Abstract
Symbiotic microbes are essential to the ecological success and evolutionary diversification of multicellular organisms. The establishment and stability of bipartite symbioses are shaped by mechanisms ensuring partner fidelity between host and symbiont. In this minireview, we demonstrate how the interface of chemical signals and host structures influences fidelity between legume root nodules and rhizobia, Hawaiian bobtail squid light organs and Allivibrio fischeri, and fungus-growing ant crypts and Pseudonocardia. Subsequently, we illustrate the morphological diversity and widespread phylogenetic distribution of specialized structures used by hosts to house microbial symbionts, indicating the importance of signal-structure interfaces across the history of multicellular life. These observations, and the insights garnered from well-studied bipartite associations, demonstrate the need to concentrate on the signal-structure interface in complex and multipartite systems, including the human microbiome.
Keywords: Allivibrio fischeri, crypt, fidelity, light organ, NCR peptides, root nodule, rhizobia, Pseudonocardia, signals, signaling, structures
Introduction
The diversity and complexity of plants and animals has been shaped, at least in part, through the formation of beneficial associations with symbiotic microbes [1]. The importance of beneficial symbioses in the evolution of plants and animals is illustrated by the virtual ubiquity of symbiotic microbes in aiding digestion across metazoans and by many plants engaging in mutualisms with mycorrhizal fungi. Despite the critical role symbiotic microbes play in shaping the biology of their hosts, our understanding of the formation, maintenance, and codiversification of symbiotic associations is limited, and largely informed by our understanding of a relatively small number of symbioses.
Symbiotic associations, by definition, are interactions that involve two or more species living together in an intimate association. Thus, the host-microbe interface—where the host and symbiont interact directly and exchange complex chemical signals—is fundamental for the establishment and maintenance of the symbiosis. The importance of this interface has been demonstrated through extensive studies of several model bipartite symbiosis, where high host specificity of single symbiont strains greatly facilitates studying how partner fidelity is shaped by chemical signaling and host physiology, behavior, and morphology. Herein, we illustrate the importance of the host-microbe interface in shaping the establishment and maintenance of symbiosis by reviewing three well-studied symbiotic systems involving elaborate structures: legume root nodules and rhizobia, Hawaiian bobtail squid light organs and Allivibrio fischeri, and fungus-farming ant crypts and Pseudonocardia. Finally, we document the widespread distribution of convergently evolved specialized structures for maintaining microbial symbionts across animals and plants and argue that deeper understanding of the general principles of symbiosis are possible through determining the chemical and physical underpinnings that shape the interface between hosts and symbiotic microbes.
Partner fidelity is maintained through cognate signal exchange in the legume root nodule-rhizobia symbiosis.
Many legumes acquire nitrogen through symbiosis with nitrogen-fixing rhizobia, which are housed in specialized structures called root nodules (Figure 1A). These structures accommodate the rhizobial symbionts in an anoxic environment suitable for nitrogenase activity [2]. During the establishment of this symbiosis, the rhizobia gain access into inner cortical root cells through infection threads and are able to directly exchange molecules with the legume host [3,4]. Consequently partner fidelity is obtained through legumes screening rhizobial symbionts from the milieu of incompatible microbes and potential pathogens occupying the rhizosphere [5].
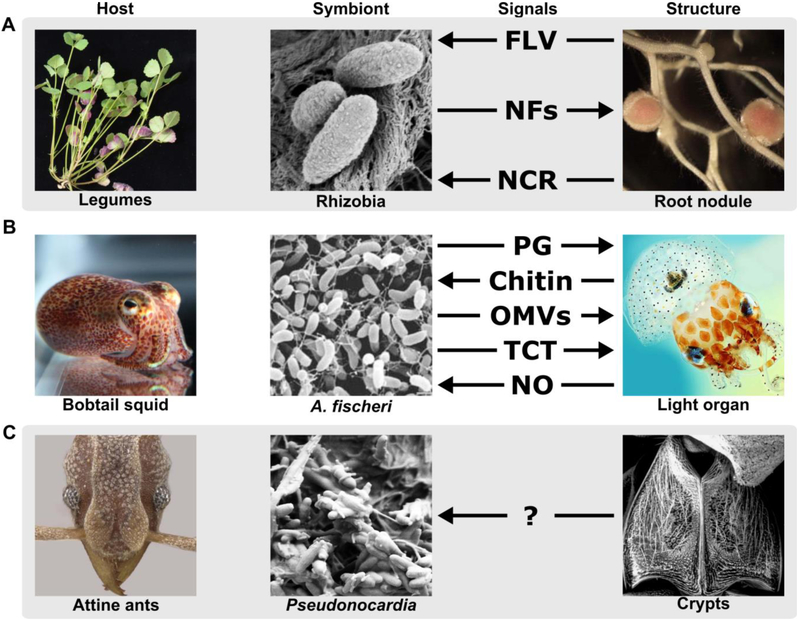
Figure 1. Signal-structure interfaces in bipartite associations between (A) legumes and rhizobia, (B) Hawaiian bobtail squid and A. fischeri, and (C) attine ants and Pseudonocardia.
The arrows indicate the direction of signals from producer to respondent and are arranged from top to bottom in each panel to indicate the order of signaling. FLV, flavonoids; NF, nodulation factors; NCR, nodule-specific cysteine-rich peptides; PG, peptidoglycan; OMVs; outer membrane vesicles; TCT, tracheal cytotoxin; NO, nitric oxide. Image credits: legume, rhizobia, and root nodule, Jean Michel Ané; bobtail squid, Mark Mandell; A. fischeri from [28]; light organ from Spencer Nyholm, under the terms of the Creative Commons Attribution License; attine ant, Ted Schultz; Pseudonocardia, Cameron Currie; ant crypt, Cameron Currie.
Nodulation begins when legume roots release flavonoids, which function as chemoattractants for rhizobia. Notably, different legume species produce distinct suites of flavonoids that attract coadapted rhizobial symbionts (recently reviewed in [6]). In turn, the coevolved rhizobia respond to the flavonoid signal by activating expression of nod genes to produce lipochitooligosaccharides called Nodulation Factors (NFs). NFs stimulate root hairs to curl and enclose the rhizobia, leading to infection thread formation, differentiation of the rhizobia into bacteroids, and ultimately to root nodulation (reviewed in [7,8]). Rhizobia produce specific NFs comprised of variable numbers of N-acetylglucosamine units that may be further modified by the addition of acetyl, carbamoyl, methyl, or variable acyl groups to the terminal non-reducing sugar and addition of acetate, D-arabinose, fucose, glycerol, or sulfate groups to the terminal reducing sugar [9]. Because NFs directly bind to legume membrane receptors called NF Receptors 1 and 5 [10,11], incompatible NF-receptor pairs will fail to initiate signal transduction in the host legume and prevents incompatible symbionts from stimulating nodule formation.
In addition to providing the rhizobia with an environment for nitrogen fixation, the root nodule facilitates the transfer of nodule-specific cysteine-rich (NCR) peptides from the host to the symbiont. These peptides are comprised of 30–50 variable amino acids with 4 or 6 conserved cysteine residues [12]. As many NCR peptides exhibit antimicrobial activity against rhizobia in vitro, it was initially thought NCR peptides function solely in rhizobia population control [13]. However, mutants of the model legume Medicago truncatula that are unable to produce specific NCR peptides form defective root nodules and are unable to fix nitrogen [14], suggesting that NCR peptides have additional functions in the symbiosis. Indeed, the antimicrobial activity of NCR peptides occurs at high concentrations [15], but at sublethal concentrations these peptides alter the bacterial transcriptional program. Specifically, NCR peptides activate regulons that are crucial for symbiosis [15,16], essential for rhizobia to adapt to the intracellular environment of their legume hosts, and necessary for their transition into differentiated bacteroids [17]. Furthermore, variation in NCR peptide sequence coupled with observations that differences in peptide regioisomeric and reduction state modulate the biological activity of these peptides [15,18]. These various modifications bestow NCR peptides with strain-specific activity toward different rhizobial species, which is exemplified by the large number of predicted NCR peptides encoded per genome (often >100) that control the resulting bacteroid’s morphology and size [19]. In summary, root nodule symbioses are formed, and partner fidelity is maintained through production of highly variable and specific chemical signals by both legume hosts and their coevolved rhizobial symbionts.
Non-specific signals and structural adaptations maintain partner fidelity in Hawaiian bobtail squid light organ symbiosis.
The nocturnal Hawaiian bobtail squid (Euprymna scolopes) masks its shadow from predators using counterillumination produced by the bioluminescent Gram-negative bacterium Aliivibrio fischeri (formerly Vibrio fischeri), which colonize a specialized structure called the light organ [20] (Figure 1B). Similar to the legume-rhizobia symbiosis, acquisition of A. fischeri by squid is horizontal and dependent upon screening to select symbionts from the greater ocean bacterial community [21]. As the bioluminescence produced by A. fischeri is essential for camouflage and bacterial colonization of the light organ is concomitant with irreversible morphological changes [22], there is strong selective pressure for squid to maintain partner fidelity and occlude non-bioluminescent bacteria from persistently colonizing the light organ.
Colonization of the light organ by A. fischeri occurs in three phases [23]. In the first phase, newly hatched squid pass microbe-laden seawater through their mantle cavity and over a pair of mucus-coated organs called the ciliated epithelial fields. In response to peptidoglycan derivatives, this appendage secretes mucus stores that facilitate bacterial aggregation on the ciliated epithelial fields [24]. During the second phase of colonization, the squid uses ciliary motions to draw aggregates toward pores at the base of the ciliated epithelial field and into portal ducts [24]. The final colonization phase occurs when A. fischeri migrate in response to a gradient of chitin oligosaccharides (chitobiose) produced in the duct and into the deep crypts of the light organ [23]. Analogous to bacteroid differentiation in the root nodule, when A. fischeri cells colonize the light organ, they proliferate and undergo transcriptional reprogramming [21], which results in the loss of flagella [25] and the release a diaminopimelic acid-type peptidoglycan fragment called tracheal cytotoxin and lipid A. Together these molecules act as synergistic signals to trigger widespread morphological changes that include apoptosis and regression of the ciliated fields [22]. However, it was recently discovered that A. fischeri shed outer membrane vesicles (OMVs) containing lipid A during proliferation in the light organ, which are devoid of tracheal cytotoxin, yet are sufficient to trigger equivalent morphological differentiation in the squid [26,27]. Moreover, exposure to OMVs shed by non-symbiotic Gram-negative bacteria induces hemocyte migration in squid, which is a hallmark for light organ morphogenesis [26]. Together, the non-specific signal exchange between A. fischeri and squid suggest that mechanisms to maintain partner fidelity must occur upstream of light organ colonization.
A major determinant of squid colonization specificity is biofilm formation by A. fischeri [28,29]. Furthermore, squid also contribute to partner fidelity with A. fischeri through specialized mucosal structures that function as intrinsic physical barriers to colonization [30] and by generating environmental conditions to select A. fischeri over non-symbiotic bacteria [31]. For instance, though peptidoglycan isolated from distantly related bacteria indiscriminately induces mucus secretion by the squid, these secretions contain galaxin protein EsGal1, which contains an antimicrobial repeat domain that inhibits the growth of Gram-positive bacteria in vitro [32]. During colonization bacteria are exposed to nitric oxide produced by squid [33]. By producing a flavohemoglobin [34] and an alternative oxidase [35] A. fischeri survive oxygen radical stress, which inhibits competing bacteria. In addition, nitric oxide may disperse A. fischeri biofilms and prompt entry of A. fischeri into the light organ ducts [36]. In specific response to small aggregates of A. fischeri (≤5 cells) the squid produce and secrete an enzyme called chitotriosidase into the mucus fields and ducts. This enzyme is an endochitinase that produces chitobiose, which primes A. fischeri for chemotaxis toward light organ ducts [37]. In conclusion, by coupling signaling to specialized structures, the squid maintains partner fidelity with A. fischeri, despite the reliance on generic microbial signals, such as lipopolysaccharide and peptidoglycan in the microbial world.
Behavioral and structural adaptations maintain partner fidelity in the Attine ant-Pseudonocardia symbiosis.
Attine ants engage in an ancient, coevolved, and obligate mutualism with fungi they cultivate for food in specialized gardens. The cultivated fungi are host to coevolved and specialized fungal parasites in the genus Escovopsis [38], which function as “crop diseases”, directly consuming the fungal cultivar [39,40]. To help defend their fungus garden from Escovopsis many attine ants engage in a mutualism with bacteria in the genus Pseudonocardia, which occur as exosymbionts, growing directly on the cuticle of worker ants and queens [41,42] (Figure 1C). Pseudonocardia produce antifungal antibiotics with potent and directed antagonistic properties towards Escovopsis [43,44].
New adults emerge from their pupal casing aposymbiotic, immediately after which Pseudonocardia-covered workers inoculate the new adult’s exoskeleton with the symbiont [45]. If a new adult worker does not acquire Pseudonocardia within the first two hours after emerging, it is no longer able to associate with the exosymbiont [45]. Thus, the specialized transmission behavior and narrow temporal window for acquisition helps ensure the specificity and maintenance of ant-Pseudonocardia symbiosis, as the chance for introduction of other microbial symbionts or opportunists is greatly reduced. Indeed, there is high partner fidelity between attine ants and Pseudonocardia: a single strain of Pseudonocardia colonizes worker ants and queens within an individual colony [46,47]. Across generations, partner fidelity is supported by maternal transmission: Pseudonocardia are transferred vertically between generations by colony-founding queens [41]. As predicted by vertical transmission, there is high relatedness of Pseudonocardia both between ant colonies within populations as well as across species within the attine ant genera [48,49], indicating that this partner fidelity is maintained over large evolutionary scales.
Just as with legumes and squid, attine ants have specialized structures and glands associated with the ant-Pseudonocardia interface. The structures can be elaborate, including large crypts or invaginations in the ant exoskeleton [42,50]. In most genera, the structures include tubercles, on which Pseudonocardia directly grow. These tubercles are connected to internal glands that provide nutrients for Pseudonocardia growth [51]. Although the chemical signals involved at the ant-Pseudonocardia interface remain to be determined, it is clear that this interface is critical to the stability and fidelity of the association. First, amino acid based stable isotope studies indicating that Pseudonocardia obtain all their nutrition directly from ants [51]. Second, by restricting the temporal window for Pseudonocardia acquisition before newly emerged adult workers leave the relatively hygienic environment of the fungus garden, it is improbable that these ants will become colonized by non-partner Pseudonocardia or other non-symbiotic microbes. Though the precise mechanisms are still under investigation, this narrow acquisition window is likely controlled by a programed disruption of gland cells that feed the Pseudonocardia. Third, ant-Pseudonocardia switching experiments have demonstrated significant reductions in Pseudonocardia acquisition in non-native pairings [52], indicating nutritional and glandular secretion coadaptation at finer phylogenetic levels. Fourth, phylogenomic analyses show that the anatomical modification to house Pseudonocardia have arisen independently at least three times throughout evolutionary history of attine ants [50]. Taken together, the above evidence indicates that the maintenance, stability, and fidelity of the ant-Pseudonocardia association is largely mediated by the chemical signaling occurring embedded within the interface of the symbiosis on the ant exoskeleton.
Conclusion
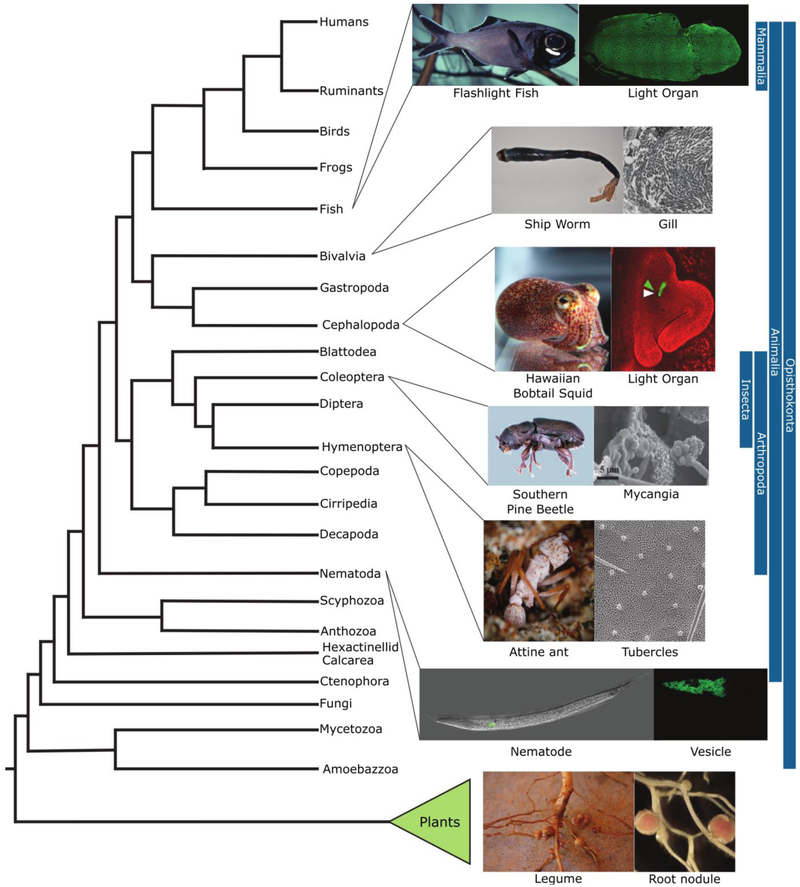
Signal-structure interfaces between hosts and their microbial symbionts are crucial for maintaining legume-rhizobia, squid-A. fischeri, and ant-Pseudonocardia partner fidelity (Figure 1). Integral to each symbiosis is the presence of modified morphological structures that are essential for maintaining the microbial symbionts and mediating host-symbiont signal exchange. Similar to these three well-studied bipartite associations highlighted above, elaborate structural modifications for housing microbial symbionts occur across plants and animals (Figure 2). For example, crypt structures that house fungal symbionts—mycangia—have evolved numerous times within beetles [53] and the crypts that house Pseudonocardia in attine ants evolved at least three independent times [50]. Similar to the light organ of bobtail squid, an analogous structure has convergently evolved in flashlight fish for harboring bioluminescent bacteria, including A. fischeri [54]. Notably, biofilm formation by A. fischeri is required for successful colonization of both squid and fish light organs. However, production of biofilm exopolysaccharide is regulated by distinct A. fischeri lineage-specific pathways, suggesting specific adaptations have arisen to modulate the signal-structure interface across different hosts [55]. In addition, there are many more examples of structural modifications that house symbiotic microbes across eukaryotes, including the gills of shipworms [56] and the vesicles of nematodes [57], . Taken together, this suggests host structural modification are critical to the host-microbe interface within bipartite symbioses.
Figure 2. Convergent evolution of structures to maintain microbial symbionts across the eukaryotic tree of life.
The phylogenetic tree shows the relationships between different eukaryotic lineages. Each inset represents an example of a eukaryotic host (left) from the specified lineage that uses a specialized structure (right) to establish and maintain a bipartite association with a microbial symbiont. Image credits: flashlight fish, Stefan Herlitze; flashlight fish light organ adapted from [69] under the terms of the Creative Commons Attribution License; ship worm and gill, Margo Haygood; bobtail squid and light organ, Mark Mandel; southern pine beetle, Erich Vallery under the terms of the Creative Commons Attribution License; mycangia, Kier Klepzig [70]; attine ant, Don Parsons; ant crypt, Cameron Currie; nematode and vesicle modified from [71] under the terms of the Creative Commons Attribution License; legume and root nodule, Jean Michel Ané.
Beyond bipartite host-microbe systems, within the widespread occurrence of microbiomes associating with diverse hosts, the presence of structural modifications at the host-microbe interface are much less recognized. These modifications may not be as well characterized in part because the associations between hosts and their microbiomes appear more plastic than bipartite associations. In animals, the majority of microbe-host interfaces occur in the gastrointestinal tract, which may harbor structural modifications to accommodate microbial symbionts that are analogous to the modifications observed in bipartite systems [58-64]. For example, both ruminants (Class Mammalia) and the hoatzin (Class Aves) have convergently evolved specialized structures, the rumen and the crop, respectively, in their foreguts that facilitate the growth of communities of fermentative microbes [65]. Further, examples showing concordance between host phylogeny and microbiome composition [66] and co-diversification of microbial symbionts with their hosts [67] suggest that there are suitable frameworks in place for the evolution of signal-structure interfaces in all animals, including mammals. It is tempting to hypothesize that the suite of antimicrobial peptides produced by the human immune system may be analogous to NCR peptides in modulating the physiology of microbes and to promote the growth of specific symbionts, as recently uncovered in amphibian systems [68]. Nevertheless, it is clear that signal-structure interfaces influence partner fidelity across the tree of life and these interfaces likely occur not only in bipartite associations, but also within more complex microbiomes. We believe more concerted efforts examining the signal-structure interfaces that occur across phylogenetically diverse host-microbe associations will help identify the general principles that govern the maintenance, stability, and coevolution of symbiotic systems.
Highlights.
Partner fidelity is essential for establishing and maintaining symbioses
Exchange of specific signals is one mechanism to ensure partner fidelity
Signal-structure interfaces evolve to ensure fidelity when using non-specific signals
Specialized structures for accommodating symbionts are widespread
Signal-structure interfaces are common and may occur within the human microbiome
Acknowledgements
We thank Caitlin Carlson for assistance in generating figures. We thank Jean-Michel Ané and Mark Mandel for providing images of legume root nodules and the bobtail squid light organ, respectively. We thank Heidi Horn, Daniel May, Mark Mandel, and Margaret Thairu for critical reading of the manuscript.
Funding
This project was supported through National Institutes of Health (NIH) U19 Al109673, NIH U19 TW009872). Reed M. Stubbendieck was supported by a National Library of Medicine training grant to the Computation and Informatics in Biology and Medicine Training Program (NLM 5T15LM007359).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kiers ET, West SA: Evolutionary biology. Evolving new organisms via symbiosis. Science 2015, 348:392–4. [DOI] [PubMed] [Google Scholar]
- 2.Ott T, van Dongen JT, Günther C, Krusell L, Desbrosses G, Vigeolas H, Bock V, Czechowski T, Geigenberger P, Udvardi MK: Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr Biol 2005, 15:531–5. [DOI] [PubMed] [Google Scholar]
- 3.Clarke VC, Loughlin PC, Day DA, Smith PMC: Transport processes of the legume symbiosome membrane. Front Plant Sci 2014, 5:699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke VC, Loughlin PC, Gavrin A, Chen C, Brear EM, Day DA, Smith PMC: Proteomic analysis of the soybean symbiosome identifies new symbiotic proteins. Mol Cell Proteomics 2015, 14:1301–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marchetti M, Capela D, Glew M, Cruveiller S, Chane-Woon-Ming B, Gris C, Timmers T, Poinsot V, Gilbert LB, Heeb P, et al. : Experimental evolution of a plant pathogen into a legume symbiont. PLoS Biol 2010, 8:e1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu C-W, Murray JD: The Role of Flavonoids in Nodulation Host-Range Specificity: An Update. Plants (Basel, Switzerland) 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desbrosses GJ, Stougaard J: Root nodulation: a paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe 2011, 10:348–58. [DOI] [PubMed] [Google Scholar]
- 8.Parniske M: Uptake of bacteria into living plant cells, the unifying and distinct feature of the nitrogen-fixing root nodule symbiosis. Curr Opin Plant Biol 2018, 44:164–174. [DOI] [PubMed] [Google Scholar]
- 9.Long SR: Rhizobium symbiosis: nod factors in perspective. Plant Cell 1996, 8:1885–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al. : A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 2003, 425:637–40. [DOI] [PubMed] [Google Scholar]
- 11.Broghammer A, Krusell L, Blaise M, Sauer J, Sullivan JT, Maolanon N, Vinther M, Lorentzen A, Madsen EB, Jensen KJ, et al. : Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc Natl Acad Sci U S A 2012, 109:13859–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alunni B, Kevei Z, Redondo-Nieto M, Kondorosi A, Mergaert P, Kondorosi E: Genomic organization and evolutionary insights on GRP and NCR genes, two large nodule-specific gene families in Medicago truncatula. Mol Plant Microbe Interact 2007, 20:1138–48. [DOI] [PubMed] [Google Scholar]
- 13.Haag AF, Baloban M, Sani M, Kerscher B, Pierre O, Farkas A, Longhi R, Boncompagni E, Hérouart D, Dall’angelo S, et al. : Protection of Sinorhizobium against host cysteine-rich antimicrobial peptides is critical for symbiosis. PLoS Biol 2011, 9:e1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim M, Chen Y, Xi J, Waters C, Chen R, Wang D: An antimicrobial peptide essential for bacterial survival in the nitrogen-fixing symbiosis. Proc Natl Acad Sci U S A 2015, 112:15238–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shabab M, Arnold MFF, Penterman J, Wommack AJ, Bocker HT, Price PA, Griffitts JS, Nolan EM, Walker GC: Disulfide cross-linking influences symbiotic activities of nodule peptide NCR247. Proc Natl Acad Sci U S A 2016, 113:10157–62.••This study takes a systematic structure-activity relationship approach to characterize the function of the conserved cysteine residues in NCR peptides and found that each peptide variant had different biological activities, suggesting that legume-rhizobia interactions may be modulated by peptide modifications.
- 16.Penterman J, Abo RP, De Nisco NJ, Arnold MFF, Longhi R, Zanda M, Walker GC: Host plant peptides elicit a transcriptional response to control the Sinorhizobium meliloti cell cycle during symbiosis. Proc Natl Acad Sci U S A 2014, 111:3561–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horváth B, Domonkos Á, Kereszt A, Szűcs A, Ábrahám E, Ayaydin F, Bóka K, Chen Y, Chen R, Murray JD, et al. : Loss of the nodule-specific cysteine rich peptide, NCR169, abolishes symbiotic nitrogen fixation in the Medicago truncatula dnf7 mutant. Proc Natl Acad Sci U S A 2015, 112:15232–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, Liu J, Li H, Yang S, Körmöczi P, Kereszt A, Zhu H: Nodule-Specific Cysteine-Rich Peptides Negatively Regulate Nitrogen-Fixing Symbiosis in a Strain-Specific Manner in Medicago truncatula. Mol Plant-Microbe Interact 2018, 31:240–248.•Using crosses of recombinant inbred lines, this study shows that legume-rhizobia partner fidelity at the level of nitrogen fixation is controlled by NCR peptides. Wang and colleagues validate these findings using chemically synthesized NCR peptides.
- 19.Herczeg R, Montiel J, Bálint B, Downie JA, Farkas A, Mergaert P, Bihari P, Kondorosi É, Kereszt A: Morphotype of bacteroids in different legumes correlates with the number and type of symbiotic NCR peptides. Proc Natl Acad Sci 2017, 114:5041–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones BW, Nishiguchi MK: Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Mar Biol 2004, 144:1151–1155. [Google Scholar]
- 21.Thompson LR, Nikolakakis K, Pan S, Reed J, Knight R, Ruby EG: Transcriptional characterization of Vibrio fischeri during colonization of juvenile Euprymna scolopes. Environ Microbiol 2017, 19:1845–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koropatnick T, Goodson MS, Heath-Heckman EAC, McFall-Ngai M: Identifying the cellular mechanisms of symbiont-induced epithelial morphogenesis in the squid-Vibrio association. Biol Bull 2014, 226:56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandel MJ, Schaefer AL, Brennan CA, Heath-Heckman EAC, Deloney-Marino CR, McFall-Ngai MJ, Ruby EG: Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Appl Environ Microbiol 2012, 78:4620–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ: Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci U S A 2000, 97:10231–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruby EG, Asato LM: Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol 1993, 159:160–7.•This is the first study to demonstrate that OMVs shed by A. fischeri during normal growth are sufficient to trigger morphological differentiation of the squid host, independent of tracheal cytotoxin.
- 26.Aschtgen M-S, Wetzel K, Goldman W, McFall-Ngai M, Ruby E: Vibrio fischeri-derived outer membrane vesicles trigger host development. Cell Microbiol 2016, 18:488–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aschtgen M-S, Lynch JB, Koch E, Schwartzman J, McFall-Ngai M, Ruby E: Rotation of Vibrio fischeri Flagella Produces Outer Membrane Vesicles That Induce Host Development. J Bacteriol 2016, 198:2156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yip ES, Grublesky BT, Hussa EA, Visick KL: A novel, conserved cluster of genes promotes symbiotic colonization and σ54-dependent biofilm formation by Vibrio fischeri. Mol Microbiol 2005, 57:1485–1498. [DOI] [PubMed] [Google Scholar]
- 29.Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG: A single regulatory gene is sufficient to alter bacterial host range. Nature 2009, 458:215–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nyholm SV, Deplancke B, Gaskins HR, Apicella MA, McFall-Ngai MJ: Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl Environ Microbiol 2002, 68:5113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nyholm SV, McFall-Ngai MJ: Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: the first site of symbiont specificity. Appl Environ Microbiol 2003, 69:3932–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heath-Heckman EAC, Gillette AA, Augustin R, Gillette MX, Goldman WE, McFall-Ngai MJ: Shaping the microenvironment: evidence for the influence of a host galaxin on symbiont acquisition and maintenance in the squid-Vibrio symbiosis. Environ Microbiol 2014, 16:3669–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ: NO means “yes” in the squid-vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell Microbiol 2004, 6:1139–51. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Dunn AK, Wilneff J, McFall-Ngai MJ, Spiro S, Ruby EG: Vibrio fischeri flavohaemoglobin protects against nitric oxide during initiation of the squid-Vibrio symbiosis. Mol Microbiol 2010, 78:903–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunn AK: Alternative Oxidase Activity Reduces Stress in Vibrio fischeri Cells Exposed to Nitric Oxide. J Bacteriol 2018, 200:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson CM, Tischler AH, Tarnowski DA, Mandel MJ, Visick KL: Nitric oxide inhibits biofilm formation by Vibrio fischeri via the nitric oxide sensor HnoX. Mol Microbiol 2019, 111:187–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kremer N, Philipp EER, Carpentier M-C, Brennan CA, Kraemer L, Altura MA, Augustin R, Häsler R, Heath-Heckman EAC, Peyer SM, et al. : Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe 2013, 14:183–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Currie CR, Wong B, Stuart AE, Schultz TR, Rehner SA, Mueller UG, Sung G-H, Spatafora JW, Straus NA: Ancient tripartite coevolution in the attine ant-microbe symbiosis. Science 2003, 299:386–8. [DOI] [PubMed] [Google Scholar]
- 39.Reynolds HT, Currie CR: Pathogenicity of Escovopsis weberi: The parasite of the attine ant-microbe symbiosis directly consumes the ant-cultivated fungus. Mycologia 2004, 96:955–9. [PubMed] [Google Scholar]
- 40.Custodio BC, Rodrigues A: Escovopsis kreiselii specialization to its native hosts in the fungiculture of the lower attine ant Mycetophylax morschi. Antonie Van Leeuwenhoek 2019, 112:305–317. [DOI] [PubMed] [Google Scholar]
- 41.Currie CR, Scott JA, Summerbell RC, Malloch D: Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 1999, 398:701–704. [Google Scholar]
- 42.Currie CR, Poulsen M, Mendenhall J, Boomsma JJ, Billen J: Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science 2006, 311:81–3. [DOI] [PubMed] [Google Scholar]
- 43.Oh DC, Scott JJ, Currie CR, Clardy J: Mycangimycin, a polyene peroxide from a mutualist Streptomyces sp. Org Lett 2009, 11:633–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Arnam EB, Ruzzini AC, Sit CS, Horn H, Pinto-Tomás AA, Currie CR, Clardy J: Selvamicin, an atypical antifungal polyene from two alternative genomic contexts. Proc Natl Acad Sci U S A 2016, 113:12940–12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marsh SE, Poulsen M, Pinto-Tomás A, Currie CR: Interaction between workers during a short time window is required for bacterial symbiont transmission in Acromyrmex leaf-cutting ants. PLoS One 2014, 9:e103269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poulsen M, Cafaro M, Boomsma JJ, Currie CR: Specificity of the mutualistic association between actinomycete bacteria and two sympatric species of Acromyrmex leaf-cutting ants. Mol Ecol 2005, 14:3597–604. [DOI] [PubMed] [Google Scholar]
- 47.Andersen SB, Hansen LH, Sapountzis P, Sørensen SJ, Boomsma JJ: Specificity and stability of the Acromyrmex-Pseudonocardia symbiosis. Mol Ecol 2013, 22:4307–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cafaro MJ, Poulsen M, Little AEF, Price SL, Gerardo NM, Wong B, Stuart AE, Larget B, Abbot P, Currie CR: Specificity in the symbiotic association between fungus-growing ants and protective Pseudonocardia bacteria. Proceedings Biol Sci 2011, 278:1814–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDonald BR, Chevrette MG, Klassen JL, Horn HA, Caldera EJ, Wendt-Pienkowski E, Cafaro MJ, Ruzzini AC, Van Arnam EB, Weinstock GM, et al. : Biogeography and Microscale Diversity Shape the Biosynthetic Potential of Fungus-growing Ant-associated Pseudonocardia. bioRxiv 2019, doi: 10.1101/545640. [DOI] [Google Scholar]
- 50.Li H, Sosa-Calvo J, Horn HA, Pupo MT, Clardy J, Rabeling C, Schultz TR, Currie CR: Convergent evolution of complex structures for ant-bacterial defensive symbiosis in fungus-farming ants. Proc Natl Acad Sci U S A 2018, doi: 10.1073/pnas.1809332115.••By reconstructing the evolutionary history of the ant-Pseudonocardia defensive symbiosis, this study reveals the convergent evolution of anatomical adaptations for maintaining beneficial symbionts in fungus-growing ants.
- 51.Steffan SA, Chikaraishi Y, Currie CR, Horn H, Gaines-Day HR, Pauli JN, Zalapa JE, Ohkouchi N: Microbes are trophic analogs of animals. Proc Natl Acad Sci U S A 2015, 112:15119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Armitage SAO, Broch JF, Marín HF, Nash DR, Boomsma JJ: Immune defense in leaf-cutting ants: a cross-fostering approach. Evolution 2011, 65:1791–9. [DOI] [PubMed] [Google Scholar]
- 53.Hulcr J, Stelinski LL: The Ambrosia Symbiosis: From Evolutionary Ecology to Practical Management. Annu Rev Entomol 2017, 62:285–303. [DOI] [PubMed] [Google Scholar]
- 54.Haygood MG: Light organ symbioses in fishes. Crit Rev Microbiol 1993, 19:191–216. [DOI] [PubMed] [Google Scholar]
- 55.Rotman ER, Bultman KM, Brooks JF, Gyllborg MC, Burgos HL, Wollenberg MS, Mandel MJ: Natural Strain Variation Reveals Diverse Biofilm Regulation in Squid-Colonizing Vibrio fischeri. J Bacteriol 2019, 201:1–20.••This study surveys A. fischeri isolates worldwide, finds that biofilm formation is a requirement for successful squid colonization, and shows that distinct lineages of A. fischeri have evolved alternative mechanisms to regulate the biofilm formation pathway.
- 56.Distel DL, Altamia MA, Lin Z, Shipway JR, Han A, Forteza I, Antemano R, Limbaco MGJP, Tebo AG, Dechavez R, et al. : Discovery of chemoautotrophic symbiosis in the giant shipworm Kuphus polythalamia (Bivalvia: Teredinidae) extends wooden-steps theory. Proc Natl Acad Sci U S A 2017, 114:E3652–E3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martens EC, Goodrich-Blair H: The Steinernema carpocapsae intestinal vesicle contains a subcellular structure with which Xenorhabdus nematophila associates during colonization initiation. Cell Microbiol 2005, 7:1723–35. [DOI] [PubMed] [Google Scholar]
- 58.Kikuchi Y, Hosokawa T, Fukatsu T: An ancient but promiscuous host-symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J 2011, 5:446–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohbayashi T, Takeshita K, Kitagawa W, Nikoh N, Koga R, Meng X-Y, Tago K, Hori T, Hayatsu M, Asano K, et al. : Insect’s intestinal organ for symbiont sorting. Proc Natl Acad Sci U S A 2015, 112:E5179–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alonso-Pernas P, Arias-Cordero E, Novoselov A, Ebert C, Rybak J, Kaltenpoth M, Westermann M, Neugebauer U, Boland W: Bacterial Community and PHB-Accumulating Bacteria Associated with the Wall and Specialized Niches of the Hindgut of the Forest Cockchafer (Melolontha hippocastani). Front Microbiol 2017, 8:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oishi S, Moriyama M, Koga R, Fukatsu T: Morphogenesis and development of midgut symbiotic organ of the stinkbug Plautia stali (Hemiptera: Pentatomidae). Zool Lett 2019, 5:16.•Examining midgut morphogenesis in the stinkbug Plautia stali, Oishi and colleagues show the close integration of microbial symbionts with host gut development.
- 62.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV: Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A 2008, 105:20858–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Earle KA, Billings G, Sigal M, Lichtman JS, Hansson GC, Elias JE, Amieva MR, Huang KC, Sonnenburg JL: Quantitative Imaging of Gut Microbiota Spatial Organization. Cell Host Microbe 2015, 18:478–88.••This study demonstrates the importance of spatial segregation of microbes within microhabitats in the gastrointestinal tract, and suggests that distinct signal-structure interfaces may exist in the gastrointestinal tract.
- 64.Mark Welch JL, Hasegawa Y, McNulty NP, Gordon JI, Borisy GG: Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc Natl Acad Sci U S A 2017, 114:E9105–E9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Godoy-Vitorino F, Goldfarb KC, Karaoz U, Leal S, Garcia-Amado MA, Hugenholtz P, Tringe SG, Brodie EL, Dominguez-Bello MG: Comparative analyses of foregut and hindgut bacterial communities in hoatzins and cows. ISME J 2012, 6:531–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brooks AW, Kohl KD, Brucker RM, van Opstal EJ, Bordenstein SR: Phylosymbiosis: Relationships and Functional Effects of Microbial Communities across Host Evolutionary History. PLoS Biol 2016, 14:e2000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moeller AH, Caro-Quintero A, Mjungu D, Georgiev AV., Lonsdorf EV, Muller MN, Pusey AE, Peeters M, Hahn BH, Ochman H: Cospeciation of gut microbiota with hominids. Science 2016, 353:380–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woodhams DC, Rollins-Smith LA, Reinert LK, Lam BA, Harris RN, Briggs CJ, Vredenburg VT, Patel BT, Caprioli RM, Chaurand P, et al. : Probiotics Modulate a Novel Amphibian Skin Defense Peptide That Is Antifungal and Facilitates Growth of Antifungal Bacteria. Microb Ecol 2019, doi: 10.1007/s00248-019-01385-9. [DOI] [PubMed] [Google Scholar]
- 69.Hellinger J, Jägers P, Donner M, Sutt F, Mark MD, Senen B, Tollrian R, Herlitze S: The Flashlight Fish Anomalops katoptron Uses Bioluminescent Light to Detect Prey in the Dark. PLoS One 2017, 12:e0170489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuceer C, Hsu C-Y, Erbilgin N, Klepzig KD: Ultrastructure of the mycangium of the southern pine beetle, Dendroctonus frontalis (Coleoptera: Curculionidae, Scolytinae): complex morphology for complex interactions. Acta Zool 2011, 92:216–224. [Google Scholar]
- 71.Stilwell MD, Cao M, Goodrich-Blair H, Weibel DB: Studying the Symbiotic Bacterium Xenorhabdus nematophila in Individual, Living Steinernema carpocapsae Nematodes Using Microfluidic Systems. mSphere 2018, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]