Abstract
Brain-derived neurotrophic factor (BDNF) signaling regulates synaptic plasticity in the hippocampus (HC) and prefrontal cortex (PFC), and has been extensively linked with fear memory expression in rodents. Notably, disrupting BDNF production from promoter IV-derived transcripts enhances fear expression in mice, and decreases fear-associated HC-PFC synchrony, suggesting that Bdnf transcription from promoter IV plays a key role in HC-PFC function during fear memory retrieval. To better understand how promoter IV-derived BDNF controls HC-PFC connectivity and fear expression, we generated a viral construct that selectively targets cells expressing promoter IV-derived Bdnf transcripts (“p4-cells”) for tamoxifen-inducible Cre-mediated recombination (AAV8-p4Bdnf-ERT2CreERT2-PEST). Using this construct, we found that ventral hippocampal (vHC) p4-cells are recruited during fear expression, and that activation of these cells causes exaggerated fear expression that co-occurs with disrupted vHC-PFC synchrony in mice. Our data highlight how this novel construct can be used to interrogate genetically defined cell types that selectively contribute to BDNF-dependent behaviors.
Subject terms: Neuroscience, Molecular biology
Introduction
In humans, a core cognitive symptom of several neuropsychiatric disorders, including post-traumatic stress disorder (PTSD) and generalized anxiety disorder, is dysregulation of learned fear [1, 2]. In rodents, freezing (cessation of movement) to a fear-conditioned cue or context (e.g., a tone or environment that has been paired with a foot-shock) is routinely used as a proxy for fear memory. Fear expression has been attributed to a network of brain areas in rodents, including the hippocampus [3, 4], medial prefrontal cortex (mPFC) [5], and amygdala [6, 7], among others. The rodent hippocampus routes information through uni-directional monosynaptic projections in ventral CA1 to both the prelimbic (PrL) and infralimbic (IL) subregions of the mPFC [8, 9]. A dissociation between PrL and IL function exists, such that the PrL contributes to expression of learned fear, while the IL contributes to extinction of learned fear [10]. The HC and mPFC functionally interact during fear expression and extinction [11, 12], suggesting a critical role for communication between these regions during fear memory expression. vHC-mPFC synapses undergo plasticity in response to paired-pulse stimulation [13], suggesting that structural changes in this circuit underlie its ability to support fear memory. Brain-derived neurotrophic factor (BDNF) is a signaling molecule that critically regulates synaptic plasticity in the developing and adult brain [14, 15], and a host of previous studies have demonstrated a relationship between BDNF gene expression and fear memory in rodents [16–19]. Release of BDNF protein in the HC-IL circuit facilitates fear extinction (decreases freezing [20]), suggesting that BDNF plays a role in regulation of fear behavior by controlling plasticity in the hippocampal-prefrontal network, which influences functional connectivity within this circuit [21]. BDNF gene expression is controlled by different promoters, which generate unique transcripts that encode the same BDNF protein [22–25]. Tight regulation at Bdnf promoters allows for restricted temporal, spatial, and activity-dependent expression, which is critical for modulating synaptic plasticity and circuit-specific behavior. Distinct expression patterns suggest that transcripts are localized to discrete circuits in defined brain regions. Supporting this notion, disruption of BDNF production from promoter-specific transcripts affects BDNF protein expression in a brain region-specific manner [26]. Since they produce the same protein, individual transcripts may serve similar molecular roles but function in cell populations that differentially contribute to distinct behavioral phenotypes [27–30]. Promoter-specific transcription of BDNF is regulated by a number of factors; for example, transcription from promoter IV (p4) is highly activity-dependent, as exon IV-containing Bdnf transcripts are upregulated following membrane depolarization [31] and kainic acid-induced seizures [32]. Recent evidence suggests that BDNF production from individual promoters differentially impacts fear memory; specifically, BDNF disruption from promoter IV (p4)-derived transcripts impairs fear expression in mice [33], and decreases fear-related hippocampal-prefrontal oscillatory synchrony [12]. These results suggest that transcription of Bdnf from p4 critically regulates hippocampal-prefrontal plasticity during fear memory recall and expression. Gaining genetic access to neurons with transcript-specific expression of Bdnf could therefore provide an avenue for interrogating and manipulating spatially defined populations of cells that selectively regulate fear behavior. To selectively tag “p4-cells” for labeling and manipulation, we developed an adeno-associated adenovirus (AAV) expressing an ERT2CreERT2 fusion protein under control of Bdnf promoter IV. Utilizing this construct with 4-hydroxy-tamoxifen (4OHT) causes expression of Cre recombinase only in p4-cells. We leveraged this construct to selectively label and manipulate p4-cells in the vHC of behaving mice during recall of both a fear-associated context and a fear-associated tone/context combination, allowing us to causally probe relationships between activity of these cells, fear-related behavior, and fear-related vHC-mPFC circuit dynamics. We found that vHC p4-cells are recruited during learned fear expression, and that synthetic activation of these cells increases fear expression while attenuating fear-related oscillatory synchrony within the vHC, as well as between the vHC and mPFC.
Materials and methods
Animals
Male wild-type (w/t) C57Bl6/J mice were group-housed (3–5 animals per cage) and maintained on a 12 h light/dark cycle in a temperature and humidity-controlled colony room. All animals had ad libitum access to food and water. Animals were 10–16 weeks of age, and weighed 25–35 g at time of surgery. All procedures were in accordance with the Institutional Animal Care and Use Committee of SoBran Biosciences Inc.
Surgeries and 4OHT injections
Mice were anesthetized with isoflurane (1–2.5% in oxygen), placed into a stereotaxic frame, and an incision was made along the midline of the scalp. The skull was leveled, bregma was identified, and small holes were drilled with a 0.9 mm burr (Fine Science Tools) above the vHC (−3.2 mm AP, ±3.1 mm ML) for viral injections. For immunohistochemistry and in situ hybridization experiments, 5 µl of AAV8-p4BDNF-ERT2CreERT2 (3.4 × 10^11 gc/ml) was mixed with 5 µl of AAV8-CAG-FLEX-tdTomato (2.9 × 10^12 vg/ml; Addgene) and 5 µl of AAV1-hSyn-eGFP-WPRE-bGH (3.7 × 10^11 gc/ml; Addgene). For behavior/electrophysiology experiments, 5 µl of AAV8-p4BDNF-ERT2CreERT2 was mixed with 5 µl of either AAV8-hSyn-DIO-mCherry (2.6 × 10^13 gc/ml; control group) or AAV8-hSyn-DIO-hM3Dq-mCherry (2.2 × 10^13 gc/ml; experimental group). A total volume of 600 nl/hemisphere was injected into the vHC (−3.4 mm DV) at a rate of 200 nl/min via a 10 µl syringe (Hamilton) controlled by an automated infusion pump (World Precision Instruments). For LFP recordings, two stereotrodes (35 µm stainless steel, California Fine Wire) were also implanted into the vHC and mPFC (+2.0 AP, ±0.3 ML, −1.8 DV). Stereotrodes were attached to a headmount, along with two wires soldered to two bone screws (Fine Science Tools). Ground screws were implanted above the frontal cortex on the hemisphere opposite the mPFC electrode, and directly above the lambda skull suture. Mice received a sub-cutaneous injection of a local anesthetic (Bupivicaine) along the incision site, as well as an i.p. injection of an analgesic (Meloxicam, 5 mg/kg) during surgery. Meloxicam injections were given for three days post-surgery for pain relief. Two weeks post-surgery, mice were given an i.p. injection of 4OHT (20 mg/kg) for three consecutive days directly prior to the onset of the dark cycle to induce recombination of Cre in promoter IV-expressing vHC neurons. 4OHT was prepared by first dissolving 10 mg powder (Sigma, catalog # H6278) in 250 µl of DMSO, and then mixing the 4OHT solution with 400 µl of 25% Tween-80 in 4.35 mL of filtered 1× PBS, for a final 4OHT concentration of 2 mg/ml [34]. Behavioral testing for immunohistochemistry/in situ hybridization was completed 2 weeks post-4OHT injection.
Immunohistochemistry/cresyl violet staining
For c-Fos immunohistochemistry, mice were trans-cardially perfused with 4% paraformaldehyde 2 h following the termination of tone/context recall. Brains were extracted and stored in 4% paraformaldehyde for 24 h, and cryoprotected in 30% sucrose in 1× PBS/sodium azide (0.05%) for 2-3 d. Coronal sections (50 μm) of the vHC were cut on a sliding microtome (Leica) with attached freezing stage (Physitemp), washed in 5% Tween-80 in 1× PBS, and incubated in blocking solution (0.5% Tween-80, 5% normal goat serum in 1× PBS) with agitation for 6–8 h. The sections were then incubated in 1:1000 anti-Fos antibody (Millipore; cat # ABE457) in blocking solution overnight at 4 °C with agitation. The following day, the sections were washed, incubated in 1:1000 goat anti-rabbit AlexaFluor 647 (Sigma) in blocking solution for 2 h with agitation, washed again, and incubated in 1:5000 DAPI (Sigma) in 1× PBS for 20 min. The sections were then mounted and coverslipped, and co-expression of either tdTomato/c-Fos (for analysis of recruitment of p4-cells during fear behavior) or mCherry/c-Fos (for verification of excitatory DREADD efficacy) was visualized at ×20 magnification on a Zeiss 700 LSM confocal microscope. c-Fos, tdTomato, and mCherry-expressing cells were quantified from maximum intensity projections created from z-stacked, tiled images. For visualization of electrode tracks in the vHC and PrL, we stained 50 μm coronal sections with 0.1% cresyl violet, and imaged on an Olympic BX51 upright light microscope.
Single-molecule in situ hybridization
For verification that exon IV-containing Bdnf and Cre were co-localized with tdTomato, we followed previously published procedures [35]. Briefly, animals were killed and brains immediately extracted and flash-frozen in 2-methylbutane (ThermoFisher). Coronal sections of vHC (16 µm) were cut, and mounted onto slides (VWR, SuperFrost Plus). The slides were quickly fixed in 10% buffered formalin at 4 °C, washed in 1× PBS, and dehydrated in ethanol. Slides were next pre-treated with a protease solution, and subsequently incubated at 40 °C for 2 h in a HybEZ oven (ACDBio) with a combination of probes for Cre, tdTomato, and exon IV-containing Bdnf mRNA. Following incubation with the probes, the slides were incubated at 40 °C with a series of fluorescent amplification buffers. DAPI was applied to the slides, and the slides were cover-slipped with Fluoro-Gold (SouthernBiotech). Transcript expression was visualized on a Zeiss LSM 700 confocal microscope with a ×40 oil-immersion lens (Cre: 555 nm, tdTomato: 488 nm, exon IV-containing Bdnf: 647 nm, DAPI: 405 nm). For quantification of transcript co-localization, ×40 z-stacks from the ventral dentate gyrus were taken, and custom MATLAB functions were used to first isolate cell nuclei from the DAPI channel using the cellsegm toolbox [36] combined with a watershed algorithm for cell splitting in 3 dimensions. Once centers and boundaries of individual cells were isolated, an intensity threshold was set for transcript detection, and watershed analysis was used to split the remaining pixels in each channel into identified transcripts. Custom MATLAB functions were then used to determine the size of each detected transcript (regionprops3 function in Image Processing toolbox), and split unusually large areas of fluorescence into multiple transcripts based on known transcript size. Each transcript was then assigned to a cell based on its position in 3 dimensions; transcripts that were detected outside of the boundaries of a cell were excluded from further analysis.
Behavior
For fear conditioning, animals were conditioned to associate a 30 s tone (4000 Hz) with a 2 s, 0.6 mA foot-shock. 4 tone-shock combinations were given, with the shock co-terminating with the last 2 s of each tone presentation. Tone-shock combinations were separated by 90 s intervals. 48 h later, mice were placed back into the conditioning chamber to measure recall of the conditioning context (in the absence of the tone) for 180 s. Following pre-tone context recall, a total of 23 tones (30 s each) were presented in the absence of the foot-shock for recall of the tone in the fear-associated context (tone/context recall). Tones were separated by 5 s intervals. Freezing (cessation of movement) during acquisition and recall sessions was quantified with automated tracking software (FreezeScan; CleverSys, Inc.). For mice with either hM3Dq or mCherry expression in the vHC, clozapine-N-oxide (CNO, Tocris; 5 mg/kg, i.p.) dissolved in 1× PBS was administered 45 min prior to context recall. For mice used to measure c-Fos expression in p4-cells following fear recall, half of the mice were given foot-shocks during acquisition, and half of the mice were presented with the tone in the absence of a foot-shock. In order to assess possible effects of hM3Dq activation on locomotor activity, home-cage activity (movement, rearing) was measured in hM3Dq and mCherry-expressing mice (HomeCageScan software; CleverSys, Inc.) both 1 h prior to and 1 h after CNO injections. We further investigated whether hM3Dq activation caused anxiogenic or anxiolytic effects by administering CNO and measuring time spent in the center and number of center crossings in an open field for 1 h. Position data for open field testing were captured with CaptureStar software (CleverSys, Inc.), and occupancy maps, time spent in the center, and number of center crossings were analyzed with custom MATLAB functions. Fear conditioning, home-cage activity, and open field testing were conducted 7 d apart.
Electrophysiology and LFP analysis
Local field potentials (LFPs) from the PL and the vHC were recorded during context and tone/context recall (Sirenia software; Pinnacle). LFPs were sampled at 2 kHz and bandpass filtered between 0 and 50 Hz. Raw traces were visually inspected for noise artifacts, and de-trended with custom MATLAB functions. Multitaper spectral analysis was used for power density estimation and phase coherence via Chronux toolbox routines in MATLAB (http://chronux.org [37]). For phase-amplitude coupling analysis, phase and amplitude values for frequency pairs were extracted via Morlet wavelet convolution. Phase and amplitude values were then binned (num. bins = 18 per cycle), and mean amplitude values were normalized by dividing each bin value by the summed value over all bins. A modulation index value was then derived by calculating the Kullback-Leibler distance between the observed phase-amplitude distribution and a uniform (null) phase-amplitude distribution [38]:
where P is equal to the observed distribution, and Q is equal to a uniform distribution. For Granger causality, custom MATLAB functions were used to calculate a univariate autoregression of the mPFC LFP at times t:
which was augmented with lagged values of the vHC LFP:
with number of lags being determined by the model order, which was set according to the Bayes’ information criterion (BIC) for each data set. A Granger causal index was then calculated by dividing the index of vHC → mPFC directionality by the total of the indices for both vHC → mPFC and mPFC → vHC directionality. Thus, if the vHC led more strongly, a Granger causal index > 0.5 would be the result, while stronger mPFC directionality would yield a Granger causal index < 0.5.
Statistical tests
Individual statistical tests and results of those tests are noted in figure legends throughout the manuscript (statistics performed with GraphPad Prism software). An alpha level of 0.05 was used to determine statistical significance for all tests. For binary classification of tdTomato expression based on number of Bdnf transcripts, we used logistic regression to predict whether or not tdTomato was expressed (cutoff of 5 transcripts) in each cell by training the classifier on transcript data from all cells in each image with the exception of five cells, which were used as a testing set. This procedure was repeated until all cells had been used as test data, and classifier accuracy was calculated by comparing the outcome predicted by the classifier with the training label for each cell. Receiver operating characteristic (ROC) curves were used to validate classifier accuracy, and area under the ROC curve was calculated with custom MATLAB functions. For comparison data, training labels and number of Bdnf transcripts were randomly shuffled relative to each other and prediction accuracy and ROC curves were calculated with the shuffled data as described above.
Results
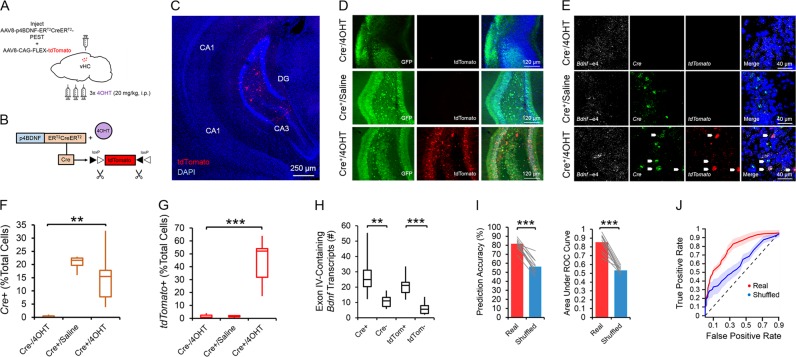
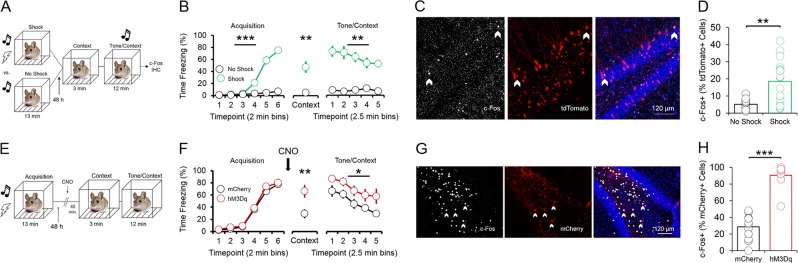
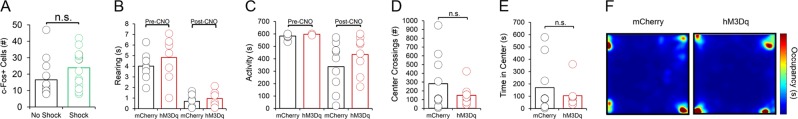
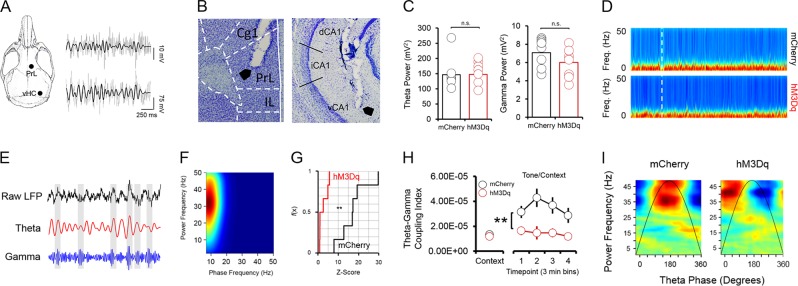
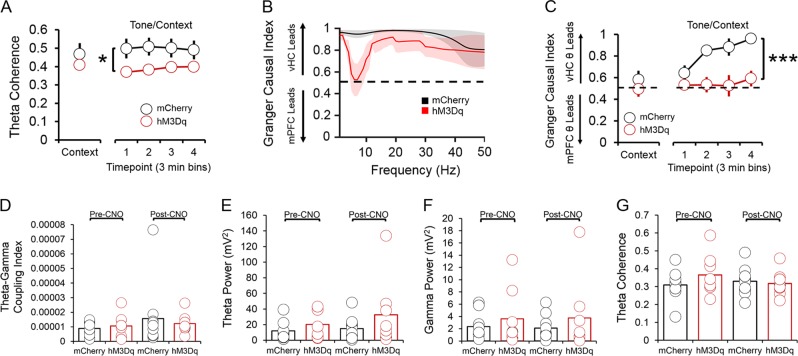
To develop a method for selectively tagging cells expressing BDNF from promoter IV (“p4-cells”) for labeling and manipulation, we first cloned a tamoxifen-inducible Cre-recombinase expression cassette (ERT2CreERT2-PEST) downstream of the proximal mouse Bdnf promoter IV sequence (initial 600 bases upstream of the exon IV transcription start site), which was then inserted into a pAAV1 backbone and packaged as high titer adeno-associated virus (AAV8-p4BDNF-ERT2CreERT2-PEST; Cyagen Biosciences). To validate that the p4BDNF-ERT2Cre-ERT2 construct undergoes recombination selectively in the presence of 4OHT, we performed intracranial injections of AAV8-p4BDNF-ERT2CreERT2-PEST with a Cre-dependent AAV8-FLEX-tdTomato into the vHC of adult mice (Fig. 1a). Previous research has shown that p4-derived Bdnf is upregulated in mice during their active (dark) cycle [39]. Following a two week incubation period, we therefore administered 4OHT (20 mg/kg, i.p.; 3×/d) immediately prior to the animals’ dark cycle for Cre/lox-mediated expression of tdTomato protein. We injected 4OHT for three consecutive days in order to maximize tagging of p4-expressing neurons with the p4BDNF-ERT2CreERT2 construct. Two weeks later, animals were killed, and tdTomato expression was compared between mice that received vehicle (saline) injections and mice that received 4OHT. Robust tdTomato expression was observed in the ventral dentate gyrus of 4OHT-injected (Cre+/4OHT+) mice (Fig. 1c), while virtually no tdTomato expression was seen in vHC sections of mice that received saline injections (Cre+/4OHT-; Fig. 1d). We found virtually no tdTomato expression in mice injected with AAV8-FLEX-tdTomato and 4OHT in the absence of p4BDNF-ERT2CreERT2 (Cre-/4OHT+; Fig. 1d), demonstrating that Cre-mediated recombination requires the presence of 4OHT [40]. To further investigate whether Cre and tdTomato transcripts were restricted to cells containing promoter IV-derived Bdnf transcripts in Cre+/4OHT+ mice, we used single-molecule fluorescence in situ hybridization (RNAscope) to visualize co-expression of virally-induced Cre, tdTomato, and exon IV-containing Bdnf transcripts within individual ventral dentate neurons in these animals (Fig. 1e). We found that Cre mRNA was expressed only in animals that received infusions of p4BDNF-ERT2CreERT2 (F(2,15) = 7.72, p = 0.005, one-way ANOVA; Fig. 1f), while tdTomato mRNA expression was limited to mice that received infusions of p4BDNF-ERT2CreERT2 and AAV8-FLEX-tdTomato with 4OHT injections (F(2,15) = 25.35, p < 0.0001, one-way ANOVA; Fig. 1g), further demonstrating that binding of 4OHT to the ERT2 fusion protein allows Cre to functionally excise the FLEX cassette for tdTomato transcription and translation. In mice that received both p4BDNF-ERT2CreERT2 and 4OHT, the number of exon IV-containing Bdnf transcripts was significantly higher in Cre+ as compared to Cre- cells (t(9) = 4.44, p = 0.0016), as well as tdTomato+ as compared to tdTomato- cells (t(9) = 6.66, p < 0.0001, paired t-tests; Fig. 1h). We further trained a binary classifier using logistic regression to determine whether number of exon IV-containing Bdnf transcripts could predict tdTomato co-expression, and found that the classifier performed with high accuracy compared with shuffled data (t(9) = 9.82, p < 0.0001 for prediction accuracy; t(9) = 9.67, p < 0.0001 for area under ROC curve; paired t-tests; Fig. 1I, j), demonstrating that the p4BDNF-ERT2CreERT2 construct targets p4-expressing cells with high precision. To investigate whether p4-cells are recruited during fear expression, we again labeled ventral dentate gyrus p4-cells with tdTomato, and compared levels of the immediate early gene (IEG) product c-Fos in tdTomato-labeled p4-cells in mice that were conditioned to freeze to a tone in the presence of a foot-shock (shock group), compared to mice that were presented with the tone in the absence of foot-shocks during the conditioning session (no shock group; Fig. 2a). As expected, mice in the shock group froze at significantly higher levels than mice in the no shock group during conditioning (F(1,5) = 73.58, p < 0.0001, group × time interaction), context recall (t(10) = 4.37, p = 0.0014, unpaired t-test), and tone/context recall (F(1,5) = 5.4, p = 0.0014, group × time interaction; Fig. 2b). Mice were killed 2 h after tone/context recall to measure c-Fos levels during fear expression. A higher percentage of p4-cells co-expressed c-Fos in the shock group compared to the no shock group (t(14) = 4.1, p = 0.0003, unpaired t-test; Fig. 2d), while total number of c-Fos + cells did not significantly differ between conditions (p > 0.05 for unpaired t-test; Fig. 3a), indicating that ventral dentate p4-cells are selectively recruited during learned fear expression. We next asked whether synthetic excitation of p4-cells is sufficient to potentiate fear expression during recall. To answer this question, we expressed the excitatory DREADD receptor hM3Dq in p4-cells by co-infusing AAV8-p4BDNF-ERT2CreERT2-PEST with AAV8-hSyn-DIO-hM3Dq-mCherry in the vHC (“hM3Dq” group). 45 min before context recall, we administered clozapine-N-oxide (CNO, 5 mg/kg, i.p.) to engage the hM3Dq receptor in p4-cells (Fig. 2e). Activation of vHC p4-cells caused increased freezing during both context (t(18) = 2.96, p = 0.01, unpaired t-test) and tone/context recall (F(1,18) = 6.08, p = 0.02, main effect of group), as compared to CNO-injected animals that expressed a control reporter (AAV8-hSyn-DIO-mCherry, “mCherry” group; Fig. 2f). In order to ensure that CNO injections reliably activated promoter IV-expressing vHC neurons, we measured co-expression of c-Fos and mCherry in hM3Dq and mCherry mice after CNO injections, and found that the percentage of mCherry cells co-expressing c-Fos was significantly higher in hM3Dq mice (t(24) = 11.75, p < 0.0001, unpaired t-test; Fig. 2h). Effects of vHC p4-cell activation on fear expression were not due to general deficits in movement or anxiety, as neither locomotor (Fig. 3b, c) nor anxiety-like behavior (Fig. 3d-f) differed between groups post-CNO injection. These results demonstrate that vHC p4-cells represent a sub-population capable of enhancing contextually-mediated fear memory recall. Systemic reductions in p4-derived BDNF drives increased fear expression, which co-occurs with decreased hippocampal-prefrontal synchrony [12], suggesting that p4-cells modulate vHC-mPFC circuit function to impact fear behavior. To test this hypothesis, we recorded simultaneous local field potentials (LFPs) from the vHC and PrL, a sub-region of the PFC that is highly implicated in fear expression, during context and tone/context recall in hM3Dq and mCherry mice after CNO injections. Activation of vHC p4-cells did not significantly affect broadband power in the vHC (Fig. 4c, d). We further found that slow gamma oscillations (30–50 Hz) preferentially coupled to theta oscillations (4–12 Hz) within the vHC (Fig. 4e, f) at higher-than-chance levels (p < 0.01, Kolmogorov-Smirnov test; Fig. 4g) during tone/context recall. Theta-gamma coupling in the vHC was significantly lower in hM3Dq mice following CNO injections (F(1,17) = 10.71, p = 0.01, main effect of group; Fig. 4h). Furthermore, we found that slow gamma also shifted its preferred theta coupling phase from the peak (~180 degrees) to the trough (~30 degrees) in hM3Dq + mice (Fig. 3i), possibly reflecting a disruption of CA3 input to CA1 following stimulation of p4-cells [41]. Decreased theta-gamma coupling in the vHC was concomitant with decreased theta phase coherence between the vHC and PrL (F(1,17) = 8.34, p = 0.01, main effect of group; Fig. 5a), suggesting that activation of vHC p4-cells disrupts vHC-PrL synchrony to promote fear expression. Disruptions in vHC-PrL theta synchrony were directionally-specific, as the vHC strongly led the PrL in all frequency bands measured in mCherry mice; this directionality was selectively disrupted in the theta frequency band in hM3Dq mice (Fig. 5b) during tone/context recall (F(1,17) = 28.57, p < 0.0001, main effect of group; Fig. 5c). CNO injections did not significantly affect theta-gamma coupling (Fig. 5d), or theta phase coherence (Fig. 5g) in the home cage.
Fig. 1.
The p4BDNF-ERT2CreERT2 construct selectively targets cells expressing exon IV-containing Bdnf transcripts in the vHC (p4-cells). a Schematic of injection strategy for labeling of p4-cells in the vHC. b 4OHT binds to the ERT2CreERT2 fusion protein to allow Cre/lox-mediated expression of tdTomato protein. c tdTomato protein is largely limited to the ventral dentate gyrus in animals that received injections of both p4BDNF-ERT2CreERT2 and 4OHT. d tdTomato protein expression is observed only in animals that received injections of p4BDNF-ERT2CreERT2 and 4OHT (GFP labeling from co-injections of AAV1-hSyn-GFP were used to verify injection accuracy). e Single-molecule in situ hybridization reveals expression of Cre mRNA in animals that received injections of p4BDNF-ERT2CreERT2, and tdTomato expression is restricted to animals that received injections of both p4BDNF-ERT2CreERT2 and 4OHT. f A significantly higher percentage of cells co-express Cre in animals that received injections of p4BDNF-ERT2CreERT2, in comparison to animals that received injections of 4OHT without p4BDNF-ERT2CreERT2. g A significantly higher percentage of cells co-express tdTomato mRNA in animals that received both p4BDNF-ERT2CreERT2 and 4OHT, as compared to both control groups. h Cells that express either Cre or tdTomato co-express a higher number of exon IV-containing Bdnf transcripts, as compared to cells that do not express Cre or tdTomato. i A logistic regression model trained to predict whether a cell expresses tdTomato based on number of exon IV-containing Bdnf transcripts performs with high accuracy for observed data, but not data in which number of Bdnf transcripts is randomly shuffled relative to tdTomato expression probability (left panel). Receiver operating characteristic (ROC) curves on classifier performance confirm model accuracy in real and shuffled data (right panel). j Regression classifier ROC curves for real and shuffled data (dashed line = chance)
Fig. 2.
p4-cells in the vHC enhance fear expression during context and tone/context recall. a Schematic of experimental design for assessing recruitment of vHC p4-cells during fear expression. All animals received injections of p4BDNF-ERT2CreERT2, FLEX-tdTomato, and 4OHT prior to fear conditioning. Half of the animals received a tone-shock combination during conditioning, and the other half received only the tone. b Animals in the shock group froze significantly more frequently than animals in the no shock group during conditioning, context recall, and tone/context recall. (c) c-Fos immunohistochemistry was used to identify p4-cells in the vHC that were recruited during recall. d A higher proportion of p4-cells (tdTomato+) in the vHC co-express fear-induced c-Fos in mice that received a shock during conditioning. e Schematic of experimental design for assessing whether vHC p4-cells enhance fear expression during recall. All animals received injections of p4BDNF-ERT2CreERT2 and 4OHT, and half of the animals additionally received injections of an AAV encoding for a Cre-dependent excitatory DREADD (hM3Dq) fused to a fluorescent reporter (mCherry), while the other half received injections of an AAV coding for the Cre-dependent expression of only mCherry. f Synthetic activation of vHC p4-cells prior to context recall significantly increases freezing during both context recall and tone/context recall. g c-Fos immunohistochemistry was used to verify that CNO injections activate hM3Dq-expressing vHC cells. h A significantly higher proportion of mCherry+cells in the vHC co-express c-Fos following CNO injections in mice that express hM3Dq-mCherry, as compared to mice that express only mCherry
Fig. 3.
Effects of vHC p4-cell activation on fear recall are fear-specific. (a) No significant difference in number of c-Fos+cells in the vHC following fear recall exists between mice that received a shock during conditioning, and mice that did not receive a shock during conditioning. (b) No significant differences in home cage total movement exist between groups either before or after CNO injections. (c) No significant differences in rearing in the home cage are observed between groups either before or after CNO injections. (d) Activation of vHC p4-cells does not affect number of center crossings, or (e) time spent in the center of an open field. (f) Open field occupancy does not differ between groups following CNO injections
Fig. 4.
Fear-related coupling in the vHC is disrupted following activation of vHC p4-cells. a Schematic of electrode placements and sample LFP recordings from the PrL and vHC (gray), with filtered theta superimposed on top (black). b Histology showing placement of an electrode tip in the vHC (left panel) and PrL (right panel). c Neither theta (4–12 Hz) power (left panel), nor slow gamma (30–50 Hz) power (right panel) significantly differ between hM3Dq and mCherry mice during context and tone/context recall. d Spectrograms from the vHC in hM3Dq and mCherry mice during fear recall. White line = start of tone/context recall. e Within the vHC, bouts of high-amplitude slow gamma oscillations tended to occur around the peaks of the theta oscillation during fear recall. f This phase-amplitude coupling occurred most strongly between hippocampal theta (phase on the x-axis) and slow gamma (amplitude on the y-axis) oscillations (heat = coupling index). g In order to determine whether observed theta-gamma coupling in the vHC during tone/context recall was greater than what would be expected by chance, phase-amplitude distributions were shuffled 500 times and a theta-gamma coupling index value was obtained for each shuffled distribution. The observed theta-gamma coupling value was compared to the shuffled distribution of coupling values to obtain a z-score. Cumulative frequency distributions of these z-scores were significantly different between hM3Dq and mCherry animals. h Theta-gamma coupling was significantly lower during tone/context recall in hM3Dq mice. (i) In addition to a decrease in theta-gamma coupling following CNO injections in hM3Dq mice, slow gamma oscillations shifted their preferred theta coupling phase from the peak to the trough in hM3Dq animals
Fig. 5.
vHC-PrL synchrony during fear recall and in the home cage. a Phase coherence between theta oscillations in the vHC and PrL was significantly lower in hM3Dq mice during tone/context recall. b The vHC LFP led the PrL LFP in all frequency bands during tone/context recall in mCherry mice, while CNO injections severely disrupted this effect selectively in the theta frequency band in hM3Dq mice (black dashed line = chance). c vHC theta strongly led PrL theta during tone/context recall in mCherry mice, and this effect was abolished in hM3Dq mice. d vHC theta-gamma coupling, e theta power, f slow gamma power, and g theta phase coherence between the vHC and PrL did not significantly differ between hM3Dq and mCherry animals in the home cage, either before or after CNO injections
Discussion
Our results describe a novel method for targeting genetically defined cell populations based on transcript-specific BDNF expression, expanding on previous research using the CreERT2 system to label and manipulate IEG-expressing neurons during behavior [34, 42]. Here, we demonstrate that p4-cells cells in the ventral dentate gyrus represent a sub-population that is capable of enhancing learned fear expression. Increased fear expression following activation of these cells co-occurred with decreased vHC-PrL synchrony, suggesting that these neurons regulate fear expression by inhibiting vHC-PrL communication, which has previously been shown to affect expression of learned fear [43]. Previous studies have shown that broad inhibition of ventral dentate granule cells does not affect contextually mediated fear expression [44], suggesting that p4-cells are a spatially, genetically, and functionally-distinct subgroup within the vHC. In the current study, we used the p4Bdnf-ERT2CreERT2 construct to broadly tag p4-cells over a 3 day period in order to assess the relationship between function of these cells and fear expression. Future studies could use this construct to label and manipulate behaviorally relevant p4-cells (i.e., cells expressing p4-derived Bdnf during conditioning vs. recall) by varying the timing of 4OHT injections relative to behavior. Results from these studies could address hypotheses about whether the same p4-cells participate in both conditioning and recall, adding to previous literature demonstrating overlap between conditioning and recall-activated ensembles in the dentate gyrus [37] and PFC [45]. The current results lay the groundwork for these studies by causally linking activity in a spatially-localized network of cells that constitutively express p4-derived Bdnf and fear recall, and provide a tool for further investigation of how these cells might contribute to distinct phases of fear behavior. Promoter-specific Bdnf transcripts are localized to distinct sub-cellular compartments in neurons [26–29], and global disruption of BDNF translation from distinct promoters results in brain region-specific patterns of BDNF expression [26]. Translation of BDNF from distinct promoters results in divergent phenotypes in mice; specifically, disruption of BDNF production from promoter I transcripts results in hyperphagia and increased aggression [26], while disruption of BDNF production from promoter IV transcripts results in increased fear expression [12, 33]. Fear expression has additionally been linked with upregulation of exon IV-containing Bdnf transcripts [17, 46], and epigenetic modulation at promoter IV of the Bdnf gene [16], pointing to a selective role for promoter IV-dependent transcription in fear regulation. Our data extend these results by showing that promoter IV-derived Bdnf affects fear behavior by virtue of spatially-selective expression in cells that are important for fear expression, supporting the theory that Bdnf splice variants differentially impact behavior by modulating function in disparate brain regions and cell types [26, 47]. All p4-cells in our data set were located in the ventral dentate gyrus, presumably because this is where endogenous expression of p4-transcripts was strong enough to drive the reporter in the p4Bdnf-ERT2CreERT2 construct. The dentate gyrus sends mossy fibers that synapse in CA3; Schaeffer collaterals from CA3 then synapse on CA1 cells, which directly project to the mPFC [8, 9]. Disruptions in vHC phase-amplitude modulation following p4-cell activation suggest that activation of these cells inhibits vHC-mPFC synchrony by de-coupling CA3-CA1 connectivity [41], which then leads to decreased hippocampal-prefrontal function, as also seen during increased fear expression in mutant mice with disrupted BDNF production from promoter IV-transcripts [12]. Taken together, our data provide insights into how sub-populations of cells defined by isoform-specific expression of Bdnf can regulate complex behavior and circuit function, and contribute to a growing understanding of how BDNF signaling impacts hippocampal-prefrontal function in fear-related behavior. Our p4Bdnf-ERT2CreERT2-PEST construct can be used in conjunction with a wide variety of Cre-dependent constructs to understand how Bdnf splice isoforms impact behavior; for example, by using a Cre-dependent inhibitory DREADD receptor to inhibit these cells, or inhibition of BDNF’s cognate receptor TrkB in conjunction with excitation of p4-cells. Our p4Bdnf-ERT2CreERT2-PEST construct provides a tool for causally relating cell-type specific expression of splice variants to behavior, which will be important for understanding how isoform-specific Bdnf expression impacts a variety of diverse behaviors in rodents [48], and humans [49].
Funding and disclosure
Funding for these studies was provided by the Lieber Institute for Brain Development and a National Institute of Mental Health R01 to KM (MH105592). The authors declare no competing interests.
Acknowledgements
We thank Richard de los Santos Abreu, John Hobbs, Madhavi Tippani, and Danisha Gallop for technical assistance. We thank Pierre Chambon (IGBMC) for use of the CreERT2 construct.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Orr SP, Roth WT. Psychophysiological assessment: clinical applications for PTSD. J Affect Disord. 2000;61:225–40. doi: 10.1016/s0165-0327(00)00340-2. [DOI] [PubMed] [Google Scholar]
- 2.Lommen MJJ, Engelhard IM, Sijbrandij M, van den Hout MA, Hermans D. Pre-trauma individual differences in extinction learning predict posttraumatic stress. Behav Res Ther. 2013;51:63–67. doi: 10.1016/j.brat.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Kjelstrup KG, Tuvnes FA, Steffenach HA, Murison R, Moser EI, Moser M-B. Reduced fear expression after lesions of the ventral hippocampus. Proc Natl Acad Sci USA. 2002;99:10825–30. doi: 10.1073/pnas.152112399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trivedi MA, Coover GD. Lesions of the ventral hippocampus, but not the dorsal hippocampus, impair conditioned fear expression and inhibitory avoidance on the elevated T-maze. Neurobiol Learn Mem. 2004;81:172–84. doi: 10.1016/j.nlm.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Morgan MA, LeDoux JE. Differential contribution of dorsal and ventral medial prefrontal cortex to the acquisition and extinction of conditioned fear in rats. Behav Neurosci. 1995;109:681–8. doi: 10.1037//0735-7044.109.4.681. [DOI] [PubMed] [Google Scholar]
- 6.Anglada-Figueroa D, Quirk GJ. Lesions of the basolateral amygdala block expression of conditioned fear but not extinction. J Neurosci. 2005;25:9680–5. doi: 10.1523/JNEUROSCI.2600-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciocchi S, Herry C, Grenier F, Wolff SBE, Letzkus J, Vlachos I, et al. Encoding of conditioned fear in central amygdala inhibitory circuits. Nature. 2010;468:277–82. doi: 10.1038/nature09559. [DOI] [PubMed] [Google Scholar]
- 8.Swanson LW. A direct projection from Ammon’s horn to prefrontal cortex in the rat. Brain Res. 1981;217:150–4. doi: 10.1016/0006-8993(81)90192-x. [DOI] [PubMed] [Google Scholar]
- 9.Jay TM, Witter MP. Distribution of hippocampal CA1 and subicular efferents in the prefrontal cortex of the rat studied by means of anterograde transport of Phaseolus vulgaris-leucoagglutinin. J Comp Neurol. 1991;313:574–86. doi: 10.1002/cne.903130404. [DOI] [PubMed] [Google Scholar]
- 10.Sierra-Mercado D, Padilla-Coreano N, Quirk GJ. Dissociable roles of prelimbic and infralimbic cortices, ventral hippocampus, and basolateral amygdala in the expression and extinction of conditioned fear. Neuropsychopharmacology. 2011;36:529–38. doi: 10.1038/npp.2010.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lesting J, Narayanan RT, Kluge C, Sangha S, Seidenbecher T, Pape H-C. Patterns of coupled theta activity in amygdala-hippocampal-prefrontal cortical circuits during fear extinction. PLoS ONE. 2011 doi: 10.1371/journal.pone.0021714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill JL, Hardy NF, Jimenez DV, Maynard KR, Kardian AS, Pollock CJ, et al. Loss of promoter IV-driven BDNF expression impacts oscillatory activity during sleep, sensory information processing and fear regulation. Transl Psychiatry. 2016;6:e873. doi: 10.1038/tp.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jay TM, Burette F, Laroche S. Plasticity of the hippocampal-prefrontal cortex synapses. J Physiol Paris. 1996;90:361–6. doi: 10.1016/s0928-4257(97)87920-x. [DOI] [PubMed] [Google Scholar]
- 14.Akaneya Y, Tsumoto T, Kinoshita S, Hatanaka H. Brain-derived neurotrophic factor enhances long-term potentiation in rat visual cortex. J Neurosci. 1997;17:6707–16. doi: 10.1523/JNEUROSCI.17-17-06707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu B. BDNF and activity-dependent synaptic modulation. Learn Mem. 2003;10:86–98. doi: 10.1101/lm.54603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn Mem. 2007;14:268–76. doi: 10.1101/lm.500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of bdnf gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–86. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soliman F, Glatt CE, Bath KG, Levita L, Jones RM, Pattwell SS, et al. A genetic variant BDNF polymorphism alters extinction learning in both mouse and human. Science. 2010;327:863–6. doi: 10.1126/science.1181886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi DC, Maguschak KA, Ye K, Jang S-W, Myers KM, Ressler KJ. Prelimbic cortical BDNF is required for memory of learned fear but not extinction or innate fear. Proc Natl Acad Sci USA. 2010;107:2675–80. doi: 10.1073/pnas.0909359107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters J, Dieppa-Perea LM, Melendez LM, Quirk GJ. Induction of fear extinction with hippocampal-infralimbic BDNF. Science. 2010;328:1288–90. doi: 10.1126/science.1186909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill JL, Martinowich K. Activity-dependent signaling: Influence on plasticity in circuits controlling fear-related behavior. Curr Opin Neurobiol. 2016;36:59–65. doi: 10.1016/j.conb.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, et al. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–89. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- 23.West AE, Chen WG, Dalva MB, Dolmetsch RE, Kornhauser JM, Shaywitz AJ, et al. Calcium regulation of neuronal gene expression. Proc Natl Acad Sci USA. 2001;98:11024–31. doi: 10.1073/pnas.191352298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2006;85:525–35. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.West AE, Pruunsild P, Timmusk T. Handb Exp Pharmacol. Berlin, Heidelberg: Springer; 2014. pp. 67–100. [DOI] [PubMed] [Google Scholar]
- 26.Maynard KR, Hill JL, Calcaterra NE, Palko ME, Kardian A, Paredes D, et al. Functional role of BDNF production from unique promoters in aggression and serotonin signaling. Neuropsychopharmacology. 2016;41:1943–55. doi: 10.1038/npp.2015.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, et al. Distinct role of long 3’ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–87. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau AG, Irier HA, Gu J, Tian D, Ku L, Liu G, et al. Distinct 3’UTRs differentially regulate activity-dependent translation of brain-derived neurotrophic factor (BDNF) Proc Nat Acad Sci USA. 2010;107:15945–50. doi: 10.1073/pnas.1002929107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baj G, Leone E, Chao MV, Tongiorgi E. Spatial segregation of BDNF transcripts enables BDNF to differentially shape distinct dendritic compartments. Proc Natl Acad Sci USA. 2011;108:16813–8. doi: 10.1073/pnas.1014168108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baj. G, D’Alessandro V, Musazzi L, Mallei A, Sartori CR, Sciancalepore M, et al. Physical exercise and antidepressants enhance BDNF targeting in hippocampal CA3 dendrites: Further evidence of a spatial code for BDNF splice variants. Neuropsychopharmacology. 2012;37:1600–11. doi: 10.1038/npp.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shieh PB, Hu S-C, Bobb K, Timmusk T, Ghosh A. Identification of a signaling pathway involved in calcium regulation of BDNF expression. Neuron. 1998;20:727–40. doi: 10.1016/s0896-6273(00)81011-9. [DOI] [PubMed] [Google Scholar]
- 32.Metsis M, Timmusk T, Arenas E, Persson H. Differential usage of multiple brain-derived neurotrophic factor promoters in the rat brain following neuronal activation. Proc Natl Acad Sci USA. 1993;90:8802–6. doi: 10.1073/pnas.90.19.8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakata K, Martinowich K, Woo NH, Schloesser RJ, Jimenez DV, Ji Y, et al. Role of activity-dependent BDNF expression in hippocampal-prefrontal cortical regulation of behavioral perseverance. Proc Natl Acad Sci USA. 2013;110:15103–8. doi: 10.1073/pnas.1222872110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye L, Allen WE, Thompson KR, Tian Q, Hsueh B, Ramakrishnan C, et al. Wiring and molecular features of prefrontal ensembles representing distinct experiences. Cell. 2016;165:1776–88. doi: 10.1016/j.cell.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colliva, A, Maynard, KR, Martinowich, K, & Tongiorgi, E. Detecting single and multiple BDNF transcripts by in situ hybridization in neuronal cultures and brain sections. Neuromethods, Humana Press: 2019.
- 36.Hodneland E, Kogel T, Frei DM, Gerdes HH, Lundervold A. CellSegm – a MATLAB toolbox for high-throughput 3D cell segmentation. Source Code Biol Med. 2013;8:16. doi: 10.1186/1751-0473-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bokil H, Andrews P, Kulkarni JE, Mehta S, Mitra PP. Chronux: A platform for analyzing neural signals. J Neurosci Methods. 2010;192:146–51. doi: 10.1016/j.jneumeth.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tort ABL, Komorowski R, Eichenbaum H, Kopell N. Measuring phase-amplitude coupling between neuronal oscillations of different frequencies. J Neurophys. 2010;104:1195–210. doi: 10.1152/jn.00106.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinowich K, Schloesser RJ, Jimenez DV, Weinberger DR, Lu B. Activity-dependent brain-derived neurotrophic factor expression regulates cortistatin-interneurons and sleep behavior. Mol Brain. 2011;4:11. doi: 10.1186/1756-6606-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, et al. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: Comparison of the recombinase activity of the tamoxifen-inducible Cre-ERT and Cre-ERT2 recombinases. Nucleic Acids Res. 1999;27:4324–7. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colgin LL, Denninger T, Fyhn M, Hafting T, Bonnevie T, Jensen O, et al. Frequency of gamma oscillations routes flow of information in the hippocampus. Nature. 2009;462:353–7. doi: 10.1038/nature08573. [DOI] [PubMed] [Google Scholar]
- 42.Denny CA, Kheirbek MA, Alba EL, Tanaka KF, Brachman RA, Laughman KB, et al. Hippocampal memory traces are differentially modulated by experience, time, and adult neurogenesis. Neuron. 2014;83:189–201. doi: 10.1016/j.neuron.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sotres-Bayon F, Sierra-Mercado D, Pardilla-Delgado E, Quirk GJ. Gating of fear in prelimbic cortex by hippocampal and amygdala inputs. Neuron. 2012;76:804–12. doi: 10.1016/j.neuron.2012.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kheirbek MA, Drew LJ, Burghardt NS, Costantini DO, Tannenholz L, Ahmari SE, et al. Differential control of learning and anxiety along the dorsoventral axis of the dentate gyrus. Neuron. 2013;77:955–68. doi: 10.1016/j.neuron.2012.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeNardo LA, Liu CD, Allen WE, Adams EL, Friedmann D, Fu L, et al. Temporal evolution of cortical ensembles promoting remote memory retrieval. Nat Neurosci. 2019;22:460–9. doi: 10.1038/s41593-018-0318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mizuno K, Dempster E, Mill J, Giese KP. Long-lasting regulation of hippocampal Bdnf gene transcription after contextual fear conditioning. Genes Brain Behav. 2012;11:651–9. doi: 10.1111/j.1601-183X.2012.00805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pruunsild P, Kazantseva A, Aid T, Timmusk T. Dissecting the human BDNF locus: Bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90:397–406. doi: 10.1016/j.ygeno.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakata K, Woo NH, Martinowich K, Greene JS, Schloesser RJ, Shen L, et al. Critical role of promoter IV-driven BDNF transcription in GABAergic transmission and synaptic plasticity in the prefrontal cortex. Proc Natl Acad Sci USA. 2009;106:5942–7. doi: 10.1073/pnas.0811431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han JC, Liu Q-R, Jones M, Levinn RL, Menzie CM, Jefferson-George KS, et al. Brain-derived neurotrophic factor and obesity in the WAGR syndrome. N Engl J Med. 2008;359:918–27. doi: 10.1056/NEJMoa0801119. [DOI] [PMC free article] [PubMed] [Google Scholar]