Abstract
Small nuclear ribonucleoprotein polypeptide G (SNRPG), often referred to as Smith protein G (SmG), is an indispensable component in the biogenesis of spliceosomal uridyl-rich small nuclear ribonucleoprotein particles (U snRNPs; U1, U2, U4 and U5), which are precursors of both the major and minor spliceosome. SNRPG has attracted significant attention because of its implicated roles in tumorigenesis and tumor development. Suggestive evidence of its varying expression levels has been reported in different types of cancers, which include breast cancer, lung cancer, prostate cancer and colon cancer. The accumulating evidence suggests that the splicing machinery component plays a significant role in the initiation and progression of cancers. SNRPG has a wide interaction network, and its functions are predominantly mediated by protein-protein interactions (PPIs), making it a promising anti-cancer therapeutic target in PPI-focused drug technology. Understanding its roles in tumorigenesis and tumor development is an indispensable arsenal in the development of molecular-targeted therapies. Several antitumor drugs linked to splicing machinery components have been reported in different types of cancers and some have already entered the clinic. However, targeting SNRPG as a drug development tool has been an overlooked and underdeveloped strategy in cancer therapy. In this article, we present a comprehensive and perspective view on the oncogenic potential of SNRPG in PPI-focused drug discovery.
Keywords: DEAD-box helicase 20, mRNA splicing, RBBP6, SNRPG, protein-protein interactions, transforming acidic coiled-coil containing protein 1, tumorigenesis
Introduction
Protein-protein interactions (PPIs) are indispensable in normative cellular processes and are tremendously important mediators in the progression of many disease states [1-3]. More than 600,000 disease-relevant PPIs have so far been reported in the human interactome [4,5], most of which remain elusive and underexplored. Optimizing the integration of PPIs with conventional and targeted cytotoxic therapies may lead to greatly protracted remissions and even curative therapies for several diseases, including cancer [6-8]. Over the years PPIs have been regarded as prototypically “intractable” and “undruggable” owing to their highly dynamic and expansive interfacial areas [2,3]. However, owing to improving technology expertise, the advent of PPI-focused smart-drug technology presents a notable advance in disease diagnostics and therapeutic studies [7-9]. PPIs have emerged as significant arsenals in the drug development armory and inhibiting PPIs using small molecules or peptides modulates biochemical pathways and has therapeutic significance [4,7,8,10].
The emergence of PPI-focused drug technology has prompted scientists to consider targeting splicing machinery components as possible targets in alleviating the existing cancer challenges [5-7]. The strategy ushered in a new dawn in the field of drug discovery. A few drugs are already on the market and some potential drug-like candidates are in clinical trials [2,3,11-13]. Nevertheless, targeting Smith (Sm) proteins as PPI drug development tools has been an overlooked and underdeveloped strategy in cancer therapy [7,14]. Varying expression levels have been reported in different types of cancers, which include breast cancer, lung cancer, prostate cancer and colon cancer [15-20]. However, very little is known about their putative interactions in cancer-cell protein networks and their roles in different types of cancers.
Understanding their functional implications may lead to new avenues to design and develop PPI-focused therapeutic drugs in cancer [2,3,5-7,21,22]. In this article, we present a comprehensive view and perspective on the oncogenic potential of SNRPG in PPI-focused drug technology.
PPI interfaces
PPIs occur over a relatively large interfacial area of approximately 1000 to 4000 Å2. The area is relatively prodigious in comparison to the mean contact area obligated for inhibition by small molecule inhibitors (300 to 1000 Å2) [23,24]. The interfacial area of PPIs harbors incontrovertible hydrophobic regions called “hot spots”. These hydrophobic regions contribute to the binding affinity and help to hold the two interacting proteins together [25,26]. Typically, hot spot density on the protein-protein interface composes 10% of the binding site residues. The amount of the structurally conserved residues (energetic hot spots) increases with the expansion of the interacting surface area [27].
Hot spot regions usually occur in clusters and within each cluster tightly packed hot spots form a network of conserved interactions called hot regions (shown in Figure 1) [28]. The cooperative contributions of hot spots within one hot region stabilize PPIs. Hot regions are networked; their energetic contributions can be additive or cooperative and contribute dominantly to the stability of PPIs [29,30]. Hot spot pockets for PPIs are distinguishable from the other regions of the protein surface owing to their concave topology, combined with a pattern of hydrophobic and polar functionality. This combination of properties confers on concave hot regions a tendency to bind other proteins and small organic compounds possessing some polar functionality decorating a largely hydrophobic scaffold [31].
Figure 1.
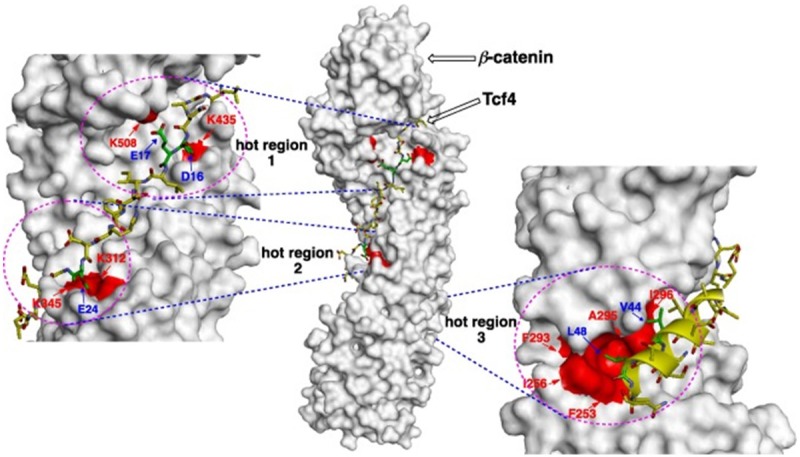
Cartoon representation of β-catenin/T-cell factor (Tcf) PPI interface showing three hot regions. Hot region 1 (K435 and K508 of β-catenin and D16 and E17 of Tcf4), hot region 2 (K312 and K345 of β-catenin and E24 and E29 of Tcf4) and hot region 3 (F253, I256, F293, A295, and I296 of β-catenin and V44 and L48 of Tcf4). The cooperative contributions of hot spots within one hot region stabilize PPIs (Figure taken from [31]).
Hot spot regions are rich in hydrogen bonding and hydrophobic amino acids (Trp, Arg and Tyr), which contribute to π-interactions and the binding of free energy [32]. Systematic alanine scanning mutagenesis has revealed that exchanging amino acid residues for alanine in these hot spot regions reduces the binding affinity by at least 2 kcal/mol [32]. Further analysis observed that hot spot regions comprise a core and a rim region. The rim region has an amino acid composition similar to the whole interfacial area, whereas the core region consists solely of aromatic residues [33,34]. The core region fosters the α-helix, β-sheet and β-turn motifs, with the α-helix having a higher ratio in most of the secondary protein structures. The α-helix actively binds into the grooves of binding partners and modulates the functioning of a large number of the disease-relevant PPIs [26,35].
Comprehensive view
SNRPG is an approximately 8.5 kDa core-splicing and cancer-implicated Sm protein whose functions are predominantly mediated by PPIs [16,36,37]. The SNRPG protein coding gene is found on chromosome 2p13.3 and is made up of 8 exons. The gene comprises 455 nucleotides with an open reading frame encoding a predicted protein of 76 amino acid residues. An important paralog of this gene is LSM7. SNRPG protein has a theoretical pI of 8.9 and is translated in vitro from a single SNRPG mRNA that migrates as a doublet on high-TEMED SDS-PAGE [38]. The two bands represent conformational isomers of the same protein. However, several transcript variants encoding different isoforms have been found for this gene. Northern blot analysis revealed that the SNRPG gene is expressed as an approximately 0.5-kb mRNA in HeLa cells [39].
SNRPG is a bona fide component of survival of motor neurons (SMN)-Sm protein complex, U1 snRNP, U2 snRNP, U12 type spliceosomal complex, U4 snRNP, U5 snRNP, spliceosomal tri-snRNP complex, catalytic step 2 spliceosome, Cytosol, methylosome, nucleoplasm, small nuclear ribonucleoprotein complex and spliceosomal complex [21]. Among its related pathways are the mRNA splicing-minor pathway and transport of the SLBP independent mature mRNA. The protein may also be a part of the U7 small nuclear ribonucleoprotein (U7 snRNP) complex, which participates in the processing of the 3’ end of histone transcripts [21]. However, it plays a yet uncharacterised role in linking core pre-mRNA splicing proteins to various cancers.
As shown in Figure 2, varying expression levels of SNRPG have been reported in different types of cancers, which include colorectal cancer, breast cancer, lung cancer, prostate cancer and liver cancer [15-20]. According to Blijlevens and co-workers, increased expression levels of SNRPG protein in different types of cancers show a positive correlation with disease initiation, progression and severity [40]. The varying expression levels of SNRPG in different types of cancers may be explained by the overexpression of the protein, the mislocalisation of unassembled protein or the mislocalisation of misassembled protein [41]. Thus, SNRPG may contribute significantly to the initiation and progression of cancers [14,16,37,42-46].
Figure 2.
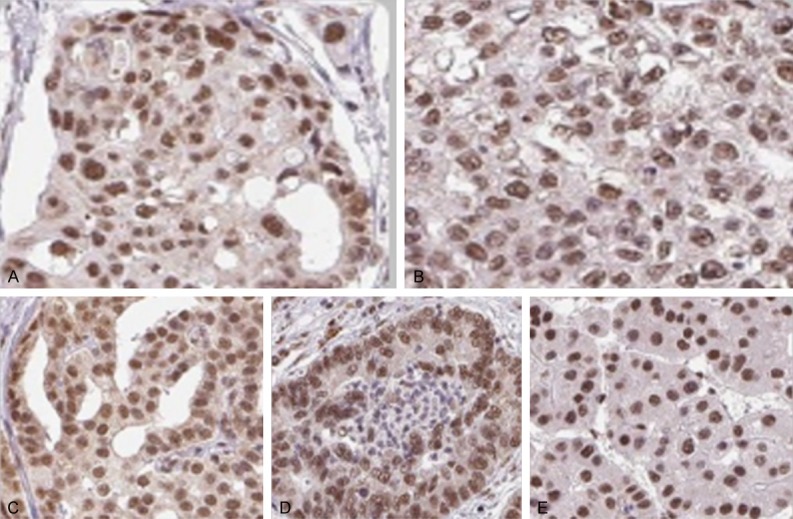
Antibody staining of five standard cancer tissues samples highlighting the localization of SNRPG in tumor cells. A. Colorectal Cancer. B. Breast Cancer. C. Prostate Cancer. D. Lung Cancer. E. Liver Cancer. Antibodies are labeled with DAB (3,3’-diaminobenzidine) and the resulting brown staining indicates where an antibody has bound to its corresponding antigen (SNRPG). Staining: Medium, Intensity: Moderate, Quantity: > 75%, Location: Nuclear, Magnification: 40 × (Figure taken from [18]).
SNRPG, like other Sm proteins, is characterised by the presence of a conserved motif called the Sm motif. As shown in Figure 3, the Sm motif consists of two conserved regions that are separated by a non-conserved linker region, Sm1 and Sm2. The conserved motif comprises an antiparallel β sheet of β5↑•β1↓•β2↑•β3↓•β4↑ topology [39]. Several of the Sm subunits are decorated by additional unstructured C terminal extensions and secondary structure elements. The Sm motif encodes for a common folding domain (Sm domain) that is responsible for mediating PPIs between Sm proteins through the antiparallel β strands [47]. Moreover, SNRPG possesses two solvent-exposed hydrophobic interaction surfaces that are prone to nonspecific interactions under physiological conditions [47-52]. According to Stark and co-workers SNRPG has a wide interaction network comprising more than 138 interactions with more than 115 identified interactors [21]. Its functions are mediated by both the specific and non-specific PPIs.
Figure 3.
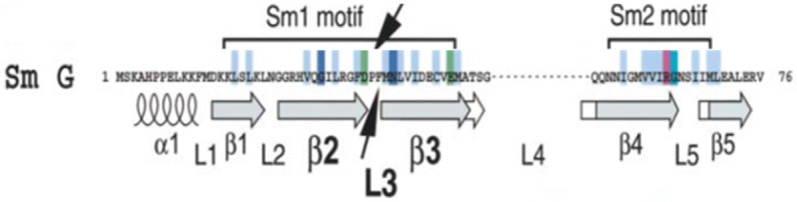
Human SNRPG protein primary structure alignment showing Sm1 and Sm2 motifs. Conserved amino acids are highlighted as follows: Light blue (uncharged hydrophobic residues), green (acidic amino acids), purple (basic amino acids), dark blue (100% conserved amino acids) and turquoise (80% conserved glycine). Arrows mark the cross-linked amino acids in the protein sequences as identified by N-terminal sequencing, for example Phe37, Met38 and Asn39. The cross-linking sites are located within loop L3 of the Sm1 motif (Figure taken from [39]).
Prior to their involvement in the splicing cycle, SNRPG together with the other Sm proteins initially undergo translation in the cytoplasm and follow a hierarchical maturation pathway in which they interact independently of snRNA (shown in Figure 5) [53]. The activity is mediated predominantly by the assembly chaperone pICln, which inhibits the pre-mature binding of Sm proteins onto U snRNA and recruits all newly synthesized Sm proteins to the protein arginine methyltransferase 5 (PRMT5) complex forming three hetero-oligomers, D3/B, D1/D2 and E/F/G [22,41,52,54]. The PRMT5-complex (comprising the Type II methyltransferase PRMT5, WD45 and pICln) promotes symmetric dimethylation of arginines on Sm proteins B/B’, D1 and D3. The SMN-complex interacts with the PRMT5-complex facilitating the assembly of Sm proteins onto the “Sm-site” of U snRNAs forming the U snRNPs, as shown in Figure 4 [41]. After additional modification and processing steps, the U snRNP is targeted to its nuclear site of function, where it ultimately accumulates in interchromatin region structures known as splicing speckles [22,41,53].
Figure 5.
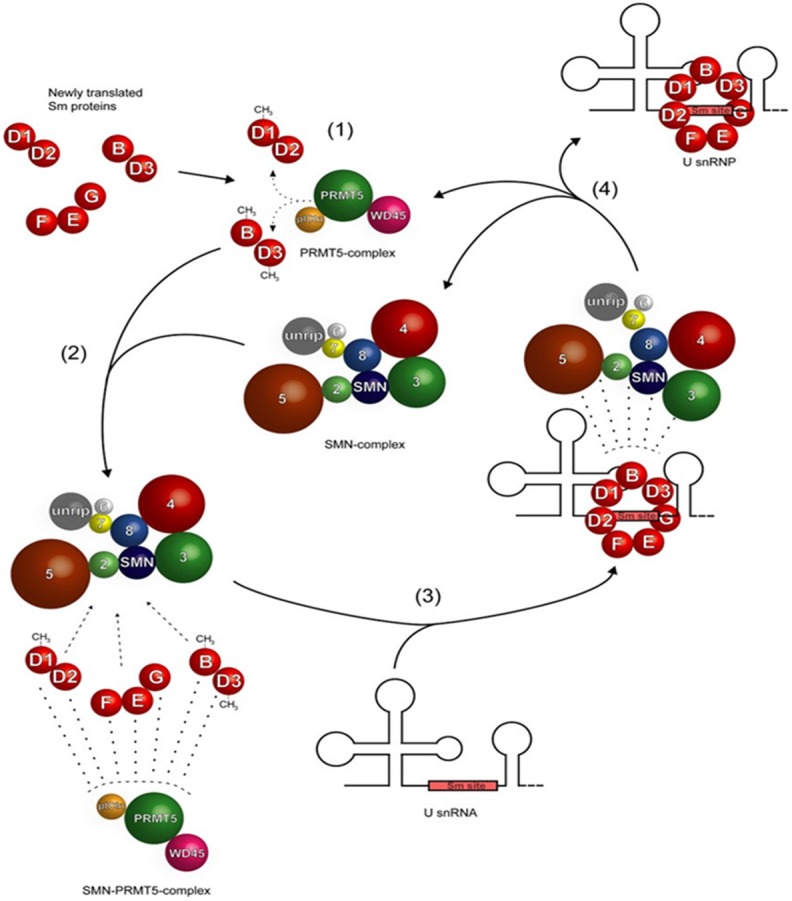
Model of assisted assembly of U snRNPs. Sm proteins are initially translated in the cytoplasm and sequestered by the PRMT5-complex, consisting of the Type II methyltransferase PRMT5, WD45 (also termed Mep50) and pICln, which promotes symmetric dimethylation of arginines on Sm proteins B/B0, D1 and D3 (step 1). Next, the SMN-complex interacts with the PRMT5-complex to form an SMN-PRMT5-complex in which the Sm proteins are transferred onto the SMN-complex (step 2). These Sm proteins are assembled onto the “Sm-site” of U snRNAs to form U snRNPs (step 3). Finally, the U snRNP, the SMN-complex and PRMT5-complex dissociate and the latter two engage in a new round of U snRNP (Figure extracted from [53]).
Figure 4.
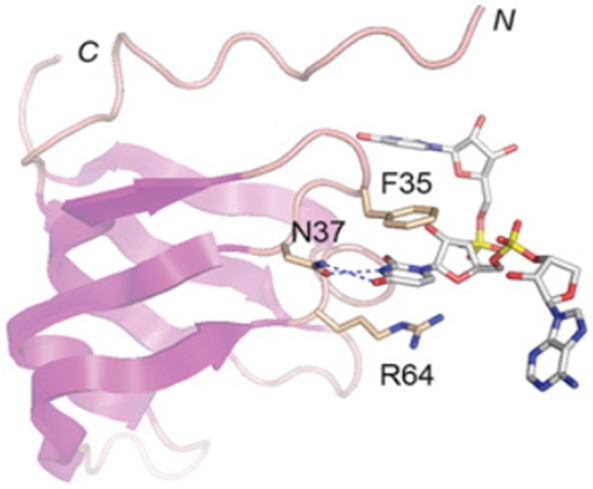
Stereo view of the human SNRPG (depicted as a cartoon trace with magenta β strands) and its interactions with the Sm site in U1 snRNA. Selected amino acids are shown as stick models and numbered according to their positions in the SNRPG polypeptide. Atomic contacts are indicated by dashed lines (Figure taken from [14]).
Very little is known about the manner in which the Sm proteins recognize and interact with the RNA-Sm site element. However, SNRPG has been highlighted to play a critical role in the direct recognition of the Sm site in the U snRNPs assembly [55]. According to Heinrichs and co-workers a direct contact between the SNRPG and the 5’ part of the Sm site element within HeLa U1 snRNP particles was demonstrated by cross-linking approaches [55]. As indicated in Figure 2, the cross-linking sites are located within loop L3 of the Sm1 motif. The cross-linked amino acids are identified in the protein sequences as identified by N-terminal sequencing: Phe37, Met38 and Asn39 [55]. The cross-linking observed for the SNRPG is an outstanding feature and provides the first line of evidence that SNRPG plays a yet uncharacterized and pivotal role in the functional activities of Sm proteins.
In this context, cells are engineered to use a plethora of PPI networks to provide a therapeutically tractable way of tweaking and manipulating the interplay of Sm proteins in order to maintain and address the normative cellular functions and progression of many disease states, including cancer [1,56]. As shown in Figure 6, deficiency of pICln due to pathophysiological cues and disease progression has been linked to mislocalization of the unassembled and/or misassembled Sm proteins and their subsequent degradation via autophagy [41]. SMN deficiency has been linked to accumulation of Sm proteins over the pICln [41,51,57]. The reduced expression of functional SMN caused by genomic mutations has been linked to the debilitating human disorder spinal muscular atrophy [58].
Figure 6.
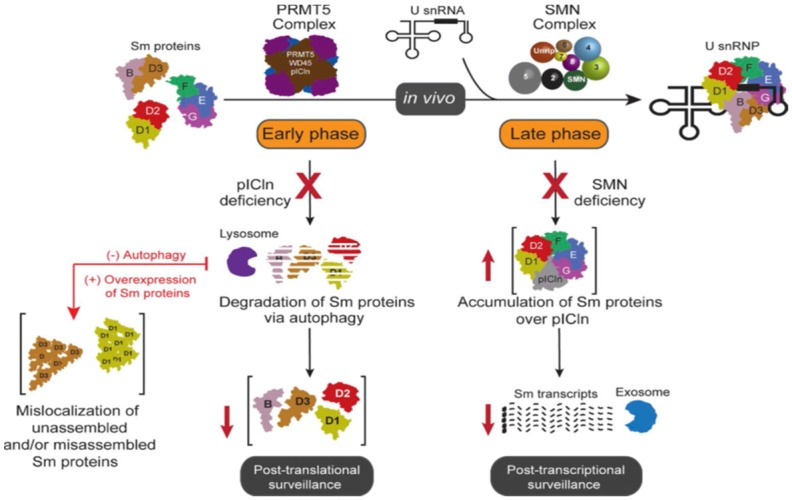
Schematic representation of the dysregulatory events in the homeostasis of U snRNPs. Dysregulation of Sm proteins during U snRNP assembly causes cellular proteotoxicity. Early phase plCln deficiency leads to degradation of Sm proteins via autophagy and mislocalisation of unassembled and/or misassembled Sm proteins. Late phase SMN deficiency leads to accumulation of Sm proteins over plCln (Figure taken from [53]).
In case of abnormality or impairment in the regulation of the assembly pathway, the cell activates fail-safe measures, including targeted autophagosome-mediated Sm protein degradation and exosome-processed Sm-encoding transcript degradation [41,51,57,58]. These measures refine cellular quality control mechanisms to prevent proteotoxicity during imbalances in UsnRNP assembly [41]. Therefore, the regulation of Sm proteins during U snRNP assembly is tremendously important to prevent cellular proteotoxicity in disease progression [22,41,53].
Perspective view
Recent studies have shown significant evidence that deregulation of spliceosomal Sm proteins is linked to pathophysiological cues and disease states, such as cancer [40,59]. The interference of Sm protein expression has been shown to induce apoptosis in non-small-cell lung cancer (NSCLC) cells [40]. According to Blijlevens and co-workers Sm proteins are frequently upregulated in NSCLC and their increased expression shows positive correlation with disease severity [40]. Despite their inability to induce apoptosis in non-malignant cells, Sm proteins represent a particularly useful novel target for selective treatment of NSCLC [40]. However, their functional basis remains elusive and yet to be fully understood.
In another study, Jin and co-workers investigated the effects of silencing SNRPN expression on cell growth using the Daoy human medulloblastoma cell line in vitro [60]. The study observed that the knockdown of SNRPN markedly reduced the proliferation and colony-forming ability of Daoy medulloblastoma cells. The results indicate that SNRPN may be a potential novel target for the development of pharmacological therapeutics in human medulloblastoma [60]. Variable methylation of SNRPN has also been linked to germ cell tumors and acute myeloid leukemia [61,62]. SNRPN depletion inhibits the proliferation and colony formation of BxPC-3 pancreatic adenocarcinoma cells, leading to S phase cell cycle arrest and cell accumulation at the sub G1 phase [63]. However, the signalling pathway of SNRPN involved in the BxPC-3 cell proliferation and tumorigenesis remains unclear and yet to be fully elucidated. The results suggest that SNRPN may promote pancreatic adenocarcinoma cell growth via regulation of the cell cycle and apoptosis, and lentivirus mediated SNRPN knockdown may be a potential therapeutic method for the treatment of pancreatic cancer [60].
Using semi-quantitative RT-PCR, Anchi and co-workers also reported the involvement of SNRPE in cell proliferation and progression of high-grade prostate cancer through the regulation of androgen receptor expression [64]. SNRPE overexpression promoted prostate cancer cell proliferation in high-grade prostate cancer cells compared with normal prostatic epithelial cells, indicating its oncogenic effects. Its knockdown expression by short interfering RNA (siRNA) resulted in the marked suppression of prostate cancer cell proliferation [64]. Furthermore, the study observed that the regulation of androgen receptor expression by SNRPE is essential for cell proliferation and progression of high-grade prostate cancer. Thus, SNRPE may present a novel molecular target for cancer drugs [64].
SNRPG is one good example of proteins that have been implicated in cancer and whose functions are predominantly mediated by PPIs [16,36,37]. According to Johnson and co-workers most cancer-implicated proteins possess structural domains that have a higher ratio of infidelity compared to non-cancer-implicated proteins, making them more prone to interaction with a wide diversity of proteins [65]. Cancer-implicated proteins have a large number of interacting proteins and occupy a central position in cancer-cell protein networks [66-68]. PPIs between cancer-implicated proteins have a higher probability of being related to the cancer processes than non-interacting proteins [1,56,65,69]. The accumulative and suggestive evidence of the varying expression levels in more than 20 different types of cancers makes SNRPG a promising anti-cancer therapeutic target in PPI-focused drug technology [16].
SNRPG and retinoblastoma binding protein 6 (RBBP6)
Putative PPIs between SNRPG and RBBP6 have been suggested. RBBP6 is a 250 kDa splicing-associated human protein initially known to bind to the retinoblastoma gene product, pRB [15,17]. The RBBP6 gene is known to possess six different domains (shown schematically in Figure 7) that have been characterized and linked with different types of cancer [15]. RBBP6 has three well-conserved N-terminal domains (“domain with no name” (DWNN), zinc knuckle and “really interesting new gene” (RING) finger domain) and three C-terminal domains (proline-rich SR domain, Rb-binding domain and p53-binding domain) [15,70,71]. Even though the functions of the N-terminal domains are not yet fully understood, it is well understood that RBBP6 is linked to tumorigenesis and tumor development, and its functions are predominantly mediated by PPIs [17]. RBBP6 has been characterized and linked with 14 different types of cancer at varying expression levels [15,19,42,44,46,71-75]. It interacts with the two-prototypical tumor suppressor proteins p53 and pRB [15,17,19].
Figure 7.
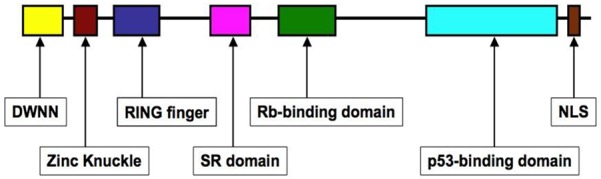
The domain organisation in human RBBP6. RBBP6 has three well-conserved N-terminal domains namely (i) “Domain with no name” (DWNN), (ii) Zinc knuckle and (iii) RING (really interesting new gene) finger domain, and three C-terminal domains, viz (i) proline-rich SR domain, (ii) Rb-binding domain and (iii) p53-binding domain (Figure taken from [76]).
Among other oncogenic functions, RBBP6 facilitates interaction between p53 and its negative regulator, MDM2, leading to enhanced p53 ubiquitination and degradation [17,19]. It also interferes with the binding of p53 to DNA and facilitates the ubiquitination of pRb [17]. RBBP6 interacts directly with the pro-proliferative transcription factor Y-box-binding protein-1 (YB-1). Its overexpression in cultured mammalian cells leads to suppression of the anti-apoptotic YB-1 in a proteasome-dependent manner [71]. However, because it down-regulates both the pro-apoptotic p53 and the anti-apoptotic YB-1, the effect of RBBP6 on tumorigenesis is likely to be highly complex [45].
Accumulative evidence has shown that RBBP6 interacts with core splicing Sm proteins, SNRPB [15] and SNRPG [43,45]. Using immunoblot analysis, Simons and co-workers observed that the N-terminal domain of RBBP6 interacts with Sm proteins [15]. The result points to a possible connection between tumor suppressor proteins and the splicing machinery components. The putative effects of the N-terminal domain of RBBP6 on Sm proteins is an interesting finding that may catapult investigations to see whether tumor suppressor proteins can directly influence Sm proteins in pre-mRNA splicing. Using a yeast 2-hybrid (Y2H) technique, Chibi and co-workers postulated that RBBP6 may perhaps interact with the core splicing SNRPG protein through its N-terminal domains, which is a crucial component of the RNA processing machinery in the cell [43]. These suggestions substantiate the possible involvement of RBBP6 in pathways linked to the pre-mRNA splicing machinery. However, the precise mechanisms involved remain elusive and yet to be characterised.
Furthermore, Kappo and co-workers identified two copies of SNRPG (conformational isomers of the same protein) as part of the five substrates that bind to the N-terminal domain of RBBP6 [45]. The Y2H findings support the results that there might be a link between RBBP6 and the Sm proteins in the initiation and progression of cancers [15,43]. Considering the critical role played by SNRPG in the formation of the hetero-oligomer E/F/G, Sm protein assembly and the subsequent assembly of Sm proteins onto the “Sm-site” of U snRNAs forming the U snRNPs, the results may suggest a possible strong link between SNRPG, pRb/p53 pathways and tumorigenesis [37]. Although many aspects of the above model remain to be proven and the mechanisms and functional basis of the links have yet to be fully understood, the findings have strong and interesting implications that prompt further investigations into the oncogenic potential of the core splicing SNRPG protein in the initiation and progression of cancers.
The possible connection between SNRPG and the N-terminal domains of RBBP6 relates to features that suggest that these proteins may be the “forgotten link” connecting the cellular pre-mRNA splicing mechanism to tumorigenesis and tumor development. First, the abundant localization of SNRPG and RBBP6 in the nucleolus and nuclear speckles in tumor cells indicates a close connection between RBBP6 and several pre-mRNA splicing components [77]. Second, the association of RBBP6 with the Sm antigens in nuclear extracts, as shown by coimmunoprecipitation, suggests that RBBP6 is associated with these splicing factors in the living cells. Third, RBBP6 cDNA encodes an SR region that has biochemical properties similar to known SR splicing proteins, mediates the ubiquitination of p53 and pRb and interacts with SNRPB and SNRPG through its DWNN. It has been speculated that RBBP6 may link mRNA 3’-end processing to pRb/p53 pathways and tumorigenesis through its DWNN [37]. However, the question of how this happens remains unanswered.
The physiological relevance of the interactions and the functional basis of the association between SNRPG and DWNN still remain obscure. Very little is known about the putative PPIs between SNRPG and the N-terminal domains of RBBP6. The two proteins play an as yet uncharacterised role in linking splicing machinery components to tumorigenesis and tumor development in various cancers. Perhaps understanding the binding events between RBBP6’s N-terminal domains and SNRPG may lead to new knowledge on how the two proteins relate in regulating splicing, tumorigenesis and tumor development. Inhibiting these PPIs may present a potential drug target in cancer diagnostics and therapeutic studies. Thus, new avenues to design and develop new therapeutic drugs may be established.
SNRPG and transforming acidic coiled coil containing protein 1 (TACC1)
The TACC1 gene is in region p12 of chromosome 8. Its mRNA is ubiquitously expressed and encodes a protein with an apparent molecular mass of 125 kDa [16]. The TACC1 protein’s subcellular localization is within the cell cytoplasm and especially concentrated in the perinuclear area [16]. TACC1 was first identified as a potential oncogene; it is sometimes included in the amplification of the 8p12 region in breast cancers and can transform fibroblasts [78]. Based on the differential expression assay of chromosome 8p11-21 genes, researchers identified the TACC1 gene, whose mRNA is reduced or absent in breast carcinomas [79]. TACC1 mRNA gene expression is downregulated in various types of tumors. Using immunohistochemistry of tumor tissue-microarrays and sections, the level of TACC1 protein is down-regulated in breast cancer [16]. Furthermore, using the two-hybrid screen in yeast, GST pull-downs and co-immunoprecipitations, Conte and co-workers identified SNRPG as one of the two potential binding partners for TACC1 in breast cancer (shown in Figure 8) [16]. The findings suggested that TACC1 might play a role in the control of mRNA metabolism. Thus, Conte and co-workers speculated that down-regulation of TACC1 may alter the control of mRNA homeostasis in polarized cells and subsequently participate in the oncogenic processes [16].
Figure 8.
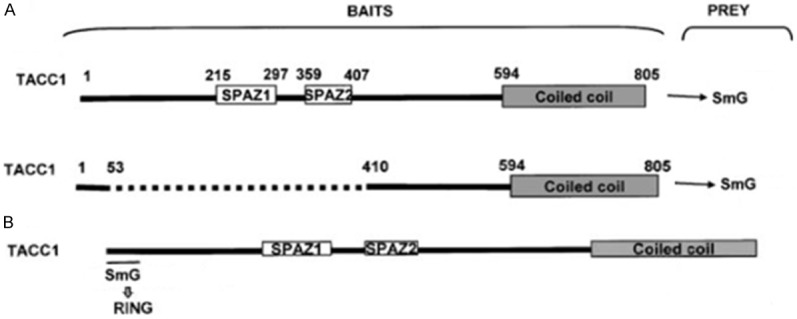
Mapping of TACC1/SNRPG interactions using the two-hybrid method in yeast. A. Schematic representation of TACC1 protein showing its three different regions: N-terminus, central serine/proline-rich region with two SPAZ motifs and coiled-coil C-terminus. Below are the different fragments generated as baits. B. Results of the mapping showing SNRPG binding to the N-terminus region of TACC1 in yeast two-hybrid experiments (Figure taken from [16]).
To delineate the region of TACC1 that interacts with SNRPG, TACC1 fragment encoded proteins fused to the LEX binding domain were used as baits in two-hybrid assays against the SNRPG prey [80]. As shown in Figure 8, the results observed that the binding region of SNRPG on TACC1 is thus restricted to the N-terminus region of TACC1. GST pull-down and co-immunoprecipitation experiments confirmed that the N-terminus region of TACC1 is indeed the site of binding for the Sm protein [80]. Inhibiting the interaction between TACC1 and SNRPG, using small molecules or peptides may modulate cancer-cell networks and have therapeutic significance. Hence investigating the binding events in the interactions between SNRPG and TACC1, and their relations in regulating splicing, tumorigenesis and tumor development may help identify new avenues to design and develop PPI-focused therapeutic drugs. Currently, the physiological relevance of the interactions and the functional basis for their association remain elusive and uncharacterized.
SNRPG and DEAD-box helicase 20 (DDX20)
DEAD-box helicase 20 (DDX20), commonly known as gem-associated protein 3 (Gemin3), is an ATP-dependent enzyme in humans that is encoded by the DDX20 gene [81,82]. It is a component of the SMN complex that is tremendously important in the assembly and reconstruction of different Sm protein complexes [83]. Cleavage of the DDX20 by the poliovirus-encoded proteinase 2Apro has been shown to result in DDX20 inactivation and reduced snRNP assembly [84]. DDX20 may act as a tumor suppressor in hepatocellular carcinoma and as a tumor promoter in breast cancer [85]. According to Chen and co-workers, DDX20 deficiency enhances NF-κB activity by impairing the NF-κB-suppressive action of microRNAs. The findings suggest that dysregulation of the microRNA machinery components may also be involved in pathogenesis in various human diseases such as cancer [85].
One good example is miRNA-140, which acts as a liver tumor suppressor. Deficiency of DDX20 leads to the impairment of miRNA-140 function. Functional impairment of miRNAs has been linked to hepatocarcinogenesis [85]. Similarly, DDX20 may promote the progression of prostate cancer through the NF-κB pathway [85]. Clinical investigations by Shin and co-workers found that a positive DP103/NF-κB feedback loop promotes constitutive NF-κB activation in invasive breast cancers [86]. The activation of this pathway is linked to cancer progression and the acquisition of chemotherapy resistance. It implies that DP103 has potential as a therapeutic target for breast cancer treatment [86].
DEAD box proteins have been found to be involved in many aspects of RNA metabolism, including Sm-Sm protein interactions, pre-mRNA splicing, mRNA transport, mRNA degradation and translation in eukaryotes and prokaryotes [87-92]. Using a biochemical approach, Charroux and co-workers observed that anti-DDX20 mAbs immunoprecipitated the spliceosomal RNPG protein, as well as several other unidentified Sm proteins. Gemin3 interacts directly with Sm core proteins, including B/B’, D2, and D3 [93]. In addition, DDX20 is uniformly distributed in the cytoplasm, where U snRNP assembly takes place, and it can be specifically co-immunoprecipitated with the cytoplasmic pool of Sm proteins [93]. Taken together, these findings suggest that DDX20 and SNRPG may play an important role linking the spliceosomal snRNP biogenesis to tumorigenesis and tumor development. Finding small-molecule or peptide inhibitors for the interaction between DDX20 and SNRPG may help modulate cancer-cell networks and open up other avenues for the designing and development of new PPI-focused smart drugs.
Conclusions
Despite the strong and interesting implications associated with SNRPG and its significant prowess as a potential smart-drug discovery target in PPI-focused diagnostics and therapeutic studies [14], the oncogenic potential of SNRPG remains to be proven. The mechanisms and functional basis of its operations in linking the splicing machinery to tumorigenesis and tumor development remain elusive and yet to be fully investigated. The findings presented in this study prompt further investigations. However, it is noteworthy that the foundational basis of the views here presented in this article is based solely on the questionable Y2H technique. Despite the popularity, relative methodical simplicity, diversity and high-throughput capacity, as well as screening method for interactomics, Y2H techniques face the problem of false positives [94]. False positives in Y2H are physical interactions detected in the screening in yeast that are not reproducible in an independent system. A list of recurrent false positives exists and often depends on the Y2H system used [94]. Nonetheless, in this study we confirm that there is no data so far reported to prove and support that the Y2H results presented in line with SNRPG are false positives.
Acknowledgements
Research reported in this article was supported by the South African National Research Foundation (NRF) through funding received via a Thuthuka Grant awarded to Abidemi Paul Kappo (Grant No: 107262) and a Doctoral Bursary received by Lloyd Mabonga from the South African Department of Science and Technology (DST). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the South African DST or the NRF.
Disclosure of conflict of interest
None.
References
- 1.Du L, Grigsby SM, Yao A, Chang Y, Johnson G, Sun H, Nikolovska-Coleska Z. Peptidomimetics for targeting protein-protein interactions between DOT1L and MLL oncofusion proteins AF9 and ENL. ACS Med Chem Lett. 2018;9:895–900. doi: 10.1021/acsmedchemlett.8b00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robertson NS, Spring DR. Using peptidomimetics and constrained peptides as valuable tools for inhibiting protein-protein interactions. Molecules. 2018;23 doi: 10.3390/molecules23040959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang G, Andersen J, Gerona-Navarro G. Peptidomimetics targeting protein-protein interactions for therapeutic development. Protein Pept Lett. 2018;25:1076–1089. doi: 10.2174/0929866525666181101100842. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez MW, Kann MG. Chapter 4: protein interactions and disease. PLoS Comput Biol. 2012;8:e1002819. doi: 10.1371/journal.pcbi.1002819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Díaz-Eufracio BI, JesúsNaveja J, Medina-Franco JL. Protein-protein interaction modulators for epigenetic therapies. Adv Protein Chem Struct Biol. 2018;110:65–84. doi: 10.1016/bs.apcsb.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kale J, Osterlund EJ, Andrews DW. BCL-2 family proteins: changing partners in the dance towards death. Cell Death Differ. 2018;25:65–80. doi: 10.1038/cdd.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor IR, Dunyak BM, Komiyama T, Shao H, Ran X, Assimon VA, Kalyanaraman C, Rauch JN, Jacobson MP, Zuiderweg ERP, Gestwicki JE. High throughput screen for inhibitors of protein-protein interactions in a reconstituted heat shock protein 70 (Hsp70) complex. J Biol Chem. 2018;293:4014–4025. doi: 10.1074/jbc.RA117.001575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pelay-Gimeno M, Glas A, Koch O, Grossmann TN. Structure-based design of inhibitors of protein-protein interactions: mimicking peptide binding epitopes. Angew Chem Int Ed Engl. 2015;54:8896–8927. doi: 10.1002/anie.201412070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keskin O, Gursoy A, Ma B. Principles of protein-protein interactions: what are the preferred ways for proteins to interact? Chem Rev. 2008;108:1225–44. doi: 10.1021/cr040409x. [DOI] [PubMed] [Google Scholar]
- 11.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. A knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuller JC, Burgoyne NJ, Jackson RM. Predicting druggable binding sites at the protein-protein interface. Drug Discov Today. 2009;14:155–161. doi: 10.1016/j.drudis.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Xu GG, Guo J, Wu Y. Chemokine receptor ccr5 antagonist maraviroc: medicinal chemistry and clinical applications. Curr Top Med Chem. 2014;14:1504–1514. doi: 10.2174/1568026614666140827143745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwer B, Kruchten J, Shuman S. Structure-function analysis and genetic interactions of the SmG, SmE, and SmF subunits of the yeast Sm protein ring. RNA. 2016;22:1320–8. doi: 10.1261/rna.057448.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simons A, Melamed-Bessudo C, Wolkowicz R, Sperling J, Sperling R, Eisenbach L, Rotter V. PACT: cloning and characterization of a cellular p53-binding protein that interacts with Rb. Oncogene. 1997;14:145–155. doi: 10.1038/sj.onc.1200825. [DOI] [PubMed] [Google Scholar]
- 16.Conte N, Charafe-Jauffret E, Delaval B, Adélaïde J, Ginestier C, Geneix J, Isnardon D, Jacquemier J, Birnbaum D. Carcinogenesis and translational controls: TACC1 is down-regulated in human cancers and associates with mRNA regulators. Oncogene. 2002;21:5619–5630. doi: 10.1038/sj.onc.1205658. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Deng B, Xing G. PACT is a negative regulator of p53 and essential for cell growth and embryonic development. Proc Natl Acad Sci U S A. 2007;104:7951–7956. doi: 10.1073/pnas.0701916104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ezkurdia I, Juan D, Rodriguez JM. Multiple evidence strands suggest that there may be as few as 19 000 human protein-coding genes. Hum Mol Genet. 2014;23:5866–5878. doi: 10.1093/hmg/ddu309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan F, Allam M, Tincho MB, Pretorius A. Implications of RBBP6 in various types of cancer; Proceedings IWBBIO 2014. Conference Paper Granada; 2014. pp. 7–9. [Google Scholar]
- 20.Hull R, Oosthuysen B, Cajee U. The drosophila retinoblastoma binding protein 6 family member has two isoforms and is potentially involved in embryonic patterning. Int J Mol Sci. 2015;16:10242–10266. doi: 10.3390/ijms160510242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohtani M. Plant snRNP biogenesis: a perspective from the nucleolus and cajal bodies. Front Plant Sci. 2018;8:2184. doi: 10.3389/fpls.2017.02184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones S, Thornton JM. Principles of protein-protein interactions. Proc Natl Acad Sci U S A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conte LL, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 25.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 26.Jochim AL, Arora PS. Systematic analysis of helical protein interfaces reveals targets for synthetic inhibitors. ACS Chem Biol. 2010;5:919–923. doi: 10.1021/cb1001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carbonell P, Nussinov R, Del Sol A. Energetic determinants of protein binding specificity: insights into protein interaction networks. Proteomics. 2009;9:1744–1753. doi: 10.1002/pmic.200800425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keskin O, Ma B, Nussinov R. Hot regions in protein-protein interactions: the organization and contribution of structurally conserved hot spot residues. J Mol Biol. 2005;345:1281–1294. doi: 10.1016/j.jmb.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 29.Reichmann D, Rahat O, Albeck S, Meged R, Dym O, Schreiber G. The modular architecture of protein-protein binding interfaces. Proc Natl Acad Sci U S A. 2005;102:57–62. doi: 10.1073/pnas.0407280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moza B, Buonpane RA, Zhu P, Herfst CA, Rahman AK, McCormick JK, Kranz DM, Sundberg EJ. Long-range cooperative binding effects in a T cell receptor variable domain. Proc Natl Acad Sci U S A. 2006;103:9867–9872. doi: 10.1073/pnas.0600220103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo W, Wisniewski JA, Ji H. Hot spot-based design of small-molecule inhibitors for protein-protein interactions. Bioorg Med Chem Lett. 2014;24:2546–2554. doi: 10.1016/j.bmcl.2014.03.095. [DOI] [PubMed] [Google Scholar]
- 32.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 33.Chakrabarti P, Janin J. Dissecting protein-protein recognition sites. Proteins. 2002;47:334–343. doi: 10.1002/prot.10085. [DOI] [PubMed] [Google Scholar]
- 34.Chene P. Drugs targeting protein-protein interactions. Chem Med Chem. 2006;1:400–411. doi: 10.1002/cmdc.200600004. [DOI] [PubMed] [Google Scholar]
- 35.Raj M, Bullock BN, Arora PS. Plucking the high hanging fruit: a systematic approach for targeting protein-protein interactions. Bioorg Med Chem. 2013;21:4051–4057. doi: 10.1016/j.bmc.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vo LT, Minet M, Schmitter JM, Lacroute F, Wyers F. Mpe1, a zinc knuckle protein, is an essential component of yeast cleavage and polyadenylation factor required for the cleavage and polyadenylation of mRNA. Mol Cell Biol. 2001;21:8346–8356. doi: 10.1128/MCB.21.24.8346-8356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR 3rd, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3’-processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPasy Server. In: Walker JM, editor. The Proteomics Protocols Handbook. Human Press; 2005. pp. 571–607. [Google Scholar]
- 39.Hermann H, Fabrizio P, Raker VA. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blijlevens M, Meulen-Muileman IH, Menezes RX, Smit EF, van Beusechem VW. High-throughput RNAi screening reveals cancer-selective lethal targets in the RNA spliceosome. Oncogene. 2019;38:4142–4153. doi: 10.1038/s41388-019-0711-z. [DOI] [PubMed] [Google Scholar]
- 41.Prusty AB, Meduri R, Prusty BK, Vanselow J, Schlosser A, Fischer U. Impaired spliceosomal UsnRNP assembly leads to Sm mRNA down-regulation and Sm protein degradation. J Cell Biol. 2017;216:2391–2407. doi: 10.1083/jcb.201611108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshitake Y, Nakatsura T, Monji M, Senju S, Matsuyoshi H, Tsukamoto H, Hosaka S, Komori H, Fukuma D, Ikuta Y, Katagiri T, Furukawa Y, Ito H, Shinohara M, Nakamura Y, Nishimura Y. Proliferation potential-related protein, an ideal esophageal cancer antigen for immunotherapy, identified using complementary DNA microarray analysis. Clin Cancer Res. 2004;10:6437–6448. doi: 10.1158/1078-0432.CCR-04-0841. [DOI] [PubMed] [Google Scholar]
- 43.Chibi M, Meyer M, Skepu A, G Rees DJ, Moolman-Smook JC, Pugh DJ. RBBP6 interacts with multifunctional protein YB-1 through its RING finger domain, leading to ubiquitination and proteosomal degradation of YB-1. J Mol Biol. 2008;384:908–916. doi: 10.1016/j.jmb.2008.09.060. [DOI] [PubMed] [Google Scholar]
- 44.Motadi LR, Bhoola KD, Dlamini Z. Expression and function of retinoblastoma binding protein 6 (RBBP6) in human lung cancer. Immunobiology. 2011;216:1065–1073. doi: 10.1016/j.imbio.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 45.Kappo MA, Ab E, Hassem F, Atkinson RA, Faro A, Muleya V, Mulaudzi T, Poole JO, McKenzie JM, Chibi M, Moolman-Smook JC, Rees DJ, Pugh DJ. Solution structure of RING finger-like domain of retinoblastoma-binding protein-6 (RBBP6) suggests it functions as a U-box. J Biol Chem. 2012;287:7146–7158. doi: 10.1074/jbc.M110.217059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye F, Song J, Wang Y. Proliferation potential-related protein promotes the esophageal cancer cell proliferation, migration and suppresses apoptosis by mediating the expression of p53 and interleukin-17. Pathobiology. 2018;85:322–331. doi: 10.1159/000492393. [DOI] [PubMed] [Google Scholar]
- 47.Kambach C, Walke S, Young R. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- 48.Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–9. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 49.Kroiss M, Schultz J, Wiesner J. Evolution of an RNP assembly system: a minimal SMN complex facilitates formation of UsnRNPs in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2008;105:10045–10050. doi: 10.1073/pnas.0802287105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang R, So BR, Li P, Yong J, Glisovic T, Wan L, Dreyfuss G. Structure of a key intermediate of the SMN complex reveals Gemin2’s crucial function in snRNP assembly. Cell. 2011;146:384–95. doi: 10.1016/j.cell.2011.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grimm C, Chari A, Pelz JP. Structural basis of assembly chaperone-mediated snRNP formation. Mol Cell. 2013;49:692–703. doi: 10.1016/j.molcel.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 52.Neuenkirchen N, Englbrecht C, Ohmer J, Ziegenhals T, Chari A, Fischer U. Reconstitution of the human U snRNP assembly machinery reveals stepwise Sm protein organization. EMBO J. 2015;34:1925–1941. doi: 10.15252/embj.201490350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neuenkirchen N, Chari A, Fischer U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett. 2008;582:1997–2003. doi: 10.1016/j.febslet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 54.Chari A, Golas MM, Klingenhäger M, Neuenkirchen N, Sander B, Englbrecht C, Sickmann A, Stark H, Fischer U. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell. 2008;135:497–509. doi: 10.1016/j.cell.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 55.Heinrichs V, Hack W, Lührmann R. Direct binding of small nuclear ribonucleoprotein G to the Sm site of small nuclear RNA. Ultraviolet light cross-linking of protein G to the AAU stretch within the Sm site (AAUUUGUGG) of U1 small nuclear ribonucleoprotein reconstituted in vitro. J Mol Biol. 1992;227:15–28. doi: 10.1016/0022-2836(92)90678-d. [DOI] [PubMed] [Google Scholar]
- 56.Du X, Li Y, Xia YL, Ai SM, Liang J, Sang P, Ji XL, Liu SQ. Insights into protein-ligand interactions: mechanisms, models and methods. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boulisfane N, Choleza M, Rage F. Impaired minor tri-snRNP assembly generates differential splicing defects of U12-type introns in lymphoblasts derived from a type I SMA patient. Hum Mol Genet. 2011;20:641–648. doi: 10.1093/hmg/ddq508. [DOI] [PubMed] [Google Scholar]
- 58.Lefebvre S, Bürglen L, Reboullet S. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 59.Marabti EE, Younis I. The cancer spliceome: reprograming of alternative splicing in cancer. Front Mol Biosci. 2018;5:80. doi: 10.3389/fmolb.2018.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jing J, Zhao Y, Wang C, Zhao Q, Liang Q, Wang S, Ma J. Effect of small nuclear ribonucleoprotein-associated polypeptide N on the proliferation of medulloblastoma cells. Mol Med Rep. 2015;11:3337–3343. doi: 10.3892/mmr.2015.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benetatos L, Hatzimichael E, Dasoula A, Dranitsaris G, Tsiara S, Syrrou M, Georgiou I, Bourantas KL. CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk Res. 2010;34:148–153. doi: 10.1016/j.leukres.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 62.Lee SH, Appleby V, Jeyapalan JN, Palmer RD, Nicholson JC, Sottile V, Gao E, Coleman N, Scotting PJ. Variable methylation of the imprinted gene, SNRPN, supports a relationship between intra-cranial germ cell tumours and neural stem cells. J Neurooncol. 2011;101:419–428. doi: 10.1007/s11060-010-0275-9. [DOI] [PubMed] [Google Scholar]
- 63.Fallini C, Bassell GJ, Rossoll W. Spinal muscular atrophy: the role of SMN in axonal mRNA regulation. Brain Res. 2012;1462:81–92. doi: 10.1016/j.brainres.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anchi T, Tamura K, Furihata M, Satake H, Sakoda H, Kawada C, Kamei M, Shimamoto T, Fukuhara H, Fukata S, Ashida S, Karashima T, Yamasaki I, Yasuda M, Kamada M, Inoue K, Shuin T. SNRPE is involved in cell proliferation and progression of high-grade prostate cancer through the regulation of androgen receptor expression. Oncol Lett. 2012;3:264–268. doi: 10.3892/ol.2011.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson PF, Cavanna T, Zicha D, Bates PA. Cluster analysis of networks generated through homology: automatic identification of important protein communities involved in cancer metastasis. BMC Bioinformatics. 2006;7:2. doi: 10.1186/1471-2105-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steinbrecher T, Labahn A. Towards accurate free energy calculations in ligand protein-binding studies. Curr Med Chem. 2010;17:767–785. doi: 10.2174/092986710790514453. [DOI] [PubMed] [Google Scholar]
- 67.Heneghan C, Blacklock C, Perera R, Davis R, Banerjee A, Gill P, Liew S, Chamas L, Hernandez J, Mahtani K, Hayward G, Harrison S, Lasserson D, Mickan S, Sellers C, Carnes D, Homer K, Steed L, Ross J, Denny N, Goyder C, Thompson M, Ward A. Evidence for non-communicable diseases: analysis of Cochrane reviews and randomised trials by World Bank classification. BMJ Open. 2013;3 doi: 10.1136/bmjopen-2013-003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bhandari GP, Angdembe MR, Dhimal M. State of non-communicable diseases in Nepal. BMC Public Health. 2014;14:23. doi: 10.1186/1471-2458-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanlon L, Avila JL, Demarest RM, Troutman S, Allen M, Ratti F, Rustgi AK, Stanger BZ, Radtke F, Adsay V, Long F, Capobianco AJ, Kissil JL. Notch1 functions as a tumor suppressor in a model of K-ras-induced pancreatic ductal adenocarcinoma. Cancer Res. 2010;70:4280–6. doi: 10.1158/0008-5472.CAN-09-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Witte MM, Scott RE. The proliferation potential protein related (P2P-R) gene with domains encoding heterogeneous nuclear ribonucleoprotein association and Rb1 binding shows repressed expression during terminal differentiation. Proc Natl Acad Sci U S A. 1997;94:1212–1217. doi: 10.1073/pnas.94.4.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pugh DJ, Eiso AB, Faro A. DWNN, a novel ubiquitin-like domain, implicates RBBP6 in mRNA processing and ubiquintin-like pathway. BMC Struct Biol. 2006;6:1–12. doi: 10.1186/1472-6807-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao S, Scott RE. P2P-R protein overexpression restricts mitotic progression at prometaphase and promotes mitotic apoptosis. J Cell Physiol. 2002;193:199–207. doi: 10.1002/jcp.10163. [DOI] [PubMed] [Google Scholar]
- 73.Gao S, Witte MM, Scott RE. P2P-R protein localizes to the nucleolus of interphase cells and the periphery of chromosomes in mitotic cells that show maximum P2P-R immunoreactivity. J Cell Physiol. 2002;191:145–154. doi: 10.1002/jcp.10084. [DOI] [PubMed] [Google Scholar]
- 74.Chen J, Tang H, Wu Z. Overexpression of RBBP6, alone or combined with mutant TP53, is predictive of poor prognosis in colon cancer. PLoS One. 2013;8:e66524. doi: 10.1371/journal.pone.0066524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moela P, Choene MM, Motadi LR. Silencing RBBP6 (retinoblastoma binding protein 6) sensitises breast cancer cells MCF7 to staurosporine and camptothecin-induced cell death. Immunobiology. 2014;219:593–601. doi: 10.1016/j.imbio.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 76.Muleya V. MSc Thesis. South Africa: Department of Biotechnology, University of Western Cape; Structural characterisation of the interaction between RBBP6 and the multifunctional protein YB-1. URI: http://hdl.handle.net/11394/2324 Accessed June 27, 2019. [Google Scholar]
- 77.Spector DL. Nuclear organisation of pre-mRNA processing. Curr Opin Cell Biol. 1993;5:442–448. doi: 10.1016/0955-0674(93)90009-f. [DOI] [PubMed] [Google Scholar]
- 78.Still IH, Vince P, Cowell JK. The third member of the transforming acidic coiled coil-containing gene family, TACC3, maps in 4p16, close to translocation breakpoints in multiple myeloma, and is upregulated in various cancer cell lines. Genomics. 1999;58:165–70. doi: 10.1006/geno.1999.5829. [DOI] [PubMed] [Google Scholar]
- 79.Ugolini F, Charafe-Jauffret E, Bardou VJ, Geneix J, Adélaïde J, Labat-Moleur F, Penault-Llorca F, Longy M, Jacquemier J, Birnbaum D, Pébusque MJ. WNT pathway and mammary carcinogenesis: loss of expression of candidate tumor suppressor gene SFRP1 in most invasive carcinomas except of the medullary type. Oncogene. 2001;20:5810–5817. doi: 10.1038/sj.onc.1204706. [DOI] [PubMed] [Google Scholar]
- 80.Conte N, Delaval B, Ginestier C, Ferrand A, Isnardon D, Larroque C, Prigent C, Séraphin B, Jacquemier J, Birnbaum D. TACC1-chTOG-Aurora A protein complex in breast cancer. Oncogene. 2003;22:8102–8116. doi: 10.1038/sj.onc.1206972. [DOI] [PubMed] [Google Scholar]
- 81.Takata A, Otsuka M, Yoshikawa T, Kishikawa T, Kudo Y, Goto T, Yoshida H, Koike K. A miRNA machinery component DDX20 controls NF-κB via microRNA-140 function. Biochem Biophys Res Commun. 2012;420:564–569. doi: 10.1016/j.bbrc.2012.03.034. [DOI] [PubMed] [Google Scholar]
- 82.Takata A, Otsuka M, Yoshikawa T. MicroRNA-140 acts as a liver tumor suppressor by controlling NF-κB activity by directly targeting DNA methyltransferase 1 (Dnmt1) expression. Hepatology. 2013;57:162–170. doi: 10.1002/hep.26011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shpargel KB, Matera AG. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc Natl Acad Sci U S A. 2005;102:17372–17377. doi: 10.1073/pnas.0508947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Almstead LL, Sarnow P. Inhibition of U snRNP assembly by a virus-encoded proteinase. Genes Dev. 2007;21:1086–1097. doi: 10.1101/gad.1535607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen W, Zhou P, Li X. High expression of DDX20 enhances the proliferation and metastatic potential of prostate cancer cells through the NF-κB pathway. Int J Mol Med. 2016;37:1551–1557. doi: 10.3892/ijmm.2016.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shin EM, Hay HS, Lee MH, Goh JN, Tan TZ, Sen YP, Lim SW, Yousef EM, Ong HT, Thike AA, Kong X, Wu Z, Mendoz E, Sun W, Salto-Tellez M, Lim CT, Lobie PE, Lim YP, Yap CT, Zeng Q, Sethi G, Lee MB, Tan P, Goh BC, Miller LD, Thiery JP, Zhu T, Gaboury L, Tan PH, Hui KM, Yip GW, Miyamoto S, Kumar AP, Tergaonkar V. DEAD-box helicase DP103 defines metastatic potential of human breast cancers. J Clin Invest. 2014;124:3807–3824. doi: 10.1172/JCI73451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Company M, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for the release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 88.Ohno M, Shimura Y. A human RNA helicase-like protein, HRH1, facilitates nuclear export of spliced mRNA by releasing the RNA from the spliceosome. Genes Dev. 1996;10:997–1007. doi: 10.1101/gad.10.8.997. [DOI] [PubMed] [Google Scholar]
- 89.Arenas JE, Abelson J. Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci U S A. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hamm J, Lamond AI. Spliceosome assembly: the unwinding role of DEAD box proteins. Curr Biol. 1998;8:532–534. doi: 10.1016/s0960-9822(07)00340-5. [DOI] [PubMed] [Google Scholar]
- 91.Staley JP, Guthrie C. Mechanical devices of the spliceosomemotors, clocks, springs and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 92.De la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisae: DEAD box proteins and related families. Trends Biochem Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- 93.Charroux B, Pellizzoni L, Perkinson RA, Shevchenko A, Mann M, Dreyfuss G. Gemin3: a novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J Cell Biol. 1999;147:1181–94. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brückner A, Polge C, Lentze N. Yeast two-hybrid, a powerful tool for systems biology. Int J Mol Sci. 2009;10:2763–2788. doi: 10.3390/ijms10062763. [DOI] [PMC free article] [PubMed] [Google Scholar]


