Abstract
Biological characterization of genetic variants identified in genome-wide association studies (GWAS) remains a substantial challenge. Here we used human induced pluripotent stem cells (hiPSC) and their neural derivatives to characterize common variants on chromosome 3p22 that have been associated by GWAS with major mental illnesses. HiPSC-derived neural progenitor cells carrying the risk allele of the single nucleotide polymorphism (SNP), rs9834970, displayed lower baseline TRANK1 expression that was rescued by chronic treatment with therapeutic dosages of valproic acid (VPA). VPA had the greatest effects on TRANK1 expression in hiPSC, NPC, and astrocytes. Although rs9834970 has no known function, we demonstrated that a nearby SNP, rs906482, strongly affects binding by the transcription factor, CTCF, and that the high-affinity allele usually occurs on haplotypes carrying the rs9834970 risk allele. Decreased expression of TRANK1 perturbed expression of many genes involved in neural development and differentiation. These findings have important implications for the pathophysiology of major mental illnesses and the development of novel therapeutics.
INTRODUCTION
Genome-wide association studies (GWASs) of bipolar disorder (BD) and schizophrenia (Scz) have identified several reproducible genetic markers, but the biological consequences of most variants remain undefined. 1, 2 Many common complex traits are mediated by expression quantitative trait loci (eQTLs) that affect expression of nearby (cis) or distant (trans) genes. 3 Altered gene expression is thus an important mechanism underlying genotype-phenotype associations. 4 However, eQTLs may act only in specific tissues 5–7 or particular stages of development. 8, 9 Such tissue and time-specific effects complicate studies of allelic variation and gene expression in the human central nervous system (CNS), where appropriate cells from living individuals are difficult to obtain. Past eQTL studies of variants implicated in brain disorders have thus depended on limited supplies of postmortem brain tissue. 10 Such studies are valuable, but cannot directly assess developmental effects and are often confounded by age, gender, medications, substance abuse, and agonal events.
Induced pluripotent stem cells (iPSC) offer an attractive alternative. 11 eQTL studies in human iPSC-derived cells exploit a renewable supply of cells largely free from the complications of postmortem tissue. iPSC can be easily generated from patients of known genetic background, further reducing variance that can obscure subtle signals. Model systems based on iPSC-derived cells can follow changes in gene expression during development and can be experimentally manipulated with drugs, toxins, or gene-editing. While iPSC-derived cells can be affected by culture conditions12 or somatic changes in DNA sequence or copy number 13 and do not reflect epigenetic marks in the donor’s brain, 4 they nevertheless offer great potential for dissecting the molecular mechanisms of complex brain disorders. Several recent studies have employed iPSC technology to model neuropsychiatric conditions in vitro, 15–18 but few studies have modeled functional effects of common risk variants.19
In this study, we used iPSC and their neural derivatives to investigate the cellular impact of common genetic polymorphisms previously associated with BD and Scz by GWAS. The polymorphisms lie on chromosome 3p22 near the gene TRANK1 (also known as LBA1). TRANK1 encodes a protein expressed in brain and other tissues, but whose function remains unknown. A common variant (rs9834970) located some 15 kb 3’ of TRANK1 has shown genome-wide significant association with BD in several GWAS,20–22 and nearby markers have been associated with Scz and other mental illnesses.23, 24 We previously showed that valproic acid (VPA), an effective treatment for BD, increases TRANK1 expression in immortalized cell lines, 20 but the relationship between VPA exposure, genetic risk variants, and expression of TRANK1 in neural cells remains unclear.
The present study had several objectives: 1) to assess the impact of common variants within the 3p22 locus on TRANK1 expression in patient-derived neural cells; 2) to test the effects of treatment with VPA and another commonly-used therapeutic agent, lithium, on TRANK1 expression; and 3) to uncover genes and gene networks whose expression correlates with that of TRANK1, thereby shedding light on its function. The findings established that genetic variants on chromosome 3p22 decreased expression of TRANK1 in developing neural cells and altered binding of the transcription factor, CTCF. Chronic treatment with therapeutic dosages of VPA, but not lithium, restored expression of TRANK1 mRNA and protein to control levels. Decreased expression of TRANK1 strongly perturbed many genes involved in neural development, differentiation, and apoptosis. These findings have implications for the pathophysiology of major mental illnesses and the development of novel therapeutics.
RESULTS
Induction and characterization of human iPSCs and neural derivatives.
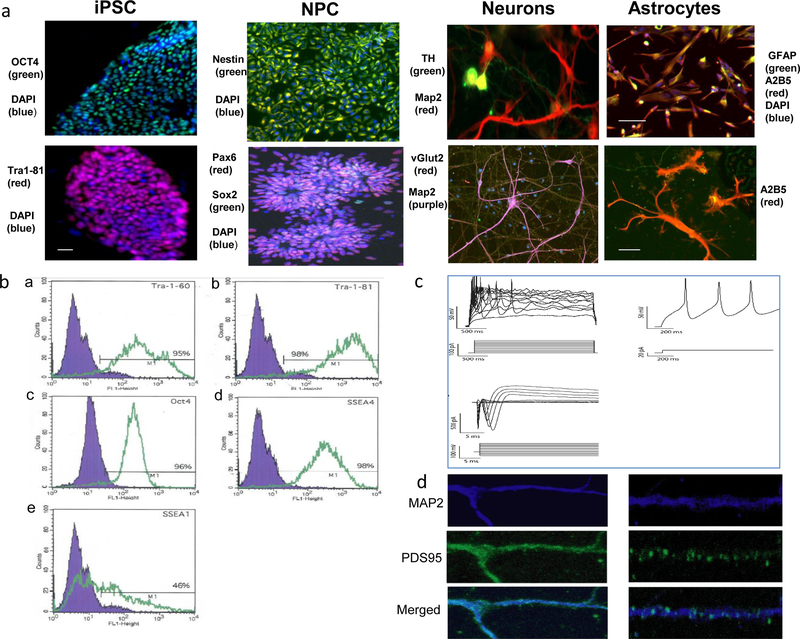
All lines exhibited typical stem cell morphology. Immunofluorescence demonstrated expression of pluripotency markers (Figure 1a). Flow cytometry was consistent with pluripotency (Figure 1b). We found good consistency of differentiation between samples: More than 96% of putative NPC were Nestin-positive; after 8 wk of neuronal differentiation, >80% of cells were positive for MAP2, and >80% of these also stained positive for vesicular glutamate transporter (vGLUT2), consistent with glutamatergic neurons. Some cells stained positive for tyrosine hydroxylase (TH), consistent with mixed dopaminergic and glutamatergic neurons. After 7 wk of astrocyte differentiation, >80% of cells stained positive for GFAP and A2B5. Representative images are shown in Figure 1a. Differentiated neurons displayed action potentials by patch-clamp (Figure 1c). These neurons also stained positive for postsynaptic density 95 (PSD95) (Figure 1d).
Figure 1. Generation of iPSCs and neural derivatives.
a, Representative images of induced pluripotent stem cells (iPSCs) and their neural derivatives subjected to immunocytochemical analysis of pluripotency and neural cell differentiation. Far left: iPSCs showed the typical morphology of human embryonic stem cells (hESCs) and positive staining for pluripotency markers OCT4 (green) and DAPI (blue) (upper panel); Tra1–81 (red) and DAPI (blue) (lower panel). Second from left: iPSC-derived neural progenitor cells (NPCs) immunostained positive for neural lineage markers Nestin (green) and DAPI (blue)(upper panel), Pax6 (red), Sox2 (green) and DAPI (blue) (lower panel) scale bar: 50 μm. Third from left: iPSC-derived neurons expressed neural markers MAP2 (red), TH (green), (upper panel), MAP2 (green) and vGlut2 (red) (lower panel). Far right: Astrocytes generated from NPCs exhibited typical astrocyte morphology and expressed astrocyte markers GFAP (green), A2B5(red) and and DAPI(blue) (upper panel) scale bar: 50 μm; and A2B5 (red) (lower panel), scale bar: 10 μm. b, Human induced pluripotent stem cells were subjected to flow cytometric to analysis with pluripotency markers. In the merged image, the blue area shows the fluorescence intensity of the IgG negative control antibody, while intensities of the antibodies of interest are shown in green. Results of negative control and iPSC lines are shown in (a) Tra-1 60 (9%, 95%), (b) Tra_1 81 (5%, 98%), (c) OCT4 (4%, 96%), (d) SSEA-4 (3%, 98%), and (e) SSEA-1(10%, 46%). c, Representative traces from whole cell-patch clamp recordings in 6-week post-differentiation neurons, demonstrating typical electrophysiological activity. d, Immunochemical staining of MAP2 (blue) and PSD95 (green) in 6-week neurons, demonstrating typical synapse staining patterns.
Gene expression profiles demonstrated distinct clusters for fibroblasts, iPSCs, NPCs and neurons, as expected (Figure S1a). All studied iPSC lines exhibited gene expression profiles typical of pluripotent stem cells (Figure S1b, Figure S1c). qRT-PCR analysis of gene expression of selected pluripotency and NPC markers further validated these cell lines (Figure S2a-b). All iPSC lines maintained normal karyotypes (Figure S2c; Supplemental Methods).
TRANK1 expression after VPA treatment in iPSCs and neuronal derivatives.
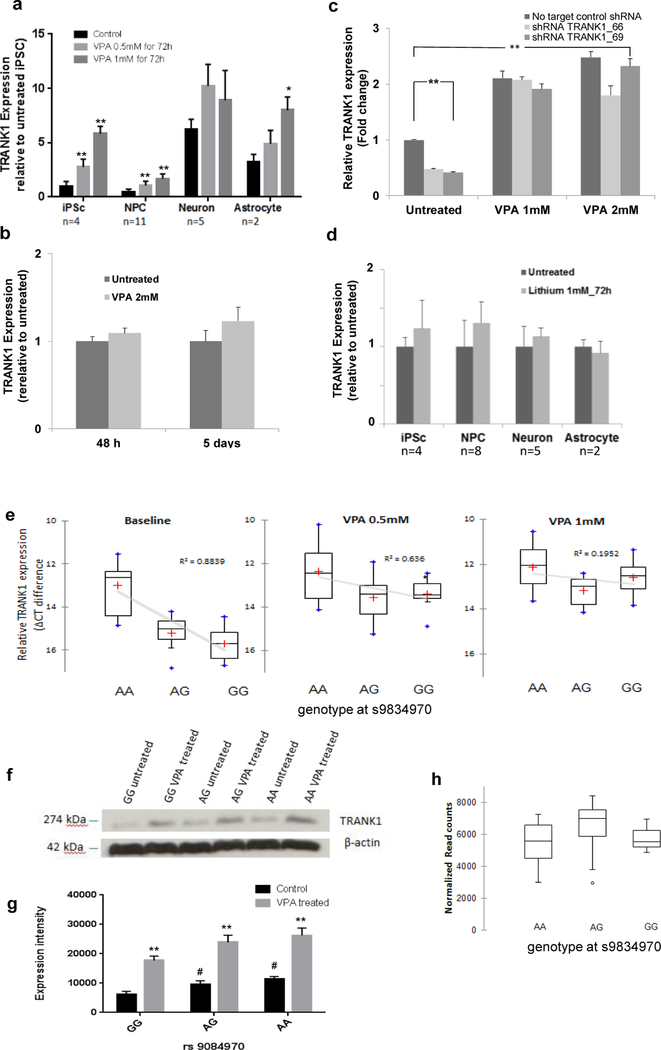
Previously, we showed that VPA increased expression of TRANK1 in immortalized, non-neural cell lines.20 To assess the effects of VPA on neural cells, monolayer-cultured iPSCs, and neural derivatives were treated with VPA. In iPSCs and NPCs, TRANK1 expression increased 3 to 6-fold after 72 h treatment with VPA (Figure 2a). Expression increased with VPA dosage over the typical therapeutic range of 0.5 to 1.0 mM. VPA also significantly increased TRANK1 expression in astrocytes, but less than in iPSC or NPCs. In contrast, VPA had no detectable effect on TRANK1 expression in neurons. A similar lack of change in TRANK1 expression was seen in rat hippocampal neurons treated for ≤5 days with VPA doses ≤ 2 mM (Figure 2b). These results show that VPA has the greatest effects on TRANK1 expression in actively growing cells, diminishing with differentiation.
Figure 2. Effects of cell type, drug treatment, and rs9834970 genotype on TRANK1 expression.
a, Human induced pluripotent stem cell (iPSC), neural progenitor cell (NPC), neuronal, and astrocyte lines were grown in independent cultures and treated with therapeutic dosages of VPA (0.5mM, 1mM) or vehicle for 72 hours. TRANK1 mRNA expression was determined by reverse transcription polymerase chain reaction(RT-PCR). VPA treatment significantly increased TRANK1 expression in iPSCs, NPCs, and astrocytes, but not in neurons (*P<0.05, **P <0.01). b, TRANK1 expression in rat E18 hippocampal neurons cultured for 8d before treatment was also unaffected by VPA, even after 48h or 5 days of treatment at 2 mM dose. c, TRANK1 expression in HeLa cells was significantly increased by treatment with 1 mM or 2 mM VPA after stable shRNA knockdown of TRANK1. **P<0.01 TRANK1 shRNA knockdown vs no target shRNA control. ## P<0.01 VPA (1mM or 2 mM) treated vs untreated condition. ^^ P<0.01 shRNA TRANK1_66 knockdown vs no target control shRNA on 2mM VPA treatment condition. d, Lithium (1 mM) had no effect on TRANK1 expression in any of the 4 cell types tested. e, Neural progenitor cells (NPCs) carrying the risk allele (G) showed reduced TRANK1 mRNA expression (left) that was rescued by 72 h of VPA treatment at dosages of 0.5 mM (middle) or 1 mM (right). Values are expressed as mean relative ΔCT difference ± S.E.M. Comparisons: baseline, GG vs AA, p<0.0001; baseline, GG vs AG, p<0.05; baseline, GG vs VPA 0.5 mM, GG, p<0.05; baseline GG vs VPA 1 mM, GG, p<0.01. n =3 for GG carries; n = 3 for AA carries; n =5 for AG carries. f, Western blot: Qualitative increase in binding of anti-TRANK1 antibody to 274 kDa protein band extracted from NPCs in A-allele carriers vs G-allele carriers at both untreated and after treatment with 1 mM VPA conditions; n =3 for each condition. g, Quantification of western blot signal intensities with image J, **P<0.01 VPA treated vs untreated, # P<0.05 allele AA and AG vs GG at rs9834970. h, No relationship between rs9834970 genotype and TRANK1 mRNA counts (n=22) in human postmortem dorsolateral prefrontal cortex from RNA seq.
VPA also reversed the effects of stable shTRANK1 knockdown in HeLa cells (n=6) (Figure 2c). This suggests that VPA affects TRANK1 expression at the level of transcription.
Unlike VPA, but consistent with our published results in immortalized cells.20 lithium chloride had no effect on TRANK1 expression in any of the iPSC-derived cells we tested (Figure 2d). Thus, subsequent experiments focused on VPA.
Genetic variation at rs9834970 and TRANK1 expression in iPSCs and neural derivatives.
Previous GWAS have consistently found that the G-allele of rs9834970 is significantly more frequent than the A-allele among people with BD or schizophrenia.2, 20–26 This SNP lies about 15 kb downstream of TRANK1, within a DNaseI hypersensitive site (ENCODE). Conditional analysis of BD GWAS data confirmed that rs9834970 fully accounted for the association signal at this locus (Figure S3). To test whether variation at rs9834970 affects expression of TRANK1, we compared TRANK1 expression in iPSCs, NPCs, and neurons.
Genotype at rs9834870 had a substantial impact on baseline expression of TRANK1 mRNA in both iPSC and NPC. Cell lines carrying the risk allele (AG or GG genotypes) showed significantly lower baseline TRANK1 expression than AA homozygotes. NPCs with the GG genotype (n=3) showed a 5.6-fold decrease in TRANK1 expression (p<0.0001) compared to the AA genotype (n=3) and a 1.35-fold decrease (p<0.05) compared to the AG genotype (n=5) (Figure 2e). Similar results were seen in iPSCs, where GG homozygotes showed significantly lower baseline TRANK1 expression than AA homozygotes (p<0.0001; Figure S4).
To test whether similar genotype effects could be detected in postmortem brain, we used RNA sequencing data obtained from human dorsolateral prefrontal cortex. 27 Baseline expression of TRANK1 was very low, and no genotype-specific effects were observed (Figure 2h). SNP rs9834970 also showed no evidence of association with TRANK1 expression in any of the 10 brain regions in BRAINEAC, 28 (Figure S5) or in any of the tissues reported by GTEX (http://www.gtexportal.org, accessed 1/12/2018). These results suggest that rs9834970 does not affect expression of TRANK1 in mature brain or exerts cell-type specific effects not easily detected in bulk tissue.
VPA Treatment Rescues Expression of TRANK1 in Risk-Allele Carriers.
Chronic treatment with therapeutic doses of VPA increased expression of TRANK1 in NPC carrying the risk allele of rs9834970, normalizing expression in those cells (Figure 2e, middle and right panels). Both 0.5 mM and 1 mM VPA significantly increased TRANK1 expression in risk allele carriers. ANOVA analysis confirmed a significant VPA by genotype interaction (F (1,46) =4.452; p<0.05). Fig 2e. shows that the increase in TRANK1 expression after VPA treatment presented in Fig 1a is driven by lines with AG or GG genotypes. Western blotting in cell lysates derived from NPC confirmed that TRANK1 protein expression was lower in risk (G) allele carriers than in AA homozygotes, and that VPA treatment increased expression of TRANK1 protein (Figure 2f-2g). We conclude that VPA treatment rescued expression of TRANK1 mRNA and protein in NPCs carrying the risk allele at rs9834970. No significant effect of genotype or VPA on TRANK1 expression was detected in neurons (Data not shown).
Predicted regulatory effects of rs9834970 and nearby SNPs.
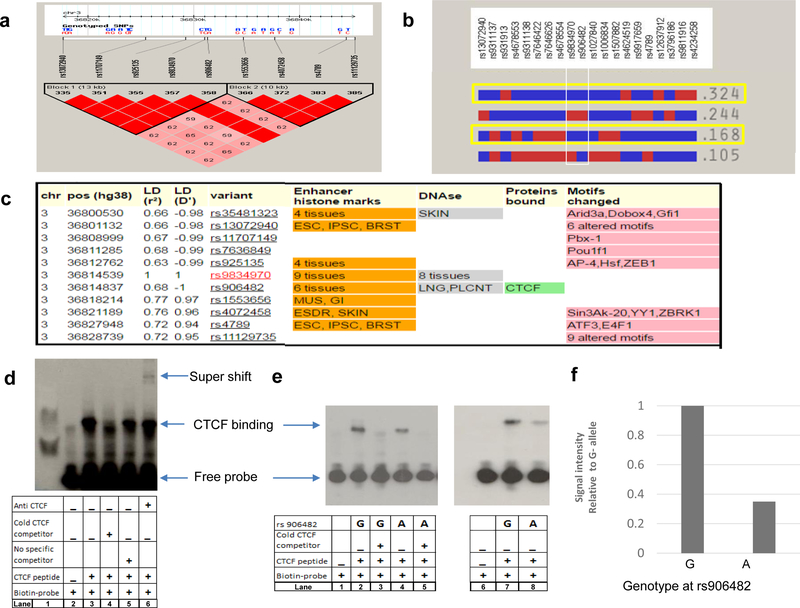
Several common SNPs are in strong linkage disequilibrium with rs9834970 (Figure 3a). Five of these SNPs have been associated with BD and/or schizophrenia in previous GWAS. 20, 22–24, 29 In individuals of European ancestry, SNPs in this region form 4 common haplotypes, two of which carry the rs9834970 risk allele (Figure 3b, yellow highlight). Both haplotypes also carry the “G” allele at rs906482 (Fig 3b, white boxed region).
Figure 3. Allele-specific effects on binding of CTCF protein to DNA.
a, Linkage disequilibrium (LD) plot. Common SNPs in the region fall into two large haplotype blocks. LD calculated as D’ in HapMAP Phase III CEU samples using Haploview 4.1; haplotype blocks based on confidence intervals 56. Red squares depict D’ values of 100%; pink squares show D’ values <100%. b, Block 1 haplotypes with CEU frequencies >5%. Both common haplotypes that carry the risk allele at rs9834970 (outlined in yellow) also carry the G-allele at of rs906482. c, Functional annotation of common SNPs in LD with rs9834970. SNPs with r2>0.6 in CEU were annotated with the core 15-state (ChromHMM) model using HaploReg v4.1. Several SNPs alter chromatin state, DNAse I sensitive sites, or regulatory motifs. Only rs906482 alters protein binding by CTCF. Abbreviations: ESC, embryonic stem cells; IPSC, induced pluripotent stem cells; BRST, mammary epithelial cells; SKIN, foreskin keratinocyte; LNG, fetal lung; PLCNT, placenta; MUS, fetal muscle leg; GI, small intestine; ESDR H1-derived neuronal progenitor cells. d, Representative EMSA plot of CTCF binding around rs906482. Lane 1 shows MW marker, lane 2 shows probe-only (negative) control, lane 3 shows probe plus CTCF peptide (positive) control, lane 4 shows that excess unlabeled (“cold”) CTCF competitor displaced binding of biotin-labeled CTCF, lane 5 shows binding of biotin-labeled CTCF with no competitor, lane 6 shows increased MW band due to binding by anti-CTCF antibody (“supershift”). e, Genotype-specific differential DNA binding by CTCF. Lane 1 shows probe-only (negative) control, lane 2 shows CTCF binding around G-allele, lane 3 shows displacement by excess unlabeled (“cold”) competitor, lane 4 shows CTCF binding around A-allele, lane 5 shows displacement by unlabeled (“cold”) competitor, lanes 6 to 8 show replicate assays corresponding to lanes 1, 4, and 2. f, Quantification of signal intensities from e., lanes 7 and 8. Statistical significance was tested by Student’s t-test.
Functional annotation by the Roadmap Epigenomics and ENCODE projects 30 indicates many SNPs in this region have an impact on chromatin state or transcription factor binding in various tissues (Figure 3c), but only rs906482 alters DNA-protein binding. This SNP alters binding by the zinc finger protein, CTCF, in multiple cell types (http://insulatordb.uthsc.edu). Since CTCF is an important gene regulatory protein in vertebrates, 31 we further investigated the impact of rs906482 on CTCF binding in NPCs. Electrophoretic mobility shift assays (EMSA) were performed using probes surrounding rs906482. CTCF peptide displayed significantly higher affinity for the (G) allele at rs906482 (Figure 3d-f). As noted above, this allele usually resides on the same haplotype as the risk (G) allele of rs9834970 (Figure 3c). These results suggest that common genetic variation near TRANK1 affects expression through a CTCF-mediated mechanism.
In light of these results, we repeated the eQTL analyses with rs906482 genotype as the independent variable. The results were similar to those observed for rs9834970, with an even stronger baseline relationship between genotype and TRANK1 expression. As with rs9834970, VPA also rescued TRANK1 expression in carriers of the G-allele at rs906842 (Figure S6).
Genome-wide gene expression profiles.
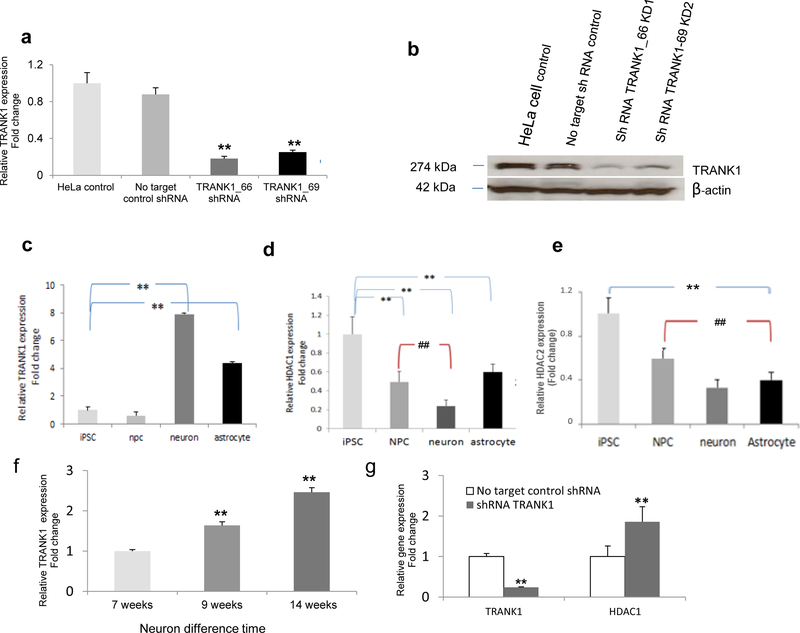
To explore the impact of decreased TRANK1 expression at the cellular level, two stable knockdowns of TRANK1 were generated with TRANK1 shRNA lentiviral constructs in HeLa cells. The extent of TRANK1 knockdown assessed at both mRNA and protein levels was substantial (Figure 4a-4b).
Figure 4. Inverse relationship between expression of TRANK1 and HDAC1 or HDAC2.
a, TRANK1 protein expression in stable knockdown Hela cell lines, shRNA TRANK1_66 and shRNA TRANK1_69, relative to no-target (scrambled) shRNA. Values based on average of three Western blots. Baseline expression in HeLa lines is shown on the left. b, Representative Western blot.
Relative mRNA expression of c, TRANK1 d, HDAC1, and e, HDAC2, in iPSC (n=4), NPC (n=11), neurons (n=5), and astrocytes (n=2). **p<0.01, all cell types compared to iPSCs; ##p<0.01, neurons, and astrocytes compared to NPCs. f, Increase in TRANK1 mRNA expression during 14 weeks of neuronal differentiation and maturation. **p<0.01, compared to 7-wk neurons. g. Relative HDAC1 and TRANK1 expression in TRANK1 knockdown HeLa cells (n=6). **p<0.01, compared to no target control shRNA.
Four gene-specific shRNA sequences designed for human TRANK1 gene; one negative construct and one “non-target” construct were transduced separately into HeLa cells (Table S7). HeLa cells were seeded in 48-well plates (50% confluent) overnight, and infected with lentivirus at multiplicity of infection (MOL) of 10, with serum-free, antibiotic-free DMEM media containing 8 μg/mL of Polybrene (Sigma Aldrich, St. Louis, MO). After six hours, the lentiviral shRNA medium was removed and replaced with DMEM-containing 20% FBS. Forty-eight hour post-transfection, the cells were seeded into a 6-well plate and propagated in complete DMEM medium with 20% FBS and 3 μg/ml puromycin until colonies were visible. The puromycin-resistant clones were picked and expanded, and the knockdown efficiency was verified by quantitative RT-PCR and western blot. The two constructs with the best knockdown efficiency (over 70%), along with non-target shRNA control, were used for gene expression array studies.
Genome-wide gene expression profiling of knockdown lines revealed many differentially expressed genes. The 214 genes that were differentially expressed with an absolute fold change >1.75 at an FDR<0.05 (Table S1) were enriched for increased cell proliferation and survival and decreased apoptosis (Table 1a). Gene set enrichment analysis (GSEA) indicated that these differentially expressed genes were strongly enriched for the Gene Ontology (GO) terms (Table S2) “regulation of cell proliferation,” “multicellular organismal development,” and “system development.”
Table 1.
Cellular growth and proliferation differences in a, TRANK1 shRNA KD in HeLa cell lines. b, VPA treated on iPSC derived neural progenitor cells.
| a | Diseases or Functions Annotation | p-Value | Predicted Activation State | Activation z-score | # Molecules |
| proliferation of cells | 1.08E-17 | Increased | 2.264 | 93 | |
| proliferation of lymphocytes | 1.14E-08 | Increased | 2.088 | 27 | |
| proliferation of neuronal cells | 4.02E-05 | Increased | 2.598 | 18 | |
| stimulation of cells | 2.04E-12 | Increased | 2.028 | 21 | |
| Attachment of cells | 3.16E-06 | Increased | 2.121 | 9 | |
| Cell survival | 1.98E-10 | Increased | 2.140 | 43 | |
| Apoptosis of muscle cell lines | 4.82E-08 | Decreased | −2.759 | 8 | |
| b | Diseases or Functions Annotation | p-Value | Predicted Activation State | Activation z-score | # Molecules |
| formation of cells | 2.76E-04 | Decreased | −2.039 | 29 | |
| proliferation of Schwann cells | 3.23E-04 | −1.948 | 4 | ||
| proliferation of cells | 8.10E-15 | −1.793 | 109 | ||
| formation of connective tissue cells | 3.08E-04 | −1.622 | 10 | ||
| growth of axons | 3.75E-07 | −1.300 | 12 | ||
To further examine the effect of TRANK1 knockdown, we assessed cellular proliferation quantitatively using the MTT assay (Suppl. Methods). As expected, cellular proliferation was significantly increased in the knockdown lines at 2–3 days, compared with the no-target scrambled controls (p<0.001, Figure S8).
Since VPA treatment increased TRANK1 expression, we expected that VPA treatment of iPSC-derived NPCs would produce a gene expression profile that contrasted with that seen in the TRANK1 knockdown. The 304 genes differentially expressed in VPA-treated NPCs with an absolute fold change >1.75 at an FDR<0.05 were analyzed (Table S3). As expected, differentially expressed genes were enriched for decreased proliferation of cells and decreased growth of axons (Table 1B). Similarly, GSEA revealed significant enrichment for GO terms related to “cell differentiation,” “nervous system development,” “neurogenesis,” and “neuron differentiation” (Table S4). Treatment of NPCs with VPA also substantially decreased cell proliferation in vitro (Figure S8.)
Increasing TRANK1 expression with differentiation and inverse relationship with expression of histone deacetylases.
Among the genes whose expression was increased most by TRANK1 knockdown (Table S1) was histone deacetylase 1 (HDAC1), which is known to be inhibited by VPA 32. To more fully characterize the relationship between TRANK1 and HDAC1, we compared relative HDAC1 and TRANK1 expression in iPSCs, NPCs, neurons, and astrocytes. TRANK1 expression was highest in neurons (Figure 4c). In contrast, HDAC1 expression was highest in iPSCs and NPCs (Figure 4d-4e). TRANK1 expression levels rose steadily during neuronal maturation (Figure 4f). In contrast, knockdown of TRANK1 in HeLa cells led to a substantial increase in HDAC1 expression (Figure 4g). These data suggest a reciprocal relationship between TRANK1 expression and HDAC1 in developing cells.
DISCUSSION
This study used human iPSCs and neural derivatives to investigate the impact of common genetic variation and drug treatment on gene expression at a locus implicated by GWAS of major mental illnesses. The results demonstrated a link between common SNPs at the locus and expression of the nearby gene, TRANK1, in neural cells. The results also showed, for the first time, that the mood stabilizer, VPA, rescues TRANK1 expression in neural cells carrying risk alleles at this locus. Although rs9834970 has no known function, an associated nearby SNP, rs906482, strongly affects binding by the transcription factor, CTCF. Decreased expression of TRANK1 perturbed many genes involved in neural development, differentiation, and apoptosis.
This study has some limitations. The number of iPSC-derived cell lines was limited, reducing power to detect subtle effects. Additional signals may emerge as sample size increases. Both rs9834970 and rs906482 reside upon a haplotype that carries many common variants related to chromatin regulation and transcription factor binding (Figure S9), so it is possible that additional SNPs contribute to the biological impact of the risk haplotype. GWAS studies have also identified other SNPs associated with BD and schizophrenia in the TRANK1 region, 2, 21, 24, 33 some of which are not in strong linkage disequilibrium with rs9834970. Further studies that use genome editing techniques such as CRISPR/Cas9 to generate isogenic controls would be needed to establish the functional impact of each SNP. Variants within the locus may affect additional genes not investigated in the present study. Some of the genome-wide expression data was based on shTRANK1 knockdown in HeLa cell lines, which are very different from NPCs. We used HeLa since it is a well-studied line with high baseline expression of TRANK1.
Drugs can be an important influence on gene expression. This study focused on VPA since it is an effective treatment for BD and demonstrated robust effects on TRANK1 expression in previous studies. 20 However, the other drug we studied, lithium, had no apparent impact on TRANK1 expression. The TRANK1 region has also been associated with schizophrenia, and VPA is not known to exert a therapeutic effect in that mental illness. Further studies will be needed to explore the full range of drugs that affect expression of TRANK1 in neural cells.
These results suggest that the therapeutic effects of VPA in BD may be due in part to expression of TRANK1. VPA normalized TRANK1 expression in cells carrying the risk haplotype. Moreover, TRANK1 regulated many of the same cellular growth and differentiation pathways that are known to be affected by VPA, highlighting a key role for histone deacetylases, known targets of VPA. VPA affected TRANK1 expression mainly in growing cells, consistent with the observation that exposure to VPA is teratogenic during early nervous system development. 33, 34 The failure of neurons to display increased TRANK1 expression after VPA treatment may reflect the high baseline expression in neurons. However, the number of neuron lines was limited and we noted considerable variation in baseline TRANK1 expression across lines, so it is possible that subtle effects were missed. It is not obvious from these data how VPA would exert a beneficial effect in the mature brain. VPA did have a significant impact on TRANK1 expression in astrocytes, suggesting one way that the therapeutic window could extend into adulthood. Recent work has highlighted the potential importance of astrocytes and other glia in the pathophysiology of neuropsychiatric disorders.35–37
This study demonstrated reciprocal effects of TRANK1 and HDAC1 on gene expression, cell proliferation, and differentiation. We observed an increase in TRANK1 expression over baseline in more-differentiated cells, where HDAC1 expression was decreased. Proof of a causal relationship between TRANK1 and HDAC1 expression would require experimental overexpression of TRANK1, beyond the scope of the present study. These data may be consistent with a model whereby HDAC1 inhibition by VPA normalizes TRANK1 expression in carriers of the risk allele. However, VPA affects many genes by inhibiting histone deacetylases, so the effect of VPA on TRANK1 expression is not specific. Future studies are needed to assess whether similar cellular phenotypes follow treatment with other HDAC inhibitors. In any case, our results are consistent with the literature implicating VPA and other histone deacetylase inhibitors in neuronal growth and differentiation. 34, 38, 39
While the precise mechanism whereby VPA and genetic variation near TRANK1 affect gene expression remains to be fully elucidated, our findings suggest an important role for CTCF. We showed that genetic variation within risk haplotypes alters CTCF binding affinity, and that decreased CTCF binding at this locus correlates with increased expressed of TRANK1. Recent CHIP-seq data from the ENCODE Project also shows substantial CTCF binding in the vicinity of rs906482. CTCF influences gene expression by acting as a transcriptional insulator, orchestrating long-range DNA-looping interactions between distal enhancers and their cognate promoters.40 Genome-wide analysis of CTCF has demonstrated its crucial role in higher-order chromatin organization that can increase or curtail enhancer-promoter interactions, depending on the relative positions of these regulatory elements. 25 Increased CTCF binding to the risk allele may block the effect of a distal enhancer that otherwise activates TRANK1 expression. Finding that enhancer is an important future goal and may suggest other therapeutic approaches to the regulation of TRANK1 expression in brain.
We speculate that CTCF may also play a role in the mechanism whereby VPA impacts TRANK1 expression. Previous studies have shown that CTCF can recruit histone deacetylases to the transcription activation complex. 41 VPA also decreases CTCF expression. 42 Thus, VPA may directly antagonize gene repression by CTCF.
This study illustrates a potential strategy to investigate GWAS findings in neuropsychiatric disorders. Genotype-specific iPSC-derived cells can be used to study allelic variation and gene expression across a range of cell types, developmental stages, and environmental conditions, offering a degree of cellular, temporal, and experimental resolution not possible with blood or post-mortem brain. We report what appears to be a cell-type specific effect of a cis-regulatory variant on gene expression, consistent with recent studies demonstrating that cis-regulation can differ between brain regions, 43 developmental stages, 44 and cell types. 45 iPSCs and neural derivatives also provide a platform for screening therapeutic drugs in patient-specific cells. We have shown how such studies can identify potentially causal genetic variants, enhance understanding of disease mechanisms, and illuminate relationships between genetic variation and treatments for neuropsychiatric disorders. More work is needed to develop scalable, high-throughput strategies for screening larger numbers of genetic risk variants, especially those whose impact is specific to mature cells or multi-cellular functions, such as neural circuits.
METHODS
Reprogramming and characterization of iPSCs
Eleven iPSC lines were used in this study (Table S5). All were generated from fibroblast cultures using lentiviral transduction with STEMCCA vector (Millipore). 46 Pluripotency was assessed by immunostaining for pluripotency markers (Supplementary Methods), flow cytometry quantification (Supplementary Methods), qPCR for specific markers, and PluriTest. 47 IPSC-derived cells were characterized by standard immunochemical analysis, qRT-PCR, and electrophysiology (Supplementary Methods). iPS clones that displayed a normal karyotype were selected for further studies.
Generation of NPCs and differentiation into neurons and astrocytes
iPSC lines were differentiated into NPCs using AggreWell™ 800 plates (StemCell Technologies) in STEMdiff™ neural induction medium. Neuronal differentiation was induced by Neurobasal (Invitrogen), supplemented with 1× B27 (Invitrogen), 10 ng/ml BDNF (Peprotech), 10 ng/ml GDNF (Peprotech), L-ascorbic acid (200 ng/ml), and cAMP (1μM) (Sigma-Aldrich). The cells were cultured in neuron medium for >2 mo. For astrocyte differentiation, NPCs were plated on Geltrex-coated culture dishes in STEMdiff™ Neural Progenitor Medium. After 2 days, medium was changed to DMEM, 1% N2 supplement (Invitrogen), 1% FBS, 20 ng/ml CTNF (R&D Systems). After 6–8 weeks, 60–80% cells were positive for astrocyte-specific markers.
Hippocampal neurons were prepared from fetal rat brain at embryonic day 18 (E18) as described. 48
Genotyping and quantitative real time PCR (qPCR)
Expression of selected genes was determined using Roche LightCycler 480 and Roche Universal ProbeLibrary System. The comparative CT method was used to quantify relative mRNA levels. Primer sequences are presented in Table S6.
All samples were genotyped on the Illumina Infinium Human OmniExpress Exome bead array. To further validate genotypes in NPCs and detect rare variants, 400 bp flanking rs9834970 was Sanger-sequenced at Macrogen. (Figure S10).
Western Blotting
Protein isolation was performed with M-PER lysis buffer (ThermoScientific, Catalog 78503). Concentration was determined using a spectrophotometer. 25ug protein was loaded onto 4–12% Tris-Glycine gels, electrophoresed, and transferred with iBlot 2 onto nitrocellulose membranes (Life Technologies). Membranes were blocked, incubated with anti-TRANK1 antibody washed 5X with TBST, and incubated with secondary antibody After washing, membranes were exposed to Amersham™ ECL™ Western blotting detection reagents (GE Healthcare) and developed. ImageJ was used for quantification.
RNA interference
MISSION™ shRNA Lentiviral Transduction Particles (Sigma Aldrich) were used for the knockdown study. Four gene-specific shRNA sequences designed for human TRANK1, one negative construct and one “non-target” construct were transduced separately into HeLa cells (Table S7). The two constructs with >70% knockdown efficiency, along with a scrambled non-target shRNA control, were used for expression array studies.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed with the Gel shift chemiluminescent kit (Active Motif). Table S8 lists sequences for synthetic double-stranded consensus CTCF and 5’ biotin-labeled oligonucleotides corresponding to rs906482. Sequence of consensus CTCF double-stranded oligonucleotide was derived from a site previously described to bind to CTCF. CTCF-associated peptide was affinity purified as described. 50 Oligonucleotide probes were designed with both alleles of rs906482, flanked by14 bp, in both a cold and 5’-biotinylated form (IDT). For each binding reaction, 5–10 pmol purified CTCF peptide was incubated for 20 min at room temperature with 20 fmol biotin-labeled duplex oligonucleotide containing the A or G allele, with or without consensus CTCF oligonucleotide in binding buffer. Super-shift was generated by anti-CTCF antibody. After incubation, the mixture was loaded on a 6% DNA retardation gel (EC6365BOX, Life Technologies), separated by electrophoresis, and transferred to a nylon membrane that was UV cross-linked and detected by chemiluminescence using stabilized streptavidin–horseradish peroxidase conjugate (Pierce). For competition assays, unlabeled consensus oligonucleotides at 100-fold molar excess were added before the biotin-labeled probe. Bands were quantified using Image J.
Microarray Analysis
Gene expression profiling was performed on RNA extracted from: a) NPC lines treated for 72 h with 0.5 mM VPA or vehicle; b) HeLa lines with stable knockdown by TRANK1 shRNA particles; and c) one HeLa line with no target shRNA control (n=8 for each condition). Groups were equally divided between array plates and hybridization batches. Total RNA, extracted with the RNeasy Mini kit (QIAGEN, Hilden, Germany) was used for cRNA amplification, Cy3 labeling, and hybridization onto Illumina HT-12_V4 beadchips. Microarray data were assembled using Genome Studio V3.0 (Illumina). Raw expression data were log2 transformed and quantile normalized using a custom R script calling Bioconductor (www.bioconductor.org). Outliers were checked based on inter-sample correlations and principal component analysis. Transcripts were considered robustly expressed and included in the analysis if the coefficient of variation lay within the linear phase of the distribution. Differential expression was tested by one-way analysis of variance. Gene-set enrichment analysis was carried out with DAVID, 51, 52 at medium stringency, with background comprising the total set of robustly-expressed probes. Differentially-expressed genes were also analyzed with GeneSpring.
Supplementary Material
ACKNOWLEDGMENTS
Supported by the Intramural Research Programs of the National Institute of Mental Health (NIMH; ZIA-MH00284311/NCT00001174), National Institute of Neurological Disease and Stroke (NINDS), and the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health. The authors have no conflicts of interest to disclose, financial or otherwise. Dr. Kevin Chen, at the NINDS Stem Cell Unit helped with Flow Cytometry Analysis; Dr. Wei Lu at the Synapse and Neural Circuit Unit (NINDS) helped with electrophysiological recordings; Dr. Mahendra Rao, formerly of the Center for Regenerative medicine (CRM), provided 2 neural progenitor lines; Dr. Manfred Boehm (Laboratory of Cardiovascular Regenerative Medicine, NHLBI) provided 5 iPSC lines; Dr. Kory Johnson (NINDS) Microarray Core helped analyze the microarray gene expression data; Drs. Amalia Dutra and Evgenia Pak of the NHGRI Cytogenetics Core performed spectral karyotyping. GM05990, GM23240, and GM23476 were obtained from Coriell Cell Repositories (Camden, NJ). Line 10593 was obtained from the Rutgers University Cell and DNA Repository (Piscataway, NJ; catalog #10C117904). Special thanks to Ioline Henter (NIMH), who provided invaluable editorial assistance.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Richards AL, Jones L, Moskvina V, Kirov G, Gejman PV, Levinson DF et al. Schizophrenia susceptibility alleles are enriched for alleles that affect gene expression in adult human brain. Mol Psychiatry 2012; 17(2): 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shinozaki G, Potash JB. New developments in the genetics of bipolar disorder. Curr Psychiatry Rep 2014; 16(11): 493. [DOI] [PubMed] [Google Scholar]
- 3.Kirsten H, Al-Hasani H, Holdt L, Gross A, Beutner F, Krohn K et al. Dissecting the genetics of the human transcriptome identifies novel trait-related trans-eQTLs and corroborates the regulatory relevance of non-protein coding loci. Hum Mol Genet 2015; 24(16): 4746–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013; 45(6): 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu F, Wang X, Hu G, Wang Y, Zhou J. The transcription factor TEAD1 represses smooth muscle-specific gene expression by abolishing myocardin function. The Journal of biological chemistry 2014; 289(6): 3308–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Gobbi M, Anguita E, Hughes J, Sloane-Stanley JA, Sharpe JA, Koch CM et al. Tissue-specific histone modification and transcription factor binding in alpha globin gene expression. Blood 2007; 110(13): 4503–4510. [DOI] [PubMed] [Google Scholar]
- 7.Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M et al. Human genomics. The human transcriptome across tissues and individuals. Science 2015; 348(6235): 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gay MH, Valenta T, Herr P, Paratore-Hari L, Basler K, Sommer L. Distinct adhesion-independent functions of beta-catenin control stage-specific sensory neurogenesis and proliferation. BMC Biol 2015; 13: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Latham KE. Stage-specific and cell type-specific aspects of genomic imprinting effects in mammals. Differentiation 1995; 59(5): 269–282. [DOI] [PubMed] [Google Scholar]
- 10.Rueckert EH, Barker D, Ruderfer D, Bergen SE, O’Dushlaine C, Luce CJ et al. Cis-acting regulation of brain-specific ANK3 gene expression by a genetic variant associated with bipolar disorder. Mol Psychiatry 2013; 18(8): 922–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korecka JA, Levy S, Isacson O. In vivo modeling of neuronal function, axonal impairment and connectivity in neurodegenerative and neuropsychiatric disorders using induced pluripotent stem cells. Mol Cell Neurosci 2016; 73: 3–12. [DOI] [PubMed] [Google Scholar]
- 12.Schwartzentruber J, Foskolou S, Kilpinen H, Rodrigues J, Alasoo K, Knights AJ et al. Molecular and functional variation in iPSC-derived sensory neurons. Nat Genet 2018; 50(1): 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C et al. Mosaic copy number variation in human neurons. Science 2013; 342(6158): 632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim DS, Lee JS, Leem JW, Huh YJ, Kim JY, Kim HS et al. Robust enhancement of neural differentiation from human ES and iPS cells regardless of their innate difference in differentiation propensity. Stem Cell Rev 2010; 6(2): 270–281. [DOI] [PubMed] [Google Scholar]
- 15.Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 2014; 515(7527): 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K et al. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry 2015; 20(3): 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mertens J, Wang QW, Kim Y, Yu DX, Pham S, Yang B et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature 2015; 527(7576): 95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bidinosti M, Botta P, Kruttner S, Proenca CC, Stoehr N, Bernhard M et al. CLK2 inhibition ameliorates autistic features associated with SHANK3 deficiency. Science 2016; 351(6278): 1199–1203. [DOI] [PubMed] [Google Scholar]
- 19.Forrest MP, Zhang H, Moy W, McGowan H, Leites C, Dionisio LE et al. Open Chromatin Profiling in hiPSC-Derived Neurons Prioritizes Functional Noncoding Psychiatric Risk Variants and Highlights Neurodevelopmental Loci. Cell Stem Cell 2017; 21(3): 305–318e308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen DT, Jiang X, Akula N, Shugart YY, Wendland JR, Steele CJ et al. Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Molecular psychiatry 2013; 18(2): 195–205. [DOI] [PubMed] [Google Scholar]
- 21.Ruderfer DM, Fanous AH, Ripke S, McQuillin A, Amdur RL, Gejman PV et al. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol Psychiatry 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goes FS, Hamshere ML, Seifuddin F, Pirooznia M, Belmonte-Mahon P, Breuer R et al. Genome-wide association of mood-incongruent psychotic bipolar disorder. Transl Psychiatry 2012; 2: e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cross-Disorder Group of the Psychiatric Genomics C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013; 381(9875): 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511(7510): 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007; 318(5858): 1917–1920. [DOI] [PubMed] [Google Scholar]
- 26.Bergen SE, O’Dushlaine CT, Ripke S, Lee PH, Ruderfer DM, Akterin S et al. Genome-wide association study in a Swedish population yields support for greater CNV and MHC involvement in schizophrenia compared with bipolar disorder. Mol Psychiatry 2012; 17(9): 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akula N, Barb J, Jiang X, Wendland JR, Choi KH, Sen SK et al. RNA-sequencing of the brain transcriptome implicates dysregulation of neuroplasticity, circadian rhythms and GTPase binding in bipolar disorder. Mol Psychiatry 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kreutzer J, Yla-Outinen L, Maki AJ, Ristola M, Narkilahti S, Kallio P. Cell culture chamber with gas supply for prolonged recording of human neuronal cells on microelectrode array. J Neurosci Methods 2017; 280: 27–35. [DOI] [PubMed] [Google Scholar]
- 29.Muhleisen TW, Leber M, Schulze TG, Strohmaier J, Degenhardt F, Treutlein J et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat Commun 2014; 5: 3339. [DOI] [PubMed] [Google Scholar]
- 30.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res 2016; 44(D1): D877–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghirlando R, Felsenfeld G. CTCF: making the right connections. Genes Dev 2016; 30(8): 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hasan MR, Kim JH, Kim YJ, Kwon KJ, Shin CY, Kim HY et al. Effect of HDAC inhibitors on neuroprotection and neurite outgrowth in primary rat cortical neurons following ischemic insult. Neurochem Res 2013; 38(9): 1921–1934. [DOI] [PubMed] [Google Scholar]
- 33.Kim BW, Yang S, Lee CH, Son H. A critical time window for the survival of neural progenitor cells by HDAC inhibitors in the hippocampus. Mol Cells 2011; 31(2): 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu IT, Park JY, Kim SH, Lee JS, Kim YS, Son H. Valproic acid promotes neuronal differentiation by induction of proneural factors in association with H4 acetylation. Neuropharmacology 2009; 56(2): 473–480. [DOI] [PubMed] [Google Scholar]
- 35.Chung WS, Welsh CA, Barres BA, Stevens B. Do glia drive synaptic and cognitive impairment in disease? Nat Neurosci 2015; 18(11): 1539–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng L, Li B, Verkhratsky A. Targeting astrocytes in bipolar disorder. Expert Rev Neurother 2016; 16(6): 649–657. [DOI] [PubMed] [Google Scholar]
- 37.Stevens B, Muthukumar AK. Cellular neuroscience. Differences among astrocytes. Science 2016; 351(6275): 813. [DOI] [PubMed] [Google Scholar]
- 38.Gottlicher M, Minucci S, Zhu P, Kramer OH, Schimpf A, Giavara S et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J 2001; 20(24): 6969–6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ganai SA, Ramadoss M, Mahadevan V. Histone Deacetylase (HDAC) Inhibitors - emerging roles in neuronal memory, learning, synaptic plasticity and neural regeneration. Curr Neuropharmacol 2016; 14(1): 55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo Y, Fu X, Jin Y, Sun J, Liu Y, Huo B et al. Histone demethylase LSD1-mediated repression of GATA-2 is critical for erythroid differentiation. Drug Des Devel Ther 2015; 9: 3153–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lutz M, Burke LJ, Barreto G, Goeman F, Greb H, Arnold R et al. Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res 2000; 28(8): 1707–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oti M, Falck J, Huynen MA, Zhou H. CTCF-mediated chromatin loops enclose inducible gene regulatory domains. BMC Genomics 2016; 17: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buonocore F, Hill MJ, Campbell CD, Oladimeji PB, Jeffries AR, Troakes C et al. Effects of cis-regulatory variation differ across regions of the adult human brain. Human molecular genetics 2010; 19(22): 4490–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burkhardt MF, Martinez FJ, Wright S, Ramos C, Volfson D, Mason M et al. A cellular model for sporadic ALS using patient-derived induced pluripotent stem cells. Molecular and Cellular Neuroscience 2013; 56: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Won H, de la Torre-Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 2016; 538(7626): 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cooper AR, Patel S, Senadheera S, Plath K, Kohn DB, Hollis RP. Highly efficient large-scale lentiviral vector concentration by tandem tangential flow filtration. J Virol Methods 2011; 177(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muller FJ, Brandl B, Loring JF. Assessment of human pluripotent stem cells with PluriTest. StemBook: Cambridge (MA), 2008. [PubMed] [Google Scholar]
- 48.Jiang X, Tian F, Du Y, Copeland NG, Jenkins NA, Tessarollo L et al. BHLHB2 controls Bdnf promoter 4 activity and neuronal excitability. J Neurosci 2008; 28(5): 1118–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao T, Wallace J, Felsenfeld G. Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex and are required for cohesin-dependent insulation activity. Mol Cell Biol 2011; 31(11): 2174–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yusufzai TM, Felsenfeld G. The 5’-HS4 chicken beta-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc Natl Acad Sci U S A 2004; 101(23): 8620–8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4(1): 44–57. [DOI] [PubMed] [Google Scholar]
- 52.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37(1): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.