Abstract
Microrchidia family CW-type zinc finger 2 (MORC2) is a newly identified chromatin remodeling enzyme with an emerging role in DNA damage response (DDR), but the underlying mechanism remains largely unknown. Here, we show that poly(ADP-ribose) polymerase 1 (PARP1), a key chromatin-associated enzyme responsible for the synthesis of poly(ADP-ribose) (PAR) polymers in mammalian cells, interacts with and PARylates MORC2 at two residues within its conserved CW-type zinc finger domain. Following DNA damage, PARP1 recruits MORC2 to DNA damage sites and catalyzes MORC2 PARylation, which stimulates its ATPase and chromatin remodeling activities. Mutation of PARylation residues in MORC2 results in reduced cell survival after DNA damage. MORC2, in turn, stabilizes PARP1 through enhancing acetyltransferase NAT10-mediated acetylation of PARP1 at lysine 949, which blocks its ubiquitination at the same residue and subsequent degradation by E3 ubiquitin ligase CHFR. Consequently, depletion of MORC2 or expression of an acetylation-defective PARP1 mutant impairs DNA damage-induced PAR production and PAR-dependent recruitment of DNA repair proteins to DNA lesions, leading to enhanced sensitivity to genotoxic stress. Collectively, these findings uncover a previously unrecognized mechanistic link between MORC2 and PARP1 in the regulation of cellular response to DNA damage.
INTRODUCTION
Cellular DNA is constantly damaged by both exogenous and endogenous genotoxic agents. Inefficient or inaccurate repair of damaged DNA could lead to genomic instability and carcinogenesis (1). To circumvent the deleterious effects of DNA damage, cells timely activate highly coordinated DNA damage response (DDR) network to repair damaged DNA (2). Eukaryotic DNA is packaged into chromatin, a highly condensed structure that intrinsically impedes the access of DNA repair machinery to DNA lesions (3,4). Consequently, dynamic remodeling of chromatin structure is essential for efficient DNA repair, which involves a concerted action of multiple chromatin-associated enzymes (5). However, how this is accomplished remains largely elusive.
Microrchidia family CW-type zinc finger 2 (MORC2) is a member of the evolutionarily conserved MORC ATPase superfamily, comprising four poorly characterized proteins including MORC1–4 (6–8). These proteins are characterized by the presence of an N-terminal catalytically active ATPase module and a central CW-type zinc finger (CW-ZF) domain (6–8). The ATPase module is composed of gyrase, Hsp90, histidine kinase, and MutL (GHKL) and S5-fold domains, which has been mechanistically linked to gene transcription and DNA repair by remodeling chromatin (7,9,10). The CW-ZF domain is present in several chromatin-associated proteins and plays a role in DNA binding and/or promoting protein–protein interactions in eukaryotic processes (8,11,12). In addition, MORC2 contains a C-terminal chromo-like domain, which is commonly found in eukaryotic chromatin proteins and can recognize methylated peptides in histones and non-histone proteins (13). These structural features indicate that MORC2 is potentially implicated in chromatin biology. Indeed, emerging evidence shows that MORC2 regulates heterochromatin formation and epigenetic gene silencing through an association with human silencing hub (HUSH) complex (14). In addition, we recently demonstrated that MORC2 is phosphorylated by p21-activated kinase 1 (PAK1) at serine 739 in response to DNA damage and facilitates ATPase-dependent chromatin remodeling and efficient DNA repair (10). However, the mechanism by which MORC2 is recruited to DNA damage sites and regulates DNA repair signaling is not completely understood.
One of the earliest events of cellular response to DNA damage is the recruitment of poly(ADP-ribose) polymerase 1 (PARP1), a highly abundant chromatin-associated enzyme, to DNA damage sites (15,16). Upon binding to DNA strand breaks, PARP1 is dramatically activated and catalyzes the synthesis of poly(ADP-ribose) (PAR) polymers at sites of DNA damage with two main consequences (15–17). First, PAR chains are covalently attached to acceptor proteins including itself and histones (a process known as PARylation), leading to chromatin relaxation that tends to increase the accessibility of DNA repair proteins to DNA lesions (17). Second, PAR serves as a chromatin-based platform for the recruitment of DNA repair factors possessing specific PAR-interacting motifs to sites of DNA lesions via non-covalent interactions, facilitating chromatin remodeling and DNA repair (15,17). PAR production is a tightly controlled process, and the rapid turnover of PAR is mainly mediated by poly(ADP-ribose) glycohydrolase (PARG), an enzyme with both endo- and exoglycosidase activities (18). Consistent with its indispensable role in DNA repair, PARP1-deficient cells are sensitive to various DNA-damaging agents (19,20). Consequently, several PARP inhibitors are being exploited clinically for the treatment of human cancers with DNA repair deficiency through the mechanism of synthetic lethality (21). In addition to DNA damage-induced auto-PARylation, the function and activity of PARP1 are tightly regulated by a variety of post-translational modifications, such as ubiquitination (22,23) and acetylation (24). Despite these advances, the upstream regulatory signals and the downstream PARylation targets of PARP1 remain largely unknown.
In this study, we report a previously unrecognized mechanistic link between MORC2 and PARP1 in the regulation of DDR. On the one hand, PARylation of MORC2 by PARP1 enhances its chromatin remodeling activities, thereby facilitating efficient DNA repair. On the other hand, MORC2 stabilizes PARP1 though a crosstalk between NAT10-mediated acetylation and CHFR-mediated ubiquitination. Consequently, depletion of MORC2 or expression of an acetylation-defective PARP1 mutant impairs PAR-dependent DNA repair signaling. These findings help to understand the mechanism of a collaborative action of chromatin-associated enzymes during cellular response to DNA damage.
MATERIALS AND METHODS
Cell culture and treatment
Human breast cancer cell lines and human embryonic kidney 293T (HEK293T) cell line were obtained from Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). All cell lines were authenticated by short tandem repeat profiling and were verified to be free of mycoplasma. Cells were cultured in DMEM or RPMI1640 media (BasalMedia, Shanghai, China) supplemented with 10% fetal bovine serum (ExCell Bio, Shanghai, China) and 1× penicillin and streptomycin (BasalMedia). For drug treatment, cells were treated with the following inhibitors for the indicated times, 5 μM TSA, 5 mM NAM, 1 mM MMS, 10 μM MG-132, 5 μM Olaparib (Selleck, Houston, USA), 10 μM ADP-HPD ammonium salt dehydrate (Santa Cruz Biotech, Santa Cruz, USA) or 100 μg/ml cycloheximide (Cell Signaling Technology, Danvers, USA). Unless otherwise stated, all reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Expression vectors
Myc-DDK-MORC2, -PARP1 and -NAT10 cDNAs were obtained from Origene (Rockville, USA) and subcloned into the lentiviral vector pCDH-CMV-MCS-EF1-Puro (System Biosciences, Mountain View, USA) to generate Flag-, HA- or Myc-tagged expression vectors. These cDNAs were also cloned into pGEX-6P-1 vector with an N-terminal GST and C-terminal His tag for purification of recombinant proteins. Flag-His-CHFR cDNA was obtained from Vigene Biosciences (Rockville, USA) and subcloned into pCDH-CMV-MCS-EF1-Puro vector to generate N-terminal Flag- tagged expression vector. Amino-acid substitutions and the deletion mutants were generated by PCR-directed mutagenesis. All construct sequences were verified by DNA sequencing. The detailed information concerning expression constructs and the primers used for molecular cloning is provided in Supplementary Tables S1 and S2. LentiCas9-Blast and lentiGuide-Puro vectors were obtained from Addgene (Cambridge, USA). The short guide RNA (sgRNA) sequences for MORC2 (5′-AGTAGACTCAGGTGTTCGCT-3) and PARP1 (5′-GTCCAACAGAAGTACGTGCA −3′) were chosen according to the Web-based CRISPR design tool from the Zhang lab (http://www.genome-engineering.org/). Indicated sgRNA sequences were cloned into lentiGuide-Puro vector following the standard protocol (25). Short hairpin RNAs (shRNA) targeting human MORC2 and NAT10 and corresponding negative control shRNAs (shNC) were obtained from Origene. Small interfering RNAs (siRNA) targeting MORC2, PARP1, PARG and MACRO domain containing 1 (MacroD1), and corresponding negative control siRNAs (siNC) were purchased form GenePharma (Shanghai, China) (Supplementary Table S3).
Plasmid transfection and lentiviral infection
Transient plasmid transfection was performed using Neofect (TengyiBio, Shanghai, China) or Lipofectamine 2000 (Invitrogen, Waltham, USA) DNA transfection reagents according to the manufacturer’s protocol. Generation of stable cell lines expressing shRNAs or cDNAs was carried out as described previously (26,27). The knockout (KO) cell lines were generated using the CRISPR/Cas9 system as described previously (25), and were validated by immunoblotting analysis and Sanger sequencing. The genomic DNA of wild-type (WT) and KO cells was extracted and used for PCR amplification of sgRNA targeting regions (primers are listed in Supplementary Table S4), and purified PCR product was subjected to Sanger sequencing. SiRNA transfection was performed using Lipofectamine 2000 transfection reagents according to the manufacturer’s instructions. The efficiency of gene silencing was assessed by immunoblotting 48 h post-transfection.
Expression and purification of recombinant proteins
The GST-tagged construct in pGEX-6P-1 was expressed in Escherichia coli strain BL21 (DE3) by addition of 1 mM IPTG (Invitrogen) into LB medium at 16°C overnight and then purified on glutathione Sepharose 4B beads (GE Healthcare, Shanghai, China). All proteins having a C-terminal His-tag were expressed in E. coli strain BL21 (DE3) by induction with 1 mM IPTG at 16°C for 16–18 h. Proteins were purified from the supernatants of lysed cells using Ni-NTA agarose (Theremofisher, Shanghai, China) according to the manufacturer’s instructions.
Antibodies, immunoblotting, immunoprecipitation and immunofluorescence
The detailed information for primary antibodies used in this study is provided in Supplementary Table S5. Secondary antibodies for immunoblotting and immunofluorescence were from Cell Signaling Technology. Immunoblotting analysis, immunoprecipitation (IP) assays and immunofluorescent staining were carried out as described previously (26,27).
Real-time quantitative PCR
Total RNA was isolated using Trizol reagent (Invitrogen) and converted to cDNA using PrimeScript RT Master Mix (Takara, Dalian, China). Real-time quantitative PCR (qPCR) analyses were performed in triplicate using SYBR Premix Ex Taq (Takara) on an Eppendorf Mastercycler ep realplex4 instrument (Eppendorf, Germany). All qPCR data were normalized to GAPDH. Primer information is described in Supplementary Table S6.
Detection of PARylated proteins
IP and detection of PARylated proteins were performed under denaturing conditions as described previously with some modifications (28). Cells were lysed in NP-40 lysis buffer (50 mM Tris-HCl, pH8, 150 mM NaCl, 0.5% NP-40, 10% glycerol, 2 mM MgCl2 and 1 mM EDTA). To sustain PAR levels throughout the experimental procedure, specific PARG inhibitor ADP-HPD (10 μM) was added to the lysis buffer for all PARylation-related experiments (29). Lysates were subjected to IP analysis with 1–3 μg antibody overnight at 4°C on a rotating platform. After washing, the beads were heated to 100°C for 5 min in 1× SDS loading buffer. Immunoblotting analysis was carried out with an anti-PAR monoclonal antibody (Trevigen, #4335-MC-100).
In vitro PARylation assay
In vitro PARylation assay was performed as described previously with some modifications (30). Briefly, purified GST-MORC2 protein (1 μg) was incubated with 100 ng recombinant full-length PARP1 protein expressed in sf9 cells (Origene) in reaction buffer containing 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 10 mM MgCl2, 10% glycerol, 1 mM DTT, 4 ng/ml sonicated salmon sperm DNA (Invitrogen) and 300 μM nicotinamide adenine dinucleotide (NAD+) in the presence or absence of 5 μM Olaparib at 37°C for 30 min. The reaction was terminated by the addition of 2× SDS loading buffer and PARylation of MORC2 was detected by immunoblotting with an anti-PAR monoclonal antibody (Trevigen, #4335-MC-100).
PLA assays
PLA assays were performed by using Duo-link In Situ-Fluorescence kits from Sigma according to the manufacturer’s instructions. Immunofluorescence staining protocol was carried out until the primary antibody incubation. Secondary antibodies conjugated to PLA probes. Ligation and amplification were performed according to the manufacturer’s instructions. The co-localization of both proteins resulted in a red PLA signal, which was visualized with a Leica SP5 confocal laser scanning microscopy (Leica Microsystems, Buffalo Grove, USA). As the PLA technique requires two specific antibodies to give a red fluorescent signal, a single primary antibody was implied as a negative signal control.
Proteomic analysis
To analyze MORC2 interacting proteins, nuclear extracts from HeLa cells were subjected to IP assays with control IgG or an anti-MORC2 antibody. After extensive washing, the bound proteins were eluted by boiling in SDS sample buffer, resolved by SDS-PAGE, visualized by Coomassie Blue (CB) staining and subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis as described previously (31). Data from LC-MS/MS analysis were searched against Swiss-Prot database by SEQUEST. Trans Proteomic Pipeline software (Institute of Systems Biology, Seattle, USA) was used to identify proteins based on the corresponding peptide sequences with ≥95% confidence. The false positive rate was <1% (31).
Chromatin isolation and micrococcal nuclease sensitivity assays
Chromatin fraction assays were carried out as described previously with some modifications (3). Briefly, 4 × 106 cells were treated with or without 1 mM methyl methanesulfonate (MMS) for 1 h and washed twice with PBS. Cells were suspended in buffer A (0.1% Triton X-100, 10 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 10% glyceral, 1 mM DTT and 300 mM sucrose) for 5 min on ice. Cytoplasmic proteins were separated from nuclei by low-speed centrifugation at 2000 rpm for 5 min at 4°C. The isolated nuclei were washed twice using buffer A, and then re-suspended in buffer B (3 mM EDTA, 0.2 Mm EGTA and 1 mM DTT) for 10 min on ice before centrifugation at 2000 rpm for 5 min. Insoluble chromatin was washed once in solution B and centrifuged at 14 000 rpm for 1 min. The final chromatin pellet was resuspended in 200 μl of SDS loading buffer and sonicated for 15 s. To digest chromatin with micrococcal nuclease (MNase), nuclei were collected by low-speed centrifugation and lysed according to the chromatin isolation protocol described above. Nuclei were resuspended in buffer A containing 20 U/μl MNase (New England Biolabs, Shanghai, China). After incubation at 37°C for 5 min, the nuclease reaction was stopped by the addition of 5 mM EGTA and then subjected to analysis using 1% native agarose gel electrophoresis.
ATPase assay
The equal amount of purified Flag-MORC2 and Flag-MORC2 2A from HEK293T cells using anti-Flag affinity gel (Bimake) was subjected to ATPase assays using the ATPase/GTPase Activity Assay Kit (Sigma) following the manufacturer’s instructions. The absorbance at 620 nm was read for all samples, standards, blanks and negative controls.
Cell viability and colony-formation assays
Cells were seeded in 96-well plates (2000 cells per well) in triplicate and treated with or without increasing doses of MMS for 3 h and then cultured in fresh media for another 24 h. Cell viability was determined by Cell Counting Kit-8 (CCK-8) (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions. For colony-formation assay, cells were seeded in 6-well plates (1000 cells per well) in triplicate and cultured under normal growth conditions in the presence or absence of MMS at the indicated doses for 2 weeks. Colonies were stained with 1% Crystal Violet and counted.
Statistical analysis
All data are presented as the mean ± standard deviation from at least three independent experiments. The Student’s t-test was used for assessing the difference between individual groups and P ≤ 0.05 was considered statistically significant.
RESULTS
MORC2 directly interacts with PARP1
To gain mechanistic insights into the biological functions of MORC2, we aimed to identify its binding partners by mass spectrometry-based proteomics. To achieve this, nuclear extracts of HeLa cells were immunoprecipitated with an anti-MORC2 antibody or control IgG, and the precipitated protein complexes were subjected to LC-MS/MS (31) (Supplementary Figure S1A). On the basis of these analyses, we identified several chromatin-associated enzymes as potential binding partners of MORC2, including PARP1 and histone acetyltransferase N-acetyltransferase 10 (NAT10) (Supplementary Figure S1B).
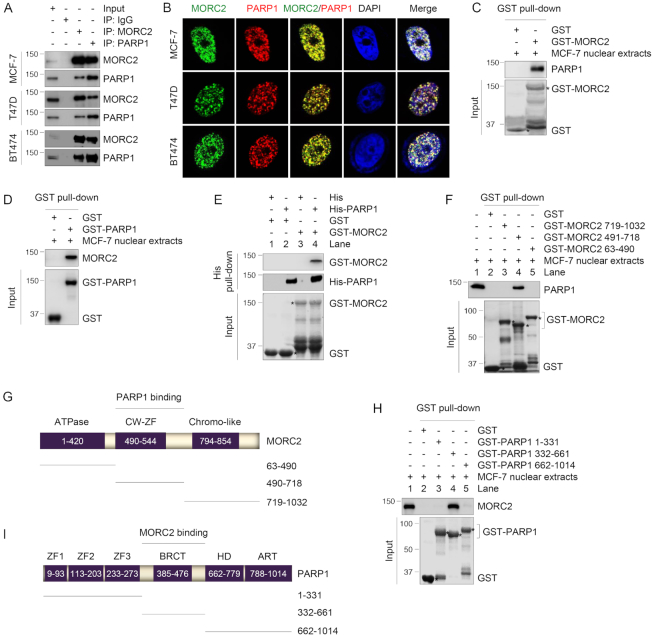
To confirm the interaction between MORC2 and PARP1, lysates from human breast cancer MCF-7, T47D and BT474 cells, which express relatively high levels of endogenous MORC2 and PARP1 (Supplementary Figure S1C), were subjected to reciprocal IP assays with either an anti-MORC2 or an anti-PARP1 antibody. Immunoblotting analysis demonstrated an interaction between MORC2 and PARP1 (Figure 1A). In contrast, MORC1 had a weaker interaction with PARP1 than MORC2, whereas MORC3 did not bind to PARP1 (Supplementary Figure S1D). In addition, the noted interaction between MORC2 and PARP1 was not mediated by DNA, as the interaction between MORC2 and PARP1 remained in the presence of either ethidium bromide (EtBr) that disrupts DNA–protein interactions or deoxyribonuclease (DNase) that degrades double- and single-stranded DNA (Supplementary Figure S1E–H). Immunofluorescent staining showed that MORC2 and PARP1 co-localized in the nucleus in MCF-7, T47D and BT474 cells (Figure 1B).
Figure 1.
MORC2 directly interacts with PARP1. (A) Lysates from MCF-7, T47D and BT474 cells were subjected to IP and immunoblotting analysis with the indicated antibodies. (B) Immunofluorescent staining of MCF-7, T47D and BT474 cells with the indicated antibodies. Nuclear counterstain was carried out using DAPI. (C and D) GST-MORC2 (C) or GST-PARP1 (D) was purified from bacteria and incubated with nuclear extracts from MCF-7 cells. The bound proteins were analyzed by immunoblotting with the indicated antibodies. GST-MORC2 or GST-PARP1 protein was stained by Coomassie blue solution as loading controls. (E) His-PARP1 was incubated with GST or GST-MORC2. The bound proteins were analyzed by immunoblotting with the indicated antibodies. GST or GST-MORC2 protein was stained by Coomassie blue solution as loading controls. (F) GST or GST-MORC2 deletion fragments were incubated with MCF-7 nuclear extracts. The pull-down protein complex was subjected to immunoblotting analysis with an anti-PARP1 antibody. GST or GST-MORC2 proteins were stained by Coomassie blue solution as loading controls. (G) Schematic diagram showing the region of MORC2 for PARP1 binding; CW-ZF, CW-type zinc finger. (H) GST or GST-PARP1 deletion fragments were incubated with MCF-7 nuclear extracts. The pull-down protein complex was analyzed by immunoblotting with an anti-MORC2 antibody. GST or GST-PARP1 proteins were stained by Coomassie blue solution as loading controls. (I) Schematic diagram showing the region of PARP1 for MORC2 binding; ART, ADP-ribosyl transferase; BRCT, BRCA1 C terminus; HD, helical subdomain; ZF, zinc finger.
To examine whether the interaction between MORC2 and PARP1 is direct, we purified GST-MORC2, GST-PARP1 and His-PARP1 from bacteria. Pull-down assay using nuclear extracts of MCF-7 cells revealed that GST-MORC2 interacted with endogenous PARP1 (Figure 1C), while GST-PARP1 bound to endogenous MORC2 (Figure 1D). Furthermore, His-PARP1 enabled to pull down GST-MORC2 under cell-free conditions (Figure 1E, lane 4), supporting a direct interaction between both proteins.
MORC2 contains an N-terminal GHKL-type ATPase domain, a central CW-ZF domain and a C-terminal chromatin-like domain (6–8). To define the domain of MORC2 that mediates its interaction with PARP1, we generated three GST-MORC2 deletion constructs and then performed GST pull-down assays using nuclear extracts from MCF-7 cells. Results showed that the central region (amino acids 491–718) containing the conserved CW-ZF domain of MORC2 was required for its interaction with PARP1 (Figure 1F and G). Interestingly, the CW-ZF domain has been proposed to mediate protein–protein interaction in eukaryotic cells (8). PARP1 has an N-terminal DNA-binding domain containing three zinc-finger (ZF) motifs, a central auto-modification domain possessing a breast cancer susceptibility protein C-terminal (BRCT) motif, and a C-terminal catalytic domain composed of an α-helical subdomain (HD) and an ADP-ribosyl transferase (ART) subdomain (32). GST pull-down assays using purified three GST-PARP1 deletion fragments and nuclear extracts from MCF-7 cells showed that the middle region (amino acids 332–661) containing the BRCT motif of PARP1 was required for interacting with MORC2 (Figure 1H and I). The BRCT motif has been shown to mediate the interaction between PARP1 and its substrates (17). Together, these results suggest that MORC2 interacts with PARP1 in vivo and in vitro.
PARP1 modifies MORC2 by PARylation
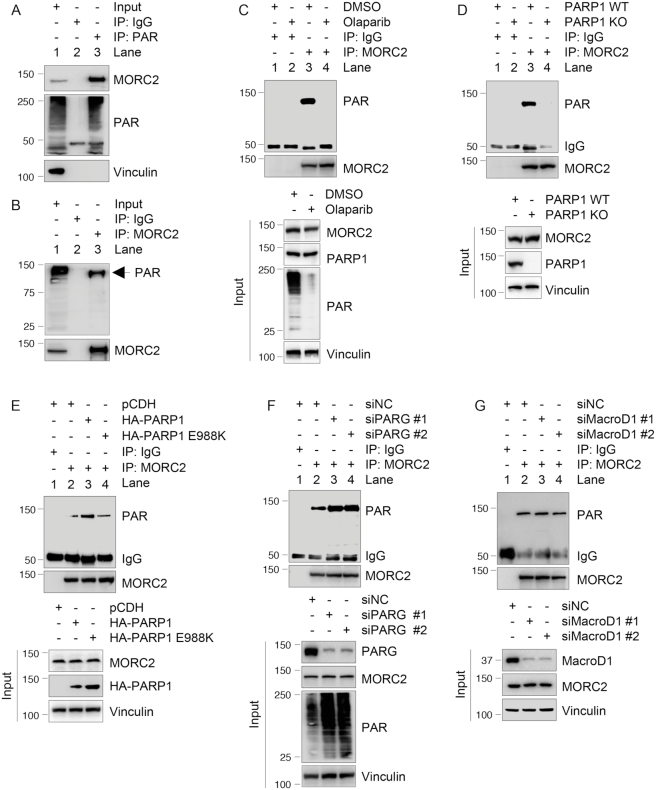
PARP1 is implicated in diverse biological processes, such as gene transcription and post-translational modification of nuclear proteins by PARylation (16). To explore the functional consequences of the noted PARP1–MORC2 interaction, we first examined whether PARP1 could modulate MORC2 expression. Interestingly, we found that knockdown of PARP1 by three independent small interfering RNAs targeting PARP1 (siPARP1 #1–3) (Supplementary Figure S2A) or knockout of PARP1 using CRISPR-Cas9 technology (Supplementary Figure S2B and C) did not significantly affect MORC2 protein levels. Immunofluorescent staining showed that depletion of PARP1 did not significantly alter nuclear localization of MORC2 (Supplementary Figure S2D and E).
To examine whether PARP1 could covalently modify MORC2 by PARylation, we used an anti-PAR antibody to pull down PARylated proteins in MCF-7 cells, which have high basal PARP1 activity (33), and then blotted the immunoprecipitates with an antibody against MORC2. Results showed that MORC2 was one of the PARylated proteins (Figure 2A, lane 3). Reciprocally, immunoprecipitated MORC2 was recognized by an anti-PAR antibody (Figure 2B, lane 3). These results demonstrated that MORC2 is a PARylated protein. Moreover, inhibition of PARP1 activity by PARP inhibitor Olaparib (Figure 2C, compare lane 4 with 3) or knockout of PARP1 (Figure 2D, compare lane 4 with 3) reduced MORC2 PARylation. To address whether the catalytic activity of PARP1 is required for MORC2 PARylation, we expressed either HA-PARP1 or catalytically inactive mutant (HA-PARP1 E988K) (34) in PARP1 KO cells and performed IP assays with an anti-MORC2 antibody. Immunoblotting analyses with an anti-PAR antibody revealed that wild-type PARP1, but not E988K mutant, effectively enhanced PARylation of MORC2 (Figure 2E, compare lane 4 with 3). Since PARG is the main PAR-degrading enzyme (18), we next knocked down PARG using two distinct siRNAs targeting PARG (siPARG #1 and #2) and then examined the PARylation levels of MORC2. As expected, depletion of PARG enhanced PARylation of MORC2 (Figure 2F, compare lanes 3–4 with 2). In contrast, knockdown of MacroD1, a mono-ADP-ribosylhydrolase (35), by two siRNAs did not significantly affect PARylation of MORC2 (Figure 2G). Together, these results indicate that MORC2 is covalently modified by PARP1-mediated PARylation.
Figure 2.
PARP1 PARylates MORC2. (A and B) Lysates from MCF-7 cells were immunoprecipitated with control IgG or an anti-PAR antibody (A) or an anti-MORC2 antibody (B), followed by immunoblotting with the indicated antibodies. (C) MCF-7 cells were treated with or without 5 μM Olaparib for 1 h. Lysates were immunoprecipitated with control IgG or an anti-MORC2 antibody, and the status of PARylation was determined by immunoblotting with an anti-PAR antibody. (D) Lysates from PARP1 WT or KO cells were immunoprecipitated with control IgG or an anti-MORC2 antibody, followed by immunoblotting with the indicated antibodies. (E) PARP1 KO cells were transfected with the indicated plasmid DNAs. After 48 h of transfection, lysates were subjected to IP and immunoblotting analysis with the indicated antibodies. (F) MCF-7 cells were transfected with non-targeting control siRNA (siNC) and two different siRNAs targeting PARG. After 48 h of transfection, lysates were subjected to IP and immunoblotting analysis with indicated antibodies. For all in vivo PARylation assays (A–F), PARG inhibitor ADP-HPD (5 μM) was added to the lysis buffer to prevent degradation of PAR. (G) MCF-7 cells were transfected with siNC and two different siRNAs targeting MacroD1. After 48 h of transfection, lysates were subjected to IP and immunoblotting analysis with indicated antibodies.
PARP1 PARylates MORC2 at two conserved residues within its CW-ZF domain
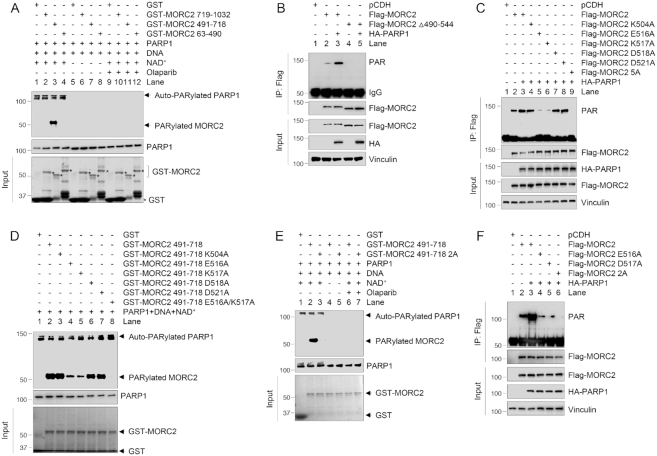
To map the PARylation sites of MORC2 by PARP1, we next carried out in vitro PARylation assays. Given that MORC2 runs with an almost similar size as PARP1 (118 versus 113 kDa), GST-MORC2 deletion fragments were used to make a distinction between the auto-PARylated PARP1 and the PARylated MORC2. As shown in Figure 3A, the fragment containing the ZF-CW domain (residues 491–718, lane 3) was PARylated in the presence of recombinant full-length PARP1, NAD+ as a donor of the ADP-ribose moiety and sheared salmon sperm DNA. The activation of PARP1 was confirmed by the detection of its auto-PARylation in the upper part of the gel. As a negative control, GST-MORC2 (resides 491–718) failed to be PARylated in the absence of NAD+ (lane 7) or in the presence of PARP inhibitor Olaparib (lane 11). In agreement with these results, ectopic expression of HA-PARP1 enhanced the PARylation of wild-type MORC2, but not its ZF-CW domain deletion mutant in vivo (Figure 3B, compare lane 5 with 3). These results suggest that MORC2 is primarily PARylated at its CW-ZF domain.
Figure 3.
Identification of the PARylation sites of MORC2. (A) In vitro PARylation assay was performed using purified deletion fragments of GST-MORC2 in the presence or absence of recombinant PARP1 enzyme, NAD+, sonicated salmon sperm DNA and PARP inhibitor Olaparib as indicated. PARylated MORC2 was detected with an anti-PAR antibody. (B) HEK293T cells were transfected with the indicated expression vectors. After 48 h of transfection, lysates were subjected to IP and immunoblotting analyses with the indicated antibodies. (C) HEK293T cells were transfected with the indicated expression vectors. After 48 h of transfection, lysates were subjected to IP and immunoblotting analyses with the indicated antibodies. (D) In vitro PARylation assay was performed using purified GST-MORC2 as indicated in the presence of recombinant PARP1 enzyme, NAD+ and sonicated salmon sperm DNA. PARylated MORC2 was detected using an anti-PAR antibody. (E) In vitro PARylation assay was performed using purified GST-MORC2 proteins in the presence or the absence of recombinant PARP1 enzyme, NAD+, sonicated salmon sperm DNA and Olaparib as indicated. PARylated MORC2 was detected using an anti-PAR antibody. (F) HEK293T cells were transfected with the indicated expression vectors. After 48 h of transfection, lysates were subjected to IP and immunoblotting analyses with the indicated antibodies.
PARylation most commonly takes place on aspartate (D), glutamate (E) and lysine (K) residues of target proteins (17). Sequence alignments of the CW-ZF domain of MORC2 from several species revealed five evolutionary conserved residues (K504, E516, K517, D518 and D521), which can be potentially PARylated (Supplementary Figure S3A). To test whether these residues are PARylated in cells, we substituted those five residues with alanine (A) alone or in combination (termed 5A) by site-directed mutagenesis, and then transfected these expression vectors into HEK293T cells with or without HA-PARP1. As shown in Figure 3C, mutation of E516 (lane 5) or K517 (lane 6) caused a significant decrease in MORC2 PARylation in the presence of HA-PARP1 as compared with its WT counterpart and other single mutants. Moreover, mutation of all 5 residues (5A) resulted in no detectable PARylation of MORC2 (lane 9). Consistently, in vitro PARylation assays showed that the PARylation levels of either GST-MORC2 E516A (lane 4) or K517A (lane 5) were significantly decreased as compared with its WT counterpart in the presence of PARP1, NAD+ and DNA (Figure 3D). Moreover, the double mutant (E516A/K517A, hereafter termed 2A) showed no detectable PARylation (lane 8). These results were further demonstrated under the conditions of inhibition of PARP1 activity by Olaparib in vitro (Figure 3E) and ectopic expression of PARP1 in vivo (Figure 3F). Collectively, these results suggest that E516 and K517 are the major PARylation sites of MORC2 by PARP1.
As the PARylation sites (E516/K517) of MORC2 reside within its interaction domain with PARP1, mutation of E516 and K517 may alter the conformation of its CW-ZF domain, resulting in disrupting its interaction with PARP1 and therefore diminishing its ability to be PARylated by PARP1. To rule out this possibility, HEK293T cells were transfected with Flag-MORC2, Flag-MORC2–2A and HA-PARP1, and treated with or without MMS for 1 h after 48 h of transfection. IP and immunoblotting analysis showed that HA-PARP1 interacted with both WT MORC2 and MORC2–2A mutant under basal condition (Supplementary Figure S3B, compare lane 4 with 2). In contrast, treatment of cells with MMS enhanced the interaction of PARP1 with WT MORC2 but not MORC2–2A mutant (Supplementary Figure S3B, compare lane 5 with 3). These results suggest that the PARylation deficiency of MORC2–2A mutant is a result of mutation of PARylation sites. Immunofluorescent staining and pulse-chase experiments using protein synthesis inhibitor cycloheximide (CHX) revealed that mutation of MORC2 PARylation sites (2A) did not significantly affect its nuclear localization (Supplementary Figure S3C) and half-live (Supplementary Figure S3D), respectively.
PARP1 recruits MORC2 to DNA damage sites and promotes MORC2 PARylation in response to DNA damage
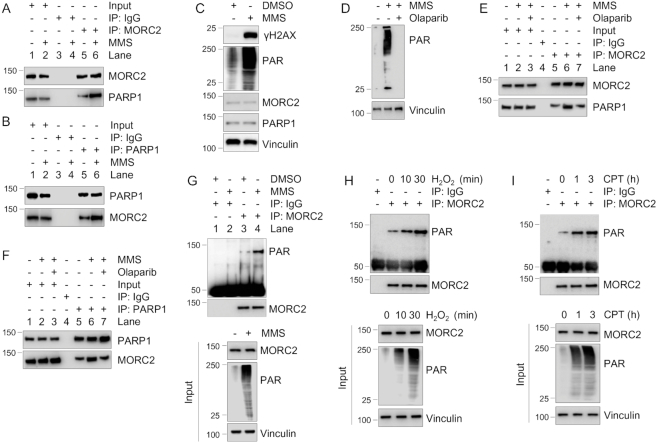
As both MORC2 and PARP1 are DNA damage-responsive proteins (10,15), we first examined whether the MORC2–PARP1 interaction is affected by DNA damage. To do this, MCF-7 cells were treated with or without DNA-damaging agent MMS in the presence or the absence of PARP inhibitor Olaparib, and then subjected to IP and immunoblotting analysis. Results showed that the association between PARP1 and MORC2 was increased after MMS treatment (Figure 4A and B, compare lane 6 with 5). As a positive control, MMS treatment significantly enhanced the levels of PAR and phosphorylated histone variant H2AX at serine 139 (termed γH2AX), two surrogate markers of DNA damage (36) (Figure 4C). Moreover, pretreatment with Olaparib decreased MMS-induced PAR activation (Figure 4D) and MMS-enhanced interaction between MORC2 and PARP1 (Figure 4E and F, compare lane 7 with 6). These results suggest that activation of PARP1 by DNA damage enhances its interaction with MORC2. To determine DNA damage affects MORC2 PARylation, we treated MCF-7 cells with DNA-damaging agents MMS, hydrogen peroxide (H2O2) and camptothecin (CPT), which have been shown to activate PARP1 enzymatic activity (29,37,38). IP and immunoblotting analyses showed that treatment with MMS (Figure 4G, compare lane 4 with 3), H2O2 (Figure 4H) and CPT (Figure 4I) enhanced MORC2 PARylation.
Figure 4.
DNA damage enhances the interaction between MORC2 and PARP1 and MORC2 PARylation. (A and B) Lysates from MCF-7 cells treated with or without 1 mM MMS for 1 h were immunoprecipitated with control IgG or an anti-MORC2 (A) or an anti-PARP1 (B) antibody, followed by immunoblotting analysis with the indicated antibodies. (C) MCF-7 cells were treated with or without 1 mM MMS for 1 h and then subjected to immunoblotting analysis with the indicated antibodies. (D-F) MCF-7 cells were pretreated with or without 5 μM Olaparib for 1 h and then treated with or without 1 μM MMS for another 1 h. Lysates were subjected to immunoblotting analysis (D) or IP assays with control IgG or an anti-MORC2 (E) or an anti-PARP1 (F) antibody. The interaction between MORC2 and PARP1 was detected by immunoblotting analysis with indicated antibodies (E and F). (G) MCF-7 cells were treated with DMSO or 1 mM MMS for 30 min. Lysates were subjected to IP and immunoblotting analysis with the indicated antibodies. (H) MCF-7 cells were treated with or without 1 mM H2O2 for the indicate times, and then subjected to IP and immunoblotting analysis with the indicted antibodies. (I) MCF-7 cells were treated with or without 1 μM CPT for the indicate times, and then subjected to IP and immunoblotting analysis with the indicted antibodies.
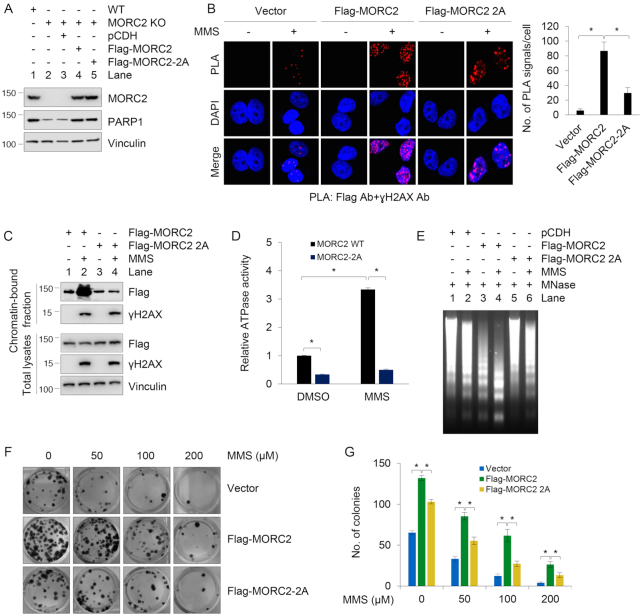
As many DNA repair factors, such as nuclear receptor NR1D1 (39), tyrosyl-DNA phosphodiesterase 1 (TDP1) (38) and histone demethylase KDM4D (30), are recruited to the sites of DNA damage in a PARP1-dependent manner, we next examined whether PARP1 is required for the recruitment of MORC2 to sites of DNA damage by the application of in situ proximity ligation assay (PLA) technique (40), which allows sensitive monitoring of the localization and interactions of DNA repair proteins at DNA breaks (41). To do this, we stably expressed Flag-MORC2 and Flag-MORC2–2A in MORC2 KO MCF-7 cells. MORC2 KO cells were verified by sequencing (Supplementary Figure S4A), and the protein levels of exogenous and endogenous MORC2 in these established cells were demonstrated by immunoblotting (Figure 5A). During eukaryotic DDR, one of the earliest events is activation of γH2AX (36), which serves as a binding platform for recruitment and/or retention of chromatin-modifying complexes and DNA repair factors in the vicinity of DNA lesions. In situ PLA assays showed that Flag-MORC2 co-localized with γH2AX following MMS treatment, but the noted co-localization was compromised in Flag-MORC2-2A expressing cells (Figure 5B). As a negative control, PLA signals failed to be detected when single antibody was used (Supplementary Figure S4B). Consistently, chromatin fractionation assays showed that MMS treatment resulted in an increase in the amount of wild-type MORC2, but not PARylation-defective mutant (2A), in the chromatin-bound fraction (Figure 5C). These results suggest that MORC2 accumulation at sites of DNA damage is dependent on PARP1, which PARylates MORC2 after damage.
Figure 5.
PARP1 recruits MORC2 to DNA damage sites and PARylation of MORC2 stimulates its chromatin remodeling activities. (A) Reconstitution of MORC2 KO MCF-7 cells with Flag-MORC2 and Flag-MORC2–2A by lentiviral infection. Expression status of exogenous and endogenous MORC2 was validated by immunoblotting. (B) MCF-7 KO cells stably expressing pCDH, Flag-MORC2 and Flag-MORC2–2A were treated with or without 1 mM MMS for 30 min, and then subjected to in vivo PLA assays with indicated antibodies. Three independent experiments were performed, and total 50 cells were counted for each experiment. (C) MCF-7 KO cells stably expressing Flag-MORC2 and Flag-MORC2–2A were treated with or without 1 mM MMS for 30 min, and subjected to cellular fractionation as described in ‘Materials and Methods’ section. Chromatin-bound fractions and total cellular lysates from undamaged and DNA damaged cells were immunoblotted with the indicated antibodies. (D) MCF-7 KO cells stably expressing Flag-MORC2 and Flag-MORC2–2A were treated with or without 1 mM MMS for 30 min. Immunoprecipitated Flag-MORC2 or Flag-MORC2–2A was subjected to ATPase assays using the ATPase/GTPase Activity Assay Kit (Sigma). Data represent mean ± s.d. from three biologically independent experiments. (E) MCF-7 KO cells stably expressing pCDH, Flag-MORC2 and Flag-MORC2–5A were treated with or without 1 mM MMS for 30 min. Isolated nuclei were subjected to MNase assays as described in ‘Materials and Methods’ section. (F and G) MCF-7 KO cells stably expressing pCDH, Flag-MORC2 and Flag-MORC2–2A were treated with or without increasing doses of MMS for 2 weeks. The representative images of Crystal Violet-stained colonies are shown in F. Quantitative results (G) are represented as mean ± s.d. as indicated from three independent experiments; *, P<0.05.
PARylation of MORC2 contributes to DNA damage response
One of our recent studies demonstrated that MORC2 exerts DNA-dependent ATPase activity to facilitate chromatin remodeling in response to DNA damage (10). Next, we sought to address whether PARylation of MORC2 affects its ATPase and chromatin remodeling activities following DNA damage. Toward this end, we set up an in vitro ATPase assay using immunoprecipitated Flag-MORC2 and Flag-MORC2-2A proteins and ATPase activity assay kit. Results showed that PARylation-defective mutant had decreased ATPase activities in response to MMS-induced DNA damage as compared with its WT counterpart (Figure 5D). To examine whether PARylation of MORC2 affects the state of chromatin relaxation, we carried out MNase sensitivity assay, which has been widely used to evaluate chromatin condensation (3). Results showed that MMS treatment of cells expressing WT MORC2 caused an increase in chromatin accessibility to MNase, but this effect was compromised in cells expressing PARylation-defective MORC2 mutant (Figure 5E, compare lane 4 with 6). One of the hallmarks of defective DNA repair is increased sensitivity to genotoxic stress. Cell survival assays revealed that expression of PARylation-defective MORC2 in MCF-7 cells led to enhanced cellular sensitivity to MMS as compared with its WT counterpart (Figure 5F and G). Together, these results suggest that PARylation is required for chromatin remodeling activity of MORC2 and for cell survival following DNA damage.
MORC2 stabilizes PARP1 through blocking ubiquitin-dependent degradation
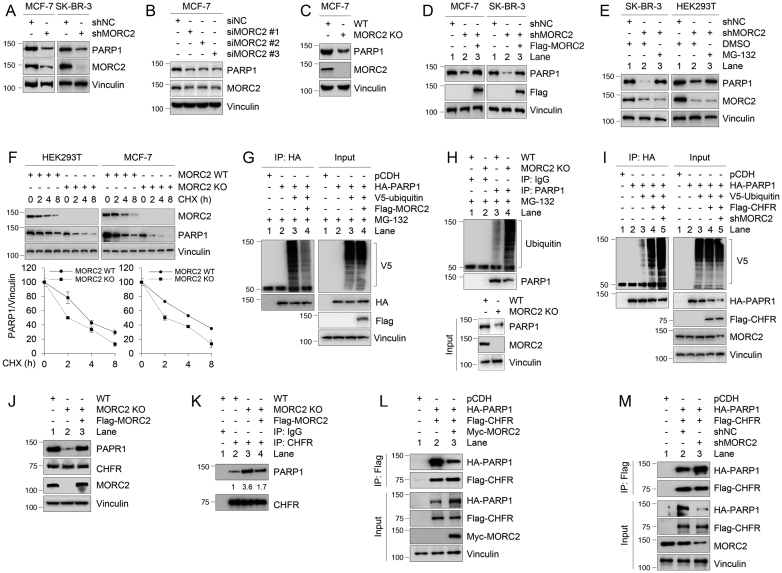
Since MORC2 has been documented to regulate gene transcription (14), we next examined whether MORC2 could affect PARP1 expression. Knockdown of MORC2 by shRNA targeting MORC2 (shMORC2) in MCF-7 and SK-BR-3 cells (Figure 6A) or by three different siRNAs against MORC2 (siMORC2 #1–3) in MCF-7 cells (Figure 6B) reduced PARP1 protein abundance. A similar effect was observed in MORC2 KO MCF-7 cells by the CRISPR-Cas9 system (Figure 6C). More importantly, reintroduction of MORC2 in MORC2-depleted MCF-7 and SK-BR-3 cells partially restored PARP1 expression (Figure 6D, compare lane 3 with 2). These results suggest that MORC2 regulates PARP1 expression.
Figure 6.
MORC2 enhances PARP1 stability through suppressing CHFR-mediated ubiquitination. (A) Lysates from MCF-7 and SK-BR-3 cells stably expressing shNC and shMORC2 were analyzed by immunoblotting with the indicated antibodies. (B) MCF-7 cells were transfected with siNC or three different siRNAs targeting MORC2 (siMORC2). After 48 h of transfection, cells were harvested for immunoblotting with the indicated antibodies. (C) Lysates from WT and MORC2 KO MCF-7 cells were analyzed by immunoblotting. (D) MCF-7 and SK-BR-3 cells stably expressing shMORC2 were transfected with Flag-MORC2. After 48 h of transfection, immunoblotting analysis was performed with the indicated antibodies. (E) MCF-7 and SK-BR-3 cells stably expressing shNC and shMORC2 were treated with DMSO or 10 μM MG-132 for 6 h. Lysates were analyzed by immunoblotting. (F) WT and MORC2 KO MCF-7 and SK-BR-3 cells were treated with 100 μg/ml CHX for the indicated times and analyzed by immunoblotting. Relative PARP1 levels (PARP1/Vinculin) are shown in lower panels. Quantitative results are represented as mean ± s.d. as indicated from three independent experiments. (G) HEK293T cells were transfected with the indicated expression vectors. After 48 h of transfection, cells were treated with 10 μM MG-132 for 6 h, and lysates were subjected to IP and immunoblotting analysis with the indicated antibodies. (H) WT and MORC2 KO MCF-7 cells were treated with 10 μM MG-132 for 6 h, and lysates were immunoprecipiated with control IgG or an anti-PARP1 antibody, followed by immunoblotting with the indicated antibodies. (I) HEK293T cells were transfected with the indicated plasmid DNAs. After 48 h of transfection, cells were treated with 10 μM MG-132 for 6 h. IP and immunoblotting analyses were performed with the indicated antibodies. In panels (G) to (I), lysis buffer was also supplemented with 10 μM MG-132 in the subsequent assays. (J and K) WT and MORC2 KO MCF-7 cells were transfected with empty vector or Flag-MORC2. After 48 h of transfection, lysates were subjected to IP and immunoblotting analyses with indicated antibodies. (L and M) HEK293T cells were transfected with the indicated expression vectors. After 48 h of transfection, lysates were subjected to IP and immunoblotting analyses with indicated antibodies.
To determine whether MORC2 regulates PARP1 at transcriptional level, we performed real-time quantitative PCR (qPCR) to measure PARP1 mRNA expression levels in control and MORC2-depleted cells. Results showed the mRNA levels of PARP1 were not significantly affected following knockdown of MORC2 by shRNA or siRNA (Supplementary Figure S5A and B), ruling out the possibility that MORC2 transcriptionally regulates PARP1 expression. Immunofluorescent staining showed that knockdown of MORC2 in MCF-7 and SK-BR-3 cells by siRNA did not significantly affect nuclear localization of PARP1 (Supplementary Figure S5C).
As PARP1 stability is controlled by ubiquitin-mediated proteolysis (22,23), we next examined whether MORC2 could affect PARP1 ubiquitination and degradation. As shown in Figure 6E, knockdown of MORC2 in SK-BR-3 and HEK293T cells led to a downregulation of PARP1, which was reversed by treatment of cells with the protease inhibitor MG-132 (compare lane 3 with 2), suggesting that MORC2 regulates PARP1 levels through a proteasome-dependent pathway. In support of this notion, knockdown of MORC2 significantly decreased half-life of endogenous PARP1 protein in the presence of eukaryote protein synthesis inhibitor CHX (Figure 6F).
To examine whether MORC2 regulates PARP1 ubiquitination levels, HEK293T cells were transfected with plasmid DNAs encoding HA-PARP1, V5-ubiquitin and Flag-MORC2 alone or in combination. Lysates were immunoprecipitated using an anti-HA antibody, followed by immunoblotting using an anti-V5 antibody. Results showed that exogenously expressed MORC2 decreased the ubiquitination levels of HA-PARP1 (Figure 6G, compare lane 4 with 3). As a control, treatment of cells with PARP1 inhibitor Olaparib did not significantly affect the appearance of a smear of polyubiquitinated PARP1 (Supplementary Figure S5D), suggesting that the observed smear of the ubiquitinated PARP1 is not from massive PAR generated by PARP1. Conversely, knockdown of endogenous MORC2 by shRNA enhanced the ubiquitination levels of endogenous PARP1 (Figure 6H, compare lane 2 with 1). These data indicate that MORC2 regulates PARP1 protein stability via the proteasome-dependent pathway.
MORC2 suppresses CHFR-mediated PARP1 ubiquitination through blocking the CHFR–PARP1 interaction
Recent several studies have reported that the E3 ligase CHFR is responsible for the ubiquitination and degradation of PARP1 (22,23). Next, we tested whether MORC2 affects CHFR-mediated PARP1 ubiquitination. As expected, knockdown of MORC2 increased CHFR-mediated PARP1 ubiquitination (Figure 6I, left panel, compare lane 5 with 4). Recently, a proteomic screen has identified the lysine 949 (K949) residue is one of ubiquitination sites of PARP1 (42). To verify this result, HEK293T cells were transfected with HA-PARP1 or HA-PARP1 K949R alone or in combination with V5-ubiquitin and Flag-CHFR. Sequential IP and immunoblotting analyses showed that the ubiquitination levels of HA-PARP1 K949R were reduced as compared with its WT counterpart (Supplementary Figure S5E, compare lane 5 with 3). We further confirmed these results by measuring PARP1 half-life in the presence of CHX. As shown in Supplementary Figure S5F, the K949R mutant increased the half-life of PARP1 as compared with its wild-type control. These results suggest that K949 is one of ubiquitination sites for PARP1.
To address the underlying mechanism by which MORC2 suppresses CHFR-mediated ubiquitination, we examined whether MORC2 affects the interaction of PARP1 with CHFR. We found that knockdown of MORC2 or re-expression of MORC2 in MORC2 KO cells affected protein expression levels of PARP1 but not CHFR (Figure 6J). IP and immunoblotting analyses showed that knockout of MORC2 in MCF-7 cells increased the interaction between CHFR and PARP1, and this effect was compromised when MORC2 was re-expressed in MORC2 KO cells (Figure 6K, compare lane 4 with 3). To confirm these results, HEK293T cells were transfected with expression vectors encoding HA-PARP1 and Flag-CHFR under the condition of overexpression of Myc-MORC2 or knockdown of MORC2 by shRNA. IP and immunoblotting analysis revealed that overexpression of MORC2 suppressed the PARP1–CHFR interaction (Figure 6L, compare lane 3 with 2), whereas knockdown of MORC2 enhanced the interaction between both proteins (Figure 6M, compare lane 3 with 2). These results suggest that MORC2 suppresses CHFR-mediated ubiquitination of PARP1 through, at least in part, compromising the interaction of PARP1 with CHFR.
NAT10 acetylates PARP1 at lysine 949 and enhances its stability
PARP1 undergoes acetylation (24), in addition to ubiquitination (22,23). To gain more insights into the mechanisms by which MORC2 stabilizes PARP1, we next examined the possibility that MORC2 enhances PARP1 stability through a crosstalk between those two PTMs. As shown in Supplementary Figure S6A, treatment of HEK293T cells with trichostatin A (TSA), an inhibitor of histone deacetylase (HDAC) classes I and II, in combination with nicotinamide (NAM), an inhibitor of the sirtuin (SIRT) family deacetylases, increased PARP1 acetylation levels. Moreover, treatment with TSA and NAM enhanced PARP1 protein levels (Supplementary Figure S6B), and PARP1 reduction following MORC2 knockdown was rescued by addition of either TSA or NAM (Supplementary Figure S6C and D, respectively). Further, TSA and NAM treatment increased the half-live of PARP1 (Supplementary Figure S6E) and decreased the ubiquitination levels of endogenous and exogenous PARP1 (Supplementary Figure S6F and G, respectively). These results indicate that MORC2 regulates PARP1 levels through, at least in part, an acetylation-dependent pathway.
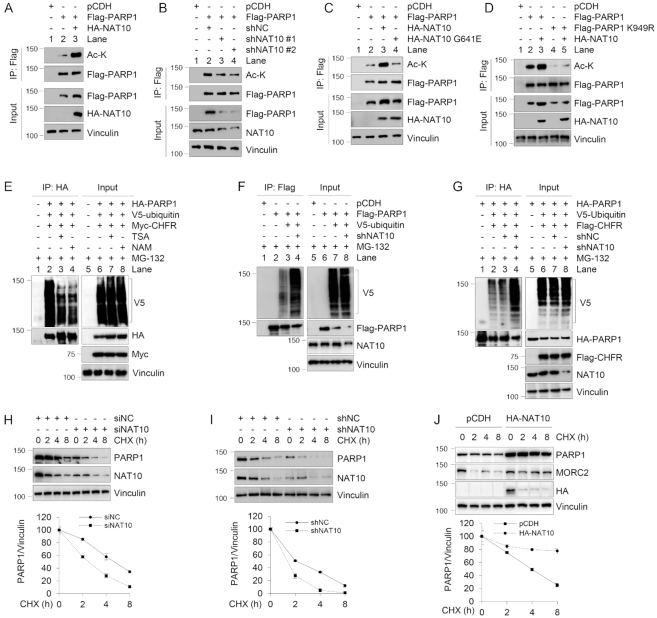
Upon examination of potential acetyltransferases that could acetylate PARP1, our attention was drawn to histone acetyltransferase NAT10, which is a potential binding partner of MORC2 revealed by our proteomic assays (Supplementary Figure S1B). NAT10 exerts acetyltransferase activity toward histones and non-histone proteins with emerging roles in DDR and human cancer development and progression (43). To test whether PARP1 is acetylated by NAT10, we ectopically expressed Flag-PARP1 alone or in combination with HA-NAT10 in HEK293T cells. IP assays with an anti-Flag antibody followed by immunoblotting using an anti-acetyl lysine (Ac-K) antibody revealed that the acetylation levels of PARP1 were increased in the presence of HA-NAT10 (Figure 7A, compare lane 3 with 2). Conversely, siRNA-mediated knockdown of NAT10 (Figure 7B, compare lanes 3 and 4 with 2) or pharmacological inhibition of NAT10 activity by small-molecule inhibitor Remodelin (44) (Supplementary Figure S6H) reduced the acetylation levels of PARP1. Moreover, expression of wild-type NAT10, but not its catalytically inactive G641E mutant (44), increased the levels of PARP1 acetylation (Figure 7C). Together, these results suggest that NAT10 acetylates PARP1 and its acetyltransferase activity is required for this event.
Figure 7.
NAT10 acetylates PARP1 at K949 and enhances its stability. (A–D) HEK293T cells were transfected with the indicated expression vectors. After 48 h of transfection, lysates were subjected to IP and immunoblotting analyses with the indicated antibodies. The immunoprecipitated PARP1 has been adjusted to be equal to make the levels of acetylated PARP1 comparable to those of immunoprecipitated PARP1. (E–G) HEK293T cells were transfected with the indicated expression vectors. After 48 h of transfection, cells were treated with 10 μM MG-132 for 6 h and lysates were subjected to IP and immunoblotting analyses with the indicated antibodies. Lysis buffer was also supplemented with 10 μM MG-132 in the subsequent assays. (H) MCF-7 cells were transfected with siNC or siNAT10. After 48 h of transfection, cells were treated with 100 μg/ml CHX for the indicated times and analyzed by immunoblotting. Relative PARP1 levels (PARP1/Vinculin) are shown in lower panel. (I) MCF-7 cells stably expressing shNC or shNAT10 were treated with 100 μg/ml CHX for the indicated times and analyzed by immunoblotting. Relative PARP1 levels (PARP1/Vinculin) are shown in lower panel. (J) HEK293T cells were transfected with pCDH or HA-NAT10. After 48 h of transfection, cells were treated with 100 μg/ml CHX for the indicated times and analyzed by immunoblotting. Relative PARP1 levels (PARP1/Vinculin) are shown in lower panel. In panels (H) to (J), quantitative results are represented as mean ± s.d. as indicated from three independent experiments.
Recent proteomic studies have identified several potential acetylation sites of PARP1 (45) (Supplementary Figure S7A). To verify these results, each lysine (K) residue was mutated into arginine (R) individually. Then, HEK293T cells were transfected with Flag-PARP1 and various mutants alone or in combination with HA-NAT10. Sequential IP and immunoblotting analyses demonstrated that ectopical expression of HA-NAT10 failed to enhance the acetylation of Flag-PARP1 K949R mutant as compared with its WT counterpart and other mutants (Supplementary Figure S7B and C). Independent experiments also demonstrated that co-expression of NAT10 increased acetylation levels of wild-type PARP1, but not PARP1 K949R mutant (Figure 7D, compare lane 3 with 5), indicating that K949 is the main acetylation residue of PARP1 by NAT10. As K949 is also one of ubiquitination sites of PARP1 (42) (Supplementary Figure S5E), it raises the possibility that PARP1 acetylation and ubiquitination might compete for the same lysine residue. As expected, CHFR-induced PARP1 ubiquitination was diminished in the presence of HDAC inhibitor TSA or NAM (Figure 7E, compare lanes 3 and 4 with 2). Moreover, knockdown of NAT10 enhanced CHFR-mediated PARP1 ubiquitination (Figure 7F and G, compare lane 4 with 3), reinforcing the idea that acetylation of PARP1 by NAT10 at K949 may positively regulate PARP1 stability by inhibiting its ubiquitination. In support of this notion, treatment of MCF-7 cells with NAT10 inhibitor Remodelin downregulated the protein but not mRNA levels of PARP1 (Supplementary Figure S7D and E, respectively). Furthermore, knockdown of NAT10 by siRNA (Figure 7H) or shRNA (Figure 7I) shortened the half-live of PARP1. Conversely, overexpression of NAT10 prolonged the half-live of PARP1 (Figure 7J). CHX chase assays showed that the K949R and K949Q mutants had enhanced stability compared to its WT counterpart (Supplementary Figure S7F). This occurs because the K949 is also one of ubiquitination sites of PARP1 (46), the K949R and K949Q mutants can not be ubiquitinated by E3 ubiquitin ligases such as CHFR, resulting in enhanced stability of PARP1. Together, these results indicate that NAT10 promotes PARP1 acetylation at K949 and enhances its stability.
MORC2 mediates the interaction of PARP1 and NAT10 and is required for NAT10-induced acetylation of PARP1
To address the mechanistic role for MORC2 in NAT10-mediated PARP1 acetylation, we next examined whether MORC2 affects the interaction between PARP1 and NAT10. Reciprocal IP assays demonstrated that MORC2, PARP1 and NAT10 formed a ternary complex at the endogenous level in human breast cancer MCF-7 and T47D cells (Supplementary Figure S8A). The noted interactions were not affected in the presence of DNase (Supplementary Figure S8B). GST pull-down assays further demonstrated that MORC2 and NAT10 bound to the central region of PARP1 (Supplementary Figure S8C), while MORC2 and PARP1 bound to the C-terminal region of NAT10 (Supplementary Figure S8D). In addition, NAT10 interacted with C-terminal region of MORC2, while PARP1 bound to the central region of MORC2 (Supplementary Figure S8E). Knockout of MORC2 in MCF-7 and HEK293T cells did not affect NAT10 protein levels (Supplementary Figure S8F). IP and immunoblotting analyses showed that knockdown of MORC2 by shRNA attenuated the interaction of exogenously expressed HA-PARP1 and GFP-NAT10 (Supplementary Figure S8G, compare lane 3 with 2). Consistently, depletion of MORC2 impaired NAT10-mediated acetylation of PARP1 (Supplementary Figure S8H, compare lane 4 with 3). More importantly, expression of NAT10 suppressed wild-type PARP1 ubiquitination (Supplementary Figure S8I, compare lane 4 with 3), and this effect was compromised under the condition of MORC2 knockdown (compare lane 5 with 4). In contrast, overexpression of NAT10 alone or in combination with MORC2 knockdown did not significantly affect the ubiquitination levels of acetylation-deficient K949R mutant (Supplementary Figure S8I, lanes 8–9). Collectively, these results indicate that MORC2 is required for NAT10-mediated acetylation of PARP1, thus enhancing PARP1 stability.
Depletion of MORC2 or expression of an acetylation-defective PARP1 mutant impairs DNA damage-induced PAR production and PAR-dependent DNA repair
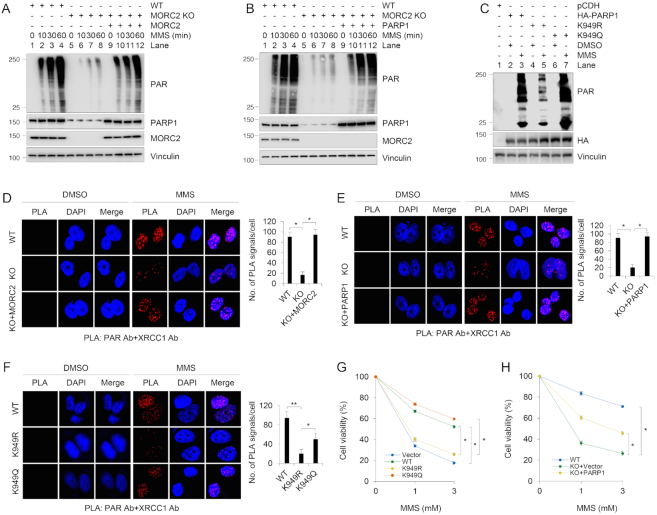
Following DNA damage, PARP1 quickly relocates to DNA damage sites and catalyzes the synthesis of PAR, which promotes the recruitment of DDR factors to the sites of DNA damage (15). To test the biological function of PARP1 stabilization by MORC2, we first examined the effects of MORC2 and PARP1 acetylation on PAR production in response to DNA damage. Immunoblotting analyses showed that MORC2 KO cells exhibited reduced PAR levels as compared with its wild-type controls following MMS treatment (Figure 8A and B). Moreover, re-expression of MORC2 or PARP1 in MORC2 KO cells partially restored the decreased PAR levels in MORC2 KO cells after MMS treatment (Figure 8A and B). It is interesting to note that K949 is located in the catalytic domain of PARP1 (residues 788–1014) and is very close to its catalytic active site E988 (34,46), acetylation of PARP1 at K949 might alter the conformation of its catalytic domain and thus regulates the catalytic activity of PARP1 in response to DNA damage. To test this notion, we examined the intrinsic PAR activity of PARP1 using purified WT, K949R or K949Q mutant of PARP1. Results showed that K949R mutant had decreased intrinsic auto-PARylation activity compared to WT or K949Q PARP1 (Supplementary Figure S9A). Consistently, PAR production was increased following MMS treatment in cells expressing wild-type PARP1 and acetylation-mimicking mutant (K949Q) as compared with acetylation-deficient mutant (K949R) (Figure 8C).
Figure 8.
Depletion of MORC2 or expression of an acetylation-defective PARP1 mutant impairs DNA damage-triggered PAR production and PAR-dependent DNA repair signaling. (A) WT and MORC2 KO MCF-7 cells stably expressing empty vector or Flag-MORC2 were treated with 1 mM MMS for 30 min. Lysates were analyzed by immunoblotting with the indicated antibodies. (B) WT and MORC2 KO MCF-7 cells stably expressing empty vector or HA-PARP1 were treated with 1 mM MMS for 30 min. Lysates were analyzed by immunoblotting with the indicated antibodies. (C) PARP1 KO MCF-7 cells stably expressing pCDH, HA-PARP1, HA-PARP1 K949R and PARP1 K949Q were treated with or without 1 mM MMS for 30 min and analyzed by immunoblotting with the indicated antibodies. (D) WT and MORC2 KO MCF-7 cells stably expressing empty vector or Flag-MORC2 were treated with or without 1 mM MMS for 30 min and then subjected to in situ PLA assays with the indicated antibodies. The representative images are shown in left. The quantitative results (right) are represented as mean ± s.d. as indicated from three independent experiments, and 50 cells were counted in each experiment. (E) WT and MORC2 KO MCF-7 cells stably expressing empty vector or HA-PARP1 were treated with or without 1 mM MMS for 30 min and then subjected to in situ PLA assays with the indicated antibodies. The representative images are shown in left. The quantitative results (right) are represented as mean ± s.d. as indicated from three independent experiments, and 50 cells were counted in each experiment. (F) PARP1 KO MCF-7 cells stably expressing HA-PARP1, HA-PARP1 K949R and PARP1 K949Q were treated with or without 1 mM MMS for 30 min. In vivo PLA assays were performed as described above. The representative images are shown in left. The quantitative results (right) are represented as mean ± s.d. as indicated from three independent experiments, and 50 cells were counted in each experiment. (G) PARP1 KO MCF-7 cells stably expressing pCDH, HA-PARP1, HA-PARP1 K949R and HA-PARP1 K949Q were treated with or without MMS at the indicated doses for 3 h and then cultured for another 24 h in the fresh media to allow cellular repair. Cell viability was determined by CCK-8 assays. Data are represented as mean ± s.d. as indicated from three independent experiments. (H) WT and MORC2 KO MCF-7 cells stably expressing empty vector or HA-PARP1 were treated with or without MMS at the indicated doses for 3 h and then cultured for another 24 h in the fresh media to allow cellular repair. Cell viability was determined by CCK-8 assays. Data are represented as mean ± s.d. as indicated from three independent experiments. **, P<0.01, *, P<0.05.
It has been well established that following DNA damage, synthesized PAR at the DNA damage sites provides the platform to recruit DNA damage response proteins to lesions (15,47). For example, it has been shown that PAR synthesis promotes X-ray cross-complementing protein 1 (XRCC1), an essential scaffold protein for base excision repair, recruitment at DNA damage sites and is important for XRCC1 function (47,48). In accordance with these results, in situ PLA assays showed that the co-localization of XRCC1 with PAR after exposure to MMS was compromised in MORC2 KO cells as compared with its WT counterparts, and re-expressed MORC2 or PARP1 in MORC2 KO cells partially restored the co-localization of XRCC1 with PAR (Figure 8D and E). Moreover, cells expressing acetylation-defective PARP1 K949R had reduced co-localization of XRCC1 with PAR as compared with cells expressing WT or acetylation-mimic (K949Q) PARP1 (Figure 8F). As a negative control, PLA signals failed to be detected when single antibody was used (Supplementary Figure S9B).
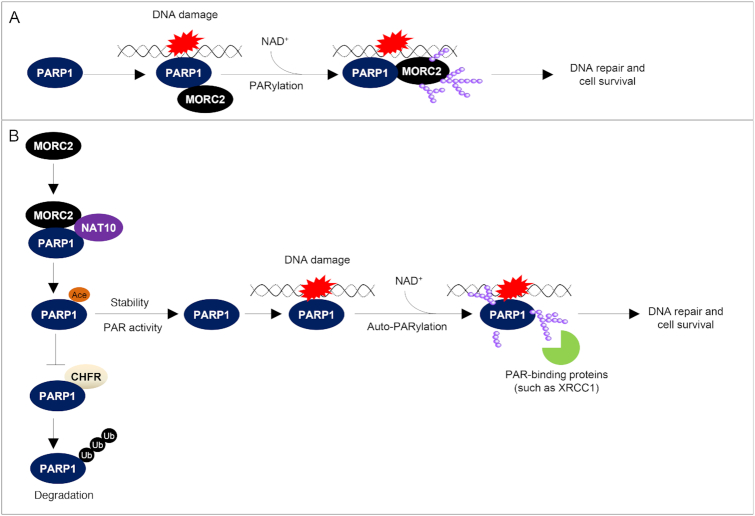
PARP1-deficient cells have reduced capacity to repair DNA damage and are therefore hypersensitive to DNA-damaging agents (19,20,37). Cell viability assays showed that cells expressing acetylation-defective PARP1 K949R were more sensitive to MMS as compared with its WT counterpart and acetylation-mimetic PARP1 K949Q mutant (Figure 8G). Moreover, cell survival was reduced in MORC2 KO cells, a phenotype that could be partially rescued by re-expression of PARP1 (Figure 8H). Together, these results suggest that PARP1 recruits MORC2 to DNA damage sites through their direct interactions and then catalyzes MORC2 PARylation to facilitate DNA damage repair (Figure 9A). Moreover, MORC2-mediated stabilization of PARP1 is implicated in DNA damage-induced PAR production and PAR-dependent DNA repair events (Figure 9B).
Figure 9.
The proposed working model. (A) Upon DNA damage, PARP1 is recruited to DNA lesions and is rapidly activated to catalyze PARylation of MORC2 using NAD+ as substrate. PARylation of MORC2 enhances its chromatin remodeling activities, thereby promoting efficient DNA repair. (B) MORC2 mediates the interaction between PARP1 and NAT10 and thereby promotes NAT10-mediated PARP1 acetylation at K949, which blocks CHFR-mediated ubiquitination and degradation of PARP1. Consequently, MORC2 regulates DNA damage-induced PAR production at the DNA damage sites and subsequent PAR-dependent recruitment of DNA repair proteins with specific PAR-binding motifs (such as XRCC1) to DNA lesions in response to DNA damage; NAD+, nicotinamide adenine dinucleotide.
DISCUSSION
In this study, we uncovered several interesting findings concerning the functional and mechanistic link between MORC2 and PARP1 in cellular response to DNA damage.
First, MORC2 is a novel PARylation target of PARP1. DDR is tightly regulated by a multitude of PTMs of chromatin or chromatin-associated proteins, which dynamically regulate protein stability, activity and protein–protein interactions (49). One of such important PTMs is PARylation, which is primarily catalyzed by PARP1, an enzyme responsible for ∼90% of the PAR synthesized under genotoxic conditions (17,29). PARylation enables to induce local chromatin restructuring through dynamic modification of chromatin-associated proteins and/or recruitment of chromatin-modifying proteins (50). Although PARylation has been extensively studied in DDR, little is known about PARylation acceptor proteins. Especially, few examples of definitive biological roles for site-specific PARylation have been reported (16). Here, we identified MORC2 as a novel binding partner and PARylation target of PARP1 (Figures 1–3). In this context, pharmacologic inhibition of PARP1, PARP1 knockdown or expression of catalytically inactive PARP1 mutant impair MORC2 PARylation. In contrast, the levels of MORC2 PARylation are elevated after genotoxic treatment and coincide with the activation of PARP1. We further showed that MORC2 is PARylated by PARP1 on two residues within the conserved CW-ZF domain. As we previously demonstrated that MORC2 is phosphorylated by PAK1 at serine 739 in response to DNA damage (10), thus PARylation is the second identified PTM of MORC2 in response to DNA damage. However, whether there is a crosstalk between phosphorylation and PARylation of MORC2 in response to DNA damage remains to be investigated in the near future.
Second, PARP1 directs MORC2 to sites of DNA damage and PARP1-mediated PARylation stimulates its chromatin remodeling activities. Emerging evidence indicates that MORC2 is a component of heterochromatin and is implicated epigenetic gene silencing by the HUSH complex (14,51). Interestingly, it has been shown that several heterochromatin-building factors, such as heterochromatin protein 1 (HP1) (52,53), SET domain bifurcated 1 (SETDB1) (54), histone deacetylase 1/2 (55) and polycomb group proteins (PcG) (56), are recruited to DNA damage sites, where they actively contribute to the DDR events. For example, HDAC1 and HDAC2, generally considered as repressive factors, accumulate at damage sites where they stimulate double-strand break repair by nonhomologous end-joining (55). In addition, heterochromatin proteins enable to undergo specific modifications to promote DNA repair in a manner that allows localized and transient chromatin relaxation (57). A case in point is KAP1, which forms heterochromatin in undamaged cells by recruiting HP1, SETDB1 and chromodomain helicase DNA binding protein 3 (CHD3) (58). Upon DNA damage, ATM-mediated phosphorylation of KAP1 at serine 824 enhances chromatin decondensation and efficient repair of damaged DNA in heterochromatin (3,59). Likewise, HP1 proteins are also phosphorylated in response to DNA damage and are important for recruiting DDR factors and dynamically reorganizing chromatin (53,60). Although one of our recent studies showed that MORC2 plays a role in the DDR through ATPase-dependent chromatin remodeling (10), how MORC2 is recruited to sites of DNA damage remains unexplored. In this study, we demonstrated that MORC2 accumulation at DNA damage sites in a PARP1-dependent manner. Furthermore, we demonstrated that PARylation, in addition to phosphorylation (10), is an important PTM for regulating the ATPase and chromatin remodeling activities of MORC2 in response to DNA damage. These findings extend our understanding of the contribution of the PARylation signaling pathway in the early chromatin remodeling at DNA lesions. In addition, previous studies have mechanistically linked PARylation at aspartate (D), glutamate (E) and lysine (K) residues of target proteins with DDR. For example, PARylation of p53 at E255, D256 and E268 has been implicated in DDR (61). Similarly, the conserved D103 is an important site of RUNX3 PARylation after DNA damage (62). As it was recently reported that serine also serves as an acceptor of PARylation upon DNA damage and this signaling is dependent on HPF1 (63,64), we cannot rule out the possibility that MORC2 PARylation at not yet identified serine residues is implicated in DDR.
Third, MORC2 stabilizes PARP1 through a crosstalk between NAT10-mediated acetylation and CHFR-mediated ubiquitination. It has been documented that the functionality and the activities of PARP1 are modulated by a variety of PTMs, such as ubiquitination and acetylation (17). In this context, E3 ubiquitin ligase CHFR has been shown to promote polyubiquitylation of PARP1 in response to DNA damage and mitotic stress (22,23). In addition, PARP1 is acetylated by the acetyltransferases p300/CBP (24) and p300/CBP-associated factor (PCAF) (65) and is deacetylated by SIRT1 (65) in addition to potential HDAC1–3 (24). Interestingly, recent two proteomic studies identified the K949 is one of ubiquitination and acetylation sites of PARP1 (42,45), thus raising the possibility that PARP1 acetylation and ubiquitination might compete for the same lysine residue. NAT10 is a member of Gcn5-related N-acetyltransferases (GNAT) family of histone acetyltransferases and exerts lysine acetyltransferase activity toward histones and nonhistone proteins (such as p53) (43). In this study, we discovered that NAT10 is a novel acetyltransferase for PARP1 acetylation at K949 and enhances PARP1 stability. Mechanistically, MORC2 forms a ternary complex with PARP1 and NAT10 and mediates the interaction between PARP1 and NAT10, thereby facilitating NAT10-mediated acetylation of PARP1 and stabilizing PARP1 (Supplementary Figure S8). In addition, we also found that MORC2 suppresses CHFR-mediated PARP1 ubiquitination (Figure 6). Expression of PARP1 K949R mutant reduced CHFR-mediated PARP1 ubiquitination, indicating that K949 could be one of ubiquitination sites for PARP1. Based on these results, we proposed that MORC2 mediates the interaction between PARP1 and NAT10 and thereby promotes PARP1 acetylation at K949 by NAT10, which blocks CHFR-mediated polyubiquitination of PARP1 at the same residue and subsequent proteasomal degradation. In support of our proposal, a crosstalk between acetylation and ubiquitination in the regulation of protein stability by competing for the same lysine residues has been documented in other proteins, such as ATP-citrate lyase (ACLY) (66).
Fourth, MORC2 is required for DNA damage-induced PAR production and PAR-dependent DNA repair signaling cascades. In response to DNA damage, PARP1 is one of the first proteins to be recruited and activated, which generates large amounts of PAR and facilitates the recruitment of DNA repair factors to DNA damage sites (15). Consequently, PARP1-defective cells have impaired DDR and are sensitive to DNA damaging agents (19,20). The role of MORC2 in controlling PARP1 stability is further supported by the evidence that DNA damage-triggered PAR production and PAR-dependent DNA repair signaling were diminished in MORC2 KO cells or cells expressing acetylation-defective PARP1 mutant. Functionally, cells depleted MORC2 or expressing acetylation-defective PARP1 mutant are sensitive to MMS-induced DNA damage. Moreover, re-expression of PARP1 in MORC2 KO cells rescued decreased cellular sensitivity to MMS treatment (Figure 8). These results indicate that MORC2 regulates DDR through, at least in part, a PARP1-dependent pathway.
In summary, findings presented here suggest that MORC2 stabilizes PARP1, which in turn PARylates MORC2, thus forming a positive feedback loop (Figure 9). This study advances our understanding of the molecular mechanisms by which multiple chromatin-remodeling enzymes cooperatively regulate chromatin remodeling and DDR to maintain genomic stability.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge the staff members of the proteomic center (Institute of Biomedical Sciences, Fudan University) for technical assistance. We are grateful to the members of the Li laboratory (especially to Hong-Yi Liu and Ying-Ying Liu) for technical assistance and to Dr Zhi-Ming Shao for funding and administrative support.
Author contributions: L.Z. performed all of the experiments and analyzed all data. D.-Q.L. conceived this study and wrote the manuscript with the input from L.Z.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China for Young Scientists [81502290 to L.Z.]; National Natural Science Foundation of China [81372847, 81572584, 81772805]; Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning [2013-06]; Science and Technology Innovation Action Plan of Shanghai Municipal Science and Technology Commission [16JC1405400]; Fudan University (to D.-Q.L.). Funding for open access charge: Lab’s money.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jeggo P.A., Pearl L.H., Carr A.M.. DNA repair, genome stability and cancer: a historical perspective. Nat. Rev. Cancer. 2016; 16:35–42. [DOI] [PubMed] [Google Scholar]
- 2. Ciccia A., Elledge S.J.. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010; 40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D.C., Lukas J., Bekker-Jensen S., Bartek J., Shiloh Y.. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 2006; 8:870–876. [DOI] [PubMed] [Google Scholar]
- 4. Murga M., Jaco I., Fan Y., Soria R., Martinez-Pastor B., Cuadrado M., Yang S.M., Blasco M.A., Skoultchi A.I., Fernandez-Capetillo O.. Global chromatin compaction limits the strength of the DNA damage response. J. Cell Biol. 2007; 178:1101–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Attikum H., Gasser S.M.. The histone code at DNA breaks: a guide to repair. Nat. Rev. Mol. Cell Biol. 2005; 6:757–765. [DOI] [PubMed] [Google Scholar]
- 6. Li D.Q., Nair S.S., Kumar R.. The MORC family: new epigenetic regulators of transcription and DNA damage response. Epigenetics. 2013; 8:685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iyer L.M., Abhiman S., Aravind L.. MutL homologs in restriction-modification systems and the origin of eukaryotic MORC ATPases. Biol. Direct. 2008; 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perry J., Zhao Y.. The CW domain, a structural module shared amongst vertebrates, vertebrate-infecting parasites and higher plants. Trends Biochem. Sci. 2003; 28:576–580. [DOI] [PubMed] [Google Scholar]
- 9. Moissiard G., Cokus S.J., Cary J., Feng S., Billi A.C., Stroud H., Husmann D., Zhan Y., Lajoie B.R., McCord R.P. et al.. MORC family ATPases required for heterochromatin condensation and gene silencing. Science. 2012; 336:1448–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li D.Q., Nair S.S., Ohshiro K., Kumar A., Nair V.S., Pakala S.B., Reddy S.D., Gajula R.P., Eswaran J., Aravind L. et al.. MORC2 signaling integrates phosphorylation-dependent, ATPase-coupled chromatin remodeling during the DNA damage response. Cell Rep. 2012; 2:1657–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He F., Umehara T., Saito K., Harada T., Watanabe S., Yabuki T., Kigawa T., Takahashi M., Kuwasako K., Tsuda K. et al.. Structural insight into the zinc finger CW domain as a histone modification reader. Structure. 2010; 18:1127–1139. [DOI] [PubMed] [Google Scholar]
- 12. Hoppmann V., Thorstensen T., Kristiansen P.E., Veiseth S.V., Rahman M.A., Finne K., Aalen R.B., Aasland R.. The CW domain, a new histone recognition module in chromatin proteins. EMBO J. 2011; 30:1939–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aravind L., Abhiman S., Iyer L.M.. Natural history of the eukaryotic chromatin protein methylation system. Prog. Mol. Biol. Transl. Sci. 2011; 101:105–176. [DOI] [PubMed] [Google Scholar]
- 14. Tchasovnikarova I.A., Timms R.T., Douse C.H., Roberts R.C., Dougan G., Kingston R.E., Modis Y., Lehner P.J.. Hyperactivation of HUSH complex function by Charcot-Marie-Tooth disease mutation in MORC2. Nat. Genet. 2017; 49:1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ray Chaudhuri A., Nussenzweig A.. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017; 18:610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gupte R., Liu Z., Kraus W.L.. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 2017; 31:101–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gibson B.A., Kraus W.L.. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012; 13:411–424. [DOI] [PubMed] [Google Scholar]
- 18. Davidovic L., Vodenicharov M., Affar E.B., Poirier G.G.. Importance of poly(ADP-ribose) glycohydrolase in the control of poly(ADP-ribose) metabolism. Exp. Cell Res. 2001; 268:7–13. [DOI] [PubMed] [Google Scholar]
- 19. Trucco C., Oliver F.J., de Murcia G., Menissier-de Murcia J.. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 1998; 26:2644–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Murcia J.M., Niedergang C., Trucco C., Ricoul M., Dutrillaux B., Mark M., Oliver F.J., Masson M., Dierich A., LeMeur M. et al.. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:7303–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lord C.J., Ashworth A.. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017; 355:1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu C., Wu J., Paudyal S.C., You Z., Yu X.. CHFR is important for the first wave of ubiquitination at DNA damage sites. Nucleic Acids Res. 2013; 41:1698–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kashima L., Idogawa M., Mita H., Shitashige M., Yamada T., Ogi K., Suzuki H., Toyota M., Ariga H., Sasaki Y. et al.. CHFR protein regulates mitotic checkpoint by targeting PARP-1 protein for ubiquitination and degradation. J. Biol. Chem. 2012; 287:12975–12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hassa P.O., Haenni S.S., Buerki C., Meier N.I., Lane W.S., Owen H., Gersbach M., Imhof R., Hottiger M.O.. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J. Biol. Chem. 2005; 280:40450–40464. [DOI] [PubMed] [Google Scholar]
- 25. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun R., Xie H.Y., Qian J.X., Huang Y.N., Yang F., Zhang F.L., Shao Z.M., Li D.Q.. FBXO22 possesses both protumorigenic and antimetastatic roles in breast cancer progression. Cancer Res. 2018; 78:5274–5286. [DOI] [PubMed] [Google Scholar]
- 27. Zhang F.L., Cao J.L., Xie H.Y., Sun R., Yang L.F., Shao Z.M., Li D.Q.. Cancer-Associated MORC2-Mutant M276I regulates an hnRNPM-Mediated CD44 splicing switch to promote invasion and metastasis in Triple-Negative breast cancer. Cancer Res. 2018; 78:5780–5792. [DOI] [PubMed] [Google Scholar]
- 28. Zhang Y., Wang J., Ding M., Yu Y.. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods. 2013; 10:981–984. [DOI] [PubMed] [Google Scholar]
- 29. Jungmichel S., Rosenthal F., Altmeyer M., Lukas J., Hottiger M.O., Nielsen M.L.. Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol. Cell. 2013; 52:272–285. [DOI] [PubMed] [Google Scholar]
- 30. Khoury-Haddad H., Guttmann-Raviv N., Ipenberg I., Huggins D., Jeyasekharan A.D., Ayoub N.. PARP1-dependent recruitment of KDM4D histone demethylase to DNA damage sites promotes double-strand break repair. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E728–E737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao J., Yu H., Lin L., Tu J., Cai L., Chen Y., Zhong F., Lin C., He F., Yang P.. Interactome study suggests multiple cellular functions of hepatoma-derived growth factor (HDGF). J. Proteomics. 2011; 75:588–602. [DOI] [PubMed] [Google Scholar]
- 32. Langelier M.F., Planck J.L., Roy S., Pascal J.M.. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012; 336:728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krukenberg K.A., Jiang R., Steen J.A., Mitchison T.J.. Basal activity of a PARP1-NuA4 complex varies dramatically across cancer cell lines. Cell Rep. 2014; 8:1808–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rolli V., O’Farrell M., Menissier-de Murcia J., de Murcia G.. Random mutagenesis of the poly(ADP-ribose) polymerase catalytic domain reveals amino acids involved in polymer branching. Biochemistry. 1997; 36:12147–12154. [DOI] [PubMed] [Google Scholar]
- 35. Feijs K.L., Forst A.H., Verheugd P., Luscher B.. Macrodomain-containing proteins: regulating new intracellular functions of mono(ADP-ribosyl)ation. Nat. Rev. Mol. Cell Biol. 2013; 14:443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M.. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998; 273:5858–5868. [DOI] [PubMed] [Google Scholar]
- 37. Rank L., Veith S., Gwosch E.C., Demgenski J., Ganz M., Jongmans M.C., Vogel C., Fischbach A., Buerger S., Fischer J.M. et al.. Analyzing structure-function relationships of artificial and cancer-associated PARP1 variants by reconstituting TALEN-generated HeLa PARP1 knock-out cells. Nucleic Acids Res. 2016; 44:10386–10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Das B.B., Huang S.Y., Murai J., Rehman I., Ame J.C., Sengupta S., Das S.K., Majumdar P., Zhang H., Biard D. et al.. PARP1-TDP1 coupling for the repair of topoisomerase I-induced DNA damage. Nucleic Acids Res. 2014; 42:4435–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ka N.L., Na T.Y., Na H., Lee M.H., Park H.S., Hwang S., Kim I.Y., Seong J.K., Lee M.O.. NR1D1 recruitment to sites of DNA damage inhibits repair and is associated with chemosensitivity of breast cancer. Cancer Res. 2017; 77:2453–2463. [DOI] [PubMed] [Google Scholar]
- 40. Soderberg O., Gullberg M., Jarvius M., Ridderstrale K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G. et al.. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 2006; 3:995–1000. [DOI] [PubMed] [Google Scholar]
- 41. Rassoolzadeh H., Coucoravas C., Farnebo M.. The proximity ligation assay reveals that at DNA double-strand breaks WRAP53beta associates with gammaH2AX and controls interactions between RNF8 and MDC1. Nucleus. 2015; 6:417–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim W., Bennett E.J., Huttlin E.L., Guo A., Li J., Possemato A., Sowa M.E., Rad R., Rush J., Comb M.J. et al.. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011; 44:325–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu X., Tan Y., Zhang C., Zhang Y., Zhang L., Ren P., Deng H., Luo J., Ke Y., Du X.. NAT10 regulates p53 activation through acetylating p53 at K120 and ubiquitinating Mdm2. EMBO Rep. 2016; 17:349–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Larrieu D., Britton S., Demir M., Rodriguez R., Jackson S.P.. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science. 2014; 344:527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lundby A., Lage K., Weinert B.T., Bekker-Jensen D.B., Secher A., Skovgaard T., Kelstrup C.D., Dmytriyev A., Choudhary C., Lundby C. et al.. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012; 2:419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marsischky G.T., Wilson B.A., Collier R.J.. Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation. Evidence for active site similarities to the ADP-ribosylating toxins. J. Biol. Chem. 1995; 270:3247–3254. [DOI] [PubMed] [Google Scholar]
- 47. Li M., Lu L.Y., Yang C.Y., Wang S., Yu X.. The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev. 2013; 27:1752–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Breslin C., Hornyak P., Ridley A., Rulten S.L., Hanzlikova H., Oliver A.W., Caldecott K.W.. The XRCC1 phosphate-binding pocket binds poly (ADP-ribose) and is required for XRCC1 function. Nucleic Acids Res. 2015; 43:6934–6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dantuma N.P., van Attikum H.. Spatiotemporal regulation of posttranslational modifications in the DNA damage response. EMBO J. 2016; 35:6–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tallis M., Morra R., Barkauskaite E., Ahel I.. Poly(ADP-ribosyl)ation in regulation of chromatin structure and the DNA damage response. Chromosoma. 2014; 123:79–90. [DOI] [PubMed] [Google Scholar]
- 51. Liu N., Lee C.H., Swigut T., Grow E., Gu B., Bassik M.C., Wysocka J.. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature. 2018; 553:228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Luijsterburg M.S., Dinant C., Lans H., Stap J., Wiernasz E., Lagerwerf S., Warmerdam D.O., Lindh M., Brink M.C., Dobrucki J.W. et al.. Heterochromatin protein 1 is recruited to various types of DNA damage. J. Cell Biol. 2009; 185:577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Baldeyron C., Soria G., Roche D., Cook A.J., Almouzni G.. HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J. Cell Biol. 2011; 193:81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Alagoz M., Katsuki Y., Ogiwara H., Ogi T., Shibata A., Kakarougkas A., Jeggo P.. SETDB1, HP1 and SUV39 promote repositioning of 53BP1 to extend resection during homologous recombination in G2 cells. Nucleic Acids Res. 2015; 43:7931–7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Miller K.M., Tjeertes J.V., Coates J., Legube G., Polo S.E., Britton S., Jackson S.P.. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 2010; 17:1144–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chou D.M., Adamson B., Dephoure N.E., Tan X., Nottke A.C., Hurov K.E., Gygi S.P., Colaiacovo M.P., Elledge S.J.. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:18475–18480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Goodarzi A.A., Noon A.T., Jeggo P.A.. The impact of heterochromatin on DSB repair. Biochem. Soc. Trans. 2009; 37:569–576. [DOI] [PubMed] [Google Scholar]
- 58. Ryan R.F., Schultz D.C., Ayyanathan K., Singh P.B., Friedman J.R., Fredericks W.J., Rauscher F.J. 3rd.. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell Biol. 1999; 19:4366–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Goodarzi A.A., Noon A.T., Deckbar D., Ziv Y., Shiloh Y., Lobrich M., Jeggo P.A.. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell. 2008; 31:167–177. [DOI] [PubMed] [Google Scholar]
- 60. Dinant C., Luijsterburg M.S.. The emerging role of HP1 in the DNA damage response. Mol. Cell Biol. 2009; 29:6335–6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kanai M., Hanashiro K., Kim S.H., Hanai S., Boulares A.H., Miwa M., Fukasawa K.. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat. Cell Biol. 2007; 9:1175–1183. [DOI] [PubMed] [Google Scholar]
- 62. Tay L.S., Krishnan V., Sankar H., Chong Y.L., Chuang L.S.H., Tan T.Z., Kolinjivadi A.M., Kappei D., Ito Y.. RUNX Poly(ADP-Ribosyl)ation and BLM interaction facilitate the fanconi anemia pathway of DNA repair. Cell Rep. 2018; 24:1747–1755. [DOI] [PubMed] [Google Scholar]
- 63. Palazzo L., Leidecker O., Prokhorova E., Dauben H., Matic I., Ahel I.. Serine is the major residue for ADP-ribosylation upon DNA damage. eLife. 2018; 7:e34334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bonfiglio J.J., Fontana P., Zhang Q., Colby T., Gibbs-Seymour I., Atanassov I., Bartlett E., Zaja R., Ahel I., Matic I.. Serine ADP-Ribosylation depends on HPF1. Mol. Cell. 2017; 65:932–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rajamohan S.B., Pillai V.B., Gupta M., Sundaresan N.R., Birukov K.G., Samant S., Hottiger M.O., Gupta M.P.. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol. Cell Biol. 2009; 29:4116–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lin R., Tao R., Gao X., Li T., Zhou X., Guan K.L., Xiong Y., Lei Q.Y.. Acetylation stabilizes ATP-citrate lyase to promote lipid biosynthesis and tumor growth. Mol. Cell. 2013; 51:506–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.