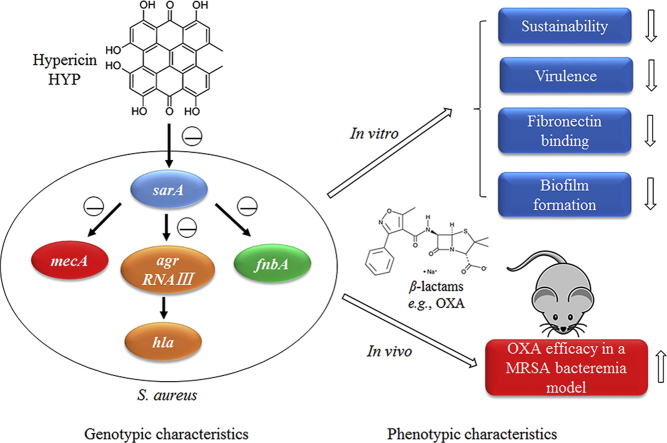
Abstract
Bacteremia is a life-threating syndrome often caused by methicillin-resistant Staphylococcus aureus (MRSA). Thus, there is an urgent need to develop novel approaches to successfully treat this infection. Staphylococcal accessory regulator A (SarA), a global virulence regulator, plays a critical role in pathogenesis and β-lactam antibiotic resistance in Staphylococcus aureus. Hypericin is believed to act as an antibiotic, antidepressant, antiviral and non-specific kinase inhibitor. In the current study, we investigated the impact of hypericin on β-lactam antibiotics susceptibility and mechanism(s) of its activity. We demonstrated that hypericin significantly decreased the minimum inhibitory concentrations of β-lactam antibiotics (e.g., oxacillin, cefazolin and nafcillin), biofilm formation and fibronectin binding in MRSA strain JE2. In addition, hypericin significantly reduced sarA expression, and subsequently decreased mecA, and virulence-related regulators (e.g., agr RNAⅢ) and genes (e.g., fnbA and hla) expression in the studied MRSA strain. Importantly, the in vitro synergistic effect of hypericin with β-lactam antibiotic (e.g., oxacillin) translated into in vivo therapeutic outcome in a murine MRSA bacteremia model. These findings suggest that hypericin plays an important role in abrogation of β-lactam resistance against MRSA through sarA inhibition, and may allow us to repurpose the use of β-lactam antibiotics, which are normally ineffective in the treatment of MRSA infections (e.g., oxacillin).
KEY WORDS: Hypericin, β-Lactams, MRSA, Synergistic effect, SarA
Graphical abstract
In this study, the authors demonstrated that hypericin, as a sarA inhibitor, had synergistic activity with β-lactams both in vitro and in a murine bacteremia model due to MRSA. The mechanisms may be related to significantly reduced biofilm formation, fibronectin binding and expression of virulence genes (e.g., fnbA and hla).
1. Introduction
Staphylococcus aureus (S. aureus), a major human pathogen, is capable of causing many infectious diseases (e.g., pneumonia, endocarditis, and bacteremia)1. Treatment of methicillin-resistant S. aureus (MRSA) infections has become challenging because this pathogen has developed resistance to almost all the standard of care (SOC) antibiotics2, 3. Therefore, there is an urgent need to develop novel therapeutic strategies against these MRSA life-threatening infections.
SarA, a key global regulator, binds on target promoters to control many virulence factors expression in S. aureus4, 5, 6, 7. As reported from prior studies, laboratory-derived sarA mutants exhibited diminished virulence in animal infection models8, 9. Importantly, we have recently reported that sarA mutants become methicillin susceptible with significantly reduced oxacillin resistance vs. their respective parental MRSA strains both in vitro and in experimental endocarditis model10. Therefore, our current studies focus on antimicrobial agents, which could inhibit sarA activity, in combination with SOC anti-S. aureus β-lactams in order to improve and/or repurpose the treatment efficacy of the antibiotics in MRSA invasive infections.
Hypericin (HYP) is a phenanthropeylene quinine pigment naturally occurring in Hypericum perforatum L. (commonly known as St. John's wort). Preclinical and clinical studies demonstrated that it possesses a variety of therapeutic activity (e.g., antidepressant11, anticancer12, and antiviral13). In addition, its antimicrobial properties have also been reported previously, especially on Gram-positive bacteria (e.g., S. aureus and Listeria monocytogenes)14, 15, 16, 17. However, little is known about its mechanism(s) of action.
In the current studies, we investigated the effect of HYP on the susceptibility of β-lactam antibiotics (e.g., oxacillin [OXA], cefazolin [CFZ] and nafcillin [NAF]) and efficacy of HYP in combination with OXA in a murine bacteremia model due to MRSA. We demonstrated that HYP significantly decreased the minimum inhibitory concentrations (MICs) of β-lactam antibiotics, biofilm formation and fibronectin binding in parallel with significantly reduced sarA, agr RNAⅢ and virulence related genes expression (e.g., mecA, fnbA and hla) in MRSA. Importantly, HYP significantly enhanced the efficacy of OXA in an experimental MRSA bacteremia model. These results suggest that the inhibition effect of HYP on sarA expression might be responsible for the synergistic effect with OXA both in vitro and in the treatment outcome in the MRSA bacteremia model.
2. Materials and methods
2.1. Bacterial strains and growth medium
JE2 strain, a plasmid-cured derivative of LAC MRSA USA300, was obtained from the National Institutes of Health Network on Antimicrobial Resistance in S. aureus (NARSA)18. A sarA deletion in MRSA strain JE2 was achieved by transducing sarA::kan mutation from ALC254319. JE2 ΔmecA is a transposon mutant with insertion in S. aureus USA 300_0032 and obtained from the Nebraska Transposon Mutant Library (NTML, Omaha, NE, USA)10. JE2 ΔsarA/pmecA is a sarA mutant strain complemented with pALC6185, which carries the entire mecA locus10. pALC 6185 is a plasmid pEPSA5 containing a 2-kb DNA fragment containing the mecA coding region20. The study strains were stored at −80 °C until thawed for use. Bacteria were routinely grown in tryptic soy broth (TSB) or TSB agar plates otherwise unless specified. All bacterial culture media were purchased from Becton Dickinson (Franklin Lakes, NJ, USA).
2.2. Determination of MICs
The MICs of HYP (Meilun Biotech, Dalian, China) and β-lactam antibiotics, including OXA, CFZ, NAF, and other SOC antibiotics on the study MRSA strains, were determined by a standard broth microdilution method21 as recommended by the Clinical and Laboratory Standards Institute (CLSI; Wayne, PA, USA). All the assays were conducted at least three times on different days and the most consistent results were presented.
2.3. Checkerboard assay
Checkerboard assays were employed to determine in vitro interactions of HYP and β-lactam antibiotics on the study MRSA strain JE2 according to the CLSI guidelines22. In brief, a final inoculum of 5 × 105 colony-forming unit/mL (CFU/mL) of MRSA cells was added into 96-well plates containing two-fold diluted HYP and β-lactam antibiotics in cation-adjusted Mueller Hinton (CAMH) broth (+2% NaCl for OXA). After incubation at 37 °C for 24 h, the combinational activity of HYP with β-lactam antibiotic was analyzed by FIC index21. The FIC was interpreted as follows: synergism, FIC ≤ 0.5; antagonism, FIC > 4.0; and indifferent when 0.5 < FIC ≤ 423.
2.4. In vitro time-kill curves
Time-killing experiments were performed using CAMH broth (+2% NaCl for OXA) with an initial inoculum of ∼5 × 105 CFU/mL of MRSA cells in the presence of sub-MICs of β-lactam antibiotics (e.g., 1/4 × MIC of OXA, CEF, and NAF) or HYP (at 1/16 × MIC or 1/8 × MIC) alone and in combination21. The concentrations of HYP and antibiotics were chosen based on the checkerboard assay results. Viability counts were performed at 0, 2, 4, 6, 8, 12 and 24 h of incubation at 37 °C. Synergistic effect was defined as a ≥2lg decrease of CFU/mL in combination vs. the most active drug alone at 24 h of incubation24.
2.5. Biofilm formation
Biofilm formation of the study strains was performed as previously described25, 26. Briefly, MRSA cells from fresh culture plates were washed and adjusted to a density of 0.5 McFarland standard and diluted 1:10 into brain heart infusion broth supplemented with 0.5% glucose. This suspension was transferred to 96-well tissue culture plates with study compound alone and in combination (1/16 × MIC or 1/8 × MIC of HYP; 1/256 × MIC of OXA, CEF, and NAF) exposure and incubated for 18 h at 37 °C. The specific concentrations of OXA and HYP were chosen based on the impact of the antibiotic alone on the biofilm formation (See Supporting Information Tables S1 and S2). After incubation, the wells were washed, air dried, and stained with safranin (0.1% in distilled water). The adhering dye was dissolved in 30% acetic acid, and absorption was measured at OD490nm to quantify biofilm formation5, 26.
2.6. Adherence to fibronectin
Fibronectin adherence assay was performed as previously described5. Briefly, 6-well tissue culture plates were coated with purified human fibronectin (50 mg/L, Sigma Chemicals, St. Louis, USA) for overnight at 4 °C and then treated with 3% bovine serum albumin (Sigma Chemicals) for 3 h at 37 °C to prevent nonspecific adhesion. Overnight cultured MRSA cells with/without HYP (1/16 × MIC or 1/8 × MIC), OXA (1/256 × MIC) alone and in combination were adjusted to OD600 nm = 1.0 (∼109 CFU/mL) and subsequently diluted 1:100 into fresh TSB with the same exposures of the compounds as for the overnight culture and incubated at 37 °C to OD600 nm ≈ 0.5. Then the MRSA cells (∼103 CFU/well) were added into the plates and incubated for 1 h. After 1 h incubation, plates were washed with PBS, TSB agar was added into each well, and incubated overnight at 37 °C. Adherence to fibronectin was expressed as the percentage (±standard deviation [SD]) of the initial inoculum bound as previously described27.
2.7. Transcription analyses by quantitative real-time PCR (qRT-PCR)
Exponential phase of MRSA cells with/without HYP and/or OXA exposure as descripted in Section 2.6 above were used for the isolation of total RNA by using a RNeasy kit (Qiagen, Los Angeles, CA, USA) as described previously10. Briefly, 2 μg of DNase treated RNA was transcribed into complementary DNA. The amplification of sarA, agr RNAⅢ, mecA, fnbA, hla and gyrB were performed using primers as described previously (see Table 1)7, 28, 29. qRT-PCR was performed using an ABI Prism 7000 instrument (Applied Biosystems, Los Angeles, CA, USA) and SYBR green PCR master kit (Applied Biosystems). gyrB was used as a control to normalize for transcript quantification. Relative quantification was calculated by the ΔΔCT method.
Table 1.
Primers used in this study.
| Primer | Sequence | Purpose |
|---|---|---|
| sarA | Forward: 5′-TCTTGTTAATGCACAACAACGTAA-3′ | RT-PCR |
| Reverse: 5′-TGTTTGCTTCAGTGATTCGTTT-3′ | ||
| fnbA | Forward: 5′-CGACACAACCTCAAGACAATAGCGG-3′ | RT-PCR |
| Reverse: 5′-CGTGGCTTACTTTCTGATGCCGTTC-3′ | ||
| mecA | Forward: 5′-TCCAGATTACAACTTCACCAGG-3′ | RT-PCR |
| Reverse: 5′-CCACTTCATATCTTGTAACG-3′ | ||
| RNAⅢ | Forward: 5′-AATTAGCAAGTGAGTAACATTTGCTAGT-3′ | RT-PCR |
| Reverse: 5′-GATGTTGTTTACGATAGCTTACATGC-3′ | ||
| hla | Forward: 5′-ACAATTTTAGAGAGCCCAACTGAT-3′ | RT-PCR |
| Reverse: 5′-TCCCCAATTTTGATTCACCAT-3′ | ||
| gyrB | Forward: 5′-CGCAGGCGATTTTACCATTA-3′ | RT-PCR |
| Reverse: 5′-GCTTTCGCTAGATCAAAGTCG-3′ | ||
| mecA promoter | Forward: 5′-ATATCGTGAGCAATGAAC TG-3′ | Gel shift |
| Reverse: 5′-TATATACCAAACCCGACAAC-3′ |
2.8. Determination the impact of HYP on SarA-mecA binding by a gel shift assay
Gel shift assay was performed to determine if SarA regulates mecA expression by directly binding to the mecA promoter, as sarA mutants exhibited increased OXA susceptibility vs. their respective isogenic MRSA parental strain10. Purified SarA protein was kindly provided by Dr. Ambrose Cheung at Dartmouth Medical School (Hanover, New Hampshire, USA)30. A 200 bp fragment encompassing the mecA promoter was generated by PCR amplification of JE2 DNA using the primer as listed in Table 131. Then, the mecA promoter DNA was incubated at room temperature for 20 min with various amounts of purified SarA protein (e.g., 0.3, 0.6 and 1.2 mg/L) in binding buffer30. To determination the impact of HYP on Sar-mecA binding, the mecA promoter DNA and 1.2 mg/L purified SarA protein were incubated at room temperature for 20 min with various concentrations of HYP (e.g., 4 and 8 mg/L) in binding buffer. The reaction mixtures were analyzed in a 6% Tris-Glycine gel (Novex, San Diego, CA, USA). The band shifts were stained by SYBR® Green Electrophoretic Mobility-Shift Assay (EMSA) Kit (Panomics, Fermont, CA, USA) and detected by exposing to UV302 nm following the manufacturer's instructions.
2.9. Cell cytotoxicity assay
Cell cytotoxicity was tested by the cell counting kit-8 (CCK-8) assay, as we previously reported32, 33. Briefly, human embryonic kidney 293 (HEK-293) cells were plated in a 96-well plate. After overnight incubation, different concentrations of HYP were added. After 24 h of incubation, the cells were treated with CCK-8 assay reagent, and OD at 450 nm was measured.
2.10. Murine bacteremia model due to MRSA strain
To further define the effect of HYP on OXA susceptibility against MRSA in vivo, a well-characterized murine bacteremia model was used34. All the animal studies complied with the ARRIVE guidelines35 and the Institutional Animal Care and Use Committee of Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center (Los Angeles, CA, USA) approved the animal study protocol. CD1 male mice (∼6 weeks) were infected by using exponential phase of MRSA JE2 cells via tail vein (108 CFU/mouse). At 24 h post-infection, animals were randomized into one of the following groups: control (without treatment); HYP alone at 5 mg/kg, iv, once daily; OXA alone at 100 mg/kg, im, three times a day (tid); or HYP-OXA combination at the doses listed above. The HYP dose was selected according to previously reported regimen in murine model36. The treatment strategy of OXA encompassed dose-regimens used in prior studies in murine sepsis model37. Treatment lasted for 3 days. The control animals were euthanized at 24 h post-infection in order to determine the MRSA density in target tissues (e.g., blood, spleen and kidney) at the beginning of treatment. Antibiotic treated animals were sacrificed at 24 h after the last treatment dose. At sacrifice, the target organs (e.g., blood, kidney and spleen) were removed and quantitatively cultured. The mean lg(CFU/g) of tissue or lg(CFU/mL) of blood (±SD) was calculated for each group for statistical comparisons.
2.11. Statistic analyses
All in vitro assays were conducted with at least two biological replicates in triplicate. Two-tailed Student's t-test was used to analyze the in vitro data and S. aureus counts in the target tissues in the murine bacteremia model between different groups. Data were expressed as Mean (±SD). P values < 0.05 were considered significant. No adjustment was made for all the P values reported in this study.
3. Results
3.1. MICs and FIC index
As expected, the MRSA strain JE2 was resistant to OXA, CFZ and NAF with MICs ranging 32–256 mg/L (Table 2). The MIC of HYP against the JE2 strain was 64 mg/L (Table 2). A synergistic activity was observed in the combination of HYP with the study β-lactam antibiotics against the JE2 stain with FIC index values ranged 0.10–0.19 (Table 2). In addition, the effect of HYP in combination with other SOC antibiotics against the study MRSA strain was tested. Except amoxicillin, ciprofloxacin, and streptomycin, all the antibiotics showed a reduction in MIC values when used in combination with 1/8 × MIC of HYP (See Supporting Information Table S3).
Table 2.
MICs of HYP, OXA, NAF and CFZ, and FIC index of the combination of HYP with the study β-lactam antibiotics against MRSA strain JE2.
| MICs (mg/L) | FIC index |
|||||
|---|---|---|---|---|---|---|
| HYP + β-lactam | ||||||
| HYP | OXA | CFZ | NAF | OXA | CFZ | NAF |
| 64 | 256 | 128 | 32 | 0.10 | 0.19 | 0.19 |
In addition, as we reported recently10, the sarA and mecA mutants in the JE2 background had significantly decreased OXA MICs, and the sarA mutant with plasmid mecA complemented strain returned the OXA MIC value to near its parental JE2 level (Table 3). In the current study, we found that the MICs of HYP on the JE2 strain set were similar with MICs ranging 32–64 mg/L. Importantly, addition of 1/8 × MIC HYP significantly reduced the MIC of OXA on the JE2 parental strain (decreased OXA MICs from 256 mg/L to 8 mg/L), and showed synergistic effect with FIC index of 0.16 (Table 3). However, this synergistic effect was not observed on the mutant strains (Table 3; FIC index >0.5). These data indicate that the synergistic effect of HYP with OXA might be due to its impact on sarA, then subsequence on mecA.
Table 3.
MICs and FIC index of OXA in the presence of HYP against MRSA JE2 wild-type, its isogenic sarA, mecA mutants and the sarA mutant complemented with mecA strains.
| Strain | MIC (mg/L) |
FIC index | ||
|---|---|---|---|---|
| HYP | OXA | OXA+1/8 × MIC HYP | ||
| JE2 wild-type | 64 | 256 | 8 | 0.16 |
| ΔsarA in JE2 | 32 | 4 | 2 | 0.56 |
| ΔmecA in JE2 | 64 | 0.5 | 0.25 | 0.63 |
| ΔsarA/pmecA in JE2 | 64 | 128 | 64 | 0.56 |
3.2. Time–killing curves
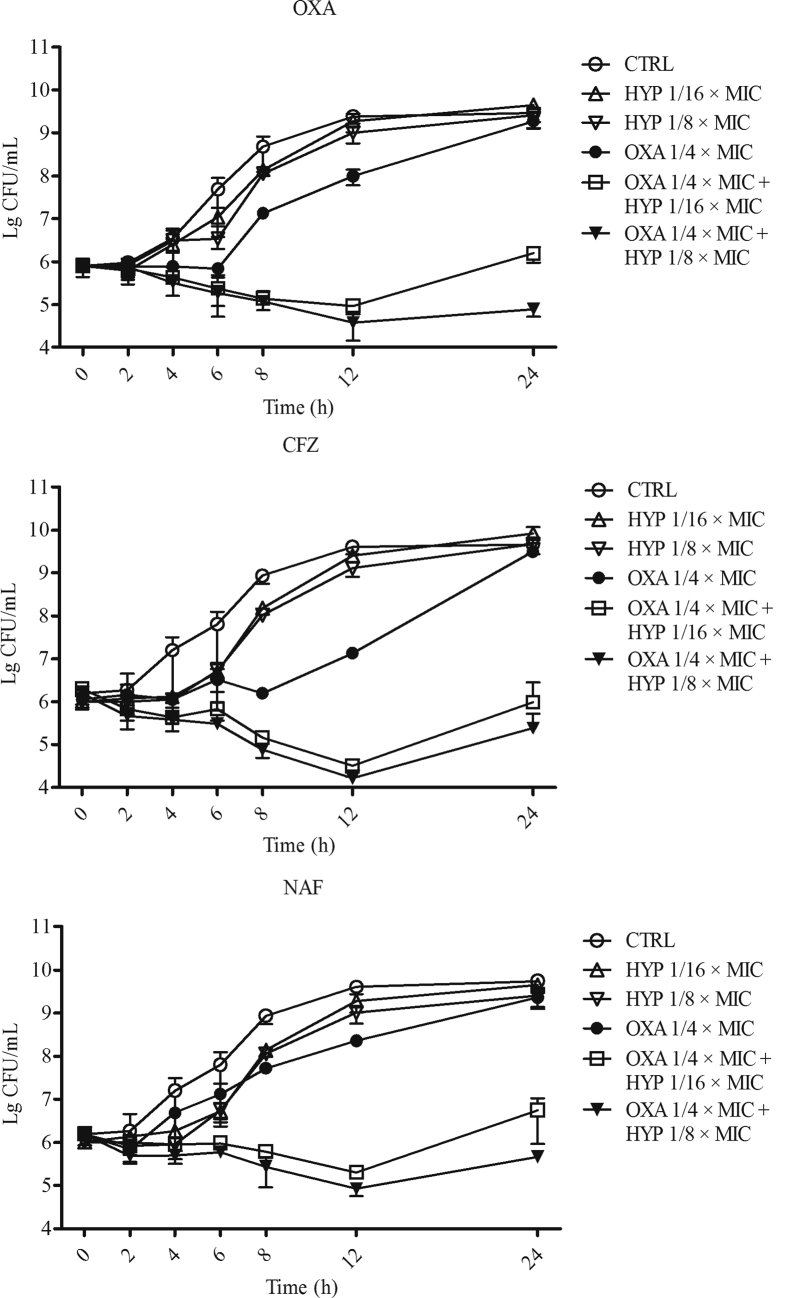
The time–killing profiles of HYP, OXA, CFZ, and NAF alone, as well as in combination against the MRSA strain JE2 are presented in Fig. 1. Control cultures without antibiotic exposure increased ∼4lg(CFU/mL) over the 24 h of incubation. HYP and β-lactam antibiotics alone at sub-MIC concentrations showed minor inhibition effect on the growth of the bacteria during 12 h of incubation, while had similar bacterial growths vs. the control groups at 24 h of incubation. Importantly, the combinations of HYP with OXA, CFZ, or NAF demonstrated synergistic effect against the MRSA strain JE2 with ∼2lg(CFU/mL) killing compared to their respective most active single drug exposure during 12 h incubation. More importantly, the combinations resulted >2lg(CFU/mL) reduction vs. HYP and β-lactams alone at 24 h time point (Fig. 1).
Figure 1.
In vitro time–killing curves of HYP, OXA, CFZ or NAF alone; and the combination of HYP with OXA, CFZ or NAF against MRSA JE2 strain. The data are the mean ± SD of MRSA counts in each group of at least two biological replicates.
3.3. The effect of HYP, β-lactam antibiotics alone and in combination on biofilm formation
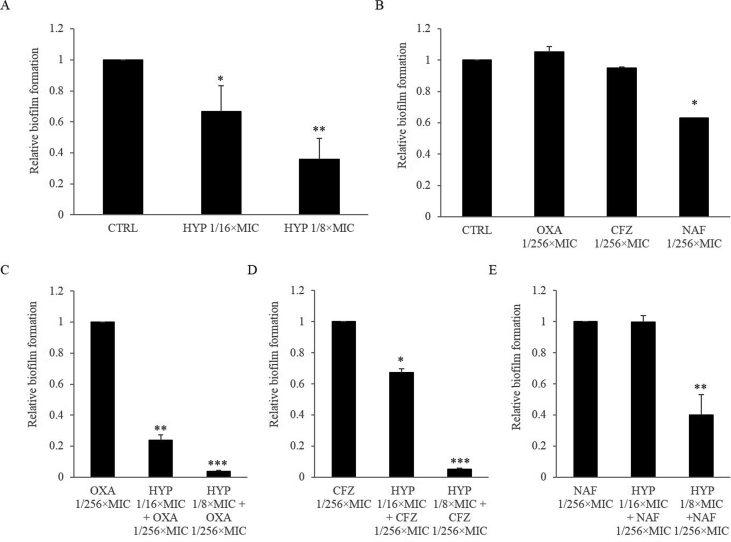
HYP alone at sub-MIC levels resulted significantly less biofilm formation than control group in a dose-dependent manner in MRSA strain JE2 (Fig. 2A). In addition, sub-MIC of NAF had significantly effect on the reduction of biofilm formation as compared to the control group, while OXA and CFZ alone had similar biofilm formation vs. the control (Fig. 2B). Of note, the combination of HYP with the study β-lactams at sub-MIC levels significantly reduced biofilm formation in a dose-dependent manner as compared to β-lactams alone (Fig. 2C–E for OXA, CFZ and NAF, respectively), except the combination of HYP at 1/16 × MIC with NAF at 1/256 × MIC (Fig. 2E).
Figure 2.
The effect of HYP (penal A), β-lactam alone (penal B), and the combination of HYP with OXA (penal C), CFZ (penal D), or NAF (penal E) on biofilm formation in MRSA. Control or β-lactam alone groups were set up as 1. Relative biofilm formation levels were represented as mean ± SD of at least two biological replicates. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001; Penals A and B vs. control; Penal C, D and E vs. β-lactam alone.
3.4. The effect of HYP, OXA alone and in combination on fibronectin binding
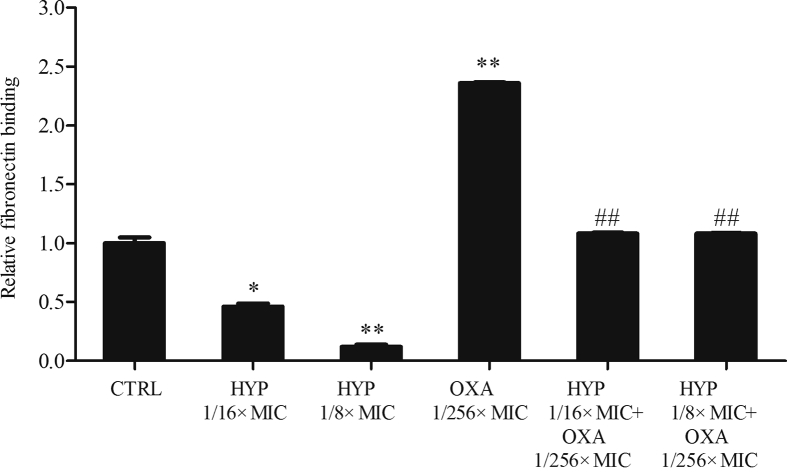
As shown in Fig. 3, in the presence of sub-MICs of HYP, the MRSA strain JE2 showed significantly decreased capability of fibronectin binding in a dose-dependent manner vs. the control group. Interestingly, sub-MIC of OXA exposure showed significantly increased binding to fibronectin as compared with the control group (Fig. 3). Of importance, the combination of HYP with OXA significantly decreased the fibronectin binding ability of the JE2 strain vs. OXA alone exposure (Fig. 3).
Figure 3.
The effect of HYP or OXA alone and in combination on fibronectin binding in MRSA strain JE2. Control group was set up as 1. Relative fibronectin binding levels were represented as mean ± SD of at least two biological replicates. ∗P < 0.01, ∗∗P < 0.001 vs. control; ##P < 0.001 vs. OXA alone.
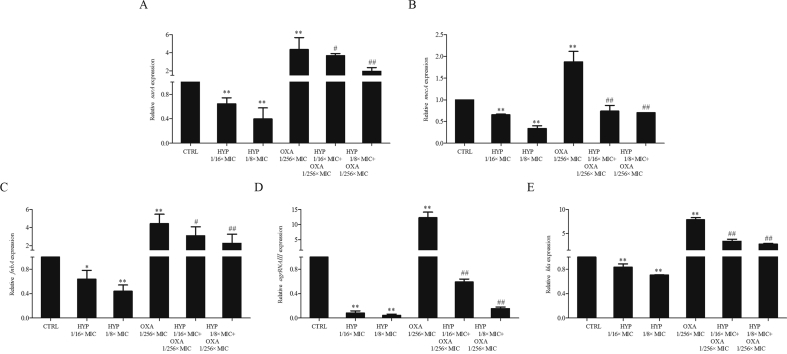
3.5. The effects of HYP, OXA alone and in combination on sarA, mecA and virulence related genes expression
It is known that mecA, encoding penicillin binding protein 2a (PBP2a), mediates the resistance to β-lactam antibiotics in MRSA35. In addition, we have recently demonstrated the important role of the global regulator, sarA, in β-lactam antibiotics resistance in MRSA10. In the current study, we found that HYP at sub-MIC levels significantly decreased sarA and mecA expressions as compared to their respective control groups (Fig. 4A and B for sarA and mecA expression, respectively). Consistent with our previous findings10, 38, sub-MICs of OXA exposure significantly induced sarA and mecA expressions vs. their respective controls (Fig. 4A and B for sarA and mecA expression, respectively). Interestingly, the combination of HYP and OXA significantly reduced the expression of sarA and mecA as compared with OXA alone (Fig. 4A and B for sarA and mecA expression, respectively). More importantly, HYP also significantly decreased agr RNAⅢ, which is a well-known sarA downstream key regulator and other virulence related genes (e.g., fnbA and hla) expression as compared to their respective control groups. Although sub-MIC of OXA exposure significantly induced these genes expression, the combination groups significantly reduced these genes expression as compared with OXA alone (Fig. 4 D and E).
Figure 4.
The effect of HYP or OXA alone; and in combinations on sarA (Penal A), mecA (Penal B), fnbA (Penal C), agrRNAⅢ (Penal D) and hla (Penal E) expressions in MRSA. Control group was set up as 1. Relative transcript levels of sarA, mecA, fnbA, agrRNAⅢ, and hla were represented as mean ± SD of at least two biological replicates. ∗P < 0.05, ∗∗P < 0.01 vs. control; #P < 0.05, ##P < 0.01 vs. OXA alone.
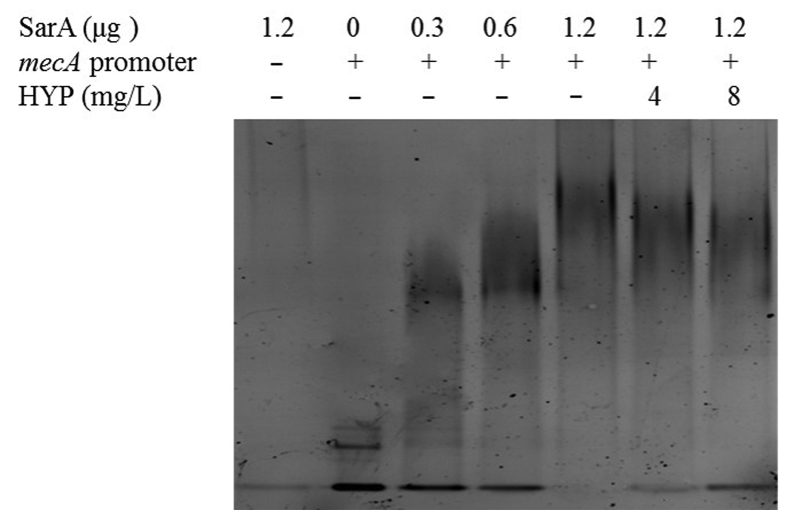
3.6. The impact of HYP on the SarA-mecA binding
A direct binding of SarA to the mecA promoter fragment was observed in a SarA protein concentration dependent manner (Fig. 5). For instance, a nearly complete shift occurred in the presence of 1.2 μg of SarA protein. Of importance, sub-MICs of HYP substantially reduced the SarA–mecA binding (Fig. 5). These results indicate that SarA positively controls mecA expression through its direct binding to mecA promoter, and HYP decreases the binding ability of SarA to mecA promoter.
Figure 5.
The effect of HYP on SarA binding ability to the mecA promoter.
3.7. The cytotoxic effect of HYP on HEK-293
The cytotoxic effect of HYP on HEK-293 cells was investigated by using CCK-8 assay. The results showed that HEK-293 cells were well tolerated to HYP at concentrations tested (Supporting Information Fig. S1).
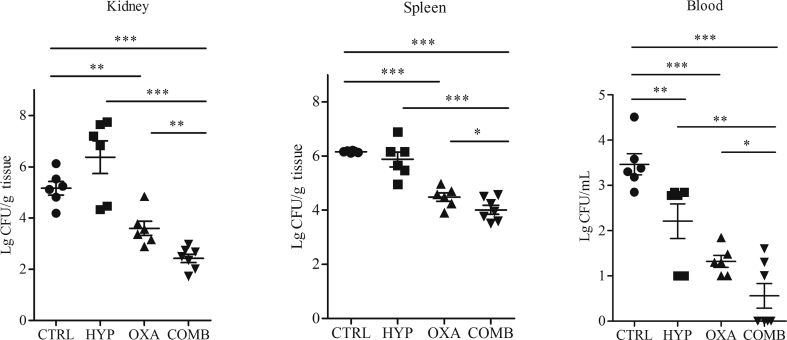
3.8. Therapeutic efficacy of HYP, OXA alone and in combination in the murine bacteremia model
HYP monotherapy showed no significant decreased in MRSA densities in kidney and spleen vs. the untreated controls, while blood samples from mice treated with HYP had significantly lower MRSA densities than those from the control animals (Fig. 6). Treatment of OXA alone resulted significant reduced MRSA density in all target tissues as compared with the control groups (Fig. 6). Of great interest, the combination of HYP and OXA exhibited significantly greater efficacy in reducing MRSA densities in all target tissues vs. all other three groups (Fig. 6).
Figure 6.
Therapeutic efficacy of HYP and OXA alone, and in combination in a mouse bacteremia model due to MRSA strain JE2. Each dot represents MRSA density in target tissues in the bacteremia model in one mouse (n = 6). Horizontal black bars indicate mean ± SD MRSA density in the target tissues. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
4. Discussion
MRSA is a major cause of invasive infections (e.g., bacteremia) with unacceptable high morbidity and mortality since its emergency in 1960s39. Vancomycin and daptomycin have been considered as the first line of antibiotics against MRSA infections. However, many MRSA strains have developed resistance to these antibiotics40, 41. Thus, an alternative treatment approach against MRSA infections is urgently needed. In our previous studies, the importance of sarA in β-lactams resistance was reported10. For instance, we demonstrated that sarA mutant strains became more susceptible to OXA as compared to their respective parental MRSA strains both in vitro and in an experimental endocarditis model. Hence, we speculated that sarA could be an optimal potential target to anti-MRSA.
In the current investigation, we observed that HYP had anti-MRSA activities with MIC of 64 mg/L. Of major importance, a synergistic effect of HYP with β-lactams (e.g., OXA, CFZ and NAF) was observed against MRSA strain JE2. In addition, the combination resulted ∼2lg(CFU/mL) reduction during 12 h incubation as compared with HYP or β-lactams alone group. However, regrowth was observed at 24 h incubation.
It is well known that mecA plays a key role in β-lactam resistance (e.g., OXA) in MRSA42, 43. In addition, we recently demonstrated that sarA regulates β-lactam antibiotic resistance in MRSA both in vitro and in an experimental endocarditis model at least in part through its effect on mecA expression10. In this study, we observed that HYP exposure significantly decreased sarA and mecA expression. Although OXA alone significantly induced sarA and mecA expression, which was consistent with others previous reported10, 38, 44, the addition of HYP significantly reduced sarA and mecA expression vs. OXA alone. To further investigate whether the synergistic effect of HYP and OXA was mediated by sarA and/or mecA, isogenic sarA, mecA and sarA/pmecA mutant strains in the JE2 background were used. Our results demonstrated that there were no synergistic effects between HYP and OXA against these mutant strains. Moreover, HYP exhibited inhibition effect on SarA–mecA promoter binding. Taken together, these data indicate that the synergistic effect at least in part was due to the impact of HYP on the inhibition of sarA activity, then subsequent on mecA expression.
Biofilm formation plays an important role in the pathogenesis of staphylococci infections45. Numerous studies have demonstrated that sarA is an important positive regulator on biofilm formation5, 46, 47, 48. The mechanism partly due to sarA negatively regulate the activity of protease and nuclease5, while positively control Bip49 and PIA/PNAG production50. Consistent with the previous studies10 , we found that JE2 sarA mutant strain formed significantly less biofilm than its isogenic parental strain. Of importance, HYP, as a sarA inhibitor, showed significantly reduced biofilm formation in a concentration-dependent manner in JE2 strain. These data were consistent with other studies showing that HYP and SarA inhibitor had anti-biofilm activity against S. aureus strains6, 15. However, the specific mechanism of HYP against MRSA biofilm formation still need further studies.
SarA is an important transcriptional regulator that interacts with other regulators (e.g., agr) and controls many virulence genes (e.g., mecA, fnbA and hla) in S. aureus51, 52. As reported from prior studies, SarA binds to fnbA promoter fragments, positively regulates FnBPs production and increases capacity of MRSA strains binding to fibronectin5, 53. HYP, as a sarA inhibitor, significantly decreased fnbA expression and fibronectin binding capacity, which were consistent with previous results related to sarA mutants and other SarA inhibitor5, 7, 54. Of importance, the combination of HYP and OXA showed significantly decreased fnbA expression and fibronectin binding capacity vs. OXA alone. In addition, Sub-MIC of HYP exposure significantly decreased agr RNAⅢ and its downstream hla genes expression6, 7, while sub-MIC of OXA exposure significantly induced these genes expression55, 56. Of importance, the combination of HYP and OXA showed significantly decreased these genes expression vs. OXA alone. Taken together, our data indicate that HYP, as a sarA inhibitor, significantly decreased agr RNAⅢ and its downstream virulence genes expression.
The most important finding in the current study was that the in vitro results were well translated into the in vivo treatment outcomes in the murine bacteremia model due to the study MRSA strain. Similar MRSA densities in kidney and spleen were observed in the HYP monotherapy group vs. the untreated controls. However, OXA treatment alone had therapeutic efficacy with significantly reduced MRSA counts in all the target tissues as compared with the control group. These results were in agreement with previous studies using similar OXA treatment regimen and bacteremia model37, 38. Of important, the combination of HYP with OXA exhibited significantly lower MRSA densities in the target tissues vs. OXA monotherapy. These combinational therapeutic effects were thought to occur by inhibition of sarA, which subsequently reduces mecA expression and OXA resistance. These data underscored the significance of HYP, as a sarA inhibitor, in the use with β-lactam antibiotic (e.g., OXA) for the treatment of bacteremia due to MRSA.
5. Conclusions
We demonstrated that HYP had inhibition effect on sarA expression, and subsequently downregulated the expression of mecA, and increased β-lactams susceptibility in MRSA. In addition, HYP, as a sarA inhibitor, significantly reduced biofilm formation, fibronectin binding and virulence-related gene expression. Notably, combination therapy regiments of HYP and OXA significantly enhanced in vivo efficacy of OXA in a murine bacteremia model due to MRSA. This combinational approach may present a novel treatment strategy against infections caused by MRSA strains by using anti-methicillin susceptible S. aureus (MSSA) β-lactam antibiotics.
Acknowledgments
This study was supported in part by CAMS Initiative for Innovative Medicine (grant numbers 2016-I2M-2-002 and 2016-I2M-3-014, China), National Mega-project for Innovative Drugs (grant number 2018ZX09721001, China), the National Science Foundation of China (grant number 81621064, China). We are very grateful to Dr. Ambrose Cheung at Dartmouth Medical School (Hanover, New Hampshire, USA) for providing the purified SarA protein.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2019.05.002.
Contributor Information
Xuefu You, Email: xuefuyou@hotmail.com.
Yan Q. Xiong, Email: yxiong@ucla.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bayer M.G., Heinrichs J.H., Cheung A.L. The molecular architecture of the sar locus in Staphylococcus aureus. J Bacteriol. 1996;178:4563–4570. doi: 10.1128/jb.178.15.4563-4570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L., Yang R., Yuan B., Liu Y., Liu C. The antiviral and antimicrobial activities of licorice, a widely-used Chinese herb. Acta Pharm Sin B. 2015;5:310–315. doi: 10.1016/j.apsb.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boucher H., Miller L.G., Razonable R.R. Serious infections caused by methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2010;51 Suppl 2:S183–S197. doi: 10.1086/653519. [DOI] [PubMed] [Google Scholar]
- 4.Cheung A.L., Nishina K.A., Trotonda M.P., Tamber S. The SarA protein family of Staphylococcus aureus. Int J Biochem Cell Biol. 2008;40:355–361. doi: 10.1016/j.biocel.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdelhady W., Bayer A.S., Seidl K., Moormeier D.E., Bayles K.W., Cheung A. Impact of vancomycin on sarA-mediated biofilm formation: role in persistent endovascular infections due to methicillin-resistant Staphylococcus aureus. J Infect Dis. 2014;209:1231–1240. doi: 10.1093/infdis/jiu007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balamurugan P., Praveen Krishna V., Bharath D., Lavanya R., Vairaprakash P., Adline Princy S. Staphylococcus aureus quorum regulator SarA targeted compound, 2-[(methylamino)methyl]phenol inhibits biofilm and down-regulates virulence genes. Front Microbiol. 2017;8:1290. doi: 10.3389/fmicb.2017.01290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arya R., Ravikumar R., Santhosh R.S., Princy S.A. SarA based novel therapeutic candidate against Staphylococcus aureus associated with vascular graft infections. Front Microbiol. 2015;6:416. doi: 10.3389/fmicb.2015.00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung A.L., Eberhardt K.J., Chung E., Yeaman M.R., Sullam P.M., Ramos M. Diminished virulence of a sar-/agr- mutant of Staphylococcus aureus in the rabbit model of endocarditis. J Clin Investig. 1994;94:1815–1822. doi: 10.1172/JCI117530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blevins J.S., Elasri M.O., Allmendinger S.D., Beenken K.E., Skinner R.A., Thomas J.R. Role of sarA in the pathogenesis of Staphylococcus aureus musculoskeletal infection. Infect Immun. 2003;71:516–523. doi: 10.1128/IAI.71.1.516-523.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L., Cheung A., Bayer A.S., Chen L., Abdelhady W., Kreiswirth B.N. The global regulon sarA regulates β-lactam antibiotic resistance in methicillin-resistant Staphylococcus aureus in vitro and in endovascular infections. J Infect Dis. 2016;214:1421–1429. doi: 10.1093/infdis/jiw386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawvere S., Mahoney M.C. St. John's wort. Am Fam Physician. 2005;72:2249–2254. [PubMed] [Google Scholar]
- 12.Zhang J., Jiang C., Figueiro Longo J.P., Azevedo R.B., Zhang H., Muehlmann L.A. An updated overview on the development of new photosensitizers for anticancer photodynamic therapy. Acta Pharm Sin B. 2018;8:137–146. doi: 10.1016/j.apsb.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobson J.M., Feinman L., Liebes L., Ostrow N., Koslowski V., Tobia A. Pharmacokinetics, safety, and antiviral effects of hypericin, a derivative of St. John's wort plant, in patients with chronic hepatitis C virus infection. Antimicrob Agents Chemother. 2001;45:517–524. doi: 10.1128/AAC.45.2.517-524.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kashef N., Karami S., Djavid G.E. Phototoxic effect of hypericin alone and in combination with acetylcysteine on Staphylococcus aureus biofilms. Photodiagn Photodyn Ther. 2015;12:186–192. doi: 10.1016/j.pdpdt.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Garcia I., Ballesta S., Gilaberte Y., Rezusta A., Pascual A. Antimicrobial photodynamic activity of hypericin against methicillin-susceptible and resistant Staphylococcus aureus biofilms. Future Microbiol. 2015;10:347–356. doi: 10.2217/fmb.14.114. [DOI] [PubMed] [Google Scholar]
- 16.Yow C.M., Tang H.M., Chu E.S., Huang Z. Hypericin-mediated photodynamic antimicrobial effect on clinically isolated pathogens. Photochem Photobiol. 2012;88:626–632. doi: 10.1111/j.1751-1097.2012.01085.x. [DOI] [PubMed] [Google Scholar]
- 17.Kairyte K., Lapinskas S., Gudelis V., Luksiene Z. Effective inactivation of food pathogens Listeria monocytogenes and Salmonella enterica by combined treatment of hypericin-based photosensitization and high power pulsed light. J Appl Microbiol. 2012;112:1144–1151. doi: 10.1111/j.1365-2672.2012.05296.x. [DOI] [PubMed] [Google Scholar]
- 18.Fey P.D., Endres J.L., Yajjala V.K., Widhelm T.J., Boissy R.J., Bose J.L. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio. 2013;4 doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trotonda M.P., Xiong Y.Q., Memmi G., Bayer A.S., Cheung A.L. Role of mgrA and sarA in methicillin-resistant Staphylococcus aureus autolysis and resistance to cell wall-active antibiotics. J Infect Dis. 2009;199:209–218. doi: 10.1086/595740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Memmi G., Filipe S.R., Pinho M.G., Fu Z., Cheung A. Staphylococcus aureus PBP4 is essential for β-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob Agents Chemother. 2008;52:3955–3966. doi: 10.1128/AAC.00049-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu X., Yang X., Li X., Lu Y., Ren Z., Zhao L. In vitro activity of sodium new houttuyfonate alone and in combination with oxacillin or netilmicin against methicillin-resistant Staphylococcus aureus. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White R.L., Burgess D.S., Manduru M., Bosso J.A. Comparison of three different in vitro methods of detecting synergy: time-kill, checkerboard, and E test. Antimicrob Agents Chemother. 1996;40:1914–1918. doi: 10.1128/aac.40.8.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Odds F.C. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother. 2003;52:1. doi: 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- 24.Belley A., Neesham-Grenon E., Arhin F.F., McKay G.A., Parr T.R., Jr., Moeck G. Assessment by time–kill methodology of the synergistic effects of oritavancin in combination with other antimicrobial agents against Staphylococcus aureus. Antimicrob Agents Chemother. 2008;52:3820–3822. doi: 10.1128/AAC.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdelhady W., Bayer A.S., Seidl K., Nast C.C., Kiedrowski M.R., Horswill A.R. Reduced vancomycin susceptibility in an in vitro catheter-related biofilm model correlates with poor therapeutic outcomes in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2013;57:1447–1454. doi: 10.1128/AAC.02073-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seidl K., Chen L., Bayer A.S., Hady W.A., Kreiswirth B.N., Xiong Y.Q. Relationship of agr expression and function with virulence and vancomycin treatment outcomes in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55:5631–5639. doi: 10.1128/AAC.05251-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlsson A., Saravia-Otten P., Tegmark K., Morfeldt E., Arvidson S. Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect Immun. 2001;69:4742–4748. doi: 10.1128/IAI.69.8.4742-4748.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveira D.C., de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ster C., Gilbert F.B., Cochard T., Poutrel B. Transcriptional profiles of regulatory and virulence factors of Staphylococcus aureus of bovine origin: oxygen impact and strain-to-strain variations. Mol Cell Probes. 2005;19:227–235. doi: 10.1016/j.mcp.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 30.Chien Y., Manna A.C., Projan S.J., Cheung A.L. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J Biol Chem. 1999;274:37169–37176. doi: 10.1074/jbc.274.52.37169. [DOI] [PubMed] [Google Scholar]
- 31.Oliveira D.C., de Lencastre H. Methicillin-resistance in Staphylococcus aureus is not affected by the overexpression in trans of the mecA gene repressor: a surprising observation. PLoS One. 2011;6 doi: 10.1371/journal.pone.0023287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang G., Pang J., Hu X., Nie T., Lu X., Li X. Daphnetin: a novel anti-Helicobacter pylori agent. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20040850. pii: E850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Z., Wang X., Zhang H., Sun J., Zheng L., Liu H. Chromopeptide A, a highly cytotoxic depsipeptide from the marine sediment-derived bacterium Chromobacterium sp. HS-13-94. Acta Pharm Sin B. 2015;5:62–66. doi: 10.1016/j.apsb.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R., Braughton K.R., Kretschmer D., Bach T.H., Queck S.Y., Li M. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 35.Wang Q., Lv Y., Pang J., Li X., Lu X., Wang X. In vitro and in vivo activity of d-serine in combination with β-lactam antibiotics against methicillin-resistant Staphylococcus aureus. Acta Pharm Sin B. 2019;9:496–504. doi: 10.1016/j.apsb.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blank M., Lavie G., Mandel M., Hazan S., Orenstein A., Meruelo D. Antimetastatic activity of the photodynamic agent hypericin in the dark. Int J Cancer. 2004;111:596–603. doi: 10.1002/ijc.20285. [DOI] [PubMed] [Google Scholar]
- 37.Jiang W., Li B., Zheng X., Liu X., Cen Y., Li J. Artesunate in combination with oxacillin protect sepsis model mice challenged with lethal live methicillin-resistant Staphylococcus aureus (MRSA) via its inhibition on proinflammatory cytokines release and enhancement on antibacterial activity of oxacillin. Int Immunopharmacol. 2011;11:1065–1073. doi: 10.1016/j.intimp.2011.02.028. [DOI] [PubMed] [Google Scholar]
- 38.Waters E.M., Rudkin J.K., Coughlan S., Clair G.C., Adkins J.N., Gore S. Redeploying β-lactam antibiotics as a novel antivirulence strategy for the treatment of methicillin-resistant Staphylococcus aureus infections. J Infect Dis. 2017;215:80–87. doi: 10.1093/infdis/jiw461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fridkin S.K., Hageman J.C., Morrison M., Sanza L.T., Como-Sabetti K., Jernigan J.A. Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med. 2005;352:1436–1444. doi: 10.1056/NEJMoa043252. [DOI] [PubMed] [Google Scholar]
- 40.CDC Vancomycin-resistant Staphylococcus aureus—Pennsylvania. MMWR Morb Mortal Wkly Rep 2002. 2002;51:902. [PubMed] [Google Scholar]
- 41.Marty F.M., Yeh W.W., Wennersten C.B., Venkataraman L., Albano E., Alyea E.P. Emergence of a clinical daptomycin-resistant Staphylococcus aureus isolate during treatment of methicillin-resistant Staphylococcus aureus bacteremia and osteomyelitis. J Clin Microbiol. 2006;44:595–597. doi: 10.1128/JCM.44.2.595-597.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He M., Shao L., Liu Q., Li J., Lin H., Jing L. Mechanism of synergy between SIPI-8294 and β-lactam antibiotics against methicillin-resistant Staphylococcus aureus. Lett Appl Microbiol. 2016;63:3–10. doi: 10.1111/lam.12583. [DOI] [PubMed] [Google Scholar]
- 43.Sakoulas G., Gold H.S., Venkataraman L., DeGirolami P.C., Eliopoulos G.M., Qian Q. Methicillin-resistant Staphylococcus aureus: comparison of susceptibility testing methods and analysis of mecA-positive susceptible strains. J Clin Microbiol. 2001;39:3946–3951. doi: 10.1128/JCM.39.11.3946-3951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mirani Z.A., Aziz M., Khan S.I. Small colony variants have a major role in stability and persistence of Staphylococcus aureus biofilms. J Antibiot (Tokyo) 2015;68:98–105. doi: 10.1038/ja.2014.115. [DOI] [PubMed] [Google Scholar]
- 45.Beenken K.E., Blevins J.S., Smeltzer M.S. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect Immun. 2003;71:4206–4211. doi: 10.1128/IAI.71.7.4206-4211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atwood D.N., Loughran A.J., Courtney A.P., Anthony A.C., Meeker D.G., Spencer H.J. Comparative impact of diverse regulatory loci on Staphylococcus aureus biofilm formation. Microbiology. 2015;4:436–451. doi: 10.1002/mbo3.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Houston P., Rowe S.E., Pozzi C., Waters E.M., O'Gara J.P. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect Immun. 2011;79:1153–1165. doi: 10.1128/IAI.00364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valle J., Toledo-Arana A., Berasain C., Ghigo J.M., Amorena B., Penades J.R. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol Microbiol. 2003;48:1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 49.Trotonda M.P., Manna A.C., Cheung A.L., Lasa I., Penades J.R. SarA positively controls bap-dependent biofilm formation in Staphylococcus aureus. J Bacteriol. 2005;187:5790–5798. doi: 10.1128/JB.187.16.5790-5798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Neill E., Pozzi C., Houston P., Smyth D., Humphreys H., Robinson D.A. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J Clin Microbiol. 2007;45:1379–1388. doi: 10.1128/JCM.02280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheung A.L., Manna A.C. Role of the distal sarA promoters in SarA expression in Staphylococcus aureus. Infect Immun. 2005;73:4391–4394. doi: 10.1128/IAI.73.7.4391-4394.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dunman P.M., Murphy E., Haney S., Palacios D., Tucker-Kellogg G., Wu S. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol. 2001;183:7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolz C., Pohlmann-Dietze P., Steinhuber A., Chien Y.T., Manna A., van Wamel W. Agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol Microbiol. 2000;36:230–243. doi: 10.1046/j.1365-2958.2000.01853.x. [DOI] [PubMed] [Google Scholar]
- 54.Xiong Y.Q., Bayer A.S., Yeaman M.R., van Wamel W., Manna A.C., Cheung A.L. Impacts of sarA and agr in Staphylococcus aureus strain Newman on fibronectin-binding protein A gene expression and fibronectin adherence capacity in vitro and in experimental infective endocarditis. Infect Immun. 2004;72:1832–1836. doi: 10.1128/IAI.72.3.1832-1836.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viedma E., Perez-Montarelo D., Villa J., Munoz-Gallego I., Larrosa N., Fernandez-Hidalgo N. Sub-inhibitory concentrations of oxacillin modify the expression of agr locus in Staphylococcus aureus clinical strains belonging to different clonal complexes. BMC Infect Dis. 2018;18:177. doi: 10.1186/s12879-018-3088-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rudkin J.K., Laabei M., Edwards A.M., Joo H.S., Otto M., Lennon K.L. Oxacillin alters the toxin expression profile of community-associated methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2014;58:1100–1107. doi: 10.1128/AAC.01618-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.