Abstract
The Drosophila embryonic central nervous system (CNS) is a complex organ consisting of ∼15,000 neurons and glia that is generated in ∼1 day of development. For the past 40 years, Drosophila developmental neuroscientists have described each step of CNS development in precise molecular genetic detail. This has led to an understanding of how an intricate nervous system emerges from a single cell. These studies have also provided important, new concepts in developmental biology, and provided an essential model for understanding similar processes in other organisms. In this article, the key genes that guide Drosophila CNS development and how they function is reviewed. Features of CNS development covered in this review are neurogenesis, gliogenesis, cell fate specification, and differentiation.
Keywords: CNS, development, Drosophila, glia, neuron, FlyBook
The fly CNS is a masterpiece of engineering and a work of art.
—with apologies to Leonardo da Vinci
Studying Drosophila CNS Development
Drosophila is a complex organism and achieves its high degree of complexity in an amazingly short developmental time. Embryonic development gives rise to a fully functional first instar larva in about a day, and after larval growth and metamorphosis (∼10 additional days), an adult fly emerges. Larvae are endowed with a sophisticated behavioral repertoire that allow them to successfully accomplish their main goals: foraging for food, eating, growing, and surviving predation. These behaviors are controlled by a CNS, consisting of a brain and ventral nerve cord (VNC), that contain ∼15,000 cells, including 1000 glia (Ito et al. 1995; Heckscher et al. 2014; Monedero Cobeta et al. 2017; Yaghmaeian Salmani et al. 2018). The embryonic CNS and its development are largely hard-wired and highly stereotyped between individuals. During larval development and metamorphosis, the far more complex adult CNS, consisting of 150,000 neurons and 15,700 glia (Jenett et al. 2012; Kremer et al. 2017), is constructed upon the embryonic CNS. Its development, while still relatively stereotyped, is significantly influenced by environmental and hormonal stimuli (Syed et al. 2017).
Understanding the genetic, molecular, and cellular bases of Drosophila embryonic CNS development has been carried out in earnest for ∼40 years (e.g., Jiménez and Campos-Ortega 1979). As with most large-scale endeavors, there were numerous intellectual antecedents and insights that drove this research, including: (1) key experimental results from related organisms, such as grasshopper (Doe and Goodman 1985); (2) the highly successful genetic screen of Nüsslein-Volhard and Wieschaus (1980) that identified embryonic patterning genes; (3) molecular studies of embryonic segmentation genes (Pankratz and Jackle 1993); (4) the utility of using multiple cytological markers to distinguish different CNS cells (Doe 1992); (5) insights from well-studied Drosophila developmental systems, such as sensory neurons (Singhania and Grueber 2014) and the visual system (Kumar 2012); and (6) and insights from vertebrate studies that led to the identification of important, new Drosophila genes (e.g., Tsuchida et al. 1994; Thor and Thomas 1997). In addition, by deconstructing CNS development into discrete cellular events, it has been possible to acquire a molecular understanding of the entire process from the postfertilization single-celled embryo to a fully functional CNS. This is a remarkable achievement of modern biology. Elucidation of Drosophila embryonic CNS development has also proven to be a useful model for studying the development of other invertebrate and vertebrate species given the strong evolutionary similarities that exist (Allan and Thor 2015). Novel insights into issues of human health have also originated from the study of Drosophila CNS development. As an example, discovery of the Drosophila single-minded (sim) gene (Crews et al. 1988; Thomas et al. 1988) led to the identification of two mouse and human sim genes: SIM1 and SIM2 (Dahmane et al. 1995; Fan et al. 1996). Human genetic studies revealed that SIM1 plays a role in appetite control and obesity (Holder et al. 2000), and is also the only known human gene associated with erectile dysfunction (Jorgenson et al. 2018). The goals of this review are to provide a comprehensive view of Drosophila embryonic CNS development while concentrating on recent studies, including neurogenesis, gliogenesis, cell fate specification, and differentiation (axon guidance mechanisms are not considered here). The focus is largely on the well-studied VNC, although aspects of brain development are included.
Structure of the Embryonic CNS
CNS segmental structure and homology
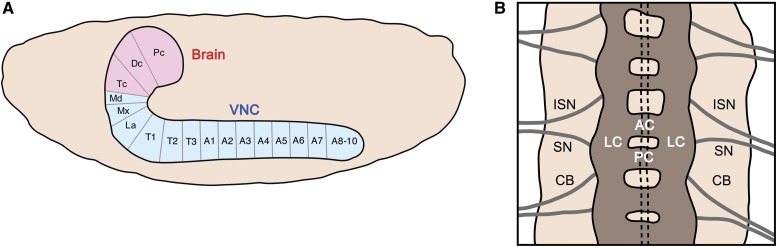
The insect CNS is a segmented organ, and each segment is referred to as a neuromere (Niven et al. 2008). The Drosophila CNS can be subdivided into the brain and VNC (Figure 1A). The embryonic brain consists of three cerebral neuromeres: protocerebrum, deutocerebrum, and tritocerebrum (Urbach and Technau 2003b). The VNC contains: (1) three subesophageal neuromeres: the mandibular, maxillary, and labial neuromeres (also referred to as S1–3), (2) three thoracic neuromeres (T1–T3), seven complete abdominal neuromeres (A1–7), and three terminal neuromeres (A8–A10) that have reduced structures (Urbach et al. 2016). Gene expression profiling of the neuroblasts (NBs) in each neuromere provides an estimate of the homology between neuromeres (Urbach et al. 2016). The T1–A7 neuromeres consist of the same pattern of 65 NBs/neuromere. The posterior abdominal neuromeres have progressively fewer NBs (A8: 63 NBs; A9: 47 NBs; A10: 23 NBs). The subesophageal neuromeres also have a reduced number of NBs (labial: ∼57 NBs; maxillary: ∼53 NBs; mandibular: 45 NBs). In the brain, 20 of 26 NBs in the tritocerebrum are homologous to VNC and subesophageal NBs, as are 18 of the 42 NBs in the deutocerebrum. In contrast, none of the 160 protocerebral NBs correspond to NBs in the VNC [144 NBs mapped by Urbach and Technau (2003a) and 16 Type II NBs identified by Walsh and Doe (2017) and Alvarez and Diaz-Benjumea (2018)]. Consequently, of the 19 neuromeres of the CNS, 18 share at least some homology with only the protocerebral neuromere divergent.
Figure 1.
Structure of the Drosophila embryonic CNS. (A) Schematic of a sagittal view of the CNS including brain (red) and ventral nerve cord (VNC; blue). Anterior is left and dorsal is top; neuromere names are listed in the text. (B) Horizontal (dorsal) view of three neuromeres of the VNC; anterior is top. The axon scaffold is shown in dark brown with the anterior commissure (AC), posterior commissure (PC), and lateral connectives (LC) indicated in one of the neuromeres. The cell bodies (CB) of the VNC are shown in tan; nerves shown include the intersegmental nerve (ISN) and segmental nerve (SN). The dotted lines represent the location of the CNS midline cells.
Cellular composition of the CNS
In the CNS, each neuromere has two bilaterally symmetric hemi-neuromeres that constitute the lateral CNS, and, in the VNC, these hemi-neuromeres are separated by a set of specialized midline cells. The numbers and types of cells derived from each embryonic NB are well-established, and, increasingly, the majority of neurons can be uniquely identified by advanced microscopic methods. Initially, the neuronal progeny of each NB was identified based on its axonal morphology by DiI labeling of NBs (Bossing et al. 1996; Schmid et al. 1999). These experiments established that each NB in a hemi-neuromere gives rise to a unique set of neurons. There are no embryonic NBs dedicated to the production of only a single cell type—instead, NBs commonly give rise to multiple cell types, including interneurons, peptidergic neurons, and motoneurons. In addition, the progeny of the same NB often have diverse axon trajectories and do not necessarily follow the same paths to their synaptic targets. DiI fills of individual CNS neurons have defined the large interneuron population of abdominal neuromeres (Rickert et al. 2011). These observations have been reinforced and expanded by detailed studies of multiple lineages using molecular markers that identify specific neurons and precursors (e.g., Karcavich and Doe 2005; Wheeler et al. 2006; Baumgardt et al. 2009). In general, neuronal migration is minimal, and the relative positions of individual neurons are similar between thoracic and abdominal neuromeres such that specific neurons can be identified by their relative position using a single marker (e.g., anti-Even-skipped staining) and computer-assisted image-acquisition and analysis (eNeuro project) (Heckscher et al. 2014). Using the eNeuro atlas data from embryonic stage 16, an A1 neuromere (including midline cells) is estimated to have a total of 713 Elav+ neurons that includes 85 motoneurons (determined by pMad staining), 602 interneurons, and 26 neurosecretory (peptidergic) cells (Dimm+ cells plus MP1 neurons). It is estimated that there are 20 glia/neuromere. There are 22 midline cells, including three midline glia, 18 neurons, and the MNB (Wheeler et al. 2006; Heckscher et al. 2014).
Going forward, characterization of embryonic CNS cells (precursors, neurons, and glia) by single-cell transcriptomic analyses will be enormously useful. It will provide a wealth of information regarding the similarities and differences among each cell type and provide a foundation for further genetic investigations of CNS development. By comparing expression profiles of CNS neurons from larvae, pupae, and adults, changes due to maturation and aging will be identified. Comparisons to neurons from other species will provide key insights into CNS evolution.
Axonal organization and peripheral nerves
CNS neurons extend axons that connect with other neurons, muscles, and the gut (Figure 1B). Within the CNS, axons assemble into one of two longitudinal connectives that run along the anterior-posterior (A–P) axis of the CNS in each neuromere. The majority of neurons (69% of interneurons; Rickert et al. 2011) extend their axons across the midline via two axon commissures: the anterior commissure and posterior commissure. Having crossed the midline, the axons join the contralateral longitudinal connective. Within, the connective, 50% of interneuronal axons turn in an anterior (ascending) direction, 20% in the posterior (descending) direction, and 30% have short axons that stay within the neuromere (Rickert et al. 2011). The preference of axons to ascend rather than descend is consistent with the need to transmit information to the brain. Axons that project the farthest tend to be born earlier than those neurons with relatively short projections. It is within the connectives that neurons synapse to neurons within the same neuromere and to neurons in other neuromeres, including the brain. Motoneurons extend their axons out of the CNS into the muscle field via three distinct nerves [segmental nerve (SN), intersegmental nerve (ISN), and transverse nerve (TN)] (Figure 1B and Figure 10) (Landgraf and Thor 2006) while sensory neurons extend axons into the CNS via the same SN and ISN (Singhania and Grueber 2014). In the near future, the entire larval CNS connectome will be determined by electron microscopic reconstruction (Eichler et al. 2017). Combined with information from expression of Gal4/split-Gal4 lines (Li et al. 2014a) and large-scale larval behavioral screens (Almeida-Carvalho et al. 2017), a sophisticated understanding of the circuitry that drives larval behavior will emerge.
Figure 10.
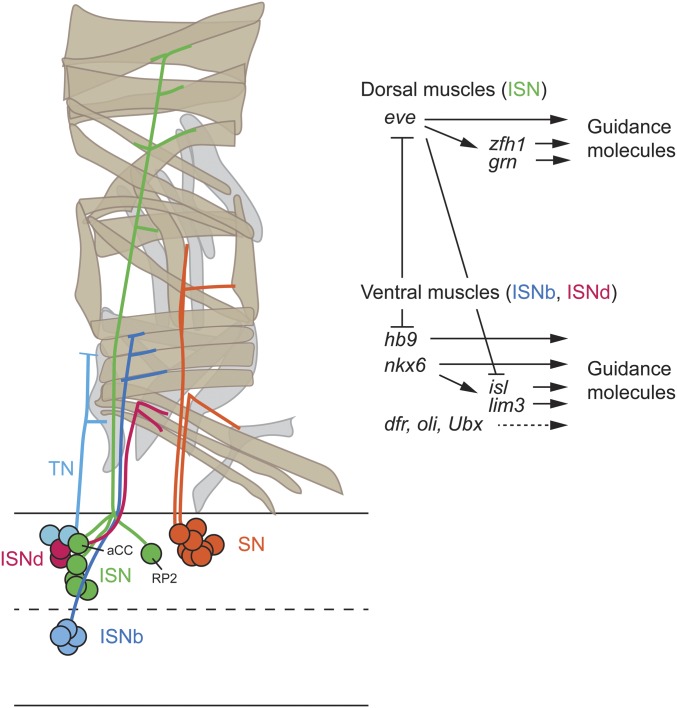
Motoneuron cell fate and axon guidance. (A) Schematic showing somatic muscles present in a hemi-segment and representative motoneurons that contribute to the transverse nerve (TN), intersegmental nerves ISN, ISNb, and ISNd, and segmental nerve (SN). The ISN aCC and RP2 motoneurons are shown. (B) Shown are TFs that control motoneuron fate, differentiation, and the guidance of ISN, ISNb, and ISNd axons that project to dorsal muscles and ventral muscles. Adapted by permission from Elsevier: Seminars in Cell & Developmental Biology (Zarin and Labrador 2017) copyright (2017).
Neural Precursor Specification
The formation of the Drosophila embryonic CNS is largely hard-wired and invariant. As the cellular blastoderm forms, the ventral-lateral region is specified to become neurogenic ectoderm, which ultimately gives rises to both the CNS and epidermis. The mesectoderm is a specialized group of neuroectodermal cells that lie at the midline and generates CNS midline neural precursors and midline glia (but not epidermis). The lateral neuroectoderm on either side of the mesectoderm comprises most of the CNS and gives rise to CNS NBs and epidermal precursors.
NB appearance and positioning
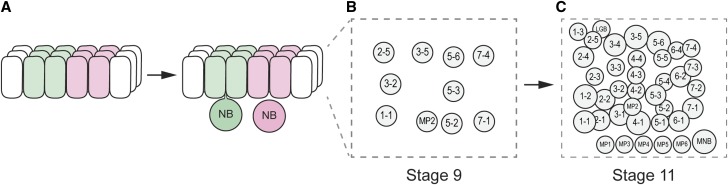
Within the neuroectoderm, NBs emerge at precise positions, and each NB/hemi-neuromere has a distinct cell fate. NBs enlarge and delaminate from the underlying ectoderm, then move internally (Figure 2A); this process occurs in five pulses over ∼4 hr of development (Doe 1992). At the beginning of NB formation (stages 8–9), 9–10 NBs are arranged in three columns/hemi-segment along the dorsal-ventral (D-V; circumferential) axis and four rows along the anterior-posterior (A-P; longitudinal) axis (Figure 2B). By the end of NB formation (late stage 11), there are 32 NBs in each hemi-neuromere (Figure 2C). Midline neuronal precursors also emerge from the mesectoderm during stages 10–11 (Wheeler et al. 2008). Two key connected questions concern the early embryonic regulatory mechanisms that direct ectodermal cells to become NBs and how these NBs acquire distinct cell fates.
Figure 2.
Neuroblast formation. (A) Neuroblasts (NB) form and delaminate from the neuroectodermal layer. Proneural clusters of neuroectodermal cells give rise to a single NB. (B and C) Shown are hemi-neuromeres (anterior to the left; midline at bottom). At stage 9 (B) ∼10 NBs have formed, and, by stage 11 (C), there are 32 NBs including MP2 and the longitudinal glioblast (LGB); midline precursors include the MNB and five MPs. Adapted by permission from Springer Nature: Nature Neuroscience Reviews (Kohwi and Doe 2013) copyright (2013), and by permission from the The Company of Biologists: Development (Urbach et al. 2016) copyright (2016).
Neural equivalence groups, proneural gene expression, and NB formation
Within the developing lateral neuroectoderm, each NB and Midline Precursor 2 (MP2) emerge from a group of ∼5–7 cells, referred to as a proneural cluster (Figure 2A) (Skeath and Carroll 1992) (MP2 is a nonstem cell neural precursor that divides into two neurons). The proneural basic helix-loop-helix (bHLH) transcription factor (TF) genes: achaete (ac), scute (sc), and lethal of scute [l(1)sc] play key roles in neural precursor formation. The l(1)sc gene is expressed in most NBs, whereas ac and sc overlap in expression and initially are expressed in a relatively small set of NBs and MP2 (Jiménez and Campos-Ortega 1990; Skeath et al. 1992). Expression of the proneural bHLH genes precedes NB formation and they are initially expressed in all cells of a proneural cluster. During neurogenesis, most cells within the cluster begin to enlarge, but, subsequently, only one cell/cluster continues to increase in size as a NB. The other surrounding cells undergo a reduction in size (Stollewerk 2000). The enlargement and formation of neural precursors are dependent on the action of proneural bHLH genes. For example, ac and sc are expressed in MP2, and an ac sc double mutant results in an absence of MP2 in >86% of segments (Skeath and Doe 1996). Similarly, the proneural bHLH genes are generally required for the enlargement and formation of NBs in the lateral CNS, although other proneural genes are required since ac sc l(sc) triple mutants result in the loss of only ∼25% of NBs (Jimenez and Campos-Ortega 1990). After NB formation, proneural bHLH gene expression remains present only in the NB; expression declines in the adjacent ectodermal cells (Skeath and Carroll 1992).
Notch signaling and NB selection
The Notch signaling pathway is required for the selection of one cell in each proneural cluster to become a NB while the other cells become epidermoblasts. Loss-of-function mutants in components of the Notch signaling pathway result in hypertrophy of the CNS at the expense of the epidermis (Lehmann et al. 1983) as all cells of the proneural cluster become NBs . This is accompanied by proneural gene expression in all cells of the cluster (Skeath and Carroll 1992). In simplified form, the Delta transmembrane protein signals from the emerging NB to the surrounding cells through the Notch transmembrane receptor (Artavanis-Tsakonas and Muskavitch 2010). Notch signaling activates the Suppressor of Hairless TF, which forms a complex with Mastermind to activate transcription of the Enhancer of split [E(spl)] bHLH genes. The E(spl) TF proteins repress proneural gene expression in the adjacent cells, which allows these cells to develop as epidermoblasts. While all cells of the proneural cluster have the potential to become a NB, how a particular cell in a cluster becomes a NB is not definitively known for the CNS. However, continued efforts in modeling and experimentation on Drosophila sensory cells and other systems provide potentially relevant models (Troost et al. 2015; Corson et al. 2017; Henrique and Schweisguth 2019).
Brain placodes and neurogenesis:
Studies on the formation of insulin-producing cells (IPCs) within the brain indicate a mode of neurogenesis distinct from the VNC (Hwang and Rulifson 2011). The IPCs arise from a NB within the dorsomedial protocerebral neuroectoderm. This NB forms, along with other NBs, from an eight-cell placode. These cells are all initially committed to become IPC NBs until Notch signaling directs seven of the eight cells to alternative NB fates. This mode of development is different from the VNC, where Notch signaling directs cells to an epidermal fate.
Dorsal-ventral patterning of the neuroectoderm: dorsal TF and neural identity genes
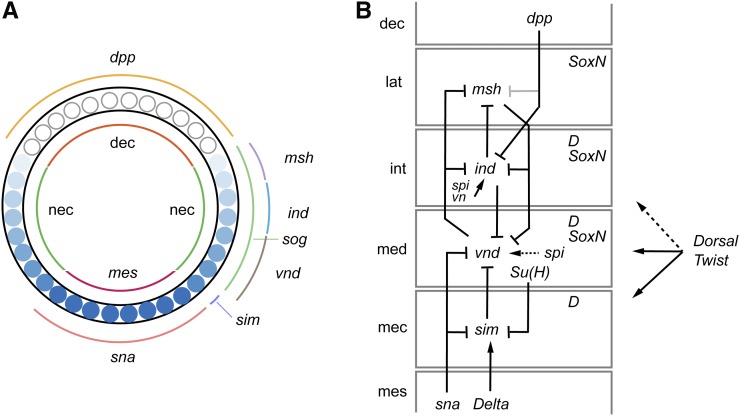
The neuroectoderm forms along both sides of the mesoderm in the blastoderm (Figure 3A). As gastrulation occurs, the mesoderm invaginates, and the two sides of the neuroectoderm converge at the ventral midline. Two key signaling pathways that govern neuroectoderm formation (commonly referred to as neural induction) are the Dorsal and Bone Morphogenetic Protein (BMP) signaling pathways. The Dorsal NF-κb-like TF forms a nuclear gradient along the dorsal–ventral (D–V) axis in the precellular blastoderm embryo with highest levels along the ventral side. The Dorsal nuclear gradient directs expression of a group of TF and signaling protein genes that subdivide the embryo along the D–V axis into mesoderm, neurogenic ectoderm, and dorsal ectoderm (Reeves and Stathopoulos 2009). The neurogenic ectoderm is further subdivided into mesectoderm (CNS midline cells) and medial, intermediate, and lateral neuroectoderm [characterized by expression of the sim, ventral nervous system defective (vnd), intermediate neuroblasts defective (ind), and muscle-specific homeobox (msh or Drop; Dr) neural identity genes, respectively] (Figure 3, A and B). These genes play important roles in directing the formation of individual neural precursor fates in the midline cells and NB columns. The precise expression of these genes is governed by the combined action of Dorsal, other TFs activated by Dorsal (Snail, Twist), multiple signaling pathways [BMP, Spitz (Spi), Notch)], and cross-regulatory inhibition as described below (reviewed in Levine and Davidson 2005; Reeves and Stathopoulos 2009) (Figure 3B).
Figure 3.
Dorsal–ventral (D–V) patterning and neural identity genes. (A) Cross-section of a blastoderm embryo showing major cell types, gradient of Dorsal protein, and expression of D–V patterning genes (ventral is bottom). Inside shows the distribution of the three main cell types: mesoderm (mes), neuroectoderm (nec), and dorsal ectoderm (dec). The blue circles represents blastoderm nuclei and indicate the levels of Dorsal protein with dark shades equivalent to high levels of nuclear protein. The domains of expression of D–V patterning genes are shown on the outside. Adapted from Hong et al. 2008, copyright (2008) National Academy of Sciences. (B) Genetic interactions and expression patterns occurring in the different neuroectodermal domains that promote neural precursor identity. Neuroectodermal domains are lateral (lat), intermediate (int), and medial (med) neuroectoderm, and mesectoderm (mec). Also shown are dorsal ectoderm (dec) and mesoderm (mes). dpp is a stronger repressor of ind expression (dark) than msh expression (gray). Maintenance of vnd expression by spi signaling is indicated by a dashed arrow. Dorsal-Twist regulation: solid lines indicate regulation by both TFs and dotted line indicates regulation by only Dorsal. Dichaete (D) and SoxN are shown in their columns of expression.
sim+ mesectodermal column:
The sim bHLH-PAS gene is a master regulator of midline cell development (Nambu et al. 1991), and is expressed in the mesectoderm (Figure 3, A and B). The sim gene is directly activated by Dorsal and Notch signaling (Kasai et al. 1998; Morel and Schweisguth 2000; Cowden and Levine 2002). It is repressed dorsally in the medial neuroectoderm by Su(H), and ventrally in the mesoderm by Snail. The Delta ligand is expressed in the mesoderm and triggers Notch signaling in the adjacent mesectoderm. This signaling converts Su(H) from a repressor to an activator, and Su(H) along with Dorsal and Twist activates sim in the mesectoderm. The action of Notch signaling limits the initial expression of sim to single cell-wide stripes. Expression of sim is maintained by autoregulation (Wharton et al. 1994). The expression of sim in single cell-wide stripes is a remarkable example of how multiple TF activators and repressors can act on a gene’s cis-regulatory elements to direct a highly specific pattern of transcription.
vnd+ medial neuroectodermal column:
Vnd is a homeobox-containing TF that functions in the medial neuroectoderm (McDonald et al. 1998) (Figure 3B). The vnd gene is directly activated by Dorsal and Twist; it is repressed by Sim and Sna ventrally and Ind and Msh dorsally. Spi signaling maintains the expression of vnd in the medial neuroectoderm.
ind+ intermediate neuroectodermal column:
Ind is also a homeobox TF that controls development of the intermediate neuroectoderm (Weiss et al. 1998) (Figure 3B). Ind is activated by Dorsal and Spi/Vein signaling, and is repressed ventrally and dorsally by Vnd and Msh, respectively. The source of Spi signaling is the medial neuroectoderm, and is dependent on rho expression (Rogers et al. 2017). The Rho intramembrane serine protease processes Spi into an active signaling factor; rho medial neuroectodermal expression is dependent on Dorsal and Twist (Ip et al. 1992).
msh+ lateral neuroectodermal column:
msh is expressed, and functions, in the lateral neuroectoderm (Isshiki et al. 1997) (Figure 3B). The positive regulation of msh is unclear, but it is repressed dorsally by Decapentaplegic (Dpp) signaling from the dorso-lateral ectoderm and ventrally by Ind and Vnd.
D–V patterning of the CNS: Dpp, Sog, and the inhibition of neurogenesis
While Dorsal is largely responsible for activating early neuroectodermal gene expression, Dpp/BMP signaling is required for formation of the dorsal ectoderm, which gives rise to the dorsal epidermis (Bier and De Robertis 2015). Dpp/BMP signaling represses neural gene expression, and multiple mechanisms exist to inhibit Dpp/BMP signaling in the neuroectoderm. However, there is also an aspect of BMP signaling that promotes distinct patterns of expression of neural identity genes in the neuroectoderm. The dpp gene is expressed at high levels in the dorsal ectoderm (Figure 3B), and the Dpp and Screw proteins act as ligands in a signaling pathway that represses neural transcription and activates epidermal transcription. Since Dpp and Scr are secreted proteins, they can migrate ventrally and repress neuronal transcription [e.g., ac and l(1)sc] in the neuroectodermal domain (Skeath et al. 1992; Biehs et al. 1996). However, the sog gene is expressed ventrally, and Sog protein binds to, and inhibits, Dpp and Scr, thus maintaining their low levels in the neuroectoderm (Biehs et al. 1996). The Brinker protein is also expressed in the neuroectoderm and acts as a transcriptional repressor to block Dpp-mediated neural transcriptional repression (Jazwinska et al. 1999). However, in the intermediate and lateral columns of the neuroectoderm, low levels of Dpp protein are present and repress ind and msh (Figure 3B). The repressive effect on ind is stronger than on msh, and this, in combination with msh ventral repression by vnd and ind, helps produce the sharp ventral border of msh expression (Mizutani et al. 2006). Enhancer affinity differences for the transcriptional effectors of Dpp signaling may contribute to the differences in repression between ind and msh (Garcia and Stathopoulos 2011; Esteves et al. 2014).
Dichaete and SoxNeuro promote neurogenesis
The High Mobility Group (HMG) SoxB family TFs, Dichaete and SoxNeuro (SoxN), play important roles along with the D–V and A–P TFs in promoting Drosophila neurogenesis (Phochanukul and Russell 2010). Within the neuroectoderm, Dichaete and SoxN are expressed in unique and overlapping stripes of cells along the D–V axis: Dichaete is expressed in the mesectodermal, medial, and intermediate columns, whereas SoxN is expressed in all three neuroectodermal columns, but not the mesectoderm (Nambu and Nambu 1996; Russell et al. 1996; Crémazy et al. 2000) (Figure 3B). Each gene shows strong mutant phenotypes in the columns where their expression does not overlap: Dichaete in the mesectoderm and SoxN in the lateral neuroectodermal column (Buescher et al. 2002; Overton et al. 2002). Dichaete mutants show strong midline glial defects and SoxN has a severe loss of lateral column NBs. SoxN also has a loss of intermediate NBs, possibly because Dichaete NB expression fades quickly (Overton et al. 2002). Not surprisingly, formation of medial and intermediate column NBs are severely affected in DichaeteSoxN double mutant embryos. Thus, Dichaete and SoxN can exhibit significant redundancy where they overlap, but they also have unique functions where they do not overlap. Genetic interaction and molecular experiments indicate that Dichaete and Ind physically interact and bind to the ac enhancer to repress ac expression in the intermediate column (Zhao and Skeath 2002; Zhao et al. 2007); Dichaete and Vnd also directly interact, but it is unknown if they function together to activate ac expression in the medial column. SoxN also genetically interacts with vnd and ind to control NB formation (Buescher et al. 2002). In the mesectoderm, Dichaete physically interacts with two TFs, Ventral veins lacking (Vvl) and Sim, to control midline gene transcription (Ma et al. 2000).
A–P patterning of the neuroectoderm
Within each hemi-segment, there exist around seven rows of NBs. Just as the epidermis is patterned intrasegmentally by the action of segmentation genes (Nusslein-Volhard and Wieschaus 1980), these genes also play a role in dictating NB formation and identity within each hemi-segment. The striped pattern of segment polarity genes derives from the action of the anterior Bicoid-Hunchback and posterior Caudal-Nanos morphogen gradients (Rosenberg et al. 2009). Key segment polarity genes involved in epidermal and CNS patterning are engrailed (en)/invected (inv), gooseberry (gsb)/gooseberry-neuro (gsb-n), hedgehog (hh), and wingless (wg) (Bhat 1999). These genes are expressed in narrow stripes within a segment: Hh and Wg are ligands for cell signaling pathways, and En/Inv and Gsb/Gsb-n are TFs. Mutant and misexpression studies revealed that these genes are required for aspects of NB identity and formation. For example, the secreted Wg morphogen gene is expressed in row five, and affects the formation of adjacent row four and row six NBs and the identity of NB 4-2 (Chu-LaGraff and Doe 1993). In another example, the interactions of segment polarity genes encoding cell signaling and transcriptional repressor proteins direct expression of huckebein (hkb) to a defined set of NBs. The hkb gene is expressed in eight NBs from rows 1, 2, 4, 5, and 7, and earlier in the corresponding neuroectodermal cells (McDonald and Doe 1997); Hkb, a TF, is required for a variety of differentiated neuronal properties, including axon guidance and neurotransmitter synthesis. Both Wg and Hh activate hkb expression in the neuroectoderm and NBs, and multiple repressors act to restrict its expression. En is expressed in neuroectodermal rows 6/7 and partially represses hkb expression in those rows; Gsb is expressed in NB rows 5 and 6, and represses hkb expression in specific NBs (e.g., NB 5-3; McDonald and Doe 1997). In principle, the combined action of D–V and A–P patterning genes not only directs the formation of a NB by activating proneural gene expression (Skeath et al. 1992, 1994), but also imparts a unique identity on each NB.
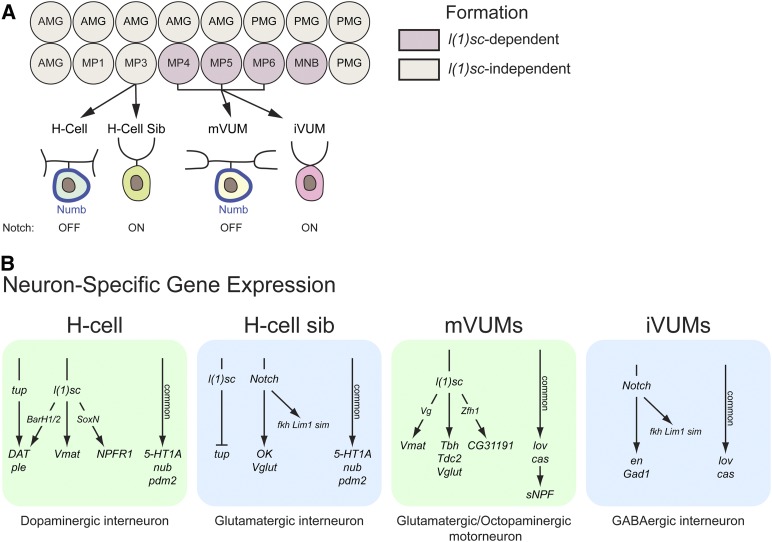
Midline precursor identity: action of segmentation genes and single-minded
The integration of D–V and A–P patterning information to generate a diverse group of neuronal precursors and glia has been studied for the CNS midline cells (Bossing and Brand 2006; Watson et al. 2011; Watson and Crews 2012). The midline neuronal precursors (arranged in order along the A–P axis) are Midline Precursors (MPs), MP1, MP3, MP4, MP5, MP6, and the median neuroblast (MNB) (Wheeler et al. 2006) (Figure 4). The MPs divide only once to generate two neurons and the MNB is a typical NB stem cell that generates ganglion mother cells (GMCs) that each divide once into two neurons (see NB Stem Cell Divisions and Asymmetric Division). The MPs are similar to GMCs, except that they are not derived from a NB. Around 10 midline glia are also generated as two discrete populations: anterior midline glia (AMG) and posterior midline glia (PMG) (Wheeler et al. 2006). The sim gene functions as a master regulator of midline cell fate: it activates the midline developmental program, including the formation of all midline neural precursors and glia, (Nambu et al. 1991) and indirectly represses the vnd+ medial CNS program (Estes et al. 2001). The Sim bHLH-PAS TF functions as a heterodimer with the broadly expressed Tango bHLH-PAS protein (Sonnenfeld et al. 1997).
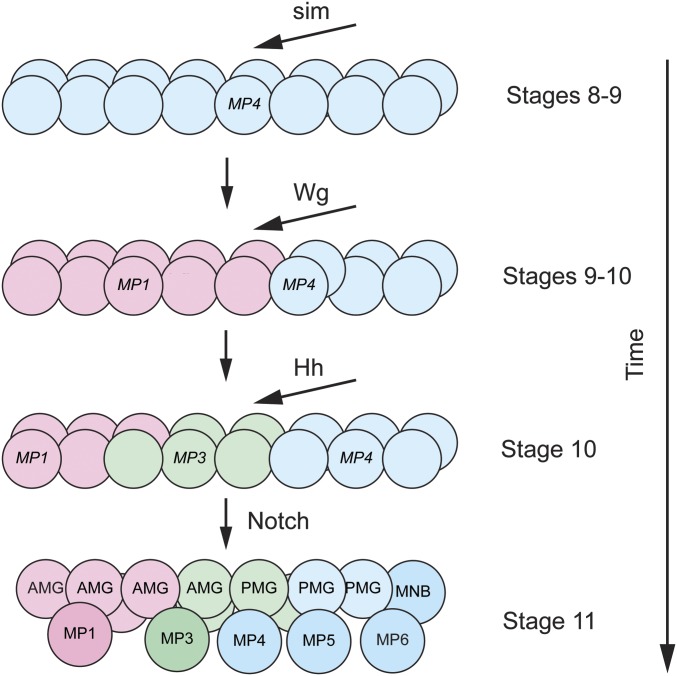
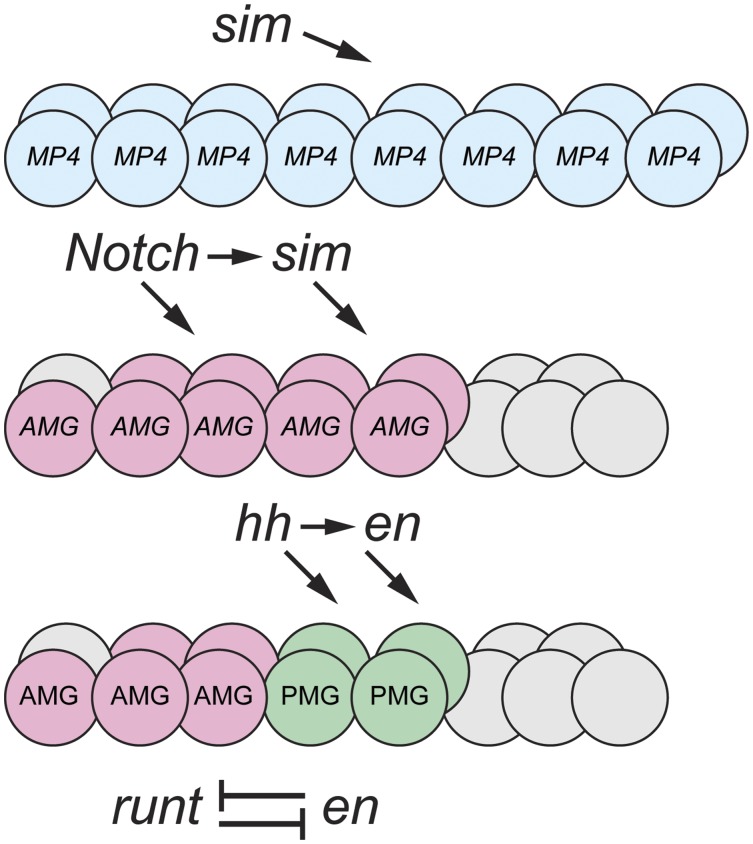
Figure 4.
Action of the Wingless, Hedgehog, and Notch signaling pathways on midline precursor identity. At stages 8–9, sim expression in the midline cells commits all midline cells to an MP4 neural fate (blue). At stages 9–10, Wg activates slp1/2 in the anterior midline compartment, and commits those cells to an MP1 fate (red). At stage 10, Hh activates gsb/gsb-n, and commits a group of cells to an MP3 fate (green). At stage 11, Notch signaling selects cells to become neural precursors and glia (AMG and PMG); the darker colors indicate the formation of neural precursors.
The formation of the neuronal precursors and midline glia are largely carried out by three signaling pathways, Wg, Hh, and Notch (Bossing and Brand 2006; Wheeler et al. 2008; Watson et al. 2011; Watson and Crews 2012) (Figure 4). Initially all midline cells are specified by sim to become a single precursor type: MP4 (Watson and Crews 2012). The Wg morphogen is secreted from lateral CNS cells in the middle of the segment, and it signals anteriorly to direct the anterior midline cells toward an MP1 fate. The Hh morphogen signals anteriorly from lateral CNS cells posterior to the wg+ cells, and directs the posterior group of MP1 cells toward an MP3 fate. Hh activates expression of gsb/gsb-n in these cells and gsb/gsb-n confers MP3 identity. At this time, there are ∼15 midline cells that constitute three equivalence groups fated to become MP1, MP3, and MP4. A third signaling pathway, Notch, carries out three functions (Wheeler et al. 2008). Notch signaling selects a single cell from each equivalence group to become the MP1, MP3, and MP4 precursors: the other cells in the MP1 and MP3 groups are directed toward a midline glial fate (the second function of Notch). In temporal sequence, the MP4 equivalence group forms MP4 → MP5 → MP6 → MNB. It is proposed that increasing levels of Notch signaling over time drive the formation of these different fates (the third function of Notch); thus, lowest levels of Notch signaling result in MP4 and the highest levels result in the MNB. Unlike the lateral CNS where Notch signaling inhibits neurogenesis and promotes epidermal cell fate; in the midline cells, Notch signaling is required for the formation of the MNB, as well as MP5 and MP6.
The role of the proneural bHLH proteins in midline cell development is complex (Stagg et al. 2011). The ac and sc genes do not play a role in neuronal precursor formation, but the l(1)sc gene is required for formation of MP4, MP5, MP6, and the MNB. However, l(1)sc does not play a role in the formation of MP1 and MP3. Instead, l(1)sc acts as a master regulator of MP3 differentiation (Stagg et al. 2011), reinforcing the idea that regulatory proteins can play different roles in different cellular contexts.
Summary
The dorsal TF directs gene expression along the D–V axis of the embryo, thus establishing the mesoderm, neuroectoderm, and dorsal ectoderm. The neuroectoderm is divided into midline, medial, intermediate, and lateral columns of gene expression. Along the A–P axis, segment polarity genes control intrasegmental patterning. Thus, each proneural cluster is defined by unique combinations of TFs, generating distinct NB and neuronal identities, while also acting on proneural genes to direct formation of NBs. Notch lateral inhibition restricts one cell in each proneural cluster to become a NB. Together, these genes are referred to as “early factors” and also regulate NB lineage progression (see Mechanisms of Neural Stem Cell Progression). Studies have shown that similar genes pattern the Drosophila embryonic brain (Urbach et al. 2006), albeit with some interesting differences. These differences include the role of A–P genes (e.g., en and empty spiracles) on D–V patterning (Seibert et al. 2009; Seibert and Urbach 2010), the roles of D–V genes in regulating Epidermal growth factor receptor (Egfr) signaling (Jussen et al. 2016), and the role of foregut-expressed sim on brain NB proliferation (Page 2003).
While the identification of these early-acting regulatory factors, and elucidation of their genetic roles in neurogenesis, are impressive, a mechanistic understanding regarding how these proteins interact and function biochemically is largely unknown. How does a cell integrate information from D–V genes (sim, vnd, ind, and msh), Sox genes, and A–P genes (en, gsb, hh, wg), to acquire a specific neural precursor identity and fate? Biochemical progress is possible; for example, using targeted DamID, it was demonstrated that Gsb opens chromatin domains in a Gsb+ NB that allows Hunchback (Hb) to bind target genes (Sen et al. 2019). In this manner, a mechanistic view of how spatial factors (Gsb) influence the function of temporal factors (Hb; see Neuroblast Temporal Cascade) is emerging. Similar genome-wide approaches can be employed to map enhancer occupancy by TFs and chromatin dynamics to identify target genes, as well as the use of traditional TF binding site mutation and transgenesis to understand how these TFs interact to control transcription and cell fate.
NB Stem Cell Divisions and Asymmetric Division
In the embryo, NBs divide in a variety of programmed ways and can give rise to: (1) another NB, (2) a GMC, (3) an intermediate neural precursor (INP), or (4) neurons (Homem and Knoblich 2012; Li et al. 2014b; Kang and Reichert 2015). GMCs divide once to give rise to two neurons. Glioblasts and glia can also emerge from NB divisions (Soustelle and Giangrande 2007) (see Glial Specification and Differentiation). In all modes, NBs divide asymmetrically in a stem cell mode to generate another NB (reviewed in Homem and Knoblich 2012; Sousa-Nunes and Somers 2013; Li et al. 2014b; Kang and Reichert 2015; Gallaud et al. 2017). This asymmetric division is characterized by: (1) the partitioning of protein complexes on the apical side of the cell that are inherited by the proliferating NB; and (2) distinct protein complexes on the basal side that inhibit the NB stem cell division mode and promote alternative cell choices, such as differentiation into neurons. This section describes the distinct modes of embryonic NB division patterns and an overview of the molecular mechanisms that guide NB asymmetric divisions. The sophisticated and comprehensive molecular and biochemical understanding of the multiple types of Drosophila neural asymmetric divisions (NB stem cells, GMCs, and sensory neuron precursors) represents one of the most important achievements and contributions of Drosophila research to the field of developmental biology.
Patterns of embryonic neural precursor divisions
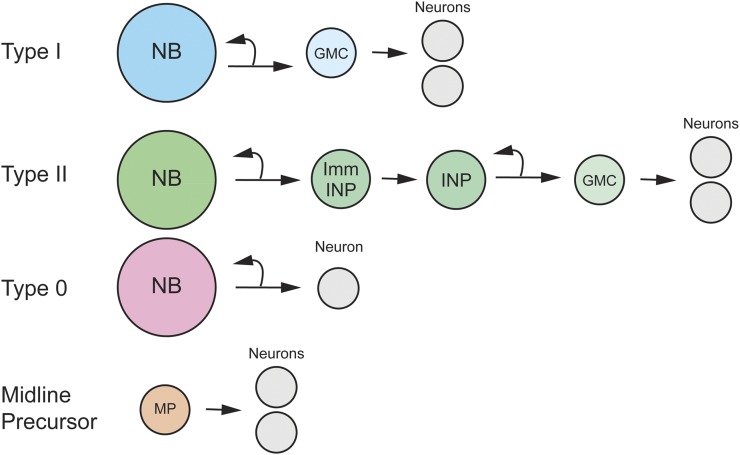
In the embryo, the CNS has five types of neural precursors that employ different patterns of division to generate neuronal progeny (Figure 5).
Figure 5.
Patterns of embryonic neural precursor divisions. Shown are Type I, Type II, Type 0 NB and Midline Precursor division modes. Type I and Type II division modes include GMCs, and Type II divisions include immature INP (Imm INP) and INP cell types.
Type I divisions (NB → GMC → neurons):
All NBs in the VNC undergo a type I division pattern at some point in their life cycle; most brain NBs also undergo Type I divisions. These NBs undergo a series of asymmetric stem cell divisions that generate a NB and GMC. Each GMC divides into two neurons.
Type II divisions (NB → INP → GMC → neurons):
There are eight NBs in each central brain hemisphere that carry out Type II divisions. This mode of division yields an INP and a NB. Division first yields an immature INP, followed by a mature INP that divides asymmetrically into another INP and a GMC—the GMC divides into two neurons. INPs divide multiple times, generating between 4–6 GMCs and 8–12 neurons (Walsh and Doe 2017). This amplification step results in lineages larger than Type I lineages. Type II divisions were first identified in the larval brain (Bello et al. 2008; Boone and Doe 2008; Bowman et al. 2008), and it was later shown that they are born during embryonic development and generate INPs during embryogenesis (Walsh and Doe 2017; Alvarez and Diaz-Benjumea 2018).
Type 0 divisions (NB → neurons):
These are NBs that switch during development from a Type I division pattern that generates NBs and GMCs to a pattern that yields a NB and a neuron (Baumgardt et al. 2014).
Midline precursor divisions (MP → neurons):
The MPs are similar to GMCs. Each divides only once to generate two neurons. However, unlike a GMC, the MPs are not derived from a NB. MP1 and MP3-6 are unpaired midline precursors that are derived from the mesectoderm (Wheeler et al. 2006). The MP2 precursors are paired, one in each hemi-neuromere adjacent to the midline (Spana et al. 1995). MP2s are derived from the medial column neuroectoderm and not the mesectoderm (Thomas et al. 1988).
NB lineages and cell number
NBs differ in their patterns of division:
Each NB is distinct within a hemi-neuromere. One of these differences is the number of neuronal progeny. For embryonic Type I NBs, these numbers can range from four neurons (NB7-3) to 36 neurons (NB7-1) (Schmid et al. 1999). Type II NBs in the brain generate even more neurons (>50 neurons/NB) (Walsh and Doe 2017). In contrast, MPs generate only two neurons. For Type I NBs, the differences in neuronal number are due, in part, to differences in the number of GMCs generated, and the timing and duration of the switch to a Type 0 division pattern. For example, NB5-6T generates nine GMCs via a Type I division mode to generate 18 neurons followed by five Type 0 divisions to generate a total of 23 neurons (Bahrampour et al. 2017). Another NB, NB3-3A has a short Type I division window of only one division followed by 11 Type 0 divisions, yielding a total of 13 neurons. In a more extreme lineage, NB7-3A has a single Type I division followed by two Type 0 divisions to generate only four neurons.
NB polarity and asymmetric divisions
Within the developing neuroectoderm, NBs delaminate and move inward. This is followed by a series of asymmetric cell divisions. For Type I divisions, GMCs emerge from the basal side of the NB (more internal) and the replenished NB forms on the apical side (Figure 6). The NB is intrinsically polarized. It inherits this polarity from the neuroectodermal cells that localize Par complex proteins on the apical side of the cell. Each division of the NB restores the apical localization of the Par proteins, and the Par complex governs the localization of basal proteins that are inherited by the GMC. The basal proteins direct the GMC division into two neurons and neuronal differentiation, while the apical cell maintains its stem cell characteristics. Understanding NB stem cell divisions and neuronal differentiation requires an appreciation of how asymmetric localization of the apical and basal components occurs.
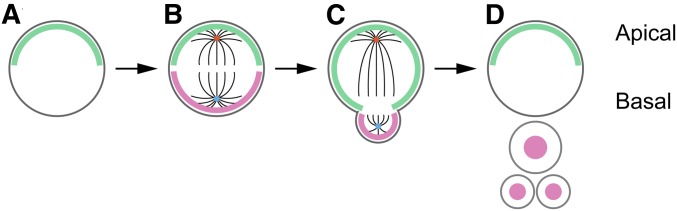
Figure 6.
Asymmetric NB division. (A) PAR complexes (green) form on the apical side of the NB cortex. (B) NB is polarized at metaphase with apical (green) and basal (red) complexes. Mother centrosome (older) is blue, and daughter centrosome (younger) is orange. (C) At telophase, the NB retains apical complexes, while the developing GMC has cortical basal factors. (D) After division, apical material again forms in the NB, whereas the basal factors enter the nucleus of GMCs and neurons. Adapted by permission from Springer Nature: Cell and Tissue Research (Kang and Reichert 2015) copyright (2014).
Apical par complex formation and function:
One key role of the apically localized Par complex is to direct the localization of the basal proteins, Miranda (Mira) and Partner of Numb (Pon), to the basal cortex (Li et al. 2014b; Gallaud et al. 2017). The functional Par complex consists of Par-6, atypical Protein Kinase C (aPKC), and Bazooka (Baz). Par complex localization is regulated by Aurora A (AurA), Lethal (2) giant larvae [L(2)gl], and Discs large (Dlg1). At prophase, the Par complex initially consists of Par-6, aPKC, and Lgl. The AurA kinase phosphorylates Par-6, which leads to the activation of aPKC. Activated aPKC phosphorylates and releases Lgl from the complex and is replaced by Baz. Additional proteins, including Cdc42, Protein phosphatase 2A (PP2A), and Dap160 also influence Par complex formation and apical localization. At metaphase, the apical complex directs the segregation of basal complex proteins to the basal cortex of the dividing NB.
Localization of basal determinants:
There are two complexes of embryonic basal proteins. One complex consists of Mira, Prospero (Pros), and Brain Tumor (Brat); the other complex consists of Numb and Pon. These proteins form a crescent along the basal cortex of the dividing NB, and are partitioned into the GMC at cytokinesis. In the embryo, the Mira-Brat-Pros complex inhibits stem cell division and promotes differentiation (Betschinger et al. 2006; Choksi et al. 2006; Lee et al. 2006). Numb inhibits Notch signaling and, in the embryo, this function occurs during neuronal cell fate acquisition after the GMC → neuron cell division step (Broadus et al. 1995; Spana and Doe 1996) (see Notch, Numb, Sanpodo, and asymmetric division). During postembryonic (but not embryonic) NB divisions, Notch promotes NB stem cell division, and this stemness function is inhibited in GMCs by Numb (Lee et al. 2006; Wang et al. 2006).
The key element in Brat and Pros basal partitioning is the localization of Mira to the basal cortex (Ikeshima-Kataoka et al. 1997; Lee et al. 2006). PP2A dephosphorylates Mira at T591, thus localizing Mira to the cell cortex (Sousa-Nunes et al. 2009). Mira is then localized to the basal side of the cortex by aPKC phosphorylation, which excludes Mira from the apical side (where aPKC resides) (Zhang et al. 2016). Mira binds Pros and Brat, and localizes the two proteins to the basal cortex.
Numb and Pon are also localized to the basal cortex. aPKC phosphorylation of Numb is required to displace Numb from the apical cortex, thus allowing its localization along the basal cortex (Knoblich et al. 1997). In addition, Polo kinase phosphorylates Pon, leading to its basal cortical localization, and Pon also contributes to proper Numb localization by directly binding to Numb (Wang et al. 2007).
Spindle orientation:
The dividing NB generates two cells (e.g., NB and GMC) with different sizes and different functional properties. While it is important that apical and basal proteins are localized to their corresponding sides of the dividing NB, it is also important that the mitotic cleavage furrow is aligned properly so that the apical and basal determinants are differentially segregated to the daughter cells (Figure 6). The importance of spindle orientation is demonstrated by genetically altering its orientation (Cabernard and Doe 2009). This results in altered cell fates, most commonly in the formation of two NBs, instead of one NB and one GMC. The apical Par complex is also important for proper spindle orientation, and key proteins that control spindle orientation are Inscuteable (Insc), Partner of Inscuteable (Pins), Mushroom body defect (Mud), and Gαi (Gallaud et al. 2017).
Insc binds to Baz in the apical Par complex, resulting in the recruitment of Insc to the apical surface (Schober et al. 1999; Wodarz et al. 1999). The G-protein, Gαi, is localized to the cortex and binds Pins; Pins, in turn binds Insc, which combines Gαi, Pins, and Insc with the Par complex (Schaefer et al. 2000; Yu et al. 2000). Pins then acts as a molecular scaffold that links the spindle to the apical surface. The interaction of Gαi with Pins results in the activation of Pins (Nipper et al. 2007); activated Pins binds to Mud, thus bringing Mud to the apical surface (Bowman et al. 2006; Izumi et al. 2006). Mud interacts with the dynein–dynactin complex. Dynein is a microtubule-associated motor protein, and the dynein–dynactin complex forces the movement of the microtubule-based spindle toward the apical side. In addition, Pins binds to Dlg, which binds a kinesin motor protein, Khc-73 (Siegrist and Doe 2005). This Pins-Dlg-Khc-73 complex anchors astral microtubules to the apical cortex, while the Pins-Mud-Dynein-Dynactin complex provides the force to move the microtubules apically (Gallaud et al. 2017).
Centrosome and spindle asymmetry:
Centrosomes serve as the microtubule organizing center (MTOC) in the cell. During cell division, the centrosome divides, and the two centrosomes exhibit unequal behavior (Rebollo et al. 2007; Rusan and Peifer 2007; Gallaud et al. 2017). The newly created (daughter) centrosome remains at the apical side. It is associated with pericentriolar matrix proteins (PCM) and retains MTOC activity, allowing the centrosome to interact with apical astral microtubules. In contrast, the mother centrosome loses its association with PCM proteins and its MTOC activity, thus removing its association with apical microtubules. During early mitosis, the mother centrosome migrates to the basal side, where it gains association with the PCM, becomes an MTOC, and interacts with basal astral microtubules. While centrosomes show asymmetry in inheritance during NB division, the consequence of the asymmetry is unclear, since mutants in which the NB missegregates the mother centrosome to the apical side undergo relatively normal asymmetric cell divisions (e.g., Singh et al. 2014; Ramdas Nair et al. 2016).
When the NB divides, it generates a new NB that is larger in size than the GMC. This size asymmetry is reflected in the positioning of the cleavage furrow along the apical-basal axis. The furrow is positioned closer to the basal cortex, and cytokinesis consequently results in a larger NB and smaller GMC. One of the key factors involved in daughter cell size asymmetry is Myosin II (Cabernard et al. 2010). Myosin II is localized uniformly at the NB cortex before mitosis. However, in response to polarity cues, Myosin II is cleared from the apical cortex by a flow directed from the basal side as mitosis begins (Roubinet et al. 2017). This lack of apical Myosin II allows the apical side to expand in comparison to the basal side. Myosin II then clears from the basal side in an apical-directed flow and accumulates in a lateral region that will become the site of the cleavage furrow. This delay in basal clearing compared to apical clearing contributes to the considerable size difference between the daughter NB and GMC. The presence of Myosin II at the future cleavage furrow directs actomyosin ring formation and subsequent cytokinesis. Multiple factors contribute to the asymmetric positioning of the cleavage site, including spindle orientation and the asymmetric localization of a Myosin II-organizing complex along the spindle (Roubinet et al. 2017).
Functions of basal determinants:
Key proteins inherited asymmetrically into the GMC (Brat, Pros) play two distinct roles: they inhibit stem cell divisions and promote neuronal differentiation. The brat gene encodes a translational repressor, and plays multiple biochemical roles. It is required for the localization of Pros into GMCs and inhibits cell cycle progression (Betschinger et al. 2006; Lee et al. 2006). Target mRNAs of brat include myc, mad, and deadpan, which encode TFs required for cellular growth and continued cell division (note that these functions of brat have largely been determined in postembryonic NB divisions). The Pros TF is tethered to the cortical cytoplasm by Mira and excluded from the nucleus in the NB (Hirata et al. 1995; Spana and Doe 1995; Ikeshima-Kataoka et al. 1997). After cytokinesis, Mira is degraded and Pros is released to enter the nucleus; the appearance of Pros in GMCs also requires brat (Betschinger et al. 2006). Within the GMC, Pros activates expression of genes that are required for neuronal differentiation and directly represses genes involved in NB fate and stem cell division (Li and Vaessin 2000; Choksi et al. 2006). Repressed NB genes include the temporal fate genes (see NB Temporal Cascade) and NB growth and cell division genes, including cyclinA, cyclinE, E2f1, and string. Pros also activates expression of the cell cycle inhibitors, dacapo (dap) and encore, and genes involved in neuronal differentiation and axonogenesis. By inhibiting NB stem cell growth, factors involved in cell division are precisely titrated so that an additional GMC → two neurons division occurs, but no further divisions.
NB Temporal Cascade
Concepts and history
NB gene expression maps and lineage analyses demonstrate that each NB in a hemi-neuromere is distinct and generates a different set of GMC and neuronal progeny (Doe 1992; Schmidt et al. 1997; Schmid et al. 1999). With each asymmetric division, a different GMC is generated, suggesting that NBs change their fate with each division cycle (Isshiki et al. 2001). In one of the most remarkable advances in the study of Drosophila neurogenesis, it was shown that the change in NB identity is due to a cascade of Temporal Transcription Factors (TTFs), which are variations of the progression: Hunchback (Hb) → Krüppel (Kr) → Pdm2/Nubbin (referred to as Pdm) → Castor (Cas) → Grainy head (Grh) (reviewed in Doe 2017) (Figure 7A). In a simple model: within a lineage, the neuronal progeny of NB1 and NB2 are different because the two NBs express different TTF profiles. This results in expression of a distinct set of target genes in each NB and their progeny. The progeny from two different NB1s that express the same TTF gene (e.g., NB3-1 and NB7-1) will be distinct because the original NBs are derived from a distinct position within the neuroectoderm and differ in their TFs. The combination of specific TTFs and developmental legacy for each NB results in different patterns of gene expression and different neuronal progeny. The original (and striking) observation was that Hb+, Pdm+, and Cas+ neurons are present in distinct layers within the CNS (reflecting their birth order), and that these regulatory genes may interact with each other to establish distinct NB sublineages (Kambadur et al. 1998). This observation led to a series of further remarkable genetic, molecular, and cellular studies describing how the TTF cascade directs NB gene expression and its consequences (Isshiki et al. 2001; Grosskortenhaus et al. 2006; Doe 2017).
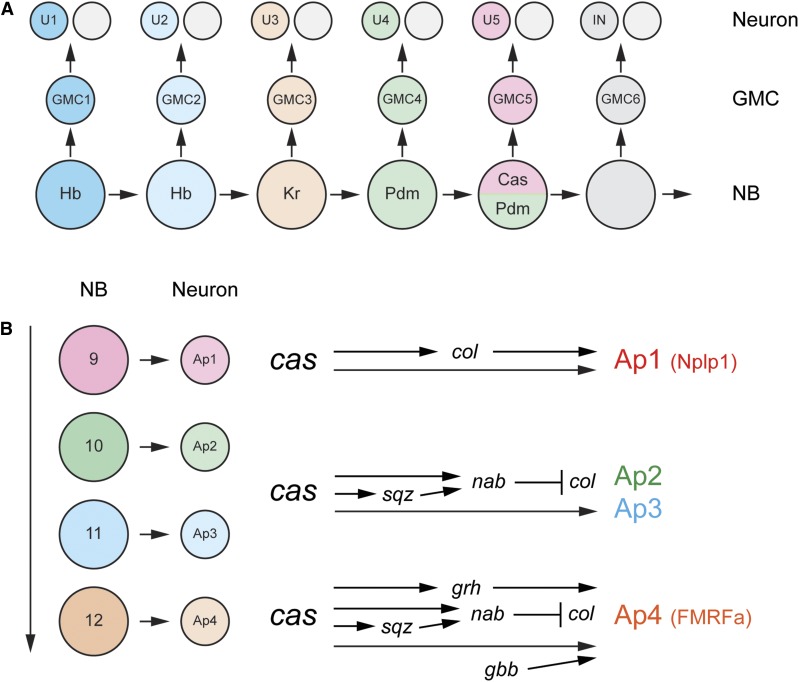
Figure 7.
Temporal transcription factor (TTF) and subtemporal transcription factor (STTF) gene cascades. (A) Progression of TTF in the NB 7-1 lineage. The U1–5 neurons are generated from GMCs 1–5. The corresponding NBs express Hb → Kr → Pdm → Cas and Pdm. Levels of Hb are higher in NB 1 (dark blue) in comparison to NB 2 (light blue). (B) In the NB 5–6T lineage, the AP1–4 neurons are generated from Type 0-dividing NBs that are present in a Cas temporal window. AP1 and AP4 are peptidergic (Nplp1 and FMRFa, respectively) and AP2/3 are not peptidergic. AP2-4 are distinct from AP1 due to the action of the Sqz and Nab STTFs that repress col. Adapted by permission from Elsevier: Cell (Baumgardt et al. 2009) copyright (2009).
The canonical Hb → Kr → Pdm → Cas → Grh cascade
Many NBs express the Hb → Kr → Pdm → Cas → Grh cascade, although variations occur in different lineages. In addition, it is clear that a number of temporal identity regulators are yet to be discovered. Several examples indicate the general principles that govern temporal NB patterning. NB7-1 generates >20 embryonic GMCs that generate >40 motoneurons and interneurons: five motoneurons, U1–U5, are distinct and are derived from GMCs 1–5, respectively; later-born GMCs give rise to interneurons (Figure 7A). GMC-1 is generated from a high Hb+ NB, GMC-2 is generated from a low Hb+ NB, GMC-3 is from a high Kr+ NB, GMC-4 from a Pdm+ NB, and GMC-5 from a Pdm+ Cas+ NB (Isshiki et al. 2001). Genetic and misexpression studies indicate that the identities of GMC-1 and the U1 motoneuron are dependent on high Hb levels; GMC-2 and U2 fates on low Hb levels; GMC-3 and U3 fates on Kr levels; GMC-4 and U4 fates on Pdm, and GMC-5 and U5 on Cas and Pdm (Pearson and Doe 2003; Grosskortenhaus et al. 2006; Seroka and Doe 2019). In other lineages (NB5-5, NB5-6T), Cas and Grh can function together to control the fate of late-born neurons (Baumgardt et al. 2009; Benito-Sipos et al. 2010). These results provide compelling evidence that TTFs act in a defined sequence to generate serial NB fates, which directly leads to distinct GMC and neuronal fates.
Variations of the TTF network
All 32 NBs in each hemi-segment and the median NB generate lineages that can differ with regards to progeny number and types of neurons and glia. Similarly, NB lineages differ in TTF gene expression: (1) while most NBs begin with expression of Hb, some late-forming NBs instead start their TTF cascade with Kr (NB3-3), Pdm (NB5-5), or Cas (NB6-1) (Tsuji et al. 2008; Benito-Sipos et al. 2010; Doe 2017). (2) In some cases, genetic experiments indicate that a TTF directs cell fate (e.g., Pdm in NB7-1) (Grosskortenhaus et al. 2006), whereas, in other lineages, its role is lacking or not apparent (e.g., Pdm in NB3-1) (Tran and Doe 2008). (3) Some TTFs span multiple NB divisions and “Subtemporal TFs (STTFs)” function within these windows to direct different cell fates. For example, in the NB5-6T lineage, NBs 9–12 divide in a Type 0 division mode to generate four neurons (Ap1-4) (Figure 7B) with three distinct fates: Ap1, Ap2/3, and Ap4. While Cas is required to generate proper Ap1-3 fates, the Squeeze and Nab STTFs function together to help distinguish Ap1 from the Ap2/3 neurons by repressing collier (col) in Ap2/3 (Baumgardt et al. 2009). Squeeze and Nab also control NB3-3 fate in the Cas expression window, and probably in additional lineages (Tsuji et al. 2008). It is clear that additional, undiscovered, TTFs and STTFs must exist to explain the full range of NB diversity.
Control of TTF timing
One important issue regarding the TTF cascade concerns how expression of TTF genes is controlled. One attractive model is that TTFs control their own expression via cross-activation and cross-repressive mechanisms (Isshiki et al. 2001). The reality is more complex, although TTF cross-repression plays a significant role (Doe 2017). Genetic studies indicate that the appearance of a TTF is not dependent on activation by prior TTFs; the TTFs that activate expression of each TTF are unknown. However, downstream TTFs can repress expression of previously expressed TTFs. Thus, pdm can repress Kr (Grosskortenhaus et al. 2006), cas can repress pdm (Grosskortenhaus et al. 2006), and grh can repress cas (Baumgardt et al. 2009), although these specific interactions may not occur in every lineage. By combining experiments with theoretical considerations, a model has been proposed that explains the sequential expression of TTFs on the decay kinetics of repressors (Averbukh et al. 2018). For example, cas expression is repressed by both Hb and Kr, and, as these proteins decay, cas expression is activated. Another important factor is the Seven-up (Svp) TF (Kanai et al. 2005). In many NB lineages, svp is expressed along with Kr and represses hb expression while promoting Kr expression (Benito-Sipos et al. 2011). Thus, it facilitates a Hb→ Kr switch. In some lineages (e.g., NB5-6), Svp also acts later in the cascade to influence neuronal cell fate (Benito-Sipos et al. 2011).
NB competence windows
One of the most fascinating developments to emerge from work on TTF patterning was the observation that misexpression of upstream NB TTFs can alter the fate of later-born NBs (Kohwi and Doe 2013). However, this cell fate transformation does not generally extend to all NBs in a lineage, but to a finite and defined number. Thus, in NB7-1, hb misexpression can change NB fate and generate additional U1/U2 neurons (the normal NB1 and NB2 progeny) in the third-to-fifth NBs in the lineage but not in later NBs (Pearson and Doe 2003). Misexpression of Kr can also extend the Kr competence window (Cleary and Doe 2006). Additional observations have provided key mechanistic insights into competence.
The first observation revealed that, during NB lineage development, the hb gene locus physically relocates from the nuclear interior (a transcriptionally active site) to the periphery in association with the nuclear lamina (a transcriptionally inactive site) (Kohwi et al. 2013). However, the relocation occurs at the end of the competence window (fifth NB in the NB7-1 lineage) well after hb expression is undetectable (after the second NB division in the NB7-1 lineage). Previous work had identified the Distal antenna (Dan) and Distal antenna-related (Danr) TFs as regulators that limit hb expression in NBs (Kohwi et al. 2011). It was further shown that misexpression of dan blocks the movement of the hb gene to the nuclear lamina, and can extend the hb competence window (Kohwi et al. 2013). However, generation of additional U1/U2 neurons requires the addition of hb expression during the extended competence window. These data indicate that Dan/Danr controls the length of the hb competence window by controlling the localization of the hb gene within the nucleus, but nuclear localization and competence is independent of hb transcription, which is required to generate U1/U2 neurons.
The second observation involved misexpression/mutant experiments indicating that the Polycomb Repressor Complexes (PRCs) also normally restrict the length of the Kr competence window in certain lineages (Touma et al. 2012). In the four NB lineages studied, only competence windows in which motoneurons were generated were PRC-dependent, suggesting that PRC establishes motoneuron competence windows that close as the lineage transitions to generating only interneurons.
These results demonstrated that NB competence involves multiple mechanisms, and that competence is independent of establishing neuronal identity. What is the purpose of competence (Kohwi et al. 2013)? It may provide a degree of stability in ensuring that a cell does not acquire an incorrect fate due to a statistical fluctuation. It may also permit the use of the same TTF to control different cell fates in different competence windows. From an evolutionary perspective, competence windows may allow an easier transition for natural selection to operate to create variations in neuronal ensembles.
The elegant genetics of TTF function raises a number of mechanistic issues. How do the TTFs interact with the early NB identity factors (Neural Precursor Specification) to generate unique NBs and progeny? Within a NB lineage (e.g., NB7-4), how does each successive NB differ with respect to gene expression (i.e., what are the gene targets of each TTF and STTFs)? Relating to the epigenetic role of Gsb (Neural Precursor Specification), Hb nuclear localization, and PRC results, how do changes in chromatin organization and accessibility influence TTF function and cell fate?
Mechanisms of Neural Stem Cell Progression
Multiple modes of embryonic NB proliferation
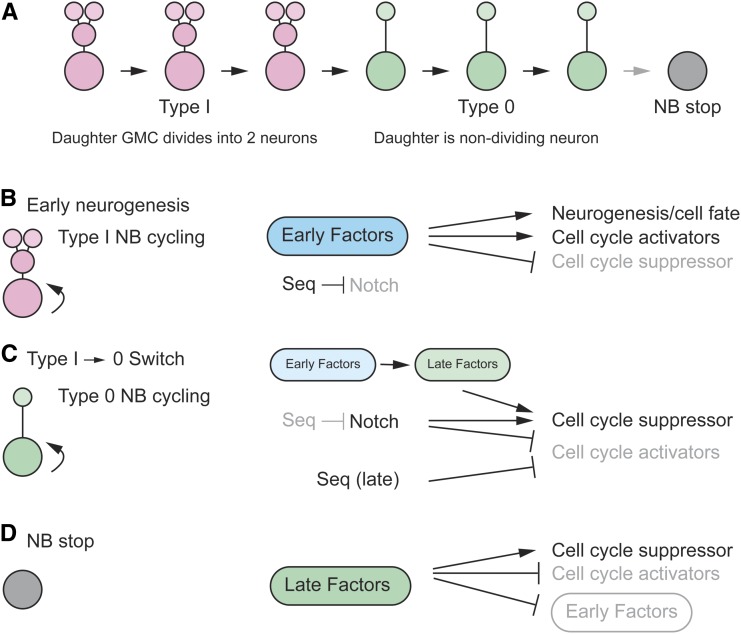
NB lineages begin as Type I NBs that generate GMCs and their two neuronal progeny. The GMC has limited proliferation potential, only dividing once. Most, if not all, NBs later transition to a Type 0 mode in which the NB stem cell division generates a single neuron in addition to a NB (Figure 8A) (Baumgardt et al. 2014). Thus, the daughter of a Type 0 NB has no proliferative potential. This is followed by a halt to NB divisions. Key issues involve understanding the factors that control Type I divisions, the switch to a Type 0 mode, and the cessation of NB division.
Figure 8.
Control of Type I cycling and the Type I→Type 0 switch. (A) In NB lineages, Type I cycling leads to a Type 0 division mode, followed by a stop in NB division. Type I GMC daughters have limited proliferative potential, dividing once, whereas type 0 NB daughters do not divide. (B) High levels of early factors (dark blue) promote neurogenesis (Type I NB cycling) by activating cell cycle factors. They also influence neural cell fate. Notch triggers the Type 0 switch but is suppressed by Seq during Type I cycling. High activity (black letters); low activity (gray letters). (C) As early factor levels decline (light blue), late factor levels rise (light green), and this promotes the Type I→0 switch in combination with Notch signaling and late-acting Seq: these genes activate expression of the dap cell cycle suppressor. Notch and late-acting Seq repress expression of cell cycle activators. (D) The stop in NB proliferation is accompanied by high levels of late factors (dark green) activating the Dap cell cycle suppressor and suppressing cell cycle activators, while also repressing early factor expression. Adapted by permission from Elsevier: Developmental Cell (Bahrampour et al. 2017) copyright (2017).
Early neurogenesis: Type I divisions
Early VNC NB divisions are Type I divisions. What factors drive this mode of division (Figure 8B)? Three groups of pan-neural TF genes with members that are broadly expressed in the early type I division stages are the Snail zinc finger family (escargot, snail, worniu), SoxB family (Dichaete, SoxN), and asense. Early TTF genes (hb, Kr, Pdm) are also present at this time. Mutants of eight of the nine genes showed reductions in NB proliferation; the exception was escargot (esg), which did not show a significant reduction in proliferation, likely due to redundancy with snail and worniu (Bahrampour et al. 2017). Misexpression experiments further indicated that these factors drive Type I NB divisions. Their downregulation is necessary for the transition to Type 0 divisions (Bahrampour et al. 2017). Mechanistically, these regulatory proteins control expression of cell cycle regulators. The precise timing of these early and late regulators is controlled by a complex web of cross-repression and cross-activation interactions among the factors. It was also shown by DamID that Asense, Snail, and Deadpan (another pan-neural expressed TF) bind to many common target genes involved in CNS development (Southall and Brand 2009). The pan-neural and TTF early factors are activated in the ectoderm and stimulate expression of the cell-cycle genes Cyclin E (CycE), string (stg), and E2f transcription factor 1 (E2f1) that promote cell division while repressing dap, a well-characterized cell cycle inhibitor gene. This results in NB formation and drives multiple rounds of Type I NB divisions while also specifying neuronal fates since both pan-neural and temporal genes influence neuronal identity.
Switch from type I → 0 divisions and cell cycle exit
Since the Type I → 0 switch normally requires alterations in cell division, prominent cell-cycle genes were tested for their effects on NB5-6T cell division and the transition from a Type I → Type 0 mode (Baumgardt et al. 2014). Mutants of dap do not influence Type I divisions but result in conversion of a Type 0 division mode to a Type I division mode; misexpression of dap in Type I NBs prematurely triggers the Type I → 0 switch. These results indicate that dap normally suppresses Type I divisions in Type 0 cells. This is consistent with its expression: absent in early NB Type I NBs but strongly expressed late in Type 0 NBs. Genetic experiments also revealed that mutants of the cell cycle G1/S regulators CycE and E2f1 resulted in a reduction in neuronal number, and misexpression led to increases in NB and GMC divisions at the expense of Type 0 divisions.
While cell cycle regulators influence cell division and the Type I → 0 switch, the question arises as to how these processes are controlled (Figure 8C). The NB5-6T lineage generates 20 neurons from stage 9 to stage 15. The first eight divisions are Type I and generate 16 neurons and the last four divisions are Type 0 generating four neurons, Ap1–4. As NB division progresses during Type I neurogenesis, Notch signaling in the NB is weak at stage 10, but progressively strengthens by stage 12, just before the transition to stage 0 occurs. Mutants in Notch signaling result in additional Ap neurons due to type 0 NBs becoming transformed to a Type I mode and generating two Ap neurons/division instead of one (Ulvklo et al. 2012). These results demonstrate that Notch signaling contributes to the Type I → 0 switch.
How is Notch signaling controlled? One key factor is the Sequoia (Seq) zinc finger TF (Gunnar et al. 2016) (Figure 8C). Seq proteins levels are relatively high during early Type I NB divisions but gradually weaken. This is the opposite of Notch activity levels and one aspect of seq function is to suppress Notch signaling in Type I NBs. As Seq levels decline, Notch activity is enhanced and the Type I → 0 switch occurs. Notch signaling activates the E(spl)HLH TF genes and E(spl)HLH represses CycE, E2f1, and stg expression. In addition, Notch signaling activates expression of the dap cell cycle inhibitor gene. These effects of Notch signaling combine to inhibit stem cell proliferation. The role of seq is even more complex: Seq is also present at late stages and directs Type 0 patterns of division (Gunnar et al. 2016). In this mode, Seq directly represses CycE and E2f1, which helps drive the Type 0 division mode (Figure 8C).
As the expression of the early Type I mode pan-neural regulators (snail, SoxB, asense family genes) declines, they activate expression of a set of late Hox and TTF genes: Antennapedia (Antp), cas, and grh. The reduction in the levels of early factors and increase in levels of the late factors drives the transition to the Type 0 division mode (Figure 8C). As the late factors increase to even higher levels, NBs exit the cell cycle (Figure 8D) (Baumgardt et al. 2014; Bahrampour et al. 2017). Mechanistically, the late factors activate expression of dap and repress expression of CycE, E2f1, and stg. Thus, the late factors promote the Type I → 0 transition and cell cycle exit, and Notch signaling also controls the Type I → 0 switch in a distinct pathway. Since TTFs, such as Cas and Grh, also control cell fate specification (see Peptidergic neuron differentiation), this is an efficient way for regulatory genes to control both the number and subtype of neurons. The concept that the early regulators promote Type I NB divisions and the late regulators promote cell cycle exit and Type 0 divisions was reinforced by misexpression experiments (Bahrampour et al. 2017). Combinations of early regulators misexpressed in the developing wing disc generate proliferating NBs and embryonic-like VNC neural lineages, whereas misexpression of late regulators result in a reduction of proliferation.
Hox genes and the neuromere-specific differences in neuronal numbers
While most neurons and glia appear homologous between abdominal, thoracic, and gnathal segments, there are significant differences. These differences are largely under the control of the Antp and Bithorax-complex (BX-C) Hox proteins and their Pbx family and Meis family cofactors: Extradenticle (Exd) and Homothorax (Hth) (Karlsson et al. 2010). The BX-C proteins include Ultrabithorax (Ubx), abdominal-A (abd-A), and Abdominal-B (Abd-B). For example, thoracic NB 5–6 (NB 5–6T) generates a set of four neurons (the Ap cluster, Ap1–4) that are not generated by abdominal NB 5–6 (NB 5–6A) (Figure 7B). In abdominal neuromeres, the BX-C Hox genes (Ubx, abd-A, and Abd-B), in combination with Exd and Hth, terminate the NB 5–6A lineage before the Ap cluster neurons are born by triggering NB cell cycle exit and apoptosis. In thoracic neuromeres, the absence of BX-C input and the presence of Antp in cooperation with exd and high levels of hth extends the progression of NB 5–6 divisions, resulting in Ap cluster neurons in NB 5–6T. Antp and Hth work largely by activating expression of col, which drives Ap cluster neuron development (Figure 7B). The presence of grh expression in this lineage is also important for formation of Ap4 (Figure 7B). In gnathal and posterior brain segments, NB 5–6 has an extended proliferation window similar, to that of NB 5–6T in thoracic segments, but the absence of Antp and weak or absent expression of grh results in an absence of Ap cluster neurons. These results, and similar work on other lineages, indicate the important role that Hox genes play in directing segment-specific neural and glial fates.
The average sizes of neural lineages differ in the embryonic CNS as a gradient, with the anterior-most NB lineages (brain) possessing the largest number of cells/lineage, and posterior-most regions (A8–10) the smallest. Difference in lineage size is controlled by the number of NB divisions and timing of the Type I → 0 switch (a few NBs also employ a Type II mode of division that yields large lineages). In the brain, NBs have a longer proliferative phase, do not undergo a Type I → 0 switch, and undergo faster daughter cell cycles, which together leads to larger lineages (Yaghmaeian Salmani et al. 2018). Lineage size is controlled by a gradient of early factors with highest levels in the brain region (Monedero Cobeta et al. 2017; Bahrampour et al. 2019). Not surprisingly, the gradient of early factors is controlled by a Hox gene gradient of (Antp - Ubx - abd-A - Abd-B; A→P) (Monedero Cobeta et al. 2017), and Hox gene expression is controlled by Polycomb (PRC2) repression (Yaghmaeian Salmani et al. 2018; Bahrampour et al. 2019). Thus, in the brain, PRC2 represses Hox gene expression allowing early factors to maintain NB proliferation and inhibit the Type I →Type 0 switch. In the VNC, increased Hox activity along the A–P axis represses early factor gene expression, limiting the sizes of NB lineages. In the most extreme case in A8–10, the Abd-B Hox gene strongly represses early factor gene expression, resulting in the smallest NB lineages.
NB quiescence and apoptosis
For most NBs, their proliferation ends during embryogenesis, and they either undergo apoptosis or enter a quiescent state. In the A3–7 abdominal hemi-neuromeres, all but three NBs undergo apoptosis; in A2, an additional NB survives; in A1, 12 NBs plus the MNB survive; in T1–3, 23 NBs plus the MNB survive (Truman and Bate 1988; Birkholz et al. 2015). The surviving NBs enter G0 or G1-like quiescence at the end of their embryonic divisions and are reactivated during larval development to continue proliferating and generating neurons (Maurange and Gould 2005). In the brain, most NBs also enter quiescence and recommence proliferation during larval development. The exceptions are four mushroom body NBs and another lateral brain NB that escape quiescence and divide throughout embryonic and postembryonic development (Ito and Hotta 1992). The timing of NB apoptosis and quiescence is idiosyncratic to each NB lineage and, thus, varies with respect to developmental time. This indicates that it is not due to a global signal (Maurange and Gould 2005).
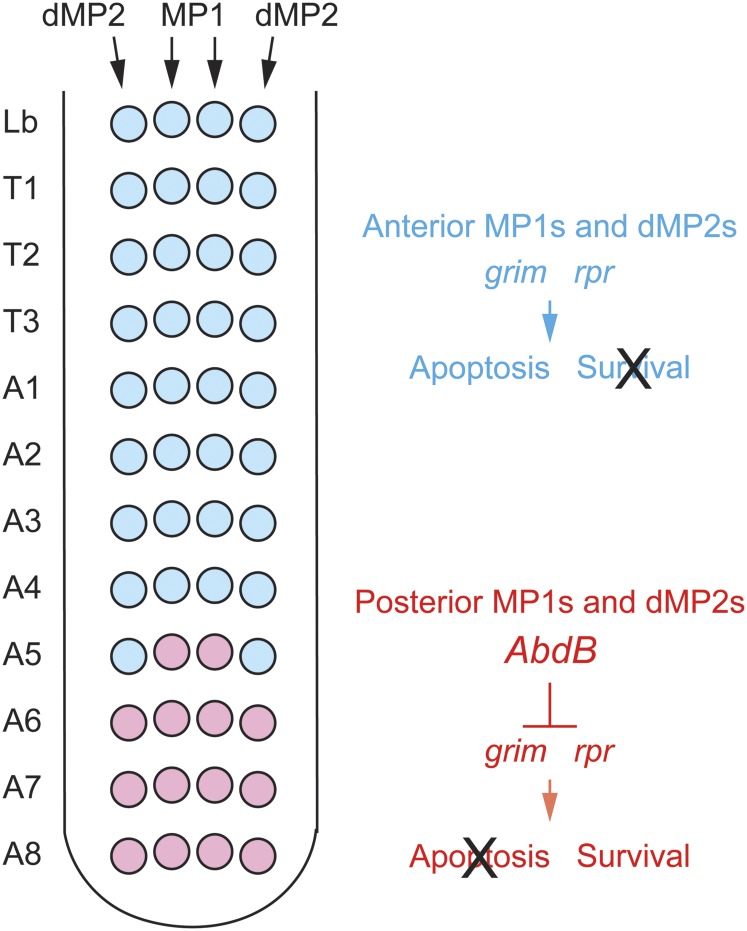
What controls whether a NB undergoes apoptosis or enters quiescence? Embryonic abdominal NBs that undergo apoptosis complete their divisions and undergo apoptosis during embryogenesis (White et al. 1994). The reaper (rpr), grim, and sickle (skl) proapoptotic genes are required for embryonic NB apoptosis, and are regulated by a common control region (White et al. 1994; Peterson et al. 2002; Tan et al. 2011). The abd-A homeotic gene, which is expressed in abdominal segments, is required for apoptosis (Prokop et al. 1998). The neuronal and glial progeny of the abdominal NBs produce Delta, which activates Notch, which is present in NBs. This triggers a pulse of abd-A expression that upregulates proapoptotic gene expression and NB apoptosis (Arya et al. 2015). As NBs age, they accumulate a repressive chromatin environment characterized by enhanced H3K27me3 histone methylation. The Cut TF opposes this increase in histone methylation, resulting in a permissive chromatin environment at the proapoptotic gene NB enhancer, allowing higher proapoptotic gene expression (Arya et al. 2019).
The alternative fate, entry into quiescence, is governed by a combination of regulatory proteins (Sousa-Nunes et al. 2010). Examining multiple lineages (Tsuji et al. 2008; Baumgardt et al. 2014; Bahrampour et al. 2017), the Hox proteins, Antp and Abd-A, were shown to influence NB quiescence: Antp promotes quiescence in thoracic segments and Abd-A prevents quiescence (promotes apoptosis) in abdominal segments.
Neuronal Formation and Differentiation
In Type I division modes, one GMC is generated in each division cycle and the GMC then divides to yield two neurons. At the midline, MPs form and also divide into two neurons. Most of the GMC and MP divisions are asymmetric, generating two nonidentical neurons (Spana and Doe 1996; Wheeler et al. 2006). After neurons are generated, they undergo differentiation into mature cells that function in larval neurotransmission. Differentiation includes the acquisition of neuronal morphology, axonal pathfinding, synaptic target recognition, synaptogenesis, dendrite formation, and the appearance of ion channels, neurotransmitters, neurotransmitter receptors, and neurotransmitter transporters. How neurons acquire their differentiated properties is challenging to study since most embryonic neurons are significantly different from each other. Consequently, differentiation is likely controlled by a large number of regulatory proteins that function combinatorially to activate specific differentiation programs in each neuron.
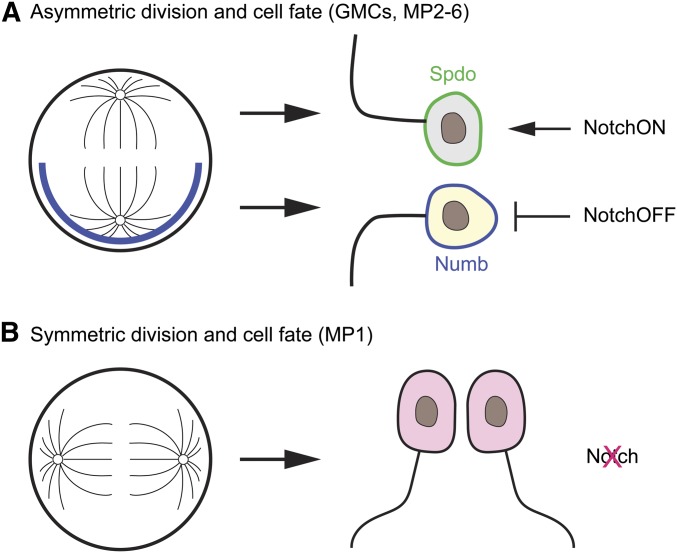
Notch, Numb, Sanpodo, and asymmetric division
During asymmetric GMC or MP divisions, the Numb protein is asymmetrically localized to just one of the two progeny (Figure 9A). The presence of Numb inhibits Delta-Notch signaling, so that differentiation of one daughter cell is dependent on Notch (NotchON), whereas the other cell that inherited Numb is independent of Notch (NotchOFF) (Spana et al. 1995; Spana and Doe 1996). Sanpodo is a transmembrane protein that is required for Notch function (Skeath and Doe 1998; O’Connor-Giles and Skeath 2003; Babaoglan et al. 2009). In the NotchOFF cell, the presence of Numb inhibits the membrane localization of Sanpodo, resulting in the loss of Notch activity. The biochemical and cell biological roles of these proteins, and a number of other interacting proteins involved in neuronal cell fate, have largely been revealed studying Drosophila sensory neuron asymmetric divisions (Derivery et al. 2015; Schweisguth 2015), but the basic mechanisms are also likely employed in GMC/MP asymmetric divisions.
Figure 9.
Asymmetric divisions of GMCs and MPs. (A) During asymmetric division of GMCs and MP2-6, Numb accumulates at the basal side of the GMC/MP and is localized in one of the daughter neurons. Spdo accumulates at the membrane of the daughter neuron without Numb and facilitates Notch signaling (NotchON). The appearance of Numb in the other daughter neuron results in the inhibition of Notch signaling (NotchOFF). (B) The MP1 cell undergoes a symmetric division to form two identical MP1 neurons. Notch signaling is not utilized to generate MP1 neuronal fates.
Motoneuron fate and axon guidance
Each hemi-neuromere of the Drosophila CNS contains 41 motoneurons, and the midline cells have three additional unpaired mVUM motoneurons (Landgraf and Thor 2006; Wheeler et al. 2006; Kohsaka et al. 2012; Heckscher et al. 2014). Drosophila motoneurons have diverse NB origins, functions, and synaptic targets; they are not generated from dedicated “motoneuron” NBs (Landgraf and Thor 2006). GMCs commonly divide into a motoneuron and an interneuron. Generally, each motoneuron innervates a distinct muscle or set of muscles. Motoneurons project to the muscle field via three main nerves: the ISN, SN, and TN (Figure 10). The branches extending from these nerves reflect individual motorneuron axons that target specific muscles. Motoneurons have distinct synaptic endings (Ib, Is, II, III) with different physiological properties. Numerous regulatory genes have been identified that control motoneuron fates. Since the axon trajectories of each motoneuron can be followed and studied, considerable success has been achieved in identifying the control genes and the cell signaling and adhesion proteins that mediate motoneuron axon guidance (Zarin and Labrador 2019).
Dorsal muscle-innervating motoneurons:
The even-skipped (eve) TF gene directs a group of ISN motoneurons to extend their projections onto dorsal muscles (Figure 10) (Landgraf et al. 1999). Furthermore, eve, Zn finger homeodomain 1 (zfh1), and grain combinatorially control the expression of genes involved in the dorsal muscle projections of the ISN aCC and RP2 motoneurons (Garces and Thor 2006; Zarin et al. 2014). Together, these TFs control expression of the Fasciclin 2 and Neuroglian cell adhesion genes and the unc-5 and beaten-path Ia guidance receptor genes (Zarin et al. 2014). These guidance molecules work in distinct ways, but together they direct the motoneuron axons to their synaptic targets. In addition, exit from the CNS to the muscle field of all motoneurons is controlled by zfh1 (Layden et al. 2006).
Ventral muscle-projecting motoneurons:
In ventrally projecting motoneurons, a group of homeobox-containing regulatory proteins, Nkx6 (HGTX), Hb9 (Extra-extra), Islet (Isl; Tailup, Tup), Lim3, Drifter (Ventral veins lacking), Ubx, and the Olig family bHLH TF work in complex ways to control the trajectories of the ISNb, ISNd, and TN axons to their ventral muscle targets (Figure 10)(Zarin and Labrador 2017). Early work indicated that the Hb9 and Nk6 regulatory proteins function together to direct motoneuronal cell fate and axon guidance (Broihier and Skeath 2002; Odden et al. 2002; Broihier et al. 2004; Cheesman et al. 2004). In addition, Dfr, Isl, and Lim3 were shown to direct motoneuron axons onto specific ventral muscles. For example, Isl is present in both ISNb and ISNd motoneurons, whereas Lim3 is present only in ISNb motoneurons. In Lim3 mutant embryos, the ISNb axons are misrouted into the ISNd tract, and misexpression of Lim3 in the ISNd neurons redirects their axons into the ISNb nerve. This establishes that Lim3 directs expression of genes involved in ISNb trajectories (Thor et al. 1999). The ISNb motoneurons express dfr in addition to Isl and Lim3, and each gene is required for ISNb pathfinding (Certel and Thor 2004). Lim3 and Isl, but not dfr, are also expressed in the TN motoneurons. Misexpression of dfr in these neurons redirects their projections to the ISNb nerve instead of the TN, establishing the role of dfr in controlling ISNb axon guidance. Olig and Ubx are also expressed in ventral motoneurons and mutants have defects in axon guidance (Oyallon et al. 2012; Hessinger et al. 2017; Zarin and Labrador 2017).
Fates of the dorsal and ventral motoneurons are maintained by cross-repressive interactions. The ventral muscle TF genes, Hb9 and Nxk6, repress eve, which controls dorsal muscle guidance, and misexpression of eve has the ability to repress expression of Hb9, isl, Nkx6, and Lim3 (Broihier and Skeath 2002).
Peptidergic neuron differentiation
The Drosophila CNS has ∼46 abdominal neurons/hemineuromere plus five midline neurons that are peptidergic (Fontana and Crews 2012; Heckscher et al. 2014). These neurons secrete a variety of neuropeptides that are generated from ∼42 precursor genes (Nässel and Winther 2010). Some neurons secrete multiple neuropeptides: for example, the midline MP1 neurons secrete Pigment-dispersing factor (Pdf), Neuropeptide-like precursor 1 (Nplp1), and Proctolin (Fontana and Crews 2012). Neuropeptides are also present in cells that secrete nonpeptide neurotransmitters: the midline iVUM5 neuron is both GABAergic and peptidergic (expresses the short neuropeptide F precursor (sNPF) gene) (Fontana and Crews 2012). Many peptidergic (or neurosecretory) cells are characterized by dense core vesicles that contain secretory peptides and the processing enzymes that generate neuropeptides. Most peptidergic neurons contain Dimmed, a bHLH TF that increases the levels of processing enzymes and neuropeptides (Hewes et al. 2003; Park et al. 2008). Dimmed has been termed a “scaling factor” since it raises the levels of peptidergic neuron-specific proteins, but does not direct a cell to become peptidergic (Mills and Taghert 2012).
Well-studied Drosophila peptidergic neurons include the Apterous (Ap)+ peptidergic neurons: the regulatory pathways that control their differentiation have been analyzed in detail (Baumgardt et al. 2009, 2014; Thor 2009). NB 5–6T gives rise to a set of four Ap+ neurons (Ap1-4) (Figure 7B), two of which, Ap1 (also called Tvb and Tv1) and Ap4 (Tv and Tv4), are peptidergic. Ap1 expresses the Nplp1 neuropeptide gene and Ap4 expresses the FMRFamide (FMRFa) neuropeptide gene. The FMRFa+ Ap4 neuron innervates the dorsal neurohemal organ—a neuroendocrine organ—whereas Ap1 axons remain within the CNS in an ipsilateral axon tract. While there are several shared regulatory pathway components, each neuron employs distinct mechanisms to carry out their differentiation programs. The Cas TTF regulates fate in all four Ap+ neurons. Cas activates expression of the col TF gene. The Ladybird TF and other early factors also activate expression of col in Ap1 (Gabilondo et al. 2016). In the Ap1 neuron, Col activates ap, dimm, and eyes absent (eya) expression, and, together, these TFs activate Nplp1 expression; Col acts in a feed-forward mode to maintain its expression while also activating Nplp1. Other aspects of Ap1 differentiation, including the expression of secretory pathway genes, axon guidance genes, as well as Nplp1, are controlled by distinct combinations of regulatory genes (Stratmann et al. 2019). Thus, differentiation of Ap1, and probably all Drosophila neurons (also see Midline neuronal differentiation), is the sum of multiple regulatory subroutines.
In the nonpeptidergic Ap2/3 neurons, Cas activates expression of the STTF genes sqz and nab, which blocks col expression leading to a nonpeptidergic fate (Figure 7B). In the Ap4 neuron, Col also activates expression of ap, dimm, and eya. However, additional factors direct Ap4 to express FMRFa. In Ap4, Cas activates grh expression, and Grh and Dachshund (Dac) function with Ap, Dimm, and Eya to activate FMRFa expression (Figure 7B). In addition, a retrograde signal, the Glass bottom boat (Gbb) BMP, from the neurohemal organ target activates the Wishful thinking (Wit) Gbb receptor on the Ap4 neuron to help activate FMRFa expression (Allan et al. 2003). The retrograde BMP signal may coordinate the precise timing of neuropeptide gene expression or impart the additional specificity required for the differentiation of individual neurons.
Midline neuron differentiation
The complexities and general features of CNS neuronal differentiation are well-illustrated by midline neuronal differentiation, given the high degree of cellular diversity in this small set of cells. There are six midline neuronal precursors: five MPs (MP1 and MP3–6) and the MNB (Figure 11A). Each MP divides into two neurons, similar to a GMC, with the exception that MPs are not derived from NBs.
Figure 11.
Diverse neuronal types are generated from midline neural precursors. (A) H-Cell and H-cell sib derive from the asymmetric division of MP3, whose formation does not require the l(1)sc proneural gene. MP4–6 require l(1)sc for formation, and each gives rise to an mVUM and iVUM. (B) H-cell and mVUMs are NotchOFF neurons that utilize l(1)sc to control cell-type specific development. The regulation of differentiation genes requires multiple intermediate TFs. H-cell sib and iVUMs are NotchON neurons. Neural gene expression common to both progeny of an MP (H-cell/H-cell sib or mVUMs/iVUMs) employ regulatory pathways distinct from the l(1)sc and Notch pathways.
MP3-6 and the nonmidline MP2s divide asymmetrically, with one daughter cell fate being dependent on Notch signaling, and the other daughter cell independent of Notch signaling (Figure 9A and Figure 11A) (Spana and Doe 1996; Wheeler et al. 2008). This distinction is due largely to the asymmetric localization of Numb, which inhibits Notch signaling. The MP1 precursor divides symmetrically to yield two identical neurons (based on the same pattern of gene expression and morphology) (Figure 9B) (Wheeler et al. 2006). Unlike MP2-6, which divides perpendicular to the longitudinal axis, MP1 divides parallel to the longitudinal axis. Notch signaling plays no apparent role in MP1 neuronal differentiation, consistent with the symmetric division of the MP1 precursor (Wheeler et al. 2008).
The progeny of MPs are diverse neuronal cell types (Figure 11B). MP3 gives rises to H-cell (dopaminergic interneuron) and H-cell sib (glutamatergic interneuron), whereas MP4–6 each gives rise to an iVUM (GABAergic interneuron) and an mVUM (octopaminergic, glutamatergic neuromodulatory motoneuron). The MP1 neurons are peptidergic motoneurons. The MNB generates a large number of local interneurons throughout embryonic and postembryonic development.
H-cell dopaminergic neuronal differentiation:
Within the VNC, there are only five dopaminergic neurons: the midline H-cell and two neurons in each lateral hemi-neuromere: the paramedial DA neuron and dorsal lateral DA neuron (Lundell and Hirsh 1994). These dopaminergic neurons are produced from the asymmetric divisions of MP3 and GMCs; in each case, the dopaminergic daughter cell is NotchOFF (Wheeler et al. 2008; Tio et al. 2011). The absence of Notch signaling on dopaminergic neuronal fate is dependent on the asymmetric localization of Numb in the dopaminergic daughter cell.
H-cell expresses a set of differentiated gene products (Figure 11B) involved in: (1) biosynthesis of dopamine [Dopa decarboxylase (Ddc) and tyrosine hydroxylase (Pale, Ple)], (2) packaging of dopamine into synaptic vesicles [Vesicular monoamine transporter (Vmat)], (3) transport through the plasma membrane [Dopamine transporter (DAT)], and (4) two neurotransmitter receptors [Neuropeptide F receptor; NPFR and 5-hydroxytryptamine (serotonin) receptor 1A (5-HT1A)]. The MP3 precursor that gives rise to H-cell expresses the related Gsb and Gsb-n TFs that are required for MP3 fate (Watson and Crews 2012). These TFs are also present in H-cell at its birth, as are a second set of downstream TFs: L(1)sc and Tup (Stagg et al. 2011). There are three distinct regulatory pathways that govern H-cell differentiation (Figure 11B) (Wheeler et al. 2008; Stagg et al. 2011): (1) the l(1)sc pathway controls expression of all genes specific to H-cell dopaminergic differentiation; (2) the tup pathway controls expression of ple, DAT, and Ddc (as does the l(1)sc pathway); and (3) a distinct pathway controls expression of differentiation genes present in both H-cell and H-cell sib (e.g., 5-HT1A serotonin receptor gene). While l(1)sc is a key regulator of H-cell differentiation, it is unlikely to control the transcription of differentiation genes directly. Instead, it activates expression of an intermediate set of TF genes that control expression of subsets of differentiated genes. These include SoxN, which controls expression of NPFR1, and BarH1, which controls expression of DAT and ple. Thus, in two distinct pathways, related homeobox proteins, Tup and BarH1, control expression of DAT and ple—they may use similar sites on target genes to control transcription since they have the same DNA binding site specificity. The gsb, l(1)sc, and tup genes also control differentiated gene expression in the paramedial and dorsal lateral dopaminergic neurons (Thor and Thomas 1997; Stagg et al. 2011; Watson and Crews 2012). One key feature is that there is a central regulator of differentiation, l(1)sc, and it controls an intermediate group of TFs that regulate different aspects of differentiation.
mVUMs and H-cell:
H cell and mVUM4-6 are the NotchOFF progeny of MP3 and MP4-6, respectively. mVUMs are glutamatergic and octopaminergic motoneurons, and only a single prominent differentiated gene, Vmat, is co-expressed with H-cell. Interestingly, mVUM differentiation gene expression [including diacyl glycerol kinase (dgk), Vmat, Tyramine β hydroxylase (Tbh), zfh1] is dependent on l(1)sc, just as l(1)sc also controls H-cell differentiation gene expression (Stagg et al. 2011). However, the role of l(1)sc in VUM4-6 development is more complex, since it also plays a proneural role and is required for MP4-6 formation (Figure 11A) (Stagg et al. 2011). The intermediate group of TFs downstream of L(1)sc is different between H-cell (BarH1/H2, SoxN) and mVUMs (Vestigial, Zfh1) (Figure 11B). Interestingly, Vmat expression is dependent on l(1)sc in both cell types, but employs different intermediate regulators: Vestigial controls Vmat expression in mVUMs, but Vestigial is not present in H-cell.
iVUMs and H-cell-sib:
The H-cell sib and iVUM4-6 neurons are the NotchON progeny of MP3 and MP4-6, respectively. These neurons are distinct: H-cell sib is a glutamatergic interneuron and the iVUMs are GABAergic interneurons. Interestingly, both have similar patterns of gene expression with respect to a suite of intermediate TFs (Fork head, Lim1, Sim), yet become distinct neurons with dissimilar patterns of terminal gene expression (Wheeler et al. 2008) (Figure 11B). Their respective MPs have distinct regulatory histories: MP3 fate is dependent on Gsb/Gsb-n (Watson and Crews 2012), whereas MP4-6 fate is dependent on Notch and l(1)sc (Wheeler et al. 2008; Stagg et al. 2011); MP4-6 cells are also En+ as are iVUMs. These data suggest a model in which Notch signaling in MP progeny direct expression of a group of related intermediate TFs that interact with different inherited regulatory proteins (such as Gsb/Gsb-n in H-cell sib and En/Inv in iVUMs) to control distinct patterns of differentiated gene expression.
Parallel pathways for genes expressed in both daughters:
There are differentiation genes that are expressed in both daughter cells of an MP that are controlled by a distinct, unknown pathway (Wheeler et al. 2008). For example, the 5-HT1A serotonin receptor, nub, and pdm are expressed in both H-cell and H-cell sib, yet their expression is not dependent on l(1)sc (H-cell) or Notch signaling (H-cell sib). Similarly, Jim Lovell (lov) and cas are expressed in iVUMs and mVUMs, yet are also not controlled by l(1)sc or Notch.
The role of cas in iVUM differentiation illustrates the complex regulation of Drosophila CNS development. Although the three iVUMs appear similar with respect to gene expression and axon trajectories, they are distinct. The sNPF neuropeptide receptor gene is expressed only in iVUM5. Interestingly, the Cas protein is present at high levels in iVUM5, lower levels in iVUM6, and is nearly absent in iVUM4. cas is required for sNPF expression in iVUM5 as expression is absent in cas mutants. An attractive model is that high levels of Cas direct sNPF expression in iVUM5, but not the other iVUMs, which have reduced cas levels. However, overexpression of cas in iVUM4 and iVUM6 fails to activate sNPF expression, indicating that specificity (expression of sNPF) requires factors in addition to Cas.
Do midline neurons regenerate?:
It is generally considered axiomatic that postmitotic neurons are incapable of further dividing or regenerating. In contrast to that view, it has been proposed that postmitotic, undifferentiated, midline cells are capable of regeneration (Bossing et al. 2012; Prehoda 2012). It was proposed that a midline neuron employs a microtubule-based system to sense when its neuronal neighbor is damaged, and then undergoes a regenerative cell division. Evidence for this assertion came from studies in which stage 10 undifferentiated midline cells were ablated; it was commonly observed that the adjacent surviving cell divided again. Previous work had proposed that mesectodermal cells undergo a synchronous division at stage 8—at this time, MPs divide into their daughter neurons and no subsequent MP divisions occur (Klämbt et al. 1991). Thus, in the case of MP-derived neurons, it was proposed that it is these postmitotic neuronal progeny of MPs that were ablated and underwent cell division (regeneration) (Bossing et al. 2012). This cell division does not occur in older differentiated neurons that have extended axons.
This model of regeneration is dependent on the assertion that MPs have divided into two neurons before stage 10, and, in the case of MPs, it is their daughter neurons and not MPs that are being ablated. However, live imaging studies revealed that MPs do not divide at stage 8, as proposed earlier, but instead divide during stage 11 (Wheeler et al. 2008); these results are consistent with published lineage data describing normal midline cell development (Bossing and Technau 1994; Wheeler et al. 2008). Consequently, an alternative interpretation of the ablation data are that it is not stage 10 undifferentiated neurons that are being ablated, but MPs that have yet to normally divide. Thus, when one MP is ablated, it is not neuronal regeneration that is being observed, but an adjacent MP normally dividing into its two daughter neurons.
Neurophysiological properties
The physiological properties of a neuron are governed by its constituent ion channels, neurotransmitter receptors, and accessory proteins. Regulating the levels of these proteins influences the characteristic physiological properties of each neuron. For example, the dorsal-projecting aCC and RP2 motoneurons express the eve TF gene. When Eve levels were increased experimentally in these cells, reduced levels of Slowpoke, a BK Ca+2-gated potassium channel, was observed (Pym et al. 2006). This resulted in a reduction in IKfast conductance. In addition, reduced levels of nAcRa-96Aa—a nicotinic acetylcholine receptor subunit—were also observed, and this led to a decrease in sensitivity for acetylcholine. In a set of ventral-projecting motoneurons, Isl represses Shaker, which encodes a K+ channel that mediates an A-type K+ current, whereas dorsal-projecting neurons that do not express isl have higher levels of Shaker (Wolfram et al. 2012). Thus, these differences in isl levels result in differences in Shaker-mediated action potential-firing between the two groups of motoneurons. Some ventral motoneurons express only isl, and some express isl and Lim3 (Thor et al. 1999). Since Lim3 can enhance repression of Shaker by Isl (Wolfram et al. 2014), this further suggests how combinations of regulatory proteins can differentially influence synaptic transmission in distinct neurons.
Summary
As described in this section, Drosophila neuronal differentiation is a complex process in which the morphological and physiological properties of each cell are controlled by multiple regulatory pathways. These pathways function in diverse ways, which demonstrates why it is important to study the differentiation of a variety of neurons. Still, there is much to be learned regarding how specific neurons differentiate. Complicating matters is the increasingly large number of neuronal-expressed genes that must be studied. Ideally, future genetic experiments will involve genome-wide assays of individual neurons to simplify the labor involved.
Glial Specification and Differentiation
Types of glia and their functions
The Drosophila embryonic CNS contains multiple types of glia that carry out a variety of functional roles (Ito et al. 1995; Crews 2010; Freeman 2015; Altenhein et al. 2016). By the larval stages, the CNS glia will broadly cover and intercalate throughout the CNS, yet they are derived from a relatively small number of cells. The CNS glia can be broadly grouped into three major classes: (1) surface glia, (2) cell body glia, and (3) neuropil and nerve glia (Figure 12). Glia also function in the embryonic CNS to phagocytose dead cells; a role analogous to microglia in vertebrates (Kurant 2011).
Figure 12.
Embryonic CNS glial cell types. Shown is a cross-section of the CNS with cell bodies in gray, and the longitudinal neuropil in white. Six types of CNS glia are shown (purple). Modified by permission from Cold Spring Harbor Laboratory Press: Cold Spring Harbor Perspectives in Biology (Freeman 2015) copyright (2015).
Surface glia together envelop the entire CNS and consist of three glial subtypes: perineurial glia, subperineurial glia, and channel glia. The perineurial glia and hemocytes secrete a lamellar barrier sheath around the outside of the CNS. Below the perineurial glia are the subperineurial glia, which cover the CNS and are the main component of the blood-brain-barrier that dictates the movement of molecules between the CNS and hemolymph (Limmer et al. 2014). The surface glia and lamella do not form completely until the late stages of larval development, but are mentioned since they originate from embryonic CNS precursors. The channel glia form a pore at the midline (Ito et al. 1995) that is lined with basement membrane material (Olson et al. 1990; Olofsson and Page 2005). Channel glia act as a pathfinding surface for trachea to innervate the CNS (Englund et al. 2002).
Cell body glia reside within the CNS and ensheath neurons extensively, thus isolating each nerve cell from adjacent cells. Analysis of cell body glial gene expression revealed expression of genes involved in metabolism, neurotransmitter uptake, and neurotransmitter recycling, indicating potential functions (Freeman et al. 2003; Altenhein et al. 2006).
Neuropil and nerve glia directly contact axons and nerves and consist of several subtypes: ensheathing glia, astrocyte-like glia, anterior midline glia, and wrapping glia. Ensheathing glia wrap the neuropil and insulate axons; this includes the longitudinal glia that ensheath the CNS axon connectives. Astrocyte-like glia have their cell bodies at the cell-body–neuropil interface, and extend their glial processes throughout the neuropil in close proximity to synapses (Stork et al. 2014). Astrocytes play a number of roles in neurotransmitter clearance and metabolism, possibly regulating neuronal physiology, and the postembryonic remodeling of synapses, phagocytosis of pruned debris, and synapse formation (Freeman 2015). Midline glia consists of two subtypes: AMG and PMG (Dong and Jacobs 1997; Wheeler et al. 2006). The AMG ensheath the axon commissures that cross the CNS midline and carry out a number of developmental roles via multiple signaling pathways (Crews 2010). AMG also secrete neurotrophic molecules that act to maintain axonal growth (Zhu et al. 2008). The PMG are a transient cell type that undergo apoptosis during embryonic development. Their function is a mystery, but is presumably developmental.
Drosophila peripheral nerves consist of motoneuron axons that exit the CNS and innervate muscles, and sensory neuron axons that originate in peripheral sensory neurons and enter the CNS. These nerves are enveloped by wrapping glia as well as by the subperineurial glia and perineurial glia that cover the CNS. The perineurial glia and subperineurial glia also infiltrate the neuromuscular junction and their morphology is sensitive to synaptic activity and growth (Brink et al. 2012). Sensory neurons are also associated with glia.
Glial lineages
All of the embryonic glia, except the midline glia, derive from cells expressing the glial master regulator, glial cells missing (gcm) (Jones 2005; Soustelle and Giangrande 2007). Despite this similarity, embryonic glia are generated in diverse ways from a relatively modest number of progenitors. While the precise division patterns of all embryonic glia are not known, there are six different lineage types that have been recognized (including midline glia) (Altenhein et al. 2016). Most glia are derived from a precursor, referred to as a neuroglioblast (NGB), that gives rise to both neurons and glia. Using the nomenclature described in Altenhein et al. (2016), there are three NGBs that constitute Type 1 lineages, and the NGB divides into a stem cell mode while generating GMCs that divide asymmetrically to yield a neuron and a glial cell. The progeny from these three NGBs all generate surface glia that are mainly subperineurial glia with one peripheral glia cell that helps form the perineurial sheath surrounding peripheral nerves. There are two Type 2 lineages (3 NGBs) that either initially generate NGBs with GMCs, giving rise to a neuron and glial cell (Type 2a) or a NGB with GMCs that generate only neurons (Type 2b). However, in both Type 2 lineages, there is a switch that occurs in which the NGB becomes a glioblast (GB) that generates GMCs producing two glial cells and no neurons. Type 2 lineages produce a variety of glial cells, even within the same lineage. For example, NB 7-4 generates surface glia, cell body glia, and neuropil glia (Beckervordersandforth et al. 2008). There are also two Type 3 lineages: in Type 3a lineages, the NGB divides to give rise to a GB and a NB that produce glia and neurons, respectively; in Type 3b lineages, there is only a GB that generates glia. The NGB Type 3a lineage (NB 6–4) generates cell body glia, whereas the Type 3b GB lineage generates neuropil-associated glia, also referred to as longitudinal glia since they lie along the longitudinal connectives. Finally, the midline glia are derived from mesectodermal cells (Bossing and Technau 1994).
Gliogenesis and Glial cell missing
They key factor in specifying embryonic glia is the GCM zinc finger TF (Hosoya et al. 1995; Jones et al. 1995; Vincent et al. 1996). In gcm loss of function mutants, glia fail to form, and presumptive glia become neurons; when gcm is ectopically expressed throughout the CNS, presumptive neurons are transformed into glia. Thus, gcm drives glial differentiation and represses neuronal differentiation (Figure 13). This master regulator role of gcm in glial development has no clear counterpart controlling neuronal development (Altenhein et al. 2016). Glial regulation of gcm transcription involves multiple cis-control elements that regulate: (1) generalized expression in the CNS, (2) glial-lineage specific expression, and (3) autoregulation (Jones et al. 2004). The generalized CNS element is required for glial expression in the CNS, and additional elements repress expression in CNS neurons. There are multiple, modular enhancers that control lineage-specific gcm transcription; for example, a distinct control element has been identified that drives expression only in the GB 6-4A lineage (Jones et al. 2004). These lineage-specific gcm control modules likely require input from the same CNS patterning TFs and signaling proteins that drive NB development (see Neural Precursor Specification) (Kim et al. 2015). Gcm activates the reversed polarity (repo) TF gene. Repo, in turn provides positive feedback along with gcm autoregulation to maintain glial expression until glial fate and differentiation occurs (Flici et al. 2014). Then, high levels of Repo result in the degradation of GCM protein (Ho et al. 2009; Flici et al. 2014), which is necessary since inappropriate expression of gcm later in development results in developmental instability.
Figure 13.
GCM control of glial development. GCM controls glial formation and differentiation (black), and inhibits neuronal differentiation (blue). Modified by permission from John Wiley and Sons: Wiley Interdisciplinary Reviews: Developmental Biology (Altenhein et al. 2016) copyright (2015).
Downstream targets of Gcm include the repo, pointed (pnt), and tramtrack (ttk) TF genes, as well as glial differentiation genes; repo is a direct Gcm target (Lee and Jones 2005), and it is likely that pnt and ttk are also direct Gcm targets. These four regulatory proteins function to define a network controlling glial differentiation (Altenhein et al. 2016) (Figure 13). Repo and Ttk control differentiation by activating the glial program and repressing the neuronal program (Giesen et al. 1997). Pnt and Repo combine to regulate genes involved in axonal ensheathment and glia–glia cell contacts (e.g., locomotion defects; loco) (Klaes et al. 1994; Granderath et al. 2000). Additional genes involved in glial differentiation, including axon ensheathment, phagocytosis, migration, and neurotransmitter metabolism, have Gcm binding sites in their regulatory DNA, and may be direct targets (Freeman et al. 2003; Altenhein et al. 2006). Another gcm-related gene is gcm2, which is expressed at relatively low levels in glia, and plays a minor role in glial differentiation in comparison to gcm (Alfonso and Jones 2002). Interestingly, both gcm and gcm2 are expressed in hemocytes, and, together, they control hemocyte migration and development of these cells into active phagocytes. Repo is not expressed in hemocytes and acts to repress hemocyte gene expression in glial cells (Trébuchet et al. 2018).
Specification of glial cell fate in individual glial precursors
Detailed investigations of several glial lineages provide insights into how Gcm is localized to GB and glial cell fate is specified. NB 6-4T is a Type 3a thoracic (T) NGB, and gives rise to a NB and GB; the GB generates three cell body glia (Akiyama-Oda et al. 2000; Freeman and Doe 2001; Ragone et al. 2001). Analysis of early division cycles revealed that Gcm protein is present in the NGB before division, and is present in both presumptive NB and GB after the first division. However, soon after division, Gcm is downregulated in the NB daughter cell but is maintained at a high level in the GB, where it drives formation of glia. After division of the NGB into NB and GB, gcm mRNA is distributed evenly between the two cells at low levels. Subsequently, gcm mRNA is downregulated in the NB and upregulated in the GB. The upregulation of gcm is controlled by Pros, which is localized asymmetrically to the GB after NGB division (Freeman and Doe 2001). Mutants of pros reveal defects in glial gene expression (repo) and a failure of NGB 6-4T progeny migration. Mira is also asymmetrically localized to the GB and is required for asymmetric localization of pros RNA and protein. In mira mutants, there is a defect in asymmetric localization of pros RNA and protein, such that it is present in both daughter cells of the NGB, and the presumptive NB is converted to an additional glial cell in some hemi-segments.
Whereas NGB 6-4T gives rise to a NB and GB by asymmetric cell division, the homologous NB in abdominal segments, GB 6-4A, is a Type 3b GB that generates only glial cells. This difference is under the control of multiple regulatory genes, including Hox, CycE, and apontic (apt). NGB 6-4T represents a ground state and the action of abd-A and Abdominal-B (Abd-B) in abdominal segments converts NGB 6-4T to GB 6-4A (Berger et al. 2005). These Hox genes carry out this conversion by downregulating expression of CycE in abdominal segments. In NGB 6-4T, CycE, acting in a role largely independent of its function in the cell cycle, is asymmetrically distributed to the NB but not GB. In NB 6-4T, CycE inhibits Pros localization, which results in a loss of gcm expression and promotes neuronal fates. In GB 6-4A, the Hox gene downregulation of CycE allows both daughter cells of the GB to become glial precursors. Another component that regulates glial-neuronal differences in NB 6-4 is the Apt TF (Shen et al. 2018). apt is expressed in NB 6-4T and activates CycE expression leading to neuronal fates, while gcm in NG 6-4A suppresses apt and CycE expression, thus supporting glial fates.
Development of midline glia
The midline glia are distinct in their developmental origins compared to lateral CNS glia. These unique cells generally have patterns of gene expression distinct from lateral glia or other cell types; most striking is the absence of gcm and repo expression in midline glia. Although it was proposed that midline glial fates are established by stage 8 (Klambt et al. 1991), later work demonstrated that this occurs later during stages 10–11 in a multi-step process (Wheeler et al. 2008). Initially, sim expression in the mesectoderm converts neuroectodermal cells to a midline neuronal precursor fate (Watson and Crews 2012) (see Midline precursor identity: action of segmentation genes and single-minded). Around stage 10, Notch signaling selects a subset of these cells to become midline glia (Wheeler et al. 2008). sim expression is maintained in midline glia due to Notch input. Molecular studies have demonstrated that Sim directly regulates expression of genes involved in glial differentiation and function, such as slit and wrapper (Wharton et al. 1994; Estes et al. 2008). It is proposed that the formation of midline glia from neuroectodermal cells is due to a sim → Notch → sim pathway (Figure 14); other TF cofactors, including Dichaete, Drifter, Zelda, and Pointed, are also involved (Ma et al. 2000; Pearson et al. 2012; Pearson and Crews 2014).
Figure 14.
Formation of midline glia. The sim gene is a master regulator of midline cell fate, and initially commits midline cells to an MP4 neural precursor fate. Notch signaling directs a group of midline cells to become AMG, in part by maintaining expression of sim in midline glia. Hh signaling directs formation of PMG, in part by activating expression of en. The Runt and En TFs cross-repress and lock in AMG and PMG fates.
Midline glia consist of two distinct populations: AMG and PMG (Dong and Jacobs 1997; Wheeler et al. 2012). AMG are the glia that ensheath the axon commissures, whereas PMG are a transient nonensheathing glia. Notch signaling triggers the AMG fate for all midline glia (Wheeler et al. 2008). In the posterior part of the segment, Hh signaling emanating from a stripe of cells adjacent, and perpendicular, to the midline converts a subset of AMG to become PMG (Figure 14) (Watson et al. 2011). This developmental switch is due, in part, to the activation of en in PMG by Hh signaling. The fates of these two distinct cell types are maintained by cross-repression between the Runt TF in AMG and En in PMG.
Midline glial migration and commissure ensheathment
The AMG initially reside in the neuroectodermal layer and migrate internally to ultimately ensheath the axon commissures. The PMG also migrate internally and some PMG will physically interact with the commissural axons, but they do not ensheath them. This process was systematically studied by live and fixed imaging (Wheeler et al. 2012) and revealed an orderly process in which the position of the AMG during their internal migration dictates how each cell will migrate and surround the axon commissures. On average, three AMG ensheath the anterior and posterior commissures. They occupy specific positions with respect to the commissures and their migratory paths are stereotyped.
During the migration process, the AMG and PMG are in close contact with the developing axon commissures (Jacobs and Goodman 1989; Wheeler et al. 2012). Two key components of this interaction are the Wrapper and Neurexin IV (Nrx-IV) proteins (Noordermeer et al. 1998; Stork et al. 2009; Wheeler et al. 2009). Wrapper is present on the surface of midline glia and Neurexin IV is present on the surface of neurons and axons. In the absence of either gene, midline glia fail to migrate, ensheath, and infiltrate the commissural axons. It was further demonstrated that Nrx-IV and Wrapper physically interact to promote midline glial-axonal interactions. Additional signaling pathways and molecules, including Egfr, PDGF- and VEGF-receptor related (Pvr), Breathless (Btl), Fear-of-intimacy (Foi), and Gliolectin (Glec), regulate specific aspects of midline glial development (Crews 2010). These processes include: (1) midline glial-axon interactions and midline glial survival (Egfr), (2) guidance of midline glia to specific midline locations (Pvr–AMG; Btl–PMG), (3) termination of midline glial migration (Foi), and (4) additional interactions between axons and midline glia (Glec). Consequently, what seems to be a relatively simple process is complex, requiring multiple mechanisms to position the midline glia so they can carry out their function. What is poorly understood is how these signaling pathways and accessory proteins interact to form the functional midline glial unit.
Longitudinal glial migration
In the embryo, multiple glial cell types are born a considerable distance from where they ultimately reside; this includes the longitudinal glia, the medialmost cell body glia (MM-CBG), and channel glia. The longitudinal glia reside on the axon connectives, whereas the MM-CBG and channel glia migrate to the midline: MM-CBG flank midline neurons, and the channel glia form a pore at the midline. Migration of longitudinal glia has been relatively well studied; in contrast, not much is known mechanistically regarding how MM-CBG and channel glia migrate to the midline. In the embryo, the longitudinal glia ultimately reside in an orderly manner along the longitudinal connectives (Jacobs et al. 1989). These glia arise from a single longitudinal glioblast (LGB, a Type 3b GB) in each hemi-neuromere that generates only glial progeny. Each LGB divides into four progeny that migrate toward the axonal connectives (Stacey et al. 2007); additional divisions contribute to the final number of longitudinal glia. The migration of the LGB is due, in part, to Netrin/Frazzled signaling (von Hilchen et al. 2010). Netrins and Frazzled are important components of a midline-derived signal (Netrins) that interacts with Frazzled on axons and contributes to their midline crossing (Brankatschk and Dickson 2006; Howard et al. 2017). In the case of glial migration, the LGB arises from a neuroectodermal region expressing frazzled; Netrin A and Netrin B are expressed in a region of the neuroectoderm between the commissures and the LGB birthplace. Acting as attractants, Netrins (Netrin A plays a more significant role) attract the longitudinal glia to a position near the connectives. Thus, guidance cues that normally attract neuronal axons to the midline are positioned in the embryo to also guide glia to their proper positions. Similarly, some peripheral nerve glia that are born in the CNS are guided by midline-derived Netrin B away from the CNS onto peripheral nerves (von Hilchen et al. 2010). These peripheral glia express the Unc5 Netrin receptor, which acts as a repulsive receptor upon interaction with Netrin. This is the same mechanism employed by motoneurons that guide their axons away from the CNS to the periphery (Keleman and Dickson 2001).
Apoptosis
Apoptosis of neurons and glia is widespread during embryonic and postembryonic CNS development (Pinto-Teixeira et al. 2016). It is estimated that 25–30% of neurons born during embryonic CNS development undergo apoptosis by the end of embryogenesis (Rogulja-Ortmann et al. 2007). Apoptosis comes in a variety of modes. Neuronal and neuroblast cell death are often intrinsically programmed (Karcavich and Doe 2005). Midline glial apoptosis or survival is dependent on appropriate cell and survival factors (Bergmann et al. 2002), as well as hormonal influences (ecdysone; Giesen et al. 2003). Neurotrophic factors are also present in the CNS, and play a role in controlling neuronal and glia survival (Hidalgo et al. 2011). Apoptotic CNS cells are phagocytosed by CNS glia and removed from the CNS (Sonnenfeld and Jacobs 1995).
Neuronal apoptosis and programmed cell death
Programmed neuronal cell death is mediated by expression of members of the proapoptotic gene family: grim, head involution defective (hid), rpr, and skl. Interestingly, different proapoptotic genes (or combinations) are used in different cell types and at different times of development (Pinto-Teixeira et al. 2016). Programmed neuronal cell death can occur soon after birth, and before the neurons differentiate. In the case of NB 7-3, GMC-2 divides asymmetrically into two neurons: EW2 and EW2sib (Karcavich and Doe 2005). Soon after birth, EW2sib, the NotchON daughter cell, undergoes apoptosis. In contrast, some neurons are generated and play developmental roles before undergoing apoptosis. In most segments, the MP1 and dMP2 neurons extend axons that act as pioneers for axon guidance (Hidalgo and Brand 1997). Other neurons extend axons along the MP1 and dMP2 tracts in the formation of specific longitudinal axon fascicles. Once this pioneering function is completed, the neurons die (Miguel-Aliaga and Thor 2004). Interestingly, MP1 and dMP2 apoptosis is segment-dependent; MP1 and dMP2 neurons persist in posterior abdominal segments while undergoing apoptosis in anterior segments (Figure 15). This is controlled by expression of the Abd-B Hox gene in posterior segments. In anterior segments, the Fkh and Hb9 TFs activate expression of the proapoptotic grim and rpr genes in MP1 and dMP2, thus triggering apoptosis. In the posterior abdominal segments, grim and rpr expression is inhibited by Abd-B, which prevents apoptosis.
Figure 15.
Hox gene control of neuronal apoptosis. In anterior neuromeres, dMP2s undergo apoptosis (Lb→A5) and MP1s undergo apoptosis in Lb→A4. In posterior neuromeres, the Abd-B Hox gene inhibits apoptosis resulting in survival of dMP2s in A6→A8 and dMP1s in A5→A8. Adapted by permission from The Company of Biologists: Development (Miguel-Aliaga and Thor 2004) copyright (2004).
Trophic control of CNS cell number and viability
Survival of both neurons and glia in the Drosophila embryonic CNS is supported by trophic factors (Hidalgo et al. 2011). Trophic systems can regulate the number of cells by directly influencing the function of proapoptotic factors or by influencing metabolic systems that maintain cell health and viability. The Drosophila CNS employs multiple types of trophic factors produced by neurons, glia, and midline cells.
Neuronal survival:
Genetic ablation of glia results in an increase in neuronal apoptosis, indicating that glia play a role in neuronal survival (Booth et al. 2000). There are several structurally related Drosophila neurotrophins involved in neuronal survival: Neurotrophin 1 (NT1), Spätzle (Spz), and Spätzle 5 (Spz5) (Zhu et al. 2008). Expression of these neurotrophins is prominent in the CNS midline cells, and genetic reduction of midline NT1 and Spz5 results in increased neuronal apoptosis. The Drosophila neurotrophins secreted at the midline could be acting directly on the proximate commissural axons, or acting more broadly. In addition, the neurotrophins are expressed in muscle cells, and their genetic reduction in muscle target tissue results in aberrant motoneuronal axon targeting. Another Drosophila neurotrophin is MANF (Drosophila mesencephalic astrocyte-derived neurotrophic factor), which is secreted by glia adjacent to dopaminergic neurons, and is required for their survival (Palgi et al. 2009).
Longitudinal glial number:
The survival of longitudinal glia is under trophic control by neurons. Vein-Egfr signaling between neurons and glia influences longitudinal glial survival (Hidalgo et al. 2001). Longitudinal glia are generated in modest excess, with the excess glia undergoing apoptosis. There is a subset of longitudinal glia that possess Egfr, and are stimulated by the Vein ligand that is produced by pioneer neurons. Genetic reduction of vein results in an increase in longitudinal glia apoptosis, indicating that Vein acts a neuronally derived glial survival factor.
Midline glial survival:
Midline glia ensheath the axon commissures, and are themselves a source of trophic and signaling factors. The midline glia represent a classic example of how cell number is controlled by neuron (axon)-glial interactions. The axon commissures are ensheathed by ∼3 AMG/segment (Wheeler et al. 2012); on average, these cells are derived from ∼10 AMG. Survival of AMG is dependent on Spi-Egfr interactions with the Spi signal emanating from the adjacent axon commissures (Bergmann et al. 2002). Those AMG in close contact with the commissures are more likely to receive sufficient Spi and survive. The Spi pathway works to inhibit apoptosis by phosphorylation and inactivation of the Hid proapoptotic protein. Midline glial numbers are also dependent on ecdysone levels (Giesen et al. 2003). In the embryo, ecdysone results in a modest reduction in midline glial number—possibly by promoting apoptosis or a reduction in proliferation.
Glial phagocytosis of dying cells
Once CNS cells undergo apoptosis, they need to be removed, since retention of corpses and unwanted cellular material may affect nervous system development and function. The CNS is ensheathed by surface glia, which are impermeable to circulating hemocytes. Consequently, it is macrophage-like CNS glia, and not hemocytes, that undertake phagocytosis of dying CNS cells (Sonnenfeld and Jacobs 1995; Kurant et al. 2008). These phagocytic cells are cell body glia that morphologically resemble astrocytes. The six microns under (simu) gene is involved in recognition and engulfment of the dying cells, and the draper gene participates in dead cell degradation (Kurant et al. 2008).
CNS condensation and apoptosis
During stages 15–17, the developing CNS undergoes a decrease in longitudinal size, referred to as CNS condensation (Olofsson and Page 2005). By the end of embryonic development, the CNS has been shortened by ∼25%. Early in embryogenesis, the neural and epidermal cell layers are adjacent, but, by stages 12–13, the layers begin to separate (Page and Olofsson 2008). Separation is complete by stage 14. Mutants that prevent neural–epidermal tissue separation (e.g., sim) also inhibit CNS condensation, suggesting that tissue separation is required for shortening (Page and Olofsson 2008). Apoptosis plays a critical role in this process, since genetically inhibiting apoptosis results in an absence of tissue separation and condensation (Page and Olofsson 2008). Apoptosis precedes tissue separation and CNS condensation, and death of epidermal cells is probably a key factor, although death of CNS neurons and glia contribute.
Interestingly, Drosophila hemocytes (phagocytes) are born in the anterior fat body cells, but migrate in a posterior direction above and below the CNS midline. Genetic and ablation studies demonstrated that hemocyte function is required for proper CNS morphogenesis, including condensation and the formation of the midline channels (Sears et al. 2003; Olofsson and Page 2005; Defaye et al. 2009; Evans et al. 2010). The phagocytic ability of migrating hemocytes and their secretion of extracellular matrix components likely both contribute to their roles in CNS morphogenesis.
Outlook
The study of Drosophila CNS development has been a remarkably successful endeavor, and much of this research is as splendid as the best developmental biology ever published. It is now possible to conceptualize how a series of developmental events can lead to the formation of the CNS and its diverse array of neurons and glia. It is also clear that many of these developmental processes are significantly more complex than are currently understood, and, in many cases, a biochemical appreciation is lacking. On a positive note, new advances in genetic technologies are well suited toward examining Drosophila CNS development, and well-planned genetic screens still provide a wealth of information (e.g., Bivik et al. 2015). Given the considerable advantages of studying a well-characterized experimental system, a dramatic leap in mechanistically grounded knowledge is possible, particularly if imbued with a collaborative spirit.
Acknowledgments
I want to acknowledge all of the members of my laboratory for their hard work, dedication, creativity, technical excellence, and keen insights. I would like to thank the many colleagues I have had the good fortune to interact with over the years, with special thanks to Corey Goodman and John Thomas. I would also like to thank FlyBook editors John Carlson and Jim Truman for the opportunity to write this review and Susan Whitfield for help with illustrations. Thanks also to the Lucille P. Markey Charitable Trust for its support.
Footnotes
Communicating editor: J. Carlson
Literature Cited
- Akiyama-Oda Y., Hotta Y., Tsukita S., and Oda H., 2000. Mechanism of glia-neuron cell-fate switch in the Drosophila thoracic neuroblast 6–4 lineage. Development 127: 3513–3522. [DOI] [PubMed] [Google Scholar]
- Alfonso T. B., and Jones B. W., 2002. gcm2 promotes glial cell differentiation and is required with glial cells missing for macrophage development in Drosophila. Dev. Biol. 248: 369–383. 10.1006/dbio.2002.0740 [DOI] [PubMed] [Google Scholar]
- Allan D. W., and Thor S., 2015. Transcriptional selectors, masters, and combinatorial codes: regulatory principles of neural subtype specification. Wiley Interdiscip. Rev. Dev. Biol. 4: 505–528. 10.1002/wdev.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan D. W., St Pierre S. E., Miguel-Aliaga I., and Thor S., 2003. Specification of neuropeptide cell identity by the integration of retrograde BMP signaling and a combinatorial transcription factor code. Cell 113: 73–86. 10.1016/S0092-8674(03)00204-6 [DOI] [PubMed] [Google Scholar]
- Almeida-Carvalho M. J., Berh D., Braun A., Chen Y. C., Eichler K. et al. , 2017. The Ol1mpiad: concordance of behavioural faculties of stage 1 and stage 3 Drosophila larvae. J. Exp. Biol. 220: 2452–2475. 10.1242/jeb.156646 [DOI] [PubMed] [Google Scholar]
- Altenhein B., Becker A., Busold C., Beckmann B., Hoheisel J. D. et al. , 2006. Expression profiling of glial genes during Drosophila embryogenesis. Dev. Biol. 296: 545–560. 10.1016/j.ydbio.2006.04.460 [DOI] [PubMed] [Google Scholar]
- Altenhein B., Cattenoz P. B., and Giangrande A., 2016. The early life of a fly glial cell. Wiley Interdiscip. Rev. Dev. Biol. 5: 67–84. 10.1002/wdev.200 [DOI] [PubMed] [Google Scholar]
- Alvarez J. A., and Diaz-Benjumea F. J., 2018. Origin and specification of type II neuroblasts in the Drosophila embryo. Development 145: pii: dev158394. 10.1242/dev.158394 [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., and Muskavitch M. A., 2010. Notch: the past, the present, and the future. Curr. Top. Dev. Biol. 92: 1–29. 10.1016/S0070-2153(10)92001-2 [DOI] [PubMed] [Google Scholar]
- Arya R., Sarkissian T., Tan Y., and White K., 2015. Neural stem cell progeny regulate stem cell death in a Notch and Hox dependent manner. Cell Death Differ. 22: 1378–1387. 10.1038/cdd.2014.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya R., Gyonjyan S., Harding K., Sarkissian T., Li Y. et al. , 2019. A Cut/cohesin axis alters the chromatin landscape to facilitate neuroblast death. Development 146:dev166603. 10.1242/dev.166603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averbukh I., Lai S. L., Doe C. Q., and Barkai N., 2018. A repressor-decay timer for robust temporal patterning in embryonic Drosophila neuroblast lineages. eLife 7: e38631 10.7554/eLife.38631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babaoglan A. B., O’Connor-Giles K. M., Mistry H., Schickedanz A., Wilson B. A. et al. , 2009. Sanpodo: a context-dependent activator and inhibitor of Notch signaling during asymmetric divisions. Development 136: 4089–4098. 10.1242/dev.040386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrampour S., Gunnar E., Jonsson C., Ekman H., and Thor S., 2017. Neural lineage progression controlled by a temporal proliferation program. Dev. Cell 43: 332–348.e4. 10.1016/j.devcel.2017.10.004 [DOI] [PubMed] [Google Scholar]
- Bahrampour S., Jonsson C., and Thor S., 2019. Brain expansion promoted by polycomb-mediated anterior enhancement of a neural stem cell proliferation program. PLoS Biol. 17: e3000163 10.1371/journal.pbio.3000163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgardt M., Karlsson D., Terriente J., Diaz-Benjumea F. J., and Thor S., 2009. Neuronal subtype specification within a lineage by opposing temporal feed-forward loops. Cell 139: 969–982. 10.1016/j.cell.2009.10.032 [DOI] [PubMed] [Google Scholar]
- Baumgardt M., Karlsson D., Salmani B. Y., Bivik C., MacDonald R. B. et al. , 2014. Global programmed switch in neural daughter cell proliferation mode triggered by a temporal gene cascade. Dev. Cell 30: 192–208. 10.1016/j.devcel.2014.06.021 [DOI] [PubMed] [Google Scholar]
- Beckervordersandforth R. M., Rickert C., Altenhein B., and Technau G. M., 2008. Subtypes of glial cells in the Drosophila embryonic ventral nerve cord as related to lineage and gene expression. Mech. Dev. 125: 542–557. 10.1016/j.mod.2007.12.004 [DOI] [PubMed] [Google Scholar]
- Bello B. C., Izergina N., Caussinus E., and Reichert H., 2008. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 3: 5 10.1186/1749-8104-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito-Sipos J., Estacio-Gomez A., Moris-Sanz M., Baumgardt M., Thor S. et al. , 2010. A genetic cascade involving klumpfuss, nab and castor specifies the abdominal leucokinergic neurons in the Drosophila CNS. Development 137: 3327–3336. 10.1242/dev.052233 [DOI] [PubMed] [Google Scholar]
- Benito-Sipos J., Ulvklo C., Gabilondo H., Baumgardt M., Angel A. et al. , 2011. Seven up acts as a temporal factor during two different stages of neuroblast 5–6 development. Development 138: 5311–5320. 10.1242/dev.070946 [DOI] [PubMed] [Google Scholar]
- Berger C., Pallavi S. K., Prasad M., Shashidhara L. S., and Technau G. M., 2005. A critical role for cyclin E in cell fate determination in the central nervous system of Drosophila melanogaster. Nat. Cell Biol. 7: 56–62. 10.1038/ncb1203 [DOI] [PubMed] [Google Scholar]
- Bergmann A., Tugentman M., Shilo B. Z., and Steller H., 2002. Regulation of cell number by MAPK-dependent control of apoptosis: a mechanism for trophic survival signaling. Dev. Cell 2: 159–170. 10.1016/S1534-5807(02)00116-8 [DOI] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., and Knoblich J. A., 2006. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124: 1241–1253. 10.1016/j.cell.2006.01.038 [DOI] [PubMed] [Google Scholar]
- Bhat K. M., 1999. Segment polarity genes in neuroblast formation and identity specification during Drosophila neurogenesis. BioEssays 21: 472–485. [DOI] [PubMed] [Google Scholar]
- Biehs B., Francois V., and Bier E., 1996. The Drosophila short gastrulation gene prevents Dpp from autoactivating and suppressing neurogenesis in the neuroectoderm. Genes Dev. 10: 2922–2934. 10.1101/gad.10.22.2922 [DOI] [PubMed] [Google Scholar]
- Bier E., and De Robertis E. M., 2015. EMBRYO DEVELOPMENT. BMP gradients: a paradigm for morphogen-mediated developmental patterning. Science 348: aaa5838 10.1126/science.aaa5838 [DOI] [PubMed] [Google Scholar]
- Birkholz O., Rickert C., Nowak J., Coban I. C., and Technau G. M., 2015. Bridging the gap between postembryonic cell lineages and identified embryonic neuroblasts in the ventral nerve cord of Drosophila melanogaster. Biol. Open 4: 420–434. 10.1242/bio.201411072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivik C., Bahrampour S., Ulvklo C., Nilsson P., Angel A. et al. , 2015. Novel genes involved in controlling specification of Drosophila FMRFamide neuropeptide cells. Genetics 200: 1229–1244. 10.1534/genetics.115.178483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone J. Q., and Doe C. Q., 2008. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev. Neurobiol. 68: 1185–1195. 10.1002/dneu.20648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth G. E., Kinrade E. F., and Hidalgo A., 2000. Glia maintain follower neuron survival during Drosophila CNS development. Development 127: 237–244. [DOI] [PubMed] [Google Scholar]
- Bossing T., Barros C. S., Fischer B., Russell S., and Shepherd D., 2012. Disruption of microtubule integrity initiates mitosis during CNS repair. Dev. Cell 23: 433–440. 10.1016/j.devcel.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossing T., and Technau G. M., 1994. The fate of the CNS midline progenitors in Drosophila as revealed by a new method for single cell labelling. Development 120: 1895–1906. [DOI] [PubMed] [Google Scholar]
- Bossing T., and Brand A. H., 2006. Determination of cell fate along the anteroposterior axis of the Drosophila ventral midline. Development 133: 1001–1012. 10.1242/dev.02288 [DOI] [PubMed] [Google Scholar]
- Bossing T., Udolph G., Doe C. Q., and Technau G. M., 1996. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev. Biol. 179: 41–64. 10.1006/dbio.1996.0240 [DOI] [PubMed] [Google Scholar]
- Bowman S. K., Neumuller R. A., Novatchkova M., Du Q., and Knoblich J. A., 2006. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev. Cell 10: 731–742. 10.1016/j.devcel.2006.05.005 [DOI] [PubMed] [Google Scholar]
- Bowman S. K., Rolland V., Betschinger J., Kinsey K. A., Emery G. et al. , 2008. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell 14: 535–546. 10.1016/j.devcel.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brankatschk M., and Dickson B. J., 2006. Netrins guide Drosophila commissural axons at short range. Nat. Neurosci. 9: 188–194. 10.1038/nn1625 [DOI] [PubMed] [Google Scholar]
- Brink D. L., Gilbert M., Xie X., Petley-Ragan L., and Auld V. J., 2012. Glial processes at the Drosophila larval neuromuscular junction match synaptic growth. PLoS One 7: e37876 10.1371/journal.pone.0037876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadus J., Skeath J. B., Spana E. P., Bossing T., Technau G. et al. , 1995. New neuroblast markers and the origin of the aCC/pCC neurons in the Drosophila central nervous system. Mech. Dev. 53: 393–402. 10.1016/0925-4773(95)00454-8 [DOI] [PubMed] [Google Scholar]
- Broihier H. T., and Skeath J. B., 2002. Drosophila homeodomain protein dHb9 directs neuronal fate via crossrepressive and cell-nonautonomous mechanisms. Neuron 35: 39–50. 10.1016/S0896-6273(02)00743-2 [DOI] [PubMed] [Google Scholar]
- Broihier H. T., Kuzin A., Zhu Y., Odenwald W., and Skeath J. B., 2004. Drosophila homeodomain protein Nkx6 coordinates motoneuron subtype identity and axonogenesis. Development 131: 5233–5242. 10.1242/dev.01394 [DOI] [PubMed] [Google Scholar]
- Buescher M., Hing F. S., and Chia W., 2002. Formation of neuroblasts in the embryonic central nervous system of Drosophila melanogaster is controlled by SoxNeuro. Development 129: 4193–4203. [DOI] [PubMed] [Google Scholar]
- Cabernard C., and Doe C. Q., 2009. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev. Cell 17: 134–141. 10.1016/j.devcel.2009.06.009 [DOI] [PubMed] [Google Scholar]
- Cabernard C., Prehoda K. E., and Doe C. Q., 2010. A spindle-independent cleavage furrow positioning pathway. Nature 467: 91–94. 10.1038/nature09334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certel S. J., and Thor S., 2004. Specification of Drosophila motoneuron identity by the combinatorial action of POU and LIM-HD factors. Development 131: 5429–5439. 10.1242/dev.01418 [DOI] [PubMed] [Google Scholar]
- Cheesman S. E., Layden M. J., Von Ohlen T., Doe C. Q., and Eisen J. S., 2004. Zebrafish and fly Nkx6 proteins have similar CNS expression patterns and regulate motoneuron formation. Development 131: 5221–5232. 10.1242/dev.01397 [DOI] [PubMed] [Google Scholar]
- Choksi S. P., Southall T. D., Bossing T., Edoff K., de Wit E. et al. , 2006. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev. Cell 11: 775–789. 10.1016/j.devcel.2006.09.015 [DOI] [PubMed] [Google Scholar]
- Chu-LaGraff Q., and Doe C. Q., 1993. Neuroblast specification and formation regulated by wingless in the Drosophila CNS. Science 261: 1594–1597. 10.1126/science.8372355 [DOI] [PubMed] [Google Scholar]
- Cleary M. D., and Doe C. Q., 2006. Regulation of neuroblast competence: multiple temporal identity factors specify distinct neuronal fates within a single early competence window. Genes Dev. 20: 429–434. 10.1101/gad.1382206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corson F., Couturier L., Rouault H., Mazouni K., and Schweisguth F., 2017. Self-organized Notch dynamics generate stereotyped sensory organ patterns in Drosophila. Science 356: pii: eaai7407. 10.1126/science.aai7407 [DOI] [PubMed] [Google Scholar]
- Cowden J., and Levine M., 2002. The Snail repressor positions Notch signaling in the Drosophila embryo. Development 129: 1785–1793. [DOI] [PubMed] [Google Scholar]
- Crémazy F., Berta P., and Girard F., 2000. Sox neuro, a new Drosophila Sox gene expressed in the developing central nervous system. Mech. Dev. 93: 215–219. 10.1016/S0925-4773(00)00268-9 [DOI] [PubMed] [Google Scholar]
- Crews S. T., 2010. Axon-glial interactions at the Drosophila CNS midline. Cell Adhes. Migr. 4: 67–71. 10.4161/cam.4.1.10208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews S. T., Thomas J. B., and Goodman C. S., 1988. The Drosophila single-minded gene encodes a nuclear protein with sequence similarity to the per gene product. Cell 52: 143–151. 10.1016/0092-8674(88)90538-7 [DOI] [PubMed] [Google Scholar]
- Dahmane N., Charron G., Lopes C., Yaspo M. L., Maunoury C. et al. , 1995. Down syndrome-critical region contains a gene homologous to Drosophila sim expressed during rat and human central nervous system development. Proc. Natl. Acad. Sci. USA 92: 9191–9195. 10.1073/pnas.92.20.9191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defaye A., Evans I., Crozatier M., Wood W., Lemaitre B. et al. , 2009. Genetic ablation of Drosophila phagocytes reveals their contribution to both development and resistance to bacterial infection. J. Innate Immun. 1: 322–334. 10.1159/000210264 [DOI] [PubMed] [Google Scholar]
- Derivery E., Seum C., Daeden A., Loubery S., Holtzer L. et al. , 2015. Polarized endosome dynamics by spindle asymmetry during asymmetric cell division. Nature 528: 280–285. 10.1038/nature16443 [DOI] [PubMed] [Google Scholar]
- Doe C. Q., 1992. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development 116: 855–863. [DOI] [PubMed] [Google Scholar]
- Doe C. Q., 2017. Temporal patterning in the Drosophila CNS. Annu. Rev. Cell Dev. Biol. 33: 219–240. 10.1146/annurev-cellbio-111315-125210 [DOI] [PubMed] [Google Scholar]
- Doe C. Q., and Goodman C. S., 1985. Early events in insect neurogenesis. II. The role of cell interactions and cell lineage in the determination of neuronal precursor cells. Dev. Biol. 111: 206–219. 10.1016/0012-1606(85)90446-4 [DOI] [PubMed] [Google Scholar]
- Dong R., and Jacobs J. R., 1997. Origin and differentiation of supernumerary midline glia in Drosophila embryos deficient for apoptosis. Dev. Biol. 190: 165–177. 10.1006/dbio.1997.8688 [DOI] [PubMed] [Google Scholar]
- Eichler K., Li F., Litwin-Kumar A., Park Y., Andrade I. et al. , 2017. The complete connectome of a learning and memory centre in an insect brain. Nature 548: 175–182. 10.1038/nature23455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C., Steneberg P., Falileeva L., Xylourgidis N., and Samakovlis C., 2002. Attractive and repulsive functions of Slit are mediated by different receptors in the Drosophila trachea. Development 129: 4941–4951. [DOI] [PubMed] [Google Scholar]
- Estes P., Mosher J., and Crews S. T., 2001. Drosophila single-minded represses gene transcription by activating the expression of repressive factors. Dev. Biol. 232: 157–175. 10.1006/dbio.2001.0174 [DOI] [PubMed] [Google Scholar]
- Estes P., Fulkerson E., and Zhang Y., 2008. Identification of motifs that are conserved in 12 Drosophila species and regulate midline glia vs. neuron expression. Genetics 178: 787–799. 10.1534/genetics.107.080440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves F. F., Springhorn A., Kague E., Taylor E., Pyrowolakis G. et al. , 2014. BMPs regulate msx gene expression in the dorsal neuroectoderm of Drosophila and vertebrates by distinct mechanisms. PLoS Genet. 10: e1004625 10.1371/journal.pgen.1004625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans I. R., Hu N., Skaer H., and Wood W., 2010. Interdependence of macrophage migration and ventral nerve cord development in Drosophila embryos. Development 137: 1625–1633. 10.1242/dev.046797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C. M., Kuwana E., Bulfone A., Fletcher C. F., Copeland N. G. et al. , 1996. Expression patterns of two murine homologs of Drosophila single-minded suggest possible roles in embryonic patterning and in the pathogenesis of Down syndrome. Mol. Cell. Neurosci. 7: 1–16. 10.1006/mcne.1996.0001 [DOI] [PubMed] [Google Scholar]
- Flici H., Cattenoz P. B., Komonyi O., Laneve P., Erkosar B. et al. , 2014. Interlocked loops trigger lineage specification and stable fates in the Drosophila nervous system. Nat. Commun. 5: 4484 10.1038/ncomms5484 [DOI] [PubMed] [Google Scholar]
- Fontana J. R., and Crews S. T., 2012. Transcriptome analysis of Drosophila CNS midline cells reveals diverse peptidergic properties and a role for castor in neuronal differentiation. Dev. Biol. 372: 131–142. 10.1016/j.ydbio.2012.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. R., 2015. Drosophila central nervous system glia. Cold Spring Harb. Perspect. Biol. 7: pii: a020552. 10.1101/cshperspect.a020552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. R., and Doe C. Q., 2001. Asymmetric Prospero localization is required to generate mixed neuronal/glial lineages in the Drosophila CNS. Development 128: 4103–4112. [DOI] [PubMed] [Google Scholar]
- Freeman M. R., Delrow J., Kim J., Johnson E., and Doe C. Q., 2003. Unwrapping glial biology: gcm target genes regulating glial development, diversification, and function. Neuron 38: 567–580. 10.1016/S0896-6273(03)00289-7 [DOI] [PubMed] [Google Scholar]
- Gabilondo H., Stratmann J., Rubio-Ferrera I., Millan-Crespo I., Contero-Garcia P. et al. , 2016. Neuronal cell fate specification by the convergence of different spatiotemporal cues on a common terminal selector cascade. PLoS Biol. 14: e1002450 (erratum: PLoS Biol. 14: e1002495). 10.1371/journal.pbio.1002450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaud E., Pham T., and Cabernard C., 2017. Drosophila melanogaster neuroblasts: a model for asymmetric stem cell divisions. Results Probl. Cell Differ. 61: 183–210. 10.1007/978-3-319-53150-2_8 [DOI] [PubMed] [Google Scholar]
- Garces A., and Thor S., 2006. Specification of Drosophila aCC motoneuron identity by a genetic cascade involving even-skipped, grain and zfh1. Development 133: 1445–1455. 10.1242/dev.02321 [DOI] [PubMed] [Google Scholar]
- Garcia M., and Stathopoulos A., 2011. Lateral gene expression in Drosophila early embryos is supported by Grainyhead-mediated activation and tiers of dorsally-localized repression. PLoS One 6: e29172 10.1371/journal.pone.0029172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesen K., Hummel T., Stollewerk A., Harrison S., Travers A. et al. , 1997. Glial development in the Drosophila CNS requires concomitant activation of glial and repression of neuronal differentiation genes. Development 124: 2307–2316. [DOI] [PubMed] [Google Scholar]
- Giesen K., Lammel U., Langehans D., Krukkert K., Bunse I. et al. , 2003. Regulation of glial cell number and differentiation by ecdysone and Fos signaling. Mech. Dev. 120: 401–413. 10.1016/S0925-4773(03)00009-1 [DOI] [PubMed] [Google Scholar]
- Granderath S., Bunse I., and Klambt C., 2000. Gcm and pointed synergistically control glial transcription of the Drosophila gene loco. Mech. Dev. 91: 197–208. 10.1016/S0925-4773(99)00304-4 [DOI] [PubMed] [Google Scholar]
- Grosskortenhaus R., Robinson K. J., and Doe C. Q., 2006. Pdm and Castor specify late-born motor neuron identity in the NB7–1 lineage. Genes Dev. 20: 2618–2627. 10.1101/gad.1445306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnar E., Bivik C., Starkenberg A., and Thor S., 2016. Sequoia controls the type I>0 daughter proliferation switch in the developing Drosophila nervous system. Development 143: 3774–3784. 10.1242/dev.139998 [DOI] [PubMed] [Google Scholar]
- Heckscher E. S., Long F., Layden M. J., Chuang C. H., Manning L. et al. , 2014. Atlas-builder software and the eNeuro atlas: resources for developmental biology and neuroscience. Development 141: 2524–2532. 10.1242/dev.108720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrique D., and Schweisguth F., 2019. Mechanisms of Notch signaling: a simple logic deployed in time and space. Development 146: pii: dev172148. 10.1242/dev.172148 [DOI] [PubMed] [Google Scholar]
- Hessinger C., Technau G. M., and Rogulja-Ortmann A., 2017. The Drosophila Hox gene Ultrabithorax acts in both muscles and motoneurons to orchestrate formation of specific neuromuscular connections. Development 144: 139–150. 10.1242/dev.143875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewes R. S., Park D., Gauthier S. A., Schaefer A. M., and Taghert P. H., 2003. The bHLH protein Dimmed controls neuroendocrine cell differentiation in Drosophila. Development 130: 1771–1781. 10.1242/dev.00404 [DOI] [PubMed] [Google Scholar]
- Hidalgo A., and Brand A. H., 1997. Targeted neuronal ablation: the role of pioneer neurons in guidance and fasciculation in the CNS of Drosophila. Development 124: 3253–3262. [DOI] [PubMed] [Google Scholar]
- Hidalgo A., Kinrade E. F., and Georgiou M., 2001. The Drosophila neuregulin vein maintains glial survival during axon guidance in the CNS. Dev. Cell 1: 679–690. 10.1016/S1534-5807(01)00074-0 [DOI] [PubMed] [Google Scholar]
- Hidalgo A., Kato K., Sutcliffe B., McIlroy G., Bishop S. et al. , 2011. Trophic neuron-glia interactions and cell number adjustments in the fruit fly. Glia 59: 1296–1303. 10.1002/glia.21092 [DOI] [PubMed] [Google Scholar]
- Hirata J., Nakagoshi H., Nabeshima Y., and Matsuzaki F., 1995. Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature 377: 627–630. 10.1038/377627a0 [DOI] [PubMed] [Google Scholar]
- Ho M. S., Chen H., Chen M., Jacques C., Giangrande A. et al. , 2009. Gcm protein degradation suppresses proliferation of glial progenitors. Proc. Natl. Acad. Sci. USA 106: 6778–6783. 10.1073/pnas.0808899106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holder J. L. Jr., Butte N. F., and Zinn A. R., 2000. Profound obesity associated with a balanced translocation that disrupts the SIM1 gene. Hum. Mol. Genet. 9: 101–108. 10.1093/hmg/9.1.101 [DOI] [PubMed] [Google Scholar]
- Homem C. C., and Knoblich J. A., 2012. Drosophila neuroblasts: a model for stem cell biology. Development 139: 4297–4310. 10.1242/dev.080515 [DOI] [PubMed] [Google Scholar]
- Hong J. W., Hendrix D. A., Papatsenko D., and Levine M. S., 2008. How the Dorsal gradient works: insights from postgenome technologies. Proc. Natl. Acad. Sci. USA 105: 20072–20076. 10.1073/pnas.0806476105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya T., Takizawa K., Nitta K., and Hotta Y., 1995. Glial cells missing: a binary switch between neuronal and glial determination in Drosophila. Cell 82: 1025–1036. 10.1016/0092-8674(95)90281-3 [DOI] [PubMed] [Google Scholar]
- Howard L. J., Brown H. E., Wadsworth B. C., and Evans T. A., 2019. Midline axon guidance in the Drosophila embryonic central nervous system. Semin. Cell Dev. Biol. 85: 13–25. 10.1016/j.semcdb.2017.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H. J., and Rulifson E., 2011. Serial specification of diverse neuroblast identities from a neurogenic placode by Notch and Egfr signaling. Development 138: 2883–2893. 10.1242/dev.055681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H., Skeath J. B., Nabeshima Y., Doe C. Q., and Matsuzaki F., 1997. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature 390: 625–629. 10.1038/37641 [DOI] [PubMed] [Google Scholar]
- Ip Y. T., Park R. E., Kosman D., Bier E., and Levine M., 1992. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. 6: 1728–1739. 10.1101/gad.6.9.1728 [DOI] [PubMed] [Google Scholar]
- Isshiki T., Takeichi M., and Nose A., 1997. The role of the msh homeobox gene during Drosophila neurogenesis: implication for the dorsoventral specification of the neuroectoderm. Development 124: 3099–3109. [DOI] [PubMed] [Google Scholar]
- Isshiki T., Pearson B., Holbrook S., and Doe C. Q., 2001. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106: 511–521. 10.1016/S0092-8674(01)00465-2 [DOI] [PubMed] [Google Scholar]
- Ito K., and Hotta Y., 1992. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev. Biol. 149: 134–148. 10.1016/0012-1606(92)90270-Q [DOI] [PubMed] [Google Scholar]
- Ito K., Urban J., and Technau G. M., 1995. Distribution, classification, and development of Drosophila glial cells in the late embryonic and early larval ventral nerve cord. Rouxs Arch. Dev. Biol. 204: 284–307. 10.1007/BF02179499 [DOI] [PubMed] [Google Scholar]
- Izumi Y., Ohta N., Hisata K., Raabe T., and Matsuzaki F., 2006. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat. Cell Biol. 8: 586–593. 10.1038/ncb1409 [DOI] [PubMed] [Google Scholar]
- Jacobs J. R., and Goodman C. S., 1989. Embryonic development of axon pathways in the Drosophila CNS. II. Behavior of pioneer growth cones. J. Neurosci. 9: 2412–2422. 10.1523/JNEUROSCI.09-07-02412.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J. R., Hiromi Y., Patel N. H., and Goodman C. S., 1989. Lineage, migration, and morphogenesis of longitudinal glia in the Drosophila CNS as revealed by a molecular lineage marker. Neuron 2: 1625–1631. 10.1016/0896-6273(89)90051-2 [DOI] [PubMed] [Google Scholar]
- Jazwinska A., Rushlow C., and Roth S., 1999. The role of brinker in mediating the graded response to Dpp in early Drosophila embryos. Development 126: 3323–3334. [DOI] [PubMed] [Google Scholar]
- Jenett A., Rubin G. M., Ngo T. T., Shepherd D., Murphy C. et al. , 2012. A GAL4-driver line resource for Drosophila neurobiology. Cell Reports 2: 991–1001. 10.1016/j.celrep.2012.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez F., and Campos-Ortega J. A., 1979. A region of the Drosophila genome necessary for CNS development. Nature 282: 310–312. 10.1038/282310a0 [DOI] [PubMed] [Google Scholar]
- Jiménez F., and Campos-Ortega J. A., 1990. Defective neuroblast commitment in mutants of the achaete-scute complex and adjacent genes of D. melanogaster. Neuron 5: 81–89. 10.1016/0896-6273(90)90036-F [DOI] [PubMed] [Google Scholar]
- Jones B. W., 2005. Transcriptional control of glial cell development in Drosophila. Dev. Biol. 278: 265–273. 10.1016/j.ydbio.2004.11.022 [DOI] [PubMed] [Google Scholar]
- Jones B. W., Fetter R. D., Tear G., and Goodman C. S., 1995. Glial cells missing: a genetic switch that controls glial vs. neuronal fate. Cell 82: 1013–1023. 10.1016/0092-8674(95)90280-5 [DOI] [PubMed] [Google Scholar]
- Jones B. W., Abeysekera M., Galinska J., and Jolicoeur E. M., 2004. Transcriptional control of glial and blood cell development in Drosophila: cis-regulatory elements of glial cells missing. Dev. Biol. 266: 374–387. 10.1016/j.ydbio.2003.10.013 [DOI] [PubMed] [Google Scholar]
- Jorgenson E., Matharu N., Palmer M. R., Yin J., Shan J. et al. , 2018. Genetic variation in the SIM1 locus is associated with erectile dysfunction. Proc. Natl. Acad. Sci. USA 115: 11018–11023. 10.1073/pnas.1809872115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jussen D., von Hilchen J., and Urbach R., 2016. Genetic regulation and function of epidermal growth factor receptor signalling in patterning of the embryonic Drosophila brain. Open Biol. 6: pii: 160202. 10.1098/rsob.160202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambadur R., Koizumi K., Stivers C., Nagle J., Poole S. J. et al. , 1998. Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 12: 246–260. 10.1101/gad.12.2.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai M. I., Okabe M., and Hiromi Y., 2005. seven-up Controls switching of transcription factors that specify temporal identities of Drosophila neuroblasts. Dev. Cell 8: 203–213. 10.1016/j.devcel.2004.12.014 [DOI] [PubMed] [Google Scholar]
- Kang K. H., and Reichert H., 2015. Control of neural stem cell self-renewal and differentiation in Drosophila. Cell Tissue Res. 359: 33–45. 10.1007/s00441-014-1914-9 [DOI] [PubMed] [Google Scholar]
- Karcavich R., and Doe C. Q., 2005. Drosophila neuroblast 7–3 cell lineage: a model system for studying programmed cell death, Notch/Numb signaling, and sequential specification of ganglion mother cell identity. J. Comp. Neurol. 481: 240–251. 10.1002/cne.20371 [DOI] [PubMed] [Google Scholar]
- Karlsson D., Baumgardt M., and Thor S., 2010. Segment-specific neuronal subtype specification by the integration of anteroposterior and temporal cues. PLoS Biol. 8: e1000368 10.1371/journal.pbio.1000368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai Y., Stahl S., and Crews S., 1998. Specification of the Drosophila CNS midline cell lineage: direct control of single-minded transcription by dorsal/ventral patterning genes. Gene Expr. 7: 171–189. [PMC free article] [PubMed] [Google Scholar]
- Keleman K., and Dickson B. J., 2001. Short- and long-range repulsion by the Drosophila Unc5 netrin receptor. Neuron 32: 605–617. 10.1016/S0896-6273(01)00505-0 [DOI] [PubMed] [Google Scholar]
- Kim H. J., Ahn H. J., Lee S., Kim J. H., Park J. et al. , 2015. Intrinsic dorsoventral patterning and extrinsic EGFR signaling genes control glial cell development in the Drosophila nervous system. Neuroscience 307: 242–252. 10.1016/j.neuroscience.2015.08.049 [DOI] [PubMed] [Google Scholar]
- Klaes A., Menne T., Stollewerk A., Scholz H., and Klambt C., 1994. The Ets transcription factors encoded by the Drosophila gene pointed direct glial cell differentiation in the embryonic CNS. Cell 78: 149–160. 10.1016/0092-8674(94)90581-9 [DOI] [PubMed] [Google Scholar]
- Klämbt C., Jacobs J. R., and Goodman C. S., 1991. The midline of the Drosophila central nervous system: a model for the genetic analysis of cell fate, cell migration, and growth cone guidance. Cell 64: 801–815. 10.1016/0092-8674(91)90509-W [DOI] [PubMed] [Google Scholar]
- Knoblich J. A., Jan L. Y., and Jan Y. N., 1997. The N terminus of the Drosophila Numb protein directs membrane association and actin-dependent asymmetric localization. Proc. Natl. Acad. Sci. USA 94: 13005–13010. 10.1073/pnas.94.24.13005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaka H., Okusawa S., Itakura Y., Fushiki A., and Nose A., 2012. Development of larval motor circuits in Drosophila. Dev. Growth Differ. 54: 408–419. 10.1111/j.1440-169X.2012.01347.x [DOI] [PubMed] [Google Scholar]
- Kohwi M., and Doe C. Q., 2013. Temporal fate specification and neural progenitor competence during development. Nat. Rev. Neurosci. 14: 823–838. 10.1038/nrn3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M., Hiebert L. S., and Doe C. Q., 2011. The pipsqueak-domain proteins Distal antenna and Distal antenna-related restrict Hunchback neuroblast expression and early-born neuronal identity. Development 138: 1727–1735. 10.1242/dev.061499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M., Lupton J. R., Lai S. L., Miller M. R., and Doe C. Q., 2013. Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell 152: 97–108. 10.1016/j.cell.2012.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer M. C., Jung C., Batelli S., Rubin G. M., and Gaul U., 2017. The glia of the adult Drosophila nervous system. Glia 65: 606–638. 10.1002/glia.23115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J. P., 2012. Building an ommatidium one cell at a time. Dev. Dyn. 241: 136–149. 10.1002/dvdy.23707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurant E., 2011. Keeping the CNS clear: glial phagocytic functions in Drosophila. Glia 59: 1304–1311. 10.1002/glia.21098 [DOI] [PubMed] [Google Scholar]
- Kurant E., Axelrod S., Leaman D., and Gaul U., 2008. Six-microns-under acts upstream of Draper in the glial phagocytosis of apoptotic neurons. Cell 133: 498–509. 10.1016/j.cell.2008.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf M., and Thor S., 2006. Development of Drosophila motoneurons: specification and morphology. Semin. Cell Dev. Biol. 17: 3–11. 10.1016/j.semcdb.2005.11.007 [DOI] [PubMed] [Google Scholar]
- Landgraf M., Roy S., Prokop A., VijayRaghavan K. and Bate M., 1999. even-skipped determines the dorsal growth of motor axons in Drosophila. Neuron 22: 43–52. 10.1016/s0896-6273(00)80677-7 [DOI] [PubMed] [Google Scholar]
- Layden M. J., Odden J. P., Schmid A., Garces A., Thor S. et al. , 2006. Zfh1, a somatic motor neuron transcription factor, regulates axon exit from the CNS. Dev. Biol. 291: 253–263. 10.1016/j.ydbio.2005.12.009 [DOI] [PubMed] [Google Scholar]
- Lee B. P., and Jones B. W., 2005. Transcriptional regulation of the Drosophila glial gene repo. Mech. Dev. 122: 849–862. 10.1016/j.mod.2005.01.002 [DOI] [PubMed] [Google Scholar]
- Lee C. Y., Andersen R. O., Cabernard C., Manning L., Tran K. D. et al. , 2006. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Dev. 20: 3464–3474. 10.1101/gad.1489406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R., Jimenez F., Dietrich U., and Campos-Ortega J. A., 1983. On the phenotype and development of mutants of early neurogenesis in Drosophila melanogaster. Wilehm Roux Arch Dev Biol 192: 62–74. 10.1007/BF00848482 [DOI] [PubMed] [Google Scholar]
- Levine M., and Davidson E. H., 2005. Gene regulatory networks for development. Proc. Natl. Acad. Sci. USA 102: 4936–4942. 10.1073/pnas.0408031102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., and Vaessin H., 2000. Pan-neural Prospero terminates cell proliferation during Drosophila neurogenesis. Genes Dev. 14: 147–151. [PMC free article] [PubMed] [Google Scholar]
- Li H. H., Kroll J. R., Lennox S. M., Ogundeyi O., Jeter J. et al. , 2014a A GAL4 driver resource for developmental and behavioral studies on the larval CNS of Drosophila. Cell Reports 8: 897–908. 10.1016/j.celrep.2014.06.065 [DOI] [PubMed] [Google Scholar]
- Li S., Wang H., and Groth C., 2014b Drosophila neuroblasts as a new model for the study of stem cell self-renewal and tumour formation. Biosci. Rep. 34: e00125. 10.1042/BSR20140008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limmer S., Weiler A., Volkenhoff A., Babatz F., and Klambt C., 2014. The Drosophila blood-brain barrier: development and function of a glial endothelium. Front. Neurosci. 8: 365 10.3389/fnins.2014.00365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundell M. J., and Hirsh J., 1994. Temporal and spatial development of serotonin and dopamine neurons in the Drosophila CNS. Dev. Biol. 165: 385–396. 10.1006/dbio.1994.1261 [DOI] [PubMed] [Google Scholar]
- Ma Y., Certel K., Gao Y., Niemitz E., Mosher J. et al. , 2000. Functional interactions between Drosophila bHLH/PAS, Sox, and POU transcription factors regulate CNS midline expression of the slit gene. J. Neurosci. 20: 4596–4605. 10.1523/JNEUROSCI.20-12-04596.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurange C., and Gould A. P., 2005. Brainy but not too brainy: starting and stopping neuroblast divisions in Drosophila. Trends Neurosci. 28: 30–36. 10.1016/j.tins.2004.10.009 [DOI] [PubMed] [Google Scholar]
- McDonald J. A., and Doe C. Q., 1997. Establishing neuroblast-specific gene expression in the Drosophila CNS: huckebein is activated by Wingless and Hedgehog and repressed by Engrailed and Gooseberry. Development 124: 1079–1087. [DOI] [PubMed] [Google Scholar]
- McDonald J. A., Holbrook S., Isshiki T., Weiss J., Doe C. Q. et al. , 1998. Dorsoventral patterning in the Drosophila central nervous system: the vnd homeobox gene specifies ventral column identity. Genes Dev. 12: 3603–3612. 10.1101/gad.12.22.3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel-Aliaga I., and Thor S., 2004. Segment-specific prevention of pioneer neuron apoptosis by cell-autonomous, postmitotic Hox gene activity. Development 131: 6093–6105. 10.1242/dev.01521 [DOI] [PubMed] [Google Scholar]
- Mills J. C., and Taghert P. H., 2012. Scaling factors: transcription factors regulating subcellular domains. BioEssays 34: 10–16. 10.1002/bies.201100089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani C. M., Meyer N., Roelink H., and Bier E., 2006. Threshold-dependent BMP-mediated repression: a model for a conserved mechanism that patterns the neuroectoderm. PLoS Biol. 4: e313 10.1371/journal.pbio.0040313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monedero Cobeta I., Salmani B. Y., and Thor S., 2017. Anterior-posterior gradient in neural stem and daughter cell proliferation governed by spatial and temporal Hox control. Curr. Biol. 27: 1161–1172. 10.1016/j.cub.2017.03.023 [DOI] [PubMed] [Google Scholar]
- Morel V., and Schweisguth F., 2000. Repression by suppressor of hairless and activation by Notch are required to define a single row of single-minded expressing cells in the Drosophila embryo. Genes Dev. 14: 377–388. [PMC free article] [PubMed] [Google Scholar]
- Nambu P. A., and Nambu J. R., 1996. The Drosophila fish-hook gene encodes a HMG domain protein essential for segmentation and CNS development. Development 122: 3467–3475. [DOI] [PubMed] [Google Scholar]
- Nambu J. R., Lewis J. O., Wharton K. A. Jr., and Crews S. T., 1991. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell 67: 1157–1167. 10.1016/0092-8674(91)90292-7 [DOI] [PubMed] [Google Scholar]
- Nässel D. R., and Winther A. M., 2010. Drosophila neuropeptides in regulation of physiology and behavior. Prog. Neurobiol. 92: 42–104. 10.1016/j.pneurobio.2010.04.010 [DOI] [PubMed] [Google Scholar]
- Nipper R. W., Siller K. H., Smith N. R., Doe C. Q., and Prehoda K. E., 2007. Galphai generates multiple Pins activation states to link cortical polarity and spindle orientation in Drosophila neuroblasts. Proc. Natl. Acad. Sci. USA 104: 14306–14311. 10.1073/pnas.0701812104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niven J. E., Graham C. M., and Burrows M., 2008. Diversity and evolution of the insect ventral nerve cord. Annu. Rev. Entomol. 53: 253–271. 10.1146/annurev.ento.52.110405.091322 [DOI] [PubMed] [Google Scholar]
- Noordermeer J. N., Kopczynski C. C., Fetter R. D., Bland K. S., Chen W. Y. et al. , 1998. Wrapper, a novel member of the Ig superfamily, is expressed by midline glia and is required for them to ensheath commissural axons in Drosophila. Neuron 21: 991–1001. 10.1016/S0896-6273(00)80618-2 [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., and Wieschaus E., 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287: 795–801. 10.1038/287795a0 [DOI] [PubMed] [Google Scholar]
- O’Connor-Giles K. M., and Skeath J. B., 2003. Numb inhibits membrane localization of Sanpodo, a four-pass transmembrane protein, to promote asymmetric divisions in Drosophila. Dev. Cell 5: 231–243. 10.1016/S1534-5807(03)00226-0 [DOI] [PubMed] [Google Scholar]
- Odden J. P., Holbrook S., and Doe C. Q., 2002. Drosophila HB9 is expressed in a subset of motoneurons and interneurons, where it regulates gene expression and axon pathfinding. J. Neurosci. 22: 9143–9149. 10.1523/JNEUROSCI.22-21-09143.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson B., and Page D. T., 2005. Condensation of the central nervous system in embryonic Drosophila is inhibited by blocking hemocyte migration or neural activity. Dev. Biol. 279: 233–243. 10.1016/j.ydbio.2004.12.020 [DOI] [PubMed] [Google Scholar]
- Olson P. F., Fessler L. I., Nelson R. E., Sterne R. E., Campbell A. G. et al. , 1990. Glutactin, a novel Drosophila basement membrane-related glycoprotein with sequence similarity to serine esterases. EMBO J. 9: 1219–1227. 10.1002/j.1460-2075.1990.tb08229.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overton P. M., Meadows L. A., Urban J., and Russell S., 2002. Evidence for differential and redundant function of the Sox genes Dichaete and SoxN during CNS development in Drosophila. Development 129: 4219–4228. [DOI] [PubMed] [Google Scholar]
- Oyallon J., Apitz H., Miguel-Aliaga I., Timofeev K., Ferreira L. et al. , 2012. Regulation of locomotion and motoneuron trajectory selection and targeting by the Drosophila homolog of Olig family transcription factors. Dev. Biol. 369: 261–276. 10.1016/j.ydbio.2012.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page D. T., 2003. A function for Egf receptor signaling in expanding the developing brain in Drosophila. Curr. Biol. 13: 474–482. 10.1016/S0960-9822(03)00094-0 [DOI] [PubMed] [Google Scholar]
- Page D. T., and Olofsson B., 2008. Multiple roles for apoptosis facilitating condensation of the Drosophila ventral nerve cord. Genesis 46: 61–68. 10.1002/dvg.20365 [DOI] [PubMed] [Google Scholar]
- Palgi M., Lindstrom R., Peranen J., Piepponen T. P., Saarma M. et al. , 2009. Evidence that DmMANF is an invertebrate neurotrophic factor supporting dopaminergic neurons. Proc. Natl. Acad. Sci. USA 106: 2429–2434. 10.1073/pnas.0810996106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz M. J., and Jackle H., 1993. Blastoderm Segmentation, pp. 467–516 in The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- Park D., Shafer O. T., Shepherd S. P., Suh H., Trigg J. S. et al. , 2008. The Drosophila basic helix-loop-helix protein DIMMED directly activates PHM, a gene encoding a neuropeptide-amidating enzyme. Mol. Cell. Biol. 28: 410–421. 10.1128/MCB.01104-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson B. J., and Doe C. Q., 2003. Regulation of neuroblast competence in Drosophila. Nature 425: 624–628. 10.1038/nature01910 [DOI] [PubMed] [Google Scholar]
- Pearson J. C., and Crews S. T., 2014. Enhancer diversity and the control of a simple pattern of Drosophila CNS midline cell expression. Dev. Biol. 392: 466–482. 10.1016/j.ydbio.2014.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C., Carney G. E., Taylor B. J., and White K., 2002. Reaper is required for neuroblast apoptosis during Drosophila development. Development 129: 1467–1476. [DOI] [PubMed] [Google Scholar]
- Pearson J. C., Watson J. D., and Crews S. T., 2012. Drosophila melanogaster Zelda and Single-minded collaborate to regulate an evolutionarily dynamic CNS midline cell enhancer. Dev. Biol. 366: 420–432. 10.1016/j.ydbio.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phochanukul N., and Russell S., 2010. No backbone but lots of Sox: invertebrate Sox genes. Int. J. Biochem. Cell Biol. 42: 453–464. 10.1016/j.biocel.2009.06.013 [DOI] [PubMed] [Google Scholar]
- Pinto-Teixeira F., Konstantinides N., and Desplan C., 2016. Programmed cell death acts at different stages of Drosophila neurodevelopment to shape the central nervous system. FEBS Lett. 590: 2435–2453. 10.1002/1873-3468.12298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prehoda K. E., 2012. Microtubules in distress release arrest. Dev. Cell 23: 233–234. 10.1016/j.devcel.2012.07.005 [DOI] [PubMed] [Google Scholar]
- Prokop A., Bray S., Harrison E., and Technau G. M., 1998. Homeotic regulation of segment-specific differences in neuroblast numbers and proliferation in the Drosophila central nervous system. Mech. Dev. 74: 99–110. 10.1016/S0925-4773(98)00068-9 [DOI] [PubMed] [Google Scholar]
- Pym E. C., Southall T. D., Mee C. J., Brand A. H., and Baines R. A., 2006. The homeobox transcription factor Even-skipped regulates acquisition of electrical properties in Drosophila neurons. Neural Dev. 1: 3 10.1186/1749-8104-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragone G., Bernardoni R., and Giangrande A., 2001. A novel mode of asymmetric division identifies the fly neuroglioblast 6–4T. Dev. Biol. 235: 74–85. 10.1006/dbio.2001.0296 [DOI] [PubMed] [Google Scholar]
- Ramdas Nair A., Singh P., Salvador Garcia D., Rodriguez-Crespo D., Egger B. et al. , 2016. The microcephaly-associated protein Wdr62/CG7337 is required to maintain centrosome asymmetry in Drosophila neuroblasts. Cell Rep. 14: 1100–1113. 10.1016/j.celrep.2015.12.097 [DOI] [PubMed] [Google Scholar]
- Rebollo E., Sampaio P., Januschke J., Llamazares S., Varmark H. et al. , 2007. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev. Cell 12: 467–474. 10.1016/j.devcel.2007.01.021 [DOI] [PubMed] [Google Scholar]
- Reeves G. T., and Stathopoulos A., 2009. Graded Dorsal and differential gene regulation in the Drosophila embryo. Cold Spring Harb. Perspect. Biol. 1: a000836 10.1101/cshperspect.a000836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert C., Kunz T., Harris K. L., Whitington P. M., and Technau G. M., 2011. Morphological characterization of the entire interneuron population reveals principles of neuromere organization in the ventral nerve cord of Drosophila. J. Neurosci. 31: 15870–15883. 10.1523/JNEUROSCI.4009-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers W. A., Goyal Y., Yamaya K., Shvartsman S. Y., and Levine M. S., 2017. Uncoupling neurogenic gene networks in the Drosophila embryo. Genes Dev. 31: 634–638. 10.1101/gad.297150.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulja-Ortmann A., Luer K., Seibert J., Rickert C., and Technau G. M., 2007. Programmed cell death in the embryonic central nervous system of Drosophila melanogaster. Development 134: 105–116. 10.1242/dev.02707 [DOI] [PubMed] [Google Scholar]
- Rosenberg M. I., Lynch J. A., and Desplan C., 2009. Heads and tails: evolution of antero-posterior patterning in insects. Biochim. Biophys. Acta 1789: 333–342. 10.1016/j.bbagrm.2008.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubinet C., Tsankova A., Pham T. T., Monnard A., Caussinus E. et al. , 2017. Spatio-temporally separated cortical flows and spindle geometry establish physical asymmetry in fly neural stem cells. Nat. Commun. 8: 1383 10.1038/s41467-017-01391-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan N. M., and Peifer M., 2007. A role for a novel centrosome cycle in asymmetric cell division. J. Cell Biol. 177: 13–20. 10.1083/jcb.200612140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S. R., Sanchez-Soriano N., Wright C. R., and Ashburner M., 1996. The Dichaete gene of Drosophila melanogaster encodes a SOX-domain protein required for embryonic segmentation. Development 122: 3669–3676. [DOI] [PubMed] [Google Scholar]
- Schaefer M., Shevchenko A., Shevchenko A., and Knoblich J. A., 2000. A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr. Biol. 10: 353–362. 10.1016/S0960-9822(00)00401-2 [DOI] [PubMed] [Google Scholar]
- Schmidt H., Rickert C., Bossing T., Vef O., Urban J. et al. , 1997. The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev. Biol. 189: 186–204. 10.1006/dbio.1997.8660 [DOI] [PubMed] [Google Scholar]
- Schmid A., Chiba A., and Doe C. Q., 1999. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development 126: 4653–4689. [DOI] [PubMed] [Google Scholar]
- Schober M., Schaefer M., and Knoblich J. A., 1999. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature 402: 548–551. 10.1038/990135 [DOI] [PubMed] [Google Scholar]
- Schweisguth F., 2015. Asymmetric cell division in the Drosophila bristle lineage: from the polarization of sensory organ precursor cells to Notch-mediated binary fate decision. Wiley Interdiscip. Rev. Dev. Biol. 4: 299–309. 10.1002/wdev.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears H. C., Kennedy C. J., and Garrity P. A., 2003. Macrophage-mediated corpse engulfment is required for normal Drosophila CNS morphogenesis. Development 130: 3557–3565. 10.1242/dev.00586 [DOI] [PubMed] [Google Scholar]
- Seibert J., and Urbach R., 2010. Role of en and novel interactions between msh, ind, and vnd in dorsoventral patterning of the Drosophila brain and ventral nerve cord. Dev. Biol. 346: 332–345. 10.1016/j.ydbio.2010.07.024 [DOI] [PubMed] [Google Scholar]
- Seibert J., Volland D., and Urbach R., 2009. Ems and Nkx6 are central regulators in dorsoventral patterning of the Drosophila brain. Development 136: 3937–3947. 10.1242/dev.041921 [DOI] [PubMed] [Google Scholar]
- Sen S. Q., Chanchani S., Southall T. D., and Doe C. Q., 2019. Neuroblast-specific open chromatin allows the temporal transcription factor, Hunchback, to bind neuroblast-specific loci. eLife 8: pii: e44036. 10.7554/eLife.44036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seroka A. Q., and Doe C. Q., 2019. The Hunchback temporal transcription factor determines motor neuron axon and dendrite targeting in Drosophila. Development 146:dev175570. 10.1242/dev.175570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Wang L., Hirose S., Zhou Z., and Liu Q., 2018. The transcriptional factor Apt regulates neuroblast differentiation through activating CycE expression. Biochem. Biophys. Res. Commun. 499: 889–894. 10.1016/j.bbrc.2018.04.012 [DOI] [PubMed] [Google Scholar]
- Siegrist S. E., and Doe C. Q., 2005. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell 123: 1323–1335. 10.1016/j.cell.2005.09.043 [DOI] [PubMed] [Google Scholar]
- Singh P., Ramdas Nair A., and Cabernard C., 2014. The centriolar protein Bld10/Cep135 is required to establish centrosome asymmetry in Drosophila neuroblasts. Curr. Biol. 24: 1548–1555. 10.1016/j.cub.2014.05.050 [DOI] [PubMed] [Google Scholar]
- Singhania A., and Grueber W. B., 2014. Development of the embryonic and larval peripheral nervous system of Drosophila. Wiley Interdiscip. Rev. Dev. Biol. 3: 193–210. 10.1002/wdev.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeath J. B., and Carroll S. B., 1992. Regulation of proneural gene expression and cell fate during neuroblast segregation in the Drosophila embryo. Development 114: 939–946. [DOI] [PubMed] [Google Scholar]
- Skeath J. B., and Doe C. Q., 1996. The achaete-scute complex proneural genes contribute to neural precursor specification in the Drosophila CNS. Curr. Biol. 6: 1146–1152. 10.1016/S0960-9822(02)70681-7 [DOI] [PubMed] [Google Scholar]
- Skeath J. B., and Doe C. Q., 1998. Sanpodo and Notch act in opposition to Numb to distinguish sibling neuron fates in the Drosophila CNS. Development 125: 1857–1865. [DOI] [PubMed] [Google Scholar]
- Skeath J. B., Panganiban G., Selegue J., and Carroll S. B., 1992. Gene regulation in two dimensions: the proneural achaete and scute genes are controlled by combinations of axis-patterning genes through a common intergenic control region. Genes Dev. 6: 2606–2619. 10.1101/gad.6.12b.2606 [DOI] [PubMed] [Google Scholar]
- Skeath J. B., Panganiban G. F., and Carroll S. B., 1994. The ventral nervous system defective gene controls proneural gene expression at two distinct steps during neuroblast formation in Drosophila. Development 120: 1517–1524. [DOI] [PubMed] [Google Scholar]
- Sonnenfeld M. J., and Jacobs J. R., 1995. Macrophages and glia participate in the removal of apoptotic neurons from the Drosophila embryonic nervous system. J. Comp. Neurol. 359: 644–652. 10.1002/cne.903590410 [DOI] [PubMed] [Google Scholar]
- Sonnenfeld M., Ward M., Nystrom G., Mosher J., Stahl S. et al. , 1997. The Drosophila tango gene encodes a bHLH-PAS protein that is orthologous to mammalian Arnt and controls CNS midline and tracheal development. Development 124: 4571–4582. [DOI] [PubMed] [Google Scholar]
- Sousa-Nunes R., Cheng L. Y., and Gould A. P., 2010. Regulating neural proliferation in the Drosophila CNS. Curr. Opin. Neurobiol. 20: 50–57. 10.1016/j.conb.2009.12.005 [DOI] [PubMed] [Google Scholar]
- Sousa-Nunes R., and Somers W. G., 2013. Mechanisms of asymmetric progenitor divisions in the Drosophila central nervous system. Adv. Exp. Med. Biol. 786: 79–102. 10.1007/978-94-007-6621-1_6 [DOI] [PubMed] [Google Scholar]
- Sousa-Nunes R., Chia W., and Somers W. G., 2009. Protein phosphatase 4 mediates localization of the Miranda complex during Drosophila neuroblast asymmetric divisions. Genes Dev. 23: 359–372. 10.1101/gad.1723609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soustelle L., and Giangrande A., 2007. Glial differentiation and the Gcm pathway. Neuron Glia Biol. 3: 5–16. 10.1017/S1740925X07000464 [DOI] [PubMed] [Google Scholar]
- Southall T. D., and Brand A. H., 2009. Neural stem cell transcriptional networks highlight genes essential for nervous system development. EMBO J. 28: 3799–3807. 10.1038/emboj.2009.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana E. P., and Doe C. Q., 1995. The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development 121: 3187–3195. [DOI] [PubMed] [Google Scholar]
- Spana E. P., and Doe C. Q., 1996. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron 17: 21–26. 10.1016/S0896-6273(00)80277-9 [DOI] [PubMed] [Google Scholar]
- Spana E. P., Kopczynski C., Goodman C. S., and Doe C. Q., 1995. Asymmetric localization of numb autonomously determines sibling neuron identity in the Drosophila CNS. Development 121: 3489–3494. [DOI] [PubMed] [Google Scholar]
- Stacey S. M., Thomas G. B., Labbe A., and Van Meyel D. J., 2007. Longitudinal glia in the fly CNS: pushing the envelope on glial diversity and neuron-glial interactions. Neuron Glia Biol. 3: 27–33. 10.1017/S1740925X07000506 [DOI] [PubMed] [Google Scholar]
- Stagg S. B., Guardiola A. R., and Crews S. T., 2011. Dual role for Drosophila lethal of scute in CNS midline precursor formation and dopaminergic neuron and motoneuron cell fate. Development 138: 2171–2183. 10.1242/dev.056507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stollewerk A., 2000. Changes in cell shape in the ventral neuroectoderm of Drosophila melanogaster depend on the activity of the achaete-scute complex genes. Dev. Genes Evol. 210: 190–199. 10.1007/s004270050303 [DOI] [PubMed] [Google Scholar]
- Stork T., Thomas S., Rodrigues F., Silies M., Naffin E. et al. , 2009. Drosophila Neurexin IV stabilizes neuron-glia interactions at the CNS midline by binding to Wrapper. Development 136: 1251–1261. 10.1242/dev.032847 [DOI] [PubMed] [Google Scholar]
- Stork T., Sheehan A., Tasdemir-Yilmaz O. E., and Freeman M. R., 2014. Neuron-glia interactions through the Heartless FGF receptor signaling pathway mediate morphogenesis of Drosophila astrocytes. Neuron 83: 388–403. 10.1016/j.neuron.2014.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann J., Ekman H., and Thor S., 2019. A branching gene regulatory network dictating different aspects of a neuronal cell identity. Development 146: dev174300. 10.1242/dev.174300 [DOI] [PubMed] [Google Scholar]
- Syed M. H., Mark B., and Doe C. Q., 2017. Steroid hormone induction of temporal gene expression in Drosophila brain neuroblasts generates neuronal and glial diversity. eLife 6: pii: e26287. 10.7554/eLife.26287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Yamada-Mabuchi M., Arya R., St Pierre S., Tang W. et al. , 2011. Coordinated expression of cell death genes regulates neuroblast apoptosis. Development 138: 2197–2206. 10.1242/dev.058826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. B., Crews S. T., and Goodman C. S., 1988. Molecular genetics of the single-minded locus: a gene involved in the development of the Drosophila nervous system. Cell 52: 133–141. 10.1016/0092-8674(88)90537-5 [DOI] [PubMed] [Google Scholar]
- Thor S., 2009. Drosophila apterous neurons: from stem cell to unique neuron, pp. 213–217 in Developmental Neurobiology, edited by Lemke G., Academic Press, London, UK. [Google Scholar]
- Thor S., and Thomas J. B., 1997. The Drosophila islet gene governs axon pathfinding and neurotransmitter identity. Neuron 18: 397–409. 10.1016/S0896-6273(00)81241-6 [DOI] [PubMed] [Google Scholar]
- Thor S., Andersson S. G., Tomlinson A., and Thomas J. B., 1999. A LIM-homeodomain combinatorial code for motor-neuron pathway selection. Nature 397: 76–80. 10.1038/16275 [DOI] [PubMed] [Google Scholar]
- Tio M., Toh J., Fang W., Blanco J., and Udolph G., 2011. Asymmetric cell division and Notch signaling specify dopaminergic neurons in Drosophila. PLoS One 6: e26879 10.1371/journal.pone.0026879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touma J. J., Weckerle F. F., and Cleary M. D., 2012. Drosophila Polycomb complexes restrict neuroblast competence to generate motoneurons. Development 139: 657–666. 10.1242/dev.071589 [DOI] [PubMed] [Google Scholar]
- Tran K. D., and Doe C. Q., 2008. Pdm and Castor close successive temporal identity windows in the NB3–1 lineage. Development 135: 3491–3499. 10.1242/dev.024349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trébuchet G., Cattenoz P. B., Zsamboki J., Mazaud D., Siekhaus D. E. et al. , 2018. The Repo homeodomain transcription factor suppresses hematopoiesis in Drosophila and preserves the glial fate. J. Neurosci. 39: 238–255. 10.1523/JNEUROSCI.1059-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troost T., Schneider M., and Klein T., 2015. A re-examination of the selection of the sensory organ precursor of the bristle sensilla of Drosophila melanogaster. PLoS Genet. 11: e1004911 10.1371/journal.pgen.1004911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman J. W., and Bate M., 1988. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev. Biol. 125: 145–157. 10.1016/0012-1606(88)90067-X [DOI] [PubMed] [Google Scholar]
- Tsuchida T., Ensini M., Morton S. B., Baldassare M., Edlund T. et al. , 1994. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell 79: 957–970. 10.1016/0092-8674(94)90027-2 [DOI] [PubMed] [Google Scholar]
- Tsuji T., Hasegawa E., and Isshiki T., 2008. Neuroblast entry into quiescence is regulated intrinsically by the combined action of spatial Hox proteins and temporal identity factors. Development 135: 3859–3869. 10.1242/dev.025189 [DOI] [PubMed] [Google Scholar]
- Ulvklo C., MacDonald R., Bivik C., Baumgardt M., Karlsson D. et al. , 2012. Control of neuronal cell fate and number by integration of distinct daughter cell proliferation modes with temporal progression. Development 139: 678–689. 10.1242/dev.074500 [DOI] [PubMed] [Google Scholar]
- Urbach R., and Technau G. M., 2003a Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development 130: 3621–3637. 10.1242/dev.00533 [DOI] [PubMed] [Google Scholar]
- Urbach R., and Technau G. M., 2003b Segment polarity and DV patterning gene expression reveals segmental organization of the Drosophila brain. Development 130: 3607–3620. 10.1242/dev.00532 [DOI] [PubMed] [Google Scholar]
- Urbach R., Volland D., Seibert J., and Technau G. M., 2006. Segment-specific requirements for dorsoventral patterning genes during early brain development in Drosophila. Development 133: 4315–4330. 10.1242/dev.02605 [DOI] [PubMed] [Google Scholar]
- Urbach R., Jussen D., and Technau G. M., 2016. Gene expression profiles uncover individual identities of gnathal neuroblasts and serial homologies in the embryonic CNS of Drosophila. Development 143: 1290–1301. 10.1242/dev.133546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent S., Vonesch J. L., and Giangrande A., 1996. glide directs glial fate commitment and cell fate switch between neurones and glia. Development 122: 131–139. [DOI] [PubMed] [Google Scholar]
- von Hilchen C. M., Hein I., Technau G. M., and Altenhein B., 2010. Netrins guide migration of distinct glial cells in the Drosophila embryo. Development 137: 1251–1262. 10.1242/dev.042853 [DOI] [PubMed] [Google Scholar]
- Walsh K. T., and Doe C. Q., 2017. Drosophila embryonic type II neuroblasts: origin, temporal patterning, and contribution to the adult central complex. Development 144: 4552–4562. 10.1242/dev.157826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Somers G. W., Bashirullah A., Heberlein U., Yu F. et al. , 2006. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 20: 3453–3463. 10.1101/gad.1487506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ouyang Y., Somers W. G., Chia W., and Lu B., 2007. Polo inhibits progenitor self-renewal and regulates Numb asymmetry by phosphorylating Pon. Nature 449: 96–100. 10.1038/nature06056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J. D., and Crews S. T., 2012. Formation and specification of a Drosophila dopaminergic precursor cell. Development 139: 3316–3325. 10.1242/dev.079525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J. D., Wheeler S. R., Stagg S. B., and Crews S. T., 2011. Drosophila hedgehog signaling and engrailed-runt mutual repression direct midline glia to alternative ensheathing and non-ensheathing fates. Development 138: 1285–1295. 10.1242/dev.056895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J. B., Von Ohlen T., Mellerick D. M., Dressler G., Doe C. Q. et al. , 1998. Dorsoventral patterning in the Drosophila central nervous system: the intermediate neuroblasts defective homeobox gene specifies intermediate column identity. Genes Dev. 12: 3591–3602. 10.1101/gad.12.22.3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton K. A. Jr., Franks R. G., Kasai Y., and Crews S. T., 1994. Control of CNS midline transcription by asymmetric E-box-like elements: similarity to xenobiotic responsive regulation. Development 120: 3563–3569. [DOI] [PubMed] [Google Scholar]
- Wheeler S. R., Kearney J. B., Guardiola A. R., and Crews S. T., 2006. Single-cell mapping of neural and glial gene expression in the developing Drosophila CNS midline cells. Dev. Biol. 294: 509–524. 10.1016/j.ydbio.2006.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler S. R., Stagg S. B., and Crews S. T., 2008. Multiple Notch signaling events control Drosophila CNS midline neurogenesis, gliogenesis and neuronal identity. Development 135: 3071–3079. 10.1242/dev.022343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler S. R., Banerjee S., Blauth K., Rogers S. L., Bhat M. A. et al. , 2009. Neurexin IV and Wrapper interactions mediate Drosophila midline glial migration and axonal ensheathment. Development 136: 1147–1157. 10.1242/dev.030254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler S. R., Pearson J. C., and Crews S. T., 2012. Time-lapse imaging reveals stereotypical patterns of Drosophila midline glial migration. Dev. Biol. 361: 232–244. 10.1016/j.ydbio.2011.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K., Grether M. E., Abrams J. M., Young L., Farrell K. et al. , 1994. Genetic control of programmed cell death in Drosophila. Science 264: 677–683. 10.1126/science.8171319 [DOI] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Kuchinke U., and Knust E., 1999. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature 402: 544–547. 10.1038/990128 [DOI] [PubMed] [Google Scholar]
- Wolfram V., Southall T. D., Brand A. H., and Baines R. A., 2012. The LIM-homeodomain protein islet dictates motor neuron electrical properties by regulating K(+) channel expression. Neuron 75: 663–674. 10.1016/j.neuron.2012.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfram V., Southall T. D., Gunay C., Prinz A. A., Brand A. H. et al. , 2014. The transcription factors islet and Lim3 combinatorially regulate ion channel gene expression. J. Neurosci. 34: 2538–2543. 10.1523/JNEUROSCI.4511-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaghmaeian Salmani B., Monedero Cobeta I., Rakar J., Bauer S., Curt J. R. et al. , 2018. Evolutionarily conserved anterior expansion of the central nervous system promoted by a common PcG-Hox program. Development 145: dev160747. 10.1242/dev.160747 [DOI] [PubMed] [Google Scholar]
- Yu F., Morin X., Cai Y., Yang X., and Chia W., 2000. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell 100: 399–409. 10.1016/S0092-8674(00)80676-5 [DOI] [PubMed] [Google Scholar]
- Zarin A. A., and Labrador J. P., 2019. Motor axon guidance in Drosophila. Semin. Cell Dev. Biol. 85: 36–47. 10.1016/j.semcdb.2017.11.013 [DOI] [PubMed] [Google Scholar]
- Zarin A. A., Asadzadeh J., Hokamp K., McCartney D., Yang L. et al. , 2014. A transcription factor network coordinates attraction, repulsion, and adhesion combinatorially to control motor axon pathway selection. Neuron 81: 1297–1311. 10.1016/j.neuron.2014.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Huang Z. X., Bao H., Cong F., Wang H. et al. , 2016. Phosphotyrosyl phosphatase activator facilitates localization of Miranda through dephosphorylation in dividing neuroblasts. Development 143: 35–44. 10.1242/dev.127233 [DOI] [PubMed] [Google Scholar]
- Zhao G., and Skeath J. B., 2002. The Sox-domain containing gene Dichaete/fish-hook acts in concert with vnd and ind to regulate cell fate in the Drosophila neuroectoderm. Development 129: 1165–1174. [DOI] [PubMed] [Google Scholar]
- Zhao G., Boekhoff-Falk G., Wilson B. A., and Skeath J. B., 2007. Linking pattern formation to cell-type specification: Dichaete and Ind directly repress achaete gene expression in the Drosophila CNS. Proc. Natl. Acad. Sci. USA 104: 3847–3852 [corrigenda: Proc. Natl. Acad. Sci. USA 105: 3658 (2008)]. 10.1073/pnas.0611700104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Pennack J. A., McQuilton P., Forero M. G., Mizuguchi K. et al. , 2008. Drosophila neurotrophins reveal a common mechanism for nervous system formation. PLoS Biol. 6: e284 10.1371/journal.pbio.0060284 [DOI] [PMC free article] [PubMed] [Google Scholar]