Abstract
Background
Well differentiated liposarcoma (WDLPS) can be difficult to distinguish from lipoma. Currently, this distinction is made by testing for MDM2 amplification, which requires a biopsy. The aim of this study was to develop a noninvasive method to predict MDM2 amplification status using radiomics features derived from MRI.
Methods
Patients with an MDM2‐negative lipoma or MDM2‐positive WDLPS and a pretreatment T1‐weighted MRI scan who were referred to Erasmus MC between 2009 and 2018 were included. When available, other MRI sequences were included in the radiomics analysis. Features describing intensity, shape and texture were extracted from the tumour region. Classification was performed using various machine learning approaches. Evaluation was performed through a 100 times random‐split cross‐validation. The performance of the models was compared with the performance of three expert radiologists.
Results
The data set included 116 tumours (58 patients with lipoma, 58 with WDLPS) and originated from 41 different MRI scanners, resulting in wide heterogeneity in imaging hardware and acquisition protocols. The radiomics model based on T1 imaging features alone resulted in a mean area under the curve (AUC) of 0·83, sensitivity of 0·68 and specificity of 0·84. Adding the T2‐weighted imaging features in an explorative analysis improved the model to a mean AUC of 0·89, sensitivity of 0·74 and specificity of 0·88. The three radiologists scored an AUC of 0·74 and 0·72 and 0·61 respectively; a sensitivity of 0·74, 0·91 and 0·64; and a specificity of 0·55, 0·36 and 0·59.
Conclusion
Radiomics is a promising, non‐invasive method for differentiating between WDLPS and lipoma, outperforming the scores of the radiologists. Further optimization and validation is needed before introduction into clinical practice.
The distinction between well differentiated liposarcomas and lipomas is needed in order to provide the correct treatment approach for patients with these tumours, but is difficult to make without a biopsy. This study shows that radiomics can be used to make this distinction based on routinely obtained MRI. The resulting radiomics model performed better in classifying these lipomatous tumours than three expert radiologists, thus showing potential as a non‐invasive diagnostic aid.
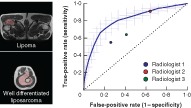
Antecedentes
Es difícil distinguir los liposarcomas bien diferenciados (well‐differentiated liposarcomas, WDLPS) de los lipomas. En la actualidad, esta distinción se realiza mediante la prueba de amplificación del gen MDM2 por biopsia. El objetivo de este estudio fue predecir de forma no invasiva el estado de amplificación del gen MDM2 para diferenciar los lipomas de los WDLPS utilizando características radiómicas a partir de la resonancia magnética.
Métodos
Se incluyeron los pacientes remitidos al instituto Erasmus MC entre 2009‐2018 por un lipoma MDM2 negativo o WDLPS MDM2 positivo y las resonancias magnéticas potenciadas en T1 correspondientes antes del tratamiento. Cuando estaban disponibles, se incluyeron otras secuencias de MRI en el análisis radiómico. Se describieron la intensidad, forma y textura de la región tumoral. Para la clasificación se utilizaron varios modelos de aprendizaje automático (machine learning). La evaluación se realizó mediante una validación cruzada aleatoria 100x. Se comparó el rendimiento de los modelos con la clasificación realizada por tres radiólogos expertos.
Resultados
Se incluyeron 116 pacientes (58 lipomas, 58 WDLPS) y 41 aparatos de MRI, con una gran heterogeneidad en las técnicas y protocolos para la adquisición de imágenes. El modelo radiómico basado únicamente en las características de las imagen en T1 dio como resultado una AUC media de 0,83, con una sensibilidad de 0,68 y una especificidad de 0,84. Un análisis adicional incorporando las imágenes ponderadas en T2 mejoró el modelo con una AUC media de 0,89, una sensibilidad de 0,74 y una especificidad de 0,88. Los tres radiólogos obtuvieron una AUC de 0,74/0,72/0,61, una sensibilidad de 0,74/0,91/0,64 y una especificidad de 0,55/0,36/0,59, respectivamente.
Conclusión
La radiómica es un método prometedor y no invasivo para diferenciar entre WDLPS y lipomas, superando la valoración de los radiólogos. Sin embargo, se necesita la optimización y validación de esta técnica antes de su introducción en la práctica clínica diaria.
Introduction
Lipomatous tumours are the most commonly observed soft tissue tumours, mostly owing to the high incidence of benign lipomas. Also within the malignant spectrum of soft tissue tumours (soft tissue sarcomas), liposarcoma is among the most frequently observed subtype1. Well differentiated liposarcoma (WDLPS) represents the largest subgroup of liposarcomas; these low‐grade, locally aggressive tumours are characterized by amplification of the MDM2 gene1. In rare cases, WDLPS can progress into a more aggressive subtype: dedifferentiated liposarcoma (DDLPS), which has a poorer prognosis1.
Several differences between lipoma and WDLPS on MRI have been described in the literature: size, location, tumour depth and intratumour heterogeneity. However, as there can be considerable overlap between these features, distinguishing between the two tumour types remains difficult, even for trained radiologists2, 3, 4, 5, 6. As the differences between lipoma/WDLPS and DDLPS are more obvious, this distinction can accurately be made solely by eye5, 7, 8, 9, 10.
An accurate diagnosis is needed to provide patients with the correct treatment and follow‐up. Whereas lipomas do not necessarily need to be excised, patients with WDLPS are generally considered candidates for surgery11. Currently, the standard way to differentiate lipoma from WDLPS is through a biopsy, which is tested for MDM2 amplification using fluorescence in situ hybridization (FISH). Amplification of the MDM2 gene is present in WDLPS, but absent in lipoma1, 12, 13. Taking a biopsy is an invasive and painful procedure for the patient, and is associated with risks, depending on tumour location, and potential sampling error.
The field of radiomics is based on the hypothesis that there is a relationship between medical imaging features and the underlying biological information, such as genetic aberrations14. Radiomics approaches have already been used in soft tissue sarcomas to predict other outcomes, such as differentiating between benign and malignant soft tissue tumours in general (not specifically lipomatous tumours)15, between intermediate‐ and high‐grade soft tissue sarcomas16, and predicting the risk of lung metastases from soft tissue sarcoma of the extremities17. Based on these results, it was hypothesized that radiomics might also be able to differentiate WDLPS from lipoma.
The aim of this study was to develop a model that predicts MDM2 amplification status using a radiomics approach, thereby differentiating WDLPS from lipoma. MRI scans obtained during routine diagnostic evaluation were used. Additionally, the performance of this model was compared with that of three trained radiologists reading the images. Finally, patients with DDLPS were included and classified by the radiologists to confirm that these tumours have distinct imaging features and can be identified without the help of additional models or tests.
Methods
Patients with a pathologically confirmed diagnosis of lipoma, WDLPS or DDLPS, a known MDM2 amplification status tested by FISH, and with at least a T1‐weighted MRI sequence available before treatment (if applicable) were included. All patients were either referred to/discussed at, or diagnosed/treated at the Erasmus MC Cancer Institute, Rotterdam, the Netherlands, between December 2009 and August 2018. As a result, some of the MRI scans were made in the referring hospitals. The study was reviewed and approved by the local medical ethics review committee (MEC‐2016‐339), and performed in accordance with national and international legislation. Need for informed consent was waived owing to the retrospective and anonymized nature of the study.
To explore the potential predictive value of different MRI sequences, several additional sequences were included, when available. Based on their use in clinical practice, the sequences were grouped into: plain T1 (T1); T1 with fat saturation (T1‐FS) including T1 inversion recovery (IR) approaches (T1‐IR; a combination of Spectral Presaturation with Inversion Recovery (SPIR), Short‐TI Inversion Recovery (STIR), Spectral Attenuated Inversion Recovery (SPAIR) and Turbo Inversion Recovery Magnitude (TIRM)); T1 with gadolinium contrast (T1‐GD); T1 with fat saturation and gadolinium contrast (T1‐FS‐GD) including T1‐IR with GD; T2 imaging (T2) including T2‐Fast Field Echo (T2FFE) and T2*; and T2‐FS including T2‐IR.
Segmentation
The lipoma and WDLPS lesions were segmented semiautomatically on the T1 images to indicate the regions of interest (ROIs)18. All images were segmented independently by either a medical masters student or a PhD candidate with an MD degree. Both were blinded to the type of lipomatous tumour. To validate segmentation accuracy, a sample set was verified by a musculoskeletal radiologist, specialized in soft tissue sarcomas. Median tumour size, defined as the maximum diameter in centimetres, and tumour volume, with corresponding i.q.r. values, were extracted from the segmentations. The DDLPS images were used only for visual classification by the radiologists, and therefore not segmented.
To transfer the segmentations to the other sequences, all sequences were spatially aligned to the T1 sequence using automated image registration (elastix software19), thereby compensating for patient movement between scans. Quality assurance was done by visual inspection.
Radiomics feature extraction
Quantitative imaging features related to intensity, shape and texture were extracted from the ROIs using PyRadiomics software20, 21. More details can be found in Appendix S1 (supporting information). The shape features quantified were morphological properties such as volume and similarity to a circle. Intensity features were quantified using first‐order statistics such as the mean and standard deviation. Texture features quantified more complex properties, such as the presence of heterogeneity and speckle patterns. When a scan type was missing for a patient, the feature values for the missing image type were imputed.
Additional features
Several additional features were selected based on the available literature and clinical relevance, including patient characteristics (age, sex and tumour location (extremity, trunk, head and neck or pelvis)) and manually scored features (tumour depth (superficial or deep), unilobular or multilobular tumour, atypical appearance on T1 image (yes or no)). These are referred to as patient and manually scored features respectively. Tumours were considered superficial when entirely located above the fascia, or as deep‐seated when located beneath the fascia, or with invasion of the fascia.
Decision model creation
To create a decision model from the features, the Workflow for Optimal Radiomics Classification (WORC) toolbox22 was used. A schematic overview of the radiomics methodology is shown in Fig. 1. In WORC, decision model creation is divided into several steps. These steps include, for example, selection of features that offer the highest predictive value and machine learning to discover the patterns in these features that distinguish between WDLPS and lipoma. For each of these steps, numerous algorithms have been proposed in the literature. WORC performs an exhaustive search amongst these algorithms, in a fully automated way, and establishes the combination of algorithms that maximizes the prediction accuracy. As the single best solution may be a coincidental finding, the 50 best performing solutions were combined into a single model, with the purpose of creating a more robust model and boosting performance. More details can be found in Appendix S2 (supporting information).
Figure 1.
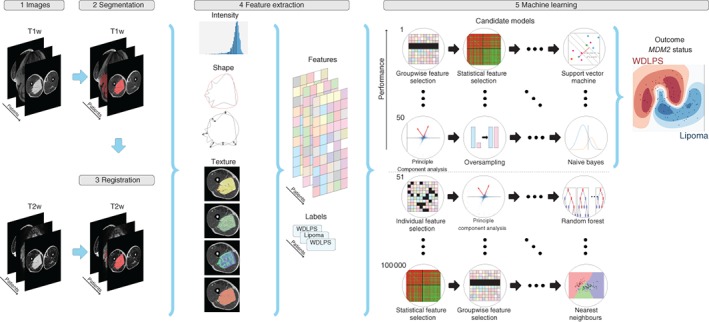
Schematic overview of the radiomics approach Inputs to the algorithm are T1‐ and T2‐weighted magnetic resonance images of well differentiated liposarcoma (WDLPS) and lipoma (1). Processing steps include segmentation of the tumour on the T1 image (2), registration of the T1 to the T2 image to transform this segmentation to the T2 image (3), feature extraction from both the T1 and T2 images (4) and the creation of a decision model from the features (5), using an ensemble of the best 50 workflows from 100 000 candidate workflows; workflows are different combinations of the different processing and analysis steps (for example the classifier used).
Experimental set‐up
To assess the predictive value of the T1 imaging features, and the additional patient and manually scored features, five models were trained and tested based on: imaging features only (model 1); patient features only (model 2); manually scored features only (model 3); a combination of imaging features and manually scored features (model 4); and volume only (model 5). The fifth model was included because WDLPS is generally larger than lipoma3. Additionally, to investigate the potential of the features independent of volume, these five models were evaluated on a volume‐matched cohort, that is a subset of the data in which the distribution of tumour volume was similar among WDLPS and lipoma. These models were trained on the full data set, but tested only on patients from the volume‐matched cohort.
Next, the potential value of other MRI sequences was explored by training and testing multiple imaging‐based radiomics models using combinations of the various MRI sequences. When a model showed more potential than the T1 imaging‐only model, it was evaluated on the volume‐matched cohort as well.
Evaluation
Model evaluation was performed through cross‐validation. The data were randomly split for 100 iterations, using 80 per cent for training and 20 per cent for testing. In each iteration, automatic workflow optimization was performed on the training set in an internal ten times random split cross‐validation (Fig. S1 , supporting information). Thus, the models were optimized solely on the training set; the test set was used only for evaluation of the final model. All splitting was done in a stratified manner to keep the balance between WDLPS and lipoma similar in all data sets.
Performance was evaluated using the area under the curve (AUC) of the receiver operating characteristic (ROC) curve, accuracy, sensitivity, specificity, negative predictive value and positive predictive value, averaged over the 100 cross‐validation iterations. Positive MDM2 amplification status (WDLPS) was defined as the positive class. Ninety‐five per cent confidence intervals for the mean performance measures were constructed using the corrected resampled t test based on all 100 cross‐validation iterations, thereby taking into account that the samples in the cross‐validation splits were not statistically independent23.
Model insights
Insight into the model was gained by ranking the patients from typical to atypical for both lipoma and WDLPS, based on the consistency of the model predictions. This was determined by the number of times (percentage) that a patient was classified correctly when included in the test set. Typical examples were patients who were always classified correctly; and atypical vice versa. In addition, to identify the individual imaging features included in the radiomics model and to assess their respective contribution to the model, univariable statistical testing of the imaging features was undertaken using the Mann–Whitney U test. P values were corrected for multiple testing using the Bonferroni correction.
Classification by radiologists
Three radiologists with expertise in soft tissue tumours classified the lipomatous tumours; radiologists 1, 2 and 3 had 3, 10 and 5 years of experience respectively. First, the radiologists had to classify the tumours as either DDLPS or WDLPS/lipoma (non‐DDLPS), to confirm that DDLPS can be recognized visually. Regardless of whether a tumour was classified as DDLPS or not, the tumours subsequently had to be classified as MDM2‐negative (lipoma) or MDM2‐positive (WDLPS/DDLPS). The classification was done using a ten‐point scale to indicate the certainty of the radiologists. The radiologists had access to all sequences that were available for each patient, as well as the age and sex.
Results
In total, 138 tumours were included: 58 patients had an MDM2‐negative lipoma, 58 had an MDM2‐positive WDLPS and 22 had an MDM2‐positive DDLPS. Most patients were men (60·1 per cent) and had a deep‐seated tumour located in a leg. Median WDLPS size was 20·4 cm and median volume was 36·3 cl, compared with 12·3 cm and 12·9 cl for lipoma (Table 1). Most of the patients underwent surgery: 32 with a lipoma, 50 with a WDLPS and 19 of those with a DDLPS. The eight patients with a WDLPS who did not have surgery were treated conservatively with an active surveillance approach, whereas the three with a DDLPS who did not have surgery had an inoperable tumour.
Table 1.
Characteristics of the patients with lipomatous tumours
| No. of patients*(n = 138) | |
|---|---|
| Age (years) † | 64 (54–71) |
| Sex ratio (M : F) | 83 : 55 |
| Diagnosis | |
| Lipoma | 58 (42·0) |
| WDLPS | 58 (42·0) |
| DDLPS | 22 (15·9) |
| Tumour location | |
| Upper extremity | 14 (10·1) |
| Lower extremity | 71 (51·4) |
| Trunk | 37 (26·8) |
| Head and neck | 6 (4·3) |
| Retroperitoneum and pelvis | 6 (4·3) |
| Paratesticular | 4 (2·9) |
| Tumour depth | |
| Superficial | 20 (14·5) |
| Deep | 118 (85·5) |
| Tumour size (cm) † | |
| Lipoma | 12·3 (9·3–15·5) |
| WDLPS | 20·4 (15·9–26·3) |
| Tumour volume (cl) † | |
| Lipoma | 12·9 (4·6–25·0) |
| WDLPS | 36·3 (22·9–85·5) |
With percentages in parentheses unless indicated otherwise;
values are median (i.q.r.).
WDLPS, well differentiated liposarcoma; DDLPS, dedifferentiated liposarcoma.
The 116 lipoma and WDLPS scans came from 41 different MRI scanners; there was wide heterogeneity in imaging hardware and acquisition protocols used, reflected in differences in magnetic field strength (1·5 T, 98 scans; 1 T, 10 scans; 3 T, 8 scans), manufacturer (Siemens, Munich, Germany, 45 scans; Philips, Amsterdam, the Netherlands, 45 scans; GE, Chicago, Illinois, USA, 26 scans), scanner model (19 different ones), slice thickness, repetition time and echo time. Additional sequences besides T1 were available in subsets of patients: T1‐FS in 55 patients (47·4 per cent), T1‐GD in 42 patients (36·2 per cent), T1‐FS‐GD in 80 patients (69·0 per cent), T2 in 76 patients (65·5 per cent) and T2‐FS in 92 patients (79·3 per cent) (Table S1 , supporting information).
Evaluation of radiomics models based on T1 imaging and additional features
The performances of models 1–5 are shown in Fig. 2 and Table S2 (supporting information). Model 1, based on the T1 imaging features, resulted in an AUC of 0·83, sensitivity of 0·68 and specificity of 0·84. Model 2, based on patient features, had a lower AUC (0·75), higher sensitivity (0·77), but lower specificity (0·59). Similarly, model 3, based on manually scored features, also had a lower AUC (0·72), higher sensitivity (0·76) and lower specificity (0·57). Model 4, combining the imaging and manually scored features, performed worse than model 1, implying that imaging features are sufficient as input. Finally, model 5, based on volume alone, performed similarly to model 1 with an AUC of 0·83, sensitivity of 0·67 and specificity of 0·84. Although the performance metrics were similar for models 1 and 5, the ROC curves in Fig. 2 show some differences. The ROC curve for the volume model (Fig. 2 e) has some sharp bends, while that for the T1 imaging model is smoother (Fig. 2 a).
Figure 2.
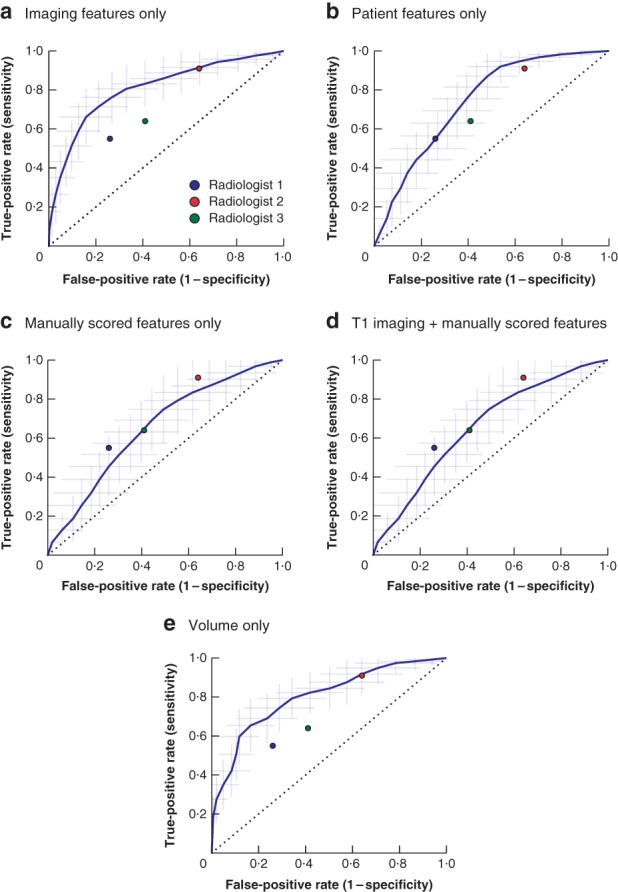
Receiver operating characteristic (ROC) curves for the radiomics models based on the T1‐weighted MRI sequence a Using imaging features only, b using patient features only, c using manually scored features only, d using T1 imaging features combined with manually scored features, and e using volume only. The shaded area indicates the 95 per cent confidence intervals of the 100 times random‐split cross‐validation; the curve is fit through their means. The performance of the three radiologists is shown.
Evaluation of the radiomics models with additional MRI sequences
Most models with an additional MRI sequence had a similar performance to the T1 imaging model (Table S3 , supporting information). However, the model combining the T1 and T2 imaging features showed a clear improvement in performance, with an AUC of 0·89, sensitivity of 0·74 and specificity of 0·88. The distribution of patient characteristics and the distribution of WDLPS and lipoma were similar across patients who had a T2 scan, indicating that the added value is within the T2 imaging features and not a result of incidental correlation with these characteristics, for example owing to selection bias.
Evaluation of models on volume‐matched cohort
Model 5, based on volume alone, illustrated that volume is indeed a strong predictive factor. The 17 tumours with a volume above 70 cl were all WDLPS, whereas the 21 tumours with a volume below 7 cl were all lipoma. In the volume‐matched cohort, consisting of the other 78 tumours with a volume between 7 and 70 cl, the volume distributions for WDLPS and lipoma were more similar. As only the T2 scans provided additional value over the T1 imaging features, the T1 + T2 imaging model was evaluated for the volume‐matched cohort as well.
The performance of both imaging‐based models (T1 and T1 + T2) was worse on the volume‐matched cohort (T1: AUC 0·69; T1 + T2: AUC 0·81) (Table 2) than on the entire cohort (AUC 0·83 and 0·89 respectively) (Table S3 , supporting information). The models based on the patient and manually scored features performed similarly to the models tested on the full cohort. The model based on volume alone still performed above chance (mean AUC 0·64), but considerably worse than on the entire data set. In this volume‐matched data set, both the T1 imaging model (AUC 0·69, sensitivity 0·60, specificity 0·74) and the T1 + T2 imaging model (AUC 0·81, sensitivity 0·66, specificity 0·84) performed considerably better than volume alone (Table 2). This showed that these models were not based solely on volume, and that other features provided additional predictive value over volume.
Table 2.
Performance of radiomics models trained on the full cohort, but evaluated in the volume‐matched cohort
| T1 imaging features only | T1 + T2 imaging features | Patient features only | Manually scored features only | Volume only | |
|---|---|---|---|---|---|
| AUC | 0·69 (0·58, 0·80) | 0·81 (0·72, 0·90) | 0·74 (0·64, 0·84) | 0·67 (0·56, 0·77) | 0·64 (0·53, 0·74) |
| Accuracy | 0·67 (0·57, 0·76) | 0·75 (0·66, 0·83) | 0·66 (0·56, 0·75) | 0·60 (0·51, 0·69) | 0·66 (0·57, 0·74) |
| Sensitivity | 0·60 (0·45, 0·75) | 0·66 (0·52, 0·79) | 0·69 (0·55, 0·83) | 0·70 (0·53, 0·87) | 0·50 (0·36, 0·64) |
| Specificity | 0·74 (0·60, 0·87) | 0·84 (0·71, 0·96) | 0·62 (0·48, 0·76) | 0·51 (0·36, 0·65) | 0·82 (0·71, 0·92) |
| NPV | 0·66 (0·54, 0·77) | 0·72 (0·60, 0·83) | 0·68 (0·56, 0·79) | 0·65 (0·49, 0·80) | 0·62 (0·53, 0·71) |
| PPV | 0·72 (0·58, 0·85) | 0·81 (0·69, 0·93) | 0·65 (0·54, 0·76) | 0·59 (0·49, 0·69) | 0·74 (0·61, 0·87) |
Values are mean (95 per cent c.i.) over the cross‐validation iterations. AUC, area under the curve; NPV, negative predictive value; PPV, positive predictive value.
Model insights
Of the 116 lipomatous tumours, 69 (26 WDLPS, 43 lipoma) were always classified correctly by model 1 in all 100 cross‐validation iterations. In contrast, 13 tumours (9 WDLPS, 4 lipoma) were always classified incorrectly. Fig. 3 shows four MRI slices of such typical and atypical examples of lipoma and WDLPS. The lesions that were always classified incorrectly were checked for possible sampling error of the biopsy. The MDM2 amplification status of eight of the 13 tumours always classified incorrectly was already determined on the resection specimen (6 WDLPS, 2 lipoma). For the other five patients, in whom it was tested on the biopsy (3 WDLPS, 2 lipoma), pathological examination of the resection specimen confirmed the diagnosis, except for one patient with a lipoma who did not undergo surgery. In the other patient with a lipoma, the resection specimen again tested negative for MDM2 amplification. The three WDLPS resection specimens were not retested.
Figure 3.
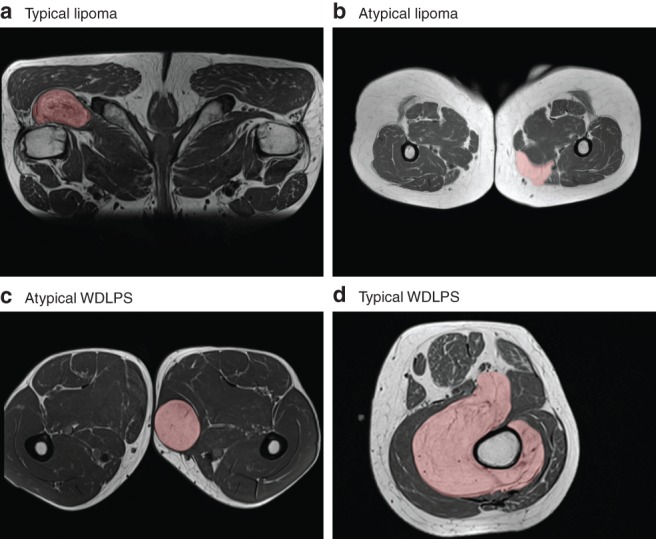
Examples of typical and atypical lipomas and well differentiated liposarcomas a Typical lipoma, b atypical lipoma, c atypical well differentiated liposarcoma (WDLPS) and d typical WDLPS. The typical examples are from two patients always classified correctly by the T1 imaging model; the atypical examples are from two patients always classified incorrectly by the T1 imaging model.
Analysis of feature importance was done for the volume‐matched cohort, as the results on the full data set were dominated by volume‐related measures. In total, 16 individual features were found to be significant after Bonferroni correction on the volume‐matched cohort (Fig. S2 , supporting information). These included 11 shape features (including several volume‐related statistics), four texture features and one intensity feature.
Radiomics models compared with radiologists
On the entire cohort, the AUCs of all three radiologists (0·74, 0·72 and 0·61 for radiologist 1, 2 and 3 respectively) (Table S4 , supporting information) were below the lower limit of the 95 per cent c.i. of the T1 imaging model (0·75 to 0·90) (Fig. 2 and Table S2, supporting information), as well as of the 95 per cent c.i. of the T1 + T2 imaging model (0·83 to 0·95) (Table S3, supporting information). The radiologists achieved sensitivity values similar to (0·64 and 0·74) or higher (0·91) than those of the radiomics models (T1: 0·68; T1 + T2: 0·74), but their specificity was much lower (radiomics: 0·84 and 0·88 respectively; radiologists 1–3: 0·55, 0·36 and 0·59 respectively). The Cohen's κ value was 0·24, 0·04 and 0·40 for all pairs of radiologists, with a mean of 0·23, indicating poor interobserver agreement.
On the volume‐matched cohort, the radiologists had a performance (AUC 0·68, 0·74 and 0·55) (Table S4 , supporting information) more similar to that of the T1 imaging model (AUC 0·69) (Table 2). On average, the T1 imaging model still performed better in terms of specificity (radiomics: 0·74; radiologists 1–3: 0·58, 0·37 and 0·50), whereas the radiologists again performed better on sensitivity (radiomics: 0·60; radiologists 1–3: 0·65, 0·88 and 0·60). However, the T1 + T2 imaging model performed much better (AUC 0·81, sensitivity 0·66, specificity 0·84) than both the T1 imaging model and the radiologists. On this cohort, the Cohen's κ values were 0·18, –0·04 and 0·34 for all pairs of radiologists, with a mean of 0·16, again indicating poor interobserver agreement.
Distinction between dedifferentiated liposarcoma and well differentiated liposarcoma/lipoma
Besides classifying lipoma and WDLPS, the radiologists also classified the scans from 22 patients with DDLPS to evaluate whether DDLPS can indeed be identified by imaging only, without the help of additional models. Radiologists 1–3 had an AUC of 0·97, 0·91 and 0·90 respectively; a sensitivity of 0·95, 0·95 and 0·91; and a specificity of 0·95, 0·56 and 0·89 in distinguishing DDLPS from non‐DDLPS (WDLPS/lipoma) (Table S4 , supporting information).
Discussion
This study shows that there is a relationship between quantitative MRI features and MDM2 amplification status, and that radiomics is a promising non‐invasive method for differentiating lipoma from WDLPS. Although the radiologists were able to distinguish between DDLPS and non‐DDLPS, they were outperformed by the T1 and T1 + T2 imaging models in differentiating WDLPS from lipoma. Moreover, the agreement between radiologists was very poor, whereas the radiomics‐based predictions were objective and reproducible (given a tumour segmentation).
Remarkably, the model trained on volume alone had a similar performance to the T1 imaging model, which included many additional features. However, in the volume‐matched data set, the T1 imaging model performed considerably better than the volume‐only model, indicating that other features do provide additional predictive value. It is already known that WDLPS is on average larger than lipoma3, and the relationship with volume (or size) in our data set was also strong; the database did not contain lipoma larger than 70 cl or WDLPS smaller than 7 cl although these do exist24, 25. However, all WDLPS lesions start as small tumours and grow over time, so the measured tumour volume depends on the moment of presentation, and a small or intermediate tumour volume is therefore not a reliable biomarker. Future research should include expansion of the data set to make the volume distributions more representative (including lipoma larger than 70 cl and WDLPS smaller than 7 cl), thereby making the radiomics model less volume‐dependent.
The models trained solely on either the patient or manually scored features performed slightly worse than the model trained on the T1 imaging features only. As the combined model did not outperform the T1 imaging model, the manually scored features did not add much in the search for the best radiomics model. Additionally, the manually scored features may be observer‐dependent, and thus prone to subjectivity. Although patient features (age, sex and tumour location) are objective, the distribution in the present data set may not be representative of clinical practice. For example, none of the patients with WDLPS were younger than 35 years, there were no lipomas among patients older than 82 years, no lipomas in the head and neck region, and no WDLPS in the pelvis or shoulder/trunk; all these might occur in daily clinical practice. Therefore, the imaging‐only models have more potential as an objective tool in clinical practice.
The results of present study are similar to those of Thornhill and colleagues26, who used a comparable approach and showed that lipomas can be distinguished from liposarcomas by texture and shape analysis. Strong points of the present study include the larger sample size (116 versus 44 in Thornhill et al.). Thornhill and co‐workers also included other liposarcoma subtypes in their model, such as DDLPS and myxoid liposarcoma (8 of 20 included liposarcomas). These other liposarcoma subtypes have distinct radiological features5, 10, which in general can be easily discriminated from lipomas by experienced radiologists. By solely including the two tumour types that are the most difficult to distinguish (WDLPS and lipoma) in the radiomics model, the present data set is more challenging and more clinically relevant. In contrast to the cases described by Thornhill et al., the diagnosis of all patients in the present data set was confirmed by verifying the MDM2 amplification status using FISH, the current standard for diagnosing and differentiating between lipoma and WDLPS1, 12, 13. The present radiomics model only requires routine MRI scans (T1, and optionally T2) without contrast injection; the other sequences did not add any predictive value to the model. As almost all standard MRI protocols include a T1 and T2 sequence, the present radiomics method is generalizable, feasible and applicable for use in daily practice. Finally, these radiomics models were developed and evaluated on a heterogeneous data set, thereby increasing the chance that the reported performance can be reproduced in a routine clinical setting when using other MRI scanners.
Advantages of using a radiomics approach over pathological assessment to differentiate between lipoma and WDLPS include sparing patients an invasive and painful biopsy, and saving the substantial costs of a radiologist performing the imaging‐guided biopsy and of the pathologist assessing it, including the costs of molecular testing by FISH. Radiomics makes use of MRI images obtained during routine diagnostic evaluation and patients do not need to undergo any additional procedure. When radiomics becomes a widely available tool, patients with WDLPS can be identified and referred to a soft tissue sarcoma expert centre at an earlier stage, with potential beneficial effects on further diagnostics, treatment and follow‐up.
Several limitations of this study should be noted, besides the volume bias already mentioned. First, segmentation of ROIs of the tumours was done manually, which inherently leads to both interobserver and intraobserver variability, as has been quantified for other cancer types27, 28, 29. Variability in segmenting the ROIs might lead to variability in the extracted imaging features and subsequently influence the classification of tumours. Additionally, manual segmentation is rather time‐consuming. This could be addressed by use of automated segmentation tools that might be available in the future. Second, variation in imaging protocols might have influenced the imaging statistics. No restrictions were put on the T1 MRI sequences regarding field strength, slice thickness, or other MRI acquisition settings, as selecting a single protocol is an unrealistic reflection of daily clinical practice and would have made the results non‐generalizable. Instead, this study shows that the present radiomics approach is robust to these variations by both training and testing the model on heterogeneous data. Third, the model is based on retrospectively collected data, which might have led to selection and information bias. This potential selection bias might have occurred particularly in the lipoma subgroup, as usually only large and atypical lipomas are referred to a sarcoma centre. However, this probably made the data set even more challenging and relevant, as these can be seen as the complex cases. Addition of the ‘small and typical’ lipomas would have made the classification easier, and radiomics is not needed to make the distinction for such lipomas.
The present radiomics model could serve as a non‐invasive, quick and low‐cost alternative to a biopsy. Although the model needs optimization to match the accuracy of a biopsy, there could be a certain patient group for whom the model may already be useful. For example, patients at high risk of complications of biopsy, or those in whom the radiomics model can predict the MDM2 amplification status with a high degree of certainty, could already be treated according to the prediction of the radiomics model. Although further research is required to identify which patients could benefit most from the present model, initial misclassification of a WDLPS as a lipoma would not harm the patient, considering that active surveillance seems a safe option in patients without (invalidating) symptoms and/or tumour growth, at least in the short term30. In addition, the performance of the radiomics model improved substantially when T2 images were added. However, only 65·5 per cent of the patients had a T2 scan available, so for a follow‐up study it is proposed to use MRI with at least both T1 and T2 sequences.
Supporting information
Supplementary Table S1 Several properties of the acquisition protocols of the 116 T1‐weighted MRI sequences of patients with lipoma or well‐differentiated liposarcoma (WDLPS) that were used to build the radiomics model.
Table S2 Performance of the radiomics models based on T1 imaging features only; patient features only; manually scored features only; the combination of T1 imaging and manually scored features; and of volume only on the full dataset. Performance for the radiomics models is reported for each experiment as mean [95% confidence interval] over the cross‐validation iterations.
Supplementary Table S3. Performance of radiomics models trained on features extracted from various MRI sequences on the full dataset. Performance is reported as mean [95% confidence interval] over the cross‐validation iterations.
Supplementary Table S4. Performance of the three radiologists in differentiating between well‐differentiated liposarcomas and lipomas on both the full and volume‐matched cohort, and in differentiating dedifferentiated liposarcoma (DDLPS) and non‐DDLPS (well‐differentiated liposarcoma [WDLPS]/lipomas).
Appendix S1: Radiomics feature extraction
Appendix S2: Technical details on decision model creation
Supplementary Figure S1 Visualization of the 100x stratified random‐split cross‐validation, including a second cross‐validation within the training set to perform the automatic workflows optimization. Optimization was done solely on the training set in order to prevent overfitting on the test set. The ensemble averages the predictions of the best 50 performing workflows to create a more robust model.
Supplementary Figure S2 P‐values of Mann–Whitney U tests of feature values for WDLPS and lipomas. Only the features that had a corrected P‐value <0·05 were included in the graph. The labels on the y‐axis correspond to the feature names: see Supplementary Materials 1 for more details.
Acknowledgements
M.V. and M.P.A.S. contributed equally to this study. The authors thank E. H. G. Oei and D. F. Hanff for classifying the lipomatous tumours. This study was financed by the Stichting Coolsingel (reference no. 567), a Dutch non‐profit foundation. M.P.A.S. acknowledges funding from the research programme STRaTeGy (project no. 14929‐14930), which is partly financed by the Netherlands Organization for Scientific Research (NWO). W.J.N. is founder, scientific lead and stock holder of Quantib.
Disclosure: The authors declare no other conflicts of interest.
References
- 1. Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F; World Health Organization, International Agency for Research on Cancer . WHO Classification of Tumours of Soft Tissue and Bone. IARC Press: Lyon, 2013. [Google Scholar]
- 2. Brisson M, Kashima T, Delaney D, Tirabosco R, Clarke A, Cro S et al MRI characteristics of lipoma and atypical lipomatous tumor/well‐differentiated liposarcoma: retrospective comparison with histology and MDM2 gene amplification. Skeletal Radiol 2013; 42: 635–647. [DOI] [PubMed] [Google Scholar]
- 3. Kransdorf MJ, Bancroft LW, Peterson JJ, Murphey MD, Foster WC, Temple HT. Imaging of fatty tumors: distinction of lipoma and well‐differentiated liposarcoma. Radiology 2002; 224: 99–104. [DOI] [PubMed] [Google Scholar]
- 4. Gupta P, Potti TA, Wuertzer SD, Lenchik L, Pacholke DA. Spectrum of fat‐containing soft‐tissue masses at MR imaging: the common, the uncommon, the characteristic, and the sometimes confusing. Radiographics 2016; 36: 753–766. [DOI] [PubMed] [Google Scholar]
- 5. Drevelegas A, Pilavaki M, Chourmouzi D. Lipomatous tumors of soft tissue: MR appearance with histological correlation. Eur J Radiol 2004; 50: 257–267. [DOI] [PubMed] [Google Scholar]
- 6. O'Donnell PW, Griffin AM, Eward WC, Sternheim A, White LM, Wunder JS et al Can experienced observers differentiate between lipoma and well‐differentiated liposarcoma using only MRI? Sarcoma 2013; 2013: 982784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kransdorf MJ, Meis JM, Jelinek JS. Dedifferentiated liposarcoma of the extremities: imaging findings in four patients. AJR Am J Roentgenol 1993; 161: 127–130. [DOI] [PubMed] [Google Scholar]
- 8. Tateishi U, Hasegawa T, Beppu Y, Satake M, Moriyama N. Primary dedifferentiated liposarcoma of the retroperitoneum. Prognostic significance of computed tomography and magnetic resonance imaging features. J Comput Assist Tomogr 2003; 27: 799–804. [DOI] [PubMed] [Google Scholar]
- 9. Yun JS, Chung HW, Song JS, Lee SH, Lee MH, Shin MJ. Dedifferentiated liposarcoma of the musculoskeletal system: expanded MR imaging spectrum from predominant fatty mass to non‐fatty mass. Acta Radiol 2019; doi: 10.1177/0284185119833060 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 10. Murphey MD, Arcara LK, Fanburg‐Smith J. From the archives of the AFIP: imaging of musculoskeletal liposarcoma with radiologic–pathologic correlation. Radiographics 2005; 25: 1371–1395. [DOI] [PubMed] [Google Scholar]
- 11. ESMO/European Sarcoma Network Working Group . Soft tissue and visceral sarcomas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow‐up. Ann Oncol 2014; 25(Suppl 3): iii102–iii112. [DOI] [PubMed] [Google Scholar]
- 12. Thway K, Wang J, Swansbury J, Min T, Fisher C. Fluorescence in situ hybridization for MDM2 amplification as a routine ancillary diagnostic tool for suspected well‐differentiated and dedifferentiated liposarcomas: experience at a tertiary center. Sarcoma 2015; 2015: 812089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kimura H, Dobashi Y, Nojima T, Nakamura H, Yamamoto N, Tsuchiya H et al Utility of fluorescence in situ hybridization to detect MDM2 amplification in liposarcomas and their morphological mimics. Int J Clin Exp Pathol 2013; 6: 1306–1316. [PMC free article] [PubMed] [Google Scholar]
- 14. Lambin P, Rios‐Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P et al Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012; 48: 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Juntu J, Sijbers J, De Backer S, Rajan J, Van Dyck D. Machine learning study of several classifiers trained with texture analysis features to differentiate benign from malignant soft‐tissue tumors in T1‐MRI images. J Magn Reson Imaging 2010; 31: 680–689. [DOI] [PubMed] [Google Scholar]
- 16. Corino VDA, Montin E, Messina A, Casali PG, Gronchi A, Marchianò A et al Radiomic analysis of soft tissues sarcomas can distinguish intermediate from high‐grade lesions. J Magn Reson Imaging 2018; 47: 829–840. [DOI] [PubMed] [Google Scholar]
- 17. Vallières M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG‐PET and MRI texture features for the prediction of lung metastases in soft‐tissue sarcomas of the extremities. Phys Med Biol 2015; 60: 5471–5496. [DOI] [PubMed] [Google Scholar]
- 18. Starmans MPA, Miclea RL, van der Voort SR, Niessen WJ, Thomeer MG, Klein S. Classification of malignant and benign liver tumors using a radiomics approach. In SPIE Medical Imaging; 2018: Image Processing; 10574D; 2018. https://www.spiedigitallibrary.org/conference-proceedings-of-spie/10574/105741D/Classification-of-malignant-and-benign-liver-tumors-using-a-radiomics/10.1117/12.2293609.short?SSO=1 [accessed 9 September 2019].
- 19. Klein S, Staring M, Murphy K, Viergever MA, Pluim JP. elastix: a toolbox for intensity‐based medical image registration. IEEE Trans Med Imaging 2010; 29: 196–205. [DOI] [PubMed] [Google Scholar]
- 20. GitHub . Predict a Radiomics Extensive Differentiable Interchangable Classification Toolkit (PREDICT) https://github.com/Svdvoort/PREDICTFastr [accessed 9 September 2019].
- 21. van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V et al Computational radiomics system to decode the radiographic phenotype. Cancer Res 2017; 77: e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. GitHub . Workflow for Optimal Radiomics Classification (WORC) https://github.com/MStarmans91/WORC [accessed 9 September 2019].
- 23. Nadeau C, Bengio Y. Inference for the generalization error. In Advances in Neural Information Processing Systems?, 2000; 307–313. http://papers.nips.cc/paper/1661-inference-for-the-generalization-error.pdf [accessed 9 September 2019].
- 24. Sanchez MR, Golomb FM, Moy JA, Potozkin JR. Giant lipoma: case report and review of the literature. J Am Acad Dermatol 1993; 28: 266–268. [DOI] [PubMed] [Google Scholar]
- 25. Smith CA, Martinez SR, Tseng WH, Tamurian RM, Bold RJ, Borys D et al Predicting survival for well‐differentiated liposarcoma: the importance of tumor location. J Surg Res 2012; 175: 12–17. [DOI] [PubMed] [Google Scholar]
- 26. Thornhill RE, Golfam M, Sheikh A, Cron GO, White EA, Werier J et al Differentiation of lipoma from liposarcoma on MRI using texture and shape analysis. Acad Radiol 2014; 21: 1185–1194. [DOI] [PubMed] [Google Scholar]
- 27. Echegaray S, Gevaert O, Shah R, Kamaya A, Louie J, Kothary N et al Core samples for radiomics features that are insensitive to tumor segmentation: method and pilot study using CT images of hepatocellular carcinoma. J Med Imaging (Bellingham) 2015; 2: 041011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu L, Lv W, Jiang J, Ma J, Feng Q, Rahmim A et al Robustness of radiomic features in [11C]choline and [18F]FDG PET/CT imaging of nasopharyngeal carcinoma: impact of segmentation and discretization. Mol Imaging Biol 2016; 18: 935–945. [DOI] [PubMed] [Google Scholar]
- 29. Parmar C, Rios Velazquez E, Leijenaar R, Jermoumi M, Carvalho S, Mak RH et al Robust radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One 2014; 9: e102107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vos M, Grünhagen DJ, Koseła‐Paterczyk H, Rutkowski P, Sleijfer S, Verhoef C. Natural history of well‐differentiated liposarcoma of the extremity compared to patients treated with surgery. Surg Oncol 2019; 29: 84–89. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1 Several properties of the acquisition protocols of the 116 T1‐weighted MRI sequences of patients with lipoma or well‐differentiated liposarcoma (WDLPS) that were used to build the radiomics model.
Table S2 Performance of the radiomics models based on T1 imaging features only; patient features only; manually scored features only; the combination of T1 imaging and manually scored features; and of volume only on the full dataset. Performance for the radiomics models is reported for each experiment as mean [95% confidence interval] over the cross‐validation iterations.
Supplementary Table S3. Performance of radiomics models trained on features extracted from various MRI sequences on the full dataset. Performance is reported as mean [95% confidence interval] over the cross‐validation iterations.
Supplementary Table S4. Performance of the three radiologists in differentiating between well‐differentiated liposarcomas and lipomas on both the full and volume‐matched cohort, and in differentiating dedifferentiated liposarcoma (DDLPS) and non‐DDLPS (well‐differentiated liposarcoma [WDLPS]/lipomas).
Appendix S1: Radiomics feature extraction
Appendix S2: Technical details on decision model creation
Supplementary Figure S1 Visualization of the 100x stratified random‐split cross‐validation, including a second cross‐validation within the training set to perform the automatic workflows optimization. Optimization was done solely on the training set in order to prevent overfitting on the test set. The ensemble averages the predictions of the best 50 performing workflows to create a more robust model.
Supplementary Figure S2 P‐values of Mann–Whitney U tests of feature values for WDLPS and lipomas. Only the features that had a corrected P‐value <0·05 were included in the graph. The labels on the y‐axis correspond to the feature names: see Supplementary Materials 1 for more details.


