Abstract
To further reveal the molecular mechanism underlying sexual differentiation of the mouse cerebral cortex and hippocampus, we reanalyzed our previous microarray study with Gene Ontology (GO) term enrichment and found that the GO term “RNA binding” was over-represented among the 89 sexually dimorphic candidate genes. Thus, we selected 16 autosomal genes annotated to the term RNA binding and profiled their mRNA expression in the developing male and female mouse cortex/hippocampus. During the first three weeks after birth, sex differences in mRNA levels of Khdrbs2, Nanos2, Rbm48, and Tdrd3 were observed in the mouse cortex/hippocampus. Of these genes, only the female-biased expression of Rbm48 in neonates was abolished by prenatal exposure to testosterone propionate (TP), while postnatal treatment of TP three weeks after birth increased Rbm48 and Tdrd3 mRNA levels in both sexes. Regardless of sex, the postnatal cortex/hippocampus also showed a marked increase in the content of androgen receptor (Ar) and estrogen receptor β (Esr2), but a decrease in estrogen receptor α (Esr1) and aromatase (Cyp19a1), which might confer the different responses of Rbm48 to prenatal and postnatal TP. Our results suggest that androgen-regulated, sexually dimorphic Rbm48 expression might present a novel molecular mechanism by which perinatal androgens control development of sexual dimorphism in cortical and hippocampal structure and function.
Keywords: Sex difference, Cerebral cortex, Hippocampus, Sex-biased gene, RNA binding protein, Androgen
1. Introduction
In humans and other mammals, sexual differentiation is essential for sex-specific development and function of both reproductive and non-reproductive organs, including the brain (Cahill 2006). This critical physiological process is dependent on not only the actions of sex hormones and other gonadal secretions following gonadal determination, but also the genetic mechanisms mediated by the genes located on the X and Y chromosomes (Arnold 2017). Besides normal neural functions, sex or gender differences are also noted in the prevalence and symptomatology of many neurological diseases and mental illnesses, such as Alzheimer’s disease, autism, depression, and schizophrenia (Bao and Swaab 2010; Pinares-Garcia et al., 2018). As sexual differentiation may confer inherent sex-specific disease vulnerability (Bao and Swaab 2011; McCarthy 2016), elucidation of the mechanisms underlying sexual dimorphism in brain structure and function will provide important insights into the etiologies of gender-biased neuropsychiatric disorders and promote the development of new therapies.
During early development, the mouse testes secrete testosterone with two perinatal peaks, the first occurring during late gestation and the second peaking on the day of birth (Motelica-Heino et al., 1988; Vom Saal and Bronson 1980). In mice and other rodent models, the developmental increase in circulating testosterone has been proven to program the expression of sex-specific reproductive and aggressive behaviors and the underlying dimorphic structural correlates in the male and female hypothalamus and amygdala (organizational effect) (Arnold 2009; Ngun et al., 2011; Quadagno et al., 1975; vom Saal 1979). To shape sexual dimorphism in the brain circuits and behavioral phenotypes, testosterone could act directly on androgen receptor (AR) and/or indirectly on estrogen receptors (ERs) via local conversion of testosterone to estradiol by aromatase (Bodo and Rissman 2007; Honda et al., 1998; Ogawa et al., 2000; Sato et al., 2004).
In addition to reproduction and aggression, many complex cognitive functions and social behaviors controlled by the cerebral cortex and hippocampus also differ between the sexes in rodent models (Gresack and Frick 2003; Isgor and Sengelaub 1998; Karlsson et al., 2015; Rizk et al., 2005). Neonatal manipulations of gonadal hormones have similarly been found to lead to sex differences in learning and cognition (Isgor and Sengelaub 1998). Associated with behavioral differences, a number of structures in these two brain regions of mice and rats have been found to be sexually dimorphic. For example, male rats show greater volumes and soma sizes in their hippocampal CA1 and CA3 pyramidal cell layer than females, and prenatal exposure of females to androgens or estrogens masculinizes the CA1 and CA3 pyramidal cell morphology (Isgor and Sengelaub 1998). These sex differences in the hippocampus may result from a greater rate of neurogenesis in males than females during the first week after birth via the activation of AR and ERs (Bowers et al., 2010; Zhang et al., 2008). Besides the hippocampus, the cerebral cortex is thicker in mice with testes but not in those with ovaries, irrespective of their sex chromosome complement (XX vs. XY), suggesting an effect of gonadal secretions on sexual differentiation of the cortex (Markham et al., 2003). AR and ERs are abundantly expressed in the developing mouse cortex and hippocampus (Kerr et al., 1995; Mogi et al., 2015; Tsai et al., 2015), and once activated, these receptors may act as transcription factors to modulate gene expression. However, the AR- or ER-specific downstream targets that are relevant to sex differences in these brain regions remain unclear.
To address this question, we previously used gene expression microarrays to identify 90 genes differentially expressed in the cortex/hippocampus of neonatal male and female mice (Armoskus et al., 2014b). By searching the Gene Ontology (GO) terms annotated to these genes, a variety of molecular functions and biological processes, including histone modifications, cell proliferation/death, androgen/estrogen signaling pathways, and synaptic organization, have been implicated in brain sexual differentiation. To further uncover meaningful associations of these genes, we performed GO enrichment analysis of our microarray data and found that the GO term, “RNA binding”, was overrepresented, suggesting that those molecular events exclusively governed by a network of sex-dimorphically expressed RNA-binding proteins might constitute a novel mechanism for controlling sexual differentiation of the developing cortex and hippocampus. In support of this possibility, an early study observed that the hippocampal mRNA levels of two splicing factors, polypyrimidine tract-binding protein-associated splicing factor (Sfpq/Psf) and splicing factor arginine/serine-rich 3 (Sfrs3/Srp20), were higher in male mice than in females, and hippocampus-dependent learning tasks upregulated expression of both splicing factors differently between the sexes (Antunes-Martins et al., 2007). Based on the growing list of neurological diseases and psychiatric disorders, in which the cortex and hippocampus are implicated, and which are linked to defects of splicing and other RNA processing mechanisms (Brinegar and Cooper 2016; Jung and Goldman 2018; Licatalosi and Darnell 2006; Manning and Cooper 2017), it has become increasingly probable that RNA binding proteins may play important roles in the regulation of neural functions served by these two brain regions. Thus, we hypothesized that during early development, sexually dimorphic expression of the genes encoding RNA binding proteins in the developing mouse cortex and hippocampus might activate a regulatory gene network to mediate the masculinizing effects of gonadal steroids on differential neural function and behavior between the sexes. To test our hypothesis, we first determined mRNA levels of 16 candidate genes annotated to RNA binding in the developing male and female mouse cortex/hippocampus. Next, we characterized expression changes of the selected genes having sexually dimorphic expression as a function of prenatal or postnatal testosterone treatment to assess their potential as downstream androgen targets that direct brain sexual differentiation.
2. Experimental Procedures
2.1. Animals
Male and female C57BL/6J mice were housed in the CSULB Animal Care Facility with a controlled 12:12h light:dark cycle (lights on at 0600 h). Food (Teklad Mouse/Rat Diet #7012; Harlan Laboratories, Inc., Indianapolis, IN) and water were provided ad libitum. Adult female mice were individually paired with a fertile male and checked daily for the presence of mating plugs. The day when a mating plug is found was designated as embryonic day 0 (E0). All experimental procedures have been approved by CSULB Institutional Animal Care and Use Committee and performed according to Association for Assessment and Accreditation of Laboratory Animal Care International guidelines.
2.2. Experimental Design
Experiment 1.
Sixteen adult female mice were individually paired with nine fertile male to produce the pups used for this experiment. Male and female pups from a total of 29 litters were sacrificed by rapid decapitation at postnatal day (PN) 0, 7, 14, and 21 (n = 8–20 per group). Their brains were immediately removed from the skulls after decapitation and coronally blocked as described previously (Armoskus et al., 2014b; Tsai et al., 2009). The cerebral cortex/hippocampus was dissected out, frozen on dry ice, and then stored at −80°C until processed for RNA extraction. The sexes of the pups were determined by examining the pigmentation in their anogenital regions as well as their gonads and reproductive tracts.
Experiment 2.
To examine if sexually dimorphic expression of selected genes was regulated by prenatal androgens, pregnant mice received a subcutaneous injection of either vehicle (0.05 ml sesame oil) or testosterone propionate (TP, 100 μg in 0.05 ml; Sigma-Aldrich) daily beginning on E16 (16 days after a mating plug was seen) until PN0 as described previously (Armoskus et al., 2014a). This treatment was to mimic the testosterone surge observed in male embryos during late gestation. The dosage and duration of TP administered were modified from previous studies showing that prenatal TP injections in pregnant dams masculinized reproductive morphology and behavior in their female offspring (Hotchkiss et al., 2007; vom Saal 1979; Wolf et al., 2002; Yucel et al., 2003). In addition, the effectiveness of our TP treatment on brain development was confirmed as it caused an increase in the density of dendritic spines in the frontal cortex of prenatally testosterone-exposed mice (Hatanaka et al., 2015). Vehicle- and TP-treated, male and female pups from nine litters (n = 10 per group) were sacrificed at PN0, and their cortex/hippocampus was similarly collected and stored as described above. The pups used in this experiment were produced by five male and six female breeders. Besides the pigmentation in the anogenital regions and the gonads, anogenital distance (AGD) was also measured using a digital caliper (Fisher) to indicate the effects of prenatal TP on genital masculinization.
Experiment 3.
To investigate the effect of testosterone on mRNA levels of sexually dimorphic genes after the critical period of brain sexual differentiation, male and female pups were treated with a subcutaneous injection of either vehicle (0.05 ml sesame oil) or TP (100 μg in 0.05 ml; Sigma-Aldrich) daily at 1000 h beginning at PN21. While our TP treatment paradigm has not been previously reported in the literature, the same dose of TP administered to newborn females was effective at masculinizing their brains and behaviors (Hisasue et al., 2010; Seney et al., 2012). The cortex/hippocampus of vehicle-and TP-treated, male and female pups from 11 litters (n = 8–11 per group) was collected 3–6 h after the last injection at PN23. AGD of individual pups was similarly measured at sacrifice, and their sexes were confirmed by examining the gonads and reproductive tracts as described above. The pups used in Experiment 3 were produced by six male and six female breeders.
2.3. RNA Extraction and cDNA Synthesis
Total RNA was extracted from the cortex/hippocampus sample using an RNeasy® Lipid Tissue kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. RNA concentration of each RNA sample was determined by measuring the absorbance at 260 nm (A260) and 280 nm (A280) using the BioRad SmartSpec™Plus spectrophotometer (Hercules, CA), and its purity was determined by calculating the A260:A280 ratio.
Using the Bio-Rad iScript™ cDNA Synthesis kit and Bio-Rad MyCycler™ thermocycler, cDNA was synthesized from 1 μg total RNA in a 20-μl reaction at 25°C for 5 min, 42°C for 30 min, and 85°C for 5 min. The cDNA was then stored at −20°C later used in PCR and qPCR to measure relative mRNA levels of selected genes.
2.4. Primer Tests
The PCR primers used in the current study were purchased from Eurofins MWG Operon LLC (Louisville, KY). The sequences of all PCR primers were first found from the Harvard Primer Bank (https://pga.mgh.harvard.edu/primerbank) and checked for specificity using NCBI’s Primer-BLAST program against RefSeq mRNA database (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). All primers (10 mM) were tested in PCR assays with 40 cycles using GoTaq® Green Master Mix (Promega Corporation, Madison, WI). All PCR primer tests were conducted with the annealing temperatures listed in Table 2. PCR products were separated on a 1.5% agarose gel containing 10 mg/ml ethidium bromide and then visualized using the FluorChem E™ System (ProteinSimple, San Jose, CA). All PCR primer sets showed a single PCR product of the expected size and no product in the negative control (without cDNA).
TABLE 2.
List of the oligonucleotide primers used for RT-PCR and RT-qPCR.
| Gene Symbol | Primer Sequence | RefSeq Accession | Annealing Temp (°C) |
PCR Product Size (bp) |
|---|---|---|---|---|
| Actb | Forward: 5’ CCAGATCATGTTTGAGACCTTCAA 3’ Reverse: 5’ CCAGAGGCATACAGGGACAGC 3’ |
NM_007393.5 | 61 | 78 |
| Ar | Forward: 5’ AGAATCCCACATCCTGCTCAA 3’ Reverse: 5’ AAGTCCACGCTCACCATATGG 3’ |
NM_013476.4 | 58 | 133 |
| Cyp19a1 | Forward: 5’ ACCTCGGGCTACGTGGATGTG 3’ Reverse: 5’ GAT GTTT GGTTT GAT GAGGAGAGC 3’ |
NM_007810.4 | 60 | 159 |
| Dhx8 | Forward: 5’ AATCCAGAGAACCAACCTGGC 3’ Reverse: 5’ GATCAGAGTTTCCATCGGTGG 3’ |
NM_144831.2 | 60 | 109 |
| Dusp11 | Forward: 5’ AACCAGCATTATGGCCGACAT 3’ Reverse: 5’ GGAGAT AGTCTTT CCACCTTT CG 3’ |
NM_028099.4 | 58 | 112 |
| Elav14 | Forward: 5’ CCAATCACCATTGACGGGATG 3’ Reverse: 5’ CAGGGGACAGGTT AT AGACGAA 3’ |
NM_010488.4 | 60 | 97 |
| Esr1 | Forward: 5’ CACTTT GATCCACCT GAT GGC 3’ Reverse: 5’ GGCACAACGTTCTTGCATTTC 3’ |
NM_007956.5 | 58 | 153 |
| Esr2 | Forward: 5’ GAAAGTGACGCGATACACCGT 3’ Reverse: 5’ CCT GT GAGGTAGGAAT GCGAA 3’ |
NM_010157.3 | 58 | 150 |
| Khdrbs2 | Forward: 5’ GGGGT GAATAT GAT GACCAGACC 3’ Reverse: 5’ CCT CGTTTACT CCAT GACCGT AG 3’ |
NM_133235.3 | 59 | 105 |
| Nanos2 | Forward: 5’ CATGGACCGTGTGAGTCTTG 3’ Reverse: 5’ TTCTTGGCAGTGACGATCAG 3’ |
NM_194064.2 | 58 | 197 |
| Ncbp2 | Forward: 5’ T GT ACT GGAAGCCACCT CT G 3’ Reverse: 5’ TGTTGCCTGGGTAGAGCTTT 3’ |
NM_026554.4 | 58 | 126 |
| Ppargc1b | Forward: 5’ TCCTGTAAAAGCCCGGAGTAT 3’ Reverse: 5’ GCT CT GGTAGGGGCAGT GA 3’ |
NM_133249.2 | 58 | 174 |
| Rbm12 | Forward: 5’ GTT AT GCCTT CGGTT GGAAA 3’ Reverse: 5’ GGCCATTT CCAAAGTT CT GA 3’ |
NM_029397.3 | 55 | 130 |
| Rbm48 | Forward: 5’ AAACCTGGTGGCGTATTCGAT 3’ Reverse: 5’ ACGCT CAACTAATT CCTT CAT GG 3’ |
NM_172991.4 | 58 | 177 |
| Rpl13a | Forward: 5’ ATGAGGTCGGGTGGAAGTACC 3’ Reverse: 5’ CAGGAGT CCGTT GGT CTT GAG 3’ |
NM_009438.5 | 62 | 179 |
| Sfpq | Forward: 5’ GGAGTTCCACCAGCAACCAT 3’ Reverse: 5’ CTGCCCAAAGCGCTCAGT 3’ |
NM_023603.3 | 60 | 69 |
| Srsf3 | Forward: 5’ TGAGGATCCCCGAGATGCT 3’ Reverse: 5’ CTTACACGGCAGCCACACAGT 3’ |
NM_013663.5 | 60 | 72 |
| Srsf10 | Forward: 5’ GCGT CAGATT GAAAT CCAGTT CG 3’ Reverse: 5’ CGAGAGCGTCTAT AT CGGT CAT 3’ |
NM_010178.3 | 60 | 126 |
| Strbp | Forward: 5’ GT GT GGT GTAAT GAGGATT GG 3’ Reverse: 5’ TTTCCGTGGGTTTGTCTTTGC 3’ |
NM_009261.4 | 58 | 101 |
| Tdrd3 | Forward: 5’ AGTGACATCAACGGTGGAAAG 3’ Reverse: 5’ GCTTGCGATTCCTCGTTATCCT 3’ |
NM_001253755.1 | 58 | 107 |
| Thoc3 | Forward: 5’ AT CT GGGATGT GAGGACT ACAA 3’ Reverse: 5’ T GT GT CTT GGCGT CAAT GAAA 3’ |
NM_028597.3 | 59 | 143 |
| Tut1 | Forward: 5’ GATT CGGACGTT GT AT CGCT G 3’ Reverse: 5’ CTCGGAGTTGCACCAAATCC 3’ |
NM_197993.3 | 59 | 130 |
| Xist | Forward: 5’ GCT GGTT CGT CTAT CTT GT GGG 3’ Reverse: 5’ GGAT CCT GCACT GGAT GAGT 3’ |
NR_001463.3 | 58 | 225 |
2.5. RT-qPCR
Serially diluted, male and female mouse cortical/hippocampal cDNA samples, ranging from 1:4 to 1: 512, were used to test for primer efficiency. Total volume of each qPCR reaction comprised of 4 μl cDNA, 1 μl forward primer (840 nM), 1 μl reverse primer (840 nM), and 6 μl ABsolute QPCR SYBR® Green Mix with ROX and MgCh (Thermo Scientific, Waltham, MA). All qPCR reactions were amplified on the StepOnePlus™ Real-Time PCR System (Life Technologies, Grand Island, NY) with an initial 15 min denaturation/polymerase-activation at 95°C, 40 cycles of denaturation at 95°C for 15 s, annealing at the optimal temperature (Table 2) for 1 min, and extension at 72° for 30 s, followed by a dissociation step at 95°C for 1 min, at 55°C for 30 s, and at 95°C for 30 s to produce a melt curve. Negative controls were also included in each assay. Ct values were analyzed by linear regression with log base 2 of the dilution as the predictor and Ct of the reaction as the response. Primers were deemed unsuitable for qPCR if the R2 value of the regression was below 0.95, or Ct-values of 1:16 dilution were above 32. The sequences, PCR product sizes, efficiencies, and annealing temperatures of all the primer used are listed in Table 2.
Using RT-qPCR with the optimal conditions as described in the dilution tests, relative mRNA levels of target genes were measured in duplicate for each cDNA sample (in 1:16 dilution) to reduce intersample variation. For each sample, relative expression of target and housekeeping genes were calculated using Pfaffl’s method against the average Ct values of the PN0 females in Experiment 1 and against the average Ct values of the vehicle-treated females in Experiments 2 and 3 (Pfaffl 2001).
Normalization and quantifications of the genes of interest was performed with the comparative Cts method relative to the housekeeping genes, Actb and Rpl113a (Armoskus et al., 2014b). For Experiments 2 and 3, target gene expression was calculated as normalized to the average Ct values of vehicle-treated females.
2.6. Statistical Analyses
The data from individual groups were first tested for equality of variances using Levene’s test. When significant deviations were detected (p<0.05), the data were log transformed and then retested for equal variance. After ensuring equal variance, the data were analyzed by two-way ANOVA to evaluate: (1) the effects of sex, age, and their interaction in Experiment 1, and (2) the effects of sex, TP treatment, and their interaction in Experiments 2 and 3. When two-way ANOVA showed significant effect or interactions, Tukey’s post-hoc test was further used for pair-wise comparison to determine which two groups were significantly different from each other. A p value <0.05 was considered statistically different.
3. Results
3.1. GO Enrichment Analysis of Microarray Data
We previously identified 90 candidate genes differentially expressed in the neonatal mouse cortex/hippocampus between male and female mice using gene expression microarrays (Armoskus et al., 2014b). In the current study, we reanalyzed those genes with GO term enrichment analysis except loc101056177 (not annotated) and found some of them to be significantly associated with the molecular function GO term, “RNA binding” (GO:0003723) (p = 0.009). Among the 89 genes, 13 were annotated with “RNA binding”, including Ddx3y, Dhx8, Eif2s3x, Eif2s3y, Elavl4, Nanos2, Ncbp2, Ppargc1b, Rbm12, Rbm48, Srsf10, Tdrd3, and Thoc3 (Table 1). Ddx3y and Eif2s3y are located on the Y chromosome and exclusively expressed in males, while Eif2s3x is an X escape gene expressed from both active and inactive X chromosomes with higher levels in females than males (Armoskus et al., 2014b). Using GO annotation search, these genes were further explored with regard to their connections to the mechanisms of post-transcriptional control regulated by RNA binding proteins. Ncbp2 was annotated with “RNA 3’-end processing” (GO:0031123) while Dhx8, Ncbp2, Srsf10, and Thoc3 were linked to “RNA splicing” (GO:0008380). Ncbp2 and Thoc3 were involved in “RNA transport” (GO:0050658). Eif2s3x, Eif2s3y, Elovl4, Nanos2, and Ncbp2 were annotated with “translation” (GO:0006412), and Elavl4, Nanos2, and Ncbp2 were linked with “RNA catabolic process” (GO:0006401). This suggests that RNA binding proteins may regulate brain sexual differentiation post-transcriptionally via mechanisms including mRNA processing (polyadenylation and splicing), export and localization, decay, and translation. Three genes, Ddx3y, Rbm12, and Rbm48, lack the GO biological process annotations.
TABLE 1.
List of the selected genes annotated to the Gene Ontology term, RNA binding and expressed in the neonatal mouse cortex/hippocampus.
| Gene Symbol | Name | RefSeq Accession | Chra | Gene Ontology Annotations |
|---|---|---|---|---|
| Sexually dimorphic genes identified by microarrays | ||||
| Ddx3yb | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | NM_012008.2 | Y | ATP binding, helicase activity, hydrolase activity, RNA binding |
| Dhx8 | DEAH (Asp-Glu-Ala-His) box polypeptide 8 | NM_144831.2 | 11 | ATP binding, ATP-dependent helicase activity, hydrolase activity, RNA binding, |
| Eif2s3xb | Eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked | NM_012010.3 | X | GTP binding, GTPase activity, translation initiation factor activity |
| Eif2s3yb | Eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked | NM_012011.2 | Y | GTP binding, GTPase activity, translation initiation factor activity |
| Elavl4 | ELAV like RNA binding protein 4 | NM_001163399.1 | 4 | AU-rich element binding, mRNA 3’-UTR binding, translation regulator activity |
| Nanos2b | Nanos C2HC-type zinc finger 2 | NM_194064.2 | 7 | mRNA binding, zinc ion binding |
| Ncbp2 | Nuclear cap binding protein subunit 2 | NM_026554.4 | 16 | RNA 7-methylguanosine cap binding, snRNA binding |
| Ppargc1b | Peroxisome proliferative activated receptor, gamma, coactivator 1 beta | NM_133249.2 | 18 | AF-2 domain binding, estrogen receptor binding, nuclear receptor transcription coactivator activity, RNA binding |
| Rbm12 | RNA binding motif protein 12 | NM_170598.2 | 2 | RNA binding |
| Rbm48b | RNA binding motif protein 48 | NM_172991.4 | 5 | RNA binding |
| Srsf10 | Serine/arginine-rich splicing factor 10 | NM_010178.3 | 4 | RNA binding, unfolded protein binding |
| Tdrd3b | Tudor domain containing 3 | NM_001253755.1 | 14 | Methylated histone binding, RNA binding, transcription coactivator activity |
| Thoc3 | THO complex 3 | NM_028597.3 | 13 | RNA binding |
| Non-dimorphic genes identified by microarrays | ||||
| Dusp11c | Dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) | NM_028099.4 | 6 | Hydrolase activity, polynucleotide 5’- phosphatase activity, protein tyrosine/serine/threonine phosphatase activity, RNA binding |
| Khdrbs2bc | KH domain containing, RNA binding, signal transduction associated 2 | NM_133235.3 | 1 | ploy(A)/poly(U) RNA binding, protein heterodimerization activity, SH3/SH2 adaptor activity |
| sfpqd | Splicing factor proline/glutamine rich | NM_023603.3 | 4 | E-box binding, histone deacetylase binding, protein homodimerization activity, RNA binding, RNA polymerase II distal enhancer sequence-specific DNA binding, transcription regulatory region sequence-specific DNA binding |
| Srsf3d | Serine/arginine-rich splicing factor 3 | NM_013663.5 | 17 | Phospholipase binding, sequence-specific mRNA binding |
| Strbpc | Spermatid perinuclear RNA binding protein | NM_009261.4 | 2 | double/single-stranded RNA binding, microtubule binding, tubulin binding |
| Tut1c | Terminal uridylyl transferase 1, U6 snRNA-specific | NM_197993.3 | 19 | ATP binding, mRNA 3’-UTR binding, polynucleotide adenylyltransferase activity, RNA uridylyltransferase activity, zinc ion binding |
Chromosome.
indicates the genes showing sex difference in expression using microarrays and RT-qPCR Armoskus et al., 2014].
indicates the genes with a 1.2 or greater fold change in expression between the sexes by microarrays, but not reaching a statistically significant difference.
indicates the genes reported by Antunes-Martins et al.(2007)
Using RT-qPCR, we confirmed the sex differences in mRNA levels of Ddx3y, Eif2s3x, and Eif2s3y in the neonatal cortex/hippocampus (Armoskus et al., 2014b), and found that their expression is independent of prenatal testosterone levels (Armoskus et al., 2014a). In this study, we continued to profile the expression of the other 10 candidates in the developing mouse cortex/hippocampus using RT-qPCR. In addition, six non-dimorphic genes, Dusp11, Khdrbs2, Sfpq, Srsf3, Strbp, and Tut1, were also included in the analysis; Dusp11, Khdrbs2, Strbp, and Tut1 displayed a 1.2 or greater fold change between the sexes in neonates with a trend towards statistical significance by microarrays while male-biased expression of Sfpq and Srsf3 was observed in the juvenile and/or adult mouse hippocampus (Antunes-Martins et al., 2007).
3.2. Effect of Age and Sex on mRNA Levels of Selected Sexually Dimorphic Candidate Genes in the Developing Mouse Cortex/Hippocampus
Body weight of each mouse used in this experiment was obtained at sacrifice (Table 3). Two-way ANOVA revealed a significant increase in body weight as age advanced (p < 0.001), but no effect of sex (p = 0.799) or interaction between age and sex (p = 0.457) on body weight.
TABLE 3.
Mean (± SE) body weights (g) of the mice used for Experiments 1, 2, and 3.
| Sex | |||||
|---|---|---|---|---|---|
| N | Female | N | Male | ||
| 0 | 16 | 1.25 ± 0.03 g | 20 | 1.26 ± 0.03 g | |
| 7 | 11 | 3.78 ± 0.14 g | 11 | 3.62 ± 0.16 g | |
| 14 | 8 | 7.64 ± 0.49 g | 8 | 7.18 ± 0.38 g | |
| 21 | 8 | 8.35 ± 0.56 g | 8 | 8.84 ± 0.24 g | |
| Experiment 2: Effect of prenatal TP | |||||
| Sex | |||||
| N | Female | N | Male | ||
| Treatment | Vehicle | 10 | 1.29 ± 0.04 g | 10 | 1.29 ± 0.03 g |
| TP | 10 | 1.24 ± 0.02 g | 10 | 1.26 ± 0.02 g | |
| Experiment 3: Effect of postnatal TP | |||||
| Sex | |||||
| N | Female | N | Male | ||
| Treatment | Vehicle | 11 | 7.59 ± 0.47 g | 8 | 8.23 ± 0.65 g |
| TP | 11 | 8.09 ± 0.20 g | 11 | 9.20 ± 0.34 g | |
N: number of mice in each group; TP: testosterone propionate;
indicates significant difference between
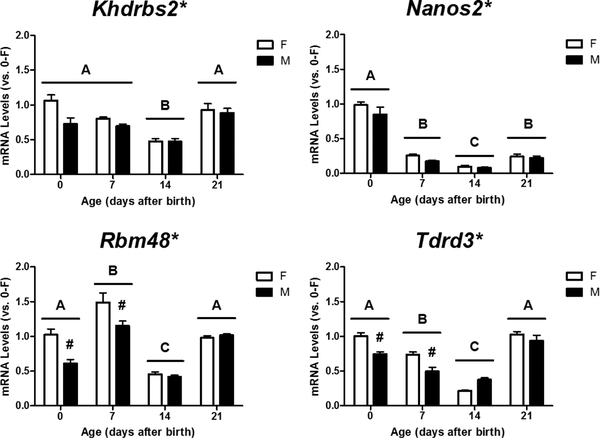
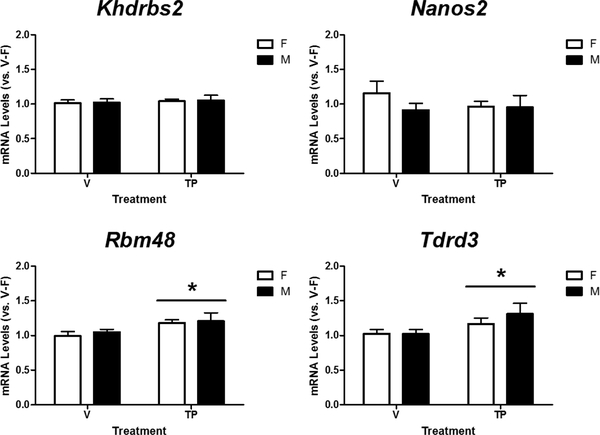
We then used RT-qPCR to determine mRNA expression of the 16 selected genes in the cortex/hippocampus of male and female mice at PN0, PN7, PN14, and PN21. Two-way ANOVA revealed a significant effect of sex on relative mRNA levels of Khdrbs2 (p = 0.015), Nanos2 (p = 0.019), Rbm48 (p < 0.001), and Tdrd3 (p = 0.003) (Figure 1). Rbm48 (p = 0.003) and Tdrd3 (p < 0.001) displayed a significant effect of interaction between age and sex, and Tukey’s post hoc analysis further revealed a femalebiased expression of both genes at PN0 and PN7, but not PN14 or PN21. Additionally, all four genes displayed a significant effect of age on their expression (p < 0.001). Khdrbs2 and Tdrd3 mRNA levels steadily declined with age after birth, but increased again at PN21. Nanos2 expression rapidly declined after birth and remained low during early development. Rbm48 mRNA levels increased at PN7, dropped to low levels at PN14, but then increased again at PN21.
Figure 1.
Sex- and age-dependent changes in relative mRNA levels of Khdrbs2, Nanos2, Rbm48, and Tdrd3 in the developing female (F) and male (M) mouse cortex/hippocampus. Expression of each gene was calculated and expressed relative to the average of neonatal females (0-F as 1-fold). * Indicates a significant difference of gene expression between the sexes (p < 0.05). # indicates a significant difference vs. females at the same age (p < 0.05). A, B, C indicate significant differences between ages (p <0.05).
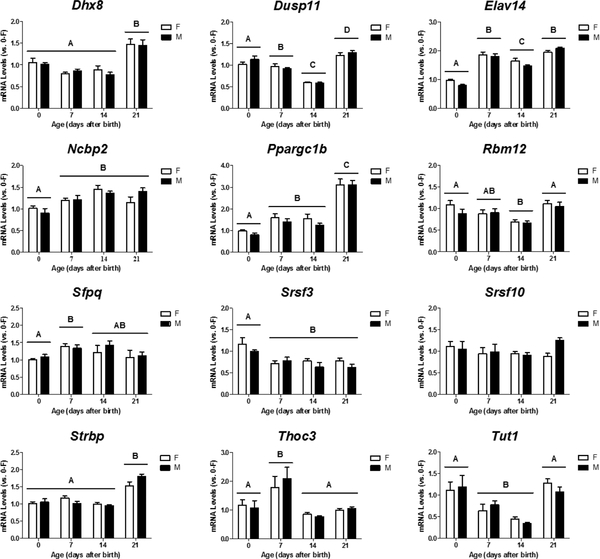
Eleven of the 12 other genes, with the exception of Srsf10, exhibited no sex-, but age-dependent changes in their mRNA levels in the developing cortex/hippocampus. A two-way ANOVA revealed a significant effect of age on relative mRNA levels of Dhx8 (p < 0.001), Dusp11 (p < 0.001), Elov14 (p < 0.001), Ncbp2 (p < 0.001), Pporgc1b (p < 0.001), Rbm12 (p = 0.001), Sfpq (p = 0.033), Srsf3 (p < 0.001), Strbp (p < 0.001), Thoc3 (p < 0.001), and Tut1 (p < 0.001) (Figure 2). Post-hoc tests further showed that expression of Dhx8, Elov14, Ncbp2, Pporgc1b, and Strbp increased with age within the first three weeks after birth whereas Srsf3 displayed a decline in expression by PN7 (p < 0.05). Dusp11, Rbm12, and Tut1 mRNA levels transiently decreased between PN7 and PN14, but Sfpq and Thoc3 displayed a brief rise in expression at PN7.
Figure 2.
Age-dependent changes in mRNA expression of Dhx8, Dusp11, Elav14, Ncbp2, Ppargc1b, Rbm12, Sfpq, Srsf3, Srsf10, Strbp, Thoc3, and Tut1 in the developing female (F) and male (M) mouse cortex/hippocampus. Relative mRNA levels of individual genes were calculated and expressed relative to the average of neonatal females (0-F as 1-fold). A, B, C, D indicate significant differences between ages (p < 0.05).
3.3. Effects of Prenatal TP Treatment on Expression of Khdrbs2, Nanos2, Rbm48, and Tdrd3 in the Cortex/Hippocampus of Neonatal Male and Female Mice
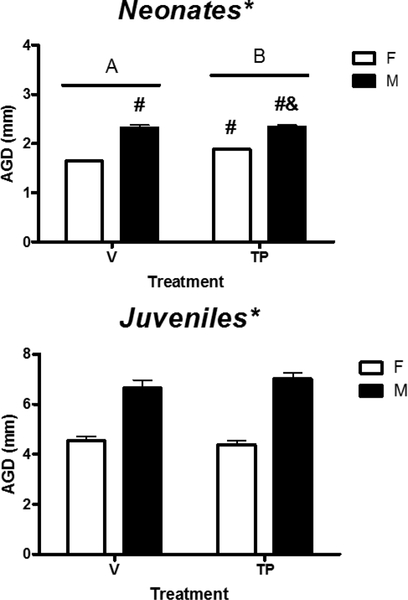
To determine if sexually dimorphic expression of Khdrbs2, Nanos2, Rbm48, and Tdrd3 in the developing cortex/hippocampus was due to prenatal testosterone secreted by the developing testes, we determined relative mRNA levels of these genes in the cortex/hippocampus of male and female neonatal mice prenatally treated with either vehicle or TP. Body weight and AGD of each mouse in this experiment were measured at sacrifice. A two-way ANOVA revealed no effect of sex, treatment, or their interaction on body weight (Table 3). In contrast, two-way ANOVA of AGD data showed significant effects of sex (p < 0.001), treatment (p = 0.004), and their interaction (p = 0.012) (Figure 3, upper panel). Tukey’s post hoc analysis further indicated that prenatal TP treatment significantly increased AGD in females, not males, but the AGD of TP-treated females was shorter than TP-treated males (p < 0.05). These observations confirmed the masculinization by fetal androgen exposure in female animals.
Figure 3.
Effects of prenatal (upper panel) and postnatal (lower panel) exposure to testosterone propionate (TP) on anogenital distance (AGD) in female (F) and male (M) neonatal and juvenile mice. * indicates a significant difference between the sexes (p < 0.05). # indicates significant difference vs. vehicle (V)-treated females (p < 0.05). & indicates significant difference vs. TP-F (p < 0.05). A, B indicate significant differences between the treatments (p < 0.05).
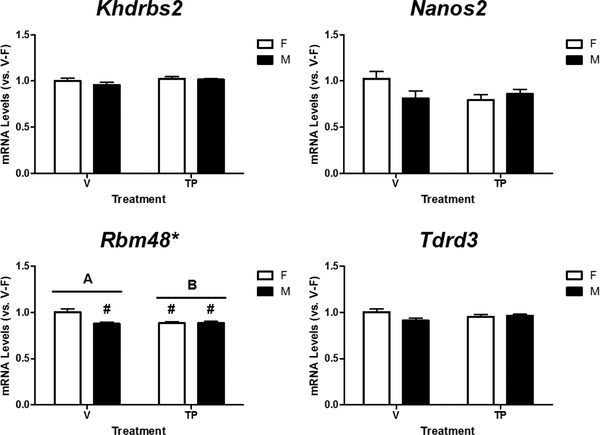
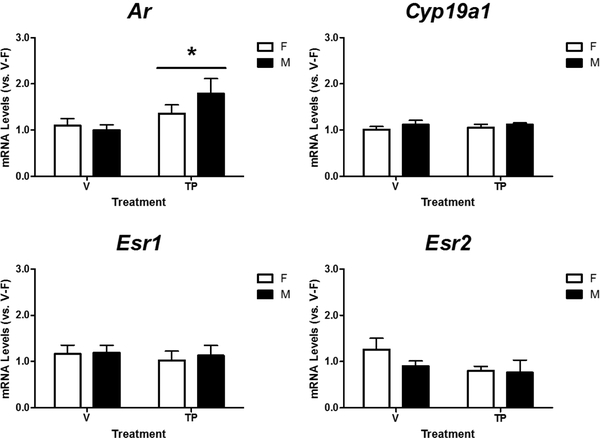
Next, we determined relative mRNA levels of Khdrbs2, Nanos2, Rbm48, and Tdrd3 in the cortex/hippocampus of male and female neonatal mice treated with either vehicle or TP during late gestation (Figure 4). A two-way ANOVA revealed a significant effect of sex (p = 0.009), treatment (p = 0.017), and their interaction (p = 0.008) on Rbm48 mRNA levels, and Tukey’s post hoc analysis further indicated a female-biased expression in vehicle-treated mice, but not in mice prenatally treated with testosterone (Figure 4, bottom left panel). Additionally, the analysis indicated no effect of sex (p = 0.274), or treatment (p = 0.196), but an interaction between sex and treatment (p = 0.048) on Nanos2 mRNA levels. However, post hoc analysis indicated no significant differences between individual groups. There was no effect of sex (p = 0.259 and p = 0.113), treatment (p = 0.104 and p = 0.970), or interaction between sex and treatment (p = 0.418 and p = 0.055) on Khdrbs2 or Tdrd3 mRNA levels in the neonatal mouse cortex/hippocampus.
Figure 4.
Effects of prenatal exposure to testosterone propionate (TP, 100 μg/0.05ml) on mRNA expression of Khdrbs2, Nanos2, Rbm48 and Tdrd3 in the female (F) and male (M) neonatal mouse cortex/hippocampus. Relative mRNA levels of each gene were calculated and expressed relative to the average of vehicle (V)-treated females (V-F as 1-fold). * indicates significant difference between the sexes (p < 0.05). # indicates significant difference versus V-F (p < 0.05). A, B indicate significant differences between the treatments (p < 0.05).
To verify the presence of sex steroid receptors and aromatase (Cyp19o1) in the cortex/hippocampus of prenatally vehicle- and TP-treated, neonatal mice, we determined relative mRNA levels of Ar, Esr1, Esr2, and Cyp19a1. There was no effect of sex, treatment, or their interaction on mRNA expression of Ar, Esr1, Esr2, and Cyp19a1 in the neonatal mouse cortex/hippocampus receiving prenatal treatments (Table 4). However, there was a trend towards a sex difference in Esr2 mRNA (p = 0.059) and a trend of interaction between sex and treatment in Esr1 mRNA (p = 0.058). Although Ar was detected in the neonatal mouse cortex/hippocampus, high cycles of threshold (Ct) values (greater than 30) observed in qPCR assays indicated low quantities of Ar mRNA.
TABLE 4.
Effect of prenatal exposure to vehicle (V) and testosterone propionate (TP, 100 μg/0.05 ml) on expression of Ar, Cyp191a, Esr1, and Esr2 mRNA in the female (F) and male (M), neonatal mouse cortex/hippocampus.
| Group | Na | Ar | N | Cyp19a1 | N | Esr1 | N | Esr2 |
|---|---|---|---|---|---|---|---|---|
| V-F | 10 | 1.03 ± 0.07b | 9 | 1.10 ± 0.16 | 10 | 1.06 ± 0.13 | 10 | 1.04 ± 0.10 |
| V-M | 10 | 0.90 ± 0.07 | 10 | 1.01 ± 0.20 | 10 | 0.82 ± 0.05 | 10 | 0.79 ± 0.05 |
| TP-F | 10 | 0.82 ± 0.04 | 10 | 1.29 ± 0.22 | 10 | 0.86 ± 0.03 | 10 | 0.81 ± 0.06 |
| TP-M | 10 | 1.06 ± 0.09 | 10 | 1.09 ± 0.20 | 10 | 0.91 ± 0.03 | 10 | 0.80 ± 0.06 |
N: number of mice in each group;
Relative mRNA levels of each gene were calculated and expressed as relative to the average of vehicle-treated females.
3.4. Effects of Postnatal TP Treatment on Expression of Khdrbs2, Nanos2, Rbm48, and Tdrd3 in the Cortex/Hippocampus of Juvenile Male and Female Mice
To determine if Khdrbs2, Nanos2, Rbm48 and Tdrd3 remained responsive to androgens after the critical period, we measured their relative mRNA levels in the cortex/hippocampus of male and female mice treated with vehicle or TP during PN21–23. Body weights of the mice used in this experiment are shown in Table 3. A two-way ANOVA revealed no effect of sex (p = 0.065), treatment (p = 0.058), or their interaction (p = 0.675) on body weight. AGD was also measured on each mouse, and a two-way ANOVA showed significant effects of sex (p < 0.001) on mean AGD, with a longer AGD in males than females (Figure 3, lower panel). On the other hand, there was no effect of treatment (p = 0.662) or interaction between sex and treatment (p = 0.252) on AGD.
A two-way ANOVA revealed that postnatal TP treatment significantly increased Rbm48 and Tdrd3 mRNA levels (p = 0.013 and p = 0.047), but there was no effect of sex (p = 0.538 and p = 0.643) or interaction between sex and treatment (p = 0.810 and p = 0.689) (Figure 5). In contrast, there was no effect of sex (p = 0.828 and p = 0.371), treatment (p = 0.575 and p = 0.596), or interaction (p = 0.980 and p = 0.427) on expression of Khdrbs2 and Nanos2.
Figure 5.
Effects of postnatal exposure to testosterone propionate (TP, 100 μg/0.05 ml) on expression of Khdrbs2, Nanos2, Rbm48, and Tdrd3 in the female (F) and male (M) mouse cortex/hippocampus at postnatal day 23. Relative mRNA levels were calculated relative to the average of vehicle (V)-treated females (V-F as 1-fold). * Indicates significant difference vs. vehicle-treated mice (p < 0.05).
We then determined relative mRNA levels of Ar, Esr1, Esr2, and Cyp19a1 in the cortex/hippocampus of vehicle- and TP-treated mice to confirm their presence in the postnatal cortex/hippocampus (Figure 6). There was no effect of TP treatment, sex, or their interaction on Esrl, Esr2, or Cyp19a1 mRNA levels in the mouse cortex/hippocampus at PN23. Interestingly, postnatal TP significantly increased Ar expression in the cortex/hippocampus (p = 0.017), but no effect of sex (p = 0.646) or interaction between treatment and sex (p = 0.358) on its mRNA levels was observed.
Figure 6.
Effects of postnatal exposure to testosterone propionate (TP, 100 μg/0.05 ml) on expression of Ar, Cyp19a1, Esr1, and Esr2 in the female (F) and male (M) mouse cortex/hippocampus at postnatal day 23. Relative mRNA levels were calculated relative to the average of vehicle (V)-treated females (V-F as 1 fold). * Indicates significant difference vs. vehicle-treated mice (p < 0.05).
3.5. Effect of Age and Sex on mRNA Levels of Ar, Cyp19a1, Esr1, and Esr2 in the Developing Mouse Cortex/Hippocampus
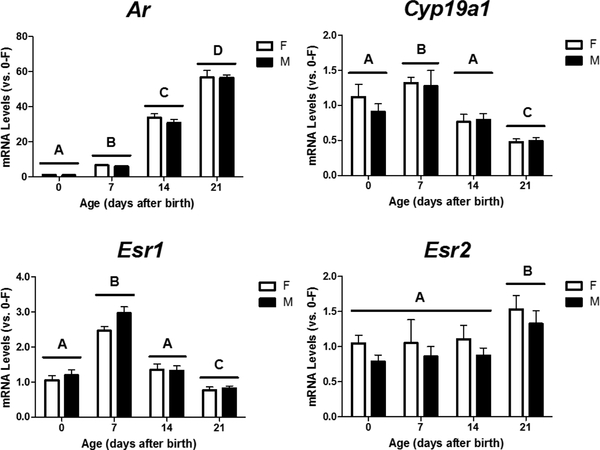
To determine if differential expression of steroid receptors and aromatase with age might play a role in conferring age-related changes in the androgenic responsiveness of the Rbm48 gene, we determined mRNA levels of Ar, Cyp19a1, Esr1, and Esr2 in the mouse cortex/hippocampus at PN0, 7, 14, and 21. Two-way ANOVA revealed a significant effect of age, but not sex with no interaction, on relative mRNA levels of Ar, Cyp19a1, Esr1, and Esr2 in the male and female mouse cortex/hippocampus during early development (Figure 7). Within the first three weeks after birth, expression of Ar (p< 0.001) and Esr2 (p = 0.033) increased with age. Post hoc analyses further demonstrated that Ar mRNA levels in the cortex/hippocampus were significantly higher on PN7 than PN0 and continued to increase at PN14 and PN21. However, the age-dependent rise in Esr2 expression didn’t occur until PN21. In contrast to Ar and Esr2, Esrl and Cyp19a1 both showed a transient rise in mRNA levels at PN7, followed by a gradual decrease with levels below those at PN0 by PN21 (p < 0.05).
Figure 7.
Developmental changes in Ar, Esr1, and Esr2 expression in the female (F) and male (M) mouse cortex/hippocampus at postnatal days 0, 7, 14, and 21. Gene expression was calculated relative to the average of neonatal females (0-F as 1 fold). A, B, C, D indicate significant differences between ages (p < 0.05).
4. Discussion
During late gestation and on the day of birth, significant elevations of circulating testosterone are secreted by the developing testes, and after these surges, testosterone levels decrease and remain low in male mice until the onset of puberty (Clarkson and Herbison 2016). The perinatal rise in testosterone establishes brain masculinization that in turn underpins sexually dimorphic behavior and physiology in male mice (Ngun et al., 2011; Quadagno et al., 1975; vom Saal 1979). Synchronously with these increases in testosterone levels during early development, Rbm48 expression is suppressed in the male compared to the female mouse cortex/hippocampus at both PN0 and PN7 specifically (Figure 1). Our finding that prenatal TP treatment in female neonates reduces Rbm48 mRNA levels to male-like levels further indicates that the sex difference in neonatal cortical and hippocampal Rbm48 expression might be due to the androgen-dependent downregulation of the Rbm48 gene (Figure 5). Testosterone has been demonstrated to exert its effects on brain masculinization through AR directly and/or ERs indirectly after local aromatization of testosterone to estradiol (Bodo et al., 2006; Bodo and Rissman 2007; Ogawa et al., 1997; Simerly et al., 1997; vom Saal 1979). In the current study, we have confirmed the presence of Ar, Cyp19a1, Esr1, and Esr2 mRNA in the neonatal mouse cortex/hippocampus (Figure 5), suggesting that both AR and ERs may be involved in the transcriptional regulation of Rbm48. While no effect of sex or TP on Ar, Cyp19a1, Esr1, and Esr2 mRNA levels in neonates was detected, the differences in circulating testosterone between the sexes during the perinatal period can still activate AR and/or ERs in a sex-dependent fashion to regulate Rbm48 transcription. Together, the differences in sex steroid milieu between male and female newborn pups appears to be the determinant of the sex difference in Rbm48 mRNA levels in the developing mouse cortex and hippocampus although the protein levels of Ar, Cyp19o1, Esr1, and Esr2 remain to be verified.
Acting as ligand-inducible transcription factors, liganded AR and ERs bind to the androgen response element (ARE) and estrogen response element (ERE), respectively, on the promoters of the genes they regulate (Denayer et al., 2010; Sanchez et al., 2002). Using a genome-wide analysis of promoter elements for estrogen receptor binding sites, Kamalakaran et al. (2005) not only confirmed the presence of ERE in the genes known to be regulated by estrogen, but also identified several new estrogen-responsive genes whose putative promoters contained EREs (Kamalakaran et al., 2005). To explore if the same molecular mechanisms may be involved in androgenic regulation of Rbm48 expression, we searched the consensus sequences of ARE and ERE in the Rbm48 promoter (−1000 to 100 bp) using the mmEPDnew, the Mus musculus (mouse) curated promoter database from Eukaryotic Promoter Database (https://epd.epfl.ch/mouse/mousedatabase.php?db=mouse). We found that its putative promoter contains one potential ARE (at position −50) and several potential EREs (at positions −802, −712, −606, −538, , −195, and −24 for ESR1, and −479, −117, −25/−24, and 44 for ESR2), which supports the Rbm48 gene as a direct transcriptional target for both liganded AR and ERs. However, our prediction cannot rule out the possibility that ligand-activated ERs may regulate Rbm48 expression via functional protein-protein interactions with other transcription factors bound to their cognate regulatory elements, such as AP-1 (at position −501), on the Rbm48 promoter in an ERE-independent manner (Kushner et al., 2000; Safe 2001). To further evaluate if the predicted ARE and EREs as well as other motifs are the functional binding sits to AR and ERs, we can perform luciferase promoter constructs containing each element and DNA-binding assays in vitro and chromatin immunoprecipitation assays in vivo.
In contrast to the neonates, male and female juvenile mice both showed increased Rbm48 mRNA levels in response to TP treatment at the same dose three weeks after birth (Figure 6), indicating that the Rbm48 gene in the developing cortex and hippocampus remains androgen-responsive during early development, but seems to be regulated by distinct mechanisms depending on age. The opposite responses of Rbm48 transcription to prenatal and postnatal testosterone might be inferred from the developmental changes in abundance of Cyp19a1 and Ar. Cyp19a1 expression in the mouse cortex/hippocampus is lower in both males and females at PN21 as compared to neonates (Figure 8). Similar to our data, Cyp19a1 mRNA expression in the mouse hippocampus remains elevated during the first two weeks after birth and then declines as adults (Ivanova and Beyer 2000). Decreased expression of Cyp19a1 might prohibit local estrogen biosynthesis in the juvenile mouse cortex/hippocampus and then dampen estrogen responsiveness by reducing transactivation of ERα and/or ERβ by ligands. On the other hand, in agreement with our previous findings (Tsai et al., 2015), Ar mRNA levels drastically increase with age during the first three weeks after birth (Figure 8), which could enhance the responsiveness of the juvenile cortex/hippocampus to androgens through augmented Ar expression. In addition, it has been reported that AR overexpression could also inhibit estradiol-stimulated ER target gene transcription by competing for transcriptional coregulators (Lanzino et al., 2005) or binding to EREs in breast cancer cells (Peters et al., 2009). Accordingly, we speculate that AR might play the major role in the androgen-dependent upregulation of Rbm48 expression three weeks after birth while the androgenic downregulation of Rbm48 transcription in neonates might be largely mediated by the activation of ERs by locally synthesized estradiol.
The Rbm48 gene encodes a 413-amino acid protein, RNA binding motif protein 48 (RBM48; 48 kDa), with a single copy of the RNA recognition motif (RRM) domain at the amino terminus (residues 78–177; RefSeq: NP_766579.3 and Ensembl: ENSMUST00000042753.13). RRM is a conserved, single-stranded RNA binding domain of 80–90 amino acids, and many RRM-containing proteins have been found to be the key regulators required for RNA biogenesis, stability, function, transport and cellular localization (Maris et al., 2005). According to the Ensembl (http://uswest.ensembl.org/Musmusculus/Info/Index), RBM48 and 270 other mouse gene products belong to the RNA-binding domain (RBD) superfamily with the presence of RRM and/or other canonical RBDs. Currently, neither RNA targets nor molecular function of RBM48 in mice have yet been identified. As proteins with structurally conserved motifs often share similar functions, we performed GO enrichment analysis of the 271 mouse RBD superfamily genes to explore the possible involvement of RBM48 in certain biological processes and signaling pathways. The results showed significant enrichment for terms related exclusively to the chemical reactions and cellular pathways that involve RNA, including RNA processing (GO:0006396), RNA splicing (GO:0008380), gene expression (GO:0010467), RNA metabolic process (GO:0016070), RNA localization (GO:0006403), RNA transport (GO:0050658), RNA stabilization (GO:0043489), and translation (GO:0006412), unsurprisingly suggesting that like many members of the RBD superfamily, RBM48 may control the fate of its bound RNAs by governing a single or multiple aspects of RNA biology, from transcription, splicing and polyadenylation to RNA modification, transport, localization, translation and turnover. Bai and colleagues (2019) have recently discovered RBM48 to function as a minor spliceosome factor in U12 splicing in plants (corn), and Rbm48 mutations cause aberrant splicing of U12-type introns and the defects that repress endosperm cell differentiation during development (Bai et al., 2019). Since RBM48 is a highly conserved protein, mouse RBM48 is predicted to also function in U12 splicing. In addition, an in vitro study has found that RBM48 may contain anti-apoptotic activity as CRISPR knockout of RBM48 in HCT116 and HeLa cells results in cleavage of both caspase 7 and PARP, the signs of apoptosis (Hart et al., 2015), which implies that RBM48 may be involved in apoptosis. In support of this, a list of RBM proteins, including RBM3, RBM4, RBM5, RBM6, RBM7, RBM8, RBM 10, RBM11, RBM12, RBM15, and RBM25, have been identified as apoptosis modulators in humans and mice (Cui and Placzek 2018; Jackson et al., 2015; Martinez-Arribas et al., 2006; Sutherland et al., 2005). Hormonally-mediated differences in cell death between the sexes are central to the sexual differentiation of many brain areas (Forger and de Vries 2010). Nunez et al. (2001) found that at the age of 6 and 7 days, male rats had a higher density of apoptotic cells in their primary visual cortex than females as the result of androgen exposure during perinatal period(Nunez et al., 2001). Based on these findings, we speculate that androgen-downregulation of Rbm48 expression might be critical for the control of apoptotic processes, resulting in sexually dimorphic neural circuits in the developing mouse cortex/hippocampus.
RT-qPCR has been commonly used as a validation tool for confirming gene expression results obtained from microarray analysis. However, of the 13 selected sexually dimorphic candidate genes identified by microarrays, only five (Ddx3y, Eif2s3x, Eif2×3y, Nanos2, and Rbm48; 38%) were confirmed with the same sex differences in expression in the developing mouse cortex/hippocampus in previous (Armoskus et al., 2014b) and current studies (Figure 1). Interestingly, Tdrd3, one of the eight false positives, had been identified as a male-biased gene by microarrays, but expressed at higher levels in females than males as measured by RT-qPCR (Figure 1). In addition, Khdrbs2 exhibited an insignificant sex bias trend in expression by microarrays, but a female-biased increase in mRNA levels by RT-qPCR (Figure 1). The majority of the selected candidates displayed a small fold change in expression, ranging from 1.16 to 1.46, and a p value of more than 0.001 between the sexes by microarrays except Ddx3y (2.8 fold, p < 0.0001), Eif2×3x (1.4 fold, p < 0.0001) and Eif2×3y (285.9 fold, p < 0.0001) (Armoskus et al., 2014b). Low correlations between microarrays and RT-qPCR have been previously reported for the measurement of mRNA expression of the genes showing less than a 1.4-fold change or a p-value greater than 0.0001 (Etienne et al., 2004; Morey et al., 2006; Wurmbach et al., 2003). The discrepancy between the results obtained from microarrays and RT-qPCR could be due to several fundamental technical differences between the two methodologies. First, microarray and RT-qPCR differ on calculation of expression data. The former requires global normalization while the latter simply utilizes the expression of one or two reference genes, such as Rpl13a and Actb, against which target gene expression is calibrated. Second, although the microarray probes and PCR primers are designed to be gene-specific, the specificity and sensitivity of array probes might be constrained by the length and sequence from allowing simultaneous hybridization of all probes and labeled cRNAs to occur at the same temperature. In contrast, the temperature at which PCR primers anneal to the cDNA template varies with the length and base composition of each primer set with optimal hybridization specificity and efficiency. Lastly, different locations on the target gene and lengths of microarray probes and PCR primers could also influence the correlation between the two methods. Even with some inconsistencies between the microarray and RT-qPCR data, microarray is still a powerful and effective tool for large-scale gene expression profiling, which helps scientists narrow the list of potential targets for further investigation of the complex molecular mechanisms underlying brain sexual differentiation.
In summary, our study has identified the Rbm48 gene as a potential downstream effector to mediate the action of perinatal testosterone in driving sexual differentiation of the cerebral cortex and hippocampus during early development. With more abundant ERs and aromatase, but low AR, expressed in the neonatal cortex/hippocampus, the perinatal rises of testosterone in male mice seem to activate ERs to suppress Rbm48 expression. As age advances, the Rbm48 gene switches from being androgen-suppressed via ERs to being androgen-stimulated through AR as AR levels rise robustly after birth. The hormonally regulated Rbm48 gene expression during early development may provide a starting point for future investigations to better understand the process of brain sexual differentiation. This will also provide new insights into the molecular mechanisms that generate gender bias in a variety of neurological diseases linked to dysfunction in these brain regions, such as Alzheimer’s disease and autism.
Highlights.
Rbm48 mRNA expression in the developing mouse cortex/hippocampus is female-biased
Prenatal testosterone reduces Rbm48 levels in the neonatal cortex/hippocampus
Postnatal testosterone increases Rbm48 levels in the juvenile cortex/hippocampus
Levels of steroid receptors and aromatase confer androgenic responsiveness of Rbm48
Acknowledgements
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award numbers SC3GM102051, 25GM0500089, and 5UL1GM118979. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We also thank Debbie Moreira, Thomas Mota, and Lauren Fisher for their technical assistance. In addition, we are also grateful to Drs. Sandra Legan and Emilie Rissman for their valuable comments and suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antunes-Martins A, Mizuno K, Irvine EE, Lepicard EM, Giese KP, 2007. Sex-dependent upregulation of two splicing factors, psf and Srp20, during hippocampal memory formation. Learn Mem. 14, 693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armoskus C, Mota T, Moreira D, Tsai HW, 2014a. Effects of prenatal testosterone exposure on sexually dimorphic gene expression in the neonatal mouse cortex and hippocampus. J Steroids Horm Sci. 5, 1000139. [PMC free article] [PubMed] [Google Scholar]
- Armoskus C, Moreira D, Bollinger K, Jimenez O, Taniguchi S, Tsai H, 2014b. Identification of sexually dimorphic genes in the neonatal mouse cortex and hippocampus. Brain Res. 1562, 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP, 2017. A general theory of sexual differentiation. J Neurosci Res. 95, 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP, 2009. The organizational-activational hypothesis as the foundation for a unified theory of sexual differentiation of all mammalian tissues. Horm Behav. 55, 570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F, Corll J, Shodja DN, Davenport R, Feng G, Mudunkothge J, Brigolin CJ, Martin F, Spielbauer G, Tseung CW, Siebert AE, Barbazuk WB, Lal S, Settles AM, 2019. RNA binding motif protein 48 is required for U12 splicing and maize endosperm differentiation. Plant Cell. 31, 715–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao AM, Swaab DF, 2011. Sexual differentiation of the human brain: Relation to gender identity, sexual orientation and neuropsychiatric disorders. Front Neuroendocrinol. 32, 214–226. [DOI] [PubMed] [Google Scholar]
- Bao AM, Swaab DF, 2010. Sex differences in the brain, behavior, and neuropsychiatric disorders. Neuroscientist. 16, 550–565. [DOI] [PubMed] [Google Scholar]
- Bodo C, Rissman EF, 2007. Androgen receptor is essential for sexual differentiation of responses to olfactory cues in mice. Eur J Neurosci. 25, 2182–2190. [DOI] [PubMed] [Google Scholar]
- Bodo C, Kudwa AE, Rissman EF, 2006. Both estrogen receptor-alpha and -beta are required for sexual differentiation of the anteroventral periventricular area in mice. Endocrinology. 147, 415–420. [DOI] [PubMed] [Google Scholar]
- Bowers JM, Waddell J, McCarthy MM, 2010. A developmental sex difference in hippocampal neurogenesis is mediated by endogenous oestradiol. Biol Sex Differ. 1, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinegar AE, Cooper TA, 2016. Roles for RNA-binding proteins in development and disease. Brain Res. 1647, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill L, 2006. Why sex matters for neuroscience. Nature Reviews Neuroscience. 7, 477–484. [DOI] [PubMed] [Google Scholar]
- Clarkson J, Herbison AE, 2016. Hypothalamic control of the male neonatal testosterone surge. Philos Trans R Soc Lond B Biol Sci. 371, 20150115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Placzek WJ, 2018. Post-transcriptional regulation of anti-apoptotic BCL2 family members. Int J Mol Sci. 19, 10.3390/ijmsl9010308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denayer S, Helsen C, Thorrez L, Haelens A, Claessens F, 2010. The rules of DNA recognition by the androgen receptor. Mol Endocrinol. 24, 898–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne W, Meyer MH, Peppers J, Meyer RA Jr, 2004. Comparison of mRNA gene expression by RT-PCR and DNA microarray. BioTechniques. 36, 618–20, 622, 624–6. [DOI] [PubMed] [Google Scholar]
- Forger NG, de Vries GJ, 2010. Cell death and sexual differentiation of behavior: Worms, flies, and mammals. Curr Opin Neurobiol. 20, 776–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresack JE, Frick KM, 2003. Male mice exhibit better spatial working and reference memory than females in a water-escape radial arm maze task. Brain Res. 982, 98–107. [DOI] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, Mero P, Dirks P, Sidhu S, Roth FP, Rissland OS, Durocher D, Angers S, Moffat J, 2015. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 163,1515–1526. [DOI] [PubMed] [Google Scholar]
- Hatanaka Y, Wada K, Kabuta T, 2015. Abnormal instability, excess density, and aberrant morphology of dendritic spines in prenatally testosterone-exposed mice. Neurochem Int. 85–86, 53–58. [DOI] [PubMed] [Google Scholar]
- Hisasue S, Seney ML, Immerman E, Forger NG, 2010. Control of cell number in the bed nucleus of the stria terminalis of mice: Role of testosterone metabolites and estrogen receptor subtypes. J Sex Med. 7, 1401–1409. [DOI] [PubMed] [Google Scholar]
- Honda S, Harada N, Ito S, Takagi Y, Maeda S, 1998. Disruption of sexual behavior in male aromatase-deficient mice lacking exons 1 and 2 of thecyp19Gene. Biochem Biophys Res Commun. 252, 445–449. [DOI] [PubMed] [Google Scholar]
- Hotchkiss AK, Lambright CS, Ostby JS, Parks-Saldutti L, Vandenbergh JG, Gray LE, 2007. Prenatal testosterone exposure permanently masculinizes anogenital distance, nipple development, and reproductive tract morphology in female sprague-dawley rats. Toxicological Sciences. 96, 335–345. [DOI] [PubMed] [Google Scholar]
- Isgor C, Sengelaub DR, 1998. Prenatal gonadal steroids affect adult spatial behavior, CA1 and CA3 pyramidal cell morphology in rats. Horm Behav. 34, 183–198. [DOI] [PubMed] [Google Scholar]
- Ivanova T, Beyer C, 2000. Ontogenetic expression and sex differences of aromatase and estrogen receptor-α/β mRNA in the mouse hippocampus. Cell Tissue Res. 300, 231–237. [DOI] [PubMed] [Google Scholar]
- Jackson TC, Du L, Janesko-Feldman K, Vagni VA, Dezfulian C, Poloyac SM, Jackson EK, Clark RS, Kochanek PM, 2015. The nuclear splicing factor RNA binding motif 5 promotes caspase activation in human neuronal cells, and increases after traumatic brain injury in mice. J Cereb Blood Flow Metab. 35, 655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y, Goldman D, 2018. Role of RNA modifications in brain and behavior. Genes Brain Behav. 17, e12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamalakaran S, Radhakrishnan SK, Beck WT, 2005. Identification of estrogen-responsive genes using a genome-wide analysis of promoter elements for transcription factor binding sites. J Biol Chem. 280, 21491–21497. [DOI] [PubMed] [Google Scholar]
- Karlsson SA, Haziri K, Hansson E, Kettunen P, Westberg L, 2015. Effects of sex and gonadectomy on social investigation and social recognition in mice. BMC Neurosci. 16, 83–015-0221-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr J, Allore R, Beck S, Handa R, 1995. Distribution and hormonal regulation of androgen receptor (AR) and AR messenger ribonucleic acid in the rat hippocampus. Endocrinology. 136, 3213–3221. [DOI] [PubMed] [Google Scholar]
- Kushner PJ, Agard DA, Greene GL, Scanlan TS, Shiau AK, Uht RM, Webb P, 2000. Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 74, 311–317. [DOI] [PubMed] [Google Scholar]
- Lanzino M, De Amicis F, McPhaul MJ, Marsico S, Panno ML, Ando S, 2005. Endogenous coactivator ARA70 interacts with estrogen receptor alpha (ERalpha) and modulates the functional ERalpha/androgen receptor interplay in MCF-7 cells. J Biol Chem. 280, 20421–20430. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB, 2006. Splicing regulation in neurologic disease. Neuron. 52, 93–101. [DOI] [PubMed] [Google Scholar]
- Manning KS, Cooper TA, 2017. The roles of RNA processing in translating genotype to phenotype. Nat Rev Mol Cell Biol. 18, 102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH, 2005. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. Febs J. 272, 2118–2131. [DOI] [PubMed] [Google Scholar]
- Markham JA, Jurgens HA, Auger CJ, De Vries GJ, Arnold AP, Juraska JM, 2003. Sex differences in mouse cortical thickness are independent of the complement of sex chromosomes. Neuroscience. 116, 71–75. [DOI] [PubMed] [Google Scholar]
- Martinez-Arribas F, Agudo D, Pollan M, Gomez-Esquer F, Diaz-Gil G, Lucas R, Schneider J, 2006. Positive correlation between the expression of X-chromosome RBM genes (RBMX, RBM3, RBM10) and the proapoptotic bax gene in human breast cancer. J Cell Biochem. 97,1275–1282. [DOI] [PubMed] [Google Scholar]
- McCarthy MM, 2016. Sex differences in the developing brain as a source of inherent risk. Dialogues Clin Neurosci. 18, 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi K, Takanashi H, Nagasawa M, Kikusui T, 2015. Sex differences in spatiotemporal expression of AR, ERα, and ERβ mRNA in the perinatal mouse brain. Neurosci Lett. 584, 88–92. [DOI] [PubMed] [Google Scholar]
- Morey JS, Ryan JC, Van Dolah FM, 2006. Microarray validation: Factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol Proced Online. 8, 175–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motelica-Heino I, Castanier M, Corbier P, Edwards DA, Roffi J, 1988. Testosterone levels in plasma and testes of neonatal mice. J Steroid Biochem. 31, 283–286. [DOI] [PubMed] [Google Scholar]
- Ngun TC, Ghahramani N, Sanchez FJ, Bocklandt S, Vilain E, 2011. The genetics of sex differences in brain and behavior. Front Neuroendocrinol. 32, 227–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JL, Lauschke DM, Juraska JM, 2001. Cell death in the development of the posterior cortex in male and female rats. J Comp Neurol. 436, 32–41. [PubMed] [Google Scholar]
- Ogawa S, Chester AE, Hewitt SC, Walker VR, Gustafsson JA, Smithies O, Korach KS, Pfaff DW, 2000. Abolition of male sexual behaviors in mice lacking estrogen receptors alpha and beta (alpha beta ERKO). Proc Natl Acad Sci U S A. 97, 14737–14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Lubahn DB, Korach KS, Pfaff DW, 1997. Behavioral effects of estrogen receptor gene disruption in male mice. Proc Natl Acad Sci U S A. 94, 1476–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AA, Buchanan G, Ricciardelli C, Bianco-Miotto T, Centenera MM, Harris JM, Jindal S, Segara D, Jia L, Moore NL, Henshall SM, Birrell SN, Coetzee GA, Sutherland RL, Butler LM, Tilley WD, 2009. Androgen receptor inhibits estrogen receptor-alpha activity and is prognostic in breast cancer. Cancer Res. 69, 6131–6140. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinares-Garcia P, Stratikopoulos M, Zagato A, Loke H, Lee J, 2018. Sex: A significant risk factor for neurodevelopmental and neurodegenerative disorders. Brain Sci. 8, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadagno DM, Wolfe HG, Kan Wha Ho G, Goldman BD, 1975. Influence of neonatal castration or neonatal anti-gonadotropin treatment on fertility, phallus development, and male sexual behavior in the mouse. Fertil Steril. 26, 939–944. [DOI] [PubMed] [Google Scholar]
- Rizk A, Robertson J, Raber J, 2005. Behavioral performance of tfm mice supports the beneficial role of androgen receptors in spatial learning and memory. Brain Res. 1034, 132–138. [DOI] [PubMed] [Google Scholar]
- Safe S, 2001. Transcriptional activation of genes by 17 beta-estradiol through estrogen receptor-Sp1 interactions. Vitam Horm. 62, 231–252. [DOI] [PubMed] [Google Scholar]
- Sanchez R, Nguyen D, Rocha W, White JH, Mader S, 2002. Diversity in the mechanisms of gene regulation by estrogen receptors. Bioessays. 24, 244–254. [DOI] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB, 2004. A functional genomics strategy reveals rora as a component of the mammalian circadian clock. Neuron. 43, 527–537. [DOI] [PubMed] [Google Scholar]
- Seney ML, Walsh C, Stolakis R, Sibille E, 2012. Neonatal testosterone partially organizes sex differences in stress-induced emotionality in mice. Neurobiol Dis. 46, 486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simerly RB, Zee MC, Pendleton JW, Lubahn DB, Korach KS, 1997. Estrogen receptor-dependent sexual differentiation of dopaminergic neurons in the preoptic region of the mouse. Proc Natl Acad Sci U S A. 94, 14077–14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland LC, Rintala-Maki ND, White RD, Morin CD, 2005. RNA binding motif (RBM) proteins: A novel family of apoptosis modulators? J Cell Biochem. 94, 5–24. [DOI] [PubMed] [Google Scholar]
- Tsai HW, Taniguchi S, Samoza J, Ridder A, 2015. Age- and sex-dependent changes in androgen receptor expression in the developing mouse cortex and hippocampus. Neurosci J. 2015, 525369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HW, Grant PA, Rissman EF, 2009. Sex differences in histone modifications in the neonatal mouse brain. Epigenetics. 4, 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vom Saal FS, Bronson FH, 1980. Sexual characteristics of adult female mice are correlated with their blood testosterone levels during prenatal development. Science. 208, 597. [DOI] [PubMed] [Google Scholar]
- vom Saal FS, 1979. Prenatal exposure to androgen influences morphology and aggressive behavior of male and female mice. Horm Behav. 12, 1–11. [DOI] [PubMed] [Google Scholar]
- Wolf CJ, Hotchkiss A, Ostby JS, LeBlanc GA, Gray LE Jr, 2002. Effects of prenatal testosterone propionate on the sexual development of male and female rats: A dose-response study. Toxicol Sci. 65, 71–86. [DOI] [PubMed] [Google Scholar]
- Wurmbach E, Yuen T, Sealfon SC, 2003. Focused microarray analysis. Methods. 31, 306–316. [DOI] [PubMed] [Google Scholar]
- Yucel S, Cavalcanti AG, Wang Z, Baskin LS, 2003. The impact of prenatal androgens on vaginal and urogenital sinus development in the female mouse. J Urol. 170, 1432–1436. [DOI] [PubMed] [Google Scholar]
- Zhang JM, Konkle AT, Zup SL, McCarthy MM, 2008. Impact of sex and hormones on new cells in the developing rat hippocampus: A novel source of sex dimorphism? Eur J Neurosci. 27, 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]