen1 selectively targets BRCA-deficient cells. Finally, we developed an MMEJ reporter and showed that Fen1 participates in MMEJ, underscoring the importance of MMEJ as a collateral repair pathway in the context of HR deficiency.
Keywords: BRCA2, synthetic lethality, MMEJ
Introduction
Damage to human genetic material frequently contributes to oncogenic transformation, and it threatens faithful transmission of genetic information across generations. In response to the wide variety of lesions in DNA caused by endogenous and exogenous sources of damage, cells have evolved a complex signal transduction pathway known as the DNA damage response (DDR). The DDR senses DNA damage, recruits a coordinated set of factors to repair the lesion appropriately, and executes a coordinated cellular response which may include immune system activation, cell cycle arrest, cellular senescence or cell death (Ciccia and Elledge, 2010).
Among the many lesions that occur in DNA, the double-strand break (DSB) is one of the most detrimental. Failure to properly repair DSBs can lead to a variety of adverse outcomes such as replication fork collapse, cell death, oncogenic translocation, or loss of telomeric DNA. Three pathways exist to repair DSB lesions: homologous recombination (HR), classical non-homologous end-joining (NHEJ), and microhomology-mediated end-joining (MMEJ). Whereas HR utilizes information from a paired sister chromatid to seamlessly repair a DSB with high fidelity, NHEJ and MMEJ can result in sequence alterations at the repaired DSB site. However, unlike NHEJ, HR and MMEJ both act on resected DSBs (Bennardo et al., 2008), directly competing for a shared substrate (Ceccaldi et al., 2015; Mateos-Gomez et al., 2015).
Several genes that play a crucial role in DSB repair are frequently mutated in cancer. Germline mutations in BRCA1 or BRCA2, both critical for the HR pathway, account for the majority of hereditary breast and ovarian cancer (Kobayashi et al., 2013; Narod, 2006; Welcsh and King, 2001). Women carrying mutations in one of these tumor suppressor genes have up to an 80% risk of developing breast cancer and up to a 50% risk of developing ovarian cancer (Narod, 2006; Petrucelli et al., 1993). Overall, nearly half of high-grade serous ovarian cancers (HGSOC) have detectable germline and somatic inactivation of HR genes, with 30% harboring alterations in BRCA1 or BRCA2 (Bitler et al., 2017). And unlike BRCA1, BRCA2 mutation also predisposes to pancreatic and prostate cancer (Roy et al., 2011).
BRCA1 and BRCA2 share a common function in facilitating HR and play additional roles in safeguarding genomic integrity. By loading Rad51 onto DNA breaks and gaps, Brca2 prevents Mre11-dependent degradation of nascent DNA at stalled replication forks (Kolinjivadi et al., 2017; Lomonosov et al., 2003; Schlacher et al., 2011; Spies et al., 2016), and with Brca1 promotes HR-mediated resolution of fork stalling (Lomonosov et al., 2003). Also, Brca2 protects telomere integrity (Doksani and de Lange, 2014) and prevents accumulation of R-loops, which can lead to replication fork stalling and interference with transcriptional elongation (Bhatia et al., 2014).
BRCA1 and BRCA2-deficient cells are exquisitely sensitive to treatment with inhibitors of poly(ADP-ribose) polymerase (PARP) (Bryant et al., 2005; Farmer et al., 2005) and this relationship has been referred to as synthetic lethality (SL). Synthetic lethality, a concept borrowed from classical genetics, describes a condition in which mutation of either of two genes is viable, but simultaneous mutation of both genes is lethal (Kaelin, 2005). Parp inhibitors have recently gained FDA approval for patients with metastatic breast cancer and a germline BRCA mutation (Robson et al., 2017a; Robson et al., 2017b), and for recurrent HGSOC (Bitler et al., 2017; Evans and Matulonis, 2017). However, dual depletion of PARP and BRCA2 by siRNA does not recapitulate the potent lethality observed upon chemical inhibition of Parp (Bryant et al., 2005). Rather than solely exploiting a genetic SL relationship, Parp inhibitors also cause lethality by physically trapping Parp onto single-strand break (SSB) intermediates, obstructing progression of replication forks (Helleday, 2011; Murai et al., 2012; Strom et al., 2011), and in that sense behaving more like classical DNA damage agents to which BRCA2-mutant tumors are also sensitive. Indeed, the toxic Parp-DNA complexes formed as a result of Parp trapping have been shown to be more cytotoxic than unresolved SSBs (Murai et al., 2012).
Targeting the intrinsic genetic vulnerabilities in BRC8A1- and BRCA2-deficient cells offers the opportunity to meet an urgent clinical need in the treatment of refractory or metastatic breast cancer in patients with a germline BRCA mutation (Narod et al., 2017) and recurrent HGSOC more broadly (Evans and Matulonis, 2017; Mirza et al., 2016). Despite recent success in clinical trials, Parp inhibitor efficacy appears to be limited by inherent and acquired resistance, underscoring the urgent need for identification of synergistic and alternative targets (Higgins et al. 2018). Therefore, we sought to explore if additional genetic synthetic lethal relationships exist with BRCA2 deficiency. We chose BRCA2 for this study because of its myriad important roles in protecting genomic integrity beyond its crucial role in HR.
To uncover novel BRCA2 synthetic lethal genes (B2SLs), we used a genetic screening approach, studying both shRNA and CRISPR-based genetic libraries in a pooled screening format, in two pairs of isogenic cell lines. We find BRCA2 mutant (B2MUT) cells to be more dependent than their wild-type counterparts (B2WT) on several pathways including base excision repair (BER), ATR activation, and MMEJ. We identify APEX2 and FEN1 as novel B2SL targets, and we show through the use of a novel cell-based reporter that FEN1 participates in MMEJ.
Results
shRNA and CRISPR screens identify B2SL Candidates
To identify novel B2SL candidates, we began by establishing a pair of cell lines that are isogenic except for the presence or absence of a BRCA2 mutation. We obtained a modified DLD-1 colon cancer cell line with a homozygous deletion of BRC repeat 6 in exon 11 that also introduces a loxP site and a stop codon between BRC repeats 5 and 6, resulting in a biallelic BRCA2 premature truncation mutation (Hucl et al., 2008). To this BRCA2 mutant (B2MUT) cell line, we introduced a full-length BRCA2 mammalian expression construct through transfection and selection for stable integrants. These add-back BRCA2 wild-type cells are a closer, though not perfect, isogenic comparison to B2MUT cells than the parental DLD-1 line, due to the genetic drift that occurs in this mismatch repair (MMR)-deficient background.
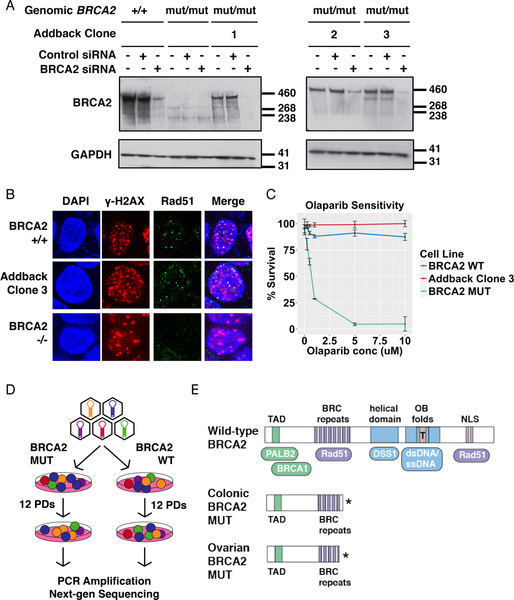
We isolated individual clones from these BRCA2 wild-type cells (B2WT) and characterized several clones to demonstrate restoration of functional BRCA2 expression. We confirmed full-length BRCA2 protein expression by Western blotting, utilizing BRCA2 siRNA to confirm the identity of the protein (Figure 1A). We observed that expression of full-length BRCA2 enhanced the growth rate of B2MUT cells (Supplemental Figure 1A) and restored their ability to form Rad51 foci in response to ionizing radiation (IR) (Figure 1B). Finally, we confirmed that expression of BRCA2 in our add-back clones restored resistance to the Parp inhibitor olaparib (Figure 1C).
Figure 1. Establishment of isogenic cell line systems for BRCA2 SL screening.
(A) Extracts from the indicated cell lines, untreated or treated with the indicated siRNAs, were immunoblotted with antibodies to BRCA2 and GAPDH. Left and right panels were run as separate gels. (B) Immunofluorescence was performed on cells of the indicated genotypes, with antibodies to γH2AX and Rad51 protein, to evaluate foci formation after 10 Gy IR. (C) The indicated cell lines were passaged in the presence of the indicated dose of olaparib or DMSO. After 3 d, cell survival was quantified using CellTiter-Blue. Error bars reflect the variability of biological triplicates. (D) Schematic diagram depicting the experimental procedure for BRCA2 SL screening in isogenic BRCA2 cell lines. (E) Schematic of wild-type BRCA2 structure, depicted with its functional domains and sites of interaction with key binding partners. Brca2 truncation mutant proteins possessed by the colonic and ovarian BRCA2 MUT cell lines used in this study are shown for comparison.
Using this pair of isogenic cell lines, we performed a series of genetic screens designed to identify novel B2SL genes. We screened a targeted library of 380 genes with a known or suspected role in the DDR; inclusion in this library was based on either a role in DNA repair described in the literature or performance of the gene in our prior screens for sensitivity to DNA damaging agents. In each screen, B2MUT and B2WT lines were separately transduced with a lentivirus-based shRNA or CRISPR gRNA library targeting these genes at a low multiplicity of infection (MOI) in triplicate (Figure 1D). Cell pellets were collected before and after passaging these cells for 12 population doublings (PDs), and the relative change of shRNAs or gRNAs during culture was determined by next-generation sequencing (NGS) of shRNA half-hairpins (Schlabach et al., 2008) or gRNAs PCR-amplified from PD0 and PD12 cell pellets.
To explore the generalizability of our results, we also obtained a second previously characterized isogenic cell line pair: ovarian PEO1 B2MUT cells, which contain a hemizygous truncation mutation in BRC repeat 5 of exon 11, and an in vitro-derived clone C4–2 selected for cisplatin resistance that contains a reversion mutation that restores full-length Brca2 expression (B2WT) (Sakai et al., 2009). Because of its evolution, the C4–2 subclone and the parental PEO1 cells are not perfectly isogenic but the use of two near isogenic cell line pairs in our screens mitigates this lack of perfect isogenicity in each case. The colonic DLD-1 B2MUT cell line and the ovarian PEO1 B2MUT cell line express similar truncation mutants of BRCA2 (Hucl et al., 2008; Sakai et al., 2009), both prematurely terminate directly in or immediately after BRC repeat 5 in exon 11 (Figure 1E). This truncation mutant retains the N-terminal transactivation domain of Brca2 (TAD) and the first four BRC repeats, which are known to bind free Rad51 (Carreira and Kowalczykowski, 2011), but lacks the DNA binding domains of Brca2 and its C-terminal Rad51 binding domain (Figure 1E). We examined all BRCA2 mutations reported from 2,433 tumors in the METABRIC breast cancer targeted exome sequencing study (Pereira et al., 2016) and found that mutations in the BRC repeat region of exon 11 are not uncommon, with about 16% of functionally impactful mutations occurring in this region (Supplemental Table 1, Supplemental Figure 1B). We screened our CRISPR-based library, but not our shRNA-based libraries, in this ovarian isogenic cell line pair (Supplemental Figure 1C).
We analyzed the results of our screens by MAGeCK (Li et al., 2014) and EdgeR (Robinson et al., 2010), relying on EdgeR analysis to calculate false discovery rates (FDRs) (Figures 2A–F, Supplemental Tables 2–6). B2SL genes deplete in the B2MUT cell line more than the B2WT cell line, and thus the fold change of B2SL genes in the B2MUT line is lower than the fold change in the B2WT line, yielding a B2MUT/B2WT ratio that is negative on a log2 scale (Figures 2A–C); this value can also be negative for genes that promote growth of B2WT cells more than B2MUT cells (Figures 2D and 2F). Overall, the magnitude of effects we observed was more potent with our CRISPR-based than our shRNA-based library, and the CRISPR library produced larger fold changes in the diploid colonic background than the aneuploid ovarian background.
Figure 2. BRCA2 Synthetic Lethal Screen Results.
(A-C) Volcano plots for the shRNA colonic, CRISPR colonic, and CRISPR ovarian B2SL screens. Significance (-log10FDR) is plotted against the genetic interaction (GI) score (average log2 fold-change for each gene in BRCA2 MUT versus BRCA2 WT cells). Genes that met a significance threshold of -log10FDR>1 are color-coded as green for relative dropout or red for relative enrichment in BRCA2 MUT vs BRCA2 WT cells. (D-F) Results from the colonic shRNA, colonic CRISPR, and ovarian CRISPR B2SL screens, plotted as the log2 fold-change in BRCA2 MUT cells against the log2 fold-change in BRCA2 WT cells. (G-H) Comparison of GI score (average log2 fold-change for each gene in BRCA2 MUT versus BRCA2 WT cells) in the ovarian CRISPR screen versus the colonic CRISPR screen (G), and the ovarian CRISPR screen versus primary shRNA screen (H).
Analysis of our screens identified POLQ as a B2SL gene (Figures 2A and 2F). POLQ encodes polymerase theta (Polθ), a large A-family DNA polymerase that plays an important role in MMEJ and is known to be synthetic lethal with HR pathway mutants (Ceccaldi et al., 2015; Mateos-Gomez et al., 2015). Its helicase domain promotes unwinding and exposure of microhomology regions on either side of a DSB (Chan et al., 2010; Ozdemir et al., 2018), while its polymerase domain extends DNA from a region of microhomology base pairing (Zahn et al., 2015). Thus, POLQ serves as a positive control for BRCA2 synthetic lethality.
SF3B2 is also a B2SL candidate. SF3B2 is part of the U2 snRNP that assembles with other snRNP components to form the spliceosome (Cretu et al., 2016). While SF3B2 loss is toxic to both B2MUT and B2WT cells, B2MUT cells appear to be more reliant on its function than B2WT cells (Figures 2A and 2D). When we re-screened 50 hits from this library in a secondary shRNA screen, we observed that SF3B2 and POLQ validated as B2SL candidates (Supplemental Figures 2A and 2B), and that our secondary shRNA screen sublibrary enriched for B2SL candidates overall (Supplemental Figures 2C and 2F).
Screening our CRISPR library in the colonic DLD-1 isogenic cell line pair confirmed POLQ as a B2SL gene and also found FEN1, APEX2, UBE2A, CLSPN, and DCLRE1C to behave as B2SLs (Figures 2B and 2E). Screening this same library in the ovarian PEO1 isogenic cell line pair revealed POLQ, FEN1, XRCC1, and RHNO1 to be among the strongest B2SL hits (Figures 2C and 2F). Because we are most interested in genes that generalize as SL broadly, we compared ovarian and colonic cancer cell lines to find POLQ, FEN1, and APEX2 as strong, consistent B2SL in both contexts (Figures 2G and 2H). While POLQ serves as a positive control, FEN1 and APEX2 represent novel B2SL genes and novel potential drug targets in BRCA-deficient tumors.
Notably, several of our B2SL hits demonstrated stronger synthetic lethality than PARP1 in both ovarian and colonic cells, including FEN1, APEX2, POLQ, XRCC1, and UBE2A. This observation underscores the well-established finding that PARP inhibitor effectiveness in BRCA2-deficient cells is due not only to a mild synthetic lethal interaction between PARP1 and BRCA2, but also because of the phenomenon of PARP trapping (Strom et al. 2011; Murai et al. 2012).
From our comparison of CRISPR screens in the ovarian and colonic cell line pairs, we noticed a striking phenomenon: every RAD51 paralog (RAD51, RAD51B, RAD51C, RAD51D, XRCC2 and XRCC3) exhibited a strongly positive genetic interaction (GI) score in both cell line contexts (Figures 2G and 2H). This positive GI score (the average log2 fold-change in B2MUT versus B2WT cells) reflects not only a detrimental effect in B2WT cells from the loss of these HR components, but also a surprising enhancement of B2MUT cell growth (Figures 2E–F). When we examined our data for the performance of BRCA2 itself we found that, like RAD51 and its paralogs, CRISPR-based knockout of these BRCA2 truncation mutants is detrimental to B2WT cells but also enhances growth of B2MUT cells, yielding a positive GI score (Figures 2E and 2F). These observations suggest that the residual BRCA2 truncation mutants present in our ovarian and colonic B2MUT cells exert a detrimental effect involving RAD51 paralogs.
To validate this result, we employed a multicolor competition assay (MCA): as previously described (Smogorzewska et al., 2007), we mixed GFP-labeled B2MUT cells with E2-Crimson-labeled B2WT cells, infected the mixture with individual gRNAs, and monitored the relative change in percent GFP+ cells by FACS (Supplemental Figure 2G). We individually tested the relative effect of two gRNAs to RAD51, RAD51C, RAD51D, and the N-terminal portion of BRCA2 in B2MUT versus B2WT cells, utilizing our colonic isogenic cell line pair. Consistent with our screen results, all of the gRNAs tested resulted in an enhanced ratio of B2MUT to B2WT cells, relative to negative control gRNAs (Supplemental Figure 2H).
It is surprising that depletion of RAD51 and its paralogs suppresses the detrimental effects of BRCA2 mutation relative to wild-type, suggesting the possibility that these two similar BRCA2 truncation mutants may behave in an unusual manner. To ask if this phenotype is specific to BRCA2 mutants truncated near BRC repeat 5, we performed MCA assays with our gRNAs in an isogenic cell line pair that possesses a different BRCA2 truncation mutant: the pancreatic CAPAN-1 cell line and its in vitro-derived clone C52 (Sakai et al., 2008). CAPAN-1 BRCA2 mutant cells have a single base pair deletion in one copy of BRCA2 that produces a frameshift and prematurely truncates the protein in BRC repeat 8 (c.[6174delT*fsTer6431]), while the second allele of BRCA2 is deleted. C5–2 cells possess a second small deletion within the mutated copy of BRCA2 that restores its correct reading frame. In an MCA assay with this second isogenic cell line pair, the same individual gRNAs targeting RAD51, RAD51C, RAD51D, and the N-terminal portion of BRCA2 no longer result in an enhancement of B2MUT cell growth relative to B2WT cells, but create the opposite effect, being more toxic in the B2MUT cells (Supplemental Figure 2H). Thus, the toxic effect of Rad51 depletion in cells expressing Brca2 mutant truncation protein terminating early in BRC repeat 5 does not extend to cells that possesses a Brca2 mutant protein with a nearly intact BRC repeat region, and is therefore likely to be an allele-specific phenomenon. We cannot rule out a possible role for other genetic or epigenetic differences in the CAPAN verses other cell line backgrounds that differentially impact the response to RAD51 paralog mutation.
BRCA2 mutant cells rely on specific repair pathways including BER
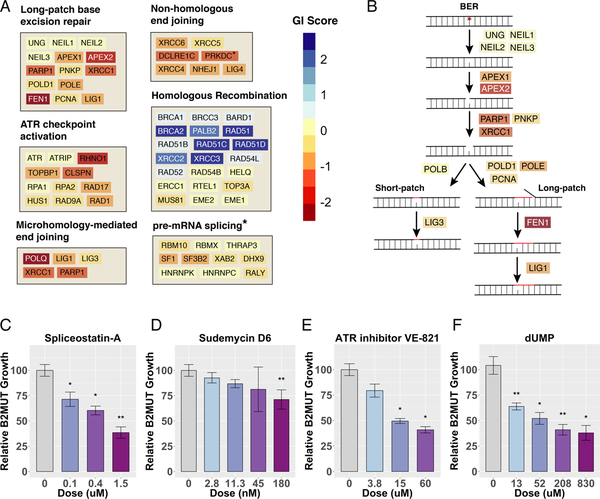
From our analysis of screens in both the ovarian and colonic isogenic cell line pairs, several pathways emerged as general themes. Overall, base excision repair (BER) scores potently as SL overall (Figure 3A). Indeed, almost every member of this pathway scores as SL except for the partially redundant glycosylases which initiate repair through this pathway by removing damaged sugars; their absence would not be expected to lead to nicks or DSBs that could result in an increased need for HR or AP sites that may produce replication stress (Figure 3B). Similarly, several members of the ATR/CHK1 signaling pathway were B2SLs, except for some of the more essential components of the ATR/ATRIP pathway such as ATR/ATRIP themselves and RPA1, which did not score using the CRISPR-based library (Figure 3A).
Figure 3. Pathway Analysis of BRCA2 Synthetic Lethality.
(A) Performance of genes in several key pathways, plotted on a color scale for genetic interaction (GI) score: the normalized average log2 fold-change across both colonic and ovarian CRISPR screens. The asterisk indicates reporting of GI score from the shRNA screen instead of combined CRISPR screens. (B) Schematic of the base excision repair (BER) pathway, showing the strength of the GI score for each gene in the pathway, plotted on the same color scale as in (A). (C-F) MCA assays in which colonic GFP-labeled BRCA2 MUT and E2-Crimson-labeled BRCA2 WT cells were mixed and co-treated with the indicated drugs. The change in percent GFP+ cells was measured by FACS after 12 d and normalized to vehicle control. Error bars reflect the variability of biological triplicates.
Known components of MMEJ scored more strongly as B2SL than NHEJ components, perhaps implying that the majority of detrimental unresolved DSBs addressed by these auxiliary pathways occurs during replication. The strikingly potent positive GI score we observe for BRCA2 and RAD51 paralogs does not extend to earlier or later steps of HR, but focally affects the components of HR involved in Rad51 loading. Finally, although SF3B2 performs as a B2SL using our shRNA library, and SF1 does as well to a lesser extent (Figure 3A, Supplemental Table 5), these essential factors do not score with the more penetrant CRISPR-based library.
To further validate some of these pathway synthetic lethalities, we employed a multicolor competition assay (MCA). As described above, we mixed GFP-labeled B2MUT cells with E2-Crimsonlabeled B2WT cells, subjected the mixture to various drug treatments, monitored the relative change in percent GFP+ cells, and normalized to vehicle control. We tested two spliceosome inhibitors, spliceostatin-A and sudemycin D6, both of which inhibit the U2 component SF3B1 (Effenberger et al., 2016). Both of these drugs caused a dose-dependent depletion of B2MUT GFP+ cells relative to B2WT E2-Crimson+ cells, supporting the hypothesis that inhibition of the U2 spliceosome component is SL with BRCA2 loss of function (Figures 3C–D).
Similarly, because several components required for ATR activation emerged as B2SL from our CRISPR screens, including RHNO1, CLSPN, and to a lesser degree the more essential TOPBP1, we tested the ATR inhibitor VE-821 for synthetic lethality in our MCA assay. This ATP-competitive inhibitor of ATR also exhibited dose-dependent selective inhibition of B2MUT cell growth versus B2WT cells (Figure 3E), confirming that B2MUT cells are more dependent on ATR activation than B2WT cells. Finally, we tested our hypothesis that B2MUT cells are more dependent on competent function of the BER pathway by increasing the load of damage that must be repaired by BER. We introduced deoxyuridine monophosphate (dUMP) directly into our cell culture medium, which has been shown to increase misincorporation of uracil into DNA (Nilsen and Krokan, 2001). dUMP also caused a dose-dependent selective depletion of B2MUT versus B2WT cells in our MCA assay, supporting our hypothesis that B2MUT cells are more dependent on BER than B2WT cells (Figure 3F).
AP endonuclease APEX2 is synthetic lethal with BRCA1 and BRCA2 loss-of-function
In our CRISPR-based screens, we identified two novel, potent, and generalizable B2SL hits: APEX2 and FEN1. APEX2 encodes Ape2, an AP endonuclease responsible for the second step of BER: after base removal by a lesion-appropriate glycosylase, Ape2 hydrolyzes the phosphodiester backbone immediately 5’ to the AP site to create a single-strand break (SSB). In addition to its AP endonuclease activity, Ape2 also possesses 3’ phosphodiesterase activity and 3’−5’ exonuclease activity, executed from the same ExoIII-like nuclease domain.
Human cells rely on two ExoIII-family type II endonucleases to generate a nick upstream of AP sites during BER, encoded by APEX1 and APEX2. APEX2 scored strongly as a B2SL gene, while APEX1 scored weakly. Ape1 and Ape2 both contain an ExoIII-like endonuclease domain with 29% sequence identity and high sequence similarity (Hadi et al., 2002), but Ape1 also contains a separate, distinct N-terminal redox domain. A critical cysteine in this redox domain reduces the heterodimeric transcription factor AP-1 (c-Jun/c-Fos), enhancing its DNA-binding activity (Xanthoudakis et al., 1994).
To validate APEX2 as a B2SL hit, we tested three individual gRNAs targeting APEX2 in an MCA assay (Figure 4A). As described above, we mixed GFP+ B2MUT colonic cells with E2-Crimson+ B2WT colonic cells and we transduced the cell mixture with each of these three individual gRNAs, separately. After selection with puromycin, we passaged the cell mixture for 7 d and quantified the change in percent GFP+ cells by FACS, normalizing to negative control gRNAs that are either non-cutting or that cut intergenic regions with low predicted off-target cutting (Supplemental Figure 3A). We verified interference with APE1 or APE2 by the relevant gRNAs with either Western blotting (Supplemental Figure 3B) or qPCR (Supplemental Figure 3C), respectively.
Figure 4. APEX2 is synthetic lethal with BRCA1 and BRCA2 deficiency.
(A-D) MCA assays were performed using the indicated gRNAs in the indicated cell backgrounds. GFP-labeled B2MUT cells were mixed with isogenic E2-Crimson-labeled B2WT cells, and the cell mixture was co-infected with the indicated individual gRNAs. After selection, the change in percent GFP+ cells was quantified by FACS before and after 7 d of culture, and normalized to negative control gRNAs. Error bars reflect the variability of biological triplicates, and the number of asterisks indicates the statistical significance for the corresponding experiment calculated by t-test (*=p<0.05,**=p<0.01,***=p<0.001). (E-F) MCA assays (as described above) in which colonic B2MUT and B2WT cells were mixed and treated for 12 d with the Ape nuclease inhibitor APEIII or the Ape1 redox inhibitor E3330. Error bars reflect the variability of biological triplicates, and asterisks indicated statistical significance as described above. (G) List of ORFs tested for complementation in cells expressing Cas9 and a gRNA to APEX2. (H) Examination of the ability of the ORFs from (F) to rescue the growth defect caused by expression of Cas9 and an APEX2 gRNA in ovarian B2MUT cells. Cells were co-infected with lentivirus expressing an APEX2 gRNA and the indicated gRNAresistant APEX2 ORF or negative control peptide. Growth was quantified after selection and growth for 8 d. (I) List of ORFs tested for complementation in cells expressing Cas9 and a gRNA to APEX2. (J) Examination of the ability of the ORFs from (I) to rescue the growth defect caused by expression of Cas9 and an APEX2 gRNA in ovarian B2MUT cells. Cells were co-infected with lentivirus expressing an APEX2 gRNA and the indicated gRNA-resistant APEX2 ORF or negative control peptide. Growth was quantified after selection and growth for 2 d.
Consistent with our screen results, gRNAs targeting APEX2 validated strongly as B2SL in both colonic and ovarian isogenic cell line pairs, while gRNAs targeting APEX1 showed weaker effects (Figures 4A–D, Supplemental Figure 3D). In addition, we asked whether inactivation of APEX1 or APEX2 exhibits synthetic lethality with BRCA1 by testing these same gRNAs in an MCA assay utilizing a BRCA1 isogenic cell line pair in the RPE background. These cell lines were generated by CRISPR-based knockout of TP53 in the RPE cell line background, followed by CRISPR-based knockout of BRCA1 in the mutant cell line. Cas9 was expressed transiently in both cases, and functional absence of BRCA1 was confirmed by Western blotting (Supplemental Figure 3E). Like its relationship to BRCA2, mutation of APEX2 is strongly SL with BRCA1 deficiency, while mutation of APEX1 exhibits a weaker genetic interaction (Supplemental Figures 3F–H).
We further assessed the synthetic lethality of APEX1 by testing two chemical inhibitors of Ape1: Ape Inhibitor III and E3330. Ape1 Inhibitor III enzymatically inhibits Ape1’s nuclease domain (Rai et al., 2012), though its potential to exert off-target effects on the structurally similar nuclease domain of Ape2 (Mol et al., 2000) has not yet been characterized. This small molecule caused selective growth inhibition of B2MUT cells relative to B2WT cells, likely through enzymatic inhibition of the nuclease domain of Ape1, and possibly also Ape2 (Figure 4E). In contrast, the small molecule inhibitor E3330 blocks the redox activity of Ape1 but does not impede its DNA repair function (Zou and Maitra, 2008). This inhibitor had no effect, suggesting that the nuclease domain, rather than the redox domain, of Ape1 contributes to its B2SL phenotype (Figure 4F).
To verify that the B2SL phenotype exhibited by APEX2 gRNAs is due to cutting of APEX2 by Cas9, rather than off-target effects, we performed a rescue experiment (Figure 4G–H), verifying expression of our rescue ORFs by Western blotting (Supplemental Figure 4A). We generated gRNA-resistant APEX2 ORFs (Figure 4G) and demonstrated that expression of gRNA-resistant wild-type Ape2 was able to rescue the growth defect in B2MUT cells caused by APEX2 gRNA cutting, relative to two negative control peptides (Figure 4H). When we introduced a point mutation in the gRNA-resistant Ape2 ORF that inactivated its nuclease domain (D277A), we found that the nuclease-deficient Ape2 mutant was no longer capable of rescuing the growth deficit caused by the APEX2 gRNA (Figure 4H). This observation suggests that the nuclease domain of Ape2 is required for the B2SL phenotype. Similarly, expression of the wild-type APE1 gene (which is not cut by our APEX2 gRNA), or its nuclease-deficient point mutant (Y171F) was not able to rescue B2MUT cell growth. The fact that Ape1 cannot complement loss of Ape2 further suggests that Ape2 possesses a distinct feature responsible for its stronger SL phenotype.
We investigated functional features that distinguish Ape2 from Ape1, in order to understand why BRCA2-deficient cells depend more heavily on Ape2 function for survival. We noted that Ape2 binds PCNA in vivo through a PIP-box motif, while Ape1 does not (Burkovics et al., 2009; Ide et al., 2003; Tsuchimoto et al., 2001). In addition, Ape2 has been found to strongly associate with replication forks, unlike Ape1 (Dungrawala et al., 2015). We hypothesized that Ape’s association with PCNA allows it to process apurinic sites at replications forks, which would otherwise block fork progression and result in breaks in the absence of BRCA2 activity. To test this model, we created a gRNA-resistant Ape2 mutant that lacks a functional PIP-box PCNA-binding motif (Figure 4I, Supplemental Figure 4B). We found that this Ape2 mutant is not capable of rescuing the growth defect caused by gRNA-mediated APEX2 depletion (Figure 4J). Thus, the PCNA-binding capability of Ape2, which promotes Ape2 localization at replication forks, is critical for its ability to promote survival of BRCA2-mutant cells.
Ape2 or Fen1 loss leads to DNA damage in BRCA2 mutant cells
Loss of BRCA2 renders cells exquisitely sensitive to DNA damaging agents that generate doublestrand breaks. PARP inhibitors generate DNA damage in BRCA2 mutant cells that imparts significant synthetic lethality. In order to investigate the lethality caused by loss of FEN1 or APEX2 in our BRCA2-mutant cells, we examined the generation of endogenous damage by measuring phosphorylated H2AX. We measured γH2AX foci, which accumulate at sites of double-strand breaks in DNA, after siRNA-mediated depletion of Ape2 or Fen1 in ovarian B2MUT cells. Depletion of either of these two proteins causes substantially more phospho-H2AX foci than a negative control siRNA (Figures 5A and 5B).
Figure 5. FEN1 and APEX2 loss leads to DNA damage in BRCA2 mutant cells.
(A) Immunofluorescence was performed on ovarian B2MUT cells transfected with the indicated siRNAs, using an antibody to γH2AX, to evaluate foci formation. (B) Quantification of the number γH2AX foci formed in (G) in 30 representative cells transfected with siRNA to the indicated genes. (C) Examination of DDR signaling in extracts of WT and BRCA2 mutant cells transfected with the indicated siRNAs and immunoblotted with the indicated antibodies. (D) Quantitation of phospho-Chk1, phospho-H2AX and phospho-RPA32 from the BRCA2 mutant cell extracts in experiment (C).
To further examine the presence of DNA damage generated by depletion of Ape2 and Fen1, we performed Western blotting of additional proteins that are phosphorylated in response to activation of the DDR kinase Atr. Depletion of Ape2 or Fen1 resulted in an increase of phospho-Chk1 and phospho-Rpa32 (Figures 5C and 5D). This effect was much more prominent in B2MUT than B2WT cells. We attribute this difference to the inability of BRCA2 mutant cells to repair the DNA damage caused by Fen1 or Ape2 depletion, resulting in a more persistent damage signal.
Flap endonuclease FEN1 is a novel B2SL that plays a role in MMEJ
FEN1, a top B2SL hit, performed strongly in both the ovarian and colonic isogenic BRCA2 mutant cell line pairs. FEN1 encodes a structure-specific endonuclease that recognizes and cleaves 5’ ended singlestranded flaps (Zou and Maitra, 2008). Several processes in DNA repair and metabolism require this enzymatic function, such as the penultimate step of BER, in which Polβ-mediated strand-displacement synthesis generates a 5’ flap that is processed by Fen1. Similarly, in lagging strand synthesis during DNA replication, Polδ extends an Okazaki fragment, encounters the subsequent 3’ fragment, and proceeds by strand-displacement synthesis to displace the downstream RNA-DNA primer, creating a 5’ flap structure that is resolved by Fen1 cleavage. In addition to this important flap endonuclease activity, Fen1’s sole active site also possesses gap endonuclease activity and 5’−3’ exonuclease activity, both of which are weaker than its flap endonuclease activity in vitro (Balakrishnan and Bambara, 2013).
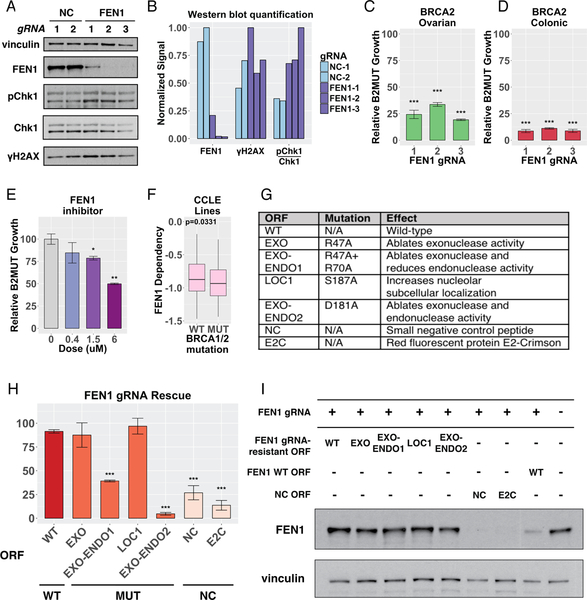
We first validated that FEN1 is a B2SL gene using the MCA assay described above. We verified that three individual gRNAs deplete the protein level of FEN1 by Western blotting (Figures 6A and 6B), and we further saw an increase in γH2AX phosphorylation and Chk1 phosphorylation at S317 in the presence of FEN1 gRNAs relative to control gRNAs, possibly reflecting an increase in DSB load (Figures 6A and 6B) as seen with siRNA depletion (Figures 5C, D).
Figure 6. FEN1 loss is synthetic lethal with BRCA1 and BRCA2 deficiency.
(A) Extracts from B2MUT ovarian cells expressing Cas9 and the indicated gRNAs were immunoblotted with the indicated antibodies. (B) Quantification of signal detected Western blotting in (A), normalized to vinculin. (C-D) MCA assays in which the indicated GFP-labeled B2MUT cells and E2-Crimson-labeled B2WT cells were mixed and co-infected with 3 individual gRNAs to FEN1. The cell mixture was cultured for 7 d, and the change in percent GFP+ cells was quantified by FACS and normalized to the average of the negative control Grna-expressing cells shown in Supplemental Figure 3A. Error bars reflect the variability of biological triplicates, and the number of asterisks indicates the statistical significance for the corresponding experiment (*=p<0.05,**=p<0.01,***=p<0.001). (E) MCA assay in which colonic GFP-labeled BRCA2 MUT cells and E2-Crimson-labeled BRCA2 WT cells were mixed and co-treated with the indicated doses of a FEN1 inhibitor. The cell mixture was cultured for 7 d, and the change in percent GFP+ cells was quantified by FACS and normalized to negative control gRNA-expressing cells. Error bars reflect the variability of biological triplicates, and asterisks indicate statistical significance as described above. (F) Dependency of BRCA1/2 MUT or BRCA1/2 WT cell lines on FEN1, determined from genome-scale CRISPR-Cas9 essentiality screens across 324 cancer cell lines from the Cancer Cell Line Encyclopedia (CCLE) (Meyers et al., 2017). (G) List of gRNA-resistant ORFs tested for complementation in BRCA2 MUT cells expressing Cas9 and a FEN1 gRNA. (H) Examination of the ability of the ORFs listed in (G) to rescue the growth defect caused by expression of Cas9 and a FEN1 gRNA. BRCA2 MUT ovarian cells were co-infected with lentivirus expressing a FEN1 gRNA and the indicated gRNA-resistant FEN1 ORF or negative control peptide. After selection and growth for 8 d, survival was quantified with a FACS-based cell counting method. (I) Extracts from BRCA2 WT cells expressing Cas9, a validated FEN1 gRNA, and the indicated gRNA-resistant ORF were immunoblotted with the indicated antibodies; all panels are derived from the same blot.
We tested these three individual gRNAs targeting FEN1 in our MCA assay and each resulted in selective growth inhibition of B2MUT versus B2WT cells, in both the ovarian and colonic backgrounds (Figures 6C–D). To ask if Fen1 depletion would induce synthetic lethality in BRCA1-mutant as well as BRCA2-mutant tumor lines, we tested three individual gRNAs to FEN1 in an MCA assay utilizing a BRCA1 isogenic cell line pair in the RPE background. Each of the gRNAs selectively inhibit growth of BRCA1 MUT cells relative to BRCA1 WT cells, indicating that FEN1 is a target applicable to patients with either a germline BRCA1 or BRCA2 mutation (Supplemental Figure 5A).
To assess whether Fen1 may represent a novel drug target in BRCA-deficient tumors, we tested a small molecule inhibitor of Fen1. This inhibitor sterically occupies the active site of Fen1, and thus impedes both its exonuclease and endonuclease functionalities (Exell et al., 2016; Ward et al., 2017). In an MCA assay, we found that this Fen1 inhibitor selectively impairs growth of B2MUT cells relative to B2WT cells, confirming that Fen1 is a viable novel B2SL drug target (Figure 6E). We also asked if we could validate FEN1 as broadly BRCA SL by utilizing the publicly available Avana CRISPR-Cas9 genome-scale dataset, analyzed by the CERES algorithm (Meyers et al., 2017). We separated 324 cell lines based on the mutation status of BRCA1 and BRCA2, and we observed that the BRCA-mutant lines are more dependent on FEN1 than the BRCA-wild-type lines (Figure 6F). This analysis provides independent confirmation of the strength of FEN1 as a B2SL hit.
Next, we asked whether the B2SL phenotype induced by our FEN1 gRNAs is directly due to ontarget cutting of FEN1 by CRISPR/Cas9. We rescued one of our validated FEN1 gRNAs with either a gRNA-resistant wild-type FEN1 ORF or negative control peptides; we found that full-length gRNAresistant wild-type FEN1 rescues the growth defect imparted by the FEN1 gRNA, confirming an on-target effect of our gRNA (Figures 6G–I). To investigate which enzymatic function of Fen1 is important for its B2SL phenotype, we made a series of point mutants, including one mutant that inactivates only the exonuclease activity of Fen1 and two different point mutants that inactivate both its endonuclease and exonuclease activity. There is no single point mutation that inactivates the endonuclease activity of Fen1 while leaving its exonuclease activity intact. We found that the exonuclease-deficient FEN1 mutant was able to rescue the effect of our FEN1 gRNA, while both endonuclease mutants were unable to rescue this effect, suggesting that BRCA2-deficient cells are selectively dependent upon the endonuclease but not the exonuclease activity of FEN1 (Figures 6G and 6H). Each FEN1 wild-type and mutant protein was expressed in the presence of the indicated FEN1 gRNA (Figure 6I) and was not toxic in the presence of a negative control gRNA (Supplemental Figure 5B).
Finally, we considered why the flap endonuclease activity of Fen1 is particularly important in BRCA2-deficient cells. We hypothesized that Fen1 may be involved in processing the 5’ flaps generated during MMEJ by Polθ-mediated strand displacement synthesis (Kent et al., 2015). In yeast, flap processing during MMEJ is accomplished by the Rad1-Rad10 endonuclease (XPF-ERCC1) (Decottignies, 2007; Ma et al., 2003). However, murine ERCC1−/− cells exhibit only a minor MMEJ defect, suggesting that an alternate nuclease may play this role in mammalian cells (Bennardo et al., 2008).
To test whether FEN1 plays a role in MMEJ flap resolution in mammalian cells, we designed a novel cell-based MMEJ reporter (Figure 7A). This reporter construct contains a red fluorescent protein (E2-Crimson) followed by a P2A peptide sequence and an in-frame downstream GFP. To create the reporter, we inserted an I-SceI cut site followed by nonsense codons in all three reading frames into a structurally and functionally critical region of E2-Crimson, and we flanked this I-SceI cut site with 9bp regions of microhomology. Because this region of E2-Crimson is functionally critical, only restoration of the wild-type sequence through utilization of the provided microhomology regions by MMEJ will result in red fluorescent signal. In contrast, end-joining repair that ablates the transcriptional stop sequence, while retaining the correct reading frame of E2-Crimson, will result in GFP fluorescence through translational restart.
Figure 7. FEN1 participates in MMEJ.
(A) Schematic of MMEJ reporter. (B-D) Negative control siRNA-transfected cells from (E) were infected with lentivirus expressing I-SceI, grown for 5 d, and were FACS-sorted for E2-Crimson+GFP+ populations (C) or GFP+ populations (D). Genomic DNA was isolated from these FACS-sorted cell populations as well as total unsorted cells (B), and the E2-Crimson construct was PCR-amplified and adapted for Illumina sequencing. The results of 500K reads were plotted for each cell population as a percent of MMEJ or NHEJ, with the indicated size (bp) of microhomology utilized for repair. Restoration of the wild-type sequence is indicated with an asterisk. (E-F) U2-OS cells containing the MMEJ reporter were transfected with the indicated siRNAs and transduced with lentiviral I-SceI. The percent of E2-Crimson+GFP+ cells was quantified by FACS after 5 d of growth. Error bars reflect variability of technical triplicates, and the number of asterisks indicates the statistical significance for the corresponding siRNA versus negative control siRNA (*=p<0.05,**=p<0.01,***=p<0.001) (F). FACS plots for a representative replicate from each experiment are shown in (E). (G) U2-OS cells containing the MMEJ reporter were treated with the indicated drugs at increasing doses and transduced with lentiviral I-SceI (DNA-PK inhibitor doses: 1=3.5 μM, 2=8.5 μM, 3=12 μM, FEN1 inhibitor doses: 1=5 μM, 2=10 μM, 3=25 μM). The percent of E2-Crimson+GFP+ cells was quantified by FACS after 5 d of growth. Error bars reflect variability of technical quadruplicates, and the number of asterisks indicates the statistical significance for the corresponding siRNA versus vehicle control, as described above.
To verify that the reporter functions as intended, we performed deep sequencing of the reporter construct following I-SceI cleavage. We introduced our MMEJ Reporter at a low MOI into U2-OS cells and reverse transfected negative control siRNA. We FACS-sorted E2-Crimson+-GFP+ cells as well as GFP+ cells and isolated genomic DNA from these sorted cell populations. After PCR-amplifying 702bp from the E2-Crimson construct and adapting the resulting PCR amplicons for Illumina sequencing, we sequenced both the sorted and unsorted populations to a read depth of 500K reads per population. We found that in the absence of any perturbation, repair of the DSB created by I-SceI, which creates a 4bp 3’ overhang, occurs more often by MMEJ than NHEJ but rarely results in restoration of the wild-type sequence (Figure 7B). In contrast, E2-Crimson+-GFP+ FACS-sorted cells predominately reflect perfect restoration of the wild-type sequence (Figure 7C). GFP+ FACS-sorted cells contain a variety of repair products that remove the transcriptional stop sequence but retain in-frame expression of GFP (Figure 7D).
After characterizing this system, we tested the effect of several perturbations on our MMEJ reporter in U2-OS cells. We compared the effect of siRNA-mediated knockdown of XRCC5 (Ku70), XRCC6 (Ku80), PARP1, FEN1, and POLθ with negative control siRNA. siRNA-mediated depletion of the core NHEJ repair proteins Ku70 and Ku80 resulted in an expected increase of MMEJ (Figures 7E–F). In contrast, siRNA-mediated depletion of POLθ or FEN1 impaired MMEJ repair (Figures 7E–F). Both POLθ and PARP1 are known to have a role in MMEJ (Sfeir and Symington, 2015) but in our case interference with PARP had only a minor effect. A defect in MMEJ repair upon siRNA-mediated depletion of Fen1 demonstrates that Fen1 plays an important functional role in MMEJ in mammalian cells.
We subsequently proceeded to test the effect of chemical inhibition of DNA-PK and Fen1 on our MMEJ reporter in U2-OS cells (Figure 7G). We observed a dose-dependent increase in signal from our MMEJ reporter through inhibition of the NHEJ mediator DNA-PK with its kinase inhibitor NU7026 (Willmore et al., 2004). Conversely, we observed a highly statistically significant dose-dependent decrease in MMEJ reporter signal through inhibition of FEN1 with its chemical inhibitor, described above (Exell et al., 2016; Ward et al., 2017). Thus, our screens for novel B2SL targets identified a second novel component of the MMEJ pathway in addition to Polθ, highlighting the dependence of B2MUT cells upon MMEJ in the absence of functional HR repair.
Discussion
It has become clear that PARP inhibitor effectiveness in BRCA-deficient cells is largely due to the phenomenon of Parp trapping, more so than exploitation of an underlying mild synthetic lethal genetic interaction between PARP and BRCA1/2 deficiencies. Furthermore, a systematic analysis of BRCA2-mutant interactions with deficiencies of other DNA repair functions has not been reported. We reasoned that unexplored SL relationships with BRCA-deficiency may exist and that these might represent valuable drug targets for metastatic or refractory BRCA-deficient breast tumors, recurrent HGSOC, and PARP inhibitor-resistant BRCA-deficient tumors. Thus, we designed a series of genetic screens to identify novel B2SL targets. We show here that there are several B2SL genes and pathways whose inactivation exhibits stronger synthetic lethality with BRCA2 loss-of-function than PARP1 inactivation. Both APEX2 and FEN1 are previously unreported, potent, and generalizable B2SL hits that represent novel drug targets.
Inactivation of APEX2 hampers the BER pathway, which overall is more important to survival in the context of BRCA2 inactivation than in counterpart BRCA2 wild-type cells. It has previously been assumed that PARP involvement in BER is SL with BRCA inactivation because unresolved SSBs create an increased DSB burden during replication. However, inactivation of APEX2 would not necessarily lead to an increase in SSBs, since Ape1 and Ape2 endonucleases are responsible for the nick-generating step of BER. Rather, in the absence of Ape2 endonuclease function, residual AP sites would persist during S phase, resulting in replication fork stalling. We hypothesize that this increase in replication stress is more toxic in the absence of functional BRCA2, since Brca2 has been shown to safeguard forks from resection (Ray Chaudhuri et al., 2016). This hypothesis is consistent with our observation that B2MUT cells are more dependent on Atr activation, given the master regulatory role of Atr activation in replication fork protection (Saldivar et al., 2017), and we found that Ape2 depletion led to more prominent phosphorylation of Chk1 and Rpa32 in B2MUT than B2WT cells, implying increased Atr activation.
Despite the fact that Ape1 is commonly believed to be the dominant BER AP endonuclease, we found that APEX2 is a stronger BRCA1 and BRCA2 SL hit than APEX1. The fact that we cannot complement Ape2 loss with overexpression of wild-type Ape1 suggests that there is a distinct feature of Ape2 that is responsible for its stronger SL phenotype. In contrast to Ape1, Ape2 binds PCNA (Burkovics et al., 2009; Ide et al., 2003; Tsuchimoto et al., 2001) and associates with replication forks (Dungrawala et al., 2015), which suggests that Ape2 may participate in replication-associated BER. Supporting this notion, Ape2 mutants with ablated PCNA-binding motifs fail to support survival of BRCA2-mutant cells. We propose that Ape2’s PCNA-association allows it to process apurinic sites at replication forks, which would otherwise block fork progression and result in breaks, requiring BRCA2 activity for repair and restoration of DNA synthesis. This model is consistent with our finding that APEX2 depletion results in an increase of γH2AX foci.
Surprisingly, we found that depletion of every human Rad51 paralog, as well as Palb2 and Brca2 itself, enhances growth of our B2MUT cell lines. We note that both the ovarian and colonic B2MUT lines possess similar BRCA2 truncation mutants, present in about ~16% of breast tumors. This truncation mutant protein is capable of binding Rad51; thus we propose that exon 11 BRCA2 truncation mutants may exert a toxic, neomorphic function involving Rad51. We did not see this same effect when we tested B2MUT cells possessing a longer truncation mutant, terminating at BRC repeat 8, suggesting that this toxic function involving Rad51 may be unique to BRCA2 truncation mutants that terminate near BRC repeat 5. This result suggests that inhibiting HR in the context of a BRCA2 exon 11 truncation mutant may actually accelerate tumor growth, implying that certain classes of HR inhibitors might exacerbate cancers if given to patients with exon 11 BRCA2 truncation mutants.
A few essential genes scored as SL in our shRNA screen, but not CRISPR-based screens, including the splicing factors SF3B2 and SF1. We validated that two inhibitors of the SF3B1 component of the U2 snRNP selectively impair growth of B2MUT cells. Since inactivation of BRCA2 leads to increased R-loop accumulation (Bhatia et al., 2014), dual inactivation of BRCA2 and SF3B1 may compound R-loop induced DNA damage to a toxic level. Alternatively, this effect may be explained by previously reported results that the U2 snRNP maintains protein levels of essential DDR components and prevents R-loop induced DNA damage (Adamson et al., 2012; Tanikawa et al., 2016). These two explanations for the effect of SF3B1 inhibition on BRCA2 mutant cells are not mutually exclusive.
Our strongest novel B2SL gene is the flap endonuclease FEN1. We validated FEN1 as a BRCA1 and BRCA2 SL target and showed that its endonuclease but not its exonuclease enzymatic activity is required to rescue the effect of FEN1 gRNA knockout in BRCA2 MUT cells. We showed using a novel cell-based reporter that FEN1 is an essential component of the MMEJ pathway; the fact that we found this phenotype through a B2SL screen underscores how important the MMEJ pathway becomes in the absence of functional HR. However, the SL phenotype of FEN1 knockout in B2MUT cells is likely multifactorial: its loss impairs BER, creates replication stress by hindering processing of Okazaki fragments, and also impairs MMEJ-mediated DSB repair. Indeed, we see that siRNA-mediated FEN1 depletion leads to increased phosphorylation of Atr substrates in B2MUT compared to B2WT cells, implying a greater degree of Atr activation. Overall, MMEJ appears to be more important in complementing loss of HR than classical NHEJ, perhaps implying that the predominant load of DSBs left unresolved by HR deficiency occurs during replication.
Finally, we showed that an existing inhibitor to FEN1 exhibits synthetic lethality in vitro, suggesting it could possibly be utilized in the treatment of BRCA-deficient tumors, subsequently or in combination with Parp inhibitors. Importantly, the toxic PARP-SSB intermediates that occur upon PARP trapping are thought to be resolved largely by BER; thus, because FEN1 executes the penultimate step of BER, it may exhibit a synergistic effect with Parp inhibition. Not only would its inhibition be SL in BRCA1deficient tumors due to the role of FEN1 in MMEJ, replication, and BER, but its inhibition should also leave toxic PARP-SSB complexes unresolved, potentially augmenting a synergistic lethality.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and request for reagents may be directed to the corresponding author Stephen J. Elledge (selledge@genetics.med.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines
Human DLD-1 cells and all clones derived from this cell line were maintained in RPMI-1640 Medium (ATCC modification) supplemented with 10% (v/v) fetal bovine serum (FBS), 100 units/mL penicillin, and 100 μg/mL streptomycin. Human PEO1 cells and their BRCA2 revertant clone C4–2 were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, 1% Pen-Strep, and 2mM glutamine. A pair of BRCA1 isogenic RPE1 cell lines was generously shared by Connor Clairmont and Alan D’Andrea; this isogenic cell line pair was created by CRISPR-based knockout of TP53 with or without CRISPR-based knockout of BRCA1 in the RPE1 background. These cells were grown in DMEM/F12 medium supplemented with 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin. HEK293T cells were maintained in DMEM supplemented with 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin.
METHOD DETAILS
Virus production and titering
To produce lentivirus, HEK293T cells were seeded in tissue culture dishes at a density equivalent to 6×105 cells per 0.9 cm2 surface area. Plasmid DNA was diluted into serum-free medium with a lentiviral packaging plasmid mixture of SV40 VSVg, Gag/Pol, Tat, and Rev, and transfected with PolyJet or MIRUS Trans-IT. After 48 h, the supernatant was harvested, filtered through a low-protein binding HT Tuffryn® membrane with 0.45 μm pores (Pall cat. #4184), and stored at −80°C. Lentiviral titer was determined by transducing the cell line of interest plated at clonogenic density with serial dilutions of virus in the presence of 4–8 μg/mL polybrene. After selecting with puromycin or NAT, colonies were counted to determine viral titer. The same process was utilized to package retrovirus when applicable, except that a retroviral packaging plasmid mixture of 1:1 Gag/Pol and VSVG was co-transfected with retroviral constructs. All transductions were performed in the presence of 4–8 μg/mL polybrene.
Generation of isogenic cell lines
We obtained a BRCA2 mutant cell line (BRCA2−/− DLD-1) from Horizon Pharma. This cell line was originally generated in the colonic pseudodiploid DLD-1 background through two successive rounds of homologous recombination introduced by adeno-associated virus, each of which created a small deletion in exon 11, leaving a residual loxP site and premature stop codon (Hucl et al., 2008). We received a pcDNA3.1-neo construct for full-length BRCA2 expression as a generous gift from Ralph Scully. We transformed this plasmid into DH5α E. coli and maintained growth of bacteria at 30°C in the presence of Ampicillin. The amplified plasmid was linearized, treated with calf alkaline phosphatase, and transfected into mutant BRCA2−/− DLD-1cell line with MIRUS. Neomycin selection was maintained for several weeks to identify stable integrants, and individual clones were isolated. Restoration of full-length BRCA2 expression was confirmed by Western blotting, and selected clones were shown to exhibit olaparib resistance and Rad51 foci formation after 10Gy IR.
shRNA screens
We designed a targeted sublibrary of 380 genes with either a known or suspected role in the DNA damage response. Inclusion of genes in this DNA damage sublibrary was based either on a known role in DNA repair described in the literature, or performance of the gene in prior screens for sensitivity to DNA damaging agents (unpublished data). We began by performing a primary shRNA screen to these 380 genes, with an shRNA library targeting each gene with 50 different shRNAs (~19K total). Pooled oligonucleotides encoding these shRNAs were cloned into the mir30 backbone context in the retroviral hairpin-expressing vector MSCV (Stegmeier et al., 2005). The library was packaged into retrovirus along with 10 negative control shRNAs in a pooled format, and titer was measured by a colony formation assay.
A pooled shRNA screen was then performed by transducing this retroviral library into both colonic DLD-1 B2MUT and B2WT cell lines at a low multiplicity of infection (0.5) in triplicate at a representation of 1000 cellular integrations per shRNA. After selection in 4 μg/mL puromycin for 3 d, cells were passaged for 12 population doubling (PDs), and cell pellets were collected both after puromycin selection (PD0) and after passaging cells for 12 population doublings (PD12). DLD-1 B2MUT cells required 35 d to reach 12 PDs, while DLD-1 B2WT cells required 19 d. Genomic DNA was isolated from these cell pellets by phenol/chloroform extraction, and the relative representation of library reagents in each sample was determined by Illumina sequencing of PCR-amplified half-hairpins (Schlabach et al., 2008).
Fifty genes that performed well in the primary shRNA screen were selected to comprise a secondary screen sublibrary. Ten hairpin sequences were designed to each of these fifty genes (~500 total) and synthesized along with 772 negative control shRNAs. Pooled oligonucleotides encoding these hairpins were cloned into the mir-E shRNA context in the lentiviral vector pHAGE pInducer10 (Meerbrey et al., 2011). A pooled shRNA screen was performed similarly to the primary shRNA screen in both colonic DLD-1 B2MUT and B2WT cell lines, with the lentiviral library pool being transduced at low multiplicity of infection (0.2) in triplicate at a representation of 1000. After selection with 4 μg/mL puromycin for 3 d, cells were passaged for 12 PDs, and cell pellets were collected from both starting PD0 and final PD12 cell populations. Again, genomic DNA was isolated from these cell pellets by phenol-chloroform extraction, and the relative representation of library reagents was determined by Illumina sequencing of PCR-amplified half-hairpins (Schlabach et al., 2008).
CRISPR screens
gRNAs were designed to 357 of the 380 DNA damage sublibrary genes targeted by the primary shRNA library. Ten gRNAs were designed to each gene, yielding ~3500 gRNAs total. A negative control library targeted to the E. coli genome with minimal predicted off-target cutting sites in the human genome was designed in parallel. These oligonucleotides were cloned in a pooled format into the LentiCRISPRv2 plasmid, which contains EFS-driven Cas9 (Sanjana et al., 2014). The DNA damage sublibrary and E. coli targeted negative control library were packaged into lentivirus and titered separately, but pooled before transduction into colonic or ovarian B2MUT or B2WT cells in triplicate at a MOI of 0.2. After puromycin selection (4 μg/mL for colonic B2MUT and B2WT lines and 1.5μg/mL for ovarian B2MUT and B2WT lines), cells were passaged for 12 PDs, and cell pellets were collected at both PD0 and PD12. The number of days required to achieve PD12 varied by cell lines (B2MUT colonic 33 d, B2WT colonic 18 d, B2MUT ovarian 17 d, B2WT ovarian 14 d). Genomic DNA was isolated from cell pellets using phenol-chloroform extraction, and gRNAs were PCR-amplified from genomic DNA and adapted for Illumina sequencing.
Multicolor Competition Assay
Individual gRNAs or small molecules were tested for synthetic lethality using a multi-color competition assay (MCA) (Luo et al., 2009). In this assay, GFP-labeled B2MUT cells are mixed with B2WT cells labeled with the DsRed-derivative red fluorescent protein E2-Crimson (E2C), at a 2:1 ratio. The percent of GFP+ cells was monitored over time using fluorescence-activated cell sorting (FACS), and unmixed GFP-labeled B2MUT and E2C-labeled B2WT cells were maintained in parallel to assure the purity of each population.
For MCA assays evaluating the effect of gRNAs, individual gRNAs were cloned into the LentiCRISPRv2 plasmid containing the previously described F+E modifications to the tracrRNA sequence (Chen et al., 2013). After sequence verification of the cloned gRNA, each gRNA to be tested was packaged separately into lentivirus (Sanjana et al., 2014; Shalem et al., 2014). Negative control gRNAs included three sequences targeted to the E. coli genome that are predicted to have no off-target cutting sites in the human genome, and several intergenic gRNA sequences predicted to have a single cutting site in the human genome, in a DNAse hypersensitive region, on chromosome 2, 15, or 16. The 2:1 mixture of pre-labeled B2MUT and B2WT cells, or B1MUT and B1WT cells, was infected with each gRNA to be tested, selected with puromycin, and the percent GFP+ cells after selection was measured as the starting percent GFP. After passaging in culture for 1 week, the percent of GFP+ cells from each sample was measured again. The change in percent GFP after passaging cells was normalized to the average of all negative control gRNAs and adjusted for differing relative growth rates as previously described (Pritchard et al., 2013), according to the following formula:
For MCA assays evaluating the effect of small molecules, serial dilutions of each drug were prepared in DMSO, with the final concentration of DMSO in culture never exceeding 0.45%. We verified that the addition of 0.45% DMSO had no effect on the change in percent GFP+ cells over time. We mixed GFPlabeled B2MUT with E2C-labeled B2WT cells as described above, and then added drug to the mixture every 3–4 days for 1 week of passaging in culture. We measured the percent of GFP+ positive cells by FACS and normalized to DMSO-treated cells using the formula above. The only exception to this procedure was for 2’-deoxyuridine 5’-monophosphate disodium salt (dUMP), which was solubilized in H2O. For this compound, change in percent GFP+ cells was normalized to the change in percent GFP+ cells with the addition of the same volume of H2O.
The drugs test in this study include: APE Inhibitor III (Calbiochem), ATR inhibitor VE-821 (Axon 1893), dUMP (Sigma D3876), E3330 (Sigma E8534), Olaparib (Selleck Chemicals S1060), and SSA (Adooq Bioscience A12700). Sudemycin 6 was a generous gift from Dr. Thomas Westbrook and FEN1 inhibitor was generously provided by Dr. Stephen Durant from AstraZeneca (Exell et al., 2016).
Western blotting
Cells were trypsinized, washed in PBS, counted in order to standardize gel loading, and lysed by resuspension in 2X NuPAGE LDS Sample Buffer (Thermo) containing Halt protease and phosphatase inhibitors (Thermo). Each sample was sonicated 4 times for 15 second intervals, with at least 15 seconds rest on ice in between successive sonication periods, before being boiled for 5 minutes at 95°C. Protein lysates were separated on 4–12% Bis-Tris gels (Thermo) and transferred to nitrocellulose membranes (Bio-Rad), which were blocked with 0.45μm-filtered 5% BSA in TBST (Cell Signaling Technology). The following primary antibodies and dilutions were used for immunoblotting: vinculin (1:1000, Sigma V9131), GAPDH (1:1000, Santa Cruz, sc-25778), BRCA2 (1:1000, Millipore OP95), phospho-Chk1 S345 (1:1000, Cell Signaling 2348), Chk1 (1:1000, Cell Signaling 2360), phospho H2AX S130 (1:1000, Millipore 05–636), FEN1 (1:500, Thermo 4E7), phospho RPA32 S4/8 (1:2000, Bethyl A300–254). Goat anti-mouse (1:5000, Jackson 115-035-003) or goat anti-rabbit (1:5000, Jackson 111-035-003) HRP-conjugated secondary antibodies were used for detection with enhanced chemiluminescence (ECL) (Perkin-Elmer NEL104001EA). For experiments involving siRNA-mediated depletion, ON-TARGET plus siRNA reagents were purchased from Dharmacon. Combined pools of 4 individual siRNAs were transfected using Lipofetamine RNAiMAX reagent (Thermo Fisher) and Western blotting was performed 48–72 hours after transfection.
Immunofluorescence
Cells grown on coverslips were fixed for 10 minutes at room temperature in 3.7% formaldehyde, freshly diluted in PBS. Cells were then permeabilized for 10 minutes at room temperature in 0.5% Triton-X in PBS and blocked for 20 minutes in 0.5% BSA and 0.2% gelatin from cold water fish skin (Sigma) in PBS. Three PBS washes occurred before each of these steps. Primary and secondary antibodies were diluted in PBS with 0.5% BSA and 0.2% gelatin from cold water fish skin (Sigma). Unless otherwise stated, primary antibody incubations were performed for 2 hours at room temperature in a dark, humidified chamber followed by a 1 h secondary antibody incubation in a dark, humidified chamber. The following antibodies and dilutions were used in this study: phospho-H2AX S139 (1:500, Millipore 05–636), Rad51 (1:200, 45 min, 37°C, Santa Cruz sc8349). Secondary antibodies included Alexa Fluor® 488 goat anti-mouse IgG (1:1000, Invitrogen A-11001), Alexa Fluor® 594 goat anti-mouse IgG (1:1000, Invitrogen A-11005), Alexa Fluor® 647 goat anti-mouse IgG (1:1000, Invitrogen A-21236), and Alexa Fluor® 488 goat anti-rabbit IgG (1:1000, Invitrogen A-11008). Coverslips were mounted on slides using Vectashield mounting medium with DAPI (Vector Laboratories), and imaging was performed on an Olympus Fluoview FV1000 Confocal Microscope. For experiments involving siRNA-mediated depletion, ON-TARGET plus siRNA reagents were purchased from Dharmacon. Combined pools of 4 individual siRNAs were transfected using Lipofetamine RNAiMAX reagent (Thermo Fisher) and immunofluorescence assays were performed 48–72 hours after transfection.
Complementation Assay
The ORF encoding full-length APEX2 was obtained from the human ORFeome 8.1 collection, and a stop codon was introduced after the last residue of the APEX2 coding sequence. A gRNA-resistant version of this construct was constructed by site-directed mutagenesis (Agilent) of the PAM and several bases in the seed sequence of the corresponding to a pre-validated APEX2 gRNA (APEX2 gRNA #3: 5’- GGTAGCATTGTCCTTACAGA-3’). This gRNA-resistant construct was used as the substrate to make several APEX2 point mutants.
The full-length ORF encoding FEN1 terminated by its natural stop codon was obtained from the Ultimate ORF collection (Thermo Fisher). A gRNA-resistant version of this construct was cloned by site-directed mutagenesis (Agilent) of the PAM and several bases in the seed sequence of a pre-validated gRNA to FEN1 (FEN1 gRNA #3: 5’- GGCTGGCAAAGTCTATGCTG-3’). This gRNA-resistant construct was used as the substrate to make several FEN1 point mutants.
All resulting constructs were cloned into a lentiviral destination vector by a Gateway LR reaction. A small, 20 amino-acid negative control peptide and the red fluorescent protein E2C were cloned in parallel to serve as negative controls. Pre-validated gRNAs to APEX2 or FEN1 were transduced into ovarian B2MUT cells or ovarian B2WT cells and selected with 1.5 μg/mL puromycin. Subsequently, ORF constructs were transduced and selected with 200 ug/mL nourseothricin sulfate (Gold Biotechnology N-500–1). Each gRNA and ORF combination was counted individually after selection and plated evenly at 20K cells/well in 24-well tissue culture plates, in triplicate. After growth for 8 d, cells were trypsinized and cell numbers were quantified using CellTiter-Glo (Promega). For the assay in Figure 4J, cell numbers were quantified using CyQUANT (Thermo Fisher) after 2 d of growth.
MMEJ Reporter Assay
The BstXI site of the DsRed-derivative red fluorescent protein E2-Crimson (E2C) was used to introduce an I-SceI cut site followed by a stop codon and flanked by 8bp microhomology regions. A construct containing E2C, a P2A sequence, and GFP was assembled using a sewing PCR reaction, with PCR primers terminating with Gateway attB sites. This PCR product was cloned by a BP reaction into pDONR221, and subsequently transferred to a lentiviral destination vector (pHAGE-EF1α-DEST) by a Gateway LR reaction. The resulting lentiviral construct was sequence-verified and packaged into lentivirus as described above. To create the reporter cell line, U2-OS osteosarcoma cells were infected with this lentiviral construct at a very low MOI of 0.1 and selected with 500 μg/mL nourseothricin sulfate (Gold Biotechnology). I-SceI was cloned into a lentiviral construct expressing BFP, and this construct was packaged into lentivirus to transduce reporter cells. Ninety-six hours after transduction with I-SceI, reporter cells were analyzed by FACS for BFP, GFP, and E2C expression. The percent of GFP+ and E2C+ cells were quantified from gated BFP+ cells that received the I-SceI virus. For sequencing of the reporter, a region of 702bp containing modified E2-Crimson was PCR-amplified from genomic DNA, isolated as described above. PCR products were adapted for Illumina sequencing with the standard Illumina primer, and each sample was sequenced to a depth of 500K reads. Reads were manually aligned to the parent cassette to characterized and quantify the percent of MMEJ and NHEJ in each sample.
QUANTIFICATION AND STATISTICAL ANALYSIS
BRCA2 mutation diagramming
All mutations in BRCA2 found in the METABRIC breast cancer mutation profiling study (Pereira et al., 2016) were downloaded from cBioPortal (Cerami et al., 2012). The functional impact of all BRCA2 mutations was assessed by batch querying Polyphen-2 (Adzhubei et al., 2010). The percent of functionally impactful BRCA2 mutations that occur in the BRC repeat region was calculated by dividing the number of damaging missense, frameshift, or nonsense mutations found in the BRC repeat region divided by the total number of damaging missense, frameshift, or nonsense mutations.
Screen Data Analysis
For the primary shRNA, secondary shRNA, CRISPR colonic, and CRISPR ovarian screens, NGS sequencing reads were aligned to the respective reference library using Bowtie (Langmead, 2010) and counts were obtained for each shRNA or sgRNA. MAGeCK (Li et al., 2014), MAGeCK-VISPR (Li et al., 2015) and edgeR (Robinson et al., 2010) were used to calculate false discovery rates (FDRs) for each gene (Supplemental Tables 2–4). For the primary shRNA screen, phenotypic scores were also computed using ATARiS computational method (Supplemental Table 5) (Shao et al., 2013). The performance of each gene was ranked according to the FDR computed for its performance by edgeR and MAGeCK (Supplemental Table 6). The overall edgeR or MAGeCK CRISPR rank for each gene by edgeR or MAGeCK was determined as the average of the gene’s performance in the colonic and ovarian CRISPR screens (Supplemental Table 6). The overall CRISPR rank for each gene was determined based on the average of each gene’s edgeR and MAGeCK CRISPR rank (Supplemental Table 6).
Supplementary Material
Acknowledgements
We are grateful to Andy Elia and Teresa Davoli for helpful discussions. We thank Eric Wooten and Qikai Xu for help in data alignment, and the members of the Elledge lab for feedback. We are grateful to Qikai Xu and Mamie Li for their work in library design and development. K.E.M. is supported by award Number T32GM007753 from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health. S.J.E. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Adamson B, Smogorzewska A, Sigoillot FD, King RW, and Elledge SJ (2012). A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol 14, 318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, and Sunyaev SR (2010). A method and server for predicting damaging missense mutations. Nat Methods 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan L, and Bambara RA (2013). Flap endonuclease 1. Annu Rev Biochem 82, 119–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennardo N, Cheng A, Huang N, and Stark JM (2008). Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet 4, e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia V, Barroso SI, Garcia-Rubio ML, Tumini E, Herrera-Moyano E, and Aguilera A (2014). BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 511, 362–365. [DOI] [PubMed] [Google Scholar]
- Bitler BG, Watson ZL, Wheeler LJ, and Behbakht K (2017). PARP inhibitors: Clinical utility and possibilities of overcoming resistance. Gynecol Oncol 147, 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, and Helleday T (2005). Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917. [DOI] [PubMed] [Google Scholar]
- Burkovics P, Hajdu I, Szukacsov V, Unk I, and Haracska L (2009). Role of PCNAdependent stimulation of 3’-phosphodiesterase and 3’−5’ exonuclease activities of human Ape2 in repair of oxidative DNA damage. Nucleic Acids Res 37, 4247–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira A, and Kowalczykowski SC (2011). Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc Natl Acad Sci U S A 108, 10448–10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MI, O’Connor KW, Konstantinopoulos PA, Elledge SJ, Boulton SJ, et al. (2015). Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature 518, 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SH, Yu AM, and McVey M (2010). Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet 6, e1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. (2013). Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, and Elledge SJ (2010). The DNA damage response: making it safe to play with knives. Mol Cell 40, 179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretu C, Schmitzova J, Ponce-Salvatierra A, Dybkov O, De Laurentiis EI, Sharma K, Will CL, Urlaub H, Luhrmann R, and Pena V (2016). Molecular Architecture of SF3b and Structural Consequences of Its Cancer-Related Mutations. Mol Cell 64, 307–319. [DOI] [PubMed] [Google Scholar]
- Decottignies A (2007). Microhomology-mediated end joining in fission yeast is repressed by pku70 and relies on genes involved in homologous recombination. Genetics 176, 1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doksani Y, and de Lange T (2014). The role of double-strand break repair pathways at functional and dysfunctional telomeres. Cold Spring Harb Perspect Biol 6, a016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungrawala H, Rose KL, Bhat KP, Mohni KN, Glick GG, Couch FB, and Cortez D (2015). The Replication Checkpoint Prevents Two Types of Fork Collapse without Regulating Replisome Stability. Mol Cell 59, 998–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effenberger KA, Urabe VK, Prichard BE, Ghosh AK, and Jurica MS (2016). Interchangeable SF3B1 inhibitors interfere with pre-mRNA splicing at multiple stages. RNA 22, 350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T, and Matulonis U (2017). PARP inhibitors in ovarian cancer: evidence, experience and clinical potential. Ther Adv Med Oncol 9, 253–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exell JC, Thompson MJ, Finger LD, Shaw SJ, Debreczeni J, Ward TA, McWhirter C, Sioberg CL, Martinez Molina D, Abbott WM, et al. (2016). Cellularly active N-hydroxyurea FEN1 inhibitors block substrate entry to the active site. Nat Chem Biol 12, 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. (2005). Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921. [DOI] [PubMed] [Google Scholar]
- Hadi MZ, Ginalski K, Nguyen LH, and Wilson DM 3rd (2002). Determinants in nuclease specificity of Ape1 and Ape2, human homologues of Escherichia coli exonuclease III. J Mol Biol 316, 853–866. [DOI] [PubMed] [Google Scholar]
- Helleday T (2011). The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol 5, 387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucl T, Rago C, Gallmeier E, Brody JR, Gorospe M, and Kern SE (2008). A syngeneic variance library for functional annotation of human variation: application to BRCA2. Cancer Res 68, 5023–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide Y, Tsuchimoto D, Tominaga Y, Iwamoto Y, and Nakabeppu Y (2003). Characterization of the genomic structure and expression of the mouse Apex2 gene. Genomics 81, 47–57. [DOI] [PubMed] [Google Scholar]
- Kaelin WG Jr. (2005). The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer 5, 689–698. [DOI] [PubMed] [Google Scholar]
- Kent T, Chandramouly G, McDevitt SM, Ozdemir AY, and Pomerantz RT (2015). Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat Struct Mol Biol 22, 230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Ohno S, Sasaki Y, and Matsuura M (2013). Hereditary breast and ovarian cancer susceptibility genes (review). Oncol Rep 30, 1019–1029. [DOI] [PubMed] [Google Scholar]
- Kolinjivadi AM, Sannino V, De Antoni A, Zadorozhny K, Kilkenny M, Techer H, Baldi G, Shen R, Ciccia A, Pellegrini L, et al. (2017). Smarcal1-Mediated Fork Reversal Triggers Mre11-Dependent Degradation of Nascent DNA in the Absence of Brca2 and Stable Rad51 Nucleofilaments. Mol Cell 67, 867–881 e867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B (2010). Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics Chapter 11, Unit 11 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Koster J, Xu H, Chen CH, Xiao T, Liu JS, Brown M, and Liu XS (2015). Quality control, modeling, and visualization of CRISPR screens with MAGeCK-VISPR. Genome Biol 16, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, and Liu XS (2014). MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15, 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonosov M, Anand S, Sangrithi M, Davies R, and Venkitaraman AR (2003). Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev 17, 3017–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, and Elledge SJ (2009). A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137, 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JL, Kim EM, Haber JE, and Lee SE (2003). Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol Cell Biol 23, 8820–8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, and Sfeir A (2015). Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, Herschkowitz JI, Burrows AE, Ciccia A, Sun T, et al. (2011). The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci U S A 108, 3665–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, et al. (2017). Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet 49, 1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza MR, Monk BJ, Herrstedt J, Oza AM, Mahner S, Redondo A, Fabbro M, Ledermann JA, Lorusso D, Vergote I, et al. (2016). Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N Engl J Med 375, 2154–2164. [DOI] [PubMed] [Google Scholar]
- Mol CD, Izumi T, Mitra S, and Tainer JA (2000). DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected]. Nature 403, 451–456. [DOI] [PubMed] [Google Scholar]
- Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, and Pommier Y (2012). Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res 72, 5588–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narod S, Booth CM, and Foulkes WD (2017). Olaparib for Metastatic Germline BRCA-Mutated Breast Cancer. N Engl J Med 377, 1792. [DOI] [PubMed] [Google Scholar]
- Narod SA (2006). Modifiers of risk of hereditary breast cancer. Oncogene 25, 5832–5836. [DOI] [PubMed] [Google Scholar]
- Nilsen H, and Krokan HE (2001). Base excision repair in a network of defence and tolerance. Carcinogenesis 22, 987–998. [DOI] [PubMed] [Google Scholar]
- Ozdemir AY, Rusanov T, Kent T, Siddique LA, and Pomerantz RT (2018). Polymerase theta-helicase efficiently unwinds DNA and RNA-DNA hybrids. J Biol Chem 293, 5259–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ, et al. (2016). The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun 7, 11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrucelli N, Daly MB, and Pal T (1993). BRCA1- and BRCA2-Associated Hereditary Breast and Ovarian Cancer In GeneReviews((R)), Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, and Amemiya A, eds. (Seattle (WA)). [Google Scholar]
- Pritchard JR, Bruno PM, Gilbert LA, Capron KL, Lauffenburger DA, and Hemann MT (2013). Defining principles of combination drug mechanisms of action. Proc Natl Acad Sci U S A 110, E170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai G, Vyjayanti VN, Dorjsuren D, Simeonov A, Jadhav A, Wilson DM 3rd, and Maloney DJ (2012). Synthesis, biological evaluation, and structure-activity relationships of a novel class of apurinic/apyrimidinic endonuclease 1 inhibitors. J Med Chem 55, 3101–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Callen E, Ding X, Gogola E, Duarte AA, Lee JE, Wong N, Lafarga V, Calvo JA, Panzarino NJ, et al. (2016). Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535, 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson M, Goessl C, and Domchek S (2017a). Olaparib for Metastatic Germline BRCA-Mutated Breast Cancer. N Engl J Med 377, 1792–1793. [DOI] [PubMed] [Google Scholar]
- Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, Delaloge S, Li W, Tung N, Armstrong A, et al. (2017b). Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N Engl J Med 377, 523–533. [DOI] [PubMed] [Google Scholar]
- Roy R, Chun J, and Powell SN (2011). BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer 12, 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai W, Swisher EM, Jacquemont C, Chandramohan KV, Couch FJ, Langdon SP, Wurz K, Higgins J, Villegas E, and Taniguchi T (2009). Functional restoration of BRCA2 protein by secondary BRCA2 mutations in BRCA2-mutated ovarian carcinoma. Cancer Res 69, 6381–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, Villegas E, Jacquemont C, Farrugia DJ, Couch FJ, et al. (2008). Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 451, 1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldivar JC, Cortez D, and Cimprich KA (2017). The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat Rev Mol Cell Biol 18, 622–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, et al. (2008). Cancer proliferation gene discovery through functional genomics. Science 319, 620–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, and Jasin M (2011). Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, and Symington LS (2015). Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem Sci 40, 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao DD, Tsherniak A, Gopal S, Weir BA, Tamayo P, Stransky N, Schumacher SE, Zack TI, Beroukhim R, Garraway LA, et al. (2013). ATARiS: computational quantification of gene suppression phenotypes from multisample RNAi screens. Genome Res 23, 665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D’Andrea AD, et al. (2007). Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129, 289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies J, Waizenegger A, Barton O, Surder M, Wright WD, Heyer WD, and Lobrich M (2016). Nek1 Regulates Rad54 to Orchestrate Homologous Recombination and Replication Fork Stability. Mol Cell 62, 903–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Hu G, Rickles RJ, Hannon GJ, and Elledge SJ (2005). A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci U S A 102, 13212–13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom CE, Johansson F, Uhlen M, Szigyarto CA, Erixon K, and Helleday T (2011). Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res 39, 3166–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanikawa M, Sanjiv K, Helleday T, Herr P, and Mortusewicz O (2016). The spliceosome U2 snRNP factors promote genome stability through distinct mechanisms; transcription of repair factors and R-loop processing. Oncogenesis 5, e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto D, Sakai Y, Sakumi K, Nishioka K, Sasaki M, Fujiwara T, and Nakabeppu Y (2001). Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res 29, 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward TA, McHugh PJ, and Durant ST (2017). Small molecule inhibitors uncover synthetic genetic interactions of human flap endonuclease 1 (FEN1) with DNA damage response genes. PLoS One 12, e0179278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcsh PL, and King MC (2001). BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum Mol Genet 10, 705–713. [DOI] [PubMed] [Google Scholar]
- Willmore E, de Caux S, Sunter NJ, Tilby MJ, Jackson GH, Austin CA, and Durkacz BW (2004). A novel DNA-dependent protein kinase inhibitor, NU7026, potentiates the cytotoxicity of topoisomerase II poisons used in the treatment of leukemia. Blood 103, 4659–4665. [DOI] [PubMed] [Google Scholar]
- Xanthoudakis S, Miao GG, and Curran T (1994). The redox and DNA-repair activities of Ref-1 are encoded by nonoverlapping domains. Proc Natl Acad Sci U S A 91, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn KE, Averill AM, Aller P, Wood RD, and Doublie S (2015). Human DNA polymerase theta grasps the primer terminus to mediate DNA repair. Nat Struct Mol Biol 22, 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou GM, and Maitra A (2008). Small-molecule inhibitor of the AP endonuclease 1/REF-1 E3330 inhibits pancreatic cancer cell growth and migration. Mol Cancer Ther 7, 2012–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.