Significance
Improved therapeutic intervention for neurodegenerative diseases, particularly early in progression of the disease course, is a critical need. Huntington’s disease (HD) is a paradigmatic disorder in this search in which vulnerable individuals can be identified early. This study focuses on the earliest stages of disease in a well-characterized animal model system. We identify early aberrant transcription and chromatin changes in affected brain regions of HD mice. The study identifies the Elk-1 transcription factor as a significant regulator of early transcriptional changes in HD. Enhanced Elk-1 levels exerted beneficial effects in an in vitro model and resulted in extensive restoration of transcriptional dysregulation in vivo. These results suggest a target for alleviating pathology in HD and other neuropsychiatric conditions.
Keywords: Huntington’s disease, neurodegeneration, transcriptional dysregulation, epigenomics, gene therapy
Abstract
Huntington’s disease (HD) is a chronic neurodegenerative disorder characterized by a late clinical onset despite ubiquitous expression of the mutant Huntingtin gene (HTT) from birth. Transcriptional dysregulation is a pivotal feature of HD. Yet, the genes that are altered in the prodromal period and their regulators, which present opportunities for therapeutic intervention, remain to be elucidated. Using transcriptional and chromatin profiling, we found aberrant transcription and changes in histone H3K27acetylation in the striatum of R6/1 mice during the presymptomatic disease stages. Integrating these data, we identified the Elk-1 transcription factor as a candidate regulator of prodromal changes in HD. Exogenous expression of Elk-1 exerted beneficial effects in a primary striatal cell culture model of HD, and adeno-associated virus-mediated Elk-1 overexpression alleviated transcriptional dysregulation in R6/1 mice. Collectively, our work demonstrates that aberrant gene expression precedes overt disease onset in HD, identifies the Elk-1 transcription factor as a key regulator linked to early epigenetic and transcriptional changes in HD, and presents evidence for Elk-1 as a target for alleviating molecular pathology in HD.
Huntington’s disease (HD), a chronic genetically based neurodegenerative disorder, is most often characterized by clinical onset in adulthood despite ubiquitous expression of the mutant Huntingtin gene (HTT) from conception. Among the most significant efforts to develop a disease-modifying therapy for HD are efforts to reduce or prevent the production of the pathological mutant form of the Huntingtin protein. However, events that cause pathological consequences in HD may already be in place at the time of therapy (1), and simply stopping the production of mutant Huntingtin protein may not effectively halt or eliminate disease progression. Of particular significance are regulatory changes that cause alterations in transcriptional programs of importance to neuronal function. In this study, we have focused on identifying key molecular control elements that are contributory to pathology in HD and designed strategies to alleviate transcriptional dysregulation encountered early in the course of disease, which may contribute to the development of effective therapeutic intervention in HD.
Transcriptional dysregulation is a central pathogenic mechanism in HD that is faithfully recapitulated in mouse models of this neurodegenerative condition. Progressive changes in transcriptional profiles comprising many neuronal genes, such as neurotransmitters, neurotrophins, and their receptors were detected in experimental models and in HD patients (2–5). In close relation to transcriptional changes, epigenetic status is also altered in HD (6, 7). Global as well as gene-specific histone hypoacetylation at the promoters of the down-regulated genes were shown in various HD models (8, 9). In previous studies, we found large changes in DNA methylation and histone H3K4trimethylation in the STHdhQ111/STHdhQ7 cell line and in the R6/2 mouse model of HD (10, 11). Collectively, these studies demonstrated aberrant expression of thousands of genes in cell and animal models of HD at progressed disease stages, as well as in postmortem brain samples from HD patients. PET studies show reduced dopaminergic D1 and D2 receptor signals as soon as a decade before disease onset in HD patients, indicating that the dysregulation of critical neuronal genes begins long before the disease symptoms (12). However, many questions still remain unanswered about the early phases of the disease, which are most amenable to treatment. What are the earliest genes whose expression are impaired in the HD brain? What are the transcriptional and epigenetic regulators affected during early HD pathogenesis? Could modulating these regulators delay or prevent onset of symptoms?
To address these questions, we chose to study the R6/1 transgenic mouse model of HD because of its delayed onset of neurological symptoms (i.e., clasping) and mild disease course with a lifespan of 7 to 9 mo, without striatal cell loss even at terminal stages of disease (13–16). This progressive and sequential onset of pathogenic events is suitable for analyses of the early disease mechanisms in HD. In addition, for cross-validation of our key findings from the R6/1 line, we used the slowly progressing full-length Huntingtin model of CHL2 [Hdh(CAG)150] heterozygous knockin mice as a second mouse model (17). For genome-wide analyses of the earliest transcriptional and regulatory changes in the HD brain, we focused on histone H3K27acetylation, which marks active enhancers and transcription start sites (TSSs) (18). Coordinated activities at TSSs and distal enhancers dictate regulation of transcription and enhancers play a key role in particular for tissue-specific gene expression (19, 20).
Here, we report genome-wide changes in gene transcription accompanied by coordinated epigenetic alterations in the striatum during presymptomatic disease stages in the R6/1 and CHL2 HD mouse models. Using chromatin immunoprecipitation followed by sequencing (ChIP-seq), and by analysis of the DNA sequences that underlie the altered epigenomic regions, we identified potential transcription factors whose DNA-binding activities are altered very early in the striatum of R6/1 and CHL2 mice. In particular, by direct ChIP-seq, we were able confirm the altered DNA binding activity of the Elk-1 transcription factor, a member of the ternary complex factor (TCF) family of erythroblast transformation specific (ETS)-domain transcription factors (21). Elk-1 shows strong expression in the central nervous system (22), where it is restricted to neuronal cells (23). Elk-1 is directly activated, phosphorylated by the prosurvival mitogen-activated protein kinases (MAPK)/extracellular-signal regulated kinase (ERK) signaling pathway (24). In the striatum, Elk-1 is activated by cortico-striatal stimulation and glutamate, and is considered to be a major transcriptional regulator known to be involved in the regulation of neuronal activity-regulated immediate-early genes (23, 24). Elk-1 was reported to play a role in reducing excitotoxic cell death in the quinolinic acid-induced model of striatal damage (25), and more recently, reduction of Elk-1 levels by small-interfering RNAs was shown to promote cell death in the STHdhQ111/Q111 cell line model of HD (26). Given its altered transcriptional activity in HD mouse striatum, as well as the evidence suggesting a positive role in an in vitro HD model, we hypothesized that enhancing Elk-1 expression would be beneficial in HD. To test this hypothesis, we overexpressed Elk-1 in a primary striatal cell culture model of HD, and found that Elk-1 exerted beneficial effects against changes associated with HD pathogenesis. In line with this result, adeno-associated virus (AAV)-mediated delivery of Elk-1 into the striatum of R6/1 mice, administered before the onset of symptoms, resulted in extensive amelioration of transcriptional dysregulation characteristic of HD. Altogether, these data reveal Elk-1 transcription factor as a promising therapeutic target in HD.
Results
Transcriptional Dysregulation Precedes Overt Disease Onset in HD Models and Is Dominated by Loss of Expression of Neuronal Genes.
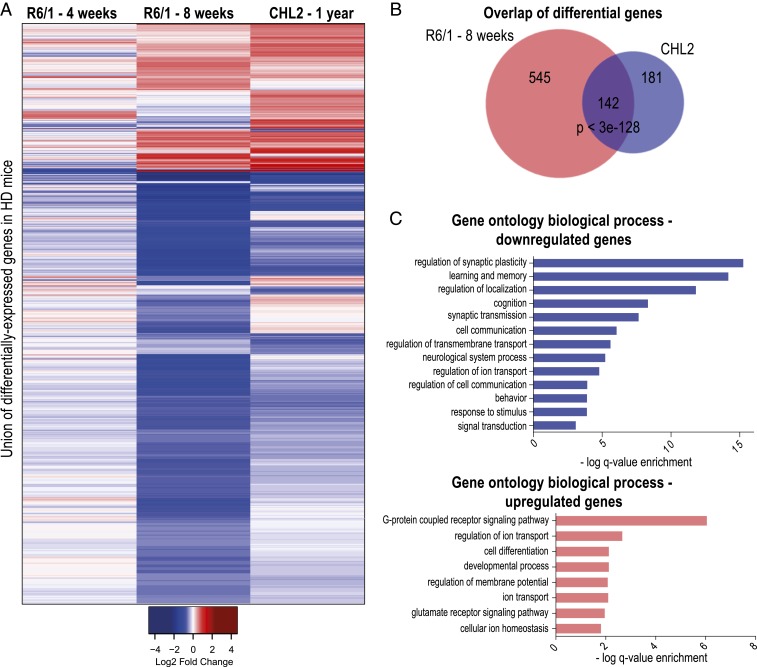
Transcriptional dysfunction is a pathogenic hallmark in HD. We and others have previously shown extensive changes in transcription comprising large number of genes at progressed disease stages in experimental models of HD and in postmortem brain tissue from HD patients (5, 10, 11). Here, our aim was to detect the earliest gene-expression changes in the brains of HD mouse models. For this purpose, we primarily used the well-established R6/1 mouse model (16) at presymptomatic disease stages, 4- and 8-wk of age, and performed RNA sequencing (RNA-seq) for genome-wide analysis of transcription in the striatum. Subsequent analysis of the data revealed that 50 genes were differentially expressed in the R6/1 striatum compared to wild-type littermates as early as 4 wk of age, and of these genes, 26 were up-regulated and 24 were down-regulated (under a false-discovery rate [FDR] of 0.05) (Dataset S1). As expected, the number of dysregulated transcripts further increased at 8 wk of age: Over 687 genes were differentially expressed in the striatum of R6/1 mice, and an overwhelming majority of these genes (609) were down-regulated, while fewer genes (78) showed increased expression in R6/1s compared to wild-types. Gene ontology (GO) analysis of the down-regulated genes revealed significant enrichments of numerous neuron-specific GO terms, such as synaptic signaling, neurological system process, and regulation of response to stimulus, indicating overall disruption of neuronal gene-expression programs early on in the striatum of R6/1 mice (Dataset S2). Key HD genes that are typically down-regulated across models and HD patients, such as dopamine receptor subtypes Drd1a and Drd2, adenosine receptor Adora2a, and many neuronal activity-regulated immediate-early genes, were among the down-regulated transcripts in the striatum of R6/1 mice at 8 wk of age. Many of the typically dysregulated HD genes that are significant at 8 wk of age already showed a similar trend at 4 wk in the R6/1 striatum, yet their expression values were below the cutoff threshold to be called statistically differential at this age. Fig. 1A shows a heatmap of differentially expressed genes in the striata of the HD mice used in our study.
Fig. 1.
R6/1 and CHL2 mice models exhibit largely overlapping transcriptional changes during prodromal disease stage of HD. (A) Heat map of differential gene expression by RNA-seq in the striata of 4- and 8-wk-old R6/1 and 1-y-old CHL2 mice, compared to respective wild-type littermates (n = 3 mice per genotype). (B) Venn diagram of differentially expressed genes in the striata of 8-wk-old R6/1 and 1-y-old CHL2 mice (significance of overlap assessed by hypergeometric test). (C) Bar graphs showing GO biological processes enriched in genes that are down-regulated and up-regulated in the striata of both R6/1 and CHL2 mice. See also SI Appendix, Fig. S1.
To further test whether these transcriptional changes are part of HD pathogenesis, we performed a second set of RNA-seq experiments in a different mouse model, CHL2 [Hdh(CAG)150], a heterozygous knockin line that carries the full-length Htt gene (17). The disease progresses more slowly in this model compared to the R6/1s. Using quantitative RT-PCR, we determined that 1 y of age was suitable for detecting early transcriptional changes in typical HD genes in the striatum of CHL2 mice. Subsequent analysis of the CHL2 RNA-seq data revealed that 324 genes were differentially expressed in the striatum compared to wild-type littermates at that age (Fig. 1 A and B). Comparison of dysregulated genes in the striata of 8-wk-old R6/1s and in the 1-y-old-CHL2s showed a highly significant overlap of 142 genes (P < 3e-128) between the two models (Fig. 1B). Genes whose disruption are central to HD pathogenesis, such as Drd2, Drd1a, Adora2a, Penk, and Ppp1r1b, as well as various neuronal activity regulated immediate-early genes like Arc and Egr1 were down-regulated in both models. Commonly down-regulated striatal genes were enriched for GO terms, such as cell communication, cognition, and signaling, while up-regulated genes were enriched in nervous system development, cell differentiation, and regulation of membrane potential (Fig. 1C).
To ascertain the concordance of the transcriptional changes in the HD mice and patients, we compared the differentially expressed genes in the striata of R6/1 and CHL2 mice with a previous HD patient study, which analyzed low-grade postmortem HD patient caudate (Vonsattel grades 0 to 2) by microarray analysis (4, 5). This revealed a significant overlap of the genes differentially expressed in the striata of the R6/1 and CHL2 mice with differentially expressed genes in the caudate nuclei of postmortem brains from HD patients (161 genes in R6/1 [P < 3e-54] and 51 genes in CHL2 [P < 1.3e-10]) (SI Appendix, Fig. S1 A and B and Dataset S3). Altogether, our results confirm that transcriptional dysregulation is a prominent disease feature in HD, and that the down-regulation of neuronal genes dominates the dysregulated HD transcriptome during the prodromal disease stage (before weight loss or appearance of clasping behavior in R6/1 and in CHL2 mice) (SI Appendix, Fig. S1 C–E).
Histone H3K27acetylation Changes in Coordination with Transcription in the Striatum of Presymptomatic HD Mice.
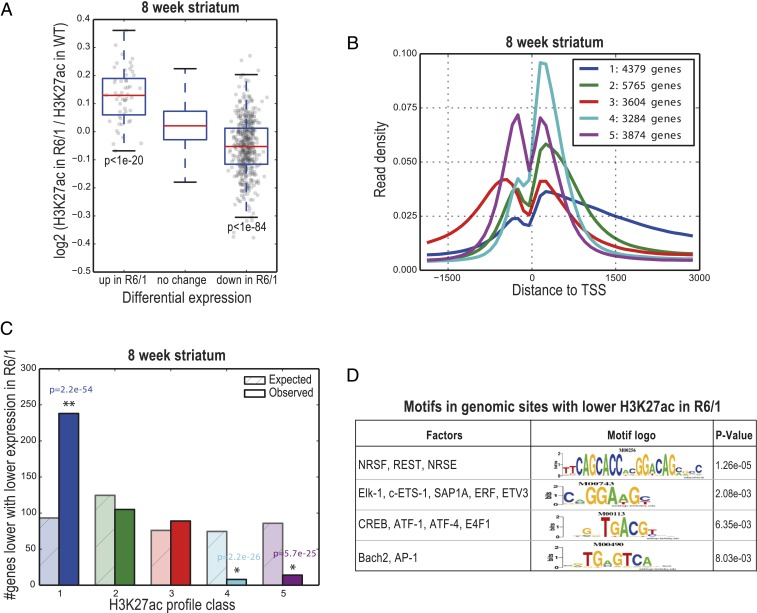
To understand the regulation of genes whose expression is altered in the striatum of HD mice during prodromal disease stages, we examined genome-wide chromatin changes. Our aim was to measure changes in activities in both proximal and distal regulatory sites, which are key players for regulation of tissue-specific gene expression, during early HD. For this purpose, we performed ChIP-seq for histone H3K27acetylation, which marks active TSSs and enhancers, in the striatum of R6/1 mice at 8 wk of age when we measured the highest number of differentially expressed genes by RNA-seq in our study. Subsequent analysis of the ChIP-seq data confirmed H3K27acetylation occupancy at TSSs and distal regulatory sites (SI Appendix, Fig. S2A). To identify genes that show changes in H3K27acetylation occupancy in HD mouse striatum, we focused our analysis on a ±10-kb window around each TSS, associating H3K27acetylation peaks to the nearest TSS within this window. Analysis of the data in wild-type mice against a nonspecific background (IgG) showed highest H3K27acetylation signal for genes (top 1,000 genes by tags) that were enriched for GO terms relevant to brain functions, such as regulation of synaptic plasticity, regulation of neuron projection development, regulation of gene expression, and modulation of synaptic transmission, indicating the active chromatin environment surrounding neuronal genes in the striatum (Dataset S4). Comparative analysis of the data for overall changes in H3K27acetylation showed that 118 genes had significantly higher and 429 genes had lower H3K27acetylation levels in the striatum of 8-wk-old R6/1 mice compared to wild-type littermates. Genes with reduced H3K27acetylation in the R6/1 striatum were associated with GO terms, such as regulation of localization and response to endogenous stimulus, while the genes with increased H3K27acetylation were enriched for dendrite morphogenesis and chromatin organization (Dataset S5).
Integrative analysis of the H3K27acetylation ChIP-seq and RNA-seq datasets revealed a highly significant overlap between genes with decreased H3K27acetylation and decreased expression and between those with increased H3K27acetylation and increased expression in the striatum of R6/1 mice at 8 wk of age (t test P < 1.5e-84 and P < 1.5e-20, respectively) (Fig. 2A), suggesting a high degree of coordination between the changes of chromatin and transcription starting from early stages of HD pathogenesis. We next analyzed our H3K27acetylation ChIP-seq data for a chromatin pattern that we previously described as marking the TSSs of neuronal genes that were down-regulated in the R6/2 strain of HD transgenic mice (11). In line with our previous findings, we detected 5 predominant patterns, or classes, of H3K27acetylation that occur in both wild-type and R6/1 mice (Fig. 2B). The class 1 pattern, which has a broad peak of H3K27acetylation downstream of the TSS in wild-type as well as in R6/1 mice, captures 47% of genes that were down-regulated in the striatum of R6/1 mice (Fig. 2C). The remaining 24%, 23%, 1%, and 4% of down-regulated genes were assigned to H3K27acetylation classes 2, 3, 4, and 5, respectively. Genes in class 1 were enriched in GO biological processes that are key for neuronal functions, such as signal transduction, regulation of nervous system development, regulation of cell communication, regulation of neurogenesis, neuron differentiation, axon guidance, behavior, and regulation of synaptic plasticity (Dataset S6). Combined with our previously published results (11), these data suggest that the neuronal genes down-regulated in HD brain present a specific H3K4trimethylation and H3K27acetylation pattern at their TSSs that is directly or indirectly affected by mutant Huntingtin. Moreover, almost none of the genes down-regulated in the striatum of R6/1 mice at 8 wk of age were in H3K27acetylation classes 4 and 5. In contrast to the down-regulated genes, those that were up-regulated in R6/1 mice did not show any significant association to a particular H3K27acetylation TSS profile (SI Appendix, Fig. S2B).
Fig. 2.
H3K27ac changes coordinately with gene-expression levels, extends into the coding region for genes that are down-regulated in HD, and predicts transcription factors with altered activity in the 8-wk-old R6/1 striatum. (A) Box-and-whisker plot for 8-wk-old R6/1 striatum show the distributions of log2 fold-change of H3K27ac reads in R6/1 compared to wild-type animals (y axis) in a 2,000-bp window around the primary TSS of up- and down-regulated genes in R6/1 mice, as measured by RNA-seq. P values were computed using t test (P < 1e-20, up-regulated vs. no change in expression; P < 1e-84, down-regulated vs. no change in expression). (B) Genes were clustered into 5 groups based on their H3K27ac profiles in the striatum of 8-wk-old wild-type animals. Plots show the density of sequence reads in window from −2 k to +3 kb of the TSS. The Inset shows the number of genes in each class. Genes in class 1 (blue) show a broad peak of H3K27ac starting at the TSS and extending into the coding region. (C) Class 1 is enriched in genes that are expressed at lower levels in R6/1. The numbers of genes with reduced expression expected and observed in each class are shown for 8-wk-old R6/1 striatum. (D) Motif enrichment in genomic regions with lower H3K27ac in R6/1 mice compared to respective wild-type littermates. See also SI Appendix, Fig. S2 and Dataset S7.
Histone H3K27acetylation Changes Reveal Transcriptional Regulators Whose Activities Are Altered in the Striatum of Presymptomatic HD Mice.
To identify transcriptional regulators whose activities may be most-proximally altered in striatum during HD pathogenesis, we analyzed the DNA sequence of regions with altered H3K27acetylation in the striatum of 8-wk-old R6/1 mice. To identify the highest-confidence transcription factor motifs, we used 3 separate methods for sequence analysis; the first based on motifs derived from TRANSFAC (27), the second based on overrepresentation of motifs using THEME (28), and the third method was a regression-based approach combining the H3K27acetylation and gene-expression datasets (29). The TRANSFAC-based approach searched for enrichment of known transcription factor binding motifs in so-called histone valleys, which have depletion of H3K27acetylation in the center and H3K27acetylation peaks on either side of the center, as the center of these chromatin structures could be accessible for transcriptional regulator binding (11). The other 2 approaches were based on motif enrichments in the vicinity of the H3K27acetylation peaks.
These analyses revealed a set of common transcriptional regulator motifs. The top motif associated with regions with lower H3K27acetylation in the R6/1 striatum at 8 wk of age was repressor element-1 transcription factor (REST)/neuron-restrictive silencer factor (NRSF)/neuron-restrictive silencer element (NRSE) (P = 1.26e-05), followed by Elk-1 binding motif (P = 2.08e-03) and cAMP-responsive element binding protein (CREB) (P = 6.35e-03) (P values derived from the TRANSFAC-based method) (Fig. 2D and Dataset S7). Both REST (30) and CREB (31) have been previously linked to HD. Our regression-based motif analysis suggested possible regulators linked to H3K27acetylation peaks in the vicinity of the down-regulated genes in R6/1, such as NF-κB, and ETS family members, including Elk-1, NEUROD1, and REST. Notably, some of these motifs, including REST and NF-κB, were also enriched in the promoters of the genes that were differentially expressed in the striatum of 4-wk-old R6/1 mice (Dataset S7), suggesting that their activities may be altered very early in the HD brain. In addition, in the CHL2 mice, H3K27acetylation ChIP-seq followed by motif analyses also showed enrichments for the ETS family, AP-1, and REST/NRSF/NRSE motifs linked to lower H3K27acetylation in the striatum of 1-y-old CHL2 mice. On the other hand, regions associated with higher H3K27acetylation in the R6/1 and CHL2 striata were associated with motifs, including members of the transcription factor OCT, ATF, and TCF families (Dataset S7).
ChIP-Seq Confirms Elk-1 Binding to Genomic Sites with Histone H3K27acetylation Signal in the Striatum of Presymptomatic HD Mice.
The preceding motif analysis of H3K27acetylation in the striatum of R6/1 mice at 8 wk of age predicted as the top-ranked motif the motif of REST, whose role in HD is well established (30), supporting the quality of our H3K27acetylation data. The next motif was that of Elk-1, which has been much less studied in the context of HD. REST is known to act as a transcriptional repressor, while Elk-1 is an activator. To understand the role of these two transcription factors in R6/1 mice, we sought to carry out genome-wide ChIP-seq. Perhaps due to the quality of the antibodies and the limiting amounts of tissue, initial ChIP-seq experiments for REST were not successful. In contrast, the Elk-1 experiments were of high quality.
Elk-1 is a master regulator of genes regulated by neuronal activity and in the striatum, where it is considered a major transcriptional regulator together with CREB (23, 24, 32). In our RNA-seq data, Elk-1 was not differentially expressed between the striata of HD and wild-type animals and, similarly, Western blotting showed that total cellular Elk-1 protein levels were not changed between the striata of R6/1 and wild-type mice at 8 wk of age (SI Appendix, Fig. S3 A and B). We used ChIP-seq to test whether Elk-1’s binding to DNA is in fact altered in HD, as predicted by our motif-analysis results. Analysis of Elk-1 ChIP-seq data showed that the top genes bound by Elk-1 in wild-type animals were associated with GO biological processes, including regulation of chromatin organization, gene expression, RNA metabolic process to cellular localization, regulation of cell differentiation, and cellular response to stimulus (Dataset S8).
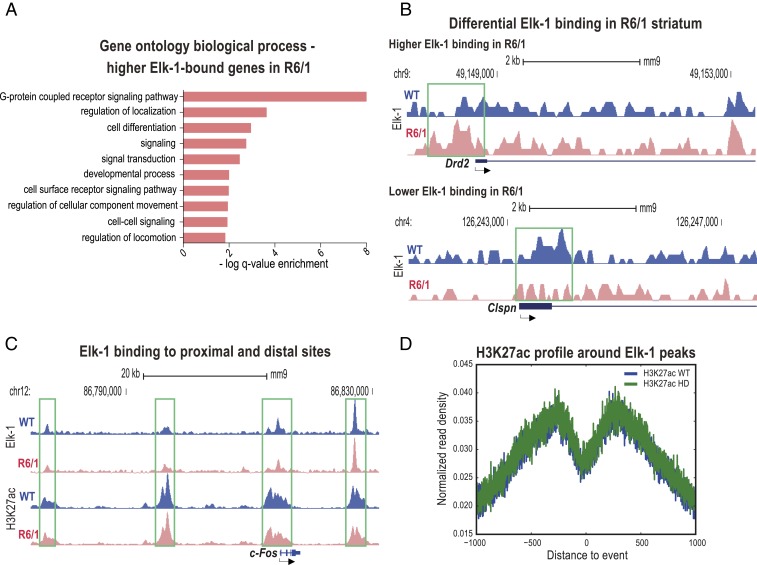
Differential analysis of the Elk-1 ChIP-seq data revealed many sites where Elk-1 binding differed significantly between the R6/1 vs. wild-type striata at 8 wk of age: 1,261 genes showed significantly higher and 193 genes showed lower Elk-1 binding in the R6/1 striatum compared to wild-type littermates (P < 1e-5). Genes with increased binding in R6/1 were enriched for the G protein-coupled receptor signaling pathway, signal transduction, cell differentiation, and regulation of localization, cellular calcium ion homeostasis GO terms, while genes with reduced binding in the R6/1 striatum were not significantly enriched for any particular categories (Fig. 3A and Dataset S9). Importantly, of 687 differentially expressed genes in our 8-wk-old R6/1 RNA-seq dataset, 101 were significantly differentially bound by Elk-1 (hypergeometric test P value for the overlap is P < 0.02) (Dataset S10). Some of these genes were Drd2, Clspn, Adora2a, Mapk4, and Rasgrp1. Fig. 3B shows genome browser views of example genes for increased and reduced Elk-1 binding around promoters of genes in R6/1, Drd2 and Clspn, respectively.
Fig. 3.
Elk-1 binds to genomic sites with H3K27ac signal, and its binding is altered in the 8-wk-old R6/1 striatum. (A) GO analysis of biological processes associated with higher Elk-1 binding in R6/1s compared to wild-type mice. (B) University of California, Santa Cruz (UCSC) Genome Browser images representing normalized Elk-1 ChIP-seq read density from 8-wk-old R6/1 and respective wild-type mice mapped at Drd2 and Claspn example gene loci. Green boxes indicate the Elk-1 occupancy around the TSSs. (C) Tracks from the UCSC Genome Browser displaying normalized Elk-1 and H3K27ac ChIP-seq read density from 8-wk-old R6/1 and respective wild-type mice mapped at a c-Fos example gene locus for proximal and distal regulatory site binding by Elk-1. Green boxes indicate the Elk-1 occupancy around the TSSs and distal sites. (D) H3K27ac profile centering on all Elk-1 peaks shows a valley-shaped chromatin pattern. See also SI Appendix, Fig. S3.
As expected, the Elk-1 motif was top among the enriched motifs within the Elk-1–bound sites (Dataset S11). However, when we focused specifically on regions that changed in binding, we were able to identify additional motifs that may provide insights into combinatorial regulation. Sites with increased Elk-1 binding in R6/1 were enriched for a number of motifs, including the Pax transcription factor family (P = 2.8e-07). On the other hand, reduced binding in R6/1 was associated with a distinct set of motifs, including the Egr transcription factor family DNA binding motif (P = 1.4e-11) (Dataset S12).
We next analyzed the localization of the Elk-1–bound sites with respect to TSSs. Notably, 45% of the Elk-1–bound sites in wild-type mice striatum were promoter regions; however, a majority of the differential Elk-1–bound sites between the R6/1 and wild-types were in distal sites (65% of all differential binding) (SI Appendix, Fig. S3C). Elk-1 binding was present both at the promoter and distal enhancer sites around c-Fos and Egr-1 genes (Fig. 3C and SI Appendix, Fig. S3D) and, as illustrated in Fig. 3C, Elk-1 peaks fell precisely within the so-called histone valleys of H3K27acetylation regions around the c-Fos gene. In line with this observation, genome-wide analysis of H3K27acetylation read distribution around all Elk-1 binding events revealed a precise valley shape with Elk-1 binding in its center (Fig. 3D). However, this H3K27acetylation valley profile did not differ in acetylation levels between the R6/1 and wild-type mice, suggesting that the differences observed in H3K27acetylation in association with Elk-1 binding were not global but specific changes linked to certain genes in R6/1 striatum.
Elk-1 Exerts Beneficial Effects Against Mutant Huntingtin-Mediated Toxicity in a Primary Striatal Culture Model of HD.
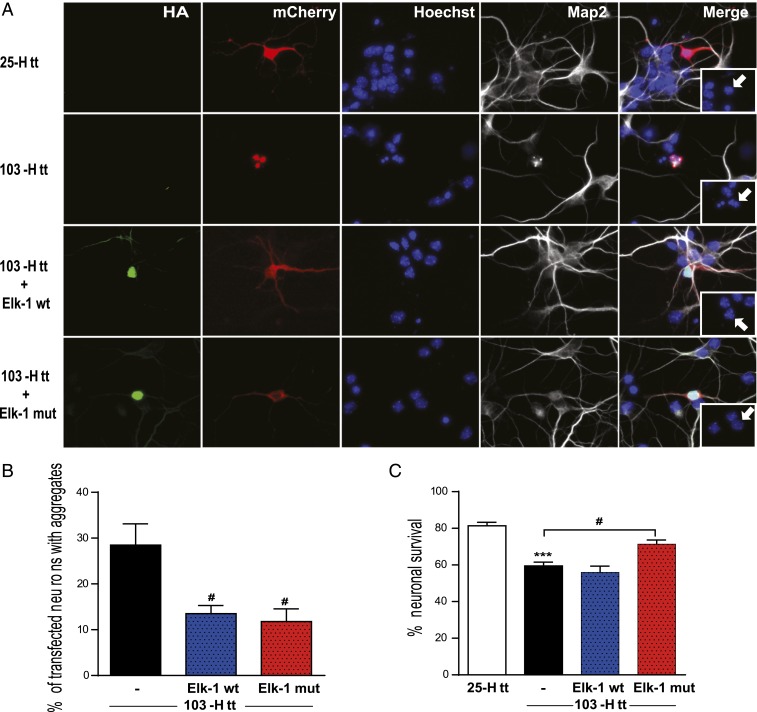
Reduction of Elk-1 expression via small-interfering RNAs was previously reported to exacerbate mutant Huntingtin toxicity in a cell line model of HD (26). To evaluate Elk-1’s beneficial potential against mutant Huntingtin toxicity, we used primary striatal neurons in culture expressing human HTT exon 1 encoding 25 (Wt-HTT) or 103 polyglutamine (Mut-HTT) stretch according to the previously established protocols (33, 34). Spontaneous degeneration and aggregate formation appear as soon as 24 h posttransfection with mutant HTT when compared to Wt-HTT transfected neurons (Fig. 4A). Because striatal neurons in culture are pure GABAergic, and hence devoid of spontaneous activity, we designed a mutated version of Elk-1 that is constitutively nuclear. It has been previously shown that ERK-induced phosphorylation of Elk-1 drives its nuclear accumulation upon glutamate stimulation. Mutation of Ser383-Ser389 phosphorylation sites of Elk-1 into Aspartate constitutively mimics phosphorylation and nuclear expression of Elk-1 (35). On the other hand, sumoylation of Elk-1 is responsible for its nuclear export (36). We therefore designed and constructed a double-mutant version of Elk-1 that is constituvely active for phosphorylation and inactive for sumoylation (3R-Asp-Elk-1 or Elk-1 mut). Cotransfection of striatal cultures with Elk-1 wild-type (Elk-1 wt) or its mutated form (Elk-1 mut) resulted in significant reduction (more than 2-fold for each) in aggregate formation, as assessed 24 h after transfection of neurons with the Elk-1 constructs (quantified based on fluorescent-protein labeling) (Fig. 4 A and B). Neuronal survival was measured 24 h after transfection and assessed based on DNA labeling with Hoechst. It showed a significant enhancement of survival by the constitutively active Elk-1 mutant expression (Fig. 4 A and C).
Fig. 4.
Elk-1 exerts beneficial effects in a primary striatal neuron culture model of HD. (A) Representative images of primary mouse striatal neurons transfected on DIV6 (in vitro day 6 in culture) with cDNAs encoding 25Q-Htt or 103Q-Htt (mCherry red) with or without HA-tagged versions of Elk-1-wt or Elk-1-mut (3R-Asp-Elk-1) (green). Immunocytochemical detection of MAP2 (grey) and Hoechst (blue) were performed to analyze the neuronal integrity. Note that 103-Htt-expressing neuron shows progressive nuclear localization of aggregates (red), neuritic retraction (grey) and DNA damage (blue). White arrows depict nuclear morphology of transfected neurons. (Magnification in the images and the Insets: 40×.) (B and C) Quantification of aggregate formation (B) and neuronal survival based on Hoechst labeling (C) in neurons were performed in transfected cells 24 h after transfection, from 3 independent experiments (100 transfected neurons per experiment) and expressed as mean ± SEM. ANOVA followed by Tukey post-hoc test. ***P < 0.001, 25Q-Htt-expressing neurons vs. 103Q-Htt-expressing neurons; #P < 0.05, 103Q-Htt-expressing neurons vs. 103Q-Htt/Elk-1-Wt or 103Q-Htt/Elk-1-mut-expressing neurons.
AAV-Mediated Elk-1 Overexpression in the Striatum Alleviates Transcriptional Dysregulation in R6/1 Mice.
Given the beneficial effects of Elk-1 in vitro, we next planned an experiment to test Elk-1’s therapeutic potential in HD in vivo. Our goal was to test the hypothesis that overexpression of Elk-1 transcription factor, delivered during prodromal disease stage, can alleviate transcriptional dysregulation, a key mechanism of pathogenesis in HD. For this purpose, we overexpressed the mouse Elk-1 gene in the striatum of R6/1 mice via an AAV-mediated gene-expression strategy. The AAV2/1 serotype was previously shown to induce long-term and widespread transduction of the neurons with no associated toxicity in mouse striatum (37, 38). AAV2/1-expressing Elk-1, along with internal ribosome entry site-based bicistronic coexpression of mCherry (Elk-1 AAV) or only the mCherry reporter (control AAV), were unilaterally delivered into the mouse dorsal striatum at 5 to 6 wk of age by stereotactic injections performed according to our previously published protocol (39). After an incubation period of 4 to 5 wk, mice were killed and brains were collected for analysis of gene expression.
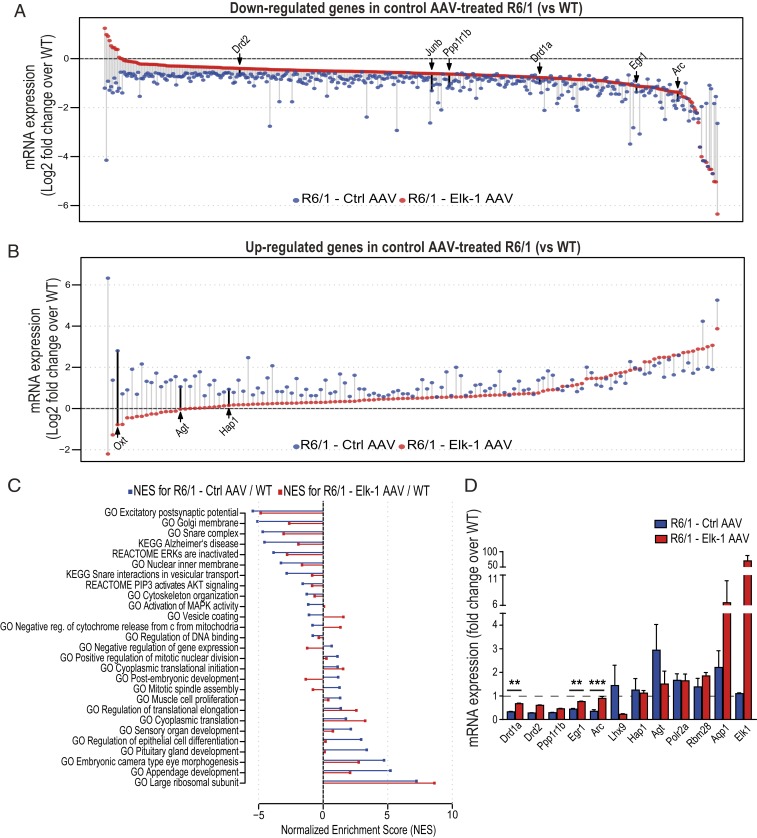
RNA-seq of striatal extracts revealed 514 differentially expressed genes between R6/1 and wild-type animals treated with control AAV, 387 of which showed lower expression in the R6/1 striatum compared to wild-type mice (at log2 fold-change cutoff of 0.5 and FDR of 0.1) (Fig. 5 A and B). As expected, dysregulated genes in the R6/1 striatum included key HD-associated genes, such as Drd1a, Drd2, Ppp1r1b (encoding the striatal marker DARPP32), Penk, and Adora2a. Next, analysis of Elk-1 AAV-injected R6/1 striatal transcriptome revealed 89 significantly differentially expressed genes compared to the control AAV-injected R6/1s (at log2 fold-change cutoff of 0.5 and FDR of 0.1); among these, 58 genes were also differentially expressed between the R6/1 and wild-type animals within the control AAV treatment group. Further analysis of the datasets, without imposing cutoff criteria on the data, revealed a much more extensive impact of Elk-1 AAV treatment on expression of the dysregulated genes in HD. Of the 387 down-regulated genes in the control AAV-injected R6/1 striatum, expression of 88% were enhanced by Elk-1 AAV treatment of R6/1s, and among these were key genes, such as Drd1a, Drd2, Ppp1r1b, and Egr1, whose expression showed a shift toward wild-type levels (Fig. 5A and Dataset S13). Similarly, expression of 78% of the genes up-regulated in the control AAV-injected R6/1 striatum were reduced by the Elk-1 AAV treatment. The effect of Elk-1 on the up-regulated genes in R6/1 is exemplified by Oxt, Agt, and Hap1 genes, which have been previously implicated in HD pathophysiology (40–42) (Fig. 5B and Dataset S13). Taken together, Fig. 5 A and B visualize a large effect of Elk-1 AAV treatment to counter transcriptional dysregulation induced by mutant Huntingtin expression in the striatum of R6/1 mice.
Fig. 5.
AAV-mediated Elk-1 overexpression alleviates dysregulation of genes in R6/1 striatum. (A and B) Dot plots showing down-regulated (A) and up-regulated (B) genes determined by RNA-seq in the striatum of R6/1 mice compared to wild-type littermates within the control AAV treatment group. Each data point represents log2 of fold-change value (HD fragments per kilobase of transcript per million mapped reads [FPKM]/wild-type FPKM) of a differentially expressed gene in R6/1 treated with Ctrl AAV (blue data points) and in R6/1 treated with Elk-1 AAV (red data points). Several previously identified HD-associated genes are labeled. n = 2 to 4 mice per genotype per group. (C) ssGSEA was performed for Elk-1 AAV- and Ctrl AAV-treated R6/1 striatal transcriptomes using log2 of fold-changes (compared to Ctrl AAV-treated wild-types) in expression of each gene in order to identify enriched gene sets from the MSigDB. A selection from the most enriched gene sets (FDR < 0.005) in Ctrl AAV-treated R6/1 mice (blue bars) and their enrichment in Elk-1 AAV-treated R6/1s (red bars) are plotted as bar graphs. Gene sets enriched in up-regulated genes in HD have positive enrichment scores, while those that are down-regulated in HD have negative enrichment scores. Standard names of the gene sets from the MSigDB are listed on the plot. NES, normalized enrichment score. (D) Quantitative RT-PCR confirmation of the expression of several key dysregulated genes (Drd1a, Drd2, Egr-1, Arc, Ppp1r1b, Lhx9, Hap1, Agt, Polr2a, Rbm28, Aqp1) in HD as well as of Elk-1 overexpression by Elk-1-expressing AAV (n = 3 to 5 mice per genotype per group). Data presented as mean ± SEM. ANOVA with Tukey post hoc test. **P < 0.01, ***P < 0.001. See also SI Appendix, Fig. S4.
Next, we explored which pathways were affected by Elk-1 overexpression. Using single-sample gene set enrichment analysis (ssGSEA) (43, 44) and gene sets from the Molecular Signatures Database (MSigDB) (45, 46), we analyzed all measured genes ranked by their fold-changes in expression in control AAV-treated R6/1 and Elk-1 AAV-treated R6/1 striatal transcriptomes compared to wild-types. This analysis associated a large number of gene sets with genes with altered expression in R6/1 striatum compared to wild-type mice within the control AAV-treated group (FDR < 0.005) (Fig. 5C and Dataset S14). Fig. 5C illustrates a selection from the most enriched gene sets in control AAV-treated R6/1 mice and their enrichments in Elk-1 AAV-treated R6/1s. The results point toward up-regulation of development-related gene sets, as well as those associated to ribosome and translation in R6/1 striatum compared to wild-types. Importantly, Elk-1 AAV treatment reduced the positive enrichment scores of the developmental gene sets while the up-regulation of translation and ribosome gene sets in R6/1s were not affected by Elk-1 overexpression. Among the negatively enriched gene sets in R6/1 striatum (lower expression in R6/1 vs. wild-type), notably the expression of gene sets related to the SNARE complex, vesicle coating, MAPK activity, and cytochrome c release from mitochondria, which is an initial step in apoptosis, were shifted toward recovery by Elk-1 overexpression in R6/1 striatum. In summary, these results indicate a global transcriptomic effect of Elk-1 overexpression that, at least to a certain extent, repairs the transcriptional effects of mutant Huntingtin gene expression in R6/1 striatum and promote a role for Elk-1 as an effective therapeutic target for rescue of aberrant transcriptional profiles in HD.
We validated the RNA-seq findings by quantitative RT-PCR, confirming the effect of Elk-1 AAV on expression of several key dysregulated genes (Drd1a, Drd2, Egr-1, Arc, Ppp1r1b, Lhx9, Hap1, Agt, Polr2a, Rbm28, Aqp1) in R6/1 striatum, as well as of Elk-1 overexpression by Elk-1–expressing AAV (Fig. 5D and SI Appendix, Fig. S4A). Additional qRT-PCR for Iba1 and mCherry expression showed that the AAVs used in our study did not induce a change in Iba1 (microglial marker) mRNA expression in mouse striatum, and confirmed robust gene expression (mCherry) from the AAVs in the ipsilateral striatum compared to the contralateral side (SI Appendix, Fig. S4B), as also shown by Western blotting for Elk-1 protein in the ipsilateral striatum (SI Appendix, Fig. S4C). Furthermore, in line with previous reports for the AAV2/1 serotype (37, 38), fluorescent microscopy demonstrated widespread transduction of striatum by the AAVs as illustrated by mCherry protein signal in coronal brain sections along the rostro-caudal axis of the mouse striatum (SI Appendix, Fig. S4D).
Discussion
An important goal of this study was to identify potential targets to alleviate transcriptional dysregulation in HD for early therapeutic intervention in the course of disease, a treatment approach that may be utilized effectively alone or in combination with the strategies, which lower the levels of toxic Htt. To accomplish this goal, we initially analyzed genome-wide transcriptional and epigenetic changes in prodromal stage Huntington’s disease using two different mouse models, the transgenic R6/1 and the mouse full-length CHL2 (CAG150) knockin mice. Transcriptional profiling revealed common dysregulation of many genes in HD striatum prior to the onset of overt disease symptoms in R6/1 and CHL2 mice and showed a large overlap with transcriptional changes in caudate nucleus from low-grade postmortem HD patient brains. Analysis of the epigenetic landscape changes via genome-wide measurement of histone H3K27acetylation led to identification of transcription factors likely to be linked to genes with altered expression during early HD. In particular, direct ChIP-seq for the Elk-1 transcription factor validated that Elk-1 binding to DNA is altered in R6/1 striatum as early as at 8 wk of age, suggesting a role for Elk-1 in transcriptional dysregulation in prodromal HD. In a primary striatal culture model of HD, enhanced Elk-1 expression resulted in a decrease in the number of Huntingtin aggregates and improved neuronal survival promoted by a constitutively active mutant form of Elk-1. AAV-mediated Elk-1 overexpression in vivo led to extensive amelioration of transcription of the genes that are dysregulated in the striatum of R6/1 mice. Given that transcriptional dysregulation is an early and central pathogenic mechanism in HD, therapies that promote Elk-1 expression and its transcriptional activity may alleviate aspects of HD progression.
Transcriptional dysregulation is a central mechanism in the cascade of pathogenic events triggered by mutant Huntingtin (3, 10, 11). Here, focusing on prodromal disease stage, we found that transcription of hundreds of key neuronal genes was altered in HD mice striatum already before the onset of overt disease symptoms. To our knowledge, this study presents the earliest transcriptional profiling data reported so far in any HD mouse model (4-wk-old R6/1). Comparison of the mice differential expression datasets with a previous HD patient postmortem caudate microarray data (up to Vonsattel grade 2) (5) showed a large overlap of dysregulated genes, suggesting that HD mice faithfully recapitulate key elements of transcriptional dysfunction in human HD and that a significant portion of the early transcriptional changes persists throughout the disease progression. Importantly, these concordant transcriptional changes in postmortem HD patient caudate and in early-stage HD mouse striatum indicate that these changes reflect true changes in transcription, and although they can be inflated by neuronal loss, they are not solely due to neuronal loss, which becomes predominant in late human HD brain with progressing disease. In this respect, information gained from early-stage mouse studies are invaluable for making a distinction between the detected gene-expression changes that are associated to neuronal dysfunction in the absence of neuronal loss, and it can be critical for prioritizing the targets for testing in preclinical mouse studies. In addition, transcriptional changes that are persistent throughout the disease course may expand the temporal window of their targeting, making them viable therapeutic candidates for different stages of the disease.
Notably, our transcriptional dataset from 8-wk-old R6/1 mice was dominated by down-regulation of genes (90% of all differentially expressed genes) and these were enriched for GO terms typical for neuronal functions, indicating that disruption of neuronal gene-expression programs predominates the early transcriptional changes in HD pathogenesis. Supporting this, our histone H3K27acetylation ChIP-seq data showed a distinctly broad chromatin pattern, which is present both in wild-type and HD mice, surrounding the promoters of neuronal genes that are down-regulated in R6/1 striatum. This is a chromatin pattern we previously described in R6/2 mouse brain, as well as in HD patient induced pluripotent stem cell-derived medium spiny neurons (11, 47).
Broad chromatin patterns at regulatory regions and superenhancers control the expression of genes that are important for maintenance of tissue identities, and therefore are strongly expressed in their respective tissue types (48), such as the expression of neuron-specific genes in brain. It is therefore conceivable that perturbations of these chromatin patterns may lead to loss of control of tissue-specific gene expression and the consequent loss of tissue identity may be associated to aberrant expression of developmental genes that are normally not expressed in differentiated cell types. In line with this, our present and previous RNA-seq datasets showed aberrant expression of genes belonging to various developmental programs in HD mice brains. Our current and previous epigenetic findings, revealing a distinct chromatin pattern around the promoters of neuronal genes that are down-regulated in the HD brain, points to a neuron-specific regulatory machinery that is selectively perturbed by mutant Huntingtin expression in neurons and that this machinery is not present or important in nonneuronal cell types. This is an exciting finding for progressing the understanding of HD pathogenesis in that it uncovers a possible key mechanism underlying the enhanced vulnerability of neurons (i.e., regulation of neuron-specific gene-expression programs) to mutant Huntingtin toxicity. These data also highlight the role of transcriptional dysregulation as a central disease mechanism in leading to neuronal dysfunction and demise in HD. Moreover, our current histone acetylation data show that expression of genes that display other chromatin patterns, which are not related to neuronal functions, are significantly less affected in the R6/1 striatum, confirming again the selectivity of mutant Huntingtin toxicity to expression of neuronal genes in HD.
DNA sequence analysis for known transcription factor DNA-binding motifs revealed strong enrichments for REST, CREB, NEUROD1, and Elk-1 transcription factors in connection with lower H3K27acetylation both in the R6/1 and CHL2 ChIP-seq datasets. Of particular relevance is the prediction that REST is the most significantly enriched motif in the regulatory regions with lower H3K27acetylation signal in HD, as the pathogenic role of REST in leading to disruption of expression of neuronal genes, such as Bdnf, is well documented (30). A role for NEUROD1 in HD pathology was implicated in relation to impaired hippocampal neurogenesis in HD and targeting NEUROD1 led to a beneficial outcome, amelioration of cognitive impairment, in R6/2 mice (47, 49). Another transcription factor with strong motif enrichment in our histone H3K27acetylation data was Elk-1, a master regulator known in particular for the control of neuronal activity-regulated immediate-early genes. Elk-1 is a member of the TCF family that is known to regulate gene expression via the serum response element DNA consensus site in association with a dimer of serum response factor (SRF) (24). However, the serum response element motif was not among the top-ranked motifs in our H3K27acetylation ChIP-seq data. In line with our finding, a previous study demonstrated that cooccupancy of promoters by SRF and Elk-1 accounted only for around 20% of the Elk-1 targets, indicating that Elk-1 can function more widely in an SRF-independent manner (50).
For triggering Elk-1’s transcriptional activity, its phosphorylation at specific residues in response to MAPKs, including ERK, is a critical event. Of note, the ERK signaling pathway plays a pivotal role in neuronal plasticity and long-term memory formation, including in response to addictive drugs in the striatum (51, 52). Elk-1 shows enriched neuronal expression in the striatum (23, 24, 32). Yet, Elk-1’s role in transcriptional dysregulation in HD has not been studied. Here, by ChIP-seq, we profiled Elk-1 binding throughout the genome and found that its binding to DNA was increased in the striatum of R6/1 mice at 8 wk of age. Previous reports showed increased Elk-1 protein expression, as well as its phosphorylation and nuclear localization in striatum at late disease stages in R6/1 (at 30 wk of age) and R6/2 mice (at 8 and 12 wk of age) (26, 53). Together with our ChIP-seq data, these findings may imply a compensatory role for Elk-1’s enhanced binding activity against ongoing transcriptional dysregulation in HD. As an example we highlighted Drd2, which displayed enhanced Elk-1 occupancy in its 5′ promoter region in R6/1. In contrast, lower Elk-1 binding can be exemplified by Clspn (Claspin), a gene related to antiapoptotic functions in neurons (54), which is down-regulated in our current RNA-seq data from the R6/1 striatum as well as in our previous expression data from R6/2 mice (11). Elk-1 has been implicated in the control of several genes dysregulated in HD, such as c-Fos, Junb, and Egr1 (53, 55, 56). Moreover, activation of Elk-1 after toxic stimuli was shown to have a beneficial effect (25), and its inhibition caused apoptosis in neuronal cultures stimulated with nerve growth factor (57). In the context of HD, inhibition of Elk-1 was shown to induce cell death in striatal STHdh cell lines (26). Here, we show that enhanced expression of a constitutively nuclear version of Elk-1 (i.e., mutated in its phosphorylation sites for MAPK/ERK and sumoylation sites) (35) reduced aggregate formation and neuronal apoptosis induced by mutant Huntingtin expression in primary striatal neurons in culture, promoting a beneficial role for Elk-1 against HD pathogenesis.
Unlike REST, which is a transcriptional repressor, Elk-1 is a transcriptional activator. As such, directly activating Elk-1 or its targets could be of therapeutic benefit. We therefore sought to map out the transcriptional program of Elk-1 and directly determine the consequences of enhancing Elk-1 protein levels in vivo. For this purpose, we performed unilateral stereotactic injections of Elk-1 AAV or control AAV into the striata of R6/1 and wild-type mice at a prodromal stage (5 to 6 wk of age). Subsequent RNA-seq after a month of viral incubation revealed a striking counteracting effect of Elk-1 AAV treatment on the dysregulated transcripts in R6/1 striatum since the expression levels of majority of the differentially expressed genes in R6/1 mice were shifted toward wild-type expression levels upon Elk-1 AAV treatment of R6/1s. GSEA analysis performed on all measured transcripts in our RNA-seq experiment identified many highly significantly enriched gene sets whose members collectively showed changes in their expression in R6/1 striatum compared to wild-type mice. Importantly, of the many negatively enriched gene sets (lower-expressed in R6/1), Elk-1 AAV treatment exhibited a repair effect on the expression of genes that are related to the SNARE complex, vesicle coating, MAPK activity, the ERK and PI3K/Akt pathways, and cytochrome c release from mitochondria, which is a first step in the cascade of events leading up to apoptotic cell death. In particular, the latter 3 gene sets, which play key roles in mitophagy and clearance of unfolded proteins (58), may contribute to Elk-1’s beneficial effects that we observed in vitro on mutant Huntingtin aggregation and striatal neuron survival. Among the positively enriched gene sets (higher-expressed in R6/1), Elk-1 exerted a normalizing effect especially on those gene sets that are related to development, such as sensory organ development, pituitary gland development, appendage development, and embryonic eye development. Our current and previous findings and reports from other laboratories showed aberrant expression of nonneuronal developmental genes as part of disease pathogenesis in HD (11, 59). In the present study, Elk-1 overexpression directly or indirectly leads to an overall normalizing net effect on the expression of these genes and this effect may contribute largely to Elk-1’s beneficial effects in the HD mouse brain. Interestingly, this effect appears to be preferential, as Elk-1 overexpression did not normalize the up-regulation of translation-related genes, which itself may be an endogenous compensatory mechanism against the ongoing transcriptional disruption in HD. Our results point toward a global transcriptomic countereffect of Elk-1 overexpression that works toward normalization of transcriptional dysregulation in HD striatum. Collectively, our findings promote a beneficial compensatory role for Elk-1 against transcriptional dysregulation in HD. Considering the large overlap of similar transcriptional changes between the striatum and cerebral cortex in HD both in mouse and human (4, 5, 11), which include common down-regulation of neuronal genes and up-regulation of developmental and translation-related gene programs, Elk-1’s therapeutic effects can be expected to be widespread, including multiple affected brain areas in HD, such as the cerebral cortex.
Further work using a slower progressing HD mouse model that matches adult onset human HD better, transduction in additional critical brain regions (e.g., cortical areas), intervention earlier in the time course of pathology, and assessment of also other key aspects of HD pathogenesis (e.g., formation of mutant Htt aggregates) may enhance the therapeutic potentials of Elk-1 transcription factor in HD and allow a more comprehensive evaluation of its therapeutic effects. Nevertheless, we have demonstrated that enhanced Elk-1 levels in the striatum results in improved molecular features in the R6/1 line of HD. Our results suggest that gene therapy for Elk-1 or its specific pharmacological activation may be a viable approach for targeting transcriptional dysregulation in HD, and perhaps for other neuropsychiatric conditions with similar molecular pathology.
Materials and Methods
RNA-Seq.
RNA-seq was performed following the Illumina mRNA Sequencing Sample Preparation Guide (Ilumina, Cat # RS-930-1001) and according to our previously published protocol (11). Details of the experimental procedures and data analyses are provided in SI Appendix, Materials and Methods.
ChIP-Seq.
Brain tissues were cross-linked with 1% (vol/vol) formaldehyde for 10 min, and the cross-linking was quenched by 0.125 M glycine. The cross-linked tissue was then homogenized, rinsed with PBS, pelleted, and frozen in liquid nitrogen. ChIP-seq assays were performed as previously described (11, 60). Details of the ChIP-seq experiments and their computational analyses are provided in SI Appendix, Materials and Methods.
Generation of AAV.
Mouse Elk-1 cDNA along with mCherry cDNA linked to an internal ribosome entry site fragment were cloned into a recombinant AAV2/1 AAV expression vector containing a CMV promoter (provided by the University of Iowa Vector Core, Iowa City). Control vector-expressed mCherry was under the control of the same promoter (AAV2/1-mCherry). Purified viruses were generated by the University of Iowa Vector Core. The resulting titers of the AAV2/1-Elk-1 and AAV2/1-mCherry control were determined to be 2.54 × 1,013 and 1.66 × 1,013 vector genomes (VG) per milliliter, respectively, using quantitative PCR.
Surgical Procedures for Stereotaxic Delivery of AAVs into Mouse Striatum.
The stereotaxic injections were carried out as previously described (39). Stereotaxic coordinates (striatum) were 0.9-mm anterior, 1.8-mm lateral, and 3.3-mm ventral to bregma. Animals were kept at a steady temperature using a heating pad throughout the operation and fed wet food for the first day postoperative. Meloxicam was used for preemptive and postoperative analgesia. All animal procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee at Massachusetts Institute of Technology (approved animal experiment proposal number; 0212-014-15).
Data Availability.
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (61).
Supplementary Material
Acknowledgments
We acknowledge generation of AAVs by the University of Iowa Vector Core; and thank the Massachusetts Institute of Technology (MIT) BioMicro Center and the Scientific Genomics Platforms at Max-Delbrück-Center for Molecular Medicine Berlin and at Berlin Institute of Health for sequencing of the RNA-seq and ChIP-Seq libraries. This work was supported by German Research Foundation (Deutsche Forschungsgemeinschaft) Exc 257/2 NeuroCure Cluster of Excellence (F.Y.); the Hereditary Disease Foundation (F.Y.); National Institute of Health Grant R01 NS089076 (to E.F. and D.E.H.); and sequencing support from the National Institutes of Health (P30-ES002109) through the MIT BioMicro Center.
Footnotes
The authors declare no competing interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE140118).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908113116/-/DCSupplemental.
References
- 1.Molero A. E., et al. , Selective expression of mutant huntingtin during development recapitulates characteristic features of Huntington’s disease. Proc. Natl. Acad. Sci. U.S.A. 113, 5736–5741 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cha J. H., Transcriptional signatures in Huntington’s disease. Prog. Neurobiol. 83, 228–248 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seredenina T., Luthi-Carter R., What have we learned from gene expression profiles in Huntington’s disease? Neurobiol. Dis. 45, 83–98 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Kuhn A., et al. , Mutant huntingtin’s effects on striatal gene expression in mice recapitulate changes observed in human Huntington’s disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum. Mol. Genet. 16, 1845–1861 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Hodges A., et al. , Regional and cellular gene expression changes in human Huntington’s disease brain. Hum. Mol. Genet. 15, 965–977 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Stack E. C., et al. , Modulation of nucleosome dynamics in Huntington’s disease. Hum. Mol. Genet. 16, 1164–1175 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Sadri-Vakili G., Cha J. H., Mechanisms of disease: Histone modifications in Huntington’s disease. Nat. Clin. Pract. Neurol. 2, 330–338 (2006). [DOI] [PubMed] [Google Scholar]
- 8.McFarland K. N., et al. , Genome-wide histone acetylation is altered in a transgenic mouse model of Huntington’s disease. PLoS One 7, e41423 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadri-Vakili G., et al. , Histones associated with downregulated genes are hypo-acetylated in Huntington’s disease models. Hum. Mol. Genet. 16, 1293–1306 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Ng C. W., et al. , Extensive changes in DNA methylation are associated with expression of mutant huntingtin. Proc. Natl. Acad. Sci. U.S.A. 110, 2354–2359 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vashishtha M., et al. , Targeting H3K4 trimethylation in Huntington disease. Proc. Natl. Acad. Sci. U.S.A. 110, E3027–E3036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross C. A., Tabrizi S. J., Huntington’s disease: From molecular pathogenesis to clinical treatment. Lancet Neurol. 10, 83–98 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Cummings D. M., et al. , Abnormal cortical synaptic plasticity in a mouse model of Huntington’s disease. Brain Res. Bull. 72, 103–107 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Nicniocaill B., Haraldsson B., Hansson O., O’Connor W. T., Brundin P., Altered striatal amino acid neurotransmitter release monitored using microdialysis in R6/1 Huntington transgenic mice. Eur. J. Neurosci. 13, 206–210 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Naver B., et al. , Molecular and behavioral analysis of the R6/1 Huntington’s disease transgenic mouse. Neuroscience 122, 1049–1057 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Mangiarini L., et al. , Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87, 493–506 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Lin C. H., et al. , Neurological abnormalities in a knock-in mouse model of Huntington’s disease. Hum. Mol. Genet. 10, 137–144 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Creyghton M. P., et al. , Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ong C. T., Corces V. G., Enhancers: Emerging roles in cell fate specification. EMBO Rep. 13, 423–430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim T. K., et al. , Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchwalter G., Gross C., Wasylyk B., Ets ternary complex transcription factors. Gene 324, 1–14 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Price M. A., Rogers A. E., Treisman R., Comparative analysis of the ternary complex factors Elk-1, SAP-1a and SAP-2 (ERP/NET). EMBO J. 14, 2589–2601 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sgambato V., et al. , In vivo expression and regulation of Elk-1, a target of the extracellular-regulated kinase signaling pathway, in the adult rat brain. J. Neurosci. 18, 214–226 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Besnard A., Galan-Rodriguez B., Vanhoutte P., Caboche J., Elk-1 a transcription factor with multiple facets in the brain. Front. Neurosci. 5, 35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrer I., Blanco R., Carmona M., Differential expression of active, phosphorylation-dependent MAP kinases, MAPK/ERK, SAPK/JNK and p38, and specific transcription factor substrates following quinolinic acid excitotoxicity in the rat. Brain Res. Mol. Brain Res. 94, 48–58 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Anglada-Huguet M., Giralt A., Perez-Navarro E., Alberch J., Xifró X., Activation of Elk-1 participates as a neuroprotective compensatory mechanism in models of Huntington’s disease. J. Neurochem. 121, 639–648 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Matys V., et al. , TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34, D108–D110 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macisaac K. D., et al. , A hypothesis-based approach for identifying the binding specificity of regulatory proteins from chromatin immunoprecipitation data. Bioinformatics 22, 423–429 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Soltis A. R., et al. , Hepatic dysfunction caused by consumption of a high-fat diet. Cell Rep. 21, 3317–3328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuccato C., et al. , Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat. Genet. 35, 76–83 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Shimohata T., et al. , Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nat. Genet. 26, 29–36 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Vanhoutte P., et al. , Glutamate induces phosphorylation of Elk-1 and CREB, along with c-fos activation, via an extracellular signal-regulated kinase-dependent pathway in brain slices. Mol. Cell. Biol. 19, 136–146 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia M., et al. , The mitochondrial toxin 3-nitropropionic acid induces striatal neurodegeneration via a c-Jun N-terminal kinase/c-Jun module. J. Neurosci. 22, 2174–2184 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charvin D., Vanhoutte P., Pagès C., Borrelli E., Caboche J., Unraveling a role for dopamine in Huntington’s disease: The dual role of reactive oxygen species and D2 receptor stimulation. Proc. Natl. Acad. Sci. U.S.A. 102, 12218–12223 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavaur J., et al. , A TAT-DEF-Elk-1 peptide regulates the cytonuclear trafficking of Elk-1 and controls cytoskeleton dynamics. J. Neurosci. 27, 14448–14458 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salinas S., et al. , SUMOylation regulates nucleo-cytoplasmic shuttling of Elk-1. J. Cell Biol. 165, 767–773 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanek L. M., et al. , Silencing mutant huntingtin by adeno-associated virus-mediated RNA interference ameliorates disease manifestations in the YAC128 mouse model of Huntington’s disease. Hum. Gene Ther. 25, 461–474 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J. H., et al. , Reinstating aberrant mTORC1 activity in Huntington’s disease mice improves disease phenotypes. Neuron 85, 303–315 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crook Z. R., Housman D. E., Dysregulation of dopamine receptor D2 as a sensitive measure for Huntington disease pathology in model mice. Proc. Natl. Acad. Sci. U.S.A. 109, 7487–7492 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Mello W. C., Gerena Y., Ayala-Peña S., Angiotensins and Huntington’s disease: A study on immortalized progenitor striatal cell lines. Front. Endocrinol. 8, 108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vercruysse P., Vieau D., Blum D., Petersén Å., Dupuis L., Hypothalamic alterations in neurodegenerative diseases and their relation to abnormal energy metabolism. Front. Mol. Neurosci. 11, 2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Metzger S., et al. , Huntingtin-associated protein-1 is a modifier of the age-at-onset of Huntington’s disease. Hum. Mol. Genet. 17, 1137–1146 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barbie D. A., et al. , Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liberzon A., A description of the molecular signatures database (MSigDB) web site. Methods Mol. Biol. 1150, 153–160 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Liberzon A., et al. ; The Molecular Signatures Database , The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.HD iPSC Consortium , Developmental alterations in Huntington’s disease neural cells and pharmacological rescue in cells and mice. Nat. Neurosci. 20, 648–660 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shin H. Y., Targeting super-enhancers for disease treatment and diagnosis. Mol. Cells 41, 506–514 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fedele V., Roybon L., Nordström U., Li J. Y., Brundin P., Neurogenesis in the R6/2 mouse model of Huntington’s disease is impaired at the level of NeuroD1. Neuroscience 173, 76–81 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Boros J., et al. , Overlapping promoter targeting by Elk-1 and other divergent ETS-domain transcription factor family members. Nucleic Acids Res. 37, 7368–7380 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pascoli V., Cahill E., Bellivier F., Caboche J., Vanhoutte P., Extracellular signal-regulated protein kinases 1 and 2 activation by addictive drugs: A signal toward pathological adaptation. Biol. Psychiatry 76, 917–926 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Cahill E., Salery M., Vanhoutte P., Caboche J., Convergence of dopamine and glutamate signaling onto striatal ERK activation in response to drugs of abuse. Front. Pharmacol. 4, 172 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roze E., et al. , Mitogen- and stress-activated protein kinase-1 deficiency is involved in expanded-huntingtin-induced transcriptional dysregulation and striatal death. FASEB J. 22, 1083–1093 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Freudenreich C. H., Lahiri M., Structure-forming CAG/CTG repeat sequences are sensitive to breakage in the absence of Mrc1 checkpoint function and S-phase checkpoint signaling: Implications for trinucleotide repeat expansion diseases. Cell Cycle 3, 1370–1374 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Luthi-Carter R., et al. , Decreased expression of striatal signaling genes in a mouse model of Huntington’s disease. Hum. Mol. Genet. 9, 1259–1271 (2000). [DOI] [PubMed] [Google Scholar]
- 56.Spektor B. S., et al. , Differential D1 and D2 receptor-mediated effects on immediate early gene induction in a transgenic mouse model of Huntington’s disease. Brain Res. Mol. Brain Res. 102, 118–128 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Demir O., et al. , ETS-domain transcription factor Elk-1 mediates neuronal survival: SMN as a potential target. Biochim. Biophys. Acta 1812, 652–662 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Shefa U., et al. , Mitophagy links oxidative stress conditions and neurodegenerative diseases. Neural Regen. Res. 14, 749–756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Labadorf A., et al. , RNA sequence analysis of human Huntington disease brain reveals an extensive increase in inflammatory and developmental gene expression. PLoS One 10, e0143563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.MacIsaac K. D., et al. , A quantitative model of transcriptional regulation reveals the influence of binding location on expression. PLoS Comput. Biol. 6, e1000773 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yildirim F., et al. , Early epigenomic and transcriptional changes reveal Elk-1 transcription factor as a therapeutic target in Huntington’s disease. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE140118. Deposited 7 November, 2019. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (61).