Abstract
p53, p63, and p73, the members of the p53 family of proteins, are structurally similar proteins that play central roles regulating cell cycle and apoptotic cell death. Alternative splicing at the carboxyl terminus and the utilization of different promoters further categorizes these proteins as having different isoforms for each. Among such isoforms, TA and ΔN versions of each protein serve as the pro and the anti-apoptotic proteins, respectively. Changes in the expression patterns of these isoforms are noted in many human cancers. Proteins of certain human herpesviruses, like Kaposi's sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus (EBV), interact with p53 family members and alter their expressions in many malignancies. Upon infections in the B cells and epithelial cells, EBV expresses different lytic or latent proteins during viral replication and latency respectively to preserve viral copy number, chromosomal integrity and viral persistence inside the host. In this review, we have surveyed and summarised the interactions of EBV gene products, known so far, with the p53 family proteins. The interactions between P53 and EBV oncoproteins are observed in stomach cancer, non-Hodgkin's lymphoma (NHL) of the head and neck, Nasopharyngeal Cancer (NPC), Gastric carcinoma (GC) and Burkitt's lymphoma (BL). EBV latent protein EBNA1, EBNA3C, LMP-1, and lytic proteins BZLF-1 can alter p53 expressions in many cancer cell lines. Interactions of p63 with EBNA-1, 2, 5, LMP-2A and BARF-1 have also been investigated in several cancers. Similarly, associations of p73 isoform with EBV latent proteins EBNA3C and LMP-1 have been reported. Methylation and single nucleotide polymorphisms in p53 have also been found to be correlated with EBV infection. Therefore, interactions and altered expression strategies of the isoforms of p53 family proteins in EBV associated cancers propose an important field for further molecular research.
Keywords: Epstein-Bar virus; p53, p63 and p73 isoforms; Malignancies; Cell biology; Cell death; Cell differentiation; Viruses; Cancer research
Epstein-Bar virus (EBV); p53, p63 and p73 isoforms; Malignancies; Cell biology; Cell death; Cell differentiation; Viruses; Cancer research
1. Introduction
Initiation of any malignancy involves a series of mutations in the progenitor cell induced by external and internal stress stimuli, leading to the uncontrolled cell division, clonal proliferation, and invasion. p53, a human tumor suppressor protein, plays a crucial role in cell cycle control and apoptosis (Pflaum et al., 2014; Sullivan et al., 2017). It is also involved in transcription independent cellular signaling, including cell death via mitochondria or through the cytosol mediated pathway (Speidel, 2010; Sullivan et al., 2017; Vaseva and Moll, 2009). Two other recently discovered proteins p63 and p73, which belongs to the P53 family, share a high degree of homology with p53 and aroused from the triplication of a common inherited gene (Murray-Zmijewski et al., 2006). N-terminal transactivation domain (TAD), DNA-binding domain (DBD) and the C-terminal oligomerization domain (OD) are the three common structural domains of p53, p63, and p73 recognized so far (Candi et al., 2014; Sullivan et al., 2017). p63 is involved in the limb, skin and craniofacial development, whereas p73 is known to facilitate neurogenesis (Yang et al., 1999; Yang et al., 2000). Several studies have shown that dysfunction in any of the p53 family proteins plays a key role in the progression of tumorigenesis.
Certain Human Herpesviruses (HHVs), like Herpes simplex virus (HSV) and Kaposi sarcoma-associated Herpesvirus (KSHV), interact with proteins of p53 family (Mohanty et al., 2015; Friborg et al., 1999; Maruzuru et al., 2013; Orosz et al., 2010; Santag et al., 2012). EBV (HHV-4) is an oncogenic virus of the Gamma herpesvirus family and was identified as the first human virus linked to malignancies among immunocompetent people in developing countries. EBV initially infects the oropharyngeal epithelial cell, followed by the establishment of the latency in B cells, epithelial cells and natural killer/T cells (Kang and Kieff, 2015). During latency, it can cause many human cancers, including Nasopharyngeal Carcinoma, Burkitt's lymphoma, Hodgkin's and non-Hodgkin's lymphomas (Thompson and Kurzrock, 2004) etc. This review has focused on the altered expression patterns of the p53 family proteins due to the involvement of EBV genes or their products during malignancies.
2. Main text
2.1. p53, p63, and p73 isoforms
Three promoter regions (two reside in exon 1 and one in intron 4) are identified in p53. Δ133p53 isoform is generated from the truncation of the amino-terminal region of intron 4 (Bourdon et al., 2005). Alternative splicing of exon 2 and alternative translational initiation of ATG40 in the TAD domain have created the Δ40p53 isoform (Murray-Zmijewski et al., 2006). Another isoform Δ160p53 has been generated from the start of translation at ATG160, which lacks the first 159 residues at intron 4 (Marcel et al., 2010). There are another three known isoforms of p53 (Full-length p53, p53β, andp53γ) which are produced from the alternative splicing of intron 9 (Murray-Zmijewski et al., 2006). To date, twelve p53 isoforms have been reported (Aoubala et al., 2010; Surget et al., 2014).
p63 and p73 proteins have both structural and functional similarities with the p53 protein. Due to alternative splicing of the carboxyl-terminal and utilization of different promoters, p53 family members are expressed in various isomeric forms. Transcription from P1 promoter has created TA isoform (i.e., TAp63 and TAp73); whereas the N-terminal truncated region or ΔN isoform (i.e., ΔNp63 and ΔNp73), which lacks TA domain, is generated from the P2 promoter (Moll and Slade, 2004; Soares and Zhou, 2018; Wei et al., 2012). It is evident that ΔN isoform acts as a critical negative inhibitor of TA isoform which further elaborates the functions of ΔN and TA isoforms as an anti-apoptotic factor and a pro-apoptotic factor respectively (Melino et al., 2003). Alternative splicing in the carboxyl-terminal region of the transcript has additionally produced variants of p63 and p73. Three such splice variants for p63 (α, β and γ) and nine for p73 (α, β, γ, δ, ε, θ, ζ, η, and η1) have been described to express TA or ΔN isomeric forms (Moll and Slade, 2004; Soares and Zhou, 2018; Wei et al., 2012). p63 and p73 is distinctively contained a motif, known as the sterile alpha motif (SAM), in the C-terminal region responsible for protein-protein interactions (Vikhreva et al., 2018; Wei et al., 2012).
2.2. EBV infection and gene expression
Inside the nucleus, the EBV genome either circularizes (episomal state) and undergoes latency, or it remains linear and goes through the lytic cycle (Tao et al., 2006). Different studies have demonstrated that the latency and lytic cycle in B cells and epithelial cells occur in somewhat different ways. In most cases, the lytic phase occurs after reactivation from latency in B cells followed by replication by viral DNA polymerase; in epithelial cells, this event often occurs after viral entry, followed by host DNA polymerase mediated replication (Odumade et al., 2011; Ragoczy et al., 1998). Following reactivation from latency, three types of temporal lytic genes are expressed, namely immediate early (IE), early, and late genes. Products of IE genes mainly encode transcriptional activators that act as a switch between lytic cycle and latency and enhance the expression of early genes. Early genes are responsible for replication and metabolism of the virus and for blocking of antigen processing. Finally, the late gene products play structural roles such as formation of viral capsid (VCA), and immune evasion of viruses. The functions of the EBV lytic genes are summarized in Table 1.
Table 1.
Function of the lytic genes of EBV or their products.
| Lytic Gene product | Utility | Reference | |
|---|---|---|---|
| Immediate Early |
BZLF 1 (ZEBRA, Z, EB 1 and Zta) |
|
(Baumann et al., 1998; Yang et al., 2015) |
| BRLF 1 (R, Rta) |
|
(Chang and Liu, 2000; Chen et al., 2009; Swenson et al., 2001) | |
| Early | BMRF 1 |
|
(Holley-Guthrie et al., 2005; Neuhierl and Delecluse, 2006; Zhang et al., 1997) |
| BRRF1 |
|
(Hagemeier et al., 2011; Hong et al., 2004; Yoshida et al., 2017) | |
| BALF5, BALF2, BBLF4, BSLF1, BBLF2/3, |
|
(Narita et al., 2015; Tsurumi et al., 1996; Yokoyama et al., 1999) | |
| BXLF1, BORF2 |
|
(Holton and Gentry, 1996; Johannsen et al., 2004) | |
| BARF 1 |
|
(Hoebe et al., 2013) | |
| SM |
|
(Ruvolo et al., 2001) | |
| BHRF 1 |
|
(Kawanishi et al., 2002; Zuo et al., 2017) | |
| BGLF 4 |
|
(Lee et al., 2007, 2008) | |
| BGLF 5 |
|
(Feederle et al., 2009a; Feederle et al., 2009b; Rowe et al., 2007) | |
| Late | BcLF1, BFRF3, BLRF2, and BdRF1 |
|
(Reischl et al., 1996; van Grunsven et al., 1993) |
| BLLF1 and BXLF2 |
|
(Heineman et al., 1988; Janz et al., 2000) | |
The latent genes of EBV and their roles are summarized in Table 2. During latency, EBNA-1, 2, 3A, 3B, 3C, LP or EBNA-5 (nuclear antigen) and LMP-1, 2A, 2B (membrane proteins) are expressed (Young and Murray, 2003). Besides latent proteins, expression of BART (Bam H1 A upright transcript) from Bam H1 A region of the viral genome and EBERs (EBV-encoded RNAs 1and 2) have been noticed in several malignancies (Iwakiri, 2014). The establishment of three distinct types of latent infection (Latency III, II, I), depending on viral gene expressions, have been demonstrated in infected B cells, particularly in memory cells. Latency III (EBNA-1, 2,3A, 3B, 3C, LP, LMP1and 2A/2B) is highly immunogenic and activates native B cells (Blast transformation); Latency II (EBNA1 and LMP1) is involved in B cell differentiation and Latency I (EBNA1) is essential for B cell proliferation (Young and Murray, 2003). It has been observed that latency II genes are expressed only in the epithelial cells (Shannon-Lowe et al., 2009).
Table 2.
Role of EBV latent genes or their products.
| Latency gene product | Utility | Reference |
|---|---|---|
| EBNA1 |
|
(Reisman et al., 1985; Sivachandran et al., 2012; Sugden et al., 1985; Yates et al., 1984; Yates et al., 1985) |
| EBNA2 |
|
(Cohen et al., 1989) |
| EBNA3A |
|
(Bazot et al., 2014; Harth-Hertle et al., 2013; Maruo et al., 2011; Paschos et al., 2009) |
| EBNA3B |
|
(White et al., 2012) |
| EBNA3C |
|
(Lin et al., 2002; Piovan et al., 2005; Saha et al, 2011a, 2011b; Saha and Robertson, 2011) |
| EBNA5 |
|
(Harada and Kieff, 1997; Mannick et al., 1991) |
| LMP1 |
|
(F Hu et al., 1993; Izumi et al., 1997; Mancao et al., 2005; Mosialos et al., 1995; Wang et al., 1985) |
| LMP2A |
|
(Allen et al., 2005; Bieging et al., 2009; Fish et al., 2014; Fukuda and Kawaguchi, 2014; Mancao and Hammerschmidt, 2007; Morrison and Raab-Traub, 2005; Swanson-Mungerson et al., 2010) |
| LMP2B |
|
(Rechsteiner et al., 2008; Rovedo and Longnecker, 2007) |
| EBERs |
|
(Fok et al., 2006; Houmani et al., 2009; Iwakiri et al., 2009; Komano et al., 1999; Samanta et al., 2006) |
| BHRF1 |
|
(Feederle et al., 2011; Seto et al., 2010; Xia et al., 2008) |
| BART |
|
(Choi et al., 2013; Haneklaus et al., 2012) |
2.3. Interaction of EBV with p53 isoform
The relationship between EBV infection and p53 expression is reported in idiopathic pulmonary fibrosis, gastric adenoma, gastric carcinoma, non-Hodgkin's lymphoma (NHL) of the head and neck, Nasopharyngeal Cancer, Burkitt's lymphoma and Gastric carcinoma (Lok et al., 2001). Moreover, the concentration of p53 is reported to determine cell cycle arrest and apoptosis in EBV infected B cells (Chen et al., 1998). Deletion of the residues 130–159 of EBNA3C open reading frame (ORF) is reported to have altered p53 expression compared to the wild type EBV, when the human PBMCs have been infected with EBNA3C construct (Shukla et al., 2016). Luciferase-based reporter assay has shown that the N-terminal domain of residue 130–190 of EBNA3C repressed the transcriptional activity of p53 by inhibiting DNA binding activity of p53 (Yi et al., 2009). Likewise, a direct association between EBNA3C and Gemim3 is observed, which stimulates the complex formation of p53 with gemim3, and thus inhibits the DNA binding activity of p53 in both B cell lymphoma and EBV transformed lymphoblastoid cells (Cai et al., 2011). Ubiquitin-specific-processing protease 7 (USP7) has a functional role in cell proliferation and apoptotic regulation through the interaction of p53 and Mdm2. USP7 is shown to interact with EBNA1 with a better affinity than p53 with the conserve DPGEGPS peptide in the osteosarcoma cell line (Saridakis et al., 2005). In Nasopharyngeal carcinoma, overexpression of LMP1 is reported and accumulated with p53 with an unknown mechanism. It has been noticed that LMP1 inhibits p53 mediated apoptosis through the activation of A20 (Shao et al., 2004, Liu et al., 2004). Transfection of LMP1 recombinant construct in human large cell lung carcinoma (with p53 deleted gene) and human osteogenic sarcoma cell line have established that the carboxyl-terminus activating regions of LMP1, CTAR1 or CTAR2 (related the region responsible for NF-κB activation) inhibit the transactivation of p53 through the influencing N-terminal transactivation domain. At the same time, p53-mediated DNA repair and transcription was repressed through the NF-κB pathway (Liu et al., 2004). LMP1 also blocked the p53 mediated apoptosis through the stimulation of the A20 gene expression in the non-small-cell lung cancer where temperature sensitive (ts) p53 and LMP1were stably expressed (Fries et al., 1996). DNA damage is shown to influence the ectopic p53 expression, which stimulated the endogenous expression of LMP1 in EBV transformed cell through the interaction with interferon regulatory factor 5 (IRF5) at the LMP1 promoter. Moreover ectopic IRF5 can increase the expression of endogenous LMP1 and blocked the p53 mediated apoptosis (Wang et al., 2017). EBV immediate-early protein BZLF1 (Z) is found to be interacting with the p53 through its C-terminus region and inhibits the p53 dependent transactivation in lymphoid cell, but overexpression of p53 restores its function (Zhang et al., 1994). Transfection of BZLF1 in HeLa cells was shown to enhance the p53 expression through the p53-DNA binding acceleration which eventually leads to the lytic gene replication (Sato et al., 2010). Transiently transfected p53 and p53 reporter genes in Jurkat T-lymphoblastoid cells expressing BZLF1 ORF encoded ZEBRA protein is shown to interact with p53 in vitro and alters the transcription of p53, thus enhanced the p53 dependent apoptosis in B lymphocytes and epithelial cells (Dreyfus et al., 2000). Complex formation between W repeats of 66 amino acid long peptides of EBNA5 (EBNA-LP) and p53 are also noted but how they affect the progression of malignancies is a bit unclear (Szekely et al., 1993). Table 3 lists the association of EBV with the expressions of p53 proteins.
Table 3.
Interaction of the P53 isoforms with EBV.
| p53 family | Interaction with the EBV genes or oncoproteins | EBV transformed cells/malignant tissue used |
|---|---|---|
| p53 | EBNA3C repress the transcriptional activity of p53 through130-190 region or by direct interaction with Gemim3 | B cell lymphoma cell |
| USP7 lowers the p53 level through the binding with EBNA1, | Osteosarcoma cell | |
| LMP-1 inhibits the transcription activation of p53 through the interaction of NF-κB pathway, stimulate A20 expression and inhibit p53, interact with IRF5. | Large cell lung carcinoma and Osteogenic sarcoma, Non-small-cell lung cells | |
| C terminal region of BZLF-1 binds with the p53 and alter its expression | Lymphoid and T-lymphoblastoid cell | |
| p63 | ΔNp63α interact with BARF1 and transactivate many folds. | Gastric Carcinoma cell |
| LMP-2A increases and stabilized the expression of ΔNp63α via the modulation of itch. | human keratinocyte cell | |
| EBNA5 has a direct interaction with the p63 in EBV positive Burkitt's lymphoma cell. | Burkitt's lymphoma cell | |
| EBNA2 interact with p63 through 310 to 336 amino acid sequence. | B cell lymphoma cell | |
| EBNA1 interact with the p63, but the mechanism of their interaction is still unknown. | Breast cancer tissue | |
| p73 | EBNA3C directly interfere with the p73 in the nucleus and stabilized ΔNp73. | B-cell |
| LMP-1 binds with p73 through the displacement of polycomb 2 complex component EZH2 and epigenetic changes via activation of JNK-1 | B cell | |
| Aberrant methylation in the exon 1 and SNPs in the p73 are associated with the EBV interaction. | Gastric Carcinoma and chronic lymphocytic leukemia |
2.4. Interaction of EBV with p63 isoform
BARF1 promoter contains multiple binding sites for p53 family proteins. Chromatin Immune Precipitation (ChIP) analysis has demonstrated that ΔNp63α binds to the immediate proximity of BARF1 promoter in NPC and Gastric carcinoma (GC). Co-transfection of various BARF1 promoter-reporter construct with the individual p53 family member in epithelial cell have revealed the p63 mediated transactivation of BARF1 with many folds (Hoebe et al., 2018). A physical association of LMP2A with ΔNp63α is observed in human keratinocyte, which has increased the expression and stabilization of ΔNp63α in cytoplasm and the nuclear membrane. This association is linked to the calcium-induced impairment of cellular differentiation with the involvement of PY and ITAM motifs. Co-immunoprecipitation studies have confirmed the LMP2A induced ΔNp63α expression through the modulation of Itch over ΔNp63α (Fotheringham et al., 2010). Another Co-immunoprecipitation study has explained the direct interaction between EBNA 5 and p63 in NPC and EBV positive Burkitt's lymphoma which may contribute to the stability of p63, but the mechanism is still unknown (Guo et al., 2006). To find out the relationship between EBV latent infection and the expression of p53 and p63 in breast cancer, immunohistochemistry study was performed in 85 formalin-fixed paraffin-embedded breast cancer using anti-EBNA-1, anti-p63, and anti-p53 antibodies. A significant correlation of EBNA-1 with p63 (p < 0.001) was found, but no significant association with p53 was detected (p = 0.10) (Ribeiro-Silva et al., 2004). The interaction of p63 with EBNA2 is reported in B cell lymphoma. P63 specifically recognizes and binds to the amino acid sequence of 310–336 in EBNA2. Mutation of the codon GTG > TCT in GTGGGA motif leads to the loss of recognition. The most common motif has been found among these 27 amino acid sequences to EBNA2 of both type1 and type2 EBV is GPPWWPP. Mutation of WW to SS or FF ablates the interaction of p63 which suggests that the hydrophobic and aromatic property of WW is essential for the interaction (Yalamanchili et al., 1994). Alterations of p63 expressions with the involvement of EBV are listed in Table 3.
2.5. Interaction of EBV with p73 isoform
The overview of the interaction between p73 and EBV in different malignancies is enumerated in Table 3. Immunoprecipitation and co-immunoprecipitation experiments have confirmed the formation of stable complexes between EBNA3C and p73 in the nucleus. Moreover, EBNA3C have down-regulated the p73 protein expression by stabilizing the ΔNp73 and inhibited doxorubicin-induced apoptosis in p53-null cell lines (Saos-2 and HCT p53 double mutant) (Sahu et al., 2014). The upregulation of ΔNp73 by LMP1 is reported in primary B cell infected recombinant EBV. Chip experiments have displayed the activation of ΔNp73 through the recruitment of p73 with the displacement of the polycomb 2 complex component EZH2 and epigenetic changes via activation of c-Jun NH2-terminal kinase 1 (JNK-1). LMP-1 mutant lacking the JNK-1 activating domain (CTAR2) did not influence the ΔNp73α expression levels (Accardi et al., 2013). DNA methylation in the CpG island of p73 was observed in EBV associated gastric carcinoma. The immunohistochemical assay has showed the loss of p73 expression in the EBV-associated GC compared to EBV negative GC. Methylation specific PCR reaction has detected the aberrant methylation patterns in p73 exon 1 in EBV associated gastric carcinoma (Tetsuo et al., 2007). It is observed that single nucleotide polymorphisms (SNPs) in p73 gene are important for the interaction with EBV in chronic lymphocytic leukemia. In the dominant model, two SNPs of p73 (rs3765701 and rs1885859) were shown to alter the association between aberrant EBV and chronic lymphocytic leukemia. A higher OR (odds ratio) was found in aberrant EBV positive patients with carriers of the wild-type homozygous genotypes compared to the EBV negative patients (Casabonne et al., 2011).
3. Conclusions
Despite the structural and functional similarities among p53, P63 and p73, there are many functional differences among them in relation to cell cycle regulation and malignancies. P53 regulates numerous gene expressions associated with G2/M and G1 cell cycle checkpoint regulation, DNA damage recognition and repair, apoptosis and cell death regulation (Levine and Oren, 2009). The roles of different p53 isoforms are depicted in many cancers. Elevated expression of Δ133p53 is reported in breast cancer, renal cell carcinoma, colon tumor but not in squamous carcinoma of the head and neck, suggesting a tissue-specific expression of this isoform (Boldrup et al., 2007; Bourdon et al., 2005; Fujita et al., 2009; Song et al., 2009). Like Δ133p53, overexpression of Δ40p53 is reported in human melanoma cell lines (Avery-Kiejda et al., 2008). Other isoforms like p53β and p53γ, have been found to be associated with the prognoses of several cancers. Association of p53β is reported with renal cell carcinoma and in ovarian cancer (Hofstetter et al., 2010; Song et al., 2009). Reduced expressions of p53β and p53γ have been detected in breast cancer (Bourdon et al., 2005). Logical functions of p53β and p53γ isoforms are still unclear.
The predominant expression of TAp63 is observed in the oocyte and epidermis. The absence of TAp63 in knock-out mice forms ulcers, hair defects, and decreased wound healing capacity (Suh et al., 2007). TAp63 also causes G1 cell cycle arrest by interacting with the p21 and p57/Kip2 (Guo et al., 2009b). p63 is found to interact with Bax and apoptotic death receptor in the intrinsic and extrinsic pathways, and increase apoptosis (Gressner et al., 2005). A significant expression of the ΔNp63 isoform in most epithelial cells has been described and is observed to have oncogenic potential. The Higher expression of p63, specifically ΔNp63 isoform, is associated with the prognosis of Nasopharyngeal Carcinoma (NPC), Head and Neck squamous cell carcinoma (HNSCC), Breast cancer, Lung and ovarian cancers with poor survival (Compérat et al., 2007; Crook et al., 2000; Dang et al., 2015; Marchini et al., 2008; Massion et al., 2003; Yamaguchi et al., 2000). ΔNp63α is observed to induce tumor-initiating stem-like proliferation by cooperating with Ras via interaction with the chromatin remodeling protein Lsh (Keyes et al., 2011). ΔNp63α also interact functionally with the transcription factor GLI2 in the hedgehog signaling pathway and induce tumorigenesis in osteosarcoma (Ram Kumar et al., 2014). Conversely, ΔNp63 has showed an opposite effect on metastasis. It has been suggested that the loss of ΔNp63 expression, p63 mediated repression of TGFβ-dependent cell migration, and ΔNp63α mediated regulation are involved in many cancer metastases (Barbieri et al., 2006; Hu et al., 2017; Koga et al., 2003). Contrariwise, TAp63 expression is associated with the induction of senescence and the inhibition of cell proliferation (Guo et al., 2009a; Wang et al., 2005). Absence or downregulation of that isoform can induce oncogenesis and metastasis, as studied in several cancers (Guo et al., 2004; Guo et al., 2009a; Lo Iacono et al., 2011; Park et al., 2000).
p73 has various utilities in cell cycle regulation, neuronal differentiation, and regulation of tumorigenesis. Like p53, it is involved in the regulation of different cell cycle checkpoint like the G1, G2/M and S phase through the interaction of several cell cycle controlling proteins (Balint et al., 2002; Innocente and Lee, 2005; Scian et al., 2007). p73 has been reported to transactivate the p75 neurotrophin receptor (have crucial functions in neurogenesis) and participate in the neuronal differentiation (Niklison-Chirou et al., 2013). The upregulation of Tp73 is found to be associated with the risk of breast, ovarian, hepatocellular, vulvar cancers and melanoma (Concin et al., 2004; Domínguez et al., 2006; O'Nions et al., 2001; Stiewe et al., 2004; Tve et al., 2004; Vikhanskaya et al., 2000; Zaika et al., 1999). Similarly, the expression of Np73 is reported with the projection of cervical cancer, gastric, esophageal, lung and colon and other cancers (Becker et al., 2006; Liu et al., 2006; Lööf et al., 2012; Uramoto et al., 2004; Vilgelm et al., 2010a,b). The expressions of p73α and p73β isoforms are predominant in normal colon and breast tissues, but different splice variants like p73γ, p73δ, p73φ, and p73ε are identified in colon and breast cancer (Vilgelm et al., 2010a,b; Zaika et al., 1999). Likewise, p73ε is reported to be expressed in Leukemic cell but is absent in mature myeloid cells (Tschan et al., 2000).
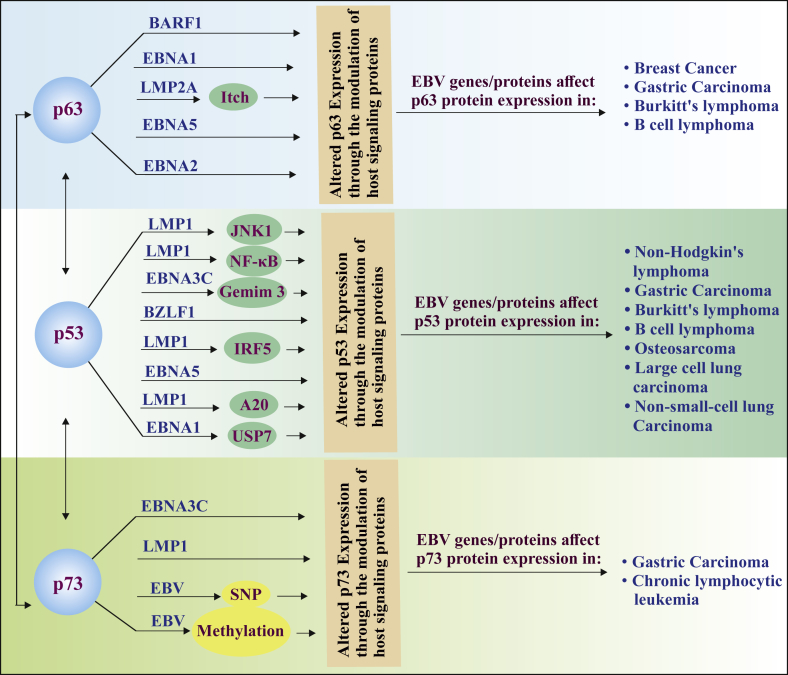
In this article, we have conceded the interaction of human herpesviruses 4 (EBV) genes or their products with the p53 family members in diverse EBV associated malignancies (Fig. 1) and highlighted the target sites of the EBV genes/proteins for future research. Furthermore, these viral-host protein interactions provide intimation for further molecular research of targeted antiviral therapy. EBV is an etiological factor for the development of NPC. A strong interaction has been reported between p63 and EBNA-5, which generally induce the expression of p63. p63 also interacts with EBNA-2, which transactivates LMP-1 and consequently induces cell proliferation and inhibits apoptosis. But it is yet to be deciphered whether p63 may have imposed any impact on LMP-1 after communicating with EBNA-2. Also, the detection of the association between p53 family members and EBV has been investigated in several cancers like carcinoma, leukemia, sarcoma, lymphoma; but the mechanisms of interactions or the expression patterns of those concerned proteins are yet to be deciphered. Despite the molecular mechanism or expression patterns, it is confirmed that the infection and expression of EBV genes are tissue specific. During infection, latency II associated genes have been found to be expressed in the epithelial cells instead of B cells (Shannon-Lowe et al., 2009). Similarly, the expression of BZLF1 variants is appeared to be cell-specific. In the same NPC patient, one variant of BZLF1 is associated with the epithelial cell and another variant with lymphocytes (Sacaze et al., 2001). This information generates curiosity about the tissue-specific interaction and expression of p53 family proteins in EBV associated cancers.
Fig. 1.
The figure has illustrated the inter-P53 family interaction and the association of the p53 family proteins with the EBV genes or their product during lytic and latency phase in several EBV associated cancer.
Moreover, the interactions among p53 family members and their isoforms have been investigated in the onset of tumorigenesis. Alteration of the TAp73 expression by ΔNp63 is reported in breast cancer (Rocco et al., 2006). Collecting data propose that the isoforms of p53, p63 and p73 interact and alter each other's expressions in the regulation of cell cycle, DNA damage, cellular stress defense and in carcinogenesis (Chen et al., 2001; Johnson et al., 2007; Wang et al., 2007; Wang and El-Deiry, 2006). Mutations in different regions of p53 have also been shown to interfere with the activation of TAp63 and TAp73 (Di Como et al., 1999; Gaiddon et al., 2001; Strano et al., 2002). It is still contradictory whether the alteration of the expression of one of the p53 family protein by EBV regulates the other protein of the same family in the same array or by any different manner. Much elaborate research is required to find out more specific mechanisms for the EBV induced regulation of p53/p63/p73 in EBV associated malignancies in particular tissues. The overall review of literature has paved a path for future studies of high importance in the virus-host interactions for oncogenic onset and in the field of molecular or clinical research to investigate the viral gene specific inhibitors.
Declarations
Author contribution statement
Koustav Chatterjee, Tathagata Choudhuri: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Piyanki Das, Nabanita Roy Chattopadhyay, Sudipa Mal: Analyzed and interpreted the data.
Funding statement
This work was supported by Visva-Bharati University, West Bengal, India. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- Accardi R., Fathallah I., Gruffat H., Mariggiò G., Le Calvez-Kelm F., Voegele C. Epstein - Barr virus transforming protein LMP-1 alters B cells gene expression by promoting accumulation of the oncoprotein ΔNp73α. PLoS Pathog. 2013;9(3) doi: 10.1371/journal.ppat.1003186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M.D., Young L.S., Dawson C.W. The Epstein-Barr virus-encoded LMP2A and LMP2B proteins promote epithelial cell spreading and motility. J. Virol. 2005;79(3):1789–1802. doi: 10.1128/JVI.79.3.1789-1802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoubala M., Murray-Zmijewski F., Khoury M.P., Fernandes K., Perrier S., Bernard H. p53 directly transactivates Δ133p53α, regulating cell fate outcome in response to DNA damage. Cell Death Differ. 2010;18:248. doi: 10.1038/cdd.2010.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery-Kiejda K.A., Zhang X.D., Adams L.J., Scott R.J., Vojtesek B., Lane D.P., Hersey P. Small molecular weight variants of p53 are expressed in human melanoma cells and are induced by the DNA-damaging agent cisplatin. Clin. Cancer Res. 2008;14(6):1659–1668. doi: 10.1158/1078-0432.CCR-07-1422. [DOI] [PubMed] [Google Scholar]
- Balint E., Phillips A., Kozlov S., L Stewart C., H Vousden K. Induction of p57KIP2 expression by p73. Proc. Natl. Acad. Sci. 2002;99(6):3529–3534. doi: 10.1073/pnas.062491899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri C.E., Tang L.J., Brown K.A., Pietenpol J.A. Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Res. 2006;66(15):7589–7597. doi: 10.1158/0008-5472.CAN-06-2020. [DOI] [PubMed] [Google Scholar]
- Baumann M., Mischak H., Dammeier S., Kolch W., Gires O., Pich D. Activation of the Epstein-Barr virus transcription factor BZLF1 by 12-O-Tetradecanoylphorbol-13-Acetate-Induced phosphorylation. J. Virol. 1998;72(10):8105–8114. doi: 10.1128/jvi.72.10.8105-8114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazot Q., Deschamps T., Tafforeau L., Siouda M., Leblanc P., Harth-Hertle M.L. Epstein–Barr virus nuclear antigen 3A protein regulates CDKN2B transcription via interaction with MIZ-1. Nucleic Acids Res. 2014;42(15):9700–9716. doi: 10.1093/nar/gku697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K., Pancoska P., Concin N., Vanden Heuvel K., Slade N., Fischer M. Patterns of p73 N-terminal isoform expression and p53 status have prognostic value in gynecological cancers. Int. J. Oncol. 2006;29(4):889–902. [PubMed] [Google Scholar]
- Bieging K.T., Amick A.C., Longnecker R. Epstein-Barr virus LMP2A bypasses p53 inactivation in a MYC model of lymphomagenesis. Proc. Natl. Acad. Sci. U.S.A. 2009;106(42):17945–17950. doi: 10.1073/pnas.0907994106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldrup L., Bourdon J.-C., Coates P.J., Sjöström B., Nylander K. Expression of p53 isoforms in squamous cell carcinoma of the head and neck. Eur. J. Cancer. 2007;43(3):617–623. doi: 10.1016/j.ejca.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon J.-C., Fernandes K., Murray-Zmijewski F., Liu G., Diot A., Xirodimas D.P. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19(18):2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Guo Y., Xiao B., Banerjee S., Saha A., Lu J. Epstein-barr virus nuclear antigen 3C stabilizes Gemin3 to block p53-mediated apoptosis. PLoS Pathog. 2011;7(12) doi: 10.1371/journal.ppat.1002418. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Candi E., Agostini M., Melino G., Bernassola F. How the TP53 family proteins TP63 and TP73 contribute to tumorigenesis: regulators and effectors. Hum. Mutat. 2014;35(6):702–714. doi: 10.1002/humu.22523. [DOI] [PubMed] [Google Scholar]
- Casabonne D., Reina O., Benavente Y., Becker N., Maynadié M., Foretová L. Single nucleotide polymorphisms of matrix metalloproteinase 9 (MMP9) and tumor protein 73 (TP73) interact with Epstein-Barr virus in chronic lymphocytic leukemia: results from the European case-control study EpiLymph. Haematologica. 2011;96(2):323–327. doi: 10.3324/haematol.2010.031161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L.-K., Liu S.-T. Activation of the BRLF1 promoter and lytic cycle of Epstein–Barr virus by histone acetylation. Nucleic Acids Res. 2000;28(20):3918–3925. doi: 10.1093/nar/28.20.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Huang S., Cooper N.R. Levels of p53 in Epstein–Barr virus-infected cells determine cell fate: apoptosis, cell cycle arrest at the G1/S boundary without apoptosis, cell cycle arrest at the G2/M boundary without apoptosis, or unrestricted proliferation. Virology. 1998;251(2):217–226. doi: 10.1006/viro.1998.9431. [DOI] [PubMed] [Google Scholar]
- Chen X., Zheng Y., Zhu J., Jiang J., Wang J. p73 is transcriptionally regulated by DNA damage, p53, and p73. Oncogene. 2001;20:769. doi: 10.1038/sj.onc.1204149. [DOI] [PubMed] [Google Scholar]
- Chen Y.-L., Chen Y.-J., Tsai W.-H., Ko Y.-C., Chen J.-Y., Lin S.-F. The Epstein-Barr virus replication and transcription activator, Rta/BRLF1, induces cellular senescence in epithelial cells. Cell Cycle. 2009;8(1):58–65. doi: 10.4161/cc.8.1.7411. [DOI] [PubMed] [Google Scholar]
- Choi H., Lee H., Kim S.R., Gho Y.S., Lee S.K. Epstein-barr virus-encoded MicroRNA BART15-3p promotes cell apoptosis partially by targeting BRUCE. J. Virol. 2013;87(14):8135–8144. doi: 10.1128/JVI.03159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J.I., Wang F., Mannick J., Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl. Acad. Sci. U.S.A. 1989;86(23):9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compérat E., Bièche I., Dargère D., Ferlicot S., Laurendeau I., Benoît G. p63 gene expression study and early bladder carcinogenesis. Urology. 2007;70(3):459–462. doi: 10.1016/j.urology.2007.04.030. [DOI] [PubMed] [Google Scholar]
- Concin N., Becker K., Slade N., Erster S., Mueller-Holzner E., Ulmer H. Transdominant ΔTAp73 isoforms are frequently up-regulated in ovarian cancer. Evidence for their role as epigenetic p53 inhibitors in vivo. Cancer Res. 2004;64(7):2449–2460. doi: 10.1158/0008-5472.can-03-1060. [DOI] [PubMed] [Google Scholar]
- Crook T., Nicholls J.M., Brooks L., O'Nions J., Allday M.J. High level expression of ΔN-p63: a mechanism for the inactivation of p53 in undifferentiated nasopharyngeal carcinoma (NPC) Oncogene. 2000;19:3439. doi: 10.1038/sj.onc.1203656. [DOI] [PubMed] [Google Scholar]
- Dang T.T., Esparza M.A., Maine E.A., Westcott J.M., Pearson G.W. ΔNp63α promotes breast cancer cell motility through the selective activation of components of the epithelial-to-mesenchymal transition program. Cancer Res. 2015;75(18):3925–3935. doi: 10.1158/0008-5472.CAN-14-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como C.J., Gaiddon C., Prives C. p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol. Cell. Biol. 1999;19(2):1438–1449. doi: 10.1128/mcb.19.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez G., García J.M., Peña C., Silva J., García V., Martínez L. ΔTAp73 upregulation correlates with poor prognosis in human tumors: putative in vivo network involving p73 isoforms, p53, and E2F-1. J. Clin. Oncol. 2006;24(5):805–815. doi: 10.1200/JCO.2005.02.2350. [DOI] [PubMed] [Google Scholar]
- Dreyfus D.H., Nagasawa M., Kelleher C.A., Gelfand E.W. Stable expression of Epstein-Barr virus BZLF-1–encoded ZEBRA protein activates p53-dependent transcription in human Jurkat T-lymphoblastoid cells. Blood. 2000;96(2):625–634. [PubMed] [Google Scholar]
- F Hu L., Chen F., Zheng X., Ernberg I., L Cao S., Christensson B. Clonability and tumorigenicity of human epithelial cells expressing the EBV encoded membrane protein LMPI. Oncogene. 1993;8(6):1575–1583. [PubMed] [Google Scholar]
- Feederle R., Bannert H., Lips H., Müller-Lantzsch N., Delecluse H.J. The Epstein-Barr virus alkaline Exonuclease BGLF5 serves pleiotropic functions in virus replication. J. Virol. 2009;83(10):4952–4962. doi: 10.1128/JVI.00170-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feederle R., Mehl-Lautscham A.M., Bannert H., Delecluse H.-J. The Epstein-Barr virus protein kinase BGLF4 and the exonuclease BGLF5 have opposite effects on the regulation of viral protein production. J. Virol. 2009;83(21):10877–10891. doi: 10.1128/JVI.00525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feederle R., Haar J., Bernhardt K., Linnstaedt S.D., Bannert H., Lips H. The members of an Epstein-Barr virus MicroRNA cluster cooperate to transform B lymphocytes. J. Virol. 2011;85(19):9801–9810. doi: 10.1128/JVI.05100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish K., Chen J., Longnecker R. Epstein-Barr virus latent membrane protein 2A enhances MYC-driven cell cycle progression in a mouse model of B lymphoma. Blood. 2014;123(4):530–540. doi: 10.1182/blood-2013-07-517649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fok V., Mitton-Fry R.M., Grech A., Steitz J.A. Multiple domains of EBER 1, an Epstein-Barr virus noncoding RNA, recruit human ribosomal protein L22. RNA. 2006;12(5):872–882. doi: 10.1261/rna.2339606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotheringham J.A., Mazzucca S., Raab-Traub N. Epstein-Barr virus latent membrane protein-2A-induced ΔNp63α expression is associated with impaired epithelial-cell differentiation. Oncogene. 2010;29:4287. doi: 10.1038/onc.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friborg J., Jr., Kong W.-p., Hottiger M.O., Nabel G.J. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- Fries K.L., Miller W.E., Raab-Traub N. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J. Virol. 1996;70(12):8653–8659. doi: 10.1128/jvi.70.12.8653-8659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K., Mondal A.M., Horikawa I., Nguyen G.H., Kumamoto K., Sohn J.J. p53 isoforms, Δ133p53 and p53β, are endogenous regulators of replicative cellular senescence. Nat. Cell Biol. 2009;11(9):1135–1142. doi: 10.1038/ncb1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., Kawaguchi Y. Role of the immunoreceptor tyrosine-based activation motif of latent membrane protein 2A (LMP2A) in Epstein-Barr virus LMP2A-induced cell transformation. J. Virol. 2014;88(9):5189–5194. doi: 10.1128/JVI.03714-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiddon C., Lokshin M., Ahn J., Zhang T., Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell. Biol. 2001;21(5):1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressner O., Schilling T., Lorenz K., Schulze Schleithoff E., Koch A., Schulze-Bergkamen H. TAp63α induces apoptosis by activating signaling via death receptors and mitochondria. EMBO J. 2005;24(13):2458–2471. doi: 10.1038/sj.emboj.7600708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Fan S., Jiang Y., Chen J., Li Z., Niu H. The expression of p63 gene in human non-small cell lung cancer. Chin. J. Lung Canc. 2004;7(1):31–34. doi: 10.3779/j.issn.1009-3419.2004.01.08. [DOI] [PubMed] [Google Scholar]
- Guo C., Pan Z.-G., Li D.-J., Yun J.-P., Zheng M.-Z., Hu Z.-Y. The expression of p63 is associated with the differential stage in nasopharyngeal carcinoma and EBV infection. J. Transl. Med. 2006;4 doi: 10.1186/1479-5876-4-23. 23-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Keyes W.M., Papazoglu C., Zuber J., Li W., Lowe S.W. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat. Cell Biol. 2009;11(12):1451–1457. doi: 10.1038/ncb1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Keyes W.M., Papazoglu C., Zuber J., Li W., Lowe S.W. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat. Cell Biol. 2009;11:1451. doi: 10.1038/ncb1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeier S.R., Barlow E.A., Kleman A.A., Kenney S.C. The Epstein-Barr virus BRRF1 protein, Na, induces lytic infection in a TRAF2- and p53-dependent manner. J. Virol. 2011;85(9):4318–4329. doi: 10.1128/JVI.01856-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haneklaus M., Gerlic M., Kurowska-Stolarska M., Rainey A.-A., Pich D., McInnes I.B. Cutting edge: mir-223 and EBV mir-BART15 regulate the NLRP3 inflammasome and IL-1β production. J. Immunol. 2012;189(8):3795–3799. doi: 10.4049/jimmunol.1200312. [DOI] [PubMed] [Google Scholar]
- Harada S., Kieff E. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J. Virol. 1997;71(9):6611–6618. doi: 10.1128/jvi.71.9.6611-6618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harth-Hertle M.L., Scholz B.A., Erhard F., Glaser L.V., Dölken L., Zimmer R., Kempkes B. Inactivation of intergenic enhancers by EBNA3A initiates and maintains polycomb signatures across a chromatin domain encoding CXCL10 and CXCL9. PLoS Pathog. 2013;9(9) doi: 10.1371/journal.ppat.1003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineman T., Gong M., Sample J., Kieff E. Identification of the Epstein-Barr virus gp85 gene. J. Virol. 1988;62(4):1101–1107. doi: 10.1128/jvi.62.4.1101-1107.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebe E.K., Le Large T.Y.S., Greijer A.E., Middeldorp J.M. BamHI-A rightward frame 1, an Epstein–Barr virus-encoded oncogene and immune modulator. Rev. Med. Virol. 2013;23(6):367–383. doi: 10.1002/rmv.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebe E., Wille C., Hagemeier S., Kenney S., Greijer A., Middeldorp J. Epstein–barr virus gene BARF1 expression is regulated by the epithelial differentiation factor ΔNp63α in undifferentiated nasopharyngeal carcinoma. Cancers. 2018;10(3):76. doi: 10.3390/cancers10030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstetter G., Berger A., Fiegl H., Slade N., Zorić A., Holzer B. Alternative splicing of p53 and p73: the novel p53 splice variant p53δ is an independent prognostic marker in ovarian cancer. Oncogene. 2010;29:1997. doi: 10.1038/onc.2009.482. [DOI] [PubMed] [Google Scholar]
- Holley-Guthrie E.A., Seaman W.T., Bhende P., Merchant J.L., Kenney S.C. The Epstein-Barr virus protein BMRF1 activates gastrin transcription. J. Virol. 2005;79(2):745–755. doi: 10.1128/JVI.79.2.745-755.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton R.H., Gentry G.A. The Epstein Barr virus genome encodes deoxythymidine kinase activity in a nested internal open reading frame. Intervirology. 1996;39(4):270–274. doi: 10.1159/000150528. [DOI] [PubMed] [Google Scholar]
- Hong G.K., Delecluse H.-J., Gruffat H., Morrison T.E., Feng W.-H., Sergeant A., Kenney S.C. The BRRF1 early gene of Epstein-Barr virus encodes a transcription factor that enhances induction of lytic infection by BRLF1. J. Virol. 2004;78(10):4983–4992. doi: 10.1128/JVI.78.10.4983-4992.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houmani J.L., Davis C.I., Ruf I.K. Growth-promoting properties of Epstein-Barr virus EBER-1 RNA correlate with ribosomal protein L22 binding. J. Virol. 2009;83(19):9844–9853. doi: 10.1128/JVI.01014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Liang S., Chen H., Lv T., Wu J., Chen D. ΔNp63α is a common inhibitory target in oncogenic PI3K/Ras/Her2-induced cell motility and tumor metastasis. Proc. Natl. Acad. Sci. 2017;114(20):E3964–E3973. doi: 10.1073/pnas.1617816114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocente S.A., Lee J.M. p73 is a p53-independent, Sp1-dependent repressor of cyclin B1 transcription. Biochem. Biophys. Res. Commun. 2005;329(2):713–718. doi: 10.1016/j.bbrc.2005.02.028. [DOI] [PubMed] [Google Scholar]
- Iwakiri D. Epstein-barr virus-encoded RNAs: key molecules in viral pathogenesis. Cancers. 2014;6(3):1615–1630. doi: 10.3390/cancers6031615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakiri D., Zhou L., Samanta M., Matsumoto M., Ebihara T., Seya T. Epstein-Barr virus (EBV)–encoded small RNA is released from EBV-infected cells and activates signaling from toll-like receptor 3. J. Exp. Med. 2009;206(10):2091–2099. doi: 10.1084/jem.20081761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi K.M., Kaye K.M., Kieff E.D. The Epstein–Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyte growth transformation. Proc. Natl. Acad. Sci. U.S.A. 1997;94(4):1447–1452. doi: 10.1073/pnas.94.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janz A., Oezel M., Kurzeder C., Mautner J., Pich D., Kost M. Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J. Virol. 2000;74(21):10142–10152. doi: 10.1128/jvi.74.21.10142-10152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannsen E., Luftig M., Chase M.R., Weicksel S., Cahir-McFarland E., Illanes D. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. U.S.A. 2004;101(46):16286–16291. doi: 10.1073/pnas.0407320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J., Lagowski J., Lawson S., Liu Y., Kulesz-Martin M. p73 expression modulates p63 and Mdm2 protein presence in complex with p53 family-specific DNA target sequence in squamous cell carcinogenesis. Oncogene. 2007;27:2780. doi: 10.1038/sj.onc.1210941. [DOI] [PubMed] [Google Scholar]
- Kang M.-S., Kieff E. Epstein–Barr virus latent genes. Exp. Mol. Med. 2015;47:e131. doi: 10.1038/emm.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanishi M., Tada-Oikawa S., Kawanishi S. Epstein–Barr virus BHRF1 functions downstream of Bid cleavage and upstream of mitochondrial dysfunction to inhibit TRAIL-induced apoptosis in BJAB cells. Biochem. Biophys. Res. Commun. 2002;297(3):682–687. doi: 10.1016/s0006-291x(02)02261-1. [DOI] [PubMed] [Google Scholar]
- Keyes W.M., Pecoraro M., Aranda V., Vernersson-Lindahl E., Li W., Vogel H. ΔNp63α is an oncogene that targets chromatin remodeler Lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell. 2011;8(2):164–176. doi: 10.1016/j.stem.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga F., Kawakami S., Fujii Y., Saito K., Ohtsuka Y., Iwai A. Impaired p63 expression associates with poor prognosis and uroplakin III expression in invasive urothelial carcinoma of the bladder. Clin. Cancer Res. 2003;9(15):5501–5507. [PubMed] [Google Scholar]
- Komano J., Maruo S., Kurozumi K., Oda T., Takada K. Oncogenic role of Epstein-Barr virus-encoded RNAs in Burkitt’s lymphoma cell line akata. J. Virol. 1999;73(12):9827–9831. doi: 10.1128/jvi.73.12.9827-9831.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-P., Chen J.-Y., Wang J.-T., Kimura K., Takemoto A., Lu C.-C., Chen M.-R. Epstein-barr virus BGLF4 kinase induces premature chromosome condensation through activation of condensin and topoisomerase II. J. Virol. 2007;81(10):5166–5180. doi: 10.1128/JVI.00120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-P., Huang Y.-H., Lin S.-F., Chang Y., Chang Y.-H., Takada K., Chen M.-R. Epstein-barr virus BGLF4 kinase induces disassembly of the nuclear lamina to facilitate virion production. J. Virol. 2008;82(23):11913–11926. doi: 10.1128/JVI.01100-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A.J., Oren M. The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer. 2009;9:749. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Johannsen E., Robertson E., Kieff E. Epstein-barr virus nuclear antigen 3C putative repression domain mediates coactivation of the LMP1 promoter with EBNA-2. J. Virol. 2002;76(1):232–242. doi: 10.1128/JVI.76.1.232-242.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.-T., Chang Y.-T., Chen S.-C., Chuang Y.-C., Chen Y.-R., Lin C.-S., Chen J.-Y. Epstein–Barr virus latent membrane protein 1 represses p53-mediated DNA repair and transcriptional activity. Oncogene. 2004;24:2635. doi: 10.1038/sj.onc.1208319. [DOI] [PubMed] [Google Scholar]
- Liu S.S., Chan K.Y.-K., Cheung A.N.-Y., Liao X.-Y., Leung T.-W., Ngan H.Y.-S. Expression of ΔNp73 and TAp73α independently associated with radiosensitivities and prognoses in cervical squamous cell carcinoma. Clin. Cancer Res. 2006;12(13):3922–3927. doi: 10.1158/1078-0432.CCR-05-2573. [DOI] [PubMed] [Google Scholar]
- Lo Iacono M., Monica V., Saviozzi S., Ceppi P., Bracco E., Papotti M., Scagliotti G.V. p63 and p73 isoform expression in non-small cell lung cancer and corresponding morphological normal lung tissue. J. Thorac. Oncol. 2011;6(3):473–481. doi: 10.1097/JTO.0b013e31820b86b0. [DOI] [PubMed] [Google Scholar]
- Lok S.S., Stewart J.P., Kelly B.G., Hasleton P.S., Egan J.J. Epstein–Barr virus and wild p53 in idiopathic pulmonary fibrosis. Respir. Med. 2001;95(10):787–791. doi: 10.1053/rmed.2001.1152. [DOI] [PubMed] [Google Scholar]
- Lööf J., Pfeifer D., Ding Z., Sun X.-F., Zhang H. Effects of ΔNp73β on cisplatin treatment in colon cancer cells. Mol. Carcinog. 2012;51(8):628–635. doi: 10.1002/mc.20835. [DOI] [PubMed] [Google Scholar]
- Mancao C., Hammerschmidt W. Epstein-Barr virus latent membrane protein 2A is a B-cell receptor mimic and essential for B-cell survival. Blood. 2007;110(10):3715–3721. doi: 10.1182/blood-2007-05-090142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancao C., Altmann M., Jungnickel B., Hammerschmidt W. Rescue of “crippled” germinal center B cells from apoptosis by Epstein-Barr virus. Blood. 2005;106(13):4339–4344. doi: 10.1182/blood-2005-06-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannick J.B., Cohen J.I., Birkenbach M., Marchini A., Kieff E. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J. Virol. 1991;65(12):6826–6837. doi: 10.1128/jvi.65.12.6826-6837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel V., Perrier S., Aoubala M., Ageorges S., Groves M.J., Diot A. Δ160p53 is a novel N-terminal p53 isoform encoded by Δ133p53 transcript. FEBS Lett. 2010;584(21):4463–4468. doi: 10.1016/j.febslet.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Marchini S., Marabese M., Marrazzo E., Mariani P., Cattaneo D., Fossati R. ΔNp63 expression is associated with poor survival in ovarian cancer. Ann. Oncol. 2008;19(3):501–507. doi: 10.1093/annonc/mdm519. [DOI] [PubMed] [Google Scholar]
- Maruo S., Zhao B., Johannsen E., Kieff E., Zou J., Takada K. Epstein-Barr virus nuclear antigens 3C and 3A maintain lymphoblastoid cell growth by repressing p16(INK4A) and p14(ARF) expression. Proc. Natl. Acad. Sci. U.S.A. 2011;108(5):1919–1924. doi: 10.1073/pnas.1019599108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruzuru Y., Fujii H., Oyama M., Kozuka-Hata H., Kato A., Kawaguchi Y. Roles of p53 in herpes simplex virus 1 replication. J. Virol. 2013;87(16):9323–9332. doi: 10.1128/JVI.01581-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massion P.P., Taflan P.M., Jamshedur Rahman S.M., Yildiz P., Shyr Y., Edgerton M.E. Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res. 2003;63(21):7113–7121. [PubMed] [Google Scholar]
- Melino G., Lu X., Gasco M., Crook T., Knight R.A. Functional regulation of p73 and p63: development and cancer. Trends Biochem. Sci. 2003;28(12):663–670. doi: 10.1016/j.tibs.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Mohanty S., Sahu S.K., Chattopadhyay N.R., Kumar A., Das P., Choudhuri T. TAp63alpha induced apoptosis inhibited by Kaposi's sarcoma herpesvirus latency nuclear antigen. J. Carcinog. Mutagen. 2015;6(2) [Google Scholar]
- Moll U.M., Slade N. p63 and p73: roles in development and tumor formation. Mol. Cancer Res. 2004;2(7):371–386. [PubMed] [Google Scholar]
- Morrison J.A., Raab-Traub N. Roles of the ITAM and PY motifs of Epstein-Barr virus latent membrane protein 2A in the inhibition of epithelial cell differentiation and activation of β-catenin signaling. J. Virol. 2005;79(4):2375–2382. doi: 10.1128/JVI.79.4.2375-2382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosialos G., Birkenbacht M., Yalamanchill R., Van Arsdale T., Ware C., Kleff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80(3):389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- Murray-Zmijewski F., Lane D.P., Bourdon J.C. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13:962. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- Narita Y., Sugimoto A., Kawashima D., Watanabe T., Kanda T., Kimura H. A herpesvirus specific motif of Epstein-Barr virus DNA polymerase is required for the efficient lytic genome synthesis. Sci. Rep. 2015;5:11767. doi: 10.1038/srep11767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhierl B., Delecluse H.-J. The Epstein-Barr virus BMRF1 gene is essential for lytic virus replication. J. Virol. 2006;80(10):5078–5081. doi: 10.1128/JVI.80.10.5078-5081.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklison-Chirou M.V., Steinert J.R., Agostini M., Knight R.A., Dinsdale D., Cattaneo A. TAp73 knockout mice show morphological and functional nervous system defects associated with loss of p75 neurotrophin receptor. Proc. Natl. Acad. Sci. U.S.A. 2013;110(47):18952–18957. doi: 10.1073/pnas.1221172110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Nions J., Brooks L.A., Sullivan A., Bell A., Dunne B., Rozycka M. p73 is over-expressed in vulval cancer principally as the Delta 2 isoform. Br. J. Canc. 2001;85(10):1551–1556. doi: 10.1054/bjoc.2001.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odumade O.A., Hogquist K.A., Balfour H.H. Progress and problems in understanding and managing primary Epstein-Barr virus infections. Clin. Microbiol. Rev. 2011;24(1):193–209. doi: 10.1128/CMR.00044-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orosz L., Gallyas É., Kemény L., Mándi Y., Facskó A., Megyeri K. Involvement of p63 in the herpes simplex virus-1-induced demise of corneal cells. J. Biomed. Sci. 2010;17(1) doi: 10.1186/1423-0127-17-47. 47-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B.-J., Lee S.-J., Kim J.I., Lee S.-J., Lee C.-H., Chang S.-G. Frequent alteration of p63 expression in human primary bladder carcinomas. Cancer Res. 2000;60(13):3370–3374. [PubMed] [Google Scholar]
- Paschos K., Smith P., Anderton E., Middeldorp J.M., White R.E., Allday M.J. Epstein-barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene Bim. PLoS Pathog. 2009;5(6) doi: 10.1371/journal.ppat.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflaum J., Schlosser S., Müller M. p53 family and cellular stress responses in cancer. Front. Oncol. 2014;4(285) doi: 10.3389/fonc.2014.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovan E., Tosello V., Indraccolo S., Cabrelle A., Baesso I., Trentin L. Chemokine receptor expression in EBV-associated lymphoproliferation in hu/SCID mice: implications for CXCL12/CXCR4 axis in lymphoma generation. Blood. 2005;105(3):931–939. doi: 10.1182/blood-2004-03-0799. [DOI] [PubMed] [Google Scholar]
- Ragoczy T., Heston L., Miller G. The Epstein-Barr virus rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 1998;72(10):7978–7984. doi: 10.1128/jvi.72.10.7978-7984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram Kumar R.M., Betz M.M., Robl B., Born W., Fuchs B. ΔNp63α enhances the oncogenic phenotype of osteosarcoma cells by inducing the expression of GLI2. BMC Canc. 2014;14(1):559. doi: 10.1186/1471-2407-14-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M.P., Berger C., Zauner L., Sigrist J.A., Weber M., Longnecker R. Latent membrane protein 2B regulates susceptibility to induction of lytic Epstein-Barr virus infection. J. Virol. 2008;82(4):1739–1747. doi: 10.1128/JVI.01723-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischl U., Gerdes C., Motz M., Wolf H. Expression and purification of an Epstein-Barr virus encoded 23-kDa protein and characterization of its immunological properties. J. Virol Methods. 1996;57(1):71–85. doi: 10.1016/0166-0934(95)01970-7. [DOI] [PubMed] [Google Scholar]
- Reisman D., Yates J., Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol. Cell. Biol. 1985;5(8):1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro-Silva A., Ramalho L.N.Z., Garcia S.B., Zucoloto S. Does the correlation between EBNA-1 and p63 expression in breast carcinomas provide a clue to tumorigenesis in Epstein-Barr virus-related breast malignancies? Braz. J. Med. Biol. Res. 2004;37:89–95. doi: 10.1590/s0100-879x2004000100013. [DOI] [PubMed] [Google Scholar]
- Rocco J.W., Leong C.-O., Kuperwasser N., DeYoung M.P., Ellisen L.W. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9(1):45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Rovedo M., Longnecker R. Epstein-barr virus latent membrane protein 2B (LMP2B) modulates LMP2A activity. J. Virol. 2007;81(1):84–94. doi: 10.1128/JVI.01302-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe M., Glaunsinger B., van Leeuwen D., Zuo J., Sweetman D., Ganem D. Host shutoff during productive Epstein–Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc. Natl. Acad. Sci. 2007;104(9):3366–3371. doi: 10.1073/pnas.0611128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvolo V., Gupta A.K., Swaminathan S. Epstein-barr virus SM protein interacts with mRNA in vivo and mediates a gene-specific increase in cytoplasmic mRNA. J. Virol. 2001;75(13):6033–6041. doi: 10.1128/JVI.75.13.6033-6041.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacaze C., Henry S., Icart J., Mariamé B. Tissue specific distribution of Epstein-Barr virus (EBV) BZLF1 gene variants in nasopharyngeal carcinoma (NPC) bearing patients. Virus Res. 2001;81(1):133–142. doi: 10.1016/s0168-1702(01)00376-8. [DOI] [PubMed] [Google Scholar]
- Saha A., Robertson E.S. Functional modulation of the metastatic suppressor Nm23-H1 by oncogenic viruses. FEBS Letters. 2011;585(20):3174–3184. doi: 10.1016/j.febslet.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A., Bamidele A., Murakami M., Robertson E.S. EBNA3C attenuates the function of p53 through interaction with inhibitor of growth family proteins 4 and 5. J. Virol. 2011;85(5):2079–2088. doi: 10.1128/JVI.02279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A., Halder S., Upadhyay S.K., Lu J., Kumar P., Murakami M. Epstein-barr virus nuclear antigen 3C facilitates G1-S transition by stabilizing and enhancing the function of cyclin D1. PLoS Pathog. 2011;7(2) doi: 10.1371/journal.ppat.1001275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu S.K., Mohanty S., Kumar A., Kundu C.N., Verma S.C., Choudhuri T. Epstein–Barr virus nuclear antigen 3C interact with p73: interplay between a viral oncoprotein and cellular tumor suppressor. Virology. 2014;448:333–343. doi: 10.1016/j.virol.2013.10.023. [DOI] [PubMed] [Google Scholar]
- Samanta M., Iwakiri D., Kanda T., Imaizumi T., Takada K. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 2006;25(18):4207–4214. doi: 10.1038/sj.emboj.7601314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santag S., Jäger W., Karsten C.B., Kati S., Pietrek M., Steinemann D. Recruitment of the tumour suppressor protein p73 by Kaposi’s Sarcoma Herpesvirus latent nuclear antigen contributes to the survival of primary effusion lymphoma cells. Oncogene. 2012;32:3676. doi: 10.1038/onc.2012.385. [DOI] [PubMed] [Google Scholar]
- Saridakis V., Sheng Y., Sarkari F., Holowaty M.N., Shire K., Nguyen T. Structure of the p53 binding domain of HAUSP/USP7 Bound to Epstein-Barr nuclear antigen 1. Mol. Cell. 2005;18(1):25–36. doi: 10.1016/j.molcel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Sato Y., Shirata N., Murata T., Nakasu S., Kudoh A., Iwahori S. Transient increases in p53-responsible gene expression at early stages of Epstein-Barr virus productive replication. Cell Cycle. 2010;9(4):807–814. doi: 10.4161/cc.9.4.10675. [DOI] [PubMed] [Google Scholar]
- Scian M.J., Carchman E.H., Mohanraj L., Stagliano K.E.R., Anderson M.A.E., Deb D. Wild-type p53 and p73 negatively regulate expression of proliferation related genes. Oncogene. 2007;27:2583. doi: 10.1038/sj.onc.1210898. [DOI] [PubMed] [Google Scholar]
- Seto E., Moosmann A., Grömminger S., Walz N., Grundhoff A., Hammerschmidt W. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 2010;6(8) doi: 10.1371/journal.ppat.1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon-Lowe C., Adland E., Bell A.I., Delecluse H.-J., Rickinson A.B., Rowe M. Features distinguishing Epstein-Barr virus infections of epithelial cells and B cells: viral genome expression, genome maintenance, and genome amplification. J. Virol. 2009;83(15):7749–7760. doi: 10.1128/JVI.00108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J.-Y., Ernberg I., Biberfeld P., Heiden T., Zeng Y.-X., Hu L.-F. Epstein-Barr virus LMP1 status in relation to apoptosis, p53 expression and leucocyte infiltration in nasopharyngeal carcinoma. Anticancer Res. 2004;24:2309–2318. [PubMed] [Google Scholar]
- Shukla S.K., Jha H.C., El-Naccache D.W., Robertson E.S. An EBV recombinant deleted for residues 130-159 in EBNA3C can deregulate p53/Mdm2 and Cyclin D1/CDK6 which results in apoptosis and reduced cell proliferation. Oncotarget. 2016;7(14):18116–18134. doi: 10.18632/oncotarget.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivachandran N., Wang X., Frappier L. Functions of the Epstein-Barr virus EBNA1 protein in viral reactivation and lytic infection. J. Virol. 2012;86(11):6146–6158. doi: 10.1128/JVI.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares E., Zhou H. Master regulatory role of p63 in epidermal development and disease. Cell. Mol. Life Sci. : CMLS. 2018;75(7):1179–1190. doi: 10.1007/s00018-017-2701-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Huo S.-w., Lü J.-j., Liu Z., Fang X.-l., Jin X.-b., Yuan M.-z. Expression of p53 isoforms in renal cell carcinoma. Chin. Med. J. 2009;122(8):921–926. [PubMed] [Google Scholar]
- Speidel D. Transcription-independent p53 apoptosis: an alternative route to death. Trends Cell Biol. 2010;20(1):14–24. doi: 10.1016/j.tcb.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Stiewe T., Tuve S., Peter M., Tannapfel A., Elmaagacli A.H., Pützer B.M. Quantitative TP73 transcript analysis in hepatocellular carcinomas. Clin. Cancer Res. 2004;10(2):626–633. doi: 10.1158/1078-0432.ccr-0153-03. [DOI] [PubMed] [Google Scholar]
- Strano S., Fontemaggi G., Costanzo A., Giulia Rizzo M., Monti O., Baccarini A. Physical interaction with human tumor-derived p53 mutants inhibits p63 activities. J. Biol. Chem. 2002;277:18817–18826. doi: 10.1074/jbc.M201405200. [DOI] [PubMed] [Google Scholar]
- Sugden B., Marsh K., Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Mol. Cell. Biol. 1985;5(2):410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh K.-Y., Lacouture M., Gerami P. p63 in primary cutaneous carcinosarcoma. Am. J. Dermatopathol. 2007;29(4):374–377. doi: 10.1097/DAD.0b013e31812f52bd. [DOI] [PubMed] [Google Scholar]
- Sullivan K.D., Galbraith M.D., Andrysik Z., Espinosa J.M. Mechanisms of transcriptional regulation by p53. Cell Death Differ. 2017;25:133. doi: 10.1038/cdd.2017.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surget S., Khoury M.P., Bourdon J.-C. Uncovering the role of p53 splice variants in human malignancy: a clinical perspective. OncoTargets Ther. 2014;7:57–68. doi: 10.2147/OTT.S53876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Mungerson M., Bultema R., Longnecker R. Epstein–Barr virus LMP2A imposes sensitivity to apoptosis. J. Gen. Virol. 2010;91(Pt 9):2197–2202. doi: 10.1099/vir.0.021444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson J.J., Holley-Guthrie E., Kenney S.C. Epstein-barr virus immediate-early protein BRLF1 interacts with CBP, promoting enhanced BRLF1 transactivation. J. Virol. 2001;75(13):6228–6234. doi: 10.1128/JVI.75.13.6228-6234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely L., Selivanova G., Magnusson K.P., Klein G., Wiman K.G. EBNA-5, an Epstein-Barr virus-encoded nuclear antigen, binds to the retinoblastoma and p53 proteins. Proc. Natl. Acad. Sci. U.S.A. 1993;90(12):5455–5459. doi: 10.1073/pnas.90.12.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q., Young L., Woodman C., G Murray P. Epstein-Barr virus (EBV) and its associated human cancers - genetics, epigenetics, pathobiology and novel therapeutics. Front. Biosci. 2006;11(1):2672–2713. doi: 10.2741/2000. [DOI] [PubMed] [Google Scholar]
- Tetsuo U., Ja-Mun C., Hiroshi U., Rumi H., Moon-Sung C., Makoto S. p73 gene promoter methylation in Epstein-Barr virus-associated gastric carcinoma. Int. J. Cancer. 2007;120(1):60–66. doi: 10.1002/ijc.22275. [DOI] [PubMed] [Google Scholar]
- Thompson M.P., Kurzrock R. Epstein-barr virus and cancer. Clin. Cancer Res. 2004;10(3):803–821. doi: 10.1158/1078-0432.ccr-0670-3. [DOI] [PubMed] [Google Scholar]
- Tschan M.P., Grob T.J., Peters U.R., Laurenzi V.D., Huegli B., Kreuzer K.A. Enhanced p73 expression during differentiation and complex p73 isoforms in myeloid leukemia. Biochem. Biophys. Res. Commun. 2000;277(1):62–65. doi: 10.1006/bbrc.2000.3627. [DOI] [PubMed] [Google Scholar]
- Tsurumi T., Kobayashi A., Tamai K., Yamada H., Daikoku T., Yamashita Y., Nishiyama Y. Epstein–barr virus single-stranded DNA-binding protein: purification, characterization, and action on DNA synthesis by the viral DNA polymerase. Virology. 1996;222(2):352–364. doi: 10.1006/viro.1996.0432. [DOI] [PubMed] [Google Scholar]
- Tve S., Wagner S., Schittek B., Pützer B. Alterations of ΔTA-p73 splice transcripts during melanoma development and progression. Int. J. Cancer. 2004;108(1):162–166. doi: 10.1002/ijc.11552. [DOI] [PubMed] [Google Scholar]
- Uramoto H., Sugio K., Oyama T., Nakata S., Ono K., Morita M. Expression of ΔNp73 predicts poor prognosis in lung cancer. Clin. Cancer Res. 2004;10(20):6905–6911. doi: 10.1158/1078-0432.CCR-04-0290. [DOI] [PubMed] [Google Scholar]
- van Grunsven W.M., vaHeerde E.C., de Haard H.J., Spaan W.J., Middeldorp J.M. Gene mapping and expression of two immunodominant Epstein-Barr virus capsid proteins. J. Virol. 1993;67(7):3908–3916. doi: 10.1128/jvi.67.7.3908-3916.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseva A.V., Moll U.M. The mitochondrial p53 pathway. Biochim. Biophys. Acta Bioenerg. 2009;1787(5):414–420. doi: 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikhanskaya F., D'Incalci M., Broggini M. p73 competes with p53 and attenuates its response in a human ovarian cancer cell line. Nucleic Acids Res. 2000;28(2):513–519. doi: 10.1093/nar/28.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikhreva P., Melino G., Amelio I. p73 alternative splicing: exploring a biological role for the C-terminal isoforms. J. Mol. Biol. 2018;430(13):1829–1838. doi: 10.1016/j.jmb.2018.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgelm A.E., Hong S.M., Washington M.K., Wei J., Chen H., El-Rifai W., Zaika A. Characterization of ΔNp73 expression and regulation in gastric and esophageal tumors. Oncogene. 2010;29:5861. doi: 10.1038/onc.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgelm A.E., Washington M.K., Wei J., Chen H., Prassolov V.S., Zaika A.I. Interactions of the p53 protein family in cellular stress response in gastrointestinal tumors. Mol. Cancer Ther. 2010;9(3):693–705. doi: 10.1158/1535-7163.MCT-09-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., El-Deiry W.S. p73 or p53 directly regulates human p53 transcription to maintain cell cycle checkpoints. Cancer Res. 2006;66(14):6982–6989. doi: 10.1158/0008-5472.CAN-06-0511. [DOI] [PubMed] [Google Scholar]
- Wang D., Liebowitz D., Kieff E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell. 1985;43(3, Part 2):831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- Wang D.-Y., Cheng C.-C., Kao M.-H., Hsueh Y.-J., Ma D.H.K., Chen J.-K. Regulation of limbal keratinocyte proliferation and differentiation by TAp63 and ΔNp63 transcription factors. Investig. Ophthalmol. Vis. Sci. 2005;46(9):3102–3108. doi: 10.1167/iovs.05-0051. [DOI] [PubMed] [Google Scholar]
- Wang J., Liu Y.-X., Hande M.P., Wong A.C., Jin Y.J., Yin Y. TAp73 is a downstream target of p53 in controlling the cellular defense against stress. J. Biol. Chem. 2007;(40):29152–29162. doi: 10.1074/jbc.M703408200. [DOI] [PubMed] [Google Scholar]
- Wang Q., Lingel A., Geiser V., Kwapnoski Z., Zhang L. Tumor suppressor p53 stimulates the expression of Epstein-Barr virus latent membrane protein 1. J. Virol. 2017;91(20):e00312–e00317. doi: 10.1128/JVI.00312-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Zaika E., Zaika A. p53 family: role of protein isoforms in human cancer. J. Nucleic Acids. 2012;2012:687359. doi: 10.1155/2012/687359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R.E., Rämer P.C., Naresh K.N., Meixlsperger S., Pinaud L., Rooney C. EBNA3B-deficient EBV promotes B cell lymphomagenesis in humanized mice and is found in human tumors. J. Clin. Investig. 2012;122(4):1487–1502. doi: 10.1172/JCI58092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T., O’Hara A., Araujo I., Barreto J., Carvalho E., Sapucaia J.B. EBV MicroRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68(5):1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalamanchili R., Tong X., Grossman S., Johannsen E., Mosialos G., Kieff E. Genetic and biochemical evidence that EBNA 2 interaction with a 63-kDa cellular GTG-binding protein is essential for B lymphocyte growth transformation by EBV. Virology. 1994;204(2):634–641. doi: 10.1006/viro.1994.1578. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Wu L., Caballero O.L., Hibi K., Trink B., Resto V. Frequent gain of the p40/p51/p63 gene locus in primary head and neck squamous cell carcinoma. Int. J. Cancer. 2000;86(5):684–689. doi: 10.1002/(sici)1097-0215(20000601)86:5<684::aid-ijc13>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R.T. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yang A., Walker N., Bronson R., Kaghad M., Oosterwegel M., Bonnin J. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- Yang J., Deng W., Hau P.M., Liu J., Lau V.M.Y., Cheung A.L.M. Epstein–Barr virus BZLF1 protein impairs accumulation of host DNA damage proteins at damage sites in response to DNA damage. Lab. Investig. 2015;95:937. doi: 10.1038/labinvest.2015.69. [DOI] [PubMed] [Google Scholar]
- Yates J., Warren N., Reisman D., Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. U.S.A. 1984;81(12):3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J.L., Warren N., Sugden B. Stable replication of plasmids derived from Epstein–Barr virus in various mammalian cells. Nature. 1985;313:812. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- Yi F., Saha A., Murakami M., Kumar P., Knight J.S., Cai Q. Epstein–Barr virus nuclear antigen 3C targets p53 and modulates its transcriptional and apoptotic activities. Virology. 2009;388(2):236–247. doi: 10.1016/j.virol.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama N., Fujii K., Hirata M., Tamai K., Kiyono T., Kuzushima K. Assembly of the Epstein–Barr virus BBLF4, BSLF1 and BBLF2/3 proteins and their interactive properties. J. Gen. Virol. 1999;80(11):2879–2887. doi: 10.1099/0022-1317-80-11-2879. [DOI] [PubMed] [Google Scholar]
- Yoshida M., Watanabe T., Narita Y., Sato Y., Goshima F., Kimura H., Murata T. The Epstein-Barr virus BRRF1 gene is dispensable for viral replication in HEK293 cells and transformation. Sci. Rep. 2017;7:6044. doi: 10.1038/s41598-017-06413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L.S., Murray P.G. Epstein–Barr virus and oncogenesis: from latent genes to tumours. Oncogene. 2003;22:5108. doi: 10.1038/sj.onc.1206556. [DOI] [PubMed] [Google Scholar]
- Zaika A.I., Kovalev S., Marchenko N.D., Moll U.M. Overexpression of the wild type <em>p73</em> gene in breast cancer tissues and cell lines. Cancer Res. 1999;59(13):3257–3263. [PubMed] [Google Scholar]
- Zhang Q., Gutsch D., Kenney S. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol. Cell. Biol. 1994;14(3):1929–1938. doi: 10.1128/mcb.14.3.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Holley-Guthrie E., Ge J.-Q., Dorsky D., Kenney S. The Epstein–Barr virus (EBV) DNA polymerase accessory protein, BMRF1, activates the essential downstream component of the EBV oriLyt. Virology. 1997;230(1):22–34. doi: 10.1006/viro.1997.8470. [DOI] [PubMed] [Google Scholar]
- Zuo L., Yue W., Du S., Xin S., Zhang J., Liu L. An update: Epstein-Barr virus and immune evasion via microRNA regulation. Virol. Sin. 2017;32(3):175–187. doi: 10.1007/s12250-017-3996-5. [DOI] [PMC free article] [PubMed] [Google Scholar]