Abstract
Rare genetic variants in yeast explain a large amount of phenotypic variation in a complex trait like growth.
Research organism: S. cerevisiae
Related research article Bloom JS, Boocock J, Treusch S, Sadhu MJ, Day L, Oates-Barker H, Kruglyak L. 2019. Rare variants contribute disproportionately to quantitative trait variation in yeast. eLife 8:e49212. doi: 10.7554/eLife.49212
Related research article Fournier T, Abou Saada O, Hou J, Peter J, Caudal E, Schacherer J. 2019. Extensive impact of low-frequency variants on the phenotypic landscape at population-scale. eLife 8:e49258. doi: 10.7554/eLife.49258
Although most of the 3000 million nucleotides in the human genome are the same in every person on the planet, there are about 90 million sites that can vary between individuals. The source of all phenotypic variation in humans lies in these 90 million genetic variants, and in their interactions with each other and with the environment. Identifying the genetic variants that are involved in a specific trait (such as height or disease status) is a long-standing goal in biology.
Today, researchers rely on genome-wide association studies (GWAS) to find the genetic variants that are relevant to a specific trait. In GWAS the genomes of individuals are analyzed to see if particular genetic variants are correlated with variation in traits of interest. GWAS results have identified hundreds of variants underlying phenotypic variation in humans, mice, fruit flies, rice, maize, and many other taxa. Yet, despite the large number of alleles that have been identified using this technique, the amount of phenotypic variation they explain is just a fraction of what twin and pedigree studies predict is heritable. For example, twin studies have shown that approximately 80% of variation in human height can be explained by genetic factors (Silventoinen et al., 2012). However, the results of the best powered GWAS only explain around 20% of such variation (Wood et al., 2014). This gap is known as the ‘missing heritability problem’.
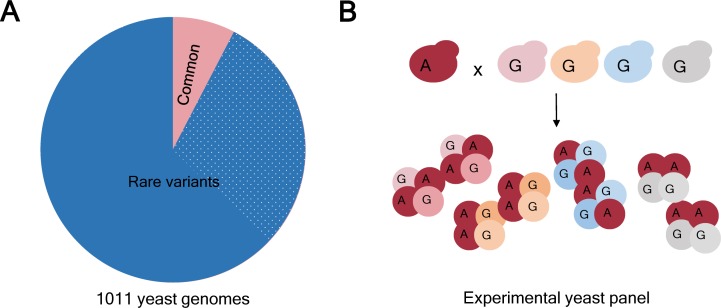
Rare and low-frequency genetic variants (which have allele frequencies of <1% and <5% respectively) have been proposed as one explanation for the missing heritability problem (reviewed in Gibson, 2012). Such variants are routinely excluded from GWAS studies because when an allele is present in few individuals, the statistical analysis used to draw correlations between traits and alleles is not powerful enough to obtain significant results. As a consequence around 90% of genetic variation in humans and other organisms like yeast has so far gone unexplored (Figure 1A; Auton et al., 2015; Peter et al., 2018). The missing heritability might be hiding in plain sight, but until now, studying the effect of rare alleles on the variation of traits influenced by more than one gene was extremely challenging. Now, in eLife, two independent groups report the results of experiments on yeast which show that rare variants have a fundamental role in phenotypic variation at the population level.
Figure 1. Allele frequency in natural isolates of yeast and in the experimental populations.
(A) Based on a study of 1011 genomes it is known that 93% of the genetic variants in the yeast Saccharomyces cerevisiae are rare (that is, they have a frequency <1%; blue). Moreover, just over 508000 variants (31% of the total; dotted blue) were found in just 1 of the 1011 genomes studied. However, genome-wide association studies (GWAS) tend to focus on the 7% of genetic variants that are common (that is, have a frequency >1%; pink). (B) The frequency of a rare allele can be increased by crossing a yeast isolate carrying the rare variant with an isolate with the alternative (more common) variant. To obtain a variety of isolates with a specific rare allele in different genetic backgrounds, the isolate carrying the rare variant (allele A, dark red) can be crossed with several different isolates with the alternative allele (allele G, pink, yellow, blue, grey). As a result, allele A is more frequent in the experimental panel than in the parental isolates, making it suitable for GWAS analysis. Importantly, regardless of the frequency that any allele reaches in the experimental panel, the real natural frequency can be looked up in the collection of 1011 yeast genomes (panel A).
In a monumental effort, the two groups independently selected a set of wild and domesticated yeast isolates from all over the world and crossed them to generate a genetically diverse panel of thousands of strains (Figure 1B). They then exposed each cross to more than 35 different media conditions and quantified their growth by measuring colony size. As a result of the crossing scheme, genetic variants that were present in just one or a few yeast isolates were now present in hundreds of samples in the experimental panels (Figure 1B). This allowed the groups to include a large number of rare variants (up to 28% of the total) in the GWAS analysis: many of these variants would have been excluded from traditional GWAS studies due to their low allele frequency.
Both groups independently identified thousands of genetic variants associated with growth, and estimated that over half of growth variance can be attributed to additive effects. To determine how variants with different frequencies contributed to phenotypic effects, variants were classified into either rare (<1%) and common (>1%) (Bloom et al., 2019), or rare (<1%), low frequency (1–5%) and common (>5%) (Fournier et al., 2019). This classification was based on 1011 yeast genomes that represent global yeast diversity (Figure 1A; Peter et al., 2018). Strikingly, rare variants contributed a disproportionate amount to phenotypic variation in both studies.
In one study Joseph Schacherer and co-workers at the University of Strasbourg – including Téo Fournier as first author – found that 16% of the GWAS results were rare alleles even when they made up just 4% of all the variants used in the experiments (Fournier et al., 2019). In the other study Joshua Bloom, Leonid Kruglyak and colleagues at UCLA estimated that over half of the observed growth variation can be explained by rare variants, even when they represented only 28% of the variants used (Bloom et al., 2019). The UCLA team also found that the rare variants detected in GWAS tend to have larger effect sizes than common variants, tend to reduce growth ability, and tend to have arisen more recently in evolutionary time.
These results join recent efforts exploring the effect of rare variants on complex traits. For human height it has been shown that rare variants have effect sizes ten times larger than common variants (Marouli et al., 2017), and that together they account for most of the missing heritability in this trait (Wainschtein et al., 2019). In parallel, it was estimated that at least a quarter of gene expression heritability in humans is accounted for by rare variants (Hernandez et al., 2019). The fact that in humans, as well as yeast, the contribution of rare variants to complex traits is now beyond doubt suggests that it may be the same in other species. However, addressing this question in organisms with larger genomes and not amenable to crossing schemes remains challenging. But rest assured, researchers will find a way.
Biography
Luisa F Pallares is at the Lewis-Sigler Institute for Integrative Genomics and the Department of Ecology and Evolutionary Biology, Princeton University, Princeton, United States
Competing interests
No competing interests declared.
References
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR, 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JS, Boocock J, Treusch S, Sadhu MJ, Day L, Oates-Barker H, Kruglyak L. Rare variants contribute disproportionately to quantitative trait variation in yeast. eLife. 2019;8:e49212. doi: 10.7554/eLife.49212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier T, Abou Saada O, Hou J, Peter J, Caudal E, Schacherer J. Extensive impact of low-frequency variants on the phenotypic landscape at population-scale. eLife. 2019;8:e49258. doi: 10.7554/eLife.49258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. Rare and common variants: twenty arguments. Nature Reviews Genetics. 2012;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RD, Uricchio LH, Hartman K, Ye C, Dahl A, Zaitlen N. Ultrarare variants drive substantial cis heritability of human gene expression. Nature Genetics. 2019;51:1349–1355. doi: 10.1038/s41588-019-0487-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marouli E, Graff M, Medina-Gomez C, Lo KS, Wood AR, Kjaer TR, Fine RS, Lu Y, Schurmann C, Highland HM, Rüeger S, Thorleifsson G, Justice AE, Lamparter D, Stirrups KE, Turcot V, Young KL, Winkler TW, Esko T, Karaderi T, Locke AE, Masca NG, Ng MC, Mudgal P, Rivas MA, Vedantam S, Mahajan A, Guo X, Abecasis G, Aben KK, Adair LS, Alam DS, Albrecht E, Allin KH, Allison M, Amouyel P, Appel EV, Arveiler D, Asselbergs FW, Auer PL, Balkau B, Banas B, Bang LE, Benn M, Bergmann S, Bielak LF, Blüher M, Boeing H, Boerwinkle E, Böger CA, Bonnycastle LL, Bork-Jensen J, Bots ML, Bottinger EP, Bowden DW, Brandslund I, Breen G, Brilliant MH, Broer L, Burt AA, Butterworth AS, Carey DJ, Caulfield MJ, Chambers JC, Chasman DI, Chen YI, Chowdhury R, Christensen C, Chu AY, Cocca M, Collins FS, Cook JP, Corley J, Galbany JC, Cox AJ, Cuellar-Partida G, Danesh J, Davies G, de Bakker PI, de Borst GJ, de Denus S, de Groot MC, de Mutsert R, Deary IJ, Dedoussis G, Demerath EW, den Hollander AI, Dennis JG, Di Angelantonio E, Drenos F, Du M, Dunning AM, Easton DF, Ebeling T, Edwards TL, Ellinor PT, Elliott P, Evangelou E, Farmaki AE, Faul JD, Feitosa MF, Feng S, Ferrannini E, Ferrario MM, Ferrieres J, Florez JC, Ford I, Fornage M, Franks PW, Frikke-Schmidt R, Galesloot TE, Gan W, Gandin I, Gasparini P, Giedraitis V, Giri A, Girotto G, Gordon SD, Gordon-Larsen P, Gorski M, Grarup N, Grove ML, Gudnason V, Gustafsson S, Hansen T, Harris KM, Harris TB, Hattersley AT, Hayward C, He L, Heid IM, Heikkilä K, Helgeland Ø, Hernesniemi J, Hewitt AW, Hocking LJ, Hollensted M, Holmen OL, Hovingh GK, Howson JM, Hoyng CB, Huang PL, Hveem K, Ikram MA, Ingelsson E, Jackson AU, Jansson JH, Jarvik GP, Jensen GB, Jhun MA, Jia Y, Jiang X, Johansson S, Jørgensen ME, Jørgensen T, Jousilahti P, Jukema JW, Kahali B, Kahn RS, Kähönen M, Kamstrup PR, Kanoni S, Kaprio J, Karaleftheri M, Kardia SL, Karpe F, Kee F, Keeman R, Kiemeney LA, Kitajima H, Kluivers KB, Kocher T, Komulainen P, Kontto J, Kooner JS, Kooperberg C, Kovacs P, Kriebel J, Kuivaniemi H, Küry S, Kuusisto J, La Bianca M, Laakso M, Lakka TA, Lange EM, Lange LA, Langefeld CD, Langenberg C, Larson EB, Lee IT, Lehtimäki T, Lewis CE, Li H, Li J, Li-Gao R, Lin H, Lin LA, Lin X, Lind L, Lindström J, Linneberg A, Liu Y, Liu Y, Lophatananon A, Luan J, Lubitz SA, Lyytikäinen LP, Mackey DA, Madden PA, Manning AK, Männistö S, Marenne G, Marten J, Martin NG, Mazul AL, Meidtner K, Metspalu A, Mitchell P, Mohlke KL, Mook-Kanamori DO, Morgan A, Morris AD, Morris AP, Müller-Nurasyid M, Munroe PB, Nalls MA, Nauck M, Nelson CP, Neville M, Nielsen SF, Nikus K, Njølstad PR, Nordestgaard BG, Ntalla I, O'Connel JR, Oksa H, Loohuis LM, Ophoff RA, Owen KR, Packard CJ, Padmanabhan S, Palmer CN, Pasterkamp G, Patel AP, Pattie A, Pedersen O, Peissig PL, Peloso GM, Pennell CE, Perola M, Perry JA, Perry JR, Person TN, Pirie A, Polasek O, Posthuma D, Raitakari OT, Rasheed A, Rauramaa R, Reilly DF, Reiner AP, Renström F, Ridker PM, Rioux JD, Robertson N, Robino A, Rolandsson O, Rudan I, Ruth KS, Saleheen D, Salomaa V, Samani NJ, Sandow K, Sapkota Y, Sattar N, Schmidt MK, Schreiner PJ, Schulze MB, Scott RA, Segura-Lepe MP, Shah S, Sim X, Sivapalaratnam S, Small KS, Smith AV, Smith JA, Southam L, Spector TD, Speliotes EK, Starr JM, Steinthorsdottir V, Stringham HM, Stumvoll M, Surendran P, 't Hart LM, Tansey KE, Tardif JC, Taylor KD, Teumer A, Thompson DJ, Thorsteinsdottir U, Thuesen BH, Tönjes A, Tromp G, Trompet S, Tsafantakis E, Tuomilehto J, Tybjaerg-Hansen A, Tyrer JP, Uher R, Uitterlinden AG, Ulivi S, van der Laan SW, Van Der Leij AR, van Duijn CM, van Schoor NM, van Setten J, Varbo A, Varga TV, Varma R, Edwards DR, Vermeulen SH, Vestergaard H, Vitart V, Vogt TF, Vozzi D, Walker M, Wang F, Wang CA, Wang S, Wang Y, Wareham NJ, Warren HR, Wessel J, Willems SM, Wilson JG, Witte DR, Woods MO, Wu Y, Yaghootkar H, Yao J, Yao P, Yerges-Armstrong LM, Young R, Zeggini E, Zhan X, Zhang W, Zhao JH, Zhao W, Zhao W, Zheng H, Zhou W, EPIC-InterAct Consortium, CHD Exome+ Consortium, ExomeBP Consortium, T2D-Genes Consortium, GoT2D Genes Consortium, Global Lipids Genetics Consortium, ReproGen Consortium, MAGIC Investigators. Rotter JI, Boehnke M, Kathiresan S, McCarthy MI, Willer CJ, Stefansson K, Borecki IB, Liu DJ, North KE, Heard-Costa NL, Pers TH, Lindgren CM, Oxvig C, Kutalik Z, Rivadeneira F, Loos RJ, Frayling TM, Hirschhorn JN, Deloukas P, Lettre G. Rare and low-frequency coding variants alter human adult height. Nature. 2017;542:186–190. doi: 10.1038/nature21039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter J, De Chiara M, Friedrich A, Yue JX, Pflieger D, Bergström A, Sigwalt A, Barre B, Freel K, Llored A, Cruaud C, Labadie K, Aury JM, Istace B, Lebrigand K, Barbry P, Engelen S, Lemainque A, Wincker P, Liti G, Schacherer J. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature. 2018;556:339–344. doi: 10.1038/s41586-018-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silventoinen K, Sammalisto S, Perola M, Boomsma DI, Cornes BK, Davis C, Dunkel L, de Lange M, Harris JR, Hjelmborg JVB, Luciano M, Martin NG, Mortensen J, Nisticò L, Pedersen NL, Skytthe A, Spector TD, Stazi MA, Willemsen G, Kaprio J. Heritability of adult body height: a comparative study of twin cohorts in eight countries. Twin Research. 2012;6:399–408. doi: 10.1375/136905203770326402. [DOI] [PubMed] [Google Scholar]
- Wainschtein P, Jain DP, Yengo L, Zheng Z, Cupples LA. Recovery of trait heritability from whole genome sequence data. bioRxiv. 2019 doi: 10.1101/588020. [DOI]
- Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Chu AY, Estrada K, Luan Jian'an, Kutalik Z, Amin N, Buchkovich ML, Croteau-Chonka DC, Day FR, Duan Y, Fall T, Fehrmann R, Ferreira T, Jackson AU, Karjalainen J, Lo KS, Locke AE, Mägi R, Mihailov E, Porcu E, Randall JC, Scherag A, Vinkhuyzen AAE, Westra H-J, Winkler TW, Workalemahu T, Zhao JH, Absher D, Albrecht E, Anderson D, Baron J, Beekman M, Demirkan A, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Fraser RM, Goel A, Gong J, Justice AE, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Lui JC, Mangino M, Leach IM, Medina-Gomez C, Nalls MA, Nyholt DR, Palmer CD, Pasko D, Pechlivanis S, Prokopenko I, Ried JS, Ripke S, Shungin D, Stancáková A, Strawbridge RJ, Sung YJ, Tanaka T, Teumer A, Trompet S, van der Laan SW, van Setten J, Van Vliet-Ostaptchouk JV, Wang Z, Yengo L, Zhang W, Afzal U, Ärnlöv J, Arscott GM, Bandinelli S, Barrett A, Bellis C, Bennett AJ, Berne C, Blüher M, Bolton JL, Böttcher Y, Boyd HA, Bruinenberg M, Buckley BM, Buyske S, Caspersen IH, Chines PS, Clarke R, Claudi-Boehm S, Cooper M, Daw EW, De Jong PA, Deelen J, Delgado G, Denny JC, Dhonukshe-Rutten R, Dimitriou M, Doney ASF, Dörr M, Eklund N, Eury E, Folkersen L, Garcia ME, Geller F, Giedraitis V, Go AS, Grallert H, Grammer TB, Gräßler J, Grönberg H, de Groot LCPGM, Groves CJ, Haessler J, Hall P, Haller T, Hallmans G, Hannemann A, Hartman CA, Hassinen M, Hayward C, Heard-Costa NL, Helmer Q, Hemani G, Henders AK, Hillege HL, Hlatky MA, Hoffmann W, Hoffmann P, Holmen O, Houwing-Duistermaat JJ, Illig T, Isaacs A, James AL, Jeff J, Johansen B, Johansson Åsa, Jolley J, Juliusdottir T, Junttila J, Kho AN, Kinnunen L, Klopp N, Kocher T, Kratzer W, Lichtner P, Lind L, Lindström J, Lobbens S, Lorentzon M, Lu Y, Lyssenko V, Magnusson PKE, Mahajan A, Maillard M, McArdle WL, McKenzie CA, McLachlan S, McLaren PJ, Menni C, Merger S, Milani L, Moayyeri A, Monda KL, Morken MA, Müller G, Müller-Nurasyid M, Musk AW, Narisu N, Nauck M, Nolte IM, Nöthen MM, Oozageer L, Pilz S, Rayner NW, Renstrom F, Robertson NR, Rose LM, Roussel R, Sanna S, Scharnagl H, Scholtens S, Schumacher FR, Schunkert H, Scott RA, Sehmi J, Seufferlein T, Shi J, Silventoinen K, Smit JH, Smith AV, Smolonska J, Stanton AV, Stirrups K, Stott DJ, Stringham HM, Sundström J, Swertz MA, Syvänen A-C, Tayo BO, Thorleifsson G, Tyrer JP, van Dijk S, van Schoor NM, van der Velde N, van Heemst D, van Oort FVA, Vermeulen SH, Verweij N, Vonk JM, Waite LL, Waldenberger M, Wennauer R, Wilkens LR, Willenborg C, Wilsgaard T, Wojczynski MK, Wong A, Wright AF, Zhang Q, Arveiler D, Bakker SJL, Beilby J, Bergman RN, Bergmann S, Biffar R, Blangero J, Boomsma DI, Bornstein SR, Bovet P, Brambilla P, Brown MJ, Campbell H, Caulfield MJ, Chakravarti A, Collins R, Collins FS, Crawford DC, Cupples LA, Danesh J, de Faire U, den Ruijter HM, Erbel R, Erdmann J, Eriksson JG, Farrall M, Ferrannini E, Ferrières J, Ford I, Forouhi NG, Forrester T, Gansevoort RT, Gejman PV, Gieger C, Golay A, Gottesman O, Gudnason V, Gyllensten U, Haas DW, Hall AS, Harris TB, Hattersley AT, Heath AC, Hengstenberg C, Hicks AA, Hindorff LA, Hingorani AD, Hofman A, Hovingh GK, Humphries SE, Hunt SC, Hypponen E, Jacobs KB, Jarvelin M-R, Jousilahti P, Jula AM, Kaprio J, Kastelein JJP, Kayser M, Kee F, Keinanen-Kiukaanniemi SM, Kiemeney LA, Kooner JS, Kooperberg C, Koskinen S, Kovacs P, Kraja AT, Kumari M, Kuusisto J, Lakka TA, Langenberg C, Le Marchand L, Lehtimäki T, Lupoli S, Madden PAF, Männistö S, Manunta P, Marette A, Matise TC, McKnight B, Meitinger T, Moll FL, Montgomery GW, Morris AD, Morris AP, Murray JC, Nelis M, Ohlsson C, Oldehinkel AJ, Ong KK, Ouwehand WH, Pasterkamp G, Peters A, Pramstaller PP, Price JF, Qi L, Raitakari OT, Rankinen T, Rao DC, Rice TK, Ritchie M, Rudan I, Salomaa V, Samani NJ, Saramies J, Sarzynski MA, Schwarz PEH, Sebert S, Sever P, Shuldiner AR, Sinisalo J, Steinthorsdottir V, Stolk RP, Tardif J-C, Tönjes A, Tremblay A, Tremoli E, Virtamo J, Vohl M-C, Electronic Medical Records and Genomics (eMEMERGEGE) Consortium, MIGen Consortium, PAGEGE Consortium, LifeLines Cohort Study. Amouyel P, Asselbergs FW, Assimes TL, Bochud M, Boehm BO, Boerwinkle E, Bottinger EP, Bouchard C, Cauchi S, Chambers JC, Chanock SJ, Cooper RS, de Bakker PIW, Dedoussis G, Ferrucci L, Franks PW, Froguel P, Groop LC, Haiman CA, Hamsten A, Hayes MG, Hui J, Hunter DJ, Hveem K, Jukema JW, Kaplan RC, Kivimaki M, Kuh D, Laakso M, Liu Y, Martin NG, März W, Melbye M, Moebus S, Munroe PB, Njølstad I, Oostra BA, Palmer CNA, Pedersen NL, Perola M, Pérusse L, Peters U, Powell JE, Power C, Quertermous T, Rauramaa R, Reinmaa E, Ridker PM, Rivadeneira F, Rotter JI, Saaristo TE, Saleheen D, Schlessinger D, Slagboom PE, Snieder H, Spector TD, Strauch K, Stumvoll M, Tuomilehto J, Uusitupa M, van der Harst P, Völzke H, Walker M, Wareham NJ, Watkins H, Wichmann H-E, Wilson JF, Zanen P, Deloukas P, Heid IM, Lindgren CM, Mohlke KL, Speliotes EK, Thorsteinsdottir U, Barroso I, Fox CS, North KE, Strachan DP, Beckmann JS, Berndt SI, Boehnke M, Borecki IB, McCarthy MI, Metspalu A, Stefansson K, Uitterlinden AG, van Duijn CM, Franke L, Willer CJ, Price AL, Lettre G, Loos RJF, Weedon MN, Ingelsson E, O'Connell JR, Abecasis GR, Chasman DI, Goddard ME, Visscher PM, Hirschhorn JN, Frayling TM. Defining the role of common variation in the genomic and biological architecture of adult human height. Nature Genetics. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]