Transvection is a form of gene regulation the depends on interactions between regulatory elements on separate chromosomes. In one form of transvection that is prevalent in Drosophila, an enhancer can act...
Keywords: chromatin, long-range enhancer, position effects, somatic homolog pairing, transvection
Abstract
Transvection is an epigenetic phenomenon wherein regulatory elements communicate between different chromosomes in trans, and is thereby dependent upon the three-dimensional organization of the genome. Transvection is best understood in Drosophila, where homologous chromosomes are closely paired in most somatic nuclei, although similar phenomena have been observed in other species. Previous data have supported that the Drosophila genome is generally permissive to enhancer action in trans, a form of transvection where an enhancer on one homolog activates gene expression from a promoter on a paired homolog. However, the capacity of different genomic positions to influence the quantitative output of transvection has yet to be addressed. To investigate this question, we employed a transgenic system that assesses and compares enhancer action in cis and in trans at defined chromosomal locations. Using the strong synthetic eye-specific enhancer GMR, we show that loci supporting strong cis-expression tend to support robust enhancer action in trans, whereas locations with weaker cis-expression show reduced transvection in a fluorescent reporter assay. Our subsequent analysis is consistent with a model wherein the chromatin state of the transgenic insertion site is a primary determinant of the degree to which enhancer action in trans will be supported, whereas other factors such as locus-specific variation in somatic homolog pairing are of less importance in influencing position effects on transvection.
IN 1954, geneticist Ed Lewis coined the term transvection to describe an unexpected case of intragenic complementation between two alleles of Ultrabithorax (Ubx) in Drosophila melanogaster (Lewis 1954). Specifically, he found that complementation via transvection is dependent upon the physical juxtaposition of homologous chromosomes—a phenomenon known as somatic homolog pairing that is found primarily in Dipteran species (reviewed by McKee 2004; Joyce et al. 2016). Chromosomal rearrangements that disrupt the ability of homologous loci to pair result in reduced complementation via transvection, demonstrating that trans-communication can take place between different chromosomes.
Since Lewis’ discovery, examples of pairing-dependent regulation of gene expression have been discovered at many other loci in the Drosophila genome, and similar phenomena have been uncovered in other organisms (reviewed by Kennison and Southworth 2002; Kassis 2012; Joyce et al. 2016). Extensive molecular analyses have shown that transvection can occur by several mechanisms, leading to either activation or repression of transcription (reviewed by Duncan 2002). In one form of transvection, which likely accounts for the original observations of Lewis (1954), an enhancer on one homolog can act in trans on a promoter on the paired homolog. This form of transvection is frequently observed between paired mutant alleles wherein one allele lacks enhancer sequences (an “enhancerless” allele) and the other lacks a functional promoter (“promoterless”); the functional enhancer acting on the intact promoter in trans restores the capacity for transcription, leading to intragenic complementation (Geyer et al. 1990).
Through primarily transgenic approaches, several studies have supported that the Drosophila genome is generally permissive to enhancer action in trans. These studies involve creating pairs of transgenic constructs that mimic the enhancerless and promoterless alleles of known mutants that participate in transvection. For example, Chen et al. (2002) modified a yellow transgene to carry FRT and loxP sites such that, following integration of the transgene into the genome, treatment with the recombinases Cre or FLP would create enhancerless and promoterless derivatives of yellow. Acting alone, neither derivative was capable of fully rescuing pigmentation in a background where the natural yellow locus was deleted; however, when enhancerless and promoterless transgenic derivatives were placed in trans to one another so they could pair, increased levels of yellow pigmentation were observed, consistent with the occurrence of enhancer action in trans. Notably, all eight genomic locations tested supported transvection by this pigmentation assay (Chen et al. 2002), and subsequent analysis employing similar yellow-based transgenes confirmed that enhancer action in trans was permitted at a further dozen genomic locations (Kravchenko et al. 2005). Similarly, fluorescent reporters placed at several common attP-based landing sites have been shown to permit transvection when they are placed in trans and allowed to pair (Bateman et al. 2012; Mellert and Truman 2012; Blick et al. 2016), further supporting that the genome is generally able to support enhancer action in trans.
Despite the growing consensus on the permissivity of the genome for transvection, little is known about the potential impact of position effects on enhancer action in trans. Conventional transgenes that rely on enhancer-promoter interactions in cis can vary greatly in their expression levels depending on the local chromatin environment surrounding an insertion (e.g., Markstein et al. 2008; Chen and Zhang 2016; Corrales et al. 2017; Wu et al. 2017). In theory, expression from transgenes that rely on enhancer action in trans may be similarly impacted by differences in local chromatin structure. Furthermore, since transvection requires that two chromosomes be closely paired, transvection could also be affected by positional differences in the degree of somatic homolog pairing (Fung et al. 1998; Williams et al. 2007; Bateman and Wu 2008; Joyce et al. 2012).
Here, we address how position effects can impact enhancer action in trans in the Drosophila genome. Using an eye tissue-specific enhancer and a green fluorescent protein (GFP) reporter, we employ a Cre/loxP- and FLP/FRT-based strategy to assess the capacity of different genomic locations to support transvection via enhancer action in trans. By quantitatively assessing GFP fluorescence, we find variation in the degree to which transvection is supported at different genomic locations.
Our analysis shows that sites supporting strong enhancer activity in cis are likely to show higher expression in trans and map to regions of open chromatin in multiple cell types, whereas sites supporting weak cis-activity tend to show lower levels of transvection. Furthermore, by employing DNA-FISH on larval eye discs using fluorescent probes targeting several transgene insertion sites, we find no relationship between expression levels resulting from transvection and variation in somatic homolog pairing. Thus, our data are consistent with a model wherein the local chromatin state is a primary determinant of transcriptional output supported by transvection.
Materials and Methods
Stocks and fly husbandry
The following stocks were obtained from the Bloomington Drosophila Stock Center: y w; Sco/ CyO, Cre-w, carrying a Cre recombinase transgene on the second chromosome balancer CyO, the stock y w; MKRS, hs-FLP/ TM6B, Cre-w, carrying a heat-shock-inducible FLP construct on the MKRS chromosome and a Cre construct on the balancer TM6B, the stock y w; Bc/CyO, HoP2.1, carrying a transgene encoding the P-element transposase on CyO, the stocks GMR-GAL4 and UAS-lamin-GFP, which express GAL4 under the control of the GMR enhancer and a GFP-tagged nuclear lamin under the control of the UAS enhancer, respectively, and a stock carrying za—a loss of function allele of z. All flies were cultured at 25° on standard Drosophila cornmeal, yeast, sugar, and agar medium with p-hydroxybenzoic acid methyl ester as a mold inhibitor (Bateman et al. 2012).
Cloning, transformation, and P-element mobilization
The plasmid pP[TV2-GMR] was constructed using the vector pWFL (Siegal and Hartl 1996), provided by Mark Siegel, GMR (Moses and Rubin 1991) sequences from pGMR, provided by Jeff Settleman, and hsp70-GFP sequences from the plasmid R1NheXho-GFP, provided by Sean Carroll. A detailed cloning strategy can be provided upon request. Transgenic flies were generated by co-injecting 0.5 mg/ml pP[TV2-GMR] with 0.1 mg/ml “wings clipped” helper plasmid in water into w1118 embryos as previously described (Rubin and Spradling 1982; Morris et al. 1998). Each independent insertion was mapped to a chromosome by segregation analysis. To determine precise genomic positions, genomic DNA flanking each insertion was amplified via inverse PCR (Ochman et al. 1988) as described by E. J. Rehm of the Berkeley Drosophila Genome Project (http:/www.fruitfly.org), and the resulting fragments were sequenced and mapped to the reference genomic sequence. One line was found to be inserted into an X-element on the third chromosome, and its precise location was not determined given the multiple X-element annotations on chromosome 3. Lines with evidence of multiple insertions and lines with homozygous lethal insertions were discarded from further analysis.
An insertion of P[TV2-GMR] on the X chromosome was remobilized to generate additional insertions on the autosomes. Briefly, P[TV2-GMR]X virgin females were crossed with w; Bc/CyO, HoP2.1 males to produce P[TV2-GMR]X/Y; +/CyO, HoP2.1 males. These males were crossed to w1118 virgin females, and male F2 progeny were screened for loss of the CyO, HoP2.1 balancer and gain of mini-white eye color, indicating a new insertion of P[TV2-GMR]. Each independent insertion was mapped as described above.
Enhancerless and promoterless derivatives of each mapped P[TV2-GMR] insertion were generated by crossing to flies carrying FLP recombinase under the control of a heat shock promoter (hs-FLP) (Golic and Lindquist 1989), or to flies carrying a weak but constitutive germline source of Cre recombinase (Cre-w) (Siegal and Hartl 1996). Successful removal of the FLP- or Cre-cassette from P[TV2-GMR] was monitored by the loss of mini-white eye color. Each enhancerless and promoterless line was established from a single Cre or FLP event by mating w− progeny singly to stocks carrying balancer chromosomes. An example of a cross scheme for a third chromosome insertion is provided in Supplemental Material, Figure S1.
Quantitative imaging of GFP fluorescence
Eye-antennal discs were dissected from wandering third-instar larvae in phosphate-buffered saline (PBS) and fixed in 4% formaldehyde (Electron Microscopy Sciences) in PBS for 20 min. Discs were rinsed three times in PBST (PBS with 0.1% Triton X-100) and mounted in-Fluoromount G (Southern Biotech). Discs were visualized on a Leica SP8 confocal microscope using a HyD detector in photon counting mode, with laser power adjusted to 45 μW as measured at the stage with an X-Cite XR2100 power meter (Lumen Dynamics). Four to six discs of each genotype were imaged in 16 µm z-stacks using 0.5 µm slices.
To quantify GFP fluorescence, each stack was summed to a single plane using the “sum slices” function of ImageJ imaging software. Then, regions of interest were defined for the GMR expression domain from the morphogenetic furrow to the posterior of the eye discs, and for the undifferentiated anterior compartment of the eye disc (see Figure 2B). Finally, the background-subtracted mean fluorescence was determined by subtracting the mean pixel intensity of the anterior compartment from that of the GMR expression domain.
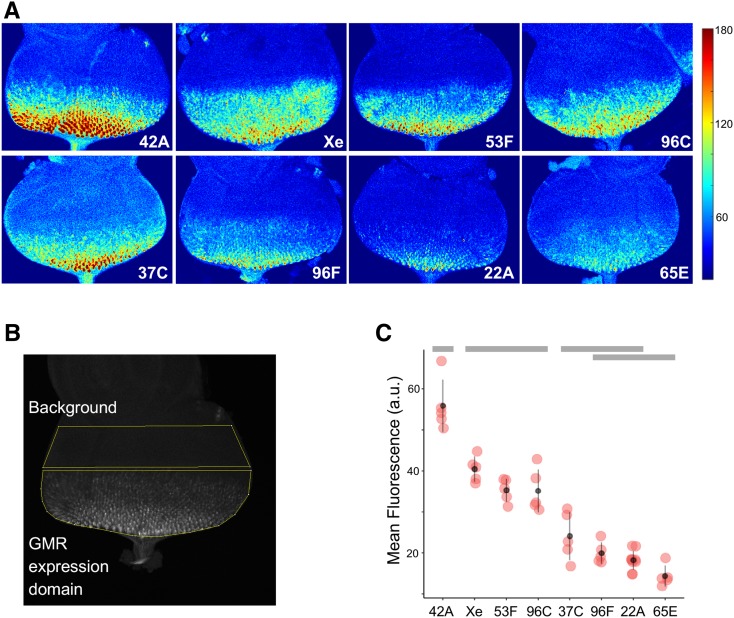
Figure 2.
GMR activation of GFP in cis varies at different genomic locations. (A) Representative images of third-instar eye disc for each location showing GFP fluorescence produced by GMR activity in cis, pseudocolored to indicate low (blue) or high (red) fluorescence. Color scale to right (arbitrary units). (B) Schematic for quantitative assessment of GFP fluorescence. Each 3D confocal image stack is flattened by summing pixel intensities in z-stacks, and the mean pixel intensity for the GMR expression domain (posterior to the morphogenetic furrow) is adjusted for background fluorescence by subtracting the mean pixel intensity of undifferentiated cells anterior to the furrow (background). See Materials and Methods for further details. (C) Quantification of GFP fluorescence resulting from GMR activity in cis at different genomic locations (a.u., arbitrary units.) ANOVA analysis shows significant differences across genotypes (P < 0.0001), gray bars indicate statistical groupings based on Tukey’s multiple comparison tests.
Reverse transcription and quantitative PCR
Reverse transcription and quantitative PCR were performed on a StepOne Real-Time PCR system (Applied Biosystems, Foster City, CA) as previously described (Bateman et al. 2012) using primers GFPRT_1 (5′-ATTCTCGTGGAACTGGATGG) and GFPRT_2 (5′-AGCTTTCCAGTGGTGCAGAT) targeting the GFP coding region, and primers RP49-58F (5′-TACAGGCCCAAGATCGTGAAG) and RP49-175R (5′-GACGCACTCTGTTGTCGATACC) targeting the housekeeping rp49 cDNA (Moon et al. 2008) as an internal reference. Relative levels of transcript were calculated via the ΔΔCt method using StepOne software.
Oligopaint design and DNA-FISH
Oligopaint (oligo) probes (Beliveau et al. 2012) were designed for genomic regions surrounding insertions of P[TV2-GMR] at 22A, 42A, and 96F, and synthesized as a combined library by CustomArray, (Bothell, WA). Each probe includes a central 32 nt sequence that is complementary to Drosophila genomic DNA flanked by two pairs of 21-mer primer binding sites, one of which allows universal amplification of the entire library, and the other amplification of a probe sublibrary that target a contiguous segment of the genome. Probed regions were between 70 and 90 kb in length, with oligos covering the regions of interest at a density of between 11.4 and 16.1 oligos/kb. Each probe consisted of between 2030 and 2411 unique oligos. Total library amplification by emulsion PCR was carried out as described (Beliveau et al. 2012), and sublibraries of probes targeting specific genomic regions were synthesized by PCR using a forward primer with a fluorescent label at its 5′ end and a reverse primer with a phosphate at its 5′ end. Each probe library was made single-stranded via lambda exonuclease treatment as described (Beliveau et al. 2015). A fourth labeled Oligopaint probe set covering 500 kb surrounding the P[TV2-GMR] insert at 96C was ordered directly from the Joyce Lab Oligopaint Production Service (University of Pennsylvania).
DNA-FISH was carried out as described (Viets et al. 2018) on tissue isolated from flies of genotype GMR-GAL4/UAS-laminB-GFP, which express a GFP-tagged nuclear lamin protein in cells where GMR is active. Eye discs from wandering third-instar larvae were dissected in PBS and incubated in heptane fixative solution (75% heptane, 1% formaldehyde, and 0.125% Tergitol NP-40 in PBS) for 10 min. After three brief rinses and three 5-min washes in PBX (PBS with 0.3% Triton X-100), discs were removed from mouth hooks and blocked for 1 hr in PBX containing 1.5% bovine serum albumin (BSA); anti-GFP antibody (Life Technologies) was then added at a 1:2000 dilution and the solution was incubated overnight at 4°. Discs were then washed three times in PBX for 20 min each, followed by overnight incubation at 4° with goat anti-rabbit secondary antibody conjugated to AlexaFluor-488 (Invitrogen) at a 1:250 dilution in PBX. Following two 20-min washes in PBX and one 20-min wash in PBS, discs were rinsed four times in 2× SSCT (2× SSC with 0.1% Tween-20), then stepped through three 10-min washes in 2× SSCT containing 20, 40, and 50% formamide. Discs were then incubated in 2× SSCT with 50% formamide at 37° for 4 hr, 92° for 3 min, and 60° for 20 min. Next, ∼30 pmol of Oligopaint probe in hybridization mix (50% formamide, 10% dextran sulfate, 250 µg/ml RNAse A in 2× SSCT) was added, and the solution was heated to 91° for 3 min, followed by overnight hybridization at 37° with agitation. Discs were then washed in 2× SSCT with 50% formamide (2× 30 min at 37°), shifted to room temperature, and washed with SSCT with 20% formamide (10 min), 2× SSCT (10 min) and 2× SSC (1 × 10 min). For the Oligopaint targeting 96C, a labeled secondary probe was used to visualize the unlabeled primary probe (Beliveau et al. 2014); in this case, prior to washing in 2× SSCT with 20% formaldehyde, discs were incubated overnight with 20 pmol of secondary probe in 2× SSCT with 50% formamide at 37° with agitation, then washed twice for 30 min in 2× SSCT. Discs were mounted in Fluoromount G with DAPI (Electron Microscopy Sciences, Hatfield, PA). Slides were stored at 4°, and images were acquired using a Leica SP8 confocal microscope. FISH signals in GFP-positive nuclei were scored manually as paired if a single signal was visible in a nucleus, or unpaired if two or more signals were visible.
Data availability
All stocks and reagents are available upon request. All data necessary for confirming the conclusions of this work are represented in the text, figures, tables, and supplemental information. Publicly available genomic data on chromatin states in cultured cells (Filion et al. 2010; Kharchenko et al. 2011) were accessed via the genome browser at flybase.org. Supplemental material available at figshare: https://doi.org/10.25386/genetics.9973646.
Results
To assess potential position effects on enhancer action in cis and in trans, we employed a recombinase-based strategy to modify transgenic constructs inserted into various positions in the genome. Specifically, we modified a P-element transformation vector carrying FRT and loxP sites (Siegal and Hartl 1996) so that, when inserted into the genome, GFP would be expressed under the control of the eye-specific synthetic enhancer GMR and the hsp70 promoter. For each insertion, subsequent exposure to Cre or FLP creates an enhancerless or promoterless derivative, respectively, that can be used to assess enhancer action in trans (Figure 1). As we established transgenic lines, mapped their genomic positions, and created enhancerless and promoterless derivatives, we selected insertions that met two conditions: first, that the insert was homozygous viable so that enhancerless and promoterless derivatives could be analyzed in trans in third instar larvae, and second, that the enhancerless and promoterless transgenes showed no evidence of GFP expression in the eye disc as hemizygotes due to trapping of enhancers near the point of insertion, such that GFP expression in enhancerless/promoterless flies could only result from transvection. In total, we selected insertions at eight genomic locations for further analysis (Table 1).
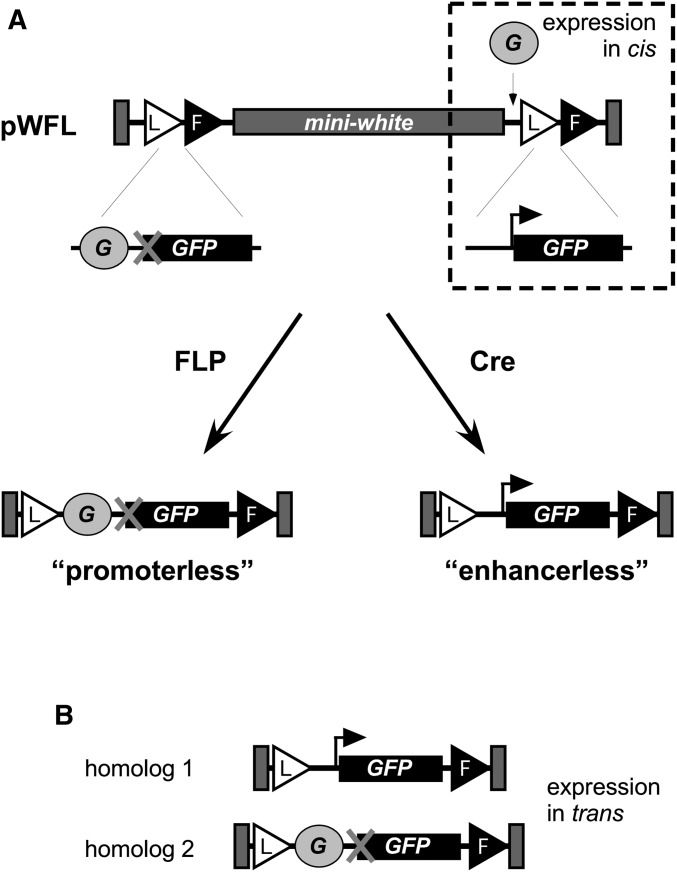
Figure 1.
Schematic for transgenic analysis. (A) The vector pWFL (Siegal and Hartl 1996) was modified to create pP[TV2-GMR], carrying transgenic fragments as indicated. G, GMR enhancer; L, loxP site; F, FRT site; bent arrow, hsp70 core promoter; X indicates a lack of a promoter. With no modification to the insertion, GMR activity in cis can be assessed. Exposure to FLP or Cre creates enhancerless or promoterless derivatives, respectively, which can be combined to assess GMR activity in trans (B).
Table 1. Genomic features of insertion sites used in this study.
| Polytene band | Position (Release 6) | Nearest gene | Position relative to nearest gene | BG3 chromatin state | S2 chromatin state | Kc chromatin state | TAD Position | Nearest Insulator |
|---|---|---|---|---|---|---|---|---|
| 42A | 2R:6,214,221 | l(2)k14710 | 103 bp upstream | active, red | active, red | active, yellow | TAD | 8 (I) |
| X-element | ND | ND | ND | ND | ND | ND | ND | ND |
| 53F | 2R:17,100,154 | intergenica | intergenic | active, choral | active, brown | active, red | boundary | 1621 (I) |
| 96C | 3R:25,208,367 | Cad96Ca | 86bp upstream | active, choral | active, choral | (gap) | TAD | 6908 (I) |
| 37C | 2L:19,158,447 | brat | intronic | active, brown | active, red | active, red | TAD | 7651 (II) |
| 96F | 3R:26,037,053 | HLHm7 | 5′ UTR | inactive, black | active, choral | inactive, blue | TAD | 1174 (I) |
| 22A | 2L:1,668,371 | Chinmo | 1st intron of A tx | inactive, black | active, brown | active, red | boundary | 3709 (II) |
| 65E | 3L:7,134,401 | melted | 117 upstream | inactive, gray | inactive, gray | inactive, blue | TAD | 252 (I) |
Genomic positions were determined by sequencing of inverse PCR products. Chromatin states for Kc cells are as defined by Filion et al. (2010) according to a five-state model, and those for BG3 and S2 cells are as defined by Kharchenko et al. (2011) according to a nine-state model; TAD boundaries are according to subkilobase resolution Hi-C data of Eagen et al. (2017); insulators are according to Nègre et al. (2010), with the distance from the insertion site to the nearest insulator and the type of insulator (I or II) given in base pairs. Class I insulators are principally bound by BEAF-32/CP190/CTCF, and Class II insulators are bound by Su(Hw).
Insertion is intergenic between GstS1 and CG30456.
For each insertion site that satisfied our criteria, we first examined GFP fluorescence resulting from the GMR enhancer acting in cis on the hsp70 promoter in a single-copy hemizygous insertion. Developing eye discs from wandering third instar larvae of each genotype were dissected, fixed, and analyzed using quantitative confocal microscopy methods (see Materials and Methods). For each sample, the intensity of fluorescence in the GMR expression domain was normalized to the background fluorescence of a domain where GMR was not active in order to account for potential differences in basal transcription from the hsp70 promoter at different genomic locations (Figure 2, A and B). Among the eight locations, we observe roughly a fourfold difference in normalized fluorescence intensity from the highest-expressing to the lowest-expressing transgenic lines, reflecting position effects on the activity of the GMR enhancer in cis (Figure 2).
To better understand the nature of the observed position effects for cis-expression, we assessed existing data regarding histone modifications for each insertion site as predicted from genome-wide analyses in cultured cells (Table 1) (Filion et al. 2010; Kharchenko et al. 2011). For the seven transgenic insertions that were uniquely mapped to the genome, we observed a strong relationship between the levels of expression observed in vivo and the chromatin states, classified as either “active” or “inactive,” around the insertion site. In particular, the transgenic insertion sites showing the highest levels of expression corresponded to regions with active chromatin marks (red, brown, or choral chromatin colors in the nine-state chromatin model) in all three cell lines tested, whereas the insertion sites showing the lowest expression were mapped to regions with inactive marks (gray, blue, or black) in some or all cell lines. Notably, each of the three lowest-expressing lines was characterized by a repressive chromatin state in BG3 cells, a neuronally derived cell line that is likely more similar to the larval photoreceptors where GMR is active than are embryonically derived S2 and Kc cells (Ui et al. 1994; Kharchenko et al. 2011). In agreement with this, prior studies of gene expression for the genes closest to each of the three lowest-expressing inserts, melted, HLHm7, and chinmo, showed no evidence of expression in third instar larval photoreceptors (de Celis et al. 1996; Mikeladze-Dvali et al. 2005; Flaherty et al. 2010). Thus, position effects on expression levels in cis are consistent with active vs. inactive chromatin profiles in cultured cells, particularly those of BG3 cells.
Positions effects influence enhancer action in trans
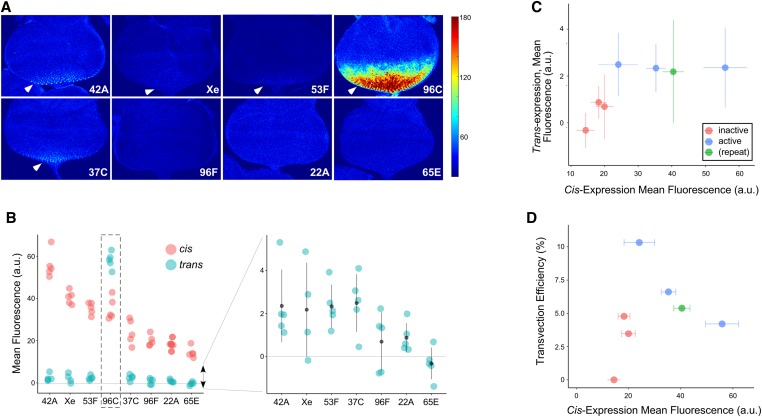
We next assessed each of the eight locations for the capacity to support transvection. Specifically, we crossed lines carrying corresponding enhancerless and promoterless transgenic derivatives at each location, and assessed transvection in the resulting progeny. In qualitatively assessing GFP fluorescence, seven of the eight lines showed strongly reduced levels of fluorescence generated via trans-expression of GFP relative to their respective cis-expressing lines, consistent with prior observations of transvection by GMR (Figure 3, A and B) (Bateman et al. 2012; Blick et al. 2016). Conversely, one location, an insert in polytene band 96C, consistently showed exceptionally bright GFP fluorescence resulting from transvection, with intensity similar to the brightest fluorescence observed in our analysis of cis-expression. Quantitative analysis of confocal images supported these observations: for position 96C, the mean intensity of GFP fluorescence resulting from transvection was 1.6-fold higher than that resulting from cis-expression at the same location, whereas the analogous comparison for all other locations showed fluorescence intensity resulting from transvection no higher than one-tenth that resulting from cis-expression (Figure 3B).
Figure 3.
GMR activation of GFP in trans varies at different genomic locations. (A) Representative images of third instar eye disc for each location showing GFP fluorescence produced by GMR activity in trans, pseudocolored to indicate low (blue) or high (red) fluorescence. Color scale to right (arbitrary units), arrowheads indicate discernable expression in posterior-most cells. (B) Quantification of GFP fluorescence resulting from GMR activity in cis (pink) or trans (blue) at different genomic locations (a.u., arbitrary units.) Dotted box highlights exceptional trans-activity at position 96C. Graph to the right shows data for trans-expression at a more detailed scale (point and whiskers = mean ± SD; data for 96C excluded; horizontal line, background subtracted fluorescence = 0.) (C) mean ± SD for GFP fluorescence resulting from GMR action in trans (y-axis) vs. cis (x-axis). Points are colored according to their chromatin type in cultured BG3 cells (Kharchenko et al. 2011) (pink, “inactive” chromatin, blue, “active” chromatin, green, insertion in repetitive element that was not precisely mapped). (D) Transvection efficiency (fluorescence from trans-activity relative to cis-activity; y-axis) vs. mean fluorescence resulting from cis-expression (x-axis). Coloring as in (C). Note that position 96C is excluded from plots in (C and D).
We were surprised by the high degree of transvection supported by the insertion at position 96C. In mapping this transgene, we discovered evidence of an aberrant insertion event that resulted in a deletion of sequences in the 5′ P end of the element and a duplication of the promoter region and first exons of the mini-white gene (Figure S2A). These rearrangements still allowed for proper Cre and FLP treatment of the construct to create enhancerless and promoterless derivatives, which would be predicted to pair as intended when placed in trans with one another with the exception of a mini-white fragment replacing the 5′ P end. The duplicated mini-white promoter carried by the aberrant insert was located downstream of the GFP transgene, and therefore would not directly contribute to transcription through the GFP transcription unit. The duplicated region also encodes two binding sites for the DNA-binding protein Zeste, a transcription factor that has previously been implicated as a positive regulator of transvection (Benson and Pirrotta 1988). We tested whether Zeste activity could be responsible for the high level of transvection associated with the insertion at 96C by assessing GFP fluorescence in larvae lacking functional zeste, but the elevated GFP levels resulting from trans- vs. cis-expression were still observed in a loss-of-function za background (Figure S2B), indicating that Zeste is unlikely to contribute to the unusual degree of transvection observed for this transgenic insertion.
We next assessed GFP fluorescence generated via transvection at the remaining seven sites. Among this set, we observed a significant correlation between the mean GFP fluorescence produced in cis vs. that produced in trans at each location (r = 0.74, P = 0.03) (Figure 3, A–C), suggesting that position effects on transcriptional output via enhancer action in cis and in trans are directly comparable. Consistent with this, the site that supported the weakest levels of cis-expression out of all tested locations (65E) was the only location to show mean GFP fluorescence resulting from transvection that was undetectable relative to background. Similarly, the combined mean fluorescence for the three sites that mapped to open chromatin in cultured cells (excluding position 96C) was significantly higher than that of the three sites that mapped to silent chromatin in BG3 cells (2.4 ± 0.3 a.u. (n = 15 discs) vs. 0.6 ± 0.2 a.u. (n = 15), P = 1 × 10−4, Student’s t-test), indicating that the observed correlation between cis-expression and chromatin structure in cultures cells is also observed for trans-expression.
As a final comparison, we assessed the efficiency of transvection, which we defined as the level of fluorescence resulting from trans-expression relative to that from cis-expression, at each location. Excluding the special case of position 96C, transvection efficiencies ranged from 0 to 10.3%, with no significant correlation to cis-expression level (Pearson’s r = 0.2, P = 0.66) or chromatin states in cultured cells (P = 0.13, Student’s t-test). In sum, our data support that “open” vs. “closed” chromatin structures have similar general effects on transcriptional output via cis- and trans-expression, but are unlikely to strongly affect the efficiency of enhancer action in trans relative to cis-output at a given genomic location.
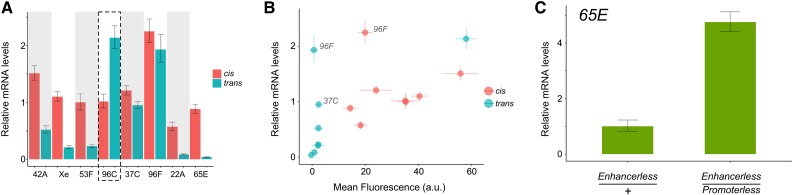
To support our analysis of GFP fluorescence, we used quantitative RT-PCR to compare GFP mRNA levels resulting from cis- and trans-based expression at each position. Note that, due to the difficulty in separating differentiated eye cells from the remainder of the eye-antennal disc, this analysis reflects mRNA expression across the entire tissue rather than specific expression in GMR-positive cells. Relative levels of GFP mRNA were consistent with our analysis of GFP fluorescence, including elevated levels of GFP expression resulting from transvection at position 96C, with the exception of data points showing higher than anticipated RNA levels for cis- and trans-expression at position 96F and for trans-expression at position 37C (Figure 4). The insertion at position 96F is in the 5′ UTR of HLHm7, a gene expressed in regions of the eye-antennal disc outside of the GMR expression domain, including a subset of developing antennal and ocellar cells and a region of eye primordial cells anterior to the morphogenetic furrow (de Celis et al. 1996). Although we did not observe GFP fluorescence in these domains in lines with inserts at 96F, in situ hybridization using a probe specific for the GFP transgene shows a distinct background of GFP RNA in the antennal and ocellar portions of the disc, which likely contributes to the higher than anticipated qRT-PCR data (Figure S3). Similarly, the insert at 37C is within an intron of the brat gene, and in situ hybridization for GFP mRNA in discs carrying the enhancerless transgenic derivative at position 37C shows a stripe of mRNA expression in primordial eye cells just anterior to the morphogenetic furrow, similar to that observed for brat expression (Figure S3) (Frank et al. 2002). Excluding these three outliers, we observed a high degree of correlation between GFP fluorescence and qRT-PCR (Pearson’s r = 0.93, P < 0.0001), indicating that the observed differences in fluorescence are transcriptionally driven. Additionally, we observed a significant correlation between mean expression levels produced by cis vs. trans expression for each line as assessed by qRT-PCR (r = 0.9, P = 0.02). Finally, we observed significant position-to-position differences in mRNA levels resulting from enhancer action in trans (P < 0.0001, ANOVA), with post hoc tests showing significant differences among all pair-wise comparisons except for lines Xe and 53A (Tukey’s HSD, P < 0.05). In sum, our qRT-PCR analysis further supports that position effects impact enhancer action in trans.
Figure 4.
Quantitative RT-PCR analysis of GFP transcript levels generated by GMR activity in cis and trans. (A) Total RNA from whole third instar larval eye-antennal discs of each genotype was subjected to quantitative RT-PCR using GFP-specific primers. All data are presented as relative to cis-expression from position 53F (= 1.0). Each bar in the figure represents three biological replicates; error bars represent 95% confidence interval. Dotted box highlights exceptional trans-activity at position 96C. (B) GFP expression by GMR as assessed by qRT-PCR (y-axis; data from (A)) vs. mean fluorescence (x-axis; data from Figure 2 and Figure 3; a.u., arbitrary units). Outliers with evidence of ectopic position-dependent GFP mRNA outside of the GMR expression domain are indicated (37C, 96F). (C) qRT-PCR using GFP-specific primers demonstrating elevated GFP RNA when GMR acts in trans at position 65E (enhancerless/promoterless) relative to the enhancerless construct in the absence of GMR at that position.
Our assessment of GFP fluorescence showed no evidence of transvection at location 65E, which could be caused by a complete absence of communication between homologous chromosomes at that site, or could result from extremely low levels of mRNA produced by enhancer action in trans that do not elevate GFP fluorescence above background levels. To differentiate between these possibilities, we used quantitative RT-PCR to compare mRNA levels resulting from transvection at position 65E to those resulting from a hemizygous insertion of the enhancerless transgenic derivative in the absence of a GMR enhancer on either homolog. We observed a 4.75-fold increase in GFP mRNA levels for the transvection genotype relative to the enhancerless control (Figure 4C), demonstrating that enhancer action in trans is indeed supported at this site, but that the resulting levels of gene expression are insufficient to detect the phenotype of GFP fluorescence above background. In sum, all eight positions that we queried support transvection to varying degrees, with a significant correlation between positions effects on cis- and trans-based expression.
Position effects on enhancer action in trans do not reflect variation in somatic homolog pairing
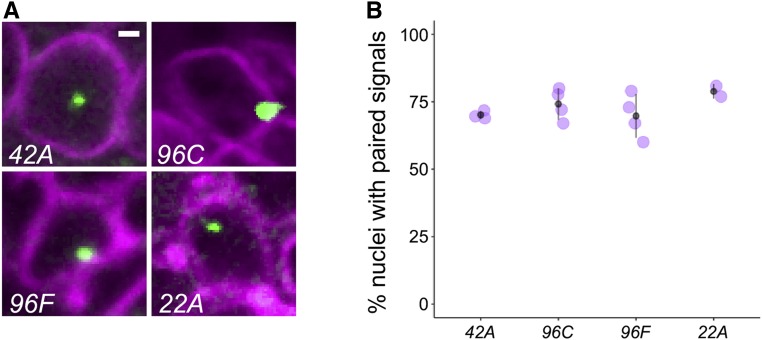
Prior analyses of somatic homolog pairing via DNA-FISH have shown potential variation in the percentage of nuclei that pair at different genomic locations and in different cell types (Fung et al. 1998; Williams et al. 2007; Bateman and Wu 2008; Joyce et al. 2012). To assess whether position effects on transvection could result from variation in somatic homolog pairing, we designed Oligopaint probes (Beliveau et al. 2015) targeting genomic regions surrounding four of our transgenic insertions, including position 96C with unusually high levels of transvection, and three other positions that support different levels of GMR activity in cis and in trans. Scoring of the percentage of nuclei with paired signals for each of these locations in GMR-positive cells from third instar larval eye discs showed a range of mean pairing levels from 69.7 ± 8.1% to 78.8 ± 2.9%, consistent with pairing levels observed for other loci (Figure 5) (Fung et al. 1998; Williams et al. 2007; Bateman and Wu 2008; Joyce et al. 2012). However, we find no significant difference among the pairing levels observed at these four sites (P = 0.32, ANOVA), indicating that variation in somatic homolog pairing is unlikely to contribute to the broad variation in enhancer activity in cis and in trans observed in our experiment.
Figure 5.
Position effects on transvection are not due to variation in somatic homolog pairing. (A) representative nuclei showing DNA-FISH in third instar retinal cells. Cells were identified as GMR positive using an anti-GFP antibody in discs expressing UAS-laminB-GFP under the control of GMR-GAL4. The insertion position targeted by the Oligopaint probe is indicated in each panel. Bar, 1 µm. (B) quantification of somatic homolog pairing. Each point represents the percentage of nuclei with a single fluorescence signal scored in ∼25–40 nuclei derived from a single eye disc. Two to four discs were scored for each genotype. Point and whiskers represent mean ± SD.
Finally, in addition to our analyses of local chromatin states and somatic homolog pairing, we also considered whether position effects on enhancer activity in cis or in trans could be influenced by proximity of a transgenic insertion site to other nearby genomic features, including local gene structures, TAD boundaries, or insulators, and found no clear patterns of correlation (Table 1). While we cannot exclude the possibility that a complex interplay of any number of features can influence transvection, our data most strongly support that the local chromatin state is a major determinant for the capacity of a locus to support transcription, with similar impacts on enhancer activity in cis and in trans.
Discussion
Here, we used a transgenic approach to quantitatively assess locus-specific variation in transvection. Our analysis demonstrates a high correlation between the transcriptional output of an enhancer in cis and in trans at the same location. In considering possible mechanisms to account for position effects on transvection, we observed a strong correlation between the degree of transvection supported in vivo and the chromatin state, active or inactive, as assessed in cultured cells. Other possible factors, including locus-specific variation in somatic homolog pairing, did not show a clear pattern when compared to variation in transvection, arguing that position effects on enhancer action in trans are largely determined by local chromatin modifications.
Our data are in general agreement with prior findings on the permissivity of the genome to transvection (Chen et al. 2002; Kravchenko et al. 2005; Bateman et al. 2012; Mellert and Truman 2012). While only seven of eight locations in our study supported enhancer action in trans as assessed by GFP fluorescence, the location that failed to show fluorescence represented the position with the weakest levels of GMR activity in cis, and quantitative analysis of mRNA levels showed that enhancer action in trans was indeed supported for the location. It is therefore likely that weak trans-activity at this position leads to mRNA levels below a threshold required to produce GFP fluorescence above background levels. This model could explain why prior studies of transvection using yellow-based transgenes did not identify loci lacking a capacity to support transvection, given that very little yellow mRNA is required to produce changes in adult cuticle pigmentation (Morris et al. 2004), although it could also be the case that yellow-based transgenes carry transvection-promoting sequences that are not present in our fluorescent reporters.
Previous observations of locus-specific variation in levels of somatic homolog pairing raised the question of whether position effects on transvection would arise from variation in interhomolog contacts. Our data show no clear difference in pairing levels for four different locations that support vastly different levels of transcriptional output via enhancer action in trans, suggesting that variation in somatic homolog pairing is not a major contributor to position effects on transvection. Prior studies of pairing-sensitive silencing—a form of transvection that results in decreased gene expression when transgenes carrying certain polycomb response elements (PREs) or other silencing motifs are paired—have also shown evidence of position effects (reviewed by Kassis 2002). Although the precise mechanisms causing position effects on pairing-sensitive silencing have not been determined, it has been postulated that flanking genomic DNA is highly influential (Kassis 2002). As a caveat in interpreting our data, we note that the optical resolution of confocal microscopy used in our assessment may not reveal subtle differences in somatic homolog pairing that could occur within ∼0.3–0.5 µm, which represents the approximate size of a DNA-FISH signal in our assay. It should also be noted that a baseline level of somatic homolog pairing must exist at each site in order for transvection to be supported, since disruption of pairing via rearrangement reduces or eliminates enhancer action in trans (Lewis 1954), and attempts to observe transvection in organisms with little to no somatic homolog pairing have frequently failed to show robust enhancer action in trans (e.g., Noordermeer et al. 2011). Thus, although we find no clear correlation between variation in pairing and position effects on transvection in this study, each site assessed has at least a minimal level of somatic homolog pairing required for enhancer action in trans.
Despite a general trend for enhancer action in trans to result in reduced levels of activity relative to action in cis, our analysis uncovered one exceptional insertion where transvection was surprisingly robust, producing GFP fluorescence and steady-state mRNA levels higher than those observed for cis-activity at that location. No activity was observed for either the enhancerless or promoterless construct alone as either a homozygous or hemizygous insertion at this location, demonstrating that the robust activity required the two constructs to interact in trans. This insert is located just upstream of the gene Cad96Ca in polytene band 96C, and we observe no obvious genomic feature that could account for enhanced transvection, nor do we observe exceptional levels of somatic homolog pairing via DNA-FISH using Oligopaint probes targeting this region. Notably, a previous study of transvection at the Malic enzyme locus demonstrated trans-activity greater than the expected cis-activity in genotypes carrying certain promoter deletions (Lum and Merritt 2011), indicating that unusually high levels of transvection may exist under special circumstances. While it is possible that the region upstream of Cad96Ca generally supports robust transvection due to unknown genomic features, we cannot exclude the possibility that the structural change observed for this P-element insertion may play a central role, perhaps through duplicated sequences within the white promoter that enhance transvection, or via the deletion of unknown elements in the 5′ end of the P element that antagonize transvection.
In sum, our analysis provides further resolution to the question of whether transvection is a general feature in the Drosophila genome. In combination with a previous study demonstrating that the strength of an enhancer’s activity is an important determinant of whether it can act in trans (Blick et al. 2016), a general picture is emerging wherein factors that contribute to higher levels of gene expression have an overall positive impact on transvection via enhancer action in trans. However, some specific sequences can positively impact enhancer action in trans via other mechanisms, including well characterized elements such as insulators (Kravchenko et al. 2005; Schoborg et al. 2013; Fujioka et al. 2016; Piwko et al. 2019), and other less characterized elements (Hopmann et al. 1995; Ronshaugen and Levine 2004; Blick et al. 2016). Thus, while a general correlation exists between gene expression levels in cis and in trans, the Drosophila genome likely harbors sequences that further regulate the capacity for regulatory elements to interact between chromosomes.
Acknowledgments
Thanks to Eric Joyce, Brian Beliveau, Hien Hoang, and Ting Wu for help with OligoPaint design and synthesis and for stimulating discussions, to Kayla Viets and Bob Johnston for help with OligoPaint FISH in eye discs, Jeff Settleman, Mark Siegel, and Sean Carroll for plasmids, and the Bloomington Drosophila Stock Center for fly stocks. This work was supported by grants from the National Institute of General Medical Sciences of the National Institutes of Health (P20 GM0103423), a Faculty Early Development (CAREER) award from the National Science Foundation to J.R.B. (1349779), and Bowdoin College.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.9973646.
Communicating editor: J. Birchler
Literature Cited
- Bateman J. R., and Wu C. T., 2008. A genomewide survey argues that every zygotic gene product is dispensable for the initiation of somatic homolog pairing in Drosophila. Genetics 180: 1329–1342. 10.1534/genetics.108.094862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman J. R., Johnson J. E., and Locke M. N., 2012. Comparing enhancer action in cis and in trans. Genetics 191: 1143–1155. 10.1534/genetics.112.140954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliveau B. J., Joyce E. F., Apostolopoulos N., Yilmaz F., Fonseka C. Y. et al. , 2012. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl. Acad. Sci. USA 109: 21301–21306. 10.1073/pnas.1213818110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliveau B. J., Apostolopoulos N., and Wu C. T., 2014. Visualizing genomes with Oligopaint FISH probes. Curr. Protoc. Mol. Biol. 105: Unit 14.23 10.1002/0471142727.mb1423s105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliveau B. J., Boettiger A. N., Avendano M. S., Jungmann R., McCole R. B. et al. , 2015. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat. Commun. 6: 7147 10.1038/ncomms8147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson M., and Pirrotta V., 1988. The Drosophila zeste protein binds cooperatively to sites in many gene regulatory regions: implications for transvection and gene regulation. EMBO J. 7: 3907–3915. 10.1002/j.1460-2075.1988.tb03277.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blick A. J., Mayer-Hirshfeld I., Malibiran B. R., Cooper M. A., Martino P. A. et al. , 2016. The capacity to act in trans varies among Drosophila enhancers. Genetics 203: 203–218. 10.1534/genetics.115.185645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. L., Huisinga K. L., Viering M. M., Ou S. A., Wu C. T. et al. , 2002. Enhancer action in trans is permitted throughout the Drosophila genome. Proc. Natl. Acad. Sci. USA 99: 3723–3728. 10.1073/pnas.062447999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., and Zhang J., 2016. The genomic landscape of position effects on protein expression level and noise in yeast. Cell Syst. 2: 347–354. 10.1016/j.cels.2016.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales M., Rosado A., Cortini R., van Arensbergen J., van Steensel B. et al. , 2017. Clustering of Drosophila housekeeping promoters facilitates their expression. Genome Res. 27: 1153–1161. 10.1101/gr.211433.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis J. F., de Celis J., Ligoxygakis P., Preiss A., Delidakis C. et al. , 1996. Functional relationships between Notch, Su(H) and the bHLH genes of the E(spl) complex: the E(spl) genes mediate only a subset of Notch activities during imaginal development. Development 122: 2719–2728. [DOI] [PubMed] [Google Scholar]
- Duncan I. W., 2002. Transvection effects in Drosophila. Annu. Rev. Genet. 36: 521–556. 10.1146/annurev.genet.36.060402.100441 [DOI] [PubMed] [Google Scholar]
- Eagen K. P., Aiden E. L., and Kornberg R. D., 2017. Polycomb-mediated chromatin loops revealed by a subkilobase-resolution chromatin interaction map. Proc. Natl. Acad. Sci. USA 114: 8764–8769. 10.1073/pnas.1701291114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion G. J., van Bemmel J. G., Braunschweig U., Talhout W., Kind J. et al. , 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224. 10.1016/j.cell.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty M. S., Salis P., Evans C. J., Ekas L. A., Marouf A. et al. , 2010. chinmo is a functional effector of the JAK/STAT pathway that regulates eye development, tumor formation, and stem cell self-renewal in Drosophila. Dev. Cell 18: 556–568. 10.1016/j.devcel.2010.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D. J., Edgar B. A., and Roth M. B., 2002. The Drosophila melanogaster gene brain tumor negatively regulates cell growth and ribosomal RNA synthesis. Development 129: 399–407. [DOI] [PubMed] [Google Scholar]
- Fujioka M., Mistry H., Schedl P., and Jaynes J. B., 2016. Determinants of chromosome architecture: insulator pairing in cis and in trans. PLoS Genet. 12: e1005889 10.1371/journal.pgen.1005889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung J. C., Marshall W. F., Dernburg A. F., Agard D. A., and Sedat J. W., 1998. Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. J. Cell Biol. 141: 5–20. 10.1083/jcb.141.1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer P. K., Green M. M., and Corces V. G., 1990. Tissue-specific transcriptional enhancers may act in trans on the gene located in the homologous chromosome: the molecular basis of transvection in Drosophila. EMBO J. 9: 2247–2256. 10.1002/j.1460-2075.1990.tb07395.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic K. G., and Lindquist S., 1989. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59: 499–509. 10.1016/0092-8674(89)90033-0 [DOI] [PubMed] [Google Scholar]
- Hopmann R., Duncan D., and Duncan I., 1995. Transvection in the iab-5,6,7 region of the bithorax complex of Drosophila: homology independent interactions in trans. Genetics 139: 815–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce E. F., Williams B. R., Xie T., and Wu C. T., 2012. Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genet. 8: e1002667 10.1371/journal.pgen.1002667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce E. F., Erceg J., and Wu C. T., 2016. Pairing and anti-pairing: a balancing act in the diploid genome. Curr. Opin. Genet. Dev. 37: 119–128. 10.1016/j.gde.2016.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis J. A., 2002. Pairing-sensitive silencing, polycomb group response elements, and transposon homing in Drosophila. Adv. Genet. 46: 421–438. [DOI] [PubMed] [Google Scholar]
- Kassis J. A., 2012. Transvection in 2012: site-specific transgenes reveal a plethora of trans-regulatory effects. Genetics 191: 1037–1039. 10.1534/genetics.112.142893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison J. A., and Southworth J. W., 2002. Transvection in Drosophila. Adv. Genet. 46: 399–420. 10.1016/S0065-2660(02)46014-2 [DOI] [PubMed] [Google Scholar]
- Kharchenko P. V., Alekseyenko A. A., Schwartz Y. B., Minoda A., Riddle N. C. et al. , 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485. 10.1038/nature09725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchenko E., Savitskaya E., Kravchuk O., Parshikov A., Georgiev P. et al. , 2005. Pairing between gypsy insulators facilitates the enhancer action in trans throughout the Drosophila genome. Mol. Cell. Biol. 25: 9283–9291. 10.1128/MCB.25.21.9283-9291.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B., 1954. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am. Nat. 88: 225–239. [Google Scholar]
- Lum T. E., and Merritt T. J., 2011. Nonclassical regulation of transcription: interchromosomal interactions at the malic enzyme locus of Drosophila melanogaster. Genetics 189: 837–849. 10.1534/genetics.111.133231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M., Pitsouli C., Villalta C., Celniker S. E., and Perrimon N., 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40: 476–483. 10.1038/ng.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee B. D., 2004. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim. Biophys. Acta 1677: 165–180. 10.1016/j.bbaexp.2003.11.017 [DOI] [PubMed] [Google Scholar]
- Mellert D. J., and Truman J. W., 2012. Transvection is common throughout the Drosophila genome. Genetics 191: 1129–1141. 10.1534/genetics.112.140475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikeladze-Dvali T., Wernet M. F., Pistillo D., Mazzoni E. O., Teleman A. A. et al. , 2005. The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell 122: 775–787. 10.1016/j.cell.2005.07.026 [DOI] [PubMed] [Google Scholar]
- Moon N. S., Di Stefano L., Morris E. J., Patel R., White K. et al. , 2008. E2F and p53 induce apoptosis independently during Drosophila development but intersect in the context of DNA damage. PLoS Genet. 4: e1000153 10.1371/journal.pgen.1000153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. R., Chen J. L., Geyer P. K., and Wu C. T., 1998. Two modes of transvection: enhancer action in trans and bypass of a chromatin insulator in cis. Proc. Natl. Acad. Sci. USA 95: 10740–10745. 10.1073/pnas.95.18.10740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. R., Petrov D. A., Lee A. M., and Wu C. T., 2004. Enhancer choice in cis and in trans in Drosophila melanogaster: role of the promoter. Genetics 167: 1739–1747. 10.1534/genetics.104.026955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses K., and Rubin G. M., 1991. Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev. 5: 583–593. 10.1101/gad.5.4.583 [DOI] [PubMed] [Google Scholar]
- Nègre N., Brown C. D., Shah P. K., Kheradpour P., Morrison C. A. et al. , 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 6: e1000814 10.1371/journal.pgen.1000814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D., de Wit E., Klous P., van de Werken H., Simonis M. et al. , 2011. Variegated gene expression caused by cell-specific long-range DNA interactions. Nat. Cell Biol. 13: 944–951. 10.1038/ncb2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Gerber A. S., and Hartl D. L., 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120: 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwko P., Vitsaki I., Livadaras I., and Delidakis C., 2019. The role of insulators in transgene transvection in Drosophila. Genetics 212: 489–508. 10.1534/genetics.119.302165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronshaugen M., and Levine M., 2004. Visualization of trans-homolog enhancer-promoter interactions at the Abd-B Hox locus in the Drosophila embryo. Dev. Cell 7: 925–932. 10.1016/j.devcel.2004.11.001 [DOI] [PubMed] [Google Scholar]
- Rubin G. M., and Spradling A. C., 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353. 10.1126/science.6289436 [DOI] [PubMed] [Google Scholar]
- Schoborg T., Kuruganti S., Rickels R., and Labrador M., 2013. The Drosophila gypsy insulator supports transvection in the presence of the vestigial enhancer. PLoS One 8: e81331 10.1371/journal.pone.0081331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal M. L., and Hartl D. L., 1996. Transgene Coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics 144: 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui K., Nishihara S., Sakuma M., Togashi S., Ueda R. et al. , 1994. Newly established cell lines from Drosophila larval CNS express neural specific characteristics. In Vitro Cell. Dev. Biol. Anim. 30A: 209–216. 10.1007/BF02632042 [DOI] [PubMed] [Google Scholar]
- Viets K., Sauria M., Chernoff C., Anderson C., Tran S. et al. , 2018. TADs pair homologous chromosomes to promote interchromosomal gene regulation. bioRxiv. doi: 10.1101/445627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B. R., Bateman J. R., Novikov N. D., and Wu C. T., 2007. Disruption of topoisomerase II perturbs pairing in drosophila cell culture. Genetics 177: 31–46. 10.1534/genetics.107.076356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. L., Li B. Z., Zhang W. Z., Song K., Qi H. et al. , 2017. Genome-wide landscape of position effects on heterogeneous gene expression in Saccharomyces cerevisiae. Biotechnol. Biofuels 10: 189 10.1186/s13068-017-0872-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All stocks and reagents are available upon request. All data necessary for confirming the conclusions of this work are represented in the text, figures, tables, and supplemental information. Publicly available genomic data on chromatin states in cultured cells (Filion et al. 2010; Kharchenko et al. 2011) were accessed via the genome browser at flybase.org. Supplemental material available at figshare: https://doi.org/10.25386/genetics.9973646.