Abstract
Background
Genome-wide association studies (GWAS) are extensively used to identify single nucleotide polymorphisms (SNP) underlying the genetic variation of complex traits. However, much uncertainly often still exists about the causal variants and genes at quantitative trait loci (QTL). The aim of this study was to identify QTL associated with residual feed intake (RFI) and genes in these regions whose expression is also associated with this trait. Angus cattle (2190 steers) with RFI records were genotyped and imputed to high density arrays (770 K) and used for a GWAS approach to identify QTL associated with RFI. RNA sequences from 126 Angus divergently selected for RFI were analyzed to identify the genes whose expression was significantly associated this trait with special attention to those genes residing in the QTL regions.
Results
The heritability for RFI estimated for this Angus population was 0.3. In a GWAS, we identified 78 SNPs associated with RFI on six QTL (on BTA1, BTA6, BTA14, BTA17, BTA20 and BTA26). The most significant SNP was found on chromosome BTA20 (rs42662073) and explained 4% of the genetic variance. The minor allele frequencies of significant SNPs ranged from 0.05 to 0.49. All regions, except on BTA17, showed a significant dominance effect. In 1 Mb windows surrounding the six significant QTL, we found 149 genes from which OAS2, STC2, SHOX, XKR4, and SGMS1 were the closest to the most significant QTL on BTA17, BTA20, BTA1, BTA14, and BTA26, respectively. In a 2 Mb windows around the six significant QTL, we identified 15 genes whose expression was significantly associated with RFI: BTA20) NEURL1B and CPEB4; BTA17) RITA1, CCDC42B, OAS2, RPL6, and ERP29; BTA26) A1CF, SGMS1, PAPSS2, and PTEN; BTA1) MFSD1 and RARRES1; BTA14) ATP6V1H and MRPL15.
Conclusions
Our results showed six QTL regions associated with RFI in a beef Angus population where five of these QTL contained genes that have expression associated with this trait. Therefore, here we show that integrating information from gene expression and GWAS studies can help to better understand the genetic mechanisms that determine variation in complex traits.
Keywords: Residual feed intake, GWAS, QTL, RNA-seq, Gene expression, Angus, Beef cattle
Background
The incorporation of genomic information in livestock breeding programs is a common strategy to improve accuracy of selection for economically important traits. This is most useful for traits in the breeding objective that are not often measured by breeders. In beef cattle, the aim of most production systems is to select for more feed efficient animals since feed costs constitute around 70% of the total expenses [1]. The measurement of feed intake is costly, usually requiring expensive equipment to determine phenotypes for growth and feed intake in a 70 days test period in a feedlot. Feed efficiency in beef cattle is often expressed as residual feed intake (RFI) which is the difference between the observed feed intake recorded over a period of time and the expected feed intake based on the animal’s growth rate and maintenance requirement [2]. RFI reflects the variation in feed intake conditional on productivity, and therefore the variation in RFI can be used to explore the underlying causes of genetic variation using genomic technologies.
Modern genomic tools can be utilised to unravel the underlying biology of genetic variability in plants and animals. Methods that examine the heritability of traits, along with genome-wide association and gene expression studies have been utilised to attempt to understand the genetic basis underlying phenotypic differences between individuals. Numerous studies have reported variance components and heritabilities for RFI in cattle and correlations with other important production traits [3–5]. More recently, genome-wide association studies (GWAS) have been used to reveal the genomic architecture of polygenic traits by finding statistical associations between the phenotype and genetic markers assumed close to putative QTL (quantitative trait loci). Several GWAS for RFI have been performed in beef and dairy cattle [6–10]. Estimates of heritability of RFI range from low to moderate (0.14 to 0.49) and GWAS point at QTL for this trait in regions on many chromosomes (BTA3, BTA5, BTA6, BTA8, BTA12, BTA13, BTA15, BTA17, BTA18, BTA20, BTA21 and BTA22, see references 6, 7, 9). Identification of the causal variants in these QTL regions could help to better understand the genetic mechanisms underlying this trait. However, GWAS results are rarely conclusive and studies often require more phenotypic data on a larger number of animals as well as denser SNP panels to precisely locate the causative mutations, and genes involved in RFI.
Additional to GWAS, a number of studies have used transcriptomic data to find the genes that are differentially expressed with contrasting phenotypes or genotypes [11, 12], e.g. some studies have identified genes significantly associated with RFI in Angus cattle [13] and others have contrasted divergent lines or extreme phenotypes in other breeds of beef cattle [14]. However, there has been little consistency among the results of these studies. Gene expression studies are challenging, and they can vary widely in describing transcriptomic differences encompassing different tissues, breeds, sex, and age. A multi-tissue transcriptome approach combined with GWAS results may allow validation and better interpretation of GWAS findings, potentially giving a better insight into the genetic mechanisms and the biology behind this trait.
The aim of the current study was to perform a GWAS for RFI with imputed high density (770 K) genotypes in Australian Angus steers to detect significant SNPs statistically associated with phenotypic variation in RFI. Additionally, results from a multi-tissue gene expression experiment (RNA-seq) in a separate Angus population were used to further strengthen evidence for particular genes being involved in the genetic regulation of feed efficiency in beef cattle.
Results
Genome-wide association study
The estimated heritability for RFI based on the 2190 steers used for the GWAS was 0.3 (±0.04) using a genomic relationship matrix and after correcting for fixed effects of the contemporary groups. Genomic inflation lambda (λ) values of 0.95 show that the resulting p-values from the GWAS follow a chi-squared distribution and there was no sign of any systematic bias, e. g. due to population structure. From a visual evaluation (Q-Q plot), the distribution of most of the observed p-values aligned with the distribution of the expected p-values except for the significant p-values from SNPs associated with RFI (Additional file 2: Figure S1).
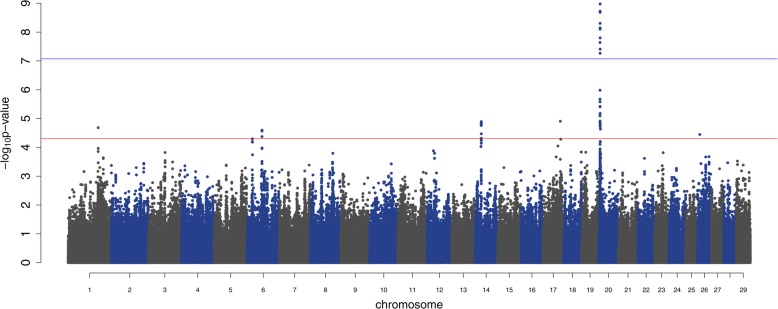
The GWAS resulted in 78 significant SNP from six QTL regions (on BTA1, BTA6, BTA14, BTA17, BTA20 and BTA26) when using as a threshold –log10(p) < 5e− 5 (Table 1; Additional file 1: Table S1), and from these, only 11 SNPs passed the more stringent threshold (−log10(p) < 8.51e− 8), with all of these located in a single QTL on BTA20 (Fig. 1).
Table 1.
Identified QTL associated with RFI in beef Angus (B. Taurus UMD3.1)
| Chromosome | Regions (Mb) | Significant SNPs |
|---|---|---|
| 1 | 11.05–11.06 | 2 |
| 6 | 55.18–55.08 | 10 |
| 14 | 24.18–24.39 | 13 |
| 17 | 63.63 | 1 |
| 20 | 4.88–6.12 | 31 |
| 26 | 8.91 | 1 |
Fig. 1.
Manhattan plot of SNP’s p-values of association with RFI. The lines represent the significant thresholds at -log10(p) > 7 (blue) and -log10(p) > 4.3 (red)
Further details on all significant SNPs are shown in the Additional file 1: Table S1), while the QTL regions with the most significant SNPs can be found in Table 1. The most significant SNP overall was found on chromosome BTA20 and this SNP explained 4% of the genetic variance, while the variance explained by the most significant SNPs in each of the other QTL was smaller (< 2%) (Table 2). From the most significant SNP per region, four of them (rs42662073, rs137349090, rs42005099, rs42322957) have MAF > 0.3 while two (rs42544395 and rs133158056) have MAF < 0.09 (Table 2). The SNPs rs42005099 and rs42322957 had a negative partial dominance (d) effect of − 0.396 and − 0.336, respectively, while rs137349090 had only an additive (a) effect with animals having the AA genotype being the more efficient (i.e. the lowest phenotypic value for RFI; Additional file 3: Table S2).
Table 2.
Association analysis of genotypes from the most significant SNP in each QTL region for RFI in Angus (B. Taurus UMD3.1)
| SNP | Position | A1a | A2b | Frc | -log10 (P) | %d | Effecte |
|---|---|---|---|---|---|---|---|
| rs42662073 | 20:4883142 | B | A | 0.45 | 8.98 | 4% | −0.26 |
| rs137349090 | 17:63630684 | A | B | 0.34 | 4.91 | 2% | −0.19 |
| rs42544395 | 14:24181858 | A | B | 0.08 | 4.89 | 2% | −0.34 |
| rs42005099 | 1:110543274 | A | B | 0.49 | 4.68 | 2% | 0.18 |
| rs42322957 | 6:55181977 | B | A | 0.38 | 4.59 | 2% | 0.19 |
| rs133158056 | 26:8907239 | A | B | 0.049 | 4.45 | 2% | −0.39 |
aA1: reference allele; bA2: other allele; cFr: MAF (minor allele frequency); dpercentage of variance explained by the genotype; eSNP effect
A window of 1 Mb surrounding the most significant SNPs was used to find candidate genes with biological importance for RFI. In total, 149 genes were found in these regions and 54 among them corresponded to uncharacterized proteins (Additional file 3: Table S2). Except for the rs137349090 SNP on BTA17, which is in an intronic section of the gene 2′-5′-Oligoadenylate Synthetase 2 (OAS2), all other SNPs were found in intergenic regions. For the most significant SNP in this study, rs42662073 on BTA20, Stanniocalcin 2 (STC2) is the closest gene. In the case of chromosomes BTA1, BTA6, BTA14, BTA26, the gene closest to the significant SNPs are Short Stature Homeobox 2 (SHOX2), LOC104968862, XK Related 4 (XKR4), and Sphingomyelin Synthase 1 (SGMS1), respectively. The most relevant genes based on the biological functions reported in other studies (related to feed efficiency and growth) are summarized in Table 3.
Table 3.
Previously reported role of the candidate genes located in the genomic regions associated with RFI in Angus
| Genomic region (1 Mb) | Candidate genes | Function |
|---|---|---|
|
BTA20 3.88–5.88 |
DUSP1-Dual Specificity Phosphatase 1 | Up-regulated gene in muscle from efficient broilers [15] and adipose tissue from obese humans [16] |
| ERGIC1 - Endoplasmic Reticulum-Golgi Intermediate Compartment 1 | Associated with MMWT in cattle [10] | |
| RPL26L1-Ribosomal Protein L26 Like 1 | Gene associated with MMWT in cattle [10] and up regulate in breast carcinoma [17] | |
| STK10-Serine threonine kinase 10 | Significantly associated with slaughter weight in beef cattle [18] | |
| ATP6V0E1- ATPase H+ Transporting V0 Subunit E1 | Involved in oxidative phosphorylation with up-regulation in rumen epithelium of low RFI cattle [19] | |
| STC2-Stanniocalcin 2 | Associated with RFI and MBW in cattle [20]; possible modulator of carcass and meat quality traits in beef cattle [21]. Overexpression resulted in postnatal growth restriction in mice [22] | |
| CPEB4- Cytoplasmic Polyadenylation Element Binding Protein 4 | Gene associated with rib eye area in Nelore [23]. Nearby a suggestive SNP (p-value 1.38e− 05) for average daily gain in pig [24] | |
| NEURL1B- Neuralized E3 Ubiquitin Protein Ligase 1B | Associated with day 2 of preadipocyte differentiation in chicken [25], nearby gene to the single nucleotide variants associated with body mass index in adult humans [26], nearby gene to significant SNP for longissimus dorsi muscle area in Hanwoo cattle [27] | |
| BOD1- Biorientation Of Chromosomes in Cell Division 1 | Inhibits PP2A-B56 regulating the function of Plk1 in mitotic cells at spindle poles and kinetochores [28] | |
|
BTA17 6.26–6.46 |
SDS- Serine Dehydratase, SDSL- Serine Dehydratase Like | Low expression in bovine jejunal epithelium tissue due to restricted dietary [29] |
| DTX1- Deltex E3 Ubiquitin Ligase 1 | Regulates transcription in the nucleus downstream the Notch receptor [30] | |
| SLC8B1- Solute Carrier Family 8 Member B1 | Up-regulation in high-efficient broiler chickens [31] | |
| OAS2–2′-5′-Oligoadenylate Synthetase 2 | Up-regulated in Blonde d’Aquitaine during embryonic muscle developmental when contrasting with Charolais [32] | |
| PTPN11- Protein Tyrosine Phosphatase, Non-Receptor Type 11 | Down regulated gene in high-RFI Holstein [33], and control cell proliferation in postnatal mice [34] | |
| RPL6- Ribosomal Protein L6 | Differentially expressed gene in divergent RFI lines of pigs [35] | |
| LHX5- LIM Homeobox 5 | Regulates the development and distribution of Cajal-Retzius cells in the developing forebrain [36] | |
| TPCN1- Two Pore Segment Channel 1 | Mice with knock down of the Tpcn1/2 had increase body mass due to faster increase in fat mass compare with the wile mice [37] | |
|
BTA14 2.31–2.51 |
XKR4- XK Related 4 | SNP associated with ADFI and ADG in cattle [38], and backfat thickness in Nelore [39] |
| SOX17- SRY-Box 17 | Transcriptional regulator of differentiation in embryonic stem cells in mouse [40]. Significant SNP associated with EBV for paternal calving ease in cattle [41] | |
|
BTA1 1.10–1.11 |
VEPH1- Ventricular Zone Expressed PH Domain Containing 1 | Candidate gene for rump fat thickness in Nellore [9] |
| PTX3- Pentraxin 3 | Up-regulated in breast muscle of high-feed efficient broilers [42] | |
| MFSD1-Major facilitator superfamily domain containing 1 | Down-regulated gene in brainstem and hypothalamus of mice raised on high-fat diet [43] | |
|
BTA6 5.41–5.61 |
LOC104968862 LOC104968863 -un characterize proteins |
Located in the region of SNPs for rump fat thickness [39] |
|
BTA26 7.90–9.90 |
MINPP1- Multiple Inositol-Polyphosphate Phosphatase 1 | Maintains the levels of InsP5 and InsP6 which are essential to normal cell growth [44] |
| A1CF- APOBEC1 Complementation Factor | Splicing regulator and the A1CF loss of function elevated triglycerides levels in mice [45] | |
| PAPSS2–3′-Phosphoadenosine 5′-Phosphosulfate Synthase 2 | Gene located nearby a SNP associated with DMI in feedlot steers [46]; After treating cartilage from bovine with TGF-β, the expression of gene PAPSS2 was up-regulated in articular chondrocytes, while the expression was down-regulated in cartilage from mice with negative mutation of the TGF-β receptor [47] | |
| SGMS1- Sphingomyelin Synthase 1 | Gene nearby a significant SNP associated with RFI in pigs [48] and average daily feed intake [49] | |
| ASAH2- N-Acylsphingosine Amidohydrolase 2 | Up-regulated in pigs with low feed conversion ratio [50] |
1Mb: 1 M base, BTA: Bos Taurus Autosome
Gene expression integration
Genes significantly associated with RFI- GSA (at p-value< 0.001, GSAp < 0.001) - in each tissue/sex were identified. The bull dataset had a higher number of GSAp < 0.001 with 23 (A-bulls_liver) genes in liver and 21 (D-bulls_muscle) genes in muscle, while H-steers_liver, H-heifers_blood, H-heifers_liver, and H-steers_blood datasets had 8, 6, 5, and 1 GSAp < 0.001 respectively (Additional file 6: Table S5). From all of the GSA, only the Eukaryotic translation initiation factor 3H (EIF3H) gene was found significantly associated with RFI, based on their expression in A-bulls_liver and D-bulls_muscle tissues. However, the expression effect was opposite in different tissues (0.101 in liver and − 0.085 in muscle).
The most significant GSAp < 0.001 in the QTL for RFI from all of the datasets are shown in Table 4 while the complete list is presented in the (Additional file 5: Table S4). The five most significant GSA, based on their p-value, were Neuronal Regeneration Related Protein (NREP), N-Acetylated Alpha-Linked Acidic Dipeptidase Like 1 (NAALADL1), Nuclear Receptor Coactivator 4 (NCOA4), CD8b Molecule (CD8B), and 7-Dehydrocholesterol Reductase (DHCR7). On the other hand, when the GSA were ranked based on the effect that gene expression had on the phenotype, the top five were Interferon gamma inducible protein 47 (IFI47), Coiled-Coil Domain Containing 38 (CCDC38), Glutathione S-Transferase Mu 2 (GSTM2), uncharacterized protein (ENSBTAG00000040281), and Retinol Binding Protein 1 (RBP1). The pathways related to the top significant GSAp < 0.001 were Cholesterol biosynthesis, Fatty acid degradation, MAPK signaling pathway, and PI3K-Akt signaling pathway (Table 4).
Table 4.
Most significant gene expression associated with RFI and their related metabolic pathway
| Symbol | Effect | p-value | Dataseta | Pathway |
|---|---|---|---|---|
| NREP-Neuronal Regeneration Related Protein | −0.21 | 4.66E-06 | BM | MECP2 and Associated Rett Syndrome |
| NAALADL1- N-Acetylated Alpha-Linked Acidic Dipeptidase Like 1 | 0.33 | 1.33E-05 | HB | – |
| NCOA4- Nuclear Receptor Coactivator 4 | 0.18 | 1.49E-05 | SL | Pathways in cancer, Thyroid cancer |
| CD8B- CD8b Molecule | 0.34 | 1.97E-05 | HB | T-Cell Receptor and Co-stimulatory Signaling, Innate Immune System |
| DHCR7–7-Dehydrocholesterol Reductase | −0.29 | 2.32E-05 | BM | Regulation of cholesterol biosynthesis by SREBP (SREBF), cholesterol biosynthesis I |
| ENSBTAG00000039588 | 0.39 | 3.81E-05 | HB | – |
| SYNE2- Spectrin Repeat Containing Nuclear Envelope Protein 2 | 0.11 | 4.01E-05 | BM | Meiosis, Ovarian Infertility Genes |
| OLFML1- Olfactomedin Like 1 | −0.51 | 4.56E-05 | BM | – |
| ANGPTL2- Angiopoietin Like 2 | −0.27 | 5.93E-05 | BM | Common Cytokine Receptor Gamma-Chain Family Signaling Pathways |
| ACADSB- Acyl-CoA Dehydrogenase Short/Branched Chain | −0.29 | 6.29E-05 | BL | Fatty acid degradation, Valine, leucine and isoleucine degradation, Metabolic pathways, Fatty acid metabolism |
| DDIT3- DNA Damage Inducible Transcript 3 | −0.22 | 7.26E-05 | BM | MAPK signaling pathway, Protein processing in endoplasmic reticulum, Non-alcoholic fatty liver disease (NAFLD), Transcriptional misregulation in cancer |
| TDRP- Testis Development Related Protein | −0.45 | 1.21E-04 | HL | – |
| CCDC38- Coiled-Coil Domain Containing 38 | 0.68 | 1.67E-04 | BL | – |
| AIM1- Absent in melanoma 1 | 0.15 | 1.91E-04 | BL | Fatty acid degradation, alpha-Linolenic acid metabolism, Metabolic pathways, Biosynthesis of secondary metabolites, Fatty acid metabolism |
| PLA2G16- Phospholipase A2 Group XVI | 0.18 | 2.32E-04 | HL | Glycerophospholipid metabolism, Ether lipid metabolism, Arachidonic acid metabolism, Linoleic acid metabolism, alpha-Linolenic acid metabolism, Metabolic pathways, Ras signaling pathway, Regulation of lipolysis in adipocytes |
| TMEM135- Transmembrane Protein 135 | −0.21 | 2.50E-04 | BM | – |
| CENPM- Centromere Protein M | 0.45 | 2.82E-04 | SL | Chromosome Maintenance, Signaling by Rho GTPases |
| EIF2A- Eukaryotic Translation Initiation Factor 2A | 0.16 | 2.83E-04 | BL | RNA transport, Protein processing in endoplasmic reticulum |
| SQLE- Squalene Epoxidas | −0.45 | 2.84E-04 | BM | Steroid biosynthesis, Metabolic pathways, Biosynthesis of antibiotics |
| GSTT3- Glutathione S-Transferase Theta 3 | 0.27 | 2.91E-04 | BL | Glutathione metabolism, Metabolism of xenobiotics by cytochrome P450, Drug metabolism - cytochrome P450, Chemical carcinogenesis |
| HMGCS1–3-Hydroxy-3-Methylglutaryl-CoA Synthase 1 | −0.24 | 2.94E-04 | BM | Synthesis and degradation of ketone bodies, Valine, leucine and isoleucine degradation, Butanoate metabolism, Terpenoid backbone biosynthesis, Metabolic pathways, Biosynthesis of antibiotics |
| GHDC- GH3 Domain Containing | 0.17 | 3.59E-04 | HL | Innate Immune System |
| EIF3H- Eukaryotic Translation Initiation Factor 3 Subunit H | 0.10 | 3.62E-04 | BL | RNA transport, Measles |
| GPATCH11- G-Patch Domain Containing 11 | 0.33 | 4.14E-04 | SL | – |
| NET1- Neuroepithelial Cell Transforming 1 | −0.14 | 4.19E-04 | HL | p75 NTR receptor-mediated signaling, fMLP Pathway |
| BCKDHB- Branched Chain Keto Acid Dehydrogenase E1 Subunit Beta | 0.10 | 4.32E-04 | SL | Valine, leucine and isoleucine degradation, Metabolic pathways, Biosynthesis of antibiotics |
| DNER- Delta/Notch Like EGF Repeat Containing | −1.13 | 4.33E-04 | BM | Signaling by NOTCH1 and NOTCH2 Activation, Transmission of Signal to the Nucleus |
| ALDH5A1- Aldehyde Dehydrogenase 5 Family Member A1 | 0.14 | 4.81E-04 | BL | Alanine, aspartate and glutamate metabolism, Butanoate metabolism, Metabolic pathways |
| ROBO2- Roundabout Guidance Receptor 2 | 0.49 | 5.03E-04 | BL | Axon guidance |
| GSTM2- Glutathione S-Transferase Mu 2 | 0.61 | 5.51E-04 | BL | Glutathione metabolism, Metabolism of xenobiotics by cytochrome P450, Drug metabolism - cytochrome P450, Chemical carcinogenesis |
| PRICKLE1-Prickle Planar Cell Polarity Protein 1 | −0.31 | 5.62E-04 | BL | Wnt signaling pathway |
| ENSBTAG00000040281 | 0.59 | 6.02E-04 | HB | – |
| IFI47- Interferon Gamma Inducible Protein 4 | 0.88 | 6.02E-04 | SL | TNF signaling pathway |
| ENSBTAG00000002786 | −1.10 | 6.42E-04 | SL | – |
| RBP1- Retinol Binding Protein 1 | 0.55 | 7.01E-04 | HB | Nicotinate and nicotinamide metabolism, Metabolic pathways |
| UBE2D2- Ubiquitin Conjugating Enzyme E2 D2 | 0.10 | 7.21E-04 | SL | Ubiquitin mediated proteolysis, Protein processing in endoplasmic reticulum |
| ENSBTAG00000001489 | −0.18 | 7.24E-04 | SB | Phagosome, Gap junction |
| GDPGP1- GDP-D-Glucose Phosphorylase 1 | 0.32 | 7.40E-04 | HB | – |
| ITGB4- Integrin Subunit Beta 4 | 0.34 | 8.59E-04 | HL | PI3K-Akt signaling pathway, Focal adhesion, ECM-receptor interaction, Regulation of actin cytoskeleton, Hypertrophic cardiomyopathy (HCM), Arrhythmogenic right ventricular cardiomyopathy (ARVC), Dilated cardiomyopathy |
| URI1- URI1, Prefoldin Like Chaperone | 0.19 | 9.32E-04 | SL | Translational Control, Apoptosis and Autophagy |
aBM: bulls_muscle, HB: H-heifers_blood, SL: H-steers_blood, BL: A-bulls_liver, HL: H-heifers_liver
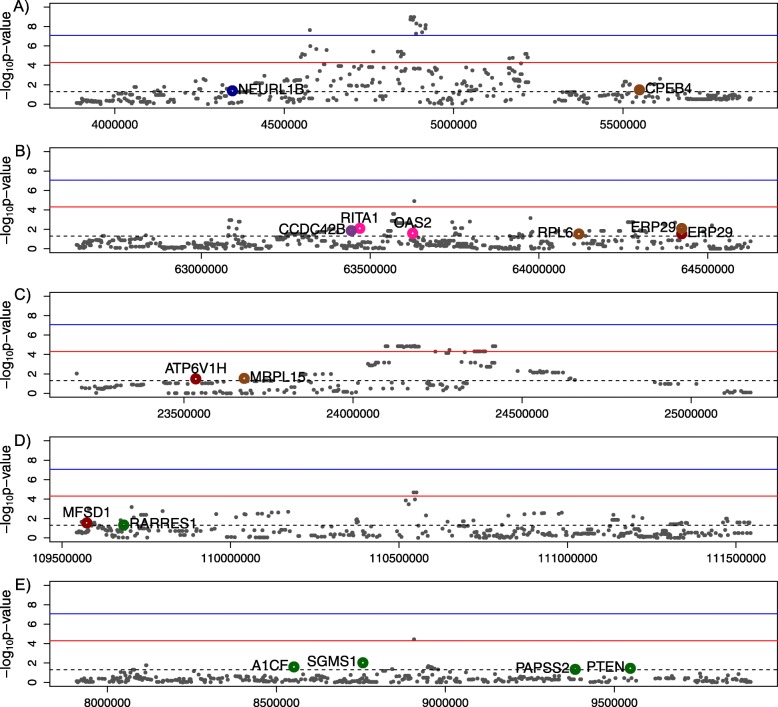
When the window was extended to 2 Mb, 15 genes whose expression was associated with RFI (p < 0.05; GSAp < 0.05) were identified around the top significant SNPs on BTA1, BTA14, BTA17 and BTA26. The region on BTA6 between 54 and 56 Mb does not code for any genes, therefore, there are no results for the gene expression in that region. For the most significant QTL on BTA20 positioned between 3.88 and 5.88 Mb, the GSA NEURL1B and CPEB4 were found (Fig. 2a), their expression had a positive effect on RFI (0.254 and 0.064 respectively). The region with most GSAp < 0.05 genes was BTA17 (Fig. 2b) with five genes (RITA1- RBPJ Interacting and Tubulin Associated 1, CCDC42B- Coiled-Coil Domain Containing 42, OAS2, RPL6- Ribosomal Protein L6, and ERP29- Endoplasmic Reticulum Protein 29). Gene ERP29 was significantly associated in two datasets (H-steers_liver and D-bulls_muscle). However, similar to gene EIF3H mentioned before, the direction of the effect was found to be opposite in different tissues, with a regression of RFI on lcpm of 0.079 in liver and − 0.12 in muscle. The QTL region with the second most GSAp < 0.05 was BTA26 (between 7.90 and 9.90 Mp) with four GSA (A1CF- APOBEC1 Complementation Factor, SGMS1, PAPSS2–3′-Phosphoadenosine 5′-Phosphosulfate Synthase 2, PTEN- Phosphatase and Tensin Homolog) (Fig. 2e). From these genes, the gene expression of A1CF was down-regulated (− 0.10), while the other GSAp < 0.05 were up-regulated. The QTL regions with a smaller number of GSAp < 0.05 were BTA14 (with ATP6V1H and MRPL15 between 2.31 and 2.51 Mb; Fig. 2c) and BTA1 (with MFSD1 and RARRES1 between 1.10 and 1.11 Mb; Fig. 2d).
Fig. 2.
Genes significantly associated with RFI in the region of 2 MB around the significant SNPs in a) BTA20, b) BTA17, c) BTA14, d) BTA1, and e) BTA26. Colors indicate the corresponding dataset: bulls_liver (green), bulls_muscle (brown), steers_liver (red), steers_blood (purple), heifers_liver (blue), and heifers_blood (pink)
Discussion
In this study, the heritability estimated for RFI (h2 = 0.3) is in agreement with other estimates reported previously for other Angus populations [10, 20, 51], an Angus-Brahman herd (0.30) [52], and Nellore (0.17) [53]. However, in some other studies in Angus and Charolais populations, the heritability has been reported as high as 0.47 and 0.68, respectively [54]. Most of those studies, however, are based on relatively small data sets.
Genome-wide association for RFI
Six QTL regions were identified to be associated with RFI on BTA1, BTA6, BTA14, BTA17, BTA20, and BTA26 (Table 1). A QTL for RFI on BTA20 has been reported in earlier studies, however, it is not the same location as in this study. The significant SNP for RFI (20_51402608) [6] was identified in Angus and is located 46.5 Mb from our most significant SNP while on chromosome 20 there was a significant QTL for ADG (BTA20_39) in SimAngus which is 34.1 Mb apart from our QTL for RFI [10]. The differences in regions found in our results compared with the regions reported in earlier studies could be due to the use of different Angus population, the number of animals used, some findings maybe false positives, or the approach applied to measure and define RFI might differ. Additionally, the fact that nearby SNPs have been previously reported as being associated for other traits (like MMWT, DMI) could be due to the pleiotropic effect of some regions. For example, the same regions have been associated for DMI-MBW, ADG-MBW, RFI-MBW [20], and RFI-DFI [6]. Although RFI and ADG and MBW had no correlation at the phenotypic level due to the conditional adjustment, there could still be a correlation at the genetic level [55], albeit relatively small. Interestingly, the gene STC2 was the closest to the QTL on BTA20 in our study, and previous studies have reported SNPs (rs133032375) in this region significantly associated with mid-test weight and RFI in Hereford [20]. This gene STC2 is a proteinase inhibitor of PAPP-A and the over-expression of STC2 in mice causes a reduction in postnatal growth compared with normal mice [22, 56]. Additionally, mice with an over-expression of human STC2 showed reducing bone and skeletal muscle growth [57].
There were five other regions identified in this study that provided further information of candidate genes with biological relevance to RFI (Table 1). On BTA1, a close QTL has been identified in BTA1_103459113 associated with RFI [6], while BTA1_106 [10], and BTA1_107 [20] were associated with feedlot dry matter intake (DMI), BTA1_108 was identified for MMWT [10]. Here we identified the nearby gene PTX3 which previously was reported as up-regulated in breast muscle of high-efficient broilers [42]. Another gene found in the 1 Mb window from the significant SNP for BTA1 is MFSD1 which is down-regulated in the brainstem and hypothalamus of mice raised on a high-fat diet [43].
On BTA14, the SNP rs42544395 was the most significant for RFI (Table 2), which is close to the SNP identified in SimAngus 14_17 for DMI, BTA14_24, BTA14_25 and BTA14_26 for MMWT, while BTA14_27 was associated with RFI in Angus [10]. In another population of Angus cattle, the SNP BovineHD1400006992 (BTA14_24114365) was significantly associated with PW_lwt, and SNP BovineHD1400007153 (BTA14_24621142) was associated with RFI [6]. The closest gene to SNP rs42544395 is XKR4 which was associated with feed intake and growth in cattle [38]. This gene was also reported as associated with rump fat thickness [58] and back fat [39]. In the Nellore breed, the XKR4 gene was associated with tenderness [59].
The SNP 17_58 was earlier reported for RFI in Angus [10] and it is close to the identified QTL on BTA17 (rs137349090). Multiple interesting genes were identified in the 1 Mb region surrounding this SNP (Table 3). The OAS2 gene seems to play an important role during muscle development [60]. Another gene, SLC8B1, was reported as up-regulated in high-efficient broiler chickens [31], while the gene PTPN11 was down regulated in high-RFI Holstein [33]. Divergent RFI lines of pigs had differential expression of RPL6 [35], another gene located close to rs137349090. The estimation of dominance effects in the most significant SNPs for each QTL showed that with the exception of SNP rs137349090, all SNPs had a significant dominance effect, with some even showing overdominance (Additional file 4: Table S3). Similarly, significant dominance and epistatic effects for carcass, growth and fertility traits were found in Angus cattle [61]. This pattern of large dominance effect is consistent with the suggestion by Jiang, et al. [62] that the contribution of non-additive effects to the total genetic variance for complex trait in Holstein cattle can be considerable. However, as pointed out by Hill et al. (2008), most of the dominance effects are captured by the additive genetic variance [63].
In our study, we found more negative than positive dominance effects, which is in agreement with those reported previously for RFI, age at puberty and postpartum anoestus interval [61]. The SNP rs137349090 identified on BTA17 had no dominance effects, which is a relatively accurate estimate as we found that this SNP has a sufficient number of observations for each of the genotypes (MAF = 0.34). There are two important GSA in this region (RPL6 and ERP29; see Fig. 2b) that were reported as differentially expressed in divergent lines for RFI in pigs [35] and chicken [64]. Altogether, these results suggest that the information on this SNP genotype might contribute to a higher accuracy of genomic prediction of phenotype or breeding value for RFI.
Finally, we identified a QTL for RFI on BTA26, and this region has not been reported previously for RFI. In this region, the gene SGMS1 is close to the most significant SNP rs133158056. The function of the SGMS1 gene has not been documented, however, significant SNPs for RFI [48] and average feed intake [49] have been identified in pig for the homologous gene region. Moreover, in the present study, the expression of SGMS1 in bulls strongly selected for RFI was significantly higher (with a positive effect 0.11) than in animals selected for low RFI.
Gene expression overlap with GWAS results
The use of RNA-seq in multiple tissues of Angus cattle allowed us to identify the genes that were significantly associated (GSAp < 0.05) with RFI inside a window of 2 MB from the most significant SNPs. The genes NEURL1B and CPEB4 were located on BTA20 (Fig. 2a) and have not been reported to be associated with feed efficiency traits in previous studies, motivating further analysis of these genes to validate these results and to determine the role of these genes in relation to RFI. On BTA17, we found a relatively high number of GSA, with gene ERP29 proximal to the SNP rs137349090. This gene has been reported previously as differentially expressed between high and low RFI lines in chickens [64]. In addition, on chromosome BTA17, the gene RITA1 was significantly associated with RFI and is close to the significant SNP rs137349090. This gene is a tubulin-binding protein that acts as a negative regulator in the Notch signaling pathway. However, there is no previous report of this gene to be associated with feed efficiency traits. On BTA14, the gene ATPase H+ Transporting V1 Subunit H (ATP6V1H) was nearest to the significant SNP (rs42544395) and was significantly associated (GSAp < 0.05) for RFI in steer liver tissue (Fig. 2c). This gene has been reported previously for traits that define puberty (age at first corpus luteum and scrotal circumference of 26 cm) in Brahman cattle [65]. The scrotal circumference was reported to be higher in young bulls with high RFI (when backfat thickness was corrected for in the model) [66]. Nonetheless, in a previous study, there was no detrimental effect of low RFI on scrotal circumference of bulls [67]. On the same chromosome, the expression of the gene Mitochondrial Ribosomal Protein L15 (MRPL15) was significantly associated (GSAp < 0.05) with RFI in muscle from D-bulls. Previous reports have shown low expression of MRPL15 in double muscle Semitendonosus in cattle [32], but a higher gene expression was found in more feed efficient broilers [68].
We used a higher significance threshold (GSAp < 0.001) for a genome wide search for differentially expressed genes, and observed that the highest number of GSAp < 0.001 was observed in A-bulls_liver followed by D-bulls_muscle, H-steers_liver, H-heifers_blood, H-heifers_liver, and H-steers_blood with 23, 21, 8, 6, 5, and 1 GSA respectively (Additional file 6: Table S5). The higher number of GSAp < 0.001 in A-bulls_liver agrees with the larger variation in phenotypes for RFI found in the bulls used in this study, while the lower number of GSA in the H-groups, maybe due to the smaller variation in RFI values observed among heifer and steers in these groups (Table 6). Additionally, the small library size used in this analysis could lead to missing observations on some genes relevant in the RFI biology.
Table 6.
Number of samples used in the GWAS and GSA analyses and summary statistics for RFI
| Dataset (analysis) | N | Mean | SD | Min | Max |
|---|---|---|---|---|---|
| Genotyped Steers (GWAS) | 2190 | −2.11 | 1.66 | −9.3 | 4.2 |
| Bull liver (A-bulls: GSA) | 27 | 0.16 | 0.99 | −1.43 | 1.89 |
| Bull muscle (D-bulls: GSA) | 47 | −0.02 | 0.96 | −1.96 | 2.62 |
| Heifers & Steers blood-liver (H-cohort: GSA) | 52 | −0.004 | 0.81 | −1.84 | 1.87 |
SD: standard deviation, Min: minimum value, Max: maximum value, GSA: gene significantly associated, GWAS: genome-wide association studies
The gene EIF3H was found significantly associated in A-bulls_liver and D-bulls_muscle. Interestingly, this gene was observed to be over-expressed in Hanwoo cattle for animals with increasing CWT and EMA [69]. In trout, the EIF3H gene has been shown to be involved in compensatory muscle growth [70]. Further studies are needed to better understand the role of EIF3H gene on RFI in liver and muscle tissues.
The top significant genes (GSAp < 0.001) seem to play a role in RFI as they were shown to be associated with phenotypic differences and their functional annotation is consistent with RFI as a biological trait. The gene NREP for example, is close to a region associated with feed efficiency in Nellore cattle [71]. Another interesting gene is GSTM2, which in this study, we found positively associated with RFI in liver tissue from bulls. GTM gene family had been reported previously to be associated with RFI, GSTM1 and GSTM3 were highly expressed in high-RFI animals in liver [11]. GSTM1 and GSTM2 were significantly correlated with RFI-EBV and were up-regulated also in liver from high RFI steers [72]. The corresponding pathways in which the top significant GSAp < 0.001 were involved is reported in Table 4. In spite of the underpowered small library size obtained in the RNA-seq data, multiple genes could be found to have a significant effect on RFI, based on their observed expression and their potential function in pathways like Cholesterol biosynthesis, Fatty acid metabolism, MAPK signaling pathway, Glycerophospholipid metabolism, Metabolism of xenobiotics by cytochrome P450 and PI3K-Akt signaling pathway (Table 4). The diversity of pathways found in our results and other studies [19] reflect many processes involved in RFI and the genetic complexity of this trait.
Limitations of the study
This study has some limitations in the analysis of gene expression (RNA-seq). The small library size obtained from sequencing the RNA reduced the chance of finding strong GSA for RFI in the transcriptome of the animals. A pathway enrichment analysis from the obtained GSA was not significant (for the Benjamini multiple test) and was not included in this study because it could be affected by the low level of deep sequencing. However, we still consider our results meaningful and they could be used to validate results from the GWAS, overall offering more evidence and a biological interpretation of their potential role in determining genetic variation in RFI. The Angus populations used for the genomic analysis came from a commercial herd while the dataset used in the transcriptomic analysis was from animals under divergent selection lines for RFI. In addition, the animals were tested for RFI at different ages in both datasets, 18 months and 13 months for the GWAS and transcriptomic dataset, respectively. Therefore, due to these differences, results in this study, obtained by combining gene expression results in RFI contrasting phenotypes and results from genetic variants found to be associated with phenotypic variation in RFI should be interpreted with care. Nevertheless, our results encourage the use of various types of “omics” information in the same population as a way to decipher the genetic and genomic architecture of complex traits and as a way to obtain a better biological interpretation of the trait.
Future implications
The approach followed in this study illustrated the benefit of combining genomic information with gene expression to obtain an enriched overview of the genes implicated in RFI. The genes identified in this study could be used as a target for further functional studies, to help further elucidate their role in other cattle breeds or with different diets.
In future studies, the access to whole-genome sequence and larger datasets are desired to confirm and refine the five suggestive QTL at BTA1, BTA6, BTA14, BTA17, and BTA26. The use of sequence data and larger more diverse or multi-breed population could alleviate the limitation due to linkage disequilibrium (LD), where the genotypes of multiples SNPs would be correlated with the causal variant. Furthermore, sequence information is more likely to uncover QTL in other regions. Finally, combining information from both GWAS and transcriptomic profiling could help to select the SNPs that can contribute to an increased accuracy of prediction of phenotype or breeding values for RFI.
Conclusion
In this study, we investigated the genome-wide association of SNPs with RFI in an Australian Angus beef cattle population. We identified six QTL regions associated with RFI (BTA1, BTA6, BTA14, BTA17, BTA20 and BTA26) reflecting the polygenic nature of this trait. Promising candidate genes were identified around the most significant SNPs in each QTL. We also revealed 15 genes in these QTL regions whose expression were significantly associated with phenotypic and genetic differences in RFI (NEURL1B, CPEB4, RITA1, CCDC42B, OAS2, RPL6, ERP29, ATP6V1H, MRPL15, MFSD1, RARRES1, A1CF, SGMS1, PAPSS2 and PTEN). Our approach demonstrates that combining GWAS and RNA-seq information improves the interpretation of GWAS results and gives it a more biological connotation.
Methods
Data used for the GWAS
Animals and phenotypes
All phenotypic data were collected on 2190 Angus steers from the Angus Sire Benchmarking Program (ASBP, also known as the Angus Beef Information Nucleus) during a feedlot testing period between 2013 and 2017. This structured dataset represented a progeny test of registered Angus sires from herds located in New South Wales and Victoria, Australia. All the procedures were managed according to the welfare guidelines established by the Australian Animal Welfare Standards and guidelines for cattle (Edition one 2013) approved by the University of New England Animal Ethics Committee (Approval No. AEC12–082). All steers, were born from fixed time AI in various herds and within each herd-year calves had a maximum variation in age of 15 days. The steers were moved from pasture feeding into a feedlot at an average of 17 months of age. Before entering the test period in the feedlot, these animals had an adjustment period of 21 days followed by a 70 d test period in the Tullimba research feedlot (30°20′S, 151°10′E, altitude 560 m) near Kingstown, NSW, Australia, as described in [73] and after the test all the animals were returned to industry owners. Daily records on feed intake and fortnightly records of body weight were used to derive average daily gain and metabolic mid-weight [1, 74] and were fitted together with age in the following model:
| 1 |
where Yi is the daily feed intake of animal i (kg/day), βo is the regression intercept, β1 is the partial regression coefficient of feed intake on average daily gain (kg/day), β2 corresponds to the partial regression coefficient of feed intake on metabolic mid weight (kg0.73), β3 is the partial regression coefficient of feed intake on the feed test management group (defined as feedlot test pen within herd of origin and year), and ei is the residual error in feed intake of animal i, therefore defined as the phenotype for residual feed intake (RFI). The mean and standard deviation for RFI are shown in Table 6, with a range in RFI from − 9.3 to 4.2 kg/day.
Genotypes
All 2190 steers with RFI phenotypes were genotyped using various lower density SNP panels (Table 5). Their genotypes were imputed to medium density (50 k) and then to high density (HD- 770 K) as part of an imputation performed on the wider Australian Angus population. The reference population for the 50 k imputation consisted of 11,226 animals from the Angus Australia genotyped with a number of 50 k arrays (see RefImp50k in Table 5). The reference population for the high density imputation consisted of 1069 animals, again from Angus Australia (See RefImpHD, Table 5). For each SNP chip listed in Table 5, quality control (QC) was applied where only autosomal SNPs and the SNPs with a call rate higher than a 0.6 GeneCall score were kept. Further QC was undertaken using Plink v1.90b3.42 [75], filtering out those SNPs with minor allele frequency (MAF) < 0.01, deviation from Hardy Weinberg equilibrium (P < 10− 6), and those SNPs with more than 5% missing genotypes. Only animals that had a valid genotype on more than 95% of the SNPs were kept in the analysis. The total number of animals and SNPs remaining after quality control are shown in Table 5. The 50 k reference consisted of 39.7 k SNPs after QC and the merging of 50 k reference chips. Similarly, 587,437 SNPs remained after QC for the HD reference chip. Imputation to 50 k and then HD was undertaken using FImpute v2.2 [76]. The final dataset consisted of 2190 phenotyped steers.
Table 5.
SNPs in each chip panel (SNPs after quality control) and samples used in the imputation
| SNP Chip | Number of SNPs (after QCa) |
Number of samples |
|---|---|---|
| Low density panel | ||
| GGP_8K | 8753 (7569) | 849 (325) |
| GGPLD-10 K-V2.0 | 9323 (7636) | 980 (424) |
| GGPLD-20 K-V1.0 | 20,701 (14,062) | 1189 (534) |
| GGPLD-20 K-V3.25 | 25,712 (18,786) | 1138 (374) |
| GGPLD-26 K-V3.0 | 26,728 (18,187) | 1344 (526) |
| GGPLD-30 K-V4.0 | 30,865 (20,560) | 1556 (1) |
| GGPLD-9 K | 8659 (6230) | 574 (6) |
| RefImp50k | ||
| LDMAX_SNPMap | 56,955 (39,736) | 3950 |
| ZM2_SNPMap | 60,911 (42,522) | 778 |
| GSTP_SNPMap | 54,609 (39,706) | 6673 |
| ZOE-50 K | 54,001 (37,231) | 231 |
| RefImpHD | ||
| GGPHD-770 K | 777,984 (587,437) | 1069 |
In low density arrays only the samples indicated in parenthesis were phenotyped for RFI. aQC: quality control
To ensure imputation accuracy was acceptable, a simple cross validation was performed. One thousand animals were extracted from the 50 k reference (RefImp50k) population, their genotypes were updated to only include the low density SNP and they were subsequently imputed back up to 50 k. The HD reference genotypes were evaluated by extracting 100 animals and keeping only the SNPs corresponding to the 50 k panel and then imputed up to HD. The accuracy of imputation was measured with the correlation between the imputed genotypes and the true genotypes. The imputation accuracy was also measured as concordance which is the proportion of SNPs with matching imputed and original genotypes. The average accuracy of imputation measured as correlation and concordance were 0.96 and 0.98, respectively, for low density imputed to 50 k; while a value of 0.99 was obtained for imputing from 50 k to high density.
Genome-wide association study
The GWAS for RFI was performed using GCTA v1.26.0 (Yang et al. 2011) fitting the genomic relationship matrix (G) [77] in a univariate linear mixed model:
| 2 |
where y is the RFI phenotypic value, μ is the mean, cgi is the effect of contemporary group i, gj is a fixed effect of the allele dosage at a single SNPj to contrast with fitting 3 genotypes (i.e. a covariate representing the number of “1” alleles), α is the regression coefficient for the allele substitution effect, ak is the random additive genetic effect of animal k, and eijk is a random residual effect [73]. A total number of 98 contemporary groups (CG) were formed by concatenating herd of origin, year of birth, age, management group prior to the feed test, feed test management group and birth type [78]. A matrix G with genomic relationships among the steers was calculated based on [77] and was fitted as a covariance matrix for the additive genetic effects, i.e. var.(a) = , in order to account for residual additive genetic effects as well as for population structure effects and family relatedness. The effect of individual SNPs was estimated each time using the same estimated value for . Therefore, model [2] was run 587,437 times, once for each SNP.
To control false-positive associations, a Bonferroni correction was applied. The SNPs were considered significant when its p-value < 0.05/587,437 giving a threshold of -log10(p) > 7. Additionally, we also used a lower threshold (−log10(5e− 5) > 4.3) to identify SNPs that were not statistically significant but that could be close to genes with a biological function that could be related to RFI. QTL regions were defined as the section of the genome that contains significant SNP (−log10(5e− 5) > 4.3) extending for 1 Mb on either side of the significant SNPs.
Genomic inflation of the GWAS was calculated as the median of the chi-squared test divided by the expected median of the chi-square distribution expressed as lambda (λ). The variance explained by the significant SNP as a proportion of genetic variance was calculated as the percentage of:
where p and q (=1-p) are the allele frequencies for the i-th SNP, α2 is the estimated additive effect of the i-th SNP and is the additive genetic variance.
Candidate genes within the QTL regions were further investigated for their function. For the most significant SNPs in each QTL region, we used the model [2] but with the three genotypes for a SNP locus as a fixed class variable in a univariate linear mixed model using MTG2 v2.09 [79] in order to estimate both additive (a) and dominance (d) effects at each SNP using the formulas [80]:
where and are the estimated effects of the homozygous genotypes, and is the effect of the heterozygous genotype.
Gene expression by RNA sequencing
Experimental design
The animals used for the gene expression section were an independent dataset to the animals used in the GWAS analysis. All the procedures involved in the experiment were approved by the University of New England Animal Ethics Committee (AEC 06/123, AEC14–002 and AEC14–036) and New South Wales Department of Primary Industries (NSW DPI) Animal Research Authority (ORA09/015, ORA 13/16/004).
Three cohorts of animals were used for gene expression studies. All animals used for the gene expression study came from the feed efficiency selection lines of Angus cattle at the Agricultural Research Centre, Trangie, NSW, Australia. Further details on the selection design can be found in Arthur, et al. [1]. The first set of animals (A-bulls) consisted of 27 young bulls born in 2005 which belonged to approximately the third generation since the start of the divergent RFI selection lines [11]. These A-bulls were selected from the highest and lowest phenotypes for RFI out of a tested cohort of 90 young bulls. All these animals were reared with their mothers on pasture, weaned at about 7 months of age, and later were reared on grazing pasture till they reached feedlot entry weight (13 months). The RFI test was conducted for 70 days in the Tullimba test station using an automated feeding system which delivers and records individual animal feed intake. Based on the performance in the RFI test, 30 animals with the lowest RFI and 30 animals with the highest RFI were chosen out of 90 for collection of liver biopsies at the end of RFI testing [11]. From those animals, only 27 animals were used in this study for the gene expression analysis. The detailed procedures of the liver biopsy were described in [11] and animals were administered by appropriate pain relief and post-operative care, as directed by the veterinary surgeon. After 2 weeks of the biopsy, the animals were returned back to Trangie Agricultural Research Centre for breeding or used for other research projects.
The second set of animals were 47 young bulls (D-bulls) which were born in 2008 and were progeny of A-bulls. The D-bulls calves were reared with their mothers on pasture till weaning (~ 230 day). After weaning, the young D-bulls were reared on grazing pasture until they reached feedlot entry weight (around 13 months). The top 25 high RFI and bottom 22 low RFI young bulls were selected to be tested for RFI at Trangie. The selection of top and bottom RFI groups were based on RFI_EBV extracted from BREEDPLAN in May 2009. Tissue biopsies were collected from the Semitendinosus muscle at the end of the RFI test [81].
The third cohort of 25 steers (H-steers) and 27 heifers (H-heifers) were one further generation down of the selection line and were progeny of D-bulls and born in 2012. The male calves were castrated at 4 months of age. The young male and female calves were reared with their mothers on pasture until weaning (~ 230 day). After weaning, 32 heifers and 32 steers were transferred to the NSW DPI Agricultural Research and Advisory Station (Glen Innes, NSW, Australia) and grown on native pastures until they reached feedlot entry weight of approximately 400 kg BW (~ 560 days). Before the RFI test, animals were given a 2 week period of adaptation to the feedlot ration. The RFI was measured with an automated recording system for 70 days. During this period, animals had ad libitum access to a pelleted diet which 75% grain, 10% sorghum hay, and 5% protein pellets, plus monensin, vitamins, and mineral supplement. This diet had an average energy content of 10.5 MJ metabolizable energy (ME) per kilogram dry matter and 15 to 17% crude protein. Straw was provided at an average of 0.5 kg per animal per day. The animals were transferred to a respiration chamber facility at University of New England with the same diet for RFI testing. Liver tissue was collected by biopsies at the second week after the end of RFI testing [11, 82]. In addition, peripheral venous blood samples were extracted from the tail (coccygeal) vein of cattle and it was directly placed into the PAXgene Blood RNA Tubes (Qiagen, BD, cat. no. 762165). The collection tubes were gently inverted 10 times, stored at 4 °C, and transferred to − 20 °C for long term storage.
Phenotypic traits
The average daily gain, net feed intake, average daily feed intake and metabolic mid-test weight (MMWT) were recorded for all the 126 animals. RFI was calculated for each animal based on a linear regression model of feed intake on metabolic mid-test live weight, ADG and fitting contemporary groups as fixed effect [1]. The mean and standard deviation for each dataset used in gene expression analysis is shown in Table 6. The average RFI of the samples with RNA sequences were centred close to zero and ranged from − 1.96 to 2.62.
RNA extraction and library preparation
Total RNA was isolated from liver and muscle tissue using TRI Reagent (Ambion, Applied Biosystem, Austing, TX, USA) and PAXgene Blood RNA Kit (Qiagen BD, cat. no. 762165) was used for blood samples according to the manufacturer’s instructions. The quantity of the RNA was determined by spectrophotometer Nanodrop ND-1000 (Nanodrop Technologies) and by electrophoresis on 1% agarose gel. We assessed the integrity of the RNA on an Angilent 2100 Bioanalyzer (Agilent Technologies, CA, USA). All samples had an RNA integrity number (RIN) larger than 7. RNA samples were purified using RNeasy mini columns with DNase I (Qiagen). RNA-seq libraries were prepared using the TruSeq RNA sample preparation kit (Illumina) according to the manufacturer’s protocol. The RNA-seq libraries for A-bulls and D-bulls used the polyadenylated fraction of RNA from each animal by using modified protocol of Illumina sample preparation for RNA-Seq protocol (Illumina Inc) at AgriBio (Biosciences Research Centre, Bundoora, Victoria). The RNA-seq libraries for H-steers and H-heifers were prepared by Beijing Genomics Institute (Shengzhgen, China). RNA-seq libraries were sequenced on the HiSeq2000 sequencer (Illumina) in a 101-cycle paired end run. One hundred base paired end reads were called with CASAVA v1.8 and output in fastq format.
RNA-seq analysis
The software FastQC v0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/ fastqc/) was used to assess the quality of the RNA sequences, while Trimmomatic v0.36 [83] was used in the pre-processing step to remove the low-quality reads and adaptors. The software TopHat 2.0.5 was used with default parameters [84] mapped the cleaned reads to the bovine reference genome (Bos taurus, Ensembl UMD3.1) and HTSeq v0.6.1 [85] was used to assemble the reads. The mapping summary for all datasets is shown in (Table 7). The following steps where done in R software [86]. We filtered the genes with no expression and normalized the gene counts with the trimmed mean of M-values normalization (TMM) using the R package edgeR v3.18.1 [87].
Table 7.
Alignment summary of reads to the B. taurus reference genome (UMD3.1 Ensembl)
| Dataset | Input reads | Pair reads Mapped | Mapping rate | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Average | Min | Max | Total | Average | Min | Max | ||
| A_Liver | 199,721,632 | 7,397,097 | 3,968,542 | 12,494,809 | 167,142,561 | 6,190,465 | 3,123,525 | 11,360,856 | 82% |
| D_Muscle | 317,358,653 | 6,752,312 | 4,039,955 | 9,423,044 | 285,957,850 | 6,084,210 | 3,686,141 | 8,662,681 | 89% |
| H_Liver | 579,132,020 | 10,724,667 | 5,173,045 | 11,536,836 | 529,820,963 | 10,188,865 | 9,397,174 | 10,820,527 | 91% |
| H_Blood | 538,841,497 | 11,225,865 | 8,976,989 | 11,547,203 | 462,519,935 | 9,635,832 | 7,610,221 | 10,495,347 | 85% |
Linear regressions for RFI on the log2 copies per million (lcpm) were performed separately for each dataset (A-bulls_liver, bulls_muscle, H-heifers_blood, H-steers_blood, H-heifers_liver, and H-steers_liver) to find the genes significantly associated (GSA p < 0.05) with RFI from the set of genes that were within a 2 Mb window from the significant QTL found in the GWAS analysis. Additionally, we used expression from all the genes in the genome to select with p < 0.001 GSA genes (GSA p < 0.001). Based on the sign of the regression coefficient, a positive value indicated that the gene is up-regulated showing high expression, while a negative value indicates less gene expression abundance (down-regulated) with higher values for RFI (less efficient animals).
Supplementary information
Additional file 1: Table S1. Mapping complete information
Additional file 2: Figure S1. Q-Q plot of p-values for the GWAS for RFI.
Additional file 3: Table S2. Significant SNPs information.
Additional file 4: Table S3. Additive and dominance effects.
Additional file 5: Table S4. Information of the genes located near the significant SNPs.
Additional file 6: Table S5. Genes significantly associated at p < 0.001 with RFI.
Acknowledgements
The authors gratefully acknowledge the Angus Society of Australia for their support to this research and the access to the data used for this study through the Angus Sire Benchmarking Project. The authors also would like to extend the acknowledgements to the Angus bull breeders, co-operator herds, technicians, supply chain partners and research organizations for the support provided to the Angus Sire Benchmarking Project.
Meat and Livestock Australia provided funding for feed intake data collection (B.SBP.0089) and co-funding support for the ASBP (PSH.0528).
The gene expression work was founded by the Department of Agriculture, Fishery and Forest and Meat & Livestock Australia.
Abbreviations
- ADG
Average Daily Gain
- bp
base pairs
- BP
Biological Process
- BTA
Bos taurus autosome
- CC
Cellular components
- G
Genomic relationship matrix
- GSA
Genes Significantly Associated
- GWAS
Genome-wide Association Studies
- lcpm
log2 copies per million
- LD
Linkage Disequilibrium
- MAF
Minor Allele Frequencies
- Mb
Mega bases
- MF
Molecular function
- MWT
Metabolic mid weight
- QTL
Quantitative Trait Loci
- REML
Restricted maximum likelihood
- RFI
Residual Feed Intake
- RNA-seq
RNA sequencing
- SNP
Single Nucleotide Polymorphism
- TMM
Trimmed mean of M-values normalization
Authors’ contributions
SDLHS carried out the analyses of the genomic (quality control, imputation) and transcriptomic (quality control, pre-processing, mapping, assembly) datasets, performed the statistical analyses and wrote the manuscript. ND and SC participated in the imputation and interpretation of the genomic analyses. CG advised on the analyses of the genomic and transcriptomic datasets. JHJVDW assisted with the genomic analyses and writing of the paper. YC designed the experiment of the divergent lines and generated the RNA-seq datasets. All authors provided a critical review and approved the final manuscript.
Funding
Meat and Livestock Australia (MLA) have provided funding support for feed intake data collection (B.SBP.0089) and co-funding support for the Angus Sire Benchmarking Project (PSH.0528). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
Summary data of the analysis are included in this published article as Additional files. The RNA sequences raw data are available from the National Centre for Biotechnology Information Sequence Read Archive (SRA) under the accession BioProject numbers: PRJNA393239 and PRJNA579776. The genotypes used in this study are not stored publicly, however, this data is available from the Angus Society of Australia on reasonable request.
Ethics approval and consent to participate
For the GWAS study, all the procedures were managed according to the welfare guidelines established by the Australian Animal Welfare Standards and guidelines for cattle (Edition one 2013) approved by the University of New England Animal Ethics Committee (Approval No. AEC12–082).
The procedures involved in the RNA-seq experiment were approved by the University of New England Animal Ethics Committee (AEC 06/123, AEC14–002 and AEC14–036) and New South Wales Department of Primary Industries (NSW DPI) Animal Research Authority (ORA09/015, ORA 13/16/004).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sara de las Heras-Saldana, Email: sdelash2@une.edu.au.
Samuel A. Clark, Email: Sam.Clark@une.edu.au
Naomi Duijvesteijn, Email: nduijves@une.edu.au.
Cedric Gondro, Email: gondroce@msu.edu, Email: cgondro2@une.edu.au.
Julius H. J. van der Werf, Email: jvanderw@une.edu.au
Yizhou Chen, Email: yizhou.chen@dpi.nsw.gov.au.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-019-6270-4.
References
- 1.Arthur P, Archer J, Johnston D, Herd R, Richardson E, Parnell P. Genetic and phenotypic variance and covariance components for feed intake, feed efficiency, and other postweaning traits in Angus cattle. J Anim Sci. 2001;79(11):2805–2811. doi: 10.2527/2001.79112805x. [DOI] [PubMed] [Google Scholar]
- 2.Koch RM, Swiger LA, Chambers D, Gregory KE. Efficiency of feed use in beef cattle 1. J Anim Sci. 1963;22(2):486–494. doi: 10.2527/jas1963.222486x. [DOI] [Google Scholar]
- 3.Veerkamp R, Emmans G, Cromie A, Simm G. Variance components for residual feed intake in dairy cows. Livest Prod Sci. 1995;41(2):111–120. doi: 10.1016/0301-6226(94)00056-D. [DOI] [Google Scholar]
- 4.Herd RM, Bishop SC. Genetic variation in residual feed intake and its association with other production traits in British Hereford cattle. Livest Prod Sci. 2000;63(2):111–119. doi: 10.1016/S0301-6226(99)00122-0. [DOI] [Google Scholar]
- 5.De Haas Y, Windig J, Calus M, Dijkstra J, De Haan M, Bannink A, Veerkamp R. Genetic parameters for predicted methane production and potential for reducing enteric emissions through genomic selection. J Dairy Sci. 2011;94(12):6122–6134. doi: 10.3168/jds.2011-4439. [DOI] [PubMed] [Google Scholar]
- 6.Bolormaa S, Hayes B, Savin K, Hawken R, Barendse W, Arthur P, Herd R, Goddard M. Genome-wide association studies for feedlot and growth traits in cattle. J Anim Sci. 2011;89(6):1684–1697. doi: 10.2527/jas.2010-3079. [DOI] [PubMed] [Google Scholar]
- 7.Rolf MM, Taylor JF, Schnabel RD, McKay SD, McClure MC, Northcutt SL, Kerley MS, Weaber RL. Genome-wide association analysis for feed efficiency in Angus cattle. Anim Genet. 2012;43(4):367–374. doi: 10.1111/j.1365-2052.2011.02273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khansefid M, Pryce J, Bolormaa S, Miller S, Wang Z, Li C, Goddard M. Estimation of genomic breeding values for residual feed intake in a multibreed cattle population. J Anim Sci. 2014;92(8):3270–3283. doi: 10.2527/jas.2014-7375. [DOI] [PubMed] [Google Scholar]
- 9.Santana M, Ventura R, Utsunomiya Y, Neves H, Alexandre P, Oliveira Junior G, Gomes R, Bonin M, Coutinho L, Garcia J. A genomewide association mapping study using ultrasound-scanned information identifies potential genomic regions and candidate genes affecting carcass traits in Nellore cattle. J Anim Breed Genet. 2015;132(6):420–427. doi: 10.1111/jbg.12167. [DOI] [PubMed] [Google Scholar]
- 10.Seabury C, Oldeschulte D, Saatchi M, Beever J, Decker J, Halley Y, Bhattarai E, Molaei M, Freetly H, Hansen S. Genome-wide association study for feed efficiency and growth traits in US beef cattle. BMC Genomics. 2017;18(1):386. doi: 10.1186/s12864-017-3754-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Gondro C, Quinn K, Herd RM, Parnell PF, Vanselow B. Global gene expression profiling reveals genes expressed differentially in cattle with high and low residual feed intake. Anim Genet. 2011;42(5):475–490. doi: 10.1111/j.1365-2052.2011.02182.x. [DOI] [PubMed] [Google Scholar]
- 12.Tizioto PC, Coutinho LL, Decker JE, Schnabel RD, Rosa KO, Oliveira PS, Souza MM, Mourão GB, Tullio RR, Chaves AS. Global liver gene expression differences in Nelore steers with divergent residual feed intake phenotypes. BMC Genomics. 2015;16(1):242. doi: 10.1186/s12864-015-1464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khansefid M, Millen C, Chen Y, Pryce J, Chamberlain A, Vander Jagt C, Gondro C, Goddard M. Gene expression analysis of blood, liver, and muscle in cattle divergently selected for high and low residual feed intake. J Anim Sci. 2017;95(11):4764–4775. doi: 10.2527/jas2016.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexandre PA, Kogelman LJ, Santana MH, Passarelli D, Pulz LH, Fantinato-Neto P, Silva PL, Leme PR, Strefezzi RF, Coutinho LL. Liver transcriptomic networks reveal main biological processes associated with feed efficiency in beef cattle. BMC Genomics. 2015;16(1):1073. doi: 10.1186/s12864-015-2292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong B, Hudson N, Seo D, Lee S, Khatri B, Lassiter K, Cook D, Piekarski A, Dridi S, Anthony N. RNA sequencing for global gene expression associated with muscle growth in a single male modern broiler line compared to a foundational barred Plymouth rock chicken line. BMC Genomics. 2017;18(1):82. doi: 10.1186/s12864-016-3471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khadir A, Tiss A, Abubaker J, Abu-Farha M, Al-Khairi I, Cherian P, John J, Kavalakatt S, Warsame S, Al-Madhoun A. MAP kinase phosphatase DUSP1 is overexpressed in obese humans and modulated by physical exercise. Am J Physiol Endocrinol Metab. 2015;308(1):E71–E83. doi: 10.1152/ajpendo.00577.2013. [DOI] [PubMed] [Google Scholar]
- 17.Guimaraes JC, Zavolan M. Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol. 2016;17(1):236. doi: 10.1186/s13059-016-1104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karisa BK, Thomson J, Wang Z, Stothard P, Moore SS, Plastow GS. Candidate genes and single nucleotide polymorphisms associated with variation in residual feed intake in beef cattle. J Anim Sci. 2013;91(8):3502–3513. doi: 10.2527/jas.2012-6170. [DOI] [PubMed] [Google Scholar]
- 19.Kong RSG, Liang G, Chen Y, Stothard P. Transcriptome profiling of the rumen epithelium of beef cattle differing in residual feed intake. BMC Genomics. 2016;17(1):592. doi: 10.1186/s12864-016-2935-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saatchi M, Beever J, Decker J, Faulkner D, Freetly H, Hansen S, Yampara-Iquise H, Johnson K, Kachman S, Kerley M. QTLs associated with dry matter intake, metabolic mid-test weight, growth and feed efficiency have little overlap across 4 beef cattle studies. BMC Genomics. 2014;15(1):1004. doi: 10.1186/1471-2164-15-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramayo-Caldas Y, Fortes M, Hudson N, Porto-Neto L, Bolormaa S, Barendse W, Kelly M, Moore S, Goddard M, Lehnert S. A marker-derived gene network reveals the regulatory role of PPARGC1A, HNF4G, and FOXP3 in intramuscular fat deposition of beef cattle. J Anim Sci. 2014;92(7):2832–2845. doi: 10.2527/jas.2013-7484. [DOI] [PubMed] [Google Scholar]
- 22.Gagliardi AD, Kuo EY, Raulic S, Wagner GF, DiMattia GE. Human stanniocalcin-2 exhibits potent growth-suppressive properties in transgenic mice independently of growth hormone and IGFs. Am J Physiol Endocrinol Metab. 2005;288(1):E92–E105. doi: 10.1152/ajpendo.00268.2004. [DOI] [PubMed] [Google Scholar]
- 23.Mudadu MA, Porto-Neto LR, Mokry FB, Tizioto PC, Oliveira PS, Tullio RR, Nassu RT, Niciura SC, Tholon P, Alencar MM. Genomic structure and marker-derived gene networks for growth and meat quality traits of Brazilian Nelore beef cattle. BMC Genomics. 2016;17(1):235. doi: 10.1186/s12864-016-2535-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng Q, Wang K, Liu X, Zhou H, Xu L, Wang Z, Fang M. Identification of growth trait related genes in a Yorkshire purebred pig population by genome-wide association studies. Asian Australas J Anim Sci. 2017;30(4):462. doi: 10.5713/ajas.16.0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang T, Zhang X, Han K, Zhang G, Wang J, Xie K, Xue Q. Genome-wide analysis of lncRNA and mRNA expression during differentiation of abdominal preadipocytes in the chicken. G3. 2017;7:953–966. doi: 10.1534/g3.116.037069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bryan MS, Argos M, Pierce B, Tong L, Rakibuz-Zaman M, Ahmed A, Rahman M, Islam T, Yunus M, Parvez F. Genome-wide association studies and heritability estimates of body mass index related phenotypes in Bangladeshi adults. PLoS One. 2014;9(8):e105062. doi: 10.1371/journal.pone.0105062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Gao Y, Kim Y, Iqbal A, Kim J. A whole genome association study to detect additive and dominant single nucleotide polymorphisms for growth and carcass traits in Korean native cattle, Hanwoo. Asian Aust J Anim Sci. 2017;30(1):8. doi: 10.5713/ajas.16.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porter IM, Schleicher K, Porter M, Swedlow JR. Bod1 regulates protein phosphatase 2A at mitotic kinetochores. Nat Commun. 2013;4:2677. doi: 10.1038/ncomms3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keogh K, Waters SM, Cormican P, Kelly AK, Kenny DA. Effect of dietary restriction and subsequent re-alimentation on the transcriptional profile of bovine jejunal epithelium. PLoS One. 2018;13(3):e0194445. doi: 10.1371/journal.pone.0194445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto N, Yamamoto S, Inagaki F, Kawaichi M, Fukamizu A, Kishi N, Matsuno K, Nakamura K, Weinmaster G, Okano H. Role of Deltex-1 as a transcriptional regulator downstream of the notch receptor. J Biol Chem. 2001;276:45031–45040. doi: 10.1074/jbc.M105245200. [DOI] [PubMed] [Google Scholar]
- 31.Zhou N, Lee WR, Abasht B. Messenger RNA sequencing and pathway analysis provide novel insights into the biological basis of chickens’ feed efficiency. BMC Genomics. 2015;16(1):195. doi: 10.1186/s12864-015-1364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cassar-Malek I, Passelaigue F, Bernard C, Léger J, Hocquette J. Target genes of myostatin loss-of-function in muscles of late bovine fetuses. BMC Genomics. 2007;8(1):63. doi: 10.1186/1471-2164-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salleh MS, Mazzoni G, Höglund JK, Olijhoek DW, Lund P, Løvendahl P, Kadarmideen HN. RNA-Seq transcriptomics and pathway analyses reveal potential regulatory genes and molecular mechanisms in high-and low-residual feed intake in Nordic dairy cattle. BMC Genomics. 2017;18(1):258. doi: 10.1186/s12864-017-3622-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griger J, Schneider R, Lahmann I, Schöwel V, Keller C, Spuler S, Nazare M, Birchmeier C. Loss of Ptpn11 (Shp2) drives satellite cells into quiescence. Elife. 2017;6:e21552. doi: 10.7554/eLife.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gondret F, Vincent A, Houée-Bigot M, Siegel A, Lagarrigue S, Causeur D, Gilbert H, Louveau I. A transcriptome multi-tissue analysis identifies biological pathways and genes associated with variations in feed efficiency of growing pigs. BMC Genomics. 2017;18(1):244. doi: 10.1186/s12864-017-3639-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miquelajáuregui A, Varela-Echavarría A, Ceci ML, García-Moreno F, Ricano I, Hoang K, Frade-Pérez D, Portera-Cailliau C, Tamariz E, De Carlos JA. LIM-homeobox gene Lhx5 is required for normal development of Cajal–Retzius cells. J Neurosci. 2010;30(31):10551–10562. doi: 10.1523/JNEUROSCI.5563-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lear PV, González-Touceda D, Porteiro Couto B, Viaño P, Guymer V, Remzova E, Tunn R, Chalasani A, García-Caballero T, Hargreaves IP. Absence of intracellular ion channels TPC1 and TPC2 leads to mature-onset obesity in male mice, due to impaired lipid availability for thermogenesis in brown adipose tissue. Endocrinology. 2015;156(3):975–986. doi: 10.1210/en.2014-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindholm-Perry A, Kuehn L, Smith T, Ferrell C, Jenkins T, Freetly H, Snelling W. A region on BTA14 that includes the positional candidate genes LYPLA1, XKR4 and TMEM68 is associated with feed intake and growth phenotypes in cattle 1. Anim Genet. 2012;43(2):216–219. doi: 10.1111/j.1365-2052.2011.02232.x. [DOI] [PubMed] [Google Scholar]
- 39.de Oliveira Silva RM, Stafuzza NB, de Oliveira FB, de Camargo GMF, Ceacero TM, Cyrillo JNDSG, Baldi F, Boligon AA, Mercadante MEZ, Lourenco DL. Genome-wide association study for carcass traits in an experimental Nelore cattle population. PLoS One. 2017;12(1):e0169860. doi: 10.1371/journal.pone.0169860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niakan KK, Ji H, Maehr R, Vokes SA, Rodolfa KT, Sherwood RI, Yamaki M, Dimos JT, Chen AE, Melton DA. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev. 2010;24(3):312–326. doi: 10.1101/gad.1833510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pausch H, Flisikowski K, Jung S, Emmerling R, Edel C, Götz KU, Fries R. Genome-wide association study identifies two major loci affecting calving ease and growth-related traits in cattle. Genetics. 2011;187(1):289–297. doi: 10.1534/genetics.110.124057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong B, Song J, Lee J, Hargis B, Wing T, Lassiter K, Bottje W. Gene expression in breast muscle associated with feed efficiency in a single male broiler line using a chicken 44K oligo microarray. I. Top differentially expressed genes. Poult Sci. 2011;90(11):2535–2547. doi: 10.3382/ps.2011-01435. [DOI] [PubMed] [Google Scholar]
- 43.Perland E, Hellsten SV, Lekholm E, Eriksson MM, Arapi V, Fredriksson R. The novel membrane-bound proteins MFSD1 and MFSD3 are putative SLC transporters affected by altered nutrient intake. J Mol Neurosci. 2017;61(2):199–214. doi: 10.1007/s12031-016-0867-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chi H, Yang X, Kingsley PD, O’Keefe RJ, Puzas JE, Rosier RN, Shears SB, Reynolds PR. Targeted deletion of Minpp1 provides new insight into the activity of multiple inositol polyphosphate phosphatase in vivo. Mol Cell Biol. 2000;20(17):6496–6507. doi: 10.1128/MCB.20.17.6496-6507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin J, Conlon DM, Wang X, Von Nostrand E, Robano I, Park Y, Strong A, Radmanesh B, Barash Y, Rader DJ. RNA-binding protein A1CF modulates plasma triglyceride levels through posttranscriptional regulation of stress-induced VLDL secretion. BioRxiv. 2018:397554. 10.1101/397554.
- 46.Serao NV, Gonzalez-Pena D, Beever JE, Bollero GA, Southey BR, Faulkner DB, Rodriguez-Zas SL. Bivariate genome-wide association analysis of the growth and intake components of feed efficiency. PLoS One. 2013;8(10):e78530. doi: 10.1371/journal.pone.0078530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramaswamy G, Sohn P, Eberhardt A, Serra R. Altered responsiveness to TGF-β results in reduced Papss2 expression and alterations in the biomechanical properties of mouse articular cartilage. Arthritis Res Ther. 2012;14(2):R49. doi: 10.1186/ar3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Do DN, Ostersen T, Strathe AB, Mark T, Jensen J, Kadarmideen HN. Genome-wide association and systems genetic analyses of residual feed intake, daily feed consumption, backfat and weight gain in pigs. BMC Genet. 2014;15(1):27. doi: 10.1186/1471-2156-15-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Onteru SK, Gorbach DM, Young JM, Garrick DJ, Dekkers JC, Rothschild MF. Whole genome association studies of residual feed intake and related traits in the pig. PLoS One. 2013;8(6):e61756. doi: 10.1371/journal.pone.0061756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Y, Hou Y, Liu F, Liu A, Jing L, Zhao C, Luan Y, Miao Y, Zhao S, Li X. Transcriptome analysis reveals that Vitamin A metabolism in the liver affects feed efficiency in pigs. G3. 2016;6:3615–3624. doi: 10.1534/g3.116.032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu D, Miller S, Sargolzaei M, Kelly M, Vander Voort G, Caldwell T, Wang Z, Plastow G, Moore S. Genome-wide association analyses for growth and feed efficiency traits in beef cattle. J Anim Sci. 2013;91(8):3612–3633. doi: 10.2527/jas.2012-5716. [DOI] [PubMed] [Google Scholar]
- 52.Elzo M, Thomas M, Martinez C, Lamb G, Johnson D, Rae D, Wasdin J, Driver J. Genomic–polygenic evaluation of multibreed Angus–Brahman cattle for postweaning feed efficiency and growth using actual and imputed Illumina50k SNP genotypes. Livest Sci. 2014;159:1–10. doi: 10.1016/j.livsci.2013.11.005. [DOI] [Google Scholar]
- 53.Silva R, Fragomeni B, Lourenco D, Magalhães A, Irano N, Carvalheiro R, Canesin R, Mercadante M, Boligon A, Baldi F. Accuracies of genomic prediction of feed efficiency traits using different prediction and validation methods in an experimental Nelore cattle population. J Anim Sci. 2016;94(9):3613–3623. doi: 10.2527/jas.2016-0401. [DOI] [PubMed] [Google Scholar]
- 54.Chen L, Schenkel F, Vinsky M, Crews D, Jr, Li C. Accuracy of predicting genomic breeding values for residual feed intake in Angus and Charolais beef cattle. J Anim Sci. 2013;91(10):4669–4678. doi: 10.2527/jas.2013-5715. [DOI] [PubMed] [Google Scholar]
- 55.Kennedy BW, Van der Werf JH, Meuwissen TH. Genetic and statistical properties of residual feed intake. J Anim Sci. 1993;71(12):3239–3250. doi: 10.2527/1993.71123239x. [DOI] [PubMed] [Google Scholar]
- 56.Jepsen MR, Kløverpris S, Mikkelsen JH, Pedersen JH, Füchtbauer EM, Laursen LS, Oxvig C. Stanniocalcin-2 inhibits mammalian growth by proteolytic inhibition of the insulin-like growth factor axis. J Biol Chem. 2015;290(6):3430–3439. doi: 10.1074/jbc.M114.611665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnston J, Ramos-Valdes Y, Stanton LA, Ladhani S, Beier F, DiMattia GE. Human stanniocalcin-1 or-2 expressed in mice reduces bone size and severely inhibits cranial intramembranous bone growth. Transgenic Res. 2010;19(6):1017–1039. doi: 10.1007/s11248-010-9376-7. [DOI] [PubMed] [Google Scholar]
- 58.Porto Neto L, Bunch R, Harrison B, Barendse W. Variation in the XKR4 gene was significantly associated with subcutaneous rump fat thickness in indicine and composite cattle. Anim Genet. 2012;43(6):785–789. doi: 10.1111/j.1365-2052.2012.02330.x. [DOI] [PubMed] [Google Scholar]
- 59.Magalhães AF, de Camargo GM, GAF J, Gordo DG, Tonussi RL, Costa RB, Espigolan R, MdO R, Bresolin T, de Andrade WB. Genome-wide association study of meat quality traits in Nellore cattle. PloS One. 2016;11(6):e0157845. doi: 10.1371/journal.pone.0157845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cassar-Malek I, Boby C, Picard B, Reverter A, Hudson N. Molecular regulation of high muscle mass in developing blonde d’Aquitaine cattle foetuses. Biol Open. 2017;6(10):1483–1492. doi: 10.1242/bio.024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bolormaa S, Pryce JE, Zhang Y, Reverter A, Barendse W, Hayes BJ, Goddard ME. Non-additive genetic variation in growth, carcass and fertility traits of beef cattle. Genet Sel Evol. 2015;47(1):26. doi: 10.1186/s12711-015-0114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang J, Shen B, O’Connell JR, VanRaden PM, Cole JB, Ma L. Dissection of additive, dominance, and imprinting effects for production and reproduction traits in Holstein cattle. BMC Genomics. 2017;18(1):425. doi: 10.1186/s12864-017-3821-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hill WG, Goddard ME, Visscher PM. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 2008;4(2):e1000008. doi: 10.1371/journal.pgen.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee J, Karnuah AB, Rekaya R, Anthony NB, Aggrey SE. Transcriptomic analysis to elucidate the molecular mechanisms that underlie feed efficiency in meat-type chickens. Mol Gen Genomics. 2015;290(5):1673–1682. doi: 10.1007/s00438-015-1025-7. [DOI] [PubMed] [Google Scholar]
- 65.Fortes M, Lehnert S, Bolormaa S, Reich C, Fordyce G, Corbet N, Whan V, Hawken R, Reverter A. Finding genes for economically important traits: Brahman cattle puberty. Anim Prod Sci. 2012;52(3):143–150. doi: 10.1071/AN11165. [DOI] [Google Scholar]
- 66.Awda B, Miller S, Montanholi Y, Voort GV, Caldwell T, Buhr M, Swanson K. The relationship between feed efficiency traits and fertility in young beef bulls. Can J Anim Sci. 2013;93(2):185–192. doi: 10.4141/cjas2012-092. [DOI] [Google Scholar]
- 67.Wang Z, Colazo M, Basarab J, Goonewardene L, Ambrose D, Marques E, Plastow G, Miller S, Moore S. Impact of selection for residual feed intake on breeding soundness and reproductive performance of bulls on pasture-based multisire mating. J Anim Sci. 2012;90(9):2963–2969. doi: 10.2527/jas.2011-4521. [DOI] [PubMed] [Google Scholar]
- 68.Bottje WG, Lassiter K, Piekarski-Welsher A, Dridi S, Reverter A, Hudson NJ, Kong B. Proteogenomics reveals enriched ribosome assembly and protein translation in Pectoralis major of high feed efficiency pedigree broiler males. Front Physiol. 2017;8:306. doi: 10.3389/fphys.2017.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee S, Van Der Werf J, Kim N, Lee S, Gondro C, Park E, Oh S, Gibson J, Thompson J. QTL and gene expression analyses identify genes affecting carcass weight and marbling on BTA14 in Hanwoo (Korean cattle) Mamm Genome. 2011;22(9–10):589. doi: 10.1007/s00335-011-9331-9. [DOI] [PubMed] [Google Scholar]
- 70.Rescan P, Cam A, Rallière C, Montfort J. Global gene expression in muscle from fasted/refed trout reveals up-regulation of genes promoting myofibre hypertrophy but not myofibre production. BMC Genomics. 2017;18(1):447. doi: 10.1186/s12864-017-3837-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Olivieri BF, Mercadante MEZ, Cyrillo JNDSG, Branco RH, Bonilha SFM, de Albuquerque LG, de Oliveira Silva RM, Baldi F. Genomic regions associated with feed efficiency indicator traits in an experimental Nellore cattle population. PLoS One. 2016;11(10):e0164390. doi: 10.1371/journal.pone.0164390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen Y, Arthur P, Herd R, Quinn K, Barchia I. Using genes differentially expressed in bulls to classify steers divergently selected for high and low residual feed intake. Anim Prod Sci. 2012;52(7):608–612. doi: 10.1071/AN11266. [DOI] [Google Scholar]
- 73.Torres-Vázquez JA, van der Werf J, Clark SA. Genetic and phenotypic associations of feed efficiency with growth and carcass traits in Australian Angus cattle. J Anim Sci. 2018;96:4521–4531. doi: 10.1093/jas/sky325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Archer J, Arthur P, Herd R, Parnell P, Pitchford W. Optimum postweaning test for measurement of growth rate, feed intake, and feed efficiency in British breed cattle. J Anim Sci. 1997;75(8):2024–2032. doi: 10.2527/1997.7582024x. [DOI] [PubMed] [Google Scholar]
- 75.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sargolzaei M, Chesnais JP, Schenkel FS. A new approach for efficient genotype imputation using information from relatives. BMC Genomics. 2014;15(1):478. doi: 10.1186/1471-2164-15-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Graser H, Tier B, Johnston D, Barwick S. Genetic evaluation for the beef industry in Australia. Aust J Exp Agric. 2005;45(8):913–921. doi: 10.1071/EA05075. [DOI] [Google Scholar]
- 79.Lee SH, Van der Werf J. MTG2: an efficient algorithm for multivariate linear mixed model analysis based on genomic information. Bioinformatics. 2016;32(9):1420–1422. doi: 10.1093/bioinformatics/btw012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Falconer DS, Mackay T. Introduction to quantitative genetics. 4. 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Donoghue K, Arthur P, Wilkins J, Herd R. Onset of puberty and early-life reproduction in Angus females divergently selected for post-weaning residual feed intake. Anim Prod Sci. 2011;51(3):183–190. doi: 10.1071/AN10097. [DOI] [Google Scholar]
- 82.Herd R, Velazco J, Arthur P, Hegarty R. Associations among methane emission traits measured in the feedlot and in respiration chambers in Angus cattle bred to vary in feed efficiency. J Anim Sci. 2016;94(11):4882–4891. doi: 10.2527/jas.2016-0613. [DOI] [PubMed] [Google Scholar]
- 83.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.R core Team . R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 87.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Mapping complete information
Additional file 2: Figure S1. Q-Q plot of p-values for the GWAS for RFI.
Additional file 3: Table S2. Significant SNPs information.
Additional file 4: Table S3. Additive and dominance effects.
Additional file 5: Table S4. Information of the genes located near the significant SNPs.
Additional file 6: Table S5. Genes significantly associated at p < 0.001 with RFI.
Data Availability Statement
Summary data of the analysis are included in this published article as Additional files. The RNA sequences raw data are available from the National Centre for Biotechnology Information Sequence Read Archive (SRA) under the accession BioProject numbers: PRJNA393239 and PRJNA579776. The genotypes used in this study are not stored publicly, however, this data is available from the Angus Society of Australia on reasonable request.